Purpose of review
Clostridioides difficile infection (CDI) is the most common cause of healthcare-associated diarrhoea in western countries, being categorized as an urgent healthcare threat. Historically, researchers have relied on the use of in vivo animal models to study CDI pathogenesis; however, differences in physiology and disease prognosis compared with humans limit their suitability to model CDI. In vitro models are increasingly being used as an alternative as they offer excellent process control, and some are able to use human ex-vivo prokaryotic and/or eukaryotic cells.
Recent findings
Simulating the colonic environment in vitro is particularly challenging. Bacterial fermentation models have been used to evaluate novel therapeutics, explore the re-modelling of the gut microbiota, and simulate disease progression. However, they lack the scalability to become more widespread. Models that co-culture human and bacterial cells are of particular interest, but the different conditions required by each cell type make these models challenging to run. Recent advancements in model design have allowed for longer culture times with more representative bacterial populations.
Summary
As in vitro models continue to evolve, they become more physiologically relevant, offering improved simulations of CDI, and extending their applicability.
Keywords: Clostridioides difficile, Clostridioides difficile infection, gut microbiota, gut model, in vitro
INTRODUCTION
Clostridioides difficile is a Gram-positive, endospore-producing anaerobe, which can colonize the intestinal microbiota. When the complex gut microbial ecosystem is perturbed, it allows the bacterium rapid expansion, leading to symptomatic C. difficile infection (CDI). Disease severity can range from mild diarrhoea to toxic megacolon, perforation of the colon and, ultimately, death. CDI poses a significant healthcare burden across the globe, as the leading cause of healthcare-associated diarrhoea [1].
Box 1.
no caption available
The pathogenesis of CDI is largely associated with the use of broad-spectrum antibiotics, which limit the competitive exclusion afforded by the endogenous microbiota, allowing C. difficile proliferation. Fluoroquinolones, clindamycin, and beta-lactams (particularly cephalosporins) are associated with high CDI risk [2,3]. The recommended antibiotic treatments for C. difficile, vancomycin and fidaxomicin, [4] may compound the factors, which caused the initial C. difficile proliferation, leading to a significant number of patients relapsing or developing recurrent CDI (rCDI).
Faecal microbiota transplant (FMT) is a relatively crude but effective restorative therapy, shown to prevent rCDI. FMT acts by reseeding the microbiota, restoring microbial and functional diversity. Autologous transplants are effective [5] but not always feasible. Due to the risk of transplanting pathogens, which may lead to further complications for the patient or potentially unknown long-term health consequences, allogenic transplants are usually reserved for patients who have suffered from multiple instances of rCDI. Alternatives to FMT are defined as restorative therapies that reseed preselected beneficial communities of the endogenous microbiota [5–7]. However, none of these novel treatments are yet authorized for use clinically [6].
With interactions between C. difficile, host, and microbiota having a key role in CDI pathogenesis, the ability to accurately model these interactions will be crucial to future C. difficile research. Current studies often rely on the use of in vivo animal models – namely hamster and mouse models [8,9]. They are particularly used when studying immunological aspects of C. difficile; however, immune responses vary greatly between species and disease progression in these models is often dissimilar to that in humans [10]. Further limitations include the differences in anatomy and microbiota composition [11], plus there is a general need to reduce and refine the use of animals in research. This has promoted the use of in vitro models to simulate CDI.
The complex multifaceted aetiologies behind CDI mean that model systems, which can maintain complex and stable microbial communities and model their interactions with host physiology will be invaluable in further understanding C. difficile/CDI and for the development of novel therapies.
CURRENT IN VITRO MODELS FOR STUDYING CLOSTRIDIOIDES DIFFICILE INFECTIONS
Bacterial fermentation models
Batch fermentation
Batch fermentation models are the simplest in vitro model and consist of a reaction vessel with controlled internal conditions [12▪]. They provide a quick and relatively inexpensive screening tool for the metabolization of specific substrates. However, depletion of nutrients, media acidification, and build-up of metabolites limit the experiment duration – typically less than 48 h – making these models unsuitable for longitudinal studies [13]. Nonetheless, batch fermentation is used to study CDI. For instance, a batch model was used to assess the efficacy of a ‘Bacteriophage Cocktail’ to clear CDI [14], while a separate study used a simple batch model consisting of six-well plates, to investigate the sporulation of C. difficile in faecal emulsions from different patients, showing that a dysbiotic microbiota is more susceptible to CDI, and this susceptibility is strain-dependent [15].
