Abstract
The repair of damaged articular cartilage is an unmet medical need. Chondrocyte-based cell therapy has been used to repair cartilage for over 20 years despite current limitations. Chondrocyte dedifferentiation upon expansion in monolayer is well known and is the main obstacle to their use as cell source for cartilage repair. Consequently, current approaches often lead to fibrocartilage, which is biomechanically different from hyaline cartilage and not effective as a long-lasting treatment. Here, we describe an innovative 3-step method to engineer hyaline-like cartilage microtissues, named Cartibeads, from high passage dedifferentiated chondrocytes. We show that WNT5A/5B/7B genes were highly expressed in dedifferentiated chondrocytes and that a decrease of the WNT signaling pathway was instrumental for full re-differentiation of chondrocytes, enabling production of hyaline matrix instead of fibrocartilage matrix. Cartibeads showed hyaline-like characteristics based on GAG quantity and type II collagen expression independently of donor age and cartilage quality. In vivo, Cartibeads were not tumorigenic when transplanted into SCID mice. This simple 3-step method allowed a standardized production of hyaline-like cartilage microtissues from a small cartilage sample, making Cartibeads a promising candidate for the treatment of cartilage lesions.
Keywords: articular cartilage, hyaline cartilage, tissue engineering, microtissues, dedifferentiation, re-differentiation, WNT signaling
Graphical Abstract
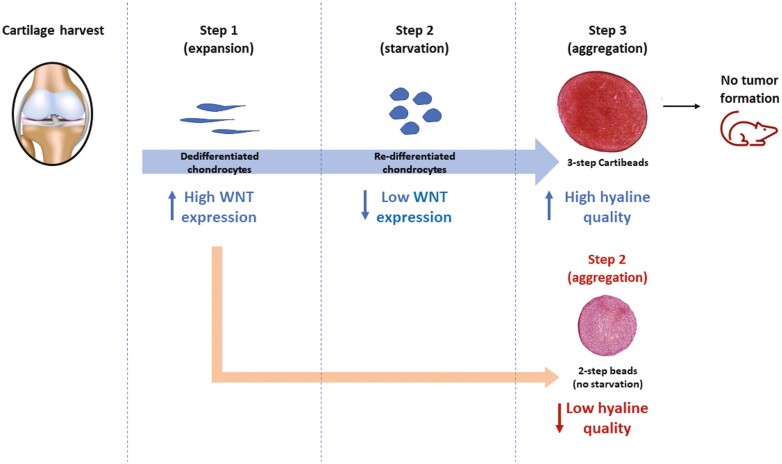
Graphical Abstract.
Significance Statement.
This article reports a novel cellular therapy for chondral lesions using three-dimensional engineered cartilage tissues of high hyaline quality, called Cartibeads. Here, the authors have characterized the produced cartilage using quantitative and qualitative analysis showing their hyaline composition. Cartibeads as a method allows the mass amplification of chondrocytes that dedifferentiate and to redifferentiate them into chondrocytes capable of producing an extracellular matrix with characteristics close to hyaline cartilage, independently of the age of the patient and the degenerative status of the donor joint.
Background
Chondrocyte-based cell therapy has been used in attempt to repair cartilage for over 20 years, but faces numerous limitations.1 Chondrocytes, as the only resident cells in cartilage, represent a key cell source for cartilage regeneration. However, cartilage has a low cell density, which decreases with age.2 Chondrocytes are responsible for the maintenance of the hyaline cartilage matrix, characterized by the presence of glycosaminoglycan (GAG) and type II collagen.3,4 A major challenge for chondrocyte-based cell therapy is that chondrocytes dedifferentiate in 2-dimensional (2D) cell culture, resulting in a rapid loss of chondrogenic phenotype5 and inability to produce type II collagen and GAG.6,7 Accordingly, chondrocytes differentiate into fibroblast-like cells that produce type I collagen, a key component of fibrocartilage. Fibrocartilage is biomechanically different from hyaline cartilage and is not an effective tissue for long-lasting cartilage repair.8
Chondrogenic phenotype can be partly maintained in 3-dimensional (3D) culture systems,9 with or without the use of external scaffolds. However, re-differentiation efficiency decreases with passage number. Consequently, most current approaches developed for cell therapy use cells up to passage 3.10,11 High cell passage can be achieved with fetal or juvenile cartilage.12 However, hyaline quality decreases with cell passage and fetal chondrocytes may evolve into hypertrophic tissue.13 For chondrocyte-based cell therapy the harvest of large cartilage biopsies is therefore required to reach sufficient cell numbers, with the risk of creating donor-site lesions. Another challenge for autologous chondrocyte-based cell therapy is the difficulty to expand and differentiate cells from elderly patients,2 excluding this population from most clinical trials as inclusion is limited to the age of 55.14
Pluripotent15,16 as well as multipotent stem cells such as mesenchymal stem cells (MSCs)17,18 have higher capacity of self-renewal than articular chondrocytes. However, pluripotent stem cells are associated with an increased risk of tumorigenicity, while MSCs tend to produce fibrocartilaginous scar tissue and hypertrophic cartilage.19 Indeed, MSCs show commitment toward osteogenesis by default. When induced to chondrogenesis, MSCs typically form hypertrophic cartilage that undergoes endochondral ossification. Consequently, cartilaginous tissue derived from MSCs typically undergoes remodeling into bone when transplanted in vivo.20 MSCs have been considered as a preferable cell source due to their ease of isolation. However, due to the heterogeneity of MSCs isolated from different sources (adipose tissue or bone marrow), their therapeutic use is unstable and limited.21 Current clinical trials show improvement in patient scores22 but have not yet shown evidence of regenerating articular cartilage.
Long-term success in the treatment of cartilage lesions requires implantation and integration of high-quality hyaline tissues resembling native cartilage.23,24 Nasal chondrocytes have been used instead of articular chondrocytes for their greater capacity to generate superior and reproducible hyaline-like cartilage tissues,25 but dedifferentiation issues remain. Currently, only 5 autologous chondrocyte-based cell therapies have been approved for human use to treat focal cartilage lesions26; MACI (Vericel,27), Spherox (Co.don,28,29), Chondron (Sewon Cellontech,30), Cartigrow (Regrow Bioscience31), and Ortho-ACI (Orthocell,32). Two of these therapies use animal-derived external scaffolds (MACI and Ortho-ACI) and none of them have yet established potency criteria based on GAG quantity and type II collagen expression.
Here, we present an innovative 3-step method allowing the use of extensively cultured adult dedifferentiated human chondrocytes to generate cartilage microtissues called Cartibeads. The Cartibeads method can reverse the loss of chondrogenic phenotype in extensively expanded chondrocytes up to passage 9 and from donors over 80 years old. Mechanistically, we identified a decrease in the WNT signaling pathway that facilitated the chondrogenic re-differentiation. Cartibeads were able to produce their own hyaline matrix without the support of an external scaffold. We used high level of GAG and type II collagen detection as potency criteria to evaluate the hyaline quality of Cartibeads. Moreover, for safety evaluation, we showed that Cartibeads were not tumorigenic upon implantation in immunodeficient SCID mice.
Methods
Production of Cartibeads
Human cartilage samples were obtained from consenting living donors (age 18 to 80 years old) following orthopedic procedures for various indications (Supplementary Table S1). The collected cartilage was transferred to the laboratory in a sterile recipient with normal saline (NaCl 0.9%) at room temperature.
The cartilage was sliced into small pieces (1 mm) to facilitate the extraction of chondrocytes by enzymatic digestion. Cartilage pieces were placed in orbital rotation at 37 °C overnight, with collagenase type II (400 U/mL, ThermoFisher) in medium E (ME; DMEM medium with high glucose (Gibco), 10% human serum (Sigma),1× L-glutamine,1× penicillin-streptomycin (P/S; ThermoFisher, 10378016), 1× non-essential amino acids (Gibco), 20 ng/mL FGF2, 10 ng/mL PDGF-BB, 5 ng/mL TGF-β3 (Cell Guidance)) containing antibiotic (gentamicin, 50 µg/mL, ThermoFisher), and antifungal (amphotericin B or fungizone , 0,25 µg/mL, ThermoFisher).
