Abstract
Phage–microbe interactions are appealing systems to study coevolution, and have also been increasingly emphasized due to their roles in human health, disease, and the development of novel therapeutics. Phage–microbe interactions leave diverse signals in bacterial and phage genomic sequences, defined as phage–host interaction signals (PHISs), which include clustered regularly interspaced short palindromic repeats (CRISPR) targeting, prophage, and protein–protein interaction signals. In the present study, we developed a novel tool phage–host interaction signal detector (PHISDetector) to predict phage–host interactions by detecting and integrating diverse in silico PHISs, and scoring the probability of phage–host interactions using machine learning models based on PHIS features. We evaluated the performance of PHISDetector on multiple benchmark datasets and application cases. When tested on a dataset of 758 annotated phage–host pairs, PHISDetector yields the prediction accuracies of 0.51 and 0.73 at the species and genus levels, respectively, outperforming other phage–host prediction tools. When applied to on 125,842 metagenomic viral contigs (mVCs) derived from 3042 geographically diverse samples, a detection rate of 54.54% could be achieved. Furthermore, PHISDetector could predict infecting phages for 85.6% of 368 multidrug-resistant (MDR) bacteria and 30% of 454 human gut bacteria obtained from the National Institutes of Health (NIH) Human Microbiome Project (HMP). The PHISDetector can be run either as a web server (http://www.microbiome-bigdata.com/PHISDetector/) for general users to study individual inputs or as a stand-alone version (https://github.com/HIT-ImmunologyLab/PHISDetector) to process massive phage contigs from virome studies.
Keywords: Phage–host interaction, Virome, CRISPR, Prophage, Machine learning
Introduction
Phages play key roles in shaping the community structure of human and environmental microbiota, and provide tools for the precise manipulation of specific microbes. Recent studies have highlighted the influence of phage–microbe interactions on mammalian health and disease, and their great potential in the development of novel therapeutics, such as phage therapy, to combat multidrug-resistant (MDR) infections. Therefore, it is critical to identify and fully understand these interactions [1]. The molecular and ecological coevolutionary processes of phages and microbes leave various signals in their genomic sequences that can be used to trace phage–host interactions [2]. In addition to experimental methods, recent advances in large-scale genomic and metagenomic sequencing and computational approaches have deepened our understanding of phage–microbe interactions, and advanced new challenges in investigating such phage–host interaction signals (PHISs).
PHISs can be grouped into five categories based on their means of detection. First, PHISs can be detected by identifying putative prophage regions in bacterial genomes, defined as integrated phages that insert their genomes into their bacterial hosts. Several in silico tools for prophage detection in sequenced genomes have been developed, including VirSorter [3], PHASTER [4], Prophinder [5], Phage_Finder [6], PhageWeb [7], and DBSCAN-SWA [8]. Recently, a microbe–phage interaction database, Microbe Versus Phage (MVP), was developed based on prophage inference [9]. Second, PHISs can be detected by sequence composition analysis, a commonly used alignment-free method, based on the observation that phages share highly similar genomic signatures (such as k-mer or codon usage) with their hosts, because phage replication is dependent on the translation machinery of its bacterial host [10]. VirHostMatcher [11], WIsH [12], and PHP [13] are such tools for predicting the hosts of viral genomes, or even short viral contigs based on k-mer signals. Third, clustered regularly interspaced short palindromic repeats (CRISPR) spacer sequences have been applied to infer phage–host interactions, given that bacterial hosts incorporate spacer sequences from phages that infect them [14], [15], [16]. Fourth, genetic homology analysis, based on the homology between phage and bacterial genes, can also be used to identify phage–bacterial relationships [17], [18], [19]. Fifth, protein–protein interactions (PPIs) have been applied to predict phage–host interactions because the interactions between phages and microbes are dependent mainly on the interactions between their encoded proteins [20]. Recently, VirHostMatcher-Net and RaFAH have been developed to predict phage–host interactions by integrating multiple PHISs including CRISPR + k-mer and CRISPR + tRNA + homolog signals, respectively [21], [22].
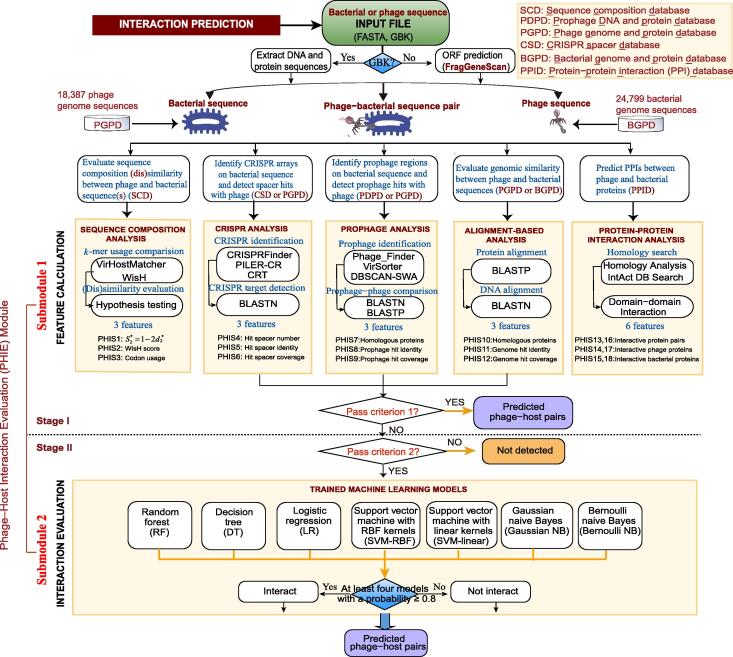
Although various methods have been proposed to predict phage–host interactions, these predictions usually use only a single or a couple of limited in silico signal(s), and therefore have limited accuracy and coverage [2]. Meanwhile, the number of viruses identified in virome studies is increasing exponentially, and there is a massive demand for a tool that is capable of incorporating all types of PHISs and conveniently predicting the microbial hosts of viruses. However, to the best of our knowledge, all currently available tools are limited to certain interaction features, and there is no published web server implementation or informed stand-alone software available to integrate all types of PHISs for comprehensive prediction of global phage–host interactions. To meet this urgent demand, we developed a novel integrative tool to predict phage–host interactions by detecting and integrating diverse in silico PHISs, and scoring the probability of phage–host interactions using machine learning models based on PHIS features. Phage–host interaction signal detector (PHISDetector) captures phage–host associations in a data-driven manner, and is available as a software pipeline for phage–host interaction identification, annotation, and analysis in a comprehensive and user-friendly manner (Figure 1). The PHISDetector can be run either as a web server (http://www.microbiome-bigdata.com/PHISDetector/) or as a stand-alone version on a standard desktop computer (https://github.com/HIT-ImmunologyLab/PHISDetector).
Figure 1.
PHISDetector pipeline for prediction and evaluation of microbe–phage interactions
For candidate phage–bacterial sequence pairs, eighteen PHIS features belonging to five categories are calculated using sequence composition similarity, CRISPR targeting, prophage, genetic homology, and PPI/DDI. Then, a two-stage procedure is performed to predict and evaluate their interactions. In stage I, phage–host pairs with high reliability are detected using criterion 1 and returned directly as final predicted results. In stage II, phage–host pairs with potential PHISs based on criterion 2 are retained and further evaluated using seven well-trained machine learning models including RF, DT, LR, SVM-RBF, SVM-linear, Gaussian NB, and Bernoulli NB, and the phage–host pairs distinguished by at least four models with a probability ≥ 0.8 were returned. PPI, protein–protein interaction; DDI, domain–domain interaction; RF, random forest; DT, decision tree; LR, logistic regression; SVM, support vector machine; RBF, radial basis function; NB, naive Bayes; GBK, GenBank; PHIS, phage–host interaction signal; CRISPR, clustered regularly interspaced short palindromic repeats; ORF, open reading frame; BLASTN, Nucleotide Basic Local Alignment Search Tool; BLASTP, Protein Basic Local Alignment Search Tool; CSD, CRISPR spacer database; PDPD, prophage DNA and protein database; PGPD, phage genome and protein database; SCD, sequence composition database; BGPD, bacterial genome and protein database; PPID, protein–protein interaction database.
Method
Creation of custom databases
Phage genome and protein database
The phage genome database contained 18,387 complete phage genome sequences collected from Millardlab (https://millardlab.org/bioinformatics/bacteriophage-genomes/), which were extracted from the GenBank (GBK) database on April 2021. Open reading frames (ORFs) were annotated using FragGeneScan. Phage sequences shorter than 1000 nt were removed from the database. Finally, 1,255,004 non-redundant phage protein sequences were clustered using CD-HIT at a clustering cutoff of 100% identity over 100% alignment of the shorter sequence [23], and were used to build the phage protein database.
Bacterial genome and protein database
The bacterial genome and protein database (BGPD) contained 24,799 completely assembled bacterial genomes downloaded from the National Center for Biotechnology Information (NCBI) FTP site (https://ftp.ncbi.nlm.nih.gov/) on May 2021 and 22,662,539 non-redundant bacterial protein sequences obtained according to the same processing steps as those used for the phage genome and protein database (PGPD).
Sequence composition database
The sequence composition database (SCD) contained k-mer (k = 6) frequency and codon usage calculated for each of 24,799 completely sequenced bacterial genomes and 18,387 phage genome sequences, as well as homogeneous Markov models trained for each of the 24,799 bacterial genomes using the WIsH method.
