Key Points
We developed a model for automated kidney and liver volumetry in ADPKD to provide assistance with time-consuming volumetry.
The model works in both coronal and axial planes and was tested in the real-life setting using large multicentric cohorts.
The trained model is published along with the code to allow for further joint development and integration into commercial software packages.
Keywords: Cystic kidney disease, ADPKD, automated, deep neural network, kidney, liver, polycystic kidney disease, segmentation, volumetry
Visual Abstract
Abstract
Background
Imaging-based total kidney volume (TKV) and total liver volume (TLV) are major prognostic factors in autosomal dominant polycystic kidney disease (ADPKD) and end points for clinical trials. However, volumetry is time consuming and reader dependent in clinical practice. Our aim was to develop a fully automated method for joint kidney and liver segmentation in magnetic resonance imaging (MRI) and to evaluate its performance in a multisequence, multicenter setting.
Methods
The convolutional neural network was trained on a large multicenter dataset consisting of 992 MRI scans of 327 patients. Manual segmentation delivered ground-truth labels. The model’s performance was evaluated in a separate test dataset of 93 patients (350 MRI scans) as well as a heterogeneous external dataset of 831 MRI scans from 323 patients.
Results
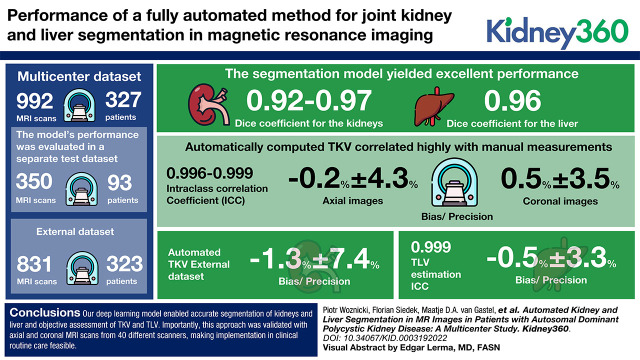
The segmentation model yielded excellent performance, achieving a median per study Dice coefficient of 0.92–0.97 for the kidneys and 0.96 for the liver. Automatically computed TKV correlated highly with manual measurements (intraclass correlation coefficient [ICC]: 0.996–0.999) with low bias and high precision (−0.2%±4% for axial images and 0.5%±4% for coronal images). TLV estimation showed an ICC of 0.999 and bias/precision of −0.5%±3%. For the external dataset, the automated TKV demonstrated bias and precision of −1%±7%.
Conclusions
Our deep learning model enabled accurate segmentation of kidneys and liver and objective assessment of TKV and TLV. Importantly, this approach was validated with axial and coronal MRI scans from 40 different scanners, making implementation in clinical routine care feasible.
Clinical Trial registry name and registration number: The German ADPKD Tolvaptan Treatment Registry (AD[H]PKD), NCT02497521
Introduction
Autosomal dominant polycystic kidney disease (ADPKD) is the most common hereditary kidney disorder, with a genetic prevalence of approximately one in 1000 (1). It is characterized by the development of multiple cysts in the kidneys, which progressively impair kidney function, leading to kidney failure in up to 75% of patients by the age of 70 (2). In parallel to the kidney function decline, ADPKD is characterized by a progressive increase in kidney volume over time.
Total kidney volume (TKV) has become an important biomarker for assessing disease severity and can be used to predict future loss of the eGFR (3,4). Assessing the risk of future kidney failure is especially important at early disease stages when renal function might not be impaired yet (5) and can be achieved using the Mayo Imaging Classification (6) on the basis of height-adjusted TKV measurement. This tool identifies patients with a risk of rapid progression for clinical trials and selects patients for treatment with disease-modifying drugs. Importantly, TKV increase over time has been widely accepted as a surrogate end point for the evaluation of the efficacy of new pharmacologic therapies (7,8).
Polycystic liver disease is the most common extrarenal manifestation of ADPKD. Liver cysts are typically present in early disease stages and become increasingly common in advanced stages, with a relative loss of liver parenchyma. Liver function is usually preserved, but an increase in total liver volume (TLV) is known to be associated with a lower quality of life (9).
The current gold standard for measuring TKV and TLV relies on manual tracing of the organ boundaries. However, this approach requires a lot of time and is subject to intra- and inter-reader variability (10). Recently, automated and semiautomated approaches to kidney volumetry (11–14) as well as joint kidney and liver volumetry (15,16) have gained increasing attention and may provide a future solution to this problem. Their practical utility is limited by the unknown generalizability of the models across imaging data from different scanner manufacturers, which is a recognized problem, especially for magnetic resonance imaging (MRI) data (17,18).
Convolutional neural networks have been responsible for recent phenomenal advances in tasks like object classification and image segmentation. U-Net (19) is arguably the most successful deep learning architecture for semantic segmentation, and its variants are currently the state of the art for biomedical image segmentation (F. Isensee et al., unpublished data) (20,21).
The aim of this study was to develop a deep neural network for fully automated volumetry of kidneys and liver and to validate it in a large longitudinal cohort as well as assess its generalizability in a multicenter, multiscanner, multisequence setting.
Materials and Methods
Study Design
We analyzed MRI data from patients with ADPKD participating in two different prospective studies: the German ADPKD Tolvaptan Treatment Registry, University of Cologne (UoC) and the Developing Intervention Strategies to Halt Progression of Autosomal Dominant Polycystic Kidney Disease (DIPAK1) study, a Dutch collaborative (8). MRI scans of these patients were obtained in multiple centers in Germany and The Netherlands. Both studies were approved by the respective local institutional review boards (the Ethics Committee of the Medical Faculty UoC and the medical ethics committees of all involved Dutch institutes). The DIPAK1 study, a randomized, controlled trial, studied whether lanreotide could ameliorate the rate of disease progression in patients with rapidly progressive ADPKD. Patients were randomized 1:1 for lanreotide or placebo treatment, which affected the natural progression of the disease, by means of kidney and liver volume growth.
