Abstract
Low temperature affects the yield and quality of crops. Inducer of CBF expression 1 (ICE1) plays a positive role in plant cold tolerance by promoting the expression of CRT binding factor (CBF) and cold-responsive (COR) genes. Several ICE1-interacting transcription factors (TFs) that regulate plant cold tolerance have been identified. However, how these TFs affect the function of ICE1 and CBF expression under cold conditions remains unclear. Here, we identified the MYC-type TF MdbHLH4, a negative regulator of cold tolerance in Arabidopsis (Arabidopsis thaliana) and apple (Malus domestica) plants. Under cold conditions, MdbHLH4 inhibits the expression of MdCBF1 and MdCBF3 by directly binding to their promoters. It also interacts with MdICE1L, a homolog of AtICE1 in apple, and inhibits the binding of MdICE1L to the promoters of MdCBF1/3 and thus their expression. We showed that MdCAX3L-2, a Ca2+/H+ exchanger (CAX) family gene that negatively regulates plant cold tolerance, is also a direct target of MdbHLH4. MdbHLH4 reduced apple cold tolerance by promoting MdCAX3L-2 expression. Moreover, overexpression of either MdCAX3L-2 or MdbHLH4 promoted the cold-induced ubiquitination and degradation of MdICE1L. Overall, our results reveal that MdbHLH4 negatively regulates plant cold tolerance by inhibiting MdCBF1/3 expression and MdICE1L promoter-binding activity, as well as by promoting MdCAX3L-2 expression and cold-induced MdICE1L degradation. These findings provide insights into the mechanisms by which ICE1-interacting TFs regulate CBF expression and ICE1 function and thus plant cold tolerance.
One-sentence summary MdbHLH4 negatively regulates plant cold tolerance by inhibiting MdCBF1/3 expression and MdICE1L promoter-binding activity and promoting MdCAX3L-2 expression and cold-induced MdICE1L degradation.
Introduction
Low temperature is one of the major factors that limits geographical distribution of plants and adversely affects crop yield and quality (Thomashow, 1999; Guo et al., 2018; Ding and Yang, 2022). To rapidly sense and respond to cold stress, plants have evolved complex and elaborate regulatory mechanisms (Kidokoro et al., 2017; Guo et al., 2018; Ding et al., 2019; Ding and Yang, 2022). The activation of cold-responsive (COR) genes plays a key role in plant cold signaling (Chinnusamy et al., 2003; Miura et al., 2011; Guo et al., 2018), and DRE binding factor 1/CRT binding factor (DREB1/CBF) transcription factors (TFs) can directly promote their expression in response to cold (Stockinger et al., 1997; Gilmour et al., 2004; Chinnusamy et al., 2007; Liu et al., 2018; Song et al., 2021). Till now, many TFs involved in the regulation of CBFs expression have been identified, such as the negative regulators MYB15 (Agarwal et al., 2006), ethylene insensitive 3 (EIN3) (Shi et al., 2012), phytochrome-interacting factors (PIFs) (Lee and Thomashow, 2012; Jiang et al., 2017), and the positive regulators inducer of CBF expression 1 (ICE1) (Chinnusamy et al., 2003) and calmodulin binding transcription activators (CAMTAs) (Doherty et al., 2009; Kim et al., 2013).
ICE1 is a member of the basic helix–loop–helix (bHLH) TF family (Lee et al., 2005). The bHLH domain consists of two parts: the basic region responsible for DNA binding and the HLH region responsible for homodimerization or heterodimerization (Pires and Dolan, 2010; Feller et al., 2011). Based on sequence alignment and 3D structure analysis, a conserved H5-E9-R13 motif and several residues crucial for DNA binding and protein interaction have been identified in this domain (Pires and Dolan, 2010; Lian et al., 2017; Su et al., 2017). Generally, bHLH proteins specifically recognize and bind to canonical E-box/MYC cis-elements (CANNTG) in the promoters of target genes to regulate their expression (Pires and Dolan, 2010; Feller et al., 2011). In Arabidopsis (Arabidopsis thaliana), ICE1 directly binds to the E-box in the CBF3 promoter and promotes its expression in response to cold stress (Chinnusamy et al., 2003). Subsequent studies reveal that the ICE1-CBF module is widespread and conserved in various plant species, such as rice (Oryza sativa), wheat (Triticum aestivum), barley (Hordeum vulgare), tomato (Solanum lycopersicum), potato (Solanum tuberosum), and apple (Malus domestica) (Pino et al., 2007; Morran et al., 2011; Feng et al., 2012, 2013; Guo et al., 2018; An et al., 2021).
In addition to ICE1, several ICE1-interacting TFs that regulate CBFs expression have been identified, such as the negative regulators MYC67 and MYC70. As MYC-type bHLH TFs, these two proteins can bind to the promoter of CBF3 and inhibit its expression under cold stress (Ohta et al., 2018). Moreover, MYC67/70 may suppress CBFs expression by inhibiting the promoter-binding activity of ICE1 via unknown mechanisms (Ohta et al., 2018). A similar situation applies to MYB15, a key negative regulator of cold-induced CBFs expression (Agarwal et al., 2006). Recent studies have revealed the molecular mechanism by which cold stress regulates the activity and stability of MYB15 protein (Kim et al., 2017; Wang et al., 2019). However, whether and how the MYB15–ICE1 interaction affects ICE function remains unclear (Agarwal et al., 2006). In apple, a BBX (B-box) TF MdBBX37 functions positively in cold tolerance by directly activating MdCBF1/4 expression. Moreover, it can enhance the binding activity of MdICE1 to the MdCBF1 promoter by interacting with MdICE1 (An et al., 2021).
Calcium signaling plays an important role in plant cold response. Following exposure to cold stress, cytoplasmic Ca2+ levels rapidly increase (Knight et al., 1996; Guo et al., 2018; Cui et al., 2020; Ding and Yang, 2022). This Ca2+ signature induces a series of downstream events, such as the expression of CBF and COR genes (Chinnusamy et al., 2007; Dodd et al., 2010). The Ca2+ signature also affects the protein stability of ICE1. Cold stress induces the degradation of ICE1 through the 26S ubiquitin-proteasome pathway, which is promoted by the phosphokinases mitogen-activated protein kinases 3/6 (MPK3/6) (Li et al., 2017). Two plasma membrane-localized Ca2+/calmodulin (CaM)-regulated receptor-like kinases (CRLKs), CRLK1 and CRLK2, can suppress the activity of MPK3/6 and ICE degradation through the MEKK1–MKK2–MPK4 cascade (Zhao et al., 2017; Guo et al., 2018; Ding and Yang, 2022). Notably, the kinase activity of CRLKs is stimulated by Ca2+/CaM binding (Yang et al., 2010; Zhao et al., 2017). Open stomata 1 (OST1), a sucrose non-fermenting-related protein kinase 2 family member, maintains ICE stability by suppressing HOS1-mediated ubiquitination of ICE1 (Dong et al., 2006; Ding et al., 2015). Since the cold-activated OST1 activity does not depend on ABA, other pathways responsible for OST1 activity have been suggested, such as cold-induced Ca2+ signaling (Ding et al., 2015; Lee and Seo, 2021; Ding and Yang, 2022). Several other key factors that affect ICE1 protein stability have also been identified, such as the SUMO E3 ligase SIZ1 (SAP and MIZ1 domain-containing ligase 1) (Miura et al., 2007) and the GSK3-like kinase brassinosteroid-insensitive 2 (BIN2) (Ye et al., 2019). However, the mechanisms of their cold activation remain elusive.
Due to the crucial role of calcium signaling in cold response, many Ca2+ channels and transporters involved in the regulation of cold-induced Ca2+ signature and plant cold tolerance have been identified, including cyclic nucleotide-gated ion channel (CNGCs) (Cui et al., 2020; Wang et al., 2021), Mid1-complementing activity (MCAs) (Yamanaka et al., 2010; Mori et al., 2018; Yoshimura et al., 2021), and ANNEXIN1 (Liu et al., 2021). The Ca2+/H+ exchangers (CAXs) are a subgroup within the Ca2+/cation antiporter superfamily (Shigaki and Hirschi, 2006; Manohar et al., 2011). In general, CAX proteins are localized to the vacuole membrane and promote the compartmentalization of cytosolic Ca2+ into vacuoles, which prevents the accumulation of excess Ca2+ in the cytoplasm (Wu et al., 2013; Pittman and Hirschi, 2016; Ma and Berkowitz, 2017). In Arabidopsis, the expression of CAX1 is significantly induced by cold stress, and cax1 mutants exhibit increased CBF expression and enhanced cold acclimation (Catala et al., 2003). Ectopic expression of AtCAX1 in tobacco (Nicotiana tabacum) leads to hypersensitivity to cold stress (Hirschi, 1999). The role of CAX proteins in regulating plant cold tolerance has also been identified in some other plant species (Xu et al., 2013; Li et al., 2017). However, the mechanisms underlying the cold-induced CAXs expression remain unclear. In apple, most CAX family genes respond significantly to cold stress (Mao et al., 2021), but no detailed functional study of these genes in response to cold stress has been conducted to date.
Apple is one of the most economically important fruit crops worldwide and cold stress is one of the major environmental factors affecting the productivity and quality of apples. Here, we identified a bHLH TF MdbHLH4 that negatively regulates apple cold tolerance. It inhibits cold-induced MdCBF1/3 expression either directly by binding to their promoters or indirectly by inhibiting the promoter-binding activity of MdICE1L. Besides, MdbHLH4 can reduce cold tolerance by promoting MdCAX3L-2 expression and MdICE1L degradation. Generally, the results of this study provide a mechanistic understanding of how the ICE1-interacting protein MdbHLH4 regulates CBF expression and ICE1 function and thus apple cold tolerance.
Results
Gene cloning and characterization of the bHLH TF MdbHLH4
In a previous study, we identified the bHLH family protein MdbHLH4, which might play an important role in the abiotic stress response (Mao et al., 2017). The full-length coding sequence (CDS) of MdbHLH4 is 1,293 bp and encoded a protein with 430 aa. A BLASTP search against the Arabidopsis proteome suggested that MdbHLH4 was a homolog of AtMYC70 (AT2G46810) in apple. Gene structure comparison showed that the exon–intron composition of MdbHLH4 and AtMYC70 was similar (Supplemental Figure S1).
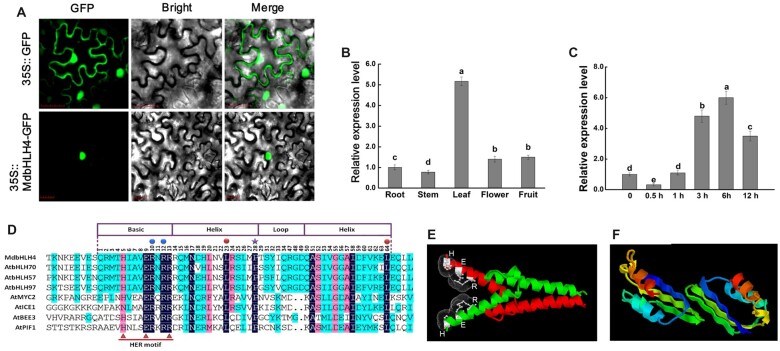
Subcellular localization analysis in Nicotiana benthamiana leaves showed that MdbHLH4 is a nuclear-localized protein (Figure 1A). Tissue-specific expression analysis showed that MdbHLH4 is constitutively expressed in all tested apple tissues. Its expression was highest in apple leaves, followed by the flowers and fruits, and its expression was lowest in stems (Figure 1B). Under cold treatment, the expression of MdbHLH4 was initially significantly down-regulated, and this was followed by rapid and significant up-regulation of its expression (Figure 1C), suggesting that MdbHLH4 may function in the later stage of the cold response (Zhao et al., 2017; Ye et al., 2019).
Figure 1.
Protein characteristics and expression analysis of MdbHLH4. A, Subcellular localization of MdbHLH4-GFP in N. benthamiana leaves. Bars = 30 μm. B, Relative expression levels of MdbHLH4 in different apple tissues. C, Expression analysis of MdbHLH4 in apple leaves under cold treatment. Different letters in B and C represent significant differences based on one-way ANOVA and Duncan’s test (P < 0.05). Error bars indicate the SD of three biological replicates. D, Sequence alignment of the bHLH domain of MdbHLH4 and bHLH proteins in Arabidopsis. Rectangular boxes represent the bHLH domain. Triangles indicate the HER motif. Dots and stars represent highly conserved amino acids (R10, R12, L23, L64, and P28) in the bHLH family. E and F, The 3D structure diagram of the dimer formed by the bHLH domain (E) or the C-terminal domain (F) of MdbHLH4.
