SUMMARY
Glutathione (GSH) functions as a major sulfur repository and hence occupies an important position in primary sulfur metabolism. GSH degradation results in sulfur reallocation and is believed to be carried out mainly by γ‐glutamyl cyclotransferases (GGCT2;1, GGCT2;2, and GGCT2;3), which, however, do not fully explain the rapid GSH turnover. Here, we discovered that γ‐glutamyl peptidase 1 (GGP1) contributes to GSH degradation through a yeast complementation assay. Recombinant proteins of GGP1, as well as GGP3, showed high degradation activity of GSH, but not of oxidized glutathione (GSSG), in vitro. Notably, the GGP1 transcripts were highly abundant in rosette leaves, in agreement with the ggp1 mutants constantly accumulating more GSH regardless of nutritional conditions. Given the lower energy requirements of the GGP‐ than the GGCT‐mediated pathway, the GGP‐mediated pathway could be a more efficient route for GSH degradation than the GGCT‐mediated pathway. Therefore, we propose a model wherein cytosolic GSH is degraded chiefly by GGP1 and likely also by GGP3. Another noteworthy fact is that GGPs are known to process GSH conjugates in glucosinolate and camalexin synthesis; indeed, we confirmed that the ggp1 mutant contained higher levels of O‐acetyl‐l‐Ser, a signaling molecule for sulfur starvation, and lower levels of glucosinolates and their degradation products. The predicted structure of GGP1 further provided a rationale for this hypothesis. In conclusion, we suggest that GGP1 and possibly GGP3 play vital roles in both primary and secondary sulfur metabolism.
Keywords: glutathione metabolism, gamma‐glutamyl peptidase, gamma‐glutamyl cyclotransferase, glucosinolate metabolism, sulfur metabolism, sulfur deficiency, cysteine, Arabidopsis thaliana
Significance Statement
Glutathione (GSH) is a major repository for organic sulfur in plants. In this study, we identified novel GSH degradation enzymes, γ‐glutamyl peptidases (GGPs), which may supply a substantial amount of cysteine in the cell. Moreover, GGPs also function in glucosinolate and camalexin synthesis, so GGPs can be regarded as enzymes that play important roles in both GSH degradation and glucosinolate and camalexin synthesis, that is, primary and secondary sulfur metabolism.
INTRODUCTION
Sulfur is a macronutrient for plants. Sulfur is taken up by plant cells mainly as sulfate, which is then reduced to sulfide. Sulfide can be incorporated into the amino acid skeleton of O‐acetyl‐l‐Ser (OAS) to produce Cys, which is the donor for all metabolites containing reduced sulfur (Koprivova & Kopriva, 2014). Cys, however, is not the storage form of reduced sulfur because extra concentrations of Cys are toxic due to their high reactivity (Deshpande et al., 2017; Romero et al., 2014). Cys is incorporated into glutathione (GSH; l‐γ‐glutamyl‐l‐cysteinyl‐glycine), which is stable and exists in high concentrations in plant tissues (Leustek et al., 2000). Thus, GSH functions as a vital Cys source. Similarly, GSH concentrations in the phloem sap are high (in the millimolar range); therefore, GSH is also considered the transport form of reduced sulfur (Kuzuhara et al., 2000). In addition, GSH plays a variety of essential roles in plants. GSH reversibly undergoes redox reactions through its thiol residue and maintains the redox status of the cells. When oxidized, two GSH molecules form dimers and become oxidized GSH (GSSG), which is returned to the reduced GSH by GSH reductase. GSH is essential for alleviating oxidative stress in plants because most GSH in the cell is in its reduced form and can reduce oxidants (Foyer & Noctor, 2011; Noctor et al., 2011).
GSH is synthesized from its constituent amino acids via a two‐step ATP‐dependent reaction. First, γ‐glutamyl‐cysteine (γ‐Glu‐Cys) is synthesized from Glu and Cys by γ‐glutamyl‐cysteine synthetase (γ‐ECS; Hell & Bergmann, 1990), which is encoded by GSH1 (May & Leaver, 1994) and considered important for the GSH synthesis rate (Noctor et al., 2012). GSH is synthesized from γ‐Glu‐Cys and Gly by GSH synthetase, which is encoded by GSH2 (Wang & Oliver, 1996). However, the GSH degradation pathways are not as straightforward as this synthesis pathway. In mammals, GSH is degraded during the γ‐glutamyl cycle (Figure S1; Orlowski & Meister, 1970; Meister & Larsson, 1995). In this cycle, GSH is degraded by an exoenzyme, γ‐glutamyl transpeptidase (GGT), which hydrolyzes GSH and transfers the Glu residue to water or other amino acids. When transferred to water, Glu is produced; when transferred to amino acids, γ‐glutamyl amino acids are produced. Glu and γ‐glutamyl amino acids are incorporated into cells, and γ‐glutamyl amino acids are processed by the cytosolic enzyme γ‐glutamyl cyclotransferase (GGCT). This enzyme detaches the γ‐glutamyl residue of γ‐glutamyl amino acids and releases it as 5‐oxoproline, which can be converted to Glu by another cytosolic enzyme, oxoprolinase. The other reaction product of GSH degradation by GGT is cysteinyl‐glycine (Cys‐Gly). This dipeptide is degraded into Cys and Gly by dipeptidases. In this way, GSH is degraded into three constituent amino acids, Glu, Cys, and Gly, and they are utilized for the biosynthesis of proteins or GSH in the cells (Figure S1).
In Arabidopsis, the turnover of GSH is as high as 80% in 1 day under normal conditions (Ohkama‐Ohtsu et al., 2008), and genes responsible for the degradation pathway have been extensively explored. There are four homologs of mammalian GGTs: GGT1, 2, 3, and 4. GGT2 is expressed mostly in young siliques and barely in roots, and GGT3 is hardly expressed and is not regarded as functional; only GGT1 and GGT4 are actively expressed in the whole plant body (Destro et al., 2010; Ohkama‐Ohtsu, Radwan, et al., 2007; Ohkama‐Ohtsu, Zhao, et al., 2007). GGT1 and GGT2 are localized to the apoplast and GGT4 to the vacuole (Ferretti et al., 2009; Grzam et al., 2007; Martin et al., 2007; Ohkama‐Ohtsu, Radwan, et al., 2007; Ohkama‐Ohtsu, Zhao, et al., 2007). Because ggt1/ggt4 knockout mutants did not show GGT activity, they were utilized for the investigation of the γ‐glutamyl cycle in plants (Ohkama‐Ohtsu et al., 2008). However, the GSH degradation rate and GSH concentration were not affected by defects in GGT activity. This strongly suggests that GGT is not involved in GSH degradation. This was also consistent with the fact that plants have two orders of magnitude less GSH in the extracellular space than in the cytoplasm (Ohkama‐Ohtsu, Radwan, et al., 2007). Instead, it was proposed that GGT1 is responsible for GSSG degradation in the apoplast (Ohkama‐Ohtsu, Radwan, et al., 2007) and GGT4 is responsible for GSH conjugate degradation in the vacuole (Grzam et al., 2007; Ohkama‐Ohtsu, Zhao, et al., 2007). Therefore, other GSH degradation pathways must be identified.
Ohkama‐Ohtsu et al. (2008) also found that 5‐oxoproline levels are correlated with GSH or γ‐Glu‐Cys levels, and they proposed that GGCT, a cytosolic enzyme that degrades γ‐glutamyl amino acids and releases 5‐oxoproline, also degrades GSH. In addition, activity assays using protein extracts suggested that 5‐oxoproline is more likely to be supplied by GSH degradation than by γ‐Glu‐Cys degradation (Ohkama‐Ohtsu et al., 2008). Later, GGCT, which has GSH degradation activity, was identified in mammals (Kumar et al., 2012), and three homologs have been identified in Arabidopsis: GGCT2;1, GGCT2;2, and GGCT2;3. All of them degrade GSH at physiological concentrations into 5‐oxoproline and Cys‐Gly in vitro and complement the GSH degradation‐defective yeast mutant (Kumar et al., 2015; Paulose et al., 2013). Among the GGCT genes, the expression patterns of GGCT2;1 have been analyzed in detail. GGCT 2;1 mRNA is rapidly accumulated under various conditions, including pollen tube growth (Wang et al., 2008), heavy metal stress (Kovalchuk et al., 2005), salinity stress (Gong et al., 2005), and sulfur starvation (Bielecka et al., 2014; Hubberten et al., 2012; Maruyama‐Nakashita et al., 2005). Paulose et al. (2013) reported that the promotion of GGCT2;1 expression under arsenite stress can contribute to heavy metal tolerance by recycling Glu in GSH and saving energy for de novo Glu synthesis. Joshi et al. (2019) also reported that the upregulated expression of GGCT2;1 under sulfur deficiency is involved in the regulation of root architecture by controlling GSH homeostasis. However, the mechanism responsible for the rapid turnover of GSH is still unclear because the expression of GGCT2;1 is very low under normal conditions. We hypothesized that enzymes other than GGCT2;1 contribute to cytosolic GSH degradation under normal conditions.
In the present study, we showed that the cytosolic enzymes γ‐glutamyl peptidase 1 (GGP1) and GGP3 have GSH degradation activity comparable to that of GGCT2;1, GGCT2;2, and GGCT2;3. Based on the results of expression analysis and mutant analysis, we suggest that cytosolic GSH is fundamentally degraded by GGP1, except that GGCT2;2 is crucial in developing organs and GGCT2;1 is vital under sulfur deficiency. Interestingly, GGP1 and GGP3 also process GSH conjugates in the biosynthesis of glucosinolates (GSLs) and camalexin (Geu‐Flores et al., 2009), so they are expected to have dual roles. This hypothesis was tested through metabolome analysis and protein structure analysis; therefore, we suggest that GGPs play important roles in both GSH degradation and GSL and camalexin synthesis, that is, primary and secondary sulfur metabolism.
RESULTS
Isolation of A. thaliana cDNA clones that complement yeast dug2Δ or dug3Δ mutants
An Arabidopsis thaliana cDNA library built in the yeast expression vector pFL61 (Minet et al., 1992) was transformed into the Saccharomyces cerevisiae yeast mutants dug2Δ or dug3Δ (DUG: Defective in Utilization of GSH; Ganguli et al., 2007). In S. cerevisiae, three mutants, dug1Δ, dug2Δ, and dug3Δ, are defective in GSH utilization (Ganguli et al., 2007). The (DUG2–DUG3)2 complex first degrades GSH into Glu and Cys‐Gly, and then the (DUG1)2 homodimer degrades Cys‐Gly into Cys and Gly (Kaur et al., 2009, 2012). As a result of this transformation, approximately 4.6 × 104 yeast transformants were obtained in dug2Δ, and two independent colonies were isolated from the minimal selection plates with GSH as the sole sulfur source. From the approximately 6.9 × 104 yeast transformants in dug3Δ, three independent colonies were isolated under the same growth conditions. Plasmids were isolated from these colonies, and cDNAs expressed from the library were sequenced. All clones that complemented the dug2Δ and dug3Δ yeast mutants corresponded to the gene GGP1, which was initially identified as an enzyme processing GSH conjugates in the biosynthesis of GSLs and camalexin (Geu‐Flores et al., 2009).
To confirm the complementation of dug2Δ and dug3Δ yeast with GGP1, the open reading frame (ORF) of GGP1 was cloned into the yeast expression vector p416TEF (Mumberg et al., 1995) and transformed into dug2Δ and dug3Δ mutants. Both yeast mutants transformed with the GGP1 ORF grew on the medium when GSH was the sole sulfur source (Figure 1a). This result suggests that GGP1 from Arabidopsis is sufficient to degrade GSH, whereas yeast DUG2 and DUG3 need to form an active complex. The genes GGCT2;1, GGCT2;2, and GGCT2;3 were also cloned into p416TEF and introduced into dug2Δ or dug3Δ mutants. As previously demonstrated, all GGCT genes complemented both yeast mutants (Figure 1a) (Kumar et al., 2015).
Figure 1.
