Summary
Extensive remodeling of host gene expression by nonstructural protein 1 (nsp1) of coronaviruses is a well-documented and conserved aspect of coronavirus-host takeover. Using comparative transcriptomics we investigated the diversity of transcriptional targets between various nsp1 proteins. Additionally, affinity purification followed by mass spectrometry was implemented to identify common interactors between the different nsp1 proteins. Although we detected widespread RNA destabilization, closely related nsp1 showed little similarities in clustering of targeted genes. We observed a partial overlap in transcriptional targeting between α-CoV 229E and MERS nsp1, which may suggest a common targeting mechanism, as MERS nsp1 preferentially targets nuclear transcripts. Our interactome data show great variability between nsp1 interactions, with 229E nsp1, the smallest nsp1 tested here, interacting with the most number of host proteins. Although nsp1 is a rather well-conserved protein with conserved functions across different coronaviruses, our data indicate that its precise effects on the host cell are virus specific.
Subject areas: Microbiology, Molecular microbiology, Virology
Graphical abstract
Highlights
-
•
nsp1s from multiple coronaviruses alter expression of host mRNA
-
•
Transcriptional targeting between nsp1 of different coronaviruses is highly varied
-
•
Host-nsp1 protein interactions are unique among different coronaviruses
Microbiology; Molecular microbiology; Virology
Introduction
The past 20 years have seen the emergence of three highly pathogenic human coronaviruses (HCoVs), including SARS-CoV, MERS-CoV, and SARS-CoV-2. Since 2002 and the first coronavirus epidemic, these viruses have jumped to the forefront of public awareness as major public health threats. Other HCoVs routinely circulate in the human population, such as HCoV-HKU1 or HCoV-229E, which cause mild to moderate upper respiratory tract infections.
Coronaviruses consist of 4 genera: α-CoV, β-CoV, gammacoronavirus (γ-CoV), and deltacoronavirus (δ-CoV).1,2,3,4 Although γ-CoVs and δ-CoVs primarily infect birds,4 α-CoV and β-CoV only infect mammals. The three highly pathogenic HCoVs, SARS-CoV (referred to as SARS1 herein), MERS-CoV, and SARS-CoV-2 (referred to as SARS2 herein), belong to the genus β-CoV and are believed to freely circulate in and originate from bats.5,6 All these viruses have a highly conserved genomic organization and are the largest known RNA viruses to date. The 5′-terminal two-thirds of their genome encodes two overlapping open reading frames (ORF1a and ORF1ab), which results in the production of two large polyproteins. Nsp1 is the most N-terminal peptide released from the 5′-most polyprotein. Nsp1’s role during β-CoV infection has long been studied, and nsp1 was revealed to be a host shutoff protein that controls anti-viral responses by globally reducing host gene expression.7,8 SARS2 nsp1 similarly to SARS1 nsp1 not only induces translational shutdown7,9 but also promotes the degradation of its target RNA by binding to the 40S ribosomal subunit.10,11,12 Furthermore, MERS-CoV nsp1 selectively targets mRNA synthesized in the host cell nucleus for degradation and thus inhibits translation in host cells.13,14 How the various nsp1 proteins mediate this extensive shutoff phenotype at the RNA level remains unknown.15,16 nsp1 shares no resemblance in its primary amino acid sequence or protein structure with any known RNases and has been hypothesized to co-opt a host endonuclease to induce endonucleolytic cleavage of template mRNA transcripts that interact with 40S ribosomes. Yet, the identity of this putative host RNase, although extensively looked for, remains unknown.
Intriguingly, only α- and β-CoV encode nsp1, whereas γ- and δ-CoV lack this protein.2,3,17,18,19,20 The sizes of nsp1 in β-CoV also differ from the α-CoVs’ nsp1, with the α-CoVs’ nsp1 being substantially smaller than their β-CoV counterparts. Although these differences may have important consequences in the role of nsp1 during infection, it appears that nsp1 proteins from HCoV-229E and HCoV-NL63 might still be able to bind the 40S ribosomal subunit to affect host mRNA stability18,21,22 reminiscing of how the β-CoV nsp1 triggers host shutoff.
The extensive study of the role of SARS2, SARS1, and MERS nsp1 has revealed a pervasive role in reshaping the host gene expression environment. In this study, we set out to compare the impact of nsp1 on the host cell not only from the highly pathogenic β-CoV but also from the α-CoVs 229E. We hypothesized that the extent of nsp1-mediated RNA decay may vary among the different HCoVs, which could account for some of the severity of these infections. To address this possibility, we generated a library of nsp1-inducible cells from 4 HCoVs and explored the extent of nsp1 effect on the host transcriptome by RNA sequencing (RNA-seq). Interestingly, we found that widespread targeting of RNA is conserved among these nsp1 but the range of targets is different. Moreover, we investigated the interactome of these nsp1 proteins by mass spectrometry to identified common and divergent interactors and identified factors that may contribute to nsp1 targeting of RNA. This work provides important insights into the fundamental differences between highly pathogenic and common coronaviruses’ nsp1 and refines our understanding of nsp1-mediated decay.
Results
Inducible Expression of coronavirus nsp1 proteins
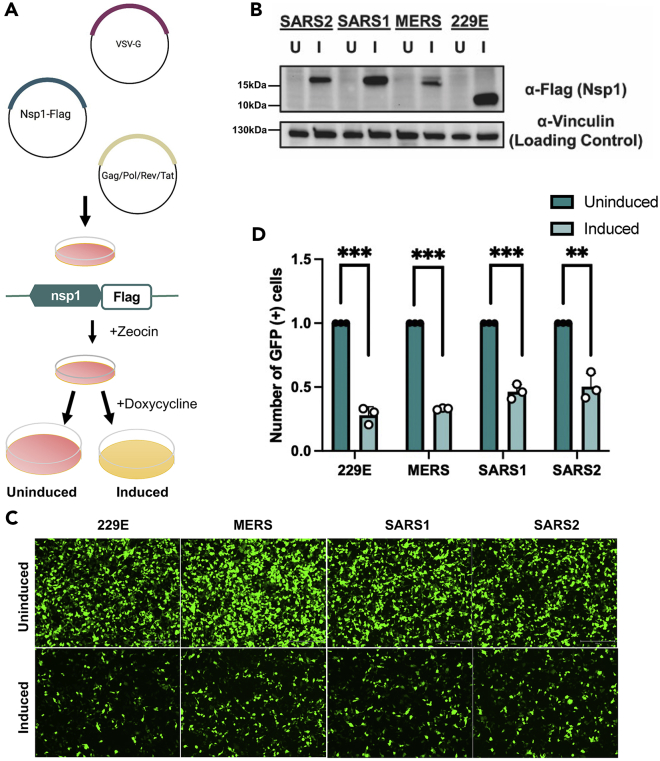
Previous work has shown conservation in the ability of α- and β-CoVs to modulate host gene expression pathways.15 However, to date, it remains unclear how diverse is the impact on the host transcriptome between α- and β-CoV nsp1 proteins. To assess the influence of different coronavirus nsp1 proteins on the host gene expression environment, we generated a library of nsp1-inducible cell lines using the nsp1 protein of 4 different HCoVs. To generate this library, along with SARS2 and MERS, we constructed 229E and SARS1 coronavirus nsp1 lentiviral plasmids derived from the pLVX-TetOne-Zeo-CoV2-nsp1-3xFLAG gifted to us by the Glaunsinger Lab. Following production of lentiviral plasmids, we performed lentiviral transduction on HEK293T cells using select pLVX CoV nsp1-Flag plasmid and pMD2.G and psPAX2 envelop and packaging plasmids (Figure 1A). After transduction, cells underwent selection using zeocin at a concentration of 325 μg/mL. Following selection, to verify that our cell lines were inducible and would produce CoV nsp1 protein, cells were induced with 1 μg/mL doxycycline. Twenty-four hours post-induction samples were collected and protein samples were western blotted with an anti-FLAG antibody (Figure 1B). All 4 nsp1 proteins express well under induction, at the expected size, and no leaky expression was observed in the uninduced samples. Transcript levels of nsp1 were also assessed by RT-qPCR (Figure S1). Once we verified that all 4 CoV nsp1 proteins could be properly expressed, we next determined if these induced nsp1-regulated gene expression as expected. As reported before, nsp1 can efficiently degrade GFP transcripts and reduce GFP expression by itself.10,12 We thus next expressed a GFP reporter; then the transduced cells were either left in an uninduced state or induced with doxycycline. Twenty-four hours post-transfection and induction, GFP expression was measured with fluorescence microscopy Figure 1C) and quantified using ImageJ (Figure 1D). As expected, induction led to a significant decrease in the intensity of GFP expression in all CoV nsp1-expressing cells. This is in line with the observation that despite being a smaller protein, the 229E nsp1 is able to regulate the expression of genes, likely due to a highly conserved domain.22 These results show that our library of nsp1-expressing cell lines do not have leaky expression, are inducible, and are able to modulate gene expression.
