SUMMARY
Intraflagellar transport (IFT) is the highly conserved process by which proteins are transported along ciliary microtubules by a train-like polymeric assembly of IFT-A and IFT-B complexes. IFT-A is sandwiched between IFT-B and the ciliary membrane, consistent with its putative role in transporting transmembrane and membrane-associated cargoes. Here, we have used single-particle analysis electron cryomicroscopy (cryo-EM) to determine structures of native IFT-A complexes. We show that subcomplex rearrangements enable IFT-A to polymerize laterally on anterograde IFT trains, revealing a cooperative assembly mechanism. Surprisingly, we discover that binding of IFT-A to IFT-B shields the preferred lipid-binding interface from the ciliary membrane but orients an interconnected network of β-propeller domains with the capacity to accommodate diverse cargoes towards the ciliary membrane. This work provides a mechanistic basis for understanding IFT-train assembly and cargo interactions.
Graphical Abstract
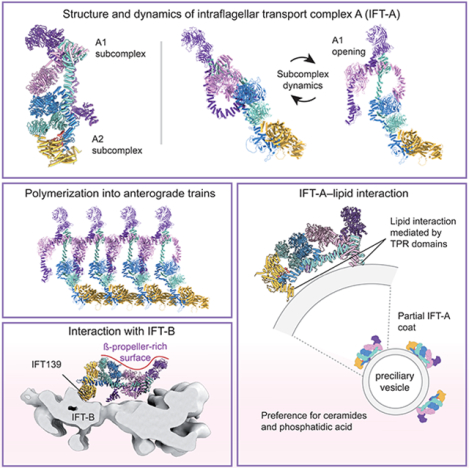
INTRODUCTION
Cilia, or flagella, are protrusions of eukaryotic cells with pleotropic functions that span from cellular motility to the sensation of physical, chemical and protein signals1. Ciliogenesis, cilia homeostasis, and the establishment of cilia signaling pathways rely on a dedicated and highly conserved trafficking mechanism called intraflagellar transport (IFT) which shuttles proteins in, out, and within cilia2. Consistent with the important role of IFT in most cilia functions, defective IFT is associated with a wide range of genetically heterogeneous human diseases including skeletal ciliopathies, polycystic kidney disease, retinal degeneration, and Bardet-Biedl syndrome (BBS)3.
Anterograde IFT toward the ciliary tip is powered by kinesin-24, whereas retrograde transport is driven by dynein-25. These two ATP-consuming motors travel processively along the outer microtubules of the axoneme at velocities between 2 to 4 μm/s4 while avoiding collisions6. The majority of IFT cargoes are coupled indirectly to the motors by IFT-A, IFT-B and the BBSome, which are multisubunit complexes of 6, 16, and 8 subunits, respectively. IFT-B is implicated in trafficking soluble proteins including tubulin7,8 and precursors of axonemal complexes9–11, whereas the BBSome and IFT-A are thought to transport transmembrane and membrane-associated proteins12–15. Cargoes are thought to be recognized by the IFT complexes by both direct interactions7,16 and through adaptor molecules10,13.
In the cilium and during assembly at the ciliary base, the IFT complexes assemble with their motors into polymeric assemblies known as “trains”4,17,18. Anterograde trains are compact structures approximately 200–500 nm in length with clearly defined periodicities, whereas retrograde trains have a less dense zig-zag pattern with a longer repeat length6. Subtomogram averaging has revealed repeating copies of IFT-B form the backbone of the anterograde train and make extensive contacts with dynein-2, which is carried as an inactive passenger19. IFT-A is situated between IFT-B and the ciliary membrane, where it presumably contacts its transmembrane cargoes. Although IFT-A is substoichiometric relative to IFT-B, it always occurs in linear arrays18,19.
Individual IFT proteins, including 4 IFT-A subunits, are rich in N-terminal β-propeller domains and C-terminal tetratricopeptide repeats (TPRs). Given that similar domain organizations occur in the subunits of vesicle coatomers (COPI, COPII and clathrin) and tethering complexes (CORVET, HOPs), it has been proposed that they share an evolutionary relationship20,21. Mounting evidence supports an extraciliary role for IFT proteins in vesicular trafficking, including localization of IFT46 to the surface of periciliary vesicles by in-situ immunogold labeling22 and the loss of densely coated periciliary vesicles in mouse cells deficient in the IFT-A subunit, IFT12123. How IFT-A binds vesicles, polymerizes into trains, and transitions between these states, are unanswered questions of fundamental importance toward a mechanistic understanding of IFT.
Unlike IFT-B7,24–30, dynein-231 and the BBSome32–34, structural data for the IFT-A complex is limited. Here, we have used single-particle cryo-EM to determine 3–4 Å resolution structures of native IFT-A complexes. In combination with spatial information from IFT trains in situ and by reconstituting lipid binding in vitro, we reveal the architecture of IFT-A, the mechanism by which IFT-A polymerizes into trains, and the surfaces capable of binding cargoes and membranes.
RESULTS
For a source of native IFT-A, we used Leishmania tarentolae, a single-celled uniflagellate of the Kinetoplastida order. Although L. tarentolae is not pathogenic to humans, it is closely related to organisms responsible for three major human diseases: leishmaniasis (various Leishmania species), African sleeping sickness (Trypanosoma brucei), and Chagas disease (Trypanosoma cruzi). Their flagella are crucial for their motility, pathogenicity, and viability35. For these reasons and their amenability to reverse genetics, the Kinetoplastida order has long been used for studies of flagella biology. Microscopy studies have demonstrated that the IFT machinery is conserved between Kinteoplastids and Chlamydomonas: both T. brucei and C. reinhardtii form anterograde trains of similar length (from ~200 nm to over 500 nm)18,36,37 and velocity (2–2.5 μm/s)36–39. Thin-section transmission electron microscopy revealed IFT trains in L. tarentolae flagella (Fig. S1A).
To purify the IFT-A complex from L. tarentolae, we created a strain expressing FLAG-tagged IFT43, the smallest of the IFT-A subunits. Anti-FLAG affinity purification recovered not only all six IFT-A subunits (IFT43, 121, 122, 139, 140 and 144), but most IFT-B subunits (Table S1A) indicating that IFT-A and -B coprecipitate. To simultaneously improve the purity and stability of the IFT-A complex, we centrifuged the sample through a sucrose gradient containing the chemical crosslinker, DTSSP (3,3’-Dithiobis (sulfosuccinimidylpropionate)). Fractions containing IFT-A were pooled from the gradient and purified further using ion-exchange chromatography (Fig. S1C). SDS-PAGE analysis (Fig. S1D) and mass spectrometry (Table S1B) revealed that the purified sample was almost exclusively IFT-A and that the interaction with IFT-B did not survive the purification process even in the presence of the crossslinker. Two-dimensional classification of particles from negative-stain electron microscopy revealed that the sample contained three major populations (Fig. 1A). One population was an elongated particle, approximately 300 Å in length and formed by two subcomplexes, one with a characteristic V shape. An almost identical class was obtained for IFT-A purified without DTSSP demonstrating that this state is not induced by the crosslinker (Fig. S2A). The other two populations were of these subcomplexes alone. Intriguingly, we did not observe IFT-A polymers in vitro, suggesting that polymerization of IFT-A is dependent on the prior oligomerization of IFT-B. This finding is consistent with the order of train assembly observed in vivo18.
Figure 1. IFT-A is formed by two interconnected subcomplexes.

A. Two-dimensional class averages from negative-stain electron microscopy showing an intact Leishmania tarentolae IFT-A complex (top), an A1 subcomplex (middle) and an A2 subcomplex (bottom).
B. Cryo-EM processing of purified IFT-A revealed three distinct classes. States 1 and 2 differ in the position of IFT139. The third class corresponded to the IFT-A2 subcomplex. Maps are sharpened using DeepEMhancer (for visualization purposes only) and colored by subunit.
C. Domain organization of the six IFT-A subunits with boundaries numbered. Abbreviations: TPR, tetratricopeptide repeat; ZnF, zinc-finger.
D. Two views showing the atomic model of IFT-A (state 2).
Structure of a monomeric IFT-A complex
To gain a better understanding of the architecture of IFT-A, we used cryo-EM single-particle analysis. Cryo-EM revealed two major populations of intact IFT-A that differed in the orientation of a large α-solenoid protein (IFT139) at one end of the complex (Fig. 1B). The structures of these populations were resolved separately to overall resolutions of 4.0 Å (class 1) and 3.6 Å (class 2). Because class 2 had more than twice the number of particles of class 1 and higher resolution, we focused our analysis on this conformational state. Even after separating the large-scale movement of IFT139, other parts of the complex showed a high degree of lability. By masking, and then combining, specific regions during processing we generated a composite map with improved local resolution (Figs. S1 and S3). The resolved sidechains of the map (Fig. S3C) allowed unambiguous modeling of all six subunits. High-resolution maps were necessary to confidently distinguish between the similar domain architectures of the IFT121/122/140/144 subunits (Fig. 1C). The final atomic model (Fig. 1D) is consistent with chemical crosslinking mass spectrometry (XL-MS) data obtained for Tetrahymena thermophila IFT-A40 (Fig. S4). The few outliers, with distances beyond the crosslinking capabilities of disuccinimidyl sulfoxide41, mostly involve residues within the TPR domains of IFT140 and IFT144, which, as we show later, are capable of large-scale movements37
Our structure revised a prior description of IFT-A as having core and peripheral subcomplexes13,42. We found that the IFT-A “core” (IFT122, 140 and 144) actually corresponded to the V-shaped subcomplex and that the “peripheral” subcomplex of IFT43, 121 and 139 also contained IFT122 as an integral component (Fig. 1D). Given that neither subcomplex is peripheral to the other, we redefine the core subcomplex as IFT-A1 and the peripheral subcomplex as IFT-A2, reflecting a similar nomenclature used for IFT-B subcomplexes28.
Both subcomplexes have distinctive architectures. The V-shape of the A1 subcomplex is generated by a heterodimer of IFT140/144 in which their tandem β-propeller domains are splayed ~50 Å apart. IFT121/122 form a similar heterodimer in the A2 subcomplex but with their β-propeller domains closed together into a tetrameric arrangement. The heterodimerization of IFT121/122 and IFT140/144 occurs through a common dimerization module that involves an antiparallel interaction between the first 5 helices of their respective TPR domains (Fig. 2A,B). This TPR–TPR interaction is different from how subunits with similar domain architectures heterodimerize in COPI coatamers43 and the HOPs complex44.
Figure 2. Inter-subunit interactions.
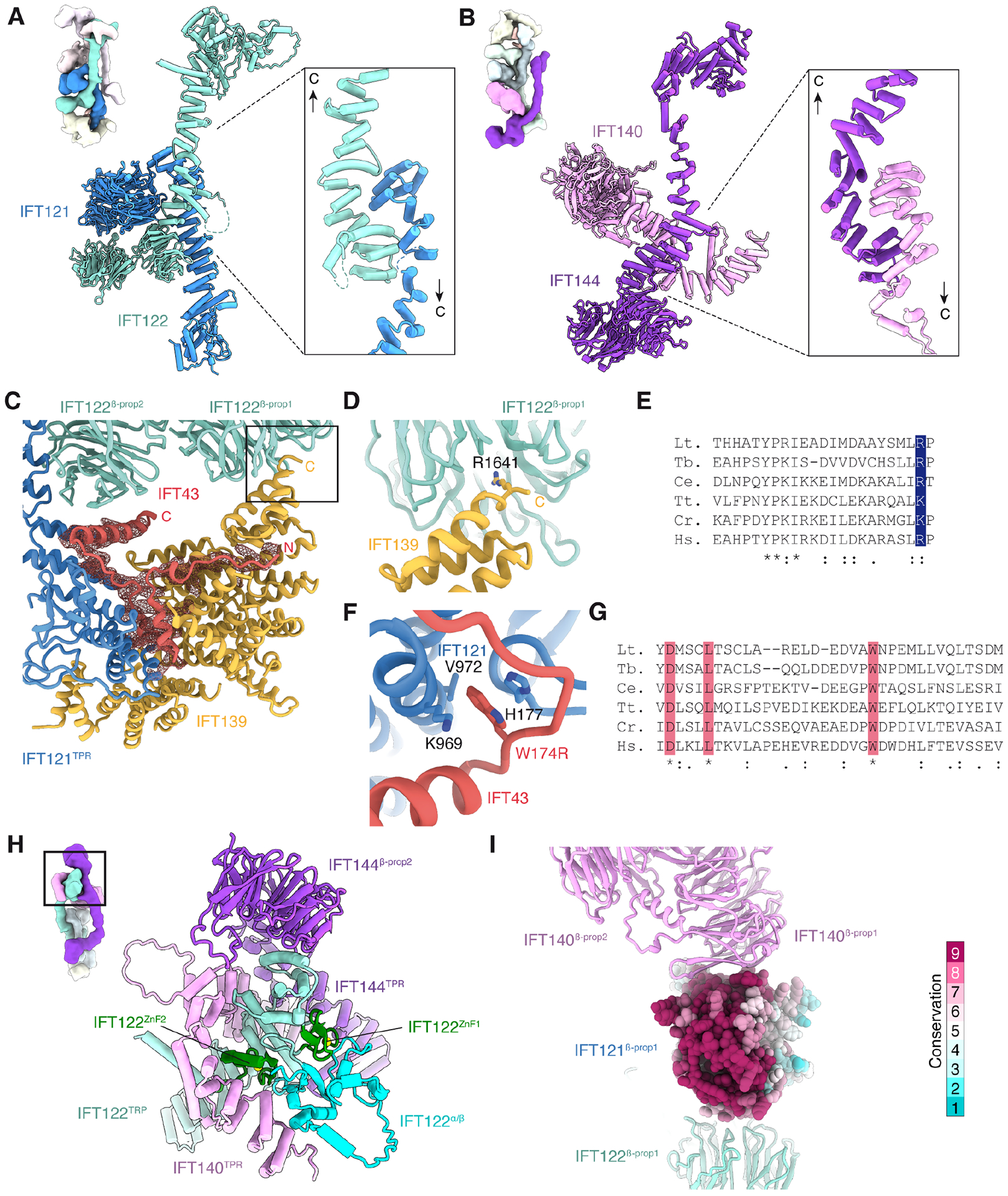
Antiparallel TPR–TPR interactions are responsible for the heterodimerization of (A) IFT121 and IFT122 in the A2 subcomplex and (B) IFT140 and IFT144 in the A1 subcomplex.