Protein sequence analysis revealed that MdbHLH4 contains the conserved bHLH domain, which contains the conserved HER (H5-E9-R13) motif and several residues that are crucial for DNA binding and dimer stabilization (Figure 1D; Pires and Dolan, 2010). 3D structure prediction showed that the bHLH domain of MdbHLH4 can interact with itself to form a homodimer (Figure 1E andSupplemental Figure S2A), and the side chains of the H5-E9-R13 residues can form a perfect clip-like structure for DNA binding (Figure 1E;Su et al., 2017). In addition to the bHLH domain, the C-terminal of MdbHLH4 may be an important region for dimer formation (Figure 1F andSupplemental Figure S2B).
MdbHLH4 negatively regulates cold tolerance in transgenic Arabidopsis and apple plants
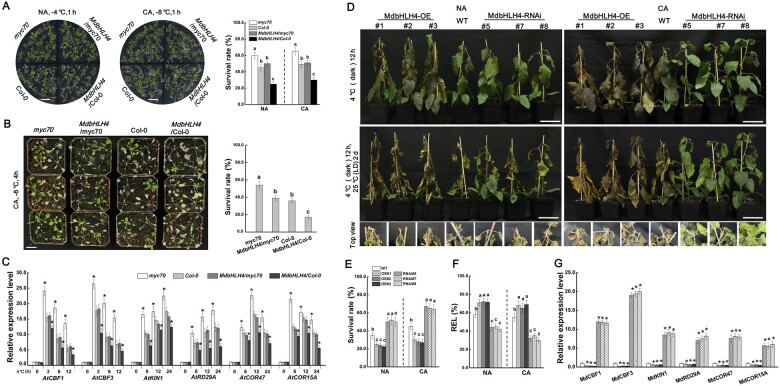
To identify the function of MdbHLH4 in regulating plant cold tolerance, MdbHLH4 was transformed into Col-0 and myc70 mutant (SALK_069605C) Arabidopsis seedlings. T3 seeds of homozygous transgenic Arabidopsis with high MdbHLH4 expression were selected for the following experiments (Supplemental Figure S3). Seedlings grown on 0.5 MS medium were divided into non-acclimated (NA) and cold-acclimated (CA) groups for cold treatment. The survival rate of myc70 mutant seedlings was highest under both NA and CA treatment conditions. Ectopic expression of MdbHLH4 in myc70 mutant could complement its cold-insensitivity phenotype. Moreover, ectopic expression of MdbHLH4 significantly decreased the survival rate of Col seedlings (Figure 2A).
Figure 2.
Functional characterization of MdbHLH4 in regulating cold tolerance in transgenic Arabidopsis and apple plants. A, Growth phenotype and survival rate of Arabidopsis seedlings after cold treatment. NA, without cold acclimation; CA, with cold acclimation (4°C for 12 h). Scale bars, 1 cm. B, The growth phenotype and survival rate of Arabidopsis plants (3 weeks old) after cold treatment. Scale bar, 2 cm. C, Expression analysis of COR genes in Arabidopsis under cold treatment. D–F, Freezing phenotype (D), survival rate (E), and leaf REL (F) of WT and MdbHLH4 transgenic apple plants under cold treatment. Scale bars, 9 cm. G, Expression analysis of COR genes in apple plants. Error bars indicate the SD of three biological replicates in all graphs. Bars labeled with * in each panel are significantly different from the Col-0 in C and WT in G (P < 0.05, Student’s t test). Different letters in A, B, E, F, and G indicate significant differences at P < 0.05 based on one-way ANOVA and Duncan’s tests.
To further determine the function of MdbHLH4 in cold tolerance regulation in Arabidopsis, 3-week-old seedlings grown in nutrition pots were subjected to freezing treatment after cold accumulation. Ectopic expression of MdbHLH4 significantly decreased the survival rate of myc70 and Col seedlings (Figure 2B). We analyzed the expression of several well-known COR genes in Arabidopsis, which contain CBF1, CBF3, KIN1, RD29A, COR47, and COR15A. Cold treatment significantly induced their expression, which was clearly inhibited by the ectopic expression of MdbHLH4 (Figure 2C). These results indicate that MdbHLH4 plays a negative role in the Arabidopsis cold response.
To identify the function of MdbHLH4 in regulating cold tolerance in apple, several transgenic apple plants overexpressing MdbHLH4 (overexpression [OE]) or with MdbHLH4 expression interfered (RNA interference [RNAi]) were generated (Supplemental Figure S4A). Three OE lines (OE-1, OE-2, and OE-3) with high MdbHLH4 expression and three RNAi lines (RNAi-5, RNAi-7, and RNAi-8) with MdbHLH4 expression significantly inhibited were used for subsequent cold treatments (Supplemental Figure S4B). Plants in the NA group were treated with −6°C for 3 h without cold acclimation. After recovering at 4°C for 12 h, all three OE lines showed obvious cold stress damage, with most leaves turning brown. In contrast, most leaves of WT and RNAi lines were still green (Figure 2D). After culture at 25°C for 2 days, the phenotypic differences among lines were more obvious. Most leaves of the OE lines withered and died, while several leaves of the RNAi plants remained alive and even appeared bright green, suggesting that stress-induced damage in OE plants was more severe (Figure 2D).
Plants in the CA group were treated with −8°C for 4 h after cold acclimation. OE plants exhibited more severe stress damage and RNAi plants experienced less damage compared with WT plants (Figure 2D). Measurements of the survival rate and leaf ion leakage were consistent with the phenotypic differences. Compared with WT plants, OE lines had a lower survival rate and higher ion leakage, whereas RNAi lines had a higher survival rate and lower ion leakage; this pattern was observed for plants in both the NA and CA groups (Figure 2, E and F). Moreover, the expression of COR genes was down-regulated in OE plants but significantly up-regulated in RNAi plants compared with WT (Figure 2G). These findings indicate that MdbHLH4 plays a negative regulatory role in the cold stress response of apple plants.
MdbHLH4 directly binds to the promoters of MdCBF1/MdCBF3 and inhibits their expression
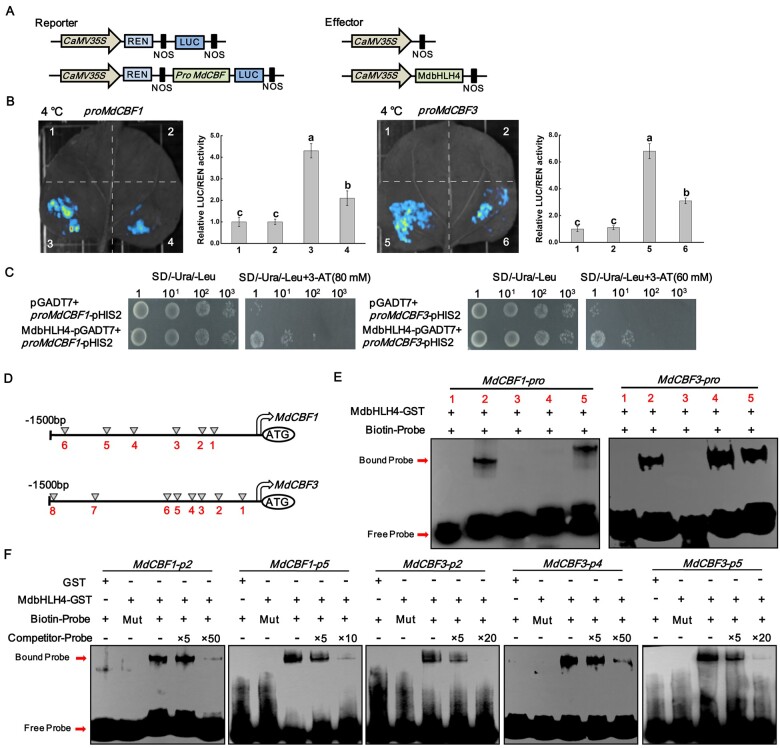
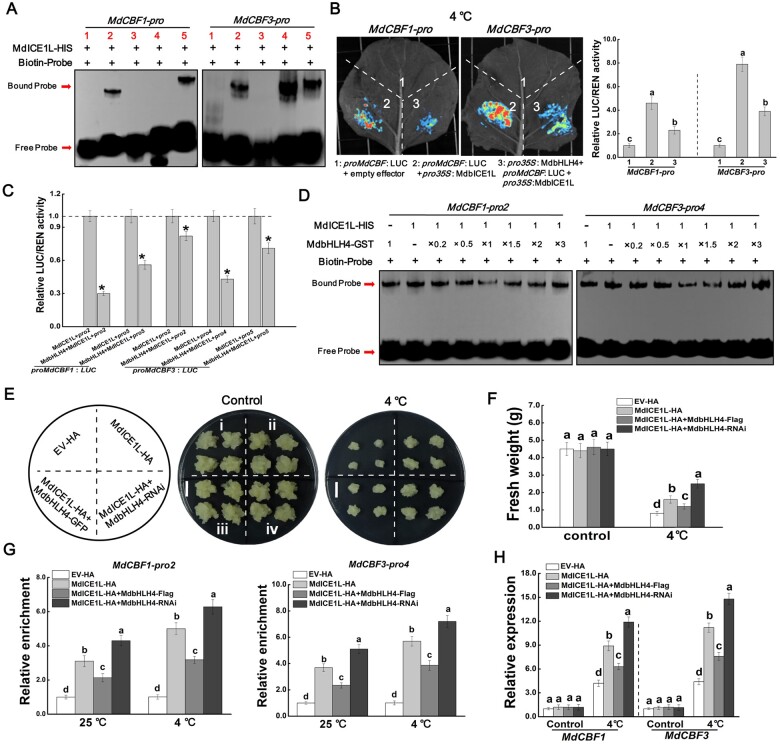
Since the expression of MdCBF1 and MdCBF3 decreased in MdbHLH4-OE plants under cold conditions, we identified the role of MdbHLH4 in regulating the expression of these two genes using the dual-luciferase (LUC) reporter system. The promoter sequences of MdCBF1 and MdCBF3 were cloned into the reporter vector, and the CDS of MdbHLH4 was cloned into the effector vector (Figure 3A). No fluorescence was observed when the two empty vectors were co-expressed. When the MdCBF1pro::LUC or MdCBF3pro::LUC construct was co-expressed with the empty effector vector, an obvious fluorescence signal was observed after cold treatment, and this signal was significantly inhibited by the co-expression of the MdbHLH4 protein (Figure 3B). Measurements of relative LUC/REN (Firefly Luciferase/Renilla Luciferase) activity were also consistent with the fluorescence observation results (Figure 3B). These findings suggest that MdbHLH4 inhibits the expression of MdCBF1 and MdCBF3 in plants under cold conditions.
Figure 3.
MdbHLH4 directly represses MdCBF1/3 expression under cold stress. A, Schematic diagram of the constructed reporter and effector vectors. B, Fluorescence observations and relative LUC/REN activity measurements in dual-LUC assays. The relative LUC/REN value is the average of three biological replicates, with each replicate containing three N. benthamiana plants. Error bars indicate the sd of three biological replicates. 1, empty reporter and effector; 2, empty reporter+35S::MdbHLH4; 3, proMdCBF1::LUC+empty effector; 4, proMdCBF1::LUC+35S::MdbHLH4; 5, proMdCBF3::LUC+empty effector; and 6, proMdCBF3::LUC+35S::MdbHLH4. C, Identification of the promoter-binding ability of MdbHLH4 on the promoters of MdCBF1/3 by Y1H assays. D, Diagram of the potential MdbHLH4-binding sites within the MdCBF1/3 promoters. E, EMSAs showing the binding of MdbHLH4 to the MdCBF1/3 promoters. Numbers indicate the potential binding sites in D. F, Verification of the binding specificity of MdbHLH4 by competing EMSAs. Competitor, probes without biotin labeling. Mut, mutant probes with core sequences within E-box altered. 5×, 10×, and 50× represent the rates of competitor probes. Sequences of different probes are provided in Supplemental Table S1.