Figure 1.
Inducible expression of 4 coronavirus nsp1s results in downregulation of GFP
(A) Diagram representing lentiviral transduction and cell induction protocols. HEK293T cells were transfected with pLVX plasmids expressing the 4 nsp1s selected (from 229E, SARS1, SARS2, MERS) under a doxycycline-inducible promotor along with the lentiviral envelope and packaging plasmids.
(B) Transduced cells were induced (I) with doxycycline for 24 hours (or left uninduced [U]). Cells were then harvested, lysed, resolved on SDS-PAGE, and immunoblotted with the indicated antibodies. Vinculin was used as the loading control.
(C and D) (C) Transduced cells were transfected with a GFP reporter for 24 hours and then nsp1 expression (as indicated) was induced with doxycycline for 24 hours. GFP expression was monitored using fluorescence microscopy and quantification of GFP-positive cells in (D). ∗∗p < 0.01, ∗∗∗p < 0.001.
Coronavirus nsp1 comparative RNA-seq shows differences in gene expression
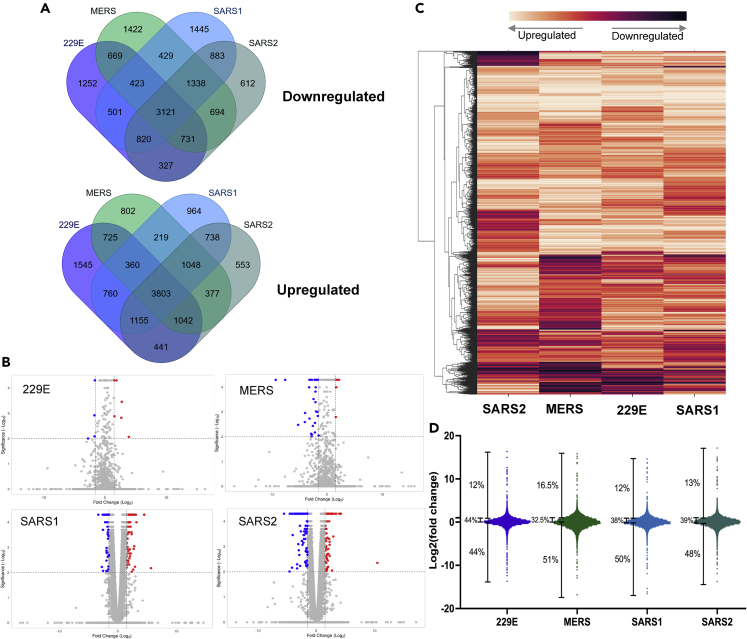
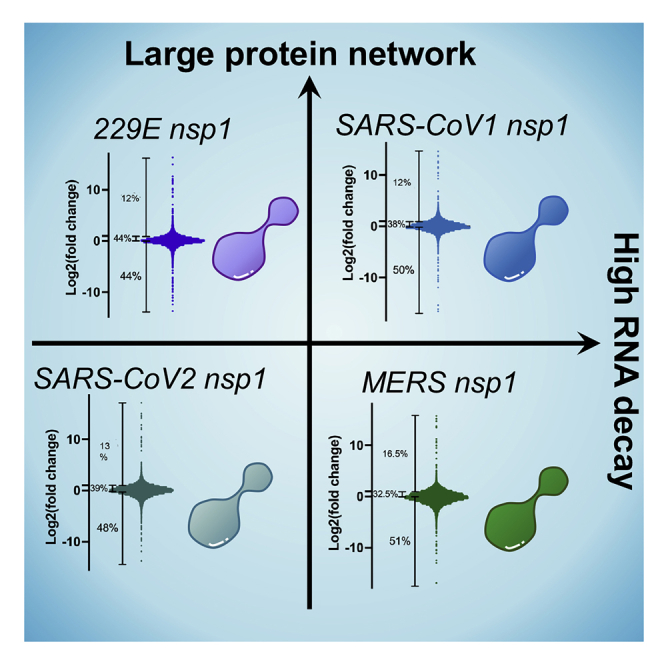
After confirming that our inducible cell lines function as expected, we sought to explore the differences in the extent of nsp1 targeting on the host transcriptome. Using the lentivirally transduced cell lines described earlier, we induced the expression of nsp1 from SARS1, SARS2, MERS, or 229E (using uninduced cells as controls). Twenty-four hours post-induction, total RNA was extracted, polyA selected, and cDNA library was prepared and sequenced. Of a total of 19,331 genes identified by RNA-seq, 15,779 genes appeared in all 4 datasets (Table S1). More specifically, we identified 3,121 genes that were consistently downregulated upon expression of all 4 nsp1 and 3,833 that were consistently upregulated (Figure 2A). Unsurprisingly, 229E nsp1, as the only representative of an α-CoV, has the most unique pattern of genes up- and downregulated, perhaps indicating that its targeting mechanism differs from that of the β-CoV nsp1. Furthermore, fold change patterns induced by 229E and MERS nsp1 as observed by volcano plots appeared different than the SARS1 and SARS2 plots (Figure 2B). This might suggest that the fold change distribution may not follow a normal distribution. Previous studies had indicated that MERS preferentially targets nuclear mRNA, whereas our data here represent whole-cell RNA pools, which could skew our data representation.13 This might suggest that 229E nsp1 similarly only targets a subset of RNA in cells. We next performed hierarchical clustering on this comparative transcriptomics dataset. Figure 2C shows a heatmap of the correlation matrix across all transcripts. In line with previous analyses, our data indicate that all the nsp1 tested trigger massive RNA degradation with >50% of the detected genes downregulated upon each nsp1 expression (Figure 2D). Based on our analysis, between 30% and 45% of genes had a fold change between 1 and 2, indicating that nsp1 had little to no effect on them. Surprisingly, we also detected close to 15% of genes that seem to be upregulated upon nsp1 induction. Overall, our data indicate that α- and β-CoV nsp1 share the ability to widely trigger RNA decay; however, the precise target of each of the nsp1 differs, suggesting that the host cell is likely differentially affected.
Figure 2.
RNA-seq analysis shows differential expression of host genes as impacted by coronavirus nsp1 expression
(A) Venn diagrams showing the extent of unique and shared genes identified by RNA-seq between each coronavirus nsp1 RNA-seq dataset.
