Abstract
The cytokine granulocyte-macrophage-colony stimulating factor (GM-CSF) possesses the capacity to differentiate monocytes into macrophages (MØs) with opposing functions, namely, proinflammatory M1-like MØs and immunosuppressive M2-like MØs. Despite the importance of these opposing biological outcomes, the intrinsic mechanism that regulates the functional polarization of MØs under GM-CSF signaling remains elusive. Here, we showed that GM-CSF-induced MØ polarization resulted in the expression of cytokine-inducible SH2-containing protein (CIS) and that CIS deficiency skewed the differentiation of monocytes toward immunosuppressive M2-like MØs. CIS deficiency resulted in hyperactivation of the JAK-STAT5 signaling pathway, consequently promoting downregulation of the transcription factor Interferon Regulatory Factor 8 (IRF8). Loss- and gain-of-function approaches highlighted IRF8 as a critical regulator of the M1-like polarization program. In vivo, CIS deficiency induced the differentiation of M2-like macrophages, which promoted strong Th2 immune responses characterized by the development of severe experimental asthma. Collectively, our results reveal a CIS-modulated mechanism that clarifies the opposing actions of GM-CSF in MØ differentiation and uncovers the role of GM-CSF in controlling allergic inflammation.
Keywords: CIS, GM-CSF, Macrophage, M2, M1
Subject terms: Autoimmunity, Cell signalling
Introduction
Macrophages (MØs) play crucial roles in immune defense against invading pathogens and have important functions in regulating and maintaining tissue homeostasis [1]. It has long been appreciated that MØs show functional plasticity/polarization and dynamically respond to different physiological situations. External cues, including cytokines and Toll-like receptor (TLR) agonists, can direct MØ functional polarization [2]. Historically, interferon (IFN)-γ and interleukin (IL)-4 have been used to induce polarization of classically activated M1 MØs with strong proinflammatory functions and alternatively activated M2 MØs with anti-inflammatory functions, respectively [2].
Differing from the aforementioned modes of MØ polarization, MØs generated from bone marrow (BM) progenitors or monocytes in the presence of granulocyte-macrophage-colony stimulating factor (GM-CSF) or macrophage-colony stimulating factor (M-CSF), respectively, can display both M1- and M2-like characteristics [3]. GM-CSF-differentiated MØs are thought to be M1-like, producing proinflammatory cytokines upon stimulation with TLR ligands [3–5]. Corroborating evidence from an autoimmune disease context supports a proinflammatory role for GM-CSF in vivo (reviewed in [6, 7]). Consistent with its proinflammatory properties, GM-CSF has long been used in the vaccination setting as a strong immune adjuvant to promote antitumor immunity [8].
Paradoxically, GM-CSF has also been associated with the development of suppressive M2-like MØs in various tumor settings [9–11] and after renal ischemia [12]. GM-CSF also has a critical role in the induction of allergic inflammation [13, 14], and consequently, its neutralization was found to dampen inflammation in certain allergic settings [13], although it is not clear that M2-like MØs are directly responsible for the induction of allergic inflammation. Nevertheless, M2-like MØs are known to support Th2 immunity [15]. A protective role for GM-CSF in murine models of dextran sulfate sodium (DSS)-induced colitis was also associated with GM-CSF-induced myeloid cells [16]. Together, these studies point to divergent functional outcomes driven by GM-CSF signaling in MØs, yet the molecular mechanism governing the functional dichotomy of the effects of GM-CSF on MØs is largely unknown.
The strength and duration of GM-CSF signaling are tightly regulated by the induction of suppressors of cytokine signaling (SOCS) proteins, which act in a negative feedback loop to limit cytokine responses [17, 18]. Among the SOCS family members, cytokine-inducible SH2 protein (CIS) is a known target of STAT5 activation [19–21] and is induced by GM-CSF in MØs [19, 22]. CIS-deficient mice housed in a specific pathogen-free environment showed no overt defects in myelopoiesis, suggestive of low GM-CSF or CIS expression in an unchallenged situation [23, 24]. In line with this, the impact of CIS deficiency in natural killer (NK) cells or T cells became apparent only following stimulation with exogenous IL-15 or T cell receptor (TCR) engagement, respectively [25–27].
Here, we explored the role of CIS in regulating MØ functional polarization following GM-CSF stimulation. We found that CIS deficiency resulted in the development of MØs with strong immunosuppressive functions, limited production of IL-12 and an M2-like MØ gene signature that ultimately induced a Th2-biased immune response in vivo. The functional skewing was largely the result of hyperactivation of the JAK-STAT5 signaling pathway, which led to suboptimal IRF8 induction and thus crippled the M1-like polarization program. Thus, our study highlights a critical role for CIS in fine-tuning GM-CSF signaling and promoting M1-like features in macrophages.
Results
CIS deficiency leads to the generation of MØs that strongly inhibit T-cell responses
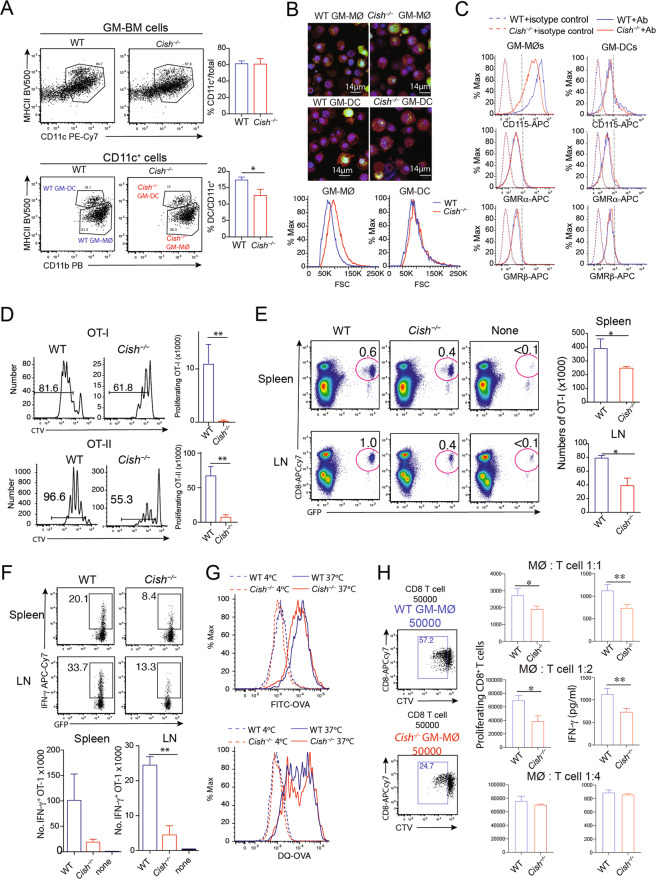
In line with previous studies [23, 26], we did not observe any conspicuous changes in the frequencies of myeloid cell types or BM hematopoietic progenitors in Cish−/− mice (Figs. S1 and S2). To investigate the role of CIS in MØ differentiation, we cultured BM progenitors from WT and Cish−/− mice [26] for 7 days with GM-CSF to generate MØs (GM-MØs; CD11c+MHCIIint CD11bhiCD115hiCD86loFlt3lo) and dendritic cells (DCs) (GM-DCs; CD11c+MHCIIhiCD11bintCD115-CD86hiFlt3hi) [28] (Fig. 1A). GM-MØs dominated in both genotypes (80–90% of total CD11c+ cells), with a small but significant reduction in the proportion of GM-DCs in Cish−/− BM cell cultures (Fig. 1A). Flow cytometry and fluorescence microscopy experiments revealed that Cish−/− GM-MØs were larger in size than WT GM-MØs (Fig. 1B). We also found that Cish−/− GM-MØs had lower cell-surface expression of CD115 (Macrophage-Colony Stimulating Factor receptor; M-CSFR) than WT GM-MØs, while GM-MØs of both genotypes expressed similar levels of the GM-CSF receptor (GM-CSFR) α and β subunits (Fig. 1C). Downregulation of CD115 by GM-CSF was dose dependent and cell intrinsic since Cish−/− GM-MØs derived from a coculture containing WT cells also had lower CD115 expression (Fig. S3A, B). Similarly, the expression of other myeloid markers, such as CD209, CCR2, F4/80 and CD14, was reduced in Cish−/− GM-MØs compared to their WT counterparts (Fig. S3B, C). In contrast, CIS deficiency in GM-DCs had a limited impact on the expression of multiple cell-surface markers (Fig. S3D).
Fig. 1.
Cish−/− GM-MØs have a reduced capacity to induce T-cell responses. A WT and Cish−/− GM-MØ BM cells were cultured with 10 ng/mL GM-CSF for 7 days. Harvested cells were analyzed to identify GM-MØs and GM-DCs. B Sorted GM-MØs and GM-DCs were stained with CellTracker violet (red), LysoTracker green (green) and SIR-DNA (blue). Histograms showing cell size measured from the forward scatter data for WT and Cish−/− GM-MØs and GM-DCs. C Histograms showing the expression of M-CSF (CD115) and GM-CSF receptors (GMRα and GMRβ) on WT and Cish−/− GM-MØs and GM-DCs. D CTV-labeled OT-I or OT-II T cells were cultured with WT or Cish−/− GM-MØs in the presence of ovalbumin (OVA). Bar graphs represent the mean number ± S.D. of proliferating OT-I T cells at 48 h (top) and OT-II T cells at 60 h (bottom). Data are representative of three independent experiments. E B6 mice that had been previously injected with GFP-OT-I cells were intravenously infused with OVA-pulsed WT (n = 3) or Cish−/− GM-MØs (n = 3). *P < 0.05, Student’s t test. Antigen-induced T-cell expansion was evaluated 5 days after MØ transfer. F IFN-γ production by GFP-OT-I cells was evaluated after 4 h of stimulation with PMA/ionomycin. **P < 0.01, Student’s t test. Data are representative of two independent experiments. G Antigen uptake and processing of soluble OVA by WT and Cish−/− GM-MØs. For antigen uptake, cells were incubated with FITC‐OVA at 37 °C or on ice for 30 min. For antigen processing, cells were incubated with DQ-OVA at 37 °C for 30 min. Then, the samples were either incubated at 37 °C for an additional 90 min or kept on ice. H Purified CD8+ T cells from B6 mice were stimulated with anti-CD3/anti-CD28 antibodies with the indicated number of WT or Cish−/− GM-MØs for 3 days. Cell proliferation and cytokine production were then determined. *P < 0.05, **P < 0.01; Student’s t test
Next, we sought to compare the functions of GM-MØs generated from WT or Cish−/− mice. First, we evaluated GM-MØs for their ability to stimulate antigen-specific T-cell proliferation. To this end, GM-MØs were pulsed with ovalbumin (OVA) and subsequently cocultured with dye-labeled MHCI- and MHCII-restricted OVA-specific CD8+ or CD4+ T cells (OT-I and OT-II cells, respectively). T-cell proliferation was significantly weaker when T cells were cocultured with OVA-pulsed Cish−/− GM-MØs than when T cells were cocultured with OVA-pulsed WT GM-MØs (Fig. 1D, Fig. S3E). A similar observation was made for GM-DCs (Fig. S3E), suggesting that CIS expression in MØs is required to promote T-cell expansion. In line with the above findings, adoptive transfer of OVA-pulsed GM-MØs into C57BL/6 mice that had previously received GFP+ OT-I cells revealed that antigen-driven T-cell expansion and IFN-γ production were weaker in mice vaccinated with OVA-pulsed Cish−/− GM-MØs than those vaccinated with WT GM-MØs (Fig. 1E, F). The difference in the T-cell responses induced by WT and Cish−/− GM-MØs was not due to antigen uptake or processing, since GM-MØs of both genotypes had a similar uptake and processing capacity (Fig. 1G). Moreover, when we bypassed antigen uptake/presentation by stimulating purified CD8+ T cells with anti-CD3/anti-CD28 antibodies, Cish−/− GM-MØs (at MØ:T-cell ratios of 1:1 and 1:2) were more potent in suppressing T-cell proliferation and IFN-γ production than were WT GM-MØs (Fig. 1H). Thus, we conclude that CIS deficiency leads to the generation of MØs that inhibit T-cell expansion.
