Graphical abstract
Keywords: Nanobody, SARS-CoV-2, COVID-19, Variants
Abstract
Global health and economy are deeply influenced by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and its newly emerging variants. Nanobodies with nanometer-scale size are promising for the detection and treatment of SARS-CoV-2 and its variants because they are superior to conventional antibodies in terms of cryptic epitope accessibility, tissue penetration, cost, formatting adaptability, and especially protein stability, which enables their aerosolized specific delivery to lung tissues. This review summarizes the progress in the prevention, detection, and treatment of SARS-CoV-2 using nanobodies, as well as strategies to combat the evolving SARS-CoV-2 variants. Generally, highly efficient generation of potent broad-spectrum nanobodies targeting conserved epitopes or further construction of multivalent formats targeting non-overlapping epitopes can promote neutralizing activity against SARS-CoV-2 variants and suppress immune escape.
1. Introduction
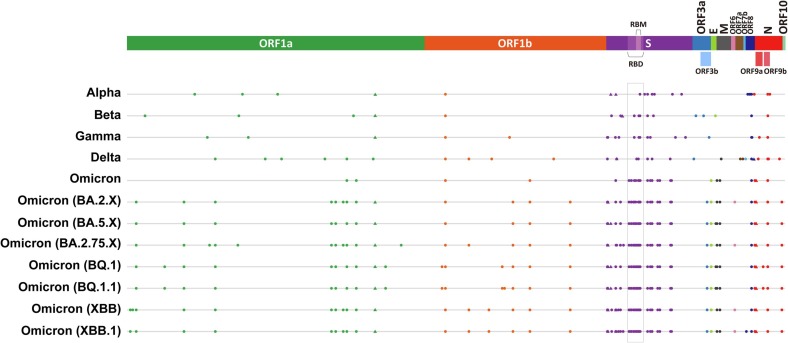
The coronavirus disease 2019 (COVID-19) pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection, has posed unprecedented influence on global health and economy, mainly due to its high transmissibility [1], high mutation rate [2], [3], asymptomatic infection [4], lack of effective prevention and treatment methods, as well as inappropriate management strategies. Moreover, patients who have recovered from COVID-19 are still affected. Recent studies have shown that about 1 out of 8 patients will experience “long-term COVID-19” symptoms after recovery [5]. In addition, the SARS-CoV-2 virus keeps evolving into various new variants, among which the variants with enhanced transmissibility or infectivity are classified as variants of concern (VOC), including Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2), and the currently circulating Omicron (B.1.1.529) variant based on the latest information of World Health Organization (WHO) (https://www.who.int/activities/tracking-SARS-CoV-2-variants/) (Fig. 1 ), and this has brought great challenges to the prevention and control of the virus [6], [7]. For the currently globally dominant Omicron variant, the coexistence of various strains has led to the emergence of Omicron XD, XE, XF, and other recombinants, which are more infectious and resistant to developed vaccines and antiviral drugs [2], [8].
Fig. 1.
Mutations of SARS-CoV-2 variants. Mutations of SARS-CoV-2 variants are depicted (amino acid substitutions are indicated as dots (●), while deletions as triangles (▲) with different colors representing different proteins of the virus. The rectangular box indicates RBD, which is the convergent region of the SARS-CoV-2 variants mutations. Generated based on sequences from GISAID (https://gisaid.org/wiv04/, and https://outbreak.info/situation-reports/).
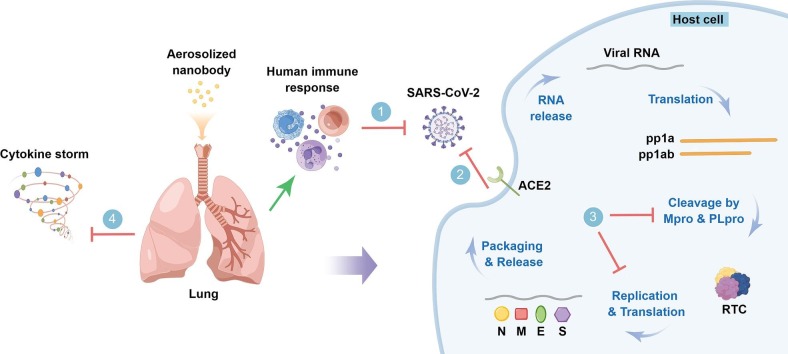
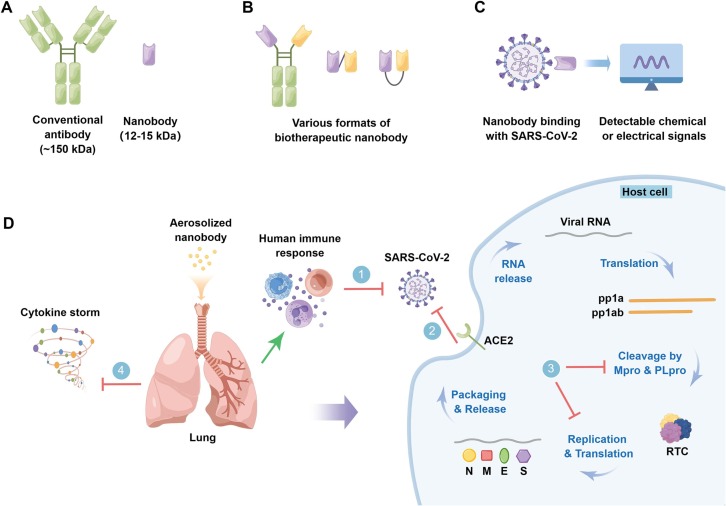
Nanobodies (Nbs), also known as VHHs or single-domain antibodies, are small antigen-binding fragments derived from unique heavy-chain-only antibodies (HCAbs) in camelids [9]. Compared with conventional antibodies, nanobodies are featured by small size (12–15 kDa, about one-tenth the size of conventional antibodies), single domain, high stability (resistance to high temperature and extreme pH conditions) and long shelf-life, high solubility, deep-tissue penetration, cryptic epitope accessibility, amenable to versatile modalities, lack of glycosylation as well as Fc-mediated immune activation, and high amount but cost-effective prokaryotic production, all of which make nanobodies excellent diagnostic and therapeutic tools [10] (Fig. 2 ). In particular, the robustness of nanobodies enables their inhaled delivery, which appears to be an ideal administration route for the treatment of pulmonary diseases including COVID-19 [11], [12], this is because nebulized nanobodies increase their lung delivery efficiency and concentration in bronchoalveolar lavage fluid, and allow the nanobodies to reach their maximum concentration (Cmax) more quickly [13] (Fig. 2D). It has been reported that multimeric nanobodies retain their function after aerosolization, lyophilization, and heat treatment, enabling direct aerosolized delivery to the nasal and lung tissues, which are two key sites of SARS-CoV-2 replication [14].
Fig. 2.
Prevention, Detection, and treatment of the SARS-CoV-2 and its variants with nanobodies. A. Schematic representation of conventional antibody and nanobody which is the variable domain of heavy chain-only antibody (HCAb) and only one tenth the size of the former. B. Nanobodies are adaptable to different multivalent formats, enabling enhanced avidity, simultaneous engagement of multiple spatially discrete non-overlapping epitopes, and mitigation of viral escape, especially when using conserved epitopes as targets. Here, multivalent formats of nanobodies are exemplified by dimerization achieved by genetic immunoglobulin (Ig) Fc fusion or C-to-N-terminal assembly, or otherwise C-to-C-terminal fusion of nanobodies through chemical approach. C. Application of robust nanobodies in SARS-CoV-2 detection, especially in point-of-care testing (POCT), during which binding events are translated into detectable chemical or electrical signals. D. Local pulmonary delivery of aerosolized nanobodies is promising in prevention and treatment of the SARS-CoV-2 and its variants by inducing human immune response (1), blocking their life cycle at different stages, such as viral entry (including viral binding and the subsequent membrane fusion) (2) and viral replication (3), or by modulating host inflammation (cytokine storm) (4). Green line with arrow indicates “Induction”, and red “T” shaped lines indicate “Inhibition”. By Figdraw.
2. Life cycle of SARS-CoV-2
SARS-CoV-2 is an enveloped virus with positive-sense single-stranded RNA (ssRNA) that encodes four structural proteins (the spike (S), nucleocapsid (N), membrane (M), and envelope (E) proteins), which are required for viral assembly and cell entry, 16 non-structural proteins (NSPs) (nsp1-16) that are essential for viral replication, and 9 accessory proteins that may interfere with host immunity [15], [16]. The coronavirus homotrimeric S glycoprotein consists of the receptor-binding domain (RBD)-containing S1 and S2 subunits that mediates viral attachment and subsequent fusion with host cell membranes [17], [18]. Interestingly, the prefusion form of each RBD is in a dynamic equilibrium switching between two conformations: the receptor-inaccessible down (or closed) conformation and the receptor-accessible up (or open) conformation which is considered to be less stable [19], [20], depending on the transient hide or exposure of the receptor binding determinants, and S protein is predominantly in the 3down or 1up2down conformation [19], [21]. S protein with at least one RBD in the up conformation is accessible to ACE2 and their binding triggers the S protein trimer dissociation as well as fusion events, whereas S protein with all three RBDs in the down conformation is inactive [14], [22], [23], [24]. Cryo-EM experiments showed that upon furin cleavage, RBD undergoes a transition from a compact inaccessible down conformation to a partially accessible form, followed by an ACE2-bound up conformation [25].
