Abstract
Acquired generalized lipodystrophy (AGL) is a rare condition characterized by massive loss of adipose tissue through the body, causing severe metabolic complications. Autoimmune destruction of adipocytes is strongly suspected based on the frequent association of AGL with autoimmune disorders. In 2018, autoantibodies against perilipin 1 (PLIN1) were identified in three patients with autoimmune-associated AGL. However, the pathogenic mechanism and clinical impact of anti-PLIN1 remain unsolved. The prevalence of anti-PLIN1 autoantibodies in an AGL cohort of 40 patients was 50% (20 of 40). Among positive patients, 10 had the autoimmune variety and 10 had panniculitis-associated AGL. The IgG isotype was predominant, although some IgM antibodies were detected. Epitope-mapping studies did not identify a single, major epitope. Instead, autoantibodies typically bound to several different peptides, among which the central (233–405) domain was detected in all antibody-positive patients, for both IgG and IgM autoantibodies. In-depth epitope mapping indicated that anti-PLIN1 autoantibodies predominantly recognize the αβ-hydrolase domain containing 5 (ABHD5) binding site (383–405). Autoantibodies dose-dependently blocked the binding of PLIN1 to ABHD5 and caused a dislocation of ABHD5 toward the cytosol, leading to an increase in lipolysis and lipase activities. Finally, anti-PLIN1 titers significantly correlated with the amount of fat loss, metabolic control impairment, and severity of liver injury. Our data strongly support that anti-PLIN1 autoantibodies are a diagnostic biomarker and a cause of lipodystrophy in patients with AGL.
Introduction
Acquired generalized lipodystrophy (AGL), also known as Lawrence syndrome (ORPHAcode 79086), is an ultra-rare disease characterized by the loss of adipose tissue in all or nearly all depots. Typically, generalized fat loss develops during childhood or adolescence. Patients with AGL may develop several health complications, particularly those associated with severe insulin resistance, including hypertriglyceridemia, diabetes, hepatic steatosis, acanthosis nigricans, menstrual irregularities, and polycystic ovary syndrome (1). These patients usually have markedly reduced levels of leptin and adiponectin resulting from low adipocyte mass (1,2). Approximately 25% of patients debut with an episode of panniculitis (type 1), and some of them have clinical evidence of autoimmunity (3). Another 25% of cases are associated with autoimmune diseases without panniculitis (type 2), such as juvenile dermatomyositis, Hashimoto thyroiditis, autoimmune hemolytic anemia, and Sjogren syndrome, among others. In the remaining patients, the underlying mechanism of fat loss is not clear (idiopathic variety or type 3) (3,4).
The diagnosis of AGL is based on medical history, body fat composition, presence of autoimmunity, and absence of family history of lipodystrophy. Although the associations with autoimmunity suggest that AGL has an autoimmune basis, the pathogenic mechanism resulting in the loss of adipose tissue remains unknown. Thus, the search for reliable clinical biomarkers is of key importance to improve diagnosis and treatment.
In 2018, our group reported the presence of a novel autoantibody in some patients with the autoimmune variety of AGL (5). This antibody was directed against perilipin 1 (PLIN1), which is required for the optimal regulation of the lipolytic pathway. Using a mouse model of preadipocytes to analyze lipolytic activity, we showed that these autoantibodies significantly increased basal lipolytic rates but did not modify stimulated lipolysis (5). PLIN1 frameshift variants have been described in patients affected by familial partial lipodystrophy type 4 (FPLD4) (6–8). Most of the FPLD4-associated PLIN1 variants disrupt the ability of PLIN1 to inhibit basal lipolysis in adipocytes because the C-terminal domain of PLIN1 (amino acids 380–427) fails to interact with αβ-hydrolase domain containing 5 (ABHD5), leading to the constitutive activation of adipocyte triglyceride lipase (6,7). Considering the similarities in the functional implications between autoantibodies against PLIN1 and PLIN1 pathogenic variants, the characterization of these autoantibodies in patients with AGL is essential.
In this study, we describe the prevalence, epitope mapping, and associated immunological features of anti-PLIN1 autoantibodies in a large cohort of patients with AGL. Additionally, functional studies were performed to analyze the blocking activity of the autoantibodies, demonstrating the pathogenic mechanism that leads to lipodystrophy development. Finally, a comparison of the clinical presentations in patients with and without anti-PLIN1 is presented, which demonstrates a significant correlation between autoantibody titers and clinical severity.
Research Design and Methods
Patients and Biological Samples
Forty patients (7 from Spain, 3 from Italy, and 30 from the U.S.) were diagnosed with AGL on the basis of fat loss during childhood or adulthood affecting large areas of the body and having ruled out other causes of weight loss. Congenital generalized lipodystrophy (CGL) was also excluded based on the natural course of the disease, clinical features, age at onset, exclusion of pathogenic variants in CGL-related genes (AGPAT2, BSCL2, CAV1, PTRF, and PPARG), and absence of familial history of lipodystrophy. The classification of patients with AGL into subtypes (autoimmune, panniculitis, and idiopathic) was performed on the basis of previously proposed criteria (3).
The study was conducted according to the Declaration of Helsinki and approved by the Clinical Research Ethics Committee of La Paz University Hospital (project code PI-3522) and the Institutional Review Board of the National Institute of Diabetes and Digestive and Kidney Diseases (ClinicalTrials.gov, NCT00001987). All participants provided written informed consent for participation in the study and the publication of their clinical and biochemical information.
Blood was drawn to obtain fresh serum and kept frozen at −80°C until further use. Additionally, sera samples were obtained from 27 patients with acquired partial lipodystrophy (APL) (85% females; mean age 33.7 years [range 11–76], and mean BMI 21.7 kg/m2 [16.3–33.8]), 50 healthy volunteers (80% females, mean age 29.9 years [6–66], and mean BMI 23.4 kg/m2 [17.6–26.5]), and 20 obese individuals (50% females, mean age 51.5 years [31–64], and mean BMI 45.9 kg/m2 [36.8–67.8]).
Recombinant Proteins and Synthetic Peptides
Recombinant full-length PLIN1 (1–522) and ABHD5 were obtained from OriGene (Rockville, MD). Central and C-terminal recombinant fragments of PLIN1 (233–405, code CSB-MP526447HU1, and 411–496, code CSB-MP526447HU2) were obtained from Cusabio (Houston, TX). NZYTech (Lisbon, Portugal) manufactured 12 overlapping peptides for sequence 233–405. All details about recombinant proteins and peptides can be found in Supplementary Tables 1 and 2.
Detection, Characterization, and Quantification of Anti-PLIN1 Autoantibodies by ELISA
Anti-PLIN1 IgG autoantibodies were detected by ELISA assay as previously described (5). Additionally, this method has been modified using different HRP-conjugated secondary antibodies to detect human IgM or IgA autoantibodies (at 1/5,000 dilution; both from Jackson ImmunoResearch Laboratories). Using this approach, anti-PLIN1 autoantibodies were quantified by ELISA using a reference patient serum (from patient AGL3) with a given arbitrary value of 1,000 arbitrary units (AU)/mL. A twofold dilution series of patients’ serum was added, and the results were interpolated to the standard curve of the positive control.
Epitope-mapping studies were performed by coating ELISA plates with 100 ng/well of each recombinant fragment or peptide and then detecting the binding of specific Ig isotype (IgG, IgM, or IgA) in the same conditions described by our group (5). Samples with absorbance two times SD greater than the mean of normal control subjects’ (n = 50) absorbance values were deemed positive.
IgG subclasses were determined by ELISA as described to detect anti-PLIN1 autoantibodies with some modifications (5). Briefly, Ig subclasses were detected using a specific mouse peroxidase-conjugated anti-human IgG1 at 1/500 (9054–05), anti-human IgG2 at 1/500 (9060–05), anti-human IgG3 at 1/250 (9210–05) (all from SouthernBiotech, Birmingham, AL), and anti-human IgG4 at 1/250 (A10654; Invitrogen, Madrid, Spain). Similarly, Ig light chains were detected using polyclonal antibodies, respectively, raised against the human κ and λ light chains (Helena Biosciences, Gateshead, U.K.) at 1/1,000, followed by their appropriate conjugated secondary antibody (Dako, Madrid, Spain).