C. Atomic model (cartoon) and cryo-EM density (mesh) showing the position of IFT43 mediating the interaction between IFT139 and IFT121TPR.
D. The penultimate residue of IFT139 (R1641) inserts into the center of IFT122β-prop1.
E. A basic residue in the ultimate or penultimate position (highlighted in blue) is a conserved feature of IFT139 sequences from Leishmania tarentolae (Lt.), Trypanosoma brucei (Tb.), Caenorhabditis elegans (Ce.), Tetrahymena thermophila (Tt.), Chlamydomonas reinhardtii (Cr.), and Homo sapiens (Hs.).
F. A model of the interaction between human IFT43 and IFT121 based on the atomic model of Lt. IFT-A. IFT43 W174, which is mutated to an arginine in skeletal ciliopathies46, inserts into a conserved pocket lined by H177, K969 and V972 (human residue numbering).
G. Sequence alignment of the conserved region of IFT43. Invariant residues, including W174, are highlighted in red and marked with an asterisk. Conserved residues are marked with one or two dots.
H. Organization of domains within the apex of the V-shaped A1 subcomplex.
I. IFT140β-prop1 binds to a conserved surface region of IFT121β-prop1.
IFT139, at the bottom of the A2 complex, interacts with IFT122β-prop2 and the C-terminal TPRs of IFT121 (Fig. 2C). The interaction with IFT122β-prop2 involves the insertion of the positively charged sidechain of the penultimate residue of IFT139, R1642, into the center of the β-propeller (Fig. 2D). This interaction is likely highly conserved, as a basic residue occurs at or near the end of most IFT139 sequences (Fig. 2E). The interaction between IFT139 and IFT121TPR is stabilized by IFT43, which contributes short α-helices to the interface (Fig. 2C). The mediatory position of IFT43 explains why Chlamydomonas ift43 mutants have barely detectable levels of IFT13945. In humans, a mutation (W174R) within the IFT121–IFT43 interface (Fig. 2F) is associated with a skeletal ciliopathy46, demonstrating the importance of this interaction for proper IFT-A integrity and function. Notably, only the conserved region of IFT43 is resolved in our maps (Fig. 2G). The N-terminus, which is dispensable for ciliogenesis45, is predicted to be unstructured.
The subcomplexes are connected by IFT122TPR, which extends from the A2 subcomplex to engage with the TPR domains of IFT140 and IFT144 at the apex of the V-shaped A1 subcomplex (Fig. 2H). This interaction explains why IFT122 can form a stable complex with IFT140/144 in a Chlamydomonas mutant lacking IFT12142 and in human cell lines overexpressing fluorescently tagged IFT subunits47. To address whether IFT122 would segregate with either the A1 or the A2 subcomplex in their separated forms, we resolved a 3.8-Å resolution structure of the A2 subcomplex alone, which revealed IFT122 together with IFT43/121/139 (Fig. 1C and Fig. S1–2). In this subcomplex, IFT122TPR is not resolved, indicating that the connecting bridge is flexible in the absence of the A1 subcomplex. The isolated A1 subcomplex therefore likely consists of just IFT140/144, consistent with their ability to co-immunoprecipitate in Chlamydomonas ift122 mutants42.
In addition to IFT122TPR, the A1 and A2 subcomplexes are associated by an inter-propeller interaction between IFT140β-prop1 and a conserved surface of IFT121β-prop1 (Fig. 2I). Another, more labile connection (based on the relative weakness of the density) involves the C-terminus of IFT144, which extends from the A1 subcomplex to loosely contact IFT121TPR in the A2 subcomplex (Fig. 1D). As described later, both these interactions change when IFT-A polymerizes into anterograde trains. The C-terminus of IFT140TPR has no discernable density, indicative of extreme flexibility.
IFT121, IFT122 and IFT144 all terminate with two Cys4 zinc-finger (ZnF) domains, suggesting they share a common evolutionary origin (Fig. S5). These ZnF domains are highly conserved, although not all species have two ZnF domains per subunit; human IFT121, for example, lacks the first ZnF domain (Fig. S5A). In IFT122, the ZnF domains are separated by a small mixed α/β domain of unknown function (Fig. 2H and Fig. S5B). Loss of a Zn-coordinating cysteine (C1267Y) in human IFT144 is associated with short-rib thoracic dysplasia48, demonstrating that correct folding of the ZnF domain is required for proper IFT-A function. Proteins with similar WD40–TPR architectures were also found to terminate in ZnFs in the membrane-binding GATOR2 complex49 where they mediate heterodimerization. There is no evidence from our structure to support a similar dimerization function for the ZnF domains of IFT-A.
An atomic model of IFT-A polymerized on an anterograde IFT train
To determine how IFT-A polymerizes during IFT, we docked our atomic model into subtomogram averages of IFT-A from anterograde trains19,40, the best of which has a resolution of 23 Å40 (Fig. 3). The A2 subcomplex can be fitted unambiguously due to its distinctive arrangement of 4 β-propellers (two each from IFT121 and IFT122) and a large α-solenoid (IFT139) (Fig. 3B–C). Unexpectedly, the A1 subcomplex shows a substantial conformational rearrangement as a result of polymerization (Fig. 3D). It has rotated relative to A2, breaking the inter-propeller contacts between IFT121β-prop1 and IFT140β-prop1 (Video S1). The TPR domain of IFT122 straightens, and potentially rigidifies based on the strength of the density, in response to the rotation (Fig. 3D, inset). The liberated IFT140 rotates to contact IFT121β-prop2 of its distal neighbor. Despite this movement, the distance between the splayed β-propeller domains of IFT140/144 remains largely unchanged (~50 Å), consistent with a rigid-body motion. By generating lateral contacts between neighboring molecules, the swiveling of the A1 domain is a crucial step in IFT-A polymerization.
Figure 3. Organization of polymerized IFT-A in anterograde intraflagellar transport (IFT) trains.
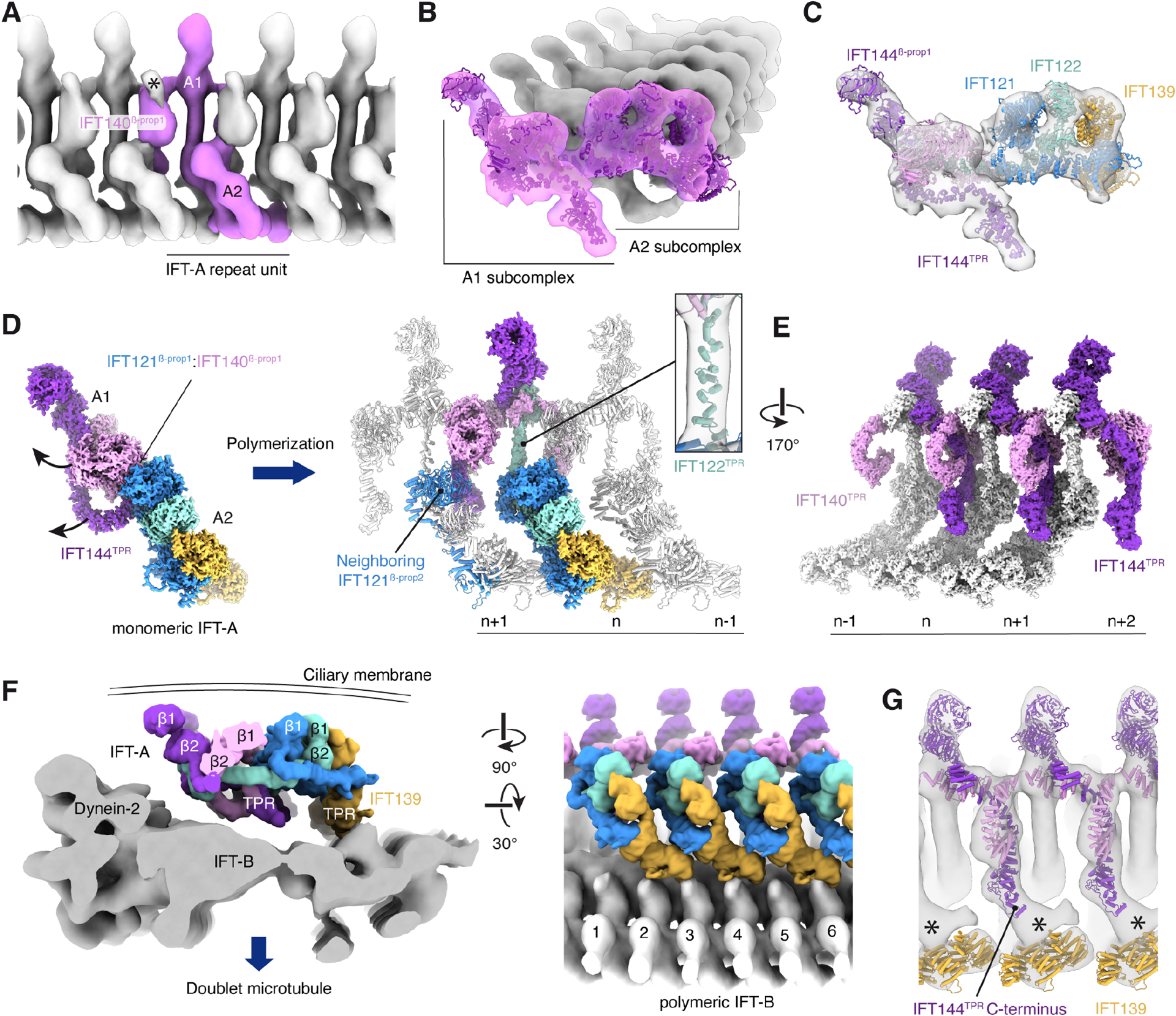
A. A composite subtomogram average (generated by merging repeating copies of EMD-2679140) showing C. reinhardtii IFT-A polymerized on an anterograde IFT train and viewed from the ciliary membrane. The repeat unit corresponding to a single IFT-A monomer is colored pink. Additional density (marked with an asterisk) not accounted for by our model is observed above the IFT140β-prop1 domain.
B. View from the distal end of a train showing the atomic model of C. reinhardtii fit into the subtomogram average.
C. IFT-A, as in panel B, but with each subunit colored.
D. The A1 subcomplex reorients from its position in the monomeric complex. The arrows show the direction of movement. Rotation of the A1 subcomplex causes the IFT121β-prop1:IFT140β-prop1 interaction to break, and IFT140β-prop1 to interact with IFT121β-prop2 of the neighboring complex. IFT122TPR straightens in response to these subcomplex rearrangements (inset).
E. Arrangement of IFT140TPR and IFT144TPR on the underside of the IFT-A complex (viewed from IFT-B). IFT144TPR interacts with the neighboring molecule where it is bound by IFT140TPR.
F. Two views showing the position of IFT-A (in surface representation) relative to the subtomogram average of IFT-B (EMD-15261)18. IFT-A is predominantly tethered to IFT-B by the IFT139 subunit.
G. View of IFT-A from IFT-B showing atomic models of IFT139/140/144 (the other subunits are hidden for clarity). Additional density (marked with an asterisk) is seen between the C-terminus of IFT144TPR and IFT139.
We also observed large changes in the positions of the TPR domains of IFT140 and IFT144 on the underside of the IFT-A complex. The C-terminus of IFT144TPR is released from its loosely bound position on IFT121TPR to engage IFT121β-prop2 of its distal neighbor (Fig. 3D–E). This appears to induce ordering of the C-terminal region of IFT140TPR, which curls to interact with IFT144TPR in its new position (Fig. 3E). It should be noted that these positions are tentative as accurate modeling of the TPRs was challenging due to the 23 Å resolution of the subtomogram average. The position of IFT144 in the train may explain some of the outlier crosslinks that were incompatible with the model of monomeric IFT-A (Fig. S4). TPR–TPR interactions also occur between IFT139 and IFT121 from neighboring complexes. In order for these interactions to occur, IFT139 has rotated further from its position in either state 1 or state 2. The need for IFT139 to be dynamic may explain why we observed multiple states. In conclusion, collective cooperativity provided by multiple lateral interactions explain why IFT-A polymerizes in linear arrays on IFT-B rather than binds sporadically18.
To visualize how these IFT-A polymers are oriented on IFT trains, we next docked our atomic model into a composite map of an IFT train assembled at the ciliary base prior to anterograde transport (EMD-15261)18 (Fig. 3F). This revealed that IFT-A adopts a slanted configuration on the train relative to IFT-B with a footprint covering approximately 3 IFT-B complexes. IFT-A is tethered to IFT-B through unassigned densities either side of the IFT139 subunit (Fig. 3G). Candidate IFT-B subunits that might mediate these contacts include IFT70, IFT88, and IFT172 which crosslink with IFT-A40, and IFT74/81, which integrative modeling with AlphaFold2 predictions have proposed to be positioned near IFT-A29. Interestingly, these five proteins are the top five IFT-B subunits that copurify with L. tarentolae IFT-A (Table S1A).
The largest unassigned density is between the C-terminus of IFT144TPR and IFT139 (Fig. 3G). A crucial role for the C-terminus of IFT144 in binding IFT-B would agree with recent biochemical evidence from IFT144 truncations50,51. The C-terminus of IFT172 has been proposed to bridge IFT144 and IFT139 on anterograde trains29, potentially explaining this unassigned density. The link with IFT172 could explain why mutations in IFT17252,53 phenocopy those caused by IFT-A mutants (Table S3).
A consequence of the orientation of IFT-A on the IFT-B polymer is that all six β-propeller domains are on the upper face, closest to the ciliary membrane and all TPR domains face the IFT-B complex (Fig. 3F). The BBSome is also thought to position multiple β-propeller domains toward the ciliary membrane32,33. In the subtomogram average of anterograde IFT-A40, we observed additional density above the interface between IFT140β-prop1 and IFT140β-prop2 that cannot be explained by our atomic model (Fig. 3A). This density may correspond to a cargo or cargo adaptor that is present in vivo but absent from the purified IFT-A used for cryo-EM analysis. A role for the IFT140 β-propeller domains in cargo binding would be consistent with loss of ciliary localization of GTPases, lipid-anchored proteins, and cell signaling proteins in Chlamydomonas mutant strains lacking the IFT140 β-propeller domains54.