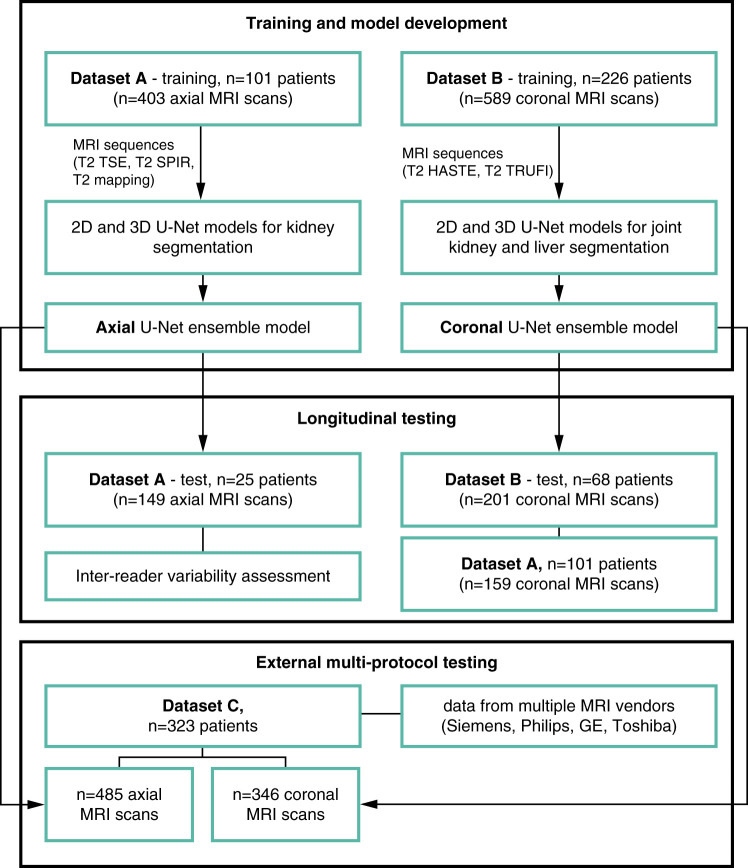
A flowchart of the procedures for patient stratification, model development, and validation is shown in Figure 1. We trained a model for axial magnetic resonance (MR) images using data from UoC (dataset A) and a model for coronal MR images with data from the DIPAK1 study (dataset B). Each model was trained on a subset of the dataset and tested on a separate subset, including longitudinal data. The models were externally validated on dataset C, which contained MRI data from numerous outpatient centers in Germany. The coronal model was additionally validated on dataset A.
Figure 1.
Flowchart of the procedures for model development and testing. 2D, two dimensional; 3D, three dimensional; HASTE, half-Fourier–acquired single-shot turbo spin echo; MRI, magnetic resonance imaging; SPIR, spectral presaturation with inversion recovery; T2, T2-weighted sequence; TRUFI, coronal T2 true fast imaging with steady-state free precession; TSE, turbo spin echo.
MRI Data
The study workflow is depicted in Figure 1. A total of 126 patients with ADPKD with varying levels of disease severity who underwent abdominal MRI in UoC between October 2015 and May 2017 were included in dataset A. A total of 71 patients had additional longitudinal data from a follow-up examination performed approximately 1 year after the baseline examination (median of 373 days; range, 297–600 days). Exclusion criteria were (1) unavailability of imaging data, (2) severe imaging artifacts, (3) other diagnoses than ADPKD, and (4) unavailability of clinical information. MR images were acquired with a 1.5-Tesla scanner (Ingenia 1.5T; Philips Healthcare, Best, The Netherlands) following a standardized scan protocol described in Supplemental Table 1A. Four sequences, including axial T2 TSE, T2 spectral presaturation with inversion recovery (SPIR), and T2 mapping as well as coronal T2 TSE, were included in the analysis. A detailed flowchart of inclusion and exclusion criteria for datasets A and C is shown in Supplemental Appendix 2.
MRI data from 294 patients with ADPKD and a high likelihood of disease progression enrolled in a randomized, multicenter, controlled clinical trial (DIPAK1 [8,22]) were included in our study as dataset B. MRI data were acquired at baseline and at two follow-ups (after 120 and 132 weeks) for longitudinal analysis. This dataset comprised data from 260 patients at baseline, 250 patients at the first follow-up, and 239 patients at the second follow-up. Two sequences were included: coronal half-Fourier–acquired single-shot turbo spin echo (HASTE) and coronal T2 true fast imaging with steady-state free precession (TRUFI). The MRI protocols as well as inclusion and exclusion criteria have been published previously (10) and are additionally described in Supplemental Table 1B.
Additionally, 323 patients with ADPKD treated at UoC who had MRI examinations performed externally in different radiology outpatient centers between January 2016 and March 2020 were used for external validation (dataset C). Patients had data from 40 different MRI scanner models and vendors acquired with a wide variety of image acquisition protocols. We decided to include all T2-weighted images. Acquisition parameters of the most common sequences included are presented in Supplemental Table 1C, and inclusion/exclusion criteria are included in Supplemental Appendix 2.
Manual Segmentation and Preprocessing
In dataset A, kidney boundaries were manually traced in axial T2 SPIR scans by an experienced board-certified radiologist using 3D-Slicer (23). Liver volume was not routinely measured in dataset A; however, a subset of 29 patients had TLV measurements in coronal T2-weighted images performed using manual tracing in 3D-Slicer. These patients as well as TKV test cases from dataset A were additionally segmented by a second reader to estimate inter-reader variability in TLV and TKV, respectively. In dataset B, the kidney and liver boundaries were manually traced in coronal images using Analyze (version 11.0; Biomedical Imaging Resource, Mayo Foundation, Rochester, NY). In dataset C, kidney boundaries were manually traced by a radiologist using Intellispace Discovery (Philips Healthcare). In each cohort, the readers were blinded to patient information as well as previous tracing, and the renal hilum for TKV and the gall bladder and main portal vein for TLV were excluded from the organ outline.
For training, each individual MR image was cropped to the region of nonzero values, and z-score normalization was applied. The median image voxel size was 1.37 mm in plane with a median slice thickness of 4 mm for training images in dataset A and 0.84 mm in plane with a median slice thickness of 5 mm for training images in dataset B. Because of relatively large anisotropy, we trained separate models for axial and coronal MRI scans and resampled the images to the median voxel spacing of their respective planes.
Training Procedure
We experimented with models on the basis of two-dimensional (2D) and three-dimensional (3D) U-Net networks and used an ensemble of them in our final model. Every single network had an encoder-decoder architecture with skip connections to effectively aggregate semantic information while retaining high-resolution spatial information. The models were trained using five-fold crossvalidation using the nnU-Net framework (F. Isensee et al., unpublished data), which is the current state of the art in abdominal organ segmentation. The models used instance normalization. Because of GPU memory limitations, training and inference were performed on patches, which were subsequently aggregated to produce the final prediction. The training procedure included the use of the Adam optimizer with an initial learning rate of 3 × 10−4 and early stopping. The loss function was a combination of Dice and crossentropy loss, as proposed by Isensee et al. (F. Isensee et al., unpublished data). The whole training took 8 days on a machine with two Nvidia Tesla V100 16-GB GPUs. At inference, image patches were chosen to overlap half of the patch size and were mirrored along the x and y axes. The final model was an ensemble of 2D and 3D U-Net architectures using the five networks trained in five-fold crossvalidation. Kidney and liver volumes were calculated from the output segmentation.
