Summary
To investigate B-cell differentiation and maturation occurring in the germinal center (GC) using in vitro culture systems, key factors and interactions of the GC reaction need to be accurately simulated. This study aims at improving in vitro GC simulation using 3D culture techniques. Human B-cells were incorporated into PEG-4MAL hydrogels, to create a synthetic extracellular matrix, supported by CD40L cells, human tonsil-derived lymphoid stromal cells, and cytokines. The differentiation and antibody production of CD19+B-cells was best supported in a 5.0%-PEG-4MAL, 2.0 mM-RGD-peptide composition. The 3D culture significantly increased plasmablast and plasma cell numbers as well as antibody production, with less B-cell death compared to 2D cultures. Class switching of naive CD19+IgD+B-cells toward IgG+ and IgA+B-cells was observed. The formation of large B-cell clusters indicates the formation of GC-like structures. In conclusion, a well-characterized and controllable hydrogel-based human 3D lymphoid model is presented that supports enhanced B-cell survival, proliferation, differentiation, and antibody production.
Subject areas: Biological sciences, Bioengineering, Cell engineering, Tissue Engineering
Graphical abstract
Highlights
-
•
Optimization of a 3D PEG-4MAL hydrogel for the culture of human B-cells in vitro
-
•
3D cultures improve survival, proliferation, and differentiation of human B-cells
-
•
Mimicking the stromal lymphoid microenvironment promotes human B-cell expansion
-
•
The in vitro human 3D lymphoid model facilitates class switching of naive B-cells
Biological sciences; Bioengineering; Cell engineering; Tissue Engineering
Introduction
During immune responses, naive B-cells are activated through the ligation of their immunoglobulin (Ig) B-cell receptor (BCR) with encountered antigen. Upon activation, secondary lymphoid tissues develop specialized microenvironments known as germinal centers (GCs).1 Within these GCs, activated B-cells intricately interact with neighboring cell types to commence a B-cell response. This B-cell response includes proliferation, differentiation, and somatic hypermutation, a process editing the antigen-binding region of the BCR, followed by the selection of B-cells based on BCR affinity, generating high-affinity antibodies.2 Examples of supporting GC cell types required for these B-cell processes are lymphoid stromal cells (LSCs) and T follicular helper cells. T-cell - B-cell interactions also initiate class-switch recombination, a process replacing the constant region of the BCR for another isotype, generating antibodies with specialized effector functions.2,3 Ultimately, GC B-cells exit the GCs either as memory B-cells or long-lived plasma cells.2,4 Upon antigen reencounter during secondary immune responses, memory B-cells within the GCs are reactivated, undergoing further BCR editing and differentiation, forming the next generation of memory B-cells and plasma cells as progeny with improved BCR affinity and function.
The GCs have cellular and non-cellular components, containing extracellular matrix (ECM) proteins such as collagens, fibronectin, laminin, and vitronectin.5,6 These proteins are essential for the formation of stromal networks, along which both B-cells and T-cells can migrate to interact with each other.5 ECM components also play a role in the binding of chemokines, creating gradients that also steer the migration of B-cells and T-cells.7 However, most factors regulating the outcome of GC reactions are still poorly understood. To investigate the complex cell-cell and cell-matrix interactions that take place in the GCs, a model is needed that can simulate these processes.
In the past decades, the study of B-cells and the GC reaction has been performed using both 2D in vitro culture models and 3D in vivo animal models.8,9,10,11,12 Animal models such as mice offer the possibility to study B-cells in their complex 3D environment; however, the translation of such results toward humans can be challenging.13 Current 2D in vitro cultures to investigate B-cells in vitro are based on the stimulation of B-cells with CD40L expressing cells and various cytokines. These methods also have challenges of their own, being faced with limitations in recapitulating the complexity of the GC reaction. The natural microenvironment provides signals to the residing B-cells through cellular adherence to the ECM.14 These cell-matrix interactions, as well as cell-cell interactions, are important for differentiation, proliferation, and cellular functions in vivo.15 This also explains why some primary (malignant) B-cell types are difficult to culture using these conventional 2D systems, such as plasma cells, multiple myeloma, and chronic lymphocytic leukemia, as they depend on these interactions for their survival and growth.16,17,18 A better representative model could aid the study of both age- and vaccine-related differences in memory B cell responses after infection or vaccination, studying expansion capacity, antibody yield, affinity and function, and the respective stimulus-or T-cell dependency of these responses.
An alternative approach for the study of GCs is the use of organoids or 3D culture methods, offering the possibility to study the B-cells in an artificial 3D environment. 3D culture models can mimic the surrounding ECM and tissue hierarchy using a 3D hydrogel.19 This offers the possibility to more accurately study organ behavior, mimicking relevant cell-matrix as well as cell-cell interactions, further bridging the gap between 2D cell cultures and in vivo processes.20
3D hydrogels and their applications in organoids or 3D in vitro models have been extensively researched. Hydrogels of natural origin can be used, mimicking the natural ECM.21 However, natural hydrogels are in general animal derived and are faced with batch-to-batch variations. Synthetic hydrogels have been developed to overcome these problems, biologically functionalizing inert polymers.22,23 As a synthetic replacement of the widely used Matrigel in organoids, PEG gels were developed that were functionalized with minimal adhesion cues.22,23,24,25,26 Various PEG formulations have been studied for the application in 3D in vitro GC mouse models, with functionalized PEG-4MAL supporting murine B cell expansion and germinal center-like induction.27
3D in vitro GC mouse models have been shown to exceed classical 2D cultures in terms of B-cell growth, survival, and the presence of antibody class switching.28,29,30 For a better translation of in vitro research outcomes when studying human processes and diseases, a human model would be preferable. Models including both human B-cells and the stromal cell fraction have recently been developed through the culture of human tonsil tissue in single cell suspensions.31 However tonsillar tissue, even though most representative for the GC reaction, is not readily available, whereas peripheral blood mononuclear cells (PBMCs) are. Human PBMC-derived B-cells and T-cells have recently been studied in an organ-on-a-chip model, which was shown to recapitulate many features of germinal centers.32 In this study, animal-derived hydrogels were used as the 3D matrix. These matrices are of complex and variable compositions,33 and can potentially elicit undesired immune reactions.22,34
The aim of this study is to develop an accessible human 3D lymphoid model as an improvement to current human 2D in vitro systems, which can be used as a platform to study B-cells in an environment mimicking the GC. The developed model is building on experience within our group, gained in a 3D bone marrow niche model for the culture of multiple myeloma cells.35,36,37,38 The model is optimized in terms of cellular composition (LSCs,39 CD40L cells (CD40LCs) mimicking T-cell co-stimulation, and bulk B-cells versus naive or activated B-cells), cytokine supplementation and 3D hydrogel composition. Added cytokines are naturally produced by GC cell types that are currently not included in the model; T follicular helper cells (IL2, IL4, IL6, IL10, and IL21), follicular dendritic cells (IL15), and myeloid cells such as macrophages (IFN-α, APRIL), and play a role in the proliferation of B-cells, GC formation and maintenance as well as antibody response.8,9,40,41 As a 3D hydrogel, a four-armed polyethylene glycol macromer with maleimide groups at each terminus (PEG-4MAL) was used.23,25,26,27 This PEG-4MAL system can be tailored in terms of stiffness and functionalized using ECM-mimicking peptides. The setup of this human 3D lymphoid model has been performed using human PBMC-derived B-cells from healthy adults. The developed human 3D lymphoid model surpasses 2D methods in supporting B-cells and mimicking the GC environment, using accessible cell types and tunable, well-characterized materials.
Results
B-cell culture optimization in 2D
Our first aim was to identify a 2D culture condition best supporting the survival, proliferation, differentiation, and antibody production of the cultured B-cells. 2D cultures were optimized in terms of medium additives and supporting cells. The compared culturing methods aim at mimicking the activation and differentiation of B-cells occurring in a GC reaction, using toll-like receptor (TLR) activation (mimicking antigen activation) and/or activation of CD40 (mimicking T-cell help) and the addition of cytokines (Table 1).
Table 1.
Medium compositions
| Timepoint | CK1 | CK2 | CK3 |
|---|---|---|---|
| Day 0–4 | CPG ODN, IL2, IL10, IL15 | CPG ODN, IL2, IL4, IL10, IL15, IL21 | IL4, IL21 |
| Day 4–7 | IL2, IL6, IL10, IL15 | IL2, IL4 IL6, IL10, IL15, IL21 | IL4, IL21 |
| Day 7–11 | IL6, IL15, IFNα | IL4, IL6, IL15, IFNα, IL21 | IL4, IL21 |
| Day 11–14 | IL6, APRIL | IL4, IL6, IL21, APRIL | IL4, IL21 |
Used additives in cytokine mix 1 (CK1), cytokine mix 2 (CK2), and cytokine mix 3 (CK3) over time. All three mixtures have IMDM containing 10% FCS (v/v), 100 units/mL of penicillin, and 100 μg/mL of streptomycin as a basis.
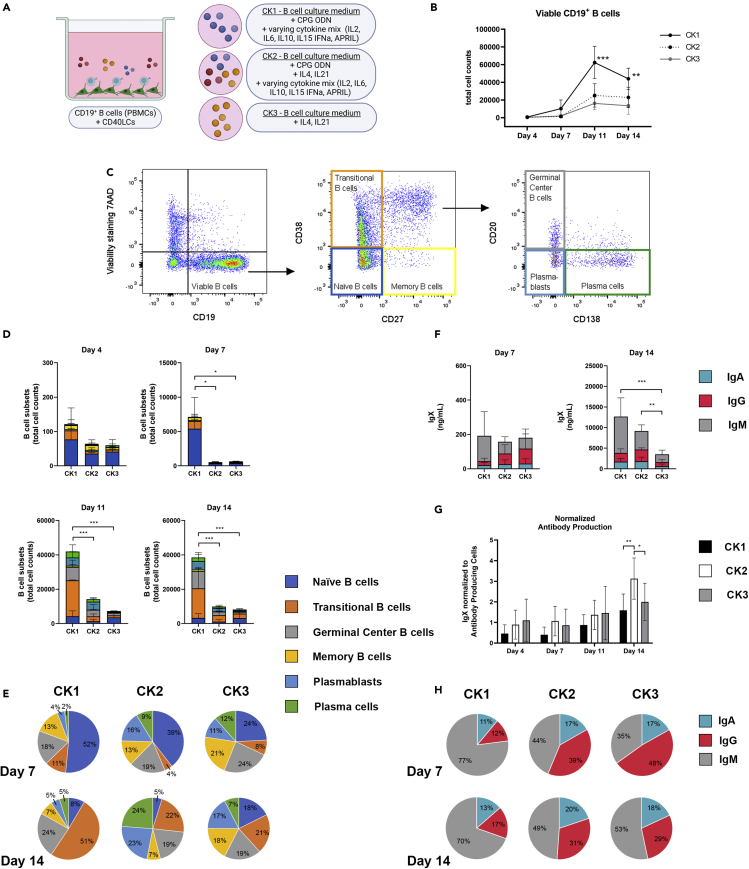
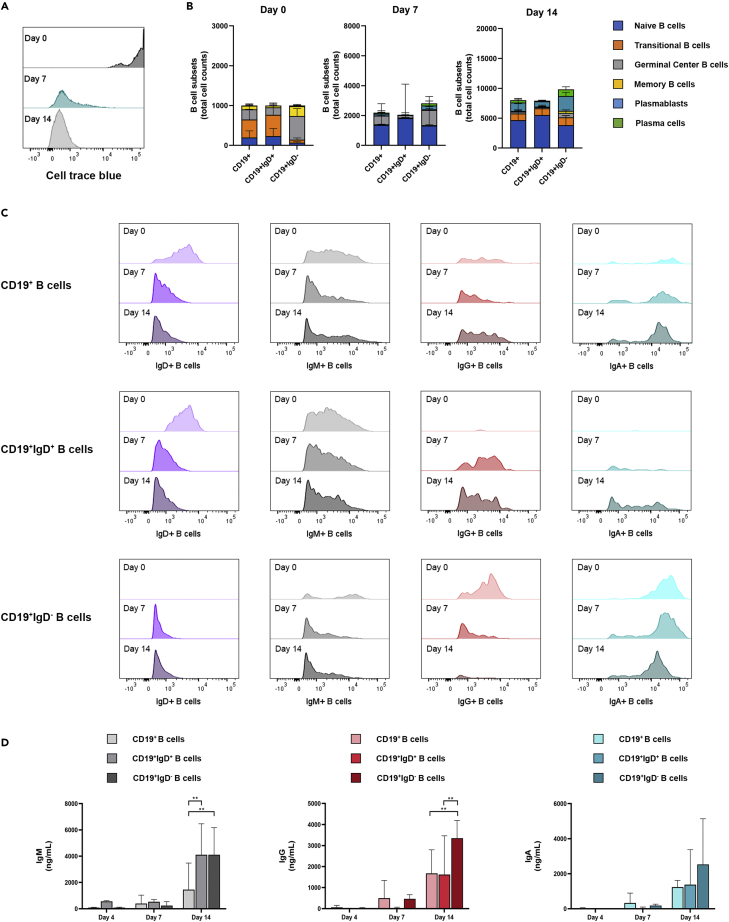
At first, human PBMC-derived CD19+ B-cells were co-cultured with irradiated murine fibroblast L-cells expressing human CD40L (CD40LCs, Figure 1A), as commonly used 2D in vitro systems.42,43 Potential culturing methods were based on previously described 2D B-cell culture systems.8,9,42 Jourdan et al. aimed at the generation and survival of plasma cells from memory B cells using CPG ODN, IL2, IL10, IL15, IL6, IFN-α and APRIL (CK1), and Marsman et al. on behalf of the T2B consortium,43 optimized protocols for T-cell-dependent activation of B cells using IL4 and IL21 (CK3). These two B-cell culture systems were combined in CK2 (Table 1), and compared by looking at the resulting quantity (Figure 1B) and differentiation status of cultured CD19+ B-cells over time. Six different B-cell subtypes were analyzed, i.e. naive B-cells (CD19+CD20+CD27−CD38−CD138-), transitional B-cells (CD19+CD20+CD27−CD38+CD138-), germinal center B-cells (CD19+CD20+CD27+CD38+CD138-), memory B-cells (CD19+CD20+CD27+CD38−CD138-), plasmablasts (CD19+CD20−CD27+CD38+CD138-) and plasma cells (CD19+CD20−CD27+CD38+CD138+) (Figure 1C). Input cells contained mainly naive, CD27− B-cell types (69.17% ± 12.72% naive B-cells and transitional B-cells) and also CD27+ memory B-cells and germinal center B-cells (30.49% ± 12.59%, Figure S1). Germinal center B-cells have low CD38 expression and still express CD20, a post-germinal center phenotype that has not transitioned toward plasmablast.44 Concentration and isotype of the produced antibodies were measured by a multiplex immunoassay.