To determine whether MdbHLH4 can directly bind to the promoters of MdCBF1/3, the yeast one-hybrid (Y1H) system was used. Compared with the control group, yeast strains transformed with the MdbHLH4-pGADT7 and proMdCBF1-pHIS2 (or proMdCBF3-pHIS2) vectors grew better in the screening medium (Figure 3C). To further confirm the direct binding of MdbHLH4 protein to the promoters of MdCBF1 and MdCBF3, electromobility shift assays (EMSAs) were carried out. Sequence analysis showed that there were six and eight putative binding sites in the promoters of MdCBF1 and MdCBF3, respectively (Figure 3D). We focused on the five sites closest to the start codons of these two genes in this study. Gene-specific probes labeled with biotin were designed and synthesized according to sequences around these sites (Supplemental Figure S5 and Supplemental Table S1). The EMSA results showed that MdbHLH4 binds directly to the second and fifth sites of the MdCBF1 promoter and to the second, fourth, and fifth sites of the MdCBF3 promoter (Figure 3E). To further confirm the binding specificity of MdbHLH4 to these sites, mutant probes with core sequences in the E-box altered were generated, and probes without biotin labels (competitor probes) were synthesized (Supplemental Figure S5 and Supplemental Table S1). MdbHLH4 could not bind to the mutant probes and the addition of competitor probes could clearly inhibit the binding of MdbHLH4 to the corresponding biotin-labeled probes (Figure 3F). These findings indicate that MdbHLH4 could directly bind to the promoters of MdCBF1 and MdCBF3 and inhibit their expression under cold conditions.
Protein interactions between MdbHLH4 and MdICE1L/MdICE1
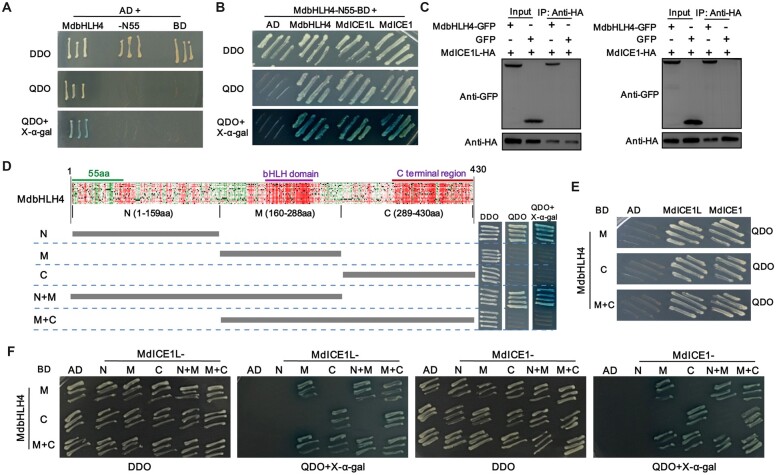
Using AtICE1 protein as a query, we conducted BLASTP searches of the apple proteome (GDDH13). The protein MD09G1003800 had the highest score and was identified as the ICE1 homolog in apple. To distinguish this protein from the recently published MdICE1 (An et al., 2021), which has also been referred to as MdCIbHLH1 (Feng et al., 2012), we refer to it as MdICE1L (Supplemental Figure S6). The full-length MdbHLH4 protein exhibited strong self-activation activity; consequently, 55 aa in its N-terminal were removed to abolish this activity (MdbHLH4-N55; Figure 4A). The yeast two-hybrid (Y2H) results showed that MdbHLH4 could either interact with itself or with MdICE1L/MdICE1 (Figure 4B). To verify the interaction between MdbHLH4 and MdICE1L/MdICE1 in vivo, MdbHLH4-GFP and MdICE1L-HA (or MdICE1-HA) proteins were transiently co-expressed in N. benthamiana leaves. The results of the co-immunoprecipitation (Co-IP) assays showed that MdbHLH4 could interact with both MdICE1L and MdICE1 in vivo (Figure 4C).
Figure 4.
MdbHLH4 interacts with MdICE1L/MdICE1. A, Identification of the self-activation activity of full-length and truncated MdbHLH4 (MdbHLH4-N55) based on Y2H assays. B and C, Protein interaction identification between MdbHLH4 and MdICE1L/MdICE1 by Y2H (B) and Co-IP (C) assays. D, Prediction of the effects of point mutations, functional region division, and self-activation activity identification of different regions of MdbHLH4. Detailed results of the effects of point mutations are provided in Supplemental Figure S7. E, Identification of crucial regions for the MdbHLH4–MdICE1L/MdICE1 interaction in MdbHLH4. AD, pGAD424 and BD, pGBT9. F, Identification of crucial regions for the MdbHLH4–MdICE1L/MdICE1 interaction in MdICE1L/MdICE1. AD, pGADT7; BD, pGBKT7; DDO, SD medium without leucine and tryptophan; and QDO, SD medium without leucine, tryptophan, histidine, and adenine.
To identify the important regions of MdbHLH4 underlying the protein interaction, we predicted the effects of point mutations in the MdbHLH4 amino acid sequence. Amino acids within the bHLH domain and the C-terminal region are most conserved (Figure 4D andSupplemental Figure S7). Consequently, we divided the MdbHLH4 protein into three regions: the N-terminal region (N, 1–159 aa), the intermediate region (M, 160–288 aa), and the C-terminal region (C, 289–430 aa). The N-terminal region showed strong self-activation activity, and no self-activation activity was observed for the other regions (Figure 4D), suggesting that this region is the transcriptional activation domain. Furthermore, the Y2H assays showed that either the M or C region of MdbHLH4 could individually interact with MdICE1L/MdICE1 (Figure 4E), indicating that the C-terminal of MdbHLH4 is a key region mediating the MdbHLH4–MdICE1L/MdICE1 interaction in addition to the bHLH domain.
To identify the crucial regions of MdICE1L/MdICE1 underlying the protein interaction, we further divided these two proteins into three regions (N, M, and C), based on previous studies on AtICE1 (Hu et al., 2013, 2019) and the prediction results of the effects of point mutations on MdICE1L/MdICE1 (Supplemental Figure S8). The Y2H results showed that the M and C regions of MdICE1L/MdICE1 proteins could interact with the corresponding regions of MdbHLH4, respectively (Figure 4F). These results further support that both the M and C regions of MdbHLH4 are important for the MdbHLH4–MdICE1L/MdICE1 interaction.
The MdbHLH4–MdbICE1L interaction inhibits the binding ability of MdICE1L on MdCBF1/MdCBF3 promoters
We identified the binding of MdICE1L to the promoters of MdCBF1 and MdCBF3 by EMSAs. As expected, MdICE1L binds directly to the promoters of these two genes. Moreover, the binding sites of MdICE1L were the same as those of the MdbHLH4 protein (Figures 3E and 5A). We identified the transcriptional activation activity of MdICE1L on MdCBF1/MdCBF3 promoters by dual-LUC assays. Compared with the combinations MdCBF1pro::LUC or MdCBF3pro::LUC plus empty effector vector (control), the fluorescence intensity and relative LUC/REN activity were significantly increased when MdICE1L was co-expressed (Figure 5B). When MdbHLH4 protein was further co-expressed in these combinations, the MdICE1L-mediated transcriptional activation was significantly inhibited (Figure 5B), indicating that MdbHLH4 inhibits the transcriptional activation of MdICE1L on MdCBF1/MdCBF3.
Figure 5.
MdbHLH4 represses the binding activity of MdICE1L on MdCBF1/3 promoters. A, EMSAs showing that MdICE1L directly binds to the promoters of MdCBF1/3. Numbers indicate the binding sites in Figure 3. B, Dual-LUC assays showing that MdbHLH4 inhibits the transcriptional activation of MdCBF1/3 by MdICE1L under cold stress. C, Identification of the MdICE1L-binding sites that are most significantly affected by MdbHLH4 through dual-LUC assays. pro2, pro4, and pro5 represent the effector vectors containing the corresponding binding sites in MdCBF1/3 promoters in A. Significant differences (*) were determined using Student’s t test: P < 0.05. D, EMSAs showing that MdbHLH4 inhibits the promoter-binding activity of MdICE1L. MdbHLH4-GST and MdICE1L-HIS proteins were added to the binding buffer at the indicated molar ratios. E and F, MdbHLH4 reduced the enhancement effect of MdICE1L on the cold tolerance of transgenic apple calli. Growth phenotype (E) and fresh weight (F) after cold treatment are shown. EV-HA indicates apple calli transformed with the empty vector pCambia35S-3HA. Bars represent 1 cm. G, ChIP-qPCR assays showing that MdbHLH4 inhibits the binding of MdICE1L to the promoters of MdCBF1/3 in apple. Apple calli treated with 4°C/25°C for 3 h were harvested for ChIP assays. Immunoprecipitation was performed with or without anti-HA antibody. The relative enrichment was calculated as the ratio of MdbHLH4/MdICE1L transgenic to control (EV-HA) samples. H, Expression analysis of MdCBF1/3 in transgenic calli. Apple calli treated with 4°C/25°C (control) for 3 h were harvested for RT-qPCR analysis. Error bars indicate the SD of three biological replicates in all graphs. Different letters in B, F, G, and H represent significant differences based on one-way ANOVA and Duncan’s tests (P < 0.05).
To characterize the effect of MdbHLH4 on the promoter-binding ability of MdICE1L, we first identified the MdICE1L-binding sites that were most significantly affected by MdbHLH4 using the dual-LUC system. Segments containing a single binding site were cloned into the reporter vector and then transiently co-expressed with the 35S::ICE1L effector vector in N. benthamiana leaves. Among these binding sites, the relative LUC/REN activity of combinations containing MdCBF1-pro2 and MdCBF3-pro4 declined most significantly (Figure 5C). Competing EMSAs also showed that MdbHLH4 should have the strongest binding activity to these two sites (Figure 3F). Thus, these two sites were selected for subsequent experiments.
EMSAs showed that the size of the bands of the probe-bound MdbHLH4-GST and MdICE1L-HIS proteins was similar and could not be distinguished (Figure 5D). To analyze the effect of MdbHLH4 on the probe-binding ability of MdICE1L, we added the MdbHLH4 protein to various content gradients. The addition of MdbHLH4 protein substantially reduced the brightness of the bands of the probe-bound MdICE1L protein (Figure 5D), suggesting that the MdbHLH4–MdICE1L interaction has a negative effect on the binding ability of MdICE1L. Furthermore, when the content of MdbHLH4 protein was more than that of MdICE1 (molar ratios > 1), the brightness of the bands increased with the MdbHLH4 content (Figure 5D), indicating that this protein interaction also inhibits the binding ability of MdbHLH4. These results suggest that the MdbHLH4–MdbICE1L interaction may block the binding ability of both MdICE1L and MdbHLH4 proteins on MdCBF1/MdCBF3 promoters.
To further confirm this result in vivo, MdICE1L-HA and MdbHLH4-Flag OE vectors were constructed, and several transgenic apple calli transformed with different combinations of these vectors were obtained (Figure 5E and Supplemental Figure S9). Under 4°C, MdICE1L OE significantly alleviated the inhibitory effect of cold stress on growth. This effect of MdICE1L was inhibited by MdbHLH4 OE but significantly enhanced when MdbHLH4 expression was interfered, suggesting that MdbHLH4 negatively regulates the function of MdICE1L in apple (Figure 5, E and F). Subsequently, these transgenic apple calli were used for chromatin immunoprecipitation-quantitative PCR (ChIP-qPCR) analysis. Results further support that MdICE1L could bind to the two sites MdCBF1-pro2 and MdCBF3-pro4 (Figure 5G). Furthermore, MdbHLH4 OE significantly inhibited the binding activity of MdICE1L to these sites, and the opposite effect was observed when MdbHLH4 expression was interfered, especially under cold treatment (Figure 5G). We also detected the expression level of MdCBF1 and MdCBF3 in these transgenic apple calli. Under cold treatment, MdICE1L OE significantly promoted their expression, which was inhibited by MdbHLH4 co-expression (Figure 5H). These results further support the negative effect of the MdbHLH4–MdbICE1L interaction on MdICE1L promoter-binding activity and thus MdCBF1/3 expression.
