ABSTRACT
Many bacterial species use the secondary messenger, c-di-GMP, to promote the production of biofilm matrix components. In Pseudomonas aeruginosa, c-di-GMP production is stimulated upon initial surface contact and generally remains high throughout biofilm growth. Transcription of several gene clusters, including the Sia signal transduction system, are induced in response to high cellular levels of c-di-GMP. The output of this system is SiaD, a diguanylate cyclase whose activity is induced in the presence of the detergent SDS. Previous studies demonstrated that Sia-mediated cellular aggregation is a key feature of P. aeruginosa growth in the presence of SDS. Here, we show that the Sia system is important for producing low levels of c-di-GMP when P. aeruginosa is growing planktonically. In addition, we show that Sia activity is important for maintaining cell-associated Psl in planktonic populations. We also demonstrate that Sia mutant strains have reduced cell-associated Psl and a surface attachment-deficient phenotype. The Sia system also appears to posttranslationally impact cell-associated Psl levels. Collectively, our findings suggest a novel role for the Sia system and c-di-GMP in planktonic populations by regulating levels of cell-associated Psl.
IMPORTANCE The biofilm matrix of the opportunistic pathogen Pseudomonas aeruginosa is composed of exopolysaccharides (EPS), proteins, and nucleic acids. In P. aeruginosa, an increase in the small molecule c-di-GMP causes an increase in biofilm matrix production both transcriptionally and posttranslationally. C-di-GMP is synthesized by diguanylate cyclases (DGCs), and P. aeruginosa encodes many DGCs which are active under different conditions and influence specific pathways. Here, we demonstrate that the DGC SiaD, specifically, posttranslationally regulates production of the cell-associated form of the EPS Psl. Since cell-associated Psl is essential for attachment to surfaces, mutants lacking SiaD activity do not attach robustly to surfaces. Our findings reveal a novel mechanism for regulating production of an EPS important for colonization of infection sites.
KEYWORDS: Pseudomonas aeruginosa, biofilm formation, c-di-GMP
INTRODUCTION
Many bacteria exist as part of large multicellular aggregates called biofilms that are encased by a self-produced extracellular matrix (1–3). Pseudomonas aeruginosa is a model organism for studying biofilms in the laboratory and is an opportunistic pathogen that causes chronic infections that resist eradication despite intensive treatment (4–6). P. aeruginosa forms a biofilm, encased by matrix components, at some infection sites (7–9).
The P. aeruginosa biofilm matrix is primarily composed of extracellular DNA, proteins, including the large adhesion protein CdrA, and up to three exopolysaccharides (EPS): alginate, Pel, and Psl (10). CdrA increases aggregate stability by binding to itself, Pel, and Psl (11, 12). In nonmucoid strains, Pel and Psl serve as the primary matrix scaffolds (13–15). Psl is a neutral EPS composed of d-mannose, d-glucose, and l-rhamnose that promotes initial attachment to surfaces and protects the bacterium from various stressors (16–20). Psl and Pel exist in both cell-free and cell-associated forms (16, 21). The mechanism by which Psl and Pel associate with the cell is unknown.
In P. aeruginosa, the intracellular signaling molecule, cyclic diguanylate monophosphate (c-di-GMP), facilitates the transition of planktonic cells to the biofilm state (22, 23). C-di-GMP is synthesized by diguanylate cyclases (DGC) which contain a GG(D/E)EF domain and is degraded by phosphodiesterases harboring a HD-GYP or EAL domain (22, 24). When a cell contacts a surface, the Wsp and Pil-Chp surface sensing systems increase c-di-GMP which promotes the transition to biofilm growth (25, 26). High levels of c-di-GMP results in transcriptional repression of flagellar and pili motility genes and increases expression of the cdrAB, pel, and psl operons at the level of transcription and translation (23, 27). Production of Pel is also induced by c-di-GMP posttranslationally (28).
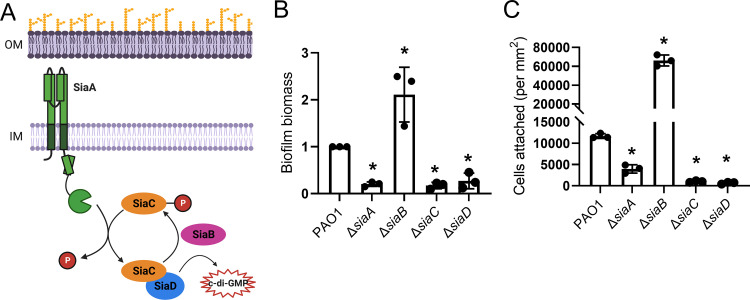
The siaABCD operon is also elevated under conditions of high c-di-GMP (23, 25). The siaABCD operon encodes a signal transduction complex that controls the activity of a DGC, SiaD (29). In response to an unknown environmental signal, the inner membrane-associated Ser/Thr phosphatase, SiaA, dephosphorylates SiaC. This allows SiaC to bind SiaD, promoting c-di-GMP synthesis. SiaB is a kinase that competitively binds SiaC and rephosphorylates it. Once rephosphorylated, SiaC will no longer bind SiaD, and DGC activity is shut off (29) (Fig. 1A).
FIG 1.
The Sia signaling network impacts attachment of P. aeruginosa to surfaces. (A) The Sia signaling network regulates SiaD diguanylate cyclase activity. In response to an unknown environmental signal, SiaA dephosphorylates SiaC. SiaC then binds SiaD, activating SiaD DGC activity. SiaB is a kinase that competitively binds SiaC and rephosphorylates it. Once rephosphorylated, SiaC will no longer bind SiaD, and DGC activity is shut off. Figure created in BioRender.com (29). (B) Static biofilm formation of sia mutant strains. Biofilm biomass produced by each strain was measured by crystal violet staining and normalized to wild type (PAO1). Presented as mean and standard deviation. N, 3 biological replicates, *P < 0.05. (C) Adherence of sia mutant strains to glass. Cells were incubated on a glass coverslip, rinsed and attached cells were immediately quantified by microscopy. Presented as mean and standard deviation. N, 3 biological replicates, *, P < 0.05.
The sia operon was first described as essential for cellular aggregation of P. aeruginosa when grown on the detergent sodium dodecyl sulfate (SDS) (30, 31). It was later determined that SiaD DGC activity is essential for cellular aggregation in response to both SDS and tellurite (TeO42−), therefore increasing P. aeruginosa survival in the presence of these stressors (30–33). Subsequent work showed that mutations in both siaA and siaD cause a decrease in psl, cdrAB, and cupA expression (31). SiaA and SiaD also affect the activity of the translational regulator RsmA, by regulating the levels of the small regulatory RNA rsmZ (32, 34). These studies show that the Sia signaling network responds to an environmental signal that stimulates P. aeruginosa aggregate formation through SiaD-mediated c-di-GMP synthesis.