Prophage DNA and protein database
The prophage DNA database contained the DNA sequences of 234,045 prophage regions identified in 21,032 bacterial genomes using Phage_Finder or DBSCAN-SWA (our in-house developed prophage detection tool). The prophage protein database contained 1,182,233 protein sequences predicted using FragGeneScan in these prophage regions.
CRISPR spacer database
We identified a total of 119,958 CRISPR arrays from bacterial genome sequences in the BGPD using CRT, CRISPRFinder [24], and PILER-CR [25], and collected 91,685 CRISPR arrays from the CRISPRminer database (https://www.microbiome-bigdata.com/CRISPRminer/) [26] and 11,767,782 spacers from CrisprOpenDB (https://crispr.genome.ulaval.ca) [27]. By merging the CRISPR spacers from the aforementioned collections, 13,183,722 spacer sequences from 578,698 bacterial and archaeal contig sequences were extracted to build the CRISPR spacer database (CSD).
PPI database
To extract the PPIs that constitute the basis for phage–host physical interaction prediction, 1) PPIs between bacterial and phage proteins were inferred by examining the PPIs of their homologous proteins in the IntAct Molecular Interaction Database (https://www.ebi.ac.uk/intact/), and 2) the frequencies at which these PPIs occur in the positive and negative training sets (occur more than twice in the positive compared with the negative training set) were compared. Finally, 912 non-redundant PPIs that were correlated with phage–host interactions were retained. In the same way, 318 non-redundant Pfam domain–domain interactions (DDIs) were selected and used for further evaluation of phage–host interactions.
Phage–host interaction datasets
We totally collected three benchmark datasets with annotated or known phage–host interactions from previous studies [2], [11], [18] (Table S2). The combined dataset included 2511 phage–host pairs and were split into three mutually exclusive sets at the strain level for model training and external validation. Briefly, the host information was extracted from the fields “isolate host=” or “host=”, which were annotated in each of the phage GBK files. The dataset from Edwards et al. [2] including 817 phage–host pairs was used as the positive training set. The 936 phage–host pairs exclusively occurred in the study of Villarroel et al. [18], which originally included 1747 pairs, were used as the positive test set. An extra of 758 phage–host pairs got from Ahlgren et al. [11] after excluding the shared phages with the aforementioned two datasets was used as an independent benchmark dataset to validate the performance of PHISDetector. The negative training and test datasets were built artificially by matching phages with bacteria from a species other than their known hosts in a degree-preserving manner (using edge swaps but only for uniquely connecting pairs).
Calculation of PHIS features
Diverse PHIS features in bacterial and phage genomes could potentially impinge on host range determination. We constructed 18 features belonging to five categories in our framework. A detailed definition of individual features is provided in Table S2. 1) Sequence composition-related features, including similarity score, WIsH score, and codon usage score, were used to evaluate the similarity of sequence composition of a pair of phage–host genome sequences. The similarity score () was calculated to measure the similarity of the oligonucleotide frequency pattern. WIsH score was calculated based on estimated k-mer frequencies [11], [12]. The codon usage score was evaluated as the dissimilarity in codon usage profiles of phage and bacterial coding regions. 2) CRISPR-related features included CRISPRnum, CRISPRidn, and CRISPRcov, representing the number of shared CRISPR spacers, the best identity, and the coverage over all the hits between the host spacers and the phage genome, respectively. 3) Prophage-related features, including PROPphp, PROPidn, and PROPcov, were defined to evaluate the similarity between bacterial prophage region(s) and phage genomes based on homologous protein comparison and nucleotide sequence similarity. 4) Genetic homology features, including ALNhpc, ALNidn, and ALNcov, represent the sequence homology between phage and bacterial genome regions. 5) PPI- or DDI-based features, including PPInum, PPIbap, PPIphp, DDInum, DDIbap, and DDIphp, were calculated to evaluate the interacting potential for each phage–host pair based on the interactions between their encoded proteins as follows: the number of PPIs or DDIs between the bacterial and phage proteins (PPInum and DDInum), the proportion of bacterial proteins involved in PPIs or DDIs (PPIbap and DDIbap), and the proportion of phage proteins involved in PPIs or DDIs (PPIphp or DDIphp).
General phage–host interaction prediction workflow
For candidate phage–bacterial sequence pairs, a two-stage procedure, phage–host interaction evaluation (PHIE) module, was performed to calculate the 18 PHIS features first (stage I), and then to predict and evaluate their interactions (stage II) (Figure 1). In stage I, phage–host pairs with high reliability were detected using criterion 1 in Table S3. Criterion 1 is defined as follows: 1) a strong prophage signal was defined as ≥ 80% overall sequence identity between the bacterial prophage region and phage genome with ≥ 75% prophage coverage, or ≥ 70% of the prophage proteomes are homology (≥ 40% overall amino acid identity and ≥ 70% overall coverage) with the phage proteomes; 2) strong genetic homology signal was defined as ≥ 80% overall sequence identity (ALNidn) between bacterial and phage genomes with ≥ 75% phage genome coverage (ALNcov), or ≥ 70% of the phage proteomes are homology (ALNhpc) (≥ 40% overall amino acid identity and ≥ 70% overall coverage) with the bacterial proteomes; and 3) a strong CRISPR signal was defined as mismatch ≤ 2, spacer coverage ≥ 95%, and e-value ≤ 1E−2 for CRISPR spacer and protospacer matching. In stage II, phage–host pairs with potential PHISs based on criterion 2 (defined in Table S3), which denoted less stringent prophage, genetic homology, and CRISPR signal requirements, were retained and further evaluated using trained machine learning models. Seven machine learning models, namely, random forest (RF), decision tree (DT), logistic regression (LR), support vector machine (SVM) with radial basis function (RBF) kernels (SVM-RBF), SVM with linear kernels (SVM-linear), Gaussian naive Bayes (NB), and Bernoulli NB, were trained on the training dataset with 18 PHIS features (Table S2). Ten-fold cross-validation was used to determine the best configuration parameters. Trained models were used to predict phage–host interactions, and the phage–host pairs discriminated by at least four models with a probability of at least 0.8 were returned (Figure 1). All analyses were carried out using the Python package ‘scikit-learn’ [28]. Briefly, criterion 1 is used to screen out the phage–host pairs with high reliability predicted only using single strong CRISPR, prophage, or genetic homology signals, but not guarantee a high overall prediction score based on machine learning models which consider the effects of all 18 PHIS features. Criterion 2 is used to screen out potential phage–host pairs with weak single signal(s). And these candidate pairs will be further evaluated using the machine learning models based on the overall combination effects of all 18 PHIS features.
Integrated analysis tools
The PHISDetector tool is composed of seven independent analysis modules that allow for 1) identification of diverse in silico PHISs, including oligonucleotide profile analysis, CRISPR analysis, prophage analysis, and PPI analysis; 2) analysis of specialty genes, including virulence factors (VFs) and antibiotic resistance genes (ARGs); and 3) performance of similarity analysis and co-occurrence/co-abundance analysis. These integrated tools can be accessed via http://www.microbiome-bigdata.com/PHISDetector/index/tools/.
Oligonucleotide profile analysis
This module predicts the bacterial host of phages by examining various oligonucleotide frequency (ONF)-based distances/dissimilarities using VirHostMatcher. For the prediction of the prokaryotic host of short viral contigs, an extra WIsH approach is provided. Note that an extra taxonomy file is required when using the VirHostMatcher approach, so we have provided a tool to generate the taxonomy file for the input bacterial genomes using NCBI accession IDs.
CRISPR analysis
CRISPR spacer sequences are computationally identifiable sequence signatures of previous phage–host infections. In this module, three scenarios of analysis are supported. 1) Users can provide their input either as spacer sequences in (multi-)FASTA format, such as CRISPRFinder, PILER-CR, or Seq2CRISPR [29] output files, or as a bacterial genome sequence for which the CRISPR spacers will be automatically identified using PILER-CR. Next, putative protospacer targets will be identified by a Nucleotide Basic Local Alignment Search Tool (BLASTN) search of the spacer input against the viral reference database. 2) Users can upload viral sequences that will undergo BLASTN searches against the spacer reference database. The spacer reference database has been built in our pipeline, including spacers predicted from the complete and draft bacterial genomes in the NCBI database. The bacterial sources of the identified spacers are predicted to be the potential hosts of the viral sequences. 3) Users can examine the phage–host links by CRISPR spacer–protospacer matching between the uploaded bacterial and phage sequences in (multi-)FASTA format. The spacer sequences will be predicted on the bacterial sequence using PILER-CR first, and will then be aligned to the phage sequences by BLASTN.