MdbHLH4 directly binds to the MdCAX3L-2 promoter and promotes its expression
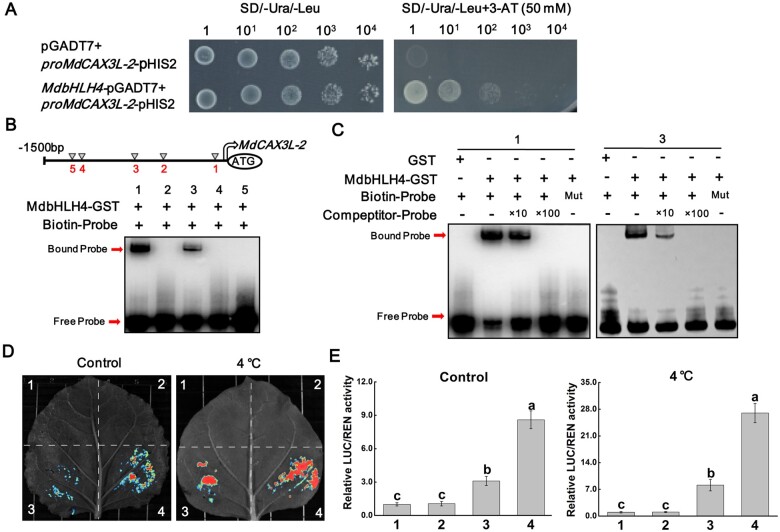
In our recent study, we identified a CAX family gene MdCAX3L-2 that significantly responds to cold stress and promotes the compartmentalization of Ca2+ into vacuoles (Mao et al., 2021). To identify the upstream TFs that regulate MdCAX3L-2 expression, we screened the apple cDNA library using Y1H assays. These assays revealed that MdbHLH4 could bind to the promoter of MdCAX3L-2 (Figure 6A). To further verify the direct binding of MdbHLH4 to the MdCAX3L-2 promoter, EMAS assays were performed. Sequence analysis showed that there were five putative binding sites in the MdCAX3L-2 promoter (Figure 6B and Supplemental Figure S5). The EMSA results showed that MdbHLH4 could directly bind to the first and third sites in the MdCAX3L-2 promoter (Figure 6B). The binding specificity of MdbHLH4 to these two sites was also verified by EMAS assays with the addition of mutant and competing probes (Figure 6C;Supplemental Figure S5 and Supplemental Table S1).
Figure 6.
MdbHLH4 directly binds to the promoter of MdCAX3L-2 and activates its expression. A, Y1H assays showing the interaction between MdbHLH4 and the MdCAX3L-2 promoter. B, EMSAs showing that MdbHLH4 directly binds to the MdCAX3L-2 promoter. Putative binding sites in the MdCAX3L-2 promoter are shown in the schematic diagram. C, Identification of MdbHLH4-binding specificity to sites 1 and 3 in B by competing EMSAs. Sequences of different probes can be found in Supplemental Table S1. D and E, Dual-LUC assays showing the transcriptional activation activity of MdbHLH4 on the MdCAX3L-2 promoter in N. benthamiana. 1, empty reporter and effector; 2, empty reporter + 35S::MdbHLH4; 3, proMdCAX3L-2::LUC + empty effector; and 4, proMdCAX3L-2::LUC + 35S::MdbHLH4. Error bars indicate the sd of three biological replicates. Different letters represent significant differences based on one-way ANOVA and Duncan’s tests (P < 0.05).
To investigate the transcriptional regulatory activity of MdbHLH4 on MdCAX3L-2 promoter, the dual-LUC system was used. Fluorescence observation indicated that the transcriptional activity of the MdCAX3L-2 promoter was low under normal conditions and could be induced by cold treatment (Figure 6D). When the MdbHLH4 protein was co-expressed, the transcriptional activity of the MdCAX3L-2 promoter increased obviously under both normal and cold treatment conditions (Figure 6D). Measurements of the relative LUC/REN activity were also consistent with the fluorescence observation results (Figure 6E). These results indicate that MdbHLH4 can directly promote MdCAX3L-2 expression in response to cold conditions.
MdCAX3L-2 enhances the negative effect of MdbHLH4 on cold tolerance
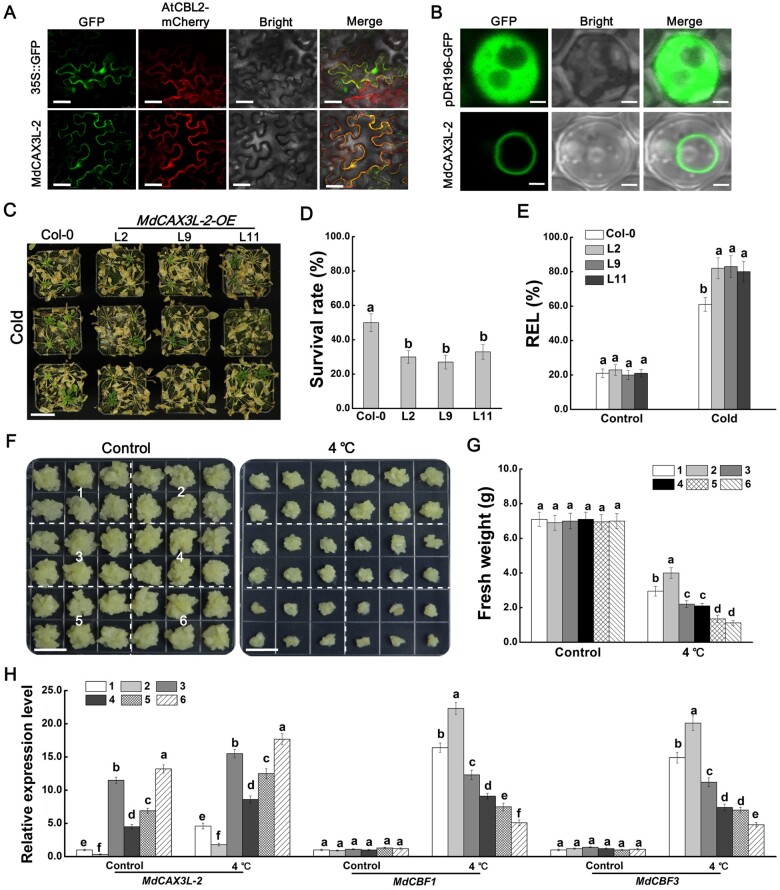
We transiently expressed the MdCAX3L-2-GFP fusion protein in N. benthamiana leaves to identify its subcellular localization. The green fluorescence of the MdCAX3L-2-GFP fusion protein was a perfect match with the red fluorescence of AtCBL2-mCherry, which made the false color of the fluorescence yellow (Figure 7A). This result indicates that MdCAX3L-2 is a tonoplast-localized protein. We also identified the subcellular localization of MdCAX3L-2 in yeast cells. The green fluorescence of the MdCAX3L-2-GFP fusion protein could only be detected in the vacuole membrane in yeast cells (Figure 7B). These results, combined with the ability of MdCAX3L-2 to strongly promote the compartmentalization of cytosolic Ca2+ into vacuoles (Mao et al., 2021), indicate that MdCAX3L-2 should play a negative role in cold-induced Ca2+ signatures.
Figure 7.
MdCAX3L-2 enhances the negative effect of MdbHLH4 on cold tolerance. A and B, Localization of MdCAX3L-2 protein in N. benthamiana epidermal cells (A) and yeast cells (B). AtCBL2-mCherry was used as a tonoplast location marker. GFP protein was used as a control. Scale bars, 30 μm in A and 3 μm in B. C–E, OE of MdCAX3L-2 reduced Arabidopsis cold tolerance. Growth phenotype (C), survival rate (D), and REL (E) of WT and MdCAX3L-2 transgenic Arabidopsis seedlings (1-mouth-old) after cold treatment. Scale bar, 4 cm. F, MdbHLH4 increased the sensitivity of MdCAX3L-2-OE transgenic apple calli to cold stress. 1, WT; 2, MdCAX3L-2-RNAi; 3, MdCAX3L-2-Flag; 4, MdbHLH4-GFP; 5, proMdCAX3L-2::MdCAX3L-2-Flag + MdbHLH4-GFP; and 6, MdCAX3L-2-Flag+MdbHLH4-GFP. Bars represent 1 cm. G, Fresh weight of calli in F. H, Expression analysis of MdCAX3L-2 and MdCBF1/3 in WT and transgenic apple calli in F. Apple calli treated at 4°C for 3 h were harvested for RT-qPCR analysis. Error bars indicate the sd of three biological replicates in all graphs. Different letters in D, E, G, and H represent significant differences based on one-way ANOVA and Duncan’s tests (P < 0.05).
To identify the role of MdCAX3L-2 in regulating plant cold tolerance, we obtained transgenic Arabidopsis seedlings with “Col” background (Supplemental Figure S10). Under normal conditions, no obvious phenotypic differences were observed. After cold treatment, the survival rate of the MdCAX3L-2 transgenic lines was significantly lower than that of the WT plants (Figure 7, C and D). Measurements of the REL also indicated that the MdCAX3L-2 transgenic seedlings experienced more severe stress damage (Figure 7E). These results indicate that MdCAX3L-2 negatively regulates cold tolerance in Arabidopsis.
To confirm the negative role of MdCAX3L-2 in regulating cold tolerance in apple, MdCAX3L-2 expression vectors driven by 35S (35S::MdCAX3L-2-Flag) or its own promoter (MdCAX3L-2pro::MdCAX3L-2-Flag) and the RNAi vector MdCAX3L-2-RNAi were constructed and transformed into apple calli (Figure 7F). After cold treatment, the growth of WT was significantly inhibited, and this inhibitory effect could be alleviated by interfering with MdCAX3L-2 expression; the inhibitory effect was strengthened when MdCAX3L-2 expression was up-regulated (Figure 7, F and G). These results demonstrate that MdCAX3L-2 plays a negative role in cold tolerance regulation in apple.
To determine whether MdbHLH4 could reduce apple cold tolerance by promoting MdCAX3L-2 expression, the MdbHLH4-GFP OE vector was transformed into these MdCAX3L-2 transgenic lines to obtain double transgenic apple calli. Apple calli transformed with MdCAX3L-2pro::MdCAX3L-2 did not show significant differences in growth and MdCAX3L-2 expression compared with WT plants (Supplemental Figure S11). However, when 35S::MdbHLH4-GFP and MdCAX3L-2pro::MdCAX3L-2-Flag vectors were co-transformed in apple calli, the growth inhibitory effect of cold on this double transgenic line was more pronounced than that on apple calli overexpressing MdbHLH4 alone. After cold treatment, the growth status of this double transgenic line was similar to that of apple calli co-transformed with 35S::MdbHLH4-GFP and 35S::MdCAX3L-2-Flag vectors (Figure 7, F and G). Expression analysis also revealed that the expression of MdCAX3L-2 was more significantly up-regulated by MdbHLH4-GFP co-transformation (Figure 7H). These findings indicate that in addition to inhibiting MdCBF1/3 expression, MdbHLH4 may also reduce apple cold tolerance by promoting MdCAX3L-2 expression.
OE of MdCAX3L-2 and MdbHLH4 promotes the cold-induced ubiquitination and protein degradation of MdICE1L in apple
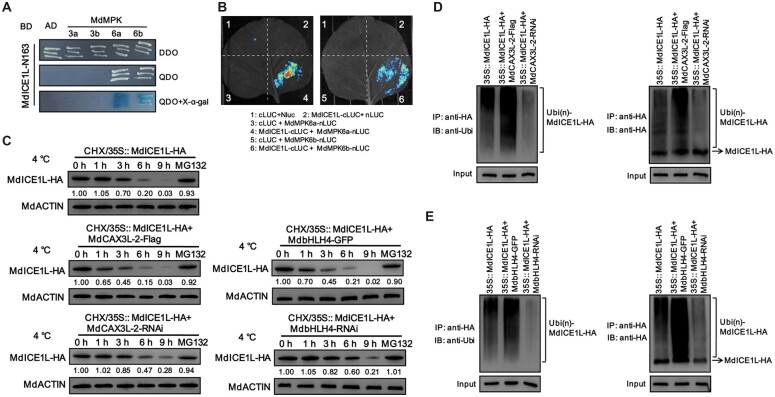
Under cold stress, MPK3/6 promotes the ubiquitination and degradation of ICE1 by phosphorylation of ICE1, and Ca2+/CAM-activated CRLKs inhibit this process to promote ICE stability (Zhao et al., 2017). Given that MdCAX3L-2 inhibits Ca2+ accumulation in the cytoplasm (Mao et al., 2021) and MdCBF1/3 expression (Figure 7H), it may negatively regulate the stability of MdICE1L under cold stress. To test this hypothesis, we first studied the interaction between MdICE1L and MdMPK3/6. Two MdMPK3 and two MdMPK6 proteins were identified from the apple genome through a BLASTP search using AtMPK3 and AtMPK6 as queries, and these were referred to as MdMPK3a/b and MdMPK6a/b, respectively. Y2H and split-LUC assays revealed that MdICE1L interacted with MdMPK6a/b (but not MdMPK3a/b) both in vitro and in vivo (Figure 8, A and B). Subsequently, MdICE1L-HA transgenic apple calli were exposed to cold conditions for different periods, and the protein level of MdICE1L-HA was detected. The protein level of MdICE1L-HA decreased noticeably under cold treatment (Figure 8C), and this process was substantially inhibited by the 26S proteasome inhibitor MG132, suggesting that the process of cold-induced ubiquitination and degradation of ICE1 is conserved in apple.
