Abstract
Diabetes has been a worldwide healthcare problem for many years. Current methods of treating diabetes are still largely directed at symptoms, aiming to control the manifestations of the pathology. This creates an overall need to find alternative measures that can impact on the causes of the disease, reverse diabetes, or make it more manageable. Understanding the role of key players in the pathogenesis of diabetes and the related β-cell functions is of great importance in combating diabetes. PDX1 is a master regulator in pancreas organogenesis, the maturation and identity preservation of β-cells, and of their role in normal insulin function. Mutations in the PDX1 gene are correlated with many pancreatic dysfunctions, including pancreatic agenesis (homozygous mutation) and MODY4 (heterozygous mutation), while in other types of diabetes, PDX1 expression is reduced. Therefore, alternative approaches to treat diabetes largely depend on knowledge of PDX1 regulation, its interaction with other transcription factors, and its role in obtaining β-cells through differentiation and transdifferentiation protocols. In this article, we review the basic functions of PDX1 and its regulation by genetic and epigenetic factors. Lastly, we summarize different variations of the differentiation protocols used to obtain β-cells from alternative cell sources, using PDX1 alone or in combination with various transcription factors and modified culture conditions. This review shows the unique position of PDX1 as a potential target in the genetic and cellular treatment of diabetes.
Keywords: PDX1, pancreas, β-cells, insulin, diabetes
1 Introduction
Diabetes mellitus (diabetes) is a chronic metabolic disease characterized by high blood glucose levels and associated with impaired insulin secretion, insulin action, or both (Weir et al., 1990; Kerner et al., 2014). According to the World Health Organization, more than 400 million people are living with diabetes worldwide, prompting global efforts to stop the rise of the disease. Many approaches are being used in the treatment of diabetes, ranging from conventional methods, like the use of pharmaceutical compounds, to more advanced approaches such as gene and cellular therapies (Ryan et al., 2005; Wong et al., 2010; Sorli and Heile, 2014; Hering et al., 2016; Loretelli et al., 2020). However, many of them fall short as substitutes for the sophistication of human β-cells. Therefore, in pursuit of understanding the pathogenesis of diabetes, we must understand the key players in the molecular mechanisms of β-cells. One of these key players is pancreatic and duodenal homeobox 1 (PDX1), also known as insulin-promoting factor 1 (IPF1).
Although PDX1 is predominantly expressed in β-cells and some δ-cells of the islets of Langerhans, it is also expressed in the gastrointestinal tract (duodenum, stomach, pancreas), and the central nervous system during development (Perez-Villamil et al., 1999; Fagerberg et al., 2014). PDX1 is one of the early-expressed genes during pancreas development and one that persists through β-cell maturation. Studies have revealed its role in the normal development of the pancreas by regulating the fate and propagation of pancreatic precursor cells (Jonsson et al., 1994). Furthermore, PDX1-expressing progenitors give rise to both exocrine and endocrine lineages, hence PDX1 loss-of-function mutations lead to arrested development of the exocrine compartment and an underdeveloped endocrine compartment, since endocrine lineage is also affected by NGN3-expressing progenitors (Oliver-Krasinski et al., 2009). Mutations in the PDX1 gene cause pancreas agenesis, maturity-onset diabetes of the young 4 (MODY4) and other pancreatic dysfunctions (Jonsson et al., 1994; Stoffers et al., 1997). PDX1 is also linked to diabetes pathogenesis; in type 1 diabetes (T1D) PDX1 autoantibodies have been detected, while in type 2 diabetes (T2D), PDX1 expression levels are compromised (Li et al., 2010; Guo et al., 2013; Abreu et al., 2021). These data solidify the role of PDX1 as a master regulator of embryonic pancreatic formation, in both exocrine and endocrine compartments, and most importantly in the maturation and development of β-cell function.
2 PDX1 gene structure and regulation of expression
PDX1 was first described as a nuclear endodermal protein expressed in the epithelium of the duodenum and the pancreas (Wright et al., 1989). Later, PDX1 was linked to β-cells’ specific expression of insulin by binding and transactivating the insulin gene promoter (Ohlsson et al., 1993). PDX1 also activates other pancreas-associated genes like those for SST (somatostatin), GCK (glucokinase), IAPP (islet amyloid polypeptide), RFX6 (regulatory factor X6), HNF1B (HNF1 homeobox B), and even PDX1 itself (Wang et al., 2018).
The PDX1 gene is highly conservative among different species. It is composed of two exons spanning a region of 6 kb on chromosome 13 and encoding a protein of 283 amino acids without any known splice forms (Stoffel et al., 1995). One exon encodes for the NH2- terminal region containing a DNA activation domain and the other exon encodes for the COOH- terminal region and the homeodomain region that contains three helixes and harbors a nuclear localization signal responsible for DNA binding (Inoue et al., 1996; Melloul et al., 2002; Schwitzgebel et al., 2003; Stanojevic et al., 2004).
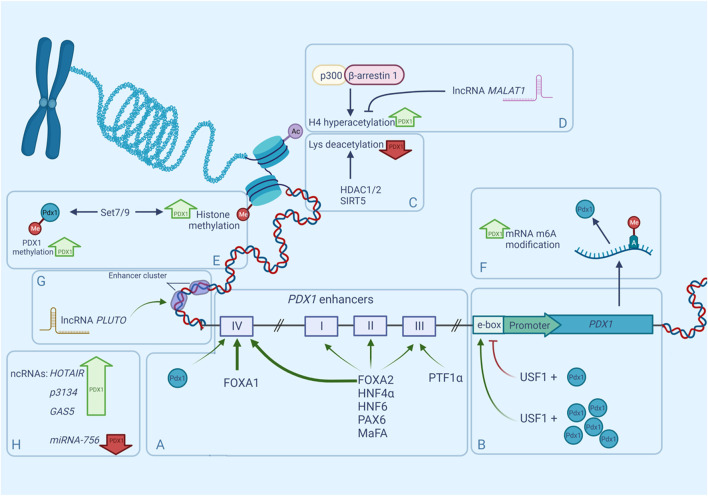
Although PDX1 has only one promoter (Figure 1), it is regulated by a large number of distant enhancers mainly located in the 5′-flanking region of the gene (Sharma et al., 1996; Wu et al., 1997; Campbell and Macfarlane, 2002). Those enhancers were first discovered and characterized by studying the nuclease hypersensitive sites (HSS). HSS are markers of transcription factor (TF) binding sites, and three of them have been identified in the approximate area between −3,000 and +180 bp of the mouse Pdx1 gene. HSS1 (−2,560 to −1880), has demonstrated an ability to control the β-cell-specific expression of PDX1 (Wu et al., 1997). Further investigation of the HSS1 site showed that it can be divided into three subdomains: area I (−2,694 to −2,561 bp), area II (−2,139 to −1958 bp), and area III (−1879 to −1799 bp). The most distant PDX1 enhancer is located near −6,000 bp in the mouse gene and near −8,300 bp in the human gene and is known as area IV (Gerrish et al., 2004). Areas I, III, and IV are very conserved between mice, humans, and chickens, sharing 78–89% similarity, whereas area II is present only in mammals (Gerrish et al., 2000; Gerrish et al., 2004). Analyses of the enhancer region have shown that areas I–III contain binding sites for important transcription factors participating in pancreatic organogeneses, such as HNF1α, FOXA2, HNF6, PAX6, and MaFA (Dassaye et al., 2016), moreover, areas I–II contain binding sites both for transcriptional activators and inhibitors of PDX1 (Gerrish et al., 2004). Further analyses of enhancer functions show that areas I, II, and IV are capable of maintaining the expression of the β-cell-specific reporter in transfection assays independently of each other (Marshak et al., 2000; Gerrish et al., 2004), and together areas I and II can induce strong PDX1 expression in β-cells (Van Velkinburgh et al., 2005). Although area III seems to be less important and does not drive β-cell-selective activity, it contributes to PDX1 expression during embryogenesis through its binding of the PTF1α transactivator (Gerrish et al., 2000; Wiebe et al., 2007). Furthermore, the removal of areas I–III in vivo leads to decreased expression of PDX1 and impairs the formation of the pancreas in the early development stages in mice (Fujitani et al., 2006)—an outcome similar to pancreatic agenesis in homozygous loss-of-function PDX1 phenotypes.
FIGURE 1.
Regulation of mammalian PDX1 gene by multiple factors. Expression of PDX1 is controlled by numerous epigenetic factors such as nucleosome positioning, histone methylation, histone acetylation, accessibility of enhancers and non-coding RNAs. PDX1 expression can be upregulated (bold green upward arrows) or downregulated (bold red downward arrows) by any of these factors. (A). Distant enhancers known as areas I-IV, participate in PDX1 transcription regulation due to their ability to bind transcription factors and boost gene expression. Same TFs can bind to multiple areas with different affinities, for example FOXA1/2 occupy area IV more efficiently (bold green arrows) than other areas (narrow green arrows). (B). regulation of PDX1 through upstream regulatory factors (USF) and E-box. (C). interaction of PDX1 and histone deacetylases. (D). interaction between β-arrestin-1, P300 and their role in PDX1 regulation. (E). PDX1 recruitment to Lys methyltransferase Set7/9. (F). RNA modification in PDX1 regulation. (G). (H). role of ncRNAs in PDX1 regulation.
As was mentioned before, Area II is unique to mammals and has multiple essential functions both in early endocrine cell specification and postnatal β-cell maturation. Deletion of this locus does not affect pancreas development or size, but it does change islet cell composition. Homozygous absence of Area II in PDX+/-pancreas (PDX1ΔII/−) results in a large decrease in endocrine progenitors, abnormal β-cell specification, and subsequent hyperglycemia in newborn mice (Yang et al., 2017). Furthermore, the PDX1 deficient condition associated with the PDX1ΔII/− state caused a significant alteration in the proportion of α- and β-cells, shifting the balance towards the α-cell type, probably because of the inhibited PDX1-mediated repression of the ARX gene expression—a main α-cell fate inductor (Collombat et al., 2007; Yang et al., 2017). These data demonstrate that area II of the PDX1 enhancer participates in maintaining a proper α- and β-cell balance in the islets. Of note, PDX1 repression of the ARX gene is mediated by co-expression of the Groucho-related gene 3 (GRG3), a member of the Groucho family of co-repressors (Metzger et al., 2014), but interactions of the GRG3, ARX, and PDX1 area II enhancers are yet to be investigated.
Like area II, area IV is not essential for normal organogenesis of the pancreas, but it regulates postnatal PDX1 expression and β-cell functions and growth. Area IV has binding sites for both FOXA1 and FOXA2 which are pioneer TFs of several foregut-derived organs, including the pancreas (Figures 1A). These TFs occupy area IV more efficiently than other areas and their bindings are heavily dependent on developmental time, and increases with age (Gao et al., 2008). Interestingly, mice with area IV mutations exhibited sexually dimorphic phenotypes: affected diabetic males with reduced PDX1 levels manifested hyperglycemia at weaning time versus phenotypically normal females (Spaeth et al., 2017). Spaeth et al. (2017) also suggested that PDX1 may autoregulate area IV during weaning which could explain why PDX1 levels increase in this period of development (Stolovich-Rain et al., 2015). Additionally, area IV of the PDX1 enhancer is tissue-specific and is upregulated by HNF3β and NEUROD/b2 strictly in islet cells (Naya et al., 1997; Kaestner, 2000). This region is also influenced by glucocorticoids, which reduce PDX1 expression by interfering with HNF3β activity (Sharma et al., 1997).
Besides its occupation of the PDX1 enhancer, transactivation of the gene by FOXA2 is also augmented by FAM3A—a mitochondrial protein that enhances ATP production. Yang et al. (2020) studied the role of mitochondria in regulating PDX1 expression in pancreatic β-cells and demonstrated that FAM3A-induced ATP production elevates cellular Ca2+ levels, which results in the release of activated calmodulin to function as a co-activator of FOXA2, thus stimulating PDX1 gene transcription. This is one of the possible mechanisms that link mitochondrial dysfunction with insulin deficiency under diabetic conditions.
After transcription, PDX1 cooperates with FOXA1 and FOXA2 (FOXA1/2) to promote downstream gene regulation. Generally, FOXA1/2 bind to the enhancers of target genes, assisting increased deposition of histone H3 lysine four mono-methylation (H3K4me1) that leads to chromatin relaxation and accessibility for other TFs, a process known as enhancer priming (Lee et al., 2019). A recent study has shown that FOXA1/2 recruitment to primed enhancers before pancreatic lineage induction is independent of PDX1, whereas, in contrast, FOXA1/2 binding with unprimed enhancers requires cooperation with PDX1 (Geusz et al., 2021). This indicates that full chromatin accessibility and enhancer activation during β-cell development are heavily reliant on PDX1.
Another group of enhancers are the E-boxes (enhancer boxes)—short DNA sequences that share a signature motif CANNTG (N for any nucleotide) and act like protein-binding sites (Massari and Murre, 2000). During pancreas genesis, PDX1 activates its promoter through the proximal (−104/−99 bp) E-box motif in the PDX1 promoter region, creating a positive autoregulatory loop (Melloul et al., 2002). Upstream stimulatory factor 1 (USF1) occupies that E-box motif and has been proven to be crucial for the autoregulation of PDX1 (Amemiya-Kudo et al., 2011). First, it was discovered that USF1 forms a complex with PDX1, and together they activate the expression of the PDX1 gene, however further investigation revealed that USF1 has a dose-dependent repressing effect on the PDX1 promoter, but is successively abrogated in a higher concentration of PDX1 (Spohrer et al., 2017) (Figure 1B).
Interestingly, both PDX1 and USF1 have been identified as substrates for protein kinase CK2 (Meng et al., 2010; Lupp et al., 2014). Spohrer et al. (2017) have shown that CK2 is a negative regulator of USF1-dependent PDX1 transcription because of the CK2 phosphorylation of USF1, which strengthened the USF1 interaction with the PDX1 protein.
All these combined findings indicate the importance of the conserved enhancer elements for PDX1 expression both in pancreas development and in the maintenance of β-cells.
3 Epigenetic control of PDX1 function
Epigenetic modifications are an important part of the gene expression machinery that operate transcription by changing the state of the chromatin. PDX1 regulates pancreas development and β-cell maintenance with the help of various recruited coregulators. Among them are numerous epigenetic modifiers that participate in DNA methylation, histone modification, chromatin remodeling, and ncRNA operation (Spaeth et al., 2016).