Prophage analysis
The prophage analysis module accepts both raw DNA sequences in FASTA format and annotated genomes in GBK format, and performs analysis using three prophage detection programs: Phage_Finder, VirSorter, and DBSCAN-SWA. DBSCAN-SWA implements an algorithm combining the DBSCAN algorithm and SWA, referring to the theory of PHASTER, a widely used web tool for prophage prediction with no available stand-alone version or source code [4]. In addition, tRNA sites are annotated using ARAGORN [30] for raw DNA sequences, and extracted for annotated sequences. The sequences of 10 upstream and downstream proteins for each cluster using integrase as the anchor are extracted to examine putative attachment (ATT) sites using BLASTN, with the parameters ‘-task blastn-short-evalue 1000’. Finally, the predictive prophage region is characterized using BLASTN against the Universal Protein (UniProt) viral genome DNA sequences, and the best hitting phage organism is returned. We also use the viral UniProt TrEML reference database to annotate the predicted ORFs in the prophage region. Annotated ORFs with taxonomy information are then subjected to a voting system, and the prophage region is assigned a taxonomy based on the most abundant ORF taxonomy annotated within the prophage. The distribution of prophage-like elements detected by different methods and their size relative to the genome of their host are shown on an interactive circular genome viewer, encoded using AngularPlasmid (https://angularplasmid.vixis.com). The corresponding prophage annotation is shown in the right panel when clicking on the regions.
PPI analysis
Interactions between bacteriophage proteins and bacterial proteins are important for efficient infection of the host cell. We assign bacterial and phage genes to homologs in the Universal Protein Knowledgebase (UniProtKB) protein database based on amino acid sequence homology via double index alignment of next-generation sequencing data (DIAMOND) searches [31]. The interactions between bacteriophage and bacterial proteins are inferred by examining the PPIs of their homologs in the IntAct Molecular Interaction Database (https://www.ebi.ac.uk/intact/). The interactions between bacteriophage proteins and bacterial proteins may contribute to understanding the infectious interactions between bacteria and phages.
Specialty gene check
As accessory genetic elements, bacteriophages play a crucial role in disseminating genes and promoting genetic diversity within bacterial populations. They can transfer genes encoding VFs, such as toxins, adhesins, and aggressions, to promote the virulence of the host bacteria. ARGs in bacterial chromosomes or plasmids can also be mobilized by phages during the infection cycle to increase antibiotic resistance. To identify specialty genes for a pair of bacteria–phage genomes, ORFs are first predicted using FragGeneScan, then further predicted using Short, Better Representative Extract Dataset (ShortBRED) [32] and Resistance Gene Identifier (RGI, v3.1.1; https://github.com/arpcard/rgi) against the virulence factor database (VFDB; http://www.mgc.ac.cn/VFs/) [33] and the Comprehensive Antibiotic Resistance Database (CARD; https://card.mcmaster.ca/) [34], respectively. This analysis facilitates our understanding of how specialty genes are transferred between bacteria and phages.
Similarity analysis
In this module, the similarity between the query phage genome and the genomes of 2196 (or 1871) reference phages with known host genera (or species) is calculated using HostPhinder [18], and the corresponding bacterial host species of similar phages is returned, using a tree viewer and a table to illustrate the prediction process. The GeneNet [19] program is also provided to predict the phage host range based on a built-in gene-based virus–host reference network.
Co-occurrence analysis
This module receives relative abundance profiles in text file format as input, and uses CoNet [35] implementation with Java to calculate the co-occurrence or co-exclusion relationships between the abundance of bacterial and phage organisms across samples. The co-occurrence analysis is mainly divided into initial network computation and assessment of significance. The network is computed by scoring the association strength between bacteria and phage, in which five metrics are calculated by default including correlation metrics (Pearson, Spearman), similarity metrics (mutual information), and distance metrics (Kullback–Leibler, Bray Curtis). Next, the significance of the associations is assessed with a permutation test and bootstraps, and multiple testing corrections are performed with the Benjamini–Hochberg procedure by default. Finally, networks obtained from diverse measures are combined through voting systems using the Sims method. We also incorporate Cytoscape.js [36], an open-source graph theory library written in JavaScript for network visualization so that the differences among the networks constructed using distinct metrics could easily be observed and compared.
Evaluation methods
One-sided t-test was used to examine whether the signal scores were significantly greater or lower (P < 0.05) in positive phage–host pairs than in negative pairs. The receiver operating characteristic (ROC) curves were used to assess the power of predictive signals by plotting the false positive rate (1 – specificity) vs. the true positive rate (sensitivity) according to the change in threshold for each signal feature. The area under the ROC curve (AUC) is a measure of the ability of the model to rank true interactions higher than non-interactions, independent of the prediction score threshold. The sensitivity (true positive rate) and specificity (true negative rate) were used as accuracy metrics to assess the prediction results.
Results
An integrated approach for predicting phage–host interactions
Phage–host interactions can be inherently traced by various signals recorded in their genomic sequences. We designed five categories of features to represent diverse in silico PHISs that contribute to the prediction of phage–host interactions. First, since temperate phages are ubiquitous in nature, with nearly half of sequenced bacteria bearing lysogens [37], we can link phages with their bacterial hosts by identifying the integrated prophages and comparing them with the phage genomes. Thus, we incorporated prophage-related features (PROPnum, PROPidn, and PROPcov) into our tool to evaluate the similarity between integrated prophage(s) and phage genomes based on homologous protein alignment using DIAMOND Protein Basic Local Alignment Search Tool (BLASTP), and nucleotide alignment using BLASTN. Second, given that phages ameliorate their nucleotide composition toward that of their bacterial hosts, we next added sequence composition features ( similarity, WIsH score, and codon usage score) to reflect highly similar patterns in codon usage or short nucleotide words (k-mers) shared between some phages and their hosts. Third, as CRISPR-Cas systems have been found in ∼ 45% of bacterial genomes [24], and approximately 7% of all detectable spacers can match known sequences, most of which originate from phage genomes [38], we incorporated CRISPR features (CRISPRnum, CRISPRidn, and CRISPRcov) into our tool to identify past infections between a phage and its hosts. Fourth, we incorporated genetic homology features (ALNhpc, ALNidn, and ALNcov) to represent genetic homologous sequences that were acquired during a past infection event. Finally, DDI scores (DDInum, DDIbap, and DDIphp) and PPI scores (PPInum, PPIbap, and PPIphp) were combined to evaluate interactions between proteins from the phage and its bacterial hosts. The combination of these categories of PHIS features increases the possibility of capturing additional interacting signals derived from different known mechanisms. Based on the aforementioned in silico PHIS features, we next carried out machine learning modeling to systematically integrate the categories of PHISs to predict phage–host interactions. The overall prediction framework is illustrated in Figure 1.
Evaluation of the predictive power of in silico PHISs
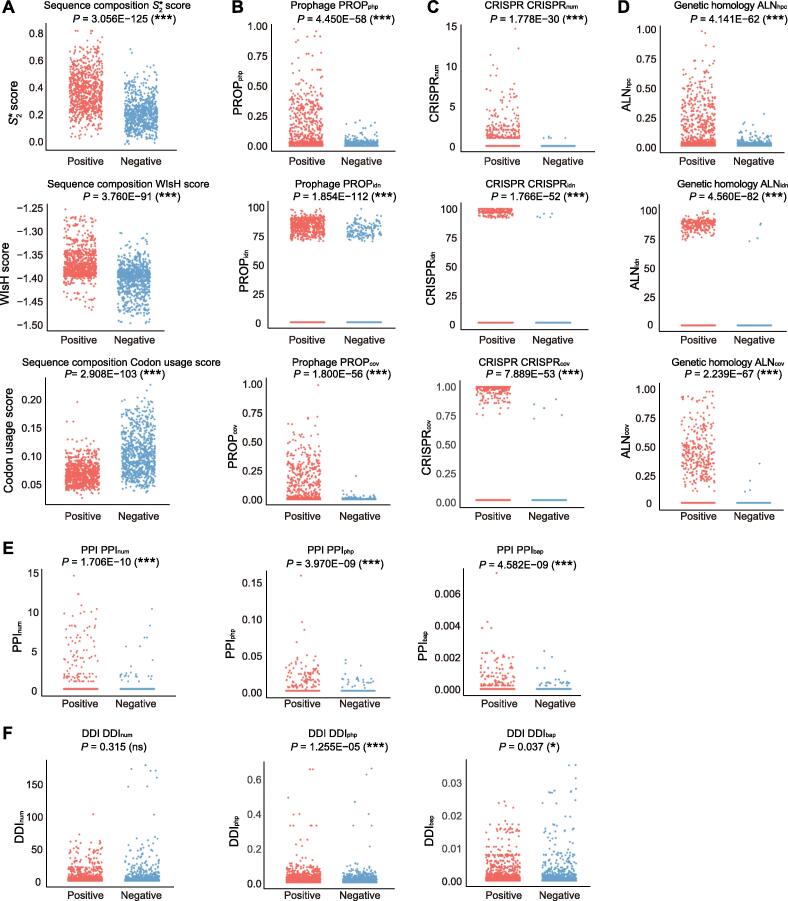
To assess the discriminatory power of each of the 18 PHIS features, one-sided t-test was used to determine the difference between the mean scores of each PHIS feature in the positive and negative phage–host pairs from a training set containing 817 phages and 143 host bacterial species. All features from the five categories showed extraordinary discriminating abilities, except for DDI-based features, which have acceptable discriminating abilities (Figure 2). For sequence composition analysis, positive phage–host pairs had significantly higher S2* similarity (P = 3.056E−125), WIsH score (P = 3.760E−91), and codon usage similarity (P = 2.908E−103) than negative pairs (Figure 2A). In terms of the three prophage-related features, significant discriminant power could be observed between the positive and negative pairs (PROPphp, P = 4.450E−58; PROPidn, P = 1.854E−112; and PROPcov, P = 1.800E−56; Figure 2B). All CRISPR scores were significantly higher for the positive phage–host pairs than for the negative ones (CRISPRnum, P = 1.778E−30; CRISPRidn, P = 1.766E−52; and CRISPRcov, P = 7.889E−53; Figure 2C). For genetic homology features, positive and negative pairs were significantly different based on homologous comparisons between phage and bacterial nucleotide and protein sequences (ALNhpc, P = 4.141E−62; ALNidn, P = 4.560E−82; and ALNcov, P = 2.239E−67; Figure 2D). All PPI-based features also showed extraordinary discriminating abilities, but the DDI-based features did not provide good discriminative abilities (Figure 2E and F). The discriminating ability of these features was also validated by calculating the AUC values. Similarly, except for DDI-based features which had weak discrimination abilities, the other features could achieve excellent discriminating ability (with AUC ≥ 0.792) (Table S4).