A function for the Sia system in the absence of added stressors has not been reported. Here, we show that mutations in the sia operon influence planktonic production of both c-di-GMP and the cell-associated form of Psl. This results in Sia-dependent changes to surface attachment. Screening of additional DGC mutant strains revealed that only the Sia system has a significant impact on the planktonic levels of cell-associated Psl. Additionally, in a Sia-deficient background, cell-associated Psl levels are impacted even when the psl operon is expressed under the control of an inducible promoter. This suggests that the Sia system affects Psl distribution posttranslationally. Collectively, our results indicate that the Sia system contributes to both planktonic levels of c-di-GMP and cell-associated Psl.
RESULTS
The Sia system promotes the attachment of planktonic cells to a surface.
We initially sought to determine the impact of the Sia signaling network on P. aeruginosa biofilm formation. Mutations that prevent SiaD DGC activity, ΔsiaA, ΔsiaC, and ΔsiaD, impede biofilm formation, in the absence of external stressors. Whereas a ΔsiaB mutation constitutively activates SiaD and increases biofilm formation (Fig. 1B, Fig. S1A) (29, 35). The observed changes were not due to changes in growth and this phenotype could be partially rescued (Fig. S1BC).
We used a microscopic approach to gain insight into how the Sia system influences surface attachment. GFP expressing, midlog-phase planktonic cells (OD600nm = 0.5) were inoculated onto a glass coverslip and allowed to attach for 10 min. Unattached cells were washed away and attached cells were quantified by microscopy (Fig. 1C). Compared with the parental strain, ΔsiaA, ΔsiaC, and ΔsiaD mutant strains exhibited an attachment defect, while a ΔsiaB mutant strain showed an increase in attached cells. This phenotype could be also be partially restored by expressing the sia genes in trans. (Fig. S1D). These data demonstrate that the Sia system impacts the surface attachment of planktonic cells.
The Sia system regulates cell-associated Psl levels during planktonic growth.
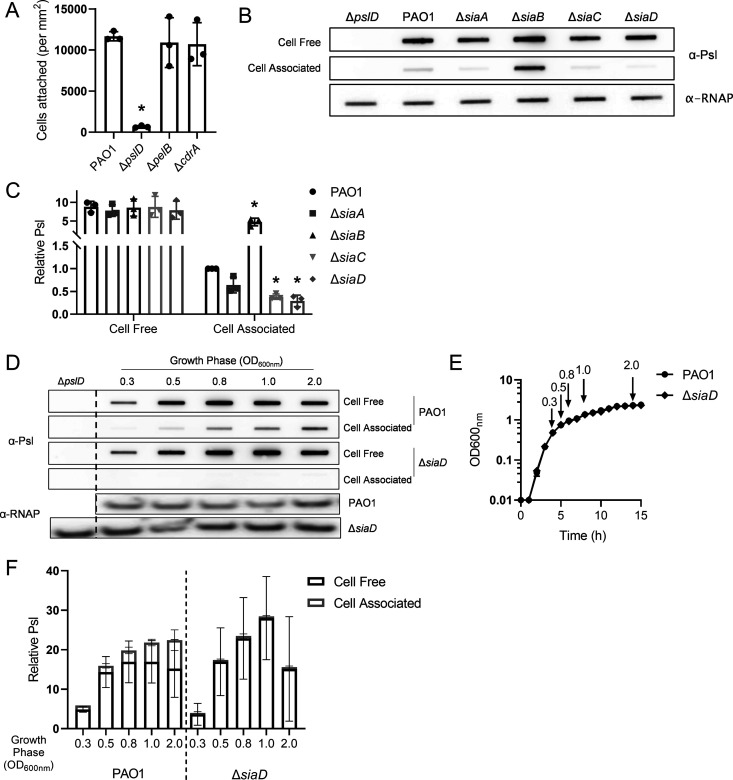
While expression of any of the major P. aeruginosa nonmucoid matrix components, Pel, Psl, or CdrA, has the potential to increase attachment, only Psl is produced during planktonic growth in the lab strain PAO1 (11, 13). As a result, only strains that cannot produce Psl exhibit impaired surface attachment. (Fig. 2A, Fig. S2A); (13, 18, 36).
FIG 2.
The Sia signaling network impacts the surface adherence of planktonic cells through control of cell-associated Psl. (A) Adherence of matrix mutant strains to glass. Cells were incubated on a glass coverslip, rinsed and attached cells were immediately quantified by microscopy. (B) Representative immunoblot for Psl produced by sia mutants, extracted from midlog planktonic cells (OD600nm = 0.5). RNAP served as a loading control. (C) Quantification of relative Psl production calculated using blots in 2B. Psl band intensity was normalized to RNAP levels and then compared to wild-type (PAO1) cell-associated Psl. (D) Representative immunoblot for Psl from PAO1 and ΔsiaD throughout planktonic growth. Negative-control samples (ΔpslD) were processed at OD600nm = 2. At each time point 1 × 109 cells were processed. RNAP served as a loading control. (E) Growth curve of strains evaluated in 2D, in Lennox Broth. The time points collected for 2D are marked on the curve. (F) Quantification of relative cell-free and cell-associated Psl production calculated using blots in 2D. Psl band intensity was normalized to RNAP levels and then compared to PAO1 at OD600nm = 0.3. Presented as mean and standard deviation. N, 3 biological replicates, *, P < 0.05.
We hypothesized that the Sia system regulates initial attachment by altering planktonic Psl production. To test this, we performed Psl immunoblots on both cell-free and cell-associated fractions. The amount of cell-free Psl made by these strains was similar. However, we observed that, compared to the wild-type strain, the ΔsiaC and ΔsiaD mutant strains have a significant decrease in cell-associated Psl levels (Fig. 2B, C; Fig. S2B, C). The ΔsiaA mutant strain also appeared to have decreased cell-associated Psl, but this was not statistically significant. Conversely, deleting siaB caused an increase in cell-associated Psl levels. Although there are changes in the levels of cell-associated Psl, only the ΔsiaB mutant strain shows a significant change in total Psl production (Fig. S2D). These results are consistent with the initial attachment data (Fig. 1C) and suggest that attachment differences between strains are due to the levels of cell-associated Psl.
To determine whether our observations are growth phase dependent, we measured cell-free and cell-associated Psl production throughout the growth curve (Fig. 2D to F). For this experiment, we compared wild type (PAO1) to ΔsiaD, since SiaD is the ultimate output of the system. There was no significant difference in the amount of cell-free Psl made between PAO1 and the ΔsiaD mutant strain at any time point (Fig. 2D, F). For PAO1, we observed that as cells enter stationary phase, the amount of cell-associated Psl gradually increases (Fig. 2D, F). However, we observed reduced levels of cell-associated Psl in the ΔsiaD mutant strain at every time point (Fig. 2D, F). These results suggest that the Sia system is important for maintaining levels of cell-associated Psl regardless of the growth phase of the cells. Depending on the nature of the sia mutation, this translates to reduced (ΔsiaA, ΔsiaC, ΔsiaD) or elevated (ΔsiaB) surface attachment phenotypes.
SiaD enzymatic activity is an important contributor to planktonic levels of c-di-GMP and Sia-dependent phenotypes.