The life cycle of SARS-CoV-2 starts with host cell entry, a critical step and a key target for COVID-19 preventive and therapeutic drugs. Upon reaching the cell surface, RBD in the up conformation binds to the peptidase domain (PD) of angiotensin-converting enzyme 2 (ACE2), resulting in a conformational change in the S1 subunit and the subsequent exposure of the S2 subunit located S2′ proteolytic site [25], which is cleaved by highly expressed host transmembrane serine protease 2 (TMPRSS2) in lung epithelial cells, and this leads to the rapid cell entry of wild-type or previous variants of SARS-CoV-2 [26]. Otherwise, for Omicron, the virus mainly infects the upper airway epithelial cells with low TMPRSS2 expression, followed by being slowly endocytosed into endolysosomes, where the S2′ site is cleaved by cathepsin L [26], [27]. Cleavage at the S2′ site further results in exposure of the fusion peptide (FP) that induces fusion between the viral and host cellular membranes [28]. Viral RNA is released through fusion pore into the cytoplasm of host cell and then hijacks the host ribosomal machinery to produce viral polypeptides (pp1a and pp1ab), which are cleaved by Mpro and PLpro proteins [15], [29]. The resulting replication-transcription complex (RTC) which contains RNA-dependent RNA polymerase (RdRp) replicates the viral ssRNA [30]. The N protein is biosynthesized in the cytoplasm, while other structural proteins including S, M, and E proteins are translated on the rough endoplasmic reticulum (RER) to confer post-translational modifications (PTMs), and the S protein is cleaved at the S1-S2 boundary by furin or furin-like proprotein convertase in the Golgi apparatus, resulting in the non-covalently associated S1 and S2 subunit [31], [32]. Viral structural proteins and genomic RNA are then transported for virion assembly to the ER-Golgi intermediate compartment (ERGIC), where the N protein is packaged internally along with ssRNA, while the S, E, and M proteins are incorporated into the viral membrane. The virions are then released from host cells through exocytosis to restart the life cycle [33] (Fig. 2D).
3. Diagnosis of COVID-19 with nanobodies
Since SARS-CoV-2 is an evolving virus which has and may further strengthen its airborne transmission [2], [34], high transmissibility, infectious asymptomatic infections [35], and other characteristics, readily and highly sensitive detection of the virus plays a pivotal role in curbing the ongoing pandemic. The current molecular detection of SARS-CoV-2 mainly utilizes real-time reverse transcription polymerase chain reaction (RT-PCR) or loop-mediated isothermal amplification (LAMP) with high sensitivity [36], [37]. Nevertheless, this gold-standard method not only relies heavily on laboratory equipment, but also is costly and time-consuming, which is not conducive to mass screening and timely interruption of viral transmission [38]. Alternatively, viral antigen proteins can be directly detected through a cost-effective and time-saving antibody-based immunoassay without sample processing [39]. The specificity, sensitivity, and stability of antibodies are critical for the viral detection performance in immunoassays [40], and nanobodies are excellent reagents mainly due to their improved shelf-life, and capacity of high-density coupling to raise detection sensitivity resulted from small size [41], especially in point-of-care testing (POCT) which provide on-the-spot results, such as lateral flow immunoassays (LFIs).
Currently, many nanobody-based immunoassays target different proteins of SARS-CoV-2 to convert binding events into detectable chemical or electrical signals (Fig. 2C) (Table 1 ). Based on the immunoreaction between an antigen and its antibody, laboratory-based sandwich enzyme-linked immunosorbent assay (ELISA) with labelled immunosensor is a commonly used quantitative method. However, its application in high-throughput mass screening and portable use is limited by time-consuming, high cost, as well as complex operation, while all of these are overcome in LFIs, in which binding events are amplified and converted into electrical signals readout through electrochemical transistors [42], [43]. Au nanoparticles (AuNPs) anchored with S protein-specific nanobodies have been used as photoelectrochemical (PEC) immunosensors with a reported limit of detection (LOD) of ∼ 5 fg/mL for S protein, in which the capture of S proteins by nanobodies increases the steric effect, resulting in impediment of photoinduced charges diffusion and reduced photocurrent signal [44]. In another research, AuNPs were coated with nanobodies targeting non-overlapping epitopes of RBD and served as multivalent antigen-binding sensors called Nano2RED, in which antigen binding signals are converted into detectable physical AuNP aggregation with a LOD for RBD as low as ∼ 1.3 pM in diluted human serum [45]. Both AuNP-based methods described above can obtain readout from a sample within 30 min. Nanobody-functionalized electrodes bioconjugated with RBD binding nanobody VHH72 fused SpyCatcher with high density and controlled orientation, were formatted into sensors by combining with solution-processable conjugated polymer in the transistor channel and give single-molecule-sensitive readouts within 15 min using unprocessed saliva or nasopharyngeal swab samples [46]. In another research, Pagneux et al. introduced a cysteine mutant in a loop between frameworks 1 and 2 of VHH72 to make it perpendicular orientated on the sensor surface, resulting in a nanobody-functionalized electrochemical platform using differential pulse voltammetry (DPV) as a readout, which detects saliva and nasopharyngeal swab samples rapidly within 10 min [47]. Nevertheless, the capture of analyte molecules by biosensors commonly relies on diffusion-dominated transport or manual mixing, which is still time-consuming and labor-intensive. To address this issue, Koklu et al. introduced alternating current electrothermal flow (ACET) with micro-stirring effects to accelerate the device operation, thereby reducing the sample-to-result time to approximately only 2 min [48]. Apart from improving intrinsic sensitivities, efforts have also been made to capture and concentrate SARS-CoV-2 virus using nanobody-functionalized cellulose to improve detection capability, which can also be used for virus sampling and virus removal [49]. It is worth noting that although the S protein is currently the most commonly used target for antigen detection, the N protein is also used [50], [51] due to its abundant expression and presence in nasopharyngeal swabs [52], [53]. More importantly, the N protein is a potential universal target because it is more conserved than the S protein with convergent mutations [54] (Fig. 1).
Table 1.
Summary of nanobody-based immunoassay methods for SARS-CoV-2 detection.
| Nanobody name | Nanobody target | Affinity to target (KD) | Method | Sample source | Specificity | Sensitivity | Sample to-result time | Features | References |
|---|---|---|---|---|---|---|---|---|---|
| NTD-E4, NTD-B6 | N protein | 23.8 ± 2.1 nM (NTD-E4), 46.5 ± 2.6 nM (NTD-B6) | ELISA | Nasal swabs | – | – | Several hours | – | [51] |
| E2, B6, C2 | N protein | 0.8 nM (E2), 1.6 nM (B6), 1.1 nM (C2) | ELISA | – | – | LOD of 50 pg/mL for N and 1.28 × 103 pfu/mL for the killed virus |
Several hours | – | [50] |
| C5, F2 | S protein | 99 pM (C5), 40 pM (F2) | ELISA | – | – | 103 ffu/ml for inactivated virus | Several hours | – | [116] |
| – | RBD of S protein | – | ELISA | Saliva, nasopharyngeal swabs | 100 % | Saliva - 93.3 % (Ct ≤ 30), 87.5 % (Ct ≤ 35), 86 % (Ct ≤ 40); Nasopharyngeal swabs - 90 % (Ct ≤ 30), 80 % (Ct ≤ 35), 88 % (Ct ≤ 40) | Several hours | – | [117] |
| – | S protein | Photoelectrochemical (PEC) immunosensor | – | – | LOD = 2.8 × 10−17 M (5 fg/mL) | 30 min | Label-free, convenient, rapid, low cost | [44] | |
| RBD8/ RBD10 |
RBD of S protein | – | Nanobody-functionalized nanoparticles for rapid, electronic detection (Nano2RED) | Serum, blood | – | LOD=∼1.3 × 10−12 M, for RBD in diluted human serum | 5–20 min | Label-free, rapid detection, low cost | [45] |
| VHH72-C13 | RBD of S protein | 12.1 nM | Nanobody-functionalized electrochemical platform | Saliva, nasopharyngeal swab | Saliva-85 %, nasopharyngeal swab-90 % | Saliva-80 %, nasopharyngeal swab-90 %, LOD = 1.2 × 104 viral RNA copies mL−1, 2 × 10−17 M |
10 min | Portable, quick, accurate, user-friendly | [47], [118] |
| VHH72 | RBD of S protein | 23 nM | Organic electrochemical transistors (OECTs) | Saliva, nasopharyngeal swab, serum | – | Single-molecule, sensitivity comparable to RT-PCR, saliva (LOD = 1.2 × 10−21 M), nasopharyngeal swab (LOD = 1.9 × 10−14 M) | <15 min | Label-free, rapid, sensitive, reusable, simple, robust | [46], [118] |
| Ty1 | RBD of S protein | 24.9 ± 10.3 nM | Alternating current electrothermal flow (ACET) integrated OECT-based immunosensor | Saliva | – | LOD = 1 × 10−16 M | ∼2 min | Label-free, partially reusable, speedy detection | [48] |
In addition to direct detection of the virus, diagnostic testing of human neutralizing antibodies (NAbs) against SARS-CoV-2 is also of great significance to monitor the spread of viral infection or the effectiveness of vaccination at individual and population scales. While currently used detection methods are usually immunoassays (ELISA, LFI, etc.) that do not distinguish NAbs from non-NAbs, or viral neutralization test (VNT) that require the use of live SARS-CoV-2 virions and biosafety level 3 (BSL-3) facility, which are often time-consuming, laborious, and relatively expensive [55]. Wagner et al. established a competitive multiplex binding assay (NeutrobodyPlex) using a biparatopic nanobody (bipNb) consisting of two RBD-specific inhibitory nanobodies and succeed in high-throughput screening and detailed analysis of NAbs against RBD [56]. Nanobodies have also been applied in POCT, enabling speedy, highly portable, semi-quantitative, and cost-effective NAb measurements. Redecke et al. constructed a bi-specific protein comprising a red blood cell (RBC) binding nanobody and RBD to mediate instantaneous hemagglutination in the presence of RBD-targeted NAbs, and they used artificial intelligence (AI) to analyze the digital images captured by cell phones, promoting its application in POCT [57]. Notably, these methods are adaptable to other pathogens.