Once extracted, cells were washed and plated (p0) onto extracellular matrix (MaxGel , Sigma) pre-coated T25 cm2 flasks and cultured for 12 to 16 days in ME with antibiotic (gentamicin, 50 µg/mL, ThermoFisher) and antifungal (amphotericin B or fungizone, 0.25 µg/mL, ThermoFisher), which were maintained for 5 days. All 2D cell culture was conducted on extracellular matrix-coated flasks and in atmospheric oxygen conditions (21% O2) with 10% CO2. At confluence, cells were passaged in 1 T75 (p1) and later split in 2× T75 (p2) until confluence. At this stage, cells could be frozen for back-up (p2). After cell expansion in ME (step 1), cells were cultured for 7 days in medium R (MR; DMEM medium with high glucose (Gibco), 10% human platelet lysate containing high concentration of PDGF (PDGF-BB ~9 ng/mL, PDGF-AB~35 ng/mL,) and TGF-β1 (~140 ng/mL) and low concentration of FGF-2 (<0.05 ng/mL) (Sexton biotech), 1× L-glutamine, 1× penicillin-streptomycin (P/S; ThermoFisher, 10378016), 1× non-essential amino acid (Gibco)). Culture in MR represents step 2, or the re-differentiation phase, where a decrease in cell growth was observed. In step 3, chondrocytes were collected and resuspended in chondrogenic medium I (MI; DMEM medium with high glucose, 1× ITS-X, 1× L-glutamine, 1× penicillin-streptomycin (P/S; ThermoFisher, 10378016), 1× non-essential amino acid (Gibco, 11140050), ascorbic acid 200 µM (Sigma), 40 ng/mL of TGF-β3, 40 ng/mL BMP2, 40 ng/mL IGF-1 (Cell Guidance)) to obtain 0.2 × 106 cells/well in conical 96-well polypropylene plates (~20 × 106 cells/plate). The 96-well plates were centrifuged 5 minutes at 300g to allow cell aggregation into Cartibeads. During step 3, Cartibeads are cultured for 15 days in 3D culture in low oxygen level (5% O2) with 10% CO2, until fully formed. Cartibeads are removed from the 96-well plates and pooled together. Chondrocytes used to generate beads or Cartibeads in the 2- and 3-step methods came from cells cultured at passages 3 to 9.
To validate the role of WNT pathway downregulation during the re-differentiation phase (step 2 in the 3-step method), we used ME supplemented with 10 µM of XAV-939 (Sigma), a WNT pathway inhibitor. Chondrocytes were cultured for 4 days in ME + XAV-939 and then collected to form beads, following the 2-step method. Chondrocytes cultured in ME were used as control for data normalization.
GAG Quantification
Glycosaminoglycan (GAG) content was evaluated by the dimethylmethylene blue assay (DMMB) (Sigma, 341088). Chondroitin sulphate A (Sigma, C9819) was used to generate 6 standards, concentrations ranging from 0 to 50 µg/mL Chondroitin sulphate C (Sigma, C4384) was used to generate low and high Internal Quality Control (IQC) solutions, concentrations 15 and 35 µg/mL, respectively. Cartibeads were digested with proteinase K (1 mg/mL, Promega, V3021) in Tris-HCl buffer (50 mM, pH 8, Sigma), for 15 ± 2 h at 56 °C. The enzymatic digestion was stopped by incubation at 97 °C for 15 minutes. The resulting sample was diluted (1:5-1:10) in Tris-HCl buffer (50 mM, pH 8) for the assay. One hundred micro liters of standard, ICQ, or sample were read in triplicates with a spectrophotometer (λ = 525 nm) after 5 minutes reaction with 1 mL of DMMB working solution.
DNA Quantification
The GAG content was normalized to the DNA content, which was measured with PicoGreen-Qubit assay. Standards and ICQ were prepared from calf thymus DNA in TE buffer (200 mM Tris-HCl, 20 mM EDTA, pH 7.5). The samples used are those obtained from the proteinase K digestion, then diluted 1:15 in TE buffer. For this assay, 100 μL of standard, IQC, or sample were taken in triplicates. Then, 100 μL of diluted PicoGreen Quant-It solution (1:200, ThermoFisher, P11496) was added. The sample was incubated for 5 minutes, during which time the intercalant is complexed with the DNA. Finally, the reading was performed in Qubit 4 Fluorometer (ThermoFisher, Q33238), with excitation peak at 485 nm.
Safranin-O Staining
Safranin-O staining of cartilage beads and native cartilage samples (formalin-fixed, paraffin-embedded) was performed to reveal glycosaminoglycans (GAG). 5µm paraffin slides were deparaffinated by xylol and rehydrated by consecutive alcohol bath (concentrations 100%, 95% and 70%) with a final 5-minutes distilled water bath. Then, haematoxylin staining was used for nucleus counterstaining, followed by a 5-minutes wash with running warm water. Fast Green 0.001% (Sigma F7252) was used to stain the cytoplasm for 5 minutes, then washed out with acetic acid (1:100). The slides were washed out immediately with distilled water. Safranin-O 0.1% (Sigma S2255) was applied for staining of GAGs for 2.5 minutes, then washed repeatedly with distilled water. Dehydration was achieved by vigorous shaking the rack 10-20 times in alcohol baths (concentration 95% and 100%), followed by a xylol bath until slides assembly with Eukitt resin (Batch A1113, KiNDO1500).
Immunohistochemical Staining
Five micrometers of paraffin slides were dried overnight at 47 °C. Slides were deparaffinated by xylol and rehydrated by consecutive alcohol baths (concentrations 100%, 95%, and 70%). Cartibeads samples were immersed in 0.01M citrate buffer bath at pH=6, heated 3 times, 5 minutes each in microwave at 620w and then cooled in ice bath for 20 minutes. The slides were then rinsed in PBS for 5 minutes. The primary antibodies were then used (Collagen I Abcam ab34710; Collagen II Abcam ab34712 and Sigma SAB4500366). Of note, the 2 type II collagen antibodies that were used showed similar results. The antibodies were diluted 1:100 in 0.3% Triton X-100 in PBS and placed on different samples for 1 hour at room temperature. After rinsing for 5 minutes with PBS, secondary antibodies were used: a biotin-conjugated anti mouse (Vector lab, BA-2000), anti-rabbit IgG (Vector lab BA-1000), and an avidin-biotin peroxidase detection system with 3,3ʹ-diaminobenzidine substrate (DAB, Vector Labs). Samples were counterstained with pure filtered haematoxylin. Dehydration was achieved by vigorous shaking of the rack 10-20 times in alcohol baths (concentration 95% and 100%), followed by a pure xylol bath until slides assembly with Eukitt resin (Batch A1113, KiNDO1500). A Nikon Eclipse C1 Confocal microscope as well as a Nikon Eclipse TE2000-E were used for imaging.
Immunoblotting
Cells were lysed for 30 minutes on ice in ice-cold RIPA buffer (Life Technologies) supplemented with phosphatase and protease inhibitors (Complete anti-protease cocktail; Roche). Protein (10μg) was separated by SDS-PAGE (Bio-Rad) and transferred to PVDF membranes (Amersham). Blots were probed with anti-phospho-β-catenin (Cell Signaling; 5651T), Axin1 (Cell Signaling; 2087), β-actin HRP (Sigma-Aldrich) and GADPH (Cell Signaling; 2118) (1:1000) followed by the HRP-Rabbit or Mouse conjugated antibodies (1:5000).
Multipotency Assessment
Human primary mesenchymal stem cells (MSCs) were derived from adipose tissue or bone marrow. Monolayer human MSC (kindly provided by Dr. Mathurin Baquié), ASCs (kindly provided by Dr. Olivier Preynat-Seauve), or chondrocytes were expanded in ME before differentiation into the chondrogenic, adipogenic, and osteogenic fate to assess their multipotency. Tri-lineage differentiation was performed as previously described.33 The osteogenic, adipogenic, and chondrogenic differentiations were assessed using Alizarin Red S staining (Merck, TMS-008-C), Oil Red O staining (Sigma, O1391) and Safranin-O (Sigma, S2255)/Fast Green staining (Sigma F7252), respectively.
Flow Cytometry
To characterize the expression of MSCs surface markers, we performed flow cytometry analysis using chondrocytes from 3 donors. MSCs/ASCs were used as positive controls. Chondrocytes and MSCs/ASCs were expanded in ME before performing FACS analysis. To characterize MSCs, cell surface markers CD90 (R&D Systems; 965663), CD73 (BioLegend; 344005), CD105 (R&D Systems; 965665), CD45 (R&D Systems; 965662), CD14 (Abcam; ab2806), and CD34 (BD Pharmingen; 55824) were used. Three samples per donor were used in biological triplicate. 100 000 cells were fixed with PFA 4% then stained for CD73-CFS, CD90-APC, and CD105-PerCP during 1 hour at RT in FACs buffer (BSA-Azide-PBS). A minimum of 10 000 living cells was acquired with a Gallios flow cytometer and the analysis was done using the FlowJo software. Results represent the average of 3 independent experiments for CD73 and CD90. Two independent experiments were performed for CD105. For the gating strategy, living cells were first selected, then single cells were identified based on FSC-W and FSC-A to remove doublets. The positive staining was defined based on the negative control IgG-CFS, IgG-APC, and IgG-PerCP for CD73-CFS, CD90-APC, and CD105-PerCP, respectively.
