Abstract
In the last two years, the world encountered the SARS-CoV-2 virus, which is still dominating the population due to the absence of a viable treatment. To eradicate the global pandemic, scientists, doctors, and researchers took an exceptionally significant initiative towards the development of effective therapeutics to save many lifes. This review discusses about the single-domain antibodies (sdAbs), also called nanobodies, their structure, and their types against the infections of dreadful SARS-CoV-2 virus. A precise description highlights the nanobodies and their therapeutic application against the other selected viruses. It aims to focus on the extraordinary features of these antibodies compared to the conventional therapeutics like mAbs, convalescent plasma therapy, and vaccines. The stable structure of these nanobodies along with the suitable mechanism of action also confers greater resistance to the evolving variants with numerous mutations. The nanobodies developed against SARS-CoV-2 and its mutant variants have shown the greater neutralization potential than the primitive ones. Engineering of these specialized antibodies by modern biotechnological approaches will surely be more beneficial in treating this COVID-19 pandemic along with certain other viral infections.
Keywords: Nanobodies, SARS-CoV-2, Neutralization, Virus
1. Introduction
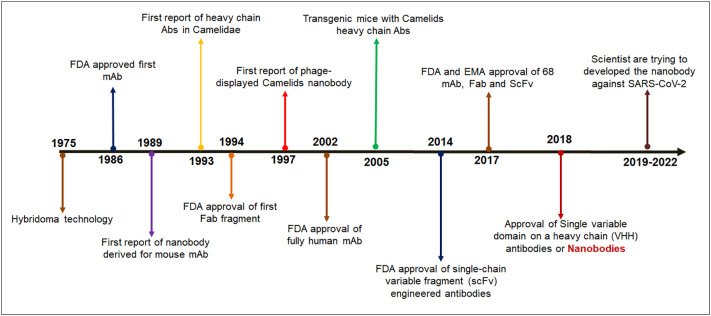
With the onset of the global infection by the SARS-CoV-2 virus, the scientific community got engaged in discovering effective therapeutics to eradicate the viral infection's detrimental effects. One of the effective treatment procedures is the use of monoclonal antibodies. [1]. It gave a promising breakthrough to the pharmaceutical industry. Monoclonal antibodies have been proven successful groups of therapeutic molecules which has can treat SARS-CoV-2 infections. Besides, it was also a ray of hope in the previous global outbreaks, namely MERS and SARS. However, due to certain limitations, the replacement of monoclonal antibodies (mAb's) becomes essential. The single-domain antibodies (sdAbs), also called nanobodies, can be a more potent weapon for treating the pandemic. These antibodies are immunoglobulins that are isolated from camelids. They possess very low molecular weight, ranging from 12 to 15 kD, as a result, they can access multiple antigenic epitopes. This is a great advantage of nanobodies compared to monoclonal antibodies [2], [3], [4]. Nanobody development events from the invention of hybridoma technology were a breakthrough, and this event has shown a new direction for the treatment of diverse diseases and COVID-19 infection (Fig. 1 ).
Fig. 1.
Timelines show different breakthrough of nanobodies development.
The receptor-binding domain (RBD) of the S-glycoprotein stimulates the attachment of the SARS-CoV-2 virus with the ACE2 receptor of the host cell. These, in turn, further stimulate an alteration in the conformation of the RBD, enhancing the virus to cleave the host cell's membrane. Several data obtained from X-Ray crystallography and cryo-EM microscopy suggest the efficiency of nanobodies in this regard. Nanobodies target the epitopes on the Spike RBD, which hinders the viral attachment to the host cell, nullifying the chances of infection [5]. For instance, Zhenlin Yang discussed a potent nanobody n3113 that binds to the RBD in the open state without involving the ACE2 receptor. The interaction of the n3113 antibody with the epitopes of the RBD makes the Spike protein extremely stable, but it does not allow the virus to fuse into the membrane. This provides strong evidence of neutralization. Moreover, n3113 binding has shown exclusive neutralization efficiencies in emerging mutant variants like Alpha (B.1.1.7), Beta (B.1.351), and Gamma (P.1) [6].
Treating several diseases with primitive antibodies might lead to serious problems and disturbing health. In order to overcome these limitations, nanobodies are employed. Due to some extraordinary physical characteristics, patients can inhale nanobodies, which have been a promising breakthrough in treating several viral infections. For example, the ALX – 0171 nanobody developed by Ablynx was found to be an effective antiviral therapeutic agent in treating the infection caused by RSV (Respiratory syncytial virus). Clinical trial studies elucidated that this nanobody responded well at all administered doses. Several methods are being adopted to minimize the immunogenic risk of administering nanobodies from camelids. The main aim of engineering these antibodies is to make them humanized. However, the process is extremely laborious and time taking. Successful preparation of humanized nanobodies carries several regions of camelid origin (like R45, F37, G47) to ensure a specific binding with the antigenic epitopes [2], [7].
Implementing nanobodies as a potent therapeutic agent has been fruitful in controlling several viral infections like Hepatitis, Rabies, HIV, Rotavirus, Ebola etc. [8], [9], [10]. The flexible structure of these antibodies ensures a large-scale production in both prokaryotic and eukaryotic organisms. Moreover, the nanobodies can target several conserved inaccessible viral epitopes, which is not possible by the primitive ones. Besides the small size, these antibodies possess a wide range of complementarity-determining regions (CDRs), stimulating their binding affinity. Owing to these antibodies' small nano-molecular size, they can penetrate through the body tissues to access those epitopes. Several reports highlight the superiority of nanobodies over conventional mAbs in treating SARS-CoV-2 infection. A major reason behind this successful breakthrough is the ability of these nanobodies to be transformed into a multimeric assembly, improving its overall binding with the antigenic epitopes to a larger extent. These nanobodies have also shown promising results in the case of the evolving SARS-CoV-2 variants. Nanobodies can take up a variety of forms in order to trigger the immune system to kill the disease-causing organism. Due to their compatible size, these antibodies can be fused with the Fc domain rendering advanced results compared to the primitive ones [11], [12] .
Thus, in this review, we broadly emphasize the nanobodies (especially the single domain antibodies) and their role in controlling the COVID-19 pandemic. We will also highlight the extraordinary features possessed by these antibodies and their superiority over the conventionally proposed ones. The pandemic have been a major breakthrough to understand the plethora of limitations of the conventionally used neutralizing antibodies in treating the SARS-CoV-2 infections. This has initiated the pharmaceutical sector to choose an alternative agent which can be extremely efficient in treating SARS-CoV-2 infections. Therefore, this review will elucidate the mechanism of action of these nanobodies and how it prevents SARS-CoV-2 infections, considering several trending significant mutations. Subsequently, special attention has also been paid to the diverse type of nanobodies and their actions against other significant viral diseases.
2. Structure of a “nanobody”
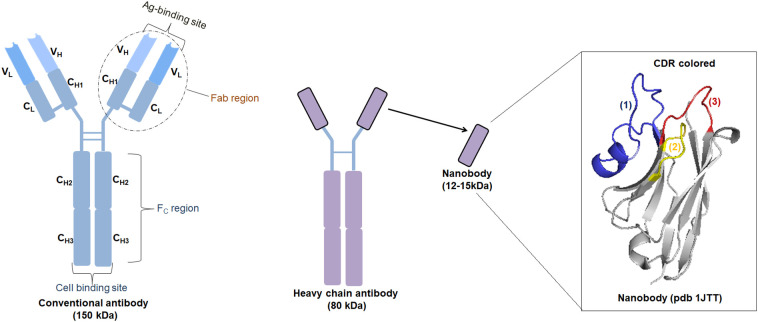
As discussed earlier, nanobodies are heavy-chain antibodies (HCAbs) derivatives. These are first isolated from camelid specifies. These antibodies are devoid of the L chain and the H chain's first constant region (also called CH1). The IgG2 region of these antibodies is greater in length compared to the primitive ones and can compensate for the lack of the CH1 region. In fact, the greater length of the IgG2 constitutes several proline and glycine repeats which widens the space for linking with the epitopes. The definitive studies on the structure of these nanobodies clearly indicated that these antibodies use only a single domain of the variable region (VHH) to access the epitopes; a completely different structure from the primitive ones (Fig. 2 ). The genetic modification of these antibodies by introducing a heavy chain and the human Fc domain makes it extremely capable of preventing the SARS-CoV-2 infection. Incorporating the Fc domain in the nanobodies accelerates their transition efficiency by rendering a longer lifespan. The introduction of this component also helps in the interaction of these nanobodies with specific antigens, other cells, and immune system organs [3], [11].
Fig. 2.
Fine structure of conventional, Heavy-Chain antibody (HCAb), and single-domain antibodies (nanobodies). The ribbon representation of a nanobody (PDB ID: 1JTT). The framework regions are in grey, the hypervariable H1, H2 and H3 antigen binding loops are in [1] blue [2] yellow, and [3] red, respectively.