4. Prevention of SARS‑CoV‑2 infection
Effective vaccines are significant in blocking the spread of COVID-19, while most of the vaccines currently in use are sophisticated to manufacture and entail specialized storage and transportation conditions, which limits their distribution, especially in remote areas. It is promising to develop vaccines based on nanobodies due to their superior stability. Pishesha et al. fused RBD with a nanobody targeting class II MHC antigens to develop a candidate vaccine, which delivers RBD directly and specifically to antigen-presenting cells (APCs), resulting in robust humoral and cellular immunity against SARS-CoV-2 as well as its variants (Alpha, Beta, and Gamma) in both young and aged mice. Moreover, this vaccine remains stable with full efficacy for room temperature storage or lyophilization [58]. Besides, innate anti-viral immune response was also used. IFN-β inhibits SARS-CoV-2 replication by upregulating the expression of interferon-stimulated genes (ISGs) and has been used to treat patients with COVID-19. Lyu et al. generated exosomes conjugated with IFN-β fused by S protein-targeting nanobody to selectively deliver IFN-β to virus-infected cells, which may also avoid the side effects caused by systemic administration of IFN-β [59].
5. Treatment of SARS‑CoV‑2
All strategies that can effectively disrupt the life cycle of SARS-CoV-2 in humans have potential therapeutic value for the treatment of COVID-19. In addition, nanobodies also have the potential to modulate host immune response in hospitalizations associated with severe COVID-19 (Fig. 2D) (Table 2 ).
Table 2.
Summary of nanobodies in treatment of SARS-CoV-2 variants.
| Nanobody name | Nanobody target | Nanobody format | Source | Screening method | Expression system | Affinity to target (KD) |
Neutralizing pseudovirus (IC50) |
Neutralizing SARS-CoV-2 (IC50) |
Mechanism | Virus variants | Features | References |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NB1A2, NB2B4 | Mpro | Monovalent | Mpro-immunized camel |
Phage display | Top10F’ cells | 2.43 nM, 0.46 nM | – | – | Targeting Mpro dimer and dissociate it into monomers, engaging the catalytic and substrate binding sites of Mpro |
– | – | [76] |
| Nb15-NbH-Nb15 | S protein | Heterotrimeric | Alpaca immunized with the extracellular domain of S protein (S1 + S2 ECD, S) | Phage display |
293F cells | 0.541 nM | 0.0004 ± 0 μg/ml | – | – | Wild type, Alpha, Delta, Epsilon, B.1.429 | Nb15-NbH-Nb15 formatting; intranasal administration |
[119] |
| See original literature | S protein | Monomeric, dimeric, trimeric | Llamas immunized with S1 and S2 Fc fusion proteins | Combining in vivo antibody affinity maturation and proteomics | Arctic Express (DE3) cells | See original literature | See original literature | See original literature | Multiple modes (see original literature) |
Alpha, Beta, Gamma | Combining in vivo antibody affinity maturation and proteomics, synergistic viral-neutralizing effects analysis | [96] |
| See original literature | S protein (RBD, S2, or NTD) |
Monovalent, bivalent (Fc fusion) | Llamas immunized with S protein (and boosted with RBD) | Phage display |
E. coli (Monovalent) HEK293-6E cells (Fc fusion) |
See original literature | See original literature | See original literature | See original literature | Wild type, Alpha, Beta, Omicron |
Targets besides RBD | [92] |
| saRBD-1 | RBD of S protein | Monovalent, bivalent | Spike RBD immunized alpaca |
Phage display | Bacteria, 293F cells | 750 pM (monovalent), 302 pM (bivalent) | 4.26 nM (monovalent), 100 pM (bivalent) | – | Competitive binding to RBD of the S protein | Wild type, Alpha, Beta, Gamma, Delta, Epsilon | – | [120] |
| Sb#15, Sb#68 | RBD of S protein | Monovalent, bivalent | Synthetic sybody libraries | Ribosome display + phage display | E. coli MC1061 cells | 12 nM (Sb#15), 9 nM (Sb#68), 0.3 nM (GS4), | 2.3 µg/ml (Sb#15), 2.3 µg/ml (Sb#68), 0.02 µg/ml (G4S), 0.01 µg/ml (Tripod-GS4r) | – | Interfering with ACE2 interaction | Wild type, Alpha, Beta, Delta, | – | [69] |
| VHH72 | RBD of S protein | Bivalent, VHH72-Fc | Llama immunized with SARS-CoV-2 S glycoprotein | Phage display | HEK293 cells, Pichia pastoris | 23 nM | 0.2 µg/mL | – | Disrupting RBD dynamics and receptor binding | Wild type, Delta, Beta, Kappa | Extracellular vesicles-targeting platform | [118], [121] |
| 1B-3F-Fc, 3F-1B-2A-Fc, 1B-3F-2A-Fc | RBD of S protein | Bispecific, trispecific | Naïve and synthetic humanized llama VHH libraries | Phage display | Expi293 cells | 0.25 nM (1B-3F-Fc), 46.8 pM (3F-1B-2A-Fc), 94.7 pM (1B-3F-2A-Fc) | 3.00 nM (3F-1B-2A-Fc), 6.44 nM (1B-3F-2A-Fc) | – | Interfering with ACE2 interaction | Wild type, Alpha, Beta, Delta, Lambda, Omicron |
– | [100], [112], [122] |
| NB1A7, NB1B11 | RBD of S protein | Monovalent, multivalent | Recombinant RBD-immunized camel |
Phage display | Periplasm of E. coli BL21 (DE3) (monovalent), HEK293F cells (multivalent) | 0.74 ± 0.003 nM (NB1A7), 6.76 ± 0.027 nM (NB1B11) |
131.3 ± 1.39 nM (NB1A7), 303.9 ± 1.46 nM (NB1B11) | 1.0 ± 1.5 nM (Format I: NB1A7-Fc + NB1B11-Fc), 0.6 ± 1.19 nM (Format II: NB1A7-NB1B11-Fc) |
Blocking the interaction between ACE2 and RBD | – | – | [61] |
| n3113v, n3130v, bn03, n3113.1-Fc (Y58L) |
RBD of S protein | Monovalent, bivalent | Grafted fully human single-domain antibody | Phage-display, in vitro affinity maturation through error-prone PCR | E. coli HB2151 | 0.81–2.54 nM (n3113v); 1.16–6.04 nM (n3130v); ≤1 nM (bn03) | 0.28 µg/mL (bn03) | – | Inducing S trimer to unstable wide-up state, inhibiting S protein mediated membrane fusion |
Wild type, Alpha, Beta, Gamma, Delta, Omicron | Human origin single-domain antibody library | [68], [99], [123] |
| RBD-1-2G | RBD of S protein | Monovalent, bivalent (RBD-1-2G-Fc), trivalent | Synthetic humanized nanobody libraries | Phage-display | Vibrio natriegens (Monovalent); | 9.4 nM (monovalent toward RBD); 1.9 nM (bivalent RBD-1-2G-Fc); 0.1 nM (trivalent) | 490 nM (RBD-1-2G); 88 nM (RBD-1-2G-Fc); 4.1 nM (RBD-1-2G-Tri) | 1211 nM (RBD-1-2G); 255 nM (RBD-1-2G-Fc); 182 nM (RBD-1-2G-Tri) | Interfering with ACE2 interaction | Wild type, Alpha | Synthetic humanized nanobody libraries | [124] |
| P2C5, P2G1, P5F8 | RBD of S protein | Monovalent, bivalent | Bactrian camel immunized with SARS-CoV-2 RBD | Phage-display | E. coli BL21 cells | 3.97 nM (P2C5), 5.36 nM (P2G1) and 1.94 nM (P5F8) | – | – | Competing with ACE2 for RBD binding | SARS-CoV-2B.1.1.1, Alpha, Beta, Gamma, Delta and Omicron | – | [125] |
| NIH-CoVnb-112 | RBD of S protein | Monovalent | Llama immunized with SARS-CoV-2 S1 protein | Phage-display | BL21 (DE3) competent E. coli strain; X-33 Pichia pastoris |
1–5 nM | – | – | Competing with ACE2 for RBD binding | Wild type, Alpha, Beta, Gamma, and Delta. | Nebulized delivery of NIH-CoVnb-112 | [126], [127] |
| Nb1, Nb2 | RBD of S protein | Bivalent (Nb1–Nb2), tetravalent (Nb1–Nb2-Fc) | Synthetic library | Phage-display | Escherichia coli (Nb1–Nb2), Expi293F cells (Nb1–Nb2-Fc) | ≤0.001 nM | Nb1-Nb2 (All variants ≤ 0.0865 nM) | Nb1-Nb2 (WT-1.207 nM, Alpha-0.8149 nM, Beta-1.776 nM, Gamma-13.01 nM, Delta-0.7317 nM); Nb1-Nb2-Fc (WT-0.0168 nM, Alpha-0.0117 nM, Beta-0.0097 nM, Gamma-0.0987 nM, Delta-0.0232 nM, Omicron-1.46 nM) |
Competing with ACE2 for RBD binding | Alpha, Beta, Gamma, Delta, Lambda (C.37), Kappa (B.1.617.1), Mu (B.1.621), and Omicron | – | [128] |
| 1.10, 1.26, 1.29, 2.15 | RBD of S protein | Monospecific and bispecific | Dromedary camels immunized with RBDmFc |
Surface Display in Bacteria | HEK- 293F cells |
Nb - 3.96 nM (1.10), 0.26 nM (1.26), 12.9 nM (1.29), 0.39 nM (2.15); HCAb - 0.77 nM (1.10), 0.08 nM (1.26), 0.63 nM (1.29), 0.08 nM (2.15) |
– | – | Competing with ACE2 for RBD binding | WA1, Alpha, Beta, Gamma, Delta, Kappa, Zeta | – | [129] |