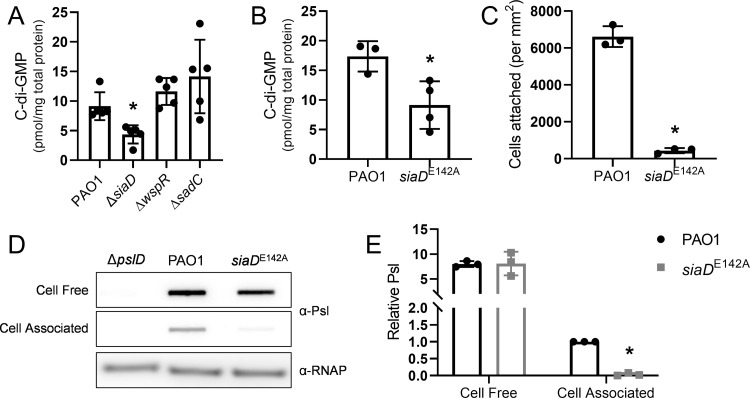
We hypothesized that the influence of the Sia system on cell-associated Psl could be through modulating intracellular levels of c-di-GMP. To directly test this hypothesis, we measured c-di-GMP levels of wild type and a ΔsiaD mutant during planktonic growth. Indeed, the ΔsiaD mutant has an approximately 2-fold decrease in c-di-GMP levels compared to wild type (Fig. 3A). This decrease can be rescued (Fig. S3A). Two other well-characterized DGCs in P. aeruginosa, WspR and SadC, were analyzed for their potential contribution to c-di-GMP pools in planktonically grown cells. We found that mutations in these DGCs did not significantly affect c-di-GMP levels. This suggests that SiaD is an important contributor to c-di-GMP in planktonic cells.
FIG 3.
The Sia system impacts levels of cell-associated Psl through SiaD diguanylate cyclase activity. (A) C-di-GMP levels of by diguanylate cyclase mutant strains. C-di-GMP was extracted from midlog planktonic cells (OD600nm = 0.5). Presented as mean and standard deviation. N = 5 biological replicates, *, P < 0.05. (B) C-di-GMP levels of the siaDE142A catalytic mutant strain. C-di-GMP was extracted from midlog planktonic cells (OD600nm = 0.5). Presented as mean and standard deviation. N = 3 biological replicates, *, P < 0.05. (C) Adherence of the siaDE142A mutant strain to glass. Cells were incubated on a glass coverslip, rinsed and attached cells were immediately quantified by microscopy. Presented as mean and standard deviation. N, 3 biological replicates, *P < 0.05. (D) Representative immunoblot for Psl produced by the siaDE142A mutant, extracted from midlog, planktonic cells (OD600nm = 0.5). RNAP served as a loading control. (E) Quantification of relative Psl production calculated using blots in 3D. Psl band intensity was normalized to RNAP levels and then compared to wild-type (PAO1) cell-associated Psl. N, 3 biological replicates, *, P < 0.05.
To further evaluate the role of SiaD DGC activity during planktonic growth, we made a targeted mutation in the GGEEF domain of SiaD and introduced this allele onto the chromosome. Western blot analysis showed that the E142A mutation did not affect steady-state levels of SiaD (Fig. S3B). We observed that the catalytic siaD mutant strain, like ΔsiaD, has a 2-fold decrease in c-di-GMP levels compared to wild type (Fig. 3B; Fig. S3A). Similarly, cells expressing the catalytically defective SiaD demonstrate an attachment defect (Fig. 3C, Fig. S3C) and a decrease in levels of cell-associated Psl (Fig. 3D and E; Fig. S3D, E). Taken together, these data support that SiaD DGC enzymatic activity is responsible regulating cell-association of Psl and initial attachment.
Planktonic levels of cell-associated Psl are not impacted by other DGCs.
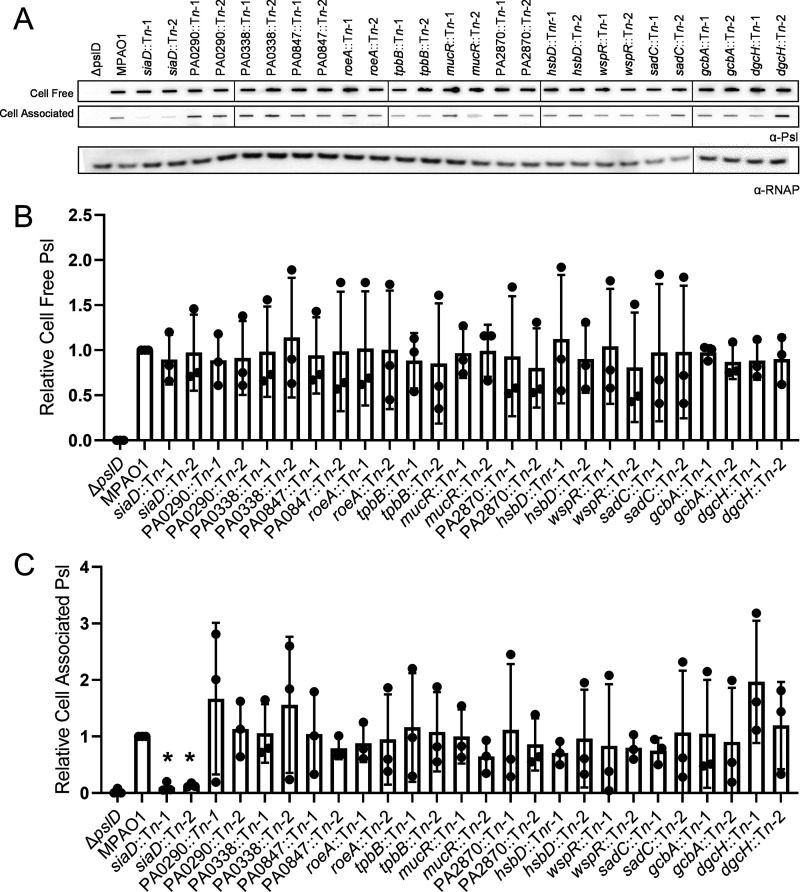
It was not known whether other DGCs contribute to Psl production during planktonic growth. Therefore, we tested a panel of mutant strains from the PAO1 transposon mutant library (37, 38). We chose 13 DGCs that have been shown to be enzymatically active (29, 39–42). There was no significant change in cell-free Psl levels in any of the transposon mutants (Fig. 4A and B). There was a significant decrease in cell-associated Psl only in the siaD transposon mutants (Fig. 4A, C). These data suggest that the Sia system specifically impacts levels of cell-associated Psl during planktonic growth.
FIG 4.
Planktonic levels of cell-associated Psl are not impacted by DGCs than SiaD. (A) Representative immunoblot for Psl produced by DGC transposon insertion mutants, extracted from midlog planktonic cells (OD600nm = 0.5). RNAP served as a loading control. (B) Quantification of cell-free and (C) cell-associated Psl production calculated using blots in 4A. Psl band intensity was normalized to RNAP levels and then compared to the wild-type control (MPAO1). Presented as mean and standard deviation. N, 3 biological replicates, *, P < 0.05.