Figure 2.
Distributions of 18 PHIS feature values in 817 interacting phage–host pairs and non-interacting phage–host pairs
A. Jitter scatter plots of sequence composition feature values, including score, WIsH score, and codon usage score. B. Jitter scatter plots of prophage-related feature values, including PROPphp, PROPidn, and PROPcov. C. Jitter scatter plots of CRISPR-related feature values, including CRISPRnum, CRISPRidn, and CRISPRcov. D. Jitter scatter plots of genetic homology feature values, including ALNhpc, ALNidn, and ALNcov. E. Jitter scatter plots of PPI-based features, including PPInum, PPIphp, and PPIbap. F. Jitter scatter plots of DDI-based features, including DDInum, DDIphp, and DDIbap. *, P < 0.05; ***, P < 0.001; ns, not significant.
Machine learning models for phage–host interaction predictions
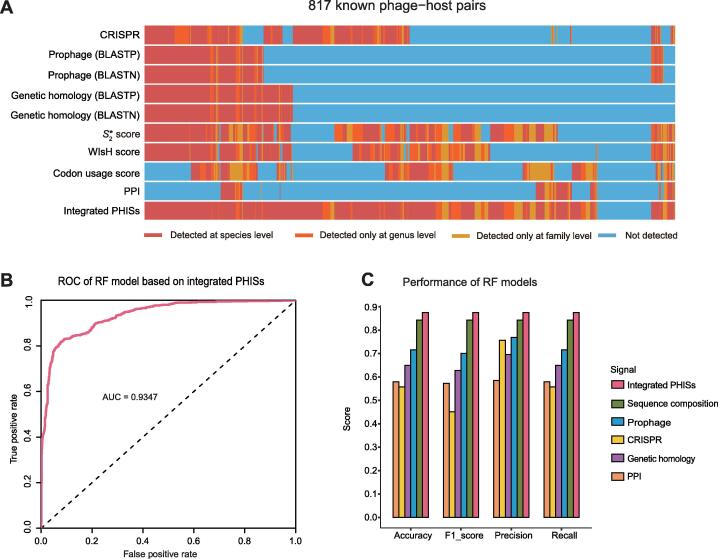
A single PHIS category could identify only a limited number of positive interactions for the training set (16.4%–41.25%) (Figure 3A), while the combination of multiple categories of PHIS features could identify many more known interactions at the species to family level (70.13%–89.84%). A phage–host pair is decided as positive or validated by a given feature using criterion 1 for the CRISPR, prophage, and genetic homology signals, and pre-determined values for sequence composition and PPI signals in Table S3. It is worth noting that, at the species level, about 30% of the known phage–host interactions did not contain any of the detectable signals defined in our study. These results indicate that different types of PHISs may reflect distinct interacting mechanisms that are requisitioned by different phage–host sub-populations and more phage–host interaction signals need to be discovered or incorporated.
Figure 3.
Comparison of the predictive power of single PHIS category or the integrated phage–host interaction signals
A. Heatmap showing whether known phage–host pairs are validated by diverse in silico PHISs. B. ROC curve showing the discriminative ability between positive and negative pairs in the test set using the integrated model combining all PHISs, with the AUC value of 0.9347. C. Bar chart displaying the performance of RF models based on integrated PHISs or single PHIS category in the test set by four evaluation indexes, namely, accuracy, F1-score, precision, and recall. ROC, receiver operating characteristic; AUC, area under the ROC curve.
Based on the aforementioned 18 in silico PHIS features, we carried out machine learning modeling to systematically integrate various categories of PHISs to predict and evaluate phage–host interactions. Seven machine learning models (RF, DT, LR, SVM-RBF, SVM-linear, Gaussian NB, and Bernoulli NB) were applied to the training dataset containing 817 phages and 143 host bacterial species (Table S1; Figure S1). Ten-fold cross-validation was performed to determine the best configuration parameters. Next, we applied these trained models to validate the test set containing 936 phages infecting 110 host species (Table S1; Figure S1), and plotted the corresponding ROCs. The AUC, which measures the discriminative ability between positive and negative pairs in the test set, was 0.5738–0.9275 for each trained model, with the RF model achieving the best performance (Figure S2).
To further prove that the machine learning model integrating all PHIS categories performs better than nonintegrated models, we also trained RF models using each individual PHIS category and integrated signals, and tested the corresponding discriminative abilities in the test set. AUCs ranged between 0.5738–0.9275 for sequence composition, CRISPR, prophage, genetic homology, and PPI individually, while the AUC of our integrated model achieved 0.9347 (Figure 3B, Figure S2). In addition, by calculating the four general evaluation indexes, including accuracy, F1-score, precision, and recall, we showed that our integrated model performed much better than each individual PHIS category, with a score approximate to 0.875 in all these indexes (Figure 3C). Although at the strain level the datasets for model training and external validation are mutually exclusive, at the phage genus level they are not completely exclusive but with 96 shared phage genera. Therefore, to further evaluate the ability of the models in predicting phage–host interactions for novel phages, we split the test dataset into 698 ‘experienced’ phages whose genera occurred in the training set and 238 ‘novel’ phages whose genera were not used in the training set. By random sampling of the ‘experienced’ and ‘novel’ phages with equal size 100 times, the median accuracy for predicting ‘novel’ phages using our trained models achieved 0.72, decreased by 0.13 than that for the ‘experienced’ phages (which achieved a prediction accuracy of 0.85) (Figure S3A). Therefore, our approach, which integrated five categories of PHIS features using machine learning models, exhibited robust predictive power for phage–host interactions even for new phages unseen in the training models. Furthermore, we evaluate the performance of PHISDetector in predicting hosts for phages at various lengths based on the predicted results for 1434 phages with known phage–host interactions from the two external test sets. As is shown in Figure S3B, PHISDetector could predict hosts for a majority of phages (with a length of 10–100 kb) with a high accuracy of 0.64–0.88 or 0.86–0.92 at the genus or family level (Figure S3B). For the phages with lengths less than 10 kb or greater than 100 kb, PHISDetector could obtain a prediction accuracy of 0.25 and 0.47 at the genus level, but achieve 0.99 and 0.76 at the family level. Therefore, predicting hosts for shorter viral contigs probably required more phage–host interaction signals to accurately predict their hosts.
Advanced features of PHISDetector
Our prophage analysis module integrated two popular programs, Phage_Finder and VirSorter, and our in-house developed tool DBSCAN-SWA, which combines the DBSCAN algorithm [39] with SWA. DBSCAN-SWA presents the best detection power based on an analysis using a controlled dataset, including 184 manually annotated prophages, with a detection rate of 85%, which is greater than that of Phage_Finder (63%) or VirSorter (74%). Combining all three methods (provided as a “merge” function in the prophage analysis module), 92 % of the reference prophages could be detected. We also added a prophage annotation step to indicate possible integrated phages in the predicted regions.
Our CRISPR analysis module facilitates two-way analysis. If a phage genome is submitted, it will be compared with our in-house collected spacer database (13,183,722 spacer sequences) to quickly detect CRISPR-targeting associations between the input phage sequence and microbial genomes in NCBI. If a bacterial genome is received, the PHISDetector will detect the CRISPR spacer automatically and compare it with an in-house collected PGPD to find the target phage. If a bacterium–phage pair is received, the PHISDetector will detect the CRISPR spacer automatically in the bacterial genome sequence and compare it against the query phage genome to predict the CRISPR-targeting association.
The sequence composition analysis module supports VirHostMatcher, WIsH, and codon usage, which are complementary because VirHostMatcher may be more suitable for complete genomes, whereas WIsH (for virus contigs shorter than 10 kb) and codon usage distance can be detected in both complete and incomplete genomes. The genetic homology module detects the sequence homology between any phage–host pair regions of genetic homology, and provides visualizations to display the degree of matching between the phage–host pair as circular genome viewers. In the co-abundance analysis module, we used the CoNet program to infer a viral and bacterial co-occurrence network. As a plug-in of Cytoscape, we adapted CoNet to a web version to better aid biologists without a computational background to use and adjust parameters. As shown in the Figure 3A, a distinct phage–host sub-population was determined by PPIs/DDIs compared with other categories of signals, with the hosts of 19 phages (2.3%) were correctly identified only by PPIs/DDIs but not supported by any other signals. Therefore, the PPIs/DDIs could reflect the phage–host interactions originated from the interactions between their encoded proteins though not supported by other categories of signals. Therefore, we also provided a functional module for the detection of PPIs and DDIs between a pair of phage–host genomes to better understand their interplay at the protein level. In addition, to assist in characterizing phage genomes for therapeutic applications, we introduced a specialty gene check module to detect VFs and ARGs.