Orientation of IFT-A on membranes
The proposed evolutionary relationship between IFT-A and vesicle-binding coatomers and tethers20,21 and its membrane-proximal location on IFT trains19 support a direct interaction between IFT-A and membranes. Previous work with a recombinant Chlamydomonas IFT-A subcomplex (IFT43/121/139) had identified preferential binding to anionic phosphoglycerates23. To test if native L. tarentolae IFT-A has similar lipid specificity, we used a lipid-protein overlay assay (Fig. 4A). Of the 15 lipids tested, only phosphatidic acid (PA) and 3-sulfogalactosylceramide showed binding to IFT-A. Ceramide binding is consistent with the ability of ceramide to pull-down IFT complexes from Chlamydomonas flagella55. Because PA was also the dominant interaction detected for Chlamydomonas IFT43/121/139 subcomplexes23, we next tested whether IFT-A could bind PA-containing liposomes. Negative-stain electron microscopy showed partial decoration of the liposome surface but not the formation of cage-like coats (Fig. 4B). IFT-A did not decorate liposomes formed without PA (Fig. S6). Furthermore, two-dimensional class averaging revealed a consistent mode of attachment of IFT-A to the liposome surface, in which the curvature of the IFT-A complex complements the convex curvature of the liposome (Fig. 4C). By overlaying our atomic model onto these class averages (Fig. 4D), we could determine the approximate surfaces that face, and potentially interact, with the membrane. The closest contact is with the apex of the A1 module, which consists of the entwined TPR domains of IFT122, IFT140 and IFT144 (Fig. 2C). At the other end of the complex, IFT139 approaches the membrane in some but not all cases (Fig. 4B). A role for IFT139 in membrane binding would be consistent with the ability of IFT43/121/139 but not IFT43/121 to bind PA-containing liposomes and PA in lipid overlap assays23. Thus, the preferred membrane binding interface of the IFT-A complex involves its TPR domains. The ability of TPR domains to bind lipids is supported by a recent crystal structure of the TPR domain of a mitochondrial protein, PTPIP51, bound to PA56. Surprisingly, the same TPR surfaces face, and even engage with, IFT-B in anterograde trains rather than the ciliary membrane (Fig. 3F), rendering these two binding modes incompatible.
Figure 4. IFT-A uses TPR domains for membrane attachment.
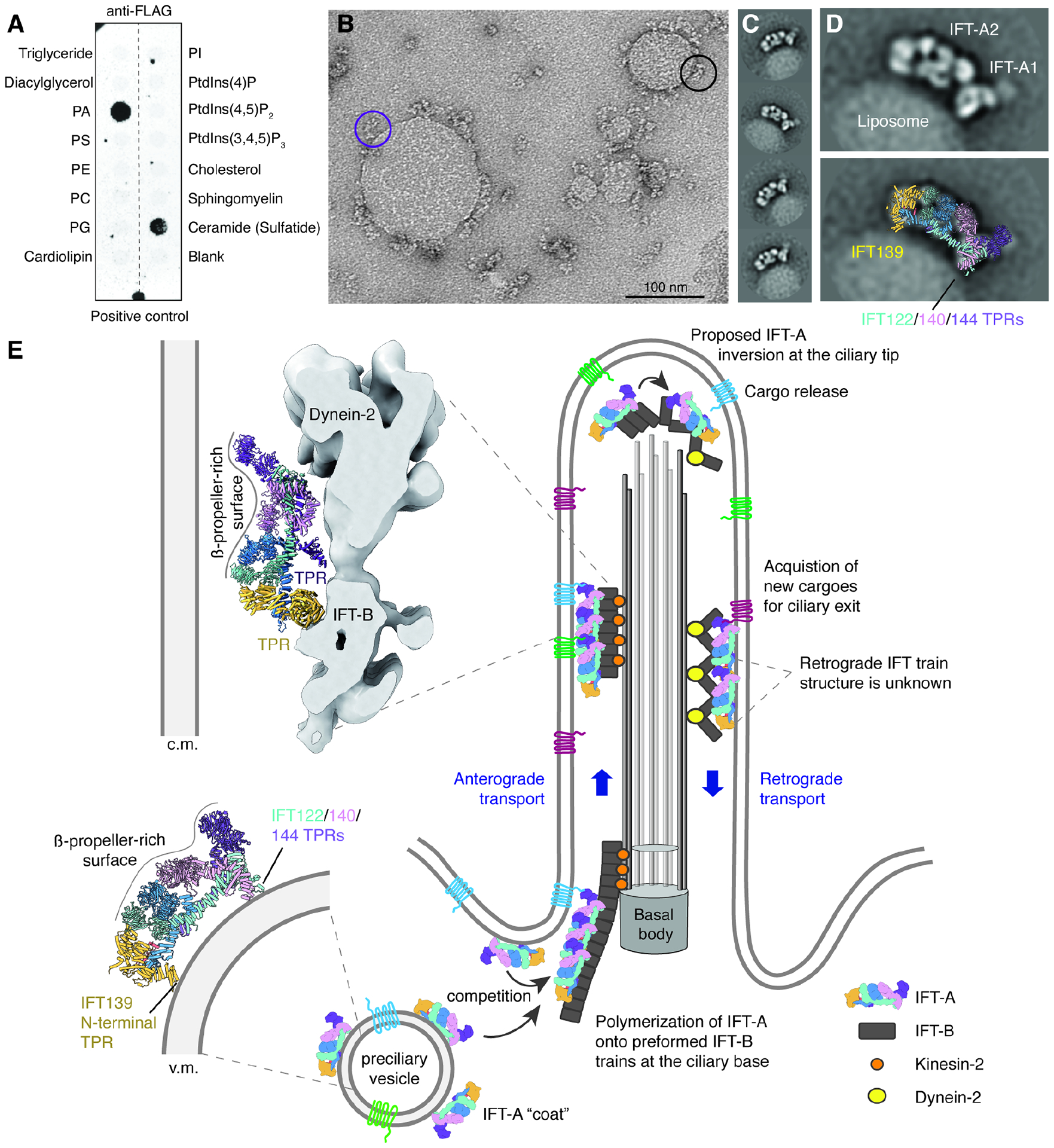
A. Results of a lipid-strip overlay assay showing that IFT-A has specificity for phosphatidic acid (PA) and 3-sulfogalactosylceramide. Identical results were obtained twice. Lipid abbreviations: PS, phosphatidylserine; PE, phosphatidylethanolamine; PC, phosphatidylcholine; PG, phosphatidylglycerol; PI, phosphatidylinositol.
B. A negative-stain electron micrograph showing PA-containing liposomes coated with IFT-A. The purple circle highlights an IFT-A complex tethered to the membrane only through the A1 subcomplex, whereas the black circle highlights an IFT-A complex tethered through its A1 and A2 subcomplexes.
C. Two-dimensional (2D) class averages showing a consistent arrangement on IFT-A on the liposome surface.
D. A 2D class average of a liposome-bound IFT-A (top) overlaid with the atomic model of IFT-A (bottom). Membrane contacts appear to involve the TPRs of IFT139 (yellow) and IFT122/140/144 (teal/pink/purple).
E. A hypothetical model for how IFT-A might convert between different orientations depending on whether it is associated with vesicles or in anterograde or retrograde trains. In this model, IFT-A binds vesicle (v.m) and ciliary membranes (c.m.) during retrograde transport through its TPR domains. These TPR domains are shielded from the membrane during anterograde transport by binding IFT-B.
Ciliopathy-associated IFT-A variants reveal potential cargo-binding sites
A structure of IFT-A allowed us to analyze mutations that cause human ciliopathies. First, we exploited the structural conservation of the IFT-A complex and the predictive power of AlphaFold2 to build an atomic model of human IFT-A. Onto this model, we mapped all missense variants in IFT-A that are annotated as either pathogenic or likely pathogenic in the ClinVar database (Fig. 5A and Table S3). The mutations are dispersed throughout the structure with all six subunits harboring variants. Most mutations occur within the hydrophobic cores of individual subunits, where they may disrupt complex formation by instigating defective folding pathways. A smaller subset, including IFT43 W174R (Fig. 2F), map to intersubunit interfaces, where they could destabilize complex formation. We also observed 18 mutations occurring at surface-exposed sites (Fig. 5B). We speculate that mutation of residues that face the ciliary membrane and occur within conserved regions, such as IFT144 D159 (Fig. 5C), may elicit their deleterious effects by disrupting cargo binding rather than IFT-A complex formation.
Figure 5. Location of disease-causing missense variants.
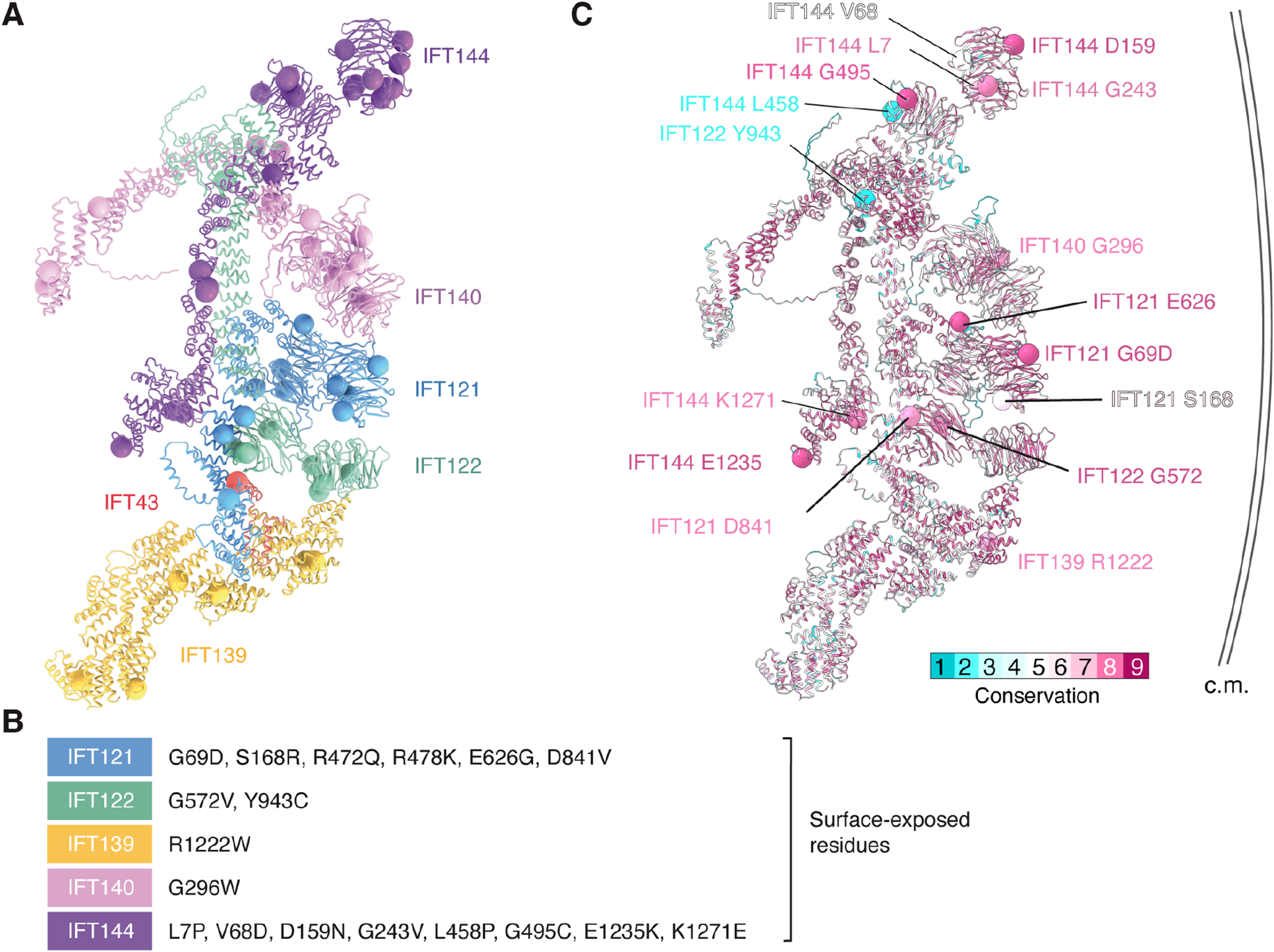
A. Human disease-causing missense variants (Table S3) mapped onto a model of human IFT-A, colored by subunit. Spheres mark the Cα atom of the mutated residue.
B. A list of surface-exposed residues mutated in human disease.
C. The atomic model and surface-facing mutation sites listed in panel B colored by conservation. Conservation scores were calculated using ConSurf61. The side facing right abuts the ciliary membrane (c.m.) in anterograde trains, whereas the surface facing left engages IFT-B and preferentially binds lipids.
DISCUSSION
Intraflagellar transport, essential for ciliogenesis and the signaling capabilities of cilia, requires the formation of megadalton polymeric assemblies that transport cargoes to and from the cilium. Here we have used cryo-EM to determine structures of native L. tarentolae IFT-A complexes. The structures revealed that IFT-A has a bilobal architecture of two subcomplexes, A1 and A2. Although the IFT-A assembly pathway remains to be elucidated, the structure supports a model where IFT121/122 and IFT140/144 independently heterodimerize through antiparallel TPR interactions, before recruiting each other and IFT43/139. Contemporary work reporting the cryo-EM structure of a human IFT-A complex, reconstituted from recombinant proteins, also shows a bilobal architecture57. However, the A1 and A2 subcomplexes are highly flexible relative to one another and are only connected through the IFT122TPR domain. No interaction between IFT140β-prop1 and IFT121β-prop1 was observed. This striking disparity in architecture and dynamics may reflect species-specific differences or a consequence of sample preparation methods (native versus recombinant). In negative-stain electron microscopy of L. tarentolae IFT-A purified without a crosslinker we observed the “closed” state and also less populated “open” states (Fig. S2A,B). A partially open state, in which the IFT140β-prop1 and IFT121β-prop1 are no longer in contact, was also observed as a minor class in our cryo-EM dataset of the crosslinked IFT-A sample (Fig. S2C). From these data, we conclude that native IFT-A exists as an equilibrium between closed the open states. The ability of the IFT140β-prop1 and IFT121β-prop1 interface to open is consistent with our mechanism for oligomerization (Fig. 3), in which this interface has to break to allow the IFT-A1 domain to swivel to contact its neighboring complex.
We show, using in-vitro reconstitution with liposomes, that the fully assembled complex can bind membranes through it TPR domains with specificity for phosphatidic acid and ceramides. Ceramides were shown recently to promote ciliogenesis, regulate retrograde IFT, and pull-down Chlamydomonas IFT subunits55. The ability of IFT-A to bind membranes provides further evidence for a shared evolutionary relationship with coatomers. Although we saw no evidence for IFT-A forming cage-like structures on liposomes, further work will be needed to discover if IFT-A can form ordered vesicle coats in cells.