CIS deficiency leads to the generation of mouse and human MØs with reduced IL-12 production
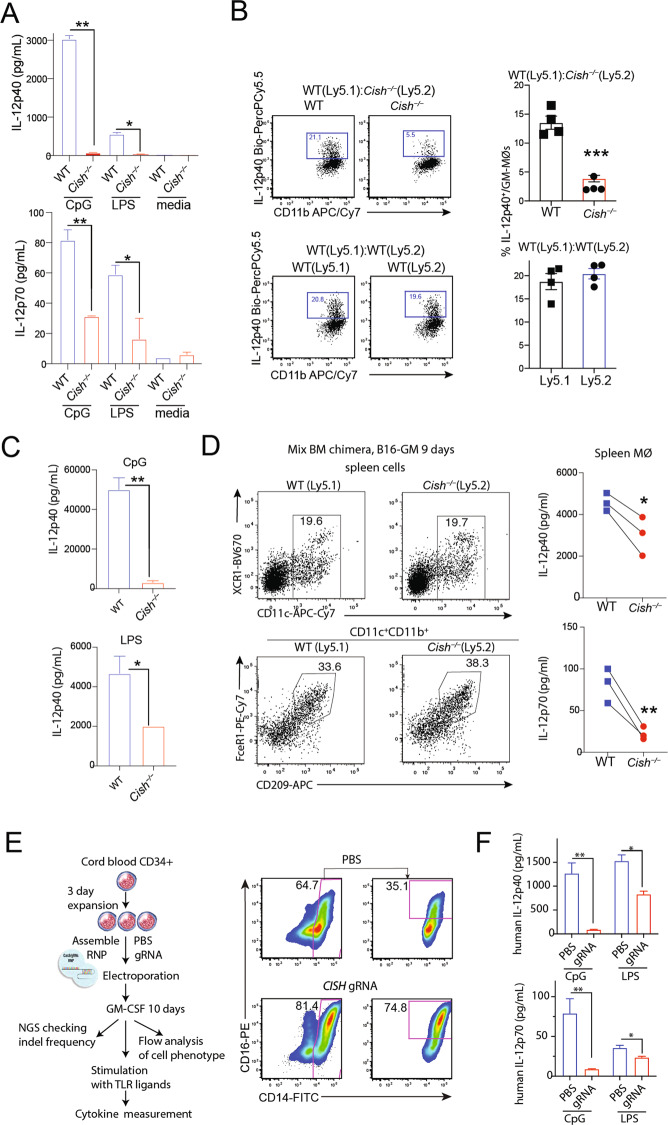
We next investigated the mechanism underlying the lack of IFN-γ production by CD8+ T cells cocultured with Cish−/− GM-MØs (Fig. 1F, H). As IL-12 is a key cytokine promoting IFN-γ production [29], we measured IL-12 production by MØs upon TLR agonism. Compared to WT GM-MØs, Cish−/− GM-MØs produced substantially less IL-12 when stimulated with CpG or LPS (Fig. 2A, Fig. S4A). WT and Cish−/− BM coculture experiments confirmed that the reduced IL-12 production of Cish−/− GM-MØs was cell intrinsic (Fig. 2B). IL-12 production was also reduced in Cish−/− GM-MØs following stimulation with either PolyI:C or an agonistic anti-CD40 antibody (Fig. S4B). The requirement for CIS was relatively selective for IL-12, as the expression of other cytokines (e.g., IL-6, IL-10 and TNF-α) was similar between WT GM-MØs and Cish−/− GM-MØs (Fig. S4C). The defective IL-12 production by Cish−/− GM-MØs was observed across a range of GM-CSF concentrations (Fig. S4D). In line with earlier reports [28, 30], GM-DCs produced less IL-12 than GM-MØs, and IL-12 production was CIS independent (Fig. S4E). These results suggest that CIS plays a critical role in promoting IL-12 production by GM-MØs.
Fig. 2.
Cish−/− MØs produce a relatively low amount of IL-12. A IL-12 production by WT and Cish−/− GM-MØs stimulated with CpG or LPS for 20 h. *P < 0.05, **P < 0.01; multiple-group ANOVA. B Production of IL-12 by WT (Ly5.1) and Cish−/− (Ly5.2) GM-MØs derived from cocultures with BM cells isolated from individual mice. The harvested cells were stimulated with CpG for 4 h. ***P < 0.001, Student’s t test. C IL-12 production by spleen cells isolated from WT or Cish−/− mice. Mice were engrafted with B16-GM cells for 9 days. Spleen cells were stimulated with CpG or LPS for 20 h. Bar graphs show the mean ± S.D. of the IL-12 concentration in culture supernatants. *P < 0.05, **P < 0.01; Student’s t test. D IL-12 production by splenic MØs isolated from WT or Cish−/− mice. Mixed bone marrow chimeric mice reconstituted with both WT (Ly5.1) and Cish−/− (Ly5.2) BM cells were engrafted with B16-GM cells for 9 days. Sorted MØs were stimulated with CpG for 20 h. Line graphs show IL-12 production by paired WT and Cish−/− splenic MØs from the same host. *P < 0.05, **P < 0.01; Student’s t test. E, F IL-12 production by human GM-MØs with CISH deletion. CD34+ cells isolated from human cord blood were transfected with Cas9 assemble RNP and CISH guide RNA. The transfected CD34+ cells were cultured with human GM-CSF (5 ng/ml) for 7 days. The cells were stimulated with CpG or LPS. E FACS plots show the resulting human GM-MØ population. F Bar graphs show IL-12 production by human GM-MØs. *P < 0.05, **P < 0.01; multiple-group ANOVA. Data are representative of three independent experiments
The limited number of splenic MØs (identified as CD11c+CD11b+CD209+FceR1+ cells) in mice, probably due to the low abundance of GM-CSF in unchallenged mice [31, 32], precluded us from corroborating the above findings in vivo. To circumvent this issue, we challenged WT or Cish−/− mice for 9 days with a B16 melanoma cell line producing GM-CSF (B16-GM) [8]. When splenocytes isolated from challenged WT or Cish−/− mice were stimulated with CpG or LPS, we found that IL-12 production was substantially reduced in the absence of CIS (Fig. 2C). To evaluate the contribution of MØs to IL-12 production, we generated mixed bone marrow chimeric mice by reconstituting lethally irradiated recipient mice (C57BL/6-Ly5.1) with mixed WT (Ly5.1) and Cish−/− (Ly5.2) BM cells. Following reconstitution, the mice were engrafted with B16-GM cells for 9 days. MØs of both WT origin and Cish−/− origin were then isolated and stimulated with CpG for 20 h. Similar to in vitro-generated GM-MØs, splenic Cish−/− MØs isolated from B16-GM cell-bearing mice produced significantly less IL-12 than WT MØs (Fig. 2D).
Finally, we investigated whether CIS deficiency also impacted the capacity of human MØs to produce IL-12. To this end, CD34+ cells derived from human cord blood were electroporated with Cas9 assembled in a ribonucleoprotein particle (RNP) with CISH guide (g)RNA. The indel frequency for donor CD34+ cells transfected with CIS guide-RNPs was 83% (based on next-generation sequencing). Transfected CD34+ cells were cultured with human GM-CSF (5 ng/mL) for 7–10 days. CISH gRNA- and mock-transfected cells differentiated into CD14+CD16+ cells. Notably, the percentage of CD14+CD16+ cells was higher in cultures with CISH gRNA than those that underwent mock transfection (Fig. 2E). When cells were stimulated with CpG or LPS, the production of IL-12 by cells targeted with CISH gRNA was substantially reduced compared to that of mock-transfected cells (Fig. 2E). Taken together, these data point to a critical role for CIS in controlling the production of IL-12 in both mouse and human MØs.
CIS deficiency imprints MØs with M2-like characteristics
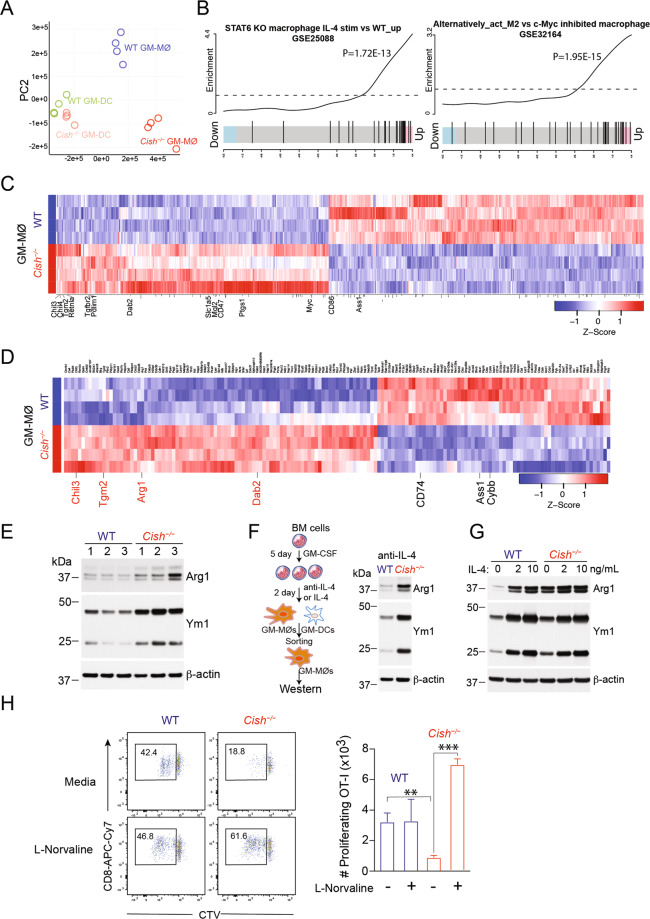
We showed above that Cish−/− MØs strongly inhibited T-cell responses and had reduced IL-12 production compared to WT MØs. To gain insights into the molecular mechanisms underlying these functional changes, we performed RNA sequencing (RNA-seq) of sorted GM-MØs and GM-DCs from WT and Cish−/− mice (n = 4). Principal component analysis (PCA) revealed that CIS and cell lineage identity accounted for the major differences between the WT and Cish−/− samples. The impact of CIS deficiency was more prominent in GM-MØs than in GM-DCs, as evidenced by the increased separation of the MØ groups in the PCA plot (Fig. 3A) and the higher number of differentially expressed genes (DEGs) between the Cish−/− and WT samples (1029 and 713 DEGs for GM-MØs and GM-DCs, respectively) (Supplementary Table 1). As MØs are the dominant cell type generated under GM-CSF culture conditions and are more profoundly impacted by CIS deficiency, we chose MØs for further detailed analysis. Gene ontology analysis highlighted many differences between WT and Cish−/− GM-MØs that correlated with their differences in function, cell cycling and metabolism (Fig. S5A).
Fig. 3.
Cish−/− MØs display characteristics of M2 MØs. A–C RNA-seq analysis of WT and Cish−/− GM-MØs and GM-DCs. A Principal component analysis (PCA) plot of TPM values over dimensions 1 and 2 with samples from 4 individual mice per group colored by group for GM-MØs and GM-DCs. B Gene set enrichment analysis barcode plot comparing the expression of upregulated genes in Cish–/– GM-MØs with two previously reported M2 MØ signature gene sets. C Heatmap showing DEGs, with M2 MØ signature genes indicated in red (log2FC>X, FDR < 0.05). D Proteomic analysis of GM-MØs derived from 4 individual WT and Cish−/− mice. Heatmap showing the expression of differentially regulated proteins by the two types of GM-MØs, with M2 proteins indicated in red (log2FC > 0.6, FDR<0.05). E Western blot analysis of Arg1 and Ym1 in WT or Cish−/− GM-MØs derived from 3 individual donors. F, G BM cells were cultured with GM-CSF for 5 days, and an anti-IL-4 antibody or recombinant IL-4 was then added. After 2 days, GM-MØs were sorted and analyzed for Arg1 and Ym1 expression by Western blotting. F Cultures ± anti-IL-4 antibody. G Cultures ± recombinant IL-4. H CTV-labeled OT-1 CD8+ T cells were cocultured with GM-MØs and stimulated with OVA ± 12 mM L-norvaline. Dot plots show the OVA-stimulated proliferation of CTV-labeled CD8+ T cells. Bar graphs show the numbers of proliferating CD8+ T cells. *P < 0.05, **P < 0.01, **P < 0.001; multiple-group ANOVA
GM-CSF is thought to bias MØ polarization toward the proinflammatory M1-like state [3, 4], and we therefore expected that CIS deficiency would strengthen GM-CSF signaling and further increase M1 MØ polarization. Instead, analysis of the RNA-seq data indicated that the genes upregulated in Cish−/− GM-MØs were positively correlated with gene signatures derived from IL-4-induced M2 MØs (GSE25088) [33] and GSE32164 [34] (Fig. 3B). Next, we analyzed genes known to be associated with MØ functional polarization among the DEGs in Cish−/− GM-MØs [35, 36] (Supplementary Table 2). Over 64% (35/56) of the known M2 MØ-associated DEGs (Supplementary Table 2) were upregulated in Cish−/− GM-MØs (Fig. 3C), including prototypic genes such as Chil3 (Ym1), Chil4, Retnla (Fizz1) and Tgm2, while over 85% (26/30) (Supplementary Table 2) of the known M1 MØ-associated DEGs were downregulated in Cish−/− GM-MØs, thus suggesting that CIS deficiency skews GM-CSF-induced MØs toward an M2-like phenotype.