The sequence of the amino acid residues in the case of nanobody illustrates similar structural attributes of the HV (hypervariable) and the VH domains. However, minor differences can be pointed out in the FR2 (framework 2) and CDR regions. The FR2 region is extremely conserved and induces some replacement of amino acids, which makes the nanobodies structurally flexible [13]. As discussed earlier, nanobodies' engineering is essential to improve their immunogenicity. Replacing these amino acid residues results in the dimerization of the nanobodies when they are humanized. Studies suggest that these replacements only make the antibodies proficient in working without the VH chain. Moreover, these antibodies do not possess any flat or deep groove surfaces in order to interact with the antigenic epitopes. A major approach to structurally modify the nanobodies is the grafting of several CDRs. Incorporating the CDR loops from conventional antibodies accelerates the efficacy of these nanobodies. The CDR grafting also improves the avidity of the antibody [3], [14].
3. Mechanism of action of nanobodies
The S-glycoprotein of the SARS-CoV-2 virus is believed to contain multiple epitopes, which are the major targets of the antibodies. The S-glycoprotein of the virus attaches to the surface of the host ACE2 receptor and other co-receptors and mediates the event of membrane fusion [15]. Due to the structural conformation, the receptor-binding domain (RBD) exists in two known conformations, namely “up” and “down”. The “up” state is preferred as it is easily accessible by the antibodies in comparison with the “down” state. According to the prior discussion, it is very clear that nanobodies have been proven to be a very efficient therapeutic agent than the normal mAbs. The rate of neutralization increases many folds as these nanobodies targets more than one epitope residing in the RBD of the Spike protein. The “up” conformation strongly favors the binding of the virus with the host receptor conferring many secondary changes, leading to the S1 dissociation. Hence, like mAbs, nanobodies too show major neutralization in the “up” state of the RBD. The nanobodies also activate the fusion cascade of the host cell upon viral interaction. The binding of the ACE2 receptor with the Spike protein activates this cascade. This binding also facilitates the cleavage of some structures, leading to conformational changes that activate these nanobodies to neutralize inside the host cells. These nanobodies are capable of stabilizing the RBD in the “up-conformation” which is another reason for these therapeutic agents to accelerate the membrane fusion [16].
The first neutralizing nanobody against SARS-CoV-2 was a potent therapeutic agent against the MERS-CoV and SARS-CoV-1 viruses. Such nanobody which targets the receptor-binding domain of these viruses have shown promising neutralizing efficacy and cross-reactivity against the SARS-CoV-2 virus [17]. These were followed by nanobodies, or “sybodies,” against SARS-CoV-2 that were found in synthetic libraries [18], [19]. To reduce the immunological reaction in humans, the framework portions of the synthetic library were somewhat humanized [2]. Other methods include humanizing the nanobody backbone, lowering the possibility of immunological recognition, and creating a platform to create single-domain antibodies that are unique to SARS-CoV-2 and are of human origin. In other investigations, nanobodies were successfully isolated and fused to the human IgG1-Fc region, enhancing their ability to bind and neutralize [20], [21], [22], [23]. Another set of nanobodies directed against the epitopes of the SARS-CoV-2 Spike protein was also identified from a yeast surface-displayed synthetic library. Bivalent and trivalent nanobodies were created by the researchers using Nb6, which resulted in a 2000-fold boost in inhibitory activity against both a fictitious virus and live SARS-CoV-2 in infection testing [24].
Nanobodies primary method of combating COVID-19 is by preventing the interaction between Spike protein and ACE2. By using yeast surface libraries, Schoof et al. discovered two kinds of neutralizing nanobodies. While class II Nbs like Nb3 linked to a distinct, unnamed epitope on Spike proteins, class I Nbs like Nb6 and Nb11 are adhered to RBD of the Spike protein. The most effective class I Nb's binding site was discovered using cryo-electron microscopy (cryo-EM). It was discovered that Nb6 and Nb11 can bind both the up and down conformations of RBD. Unusually, two neighboring RBD in the down conformation were stabilized by the binding of Nb6. This was probably going to make it easier for other Nb6 molecules to bind [24].
Dong et al. used a humanized llama VHH to test the effectiveness of nanobodies against COVID-19. The binding site of the Spike protein was targeted by 91 high-affinity nanobodies, 69 of which had a unique sequence. Further research on the 69 distinct nanobodies revealed that 15S protein binders inhibit the interaction between the Spike protein and the ACE2 receptor, strengthening the neutralizing effect against COVID-19 infection [25]. Additionally, five single domain nanobodies (sdNbs) that act against COVID-19 Spikes were identified in a subsequent study by Chi et al. The pseudotyped COVID-19 particle displayed a low affinity for these monovalent nanobodies. Upon fusing with human IgG Fc-domain, it was discovered that an attempt had been made to enhance the neutralizing activity of these single domain nanobodies. When compared to standard single domain nanobodies, the activity of fc-fused single domain nanobodies increased tenfold [26].
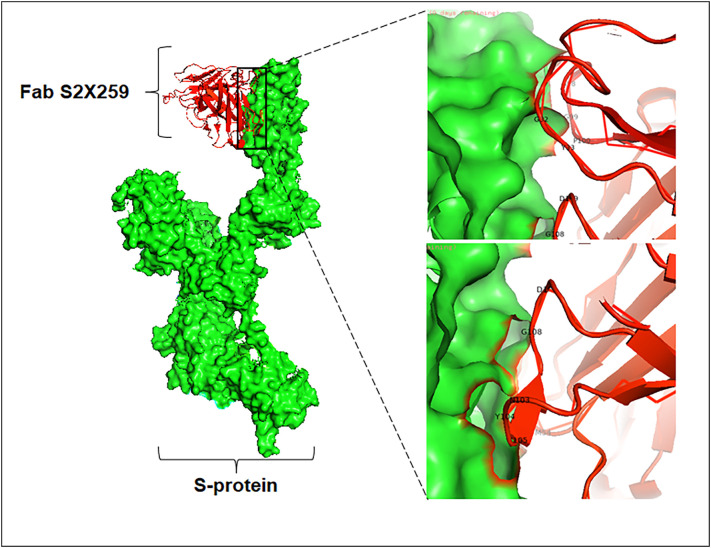
Several studies have proved the efficacy of using antibody cocktails in treating COVID-19. The nanobodies are also not an exception. Koenig and his colleagues concluded that the administration of combined nanobodies has been extremely proficient in treating the SARS-CoV-2 infection. They investigated that incorporating four nanobodies was extremely potent in inhibiting viral replication inside the host. In order to make them capable of recognizing a wide range of antigenic epitopes, the nanobodies are engineered by changing some of the domain constituents and joining them with suitable linker peptides [16]. Moreover, recognizing more than one epitope can also prevent emerging variants from escaping the immune system. Koenig et al. also highlighted several neutralization strategies adopted by these nanobodies against this virus. Interaction of the two paratopes of the nanobodies or the preparation of multivalent nanobody complexes mediates neutralization to a greater extent. The nanobodies generally recognize the epitopic regions with the help of a loop-like structure formed using CDR1, CDR2, and CDR3, a major part constituting the antigen-binding site. The incorporation of the human Fc region also facilitates the neutralization activity against the SARS-CoV-2 virus. Due to the presence of this Fc domain, the nanobodies can overcome the ACE2 binding and are able to recognize the antigenic epitopes present in the RBD (Fig. 3 ) [16], [27].
Fig. 3.
Structure of SARS-CoV-2 Spike monomer bound to nanobody (Fab S2X259) [PDB ID: 7RA8]. The interacting residues have been marked in the bounding region.
4. Special characteristics of a “nanobody”
A more potent neutralization approach is observed in the case of nanobodies than in conventional monoclonal antibodies. A good therapeutic agent should possess unique characteristics to treat several widespread pandemics effectively. One of the most important characteristics is the easy production process for the molecules. According to the data obtained from X-Ray crystallography and cryo-EM, nanobodies have a quite simple structure in comparison to the other classes of antibodies. Hence, these nanobodies can be easily produced in all host organisms, namely the prokaryotes and eukaryotes. Besides, it possesses several unique biophysical properties, making it distinct from the other different classes of antibodies. The conventional antibodies employed for treating many diseases are immunogenic in nature; however, nanobodies are an exception. The suitable structural architecture allows these antibodies to possess this unique property. Moreover, these nanobodies can be easily eliminated from the blood owing to their nanomolar size [3], [28].
The nanobodies are extremely capable of tolerating high temperatures. Additionally, these antibodies can be engineered easily and made more resistant to a greater temperature scale by introducing several amino acids. The resistive disulfide bonds in the nanobodies are unlikely to be degraded by proteases like chymotrypsin. Due to the presence of extremely conserved sequences of the nanobody, these can be used to graft antigen-binding loops to unstable nanobodies, which are not very potent for recognizing antigens. The mechanism of action of these “nanobodies” somewhat mimics the working of an enzyme. The complementarity of the concave architecture of the paratope with the convex epitope induces a catalytic effect. This makes the nanobodies capable of binding and accessing a wider range of epitopes [3]. The nanobodies are also very efficient in deeply penetrating the host tissues once they are incorporated into the body. They are more soluble than conventional antibodies and can withstand extreme environmental conditions without hampering their function [28]. The significant characteristics of the nanobodies, making them extremely good therapeutic agents compared to conventional antibodies, are elucidated in Table 1 .