Surprisingly, we discovered that IFT-A uses a common interface to engage both liposomes and IFT-B, with the N-terminal TPRs of IFT139 integral to both interactions. This contradicted our expectation that IFT-A would engage both membranes and IFT-B through opposing interfaces given that it is sandwiched between the two in anterograde trains. We hypothesize that our membrane-associated form, with lipid contacts mediated by TPR domains, corresponds to IFT-A on preciliary vesicles and/or retrograde trains (Fig. 4E). If our preciliary vesicle hypothesis is correct, competition between binding membranes or IFT-B at the ciliary base might explain how IFT-A transitions from a vesicle- or membrane-bound state to one that engages IFT-B on assembling trains in the cytosol. This is not to say that IFT-A in anterograde trains cannot bind membranes. Membrane binding could still be achieved through a lower affinity interface or through adaptors such as TULP3, which associate with IFT-A during anterograde transport13,57,58 and have affinity for phosphoinositides13.
If our retrograde train hypothesis is correct, it would require IFT-A to invert during the conversion from anterograde to retrograde trains at the ciliary tip (Fig. 4E). Rotation on a hinge provided by the flexible IFT139 subunit could allow inversion to occur without breaking contact with IFT-B, which is thought to be preserved during the transition59. A consequence of the inversion model is that it would physically break interactions with anterograde cargoes, releasing them from futile recycling back to the cytosol. By exposing a different interface to the ciliary membrane, the inverted IFT-A could recognize a different set of cargoes from those delivered to the cilium. A role for IFT-A in binding lipids specifically in retrograde trains may explain why ceramide biosynthesis inhibitors slow retrograde but not anterograde trains in Chlamydomonas flagella55. In-situ structures of IFT-A-coated periciliary vesicles and retrograde IFT trains will be needed to test these hypotheses. How and when IFT-A converts between monomeric, polymeric, and membrane-bound forms has fundamental implications for the assembly and conversion of IFT trains, and the pickup and release of ciliary cargoes.
Implications for cargo binding
Our structures and structural analysis suggest that both the outer edges and upper surface of the β-propeller domains may be involved in cargo recognition, and that the β-propeller domains of IFT140 and IFT144 in the IFT-A1 subcomplex may be particularly important. First, surface-exposed residues that are mutated in ciliopathies, and which may correspond to cargo binding sites, map to both the edges and upper surfaces of the β-propeller domains (Fig. 5). Second, density in the subtomogram average of an anterograde IFT-A train that cannot be explained by our model (Fig. 3A) is associated with the edges of the IFT140 β-propeller domains. This region includes G296, one of residues mutated in short-rib thoracic dysplasia (Table S3). Although we cannot identity the origin of this additional density, it indicates that factors may be able to bind this interface. Finally, a role for IFT140 in cargo binding is consistent with biochemical evidence showing a direct association between IFT140 and the ciliary targeting sequence of the ciliary G-protein coupled receptor, somatostatin receptor 3 (SSTR3)16.
A recent study has mapped the TULP3 binding site on human IFT-A to an acidic patch on the outside of IFT122 ZnF domain57. Using AlphaFold2 multimer, this region was predicted to bind the N-terminal helix of TULP3, which had previously been shown to be required for IFT-A binding13. Mutation of five conserved IFT122 residues within this interface, including charge reversal of three acidic residues, abolished TULP3 binding without disrupting the incorporation of IFT122 into IFT-A57. Binding to the IFT122 ZnF domain would place the phosphoinositide-binding TUBBY domain of TULP313 in the vicinity of the A1 subcomplex β-propeller domains on the end of a flexible tether. Our structure of the L. tarentolae IFT-A demonstrates that the putative binding site for the TULP3 N-terminal helix is conserved, including the acidic residues that are proposed to interact electrostatically with the basic residues of the helix. However, the L. tarentolae genome does not encode an obvious homolog of TULP3, and no TUBBY domain-containing proteins copurified with IFT-A. Whether L. tarentolae has an adaptor equivalent of TULP3 awaits investigation.
Limitations of the study
Here, we have shown that IFT-A displays lipid specificity and can partially coat liposomes in vitro, consistent with the in-vivo visualization of IFT121-dependent densely coated periciliary vesicles in mammalian cells23. However, further work will be needed to demonstrate exactly how IFT-A binds native vesicles, and whether it co-migrates with IFT-B during vesicular trafficking. Are they delivered to the ciliary base separately or as a preformed complex? How IFT-A transitions from its vesicle-bound form to the IFT train also needs further clarification. Does uncoating require membrane fusion, or is uncoating a prerequisite for membrane fusion? To aid these studies, our high-resolution structures will provide templates to identify IFT-A at different stages of trafficking in electron micrographs and tomograms60. While we show that lateral polymerization of IFT-A into IFT trains requires large subcomplex rearrangements, whether IFT-B polymerization follows similar principles will require structures of IFT-B before train formation. The ability to determine structures of native IFT complexes provides a viable avenue to achieve this aim. Similarly, although our structures reveal potential cargo-binding sites, the molecular basis for substrate recognition will require structures of co-complexes with membrane-protein cargoes and their adaptors.
STAR★Methods
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Alan Brown (alan_brown@hms.harvard.edu).
Materials availability
A pLEXSY-hyg2.1 plasmid expressing Flag-tagged IFT43 has been deposited with Addgene (https://www.addgene.org/) with ID #194433.
Data and Code Availability
Composite cryo-EM maps of L. tarentolae IFT-A state 1 and 2 have deposited in the Electron Microscopy Data Bank (EMDB) with accession numbers EMD-28866 and EMD-28867. Half maps and masks are deposited as additional maps associated with these entries. Atomic models of IFT-A state 1 and state 2 have been deposited in the Protein Data Bank (PDB) with accession numbers 8F5O and 8F5P.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Leishmania tarentolae
Leishmania tarentolae strain P10 (Jena Bioscience, #LT-101) cells were grown in BHI medium (HIMEDIA, #N210) containing 5 μg/mL hemin chloride (Sigma, #3741) and 10 U/mL Penicillin-Streptomycin (Gibco, #15070063) at 26°C in the dark. Cells were maintained as static suspension cultures as described in the LEXSY expression kit manual (Jena Bioscience).
METHOD DETAILS
Sample preparation for thin-section transmission electron microscopy
Cells in late logarithmic phase were diluted 1:10 and left overnight. The next day, the cells were fixed by a 5-min incubation with microscopy-grade glutaraldehyde at a final concentration of 2.5%. The cells were collected by centrifugation at 800 × g for 5 min. The pellet was resuspended in 1.5 mL fresh BHI medium without hemin and antibiotics. To this, an equal volume of fixation reagent containing 2% formaldehyde, 2.5% glutaraldehyde and 0.3% picric acid in 0.1 M sodium cacodylate buffer (pH 7.4) was added and left for 1 hr on a rocking platform. 200 μL of cell suspension (containing ~20–50 million cells) was then loaded onto 12.7 mm Aclar coverslips (Electron Microscopy Sciences, #50425–25) precoated with poly-L-lysine (Sigma-Aldrich, #P8920) and allowed to settle for 30 min. Excess liquid was aspirated, and the coverslip was spun at 4680 × g on a plate spinner for 3 min. The coverslip was then washed three times with 0.1 M sodium cacodylate buffer followed by fixation with 1% osmium tetroxide. After rinsing with 50 mM maleate buffer (pH 5.2), the sample was further incubated with 1% aqueous uranyl acetate for 1 hr at room temperature. The fixed sample was dehydrated by washing with escalating ethanol concentrations (70, 90 and 100%). Following dehydration, the sample was treated with propyleneoxide for 1 hr, infiltrated with epoxy resin and finally embedded in freshly mixed Epon. 50 nm sections were collected from the Epon blocks and mounted on the specimen grid. For contrast, the grid was stained with lead citrate and uranyl acetate. The images were collected on a 120 kV Tecnai T12 microscope (Thermo Fisher Scientific).
Construct design
To constitutively express IFT43-FLAG in L. tarentolae from a genome-integrated position, we modified expression vector pLEXSY-hyg2.1 (Jena Bioscience, #EGE-1310hyg) to insert a linker, 3X FLAG peptide sequence, and a XbaI restriction site into the KpnI site of the vector. The L. tarentolae gene encoding IFT43 was amplified from genomic DNA using primer oligonucleotides (Key Resources Table) and Phusion High-Fidelity DNA polymerase (Thermo Fisher Scientific, #F-530S). The amplified gene was inserted into the modified pLEXSY-hyg2.1 vector between the 5´ BglII or NcoI and 3´ XbaI sites. The ligated vector was transformed into E. coli DH5α cells and plated onto agar plates containing ampicillin. Bacterial colonies were screened by colony PCR using the gene-specific primers (Key Resources Table). The plasmids were purified and sequence verified before transfection.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rat anti-FLAG antibody | BioLegend | Cat #637301; RRID:AB_1134266 |
| Horseradish peroxidase-conjugated anti-rat secondary antibody | Cell Signaling Technology | Cat #7077S |
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| 1,2-dipalmitoyl-sn-glycero-3-phosphate (PA) | Avanti Polar Lipids | Cat #830855P |
| 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE) | Avanti Polar Lipids | Cat #850757 |
| 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1’-rac-glycerol) (POPG( | Avanti Polar Lipids | Cat #840457 |
| 3,3’-dithiobis(sulfosuccinimidyl propionate) (DTSSP) | Thermo Fisher Scientific | Cat #21578 |
| 3X FLAG peptide | Pepmic | Custom synthesis, no identifier |
| Anti-FLAG M2 resin | Sigma-Aldrich | Cat #A2220 |
| BHI medium | HIMEDIA | Cat #N210 |
| Hemin chloride | Sigma | Cat #3741 |
| Penicillin-Streptomycin | Gibco | Cat #15070063 |
| Poly-L-lysine | Sigma-Aldrich | Cat #P8920 |
| Phusion High-Fidelity DNA polymerase | Thermo Fisher Scientific | Cat #F-530S |
| Polycarbonate membrane | Avanti Polar Lipids | Cat #610005 |
| Trypsin | Promega | Cat #90057 |
| Critical Commercial Assays | ||
| Novex ECL chemiluminescence kit | Thermo Fisher Scientific | Cat #WP20005 |
| Membrane lipid strips | Echelon Bioscience | Cat #P-6002 |
| Deposited Data | ||
| Subtomogram average of C. reinhardtii IFT-A obtained from anterograde trains | McCafferty et al., 202240 | EMDB: EMD-26791 |
| Subtomogram average of C. reinhardtii IFT-A obtained from anterograde trains | Jordan et al., 201819 | EMDB: EMD-4304 |
| Composite subtomogram average of an assembled C. reinhardtii IFT train at the ciliary transition zone | van den Hoek et al., 202218 | EMDB: EMD-15261 |
| Composite cryo-EM map of Leishmania tarentolae IFT-A state 1 | This paper | EMDB: EMD-28866 |
| Composite cryo-EM map of Leishmania tarentolae IFT-A state 2 | This paper | EMDB: EMD-28867 |
| Atomic model of Leishmania tarentolae IFT-A state 1 | This paper | PDB: PDB 8F5O |
| Atomic model of Leishmania tarentolae IFT-A state 2 | This paper | PDB: PDB 8F5P |
| Chemical crosslinking mass spectrometry data obtained for Tetrahymena thermophila IFT-A | McCafferty et al., 202240 | MassIVE/ProteomeXchange: PXD032818 |
| Experimental Models: Cell Lines | ||
| Experimental Models: Organisms/Strains | ||
| Leishmania tarentolae strain P10 | Jena Bioscience | Cat #LT-101 |
| Bacteria: Escherichia coli DH5α | New England Biolabs | Cat #C2987H |
| Oligonucleotides | ||
|
LtIFT43_Fw1 CCTTGCCACCAGATCTGCCATGTCCACGACTTCTATGACTGCC |
Sigma | N/A |
|
LtIFT43_Rv1 CCGGAGCCTCTAGACTTCGAAACCTGCTTGGATG |
Sigma | N/A |
| Recombinant DNA | ||
| Plasmid: pLEXSY-hyg2.1 | Jena Bioscience | Cat #EGE-1310hyg |
| Plasmid: pLEXSY-hyg2.1_LtIFT43_Flag | This paper | Addgene: Cat #194433 |
| Software and Algorithms | ||
| AlphaFold2 | Jumper et al., 202170 | https://www.deepmind.com/ |
| ChimeraX v1.4 | Pettersen et al., 202169 | https://www.rbvi.ucsf.edu/chimerax/ |
| Clustal Omega v1.2.2 | Sievers et al., 201174 | http://www.clustal.org/omega/ |
| Coot v.0.9.8.3 | Brown et al., 201571 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| cryoSPARC v3.3.2 | Punjani et al., 201766 | https://cryosparc.com/ |
| DeepEMhancer | Sanchez-Garcia et al., 202162 | https://github.com/rsanchezgarc/deepEMhancer |
| Prism v9 | Graphpad Software, USA | https://www.graphpad.com/ |
| phenix.combine_focused_maps v1.18.2–3874 | Liebschner et al, 201968 | https://www.phenix-online.org/ |
| phenix.molprobity v1.18.2–3874 | Chen et al., 201073 | https://www.phenix-online.org/ |
| phenix.real_space_refine v1.18.2–3874 | Afonine et al., 201872 | https://www.phenix-online.org/ |
| RELION 4.0 | Zivanov et al., 201867 | https://www3.mrc-lmb.cam.ac.uk/relion/ |
| SBGrid | Morin et al., 201365 | https://sbgrid.org/ |
| SerialEM v.3.8.5 | Schorb et al., 201964 | https://bio3d.colorado.edu/SerialEM/ |
| SEQUEST | Thermo Fisher Scientific | https://www.thermofisher.com |
| Other | ||
| Aclar coverslips | Electron Microscopy Sciences | Cat #50425–25 |
| Carbon-coated copper grids | Electron Microscopy Sciences | Cat #CF200-Cu |
| R2/1, 200 mesh gold, QUANTIFOIL grids | Electron Microscopy Sciences | Cat #220210 |
Transfection
For transfection, L. tarentolae cell density was adjusted to 108 cells/mL, followed by incubation on ice for 10 min. 380 μL of this culture was mixed with 2 μg of linearized (SwaI digested and gel purified) plasmid. This mixture was immediately transferred to a pre-cooled electroporation cuvette (d=2 mm) and electroporated using a Gene Pulser Xcell Eukaryotic System (Bio-Rad) using time constant protocol at 450 V for 3.5 ms. The cuvette was then put back on ice for 10 min. The cells were allowed to recover overnight in fresh BHI-hemin medium as a static suspension culture. The cells were harvested by centrifugation at 2,000 × g for 3 min, resuspended in 200 μL and plated onto BHI agar containing 100 μg/mL hygromycin. The plate was incubated for 4–5 days until colonies were visible. Individual colonies were transferred to 150 μL BHI-hemin medium in a 96-well plate. For protein production, the cells were grown at scale as agitated suspension cultures.