Figure 8.
MdbHLH4 and MdCAX3L-2 promote the cold-induced ubiquitination and degradation of MdICE1L. A, Identification of the MdICE1L-MdMPK6a/b interaction using Y2H assays. B, Identification of the MdICE1L–MdMPK6a/b interaction through split-LUC assays. C, MdCAX3L-2 and MdbHLH4 promoted the cold-induced degradation of MdICE1L protein. The protein levels of MdICE1L-HA in transgenic calli (incubated with CHX) were detected at specified time points under 4°C treatment with the anti-HA antibody. Apple calli treated with CHX plus MG132 were sampled at 9 h of cold treatment. Numbers under the upper panel represent the band intensity, which was determined using ImageJ software. D and E, OE of either MdCAX3L-2 (D) or MdbHLH4 (E) promotes the ubiquitination of MdICE1L under cold stress. The ubiquitination level of MdICE1L-HA was detected using anti-ubiquitin (left) and anti-HA (right) antibodies.
Using MdICE1L-HA transgenic apple callus as background, double transgenic apple calli overexpressing MdbHLH4 or MdCAX3L-2 and calli with the expression of these two genes interfered were generated. MdCAX3L-2-OE promoted the cold-induced degradation of MdICE1L-HA, and interference of MdCAX3L-2 expression delayed the degradation process (Figure 8C). Besides, OE of MdbHLH4 also promoted the degradation of MdICE1L-HA, which could be delayed by interfering MdbHLH4 expression (Figure 8C). These results indicate that the MdbHLH4–MdCAX3L-2 module negatively affects MdICE1L stability under cold stress.
To investigate whether MdbHLH4 and MdCAX3L-2 promote MdICE1L degradation by promoting its ubiquitination, we detected the ubiquitination level of MdICE1L-HA in transgenic apple calli. As expected, interference of MdCAX3L-2 expression inhibited the cold-induced ubiquitination of MdICE1L, and OE of MdCAX3L-2 promoted its ubiquitination (Figure 8D). Similar patterns were observed in MdbHLH4/MdICE1L-HA double transgenic apple calli (Figure 8E). These findings suggest that the MdbHLH4–MdCAX3L-2 module can reduce apple cold tolerance by promoting the cold-induced ubiquitination and degradation of MdICE1L.
Discussion
Cold stress is one of the major environmental factors adversely affecting crop yield and quality. The ability of plants to sense cold stress and acclimate to freezing conditions has been studied extensively in the last several decades (Guo et al., 2018; Ding and Yang, 2022). In China, cold stress seriously restricts the development of the apple industry, especially early spring chilling and late spring frosts (Feng et al., 2012; An et al., 2021). Elucidating the mechanisms underlying the response of plants to cold stress could facilitate the breeding of cold-tolerant varieties.
To balance the allocation of resources to growth, development, and the cold stress response, plants have evolved a series of regulatory mechanisms to mediate the expression of CBF genes (Ding et al., 2019; Chen et al., 2021; Ding and Yang, 2022). In Arabidopsis, there are three COR CBF genes (CBF1–CBF3) (Chinnusamy et al., 2003; Gilmour et al., 2004). ICE1 and its homolog ICE2 regulate the expression of downstream COR genes and plant cold tolerance by binding to the promoters of CBF3 and CBF1, respectively (Chinnusamy et al., 2003; Fursova et al., 2009). Besides, the increased cold tolerance and expression of CBF1/3 in the cbf2 knock-out mutant suggest that CBF2 is a negative regulator of CBF1/3 expression (Novillo et al., 2004). These results indicate that CBF1 and CBF3 are key factors in the cold signaling pathway mediated by ICE1 and its homologs (Guo et al., 2018; Ding and Yang, 2022). MYC70 is an ICE1-interacting protein that negatively regulates CBF expression by directly binding to the CBF3 promoter, while MdICE1 enhances apple cold tolerance by directly regulating MdCBF1 expression (An et al., 2021). Since MdbHLH4 is a homolog of MYC70 and interacts with MdICE1L/MdICE1 (Figures 1, 2, and 4 and Supplemental Figure S1), we focused on the regulatory function of MdbHLH4 on both MdCBF1 and MdCBF3 in this study. Our findings indicate that MdbHLH4 negatively regulates apple cold tolerance by directly inhibiting the expression of MdCBF1 and MdCBF3 under cold stress (Figures 2 and 3), suggesting a key negative role of MdbHLH4 in cold-induced CBFs expression in apple. This also suggests that the function and regulatory mechanism of MYC70 in cold response may be conserved in different plant species.
In addition to directly inhibiting CBF3 expression, MYC70 may also inhibit the binding activity of ICE1 to the CBF3 promoter via unknown mechanisms (Ohta et al., 2018). It has been proposed that MYC70 homodimer or MYC67–MYC70 heterodimer may inhibit the binding activity of ICE1 by covering the E-box/MYC cis-elements in the CBF3 promoter (Ohta et al., 2018). Besides, MYC67/MYC70 might affect the transcriptional activation activity of ICE1 by interacting with ICE1 (Ohta et al., 2018). For example, EIN3 and EIN3-like 1 antagonize the transcriptional activation activity of MYC2 by interacting with MYC2 (Song et al., 2014). In this study, we found that MdICE1L could bind to the promoters of MdCBF1/3 to enhance their expression and thus apple cold tolerance (Figure 5). The inhibitory effect of MdbHLH4 on the transcriptional activation of MdCBF1/3 by MdICE1L (Figure 3, A and B) and the MdbHLH4–MdICE1L interaction (Figure 4) suggests that these two hypotheses may also apply to MdbHLH4, especially the binding sites of MdbHLH4 are identical to those of MdICE1L (Figures 3E and 5A). To determine how MdbHLH4 affects the function of MdICE1L, EMSAs with the MdbHLH4 protein added to various content gradients were carried out (Figure 5D). The results revealed that the MdbHLH4–MdICE1L interaction inhibits the binding activity of both proteins on CBF promoters. ChIP assays also indicated that MdbHLH4 inhibits MdICE1L promoter-binding activity (Figure 5G). These results indicate that MdbHLH4 more likely abolishes the binding activity of MdICE1L (and itself) by interacting with MdICE1L, rather than via the routes proposed in the two aforementioned hypotheses. These findings provide insights into the mechanisms by which ICE1-interacting proteins affect ICE1 function; this information could be used to guide studies of other key negative regulators in cold response, such as MYB15, MYC67, and MYC70 (Agarwal et al., 2006; Ohta et al., 2018).
Calcium signaling is crucial for plant development and stress responses. Upon exposure to cold, a rapid Ca2+ influx into the cytosol occurs, which is achieved via several types of Ca2+ channels and transporters (Demidchik et al., 2018; Guo et al., 2018; Kudla et al., 2018; Lee and Seo, 2021). Recent studies in Arabidopsis and rice have shown that loss of function of these Ca2+ channels or transporters, which contain AtCNGC2, AtCNGC4 (Cui et al., 2020), OsCNGC9 (Wang et al., 2021), OsCNGC14, OsCNGC16 (Cui et al., 2020), AtANNEXIN1 (Liu et al., 2021), AtMCA1, and AtMCA2 (Mori et al., 2018), reduces the cold-induced Ca2+ signature and results in decreased cold tolerance. CAX proteins are Ca2+/H+ antiporters that inhibit Ca2+ accumulation in the cytoplasm by mediating the entry of cytosolic Ca2+ into vacuoles to maintain a basal cytosolic Ca2+ concentration (Lee and Seo, 2021). This suggests that CAXs may negatively regulate cold-induced Ca2+ signatures and thus plant cold tolerance, such as AtCAX1 (Hirschi, 1999; Catala et al., 2003). To date, the mechanisms underlying the cold-induced expression of CAX genes remain unclear. In apple, cold stress significantly induces MdCAX3L-2 expression. Ectopic expression of MdCAX3L-2 in yeast cells strongly promotes the compartmentalization of Ca2+ into vacuoles (Mao et al., 2021), suggesting a potential role of MdCAX3L-2 in the regulation of cold-induced Ca2+ signature. Here, we proved that MdCAX3L-2 had a negative effect on cold tolerance and CBFs expression in Arabidopsis and apple calli (Figure 7). Moreover, MdbHLH4 can directly bind to the MdCAX3L-2 promoter to enhance its expression (Figure 6) and thus negatively regulate apple cold tolerance (Figure 7, F–H). These results provide insights into the response of CAX genes to cold stress, and suggest that MdbHLH4 may negatively affect apple cold tolerance by inhibiting cold-induced Ca2+ signatures via the MdbHLH4–MdCAX3L-2 module.
The cold-induced Ca2+ signature positively regulates the protein stability of ICE1. Phosphorylation of ICE1 mediated by the MKK4/5-MPK3/6 cascade has been shown to promote ubiquitination and thus the degradation of ICE1 in response to cold (Li et al., 2017; Zhao et al., 2017). Two plasma membrane (PM)-localized kinases CRLK1 and CRLK2, whose activity is stimulated by binding to Ca2+/CaM (Yang et al., 2010), positively regulate ICE1 stability and freezing tolerance by suppressing the cold-induced activity of MPK3/MPK6 (Zhao et al., 2017). These results, combined with the MdICE1L–MdMPK6 interaction and cold-induced MdICE1L degradation (Figure 8, A–C), suggest that the MPK6-promoted ICE1 ubiquitination and degradation pathway may be conserved in apple, and that the MdbHLH4–MdCAX3L-2 module may negatively regulate MdICE1L stability by inhibiting cold-induced Ca2+ signatures. To test this hypothesis, we compared the degradation rate and ubiquitination level of MdICE1L in various transgenic apple calli under cold treatment. As expected, OE of either MdbHLH4 or MdCAX3L-2 promoted the cold-induced ubiquitination and degradation of MdICE1L (Figure 8, C and D). These findings suggest that the MdbHLH4–MdCAX3L-2 module negatively regulates MdICE1L stability under cold stress, and provide a basis for further studies on the function and regulatory mechanisms of CAX proteins in plant cold response.