Genome sequencing of the islets of T2D donors has revealed that the PDX1 gene is heavily methylated in diabetes (Volkov et al., 2017). Interestingly, high levels of glucose promote methylation of PDX1 in isolated T2D islets, decreasing the expression of the gene even further (Yang et al., 2012). Different levels of glucose also affect the chromatin landscape shifting of PDX1 in β-cells. According to Mosley and Ozcan (2004) in the presence of low glucose levels, PDX1 is located in the nuclear periphery, where it interacts with the histone deacetylases HDAC1 and HDAC2, allowing them to target the insulin promoter, producing condensed chromatin and consequently reducing insulin gene expression (Figure 1C). When the concentration of glucose becomes high, PDX1 translocates to the nucleoplasm where it interacts with histone acetyltransferase p300. P300 causes the hyperacetylation of histone H4 in the insulin promoter, thus stimulating insulin gene expression (Mosley et al., 2004) (Figure 1D). However, in contrast to the studies of Mosley and others (Rafiq et al., 1998; Elrick and Docherty, 2001), later experiments did not find any evidence of glucose-dependent changes in the localization of PDX1 (Spohrer et al., 2017).
Recently it has been shown that β-arrestin-1 participates in the engagement of p300 to PDX1 (Figure 1D). β-Arrestin-1 and β-arrestin-2 are intracellular signaling proteins that participate in the sensitization of many G protein-coupled receptors (GPCRs) (Pierce and Lefkowitz, 2001) or act independently (Shukla et al., 2011). Tissue-specific knockout of β-arrestin-2 in mouse β-cells causes metabolic deficits, including impaired insulin secretion and reduced glucose tolerance (Zhu L. et al., 2017). Surprisingly, β-cell-specific β-arrestin-1 knockout in mice fed a standard chow did not show similar deficits (Barella et al., 2019), but severe impairment in glucose tolerance in mice on an obesogenic diet was reported (Barella et al., 2021). Further investigation showed that β-arrestin-1 deficiency in β-cells leads to reduced PDX1 expression because of the lack of β-arrestin-1 complexes with p300, which normally promote PDX1 transcription (Barella et al., 2021).
The nucleosome state heavily affects the availability of chromatin for TFs. The SWI/SNF family members forming part of the ATP-dependent chromatin remodeling complex are key regulators of nucleosome positioning (Euskirchen et al., 2012). Like HDACs, the SWI/SNF complex interacts with PDX1 in a glucose-dependent manner (McKenna et al., 2015). The mammalian SWI/SNF complex contains ATPase subunits, either BRG1 or BRM. In low glucose, PDX1 interacts with the BRM:SWI/SNF complex, allowing it to repress its target genes INS1, SLC2A2, and UCN3. Whereas in high glucose, PDX1 binds to the BRG1:SWI/SNF complex, which, in contrast, enhances the expression of these genes (Kim and Kulkarni, 2020).
Class III HDACs also known as sirtuins (SIRTs) are involved in the regulation of pancreas development and glucose homeostasis. SIRT1 promotes β-cell formation by boosting the transcription of PDX1 through the deacetylation of FOXA2 on the promoter of the PDX1 gene (Wang et al., 2013). Oppositely, SIRT5 downregulates the transcription of PDX1 through H4K16 deacetylation of its promoter region (Ma and Fei, 2018).
PDX1 co-modifiers are also involved in histone methylation and demethylation. For example, PDX1 recruits Lys methyltransferase Set7/9 to the INS gene, where it performs H3-K4 methylation and thus activates transcription of the gene in mouse β-cells (Deering et al., 2009). Set7/9 also takes part in PDX1 expression regulation (Figure 1E). The β-cells specific knockout of Set7/9 results in the downregulation of PDX1 and other important β-cell genes like MAFA, GCK, and GLUT2, causing a shift from insulin production to increased proliferation (Maganti et al., 2015; Jetton et al., 2021). Furthermore, Set7/9 seems to be important for the PDX1 protein itself: it has been shown that Set7/9 methylates the N-terminal residue Lys-131 of PDX1, augmenting PDX1 transcriptional activity, which is important for the maintenance of normal β-cell function and glucose homeostasis (Maganti et al., 2015).
RNA modification is a relatively newly discovered mechanism of gene expression regulation that also takes part in PDX1 regulation. One of the most abundant RNA modifications is the methylation of the adenosine in N6-position-m6A (Figure 1F) (Frye et al., 2018). It was shown that the m6A landscape of T2D islets significantly differs from the landscape of healthy islets—the mRNA pool in T2D islets is hypomethylated. Artificial depletion of m6A levels in EndoC-βH1 (immortalized human β-cell line) cells results in G0-G1 cell cycle arrest and impaired insulin secretion due to downregulation of the insulin/IGF1–AKT–PDX1 pathway, decreasing the AKT phosphorylation and PDX1 protein levels (De Jesus et al., 2019).
Non-coding RNAs (ncRNA) are other key players in gene regulation. More than 1,000 cell-type-specific long non-coding RNAs (lncRNA) have been identified both in human and murine pancreatic islets (Moran et al., 2012; Benner et al., 2014), most of which are located outside of genes, but near the islet-specific chromatin domains and protein-coding regions (Moran et al., 2012).
Frequently, enhancer clusters are targets of lncRNAs. An enhancer cluster, otherwise defined as a superenhancer, is a group of enhancers in close genomic proximity that are bound by multiple TFs (Pott and Lieb, 2015). LncRNAs affect the binding of TFs with pancreatic-enhancer-cluster-associated genes, changing their affinity for targets (Pasquali et al., 2014; Akerman et al., 2017). Knockdown of islet-specific lncRNAs identified their ability to modulate gene expression and, consequently, insulin secretion in human β-cells. It was shown that in T2D islets several lncRNAs were significantly altered compared to healthy islets (Akerman et al., 2017; Sathishkumar et al., 2018).
PDX1 is regulated by many lncRNAs, but only a few are well-characterized. One of them is HI-LNC71, also known as PLUTO (PDX1 locus upstream transcript). PLUTO regulates PDX1 transcription by affecting the 3D contacts between the enhancer cluster and the PDX1 promoter (Figures 1G). Although lncRNA sequences generally are not conserved across different species, PLUTO regulation of PDX1 has been confirmed for both mouse and human orthologs, underlining the possible importance of that lncRNA in the modulation of PDX1 expression (Akerman et al., 2017).
Circulating lncRNA-p3134 also participates in the regulation of PDX1 and other β-cell-associated TFs (Figure 1H). It has been shown that overexpression of lncRNA-p3134 in the mouse pancreatic β-cell line MIN6 upregulates Pdx1, MAFA, and Glut2 expression levels and increases glucose-stimulated insulin secretion consistent with the upregulation of insulin-associated TFs. Moreover, in the condition of high glucose exposure this overexpression partially reversed the inhibitory effect of glucotoxicity on PDX1 expression, and, as a consequence, the glucose-stimulated insulin secretion (GSIS) function was restored (Ruan et al., 2018). PDX1 and MAFA expression are also affected by GAS5 (growth arrest-specific transcript 5) (Jin et al., 2017; Esguerra et al., 2020) and HOTAIR lncRNAs (Zhu, 2020). GAS5 is a key regulatory factor in mammalian cell growth, proliferation, and apoptosis (Lander et al., 2001). It is known for repressing the glucocorticoid receptor function (Kino et al., 2010) and its low levels in human serum are associated with T2D (Carter et al., 2015). Knockdown of lncRNA GAS5 expression resulted in decreased expression of PDX1 and MAFA (Jin et al., 2017), and a similar effect was observed for HOTAIR knockdown (Zhu, 2020). Under glucocorticoid-caused GAS5 downregulation, both PDX1 and NKX6-1 levels were affected (Esguerra et al., 2020).
Although the ncRNA MALAT1 is associated with the regulation of alternative splicing (Tripathi et al., 2010) (Figure 1D), recently it has been reported that MALAT1 decreases the expression of PDX1 by suppressing histone acetylation of the Pdx1 promoter in MIN6 cells (Ding et al., 2020).
RNA-dependent regulation of PDX1 expression also involves short non-coding RNAs. For example, miRNA-765 targets the PDX1 gene and reduces its products on both the mRNA and protein levels, which results in impaired survival and function of pancreatic β-cells (Zheng et al., 2021).
Taken together, this data indicates that the participation of ncRNAs in PDX1 regulation makes them a potential target for T2D treatment procedures and use in protocols directing transdifferentiation into β-cells.
4 Role of PDX1 in pancreas organogenesis and β-cells maturity
The human pancreas is composed of exocrine and endocrine compartments. The orchestration of differentiation and acquisition of cell identity requires the intricate and coordinated expression of different transcription factors during pancreas development.
4.1 Role of PDX1 during early pancreatic development
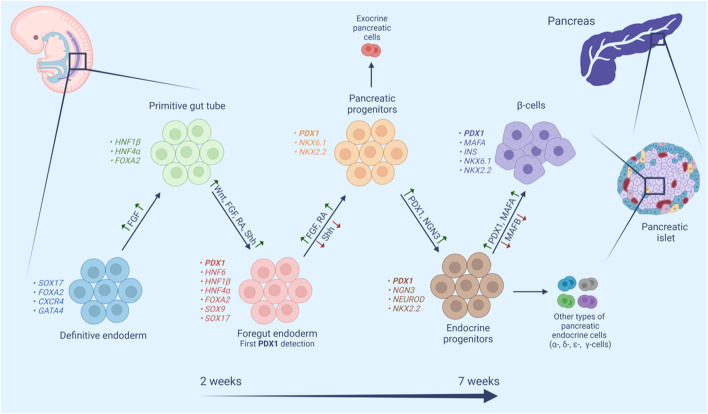
The pancreas originates from a flat sheet of cells known as the definitive endoderm (Figure 2). The definitive endoderm shares a common progenitor with mesoderm called mesendoderm. Specification of the endoderm is affected by many factors including Wnt and Nodal signaling, both of which favor endoderm formation at high levels (Lickert et al., 2002; Vincent et al., 2003). The definitive endoderm then folds into a primitive gut tube, which in turn develops into foregut endoderm (Wells and Melton, 1999). Fibroblast growth factor (FGF), retinoic acid (RA), and the Wnt, and Sonic hedgehog signaling (Shh) pathways have been implicated in foregut endoderm formation (Spence and Wells, 2007). At this stage, the expression of PDX1 is detected making it one of the earliest transcription factors to be expressed in the developing pancreas (Jennings et al., 2013). The fate of the foregut endoderm is determined by Shh signaling. Repression of Shh is necessary for the development of the foregut endoderm into pancreatic progenitor, meanwhile, expression of Shh causes loss of pancreatic gene expression (Apelqvist et al., 1997; Kim et al., 1997). Repression of Shh marks the primary pancreas specification at around 29 days post-conception in humans. The molecular events underlying the primary specification are still not clear, one study has suggested that secretion of FGF2 by the notochord represses Shh signaling and thereby induces expression of pancreatic genes including PDX1 (Hebrok et al., 1998). Moreover, RA signaling at this stage induces several transcription factors including PDX1 (Kumar et al., 2003). The early expression of PDX1 is accompanied by the expression of SOX9, GATA4, FOXA2, and SOX17, however, approximately 30–33 days post-conception, the expression of SOX17 is later lost and replaced with that of NKX6.1. This expression profile of PDX1, SOX9, NKX6.1, and FOXA2 is a hallmark of multipotent pancreatic progenitors (Jennings et al., 2013). Pancreatic progenitors can give rise to a variety of pancreatic cells including exocrine and endocrine cells (Aydin et al., 2020). NKX6.1 is a critical transcription factor in pancreatic β-cell function and proliferation. NKX6.1 expression increases throughout pancreas development where it engages in endocrine commitment and later become restricted to β-cell (Al-Khawaga et al., 2018). The balance between NKX6.1 and PTF1A determines the fate of pancreatic progenitors into exocrine and endocrine commitment. Studies have suggested an antagonistic mechanism between these 2 TFs, in which overexpression of NKX6.1 reduced PTF1A expression and subsequently acinar cell generation, while reduced expression of NKX6.1 showed high expression of PTF1A and a substantial reduction in endocrine progenitors (Schaffer et al., 2010). The simultaneous co-expression of PDX1 and NKX6.1 in pancreatic progenitors warrants their commitment to mono-hormonal, glucose-responsive β-cells (Aigha and Abdelalim, 2020; Memon and Abdelalim, 2020). On the other hand, pancreatic progenitors expressing PDX1 without NKX6.1 (PDX1+/NKX6.1-) develop into poly-hormonal β-cell that fail to function properly in vivo (Aigha and Abdelalim, 2020). Interestingly, Memon et al. (2021) have demonstrated that PDX1-/NKX6.1+ progenitors can give rise to insulin producing glucose-responsive β-cells.
FIGURE 2.
Schematic representation of human β-cell maturation stages. β-Cell differentiation is a complex multistage process. In each stage cell populations have distinctive markers and require both activation (green upward arrows) and repression (red downward arrows) of different signaling pathways, transcription factors, hormones and small molecules.
By the end of the embryonic period, NGN3 detection marks the endocrine commitment and the appearance of pancreatic endocrine cells including fetal β-cells with subsequent detection of nuclear NKX2.2, NKX6.1, PDX1, FOXA2, and ISL1 (Jennings et al., 2013). NGN3 is downregulated by the Notch signaling pathway and upregulated by a cross-regulatory transcription factor network composed of four transcription factors (SOX9, HNF6, HNF1b, FOXa2) (Apelqvist et al., 1999; Lynn et al., 2007). Studies have shown that PDX1 is a member of this cross-regulatory network and it participates directly in the expression of NGN3, which leads to the upregulation of NEUROD1 pushing the cells toward endocrine differentiation (Oliver-Krasinski et al., 2009). A different study has shown a cooperative connection between PDX1 and HNF6 in the early pancreatic bud and their role in the activation of NGN3, subsequent endocrine specification, and the functional maturation of β-cells (Henley et al., 2016). On the other hand, activation of NGN3 leads to the activation of HES-1 in neighboring cells, which inhibits endocrine differentiation through its actions on NGN3 (Jensen et al., 2000). This lateral inhibition model ensures that some progenitors undergo a programmed downregulation of NGN3 and PDX1, which is required for the formation of the exocrine compartment of the pancreas (Kaneto et al., 2007).