A large variety of 2D data augmentation techniques were used to prevent overfitting, including mirroring, random rotation, random scaling, and random elastic deformations. The augmentations were applied slice-wise in plane for each sample during training for 2D and 3D U-Nets. Additional details on the training and experiments can be found in Supplemental Appendix 3, including selected training hyperparameters in Supplemental Table 3. The segmentation model and code can be found online at https://github.com/pwoznicki/ADPKD.
Statistical Analyses and Evaluation
Baseline patient characteristics are reported as mean ± SD for normal distributions and median (interquartile range) for skewed distributions. Data were tested for normality using the Shapiro–Wilk test. Spatial overlap between predictions and reference outlines was assessed using the Dice coefficient, the Jaccard index, sensitivity, precision, the Hausdorff distance, and mean surface distance, reported as median and 10–90 percentile range. The segmentations were evaluated separately for each MRI sequence as well as for each organ (right kidney, left kidney, and liver). The P values for the statistical difference between our model and the human reader were computed using the paired t test for normally distributed data and the Wilcoxon signed-rank test for non-normally distributed data. Agreement between TKV and TLV obtained from automated segmentation and manual tracing was assessed using linear regression and Bland–Altman analyses. Both actual and percentage differences were evaluated. We compared the Mayo risk classes (6) calculated with the automated method with the reference method. Statistical analyses were implemented in Python (version 3.7.9; Python Software Foundation, Wilmington, DE).
Results
Cohort Characteristics
Characteristics of the analyzed study cohorts are described in Table 1. Dataset A consisted of 126 patients with a mean age of 43±12 years and a mean eGFR of 73±28 ml/min per 1.73 m2. A total of 294 patients with a mean age of 48±7 years and a mean eGFR of 52±12 ml/min per 1.73 m2 represented dataset B. Dataset C included 323 patients with a mean age of 44±13 years and a mean eGFR of 67±32 ml/min per 1.73 m2. Supplemental Table 4 describes detailed characteristics for training and test splits of datasets A and B.
Table 1.
Patient cohort baseline characteristics
| Parameter | Dataset Aa | Dataset Ba | Dataset C |
|---|---|---|---|
| Patients in total | 126 | 294 | 323 |
| Training | 101 | 226 | 0 |
| Test | 25 | 68 | 323 |
| Patients with a single examination | 56 | 30 | 323 |
| Patients with one follow-up examination | 71 | 73 | 0 |
| Patients with two follow-up examinations | 0 | 191 | 0 |
| Age, yr | 43.1±12.2 | 48.3±7.4 | 44.3±13.1 |
| Men | 55 (45%) | 140 (48%) | 134 (41%) |
| Women | 71 (55%) | 154 (52%) | 192 (59%) |
| Height, m | 1.75±0.11 | 1.77±0.10 | 1.74±0.09 |
| Weight, kg | 83.5±19.9 | 84.1±16.7 | 80.1±17.4 |
| BMI, kg/m2 | 27.1±5.4 | 26.9±4.6 | 26.2±5.0 |
| eGFR, CKD-EPI, ml/min per 1.73 m2 | 72.8±28.3 | 51.6±11.6 | 66.8±31.6 |
| Mayo Risk class | |||
| 1A and 1B | 37 (29%) | 60 (20%) | 81 (25%) |
| 1C–1E | 89 (71%) | 234 (80%) | 245 (75%) |
| Kidney volume, ml | |||
| Total | 1602±982 | 2399±1600 | 1917±1755 |
| Left | 812±507 | 1238±827 | 930±944 |
| Right | 790±580 | 1161±806 | 842±899 |
| Liver volume,b ml | 2457±1471 | 2617±1607 | Not measured |
BMI, body masss index; CKD-EPI, Chronic Kidney Disease Epidemiology Collaboration.
Parameters measured at baseline.
In dataset A measured in a subset of 28 patients.
Segmentation Performance
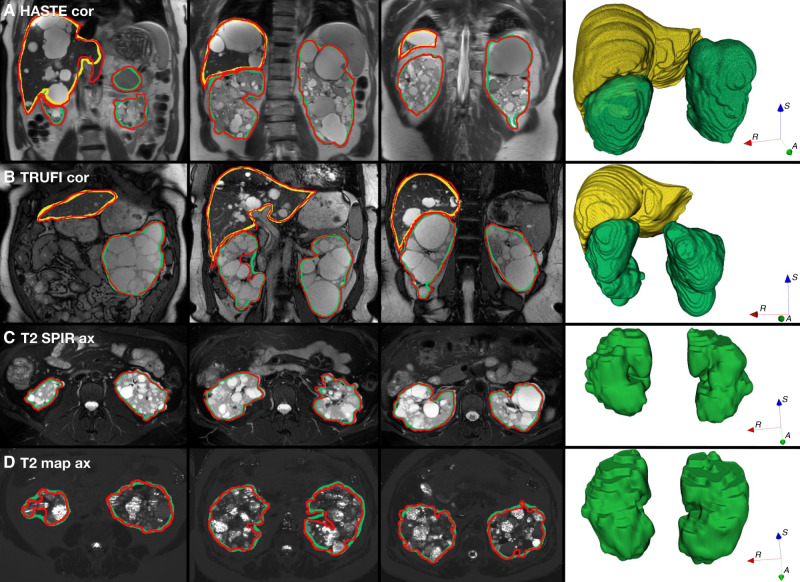
Automated segmentation results are summarized in Table 2 and Supplemental Table 5. In test set A, automated segmentation of kidneys achieved a median Dice coefficient of 0.94. Results were best for axial T2 SPIR images (median Dice, 0.96) compared with axial T2 images (Dice, 0.94) and axial T2 mapping (Dice, 0.92). In test set B, median Dice coefficients of 0.97 and 0.96 were achieved for kidneys and the liver, respectively. Results were similar for coronal HASTE images (median Dice, 0.97 for kidneys and 0.96 for liver) and coronal TRUFI scans (median Dice, 0.96 for kidneys and the liver). Segmentation results were consistent for both right and left kidneys, with a median Dice coefficient of 0.94 for both kidneys in dataset A and of 0.97 for the right and left kidneys in dataset B. Our model required between 2 and 4 minutes to segment a single MR scan from test datasets. Examples of automated segmentation of kidneys and liver using different MRI sequences are shown in Figure 2.
Table 2.