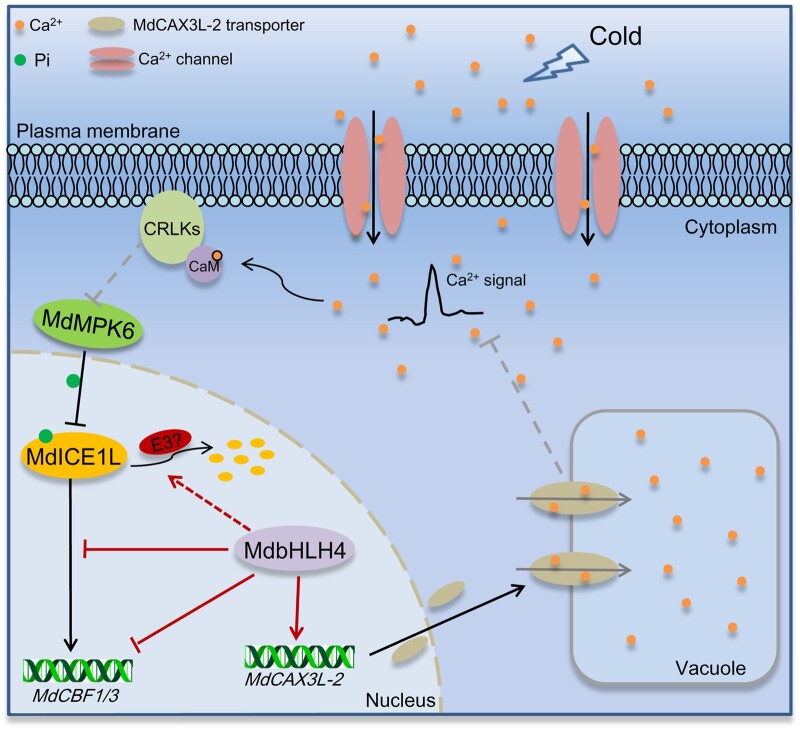
The temporal expression patterns of CBF genes can be divided into the off, on, and attenuated stages during the cold response (Zhao et al., 2017; Ye et al., 2019). Upon exposure to cold stress, CBF expression is strongly induced within 15–30 min and peaks at 2–3 h, and the protein level of ICE1 begins to decrease at 3 h after cold treatment (Novillo et al., 2004; Medina et al., 2011; Ye et al., 2019). These results, combined with the expression pattern of MdbHLH4 and its negative role on MdICE1L stability under cold stress (Figures 1C and 8), suggest that MdbHLH4 should function in the attenuated stage of cold signaling. We propose a working model for the molecular mechanism by which MdbHLH4 regulates the cold response in apple (Figure 9). Upon exposure to cold stress, MdICE1L is activated rapidly and initials the expression of MdCBF1/3 to activate the cold response. MdbHLH4 expression is then induced, and MdbHLH4 protein accumulates. MdbHLH4 directly inhibits MdCBF1/3 expression by binding to their promoters. MdbHLH4 also interacts with MdICE1L to inhibit its promoter-binding activity and thus MdCBF1/3 expression. Besides, MdbHLH4 helps attenuate cold-induced Ca2+ signatures by promoting MdCAX3L-2 expression to inhibit overreactions, which lead to the enhanced ubiquitination and degradation of MdICE1L. The multiple roles of MdbHLH4 in regulating CBF expression and ICE1 function indicate that it is a key negative regulator in the apple cold response. The results of this study provide insights that will aid future studies of the mechanisms underlying the ICE1-mediated cold stress response as well as plant growth and development.
Figure 9.
A working model showing the roles of MdbHLH4 in cold stress signaling. When plants are exposed to cold stress, MdICE1L is activated rapidly, and the expression of MdCBF1/3 is initiated, a process that as least partially depends on cold-induced Ca2+ signaling. Subsequently, cold signaling activates the expression of MdbHLH4. The accumulated MdbHLH4 protein directly inhibits MdCBF1/3 expression by binding to their promoters. MdbHLH4 also inhibits their expression by inhibiting the promoter-binding activity of MdICE1L through the MdbHLH4–MdICE1L interaction. Besides, MdbHLH4 activates the expression of MdCAX3L-2 to promote the compartmentalization of cytosolic Ca2+ into vacuoles and thus attenuate the cold-induced Ca2+ signature. This reduces the cold-induced expression of CBF and COR genes and thus plant cold tolerance. This also leads to the enhanced ubiquitination and degradation of MdICE1L in response to cold, a process that may be related to MdMPK6a/b in apple.
Materials and methods
Plant materials and growth conditions
Plant materials used in this study included “GL-3” (a seedling clone from open-pollinated “Royal Gala”) apple (M. domestica) plants (Dai et al., 2013), apple calli (“Orin”), and A. thaliana seedlings (Col-0 and myc70 mutant). Before cold treatment, tissue-cultured apple plants were transferred to rooting medium (MS medium supplemented with 0.5 mg/L IAA (3-Indoleacetic acid) and 0.5 mg/L IBA (Indolebutyric acid)). Thirty-five days later, plants with roots were transplanted into pots containing a mixture of nutrient soil and perlite (1:1, v/v) and grown in a growth chamber under long-day (LD; 16-h light/8-h dark) conditions. “Orin” calli were grown on MS medium containing 1.5 mg/L 2,4-d and 0.4 mg/L 6-BA (6-benzyl aminopurine) in the dark at 25°C unless stated otherwise. Arabidopsis seedlings were grown at 23°C under short-day (SD; 8-h light/16-h dark) conditions before cold treatment.
Bioinformatics analysis
The intron–exon schematic structures of MdbHLH4 and AtMYC70 were drawn using GSDS software (http://gsds.gao-lab.org/). The conserved bHLH domain in MdbHLH4 was identified using CD-Search (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi), SMART (http://smart.embl-heidelberg.de/), and HMMER (https://www.ebi.ac.uk/Tools/hmmer/). Multiple sequence alignment of bHLH proteins from Arabidopsis and apple was performed using DNAMAN software (Version 6.0.3.99). The 3D structures of the bHLH and C-terminal domains of MdbHLH4 were predicted using SWISS-MODEL (https://swissmodel.expasy.org/) and visualized using RsaMol software (Version 2.7.5.2). Point mutation prediction of the MdbHLH4 protein was performed using PredictProtein (https://predictprotein.org/).
Vector construction and genetic transformation
The CDSs of MdbHLH4 and MdCAX3L-2 were cloned into the pCambia2300-GFP and pCambia35S-4MYC-3FLAG vectors, respectively, to construct the OE vectors (MdbHLH4-GFP/-Flag and MdCAX3L-2-GFP/-Flag). Selected gene-specific fragments of MdbHLH4 and MdCAX3L-2 were cloned into the RNAi vector pK7GWIWG2D (with a GFP selection marker) to generate MdbHLH4-RNAi and MdCAX3L-2-RNAi vectors, respectively. The CDS of MdICE1L was inserted into pCambia35S-3HA to generate MdICE1L-HA. To construct a MdCAX3L-2 OE vector driven by its own promoter, the MdCAX3L-2 promoter (1500 bp upstream of the start codon) was cloned from apple and used to replace the 35S promoter of the constructed MdCAX3L-2-Flag vector. The primers are listed in Supplemental Table S1.
Transgenic Arabidopsis and apple materials were obtained using the Agrobacterium-mediated transformation method. Six-week-old wild-type (Col-0) and myc70 mutant Arabidopsis plants were used for genetic transformation, which was conducted via the floral dip method (Clough and Bent, 1998). Twenty-day-old liquid apple calli were used for genetic transformation as described in a previous study (Yang et al., 2021). Thirty-day-old tissue-cultured “GL-3” plants were used to obtain MdbHLH4-OE and MdbHLH4-RNAi transgenic apple plants as previously described (Dai et al., 2013; Zhao et al., 2020).
Cold treatment
Both Arabidopsis and apple plants were subjected to two types of cold treatments: with cold acclimation (4°C for 12 h; CA) or without cold acclimation (NA). For Arabidopsis seedlings (7 days old) grown in 0.5 MS medium, seedlings in the NA group were treated with −4°C for 1 h, whereas seedlings in the CA group were treated with −8°C for 1 h. After cold treatment, seedlings were placed at 4°C for 12 h under dark conditions for recovery. After 2 days of culture at 23°C under LD conditions, the survival rate was calculated. One replicate comprised five plates and there were a total of three biological replicates. Arabidopsis plants grown in nutrition pots were treated with −8°C for 4 h after cold acclimation. Other treatments after cold treatment were consistent with those of Arabidopsis seedlings cultured in plates. Three independent biological replications were performed, with 20 seedlings of each line in each biological replicate.
For apple plants, 6-week-old WT and MdbHLH4 transgenic (OE/RNAi) plants with a consistent growth state were used in cold treatments (CA/NA). Plants in the NA group were treated with −6°C for 3 h, whereas plants in the CA group were treated with −8°C for 4 h after cold acclimation (4°C for 12 h). For the NA and CA treatments, freezing began at 0°C, the temperature dropped by 2°C per hour until reaching the desired temperature, and then the plants were held at this temperature for specified time period. After cold treatment, all plants were exposed to 4°C under dark conditions for 12 h for recovery. Subsequently, these plants were cultured at 25°C for 2 days under LD conditions. Three independent biological replications were performed, with 15 plants of each line in each biological replicate.
For apple calli, WT and transgenic calli were placed on new medium plates and pre-cultured for 5 days under normal conditions (25°C, dark). These plates were then subjected to 4°C under dark conditions for cold treatment. After 20 days of growth, images were taken and the fresh weight was measured. WT and calli transformed with the empty vector pCambia35S-3HA were used as controls. One replicate comprised three plates and there were a total of three biological replicates.
Subcellular localization
To determine the subcellular localization of proteins in plants, MdbHLH4-GFP and MdCAX3L-2-GFP fusion proteins were transiently expressed in N. benthamiana leaves as previously described (Yang et al., 2021). AtCBL2-mCherry was used as a tonoplast location marker. To identify the subcellular localization of MdCAX3L-2 in yeast, the CDS of MdCAX3L-2 was cloned into the pDR196-GFP vector. The recombinant vector was then transformed into the K667 yeast strain using the LiAc/PEG method. The fluorescence was captured by a confocal laser scanning microscope (TCS-SP8 SR, Leica). GFP fluorescence signals were detected at 500–535 nm after excitation at 488 nm, while mCherry was excited at 543 nm and scanned at 600–630 nm.
RNA extraction, gene expression analysis, and REL measurement
Total RNA of different plant materials was isolated using an RNAprep pure Plant Kit (Tiangen, Beijing, China) according to the manufacturer’s instructions. Reverse transcription was conducted for single-stranded DNA synthesis using the PrimeScript RT Reagent Kit (TaKaRa, Shiga, Japan). Reverse transcription qPCR (RT-qPCR) analysis was performed as previously described (Mao et al., 2017) using MdMDH as the reference gene. Relative electrolyte leakage (REL) was determined according to the following formula: REL = (D1 − D0)/(D2 − D0)×100% (Hang et al., 2021; Yang et al., 2021).
Y1H assays
The promoter fragments of MdCBF1 (1,200 bp), MdCBF3 (1,000 bp), and MdCAX3L-2 (1,500 bp) were cloned into the pHIS2 vector, and the CDSs of MdbHLH4 and MdICE1L were cloned into the pGADT7 vector. These constructs were transformed into the Y187 yeast strain with specified combinations, with the empty pGADT7 as a negative control. Positive transformants were inoculated to the screening medium (SD base/-Ura-Trp supplemented with 3-AT) to test for possible interactions.
Dual-LUC assays
The designed promoter fragments (1,500 bp) were cloned into the reporter vector pGreenII 0800-LUC, and the CDSs of MdbHLH4 and MdICE1L were cloned into the effector vector pGreenII 62-SK. The recombinant vectors were injected into N. benthamiana leaves with specified combinations by Agrobacterium-mediated transformation for transient expression. After 3 days of culture under normal conditions (25°C, LD) and treatment with 4°C for 6 h, the parts surrounding the injection site were collected for luminescence detection with a detection kit (Promega, Madison, Wisconsin, USA). LUC fluorescence was photographed with a Lumazone Pylon 2048B imaging system (Princeton, New Jersey, USA).
Electromobility shift assays
MdbHLH4-GST and MdICE1L-HIS fusion proteins were expressed in Escherchia coli BL21 and purified with GST and HIS purification columns (Beyotime, Shanghai, China), respectively. EMSAs were performed using the LightShift Chemiluminescent EMSA kit (Thermo Scientific, Waltham, Massachusetts, USA) according to the manufacturer’s instructions. Sequences of various probes are listed in Supplemental Table S1.
ChIP-qPCR assays
MdICE1L-HA and MdICE1-HA/MdbHLH4 transgenic apple calli were treated at 4°C for 3 h and then were harvested and immersed in formaldehyde solution for cross-linking. After sonication, chromatin from control (EV-HA) and MdICE1L-HA transgenic calli was immunoprecipitated with or without anti-HA antibody (Yeasen, Shanghai, China). Relative enrichment values of the promoter fragments were detected by qPCR, with the enrichment of control (EV-HA) samples used as the reference, which was set to 1.0. Three biological replicates were performed, with each containing four technical replicates. The primers are listed in Supplemental Table S1.