CGH Array
Comparative genomic hybridization (CGH) array analysis was performed on chondrocytes at different passages (Supplementary Table S3). DNA was extracted using the QIAGEN QIAamp DNA Mini Kit (Qiagen, Hilden) according to the manufacturer’s protocol. Array CGH was performed using the Agilent SurePrint G3 Human CGH Microarray kit 4_44K (design ID 014950) with 43 kb overall median probe spacing (Agilent Technologies). Practical resolution was approximately 200 kb. Labeling and hybridization were performed following the protocols provided by the manufacturers. Briefly, sample DNA and DNA of a sex-matched control (1 µg of each) was labeled with Cy3-dUTP and Cy5-dUTP, respectively (Sure Tag Labelling Kit, Agilent Technologies). Labeled products were purified by Amicon Ultra 30 K filters (Millipore). Hybridization was performed according to the protocol provided by Agilent. Sample and control DNA was pooled and hybridized with 2 mg of Human Cot-I DNA at 65 °C with rotation for 24 h. Arrays were analyzed using an Agilent SureScan Microarray scanner and the Agilent Feat. A graphical overview was obtained using Cytogenomics 5.1.2.1 software and used to determine genetic stability (absence of deletions and duplications). Data analysis was done on UCSC Genome Browser Human Genome build19.
RNA Extraction and RT-PCR
RNA extraction was performed after 2D (chondrocytes) and 3D culture (Cartibeads). Isolation of total RNA was performed using the RNeasy kit from Qiagen according to the manufacturer’s instructions and was measured using a spectrometer. Five hundred nanograms of total RNA were used to synthesize cDNA using the PrimeScript RT Reagent Kit (Takara) according to manufacturer’s protocol. Real-time PCR was performed with PowerUp SYBR Green Master Mix (Applied Biosystems) using the QuantStudio 12K Flex Real-Time PCR System (Thermo Fisher Scientific) at the Genomic platform core facilities (University of Geneva). Primer sequences are described in Supplementary Table S5. First, efficacy tests were performed for all primers for validation prior utilization. The relative level of each sample was normalized to 2 housekeeping genes (ALAS1 and EEF1). RT-PCR reactions were carried out, at least, in technical and biological triplicates, and the average cycle threshold (CT) values were determined.
RNASeq
We performed RNA sequencing (RNAseq) analysis using 3 donor samples: donor 14 (cells in passage p3 in 2D culture, and p4/p5 in 3D culture for 2-step and 3-step method, respectively), donor 18 (p5 in 2D and p6/p7 in 3D) and donor 27 (p4 in 2D and p5/p6 in 3D). One biological sample per donor was used. RNA was extracted at different time points for the 3-step Cartibeads method: (i) end of step 1 (dedifferentiated chondrocytes, ME), (ii) end of step 2 (re-differentiated chondrocytes, MR), and (iii) end of step 3 (Cartibeads, MI). For the 2-step method, 2 time points were selected: (i) end of step 1 (dedifferentiated chondrocytes, ME) and (ii) end of step 2 (beads, MI). Of note, the starting material corresponding to the expansion in ME (step 1) is identical for the 2- and 3-step methods.
As previously described, the Agilent 2100 Bioanalyzer was used for RNA quality control prior sequencing, the SR100—libraries TruSeqHT stranded—Illumina HiSeq 4000 were used, and the sequencing quality control was performed with FastQC v.0.11.5.34 The quality distribution along the reads were evaluated and validated for all samples. The UCSC human hg38 reference was used to map the reads with STAR aligner v.2.5.3a to the reference genome. The average mapping rate was 93.54%. The transcriptome metrics were evaluated with the Picard tools v.1.141 and the differential expression analysis was performed with the statistical analysis R/Bioconductor package edgeR v. 3.18.1.35,36 Briefly, the counts were normalized according to the library size and filtered. The genes having a count above 1 count per million reads (cpm) in at least 3 samples were kept for the analysis. The raw gene number of the set was 26 485. The poorly or not expressed genes were filtered out. The final data set consisted of 13 884 genes. The differentially expressed genes tests were done with exact Test using a negative binomial distribution. The P-value of differentially expressed genes was corrected for multiple testing error using Benjamini-Hochberg with a 5% FDR (false discovery rate). The differentially expressed genes tests were done with edgeR using a negative binomial distribution. Panther analysis was used for determining the enrichment for each family of genes (number of entities).37,38
Implantation of Human Cartibeads in SCID/NOD Mice for Safety Study
Sixty-two male SCID/NOD mice were used to test the safety of Cartibeads. Fifty-four mice received human Cartibeads by subcutaneous implantation. One control group was used, where 8 animals received aggregated A549 adenocarcinoma cells (the number of mice per group is summarized in Supplementary Table S4). All implanted human Cartibeads were cultured according to the standardized 3-step method and implanted in the same manner. General anaesthesia was achieved with 4% isoflurane followed by 2% isoflurane under mask with 5% oxygen. Local disinfection of the skin was performed with 70% alcohol after shaving the back. Skin incision of 0.5 cm caudally from the occipital pole was performed. The produced tissues (Cartibeads and A549 adenocarcinoma beads), of 200 000 cells each, were implanted subcutaneously with a sterile pipet (1 Cartibead, or 1 or 5 A549 beads/animal). The skin was then closed with surgical glue (Histoacryl, B. Braun Surgical S.A.). In order to locate the site and orientation of the sample of skin, we performed a 4 cardinal point tattoo with a sterile 26-gauge needle and green tattoo ink. Mice in the control adenocarcinoma group were euthanized at 4 to 6 weeks, when tumor size and/ or its systemic effects compromised animal wellbeing beyond the needs of the study. Mice in the Cartibeads group were followed up to 6 months. Skin and organs were harvested and examined by 2 independent study members at each stage. Samples were all conserved 1/3 in formol 4% and 2/3 dry at −2 0 °C. No blood work was performed on the study group nor the controls as it was not required by the medical authorities for such product, and it would have generated undue stress to the study animals.
All work was performed in accordance with the animal research committee of Geneva under the approved protocol (GE/12/18). Sixty-two male SCID/NOD mice aging from 7- to 10 weeks and weighing approximately 30 g were purchased from Charles Rivers and housed 4 per cage. Housing was at a standard husbandry for specific pathogen-free provided by animal facility at the university medical centre. Mice were allowed to acclimate for at least 2 weeks before any manipulation. Due to abusive behavior, 8 mice had to be euthanized early on.
Quantification and Statistical Analysis
Sample size and statistics for each experiment were provided in the corresponding results section and/or in the figure legends. One-way ANOVA was used for the statistical analyses (except when Student’s t test is noticed), with P < .05 considered significant.
Availability of Data and Materials
The Cartibeads production method and the composition of mediums E, R, and I that are described in the Methods section are patented under European patent number WO2021028335. Moreover, raw data will be available upon request.
Results
Engineering and Characterization of Cartibeads
In this study, we generated cartilage microtissues named Cartibeads. The Cartibeads method is an innovative 3-step protocol (Fig. 1a) consisting of chondrocyte expansion in 2-dimensional culture (2D; step 1), followed by cell re-differentiation (2D; step 2), and finally by 3-dimensional culture (3D; step 3). Step 1 is characterized by dedifferentiation of native chondrocytes while step 2 is defined by chondrogenic commitment. Step 3 leads to Cartibeads formation by chondrocyte aggregation. We were able to engineer Cartibeads of high hyaline quality from adult dedifferentiated chondrocytes cultured at passages 5 to 9. Donors were up to 80 years of age and included patients with osteoarthritis (Supplementary Table S1).
Figure 1.
Generation and characterization of Cartibeads engineered by a 3-step method. (a) Cartibeads generation and study plan. 1. Harvesting: A cartilage sample is collected to extract chondrocytes. 2. Production: Scheme of the 3-step method for the generation of Cartibeads derived from dedifferentiated chondrocytes; step 1 (expansion) and step 2 (re-differentiation) were performed in 2D and in atmospheric oxygen conditions (21% oxygen), step 3 (Cartibeads formation) was performed in 3D culture and in hypoxic conditions (5% oxygen). (b) Quantification of GAG content (µg GAG/Cartibeads) using the DMMB assay. Cartibeads used for GAG quantification were generated from cells at passages P4 to P6. For 4 donors, the lines represent the error bars from 3 independent experiments. (c) Quantification of GAG content in Cartibeads normalized to DNA content, expressed as a GAG/DNA ratio (µg GAG/µg DNA). (d) Histological qualitative analysis of Cartibeads. Cartibeads from 3 donors were stained for Safranin-O staining (GAG in red, top panel), type II collagen (brown DAB staining, middle panel), and type I collagen (brown DAB staining, lower panel) (for colour figure refer to online version). Scale bars 100 µm.
