Abstract
Acquired lipodystrophy is often characterized as an idiopathic subtype of lipodystrophy. Despite suspicion of an immune-mediated pathology, biomarkers such as autoantibodies are generally lacking. Here, we used an unbiased proteome-wide screening approach to identify autoantibodies to the adipocyte-specific lipid droplet protein perilipin 1 (PLIN1) in a murine model of autoimmune polyendocrine syndrome type 1 (APS1). We then tested for PLIN1 autoantibodies in human subjects with acquired lipodystrophy with two independent severe breaks in immune tolerance (including APS1) along with control subjects using a specific radioligand binding assay and indirect immunofluorescence on fat tissue. We identified autoantibodies to PLIN1 in these two cases, including the first reported case of APS1 with acquired lipodystrophy and a second patient who acquired lipodystrophy as an immune-related adverse event following cancer immunotherapy. Lastly, we also found PLIN1 autoantibodies to be specifically enriched in a subset of patients with acquired generalized lipodystrophy (17 of 46 [37%]), particularly those with panniculitis and other features of autoimmunity. These data lend additional support to new literature that suggests that PLIN1 autoantibodies represent a marker of acquired autoimmune lipodystrophies and further link them to a break in immune tolerance.
Introduction
Lipodystrophy is a clinical syndrome defined by progressive loss of adipose tissue in some parts of the body (partial) or throughout (generalized) and may be inherited (genetic/familial) or acquired (1). The excessive loss of adipocytes can lead to clinically severe outcomes including insulin resistance, metabolic abnormalities (diabetes, hypertriglyceridemia, and hepatic steatosis) or even death (1–3). Lipodystrophy is diagnosed largely by clinical phenotype, but heterogeneous presentation, clinical mimicry, and frequent association with complex disorders underscore the need for biomarkers in this disorder. Mutations in several lipid droplet–associated genes explain a genetic etiology for a subset of lipodystrophies; however, the acquired lipodystrophy subtype is largely idiopathic and biomarkers are lacking (2,3).
Frequent co-occurrence of acquired lipodystrophy with autoimmune disease has fueled suspicion of an immune-mediated pathology (4–6). We hypothesized that autoimmune-associated acquired lipodystrophy may be associated with a common autoantibody that is adipocyte specific. Here, we identify autoantibodies to perilipin 1 (PLIN1), a gene mutated in a subset of inherited lipodystrophies, in two clinical cases of autoimmune-associated acquired lipodystrophy. One patient had a mutation in the autoimmune regulator gene, AIRE (7). Mutations in AIRE lead to the development of autoimmune polyendrocine syndrome type 1 (APS1) (or autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy [APECED]; OMIM no. 240300), which is characterized by breakdown in central tolerance and production of high-affinity autoantibodies to tissue-restricted antigens (8,9). The second patient developed lipodystrophy as an immune-related adverse event (irAE) following cancer immunotherapy (10). Finally, we broadly extended these findings and found PLIN1 autoantibodies to be specifically enriched in a subset of patients with acquired generalized lipodystrophy (AGL), particularly those with panniculitis along with other features of autoimmunity. The disparate nature of these cases, bound by a common autoantibody and clinical phenotype, suggests that anti-PLIN1 may be a molecular indicator of autoimmune-associated acquired lipodystrophy in multiple clinical contexts.
Research Design and Methods
Mouse Sera Collection
Aire−/− mice were maintained on a Balb/C background under SPF barrier conditions at University of California, San Francisco. Sera from wild-type and Aire−/− mice were prepared from tail vein bleeds (100 μL) as previously described (11). Sera were flash frozen and stored at −80°C until use. Tissue for immunohistochemistry was prepared from adult wild-type C57BL/6J mice (The Jackson Laboratory).
Phage Display and Immunoprecipitation (Phage Immunoprecipitation Sequencing)
The T7 Phage Display library (phage immunoprecipitation sequencing [PhIP-Seq] library) used here and experimental protocol were previously validated (12,13). Briefly, 1 μL mouse sera is incubated with 1 mL PhIP-Seq library (1010 pfu/mL) and incubated for 12–18 h at 4°C. Antibody-bound phages are immunoprecipitated with a mix of protein A and protein G magnetic beads (Thermo Fisher Scientific), eluted, and sequenced for identification of the unknown phage antigen(s). Peptide counts were summed with respect to annotated proteins, and total reads were normalized to 100,000 (RP100K). Candidate antigens were called based on read count >50 RP100K in all Aire−/− mice and absent in Aire+/+ controls.
Immunoprecipitation From 293T Overexpression Lysates
Full-length mouse pCMV6-Plin1-myc-flag (RC206292; OriGene Technologies) was sequenced verified and used for transfection in 293T cells. Whole cell lysates containing overexpressed PLIN1 were made to 1 mg/mL and 10 μL was set for input for Western blotting. One microliter of mouse sera or mock was added to 500 µL whole cell lysate, incubated for 12 h at 4°C. Antibodies were immunoprecipitated with protein A and protein G beads, washed three times with radioimmunoprecipitation assay buffer (140 mmol/L NaCl, 10 mmol/L Tris-HCL, 1.0% Triton-X, 0.1% SDS), and boiled in 2× Laemmli with β-mercaptoethanol. Immunoprecipitation (IP) elutions were run on a reducing SDS-PAGE 4–12% Bis-Tris gel (NP0349BOX; Thermo Fisher Scientific), transferred to nitrocellulose, and immunoblotted with primary anti-Flag antibody (1:5,000, rabbit anti-Flag; Cell Signaling Technology). To visualize anti-Flag we used infrared anti-rabbit secondary antibody (926-68703; LI-COR).
Indirect Immunofluorescence on Mouse Tissue
Adult wild-type mice were sacrificed and then perfused with 4% paraformaldehyde, and stomach was postfixed for 1 h, following sucrose/optimal cutting temperature compound (OCT) embedding for cryosectioning. Sections were cut 12 μm thick. Indirect immunofluorescent stains were obtained through incubating serum from anti-PLIN1 autoantibody–positive sample or control sample or commercial antibody to PLIN1 (Sigma-Aldrich) at a dilution of 1:1,000 on tissue section. Samples were washed and then developed with an FITC-conjugated secondary anti-human IgG antibody (Abcam). Representative images were captured on Nikon Ti confocal microscope at the University of California, San Francisco, imaging core.
Radioligand Binding Assay
An expression plasmid containing full-length human PLIN1 coding sequence under the control of a T7 promoter (RC206292; OriGene Technologies) was sequence verified and used as a DNA template for in vitro translation of the PLIN1 protein. PLIN1 was synthesized in the presence of [35S]methionine to radiolabel the protein as previously described (14). Individual serum samples from both index patient cases, as well as a cohort of control subjects without evidence of lipodystrophy (including normal control patients, patients with identified APS1, and checkpoint blockade–treated patients) were incubated with radiolabeled PLIN1 at 4°C overnight. Antibody-bound protein was immunoprecipitated with protein A/G beads and washed and total radioactive counts were obtained by scintillation. A commercial PLIN1-specific antibody (no. HPA024299; Sigma-Aldrich) was used as a positive control. The antibody index was calculated as follows: (sample value − mean blank value)/(positive control antibody value − mean blank value). A threshold for a positive result was set as 3 SD above the mean for normal control subjects (indicated by dotted line).