Figure 1.
Three varying cytokine mixes were tested for their potential to support different B-cell subsets over time in vitro
CD19+ B-cells (n = 3, see also Figure S1) were co-cultured with CD40L cells (CD40LCs) in 2D in either cytokine mix 1 (CK1), cytokine mix 2 (CK2) or cytokine mix 3 (CK3) (Table 1).
(A) Schematic overview of the studied 2D co-culture conditions.
(B) Absolute number of viable CD19+ B cells over time.
(C) Gating strategy for six B-cell subsets.
(D) B cell subsets (total cell counts) of the absolute number of cells within each culture well and (E) relative frequencies on days 7 and 14, of the six B-cell subsets.
(F) Production of IgX (IgM, IgG, and IgA) in the culture supernatant on days 7 and 14.
(G) Total produced IgX normalized to the total amount of antibody-producing cells (plasmablasts and plasma cells) present in the cultures over time.
(H) Relative frequency of the production of IgX (IgA, IgM, and IgG) in the culture supernatant on days 7 and 14.
(B, D-H) All data compares B-cells cultured using CK1, CK2, or CK3, showing the mean (n = 3). Error bars indicate the SD. ∗ = p > 0.05, ∗∗ = p > 0.01, ∗∗∗ = p > 0.001.
In all three medium compositions (CK1-3, Table 1), cells survived during 14 days of culture and differentiated toward mature plasmablast and plasma cell phenotypes (Figure 1D). CK1 supported more extensive proliferation of B-cells compared to CK2 and CK3. This CK1 proliferation was not evenly distributed, but mainly occurred in the naive B-cell population during the first 7 days of culture, and in the transitional B-cell population during the subsequent 7 days of culture (Figures 1D and 1E).
The significantly increased number of cells in CK1 at day 7 did not result in enhanced cumulative antibody production. At day 14, a significant increase in the total IgX antibody production measured (the sum of IgA, IgG, and IgM) was observed in CK1 and CK 2 compared to CK3 (Figure 1F). When normalizing this antibody production to the antibody-producing cells (plasmablasts and plasma cells) in each condition, CK2 had a significantly higher antibody production (Figure 1G). Looking at the relative frequency of IgA, IgG, and IgM, CK1 had a higher frequency of IgM compared to CK2 and CK3, with a more even distribution of the three antibody isotypes in the latter two (Figure 1H).
Altogether, CK1 outperformed CK2 and CK3 in terms of cellular proliferation; however, this increase in B-cells could mainly be attributed to unactivated, non–antibody-producing B-cell subtypes. CK2 and CK3 resulted in more stable relative frequencies of B-cell subtypes over time, with CK2 having the highest relative frequency of antibody-producing cells, as well as normalized antibody production. Therefore CK2 was selected of the three for all subsequent experiments.
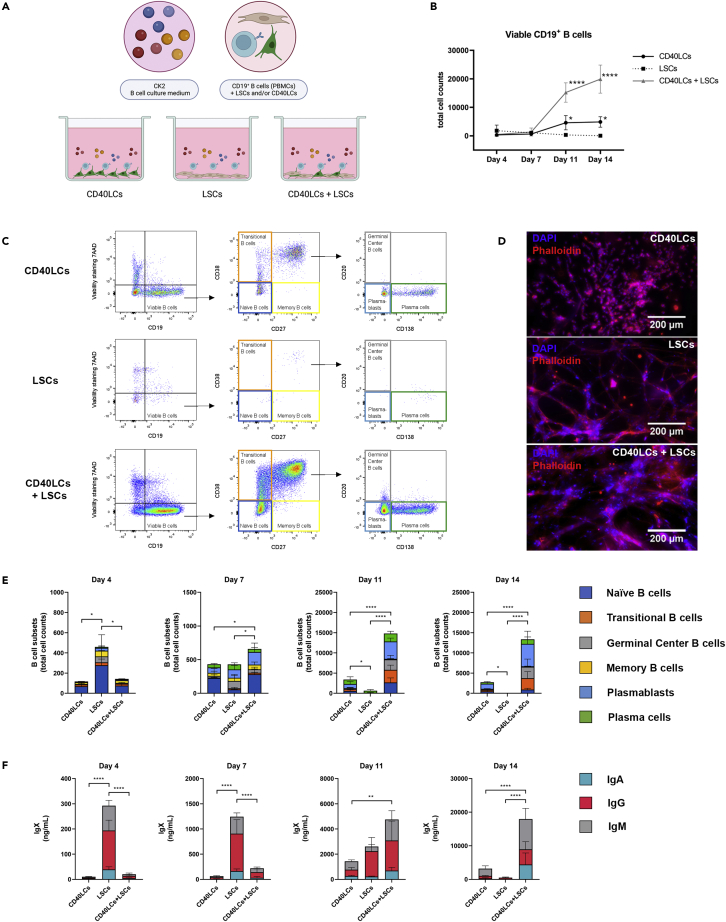
Next, the use of supporting cells was optimized. Human PBMC-derived CD19+ B-cells were co-cultured with irradiated murine fibroblast L-cells expressing human CD40L (CD40LCs) as a golden standard. Alternative methods tested were human tonsil-derived stromal cells (LSCs) and a combination of both CD40LCs and LSCs (Figure 2A). Again quantity and differentiation status of cultured CD19+ B-cell subsets were assessed over time (Figures 2B and 2C). The supporting cells were present during all time points, as confirmed by fluorescent microscopy, providing a varying supportive environment for the cultured B-cells per condition (Figure 2D). CD40LCs supported B-cell survival, proliferation, differentiation, and antibody production for 14 days. LSCs showed significantly higher B-cell numbers at day 4 compared to CD40LCs (Figure 2E). These were accompanied by significantly higher antibody levels at day 4 (Figure 2F). These higher antibody levels were still present at day 7; however, the number of B-cells did not further increase, whereas CD40LCs cultured B-cells did increase in number on day 7. LSCs were not capable of supporting viable, proliferating, and differentiating B-cells during 14 days of culture, with rapidly declining B-cell numbers at days 11 and 14. The triple culture of CD19+ B-cells, CD40LCs, and LSCs significantly increased B-cell numbers from day 7 onwards, also followed by increased antibody production (Figures 2E and 2F). Therefore the culture of CD19+ B-cells, supported by CD40LCs and LSCs, was selected as the most optimal culture condition.
Figure 2.
CD40L cells (CDLCs), lymphoid stromal cells (LSCs), or CD40LCs + LSCs were co-cultured with CD19+ B-cells in CK2 (Table 1) and tested for their potential to support different B-cell subsets over time in vitro (2D)
(A) Schematic overview of the studied 2D co-culture conditions.
(B) Absolute number of viable CD19+ B cells over time.
(C) Flow cytometry results of one of the analyzed donors, illustrating the differences between the tested supporting cell combinations on the co-cultured B-cells at day 14.
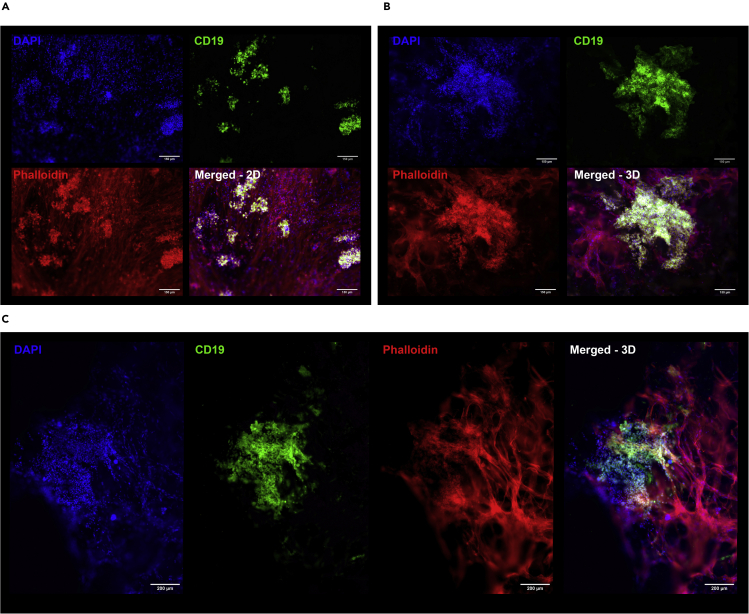
(D) Fluorescent images of the added supporting cell types in the co-cultures (blue = DAPI, red = phalloidin, scale bars represent 200 μm).
(E) B cell subsets (total cell counts) within each culture well of the absolute number of cells on days 4, 7, 11 and 14, of the six B-cell subsets.
(F) Production of IgX (IgM, IgG, and IgA) in the culture supernatant on days 4, 7, 11 and 14.
(E and F) Data showing the mean ± SD (n = 3). ∗ = p > 0.05, ∗∗ = p > 0.01, ∗∗∗ = p > 0.001, ∗∗∗∗ = p > 0.0001.