We also compared the DEGs to the gene signature of M-CSF-derived M2-like macrophages [37]. We found that the genes downregulated in Cish−/− GM-MØs were more strongly positively correlated with the gene signatures decreased in M-CSF-derived M2-like MØs compared with those in GM-CSF-derived M1-like macrophages (Fig. S5C). On the other hand, the genes upregulated in Cish−/− GM-MØs did not correlate with those upregulated in M-CSF-derived MØs (Fig. S5C) but instead positively correlated with gene signatures derived from IL-4-induced M2 MØs (Fig. 3B). Thus, Cish−/− GM-MØs resemble M-CSF-derived MØs in certain aspects but differ from them in other key parameters.
To support our transcriptomic analysis, we also performed label-free quantitative proteomic analysis of GM-MØs derived from WT and Cish−/− mice (n = 4) (Supplementary Table 3). In accordance with the transcriptional data, we found that several typical M2 MØ-associated proteins were upregulated in Cish−/− GM-MØs, including Arg1, Chil3, Tgm2 and Dab2 (Fig. 3D). In contrast, the proteins that were downregulated in Cish−/− GM-MØs included M1 MØ-associated proteins, such as CD74, Ass1 and Cybb (Fig. 3D). Overall, both the RNA-seq and proteomic analyses revealed that loss of CIS resulted in the development of GM-MØs sharing some features characteristic of M2-like MØs.
Next, we selected two commonly used M2 markers, Arg1 and Ym1, for further validation by western blotting. Cish−/− GM-MØs sorted from BM cultures derived from 3 individual mice all expressed higher levels of Arg1 and Ym1 than WT GM-MØs (Fig. 3E), thus corroborating the above results. To convert Cish−/− GM-MØs into M1-like MØs, we stimulated WT and Cish−/− GM-MØs with IFN-γ and LPS, potent M1 MØ-inducing cytokines. While IFN-γ and LPS treatment reduced the expression of Arg1 and Ym1 in both WT GM-MØs and Cish−/− GM-MØs, the latter retained substantially higher levels of these M2 MØ markers (Fig. S5B).
IL-4 is a prototypical cytokine that drives the differentiation of M2 MØs [36]. To investigate whether the M2-like phenotype of Cish−/− GM-MØs was influenced by endogenous IL-4, a neutralizing anti-IL-4 antibody was added to GM-CSF BM cell cultures on Day 5. Cish−/− GM-MØs maintained higher expression of Ym1 and Arg1 (Fig. 3F). Furthermore, addition of exogenous IL-4 in the late stage of cell differentiation increased the expression of Arg1 and Ym1 in GM-MØs, but these increases were comparable between the WT and Cish−/− genotypes (Fig. 3G). This observation suggests that the induction of the M2-like phenotype in Cish−/− GM-MØs is unlikely to be directly due to IL-4.
Finally, we examined the functional consequence of the high level of Arg1 in Cish−/− GM-MØs. Arginine availability is key to an optimal T-cell immune response [38]. As the Arginase 1 inhibitor L-norvaline enhances T-cell proliferation in the presence of antigen-presenting cells [39], we investigated whether inhibition of Arg-1 activity in Cish−/− GM-MØs could relieve the suppressive activity of Cish−/− GM-MØs (Fig. 1D–H). The addition of L-norvaline greatly enhanced the proliferation of OT-I T cells stimulated in the presence of Cish−/− GM-MØs but not that of those stimulated in the presence of WT GM-MØs (Fig. 3H). Thus, we contend that M2-phenotype molecular imprinting in the absence of CIS indeed leads to a gain of suppressive function, which can be partially relieved by inhibiting Arginase 1.
Enhanced STAT5 activation in the absence of CIS contributes to the development of M2-like MØs
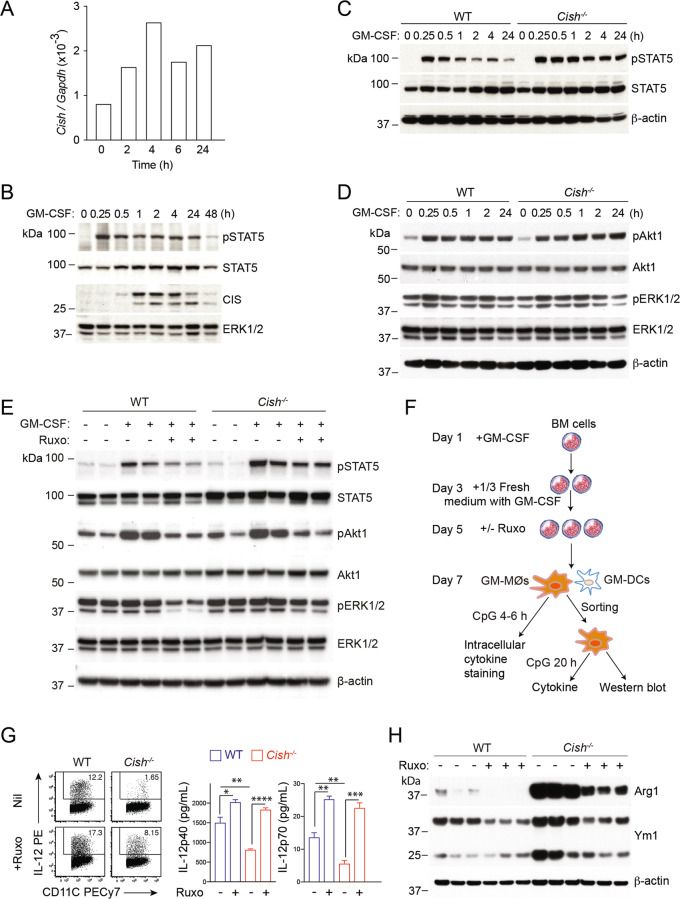
Next, we investigated the signaling events leading to the development of M2-like features in the absence of CIS. Isolated GM-MØs that were rested and then restimulated with GM-CSF induced the mRNA and protein expression of CIS and STAT5 activation (measured as STAT5 tyrosine phosphorylation) in GM-MØs (Fig. 4A and B). Furthermore, CIS deficiency in GM-MØs led to enhanced and prolonged STAT5 activation after GM-CSF restimulation (Fig. 4C) but no differences in activation of the PI3K/Akt or ERK1/2 MAPK pathway (Fig. 4D).
Fig. 4.
Cish−/− GM-MØs have enhanced STAT5 activation, contributing to an M2-like phenotype. A, B Purified GM-MØs, generated on Day 7 in the presence of GM-CSF, were restimulated in vitro with recombinant GM-CSF (50 ng/ml) for the indicated times. A The induction of Cish mRNA expression relative to that of the housekeeping gene Gapdh in GM-MØs upon GM-CSF stimulation was determined by RT‒qPCR analysis of extracted RNA samples. B The induction of CIS protein expression and STAT5 activation in GM-MØs upon GM-CSF restimulation were determined by immunoblotting. C, D Purified WT and Cish−/− GM-MØs, generated on Day 7 in the presence of GM-CSF, were stimulated in vitro with recombinant GM-CSF (50 ng/ml) for the indicated times. Cell lysates were analyzed by western blotting to assess (C) STAT5 activation and (D) Akt1 and ERK 1/2 activation. E BM cells from WT or Cish−/− mice were cultured with GM-CSF for 5 days. GM-MØs were harvested and stimulated ± GM-CSF (50 ng/ml) or the JAK inhibitor ruxolitnib (Ruxo) (0.2 μM) for 4 h. The cells were lysed for Western blotting to assess STAT5, Akt1 and ERK1/2 activation. Data were derived from 2 individual mice. F-G BM cells were cultured with GM-CSF for 5 days and supplemented ± RUXO (0.2 μM) for 2 days. G The IL-12 production of GM-MØs upon CpG stimulation was evaluated by intracellular cytokine staining and secretion. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; multiple-group ANOVA. Data are representative of two independent experiments. H Purified WT and Cish−/− GM-MØs were analyzed for Ym1 and Arg1 expression by western blotting. In each western blot, β-actin was used as a loading control, with the exception of (B), in which ERK1/2 was used as the loading control. Data were derived from 3 individual mice
To investigate whether enhanced JAK/STAT5 signaling is responsible for some of the functional changes observed in Cish−/− GM-MØs, we tuned down GM-CSF signaling using the JAK inhibitor ruxolitinib (RUXO, 0.2 μM). Hyperactivation of STAT5, but not Akt1 or ERK1/2, upon GM-CSF stimulation was also observed on Day 5 in BM-derived Cish−/− GM-MØs (Fig. 4E). The addition of RUXO to the medium substantially reduced the phosphorylation of STAT5 in both WT GM-MØs and Cish−/− GM-MØs (Fig. 4E). As expected, RUXO-treated GM-MØs also showed reduced phosphorylation of Akt1 and ERK1/2, whose activation depends on JAK kinase activation (Fig. 4E). Thus, these observations suggested that the impaired IL-12 production observed in Cish−/− GM-MØs (Fig. 2B and C) was independent of the activation statuses of Akt1 and ERK1/2. Importantly, the RUXO-mediated downmodulation of pSTAT5 restored the capacity of Cish−/− GM-MØs to produce IL-12 following CpG stimulation, and IL-12 production by WT GM-MØs was further enhanced by RUXO (Fig. 4F and G). This suggests that decreasing JAK/STAT5 signaling output in GM-MØs, which is normally mediated by CIS, is critical for optimal IL-12 production. The higher expression of Arg1 and Ym1 observed in Cish−/− GM-MØs was greatly reduced following RUXO addition to the cell culture (Fig. 4H). Taken together, these data suggest that CIS deficiency in GM-MØs results in increased activity of the JAK/STAT5 signaling pathway leading to the development of M2-like MØ features and reduced IL-12 production.