(B) Volcano plot of all genes differentially expressed between uninduced samples versus coronavirus nsp1-expressing cells. Data points are shown as a representation of the log2(fold change) versus the -log10(p value) plotted by VolcaNoseR2. Significance cutoff was set to the programs default setting, with significantly downregulated genes being labeled blue and significantly upregulated genes being labeled red.
(C) Hierachical clustering and heatmap of RNA-seq data. Data were clustered based on fold change with columns representing expression of different coronavirus nsp1 based on relatedness (MERS, 229E, SARS1, SARS2). Expression levels were normalized relative to uninduced sample expression and represented as a heatmap. Transcripts are clustered based on complete linkage method to place genes with high similarities together with dendrogram on the left.
(D) Distribution of fold change per nsp1 tested over uninduced sample and corresponding percentages on degrading transcripts.
Validation of coronavirus nsp1 RNA-seq gene expression patterns by RT-qPCR
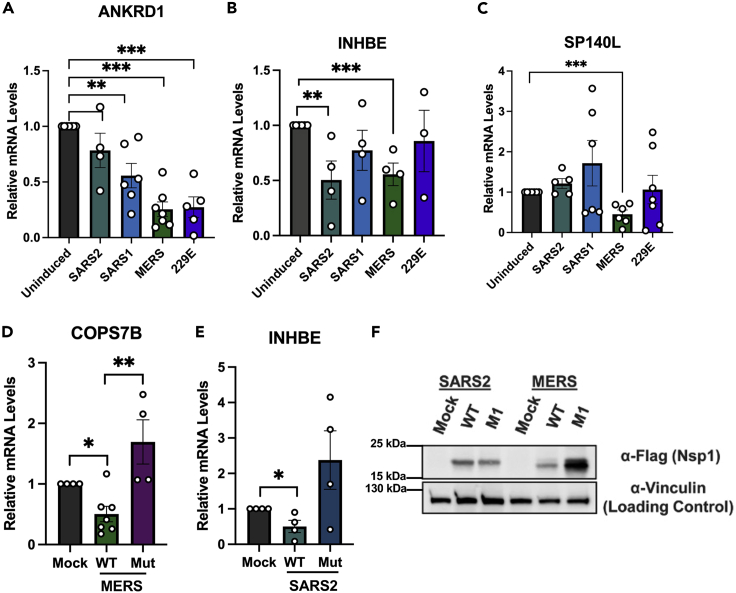
RNA-seq provided an extensive range of data about the effects of expression of the different coronavirus nsp1s on host gene expression. After data sorting and processing, we noticed some genes with particular patterns of degradation upon expression of the nsp1. We thus next wanted to validate these patterns by RT-qPCR (Figure 3). The first gene that we tested was ANKRD1. There are multiples links between ANKRD1 and coronaviruses; for example, upon porcine epidemic diarrhea virus (PEDV)—an α-CoV—infection ANKRD1 is downregulated23 or viral load in SARS-CoV-2 infections is increased if ANKRD1 is knocked down.24 Our RNA-seq data showed that ANKRD1 was downregulated upon expression of all tested β-CoV nsp1s. We thus induced the expression of each nsp1 in cells and collected total RNA to examine ANKRD1 expression by RT-qPCR. Our results recapitulate the RNA-seq data. We next tested the gene INHBE, encoding a protein associated with cell proliferation and apoptosis, which showed higher downregulation in SARS2 and MERS, with slight downregulation in SARS1 and 229E in our RNA-seq data. This pattern was again reflected in the RT-qPCR results (Figure 3B). We next investigated a gene (SP140L) that encodes a protein that binds zinc and acts as a transcription regulator. SP140L appears to be slightly upregulated upon expression of SARS2 and SARS1 nsp1, but downregulated with MERS and 229E nsp1 expression. Although our 229E nsp1 in this case appeared to have no effect on SP140L expression, MERS nsp1 expression led to strong downregulation as would be expected from our RNA-seq data (Figure 3C).
Figure 3.
RNA-seq validation
(A–D) 293T cells were either left uninduced or induced for 24 hours as described above, total RNA was harvested, and RT-qPCR was performed to measure expression of the endogenous genes ANKRD1 (A), INHBE (B), and SP140L (C).
(D and E) 293T cells were transfected with a FLAG-tagged WT or mutant version of nsp1 from MERS (D) or SARS2 (E) as indicated (or mock transfected as a control). Total RNA was harvested and RT-qPCR performed to measure expression of the endogenous genes COP17S or INHBE.
(F) 293T cells were transfected with a FLAG-tagged WT or mutant version of nsp1 as indicated. After 24 hours, cells were harvested, lysed, resolved on SDS-PAGE, and immunoblotted with the indicated antibodies. Vinculin was used as the loading control. n.s., not significant; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
Our understanding of how nsp1 triggers RNA decay remains mostly vague; however, some nsp1 mutants have been identified to specifically have no RNA decay activity. Residues R146 and K147 in MERS and R124 and K125 in SARS2 have been shown to be required for 40S binding, and mutating these residues has been shown to affect RNA turnover.7,12,25,26 We thus next wanted to validate our RNA-seq data using these mutants. We thus constructed FLAG-tagged versions of an MERS nsp1 mutant (R146A + K147A here referred to as MERS nsp1_mut) and a SARS2 nsp1 mutant (R124A + K125A here referred to as SARS2 nsp1_mut), which are both expected to abrogate nsp1-mediated RNA decay. For MERS nsp1_mut we looked at COPS7B, a member of the COP9 signalosome complex involved in ubiquitination, which we found to be downregulated by MERS nsp1 expression. To compare nsp1 wild-type (WT) and mutant version, 293T cells were transfected with a FLAG-tagged WT MERS nsp1, or MERS nsp1_mut (or mock transfected). After 24 hours, total RNA was extracted and used to assess COPS7B expression by RT-qPCR. WT nsp1 induced a strong reduction in COPS7B expression as expected from the RNA-seq data, whereas the nsp1 mutant failed to induce degradation (Figure 3D). Similarly, for SARS2 nsp1, we compared the expression of INHBE upon WT or mutant expression and saw that nsp1_mut failed to induce INHBE mRNA degradation (Figure 3E). We verified that this loss of decay was not due to a lower expression of the mutant compared with the WT by western blotting (Figure 3F). Combined together, these results show that all nsp1 tested specifically induce mRNA decay. Additionally, we show that the mutant variants of the SARS2 and MERS nsp1 proteins we have are able to ablate mRNA degradation.