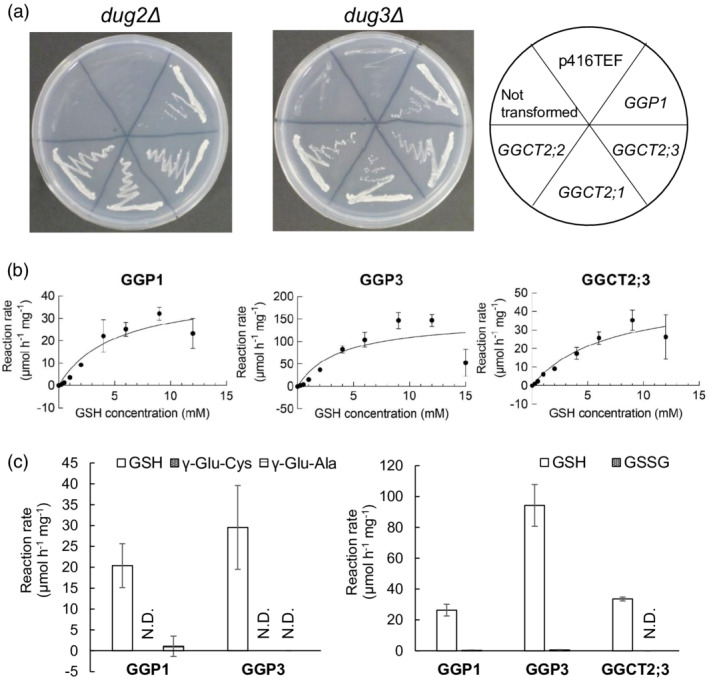
GSH degradation activity of γ‐glutamyl peptidases (GGPs). (a) Functional complementation assay of yeast mutants with Arabidopsis GGP1 together with γ‐glutamyl cyclotransferase 2;1 (GGCT2;1), GGCT2;2, and GGCT2;3. The Saccharomyces cerevisiae dug2Δ and dug3Δ mutants (DUG: Defective in Utilization of Glutathione) were transformed with plasmids carrying GGP1, GGCT2;1, GGCT2;2, or GGCT2;3 driven by the TEF promoter in the p416TEF vector and grown in SD medium supplemented with glutathione (GSH). The vector p416TEF was used as a control. (b) Michaelis–Menten plot of GSH degradation activity of recombinant GGP1, GGP3, and GGCT2;3. Recombinant GGP1, GGP3, and GGCT2;3 were incubated in 50 μl of reaction mixture containing 0.25 to 15.0 mm GSH for 30 min at 37°C. (c) Degradation activity of recombinant enzymes toward other γ‐glutamyl compounds, γ‐Glu‐Cys, γ‐Glu‐Ala (left), or GSSG (right). The values and error bars represent the mean and standard deviation of three biological replicates. Kinetic parameters were calculated using GraphPad Prism 9.
Degradation activities of recombinant proteins from E. coli
We investigated the enzymatic properties of GGPs and GGCTs using recombinant proteins. There are five GGP genes in Arabidopsis, but the expression of GGP2, GGP4, and GGP5 was very low, and only GGP1 and GGP3 were actively expressed across all tissues (Geu‐Flores et al., 2011); therefore, in addition to GGP1, GGP3 was analyzed in this experiment.
GGP1, GGP3, GGCT2;1, GGCT2;2, and GGCT2;3 were expressed in Escherichia coli. GGP1, GGP3, and GGCT2;3 collected from the soluble fraction were utilized for the subsequent measurement of enzyme kinetics and analyzed together with the reported properties of GGCT2;1 and GGCT2;2 (Kumar et al., 2015; Paulose et al., 2013). The activity and affinity of the proteins were analyzed with various GSH concentrations ranging from 0.25 to 15.0 mm (Figure 1b; Table 1). The K m values of GGP1, GGP3, and GGCT2;3 were 5.0, 3.7, and 6.7 mm, respectively; their k cat values were 0.33, 1.17, and 0.31 sec−1, respectively.
Table 1.
Kinetic parameters of GGP1, GGP3, and GGCT2;3
| Enzyme | k cat (sec−1) | K m (mm) | k cat/K m (sec−1 m −1) |
|---|---|---|---|
| GGP1 | 0.33 ± 0.06 | 5.0 ± 2.0 | 66 |
| GGP3 | 1.17 ± 0.22 | 3.7 ± 2.0 | 297 |
| GGCT2;3 | 0.31 ± 0.07 | 6.7 ± 3.0 | 46 |
In addition to GSH, the degradation activity of these enzymes toward other γ‐glutamyl compounds, namely, GSSG, γ‐glutamyl‐cysteine (γ‐Glu‐Cys), and γ‐glutamyl‐alanine (γ‐Glu‐Ala), was also analyzed (Figure 1c). However, the activity of GGCT2;3 toward γ‐Glu‐Cys and γ‐Glu‐Ala was excluded from the analysis because it is known to be much lower than that toward GSH (Kumar et al., 2015). The results showed that GGP1, GGP3, and GGCT2;3 hardly degraded γ‐Glu‐Cys, γ‐Glu‐Ala, or GSSG.
Comparison of gene expression levels of GGP and GGCT among organs
The distribution of GGP1, GGP3, GGCT2;1, GGCT2;2, and GGCT2;3 expression was investigated by dividing plants at the vegetative and reproductive stages into 10 parts and analyzing the expression levels of these genes by quantitative reverse transcription PCR (RT‐PCR) using the absolute quantification method. The 10 plant parts were the mature and young leaves of vegetative‐stage plants and the flowers/buds, siliques, stems, cauline and rosette leaves, and the upper, middle, and lower roots of reproductive‐stage plants. In general, all GGP and GGCT genes, except GGCT2;1, were constitutively expressed in all organs (Figure 2). In particular, GGP1 transcripts were abundant in the rosette leaves. In contrast, the expression of GGCT2;1 was much lower than that of the other four genes in all organs.
Figure 2.
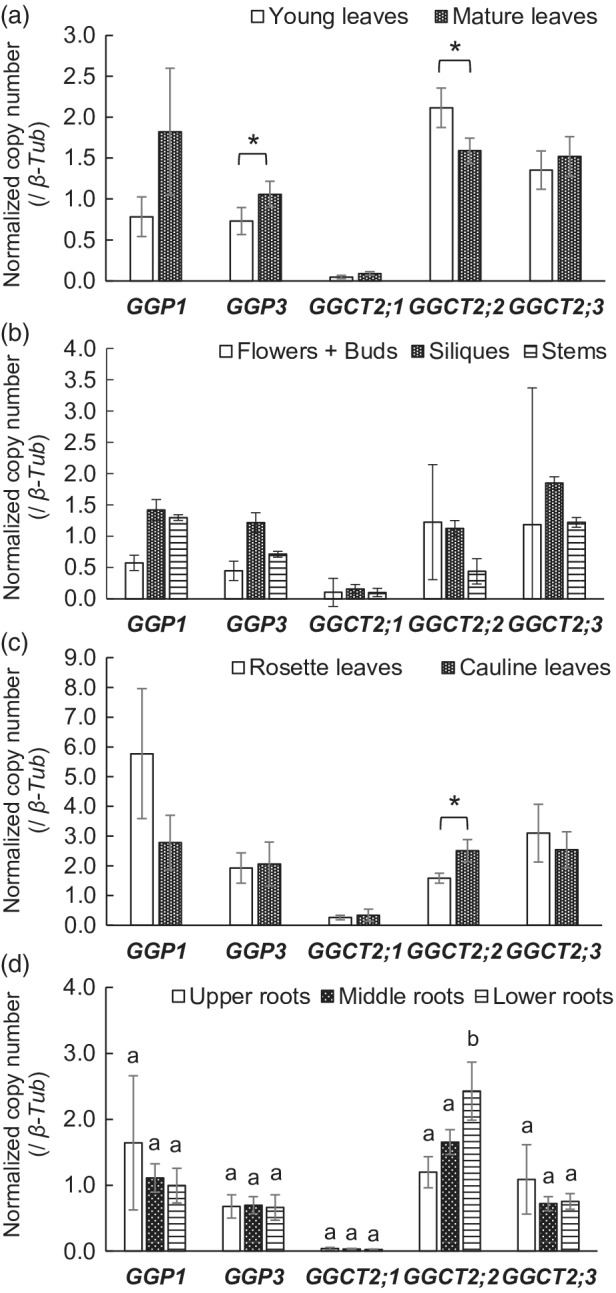
Normalized copy numbers of GGP1, GGP3, GGCT2;1, GGCT2;2, and GGCT2;3 transcripts in various organs of wild‐type (WT) plants. Plants were grown in hydroponic culture. Transcript copy numbers of GGP1, GGP3, GGCT2;1, GGCT2;2, and GGCT2;3 were analyzed by quantitative RT‐PCR using the absolute quantification method and normalized to β‐tubulin. (a) Comparison of young and old leaves from the vegetative‐stage samples. (b–d) Results of the reproductive‐stage samples. (b) Results of flowers/buds, siliques, and stems. (c) Comparison of the rosette and cauline leaves of reproductive‐stage samples. (d) Comparison of the upper, middle, and lower roots of the reproductive‐stage samples. The values and error bars represent the mean and standard deviation of four or five biological replicates. In (a) and (c), asterisks indicate significant differences (Student's t‐test, P < 0.05). In (d), different letters indicate significant differences (Tukey's test, P < 0.05).
It was also notable that the amount of GGP3 transcript was significantly higher in mature leaves than in young leaves at the vegetative stage (Figure 2a), and a similar tendency was observed for GGP1 (P < 0.1). As mentioned above, GGP1 transcripts were highly abundant when the leaves achieved a rosette arrangement at the reproductive stage (Figure 2b,c). In contrast, the amount of GGCT2;2 transcripts was significantly higher in young leaves than in mature leaves at the vegetative stage (Figure 2a). A similar trait was confirmed for the reproductive stage, wherein the GGCT2;2 transcript in cauline leaves was significantly higher than that in rosette leaves (Figure 2c). Furthermore, GGCT2;2 transcript levels in the lower roots, which are considered to be the youngest roots, were significantly higher than in the middle and upper roots (Figure 2d).
Changes in GSH metabolism in the ggp1 and ggct2;1 knockout mutants
To confirm the role of GGP1 in GSH degradation in planta, GSH concentrations in the ggp1‐1 (GK‐319F10) and ggp1‐2 (SALK_089634) knockout mutants (Geu‐Flores et al., 2011) were measured together with the ggct2;1–2 knockout mutant (SALK_56007, Joshi et al., 2019) and their corresponding wild‐type plants (Columbia‐0 [Col‐0]) using 14‐day‐old liquid‐cultured seedlings. Prior to this experiment, these plants were grown on rockwool under normal conditions until they set seeds, but they did not show any evident phenotypic changes. GSH concentrations in the ggp1‐1 and ggp1‐2 mutants were significantly higher than those in the wild‐type plants when grown under normal conditions, whereas the ggct2;1–2 mutant did not show any significant difference (Figure 3a), suggesting that GGP1 contributes to GSH degradation under normal conditions. We also measured GSH concentrations in mutants under sulfur‐ or nitrogen‐deficient conditions. Under sulfur deficiency, the ggp1‐1, ggp1‐2, and ggct2;1–2 mutants showed significantly higher GSH levels than wild‐type plants (Figure 3a). Higher GSH levels under sulfur deficiency have also been reported for ggct2;1–1 (SALK_117578), another T‐DNA insertion mutant of GGCT2;1 (Joshi et al., 2019). However, under nitrogen deficiency, the elevation of GSH levels was only observed in ggp1‐1 and ggp1‐2, and not in ggct2;1–2, which was similar to normal conditions. These results suggest that GGP1 constantly degrades GSH, whereas GGCT2;1 breaks down only under sulfur deficiency. Considering that the perturbed GSH degradation may affect Cys supply, Cys contents in the mutants were also quantified; however, they were not decreased, and rather increased (Figure 3b). Because Cys is a vital metabolite for the synthesis of proteins or compounds containing reduced sulfur, the Cys concentration is presumably to be tightly regulated.
Figure 3.