Downregulation of IRF8 by enhanced STAT5 activation contributes to the development of M2-like MØs
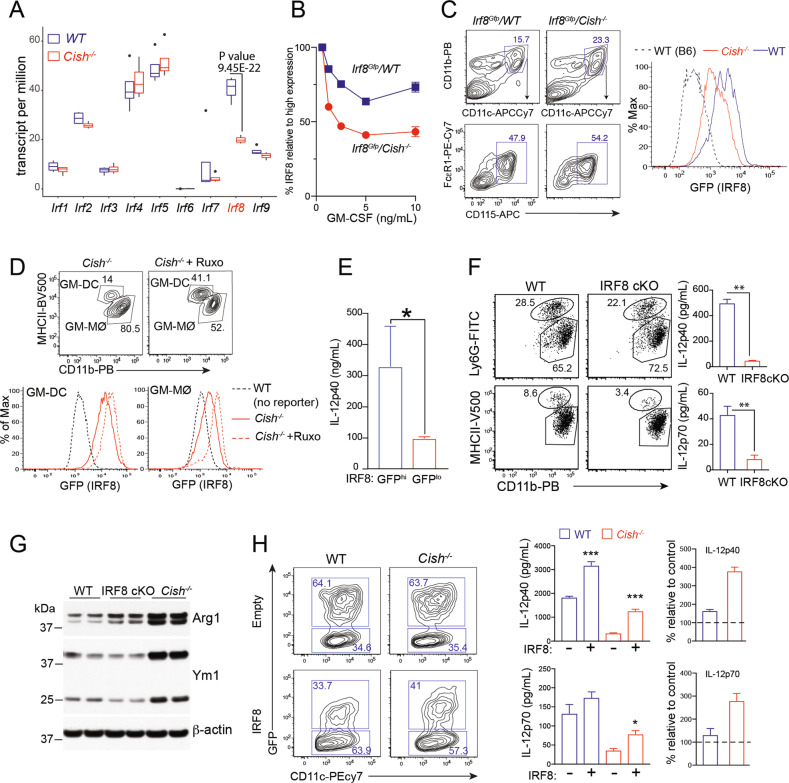
To define the molecular consequences of the sustained JAK/STAT5 activation observed in Cish−/− GM-MØs, we further analyzed our transcriptomic dataset. We focused our analysis on transcription factors known to regulate M1/M2 cell fate decisions, particularly the interferon responsive factor (IRF) family members known to be critical for MØ polarization [40]. Quantitation of the mRNA levels for the 9 members of this family revealed that only Irf8 mRNA transcript levels were substantially reduced in Cish−/− GM-MØs compared to WT GM-MØs (Fig. 5A). To substantiate these findings, we crossed Cish−/− mice with Irf8Gfp reporter mice [41], allowing us to measure IRF8 expression at the single-cell level. GM-CSF reduced the expression of IRF8 in GM-MØs in a dose-dependent manner (Fig. 5B). Comparison of GM-MØs derived from Irf8Gfp/WT and Irf8Gfp/Cish−/− mice confirmed the reduced expression of IRF8 in GM-MØs lacking CIS (Fig. 5B). Consistent with this observation, the expression of IRF8 was substantially reduced in MØs isolated from Irf8Gfp/Cish−/− mice that had been challenged with B16-GM tumors for 9 days compared to those isolated from similarly treated Irf8Gfp/WT mice (Fig. 5C). We next investigated whether attenuation of JAK/STAT signaling could rescue Irf8 transcription in Cish−/− GM-MØs. Irf8Gfp/Cish−/− BM progenitors were cultured in the presence of GM-CSF with/without the addition of RUXO on Day 5 of culture. On Day 7, we noted that the Irf8Gfp/Cish−/− GM-MØs treated with RUXO exhibited higher IRF8-GFP expression than the untreated Cish−/− GM-MØs (Fig. 5D). Similar observations were made for GM-DCs (Fig. 5D). These results suggest that the sustained JAK/STAT5 activation observed in Cish−/− GM-MØs inhibits IRF8 expression.
Fig. 5.
Cish−/− MØs have reduced expression of IRF8, resulting in an M2-like phenotype. A Interferon regulatory factor (IRF) expression in WT and Cish−/− GM-MØs was analyzed by RNA-seq. Boxplot showing IRF expression in WT and Cish−/− GM-MØs (n = 4). B BM cells from Irg8Gfp mice ± Cish deficiency were cultured with different concentrations of GM-CSF. Heatmap showing the MFI of Irf8Gfp in GM-MØs. C Irf8Gfp mice ± Cish deficiency were engrafted with B16-GM cells. Isolated spleen MØs were evaluated for the expression of IRF8-GFP. Negative controls (dotted line) were MØs from B6 mice. D Irf8Gfp/Cish−/− BM cells were cultured in the presence of GM-CSF for 5 days. The cells were then cultured with RUXO for an additional 2 days, and IRF8-GFP expression by GM-MØs and GM-DCs was evaluated. E GM-MØs from Irf8Gfp mice were sorted into IRF8-GFPhi (top 50%) and IRF8-GFPlo (bottom 50%) populations. The sorted GM-MØs were then stimulated with CpG for 20 h, and the IL-12 concentration in the culture supernatant was measured by ELISA. *P < 0.05, Student’s t test. F, G GM-MØs from CD11c-cre-IRF8fl/fl (IRF8cKO) and WT mice were stimulated with CpG for 20 h. F IL-12 production was then measured by ELISA. **P < 0.01, Student’s t test. G Sorted GM-MØs cultured from BM cells isolated from IRF8cKO, WT or Cish−/− mice were also analyzed for the expression of Arg1 and Ym1 by western blotting. H BM progenitor cells from WT and Cish−/− mice were transduced with either an empty/GFP- or IRF8/GFP-encoding retrovirus and cultured with 10 ng/ml GM-CSF for 5 days. The gated population represents the frequency of transduced (GFP+) GM-MØs. Sorted transduced GM-MØs were stimulated with CpG for 20 h. The IL-12 concentration in the culture supernatant is presented in bar graphs. The right panels show the % increase in IL-12 production by cells transduced with the IRF8/GFP-encoding vector relative to that of cells of the same genotype transduced with the empty/GFP-encoding vector. Data are representative of 3 independent experiments. *P < 0.05, ***P < 0.001; multiple-group ANOVA
We then explored whether the downregulation of IRF8 contributed to the impaired IL-12 production observed in Cish−/− GM-MØs. The first supporting evidence came from the experiment performed with Irf8Gfp reporter mouse, in which we found that IRF8-GFPhi GM-MØs produced significantly higher amounts of IL-12 than IRF8-GFPlo GM-MØs (Fig. 5E). We then tested whether a lack of IRF8 impacted the capacity of GM-MØs to produce IL-12. To circumvent the defective myelopoiesis observed in germline IRF8-KO mice [42], we used CD11c-Cre/Irf8fl/fl (IRF8cKO) mice to generate GM-MØs. In this system, the compartment of Ly6G+ granulocytes was similar between WT and IRF8cKO mice, while that of GM-DCs were slightly reduced in IRF8cKO mice (Fig. 5F). Consistent with our earlier observations, GM-MØs derived from IRF8cKO mice exhibited reduced IL-12 production following CpG stimulation compared to their WT counterparts (Fig. 5F). IRF8cKO GM-MØs also showed increased expression of the M2 MØ marker Arg1 compared to WT GM-MØs. However, the phenotype of IRF8cKO GM-MØs was less pronounced than that of Cish−/− GM-MØs (Fig. 5G). Crucially, IRF8 overexpression in Cish−/− GM-MØs substantially increased IL-12 production following CpG stimulation, suggesting that the reduced Irf8 expression observed in Cish−/− GM-MØs impaired their capacity to produce IL-12 (Fig. 5H). Collectively, our data suggest that CIS deficiency leads to increased STAT5 activation, resulting in the downregulation of IRF8 and hindering GM-MØ polarization into M1-like MØs.
IRF8 regulates the gene program involved in MØ polarization
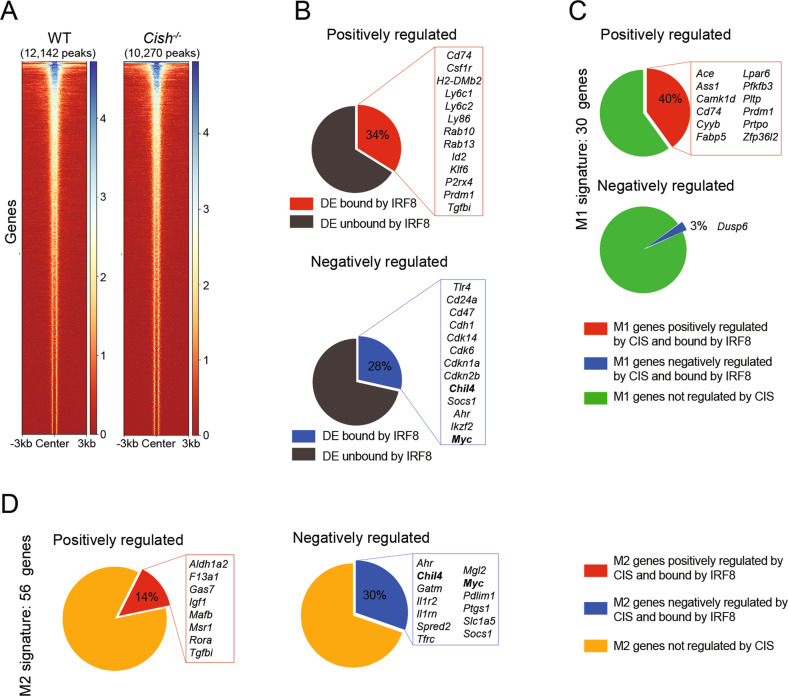
Our results point to a critical role for CIS in maintaining an adequate IRF8 concentration, which ultimately controls MØ polarization. To gain insight into the potential instructive role of IRF8 regulation in controlling GM-MØ polarization, we performed CUT&Tag sequencing [43] to identify the genes directly targeted by IRF8 in GM-MØs. To facilitate our pull-down strategy, we derived GM-MØs from Irf8Gfp/WT and Irf8Gfp/Cish−/− mice, in which IRF8 is expressed as a fusion protein with GFP, and the pull-down step was performed with an anti-GFP Ab (Fig. S6A and B). Using this strategy, we identified 12,142 binding sites occupied by IRF8 in the genome of WT GM-MØs and 10,270 binding sites in that of Irf8Gfp/Cish−/− GM-MØs (Fig. 6A). Comparison of our genome-wide IRF8 binding dataset with our RNA-seq data allowed us to identify putative IRF8-regulated genes. For genes positively regulated by CIS (downregulated in Cish−/− GM-MØs), 34% of them displayed at least one IRF8 binding site, while for genes negatively regulated by CIS (upregulated in Cish−/− GM-MØs), 28% of them displayed at least one IRF8 binding site (Fig. 6B). As we reasoned that CIS deficiency in GM-MØs led to a perturbed polarization potential, in part due to the lack of IRF8 upregulation, we focused on the genes associated with MØ polarization (Supplemental Table 1). For M1 MØ-associated genes, 40% of the M1 MØ genes positively regulated by CIS (Ace, Cd74 and Cyyb) were bound by IRF8, while only 3% of the M1 MØ genes negatively regulated by CIS (Dusp6) were bound by IRF8 (Fig. 6C). On the other hand, of the 56 M2 MØ signature genes bound by IRF8, 30% were negatively regulated by CIS (Ahr, Chil4 and Myc), and 14% were positively regulated by CIS (F13a1 and Msr1) (Fig. 6D). Taken together, these observations support that the CIS-mediated modulation of IRF8 plays a key role in the regulation of the M1-like state.
Fig. 6.