Results of automatic kidney and liver segmentation for different magnetic resonance imaging sequences
| Cohort, Organ, and Magnetic Resonance Imaging Sequence | No. of Scans | Dice | Jaccard | Sensitivity | Precision | Hausdorff | Mean Surface Distance |
|---|---|---|---|---|---|---|---|
| Dataset A test | |||||||
| Kidneys | |||||||
| T2 SPIR ax | 43 | 0.96 [0.90–0.97] | 0.92 [0.82–0.95] | 0.95 [0.91–0.97] | 0.97 [0.89–0.97] | 11.0 [7.0–18.8] | 0.51 [0.28–1.33] |
| T2 ax | 43 | 0.94 [0.89–0.95] | 0.88 [0.80–0.91] | 0.92 [0.89–0.95] | 0.95 [0.89–0.97] | 11.5 [8.0–15.9] | 0.78 [0.52–1.39] |
| T2 map ax | 38 | 0.92 [0.89–0.94] | 0.85 [0.80–0.89] | 0.92 [0.88–0.95] | 0.93 [0.87–0.96] | 7.2 [5.0–20.0] | 0.46 [0.35–1.0] |
| Dataset B test | |||||||
| Kidneys | |||||||
| HASTE cor | 182 | 0.97 [0.94–0.98] | 0.93 [0.88–0.95] | 0.97 [0.94–0.98] | 0.96 [0.94–0.98] | 10.0 [6.2–18.8] | 0.36 [0.26–0.56] |
| TRUFI cor | 18 | 0.96 [0.94–0.98] | 0.93 [0.89–0.95] | 0.96 [0.94–0.98] | 0.97 [0.94–0.98] | 10.3 [7.3–15.8] | 0.35 [0.28–0.51] |
| Liver | |||||||
| HASTE cor | 165 | 0.96 [0.94–0.97] | 0.93 [0.88–0.94] | 0.96 [0.93–0.98] | 0.96 [0.93–0.98] | 14.0 [8.6–22.6] | 0.46 [0.36–0.72] |
| TRUFI cor | 16 | 0.96 [0.95–0.97] | 0.93 [0.90–0.95] | 0.96 [0.92–0.97] | 0.97 [0.95–0.98] | 16.3 [5.9–29.3] | 0.48 [0.39–0.68] |
Values are reported as median and 10–90 percentile range (in square brackets). Hausdorff distance and mean surface distance are measured in voxels. T2, T2-weighted sequence; SPIR, spectral presaturation with inversion recovery; ax, axial; T2 map, T2 mapping; HASTE, half-Fourier–acquired single-shot turbo spin echo; cor, coronal; TRUFI, coronal T2 true fast imaging with steady-state free precession.
Figure 2.
Examples comparing the automated segmentation of the kidneys and the liver with the ground truth, showing very high precision of the automated method. (A) and (B) correspond to patients from the dataset B test: (A) HASTE coronal and (B) TRUFI. (C) and (D) correspond to patients from the dataset A test: (C) T2 SPIR axial and (D) T2 map axial. Red contours indicate the reference segmentation (manual), and green and yellow contours indicate automated segmentation of the kidneys and liver, respectively. ax, axial; cor, coronal.
Internal Validation
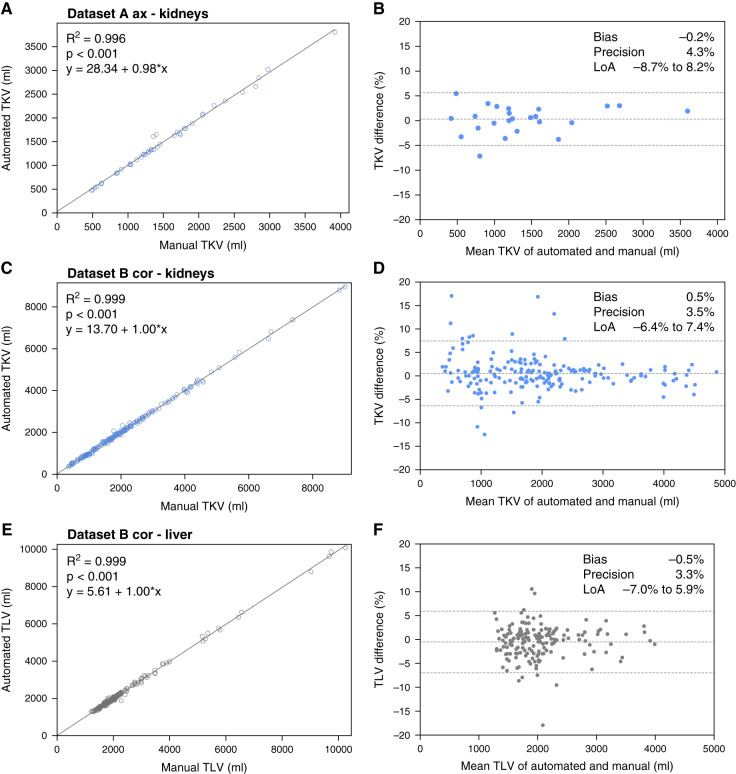
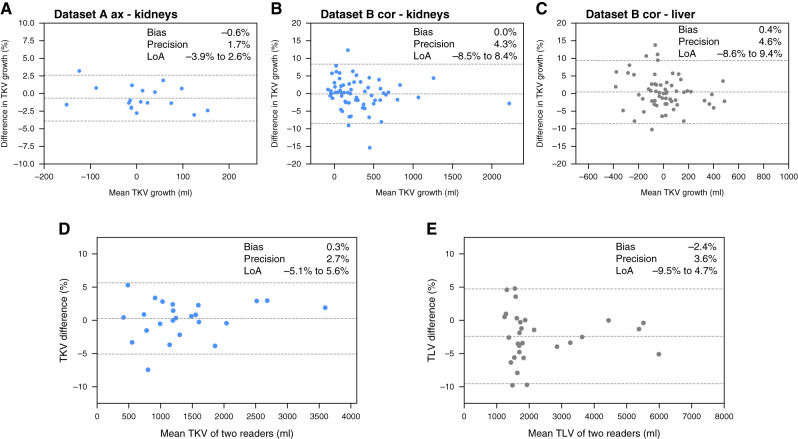
Linear regression and Bland–Altman plots comparing our method with manual tracing for TKV and TLV measurement are shown in Figure 3. The results are compared with the inter-reader variability of the reference method, as presented in Figure 4. For TKV estimation, the bias of our method was −0.2% in dataset A and 0.5% in dataset B, and precision values were 4% and 4% for datasets A and B, respectively. For TLV estimation, which was evaluated in dataset B, bias was −0.5%, and precision was 3%. These results were comparable with inter-reader variability, with bias and precision of 0.3% and 3%, respectively, for kidneys and of −2% and 4%, respectively, for liver volume estimation. In test set A, only one of 25 patients was reclassified into another Mayo risk group (Supplemental Table 6).
Figure 3.
Internal validation of the model, showing high correlation with the ground truth as well as high precision and low bias of total kidney and liver volumetry. (A, C, and E) High correlation and (B, D, and F) agreement assessed with Bland–Altman plots for (A–D) total kidney volume (TKV) measurements and (E and F) total liver volume (TLV) measurements. LoA, limit of agreement.
Figure 4.