Study Approval: Human Studies
Figure 2 involves our index APS1 patient with lipodystrophy (case report 1) as well as APS1 patients without lipodystrophy and healthy controls. Ethics approval for the study of the index APS1 patient with lipodystrophy was granted by the ethics committee at Endocrinology Research Center of Russia, and all patients or their parents or guardians provided written informed consent. Information regarding ethics approval for the study of APS1 patients without lipodystrophy in Fig. 2 was previously published (15); and approval was granted by the National Insitute of Allergy and Infectious Disease Institutional Review Board, Bethesda, MD, and patients provided written informed consent. Healthy control plasma for Fig. 2 was obtained from the New York Blood Center, where they were collected under informed consent, including consent for usage for research and publication. Figure 3 involves our index cancer immunotherapy patient with lipodystrophy (case report 2) as well as checkpoint immunotherapy patient control subjects. Ethics approval and signed and written informed consent for case report 2 in Fig. 3 was granted by Hospices Civils de Lyon as part of the study Immune modulation Study in Patients with Metastatic Melanoma Treated with anti-PD1 Monoclonal Antibodies (PAIR) (clinical trial reg. no. NCT02626065, ClinicalTrials.gov). Ethics approval for the study of checkpoint immunotherapy patient control subjects for Fig. 3 was provided by the Human Research Protection Program International Review Board, and all participants gave written informed consent. Supplementary Fig. 1 contains a study of additional APS1 patients without lipodystrophy and healthy control subjects. Ethics approval for the study of all patients in Supplementary Fig. 1 was granted by the Regional Ethical Committee of Western Norway (approval nos. 2009/2555 and 2018/1417), and all participants gave written informed consent for participation. Ethics approval for mouse studies was granted by University of San Francisco International Review Board (protocol AN177913). The research study for control subjects with diabetes (Fig. 4) was approved by the Colorado Multiple Institutional Review Board (IRB no. 92-292). Study subjects with new- and recent-onset T1D were recruited from the Barbara Davis Center for Diabetes clinics. Peripheral blood was obtained for islet autoantibody measurements and HLA typing. Written informed consent was obtained from each participant and guardian when the participant was <18 years of age, and the Colorado Multiple Institutional Review Board approved the study.
Figure 2.
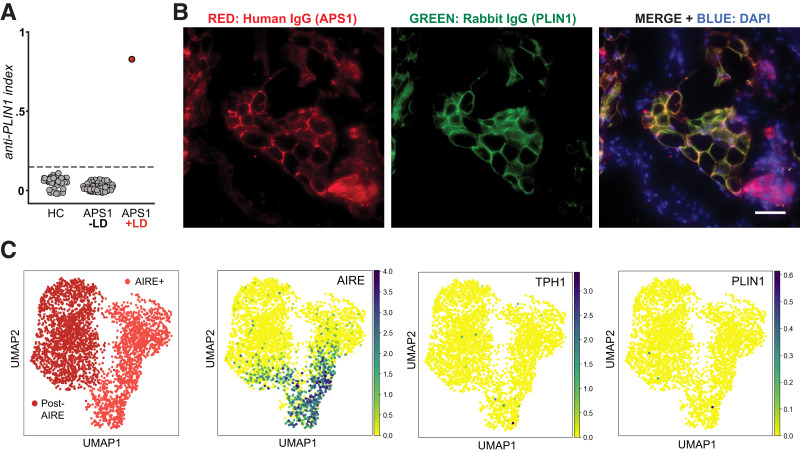
Autoantibodies to PLIN1 in sera from a patient with APS1 and acquired lipodystrophy. A: RLBA for detection of anti-PLIN1 antibodies. Radiolabeled PLIN1 protein was incubated with sera from healthy control subjects (HC) (n = 54) or from APS1 patients with (n = 1) or without (n = 68) lipodystrophy (LD). Dotted line indicates mean ± 3 SD healthy control subjects. B: Validation of autoantibodies to PLIN1 in orthogonal cell–based assay. Fixed stomach tissue of mice was mounted and immunostained with sera from case report patient 1 and commercial antibody to PLIN1. A mix of secondary antibodies anti-human IgG Alexa Fluor 547 and anti-rabbit Alexa Fluor 488 at 1:2,000 was used to visualize human IgG and PLIN1 antibodies, respectively. A merge is provided on the right of individual images. Images taken at ×20 magnification. Note yellow in merge indicating colocalization of PLIN1 (green) and human IgG (red). DAPI is blue and indicates nuclei. Scale bar indicates 100 μm. C: Examination of RNA expression of AIRE, PLIN1,and TPH1 genes in human mTECs using a publicly available human thymic RNA sequencing data set (Park et al. [16] and Bautista et al. [17]).
Figure 3.
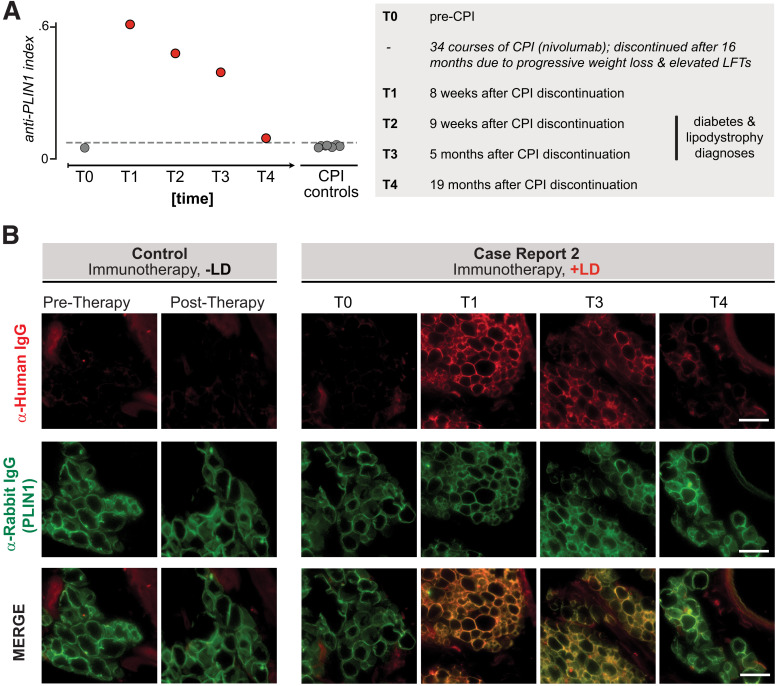
Autoantibodies to PLIN1 in sera from a patient with autoimmune AGL following cancer immunotherapy. A: RLBA screening for PLIN1 antibodies in sera from all time points from index patient 2 as well as checkpoint-treated control patients without lipodystrophy (n = 7), as done for case report 1. Dotted line indicates mean ± 3 SD healthy control subjects (n = 11). Numbers to the right of circles indicate the time point series, for reference in panel B. The left panel describes the clinical timeline corresponding to time points T0–T4. Checkpoint inhibitor therapy (CPI) was discontinued at 34 cycles (16 months) due to progressive weight loss and elevated liver function tests (LFTs), with further workup over subsequent months. B: Validation of autoantibodies to PLIN1 with use of immunohistochemistry on mouse omental adipose tissue, as in Fig. 2B. Sera were used from either a control subject (immunotherapy but no AGL) or case report 2 patient (immunotherapy with AGL), with various time points. Each column represents an individual sample; from left to right: sera from checkpoint control pretreatment, sera from checkpoint control posttreatment with no autoimmunity, case report 1 pretherapy, case report 1 posttherapy 1, case report 1 posttherapy 3, case report 1 posttherapy 4. Images are 600 × 600 pixel insets from original ×40 image. Scale bar represents 100 μm. LD, lipodystrophy.
