Abstract
The presence of mineral fluoride (F−) in the environment has both a geogenic and anthropogenic origin, and the halide has been described to be toxic in virtually all living organisms. While the evidence gathered in different microbial species supports this notion, a systematic exploration of the effects of F− salts on the metabolism and physiology of environmental bacteria remained underexplored thus far. In this work, we studied and characterized tolerance mechanisms deployed by the model soil bacterium Pseudomonas putida KT2440 against NaF. By adopting systems‐level omic approaches, including functional genomics and metabolomics, we gauged the impact of this anion at different regulatory levels under conditions that impair bacterial growth. Several genes involved in halide tolerance were isolated in a genome‐wide Tn‐Seq screening—among which crcB, encoding an F−‐specific exporter, was shown to play the predominant role in detoxification. High‐resolution metabolomics, combined with the assessment of intracellular and extracellular pH values and quantitative physiology experiments, underscored the key nodes in central carbon metabolism affected by the presence of F−. Taken together, our results indicate that P. putida undergoes a general, multi‐level stress response when challenged with NaF that significantly differs from that caused by other saline stressors. While microbial stress responses to saline and oxidative challenges have been extensively studied and described in the literature, very little is known about the impact of fluoride (F−) on bacterial physiology and metabolism. This state of affairs contrasts with the fact that F− is more abundant than other halides in the Earth crust (e.g. in some soils, the F− concentration can reach up to 1 mg gsoil −1). Understanding the global effects of NaF treatment on bacterial physiology is not only relevant to unveil distinct mechanisms of detoxification but it could also guide microbial engineering approaches for the target incorporation of fluorine into value‐added organofluorine molecules. In this regard, the soil bacterium P. putida constitutes an ideal model to explore such scenarios, since this species is particularly known for its high level of stress resistance against a variety of physicochemical perturbations.
INTRODUCTION
Fluorine (F), the most electronegative and reactive element of the periodic table, is widely found either in the form of mineral fluoride (F−) salts, including the free F− anion itself, or as organofluorine compounds (Cheng & Ma, 2021; Greenwood & Earnshaw, 1984; Gribble, 2002). Estimated to be the 13th most abundant element in the Earth crust, F is widespread in soils and water (Chahine et al., 2022; Mason & Moore, 1982). Although the concentration of F− tends to be rather low in unpolluted areas, its abundance can locally increase to rather significant levels as a result of natural phenomena, e.g. volcanic or geothermal activity. Moreover, the presence of organofluorine compounds in multiple industrial applications had led to human activity‐related increases of the F− concentration in natural environments (Harsanyi & Sandford, 2015; Wackett, 2021a). This is the case of polluted waters derived from untreated industrial waste that comprises, among other examples, effluents from chemical plants manufacturing organofluorine compounds or aluminium smelters (Camargo, 2003; Muini et al., 2012). F− is likewise used as an additive in dental hygiene products due to its multiple (yet controversial) beneficial effects—which include reducing the incidence of cavities, promoting enamel re‐mineralization and the general antimicrobial effect of the anion on the oral microbiota (Bradshaw et al., 1990; Johnston & Strobel, 2020).
Over the years, the toxicity of F− has been studied in a variety of organisms—from algae and other aquatic species (Bhatnagar & Bhatnagar, 2004; Camargo, 2003) to mixed bacterial populations, e.g. dental biofilms, wastewater communities or even in the rat microbiome (Bradshaw et al., 2002; Dionizio et al., 2021; Hamilton, 1990; Marquis, 1995; Ochoa‐Herrera et al., 2009). The impact of F− on microbial physiology has been likewise assessed in individual bacterial species that form part of the oral and skin microbiota, e.g. Streptococcus mutans (Cai et al., 2017; Damé‐Teixeira et al., 2019; Liao et al., 2017), Bifidobacterium sp. (Manome et al., 2019) and Streptococcus pyogenes (Thongboonkerd et al., 2002), as well as in natural isolates of Enterobacter cloacae (Liu et al., 2017) endowed with high tolerance towards F− salts. A conclusion shared by these studies is that F− negatively affects the performance of a number of key enzymes in central carbon metabolism. A critical example of this sort is the inhibition of both enolase and F‐ATPase, although F− salts also exert an inhibitory impact on pyrophosphatases and catalases (Baykov et al., 2000; Li, 2003; Ma et al., 2014; Marquis et al., 2003; Qin et al., 2006; Samygina et al., 2007; Wackett, 2021b, 2021c). Considering this circumstance, it does not come as a surprise that bacteria evolved F− tolerance mechanisms, the most widespread of which is represented by specific membrane exporters that keep low intracellular F− levels (Baker et al., 2012; McIlwain et al., 2021). Two unrelated classes of F−‐detoxifying transporters have been described thus far: (i) the CLCF‐type F−/H+ antiporter, a subclass of the widespread CLC (voltage‐gated chloride channel) superfamily of anion‐transport proteins (Stockbridge et al., 2012) that includes the EriCF transporter of Pseudomonas syringae and (ii) a group of small membrane proteins generically known as the ‘Fluc’ family, which comprises the CrcB F− channel of Escherichia coli (Last et al., 2016; Stockbridge et al., 2013). Additionally, mineral F− is released during microbial biodegradation of organofluorines (Bygd et al., 2021; Neilson & Allard, 2002; Wackett, 2021a; Wackett, 2022; Wirth & Nikel, 2021), and anion export seems to play a role in mitigating its toxic effects under biodegradation conditions.
Recent developments in synthetic biology have enlarged the portfolio of building blocks produced by engineered bacteria to include organofluorines (Cheng & Ma, 2021; Cros et al., 2022; Wu et al., 2020). We have recently engineered the model environmental bacterium Pseudomonas putida towards production of fluorometabolites from mineral F− (Calero et al., 2020; Pardo et al., 2022)—yet the effects of these salts on the bacterial physiology remain unstudied. Pseudomonas putida is well known for its tolerance towards different stress conditions, e.g. solvents (Bitzenhofer et al., 2021; Kieboom et al., 1998; Mosqueda et al., 1999) and toxic aromatic compounds (Calero et al., 2018; Fernández‐Cabezón et al., 2022), and has garnered recognition as a robust bacterial cell factory (Nikel et al., 2016a; Volke et al., 2020a, 2022; Weimer et al., 2020). Thus, understanding the impact of F− on the metabolism of P. putida has broad interest not only from a fundamental perspective but also in terms of metabolic engineering aimed at organofluorine biosynthesis. Hence, in this work, we attempted to elucidate the effects of F− on the physiology and metabolism of P. putida KT2440 using a multi‐omic approach that includes transposon insertion sequencing (Tn‐Seq), genetic dissection of key functions relevant for salt tolerance, quantitative physiology experiments and network‐wide, high‐resolution metabolomic profiling. Our results indicate that the F− ion exerts a multi‐level effect on P. putida, unique and different when compared to that brought about by other halides, which leads to a significant restructuring of central carbon metabolism towards counteracting the toxic effects of fluoride salts.
EXPERIMENTAL PROCEDURES
Bacterial strains, plasmids and culture conditions
The bacterial strains and plasmids employed in this study are listed in Table 1. Unless otherwise stated, P. putida KT2440 was routinely grown at 30°C in lysogeny broth (LB) medium or onto LB agar plates according to standard protocols (Sambrook & Russell, 2001). M9 minimal medium containing 5 g L−1 glucose as the only carbon source (Abril et al., 1989; Nikel et al., 2016b; Sánchez‐Pascuala et al., 2019) was used for all the gene expression and microbial physiology assays. Escherichia coli DH5α was used as a host for cloning and plasmid maintenance and was grown in LB at 37°C. When relevant, antibiotics were used at the following final concentrations: chloramphenicol (Cm), 30 μg ml−1; kanamycin (Km), 50 μg ml−1; gentamicin (Gm), 10 μg ml−1; and streptomycin (Str), 100 μg ml−1. Electrocompetent P. putida cells were prepared by washing the biomass from an overnight culture (in LB medium) with 300 mM sucrose (Volke et al., 2020b, 2020c). Sodium fluoride (NaF) was purchased from Sigma‐Aldrich (St. Louis, MO, USA; cat. # 201154) and used as stressor and inducer of the fluoride‐responsive riboswitch (FRS) as indicated in the text.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics a | References |
|---|---|---|
| Strains | ||
| E. coli DH5α | Cloning host; F− λ− endA1 glnX44(AS) thiE1 recA1 relA1 spoT1 gyrA96(NalR) rfbC1 deoR nupG Φ80(lacZΔM15) Δ(argF‐lac)U169 hsdR17(r K − m K +) | Meselson and Yuan (1968) |
| P. putida KT2440 | Wild‐type strain; derivative of P. putida mt‐2 cured of the TOL plasmid pWW0, hsdR1 | Bagdasarian et al. (1981) |
| P. putida KT2440::mini‐Tn7(Gm)::mini‐Tn5(Km)::GFP | P. putida KT2440 derivative, pooled library of mini‐Tn5 insertions in a strain inserted with a mini‐Tn7(Gm) and GFP+ background, KmR GmR | Calero et al. (2018) |
| P. putida ΔcrcB | P. putida KT2440 derivative, in‐frame deletion of crcB (PP_4001) | |
| P. putida ΔrarA | P. putida KT2440 derivative, in‐frame deletion of rarA (PP_4002) | This study |
| P. putida ΔprtR | P. putida KT2440 derivative, in‐frame deletion of prtR (PP_2889) | This study |
| P. putida ΔlapA | P. putida KT2440 derivative, in‐frame deletion of lapA (PP_0168) | This study |
| P. putida ΔlapD | P. putida KT2440 derivative, in‐frame deletion of lapD (PP_0165) | This study |
| P. putida ΔflhA | P. putida KT2440 derivative, in‐frame deletion of flhA (PP_4344) | This study |
| P. putida ΔyijP | P. putida KT2440 derivative, in‐frame deletion of yijP (PP_2579) | This study |
| P. putida Δorn | P. putida KT2440 derivative, in‐frame deletion of orn (PP_4902) | This study |
| P. putida ΔdusB | P. putida KT2440 derivative, in‐frame deletion of dusB (PP_4820) | This study |
| P. putida ΔvacB | P. putida KT2440 derivative, in‐frame deletion of vacB (PP_4880) | This study |
| Plasmids | ||
|---|---|---|
| pS441·FRSv1 | Plasmid‐borne fluoride biosensor based on a fluoride‐responsive riboswitch (FRS); derivative of vector pSEVA441 (Silva‐Rocha et al., 2013), FRSv1 → msfGFP; StrR | Calero et al. (2020) |
| pGNW2 | Plasmid for introducing genome deletions in Gram‐negative bacteria; derivative of vector pEMG (Martínez‐García & de Lorenzo, 2011) carrying P 14g → msfGFP; KmR | Wirth et al. (2020) |
| pGNW2·ΔrarA | Derivative of vector pGNW2 for deletion of rarA (PP_4002) in P. putida KT2440 | This study |
| pGNW2·ΔprtR | Derivative of vector pGNW2 for deletion of prtR (PP_2889) in P. putida KT2440 | This study |
| pGNW2·ΔlapA | Derivative of vector pGNW2 for deletion of lapA (PP_0168) in P. putida KT2440 | This study |
| pGNW2·ΔlapD | Derivative of vector pGNW2 for deletion of lapD (PP_0165) in P. putida KT2440 | This study |
| pGNW2·ΔflhA | Derivative of vector pGNW2 for deletion of flhA (PP_434) in P. putida KT2440 | This study |
| pGNW2·ΔyijP | Derivative of vector pGNW2 for deletion of yijP (PP_2579) in P. putida KT2440 | This study |
| pGNW2·Δorn | Derivative of vector pGNW2 for deletion of orn (PP_4902) in P. putida KT2440 | This study |
| pGNW2·ΔdusB | Derivative of vector pGNW2 for deletion of dusB (PP_4820) in P. putida KT2440 | This study |
| pGNW2·ΔvacB | Derivative of vector pGNW2 for deletion of vacB (PP_4880) in P. putida KT2440 | This study |
| pSEVA628S | Helper plasmid for genomic deletions; oriV(RK2), xylS, Pm → I‐SceI; GmR | Martínez‐García et al. (2015) |
| pS2513·PHP | Plasmid‐borne intracellular pH biosensor; derivative of vector pSEVA2513 (Silva‐Rocha et al., 2013), P EM7 → msfGFP; KmR | Arce‐Rodríguez et al. (2019) |
Antibiotic markers: Gm, gentamicin; Km, kanamycin; Nal, nalidixic acid; Rif, rifampicin; Str, Streptomycin.
General DNA manipulations and construction of plasmids and mutant strains
Plasmids for gene deletion of targets identified in the Tn‐Seq experiments were constructed by amplifying the upstream and downstream regions of the corresponding genes using the primers described in Table S1, and assembling the homology regions by USER cloning (Geu‐Flores et al., 2007) into the suicide plasmid pGNW2 (Wirth et al., 2020). Clean gene deletions in P. putida were constructed by I‐SceI meganuclease‐mediated homologous recombination as described by Volke et al. (2020b). The correctness of gene deletions was verified by DNA sequencing.
Fluoride toxicity assays
Overnight cultures of P. putida KT2440 and P. putida ΔcrcB were diluted 20 times in 10 ml of modified M9 minimal medium containing different NaF concentrations. Cells were grown in 96‐well microtiter plates (flat bottom; Greiner Bio‐One, Fredensborg, Denmark) at 30°C and medium agitation in an ELx808 microtiter plate reader (BioTek Instruments Inc., Winooski, VT, USA). Cells exposed to NaF at various concentrations were incubated in the microtiter plate reader for 24 h, and bacterial growth was followed by taking optical density at 600 nm (OD600) readings every 30 min. OD600 values after 20 h were selected to calculate and plot the minimal inhibitory concentrations (MICs) in connection to NaF treatments. The NaF concentrations used to this end were 0, 25, 50, 75, 125, 200 and 250 mM for P. putida KT2440, and 0, 0.1, 0.5, 1 and 5 mM for P. putida ΔcrcB. To analyse the growth profile of mutants identified in the Tn‐Seq experiments, overnight cultures of P. putida KT2440 and the corresponding mutants were diluted 20 times in 10 ml of modified M9 minimal medium containing NaF at 25 mM. Cultures were then incubated in 96‐well microtiter plates (flat bottom, Greiner Bio‐one) at 30°C and medium agitation in an ELx808 microtiter plate reader (BioTek Instruments Inc.), and growth was followed by measuring the OD600 every 30 min. OD600 values after 20 h of incubation were then selected to be plotted and to calculate MICs as indicated previously (Calero et al., 2020).