IRF8 regulates genes associated with MØ polarization. A Heatmap showing IRF8 binding in WT and Cish−/− GM-MØs. The number of IRF8 peaks identified for each genotype is shown. B Differentially expressed genes (DEGs) between WT and Cish–/– GM-MØs displaying at least one IRF8 binding site in the locus. Peaks were assigned to the closest gene. Top: genes positively regulated by CIS (downregulated in Cish−/− GM-MØs), bottom: genes negatively regulated by CIS (upregulated in Cish−/− GM-MØs). C M1 MØ-associated DEGs displaying an IRF8 binding site in the locus. Top: M1MØ-associated genes positively regulated by CIS, bottom: M1 MØ-associated genes negatively regulated by CIS. D M2 MØ-associated DEGs displaying an IRF8 binding site in the locus. Top: M2 MØ-associated genes positively regulated by CIS, bottom: M2 MØ-associated genes negatively regulated by CIS
Cish−/− MØs increase the Th2 response and the severity of allergic asthma
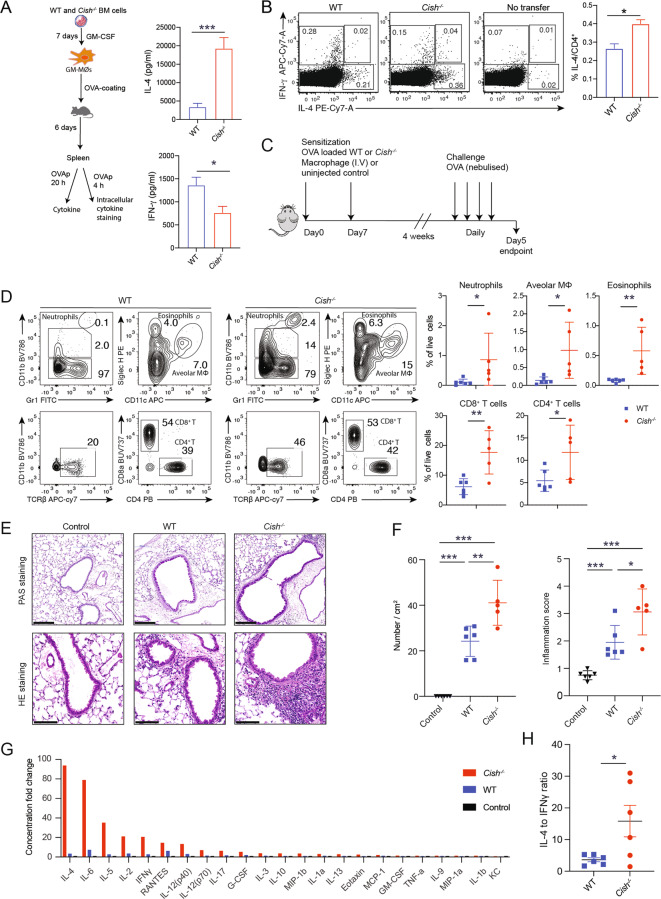
To determine if the M2-like MØ phenotype observed in the absence of CIS evoked a Th2 response in vivo, we transferred OVA-pulsed WT and Cish−/− GM-MØs into C57BL/6 mice and measured the production of IL-4 and IFN-γ by restimulated splenocytes after 6 days. IL-4 production was substantially increased in cells from mice that received OVA-pulsed Cish−/− GM-MØs compared to those from mice immunized with OVA-pulsed WT GM-MØs, while IFN-γ levels were reduced (Fig. 7A). Ex vivo evaluation of OVA-stimulated CD4+ T cells confirmed that Cish−/− GM-MØs induced more IL-4-producing cells (Fig. 7B). Next, we compared the impacts of transferred OVA-pulsed GM-MØs of WT or Cish−/− origin on the induction of Th2 allergic inflammation [44]. To this end, we immunized WT mice with OVA-pulsed WT or Cish−/− MØs and then challenged them with nebulized OVA 4 weeks later (Fig. 7C). Following the challenge, the lungs of mice that were preimmunized with OVA-pulsed Cish−/− MØs contained significantly increased immune infiltrates compared to those of mice that received OVA-pulsed WT MØs (Fig. 7D). These immune infiltrates were accompanied by an increased prevalence of PAS+ mucus-producing cells, enhanced lung tissue inflammation intensity and pronounced increases in the concentrations of a wide array of cytokines in the bronchoalveolar lavage fluid (BALF) in mice preimmunized with OVA-pulsed Cish−/− MØs (Fig. 7E–G). Notably, the IL-4:IFN-γ ratio from the same samples was calculated to reflect a Th2 bias, and the BALF of mice immunized with OVA-pulsed Cish−/− MØs preferentially contained increased levels of the Th2 cytokine IL-4 (Fig. 7H). Taken together, these data suggested that CIS deficiency resulted in the polarization of MØs featuring a strong capacity to induce a robust Th2 response that exacerbated the allergic asthma response. Thus, CIS modulated Th differentiation through control of MØ polarization independent of the reported cell-intrinsic role of CIS in controlling T-cell polarization [45].
Fig. 7.
Cish-/- MØs promote Th2 responses and exacerbate allergic asthma. A Schematic showing the experimental approach for assessing the immune response induced by WT or Cish−/− GM-MØs. Six days after the transfer of OVA-coated GM-MØs, harvested spleen cells from 3 individual mice were cultured with OVA for 20 h. The supernatant was harvested for a cytokine assay. The bar graph shows the concentrations of IL-4, IFN-γ and IL-17A. B Spleen cells from mice treated as in (A) were pulsed with OVA323-339 for 4 h. The proportions of IL-4- and IFN-γ-producing CD4+ T cells were tested via intracellular staining. Dot plots show IL-4 and IFN-γ expression in CD4+ T cells. The bar graph shows the percentage of IL-4+ CD4+ T cells. *P < 0.05, Student’s t test. C Experimental protocol for OVA-induced inflammation. D Contour plots show the gating strategies for neutrophils, alveolar macrophages, eosinophils and T cells in the bronchoalveolar lavage fluid. Scatter plots show the mean ± SEM of different immune cell populations from individual mice treated as in (C). E Representative PAS and HE staining of the airways of mice treated as in (C). Scale bar = 200 μm. F Quantification of PAS+ cells and the inflammation score. Bar graphs show the mean ± SEM of the indicated populations. Each dot represents one mouse. G The bronchoalveolar lavage fluid of mice treated as in (C) was analyzed for cytokine and chemokine expression by a BioPlex assay. The bar graph shows the fold change over the mean level in control mice. H Ratio of the IL-4 and IFN-γ concentrations in the same sample. Each dot represents one mouse. **P < 0.01, ***P < 0.001; Student’s t test
Discussion
A central finding from our study is that CIS has a marked impact on GM-CSF-induced MØ polarization. In its absence and in microenvironments where GM-CSF was abundant, MØs became immunosuppressive, producing less IL-12, which ultimately resulted in the induction of a Th2 immune response. In contrast to IL-4-induced M2 polarization, which depends on the STAT6/IRF4 signaling axis [46], CIS deficiency induced M2-like MØ polarization following GM-CSF exposure through sustained activation of STAT5 and consequent downregulation of IRF8. Thus, we propose that CIS activity may represent a key intrinsic regulatory mechanism responsible for the functional polarization of MØs.
At the cellular level, the development of proinflammatory M1-like MØs in vitro upon GM-CSF stimulation has been documented for both humans and mice [3, 5]. Similarly, in murine models of autoimmunity, GM-CSF has been shown to promote MØ pathogenicity associated with the production of proinflammatory cytokines [47, 48]. However, GM-CSF has also been associated with the development of suppressive M2-like MØs in various settings [11, 12, 16]. How GM-CSF drives these contrasting cellular fates of MØs is still largely unknown. A simple interpretation of the dichotomy of MØ polarization under GM-CSF stimulation is a model of ligand abundance-mediated polarization, as the development of suppressive M2-like MØs is largely associated with high levels of GM-CSF [11, 12]. At the molecular level, heightened STAT5 activation under GM-CSF stimulation has been linked to the development of M2-like MØs [12, 49]. Accordingly, neutralization of GM-CSF or STAT5 inhibition can reverse M2 MØ polarization [50]. Our study identified another layer of control independent of GM-CSF abundance. Here, we report that CIS acts as a cell-intrinsic rheostat in controlling STAT5 activation independent of the activation of PI3K or MAPK. CIS deficiency in GM-MØs led to heightened and sustained STAT5 activation that favored M2-like MØ differentiation. Fittingly, attenuation of upstream signaling with a JAK inhibitor increased IL-12 production and reduced the expression of Arg1 and Ym1 in Cish−/− MØs.
Here, we revealed the CIS-modulated GM-CSF/STAT5/IRF8 signaling axis that acts in MØ polarization regulation. In our study, GM-CSF signaling in Cish−/− MØs resulted in suboptimal expression of IRF8, in part due to the hyperactivation of STAT5. Accordingly, JAK inhibition in Cish−/− MØs prevented STAT5 phosphorylation and restored normal IRF8 expression. STAT5 activation has been shown to suppress the induction of Irf8 transcription in plasmacytoid DCs [51]. IRF8 has also been reported to be a critical positive regulator of IL-12 expression [52, 53]. This strongly indicates that Irf8 downregulation is functionally linked to the reduced IL-12 production in Cish−/− MØs. Earlier studies provided circumstantial evidence supporting the idea that downregulation/silencing of IRF8 could represent a defining event favoring the development of M2-like MØs through increased expression of M2 MØ genes [54–56]. Our study demonstrated the extent of IRF8 binding in MØs, particularly in the vicinity of many genes that have been associated with MØ polarization, suggesting a critical instructive role in driving MØ polarization. Despite its importance, the downmodulation of IRF8 is unlikely to be the sole downstream event of the GM-CSF/STAT5 pathway that leads to M2 MØ polarization in Cish−/− MØs, although the identity of any other contributing signaling pathways will require future studies.
Although M2-like MØs are known to promote Th2-cell differentiation [15], the molecular mechanisms responsible for this process are not well understood. It has previously been reported that the M2 MØ signature genes Ym1/2 can directly influence Th2-cell development [57, 58]. Our transcriptomic and proteomic approaches highlighted the elevated expression of TGM2, Arg1 and FIZZ1 in Cish−/− GM-MØs (Fig. 3), all of which have been shown to play roles in allergic asthma [59]. Thus, we contend that CIS expression in myeloid cells has a prominent role in regulating Th2 immunity, independent of the reported role for CIS in T cells involved in promoting Th2-cell differentiation [45].
Overall, we reveal that CIS acts as a brake on GM-CSF signaling and is critical for MØ polarization. Through dissection of the molecular and functional nature of Cish−/− GM-MØs, we provide significant insights into how GM-CSF can shape MØ polarization. We argue that CIS expression in myeloid cells may be beneficial in allergic responses and antitumor immunity by curtailing GM-CSF signaling to prevent M2 MØ polarization and pathogenic Th2 responses.
Materials and methods
Mice
Mice were housed under specific pathogen-free conditions at The Walter & Eliza Hall Institute of Medical Research. The strains are listed in Supplementary Table 4. Cish−/− mice were kindly provided by J. Ihle and E. Parganas (St. Jude Children’s Research Hospital). Chimeric mice were from the F1 generation (Ly5.1/Ly5.2); these recipients were irradiated with 2 doses of 550 rads and reconstituted with 2 × 106 BM cells from Ly5.1 and Cish−/− mice. All experiments were performed following relevant guidelines and regulations approved by the Walter & Eliza Hall Institute of Medical Research animal ethics committee (Projects #2016.014, #2017.008, and #2018.040).
Cell preparation, antibodies, and flow cytometry
Cells were isolated from the spleen, lungs and tumors by digestion in collagenase/DNase I. The antibodies used in this study are listed in Supplementary Table 4. Cell numbers were determined by the addition of fluorochrome-conjugated calibration beads (BD Biosciences, San Jose, CA) directly to the samples. To evaluate expression levels, a fluorescence minus one (FMO) control was included. Data were collected using a FACSVerse (BD Biosciences) and analyzed using FlowJo software (Tree Star, Ashland, OR). Cell sorting was performed using a FACSAria or an Influx cell sorter (BD Biosciences).
BM cell culture
BM cells from mice were isolated by flushing femurs and tibias with 5 ml PBS supplemented with 2% heat-inactivated fetal bovine serum (FBS) (Sigma Aldrich, Lenexa, KS, USA). The BM cells were centrifuged once and then resuspended in tris-ammonium chloride at 37 °C for 30 s to lyse red blood cells. The cells were centrifuged again and then strained through a 70-μm filter before being resuspended in RPMI-1640 medium supplemented with 10% FBS. For GM-CSF-stimulated cultures, BM cells were resuspended at 0.5 × 106/ml in medium containing titrated doses of GM-CSF in 12-well plates. After 3–4 days, fresh medium with cytokines was added to the cultures. Cell cultures were maintained for up to 7 days. For further analysis, MØs and DCs were sorted on a FACSAria (BD Biosciences).
T-cell proliferation assays
CD4+ T cells (OT-II) and CD8+ T cells (OT-I) were purified from TCR-transgenic mice by sorting and labeled with CTV (Invitrogen, Thermo Fisher, Waltham; MA) according to the manufacturer’s protocol. Labeled cells were cultured at 1 × 105 cells in 200 μL RPMI-1640 medium supplemented with 10% FBS in flat-bottom 96-well plates in the absence or presence of antigen-presenting cells and defined antigens. The cell cultures were harvested 3 days later. T-cell proliferation was evaluated by monitoring CTV dilution. The arginase inhibitor 1 L-norvaline (12 mM, Sigma‒Aldrich) was included in some cultures. The cytokine levels in culture supernatants were analyzed using a BioPlex kit (Bio-Rad). In some experiments, CTV-labeled CD8+ T cells were also stimulated with 5 μg/ml anti-CD3 and 2 μg/ml anti-CD28 for 3 days. Cell proliferation and cytokine production were evaluated as described above.