B-cell culture optimization in 3D
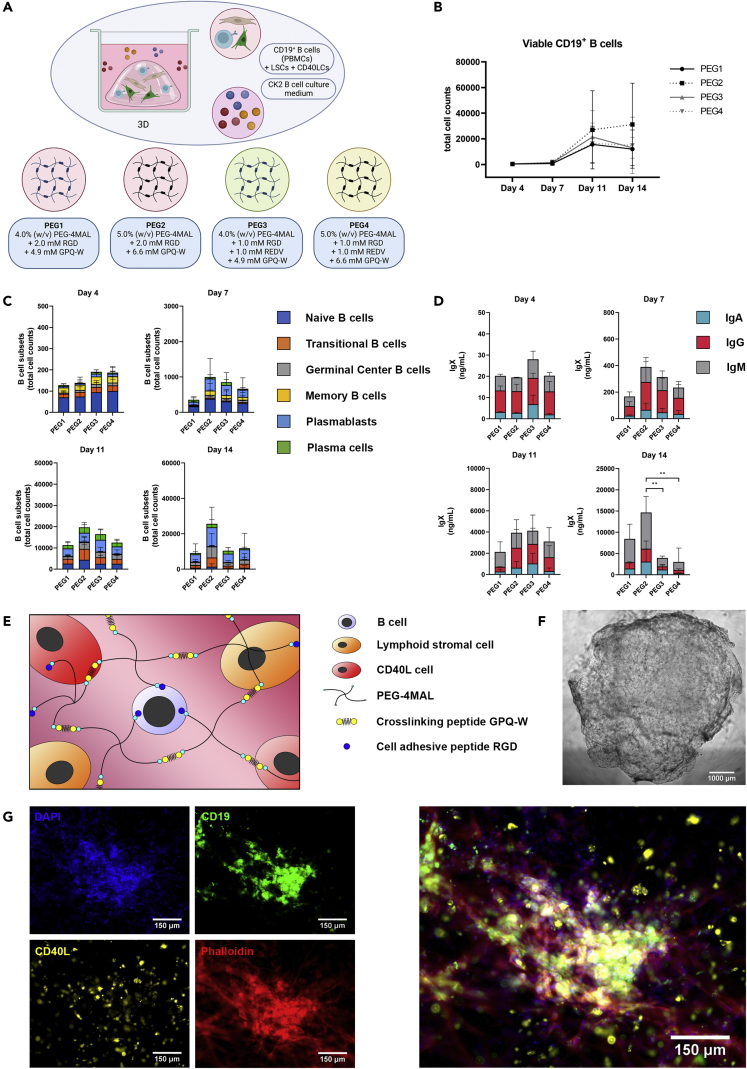
After optimizing the 2D culture of CD19+ B-cells, a 3D culture was developed. Six hydrogel conditions (PEG1-PEG6, Table 2) were studied for their effect on CD40LCs- and LSCs-co-cultured CD19+ B-cells. Hydrogels were varied in polymer content, (4.0% (w/v) and 5.0% (w/v) PEG-4MAL), and peptide composition (2.0 mM RGD, 1.0 mM RGD and 1.0 mM REDV, or 2.0 mM REDV), mimicking either the minimal integrin-binding motif in fibronectin (RGD),45 and/or the motif in fibronectin that is recognized by the integrin α4β1 or VLA-4 (REDV). RGD peptides facilitate integrin-stimulated adhesion and spreading of cells in 3D environments,46 REDV facilitates BCR-mediated adhesion of B-cells to fibronectin by VLA-4.47
Table 2.
Hydrogel compositions
| Hydrogel name | PEG-4MAL concentration | RGD concentration | REDV concentration | GPQ-W concentration |
|---|---|---|---|---|
| Hydrogel 1 (PEG1) | 4.0% (w/v) | 2.0 mM | – | 4.9 mM |
| Hydrogel 2 (PEG2) | 5.0% (w/v) | 2.0 mM | – | 6.6 mM |
| Hydrogel 3 (PEG3) | 4.0% (w/v) | 1.0 mM | 1.0 mM | 4.9 mM |
| Hydrogel 4 (PEG4) | 5.0% (w/v) | 1.0 mM | 1.0 mM | 6.6 mM |
| Hydrogel 5 (PEG5) | 4.0% (w/v) | – | 2.0 mM | 4.9 mM |
| Hydrogel 6 (PEG6) | 5.0% (w/v) | – | 2.0 mM | 6.6 mM |
PEG4-MAL hydrogels were tested in 2 different concentrations and 3 different peptide functionalizations using RGD, REDV, or a combination of the two. The concentration of cross-linking peptide GPQ-W was adjusted according to the amount of PEG-4MAL added in the hydrogel.
PEG5 and PEG6, containing only the REDV peptide, formed a less stable gel over time compared to PEG1-PEG4, containing either RGD or a combination of RGD and REDV. Overall cellular viability, including LSC viability, was decreased in PEG5 and PEG6 (Figures S2A-S2C). PEG1-PEG4 formed a stable hydrogel, that did not affect cellular viability over time (Figures S2A-S2G). These four hydrogel conditions were further selected for analysis of their potential as a 3D B-cell environment (Figure 3A). Of these, the 5% (w/v) PEG-4MAL gels showed a trend of a higher compressive modulus compared to the 4% (w/v) PEG-4MAL gels (Figure S2D). In line with these results, the 5% (w/v) PEG-4MAL gels showed a less steep increase in swelling ratio in the first hours and days after hydrogel fabrication, resulting in less hydrogel degradation compared to the 4% (w/v) PEG-4MAL gels, as shown by a higher stabilized swelling ratio (Figures S2F and S2G).
Figure 3.
Optimization of the functionalized hydrogel that is used as a 3D matrix for the 3D in vitro B-cell co-cultures (see also Figure S2)
CD19+ B-cells (n = 3), CD40LCs and LSCs were incorporated in the varying hydrogel compositions (3D, Table 2) and cultured in CK2 (Table 1).
(A) Schematic overview of the studied 3D co-culture conditions.
(B) Absolute number of viable CD19+ B cells over time.
(C) B cell subsets (total cell counts) within each 3D culture of the absolute number of cells per well on days 4, 7, 11 and 14, of the six B-cell subsets.
(D) Production of IgX (IgM, IgG, and IgA) in the culture supernatant on days 7 and 14.
(B-D) Data showing the mean ± SD (n = 3). ∗∗ = p > 0.01.
(E) Schematic representation of the selected PEG2 5.0% (w/v) PEG-4MAL and 2.0 mM RGD functionalized hydrogel, forming a stable 3D hydrogel when crosslinked using a GPQ-W cross-linking peptide. CD19+ B-cells, CD40LCs, as well as LSCs are homogeneously distributed throughout the hydrogel upon cross-linking.
(F) Brightfield image of a 40 μL crosslinked PEG2 (5.0% (w/v) PEG-4MAL and 2.0 mM RGD) 3D co-culture, scalebar represents 1000 μm.
(G) Fluorescent images of the PEG2 3D co-cultures, illustrating the possibility of cellular tracing and staining. Blue + green + red cells are CD19+ B-cells, blue + yellow + red cells are CD40LCs, blue + red cells are LSCs (blue = DAPI, green = CD19 immunostaining, yellow = DiI labeling, red = phalloidin, scale bars represent 150 μm).
When comparing B-cell survival, proliferation, and differentiation, no significant differences were found between the four stable hydrogels analyzed (Figures 3B and 3C). However, a trend was visible, more viable B-cells were present in PEG2, accompanied by higher antibody levels. This difference was found to be significant on day 14 when comparing PEG2 with PEG3 and PEG4 (Figure 3D). The inclusion of cells within PEG2 did not result in an enhanced hydrogel degradation during 14 days of culture (Figure S2E). The PEG2 hydrogel was therefore selected, resulting in an optimized 3D culture containing 5.0% (w/v) PEG-4MAL and 2.0 mM RGD, schematically illustrated in Figure 3E. Next to flowcytometric analysis of the cultured cell types, also brightfield microscopy (Figure 3F), and fluorescent microscopy were possible on the generated 3D cultures, discriminating the various cell types within their tissue-engineered environment (Figure 3G).
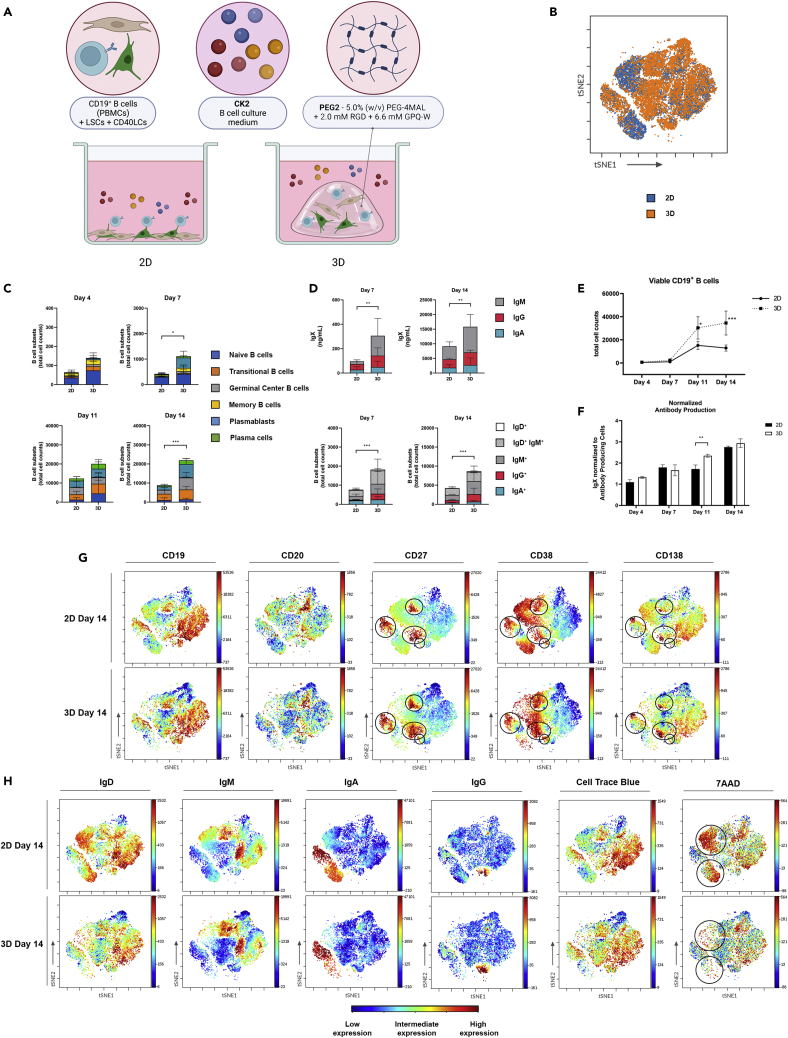
3D cultures of CD19+ B-cells induce a significant increase in B-cells and antibody production
Using all optimized culture conditions, 2D and 3D cultures were compared (Figure 4A, Table S1). The addition of the PEG2 3D matrix did not result in a preferential outgrowth of specific B-cell subsets or isotypes, illustrated by comparable clustering in ViSNE plots (Figure 4B). The 3D culture condition did result in a significant increase in B-cells on days 7 and 14, with similar trends on days 4 and 11, comparing the absolute number of cells at each time point (Figure 4C). Antibody production was increased in the 3D cultures, in line with the increased numbers of B-cells (Figures 4D and 4E). When normalizing this antibody production to the antibody-producing cells (plasmablasts and plasma cells) in each condition, the 3D cultures had a significantly higher antibody production on day 11. This difference was not observed at the other time points (Figure 4F). The 2D culture of B-cells did result in increased cell death (large 7AAD-positive cell clusters), not present in 3D cultured B-cells (Figures 4G and 4H). In both conditions, plasmablasts and plasma cells were present in the IgM, IgG, and IgA clusters, with the largest cluster of antibody-producing cells no longer expressing membrane-bound immunoglobulins (Figure 4G). The 3D culture condition did not result in a preferential outgrowth of specific B-cell subsets or isotypes, illustrated by comparable clustering in ViSNE plots and the coinciding surface marker expressions (Figures 4G and 4H).