Analysis of growth curves and calculation of kinetic parameters
Growth curves of P. putida KT2440 for quantitative physiology experiments were assessed in batch cultures performed in 50 ml of M9 minimal medium in 250‐ml Erlenmeyer flasks at 30°C and with continuous shaking at 180 rpm. Overnight cultures, previously grown in M9 minimal medium with the corresponding carbon source, were diluted to obtain an initial OD600 of 0.1 in fresh M9 minimal medium or in the same medium containing NaF 50 mM. Cultures were incubated under the same conditions indicated above. Growth was followed by measuring the OD600 every hour. Specific growth rates [μ (h−1)] and lag phases [λ (h)] were calculated as described by Nikel et al. (2015a) using the following formulae:
where t ΔOD is the time t when an increase of OD600 up to 0.05 is first noticed in any given culture.
Tn5 transposon mutant library screening by growth‐based selection
A Tn5 mutant library derived from P. putida KT2440::mini‐Tn7(Gm), i.e. P. putida KT2440::mini‐Tn7(Gm)::mini‐Tn5(Km)::GFP (Calero et al., 2018), was used in growth‐based, Tn‐Seq selection experiments under NaF stressful conditions. Bacterial cryostocks, containing approximately individual 100,000 mutants (as determined by colony counting on LB agar plates), were thawed on ice, washed twice with fresh LB medium and grown overnight in 50 ml of LB with proper antibiotics at 30°C in 250‐ml Erlenmeyer flasks as indicated in the preceding section. A fraction of this overnight LB medium culture was used to inoculate 50 ml of M9 minimal medium with adequate antibiotics to a starting OD600 of 0.1 after washing the cells with M9 minimal medium or M9 minimal medium supplemented with NaF at 50 mM. Cells were incubated at 30°C and 200 rpm and bacterial growth was followed by measuring OD600 roughly every 2 h. When the cultures reached late exponential phase (at an OD600 of approximately 2), the cells were harvested and stored at −20°C prior to genome DNA (gDNA) extraction. Three independent biological replicates were carried out for each condition.
Library preparation for Tn‐Seq screening experiments
The gDNA libraries prepared from the control and NaF‐stressed cultures were obtained essentially as described by Lennen and Herrgård (2014). In brief, P. putida gDNA was extracted using the PureLink Genomic DNA kit (Life Technologies Corp., Carlsbad, CA, USA), and 3 μg of total gDNA was sheared in 300‐bp fragments using an E220‐focused ultrasonicator (Covaris LLC, Woburn, MA, USA). End‐repair (NEBNext End repair module; New England BioLabs, Ipswich, MA, USA) and A‐tailing using the Klenow fragment of the E. coli DNA polymerase I (Thermo Scientific, Waltham, MA, USA) was performed on the fragments prior to adding the adapters. Next, Illumina TruSeq adapters were ligated using the NEBNext Quick Ligation Module (New England BioLabs) and the transposon‐chromosome junctions were amplified by PCR with a primer specific for the adapter and a biotinylated primer specific for the corresponding mini‐Tn5 inserts. Biotinylated PCR products were subsequently purified by affinity capture using the Dyna MyOne streptavidin C1 beads (Thermo Scientific). Library size distribution was validated by qPCR, using the KAPA qPCR quantification kit, the Agilent RNA 6000 Pico kit and the Agilent 2100 Bioanalyzer (Agilent Technologies Inc., Santa Clara, CA, USA). Finally, the library was sequenced in an Illumina Inc. (San Diego, CA, USA) MiSeq™ benchtop sequencer.
Time‐course, high‐resolution metabolomic analysis of NaF‐treated P. putida cultures
Pre‐cultures of P. putida KT2440 and the ΔcrcB mutant were incubated in 50‐ml Falcon tubes containing 10 ml of M9 minimal medium supplemented with glucose as the carbon source. The experiments were set in a 100‐ml glass mini‐bioreactor with four lateral baffles to increase aeration. The temperature was maintained at 30°C by a water‐thermostatic double jacket, and the reactor setup was placed onto a magnetic agitator, keeping the stirrer speed at 800 rpm. An aliquot of the pre‐culture was inoculated in 50‐ml of M9 minimal medium (with the relevant additives) to an initial OD600 of 0.05. The biomass in the sample used as inoculum was centrifuged for 5 min at 5000g and washed twice with M9 minimal medium without any carbon source to remove carry‐over metabolites. When the cultures reached an OD600 = 0.5, three 1‐ml samples were taken as reference (termed t C in these experiments). At t C, NaF was added to the reactor at 50 mM for cultures of P. putida KT2440 and 1 mM for the ΔcrcB mutant. Next, samples were taken at 0 (i.e. immediately after NaF addition), 10, 20, 30, 45, 90, 180, 300, 600, 1800 and 5400 s. Each sample was rapidly filtrated in MF Millipore™ membrane filter (0.45‐μm pore size; Sigma‐Aldrich Co.) and the filter containing the bacterial biomass was placed onto a mini‐Petri dish with 1 ml of quenching solution [40% (vol./vol.) acetonitrile, 40% (vol./vol.) methanol and 100 mM formic acid] precooled to −20°C. The solution containing the quenched biomass was transferred into a 2‐ml tube; the filter was rinsed with an extra 1‐ml of quenching solution and collected in the same tube. Samples were promptly placed in a dry ice bath and kept under these conditions for 30 min. Next, the samples were thawed and centrifuged at 13,000g for 5 min and the supernatants were transferred to a new tube, where the solvents were evaporated in a SpeedVac centrifuge for ca. 2 h at 45°C until they were fully dried. Finally, the samples were stored at −80°C until analysis. Just prior to metabolite analysis, the samples were resuspended in 100 μl of LC–MS grade water. Two biological replicates were carried out as indicated above, with technical duplicates for each experiment.
Chromatographic separation of metabolites was done with an ACQUITY UPLC™ high‐strength silica T3 column (1.8 μm × 2.1 mm × 30 mm, Agilent Technologies Inc.) in an HPLC apparatus (Shimadzu; Columbia, MD, USA). A gradient of eluent A [10 mM acetic acid, 5% (vol./vol.) methanol, 10 mM tributylamine and 2% (vol./vol.) 2‐propanol] and 2‐propanol was implemented for metabolite separation as described by McCloskey et al. (2018). The flow rate was set to 0.5 ml min−1 with a total run time of 4.4 min; the autosampler was kept at 10°C and the column oven was set at 40°C with an injection volume of 10 μl. For metabolite identification and determination, a mass spectrometer (QTrap™ AB SCIEX mass spectrometer 5500) was operated in negative ion mode with the following settings: ionization set, −4500; temperature, 500°C; curtain gas, 45; collision gas, high; ion source gas, 1; and ion source gas pressure, 250 pound square inch−1. Peak intensities for metabolites within central carbon metabolism were obtained from this analysis by processing the raw mass spectrometry data through the SmartPeak workflow (Kozaeva et al., 2021; Kutuzova et al., 2020; Nikel et al., 2009). Metabolomics data analysis was carried out in Excel and the analysed data—metabolite fold change (FC)—were used as input to visualize the data in heatmap by using Seaborn Python package.
Assessing the intracellular F− concentration with a fluoride‐responsive fluorescent biosensor
Plasmid pS441·FRSv1 (Calero et al., 2020), carrying a monomeric superfolder gfp (msfGFP) gene under control of a synthetic riboswitch‐promoter element, was transformed into both the wild‐type strain and the P. putida deletion mutants constructed in this work and selected according to the Tn‐Seq analysis. The intracellular F− concentration in these strains was correlated to the msfGFP fluorescence output. To this end, overnight pre‐cultures of strains carrying plasmid pS441·FRSv1 were diluted 20 times in 5 ml of M9 minimal medium and the cell suspension was distributed in 96‐well microtiter plates (flat bottom; Greiner Bio‐One, Kremsmünster, Austria). Cells were grown for 3 h at 30°C with agitation at 300 rpm, after which the cultures were added with NaF at 0, 0.25, 1 and 15 mM. The kinetics of bacterial growth (estimated as the OD600) and msfGFP fluorescence, using wavelengths of excitation and emission of 485 and 528 nm, respectively, were followed in a SynergyMX microtiter plate reader (BioTek Instruments Inc.) for 20 h. Measurements were carried out every 10 min on three independent biological replicates, and the normalized fluorescence values at 10 and 15 h after NaF addition were plotted and compared across conditions.
Evaluating the impact of NaF on the intracellular pH homeostasis
Changes in the intracellular change of pH in the presence of NaF were studied by using a non‐invasive, pH‐sensitive fluorescence biosensor (an engineered variant of GFP) borne by plasmid pS2513·PHP (Arce‐Rodríguez et al., 2019). This plasmid was transformed into the wild‐type strain P. putida KT2440 and its ΔcrcB mutant derivative. Overnight pre‐cultures of these transformed strains were diluted 20 times in 10 ml of M9 minimal medium and distributed in 96‐well microtiter plates (flat bottom; Greiner Bio‐One). Cells were grown for 4 h at 30°C and 300 rpm, after which cultures of P. putida KT2440 were added with NaF at 0, 25 and 50 mM, whereas cultures of P. putida ΔcrcB were supplemented with NaF at 0, 0.5 and 1 mM. The kinetics of bacterial growth (estimated as the OD600) and ratiometric fluorescence determinations, using wavelengths of excitation and emission of 485 and 405 nm, respectively, were assessed in a SynergyMX microtiter plate reader (BioTek Instruments Inc.) for 20 h. Measurements were carried out every 10 min on three independent biological replicates, and the fluorescence ratios at 405 nm/485 nm were plotted and compared across conditions.
Drops assays were also performed to study the interplay between changes in the pH of the surrounding medium and F− toxicity. In this case, overnight cultures of P. putida KT2440 and the ΔcrcB mutant, grown in either LB medium or in M9 minimal medium, were serially diluted in the same fresh medium, and 10‐μl of the corresponding dilutions were spotted onto LB or M9 minimal medium agar plates supplemented with 10 μg ml−1 of phenol red (phenolsulfonphthalein) and, in some cases, NaF at 1 mM. Droplets were let dry out before placing the plates at 30°C for 24 or 48 h. Individual and combined cultures of the two strains were assayed in these experiments as indicated in the text.
Data and statistical analysis
All the experiments reported in this article were independently repeated at least twice (as indicated in the corresponding figure or table legend), and the mean value of the corresponding parameter ± standard deviation is presented. When relevant, the level of significance of the differences when comparing results was evaluated by means of the Student's t‐test with α = 0.01 or α = 0.05 as indicated in the figure legends.
Reads obtained from Illumina sequencing were initially processed to discard unspecific sequences and trim the length of the correct ones to 120 bp, leaving 25 bp of gDNA sequence that can be used for alignment and comparison to the P. putida KT2440 genome. Genes that were conditionally more abundant (or underrepresented) under a given experimental condition were identified using the ESSENTIALS pipeline (Zomer et al., 2012). Up to three mismatched nt were tolerated in the Tn barcode sequence, and a minimum 20‐nt sequence match was required for alignment. Insertions were ignored near the 3′‐end of genes, and gene‐level insertion counts were normalized across samples using trimmed mean of M values (TMM). A LOESS (i.e. locally weighted scatterplot smoothing) regression was also applied to correct the bias due to genomic location insertion. p‐values were calculated using the quantile‐adjusted conditional maximum likelihood method and adjusted with the Benjamini–Hochberg procedure (Subramaniyam et al., 2019). The P. putida KT2440 reference genome was obtained from the GenBank entry NC_002947.3 (Belda et al., 2016). Read counts per insertion site of the control and target samples were tested for significant differences, and those with a binary logarithm (log2) of the FC lower than −2 or higher than 2, as well as a p‐value <0.05, were selected as conditionally essential genes (Burby et al., 2017).
RESULTS AND DISCUSSION
Assessing the impact of F− on the growth of wild‐type P. putida KT2440
The first step to evaluate the tolerance of P. putida towards F− entailed a quantitative assessment of the MIC of this ion under different growth conditions—both in rich medium (LB) and in M9 minimal medium added with 5 g L−1 glucose as the only carbon source. Thus, wild‐type P. putida KT2440 was cultivated in these culture media added with NaF at different concentrations (up to 250 mM) in microtiter plates, and the optical density at 600 nm (OD600) was followed over time to estimate bacterial growth. After 20 h of incubation, we found that P. putida tolerated NaF within different concentration ranges depending on the culture medium used for the experiments [Figure 1(A)]. Cells could grow in the presence of 75 mM NaF in LB medium up to final OD600 values >70% of that registered in control experiments (i.e. no F− in the medium)—yet bacterial growth in M9 minimal medium with glucose was impaired even at the lowest NaF concentration tested. When the halide was added to the media at 125 mM, bacterial growth was similarly (and dramatically) affected in both culture conditions [Figure 1(A)], with a >90% reduction in OD600 values. Hence, the MIC of NaF lies in the range of 75 mM in M9 minimal medium and 125 mM in LB medium. We also reasoned that Na+, the counterion of F− in these experiments, could exert a deleterious effect on growth when added at high concentrations as the ones used in our experiments. However, a similar impact on the growth patterns of strain KT2440 was observed when the cells were incubated in the presence of different KF concentrations in M9 minimal medium with glucose (Figure S1 in the Supporting Information). Furthermore, in order to discriminate whether the effects seen in these cultures were simply due to changes in the ionic strength brought about by the addition of salts to the culture media, MIC assays were performed using NaCl, widely used as a saline stress agent (Hachicho et al., 2017). In this case, cells could tolerate the salt up to 0.5 M, and some growth could be detected even when LB medium was supplemented with 1 M NaCl [Figure 1(B)]. These observations indicate that F− exerts an impact on bacterial growth unique to this halide.
FIGURE 1.