Physical Interaction of PLIN1 and ABHD5 by ELISA
For the detection of the interaction between PLIN1 and ABHD5, ELISA plates (Costar Medium binding) were coated with 100 ng of recombinant human PLIN1 (OriGene) in carbonate/bicarbonate (pH 9.3) overnight at 4°C. The next day, wells were blocked with assay buffer (PBS containing 0.1% Tween 20 and 5% BSA) for 1 h at 37°C. Then, 100 ng protein/well of recombinant human ABHD5 in assay buffer was added and incubated 1 h at 37°C. After washing the plates five times with PBS-0.1% Tween 20, the complex was detected using a 1/1,000 dilution of a monoclonal anti-ABHD5 antibody (AM31371PU-N; provided by OriGene). After five further washes, horseradish peroxidase–conjugated secondary antibody (Jackson ImmunoResearch Laboratories) diluted in the same buffer (1/1,000) was added. The absorbance of ABTS was determined at 405 nm using 620 nm as a reference wavelength. To detect the interacting domain of PLIN1 with ABHD5, we performed an ELISA assay under the same conditions, but the plates were coated with 100 ng/well of each PLIN1 fragment.
Detection of Blocking Activity of Anti-PLIN1 Autoantibodies by ELISA
For blocking assays, ELISA plates (Costar Medium binding) were coated with 100 ng/well of recombinant ABHD5 (OriGene) in carbonate/bicarbonate (pH 9.3) overnight at 4°C. The plates were blocked with assay buffer (PBS supplemented with 0.1% Tween 20 and 5% BSA) for 1 h at 37°C and washed five times with PBS-0.1% Tween 20. On the same plate, a twofold dilution series (1/250, 1/500, 1/1,000, and 1/2,000) of patients’ serum was mixed with a fixed amount (100 ng) of recombinant PLIN1 (OriGene) and incubated for 1 h at 37°C. After washing the plates, bound PLIN1 was detected using a monoclonal rabbit anti-human PLIN1 (no. 9349; Cell Signaling Technology) at 1/1,000 dilution. After five more washes, 100 µL of a 1/2,000 dilution of goat anti-rabbit IgG antibody coupled with HRP (AP132P; Merck) was added, and the plates were kept at 37°C for 1 h. The enzymatic reaction was developed with ABTS (Merck) and measured at 405 nm using 620 nm as a reference wavelength. The absorbance determined was proportional to the amount of bound PLIN1, and it was the maximal from the well in the absence of serum. The reduction of the maximal signal (blocking capacity) by the anti-PLIN1 blocking antibodies was proportional to the blocking activity of the sample, calculated as a percentage: blocking capacity (%) = 100 × (1 − [A405 assay sample/A405 total PLIN1-ABHD5 binding]).
Measurement of Lipolytic and Lipase Activities
Basal and stimulated lipolysis were measured on differentiated mouse 3T3-L1 preadipocytes under the same conditions previously described (5), but using the human serum at 1:10 dilutions instead of purified IgG. For measurement of lipase activity, once preadipocytes were incubated with human serum, the cells were lysed using RIPA buffer, and lipase activity was analyzed using the Lipase Activity Assay kit (MAK046; Sigma-Aldrich) following the manufacturer’s instructions.
Immunocytochemistry
Differentiated mouse 3T3-L1 preadipocytes were incubated with human serum at 1:10 dilution during 180 min at 37°C, 5% CO2. Fixed (4% paraformaldehyde) and permeabilized cells were incubated overnight at 4°C with an FITC-conjugated polyclonal rabbit F(ab)2 anti-human IgG (1:250; Dako) and a rabbit anti-human PLIN1 antibody (no. 3470; from Cell Signaling Technology) or a rabbit ABHD5 polyclonal antibody (no. PA5–78704; Thermo Fisher Scientific), both at 1:100. The reaction was developed using a convenient biotin-conjugated secondary antibody (1:1,000; Dako) followed by avidin-Texas Red–conjugated (1:1,000; A-2006; Vector Laboratories). Cells were then mounted with DAPI-containing Vectashield (Vector Laboratories). Sections were examined using a Leica TCS SPE spectral confocal microscope (Leica Microsystems, Heidelberg, Germany).
Statistical Analysis
Statistical analyses were conducted using Prism 8 (GraphPad Software, La Jolla, CA). The results are shown as either mean and SD (or median and interquartile range [IQR]) for continuous variables or absolute numbers and percentage for categorical variables. Frequency data were compared using the χ2 test (or Fisher exact tests, where appropriate). Differences between groups were assessed using the Mann-Whitney test for nonparametric data, while for parametric data, unpaired Student t tests were applied. Pearson or Spearman correlation coefficients were performed to test associations. The Bonferroni multiple-comparison test was used for comparisons of all of the groups for lipolysis and lipase activity assays. Statistical significance was considered when P < 0.05.
Data and Resource Availability
All data generated or analyzed during this study are included in the published article (and its supplementary files online). The mouse lines analyzed during the current study are available from the corresponding author upon reasonable request.
Results
Cohort Description
Physical, anthropometric, and clinical features of the patients’ cohort are summarized in Table 1. Some patients were treated with recombinant human leptin (metreleptin; n = 23) at the time of data and sample collection. Twenty-two patients (55%) were classified as having the autoimmune variety of AGL and 15 patients (37.5%) as the panniculitis variety, of whom 10 had both panniculitis and autoimmune disease or autoantibody positivity and 5 had panniculitis without autoimmunity. The remaining three patients (7.5%) had the idiopathic variety. Females were affected three times more often than males. Most patients developed lipodystrophy in childhood and adolescence and rarely after 30 years of age. The extent of fat loss was substantial in all patients. The prevalence of metabolic derangements (acanthosis nigricans, hyperglycemia, diabetes, hyperinsulinemia, hypertriglyceridemia, steatohepatitis, hypercholesterolemia, and low levels of HDL cholesterol) was higher at first clinical evaluation. Circulating leptin levels at first clinical visit were lower than normal in most of the patients, and more than half of the cohort required treatment with recombinant leptin. Mild to moderate elevation of serum transaminases (AST and ALT) was frequently seen. There were no abnormalities in Ig levels. Some patients showed a reduction of complement C4 levels, while C3 was found within normal range.
Table 1.
Clinical and demographical characteristics of patients with AGL included in the study
| All patients with AGL (n = 40) | Reference range | |
|---|---|---|
| AGL variety, n (%) | — | |
| Autoimmune | 22 (55) | |
| Panniculitis | 5 (12.5) | — |
| Panniculitis plus autoimmune | 10 (25) | — |
| Idiopathic | 3 (7.5) | — |
| Female, n (%) | 30 (75) | — |
| Age (years), mean (SD) | 28.5 (18.2) | — |
| Lipodystrophy onset (years), median (IQR) | 6 (3–13.5) | — |
| Time with lipodystrophy (years), median (IQR) | 11.5 (6–17.2) | — |
| BMI (kg/m2), mean (SD) | 19.6 (3.5) | 18–25 |
| Percent total fat, mean (SD) | 11.6 (5.3) | 27.2–33.3 |
| Acanthosis, n (%) | 16 (40) | — |
| Glucose (mg/dL), median (IQR) | 106 (85.9–148.3) | 74–106 |
| Insulin (μU/mL), median (IQR) | 43.2 (13.3–95.5) | 3–25 |
| Diabetes, n (%) | 28 (70) | — |
| HbA1c (mmol/mol), median (IQR) | 42 (36–63) | 20–42 |
| HOMA-IR, median (IQR) | 12.4 (2.9–32.2) | 0–2.6 |
| Triglycerides (mg/dL), median (IQR) | 163.5 (122–352) | <150 |
| Total cholesterol (mg/dL), median (IQR) | 148 (128–193) | <200 |
| HDL (mg/dL), median (IQR) | 28 (22–33.5) | >40 |
| Leptin at baseline (ng/mL), median (IQR) | 1.3 (0.5–2.7) | <12 ng/mL in women, <8 ng/mL in men |
| Metreleptin treatment, n (%) | 23 (57.5) | — |
| Leptin after metreleptin treatment (ng/mL), median (IQR) | 9.6 (1.9–69.2) | <12 ng/mL in women, <8 ng/mL in men |
| Fatty liver, n (%) | 31 (77.5) | — |
| ALT (IU/L), median (IQR) | 39.5 (25.3–86.5) | <35 |
| AST (IU/L), median (IQR) | 30.5 (21–53.3) | <40 |
| IgG (mg/dL), median (IQR) | 1,077 (948.5–1,333) | 725–1,900 |
| IgA (mg/dL), median (IQR) | 151.5 (109–232.5) | 50–340 |
| IgM (mg/dL), median (IQR) | 89.3 (52.5–133.5) | 45–280 |
| C3 (mg/dL), median (IQR) | 125.4 (109.2–139) | 75–135 |
| C4 (mg/dL), median (IQR) | 17.4 (12.6–23.9) | 14–60 |
| Autoimmunity*, n (%) | 32 (80) | — |
| Immunosuppressant treatment, n (%) | 10 (25) | — |
Data are reported as mean ± SD, median (IQR), or absolute number (percentage).
