Abstract
The mucosal origins hypothesis of rheumatoid arthritis (RA) proposes a central role for mucosal immune responses in the initiation or perpetuation of the systemic autoimmunity that occurs with disease. However, the connection between the mucosa and systemic autoimmunity in RA remains unclear. Using dual IgA and IgG family plasmablast-derived monoclonal autoantibodies obtained from peripheral blood of individuals at-risk for RA, we identified cross-reactivity between RA-relevant autoantigens and bacterial taxa in the closely related families Lachnospiraceae and Ruminococcaceae. After generating bacterial isolates within the Lachnospiraceae/Ruminococcaceae genus Subdoligranulum from the feces of an individual, we confirmed monoclonal antibody binding, as well as CD4+ T cell activation in individuals with RA compared to control individuals. Additionally, when Subdoligranulum isolate 7 but not isolate 1 colonized germ-free mice, it stimulated Th17 cell expansion, serum RA-relevant IgG autoantibodies, and joint swelling reminiscent of early RA, with histopathology characterized by antibody deposition and complement activation. Systemic immune responses were likely due to mucosal invasion along with the generation of colon isolated lymphoid follicles (ILFs) driving increased fecal and serum IgA by isolate 7, as B and CD4+ T cell depletion not only halted intestinal immune responses but also eliminated detectable clinical disease. In aggregate, these findings demonstrate a mechanism of RA pathogenesis through which a specific intestinal strain of bacteria can drive systemic autoantibody generation and joint-centered antibody deposition and immune activation.
One-sentence summary:
An autoantibody-targeted Subdoligranulum strain activates T cells in individuals with rheumatoid arthritis and drives arthritis in mice.
Editor’s Summary:
A Bacterial Driver of Arthritis. Autoantibodies can be detected in individuals at-risk for developing rheumatoid arthritis (RA) prior to development of clinical disease. The source of these autoantibodies, however, remains unclear. Here, Chriswell et al. identified that IgG and IgA autoantibodies from individuals who are at-risk for RA cross-react against gut bacteria in the Lachnospiraceae and Ruminococcaceae families. Further analysis identified a bacterial strain from the Subdoligranulum genus that was associated with autoantibody development. Mice colonized with this Subdoligranulum isolate developed arthritis with pathology similar to human RA. These findings suggest that this Subdoligranulum strain may be a major contributor to RA autoantibody development.
Introduction
The natural history of rheumatoid arthritis (RA) has been the focus of study for years, yet the causal triggers of RA remain unclear. Biomarkers that provide insights into disease mechanisms include two types of autoantibodies – anti-citrullinated protein antibodies (ACPA) and rheumatoid factor (RF) – that develop years before disease onset and are predictive for future joint disease onset and severity(1–4). During this preclinical period, individuals are considered “at-risk” for RA(5).
ACPA preferentially bind citrulline-containing epitopes on an array of proteins and can contribute to joint damage in a murine model of RA(6). RF targets the constant region of immunoglobulin, and persistently elevated titers correlate with clinical disease activity(7). Notably, these antibodies arise from separate B cell lineages and mechanisms(8), suggesting that they function differentially in the development of RA. ACPA+ B cells undergo upregulation of genes that promote T cell-dependent responses, whereas RF+ B cells upregulate transcriptional programs that invoke innate immunological memory reactivation(8).
With regard to the initial development of RA-related autoantibodies, the mucosal origins hypothesis suggests that environmental interactions and chronic inflammation at mucosal surfaces may be important early drivers of RA pathogenesis(9). In support of this paradigm, ACPA are detected in the lungs of individuals with longstanding RA(10), as well as in the sputum of individuals at-risk for RA(11), and this is associated with the presence of elevated cytokines, complement activation, and neutrophil extracellular trap (NET) formation(12). The oral mucosa is also implicated, as supported by the finding that Porphyromonas gingivalis, a bacteria causally linked to chronic periodontitis, encodes an enzyme capable of citrullinating proteins(13), and ACPA+ individuals exhibit a higher relative abundance of P. gingivalis in their oral microbiome(14). Another potential oral pathway links Aggregatibacter actinomycetemcomitans that are expanded in the periodontium of individuals with chronic RA(15), and have the capacity to induce host cell hyper-citrullination(16) rather than direct enzymatic citrullination of proteins. Furthermore, an orally-derived strain of Streptococcus has been found to be capable of inducing arthritis in the autoimmune arthritis-prone SKG mice(17). Additional data from both human and murine studies implicate the intestinal mucosa. Expansion of Prevotella copri in the gut of individuals with new-onset RA(18) and during the preclinical phase(19) has been described, and other groups demonstrate perturbations in Lactobacilli(20) and rare lineages of bacteria(21) in RA that can modulate experimental arthritis models. A further relationship to Prevotella is suggested by the finding that human leukocyte antigen (HLA)-DR-containing peptides, eluted from antigen-presenting cells of patients with chronic RA, derived from these bacteria share homology with two additional RA-specific autoantigens(22–24). Thus, certain lines of evidence support immune responses to bacteria at mucosal surfaces as modifiers of RA relevant autoantibodies and amplifiers of underlying joint inflammation. However, despite these clinical associations, none of the proposed bacterial lineages have demonstrated an arthritogenic effect in wild type mice in the absence of other strongly pro-inflammatory factors.
Plasmablasts are a potentially highly informative subset of B cells, as they are circulating components of ongoing local immune responses(25–27) and could link immune responses at the mucosa with the joint. Recently, Kinslow et al. described a substantial expansion of IgA+ plasmablasts in the circulation of individuals at-risk for RA(28). Following variable region sequencing, a large number of the isolated plasmablasts were found to arise in shared clonal families with both IgA+ and IgG+ members. These findings suggested a shared mucosal and systemic immune response evolution, although the specific triggers and mechanisms by which this conversion might happen remains unknown. We hypothesized that the antibodies produced by these dual IgA/IgG-containing family plasmablasts would recognize a mucosa-associated bacterium that could stimulate the development of autoimmunity. In this report, we utilized informative monoclonal antibodies (mAbs) derived from circulating dual IgA/IgG-containing plasmablast families to identify candidate intestinal bacteria in feces from at-risk individuals. From that source, we isolated a bacterial strain that is targeted by both dual IgA/IgG family plasmablasts and CD4+ T cells from patients with RA. We found that colonizing mice with this bacterial strain stimulated RA-related autoantibodies and joint swelling with both IgG/IgA deposition and complement activation; we further demonstrated a B- and T-cell dependent pathway by which these pathologic changes arise. This human bacterial species, provisionally classified as a Subdoligranulum didolesgii, provides a causal link between the mucosal immune system, RA autoimmunity, and joint-centered pathology. Studies of this strain will provide a clinically relevant model for understanding of the drivers of microbially-driven RA-related autoimmunity.
Results
A subset of dual IgA/IgG family plasmablast-derived monoclonal antibodies target RA-relevant antigens as well as bacteria in families Lachnospiraceae and Ruminococcaceae.
We previously identified a population of circulating plasmablasts that belong to dual IgA/IgG clonal families in individuals at risk for RA(28). Hypothesizing that these plasmablasts may inform a mucosal to systemic immune response conversion leading to targeting of RA-relevant antigens, we established mAbs derived from plasmablasts (PB-mAbs) from shared IgA/IgG clonal families. Plasmablasts were isolated from individuals at-risk for the development of RA (n=4) and individuals with early RA (<1 year from diagnosis; n=2; tables S1 and S2), and mAbs were selected for further study due to their ability to bind RA-relevant citrullinated antigens(28). Sequences from the variable regions were cloned onto a mouse IgG2a framework and expressed. A total of 94 successfully generated PB-mAbs confirmed binding of numerous RA-relevant autoantigens by multiplex array (Fig. 1A, data file S1 and S2).
Figure 1: A subset of dual IgA/IgG family plasmablast-derived monoclonal antibodies cross-react with RA-relevant antigens and predominantly bind families Lachnospiraceae and Ruminococcaceae.
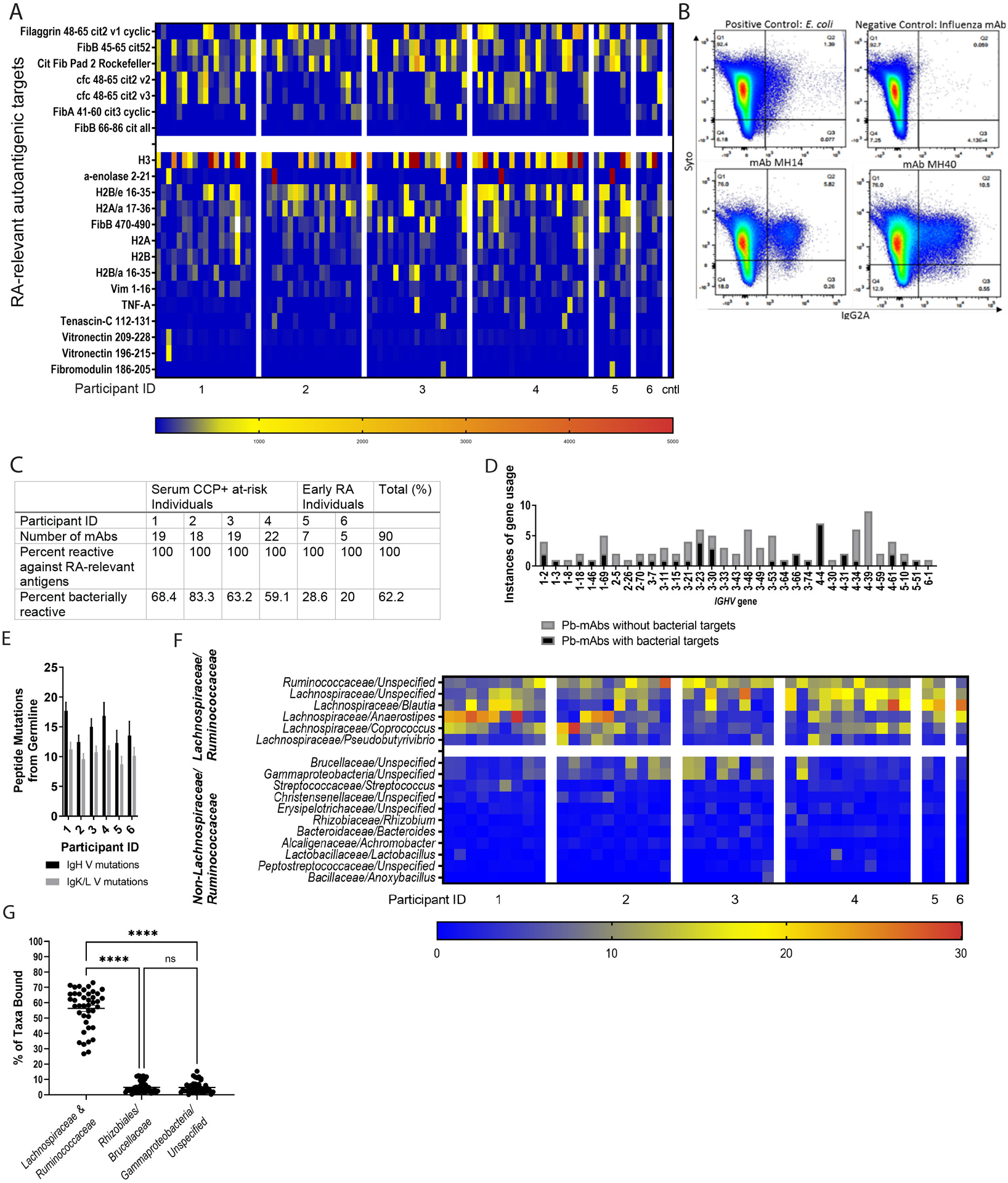
(A) 94 plasmablast-derived mAbs (PB-mAbs) from at-risk (n=4) and early RA (n=2) individuals belonging to shared IgG and IgA clonal families were applied to a planar array containing 346 different citrullinated and native peptide targets. The heatmap demonstrates degree of reactivity between individual PB-mAbs (columns, x-axis) with specific antigens (rows, y-axis). Cntl, control. (B and C) These PB-mAbs were screened for cross-reactivity against a broadly representative pool of fecal bacteria created from feces from individuals with early RA (n=5), at-risk (n=8) and healthy controls (n=5). The fecal pool was exposed to each antibody and analyzed by flow cytometry. The samples that were greater than two standard deviations above background staining were considered to be positive for bacterial binding. (B) Representative flow plots demonstrating binding to Syto9Green (a nucleic acid stain, y-axis) and IgG2A-PE (representing mAb binding, x-axis) are shown. The top left displays binding to E. coli (positive control), the top right to an influenza mAb (negative control). The bottom two graphs are representative binding plots of two of the positive mAbs (14 and 40). (C) The table summarizes PB-mAb reactivity against RA-relevant antigens and bacteria, separated by individual. (D) PB-mAbs with and without bacterial targets were analyzed for IGHV gene usage. The count of mAbs expressing each IGHV gene is displayed (y-axis) against represented IGHV gene segments (x-axis). mAbs with bacterial targets are displayed in gray and mAbs without bacterial targets are displayed in black. (E) The amino acid mutations from germline (y-axis) are demonstrated for each individual (x-axis). The number of IgH V mutations are displayed in black and the number of IgL/K mutations are displayed in gray. (F) The mAb-bound bacterial fraction underwent 16S rRNA sequencing and taxonomic identification. The heatmap displays percentage of total bacteria bound represented by each taxa displayed; the top taxa bound for all mAbs are shown. Each mAb is shown (x-axis), segregated by the individual from whom it was derived. Bacteria are segregated by either belonging to families Lachnospiraceae or Ruminococcaceae (top of y-axis) or not belonging to either Lachnospiraceae or Ruminococcaceae (bottom of y-axis). (G) The three most bound taxa from each PB-mAb are represented out of the total bacteria bound (x-axis). The percent of total bacteria bound is quantified for each mAb (y-axis) and shown as round symbols for the individual mAbs and mean ± SEM as bars. P<0.0001; ns, not significant, by one-way ANOVA with Tukey’s post-test.