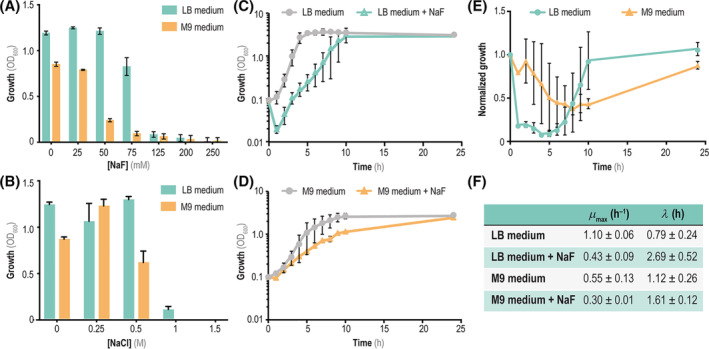
Impact of mineral F− on growth profiles of P. putida KT2440. (A) Minimal inhibitory concentration (MIC) assays with different NaF concentrations added to either LB medium (green) or M9 minimal medium containing 5 g L−1 glucose as the only carbon source (orange). Growth was estimated as the optical density measured at 600 nm (OD600) after 20 h of incubation. (B) MIC assays carried out by adding NaCl to the culture media instead of NaF. (C) Growth curves of P. putida KT2440 in shaken‐flask cultures in LB medium (grey circles) or LB medium supplemented with 50 mM NaF (green triangles). (D) Time‐course growth curves in Erlenmeyer flasks of P. putida KT2440 in M9 minimal medium (grey circles) and M9 supplemented with 50 mM of NaF (orange triangles). In all cases, 5 g L−1 glucose was added as the only carbon source. (E) Tolerance of P. putida KT2440 to F− in both rich and defined culture media. Time‐resolved tolerance curves represent the ratio between the bacterial growth (estimated as the OD600) in the presence of 50 mM NaF and the growth in the control condition in LB medium (green circles) and M9 minimal medium containing 5 g L−1 glucose (orange triangles). (F) Quantitative physiology parameters (maximum specific growth rate, μ max and duration of the lag phase, λ) for cultures of P. putida KT2440 in LB medium or M9 minimal medium containing 5 g L−1 glucose, supplemented or not with 50 mM NaF. In all cases, the error bars represent standard deviations of average values calculated from at least three independent experiments. Abbreviation: LB, lysogeny broth
Based on these preliminary experiments, we carried out shaken‐flask cultures of P. putida KT2440 incubated in both media supplemented with 50 mM NaF with the purpose of analysing the effect of F− on quantitative physiology parameters, e.g. the extension of the lag phase (λ) and the maximum specific growth rate (μ max). Both kinetic parameters were affected by the presence of the halide in the medium, although the final OD600 values (upon 24 h of incubation) in these experiments were remarkably similar to those reached in control cultures, without any added F− [Figure 1(C,D)]. These observations would suggest that a recovery effect (e.g. halide detoxification) occurs during exponential growth, enabling the cells to cope with the stressful conditions to reach a final biomass density within the range of untreated cultures. This aspect was evidenced by plotting the normalized growth of P. putida (i.e. OD600 in the presence of NaF/OD600 in untreated cultures) over time [Figure 1(E)]. Cultures grown in M9 minimal medium with glucose had a relatively stable behaviour, as judged by normalized growth values that gradually decreased over the first 10 h. LB medium cultures, in contrast, were strongly affected by the presence of NaF during early exponential growth (with normalized growth values immediately dropping by >90%)—yet P. putida recovered faster in rich medium than in M9 minimal medium. Figure 1(F) shows the actual λ and μ max values for all shaken‐flask experiments. Accordingly, μ max was reduced by 62% and 43% upon NaF treatment in LB and M9 minimal medium, respectively. In contrast, λ increased by 3.5‐fold in LB medium added with F−, while this growth parameter was essentially unaffected in M9 minimal medium containing glucose as the carbon substrate [Figure 1(F)]. Prompted by these results, which point to the existence of a potential F− detoxification mechanism, we investigated the genome‐wide genetic basis of the observed phenotypes as indicated in the next section.
Tn‐Seq identifies the pool of genes involved in the tolerance of P. putida towards F−
With the aim of identifying genes involved in the response of P. putida KT2440 to F−, we adopted a library of random mini‐Tn5 insertions (Calero et al., 2018) to perform a genome‐wide Tn‐Seq analysis of cells exposed to NaF stress. This transposon‐insertion library consists of a pooled mixture of individual P. putida clones carrying two independent (mini‐Tn7 and mini‐Tn5) transposons (Calero et al., 2018). The library was constructed in two steps. First, the mini‐Tn7 transposon, bearing a gentamicin (Gm) resistance cassette, was integrated in the genome to facilitate positive selection in subsequent steps. Afterwards, the mini‐Tn5 module, endowed with a kanamycin (Km) resistance determinant, was randomly inserted throughout the bacterial genome (Table 1). The mini‐Tn5 transposon module interrupts genes at different positions of the open reading frame (ORF) in a random fashion (Martínez‐García et al., 2014), therefore part of the (non‐lethal) insertion mutants will display a phenotype under specific growth conditions for which the cognate function is relevant. The experimental pipeline used in the Tn‐Seq experiments involved four main steps [Figure 2(A)]: (i) growth‐based selection and clonal enrichment of P. putida clones under control and stressful (i.e. in the presence of NaF) conditions, (ii) extraction and preparation of gDNA libraries from these cultures, (iii) high‐resolution DNA sequencing of these gDNA libraries and (iv) phenotypic characterization of the impact of each isolated ORF, identified as to be involved in NaF tolerance, by constructing in‐frame deletion mutants of strain KT2440.
FIGURE 2.
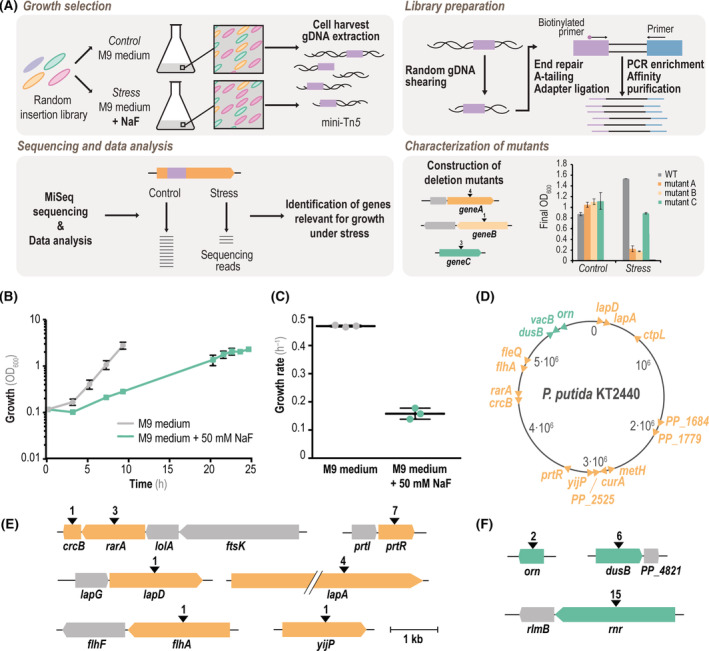
Tn‐Seq‐guided identification of genes relevant for bacterial growth in the presence of mineral F−. (A) Workflow of the experimental procedure for Tn‐Seq experiments. A Tn5 insertion library (transposons depicted as purple boxes) was used in growth assays under control and stress (+NaF) conditions in order to assess differential growth of Tn5‐insertion mutants of wild‐type (WT) P. putida KT2440. After harvesting the bacterial biomass, genome DNA (gDNA) was extracted and processed by mechanical shearing, A‐tailing and ligation of adapters (blue boxes), followed by PCR enrichment as indicated in Experimental procedures. The resulting library was subsequently analysed by next‐generation sequencing (MiSeq), and the number of readings under different experimental conditions were compared to assess the differential abundance of individual insertions. Genes relevant for growth of strain KT2440 in the presence of NaF were identified, and the deletion mutants in the corresponding genes (listed in Table 1) were constructed and characterized as described in the text. (B) Growth of the library of Tn5‐insertion mutants of P. putida KT2440 in M9 minimal medium containing 5 g L−1 glucose as the only carbon source (grey) and the same medium supplemented with 50 mM NaF (green). Growth was estimated as the optical density measured at 600 nm (OD600), and the biomass was harvested for gDNA extraction and processing at the end‐point (i.e. no further changes in OD600) for each culture condition. (C) Specific growth rates of P. putida KT2440 grown under control conditions (M9 minimal medium containing 5 g L−1 glucose, grey) and stress conditions (+50 mM NaF, green). (D) Physical map of the genome of strain KT2440 identifying the insertion sites in relevant genes found in the Tn‐Seq assay. Insertions with a log2(fold change, FC) < −2 (genes relevant for survival in the presence of NaF) are shown in orange. Insertions with a log2(FC) > 2 (mutations positively affecting growth with NaF) are indicated in green. (E) Genetic context of genes identified to be important for bacterial growth in the presence of 50 mM NaF (orange) by Tn‐Seq. The number of insertions is indicated with black arrowheads, and genes highlighted in grey represent the genetic context of predicted operons. (F) Same as panel (E), but showing genes with positively affecting insertions (green) and their genetic context. The same scale is used in panels (E) and (F)
As described in the first section of the article, the addition of 50 mM NaF to M9 minimal medium with glucose as the carbon source had a significant impact on the cell physiology, but it still permits bacterial growth to significant levels. Therefore, we selected this F− concentration as the standard stress condition for library enrichment and selection, using M9 minimal medium with glucose as control. The growth of the mini‐Tn5 libraries was monitored over time until they reached an OD600 of ~2 [Figure 2(B)], when they were harvested for gDNA extraction. Under these culture conditions, μ max dropped from 0.46 h−1 in the control experiment to 0.15 h−1 in the presence of NaF [Figure 2(C)], consistent with our previous observations in wild‐type P. putida cultures (Figure 1). Sequencing of the junctions between the transposon and the bacterial genome (i.e. at the landing spot) was achieved by generating an enriched amplification library using a pUT‐Km transposon‐specific oligonucleotide in order to enrich the junctions (Calero et al., 2018; Lennen & Herrgård, 2014). Using these generated libraries, we sought to identify genes involved in tolerance towards F− by comparing the abundance of sequencing reads of each insertion found in the target experimental conditions and control experiments. Both (i) the binary logarithm of the fold change [log2(FC)] of the reads‐per‐insertion parameter, which indicates the difference in the abundance of readings for each insertion in both conditions, and (ii) the p‐values, which assess the significance of that difference, were used as the criteria for selection of candidate genes (Burby et al., 2017).
Several genes were found to be important for the growth of P. putida KT2440 when exposed to high F− concentrations [Figure 2(D); Table 1 and Table S2 in the Supporting Information]. The ORFs identified in this screening experiment are scattered throughout the genome of P. putida KT2440 [Figure 2(D)], which indicates that there was no significant positional bias in the Tn‐Seq analysis. The top‐scoring genes predicted to confer either resistance to F− are indicated in Figure 2(E), together with the physical map of the chromosome context and the actual number of mini‐Tn5 insertions found therein. The gene encoding the F− exporter CrcB had a top score [log2(FC) = −2.79 and p‐value = 0.0151; Table 2] among the functions expected to confer NaF resistance. This observation does not come as a surprise, given the known involvement of CrcB in F− detoxification in several bacterial species (McIlwain et al., 2021). The identification of such a short length (374 bp) gene also served as an internal quality control feature—indicating that the insertion library had enough quality to be used to identify other genes related to F− toxicity. Interestingly, the upstream gene in the operon where crcB is located, rarA (PP_4002), was also identified as important for F− tolerance. Three independent mini‐Tn5 insertions were found to be more represented in the control experiment than in the stress (+NaF) condition. RarA has been previously annotated as a recombination factor, and it has been described as having a role in the tolerance of P. putida towards the acid stress brought about by HCl treatment (Reva et al., 2006). We also discovered that prtR (PP_2889) was involved in the resistance phenotype in these experiments. PrtR is annotated as a transmembrane σ factor, and it has been previously described to display a role in P. fluorescens in controlling the formation of extracellular proteases and siderophores, as well as temperature‐dependent lipases and proteases (Burger et al., 2000; Llamas et al., 2014; Song et al., 2014). This gene was also identified in a high‐throughput screening towards enhanced fitness for aromatic substrates conversion by engineered P. putida strains (Eng et al., 2021).
TABLE 2.