Finally, a consensus analysis using machine learning models was performed to indicate the possible integrity of the predicted interactions and the interplay among different PHISs. Based on PHIS detection for the training set, consisting of 817 known phage–host interactions, more than 85% of the phage hosts were correctly identified at the species level by combining all categories of signals. The integrated RF model trained on the training set attained the best performance, with an AUC value of 0.9347 and an accuracy of 0.875 for the test set (936 known phage–host pairs). Therefore, the PHISDetector can predict interactions that could not have been detected using a single category of signals, and can precisely calculate the possibility of novel phage–microbe pairs.
Case study 1: identification of hosts of metagenomic viral contigs using PHISDetector
As a large number of new viral genomes or sequence fragments are being unveiled by viral metagenomics, predicting the microbial hosts for these metagenomic phage contigs remains one of the most fundamental challenges in understanding the ecological roles of phages [40]. The stand-alone version of the PHISDetector is particularly powerful for expanding our framework for large viral metagenomics dataset analysis. Users can submit high-throughput sequencing-derived phage sequences as the input, and the predicted bacterial hosts of these phages are returned.
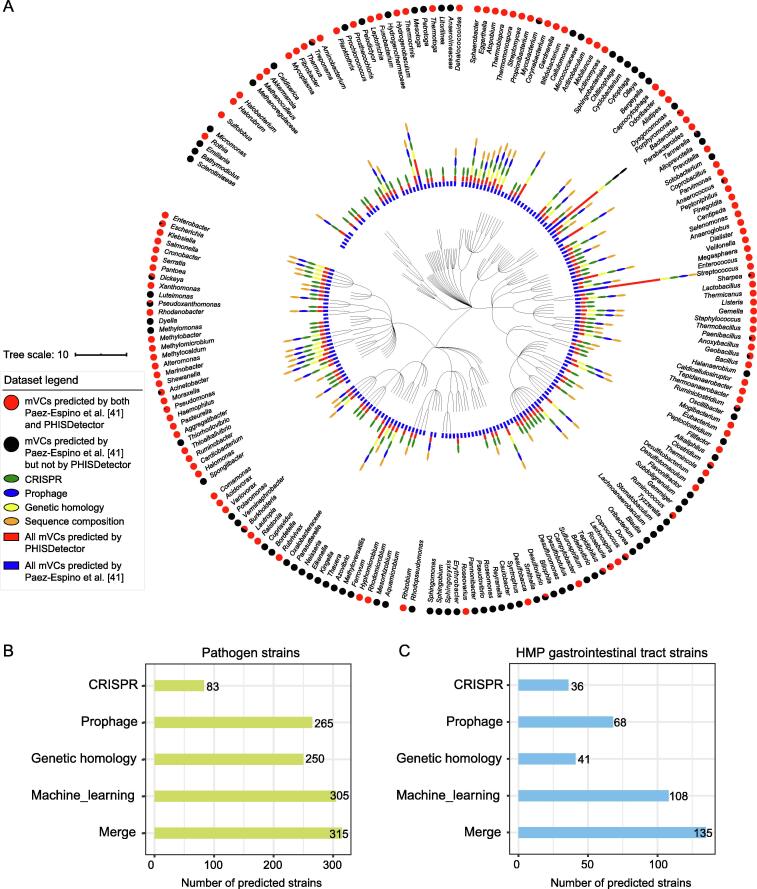
We tested a set of 125,842 metagenomic viral contigs (mVCs) from 3042 geographically diverse samples [41] and predicted their bacterial hosts using PHISDetector. First, using criterion 1, we could predict the bacterial hosts of 13,304 (10.57%), 2221 (1.76%), and 276 (0.22%) mVCs by matching CRISPR spacers, genetic homology of bacterial genomes, and microbial prophages with mVCs, respectively. Second, using criterion 2, 111,058 (88.25%) mVCs were retained for further evaluation by machine learning models (Figure 1). Finally, 64,957 mVCs (51.62%) were returned with predicted hosts at the genus level, supported by at least two trained machine learning models with a probability ≥ 0.8. Compared with the original study, in which only 9607 (7.7%) of the mVCs whose hosts were bacteria were predicted mainly through CRISPR spacers and transfer RNA matches, PHISDetector annotated hosts for 69,257 (55.03%) of all mVCs, and the predicted hosts matched the previous annotation in 62.34% of cases at the genus level (Figure 4A, Figure S4; Tables S5 and S6). In summary, PHISDetector can successfully predict bacterial hosts for virome contigs in large datasets.
Figure 4.
Performance of PHISDetector in predicting bacterial hosts for mVCs and the infecting phages for MDR bacteria and human gut bacteria
A. Phylogenetic distribution of the bacterial hosts for mVCs. In total, 205 genera are shown in the phylogenetic tree. The innermost circle (blue rectangles) shows the number of mVCs assigned to a genus by Paez-Espino et al. [41] and the adjacent circle (red rectangles) shows the number of mVCs assigned to a genus by PHISDetector. In the middle circles, various signals detected for a genus are marked: CRISPR (green ovals), prophage (blue ovals), genetic homology (yellow ovals), and sequence composition (orange ovals). In the outermost circle, the pie charts indicate the consistency of host prediction at genus level between Paez-Espino et al. [41] and PHISDetector. Red indicates the fraction of mVCs assigned to a genus by Paez-Espino et al. [41] that is also correctly predicted by PHISDetector. Black indicates the fraction of mVCs assigned to a genus by Paez-Espino et al. [41] but not predicted by PHISDetector. B. and C. The number of strains with reliable phages predicted using strong CRISPR, prophage, or genetic homology signals using criterion 1 or using the trained machine learning model, respectively, for 315 MDR bacteria (B) and 454 human gut bacteria (C). mVC, metagenomic viral contig; MDR, multidrug-resistant; HMP, Human Microbiome Project.
Case study 2: prediction of infecting phages for MDR bacteria and human gut bacteria
Antibiotic resistance in bacteria, especially a dramatic increase in MDR bacteria, has emerged as a global challenge over the past century. As viruses bear the ability to kill or inhibit bacteria, bacteriophages may provide a therapeutic opportunity to combat MDR bacteria. To demonstrate this application, we extracted 368 clinical bacterial pathogen isolates from the NCBI Pathogen Detection database (https://www.ncbi.nlm.nih.gov/pathogens/isolates/, using the query “host: Homo sapiens && epi_type: clinical && asm_acc: GCA* && creation_date: 2020 && AMR_genotypes: *”). These bacterial isolates belong to 31 species, and have complete bacterial genome sequences and predicted antimicrobial resistance (AMR) genotypes. We applied PHISDetector to predict potential infecting phages for these pathogens, and obtained a total of 927 reliable infecting phages for 315 bacterial isolates (85.6%) from 21 species (67.7%). Among these bacterial isolates, 83 (26%), 265 (84%), and 250 (79%) strains were detected based on strong CRISPR, prophage, and genetic homology signals using criterion 1, respectively. PHISs could be detected in most of the strains (305, 96.8%) using a machine learning model (Figure 4B). Reliable prediction of infecting phages is considered when there is a match between the genus of the bacterial host for a phage and the genus of the query bacteria. These predicted interactions may provide vital information for the development of novel phage therapies for the treatment of MDR bacterial infections. Detailed information can be accessed and browsed via the PHISDetector webserver (http://www.microbiome-bigdata.com/PHISDetector/index/case_study).
Although a growing body of research has emphasized the role of the human gut microbiome in human health and disease, knowledge of phages that can infect major human gut commensal bacteria remains limited. The discovery of their interactions will provide potential tools for precise manipulation of specific microbes in human gut and be of great benefit to studies about the function of intestinal symbiotic bacteria as well as development of therapies for treating pathogenic bacteria. Therefore, we collected 454 bacterial isolates (representing 222 species) from the human gastrointestinal tract that have complete sequences and annotation via https://www.hmpdacc.org/hmp/HMRGD/. In total, 416 candidate phages for 135 (30%) bacteria from 55 (25%) species were predicted with high reliability using a PHISDetector, with 95 (70.4%) strains detected by strong signals in criterion 1, and 108 (80%) using a machine learning model (Figure 4C). PHISDetector performed dependably for major gut bacteria; for example, as one of the most abundant bacterial species in the human gut microbiota, a reduction in the abundance of Faecalibacterium prausnitzii is relevant to the pathogenesis of inflammatory bowel disease (IBD) [42]. PHISDetector could identify the exact infecting phages for several F. prausnitzii strains, whose phages were first isolated in 2018 [43] (Table 1). It should also be noted that only a limited number of phages could be isolated because most human gut bacteria are anaerobic, and are difficult to isolate or culture. Meanwhile, among the 319 bacterial isolates (182 species) for which PHISDetector failed to predict any reliable phages, in 88 species (∼ 48%) this was due to the lack of sequenced phages infecting bacteria within the same genus in our PGPD. With more human gut virome studies conducted, more gut phage DNA will be isolated, sequenced, and added into viral databases, which will notably enhance the detection ability of PHISDetector in the future.
Table 1.