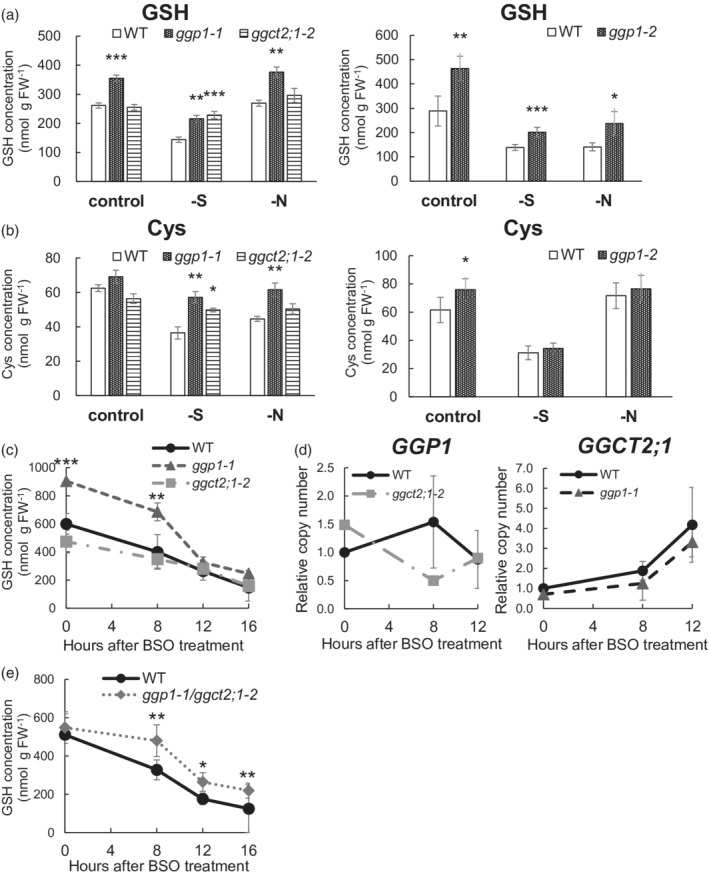
Impact of GGP1 or GGCT2;1 perturbation on GSH metabolism. (a) GSH concentration of ggp1‐1 and ggct2;1–2 (left) and ggp1‐2 (right) mutants. (b) Cys concentration of ggp1‐1 and ggct2;1–2 (left) and ggp1‐2 (right) mutants. (c) Time‐dependent changes in GSH concentration after buthionine sulfoximine (BSO) treatment in the ggp1 and ggct2;1 mutants. (d) Time‐dependent changes in the relative expression of GGP1 (left) and GGCT2;1 (right) after BSO treatment. (e) Time‐dependent changes in GSH concentration after BSO treatment in the ggp1/ggct2;1 double mutant. The values and error bars represent the mean and standard deviation of four to five biological replicates. Asterisks indicate significant differences from the wild type; *P < 0.05, **P < 0.01, Dunnett's test (left figures of (a), (b), and (c)) or Student's t‐test (right figures of (a), (b), and (e)).
We also monitored the GSH degradation rate after treating plants with buthionine sulfoximine (BSO), an established inhibitor of GSH synthesis (Griffith & Meister, 1979). However, the ggp1 mutant did not show a significant decrease in the rate of GSH degradation (Figure 3c). A possible reason was that high GSH concentrations in the ggp1 mutant led to active GSH degradation by the remaining GSH degradation enzymes because an enzymatic reaction is generally catalyzed at a higher rate under higher substrate concentrations even when there is the same amount of an enzyme. Correspondence between active GSH degradation and increased GGCT2;1 expression after 8 h (Figure 3c,d) also implied that GGCT2;1 worked more effectively in the ggp1 mutant under BSO treatment.
Therefore, we created the ggp1‐1/ggct2;1–2 double knockout mutant and analyzed its GSH degradation rate using the same BSO treatment (Figure 3e). Although the initial GSH concentration was similar between genotypes in this trial, the double mutant showed a significantly higher GSH level than wild‐type plants after 8 h of BSO treatment, which was maintained until 16 h. Therefore, the double mutant degraded GSH more slowly than wild‐type plants. This suggests that GGP1 and GGCT2;1 redundantly contribute to GSH degradation in Arabidopsis plants.
We also checked the expression levels of other genes in wild‐type plants and the ggp1‐1/ggct2;1–2 mutant after BSO treatment to gain more insight into the underlying mechanisms. The time‐dependent expression levels of GGP3, GGCT2;2, and GGCT2;3 were hardly affected (Figure S3a), suggesting that the observed changes in GSH degradation were caused primarily by the disruption of GGP1 and GGCT2;1. We also suspected that the decreased accumulation of GSH might be caused by the weakened expression of GSH1 or GSH2, involved in GSH synthesis (May & Leaver, 1994; Wang & Oliver, 1996); however, no significant differences were detected between wild‐type plants and the mutant (Figure S3b). Another mechanism such as the post‐translational regulation of the activity of γ‐ECS, a product of GSH1, might also be involved (Jez et al., 2004).
Changes in metabolites in the ggp1 and ggct2;1 mutants
GSH degradation supplies three amino acids, Cys, Glu, and Gly, so this reaction is considered to have a broad impact on plant metabolism, especially sulfur metabolism. To find further evidence of GSH degradation by GGP1 and analyze the effects of GSH degradation on whole plant metabolism, we conducted a widely targeted metabolome analysis (Sawada et al., 2009; Uchida et al., 2020) (Figure 4; Table S6; Data S1). Based on our hypothesis that GGP1 fundamentally degrades GSH and GGCT2;1 accelerates GSH degradation under sulfur deficiency, ggp1‐1 and ggct2;1–2 single mutants were used for the analysis. These plants were grown under normal, sulfur‐deficient, and nitrogen‐deficient conditions because amino acid supply from GSH may play a more important role under these stresses and because the expression of GGCT2;1 and GGP1 is promoted under sulfur and nitrogen deficiency, respectively, as shown below. Out of 443 compounds examined, 223 compounds were detected and 134 compounds showed significant differences between wild‐type plants and any of the mutants. In the metabolites involved in sulfur assimilation, higher OAS levels were observed in the ggp1 and ggct2;1 mutants grown under sulfur deficiency (Figure 4a). OAS, a substrate for Cys synthesis together with sulfide, accumulates under sulfur deficiency and functions as a signaling molecule (Aarabi et al., 2020; Ohkama‐Ohtsu et al., 2004; Watanabe et al., 2021). In our experiments, OAS levels increased 9.3‐fold under sulfur deficiency in wild‐type plants, and this accumulation was significantly higher in the ggp1 and ggct2;1 mutants, suggesting that these mutants might sense stronger sulfur deficiency. The levels of 5‐oxoproline, a product of the GGCT pathway, were increased in the ggp1 mutant under control or sulfur‐deficient conditions, but decreased under nitrogen deficiency; therefore, it remains unclear whether the GGCT pathway was activated in the ggp1 mutant.
Figure 4.
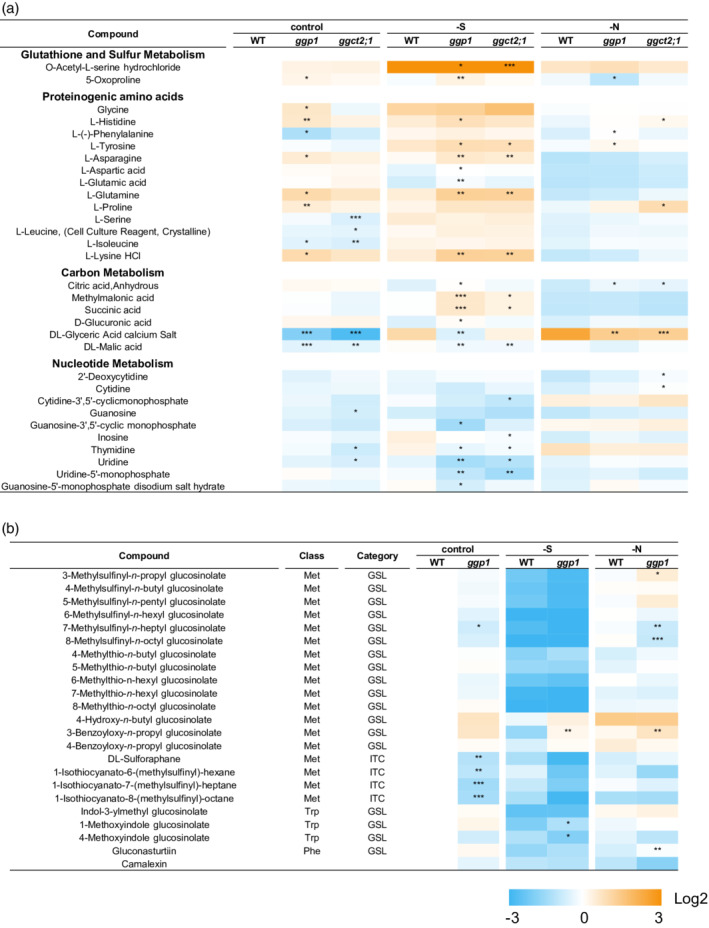
Changes in metabolite contents in wild‐type (WT), ggp1, and ggct2;1 plants. Plants were grown in liquid culture and utilized for widely targeted metabolomics. (a) Changes in metabolites involved in glutathione and sulfur, amino acid, carbon, or nucleotide metabolism. (b) Changes in glucosinolate, isothiocyanate, and camalexin levels in the ggp1‐1 mutant. Asterisks indicate significant differences from the WT grown under the same conditions; *P < 0.05, **P < 0.01, ***P < 0.001, Dunnett's test.
Several amino acids showed higher concentrations in the ggp1 mutant, but some showed lower concentrations (Figure 4a). These changes were only slightly observed under nitrogen starvation. This was different from our expectation that amino acid concentrations would decrease because GSH catabolism functions also as nitrogen distribution and because the GGP1 transcript levels were even increased under nitrogen deficiency. This possibly means that the perturbation of GSH degradation induced some change in nitrogen metabolism. Lack of GGP1 or GGCT2;1 also led to different concentrations of compounds related to carbon and nucleotide metabolism, especially under sulfur deficiency. Besides these compounds, many metabolites including the ones related to flavonoid or lysine metabolisms showed different concentrations in the ggp1 or ggct2;1 mutant (Table S6). The broader relevance of GSH degradation to plant metabolism is of particular interest.
The contents of GSLs and their degradation products, isothiocyanates (ITCs), were also affected by GGP1 disruption (Figure 4b). This was expected because GGP1 also functions in GSL biosynthesis (Geu‐Flores et al., 2011). In the ggp1 mutants, many methylsulfinyl‐type GSLs, especially those with long side chains, showed significant decreases or a tendency to decrease: 6‐methylsulfinyl‐n‐hexyl‐glucosinolate (6MSH), 7‐methylsulfinyl‐n‐heptyl‐glu‐cosinolate (7MSH), and 8‐methylsulfinyl‐n‐octyl‐glucosinolate (8MSO). This result, too, was consistent with the report by Geu‐Flores et al. (2011), showing that these compounds decreased more significantly in the ggp1/ggp3 double mutant. In this experiment, we further found that the levels of sulforaphane, 1‐isothiocyanato‐6‐(methylsulfinyl)‐hexane, 1‐isothiocyanato‐7‐(methylsulfinyl)‐heptane, and 1‐isothio‐cyanato‐8‐(methylsulfinyl)‐octane, which are ITCs produced from 4MSB, 6MSH, 7MSH, and 8MSO, respectively, significantly decreased in the ggp1 mutant. Lastly, GGP1 appears to be also involved in camalexin synthesis, as the camalexin levels in the ggp1 mutants showed a tendency to decrease, although this decrease was not statistically significant, probably due to a lack of biological stress.
Gene expression levels in wild‐type plants and the ggp1 and ggct2;1 mutants under normal, sulfur‐deficient, and nitrogen‐deficient conditions
To gain a deeper understanding of the causes and effects of the metabolite changes in the ggp1 and ggct2;1 mutants, we investigated expression levels of genes involved in sulfur or GSH metabolism under control, sulfur‐deficient, and nitrogen‐deficient conditions (Figure 5).
Figure 5.