PDX1 also plays a crucial role in cell fate determination, this is manifested in several aspects. The relationship between PDX1 and MAFA is one example, MAFA upregulates PDX1 expression in adult islets (Samaras et al., 2003), however, it is not necessary for primary PDX1 induction. Meanwhile, PDX1 is expressed in cells before MAFA and it induces MAFA−MAFB+ to MAFA+MAFB− transition in the late stages of embryogenesis (Nishimura et al., 2006). This transition contributes to the differentiation and maturation of pancreatic β-cells while concurrently pushing cells away from α-cell commitment, where MAFB plays a pivotal role (Nishimura et al., 2006; Hang and Stein, 2011; Conrad et al., 2016).
The role of PDX1 in pancreatic fate determination is also seen by its repression of the intestinal progenitors in the gut tube. PDX1 in cooperation with SOX9 represses the intestinal master transcription factor CDX2 in the pancreatic domain of the gut tube, thus preventing intestinal fate conversion while upregulating pancreas-restricted TFs like PTF1α and NKX6.1 (Shih et al., 2013; Shih et al., 2015). Moreover, PDX1 also downregulates hepatic-specific genes by its actions on HNF1α. The human HNF4α gene contains two promoters P1 and P2 that drive the expression of two sets of isoforms, HNF4α one to six and HNF4α seven to nine, respectively. The P1- and P2-derived isoforms have different functions and different affinity to HNF4α targets (Thomas et al., 2001). PDX1 upregulates HNF1α P2 transcripts with weaker transactivation potential than P1 transcripts, causing them to compete for targets and eventually reducing the activation of target liver-specific genes by the P1 transcripts (Donelan et al., 2015).
4.2 Role of PDX1 in pancreatic islets
During embryogenesis, β-cells are multihormonal immature cells with high proliferative ability; the features of functional β-cells are gained after birth, particularly around weaning time, as demonstrated by their robust GSIS with simultaneous loss of proliferative ability (Sun et al., 2021). The maturation of β-cells is controlled by many transcription factors, the most important of which is MAFA—a TF that plays a role in insulin granule synthesis and secretion (Hang et al., 2014). PDX1 induces MAFB to MAFA transition in later stages of embryogenesis (discussed above). Furthermore, PDX1 plays a significant role in the maturation and identity perseveration of β-cells after birth as it controls the activation of insulin and other genes responsible for glucose sensing and metabolism like GLUT2 and glucokinase (Kaneto et al., 2008). In addition, Kropp et al. (2018) showed that the cooperative function of PDX1 and osteocalcin 1 (OC1) is necessary for the specification and differentiation of pancreatic endocrine cells and postnatal islet maturation.
Interestingly, some degree of β-cell heterogeneity has been reported in normal adult islets (Szabat et al., 2009). Nasteska et al. (2021) have shown that β-cells exist in different stages of maturation in adult islets. Adult islets house highly plastic, immature β-cells with low expression of PDX1 and MAFA alongside highly mature β-cells with high expression of both PDX1 and MAFA. Presumably low levels of PDX1 promote insulin expression during the maturation process, while higher PDX1 levels in mature cells maintain β-cells in differentiated state promoting increased expression of GLUT2 and glucokinase (Szabat et al., 2009). This heterogeneity of β-cells is crucial for proper islet function since it constitutes a reservoir of cells that can be mobilized under stress conditions (Nasteska et al., 2021).
There’s combinatorial effect of PDX1 and FOXA2 in postnatal maturation of β-cells, we mentioned before that both TFs cooperate to promote downstream gene regulation (Lee et al., 2019). MAFA, Ins1, and Slc2a2 are among the genes activated by both TFs in the islets and they are involved β-cells maturation, function, insulin secretion and MODY (Bastidas-Ponce et al., 2017). One study generated mice with reduced expression of both TFs which resulted in upregulation of genes responsible for α-cell fate like MAFB with alterations in β-cells numbers and developed hyperglycemia at weaning time. Moreover, the deletion of PDX1 in mature β-cells led to them losing multiple β-cell markers, combined with either loss of hormone expression or to their adapting of a glucagon-secreting ⍺-cell phenotype since PDX1 binds and inhibits several ⍺-cell genes like MAFB, as was mentioned before (Ritz-Laser et al., 2003; Gao et al., 2014).
5 PDX1 and diabetes
Mutations of the PDX1 gene lead to different outcomes that affect pancreas structure, functions or both. Homozygous mutations in the PDX1 gene and other mutations that impair the functionality of the PDX1 protein during embryonic development cause pancreas agenesis in mice and humans and eventually lead to fatal perinatal hyperglycemia (Jonsson et al., 1994; Stoffers et al., 1997; Hui and Perfetti, 2002). While heterozygous mutations in PDX1 cause maturity-onset diabetes of the young 4 (MODY4) (Abreu et al., 2021). MODY four is a rare monogenic subtype of diabetes mellitus, it is caused by different various mutations in PDX1 gene and its transactivation domain (Anik et al., 2015; Deng et al., 2019; Abreu et al., 2021; Yoshiji et al., 2022).
Studies have shown that a decrease in PDX1 expression in PDX1 knockout mice concurred with poor maturation of β-cells after birth, impaired expression of several β-cells genes, and the appearance of poly-hormonal cells within the islets (Bastidas-Ponce et al., 2017; Spaeth et al., 2017; Jara et al., 2020). In mature β-cells, pdx1 haploinsufficiency leads to diabetes in mice and humans through adapting ⍺-cell phenotype (discussed above).
In fact, a lot of diabetes phenotypes are associated with mutations in the PDX1 gene. The nature of the phenotype and severity of the condition depends on the type and location of the mutation. Missense mutations in the transactivator domain of PDX1 can reduce the ability of the PDX1 protein to activate the expression of its target genes during β-cell development and maturation. As an example of the severity of different mutations, the substitution Pro-Thr in the 33 position (P33 T) causes a greater impact on β-cell formation and function than Cys-Arg in the 18 position (Wang et al., 2019). The P33 T mutation not only impairs binding with DNA targets and transcriptional activation functions, but also predisposes to reduced birth weight, miscarriage, and early postnatal death (Gragnoli et al., 2005). Proline insertions have also been shown to be pathogenic and associated with MODY (Karim et al., 2005; Elbein et al., 2006). Several cases of the PDX1-mutant MODY are reported, where glutamic acid in position 178 of PDX1 was substituted with other amino acids (Schwitzgebel et al., 2003; Nicolino et al., 2010; Abreu et al., 2021). Glutamic acid in that position in the homeodomain is evolutionarily conserved among several species, and this mutation seems to decrease PDX1 half-life, which could prevent the proper self-activation of PDX1 and consequently decreases protein levels (Schwitzgebel et al., 2003; Abreu et al., 2021). In some cases, different point mutations in the PDX1 gene impair endocrine progenitor and β-cell development, leading to the downregulation of several PDX1 target genes responsible for insulin synthesis and secretion, which gives rise to non-functional differentiated β-cells with poor responses to glucose changes (Wang et al., 2019).
PDX1 is also linked to both type 1 and type 2 diabetes pathogenesis; in type 1 diabetes (T1D) PDX1 autoantibodies have been detected (Li et al., 2010). Interestingly, these autoantibodies can be used in screening high-risk population susceptible for developing T1D (Donelan et al., 2013). While in type 2 diabetes (T2D), PDX1 expression levels are compromised (Li et al., 2010; Guo et al., 2013; Abreu et al., 2021). One study found an enrichment of T2D-associated SNPs in PDX1 occupied sites located in the intronic regions of TCF7L2 and HNF1B. Hnf1β is involved in controlling proliferation and survival of multipotent pancreatic progenitors and deletion of the gene causes pancreatic hypoplasia (De Vas et al., 2015), while TCF7L2 has been identified as the locus conveying the highest risk for developing T2DM (McCarthy and Zeggini, 2009). Mutations in their cis-regulatory regions of these two genes predispose to diabetes (Wang et al., 2018). A recent study has found that selected genetic SNPs in PDX1 and MC4R could modify the risk of T2D (Wang et al., 2021).
A number of studies have found a connection between DNA methylation of PDX1 and reduced activity in T2D islets (Liu et al., 2021). Yang et al. (2012) found out that PDX1 was one of 15 genes with CpG islands within the promoter that were methylation-susceptible in T2D (Yang et al., 2012). DNA methylation leads to reduced levels of PDX1 protein and mRNA, resulting in impaired expression of both GLUT2 and insulin and causing development of hyperglycemia (Ahlgren et al., 1998). Under high glucose concentration, DNA methylation level may increase abnormally which results in decreased insulin secretion and subsequently leads to diabetes (Pinzon-Cortes et al., 2017).
Normal pancreas development with β-cell formation and maturation is a complex multilayered process that can provide a guideline for the induction of insulin-producing cells from non-β cells using various transcription factors in a way mimicking the natural process.
6 Role of PDX1 in reprogramming different cell types into pancreatic β-cells
The most promising way to synthesize β-cells from non-β-cells appears to be by mimicking the natural process in which β-cells are developed during pancreas genesis. Based on evidence from years of studies of developmental biology, β-like-cells or at least insulin-producing cells, can be obtained from pluripotent cells, multipotent cells, or mature cell types. Since PDX1 is a master regulator of β-cell differentiation and maturation, a lot of differentiation approaches use PDX1 as the main factor or as a part of multifactorial protocols. In this section, we will review the research in which PDX1 has been used to obtain insulin-producing β-like cells from non-β-cells (Table 1).
TABLE 1.
Summary of studies using PDX1 in obtaining β-cells (full version Supplementary Material).
| Study | Cell source | Transcription factors | Small molecules | Outcome |
|---|---|---|---|---|
| Differentiated adult cells | ||||
| (Ferber et al. 2000) | mouse liver cells | PDX1 | N/A | Insulin secretion |
| (Ber et al. 2003) | mouse liver cells | PDX1 | N/A | Induction of pancreatic Exocrine and endocrine genes, insulin secretion |
| (Imai et al., 2005, Kaneto et al., 2005, Cao et al., 2004, Tang et al., 2006) | mouse liver cells | PDX1-VP16 PDX1 | N/A | Expression of pancreatic markers, insulin secretion |
| (Wang et al. 2007) | mouse liver cells | PDX1-NGN3 | N/A | Insulin secretion, near normal GSIT function |
| (Zhou et al. 2008) | mouse pancreatic exocrine cells | PDX1- MAFA-NGN3 | N/A | Adoption of β-cells morphology and marker expression, insulin secretion |
| (Banga et al. 2012) | mouse hepatic duct-like cells | PDX1-MAFA- NGN3 | N/A | Adoption of β-cells morphology and marker expression, insulin secretion |
| (Cim et al. 2012) | rat liver cells | PDX1-NGN3-MAFA | N/A | Expression of insulin mRNA |
| (Hickey et al. 2013) | mouse gall bladder epithelial cells | PDX1-MAFA-NGN3 | retinoic acid, dibenzazepine | Upregulation of β-cells genes, insulin secretion |
| (Akinci et al. 2013) | rat pancreatic exocrine cells and hepatocytes mouse hepatocyte-derived small cells | PDX1-MAFA-NGN3 | DAPT, BIX-01294, NECA | Increased expression of β-cell markers, insulin secretion |
| (Chen et al. 2014) | mouse intestine crypts cells | PDX1-MAFA-NGN3 | N/A | Adoption of β-cells morphology, insulin secretion |
| (Cardinale et al. 2015) | human biliary tree stem cells | PDX1 | bFGF, PDX-1 peptide | Increased expression of β-cells markers, insulin and C-peptide secretion |
| (Lima et al. 2016) | human exocrine pancreatic cells | PDX1-NGN3-MAFA-PAX4 | ITS, 5-aza-2′-deoxycytidine, sodium butyrate, SB431542, Y27632, betacellulin, exendin-4, nicotinamide | Insulin packaging and secretion |
| (Xiao et al. 2018) | mouse pancreatic alpha cells | PDX1-MAFA | N/A | Increased β-cell mass, insulin secretion |
| Embryonic stem cells | ||||
| (Xu et al. 2013) | mouse ESCs | PDX1-MAFA- NEUROD-NGN3 | β-mercaptoethanol, activin A, retinoic acid, ITS, bFGF, EGF, N2, B27, nicotinamide | Increased expression of β-cells markers, insulin secretion |
| (Salguero-Aranda et al. 2016) | mouse ESCs | increased PDX1 expression by small molecules | DETA-NO, valproic acid, P300 inhibitor C646, β-mercaptoethanol | Increased expression of β-cells markers, glucose responsivity, insulin secretion |
| Induced pluripotent stem cells | ||||
| (Saxena et al. 2016) | human iPSCs | PDX1-MAFA- NGN3 | Activin A, wnt3A, bFGF, BMP4, VEGF, noggin, FGF10, KGF, EGF, B27, ascorbic acid, KAAD-cyclopamine, retinoic acid, Y-27632, vitamin A, T3, Alk5 inhibitor, dibenzazepine | Adoption of β-cells morphology, insulin secretion |
| (Rajaei et al. 2018) | human iPSCs | PDX1 | activin A, Wnt3a, KGF, EGF, SB431542, B27, KAAD cyclopamine, retinoic acid, noggin, IBMX | Insulin and C-peptide secretion, adoption of β-cells morphology |
| Mesenchymal stem cells | ||||
| (Yuan et al. 2010) | rat bone marrow-derived MSCs | PDX1 | N/A | Acquisition of β-cells phenotype, insulin secretion |
| (He et al. 2011) | human umbilical cord MSCs | PDX1 | EGF, B27, GLP-1, betacellulin, HGF, nicotinamide, β-mercaptoethanol | Adoption of β-cells morphology and marker expression, insulin and C-peptide secretion |
| (Lima et al. 2013) | human exocrine pancreas-derived MSCs | PDX1-MAFA-NGN3-PAX4 | betacellulin, exendin-4, nicotinamide | Insulin secretion |
| (Chun et al. 2015) | human amniotic fluid-derived MSCs | PDX1 | activin A, β-mercaptoethanol, N2, B27, bFGF, nicotinamide | Expression of β-cells markers, insulin, and C-peptide secretion |
| (Xu et al. 2017) | human umbilical cord MSCs | PDX1-PAX4 | B27 | Acquisition of β-cells phenotype, expression of endocrine markers, insulin secretion |
| (Gao et al. 2018) | human adipose tissue-derived MSCs | PDX1 | N/A | Insulin secretion |
Early efforts directed towards obtaining β-cells, focused on embryonic stem cells (ESCs), taking advantage of the robust pluripotency of ECSs and established knowledge about natural β-cell genesis (Lumelsky et al., 2001). ESCs are guided towards definitive endoderm, then pancreatic progenitors, followed by endocrine progenitors, and finally mature β-cells. These protocols exploit culture conditions to guide the differentiation process into insulin-producing cells while concurrently using forced expression of the transcriptional factors regulating β-cell identity (Hebrok, 2012). D'Amour et al. (2005) was one of the first groups that differentiated ESCs into endoderm derivatives using Activin A in low serum content. Since then, various protocols have emerged with different modifications, changing signaling molecules, transcription factors, and culture conditions. Cho et al. (2008) have shown that exposure of human ESCs to betacellulin and nicotinamide alongside other factors sustains PDX1 expression and further induces β-cell differentiation. This approach of stimulating PDX1 expression indirectly was also adapted by Salguero-Aranda et al. (2016). Exposing mouse ESCs to diethylenetriamine nitric oxide adduct leads to enhanced expression of Pdx1 when combined with valproic acid, and p300 inhibitor, enhanced pancreatic lineage specification and the generation of glucose-responsive insulin-producing cells (Salguero-Aranda et al., 2016). Another approach is the forced expression of PDX1 by viral transduction, using adenoviral transduction of PDX1+MAFA and either NGN3 or NEUROD alongside small molecules in a three-step protocol that guided the differentiation of mouse ESCs into insulin- and C-peptide—producing cells with elevated expression of β-cell markers (Xu et al., 2013). However, many of the obtained cells are not considered “true” β-cells mainly because they are immature, multihormonal, and unable to regulate glucose levels.