Continuous single-stage models
Continuous single-stage models (CSS) models also consist of a single reaction vessel, but the continuous or semi-continuous influx of nutrient-rich media and efflux of waste products allows for longer culture times, where bacterial populations can stabilize and form trophic chains [16]. Their main disadvantage is that they only simulate a single colonic region, so microbial dynamics across the entire gastrointestinal tract cannot be characterized. However, their simplicity and low cost compared with multistage models make repeats more feasible. For example, the Mini BioReactor Arrays (MBRAs) allows for 24 CSS to be run simultaneously, promoting the growth of stable microbial communities [17]. The MBRAs have been used to demonstrate that an in vitro gut microbiota modulated with polyphenols has decreased C. difficile colonization resistance [18▪], and that Fusobacterium nucleatum acts synergistically with C. difficile in the formation of biofilms [19▪].
Continuous multistage models
Continuous multistage (CMS) models were first devised in the 1980s and, subsequently, validated against the colonic contents of sudden death victims [20]. In brief, the original model consisted of three vessels arranged sequentially to simulate the proximal, medial, and distal colonic regions. Each vessel is maintained at conditions (pH, temperature and %O2) specifically designed to mimic each colonic region [21▪▪,22]. This arrangement has become the standard reference from which other models have been developed.
A variation of this triple-stage model has been extensively used for studying CDI pathogenesis. Recent work includes evaluating the propensity of oral antibiotics to induce CDI: omadacycline, first-generation cephalosporins and eravacycline are among those that showed a low CDI association [22–24]. These models have also been used to evaluate the potential of a novel antibody therapy to prevent CDI [25], with results showing good efficacy at neutralizing toxin production and rCDI prevention when combined with vancomycin. A further study showed that trehalose-induced remodelling of the gut microbiota can lead to colonization resistance against C. difficile[21▪▪]. Furthermore, it is possible to modify the standard CMS model to support biofilm growth and study sessile populations – using this model, it was demonstrated that biofilms can harbour C. difficile and cause rCDI [26▪▪].
Although the majority of CDI studies using CMS models used the described triple-stage setup, other in vitro systems are available. The Simulator of the Human Intestinal Microbial Ecosystem (SHIME) [27,28] is a commercial system, which builds upon the triple-stage model by adding two fermentation vessels to simulate the stomach and small intestine. In doing so, it is one of the few models, which simulates the entire gastrointestinal tract; however, the entire system is initiated with derived faecal matter, which is unlikely representative of the physiological conditions of the upper gastrointestinal tract.
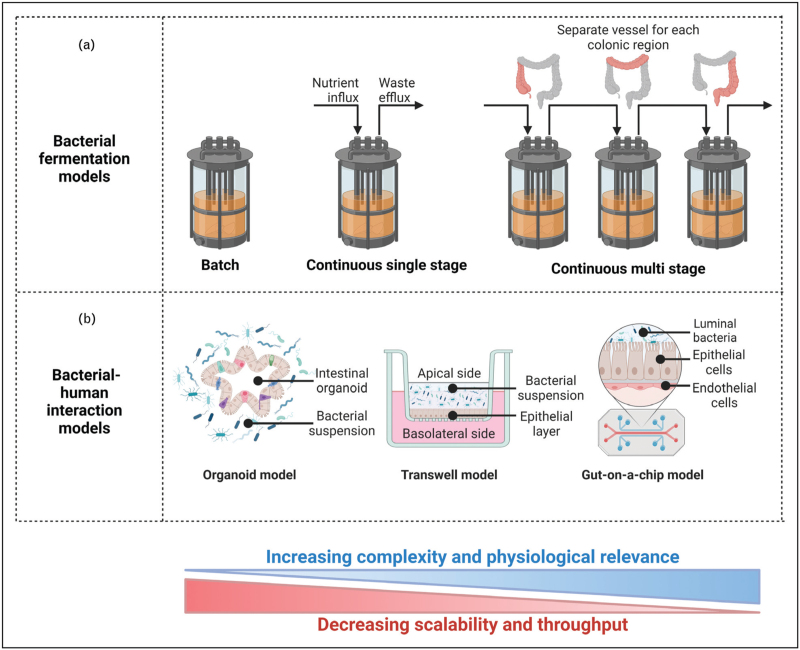
CMS models have been extensively used for preclinical research and correlate well with patient outcomes [22,24]. They allow for long-lasting longitudinal studies and sampling of different colonic regions to study microbiota spatial variation, which can only be achieved through invasive surgery in in vivo models. Despite showing greater control and fewer ethical restraints than animal models, CMS are resource-intensive and thus impractical to run in large numbers, limiting the possibility of repeats. A summary of bacterial fermentation set-ups is shown in Fig. 1a.