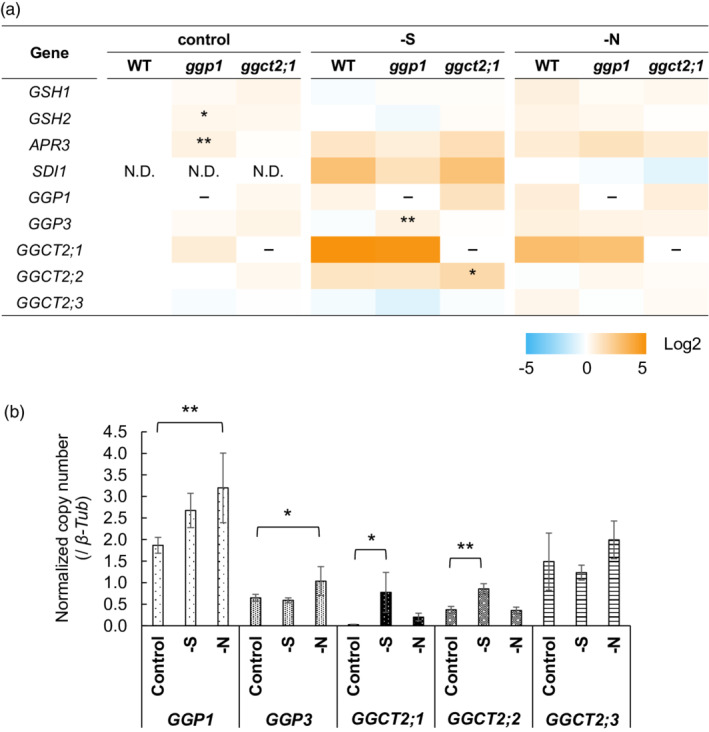
Gene expression levels in the wild type (WT) and the ggp1 and ggct2;1 mutants under normal, sulfur‐deficient, or nitrogen‐deficient conditions. Plants were grown by liquid culture for 14 days, and gene expression levels were analyzed by quantitative RT‐PCR using the absolute quantification method (n = 4–6). Expression levels were normalized to β‐tubulin. (a) Relative gene expression compared with the WT grown under control conditions (or nitrogen deficiency for SDI1) (logarithmic scale). Asterisks indicate significant differences from the WT grown under the same conditions; *P < 0.05, **P < 0.01, Dunnett's test. (b) Detailed results of GGP and GGCT expression levels in the WT (linear scale). The values and error bars represent the mean and standard deviation. Asterisks indicate significant differences from the WT grown under control conditions; *P < 0.05, **P < 0.01, Dunnett's test.
We first confirmed that GSH accumulation in the ggp1 and ggct2;1 mutants was not caused by promoted GSH synthesis but by disturbed GSH degradation. The expression levels of GSH1 and GSH2 were mostly similar between wild‐type plants and the ggp1 and ggct2;1 mutants, except for the slight increase in GSH2 expression (1.3‐fold) in the ggp1 mutant (Figure 5a), suggesting that GSH accumulation in the mutants was not caused by accelerated GSH synthesis.
We further analyzed the SDI1 and APR3 expression levels, which respond to OAS accumulation (Aarabi et al., 2020; Watanabe et al., 2021), because OAS is more greatly accumulated in the ggp1 and ggct2;1 mutants than in wild‐type plants under sulfur deficiency (Figure 4a). The SDI1 and APR3 expression levels were not significantly elevated in the mutants under sulfur deficiency (Figure 5a); however, this does not necessarily contradict the observed OAS accumulation as discussed later. On the other hand, under normal conditions, the expression of APR3, which encodes the rate‐limiting enzyme in the assimilatory reduction of sulfate (Watanabe et al., 2021), was significantly higher in the ggp1 mutant, suggesting that sulfur assimilation was possibly enhanced in the ggp1 mutant to complement the perturbed GSH degradation.
We also analyzed the expression levels of GGPs and GGCTs, including their responses to sulfur or nitrogen deficiency in wild‐type plants (Figure 5a,b), because the amino acid supply from GSH degradation may be facilitated under these conditions. Joshi et al. (2019) reported that GGCT2;1 expression increased 31‐fold under sulfur deficiency compared with control conditions (Figure 5b). Interestingly, the GGCT2;2 transcript levels significantly increased 2.3‐fold in the same comparison. On the other hand, GGP1 and GGP3 expression levels in wild‐type plants were slightly upregulated under nitrogen deficiency. In a comparison of wild‐type plants and mutants, the expression levels of GGP3 and GGCT2;2 were increased in the ggp1 and ggct2;1 mutants under sulfur deficiency, respectively (Figure 5a), suggesting that GGP3 and GGCT2;2 might complement the functions of GGP1 and GGCT2;1 under these conditions.
DISCUSSION
GSH degradation activity of GGP1 and GGP3
In the present study, we found that GGPs, enzymes that were originally reported to process GSH conjugates in the biosynthesis of GSLs and camalexin (Geu‐Flores et al., 2009), also degrade GSH. The yeast mutants defective in GSH degradation, dug2Δ or dug3Δ, were complemented by Arabidopsis GGP1 (Figure 1a) and grew on medium where GSH was the sole sulfur source. In addition, recombinant GGP1 and GGP3 proteins degraded GSH (Figure 1b; Table 1). The K m values of GGP1 and GGP3 for GSH were 5.0 mm and 3.7 mm, which are reasonable because the cytosolic GSH concentration ranges from 2.8 mm to 4.5 mm (Koffler et al., 2013). This reasonability is confirmed by the fact that the K m values of GGCTs are 1.9 mm for GGCT2;1 (Paulose et al., 2013), 1.7 mm for GGCT2;2 (Kumar et al., 2015), and 6.7 mm (Table 1) or 4.9 mm (Kumar et al., 2015) for GGCT2;3. The activities of GGP1 and GGP3 were moderate because their k cat values were 1.1‐fold and 3.8‐fold higher than that of GGCT2;3 (Table 1), respectively, whereas the k cat value of GGCT2;2 was 5.7‐fold higher than that of GGCT2;3 (Kumar et al., 2015).
Another remarkable characteristic of GGPs is the abundance of GGP1 transcripts. The GGP1 transcript level was the highest among GGPs and GGCTs in rosette leaves of plants in the reproductive stage (Figure 2), which occupied the highest volume in the plant body. The GGP1 transcript was also the most abundant in 2‐week‐old liquid‐cultured plants (Figure 5b). Additionally, in The Arabidopsis Information Resource (TAIR) database (http://www.arabidopsis.org/), the numbers of expressed sequence tags associated with GGP1, GGP3, GGCT2;1, GGCT2;2, and GGCT2;3 were 335, 66, 13, 118, and 52, respectively, and five clones that complemented the dug2Δ or dug3Δ mutants in our screening all corresponded to GGP1.
Considering these facts, it is highly possible that GGPs, especially GGP1, substantially degrade GSH in planta. This hypothesis was verified using ggp1 knockout mutant plants. ggp1 mutants accumulated significantly more GSH than wild‐type plants (Figure 3a), indicating that GGP1 contributes to GSH degradation in plants. This cannot be attributed to GSH synthesis because expression levels of the genes involved in GSH synthesis, GSH1 and GSH2, were not affected in the ggp1 and ggct2;1 mutants in most cases (Figure 5a); in particular, there was no significant difference in the expression levels of GSH1, which is considered to be important for the GSH synthesis rate (Noctor et al., 2012). Additionally, a decreased GSH degradation rate was actually observed in the ggp1/ggct2;1 mutant (Figure 3e). Our metabolome data also support the involvement of GGP1 in GSH degradation (Figure 4). Although many GSLs and ITCs showed a significant decrease or a tendency to decrease in ggp1 mutant plants (Figure 4a), which is consistent with the previous findings that GGPs are involved in GSL biosynthesis (Geu‐Flores et al., 2011), some results cannot be explained in the context of GSL biosynthesis; rather, they can be well explained in terms of GSH degradation. OAS, a signaling molecule for sulfur starvation (Aarabi et al., 2020; Ohkama‐Ohtsu et al., 2004; Watanabe et al., 2021), was accumulated under sulfur deficiency, and this accumulation was more conspicuous in the ggp1 mutant (Figure 4a), suggesting that it might sense stronger sulfur starvation. This result cannot be explained by the impaired GSL synthesis because OAS levels did not differ between wild‐type plants and the myb28myb29 mutant, whose Met‐derived GSL level was below 0.1% of that of wild‐type plants (Li et al., 2013). Instead, it is more likely that this OAS accumulation was caused by perturbed GSH degradation, considering that the ggct2;1 mutant similarly showed higher OAS levels under sulfur deficiency. Although the expression levels of OAS‐responsive genes such as APR3, SDI1, and GGCT2;1 (Aarabi et al., 2020; Watanabe et al., 2021) were not significantly affected by this OAS accumulation (Figure 5a), this can be well explained according to the report by Sogawa et al. (2005). They reported that additional OAS application does not induce sulfur‐responsive gene expression when plants are already exposed to sulfur deficiency, and also found that GSH negatively regulates sulfur‐responsive gene expression. Therefore, it is not surprising that the ggp1 mutant with high GSH concentration did not show increased expression of OAS‐responsive genes under sulfur starvation, even if it sensed stronger sulfur deficiency and accumulated OAS.
Overview of GSH degradation in Arabidopsis
Our findings suggest that Arabidopsis has five cytosolic GSH degradation enzymes, GGP1, GGP3, GGCT2;1, GGCT2;2, and GGCT2;3. Their functional differences need to be elucidated. We conducted expression analysis and found that the expression patterns of GGPs and GGCTs differed depending on the organ type (Figure 2). The expression of GGP1 and GGP3 was higher in mature organs than in young ones (Figure 2a,c). In particular, GGP1 has the highest transcript levels among GGPs and GGCTs, and is considered to substantially degrade GSH in plants, as discussed in the previous section. GGPs are localized to the vascular tissues of leaves (Geu‐Flores et al., 2011), so their physiological role is to degrade GSH and distribute constituent amino acids for protein and metabolite synthesis in the vascular tissue of leaves. Cys is the precursor molecule of numerous sulfur‐containing metabolites such as Met, vitamins, cofactors, and Fe‐S clusters, and Met can be further processed into S‐adenosyl‐l‐Met or ethylene (Romero et al., 2014).
In contrast, the expression of GGCT2;2 was higher in young organs than in old ones; for example, it was higher in young leaves than in mature leaves (Figure 2a), in cauline leaves than in rosette leaves (Figure 2c), and in lower roots than in middle or upper roots (Figure 2d). These data mostly coincide with public microarray data visualized by Genevestigator (https://www.genevestigator.com/) (Figure S4). GGCT2;2 has a high affinity and high activity compared to other GGPs and GGCTs. The K m of GGCT2;2 is 1.7 mm (Kumar et al., 2015), which is lower than those of GGP1 (5.0 mm, Table 1), GGP3 (3.7 mm, Table 1), and GGCT2;3 (6.7 mm, Table 1; 4.9 mm, Kumar et al., 2015). The k cat of GGCT2;2 was approximately 5.7‐fold higher than that of GGCT2;3 (Kumar et al., 2015), whereas the k cat values of GGP1 and GGP3 were 1.1‐fold and 3.8‐fold higher than that of GGCT2;3, respectively (Table 1). Therefore, GGCT2;2 may actively degrade GSH in developing organs and provide constituent amino acids to support plant growth. The involvement of GGCT2;2 in plant growth can be inferred from the fact that GGCT2;1 is explicitly expressed under sulfur deficiency and affects root architecture (Joshi et al., 2019). It would be interesting to investigate whether GGCT2;2 is related to root architecture under normal conditions.
Under sulfur deficiency, GGCT2;1 and GGCT2;2 transcript levels increased 31‐fold and 2.3‐fold, respectively (Figure 5), so their contribution, especially that of GGCT2;1, will increase. Because the GSH concentration is low under sulfur deficiency, GGCT2;1 and GGCT2;2, which have a higher affinity for GSH than GGP1, GGP3, and GGCT2;3 (Table 1; Paulose et al., 2013; Kumar et al., 2015), may be more efficient under these conditions.
There is another crucial difference between GGPs and GGCTs. Whereas GGPs directly produce Glu without ATP consumption (Geu‐Flores et al., 2011), GGCTs produce 5‐oxoproline, which requires ATP to be converted to Glu (Ohkama‐Ohtsu et al., 2008). Therefore, the degradation pathway through GGPs is more energy‐efficient than the pathway through GGCTs. It is conceivable that plants fundamentally degrade GSH with GGPs and utilize GGCTs only when the demand for GSH degradation is high, for example, in developing organs or under sulfur deficiency. Notably, our findings suggest that plants utilize two types of GSH degradation pathways to meet the demand for constituent amino acids with less energy.