Longitudinal validation shows high agreement between the growth of total kidney and liver volumes measured with our method and the reference method. (A)–(C) show Bland–Altman plots for the agreement between the growth rate of TKV and TLV between baseline and follow-up for (A) kidney volume growth in the axial MRI series in dataset A, (B) kidney volume growth in the coronal MRI series in dataset B, and (C) liver volume growth in the coronal MRI series in dataset B. (D) and (E) show Bland–Altman plots for the agreement of the reference method of (D) TKV and (E) TLV measurement with manual tracing between two different readers.
Longitudinal Validation
Supplemental Table 7 presents the absolute and percentage growth in TKV and TLV during follow-up. No significant differences were found between the automated and manual volumetric methods for kidneys and liver. Figure 4 shows Bland–Altman plots for agreement between the growth rate of TKV and TLV from baseline to follow-up. For TKV growth estimation, bias and precision were −0.6% and 2% for dataset A (Figure 4A), respectively, and 0% and 4% for dataset B (Figure 4B), respectively. TLV growth demonstrated bias of 0.4% and precision of 5% in dataset B (Figure 4C).
External Validation
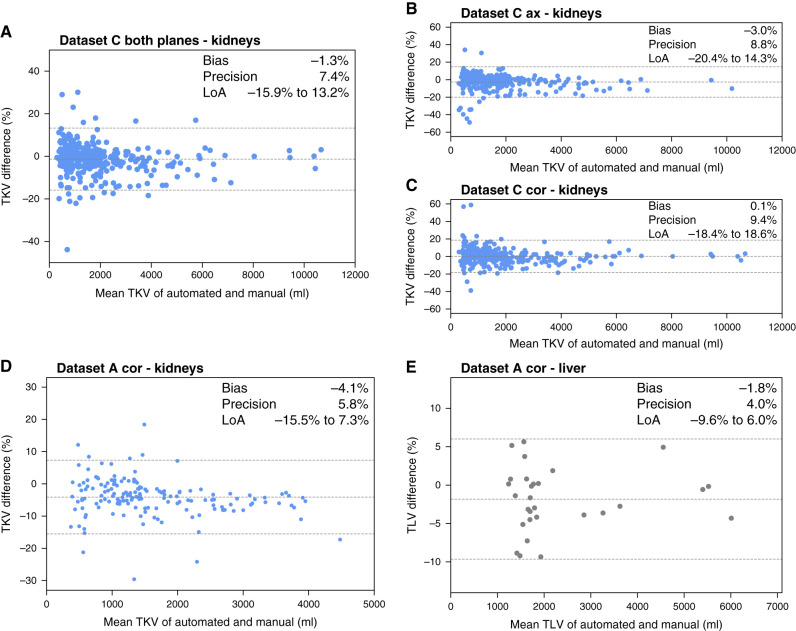
Figure 5 shows Bland–Altman plots comparing the difference between TKV and TLV for external data, which included dataset C and coronal images from dataset A. Because of high variability in acquisition protocols and scanners in dataset C, we calculate TKV in this cohort as a median of TKVs calculated across all included MRI sequences. We observed that in dataset A (Figure 5, D and E), the results were relatively good, with bias and precision of −4% and 6%, respectively, for TKV and −2% and 4%, respectively, for TLV. In dataset C (Figure 5, A–C), bias was −1% and precision was 7% when sequences in both axial and coronal orientations were included, and it was more precise than TKV calculated from images in either orientation separately.
Figure 5.
External validation of the automated model shows that it generalizes well across various MRI scanners and protocols. Bland–Altman plots for assessing the difference between TKV and TLV calculated from the automated segmentation and reference volumetry method of manual tracing for the external data. (A)–(C) present results for kidney volumetry in dataset C, and (D) and (E) present results for kidney and liver volumetry, respectively, in the coronal MRI series of dataset A.
Multiscanner Validation
Performance of the automated TKV estimation in MR images from dataset C, grouped by scanner manufacturers and models, is evaluated in Table 3. We report the total number of patients and scans as well as correlation and TKV difference from the reference standard. The correlation between automated and reference TKV varied from 0.990 for Siemens Amira to 0.999 for Philips Intera, Philips Ingenia S, and Toshiba scanners and >0.999 for Philips Achieva and Achieva dStream. Across the scanners, bias was in the range of −2% to 3%, and precision was in the range of 3%–11%.
Table 3.
Performance of our model in total kidney volume estimation in magnetic resonance images from dataset C, grouped by scanner manufacturer and model, as compared with reference manual tracing
| Manufacturer and Model | No. of Patients | No. of Scans | Correlationa | Total Kidney Volume Difference,b ml | Total Kidney Volume Difference, % | P Valuec |
|---|---|---|---|---|---|---|
| Siemens | ||||||
| Aera | 43 | 109 | 0.991 | −67±185 | −1.9±9.9 | 0.16 |
| Avanto | 41 | 113 | 0.992 | −47±160 | −1.9±7.8 | 0.07 |
| SymphonyTim | 29 | 72 | 0.991 | −52±200 | −1.3±8.7 | 0.58 |
| Avanto fit | 12 | 23 | 0.995 | −29±306 | 1.3±5.8 | 0.37 |
| Amira | 8 | 25 | 0.990 | 2±81 | 0.5±5.0 | 0.84 |
| Other models | 43 | 92 | 0.994 | −63±167 | −1.9±7.1 | 0.01 |
| Philips | ||||||
| Ingenia | 26 | 68 | 0.998 | −41±105 | 1.4±5.8 | 0.37 |
| Intera | 31 | 80 | 0.995 | −116±550 | −2.3±6.3 | 0.07 |
| Achieva | 24 | 66 | >0.999 | −56±134 | −1.1±3.9 | 0.05 |
| Ingenia S | 14 | 40 | 0.999 | −39±75 | 2.0±4.6 | 0.17 |
| Achieva dStream | 7 | 20 | >0.999 | −10±27 | 0.3±3.1 | 0.69 |
| Panorama HFO | 8 | 20 | 0.997 | −52±281 | 2.5±11.1 | >0.99 |
| GE | ||||||
| Multiple models | 29 | 73 | 0.997 | −31±108 | −0.9±5.8 | 0.001 |
| TOSHIBA | ||||||
| Multiple models | 5 | 12 | 0.999 | 149±378 | −0.1±11.0 | 0.81 |
High correlations and consistently small differences from reference volumetry can be noticed for the automated method across all of the models.
Pearson correlation coefficient.
Total kidney volume difference was calculated between automated and manual methods.
Calculated with the Wilcoxon signed-rank test.
Discussion
In this study, we developed a deep neural network for fully automated joint kidney and liver segmentation and validated it on multiple MRI sequences in longitudinal as well as external datasets. We obtained high accuracy for automated kidney and liver segmentation as well as high agreement between automatically calculated TKV and TLV and human readers. Importantly, our method enables accurate kidney segmentation in both axial and coronal image orientation.