HOMA-IR, HOMA of insulin resistance.
The presence of autoantibody positivity and/or autoimmune disease.
Eighty percent of patients with AGL from our cohort had evidence of autoimmune disease and/or autoantibody positivity. Nine of 40 patients (22.5%) had type 1 diabetes with anti-GAD65 and islet cell autoantibodies. Twelve of 40 patients (30%) had Hashimoto thyroiditis with detectable autoantibodies against thyroid peroxidase and/or thyroglobulin. Six of 40 patients (15%) had autoimmune hepatitis (AIH), and one of them was positive for antinuclear antibodies and anti–smooth muscle. Two of 40 patients (5%) had celiac disease, associated with IgA deficiency in 1 of them; 4 of 40 (10%) had juvenile dermatomyositis; 2 of 40 (5%) had autoimmune hemolytic anemia; 2 (5%) had vitiligo; and 2 (5%) had autoimmune thrombocytopenia. Other autoimmune diseases seen in single patients in the cohort were: arthritis, Addison disease, membranoproliferative glomerulonephritis type 1, immune complex–mediated glomerulonephritis, idiopathic neutropenia, systemic lupus erythematosus, autoimmune pancytopenia, and chronic inflammatory demyelinating polyradiculoneuropathy. Autoantibody positivity without clinical evidence of a related autoimmune disorder was reported in many patients, including antinuclear antibodies, double-stranded DNA antibody, PM/Scl100 antibody, anti-RNP, anti–parietal cell, anti-testis, and 21-hydroxylase autoantibodies, and Coombs positivity.
Other immunological features observed in our cohort were peripheral T-cell lymphoma (3 of 40; 7.5%), psoriasis (3 of 40; 7.5%), immunodeficiency secondary to LRBA deficiency (1 of 40; 2.5%), and common variable immunodeficiency (1 of 40; 2.5%). In addition, 1 patient of 40 (2.5%) developed generalized lipodystrophy as an immune-related adverse event (irAE) after pembrolizumab treatment for cutaneous melanoma.
Occurrence of Anti-PLIN1 Autoantibodies
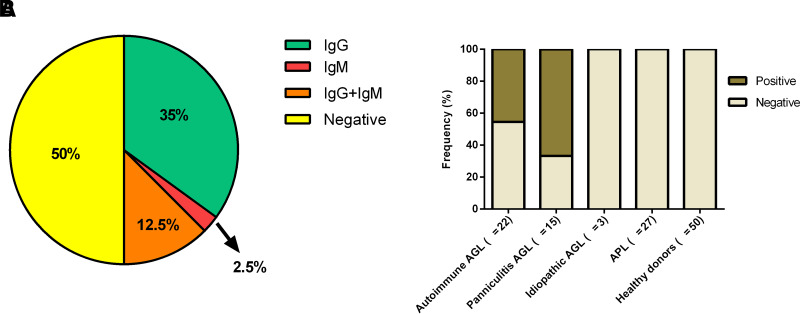
The prevalence of anti-PLIN1 antibodies in patients with AGL was 50% (20 of 40) (Fig. 1A). Among the anti-PLIN1–positive patients, 14 (35%) were positive for IgG, 5 (12.5%) were copositive for IgG and IgM, 1 (2.5%) was positive for IgM only, and no IgA was found.
Figure 1.
Frequency of anti-PLIN1 autoantibodies. A: Distribution of patients AGL positive for isolated IgG or IgM autoantibodies, copositive for IgG and IgM, and negative for anti-PLIN1 antibodies. B: Frequency of anti-PLIN1 in patients with different AGL types (autoimmune, panniculitis, and idiopathic) or APL and healthy donors.
Anti-PLIN1 antibodies were detected in 10 of 22 (45.5%) patients with the autoimmune variety, and 10 of 15 (66.7%) with the panniculitis variety (Fig. 1B). Among the PLIN1 positive panniculitis-associated patients, 7 of 10 (70%) also presented with autoimmune disease or autoantibody positivity. Anti-PLIN1 autoantibodies were not found in idiopathic AGL, healthy control subjects, obese individuals, and patients with APL (Fig. 1B).
Characterization of Autoantibodies Against PLIN1
In our AGL cohort, the predominant subclass was IgG1 (15 of 19; 78.9%), followed by IgG3 (12 of 19; 63.2%), IgG2 (5 of 19; 26.3%), and IgG4 (1 of 19; 5.2%). Four of 19 patients (21%) were copositive for IgG1 and IgG3; another 4 of 19 patients (21%) were copositive for IgG1, IgG2, and IgG3; 1 patient (1 of 19; 5.2%) was copositive for IgG1 and IgG2; and finally, 1 patient (5.2%) was copositive for IgG1 and IgG4 (Table 2). Concerning Ig light chains, 5 of 20 patients (25%) were positive for κ, 15 of 20 (75%) for both κ and λ, and none for λ only (Table 2). We observed a wide range of anti-PLIN1 titers, particularly in patients with IgG-type antibodies (Table 2).
Table 2.
Immunological features of autoantibodies against PLIN1
| Code | AGL variety | Autoantibody isotype | IgG autoantibody subclass | Autoantibody light chains | IgG autoantibody titers (AU/mL) | IgM autoantibody titers (AU/mL) |
|---|---|---|---|---|---|---|
| AGL1 | Autoimmune | IgG, IgM | IgG1, IgG2, IgG3 | κ | 323.2 | 1,513.4 |
| AGL2 | Autoimmune | IgG | IgG1 | κ | 110.7 | — |
| AGL3 | Autoimmune | IgG, IgM | IgG1 | κ+λ | 1,000* | 1,000* |
| AGL9 | Autoimmune | IgG | IgG3 | κ+λ | 7,998.2 | — |
| AGL10 | Panniculitis plus autoimmune | IgG | IgG3 | κ+λ | 102 | — |
| AGL11 | Panniculitis | IgG | IgG1, IgG2, IgG3 | κ | 9,763.6 | — |
| AGL13 | Panniculitis plus autoimmune | IgG | IgG1, IgG4 | κ+λ | 523.1 | — |
| AGL14 | Panniculitis | IgG | IgG3 | κ+λ | 72.1 | |
| AGL16 | Panniculitis plus autoimmune | IgG | IgG1, IgG2, IgG3 | κ+λ | 348.2 | — |
| AGL18 | Autoimmune | IgG | IgG1, IgG2, IgG3 | κ+λ | 1,026.4 | — |
| AGL20 | Autoimmune | IgG | IgG1, IgG2 | κ+λ | 1,080.3 | |
| AGL21 | Autoimmune | IgM | — | κ+λ | — | 1,386.5 |
| AGL23 | Panniculitis plus autoimmune | IgG | IgG1, IgG3 | κ+λ | 1,047.1 | — |
| AGL24 | Autoimmune | IgG | IgG1 | κ+λ | 1,356.1 | |
| AGL28 | Autoimmune | IgG | IgG3 | κ+λ | 421.4 | |
| AGL32 | Panniculitis | IgG, IgM | IgG1, IgG3 | κ+λ | 4,039.1 | 4,599.7 |
| AGL33 | Panniculitis plus autoimmune | IgG | IgG1, IgG3 | κ+λ | 33,640 | — |
| AGL37 | Panniculitis plus autoimmune | IgG, IgM | IgG1, IgG3 | κ | 909.8 | 4,929.7 |
| AGL38 | Autoimmune | IgG | IgG1 | κ | 4,842.7 | — |
| AGL39 | Panniculitis plus autoimmune | IgG, IgM | IgG1 | κ+λ | 198,200 | 3,899.7 |
A reference patient serum (AGL3) was used to calibrate the assay and to calculate anti-PLIN1 autoantibody titers in AU per milliliter.