Based on the above discussion, we developed a model of GSH degradation in mature organs (Figure 6). In this model, cytosolic GSH is considered to be degraded mainly by GGP1 (Figure 6a), and the degradation is boosted by GGCT2;1 under sulfur deficiency (Figure 6b). This seems rational because the GGP pathway is energy‐efficient yet its affinity to GSH is low, whereas the GGCT pathway is energy consuming yet its affinity is high. This model also takes into account GSH conjugate processing by GGPs, which will be discussed in the next section.
Figure 6.
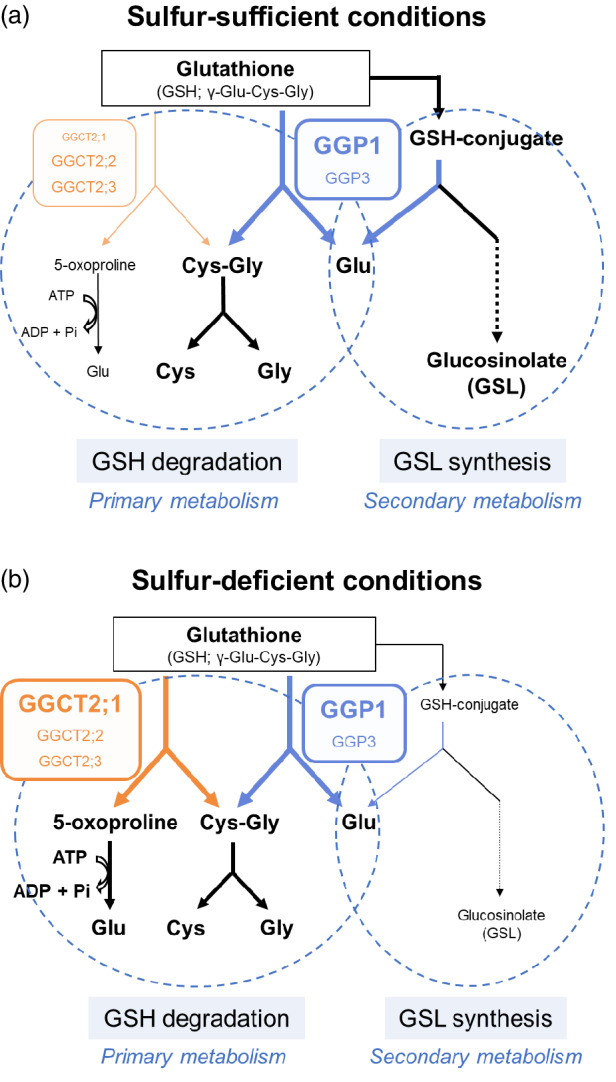
Models of GSH degradation and GSL synthesis in Arabidopsis. (a) Under sulfur‐sufficient conditions, GGP1 is estimated to play a central role in GSH degradation and to process GSH conjugates in the GSL synthesis pathway. (b) Under sulfur‐deficient conditions, the expression of GGCT2;1 is promoted, and GGCT2;1 accelerates GSH degradation. GSL synthesis is reduced under these conditions.
GSH degradation may have a broad impact on plant metabolism. Our metabolome analysis showed that amino acid concentrations were perturbed in the ggp1 mutant under control and sulfur‐deficient conditions, and metabolites involved in carbon and nucleotide metabolism were also affected under sulfur deficiency (Figure 4a). These changes were similar to those of the ggct2;1 mutant under sulfur deprivation, suggesting that these changes were caused by disturbance of GSH degradation, not GSL synthesis. In other words, it seems that plants had to deal with the perturbed GSH degradation by modifying their metabolism. Nonetheless, it is not easy to identify what brings about these changes in the ggp1 or ggct2;1 mutant: it may be altered amino acid supply or an altered redox environment caused by GSH accumulation. Considering that some changes were observed only under sulfur starvation, one possibility is that sulfur metabolism was affected by perturbed GSH degradation, and this further influenced nitrogen or carbon metabolism because they are cooperatively regulated (Courbet et al., 2019; Li et al., 2020). Decreased nucleoside and nucleotide levels possibly mean that replication or transcription activity was changed in the ggp1 and ggct2;1 mutants under sulfur deficiency. It is also possible that other factors such as other constituent amino acids, Glu and Gly, or the redox environment affected metabolisms, but this is not clear at this time. It is of great interest to investigate how far, how much, and through which pathway GSH degradation has an influence on plant physiology.
Dual roles of GGP1 and GGP3 in GSH degradation and GSL synthesis
GGP1 and GGP3 were initially reported as enzymes that process GSH conjugates in the biosynthesis of GSLs and camalexin (Geu‐Flores et al., 2009) and therefore considered to have dual roles in both GSH degradation and GSL and camalexin synthesis, that is, primary and secondary sulfur metabolism. The K m value of GGP1 is appropriate for both GSH (Table 1) and a GSH conjugate (37 ± 8 μm for S‐[(Z)‐phenylaceto‐hydroximoyl]‐l‐glutathione (GS‐B); Geu‐Flores et al., 2009). According to the ATTED‐II database (Obayashi et al., 2018, https://atted.jp/), GGP1 is co‐expressed with genes involved in sulfur assimilation as well as genes involved in GSL synthesis: ATP sulfurylase 1 (APS1, AT3G22890), APS reductase 1 (APR1, AT4G04610), APS kinase 1 (APK1, AT2G14750), APS kinase 2 (APK2, AT4G39940), ATP sulfurylase 3 (APS3, AT4G14680), and serine acetyltransferase2;2 (SERAT2;2, AT3G13110). Our metabolome data also support the hypothesis of the dual roles of GGPs (Figure 4). As discussed in the previous section, many GSLs and ITCs showed significant decreases or a tendency to decrease in the ggp1 mutant, as already reported by Geu‐Flores et al. (2011); however, some results, such as the accumulation of OAS, a signaling molecule for sulfur starvation, are better explained by the lack of GSH degradation. Finally, but importantly, the ggp1 mutants showed higher GSH levels (Figure 3a). Analyzing these facts, GGPs are indeed involved in the degradation of both GSH and GSH conjugates in plants.
Therefore, we further evaluated the feasibility of the acceptance of GSH and GSH conjugates by GGP1 by analyzing its three‐dimensional structure and substrate recognition mechanism. The predicted protein structure of GGP1 is available in the AlphaFold Protein Structure Database (Figure 7, left) (Varadi et al., 2021). GGP1 belongs to the peptidase family C26 (γ‐glutamyl hydrolase family) in the MEROPS database with an ID of C26.A05 (Rawlings et al., 2014). GGP1 has a catalytic triad consisting of C100, H192, and E194 (magenta in Figure 7), which are completely conserved in enzymes belonging to the C26 family (Figure S5). In the crystal structure of anthranilate synthase from Serratia marcescens, a covalently bound glutamyl thioester to the catalytic cysteine residue was observed in the TrpG (glutamine amidotransferase) subunit (cyan in Figure 7, right) (Spraggon et al., 2001). Based on this catalytic intermediate structure, we modeled the GSH conjugate GS‐B into the active site of GGP1 (yellow in Figure 7, right). The γ‐Glu moiety of GS‐B was recognized by the side chain of Q104 and the main chain amides of Q156 and D157. Q104 is also completely conserved in the C26 family, which consists of enzymes acting on substrates with a γ‐Glu moiety (Figure S5). The Gly moiety of the GSH conjugate was placed near R206, which can form a salt bridge with the carboxylate group (Figure 7, right). The phenyl group of GS‐B was placed near the side chain of Y212. The presence or absence of this π‐π interaction may account for the different K m values of GGP1 toward GS‐B or GSH. R206 is conserved in GGP3 and the homologs in soybean (Glycine max) and rice (Oryza sativa) but not in moss (Physcomitrium patens) (see Figures S5 and S6), suggesting that these plant homologs of GGP1 can also hydrolyze GSH, except in moss. This may further imply that their ancestral enzyme originally evolved for GSH degradation after the appearance of vascular plants, as GSLs are metabolites specifically found in and around Brassicales. In contrast, Y212 was only present in GGP1. However, GGP3 has a Leu residue (L216) at this position, and hydrophobic interactions with GSLs derived from hydrophobic amino acids (Phe, Trp, and Met) will be retained. In summary, it was presumed based on the predicted three‐dimensional structure that GGP1 and GGP3 are able to hydrolyze the γ‐glutamyl peptide bond of GSH as well as GSH conjugates.
Figure 7.
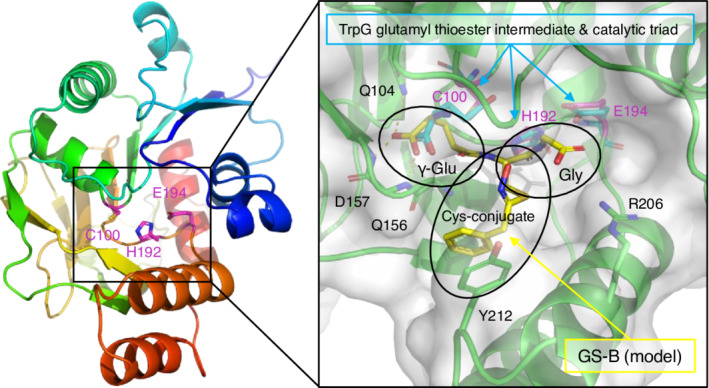
Predicted structure of GGP1 and interaction with GS‐B. The protein structure of GGP1 was obtained from the AlphaFold Protein Structure Database. Left, the overall structure of GGP1 in rainbow colors (blue and red for the N‐ and C‐termini, respectively). The catalytic triad residues are shown as magenta sticks. Right, the active site of GGP1 (green). The molecular surface of GGP1 (white) is transparently shown. A GSH conjugate (GS‐B, yellow sticks) was manually modeled with a reference to the covalently bound glutamyl thioester intermediate of the glutamine amidotransferase (TrpG) subunit of anthranilate synthase from S. marcescens (PDB ID: 1I7Q).
Based on these results, we conclude that GGPs catalyze two reactions in plants (Figure 6). In this case, the priority of the reactions likely varies depending on the substrate concentration. The affinity of GGPs for GSH conjugates is much higher than that for GSH, so when GSH conjugates accumulate, processing of GSH conjugates will be prioritized over GSH degradation, leading to an automatic reduction in GSH degradation. Although it is well known that GSLs and camalexin function in alleviating biotic stresses, high concentrations of GSH are also crucial for resistance to viruses, bacteria, and fungi because GSH functions in ROS scavenging and signaling (Hernández et al., 2017). Therefore, it is very reasonable for plants experiencing biotic stress to synthesize GSLs and camalexin and simultaneously inhibit GSH degradation. Additionally, it is notable that Ralstonia solanacearum, a pathogenic bacterium, produces the virulence effector protein RipAY, which has GGCT activity (Mukaihara et al., 2016). GSH degradation by RipAY causes GSH depletion in plant cells and increases susceptibility in the early stages of infection.
In contrast, GSH degradation may be prioritized over GSH conjugate degradation under sulfur starvation (Figure 6b) because the expression of upstream genes in GSL synthesis pathways is downregulated under sulfur‐deficient conditions (Aarabi et al., 2016), which probably leads to the decrease in GSH conjugates. It is reasonable that GSH degradation is activated under sulfur deficiency because GSH degradation functions as the distribution of stored organic sulfur. In addition, it has recently been shown that GSLs are degraded under sulfur deficiency to reallocate sulfur atoms, and that GSH is required for degradation (Sugiyama et al., 2021). Therefore, GSH consumption for GSL synthesis may decrease under sulfur deficiency, and GSH may be utilized for GSL degradation. Overall, the dual functions of GGPs potentially contribute to the sulfur flow shift from sulfur consumption in GSL synthesis to sulfur reallocation through GSH and GSL degradation under sulfur deficiency.
EXPERIMENTAL PROCEDURES
Yeast strains and screening
The S. cerevisiae strains used in this study, listed in Table S1, were obtained from EUROSCARF (http://www.euroscarf.de/). Strains Y05729 (dug2Δ) and Y05729 (dug3Δ) were constructed from BY4741 by insertional mutagenesis (Winzeler et al., 1999). All strains were missing MET15 and were unable to assimilate inorganic sulfur.