The emergence of induced pluripotent stem cells (iPSCs), circumvented ethical considerations related to ESCs and opened doors to generate patient-specific therapeutic cells. As with ESCs, differentiation protocols for iPSCs follow the same developmental stages as β-cells, beginning with definitive endoderm and moving towards pancreatic progenitors and differentiated β-cells. PDX1 plays a significant role in the differentiation of iPSCs into islet β cells, manifested by its activation of NGN3 and PAX6 expression after binding to their promoter regions and activating downstream gene networks (Qin et al., 2015). (Walczak et al., 2016) showed that lentiviral transduction of iPSCs with PDX1 and NKX6.1 induced the formation of insulin-producing cells with a morphology resembling β-cells, moreover, the derived cells secreted C-peptide in a glucose-responsive manner (Rajaei et al., 2018). Alongside PDX1, two other factors are usually used to push cells into β-cell differentiation. These are MAFA and NGN3. Together all three factors: PDX1, MAFA, and NGN3 are known as the PMN factors and play a critical role in β-cell differentiation and maturation. They have been used together in many research projects to obtain β-cells (Zhu Y. et al., 2017). For example, one study used a system to express the PMN factors in iPSCs under the control of a vanillic acid-dependent switch, coupled with a synthetic signaling cascade. Doing so allowed precise replication of the dynamics of endogenous expression and resulted in β-like cells comparable to human pancreatic islets in GSIS (Saxena et al., 2016). Differentiation protocols have witnessed many variations allowing the generation of cell populations called stem cell-derived islets (SC-islets) (Hogrebe et al., 2020; Maxwell and Millman, 2021). Although promising, generating human iPSCs remains inefficient and expensive, thus hindering the generation of large numbers of patient-specific lines.
Another promising source from which to obtain β-cells is mesenchymal stem cells (MSCs). MSCs have manifold advantages, like immunomodulation, which reduce immune system activation and anti-inflammatory factor release after transplantation (Scuteri and Monfrini, 2018). Transfection of bone marrow-derived MSCs with a recombinant plasmid harboring PDX1 prompted islet-like structure formation, insulin secretion, and the adoption of β-cell morphology (Yuan et al., 2010). Furthermore, the level of PDX1 expression is closely correlated with the level of insulin mRNA and the level of insulin secretion in differentiated, stable MSC cell lines (Yuan et al., 2012). Similar results have been shown in amniotic-fluid-derived MSCs upon adenoviral transduction with PDX1 in controlled culture conditions. Such MSCs were able to express β-cell markers, and secrete insulin and C-peptide (Chun et al., 2015). Similarly, umbilical-cord-derived-MSCs formed islet-like structures containing β-like cells that produced insulin, C-peptide, and other endocrine markers after delivery of PDX1 (He et al., 2011) or the co-delivery of PDX1 and PAX4 using recombinant adenovirus under controlled media conditions (Xu et al., 2017).
Using small molecules and controlled culture conditions are of great importance in obtaining β-cells from other cell types. Lima et al. (2013) obtained functional β-like-cells from pancreatic-exocrine-tissue-derived MSCs after transduction with adenovirus harboring PMN + PAX4. Interestingly, the removal of serum from the media, stopping epithelial to mesenchymal transition (EMT), with Rho-associated kinase (ROCK), and the addition of small molecules like betacellulin, exendin-4 and nicotinamide resulted in better insulin expression and glucose regulation (Lima et al., 2013). Human adipose-tissue-derived-MSCs could also be differentiated into functional islet-like cells after transduction with adenovirus harboring PDX1. Differentiated cells secreted insulin and could regulate glucose in diabetic mice (Gao et al., 2018). All things considered, MSCs are great candidates for the treatment of diabetes, however, more studies are needed to evaluate the long-term differentiative capabilities of MSCs and their safety in application.
Soon after breaking the rigidity of adult differentiated cell types, new research focused on reprogramming mature cells directly into insulin-producing cells without inducing pluripotency. Obtaining β-cells from terminally differentiated cells might be a well-rounded alternative to the use of stem cells, considering their abundance and low risk of tumorigenesis, especially when sharing common progenitors with β-cells. Akinci et al. (2012) introduced the PMN factors using an adenoviral vector into 8 cell types, and demonstrated a bigger likelihood of developmentally related cells to transdifferentiate into β-cells. Of the 8 cell types studied, pancreatic exocrine and SOX9+ hepatic cells upregulated β-cell markers and secreted insulin (Akinci et al., 2012; Akinci et al., 2013).
Liver cells are developmentally related to β-cells and they could be reprogrammed to adopt β-cell morphology and markers. Much research has attempted to carry out in vivo differentiation in animal subjects starting from liver cells. For example, Ferber et al. (2000) identified the role of PDX1 in transforming murine liver cells into insulin-secreting cells after adenoviral transduction. The same results were repeated by Wang et al. (2007) who used plasmids harboring PDX1 or NGN3 with an unrelated adenovirus expressing the human coagulation factor IX gene (AdVhFIX). Diabetic mice treated with either of the two plasmids showed reduced hyperglycemia. Interestingly, when either of the two transcription factors was administrated in adeno-associated viral construct (AAV) there was no response in glucose levels. Later, when another group evaluated the effect of the hydrodynamic delivery of different plasmids carrying PMN factors in the livers of rats, they found that some plasmids like CpG-depleted plasmid (pCpG) and increased the levels of insulin mRNA up to 50 fold. Moreover, they detected signs of differentiation towards β-cells that were able to control glucose in hyperglycemic rats for 1 week (Cim et al., 2012). Similar results were obtained by Banga et al. (2012), who delivered PMN factors into murine livers by adenoviral transduction and obtained glucose-sensitive cells that ameliorated diabetes in mice. Hepatic to pancreatic differentiation could also be achieved by ectopic expression of PDX1 by recombinant adenovirus (Ber et al., 2003).
Some studies have used the active form of PDX1 (PDX1-VP16) in which the activation domain from herpes simplex virus (VP16) is fused to the C-terminus of PDX1, which allows PDX1 to activate target genes without association with other co-factors (Horb et al., 2003). Compared to PDX1, PDX1-VP16 is more efficient in initiating liver-to-endocrine pancreas differentiation, however, both of them upregulated β-cell genes and reversed hyperglycemia in diabetic mice upon lentiviral transduction (Tang et al., 2006). Imai et al. (2005) showed that adenoviral transduction with PDX1-VP16 induced murine hepatocytes to produce insulin and to regulate hyperglycemia in diabetic mice, however, the expression pattern of the hepatocyte was maintained. Kaneto et al. (2005) also showed that PDX1-VP16 increased insulin and other pancreatic factors in the livers of mice and controlled hyperglycemia, especially when combined with NEUROD or NGN3. Cao et al. (2004) showed that the complete transdifferentiation of hepatic cells into insulin-producing cells by PDX1-VP16 requires additional external factors like high glucose and hyperglycemia. Introducing exogenous PDX1-VP16 protein into definitive endoderm cells generated from ESCs, mimicking the natural pattern of PDX1 expression, induced an endocrine pancreas-like cell phenotype, in which 30% of the cells were β-like cells (Bernardo et al., 2009). Taking into account that the PDX1-VP16 approach showed convincing results in the conversion of ESCs and hepatic cells into β-like cells, perhaps this approach can be also adapted for the transdifferentiation of other adult cells into β-cells.
In vivo studies have also been performed on other cell types. Zhou et al. (2008) used adenovirus to deliver PMN into differentiated pancreatic exocrine cells. The reprogrammed cells resembled β-cells morphologically and functionally. Another study screened the effect of PMN factors in a variety of tissues. They found that expression of PMN in the intestinal crypts gives them β-like features including glucose sensitivity and insulin secretion ability (Chen et al., 2014).
α-cells have been particularly well-studied as sources for β-cell compensation due to their sharing of a close progenitor. The differential plasticity between alpha and β-cells was proven to be subject to the effects of PDX1. Introducing PDX1 in NGN3+ endocrine progenitors in the embryonic period in mice resulted in rapid postnatal reprogramming of α-cells to insulin-positive cells resembling β-cells (Yang et al., 2011). Transduction of AAV carrying PDX1 and MAFA into the pancreas of diabetic mice leads to increased α-to β-cell transdifferentiation, increased glucose responsiveness, and normalized blood sugar (Xiao et al., 2018). Moreover, repression of α-cell genes like ARX while upregulating PAX4 is another approach to guide α-to β-cells differentiation. Lima et al. (2016) used PMN factors alongside PAX4 and siRNA against ARX in the exocrine pancreas to favor the formation of β-over α-cells. The obtained β-like cells efficiently processed and secreted insulin and were able to respond to glucose and normalize its levels in diabetic mice.
Gall bladder epithelial cells have also been studied in this regard, after adenoviral transduction with PMN factors. The reprogrammed cells upregulated β-cell genes while downregulating epithelial genes, however, they were not true functional β-cells because they were non-responsive to glucose despite their insulin secretion ability (Hickey et al., 2013). Cardinale et al. (2015) identified a heterogeneous stem/progenitor cell population in the human biliary tree that, when exposed to PDX1 protein in the media, internalized the protein and started insulin and C-peptide production, indicating endocrine differentiation. Another group upregulated the expression of PDX1 indirectly by identifying an andrographolide named C1037 that can stimulate PDX1 expression in both its resulting mRNA and protein levels. Pancreatic duct cells treated with C1307 increased expression of insulin while decreasing glucagon levels compared to control groups (Zhang et al., 2020).
The numerous variations of differentiation protocols rule out the existence of a unified method and source for obtaining β-cells. Although promising, this numerosity indicates the need for more studies and clinical trials to find a unified protocol involving the least amount of genetic and cellular manipulation. All the protocols summarized in this review have focused on the activation of important β-cell genes, primarily PDX1. The methods of activation alongside culture conditions and added small molecules can vary and affect the differentiation outcome. Some protocols rely solely on small molecules to provide conditions similar to β-cell formation in vivo, which avoids problems associated with viral transduction. Choosing the right cell of origin can mean the difference between a complex or a simple protocol. As mentioned before, cells that share a common progenitor with β-cells are easier to transform. Stem cells have great pluripotency potential but this raises the risk of tumorigenesis. In conclusion, this data shows the importance of PDX1 in obtaining β-cells considering its role as a master regulator of β-cell function and identity.
7 Conclusion
Numerous studies on PDX1 have demonstrated its critical role in organogenesis, differentiation, maturation, and in maintaining, and preserving β-cell identity. Moreover, much research has accomplished the differentiation of non-β-cells into insulin-producing cells with the aid of PDX1 as a master regulator in the differentiation protocols, or alongside other factors and soluble molecules. This property of PDX1 makes it a particularly important target for gene or replacement therapy approaches to the treatment of diabetes. Our increased knowledge of PDX1 and other pancreatic endocrine factors provides the cornerstone for optimizing differentiation protocols. Through the years, these protocols have witnessed many advances and variations, including various combinations of transcription factors, culture media components, and different source cells. However, we are still far from finding a bona fide alternative to human β-cells, but our understanding of the role of transcription factors, their interactions and intricate regulation by each other together with the further impact of small molecules and epigenetic factors should eventually allow us to obtain functional β-cells that would be suitable for transplantation therapy in the treatment of diabetes.
Author contributions
NE, KS, and ED: collection of data on the role of PDX1 in β-cell functions and identity, compilation of article content. NE: compilation of Table 1 content. KS: drawing Figures 1, 2.