IFT-A purification (with crosslinker)
L. tarentolae cells from 32 L culture volumes were harvested by centrifugation at 5,422 × g for 10 min. The cell pellets were resuspended in ~450 mL ice-cold lysis buffer (35 mM HEPES pH 7.4, 100 mM KCl, 5 mM MgSO4, 1 mM CaCl2, 1 mM DTT, Protease inhibitor cocktail, 20 mM NaF, 50 mM β-glycerophosphate, 1% Triton X100, and DNAse) and incubated at 4°C for 15 min with continuous stirring. The lysate was centrifuged at 42,510 × g for 30 min. The clarified lysate was applied to 4 mL anti-FLAG M2 resin (Sigma-Aldrich, #A2220) preequilibrated with wash buffer (35 mM HEPES pH 7.4, 100 mM KCl, 5 mM MgSO4, 1 mM CaCl2, 1 mM DTT). The column was washed with 200 mL wash buffer and eluted with wash buffer containing 100 μg/mL 3X FLAG peptide (Pepmic). The eluate was concentrated using a 15 mL concentrator with a 100 kDa cutoff Amicon Ultra filter (Millipore) to less than 1 mL and loaded onto a 13 mL 10–40% continuous sucrose gradient prepared in wash buffer with a gradient maker (Gradient Master, BioComp). Crosslinking reagent 3,3’-dithiobis(sulfosuccinimidyl propionate) (DTSSP; Thermo Fisher Scientific, #21578) was added to the 40% sucrose solution to a final concentration of 1 mM prior to making the gradient. The sample was centrifuged at 200,000 × g for 16 hrs using a SW40Ti rotor in an Optima LE-80K ultracentrifuge (Beckman Coutler) at 4°C. The gradient was fractionated into 600 μL aliquots with the crosslinking reaction quenched by adding 40 mM Tris pH 7.4. The fractions containing the complex were pooled and concentrated with a 4 mL Amicon Ultra concentrator with a 100 kDa cutoff (Millipore). The sample was loaded onto a MonoQ5/50 GL column (Cytiva, #17516601) preequilibrated with buffer A (20 mM HEPES pH 7.4, 5 mM MgSO4, 1 mM CaCl2, 50 mM KCl, and 1 mM DTT) and eluted using a gradient up to buffer B (20 mM HEPES pH 7.4, 5 mM MgSO4, 1 mM CaCl2, 1 M KCl, and 1 mM DTT). Peak fractions were analyzed by SDS-PAGE, mass spectrometry, and negative-stain electron microscopy.
IFT-A purification (without detergent or crosslinker)
Cell pellets were resuspended in ~300 mL ice-cold lysis buffer without detergent (35 mM HEPES pH 7.4, 100 mM KCl, 5 mM MgSO4, 1 mM CaCl2, 1 mM DTT, Protease inhibitor cocktail, 20 mM NaF, 50 mM β-glycerophosphate, 8% sucrose, and DNAse). The cells were lysed with a high-pressure homogenizer-EmulsiFlex-C3 (Avestin) with an applied pressure of 50–60 psi. The lysate was centrifuged at 42,510 × g for 30 min. The clarified lysate was applied to, and eluted from, a 4 mL anti-FLAG M2 resin as described above. The eluate was concentrated to ~0.5 mL and loaded onto Superpose 6 Increase 10/300 GL column (Cytiva, #29091596) preequilibrated in 20 mM HEPES pH 7.4, 100 mM KCl, 5 mM MgSO4, 1 mM CaCl2, and 1 mM DTT. Peak fractions were analyzed immediately by negative-stain electron microscopy.
Mass spectrometry
Mass spectrometry was performed at the Taplin Mass Spectrometry Facility at Harvard Medical School. In brief, the IFT-A samples were provided to the facility as dehydrated SDS-PAGE gel pieces. The gel pieces were rehydrated with 50 mM ammonium bicarbonate solution containing 12.5 ng/mL trypsin (Promega, Cat. #90057). After 45 min at 4°C, the trypsin solution was replaced with 50 mM ammonium bicarbonate solution and left at 37°C overnight. Peptides were extracted by removing the ammonium bicarbonate solution, followed by a wash with a solution containing 50% acetonitrile and 1% formic acid. The extracts were then dried using a vacuum concentrator for one hour and stored at 4°C. For mass spectrometry, the samples were reconstituted in 5–10 mL of solvent A (2.5% acetonitrile, 0.1% formic acid) and loaded onto a pre-equilibrated reverse-phase capillary column (100 mm inner diameter, and ~30 cm length) containing 2.6 mm C18 spherical silica beads using a Famos auto sampler (LC Packings). A gradient was formed, and peptides were eluted with increasing concentrations of solvent B (97.5% acetonitrile, 0.1% formic acid). As peptides eluted, they were subjected to electrospray ionization and entered into an LTQ Orbitrap Velos Pro ion-trap mass spectrometer (Thermo Fisher Scientific). Peptides were detected, isolated, and fragmented to produce a tandem mass spectrum of fragment ions for each peptide. Protein identity was determined using Sequest (Thermo Fisher Scientific). The data were filtered to between a one and two percent peptide false discovery rate.
Lipid-binding assays
Membrane lipid strips (Echelon Bioscience, #P-6002) were incubated with 5 mL Tris-buffered saline (TBS) containing 3% bovine serum albumin (BSA) overnight on an orbital shaker at 4°C. The next day, the buffer was renewed, and the strip incubated for another hour. The strip was then incubated with a 5 mL solution of ~0.05 mg/mL IFT-A in buffer containing 20 mM HEPES pH 7.4, 5 mM MgSO4, 1 mM CaCl2, 50 mM KCl, and 1 mM DTT at room temperature with constant shaking. After 2 hrs, the protein solution was discarded, and the strip washed twice (5 min per wash) with a total of 10 mL blocking buffer (TBS with 0.001% tween-20 (TBST) and 5% milk). The strip was then incubated with 5 mL blocking buffer containing a 1:500 dilution of anti-FLAG antibody (BioLegend, #637301) overnight with constant shaking at 4°C. Next, the strip was washed three times with 10 mL TBST for a total of 30 min before incubating with a horseradish peroxidase-conjugated anti-rat secondary antibody (Cell Signaling Technology, #7077S) for 1 hr at room temperature. Finally, the strip was washed three times with TBST and once with TBS (10 mL per wash, each for 10 min). Signal was detected using the Novex ECL chemiluminescence kit (Thermo Fisher Scientific, #WP20005).
Liposome-binding assay
Liposomes were generated using a lipid composition (11.4% PA, 63.2% POPE, 25.4% POPG) that replicates the C. reinhardtii ciliary membrane23,63. 1,2-dipalmitoyl-sn-glycero-3-phosphate (16:0 PA, #830855P), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (16:0–18:1 POPE, #850757) and 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1’-rac-glycerol) (16:1–18:1 POPG, #840457) were purchased from Avanti Polar Lipids. Liposomes were prepared using thin-film hydration followed by extrusion. Briefly, lipid stock solutions in chloroform were mixed to get 100 nM total lipid in the desired composition in a glass tube. The mixture was dried using N2 gas to form a thin layer followed by drying overnight in a vacuum desiccator. The dried film was hydrated with 1 mL buffer containing 30 mM HEPES pH 7.4 and 100 mM KCl. The buffer was first warmed to the phase transition temperature of PA (65°C) to help liposome formation. The solution was sonicated in a water bath sonicator for 5 min. The liposome solution was then extruded 50–100 times through a 100 nM polycarbonate membrane (Avanti Polar Lipids, #610005). Liposomes containing POPE (63.2%) and POPG (36.8%) were prepared using same method. Purified IFT-A was mixed with liposomes in molar ratios of 10:1, 20:1, 40:1 and 60:1 and incubated 30 on ice and analyzed by negative stain electron microscopy.
Negative-stain electron microscopy
Carbon-coated copper grids (Electron Microscopy Sciences, #CF200-Cu) were glow discharged at 30 mA for 30 s. A 4 μl aliquot of IFT-A sample (0.02 mg/mL) was applied to the grid. After incubating for 1 min, the grid was washed twice with 10 mM HEPES pH 7.4 and twice with 1.5% Uranyl formate followed by staining with 1.5% Uranyl formate for 2 min. The grids were imaged using either a 100 kV CM10 (Philips) or a 120 kV Tecnai T12 (Thermo Fisher Scientific) microscope. Images were recorded using an Ultrascan 895 CCD camera (Gatan).
Cryo-grid preparation
Cryo-EM grids of IFT-A were prepared using a Vitrobot Mark IV (Thermo Fisher Scientific). 3 μL aliquots of purified complex at concentrations of 1.1 mg/mL were applied onto glow-discharged 2-nm thick carbon coated QUANTIFOIL grids (R2/1, 200 mesh gold, Electron Microscopy Sciences, #220210). The grids were blotted for 5.5 s with a blot force of 12 and 100% humidity before being plunged into liquid ethane cooled by liquid nitrogen.
Cryo-EM data collection
Images were acquired on a Titan Krios microscope equipped with a BioQuantum K3 Imaging Filter (slit width 20 eV) and a K3 direct electron detector (Gatan) and operating at an acceleration voltage of 300 kV. Images were recorded at a defocus range of −1.5 μm to −2.5 μm with a nominal magnification of 105,000 ×, resulting in a pixel size of 0.83 Å. Each image was dose-fractionated into 50 movie frames with a total exposure time of 2.963 s, resulting in a total dose of ~57.8 electrons per Å2. SerialEM v.3.8.5 was used for data collection64.
Cryo-EM data processing
Software used for cryo-EM data processing and model building were installed and managed by SBGrid65. Data were processed using cryoSPARC v3.3.266 and RELION 4.067. Individual movies were motion corrected in RELION before being exported to cryoSPARC for Patch-based CTF estimation. Curate Exposures was used to select 25,415 good micrographs. Next, we used a template-free blob picker to pick particles on a subset of 5,492 micrographs from which we generated 14 distinct two-dimensional classes with well-resolved features. Using these 2D classes as templates for picking, we obtained 1,203,589 “large” and 696,079 “small” particles from the full dataset. Large particles corresponded to the intact IFT-A complex, whereas the small particles corresponded to individual IFT-A2 subcomplexes. After alignment in cryoSPARC, the large and small particles were transferred to RELION 4.0 for further processing. The large particles were classified with or without alignment into 4 to 6 classes. The classes were then combined into two states that differ in the position of IFT139. State 1 particles (239,280 in total) were reextracted without binning, polished and refined to a final resolution of 4.0 Å. Next, we performed signal subtraction for the A1 and A2 subcomplexes followed by 3D classification. Mask-focused refinement was performed on five local regions (shown in Fig. S1E) to obtain higher-resolution maps. Unbinned state 2 particles (563,466 in total) were polished and refined to a final resolution of 3.6 Å. To improve local resolution, mask-focused refinement was performed on the A2 subcomplex. In addition, mask-focused classification and refinement was performed on the A1 subcomplex and IFT139 to improve the resolution of these dynamic regions.
Mask-focused refinement maps were sharpened by RELION postprocessing and merged into a composite map using PHENIX combine_focused_maps68 with default settings. These composite maps were used for model building, refinement, and analysis. Maps sharpened using DeepEMhancer62 were used to make figures in ChimeraX v1.469.
Model building and refinement
Atomic models of the L. tarentolae IFT-A subunits were predicted using AlphaFold270 and docked into the cryo-EM maps using Coot v.0.9.8.371. The models were then adjusted in Coot to better fit the maps using real-space refinement. Zinc ions were added to the zinc-finger domains. The final model was refined in real space using Phenix72 with Ramachandran and secondary structure restraints applied. Model statistics were calculated with MolProbity73 implemented within Phenix. In general, only protein regions with resolved sidechains or secondary structure are included in the final, deposited models. Flexible loops and domains have been removed including the C-terminal zinc finger domain of IFT144 and the N-terminal TPRs of IFT139. More of the IFT139 N-terminus could be modeled in state 1 than state 2. An exception was made for IFT144β-prop1, which has relatively weak density due to flexibility but was left in the model.
Atomic models of IFT-A from Tetrahymena thermophila (used for validation against crosslinking data; Fig. S4), Chlamydomonas reinhardtii (used for modeling IFT-A in anterograde trains; Fig. 3) and Homo sapiens (used for interpreting human mutation data; Fig. 2F and Fig. 5) were built by superposing sections of AlphaFold2 models onto the atomic model of L. tarentolae IFT-A. Sequence alignments were performed with Clustal Omega v1.2.274.
The atomic model of IFT-A polymerized on IFT trains was generated by docking the homology model of the C. reinhardtii IFT-A into the subtomogram average of IFT-A obtained from anterograde trains (EMD-26791)40. Docking revealed that the segmented density did not correspond with the monomeric unit of IFT-A. We therefore used a lower resolution subtomogram average that contains approximately 3 copies of IFT-A (EMD-4304)19 to proliferate EMD-26791 into a linear array. Docking was guided by the A2 subcomplex, where the shape and curvature allowed unambiguous positioning. The A1 domain was then rotated into position as a rigid body. The IFT140TPR and IFT144TPR domains were then manually adjusted in Coot to fit the density. To resolve the position of IFT-A relative to IFT-B, the atomic model was docked into the composite subtomogram average of an assembled C. reinhardtii IFT train at the ciliary transition zone (EMD-15261)18.
QUANTIFICATION AND STATISTICAL ANALYSIS
Estimations of the resolution of the cryo-EM density maps (reported in Figures S1 and S3) are based on the 0.143 FSC criterion75. All statistical validation performed on the deposited models (PDB: 8F5O and PDB: 8F5P) was done using the PHENIX package73 (Table S2).