We used high level of GAG and type II collagen detection as potency criteria to evaluate the hyaline quality of Cartibeads. Quantitative analysis of Cartibeads revealed high GAG content, which was independent of cell passage number, donor age, and osteoarthritic status of the joint (Fig. 1b). We measured an average of 40 µg of GAG/Cartibead and an average ratio of 50 µg GAG/µg DNA (SD = 45) (Fig. 1b, c). GAG presence in Cartibeads was qualitatively confirmed by staining with Safranin-O (Fig. 1d, top panel). The hyaline quality of Cartibeads was further confirmed by immunohistochemical staining, which showed a strong detection of type II collagen (Fig. 1d, middle panel) and weak detection of type I collagen (Fig. 1d, bottom panel).
Transcriptomic Analyses of the 3-step Cartibeads Method
To identify the key molecular pathways allowing the production of Cartibeads, the 3-step Cartibeads method was compared to a 2-step method, classically used in tissue engineering (Fig. 2a). In both methods, chondrocytes were extensively expanded with medium E (ME), containing FGF-2 supplementation. In the 3-step method we introduced an extra step, corresponding to a starvation phase, using medium R (MR), which does not contain additional FGF-2. In both methods, the final step consists in 3D culture for 2 weeks in maturation medium I (MI), containing supplementation with TGF-β3, BMP-2, and IGF-1 (Figs 1a and 2a). In terms of morphology, the 3-step method resulted in larger, white-coloured beads (diameter of 1-2 mm) than the 2-step method (diameter 0.5-1 mm) and a higher quantity of GAG/bead (Supplementary Fig. S1a, b, c). Increased levels of GAG confirmed the improved chondrogenic re-differentiation in the 3-step Cartibeads method. This was attributed to the introduction of the starvation step with MR, which facilitates the re-differentiation of chondrocytes and hyaline matrix production.
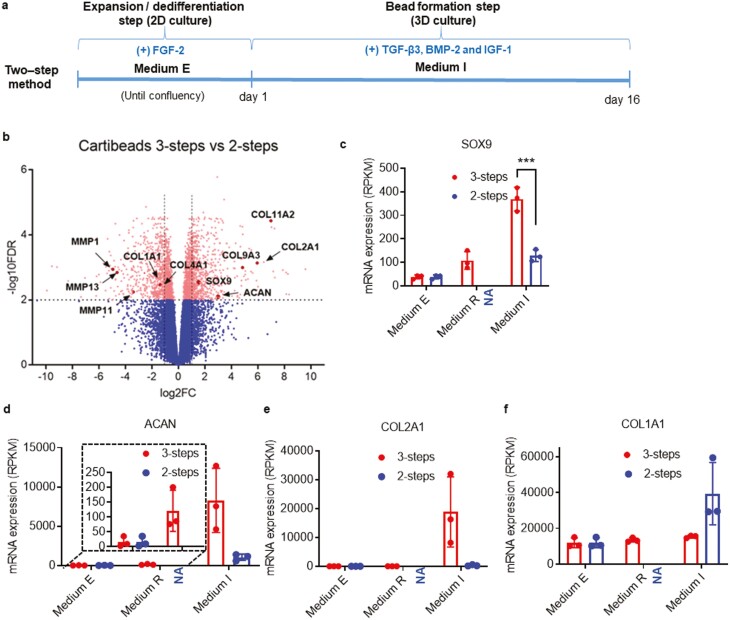
Figure 2.
Improvement of the hyaline characteristics of Cartibeads engineered from the 3-step method compared to the 2-step method. (a) Scheme of the 2-step method for the generation of cartilage beads. Chondrocytes were expanded in step 1 in ME and directly used in step 2 in MI for beads generation. (b) Visualization of RNAseq results from chondrocytes in 3-step versus 2-step method. The Volcano plot shows results with statistical significance (FDR<.01) versus magnitude of change (>2-fold change). The plot highlights genes that are significantly (scatter above the dotted line) upregulated (right-hand side) and downregulated (left-hand side). In this plot, ACAN and COL2A1 are increased in the 3-step method compared to the 2-step method, while COL1A1 is decreased. c, d, e, f) Data from RNAseq analysis comparing mRNA expression levels (in RPKM) for SOX9 (c), ACAN (d), COL2A1 (e), and COL1A1 (f) genes in the 3-step and 2-step method. RNAseq analysis was made from 3 donor samples. One biological sample per donor was used. In all the study, chondrocytes are used between passages 3 and 7. Of note, the starting material corresponding to the expansion step (step 1) in ME is identical for the 2-step (right-hand side bars) and 3-step (left-hand side bars) method. NA (not applicable) refers to the lack of this step in the 2-step method, thus resulting in an absence of data. ***P < .001.
To understand the molecular mechanisms involved in increased production of hyaline cartilage, we performed RNA sequencing (RNAseq) analysis using 3 donor samples at each critical step of both methods. RNA was extracted at different time points for the 3-step Cartibeads method: (i) end of step 1 (dedifferentiated chondrocytes, ME), (ii) end of step 2 (re-differentiated chondrocytes, MR), and (iii) after 15 days for step 3 (Cartibeads, MI). For the 2-step method, 2 time points were selected: (i) end of step 1 (dedifferentiated chondrocytes, ME), and (ii) end of step 2 (beads, MI). Of note, the end of expansion (step 1) is identical in both methods. We first compared the cartilage microtissues generated by both methods (Fig 2b, S1d), and observed significantly changed levels of genes involved in collagen and ECM degradation/ formation along with ECM organization (COL4A1, COL9A3, COL11A2, MMP1, MMP11, MMP13) in Cartibeads as opposed to the 2-step method beads. Differential gene expression analysis showed that the 3-step method induced higher levels of SOX 9 (a transcription factor inducing chondrogenic commitment), COL2A1 (type II collagen), and ACAN (aggrecan, a GAG-associated protein) in Cartibeads, while COL1A1 (type I collagen, specific of fibrocartilage), was higher in the 2-step method beads (Fig 2b, c, d, e, f).
Our data showed the dedifferentiation of chondrocytes expanded in ME by the detection of COL1A1 expression and the absence of hyaline cartilage markers such as type COL2A1 and ACAN (Fig. 2d, e, f). We confirmed the enrichment of dedifferentiated cells in ME by flow cytometry, indicating that the proportion of cells expressing mesenchymal stem cell (MSCs) markers CD73, CD90, and CD105 (as defined by the International Society for Cell Therapy (ISCT)) was more than 90% of the total cell population at the end of expansion (Supplementary Fig. S2a and Table S2). Indeed, MSCs are expected to be positive (>95%) for CD90, CD73, and CD105, and negative (<2%) for CD45, CD14, and CD34, as shown in Fig S2a.39 Furthermore, multipotency of this population of dedifferentiated chondrocytes was evaluated by their capacity to differentiate toward other MSCs derivative cells (osteocytes, adipocytes, and chondrocytes). Although dedifferentiated chondrocytes were able to differentiate into the 3 cell-types, their multipotency potential was lower compared to MSCs/ASCs (Supplementary Fig. S2b).