Antigen uptake and processing
For antigen uptake, 1 × 106 cells were incubated with 100 μg/mL FITC‐OVA at 37 °C or kept on ice for 30 min. For antigen processing, 1 × 106 cells were incubated with 100 μg/mL DQ-OVA (ThermoFisher) at 37 °C for 30 min. Then, the samples were either incubated at 37 °C for an additional 90 min or kept on ice. After the incubation, the cells were washed with cold PBS containing 2% FBS‐EDTA (0.02 mM) and analyzed by flow cytometry.
Cell signaling analysis by immunoblotting
GM-MØs were isolated by FACS sorting. The purity of enriched cells was consistently >95%. For cell signaling assessment, cells were washed free of GM-CSF, rested for 4 h and then restimulated in vitro with recombinant GM-CSF (50 ng/ml) for the indicated times in RPMI-1640 medium supplemented with antibiotics and 10% heat-inactivated FBS. The cells were washed in cold PBS and pelleted, followed by lysis. Approximately 5 × 105 sorted cells were collected per sample and lysed in 100 μl lysis buffer (50 mM Tris-HCl, pH 7.4; 150 mM NaCl; 0.25% deoxycholic acid; 1% NP-40; and 1 mM EDTA) supplemented with protease inhibitors (Complete Cocktail tablets, Roche), 1 mM PMSF, 1 mM Na3VO4 and 1 mM NaF for 30 min on ice. The lysates were clarified by centrifugation at 13,000 rpm for 15 min at 4 °C. Gel electrophoresis was carried out with 4–12% NuPAGE Bis-Tris gels, followed by transfer to nitrocellulose membranes (Amersham) and immunoblotting. The primary antibodies are listed in Supplementary Table 4.
Cell stimulation and cytokine assay
MØs and moDCs derived from 7-day BM cultures were purified by sorting. Then, the cells were cultured at 5 × 104 cells in 200 μl RPMI-1640 medium supplemented with 10% FBS in U-bottom 96-well plates in the absence or presence of LPS (1 μg/mL, Sigma), CpG ODN 1668 (1 μM, InvivoGen), an agonistic anti-CD40 Ab (5 μg/ml, clone FGK4.5) or PolyI:C (5 μg/ml, InvivoGen) for 20 h. For detection of cytokines in supernatants in in vitro assays, the indicated cytokines were detected using a BioPlex kit (Bio-Rad) or ELISA kit (Invitrogen) following the manufacturer’s instructions. For intracellular cytokine staining, cells were stimulated for 4–6 h with 0.5 μM CpG or 50 ng/ml PMA/1 mM ionomycin with GolgiStop (BD Biosciences), stained for cell-surface markers, fixed/permeabilized using a Cytofix/Cytoperm kit (BD Biosciences), and stained with anti-cytokine antibodies. The antibodies are listed in Supplementary Table 4.
JAK inhibition and IL-4 neutralization during in vitro MØ differentiation
For JAK inhibition, BM cells were cultured at 0.5 × 106 cells/ml in medium containing 10 ng/mL GM-CSF in 12-well plates for 5 days. Ruxolitinib (RUXO) (0.2 μM, Invitrogen) was added, and the cells were harvested on Day 7. For IL-4 neutralization, BM cells were cultured at 0.5 × 106 cells/ml in medium containing 10 ng/mL GM-CSF in 12-well plates in the presence of an anti-IL-4 antibody (50 μg/ml, clone 11B11) for 7 days.
Tumor induction
A total of 1–5 × 105 GM-CSF-transduced mouse melanoma B16F10 cells (B16-GM cells, provided by Prof. Jose Villadangos with permission from Prof Glenn Dranoff [8]) were injected s.c. into the flank of test mice. The mice were evaluated after 1–2 weeks.
RNA sequencing
GM-DCs and GM-MØs from 4 individual mice were enriched by sorting. RNA was isolated independently from biological replicates with RNeasyPlus Mini kits (Qiagen). mRNA reverse transcription and cDNA libraries were prepared using a TruSeq RNA Sample preparation kit (Illumina) following the manufacturer’s instructions. Samples were sequenced with an Illumina NextSeq 500 sequencing system, producing between 14–20 x107 single-end 85-bp reads per sample.
Bioinformatic analysis of RNA-seq data
RNA-seq reads were quantified against the Ensembl [60] v31 GRCm38 transcriptome using Kallisto [61]. Differential expression analysis was conducted using Sleuth [62] at the gene level via the method of Pimentel et al. [63]. Q-values were calculated using the Benjamini–Hochberg [64] adjustment, and log-fold changes at the gene level were estimated by combining transcript level abundances normalized to transcripts per million reads (TPM) [65]. Heatmaps of TPM expression values were plotted with the coolmap function of the limma package [66].
Sample preparation for mass spectrometry-based proteomics
GM-MØs (1–1.5 x 106 cells/replicate) from 4 individual WT or Cish−/− mice were sorted on a FACSAria to achieve a final purity of 99–100%. The cells were washed three times with ice-cold PBS prior to dry cell pellet storage at –80 °C. The cells were lysed in preheated (95 °C) 5% SDS/10 mM Tris/10 mM Tris (2-carboxyethyl) phosphine/5.5 mM 2-chloroacetamide and heated at 95 °C for 10 min. Neat trifluoracetic acid (Sigma‒Aldrich) was added to hydrolyze the DNA, resulting in a final concentration of 1%. The lysates were quenched with 4 M Tris (pH 10), resulting in a final concentration of ~140 mM Tris (pH 7). The myeloid cell protein lysates (~50 μg) were prepared for mass spectrometry (MS) analysis as previously described [67]. Acidified peptide mixtures were analyzed by nanoflow reversed-phase liquid chromatography–tandem mass spectrometry (LC–MS/MS) on an Easy-nLC 1000 system (Thermo Fisher Scientific) coupled to a Q-Exactive HF (QE-HF) mass spectrometer equipped with a nanoelectrospray ion source and in-source column heater (Sonation) at 40 °C for automated MS/MS (Thermo Fisher Scientific). The peptide mixtures were loaded in buffer A (0.1% formic acid, 2% acetonitrile, and Milli-Q water) and separated by reverse-phase chromatography using a C18 fused silica column (packed emitter, internal diameter: 75 μm, outer diameter: 360 μm × 25 cm length, IonOpticks) using flow rates and data-dependent methods previously described [25]. Raw files consisting of high-resolution MS/MS spectra were processed with MaxQuant (version 1.5.8.30) for feature detection and protein identification using the Andromeda search engine as previously described [67]. LFQ quantification was selected, with a minimum ratio count of 2. PSM and protein identifications were filtered using a target-decoy approach at an FDR of 1%. Only unique and razor peptides were considered for quantification, with intensity values present in at least 2 out of the 3 replicates per group. Statistical analyses were performed using LFQAnalyst (https://bioinformatics.erc.monash.edu/apps/LFQ-Analyst/), whereby the LFQ intensity values were used for protein quantification. Missing values were replaced by values drawn from a normal distribution of 1.8 standard deviations and a width of 0.3 for each sample (Perseus-type). Proteinwise linear models combined with empirical Bayes statistics were used for differential expression analysis using the Bioconductor package limma, whereby the adjusted p value cutoff was set at 0.05 and the log2-fold change cutoff was set at 1. The Benjamini‒Hochberg (BH) method for FDR correction was used.
CRISPR/CAS9-mediated deletion of CISH in human CD34+ cord blood cells
CD34+ cells were isolated from human cord blood (Stemcell Technologies) and cultured for 2 days in expansion medium (Miltenyi). CIS RNPs were assembled by incubating 1 ml of 30 mM CISH gRNA (5ʹ-CTCACCAGATTCCCGAAGGT-3ʹ; Synthego), 1.7 ml of 67 nM Cas9 (Integrated DNA Technologies), 1 ml of electroporation enhancer (Integrated DNA Technologies) and 1.3 ml of PBS for 10 min at room temperature. A total of 5 × 105 cells were pelleted, resuspended in RNP solution and 20 ml of primary cell P3 buffer (Lonza) and transferred to an electroporation cuvette (Lonza). The cells were electroporated using a 4D-Nucleofector (Lonza) with pulse code CM-137 and rested in complete medium for 10 min before being transferred to a cell culture dish. The cells were cultured with 5 ng/mL huGM-CSF (R&D) for 7 days. A small pellet of cells was collected for sequencing to determine the CIS indel frequency.
Cell transfer
WT and Cish−/− GM-MØs were incubated with 10 mg/ml OVA in serum-free medium at 37 °C for 30 min. After washing with cold PBS, OVA-pulsed WT and Cish−/− GM-MØs were injected i.v. into B6 mice or Rag1−/−/Il2rg−/− mice infused with 106 Ly5.1-OT-1 cells 2 weeks prior. OT-1 cell expansion was evaluated 3–7 days after GM-MØ injection.
CUT&Tag sequencing
CUT&Tag was performed with the Hyperactive in situ ChIP Library Prep Kit (Vazyme) following the manufacturer’s recommendations. Briefly, Cish−/− GM-MØs were sorted from GM-CSF-supplemented cultures of BM cells isolated from WT Irf8Gfp or Cish−/− Irf8Gfp mice. GFP+ GM-MØs (1 × 105 cells) were bound to concanavalin A-coated magnetic beads (Bags laboratories) and subjected to immunoprecipitation with 0.5 μg of primary antibody (rabbit anti-GFP, ab290; rabbit anti-H3K27me3, CST: 9733; or rabbit anti-mouse IgG control). Following the primary antibody incubation and washing using a magnetic stand, a secondary anti-rabbit antibody was added and incubated under gentle agitation for 1 h (Antibodies online ABIN101961). The cells were washed and incubated for 1 h in a mix of Hyperactive pG-Tn5/pA-Tn5 Transposon with Dig-300 Buffer at a final concentration of 0.04 μM. Excess reagents were washed out using the magnetic stand, and the cells were resuspended in 100 μl of Tagmentation buffer and incubated at 37 °C for 1 h. The reaction was stopped by heat inactivation (55 °C for 10 min). DNA was purified using Ampure XP beads (Beckman Coulter). For library amplification, 24 μl of DNA was mixed with 10 μl of 5x TAB, 1 μl of TAE, and 5 μl of uniquely barcoded i5 and i7 primers [68] and amplified for 14 cycles (72 °C for 5 min; 98 °C for 30 s; 14 cycles of 98 °C for 10 s, 63 °C for 10 s and 72 °C for 1 min; and hold at 4 °C). The PCR products were purified with Ampure XP beads and eluted in 25 µl of water. The eluted DNA was checked for region molarity via a high-sensitivity D5000 TapeStation (Fig. S6B). The libraries were sequenced on an Illumina NextSeq platform, and 150-bp paired-end reads were generated.
CUT&Tag data analysis
Raw data were uploaded to the Galaxy Australia website (https://usegalaxy.org.au/). The sequence quality was checked via FastQC and MultiQC. Then, the raw data were aligned to mm10 via Bowtie2 version 2.3.4.3 with the following options: --local --very-sensitive-local --no-unal --no-mixed --no-discordant -- phred33 -I 10 -X 700 [69, 70]. Duplicate reads were removed via a SAM/BAM filter. The biological duplicate sample reads were pooled together via the Merge BAM files package. Then, peaks were called via MACS2 (q < 0.01) [71, 72]. The binding peaks were viewed using the Integrated Genome Brower (IGB) [73]. The peaks were annotated via ChIPseeker [74]. The peak intersection between WT and Cis–/– GM-MØs was calculated via Bedtool [75].
Retroviral transduction
Retroviral supernatants were generated by transient transfection of 293T cells with plasmids encoding viral envelope proteins (pMD1-gag-pol and pCAG-Eco) and the expression vector pMSCV-iresGFP or pMSCV-IRF8iresGFP using FuGeneHD (Promega). The retroviral supernatants were centrifuged onto RetroNectin (Takara)-coated plates for 45 min at 4000 rpm and 32 °C. Cells were then cultured with the virus in the presence of 4 μg/ml polybrene (Sigma‒Aldrich) for 12 h.