| VHH72 | RBD of S protein | Multivalent (bivalent, tetravalent, hexavalent) | Llama immunized with SARS-CoV-2 S glycoprotein | Phage display | HEK293-6E cells | 23 nM | 3.3 ± 1.8 nM (bivalent), 0.34 ± 0.072 nM (tetravalent), 0.035 ± 0.003 nM (hexavalent) | – | Competing with ACE2 for RBD binding | Wild-type, Alpha, Beta | High valencies formatting of nanobody (hexavalent) | [103], [118] |
| Nb12, Nb15, Nb17, Nb19, Nb30, and Nb56 | RBD of S protein | Monomer, bivalent, trimer | Llama or nanomice immunized with RBD and/or S protein | Phage display |
WK6 cells (ATCC, 47078), Expi293 cells (Fc conjugated nanobodies) | Monomer - 8.15 nM (Nb15), 5.59 nM (Nb17), 4.72 nM (Nb19), 3.26 nM (Nb56), 30.0 nM (Nb12), 6.55 nM (Nb30); Trimer - 16.4 pM (Nb15), <1 pM (Nb17), 35.1 pM (Nb19), 38.2 pM (Nb56), <1 pM (Nb12), 820 pM (Nb30) |
Monomer - 11.7 nM (Nb12), 0.4 nM (Nb15), 0.6 nM (Nb17), 0.3 nM (Nb19), 6.9 nM (Nb30), 0.9 nM (Nb56); Bivalent - 0.7 nM (Nb12-llama), 0.5 nM (Nb12-human), 0.3 nM (Nb15-human), 79 pM (Nb17-human), 66 pM (Nb19-human), 1.9 nM (Nb30-llama), 1.8 nM (Nb30-human), 0.1 nM (Nb56); Trimer - 0.1 nM (Nb12-human), 65 pM (Nb12-llama), 0.1 nM (Nb15-human), 18 pM (Nb17-human), 9 pM (Nb19-human), 55 pM (Nb56-human) |
– | Interfering with ACE2–RBD associations | WA1, Alpha, Beta, Gamma | Creation of nanobody-producing mice - nanomice | [91] |
| RBD1i13, RBD3i17, RBD6id, RBD10i10, RBD10i14, RBD11i12 | RBD of S protein | Monovalent, bivalent (Fc fusion) | A synthetic yeast-displayed library of nanobodies |
Autonomous hypermutation yeast surface display | Expi293 cells | 32.2 nM (RBD1i13), 230 nM (RBD3i17), 263 nM (RBD6id), 2.14 nM (RBD10i10), 0.72 nM (RBD10i14), 316 nM (RBD11i12) | 0.05 μg/ml (RBD1i13), 0.116 μg/ml (RBD3i17), 0.056 μg/ml (RBD6id), 0.19 μg/ml (RBD10i10), 0.42 μg/ml (RBD10i14), 0.04 μg/ml (RBD11i12) | – | – | – | Autonomous hypermutation yeast surface display (AHEAD) | [109] |
| W25 | RBD of S protein | Monovalent (nanobody, monomeric Fc fusion), bivalent (Fc fusion) | Alpaca immunized with full S protein | bacterial display | Priplasmic expression the E. coli wk6 strain (W25) ExpiCHO cells (W25Fc, W25FcM) |
∼ 0.3 nM (RBD) | – | W25 - 9.82 ± 1.92 nM (D614), 5.09 ± 1.09 nM (G614 variant); W25FcM - 27.40 ± 8.38 nM (D614), 12.36 ± 2.84 nM (G614); W25Fc - 7.39 ± 2.39 nM (D614), 3.69 ± 0.96 nM (G614) |
Competing with ACE2 for RBD binding | D614 variant, G614 variant | – | [109], [130] |
| Nb20, Nb21, Nb34, Nb105, Nb95, Nb17, Nb36 |
RBD of S protein | Monovalent, homodimeric, homotrimeric, heterodimeric | Llama immunized with RBD-Fc fusion protein | Proteomic identification of high-affinity RBD-Nbs | BL21 (DE3) cells | 10.4 pM (Nb20), <1 pM (Nb21) | 0.102 nM (Nb20), 0.045 nM (Nb21), 1.991 nM (Nb34), 5.105 nM (Nb105), 10.05 nM (Nb95), 1.106 nM (Nb17), 1.563 nM (Nb36) | 0.048 nM (Nb20), 0.022 nM (Nb21), 1.125 nM (Nb34), 5.070 nM (Nb105), 5.105 nM (Nb95), 1.500 nM (Nb17) | Sterically interfering with ACE2 binding (directly for class I nanobodies, or indirectly for class II nanobodies), lock the spike in an all-RBD-up conformation or destabilize it (for class III nanobodies) | Alpha, Delta | Proteomic identification of high-affinity RBD-Nbs, resist resistance of nanobody to lyophilization and aerosolization | [101], [105] |
| WNb 2, WNb 7, WNb 15, WNb 36 | RBD of S protein | Monovalent, bivalent (Fc fusion) | Alpacas immunized with coronavirus spike and receptor-binding domains (RBD) | Phage display | – | 0.14 - 19.49 nM | Wild type - 0.33 nM (WNbFc 2), 3.18 nM (WNbFc 7), 2.55 nM (WNbFc 15), 0.10 nM (WNbFc 36); N501Y D614G variant - 0.30 nM (WNbFc 2), 5.04 nM (WNbFc 7), 4.91 nM (WNbFc 15), 0.11 nM (WNbFc 36) |
Interfering with ACE2–RBD associations | Wild type and the N501Y D614G variant |
– | [131] | |
| C5, H3, C1, F2 | RBD of S protein | Monomeric, dimeric (Fc fusion), trimeric | Llama immunized with RBD, RBDFc, and/or S protein | Phage display | WK6 E. coli strain (monomeric), Expi293® cells (dimeric, trimeric) | RBD - 615 pM (C1), 99 pM (C5), 25 pM (H3), 40 pM (F2), 18 pM (C5 trimer), 0.3 pM (H3 trimer), 53 pM (C1 trimer) |
– | 18 pM (C5 with Victoria), 25 pM (C5 with Alpha) | Interfering with ACE2–RBD associations | Victoria, Alpha, Beta | – | [65] |
| DL28 | RBD of S protein | Monomeric, dimeric (Fc fusion) | Alpaca immunized with RBD | Phage display | E. coli (monomeric), Expi293 suspension cells | 1.56 nM | – | 5.39 nM (Wild type), 4.61 nM (Alpha), 13.95 nM (Beta), 17.16 nM (Gamma), 21.88 nM (Delta), 8.68 nM (Omicron) | Keeping an RBD loop in a conformation incompatible with ACE2-binding |
Wild type, Alpha, Beta, Gamma, Delta, Omicron | – | [65], [66] |
| H7, G12 | RBD of S protein | Dimeric (Fc fusion) | Synthetic humanized VHH library | Phage display, affinity maturation by CDR1 and CDR2 shuffling |
FreeStyle™ 293-F cells |
4.485 nM (H7-Fc), <1 pM (G12 × 3-Fc) |
– | D614G variant - 133.8 ng/ml (H7-Fc), 13.1 ng/ml (G12 × 3-Fc); Delta - 12.3 ng/ml (H7-Fc), 0.9 ng/ml (G12 × 3-Fc); Omicron BA.1 - 106 ng/ml (H7-Fc), 9.6 ng/ml (G12 × 3-Fc) |
– | Wild type, D614G variant, Delta, Omicron BA.1 | – | [66], [132] |
| C5G2 | RBD of S protein | Monovalent | Synthetic nanobody library |
Phage display-screening and affinity maturation |
E. coli BL21 (DE3) |
1.62 nM | 1.59 nM (Wild type), 0.95 nM (Alpha), 0.56 nM (Beta), 1.06 nM (Gamma), 0.3 nM (Omicron) | – | Binding to RBD at a conserved region and the neighboring NTD domain through its CDR3, and inhibit ACE2 binding by FR2 due to steric hindrance | Wild type, Alpha, Beta, Gamma,Omicron | One nanobody wit triple functions | [132], [133] |
| Nb-007 | RBD of S protein | Monomeric, dimeric (Fc fusion) | Alpaca immunized subcutaneously with RBD | Phage Display |
Brevibacillus choshinensis SP3 cells/ E. coli BL21 (DE3) (monomeric), 293 T cells (Fc fusion) |
67.4 pM | Nb-007 - 37.6 nM (Wild type), 8.13 mM (Beta), 1.07 mM (Delta); Nb-007-Fc - 1.64 nM (Wild type), 42.6 nM (Delta) |
– | Competing with ACE2 for RBD binding | Wild type, Beta, Delta | – | [133], [134] |
5.1. Blocking of viral invasion
Currently, S protein is the most prominent target of antibody-based targeted therapy for COVID-19 [60], mainly due to its vital role in viral recognition and fusion with host membranes, as previously described. Neutralizing nanobodies block viral invasion in several ways.
First, interfering with ACE binding. Most of the identified nanobodies showed efficacy by directly engaging epitopes of RBD. Another class of nanobodies, such as NB1A7 [61], do not bind to receptor-binding motif (RBM) of RBD, but they sterically hinder ACE2 binding. NB1B11 was reported to bind to RBM in both its up and down conformation and buries the ACE2-binding site, thereby blocking RBD/ACE2 attachment [61]. Apart from NB1B11, several nanobodies such as Ty1 [62], Sb23 [63], P17 and P86 [64]), have similar performance of binding both conformations, demonstrating their excellent antiviral efficacy. Second, stabilizing the epitope-inaccessible/inactive form of S protein. These nanobodies recognize RBD epitopes overlapping with the ACE2 binding site, and they bind the S protein in its fully inactive conformation (3downs), locking its receptor binding domains into an inaccessible state [14], [65], [66]. Third, inhibiting SARS-CoV-2 S protein-mediated membrane fusion with target cells or premature inducing of the post-fusion conformation to preclude bona fide fusion events. It has been reported that RBD bound to nanobody K-874A can still interact with ACE2, resulting in blocking viral membrane fusion but not viral attachment [67]. Growing evidence suggests that neutralizing nanobody binding can stabilize S protein in its up conformation, leading to transitions from premature to an irreversible post-fusion state and inactivation [61], [68] or aberrant activation of the fusion machinery that renders virions noninfectious [69], [70], probably due to exposure of proteolysis site and the subsequent shedding of S1.