An A. thaliana cDNA library was cloned into the expression plasmid pFL61 and transformed into competent yeast cells of dug2Δ or dug3Δ strains (Minet et al., 1992) using the lithium acetate method (Ito et al., 1983). The transformants were screened on minimal selection plates (SD‐Ura), in which all sulfate ions were replaced with chloride ions and supplemented with 200 μm GSH. All plates were incubated at 28°C for 3–5 days. Transformation efficiency was calculated from yeast grown on SD‐Ura plates supplemented with 200 μm Met. The plasmids transformed into yeast were extracted using a Zymoprep™ Yeast Plasmid Miniprep I kit (Zymo Research, http://www.zymoresearch.com/).
The inserts in the clones were sequenced with ABI PRISM 3130 and 3500 Genetic Analyzers (Applied Biosystems, Waltham, MA, USA) and primers pGK5 and pGK3 (Table S2) (Nozawa et al., 2006).
Construction of the plasmids for transformation into yeast
Total RNA was extracted from A. thaliana using an RNeasy Plant Mini Kit (Qiagen, Hilden, Germany), followed by cDNA synthesis with SuperScript III reverse transcriptase (Invitrogen, Waltham, MA, USA, https://www.thermofisher.com/) and the Oligo (dT)16 primer. The ORF sequences for GGP1 (AT4G30530), GGCT2;1 (AT5G26220), GGCT2;2 (AT4G31290), and GGCT2;3 (AT1G44790) were amplified from the A. thaliana cDNA template using the primer sets listed in Table S2. The amplified product for GGP1 was subcloned between the EcoRI and XhoI sites, and those for GGCT2;1, GGCT2;2, and GGCT2;3 were subcloned between the BamHI and EcoRI sites of the p416TEF yeast expression vector (Mumberg et al., 1995). These plasmids were transformed into dug2Δ and dug3Δ yeast strains using the lithium acetate method (Ito et al., 1983).
Construction of E. coli expression strains
The coding sequences (CDSs) of GGP1, GGP3 (AT4G30550), GGCT2;1, GGCT2;2, and GGCT2;3 were obtained from TAIR (https://www.arabidopsis.org/), and they were amplified by PCR using KOD‐Plus‐Neo (TOYOBO, Osaka, Japan) with the primers listed in Table S3. The PCR products were cloned into the entry vector pENTR/D‐TOPO (Invitrogen), and the inserts were confirmed by Eurofins Genomics (Tokyo, Japan) with M13 primers (Table S3). Subsequently, each CDS was cloned into the destination vector pDEST17 (Invitrogen) via LR recombination. Finally, the constructed plasmids were transformed into E. coli expression strain BL21‐AI (Invitrogen). All operations were performed according to the manufacturer's protocols.
Expression and purification of the recombinant proteins
BL21‐AI possessing each construct was cultured in LB medium containing 100 μg ml−1 ampicillin up to an OD600 value of approximately 0.6 at 37°C and 150 rpm. l‐Arabinose was added to a final concentration of 0.02% (w/v), and the cells were cultured for another 17 h at 18°C at 120 rpm. Cells were harvested by centrifugation at 2500 g for 5 min at 4°C. The target proteins were obtained as N‐terminal His‐tagged proteins, followed by purification using a His60 Ni Gravity Column Purification Kit (Clontech Laboratories, Mountain View, CA, USA), according to the manufacturer's instructions, except that 1 mm phenylmethanesulfonyl fluoride was added to the extraction and wash buffers of the kit. Of the five proteins expressed in E. coli, GGP1, GGP3, and GGCT2;3 were collected from the soluble fraction and used for subsequent experiments. The proteins were dialyzed overnight in a buffer containing 50 mm Tris–HCl (pH 8.0) and 150 mm NaCl at 4°C. The results were confirmed using SDS‐PAGE (Figure S2). Protein concentrations were determined using Bradford reagent (Bio‐Rad, Hercules, CA, USA), with bovine serum albumin as the standard.
Activity assay of recombinant proteins for GSH or other γ‐glutamyl compounds
Kinetic parameters were determined using 0.5 μg of GGP1, 0.25 μg of GGP3, or 2.2 μg of GGCT2;3. The proteins were incubated in 50 μl of reaction mixture containing 50 mm Tris–HCl (pH 8.0) and 0.25 to 15.0 mm GSH for 30 min at 37°C, and the reaction was terminated by adding 10 μl of 1.5 m HCl. The solution was centrifuged at 11 000 g for 15 min at 4°C, and the Cys‐Gly released in the supernatant was quantified by HPLC, as described below. Kinetic parameters were calculated using GraphPad Prism 9 software (GraphPad Software, San Diego, CA, USA). For degradation activity for other γ‐glutamyl compounds, 0.5 μg of GGP1 or 0.25 μg of GGP3 was incubated in 50 μl of reaction mixture containing 50 mm Tris–HCl (pH 8.0) and 5.0 mm GSH, 5.0 mm γ‐Glu‐Cys (Sigma‐Aldrich), 5.0 mm γ‐Glu‐Ala (Sigma‐Aldrich, St. Louis, MO, USA), or 2.5 mm GSSG. The reaction was conducted as described previously. As described below, the released Cys‐Gly was quantified for GSH, Cys was quantified for γ‐Glu‐Cys, and Ala was quantified for γ‐Glu‐Ala.
Analysis of Ala
Ala in the reaction solutions of activity assays was derivatized with the AccQ‐Fluor Reagent Kit (Waters, Milford, MA, USA, http://www.waters.com/waters/home) and measured by HPLC using an AccQ‐Tag column (Waters) according to the manufacturer's instructions.
Thiol analysis with HPLC
Thiols extracted from plant tissues with 0.01 m HCl or thiols in the reaction solutions of activity assays were quantified by HPLC after derivatization with monobromobimane (Minocha et al., 2008; Nishida et al., 2016). The HPLC system (Shimadzu, Japan) consisted of a system controller (CBM‐20A), two pumps (LC‐20 AD), a degasser (DGU‐20A3), an autosampler (SIL‐20A), a column oven (CTO‐20 AC), and a fluorescence detector (RF‐20A). The column was a Shim‐pack FC‐ODS (4.6 mm × 150 mm; Shimadzu, Japan).
Hydroponic culture of plants for sampling at vegetative and reproductive stages
Arabidopsis thaliana Col‐0 wild‐type plants were grown hydroponically in 50‐ml conical tubes at 22°C under 16 h light (100 μmol−1 m−2 sec−1)/8 h dark as described by Ohkama‐Ohtsu, Zhao, et al. (2007) using MGRL medium (Fujiwara et al., 1992; Hirai et al., 1995). The vegetative‐stage samples were grown for 22 days. The three oldest leaves from the first true leaf were collected as mature leaves, and the three youngest leaves were collected as young leaves. For the reproductive stage, samples were grown for 33 days, and flowers + buds, siliques 1–2 cm in length, stems, and all cauline and rosette leaves were collected separately. Roots were divided into three equally long parts, and the upper, middle, and lower roots were collected separately. Samples were immediately frozen in liquid nitrogen and stored at −80°C until RNA extraction.
Liquid culture of plants in flasks
Plants were grown in flasks, as described by Xiang and Oliver (1998), using MGRL medium (Fujiwara et al., 1992). For sulfur‐deficient treatments, all sulfate ions in the MGRL medium were replaced with an equal molar concentration of chloride ions (Hirai et al., 1995). In nitrogen‐deficient treatments, all nitrate ions were replaced with an equal molar concentration of chloride ions. In the nitrogen‐deficient experiment, 12‐day‐old plants grown in standard medium were transferred to nitrogen‐deficient medium and grown for an additional 2 days. In sulfur‐deficient experiments, 10‐day‐old plants grown in the standard medium were transferred to sulfur‐deficient medium and grown for an additional 4 days. Control plants were grown for 14 days in standard MGRL medium, and the medium was replaced after 10 days. The entire bodies of plants in each flask were pooled, immediately frozen with liquid nitrogen, and stored at −80°C until analysis.
RNA extraction and quantitative PCR analysis
RNA was extracted from frozen samples using the RNeasy Plant Mini Kit (Qiagen, Hilden, Germany), followed by reverse transcription using the PrimeScript™ RT Reagent Kit with gDNA Eraser (Perfect Real Time) (Takara Bio, Shiga, Japan). Quantitative PCR was conducted using the KAPA SYBR FAST qPCR Master Mix (2×) Kit (KAPA Biosystems, Wilmington, MA, USA) and a LightCycler 96 (Roche Diagnostics, Basel, Switzerland) following the manufacturer's protocols. Basically, the absolute quantification method was adopted using serial dilutions of DNA with known copy numbers for the standard curves; only for the expression analysis in BSO treatment, the relative quantification method was adopted. The copy number of β‐tubulin (TUB4, AT5G44340) was used as the internal standard. The primers used are shown in Table S4. The normalized copy number was determined by dividing the copy number of each gene by that of β‐tubulin.
BSO treatment and time‐course analysis
Plants were grown in flasks containing 1/2 Murashige and Skoog medium under constant light at 22°C for 10 days (Xiang & Oliver, 1998). To inhibit GSH synthesis, 1 mm BSO (final concentration) was added, and plants were sampled at each time point and used for further thiol analysis or expression analysis as described above.
Screening of Arabidopsis T‐DNA knockout mutants for ggp1 and ggct2;1
Unless otherwise indicated, A. thaliana plants were grown on rockwool in a plastic container at 22°C under a 16 h light (150 μmol m−2 sec−1)/8 h dark photoperiod to screen mutant lines harboring T‐DNA homozygously. The primers used for screening the ggp1 and ggct2;1 mutant plants are shown in Table S5. The ggp1‐1 mutant, which was used by Geu‐Flores et al. (2011), was provided by GABI‐kat (http://www.gabi‐kat.de/, stock no. GK‐319F10) (Rosso et al., 2003). Homozygous plants were screened by PCR using the gene‐specific primers GK‐319‐F and GK‐319‐R and the T‐DNA left‐border primer GABI‐8474 (http://www.gabi‐kat.de/). A lack of amplifiable transcripts of AT4G30530 in the ggp1‐1 mutant was reported by Geu‐Flores et al. (2011), and it was confirmed by quantitative RT‐PCR using primers At4g30530F and At4g30530R. The ggp1‐2 mutant (SALK_089634) was provided by the Salk Institute (Alonso et al., 2003) and was obtained from the Arabidopsis Biological Resource Center (ABRC, https://abrc.osu.edu/). Homozygous plants were screened using the gene‐specific primers SALK_089634F and SALK_089634R and the T‐DNA left‐border primer pROKr3 (Lin & Oliver, 2008). The lack of an amplifiable transcript of AT4G30530 in the ggp1‐1 mutant was confirmed by quantitative RT‐PCR using primers At4g30530F and At4g30530R. The ggct2;1–2 mutant (SALK_56007), which was used by Joshi et al. (2019), was provided by the Salk Institute (Alonso et al., 2003) and was obtained from the ABRC (https://abrc.osu.edu/). Homozygous plants were screened by PCR using the gene‐specific primers GGCT2,1F and GGCT2,1R and the T‐DNA left‐border primer pROKr3 (Lin & Oliver, 2008). A lack of an amplifiable transcript of AT5G26220 in the ggct2;1–2 mutant was reported by Joshi et al. (2019), and we confirmed this by RT‐PCR using the primers At5g26220F and At5g26220R. The ggp1‐1/ggct2;1–2 double knockout mutant was created by cross‐pollination and selection by PCR and RT‐PCR using the above primers.
Widely targeted metabolomics
Plants were grown in liquid culture under normal conditions, sulfur deficiency, and nitrogen deficiency, as described above (n = 4). A widely targeted metabolome analysis was conducted according to Uchida et al. (2020) with some modifications. Plant shoots and roots were lyophilized and used for extraction with 1 ml of extraction solvent (80% methanol [v/v], 0.1% formic acid [v/v]) per 4 mg of powdered tissue. Two internal standards were used: 8.4 nm lidocaine (positive ion mode) and 210 nm 10‐camphorsulfonic acid (negative ion mode). The extracted solution (25 μl) was further diluted using 75 μl of extraction solvent, and 25 μl of this diluted solution was dried and dissolved in 250 μl of LC–MS grade water. In the end, 100 μg ml−1 sample solution was obtained, and 1 μl of the solution was subjected to LC‐QqQ‐MS/MS analysis (LCMS‐8050, Shimadzu, Japan). The detection conditions of the metabolites were the same as described previously (Uchida et al., 2020). Metabolite signals normalized by those of respective internal standards were compared between wild‐type plants and the ggp1‐1 or ggct2;1–2 mutants using Dunnett's test at each nutrient condition. Metabolites showing a low signal‐to‐noise ratio (<3 compared to the signals in solvent control) or a high variance (relative standard deviation > 50%) in wild‐type plants at normal conditions were excluded. For the analysis of GSL‐related metabolites, 20 GSL species previously detected in Col‐0 (Brown et al., 2003) were verified.