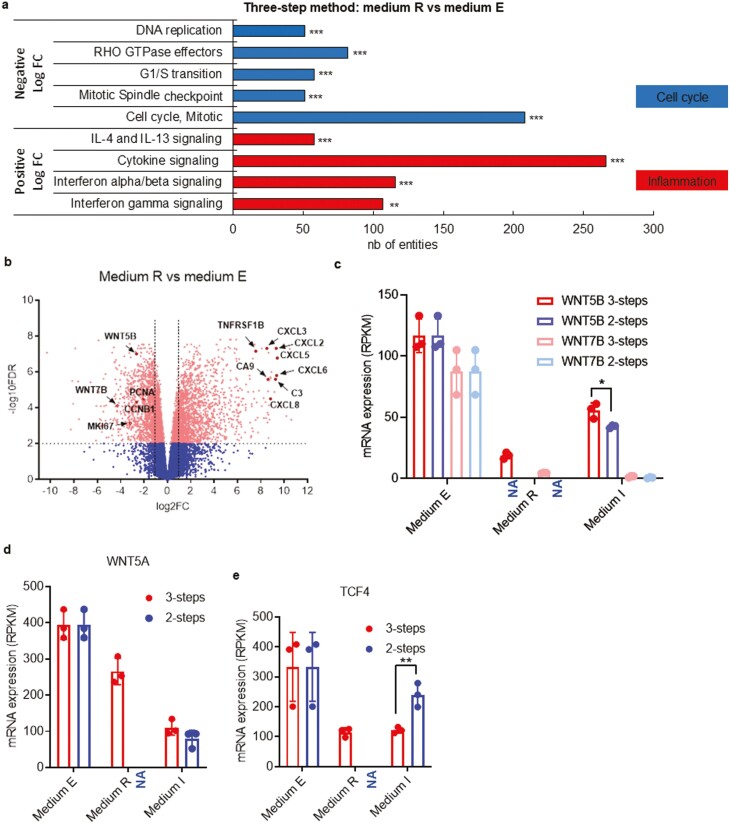
When we compared the gene expression pattern of cells in MR (step 2, re-differentiation step) to cells in ME (step 1, expansion step), we found higher levels of expression of genes involved in inflammation processes (interleukin and cytokine signaling along with interferon signaling) during redifferentiation. In parallel, we found lower levels of expression of genes implicated in the cell cycle (CCNB1, MKI67, PCNA) (Fig. 3a, b). Transcriptomic analyses of the 3-step Cartibeads method identified low level expression of WNT genes as a key pathway involved in the high quality of hyaline matrix. By comparing the expansion step with the re-differentiation step, we observed high expression levels of WNT5A, WNT5B, and WNT7B genes during the expansion step (step 1, ME, both methods) (Fig. 3b, c, d). These genes were strongly downregulated during the re-differentiation step (MR, 3-step method) and during 3D culture (MI, both methods), except for WNT5B, which showed intermediate expression in 3D culture (MI, both methods) (Fig. 3c). These results are in line with previous studies reporting an upregulation of WNT upon FGF-2 treatment.40 Indeed, WNT signaling is involved in stem-like phenotype. Accordingly, the lack of FGF-2 supplementation in MR led to WNT signaling downregulation during the re-differentiation phase (Fig. 3c, d). WNT downregulation was in correlation with the decrease of TCF4 in the 3-step method, a transcription factor well-known as downstream effector of the canonical WNT signaling pathway (Fig.3e). We observed a comparable decrease of WNT5A, WNT5B and WNT7B genes in the 2-step method during the 3D phase, but significantly higher expression of TCF4 (Fig. 3b, c, d, e).
Figure 3.
Inhibition of WNT signalling enhances the production of hyaline matrix components. (a) Gene set enrichment analysis and functional annotation clustering are based on the differential expression levels of MR versus ME in the 3-step method. Histograms show the number of genes for each family of genes for which a significant enrichment was found (number of genes differentially expressed). Top gene sets had lower expression in MR than in ME (negative FC), while bottom gene sets had higher expression in MR (positive FC). (b) Visualization of RNAseq results from chondrocytes in MR versus ME in the 3-step method. The Volcano plot shows results with statistical significance (FDR < .01) versus magnitude of change (>2-fold change). The plot highlights genes that are significantly (scatter above the dotted line) upregulated (right-hand side) and downregulated (left-hand side). In this plot, WNT5B, WNT7B, and MKI67 are decreased in MR versus ME in the 3-step method. Data from RNAseq analysis comparing mRNA expression levels (in RPKM) for WNT5B and WNT7B (c), WNT5A (d), TCF4 (e) genes in the 3-step and 2-step method. RNAseq analysis was made from 3 donor samples. One biological sample per donor was used. In all the study, chondrocytes are used between passage 3 to 7. Of note, the starting material corresponding to the expansion step (step 1) in ME is identical for the 2-step (right-hand side bars) and 3-step (left-hand side bars) method. NA (Not Applicable) refers to the lack of this step in the 2-step method, thus resulting in an absence of data. *P < .05, **P < .01; ***P < .001.
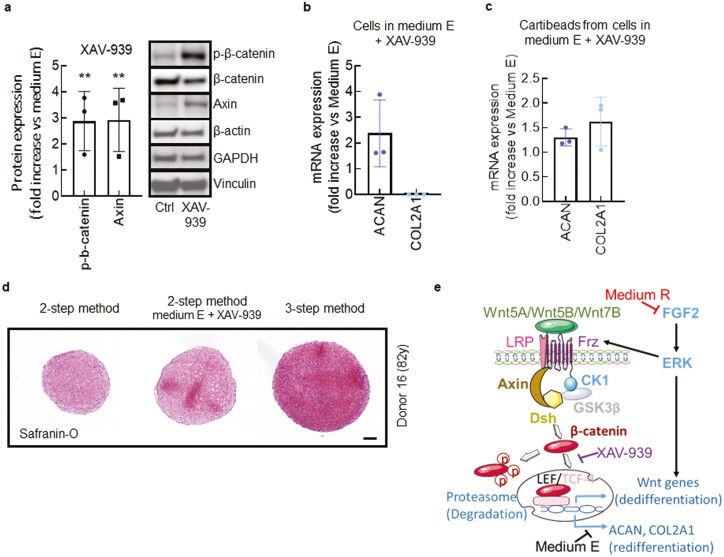
To validate the role of WNT pathway downregulation during the re-differentiation phase (MR) (Fig. 1a), we used ME supplemented with 10 µM of XAV-939, a WNT pathway inhibitor, for 4 days (Fig. 4a). Upon XAV-939 treatment, we observed an increase of phospho-β-catenin and axin (Fig. 4a), known indicators of WNT signaling blockade.41 Pharmacological inhibition of WNT induced an increase of the ACAN and COL2A1 expression in the 2-step method after 2D and 3D culture (Fig. 4b, c), similarly to MR in the 3-step method. As a result, pharmacological inhibition of WNT by XAV-939 resulted in the presence of hyaline features of beads obtained in the 2-step method, as confirmed by Safranin-O staining (Fig. 4d).
Figure 4.
Pharmacological inhibition of WNT signalling by XAV-939. (a) Immunoblots showing expression of the indicated proteins in chondrocytes exposed to the WNT signalling inhibitor XAV-939 (10 µM). Histogram shows the densitometry analysis of β-catenin and Axin in chondrocytes after 4 days of culture in ME (control) versus ME + XAV-939. Data are represented as mean ± SEM. A Student’s t test was used for the statistical analysis. (b, c) mRNA expression of ACAN and COL2A1 determined by qPCR. We compared chondrocytes after 4 days of culture in ME versus ME + XAV-939 (b), and beads (2-step method) from chondrocytes cultured in ME versus ME + XAV-939 (c). (d) Safranin-O staining of GAG in beads produced from the 2-step method without (left panel) or with XAV-939 (middle panel), and in Cartibeads from the 3-step method (right panel). Scale bar 100µm. (e) Scheme summarizing the molecular basis for WNT signalling pathway in ME, and WNT inhibition when chondrocytes are cultured in MR or in ME + XAV-939, allowing the production of a hyaline matrix containing ACAN and COL2A1. **P < .01.
Altogether, transcriptomic analysis identified the involvement of WNT signaling pathway and its modulation in chondrocyte dedifferentiation and re-differentiation, together with WNT5A, WNT5B, and WNT7B acting as potential main mediators of this response (Fig. 3c, d). ME supplemented with XAV-939 partially mimicked the effect of MR, since FGF-2 supplementation in ME may act on other pathways to prevent cell differentiation (Fig 4e). Consequently, due to the use of MR lacking FGF-2 supplementation, the 3-step method naturally induced the downregulation of WNT genes in 7 days, without the use of a costly pharmacological molecule.