Figure 4.
Comparison of 2D versus 3D co-cultures of CD19+ B-cells
CD19+ B-cells (n = 3) were cultured either on top of CD40LCs and LSCs (2D), or were incorporated together with CD40LCs and LSCs in PEG2 (3D) in CK2 (see also Table S1).
(A) Schematic overview of the studied 2D vs 3D co-culture conditions.
(B) viSNE dot plots of the concatenated (n = 3) 2D and 3D data at day 14 showing the overlaid clustering of B cells.
(C) B cell subsets (total cell counts) within each 2D or 3D culture of the absolute number of cells per well on days 4, 7, 11, and 14, of the six B-cell subsets.
(D) Production of IgX (IgM, IgG, and IgA) in the culture supernatant on days 7 and 14 (top graphs) and expression of membrane-bound immunoglobulins (IgD, IgM, IgG, and IgA) on days 7 and 14 (bottom graphs).
(E) Absolute number of viable CD19+ B cells over time.
(F) Total produced IgX normalized to the total amount of antibody-producing cells (plasmablasts and plasma cells) present in the cultures over time.
(G and H) viSNE plots showing the expression of surface markers (CD19, CD20, CD27, CD38, CD138), membrane-bound immunoglobulins (IgD, IgM, IgA, IgG), and both viability and proliferation (7AAD and cell trace blue) on 2D versus 3D cultured B cells at day 14, blue indicating low expression, yellow intermediate expression and red high expression.
(C-F) Data showing the mean ± SD (n = 3). ∗ = p > 0.05, ∗∗ = p > 0.01, ∗∗∗ = p > 0.001.
Characterization of the cultured B-cells in the 3D model in terms of proliferation, class switching, and morphology
To study the proliferation of B-cells, all CD19+ B-cells were traced over time using cell trace blue. At day 7, a steep decline in positive cells is visible, with the majority of CD19+ B-cells being negative for the cell tracer. At day 14, all viable CD19+ B-cells are negative for cell trace blue, indicating the proliferation of all CD19+ B-cells, resulting in a loss of cell trace blue over time (Figure 5A).
Figure 5.
Proliferation and class switching of B-cells in the 3D model
CD19+ B-cells (n = 3) were further sorted into CD19+IgD+ and CD19+IgD− B-cells (see also Figures S3 and S4).
All three B-cell conditions were incorporated together with CD40LCs and LSCs in PEG2 (3D) and cultured in the CK2 medium mix (Table 1).
(A) Cell trace blue proliferation staining on the 3D cultured CD19+ B-cells on days 0, 7, and 14. (B) Culture dynamics of CD19+ bulk, CD19+IgD+ or CD19+IgD− sorted B-cells over time showing B cell subsets (total cell counts) of B-cell subsets on days 0, 7, and 14 (see also Figure S4). (C) Expression of membrane-bound immunoglobulins (IgD, IgM, IgG, and IgA) of the absolute number of B-cells per well on days 0, 7, and 14. (D) Production of IgM, IgG and IgA in the culture supernatant on day 4, day 7, and day 14. Data showing the mean ± SD (n = 3). ∗∗ = p > 0.01.
To study potential class switching of the cultured B-cells in the 3D model, B-cells were sorted into a naive population (CD19+IgD+) and maturated population (CD19+IgD−, Figure S3) and compared to unsorted CD19+ B-cells. The three compared conditions have equal amounts of B-cells over time. In the CD19+IgD+ cultures, less plasmablasts and plasma cells develop over time, as opposed to CD19+IgD− cultures, where more plasmablasts and plasma cells develop over time, both compared to unsorted CD19+ B-cells (Figure 5B). When analyzing B-cell isotypes using flow cytometry, unsorted CD19+ B-cell cultures show diminishing IgD and IgM populations, and increasing IgG and IgA populations over time. Naive CD19+IgD+ B-cells also show a diminishing IgD population, with a larger, more stable IgM population and an increasing IgG and IgA population over time (Figure 5C). As no mature IgG and IgA cells were present in this condition at day 0, with no production of IgG and IgA during the first 4 days of culture (Figure 5D), this increase can be attributed to the class switching of naive CD19+IgD+ B-cells.
The cultured mature CD19+IgD− B-cells show a stable IgA population over time. However, the IgG population diminishes over time, as opposed to both unsorted CD19+ and sorted CD19+IgD+ B-cells cultures (Figure 5C). In the CD19+IgD− sorted cultures, a higher presence of a CD27+CD38+CD138+ population, that is IgD−IgM-IgA-IgG-, was found at day 14. This population is diminished in CD19+IgD+ sorted cultures (Figure S4). The loss of IgG+ B-cells could therefore be attributed to the maturation of IgG+ B-cells toward CD27+CD38+CD138+ B-cells, losing their membrane-bound immunoglobulins. This is supported by a significantly higher IgG production in the supernatant of CD19+IgD− sorted cultures at day 14 (Figure 5D).
Microscopy of the 3D B-cell co-cultures shows the formation of larger GC-like B-cell clusters with an average diameter of 482 μm ± 223 μm, which are significantly larger than those formed in 2D B-cell co-cultures (91 μm ± 39 μm) (Figure 6). B-cell clusters can be discriminated either morphologically as rounded cells from their stretched, adherent stromal counterpart, or by immunocytochemistry staining for CD19 expression. All 3D B-cell outgrowth show intricate connections to the surrounding stromal cell compartment (Figure 6).
Figure 6.
Fluorescent imaging of 2D and 3D CD19+ B-cell co-cultures
(A) Fluorescent images of 2D and (B and C) 3D co-cultures. Blue = DAPI, green = CD19 immunostaining, red = phalloidin. Scale bars represent 150 μm (A and B) or 200 μm (C).
Discussion
In this study, a human 3D lymphoid model based on synthetic polymers was developed and optimized for the culture of human B-cells. The developed model exceeded a 2D human B-cell culture system, better recapitulating the tissue complexity of the GC reaction using 3D culture techniques. The added well-characterized hydrogel resembles the tissue hierarchy and natural surrounding ECM of the GC. This more accurately mimics relevant cell-cell as well as cell-matrix interactions, further bridging the gap between 2D cell cultures and in vivo GC processes. The addition of a 3D environment enhanced in vitro B-cell survival, proliferation, differentiation, and antibody production compared to 2D co-cultures. This more representative model could be used to study human memory B cell responses during aging, after infection or vaccination, or to assess hallmarks of B-cell functionality or anomality in health and disease.
The study of PBMC-isolated human B-cells via in vitro culture represents an invaluable resource for immunophenotyping, functional, and repertoire studies. By culturing B-cells in vitro, new challenges arise in cellular survival over time, and the mimicking of in vivo occurring processes such as the complex GC reaction. The proliferation and survival of activated B-cells require sustained receptor stimulation through the mimicking of T-cell interactions using stimulation via CD40L and IL4.48 Mature plasma cells are known to die rapidly when cultured in vitro without a supportive stromal environment.49 Through the supplementation of cytokines and co-cultures with stromal cells (either as an irradiated CD40L expressing cell source or as a functional human LSC), the natural cellular B-cell environment can be mimicked. However, adding these factors still does not take into account the effect of the natural ECM and 3D environment. ECM proteins have been shown to be critical in GC B-cell reactions and antigen-specific antibody production.50 Also for many signaling molecules, the ability to bind to ECM components is essential for their functionality. The key regulator of B-cell migration and lymphoid tissue architecture, CXCL13, binds to the ECM after being secreted by fibroblastic reticular cells (FRCs). This binding and thus immobilization constrains its diffusion, forming concentration gradients in 3D.7 Only in a 3D culture model, GC functionality can be studied in an environment capable of supporting such a structural organization.
To develop a functional 3D human in vitro lymphoid model, we validated its added value over a traditional golden standard 2D model. Previously developed 2D B-cell culture systems were taken as a basis, and compared through the variation of cytokine supplementation8,9,42,51 and supporting cell source. The supportive function of bone marrow and LSCs on B-cell survival in health and pathogenesis has been described previously.49,52 However when co-culturing human tonsil LSCs with CD40LCs, B-cell survival, proliferation, and differentiation was supported significantly better than single co-cultures of B-cells using either CD40LCs or LSCs as supporting cells, clearly indicating the benefit of a lymphoid tissue-like embedding. The supernatants of B-cell co-cultures with both CD40LCs and LSCs contained higher levels of APRIL, BAFF, and IL-10, compared to B-cell co-cultures containing only LSCs or CD40LCs (data not shown). Future studies will focus on the kinetics of the produced cytokines, as well as the discrimination between the multiple co-cultured cell types as potential producers.
Ideally, a hydrogel mimicking this ECM does not only reflect the 3D spatial structure of the lymphoid tissue, but also the biological composition of the ECM. Matrigel, a natural ECM-based hydrogel, has been used in a previous 3D bone marrow niche model for the culture of multiple myeloma cells,35,36,37,38 a human lymphoid follicle organ-on-a-chip model,32 and is also used extensively for the generation of organoids.53,54,55 However as a natural product produced by Engelbreth-Holm-Swarm mouse sarcoma cells, this matrix is not well-defined and batch to batch variations exist.33,56 Other natural ECM mimics used in tissue engineering applications, such as collagen, hyaluronic acid, and chondroitin sulfate hydrogels, are mainly bovine or porcine derived,21,57 and therefore faced with batch-to-batch variations. Moreover, the presence of endotoxins could elicit undesired immune reactions.34 In our immunological application, a hydrogel with a low risk of unwanted immune reactions is desired. As an alternative, we, therefore, used a well-defined, non-animal derived matrix, with a synthetic PEG polymer backbone that can be functionalized with biological components of choice.22,24,26 PEG polymers with various terminal groups have previously been optimized for their culture with murine B cells,27 showing the best results using PEG-4MAL, and subsequently used in the formation of 3D mouse immune tissues, for the study of GC B-cells.27,30 In our study, synthetic PEG-4MAL hydrogels were explored for their effect on the human instead of mouse B-cell cultures, at varying polymer concentrations and peptide compositions. The addition of REDV, facilitating BCR-mediated adhesion of B-cells to fibronectin by VLA-4,47 next to RGD did not lead to enhanced B-cell survival and antibody production. The increased presence of RGD peptides (PEG2), supporting a more general integrin-stimulated adhesion and spreading of human stromal cells in the 3D environment,46 was shown to more enhance B-cell survival. The stiffer (Figure S2D) 5.0% (w/v) PEG-4MAL and 2.0 mM RGD functionalized hydrogel (PEG2) best supported B-cell survival and antibody production, thus with a higher polymer content than the described 4.0% (w/v) PEG-4MAL and 2.0 mM RGD hydrogel for organoid and spheroid cultures,26 but with lower polymer content that the 7.5% (w/v) PEG-4MAL gels used for the murine 3D B-cell model,29,30 emphasizing the need for the optimization of ECM-mimicking hydrogels for each desired application.
When comparing the 3D-optimized culture system to the 2D culture system, it was shown that the addition of a biologically active 3D matrix resulted in a significant increase in B-cells after 14 days of culture and increased antibody production. These enhanced processes are linked to each other, as the increase in antibody production could be a result of increased B cell proliferation resulting in more antibody-secreting cells, or could be due to altered differentiation and enhanced antibody secretion per cell. The increased numbers of mature B-cells present in the 3D cultures could be attributed to the presence of cell-matrix interactions in 3D, or to the addition of an hypoxic 3D environment,58 as it has been shown that hypoxic conditions enhance the survival of human plasma cells in vitro.59 Most likely it is a combination of processes, which needs further investigation to be unraveled.