Supplementary Material
Figure S1. Purification and structure determination of Leishmania tarentolae IFT-A, Related to Figure 1. A. Left, thin-section transmission electron micrograph showing a L. tarentolae flagellum with an IFT train (marked with an arrowhead) between the axonemal doublet microtubules (DMT) and the ciliary membrane (c.m.). Also indicated is the central apparatus (CA) in the middle of the axoneme. Middle and right, electron micrographs showing the cross-sections of Leishmania flagella with putative IFT trains labeled with arrowheads.
B. Chromatogram showing the IFT-A peak following ion-exchange chromatography.
C. Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) of purified IFT-A.
D. Representative electron micrograph of IFT-A taken under cryogenic conditions.
E. Flow diagram, starting with selected two-dimensional class averages, of the steps taken to obtain structures of IFT-A complexes and the IFT-A2 subcomplex.
Figure S2. Two-dimensional (2D) class averages of Leishmania tarentolae IFT-A, Related to Figure 1. A. Comparison of 2D class averages from negative-stain electron microscopy of IFT-A purified with (+) and without (Δ) crosslinker. In both instances, the class shown is the most abundant class observed.
B. Selected 2D classes of IFT-A purified without crosslinker showing a progressive opening of a gap between the A1 and A2 subdomains.
C. 2D class averages from cryo-EM data showing that subcomplex movement is observed even with crosslinked IFT-A. This movement breaks the interaction between the N-terminal β-propeller domains of IFT121 and IFT140.
Figure S3. Resolution and map quality, Related to Figure 1. A. Fourier shell correlation (FSC) curves for IFT-A state 1 (left), IFT-A state 2 (middle) and IFT-A2 (right). The corresponding maps are shown in Fig. S1 together with their resolution determined using the FSC=0.143 criterion. Map1 corresponds to the consensus refinement before mask-focused classification/refinement.
B. Consensus maps colored by local resolution. Local map quality was improved further by mask-focused refinement.
C. Examples of map density for each of the six IFT-A subunits with landmark residues labeled. Maps were sharpened using DeepEMhancer for visualization purposes only62.
Figure S4. IFT-A structure validation using chemical crosslinking mass spectrometry, Related to Figure 1. A. Atomic model of Tetrahymena thermophila IFT-A, built using the model of Leishmania tarentolae IFT-A as a template. The Cα atoms of residues crosslinked by disuccinimidyl sulfoxide are connected with a line. Green lines indicate crosslinks within a subunit, and purple lines indicate crosslinks between two different subunits. Orange spheres represent residues that form crosslinks that exceed the distance (>45 Å) capable of being crosslinked by disuccinimidyl sulfoxide.
B. Plot of distances between Cα positions of crosslinked residues. Data were plotted using GraphPad Prism v9 (GraphPad Software).
Figure S5. IFT121, IFT122 and IFT144 have conserved C-terminal zinc-finger (ZnF) domains, Related to Figure 2. A. The C-terminus of Leishmania tarentolae (Lt.) IFT121 superposed with the AlphaFold2 model of human (Hs.) IFT121. In humans, the first ZnF domain (ZnF1) is absent but the second (ZnF2) is highly conserved. The Zn ion is shown as a green sphere and the coordinating cysteine residues are shown in stick representation.
B. Superposition of the C-terminal domains of Lt. and Hs. IFT122, each featuring two conserved ZnFs and a small mixed α/β domain.
C. Superposition of the C-termini of Lt. and Hs. IFT144 featuring tandem ZnF domains.
D. Sequence alignment of the regions shown in panels A–C. Conserved residues are marked with an asterisk. Conserved cysteines are further highlighted in yellow. Sequences were obtained from Leishmania tarentolae (Lt.), Trypanosoma brucei (Tb.), Chlamydomonas reinhardtii (Cr.), Tetrahymena thermophila (Tt.), Caenorhabditis elegans (Ce.) and Homo sapiens (Hs.). Accession numbers for the sequences are provided in Table S4.
Figure S6. IFT-A specifically decorates liposomes containing phosphatidic acid (PA), Related to Figure 4. A. Representative negative-stain electron micrograph showing IFT-A particles bound to the surface of liposomes generated with PA.
B. Representative negative-stain electron micrograph of IFT-A incubated with liposomes lacking PA. Most IFT-A particles and liposomes remain separate.
Table S2. Statistics for data collection, data processing, model refinement and validation, Related to Figure 1.
Table S4. Sequence information for IFT-A subunits, Related to STAR Methods. Sequences for L. tarentolae and T. brucei proteins were obtained from TriTrypDB105. Sequences for C. reinhardtii proteins were obtained from Phytozome106. Sequences for C. elegans proteins were obtained from WormBase107. Sequences for T. thermophila and H. sapiens proteins were obtained from UniProt108. These sequences were used for protein sequence alignments and/or to build AlphaFold2 models.
Table S1 (Provided as an Excel file). Mass-spectrometry data, Related to Figure 1. A. List of proteins identified in the elution from an anti-FLAG pulldown of lysate from Leishmania tarentolae expressing FLAG-IFT43. IFT-A subunits are highlighted in yellow and IFT-B subunits in blue. B. List of proteins surviving gradient centrifugation and ion-exchange chromatography. IFT-A subunits are highlighted in yellow. No IFT-B subunits were detected. C. Mass-spectrometry identification of phosphorylated IFT-A residues. Most phosphorylated residues occur in non-conserved surface-exposed loops.
Table S3 (Provided as an Excel file). Mutations in human IFT-A genes associated with ciliopathies, Related to Figure 5. Missense variants annotated as pathogenic or likely pathogenic obtained from the ClinVar database76. References found in the table:46,48,77–104.
Video S1. Polymerization of IFT-A into anterograde IFT trains, Related to Figure 3. Video showing the docking of an atomic model of monomeric C. reinhardtii IFT-A into a subtomogram average of polymeric IFT-A (EMD-26791) and the rotation of the A1 subcomplex that allows IFT-A to polymerize.
Highlights.
Cryo-EM structures of native Leishmania tarentolae IFT-A complexes
Subcomplex rearrangements enable IFT-A to polymerize into lateral IFT trains
IFT-A has lipid specificity and can coat liposomes
Anterograde IFT-A positions cargo-binding beta-propellers towards the ciliary membrane
ACKNOWLEDGMENTS
We thank S. Sterling, R. Walsh, and M. Mayer for assistance with electron microscopy at the Harvard Cryo-EM Center for Structural Biology. We thank R. Tomaino for mass spectrometry analysis, P. Aschauer for advice on preparing liposomes, J.R. Anderson for testing postprocessing strategies, and SBGrid for computing support. M.G. was supported by a Charles A. King Trust Postdoctoral Research Fellowship. A.B. is supported by NIGMS (GM141109 and GM143183), the Smith Family Foundation, and the Pew Charitable Trusts.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Nachury MV, and Mick DU (2019). Establishing and regulating the composition of cilia for signal transduction. Nat. Rev. Mol. Cell Biol 20, 389–405. 10.1038/s41580-019-0116-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taschner M, and Lorentzen E (2016). The Intraflagellar Transport Machinery. Cold Spring Harbor Perspectives in Biology 8, a028092. 10.1101/cshperspect.a028092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reiter JF, and Leroux MR (2017). Genes and molecular pathways underpinning ciliopathies. Nat. Rev. Mol. Cell Biol 18, 533–547. 10.1038/nrm.2017.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kozminski KG, Johnson KA, Forscher P, and Rosenbaum JL (1993). A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc National Acad Sci 90, 5519–5523. 10.1073/pnas.90.12.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pazour GJ, Wilkerson CG, and Witman GB (1998). A dynein light chain is essential for the retrograde particle movement of intraflagellar transport (IFT). J. Cell Biol 141, 979–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stepanek L, and Pigino G (2016). Microtubule doublets are double-track railways for intraflagellar transport trains. Science 352, 721–724. 10.1126/science.aaf4594. [DOI] [PubMed] [Google Scholar]
- 7.Bhogaraju S, Cajanek L, Fort C, Blisnick T, Weber K, Taschner M, Mizuno N, Lamla S, Bastin P, Nigg EA, et al. (2013). Molecular basis of tubulin transport within the cilium by IFT74 and IFT81. Science 341, 1009–1012. 10.1126/science.1240985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Craft JM, Harris JA, Hyman S, Kner P, and Lechtreck KF (2015). Tubulin transport by IFT is upregulated during ciliary growth by a cilium-autonomous mechanism. J. Cell Biol 208, 223–237. 10.1083/jcb.201409036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wren KN, Craft JM, Tritschler D, Schauer A, Patel DK, Smith EF, Porter ME, Kner P, and Lechtreck KF (2013). A differential cargo-loading model of ciliary length regulation by IFT. Curr. Biol 23, 2463–2471. 10.1016/j.cub.2013.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lechtreck KF, Liu Y, Dai J, Alkhofash RA, Butler J, Alford L, and Yang P (2022). Chlamydomonas ARMC2/PF27 is an obligate cargo adapter for intraflagellar transport of radial spokes. elife 11. 10.7554/elife.74993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dai J, Barbieri F, Mitchell DR, and Lechtreck KF (2018). In vivo analysis of outer arm dynein transport reveals cargo-specific intraflagellar transport properties. Mol. Biol. Cell 29, 2553–2565. 10.1091/mbc.e18-05-0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liem KF, Ashe A, He M, Satir P, Moran J, Beier D, Wicking C, and Anderson KV (2012). The IFT-A complex regulates Shh signaling through cilia structure and membrane protein trafficking. J Cell Biol 197, 789–800. 10.1083/jcb.201110049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mukhopadhyay S, Wen X, Chih B, Nelson CD, Lane WS, Scales SJ, and Jackson PK (2010). TULP3 bridges the IFT-A complex and membrane phosphoinositides to promote trafficking of G protein-coupled receptors into primary cilia. Gene Dev 24, 2180–2193. 10.1101/gad.1966210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lechtreck K-F, Johnson EC, Sakai T, Cochran D, Ballif BA, Rush J, Pazour GJ, Ikebe M, and Witman GB (2009). The Chlamydomonas reinhardtii BBSome is an IFT cargo required for export of specific signaling proteins from flagella. J. Cell Biol 187, 1117–1132. 10.1083/jcb.200909183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nachury MV, Loktev AV, Zhang Q, Westlake CJ, Peränen J, Merdes A, Slusarski DC, Scheller RH, Bazan JF, Sheffield VC, et al. (2007). A Core Complex of BBS Proteins Cooperates with the GTPase Rab8 to Promote Ciliary Membrane Biogenesis. Cell 129, 1201–1213. 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- 16.Ye F, Nager AR, and Nachury MV (2018). BBSome trains remove activated GPCRs from cilia by enabling passage through the transition zone. J. Cell Biol 217, 1847–1868. 10.1083/jcb.201709041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kozminski KG, Beech PL, and Rosenbaum JL (1995). The Chlamydomonas kinesin-like protein FLA10 is involved in motility associated with the flagellar membrane. J Cell Biology 131, 1517–1527. 10.1083/jcb.131.6.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van den Hoek H, Klena N, Jordan MA, Viar GA, Righetto RD, Schaffer M, Erdmann PS, Wan W, Geimer S, Plitzko JM, et al. (2022). In situ architecture of the ciliary base reveals the stepwise assembly of intraflagellar transport trains. Science 377, 543–548. 10.1126/science.abm6704. [DOI] [PubMed] [Google Scholar]
- 19.Jordan MA, Diener DR, Stepanek L, and Pigino G (2018). The cryo-EM structure of intraflagellar transport trains reveals how dynein is inactivated to ensure unidirectional anterograde movement in cilia. Nat Cell Biol 20, 1250–1255. 10.1038/s41556-018-0213-1. [DOI] [PubMed] [Google Scholar]
- 20.Jékely G, and Arendt D (2006). Evolution of intraflagellar transport from coated vesicles and autogenous origin of the eukaryotic cilium. Bioessays 28, 191–198. 10.1002/bies.20369. [DOI] [PubMed] [Google Scholar]