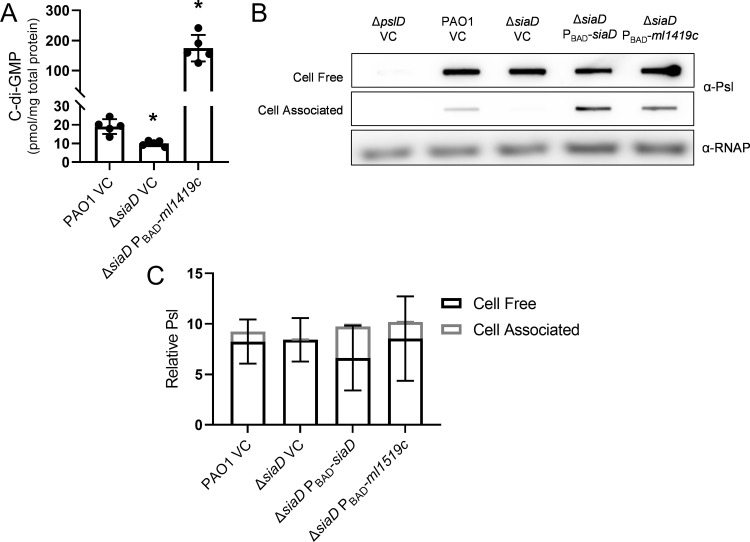
Although SiaD produced c-di-GMP is responsible for these phenotypes during planktonic growth, we hypothesized that artificially elevating c-di-GMP levels could restore cell-associated Psl levels in sia mutant strains. We already observed that strains with high c-di-GMP, ΔsiaB (29) and ΔsiaD PBAD-siaD (Fig. S3A), demonstrate an increase in cell-associated Psl levels (Fig. 2B, C, Fig. S2B, C). However, in both strains c-di-GMP increases are due to increased SiaD activity. We next determined if cell-associated levels of Psl could be rescued by expressing a nonnative, heterologously expressed DGC. When we induced expression of the Mycobacterium leprae DGC Ml1419c we observed 10-fold higher c-di-GMP levels than wild type (Fig. 5A) and a corresponding increase in cell-associated, but not cell-free Psl (Fig. 5B and C) (43). This suggests that while SiaD is responsible for levels of cell-associated Psl during planktonic growth, cell-associated Psl can be restored in a siaD mutant strain by artificially elevating intracellular c-di-GMP levels.
FIG 5.
Overproduction of c-di-GMP causes an increase in cell-associated Psl levels. (A) C-di-GMP levels of cells overexpressing the heterologous cyclase Ml1419c in P. aeruginosa. C-di-GMP was extracted from midlog planktonic cells (OD600nm = 0.5). Presented as mean and standard deviation. N, 5 biological replicates, *, P < 0.05. (B) Representative immunoblot for Psl produced by cells overexpressing siaD or ml1419c, extracted from midlog, planktonic cells (OD600nm = 0.5). RNAP served as a loading control. (C) Quantification of relative Psl production calculated using blots in 5B. Psl band intensity was normalized to RNAP levels and then compared to the wild-type vector control (PAO1 VC) cell-associated Psl. Presented as mean and standard deviation. N, 3 biological replicates, *, P < 0.05, VC=vector control.
The Sia system’s impact on cell-associated Psl levels appear to occur posttranslationally.
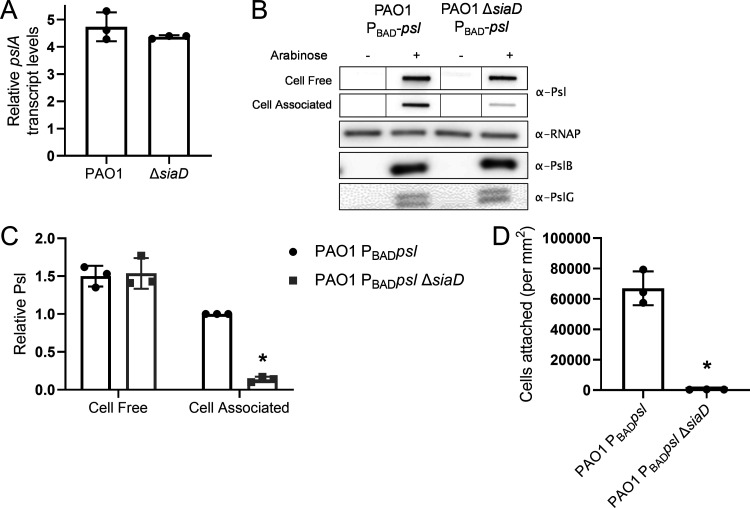
We hypothesized that the effect of SiaD on cell-associated Psl levels is posttranscriptional since a majority of Psl (the cell-free fraction) is still produced by a ΔsiaD mutant strain (Fig. 2F, G, Fig. S2D). However, since transcription of the psl operon is regulated by c-di-GMP (27), we wanted to determine whether uncoupling c-di-GMP-mediated transcriptional control of the psl operon negates the effect of siaD on Psl distribution. We used qRT-PCR to measure pslA transcript levels during exponential growth and observed no difference between PAO1 and a ΔsiaD mutant (Fig. 6A). We also utilized a strain where the native, c-di-GMP regulated promoter has been replaced with an arabinose inducible promoter, on the chromosome (PBAD-ara) (20). In this strain, psl transcription is no longer regulated by c-di-GMP. We found that a ΔsiaD mutation in this background still results in a decrease in levels of cell-associated Psl (Fig. 6B and C). This strain also demonstrated the expected corresponding decrease in initial attachment (Fig. 6D). These data suggest that the effect of the Sia system on cell-association of Psl occurs posttranscriptionally.
FIG 6.
The Sia system regulates cell-associated Psl production posttranslationally. (A) q-RT-PCR of pslA-containing transcripts relative to rpoD transcript levels. RNA was extracted from midlog planktonic cultures (OD600nm = 0.5). (B) Expression of the psl operon was driven by the PBAD promoter to uncouple any potential transcriptional control of the Psl promoter. These strains were analyzed for potential effects on Psl protein translation. Representative immunoblots are shown for Psl, PslB, and PslG. RNAP served as a loading control. (C) Quantification of relative Psl production calculated using blots in 6B. Psl band intensity was normalized to RNAP levels and then compared to wild-type (PAO1 PBADpsl) cell-associated Psl. (D) Adherence of psl overexpression strains to glass. Cells were incubated on a glass coverslip, rinsed and attached cells were immediately quantified by microscopy. Presented as mean and standard deviation. N, 3 biological replicates, *, P < 0.05.
Psl translation is repressed by the mRNA-binding regulatory protein RsmA (44). Past work found that a ΔsiaD mutant had lower levels of the small RNA rsmZ, one of the regulators of RsmA activity (32). We hypothesized that decreased levels of rsmZ could lead to an increase in RsmA binding to the psl transcript and cause a decrease in overall Psl protein translation. However, using the same PBAD-psl strain described above, we probed for two of the Psl proteins, PslB and PslG, and found no difference in the amount of Psl protein made between wild type and the ΔsiaD mutant (Fig. 6B). Together, these data suggest that the Sia system affects cell-association of Psl through a posttranslational mechanism, independent of the Rsm pathway effects on Psl.