Cell imaging
In total, 2 × 105/ml cells were suspended in 100 μl PBS, stained using a 1:1000 dye mixture (CellTracker violet LysoTracker green and DAPI, from Thermo Fisher) at 37 °C for 15 min, washed twice in PBS and plated in 96-well PerkinElmer Cell Carrier plates for imaging with a Leica SP8 confocal microscope. The volume of each cell was calculated using Imaris 9.1.2 by applying a surface to each cell using the CellTracker violet channel. Images at higher resolution were acquired using the same microscopy settings and processed on a FIJI platform.
Allergic asthma model
To establish an allergic asthma model, 5 × 106 WT or Cish−/− GM-MØs were generated, loaded with OVA and then injected into recipient mice on Day 0 and Day 7. The allergic response to OVA was induced as described previously [76].
Statistical analysis
Statistical comparisons of the mean difference between two groups from independent experiments were made using a two-tailed Student’s t test, and data for multiple groups, dose–response curves or time courses were analyzed using ANOVA. Analyses were performed with Prism v.5.0 software (GraphPad, San Diego, CA). P < 0.05 was considered statistically significant.
Supplementary information
Acknowledgements
We thank Lisa Reid, Marina Patsis, Manuela Hancock, Rhiannan Crawley, Rebekah Meeny, Tania Camilleri, and the institute flow cytometry facility for excellent technical assistance. We acknowledge the Wurundjeri people of the Kulin nation as the traditional owners and guardians of the land on which most of the work was performed. This work was supported by National Health and Medical Research Council of Australia (NHMRC) grants (1037321, 1105209, 1143976, 1150425, 1080321, 1196335, 5575500, 1054925, and 1048278), an NHMRC Independent Research Institutes Infrastructure Support Scheme grant (361646) and a Victorian State Government Operational Infrastructure Support grant. JB was supported by the Stafford Fox Medical Research Foundation.
Author contributions
Conceptualization, YZ; Methodology: SBZ, JR, MC, NGB, LFD, JB, YY, and YX; Investigation: YZ, SBZ, TBK, JR, MC, QKW, HQW, LS, RS, LFD, FSFG, YY, YX, RA, NI, and JLN; Writing—Original Draft, YZ with input from SBZ, AML, SN, SEN, MC, NGB, and YY; Review & Editing: SBZ, MC, NGB, LFD, YKX, SN, NDH, SEN, and AML; and Resources: JR, NDH, SN, and SEN.
Data availability
The RNA-seq data have been deposited in the European Nucleotide Archive (ENA) under dataset identifier PRJEB40745. The mass spectrometry proteomic data have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository under the dataset identifier PXD018390 (reviewer token: Username: reviewer98346@ebi.ac.uk Password:2MzhUnon). The CUT&Tag data will be made available upon request and will be publicly available after publication.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Shengbo Zhang, Jai Rautela, Naiara G Bediaga, Tatiana B Kolesnik.
These authors jointly supervised this work: Nicholas D Huntington, Sandra E Nicholson, Michaël Chopin, Yifan Zhan.
Contributor Information
Michaël Chopin, Email: Michael.chopin@monash.edu.
Yifan Zhan, Email: yifan.zhan@huabobio.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41423-022-00957-z.
References
- 1.Lavin Y, Mortha A, Rahman A, Merad M. Regulation of macrophage development and function in peripheral tissues. Nat Rev Immunol. 2015;15:731–44. doi: 10.1038/nri3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, Goerdt S, et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41:14–20. doi: 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fleetwood AJ, Lawrence T, Hamilton JA, Cook AD. Granulocyte-macrophage colony-stimulating factor (CSF) and macrophage CSF-dependent macrophage phenotypes display differences in cytokine profiles and transcription factor activities: implications for CSF blockade in inflammation. J Immunol (Baltim, Md: 1950) 2007;178:5245–52. doi: 10.4049/jimmunol.178.8.5245. [DOI] [PubMed] [Google Scholar]
- 4.Martinez FO, Gordon S. The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000Prime Rep. 2014;6:13. doi: 10.12703/P6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Verreck FA, de Boer T, Langenberg DM, Hoeve MA, Kramer M, Vaisberg E, et al. Human IL-23-producing type 1 macrophages promote but IL-10-producing type 2 macrophages subvert immunity to (myco)bacteria. Proc Natl Acad Sci USA. 2004;101:4560–5. doi: 10.1073/pnas.0400983101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamilton JA. Colony-stimulating factors in inflammation and autoimmunity. Nat Rev Immunol. 2008;8:533–44. doi: 10.1038/nri2356. [DOI] [PubMed] [Google Scholar]
- 7.Becher B, Tugues S, Greter M. GM-CSF: from growth factor to central mediator of tissue inflammation. Immunity. 2016;45:963–73. doi: 10.1016/j.immuni.2016.10.026. [DOI] [PubMed] [Google Scholar]
- 8.Dranoff G, Jaffee E, Lazenby A, Golumbek P, Levitsky H, Brose K, et al. Vaccination with irradiated tumor cells engineered to secrete murine granulocyte-macrophage colony-stimulating factor stimulates potent, specific, and long-lasting anti-tumor immunity. Proc Natl Acad Sci USA. 1993;90:3539–43. doi: 10.1073/pnas.90.8.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bayne LJ, Beatty GL, Jhala N, Clark CE, Rhim AD, Stanger BZ, et al. Tumor-derived granulocyte-macrophage colony-stimulating factor regulates myeloid inflammation and T cell immunity in pancreatic cancer. Cancer cell. 2012;21:822–35. doi: 10.1016/j.ccr.2012.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sielska M, Przanowski P, Wylot B, Gabrusiewicz K, Maleszewska M, Kijewska M, et al. Distinct roles of CSF family cytokines in macrophage infiltration and activation in glioma progression and injury response. J Pathol. 2013;230:310–21. doi: 10.1002/path.4192. [DOI] [PubMed] [Google Scholar]
- 11.Bronte V, Chappell DB, Apolloni E, Cabrelle A, Wang M, Hwu P, et al. Unopposed production of granulocyte-macrophage colony-stimulating factor by tumors inhibits CD8+ T cell responses by dysregulating antigen-presenting cell maturation. J Immunol (Baltim, Md: 1950) 1999;162:5728–37. [PMC free article] [PubMed] [Google Scholar]
- 12.Huen SC, Huynh L, Marlier A, Lee Y, Moeckel GW, Cantley LG. GM-CSF promotes macrophage alternative activation after renal ischemia/reperfusion injury. J Am Soc Nephrol. 2015;26:1334–45. doi: 10.1681/ASN.2014060612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cates EC, Fattouh R, Wattie J, Inman MD, Goncharova S, Coyle AJ, et al. Intranasal exposure of mice to house dust mite elicits allergic airway inflammation via a GM-CSF-mediated mechanism. J Immunol (Baltim, Md: 1950) 2004;173:6384–92. doi: 10.4049/jimmunol.173.10.6384. [DOI] [PubMed] [Google Scholar]
- 14.Willart MA, Deswarte K, Pouliot P, Braun H, Beyaert R, Lambrecht BN, et al. Interleukin-1alpha controls allergic sensitization to inhaled house dust mite via the epithelial release of GM-CSF and IL-33. J Exp Med. 2012;209:1505–17. doi: 10.1084/jem.20112691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mills CD, Kincaid K, Alt JM, Heilman MJ, Hill AM. M-1/M-2 macrophages and the Th1/Th2 paradigm. J Immunol (Baltim, Md: 1950) 2000;164:6166–73. doi: 10.4049/jimmunol.164.12.6166. [DOI] [PubMed] [Google Scholar]
- 16.Bernasconi E, Favre L, Maillard MH, Bachmann D, Pythoud C, Bouzourene H, et al. Granulocyte-macrophage colony-stimulating factor elicits bone marrow-derived cells that promote efficient colonic mucosal healing. Inflamm Bowel Dis. 2010;16:428–41. doi: 10.1002/ibd.21072. [DOI] [PubMed] [Google Scholar]
- 17.Alexander WS, Hilton DJ. The role of suppressors of cytokine signaling (SOCS) proteins in regulation of the immune response. Annu Rev Immunol. 2004;22:503–29. doi: 10.1146/annurev.immunol.22.091003.090312. [DOI] [PubMed] [Google Scholar]
- 18.Louis C, Souza-Fonseca-Guimaraes F, Yang Y, D’silva D, Kratina T, Dagley L, et al. 2020. NK cell-derived GM-CSF potentiates inflammatory arthritis and is negatively regulated by CIS. J Exp Med. 217:e20191421 [DOI] [PMC free article] [PubMed]
- 19.Feldman GM, Rosenthal LA, Liu X, Hayes MP, Wynshaw-Boris A, Leonard WJ, et al. STAT5A-deficient mice demonstrate a defect in granulocyte-macrophage colony-stimulating factor-induced proliferation and gene expression. Blood. 1997;90:1768–76. doi: 10.1182/blood.V90.5.1768. [DOI] [PubMed] [Google Scholar]
- 20.Matsumoto A, Masuhara M, Mitsui K, Yokouchi M, Ohtsubo M, Misawa H, et al. CIS, a cytokine inducible SH2 protein, is a target of the JAK-STAT5 pathway and modulates STAT5 activation. Blood. 1997;89:3148–54. doi: 10.1182/blood.V89.9.3148. [DOI] [PubMed] [Google Scholar]
- 21.Yoshimura A, Ohkubo T, Kiguchi T, Jenkins NA, Gilbert DJ, Copeland NG, et al. A novel cytokine-inducible gene CIS encodes an SH2-containing protein that binds to tyrosine-phosphorylated interleukin 3 and erythropoietin receptors. EMBO J. 1995;14:2816–26. doi: 10.1002/j.1460-2075.1995.tb07281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lehtonen A, Matikainen S, Miettinen M, Julkunen I. Granulocyte-macrophage colony-stimulating factor (GM-CSF)-induced STAT5 activation and target-gene expression during human monocyte/macrophage differentiation. J Leukoc Biol. 2002;71:511–9. doi: 10.1189/jlb.71.3.511. [DOI] [PubMed] [Google Scholar]
- 23.Marine JC, McKay C, Wang D, Topham DJ, Parganas E, Nakajima H, et al. SOCS3 is essential in the regulation of fetal liver erythropoiesis. Cell. 1999;98:617–27. doi: 10.1016/S0092-8674(00)80049-5. [DOI] [PubMed] [Google Scholar]
- 24.Matsumoto A, Seki Y, Kubo M, Ohtsuka S, Suzuki A, Hayashi I, et al. Suppression of STAT5 functions in liver, mammary glands, and T cells in cytokine-inducible SH2-containing protein 1 transgenic mice. Mol Cell Biol. 1999;19:6396–407. doi: 10.1128/MCB.19.9.6396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delconte RB, Kolesnik TB, Dagley LF, Rautela J, Shi W, Putz EM, et al. CIS is a potent checkpoint in NK cell-mediated tumor immunity. Nat Immunol. 2016;17:816–24. doi: 10.1038/ni.3470. [DOI] [PubMed] [Google Scholar]
- 26.Palmer DC, Guittard GC, Franco Z, Crompton JG, Eil RL, Patel SJ, et al. Cish actively silences TCR signaling in CD8+ T cells to maintain tumor tolerance. J Exp Med. 2015;212:2095–113. doi: 10.1084/jem.20150304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang XP, Ghoreschi K, Steward-Tharp SM, Rodriguez-Canales J, Zhu J, Grainger JR, et al. Opposing regulation of the locus encoding IL-17 through direct, reciprocal actions of STAT3 and STAT5. Nat Immunol. 2011;12:247–54. doi: 10.1038/ni.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Helft J, Bottcher J, Chakravarty P, Zelenay S, Huotari J, Schraml BU, et al. GM-CSF mouse bone marrow cultures comprise a heterogeneous population of CD11c(+)MHCII(+) macrophages and dendritic cells. Immunity. 2015;42:1197–211. doi: 10.1016/j.immuni.2015.05.018. [DOI] [PubMed] [Google Scholar]