Table 1.
Features of nanobodies compare to conventional antibodies.
| a | Nanobodies are able to recognize native epitopes, which are unusual for the classical antibodies. |
| b | Lesser amount of immunogenicity. |
| c | High stability and solubility in the harsh environments (denaturing conditions or high temperatures). |
| d | Small size facilitated to higher tissue penetration. |
| e | Nanobodies can be produced easily in yeast and bacteria. Its lack the Fc domain (glycan-harboring), and are easy to make than the standard mAbs. |
| f | Due to the absence of Fc regions, nanobodies have low risk of antibody-dependent enhancement of infection. |
| g | Life span of nanobodies can be enhanced by adding human albumin or polyethylene glycol. |
| h | Nanobodies can be used for new immunobiotechnological purposes and are very easy to manipulate. |
| i | Effortless for the selection by phage display. |
5. Several nanobodies employed for treating COVID-19
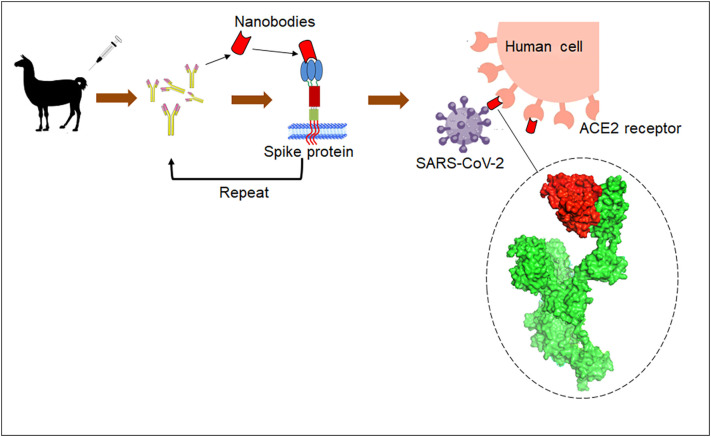
Due to some special characteristics, several nanobodies are employed for treating the COVID-19 pandemic (Table 2 ). The specialized mechanism for the production and mode of action of nanobody is illustrated in Fig. 4 . Some nanobodies are listed below:
Table 2.
Various nanobodies, their sources, and mechanism of action against SARS-CoV-2.
| Sl. No. | Nanobodies | Source | Dissociation constant (KD) value (nM) against protein of SARS-CoV-2 | Mechanism of action | Reference |
|---|---|---|---|---|---|
| 1. | Ty1 | Alpaca | Targets the RBD of the Spike protein with a greater affinity. | [74], [75] | |
| 2. | H-11 D4 | Ilama | 39 (±2) | Blocks the attachment of the ACE2 receptor with the Spike protein | [74], [76], [77] |
| 3. | H-11 H4 | Ilama | 12 (±1.5) | Blocks the attachment of the ACE2 receptor with the Spike protein. | [74], [76], [77] |
| 4. | Sb23 | Library of synbodies | 10.6 (± 2) | Blocks the interaction of the ACE2 receptor and the RBD | [78], [79] |
| 5. | K-874A | Camels (no modifications are included) | 1.4 | Inhibits the fusion of the viral membrane and prevents viral replication | [80] |
| 6. | Nb11–59 | Pichia pastoris | 21.6 | Blocks the interaction of the ACE2 receptor and the RBD | [28], [81] |
| 7. | Nb16–68 | 36 | |||
| 8. | VNAR | Sharks | 38.5–60.3 | Targets the S1-RBD and breaks the salt bridge between ACE2 and RBD | [82], [83] |
| 9. | n3113.1-Fc | – | 5.3 | Binds to the RBD of the Spike protein without involving ACE2 receptor | [84] |
Fig. 4.
A illustration about the workflow for isolating nanobody against SARS-CoV-2 Spike protein and its mode of action in human cells.
5.1. Ty1
Ty1 is a nanobody that has been isolated from Alpaca, a species belonging to camelid origin. The nanobody functions by targeting the receptor-binding domain of the S-glycoprotein. It facilitates the Ty1 to bind the RBD with a very high affinity. It can bind with the RBD in both conformations [29]. This antibody can be yielded in large amounts in bacteria, decreasing the production cost to a greater extent. Several CDRs bring about the interaction of the antibody with the RBD. For instance, the CDR1 and CDR3 are engaged in the interaction of the RBD with Ty1, whereas the CDR2 stabilizes these interactions without involving in interactions. This is the first nanobody isolated and found to be efficient in neutralizing the SARS-CoV-2 virus [30].
5.2. H11-D4 and H11-H4
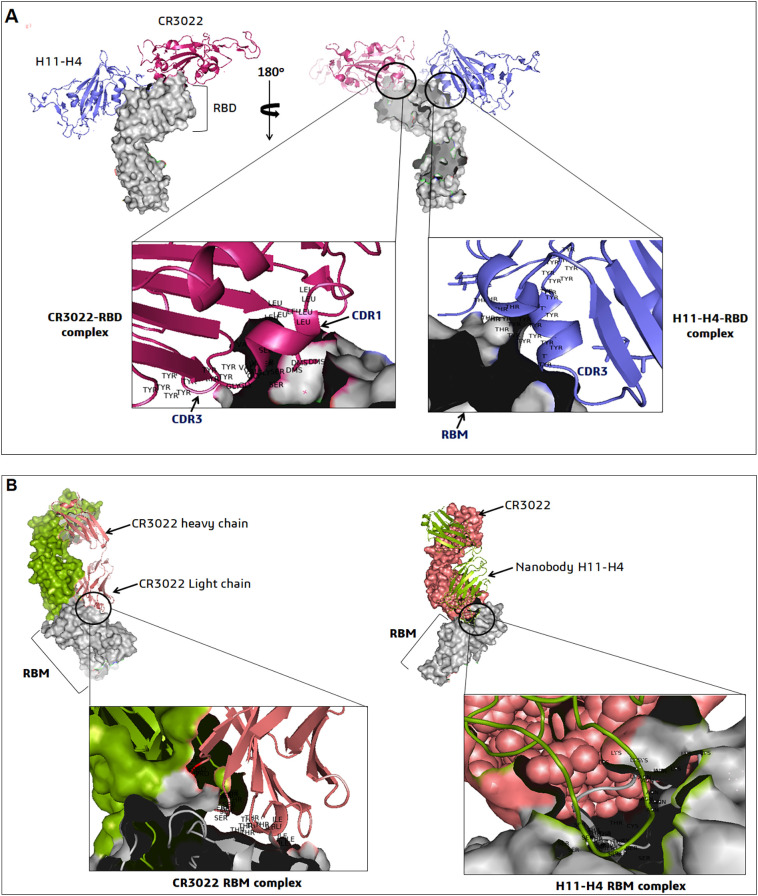
This nanobody has been derived from the Ilama's belonging to the camelid origin. The H11-D4 and H11-H4 more or less have a conserved sequence and similar neutralization mechanism, and they just differ in five positions within the CDR3 loop. This is extremely proficient in hindering the SARS-CoV-2 infection. These antibodies prevent the attachment of the S-glycoprotein with the ACE2 receptor, blocking the virus's entry. It targets a conserved epitope beside the ACE2 receptor and makes it possible to neutralize a live virus, as highlighted in the in-vitro assays [27]. Due to this feature, researchers find a ray of hope that this antibody or its combination can be extremely potent in treating COVID-19 patients facing severe symptoms. The fusion of the Fc region with this nanobody alters the binding capability with the ACE2 receptor, which can be even more functional in blocking the entry of the virus [30]. The interaction between the H11-H4 nanobody, CR3022 antibody and the epitopes containing the RBD region of SARS-CoV-2 S-protein is shown in Fig. 5 .
Fig. 5.
The nanobodies (H11-H4 and CR3022) binding epitopes interactions on RBD of SARS-CoV-2. (A) The structure of H11-H4–RBD–CR3022 complex shows that the nanobody and antibody recognized sites (CDR1, CDR3) of different epitopes [PDB ID: 6YLA]. (B) The structure of CR3022 (light chain)-RBM-H11-H4 complex showing the interacting regions of RBD-ACE2 receptor sites (PDB ID: 6ZH9). Both two figures show the different interacting residues in the nanobody interaction region.