Genes identified to affect the tolerance of P. putida towards F− stress a
| Gene | Locus | Insertions | Log2(FC) | p‐value | Description | References | Comments |
|---|---|---|---|---|---|---|---|
| rarA | PP_4002 | 3 | −4.14 | 2.13 × 10−7 | Recombination factor protein RarA | Reva et al. (2006) | Part of the PP_3999‐PP_4004 operon together with crcB; relevant under acid (HCl) stress (pH = 4.5). |
| −3.46 | 0.0047 | ||||||
| −2.33 | 0.0077 | ||||||
| crcB | PP_4001 | 1 | −2.79 | 0.0151 | Fluoride channel | — | Fluoride‐specific exporter |
| prtR | PP_2889 | 7 | −2.63 | 8.16 × 10−5 | Transmembrane anti‐σ factor PrtR | Burger et al. (2000); Llamas et al. (2014); Song et al. (2014); Calero et al. (2018) | Forms an operon with the stationary‐phase σ factor PrtI gene (PP_2888). PrtR controls the formation of external proteases and siderophores in P. fluorescens, as well as temperature‐dependent lipases and proteases. Also involved in tolerance towards p‐coumaric acid |
| −2.91 | 0.0111 | ||||||
| −2.45 | 7.21 × 10−5 | ||||||
| −2.14 | 0.0044 | ||||||
| −3.00 | 4.79 × 10−5 | ||||||
| −3.57 | 0.0002 | ||||||
| −2.60 | 0.0231 | ||||||
| lapA | PP_0168 | 4 | −2.50 | 0.0009 | Putative surface adhesion protein | Reva et al. (2006) | Involved in biofilm formation; plays a role in tolerance towards urea |
| −2.77 | 0.0392 | ||||||
| −2.21 | 6.22 × 10−8 | ||||||
| −2.18 | 0.0006 | ||||||
| PP_1779 | 2 | −4.13 | 0.0046 |
Lipopolysaccharide ABC export system, ATP‐binding protein |
Cuthbertson et al. (2010) | Mediates polysaccharides export for the formation of glycosylated macromolecules | |
| −4.57 | 0.0009 | ||||||
| lapD | PP_0165 | 1 | −2.21 | 0.0471 | Transmembrane protein | — | Bis‐(3′,5′)‐cyclic dimeric GMP‐binding protein; involved in surface attachment |
| fleQ | PP_4373 | 1 | −2.32 | 0.0341 | Transcriptional regulator | — | Bis‐(3′,5′)‐cyclic dimeric GMP‐responsive transcriptional regulator |
| flhA | PP_4344 | 1 | −2.59 | 0.0121 | Flagellar biosynthesis protein FlhA | — | Flagella assembly |
| yijP | PP_2579 | 1 | −2.33 | 0.0018 |
Phosphoethanolamine transferase CptA |
Mumm et al. (2016) | Involved in biosynthesis and modification of lipopolysaccharides. Subjected to regulation by ColRS |
| PP_2525 | 1 | −2.03 | 4.68 × 10−7 | Hypothetical protein | — | ATP‐binding protein, ATPase; 59% identity with SufC, a Fe‐S assembly ATPase, in Shewanella oneidensis MR‐1 | |
| metH | PP_2375 | 3 | −2.21 | 0.0221 | B12‐dependent methionine synthase | Domínguez‐Cuevas et al. (2006); Molina‐Henares et al. (2010) | Last step of l‐methionine biosynthesis; upregulated in presence of toluene and o‐xylene in P. putida KT2440 |
| −2.07 | 0.0431 | ||||||
| −2.08 | 0.1211 | ||||||
| curA | PP_2476 | 1 | −2.48 | 0.0044 | NADPH‐dependent curcumin/dihydrocurcumin reductase | Follonier et al. (2011) | Zinc‐containing alcohol dehydrogenase. Overexpressed when cells are stressed in pressurized cultures |
| ctpL | PP_0562 | 1 | −2.08 | 0.0191 | Methyl‐accepting chemotaxis sensory transducer | — | Chemoreceptor for 4‐chloroaniline and catechol in P. aeruginosa |
| PP_1684 | 1 | −2.17 | 0.0111 | Major facilitator superfamily transporter | Follonier et al. (2013); Molina‐Santiago et al. (2016) | Efflux pump. Overexpressed when cells are stressed in pressurized cultures. Differentially expressed in toluene‐containing cultures | |
| PP_2411 | 1 | −2.08 | 0.027 | Major facilitator family transporter | — | Unknown | |
| PP_1922 | 1 | −2.51 | 0.0022 | Hypothetical protein | — | DUF2726 domain‐containing protein (unknown) | |
| PP_2857 | 1 | −2.48 | 0.0028 | Hypothetical protein | — | Orthologue to P. fluorescens F113 PSF113_2684 and P. putida H8234 L483_12195 (autotransporter adhesin) | |
| PP_1882 | 1 | −2.19 | 0.014 | Hypothetical protein | — | Predicted to be a membrane protein; contains a YD (tyrosine‐aspartate) repeat. Similar to Rhs proteins | |
| PP_2298 | 1 | −2.63 | 0.014 | Hypothetical protein | — | Unknown | |
| PP_2398 | 1 | −2.08 | 0.017 | Hypothetical protein | — | Small peptide | |
| PP_1631 | 1 | −2.27 | 0.020 | Hypothetical protein | Seo and Kim (2018) | Putative decarboxylase. Similar to LOG family proteins, which are not very well known in prokaryotic organisms but apparently related to microbe–plant interactions | |
| PP_0925 | 1 | −2.14 | 0.024 | Hypothetical protein | — | Orthologue to an acyl‐CoA dehydrogenase present in other Pseudomonas species |
FC, fold change; LOG proteins, lonely guy proteins; CoA, coenzyme A. The references in the table indicate reports of the corresponding gene or function in either P. putida KT2440 or related species.
Furthermore, two genes related to biofilm formation were recognized in the experiment, lapA (PP_0168) and lapD (PP_0165). LapA is a massive surface adhesion protein (the largest polypeptide in strain KT2440) that participates in assembling cells during biofilm formation (Benedetti et al., 2016; López‐Sánchez et al., 2016). LapA modifies the hydrophobicity of cell membranes and is necessary for both initial attachment of P. putida and mature biofilm formation (El‐Kirat‐Chatel et al., 2014). This protein seems to be also involved in the tolerance of P. putida KT2440 towards urea (Reva et al., 2006). LapD was also isolated in our screening, probably due to its functional relationship with LapA. LapD has been previously described as a transmembrane protein that prevents LapA hydrolysis (which would trigger biofilm dispersal) by inhibiting the activity of the LapA‐specific periplasmic protease LapG (Gjermansen et al., 2010; Yousef‐Coronado et al., 2011). Two other genes selected in the Tn‐Seq experiments were involved in the regulation and synthesis of flagella, i.e. fleQ (PP_4373) and flhA (PP_4344). Finally, two genes recognized as being involved in the synthesis and export of lipopolysaccharides, i.e. yijP (PP_2579) and PP_1779 (Cuthbertson et al., 2010; Mumm et al., 2016), were likewise isolated in this experiment. The identification of such a significant number of genes involved in cell membrane structure and integrity suggests that membrane stability is of vital importance for the survival of P. putida under F− stress conditions. All the identified genes, together with further explanations of their biological role gathered from the literature, can be found in Table 2. Taken together, these results indicate that F− treatment is connected to diverse physiological and regulatory functions in P. putida KT2440—even when the actual number of genes involved in the tolerance phenotype is relatively small, as is the type and number of functional categories to which these genes belong. Additionally, the loss of function of five genes was identified to increase the growth of P. putida KT2440 in the presence of F−, as Tn5 insertions in these sequences were more represented in cultures added with NaF than in control experiments [Figure 2(F) and Table S2 in the Supporting Information]. These genes included orn, dusB and vacB, in which several independent insertions (up to 15 in the case of vacB) were enriched in the presence of F−. Clean, in‐frame deletion mutants of strain KT2440 were constructed and characterized as explained in the next section in order to elucidate the role of the genes identified in the Tn‐Seq experiments.
Phenotypic characterization of mutants of P. putida KT2440 in the presence of F−
We selected some of the genes identified to play an important role in F− homeostasis (Table 2), and we constructed in‐frame, scar‐less deletion mutants of P. putida KT2440—to circumvent any potential polar effect brought about by transposon insertion (Hutchison et al., 2019). Three of the genes identified as ‘detrimental’ for bacterial growth in the presence of F− (Table S2 in the Supporting Information) were likewise selected for quantitative physiology experiments. Hence, deletion mutants of P. putida KT2440 in crcB, rarA, prtR, lapA, lapD, flhA and yijP (i.e. important genes for growth in F−), and orn, dusB and vacB (i.e. insertions potentially affecting positively growth in the presence of the halide) were constructed by allelic replacement as per the procedure described by Wirth et al. (2020). The resulting mutant strains were individually grown in microtiter plate cultures in M9 minimal medium added with 5 g L−1 glucose, and in the same medium supplemented with NaF at 25 mM. Note that this NaF concentration (half the amount used in the Tn‐Seq experiments) was selected as to still observe the negative impact of F− on the cell physiology without completely suppressing bacterial growth according to the preliminary experiments shown in Figure 1—these conditions are especially relevant for characterizing mutants affected in tolerance against NaF. Specific growth rates [Figure 3(A)] and the cell density after 20 h of incubation [Figure 3(B)] were compared in these conditions as described in Experimental procedures, using wild‐type P. putida KT2440 as the reference. As expected, both μ max and the final cell density (OD600, optical density measured at 600 nm) were severely affected in cultures of P. putida ΔcrcB when the medium was supplemented with 25 mM NaF—which essentially inhibited bacterial growth under these conditions. Intermediate growth phenotypes between those of the wild‐type strain and the ΔcrcB mutant could be detected in cultures of the remaining mutants. Pseudomonas putida ΔrarA, for instance, displayed a reduced (ca. 50%) μ max value in the presence of NaF compared to the control experiment. F− treatment did not affect μ max of P. putida ΔlapA and P. putida ΔlapD, although the final OD600 was reduced by >30% in comparison to that in NaF‐free control experiments. Taken together, these experiments show that the genes isolated in the Tn‐Seq experiments are indeed involved in the response of P. putida KT2440 to F− treatment. The P. putida strains carrying mutations in the genes orn, dusB and vacB, which were identified in the Tn‐seq screening to have a better growth when disrupted, also showed reduced‐fitness phenotypes in the tested conditions of F− when compared to cultures of the wild‐type strain [Figure 2(F)]. Two of these genes (orn and vacB) encode ribonucleases (i.e. a broad‐range oligoribonuclease and the exoribonuclease R, respectively), whereas DusB is identified as a tRNA dihydrouridine synthase (Belda et al., 2016). Since these functions could exert a pleiotropic effect, we cannot discard a potential role of these genes in the general homeostasis of P. putida—rather than a specific response to NaF—or that the effect of these functions is restricted to very specific growth conditions. The identification of general response functions in high‐throughput screens is a common feature of functional‐genomic procedures, and it has been described in several articles where P. putida mutants are selected by enhanced fitness under specific culture conditions (Eng et al., 2021; Thompson et al., 2019, 2020).
FIGURE 3.
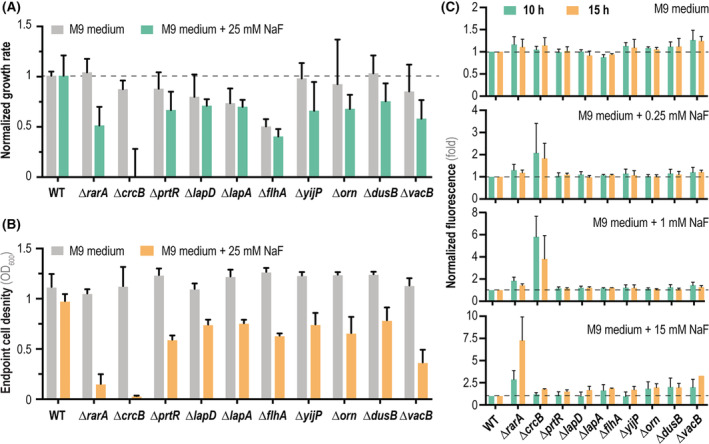
Phenotypic characterization of individual P. putida mutants in genes identified by the Tn‐Seq screening. (A) Normalized growth rates of mutant strains compared to wild‐type P. putida KT2440 (WT) grown either under control conditions (M9 minimal medium containing 5 g L−1 glucose as the only carbon source, grey bars) or stressful conditions (+25 mM NaF, green bars). Dashed lines indicate a growth rate identical to that of the WT strain. (B) Final cell density (estimated as the optical density measured at 600 nm, OD600) of the WT strain and the individual P. putida mutants after 15 h of incubation (i.e. onset of the stationary phase) under control conditions (M9 minimal medium containing 5 g L−1 glucose, grey bars) and stressful conditions (+25 mM NaF, orange bars). (C) Intracellular F− concentration estimated with a biosensor based on the fluoride‐sensitive riboswitch (FRS), which couples the presence of F− with a fluorescent (msfGFP) output. Normalized msfGFP fluorescence (indicated as the fold change) of the individual P. putida mutants carrying the FRS biosensor compared to the WT strain incubated in the presence of different NaF concentrations (0, 0.25, 1 and 15 mM). Measurements were done after 10 or 15 h of incubation, and the dashed line indicates fluorescence levels within the range of the WT strain. In all cases, error bars represent standard deviations of average values calculated from at least three independent biological replicates
Exploring the intracellular F− concentration in wild‐type and mutant P. putida strains with a non‐invasive fluoride‐responsive biosensor
The results so far indicate a general impact of F− on the cell physiology, and prompted the question of how are the genes identified in the Tn‐Seq experiment involved at the molecular level. Thus, the next step in this study was to assess the role of these genes in the intracellular F− homeostasis—especially at the light that several genes isolated in the screening seem to be involved in cell membrane‐related processes, including metabolite transport. Hence, the non‐invasive F− biosensor based on a FRS (Calero et al., 2020) was adopted to study the intracellular F− levels in all relevant strains. The FRS can be indirectly used as a means to assess F− concentrations owing to the conformation change in the secondary mRNA structure in the presence of the anion. This conformational transition exposes a (buried) ribosome binding site, leading to the translation of the reporter (i.e. msfGFP, a monomeric superfolder GFP). Plasmid pS441·FRSv1 contains a synthetic element composed of the FRS of Pseudomonas syringae controlling the expression of a fusion protein construct. This fragment consists of the leading sequence of eriC F (i.e. the gene regulated by the FRS in its original context) and msfGFP, as well as its original P eriC F promoter. We introduced plasmid pS441·FRSv1, containing the FRS‐based biosensor, in the P. putida mutants constructed herein (Table 1). Fluorescence levels were measured in M9 minimal medium cultures added with glucose in the presence of different F− concentrations during mid‐ to late‐exponential growth (10 and 15 h of incubation). In these experiments, P. putida KT2440 and its ΔcrcB knock‐out derivative were used as negative and positive controls of intracellular F− accumulation, respectively, and the fluorescence readout was normalized to that of the wild‐type strain under each culture condition. Expectedly, a clear msfGFP signal corresponding to a significant intracellular F− level could be observed in cultures of P. putida ΔcrcB/pS441·FRSv1 exposed to the salt—even when NaF was added at concentrations as low as 0.25 mM [Figure 3(C)]. The wild‐type strain, in contrast, tolerated the NaF challenge even at 15 mM, displaying a low msfGFP output that indicates efficient F− secretion via CrcB. The ΔcrcB cells could not grow at 15 mM NaF, and as a consequence, the biosensor output could not be reliably assessed in these conditions. The rest of the mutants in genes identified in our screening, in contrast, could stably maintain low intracellular F− levels when NaF was fed at concentrations up to 1 mM. The only exception was P. putida ΔrarA, which already showed a twofold increase in the fluorescence output compared to the wild‐type strain after 10 h of incubation with 1 mM NaF. At the highest NaF concentration tested, this mutant showed a fluorescence signal sevenfold higher than the control strain after 15 h of F− exposure. A slight increase in the msfGFP readout could also be detected for the rest of the P. putida mutants [Figure 3(C)]—yet the differences were not significant compared to control experiments. Taken together, these results indicate that CrcB‐dependent efflux of F− ions is by far the predominant molecular mechanism against toxicity. Building on this information, we decided to explore the basis for the observed phenotype of P. putida ΔcrcB when exposed to NaF.