Figure 4.
Autoantibodies to PLIN1 are specific to the generalized subtype of acquired lipodystrophy. RLBA screening for PLIN1 antibodies in sera from healthy control subjects (HC) (n = 63) or individuals with either lipodystrophy (AGL n = 46, APL n = 52) or autoimmune metabolic disorders not involving lipodystrophy (type 1 diabetes [T1D] n = 110, type 2 diabetes [T2D] n = 24). Dotted line represents 3 SD above the mean from healthy control subjects. Red dots depict 17 of 46 (37%) AGL patients scoring positive for PLIN1 autoantibodies.
The study protocol for control subjects and lipodystrophy patients in Fig. 4 and Supplementary Fig. 2 was approved by the Institutional Review Board of UT Southwestern Medical Center (UTSW), the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), Bethesda, MD, and the University of Michigan. Adult patients, normal healthy control subjects, and legal guardians of patients <18 years of age gave written informed consent; minors provided assent if age appropriate.
Subjects for Expanded Lipodystrophy Cohort
Serum from 13 subjects with AGL and 5 normal subjects from UTSW was studied for autoantibodies with a HuProt chip assay at the UT Southwestern Microarray and Immune Phenotyping Core laboratory. In addition, serum/plasma from subjects with AGL (UTSW n = 36, NIDDK n = 8, and University of Michigan n = 2), acquired partial lipodystrophy (APL) (UTSW n = 52) and normal healthy control subjects (UTSW n = 19) was assessed for PLIN1 autoantibody with radioligand binding assay (RLBA).
Questionnaire
Demographic data and health history were collected from AGL patients either during physician interview or with a lipodystrophy questionnaire. Presence of autoimmune disorders, such as panniculitis, rheumatoid arthritis, systemic lupus erythematosus, polymyositis or dermatomyositis, idiopathic arthritis, and Hashimoto thyroiditis or others, and metabolic disorders, such as diabetes, hypertriglyceridemia, hepatic steatosis, fatty liver, and polycystic ovarian syndrome and others, were self-reported by the patients. Height and body weight were measured with standard procedures. Patients with APL (47 female and 4 male, median age 33.5 years [minimum–maximum 7–67]) presented with gradual onset of bilaterally symmetrical subcutaneous fat loss from the face, neck, upper extremities, thorax, and abdomen but sparing the lower extremities (4). Supportive clinical criteria included 1) onset of subcutaneous fat loss during childhood and adolescence, 2) absence of family history of lipodystrophy, and 3) presence of autoimmune diseases, and laboratory criteria included 1) low serum levels of complement 3, 2) presence of serum C3 nephritic factor autoantibody, 3) proteinuria, 4) membranoproliferative glomerulonephritis, and 5) characteristic body fat distribution as documented by skinfold thickness measurements (4). Normal healthy subjects (14 female and 5 male, age 27 years [23–53]) had no medical problems and had normal complete blood counts, serum chemistry, thyroid function tests, and urinalysis.
Biochemical Analyses and Procedures
Fasting blood samples were collected and were analyzed for biochemical variables. Blood samples for the UTSW patients were sent to Quest Diagnostics (Irving, TX) for the analysis. Serum glucose, lipids, lipoproteins, and liver enzymes of UTSW patients were measured with the photometric method (Beckman Coulter AU clinical analyzer). Blood hemoglobin A1c (HbA1c) was measured with the immunoturbidimetric method (Roche Integra 800 chemistry analyzer). Serum glucose, HbA1c, lipids, and hepatic function tests for the NIDDK samples were conducted in the National Institutes of Health Clinical Center laboratory according to standard methodology. Blood samples at the University of Michigan were analyzed by the University of Michigan Health System Clinical Pathology Laboratory.
Serum Antibody Profiling With a Human Proteome Microarray
The HuProt version 3.1 array consists of ∼19,500 unique full-length proteins (recombinant proteins expressed in the saccharomyces cerevisiae and purified) printed on glass slides in duplicates. This protein panel represents 16,152 unique human genes (∼81% of the proteome) and 124 unique mouse gene. Positive control proteins are H1, H2A, H2B, H3, H4, IgG488/594, rhodamine + IgG 647, hMDM2, and Era along with human IgG and anti-human IgG in various concentrations and negative control proteins BSA (bovine serum albumin and GST in various concentrations) and printed in duplicates as well. The following procedure was used for the assay.
Human Thymic Expression Analysis
To assess expression of PLIN1 in human thymic epithelial cells (TECs), we used published single-cell RNA sequencing data sets (16,17). We analyzed expression in the Aire+ medullary TEC (mTEC) hi/mTEC(II) and Corneo-like mTEC/mTEC(III)/post-AIRE subsets from the Park et al. (16) and Bautista et al. (17) combined data set. Cells were subclustered with use of the Leiden algorithm, and expression of genes of interest was visualized with Uniform Manifold Approximation and Projection (UMAP).
Data and Resource Availability
The data sets generated during or analyzed during the current study are included in the study or Supplementary Material and/or are available from the corresponding author on reasonable request. No novel applicable resources were generated or analyzed during the current study.