Because the PB-mAbs and many circulating autoantibodies include the mucosal IgA isotype, we queried if the PB-mAbs could target intestinal bacteria. For broad representation of fecal bacteria, we pooled feces from healthy individuals (n=5), individuals at-risk for RA (n=8), and individuals with early RA (n=5; table S3, fig. S1A). By 16S rRNA gene sequencing we verified high bacterial diversity in this pool (Shannon-H Index Value 2.524; fig. S1B). Using a negative control PB-mAb targeting influenza and a positive control mAb to E. coli, we developed a flow cytometry assay to identify PB-mAb binding to fecal bacteria (Fig. 1B). Staining greater than two standard deviations above the mean fluorescence intensity of the negative controls was considered positive binding. Using this cut-off value, a total of 61.7% (58/94) of the PB-mAbs targeted intestinal bacteria (Fig. 1C), although there was no association between isotype (IgA versus IgG) and bacterial binding (fig. S2A). There was no association between autoantigen binding preference and binding of bacterial targets (fig. S2B). However, mAbs that bound bacteria utilized a subset of heavy chain variable region (IGHV) genes compared to the non-bacteria binding mAbs (Fig. 1D, fig. S2C). Additionally, the PB-mAbs were substantially mutated from germline, although the number of mutations in the bacteria binding versus non-binding PB-mAbs did not differ, indicating that these plasmablasts are likely not from a natural antibody pool (Fig. 1E, fig. S2D).
To define the bacterial targets of the PB-mAbs, we sorted the mAb-bound bacterial fraction and sequenced bacterial 16S rRNA genes using fluorescence activated cell sorting (FACS). Predominantly targeted taxa included the closely related families Lachnospiraceae and Ruminococcaceae (Fig. 1F and fig. S3A). Indeed, greater than 50% of the total bacteria bound by the bacteria-binding mAbs combined were from families Lachnospiraceae and Ruminococcaceae (Fig. 1G). When PB-mAbs binding Lachnospiraceae and Ruminococcaceae (MH 4, 28, 58, and 91) were matched against individual fecal samples from humans with early RA or healthy controls, there was no difference in binding to Lachnospiraceae or Ruminococcaceae between groups (fig. S3B), suggesting as anticipated a broad range of these overall families. These data demonstrate an interesting antibody cross-reactivity between bacterial targets, specifically Lachnospiraceae and Ruminococcaceae, among a subset of mAbs that bind RA-relevant autoantigenic targets.
Ruminococcaceae Subdoligranulum strains isolated from a human sample are targeted by PB-mAbs.
In order to investigate immune responses to specific species within Lachnospiraceae and Ruminococcaceae in RA, we first established primary bacterial isolates from an individual with greater than 40% abundance of these taxa in their feces (fig. S3C). A total of 50 isolates were established, seven identified as Lachnospiraceae/Ruminococcaceae by quantitative polymerase chain reaction (qPCR) (isolates 1 to 7), and five confirmed as pure isolates by 16S rRNA sequencing (isolates 1, 3, 4, 5, and 7). These five isolates underwent whole genome sequencing. Sequences were assembled, scaffolded, and cleaned using Abyss(29), input into the Biobakery Workflow(30), and categorized as unidentified species within the family Ruminococcaceae and genus Subdoligranulum (Fig. 2A), which we provisionally designate as a species named Subdoligranulum didolesgii. We identified this bacterium as a distinct species by aligning the genome and inserting the full-length sequence into both the SILVA Ribosomal RNA Database as well as NCBI Blast without substantial alignment to other entries into these databases. We narrowed down the five isolates to two candidates (isolates 1 and 7) for further study due to their differing abilities to induce joint swelling in mono-colonized mice (fig. S4). We confirmed that a subset of PB-mAbs (numbers 4, 28, 58, and 91), selected based on their binding of both Lachnospiraceae and Ruminococcaceae, bound both bacterial isolates 1 and 7, whereas mAb 7 that did not bind Lachnospiraceae or Ruminococcaceae did not bind isolates 1 or 7 (Fig. 2B).
Figure 2: Ruminococcaceae Subdoligranulum strains isolated from a human sample are targeted by PB-mAbs and stimulate CD4+ T cells from patients with RA.
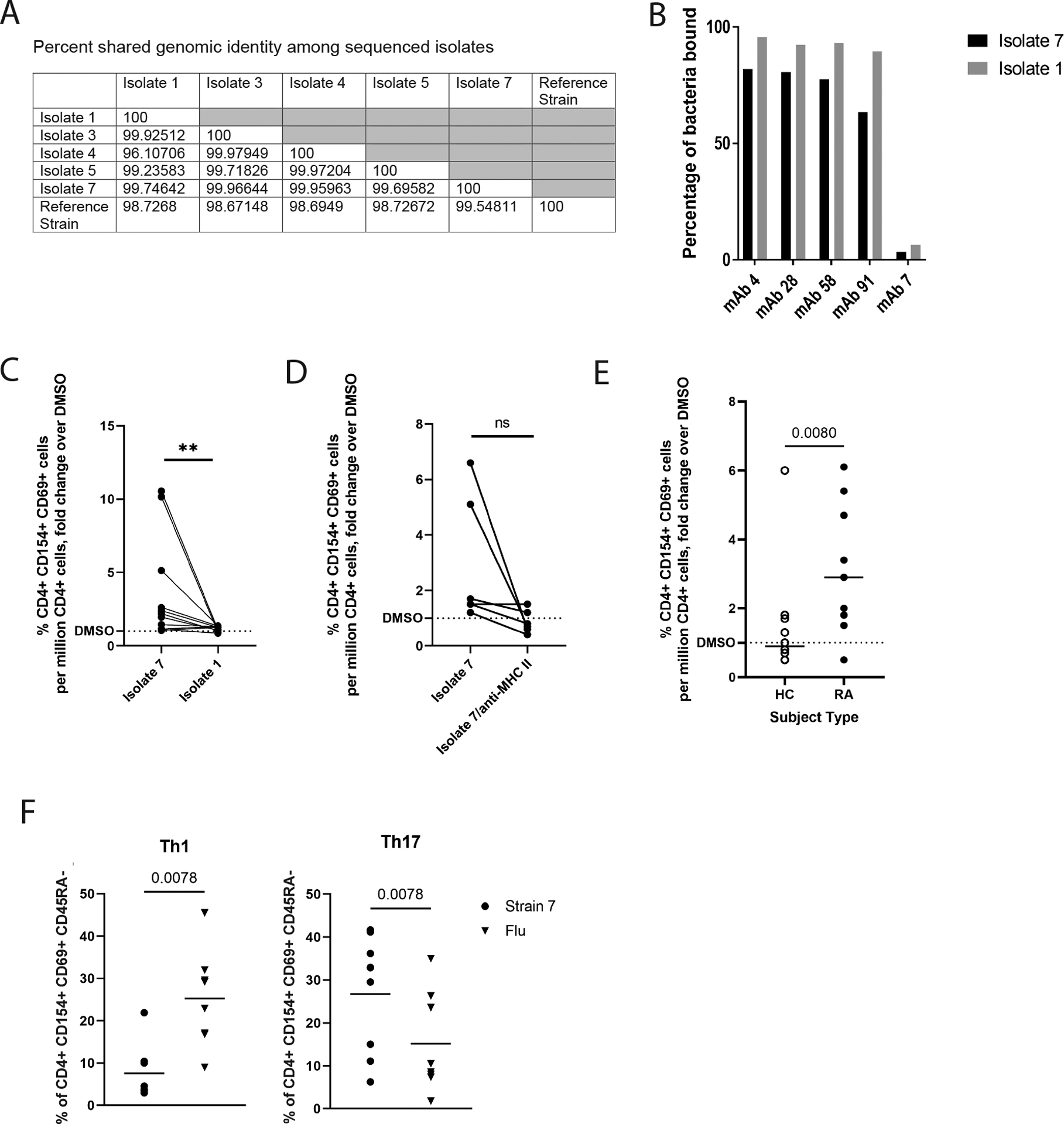
(A) Seven primary strains of Ruminococcaceae Subdoligranulum were isolated from the feces of an individual. Five isolates were selected for short read genome sequencing based on taxonomic identification by 16S rRNA sequencing. The table represents percent genomic shared identity among the 5 isolates as well as against a reference genome found to be genetically aligned (MGYG-HGUT02424; unidentified genus in Order Clostridiales, which includes Lachnospiraceae and Ruminococcaceae). (B) Isolates 1 and 7 were matched against four selected PB-mAbs (numbers 4, 28, 58, and 91) that bound highly to other various patterns of Ruminococcaceae and Lachnospiraceae species to verify that they targeted the strains. They were also matched against a control mAb (number 7) that was previously found to not bind bacteria. The percent of bacteria bound to mAb is displayed (y-axis) against each selected mAb (x-axis). Binding by isolate 7 is shown in black, and binding to isolate 1 is shown in gray. (C) Human peripheral blood mononuclear cells (PBMCs) from individuals with RA (n=11) were stimulated with 50ng/ml isolates 1 or 7. Fold change of the CD4+ T cell response relative to DMSO (horizontal dotted line) is displayed against binding to isolates 1 and 7 (x-axis). **P<0.01, non-parametric Wilcoxon matched-pairs signed rank test. (D) A Class II HLA-DR (clone L243) blocking antibody or an equal volume of phosphate-buffered saline was applied at 20μg/mL for 30 minutes prior to stimulation of PBMCs from individuals with RA (n=5) with isolate 7. Fold change of the CD4+ T cell response relative to DMSO (horizontal dotted line) is displayed against binding to isolate 7 versus isolate 7 blocked with L243 (x-axis). ns, not significant by non-parametric Wilcoxon matched-pairs signed rank test. (E) Isolate 7 specific responses among CD4+ T cells was tested comparing CD4+ T cells isolated from individuals with RA (n=11) to CD4+ T cells isolated from healthy controls (n=12). Fold change of the CD4+ T cell response relative to DMSO (y-axis) is displayed (x-axis). Data were analyzed using a nonparametric Mann-Whitney test. (F) Left: using CD45RA−/CXCR3+/CCR4−/CCR6− as a defining marker combination, we compared the relative proportion of Th1-like cells for Isolate 7-specific (circles) or influenza-specific (inverted triangles) memory CD4+ T cell responses in CD4+ T cells from individuals with RA, observing a higher proportion of Th1-like influenza specific cells (p=0.0078, nonparametric Mann-Whitney test). Right: using CD45RA-/CXCR3-/CCR4+/CCR6+ as a defining marker combination, we compared the relative proportion of Th17-like cells for Isolate 7-specific (circles) or influenza-specific (inverted triangles) memory CD4+ T cell responses, observing a significantly higher proportion of Th17-like isolate 7-specific cells (p=0.0078, nonparametric Mann-Whitney test). Horizontal bars indicate mean.