5.2. Inhibition of viral replication
After viral entry, an effective strategy is to block viral replication mediated by RTC, which is composed of a number of NSPs [71]. For example, it is recently reported that a novel mechanism for nsp9-mediated SARS-CoV-2 RNA capping, which plays essential roles in viral translation and proliferation [72], suggesting that nsp9 is a promising target. Nanobodies targeting and inactivating key proteins in viral translation are potential anti-coronaviral targeted drugs. Recently, several studies have developed nanobodies against nsp3 [73], nsp7 [74], nsp8 [74], nsp9 [75], RdRp [74], and Mpro [76]. Unfortunately, these studies did not provide direct evidence of inhibition of viral replication and further research is needed. In fact, as opposed to preventing viral entry outside the cell, interfering with viral replication likely requires delivery of nanobodies into host cells, which may be critical to their efficacy.
5.3. Modulation of host immune response
Direct antiviral strategies including interventions of blocking SARS-CoV-2 entry or replication show efficacy only in the early stage, but relatively ineffective in hospitalizations associated with severe COVID-19, which may be largely driven by hyperactive immunopathology resulted from uncontrolled systemic inflammation including cytokine storm [77]. Studies have begun using nanobodies to inhibit proinflammatory responses by blocking the interaction of proinflammatory mediators with their receptors. A report focused on myeloid cells that produce proinflammatory cytokines and chemokines (IL-1, IL-6, IL-8, CXCL10, etc.), and constructed a bispecific nanobody A8-G11-Fc using two S protein-specific nanobodies (A8 and G11) with complementary capacity to block interactions between the S1 protein and host ACE2 as well as myeloid cell receptors, and then demonstrating its capacity of blocking ACE2-mediated infection as well as the myeloid receptor-mediated proinflammatory responses [78]. In another research, Ettich et al. developed a bispecific inhibitor, c19s130Fc, involving an RBD-targeting nanobody VHH72 to block viral infection, and a nanobody VHH6 targeting IL6:sIL-6R complexes to block IL-6 trans-signaling, which has been demonstrated to be a primary driver of ongoing inflammatory reactions [79].
6. Strategies for coping with the emerging SARS-CoV-2 variants
SARS-CoV-2 is an RNA virus, which is featured by large population size and short viral generation time [80], and it has a high mutation rate despite the proofreading activity conferred by nsp14 [81]. Moreover, recombination occurs between different circulating variants when they infect the same host [2], and RNA editing is also induced by human immune system [82]. Besides, human NAbs, as well as other drugs including vaccines, may act as an additive selective pressure, supported by evidence that mutations converge in RBD, especially the RBM region, which is a major target of NAbs [61], [83], [84] (Fig. 1). All of the above result in mutations accumulation, viral evolution, and finally the emergence of new variants [2], [85]. These variants, especially the Omicron variant including its subvariants, confer S protein mutations, among which R346T, K444T, N460K, and F486S represent key neutralization escape mutations [86], [87], [88], while the N460K and F486 mutations enhance fusogenicity [88], and some of the convergent hotspots render initially developed therapeutic antibodies ineffective [6], [7], [84], [89], [90].
6.1. Enhancing avidity of nanobodies
Since mutations in variants reduce their binding to neutralizing nanobodies, increasing affinity of nanobodies reversely may be a strategy for dealing with variants. Fc region fusion has been used to enhance their binding activity as well as neutralizing activity [91], [92], and interestingly, it is reported that nanobodies fused to the Fc region of human mucosal IgA may be more potent against Omicron variants compared to the IgG equivalent, possibly due to the longer hinge region of IgA-Fc [93]. In an excellent recent study, Xu et al. isolated a set of RBD-ACE2 interface-targeting nanobodies that are ineffective against E484 or N501 substitution-carrying variants, but their homotrimeric versions showed fully neutralization activity against these variants, probably due to approximately-two orders of magnitude improvement in affinity [91]. However, high affinity nanobodies will still undergo dissociation from the targets because of their reversible interaction. To address this issue, Yu et al. designed latent bioreactive amino acid (FFY)-incorporated covalent nanobodies, which bind irreversibly to viral S protein due to the FFY-mediated proximity-enabled reactive therapeutic (PERx) mechanism, and their viral neutralization potency was enhanced drastically by the FFY incorporation [94].
6.2. Simultaneous engagement of multiple spatially discrete non-overlapping epitopes
As mentioned above, nanobodies targeting different epitopes block the SARS-CoV-2 invasion through different mechanisms. The simultaneous application of nanobodies with different epitopes as multivalent formatting or cocktails can exert a synergistic effect, resulting in greatly improved avidity and neutralization potency, as well as broad-spectrum protection against SARS-CoV-2 variants, and ultimately mitigates viral escape [14], [69], [70], [95]. It is known that single-domain nanobodies, which are only one tenth the size of conventional antibodies, are superior in simultaneously binding discrete non-overlapping epitopes because of the lower chance of steric clash [96]. In fact, it is rare to see conventional antibodies binding to the same RBD simultaneously, as they will clash with each other and/or with other RBDs [96]. Notably, multivalent nanobodies are typically generated by genetic fusions that allow only C-to-N-terminal assembly, which may interfere with the function of their complementary determining regions (CDRs). Site-specific C-to-C-terminal fusion of nanobodies can be achieved by chemical approaches mediated by π‑Clamp [97], or 4-arm PEG [98], etc. (Fig. 2B). It is worth noting that although both multivalent nanobodies and cocktails can be effective in suppressing mutational escape, the neutralization efficacy of multivalent nanobodies is generally more potent than the latter, and in one report they differ by as much as fourfold [99], [100]. Moreover, the order of the nanobodies in the multivalent construct is also critical for efficacy, as this may affect how it binds to the trimeric S protein [70].
6.3. Targeting evolutionarily conserved epitopes of SARS-CoV-2 variants
As previously mentioned, under the selective pressure of NAbs, the ACE2-binding region RBM is the primary region for convergent mutations in current circulating variants and likely to be the case for future new variants [61], [83], [101]. Therefore, nanobodies targeting this region may easily lose their neutralizing activity, suggesting the importance of isolating nanobodies targeting conserved epitopes. Epitopes of SARS-CoV-2 NAbs can be classified into three types: ACE2 RBM, cryptic epitopes in the trimeric interface, and lateral surface epitopes [99], [102]. Binding of non-RBM-targeting NAbs was reported to be highly correlated with Omicron neutralizing potency, suggesting conservatism of non-RBM epitopes. Indeed, the potency of the lateral surface epitope targeting nanobody n3113v and the trimeric interface cryptic epitopes targeting nanobody n3130v were less affected by the Omicron mutations [99]. In addition, several other studies have also confirmed that inhibition of mutational escape and the broadening of neutralizing potency against variants can be achieved by nanobodies targeting conserved epitopes [61], [91], [99], [101], [103], [104], some of which are inaccessible to conventional antibodies [91], [101].
6.4. Convenient and effective methods for generating potent neutralizing nanobodies
Typically, nanobody generation entails immunizing camelids and the subsequent isolation of peripheral blood lymphocytes, from which the variable regions of nanobodies are cloned to generate a phage display immune library, which involves large animal rearing. Moreover, all the above steps will take approximately 9–15 weeks, which is time consuming [10]. Therefore, it is challenging to cope with the rapidly evolving SARS-CoV-2 variants using traditional ways of nanobodies generation. Because of the intense arms race between virus evolution and the development of effective vaccines and targeted drugs, it is crucial to develop reliable new methods for rapid screening and mass production of effective nanobodies against the evolving SARS-CoV-2 variants.
In addition to typical display-based library screening methods, proteomics has also been used to identify high-affinity nanobodies. Nanobody-containing HCAbs were purified from RBD-immunized llama plasma using protein G and protein A Sepharose beads and analyzed by mass spectrometry after proteolysis. Finally, thousands of RBD specific candidate nanobodies were identified, of which 109 nanobodies were characterized with picomolar to femtomolar affinities [105]. Similarly, Mast et al. generated a large repertoire of nanobodies against SARS-CoV-2 as well as the emerging VOCs using proteomics combined with in vivo antibody affinity maturation, that is, the LC-MS/MS results were used to search against the sequence library generated by amplifying nanobody domain sequence using lymphocyte RNA derived from bone marrow aspirates [96].
To avoid the use of large animals that are expensive and potentially involved in animal welfare, Xu et al. created a nanobody-producing mouse (referred to as nanomice) by replacing the VH domain with 30 VHH genes from alpacas, dromedary, and Bactrian camel in mouse embryonic stem cells, which were transferred into the uteri of pseudopregnant C56BL/6 recipients, followed by mating and isolation of VHH-homozygous nanomice, from which they isolated high-affinity RBD-targeting nanobodies [91]. Alternatively, synthetic nanobodies (sybodies) libraries allow for faster and cheaper animal-free isolation of nanobodies without using animals with a higher degree of control [106]. Several sybodies with high neutralization activity [63], [107], some of which are humanized, have been selected against RBD [108].
Commonly, synthetic nanobody library screening requires large library size and well-established screening methods, mainly due to the lack of in vivo affinity maturation mediated by somatic hypermutation. To overcome these limitations while maintaining the animal-free advantage, researchers reported autonomous hypermutation yeast surface display (AHEAD), a highly accessible synthetic recombinant nanobody generation technology that emulates hypermutation using yeast. The core component - an orthogonal error-prone DNA polymerase, replicated nanobody fragments containing cytosolic plasmid (p1) to produce durable continuous hypermutation of p1-encoded nanobody, and the resulting nanobodies were surface displayed on yeast for antigen binding screening. High affinity nanobodies against S protein were rapidly generated within two weeks using AHEAD [109]. Notably, this technology not only facilitates the rapid evolution of previously discovered nanobodies, but also enables isolation of brand-new nanobodies against a given antigen.