Isolated phages reportedbyCornuault et al.[43]and predicted phages using PHISDetector for Faecalibacterium prausnitzii strains
| Strain | Isolated phage name | Isolated phage GenBank ID | Predicted phage GenBank ID |
PHIS |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
CRISPR |
Prophage |
Genetic homology |
Machine learning | |||||||
| C1 | C2 | C1 | C2 | C1 | C2 | |||||
| Faecalibacterium prausnitzii M21/2 | FP_Epona | MG711462 | NA | × | × | × | √ | × | × | × |
| Faecalibacterium prausnitzii A2-165 | FP_Mushu | MG711460 | MG711460 | × | × | √ | √ | √ | √ | × |
| FP_Lagaffe | MG711461 | MG711461 | × | × | √ | √ | √ | √ | × | |
| Faecalibacterium prausnitzii SL3/3 | FP_Toutatis | MG711466 | MG711466 | × | × | × | √ | × | √ | √ |
| Faecalibacterium prausnitzii KLE1255 | FP_Lugh | MG711464 | NA | × | × | × | × | × | × | × |
| FP_Toutatis | MG711466 | MG711466 | × | × | √ | √ | × | √ | × | |
| Faecalibacterium prausnitzii L2-6 | FP_Toutatis | MG711466 | MG711466 | × | × | √ | √ | √ | √ | × |
| FP_Lugh | MG711464 | NA | × | × | × | × | × | × | × | |
| FP_Taranis | MG711467 | MG711467 | √ | √ | √ | √ | × | √ | × | |
Note: “√” denotes that the corresponding signal could be detected for the phage–host pair; “×” denotes that the corresponding signal could not be detected for the phage–host pair. C1 and C2 denotes criterion 1 and criterion 2 defined in Table S3, respectively. PHIS, phage–host interaction signal; NA, not available.
Case study 3: making predictions and annotations using the PHISDetector webserver
The PHISDetector webserver receives bacterial or virus genomic sequences in GBK or FASTA format as input, and provides graphical results and data tables with details to download. For a FASTA input file, ORFs will be first predicted on the input genome using FragGeneScan [44], while for a GBK file, DNA sequences and ORF amino acid sequences of the genome will be extracted directly from the input GBK file (Figure 1). The PHISDetector webserver supports three types of analysis. 1) Evaluation of the interacting probability for a pair of phage and prokaryotic genomes. If a pair of phage–microbe genome sequences has been submitted, the PHIE module will be applied to indicate the possibility of the interaction (Figure 1). 2) Prediction of infecting phages for a query prokaryotic genome. If a bacterial sequence has been submitted (Figure 1, upper left), the ORFs, prophage regions, and CRISPR arrays will be initially detected. Then, the PHIE module will be performed to evaluate the interacting potential for each of the 18,387 phages in the PGPD with the input bacterial sequence. 3) Prediction of bacterial hosts for the query phage genome. If a phage sequence has been submitted (Figure 1, upper right), the PHIE module will be used to evaluate the interacting potential of the query phage with each of the 24,799 bacterial genomes in the BGPD or 13,183,722 spacers in the CSD based on CRISPR spacer matching information.
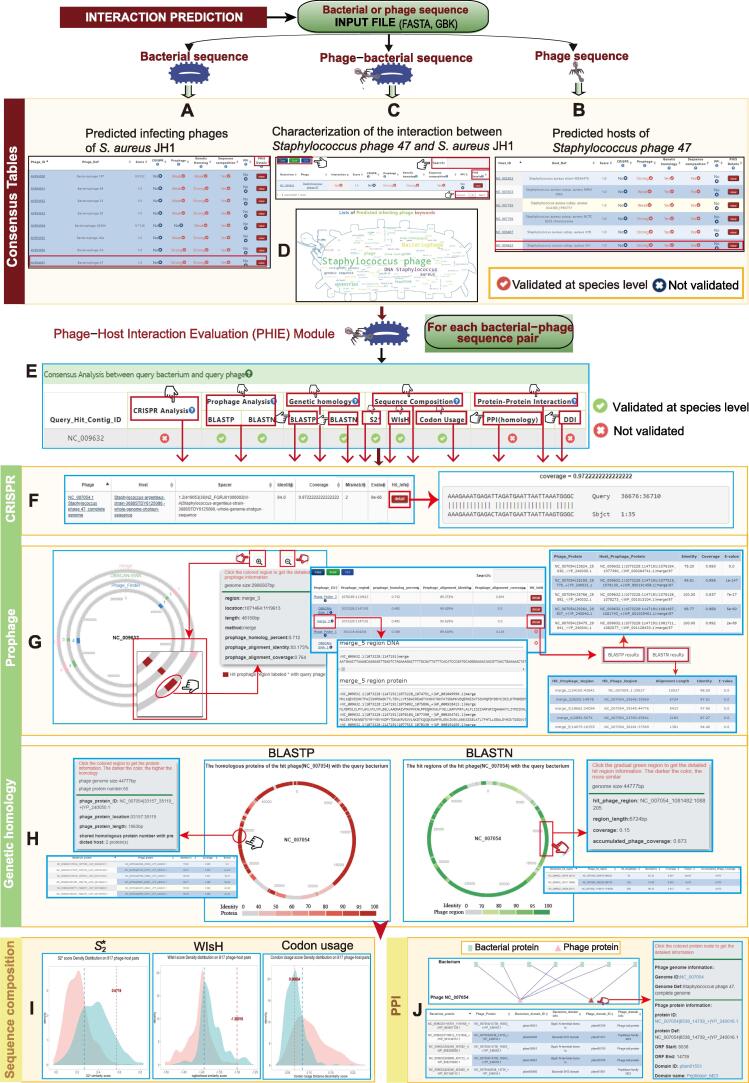
We illustrated the output results using the predictions for infecting phages of Staphylococcus aureus subsp. aureus JH1 (NC_009632) (Figure 5A), and for bacterial hosts of Staphylococcus phage 47 (NC_007054) (Figure 5B), as well as the characterization of interactions between them (Figure 5C). A word cloud plot showing the frequency of keywords for all predicted phages for S. aureus JH1 was used to visualize the dominant phages (Figure 5D). For each candidate phage–host pair, a consensus table displaying different PHISs was used to give an overview of all detected signals supporting the interaction and consistency among signals (Figure 5E). Interactive DataTables were used to display the prediction results with details for CRISPR, prophage, genetic homology, sequence composition, and PPI signals (Figure 5F–J). In addition, several kinds of interactive graphics are provided to facilitate browsing, analysis, and interpretation of the prediction results. For the CRISPR signal, a table showing matching between the bacterial host CRISPR spacer and phage protospacer is provided (Figure 5F). For the prophage signal, an interactive circular genome viewer is provided to illustrate the prophage regions in host genome with detailed information for homologous comparison between the phage and the matching prophage (Figure 5G). For genetic homology analysis, a circular genome viewer can be used to evaluate the similarity between host and phage based on homologous protein alignment by DIAMOND BLASTP and nucleotide alignment by BLASTN (Figure 5H). For sequence composition signals, , WIsH, and codon usage scores are plotted on the density curves, with red and blue curves representing the distribution of scores calculated using the positive and negative training sets, respectively (Figure 5I). For PPIs, the interactive bipartite network shows the PPIs between the phage and bacterial proteins, with DataTables providing detailed information about proteins. In addition, PHISDetector also provides seven independent analysis modules: oligonucleotide profile analysis, CRISPR analysis, prophage analysis, protein interaction, specialty gene check, similarity analysis, and co-occurrence analysis, to provide a flexible and convenient one-stop web service for oriented phage–host interaction analyses (Figure S5).
Figure 5.
Illustrations for making predictions and annotations using PHISDetector web server
A. Infecting phages identified from PGPD for bacterial strain Staphylococcus aureus subsp. aureus JH1 (NC_009632). B. Bacterial hosts for Staphylococcus phage 47 (NC_007054 or AY954957) identified from BGPD or CSD.C. Characterization of the phage–host pair (S. aureus JH1 vs. Staphylococcus phage 47). D. Word cloud showing the frequency of keywords of all predicted phages for S. aureus JH1. E. A consensus table displaying diverse PHISs detected for the phage–host interaction. F. CRISPR panel showing matching information between host CRISPR spacer and phage protospacer sequences. G. Circular genome viewers illustrating the sequence homology between the prophage regions of S. aureus JH1 and Staphylococcus phage 47, based on BLASTP or BLASTN homologous alignment. H. Circular genome viewers displaying the sequence homology between S. aureus JH1 and Staphylococcus phage 47, based on BLASTP or BLASTN homologous alignment, with color shade representing the level of similarity with detailed information shown when clicking on a region. I.S2*, WIsH, and codon usage scores (red lines) evaluating the sequence composition similarity between S. aureus JH1 and Staphylococcus phage 47 were plotted on background density curves, with red and blue curves representing the distribution of scores calculated using positive and negative training sets, respectively. J. Interactive bipartite network and tables giving the PPIs between proteins of S. aureus JH1 and its predicted phage Staphylococcus phage 47 (NC_007054).