Next, in order to evaluate the immunologic relevance of the Subdoligranulum isolates, we assessed if the bacteria were recognized by circulating T cells from individuals with RA. Peripheral blood mononuclear cells (PBMCs) from 11 individuals with RA were stimulated with 50 ng/ml oxygen-killed Subdoligranulum isolate 1 or isolate 7 for 14 hours. Compared to isolate 1, isolate 7 significantly (p<0.01) activated CD4+ T cells in the PBMCs as measured by accumulated surface CD69 and CD154 expression (Fig. 2C, fig. S5, table S4). Blocking MHC class II with an anti-DR antibody did not result in significantly (p>0.05) reduced CD69+ CD154+ expression (Fig. 2D). Furthermore, utilizing previously unexamined individuals, isolate 7 specific responses were more prevalent in CD4+ T cells derived from RA cases (N=11) as compared to healthy controls (N=12) (Fig. 2E, table S5). Finally, we examined the cell surface phenotype of isolate 7- and influenza-specific CD4+ T cells in the same RA cases (n=11) using accepted definitions for Th1-like (CD45RA−/CXCR3+/CCR4−/CCR6−) and Th17-like (CD45RA−/CXCR3−/CCR4+/CCR6+) cells(31), observing a significantly (p<0.01) higher proportion of Th1-like influenza specific CD4+ T cells and a significantly (p<0.01) higher proportion of Th17-like isolate CD4+ T specific T cells (Fig. 2F). Our findings support the hypothesis that the Subdoligranulum isolate 7 elicits immunologically-relevant CD4+ T cell responses and suggest that strain variations may be of importance to understand the pathogenesis of RA.
Subdoligranulum isolates stimulate joint swelling characterized by antibody and complement deposition in mono-colonized mice.
Having associated immunity towards Subdoligranulum isolates with RA and deriving them from a human sample, we then investigated if the isolates were capable of inducing RA-relevant autoimmunity in mice. Subdoligranulum isolates 1, 3, 4, 5, and 7 were gavaged at 5×106 colony-forming units (CFU) into germ-free DBA/1 mice. Given the previously published association of Prevotalla copri with new-onset RA(18) and the homology between proteins in this bacteria and RA-relevant autoantigens(22–24), we also mono-colonized a group of germ-free DBA/1 mice with P. copri (DSMZ 18205(32)) as a control in addition to a sterile media gavage. Four of the five Subdoligranulum strains, and neither control, caused joint swelling spontaneously starting about 14 days after bacterial gavage and persisted until 35 days when most mice were euthanized (Fig. 3, A and B and fig. S4A), although when followed through 63 days, arthritis was still observed (fig. S4B). Germ-free C57BL/6J mice also developed very mild but detectable joint swelling after mono-colonization with isolate 7 (fig. S4C). We confirmed equal, stable bacterial colonization across groups (fig. S4, D to F). In order to confirm that our findings were not due to immunologic impairments of the germ-free state of the mice(33), we colonized specific-pathogen-free (SPF) DBA/1j mice that had been pre-treated with broad spectrum antibiotics orally to deplete the microbiome. We then gavaged these mice with either isolate 7, isolate 1, P. copri, or left them untreated, and were able to replicate the paw swelling phenotype under these conditions (Fig. 3C). The finding of arthritis development with introduction of a single strain alone was unexpected, as non-transgenic murine models of joint disease typically require intradermal injection of antigen with adjuvant, adjuvant alone, or intravenous transfer of pathogenic antibodies to develop disease(34, 35).
Figure 3: A specific Subdoligranulum strain stimulates joint swelling and inflammation in mono-colonized mice that is characterized by IgG, IgA, and complement C3 deposition in joints.
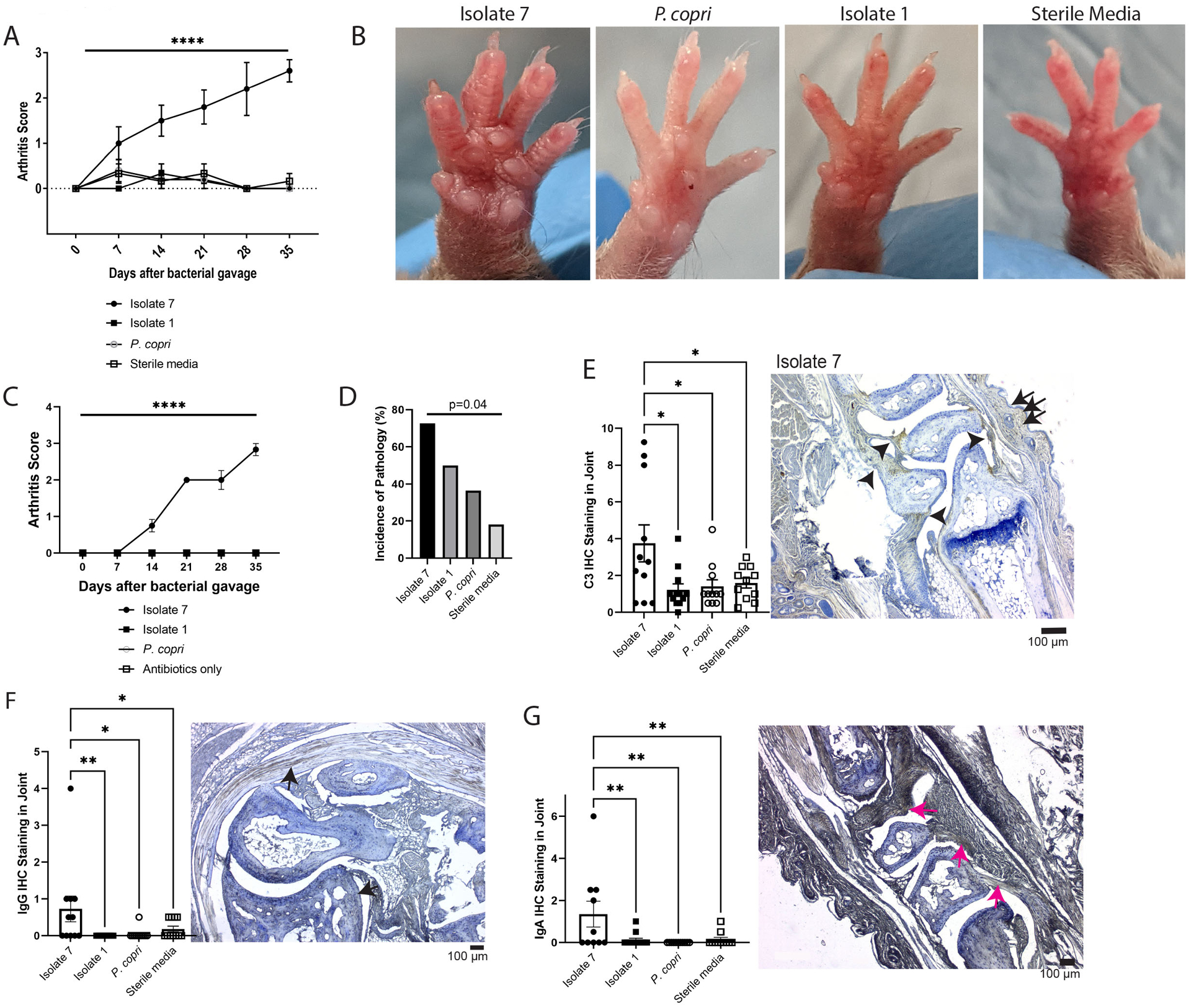
(A and B) Subdoligranulum isolates 1 and 7 as well as Prevotella copri or sterile media were gavaged separately into germ-free DBA/1 mice (n=6 isolate 1, n=6 isolate 7, n=5 P. copri, and n=6 sterile media). Mice were observed weekly for 35 days for the development of joint swelling and assessed a score based on the number of joints affected. (A) The mean ± SEM score is shown (y-axis) over time after bacterial gavage (x-axis). ****P<0.0001, repeated measures ANOVA. (B) Representative photographs of paws from the treatment groups are shown to demonstrate the swelling observed in mice mono-colonized with isolate 7. (C) SPF DBA/1j mice were treated with oral broad-spectrum antibiotics (neomycin, ampicillin, metronidazole, and vancomycin) for 5 days to deplete the microbiome. After antibiotic treatment, mice were gavaged with either Subdoligranulum isolates 1 and 7 or P. copri (n=6 isolate 7, n=6 isolate 1, n=6 P. copri, n=5 antibiotics only). Mice were observed weekly for 35 days for the development of joint swelling and assessed a score based on the number of joints affected. The mean ± SEM score is shown (y-axis) over time after bacterial gavage (x-axis). ****P<0.0001, repeated measures ANOVA. (D) Paw histology was assessed by a pathologist in a blinded fashion. Displayed is the incidence of pathology (y-axis) separated by treatment group (x-axis). Data were analyzed using Fisher’s exact test. (E) IHC for the C3 component of complement was performed on decalcified paw sections. C3 staining intensity scores are displayed (y-axis) separated by treatment group (x-axis). Symbols represent individual mice and bars indicate the mean ± SEM. *P<0.05, one-way ANOVA with Tukey’s post-test. A representative section for isolate 7 is displayed, with deposition indicated by arrows; the scale bar represents 100μm. (F) IgG deposition in decalcified paw sections was assessed by IHC. Intensity scores with symbols as individual mice and bars as mean ± SEM are displayed (y-axis), separated by treatment group (x-axis). *P≤0.05 and **P<0.01, one-way ANOVA with Tukey’s post-test. A representative section is displayed, with deposition indicated by arrows; the scale bar represents 100μm. (G) IgA deposition in decalcified paw sections was assessed by IHC and intensity scores are displayed (y-axis), separated by treatment group (x-axis). Symbols represent individual mice and bars indicate the mean ± SEM. **P≤0.01, one-way ANOVA with Tukey’s post-test. A representative section is displayed, with deposition indicated by arrows; the scale bar represents 100μm. Data are from n=12 isolate 1-gavaged, n=11 isolate 7-gavaged, n=11 P. copri-gavaged, and n=11 sterile media-gavaged animals across two experiments.
Evaluation of pathology in germ-free mono-colonized mice demonstrated a range of synovitis, osteitis, vasculitis, and soft tissue inflammation, all mild in severity (fig. S6). Yet all-combined, mice mono-colonized with isolate 7 had the highest incidence of pathology compared to the other treatment groups (Fig. 3D). Despite mild immune cell infiltrate, immunohistochemistry of the joints for immunoglobulin deposition and complement protein C3 demonstrated marked deposition both in the joint space and intradermally (Fig. 3E, fig. S7A). Additionally, immunohistochemistry demonstrated IgG and IgA deposition in the joint (Fig. 3, F and G, fig. S7, B and C). These findings suggest that an antibody mediated process may be a key factor in driving the joint swelling in this murine phenotype.
Serum IgA, RA-related autoantibodies, and splenic Th17 cells expand in Subdoligranulum isolate 7 mono-colonized mice.
In order to understand the systemic immune response following gavage with isolate 1, isolate 7, P. copri, or sterile media, serum from mice was collected on days 14 and 35 after gavage. The total serum IgA was significantly (p<0.0001 compared to isolate 1, p<0.05 compared to P. copri, and p<0.05 compared to sterile media) increased in isolate 7 mono-colonized mice at day 14 as compared to the other groups (Fig. 4A), but this normalized by day 35 (fig. S8A) and was not observed for IgG (fig. S8B). In SPF mice mono-colonized with isolate 7, both IgA and IgG were increased in the serum at days 14 and persisted at day 35 (fig. S9).
Figure 4: Subdoligranulum isolate 7 causes development of increased serum IgA, systemic RA-related autoantibodies, and expanded splenic Th17 populations.

(A to C) Serum from mice mono-colonized with either isolate 1 (n=12), isolate 7 (n=12), or P. copri (n=10), or given sterile media (n=10) was collected at days 14 and 35 after gavage. (A) The total serum IgA at 14 days after gavage was determined by ELISA; serum IgA is displayed (y-axis) against treatment group (x-axis). Symbols represent individual mice and bars indicate the mean ± SEM. *P<0.05 and ****P<0.0001, Kruskal-Wallis test with Dunn’s post-test. (B) Serum was analyzed on a planar array containing about 350 citrullinated and native peptides for autoantigens relevant in RA. A cutoff for positivity was established at the 80th percentile of autoantibody reactivity for individual samples on this assay (1.5 relative units) and the proportion of murine samples meeting or exceeding this threshold (11 antigens as displayed) at each timepoint is shown (y-axis). Each treatment group is shown from left to right. (C) Mean ± SD values for serum reactivity from each treatment group with specific RA-relevant autoantigens shown in panel (B) are shown in the table, comparing the four groups. P-values were determined by Kruskal-Wallis test with Dunn’s post-test. (D) Serum was analyzed at days 14 and 35 after bacterial gavage for the presence of murine collagen type II (CII) autoantibodies by ELISA. Mean ± SEM relative values are shown (y-axis) comparing groups (x-axis), including mice with collagen-induced arthritis (CIA). *P<0.05, **P<0.01 by Kruskal-Wallis test with Dunn’s post-test. n=6 CIA mice, n=6 isolate 7-gavaged mice, n=6 isolate 1-gavaged mice, n=6 P. copri-gavaged mice, n=6 sterile media-gavaged mice. (E) Spleens were collected from each mouse at 35 days post gavage and CD4+ T cell populations were analyzed by flow cytometry (n=7 isolate 7, n=7 isolate 1, n=6 P. copri, n=8 sterile media). The percentage (left) and absolute number (middle) of Roryt+ Th17 cells, as well as the Th17 to Treg ratio (right) is displayed. Symbols represent individual mice and bars indicate the mean ± SEM. *P<0.05 and **P<0.01 using Kruskal-Wallis test with Dunn’s post-test. (F and G) Splenocytes were collected from mice gavaged with Subdoligranulum isolate 1 or isolate 7 at 35 days after gavage and CD4+ T cells were isolated. CD4+ T cells were cocultured with bone marrow dendritic cells (BMDCs) loaded with either isolate 1, isolate 7, or no bacterial antigen for 14 hours. (F) The percentage of CD154+ CD69+ CD4+ cells is shown (y-axis) compared by treatment group (x-axis). (G) The percentage of Rorγt+ CD4+ T cells is shown (y-axis) compared by treatment group (x-axis). *P<0.05, **P<0.01 using Kruskal-Wallis test with Dunn’s post-test. n=6 isolate 7-gavaged mice, n=6 isolate 1-gavaged mice. The CD4+ T cell stimulation assay was performed in technical triplicate for each mouse and condition in vitro.