Nsp1 interactome varies among coronaviruses
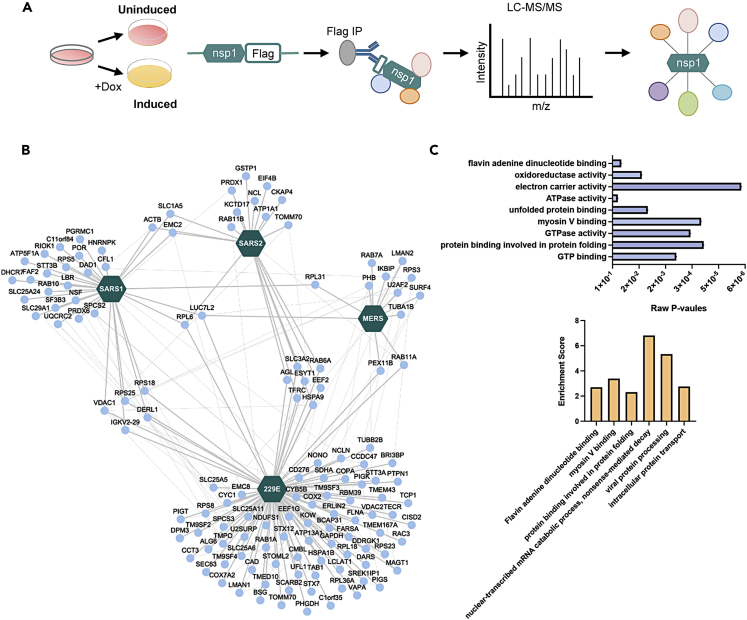
Nsp1 contribution to the widespread changes in host mRNA stability is believed to be mediated through protein-protein interactions since nsp1 itself does not appear to act as a nuclease. Yet, very little is known about nsp1 interactions, in particular in the less studied α-CoV. We first wanted to ensure that our constructs accurately recapitulate known nsp1 interactions. We thus first verified interactions with select ribosomal proteins and confirmed that nsp1 from SARS1 expressed from our induced cell line interacts with RPS2, RPS3, and RACK1 (Figure S2). To better decipher the contribution of nsp1 to the regulation of gene expression, we next compared the interactome of the 4 selected nsp1 using affinity purification coupled with liquid chromatography-tandem mass spectrometry to map the nsp1-host protein interaction network (Figure 4A). After stringent filtering, 128 unique proteins were identified in this interactome, of which 74 were unique to 229E nsp1, 8 to MERS nsp1, 21 to SARS1 nsp1, and 9 to SARS2 nsp1 (Table S2). Notably, among the interactors of nsp1, ribosomal proteins and translation initiation factors were very prominent, which is in line with previous studies investigating nsp1’s interactions as well as consistent with what is known about nsp1 function during infection.27,28 Furthermore, comparison of the interaction networks highlighted that of the 4 nsp1 tested, nsp1 from MERS was the most divergent (Figure 4B). We next performed a functional analysis of this interactome using Gene Ontology (GO) on the combined list of interactors, which revealed 6 functional groups involved in nsp1-mediated RNA decay and essential cellular biological processes that could promote viral progression (Figure 4C). These results demonstrate the differences between coronavirus nsp1-host proteins interactions and provide insight into how host proteins may contribute to nsp1 role for progression of viral infection.
Figure 4.
Affinity purification followed by mass spectrometry (AP-MS) of nsp1-host protein interactions shows few interactors shared between coronaviruses
(A) Schematic representation of the AP-MS strategy used to identify the interactors of nsp1 from 4 different coronaviruses. FLAG-tagged nsp1 expression was induced in HEK293T as described above.
(B) Cytoscape network representation of nsp1-host protein interactions identified by MS. Host proteins (nodes) and the 4 nsp1s (hexagon) are represented. Intra-network interactions among host proteins (thin gray lines) were manually curated from the STRING and IntAct databases.
(C) Gene Ontology (GO) enrichment analysis was performed on the interacting proteins for each coronavirus nsp1 using DAVID bioinformatic database. The top (blue) histogram shows the raw p values of most enriched molecular function GO terms, and the bottom histogram (orange) represents enrichment scores for the 6 clusters found in the GO term analysis.
Discussion
Destabilization and degradation of host mRNA is a prevalent strategy employed by numerous viruses as a means of usurping the host gene expression machinery and to dampen the host immune responses.29,30,31,32,33,34,35,36,37 Prior research has highlighted the ability of SARS1, SARS2, and MERS coronavirus protein nsp1 to disrupt host gene expression through two prominent methods: modulation of host gene expression through mRNA degradation as well as binding of the 40S ribosomal subunit and subsequent translational arrest.18,22,23 Moreover, some work also explored the ability of the α-CoV nsp1, such as the one from HCoV-229E, to recapitulate some of the β-CoV nsp1 function.18,22 However, the extent and how conserved nsp1-mediated RNA decay is on the host transcriptome between the different coronavirus nsp1 remains poorly understood. Yet, we know that different nsp1s have different effect on the infected cells. For example, it has recently been shown that SARS2 nsp1 better suppresses STAT1 and STAT2 phosphorylation than the nsp1 protein of SARS1 and MERS thereby more strongly repressing interferon (IFN)-1 expression.38 This supports the notion that these proteins may function differently and may show different preferences toward decay and regulation of gene expression. Another example is MERS nsp1 that was shown to preferentially target transcripts of nuclear origin, while forgoing those transcribed in the cytoplasm.13 This again highlights how despite having a similar outcome on the host regulation, the mode of action of the various nsp1 may be coronavirus specific.
Here, we sought to explore the transcriptional landscape of cells expressing the nsp1 of different coronaviruses to better understand the impact of nsp1 between the β-CoVs and the α-CoV 229E. One caveat of our approach is that that nsp1-mediated effect on the transcriptome is known to depend on the IFN state of the cell, which is not addressed here. In fact, the overall state of the cell and timing of expression may be major drivers of nsp1-dependent decay. Fisher et al., for example observed a higher level of degradation than detected here at this 24-hour time point despite also performing their study in 293T cells.39 It would thus be interesting to have a dynamic view of RNA decay in cell and to quantify nsp1 role over time and under several immune states. Here, we observed that about half of the transcripts detected by RNA-seq were downregulated upon nsp1 induction, regardless of the origin of nsp1. This suggests that all nsp1s tested in our study have a conserved and likely profound impact on their host cells via the regulation of RNA stability. To our surprise, numerous transcripts detected in our sequencing data were non-coding RNAs (ncRNAs). Given the prominent association between nsp1 and the ribosome, it has been hypothesized that nsp1’s direct role in RNA decay was likely either associated with the host ribosome stalling pathways or alternatively that the endonuclease necessary to mediate nsp1 decay was recruited at the site of nsp1-ribosome interaction. The association of nsp1 and ncRNA would suggest that maybe nsp1’s role in RNA decay may be more extensive than previously thought or perhaps that nsp1 can target multiple decay pathways. MERS nsp1 has already been suggested to lead to RNA decay by targeting transcripts independently of ribosome interactions.13,40 Based on our observation that all the nsp1 tested here target ncRNAs, it could mean that they share this ribosome-independent RNA decay function. Furthermore, it would be interesting to explore this link between nsp1 and host ncRNA, as this could have widespread consequences on the regulation of the host cell.
Although SARS1 and SARS2 nsp1’s effect on the host transcriptome appear to adopt a “classic” volcano plot shape, indicative of widespread, uniform RNA destabilization on the host transcriptome, MERS and 229E nsp1 expression datasets do not follow this pattern. When we examine the MERS plot, we see not only many genes undergoing significant negative fold changes but also high representation of genes that have significance with little to no fold change. This could reflect the fact that MERS nsp1 has been suggested to only target nuclear transcribed RNAs, and our RNA-seq data represents total cellular transcripts. Tight clustering of the data would indeed result in this with few genes significantly represented in the plot. As for 229E, however, the vast majority of identified genes represented in the volcano plot showed little significance in fold change in totality. It could suggest that 229E preferentially target a subset of transcript, but has only mild effect on the rest of the transcriptome. Alternatively, it has been suggested by Wang et al.18 that nsp1’s greatest contribution to host shutoff is not in fact due to RNA decay but more likely because of the translational arrest induced by nsp1 binding to ribosomes. Our data here would indicate that this could be true for 229E nsp1, where we observe less marked RNA decay than its β-CoV counterparts. One alternative would be possibly a technical difference between these nsp1s, perhaps due to their optimal timing of expression and/or action. Finally, recently, it was discovered that SARS-CoV2 nsp1 can disrupt nuclear export pathways,41 which could also differentially play a role in the breadth of transcripts targeted by the various nsp1s.