Identification of the Epitope Recognized by Anti-PLIN1 Autoantibodies
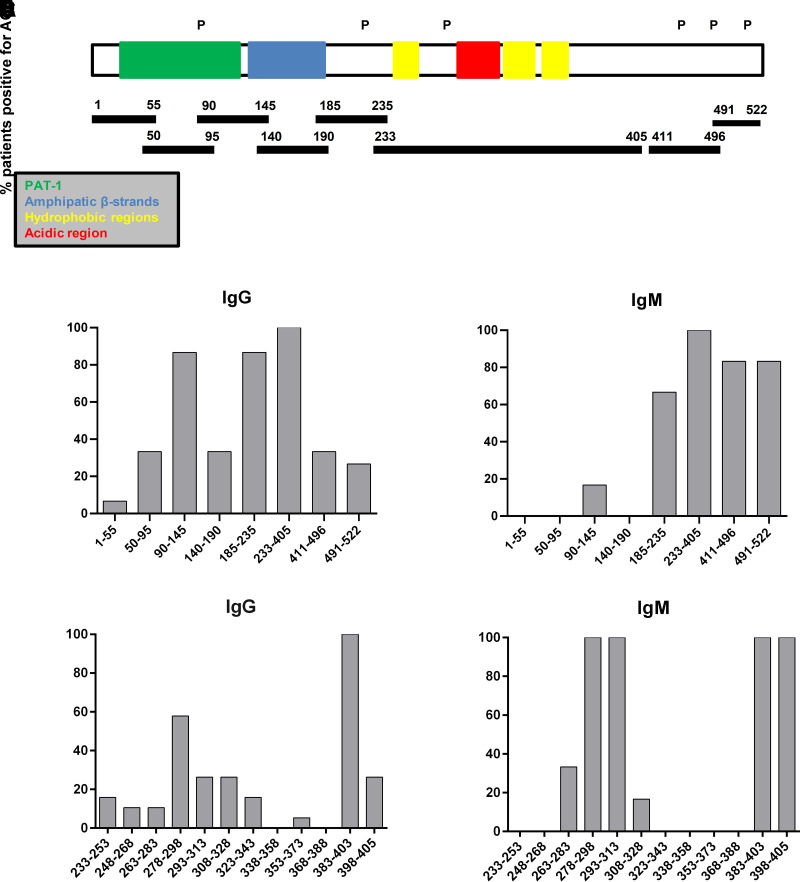
In the first set of experiments, epitope mapping was evaluated using eight PLIN1 overlapping fragments spanning the full PLIN1 protein sequence (Fig. 2A and Supplementary Table 1) to test for their suitability as target antigens in ELISA assays. Concerning IgG reactivity, autoantibodies recognized the fragment 233–405 in 19 of 19 (100%) patients. Additionally, 16 (84.2%) patients had reactivity against fragments 90–145 and 185–235; 6 (31.6%) against fragment 50–95; 5 (26.3%) against 140–190; 7 (36.8%) against fragment 411–496; 4 (21.1%) against the C-terminal domain 491–522; and 3 (15.8%) against the N-terminal domain 1–55 (Fig. 2B). IgM autoantibodies exhibited reactivity against fragment 233–405; 3 (50%) against fragment 411–496; 4 (66.7%) against fragments 491–522; 2 (33.3%) against fragment 185–235; and 1 (16.6%) against fragment 90–145. None of the patients analyzed showed IgM reactivity against fragments 1–55, 50–95, or 140–190 (Fig. 2C). Individual absorbance values for each patient and each fragment are summarized in Supplementary Figs. 1 and 2.
Figure 2.
Epitope-mapping studies. A: Schematic representation of PLIN1 domain distribution and location of PLIN1 fragments used for epitope mapping. The ∼100-amino-acid hydrophobic PAT domain is located at the extreme N terminus, followed by amphipathic β-strands. The C terminus domain is composed of three hydrophobic sequences and one acidic region comprised of amino acids 291–318. The positions of the phosphorylation sites are indicated by a “P.” The reactivity of IgG (B) or IgM (C) autoantibodies from 20 patients with AGL was tested with recombinant PLIN1 fragments, covering almost all domains of PLIN1. IgG (D) and IgM (E) anti-PLIN1 reactivity against 12 overlapping peptides spanning amino acids 233–405. Results are expressed as the percentage of total positive patients for each PLIN1 peptide.
Because anti-PLIN1 antibodies from all patients recognized the fragment 233–405 with both IgG and IgM isotype autoantibodies, we finely mapped the main epitopes present in this region by using 12 overlapping peptides spanning the 233–405 fragment (see peptide details in Supplementary Table 2). Using this approach, we found that 100% of the patients with IgG and IgM autoantibodies shared immunoreactivity against the 383–403 peptide (Fig. 2D and E).
Within the same 233–405 sequence, a second region spanning amino acids 278–298 showed substantial binding of IgG autoantibodies (11 of 19 patients; 57.9%) (Fig. 2D). Interestingly, in addition to the 383–403 fragment, 6 of 6 (100%) patients with IgM antibodies also recognized the peptides 278–298, 293–313, and 398–405 (Fig. 2E). Peptide-specific immunoreactivity data for each patient can be found in Supplementary Figs. 3 and 4.
Anti-PLIN1 Autoantibodies Block ABHD5 Binding to PLIN1
The C-terminal domain of PLIN1 is essential for maintaining basal lipolysis homeostasis. Through amino acids 380–427 in this domain, PLIN1 interacts with ABHD5 to inhibit basal lipolysis, and indeed, our results showed that this is the foremost region recognized by anti-PLIN1 autoantibodies (Fig. 2). Using two different online software approaches to predict linear antigenic epitopes (EMBOSS at https://www.bioinformatics.nl/cgi-bin/emboss/antigenic; and SVMTriP at http://sysbio.unl.edu/SVMTriP/prediction.php) in the PLIN1 sequence, we confirmed that the epitope spanning between amino acids 390–410 showed the highest antigenicity. This prompted us to analyze whether anti-PLIN1 autoantibodies could disturb the interaction between PLIN1 and ABHD5. For that, we performed an ELISA assay with either full-length PLIN1 or the recombinant fragments confirming that ABHD5 interacted with full-length PLIN1, and the binding site for this interaction was predominantly located in the region 233–405 (Supplementary Fig. 5A). Using 12 overlapping peptides spanning the 233–405 fragment, we were able to restrict the binding site of ABHD5 to amino acids 383–405 in the PLIN1 C-terminal domain (Supplementary Fig. 5B).