The detoxifying role of CrcB is specific for F−
At this point, it was clear that CrcB plays a central role in F− detoxification, as knocking out the gene encoding this exporter led to a significant intracellular accumulation of F− that impaired growth even at very low NaF concentrations. We decided to further explore the relevance of CrcB by testing the phenotype of P. putida ΔcrcB under different cultivation conditions. In the first place, we assessed the MIC for F− in 20‐h microtiter plate cultures of this mutant strain, finding that bacterial growth was impaired when M9 minimal medium is supplemented with as little as 0.5 mM NaF [Figure 4(A)]. This is an F− concentration ca. 100‐fold lower than the amount needed to inhibit the growth of wild‐type P. putida KT2440. When using rich LB medium, the MIC shifted to a 10‐fold higher F− concentration—still significantly lower than the NaF that would impair growth of strain KT2440. Also, the growth of the ΔcrcB strain in the presence of NaCl was slightly impaired compared to wild‐type P. putida, with a significant decrease in the final OD600 values when the M9 minimal medium was supplemented with 0.5 M NaCl. After a 20‐h incubation under these conditions, the cultures of P. putida KT2440 reached an OD600 of ~0.6, whereas the ΔcrcB mutant could only grow to an OD600 ~ 0.25 [Figure 4(B)]. Furthermore, we were interested in exploring if the sensitivity phenotype of the mutant strain is only evidenced in the presence of NaF or whether it extends to other types of chemical insults beyond saline stress. To this end, the growth of the wild‐type P. putida strain was compared to that of the mutant under acid stress [HCl, Figure 4(C)] and oxidative stress [H2O2, Figure 4(D), or N,N′‐dimethyl‐4,4′‐bipyridinium dichloride (paraquat), Figure 4(E)]. These stressful conditions are known to trigger a general metabolic and physiological response in P. putida (Nikel et al., 2016b, 2021). The growth pattern of both strains was similar under all three stressful conditions (i.e. both the wild‐type strain and the ΔcrcB strain displayed a similar sensitivity to the addition of HCl, H2O2 or paraquat), which indicates that CrcB is not directly involved in the tolerance towards these stressors. The next step was to investigate sensitivity to salts and physicochemical conditions that could facilitate NaF diffusion into the cells.
FIGURE 4.
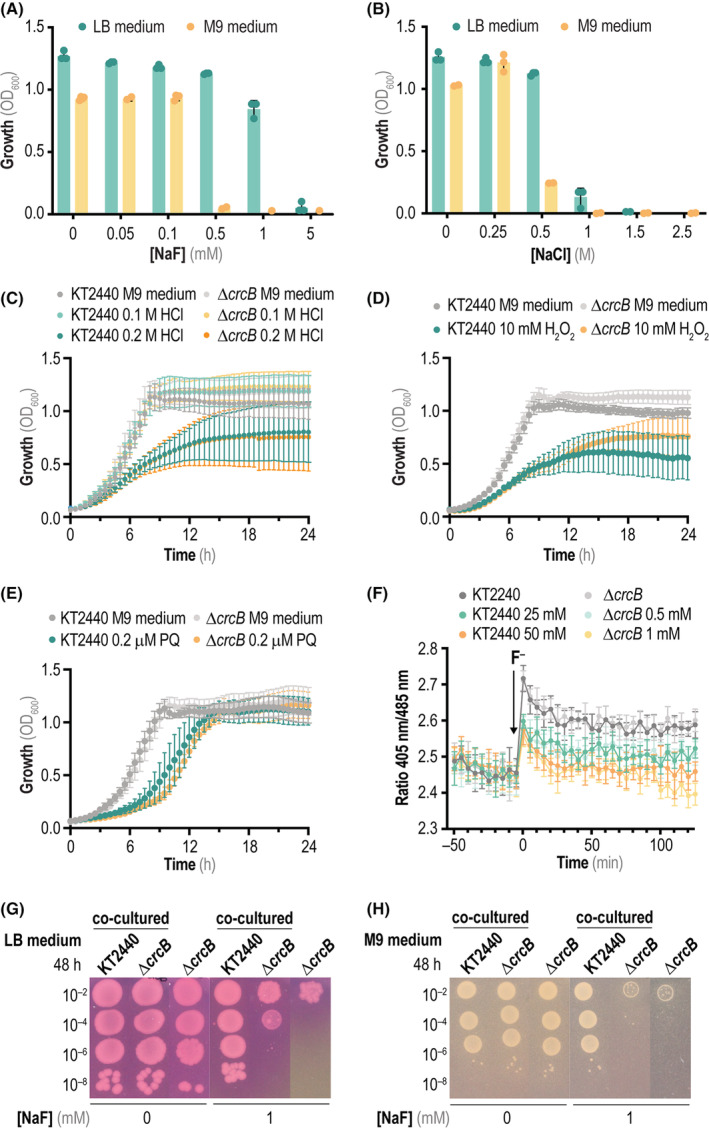
Physiological characterization of P. putida ΔcrcB in the presence of mineral F−. (A) Minimal inhibitory concentration (MIC) assay of P. putida ΔcrcB exposed to increasing NaF concentrations after 20 h of incubation in either LB medium or M9 minimal medium containing 5 g L−1 glucose as the only carbon source. Growth was estimated as the optical density measured at 600 nm (OD600); dots represent individual data per experiment with at least three independent cultures analysed per condition. (B) Same as in panel (A) but applying increasing NaCl concentrations. (C) Growth curves of wild‐type P. putida KT2440 and P. putida ΔcrcB in M9 minimal medium added with 5 g L−1 glucose as the only carbon source and HCl at different concentrations. (D) and (E) Same as in panel (C), but in this case cells were cultured in M9 minimal medium (containing 5 g L−1 glucose) added with either H2O2 or paraquat (PQ, N,N′‐dimethyl‐4,4′‐bipyridinium dichloride), respectively. In all cases, error bars represent standard deviations of average values calculated from at least three independent biological replicates. (F) Changes in the intracellular pH of P. putida KT2440 and P. putida ΔcrcB using a ratiometric, non‐invasive pH biosensor (PHP). Cells were grown in M9 minimal medium containing 5 g L−1 glucose as the only carbon source, and the intracellular pH was followed over 2 h upon addition of NaF (black arrow, at t = 0 min) by plotting the fluorescence ratio 405 nm/485 nm over time. The error bars represent standard deviations of at least four technical replicates, and a representative experiment out of three independent biological replicates is shown. (G) Survival drop assays of P. putida KT2440 and P. putida ΔcrcB in LB medium plates in the presence of the pH indicator phenol red. In some experiments, the culture medium was added with 1 mM NaF, and the plates were incubated for 48 h at 30°C. Co‐cultured strains indicate drops of the wild‐type and mutant strains that were incubated next to each other in the same plate. (H) Same as in panel (G), but the experiment was carried out in M9 minimal medium plates containing 5 g L−1 glucose as the only carbon source. Abbreviation: LB, lysogeny broth
Interplay between F− toxicity and the intracellular and extracellular pH
F− toxicity has been described to be dependent on the pH of the surrounding medium, due to the fact that hydrogen fluoride (HF) is probably the form that diffuses across cell membranes. HF is such a small molecule that it would cross biological membranes through water channels, including aquaporins (Marquis et al., 2003). Additionally, some reports indicate that HF hydrolyzes to H+ + F− upon the acid form is transported to the cytoplasm, thereby decreasing the intracellular pH and mediating acid stress due to the resulting HF ↔ H+ + F− equilibrium (Johnston & Strobel, 2020). If the equilibrium is displaced towards HF dissociation, this phenomenon would partially collapse the H+ gradient and uncouple energy metabolism—and probably other ion homeostatic processes as well (Danchin & Nikel, 2019). In order to examine whether the intracellular pH was affected in P. putida KT2440 upon exposure to NaF, we used a genetically encoded, non‐invasive pH biosensor that we have previously tailored for Pseudomonas species and other Gram‐negative bacteria (Arce‐Rodríguez et al., 2019). This pH biosensor (termed PHP) is based on a codon‐optimized gene variant of the pH‐sensitive pHluorin2 indicator protein, which displays changes in the ratio between its two excitation peaks (λ excitation = 405 and 485 nm) upon pH variations occur. This ratiometric change can be adopted to determine variations in the intracellular pH. Here, we introduced the PHP pH biosensor in a plasmid‐based format into P. putida KT2440 and the ΔcrcB strain, and grew the resulting cells in M9 minimal medium to a similar cell density whereupon NaF was added at different concentrations. The ratio in the excitation peaks at 405 and 485 nm was continuously followed over time to assess changes in the intracellular pH [Figure 4(F)]. Expectedly, no significant changes in the fluorescence ratio were detected in the absence of NaF. After spiking the cultures with the salt, however, an almost immediate increase in the pH was observed in all cultures, with a clear difference observed between the experimental conditions. The general trend observed was a lower intracellular pH in cells exposed to higher NaF concentrations (i.e. we detected an inverse relationship between the two parameters studied, which may indicate that the HF ↔ H+ + F− equilibrium is displaced towards dissociation in the cytoplasmic milieu). The qualitative differences in intracellular pH before and after addition of NaF were similar in both strains, even though the F− concentrations that elicited such response in P. putida ΔcrcB were much lower than those used for the wild‐type. This observation suggests that, although F− affected the intracellular pH in both bacterial strains, the transporter mutant suffered acidification of the cytoplasm even at low salt concentrations [Figure 4(F)].
The effect of the extracellular pH on F− toxicity was likewise addressed. The growth of P. putida KT2440 and its ΔcrcB derivative was evaluated by spotting serial dilutions of bacterial suspensions onto LB or M9 minimal medium plates in the presence or absence of 1 mM NaF. In these experiments, the culture media were supplemented with phenol red (phenolsulfonphthalein), a pH indicator that undergoes a gradual transition from yellow (λ emission = 443 nm) to red (λ excitation = 570 nm) over a pH range from 6.8 to 8.2 (Yamaguchi et al., 1997). Above pH = 8.2, phenol red turns a bright pink (fuchsia) colour. In a rich medium, such as LB broth, the growth of P. putida mediated a change in the pH towards basicity, which could be easily detected as phenol red turned fuchsia under these conditions [Figure 4(G)]—probably since amino acids and small peptides are the main source of carbon, thereby releasing ammonia (Sánchez‐Clemente et al., 2018). On the contrary, in M9 minimal medium with glucose as a carbon source, the growth of cells acidified the medium as a consequence of gluconate and 2‐ketogluconate formation (Nikel et al., 2015b), thus turning the pH indicator into a yellow hue. Changes in the medium pH, in turn, affect bacterial growth in the presence of F− because HF enters the cells more efficiently than the anionic form. As shown above, the deletion of fluoride transporter in bacterial cells leads to a higher concentration of F− in the cytoplasm than outside of the cell—the so‐called weak acid accumulation or ion trapping effect, which affects the ability of cells lacking the transporter to grow in the presence of NaF under acidic pH conditions (Ji et al., 2014). This feature could be observed when wild‐type P. putida KT2440 and the ΔcrcB strain were co‐cultured next to each other [Figure 4(G)]. In the media containing 1 mM NaF, P. putida ΔcrcB was able to grow only when the plates were basified by the growth of the wild‐type strain. Conversely, the growth of the crcB mutant was impaired in M9 minimal medium with glucose, either co‐cultured with the wild‐type or by itself [Figure 4(H)], highlighting that HF is the active, toxic form of fluorine that affects bacterial cells probably by direct permeation through the cell membrane. Considering the results from bacterial physiology experiments, the next relevant question in this study was to investigate the metabolic impact of adding F− to cultures of P. putida using a systematic metabolomic approach.
Network‐wide metabolome re‐organization upon exposure of P. putida to F−
We assessed the effect of F− exposure on intracellular metabolite concentrations both in wild‐type P. putida KT2440 and the mutant ΔcrcB strain when the cells grown on M9 minimal medium with glucose as the only carbon source. Pseudomonas putida KT2440 runs a cyclic glycolytic metabolism, composed by catabolic and anabolic activities of the Entner–Doudoroff pathway and the Embden–Meyerhof–Parnas (EMP) pathway (in a gluconeogenic fashion), together with the pentose phosphate (PP) pathway (Nikel et al., 2015b; Volke et al., 2021). Thus, the architecture of the so‐called EDEMP cycle regenerates hexose phosphates and favours catabolic NADPH formation [Figure 5(A)]. Here, we analysed the concentration of metabolites within the EDEMP and the tricarboxylic acid (TCA) cycles, together with several molecules within amino acid biosynthesis pathway and redox metabolism, using high‐resolution, targeted LC–MS/MS analysis (Figures S2–S7 in the Supporting Information). In order to provide a complete picture of the impact of NaF on the metabolome, samples were taken at different time points upon adding F− to the culture medium, and the results were represented as fold changes compared to the metabolite concentration prior to the F− challenge. Due to the significant differences in F− sensitivity of either strain, in these experiments we used a NaF concentration that would cause a similar reduction in the specific growth rate for P. putida KT2440 and the ΔcrcB mutant (50 and 1 mM, respectively; see Figures 1 and 4).
FIGURE 5.