Another interesting observation was that a fraction of transcripts appears to be stabilized and/or have increased expression upon expression of the various nsp1s. These transcripts could have function important for the regulation of the infected cell. In fact, GO term analysis of the “upregulated” transcripts for SARS2 nsp1 revealed an enrichment for genes involved in “cytokine activity.” However, it remains to be assessed whether these transcripts are truly refractory to nsp1 or whether this is a secondary effect of nsp1-mediated widespread decay and translational arrest.
Hierarchical clustering of our transcriptomic data highlight that although all nsp1 proteins induce large-scale RNA decay, there is limited overlap in their specific targets, especially between SARS1 and SARS2 nsp1. Moreover, genes that are heavily downregulated by MERS nsp1 are clustered to the lower half of the heatmap, likely representing nuclear transcripts. This is very similar to the clustered gene degraded by 229E nsp1, suggesting that 229E nsp1 may also preferentially target nuclear transcripts.
We also verified our results using the SARS2 (R124A + K125A) and MERS (R146A + K147A) nsp1 mutants, confirming that these mutants were not able to induce RNA decay like their WT counterparts. As it has been suggested before, although these mutations lead to a loss of mRNA decay potential in the nsp1 protein, this is likely not linked directly to nsp1 containing RNAse activity. As no nsp1 sequence contains amino acid sequences associated with RNA catalysis, the mutations could potentially disrupt interactions with a protein that may play this role for nsp1.
Another challenging aspect of studying nsp1 biology is that it has been difficult to identify robust interactions between nsp1 and host proteins. Small and large screens looking either specifically at nsp1 interactome or more globally at the interactome of all CoV proteins have identified only few, rarely confirmed interactors.27,42,43 Here, to increase the robustness of our results, we continued with our comparative approach and sought to identify interactors that were shared by these 4 nsp1s. Intriguingly, out of this interactome, 229E nsp1 had by far the highest number of unique interactors and MERS nsp1 had the fewest. 229E nsp1, like other α-CoV nsp1 are drastically smaller than those found in β-CoVs. We can thus speculate that it would require more interacting partners to achieve its primary functions during viral infection. It is also possible that 229E nsp1 possesses more disordered regions—regions that are known to facilitate protein-protein interactions—as it might be more difficult for a protein of only 111 amino acids to fold into complex structures. However, studies have highlighted that nsp1 from the α-CoV TGEV (transmissible gastroenteritis virus) and PEDV lack disordered regions.23,44 Therefore, the increased number of potential interactors for 229E nsp1 could be conferred by some other yet uncharacterized intrinsic domain.
Given that MERS nsp1 is known to operate very differently than the nsp1 from SARS1 and SARS2, it is perhaps less surprising to see that its interactome is also very different. Although we did not identify in this interactome any host protein with direct RNAse activity that could account for nsp1’s role in RNA decay, our GO revealed that many of the interactors that we detected were associated with mRNA catabolic process. It would thus be interesting to explore these interactions and assess whether any of these factors are essential for nsp1 activity.
In this work we sought to explore the similarities and differences between key RNA regulatory proteins during coronavirus infection, to better understand how their expression leads to different viral pathogenic conditions. We observed that even between closely related CoVs, there seems to be little similarities in clustering of genes targeted for decay and that there are few protein interactors shared between these different proteins. However, gene targeting for these proteins does not appear to be random; based on hierarchical clustering the nsp1s seem to target different gene clusters.
Limitation of study
A limitation of our study is that the nsp1 proteins were expressed from lentiviral constructs in uninfected cells. In an infection model, there could be additional viral factors impacting nsp1 interactions altering the transcriptome in a way unobserved in our study. Additionally, our nsp1s were expressed without the presence of the 5′ leader, which in the context of infection, prevents nsp1 self-cleaving. Finally, our RNA-seq was performed on total mRNA: given that MERS nsp1 is known to specifically target nucleartranscripts, nuclear fractionation may be more suited to assess MERS nsp1 impact on the transcriptome.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-Flag | Fisher Scientific | MA191878; RRID:AB_1957945 |
| Rabbit anti-Vinculin | Invitrogen | Cat#PI700062 AB_700062; RRID: AB_2532280 |
| HRP Goat anti-Rabbit IgG | Southern Biotechnology Associates | Cat#OB4030-05; RRID:AB_2687483 |
| HRP Goat anti-Mouse IgG | Southern Biotechnology Associates | Cat#OB1031-05 AB_103105 |
| Anti-FLAG® M2 Magnetic Beads | Sigma-Aldrich | M8823-1ML; RRID:AB_2637089 |
| Rabbit anti-RPS2 | Bethyl Labs | A303-794A; RRID:AB_11218192 |
| Rabbit anti-RACK1 | Bethyl Labs | A302-545A; RRID:AB_1999012 |
| Rabbit anti-RPS3 | Proteintech | 11990-1-AP; RRID:AB_2180758 |
| Bacterial and virus strains | ||
| MAX Efficiency™ DH5α Competent Cells | Invitrogen, Fisher | 18258012 |
| Chemicals, peptides, and recombinant proteins | ||
| EcoRI | New England Biolabs | R0101S |
| NotI-HF | New England Biolabs | R3189S |
| Zeocin | Invitrogen | R25001 |
| TRIzol Reagent | ThermoFisher | 15596026 |
| Turbo DNase | Invitrogen | AM2238 |
| AMV Reverse Transcriptase | Promega | PR-M5108 |
| Q5® High-Fidelity DNA Polymerase | New England Biolabs | M0491S |
| Doxycycline | BD Biosciences | NC0424034 |
| Polybrene | FISHER | Cat# NC9840454 |
| C18 tips | Pierce | UJ291680 |
| Critical commercial assays | ||
| PolyJet (transfection reagent) | SignaGen Laboratories | SL100688 |
| iTaq™ Universal SYBR® Green Supermix | Bio-Rad | 1725124 |
| In-Fusion HD Cloning Kit | TaKaRa Bio | 639650 |
| T4 DNA Ligase | New England Biolabs | M0202S |
| Deposited data | ||
| RNA-seq files | NCBI GEO | GSE217848 |
| Experimental models: Cell lines | ||
| Human: HEK 293T Cells | ATCC | CRL-3216 |
| Oligonucleotides | ||
| Table S4 | This paper | N/A |
| Recombinant DNA | ||
| pLVX TetOne-Zeo-MERS-nsp1-3xFlag | Britt Glaunsinger | Derived from Addgene #129746 |
| pLVX TetOne-Zeo-COV2-nsp1-3xFlag | Britt Glaunsinger | Derived from Addgene #129746 |
| pMD2.G | Britt Glaunsinger | Plasmid #12259 (Addgene) |
| psPAX2 | Britt Glaunsinger | Plasmid#12260 (Addgene) |
| pLVX TetOne-Zeo-CoV-nsp1-3xFlag | This Paper | N/A |
| pLVX TetOne-Zeo-229E-nsp1-3xFlag | This Paper | N/A |
| pcDNA4.TO-MERS-nsp1 | Britt Glaunsinger | N/A |
| pcDNA4.TO-CoV2-nsp1 | Britt Glaunsinger | N/A |
| Software and algorithms | ||
| Galaxy Plat-form | Afgan et al., 201845 | https://usegalaxy.org/ |
| Bowtie2 (Galaxy Plat-form) | Langmead and Salzberg |
https://usegalaxy.org/ http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| Cufflinks (Galaxy Plat-form) | Trapnell et. al., 201346 | https://usegalaxy.org/ |
| Cuffdiff (Galaxy Plat-form) | Trapnell et al., 201346 | https://usegalaxy.org/ |
| Multiple List Comparator | https://molbiotools.com/listcompare.php | |
| VolcaNoseR | Goedhart and Luijsterburg 202047 | https://huygens.science.uva.nl/VolcaNoseR2/ |
| DAVID | Nature Protocols 2009; 4(1):44 & Nucleic Acids Res. 2009; 37(1):1 | https://david.ncifcrf.gov/ |
| CRAPpome | Mellacheruvu et al & Nature Methods 2013. | https://reprint-apms.org/?q = chooseworkflow |
| Cytoscape | Genome Res. 2003. 13: 2498-2504 | https://cytoscape.org/ |
| Python | N/A | Version 3.9.1 |
| Seaborn | N/A | Version 0.11.2 |
| Matplot | N/A | Version 3.5.1 |
| Prism | N/A | Version 9.4.1 |
Resource availability
Lead contact
Requests for resources and reagents should be directed to the Lead Contact Author Mandy Muller (mandymuller@umass.edu).