5.3. Sb23
Similar to the other nanobodies, Sb23 also functions in a similar pattern by targeting the RBD. Sb23 is a nanobody that is isolated from the synthetic library according to its neutralization efficiency. The selection of these nanobodies from libraries accelerates the process of developing therapeutics. The attachment to the ACE2 receptor, in turn, blocks the entry of the virus inside the host cell. The strongest attachment of the ACE2 receptor is observed in the case of Sb23. The Sb23 binds to the inner surface of the ACE2-RBD attachment site conferring an extremely high affinity of Sb23 to block this interaction. This orientation exists in the RBD's “up” and “down” conformations. Custódio et al. reported that Sb23 also increases the stability of the “up” conformation of the RBD, making the epitopes easily accessible. Thus, the core central region of the Spike could be studied properly [19]. It will be a major breakthrough for researchers to develop efficient therapeutics targeting these epitopes.
5.4. K-874A
This nanobody follows a different neutralization mechanism from Ty1, Sb23, H11-D4, and H11-H4. These antibodies hinder SARS-CoV-2 infection by interfering with the ACE2-RBD interaction. Additionally, K-874A disrupts the fusion of the viral membrane by TMPRSS2. In the case of Syrian hamsters, introducing this nanobody through the nose improved the infection that prevailed in the animal's lungs. K-874A mainly accesses the epitopes amidst the RBD of the Spike protein and the N terminal domain. The administration of these antibodies decreased the severe symptoms of the Syrian hamsters. Upon successful administration, K-874A did not result in a massive cytokine storm, a life-threatening condition that generally occurs after SARS-CoV-2 infection. These nanobodies do not need modifications, including fusions, to work efficiently as therapeutic agents. Besides, it also inhibits the replication of the SARS-CoV-2 virus inside the host cell. Thus, the direct administration of the K-874A nanobody will prevent detrimental infection in the lungs of the SARS-CoV-2-affected patients [31].
5.5. Nb11-59 and Nb16-68
Nb11-59 is a potent neutralizing nanobody that shows a neutralizing efficiency near 50%. It is extremely stable and can be produced in large quantities in Pichia pastoris. Due to these special characteristics, this nanobody can be used as a good therapeutic agent by direct inhalation. It blocks the SARS-CoV-2 infection by competing with the ACE2 attachment with the RBD of the Spike protein. According to several health reports updated by WHO and CDC, it is very clear that the SARS-CoV-2 virus can infect the lungs, causing severe respiratory trouble. Among several therapeutics, the exceptional features of nanobodies can be a marvelous therapeutic agent to hinder such kinds of infections. Nb11-59 and Nb16-68 can be good targets for this. Moreover, Junwei Gai and his colleagues have already proved that these nanobodies possess the capability to neutralize the mutant variants, a major concern for the entire world. Owing to their size, these nanobodies can reach directly to the site of infection to disrupt viral replication [32].
5.6. VNAR (variable new antigen receptor domain)
VNAR antibodies are isolated from sharks. These extremely small and flexible antibodies make them excellent therapeutic agents for targeting unique epitopes. VNAR antibodies target the S1 domain, especially the S1-RBD. One of the most common mutations prevalent among the emerging variants is the E484K. This mutation disrupts the formation of the salt bridge between the RBD of the S1 subunit and the ACE2 receptor [33]. Other than E484K, another mutation that increases the transmissibility and virulence of the SARS-CoV-2 virus is the N501Y [34]. In vitro studies proved that the VNAR nanobodies are functional in both cases. Thus, it can be anticipated that the successful administration of these nanobodies can also retain their neutralization capacities in the emerging variants. This additional feature possessed by these nanobodies makes it extremely efficient to neutralize the mutant variants by lowering the chances of immune escape. Conventional treatments with the help of neutralizing mAbs and vaccines are somehow being escaped by these mutant variants [35].
5.7. n3113.1-Fc
The nanobody n3113 alone is not considered to be a potential target against the RBD of the Spike protein. However, slight modifications and fusion with the Fc region make these nanobodies outstanding targets to neutralize the SARS-CoV-2 virus. It can neutralize the wild-type strain and hinder the infection of the Alpha, Beta, Delta, and the other circulating variants. This nanobodies disrupts the entry of the virus by interfering with membrane fusion. The notable fact observed for these nanobodies is that it does not involve any interaction with the ACE2 receptor. Owing to this fact, it is very hard to conclude the exact mechanism of neutralization that these nanobodies follow. It retains an anti-parallel beta-strand with a paratope that somewhat resembles a convex shape facing the inner regions of the RBD. Like VNAR and K-874A, these nanobodies are also believed to be more potent neutralizing agents because of their smaller size, high stability, and lesser immunogenicity. Additionally, they can be targeted directly to the lungs, preventing infection. These features make them more resistant to the mutant variants [35].
5.8. aRBD-2-5-Fc and aRBD-2-7-Fc
The two nanobodies, namely aRBD-2-7-Fc and aRBD-2-5-Fc, have shown extremely high neutralization efficiency against the evolving mutant variants of the SARS-CoV-2 virus, especially the Kappa, Alpha, and Gamma variants. It's neutralization potency is many folds higher than the Nb21 nanobody, which has shown promising results against the wild-type SARS-CoV-2 strain [36]. Additionally, aRBD-2-5-Fc has also shown promising neutralization results in combating the heavily mutated Omicron variant. Treatment of aRBD-2-5-Fc in a hamster has resulted in the elimination of the SARS-CoV-2 infection caused by the Omicron BA.1 subvariant. Furthermore, this nanobody has been found to possess a high pharmacokinetic profile in several animal models like hamsters and mice [37].
Different significant nanobodies specific for viral diseases and their current clinical status are enlisted in Table 3 .
Table 3.
Different nanobodies that are specific for viral diseases and their current clinical status.
| Sl. No. | Nanobodies | Virus name | Status | Reference |
|---|---|---|---|---|
| 1 | Palivizumab | Respiratory syncytial virus | FDA approved | [47] |
| 2 | VHH nanobody | Dengue virus | Pre-clinical | [85] |
| 3 | Multiple VHH monovalent | Polio virus | Pre-clinical | [86] |
| 4 | VHH bivalent/albumin-linked | Rabies virus | Pre-clinical | [87] |
| 5 | MD3606-human Fc trivalent nanobody | Influenza virus | Pre-clinical | [88] |
| 6 | VHH nanobody | Hepatitis C virus | Pre-clinical | [67] |
| 7 | Anti-CXCR4 monovalent and bivalent | HIV | Pre-clinical | [89] |
| 8 | Multiple VHH monovalent | Norovirus | Pre-clinical | [90] |
| 9 | VHH /human Fc hybrid | MERS-CoV | Pre-clinical | [91] |
| 10 | H5N1-HA bivalent nanobody | Influenza virus | Pre-clinical | [43] |
6. Nanobodies employed as a therapeutic agents for other viruses
Besides SARS-CoV-2, nanobodies have shown a greater range of neutralization against many other viruses. The specific nanobodies used against different viruses are elucidated in Table 4 . Some of them are discussed below:
Table 4.
Nanobodies and their sources used against different viruses.
| Sl. no. | Name of the virus | Nanobodies neutralize the infection | Source | Reference |
|---|---|---|---|---|
| 1. | Influenza virus (Type A) | R1a-B6 | Alpaca | [42] |
| M2–7A | – | [92] | ||
| 2. | HIV-1 virus | m36 | Llama | [48] |
| JM4 | Llama | [93] | ||
| J3 | Llama | [94] | ||
| 3. | Enteric virus | ARP1 | Llama | [47] |
| 3B2 | – | [55] | ||
| Nano-85, Nano-25 | Alpaca | [57] | ||
| 4. | Herpes simplex virus 2 (HSV-2) | R33 | Llama | [59] |
| 5. | Respiratory syncytial virus (RSV) | Nb017 | Llama | [62] |
| ALX-0171 | – | [62] | ||
| 6. | Ebola virus | M24 | – | [63] |
| 7. | Hepatitis C virus | D03 | Alpaca | [67] |
| Anti-NS3, anti-NS5B and antiprotease nanobodies | Humanized-camel phage library | [47] |
6.1. Influenza virus
On the basis of varying antigenicity, the influenza virus has been segregated into four types: A, B, C, and D [38]. Owing to the common feature of viral mutation, several therapeutics employed for curing the influenza-affected subjects are somehow deviating from their potential of treating the infection. Considering the type A strain, two glycoproteins in the viral membrane are responsible for enhancing the entry of the virus inside the host cell. They are hemagglutinin and neuraminidase [39], [40]. Yep et al. have elucidated that these membrane proteins can be targeted to prepare several neutralizing mAbs. A closer look at the structural conformation of these proteins proves the presence of several concave surfaces makes it extremely suitable for preparing nanobodies [41]. The R1a-B6 is one of the nanobody that exhibits a cross-neutralizing action providing a broader aspect to cure the viral infection. These antibodies have been isolated from Alpaca. The main mechanism of these antibodies is based on inhibiting the function of the hemagglutinin protein [42]. The influenza type A strain has many of the developed nanobodies, mainly isolated from camels and llamas. In-vitro studies have demonstrated that these antibodies function by blocking the neuraminidase protein [43], [44], [45].