FIGURE 1.
Overview of the in vitro models currently available for studying C. difficile and CDI. (a) Bacterial fermentation models and (b) human interaction models. Left to right indicates models of increasing complexity and corresponding decreasing scalability. Figure created using BioRender.com.
Other in vitro models
Variations of the multicompartmental model TNO gastro-intestinal model (TIM), which simulates the gastrointestinal tract lumen conditions, have been used to study the fermentation of foods and supplements [12▪,29,30]. TIM focusses on the meal transit time, and it controls physiological parameters to reflect the conditions of the upper gastrointestine; thus, experiments have a short timeframe (>72 h). Similarly, models such as EnteroMix [31] and ECSIM [32] also have reduced run times when compared to the triple-stage model and SHIME. As these models aim at investigating the digestion process, they do not offer the experimental durability of weeks/months that CDI studies often require.
Human interaction models
Bacterial fermentation models support solely microbial growth. Although suitable to investigate microbe–microbe and drug–microbe interactions, these models lack information on how microbial composition relates to the host physiology. Human interaction models aim to bridge this gap (Fig. 1b) but are limited by the different culture requirements of prokaryotic and eukaryotic cells. Many fastidious anaerobes composing the intestinal microbiota are sensitive to low oxygen concentrations, which contrasts with the high oxygen requirements of human cells.
Three-dimensional models
Three-dimensional (3D) organoids consist of ex vivo culture of organ cells, which mimic the source tissue architecture. Intestinal organoids developed through the culture of intestinal stem cells can accurately portray the physiological composition of the human intestinal epithelium. Intestinal organoids vary with the nature of the stem cell origin; enteroids derive from adult stem cells isolated from tissue biopsies, while induced human intestinal organoids are developed from pluripotent stem cells. The latter provide a more robust representation of the intestinal epithelium and remove the need for biopsy samples, but have increased culture times (months rather than days).
Organoids have proved useful in exploring the interactions between C. difficile and the intestinal epithelium [33▪,34]. The pathogenicity of CDI is driven largely by toxin A (TcdA) and toxin B (TcdB). These are known to alter the cytoskeletal structure of the intestinal epithelium causing cell death and disrupting the epithelial barrier. Exposing organoids to C. difficile toxins demonstrated that microRNAs suppression in colonic tissues during CDI may be attributable to the actions of C. difficile TcdB [34].
However, the use of spheroid organoids for C. difficile research is not very common. Intestinal organoids form closed 3D spheres with an inner apical side representing the intestinal lumen and an outer basal side representing the submucosal side of the intestinal epithelium. This orientation reduces access to the luminal surface, impacting cell line consistency and physiology, and limiting 3D models suitable for C. difficile-related studies.
Two-dimensional monolayers and ‘gut-on-a-chip’ models
Two-dimensional (2D) monolayers derive from fragmented 3D organoids, as patient-derived cell lines are plated onto extracellular matrix-coated wells known as Transwell plates [35]. The 2D organoid-derived monolayer allows access to the apical side and provides a simple and inexpensive method of co-culturing epithelial and bacterial cells. Jejunal human intestinal organoids have been used to study the action of TcdA and TcdB, revealing mucin might reduce toxin binding in the human epithelium [33▪]. Transwell models have recently been developed, which allow extended co-culture of human epithelial cells with oxygen-sensitive bacterial species [36▪]. Although these models allow for longer culture times compared with 3D organoids and promote bacterial differentiation to form villus-like structures, they lack fluid flow and peristalsis-like motion, important in vivo characteristics of the intestinal environment.