Preclinical Safety Studies of Cartibeads In Vitro and In Vivo
Implantation of cells expanded in vitro raises a problem of potential uncontrolled proliferation. Cartibeads safety was first evaluated through in vitro preclinical studies. The CGH (comparative genomic hybridization) array analysis showed genetic stability of chondrocytes during cell amplification up to passage 11 (Supplementary Table S3). Indeed, no duplications or deletions were noted, except for loss of chromosome Y in 3 older male donors (>55 years old). These findings are consistent with previous studies stating loss of chromosome Y as a normal age-related acquired mutation in men, with no pathological implications.42-44
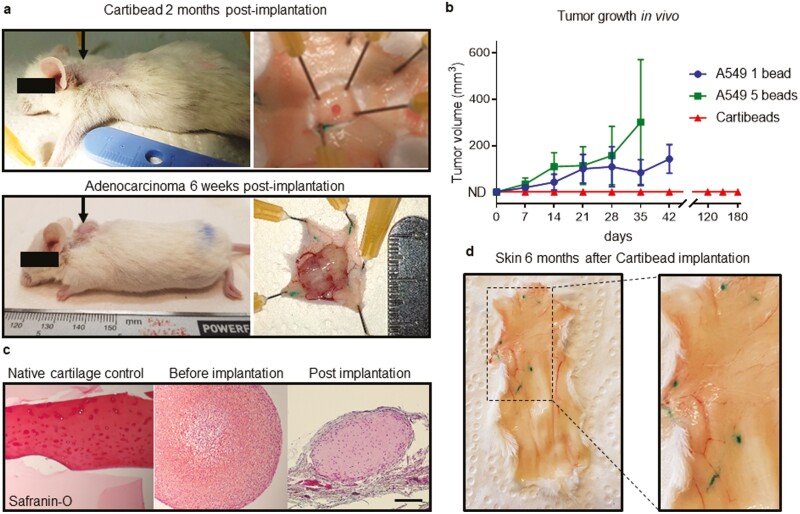
In the in vivo safety study, we evaluated a potential tumorigenic effect of implanted human Cartibeads in SCID mice. A549 human alveolar basal epithelial adenocarcinoma cells were used as positive controls (Fig. 5a, b, Supplementary Table S4). Indeed, we observed tumor development with 1 and 5 beads of A549 at 6 weeks post-implantation (Fig. 5a, b). Cartibeads size remained the same at 2 months post-implantation, and we observed a weak staining of GAG by Safranin-O, indicating degeneration (Fig. 5a, c). The Cartibeads did not expand and were mostly undetectable 6 months after implantation, thereby confirming absence of potential tumorigenicity over 6 months follow-up (Fig. 5b, d). Complete necropsy performed by 2 independent observers demonstrated the absences of local and off-site malignant changes or degenerations. All the harvested dorsal skin and 5 internal organs were macroscopically clear from any suspicious pathological changes (Fig. 5d, Supplementary Table S4).
Figure 5.
The absence of tumorigenicity of human Cartibeads following implantation in immunodeficient mice. (a) Beads were subcutaneously implanted in the back of SCID mice (black arrows). Representative pictures of a Cartibead (0.2 × 106 chondrocytes/Cartibead) at 2 months post-implantation (top panel) and a tumour derived from 5 beads of A549 adenocarcinoma cells (0.2 × 106 A549 cells/bead) at 6 weeks post-implantation (bottom panel). (b) Tumour development (mm3) after subcutaneous implantation of 1 Cartibead or 1 or 5 A549 beads per mice (n = 10 to 14 mice for Cartibeads and 2× n = 4 mice per A549 beads). ND = not detected. (c) Safranin-O staining of GAG in native cartilage (left panel), Cartibeads before implantation (middle panel) and Cartibeads 2 months post-implantation (right panel). Scale bar 200 µm. (d) Skin 6 months after Cartibeads implantation showed no alterations or residues of Cartibeads.
Discussion
The use of adult chondrocytes as starting material to regenerate cartilage is limited by culture-induced dedifferentiation. In the present study, we demonstrated that our novel 3-step method can reverse the loss of chondrogenic phenotype, solving a critical issue encountered during cell therapy using adult chondrocytes. Our data demonstrate that our method produced Cartibeads with a high-quality cartilage with hyaline features, regardless of the donor age and cartilage harvest quality (from osteoarthritic joint), using dedifferentiated chondrocytes up to passage 9. Since our patented 3-step method can overcome culture-induced dedifferentiation, it allows the extensive expansion of chondrocytes before aggregation. Therefore, Cartibeads can be generated from a small cartilage biopsy, as opposed to 200 mg on average in conventional human chondrocyte-based cell therapy.45 In a clinical setting, this would greatly reduce the risk of creating donor-site lesions.
Our method showed a ratio of GAG/Cartibead that is 20-fold higher than previously reported studies, suggesting that our method produced more hyaline cartilage and less fibrocartilage.28 Indeed, Spherox report 0.56 to 2.4 µg GAG/spheroid as opposed to our average of 40 µg GAG/Cartibead. Consistent with this, we obtained on average a ratio of GAG/DNA at least 3 times higher than other published methods.25,46 Cartilage constructs from nasal chondrocytes showed 13.3 ± 9.5 µg GAG/µg DNA, and construct from articular chondrocytes 6.3 ± 0.4 µg GAG/µg DNA, while Cartibeads showed an average of 50 µg GAG/µg DNA. We also showed that Cartibeads matrix contains abundant type II collagen and scarce type I collagen, with an intense Safranin-O glycosaminoglycan staining. Moreover, we were able to produce Cartibeads of similar characteristics from 15 donors up to 80 years old, while other methods are restricted to only 3 young donors when using high passage cells.47
In this work, we identified the pathway to re-differentiate adult dedifferentiated chondrocytes. We hypothesized that re-differentiation may be induced by the use of “re-differentiation medium” MR, without supplementation of FGF-2, while ME used during cell expansion contains FGF-2. However, FGF-2 absence in 3D culture was not sufficient to induce hyaline matrix synthesis in the 2-step method. Indeed, re-differentiation of chondrocytes most likely requires cell adhesion to a matrix-coated flask and activation of specific signaing pathways in 2D culture. Starvation of FGF-2 led to downregulation of genes involved in the WNT signaling pathway (WNT, TCF4). To the best of our knowledge, we are the first to emphasize the importance of WNT pathway inhibition in the production of high hyaline quality cartilage using dedifferentiated chondrocytes. It is well documented that WNT signaling is involved in both inhibition and stimulation of chondrogenic differentiation of adult progenitor cells.48-50 As shown in embryos, high levels of WNT/β-catenin signaling inhibit chondrogenic differentiation of stem cells, while downregulation of this pathway induces chondrogenesis.51-53 In our study, we identified WNT5A, WNT5B, and WNT7B as potential WNT isoforms involved in this mechanism. In parallel with the downregulation of the WNT pathway, we observed an increased expression of genes involved in the inflammatory pathway (interleukins, cytokines). In fact, in wound healing and tissue repair, a dynamic balance between pro- and anti-inflammatory factors is essential for effective healing.54,55 Accordingly, our data suggest that hyaline matrix production might require a dynamic balance between cellular inflammation and tissue remodeling associated with an exit of the cell cycle.56
Cartibeads certainly hold potential as a chondrocyte-based treatment for cartilage repair, which requires safety and efficacy to be demonstrated. We have evaluated Cartibeads safety in the present work. Human Cartibeads did not proliferate when implanted in SCID mice and even disappeared after 6 months, which is consistent with similar studies.57 Additionally, no other pathological changes were noticed locally at the skin level nor at a distance in other organs at the time of euthanasia in all the study group. Cartibeads efficacy up to 6 months of follow-up was evaluated in a preclinical study in the minipig model and its results are currently being processed. The local ethics committee and medical authority have consequently authorized a first-in-human phase I clinical study using autologous Cartibeads implantation in patients with cartilage damage.
Conclusion
Cartibeads are the first high-quality hyaline cartilage tissue engineered from adult dedifferentiated chondrocytes with a proven safety profile. We described the signaling pathway of chondrocytes dedifferentiation and re-differentiation, which allowed them to recover their phenotype and secrete their hyaline extracellular matrix. Collectively, Cartibeads represent a breakthrough in the field of cartilage repair and have been approved by a local ethics committee and medical authority for a first-in-human study using autologous Cartibeads implantation in patients with cartilage damage.
Supplementary Material
Acknowledgments
The authors thank the personnel of the Genomic Core Facility of the Faculty of Medicine (University of Geneva), in particular Christelle Barraclough and Didier Chollet, for their help in sample preparation, qRT-PCR, and RNAseq, as well as Natacha Civic for RNASeq analysis. We also thank the teams of the laboratory of Vincent Braunersreuther and Prof. Karl-Heinz Krause for their help.
Contributor Information
Halah Kutaish, Department of Pathology and Immunology, Medical School, University of Geneva, Geneva, Switzerland; University Medical Center, University of Geneva, Geneva, Switzerland; Foot and Ankle Surgery Centre, Centre Assal, Clinique La Colline, Hirslanden Geneva, Switzerland.
Laura Bengtsson, Vanarix SA, Lausanne, Switzerland.
Philippe Matthias Tscholl, University Medical Center, University of Geneva, Geneva, Switzerland; Department of Orthopaedics Surgery, Geneva University Hospital, Geneva, Switzerland.
Antoine Marteyn, Department of Pathology and Immunology, Medical School, University of Geneva, Geneva, Switzerland; University Medical Center, University of Geneva, Geneva, Switzerland.
Vincent Braunersreuther, Service of Clinical Pathology, Diagnostic Department, Geneva University Hospitals, Geneva, Switzerland.
Alexandre Guérin, Department of Pathology and Immunology, Medical School, University of Geneva, Geneva, Switzerland; University Medical Center, University of Geneva, Geneva, Switzerland.