Importantly, our human 3D B-cell model supported major processes in B-cell differentiation, i.e. class switching to IgG and IgA isotypes and differentiation of memory B-cells into IgM, IgG, or IgA-secreting cells. Notably, when studying the presence of class switching of CD19+IgD− sorted cells in the 3D model, loss of B-cells expressing surface IgG, but not surface IgA, was observed over time. Together with enhanced IgG production in these cultures, this suggests that the current 3D model and cytokine conditions facilitated rapid differentiation of IgG memory B-cells toward IgG-producing plasmablasts or plasma cells. IgG-expressing B-cells remained present in the total CD19+ B-cell cultures, which could be explained by simultaneous class switching of naive B-cells to IgG+ B-cells, as observed in the CD19+IgD+ sorted cultures. In future studies, the human 3D B-cell model could help to further disentangle factors and cellular interactions driving isotype-specific class-switching and plasma cell differentiation. Previously, class-switch recombination was considered a process occurring during the GC response, after the onset of somatic hypermutation, and thus an indicator of a functional GC reaction.60 More recently, the timing of class-switch recombination was revisited, as it was shown to occur prior to GC formation.3 The class-switch recombination taking place in our 3D model should therefore be considered as an indicator of functional B-cell activation and B-T-cell mimicking interactions taking place in lymphoid B-cell zones near, rather than in the GCs.
The developed human 3D in vitro lymphoid model offers the opportunity to study B-cells in the context of a mimicked 3D lymphoid environment. The use of PBMCs provides the opportunity to study and compare B-cell functionality in both health and disease, as PBMCs are widely collected and stored in clinical studies. The study of B-cell functionality in both health and disease could include pathogen-specific immune responses, B-cell mediated autoimmunity, and B-cell malignancies. The improved survival of mature B-cells in the 3D model aids the study of autoreactive plasma cells, which can play an important role in the study of autoimmune diseases.61 Also primary malignant B-cell types that cannot be cultured using conventional 2D systems, such as chronic lymphocytic leukemia, can potentially be studied using the developed 3D lymphoid model. Comparison of B cell functionality within this in vitro 3D model with in vivo data, especially in B cell-related disease, could further illuminate the relevance of this 3D model.
In the current set-up of the 3D model, available 2D models served as a basis for comparison and optimization, where extensive cytokine supplementation is necessary for both B-cell survival and maturation. For the development of the 3D model, conditions were kept equal and compared, so the added effect of the synthetic 3D matrix could be established. This 3D model could be even further optimized, including for example human T-cells, to replace the current murine CD40L cell source expressing human CD40L, or follicular DCs. By having both primary human LSCs and human T-cells present in the model, which both can contribute to the survival and maturation of B-cells through cellular interactions and cytokine production, supplemented cytokines need to be reevaluated. A further optimized 3D system could potentially function as a self-supporting system that no longer requires cytokine supplementation, with essential cell types and interactions being present, providing crucial signals and support to the cultured B-cells. Activation of B-cells will still be essential, but could include antigen-specific stimulation of B-cells via BCR, preferably via human T-cells, thus mimicking the cognate T-B interactions. By doing so, the maturation of antigen-specific antibodies can be further investigated (e.g. hypermutation, avidity maturation), as opposed to the currently used polyclonal activation of B cells via TLRs. Future plans may also include the analysis of the chemokine expression within the formed B-cell clusters, and the potential formation of light and dark zones within the currently observed outgrowths of B-cells.
In conclusion, the human 3D in vitro lymphoid model developed in this study enhanced B-cell survival, proliferation, maturation, and class switching compared to traditional 2D culture techniques. The model contains a well-defined, 3D ECM-mimicking hydrogel, T-cell mimicking CD40LCs, and human tonsil-derived LSCs. The latter adds a human stromal compartment to the model, expressing markers of characteristics of FRCs.39 The formation of large B-cell clusters within the 3D model during 14 days of culture, indicates the formation of GC-like structures, which can be followed over time using microscopy. The developed 3D lymphoid model provides an accessible human platform that can be used to study healthy and diseased B-cells in a 3D environment mimicking the natural surrounding microenvironment.
Limitations of the study
A human 3D in vitro lymphoid model offers the opportunity to study B-cell features in the context of a mimicked 3D lymphoid environment. However, the currently developed model is still a simplified version of the natural lymphoid environment, mimicking only selected aspects. The first limitation is the present choice of cell source, studying healthy B-cells isolated from PBMCs, as opposed to lymphoid tissue. This latter cell source is more representative of the cells involved in a natural GC reaction. Primary lymphoid stromal cells (LSC) were included, but from an allogeneic source. Using autologous LSCs is technically more challenging, but preferable. A second limitation is that T cell help is currently mimicked using a murine CD40L cell source expressing human CD40L. Even though irradiated before inclusion in the model, a fully human model, with autologous primary T-cells providing the essential T-cell help for the studied human B-cells, could further improve this model. Another limitation is that the survival and differentiation of B-cells are currently supported through artificial cytokine supplementation over time, as is done in traditional 2D cultures. The supplemented cytokines could dominate any natural cytokine production occurring in the system through the interaction of the B-cells with the LSCs. A self-supporting 3D model, consisting of all essential autologous cell types, would be most optimal to investigate germinal center processes.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| BUV395 Mouse Anti-Human CD19 | BD biosciences | Cat# 563549; RRID:AB_2738272 |
| Brilliant Violet 605™ anti-human CD20 Antibody | Biolegend | Cat# 302334; RRID:AB_2563398 |
| BV421 Mouse Anti-Human CD27 | BD biosciences | Cat# 562513; RRID:AB_11153497 |
| BV786 Mouse Anti-Human CD38 | BD biosciences | Cat# 563964; RRID:AB_2738515 |
| PE anti-human CD138 (Syndecan-1) Antibody | Biolegend | Cat# 356504; RRID:AB_256187 |
| Brilliant Violet 510™ anti-human IgD Antibody | Biolegend | Cat# 348220; RRID:AB_2561945 |
| FITC anti-human IgM Antibody | Biolegend | Cat# 314506; RRID:AB_493009 |
| APC/Fire™ 750 anti-human IgG Fc Antibody | Biolegend | Cat# 410724; RRID:AB_275022 |
| IgA Antibody, anti-human, APC | Miltenyi Biotec | Cat# 130-113-472; RRID:AB_2733422 |
| Purified anti-human CD19 Antibody | Biolegend | Cat# 363002; RRID:AB_2563989 |
| Mouse Ig (Whole Ab), biotinylated | GE Healthcare Life sciences | GERPN1001-2ML |
| Mouse Anti-Human Lambda-PE | Southern Biotech | Cat# 9180-09; RRID:AB_279667 |
| Mouse Anti-Human Kappa-PE | Southern Biotech | Cat# 9230-09; RRID:AB_2796710 |
| IgA from human serum | Sigma Aldrich | Cat# I4036 |
| IgG from human serum | Sigma Aldrich | Cat# l2511 |
| IgM from human serum | Sigma Aldrich | Cat# I8260 |
| Biological samples | ||
| Immunoglobulins G, A & M Serum Human | National Institute for Biological Standards and Control (NIBSC) | 67/099 |
| Chemicals, peptides, and recombinant proteins | ||
| GCRDGPQGIWGQDRCG | AAPPTec LLC | Custom made |
| GRGDSPC | AAPPTec LLC | Custom made |
| GREDVGC | AAPPTec LLC | Custom made |
| 4arm-PEG-MAL, MW 20 kDa | Laysan Bio | Cat# 4arm-PEG-MAL-20K |
| Recombinant Human IL-2 | Peprotech | Cat# 200-02 |
| Recombinant Human IL-4 | Peprotech | Cat# 200-04 |
| Recombinant Human IL-6 | Peprotech | Cat# 200-06 |
| Recombinant Human IL-10 | Peprotech | Cat# 200-10 |
| Recombinant Human IL-15 | Peprotech | Cat# 200-15 |
| Recombinant Human IL-21 | Peprotech | Cat# 200-21 |
| Recombinant Human APRIL | Peprotech | Cat# 310-10C |
| Recombinant Human IFN-alpha A (alpha 2a) Protein | R&D Systems | Cat# 11100-1 |
| Collagenase, Type I, powder | Thermo Fisher | Cat# 17018029 |
| DAPI | Sigma Aldrich | Cat# D9542 |
| Phalloidin–Atto 700 | Sigma Aldrich | Cat# 79286 |
| Streptavidin, Alexa Fluor™ 488 Conjugate | Invitrogen | Cat# S32354 |
| VECTASHIELD® Antifade Mounting Medium | Vector Labs | Cat# H-1000 |
| Critical commercial assays | ||
| EasySep™ Human CD19 Positive Selection Kit II | Stemcell technologies | Cat# 17854 |
| CellTrace™ Blue Cell Proliferation Kit, for flow cytometry | Thermo Fisher | Cat# C34568 |
| LIVE/DEAD™ Viability/Cytotoxicity Kit, for mammalian cells | Invitrogen | Cat# L3224 |
| Experimental models: Cell lines | ||
| Murine fibroblast L-cells expressing human CD40L | generated by Garrone et al. | Garrone et al., 1995 https://doi.org/10.1084/jem.182.5.1265. |
| Oligonucleotides | ||
| CpG ODN2006 | Invivogen | Cat# tlrl-2006 |
| Software and algorithms | ||
| FlowJo | FlowJo, LLC and Illumina, Inc. | RRID:SCR_008520 |
| Cytobank Premium | Beckman Coulter, Inc. | https://premium.cytobank.org/cytobank; RRID:SCR_014043 |
| GraphPad Prism 9.3.1. | GraphPad Software, Inc. | https://www.graphpad.com/; RRID:SCR_002798 |
| Leica Application Suite X | Leica Microsystems | https://www.leica-microsystems.com/products/microscope-software/p/leica-las-x-ls/; RRID:SCR_013673 |
| ImageJ 1.53c | open source | https://imagej.nih.gov/ij/; RRID:SCR_003070 |
| Bio-Plex Manager | Bio-Rad | https://www.bio-rad.com/; RRID:SCR_014330 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Cécile van Els (cécile.van.els@rivm.nl).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Primary cells and cell lines
PBMCs were isolated from healthy adult blood donors’ buffy coats (Sanquin Blood Supply, Amsterdam, The Netherlands) by density gradient centrifugation on a Ficoll-Hypaque gradient (Pharmacia Biotech). PBMCs were frozen in 50% RPMI, 40% fetal calf serum (FCS; Greiner Bio-One) and 10% dimethyl sulfoxide (DMSO; Sigma-Aldrich) and stored at −135°C until use.
Human tonsil-derived LSCs, were previously isolated and characterized.39 Human tonsil-derived LSCs expressed the FRC marker podoplanin (PDPN), while lacking the endothelial marker CD31, and were thus classified as FRCs. Murine fibroblast L-cells expressing human CD40L (CD40LCs), generated by Garrone et al.,62,63 were used in the cultures as a source for CD40 ligation. CD40LCs were gamma irradiated (2,000 rad/20 Gy) before use.
All buffy coat donations were obtained after written informed consent, and used protocols were approved by the local medical ethical committee in accordance with the Declaration of Helsinki.
Isolation of CD19+ B-cells and subsets
B-cells were isolated from the cryopreserved PBMCs via CD19 positive selection according to the manufacturers protocol (Easysep, Stemcell technologies). For further sorting of CD19+ B-cell subsets, cells were stained with anti-CD19 (1:300, PE-Cy7, J3-119, Beckman Coulter), anti-IgD (1:100, BV510, IA6-2, Biolegend), and 7AAD (1:200, 559,925, BD Biosciences) in FACS buffer (PBS (Gibco), 5 mg/mL BSA (Sigma) and 2 mM EDTA (Sigma)) and incubated at 4°C in the dark for 30 min. After incubation, cells were washed 2 times using FACS buffer and then re-suspended in sorting buffer (FACS buffer plus 25 mM HEPES (Sigma)). CD19+ B-cells were sorted into CD19+IgD+7AAD− and CD19+IgD-7AAD− populations using the purity sort mode on a FACSMelody Cell Sorter (Becton Dickinson) using the BD FACSChorus application (Becton Dickinson).