We next evaluated serum autoantibodies using a planar array containing about 350 RA-relevant autoantigens. Isolate 7-gavaged mice developed and maintained serum autoantibodies against RA-relevant autoantigens at higher proportions 14 and 35 days after gavage (p<0.05) than mice in the other groups (Fig. 4B). Furthermore, isolate 7 mono-colonized mice developed several autoantibodies targeting RA-relevant antigens, such as fibromodulin, at greater titers in comparison to the other groups (Fig. 4C). There was reactivity to both citrullinated and native peptides, analogous to the reactivity profiles of the human PB-mAbs (Fig. 1A). These data indicate that mono-colonization with isolate 7 allows for the establishment of persistent RA-relevant autoantibodies in circulation.
DBA/1 mice are prone to generate anti-murine type II collagen (anti-CII) antibodies in the collagen induced arthritis (CIA) model. Thus, we specifically assayed these by enzyme-linked immunosorbent assay (ELISA) in day 14 and 35 serum from our mono-colonized mice. Mice mono-colonized with isolate 7 developed anti-CII autoantibodies comparable to mice with CIA (Fig. 4D).
Given CD4+ T cell reactivity to isolate 7 in patients with RA, we evaluated splenic CD4+ T cell populations at days 14 and 35 in mice mono-colonized with isolate 1, isolate 7, or P. copri, or gavaged with sterile media (fig. S10A). We focused on Th17 and activated Treg populations due to the demonstrated role of intestinal microbiota in their development(36, 37), and on Tfh because of our observed changes in autoantibodies. At day 14, splenic Th17 cells were significantly increased in percentage (p<0.05 compared to P. copri and p<0.01 compared to sterile media), absolute number (p<0.05 compared to P. copri and sterile media), and Th17/Treg ratio (p<0.05 compared to P.copri and p<0.01 compared to sterile media) in isolate 7 mono-colonized mice (Fig. 4E). However, we did not observe differences in activated Treg or Tfh subsets in the isolate 7 group (fig. S10, B and C). By day 35, the expansion of Th17 cells remained in isolate 7 mono-colonized mice compared to sterile media-gavaged mice (fig. S11). At day 35, a similar expansion of Th17 cells was observed in SPF DBA/1j mice that were gavaged with isolate 7 (fig. S12, A and B), and is accompanied by a significant decrease in absolute number (p<0.05 compared to isolate 1 and P. copri, p<0.01 compared to antibiotics only) and percentage (p<0.05 compared to isolate 1, p<0.01 compared to P. copri) of splenic activated Treg populations (fig. S12C). However, similar to germ free mono-colonized mice, there was not an observable difference in Tfh frequencies between groups (fig. S12D).
To further understand this splenic Th17 skewing in vitro, splenic CD4+ T cells were isolated from germ-free DBA/1 mice mono-colonized with either isolate 1 or isolate 7 and cocultured with bone marrow-derived dendritic cells (BMDCs) loaded with either isolate 1 or isolate 7. Markers of T cell activation CD69 and CD154 were increased in CD4+ T cells from isolate 7 mono-colonized mice and stimulated with isolate 7 after 14 hours of coculture as compared to CD4+ T cells stimulated with isolate 1 (Fig. 4F, fig. S13), as well as markers of Th17 proliferation and function (Fig. 4G). Our data are consistent with others’ findings of Th17 expansion aiding in the development of autoantibody-mediated arthritis in mice(38) and in the evolution of human RA(39, 40). These findings in aggregate suggest a possible mucosal-to-systemic immune system response driven by IgA, resulting in the targeting of autoantigens by the immunoglobulins that reach the systemic circulation after being educated in the mucosa.
Subdoligranulum isolate 7 generates intestinal isolated lymphoid follicles, increased IgA, and Th17 skewing.
We next investigated the effects of Subdoligranulum isolate 7 on intestinal immunity. First, to determine if isolate 7 relative to isolate 1, P. copri, or sterile media affected intestinal permeability, fluorescein isothiocyanate (FITC)-dextran was administered orally to mice four hours prior to euthanasia, at which time serum was collected and the concentrations of FITC-dextran measured. All three mono-colonizations resulted in improved barrier compared to sterile media (Fig. 5A), indicating that the two Subdoligranulum isolates and P. copri were capable of at least partially restoring the barrier defect of germ-free mice(41). Intestinal histology revealed significantly increased numbers of isolated lymphoid follicles (ILFs) in isolate 7 mono-colonized mice as compared to P. copri and isolate 1 mono-colonized mice (p<0.05 compared to P. copri and sterile media); no difference in mean ILF size was observed (Fig. 5, B and C). We did not observe changes in colonic crypt depth or small intestinal villus morphology among isolate 7-gavaged mice (fig. S14). There was an increase in villus width and crypt depth among isolate 1-gavaged mice, which could suggest a mild injurious intestinal effect among mice gavaged with this strain (fig. S14). The ILFs in isolate 7 mono-colonized mice more closely resembled classical mature ILF morphology(42) (Fig. 5C, fig. S15), suggesting increased mucosal IgA generation(43). Indeed, luminal IgA secretion was significantly increased in isolate 7 mono-colonized mice (p<0.05 compared to sterile media, p<0.0001 compared to isolate 1)(Fig. 5D), although, similar to the serum, this difference resolved by day 35 (fig. S16A). There was no difference (p>0.05) in fecal IgG among groups (fig. S16B). An increase in fecal IgA was similarly seen in SPF mice that were gavaged with isolate 7 compared to mice gavaged with isolate 1 and P. copri at day 14 and compared to P. copri and antibiotics at day 35 (fig.S17). This indicates the possibility that T-dependent IgA is being produced and is coating the bacteria in the lumen, thereby reducing the total free IgA in the lumen. This hypothesis is supported by the presence of IgA+ B220+ B cells and IgA+ B220- plasma cells being generated in the mature ILFs seen in isolate 7 colonized mice (Fig. 5E, fig. S18). In associated mucosal lymphoid tissues mesenteric lymph nodes (MLNs) and Peyer’s patches (PPs), the ratio of Th17/Treg T cell subsets increased significantly (p<0.05) in the isolate 7 mono-colonized mice compared to sterile media gavaged-mice at day 14 following gavage (Fig. 5F), although the percentages and absolute numbers within the Th17, Treg, and Tfh populations individually were not different across groups (fig. S19, A to C). There is evidence of increased host access by isolate 7 mono-colonized mice compared to isolate 1- and P. copri-gavaged mice as demonstrated by bacterial presence in the host epithelium (Fig. 5, G and H), and mucus area in square millimeters, was reduced in isolate 7 mono-colonized groups compared to the P. copri mono-colonized groups, but greater than germ-free mice (fig. S20), suggesting that isolate 7 potentially affects mucus thickness. In aggregate, our observations suggest that Subdoligranulum isolate 7 can directly access the host to stimulate a robust intestinal immune response characterized by the formation of ILFs functioning to secrete IgA.
Figure 5: Subdoligranulum isolate 7 promotes development of intestinal isolated lymphoid follicles, increased mucosal IgA, and Th17 skewing in mucosal lymphoid tissues.

(A) FITC-Dextran was orally gavaged into mice mono-colonized with isolate 1 (n=5), isolate 7 (n=7), P. copri (n=6) or sterile media (n=6) 4 hours before euthanasia. At the time of euthanasia, serum was collected and tested for the presence of FITC-Dextran. Concentration of serum FITC-dextran is displayed (y=axis) against each treatment group (x-axis). Symbols represent individual mice and bars indicate the mean ± SEM.*P<0.05, one-way ANOVA with Tukey’s post-test. (B and C) Colon histology from each group was analyzed. (B) The number (left) and size in area (right) of isolated lymphoid follicles (ILFs) per colon (y-axis) separated by treatment group (x-axis) is shown. Symbols represent individual mice and bars indicate the mean ± SEM. *P<0.05, one-way ANOVA with Tukey’s post-test. (C) Representative colon histology containing ILFs in each group are shown. Scale bars represent 100μm. (D) Feces from mono-colonized mice at day 14 after gavage were tested for total IgA by ELISA (n=11 isolate 7-gavaged, n=12 isolate 1-gavaged, n=6 P. copri-gavaged, n=12 sterile media-gavaged). Symbols represent individual mice and bars indicate the mean ± SEM. *P<0.05; ****P<0.0001; and ns, not significant by Kruskal-Wallis test with Dunn’s post-test. (E) Representative colon immunofluorescence containing an ILF in an isolate 7-gavaged mouse is shown at 200x magnification. In red is IgA staining, in green is B220 staining, and in blue is DAPI. The arrows point to representative B220+ IgA+ B cells. (F) Mesenteric lymph nodes (MLNs) and Peyer’s patches (PPs) were evaluated at 14 days post-gavage for Th17 and activated Treg subpopulations by flow cytometry (n=7 isolate 7-gavaged, n=7 isolate 1-gavaged, n=7 P. copri-gavaged, n=8 sterile media-gavaged). The ratio of Th17:Treg cells is displayed with individual mice shown as symbols and bars indicate the mean ± SEM.. *P<0.05by Kruskal-Wallis test with Dunn’s post-test. (G and H) Bacterial presence in the host colonic epithelium was evaluated by fluorescence in situ hybridization (FISH) at 35 days post-gavage. (G) The total amount of bacteria in the epithelium across 10 fields of view is shown (y-axis), compared to treatment group (x-axis). *P<0.05, **P<0.01, ***P<0.001 by Kruskal-Wallis test with Dunn’s post-test (n=6 isolate 7-gavaged mice, n=5 isolate 1-gavaged mice, n=5 P. copri-gavaged mice, n=6 sterile media-gavaged mice). (H) Representative colon FISH images are shown at 200x magnification demonstrating bacteria (red, arrows), host mucus (green) and nuclei (blue) for each group. Dotted white lines highlight the mucosal layer as well as the colonic crypts in each image.
Joint swelling in Subdoligranulum isolate 7 colonized mice is dependent on T and B cells but not granulocytes.
To determine if the observed paw swelling was truly mediated by adaptive immunity, we selectively depleted B cells, CD4+ T cells, or granulocytes using mAbs (fig. S21) two days prior to mono-colonizing mice with isolate 7. The depletions were efficacious at depleting the B cells by two-fold, the CD4+ T cells by ten-fold, and granulocytes by five-fold on average. Mice depleted of CD4+ T or B cells did not develop paw swelling, whereas control antibody treated mice did (Fig. 6A), indicating that adaptive immunity is required for our phenotype. Though mice depleted of granulocytes developed paw swelling equal to treatment with control antibody, the onset of swelling was delayed by about one week (Fig. 6A), suggesting that, although granulocytes aid in the phenotype, they are not essential. ILFs were not affected in the CD4+ T cell-, B cell-, or granulocyte-depleted mice (Fig. 6B). Circulating and fecal IgA was significantly decreased in the B and CD4+ T cell-depleted mice at days 14 (p<0.01) and 35 (p<0.05 for B cell- and p<0.01 for CD4+ T cell-depletion) after bacterial gavage, and circulating and fecal IgG was decreased at day 35 after bacterial gavage (Fig. 6, C to F). Circulating IgA was significantly reduced in the granulocyte-depleted mice at day 14 (p<0.05) and day 35 (p=0.05), and circulating IgG was reduced in granulocyte-depleted mice at day 35 (p<0.05). These findings suggest that granulocyte-dependent IgA and IgG synthesis at the mucosal surface that spreads systemically may be important in this model.
Figure 6: Joint swelling is dependent on T and B cells but not granulocytes.