Molecular modeling of GGP1
The predicted protein structures of GGP1 and GGP3 (version 2) were obtained from the AlphaFold Protein Structure Database (https://alphafold.ebi.ac.uk/entry/Q9M0A7 and https://alphafold.ebi.ac.uk/entry/Q9M0A5). The initial molecular model of GS‐B was generated using PCMODEL (Serena Software, Bloomington, IN, USA). The crystal structure of TrpG (PDB ID: 1I7Q, chain B) was superimposed on the GGP1 structure with catalytic Cys and His residues. The γ‐Glu moiety of the molecular model of GS‐B was superimposed on the glutamyl thioester group of TrpG with carboxylate carbon (C), amino nitrogen (N), Cα, Cβ, and Cγ atoms. The conformation of the remaining part of GS‐B was manually changed using PyMOL (Schrödinger LLC, New York, NY, USA). The dihedral angles of the Cγ‐Cε bond of γ‐Glu and rotatable bonds of the Cys conjugate and Gly moieties were rotated to minimize the steric hindrance with the protein atoms by visually inspecting the molecular surface of GGP1. The modeled GS‐B structure was energy‐minimized using the optimizy.py plug‐in of PyMOL with the molecular mechanics features of Open Babel (O'Boyle et al., 2011).
AUTHOR CONTRIBUTIONS
TI, TK, KN, MU, and NO‐O designed the experiments and analyzed the data; TI, TK, KN, MU, SM, SA, KK, MS, and JI performed the experiments; AM‐N, RS, MYH, TY, and NO‐O supervised the experiments; SF performed protein structure analysis; TI, SF, AM‐N, RS, MYH, and NO‐O contributed to the writing of the manuscript.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supporting information
Data S1. Peak area matrix of all detected compounds in metabolome analysis.
Data S2. Annotation list for metabolome analysis.
Figure S1. The γ‐glutamyl cycle proposed in mammals.
Figure S2. Confirmation of the result of protein purification by SDS‐PAGE.
Figure S3. Gene expression in the ggp1/ggct2;1 mutant after buthionine sulfoximine (BSO) treatment.
Figure S4. Expression analysis based on a public microarray database.
Figure S5. Amino acid sequence alignment of GGP1 and its homologs.
Figure S6. Superimposition of the predicted protein structures of GGP1 (green) and GGP3 (cyan).
Table S1. List of yeast strains used in this study.
Table S2. Primers used for constructing and sequencing expression plasmids for yeast.
Table S3. Primers used for the construction of E. coli expression strains.
Table S4. Primers used for quantitative PCR.
Table S5. Primers used for screening ggp1 and ggct2;1 mutants and mRNA analysis.
Table S6. All significant changes in metabolite contents in wild‐type (WT), ggp1, and ggct2;1 plants.
ACKNOWLEDGMENTS
We thank Dr. Akira Nozawa (Ehime University) and Dr. Toru Fujiwara (The University of Tokyo) for kindly providing pFL61 and the method for yeast transformation. This work was partly supported by JSPS KAKENHI (grant numbers 15KT0028, 16K07639, and 19H02859 to NO‐O and 22H05573 to AM‐N).
DATA AVAILABILITY STATEMENT
The data supporting the findings of this study are available in this article and its supplementary materials.
REFERENCES
- Aarabi, F. , Kusajima, M. , Tohge, T. , Konishi, T. , Gigolashvili, T. , Takamune, M. et al. (2016) Sulfur deficiency‐induced repressor proteins optimize glucosinolate biosynthesis in plants. Science Advances, 2, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aarabi, F. , Naake, T. , Fernie, A.R. & Hoefgen, R. (2020) Coordinating sulfur pools under sulfate deprivation. Trends in Plant Science, 25, 1227–1239. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M. , Stepanova, A.N. , Leisse, T.J. , Kim, C.J. , Chen, H. , Shinn, P. et al. (2003) Genome‐wide insertional mutagenesis of Arabidopsis thaliana. Science, 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Bielecka, M. , Watanabe, M. , Morcuende, R. , Scheible, W.‐R. , Hawkesford, M.J. , Hesse, H. et al. (2014) Transcriptome and metabolome analysis of plant sulfate starvation and resupply provides novel information on transcriptional regulation of metabolism associated with sulfur, nitrogen and phosphorus nutritional responses in Arabidopsis. Frontiers in Plant Science, 5, 805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, P.D. , Tokuhisa, J.G. , Reichelt, M. & Gershenzon, J. (2003) Variation of glucosinolate accumulation among different organs and developmental stages of Arabidopsis thaliana. Phytochemistry, 62, 471–481. [DOI] [PubMed] [Google Scholar]
- Courbet, G. , Gallardo, K. , Vigani, G. , Brunel‐Muguet, S. , Trouverie, J. , Salon, C. et al. (2019) Disentangling the complexity and diversity of crosstalk between sulfur and other mineral nutrients in cultivated plants. Journal of Experimental Botany, 70, 4183–4196. [DOI] [PubMed] [Google Scholar]
- Deshpande, A.A. , Bhatia, M. , Laxman, S. & Bachhawat, A.K. (2017) Thiol trapping and metabolic redistribution of sulfur metabolites enable cells to overcome cysteine overload. Microbial Cell Factories, 4, 112–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Destro, T. , Prasad, D. , Martignago, D. , Lliso Bernet, I. , Trentin, A.R. , Renu, I.K. et al. (2010) Compensatory expression and substrate inducibility of γ‐glutamyl transferase GGT2 isoform in Arabidopsis thaliana. Journal of Experimental Botany, 62, 805–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti, M. , Destro, T. , Tosatto, S.C.E. , La Rocca, N. , Rascio, N. & Masi, A. (2009) Gamma‐glutamyl transferase in the cell wall participates in extracellular glutathione salvage from the root apoplast. The New Phytologist, 181, 115–126. [DOI] [PubMed] [Google Scholar]
- Foyer, C.H. & Noctor, G. (2011) Ascorbate and glutathione: the heart of the redox hub. Plant Physiology, 155, 2–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara, T. , Hirai, M.Y. , Chino, M. , Komeda, Y. & Naito, S. (1992) Effects of sulfur nutrition on expression of the soybean seed storage protein genes in transgenic petunia. Plant Physiology, 99, 263–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguli, D. , Kumar, C. & Bachhawat, A.K. (2007) The alternative pathway of glutathione degradation is mediated by a novel protein complex involving three new genes in Saccharomyces cerevisiae. Genetics, 175, 1137–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geu‐Flores, F. , Møldrup, M.E. , Böttcher, C. , Olsen, C.E. , Scheel, D. & Halkier, B.A. (2011) Cytosolic γ‐glutamyl peptidases process glutathione conjugates in the biosynthesis of glucosinolates and camalexin in Arabidopsis. Plant Cell, 23, 2456–2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geu‐Flores, F. , Nielsen, M.T. , Nafisi, M. , Møldrup, M.E. , Olsen, C.E. , Motawia, M.S. et al. (2009) Glucosinolate engineering identifies a γ‐glutamyl peptidase. Nature Chemical Biology, 5, 575–577. [DOI] [PubMed] [Google Scholar]
- Gong, Q. , Li, P. , Ma, S. , Indu Rupassara, S. & Bohnert, H.J. (2005) Salinity stress adaptation competence in the extremophile Thellungiella halophila in comparison with its relative Arabidopsis thaliana. The Plant Journal, 44, 826–839. [DOI] [PubMed] [Google Scholar]
- Griffith, O.W. & Meister, A. (1979) Potent and specific inhibition of glutathione synthesis by buthionine sulfoximine (S‐n‐butyl homocysteine sulfoximine). The Journal of Biological Chemistry, 254, 7558–7560. [PubMed] [Google Scholar]
- Grzam, A. , Martin, M.N. , Hell, R. & Meyer, A.J. (2007) γ‐Glutamyl transpeptidase GGT4 initiates vacuolar degradation of glutathione S‐conjugates in Arabidopsis. FEBS Letters, 581, 3131–3138. [DOI] [PubMed] [Google Scholar]
- Hell, R. & Bergmann, L. (1990) γ‐Glutamylcysteine synthetase in higher plants: catalytic properties and subcellular localization. Planta, 180, 603–612. [DOI] [PubMed] [Google Scholar]
- Hernández, J.A. , Barba‐Espín, G. & Diaz‐Vivancos, P. (2017) Glutathione‐mediated biotic stress tolerance in plants. In: Hossain, M.A. , Mostofa, M.G. , Diaz‐Vivancos, P. , Burritt, D.J. , Fujita, M. & Tran, L.‐S.P. (Eds.) Glutathione in plant growth, development, and stress tolerance. Cham: Springer International Publishing, pp. 309–329. [Google Scholar]
- Hirai, M.Y. , Fujiwara, T. , Chino, M. & Naito, S. (1995) Effects of sulfate concentrations on the expression of a soybean seed storage protein gene and its reversibility in transgenic Arabidopsis thaliana. Plant & Cell Physiology, 36, 1331–1339. [PubMed] [Google Scholar]
- Hubberten, H.M. , Drozd, A. , Tran, B.V. , Hesse, H. & Hoefgen, R. (2012) Local and systemic regulation of sulfur homeostasis in roots of Arabidopsis thaliana. The Plant Journal, 72, 625–635. [DOI] [PubMed] [Google Scholar]
- Ito, H. , Fukuda, Y. , Murata, K. & Kimura, A. (1983) Transformation of intact yeast cells treated with alkali cations. Journal of Bacteriology, 153, 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jez, J.M. , Cahoon, R.E. & Chen, S. (2004) Arabidopsis thaliana glutamate‐cysteine ligase: functional properties, kinetic mechanism, and regulation of activity. The Journal of Biological Chemistry, 279, 33463–33470. [DOI] [PubMed] [Google Scholar]
- Joshi, N.C. , Meyer, A.J. , Bangash, S.A.K. , Zheng, Z.L. & Leustek, T. (2019) Arabidopsis γ‐glutamylcyclotransferase affects glutathione content and root system architecture during sulfur starvation. The New Phytologist, 221, 1387–1397. [DOI] [PubMed] [Google Scholar]
- Kaur, H. , Ganguli, D. & Bachhawat, A.K. (2012) Glutathione degradation by the alternative pathway (DUG pathway) in Saccharomyces cerevisiae is initiated by (Dug2p‐Dug3p)2 complex, a novel glutamine amidotransferase (GATase) enzyme acting on glutathione. The Journal of Biological Chemistry, 287, 8920–8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur, H. , Kumar, C. , Junot, C. , Tolendano, M.B. & Bachhawat, A.K. (2009) Dug1p is a Cys‐Gly peptidase of the γ‐glutamyl cycle of Saccharomyces cerevisiae and represents a novel family of Cys‐Gly peptidases. The Journal of Biological Chemistry, 284, 14493–14502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koffler, B.E. , Bloem, E. , Zellnig, G. & Zechmann, B. (2013) High resolution imaging of subcellular glutathione concentrations by quantitative immunoelectron microscopy in different leaf areas of Arabidopsis. Micron, 45, 119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koprivova, A. & Kopriva, S. (2014) Molecular mechanisms of regulation of sulfate assimilation: first steps on a long road. Frontiers in Plant Science, 5, 589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalchuk, I. , Titov, V. , Hohn, B. & Kovalchuk, O. (2005) Transcriptome profiling reveals similarities and differences in plant responses to cadmium and lead. Mutation Research, 570, 149–161. [DOI] [PubMed] [Google Scholar]
- Kumar, A. , Tikoo, S. , Maity, S. , Sengupta, S. , Sengupta, S. , Kaur, A. et al. (2012) Mammalian proapoptotic factor ChaC1 and its homologues function as γ‐glutamyl cyclotransferases acting specifically on glutathione. EMBO Reports, 13, 1095–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S. , Kaur, A. , Chattopadhyay, B. & Bachhawat, A.K. (2015) Defining the cytosolic pathway of glutathione degradation in Arabidopsis thaliana: role of the ChaC/GCG family of γ‐glutamyl cyclotransferases as glutathione‐degrading enzymes and AtLAP1 as the Cys‐Gly peptidase. The Biochemical Journal, 468, 73–85. [DOI] [PubMed] [Google Scholar]
- Kuzuhara, Y. , Isobe, A. , Awazuhara, M. , Fujiwara, T. & Hayashi, H. (2000) Glutathione levels in phloem sap of rice plants under sulfur deficient conditions. Soil Science & Plant Nutrition, 46, 265–270. [Google Scholar]