Y2H assays
The CDSs or truncated fragments of MdbHLH4, MdICE1L, and MdICE1 were cloned into the prey vector pGAD424 (or pGADT7) and the bait vector pGBT9 (or pGBKT7), respectively. The recombinant vectors were co-transformed into the yeast strain Y2H-Gold with specified combinations. Positive transformants grown on SD-Trp/-Leu (DDO) medium were inoculated to the screening medium (SD-Trp/-Leu/-His/-Ade, QDO) for identification of possible interactions.
Co-IP assays
The MdbHLH4-GFP, MdICE1L-HA, MdICE1-HA, and pCambia2300-GFP vectors were transiently co-expressed in N. benthamiana leaves with specified combinations. After 3 days of culture under normal conditions, total protein was extracted from leaves and incubated with anti-HA magnetic beads (Beyotime) at 4°C overnight. The eluted solution was detected with anti-HA and anti-GFP antibodies (Yeasen).
LUC complementation imaging assays
The CDSs of MdICE1L and MdMPK6a/b were cloned into the pRI101-nLUC and pRI101-cLUC vectors, respectively. The recombinant vectors were injected into N. benthamiana leaves with specified combinations by Agrobacterium-mediated transformation for transient expression. Combinations containing empty nLUC or cLUC vectors were used as negative controls. After 3 days of culture under normal conditions (25°C, LD), the parts surrounding the injection site were collected for luminescence detection with a detection kit (Promega). The LUC fluorescence signal was captured using a Lumazone Pylon 2048B imaging system.
Protein degradation and ubiquitination analysis
For in vivo protein degradation assays, various transgenic calli showing consistent growth were placed on MS medium supplemented with 250 μM cycloheximide (CHX) (or CHX plus 100 μM MG132) and treated at 4°C. Samples were taken at specified time points. The protein level of MdICE1L-HA was detected using anti-HA antibody (Yeasen).
For ubiquitination detection, transgenic calli were treated at 4°C for 6 h in MS medium supplemented with 250 μM CHX and 100 μM MG132. The protein extracts were immunoprecipitated using anti-HA magnetic beads. The eluted solution was detected with anti-HA and anti-Ubi (Cell Signaling Technology, USA) antibodies.
Statistical analysis
IBM SPSS Statistics software (version 26; SPSS Inc., Chicago, IL, USA) was used for statistical analysis. Error bars in the bar graphs show means ± sd of three biological replicates. Significant differences (P < 0.05) were determined by one-way ANOVA followed by Duncan’s test or Student’s t test.
Accession numbers
Arabidopsis sequence data in this article can be found in the Arabidopsis Information Resource (TAIR, http://www.Arabidopsis.org/index.jsp) under accession nos. AtMYC70 (AT2G46810), AtCBF1 (AT4G25490), AtCBF3 (AT4G25480), AtKIN1 (AT5G15960), AtRD29A (AT5G52310), AtCOR47 (AT1G20440), AtCOR15A (AT2G42540), AtMPK3 (AT3G45640), and AtMPK6 (AT2G43790). Apple sequence data can be found in the Genome Database for Rosaceae (GDR; https://www.rosaceae.org) data libraries under accession nos. MdbHLH4 (MD01G1089300), MdCAX3L-2 (MD12G1165800), MdICE1 (MDP0000662999), MdICE1L (MD09G1003800), MdCBF1 (MD07G1262900), MdCBF3 (MD01G1196100), MdKIN1 (MD09G1079600), MdRD29A (MD01G1201000), MdCOR47 (MD08G1004500), MdCOR15A (MD10G1035600), MdMPK3a (MD11G1121500), MdMPK3b (MD03G1108500), MdMPK6a (MD15G1147300), and MdMPK6b (MD02G1004000).
Supplementary Material
Acknowledgments
We thank Prof. Zhihong Zhang (Shenyang Agricultural University) for providing us with the “Gala” (GL-3) apple plants. We also thank the Horticulture Science Research Center at College of Horticulture, NWAFU, for their technical support in this work.
Funding
This work was financially supported by grants from the National Key Research and Development Program of China (2019YFD1000102), the National Natural Science Foundation of China (31701894), the Key S&T Special Projects of Shaanxi Province (2020zdzx03-01-02), and the China Agriculture Research System of MOF and MARA (CARS-27).
Conflict of interest statement. The authors declare that there is no conflict of interest.
Contributor Information
Jie Yang, State Key Laboratory of Crop Stress Biology for Arid Areas/Shaanxi Key Laboratory of Apple, College of Horticulture, Northwest A&F University, Yangling 712100, China.
Xin Guo, State Key Laboratory of Crop Stress Biology for Arid Areas/Shaanxi Key Laboratory of Apple, College of Horticulture, Northwest A&F University, Yangling 712100, China.
Quanlin Mei, State Key Laboratory of Crop Stress Biology for Arid Areas/Shaanxi Key Laboratory of Apple, College of Horticulture, Northwest A&F University, Yangling 712100, China.
Lina Qiu, State Key Laboratory of Crop Stress Biology for Arid Areas/Shaanxi Key Laboratory of Apple, College of Horticulture, Northwest A&F University, Yangling 712100, China.
Peihong Chen, State Key Laboratory of Crop Stress Biology for Arid Areas/Shaanxi Key Laboratory of Apple, College of Horticulture, Northwest A&F University, Yangling 712100, China.
Weihan Li, State Key Laboratory of Crop Stress Biology for Arid Areas/Shaanxi Key Laboratory of Apple, College of Horticulture, Northwest A&F University, Yangling 712100, China.
Ke Mao, State Key Laboratory of Crop Stress Biology for Arid Areas/Shaanxi Key Laboratory of Apple, College of Horticulture, Northwest A&F University, Yangling 712100, China.
Fengwang Ma, State Key Laboratory of Crop Stress Biology for Arid Areas/Shaanxi Key Laboratory of Apple, College of Horticulture, Northwest A&F University, Yangling 712100, China.
Data availability
The authors confirm that all experimental data are available and accessible via the main text and/or the supplemental data.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Schematic diagram of the gene structures of MdbHLH4 and AtMYC70.
Supplemental Figure S2. Details of the 3D structure prediction results of MdbHLH4 protein.
Supplemental Figure S3. Identification of MdbHLH4 transgenic Arabidopsis seedlings.
Supplemental Figure S4. Identification of MdbHLH4 transgenic apple plants.
Supplemental Figure S5. Sequences of probes used in EMSAs.
Supplemental Figure S6. Sequence alignment of MdCIbHLH1, MdICE1, and MdICE1L.
Supplemental Figure S7. Prediction of the effects of point mutations on the MdbHLH4 protein.
Supplemental Figure S8. Prediction of the effects of point mutations and functional region division of MdICE1L and MdICE1 proteins.
Supplemental Figure S9. Identification of MdICE1L-HA and MdbHLH4-GFP protein expression in transgenic calli.
Supplemental Figure S10. Identification of MdCAX3L-2 transgenic Arabidopsis seedlings.
Supplemental Figure S11. Comparison of the growth phenotype, fresh weight, and MdCAX3L-2 expression between WT and proMdCAX3L-2::MdCAX3L-2 transgenic apple calli.
Supplemental Table S1. The primers used in this study.
K.M. and F.M. conceived and designed the experiments. J.Y., X.G., and Q.M. performed most of the experiments. L.Q., P.C., and W.L. provided the technical assistance. J.Y., K.M., and F.M. analyzed the data and wrote the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/General-Instructions) is: Fengwang Ma (fwm64@sina.com).
References
- Agarwal M, Hao YJ, Kapoor A, Dong CH, Fujii H, Zheng XW, Zhu JK (2006) A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J Biol Chem 281: 37636–37645 [DOI] [PubMed] [Google Scholar]
- An JP, Wang XF, Zhang XW, You CX, Hao YJ (2021) Apple B-box protein BBX37 regulates jasmonic acid mediated cold tolerance through the JAZ-BBX37-ICE1-CBF pathway and undergoes MIEL1-mediated ubiquitination and degradation. New Phytol 229: 2707–2729 [DOI] [PubMed] [Google Scholar]
- Catala R, Santos E, Alonso JM, Ecker JR, Martinez-Zapater JM, Salinas J (2003) Mutations in the Ca2+/H+ transporter CAX1 increase CBF/DREB1 expression and the cold-acclimation response in Arabidopsis. Plant Cell 15: 2940–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XX, Ding YL, Yang YQ, Song CP, Wang BS, Yang SH, Guo Y, Gong ZZ (2021) Protein kinases in plant responses to drought, salt, and cold stress. J Integr Plant Biol 63: 53–78 [DOI] [PubMed] [Google Scholar]
- Chinnusamy V, Ohta M, Kanrar S, Lee BH, Hong XH, Agarwal M, Zhu JK (2003) ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev 17: 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Zhu J, Zhu JK (2007) Cold stress regulation of gene expression in plants. Trends Plant Sci 12: 444–451 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cui YM, Lu S, Li Z, Cheng JW, Hu P, Zhu TQ, Wang X, Jin M, Wang XX, Li LQ, et al. (2020) CYCLIC NUCLEOTIDE-GATED ION CHANNELs 14 and 16 promote tolerance to heat and chilling in rice. Plant Physiol 183: 1794–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai HY, Li WR, Han GF, Yang Y, Ma Y, Li H, Zhang ZH (2013) Development of a seedling clone with high regeneration capacity and susceptibility to Agrobacterium in apple. Sci Horticult 164: 202–208 [Google Scholar]
- Demidchik V, Shabala S, Isayenkov S, Cuin TA, Pottosin I (2018) Calcium transport across plant membranes: mechanisms and functions. New Phytol 220: 49–69 [DOI] [PubMed] [Google Scholar]
- Ding Y, Yang S (2022) Surviving and thriving: how plants perceive and respond to temperature stress. Dev Cell 57: 947–958 [DOI] [PubMed] [Google Scholar]
- Ding YL, Li H, Zhang XY, Xie Q, Gong ZZ, Yang SH (2015) OST1 kinase modulates freezing tolerance by enhancing ICE1 stability in Arabidopsis. Dev Cell 32: 278–289 [DOI] [PubMed] [Google Scholar]
- Ding YL, Shi YT, Yang SH (2019) Advances and challenges in uncovering cold tolerance regulatory mechanisms in plants. New Phytol 222: 1690–1704 [DOI] [PubMed] [Google Scholar]
- Dodd AN, Kudla J, Sanders D (2010) The language of calcium signaling. Annu Rev Plant Biol 61: 593–620 [DOI] [PubMed] [Google Scholar]
- Doherty CJ, Van Buskirk HA, Myers SJ, Thomashow MF (2009) Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 21: 972–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong CH, Agarwal M, Zhang YY, Xie Q, Zhu JK (2006) The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc Natl Acad Sci USA 103: 8281–8286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feller A, Machemer K, Braun EL, Grotewold E (2011) Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J 66: 94–116 [DOI] [PubMed] [Google Scholar]
- Feng HL, Ma NN, Meng X, Zhang S, Wang JR, Chai S, Meng QW (2013) A novel tomato MYC-type ICE1-like transcription factor, SlICE1a, confers cold, osmotic and salt tolerance in transgenic tobacco. Plant Physiol Biochem 73: 309–320 [DOI] [PubMed] [Google Scholar]
- Feng XM, Zhao Q, Zhao LL, Qiao Y, Xie XB, Li HF, Yao YX, You CX, Hao YJ (2012) The cold-induced basic helix–loop–helix transcription factor gene MdCIbHLH1 encodes an ICE-like protein in apple. BMC Plant Biol 12: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fursova OV, Pogorelko GV, Tarasov VA (2009) Identification of ICE2, a gene involved in cold acclimation which determines freezing tolerance in Arabidopsis thaliana. Gene 429: 98–103 [DOI] [PubMed] [Google Scholar]
- Gilmour SJ, Fowler SG, Thomashow MF (2004) Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol Biol 54: 767–781 [DOI] [PubMed] [Google Scholar]
- Guo XY, Liu DF, Chong K (2018) Cold signaling in plants: insights into mechanisms and regulation. J Integr Plant Biol 60: 745–756 [DOI] [PubMed] [Google Scholar]
- Hang N, Shi TR, Liu YR, Ye WX, Taier G, Sun Y, Wang KH, Zhang WJ (2021) Overexpression of Os-microRNA408 enhances drought tolerance in perennial ryegrass. Physiol Plant 172: 733–747 [DOI] [PubMed] [Google Scholar]