- 29.Henry CJ, Ornelles DA, Mitchell LM, Brzoza-Lewis KL, Hiltbold EM. IL-12 produced by dendritic cells augments CD8+ T cell activation through the production of the chemokines CCL1 and CCL17. J Immunol (Baltim, Md: 1950) 2008;181:8576–84. doi: 10.4049/jimmunol.181.12.8576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun L, Rautela J, Delconte RB, Souza-Fonseca-Guimaraes F, Carrington EM, Schenk RL, et al. GM-CSF Quantity Has a Selective Effect on Granulocytic vs. Monocytic Myeloid Development and Function. Front Immunol. 2018;9:1922. doi: 10.3389/fimmu.2018.01922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chow KV, Lew AM, Sutherland RM, Zhan Y. Monocyte-derived dendritic cells promote th polarization, whereas conventional dendritic cells promote Th proliferation. J Immunol (Baltim, Md: 1950) 2016;196:624–36. doi: 10.4049/jimmunol.1501202. [DOI] [PubMed] [Google Scholar]
- 32.Erlich Z, Shlomovitz I, Edry-Botzer L, Cohen H, Frank D, Wang H, et al. Macrophages, rather than DCs, are responsible for inflammasome activity in the GM-CSF BMDC model. Nat Immunol. 2019;20:397–406. doi: 10.1038/s41590-019-0313-5. [DOI] [PubMed] [Google Scholar]
- 33.Szanto A, Balint BL, Nagy ZS, Barta E, Dezso B, Pap A, et al. STAT6 transcription factor is a facilitator of the nuclear receptor PPARgamma-regulated gene expression in macrophages and dendritic cells. Immunity. 2010;33:699–712. doi: 10.1016/j.immuni.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pello OM, De Pizzol M, Mirolo M, Soucek L, Zammataro L, Amabile A, et al. Role of c-MYC in alternative activation of human macrophages and tumor-associated macrophage biology. Blood. 2012;119:411–21. doi: 10.1182/blood-2011-02-339911. [DOI] [PubMed] [Google Scholar]
- 35.Martinez FO, Gordon S, Locati M, Mantovani A. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J Immunol (Baltim, Md: 1950) 2006;177:7303–11. doi: 10.4049/jimmunol.177.10.7303. [DOI] [PubMed] [Google Scholar]
- 36.Martinez FO, Helming L, Milde R, Varin A, Melgert BN, Draijer C, et al. Genetic programs expressed in resting and IL-4 alternatively activated mouse and human macrophages: similarities and differences. Blood. 2013;121:e57–69. doi: 10.1182/blood-2012-06-436212. [DOI] [PubMed] [Google Scholar]
- 37.Lacey DC, Achuthan A, Fleetwood AJ, Dinh H, Roiniotis J, Scholz GM, et al. Defining GM-CSF- and macrophage-CSF-dependent macrophage responses by in vitro models. J Immunol (Baltim, Md: 1950) 2012;188:5752–65. doi: 10.4049/jimmunol.1103426. [DOI] [PubMed] [Google Scholar]
- 38.Rodriguez PC, Quiceno DG, Ochoa AC. L-arginine availability regulates T-lymphocyte cell-cycle progression. Blood. 2007;109:1568–73. doi: 10.1182/blood-2006-06-031856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bronte V, Serafini P, Mazzoni A, Segal DM, Zanovello P. L-arginine metabolism in myeloid cells controls T-lymphocyte functions. Trends Immunol. 2003;24:302–6. doi: 10.1016/S1471-4906(03)00132-7. [DOI] [PubMed] [Google Scholar]
- 40.Gunthner R, Anders HJ. Interferon-regulatory factors determine macrophage phenotype polarization. Mediators Inflamm. 2013;2013:731023. doi: 10.1155/2013/731023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang H, Yan M, Sun J, Jain S, Yoshimi R, Abolfath SM, et al. A reporter mouse reveals lineage-specific and heterogeneous expression of IRF8 during lymphoid and myeloid cell differentiation. J Immunol (Baltim, Md: 1950) 2014;193:1766–77. doi: 10.4049/jimmunol.1301939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kurotaki D, Yamamoto M, Nishiyama A, Uno K, Ban T, Ichino M, et al. IRF8 inhibits C/EBPalpha activity to restrain mononuclear phagocyte progenitors from differentiating into neutrophils. Nat Commun. 2014;5:4978. doi: 10.1038/ncomms5978. [DOI] [PubMed] [Google Scholar]
- 43.Kaya-Okur HS, Wu SJ, Codomo CA, Pledger ES, Bryson TD, Henikoff JG, et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat Commun. 2019;10:1930. doi: 10.1038/s41467-019-09982-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chávez-Galán L, Olleros ML, Vesin D, Garcia I. Much More than M1 and M2 Macrophages, There are also CD169(+) and TCR(+) Macrophages. Front Immunol. 2015;6:263. [DOI] [PMC free article] [PubMed]
- 45.Yang XO, Zhang H, Kim BS, Niu X, Peng J, Chen Y, et al. The signaling suppressor CIS controls proallergic T cell development and allergic airway inflammation. Nat Immunol. 2013;14:732–40. doi: 10.1038/ni.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Satoh T, Takeuchi O, Vandenbon A, Yasuda K, Tanaka Y, Kumagai Y, et al. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat Immunol. 2010;11:936–44. doi: 10.1038/ni.1920. [DOI] [PubMed] [Google Scholar]
- 47.Croxford AL, Lanzinger M, Hartmann FJ, Schreiner B, Mair F, Pelczar P, et al. The cytokine GM-CSF drives the inflammatory signature of CCR2+ monocytes and licenses autoimmunity. Immunity. 2015;43:502–14. doi: 10.1016/j.immuni.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 48.Ko HJ, Brady JL, Ryg-Cornejo V, Hansen DS, Vremec D, Shortman K, et al. GM-CSF-responsive monocyte-derived dendritic cells are pivotal in Th17 pathogenesis. J Immunol (Baltim, Md: 1950) 2014;192:2202–9. doi: 10.4049/jimmunol.1302040. [DOI] [PubMed] [Google Scholar]
- 49.Zhang Y, Li X, Luo Z, Ma L, Zhu S, Wang Z, et al. ECM1 is an essential factor for the determination of M1 macrophage polarization in IBD in response to LPS stimulation. Proc Natl Acad Sci USA. 2020;117:3083–92. doi: 10.1073/pnas.1912774117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bhattacharya P, Thiruppathi M, Elshabrawy HA, Alharshawi K, Kumar P, Prabhakar BS. GM-CSF: An immune modulatory cytokine that can suppress autoimmunity. Cytokine. 2015;75:261–71. doi: 10.1016/j.cyto.2015.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Esashi E, Wang YH, Perng O, Qin XF, Liu YJ, Watowich SS. The signal transducer STAT5 inhibits plasmacytoid dendritic cell development by suppressing transcription factor IRF8. Immunity. 2008;28:509–20. doi: 10.1016/j.immuni.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Holtschke T, Lohler J, Kanno Y, Fehr T, Giese N, Rosenbauer F, et al. Immunodeficiency and chronic myelogenous leukemia-like syndrome in mice with a targeted mutation of the ICSBP gene. Cell. 1996;87:307–17. doi: 10.1016/S0092-8674(00)81348-3. [DOI] [PubMed] [Google Scholar]
- 53.Masumi A, Tamaoki S, Wang IM, Ozato K, Komuro K. IRF-8/ICSBP and IRF-1 cooperatively stimulate mouse IL-12 promoter activity in macrophages. FEBS Lett. 2002;531:348–53. doi: 10.1016/S0014-5793(02)03556-1. [DOI] [PubMed] [Google Scholar]
- 54.Ototake Y, Yamaguchi Y, Asami M, Komitsu N, Akita A, Watanabe T, et al. Downregulated IRF8 in Monocytes and Macrophages of Patients with Systemic Sclerosis May Aggravate the Fibrotic Phenotype. J investigative Dermatol. 2021;141:1954–63. doi: 10.1016/j.jid.2021.02.015. [DOI] [PubMed] [Google Scholar]
- 55.Twum DY, Colligan SH, Hoffend NC, Katsuta E, Gomez EC, Hensen ML, et al. IFN regulatory factor-8 expression in macrophages governs an antimetastatic program. JCI Insight. 2019;4:e124267 [DOI] [PMC free article] [PubMed]
- 56.Waight JD, Netherby C, Hensen ML, Miller A, Hu Q, Liu S, et al. Myeloid-derived suppressor cell development is regulated by a STAT/IRF-8 axis. J Clin Investig. 2013;123:4464–78. doi: 10.1172/JCI68189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cai Y, Kumar RK, Zhou J, Foster PS, Webb DC. Ym1/2 promotes Th2 cytokine expression by inhibiting 12/15(S)-lipoxygenase: identification of a novel pathway for regulating allergic inflammation. J Immunol (Baltim, Md: 1950) 2009;182:5393–9. doi: 10.4049/jimmunol.0803874. [DOI] [PubMed] [Google Scholar]
- 58.Arora M, Chen L, Paglia M, Gallagher I, Allen JE, Vyas YM, et al. Simvastatin promotes Th2-type responses through the induction of the chitinase family member Ym1 in dendritic cells. Proc Natl Acad Sci USA. 2006;103:7777–82. doi: 10.1073/pnas.0508492103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abdelaziz MH, Abdelwahab SF, Wan J, Cai W, Huixuan W, Jianjun C, et al. Alternatively activated macrophages; a double-edged sword in allergic asthma. J Transl Med. 2020;18:58. [DOI] [PMC free article] [PubMed]
- 60.Yates AD, Achuthan P, Akanni W, Allen J, Allen J, Alvarez-Jarreta J, et al. Ensembl 2020. Nucleic Acids Res. 2020;48:D682–D688. doi: 10.1093/nar/gkz966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:525–7. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 62.Roguev A, Talbot D, Negri GL, Shales M, Cagney G, Bandyopadhyay S, et al. Quantitative genetic-interaction mapping in mammalian cells. Nat Methods. 2013;10:432–7. doi: 10.1038/nmeth.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yi L, Pimentel H, Bray NL, Pachter L. Gene-level differential analysis at transcript-level resolution. Genome Biol. 2018;19:53. doi: 10.1186/s13059-018-1419-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc: Ser B (Methodol) 1995;57:289–300. [Google Scholar]
- 65.Wagner GP, Kin K, Lynch VJ. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 2012;131:281–5. doi: 10.1007/s12064-012-0162-3. [DOI] [PubMed] [Google Scholar]
- 66.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rautela J, Dagley LF, De Oliveira CC, Schuster IS, Hediyeh-Zadeh S, Delconte RB, et al. Therapeutic blockade of activin-A improves NK cell function and antitumor immunity. Sci Signal. 2019;12:eaat7527 [DOI] [PubMed]
- 68.Mezger A, Klemm S, Mann I, Brower K, Mir A, Bostick M, et al. High-throughput chromatin accessibility profiling at single-cell resolution. Nat Commun. 2018;9:3647. doi: 10.1038/s41467-018-05887-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Feng J, Liu T, Qin B, Zhang Y, Liu XS. Identifying ChIP-seq enrichment using MACS. Nat Protoc. 2012;7:1728–40. doi: 10.1038/nprot.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008;9:R137. [DOI] [PMC free article] [PubMed]
- 73.Nicol JW, Helt GA, Blanchard SG, Jr., Raja A, Loraine AE. The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinforma (Oxf, Engl) 2009;25:2730–1. doi: 10.1093/bioinformatics/btp472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yu G, Wang LG, He QY. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinforma (Oxf, Engl) 2015;31:2382–3. doi: 10.1093/bioinformatics/btv145. [DOI] [PubMed] [Google Scholar]
- 75.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinforma (Oxf, Engl) 2010;26:841–2. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Keenan CR, Iannarella N, Garnham AL, Brown AC, Kim RY, Horvat JC, et al. Polycomb repressive complex 2 is a critical mediator of allergic inflammation. JCI Insight. 2019;4:e127745. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data have been deposited in the European Nucleotide Archive (ENA) under dataset identifier PRJEB40745. The mass spectrometry proteomic data have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository under the dataset identifier PXD018390 (reviewer token: Username: reviewer98346@ebi.ac.uk Password:2MzhUnon). The CUT&Tag data will be made available upon request and will be publicly available after publication.