Material availability
All unique reagents generated in this study are available from the lead contact.
Experimental model and subject details
Plasmids and plasmid construction
The pLVX-TetOne-Zeo-SARS2-NSP1-3xFlag and pLVX-TetOne-Zeo-MERS-NSP1-3xFlag were a kind gift from the Glaunsinger Lab and were derived from the pLVX ORF18 plasmid (addgene #129746). SARS1 and 229E nsp1 were cloned into the pLVX backbone (Table S4 - List of primers - related to STAR Methods). Briefly, MERS nsp1 was removed from the vector using EcoRI and NotI. SARS1 and 229E sequences were obtained as gBlocks (IDT), PCR amplified, ligated into the same EcoRI and NotI. All plasmid constructions were verified by Sanger sequencing. Transient expression vectors pcDNA4 SARS2 nsp1 Mut 3xFlag and pcDNA MERS nsp1 mut 3xFlag were also a gift from the Glaunsinger lab.
Cells, transfections, and lentiviral transduction
HEK293T cells (ATCC) were grown in Dulbecco’s modified Eagle’s medium (DMEM – Invitrogen) supplemented with 10% fetal bovine serum (FBS). To establish the lentiviral cell lines, HEK293T cells were co-transfected with their respective zeocine-resistant, lentiviral vector along with pMD2.G and psPAX2 the envelop, packaging, and accessory plasmids in DMEM 0% FBS with using Polyjet (SignaGen). 48 h post transfection the supernatant from transfected cells was collected and filtered through a 0.45 μM filter diluted with serum free media with polybrene at a concentration of 8 μg/ml. Mixture was added to fresh HEK293T cells in a 6-well plate and underwent spinfection at 1500rpm for 1.5 h. Cells were incubated overnight then split the following day into a 10 cm plate with media containing 325 μg/ml of zeocin. After selection the established lentivirally infected cell lines were maintained in 162.5 μg/ml zeocin. For DNA transfection, HEK293T cells were plated and transfected after 24h when 70% confluent using PolyJet (SignaGen).
Western blotting
Cell lysates were prepared in TP150 lysis buffer (NaCl, 150 mM; Tris, 50 mM; NP-40, 0.5%; dithiothreitol [DTT]. 1 mM; and protease inhibitor tablets) and quantified by Bradford assay. 20ug of each sample were resolved by SDS-PAGE and Western blotted with following antibodies in TBST (Tris-buffered saline, 0.1% Tween 20): mouse anti-Flag at 1:1000 (Invitrogen) and rabbit anti-Vinculin at 1:2000 (Invitrogen). Primary antibody incubations were followed by horseradish peroxidase (HRP)-conjugated goat anti-mouse or goat anti-rabbit secondary antibodies (1:5000; Southern Biotechnology)
RT-qPCR
Total RNA was harvested using TRIzol according to the manufacture’s protocol. cDNAs were synthesized from 1 μg of total RNA using AMV reverse transcriptase (Promega) and used directly for qPCR analysis with the SYBR green qPCR kit (Bio-Rad). Signals obtained by qPCR were normalized to those for 18S.
RNA-seq
6 biological replicates of each lentiviral transduced cells (SARS1, SARS2, MERS, 229E and uninduced sample as a control) were grown to 80% confluency then induced with 1ug/ml of Doxycycline (BD Biosciences). After 24h cells were collected in TRIzol Reagent and RNA was harvested following the manufacturer’s protocol. Purity of samples were analyzed via bioanalyzer. Following poly(A) selection, libraries underwent 76-base paired-end sequencing using the NextSeq500 Mid-150 cycle kit on a NextSeq 500. Read quality was assessed using fastqc. Using Galaxy45 reads were then aligned to the human genome (hg38) by Bowtie2 and differential expression analysis were performed using Cufflink and Cuffdiff.46,48 Data was normalized using FPKM methodology based on gene length and total number of mapped reads. This analysis was performed between the induced condition to the uninduced condition for each nsp1 dataset to get differential expression of genes. For graphical representation in the heatmap, fold change values were saturated by a hyperbolic tan function with a cutoff set at 10. Hierarchical clustering was generated in Python using the SciPy package with complete linkage and Euclidian distance. Volcano plots were developed using the program VolcaNoseR47 (LCAM) by plotting the log2Fold Change vs -log10p value, with significance thresholds left at the default settings and the top 10 most significant genes highlighted and labeled. Comparison of overlapping genes and generation of Venn diagrams were generated using Multiple List Comparator (Molbiotools).
Immunoprecipitation
Cells were lysed in a low-salt lysis buffer (150 mM NaCl, 0.5% NP-40, 50 mM Tris [pH 8], 1 mM DTT, and protease inhibitor cocktail), and protein concentrations were determined by Bradford assay. For FLAG construct pull-downs, 400 μg of total protein lysates were incubated overnight with Anti-FLAG M2 Magnetic Beads (Sigma) or control G-coupled magnetic beads. Beads were then washed extensively with lysis buffer. Lastly, samples were resuspended in 4X laemmli loading dye before resolution by SDS-PAGE and further Western blotting.
Mass spectrometry
Lentiviral transduced cells were grown in 10 cm plates to an 80% confluency and then induced with 1 μg/ml of Doxycycline (BD Biosciences). 24 h post-induction, cells were harvested and lysed, and immunoprecipitation (as mentioned above) was performed overnight at 4C. Samples were extensively washed, and trypsin digested overnight. Samples were then cleaned up using a C18 column and sent to the University of Massachusetts Mass Spectrometry Center where peptides were separated by reverse-phase chromatography using nano-flow EASY-nLC 1000 UHPLC coupled to Orbitrap Fusion mass spectrometer (Thermo Scientific) with PepMap RSLCnano column (75 μm ID, 15 cm). Peptides were eluted over a 90 min 5–35% ACN gradient at 300 nL/min. Survey scans were measured in the Orbitrap analyzer at 60,000 resolutions. Data were collected in the linear ion trap using a 2-s cycle time with a full MS mass range from 400 to 100 m/z. Peptides (charge state 2–6) were fragmented using higher energy collisional dissociation using a normalized collision energy setting of 27. A dynamic exclusion time of 5 s was used and the peptide match setting was enabled. Raw files were analyzed in Proteome Discoverer 2.4 (Thermo Scientific) using the SEQUEST search algorithm using the Human Proteome database. The search parameters used were as follows: 10 ppm precursor ion tolerance and 0.4 Da fragment ion tolerance; up two missed cleavages were allowed; dynamic modifications of methionine oxidation and N-terminal acetylation. Peptide matches were filtered to a protein false discovery rate of 5% using Percolator algorithm. Data was then filtered based on the number of peptides present for each hit between 3 biological replicates against control samples. Unique hits in the experimental sample were then filtered through the CRAPome database49 to determine false positive rates. Gene Ontology (GO) enrichment analysis was then performed on the human interacting proteins for each coronavirus using DAVID bioinformatic database. Top enriched and shared clusters are identified on the network using the Cytoscape software.