Comparison with other methods
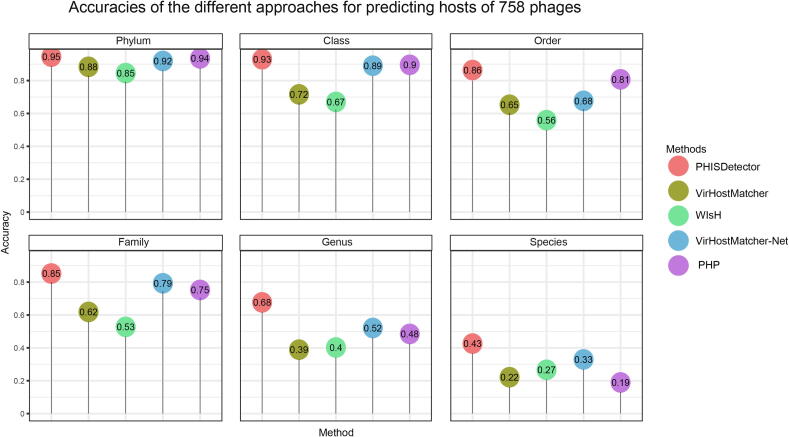
We compared PHISDetector with VirHostMatcher [11], WIsH [12], VirHostMatcher-Net [21], and PHP [13] on an independent benchmark dataset including 758 annotated phage–host pairs (see Method). Since all the four published methods and PHISDetector calculate a score to indicate the reliability of a predicted phage–host pair, we return the one with the highest score (or probability) as the predicted hosts for each phage and calculated a host prediction accuracy as the percentage of phages whose representative hosts predicted by these methods belong to the same taxonomic affiliation as their annotated hosts (Table S7). As shown in Figure 6, PHISDetector outperformed the other tools at all taxonomic levels, especially at the species and genus levels. In addition, we compared the predicted results of PHISDetector and RaFAH for 125,842 mVCs from 3042 geographically diverse samples [41]. Compared with the original study, in which only 9607 (7.7%) of the mVCs were predicted mainly through CRISPR spacers and transfer RNA matches, PHISDetector annotated hosts for 69,257 (55.03 %) of all mVCs, and the predicted hosts at the genus level matched the previous annotation in 62.34% of cases. Comparatively, with a P value threshold of 0.1, WIsH annotated 59% of the mVCs and the predicted hosts matched the previous annotation in 70% of the cases just at the family level; RaFAH just annotated hosts for 20,409 contigs (16.22%) of all mVCs with a probability ≥ 0.5, and matched the previous annotation in 47.05% of cases with a probability ≥ 0.8 and 61.5% of cases with a probability ≥ 0.5. Moreover, PHISDetector showed significantly better performance than RaFAH (bootstrap test on ROC with P = 0.0025; Figure S4; Table S6). In summary, PHISDetector presented better performance in predicting hosts for complete phage genomes or virome contigs than other available tools.
Figure 6.
Comparisonof the performance of PHISDetector with VirHostMatcher, WIsH, VirHostMatcher-Net,and PHP on 758 annotated phage–host pairs
Lollipop chart showing the prediction accuracies of the different approaches for 758 phages. Prediction accuracies were compared between PHISDetector and other four published phage–host interaction prediction tools, including VirHostMatcher, WIsH, VirHostMatcher-Net, and PHP, at different taxonomic levels, including species, genus, family, order, class, and phylum. The principle for assigning the host is that the one with the highest score (or probability) is predicted as the host for each phage.
Discussion
In the present study, we applied an integrated approach to develop PHISDetector for phage–host interaction predictions. Compared with prior tools, the PHISDetector pipeline is uniquely comprehensive, because it integrates various types of PHISs, reflecting possible phage–microbe interacting mechanisms into one tool, and adds valuable novel functionalities. Consequently, PHISDetector can predict additional interactions that cannot be detected using a single category, and can calculate the possibility of a novel phage–microbe pair using trained machine learning models. Users can choose to use the web server or stand-alone version flexibly, according to their research and resources, both of which provide well-designed, interactive visualization outputs for improved interpretation. The PHISDetector will continue to develop to incorporate additional in silico phage–host signals, and to evaluate the consistency of the association between different signals upon extensive analysis of large datasets. We hope that PHISDetector can promote research on the role of phage–host interactions from ecological and evolutionary perspectives, facilitate our understanding of their roles in human health and disease, and accelerate the development of novel therapeutic strategies, such as modulating specific microbes in a microbial community and treating MDR infections.
Code availability
The source code of PHISDetector in this work can be available at https://github.com/HIT-ImmunologyLab/PHISDetector.
Data availability
All other relevant data in this work can be available at http://www.microbiome-bigdata.com/PHISDetector/index/download.
CRediT authorship statement
Fengxia Zhou: Conceptualization, Methodology, Software, Writing - original draft. Rui Gan: Conceptualization, Methodology, Software, Writing - original draft. Fan Zhang: Conceptualization, Methodology, Software, Writing - original draft, Funding acquisition. Chunyan Ren: Writing - original draft, Validation. Ling Yu: Investigation, Validation. Yu Si: Investigation, Validation. Zhiwei Huang: Conceptualization, Writing - review & editing, Supervision, Funding acquisition. All authors have read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Acknowledgments
We thank Zeguo Sun and Weijia Zhang from Icahn School of Medicine at Mount Sinai, Jiqiu Wu from Imperial College, and Fang Wang from MD Anderson Cancer Center for helpful comments and for testing the webserver software. We thank Yunfei Ji, Jiale Zhang, and Yongkui Lai for their help in the developments of PHISDetector. This work is supported by the National Natural Science Foundation of China (Grant Nos. 31825008, 31422014, and 61872117).
Handled by Feng Gao
Footnotes
Peer review under responsibility of Beijing Institute of Genomics, Chinese Academy of Sciences / China National Center for Bioinformation and Genetics Society of China.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gpb.2022.02.003.
Supplementary material
The following are the Supplementary data to this article:
Top 20 genera of the phage and their hosts in the training and test datasets A. Fraction of top 20 phage genera in the training and test datasets. B. Fraction of top 20 host genera in the training and test datasets.
Performance of the trained random forest models based on five single phage–host interaction signals ROC of the trained RF models based on five single PHISs including CRISPR, Prophage, Genetic homology, Sequence composition, and PPI. The AUC values were shown for each signal respectively.
The performance of machine learning models for novel phages and predicting the hosts for phages at various lengths using PHISDetector A. Predicted accuracies of 100 experienced and novel phages at the species level, which are separated based on the phage genera by random sampling of the validation set 100 times. B. The left bar chart represented the fractions of phages in PGPD at various lengths. (1 kb, 10 kb] means > 1 kb and ≤ 10 kb. Other regions are similar. The right subplot showed the prediction accuracies (shown in the circles) of PHISDetector that predict hosts for 1434 phages with known phage–host interactions at various lengths, at the genus and family levels (right).
Comparison of the host prediction performance between PHISDetector and RaFAH for mVCs ROC showing the host prediction performance of PHISDetector and RaFAH for mVCs. The AUC values were shown for each approach, respectively. P value was calculated by bootstrap test to assess the significance of the difference between two ROC curves.
Illustrations for seven independent phage–host interaction related modules using the PHISDetector web server Various graphics for visualizations of seven independent analysis modules including interactive DataTables for oligonucleotide profile analysis and CRISPR analysis, heatmap for oligonucleotide profile analysis, interactive tree for similarity analysis, circular genome viewer for prophage analysis and specialty gene check, and interactive bipartite networks for PPI analysis with co-occurrence analysis.