Results
Identification of Anti-PLIN1 Autoantibodies in the Murine Model of APS1
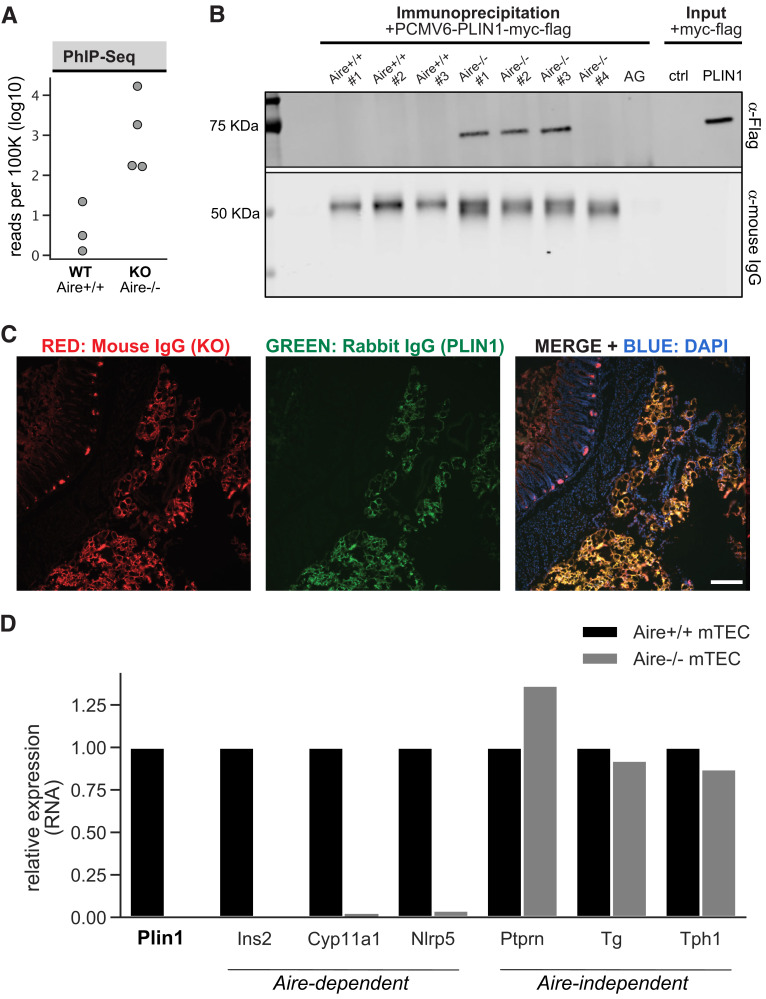
APS1 is a monogenic disorder linked to a defect in the AIRE gene that is characterized by multiple tissue-specific autoimmune diseases (18,19). Here we leveraged sera from a cohort of Aire knockout mice (Aire−/−) (n = 4) that were archived previously due to evidence of an autoantibody to an approximately 60-kDa antigen present in fat (data not shown). To identify the specificity of this autoantibody we tested sera from these four Aire−/− mice as well as three age-matched wild-type controls (Aire+/+) (n = 3) with PhIP-Seq, a proteome-wide screening approach for autoantibody discovery (20). Although each sample exhibited a distinct pattern of PhIP-Seq reactivity, phage-displayed peptides derived from the PLIN1 gene were the only shared targets enriched by all four Aire−/− mice and absent from all wild-type controls (Fig. 1A). We validated antibody reactivity in Aire−/− mice in two orthogonal assays. First, we demonstrate IP of recombinantly expressed mouse full-length Plin1-myc-flag from cell lysates using Aire−/− sera (Fig. 1B). Second, we show colocalization of Aire−/− sera with commercial antibody to PLIN1 in mouse omental adipose tissue (Fig. 1C). Aire promotes immune tolerance by driving the expression of a wide array of tissue specific antigens in thymic epithelial cells and, consistent with this activity, we found the relative amounts of Plin1 RNA in mTECs to be dependent on proper Aire expression (21) (Fig. 1D). The reduction in Plin1 expression was consistent with reductions of other known Aire-activated target genes, including Ins2, Cyp11a1, and Nlrp5, suggesting a possible mechanism contributing to autoimmunity to Plin1. These data in mice, together with prior reports linking PLIN1 mutations to inherited lipodystrophy subtypes, suggest that anti-PLIN1 immune response may also be present in human lipodystrophy associated either with APS1 or with alternative autoimmune-associated acquired lipodystrophies (22,23).
Figure 1.
Discovery and validation of autoantibodies to perilipin 1 in Aire−/− mouse sera. A: PhIP-Seq analysis in Aire−/− and Aire+/+ mice. Aggregated Plin1 PhIP-Seq data from Aire−/− (n = 4) and Aire+/+ (n = 3) mice. B: Whole cell lysates generated from 293T cells expressing full-length mouse PLIN1 were incubated with sera from Aire−/− (n = 4) or Aire+/+ (n = 3) mice. Antibodies were immunoprecipitated with use of AG beads, and IP elutions were subject to SDS-PAGE immunoblotting. AG lane indicates an IP with use of protein A/G beads only—no sera. Input lane indicates loading of whole cell lysate with and without (−) transfection of PLIN1-myc-flag plasmid. IP elutions and input were immunostained with either anti-Flag IgG to identify positive anti-Plin1 signal, or anti-mouse IgG to show qualitative capture of IgG from sera. C: Representative image of immunohistochemistry on mouse omental adipose tissue showing positive colocalization of antibodies from Aire−/− sera and commercial anti-Plin1 IgG. Primary antibodies from Aire−/− mouse sera and commercial antibody to PLIN1 were visualized with secondaries to mouse IgG (Alexa Fluor 567 [red]) and rabbit IgG (Alexa Fluor 488 [green]), respectively. DAPI (blue) stains nuclei. Scale bar indicates 150 μm. D: Examination of RNA expression of Plin1 and genes with known Aire-dependent thymic expression using a publicly available murine thymic RNA sequencing data from Aire−/− and Aire+/+ mTECs (Sansom et al. [21]). ctrl, control; KO, knockout.
Case Report No. 1: APS1 Patient
The first case of AGL in a patient with APS1 was recently reported (7). Briefly, the patient developed recurrent stomatitis at age 7 months and signs of lipodystrophy at 18 months. At 3 years of age autoimmune hepatitis developed (Child-Pugh class C) and lipodystrophy progressed. Clinical testing for lipodystrophy-associated genetic variants was negative (ZMPSTE24, LMNA, BSCL2, PLIN1, PTRF, LMNB2, POLD1, AKT2, CIDEC, PIK3CA, PPARG, PSMB8, CAV1, PPP1R3A, AGPAT2). Candidiasis in the oral cavity and esophagus was diagnosed at 4 years of age followed by primary adrenal insufficiency at age 4.5 years. At that point the diagnosis of APS1 was made, confirmed by homozygocity for the most common AIRE mutation, R257X. He died of viral H1N1 pneumonia with respiratory and adrenal failures at age 5 years and 9 months. Serum from the patient for autoantibody testing was obtained at 5 years of age.