Our method was validated on independent test sets and a large heterogeneous external dataset, which represented variable organ morphology, intensity distribution, and extensive range of TKV (range, 296–18,153 ml). Predicted segmentation masks were slightly more accurate for the coronal model, which was trained to jointly segment the kidneys and the liver, than for the axial model, which only segmented the kidneys (median Dice coefficient, 0.97 versus 0.94). Liver cysts can be challenging when measuring TKV because the distribution of cysts between the liver and the right kidney can be indistinguishable. It is possible that labeling the liver helps in differentiating the origin of unclear cysts in the MR image. We also observed that ensembling 2D U-Nets with a larger field of view and 3D U-Nets, which aggregate information along the z axis, was beneficial for segmentation performance.
The average time of manual kidney volumetry in MRI was reported to take between 32 and 90 minutes (5,10,11,13,24). Several different methods have been suggested for estimating TKV, which require less time than manual tracing, including midslice and ellipsoid methods (5). These approaches use standardized kidney measurements to calculate the estimated volume using approximation equations. Consequently, these methods are relatively imprecise, achieving bias and precision of 5% and 8%, respectively, for the midslice method and 1% and 9%, respectively, for the ellipsoid method (10).
To address this problem, more recent studies introduced automated or semiautomated polycystic kidney segmentation in MRI. The semiautomated Sheffield TKV Tool (11) showed high accuracy, with bias and precision of −0.3% and 4%, respectively, but it was subject to inter-reader variability and took around 4–7 minutes per measurement. These issues were eliminated by the fully automated approaches of Kline et al. (13), externally validated by van Gastel et al. (15), which reported performance at a level comparable with inter-reader variability. Another recent study (14) has reported automated TKV in T2-weighted MRI studies with bias and precision of 3% and 3%, respectively, for external validation and 4% and 6%, respectively, for prospective validation. However, this analysis was externally validated only on a dataset of 20 patients. We further expand on these studies, proposing a new architecture and extensive multisequence validation for TLV measurement in particular, as there have been few publications to date about the automated TLV estimation in the literature (15,25). With our method, kidney and liver volumetry takes around 2–5 minutes on a computer with a single Nvidia 1080Ti GPU.
Our established deep learning model adds important new evidence and features to currently available approaches. First, the model works on both axial and coronal sequences for TKV, and the coronal model was also validated for TLV. Second, the approach shows excellent bias and precision in several cohorts, including images from many different scanners without using standardized MRI sequences. In the internal validation, our method performs on par with the radiologist for estimating TKV (automated: 0.2%±4% and 0.5%±4% versus radiologist: 0.3%±3%) and TLV (automated: −0.5%±3% versus radiologist: −2%±4%). Similarly, high accuracy is achieved for the estimation of TKV and TLV growth in time. The highest Dice coefficient, signifying the highest segmentation accuracy, is achieved for coronal HASTE and TRUFI as well as axial T2 SPIR sequences, which were more accurate than axial T2 and T2 map sequences. We believe that this could be explained by the high robustness of HASTE and TRUFI regarding imaging of the upper abdomen with the known risk of motion artifacts due to insufficient breath holding as well as the higher contrast between the hyperintense kidney parenchyma and the surrounding hypointense perirenal fat tissue for the fat-saturated T2 SPIR compared with the regular T2 sequence. The T2 map sequence is especially sensitive to motion artifacts, which might have affected the distinction of the renal parenchyma and the surrounding tissue slightly. We suggest that the minimal acquisition mode for an accurate kidney and liver volumetry could include a single fat-saturated T2 sequence in the coronal or axial plane.
The external TKV validation in the highly heterogeneous dataset C was visibly less accurate (−1%±7%), probably owing to varying MRI protocols and hence, the ensuing lack of standardization (an overview of performance in subgroups of scanners/sequences is provided in Table 3). In this regard, bias and precision are only slightly better than the reported values for the ellipsoid equation. However, it is highly likely that the past studies strongly overestimated the clinical performance of the ellipsoid equation due to the availability of standardized MRI sequences and limited numbers of trained readers. This does not reflect the current clinical standard of care in most settings, in which MRI data are much more heterogeneous and measurements are obtained by many different radiologists not specifically trained. Also, even the ellipsoid equation requires manual measurements and is not provided on a routine basis by all radiologic practices. Both concerns are addressed using an automated model.
This study has a few limitations. First of all, the worse performance in the external dataset C, compared with datasets A and B with standardized MRI protocols, warrants further investigation into possible causes and ways to bridge this important gap. Furthermore, the subgroups of individual scanner types of different manufacturers are too small to allow for a final conclusion regarding the performance in direct comparison. The axial model does not perform liver segmentation because the manual segmentation data for the liver were not available for axial MRI studies in dataset A. Another limitation is the short time between baseline and follow-up for dataset A. Observation over a longer time period would enable quantifying more substantial disease progression. Our algorithm would not be able to distinguish patients in Mayo class 2 from patients in class 1. This step still requires the assessment by a radiologist/nephrologist but is much less time consuming than volumetry itself. Arguably, volumetry is much less important in patients in Mayo class 2 considering that this class is associated with slow disease progression. Finally, the proximity between the kidneys and the liver and the ambiguities inherent to MRI may lead to misclassification of cysts at the organ border. That is why the visual inspection is needed.
In conclusion, we developed a deep learning model for fully automated joint kidney and liver segmentation in patients with ADPKD and evaluated it in the largest multicenter cohort to date, including longitudinal and external data. We proved that our method is an accurate and robust tool for TKV and TLV estimation as well as their growth rates. Our approach was evaluated extensively on both axial and coronal MR scans obtained from 40 different MR scanners, promising future applicability in the routine clinical setting. The trained model is published along with the code used for development to allow for further joint development and usage by other centers. We hope that this may help enable the translation of the tool to routine clinical care and facilitate its incorporation in established medical imaging software solutions.
Disclosures
B. Baessler reports ownership interest in Lernrad GmbH. D.P. dos Santos reports consultancy agreements with Cook Medical. R.T. Gansevoort reports consultancy agreements with AstraZeneca, Bayer, Galapagos, Otsuka Pharmaceutical, and Sanofi-Genzyme; research funding from AstraZeneca, Bayer, Galapagos, Healthy.io, Otsuka Pharmaceuticals, and Sanofi-Genzyme; honoraria from Bayer, Galapagos, Mironid, Otsuka Pharmaceuticals, and Sanofi-Genzyme; and advisory or leadership roles as an editor of American Journal of Kidney Diseases, CJASN, Journal of Nephrology, Kidney360, Nephrology Dialysis Transplantation, and Nephron Clinical Practice and a member of the Council of the European Renal Association. R.-U. Müller reports consultancy agreements with Alnylam and Sanofi; ownership interest in Bayer, Chemocentryx, Novartis, Pfizer, Roche, and Santa Barbara Nutrients; and research funding from Otsuka Pharmaceuticals and Thermo Fisher Scientific. All research funding was paid to the employer (Department II of Internal Medicine). R.-U. Müller also reports honoraria from Alnylam and Sanofi (consultancy agreements) and advisory or leadership roles on the editorial board of Kidney and Dialysis, on the scientific advisory board of Santa Barbara Nutrients, and as chair of the board of the Working Group "Genes & Kidney" (European Renal Association). F. Grundmann is supported by funding from the German Research Foundation (GR 3932/2-2). All remaining authors have nothing to disclose.