DISCUSSION
C-di-GMP is a major determinant that promotes P. aeruginosa biofilm formation. The results of our study reveal that c-di-GMP produced by the Sia signaling network plays a key role in P. aeruginosa biology in planktonic populations. We observed that SiaD DGC activity is responsible for regulating cell-associated Psl produced during planktonic growth, and this affects the ability of cells to attach to a surface. SiaD was the only DGC of the 13 examined in this study that showed evidence of affecting Psl cell-association, suggesting that this is a specific role of the Sia system. Our data also show that cell-free Psl is the major form of Psl detected in planktonic cultures. We further demonstrate that, at later stages of planktonic growth, the relative amount of cell-associated Psl increases in a Sia-dependent manner. While specific to SiaD DGC activity during planktonic growth, cell-associated Psl in a ΔsiaD mutant strain can be restored by artificially elevating c-di-GMP. We also show that the impact of the Sia system on cell-associated Psl does not involve either transcriptional or translational control of the Psl biosynthetic machinery. These findings reveal a role for the Sia signaling network and c-di-GMP in planktonic populations and point to a novel mechanism for regulating the association of Psl to the cell.
While the output of the Sia system, c-di-GMP, has been well established, the environmental cues that stimulate the system remain a mystery. The SiaA periplasmic domain has predicted structural homology to a Cache domain (data not shown) (45). Cache domains can bind a diversity of ligands, though these ligands are typically small organic molecules, including amino acids, purines, monosaccharides, and 4-carbon dicarboxylates (46, 47). Since the Sia signaling network is essential for the SDS induced aggregation response it has been largely assumed that SiaA binds to a product of that response, e.g., a molecule released upon cell membrane permeabilization or SDS itself. However, since we show that the Sia signaling network is active during planktonic growth in the absence of stressors, we suspect that SiaA binds to either a self-produced molecule commonly released by P. aeruginosa during growth, or that low levels of a stressor are present in our liquid culture growth conditions. Since it was previously shown that exposure to exogenous, cell-free Psl causes a SiaD dependent increase in c-di-GMP levels it is also possible that some chemical feature of Psl or envelope stress caused by Psl is sensed by the Sia system (48).
The mechanism by which the Sia system controls cell-associated Psl is unclear. One hypothesis is that a minimum threshold level of cellular c-di-GMP is required to produce cell-associated Psl. Thus, the slightly lower c-di-GMP levels present in the siaD mutant strain during midlog planktonic growth might be insufficient to promote whatever mechanism is necessary to produce the cell-associated form of Psl. It appears that overexpression of DGC activity can rescue the Sia defect, but this is observed at high levels of c-di-GMP (Fig. 5A). However, the Sia system appears to be fairly specific in its ability to control cell-associated Psl. No other DGC mutant strain tested influenced levels of cell-associated Psl (Fig. 4A, C). It is possible that other DGC may play a minor role in this phenotype; however, when SiaD is present this effect is masked and so would not have been detected in this screen. In addition, the ΔsiaD mutant strain exhibited a defect in producing cell-associated Psl even in stationary phase (Fig. 2D, F), where other DGCs become active and c-d-GMP levels are known to rise, which does not support this hypothesis. An alternative hypothesis is that the Sia system directly interfaces with the mechanism responsible for cell-association of Psl. One possibility here is that SiaD interacts with the Psl biosynthetic machinery, influencing Psl synthesis and potentially the activity of the Psl hydrolase, PslG. However, none of the Psl biosynthetic proteins are known to bind c-di-GMP; however, this type of regulation has been observed for both Pel and alginate (28, 49). This would allow SiaD-generated c-di-GMP to act as an effector for these processes. An alternative hypothesis is that the Sia system may regulate the expression and/or function of an outer membrane capable of interacting with Psl. Further experimentation is required to distinguish between these different hypotheses.
Finally, this work demonstrates the biological significance of the two different forms of Psl. Cell-free Psl is known to bind complement components and impact other features of innate immunity (17, 18). Here, we show that cell-associated Psl promotes surface attachment and likely promotes aggregate formation when grown in the presence of certain stressors like SDS. This might allow the cell to use Psl as an adhesive and under the right conditions promote a protective biofilm lifestyle. A significant remaining question pertains to the molecular mechanism by which Psl is cell-associated. These findings could be significantly relevant to human disease, where activity of the Sia system and the resulting cell-associated Psl might assist in establishing biofilm infections.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains and plasmids used are listed in Table S1. E. coli and P. aeruginosa were maintained on Lennox Broth agar and grown at 37°C. For all experiments P. aeruginosa was grown in Lennox Broth. For experiments, overnight cultures were diluted to OD600nm = 0.05 and grown at 37°C with shaking to exponential phase (OD600nm = 0.5), unless otherwise stated. Plasmids were maintained with 100 μg/mL gentamicin for P. aeruginosa and 10 μg/mL gentamicin, 50 μg/mL kanamycin, or 100 μg carbenicillin for E. coli. One percent arabinose was used to induce express of the PBAD promoter.
Strain construction.
Markerless, nonpolar deletions were made using allelic exchange as previously described (50). Regions flanking the genes were amplified and sewn together using the primers listed in Table S2 and cloned into pDONRPEX18GmGW. The resulting plasmids were transformed into DH5a and verified by sequencing. The plasmid was then mobilized into P. aeruginosa using the helper plasmid pRK2013. Transconjugants were selected for on VBMM agar containing 60 μg/mL gentamicin. Counter selection was performed on NSLB containing 10% sucrose and Pseudomonas Isolation Agar. Colonies were screened by colony PCR and confirmed by sequencing. For overexpression constructs, genes were amplified using the primers listed in Table S2 and cloned into pJN105. Plasmids were confirmed by sequencing.
Growth curves.
Overnight cultures were diluted to OD600nm = 0.01 in 50 mL Lennox Broth. Cultures were incubated at 37°C with shaking at 225 rpm and the OD600nm was measured every hour for 15 h using a ThermoSpectrophic Spectrophotometer (Thermo Scientific). For hyperaggregating strains, including ΔsiaB, the sample taken was first passed through a 22-guage syringe to break up aggregates measuring the OD600nm.
Initial attachment of P. aeruginosa to glass.
GFP expressing, exponential-phase cells were diluted 1:10 and 100 μL of the mixture was incubated on a glass coverslip for 10 min at room temperature. The coverslip was washed three times with sterile distilled water, placed on a slide, and cells were visualized on a Zeiss LSM 800 confocal laser scanning microscope. Images were analyzed with Volocity software (Quorum Technologies).
Western blotting.