Funding
This research was funded by grant no. 075-15-2019-1789 from the Ministry of Science and Higher Education of the Russian Federation, allocated to the Center for Precision Genome Editing and Genetic Technologies for Biomedicine.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2022.1091757/full#supplementary-material
Nomenclature
Abbreviations
- AAV
Adeno-associated virus
- bFGF
basic fibroblast growth factor
- EMT
Epithelial–mesenchymal transition
- ESC
embryonic stem cells
- GPCR
G protein-coupled receptor
- HSS
Nuclease hypersensitive site
- iPSCs
induced pluripotent stem cells
- ITS
insulin–transferrin–selenium
- lncRNA
long non-coding RNA
- MODY
Maturity-onset diabetes of the young
- MSC
mesenchymal stem cells
- ncRNA
non-coding RNA
- PMN
PDX1, MAFA, NGN3
- RA
retinoic acid
- ROCK
Rho-associated kinase
- SC-islets
stem cells–derived islets
- Shh
Sonic hedgehog
- siRNA
Small interfering RNA
- SIRT
sirtuin
- T1D
Type 1 diabetes
- T2D
Type 2 diabetes
- TF
Transcription factor
Genes and proteins
- AdVhFIX
Human coagulation factor IX gene
- AKT
RAC-alpha serine/threonine-protein kinase
- ARX
Aristaless related homeobox
- CDX2
Caudal Type Homeobox 2
- CK2
Casein kinase II
- FAM3A
FAM3 metabolism regulating signaling molecule A
- FGF
Fibroblast growth factor
- FGF2
Fibroblast growth factor 2
- FOXA1
Forkhead box A1
- FOXA2
Forkhead box A2
- GAS5
growth arrest-specific transcript 5
- GATA4
GATA Binding Protein 4
- GCK
Glucokinase
- GLUT2
Glucose transporter 2
- GRG3
Groucho-related gene 3
- GSIS
Glucose-stimulated insulin secretion
- HDAC1
Histone deacetylase 1
- HDAC2
Histone deacetylases 2
- HES1
hes family bHLH transcription factor 1
- HNF1α
Hepatocyte nuclear factor 1 α
- HNF3β
Hepatocyte nuclear factor 3 β
- HNF4β
Hepatocyte nuclear factor 4 β
- HNF6
Hepatocyte nuclear factor 6
- IAPP
Islet amyloid polypeptide
- IGF1
Insulin-like growth factor 1
- INS1
Insulin I
- IPF1
Insulin-promoting factor 1
- MAFA
MAF bZIP transcription factor A
- MAFB
MAF bZIP transcription factor B
- NEUROD
Neuronal differentiation 1
- NGN3
Neurogenin-3
- NKX2.2
NK2 Homeobox 2
- NKX6.1
NK6 Homeobox 1
- OC1
One Cut Homeobox 1
- PAX4
Paired box 4
- PAX6
Paired box 6
- PDX1
Pancreatic and duodenal homeobox 1
- PLUTO
PDX1 locus upstream transcript
- PTF1α
Pancreas-specific transcription factor 1α
- RFX6
Regulatory factor X6
- SLC2A
Solute carrier family 2 member 9
- SOX17
SRY-Box transcription factor 17
- SOX9
SRY-Box transcription factor 9
- SST
Somatostatin
- UCN3
Urocortin 3
- USF1
Upstream stimulatory factor 1
References
- Abreu G. M. M., Tarantino R. M., da Fonseca A. C. P., de Souza R. B., Soares C., Cabello P. H., et al. (2021). PDX1-MODY: A rare missense mutation as a cause of monogenic diabetes. Eur. J. Med. Genet. 64 (5), 104194. 10.1016/j.ejmg.2021.104194 [DOI] [PubMed] [Google Scholar]
- Ahlgren U., Jonsson J., Jonsson L., Simu K., Edlund H. (1998). beta-cell-specific inactivation of the mouse Ipf1/Pdx1 gene results in loss of the beta-cell phenotype and maturity onset diabetes. Genes Dev. 12 (12), 1763–1768. 10.1101/gad.12.12.1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aigha, Abdelalim E. M. (2020). NKX6.1 transcription factor: A crucial regulator of pancreatic beta cell development, identity, and proliferation. Stem Cell Res. Ther. 11 (1), 459. 10.1186/s13287-020-01977-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akerman I., Tu Z., Beucher A., Rolando D. M. Y., Sauty-Colace C., Benazra M., et al. (2017). Human pancreatic beta cell lncRNAs control cell-specific regulatory networks. Cell Metab. 25 (2), 400–411. 10.1016/j.cmet.2016.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akinci E., Banga A., Greder L. V., Dutton J. R., Slack J. M. (2012). Reprogramming of pancreatic exocrine cells towards a beta (β) cell character using Pdx1, Ngn3 and MafA. Biochem. J. 442 (3), 539–550. 10.1042/BJ20111678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akinci E., Banga A., Tungatt K., Segal J., Eberhard D., Dutton J. R., et al. (2013). Reprogramming of various cell types to a beta-like state by Pdx1, Ngn3 and MafA. PLoS One 8 (11), e82424. 10.1371/journal.pone.0082424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Khawaga S., Memon B., Butler A. E., Taheri S., Abou-Samra A. B., Abdelalim E. M. (2018). Pathways governing development of stem cell-derived pancreatic beta cells: Lessons from embryogenesis. Biol. Rev. Camb. Philos. Soc. 93 (1), 364–389. 10.1111/brv.12349 [DOI] [PubMed] [Google Scholar]
- Amemiya-Kudo M., Oka J., Takeuchi Y., Okazaki H., Yamamoto T., Yahagi N., et al. (2011). Suppression of the pancreatic duodenal homeodomain transcription factor-1 (Pdx-1) promoter by sterol regulatory element-binding protein-1c (SREBP-1c). J. Biol. Chem. 286 (32), 27902–27914. 10.1074/jbc.M110.186221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anik A., Catli G., Abaci A., Sari E., Yesilkaya E., Korkmaz H. A., et al. (2015). Molecular diagnosis of maturity-onset diabetes of the young (MODY) in Turkish children by using targeted next-generation sequencing. J. Pediatr. Endocrinol. Metab. 28 (11-12), 1265–1271. 10.1515/jpem-2014-0430 [DOI] [PubMed] [Google Scholar]
- Apelqvist A., Ahlgren U., Edlund H., Ahlgren U. (1997). Sonic hedgehog directs specialised mesoderm differentiation in the intestine and pancreas. Curr. Biol. 7 (10), 801–804. 10.1016/s0960-9822(06)00340-x [DOI] [PubMed] [Google Scholar]
- Apelqvist A., Li H., Sommer L., Beatus P., Anderson D. J., Honjo T., et al. (1999). Notch signalling controls pancreatic cell differentiation. Nature 400 (6747), 877–881. 10.1038/23716 [DOI] [PubMed] [Google Scholar]
- Aydin S., Sagrac D., Sahin F. (2020). Differentiation potential of mesenchymal stem cells into pancreatic beta-cells. Adv. Exp. Med. Biol. 1247, 135–156. 10.1007/5584_2019_476 [DOI] [PubMed] [Google Scholar]
- Banga A., Akinci E., Greder L. V., Dutton J. R., Slack J. M. (2012). In vivo reprogramming of Sox9+ cells in the liver to insulin-secreting ducts. Proc. Natl. Acad. Sci. U. S. A. 109 (38), 15336–15341. 10.1073/pnas.1201701109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barella L. F., Rossi M., Pydi S. P., Meister J., Jain S., Cui Y., et al. (2021). β-Arrestin-1 is required for adaptive β-cell mass expansion during obesity. Nat. Commun. 12 (1), 3385. 10.1038/s41467-021-23656-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barella L. F., Rossi M., Zhu L., Cui Y., Mei F. C., Cheng X., et al. (2019). β-Cell-intrinsic β-arrestin 1 signaling enhances sulfonylurea-induced insulin secretion. J. Clin. Invest. 129 (9), 3732–3737. 10.1172/JCI126309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastidas-Ponce A., Roscioni S. S., Burtscher I., Bader E., Sterr M., Bakhti M., et al. (2017). Foxa2 and Pdx1 cooperatively regulate postnatal maturation of pancreatic beta-cells. Mol. Metab. 6 (6), 524–534. 10.1016/j.molmet.2017.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benner C., van der Meulen T., Caceres E., Tigyi K., Donaldson C. J., Huising M. O. (2014). The transcriptional landscape of mouse beta cells compared to human beta cells reveals notable species differences in long non-coding RNA and protein-coding gene expression. BMC Genomics 15, 620. 10.1186/1471-2164-15-620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ber I., Shternhall K., Perl S., Ohanuna Z., Goldberg I., Barshack I., et al. (2003). Functional, persistent, and extended liver to pancreas transdifferentiation. J. Biol. Chem. 278 (34), 31950–31957. 10.1074/jbc.M303127200 [DOI] [PubMed] [Google Scholar]
- Bernardo A. S., Cho C. H., Mason S., Docherty H. M., Pedersen R. A., Vallier L., et al. (2009). Biphasic induction of Pdx1 in mouse and human embryonic stem cells can mimic development of pancreatic beta-cells. Stem Cells 27 (2), 341–351. 10.1634/stemcells.2008-0310 [DOI] [PubMed] [Google Scholar]
- Campbell S. C., Macfarlane W. M. (2002). Regulation of the pdx1 gene promoter in pancreatic beta-cells. Biochem. Biophys. Res. Commun. 299 (2), 277–284. 10.1016/s0006-291x(02)02633-5 [DOI] [PubMed] [Google Scholar]
- Cao L. Z., Tang D. Q., Horb M. E., Li S. W., Yang L. J. (2004). High glucose is necessary for complete maturation of Pdx1-VP16-expressing hepatic cells into functional insulin-producing cells. Diabetes 53 (12), 3168–3178. 10.2337/diabetes.53.12.3168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinale V., Puca R., Carpino G., Scafetta G., Renzi A., De Canio M., et al. (2015). Adult human biliary tree stem cells differentiate to beta-pancreatic islet cells by treatment with a recombinant human Pdx1 peptide. PLoS One 10 (8), e0134677. 10.1371/journal.pone.0134677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter G., Miladinovic B., Patel A. A., Deland L., Mastorides S., Patel N. A. (2015). Circulating long noncoding RNA GAS5 levels are correlated to prevalence of type 2 diabetes mellitus. BBA Clin. 4, 102–107. 10.1016/j.bbacli.2015.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. J., Finkbeiner S. R., Weinblatt D., Emmett M. J., Tameire F., Yousefi M., et al. (2014). De novo formation of insulin-producing "neo-beta cell islets" from intestinal crypts. Cell Rep. 6 (6), 1046–1058. 10.1016/j.celrep.2014.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y. M., Lim J. M., Yoo D. H., Kim J. H., Chung S. S., Park S. G., et al. (2008). Betacellulin and nicotinamide sustain PDX1 expression and induce pancreatic beta-cell differentiation in human embryonic stem cells. Biochem. Biophys. Res. Commun. 366 (1), 129–134. 10.1016/j.bbrc.2007.11.112 [DOI] [PubMed] [Google Scholar]
- Chun S. Y., Mack D. L., Moorefield E., Oh S. H., Kwon T. G., Pettenati M. J., et al. (2015). Pdx1 and controlled culture conditions induced differentiation of human amniotic fluid-derived stem cells to insulin-producing clusters. J. Tissue Eng. Regen. Med. 9 (5), 540–549. 10.1002/term.1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cim A., Sawyer G. J., Zhang X., Su H., Collins L., Jones P., et al. (2012). In vivo studies on non-viral transdifferentiation of liver cells towards pancreatic beta cells. J. Endocrinol. 214 (3), 277–288. 10.1530/JOE-12-0033 [DOI] [PubMed] [Google Scholar]
- Collombat P., Hecksher-Sorensen J., Krull J., Berger J., Riedel D., Herrera P. L., et al. (2007). Embryonic endocrine pancreas and mature beta cells acquire alpha and PP cell phenotypes upon Arx misexpression. J. Clin. Invest. 117 (4), 961–970. 10.1172/JCI29115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad E., Dai C., Spaeth J., Guo M., Cyphert H. A., Scoville D., et al. (2016). The MAFB transcription factor impacts islet alpha-cell function in rodents and represents a unique signature of primate islet beta-cells. Am. J. Physiol. Endocrinol. Metab. 310 (1), E91–E102. 10.1152/ajpendo.00285.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amour K. A., Agulnick A. D., Eliazer S., Kelly O. G., Kroon E., Baetge E. E. (2005). Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat. Biotechnol. 23 (12), 1534–1541. 10.1038/nbt1163 [DOI] [PubMed] [Google Scholar]
- Dassaye R., Naidoo S., Cerf M. E. (2016). Transcription factor regulation of pancreatic organogenesis, differentiation and maturation. Islets 8 (1), 13–34. 10.1080/19382014.2015.1075687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jesus D. F., Zhang Z., Kahraman S., Brown N. K., Chen M., Hu J., et al. (2019). m(6)A mRNA methylation regulates human beta-cell biology in physiological states and in type 2 diabetes. Nat. Metab. 1 (8), 765–774. 10.1038/s42255-019-0089-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vas M. G., Kopp J. L., Heliot C., Sander M., Cereghini S., Haumaitre C. (2015). Hnf1b controls pancreas morphogenesis and the generation of Ngn3+ endocrine progenitors. Development 142 (5), 871–882. 10.1242/dev.110759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deering T. G., Ogihara T., Trace A. P., Maier B., Mirmira R. G. (2009). Methyltransferase Set7/9 maintains transcription and euchromatin structure at islet-enriched genes. Diabetes 58 (1), 185–193. 10.2337/db08-1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng M., Xiao X., Zhou L., Wang T. (2019). First case report of maturity-onset diabetes of the young type 4 pedigree in a Chinese family. Front. Endocrinol. 10, 406. 10.3389/fendo.2019.00406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding H., Wang F., Shi X., Ma H., Du Y., Hou L., et al. (2020). LncRNA MALAT1 induces the dysfunction of beta cells via reducing the histone acetylation of the PDX-1 promoter in type 1 diabetes. Exp. Mol. Pathol. 114, 104432. 10.1016/j.yexmp.2020.104432 [DOI] [PubMed] [Google Scholar]
- Donelan W., Li S., Wang H., Lu S., Xie C., Tang D., et al. (2015). Pancreatic and duodenal homeobox gene 1 (Pdx1) down-regulates hepatic transcription factor 1 alpha (HNF1α) expression during reprogramming of human hepatic cells into insulin-producing cells. Am. J. Transl. Res. 7 (6), 995–1008. [PMC free article] [PubMed] [Google Scholar]