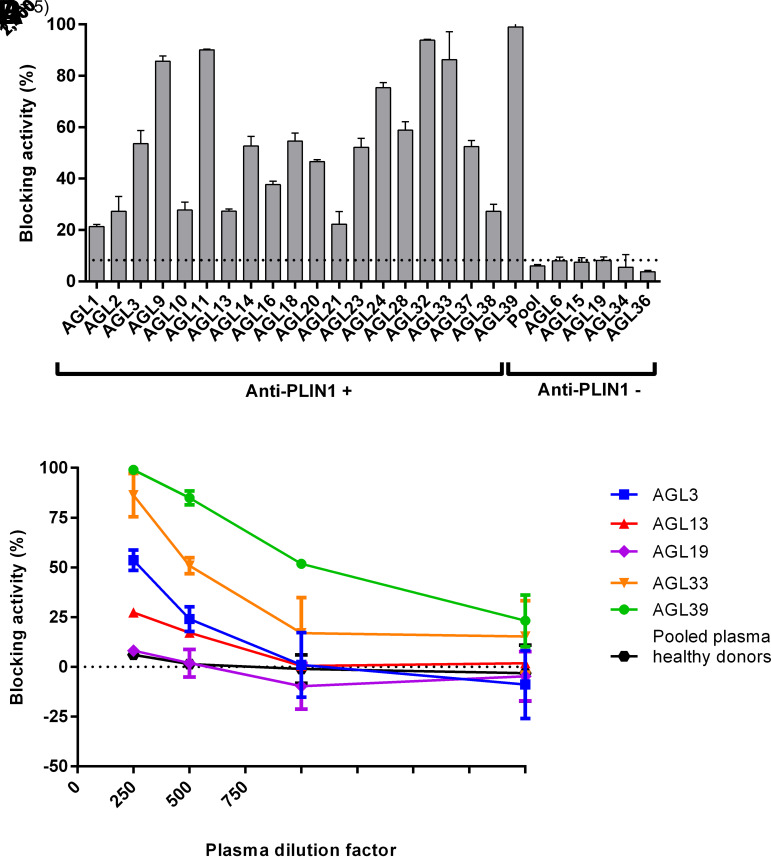
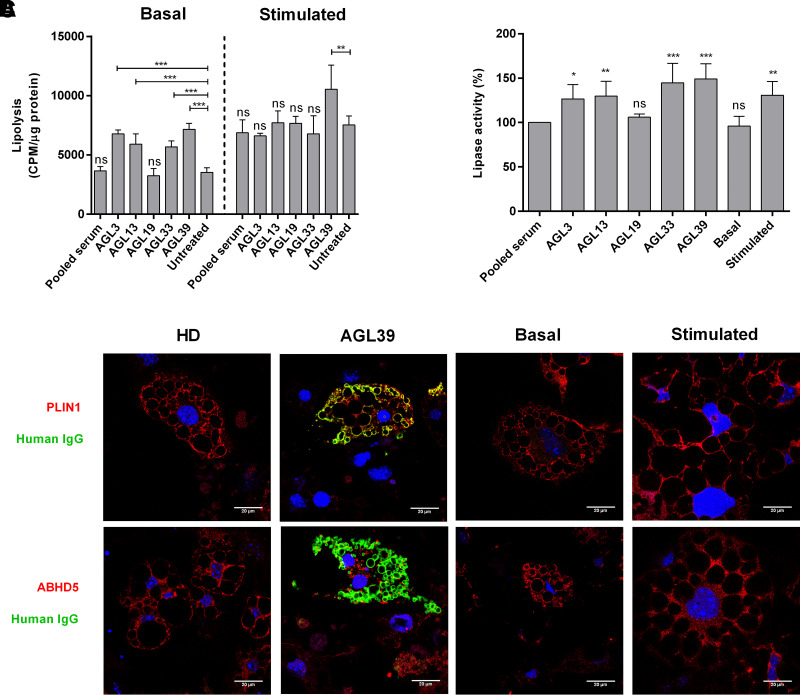
We then evaluated the ability of anti-PLIN1–positive sera to interfere with this interaction in competition ELISA assays and observed that the presence of anti-PLIN1 antibodies significantly and dose-dependently abolished ABHD5 binding as compared with antibody-negative patients with AGL and healthy donors (Fig. 3). Furthermore, anti-PLIN1 blocking activity had a strong association with IgG autoantibody titers (r = 0.61; P = 0.005), but not with IgM autoantibody titers (P = 0.66). To analyze the blocking activity observed in vitro, we performed assays in mouse 3T3-L1 preadipocytes. First, we determined the lipolysis activity (basal and stimulated) in the presence of serum from healthy donors or patients with anti-PLIN1 autoantibodies. Using this approach, we confirmed that anti-PLIN1 increased basal lipolysis (Fig. 4A) in accordance with previous results from our group (5). Interestingly, supplementation with serum from AGL39 (the patient with the highest anti-PLIN1 titer) not only affected the rates of basal lipolysis, but also produced a boost in stimulated lipolysis (Fig. 4A). Using the same approach, we found a parallel increase in lipase activity in the presence of anti-PLIN1 autoantibodies as compared with those cells untreated or treated with serum from healthy donors (Fig. 4B). Finally, we performed imaging studies to confirm subcellular localization of anti-PLIN1 and binding overlapping with ABHD5. Untreated cells or those supplemented with serum from healthy donors demonstrated intense labeling of ABHD5 and PLIN1 around the lipid droplet (Fig. 4C), while under stimulated conditions, ABHD5 was mainly located in the cytosol and PLIN1 remained on the surface of the lipid droplets (Fig. 4C). However, in the presence of serum from a patient with anti-PLIN1 autoantibodies, we observed colocalization of human IgG with PLIN1, but not with ABHD5, indicating that autoantibodies block the binding site of ABHD5 in PLIN1, thus displacing ABHD5 toward the cytosol (Fig. 4C).
Figure 3.
Blocking activity of anti-PLIN1 autoantibodies. A: Detection of blocking activity of anti-PLIN1 autoantibodies using serum samples at 1/250 dilution incubated with a fixed amount (100 ng) of recombinant PLIN1. The unspecific blocking activity (8.31%) was established using the mean value obtained for 50 healthy donors. B: Dose-dependent inhibition of the interaction of PLIN1 and ABHD5 in the presence of sera samples from anti-PLIN1–positive patients (AGL3, AGL13, AGL33, and AGL39). Serum from AGL19 and pooled serum from five healthy donors were negative for anti-PLIN1 autoantibodies. Serially diluted serum samples were mixed with a fixed amount (100 ng) of recombinant PLIN1. For both images, results were expressed as mean ± SD from two independent experiments.
Figure 4.
Functional effects of anti-PLIN1 autoantibodies on lipolysis and lipase activity. Preadipocytes (3T3-L1) were incubated for 180 min with human serum at 1/10 dilution from four patients with AGL (AGL3, AGL13, AGL33, and AGL39) with anti-PLIN1 autoantibodies, one patient without antibodies (AGL19), and a pool of healthy donors. A: Results of the radiometric assessment of basal and stimulated lipolysis. Data represent mean ± SD for triplicates of two independent experiments for each sample. Statistical significance was assessed with one-way ANOVA assay comparing the mean of each column with the mean from untreated cells. B: Results of lipase activity (nanomoles of glycerol), represented as the percentage over maximum activity from cells treated with healthy donor serum. Data represent mean ± SD for triplicates of two independent experiments for each sample. C: Confocal microscopic analysis of mouse preadipocytes revealed colocalization of PLIN1 and IgG from patient AGL39 on the lipid droplet surface (merged image, in yellow). However, no colocalization was observed when using serum from healthy donors (HD). ABHD5 was localized mainly on lipid droplets under treatment with serum from HD or untreated (basal conditions). Under stimulated conditions or in the presence of serum from AGL39, ABHD5 staining was localized on cytosol. DNA was stained with DAPI (blue); IgG binding was detected using FITC-conjugated rabbit anti-human IgG (green); and PLIN1 or ABHD5 was detected with biotin-labeled rabbit IgG followed by Texas Red–labeled streptavidin (red). Scale bars, 20 μm. *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant.
Clinical Significance of Anti-PLIN1 Autoantibodies
Patients positive for anti-PLIN1 autoantibodies were significantly younger than those without autoantibodies (P = 0.015) and had lower percent body fat (P = 0.037) (Table 3). Although metabolic alterations were common in both groups, only total cholesterol was significantly higher in patients without anti-PLIN1 autoantibodies (P = 0.013), while serum transaminases (AST and ALT) were higher in patients with anti-PLIN1 autoantibodies (P = 0.017 for AST; P = 0.006 for ALT) despite similar prevalence of fatty liver disease (Table 3). Finally, C4 complement levels were lower in patients with anti-PLIN1 compared with patients negative for this autoantibody (P = 0.011 for C4), suggestive of a mild to moderate activation of the classical pathway of the complement system (Table 3).
Table 3.