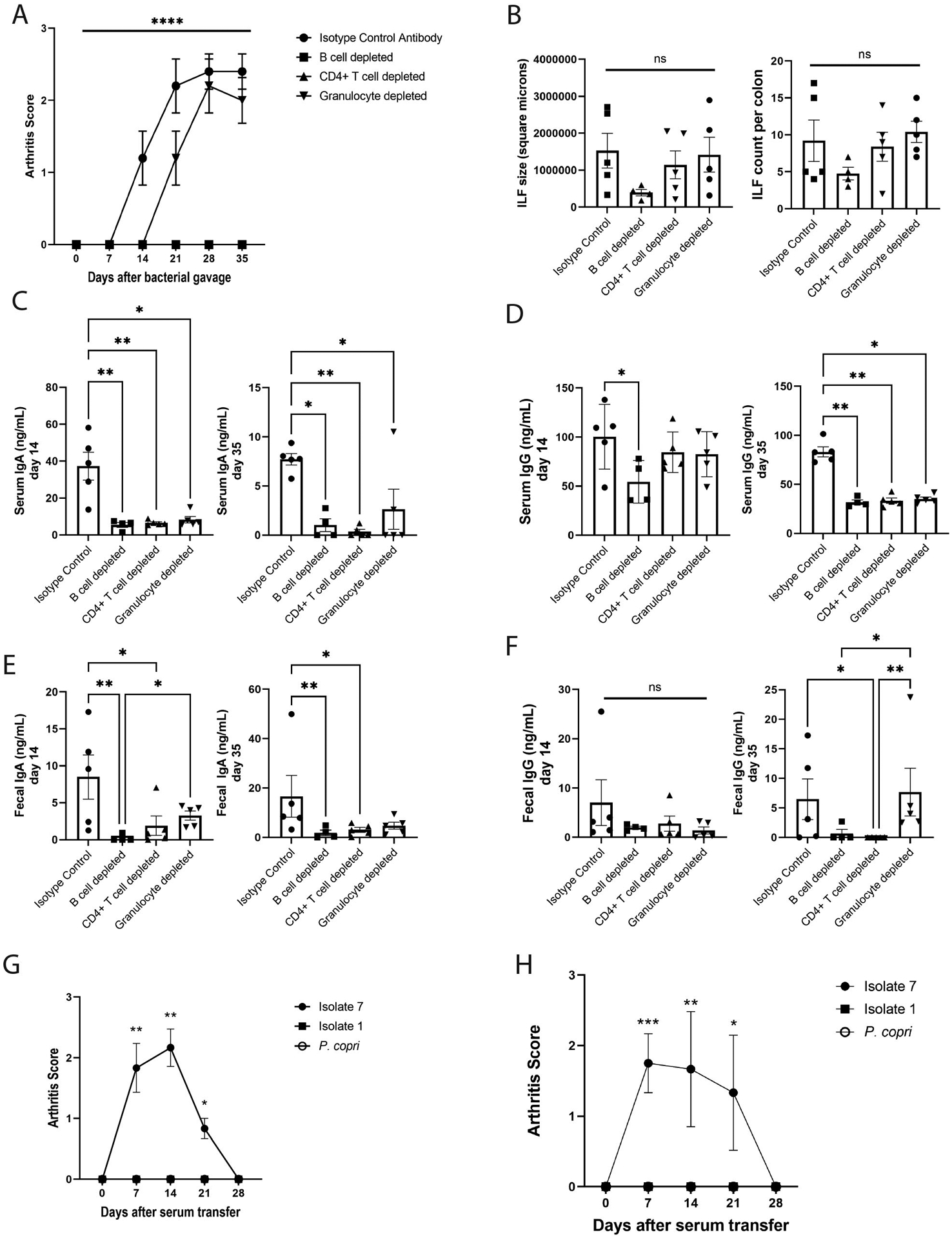
(A) Mice were selectively depleted of their B cells (n=4), CD4+ T cells (n=5), or granulocytes (n=5) through administration of depleting mAbs (n=5 isotype control). Joint swelling was assessed as previously and the mean ± SEM score (y-axis) is shown over time (x-axis). ****P<0.0001 by repeated measures ANOVA. (B) ILF area in square microns (left) and numbers (right) was assessed in colon histology from isotype control and cell-depleted mice at day 35 after bacterial gavage. Symbols represent individual mice and bars indicate the mean ± SEM. P-values were determined by one-way ANOVA with Tukey’s post-test; ns, not significant. (C to F) Total IgA and IgG in serum (C and D) and feces (E and F) at days 14 and 35 post-bacterial gavage were determined by ELISA. Symbols represent individual mice and bars indicate the mean ± SEM. *P<0.05; **P<0.01; and ns, not significant as determined by Kruskal-Wallis test with Dunn’s post-test. (G and H) Pooled day 35 serum from mice mono-colonized with isolate 1, isolate 7, and P. copri was injected into healthy germ-free DBA/1 mice or SPF DBA/1j mice, and these mice were monitored for the development of joint swelling. (G) The mean clinical score ± SEM (n=6 isolate 7 serum transfer, n=6 isolate 1 serum transfer, n=7 P. copri serum transfer) is shown relative to time after-serum transfer (x-axis) for germ-free mice. *P<0.05; **P<0.01, Kruskal-Wallis test with Dunn’s post-test. (H) The mean clinical score ± SEM (n=6 isolate 7 serum transfer, n=6 isolate 1 serum transfer, n=6 P. copri serum transfer) is shown relative to time after-serum transfer (x-axis) for SPF mice. *P<0.05; **P<0.01; ***P<0.001, Kruskal-Wallis test with Dunn’s post-test.
As the isolate 7 colonized mice develop serum autoantibodies to RA-relevant antigens, we queried if they were pathogenic. Serum was collected from mice 35 days after colonization with isolate 1, isolate 7, or P. copri mono-colonized mice and pooled by colonization group. Intraperitoneal injection of serum from isolate 7 colonized mice into healthy germ-free DBA/1 mice (Fig. 6G, fig. S22) as well as SPF DBA/1j mice (Fig. 6H) resulted in paw swelling observed within days and similar to other serum transfer studies(44, 45), resolving by 28 days following transfer, whereas serum from isolate 1 and P. copri colonized mice were unable to stimulate this phenotype. In aggregate, these findings support a potential mechanism for Subdoligranulum isolate 7 inciting B cell autoimmunity through a local intestinal-to-systemic immune response.
Subdoligranulum isolate 7 is detectable in the feces of individuals in the at-risk period and early stages of RA.
In order to determine the potential biological relevance of Subdoligranulum isolate 7 in humans at-risk for and early RA (less than 1 year from diagnosis), we utilized regions of the genome unique to isolate 7 and not present in isolate 1, to design a specific qPCR assay testing for the presence of isolate 7. The qPCR assay demonstrates specificity towards isolate 7 and sensitivity down to 10,000 bacteria per 100 mg of fecal bacteria (Fig. 7A). We then screened the feces of healthy controls (n=12), individuals at-risk for RA (n=12), and with early RA (n=12, table S6). The prevalence of isolate 7 above the limit of detection was 16.7% in the at-risk and early RA groups, but 0% in healthy controls (P<0.001, Fig. 7B). The abundance of isolate 7 varied among the positive samples, with an average of 1.637×107 ± 3.059×107 CFUs per positive sample (Fig. 7C). Previous studies have estimated that 100 mg of fecal weight contains roughly 4.1×1010 bacteria (46), indicating that our positive samples have an average of 0.04 ± 0.07% abundance of isolate 7 in their microbiomes (table S7). We similarly tested 12 unmanipulated mice from our SPF colony for the presence of isolate 7. Isolate 7 was undetectable in the feces of all of the mice (Fig. 7B). Thus, the presence of isolate 7 appears to be restricted to a subgroup of individuals at-risk for and with RA.
Figure 7: Subdoligranulum isolate 7 is detectable in the feces of individuals in the at-risk period and early stages of RA.
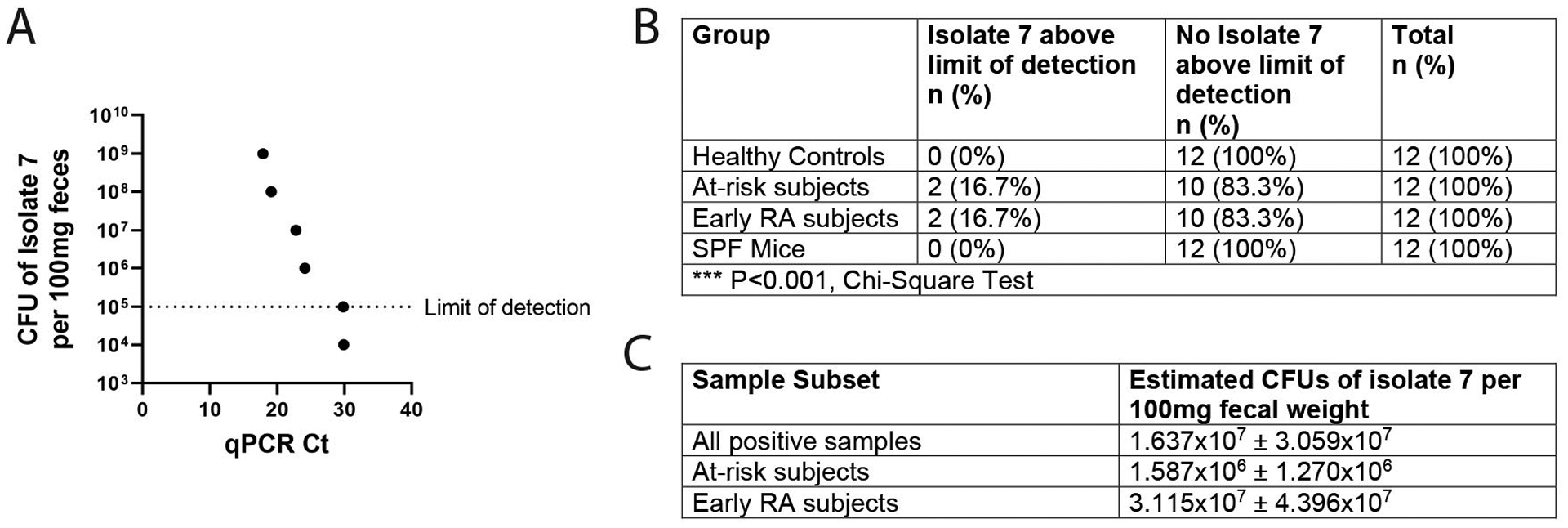
(A) Known quantities (CFU/ml) of Subdoligranulum isolate 7 were spiked into a human fecal sample to create a standard curve. The curve ranges from 1×109 CFU of isolate 7 per 100 mg of feces to 0 CFU of isolate 7, with a limit of detection set at 1×105 CFU per 100 mg of feces. A line of best fit was set for regression analysis (R2=0.9982). (B) Feces from healthy controls (n=12), individuals at-risk for RA (n=12), individuals with early RA (n=12), or SPF mice (n=12) were analyzed by qPCR for presence of isolate 7. The number and percentage of samples above the limit of detection for the assay is displayed by group. ***P<0.001, Chi-square test. (C) Regression analysis was performed utilizing the line of best fit for the assay to determine the estimated CFUs of isolate 7 in the positive samples. The mean CFUs per 100mg feces is displayed ± SEM for all positive samples (n=4), for positive samples from individuals at-risk for RA (n=2), and for positive samples for individuals with early RA (n=2).
Discussion
The mucosal origins hypothesis for the development of RA is based on compelling human immunologic and phenotypic data: IgA plasmablasts and dual IgA/IgG clonal families are expanded in the circulation during the at-risk period preceding clinically apparent RA(28); ACPA have been detected at several mucosal surfaces throughout the human body(9–11, 47) and are often of an IgA isotype(28); periodontitis, P. gingivalis, and A. actinomycetemcomitans in the oral mucosa link to ACPA generation(13, 14, 16); and cross-reactivity between proteins in P. copri and RA-relevant autoantigens has been suggested(22–24). Nevertheless, no specific microbiota-derived organism has to date been shown to be both recognized by patients with RA and able to singularly cause joint disease in experimental models. Other studies have demonstrated that strains can modulate existing arthritis(17, 48, 49), but the demonstration of the ability of a strain to incite pathology independently in genetically unmanipulated and non-mutant mice is unique. Utilizing dual IgA/IgG plasmablast derived monoclonal antibodies from human participants at risk for RA as a tool to probe the mucosal origins hypothesis, our findings establish a line of data linking a specific intestinal bacteria in the genus Subdoligranulum with local intestinal ILF formation, T cell and RA-related autoantibody development, and ultimately paw swelling associated with IgG, IgA, and C3 deposition in mice.