References
- 1.Chatterjee A., Duerkop B.A. Beyond bacteria: bacteriophage–eukaryotic host interactions reveal emerging paradigms of health and disease. Front Microbiol. 2018;9:1394. doi: 10.3389/fmicb.2018.01394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Edwards R.A., McNair K., Faust K., Raes J., Dutilh B.E. Computational approaches to predict bacteriophage–host relationships. FEMS Microbiol Rev. 2016;40:258–272. doi: 10.1093/femsre/fuv048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roux S., Enault F., Hurwitz B.L., Sullivan M.B. VirSorter: mining viral signal from microbial genomic data. PeerJ. 2015;3:e985. doi: 10.7717/peerj.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arndt D., Grant J.R., Marcu A., Sajed T., Pon A., Liang Y., et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44:W16–W21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lima-Mendez G., Van Helden J., Toussaint A., Leplae R. Prophinder: a computational tool for prophage prediction in prokaryotic genomes. Bioinformatics. 2008;24:863–865. doi: 10.1093/bioinformatics/btn043. [DOI] [PubMed] [Google Scholar]
- 6.Fouts D.E. Phage_Finder: automated identification and classification of prophage regions in complete bacterial genome sequences. Nucleic Acids Res. 2006;34:5839–5851. doi: 10.1093/nar/gkl732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Sousa A.L., Maues D., Lobato A., Franco E.F., Pinheiro K., Araujo F., et al. PhageWeb - web interface for rapid identification and characterization of prophages in bacterial genomes. Front Genet. 2018;9:644. doi: 10.3389/fgene.2018.00644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gan R., Zhou F., Si Y., Yang H., Chen C., Wu J., et al. DBSCAN-SWA: an integrated tool for rapid prophage detection and annotation. Front Genet. 2022;13:885048. doi: 10.3389/fgene.2022.885048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao N.L., Zhang C., Zhang Z., Hu S., Lercher M.J., Zhao X.M., et al. MVP: a microbe-phage interaction database. Nucleic Acids Res. 2018;46:D700–D707. doi: 10.1093/nar/gkx1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pride D.T., Wassenaar T.M., Ghose C., Blaser M.J. Evidence of host-virus co-evolution in tetranucleotide usage patterns of bacteriophages and eukaryotic viruses. BMC Genomics. 2006;7:8. doi: 10.1186/1471-2164-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahlgren N.A., Ren J., Lu Y.Y., Fuhrman J.A., Sun F. Alignment-free oligonucleotide frequency dissimilarity measure improves prediction of hosts from metagenomically-derived viral sequences. Nucleic Acids Res. 2017;45:39–53. doi: 10.1093/nar/gkw1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galiez C., Siebert M., Enault F., Vincent J., Soding J. WIsH: who is the host? Predicting prokaryotic hosts from metagenomic phage contigs. Bioinformatics. 2017;33:3113–3114. doi: 10.1093/bioinformatics/btx383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu C., Zhang Z., Cai Z., Zhu Z., Qiu Y., Wu A., et al. Prokaryotic virus host predictor: a Gaussian model for host prediction of prokaryotic viruses in metagenomics. BMC Biol. 2021;19:5. doi: 10.1186/s12915-020-00938-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stern A., Mick E., Tirosh I., Sagy O., Sorek R. CRISPR targeting reveals a reservoir of common phages associated with the human gut microbiome. Genome Res. 2012;22:1985–1994. doi: 10.1101/gr.138297.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J., Gao Y., Zhao F. Phage–bacteria interaction network in human oral microbiome. Environ Microbiol. 2016;18:2143–2158. doi: 10.1111/1462-2920.12923. [DOI] [PubMed] [Google Scholar]
- 16.Biswas A., Gagnon J.N., Brouns S.J., Fineran P.C., Brown C.M. CRISPRTarget: bioinformatic prediction and analysis of crRNA targets. RNA Biol. 2013;10:817–827. doi: 10.4161/rna.24046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dutilh B.E., Cassman N., McNair K., Sanchez S.E., Silva G.G., Boling L., et al. A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nat Commun. 2014;5:4498. doi: 10.1038/ncomms5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Villarroel J., Kleinheinz K.A., Jurtz V.I., Zschach H., Lund O., Nielsen M., et al. HostPhinder: a phage host prediction tool. Viruses. 2016;8:116. doi: 10.3390/v8050116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shapiro J.W., Putonti C. Gene co-occurrence networks reflect bacteriophage ecology and evolution. mBio. 2018;9:e01870–17. doi: 10.1128/mBio.01870-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leite D.M.C., Brochet X., Resch G., Que Y.A., Neves A., Pena-Reyes C. Computational prediction of inter-species relationships through omics data analysis and machine learning. BMC Bioinformatics. 2018;19:420. doi: 10.1186/s12859-018-2388-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang W., Ren J., Tang K., Dart E., Ignacio-Espinoza J.C., Fuhrman J.A., et al. A network-based integrated framework for predicting virus–prokaryote interactions. NAR Genom Bioinform. 2020;2:lqaa044. doi: 10.1093/nargab/lqaa044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coutinho F.H., Zaragoza-Solas A., Lopez-Perez M., Barylski J., Zielezinski A., Dutilh B.E., et al. RaFAH: host prediction for viruses of Bacteria and Archaea based on protein content. Patterns (N Y) 2021;2 doi: 10.1016/j.patter.2021.100274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fu L., Niu B., Zhu Z., Wu S., Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grissa I., Vergnaud G., Pourcel C. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007;35:W52–W57. doi: 10.1093/nar/gkm360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edgar R.C. PILER-CR: fast and accurate identification of CRISPR repeats. BMC Bioinformatics. 2007;8:18. doi: 10.1186/1471-2105-8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang F., Zhao S., Ren C., Zhu Y., Zhou H., Lai Y., et al. CRISPRminer is a knowledge base for exploring CRISPR-Cas systems in microbe and phage interactions. Commun Biol. 2018;1:180. doi: 10.1038/s42003-018-0184-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dion M.B., Plante P.L., Zufferey E., Shah S.A., Corbeil J., Moineau S. Streamlining CRISPR spacer-based bacterial host predictions to decipher the viral dark matter. Nucleic Acids Res. 2021;49:3127–3138. doi: 10.1093/nar/gkab133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., et al. Scikit-learn: machine learning in Python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]
- 29.Ye Y., Zhang Q. Characterization of CRISPR RNA transcription by exploiting stranded metatranscriptomic data. RNA. 2016;22:945–956. doi: 10.1261/rna.055988.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laslett D., Canback B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004;32:11–16. doi: 10.1093/nar/gkh152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buchfink B., Xie C., Huson D.H. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 32.Kaminski J., Gibson M.K., Franzosa E.A., Segata N., Dantas G., Huttenhower C. High-specificity targeted functional profiling in microbial communities with ShortBRED. PLoS Comput Biol. 2015;11:e1004557. doi: 10.1371/journal.pcbi.1004557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen L., Zheng D., Liu B., Yang J., Jin Q. VFDB 2016: hierarchical and refined dataset for big data analysis–10 years on. Nucleic Acids Res. 2016;44:D694–D697. doi: 10.1093/nar/gkv1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jia B., Raphenya A.R., Alcock B., Waglechner N., Guo P., Tsang K.K., et al. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017;45:D566–D573. doi: 10.1093/nar/gkw1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Faust K, Raes J. CoNet app: inference of biological association networks using Cytoscape. F1000Res 2016;5:1519. [DOI] [PMC free article] [PubMed]
- 36.Franz M., Lopes C.T., Huck G., Dong Y., Sumer O., Bader G.D. Cytoscape.js: a graph theory library for visualisation and analysis. Bioinformatics. 2016;32:309–311. doi: 10.1093/bioinformatics/btv557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Touchon M., Bernheim A., Rocha E.P. Genetic and life-history traits associated with the distribution of prophages in bacteria. ISME J. 2016;10:2744–2754. doi: 10.1038/ismej.2016.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shmakov S.A., Sitnik V., Makarova K.S., Wolf Y.I., Severinov K.V., Koonin E.V. The CRISPR spacer space is dominated by sequences from species-specific mobilomes. mBio. 2017;8:e01397–17. doi: 10.1128/mBio.01397-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ester M., Kriegel H.P., Sander J., Xu X. A density-based algorithm for discovering clusters a density-based algorithm for discovering clusters in large spatial databases with noise. Proc 2nd Int Conf Knowl Discov Data Min. 1996:226–231. [Google Scholar]
- 40.Roux S., Brum J.R., Dutilh B.E., Sunagawa S., Duhaime M.B., Loy A., et al. Ecogenomics and potential biogeochemical impacts of globally abundant ocean viruses. Nature. 2016;537:689–693. doi: 10.1038/nature19366. [DOI] [PubMed] [Google Scholar]
- 41.Paez-Espino D., Eloe-Fadrosh E.A., Pavlopoulos G.A., Thomas A.D., Huntemann M., Mikhailova N., et al. Uncovering Earth's virome. Nature. 2016;536:425–430. doi: 10.1038/nature19094. [DOI] [PubMed] [Google Scholar]
- 42.Cao Y., Shen J., Ran Z.H. Association between Faecalibacterium prausnitzii reduction and inflammatory bowel disease: a meta-analysis and systematic review of the literature. Gastroenterol Res Pract. 2014;2014 doi: 10.1155/2014/872725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cornuault J.K., Petit M.A., Mariadassou M., Benevides L., Moncaut E., Langella P., et al. Phages infecting Faecalibacterium prausnitzii belong to novel viral genera that help to decipher intestinal viromes. Microbiome. 2018;6:65. doi: 10.1186/s40168-018-0452-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rho M., Tang H., Ye Y. FragGeneScan: predicting genes in short and error-prone reads. Nucleic Acids Res. 2010;38:e191. doi: 10.1093/nar/gkq747. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Top 20 genera of the phage and their hosts in the training and test datasets A. Fraction of top 20 phage genera in the training and test datasets. B. Fraction of top 20 host genera in the training and test datasets.
Performance of the trained random forest models based on five single phage–host interaction signals ROC of the trained RF models based on five single PHISs including CRISPR, Prophage, Genetic homology, Sequence composition, and PPI. The AUC values were shown for each signal respectively.
The performance of machine learning models for novel phages and predicting the hosts for phages at various lengths using PHISDetector A. Predicted accuracies of 100 experienced and novel phages at the species level, which are separated based on the phage genera by random sampling of the validation set 100 times. B. The left bar chart represented the fractions of phages in PGPD at various lengths. (1 kb, 10 kb] means > 1 kb and ≤ 10 kb. Other regions are similar. The right subplot showed the prediction accuracies (shown in the circles) of PHISDetector that predict hosts for 1434 phages with known phage–host interactions at various lengths, at the genus and family levels (right).
Comparison of the host prediction performance between PHISDetector and RaFAH for mVCs ROC showing the host prediction performance of PHISDetector and RaFAH for mVCs. The AUC values were shown for each approach, respectively. P value was calculated by bootstrap test to assess the significance of the difference between two ROC curves.
Illustrations for seven independent phage–host interaction related modules using the PHISDetector web server Various graphics for visualizations of seven independent analysis modules including interactive DataTables for oligonucleotide profile analysis and CRISPR analysis, heatmap for oligonucleotide profile analysis, interactive tree for similarity analysis, circular genome viewer for prophage analysis and specialty gene check, and interactive bipartite networks for PPI analysis with co-occurrence analysis.
Data Availability Statement
All other relevant data in this work can be available at http://www.microbiome-bigdata.com/PHISDetector/index/download.