Funding
This research was carried out with the support of Interdisciplinary Centre for Mathematical and Computational Modelling at the University of Warsaw grant G77-36. The DIPAK Consortium is sponsored by Dutch Government grant LSHM15018 and Dutch Kidney Foundation grants CP10.12 and CP15.01. R.-U. Müller is supported by funding from German Research Foundation grants DFG MU 3629/6-1 and DFG DI 1501/9, the Marga and Walter Boll-Stiftung, Ministry of Science North Rhine-Westphalia grant NRW.Nachwuchsgruppen 2015-2021, and the PKD Foundation (the KETO-ADPKD trial).
Supplementary Material
Acknowledgments
We thank Cornelia Böhme and Polina Todorova for the excellent support of the Clinical Study Center (Department II of Internal Medicine, University Hospital Cologne) as well as Mehrdad Bahadori, Miriam Rinneburger, Katharina Lettenmeier, and Lena Merkel for their help with manual segmentations. The DIPAK Consortium is an interuniversity collaboration in The Netherlands that was established to study ADPKD and develop rational treatment strategies for this disease.
Author Contributions
B. Baessler and R.-U. Müller conceptualized the study; S. Arjune, B. Baessler, R.T. Gansevoort, F. Grundmann, L. Karner, F. Meyer, T. Persigehl, F. Siedek, and M.D.A. van Gastel were responsible for data curation; F. Meyer and P. Woznicki were responsible for investigation; D.P. dos Santos, F. Siedek, L. Karner and P. Woznicki were responsible for formal analysis; D.P. dos Santos, F. Grundmann, R.-U. Müller, and P. Woznicki were responsible for methodology; B. Baessler and R.-U. Müller were responsible for project administration; S. Arjune, L. Karner, and F. Siedek were responsible for resources; P. Woznicki was responsible for software; D.P. dos Santos and F. Meyer were responsible for validation; B. Baessler, R.T. Gansevoort, R.-U. Müller, T. Persigehl, and M.D.A. van Gastel provided supervision; F. Siedek and P. Woznicki wrote the original draft; and S. Arjune, B. Baessler, L.L. Caldeira, D.P. dos Santos, R.T. Gansevoort, F. Grundmann, L. Karner, F. Meyer, R.-U. Müller, T. Persigehl, F. Siedek, M.D.A. van Gastel, and P. Woznicki reviewed and edited the manuscript.
Supplemental Material
This article contains the following supplemental material online at http://kidney360.asnjournals.org/lookup/suppl/doi:10.34067/KID.0003192022/-/DCSupplemental.
Supplemental Appendix 1. MRI acquisition parameters.
Supplemental Appendix 2. Flowchart of patient inclusion/exclusion.
Supplemental Appendix 3. Details on model training and performed experiments.
Supplemental Table 1. MRI acquisition parameters for sequences from datasets A–C.
Supplemental Table 2. Flowchart of patient inclusion/exclusion.
Supplemental Table 3. Network training hyperparameters selected for the segmentation task.
Supplemental Table 4. Detailed cohort characteristics for the training/test split of datasets A and B.
Supplemental Table 5. Results of automated kidney and liver segmentation in test datasets A and B for all of the sequences.
Supplemental Table 6. Patient stratification according to Mayo height-adjusted TKV risk classes for automated versus reference methods of kidney volumetry in test set A.
Supplemental Table 7. Comparison of the differences in kidney and liver volume between baseline and follow-up for manual tracing and automated methods.
Supplemental Table 8. TKV growth by the Mayo imaging class for patients with longitudinal data. (n – number of subjects, TKV – total kidney volume).
References
- 1.Lanktree MB, Haghighi A, Guiard E, Iliuta I-A, Song X, Harris PC, Paterson AD, Pei Y: Prevalence estimates of polycystic kidney and liver disease by population sequencing. J Am Soc Nephrol 29: 2593–2600, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Torres VE, Harris PC, Pirson Y: Autosomal dominant polycystic kidney disease. Lancet 369: 1287–1301, 2007 [DOI] [PubMed] [Google Scholar]
- 3.Lavu S, Vaughan LE, Senum SR, Kline TL, Chapman AB, Perrone RD, Mrug M, Braun WE, Steinman TI, Rahbari-Oskoui FF, Brosnahan GM, Bae KT, Landsittel D, Chebib FT, Yu ASL, Torres VE, Harris PC; the HALT PKD and CRISP Study Investigators : The value of genotypic and imaging information to predict functional and structural outcomes in ADPKD. JCI Insight 5: 138724, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perrone RD, Mouksassi M-S, Romero K, Czerwiec FS, Chapman AB, Gitomer BY, Torres VE, Miskulin DC, Broadbent S, Marier JF: Total kidney volume is a prognostic biomarker of renal function decline and progression to end-stage renal disease in patients with autosomal dominant polycystic kidney disease. Kidney Int Rep 2: 442–450, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Magistroni R, Corsi C, Martí T, Torra R: A review of the imaging techniques for measuring kidney and cyst volume in establishing autosomal dominant polycystic kidney disease progression. Am J Nephrol 48: 67–78, 2018 [DOI] [PubMed] [Google Scholar]
- 6.Irazabal MV, Rangel LJ, Bergstralh EJ, Osborn SL, Harmon AJ, Sundsbak JL, Bae KT, Chapman AB, Grantham JJ, Mrug M, Hogan MC, El-Zoghby ZM, Harris PC, Erickson BJ, King BF, Torres VE; CRISP Investigators : Imaging classification of autosomal dominant polycystic kidney disease: A simple model for selecting patients for clinical trials. J Am Soc Nephrol 26: 160–172, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Torres VE, Meijer E, Bae KT, Chapman AB, Devuyst O, Gansevoort RT, Grantham JJ, Higashihara E, Perrone RD, Krasa HB, Ouyang JJ, Czerwiec FS: Rationale and design of the TEMPO (Tolvaptan Efficacy and Safety in Management of Autosomal Dominant Polycystic Kidney Disease and its Outcomes) 3-4 Study. Am J Kidney Dis 57: 692–699, 2011 [DOI] [PubMed] [Google Scholar]