Not only do the presence of IgA plasmablasts and shared clonality with IgG plasmablasts during the at-risk period in RA suggest a mucosal trigger for ACPA generation and disease(28), but they also provide an important tool to probe for potential antigens driving their clonal expansion. Although the hallmark of ACPA is the citrulline specificity, recent studies of PB-mAbs from the peripheral blood of patients with RA challenge this concept, often finding cross-reactivity with antigens containing and lacking posttranslational modifications(50, 51). Finding that poly-autoreactive PB-mAbs from at-risk individuals similarly reacted with Lachnospiraceae and Ruminococcaceae further supports that bacteria could potentially drive a polyreactive response with RA-related antigens, as suggested for ACPA. We are aware of the intricacies of the terms polyreactive and cross-reactive, and have chosen to term these PB-mAbs as cross-reactive, as we hypothesize that there may be a discrete antigen(s) on the Subdoligranulum isolates that is a binding target for these antibodies. However, further studies are necessary to confirm the identity of this putative antigen(s) and determine whether molecular mimicry is relevant to the interaction.
Our initial cohort of PB-mAbs was derived from a relatively small sample size of individuals at-risk for and with RA potentially limiting the generalizability of the results. We utilized this initial small cohort as a probe to identify potential intestinal bacteria that could stimulate systemic autoantibody responses. Future directions are aimed at elucidating the antigenic properties of Subdoligranulum isolate 7 to which a larger population of individuals at-risk for and with RA can be screened for serum antibody reactivity by ELISA. Given that antibodies derived from each of the initial cohort of six individuals were reactive with families Lachnospiraceae and Ruminococcaceae, we hypothesize that many individuals at-risk for and with RA will have serum reactivity, but acknowledge that there are multiple other likely relevant bacterial candidates within the intestine, as well as at other mucosal sites.
The bacterium of interest in this study, Subdoligranulum isolate 7, is a member of a highly heterogeneous group of bacteria belonging to the family Ruminococcaceae that is phylogenetically interlinked with the family Lachnospiraceae(52, 53), both within order Clostridiales. We propose to name this bacterial isolate Subdoligranulum didolesgii, as Didolesgi is the Cherokee word for arthritis or rheumatism, and the first author is an enrolled member of the Cherokee Nation of Oklahoma (54). As we have not yet determined whether this bacterium is a commensal organism or a pathobiont, we have avoided using these terms in this manuscript. Ruminococcaceae and Lachnospiraceae are two of the most abundant families in both the human and murine gut microbiome(55, 56), raising the question of why certain strains could lead to the development of arthritis without RA being a ubiquitous disease in the population. Several potential mechanisms could explain this observation. Both Lachnospiraceae and Ruminococcaceae live deep in the mucosa, occupying a niche that allows them close access to their host organism and subsequently greater immunomodulatory potential compared to bacteria localized in the lumen(57). Furthermore, heterogeneity in families Lachnospiraceae/Ruminococcaceae may be due in part to their engagement in lateral gene transfer, or alternatively infection by bacteriophages, leading to the expression of distinct proteins and gain of functions that could lead to host immunomodulation. Interestingly, at risk individuals who are serum ACPA+ harbor increased abundances of Lachnospiraceae and Ruminococcaceae phages in their feces compared to ACPA− at risk individuals and controls(58). Molecular mimicry between bacterial and host proteins may lead to the development of autoimmunity, or bacterial products may alter mucosal immune system dynamics. In support of molecular mimicry is the recent discovery of T and B cell-targeted autoantigens in RA with shared homology between synovial and P. copri proteins(22). Alternatively, murine models of autoimmune arthritis, as evidenced by the K/BxN, collagen-induced arthritis, SKG, and HLA-B27/β2m models, demonstrate that microbial stimulation of the Th17 pathway is required for the development of arthritis(59–63). Finally, community dynamics within microbial populations under different environmental pressures could lead to altered metabolite generation, which affect immune responses within the host. For example, bacterial metabolism generates short chain fatty acids like butyrate and propionate that promote Treg development and function, protecting from collagen-induced arthritis and HLA-B27/β2m-mediated arthritis(64, 65). Thus, Subdoligranulum didolesgii may be able to promote RA-relevant autoimmunity through multiple pathways.
Mice gavaged with Subdoligranulum isolate 7 developed paw swelling in a manner that is highly reproducible through blinded scoring. Although a profound immune infiltrate into the joints was absent, there was an increase in IgG, IgA, and C3 deposition. These findings could resemble the stage of RA development in humans known as tenosynovitis, which is tendon sheath inflammation that can be associated with microscopic synovitis in the hands and feet during the at-risk and early RA periods(66, 67). We see evidence of mild synovial inflammation in the joints of mice colonized with Subdoligranulum isolate 7. If we are indeed capturing a stage of joint swelling similar to tenosynovitis in our mice, this model may be more aligned with the typical pattern of human RA development than previous murine arthritis models. Additionally, this model may be more aligned with the current two-hit hypothesis for the development of RA, the first hit being an environmental stimulus that catalyzes the development of circulating RA-relevant autoantibodies and tenosynovitis, and the second hit being an additional factor, whether genetic or environmental, that triggers the overt synovitis in classifiable RA(9, 68).
We were also able to appreciate substantial local histopathologic and immune changes in the gut of the mice gavaged with Subdoligranulum isolate 7. One key finding was the generation of mature ILFs in the colon, which are known to develop dynamically in response to bacteria in the intestine(69). They are comprised mostly of B cells with T cells and CD11c+ dendritic cells interspersed(70). Mature ILFs can form germinal centers, and the B cell repertoire inside of them has been shown to resemble systemic B cell populations(71). Clinically, ILF hyperplasia is linked with IgA dysfunction(72), and has been seen in children with circulating IgA and IgG against milk proteins(73), signaling the potential for mature ILFs to stimulate systemic immune responses. Subdoligranulum isolate 7 seems to stimulate mature ILFs, likely after direct interactions with immune cells after penetrating the epithelial barrier. This finding is corroborated with our observation that Subdoligranulum isolate 7 gains increased access to the host epithelium as compared to isolate 1. In aggregate, these findings could suggest a mechanism by which isolate 7 produces an immune response that triggers a mucosal to systemic immune response conversion. Although we observed indications of inflammation at the gut, spleen, and accompanying arthritis, other sites of potential inflammation have not been studied and could provide more mechanistic clues for this observed phenotype. We propose that similar patterns of mucosal and systemic immune responses may occur in humans, as evidenced by the targeting of isolate 7 by plasmablast-associated antibodies, CD4+ T cell responses against isolate 7, and our ability to detect isolate 7 in the feces of individuals at-risk for and with early rheumatoid arthritis.
Our study has limitations. First, as discussed above, our initial plasmablast cohort was developed from six individuals at risk for or with early RA, and our conclusions would be strengthened by additional PB-mAbs from a larger group of individuals as well as from individuals without RA or risk thereof. Next, our data do not identify differences in PB-mAb binding to Lachnospiraceae and Ruminococcaceae in the feces of healthy controls versus individuals with RA. This does not necessarily indicate that there are no intrinsic differences in the microbial composition between groups and could indicate a degree of non-specificity in the binding to the bacteria of interest. PB-mAbs could be binding both pathogenic and non-pathogenic Ruminococcaceae and may do so at varying degrees of affinity. Future studies aimed at examining the “pathogenic” properties of isolate 7 and identifying target antigens may elucidate this limitation. Finally, although our qPCR data can detect the presence of Ruminococcaceae Subdoligranulum isolate 7 in the feces of individuals at-risk for and with early RA, but not in healthy controls, the tested sample size was very limited; stronger conclusions regarding the prevalence of this isolate 7 in humans at-risk for and with RA will require a larger and longitudinal population study.
Altogether our data suggest one pathway by which the intestinal microbiome and mucosal immune responses can lead to systemic autoreactivity and joint pathology that is potentially a pathway in human RA. These data are strengthened by our ability to detect Subdoligranulum isolate 7 in the feces of individuals at-risk for and with early RA. Numerous outstanding uncertainties remain: the prevalence of our specific Subdoligranulum strain in the general population versus those with RA, and strain variations that could explain our findings; specific interactions between Subdoligranulum and the host, and the mechanism for T cell and B cell responses towards it; the necessity of epithelial invasion for the observed phenotypes; the relationship between Subdoligranulum itself and the rest of the microbial community; and finally, the role of host genetics, especially shared epitope, during the immune responses to Subdoligranulum. Nevertheless, our results highlight the utility of using host immune responses, here the IgA/IgG response in plasmablasts, to identify relevant microbiota at mucosal surfaces that contribute to RA.
Materials and Methods
Study Design
The objective of this study is to understand the bacterial targets of human autoantibodies in the context of RA, to isolate these bacteria, and to understand their role as drivers of murine joint disease. For all experiments, the number of replicates, statistical test used, and P values are reported in the figure legends. The reported replicates refer to biological replicates, either murine or human. For human studies, individuals were recruited at either the University of Colorado or Benaroya Research Institute under institutional review board approval. Full details of the human cohorts utilized can be found in the supplementary materials and methods. For murine studies, mice were derived germ-free and housed at the University of Colorado gnotobiotic facility under institutional animal care and use committee approval. Cages of mice were randomly assigned to different treatment groups, and the assessor of joint swelling was blinded to treatment group. To ensure animal welfare, weight loss and signs of pain and distress were utilized as rationale for the premature endpoint of any study. For all histopathological analysis, the scorer was blinded to treatment group. Full experimental details can be found in the supplementary materials and methods.
Statistical Analysis
All raw, individual-level data for experiments where n<20 are found in data file S3. Unless noted elsewhere, normally-distributed data as determined by D’Agostino test were evaluated by analysis of variance (ANOVA) and a post-hoc t-test with Tukey’s correction. Non-parametric data were analyzed using Mann-Whitney or Kruskal Wallis with Dunn’s correction. Qualitative data were compared using Chi-squared and Fisher’s exact tests where noted. Data were analyzed using GraphPad Prism 9 Software.
Supplementary Material
Acknowledgements:
The authors would like the acknowledge Kadin E. Brooks for technical assistance in preparing this manuscript.
Funding:
The work by the authors is supported through U01 AI101981 (to VMH, WHR, JHB, KDD, KAK), P30 AR079369 (to VMH, KDD, and KAK), Pfizer ASPIRE (to KAK), T32 AR07534 (to MC and ARL), R01 AR075033 (to KAK), and the Rheumatology Research Foundation Future Physician Scientist Award (to MC).
Footnotes
List of Supplementary Materials
Supplemental materials and methods
MDAR reproducibility checklist
Data file S1 to S3
Competing Interests: The authors have no conflicts of interests to declare.
Data Availability:
All data associated with this study are in the paper or supplementary materials. Data generated from PB-mAbs, 16S rRNA, and bacterial whole genome sequencing are publicly accessible in accordance with NIH guidelines. Sequencing data are publicly available through the NIH Sequence Read Archive (SRA) accession numbers SUB10802055 (16S sequencing of human fecal samples), SUB 10768018 (16S sequencing of bacteria bound by PB-mAbs), SUB10785477 (Subdoligranulum isolates from human feces), and SUB10768012 (Subdoligranulum didolesgii strain sequences).