P. aeruginosa cultures were pelleted, resuspended in phosphate-buffered saline (NaPO4 pH 7.4, 150 mM NaCl) and sonicated on ice. The lysate was pelleted, and the concentration of protein in the supernatant was quantified using a Qubit 3.0 fluorometer (Invitrogen). Samples were normalized by protein concentration and were boiled in Laemmli buffer. For SDS-PAGE, proteins were separated on a 4–20% tris glycine gel and transferred to a 0.22 μm nitrocellulose membrane for immunoblotting. Membranes were incubated with the relevant primary antisera: anti-LasB diluted 1:6,000 (raised against AEAGGPGGNQKIGKC; Genscript), anti-6xhis diluted 1:6,000 (Lifetein; LT0426), anti-PslB diluted 1:5,000 (raised against CAPDISVDYALMERS; Genscript), or anti-PslG diluted 1:6,000 (51). All were diluted in 1% milk in tris buffered saline (50 mM tris pH 7.6, 150 mM NaCl) with 0.1% Tween 20. HRP-conjugated goat anti-rabbit (Invitogen; 32460) diluted 1:20,000 was used as the secondary antibody. Super Signal West Pico chemiluminescent substrate was used to detect signal (Thermo Fisher) and signal was visualized using an Azure Biosystems c600 imager.
Crude exopolysaccharide extractions and Psl immunoblots.
At the desired OD600nm, cells were spun down at 16,000 × g for 5 min. The supernatant contained the cell-free fraction. For the Psl growth curve, 109 cells were processed at each time point. The pellet was resuspended in an equivalent volume of 0.5 M EDTA (pH 8.0; Promega), boiled at 95°C for 20 min, with occasional vortexing, and spun down again. The supernatant contained the cell-associated fraction (Fig. S4). Both fractions were treated with final concentration of 200 μg/mL proteinase K (Sigma). For the RNA polymerase loading control, proteins were isolated from the resulting pellet. The pellet was resuspended in phosphate-buffered saline (NaPO4 pH 7.4, 150 mM NaCl) and sonicated on ice. The lysate was pelleted, and the supernatant was boiled with XT reducing buffer (Bio-Rad). Crude exopolysaccharide samples were applied to a 0.45 μm nitrocellulose membrane using a Bio-dot vacuum manifold (Bio-Rad; 1706542). Anti-Psl antiserum was diluted 1:3,000 in 1% milk in tris buffered saline (50 mM tris pH 7.6, 150 mM NaCl) with 0.1% Tween 20 (TBST) (52). HRP-conjugated anti-human (Abcam; ab98624) diluted to 1:6,000 was used as the secondary antibody. For RNA polymerase immunoblots proteins were separated on a 3–8% XT Tris-acetate gel and transferred to a PDVF membrane for immunoblotting. Anti-RNA polymerase antiserum was diluted 1:6,000 in 1% milk in TBST (Abcam; ab191598). HRP conjugated goat anti-rabbit (Invitogen; 32460) diluted 1:20,000 was used as the secondary antibody. Super Signal West Pico chemiluminescent substrate was used to detect signal (Thermo Fisher) and signal was visualized using an Azure Biosystems c600 imager. Densitometry was performed on cell-free and cell-associated samples applied to the same membrane and imagined using the same exposure, so that samples can be directly compared. Densitometry measurements were made using ImageJ. Psl band intensity was normalized to RNAP band intensity. Immunoblots which are joined together from noncontiguous portions of the same blot or combined from multiple blots (not all samples fit on a single membrane) are delineated with black lines. When samples needed to be spread over multiple membranes, controls samples were rerun with the additional samples to used for normalization.
C-di-GMP quantification.
C-di-GMP was extracted as previously described (25, 48) using 2-chloro AMP as an internal standard. Briefly, all experiments were performed on independent cultures for a total of 5 biological replicates. C-di-GMP was extracted from pelleted cells by incubating with 70% perchloric acid on ice for 1 h. The supernatant was retained and neutralized using potassium bicarbonate. Liquid chromatography MS/MS measurements were performed using an Acuity UPLC with a Synergi 4 μ Hydro RP 80A column and a C18 Guard Cartridge (Phenomenex) on a Premier XL triple-quadrupole electrospray mass spectrometer (Waters). The m/z 691 > 152 transition was used for c-di-GMP and 382 > 170 for 2-chloro-AMP. The cone voltages and collision energies were 40 V/30 eV and 35 V/20 eV, respectively. Fifteen μL of each sample were injected and the ratio of area under the curve of the c-di-GMP channel signal (retention time = 1.6 min) was divided by the area under the curve of the 2-chloroAMP signal (retention time = 2.1 min). A standard curve of 0 nM to 100 nM c-di-GMP containing 2-chloroAMP was used to quantify c-di-GMP for all samples. All c-di-GMP quantifications were normalized to protein concentration.
Crystal violet static biofilm formation assay.
200 μL of exponential-phase culture was added to wells of an untreated microtiter dish (Fisher Scientific 260860) and incubated for 20 h at 37°C. Biofilms were washed with sterile distilled water three times to remove unattached cells. Biofilms were stained with 0.1% crystal violet, solubilized with 30% acetic acid and biomass was measured (A595nm) as described previously (13). For each experiment, data were normalized to PAO1 or the PAO1 vector control (VC) as applicable.
RNA purification and quantitative real-time PCR.
RNA was extracted as previously described (13). Quantitative real-time PCR was performed with SsoAdvanced Universal SYBR green Supermix in a CFX384 touch deep well thermocycler (Bio-Rad). Primers used are listed in Table S2. pslA transcript levels are normalized to rpoD transcript levels.
Statistical analysis.
GraphPad Prism 9 was used for all statistical analyses. All experiments were analyzed using a Student's t test and a P-value <0.05 was considered statistically significant.
Footnotes
Supplemental material is available online only.
Contributor Information
Matthew R. Parsek, Email: parsem@uw.edu.
George O'Toole, Geisel School of Medicine at Dartmouth.