Quantification and statistical analysis
All results are expressed as means ± standard errors of the means (SEMs) of experiments independently repeated at least three times (individual replicate points are shown on bar graphs) using Prism9. Unpaired Student’s test was used to evaluate the statistical difference between samples. Significance was evaluated with p values as indicated in figure legends.
Acknowledgements
We thank all members of the Muller lab for helpful discussions and suggestions. Special thanks to Dr. Britt Glaunsinger and Dr. Ella Hartenian for the pLVX plasmids. We would also like to thank Dr. Ravi Ranjan at the genomics facility for guidance with sequencing and Dr. Stephen Eyles at the UMASS IALS Mass Spectrometry facility for help with protocol development and data acquisition. This research was supported a Smith-Spaulding fellowship to Y.B., training grant support (T32 GM139789) to Y.B., and an IALS midigrant to M.M.
Author contributions
Performed experiments related to RNA and protein work including cloning, western blotting, RT-qPCR, immunoprecipitations, and sample preparation for RNA-seq and mass spectrometry, Y.B. and J.M.; experimental design and analysis of data, Y.B., J.M., and M.M.; drafting and editing of manuscript, Y.B., J.M., and M.M.; supervising project, M.M.
Declaration of interests
The authors declare no competing interests.
Published: January 20, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105887.
Supplemental information
Data and code availability
-
•
The raw sequence data generated in this paper were deposited on the NCBI Gene Expression Omnibus (GEO) server and can be found here: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE217848 under the identifier GSE217848
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request
References
- 1.Gorbalenya A.E., Snijder E.J., Spaan W.J.M. Severe acute respiratory syndrome coronavirus phylogeny: toward consensus. J. Virol. 2004;78:7863–7866. doi: 10.1128/JVI.78.15.7863-7866.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J., Poon L.L.M., Guan Y., Rozanov M., Spaan W.J.M., Gorbalenya A.E. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331:991–1004. doi: 10.1016/s0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Woo P.C.Y., Huang Y., Lau S.K.P., Yuen K.Y. Coronavirus genomics and bioinformatics analysis. Viruses. 2010;2:1804–1820. doi: 10.3390/v2081803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woo P.C.Y., Lau S.K.P., Lam C.S.F., Lau C.C.Y., Tsang A.K.L., Lau J.H.N., Bai R., Teng J.L.L., Tsang C.C.C., Wang M., et al. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamitani W., Huang C., Narayanan K., Lokugamage K.G., Makino S. A two-pronged strategy to suppress host protein synthesis by SARS coronavirus Nsp1 protein. Nat. Struct. Mol. Biol. 2009;16:1134–1140. doi: 10.1038/nsmb.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Narayanan K., Huang C., Lokugamage K., Kamitani W., Ikegami T., Tseng C.T.K., Makino S. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J. Virol. 2008;82:4471–4479. doi: 10.1128/JVI.02472-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finkel Y., Mizrahi O., Nachshon A., Weingarten-Gabbay S., Morgenstern D., Yahalom-Ronen Y., Tamir H., Achdout H., Stein D., Israeli O., et al. The coding capacity of SARS-CoV-2. Nature. 2021;589:125–130. doi: 10.1038/s41586-020-2739-1. [DOI] [PubMed] [Google Scholar]
- 10.Finkel Y., Gluck A., Nachshon A., Winkler R., Fisher T., Rozman B., Mizrahi O., Lubelsky Y., Zuckerman B., Slobodin B., et al. SARS-CoV-2 uses a multipronged strategy to impede host protein synthesis. Nature. 2021;594:240–245. doi: 10.1038/s41586-021-03610-3. [DOI] [PubMed] [Google Scholar]
- 11.Lapointe C.P., Grosely R., Johnson A.G., Wang J., Fernández I.S., Puglisi J.D. Dynamic competition between SARS-CoV-2 NSP1 and mRNA on the human ribosome inhibits translation initiation. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2017715118. e2017715118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mendez A.S., Ly M., González-Sánchez A.M., Hartenian E., Ingolia N.T., Cate J.H., Glaunsinger B.A. The N-terminal domain of SARS-CoV-2 nsp1 plays key roles in suppression of cellular gene expression and preservation of viral gene expression. Cell Rep. 2021;37:109841. doi: 10.1016/j.celrep.2021.109841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lokugamage K.G., Narayanan K., Nakagawa K., Terasaki K., Ramirez S.I., Tseng C.T.K., Makino S. Middle East respiratory syndrome coronavirus nsp1 inhibits host gene expression by selectively targeting mRNAs transcribed in the nucleus while sparing mRNAs of cytoplasmic origin. J. Virol. 2015;89:10970–10981. doi: 10.1128/JVI.01352-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakagawa K., Narayanan K., Wada M., Popov V.L., Cajimat M., Baric R.S., Makino S. The endonucleolytic RNA cleavage function of nsp1 of Middle East respiratory syndrome coronavirus promotes the production of infectious virus particles in specific human cell lines. J. Virol. 2018;92 doi: 10.1128/JVI.01157-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Narayanan K., Ramirez S.I., Lokugamage K.G., Makino S. Coronavirus nonstructural protein 1: common and distinct functions in the regulation of host and viral gene expression. Virus Res. 2015;202:89–100. doi: 10.1016/j.virusres.2014.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Setaro A.C., Gaglia M.M. All hands on deck: SARS-CoV-2 proteins that block early anti-viral interferon responses. Curr. Res. Virol. Sci. 2021;2:100015. doi: 10.1016/j.crviro.2021.100015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao J., Wu C.C., Lin T.L. Complete nucleotide sequence of polyprotein gene 1 and genome organization of Turkey coronavirus. Virus Res. 2008;136:43–49. doi: 10.1016/j.virusres.2008.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y., Shi H., Rigolet P., Wu N., Zhu L., Xi X.G., Vabret A., Wang X., Wang T. Nsp1 proteins of group I and SARS coronaviruses share structural and functional similarities. Infect. Genet. Evol. 2010;10:919–924. doi: 10.1016/j.meegid.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ziebuhr J. The coronavirus replicase. Curr. Top. Microbiol. Immunol. 2005;287:57–94. doi: 10.1007/3-540-26765-4_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ziebuhr J., Schelle B., Karl N., Minskaia E., Bayer S., Siddell S.G., Gorbalenya A.E., Thiel V. Human coronavirus 229E papain-like proteases have overlapping specificities but distinct functions in viral replication. J. Virol. 2007;81:3922–3932. doi: 10.1128/JVI.02091-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang C., Lokugamage K.G., Rozovics J.M., Narayanan K., Semler B.L., Makino S. Alphacoronavirus transmissible gastroenteritis virus nsp1 protein suppresses protein translation in mammalian cells and in cell-free HeLa cell extracts but not in rabbit reticulocyte lysate. J. Virol. 2011;85:638–643. doi: 10.1128/JVI.01806-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen Z., Wang G., Yang Y., Shi J., Fang L., Li F., Xiao S., Fu Z.F., Peng G. A conserved region of nonstructural protein 1 from alphacoronaviruses inhibits host gene expression and is critical for viral virulence. J. Biol. Chem. 2019;294:13606–13618. doi: 10.1074/jbc.RA119.009713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen Z., Ye G., Deng F., Wang G., Cui M., Fang L., Xiao S., Fu Z.F., Peng G. Structural basis for the inhibition of host gene expression by porcine epidemic diarrhea virus nsp1. J. Virol. 