- 8.Meijer E, Visser FW, van Aerts RMM, Blijdorp CJ, Casteleijn NF, D’Agnolo HMA, Dekker SEI, Drenth JPH, de Fijter JW, van Gastel MDA, Gevers TJ, Lantinga MA, Losekoot M, Messchendorp AL, Neijenhuis MK, Pena MJ, Peters DJM, Salih M, Soonawala D, Spithoven EM, Wetzels JF, Zietse R, Gansevoort RT; DIPAK-1 Investigators : Effect of lanreotide on kidney function in patients with autosomal dominant polycystic kidney disease: The DIPAK 1 randomized clinical trial. JAMA 320: 2010–2019, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hogan MC, Abebe K, Torres VE, Chapman AB, Bae KT, Tao C, Sun H, Perrone RD, Steinman TI, Braun W, Winklhofer FT, Miskulin DC, Rahbari-Oskoui F, Brosnahan G, Masoumi A, Karpov IO, Spillane S, Flessner M, Moore CG, Schrier RW: Liver involvement in early autosomal-dominant polycystic kidney disease. Clin Gastroenterol Hepatol 13: 155–64.e6, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spithoven EM, van Gastel MDA, Messchendorp AL, Casteleijn NF, Drenth JPH, Gaillard CA, de Fijter JW, Meijer E, Peters DJ, Kappert P, Renken RJ, Visser FW, Wetzels JF, Zietse R, Gansevoort RT; DIPAK Consortium; DIPAK Consortium : Estimation of total kidney volume in autosomal dominant polycystic kidney disease. Am J Kidney Dis 66: 792–801, 2015 [DOI] [PubMed] [Google Scholar]
- 11.Simms RJ, Doshi T, Metherall P, Ryan D, Wright P, Gruel N, van Gastel MDA, Gansevoort RT, Tindale W, Ong ACM: A rapid high-performance semi-automated tool to measure total kidney volume from MRI in autosomal dominant polycystic kidney disease. Eur Radiol 29: 4188–4197, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma K, Rupprecht C, Caroli A, Aparicio MC, Remuzzi A, Baust M, Navab N: Automatic segmentation of kidneys using deep learning for total kidney volume quantification in autosomal dominant polycystic kidney disease. Sci Rep 7: 2049, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kline TL, Korfiatis P, Edwards ME, Blais JD, Czerwiec FS, Harris PC, King BF, Torres VE, Erickson BJ: Performance of an artificial multi-observer deep neural network for fully automated segmentation of polycystic kidneys. J Digit Imaging 30: 442–448, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goel A, Shih G, Riyahi S, Jeph S, Dev H, Hu R, Romano D, Teichman K, Blumenfeld JD, Barash I, Chicos I, Rennert H, Prince MR: Deployed deep learning kidney segmentation for polycystic kidney disease MRI. Radiol Artif Intell 4: 210205, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Gastel MDA, Edwards ME, Torres VE, Erickson BJ, Gansevoort RT, Kline TL: Automatic measurement of kidney and liver volumes from MR images of patients affected by autosomal dominant polycystic kidney disease. J Am Soc Nephrol 30: 1514–1522, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shin TY, Kim H, Lee J-H, Choi J-S, Min H-S, Cho H, Kim K, Kang G, Kim J, Yoon S, Park H, Hwang YU, Kim HJ, Han M, Bae E, Yoon JW, Rha KH, Lee YS: Expert-level segmentation using deep learning for volumetry of polycystic kidney and liver. Investig Clin Urol 61: 555–564, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan W, Huang L, Xia L, Gu S, Yan F, Wang Y, Tao Q: MRI manufacturer shift and adaptation: Increasing the generalizability of deep learning segmentation for MR images acquired with different scanners. Radiol Artif Intell 2: 190195, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bluemke DA, Moy L, Bredella MA, Ertl-Wagner BB, Fowler KJ, Goh VJ, Halpern EF, Hess CP, Schiebler ML, Weiss CR: Assessing radiology research on artificial intelligence: A brief guide for authors, reviewers, and readers-from the Radiology Editorial Board. Radiology 294: 487–489, 2020 [DOI] [PubMed] [Google Scholar]
- 19.Ronneberger O, Fischer P, Brox T: U-Net: Convolutional networks for biomedical image segmentation. In: Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015, edited by Navab N, Hornegger J, Wells WM, Frangi AF, Berlin, Springer, 2015, pp 234–241 [Google Scholar]
- 20.Kickingereder P, Isensee F, Tursunova I, Petersen J, Neuberger U, Bonekamp D, Brugnara G, Schell M, Kessler T, Foltyn M, Harting I, Sahm F, Prager M, Nowosielski M, Wick A, Nolden M, Radbruch A, Debus J, Schlemmer HP, Heiland S, Platten M, von Deimling A, van den Bent MJ, Gorlia T, Wick W, Bendszus M, Maier-Hein KH: Automated quantitative tumour response assessment of MRI in neuro-oncology with artificial neural networks: A multicentre, retrospective study. Lancet Oncol 20: 728–740, 2019 [DOI] [PubMed] [Google Scholar]
- 21.Ibtehaz N, Rahman MS: MultiResUNet: Rethinking the U-Net architecture for multimodal biomedical image segmentation. Neural Netw 121: 74–87, 2020 [DOI] [PubMed] [Google Scholar]
- 22.van Aerts RMM, Kievit W, D’Agnolo HMA, Blijdorp CJ, Casteleijn NF, Dekker SEI, de Fijter JW, van Gastel M, Gevers TJ, van de Laarschot LFM, Lantinga MA, Losekoot M, Meijer E, Messchendorp AL, Neijenhuis MK, Pena MJ, Peters DJM, Salih M, Soonawala D, Spithoven EM, Visser FW, Wetzels JF, Zietse R, Gansevoort RT, Drenth JPH; DIPAK-1 Investigators : Lanreotide reduces liver growth in patients with autosomal dominant polycystic liver and kidney disease. Gastroenterology 157: 481–491.e7, 2019 [DOI] [PubMed] [Google Scholar]
- 23.Fedorov A, Beichel R, Kalpathy-Cramer J, Finet J, Fillion-Robin J-C, Pujol S, Bauer C, Jennings D, Fennessy F, Sonka M, Buatti J, Aylward S, Miller JV, Pieper S, Kikinis R: 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn Reson Imaging 30: 1323–1341, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Turco D, Valinoti M, Martin EM, Tagliaferri C, Scolari F, Corsi C: Fully Automated segmentation of polycystic kidneys from noncontrast computed tomography: A feasibility study and preliminary results. Acad Radiol 25: 850–855, 2018 [DOI] [PubMed] [Google Scholar]
- 25.Kim Y, Bae SK, Cheng T, Tao C, Ge Y, Chapman AB, Torres VE, Yu AS, Mrug M, Bennett WM, Flessner MF, Landsittel DP, Bae KT: Automated segmentation of liver and liver cysts from bounded abdominal MR images in patients with autosomal dominant polycystic kidney disease. Phys Med Biol 61: 7864–7880, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.