REFERENCES
- 1.Hall-Stoodley L, Costerton JW, Stoodley P. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol 2:95–108. 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 2.Lebeaux D, Chauhan A, Rendueles O, Beloin C. 2013. From in vitro to in vivo models of bacterial biofilm-related infections. Pathogens 2:288–356. 10.3390/pathogens2020288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–13231322. 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 4.Høiby N, Ciofu O, Bjarnsholt T. 2010. Pseudomonas aeruginosa biofilms in cystic fibrosis. Future Microbiol 5:1663–1674. 10.2217/fmb.10.125. [DOI] [PubMed] [Google Scholar]
- 5.Serra R, Grande R, Butrico L, Rossi A, Settimio UF, Caroleo B, Amato B, Gallelli L, de Franciscis S. 2015. Chronic wound infections: the role of Pseudomonas aeruginosa and Staphylococcus aureus. Expert Rev Anti Infect Ther 13:605–613. 10.1586/14787210.2015.1023291. [DOI] [PubMed] [Google Scholar]
- 6.Roy S, Elgharably H, Sinha M, Ganesh K, Chaney S, Mann E, Miller C, Khanna S, Bergdall VK, Powell HM, Cook CH, Gordillo GM, Wozniak DJ, Sen CK. 2014. Mixed-species biofilm compromises wound healing by disrupting epidermal barrier function. J Pathol 233:331–343. 10.1002/path.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parsek MR, Singh PK. 2003. Bacterial biofilms: an emerging link to disease pathogenesis. Annu Rev Microbiol 57:677–701. 10.1146/annurev.micro.57.030502.090720. [DOI] [PubMed] [Google Scholar]
- 8.Jennings LK, Dreifus JE, Reichhardt C, Storek KM, Secor PR, Wozniak DJ, Hisert KB, Parsek MR. 2021. Pseudomonas aeruginosa aggregates in cystic fibrosis sputum produce exopolysaccharides that likely impede current therapies. Cell Rep 34:108782. 10.1016/j.celrep.2021.108782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirketerp-Møller K, Jensen P, Fazli M, Madsen KG, Pedersen J, Moser C, Tolker-Nielsen T, Høiby N, Givskov M, Bjarnsholt T. 2008. Distribution, organization, and ecology of bacteria in chronic wounds. J Clin Microbiol 46:2717–2722. 10.1128/JCM.00501-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mann EE, Wozniak DJ. 2012. Pseudomonas biofilm matrix composition and niche biology. FEMS Microbiol Rev 36:893–916. 10.1111/j.1574-6976.2011.00322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borlee BR, Goldman AD, Murakami K, Samudrala R, Wozniak DJ, Parsek MR. 2010. Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol Microbiol 75:827–842. 10.1111/j.1365-2958.2009.06991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reichhardt C, Jacobs HM, Matwichuk M, Wong C, Wozniak DJ, Parsek MR. 2020. The versatile pseudomonas aeruginosa biofilm matrix protein CdrA promotes aggregation through different extracellular exopolysaccharide interactions. J Bacteriol 202:1–9. 10.1128/JB.00216-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colvin KM, Irie Y, Tart CS, Urbano R, Whitney JC, Ryder C, Howell PL, Wozniak DJ, Parsek MR. 2012. The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environ Microbiol 14:1913–1928. 10.1111/j.1462-2920.2011.02657.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghafoor A, Hay ID, Rehm BHA. 2011. Role of exopolysaccharides in Pseudomonas aeruginosa biofilm formation and architecture. Appl Environ Microbiol 77:5238–5246. 10.1128/AEM.00637-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma L, Conover M, Lu H, Parsek MR, Bayles K, Wozniak DJ. 2009. Assembly and development of the Pseudomonas aeruginosa biofilm matrix. PLoS Pathog 5:e1000354. 10.1371/journal.ppat.1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Byrd MS, Sadovskaya I, Vinogradov E, Lu H, Sprinkle AB, Richardson SH, Ma L, Ralston B, Parsek MR, Anderson EM, Lam JS, Wozniak DJ. 2009. Genetic and biochemical analyses of the Pseudomonas aeruginosa Psl exopolysaccharide reveal overlapping roles for polysaccharide synthesis enzymes in Psl and LPS production. Mol Microbiol 176:139–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Byrd MS, Pang B, Mishra M, Edward Swords W, Wozniak DJ. 2010. The Pseudomonas aeruginosa exopolysaccharide Psl facilitates surface adherence and NF-κB activation in A549 cells. mBio 1:3–6. 10.1128/mBio.00140-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mishra M, Byrd MS, Sergeant S, Azad AK, Parsek MR, McPhail L, Schlesinger LS, Wozniak DJ. 2012. Pseudomonas aeruginosa Psl polysaccharide reduces neutrophil phagocytosis and the oxidative response by limiting complement-mediated opsonization. Cell Microbiol 14:95–106. 10.1111/j.1462-5822.2011.01704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao K, Tseng BS, Beckerman B, Jin F, Gibiansky ML, Harrison JJ, Luijten E, Parsek MR, Wong GCL. 2013. Psl trails guide exploration and microcolony formation in early P aeruginosa biofilms. Nature 497:388–391. 10.1038/nature12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma L, Jackson KD, Landry RM, Parsek MR, Wozniak DJ. 2006. Analysis of Pseudomonas aeruginosa conditional psl variants reveals roles for the psl polysaccharide in adhesion and maintaining biofilm structure postattachment. J Bacteriol 188:8213–8221. 10.1128/JB.01202-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jennings LK, Storek KM, Ledvina HE, Coulon C, Marmont LS, Sadovskaya I, Secor PR, Tseng BS, Scian M, Filloux A, Wozniak DJ, Howell PL, Parsek MR. 2015. Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix. Proc Natl Acad Sci USA 112:11353–11358. 10.1073/pnas.1503058112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simm R, Morr M, Kader A, Nimtz M, Römling U. 2004. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessibility to motility. Mol Microbiol 53:1123–1134. 10.1111/j.1365-2958.2004.04206.x. [DOI] [PubMed] [Google Scholar]
- 23.Starkey M, Hickman JH, Ma L, Zhang N, de Long S, Hinz A, Palacios S, Manoil C, Kirisits MJ, Starner TD, Wozniak DJ, Harwood CS, Parsek MR. 2009. Pseudomonas aeruginosa Rugose small-colony variants have adaptations that likely promote persistence in the cystic fibrosis lung. J Bacteriol 191:3492–3503. 10.1128/JB.00119-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmidt AJ, Ryjenkov DA, Gomelsky M. 2005. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J Bacteriol 187:4774–4781. 10.1128/JB.187.14.4774-4781.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hickman JW, Tifrea DF, Harwood CS. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci USA 102:14422–14427. 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo Y, Zhao K, Baker AE, Kuchma SL, Coggan KA, Wolfgang MC, Wong GCL, O’Toole GA. 2015. A hierarchical cascade of second messengers regulates Pseudomonas aeruginosa surface behaviors. mBio 6:1–11. 10.1128/mBio.02456-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baraquet C, Harwood CS. 2016. FleQ DNA binding consensus sequence revealed by studies of FleQ-dependent regulation of biofilm gene expression in Pseudomonas aeruginosa. J Bacteriol 198:178–186. 10.1128/JB.00539-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee VT, Matewish JM, Kessler JL, Hyodo M, Hayakawa Y, Lory S. 2007. A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol Microbiol 65:1474–1484. 10.1111/j.1365-2958.2007.05879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen G, Gan J, Yang C, Zuo Y, Peng J, Li M, Huo W, Xie Y, Zhang Y, Wang T, Den X, Liang H. 2020. The SiaA/B/C/D signaling network regulates biofilm formation in Pseudomonas aeruginosa. EMBO J 39:160–161. 10.15252/embj.2020105997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klebensberger J, Rui O, Fritz E, Schink B, Philipp B. 2006. Cell aggregation of Pseudomonas aeruginosa strain PAO1 as an energy-dependent stress response during growth with sodium dodecyl sulfate. Arch Microbiol 185:417–427. 10.1007/s00203-006-0111-y. [DOI] [PubMed] [Google Scholar]