To address this, various so-called ‘gut-on-a-chip’ models have been developed. They typically consist of two channels representing the lumen and blood vessels, separated by a semi-permeable membrane on which epithelial cells can be grown [37]. Each channel can be perfused with separate culture media to support each cell type, and recent developments have transluminal hypoxia gradients, allowing the culture of strict anaerobes [36▪] and complex faecal-derived microbiota [38]. The flexible construction of these models allows the application of fluid flow to mimic in vivo environment [39], and the co-culture of human cells [40].
CURRENT CHALLENGES AND FUTURE ADVANCES
The biggest challenges with all the above models remain the culture of a stable bacterial community, which is representative of the original inoculum, while maintaining epithelial and immune cell viability over extended periods [41]. For the study of CDI, the host–microbe interaction is of critical importance as it links bacterial composition and/or the presence of C. difficile toxins to host outcomes. Incorporating relevant human cells rather than relying on animal-derived or immortalized cell lines would simulate a more physiologically accurate response to CDI in vitro[37]. Although co-culture of human cells with selected bacterial species may be beneficial to investigate individual mechanisms of action, cell culture with a ‘complete’ microbiota would be more relevant to simulate host–microbiota interactions.
The currently available systems often compromise high throughput – which enable repeats – with limited complexity, and vice-versa. Therefore, the complex gastrointestinal models, which most closely mimic the colonic environment, are difficult to scale-up; while the most easily scalable assay-based methods have little relation to host physiology (Table 1). Future models should aim to address this issue by reducing complexity and resource requirements, and increasing automation, while maintaining clinical reflectiveness.
Table 1.
Summary of in vitro models used for studying Clostridioides difficile infection and the human gut microbiota
| In vitro model | Starting material | Advantages | Disadvantages | Ref. |
| Bacterial fermentation models | ||||
| Batch fermentation | Human faecal matter or bacterial suspension | Quick and simple to set up Relatively inexpensive Suitable for initial screening Scalable compared with more complex systems | Limited run time because of build-up of metabolites and media acidification Only one colonic region simulated No host interaction | [12▪] [13] [14] |
| Continuous single stage | Human faecal matter or bacterial suspension | Longer run times possible as media is continuously added Scalable compared with multistage models | Only one colonic region simulated No host interaction | [15] [16] [18▪] |
| Continuous multistage | Human faecal matter | Multiple colonic regions simulated, complex trophic chains established Straightforward sampling from each vessel Good correlation with clinical data | Complex systems are difficult to scale No host interaction with standard setup, although some are compatible with modules addition | [19▪] [27] [47] |
| Human interaction models | ||||
| 3D organoids | Ex vivo organ cells Bacterial suspension of selected species or metabolites | Crypt-villi structure Low equipment cost and wide availability Use of metabolites allows isolated study of response mechanisms | Short run times Microbiota cannot be profiled or sampled during co-culture | [32] |
| 2D monolayers | Ex vivo organ cells Bacterial/faecal suspension or effluent from bacterial fermentation model | Easy access to both apical and basolateral for sampling throughout culture | Difficult to maintain anaerobic conditions Peristalsis cannot be simulated | [34] [35] |
| Gut-on-a-chip | Ex vivo organ cells Bacterial/faecal suspension or effluent from bacterial fermentation model | Continuous flow and oxygen gradient extend culture times past 72 h Some models include peristalsis-like motion Possible additional chamber for immune cells Interfacing with a bacterial fermentation model | Specialized equipment and low availability Susceptible to bacterial overgrowth | [36▪] [37] |
Many of the above in vitro models have been developed within single research groups and institutions, using bespoke equipment and custom culture media, which limits standardization and poses financial restrictions as development costs are high. These differences can make it difficult to draw comparisons between studies, and so it would be beneficial to reduce method variability going forward.