- Leustek, T. , Martin, M.N. , Bick, J.‐A. & Davies, J.P. (2000) Pathways and regulation of sulfur metabolism revealed through molecular and genetic studies. Annual Review of Plant Physiology and Plant Molecular Biology, 51, 141–165. [DOI] [PubMed] [Google Scholar]
- Li, Q. , Gao, Y. & Yang, A. (2020) Sulfur homeostasis in plants. International Journal of Molecular Sciences, 21, 8926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Sawada, Y. , Hirai, A. , Sato, M. , Kuwahara, A. , Yan, X. et al. (2013) Novel insights into the function of Arabidopsis R2R3‐MYB transcription factors regulating aliphatic glucosinolate biosynthesis. Plant & Cell Physiology, 54, 1335–1344. [DOI] [PubMed] [Google Scholar]
- Lin, M. & Oliver, D.J. (2008) The role of acetyl‐coenzyme A synthetase in Arabidopsis. Plant Physiology, 147, 1822–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeira, F. , Park, Y.M. , Lee, J. , Buso, N. , Gur, T. , Madhusoodanan, N. et al. (2019) The EMBL‐EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Research, 47, W636–W641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, M.N. , Saladores, P.H. , Lambert, E. , Hudson, A.O. & Leustek, T. (2007) Localization of members of the γ‐glutamyl transpeptidase family identifies sites of glutathione and glutathione S‐conjugate hydrolysis. Plant Physiology, 144, 1715–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama‐Nakashita, A. , Nakamura, Y. , Watanabe‐Takahashi, A. , Inoue, E. , Yamaya, T. & Takahashi, H. (2005) Identification of a novel cis‐acting element conferring sulfur deficiency response in Arabidopsis roots. The Plant Journal, 42, 305–314. [DOI] [PubMed] [Google Scholar]
- May, M.J. & Leaver, C.J. (1994) Arabidopsis thaliana gamma‐glutamylcysteine synthetase is structurally unrelated to mammalian, yeast, and Escherichia coli homologs. Proceedings of the National Academy of Sciences of the United States of America, 91, 10059–10063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister, A. & Larsson, A. (1995) Glutathione synthetase deficiency and other disorders of the gamma‐glutamyl cycle. In: Scriver, C.R. , Beaudet, A.L. , Sly, W.S. & Valle, D. (Eds.) The metabolic and molecular bases of inherited disease. New York: McGraw Hill, pp. 1461–1495. [Google Scholar]
- Minet, M. , Dufour, M.E. & Lacroute, F. (1992) Complementation of Saccharomyces cerevisiae auxotrophic mutants by Arabidopsis thaliana cDNAs. The Plant Journal, 2, 417–422. [DOI] [PubMed] [Google Scholar]
- Minocha, R. , Thangavel, P. , Dhankher, O.P. & Long, S. (2008) Separation and quantification of monothiols and phytochelatins from a wide variety of cell cultures and tissues of trees and other plants using high performance liquid chromatography. Journal of Chromatography. A, 1207, 72–83. [DOI] [PubMed] [Google Scholar]
- Mukaihara, T. , Hatanaka, T. , Nakano, M. & Oda, K. (2016) Ralstonia solanacearum type III effector RipAY is a glutathione‐degrading enzyme that is activated by plant cytosolic thioredoxins and suppresses plant immunity. MBio, 7, e00359–e00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg, D. , Müller, R. & Funk, M. (1995) Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene, 156, 119–122. [DOI] [PubMed] [Google Scholar]
- Nishida, S. , Duan, G. , Ohkama‐Ohtsu, N. , Uraguchi, S. & Fujiwara, T. (2016) Enhanced arsenic sensitivity with excess phytochelatin accumulation in shoots of a SULTR1;2 knockout mutant of Arabidopsis thaliana (L.) Heynh. Soil Science & Plant Nutrition, 62, 367–372. [Google Scholar]
- Noctor, G. , Mhamdi, A. , Chaouch, S. , Han, Y. , Neukermans, J. , Marquez‐Garcia, B. et al. (2012) Glutathione in plants: an integrated overview. Plant, Cell & Environment, 35, 454–484. [DOI] [PubMed] [Google Scholar]
- Noctor, G. , Queval, G. , Mhamdi, A. , Chaouch, S. & Foyer, C.H. (2011) Glutathione. Arabidopsis Book, 9, e0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa, A. , Miwa, K. , Kobayashi, M. & Fujiwara, T. (2006) Isolation of Arabidopsis thaliana cDNAs that confer yeast boric acid tolerance. Bioscience, Biotechnology, and Biochemistry, 70, 1724–1730. [DOI] [PubMed] [Google Scholar]
- Obayashi, T. , Aoki, Y. , Tadaka, S. , Kagaya, Y. & Kinoshita, K. (2018) ATTED‐II in 2018: a plant coexpression database based on investigation of the statistical property of the mutual rank index. Plant & Cell Physiology, 59, 440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Boyle, N.M. , Banck, M. , James, C.A. , Morley, C. , Vandermeersch, T. & Hutchison, G.R. (2011) Open babel: an open chemical toolbox. Journal of Cheminformatics, 3, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkama‐Ohtsu, N. , Kasajima, I. , Fujiwara, T. & Naito, S. (2004) Isolation and characterization of an Arabidopsis mutant that overaccumulates O‐acetyl‐L‐Ser. Plant Physiology, 136, 3209–3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkama‐Ohtsu, N. , Oikawa, A. , Zhao, P. , Xiang, C. , Saito, K. & Oliver, D.J. (2008) A γ‐glutamyl transpeptidase‐independent pathway of glutathione catabolism to glutamate via 5‐oxoproline in Arabidopsis. Plant Physiology, 148, 1603–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkama‐Ohtsu, N. , Radwan, S. , Peterson, A. , Zhao, P. , Badr, A.F. , Xiang, C. et al. (2007) Characterization of the extracellular γ‐glutamyl transpeptidases, GGT1 and GGT2, in Arabidopsis. The Plant Journal, 49, 865–877. [DOI] [PubMed] [Google Scholar]
- Ohkama‐Ohtsu, N. , Zhao, P. , Xiang, C. & Oliver, D.J. (2007) Glutathione conjugates in the vacuole are degraded by γ‐glutamyl transpeptidase GGT3 in Arabidopsis. The Plant Journal, 49, 878–888. [DOI] [PubMed] [Google Scholar]
- Orlowski, M. & Meister, A. (1970) The γ‐glutamyl cycle: a possible transport system for amino acids. Proceedings of the National Academy of Sciences of the United States of America, 67, 1248–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulose, B. , Chhikara, S. , Coomey, J. , Jung, H.I. , Vatamaniuk, O. & Dhankher, O.P. (2013) A γ‐glutamyl cyclotransferase protects Arabidopsis plants from heavy metal toxicity by recycling glutamate to maintain glutathione homeostasis. Plant Cell, 25, 4580–4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings, N.D. , Waller, M. , Barrett, A.J. & Bateman, A. (2014) MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Research, 42, D503–D509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert, X. & Gouet, P. (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Research, 42, W320–W324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero, L.C. , Aroca, M.Á. , Laureano‐Marín, A.M. , Moreno, I. , García, I. & Gotor, C. (2014) Cysteine and cysteine‐related signaling pathways in Arabidopsis thaliana. Molecular Plant, 7, 264–276. [DOI] [PubMed] [Google Scholar]
- Rosso, M.G. , Li, Y. , Strizhov, N. , Reiss, B. , Dekker, K. & Weisshaar, B. (2003) An Arabidopsis thaliana T‐DNA mutagenized population (GABI‐Kat) for flanking sequence tag‐based reverse genetics. Plant Molecular Biology, 53, 247–259. [DOI] [PubMed] [Google Scholar]
- Sawada, Y. , Akiyama, K. , Sakata, A. , Kuwahara, A. , Otsuki, H. , Sakurai, T. et al. (2009) Widely targeted metabolomics based on large‐scale MS/MS data for elucidating metabolite accumulation patterns in plants. Plant & Cell Physiology, 50, 37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogawa, Y. , Ohkama‐Ohtsu, N. , Hayashi, H. , Yoneyama, T. & Fujiwara, T. (2005) Independent roles of glutathione and O‐acetyl‐L‐serine in regulation of sulfur‐responsive gene expression in Arabidopsis thaliana. Plant Biotechnology, 22, 51–54. [Google Scholar]
- Spraggon, G. , Kim, C. , Nguyen‐Huu, X. , Yee, M.C. , Yanofsky, C. & Mills, S.E. (2001) The structures of anthranilate synthase of Serratia marcescens crystallized in the presence of (i) its substrates, chorismate and glutamine, and a product, glutamate, and (ii) its end‐product inhibitor, L‐tryptophan. Proceedings of the National Academy of Sciences of the United States of America, 98, 6021–6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama, R. , Li, R. , Kuwahara, A. , Nakabayashi, R. , Sotta, N. , Mori, T. et al. (2021) Retrograde sulfur flow from glucosinolates to cysteine in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America, 118, e2017890118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida, K. , Sawada, Y. , Ochiai, K. , Sato, M. , Inaba, J. & Hirai, M.Y. (2020) Identification of a unique type of isoflavone O‐methyltransferase, GmIOMT1, based on multi‐omics analysis of soybean under biotic stress. Plant & Cell Physiology, 61, 1974–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadi, M. , Anyango, S. , Deshpande, M. , Nair, S. , Natassia, C. , Yordanova, G. et al. (2021) AlphaFold protein structure database: massively expanding the structural coverage of protein‐sequence space with high‐accuracy models. Nucleic Acids Research, 50, D439–D444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C.‐L. & Oliver, D.J. (1996) Cloning of the cDNA and genomic clones for glutathione synthetase from Arabidopsis thaliana and complementation of agsh2 mutant in fission yeast. Plant Molecular Biology, 31, 1093–1104. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Zhang, W.Z. , Song, L.F. , Zou, J.J. , Su, Z. & Wu, W.H. (2008) Transcriptome analyses show changes in gene expression to accompany pollen germination and tube growth in Arabidopsis. Plant Physiology, 148, 1201–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, M. , Chiba, Y. & Hirai, M.Y. (2021) Metabolism and regulatory functions of O‐acetylserine, S‐adenosylmethionine, homocysteine, and serine in plant development and environmental responses. Frontiers in Plant Science, 12, 643403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler, E.A. , Shoemaker, D.D. , Astromoff, A. , Liang, H. , Anderson, K. , Andre, B. et al. (1999) Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science, 285, 901–906. [DOI] [PubMed] [Google Scholar]
- Xiang, C. & Oliver, D.J. (1998) Glutathione metabolic genes coordinately respond to heavy metals and jasmonic acid in Arabidopsis. Plant Cell, 10, 1539–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Peak area matrix of all detected compounds in metabolome analysis.
Data S2. Annotation list for metabolome analysis.
Figure S1. The γ‐glutamyl cycle proposed in mammals.
Figure S2. Confirmation of the result of protein purification by SDS‐PAGE.
Figure S3. Gene expression in the ggp1/ggct2;1 mutant after buthionine sulfoximine (BSO) treatment.
Figure S4. Expression analysis based on a public microarray database.
Figure S5. Amino acid sequence alignment of GGP1 and its homologs.
Figure S6. Superimposition of the predicted protein structures of GGP1 (green) and GGP3 (cyan).
Table S1. List of yeast strains used in this study.
Table S2. Primers used for constructing and sequencing expression plasmids for yeast.
Table S3. Primers used for the construction of E. coli expression strains.
Table S4. Primers used for quantitative PCR.
Table S5. Primers used for screening ggp1 and ggct2;1 mutants and mRNA analysis.
Table S6. All significant changes in metabolite contents in wild‐type (WT), ggp1, and ggct2;1 plants.
Data Availability Statement
The data supporting the findings of this study are available in this article and its supplementary materials.