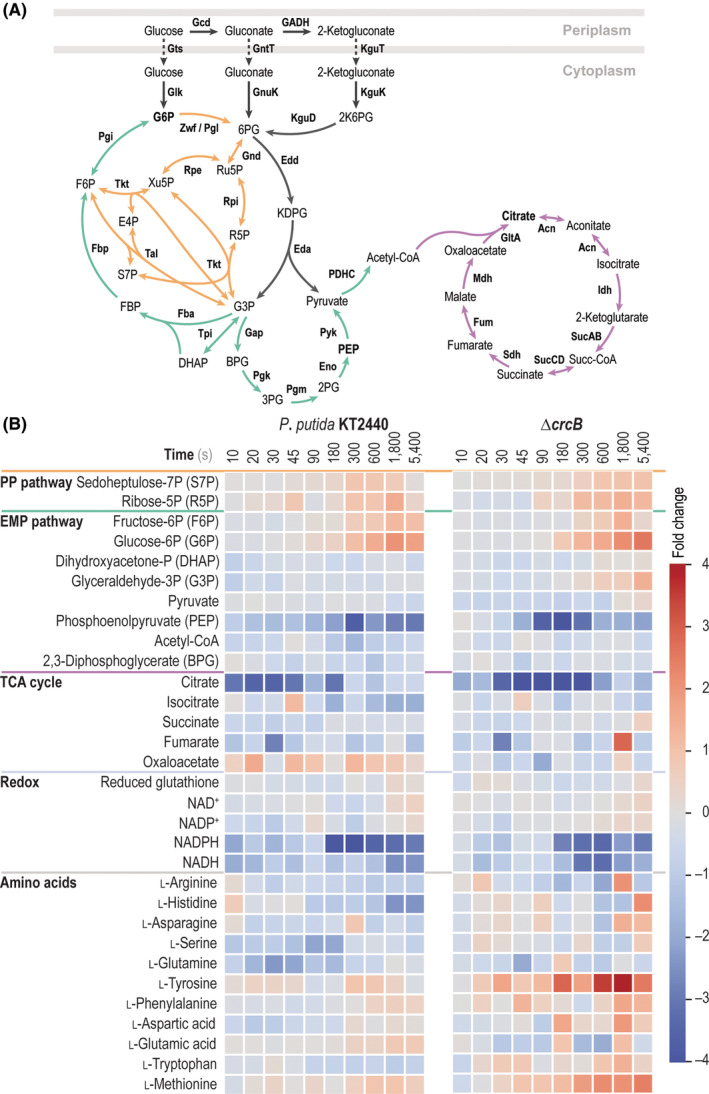
Impact of mineral F− on central carbon metabolism of P. putida KT2440 and the ΔcrcB mutant. (A) Simplified metabolic map of central metabolism in P. putida KT2440. Peripheral reactions of glucose processing and the Entner–Doudoroff pathway are highlighted in grey (transporters are indicated with dashed arrows); reactions within the pentose phosphate (PP) pathway are shown in orange. Green arrows identify the incomplete Embden–Meyerhof–Parnas (EMP) pathway, and purple arrows are used for reactions within the tricarboxylic acid (TCA) cycle. Metabolites that show a concentration difference in the presence of mineral F− are pinpointed in bold face. Abbreviations of metabolites and enzymes are as indicated by Nikel et al. (2015b, 2021). (B) Metabolomic analysis of the impact of NaF on P. putida strains grown in M9 minimal medium containing 5 g L−1 glucose as the only carbon source. The heat maps show the time‐resolved (at 10, 20, 30, 45, 90, 180, 300, 600, 1800 and 5400 s after NaF addition) averaged fold‐change in metabolite concentrations. Individual metabolites are categorized and grouped according to the metabolic blocks defined in panel (A), with the addition of cofactors relevant for redox balance and individual amino acids. All values shown in the plots were normalized to the abundance of the metabolites in cells grown under control conditions (i.e. without F−) for both wild‐type P. putida KT2440 and the ΔcrcB mutant
Some key glycolytic metabolites were affected by the F− treatment [Figure 5(B)], e.g. glucose 6‐phosphate (G6P, Figure S2) and phosphoenolpyruvate (PEP, Figure S3). In general, we detected a trend where metabolites within upper glycolysis (e.g. G6P, sedoheptulose 7‐phosphate, fructose 6‐phosphate and ribose 5‐phosphate) accumulated over time upon NaF addition in both P. putida strains. This accumulation of sugar phosphates can be explained by a general inhibitory effect of glycolytic enzymes by the halide. Several of these enzymes require Mg2+ and Mn2+ for their activity, and it has been shown that F− sequesters the cofactors and impairs enzyme activity in a rather unspecific fashion (Adamek et al., 2005; Johnston & Strobel, 2020). Moreover, we observed depletion of PEP over time, highlighting the inhibition of the enolase enzyme (which catalyses the penultimate step of the EMP pathway, converting 3‐phosphoglycerate to PEP), one of the targets of NaF toxicity previously identified in in vitro studies (Qin et al., 2006). F− also affected the metabolites of the TCA cycle, with a threefold and fourfold change citrate depletion in P. putida KT2440 and the ΔcrcB mutant, respectively (Figure S4). In this sense, some enzymes of the TCA cycle are known to be inhibited by F− (e.g. aconitase and succinate dehydrogenase). Isocitrate dehydrogenase is a rather peculiar case, as this enzyme has been shown to be both inhibited and activated by F− depending on the conditions (Adamek et al., 2005). In any case, an imbalance of the TCA cycle intermediates could be seen in the two P. putida strains tested when F− was present in the culture medium.
Redox power was likewise affected by the halide, especially at the level of NADPH and NADH, as both seemed to be depleted in the presence of F− (Figure S5). This observation could be explained either by the partial inhibition of redox‐active dehydrogenases, e.g. G6P dehydrogenase or the aforementioned isocitrate dehydrogenase, or because of higher rates of consumption of NAD(P)H to counterbalance the oxidative stress imposed by NaF (Johnston & Strobel, 2019). Interestingly, the gene encoding an NADP+‐dependent alcohol dehydrogenase (CurA, PP_2476) was also identified in the Tn‐Seq screening as to be relevant for survival in the presence of F−, which, together with the evidence gathered in physiology experiments, could point to the fact that the F− causes an effect in the redox balance (Table 2). It is worth noticing that the potential collapse of the H+ gradient due to an increase in the intracellular F− concentration could rob the cell of its capacity to couple energy metabolism to ATP synthesis—as indicated by the imbalance in the catabolic redox potential—which would directly impact growth patterns.
Lastly, two amino acids accumulated when F− was present in the medium, i.e. l‐tyrosine and l‐methionine—and this effect was especially evident when the F− transporter CrcB was removed (Figures S6 and S7). We found that the gene encoding the last enzyme involved in l‐methionine synthesis, the B12‐dependent methionine synthase MetH, seemed to be relevant for F− tolerance in the Tn‐Seq experiment (Table 2), providing evidence that l‐methionine levels play a role in the response to the halide. Importantly, the expression of metH was also found to be upregulated in P. putida KT2440 cultivated under other stress conditions, e.g. addition of toluene and o‐xylene (Domínguez‐Cuevas et al., 2006). l‐Methionine biosynthesis is also regulated as part of the general stress response in E. coli (Hondorp & Matthews, 2004). Moreover, and relevant for the conditions analysed herein, l‐methionine has been demonstrated to exert a protective role in rat tissues in the presence of F−, probably mediated by an activation of anti‐oxidative enzymes (Błaszczyk et al., 2010). The time‐dependent accumulation of l‐tyrosine in the ΔcrcB mutant may also be explained as a mechanism of defence against oxidative stress as pointed previously (Liu et al., 2020; Zhang et al., 2021). This aromatic amino acid was accumulated at elevated levels that inversely correlate with the content of its metabolic precursor, PEP. Taken together, these network‐wide metabolomic results indicate that F− mediates a general response in the distribution of metabolites as the enzymes underlying their turnover are affected by the halide (Adamek et al., 2005; Schenk et al., 2008)—either because F− binds to an essential metal in the active site of a metalloprotein or due to the complexing of F− with metals, thus acting as a substrate mimic (e.g. AlF4 − and BeF3 −·H2O).
CONCLUSION
Ever since F− became widespread in a number of environmental niches, both due to geogenic process and human‐related activities (e.g. fluoridation of drinking water, which started just after World War II), bacteria evolved specific tolerance mechanisms against this halide. However, a most interesting finding on the capacities of bacteria to adapt to F− salts is that the main mechanism to counteract toxicity seems to be active export. Furthermore, microbial organisms naturally exposed to toxic F− levels have developed fluoride‐sensing RNA devices (e.g. riboswitches, harnessed as biosensors in this study) to control the expression of genes encoding proteins that relieve the deleterious effects of this anion (Baker et al., 2012). We have extended the analysis of toxicity tolerance by screening a genome‐wide Tn‐Seq library in P. putida, which singled out CrcB (the widespread F− exporter) as the predominant mechanism for alleviating the effects of the halide. This finding is interesting since Pseudomonas is a soil bacterium that thrives across a range of environmental conditions (Volke et al., 2020a)—some of them probably involving exposure to F− salts. These observations also relate to biogeochemical processes comprising F− salts and other elements in groundwater and soil, suggesting that F− concentration should be considered when evaluating microbial response to environmental conditions. Thus, the commonplace use of NaCl as a stressor in microbial physiology studies may not be the most representative chemical insult that soil bacteria have to face in their natural environment. Moreover, Cl− cannot coordinate metal ions in the way that F− does, adding an extra layer to the complex metabolic and chemical response elicited by NaF compared to other halide salts.
Since many of the actions of F− are connected to its weak‐acid (or strong‐base) character, the mechanism of action of this anion is comparable to that of organic weak acids, e.g. metabolic acids (acetate), food preservatives, non‐steroidal anti‐inflammatory agents and fatty acids—all of which act to de‐energize the cell membrane by dissipating ΔpH (Marquis et al., 2003). These effects are further amplified by an acidic extracellular pH and involve a significant impact on several biochemical processes. Indeed, our findings indicate that the metabolic response of P. putida to a challenge with sub‐lethal concentrations of NaF invokes a network‐wide re‐arrangement of key metabolites within central carbon metabolism that, in some cases, resembles the alterations provoked by acid stress (Akkaya et al., 2018; Corona et al., 2019; Reva et al., 2006).
Besides the body of experimental evidence related to the treatment of bacterial cells with F−, as presented here, an emerging prospect of our results is their potential application for biodegradation and biosynthesis of organofluorines. As summarized by Wackett (2022) in a recent report, efficient defluorination activities (either native or engineered) will call for high levels of tolerance to increased intracellular F− concentrations by the microbial host. Pseudomonas genomes encode various enzyme types that could potentially catalyse defluorination, both through oxygenase‐mediated biodegradation processes and activities typically involved in anaerobic metabolism, e.g. radical S‐adenosyl‐l‐methionine class, vitamin B12‐mediated reductions, tyrosyl radical enzymes and aromatic dehydroxylating reductases, among many others. While the actual activity of these enzymes in defluorination processes awaits to be fully unveiled, it seems plausible that such degradative enzymes could also had a role in shaping the response of P. putida to F− salts by releasing the halide intracellularly. Moreover, the results discussed herein will also guide future engineering approaches for organofluorine biosynthesis (Nieto‐Domínguez & Nikel, 2020), where high F− levels are required to both trigger gene expression and ensure a high‐enough substrate availability for fluorinating enzymes.
CONFLICT OF INTEREST
The authors declare that there are no competing interests associated with the contents of this article.
Supporting information
TABLE S1 Oligonucleotides used in this study
TABLE S2 Beneficial insertions in the tolerance of P. putida towards F− stress.
FIGURE S1 Effect of KF on bacterial growth.
FIGURE S2 Metabolomic analysis of sugar phosphates.
FIGURE S3 Metabolomic analysis of intermediates of the lower glycolysis.
FIGURE S4 Metabolomic analysis of intermediates of the tricarboxylic acid cycle.
FIGURE S5 Metabolomic analysis of redox cofactors and related metabolites.
FIGURE S6 Metabolomic analysis of intracellular amino acids.
FIGURE S7 Metabolomic analysis of intracellular (aromatic and sulphur‐containing) amino acids.
ACKNOWLEDGEMENTS
We would like to thank Prof. Antoine Danchin for enlightening discussions on the biochemistry of fluorine. The financial support from The Novo Nordisk Foundation through grants NNF20CC0035580, LiFe (NNF18OC0034818) and TARGET (NNF21OC0067996), the Danish Council for Independent Research (SWEET, DFF‐Research Project 8021‐00039B), the European Union's Horizon 2020 Research and Innovation Programme under grant agreement No. 814418 (SinFonia) and the Cystic Fibrosis Trust, Strategic Research Centre Award–2019–SRC017 (UK) to P.I.N. is gratefully acknowledged.