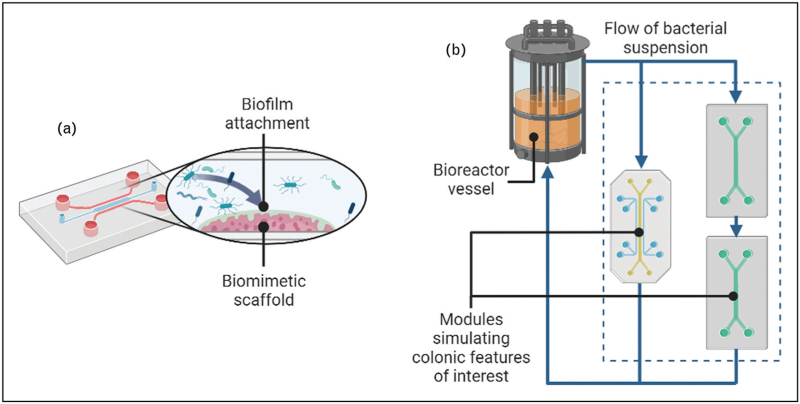
It has been shown that biofilms play a critical role in the pathogenesis of CDI, particularly in recurrent infections [26▪▪,42]. However, methods for growing and sampling intestinal biofilms in vitro are poorly standardized and vary greatly between studies. The SHIME model can be modified by the addition of mucin-coated microcosms [43], which facilitate biofilm growth, while other studies have used mucin-coated coverslips suspended within a bioreactor or bacterial suspension [19▪,44,45]. Recent studies have also shown that specially fabricated structures, more similar to the in vivo environment, can provide a greater surface area for biofilm attachment and growth [46,47]. Models with defined flow characteristics and a more representative luminal environment will be key for studying biofilm formation and its role in CDI (Fig. 2a).
FIGURE 2.
Future advances that would benefit in vitro modelling: (a) shows a gut-on-a-chip device with the inclusion of a functional surface for biofilm attachment; (b) shows how a bacterial fermentation bioreactor could be coupled with multiple organ-on-chip devices to study host interactions. Figure created using BioRender.com.
Other advances such as integrated sensing, inclusion of 3D structures, and extending culture times past 48 h will all aid in the wide-scale adoption of in vitro models, as no device can currently simulate all the characteristics of the human colonic environment in vitro[12▪]. A more pragmatic approach would be to develop a modular system whereby different in vitro ‘modules’, each with their own set of features, can be used as required (Fig. 2b). This is exemplified in the SHIME model where a flow cell can be coupled to study host–bacteria interactions [48].
FUTURE APPLICATIONS
Popularity of in vitro models as an ethical alternative to in vivo models is increasing. Their excellent process control and rapidly evolving ability to model both microbial and human components of the gastrointestinal tract have the potential to revolutionize research of gastrointestinal diseases.
As our understanding of the gut microbiota increases, the systemic implications of intestinal disease are becoming more relevant. To better support health-related studies and characterize mechanisms of disease, in vitro platforms must also evolve to accurately model inter-organ interactions. Multiorgan platforms are in development and will likely grow in popularity and relevance as they are refined [49,50].
The relationship between CDI and intestinal dysbiosis is well established [21▪▪,24,25,26▪▪]; however, dysbiosis is still poorly defined. Variation in the microbiota of individuals means there is likely a significant amount of redundancy in characterizing dysbiosis, as numerous organisms can fill similar roles. In vitro models offer the capacity to monitor the intestinal environment throughout disease progression, thus they provide means for studying unique microbial activities. Exploring the microbiota from a functional perspective through the implementation of omics technologies and defining dysbiosis as a family of functional disorders may redefine critical healthcare approaches. Understanding the functional capacity of a patient's microbiota may also provide valuable insights when curating treatment plans, by minimizing microbiota disruption and reducing the risk of CDI.
In vitro models are well suited to longitudinal studies examining how the intestinal microbiota can be reseeded using restorative therapies to re-introduce functional diversity and increase ecological robustness, in patients at risk of CDI. In vitro models can also be used to investigate the transfer of mobile genetic elements within bacterial populations, and the microbial metabolization of therapeutic agents, particularly those delivered enterally.
The final hurdle in the development of in vitro models will likely be the integration of functional immune systems for vaccine development. In vivo models remain the only viable system for studies involving adaptive immunity.