- Hirschi KD (1999) Expression of Arabidopsis CAX1 in tobacco: altered calcium homeostasis and increased stress sensitivity. Plant Cell 11: 2113–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu YR, Han X, Yang ML, Zhang MH, Pan JJ, Yu DQ (2019) The transcription factor INDUCER OF CBF EXPRESSION1 interacts with ABSCISIC ACID INSENSITIVE5 and DELLA proteins to fine-tune abscisic acid signaling during seed germination in Arabidopsis. Plant Cell 31: 1520–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu YR, Jiang LQ, Wang F, Yu DQ (2013) Jasmonate regulates the INDUCER OF CBF EXPRESSION-C-REPEAT BINDING FACTOR/DRE BINDING FACTOR1 cascade and freezing tolerance in Arabidopsis. Plant Cell 25: 2907–2924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang BC, Shi YT, Zhang XY, Xin XY, Qi LJ, Guo HW, Li JG, Yang SH (2017) PIF3 is a negative regulator of the CBF pathway and freezing tolerance in Arabidopsis. Proc Natl Acad Sci USA 114: E6695–E6702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidokoro S, Yoneda K, Takasaki H, Takahashi F, Shinozaki K, Yamaguchi-Shinozaki K (2017) Different cold-signaling pathways function in the responses to rapid and gradual decreases in temperature. Plant Cell 29: 760–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Kim HS, Bahk S, An J, Yoo Y, Kim JY, Chung WS (2017) Phosphorylation of the transcriptional repressor MYB15 by mitogen-activated protein kinase 6 is required for freezing tolerance in Arabidopsis. Nucleic Acids Res 45: 6613–6627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Park S, Gilmour SJ, Thomashow MF (2013) Roles of CAMTA transcription factors and salicylic acid in configuring the low-temperature transcriptome and freezing tolerance of Arabidopsis. Plant J 75: 364–376 [DOI] [PubMed] [Google Scholar]
- Knight H, Trewavas AJ, Knight MR (1996) Cold calcium signaling in Arabidopsis involves two cellular pools and a change in calcium signature after acclimation. Plant Cell 8: 489–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla J, Becker D, Grill E, Hedrich R, Hippler M, Kummer U, Parniske M, Romeis T, Schumacher K (2018) Advances and current challenges in calcium signaling. New Phytol 218: 414–431 [DOI] [PubMed] [Google Scholar]
- Lee BH, Henderson DA, Zhu JK (2005) The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell 17: 3155–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CM, Thomashow MF (2012) Photoperiodic regulation of the C-repeat binding factor (CBF) cold acclimation pathway and freezing tolerance in Arabidopsis thaliana. Proc Natl Acad Sci USA 109: 15054–15059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Seo PJ (2021) Ca2+talyzing initial responses to environmental stresses. Trends Plant Sci 26: 849–870 [DOI] [PubMed] [Google Scholar]
- Li H, Ding YL, Shi YT, Zhang XY, Zhang SQ, Gong ZZ, Yang SH (2017) MPK3- and MPK6-mediated ICE1 phosphorylation negatively regulates ICE1 stability and freezing tolerance in Arabidopsis. Dev Cell 43: 630–642 [DOI] [PubMed] [Google Scholar]
- Li WW, Zhou MQ, Zheng Y, Lin P, Yao XH, Lin J (2017) Characterization of CbCAX51, a cold responsive Ca2+/H+ exchanger from Capsella bursa-pastoris modulating cold tolerance in plants. Int J Agric Biol 19: 817–824 [Google Scholar]
- Lian TF, Xu YP, Li LF, Su XD (2017) Crystal structure of tetrameric Arabidopsis MYC2 reveals the mechanism of enhanced interaction with DNA. Cell Rep 19: 1334–1342 [DOI] [PubMed] [Google Scholar]
- Liu JY, Shi YT, Yang SH (2018) Insights into the regulation of C-repeat binding factors in plant cold signaling. J Integr Plant Biol 60: 780–795 [DOI] [PubMed] [Google Scholar]
- Liu QB, Ding YL, Shi YT, Ma L, Wang Y, Song CP, Wilkins KA, Davies JM, Knight H, Knight MR, et al. (2021) The calcium transporter ANNEXIN1 mediates cold-induced calcium signaling and freezing tolerance in plants. EMBO J 40: e104559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Berkowitz GA (2017) Multimeric CAX complexes and Ca2+ signaling—beyond humdrum housekeeping. J Exp Bot 68: 3997–3999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manohar M, Shigaki T, Hirschi KD (2011) Plant cation/H+ exchangers (CAXs): biological functions and genetic manipulations. Plant Biol (Stuttg) 13: 561–569 [DOI] [PubMed] [Google Scholar]
- Mao K, Dong QL, Li C, Liu CH, Ma FW (2017) Genome wide identification and characterization of apple bHLH transcription factors and expression analysis in response to drought and salt stress. Front Plant Sci 8: 480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao K, Yang J, Wang M, Liu HY, Guo X, Zhao S, Dong QL, Ma FW (2021) Genome-wide analysis of the apple CaCA superfamily reveals that MdCAX proteins are involved in the abiotic stress response as calcium transporters. BMC Plant Biol 21: 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina J, Catala R, Salinas J (2011) The CBFs: three Arabidopsis transcription factors to cold acclimate. Plant Sci 180: 3–11 [DOI] [PubMed] [Google Scholar]
- Miura K, Jin JB, Lee J, Yoo CY, Stirm V, Miura T, Ashworth EN, Bressan RA, Yun DJ, Hasegawa PM (2007) SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 19: 1403–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Ohta M, Nakazawa M, Ono M, Hasegawa PM (2011) ICE1 Ser403 is necessary for protein stabilization and regulation of cold signaling and tolerance. Plant J 67: 269–279 [DOI] [PubMed] [Google Scholar]
- Mori K, Renhu N, Naito M, Nakamura A, Shiba H, Yamamoto T, Suzaki T, Iida H, Miura K (2018) Ca2+-permeable mechanosensitive channels MCA1 and MCA2 mediate cold-induced cytosolic Ca2+ increase and cold tolerance in Arabidopsis. Sci Rep 8: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morran S, Eini O, Pyvovarenko T, Parent B, Singh R, Ismagul A, Eliby S, Shirley N, Langridge P, Lopato S (2011) Improvement of stress tolerance of wheat and barley by modulation of expression of DREB/CBF factors. Plant Biotechnol J 9: 230–249 [DOI] [PubMed] [Google Scholar]
- Novillo F, Alonso JM, Ecker JR, Salinas J (2004) CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in, stress tolerance in Arabidopsis. Proc Natl Acad Sci USA 101: 3985–3990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M, Sato A, Renhu N, Yamamoto T, Oka N, Zhu JK, Tada Y, Suzaki T, Miura K (2018) MYC-type transcription factors, MYC67 and MYC70, interact with ICE1 and negatively regulate cold tolerance in Arabidopsis. Sci Rep 8: 11622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pino MT, Skinner JS, Park EJ, Jeknic Z, Hayes PM, Thornashow MF, Chen THH (2007) Use of a stress inducible promoter to drive ectopic AtCBF expression improves potato freezing tolerance while minimizing negative effects on tuber yield. Plant Biotechnol J 5: 591–604 [DOI] [PubMed] [Google Scholar]
- Pires N, Dolan L (2010) Origin and diversification of basic–helix–loop–helix proteins in plants. Mol Biol Evol 27: 862–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman JK, Hirschi KD (2016) CAX-ing a wide net: cation/H+ transporters in metal remediation and abiotic stress signalling. Plant Biol 18: 741–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi YT, Tian SW, Hou LY, Huang XZ, Zhang XY, Guo HW, Yang SH (2012) Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and type-A ARR genes in Arabidopsis. Plant Cell 24: 2578–2595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigaki T, Hirschi KD (2006) Diverse functions and molecular properties emerging for CAX cation/H+ exchangers in plants. Plant Biol 8: 419–429 [DOI] [PubMed] [Google Scholar]
- Song S, Huang H, Gao H, Wang J, Wu D, Liu X, Yang S, Zhai Q, Li C, Qi T, et al. (2014) Interaction between MYC2 and ETHYLENE INSENSITIVE3 modulates antagonism between jasmonate and ethylene signaling in Arabidopsis. Plant Cell 26: 263–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Zhang XY, Li MZ, Yang H, Fu DY, Lv J, Ding YL, Gong ZZ, Shi YT, Yang SH (2021) The direct targets of CBFs: in cold stress response and beyond. J Integr Plant Biol 63: 1868–1887 [DOI] [PubMed] [Google Scholar]
- Stockinger EJ, Gilmour SJ, Thomashow MF (1997) Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci USA 94: 1035–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su XD, Lian TF, Xu YP (2017) Crystal structure of tetrameric Arabidopsis MYC2–DNA complex. Acta Crystallogr Found Adv 73: C300–C300 [Google Scholar]
- Thomashow MF (1999) Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol 50: 571–599 [DOI] [PubMed] [Google Scholar]
- Wang JC, Ren YL, Liu X, Luo S, Zhang X, Liu X, Lin QB, Zhu SS, Wan H, Yang Y, et al. (2021) Transcriptional activation and phosphorylation of OsCNGC9 confer enhanced chilling tolerance in rice. Mol Plant 14: 315–329 [DOI] [PubMed] [Google Scholar]
- Wang X, Ding YL, Li ZY, Shi YT, Wang JL, Hua J, Gong ZZ, Zhou JM, Yang SH (2019) PUB25 and PUB26 promote plant freezing tolerance by degrading the cold signaling negative regulator MYB15. Dev Cell 51: 222–235 [DOI] [PubMed] [Google Scholar]
- Wu M, Tong S, Waltersperger S, Diederichs K, Wang M, Zheng L (2013) Crystal structure of Ca2+/H+ antiporter protein YfkE reveals the mechanisms of Ca2+ efflux and its pH regulation. Proc Natl Acad Sci USA 110: 11367–11372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Zahid KR, He LR, Zhang WW, He X, Zhang XL, Yang XY, Zhu LF (2013) GhCAX3 gene, a novel Ca2+/H+ exchanger from cotton, confers regulation of cold response and ABA induced signal transduction. PLoS ONE 8: e66303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka T, Nakagawa Y, Mori K, Nakano M, Imamura T, Kataoka H, Terashima A, Iida K, Kojima I, Katagiri T, et al. (2010) MCA1 and MCA2 that mediate Ca2+ uptake have distinct and overlapping roles in Arabidopsis. Plant Physiol 152: 1284–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Guo X, Li WH, Chen PH, Cheng YP, Ma FW, Mao K (2021) MdCCX2 of apple functions positively in modulation of salt tolerance. Environ Exp Bot 192: 104663 [Google Scholar]
- Yang J, Li WH, Guo X, Chen PH, Cheng YP, Mao K, Ma FW (2021) Cation/Ca2+ exchanger 1 (MdCCX1), a plasma membrane-localized Na+ transporter, enhances plant salt tolerance by inhibiting excessive accumulation of Na+ and reactive oxygen species. Front Plant Sci 12: 746189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang TB, Chaudhuri S, Yang LH, Du LQ, Poovaiah BW (2010) A calcium/calmodulin-regulated member of the receptor-like kinase family confers cold tolerance in plants. J Biol Chem 285: 7119–7126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye KY, Li H, Ding YL, Shi YT, Song CP, Gong ZZ, Yang SH (2019) BRASSINOSTEROID-INSENSITIVE2 negatively regulates the stability of transcription factor ICE1 in response to cold stress in Arabidopsis. Plant Cell 31: 2682–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura K, Iida K, Iida H (2021) MCAs in Arabidopsis are Ca2+-permeable mechanosensitive channels inherently sensitive to membrane tension. Nat Commun 12: 6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao CZ, Wang PC, Si T, Hsu CC, Wang L, Zayed O, Yu ZP, Zhu YF, Dong J, Tao WA, et al. (2017) MAP kinase cascades regulate the cold response by modulating ICE1 protein stability. Dev Cell 43: 618–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Gao HB, Jia XM, Wang HB, Ke M, Ma FW (2020) The HD-Zip I transcription factor MdHB-7 regulates drought tolerance in transgenic apple (Malus domestica). Environ Exp Bot 180: 104246 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that all experimental data are available and accessible via the main text and/or the supplemental data.