An RLBA was adapted for anti-PLIN1 autoantibody screening in humans. By this assay, autoantibodies to PLIN1 were present in patient 1 but absent from a large cohort of APS1 patients without lipodystrophy (n = 68) and healthy control subjects (n = 54) (Fig. 2A). PLIN1 autoreactivity in patient 1 was further confirmed with immunohistochemistry using mouse omental adipose tissue containing fat, which showed unambiguous colocalization of human sera and commercial antibody to PLIN1 (Fig. 2B). Interestingly, with use of the same RLBA assay, PLIN1 autoantibodies were detected in several additional APS1 patients without lipodystrophy in an independent analysis involving a second, larger cohort of patients (Supplementary Fig. 1). Absence of PLIN1 autoantibodies among healthy individuals was consistent across both independent analyses.
Presence of PLIN1 autoantibodies in human APS1 is consistent with our findings in the APS1 mouse model, suggesting that tolerance to PLIN1 may be regulated by AIRE-mediated mechanisms. To further test the possibility that PLIN1 is regulated by AIRE, we mined a previously published data set representing single-cell transcriptomes of healthy human TECs to examine PLIN1 expression (17). We demonstrate that PLIN1 transcripts can be detected in AIRE-expressing and post–AIRE expressing cells in the human thymus, in a magnitude similar to that of an established AIRE target, tryptophan hydroxylase 1 (TPH1) (Fig. 2C).
Case Report No. 2: Checkpoint Blockade–Induced Lipodystrophy
Rare instances of generalized lipodystrophy as an irAE following immune checkpoint inhibitor administration have recently been described (10,24,25). Patient 2 is a 62-year-old woman with metastatic melanoma detected in her brain, lung, and liver (10). The immune checkpoint inhibitor nivolumab (an anti-PD1 monoclonal antibody) was administered for 34 cycles over 16 months, at which time the patient presented with progressive weight loss and elevated liver function tests. Workup included a liver biopsy that revealed severe steatosis, and nivolumab treatment was halted due to concern for irAE. Over the next month, the patient developed further weight loss, hyperphagia, polydipsia, and polyuria. Type 1 diabetes–related autoantibodies (to GAD, IA2, and ZnT8) were negative, and results of fasting plasma insulin, C-peptide, and HOMA of insulin resistance tests were all consistent with insulin-resistant diabetes. Her physical exam was notable for severe loss of subcutaneous fat in a broad distribution with muscle prominence consistent with lipodystrophy. Laboratory testing was notable for an undetectable leptin level and elevated triglycerides. Clinical testing for 23 lipodystrophy-associated genetic variants was also negative (AKT2, BSCL1, BSCL2, CAV1, CIDEC, DYRK1B, INSR, LIPE, LMF1, LMNA, LMNB2, NSMCE2, PCYT1A, PIK3R1, PLD3, PLIN1, POC1A, POLD1, PPARG, PSMB8, PTRF, TBC1D4, ZMPSTE24). A diagnosis of AGL was made, and her diabetes was treated with metformin, dietary changes, and high-dose basal-bolus insulin. Four months following cessation of nivolumab, there was improvement in the patient’s diabetes as noted by decreased insulin requirements and partial improvement in hypertriglyceridemia. Serum samples for patient 2 were collected at different time points throughout her clinical course under informed consent.
Longitudinal sera samples from patient 2 were tested for autoantibodies to PLIN1 with RLBA. Patient no. 2 was negative for autoantibodies to PLIN1 prior to treatment with nivolumab but was positive after 34 courses of nivolumab treatment, when she was noted to have weight loss and steatosis (Fig. 3A). To validate the immunoreactivity, immuno-labeling of mouse omental adipose tissue with patient sera and commercial antibody to PLIN1 colocalized in samples corresponding to posttreatment and lipodystrophy diagnosis time points but not pretreatment samples (Fig. 3B). Reactivity to PLIN1 appeared to decrease by 19 months after cessation of nivolumab, and although the patient’s generalized lipodystrophy phenotype did not resolve, metabolic profile did improve.
PLIN1 Autoantibodies Are Enriched in a Subset of Patients With AGL and Autoimmunity
Given the data for presence of PLIN1 autoantibodies in our two clinical cases, and extremely high signal intensity of PLIN1 autoantibodies in the HuProt array data for a small cohort of patients with AGL (Supplementary Fig. 2), we next sought to determine whether PLIN1 autoantibodies were more broadly present in subjects with acquired generalized and APL. Here, we obtained serum from cohorts of patients with AGL (n = 46) and APL (n = 52). For control subjects we included cohorts of healthy control subjects and subjects with type 1 diabetes and type 2 diabetes. On RLBA, 17 of 46 patients (37%) with AGL were positive for PLIN1 antibody and 29 were negative, whereas none of the patients with APL or any of the control populations were positive (Fig. 4).
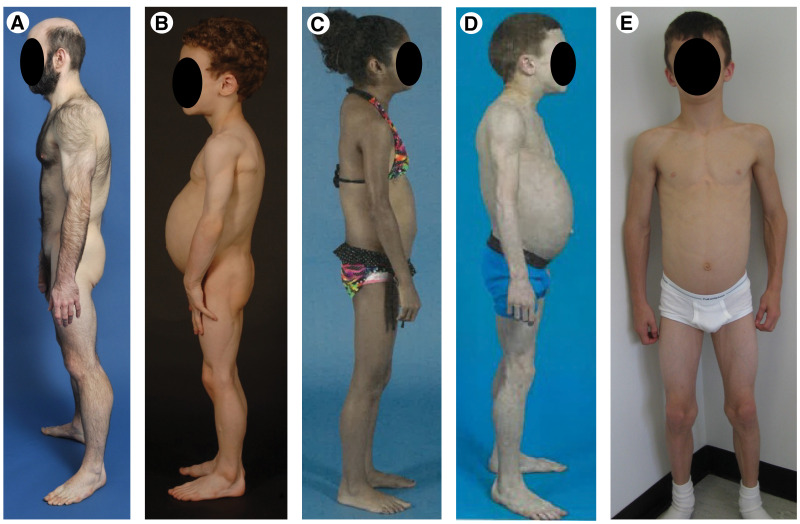
Clinical features of some of the patients with AGL are shown in Fig. 5 and were previously published (26). All patients with AGL (35 female 11 male, ages 3–59 years) presented with selective loss of body fat affecting large regions of the body beginning after birth, usually before adolescence (26). In addition, they had some of the following supportive clinical features: 1) loss of subcutaneous fat from the palms and soles, 2) acanthosis nigricans, 3) hepatosplenomegaly, 4) history of subcutaneous tender nodular swellings suggestive of panniculitis preceding onset of lipodystrophy, or 5) presence of other autoimmune diseases such as dermatomyositis, systemic lupus erythematosus, rheumatoid arthritis, and Sjögren syndrome. Common laboratory abnormalities included 1) impaired glucose tolerance or diabetes, 2) severe fasting and/or postprandial hyperinsulinemia, 3) hypertriglyceridemia and/or low levels of serum HDL cholesterol, 4) reduced serum concentrations of leptin and/or adiponectin, or 5) documentation of loss of fat from large regions of the body by anthropometry (26).