- 31.Klebensberger J, Birkenmaier A, Geffers R, Kjelleberg S, Philipp B. 2009. SiaA and SiaD are essential for inducing autoaggregation as a specific response to detergent stress in Pseudomonas aeruginosa. Environ Microbiol 11:3073–3086. 10.1111/j.1462-2920.2009.02012.x. [DOI] [PubMed] [Google Scholar]
- 32.Colley B, Dederer V, Carnell M, Kjelleberg S, Rice SA, Klebensberger J. 2016. SiaA/D interconnects c-di-GMP and RsmA signaling to coordinate cellular aggregation of Pseudomonas aeruginosa in response to environmental conditions. Front Microbiol 7:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chua SL, Sivakumar K, Rybtke M, Yuan M, Andersen JB, Nielsen TE, Givskov M, Tolker-Nielsen T, Cao B, Kjelleberg S, Yang L. 2015. C-di-GMP regulates Pseudomonas aeruginosa stress response to tellurite during both planktonic and biofilm modes of growth. Sci Rep 5:10052. 10.1038/srep10052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heurlier K, Williams F, Heeb S, Dormond C, Pessi G, Singer D, Cámara M, Williams P, Haas D. 2004. Positive control of swarming, rhamnolipid synthesis, and lipase production by the posttranscriptional RsmA/RsmZ system in Pseudomonas aeruginosa PAO1. J Bacteriol 186:2936–2945. 10.1128/JB.186.10.2936-2945.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poh WH, Lin J, Colley B, Muller N, Goh BC, Schleheck D, el Sahili A, Marquardt A, Liang Y, Kjelleberg S, Lescar J, Rice SA, Klebensberger J. 2020. The SiaABC threonine phosphorylation pathway controls biofilm formation in response to carbon availability in Pseudomonas aeruginosa. PLoS One 15:e0241019. 10.1371/journal.pone.0241019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vasseur P, Vallet-Gely I, Soscia C, Genin S, Filloux A. 2005. The pel genes of the Pseudomonas aeruginosa PAK strain are involved at early and late stages of biofilm formation. Microbiology (Reading) 151:985–997. 10.1099/mic.0.27410-0. [DOI] [PubMed] [Google Scholar]
- 37.Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, Will O, Kaul R, Raymond C, Levy R, Chun-Rong L, Guenthner D, Bovee D, Olson Mv, Manoil C. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc Natl Acad Sci USA 100:14339–14344. 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Held K, Ramage E, Jacobs M, Gallagher L, Manoil C. 2012. Sequence-verified two-allele transposon mutant library for Pseudomonas aeruginosa PAO1. J Bacteriol 194:6387–6389. 10.1128/JB.01479-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kulasakara HD, Lee V, Brencic A, Liberati N, Urbach J, Miyata S, Lee DG, Neely AN, Hyodo M, Hayakawa Y, Ausubel FM, Lory S. 2006. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3†-5†)-cyclic-GMP in virulence. Proc Natl Acad Sci USA 103:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bhasme P, Wei Q, Xu A, Naqvi STA, Wang D, Ma LZ. 2020. Evaluation and characterization of the predicted diguanylate cyclase-encoding genes in Pseudomonas aeruginosa. Microbiology Open 9:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ueda A, Wood TK. 2009. Connecting quorum sensing, c-di-GMP, pel polysaccharide, and biofilm formation in Pseudomonas aeruginosa through tyrosine phosphatase TpbA (PA3885). PLoS Pathog 5:e1000483. 10.1371/journal.ppat.1000483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De N, Pirruccello M, Krasteva PV, Bae N, Raghavan RV, Sondermann H. 2008. Phosphorylation-independent regulation of the diguanylate cyclase WspR. PLoS Biol 6:e67. 10.1371/journal.pbio.0060067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rotcheewaphan S, Belisle JT, Webb KJ, Kim HJ, Spencer JS, Borlee BR. 2016. Diguanylate cyclase activity of the Mycobacterium leprae T cell antigen ML1419c. Microbiology (Reading) 162:1651–1661. 10.1099/mic.0.000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Irie Y, Starkey M, Edwards AN, Wozniak DJ, Romeo T, Parsek MR. 2010. Pseudomonas aeruginosa biofilm matrix polysaccharide Psl is regulated transcriptionally by RpoS and post-transcriptionally by RsmA. Mol Microbiol 78:158–172. 10.1111/j.1365-2958.2010.07320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng W, Zhang C, Li Y, Pearce R, Bell EW, Zhang Y. 2021. Folding non-homologous proteins by coupling deep-learning contact maps with I-TASSER assembly simulations. Cell Rep Methods 1:100014. 10.1016/j.crmeth.2021.100014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ortega Á, Zhulin IB, Krell T. 2017. Sensory repertoire of bacterial chemoreceptors. Microbiol Mol Biol Rev 81:1–28. 10.1128/MMBR.00033-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Upadhyay AA, Fleetwood AD, Adebali O, Finn RD, Zhulin IB. 2016. Cache domains that are homologous to, but different from PAS domains comprise the largest superfamily of extracellular sensors in prokaryotes. PLoS Comput Biol 12:e1004862–21.. 10.1371/journal.pcbi.1004862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Irie Y, Borlee BR, O'Connor JR, Hill PJ, Harwood CS, Wozniak DJ, Parsek MR. 2012. Self-produced exopolysaccharide is a signal that stimulates biofilm formation in Pseudomonas aeruginosa. Proc Natl Acad Sci USA 109:20632–20636. 10.1073/pnas.1217993109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Merighi M, Lee VT, Hyodo M, Hayakawa Y, Lory S. 2007. The second messenger bis-(3′-5′)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol Microbiol 65:876–895. 10.1111/j.1365-2958.2007.05817.x. [DOI] [PubMed] [Google Scholar]
- 50.Hmelo LR, Borlee BR, Almblad H, Love ME, Trevor E, Tseng BS, Lin C, Irie Y, Storek KM, Jane J, Siehnel RJ, Howell PL, Singh PK, Tolker-Nielsen T, Parsek MR, Schweizer HP, Harrison JJ. 2015. Precision-engineering the Pseudomonas aeruginosa genome with two-step allelic exchange. Nat Protoc 10:1820–1841. 10.1038/nprot.2015.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baker P, Whitfield GB, Hill PJ, Little DJ, Pestrak MJ, Robinson H, Wozniak DJ, Howell PL. 2015. Characterization of the Pseudomonas aeruginosa glycoside hydrolase PslG reveals that its levels are critical for Psl polysaccharide biosynthesis and biofilm formation. J Biol Chem 290:28374–28387. 10.1074/jbc.M115.674929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.DiGiandomenico A, Warrener P, Hamilton M, Guillard S, Ravn P, Minter R, Camara MM, Venkatraman V, MacGill RS, Lin J, Wang Q, Keller AE, Bonnell JC, Tomich M, Jermutus L, McCarthy MP, Melnick DA, Suzich JAA, Stover CK. 2012. Identification of broadly protective human antibodies to Pseudomonas aeruginosa exopolysaccharide Psl by phenotypic screening. J Exp Med 209:1273–1287. 10.1084/jem.20120033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 and S2 and Fig. S1 to S4. Download jb.00335-22-s0001.pdf, PDF file, 1.1 MB (1.1MB, pdf)