Cell culture conditions
LSCs and CD40LCs were expanded using Dulbecco’s Modified Eagle Medium (DMEM), 10% (v/v) FCS, 100 units/mL of penicillin and 100 μg/mL of streptomycin (Gibco, Life Technologies). B-cells and B-cell co-cultures were cultured using Iscove’s modified defined medium (IMDM; Gibco, Life Technologies) containing 10% FCS (v/v), 100 units/mL of penicillin and 100 μg/mL of streptomycin (Gibco, Life Technologies). To these cultures, varying combinations of cytokines and molecules were added (cytokine mix 1 (CK1), cytokine mix 2 (CK2) or cytokine mix 3 (CK3); Table 1) in a final concentration of 10 ng/mL recombinant human IL-2 (Peprotech, 200-02), 50 ng/mL recombinant human IL-4 (Peprotech, 200-04), 50 ng/mL recombinant human IL-6 (Peprotech, 200-06), 50 ng/mL recombinant human IL-10 (Peprotech, 200-10), 10 ng/mL recombinant human IL-15 (Peprotech, 200-15), 50 ng/mL recombinant human IL-21 (Peprotech, 200-21), 50 ng/mL recombinant human APRIL (Peprotech, 310-10C), 10 μg/mL CpG ODN2006 (Invivogen, tlrl-2006-1), and 500 U/mL recombinant human IFN-alpha A (alpha 2a) protein (R&D Systems, 11,100-1).
2D and 3D cultures
Isolated CD19+ B-cells or sorted subsets were cultured in a 1:5 ratio with supporting cells (CD19+ B-cells:CD40LCs and CD19+ B-cells:LSCs) or 1:2.5:2.5 ratio (CD19+ B-cells:CD40LCs:LSCs) in the presence of an indicated cytokine and molecule mix (Table 1). In 2D, supporting cells formed a confluent layer at the bottom of a tissue culture treated 48-well plate (Cellstar Cell Culture Multiwell Plates, Greiner Bio-One), on top of which the CD19+ B-cells were cultured. In 3D, CD19+ B-cells, CD40LCs and/or LSCs were homogenously incorporated throughout an indicated hydrogel composition (Table 2) by resuspension, 20 μL gels were casted in the center of a non-tissue culture treated 48-well plate (Nunc Non-Treated Multidishes, ThermoFisher) to prevent cellular outgrowth from the 3D hydrogel. All co-cultures were cultured for 4, 7, 11 and/or 14 days in an humidified incubator at 37 °C at 5% CO2. The number of 30.000 cells per well and added medium volumes were kept equal in both 2D versus 3D. On each time point, all cells from each individual culture well were collected and measured in its entirety, resulting in an absolute total cells count that could be compared over time per condition. Medium was refreshed twice a week, adding 250 μL per well, the supernatant of each time point was stored at −20°C for further analysis.
Method details
Hydrogels
The method for the creation of a modular, cell encapsulating hydrogel, containing adhesive peptides and protease degradable crosslinking peptides, is based on the protocol of Cruz-Acuña et al.26 In short, a four-armed polyethylene glycol macromer with maleimide groups at each terminus (PEG-4MAL, MW 20 kDa, Laysan Bio) is conjugated to cysteine-containing adhesive peptides RGD (GRGDSPC, AAPPTec, custom synthesis, purity: >95%, trifluoroacetic acid (TFA) removal) and REDV (GREDVGC, AAPPTec, custom synthesis, purity: >95%, (TFA) removal) and cross-linked using GPQ-W (GCRDGPQGIWGQDRCG, AAPPTec, custom synthesis, purity: >95%, (TFA) removal). An aliquot of the PEG-4MAL macromer, adhesive RGD peptide, and cross-linker GPQ-W where allowed to reach room temperature. The needed amount of GPQ-W and RGD was weighed out and dissolved using 20 mM HEPES buffer (Sigma), after which the PH was adjusted to 7.4. Both solutions where filtered by transferring them to a separate Costar Spin-X centrifuge tube (Corning). The PEG-4MAL macromer was dissolved in sterile, prefiltered 20 mM HEPES buffer. The PEG-4MAL and RGD solution where combined in a 2:1 ratio and incubated for 15 min at 37°C, to generate the functionalized PEG-4MAL precursor solution. The cells to include in the hydrogel where combined, and spun down, after which the supernatant was removed. Cells were re-suspended in B-cell medium (20% of the desired final hydrogel volume) after which the functionalized PEG-4MAL precursor was added (60% of the desired final hydrogel volume). The cross-linking peptide solution (GPQ-W) was added to the bottom of each well, in the center of a 48-well multiwell plate (20% of the desired final hydrogel volume). The mixture comprising the functionalized PEG-4MAL precursor and the cell mixture (80% of the desired final hydrogel volume) was pipetted on top of the cross-linking peptide solution and re-suspended quickly. After casting of the hydrogel, it was left to cross-link by incubating the plate at 37°C for 20 min, after which medium was added to each well. Six different hydrogel compositions were tested during hydrogel optimization for our human 3D lymphoid model (Table 2).
Mechanical analysis
The compressive modulus (or stiffness) of PEG-4MAL hydrogels with varying PEG-4MAL content and peptide functionalization (PEG1: 4% (w/v) PEG-4MAL, 2.0 mM RGD, 4.9 mM GPQ-W, PEG2: 5% (w/v) PEG-4MAL, 2.0 mM RGD, 6.6 mM GPQ-W, PEG3: 4% (w/v) PEG-4MAL, 1.0 mM RGD, 1.0 mM REDV, 4.9 mM GPQ-W, PEG4: 5% (w/v) PEG-4MAL, 1.0 mM RGD, 1.0 mM REDV, 6.6 mM GPQ-W) was determined by unconfined compression using dynamical mechanical analysis (DMA, Q800, TA Instruments). Samples were measured (n = 3 per condition) following swelling in Iscove’s modified defined medium (IMDM; Gibco, Life Technologies) containing 10% FCS (v/v), 100 units/mL of penicillin and 100 μg/mL of streptomycin (Gibco, Life Technologies), pH 7.4 at 37°C for 24 h. The compressive modulus was calculated from the linear derivative of the stress–strain curve at 5–15% strain.
Swelling ratio
The swelling ratio (Qm) of PEG-4MAL hydrogels with varying PEG-4MAL content and peptide functionalization (PEG1: 4% (w/v) PEG-4MAL, 2.0 mM RGD, 4.9 mM GPQ-W, PEG2: 5% (w/v) PEG-4MAL, 2.0 mM RGD, 6.6 mM GPQ-W, PEG3: 4% (w/v) PEG-4MAL, 1.0 mM RGD, 1.0 mM REDV, 4.9 mM GPQ-W, PEG4: 5% (w/v) PEG-4MAL, 1.0 mM RGD, 1.0 mM REDV, 6.6 mM GPQ-W) was determined over time. Hydrogels (40 μL) were incubated in IMDM containing 10% FCS (v/v), 100 units/mL of penicillin and 100 μg/mL of streptomycin, pH 7.4 at 37°C. After carefully removing the medium, hydrogel samples were collected after 1, 2, 3 and 4 h, and at day 1, 2, 3, 4, 7, 11 and 14, measuring their mass after swelling (MS). The hydrogels were then dried in an oven at 70°C for 24 h, after which their dry mass (MD) was measured. Three hydrogels of each type were measured for each time point. The swelling ratio based on hydrogel mass was calculated using Qm = MS/MD.
Rheological analysis
The storage modulus (G′) of 5% (w/v) PEG-4MAL, 2.0 mM RGD, 6.6 mM GPQ-W (PEG2) hydrogels were measured with a discovery HR-2 hybrid rheometer (TA Instruments) in parallel plate geometry with a 20-mm diameter acrylic upper plate, at 21°C, a frequency of 1–10 rad/s, and a constant strain of 2%. Hydrogel samples, with and without cells (CD19+ B-cells,CD40LCs,LSCs) were prepared. Hydrogel samples had a diameter of 5 mm and a thickness of 1 mm. Samples were measured following swelling in IMDM containing 10% FCS (v/v), 100 units/mL of penicillin and 100 μg/mL of streptomycin, pH 7.4 at 37°C. The medium surrounding the hydrogels was carefully removed before measurement on day 1, 4, 7, 11 and 14. G′ was measured at regular intervals, rheological analysis was used as an indirect measurement of the degradation of PEG2 over time, with and without the presence of cells. Three hydrogels of each type were measured for each time point.
Flow cytometric characterization
Non-adherent cells were collected from the 2D B-cells co-cultures. 3D B-cell co-cultures were decrosslinked at day 4, 7, 11 and 14 using a 250 U/mL Collagenase Type 1 (17,018,029, Thermo Fisher) dissolved in B-cell medium, and incubated for 1 h at 37°C. Cells were re-suspended in FACS buffer containing antibodies and incubated at 4°C in the dark for 30 min. After incubation, cells were washed 2 times and then re-suspended using FACS buffer. Flow cytometric data was acquired using a FACS LSR Fortessa (Becton Dickinson).
Anti-human antibodies used for flow cytometry are: anti-CD19 (1:300, PE-Cy7, J3-119, Beckman Coulter), anti-CD19 (1:250, BUV395, SJ25C1, BD biosciences), anti-CD20 (1:150, BV605, 2H7, Biolegend), anti-CD27 (1:100, BV421, M-T271, BD Biosciences), anti-CD38 (1:150, BV786, HIT2, BD Biosciences), anti-CD45 (1:150, APC-R700, HI30, BD Biosciences), anti-CD138 (1:150, PE, MI15, Biolegend), anti-CD154 (1:200, AF647, 24–31, Biolegend), anti-IgA (1:100, APC, IS11-8E10, Miltenyi Biotec), anti-IgD (1:100, BV510, IA6-2, Biolegend), anti-IgG Fc (1:150, APC-Fire 750, M1310G05, Biolegend), anti-IgM (1:100, FITC, MHM-88, Biolegend). To exclude dead cells, 7AAD (1:200, 559,925, BD Biosciences) or Fixable Viability Stain 700 (1:4000, 564,997, BD Horizon, BD Biosciences) was added. Proliferating cells were identified using 5 μM Cell Trace Blue according to manufacturer’s protocol (C34568, Thermo Fisher).
ViSNE analyses
Biological and technical triplicates were pooled per condition, 10,000 cells were uniformly subsampled from this data. Generated figures show color coding by marker expression levels, the overall dimensionally reduced single-cell viSNE data show clustering of naive, activated and differentiated B-cell subsets as well as B-cell subclasses in the three pooled donors at different time points comparing 2D versus 3D cultures, or naive versus activated B-cells.
Immunocytochemistry
At day 14, 2D and 3D cultures were fixated in 4% (v/v) buffered formaldehyde (VWR Chemicals) and washed with PBS. Heat-mediated antigen retrieval was performed on all samples, for 15 min at 90°C in 0.5M EDTA buffer pH 8.0 (Invitrogen, ThermoFisher). Cells were permeabilized using 0.2% (v/v) Triton X-100 (Merck Millipore) in PBS for 10 min and washed with PBS. Samples were then blocked with 50 mg/mL BSA in PBS for 1 h at room temperature (RT) and subsequently washed in PBS. Immunodetection was performed by incubation with purified anti-human CD19 antibody (10 μg/mL, SJ25C1, Biolegend) or purified anti-human CD138 antibody (10 μg/mL, MI15, Biolegend) overnight at RT, followed by biotinylated sheep anti-mouse (1:200, RPN1001; GE Healthcare) for 1 h at RT, and Alexa Fluor 488 anti-streptavidin (4 μg/mL, S32354; Thermo Fisher Scientific) for 1 h at RT, all in PBS containing 50 mg/mL BSA and incubated on a plate shaker (150 rpm). All antibodies were centrifuged before use (13,000 rpm for 5 min). Between staining steps, samples were washed in 0.1% Tween 20 (v/v, Merck Millipore) in PBS. Samples were then stained for Phalloidin–Atto 700 (0.4 nmol/mL, 79,286, Sigma Aldrich) and DAPI (100 ng/mL, D9542, Sigma Aldrich), washed in PBS and mounted using Vectashield (Vector Laboratories).