2018;92:018966–e1917. doi: 10.1128/JVI.01896-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma S., Sun S., Li J., Fan Y., Qu J., Sun. L., Wang S., Zhang Y., Yang S., Liu Z., et al. Single-cell transcriptomic atlas of primate cardiopulmonary aging. Cell Res. 2021;31:415–432. doi: 10.1038/s41422-020-00412-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lokugamage K.G., Narayanan K., Huang C., Makino S. Severe acute respiratory syndrome coronavirus protein nsp1 is a novel eukaryotic translation inhibitor that represses multiple steps of translation initiation. J. Virol. 2012;86:13598–13608. doi: 10.1128/JVI.01958-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Min Y.Q., Mo Q., Wang J., Deng F., Wang H., Ning Y.J. SARS-CoV-2 nsp1: bioinformatics, potential structural and functional features, and implications for drug/vaccine designs. Front. Microbiol. 2020;11:587317. doi: 10.3389/fmicb.2020.587317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O'Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim D.-K., Weller B., Lin C.-W., Sheykhkarimli D., Knapp J.J., Kishore N., Sauer M., Rayhan A., Young V., Marin-de la Rosa N., et al. A map of binary SARS-CoV-2 protein interactions implicates host immune regulation and ubiquitination. bioRxiv. 2021 doi: 10.1101/2021.03.15.433877. Preprint at. [DOI] [Google Scholar]
- 29.Abernathy E., Glaunsinger B. Emerging roles for RNA degradation in viral replication and antiviral defense. Virology. 2015;479–480:600–608. doi: 10.1016/j.virol.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Esclatine A., Taddeo B., Evans L., Roizman B. The herpes simplex virus 1 UL41 gene-dependent destabilization of cellular RNAs is selective and may be sequence-specific. Proc. Natl. Acad. Sci. USA. 2004;101:3603–3608. doi: 10.1073/pnas.0400354101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Everly D.N., Jr., Feng P., Mian I.S., Read G.S. mRNA degradation by the virion host shutoff (Vhs) protein of herpes simplex virus: genetic and biochemical evidence that Vhs is a nuclease. J. Virol. 2002;76:8560–8571. doi: 10.1128/jvi.76.17.8560-8571.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gaglia M.M., Covarrubias S., Wong W., Glaunsinger B.A. A common strategy for host RNA degradation by divergent viruses. J. Virol. 2012;86:9527–9530. doi: 10.1128/JVI.01230-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gaucherand L., Porter B.K., Levene R.E., Price E.L., Schmaling S.K., Rycroft C.H., Kevorkian Y., McCormick C., Khaperskyy D.A., Gaglia M.M. The influenza A virus endoribonuclease PA-X usurps host mRNA processing machinery to limit host gene expression. Cell Rep. 2019;27:776–792.e7. doi: 10.1016/j.celrep.2019.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Glaunsinger B., Ganem D. Lytic KSHV infection inhibits host gene expression by accelerating global mRNA turnover. Mol. Cell. 2004;13:713–723. doi: 10.1016/s1097-2765(04)00091-7. [DOI] [PubMed] [Google Scholar]
- 35.Khaperskyy D.A., Emara M.M., Johnston B.P., Anderson P., Hatchette T.F., McCormick C. Influenza a virus host shutoff disables antiviral stress-induced translation arrest. PLoS Pathog. 2014;10:e1004217. doi: 10.1371/journal.ppat.1004217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ly M., Burgess H.M., Shah S.B., Mohr I., Glaunsinger B.A. Vaccinia virus D10 has broad decapping activity that is regulated by mRNA splicing. PLoS Pathog. 2022;18:e1010099. doi: 10.1371/journal.ppat.1010099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rowe M., Glaunsinger B., van Leeuwen D., Zuo J., Sweetman D., Ganem D., Middeldorp J., Wiertz E.J.H.J., Ressing M.E. Host shutoff during productive Epstein-Barr virus infection is mediated by BGLF5 and may contribute to immune evasion. Proc. Natl. Acad. Sci. USA. 2007;104:3366–3371. doi: 10.1073/pnas.0611128104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xia H., Cao Z., Xie X., Zhang X., Chen J.Y.C., Wang. H., Menachery V.D., Rajsbaum R., Shi P.Y. Evasion of type I interferon by SARS-CoV-2. Cell Rep. 2020;33:108234. doi: 10.1016/j.celrep.2020.108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fisher T., Gluck A., Narayanan K., Kuroda M., Nachshon A., Hsu J.C., Halfmann P.J., Yahalom-Ronen Y., Tamir H., Finkel Y., et al. Parsing the role of NSP1 in SARS-CoV-2 infection. Cell Rep. 2022;39:110954. doi: 10.1016/j.celrep.2022.110954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakagawa K., Makino S. Mechanisms of coronavirus nsp1-mediated control of host and viral gene expression. Cells. 2021;10:300. doi: 10.3390/cells10020300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang K., Miorin L., Makio T., Dehghan I., Gao S., Xie Y., Zhong H., Esparza M., Kehrer T., Kumar A., et al. Nsp1 protein of SARS-CoV-2 disrupts the mRNA export machinery to inhibit host gene expression. Sci. Adv. 2021;7:eabe7386. doi: 10.1126/sciadv.abe7386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen Z., Wang C., Feng X., Nie L., Tang M., Zhang H., Xiong Y., Swisher S.K., Srivastava M., Chen J. Interactomes of SARS-CoV-2 and human coronaviruses reveal host factors potentially affecting pathogenesis. EMBO J. 2021;40:e107776. doi: 10.15252/embj.2021107776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stukalov A., Girault V., Grass V., Karayel O., Bergant V., Urban C., Haas D.A., Huang Y., Oubraham L., Wang A., et al. Multilevel proteomics reveals host perturbations by SARS-CoV-2 and SARS-CoV. Nature. 2021;594:246–252. doi: 10.1038/s41586-021-03493-4. [DOI] [PubMed] [Google Scholar]
- 44.Jansson A.M. Structure of alphacoronavirus transmissible gastroenteritis virus nsp1 has implications for coronavirus nsp1 function and evolution. J. Virol. 2013;87:2949–2955. doi: 10.1128/JVI.03163-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Afgan E., Baker D., Batut B., van den Beek M., Bouvier D., Cech M., Chilton J., Clements D., Coraor N., Grüning B.A., et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018;46:W537–W544. doi: 10.1093/nar/gky379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trapnell C., Hendrickson D.G., Sauvageau M., Goff L., Rinn J.L., Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goedhart J., Luijsterburg M.S. VolcaNoseR is a web app for creating, exploring, labeling and sharing volcano plots. Sci. Rep. 2020;10:20560. doi: 10.1038/s41598-020-76603-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mellacheruvu D., Wright Z., Couzens A.L., Lambert J.P., St-Denis N.A., Li T., Miteva Y.V., Hauri S., Sardiu M.E., Low T.Y., et al. The CRAPome: a contaminant repository for affinity purification–mass spectrometry data. Nat. Methods. 2013;10:730–736. doi: 10.1038/nmeth.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The raw sequence data generated in this paper were deposited on the NCBI Gene Expression Omnibus (GEO) server and can be found here: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE217848 under the identifier GSE217848
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request