CONCLUSION
There are currently several in vitro platforms, which are capable of modelling microbial and human mechanisms of CDI. Simpler bacterial fermentation models are highly controllable and scalable systems suited for screening of potential therapeutic agents, whereas CMS models are fit for longitudinal studies examining remodelling of the microbiota and rCDI studies. Advances in tissue culture techniques and evolution of 3D and 2D organoid systems offer the possibility to investigate CDI mechanisms in a controllable host environment but are still limited in their potential. A likely evolution of in vitro models will be the integration of organoids into bacterial fermentation models, providing insight into how changes in microbiota composition and function may impact host cells, leading to intestinal disease.
Acknowledgements
None.
Financial support and sponsorship
None.
Conflicts of interest
There are no conflicts of interest.
Footnotes
D.E. and W.D.B. contributed equally.
REFERENCES AND RECOMMENDED READING
Papers of particular interest, published within the annual period of review, have been highlighted as:
▪ of special interest
▪▪ of outstanding interest
REFERENCES
- 1.CDC. Antibiotic resistance threats in the United States. Atlanta, GA: U.S. Department of Health and Human Services, CDC; 2019. [Google Scholar]
- 2.Czepiel J, Krutova M, Mizrahi A, et al. Mortality following clostridioides difficile infection in europe: a retrospective multicenter case-control study. Antibiotics 2021; 10:299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown KA, Langford B, Schwartz KL, et al. Antibiotic prescribing choices and their comparative C. difficile infection risks: a longitudinal case-cohort study. Clin Infect Dis 2021; 72:836–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. (NICE) NIfHaCE. Clostridioides difficile infection: antimicrobial prescribing [Online]. 2021. Available at: https://www.nice.org.uk/guidance/ng199/resources/clostridioides-difficile-infection-antimicrobial-prescribing-pdf-66142090546117. [Accessed 21 June 2022]. [Google Scholar]
- 5.Satokari R, Pietilä L, Mattila E, et al. Faecal banking at –20°C facilitates faecal microbiota transplantation for recurrent Clostridioides difficile infection in clinical practice. Infect Dis 2020; 52:662–665. [DOI] [PubMed] [Google Scholar]
- 6.Buckley AM, Moura IB, Wilcox MH. The potential of microbiome replacement therapies for Clostridium difficile infection. Curr Opin Gastroenterol 2022; 38:1–6. [DOI] [PubMed] [Google Scholar]
- 7.Feuerstadt P, Louie TJ, Lashner B, et al. SER-109, an oral microbiome therapy for recurrent Clostridioides difficile infection. N Engl J Med 2022; 386:220–229. [DOI] [PubMed] [Google Scholar]
- 8.Wexler AG, Guiberson ER, Beavers WN, et al. Clostridioides difficile infection induces a rapid influx of bile acids into the gut during colonization of the host. Cell Rep 2021; 36:109683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abutaleb NS, Seleem MN. In vivo efficacy of auranofin in a hamster model of Clostridioides difficile infection. Sci Rep 2021; 11:7093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosselot AE, Park M, Kim M, et al. Ontogeny and function of the circadian clock in intestinal organoids. Embo J 2022; 41:e106973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kieser S, Zdobnov EM, Trajkovski M. Comprehensive mouse microbiota genome catalog reveals major difference to its human counterpart. PLoS Comput Biol 2022; 18:e1009947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12▪.Roupar D, Berni P, Martins JT, et al. Bioengineering approaches to simulate human colon microbiome ecosystem. Trends Food Sci Technol 2021; 112:808–822. [Google Scholar]; The article provides an overview of the in-vitro models available to mimic the human colon.