- Donelan W., Wang H., Li S. W., Pittman D., Li Y., Han S., et al. (2013). Novel detection of pancreatic and duodenal homeobox 1 autoantibodies (PAA) in human sera using luciferase immunoprecipitation systems (LIPS) assay. Int. J. Clin. Exp. Pathol. 6 (6), 1202–1210. [PMC free article] [PubMed] [Google Scholar]
- Elbein S. C., Wang X., Karim M. A., Freedman B. I., Bowden D. W., Shuldiner A. R., et al. (2006). Role of a proline insertion in the insulin promoter factor 1 (IPF1) gene in African Americans with type 2 diabetes. Diabetes 55 (10), 2909–2914. 10.2337/db06-0434 [DOI] [PubMed] [Google Scholar]
- Elrick L. J., Docherty K. (2001). Phosphorylation-dependent nucleocytoplasmic shuttling of pancreatic duodenal homeobox-1. Diabetes 50 (10), 2244–2252. 10.2337/diabetes.50.10.2244 [DOI] [PubMed] [Google Scholar]
- Esguerra J. L. S., Ofori J. K., Nagao M., Shuto Y., Karagiannopoulos A., Fadista J., et al. (2020). Glucocorticoid induces human beta cell dysfunction by involving riborepressor GAS5 LincRNA. Mol. Metab. 32, 160–167. 10.1016/j.molmet.2019.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euskirchen G., Auerbach R. K., Snyder M. (2012). SWI/SNF chromatin-remodeling factors: Multiscale analyses and diverse functions. J. Biol. Chem. 287 (37), 30897–30905. 10.1074/jbc.R111.309302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagerberg L., Hallstrom B. M., Oksvold P., Kampf C., Djureinovic D., Odeberg J., et al. (2014). Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteomics 13 (2), 397–406. 10.1074/mcp.M113.035600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferber S., Halkin A., Cohen H., Ber I., Einav Y., Goldberg I., et al. (2000). Pancreatic and duodenal homeobox gene 1 induces expression of insulin genes in liver and ameliorates streptozotocin-induced hyperglycemia. Nat. Med. 6 (5), 568–572. 10.1038/75050 [DOI] [PubMed] [Google Scholar]
- Frye M., Harada B. T., Behm M., He C. (2018). RNA modifications modulate gene expression during development. Science 361 (6409), 1346–1349. 10.1126/science.aau1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujitani Y., Fujitani S., Boyer D. F., Gannon M., Kawaguchi Y., Ray M., et al. (2006). Targeted deletion of a cis-regulatory region reveals differential gene dosage requirements for Pdx1 in foregut organ differentiation and pancreas formation. Genes Dev. 20 (2), 253–266. 10.1101/gad.1360106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Yuan Y., Chen Y. (2018). PDX1 transfection induces human adipose derived stem cells differentiation into islet-like cells: What is the benefit for diabetic rats? Pharmazie 73 (4), 213–217. 10.1691/ph.2018.7993 [DOI] [PubMed] [Google Scholar]
- Gao N., LeLay J., Vatamaniuk M. Z., Rieck S., Friedman J. R., Kaestner K. H. (2008). Dynamic regulation of Pdx1 enhancers by Foxa1 and Foxa2 is essential for pancreas development. Genes Dev. 22 (24), 3435–3448. 10.1101/gad.1752608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao T., McKenna B., Li C., Reichert M., Nguyen J., Singh T., et al. (2014). Pdx1 maintains beta cell identity and function by repressing an alpha cell program. Cell Metab. 19 (2), 259–271. 10.1016/j.cmet.2013.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrish K., Gannon M., Shih D., Henderson E., Stoffel M., Wright C. V., et al. (2000). Pancreatic beta cell-specific transcription of the pdx-1 gene. The role of conserved upstream control regions and their hepatic nuclear factor 3beta sites. J. Biol. Chem. 275 (5), 3485–3492. 10.1074/jbc.275.5.3485 [DOI] [PubMed] [Google Scholar]
- Gerrish K., Van Velkinburgh J. C., Stein R. (2004). Conserved transcriptional regulatory domains of the pdx-1 gene. Mol. Endocrinol. 18 (3), 533–548. 10.1210/me.2003-0371 [DOI] [PubMed] [Google Scholar]
- Geusz R. J., Wang A., Lam D. K., Vinckier N. K., Alysandratos K. D., Roberts D. A., et al. (2021). Sequence logic at enhancers governs a dual mechanism of endodermal organ fate induction by FOXA pioneer factors. Nat. Commun. 12 (1), 6636. 10.1038/s41467-021-26950-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gragnoli C., Stanojevic V., Gorini A., Von Preussenthal G. M., Thomas M. K., Habener J. F. (2005). IPF-1/MODY4 gene missense mutation in an Italian family with type 2 and gestational diabetes. Metabolism. 54 (8), 983–988. 10.1016/j.metabol.2005.01.037 [DOI] [PubMed] [Google Scholar]
- Guo S., Dai C., Guo M., Taylor B., Harmon J. S., Sander M., et al. (2013). Inactivation of specific beta cell transcription factors in type 2 diabetes. J. Clin. Invest. 123 (8), 3305–3316. 10.1172/JCI65390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hang Y., Stein R. (2011). MafA and MafB activity in pancreatic beta cells. Trends Endocrinol. Metab. 22 (9), 364–373. 10.1016/j.tem.2011.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hang Y., Yamamoto T., Benninger R. K., Brissova M., Guo M., Bush W., et al. (2014). The MafA transcription factor becomes essential to islet beta-cells soon after birth. Diabetes 63 (6), 1994–2005. 10.2337/db13-1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He D., Wang J., Gao Y., Zhang Y. (2011). Differentiation of PDX1 gene-modified human umbilical cord mesenchymal stem cells into insulin-producing cells in vitro . Int. J. Mol. Med. 28 (6), 1019–1024. 10.3892/ijmm.2011.774 [DOI] [PubMed] [Google Scholar]
- Hebrok M. (2012). Generating beta cells from stem cells-the story so far. Cold Spring Harb. Perspect. Med. 2 (6), a007674. 10.1101/cshperspect.a007674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebrok M., Kim S. K., Melton D. A. (1998). Notochord repression of endodermal Sonic hedgehog permits pancreas development. Genes Dev. 12 (11), 1705–1713. 10.1101/gad.12.11.1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henley K. D., Stanescu D. E., Kropp P. A., Wright C. V. E., Won K. J., Stoffers D. A., et al. (2016). Threshold-dependent cooperativity of Pdx1 and Oc1 in pancreatic progenitors establishes competency for endocrine differentiation and beta-cell function. Cell Rep. 15 (12), 2637–2650. 10.1016/j.celrep.2016.05.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hering B. J., Clarke W. R., Bridges N. D., Eggerman T. L., Alejandro R., Bellin M. D., et al. (2016). Phase 3 trial of transplantation of human islets in type 1 diabetes complicated by severe hypoglycemia. Diabetes Care 39 (7), 1230–1240. 10.2337/dc15-1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey R. D., Galivo F., Schug J., Brehm M. A., Haft A., Wang Y., et al. (2013). Generation of islet-like cells from mouse gall bladder by direct ex vivo reprogramming. Stem Cell Res. 11 (1), 503–515. 10.1016/j.scr.2013.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogrebe N. J., Augsornworawat P., Maxwell K. G., Velazco-Cruz L., Millman J. R. (2020). Targeting the cytoskeleton to direct pancreatic differentiation of human pluripotent stem cells. Nat. Biotechnol. 38 (4), 460–470. 10.1038/s41587-020-0430-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horb M. E., Shen C. N., Tosh D., Slack J. M. (2003). Experimental conversion of liver to pancreas. Curr. Biol. 13 (2), 105–115. 10.1016/s0960-9822(02)01434-3 [DOI] [PubMed] [Google Scholar]
- Hui H., Perfetti R. (2002). Pancreas duodenum homeobox-1 regulates pancreas development during embryogenesis and islet cell function in adulthood. Eur. J. Endocrinol. 146 (2), 129–141. 10.1530/eje.0.1460129 [DOI] [PubMed] [Google Scholar]
- Imai J., Katagiri H., Yamada T., Ishigaki Y., Ogihara T., Uno K., et al. (2005). Constitutively active PDX1 induced efficient insulin production in adult murine liver. Biochem. Biophys. Res. Commun. 326 (2), 402–409. 10.1016/j.bbrc.2004.11.047 [DOI] [PubMed] [Google Scholar]
- Inoue H., Riggs A. C., Tanizawa Y., Ueda K., Kuwano A., Liu L., et al. (1996). Isolation, characterization, and chromosomal mapping of the human insulin promoter factor 1 (IPF-1) gene. Diabetes 45 (6), 789–794. 10.2337/diab.45.6.789 [DOI] [PubMed] [Google Scholar]
- Jara M. A., Werneck-De-Castro J. P., Lubaczeuski C., Johnson J. D., Bernal-Mizrachi E. (2020). Pancreatic and duodenal homeobox-1 (PDX1) contributes to beta-cell mass expansion and proliferation induced by Akt/PKB pathway. Islets 12 (2), 32–40. 10.1080/19382014.2020.1762471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings R. E., Berry A. A., Kirkwood-Wilson R., Roberts N. A., Hearn T., Salisbury R. J., et al. (2013). Development of the human pancreas from foregut to endocrine commitment. Diabetes 62 (10), 3514–3522. 10.2337/db12-1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen J., Pedersen E. E., Galante P., Hald J., Heller R. S., Ishibashi M., et al. (2000). Control of endodermal endocrine development by Hes-1. Nat. Genet. 24 (1), 36–44. 10.1038/71657 [DOI] [PubMed] [Google Scholar]
- Jetton T. L., Flores-Bringas P., Leahy J. L., Gupta D. (2021). SetD7 (Set7/9) is a novel target of PPARγ that promotes the adaptive pancreatic β-cell glycemic response. J. Biol. Chem. 297 (5), 101250. 10.1016/j.jbc.2021.101250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin F., Wang N., Zhu Y., You L., Wang L., De W., et al. (2017). Downregulation of long noncoding RNA Gas5 affects cell cycle and insulin secretion in mouse pancreatic beta cells. Cell. Physiol. biochem. 43 (5), 2062–2073. 10.1159/000484191 [DOI] [PubMed] [Google Scholar]
- Jonsson J., Carlsson L., Edlund T., Edlund H. (1994). Insulin-promoter-factor 1 is required for pancreas development in mice. Nature 371 (6498), 606–609. 10.1038/371606a0 [DOI] [PubMed] [Google Scholar]
- Kaestner K. H. (2000). The hepatocyte nuclear factor 3 (HNF3 or FOXA) family in metabolism. Trends Endocrinol. Metab. 11 (7), 281–285. 10.1016/s1043-2760(00)00271-x [DOI] [PubMed] [Google Scholar]
- Kaneto H., Miyatsuka T., Kawamori D., Yamamoto K., Kato K., Shiraiwa T., et al. (2008). PDX-1 and MafA play a crucial role in pancreatic beta-cell differentiation and maintenance of mature beta-cell function. Endocr. J. 55 (2), 235–252. 10.1507/endocrj.k07e-041 [DOI] [PubMed] [Google Scholar]
- Kaneto H., Miyatsuka T., Shiraiwa T., Yamamoto K., Kato K., Fujitani Y., et al. (2007). Crucial role of PDX-1 in pancreas development, beta-cell differentiation, and induction of surrogate beta-cells. Curr. Med. Chem. 14 (16), 1745–1752. 10.2174/092986707781058887 [DOI] [PubMed] [Google Scholar]
- Kaneto H., Nakatani Y., Miyatsuka T., Matsuoka T. A., Matsuhisa M., Hori M., et al. (2005). PDX-1/VP16 fusion protein, together with NeuroD or Ngn3, markedly induces insulin gene transcription and ameliorates glucose tolerance. Diabetes 54 (4), 1009–1022. 10.2337/diabetes.54.4.1009 [DOI] [PubMed] [Google Scholar]
- Karim M. A., Wang X., Hale T. C., Elbein S. C. (2005). Insulin Promoter Factor 1 variation is associated with type 2 diabetes in African Americans. BMC Med. Genet. 6, 37. 10.1186/1471-2350-6-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerner W., Bruckel J., German Diabetes A. (2014). Definition, classification and diagnosis of diabetes mellitus. Exp. Clin. Endocrinol. Diabetes 122 (7), 384–386. 10.1055/s-0034-1366278 [DOI] [PubMed] [Google Scholar]
- Kim H., Kulkarni R. N. (2020). Epigenetics in beta-cell adaptation and type 2 diabetes. Curr. Opin. Pharmacol. 55, 125–131. 10.1016/j.coph.2020.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. K., Hebrok M., Melton D. A. (1997). Notochord to endoderm signaling is required for pancreas development. Development 124 (21), 4243–4252. 10.1242/dev.124.21.4243 [DOI] [PubMed] [Google Scholar]
- Kino T., Hurt D. E., Ichijo T., Nader N., Chrousos G. P. (2010). Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 3 (107). 10.1126/scisignal.2000568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropp P. A., Dunn J. C., Carboneau B. A., Stoffers D. A., Gannon M. (2018). Cooperative function of Pdx1 and Oc1 in multipotent pancreatic progenitors impacts postnatal islet maturation and adaptability. Am. J. Physiol. Endocrinol. Metab. 314 (4), E308–E321. 10.1152/ajpendo.00260.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Jordan N., Melton D., Grapin-Botton A. (2003). Signals from lateral plate mesoderm instruct endoderm toward a pancreatic fate. Dev. Biol. 259 (1), 109–122. 10.1016/s0012-1606(03)00183-0 [DOI] [PubMed] [Google Scholar]
- Lander E. S., Linton L. M., Birren B., Nusbaum C., Zody M. C., Baldwin J., et al. (2001). Initial sequencing and analysis of the human genome. Nature 409 (6822), 860–921. 10.1038/35057062 [DOI] [PubMed] [Google Scholar]
- Lee K., Cho H., Rickert R. W., Li Q. V., Pulecio J., Leslie C. S., et al. (2019). FOXA2 is required for enhancer priming during pancreatic differentiation. Cell Rep. 28 (2), 382–393. 10.1016/j.celrep.2019.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. W., Koya V., Li Y., Donelan W., Lin P., Reeves W. H., et al. (2010). Pancreatic duodenal homeobox 1 protein is a novel beta-cell-specific autoantigen for type I diabetes. Lab. Invest. 90 (1), 31–39. 10.1038/labinvest.2009.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lickert H., Kutsch S., Kanzler B., Tamai Y., Taketo M. M., Kemler R. (2002). Formation of multiple hearts in mice following deletion of beta-catenin in the embryonic endoderm. Dev. Cell 3 (2), 171–181. 10.1016/s1534-5807(02)00206-x [DOI] [PubMed] [Google Scholar]