6.2. HIV-1 virus
The HIV-1 envelope protein takes the shape of a trimer and consists of gp120 and gp41. These are the main points of the target for the antibodies employed for hindering HIV-1 infection [46]. These epitopes are mainly located on the CD4 binding region and the site for interacting with the co-receptor [47]. One of the best therapeutic agents employed to elicit the immune response and inhibit viral replication is the m36 antibody. This nanobody targets a conserved sequence residing within this envelope protein, i.e., gp120. They provide a superior neutralizing capacity compared to the other bivalent antibodies [48]. When administered in patients, these nanobodies produce certain cells capable of inhibiting the HIV-1 infection. These antibodies can trigger the immune system alone or may be coupled with other antibodies [49]. The gp140 trimeric complex can act as an immunogen, and the immune response it elicits in the llama has provided evidence of several nanobodies which bind to the epitopes, especially in the sites targeted by the CD4 or the parts engaged in the interaction with other co-receptors [50]. Another protein, namely the Vpr, mediates the function of the viruses in many ways, especially in vital processes like replication and transcription. These nanobodies are developed from llamas that are immunized by either the Vpr peptide or the viral capsid protein [51].
6.3. Enteric virus
The enteric viruses are the main agents responsible for concurrent diarrhea in adults and babies. Moreover, this is a potential agent behind certain gastrointestinal complications. Some of these enteric virus family members include the norovirus and the rotavirus found in the isolates of adults and neonates. One of the nanobodies that have been isolated from the llama has shown promising results in curing these ailments. These antibodies are made from fragments of anti-rotavirus protein [52], [53], [54]. Another nanobody employed for treating this gastrointestinal ailment is 3B2, which binds with VP6, one of the capsid proteins. This provides a greater prospect of neutralization even when administered in individuals in lower doses [55]. Furthermore, noroviruses are again a serious hindrance in developing effective therapeutics because of their lower cultivation due to the frequent mutations. It comprises a shell and a P domain interconnected by a hinge [56]. Two nanobodies, namely the Nano-85 and Nano-25, target the extremely conserved residues prevailing at the lower parts of the P domain [57].
6.4. Herpes simplex virus 2 (HSV-2)
The absence of effective therapeutics against the HSV-2 virus has led to the widespread transmission of this disease. The avenue of these nanobodies has been a savior in this case too. The viral glycoprotein D is considered the first protein that initiates the process of HSV-2 infection by involving the fusion of the membrane and the interaction with the host receptors [58]. This glycoprotein serves as the binding site for the nanobody R33, isolated from the llama. However, the functional activity of the R33 nanobody can be accelerated by using an exotoxin, specifically exotoxin A. The combination of the R33 antibody along with the exotoxin serves as a potential inhibitor that restricts the production of cells essential for the process of reactivation [59]. R33 antibody can also be employed for delivering certain components to the infected cells to reduce their side effects on the body's normal cells [59]. Additionally, site-specific conjugation of medicines like immunotoxins or cytotoxins to a C-terminal cysteine not only retains the binding of nanobodies but also boosts their ability to kill virus-infected cells [47].
6.5. Respiratory syncytial virus (RSV)
Respiratory syncytial virus, or RSV, poses a severe threat to the population and is a major causative agent against acute pneumonia prevalent among kids. Like the HSV infection, glycoprotein F plays a predominant role in mediating this infection. Two humanized nanobodies, namely m35 and m17, have shown potent neutralization ability in both subgroups of the RSV virus [60]. This F glycoprotein illustrates the greater similarity between RSV type A and type B groups. Moreover, this F protein confers reduced glycosylation than the G protein. Although both of these proteins have epitopes that can be targeted by a therapeutic agent for efficient inhibition of the viral infection yet, F is chosen over G due to some additional benefits [61]. The administration of these nanobodies can also hinder the stable structural conformation of the F glycoprotein, restricting it from entering the host cell [60]. Another nanobody named Nb017 isolated from the llama has also shown considerable neutralization against the RSV [62]. A complex prepared by linking three Nb017 using a linker peptide has highlighted a broader aspect of cross-neutralization efficiency that is even higher than the licensed therapeutic palivizumab. This nanobody derived from the integration of three Nb017 is named ALX-0171. The use of this nanobody directly to the sites of the infection has considerably inhibited the process of replication [62].
6.6. Ebola virus
The Ebola virus is the causative against hemorrhagic fever that raises the fatality rate to a greater extent. The CH2 fragment of the antibody is employed to serve as the scaffold for constructing a library of the nanobodies known as the c-nanobodies. These often serve as efficient therapeutic agents. Some such as c-nanobody is M24. The structural studies involving the epitopes have implicated that this M24 nanobody can disrupt a fusion loop in the transmembrane domain of the glycoprotein responsible for the viral infection [63]. One of the interesting facts is the extremely high thermal stability of the nanobodies. These nanobodies can be used very conveniently in regions lacking refrigeration. Besides their therapeutic potential, the nanobodies can even be used to diagnose Ebola-positive subjects [64].
6.7. Hepatitis C virus
The Hepatitis C virus (HCV) is responsible for causing severe hepatitis and hepatocellular carcinoma [65]. The E2 glycoprotein present in the core of the HCV virus is responsible for mediating cellular entry and the interaction with several host-cell receptors [66]. The nanobodies have also been functional in this case. The D03 nanobody derived from alpacas targets one of the highly conserved epitopes overlapping the binding site of CD81. This nanobody consists of 20 amino acid-long complementarity-determining regions (CDR3) and can hinder cellular entry [67]. Moreover, several other nanobodies derived from camels are also capable of creating hindrances in the replication process. The anti-NS3, anti-NS5B, and some antiprotease nanobodies are linked to obtaining a greater aspect of neutralization [68], [69], [70]. Evidently, the nanobodies also restrict the production of certain cytokines, like the NFκB mediated release of cytokines, and interfere with the propagation of the virus [71].
7. Future perspective in nanobodies
The COVID-19 pandemic has sparked biotechnological advancements that have produced fresh and improved prophylactic, diagnostic, and therapeutic methods. Monoclonal antibodies have historically proven useful tools, but the pandemic has shown significant flaws, such as worldwide production restrictions. Nanobodies, produced primarily from the Camelidae family, offer an alternative to traditional monoclonal antibodies. Due to their extraordinary refolding ability, small (15 KDa) size, and recombinant nature, nanobodies undergo limitless number of potential changes [72]. The exceptional qualities of nanobodies like increased hydrophobicity, enhanced stability especially in the framework regions have increased their ability to interact with the antigenic epitopes accelerating their potency to penetrate through the tissues which can employ nanobodies to be used as nano-biodrugs for the future generations [73].
8. Conclusion
The nanobodies can be a potent therapeutic target against the SARS-CoV-2 virus and other viruses. Despite the exclusive features possessed by these nanobodies, they can be used to prevent infections in the pulmonary organs. Several reports suggested that SARS-CoV-2 infection in the lungs is extremely detrimental. Besides, the numerous amino acid substitutions also raise questions about the efficacy of the developed vaccines. These mutations alter several structural features of the virus, making it more virulent and transmissible. Properties of “immune escape” and “vaccine escape” are also a concern. Conventional mAbs, convalescent plasma therapy, and vaccines do not confer greater neutralization than nanobodies. These antibodies can be prepared very quickly and target specific inaccessible epitopes. Engineering these antibodies with the Fc fragment has also proven more efficient than the antibody alone. Thus, it is believed that these nanobodies can be used as a potential therapeutic agent to stop this alarming pandemic. Several other modifications can, however, lead to the development of a more suitable product, saving many lives.
CRediT authorship contribution statement
Manojit Bhattacharya: Validation, Formal analysis, Investigation, Figure development, Writing – Original Draft.
Srijan Chatterjee: Data curation, Resources, Investigation, Writing – Original Draft.
Sang-Soo-Lee: Validation & formal analysis.
Chiranjib Chakraborty: Conceptualization, Methodology, Project administration, Writing - Review & editing.
Ethics approval and consent to participate
Not required.
Consent for publication
Not required.
Funding
No fund received.
Declaration of competing interest
The authors declare no competing interests.
Acknowledgments
Authors are thankful to their respective University/Institute authorities.
Data availability
All data and materials included within the manuscript.