- 21.Dam T.J.P. van, Townsend MJ, Turk M, Schlessinger A, Sali A, Field MC, and Huynen MA (2013). Evolution of modular intraflagellar transport from a coatomer-like progenitor. Proc. Natl. Acad. Sci. U.S.A 110, 6943–6948. 10.1073/pnas.1221011110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wood CR, and Rosenbaum JL (2014). Proteins of the Ciliary Axoneme Are Found on Cytoplasmic Membrane Vesicles during Growth of Cilia. Curr Biol 24, 1114–1120. 10.1016/j.cub.2014.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quidwai T, Wang J, Hall EA, Petriman NA, Leng W, Kiesel P, Wells JN, Murphy LC, Keighren MA, Marsh JA, et al. (2021). A WDR35-dependent coat protein complex transports ciliary membrane cargo vesicles to cilia. elife 10. 10.7554/elife.69786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wachter S, Jung J, Shafiq S, Basquin J, Fort C, Bastin P, and Lorentzen E (2019). Binding of IFT22 to the intraflagellar transport complex is essential for flagellum assembly. Embo J 38. 10.15252/embj.2018101251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taschner M, Lorentzen A, Mourão A, Collins T, Freke GM, Moulding D, Basquin J, Jenkins D, and Lorentzen E (2018). Crystal structure of intraflagellar transport protein 80 reveals a homo-dimer required for ciliogenesis. elife 7, 929. 10.7554/elife.33067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taschner M, Kotsis F, Braeuer P, Kuehn EW, and Lorentzen E (2014). Crystal structures of IFT70/52 and IFT52/46 provide insight into intraflagellar transport B core complex assembly. J. Cell Biol 207, 269–282. 10.1083/jcb.201408002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhogaraju S, Taschner M, Morawetz M, Basquin C, and Lorentzen E (2011). Crystal structure of the intraflagellar transport complex 25/27. EMBO J. 30, 1907–1918. 10.1038/emboj.2011.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taschner M, Weber K, Mourão A, Vetter M, Awasthi M, Stiegler M, Bhogaraju S, and Lorentzen E (2016). Intraflagellar transport proteins 172, 80, 57, 54, 38, and 20 form a stable tubulin-binding IFT-B2 complex. EMBO J. 35, 773–790. 10.15252/embj.201593164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lacey SE, Foster HE, and Pigino G (2022). The Molecular Structure of Anterograde Intraflagellar transport trains. Biorxiv, 2022.08.01.502329. 10.1101/2022.08.01.502329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petriman NA, Loureiro-López M, Taschner M, Zacharia NK, Georgieva MM, Boegholm N, Wang J, Mourão A, Russell RB, Andersen JS, et al. (2022). Biochemically validated structural model of the 15-subunit intraflagellar transport complex IFT-B. Embo J, e112440. 10.15252/embj.2022112440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toropova K, Zalyte R, Mukhopadhyay AG, Mladenov M, Carter AP, and Roberts AJ (2019). Structure of the dynein-2 complex and its assembly with intraflagellar transport trains. Nat. Struct. Mol. Biol 26, 823–829. 10.1038/s41594-019-0286-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh SK, Gui M, Koh F, Yip MC, and Brown A (2020). Structure and activation mechanism of the BBSome membrane protein trafficking complex. elife 9, 3394. 10.7554/elife.53322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang S, Bahl K, Chou H-T, Woodsmith J, Stelzl U, Walz T, and Nachury MV (2020). Near-atomic structures of the BBSome reveal the basis for BBSome activation and binding to GPCR cargoes. elife 9, 213. 10.7554/elife.55954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klink BU, Gatsogiannis C, Hofnagel O, Wittinghofer A, and Raunser S (2020). Structure of the human BBSome core complex. elife 9, 213. 10.7554/elife.53910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langousis G, and Hill KL (2014). Motility and more: the flagellum of Trypanosoma brucei. Nat Rev Microbiol 12, 505–518. 10.1038/nrmicro3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buisson J, Chenouard N, Lagache T, Blisnick T, Olivo-Marin J-C, and Bastin P (2013). Intraflagellar transport proteins cycle between the flagellum and its base. J Cell Sci 126, 327–338. 10.1242/jcs.117069. [DOI] [PubMed] [Google Scholar]
- 37.Bertiaux E, Mallet A, Fort C, Blisnick T, Bonnefoy S, Jung J, Lemos M, Marco S, Vaughan S, Trépout S, et al. (2018). Bidirectional intraflagellar transport is restricted to two sets of microtubule doublets in the trypanosome flagellum. J. Cell Biol 217, 4284–4297. 10.1083/jcb.201805030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Engel BD, Ludington WB, and Marshall WF (2009). Intraflagellar transport particle size scales inversely with flagellar length: revisiting the balance-point length control model. J. Cell Biol 187, 81–89. 10.1083/jcb.200812084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iomini C, Babaev-Khaimov V, Sassaroli M, and Piperno G (2001). Protein Particles in Chlamydomonas Flagella Undergo a Transport Cycle Consisting of Four Phases. J Cell Biology 153, 13–24. 10.1083/jcb.153.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCafferty CL, Papoulas O, Jordan MA, Hoogerbrugge G, Nichols C, Pigino G, Taylor DW, Wallingford JB, and Marcotte EM (2022). Integrative modeling reveals the molecular architecture of the Intraflagellar Transport A (IFT-A) complex. eLife, 10.7554/eLife.81977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kao A, Chiu C, Vellucci D, Yang Y, Patel VR, Guan S, Randall A, Baldi P, Rychnovsky SD, and Huang L (2011). Development of a Novel Cross-linking Strategy for Fast and Accurate Identification of Cross-linked Peptides of Protein Complexes*. Mol Cell Proteomics 10, M110.002170. 10.1074/mcp.m110.002212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Behal RH, Miller MS, Qin H, Lucker BF, Jones A, and Cole DG (2012). Subunit Interactions and Organization of the Chlamydomonas reinhardtii Intraflagellar Transport Complex A Proteins*. J Biol Chem 287, 11689–11703. 10.1074/jbc.m111.287102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dodonova SO, Diestelkoetter-Bachert P, Appen A. von, Hagen WJH, Beck R, Beck M, Wieland F, and Briggs JAG (2015). A structure of the COPI coat and the role of coat proteins in membrane vesicle assembly. Science 349, 195–198. 10.1126/science.aab1121. [DOI] [PubMed] [Google Scholar]
- 44.Shvarev D, Schoppe J, König C, Perz A, Füllbrunn N, Kiontke S, Langemeyer L, Januliene D, Schnelle K, Kümmel D, et al. (2022). Structure of the HOPS tethering complex, a lysosomal membrane fusion machinery. Elife 11. 10.7554/elife.80901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu B, Zhu X, Wang L, Liang Y, Feng Q, and Pan J (2017). Functional exploration of the IFT-A complex in intraflagellar transport and ciliogenesis. PLoS Genet. 13, e1006627. 10.1371/journal.pgen.1006627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Duran I, Taylor SP, Zhang W, Martin J, Qureshi F, Jacques SM, Wallerstein R, Lachman RS, Nickerson DA, Bamshad M, et al. (2017). Mutations in IFT-A satellite core component genes IFT43 and IFT121 produce short rib polydactyly syndrome with distinctive campomelia. Cilia 6, 7. 10.1186/s13630-017-0051-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hirano T, Katoh Y, and Nakayama K (2017). Intraflagellar transport-A complex mediates ciliary entry and retrograde trafficking of ciliary G protein–coupled receptors. Mol. Biol. Cell 28, 429–439. 10.1091/mbc.e16-11-0813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang W, Taylor SP, Ennis HA, Forlenza KN, Duran I, Li B, Sanchez JAO, Nevarez L, Nickerson DA, Bamshad M, et al. (2018). Expanding the genetic architecture and phenotypic spectrum in the skeletal ciliopathies. Hum Mutat 39, 152–166. 10.1002/humu.23362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Valenstein ML, Rogala KB, Lalgudi PV, Brignole EJ, Gu X, Saxton RA, Chantranupong L, Kolibius J, Quast J-P, and Sabatini DM (2022). Structure of the nutrient-sensing hub GATOR2. Nature 607, 610–616. 10.1038/s41586-022-04939-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ishida Y, Kobayashi T, Chiba S, Katoh Y, and Nakayama K (2021). Molecular basis of ciliary defects caused by compound heterozygous IFT144/WDR19 mutations found in cranioectodermal dysplasia. Hum. Mol. Genet 10.1093/hmg/ddab034. [DOI] [PubMed] [Google Scholar]
- 51.Kobayashi T, Ishida Y, Hirano T, Katoh Y, and Nakayama K (2021). Cooperation of the IFT-A complex with the IFT-B complex is required for ciliary retrograde protein trafficking and GPCR import. Mol Biol Cell 32, 45–56. 10.1091/mbc.e20-08-0556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Halbritter J, Bizet AA, Schmidts M, Porath JD, Braun DA, Gee HY, McInerney-Leo AM, Krug P, Filhol E, Davis EE, et al. (2013). Defects in the IFT-B Component IFT172 Cause Jeune and Mainzer-Saldino Syndromes in Humans. Am J Hum Genetics 93, 915–925. 10.1016/j.ajhg.2013.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bujakowska KM, Zhang Q, Siemiatkowska AM, Liu Q, Place E, Falk MJ, Consugar M, Lancelot M-E, Antonio A, Lonjou C, et al. (2015). Mutations in IFT172 cause isolated retinal degeneration and Bardet–Biedl syndrome. Hum Mol Genet 24, 230–242. 10.1093/hmg/ddu441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Picariello T, Brown JM, Hou Y, Swank G, Cochran DA, King OD, Lechtreck K, Pazour GJ, and Witman GB (2019). A global analysis of IFT-A function reveals specialization for transport of membrane-associated proteins into cilia. J. Cell. Sci 132, jcs220749. 10.1242/jcs.220749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu D, Huang J, Zhu H, Chen Z, Chai Y, Ke J, Lei K, Peng Z, Zhang R, Li X, et al. (2022). Ciliogenesis requires sphingolipid-dependent membrane and axoneme interaction. P Natl Acad Sci Usa 119, e2201096119. 10.1073/pnas.2201096119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yeo HK, Park TH, Kim HY, Jang H, Lee J, Hwang G, Ryu SE, Park SH, Song HK, Ban HS, et al. (2021). Phospholipid transfer function of PTPIP51 at mitochondria-associated ER membranes. Embo Rep 22, e51323. 10.15252/embr.202051323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hesketh SJ, Mukhopadhyay AG, Nakamura D, Toropova K, and Roberts AJ (2022). IFT-A Structure Reveals Carriages for Membrane Protein Transport into Cilia. Biorxiv, 2022.08.09.503213. 10.1101/2022.08.09.503213. [DOI] [PubMed] [Google Scholar]
- 58.Badgandi HB, Hwang S, Shimada IS, Loriot E, and Mukhopadhyay S (2017). Tubby family proteins are adapters for ciliary trafficking of integral membrane proteins. J Cell Biology 216, 743–760. 10.1083/jcb.201607095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wingfield JL, Mekonnen B, Mengoni I, Liu P, Jordan M, Diener D, Pigino G, and Lechtreck K (2021). In vivo imaging shows continued association of several IFT A, B and dynein complexes while IFT trains U-turn at the tip. J Cell Sci 134. 10.1242/jcs.259010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lucas BA, Himes BA, Xue L, Grant T, Mahamid J, and Grigorieff N (2021). Locating macromolecular assemblies in cells by 2D template matching with cisTEM. Elife 10, e68946. 10.7554/elife.68946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ashkenazy H, Abadi S, Martz E, Chay O, Mayrose I, Pupko T, and Ben-Tal N (2016). ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Research 44, W344–50. 10.1093/nar/gkw408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sánchez-García R, Gómez-Blanco J, Cuervo A, Carazo JM, Sorzano C-OS, and Vargas J (2021). DeepEMhancer: a deep learning solution for cryo-EM volume post-processing. Commun Biol 4, 874. 10.1038/s42003-021-02399-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lechtreck KF, Brown JM, Sampaio JL, Craft JM, Shevchenko A, Evans JE, and Witman GB (2013). Cycling of the signaling protein phospholipase D through cilia requires the BBSome only for the export phase. J. Cell Biol 201, 249–261. 10.1083/jcb.201207139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schorb M, Haberbosch I, Hagen WJH, Schwab Y, and Mastronarde DN (2019). Software tools for automated transmission electron microscopy. Nat. Methods 16, 471–477. 10.1038/s41592-019-0396-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Morin A, Eisenbraun B, Key J, Sanschagrin PC, Timony MA, Ottaviano M, and Sliz P (2013). Collaboration gets the most out of software. elife 2, e01456. 10.7554/elife.01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Punjani A, Rubinstein JL, Fleet DJ, and Brubaker MA (2017). cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296. 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 67.Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJ, Lindahl E, and Scheres SH (2018). New tools for automated high-resolution cryo-EM structure determination in RELION-3. elife 7, 163. 10.7554/elife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liebschner D, Afonine PV, Baker ML, Bunkóczi G, Chen VB, Croll TI, Hintze B, Hung LW, Jain S, McCoy AJ, et al. (2019). Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr D Struct Biol 75, 861–877. 10.1107/s2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, Morris JH, and Ferrin TE (2021). UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82. 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brown A, Long F, Nicholls RA, Toots J, Emsley P, and Murshudov G (2015). Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr. D Biol. Crystallogr 71, 136–153. 10.1107/s1399004714021683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Afonine PV, Poon BK, Read RJ, Sobolev OV, Terwilliger TC, Urzhumtsev A, and Adams PD (2018). Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr D Struct Biol 74, 531–544. 10.1107/s2059798318006551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, and Richardson DC (2010). MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr 66, 12–21. 10.1107/s0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Molecular systems biology 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rosenthal PB, and Henderson R (2003). Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. Journal of Molecular Biology 333, 721–745. 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 76.Landrum MJ, Lee JM, Benson M, Brown GR, Chao C, Chitipiralla S, Gu B, Hart J, Hoffman D, Jang W, et al. (2018). ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res 46, D1062–D1067. 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Arts HH, Bongers EMHF, Mans DA, Beersum S.E.C. van, Oud MM, Bolat E, Spruijt L, Cornelissen EAM, Schuurs-Hoeijmakers JHM, Leeuw N. de, et al. (2011). C14ORF179 encoding IFT43 is mutated in Sensenbrenner syndrome. J Med Genet 48, 390. 10.1136/jmg.2011.088864. [DOI] [PubMed] [Google Scholar]
- 78.Mill P, Lockhart PJ, Fitzpatrick E, Mountford HS, Hall EA, Reijns MAM, Keighren M, Bahlo M, Bromhead CJ, Budd P, et al. (2011). Human and Mouse Mutations in WDR35 Cause Short-Rib Polydactyly Syndromes Due to Abnormal Ciliogenesis. Am J Hum Genetics 88, 508–515. 10.1016/j.ajhg.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bacino CA, Dhar SU, Brunetti-Pierri N, Lee B, and Bonnen PE (2012). WDR35 mutation in siblings with Sensenbrenner syndrome: A ciliopathy with variable phenotype. Am J Med Genet A 158A, 2917–2924. 10.1002/ajmg.a.35608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gilissen C, Arts HH, Hoischen A, Spruijt L, Mans DA, Arts P, Lier B. van, Steehouwer M, Reeuwijk J. van, Kant SG, et al. (2010). Exome Sequencing Identifies WDR35 Variants Involved in Sensenbrenner Syndrome. Am J Hum Genetics 87, 418–423. 10.1016/j.ajhg.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Walczak-Sztulpa J, Eggenschwiler J, Osborn D, Brown DA, Emma F, Klingenberg C, Hennekam RC, Torre G, Garshasbi M, Tzschach A, et al. (2010). Cranioectodermal Dysplasia, Sensenbrenner Syndrome, Is a Ciliopathy Caused by Mutations in the IFT122 Gene. Am J Hum Genetics 86, 949–956. 10.1016/j.ajhg.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Domingo-Gallego A, Pybus M, Bullich G, Furlano M, Ejarque-Vila L, Lorente-Grandoso L, Ruiz P, Fraga G, González ML, Piñero-Fernández JA, et al. (2021). Clinical utility of genetic testing in early-onset kidney disease: seven genes are the main players. Nephrol Dial Transpl 37, gfab019. 10.1093/ndt/gfab019. [DOI] [PubMed] [Google Scholar]