- Lima M. J., Muir K. R., Docherty H. M., Drummond R., McGowan N. W., Forbes S., et al. (2013). Suppression of epithelial-to-mesenchymal transitioning enhances ex vivo reprogramming of human exocrine pancreatic tissue toward functional insulin-producing beta-like cells. Diabetes 62 (8), 2821–2833. 10.2337/db12-1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima M. J., Muir K. R., Docherty H. M., McGowan N. W., Forbes S., Heremans Y., et al. (2016). Generation of functional beta-like cells from human exocrine pancreas. PLoS One 11 (5), e0156204. 10.1371/journal.pone.0156204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Lang G., Shi J. (2021). Epigenetic regulation of PDX-1 in type 2 diabetes mellitus. Diabetes Metab. Syndr. Obes. 14, 431–442. 10.2147/DMSO.S291932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loretelli C., Assi E., Seelam A. J., Ben Nasr M., Fiorina P. (2020). Cell therapy for type 1 diabetes. Expert Opin. Biol. Ther. 20 (8), 887–897. 10.1080/14712598.2020.1748596 [DOI] [PubMed] [Google Scholar]
- Lumelsky N., Blondel O., Laeng P., Velasco I., Ravin R., McKay R. (2001). Differentiation of embryonic stem cells to insulin-secreting structures similar to pancreatic islets. Science 292 (5520), 1389–1394. 10.1126/science.1058866 [DOI] [PubMed] [Google Scholar]
- Lupp S., Gotz C., Khadouma S., Horbach T., Dimova E. Y., Bohrer A. M., et al. (2014). The upstream stimulatory factor USF1 is regulated by protein kinase CK2 phosphorylation. Cell. Signal. 26 (12), 2809–2817. 10.1016/j.cellsig.2014.08.028 [DOI] [PubMed] [Google Scholar]
- Lynn F. C., Smith S. B., Wilson M. E., Yang K. Y., Nekrep N., German M. S. (2007). Sox9 coordinates a transcriptional network in pancreatic progenitor cells. Proc. Natl. Acad. Sci. U. S. A. 104 (25), 10500–10505. 10.1073/pnas.0704054104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Fei X. (2018). SIRT5 regulates pancreatic beta-cell proliferation and insulin secretion in type 2 diabetes. Exp. Ther. Med. 16 (2), 1417–1425. 10.3892/etm.2018.6301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maganti A. V., Maier B., Tersey S. A., Sampley M. L., Mosley A. L., Ozcan S., et al. (2015). Transcriptional activity of the islet beta cell factor Pdx1 is augmented by lysine methylation catalyzed by the methyltransferase Set7/9. J. Biol. Chem. 290 (15), 9812–9822. 10.1074/jbc.M114.616219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshak S., Benshushan E., Shoshkes M., Havin L., Cerasi E., Melloul D. (2000). Functional conservation of regulatory elements in the pdx-1 gene: PDX-1 and hepatocyte nuclear factor 3beta transcription factors mediate beta-cell-specific expression. Mol. Cell. Biol. 20 (20), 7583–7590. 10.1128/MCB.20.20.7583-7590.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massari M. E., Murre C. (2000). Helix-loop-helix proteins: Regulators of transcription in eucaryotic organisms. Mol. Cell. Biol. 20 (2), 429–440. 10.1128/MCB.20.2.429-440.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell K. G., Millman J. R. (2021). Applications of iPSC-derived beta cells from patients with diabetes. Cell Rep. Med. 2 (4), 100238. 10.1016/j.xcrm.2021.100238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy M. I., Zeggini E. (2009). Genome-wide association studies in type 2 diabetes. Curr. Diab. Rep. 9 (2), 164–171. 10.1007/s11892-009-0027-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna B., Guo M., Reynolds A., Hara M., Stein R. (2015). Dynamic recruitment of functionally distinct Swi/Snf chromatin remodeling complexes modulates Pdx1 activity in islet beta cells. Cell Rep. 10 (12), 2032–2042. 10.1016/j.celrep.2015.02.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melloul D., Marshak S., Cerasi E. (2002). Regulation of pdx-1 gene expression. Diabetes 51 (3), S320–S325. 10.2337/diabetes.51.2007.s320 [DOI] [PubMed] [Google Scholar]
- Memon B., Abdelalim E. M. (2020). Stem cell therapy for diabetes: Beta cells versus pancreatic progenitors. Cells 9 (2), 283. 10.3390/cells9020283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memon B., Younis I., Abubaker F., Abdelalim E. M. (2021). PDX1(-)/NKX6.1(+) progenitors derived from human pluripotent stem cells as a novel source of insulin-secreting cells. Diabetes. Metab. Res. Rev. 37 (5), e3400. 10.1002/dmrr.3400 [DOI] [PubMed] [Google Scholar]
- Meng R., Al-Quobaili F., Muller I., Gotz C., Thiel G., Montenarh M. (2010). CK2 phosphorylation of Pdx-1 regulates its transcription factor activity. Cell. Mol. Life Sci. 67 (14), 2481–2489. 10.1007/s00018-010-0348-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger D. E., Liu C., Ziaie A. S., Naji A., Zaret K. S. (2014). Grg3/TLE3 and Grg1/TLE1 induce monohormonal pancreatic beta-cells while repressing alpha-cell functions. Diabetes 63 (5), 1804–1816. 10.2337/db13-0867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran I., Akerman I., van de Bunt M., Xie R., Benazra M., Nammo T., et al. (2012). Human beta cell transcriptome analysis uncovers lncRNAs that are tissue-specific, dynamically regulated, and abnormally expressed in type 2 diabetes. Cell Metab. 16 (4), 435–448. 10.1016/j.cmet.2012.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosley A. L., Corbett J. A., Ozcan S. (2004). Glucose regulation of insulin gene expression requires the recruitment of p300 by the beta-cell-specific transcription factor Pdx-1. Mol. Endocrinol. 18 (9), 2279–2290. 10.1210/me.2003-0463 [DOI] [PubMed] [Google Scholar]
- Mosley A. L., Ozcan S. (2004). The pancreatic duodenal homeobox-1 protein (Pdx-1) interacts with histone deacetylases Hdac-1 and Hdac-2 on low levels of glucose. J. Biol. Chem. 279 (52), 54241–54247. 10.1074/jbc.M410379200 [DOI] [PubMed] [Google Scholar]
- Nasteska D., Fine N. H. F., Ashford F. B., Cuozzo F., Viloria K., Smith G., et al. (2021). PDX1(LOW) MAFA(LOW) beta-cells contribute to islet function and insulin release. Nat. Commun. 12 (1), 674. 10.1038/s41467-020-20632-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naya F. J., Huang H. P., Qiu Y., Mutoh H., DeMayo F. J., Leiter A. B., et al. (1997). Diabetes, defective pancreatic morphogenesis, and abnormal enteroendocrine differentiation in BETA2/neuroD-deficient mice. Genes Dev. 11 (18), 2323–2334. 10.1101/gad.11.18.2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolino M., Claiborn K. C., Senee V., Boland A., Stoffers D. A., Julier C. (2010). A novel hypomorphic PDX1 mutation responsible for permanent neonatal diabetes with subclinical exocrine deficiency. Diabetes 59 (3), 733–740. 10.2337/db09-1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura W., Kondo T., Salameh T., El Khattabi I., Dodge R., Bonner-Weir S., et al. (2006). A switch from MafB to MafA expression accompanies differentiation to pancreatic beta-cells. Dev. Biol. 293 (2), 526–539. 10.1016/j.ydbio.2006.02.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlsson H., Karlsson K., Edlund T. (1993). IPF1, a homeodomain-containing transactivator of the insulin gene. EMBO J. 12 (11), 4251–4259. 10.1002/j.1460-2075.1993.tb06109.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver-Krasinski J. M., Kasner M. T., Yang J., Crutchlow M. F., Rustgi A. K., Kaestner K. H., et al. (2009). The diabetes gene Pdx1 regulates the transcriptional network of pancreatic endocrine progenitor cells in mice. J. Clin. Invest. 119 (7), 1888–1898. 10.1172/JCI37028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquali L., Gaulton K. J., Rodriguez-Segui S. A., Mularoni L., Miguel-Escalada I., Akerman I., et al. (2014). Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants. Nat. Genet. 46 (2), 136–143. 10.1038/ng.2870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Villamil B., Schwartz P. T., Vallejo M. (1999). The pancreatic homeodomain transcription factor IDX1/IPF1 is expressed in neural cells during brain development. Endocrinology 140 (8), 3857–3860. 10.1210/endo.140.8.7048 [DOI] [PubMed] [Google Scholar]
- Pierce K. L., Lefkowitz R. J. (2001). Classical and new roles of beta-arrestins in the regulation of G-protein-coupled receptors. Nat. Rev. Neurosci. 2 (10), 727–733. 10.1038/35094577 [DOI] [PubMed] [Google Scholar]
- Pinzon-Cortes J. A., Perna-Chaux A., Rojas-Villamizar N. S., Diaz-Basabe A., Polania-Villanueva D. C., Jacome M. F., et al. (2017). Effect of diabetes status and hyperglycemia on global DNA methylation and hydroxymethylation. Endocr. Connect. 6 (8), 708–725. 10.1530/EC-17-0199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pott S., Lieb J. D. (2015). What are super-enhancers? Nat. Genet. 47 (1), 8–12. 10.1038/ng.3167 [DOI] [PubMed] [Google Scholar]
- Qin Y., Xiao L., Zhan X. B., Zhou H. X. (2015). Pdxl and its role in activating Ngn3 and Pax6 to induce differentiation of iPSCs into islet beta cells. Genet. Mol. Res. 14 (3), 8892–8900. 10.4238/2015.August.3.12 [DOI] [PubMed] [Google Scholar]
- Rafiq I., Kennedy H. J., Rutter G. A. (1998). Glucose-dependent translocation of insulin promoter factor-1 (IPF-1) between the nuclear periphery and the nucleoplasm of single MIN6 beta-cells. J. Biol. Chem. 273 (36), 23241–23247. 10.1074/jbc.273.36.23241 [DOI] [PubMed] [Google Scholar]
- Rajaei B., Shamsara M., Sanati M. H. (2018). In vitro generation of glucose-responsive insulin-secreting cells from PDX1-overexpressing human-induced pluripotent stem cell derived from diabetic patient. ASAIO J. 64 (6), 819–826. 10.1097/MAT.0000000000000728 [DOI] [PubMed] [Google Scholar]
- Ritz-Laser B., Gauthier B. R., Estreicher A., Mamin A., Brun T., Ris F., et al. (2003). Ectopic expression of the beta-cell specific transcription factor Pdx1 inhibits glucagon gene transcription. Diabetologia 46 (6), 810–821. 10.1007/s00125-003-1115-7 [DOI] [PubMed] [Google Scholar]
- Ruan Y., Lin N., Ma Q., Chen R., Zhang Z., Wen W., et al. (2018). Circulating LncRNAs analysis in patients with type 2 diabetes reveals novel genes influencing glucose metabolism and islet beta-cell function. Cell. Physiol. biochem. 46 (1), 335–350. 10.1159/000488434 [DOI] [PubMed] [Google Scholar]
- Ryan E. A., Paty B. W., Senior P. A., Bigam D., Alfadhli E., Kneteman N. M., et al. (2005). Five-year follow-up after clinical islet transplantation. Diabetes 54 (7), 2060–2069. 10.2337/diabetes.54.7.2060 [DOI] [PubMed] [Google Scholar]
- Salguero-Aranda C., Tapia-Limonchi R., Cahuana G. M., Hitos A. B., Diaz I., Hmadcha A., et al. (2016). Differentiation of mouse embryonic stem cells toward functional pancreatic beta-cell surrogates through epigenetic regulation of pdx1 by nitric oxide. Cell Transpl. 25 (10), 1879–1892. 10.3727/096368916X691178 [DOI] [PubMed] [Google Scholar]
- Samaras S. E., Zhao L., Means A., Henderson E., Matsuoka T. A., Stein R. (2003). The islet beta cell-enriched RIPE3b1/Maf transcription factor regulates pdx-1 expression. J. Biol. Chem. 278 (14), 12263–12270. 10.1074/jbc.M210801200 [DOI] [PubMed] [Google Scholar]
- Sathishkumar C., Prabu P., Mohan V., Balasubramanyam M. (2018). Linking a role of lncRNAs (long non-coding RNAs) with insulin resistance, accelerated senescence, and inflammation in patients with type 2 diabetes. Hum. Genomics 12 (1), 41. 10.1186/s40246-018-0173-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena P., Heng B. C., Bai P., Folcher M., Zulewski H., Fussenegger M. (2016). A programmable synthetic lineage-control network that differentiates human IPSCs into glucose-sensitive insulin-secreting beta-like cells. Nat. Commun. 7, 11247. 10.1038/ncomms11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer A. E., Freude K. K., Nelson S. B., Sander M. (2010). Nkx6 transcription factors and Ptf1a function as antagonistic lineage determinants in multipotent pancreatic progenitors. Dev. Cell 18 (6), 1022–1029. 10.1016/j.devcel.2010.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwitzgebel V. M., Mamin A., Brun T., Ritz-Laser B., Zaiko M., Maret A., et al. (2003). Agenesis of human pancreas due to decreased half-life of insulin promoter factor 1. J. Clin. Endocrinol. Metab. 88 (9), 4398–4406. 10.1210/jc.2003-030046 [DOI] [PubMed] [Google Scholar]
- Scuteri A., Monfrini M. (2018). Mesenchymal stem cells as new therapeutic approach for diabetes and pancreatic disorders. Int. J. Mol. Sci. 19 (9), 2783. 10.3390/ijms19092783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S., Jhala U. S., Johnson T., Ferreri K., Leonard J., Montminy M. (1997). Hormonal regulation of an islet-specific enhancer in the pancreatic homeobox gene STF-1. Mol. Cell. Biol. 17 (5), 2598–2604. 10.1128/MCB.17.5.2598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S., Leonard J., Lee S., Chapman H. D., Leiter E. H., Montminy M. R. (1996). Pancreatic islet expression of the homeobox factor STF-1 relies on an E-box motif that binds USF. J. Biol. Chem. 271 (4), 2294–2299. 10.1074/jbc.271.4.2294 [DOI] [PubMed] [Google Scholar]