References
- 1.Zebardast A., Hosseini P., Hasanzadeh A. The role of single-domain antibodies (or nanobodies) in SARS-CoV-2 neutralization. Mol. Biol. Rep. 2021;1–10 doi: 10.1007/s11033-021-06819-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu Y., Li C., Xia S., Tian X., Kong Y., Wang Z., et al. Identification of human single-domain antibodies against SARS-CoV-2. Cell Host Microbe. 2020;27(6):891–898. doi: 10.1016/j.chom.2020.04.023. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muyldermans S. Nanobodies: natural single-domain antibodies. Annu. Rev. Biochem. 2013;82(1):775–797. doi: 10.1146/annurev-biochem-063011-092449. [DOI] [PubMed] [Google Scholar]
- 4.Wu Y., Jiang S., Ying T. Single-domain antibodies as therapeutics against human viral diseases. Front. Immunol. 2017;8:1802. doi: 10.3389/fimmu.2017.01802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Czajka T.F., Vance D.J., Mantis N.J. Slaying SARS-CoV-2 one (single-domain) antibody at a time. Trends Microbiol. 2021;29(3):195–203. doi: 10.1016/j.tim.2020.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang Z., Wang Y., Jin Y., Zhu Y., Wu Y., Li C., et al. A non-ACE2 competing human single-domain antibody confers broad neutralization against SARS-CoV-2 and circulating variants. Signal Transduct. Target. Ther. 2021;6(1):1–8. doi: 10.1038/s41392-021-00810-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Czajka T.F., Vance D.J., Mantis N.J. Slaying SARS-CoV-2 one (Single-domain) antibody at a time. Trends Microbiol. 2021;29(3):195–203. doi: 10.1016/j.tim.2020.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esmagambetov I.B., Shcheblyakov D.V., Egorova D.A., Voronina O.L., Derkaev A.A., Voronina D.V., et al. Nanobodies are potential therapeutic agents for the ebola virus infection. Acta Nat. 2021;13(4):53–63. doi: 10.32607/actanaturae.11487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weiss R.A., Verrips C.T. Nanobodies that neutralize HIV. Vaccines (Basel) 2019;7(3) doi: 10.3390/vaccines7030077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao B., Qu M. In: Current Laboratory Techniques in Rabies Diagnosis, Research and Prevention. Rupprecht C., Nagarajan T., editors. Vol. 2. Academic Press; 2015. Chapter four - a rat basophilic leukemia cell sensor for the detection of rabies viruses; pp. 33–43. [Google Scholar]
- 11.Voss J.E. EngineeredSingle-domain Antibodies Tackle COVID Variants. 2021;595(7866):176–178. doi: 10.1038/d41586-021-01721-5. Nature Publishing Group. [DOI] [PubMed] [Google Scholar]
- 12.Xu J., Xu K., Jung S., Conte A., Lieberman J., Muecksch F., et al. Nanobodies from camelid mice and llamas neutralize SARS-CoV-2 variants. Nature. 2021;595(7866):278–282. doi: 10.1038/s41586-021-03676-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jovcevska I., Muyldermans S. The therapeutic potential of nanobodies. BioDrugs. 2020;34(1):11–26. doi: 10.1007/s40259-019-00392-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoey R.J., Eom H., Horn J.R. Structure and development of single domain antibodies as modules for therapeutics and diagnostics. Exp. Biol. Med. 2019;244(17):1568–1576. doi: 10.1177/1535370219881129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhattacharya M., Chatterjee S., Mallik B., Sharma A.R., Chakraborty C. Therapeutic role of neutralizing antibody for the treatment against SARS-CoV-2 and its emerging variants: a clinical and pre-clinical perspective. Vaccines. 2022;10(10):1612. doi: 10.3390/vaccines10101612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koenig P.-A., Das H., Liu H., Kümmerer B.M., Gohr F.N., Jenster L.-M., et al. Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape. Science. 2021;371(6530) doi: 10.1126/science.abe6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wrapp D., De Vlieger D., Corbett K.S., Torres G.M., Wang N., Van Breedam W., et al. Hoffmann M., Pöhlmann S., Graham B.S., Callewaert N., Schepens B., Saelens X., JS McLellan, VIB-CMB COVID-19 Response Team Structural basis for potent neutralization of betacoronaviruses by single-domain camelid antibodies. Cell. 2020;181(5) doi: 10.1016/j.cell.2020.04.031. 1004-1015.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong J., Huang B., Wang B., Titong A., Gallolu Kankanamalage S., Jia Z., et al. Development of humanized tri-specific nanobodies with potent neutralization for SARS-CoV-2. Sci. Rep. 2020;10(1):17806. doi: 10.1038/s41598-020-74761-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Custódio T.F., Das H., Sheward D.J., Hanke L., Pazicky S., Pieprzyk J., et al. Selection, biophysical and structural analysis of synthetic nanobodies that effectively neutralize SARS-CoV-2. Nat. Commun. 2020;11(1):1–11. doi: 10.1038/s41467-020-19204-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chi X., Zhang X., Pan S., Yu Y., Shi Y., Lin T., et al. An ultrapotent RBD-targeted biparatopic nanobody neutralizes broad SARS-CoV-2 variants. Signal Transduct. Target. Ther. 2022;7(1):44. doi: 10.1038/s41392-022-00912-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiaojie S., Yu L., Lei Y., Guang Y., Min Q. Neutralizing antibodies targeting SARS-CoV-2 spike protein. Stem Cell Res. 2020;50 doi: 10.1016/j.scr.2020.102125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu Q., Liu J., Zhao S., Gomez Castro M.F., Laurent-Rolle M., Dong J., et al. SARS-CoV-2 exacerbates proinflammatory responses in myeloid cells through C-type lectin receptors and tweety family member 2. Immunity. 2021;54(6):1304–1319.e9. doi: 10.1016/j.immuni.2021.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valenzuela Nieto G., Jara R., Watterson D., Modhiran N., Amarilla A.A., Himelreichs J., et al. Potent neutralization of clinical isolates of SARS-CoV-2 D614 and G614 variants by a monomeric, sub-nanomolar affinity nanobody. Sci. Rep. 2021;11(1):3318. doi: 10.1038/s41598-021-82833-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schoof M., Faust B., Saunders R.A., Sangwan S., Rezelj V., Hoppe N., et al. An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive spike. Science. 2020;370(6523):1473–1479. doi: 10.1126/science.abe3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dong J., Huang B., Jia Z., Wang B., Gallolu Kankanamalage S., Titong A., et al. Development of multi-specific humanized llama antibodies blocking SARS-CoV-2/ACE2 interaction with high affinity and avidity. Emerg. Microbes Infect. 2020;9(1):1034–1036. doi: 10.1080/22221751.2020.1768806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chi X., Liu X., Wang C., Zhang X., Li X., Hou J., et al. Humanized single domain antibodies neutralize SARS-CoV-2 by targeting the spike receptor binding domain. Nat. Commun. 2020;11(1):4528. doi: 10.1038/s41467-020-18387-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huo J., Le Bas A., Ruza R.R., Duyvesteyn H.M., Mikolajek H., Malinauskas T., et al. Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2. Nat. Struct. Mol. Biol. 2020;27(9):846–854. doi: 10.1038/s41594-020-0469-6. [DOI] [PubMed] [Google Scholar]
- 28.Zare H., Aghamollaei H., Hosseindokht M., Heiat M., Razei A., Bakherad H. Nanobodies, the potent agents to detect and treat the coronavirus infections: a systematic review. Mol. Cell. Probes. 2021;55 doi: 10.1016/j.mcp.2020.101692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hanke L., Vidakovics Perez L., Sheward D.J., Das H., Schulte T., Moliner-Morro A., et al. An alpaca nanobody neutralizes SARS-CoV-2 by blocking receptor interaction. Nat. Commun. 2020;11(1):1–9. doi: 10.1038/s41467-020-18174-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chakraborty C., Bhattacharya M., Sharma A.R. Emerging mutations in the SARS-CoV-2 variants and their role in antibody escape to small molecule-based therapeutic resistance. Curr. Opin. Pharmacol. 2022;62:64–73. doi: 10.1016/j.coph.2021.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haga K., Takai-Todaka R., Matsumura Y., Song C., Takano T., Tojo T., et al. Nasal delivery of single-domain antibody improves symptoms of SARS-CoV-2 infection in an animal model. PLoS Pathog. 2021;17(10) doi: 10.1371/journal.ppat.1009542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gai J., Ma L., Li G., Zhu M., Qiao P., Li X., et al. A potent neutralizing nanobody against SARS-CoV-2 with inhaled delivery potential. MedComm. 2021;2(1):101–113. doi: 10.1002/mco2.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chakraborty C., Sharma A.R., Bhattacharya M., Lee S.S. A detailed overview of immune escape, antibody escape, partial vaccine escape of SARS-CoV-2 and their emerging variants with escape mutations. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.801522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chakraborty C., Sharma A.R., Bhattacharya M., Agoramoorthy G., Evolution Lee S.S. Mode of transmission, and mutational landscape of newly emerging SARS-CoV-2 variants. mBio. 2021;12(4) doi: 10.1128/mBio.01140-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gauhar A., Privezentzev C.V., Demydchuk M., Gerlza T., Rieger J., Kungl A.J., et al. Single domain shark VNAR antibodies neutralize SARS-CoV-2 infection in vitro. FASEB J. 2021;35(11) doi: 10.1096/fj.202100986RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiang Y., Nambulli S., Xiao Z., Liu H., Sang Z., Duprex W.P., et al. Versatile and multivalent nanobodies efficiently neutralize SARS-CoV-2. Science. 2020;370(6523):1479–1484. doi: 10.1126/science.abe4747. Epub 2020/11/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma H., Zhang X., Zheng P., Dube P.H., Zeng W., Chen S., et al. Hetero-bivalent nanobodies provide broad-spectrum protection against SARS-CoV-2 variants of concern including omicron. Cell Res. 2022;32(9):831–842. doi: 10.1038/s41422-022-00700-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hause B.M., Collin E.A., Liu R., Huang B., Sheng Z., Lu W., et al. Characterization of a novel influenza virus in cattle and Swine: proposal for a new genus in the Orthomyxoviridae family. mBio. 2014;5(2) doi: 10.1128/mBio.00031-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Basu A., Antanasijevic A., Wang M., Li B., Mills D.M., Ames J.A., et al. New small molecule entry inhibitors targeting hemagglutinin-mediated influenza a virus fusion. J. Virol. 2014;88(3):1447–1460. doi: 10.1128/JVI.01225-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Clercq E. Antiviral agents active against influenza a viruses. Nat. Rev. Drug Discov. 2006;5(12):1015–1025. doi: 10.1038/nrd2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tung Yep A., Takeuchi Y., Engelhardt O.G., Hufton S.E. Broad reactivity single domain antibodies against influenza virus and their applications to vaccine potency testing and immunotherapy. Biomolecules. 2021;11(3) doi: 10.3390/biom11030407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gaiotto T., Hufton S.E. Cross-neutralising nanobodies bind to a conserved pocket in the hemagglutinin stem region identified using yeast display and deep mutational scanning. PloS one. 2016;11(10) doi: 10.1371/journal.pone.0164296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ibanez L.I., De Filette M., Hultberg A., Verrips T., Temperton N., Weiss R.A., et al. Nanobodies with in vitro neutralizing activity protect mice against H5N1 influenza virus infection. J. Infect. Dis. 2011;203(8):1063–1072. doi: 10.1093/infdis/jiq168. [DOI] [PubMed] [Google Scholar]
- 44.Tillib S.V., Ivanova T.I., Vasilev L.A., Rutovskaya M.V., Saakyan S.A., Gribova I.Y., et al. Formatted single-domain antibodies can protect mice against infection with influenza virus (H5N2) Antivir. Res. 2013;97(3):245–254. doi: 10.1016/j.antiviral.2012.12.014. [DOI] [PubMed] [Google Scholar]
- 45.Cardoso F.M., Ibanez L.I., Van den Hoecke S., De Baets S., Smet A., Roose K., et al. Single-domain antibodies targeting neuraminidase protect against an H5N1 influenza virus challenge. J. Virol. 2014;88(15):8278–8296. doi: 10.1128/JVI.03178-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koch K., Kalusche S., Torres J.L., Stanfield R.L., Danquah W., Khazanehdari K., et al. Selection of nanobodies with broad neutralizing potential against primary HIV-1 strains using soluble subtype C gp140 envelope trimers. Sci. Rep. 2017;7(1):8390. doi: 10.1038/s41598-017-08273-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu Y., Jiang S., Ying T. Single-domain antibodies as therapeutics against human viral diseases. Front. Immunol. 2017;8:1802. doi: 10.3389/fimmu.2017.01802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Badrane H., Nguyen M.H., Cheng S., Kumar V., Derendorf H., Iczkowski K.A., et al. The Candida albicans phosphatase Inp51p interacts with the EH domain protein Irs4p, regulates phosphatidylinositol-4,5-bisphosphate levels and influences hyphal formation, the cell integrity pathway and virulence. Microbiology (Reading) 2008;154(Pt 11):3296–3308. doi: 10.1099/mic.0.2008/018002-0. [DOI] [PubMed] [Google Scholar]
- 49.Jin H., Tang X., Li L., Chen Y., Zhu Y., Chong H., et al. Generation of HIV-resistant cells with a single-domain antibody: implications for HIV-1 gene therapy. Cell. Mol. Immunol. 2021;18(3):660–674. doi: 10.1038/s41423-020-00627-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matz J., Kessler P., Bouchet J., Combes O., Ramos O.H., Barin F., et al. Straightforward selection of broadly neutralizing single-domain antibodies targeting the conserved CD4 and coreceptor binding sites of HIV-1 gp120. J. Virol. 2013;87(2):1137–1149. doi: 10.1128/JVI.00461-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matz J., Herate C., Bouchet J., Dusetti N., Gayet O., Baty D., et al. Selection of intracellular single-domain antibodies targeting the HIV-1 vpr protein by cytoplasmic yeast two-hybrid system. PloS one. 2014;9(12) doi: 10.1371/journal.pone.0113729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van der Vaart J.M., Pant N., Wolvers D., Bezemer S., Hermans P.W., Bellamy K., et al. Reduction in morbidity of rotavirus induced diarrhoea in mice by yeast produced monovalent llama-derived antibody fragments. Vaccine. 2006;24(19):4130–4137. doi: 10.1016/j.vaccine.2006.02.045. [DOI] [PubMed] [Google Scholar]
- 53.Pant N., Hultberg A., Zhao Y., Svensson L., Pan-Hammarstrom Q., Johansen K., et al. Lactobacilli expressing variable domain of llama heavy-chain antibody fragments (lactobodies) confer protection against rotavirus-induced diarrhea. The Journal of infectious diseases. 2006;194(11):1580–1588. doi: 10.1086/508747. [DOI] [PubMed] [Google Scholar]
- 54.Pant N., Marcotte H., Hermans P., Bezemer S., Frenken L., Johansen K., et al. Lactobacilli producing bispecific llama-derived anti-rotavirus proteins in vivo for rotavirus-induced diarrhea. Future Microbiol. 2011;6(5):583–593. doi: 10.2217/fmb.11.32. [DOI] [PubMed] [Google Scholar]
- 55.Garaicoechea L., Olichon A., Marcoppido G., Wigdorovitz A., Mozgovoj M., Saif L., et al. Llama-derived single-chain antibody fragments directed to rotavirus VP6 protein possess broad neutralizing activity in vitro and confer protection against diarrhea in mice. J. Virol. 2008;82(19):9753–9764. doi: 10.1128/JVI.00436-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hansman G.S., Natori K., Shirato-Horikoshi H., Ogawa S., Oka T., Katayama K., et al. Genetic and antigenic diversity among noroviruses. The Journal of general virology. 2006;87(Pt 4):909–919. doi: 10.1099/vir.0.81532-0. [DOI] [PubMed] [Google Scholar]
- 57.Koromyslova A.D., Hansman G.S. Nanobody binding to a conserved epitope promotes norovirus particle disassembly. J. Virol. 2015;89(5):2718–2730. doi: 10.1128/JVI.03176-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Atanasiu D., Saw W.T., Cohen G.H., Eisenberg R.J. Cascade of events governing cell-cell fusion induced by herpes simplex virus glycoproteins gD, gH/gL, and gB. J. Virol. 2010;84(23):12292–12299. doi: 10.1128/JVI.01700-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Geoghegan E.M., Zhang H., Desai P.J., Biragyn A., Markham R.B. Antiviral activity of a single-domain antibody immunotoxin binding to glycoprotein D of herpes simplex virus 2. Antimicrob. Agents Chemother. 2015;59(1):527–535. doi: 10.1128/AAC.03818-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xun G., Song X., Hu J., Zhang H., Liu L., Zhang Z., et al. Potent human single-domain antibodies specific for a novel prefusion epitope of respiratory syncytial virus F glycoprotein. J. Virol. 2021;95(18) doi: 10.1128/JVI.00485-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Johnson S., Oliver C., Prince G.A., Hemming V.G., Pfarr D.S., Wang S.C., et al. Development of a humanized monoclonal antibody (MEDI-493) with potent in vitro and in vivo activity against respiratory syncytial virus. J. Infect. Dis. 1997;176(5):1215–1224. doi: 10.1086/514115. [DOI] [PubMed] [Google Scholar]
- 62.Detalle L., Stohr T., Palomo C., Piedra P.A., Gilbert B.E., Mas V., et al. Generation and characterization of ALX-0171, a potent novel therapeutic nanobody for the treatment of respiratory syncytial virus infection. Antimicrob. Agents Chemother. 2016;60(1):6–13. doi: 10.1128/AAC.01802-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang R., Zhang H., Peng C., Shi J., Gong R. Identification and characterization of a novel single domain antibody against ebola virus. Virol. Sin. 2021;36(6):1600–1610. doi: 10.1007/s12250-021-00454-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu J.L., Shriver-Lake L.C., Anderson G.P., Zabetakis D., Goldman E.R. Selection, characterization, and thermal stabilization of llama single domain antibodies towards ebola virus glycoprotein. Microb. Cell Factories. 2017;16(1):223. doi: 10.1186/s12934-017-0837-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gentile I., Maraolo A.E., Buonomo A.R., Zappulo E., Borgia G. The discovery of sofosbuvir: a revolution for therapy of chronic hepatitis C. Expert Opin. Drug Discovery. 2015;10(12):1363–1377. doi: 10.1517/17460441.2015.1094051. [DOI] [PubMed] [Google Scholar]