Calero, P. , Gurdo, N. & Nikel, P.I. (2022) Role of the CrcB transporter of Pseudomonas putida in the multi‐level stress response elicited by mineral fluoride. Environmental Microbiology, 24(11), 5082–5104. Available from: 10.1111/1462-2920.16110
Funding information H2020 Environment, Grant/Award Number: 814418; Novo Nordisk Fonden, Grant/Award Numbers: NNF18OC0034818, NNF20CC0035580, NNF21OC0067996; Cystic Fibrosis Trust, Strategic Research Centre Award–2019–SRC017; Danish Council for Independent Research Natural Sciences, Grant/Award Number: 8021‐00039B
REFERENCES
- Abril, M.A. , Michan, C. , Timmis, K.N. & Ramos, J.L. (1989) Regulator and enzyme specificities of the TOL plasmid‐encoded upper pathway for degradation of aromatic hydrocarbons and expansion of the substrate range of the pathway. Journal of Bacteriology, 171, 6782–6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamek, E. , Pawłowska‐Góral, K. & Bober, K. (2005) In vitro and in vivo effects of fluoride ions on enzyme activity. Annales Academiae Medicae Stetinensis, 51, 69–85. [PubMed] [Google Scholar]
- Akkaya, Ö. , Pérez‐Pantoja, D. , Calles, B. , Nikel, P.I. & de Lorenzo, V. (2018) The metabolic redox regime of Pseudomonas putida tunes its evolvability toward novel xenobiotic substrates. mBio, 9, e01512‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arce‐Rodríguez, A. , Volke, D.C. , Bense, S. , Häussler, S. & Nikel, P.I. (2019) Non‐invasive, ratiometric determination of intracellular pH in Pseudomonas species using a novel genetically encoded indicator. Microbial Biotechnology, 12, 799–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagdasarian, M. , Lurz, R. , Rückert, B. , Franklin, F.C.H. , Bagdasarian, M.M. , Frey, J. et al. (1981) Specific purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010‐derived vectors, and a host‐vector system for gene cloning in Pseudomonas . Gene, 16, 237–247. [DOI] [PubMed] [Google Scholar]
- Baker, J.L. , Sudarsan, N. , Weinberg, Z. , Roth, A. , Stockbridge, R.B. & Breaker, R.R. (2012) Widespread genetic switches and toxicity resistance proteins for fluoride. Science, 335, 233–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baykov, A.A. , Fabrichniy, I.P. , Pohjanjoki, P. , Zyryanov, A.B. & Lahti, R. (2000) Fluoride effects along the reaction pathway of pyrophosphatase: evidence for a second enzyme·pyrophosphate intermediate. Biochemistry, 39, 11939–11947. [DOI] [PubMed] [Google Scholar]
- Belda, E. , van Heck, R.G.A. , López‐Sánchez, M.J. , Cruveiller, S. , Barbe, V. , Fraser, C. et al. (2016) The revisited genome of Pseudomonas putida KT2440 enlightens its value as a robust metabolic chassis . Environmental Microbiology, 18, 3403–3424. [DOI] [PubMed] [Google Scholar]
- Benedetti, I. , de Lorenzo, V. & Nikel, P.I. (2016) Genetic programming of catalytic Pseudomonas putida biofilms for boosting biodegradation of haloalkanes. Metabolic Engineering, 33, 109–118. [DOI] [PubMed] [Google Scholar]
- Bhatnagar, M. & Bhatnagar, A. (2004) Physiology of Anabaena khannae and Chlorococcum humicola under fluoride stress. Folia Microbiologica, 49, 291–296. [DOI] [PubMed] [Google Scholar]
- Bitzenhofer, N.L. , Kruse, L. , Thies, S. , Wynands, B. , Lechtenberg, T. , Rönitz, J. et al. (2021) Towards robust Pseudomonas cell factories to harbour novel biosynthetic pathways. Essays in Biochemistry, 65, 319–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Błaszczyk, I. , Grucka‐Mamczar, E. , Kasperczyk, S. & Birkner, E. (2010) Influence of methionine upon the activity of antioxidative enzymes in the kidney of rats exposed to sodium fluoride. Biological Trace Element Research, 133, 60–70. [DOI] [PubMed] [Google Scholar]
- Bradshaw, D.J. , McKee, A.S. & Marsh, P.D. (1990) Prevention of population shifts in oral microbial communities in vitro by low fluoride concentrations. Journal of Dental Research, 69, 436–441. [DOI] [PubMed] [Google Scholar]
- Bradshaw, D.J. , Marsh, P.D. , Hodgson, R.J. & Visser, J.M. (2002) Effects of glucose and fluoride on competition and metabolism within in vitro dental bacterial communities and biofilms. Caries Research, 36, 81–86. [DOI] [PubMed] [Google Scholar]
- Burby, P.E. , Nye, T.M. , Schroeder, J.W. , Simmons, L.A. & Margolin, W. (2017) Implementation and data analysis of Tn‐seq, whole‐genome resequencing, and single‐molecule real‐time sequencing for bacterial genetics. Journal of Bacteriology, 199, e00560‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger, M. , Woods, R.G. , McCarthy, C. & Beacham, I.R. (2000) Temperature regulation of protease in Pseudomonas fluorescens LS107d2 by an ECF sigma factor and a transmembrane activator. Microbiology, 146, 3149–3155. [DOI] [PubMed] [Google Scholar]
- Bygd, M.D. , Aukema, K.G. , Richman, J.E. & Wackett, L.P. (2021) Unexpected mechanism of biodegradation and defluorination of 2,2‐difluoro‐1,3‐benzodioxole by Pseudomonas putida F1. MBio, 12, e03001‐21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, Y. , Liao, Y. , Brandt, B.W. , Wei, X. , Liu, H. , Crielaard, W. et al. (2017) The fitness cost of fluoride resistance for different Streptococcus mutans strains in biofilms. Frontiers in Microbiology, 8, 1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calero, P. , Jensen, S.I. , Bojanovič, K. , Lennen, R.M. , Koza, A. & Nielsen, A.T. (2018) Genome‐wide identification of tolerance mechanisms toward p‐coumaric acid in Pseudomonas putida . Biotechnology and Bioengineering, 115, 762–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calero, P. , Volke, D.C. , Lowe, P.T. , Gotfredsen, C.H. , O'Hagan, D. & Nikel, P.I. (2020) A fluoride‐responsive genetic circuit enables in vivo biofluorination in engineered Pseudomonas putida . Nature Communications, 11, 5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo, J.A. (2003) Fluoride toxicity to aquatic organisms: a review. Chemosphere, 50, 251–264. [DOI] [PubMed] [Google Scholar]
- Chahine, S. , Garau, G. , Castaldi, P. , Pinna, M.V. , Melito, S. , Seddaiu, G. et al. (2022) Stabilising fluoride in contaminated soils with monocalcium phosphate and municipal solid waste compost: microbial, biochemical and plant growth impact. Environmental Science and Pollution Research International, 29, 41820–41833. [DOI] [PubMed] [Google Scholar]
- Cheng, X. & Ma, L. (2021) Enzymatic synthesis of fluorinated compounds. Applied Microbiology and Biotechnology, 105, 8033–8058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corona, F. , Martínez, J.L. & Nikel, P.I. (2019) The global regulator Crc orchestrates the metabolic robustness underlying oxidative stress resistance in Pseudomonas aeruginosa . Environmental Microbiology, 21, 898–912. [DOI] [PubMed] [Google Scholar]
- Cros, A. , Alfaro‐Espinoza, G. , de Maria, A. , Wirth, N.T. & Nikel, P.I. (2022) Synthetic metabolism for biohalogenation. Current Opinion in Biotechnology, 74, 180–193. [DOI] [PubMed] [Google Scholar]
- Cuthbertson, L. , Kos, V. & Whitfield, C. (2010) ABC transporters involved in export of cell surface glycoconjugates. Microbiology and Molecular Biology Reviews, 74, 341–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damé‐Teixeira, N. , Deng, D. & Do, T. (2019) Streptococcus mutans transcriptome in the presence of sodium fluoride and sucrose. Archives of Oral Biology, 102, 186–192. [DOI] [PubMed] [Google Scholar]
- Danchin, A. & Nikel, P.I. (2019) Why nature chose potassium. Journal of Molecular Evolution, 87, 271–288. [DOI] [PubMed] [Google Scholar]
- Dionizio, A. , Uyghurturk, D.A. , Souza Melo, C.G. , Sabino‐Arias, I.T. , Araujo, T.T. , Silva Ventura, T.M. et al. (2021) Intestinal changes associated with fluoride exposure in rats: integrative morphological, proteomic and microbiome analyses. Chemosphere, 273, 129607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez‐Cuevas, P. , González‐Pastor, J.E. , Marqués, S. , Ramos, J.L. & de Lorenzo, V. (2006) Transcriptional tradeoff between metabolic and stress‐response programs in Pseudomonas putida KT2440 cells exposed to toluene. The Journal of Biological Chemistry, 281, 11981–11991. [DOI] [PubMed] [Google Scholar]
- El‐Kirat‐Chatel, S. , Beaussart, A. , Boyd, C.D. , O'Toole, G.A. & Dufrêne, Y.F. (2014) Single‐cell and single‐molecule analysis deciphers the localization, adhesion, and mechanics of the biofilm adhesin LapA. ACS Chemical Biology, 9, 485–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng, T. , Banerjee, D. , Lau, A.K. , Bowden, E. , Herbert, R.A. , Trinh, J. et al. (2021) Engineering Pseudomonas putida for efficient aromatic conversion to bioproduct using high throughput screening in a bioreactor. Metabolic Engineering, 66, 229–238. [DOI] [PubMed] [Google Scholar]
- Fernández‐Cabezón, L. , Rosich i Bosch, B. , Kozaeva, E. , Gurdo, N. & Nikel, P.I. (2022) Dynamic flux regulation for high‐titer anthranilate production by plasmid‐free, conditionally‐auxotrophic strains of Pseudomonas putida . Metabolic Engineering, 73, 11–25. [DOI] [PubMed] [Google Scholar]
- Follonier, S. , Panke, S. & Zinn, M. (2011) A reduction in growth rate of Pseudomonas putida KT2442 counteracts productivity advances in medium‐chain‐length polyhydroxyalkanoate production from gluconate. Microbial Cell Factories, 10, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follonier, S. , Escapa, I.F. , Fonseca, P.M. , Henes, B. , Panke, S. , Zinn, M. et al. (2013) New insights on the reorganization of gene transcription in Pseudomonas putida KT2440 at elevated pressure. Microbial Cell Factories, 12, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geu‐Flores, F. , Nour‐Eldin, H.H. , Nielsen, M.T. & Halkier, B.A. (2007) USER fusion: a rapid and efficient method for simultaneous fusion and cloning of multiple PCR products. Nucleic Acids Research, 35, e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjermansen, M. , Nilsson, M. , Yang, L. & Tolker‐Nielsen, T. (2010) Characterization of starvation‐induced dispersion in Pseudomonas putida biofilms: genetic elements and molecular mechanisms. Molecular Microbiology, 75, 815–826. [DOI] [PubMed] [Google Scholar]
- Greenwood, N.N. & Earnshaw, A. (1984) Chemistry of the elements. Oxford: Pergamon Press. [Google Scholar]
- Gribble, W.G. (2002) Naturally occurring organofluorines. In: Neilson, A.H. (Ed.) Organofluorines. The handbook of environmental chemistry. Berlin, Germany: Springer, pp. 121–136. [Google Scholar]
- Hachicho, N. , Birnbaum, A. & Heipieper, H.J. (2017) Osmotic stress in colony and planktonic cells of Pseudomonas putida mt‐2 revealed significant differences in adaptive response mechanisms. AMB Express, 7, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton, I.R. (1990) Biochemical effects of fluoride on oral bacteria. Journal of Dental Research, 69, 660–667. [DOI] [PubMed] [Google Scholar]
- Harsanyi, A. & Sandford, G. (2015) Organofluorine chemistry: applications, sources and sustainability. Green Chemistry, 17, 2081–2086. [Google Scholar]
- Hondorp, E.R. & Matthews, R.G. (2004) Oxidative stress inactivates cobalamin‐independent methionine synthase (MetE) in Escherichia coli . PLoS Biology, 2, e336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison, C.A. , Merryman, C. , Sun, L. , Assad‐García, N. , Richter, R.A. , Smith, H.O. et al. (2019) Polar effects of transposon insertion into a minimal bacterial genome. Journal of Bacteriology, 201, e00185‐019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, C. , Stockbridge, R.B. & Miller, C. (2014) Bacterial fluoride resistance, Fluc channels, and the weak acid accumulation effect. The Journal of General Physiology, 144, 257–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston, N.R. & Strobel, S.A. (2019) Nitrate and phosphate transporters rescue fluoride toxicity in yeast. Chemical Research in Toxicology, 32, 2305–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston, N.R. & Strobel, S.A. (2020) Principles of fluoride toxicity and the cellular response: a review. Archives of Toxicology, 94, 1051–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieboom, J. , Dennis, J.J. , Zylstra, G.J. & de Bont, J.A.M. (1998) Active efflux of organic solvents by Pseudomonas putida S12 is induced by solvents. Journal of Bacteriology, 180, 6769–6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozaeva, E. , Volkova, S. , Matos, M.R.A. , Mezzina, M.P. , Wulff, T. , Volke, D.C. et al. (2021) Model‐guided dynamic control of essential metabolic nodes boosts acetyl‐coenzyme A–dependent bioproduction in rewired Pseudomonas putida . Metabolic Engineering, 67, 373–386. [DOI] [PubMed] [Google Scholar]
- Kutuzova, S. , Colaianni, P. , Röst, H. , Sachsenberg, T. , Alka, O. , Kohlbacher, O. et al. (2020) SmartPeak automates targeted and quantitative metabolomics data processing. Analytical Chemistry, 92, 15968–15974. [DOI] [PubMed] [Google Scholar]
- Last, N.B. , Kolmakova‐Partensky, L. , Shane, T. & Miller, C. (2016) Mechanistic signs of double‐barreled structure in a fluoride ion channel. eLife, 5, e18767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennen, R. & Herrgård, M.J. (2014) Combinatorial strategies for improving multiple‐stress resistance in industrially relevant Escherichia coli strains. Applied and Environmental Microbiology, 80, 6223–6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L. (2003) The biochemistry and physiology of metallic fluoride: action, mechanism, and implications. Critical Reviews in Oral Biology and Medicine, 14, 100–114. [DOI] [PubMed] [Google Scholar]
- Liao, Y. , Brandt, B.W. , Li, J. , Crielaard, W. , van Loveren, C. & Deng, D.M. (2017) Fluoride resistance in Streptococcus mutans: a mini review. Journal of Oral Microbiology, 9, 1344509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X. , Tian, J. , Liu, L. , Zhu, T. , Yu, X. , Chu, X. et al. (2017) Identification of an operon involved in fluoride resistance in Enterobacter cloacae FRM. Scientific Reports, 7, 6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Liu, J. , Liu, M. , Strappe, P. , Sun, H. & Zhou, Z. (2020) Comparative non‐targeted metabolomic analysis reveals insights into the mechanism of rice yellowing. Food Chemistry, 308, 125621. [DOI] [PubMed] [Google Scholar]
- Llamas, M.A. , Imperi, F. , Visca, P. & Lamont, I.L. (2014) Cell‐surface signaling in Pseudomonas: stress responses, iron transport, and pathogenicity. FEMS Microbiology Reviews, 38, 569–597. [DOI] [PubMed] [Google Scholar]
- López‐Sánchez, A. , Leal‐Morales, A. , Jiménez‐Díaz, L. , Platero, A.I. , Bardallo‐Pérez, J. , Díaz‐Romero, A. et al. (2016) Biofilm formation‐defective mutants in Pseudomonas putida . FEMS Microbiology Letters, 363, fnw127. [DOI] [PubMed] [Google Scholar]
- Ma, H. , Wu, X. , Yang, M. , Wang, J. , Wang, J. & Wang, J. (2014) Effects of fluoride on bacterial growth and its gene/protein expression. Chemosphere, 100, 190–193. [DOI] [PubMed] [Google Scholar]