Frédérique Béna, Service of Genetic Medicine, Diagnostic Department, Geneva University Hospitals, Geneva, Switzerland.
Stefania Gimelli, Service of Genetic Medicine, Diagnostic Department, Geneva University Hospitals, Geneva, Switzerland.
David Longet, Department of Pathology and Immunology, Medical School, University of Geneva, Geneva, Switzerland; University Medical Center, University of Geneva, Geneva, Switzerland.
Sten Ilmjärv, Department of Pathology and Immunology, Medical School, University of Geneva, Geneva, Switzerland; University Medical Center, University of Geneva, Geneva, Switzerland.
Pierre-Yves Dietrich, Laboratory of Tumor Immunology, Oncology Department, Center for Translational Research in Onco-Hematology, Geneva University Hospitals, University of Geneva, Geneva, Switzerland.
Eric Gerstel, University Medical Center, University of Geneva, Geneva, Switzerland; Clinique la Colline, Hirslanden, Geneva, Switzerland.
Vincent Jaquet, Department of Pathology and Immunology, Medical School, University of Geneva, Geneva, Switzerland; University Medical Center, University of Geneva, Geneva, Switzerland; READS Unit, Medical School, University of Geneva, Geneva, Switzerland.
Didier Hannouche, University Medical Center, University of Geneva, Geneva, Switzerland; Department of Orthopaedics Surgery, Geneva University Hospital, Geneva, Switzerland.
Jacques Menetrey, University Medical Center, University of Geneva, Geneva, Switzerland; Centre for Sports Medicine and Exercise, Clinique la Colline, Hirslanden, Geneva, Switzerland.
Mathieu Assal, University Medical Center, University of Geneva, Geneva, Switzerland; Foot and Ankle Surgery Centre, Centre Assal, Clinique La Colline, Hirslanden Geneva, Switzerland.
Karl-Heinz Krause, Department of Pathology and Immunology, Medical School, University of Geneva, Geneva, Switzerland; University Medical Center, University of Geneva, Geneva, Switzerland.
Erika Cosset, Laboratory of Tumor Immunology, Oncology Department, Center for Translational Research in Onco-Hematology, Geneva University Hospitals, University of Geneva, Geneva, Switzerland; Team GLIMMER Of lIght (GLIoblastoma MetabolisM, HetERogeneity, and OrganoIds), Cancer Research Centre of Lyon – CRCL, Lyon, France.
Vannary Tieng, Vanarix SA, Lausanne, Switzerland.
Funding
This work was sponsored by Vanarix SA.
Conflict of Interest
H.K., L.B. disclosed leadership position and commercial research support from Vanarix SA. E.G. is a shareholder of Vanarix SA. V.T. is a co-founder and C.E.O of Vanarix SA. The other authors declared no potential conflicts of interest.
Author Contributions
H.K., E.C., P.M.T., L.B., A.M., V.B., F.M.M., A.G., F.B., S.G., J.L., I.F., D.L., D.S., and V.T. carried out experiments. V.T., H.K., and E.C. designed and supervised the study. V.T., H.K., A.M., F.M.M., and F.B. analyzed and interpreted data. E.C., H.K., L.B., and V.T. wrote the paper. E.C. and H.K. performed the in vivo mice experiments. H.K. and P.M.T. performed the in vivo minipig experiments. F.M.M., J.L., and I.F. performed the biomechanical experiment and analysis. S.I. analyzed the RNASeq data. P-Y.D., D.H., E.G., V.J., M.A., J.M., and K-H.K. provided critical feedback on the study and the manuscript. All co-authors proofread the manuscript.
Data Availability
The exact composition of the medium E, R and I is described in the patent (WO2021028335). All raw data will be published in data repository upon acceptance for publication. Moreover, raw data will be available for reviewers upon request.
Ethics Approval and Consent to Participate
Informed written consent for MSC derivation was obtained from the donors according to the local ethic committee for research 2020-01102 and NAC 14-183. Human cartilage sample collection was approved by the Swiss Ethics Committee (BASEC, 2016-00656). All mice work was performed in accordance with the animal research committee of Geneva under the approved protocol (GE/12/18).
References
- 1. Davies R. Regenerative medicine: a review of the evolution of autologous chondrocyte implantation (ACI) therapy. Bioengineering. 2019;6(1):22. 10.3390/bioengineering6010022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lotz M, Loeser RF. Effects of aging on articular cartilage homeostasis. Bone. 2012;51(2):241-248. 10.1016/j.bone.2012.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sophia Fox AJ, Bedi A, Rodeo SA. The basic science of articular cartilage: structure, composition, and function. Sports Health. 2009;1(6):461-468. 10.1177/1941738109350438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Muir H. The chondrocyte, architect of cartilage. Biomechanics, structure, function and molecular biology of cartilage matrix macromolecules. Bioessays. 1995;17(12):1039-1048. 10.1002/bies.950171208 [DOI] [PubMed] [Google Scholar]
- 5. Darling EM, Athanasiou KA. Rapid phenotypic changes in passaged articular chondrocyte subpopulations. J Orthop Res. 2005;23(2):425-432. 10.1016/j.orthres.2004.08.008 [DOI] [PubMed] [Google Scholar]
- 6. Wu L, Gonzalez S, Shah S, et al. ., Extracellular matrix domain formation as an indicator of chondrocyte dedifferentiation and hypertrophy. Tissue Eng Part C Methods. 2014;20(2):160-168. 10.1089/ten.TEC.2013.0056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Benya PD, Padilla SR, Nimni ME. Independent regulation of collagen types by chondrocytes during the loss of differentiated function in culture. Cell. 1978;15(4):1313-1321. 10.1016/0092-8674(78)90056-9 [DOI] [PubMed] [Google Scholar]
- 8. Gobbi A, Karnatzikos G, Kumar A. Long-term results after microfracture treatment for full-thickness knee chondral lesions in athletes. Knee Surg Sports Traumatol Arthrosc. 2014;22(9):1986-1996. 10.1007/s00167-013-2676-8 [DOI] [PubMed] [Google Scholar]
- 9. Benya PD, Shaffer JD. Dedifferentiated chondrocytes reexpress the differentiated collagen phenotype when cultured in agarose gels. Cell. 1982;30(1):215-224. 10.1016/0092-8674(82)90027-7 [DOI] [PubMed] [Google Scholar]
- 10. Huang BJ, Hu JC, Athanasiou KA. Cell-based tissue engineering strategies used in the clinical repair of articular cartilage. Biomaterials. 2016;98:1-22. 10.1016/j.biomaterials.2016.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Munirah S, Samsudin OC, Aminuddin BS, Ruszymah BHI. Expansion of human articular chondrocytes and formation of tissue-engineered cartilage: a step towards exploring a potential use of matrix-induced cell therapy. Tissue Cell. 2010;42(5):282-292. 10.1016/j.tice.2010.07.002 [DOI] [PubMed] [Google Scholar]
- 12. Kondo M, Kameishi S, Kim K, et al. Safety and efficacy of human juvenile chondrocyte-derived cell sheets for osteochondral defect treatment. Regen Med. 2021;6(65) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Park DY, Min B, Park SR, et al. Engineered cartilage utilizing fetal cartilage-derived progenitor cells for cartilage repair. Sci Rep. 2020;10(1):5722. 10.1038/s41598-020-62580-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Martin AR, Patel J, Zlotnick HM, Carey JL, Mauck RL. Emerging therapies for cartilage regeneration in currently excluded “red knee” populations. Regen Med. 2019;4(12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hwang NS, Elisseeff J. Application of stem cells for articular cartilage regeneration. J Knee Surg. 2009;22(1):60-71. 10.1055/s-0030-1247728 [DOI] [PubMed] [Google Scholar]
- 16. Vonk LA, De Windt TS, Slaper-Cortenbach I, Saris DB. Autologous, allogeneic, induced pluripotent stem cell or a combination stem cell therapy? Where are we headed in cartilage repair and why: a concise review. Stem Cell Res Ther. 2015;6:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Park MS, Kim YH, Jung Y, et al. In situ recruitment of human bone marrow-derived mesenchymal stem cells using chemokines for articular cartilage regeneration. Cell Transplant. 2015;24(6):1067-1083. [DOI] [PubMed] [Google Scholar]
- 18. Richter W. Mesenchymal stem cells and cartilage in situ regeneration. J Intern Med. 2009;266(4):390-405. 10.1111/j.1365-2796.2009.02153.x [DOI] [PubMed] [Google Scholar]
- 19. Mueller MB, Tuan R. Functional characterization of hypertrophy in chondrogenesis of human mesenchymal stem cells. Arthritis Rheum. 2008;58(5):1377-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pelttari K, Winter A, Steck E, et al. Premature induction of hypertrophy during in vitro chondrogenesis of human mesenchymal stem cells correlates with calcification and vascular invasion after ectopic transplantation in SCID mice. Arthritis Rheum. 2006;54(10):3254-3266. 10.1002/art.22136 [DOI] [PubMed] [Google Scholar]