Most cell-based screening methods are often restricted by the efficiency of DNA library transformation, which is not present in cell-free approaches such as ribosome display. Chen et al. reported CeVICA, a ribosome display based cell-free nanobody engineering platform, which enables the rapid isolating diverse nanobodies with favorable biophysical properties, especially high refolding capability, probably due to the lack of factors like chaperones in the cell-free environment, which favors the selection of antigen-binding nanobodies with strong inherent folding stability [110].
To accelerate the generation of potent neutralizing nanobodies, affinity maturation can be applied based on isolated nanobodies with low or moderate affinity, primarily by error-prone PCR-mediated random mutagenesis [110], or site saturation mutagenesis generated by incorporation of degenerate base combinations [14], or a newly reported CDR-swapping mutagenesis [111] through which high-affinity nanobodies can be generated simply using common-framework, non-immune nanobody libraries. In addition, computational methods have been used to predict, refine, or design nanobodies with high efficacy based on structural information, however, there is still a long way to go [100], [104], [112], [113].
7. Concluding remarks
Nanobodies are promising for detection and treatment of SARS-CoV-2 due to their small size, high stability, cryptic epitope accessibility, and low production costs. At present, a large number of nanobodies with excellent performance have been isolated against SARS-CoV-2, and studies have shown that strategies such as targeting cryptic and conserved epitopes, simultaneous targeting different epitopes, and nanobody multimerization can enhance their efficacy and reduce mutational escape. However, to date, the vast majority of nanobodies are directed against the same target - RBD, which makes it imperative to explore the details of the SARS-CoV-2 life cycle, especially for the emerging variants, including their invasion, replication, packaging, etc., and to discover new target proteins. An excellent recent study probed into SARS-CoV-2–human protein–protein interactome network using high-throughput yeast two-hybrid and mass spectrometry, and obtained high-confidence interactions which provides potential therapeutic targets for nanobodies [114].
This paper summarizes the strategies currently used, and it is worth noting that efficacy can often be maximized by combining different strategies, including new methods and technologies that are constantly emerging. For example, proteolysis-targeting chimeric (PROTAC) technology has been reported to be used to generate a live attenuated influenza A vaccine with high efficacy [115], and PROTAC-mediated attenuation of SARS-CoV-2 and its variants may also be achieved by conserved epitopes-targeting nanobodies. All in all, although not an easy way to go, the dawn has come, and we believe the conquest of COVID-19 is near in the future.
CRediT authorship contribution statement
Wenyi Wang: Supervision, Funding acquisition, Conceptualization, Investigation, Visualization, Methodology, Writing - original draft, Writing - review & editing. Yue Hu: Investigation, Visualization, Writing - original draft. Bohan Li: Investigation, Visualization, Writing - original draft. Huanan Wang: Methodology, Writing - review & editing. Jinhua Shen: Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the Fundamental Research Funds for the Central Universities, South-Central Minzu University (No. CZQ21001), and the Scientific Research Fund from South-Central Minzu University (No. YZZ20010). In particular, Dr. Wenyi Wang is grateful to Ms. Jie Dong for her full support of this work.
References
- 1.Campbell F., et al. Increased transmissibility and global spread of SARS-CoV-2 variants of concern as at June 2021. Euro Surveill. 2021;26(24):2100509. doi: 10.2807/1560-7917.ES.2021.26.24.2100509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ma K., Chen J. Omicron XE emerges as SARS-CoV-2 keeps evolving. Innovation (Camb) 2022;3(3) doi: 10.1016/j.xinn.2022.100248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bakhshandeh B., et al. Mutations in SARS-CoV-2; Consequences in structure, function, and pathogenicity of the virus. Microb. Pathog. 2021;154 doi: 10.1016/j.micpath.2021.104831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao Z., et al. A systematic review of asymptomatic infections with COVID-19. J. Microbiol. Immunol. Infect. 2021;54(1):12–16. doi: 10.1016/j.jmii.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ballering A.V., et al. Persistence of somatic symptoms after COVID-19 in the Netherlands: an observational cohort study. Lancet. 2022;400(10350):452–461. doi: 10.1016/S0140-6736(22)01214-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang W.B., et al. E484K mutation in SARS-CoV-2 RBD enhances binding affinity with hACE2 but reduces interactions with neutralizing antibodies and nanobodies: Binding free energy calculation studies. J. Mol. Graph. Model. 2021;109 doi: 10.1016/j.jmgm.2021.108035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hong J., et al. Dromedary camel nanobodies broadly neutralize SARS-CoV-2 variants. PNAS. 2022;119(18) doi: 10.1073/pnas.2201433119. e2201433119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shrestha L.B., et al. Evolution of the SARS-CoV-2 omicron variants BA.1 to BA.5: Implications for immune escape and transmission. Rev. Med. Virol. 2022;32(5):e2381. doi: 10.1002/rmv.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamers-Casterman C., et al. Naturally occurring antibodies devoid of light chains. Nature. 1993;363(6428):446–448. doi: 10.1038/363446a0. [DOI] [PubMed] [Google Scholar]
- 10.Muyldermans S. Applications of Nanobodies. Annu. Rev. Anim. Biosci. 2021;9:401–421. doi: 10.1146/annurev-animal-021419-083831. [DOI] [PubMed] [Google Scholar]
- 11.Najmeddin A., et al. Nanobodies as powerful pulmonary targeted biotherapeutics against SARS-CoV-2, pharmaceutical point of view. Biochim. Biophys. Acta Gen. Subj. 2021;1865(11) doi: 10.1016/j.bbagen.2021.129974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Heeke G., et al. Nanobodies® as inhaled biotherapeutics for lung diseases. Pharmacol. Ther. 2017;169:47–56. doi: 10.1016/j.pharmthera.2016.06.012. [DOI] [PubMed] [Google Scholar]
- 13.Yang Z., et al. Inhalable antibodies for the treatment of COVID-19. Innovation (Camb) 2022;3(6) doi: 10.1016/j.xinn.2022.100328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schoof M., et al. An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike. Science. 2020;370(6523):1473–1479. doi: 10.1126/science.abe3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu F., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bai C., Zhong Q., Gao G.F. Overview of SARS-CoV-2 genome-encoded proteins. Sci. China Life Sci. 2022;65(2):280–294. doi: 10.1007/s11427-021-1964-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jackson C.B., et al. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022;23(1):3–20. doi: 10.1038/s41580-021-00418-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peng R., et al. Cell entry by SARS-CoV-2. Trends Biochem. Sci. 2021;46(10):848–860. doi: 10.1016/j.tibs.2021.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walls A.C., et al. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181(2):281–292 e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lv Z., et al. Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody. Science. 2020;369(6510):1505–1509. doi: 10.1126/science.abc5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wrapp D., et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Labroussaa F., Jores J. SARS-CoV-2 nanobodies 2.0. Signal Transduct. Target. Ther. 2021;6(1):202. doi: 10.1038/s41392-021-00632-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan R., et al. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huo J., et al. Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2. Nat. Struct. Mol. Biol. 2020;27(9):846–854. doi: 10.1038/s41594-020-0469-6. [DOI] [PubMed] [Google Scholar]
- 25.Benton D.J., et al. Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion. Nature. 2020;588(7837):327–330. doi: 10.1038/s41586-020-2772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koch J., et al. TMPRSS2 expression dictates the entry route used by SARS-CoV-2 to infect host cells. EMBO J. 2021;40(16):e107821. doi: 10.15252/embj.2021107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fan Y., et al. SARS-CoV-2 Omicron variant: recent progress and future perspectives. Signal Transduct. Target. Ther. 2022;7(1):141. doi: 10.1038/s41392-022-00997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bestle D., et al. TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life Sci. Alliance. 2020;3(9) doi: 10.26508/lsa.202000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou P., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Q., et al. Structural Basis for RNA Replication by the SARS-CoV-2 Polymerase. Cell. 2020;182(2):417–428 e13. doi: 10.1016/j.cell.2020.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shang J., et al. Cell entry mechanisms of SARS-CoV-2. PNAS. 2020;117(21):11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoffmann M., Kleine-Weber H., Pohlmann S., A, Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 Is Essential for Infection of Human Lung Cells. Mol. Cell. 2020;78(4):779–784 e5. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shereen M.A., et al. COVID-19 infection: Origin, transmission, and characteristics of human coronaviruses. J. Adv. Res. 2020;24:91–98. doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ji S., et al. Increasing contributions of airborne route in SARS-CoV-2 omicron variant transmission compared with the ancestral strain. Build. Environ. 2022;221 doi: 10.1016/j.buildenv.2022.109328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oran D.P., Topol E.J. Prevalence of Asymptomatic SARS-CoV-2 Infection: A Narrative Review. Ann. Intern. Med. 2020;173(5):362–367. doi: 10.7326/M20-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vogels C.B.F., et al. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT-qPCR primer-probe sets. Nat. Microbiol. 2020;5(10):1299–1305. doi: 10.1038/s41564-020-0761-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ganguli A., et al. Rapid isothermal amplification and portable detection system for SARS-CoV-2. PNAS. 2020;117(37):22727–22735. doi: 10.1073/pnas.2014739117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ferretti L., et al. Quantifying SARS-CoV-2 transmission suggests epidemic control with digital contact tracing. Science. 2020;368(6491):eabb6936. doi: 10.1126/science.abb6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamayoshi S., et al. Comparison of Rapid Antigen Tests for COVID-19. Viruses. 2020;12(12):1420. doi: 10.3390/v12121420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tollånes M.C., et al. Evaluation of 32 rapid tests for detection of antibodies against SARS-CoV-2. Clin. Chim. Acta. 2021;519:133–139. doi: 10.1016/j.cca.2021.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saerens D., et al. Engineering camel single-domain antibodies and immobilization chemistry for human prostate-specific antigen sensing. Anal. Chem. 2005;77(23):7547–7555. doi: 10.1021/ac051092j. [DOI] [PubMed] [Google Scholar]