Comparative analysis between patients with and without anti-PLIN1 autoantibodies
| Anti-PLIN1–positive (n = 20) | Anti-PLIN1–negative (n = 20) | P value | |
|---|---|---|---|
| Female, n (%) | 14 (70) | 16 (80) | 0.72 |
| Age (years), mean (SD) | 17.8 (13.1) | 32.5 (20.3) | 0.015 |
| Lipodystrophy onset (years), median (IQR) | 5 (3–7) | 7 (3.5–28.5) | 0.11 |
| Time with lipodystrophy (years), median (IQR) | 11.9 (6.5–16.7) | 10.7 (6–32.1) | 0.64 |
| BMI (kg/m2), mean (SD) | 19.3 (3.4) | 19.9 (3.6) | 0.59 |
| Percent total fat, mean (SD) | 9.9 (3.8) | 13.7 (6.2) | 0.037 |
| Acanthosis, n (%) | 10 (50) | 6 (30) | 0.33 |
| Glucose (mg/dL), median (IQR) | 106 (84.4–140.4) | 107.5 (87.1–192.4) | 0.57 |
| Insulin (μU/mL), median (IQR) | 28.5 (13.2–95.8) | 49.5 (11.8–99.9) | 0.98 |
| Diabetes, n (%) | 15 (75) | 13 (65) | 0.73 |
| HbA1c (mmol/mol), median (IQR) | 42 (35–56) | 48 (36–64.3) | 0.48 |
| HOMA-IR, median (IQR) | 8.8 (3–27.1) | 13.7 (2.6–40.1) | 0.79 |
| Leptin at baseline (ng/mL), median (IQR) | 1.3 (0.45–2) | 1.2 (0.43–3.8) | 0.47 |
| Metreleptin treatment, n (%) | 14 (70) | 9 (45) | 0.20 |
| Leptin after metreleptin treatment (ng/mL), median (IQR) | 11.6 (3.5–82.5) | 5.9 (0.5–62.9) | 0.39 |
| Triglycerides (mg/dL), median (IQR) | 156.5 (112.8–379.8) | 191.5 (132–312.3) | 0.68 |
| Total Cholesterol (mg/dL), median (IQR) | 145 (118.5–165.5) | 177 (142–210.3) | 0.013 |
| HDL (mg/dL), median (IQR) | 25 (22–29.5) | 30 (22–47) | 0.09 |
| Fatty liver, n (%) | 17 (85) | 14 (70) | 0.45 |
| ALT (IU/L), median (IQR) | 64.4 (30–121.3) | 31 (18.5–49.8) | 0.017 |
| AST (IU/L), median (IQR) | 43.5 (26.3–80.8) | 24.5 (19.3–36) | 0.006 |
| Autoimmunity, n (%) | 16 (80) | 13 (65) | 0.48 |
| Immunosuppressant treatment, n (%) | 4 (20) | 6 (30) | 0.72 |
| IgG (mg/dL), median (IQR) | 1,180 (980–1,510) | 1,013 (907–1,180) | 0.09 |
| IgA (mg/dL), median (IQR) | 139 (97–245) | 192 (122–230) | 0.73 |
| IgM (mg/dL), median (IQR) | 92.2 (60–135) | 86.4 (46–133) | 0.61 |
| C3 (mg/dL), median (IQR) | 111.5 (98.8–136.1) | 133.5 (119.5–142.9) | 0.06 |
| C4 (mg/dL), median (IQR) | 14.2 (10.2–17.8) | 19.9 (16.7–26.6) | 0.011 |
Normally distributed data are reported as mean ± SD and compared with a t test; asymmetrically distributed data are reported as median (IQR) and compared with the Mann–Whitney U test. Categorical variables are reported as absolute number (percentage) and compared with Fisher exact test. P values <0.05 were considered statistically significant and are indicated in italics.
Correlation studies were performed to assess the associations of autoantibody titers with physical, demographical, and laboratory parameters (Table 4). A higher concentration of IgG anti-PLIN1 autoantibodies was associated with worse metabolic control, including higher glucose (r = 0.57; P = 0.011), insulin (r = 0.73; P = 0.002), and triglycerides (r = 0.51; P = 0.035), lower HDL-cholesterol (r = −0.50; P = 0.046), higher AST (r = 0.59; P = 0.011), and a trend toward higher ALT (P = 0.08). Elevated IgG autoantibody titers were also associated with higher circulating total IgM levels (r = 0.58; P = 0.029). There were no correlations of IgM autoantibodies titers with any physical, demographical, or laboratory parameter studied.
Table 4.
Correlation of IgG and IgM anti-PLIN1 autoantibody titers with physical, demographical, and laboratory parameters
| IgG autoantibodies | IgM autoantibodies | |||
|---|---|---|---|---|
| r | P value | r | P value | |
| Age (years) | −0.11 | 0.66 | −0.76 | 0.08 |
| Lipodystrophy onset (years) | 0.33 | 0.20 | 0.24 | 0.70 |
| Time with lipodystrophy (years) | −0.16 | 0.52 | −0.47 | 0.34 |
| BMI (kg/m2) | 0.05 | 0.84 | −0.50 | 0.30 |
| Percent total fat | −0.29 | 0.27 | −0.71 | 0.18 |
| Glucose (mg/dL) | 0.57 | 0.011 | 0.37 | 0.47 |
| Insulin (μU/mL) | 0.73 | 0.002 | −0.22 | 0.72 |
| HbA1c (mmol/mol) | 0.17 | 0.48 | 0.23 | 0.67 |
| HOMA-IR | −0.19 | 0.49 | 0.09 | 0.90 |
| Leptin (ng/mL) | 0.45 | 0.11 | 0.48 | 0.52 |
| Triglycerides (mg/dL) | 0.51 | 0.035 | 0.01 | 0.98 |
| Total cholesterol (mg/dL) | −0.17 | 0.52 | −0.70 | 0.19 |
| HDL-cholesterol (mg/dL) | −0.50 | 0.046 | 0.20 | 0.74 |
| ALT (IU/L) | 0.42 | 0.08 | 0.34 | 0.51 |
| AST (IU/L) | 0.59 | 0.011 | −0.15 | 0.80 |
| IgG (mg/dL) | −0.20 | 0.48 | −0.86 | 0.14 |
| IgA (mg/dL) | 0.06 | 0.84 | 0.08 | 0.92 |
| IgM (mg/dL) | 0.58 | 0.029 | 0.21 | 0.80 |
| C3 (mg/dL) | −0.28 | 0.26 | 0.60 | 0.20 |
| C4 (mg/dL) | −0.40 | 0.11 | −0.40 | 0.52 |
Association analyses were performed using Pearson or Spearman test. P value <0.05 was considered statistically significant and are indicated in italics.
Discussion
The rarity of generalized lipodystrophy, combined with the wide heterogeneity of the phenotype and genotype, complicates and delays diagnosis and treatment of the associated complications (9). According to the most recent consensus published, the diagnosis of generalized lipodystrophy relies on medical history, clinical features, body fat composition, and metabolic status. While the presence of a pathogenic variant in a known CGL gene provides confirmation of the diagnosis, the absence of a known pathogenic variant does not rule out a genetic form of lipodystrophy, and, at times, it may be challenging to distinguish between acquired and genetic forms of lipodystrophy (1,9,10). Thus, the presence of specific biomarkers in AGL would be helpful for a reliable and early diagnosis and may reduce the need for unnecessary genetic testing.
The etiopathogenesis of AGL remains unresolved, but based on its frequent association with autoimmune disorders, autoimmune destruction of adipocytes is strongly suspected (11–15). In 2018, our group reported the finding of autoantibodies against PLIN1 in three patients with the autoimmune variety of AGL and showed that these antibodies cause dysregulation of basal lipolysis (5). Recently, anti-PLIN1 autoantibodies have been linked to metabolic abnormalities or fat loss in two distinct situations characterized by loss of immune tolerance, such as autoimmune polyglandular syndrome type 1, and as an irAE following cancer immunotherapy with nivolumab (16). Interestingly, the patient undergoing nivolumab developed both anti-PLIN1 autoantibodies and AGL features, both of which disappeared upon the discontinuation of immunotherapy (16). All of these findings support that anti-PLIN1 autoantibodies could be a cause of generalized lipodystrophy.