References
- 1.Nielen MM, van Schaardenburg D, Reesink HW, van de Stadt RJ, van der Horst-Bruinsma IE, de Koning MH, Habibuw MR, Vandenbroucke JP, Dijkmans BA, Specific autoantibodies precede the symptoms of rheumatoid arthritis: a study of serial measurements in blood donors. Arthritis Rheum 50, 380386 (2004); published online Epub Feb ( 10.1002/art.20018). [DOI] [PubMed] [Google Scholar]
- 2.Arlestig L, Mullazehi M, Kokkonen H, Rocklov J, Ronnelid J, Dahlqvist SR, Antibodies against cyclic citrullinated peptides of IgG, IgA and IgM isotype and rheumatoid factor of IgM and IgA isotype are increased in unaffected members of multicase rheumatoid arthritis families from northern Sweden. Ann Rheum Dis 71, 825–829 (2012); published online Epub Jun ( 10.1136/annrheumdis-2011-200668). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sokolove J, Bromberg R, Deane KD, Lahey LJ, Derber LA, Chandra PE, Edison JD, Gilliland WR, Tibshirani RJ, Norris JM, Holers VM, Robinson WH, Autoantibody epitope spreading in the pre-clinical phase predicts progression to rheumatoid arthritis. PLoS One 7, e35296 (2012) 10.1371/journal.pone.0035296). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kelmenson LB, Wagner BD, McNair BK, Frazer-Abel A, Demoruelle MK, Bergstedt DT, Feser ML, Moss LK, Parish MC, Mewshaw EA, Mikuls TR, Edison JD, Holers VM, Deane KD, Timing of Elevations of Autoantibody Isotypes Prior to Diagnosis of Rheumatoid Arthritis. Arthritis Rheumatol 72, 251–261 (2020); published online Epub Feb ( 10.1002/art.41091). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerlag DM, Raza K, van Baarsen LG, Brouwer E, Buckley CD, Burmester GR, Gabay C, Catrina AI, Cope AP, Cornelis F, Dahlqvist SR, Emery P, Eyre S, Finckh A, Gay S, Hazes JM, van der Helm-van Mil A, Huizinga TW, Klareskog L, Kvien TK, Lewis C, Machold KP, Ronnelid J, van Schaardenburg D, Schett G, Smolen JS, Thomas S, Worthington J, Tak PP, EULAR recommendations for terminology and research in individuals at risk of rheumatoid arthritis: report from the Study Group for Risk Factors for Rheumatoid Arthritis. Ann Rheum Dis 71, 6382013641 (2012); published online Epub May ( 10.1136/annrheumdis-2011-200990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuhn KA, Kulik L, Tomooka B, Braschler KJ, Arend WP, Robinson WH, Holers VM, Antibodies against citrullinated proteins enhance tissue injury in experimental autoimmune arthritis. J Clin Invest 116, 961–973 (2006); published online Epub Apr ( 10.1172/JCI25422). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Zeben D, Hazes JM, Zwinderman AH, Cats A, van der Voort EA, Breedveld FC, Clinical significance of rheumatoid factors in early rheumatoid arthritis: results of a follow up study. Ann Rheum Dis 51, 1029–1035 (1992); published online Epub Sep ( 10.1136/ard.51.9.1029). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu DR, McDavid AN, Kongpachith S, Lingampalli N, Glanville J, Ju CH, Gottardo R, Robinson WH, T Cell-Dependent Affinity Maturation and Innate Immune Pathways Differentially Drive Autoreactive B Cell Responses in Rheumatoid Arthritis. Arthritis Rheumatol 70, 1732–1744 (2018); published online Epub Nov ( 10.1002/art.40578). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holers VM, Demoruelle MK, Kuhn KA, Buckner JH, Robinson WH, Okamoto Y, Norris JM, Deane KD, Rheumatoid arthritis and the mucosal origins hypothesis: protection turns to destruction. Nat Rev Rheumatol 14, 542–557 (2018); published online Epub Sep ( 10.1038/s41584-018-0070-0). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rangel-Moreno J, Hartson L, Navarro C, Gaxiola M, Selman M, Randall TD, Inducible bronchus-associated lymphoid tissue (iBALT) in patients with pulmonary complications of rheumatoid arthritis. J Clin Invest 116, 3183–3194 (2006); published online Epub Dec ( 10.1172/JCI28756). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Willis VC, Demoruelle MK, Derber LA, Chartier-Logan CJ, Parish MC, Pedraza IF, Weisman MH, Norris JM, Holers VM, Deane KD, Sputum autoantibodies in patients with established rheumatoid arthritis and subjects at risk of future clinically apparent disease. Arthritis Rheum 65, 2545–2554 (2013); published online Epub Oct ( 10.1002/art.38066). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Demoruelle MK, Harrall KK, Ho L, Purmalek MM, Seto NL, Rothfuss HM, Weisman MH, Solomon JJ, Fischer A, Okamoto Y, Kelmenson LB, Parish MC, Feser M, Fleischer C, Anderson C, Mahler M, Norris JM, Kaplan MJ, Cherrington BD, Holers VM, Deane KD, Anti-Citrullinated Protein Antibodies Are Associated With Neutrophil Extracellular Traps in the Sputum in Relatives of Rheumatoid Arthritis Patients. Arthritis Rheumatol 69, 1165–1175 (2017); published online Epub Jun ( 10.1002/art.40066). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lundberg K, Wegner N, Yucel-Lindberg T, Venables PJ, Periodontitis in RA-the citrullinated enolase connection. Nat Rev Rheumatol 6, 727–730 (2010); published online Epub Dec ( 10.1038/nrrheum.2010.139). [DOI] [PubMed] [Google Scholar]
- 14.Cheng Z, Do T, Mankia K, Meade J, Hunt L, Clerehugh V, Speirs A, Tugnait A, Emery P, Devine D, Dysbiosis in the oral microbiomes of anti-CCP positive individuals at risk of developing rheumatoid arthritis. Ann Rheum Dis 80, 162–168 (2021); published online Epub Feb ( 10.1136/annrheumdis-2020-216972). [DOI] [PubMed] [Google Scholar]
- 15.Scher JU, Abramson SB, Periodontal disease, Porphyromonas gingivalis, and rheumatoid arthritis: what triggers autoimmunity and clinical disease? Arthritis Res Ther 15, 122 (2013) 10.1186/ar4360). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Konig MF, Abusleme L, Reinholdt J, Palmer RJ, Teles RP, Sampson K, Rosen A, Nigrovic PA, Sokolove J, Giles JT, Moutsopoulos NM, Andrade F, Aggregatibacter actinomycetemcomitans-induced hypercitrullination links periodontal infection to autoimmunity in rheumatoid arthritis. Sci Transl Med 8, 369ra176 (2016); published online Epub Dec 14 ( 10.1126/scitranslmed.aaj1921). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moentadj R, Wang Y, Bowerman K, Rehaume L, Nel H, P OC, Stephens J, Baharom A, Maradana M, Lakis V, Morrison M, Wells T, Hugenholtz P, Benham H, Le Cao KA, Thomas R, Streptococcus species enriched in the oral cavity of patients with RA are a source of peptidoglycan-polysaccharide polymers that can induce arthritis in mice. Ann Rheum Dis 80, 573–581 (2021); published online Epub May ( 10.1136/annrheumdis-2020-219009). [DOI] [PubMed] [Google Scholar]
- 18.Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, Rostron T, Cerundolo V, Pamer EG, Abramson SB, Huttenhower C, Littman DR, Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife 2, e01202 (2013); published online Epub Nov 5 ( 10.7554/eLife.01202). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alpizar-Rodriguez D, Lesker TR, Gronow A, Gilbert B, Raemy E, Lamacchia C, Gabay C, Finckh A, Strowig T, Prevotella copri in individuals at risk for rheumatoid arthritis. Ann Rheum Dis 78, 590–593 (2019); published online Epub May ( 10.1136/annrheumdis-2018-214514). [DOI] [PubMed] [Google Scholar]
- 20.Zhang X, Zhang D, Jia H, Feng Q, Wang D, Liang D, Wu X, Li J, Tang L, Li Y, Lan Z, Chen B, Li Y, Zhong H, Xie H, Jie Z, Chen W, Tang S, Xu X, Wang X, Cai X, Liu S, Xia Y, Li J, Qiao X, Al-Aama JY, Chen H, Wang L, Wu QJ, Zhang F, Zheng W, Li Y, Zhang M, Luo G, Xue W, Xiao L, Li, Chen W, Xu X, Yin Y, Yang H, Wang J, Kristiansen K, Liu L, Li T, Huang Q, Li Y, Wang J, The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med 21, 895–905 (2015); published online Epub Aug ( 10.1038/nm.3914). [DOI] [PubMed] [Google Scholar]
- 21.Chen J, Wright K, Davis JM, Jeraldo P, Marietta EV, Murray J, Nelson H, Matteson EL, Taneja V, An expansion of rare lineage intestinal microbes characterizes rheumatoid arthritis. Genome Med 8, 43 (2016); published online Epub Apr 21 ( 10.1186/s13073-016-0299-7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pianta A, Arvikar SL, Strle K, Drouin EE, Wang Q, Costello CE, Steere AC, Two rheumatoid arthritis-specific autoantigens correlate microbial immunity with autoimmune responses in joints. The Journal of clinical investigation 127, 2946–2956 (2017); published online Epub Aug 1 ( 10.1172/JCI93450). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pianta A, Arvikar S, Strle K, Drouin EE, Wang Q, Costello CE, Steere AC, Evidence of the Immune Relevance of Prevotella copri, a Gut Microbe, in Patients With Rheumatoid Arthritis. Arthritis Rheumatol 69, 964–975 (2017); published online Epub May ( 10.1002/art.40003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pianta A, Chiumento G, Ramsden K, Wang Q, Strle K, Arvikar S, Costello CE, Steere AC, Identification of Novel, Immunogenic HLA-DR-Presented Prevotella copri Peptides in Patients with Rheumatoid Arthritis: Patients with Rheumatoid Arthritis. Arthritis Rheumatol, (2021); published online Epub May 27 ( 10.1002/art.41807). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kerkman PF, Fabre E, van der Voort EI, Zaldumbide A, Rombouts Y, Rispens T, Wolbink G, Hoeben RC, Spits H, Baeten DL, Huizinga TW, Toes RE, Scherer HU, Identification and characterisation of citrullinated antigen-specific B cells in peripheral blood of patients with rheumatoid arthritis. Annals of the rheumatic diseases 75, 1170–1176 (2016); published online Epub Jun ( 10.1136/annrheumdis-2014-207182). [DOI] [PubMed] [Google Scholar]
- 26.Odendahl M, Mei H, Hoyer BF, Jacobi AM, Hansen A, Muehlinghaus G, Berek C, Hiepe F, Manz R, Radbruch A, Dorner T, Generation of migratory antigen-specific plasma blasts and mobilization of resident plasma cells in a secondary immune response. Blood 105, 1614–1621 (2005); published online Epub Feb 15 ( 10.1182/blood-2004-07-2507). [DOI] [PubMed] [Google Scholar]
- 27.Radbruch A, Muehlinghaus G, Luger EO, Inamine A, Smith KG, Dorner T, Hiepe F, Competence and competition: the challenge of becoming a long-lived plasma cell. Nat Rev Immunol 6, 741–750 (2006); published online Epub Oct ( 10.1038/nri1886). [DOI] [PubMed] [Google Scholar]
- 28.Kinslow JD, Blum LK, Deane KD, Demoruelle MK, Okamoto Y, Parish MC, Kongpachith S, Lahey LJ, Norris JM, Robinson WH, Holers VM, Elevated IgA Plasmablast Levels in Subjects at Risk of Developing Rheumatoid Arthritis. Arthritis Rheumatol 68, 2372–2383 (2016); published online Epub Oct ( 10.1002/art.39771). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I, ABySS: a parallel assembler for short read sequence data. Genome Res 19, 1117–1123 (2009); published online Epub Jun ( 10.1101/gr.089532.108). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McIver LJ, Abu-Ali G, Franzosa EA, Schwager R, Morgan XC, Waldron L, Segata N, Huttenhower C, bioBakery: a meta’omic analysis environment. Bioinformatics 34, 1235–1237 (2018); published online Epub Apr 1 ( 10.1093/bioinformatics/btx754). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lubberts E, The IL-23-IL-17 axis in inflammatory arthritis. Nat Rev Rheumatol 11, 415–429 (2015); published online Epub Jul ( 10.1038/nrrheum.2015.53). [DOI] [PubMed] [Google Scholar]
- 32.Li SX, Sen S, Schneider JM, Xiong KN, Nusbacher NM, Moreno-Huizar N, Shaffer M, Armstrong AJS, Severs E, Kuhn K, Neff CP, McCarter M, Campbell T, Lozupone CA, Palmer BE, Gut microbiota from high-risk men who have sex with men drive immune activation in gnotobiotic mice and in vitro HIV infection. PLoS Pathog 15, e1007611 (2019); published online Epub Apr ( 10.1371/journal.ppat.1007611). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuhn KA, Stappenbeck TS, Peripheral education of the immune system by the colonic microbiota. Semin Immunol 25, 364–369 (2013); published online Epub Nov 30 ( 10.1016/j.smim.2013.10.002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brand DD, Latham KA, Rosloniec EF, Collagen-induced arthritis. Nat Protoc 2, 1269–1275 (2007) 10.1038/nprot.2007.173). [DOI] [PubMed] [Google Scholar]
- 35.Khachigian LM, Collagen antibody-induced arthritis. Nat Protoc 1, 2512–2516 (2006) 10.1038/nprot.2006.393). [DOI] [PubMed] [Google Scholar]