- 42.Wang W.U., et al. Label-free detection of small-molecule-protein interactions by using nanowire nanosensors. PNAS. 2005;102(9):3208–3212. doi: 10.1073/pnas.0406368102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee M.Y., et al. Organic Transistor-Based Chemical Sensors for Wearable Bioelectronics. Acc. Chem. Res. 2018;51(11):2829–2838. doi: 10.1021/acs.accounts.8b00465. [DOI] [PubMed] [Google Scholar]
- 44.Chen Y., et al. Nanobody-based label-free photoelectrochemical immunoassay for highly sensitive detection of SARS-CoV-2 spike protein. Anal. Chim. Acta. 2022;1211 doi: 10.1016/j.aca.2022.339904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen X., et al. Synthetic nanobody-functionalized nanoparticles for accelerated development of rapid, accessible detection of viral antigens. Biosens. Bioelectron. 2022;202 doi: 10.1016/j.bios.2022.113971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo K., et al. Rapid single-molecule detection of COVID-19 and MERS antigens via nanobody-functionalized organic electrochemical transistors. Nat. Biomed. Eng. 2021;5(7):666–677. doi: 10.1038/s41551-021-00734-9. [DOI] [PubMed] [Google Scholar]
- 47.Pagneux Q., et al. SARS-CoV-2 detection using a nanobody-functionalized voltammetric device. Commun. Med. (Lond.) 2022;2:56. doi: 10.1038/s43856-022-00113-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koklu A., et al. Convection Driven Ultrarapid Protein Detection via Nanobody-Functionalized Organic Electrochemical Transistors. Adv. Mater. 2022;34(35):e2202972. doi: 10.1002/adma.202202972. [DOI] [PubMed] [Google Scholar]
- 49.Sun X., et al. Nanobody-Functionalized Cellulose for Capturing SARS-CoV-2. Appl. Environ. Microbiol. 2022;88(5):e0230321. doi: 10.1128/aem.02303-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anderson G.P., et al. Single-Domain Antibodies for the Detection of SARS-CoV-2 Nucleocapsid Protein. Anal. Chem. 2021;93(19):7283–7291. doi: 10.1021/acs.analchem.1c00677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gransagne M., et al. Development of a highly specific and sensitive VHH-based sandwich immunoassay for the detection of the SARS-CoV-2 nucleoprotein. J. Biol. Chem. 2022;298(1) doi: 10.1016/j.jbc.2021.101290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gouveia D., et al. Proteotyping SARS-CoV-2 Virus from Nasopharyngeal Swabs: A Proof-of-Concept Focused on a 3 Min Mass Spectrometry Window. J. Proteome Res. 2020;19(11):4407–4416. doi: 10.1021/acs.jproteome.0c00535. [DOI] [PubMed] [Google Scholar]
- 53.Nikolaev E.N., et al. Mass-Spectrometric Detection of SARS-CoV-2 Virus in Scrapings of the Epithelium of the Nasopharynx of Infected Patients via Nucleocapsid N Protein. J. Proteome Res. 2020;19(11):4393–4397. doi: 10.1021/acs.jproteome.0c00412. [DOI] [PubMed] [Google Scholar]
- 54.Isaacs A., et al. Nucleocapsid Specific Diagnostics for the Detection of Divergent SARS-CoV-2 Variants. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.926262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Banga Ndzouboukou J.L., Zhang Y.D., Fan X.L. Recent Developments in SARS-CoV-2 Neutralizing Antibody Detection Methods. Curr Med Sci. 2021;41(6):1052–1064. doi: 10.1007/s11596-021-2470-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wagner T.R., et al. NeutrobodyPlex-monitoring SARS-CoV-2 neutralizing immune responses using nanobodies. EMBO Rep. 2021;22(5):e52325. doi: 10.15252/embr.202052325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Redecke V., et al. A rapid and affordable point of care test for antibodies against SARS-CoV-2 based on hemagglutination and artificial intelligence interpretation. Sci. Rep. 2021;11(1):24507. doi: 10.1038/s41598-021-04298-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pishesha N., et al. A class II MHC-targeted vaccine elicits immunity against SARS-CoV-2 and its variants. PNAS. 2021;118(44) doi: 10.1073/pnas.2116147118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lyu X., et al. Preventing SARS-CoV-2 Infection Using Anti-spike Nanobody-IFN-beta Conjugated Exosomes. Pharm. Res. 2022:1–9. doi: 10.1007/s11095-022-03400-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Taylor P.C., et al. Neutralizing monoclonal antibodies for treatment of COVID-19. Nat. Rev. Immunol. 2021;21(6):382–393. doi: 10.1038/s41577-021-00542-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shi Z., et al. Structural basis of nanobodies neutralizing SARS-CoV-2 variants. Structure. 2022;30(5):707–720 e5. doi: 10.1016/j.str.2022.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hanke L., et al. An alpaca nanobody neutralizes SARS-CoV-2 by blocking receptor interaction. Nat Commun. 2020;11(1):4420. doi: 10.1038/s41467-020-18174-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Custódio T.F., et al. Selection, biophysical and structural analysis of synthetic nanobodies that effectively neutralize SARS-CoV-2. Nat. Commun. 2020;11(1):5588. doi: 10.1038/s41467-020-19204-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maeda R., et al. A panel of nanobodies recognizing conserved hidden clefts of all SARS-CoV-2 spike variants including Omicron. Commun Biol. 2022;5(1):669. doi: 10.1038/s42003-022-03630-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huo J., et al. A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19. Nat. Commun. 2021;12(1):5469. doi: 10.1038/s41467-021-25480-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li T., et al. Structural Characterization of a Neutralizing Nanobody With Broad Activity Against SARS-CoV-2 Variants. Front. Microbiol. 2022;13 doi: 10.3389/fmicb.2022.875840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Haga K., et al. Nasal delivery of single-domain antibody improves symptoms of SARS-CoV-2 infection in an animal model. PLoS Pathog. 2021;17(10):e1009542. doi: 10.1371/journal.ppat.1009542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang Z., et al. A non-ACE2 competing human single-domain antibody confers broad neutralization against SARS-CoV-2 and circulating variants. Signal Transduct. Target. Ther. 2021;6(1):378. doi: 10.1038/s41392-021-00810-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Walter J.D., et al. Biparatopic sybodies neutralize SARS-CoV-2 variants of concern and mitigate drug resistance. EMBO Rep. 2022;23(4):e54199. doi: 10.15252/embr.202154199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Koenig P.A., et al. Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape. Science. 2021;371(6530):eabe6230. doi: 10.1126/science.abe6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yan L., et al. Cryo-EM Structure of an Extended SARS-CoV-2 Replication and Transcription Complex Reveals an Intermediate State in Cap Synthesis. Cell. 2021;184(1):184–193 e10. doi: 10.1016/j.cell.2020.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yan L., et al. A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors. Cell. 2022;185(23):4347–4360 e17. doi: 10.1016/j.cell.2022.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Armstrong L.A., et al. Biochemical characterization of protease activity of Nsp3 from SARS-CoV-2 and its inhibition by nanobodies. PLoS One. 2021;16(7):e0253364. doi: 10.1371/journal.pone.0253364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chouchane L., et al. Dromedary camels as a natural source of neutralizing nanobodies against SARS-CoV-2. JCI Insight. 2021;6(5) doi: 10.1172/jci.insight.145785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Esposito G., et al. NMR-Based Analysis of Nanobodies to SARS-CoV-2 Nsp9 Reveals a Possible Antiviral Strategy Against COVID-19. Adv Biol (Weinh) 2021;5(12):e2101113. doi: 10.1002/adbi.202101113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sun Z., et al. An extended conformation of SARS-CoV-2 main protease reveals allosteric targets. PNAS. 2022;119(15) doi: 10.1073/pnas.2120913119. e2120913119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lamers M.M., Haagmans B.L. SARS-CoV-2 pathogenesis. Nat. Rev. Microbiol. 2022;20(5):270–284. doi: 10.1038/s41579-022-00713-0. [DOI] [PubMed] [Google Scholar]
- 78.Lu Q., et al. SARS-CoV-2 exacerbates proinflammatory responses in myeloid cells through C-type lectin receptors and Tweety family member 2. Immunity. 2021;54(6):1304–1319 e9. doi: 10.1016/j.immuni.2021.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ettich J., et al. A Hybrid Soluble gp130/Spike-Nanobody Fusion Protein Simultaneously Blocks Interleukin-6 trans-Signaling and Cellular Infection with SARS-CoV-2. J. Virol. 2022;96(4):e0162221. doi: 10.1128/jvi.01622-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Moya A., et al. The evolution of RNA viruses: A population genetics view. PNAS. 2000;97(13):6967–6973. doi: 10.1073/pnas.97.13.6967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Romano M., et al. A Structural View of SARS-CoV-2 RNA Replication Machinery: RNA Synthesis, Proofreading and Final Capping. Cells. 2020;9(5):1267. doi: 10.3390/cells9051267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.van Dorp L., et al. No evidence for increased transmissibility from recurrent mutations in SARS-CoV-2. Nat. Commun. 2020;11(1):5986. doi: 10.1038/s41467-020-19818-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang P., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593(7857):130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 84.Cao Y., et al. Imprinted SARS-CoV-2 humoral immunity induces convergent Omicron RBD evolution. Nature. 2022 doi: 10.1038/s41586-022-05644-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Han P., et al. Receptor binding and complex structures of human ACE2 to spike RBD from omicron and delta SARS-CoV-2. Cell. 2022;185(4):630–640 e10. doi: 10.1016/j.cell.2022.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Harvey W.T., et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19(7):409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Souza P.F.N., et al. The spike glycoprotein of SARS-CoV-2: A review of how mutations of spike glycoproteins have driven the emergence of variants with high transmissibility and immune escape. Int. J. Biol. Macromol. 2022;208:105–125. doi: 10.1016/j.ijbiomac.2022.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Qu P., et al. bioRxiv; 2022. Distinct Neutralizing Antibody Escape of SARS-CoV-2 Omicron Subvariants BQ.1, BQ.1.1, BA.4.6, BF.7 and BA.2.75.2. [Google Scholar]