Fluorescent imaging
Fluorescent images were taken with a Leica DMi8 Fluorescence Microscope using a Leica EL6000 light source and Leica LASX acquisition software. Four detection channels collected the fluorescent signal from the used fluorochromes at the following wavelengths: calcein (527/30) and ethidium homodimer-1 (605/75), which were given the pseudocolors green and red, or DAPI (460/50), Alexa Fluor 488 (527/30), DiI (605/75), and Phalloidin–Atto 700 (700/75), respectively, which were given the pseudocolors blue, green, yellow, and red.
Antibody measurements
IgA, IgM and total IgG antibody concentrations in the culture supernatant were determined using a fluorescent bead-based multiplex immunoassay using Luminex technology. Culture supernatant was diluted to 1:5 (day 4 and 7) and 1:15 (day 11 and 14) in assay buffer. Dilutions were made in PBS containing 0.1% (v/v) Tween 20 and 3% (w/v) BSA. A standard curve, controls (67/099, the National Institute for Biological Standards and Control (NIBSC)) and blanks were included on each plate next to supernatants (all measured in duplicate). Beads were incubated for 45 min at RT in the dark under constant rotation at 25 rpm and subsequently washed 3 times with PBS. After the final washing step, beads were re-suspended in 100 μL PBS and shaken before analysis in a Bio-Plex 200 instrument (Bio-Rad Laboratories).
Quantification and statistical analysis
Flow cytometric data was analyzed with FlowJo v10.6.2 software. viSNE analyses of flow cytometric data were performed in cytobank premium (https://premium.cytobank.org).64 All runs used an identical random seed and the default t-SNE parameters (iterations = 1000, perplexity = 30, theta = 0.5). Fluorescent z stack images were processed using ImageJ 1.53c software to create single maximum projections. Image overviews of the 3D cultures were merged using the mosaic function of the Leica LASX software, stitching the images together using smooth and linear blending. Multiplex immunoassay data was analyzed using Bio-Plex Manager (version 4.1.1) software (Bio-Rad) with a 5-parameter fit.
All experimental groups were performed in technical triplicates. Results are presented as mean ± SD for the indicated number of donors (n) in the figure legends. p values are based on an ordinary two-way ANOVA (two-way ANOVA) for multiple hypothesis, using either Tukey’s multiple comparisons post hoc test when comparing multiple outcomes in 3 experimental conditions or more, or Sidak’s multiple comparisons post hoc test when comparing multiple outcomes in 2 experimental conditions. Data analysis was performed using GraphPad Prism 9.3.1. software. In all tests, p values <0.05 were considered statistically significant. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Acknowledgments
We would like to thank: Mylène de Ruijter (Department of Orthopedics, University Medical Center Utrecht, the Netherlands) for her help measuring the mechanical properties of the PEG-4MAL gels. Reza Nadafi (Department of Molecular Cell Biology and Immunology, Amsterdam University Medical Center, the Netherlands) for the isolation of LSCs. Martien Poelen and Maarten Emmelot for the radiation of CD40LCs. Pieter Dingemanse, Gaby Smits, and Pieter van Gageldonk for their assistance in the set up and usage of the fluorescent bead-based multiplex immunoassay. Nening Nanlohy for her help measuring culture supernatants and the subsequent analysis (all Center for Infectious Disease Control, National Institute for Public Health and the Environment, the Netherlands).
The graphical abstract and cartoons shown in Figures 1, 2, 3, and 4 were created with BioRender.com
This research is part of the Target-to-B consortium, a collaboration project that is financed by the PPP Allowance made available by Top Sector Life Sciences & Health to Samenwerkende Gezondheidsfondsen (SGF) in the Netherlands under projectnumber LSHM18055-SGF to stimulate public-private partnerships and co-financing by health foundations that are part of the SGF. We acknowledge the support of patient partners, private partners, and active colleagues of the Target-to-B consortium (see website: www.target-to-b.nl). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the article.
Author contributions
M.V.J.B. designed and carried out the experiments and analyzed the data. R.S.V.B., A.M.M.B., R.E.M., J.D.W., and C.A.C.M.V.E. supported data analyzation. M.V.J.B. wrote the article. R.S.V.B., A.M.M.B., R.E.M., J.D.W., and C.A.C.M.V.E. revised the article. J.D.W. and C.A.C.M.V.E. supervised the experiments.
Declaration of interests
The authors declare no competing interests.
Published: January 20, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105741.
Contributor Information
Jelle de Wit, Email: jelle.de.wit@rivm.nl.
Cécile A.C.M. van Els, Email: cecile.van.els@rivm.nl.
Supplemental information
Data and code availability
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This article does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Stebegg M., Kumar S.D., Silva-Cayetano A., Fonseca V.R., Linterman M.A., Graca L. Regulation of the germinal center response. Front. Immunol. 2018;9:2469. doi: 10.3389/fimmu.2018.02469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klein U., Dalla-Favera R. Germinal centres: role in B-cell physiology and malignancy. Nat. Rev. Immunol. 2008;8:22–33. doi: 10.1038/nri2217. [DOI] [PubMed] [Google Scholar]
- 3.Roco J.A., Mesin L., Binder S.C., Nefzger C., Gonzalez-Figueroa P., Canete P.F., Ellyard J., Shen Q., Robert P.A., Cappello J., et al. Class-switch recombination occurs infrequently in germinal centers. Immunity. 2019;51:337–350.e7. doi: 10.1016/j.immuni.2019.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gatto D., Brink R. The germinal center reaction. J. Allergy Clin. Immunol. 2010;126:898–907. doi: 10.1016/j.jaci.2010.09.007. quiz 908-909. [DOI] [PubMed] [Google Scholar]
- 5.Mueller S.N., Germain R.N. Stromal cell contributions to the homeostasis and functionality of the immune system. Nat. Rev. Immunol. 2009;9:618–629. doi: 10.1038/nri2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castaños-Velez E., Biberfeld P., Patarroyo M. Extracellular matrix proteins and integrin receptors in reactive and non-reactive lymph nodes. Immunology. 1995;86:270–278. [PMC free article] [PubMed] [Google Scholar]
- 7.Cosgrove J., Novkovic M., Albrecht S., Pikor N.B., Zhou Z., Onder L., Mörbe U., Cupovic J., Miller H., Alden K., et al. B cell zone reticular cell microenvironments shape CXCL13 gradient formation. Nat. Commun. 2020;11:3677. doi: 10.1038/s41467-020-17135-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jourdan M., Caraux A., De Vos J., Fiol G., Larroque M., Cognot C., Bret C., Duperray C., Hose D., Klein B. An in vitro model of differentiation of memory B cells into plasmablasts and plasma cells including detailed phenotypic and molecular characterization. Blood. 2009;114:5173–5181. doi: 10.1182/blood-2009-07-235960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jourdan M., Cren M., Robert N., Bolloré K., Fest T., Duperray C., Guilloton F., Hose D., Tarte K., Klein B. IL-6 supports the generation of human long-lived plasma cells in combination with either APRIL or stromal cell-soluble factors. Leukemia. 2014;28:1647–1656. doi: 10.1038/leu.2014.61. [DOI] [PubMed] [Google Scholar]
- 10.Cocco M., Stephenson S., Care M.A., Newton D., Barnes N.A., Davison A., Rawstron A., Westhead D.R., Doody G.M., Tooze R.M. In vitro generation of long-lived human plasma cells. J. Immunol. 2012;189:5773–5785. doi: 10.4049/jimmunol.1103720. [DOI] [PubMed] [Google Scholar]
- 11.Brynjolfsson S.F., Persson Berg L., Olsen Ekerhult T., Rimkute I., Wick M.J., Mårtensson I.L., Grimsholm O. Long-lived plasma cells in mice and men. Front. Immunol. 2018;9:2673. doi: 10.3389/fimmu.2018.02673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bergmann B., Grimsholm O., Thorarinsdottir K., Ren W., Jirholt P., Gjertsson I., Mårtensson I.L. Memory B cells in mouse models. Scand. J. Immunol. 2013;78:149–156. doi: 10.1111/sji.12073. [DOI] [PubMed] [Google Scholar]
- 13.Steiniger B.S. Human spleen microanatomy: why mice do not suffice. Immunology. 2015;145:334–346. doi: 10.1111/imm.12469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ravi M., Paramesh V., Kaviya S.R., Anuradha E., Solomon F.D.P. 3D cell culture systems: advantages and applications. J. Cell. Physiol. 2015;230:16–26. doi: 10.1002/jcp.24683. [DOI] [PubMed] [Google Scholar]
- 15.Mazzoleni G., Di Lorenzo D., Steimberg N. Modelling tissues in 3D: the next future of pharmaco-toxicology and food research? Genes Nutr. 2009;4:13–22. doi: 10.1007/s12263-008-0107-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burger J.A., Gribben J.G. The microenvironment in chronic lymphocytic leukemia (CLL) and other B cell malignancies: insight into disease biology and new targeted therapies. Semin. Cancer Biol. 2014;24:71–81. doi: 10.1016/j.semcancer.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 17.ten Hacken E., Burger J.A. Microenvironment dependency in Chronic Lymphocytic Leukemia: the basis for new targeted therapies. Pharmacol. Ther. 2014;144:338–348. doi: 10.1016/j.pharmthera.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 18.Nair J.R., Rozanski C.H., Lee K.P. Under one roof: the bone marrow survival niche for multiple myeloma and normal plasma cells. OncoImmunology. 2012;1:388–389. doi: 10.4161/onci.18746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu H., Wang Y., Cui K., Guo Y., Zhang X., Qin J. Advances in hydrogels in organoids and organs-on-a-chip. Adv. Mater. 2019;31:e1902042. doi: 10.1002/adma.201902042. [DOI] [PubMed] [Google Scholar]
- 20.Jensen C., Teng Y. Is it time to start transitioning from 2D to 3D cell culture? Front. Mol. Biosci. 2020;7:33. doi: 10.3389/fmolb.2020.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gomez-Florit M., Pardo A., Domingues R.M.A., Graça A.L., Babo P.S., Reis R.L., Gomes M.E. Natural-based hydrogels for tissue engineering applications. Molecules. 2020;25:5858. doi: 10.3390/molecules25245858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gjorevski N., Sachs N., Manfrin A., Giger S., Bragina M.E., Ordóñez-Morán P., Clevers H., Lutolf M.P. Designer matrices for intestinal stem cell and organoid culture. Nature. 2016;539:560–564. doi: 10.1038/nature20168. [DOI] [PubMed] [Google Scholar]
- 23.Cruz-Acuña R., García A.J. Synthetic hydrogels mimicking basement membrane matrices to promote cell-matrix interactions. Matrix Biol. 2017;57–58:324–333. doi: 10.1016/j.matbio.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gjorevski N., Lutolf M.P. Synthesis and characterization of well-defined hydrogel matrices and their application to intestinal stem cell and organoid culture. Nat. Protoc. 2017;12:2263–2274. doi: 10.1038/nprot.2017.095. [DOI] [PubMed] [Google Scholar]