Figure 5.
Clinical features of patients with AGL. A: A 20-year-old male with panniculitis diagnosed in early childhood. B: A 5-year-old male with hepatomegaly, hepatic steatosis, splenomegaly, diabetes, and celiac disease. C: A 12-year-old female with diabetes, dermatomyositis, hepatomegaly, and celiac disease. D: An 8-year-old male with panniculitis, fatty liver, hypothyroidism, and vitiligo. E: A 9-year-old male with IgA deficiency, hepatomegaly, and splenomegaly.
We then compared the demographics and clinical features of AGL patients with PLIN1 autoantibody with those of subjects who were negative for the antibody (Table 1). As compared with AGL patients negative for PLIN1 antibody, among those who were positive a higher proportion was male (P = 0.036) and younger in age (P = 0.011) and there was a higher prevalence among those who were positive of panniculitis (P = 0.003) and lymphadenopathy (P = 0.038) and borderline higher prevalence of autoimmune hepatitis (P = 0.055) (Table 1). There was no difference in the prevalence of arthritis, rheumatoid arthritis, polymyositis/dermatomyositis, systemic lupus erythematosus, or Hashimoto thyroiditis; however, there was a clear enrichment of multiple autoimmune features in PLIN1 antibody–positive subjects (P = 0.003). On examination of metabolic parameters there was no difference in the prevalence of diabetes, hypertriglyceridemia, hypertension, or acute pancreatitis between the PLIN1 autoantibody–positive and –negative AGL patients (Table 2). The fasting serum glucose, triglycerides, HDL cholesterol, and blood HbA1c values were also not significantly different between the two groups (Table 2). Taken together, these data support that PLIN1 autoantibodies identify a subset of patients with AGL that are enriched for having features of autoimmunity and immune dysregulation.
Table 1.
Demographics and prevalence of autoimmune diseases among PLIN1 autoantibody–positive and –negative patients with AGL
| Positive | Negative | P | |
|---|---|---|---|
| N | 17 | 29 | |
| n male/n female | 7/10 | 4/25 | 0.036 |
| Age (years) | 12 (3–59) | 31 (7–59) | 0.011 |
| Panniculitis | 65 | 17 | 0.003 |
| Lymphadenopathy | 35 | 7 | 0.038 |
| Autoimmune hepatitis | 24 | 3 | 0.055 |
| Arthritis | 35 | 17 | 0.3 |
| Hashimoto thyroiditis | 18 | 7 | 0.3 |
| Polymyositis/dermatomyositis | 12 | 14 | 1.0 |
| Rheumatoid arthritis | 19 | 7 | 0.3 |
| Systemic lupus erythematosus | 0 | 7 | 0.5 |
| Autoimmune diseases per patient | |||
| 0 | 18 | 55 | 0.003 |
| 1 | 35 | 35 | |
| 2 | 18 | 10 | |
| 3 | 29 | 0 |
Data are percentages or median (minimum–maximum) unless otherwise indicated.
Table 2.
Prevalence of metabolic diseases and metabolic variables among PLIN1 autoantibody–positive and –negative patients with AGL
| Positive | Negative | P | |
|---|---|---|---|
| N | 17 | 29 | |
| Diabetes | 71 | 52 | 0.24 |
| Hypertriglyceridemia | 80 | 75 | 1.0 |
| Hypertension | 12 | 39 | 0.09 |
| Acute pancreatitis | 12 | 14 | 1.0 |
| Serum glucose (mg/dL) | 88 (62–332) | 103 (59–316) | 0.62 |
| Serum triglycerides (mg/dL) | 343 (64–2,693) | 278 (116–5,436) | 0.68 |
| Serum HDL cholesterol (mg/dL) | 23 (11–51) | 30 (10–87) | 0.16 |
| Blood HbA1c (%) | 7.2 (5–13) | 5.1 (4.5–10.3) | 0.051 |
Data are percentages or median (minimum–maximum) unless otherwise indicated.
Discussion
Acquired lipodystrophy syndromes are an idiopathic clinical phenotype with a strong suspicion of autoimmune etiology (1). Autoimmune disease can give rise to a wide variety of clinical phenotypes, which are often related to the expression patterns of the respective targeted protein autoantigens (27). Here, the study of human patients, together with examination of mouse Aire−/− samples, suggests that acquired lipodystrophy may be marked by autoreactivity to the protein PLIN1. PLIN1 has restricted expression in adipocytes and has a well-established role in regulating lipid droplet size and maintaining lipid homeostasis throughout the body (28). Furthermore, loss of Plin1 expression in mice and humans leads to a lipodystrophy phenotype (23,29). Our unbiased discovery of PLIN1 autoantibodies in an autoimmune-prone mouse model and as well as unbiased HuProt array data revealing increased prevalence of PLIN1 autoantibodies in patients with AGL add to the growing evidence supporting an autoimmune etiology for acquired lipodystrophy and a potential role for anti-PLIN1 autoantibodies as a biomarker.
Acquired lipodystrophy is a heterogenous form of lipodystrophy that manifests as two major subtypes, AGL or APL, depending on the pattern of fat loss throughout the body (1). The etiology for both AGL and APL has generally been unclear, but clinical evidence suggests the pathophysiologies may be distinct. For instance, AGL has low serum levels of adipocytokines and is more frequently associated with autoimmune disease compared with APL (30). Indeed, in our large cohort of subjects with AGL and APL we found that PLIN1 autoantibodies were a specific marker within a subset of AGL patients (36%). Our data also reveal that PLIN1 autoantibodies are more likely to be present in AGL patients with panniculitis (type 1) as compared with those with autoimmune and idiopathic variety of AGL (types 2 and 3) (26). AGL patients with PLIN1 autoantibodies are likely to be young and male, with increased prevalence of lymphadenopathy, autoimmune hepatitis, and other autoimmune diseases. Although further work will be needed to examine this association, it may have some link to the disordered storage of fatty acids in AGL with increased fat deposition in the liver that may serve as a trigger for the autoimmune hepatitis. Overall, our findings in the AGL cohort show that a significant fraction of these patients harbor PLIN1 autoantibodies and this serves as a biomarker linking the potential etiology to an autoimmune process in the subcutaneous fat that is directed by a PLIN1-specific immune response.