- 13.Pérez-Burillo S, Molino S, Navajas-Porras B, et al. An in vitro batch fermentation protocol for studying the contribution of food to gut microbiota composition and functionality. Nat Protoc 2021; 16:3186–3209. [DOI] [PubMed] [Google Scholar]
- 14.Nale J, Redgwell T, Millard A, Clokie M. Efficacy of an optimised bacteriophage cocktail to clear Clostridium difficile in a batch fermentation model. Antibiotics 2018; 7:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horvat S, Rupnik M. Interactions between Clostridioides difficile and fecal microbiota in in vitro batch model: growth, sporulation, and microbiota changes. Front Microbiol 2018; 9:1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pham V, Mohajeri H. The application of in vitro human intestinal models on the screening and development of pre-And probiotics. Benef Microbes 2018; 9:1–18. [DOI] [PubMed] [Google Scholar]
- 17.Hobson CA, Vigue L, Naimi S, et al. MiniBioReactor Array (MBRA) in vitro gut model: a reliable system to study microbiota-dependent response to antibiotic treatment. J Antimicrob Chemother 2022; 4:dlac077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18▪.Mahnic A, Auchtung JM, Poklar Ulrih N, et al. Microbiota in vitro modulated with polyphenols shows decreased colonization resistance against Clostridioides difficile but can neutralize cytotoxicity. Sci Rep 2020; 10:8358. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study investigated C. difficile and the effects of food supplements and clindamycin using minibioreactor arrays (MBRA).
- 19▪.Engevik MA, Danhof HA, Auchtung J, et al. Fusobacterium nucleatum adheres to Clostridioides difficile via the RadD adhesin to enhance biofilm formation in intestinal mucus. Gastroenterology 2021; 160:1301.e8–1314.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study investigated C. difficile biofilm formation and its interaction with colonic MUC2 mucus layer using in vitro bioreactors.
- 20.Macfarlane GT, Macfarlane S, Gibson GR. Validation of a three-stage compound continuous culture system for investigating the effect of retention time on the ecology and metabolism of bacteria in the human colon. Microb Ecol 1998; 35:180–187. [DOI] [PubMed] [Google Scholar]
- 21▪▪.Buckley AM, Moura IB, Arai N, et al. Trehalose-induced remodelling of the human microbiota affects Clostridioides difficile infection outcome in an in vitro colonic model: a pilot study. Front Cell Infect Microbiol 2021; 11:670935. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study investigates the gut microbiota and C. difficile adaptation in response to sugar supplements.
- 22.Moura IB, Buckley AM, Ewin D, et al. Omadacycline gut microbiome exposure does not induce Clostridium difficile proliferation or toxin production in a model that simulates the proximal, medial, and distal human colon. Antimicrob Agents Chemother 2019; 63:e01581–e01518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buckley AM, Moura IB, Altringham J, et al. The use of first-generation cephalosporin antibiotics, cefalexin and cefradine, is not associated with induction of simulated Clostridioides difficile infection. J Antimicrob Chemother 2021; 77:148–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buckley AM, Altringham J, Clark E, et al. Eravacycline, a novel tetracycline derivative, does not induce Clostridioides difficile infection in an in vitro human gut model. J Antimicrob Chemother 2021; 76:171–178. [DOI] [PubMed] [Google Scholar]
- 25.Roberts AK, Harris HC, Smith M, et al. A novel, orally delivered antibody therapy and its potential to prevent Clostridioides difficile infection in preclinical models. Front Microbiol 2020; 11:578903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26▪▪.Normington C, Moura IB, Bryant JA, et al. Biofilms harbour Clostridioides difficile, serving as a reservoir for recurrent infection. NPJ biofilms microbiomes 2021; 7:16. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study reported how intestinal biofilms can act as reservoirs for C. difficile spores that can cause recurrent CDI.
- 27.Gnanasekaran T, Assis Geraldo J, Ahrenkiel DW, et al. Ecological adaptation and succession of human fecal microbial communities in an automated in vitro fermentation system. mSystems 2021; 6:e0023221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van de Wiele T, Van den Abbeele P, Ossieur W. Verhoeckx K, et al. The simulator of the human intestinal microbial ecosystem (SHIME ®). The impact of food bioactives on health: in vitro and ex vivo models. Cham (CH): Springer; 2015. 305–317. [Google Scholar]
- 29.Minekus M. Verhoeckx K, et al. The TNO gastro-intestinal model (TIM). The impact of food bioactives on health: in vitro and ex vivo models. Cham (CH): Springer; 2015. 37–46. [PubMed] [Google Scholar]
- 30.Hall AE, Moraru CI. Comparative effects of high pressure processing and heat treatment on in vitro digestibility of pea protein and starch. NPJ Sci Food 2022; 6:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salli K, Anglenius H, Hirvonen J, et al. The effect of 2’-fucosyllactose on simulated infant gut microbiome and metabolites; a pilot study in comparison to GOS and lactose. Sci Rep 2019; 9:13232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brugère J-F, Féria-Gervasio D, Popse Z, et al. The ECSIM concept (Environmental Control System for Intestinal Microbiota) and its derivative versions to help better understand human gut biology. Appl Biomed Eng 2011; 4:63–82. [Google Scholar]
- 33▪.Engevik MA, Danhof HA, Chang-Graham AL, et al. Human intestinal enteroids as a model of Clostridioides difficile-induced enteritis. Am J Physiol Gastrointest Liver Physiol 2020; 318:G870–G888. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study developed a model of C. difficile to investigate the interaction between toxins and the human intestinal epithelium.