- 36.Omenetti S, Pizarro TT, The Treg/Th17 Axis: A Dynamic Balance Regulated by the Gut Microbiome. Front Immunol 6, 639 (2015) 10.3389/fimmu.2015.00639). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang Y, Torchinsky MB, Gobert M, Xiong H, Xu M, Linehan JL, Alonzo F, Ng C, Chen A, Lin X, Sczesnak A, Liao JJ, Torres VJ, Jenkins MK, Lafaille JJ, Littman DR, Focused specificity of intestinal TH17 cells towards commensal bacterial antigens. Nature 510, 152–156 (2014); published online Epub Jun 5 ( 10.1038/nature13279). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hickman-Brecks CL, Racz JL, Meyer DM, LaBranche TP, Allen PM, Th17 cells can provide B cell help in autoantibody induced arthritis. J Autoimmun 36, 65–75 (2011); published online Epub Feb ( 10.1016/j.jaut.2010.10.007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang W, Shao S, Jiao Z, Guo M, Xu H, Wang S, The Th17/Treg imbalance and cytokine environment in peripheral blood of patients with rheumatoid arthritis. Rheumatol Int 32, 887–893 (2012); published online Epub Apr ( 10.1007/s00296-010-1710-0). [DOI] [PubMed] [Google Scholar]
- 40.Chang HH, Liu GY, Dwivedi N, Sun B, Okamoto Y, Kinslow JD, Deane KD, Demoruelle MK, Norris JM, Thompson PR, Sparks JA, Rao DA, Karlson EW, Hung HC, Holers VM, Ho IC, A molecular signature of preclinical rheumatoid arthritis triggered by dysregulated PTPN22. JCI Insight 1, e90045 (2016); published online Epub Oct 20 ( 10.1172/jci.insight.90045). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johansson ME, Jakobsson HE, Holmen-Larsson J, Schutte A, Ermund A, Rodriguez-Pineiro AM, Arike L, Wising C, Svensson F, Backhed F, Hansson GC, Normalization of Host Intestinal Mucus Layers Requires Long-Term Microbial Colonization. Cell Host Microbe 18, 582–592 (2015); published online Epub Nov 11 ( 10.1016/j.chom.2015.10.007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Glaysher BR, Mabbott NA, Isolated lymphoid follicle maturation induces the development of follicular dendritic cells. Immunology 120, 336–344 (2007); published online Epub Mar ( 10.1111/j.1365-2567.2006.02508.x). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lorenz RG, Newberry RD, Isolated lymphoid follicles can function as sites for induction of mucosal immune responses. Ann N Y Acad Sci 1029, 44–57 (2004); published online Epub Dec ( 10.1196/annals.1309.006). [DOI] [PubMed] [Google Scholar]
- 44.Corr M, Crain B, The role of FcgammaR signaling in the K/B × N serum transfer model of arthritis. J Immunol 169, 6604–6609 (2002); published online Epub Dec 1 ( 10.4049/jimmunol.169.11.6604). [DOI] [PubMed] [Google Scholar]
- 45.Christianson CA, Corr M, Yaksh TL, Svensson CI, K/BxN serum transfer arthritis as a model of inflammatory joint pain. Methods Mol Biol 851, 249–260 (2012) 10.1007/978-1-61779-561-9_19). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stephen AM, Cummings JH, The microbial contribution to human faecal mass. J Med Microbiol 13, 45–56 (1980); published online Epub Feb ( 10.1099/00222615-13-1-45). [DOI] [PubMed] [Google Scholar]
- 47.Demoruelle MK, Bowers E, Lahey LJ, Sokolove J, Purmalek M, Seto NL, Weisman MH, Norris JM, Kaplan MJ, Holers VM, Robinson WH, Deane KD, Antibody Responses to Citrullinated and Noncitrullinated Antigens in the Sputum of Subjects With Rheumatoid Arthritis and Subjects at Risk for Development of Rheumatoid Arthritis. Arthritis Rheumatol 70, 516–527 (2018); published online Epub Apr ( 10.1002/art.40401). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gupta VK, Cunningham KY, Hur B, Bakshi U, Huang H, Warrington KJ, Taneja V, Myasoedova E, Davis JM 3rd, Sung J, Gut microbial determinants of clinically important improvement in patients with rheumatoid arthritis. Genome Med 13, 149 (2021); published online Epub Sep 14 ( 10.1186/s13073-021-00957-0). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu X, Zeng B, Zhang J, Li W, Mou F, Wang H, Zou Q, Zhong B, Wu L, Wei H, Fang Y, Role of the Gut Microbiome in Modulating Arthritis Progression in Mice. Sci Rep 6, 30594 (2016); published online Epub Aug 2 ( 10.1038/srep30594). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Corsiero E, Bombardieri M, Carlotti E, Pratesi F, Robinson W, Migliorini P, Pitzalis C, Single cell cloning and recombinant monoclonal antibodies generation from RA synovial B cells reveal frequent targeting of citrullinated histones of NETs. Ann Rheum Dis 75, 1866–1875 (2016); published online Epub Oct ( 10.1136/annrheumdis-2015-208356). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Steen J, Forsstrom B, Sahlstrom P, Odowd V, Israelsson L, Krishnamurthy A, Badreh S, Mathsson Alm L, Compson J, Ramskold D, Ndlovu W, Rapecki S, Hansson M, Titcombe PJ, Bang H, Mueller DL, Catrina AI, Gronwall C, Skriner K, Nilsson P, Lightwood D, Klareskog L, Malmstrom V, Recognition of Amino Acid Motifs, Rather Than Specific Proteins, by Human Plasma Cell-Derived Monoclonal Antibodies to Posttranslationally Modified Proteins in Rheumatoid Arthritis. Arthritis Rheumatol 71, 196–209 (2019); published online Epub Feb ( 10.1002/art.40699). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.La Reau AJ, Meier-Kolthoff JP, Suen G, Sequence-based analysis of the genus Ruminococcus resolves its phylogeny and reveals strong host association. Microb Genom 2, e000099 (2016); published online Epub Dec ( 10.1099/mgen.0.000099). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meehan CJ, Beiko RG, A phylogenomic view of ecological specialization in the Lachnospiraceae, a family of digestive tract-associated bacteria. Genome Biol Evol 6, 703–713 (2014); published online Epub Mar ( 10.1093/gbe/evu050). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.We chose this name for a few significant reasons. First, we recognize that the indigenous peoples of the Americas are affected by arthritis at a disproportionate rate than other racial and ethnic groups. Secondly, we acknowledge that there is a history of erasure of the advances made by indigenous scientists and knowledge seekers. Third, because we are taught that by speaking the language of our ancestors that we breathe life into it and strengthen all our relations.
- 55.Sorbara MT, Littmann ER, Fontana E, Moody TU, Kohout CE, Gjonbalaj M, Eaton V, Seok R, Leiner IM, Pamer EG, Functional and Genomic Variation between Human-Derived Isolates of Lachnospiraceae Reveals Inter- and Intra-Species Diversity. Cell Host Microbe 28, 134–146 e134 (2020); published online Epub Jul 8 ( 10.1016/j.chom.2020.05.005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rajilic-Stojanovic M, de Vos WM, The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol Rev 38, 996–1047 (2014); published online Epub Sep ( 10.1111/1574-6976.12075). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schroeder BO, Fight them or feed them: how the intestinal mucus layer manages the gut microbiota. Gastroenterol Rep 7, 3–12 (2019); published online Epub Feb ( 10.1093/gastro/goy052). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mangalea MR, Paez-Espino D, Kieft K, Chatterjee A, Chriswell ME, Seifert JA, Feser ML, Demoruelle MK, Sakatos A, Anantharaman K, Deane KD, Kuhn KA, Holers VM, Duerkop BA, Individuals at risk for rheumatoid arthritis harbor differential intestinal bacteriophage communities with distinct metabolic potential. Cell Host & Microbe 29, 726-+ (2021); published online Epub May 12 ( 10.1016/j.chom.2021.03.020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jubair WK, Hendrickson JD, Severs EL, Schulz HM, Adhikari S, Ir D, Pagan JD, Anthony RM, Robertson CE, Frank DN, Banda NK, Kuhn KA, Modulation of Inflammatory Arthritis in Mice by Gut Microbiota Through Mucosal Inflammation and Autoantibody Generation. Arthritis Rheumatol 70, 1220–1233 (2018); published online Epub Aug ( 10.1002/art.40490). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Teng F, Felix KM, Bradley CP, Naskar D, Ma H, Raslan WA, Wu HJ, The impact of age and gut microbiota on Th17 and Tfh cells in K/BxN autoimmune arthritis. Arthritis Res Ther 19, 188 (2017); published online Epub Aug 15 ( 10.1186/s13075-017-1398-6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Asquith MJ, Stauffer P, Davin S, Mitchell C, Lin P, Rosenbaum JT, Perturbed Mucosal Immunity and Dysbiosis Accompany Clinical Disease in a Rat Model of Spondyloarthritis. Arthritis Rheumatol 68, 2151–2162 (2016); published online Epub Sep ( 10.1002/art.39681). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scher JU, Ubeda C, Artacho A, Attur M, Isaac S, Reddy SM, Marmon S, Neimann A, Brusca S, Patel T, Manasson J, Pamer EG, Littman DR, Abramson SB, Decreased bacterial diversity characterizes the altered gut microbiota in patients with psoriatic arthritis, resembling dysbiosis in inflammatory bowel disease. Arthritis Rheumatol 67, 128–139 (2015); published online Epub Jan ( 10.1002/art.38892). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Asquith M, Sternes PR, Costello ME, Karstens L, Diamond S, Martin TM, Li Z, Marshall MS, Spector TD, le Cao KA, Rosenbaum JT, Brown MA, HLA Alleles Associated With Risk of Ankylosing Spondylitis and Rheumatoid Arthritis Influence the Gut Microbiome. Arthritis Rheumatol 71, 1642–1650 (2019); published online Epub Oct ( 10.1002/art.40917). [DOI] [PubMed] [Google Scholar]
- 64.Kim DS, Kwon JE, Lee SH, Kim EK, Ryu JG, Jung KA, Choi JW, Park MJ, Moon YM, Park SH, Cho ML, Kwok SK, Attenuation of Rheumatoid Inflammation by Sodium Butyrate Through Reciprocal Targeting of HDAC2 in Osteoclasts and HDAC8 in T Cells. Front Immunol 9, 1525 (2018) 10.3389/fimmu.2018.01525). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Asquith M, Davin S, Stauffer P, Michell C, Janowitz C, Lin P, Ensign-Lewis J, Kinchen JM, Koop DR, Rosenbaum JT, Intestinal Metabolites Are Profoundly Altered in the Context of HLA-B27 Expression and Functionally Modulate Disease in a Rat Model of Spondyloarthritis. Arthritis Rheumatol 69, 1984–1995 (2017); published online Epub Oct ( 10.1002/art.40183). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ray G, Sandean DP, Tall MA, in StatPearls. (Treasure Island (FL), 2021). [Google Scholar]
- 67.Hitchon CA, El-Gabalawy HS, The synovium in rheumatoid arthritis. Open Rheumatol J 5, 107–114 (2011) 10.2174/1874312901105010107). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Klareskog L, Ronnelid J, Saevarsdottir S, Padyukov L, Alfredsson L, The importance of differences; On environment and its interactions with genes and immunity in the causation of rheumatoid arthritis. J Intern Med 287, 514–533 (2020); published online Epub May ( 10.1111/joim.13058). [DOI] [PubMed] [Google Scholar]
- 69.Knoop KA, Newberry RD, Isolated Lymphoid Follicles are Dynamic Reservoirs for the Induction of Intestinal IgA. Front Immunol 3, 84 (2012) 10.3389/fimmu.2012.00084). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hamada H, Hiroi T, Nishiyama Y, Takahashi H, Masunaga Y, Hachimura S, Kaminogawa S, Takahashi-Iwanaga H, Iwanaga T, Kiyono H, Yamamoto H, Ishikawa H, Identification of multiple isolated lymphoid follicles on the antimesenteric wall of the mouse small intestine. J Immunol 168, 57–64 (2002); published online Epub Jan 1 ( 10.4049/jimmunol.168.1.57). [DOI] [PubMed] [Google Scholar]
- 71.Wang C, McDonald KG, McDonough JS, Newberry RD, Murine isolated lymphoid follicles contain follicular B lymphocytes with a mucosal phenotype. Am J Physiol Gastrointest Liver Physiol 291, G595–604 (2006); published online Epub Oct ( 10.1152/ajpgi.00525.2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bastlein C, Burlefinger R, Holzberg E, Voeth C, Garbrecht M, Ottenjann R, Common variable immunodeficiency syndrome and nodular lymphoid hyperplasia in the small intestine. Endoscopy 20, 272–275 (1988); published online Epub Sep ( 10.1055/s-2007-1018192). [DOI] [PubMed] [Google Scholar]
- 73.Kokkonen J, Tikkanen S, Karttunen TJ, Savilahti E, A similar high level of immunoglobulin A and immunoglobulin G class milk antibodies and increment of local lymphoid tissue on the duodenal mucosa in subjects with cow’s milk allergy and recurrent abdominal pains. Pediatr Allergy Immunol 13, 129–136 (2002); published online Epub Apr ( 10.1034/j.1399-3038.2002.00090.x). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data associated with this study are in the paper or supplementary materials. Data generated from PB-mAbs, 16S rRNA, and bacterial whole genome sequencing are publicly accessible in accordance with NIH guidelines. Sequencing data are publicly available through the NIH Sequence Read Archive (SRA) accession numbers SUB10802055 (16S sequencing of human fecal samples), SUB 10768018 (16S sequencing of bacteria bound by PB-mAbs), SUB10785477 (Subdoligranulum isolates from human feces), and SUB10768012 (Subdoligranulum didolesgii strain sequences).