Interestingly, in a recent report Corvillo et al. (31) used a candidate-based approach in humans to identify PLIN1 as a putative autoantigen in autoimmune-associated AGL. Our study is complementary to this preliminary report and advances knowledge in several critical ways. First, the previous study examined a total of five patients with autoimmune-associated AGL, of whom only three were positive for PLIN1 autoantibodies. Our study confirms the very specific association of PLIN1 autoantibodies with AGL, with a large cohort of AGL patients as well as APL patients for comparison, bolstering confidence in PLIN1 autoantibodies as a potential biomarker. Second, in addition to the autoimmune variety of AGL, we show that PLIN1 autoantibodies are present in the panniculitis variety of AGL, a clinical cohort not represented in the previous study. Third, Corvillo et al. propose a functional role of PLIN1 autoantibodies in regulating lipolysis in vitro, but further investigation will be required for understanding of whether these autoantibodies have a causal relationship in vivo. Our novel association of PLIN1 autoantibodies in the Aire−/− mouse provides a model system to recapitulate autoantibodies in vivo and rigorously test for a potential function. Lastly, our extended findings of PLIN1 autoantibodies associated with AGL in two distinct disease settings provide clues about potential pathological mechanisms underlying this poorly understood disorder.
The mechanisms that trigger production of PLIN1 autoantibodies in the context of AGL are unclear. However, the two case reports presented here are instructive in that they occurred in the setting of a breakdown in two key checkpoints on immune tolerance: 1) abnormal central tolerance mediated by AIRE mutations and 2) inhibition of PD1 on peripheral T cells through exposure to immune checkpoint inhibitors (7,10). The co-occurrence of additional autoimmune responses in both patients, including autoimmune hepatitis and Addison disease in the APS1 patient and an anticancer response in the nivolumab-treated patient, provides strong circumstantial evidence that the lipodystrophy may also be of autoimmune origin. Further bolstering this notion is the presence of T cell infiltrates in the fat biopsy of the nivolumab-treated patient (10). Lastly, presence of PLIN1 autoantibodies in these two contexts points to peripheral immune tolerance mechanisms playing a role in maintaining tolerance to PLIN1.
Here we identify autoantibodies to PLIN1 in the first reported case of lipodystrophy associated with APS1 in humans (7). In the setting of APS1, loss of immune tolerance to peripheral organs is thought to occur through a loss of proper display of tissue-specific antigens in the thymus. This loss in T cell tolerance is also associated with the development of autoantibodies that reflect the T cell specificity (19). Interestingly, in mining thymic gene expression data from mice lacking Aire expression, we found evidence that Plin1 is expressed in the thymus in an Aire-dependent fashion (Fig. 1D) similar to other known self-antigens like Ins2 (insulin) (21). The marked decrease in PLIN1 self-antigen expression in the Aire-deficient thymus suggests that individuals with a deficiency in AIRE may harbor a greater risk for the development of an immune response to PLIN1 and therefore also a greater risk for developing autoimmunity to peripheral adipose tissue that expresses PLIN1. Although lipodystrophy is rare in APS1, we suggest that additional screening should be explored in this patient group and, conversely, that genetic testing for AIRE should be considered in young patients presenting with unexplained lipodystrophy or metabolic abnormalities. Although a single prior study suggests a functional role of PLIN1 autoantibodies in regulating lipolysis in vitro, further investigation will be required to understand whether these autoantibodies have a causal relationship in vivo (31). Future studies should include phenotyping of Aire−/− mice with a focus on acquired LD, which has not been examined previously due to lack of precedence.
As in case report no. 2, we also identified PLIN1 autoantibodies in association with AGL in the setting of cancer immunotherapy. As the deployment of immune checkpoint blockade therapy across many types of cancers has increased, so has the prevalence of associated irAEs (32). Endocrine adverse events can appear suddenly in individual patients and may manifest as common autoimmune diseases such as hypothyroidism and type 1 diabetes or rare autoimmune presentations such as hypophysitis and, more recently, acquired lipodystrophy (10,24,25). Given the severe consequences of some irAE, developing new and improved markers for detecting those at risk is a clinical priority. Due to the fact that autoantibodies commonly precede detection of symptoms, autoantibodies to PLIN1 may provide a useful tool toward this goal. Future studies are necessary to determine whether anti-PLIN1 antibody testing may be useful for identifying patients at risk even prior to the onset of clinically apparent symptoms.
Taken together, our results provide new insight into the specificity of the immune response in a subset of cases of idiopathic acquired lipodystrophy, adding to the growing evidence for PLIN1 autoantibodies as a possible biomarker for this syndrome. In addition, our findings of PLIN1 autoantibodies in two distinct clinical settings of a break in immune tolerance inform new mechanisms of the pathophysiology of this poorly understood disease.
Article Information
Acknowledgments. The authors thank New York Blood Center for donation of healthy plasmas for RLBA analysis. The authors thank the patients for participation in the study; Elaine Cochrane from NIDDK, National Institutes of Health (NIH), and Claudia Quittner from UT Southwestern for help with evaluation of patients; and Phoebe Ellis and Tea Huseinbegovic of UT Southwestern for illustrations.
Funding. Z.Q. was supported by American Diabetes Association grant 1-19-PDF-131. J.L.D. is funded by a grant from Chan Zuckerberg Biohub. J.L.D. and C.M.-B. are funded by the National Institute of Mental Health of the NIH (award 1R01MH122471-01). C.M.-B. is funded by The Emiko Terasaki Foundation (Project 7027742/Fund B73335) and by the National Institute of Neurological Disorders and Stroke of the NIH (award 1K99NS117800-01). S.E.V. is funded by the National Institute of Diabetes and Digestive and Kidney Diseases of the NIH (award 1F30DK123915-01). M.S.L. is funded by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases (ZIA AI001175). M.S.A. is funded by the National Institute of Allergy and Infectious Diseases (5P01AI118688), The Leona M. and Harry B. Helmsley Charitable Trust, and the Chan Zuckerberg Biohub. E.S.H. is funded by Stiftelsen Kristian Gerhard Jebsen, the Novo Nordisk Foundation, the Research Council of Norway, and the Regional Health Authorities of Western Norway. A.K.A. and A.G. are funded by NIH grant R01-DK105448 and the Southwestern Medical Foundation. R.B. is funded by the intramural research program of the National Institute of Diabetes and Digestive and Kidney Diseases.
Duality of Interest. S.D. received institutional research grants and had travel cost covered by Bristol-Myers Squibb and institutional research grants and had travel cost covered by Merck Sharp & Dohme. No other potential conflicts of interest relevant to this article were reported.
Author Contributions. C.M.-B. and S.E.V. contributed equally to the study including design of the experiments, generation of raw data, data analyses, and editing of the manuscript. C.M.-B., E.Ora., M.S.A., A.G., and J.L.D. conceptualized the study. C.M.-B., M.S.A., A.G., and J.L.D. wrote the manuscript. C.L. and M.C. assisted in immunohistochemistry experiments. Z.Q. and B.M. assisted in RLBA analysis. A.K.A., X.L., C.Z., Q.L., E. Orl., E.Ora., R.B., and A.G. contributed patient samples and metadata, clinical expertise, and HuProt analysis. A.F.K. contributed the analysis pipeline for PhIP-Seq. A.P. contributed analysis of human TEC single-cell RNA-sequencing. E.D., C.C.-A., S.D., E.F., D.A., A.M., B.E.O., M.S.L., and E.S.H. contributed samples for analysis and clinical expertise and edited the manuscript. J.L.D. and M.S.A. are the guarantors of this work and, as such, had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains supplementary material online at https://doi.org/10.2337/figshare.20060174.