Anti-PLIN1 autoantibodies were detected in 50% of patients from a large AGL cohort (Fig. 1). Several reasons may explain their absence in a proportion of AGL patients. First, despite our Western blot screening failing to detect further consistent candidates, additional autoantigens may exist other than PLIN1. Second, experimental handling and processing of samples might conceal some additional epitopes (on either PLIN1 or elsewhere). Third, the idiopathic variety of the disease (characterized in our cohort by the absence of anti-PLIN1) could hypothetically represent either early stages of disease progression in which autoimmunity has not yet manifested or a genetic-driven variety due to as-yet-unidentified genetic causes. In any event, autoantibodies are undetectable in a remarkable number of patients despite the strong association between lipodystrophy and autoimmunity, and novel experimental approaches will be necessary to identify additional adipose tissue-specific antigens.
Our present results raise the fundamental question of the origin and significance of anti-PLIN1 autoantibodies. More specifically, how did antibodies against an intracellular antigen such as PLIN1 originate, and which is the mechanistic rationale for their biological effects? A possible explanation would be that these autoantibodies arise in response to the extracellular exposure of the antigen in situations of uncontrolled cell death. We tested this hypothesis by analyzing the presence of anti-PLIN1 in a cohort of obese individuals in whom extensive adipocyte death is well documented (17), but the autoantibodies were not detected. However, a recent study has showed reduced Plin1 expression in the thymus of Aire−/− mice with anti-PLIN1 autoantibodies, which supports that Plin1 is expressed in the thymus in an Aire-dependent manner (16). This result suggests that loss of T-cell tolerance could be a possible mechanism contributing to the autoimmune response to PLIN1, as proposed in patients with autoimmune polyglandular syndrome type 1 and those under treatment with checkpoint inhibitors (16). In addition, a recent report provided the novel association of CTLA-4 haploinsufficiency and the simultaneous presence of AGL (18). Therefore, although we cannot rule out the contribution of cell death–derived extracellular exposure of PLIN1 to the emergence of these autoantibodies, we consider that there may be a genetic predisposition to develop autoimmunity against PLIN1 in patients with AGL.
In contrast, an objection may be raised regarding the epitope-mapping ELISAs presented in this study. Considering that PLIN1 is an intracellular, mostly transmembrane protein, it can be argued that the conformation when attached to a plate is very likely to be different to that of its true in vivo structure. With our results in hand though, we interpret that at least some major epitopes are indeed conserved; this is primarily supported by the fact that mutations at the identified region in the C-terminal end of PLIN1 cause FPLD4, in which the control of basal lipolysis is similarly affected. Moreover, this region (233–405 amino acids) is predicted to be cytosolic and to contain a large disordered sequence (380–427 amino acids), not having a well-defined tertiary structure, that partly encompasses with the ABHD5 binding site (https://www.uniprot.org/uniprot/O60240). These intrinsically disordered patches, which account for at least 40% of the proteome (19), are known to generally adopt an ensemble of different conformations and still carry out their function in an unstructured state. Moreover, it has been shown that disordered epitopes are as likely to be recognized by antibodies as ordered epitopes and that a significant proportion of them contain linear epitopes, which are only very weakly sensitive to the degree of disorder or to the folding status (20).
Characterization studies showed that anti-PLIN1 autoantibodies from patients with AGL were mainly of the IgG type, with the IgG1 subclass predominating, though with minor amounts of IgG2, IgG3, IgG4, and IgM also present. The presence of κ and λ light chains, as well as of all four IgG subclasses, indicates a polyclonal origin of these autoantibodies (Table 2). IgM antibodies are the first antibody class produced in response to infections and during the development of some autoimmune diseases (21). In line with this, we observed that patients with IgM autoantibodies in our cohort had shorter times from the onset of lipodystrophy than patients with IgG (8.82 vs. 13.43 years, respectively). We had only two patients with disease courses <1 year (AGL21 and AGL32), and both had IgM antibodies. Interestingly, AGL21 developed lipodystrophy as an irAE after treatment with pembrolizumab at 54 years of age, and autoantibodies were detected a few months after the onset. These results prompt us to suggest that IgM anti-PLIN1 may be biomarker of early-stage AGL that should be determined in combination with IgG in all patients with suspicion of the disease.
The polyclonal origin of anti-PLIN1 autoantibodies is consistent with the results obtained by epitope-mapping studies since immunoreactivity was not restricted to one epitope in PLIN1 (Fig. 2). However, all of the patients showed reactivity to the central domain (233–405), both for IgG and IgM autoantibodies (Fig. 2B and C). This central domain contains three hydrophobic sequences required to target and anchor PLIN1 to lipid droplets (22) (Fig. 2A). Moreover, this domain also bears the binding site of ABHD5 (amino acids 380–427), and this interaction has proved crucial for the control of basal lipolysis in vitro (23,24) and in patients with FPLD4 (6–8). Despite this significant sequence overlap, a detailed examination of the data indicated a predominant binding to the ABHD5 binding site (Fig. 2D and E). Interestingly, this epitope overlapped with the highest rank predictions of epitope antigenicity by EMBOSS and SVMTriP software suits. In view of this and considering our previous findings of impaired basal lipolysis regulation using mouse preadipocytes (5), we then studied whether anti-PLIN1 autoantibodies were able to block the binding of ABHD5. Using in vitro experimental approaches, we confirmed that anti-PLIN1 autoantibodies disrupt the interaction of PLIN1 with ABHD5 (Fig. 3) and that the blocking activity strongly correlated with autoantibody titers. This was in good agreement with the results of experiments in 3T3-L1 preadipocytes, in which supplementation with patients’ sera resulted in a significant increase in lipolysis and lipase activity (Fig. 4A and B). Similarly, we showed that anti-PLIN1 autoantibodies penetrate the cell, bind to its antigen on the surface of the lipid droplet, and induce a delocalization of ABHD5 toward the cytosol (Fig. 4C). We thus conclude that anti-PLIN1 autoantibodies mimic the pathogenic effect of FPLD4-associated PLIN1 variants and cause generalized lipodystrophy in patients with AGL.
Patients with anti-PLIN1 autoantibodies exhibited earlier onset of lipodystrophy and more extensive loss of adipose tissue (Table 3). Interestingly, autoantibody titers showed a strong association with worse glycemic and lipid control (Table 4), which could be linked to the greater loss of adipose tissue in patients with autoantibodies (Table 3). Hepatic steatosis is a frequent comorbidity in patients with AGL (1,4). Surprisingly, although a similar percentage of patients with and without antibodies had fatty liver disease, hepatocyte injury as evidenced by higher ALT and AST was significantly higher in patients with autoantibodies (Table 3) and correlated with autoantibody titers (Table 4). This difference could be explained by the predominance of AIH in patients with anti-PLIN1 (25% vs. 5% in negative patients). Similarly, the lower levels of complement C4 observed in our patients (Table 3) could be associated with AIH, in agreement with previous works (14,15). To clarify the association between anti-PLIN1 antibodies and impaired liver function, we screened for antibodies against PLIN2, the predominant perilipin expressed in the liver (25,26), but no patients showed reactivity (data not shown). In the course of liver steatogenesis, PLIN1 is the most upregulated lipid droplet protein that is linked to the degree of fatty liver and the size of liver lipid droplets (25–28). A possible explanation is that the higher presence of PLIN1 in fatty liver makes this organ a nonadipose target for anti-PLIN1 autoantibodies. Additional studies are being performed to clarify the pathogenic role of anti-PLIN1 in liver injury.
In conclusion, our data provide strong support for the conclusion that anti-PLIN1 autoantibodies are a useful biomarker for diagnosis of AGL. The association anti-PLIN1 autoantibodies with metabolic worsening suggest that they may also be a prognostic biomarker. Controlled studies may be needed to assess the effect of immunosuppressive treatments on antibody levels and the association of the latter with clinical improvement. Finally, the capacity anti-PLIN1 autoantibodies to block PLIN1/ABHD5 interactions provides a mechanistic explanation to the pathophysiology of the disease and opens the possibility for novel therapeutic strategies.
Article Information
Acknowledgments. The authors thank the patients, families, and clinicians for being part of these studies; Drs. David Vincent López and Teresa Bellón Heredia (Hospital La Paz Institute for Health Research, La Paz University Hospital, Madrid, Spain) for providing 3T3-L1 line cells; and Prof. Francesc Villarroya Gombau (Departament de Bioquimica I Biomedicina Molecular and Institut de Biomedicina de la Universitat de Barcelona, Barcelona, Spain); and David Sánchez Infantes (Department of Endocrinology and Nutrition, Institut de Recerca en Ciènces de la Salut Germans Trias i Pujol, Barcelona, Spain) for providing serum samples from obese individuals.