- Shih H. P., Seymour P. A., Patel N. A., Xie R., Wang A., Liu P. P., et al. (2015). A gene regulatory network cooperatively controlled by Pdx1 and Sox9 governs lineage allocation of foregut progenitor cells. Cell Rep. 13 (2), 326–336. 10.1016/j.celrep.2015.08.082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih H. P., Wang A., Sander M. (2013). Pancreas organogenesis: From lineage determination to morphogenesis. Annu. Rev. Cell Dev. Biol. 29, 81–105. 10.1146/annurev-cellbio-101512-122405 [DOI] [PubMed] [Google Scholar]
- Shukla A. K., Xiao K., Lefkowitz R. J. (2011). Emerging paradigms of beta-arrestin-dependent seven transmembrane receptor signaling. Trends biochem. Sci. 36 (9), 457–469. 10.1016/j.tibs.2011.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorli C., Heile M. K. (2014). Identifying and meeting the challenges of insulin therapy in type 2 diabetes. J. Multidiscip. Healthc. 7, 267–282. 10.2147/JMDH.S64084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaeth J. M., Gupte M., Perelis M., Yang Y. P., Cyphert H., Guo S., et al. (2017). Defining a novel role for the Pdx1 transcription factor in islet beta-cell maturation and proliferation during weaning. Diabetes 66 (11), 2830–2839. 10.2337/db16-1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaeth J. M., Walker E. M., Stein R. (2016). Impact of Pdx1-associated chromatin modifiers on islet beta-cells. Diabetes Obes. Metab. 18 (1), 123–127. 10.1111/dom.12730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence J. R., Wells J. M. (2007). Translational embryology: Using embryonic principles to generate pancreatic endocrine cells from embryonic stem cells. Dev. Dyn. 236 (12), 3218–3227. 10.1002/dvdy.21366 [DOI] [PubMed] [Google Scholar]
- Spohrer S., Gross R., Nalbach L., Schwind L., Stumpf H., Menger M. D., et al. (2017). Functional interplay between the transcription factors USF1 and PDX-1 and protein kinase CK2 in pancreatic beta-cells. Sci. Rep. 7 (1), 16367. 10.1038/s41598-017-16590-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanojevic V., Habener J. F., Thomas M. K. (2004). Pancreas duodenum homeobox-1 transcriptional activation requires interactions with p300. Endocrinology 145 (6), 2918–2928. 10.1210/en.2003-1188 [DOI] [PubMed] [Google Scholar]
- Stoffel M., Stein R., Wright C. V., Espinosa R., 3rd, Le Beau M. M., Bell G. I. (1995). Localization of human homeodomain transcription factor insulin promoter factor 1 (IPF1) to chromosome band 13q12.1. Genomics 28 (1), 125–126. 10.1006/geno.1995.1120 [DOI] [PubMed] [Google Scholar]
- Stoffers D. A., Zinkin N. T., Stanojevic V., Clarke W. L., Habener J. F. (1997). Pancreatic agenesis attributable to a single nucleotide deletion in the human IPF1 gene coding sequence. Nat. Genet. 15 (1), 106–110. 10.1038/ng0197-106 [DOI] [PubMed] [Google Scholar]
- Stolovich-Rain M., Enk J., Vikesa J., Nielsen F. C., Saada A., Glaser B., et al. (2015). Weaning triggers a maturation step of pancreatic beta cells. Dev. Cell 32 (5), 535–545. 10.1016/j.devcel.2015.01.002 [DOI] [PubMed] [Google Scholar]
- Sun Z. Y., Yu T. Y., Jiang F. X., Wang W. (2021). Functional maturation of immature beta cells: A roadblock for stem cell therapy for type 1 diabetes. World J. Stem Cells 13 (3), 193–207. 10.4252/wjsc.v13.i3.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabat M., Luciani D. S., Piret J. M., Johnson J. D. (2009). Maturation of adult beta-cells revealed using a Pdx1/insulin dual-reporter lentivirus. Endocrinology 150 (4), 1627–1635. 10.1210/en.2008-1224 [DOI] [PubMed] [Google Scholar]
- Tang D. Q., Lu S., Sun Y. P., Rodrigues E., Chou W., Yang C., et al. (2006). Reprogramming liver-stem WB cells into functional insulin-producing cells by persistent expression of Pdx1- and Pdx1-VP16 mediated by lentiviral vectors. Lab. Invest. 86 (1), 83–93. 10.1038/labinvest.3700368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas H., Jaschkowitz K., Bulman M., Frayling T. M., Mitchell S. M., Roosen S., et al. (2001). A distant upstream promoter of the HNF-4alpha gene connects the transcription factors involved in maturity-onset diabetes of the young. Hum. Mol. Genet. 10 (19), 2089–2097. 10.1093/hmg/10.19.2089 [DOI] [PubMed] [Google Scholar]
- Tripathi V., Ellis J. D., Shen Z., Song D. Y., Pan Q., Watt A. T., et al. (2010). The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 39 (6), 925–938. 10.1016/j.molcel.2010.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Velkinburgh J. C., Samaras S. E., Gerrish K., Artner I., Stein R. (2005). Interactions between areas I and II direct pdx-1 expression specifically to islet cell types of the mature and developing pancreas. J. Biol. Chem. 280 (46), 38438–38444. 10.1074/jbc.M508594200 [DOI] [PubMed] [Google Scholar]
- Vincent S. D., Dunn N. R., Hayashi S., Norris D. P., Robertson E. J. (2003). Cell fate decisions within the mouse organizer are governed by graded Nodal signals. Genes Dev. 17 (13), 1646–1662. 10.1101/gad.1100503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkov P., Bacos K., Ofori J. K., Esguerra J. L., Eliasson L., Ronn T., et al. (2017). Whole-Genome bisulfite sequencing of human pancreatic islets reveals novel differentially methylated regions in type 2 diabetes pathogenesis. Diabetes 66 (4), 1074–1085. 10.2337/db16-0996 [DOI] [PubMed] [Google Scholar]
- Walczak M. P., Drozd A. M., Stoczynska-Fidelus E., Rieske P., Grzela D. P. (2016). Directed differentiation of human iPSC into insulin producing cells is improved by induced expression of PDX1 and NKX6.1 factors in IPC progenitors. J. Transl. Med. 14 (1), 341. 10.1186/s12967-016-1097-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A. Y., Ehrhardt A., Xu H., Kay M. A. (2007). Adenovirus transduction is required for the correction of diabetes using Pdx-1 or Neurogenin-3 in the liver. Mol. Ther. 15 (2), 255–263. 10.1038/sj.mt.6300032 [DOI] [PubMed] [Google Scholar]
- Wang N., Tong R., Xu J., Tian Y., Pan J., Cui J., et al. (2021). PDX1 and MC4R genetic polymorphisms are associated with type 2 diabetes mellitus risk in the Chinese Han population. BMC Med. Genomics 14 (1), 249. 10.1186/s12920-021-01037-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R. H., Xu X., Kim H. S., Xiao Z., Deng C. X. (2013). SIRT1 deacetylates FOXA2 and is critical for Pdx1 transcription and beta-cell formation. Int. J. Biol. Sci. 9 (9), 934–946. 10.7150/ijbs.7529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Sterr M., AnsarullahBurtscher I., Bottcher A., Beckenbauer J., et al. (2019). Point mutations in the PDX1 transactivation domain impair human beta-cell development and function. Mol. Metab. 24, 80–97. 10.1016/j.molmet.2019.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Sterr M., Burtscher I., Chen S., Hieronimus A., Machicao F., et al. (2018). Genome-wide analysis of PDX1 target genes in human pancreatic progenitors. Mol. Metab. 9, 57–68. 10.1016/j.molmet.2018.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir G. C., Bonner-Weir S., Leahy J. L. (1990). Islet mass and function in diabetes and transplantation. Diabetes 39 (4), 401–405. 10.2337/diab.39.4.401 [DOI] [PubMed] [Google Scholar]
- Wells J. M., Melton D. A. (1999). Vertebrate endoderm development. Annu. Rev. Cell Dev. Biol. 15, 393–410. 10.1146/annurev.cellbio.15.1.393 [DOI] [PubMed] [Google Scholar]
- Wiebe P. O., Kormish J. D., Roper V. T., Fujitani Y., Alston N. I., Zaret K. S., et al. (2007). Ptf1a binds to and activates area III, a highly conserved region of the Pdx1 promoter that mediates early pancreas-wide Pdx1 expression. Mol. Cell. Biol. 27 (11), 4093–4104. 10.1128/MCB.01978-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong M. S., Hawthorne W. J., Manolios N. (2010). Gene therapy in diabetes. Self. Nonself. 1 (3), 165–175. 10.4161/self.1.3.12643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright C. V., Schnegelsberg P., De Robertis E. M. (1989). XlHbox 8: A novel Xenopus homeo protein restricted to a narrow band of endoderm. Development 105 (4), 787–794. 10.1242/dev.105.4.787 [DOI] [PubMed] [Google Scholar]
- Wu K. L., Gannon M., Peshavaria M., Offield M. F., Henderson E., Ray M., et al. (1997). Hepatocyte nuclear factor 3beta is involved in pancreatic beta-cell-specific transcription of the pdx-1 gene. Mol. Cell. Biol. 17 (10), 6002–6013. 10.1128/MCB.17.10.6002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X., Guo P., Shiota C., Zhang T., Coudriet G. M., Fischbach S., et al. (2018). Endogenous reprogramming of alpha cells into beta cells, induced by viral gene therapy, reverses autoimmune diabetes. Cell Stem Cell 22 (1), 78–90. 10.1016/j.stem.2017.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Tsang K. S., Chan J. C., Yuan P., Fan R., Kaneto H., et al. (2013). The combined expression of Pdx1 and MafA with either Ngn3 or NeuroD improves the differentiation efficiency of mouse embryonic stem cells into insulin-producing cells. Cell Transpl. 22 (1), 147–158. 10.3727/096368912X653057 [DOI] [PubMed] [Google Scholar]
- Xu L., Xu C., Zhou S., Liu X., Wang J., Liu X., et al. (2017). PAX4 promotes PDX1-induced differentiation of mesenchymal stem cells into insulin-secreting cells. Am. J. Transl. Res. 9 (3), 874–886. [PMC free article] [PubMed] [Google Scholar]
- Yang B. T., Dayeh T. A., Volkov P. A., Kirkpatrick C. L., Malmgren S., Jing X., et al. (2012). Increased DNA methylation and decreased expression of PDX-1 in pancreatic islets from patients with type 2 diabetes. Mol. Endocrinol. 26 (7), 1203–1212. 10.1210/me.2012-1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Chi Y., Meng Y., Chen Z., Xiang R., Yan H., et al. (2020). FAM3A plays crucial roles in controlling PDX1 and insulin expressions in pancreatic beta cells. FASEB J. 34 (3), 3915–3931. 10.1096/fj.201902368RR [DOI] [PubMed] [Google Scholar]
- Yang Y. P., Magnuson M. A., Stein R., Wright C. V. (2017). The mammal-specific Pdx1 Area II enhancer has multiple essential functions in early endocrine cell specification and postnatal beta-cell maturation. Development 144 (2), 248–257. 10.1242/dev.143123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y. P., Thorel F., Boyer D. F., Herrera P. L., Wright C. V. (2011). Context-specific alpha- to-beta-cell reprogramming by forced Pdx1 expression. Genes Dev. 25 (16), 1680–1685. 10.1101/gad.16875711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiji S., Horikawa Y., Kubota S., Enya M., Iwasaki Y., Keidai Y., et al. (2022). First Japanese family with PDX1-MODY (MODY4): A novel PDX1 frameshift mutation, clinical characteristics, and implications. J. Endocr. Soc. 6 (1), bvab159. 10.1210/jendso/bvab159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H., Li J., Xin N., Zhao Z., Qin G. (2010). Expression of Pdx1 mediates differentiation from mesenchymal stem cells into insulin-producing cells. Mol. Biol. Rep. 37 (8), 4023–4031. 10.1007/s11033-010-0061-y [DOI] [PubMed] [Google Scholar]
- Yuan H., Liu H., Tian R., Li J., Zhao Z. (2012). Regulation of mesenchymal stem cell differentiation and insulin secretion by differential expression of Pdx-1. Mol. Biol. Rep. 39 (7), 7777–7783. 10.1007/s11033-012-1619-7 [DOI] [PubMed] [Google Scholar]
- Zhang S., Huang F., Tian W., Lai J., Qian L., Hong W., et al. (2020). Andrographolide promotes pancreatic duct cells differentiation into insulin-producing cells by targeting PDX-1. Biochem. Pharmacol. 174, 113785. 10.1016/j.bcp.2019.113785 [DOI] [PubMed] [Google Scholar]
- Zheng L., Wang Y., Li Y., Li L., Wang X., Li Y. (2021). miR-765 targeting PDX1 impairs pancreatic beta-cell function to induce type 2 diabetes. Arch. Physiol. Biochem., 1–10. 10.1080/13813455.2021.1946561 [DOI] [PubMed] [Google Scholar]
- Zhou Q., Brown J., Kanarek A., Rajagopal J., Melton D. A. (2008). In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature 455 (7213), 627–632. 10.1038/nature07314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H. P. (2020). Silence of HOTAIR inhibits insulin secretion and proliferation in pancreatic beta cells. Eur. Rev. Med. Pharmacol. Sci. 24 (2), 784–792. 10.26355/eurrev_202001_20061 [DOI] [PubMed] [Google Scholar]
- Zhu L., Almaca J., Dadi P. K., Hong H., Sakamoto W., Rossi M., et al. (2017a). β-arrestin-2 is an essential regulator of pancreatic β-cell function under physiological and pathophysiological conditions. Nat. Commun. 8, 14295. 10.1038/ncomms14295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Liu Q., Zhou Z., Ikeda Y. (2017b). PDX1, neurogenin-3, and MAFA: Critical transcription regulators for beta cell development and regeneration. Stem Cell Res. Ther. 8 (1), 240. 10.1186/s13287-017-0694-z [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.