- 21. Du WJ, Chi Y, Yang ZX, et al. Heterogeneity of proangiogenic features in mesenchymal stem cells derived from bone marrow, adipose tissue, umbilical cord, and placenta. Stem Cell Res Ther. 2016;7(1):163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Le H, Xu W, Zhuang X, et al. Mesenchymal stem cells for cartilage regeneration. J Tissue Eng. 2020;11:2041731420943839. 10.1177/2041731420943839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Madeira C, Santhagunam A, Salgueiro JB, Cabral JM. Advanced cell therapies for articular cartilage regeneration. Trends Biotechnol. 2015;33(1):35-42. [DOI] [PubMed] [Google Scholar]
- 24. Negoro T, Takagaki Y, Okura H, Matsuyama A. Trends in clinical trials for articular cartilage repair by cell therapy. NPJ Regen Med. 2018;3:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mumme M, Barbero A, Miot S, et al. Nasal chondrocyte-based engineered autologous cartilage tissue for repair of articular cartilage defects: an observational first-in-human trial. Lancet. 2016;388(10055):1985-1994. [DOI] [PubMed] [Google Scholar]
- 26. Ramezankhani R. Two decades of global progress in authorized advanced therapy medicinal products: an emerging revolution in therapeutic strategies. Front Cell Dev Biol. 2020;17(8):547653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vericel Denmark ApS. MACI -matrix-applied characterised autologous cultured chondrocytes: EMEA/H/C/002522. European Medicines Agency (EMA).2013; Available from: https://www.ema.europa.eu/en/medicines/human/EPAR/maci. [Google Scholar]
- 28. Bartz C, Meixner M, Giesemann P, et al. An ex vivo human cartilage repair model to evaluate the potency of a cartilage cell transplant. J Transl Med. 2016;14(1):317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. CO.DON AG. Spherox: EMEA/H/C/002736. European Medicines Agency (EMA).2017; Available from: https://www.ema.europa.eu/en/medicines/human/EPAR/spherox. [Google Scholar]
- 30. Sewon Collontech CO., LTD. Chondron. Korean Ministry of Food and Drug Safety (MFDS).2016; https://www.mfds.go.kr/eng/brd/m_30/view.do?seq=70954. [Google Scholar]
- 31. Pathak S, Chaudhary D, Reddy KR, Acharya KK, Desai SM. Efficacy and safety of CARTIGROW® in patients with articular cartilage defects of the knee joint: a four year prospective studys. Int Orthop. 2022;46(6):1313-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Orthocell LTD. OrthoACI. 2022; Available from: https://orthocell.com/products/. [Google Scholar]
- 33. Liu Y, Goldberg AJ, Dennis JE, Gronowicz GA, Kuhn LT. One-step derivation of mesenchymal stem cell (MSC)-like cells from human pluripotent stem cells on a fibrillar collagen coating. PLoS One. 2012;7(3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cosset E, Petty T, Dutoit V, et al. Human tissue engineering allows the identification of active miRNA regulators of glioblastoma aggressiveness. Biomaterials. 2016;107:74-87. [DOI] [PubMed] [Google Scholar]
- 35. Huber W, Kemnitz V, Phillip V, Schmid RM, Faltlhauser A. Outcome prediction, fluid resuscitation, pain management, and antibiotic prophylaxis in severe acute pancreatitis. Intensive Care Med. 2015;41(11):2034-2035. [DOI] [PubMed] [Google Scholar]
- 36. Gentleman RC, Carey VJ, Bates DM. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mi H, Huang X, Muruganujan A, et al. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017;45(D1):D183-D189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen Y, Lun A, Smyth GK. From reads to genes to pathways: differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000Res. 2016;20(5):1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Merimi M, El-Majzoub R, Lagneaux L, et al. The therapeutic potential of mesenchymal stromal cells for regenerative medicine: current knowledge and future understandings. Front Cell Dev Biol. 2021;9:661532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Buchtova M, Oralova V, Aklian A, et al. Fibroblast growth factor and canonical WNT/beta-catenin signaling cooperate in suppression of chondrocyte differentiation in experimental models of FGFR signaling in cartilage. Biochim Biophys Acta. 2015;1852(5):839-850. [DOI] [PubMed] [Google Scholar]
- 41. Huang SM, Mishina YM, Liu S, et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461(7264):614-620. [DOI] [PubMed] [Google Scholar]
- 42. Stumm M, Boger E, Gaissmaier CG, et al. Genomic chondrocyte culture profiling by array-CGH, interphase-FISH and RT-PCR. Osteoarthritis Cartilage. 2012;20(9):1039-1045. [DOI] [PubMed] [Google Scholar]
- 43. Thompson DJ, Genovese G, Halvardson J, et al. Genetic predisposition to mosaic Y chromosome loss in blood. Nature. 2019;575(7784):652-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Guttenbach M, Koschorz B, Bernthaler U, Grimm T, Schmid M. Sex chromosome loss and aging: in situ hybridization studies on human interphase nuclei. Am J Hum Genet. 1995;57(5):1143-1150. [PMC free article] [PubMed] [Google Scholar]
- 45. Brittberg M, Recker D, Ilgenfritz J, Saris DB; SUMMIT Extension Study Group. Matrix-applied characterized autologous cultured chondrocytes versus microfracture: five-year follow-up of a prospective randomized trial. Am J Sports Med. 2018;46(6):1343-1351. [DOI] [PubMed] [Google Scholar]
- 46. Scotti C, Osmokrovic A, Wolf F, et al. . Response of human engineered cartilage based on articular or nasal chondrocytes to interleukin-1β and low oxygen. Tissue Eng Part A. 2012;18(3-4):362-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kwon H, Brown WE, O’Leary SA, Hu JC, Athanasiou KA. Rejuvenation of extensively passaged human chondrocytes to engineer functional articular cartilage. Biofabrication. 2021;13(03):5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Day TF, Guo X, Garrett-Beal L, Yang Y. Wnt/beta-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev Cell. 2005;8(5):739-750. [DOI] [PubMed] [Google Scholar]
- 49. Hill TP, Später D, Taketo MM, Birchmeier W, Hartmann C. Canonical Wnt/beta-catenin signaling prevents osteoblasts from differentiating into chondrocytes. Dev Cell. 2005;8(5):727-738. [DOI] [PubMed] [Google Scholar]
- 50. Hu H, Hilton MJ, Tu X, et al. Sequential roles of Hedgehog and Wnt signaling in osteoblast development. Development. 2005;132(1):49-60. [DOI] [PubMed] [Google Scholar]
- 51. Johnson CI, Argyle DJ, Clements DN. In vitro models for the study of osteoarthritis. Vet J. 2016;209:40-49. [DOI] [PubMed] [Google Scholar]
- 52. Westendorf JJ, Kahler RA, Schroeder TM. Wnt signaling in osteoblasts and bone diseases. Gene. 2004;341:19-39. [DOI] [PubMed] [Google Scholar]
- 53. Hartmann C. Skeletal development--Wnts are in control. Mol Cells. 2007;24(2):177-184. [PubMed] [Google Scholar]
- 54. Bosurgi L, Cao YG, Cabeza-Cabrerizo M, et al. Macrophage function in tissue repair and remodeling requires IL-4 or IL-13 with apoptotic cells. Science. 2017;356(6342):1072-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Laschober GT, Brunauer R, Jamnig A, et al. Age-specific changes of mesenchymal stem cells are paralleled by upregulation of CD106 expression as a response to an inflammatory environment. Rejuvenation Res. 2011;14(2):119-131. [DOI] [PubMed] [Google Scholar]
- 56. Karin M, Clevers H. Reparative inflammation takes charge of tissue regeneration. Nature. 2016;529(7586):307-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zscharnack M, Krause C, Aust G, et al. Preclinical good laboratory practice-compliant safety study to evaluate biodistribution and tumorigenicity of a cartilage advanced therapy medicinal product (ATMP). J Transl Med. 2015;13:160. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Cartibeads production method and the composition of mediums E, R, and I that are described in the Methods section are patented under European patent number WO2021028335. Moreover, raw data will be available upon request.
The exact composition of the medium E, R and I is described in the patent (WO2021028335). All raw data will be published in data repository upon acceptance for publication. Moreover, raw data will be available for reviewers upon request.