- 34.Monaghan TM, Seekatz AM, Markham NO, et al. Fecal microbiota transplantation for recurrent Clostridioides difficile infection associates with functional alterations in circulating microRNAs. Gastroenterology 2021; 161:255.e4–270.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y, Disalvo M, Gunasekara DB, et al. Self-renewing monolayer of primary colonic or rectal epithelial cells. Cell Mol Gastroenterol Hepatol 2017; 4:165.e7–182.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36▪.Zhang J, Huang Y-J, Yoon JY, et al. Primary human colonic mucosal barrier crosstalk with super oxygen-sensitive Faecalibacterium prausnitzii in continuous culture. Med 2021; 2:74.e9–98.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study reported a model for culture human epithelial cells alongside fastidious bacterial species.
- 37.Ashammakhi N, Nasiri R, Barros NRD, et al. Gut-on-a-chip: current progress and future opportunities. Biomaterials 2020; 255:120196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jalili-Firoozinezhad S, Gazzaniga FS, Calamari EL, et al. A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip. Nat Biomed Eng 2019; 3:520–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kulthong K, Duivenvoorde L, Mizera BZ, et al. Implementation of a dynamic intestinal gut-on-a-chip barrier model for transport studies of lipophilic dioxin congeners. RSC Adv 2018; 8:32440–32453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shah P, Fritz JV, Glaab E, et al. A microfluidics-based in vitro model of the gastrointestinal human–microbe interface. Nat Commun 2016; 7:11535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poletti M, Arnauts K, Ferrante M, Korcsmaros T. Organoid-based Models to Study the Role of Host-microbiota Interactions in IBD. J Crohns Colitis 2021; 15:1222–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frost LR, Cheng JKJ, Unnikrishnan M. Clostridioides difficile biofilms: a mechanism of persistence in the gut? PLoS Pathog 2021; 17:e1009348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van den Abbeele P, Duysburgh C, Cleenwerck I, et al. Consistent prebiotic effects of carrot RG-I on the gut microbiota of four human adult donors in the SHIME(®) Model despite baseline individual variability. Microorganisms 2021; 9:2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Engevik MA, Engevik AC, Engevik KA, et al. Mucin-degrading microbes release monosaccharides that chemoattract Clostridioides difficil and facilitate colonization of the human intestinal mucus layer. ACS Infect Dis 2021; 7:1126–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Engevik MA, Luk B, Chang-Graham AL, et al. Bifidobacterium dentium fortifies the intestinal mucus layer via autophagy and calcium signaling pathways. MBio 2019; 10:e01087-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Biagini F, Calvigioni M, De Maria C, et al. Study of the adhesion of the human gut microbiota on electrospun structures. Bioengineering 2022; 9:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Biagini F, Calvigioni M, Lapomarda A, et al. A novel 3D in vitro model of the human gut microbiota. Sci Rep 2020; 10:21499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marzorati M, Vanhoecke B, De Ryck T, et al. The HMI™ module: a new tool to study the host-microbiota interaction in the human gastrointestinal tract in vitro. BMC Microbiol 2014; 14:133–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ronaldson-Bouchard K, Teles D, Yeager K, et al. A multiorgan chip with matured tissue niches linked by vascular flow. Nat Biomed Eng 2022; 6:351–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rajan SAP, Aleman J, Wan M, et al. Probing prodrug metabolism and reciprocal toxicity with an integrated and humanized multitissue organ-on-a-chip platform. Acta Biomater 2020; 106:124–135. [DOI] [PMC free article] [PubMed] [Google Scholar]