- 25.Cruz-Acuña R., Quirós M., Farkas A.E., Dedhia P.H., Huang S., Siuda D., García-Hernández V., Miller A.J., Spence J.R., Nusrat A., García A.J. Synthetic hydrogels for human intestinal organoid generation and colonic wound repair. Nat. Cell Biol. 2017;19:1326–1335. doi: 10.1038/ncb3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cruz-Acuña R., Quirós M., Huang S., Siuda D., Spence J.R., Nusrat A., García A.J. PEG-4MAL hydrogels for human organoid generation, culture, and in vivo delivery. Nat. Protoc. 2018;13:2102–2119. doi: 10.1038/s41596-018-0036-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Graney P.L., Lai K., Post S., Brito I., Cyster J., Singh A. Organoid polymer functionality and mode of Klebsiella pneumoniae membrane antigen presentation regulates ex vivo germinal center epigenetics in young and aged B cells. Adv. Funct. Mater. 2020;30:2001232. doi: 10.1002/adfm.202001232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Purwada A., Jaiswal M.K., Ahn H., Nojima T., Kitamura D., Gaharwar A.K., Cerchietti L., Singh A. Ex vivo engineered immune organoids for controlled germinal center reactions. Biomaterials. 2015;63:24–34. doi: 10.1016/j.biomaterials.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Purwada A., Singh A. Immuno-engineered organoids for regulating the kinetics of B-cell development and antibody production. Nat. Protoc. 2017;12:168–182. doi: 10.1038/nprot.2016.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Purwada A., Shah S.B., Béguelin W., August A., Melnick A.M., Singh A. Ex vivo synthetic immune tissues with T cell signals for differentiating antigen-specific, high affinity germinal center B cells. Biomaterials. 2019;198:27–36. doi: 10.1016/j.biomaterials.2018.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wagar L.E., Salahudeen A., Constantz C.M., Wendel B.S., Lyons M.M., Mallajosyula V., Jatt L.P., Adamska J.Z., Blum L.K., Gupta N., et al. Modeling human adaptive immune responses with tonsil organoids. Nat. Med. 2021;27:125–135. doi: 10.1038/s41591-020-01145-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goyal G., Prabhala P., Mahajan G., Bausk B., Gilboa T., Xie L., Zhai Y., Lazarovits R., Mansour A., Kim M.S., et al. Ectopic lymphoid follicle formation and human seasonal influenza vaccination responses recapitulated in an organ-on-a-chip. Adv. Sci. 2022;9:e2103241. doi: 10.1002/advs.202103241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hughes C.S., Postovit L.M., Lajoie G.A. Matrigel: a complex protein mixture required for optimal growth of cell culture. Proteomics. 2010;10:1886–1890. doi: 10.1002/pmic.200900758. [DOI] [PubMed] [Google Scholar]
- 34.Heinrich M.A., Mangia M., Prakash J. Impact of endotoxins on bioengineered tissues and models. Trends Biotechnol. 2022;40:532–534. doi: 10.1016/j.tibtech.2021.12.001. [DOI] [PubMed] [Google Scholar]
- 35.Braham M.V.J., Minnema M.C., Aarts T., Sebestyen Z., Straetemans T., Vyborova A., Kuball J., Öner F.C., Robin C., Alblas J. Cellular immunotherapy on primary multiple myeloma expanded in a 3D bone marrow niche model. OncoImmunology. 2018;7:e1434465. doi: 10.1080/2162402X.2018.1434465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Braham M.V.J., Ahlfeld T., Akkineni A.R., Minnema M.C., Dhert W.J.A., Öner F.C., Robin C., Lode A., Gelinsky M., Alblas J. Endosteal and perivascular subniches in a 3D bone marrow model for multiple myeloma. Tissue Eng. Part C Methods. 2018;24:300–312. doi: 10.1089/ten.TEC.2017.0467. [DOI] [PubMed] [Google Scholar]
- 37.Braham M.V., Deshantri A.K., Minnema M.C., Öner F.C., Schiffelers R.M., Fens M.H., Alblas J. Liposomal drug delivery in an in vitro 3D bone marrow model for multiple myeloma. Int. J. Nanomed. 2018;13:8105–8118. doi: 10.2147/IJN.S184262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Braham M.V.J., Alblas J., Dhert W.J.A., Öner F.C., Minnema M.C. Possibilities and limitations of an in vitro 3D bone marrow model for the prediction of clinical responses in patients with relapsed multiple myeloma. Haematologica. 2019;104:e523–e526. doi: 10.3324/haematol.2018.213355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bar-Ephraim Y.E., Konijn T., Gönültas M., Mebius R.E., Reijmers R.M. A reproducible method for isolation and in vitro culture of functional human lymphoid stromal cells from tonsils. PLoS One. 2016;11:e0167555. doi: 10.1371/journal.pone.0167555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chu V.T., Berek C. The establishment of the plasma cell survival niche in the bone marrow. Immunol. Rev. 2013;251:177–188. doi: 10.1111/imr.12011. [DOI] [PubMed] [Google Scholar]
- 41.Ding C., Chen X., Dascani P., Hu X., Bolli R., Zhang H.G., McLeish K.R., Yan J. STAT3 signaling in B cells is critical for germinal center maintenance and contributes to the pathogenesis of murine models of lupus. J. Immunol. 2016;196:4477–4486. doi: 10.4049/jimmunol.1502043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Westerhuis B., Ten Hulscher H., Jacobi R., van Beek J., Koopmans M., Rimmelzwaan G., Meijer A., van Binnendijk R. Specific memory B cell response in humans upon infection with highly pathogenic H7N7 avian influenza virus. Sci. Rep. 2020;10:3152. doi: 10.1038/s41598-020-60048-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marsman C., Verhoeven D., Koers J., Rispens T., Ten Brinke A., van Ham S.M., Kuijpers T.W. Optimized protocols for in-vitro T-cell-dependent and T-cell-independent activation for B-cell differentiation studies using limited cells. Front. Immunol. 2022;13:815449. doi: 10.3389/fimmu.2022.815449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jackson S.M., Harp N., Patel D., Wulf J., Spaeth E.D., Dike U.K., James J.A., Capra J.D. Key developmental transitions in human germinal center B cells are revealed by differential CD45RB expression. Blood. 2009;113:3999–4007. doi: 10.1182/blood-2008-03-145979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kapp T.G., Rechenmacher F., Neubauer S., Maltsev O.V., Cavalcanti-Adam E.A., Zarka R., Reuning U., Notni J., Wester H.J., Mas-Moruno C., et al. A comprehensive evaluation of the activity and selectivity profile of ligands for RGD-binding integrins. Sci. Rep. 2017;7:39805. doi: 10.1038/srep39805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hersel U., Dahmen C., Kessler H. RGD modified polymers: biomaterials for stimulated cell adhesion and beyond. Biomaterials. 2003;24:4385–4415. doi: 10.1016/s0142-9612(03)00343-0. [DOI] [PubMed] [Google Scholar]
- 47.Spaargaren M., Beuling E.A., Rurup M.L., Meijer H.P., Klok M.D., Middendorp S., Hendriks R.W., Pals S.T. The B cell antigen receptor controls integrin activity through Btk and PLCgamma2. J. Exp. Med. 2003;198:1539–1550. doi: 10.1084/jem.20011866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Donahue A.C., Fruman D.A. Proliferation and survival of activated B cells requires sustained antigen receptor engagement and phosphoinositide 3-kinase activation. J. Immunol. 2003;170:5851–5860. doi: 10.4049/jimmunol.170.12.5851. [DOI] [PubMed] [Google Scholar]
- 49.Minges Wols H.A., Underhill G.H., Kansas G.S., Witte P.L. The role of bone marrow-derived stromal cells in the maintenance of plasma cell longevity. J. Immunol. 2002;169:4213–4221. doi: 10.4049/jimmunol.169.8.4213. [DOI] [PubMed] [Google Scholar]
- 50.He L., Gu W., Wang M., Chang X., Sun X., Zhang Y., Lin X., Yan C., Fan W., Su P., et al. Extracellular matrix protein 1 promotes follicular helper T cell differentiation and antibody production. Proc. Natl. Acad. Sci. USA. 2018;115:8621–8626. doi: 10.1073/pnas.1801196115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marsman C., Jorritsma T., Ten Brinke A., van Ham S.M. Flow cytometric methods for the detection of intracellular signaling proteins and transcription factors reveal heterogeneity in differentiating human B cell subsets. Cells. 2020;9:2633. doi: 10.3390/cells9122633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Amé-Thomas P., Maby-El Hajjami H., Monvoisin C., Jean R., Monnier D., Caulet-Maugendre S., Guillaudeux T., Lamy T., Fest T., Tarte K. Human mesenchymal stem cells isolated from bone marrow and lymphoid organs support tumor B-cell growth: role of stromal cells in follicular lymphoma pathogenesis. Blood. 2007;109:693–702. doi: 10.1182/blood-2006-05-020800. [DOI] [PubMed] [Google Scholar]
- 53.Bar-Ephraim Y.E., Kretzschmar K., Clevers H. Organoids in immunological research. Nat. Rev. Immunol. 2020;20:279–293. doi: 10.1038/s41577-019-0248-y. [DOI] [PubMed] [Google Scholar]
- 54.Clevers H. Modeling development and disease with organoids. Cell. 2016;165:1586–1597. doi: 10.1016/j.cell.2016.05.082. [DOI] [PubMed] [Google Scholar]
- 55.Drost J., Clevers H. Organoids in cancer research. Nat. Rev. Cancer. 2018;18:407–418. doi: 10.1038/s41568-018-0007-6. [DOI] [PubMed] [Google Scholar]
- 56.Kleinman H.K., Martin G.R. Matrigel: basement membrane matrix with biological activity. Semin. Cancer Biol. 2005;15:378–386. doi: 10.1016/j.semcancer.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 57.Schuurmans C.C.L., Mihajlovic M., Hiemstra C., Ito K., Hennink W.E., Vermonden T. Hyaluronic acid and chondroitin sulfate (meth)acrylate-based hydrogels for tissue engineering: synthesis, characteristics and pre-clinical evaluation. Biomaterials. 2021;268:120602. doi: 10.1016/j.biomaterials.2020.120602. [DOI] [PubMed] [Google Scholar]
- 58.Braham M.V.J., Li Yim A.S.P., Garcia Mateos J., Minnema M.C., Dhert W.J.A., Öner F.C., Robin C., Alblas J. A human hematopoietic niche model supporting hematopoietic stem and progenitor cells in vitro. Adv. Healthc. Mater. 2019;8:e1801444. doi: 10.1002/adhm.201801444. [DOI] [PubMed] [Google Scholar]
- 59.Nguyen D.C., Garimalla S., Xiao H., Kyu S., Albizua I., Galipeau J., Chiang K.Y., Waller E.K., Wu R., Gibson G., et al. Factors of the bone marrow microniche that support human plasma cell survival and immunoglobulin secretion. Nat. Commun. 2018;9:3698. doi: 10.1038/s41467-018-05853-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu Y.J., Malisan F., de Bouteiller O., Guret C., Lebecque S., Banchereau J., Mills F.C., Max E.E., Martinez-Valdez H. Within germinal centers, isotype switching of immunoglobulin genes occurs after the onset of somatic mutation. Immunity. 1996;4:241–250. doi: 10.1016/s1074-7613(00)80432-x. [DOI] [PubMed] [Google Scholar]
- 61.Hiepe F., Dörner T., Hauser A.E., Hoyer B.F., Mei H., Radbruch A. Long-lived autoreactive plasma cells drive persistent autoimmune inflammation. Nat. Rev. Rheumatol. 2011;7:170–178. doi: 10.1038/nrrheum.2011.1. [DOI] [PubMed] [Google Scholar]
- 62.Garrone P., Neidhardt E.M., Garcia E., Galibert L., van Kooten C., Banchereau J. Fas ligation induces apoptosis of CD40-activated human B lymphocytes. J. Exp. Med. 1995;182:1265–1273. doi: 10.1084/jem.182.5.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baas D.C., Koopmans M.P., de Bruin E., ten Hulscher H.I., Buisman A.M., Hendrikx L.H., van Beek J., Godeke G.J., Reimerink J., van Binnendijk R.S. Detection of influenza A virus homo- and heterosubtype-specific memory B-cells using a novel protein microarray-based analysis tool. J. Med. Virol. 2013;85:899–909. doi: 10.1002/jmv.23535. [DOI] [PubMed] [Google Scholar]
- 64.Amir E.a.D., Davis K.L., Tadmor M.D., Simonds E.F., Levine J.H., Bendall S.C., Shenfeld D.K., Krishnaswamy S., Nolan G.P., Pe'er D. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat. Biotechnol. 2013;31:545–552. doi: 10.1038/nbt.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This article does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.