- 83.Fry AE, Klingenberg C, Matthes J, Heimdal K, Hennekam RCM, and Pilz DT (2009). Connective tissue involvement in two patients with features of cranioectodermal dysplasia. Am J Med Genet A 149A, 2212–2215. 10.1002/ajmg.a.33027. [DOI] [PubMed] [Google Scholar]
- 84.Yang Q, Zhang Q, Chen F, Yi S, Li M, Yi S, Xu X, and Luo J (2021). A novel combination of biallelic IFT122 variants associated with cranioectodermal dysplasia: A case report. Exp Ther Med 21, 311. 10.3892/etm.2021.9742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu W, He X, Yang S, Zouari R, Wang J, Wu H, Kherraf Z-E, Liu C, Coutton C, Zhao R, et al. (2019). Bi-allelic Mutations in TTC21A Induce Asthenoteratospermia in Humans and Mice. Am J Hum Genetics 104, 738–748. 10.1016/j.ajhg.2019.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Davis EE, Zhang Q, Liu Q, Diplas BH, Davey LM, Hartley J, Stoetzel C, Szymanska K, Ramaswami G, Logan CV, et al. (2011). TTC21B contributes both causal and modifying alleles across the ciliopathy spectrum. Nat Genet 43, 189–196. 10.1038/ng.756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McInerney-Leo AM, Harris JE, Leo PJ, Marshall MS, Gardiner B, Kinning E, Leong HY, McKenzie F, Ong WP, Vodopiutz J, et al. (2015). Whole exome sequencing is an efficient, sensitive and specific method for determining the genetic cause of short-rib thoracic dystrophies. Clin Genet 88, 550–557. 10.1111/cge.12550. [DOI] [PubMed] [Google Scholar]
- 88.Geoffroy V, Stoetzel C, Scheidecker S, Schaefer E, Perrault I, Bär S, Kröll A, Delbarre M, Antin M, Leuvrey A, et al. (2018). Whole-genome sequencing in patients with ciliopathies uncovers a novel recurrent tandem duplication in IFT140. Hum Mutat 39, 983–992. 10.1002/humu.23539. [DOI] [PubMed] [Google Scholar]
- 89.Perrault I, Saunier S, Hanein S, Filhol E, Bizet AA, Collins F, Salih MAM, Gerber S, Delphin N, Bigot K, et al. (2012). Mainzer-Saldino Syndrome Is a Ciliopathy Caused by IFT140 Mutations. Am J Hum Genetics 90, 864–870. 10.1016/j.ajhg.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xu M, Yang L, Wang F, Li H, Wang X, Wang W, Ge Z, Wang K, Zhao L, Li H, et al. (2015). Mutations in human IFT140 cause non-syndromic retinal degeneration. Hum Genet 134, 1069–1078. 10.1007/s00439-015-1586-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Carss KJ, Arno G, Erwood M, Stephens J, Sanchis-Juan A, Hull S, Megy K, Grozeva D, Dewhurst E, Malka S, et al. (2017). Comprehensive Rare Variant Analysis via Whole-Genome Sequencing to Determine the Molecular Pathology of Inherited Retinal Disease. Am J Hum Genetics 100, 75–90. 10.1016/j.ajhg.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zenteno JC, García-Montaño LA, Cruz-Aguilar M, Ronquillo J, Rodas-Serrano A, Aguilar-Castul L, Matsui R, Vencedor-Meraz CI, Arce-González R, Graue-Wiechers F, et al. (2020). Extensive genic and allelic heterogeneity underlying inherited retinal dystrophies in Mexican patients molecularly analyzed by next-generation sequencing. Mol Genetics Genom Medicine 8, 10.1002/mgg3.1044. 10.1002/mgg3.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hull S, Owen N, Islam F, Tracey-White D, Plagnol V, Holder GE, Michaelides M, Carss K, Raymond FL, Rozet J-M, et al. (2016). Nonsyndromic Retinal Dystrophy due to Bi-Allelic Mutations in the Ciliary Transport Gene IFT140. Invest Ophth Vis Sci 57, 1053–1062. 10.1167/iovs.15-17976. [DOI] [PubMed] [Google Scholar]
- 94.Beheshtian M, Rad SS, Babanejad M, Mohseni M, Hashemi H, Eshghabadi A, Hajizadeh F, Akbari MR, Kahrizi K, Esfahani MR, et al. (2015). Impact of whole exome sequencing among Iranian patients with autosomal recessive retinitis pigmentosa. Arch Iran Med 18, 776–785. 0151811/aim.009. [PubMed] [Google Scholar]
- 95.Schmidts M, Frank V, Eisenberger T, Turki S. al, Bizet AA, Antony D, Rix S, Decker C, Bachmann N, Bald M, et al. (2013). Combined NGS Approaches Identify Mutations in the Intraflagellar Transport Gene IFT140 in Skeletal Ciliopathies with Early Progressive Kidney Disease. Hum Mutat 34, 714–724. 10.1002/humu.22294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vries J. de, Yntema JL, Die C.E. van, Crama N, Cornelissen EAM, and Hamel BCJ (2009). Jeune syndrome: description of 13 cases and a proposal for follow-up protocol. Eur J Pediatr 169, 77. 10.1007/s00431-009-0991-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Coussa RG, Otto EA, Gee H, Arthurs P, Ren H, Lopez I, Keser V, Fu Q, Faingold R, Khan A, et al. (2013). WDR19: An ancient, retrograde, intraflagellar ciliary protein is mutated in autosomal recessive retinitis pigmentosa and in Senior-Loken syndrome. Clin Genet 84, 150–159. 10.1111/cge.12196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bredrup C, Saunier S, Oud MM, Fiskerstrand T, Hoischen A, Brackman D, Leh SM, Midtbø M, Filhol E, Bole-Feysot C, et al. (2011). Ciliopathies with Skeletal Anomalies and Renal Insufficiency due to Mutations in the IFT-A Gene WDR19. Am J Hum Genetics 89, 634–643. 10.1016/j.ajhg.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shaheen R, Szymanska K, Basu B, Patel N, Ewida N, Faqeih E, Hashem AA, Derar N, Alsharif H, et al. (2016). Characterizing the morbid genome of ciliopathies. Genome Biol 17, 242. 10.1186/s13059-016-1099-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lee JM, Ahn YH, Kang HG, Ha IS, Lee K, Moon KC, Lee JH, Park YS, Cho YM, Bae J-S, et al. (2015). Nephronophthisis 13: implications of its association with Caroli disease and altered intracellular localization of WDR19 in the kidney. Pediatr Nephrol 30, 1451–1458. 10.1007/s00467-015-3068-8. [DOI] [PubMed] [Google Scholar]
- 101.Halbritter J, Porath JD, Diaz KA, Braun DA, Kohl S, Chaki M, Allen SJ, Soliman NA, Hildebrandt F, et al. (2013). Identification of 99 novel mutations in a worldwide cohort of 1,056 patients with a nephronophthisis-related ciliopathy. Hum Genet 132, 865–884. 10.1007/s00439-013-1297-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ni X, Wang J, Lv M, Liu C, Zhong Y, Tian S, Wu H, Cheng H, Gao Y, Tan Q, et al. (2020). A novel homozygous mutation in WDR19 induces disorganization of microtubules in sperm flagella and nonsyndromic asthenoteratospermia. J Assist Reprod Gen 37, 1431–1439. 10.1007/s10815-020-01770-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Montolío-Marzo S, Català-Mora J, Madrid-Aris Á, Armstrong J, Díaz-Carcajosa J, and Carreras E (2020). IFT144 and mild retinitis pigmentosa in Mainzer-Saldino syndrome: A new association. Eur J Med Genet 63, 104073. 10.1016/j.ejmg.2020.104073. [DOI] [PubMed] [Google Scholar]
- 104.Smith C, Lamont RE, Wade A, Bernier FP, Parboosingh JS, and Innes AM (2015). A relatively mild skeletal ciliopathy phenotype consistent with cranioectodermal dysplasia is associated with a homozygous nonsynonymous mutation in WDR35. Am J Medical Genetics Part 170, 760–765. 10.1002/ajmg.a.37514. [DOI] [PubMed] [Google Scholar]
- 105.Aslett M, Aurrecoechea C, Berriman M, Brestelli J, Brunk BP, Carrington M, Depledge DP, Fischer S, Gajria B, Gao X, et al. (2010). TriTrypDB: a functional genomic resource for the Trypanosomatidae. Nucleic Acids Res 38, D457–D462. 10.1093/nar/gkp851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Research 40, D1178–86. 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Harris TW, Arnaboldi V, Cain S, Chan J, Chen WJ, Cho J, Davis P, Gao S, Grove CA, Kishore R, et al. (2020). WormBase: a modern Model Organism Information Resource. Nucleic Acids Res 48, D762–D767. 10.1093/nar/gkz920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.UniProt Consortium (2021). UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Research 49, D480–D489. 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Purification and structure determination of Leishmania tarentolae IFT-A, Related to Figure 1. A. Left, thin-section transmission electron micrograph showing a L. tarentolae flagellum with an IFT train (marked with an arrowhead) between the axonemal doublet microtubules (DMT) and the ciliary membrane (c.m.). Also indicated is the central apparatus (CA) in the middle of the axoneme. Middle and right, electron micrographs showing the cross-sections of Leishmania flagella with putative IFT trains labeled with arrowheads.
B. Chromatogram showing the IFT-A peak following ion-exchange chromatography.
C. Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) of purified IFT-A.
D. Representative electron micrograph of IFT-A taken under cryogenic conditions.
E. Flow diagram, starting with selected two-dimensional class averages, of the steps taken to obtain structures of IFT-A complexes and the IFT-A2 subcomplex.
Figure S2. Two-dimensional (2D) class averages of Leishmania tarentolae IFT-A, Related to Figure 1. A. Comparison of 2D class averages from negative-stain electron microscopy of IFT-A purified with (+) and without (Δ) crosslinker. In both instances, the class shown is the most abundant class observed.
B. Selected 2D classes of IFT-A purified without crosslinker showing a progressive opening of a gap between the A1 and A2 subdomains.
C. 2D class averages from cryo-EM data showing that subcomplex movement is observed even with crosslinked IFT-A. This movement breaks the interaction between the N-terminal β-propeller domains of IFT121 and IFT140.
Figure S3. Resolution and map quality, Related to Figure 1. A. Fourier shell correlation (FSC) curves for IFT-A state 1 (left), IFT-A state 2 (middle) and IFT-A2 (right). The corresponding maps are shown in Fig. S1 together with their resolution determined using the FSC=0.143 criterion. Map1 corresponds to the consensus refinement before mask-focused classification/refinement.
B. Consensus maps colored by local resolution. Local map quality was improved further by mask-focused refinement.
C. Examples of map density for each of the six IFT-A subunits with landmark residues labeled. Maps were sharpened using DeepEMhancer for visualization purposes only62.
Figure S4. IFT-A structure validation using chemical crosslinking mass spectrometry, Related to Figure 1. A. Atomic model of Tetrahymena thermophila IFT-A, built using the model of Leishmania tarentolae IFT-A as a template. The Cα atoms of residues crosslinked by disuccinimidyl sulfoxide are connected with a line. Green lines indicate crosslinks within a subunit, and purple lines indicate crosslinks between two different subunits. Orange spheres represent residues that form crosslinks that exceed the distance (>45 Å) capable of being crosslinked by disuccinimidyl sulfoxide.
B. Plot of distances between Cα positions of crosslinked residues. Data were plotted using GraphPad Prism v9 (GraphPad Software).
Figure S5. IFT121, IFT122 and IFT144 have conserved C-terminal zinc-finger (ZnF) domains, Related to Figure 2. A. The C-terminus of Leishmania tarentolae (Lt.) IFT121 superposed with the AlphaFold2 model of human (Hs.) IFT121. In humans, the first ZnF domain (ZnF1) is absent but the second (ZnF2) is highly conserved. The Zn ion is shown as a green sphere and the coordinating cysteine residues are shown in stick representation.
B. Superposition of the C-terminal domains of Lt. and Hs. IFT122, each featuring two conserved ZnFs and a small mixed α/β domain.
C. Superposition of the C-termini of Lt. and Hs. IFT144 featuring tandem ZnF domains.
D. Sequence alignment of the regions shown in panels A–C. Conserved residues are marked with an asterisk. Conserved cysteines are further highlighted in yellow. Sequences were obtained from Leishmania tarentolae (Lt.), Trypanosoma brucei (Tb.), Chlamydomonas reinhardtii (Cr.), Tetrahymena thermophila (Tt.), Caenorhabditis elegans (Ce.) and Homo sapiens (Hs.). Accession numbers for the sequences are provided in Table S4.
Figure S6. IFT-A specifically decorates liposomes containing phosphatidic acid (PA), Related to Figure 4. A. Representative negative-stain electron micrograph showing IFT-A particles bound to the surface of liposomes generated with PA.
B. Representative negative-stain electron micrograph of IFT-A incubated with liposomes lacking PA. Most IFT-A particles and liposomes remain separate.
Table S2. Statistics for data collection, data processing, model refinement and validation, Related to Figure 1.
Table S4. Sequence information for IFT-A subunits, Related to STAR Methods. Sequences for L. tarentolae and T. brucei proteins were obtained from TriTrypDB105. Sequences for C. reinhardtii proteins were obtained from Phytozome106. Sequences for C. elegans proteins were obtained from WormBase107. Sequences for T. thermophila and H. sapiens proteins were obtained from UniProt108. These sequences were used for protein sequence alignments and/or to build AlphaFold2 models.
Table S1 (Provided as an Excel file). Mass-spectrometry data, Related to Figure 1. A. List of proteins identified in the elution from an anti-FLAG pulldown of lysate from Leishmania tarentolae expressing FLAG-IFT43. IFT-A subunits are highlighted in yellow and IFT-B subunits in blue. B. List of proteins surviving gradient centrifugation and ion-exchange chromatography. IFT-A subunits are highlighted in yellow. No IFT-B subunits were detected. C. Mass-spectrometry identification of phosphorylated IFT-A residues. Most phosphorylated residues occur in non-conserved surface-exposed loops.
Table S3 (Provided as an Excel file). Mutations in human IFT-A genes associated with ciliopathies, Related to Figure 5. Missense variants annotated as pathogenic or likely pathogenic obtained from the ClinVar database76. References found in the table:46,48,77–104.
Video S1. Polymerization of IFT-A into anterograde IFT trains, Related to Figure 3. Video showing the docking of an atomic model of monomeric C. reinhardtii IFT-A into a subtomogram average of polymeric IFT-A (EMD-26791) and the rotation of the A1 subcomplex that allows IFT-A to polymerize.
Data Availability Statement
Composite cryo-EM maps of L. tarentolae IFT-A state 1 and 2 have deposited in the Electron Microscopy Data Bank (EMDB) with accession numbers EMD-28866 and EMD-28867. Half maps and masks are deposited as additional maps associated with these entries. Atomic models of IFT-A state 1 and state 2 have been deposited in the Protein Data Bank (PDB) with accession numbers 8F5O and 8F5P.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