- 89.Miller N.L., et al. Insights on the mutational landscape of the SARS-CoV-2 Omicron variant receptor-binding domain. Cell Rep. Med. 2022;3(2) doi: 10.1016/j.xcrm.2022.100527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cao Y., et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. 2022;602(7898):657–663. doi: 10.1038/s41586-021-04385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xu J., et al. Nanobodies from camelid mice and llamas neutralize SARS-CoV-2 variants. Nature. 2021;595(7866):278–282. doi: 10.1038/s41586-021-03676-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rossotti M.A., et al. Arsenal of nanobodies shows broad-spectrum neutralization against SARS-CoV-2 variants of concern in vitro and in vivo in hamster models. Commun. Biol. 2022;5(1):933. doi: 10.1038/s42003-022-03866-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li Q., et al. Mucosal nanobody IgA as inhalable and affordable prophylactic and therapeutic treatment against SARS-CoV-2 and emerging variants. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.995412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yu B., et al. Accelerating PERx reaction enables covalent nanobodies for potent neutralization of SARS-CoV-2 and variants. Chem. 2022;8(10):2766–2783. doi: 10.1016/j.chempr.2022.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ma H., et al. Hetero-bivalent nanobodies provide broad-spectrum protection against SARS-CoV-2 variants of concern including Omicron. Cell Res. 2022;32(9):831–842. doi: 10.1038/s41422-022-00700-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mast F.D., et al. Highly synergistic combinations of nanobodies that target SARS-CoV-2 and are resistant to escape. Elife. 2021;10 doi: 10.7554/eLife.73027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Taylor R.J., et al. π-Clamp-Mediated Homo- and Heterodimerization of Single-Domain Antibodies via Site-Specific Homobifunctional Conjugation. J. Am. Chem. Soc. 2022;144(29):13026–13031. doi: 10.1021/jacs.2c04747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Moliner-Morro A., et al. Picomolar SARS-CoV-2 Neutralization Using Multi-Arm PEG Nanobody Constructs. Biomolecules. 2020;10(12):1661. doi: 10.3390/biom10121661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li C., et al. Broad neutralization of SARS-CoV-2 variants by an inhalable bispecific single-domain antibody. Cell. 2022;185(8):1389–1401 e18. doi: 10.1016/j.cell.2022.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dong J., et al. Development of humanized tri-specific nanobodies with potent neutralization for SARS-CoV-2. Sci. Rep. 2020;10(1):17806. doi: 10.1038/s41598-020-74761-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sun D., et al. Potent neutralizing nanobodies resist convergent circulating variants of SARS-CoV-2 by targeting diverse and conserved epitopes. Nat. Commun. 2021;12(1):4676. doi: 10.1038/s41467-021-24963-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hastie K.M., et al. Defining variant-resistant epitopes targeted by SARS-CoV-2 antibodies: A global consortium study. Science. 2021;374(6566):472–478. doi: 10.1126/science.abh2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zupancic J.M., et al. Engineered Multivalent Nanobodies Potently and Broadly Neutralize SARS-CoV-2 Variants. Adv. Ther. (Weinh) 2021;4(8):2100099. doi: 10.1002/adtp.202100099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Schepens B., et al. An affinity-enhanced, broadly neutralizing heavy chain-only antibody protects against SARS-CoV-2 infection in animal models. Sci. Transl. Med. 2021;13(621):eabi7826. doi: 10.1126/scitranslmed.abi7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Xiang Y., et al. Versatile and multivalent nanobodies efficiently neutralize SARS-CoV-2. Science. 2020;370(6523):1479–1484. doi: 10.1126/science.abe4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zimmermann I., et al. Generation of synthetic nanobodies against delicate proteins. Nat. Protoc. 2020;15(5):1707–1741. doi: 10.1038/s41596-020-0304-x. [DOI] [PubMed] [Google Scholar]
- 107.Li T., et al. A synthetic nanobody targeting RBD protects hamsters from SARS-CoV-2 infection. Nat. Commun. 2021;12(1):4635. doi: 10.1038/s41467-021-24905-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chi X., et al. Humanized single domain antibodies neutralize SARS-CoV-2 by targeting the spike receptor binding domain. Nat. Commun. 2020;11(1):4528. doi: 10.1038/s41467-020-18387-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wellner A., et al. Rapid generation of potent antibodies by autonomous hypermutation in yeast. Nat. Chem. Biol. 2021;17(10):1057–1064. doi: 10.1038/s41589-021-00832-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chen X., et al. A cell-free nanobody engineering platform rapidly generates SARS-CoV-2 neutralizing nanobodies. Nat. Commun. 2021;12(1):5506. doi: 10.1038/s41467-021-25777-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zupancic J.M., et al. Directed evolution of potent neutralizing nanobodies against SARS-CoV-2 using CDR-swapping mutagenesis. Cell Chem. Biol. 2021;28(9):1379–1388 e7. doi: 10.1016/j.chembiol.2021.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dong J., et al. Development of multi-specific humanized llama antibodies blocking SARS-CoV-2/ACE2 interaction with high affinity and avidity. Emerg. Microbes. Infect. 2020;9(1):1034–1036. doi: 10.1080/22221751.2020.1768806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yang J., et al. Computational design and modeling of nanobodies toward SARS-CoV-2 receptor binding domain. Chem. Biol. Drug Des. 2021;98(1):1–18. doi: 10.1111/cbdd.13847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhou Y., et al. A comprehensive SARS-CoV-2-human protein-protein interactome reveals COVID-19 pathobiology and potential host therapeutic targets. Nat. Biotechnol. 2022 doi: 10.1038/s41587-022-01474-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Si L., et al. Generation of a live attenuated influenza A vaccine by proteolysis targeting. Nat. Biotechnol. 2022;40(9):1370–1377. doi: 10.1038/s41587-022-01381-4. [DOI] [PubMed] [Google Scholar]
- 116.Girt G.C., et al. The use of nanobodies in a sensitive ELISA test for SARS-CoV-2 Spike 1 protein. R. Soc. Open Sci. 2021;8(9) doi: 10.1098/rsos.211016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kamel M., et al. Non-Invasive Detection of SARS-CoV-2 Antigen in Saliva versus Nasopharyngeal Swabs Using Nanobodies Conjugated Gold Nanoparticles. Trop. Med. Infect. Dis. 2022;7(6):102. doi: 10.3390/tropicalmed7060102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wrapp D., et al. Structural Basis for Potent Neutralization of Betacoronaviruses by Single-Domain Camelid Antibodies. Cell. 2020;181(5):1004–1015 e15. doi: 10.1016/j.cell.2020.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wu X., et al. A potent bispecific nanobody protects hACE2 mice against SARS-CoV-2 infection via intranasal administration. Cell Rep. 2021;37(3) doi: 10.1016/j.celrep.2021.109869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Weinstein J.B., et al. A potent alpaca-derived nanobody that neutralizes SARS-CoV-2 variants. iScience. 2022;25(3) doi: 10.1016/j.isci.2022.103960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Scott T.A., et al. Engineered extracellular vesicles directed to the spike protein inhibit SARS-CoV-2. Mol. Ther. Methods Clin. Dev. 2022;24:355–366. doi: 10.1016/j.omtm.2022.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Titong A., et al. First-in-class trispecific VHH-Fc based antibody with potent prophylactic and therapeutic efficacy against SARS-CoV-2 and variants. Sci. Rep. 2022;12(1):4163. doi: 10.1038/s41598-022-07952-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wu Y., et al. Identification of Human Single-Domain Antibodies against SARS-CoV-2. Cell Host Microbe. 2020;27(6):891–898 e5. doi: 10.1016/j.chom.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Fu Y., et al. A humanized nanobody phage display library yields potent binders of SARS CoV-2 spike. PLoS One. 2022;17(8):e0272364. doi: 10.1371/journal.pone.0272364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Favorskaya I.A., et al. Single-Domain Antibodies Efficiently Neutralize SARS-CoV-2 Variants of Concern. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.822159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Esparza T.J., et al. Nebulized delivery of a broadly neutralizing SARS-CoV-2 RBD-specific nanobody prevents clinical, virological, and pathological disease in a Syrian hamster model of COVID-19. MAbs. 2022;14(1):2047144. doi: 10.1080/19420862.2022.2047144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Esparza T.J., et al. High affinity nanobodies block SARS-CoV-2 spike receptor binding domain interaction with human angiotensin converting enzyme. Sci. Rep. 2020;10(1):22370. doi: 10.1038/s41598-020-79036-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chi X., et al. An ultrapotent RBD-targeted biparatopic nanobody neutralizes broad SARS-CoV-2 variants. Signal Transduct. Target. Ther. 2022;7(1):44. doi: 10.1038/s41392-022-00912-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Casasnovas J.M., et al. Nanobodies Protecting From Lethal SARS-CoV-2 Infection Target Receptor Binding Epitopes Preserved in Virus Variants Other Than Omicron. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.863831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Valenzuela N.G., et al. Potent neutralization of clinical isolates of SARS-CoV-2 D614 and G614 variants by a monomeric, sub-nanomolar affinity nanobody. Sci. Rep. 2021;11(1):3318. doi: 10.1038/s41598-021-82833-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Pymm P., et al. Nanobody cocktails potently neutralize SARS-CoV-2 D614G N501Y variant and protect mice. PNAS. 2021;118(19) doi: 10.1073/pnas.2101918118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Dormeshkin D., et al. Isolation of an escape-resistant SARS-CoV-2 neutralizing nanobody from a novel synthetic nanobody library. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.965446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhao D., et al. A potent synthetic nanobody with broad-spectrum activity neutralizes SARS-CoV-2 virus and the Omicron variant BA.1 through a unique binding mode. J. Nanobiotechnol. 2022;20(1):411. doi: 10.1186/s12951-022-01619-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yang J., et al. A Potent Neutralizing Nanobody Targeting the Spike Receptor-Binding Domain of SARS-CoV-2 and the Structural Basis of Its Intimate Binding. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.820336. [DOI] [PMC free article] [PubMed] [Google Scholar]