C.M.-B. and S.E.V. share first authorship.
M.S.A., A.G., and J.L.D. share senior authorship.
References
- 1. Garg A. Clinical review#: lipodystrophies: genetic and acquired body fat disorders. J Clin Endocrinol Metab 2011;96:3313–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Araújo-Vilar D, Santini F. Diagnosis and treatment of lipodystrophy: a step-by-step approach. J Endocrinol Invest 2019;42:61–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Özen S, Akıncı B, Oral EA. Current diagnosis, treatment and clinical challenges in the management of lipodystrophy syndromes in children and young people. J Clin Res Pediatr Endocrinol 2020;12:17–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Misra A, Peethambaram A, Garg A. Clinical features and metabolic and autoimmune derangements in acquired partial lipodystrophy: report of 35 cases and review of the literature. Medicine (Baltimore) 2004;83:18–34 [DOI] [PubMed] [Google Scholar]
- 5. Pope E, Janson A, Khambalia A, Feldman B. Childhood acquired lipodystrophy: a retrospective study. J Am Acad Dermatol 2006;55:947–950 [DOI] [PubMed] [Google Scholar]
- 6. Brown RJ, Araujo-Vilar D, Cheung PT, et al. The diagnosis and management of lipodystrophy syndromes: a multi-society practice guideline. J Clin Endocrinol Metab 2016;101:4500–4511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sorkina E, Frolova E, Rusinova D, et al. Progressive generalized lipodystrophy as a manifestation of autoimmune polyglandular syndrome type 1. J Clin Endocrinol Metab 2016;101:1344–1347 [DOI] [PubMed] [Google Scholar]
- 8. Anderson MS, Su MA. AIRE expands: new roles in immune tolerance and beyond. Nat Rev Immunol 2016;16:247–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nagamine K, Peterson P, Scott HS, et al. Positional cloning of the APECED gene. Nat Genet 1997;17:393–398 [DOI] [PubMed] [Google Scholar]
- 10. Jehl A, Cugnet-Anceau C, Vigouroux C, et al. Acquired generalized lipodystrophy: a new cause of anti–PD-1 immune-related diabetes. Diabetes Care 2019;42:2008–2010 [DOI] [PubMed] [Google Scholar]
- 11. Anderson MS, Venanzi ES, Klein L, et al. Projection of an immunological self shadow within the thymus by the aire protein. Science 2002;298:1395–1401 [DOI] [PubMed] [Google Scholar]
- 12. O’Donovan B, Mandel-Brehm C, Vazquez SE, et al. High-resolution epitope mapping of anti-Hu and anti-Yo autoimmunity by programmable phage display. Brain Commun 2020;2:fcaa059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mandel-Brehm C, Dubey D, Kryzer TJ, et al. Kelch-like protein 11 antibodies in seminoma-associated paraneoplastic encephalitis. N Engl J Med 2019;381:47–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vazquez SE, Ferré EM, Scheel DW, et al. Identification of novel, clinically correlated autoantigens in the monogenic autoimmune syndrome APS1 by proteome-wide PhIP-Seq. eLife 2020;9:e55053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ferre EM, Rose SR, Rosenzweig SD, et al. Redefined clinical features and diagnostic criteria in autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy. JCI Insight 2016;1:e88782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Park JE, Botting RA, Domínguez Conde C, et al. A cell atlas of human thymic development defines T cell repertoire formation. Science 2020;367:eaay3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bautista JL, Cramer NT, Miller CN, et al. Single-cell transcriptional profiling of human thymic stroma uncovers novel cellular heterogeneity in the thymic medulla. Nat Commun 2021;12:1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. DeVoss JJ, Anderson MS. Lessons on immune tolerance from the monogenic disease APS1. Curr Opin Genet Dev 2007;17:193–200 [DOI] [PubMed] [Google Scholar]
- 19. Husebye ES, Anderson MS, Kämpe O. Autoimmune polyendocrine syndromes. N Engl J Med 2018;378:1132–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Deutscher S. Phage display to detect and identify autoantibodies in disease. N Engl J Med 2019;381:89–91 [DOI] [PubMed] [Google Scholar]
- 21. Sansom SN, Shikama-Dorn N, Zhanybekova S, et al. Population and single-cell genomics reveal the Aire dependency, relief from Polycomb silencing, and distribution of self-antigen expression in thymic epithelia. Genome Res 2014;24:1918–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kozusko K, Tsang V, Bottomley W, et al. Clinical and molecular characterization of a novel PLIN1 frameshift mutation identified in patients with familial partial lipodystrophy. Diabetes 2015;64:299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gandotra S, Le Dour C, Bottomley W, et al. Perilipin deficiency and autosomal dominant partial lipodystrophy. N Engl J Med 2011;364:740–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Falcao CK, Cabral MCS, Mota JM, et al. Acquired lipodystrophy associated with nivolumab in a patient with advanced renal cell carcinoma. J Clin Endocrinol Metab 2019;104:3245–3248 [DOI] [PubMed] [Google Scholar]
- 25. Haddad N, Vidal-Trecan T, Baroudjian B, et al.; PATIO group . Acquired generalized lipodystrophy under immune checkpoint inhibition. Br J Dermatol 2020;182:477–480 [DOI] [PubMed] [Google Scholar]
- 26. Misra A, Garg A. Clinical features and metabolic derangements in acquired generalized lipodystrophy: case reports and review of the literature. Medicine (Baltimore) 2003;82:129–146 [DOI] [PubMed] [Google Scholar]
- 27. Rose NR. Autoimmune diseases. In International Encyclopedia of Public Health. Oxford, U.K., Elsevier, 2008, pp. 267–271 [Google Scholar]
- 28. Brasaemle DL, Subramanian V, Garcia A, Marcinkiewicz A, Rothenberg A. Perilipin A and the control of triacylglycerol metabolism. Mol Cell Biochem 2009;326:15–21 [DOI] [PubMed] [Google Scholar]
- 29. Tansey JT, Sztalryd C, Gruia-Gray J, et al. Perilipin ablation results in a lean mouse with aberrant adipocyte lipolysis, enhanced leptin production, and resistance to diet-induced obesity. Proc Natl Acad Sci U S A 2001;98:6494–6499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hussain I, Garg A. Lipodystrophy syndromes. Endocrinol Metab Clin North Am 2016;45:783–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Corvillo F, Aparicio V, López-Lera A, et al. Autoantibodies against perilipin 1 as a cause of acquired generalized lipodystrophy. Front Immunol 2018;9:2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Postow MA, Sidlow R, Hellmann MD. Immune-related adverse events associated with immune checkpoint blockade. N Engl J Med 2018;378:158–168 [DOI] [PubMed] [Google Scholar]