- 66.Khan A.G., Whidby J., Miller M.T., Scarborough H., Zatorski A.V., Cygan A., et al. Structure of the core ectodomain of the hepatitis C virus envelope glycoprotein 2. Nature. 2014;509(7500):381–384. doi: 10.1038/nature13117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tarr A.W., Lafaye P., Meredith L., Damier-Piolle L., Urbanowicz R.A., Meola A., et al. An alpaca nanobody inhibits hepatitis C virus entry and cell-to-cell transmission. Hepatology. 2013;58(3):932–939. doi: 10.1002/hep.26430. [DOI] [PubMed] [Google Scholar]
- 68.Thueng-in K., Thanongsaksrikul J., Srimanote P., Bangphoomi K., Poungpair O., Maneewatch S., et al. Cell penetrable humanized-VH/V(H)H that inhibit RNA dependent RNA polymerase (NS5B) of HCV. PloS one. 2012;7(11) doi: 10.1371/journal.pone.0049254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Phalaphol A., Thueng-In K., Thanongsaksrikul J., Poungpair O., Bangphoomi K., Sookrung N., et al. Humanized-VH/VHH that inhibit HCV replication by interfering with the virus helicase activity. J. Virol. Methods. 2013;194(1–2):289–299. doi: 10.1016/j.jviromet.2013.08.032. [DOI] [PubMed] [Google Scholar]
- 70.Jittavisutthikul S., Thanongsaksrikul J., Thueng-In K., Chulanetra M., Srimanote P., Seesuay W., et al. Humanized-VHH transbodies that inhibit HCV protease and replication. Viruses. 2015;7(4):2030–2056. doi: 10.3390/v7042030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Suzuki R., Saito K., Matsuda M., Sato M., Kanegae Y., Shi G., et al. Single-domain intrabodies against hepatitis C virus core inhibit viral propagation and core-induced NFkappaB activation. J. Gen. Virol. 2016;97(4):887–892. doi: 10.1099/jgv.0.000423. [DOI] [PubMed] [Google Scholar]
- 72.Ho M. Perspectives on the development of neutralizing antibodies against SARS-CoV-2. <span/><span>Antib. Ther</span>. 2020;3(2):109–114. doi: 10.1093/abt/tbaa009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bathula N.V., Bommadevara H., Hayes J.M. Nanobodies: the future of antibody-based immune therapeutics. Cancer Biother. Radiopharm. 2021;36(2):109–122. doi: 10.1089/cbr.2020.3941. [DOI] [PubMed] [Google Scholar]
- 74.Chakraborty C., Bhattacharya M., Sharma A.R. Emerging mutations in the SARS-CoV-2 variants and their role in antibody escape to small molecule-based therapeutic resistance. Curr. Opin. Pharmacol. 2022;62:64–73. doi: 10.1016/j.coph.2021.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hanke L., Vidakovics Perez L., Sheward D.J., Das H., Schulte T., Moliner-Morro A., et al. An alpaca nanobody neutralizes SARS-CoV-2 by blocking receptor interaction. Nat. Commun. 2020;11(1):4420. doi: 10.1038/s41467-020-18174-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huo J., Le Bas A., Ruza R.R., Duyvesteyn H.M.E., Mikolajek H., Malinauskas T., et al. Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2. Nat. Struct. Mol. Biol. 2020;27(9):846–854. doi: 10.1038/s41594-020-0469-6. [DOI] [PubMed] [Google Scholar]
- 77.Mikolajek H., Weckener M., Brotzakis Z.F., Huo J., Dalietou E.V., Le Bas A., et al. Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes. Proc. Natl. Acad. Sci. U. S. A. 2022;119(31) doi: 10.1073/pnas.2205412119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Custodio T.F., Das H., Sheward D.J., Hanke L., Pazicky S., Pieprzyk J., et al. Selection, biophysical and structural analysis of synthetic nanobodies that effectively neutralize SARS-CoV-2. Nat. Commun. 2020;11(1):5588. doi: 10.1038/s41467-020-19204-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Aria H., Mahmoodi F., Ghaheh H.S., Mavandadnejad Faranak, Zare H., Heiat M., Bakherad H. Outlook of therapeutic and diagnostic competency of nanobodies against SARS-CoV-2: a systematic review. Anal. Biochem. 2022;640 doi: 10.1016/j.ab.2022.114546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Haga K., Takai-Todaka R., Matsumura Y., Song C., Takano T., Tojo T., et al. Nasal delivery of single-domain antibody improves symptoms of SARS-CoV-2 infection in an animal model. PLoS Pathog. 2021;17(10) doi: 10.1371/journal.ppat.1009542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gai J., Ma L., Li G., Zhu M., Qiao P., Li X., et al. A potent neutralizing nanobody against SARS-CoV-2 with inhaled delivery potential. MedComm. 2020;2(1):101–113. doi: 10.1002/mco2.60. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gauhar A., Privezentzev C.V., Demydchuk M., Gerlza T., Rieger J., Kungl A.J., et al. Single domain shark VNAR antibodies neutralize SARS-CoV-2 infection in vitro. FASEB Journal : official publication of the Federation of American Societies for Experimental Biology. 2021;35(11) doi: 10.1096/fj.202100986RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen Y.L., Lin J.J., Ma H., Zhong N., Xie X.X., Yang Y., Zheng P., Zhang L.J., Jin T., Cao M.J. Screening and Characterization of Shark-Derived VNARs against SARS-CoV-2 Spike RBD Protein. Int. J. Mol. Sci. 2022;23(18):10904. doi: 10.3390/ijms231810904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang Z., Wang Y., Jin Y., Zhu Y., Wu Y., Li C., et al. A non-ACE2 competing human single-domain antibody confers broad neutralization against SARS-CoV-2 and circulating variants. Signal Transduct. Target. Ther. 2021;6(1):378. doi: 10.1038/s41392-021-00810-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fatima A., Wang H., Kang K., Xia L., Wang Y., Ye W., et al. Development of VHH antibodies against dengue virus type 2 NS1 and comparison with monoclonal antibodies for use in immunological diagnosis. PloS one. 2014;9(4) doi: 10.1371/journal.pone.0095263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Thys B., Schotte L., Muyldermans S., Wernery U., Hassanzadeh-Ghassabeh G., Rombaut B. In vitro antiviral activity of single domain antibody fragments against poliovirus. Antivir. Res. 2010;87(2):257–264. doi: 10.1016/j.antiviral.2010.05.012. [DOI] [PubMed] [Google Scholar]
- 87.Terryn S., Francart A., Lamoral S., Hultberg A., Rommelaere H., Wittelsberger A., et al. Protective effect of different anti-rabies virus VHH constructs against rabies disease in mice. PloS one. 2014;9(10) doi: 10.1371/journal.pone.0109367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Laursen N.S., Friesen R.H.E., Zhu X., Jongeneelen M., Blokland S., Vermond J., et al. Universal protection against influenza infection by a multidomain antibody to influenza hemagglutinin. Science. 2018;362(6414):598–602. doi: 10.1126/science.aaq0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jahnichen S., Blanchetot C., Maussang D., Gonzalez-Pajuelo M., Chow K.Y., Bosch L., et al. CXCR4 nanobodies (VHH-based single variable domains) potently inhibit chemotaxis and HIV-1 replication and mobilize stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(47):20565–20570. doi: 10.1073/pnas.1012865107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Koromyslova A.D., Hansman G.S. Nanobodies targeting norovirus capsid reveal functional epitopes and potential mechanisms of neutralization. PLoS Pathog. 2017;13(11) doi: 10.1371/journal.ppat.1006636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stalin Raj V., NMA Okba, Gutierrez-Alvarez J., Drabek D., van Dieren B., Widagdo W., et al. Chimeric camel/human heavy-chain antibodies protect against MERS-CoV infection. Science advances. 2018;4(8) doi: 10.1126/sciadv.aas9667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wei G., Meng W., Guo H., Pan W., Liu J., Peng T., et al. Potent neutralization of influenza A virus by a single-domain antibody blocking M2 ion channel protein. PloS one. 2011;6(12) doi: 10.1371/journal.pone.0028309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Acharya P., Luongo T.S., Georgiev I.S., Matz J., Schmidt S.D., Louder M.K., et al. Heavy chain-only IgG2b llama antibody effects near-pan HIV-1 neutralization by recognizing a CD4-induced epitope that includes elements of coreceptor- and CD4-binding sites. J. Virol. 2013;87(18):10173–10181. doi: 10.1128/JVI.01332-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.McCoy L.E., Quigley A.F., Strokappe N.M., Bulmer-Thomas B., Seaman M.S., Mortier D., et al. Potent and broad neutralization of HIV-1 by a llama antibody elicited by immunization. J. Exp. Med. 2012;209(6):1091–1103. doi: 10.1084/jem.20112655. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data and materials included within the manuscript.