Funding. This study was funded by the Instituto de Salud Carlos III and the European Regional Development Fund from the European Union (grant PI15-00255), the Spanish Autonomous Region of Madrid (Complement II-CM network, S2017/BMD-3673), the Asociación Española de Familiares y Afectados de Lipodistrofias, an intramural grant from the Xunta de Galicia (grant ED431B 2020/37), the intramural research program of the National Institute of Diabetes and Digestive and Kidney Diseases, and the Italian Ministry of Education, University and Research (project code 2017L8Z2EM). The Obesity and Lipodystrophy Center at the University Hospital of Pisa is part of the European Consortium of Lipodystrophies, the European Reference Network for Rare Endocrine Conditions (Endo-ERN), and the European Reference Network for Rare Hereditary Metabolic Disorders (MetabERN; project number 739543).
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. F.C., P.N., and M.L.-T. conceived, designed, and supervised the studies. F.C., A.L.-L., and P.N. performed the experimental works. B.S.A., G.C., S.M., F.S., D.A.-V., and R.J.B. gathered the clinical data. F.C. wrote the first draft of the manuscript and revised it with considerable input from B.S.A., A.L.-L., G.C., S.M., F.S., D.A.-V., R.J.B., P.N., M.L.-T., and F.C. is the guarantor of this work and, as such, had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains supplementary material online at https://doi.org/10.2337/figshare.20163935.
References
- 1. Brown RJ, Araujo-Vilar D, Cheung PT, et al. The diagnosis and management of lipodystrophy syndromes: a multi-society practice guideline. J Clin Endocrinol Metab 2016;101:4500–4511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Haque WA, Shimomura I, Matsuzawa Y, Garg A. Serum adiponectin and leptin levels in patients with lipodystrophies. J Clin Endocrinol Metab 2002;87:2395. [DOI] [PubMed] [Google Scholar]
- 3. Misra A, Garg A. Clinical features and metabolic derangements in acquired generalized lipodystrophy: case reports and review of the literature. Medicine (Baltimore) 2003;82:129–146 [DOI] [PubMed] [Google Scholar]
- 4. Araújo-Vilar D, Santini F. Diagnosis and treatment of lipodystrophy: a step-by-step approach. J Endocrinol Invest 2019;42:61–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Corvillo F, Aparicio V, López-Lera A, et al. Autoantibodies against perilipin 1 as a cause of acquired generalized lipodystrophy. Front Immunol 2018;9:2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gandotra S, Le Dour C, Bottomley W, et al. Perilipin deficiency and autosomal dominant partial lipodystrophy. N Engl J Med 2011;364:740–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gandotra S, Lim K, Girousse A, Saudek V, O’Rahilly S, Savage DB. Human frame shift mutations affecting the carboxyl terminus of perilipin increase lipolysis by failing to sequester the adipose triglyceride lipase (ATGL) coactivator AB-hydrolase-containing 5 (ABHD5). J Biol Chem 2011;286:34998–35006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kozusko K, Tsang V, Bottomley W, et al. Clinical and molecular characterization of a novel PLIN1 frameshift mutation identified in patients with familial partial lipodystrophy. Diabetes 2015;64:299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sorkina E, Chichkova V. Generalized lipoatrophy syndromes. Presse Med 2021;50:104075. [DOI] [PubMed] [Google Scholar]
- 10. Jéru I. Genetics of lipodystrophy syndromes. Presse Med 2021;50:104074. [DOI] [PubMed] [Google Scholar]
- 11. Ceccarini G, Magno S, Gilio D, Pelosini C, Santini F. Autoimmunity in lipodystrophy syndromes. Presse Med 2021;50:104073. [DOI] [PubMed] [Google Scholar]
- 12. Hübler A, Abendroth K, Keiner T, et al. Dysregulation of insulin-like growth factors in a case of generalized acquired lipoatrophic diabetes mellitus (Lawrence Syndrome) connected with autoantibodies against adipocyte membranes. Exp Clin Endocrinol Diabetes 1998;106:79–84 [DOI] [PubMed] [Google Scholar]
- 13. Fischer-Posovszky P, Hebestreit H, Hofmann AK, et al. Role of CD95-mediated adipocyte loss in autoimmune lipodystrophy. J Clin Endocrinol Metab 2006;91:1129–1135 [DOI] [PubMed] [Google Scholar]
- 14. Savage DB, Semple RK, Clatworthy MR, et al. Complement abnormalities in acquired lipodystrophy revisited. J Clin Endocrinol Metab 2009;94:10–16 [DOI] [PubMed] [Google Scholar]
- 15. Eren E, Özkan TB, Çakır EDP, Sağlam H, Tarım Ö. Acquired generalized lipodystrophy associated with autoimmune hepatitis and low serum C4 level. J Clin Res Pediatr Endocrinol 2010;2:39–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mandel-Brehm C, Vazquez SE, Liverman C, et al. Perilipin-1 autoantibodies linked to idiopathic lipodystrophy in the setting of two distinct breaks in immune tolerance. 28 September 2021 [preprint]. medRxiv:2021.09.24.21263657 [Google Scholar]
- 17. Cinti S, Mitchell G, Barbatelli G, et al. Adipocyte death defines macrophage localization and function in adipose tissue of obese mice and humans. J Lipid Res 2005;46:2347–2355 [DOI] [PubMed] [Google Scholar]
- 18. Ozer M, Hannibal M, Frame D, et al. MON-155 a patient with acquired generalized lipodystrophy (AGL, Lawrence syndrome) in association with CTLA-4 haploinsufficiency. J Endocr Soc 2019;3(Suppl. 1):MON-155 [Google Scholar]
- 19. Oates ME, Romero P, Ishida T, et al. D2P2: database of disordered protein predictions. Nucleic Acids Res 2013;41:D508–D516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. MacRaild CA, Richards JS, Anders RF, Norton RS. Antibody recognition of disordered antigens. Structure 2016;24:148–157 [DOI] [PubMed] [Google Scholar]
- 21. Duarte-Rey C, Bogdanos DP, Leung PSC, Anaya JM, Gershwin ME. IgM predominance in autoimmune disease: genetics and gender. Autoimmun Rev 2012;11:A404–A412 [DOI] [PubMed] [Google Scholar]
- 22. Garcia A, Sekowski A, Subramanian V, Brasaemle DL. The central domain is required to target and anchor perilipin A to lipid droplets. J Biol Chem 2003;278:625–635 [DOI] [PubMed] [Google Scholar]
- 23. Subramanian V, Rothenberg A, Gomez C, et al. Perilipin A mediates the reversible binding of CGI-58 to lipid droplets in 3T3-L1 adipocytes. J Biol Chem 2004;279:42062–42071 [DOI] [PubMed] [Google Scholar]
- 24. Patel S, Yang W, Kozusko K, Saudek V, Savage DB. Perilipins 2 and 3 lack a carboxy-terminal domain present in perilipin 1 involved in sequestering ABHD5 and suppressing basal lipolysis. Proc Natl Acad Sci USA 2014;111:9163–9168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mashek DG. Hepatic lipid droplets: a balancing act between energy storage and metabolic dysfunction in NAFLD. Mol Metab 2021;50:101115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Carr RM, Dhir R, Mahadev K, Comerford M, Chalasani NP, Ahima RS. Perilipin staining distinguishes between steatosis and non-alcoholic steatohepatitis in adults and children. Clin Gastroenterol Hepatol 2017;15:145–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Straub BK, Stoeffel P, Heid H, Zimbelmann R, Schirmacher P. Differential pattern of lipid droplet-associated proteins and de novo perilipin expression in hepatocyte steatogenesis. Hepatology 2008;47:1936–1946 [DOI] [PubMed] [Google Scholar]
- 28. Fujii H, Ikura Y, Arimoto J, et al. Expression of perilipin and adipophilin in nonalcoholic fatty liver disease; relevance to oxidative injury and hepatocyte ballooning. J Atheroscler Thromb 2009;16:893–901 [DOI] [PubMed] [Google Scholar]