- Manome, A. , Abiko, Y. , Kawashima, J. , Washio, J. , Fukumoto, S. & Takahashi, N. (2019) Acidogenic potential of oral Bifidobacterium and its high fluoride tolerance. Frontiers in Microbiology, 10, 1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquis, R.E. (1995) Antimicrobial actions of fluoride for oral bacteria. Canadian Journal of Microbiology, 41, 955–964. [DOI] [PubMed] [Google Scholar]
- Marquis, R.E. , Clock, S.A. & Mota‐Meira, M. (2003) Fluoride and organic weak acids as modulators of microbial physiology. FEMS Microbiology Reviews, 26, 493–510. [DOI] [PubMed] [Google Scholar]
- Martínez‐García, E. & de Lorenzo, V. (2011) Engineering multiple genomic deletions in Gram‐negative bacteria: analysis of the multi‐resistant antibiotic profile of Pseudomonas putida KT2440. Environmental Microbiology, 13, 2702–2716. [DOI] [PubMed] [Google Scholar]
- Martínez‐García, E. , Aparicio, T. , de Lorenzo, V. & Nikel, P.I. (2014) New transposon tools tailored for metabolic engineering of Gram‐negative microbial cell factories. Frontiers in Bioengineering and Biotechnology, 2, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez‐García, E. , Aparicio, T. , Goñi‐Moreno, A. , Fraile, S. & de Lorenzo, V. (2015) SEVA 2.0: an update of the Standard European Vector Architecture for de‐/re‐construction of bacterial functionalities. Nucleic Acids Research, 43, D1183–D1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason, B. & Moore, C. (1982) Principles of geochemistry. New York: Wiley. [Google Scholar]
- McCloskey, D. , Xu, J. , Schrübbers, L. , Christensen, H.B. & Herrgård, M.J. (2018) RapidRIP quantifies the intracellular metabolome of 7 industrial strains of E. coli . Metabolic Engineering, 47, 383–392. [DOI] [PubMed] [Google Scholar]
- McIlwain, B.C. , Ruprecht, M.T. & Stockbridge, R.B. (2021) Membrane exporters of fluoride ion. Annual Review of Biochemistry, 90, 559–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meselson, M. & Yuan, R. (1968) DNA restriction enzyme from E. coli . Nature, 217, 1110–1114. [DOI] [PubMed] [Google Scholar]
- Molina‐Henares, M.A. , de la Torre, J. , García‐Salamanca, A. , Molina‐Henares, A.J. , Herrera, M.C. , Ramos, J.L. et al. (2010) Identification of conditionally essential genes for growth of Pseudomonas putida KT2440 on minimal medium through the screening of a genome‐wide mutant library. Environmental Microbiology, 12, 1468–1485. [DOI] [PubMed] [Google Scholar]
- Molina‐Santiago, C. , Cordero, B.F. , Daddaoua, A. , Udaondo, Z. , Manzano, J. , Valdivia, M. et al. (2016) Pseudomonas putida as a platform for the synthesis of aromatic compounds. Microbiology, 162, 1535–1543. [DOI] [PubMed] [Google Scholar]
- Mosqueda, G. , Ramos‐González, M.I. & Ramos, J.L. (1999) Toluene metabolism by the solvent‐tolerant Pseudomonas putida DOT‐T1 strain, and its role in solvent impermeabilization. Gene, 232, 69–76. [DOI] [PubMed] [Google Scholar]
- Muini, D. , Vrek, D. , Mali, A.I. , Matijevic, J. , Grget, K.R. & Krmek, S.J. (2012) The concentration of fluorides in tap water and commercial bottled beverages. Acta Stomatologica Croatica, 46, 23–30. [Google Scholar]
- Mumm, K. , Ainsaar, K. , Kasvandik, S. , Tenson, T. & Hõrak, R. (2016) Responses of Pseudomonas putida to zinc excess determined at the proteome level: pathways dependent and independent of ColRS. Journal of Proteome Research, 15, 4349–4368. [DOI] [PubMed] [Google Scholar]
- Neilson, A.H. & Allard, A.S. (2002) Degradation and transformation of organic fluorine compounds. In: Neilson, A.H. (Ed.) Organofluorines. Berlin, Heidelberg: Springer, pp. 137–202. [Google Scholar]
- Nieto‐Domínguez, M. & Nikel, P.I. (2020) Intersecting xenobiology and neo‐metabolism to bring novel chemistries to life. ChemBioChem, 21, 2551–2571. [DOI] [PubMed] [Google Scholar]
- Nikel, P.I. , Zhu, J. , San, K.Y. , Méndez, B.S. & Bennett, G.N. (2009) Metabolic flux analysis of Escherichia coli creB and arcA mutants reveals shared control of carbon catabolism under microaerobic growth conditions. Journal of Bacteriology, 191, 5538–5548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikel, P.I. , Romero‐Campero, F.J. , Zeidman, J.A. , Goñi‐Moreno, A. & de Lorenzo, V. (2015a) The glycerol‐dependent metabolic persistence of Pseudomonas putida KT2440 reflects the regulatory logic of the GlpR repressor. mBio, 6, e00340‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikel, P.I. , Chavarría, M. , Fuhrer, T. , Sauer, U. & de Lorenzo, V. (2015b) Pseudomonas putida KT2440 strain metabolizes glucose through a cycle formed by enzymes of the Entner‐Doudoroff, Embden‐Meyerhof‐Parnas, and pentose phosphate pathways. The Journal of Biological Chemistry, 290, 25920–25932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikel, P.I. , Chavarría, M. , Danchin, A. & de Lorenzo, V. (2016a) From dirt to industrial applications: Pseudomonas putida as a synthetic biology chassis for hosting harsh biochemical reactions. Current Opinion in Chemical Biology, 34, 20–29. [DOI] [PubMed] [Google Scholar]
- Nikel, P.I. , Pérez‐Pantoja, D. & de Lorenzo, V. (2016b) Pyridine nucleotide transhydrogenases enable redox balance of Pseudomonas putida during biodegradation of aromatic compounds. Environmental Microbiology, 18, 3565–3582. [DOI] [PubMed] [Google Scholar]
- Nikel, P.I. , Fuhrer, T. , Chavarría, M. , Sánchez‐Pascuala, A. , Sauer, U. & de Lorenzo, V. (2021) Reconfiguration of metabolic fluxes in Pseudomonas putida as a response to sub‐lethal oxidative stress. The ISME Journal, 15, 1751–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochoa‐Herrera, V. , Banihani, Q. , León, G. , Khatri, C. , Field, J.A. & Sierra‐Álvarez, R. (2009) Toxicity of fluoride to microorganisms in biological wastewater treatment systems. Water Research, 43, 3177–3186. [DOI] [PubMed] [Google Scholar]
- Pardo, I. , Bednar, D. , Calero, P. , Volke, D.C. , Damborský, J. & Nikel, P.I. (2022) A nonconventional archaeal fluorinase identified by in silico mining for enhanced fluorine biocatalysis. ACS Catalysis, 12, 6570–6577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, J. , Chai, G. , Brewer, J.M. , Lovelace, L.L. & Lebioda, L. (2006) Fluoride inhibition of enolase: crystal structure and thermodynamics. Biochemistry, 45, 793–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reva, O.N. , Weinel, C. , Weinel, M. , Böhm, K. , Stjepandic, D. , Hoheisel, J.D. et al. (2006) Functional genomics of stress response in Pseudomonas putida KT2440. Journal of Bacteriology, 188, 4079–4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J. & Russell, D.W. (2001) Molecular cloning: a laboratory manual. Cold Spring Harbor: Cold Spring Harbor Laboratory. [Google Scholar]
- Samygina, V.R. , Moiseev, V.M. , Rodina, E.V. , Vorobyeva, N.N. , Popov, A.N. , Kurilova, S.A. et al. (2007) Reversible inhibition of Escherichia coli inorganic pyrophosphatase by fluoride: trapped catalytic intermediates in cryo‐crystallographic studies. Journal of Molecular Biology, 366, 1305–1317. [DOI] [PubMed] [Google Scholar]
- Sánchez‐Clemente, R. , Igeño, M.I. , Población, A.G. , Guijo, M.I. , Merchán, F. & Blasco, R. (2018) Study of pH changes in media during bacterial growth of several environmental strains. Proceedings, 2, 1297. [Google Scholar]
- Sánchez‐Pascuala, A. , Fernández‐Cabezón, L. , de Lorenzo, V. & Nikel, P.I. (2019) Functional implementation of a linear glycolysis for sugar catabolism in Pseudomonas putida . Metabolic Engineering, 54, 200–211. [DOI] [PubMed] [Google Scholar]
- Schenk, G. , Elliott, T.W. , Leung, E. , Carrington, L.E. , Mitić, N. , Gahan, L.R. et al. (2008) Crystal structures of a purple acid phosphatase, representing different steps of this enzyme's catalytic cycle. BMC Structural Biology, 8, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, H. & Kim, K.J. (2018) Structural insight into molecular mechanism of cytokinin activating protein from Pseudomonas aeruginosa PAO1. Environmental Microbiology, 20, 3214–3223. [DOI] [PubMed] [Google Scholar]
- Silva‐Rocha, R. , Martínez‐García, E. , Calles, B. , Chavarría, M. , Arce‐Rodríguez, A. , de las Heras, A. et al. (2013) The Standard European Vector Architecture (SEVA): a coherent platform for the analysis and deployment of complex prokaryotic phenotypes. Nucleic Acids Research, 41, D666–D675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, C. , Aundy, K. , van de Mortel, J. & Raaijmakers, J.M. (2014) Discovery of new regulatory genes of lipopeptide biosynthesis in Pseudomonas fluorescens . FEMS Microbiology Letters, 356, 166–175. [DOI] [PubMed] [Google Scholar]
- Stockbridge, R.B. , Lim, H.H. , Otten, R. , Williams, C. , Shane, T. , Weinberg, Z. et al. (2012) Fluoride resistance and transport by riboswitch‐controlled CLC antiporters. Proceedings of the National Academy of Sciences of the United States of America, 109, 15289–15294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockbridge, R.B. , Robertson, J.L. , Kolmakova‐Partensky, L. & Miller, C. (2013) A family of fluoride‐specific ion channels with dual‐topology architecture. eLife, 2, e01084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniyam, S. , DeJesus, M.A. , Zaveri, A. , Smith, C.M. , Baker, R.E. , Ehrt, S. et al. (2019) Statistical analysis of variability in TnSeq data across conditions using zero‐inflated negative binomial regression. BMC Bioinformatics, 20, 603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, M.G. , Valencia, L.E. , Blake‐Hedges, J.M. , Cruz‐Morales, P. , Velasquez, A.E. , Pearson, A.N. et al. (2019) Omics‐driven identification and elimination of valerolactam catabolism in Pseudomonas putida KT2440 for increased product titer. Metabolic Engineering Communications, 9, e00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, M.G. , Incha, M.R. , Pearson, A.N. , Schmidt, M. , Sharpless, W.A. , Eiben, C.B. et al. (2020) Fatty acid and alcohol metabolism in Pseudomonas putida: functional analysis using random barcode transposon sequencing. Applied and Environmental Microbiology, 86, e01665‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thongboonkerd, V. , Luengpailin, J. , Cao, J. , Pierce, W.M. , Cai, J. , Klein, J.B. et al. (2002) Fluoride exposure attenuates expression of Streptococcus pyogenes virulence factors. The Journal of Biological Chemistry, 277, 16599–16605. [DOI] [PubMed] [Google Scholar]
- Volke, D.C. , Calero, P. & Nikel, P.I. (2020a) Pseudomonas putida . Trends in Microbiology, 28, 512–513. [DOI] [PubMed] [Google Scholar]
- Volke, D.C. , Friis, L. , Wirth, N.T. , Turlin, J. & Nikel, P.I. (2020b) Synthetic control of plasmid replication enables target‐ and self‐curing of vectors and expedites genome engineering of Pseudomonas putida . Metabolic Engineering Communications, 10, e00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volke, D.C. , Turlin, J. , Mol, V. & Nikel, P.I. (2020c) Physical decoupling of XylS/Pm regulatory elements and conditional proteolysis enable precise control of gene expression in Pseudomonas putida . Microbial Biotechnology, 13, 222–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volke, D.C. , Olavarría, K. & Nikel, P.I. (2021) Cofactor specificity of glucose‐6‐phosphate dehydrogenase isozymes in Pseudomonas putida reveals a general principle underlying glycolytic strategies in bacteria. mSystems, 6, e00014‐21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volke, D.C. , Martino, R.A. , Kozaeva, E. , Smania, A.M. & Nikel, P.I. (2022) Modular (de)construction of complex bacterial phenotypes by CRISPR/nCas9‐assisted, multiplex cytidine base‐editing. Nature Communications, 13, 3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wackett, L.P. (2021a) Microbial responses to fluoride in the environment: an annotated selection of World Wide Web sites relevant to the topics in environmental microbiology. Environmental Microbiology Reports, 13, 955–956. [DOI] [PubMed] [Google Scholar]
- Wackett, L.P. (2021b) Why is the biodegradation of polyfluorinated compounds so rare? mSphere, 6, e00721‐21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wackett, L.P. (2021c) Nothing lasts forever: understanding microbial biodegradation of polyfluorinated compounds and perfluorinated alkyl substances. Microbial Biotechnology, 15, 773–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wackett, L.P. (2022) Pseudomonas: versatile biocatalysts for PFAS. Environmental Microbiology (in press). 10.1111/1462-2920.15990 [DOI] [PubMed] [Google Scholar]
- Weimer, A. , Kohlstedt, M. , Volke, D.C. , Nikel, P.I. & Wittmann, C. (2020) Industrial biotechnology of Pseudomonas putida: advances and prospects. Applied Microbiology and Biotechnology, 104, 7745–7766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth, N.T. & Nikel, P.I. (2021) Combinatorial pathway balancing provides biosynthetic access to 2‐fluoro‐cis,cis‐muconate in engineered Pseudomonas putida . Chem Catalysis, 1, 1234–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth, N.T. , Kozaeva, E. & Nikel, P.I. (2020) Accelerated genome engineering of Pseudomonas putida by I‐SceI—mediated recombination and CRISPR‐Cas9 counterselection. Microbial Biotechnology, 13, 233–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, L. , Maglangit, F. & Deng, H. (2020) Fluorine biocatalysis. Current Opinion in Chemical Biology, 55, 119–126. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, K. , Tamura, Z. & Maeda, M. (1997) Molecular structure of the zwitterionic form of phenolsulfonphthalein. Analytical Sciences, 13, 521–522. [Google Scholar]
- Yousef‐Coronado, F. , Soriano, M.I. , Yang, L. , Molin, S. & Espinosa‐Urgel, M. (2011) Selection of hyperadherent mutants in Pseudomonas putida biofilms. Microbiology, 157, 2257–2265. [DOI] [PubMed] [Google Scholar]
- Zhang, H. , Liu, J. , Wen, R. , Chen, Q. & Kong, B. (2021) Metabolomics profiling reveals defense strategies of Pediococcus pentosaceus R1 isolated from Harbin dry sausages under oxidative stress. LWT, 135, 110041. [Google Scholar]
- Zomer, A. , Burghout, P. , Bootsma, H.J. , Hermans, P.W.M. & van Hijum, S.A.F.T. (2012) ESSENTIALS: software for rapid analysis of high throughput transposon insertion sequencing data. PLoS One, 7, e43012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
TABLE S1 Oligonucleotides used in this study
TABLE S2 Beneficial insertions in the tolerance of P. putida towards F− stress.
FIGURE S1 Effect of KF on bacterial growth.
FIGURE S2 Metabolomic analysis of sugar phosphates.
FIGURE S3 Metabolomic analysis of intermediates of the lower glycolysis.
FIGURE S4 Metabolomic analysis of intermediates of the tricarboxylic acid cycle.
FIGURE S5 Metabolomic analysis of redox cofactors and related metabolites.
FIGURE S6 Metabolomic analysis of intracellular amino acids.
FIGURE S7 Metabolomic analysis of intracellular (aromatic and sulphur‐containing) amino acids.


