Abstract
Carotenoid cleavage, catalyzed by CAROTENOID CLEAVAGE DIOXYGENASEs (CCDs), provides signaling molecules and precursors of plant hormones. Recently, we showed that zaxinone, a apocarotenoid metabolite formed by the CCD ZAXINONE SYNTHASE (ZAS), is a growth regulator required for normal rice (Oryza sativa) growth and development. The rice genome encodes three OsZAS homologs, called here OsZAS1b, OsZAS1c, and OsZAS2, with unknown functions. Here, we investigated the enzymatic activity, expression pattern, and subcellular localization of OsZAS2 and generated and characterized loss-of-function CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats and associated protein 9)-Oszas2 mutants. We show that OsZAS2 formed zaxinone in vitro. OsZAS2 was predominantly localized in plastids and mainly expressed under phosphate starvation. Moreover, OsZAS2 expression increased during mycorrhization, specifically in arbuscule-containing cells. Oszas2 mutants contained lower zaxinone content in roots and exhibited reduced root and shoot biomass, fewer tillers, and higher strigolactone (SL) levels. Exogenous zaxinone application repressed SL biosynthesis and partially rescued the growth retardation of the Oszas2 mutant. Consistent with the OsZAS2 expression pattern, Oszas2 mutants displayed a lower frequency of arbuscular mycorrhizal colonization. In conclusion, OsZAS2 is a zaxinone-forming enzyme that, similar to the previously reported OsZAS, determines rice growth, architecture, and SL content, and is required for optimal mycorrhization.
OsZAS2 is a root- and arbusculated cell-localized zaxinone-forming enzyme required for proper rice growth and normal mycorrhization and SL homeostasis.
Introduction
Carotenoids are tetraterpene (C40) pigments consisting of long hydrocarbon chains with a conjugated double-bond system. In plants, carotenoids serve as a crucial component of photosynthesis, colorants, and antioxidants (Bouvier et al., 2003; Fraser and Bramley, 2004; Ballottari et al., 2014; Nisar et al., 2015; Hashimoto et al., 2016; Rodriguez et al., 2018; Zheng et al., 2020). In addition, the breakdown of carotenoids gives rise to a diverse group of metabolites called apocarotenoids, which includes pigments, scents, signaling molecules, growth regulators, and the precursors of the phytohormones strigolactone (SL) and abscisic acid (ABA) (Felemban et al., 2019; Moreno et al., 2021; Wang et al, 2021a; Liang et al., 2021; Zheng et al., 2021). ABA is the most-studied plant apocarotenoid hormone and a key player in plant response to abiotic and biotic stress (Peleg and Blumwald, 2011), regulation of seed maturation, dormancy, and shoot and root growth (Nambara and Marion-Poll, 2005; Moreno et al., 2021). SLs regulate a series of developmental processes. They are best known for inhibiting shoot branching/tillering, regulating root architecture, secondary growth, and senescence, and for their contribution to biotic and abiotic stress responses (Gomez-Roldan et al., 2008; Umehara et al., 2008; Ha et al., 2014; Al-Babili and Bouwmeester, 2015; Decker et al., 2017; Waters et al., 2017; Jia et al., 2019). However, SLs were originally discovered as host root-released germination stimulants for seeds of root parasitic weeds (Xie et al., 2010). Later on, they were identified as the plant-released hyphal branching factor for arbuscular mycorrhizal (AM) fungi, which paved the way for establishing plant–AM symbiosis (Akiyama et al., 2005). AM fungi symbiotic association provides the host plant with minerals, mainly phosphorus (P) and nitrogen (N), and the AM fungi with carbohydrates and lipids (Wang et al., 2017). AM symbiosis is widely distributed and formed by most of the land plants, mirroring its importance for their growth and survival (Gutjahr and Parniske, 2013; Wang et al., 2017; Fiorilli et al., 2019).
Recent studies unraveled the presence of several apocarotenoid signaling molecules, such as anchorene, iso-anchorene, β-cyclocitral, and zaxinone. Anchorene is a carotenoid-derived dialdehyde responsible for anchor root formation in Arabidopsis (Arabidopsis thaliana) (Jia et al., 2019), while its structural isomer iso-anchorene inhibits Arabidopsis root growth (Jia et al., 2021). β-Cyclocitral regulates root growth and is a retrograde signaling molecule that mediates singlet oxygen response and improves the high light tolerance by modulating the expression of oxidative stress-responsive genes (Ramel et al., 2012; Dickinson et al., 2019). Zaxinone is a regulatory metabolite, which is required for normal rice (Oryza sativa) growth and development and negatively regulates SL biosynthesis (Wang et al., 2019, 2020). Multi-omics study revealed that zaxinone also modulates cytokinin homeostasis and that its growth-promoting effect is likely caused by increased sugar metabolism in rice roots (Wang et al., 2021a, 2021b). However, exogenous application of zaxinone to Arabidopsis simultaneously increased both SL and ABA content (Ablazov et al., 2020), suggesting that it might act as a stress signal in Arabidopsis (Ablazov et al., 2020).
Apocarotenoid production in plants is mediated by carotenoid cleavage dioxygenases (CCDs), which cleave double bonds in carotenoid backbones and exhibit different substrate and cleavage site specificities (Moise et al., 2014; Dhar et al., 2020). The diversity of CCDs gives rise to a wide spectrum of apocarotenoids with unique features and functions (Giuliano et al., 2003; Auldridge et al., 2006; Ahrazem et al., 2016). Based on phylogenetic analysis and enzymatic activity, plant CCDs build six subfamilies; NINE-CIS-EPOXY CAROTENOID DIOXYGENASEs (NCEDs), CCD1, CCD4, CCD7, CCD8, and ZAXINONE SYNTHASE (ZAS) (Wang et al., 2019). NCEDs are involved in ABA biosynthesis and catalyze the cleavage of the C11, C12 (or C11′, C12′) double bond in 9-cis-epoxy carotenoids to yield the ABA precursor xanthoxin (Schwartz et al., 1997; Chernys and Zeevart, 2000). In contrast to other CCD types investigated so far, CCD1 enzymes are localized in the cytosol. Moreover, they are characterized by wide substrate and regio-specificity, cleaving many carotenoid and apocarotenoid substrates and producing dialdehyde products and volatiles that contribute to the flavor and aroma in many plants (Vogel et al., 2008; Ilg et al., 2009, 2014). CCD4 enzymes cleave the C9–C10 or C9′–C10′ double bond in bicyclic carotenoids, giving rise to C13 volatiles and C27-apocarotenoids (Bruno et al., 2015, 2016). CCD7 and CCD8 act sequentially on 9-cis-β-carotene to produce carlactone, the central intermediate of SL biosynthesis, via the intermediate 9-cis-β-apo-10′-carotenal formed by CCD7 along with the volatile β-ionone (Alder et al., 2012; Bruno et al., 2014). Carlactone is further modified by cytochrome P450s (711 clades), such as the Arabidopsis MORE AXILLARY GROWTH1 or the rice carlactone oxidase, leading to the formation of canonical and noncanonical SLs (Abe et al., 2014; Zhang et al., 2014; Jia et al., 2018; Ito et al., 2022).
ZAS is a recently discovered member of the CCD family, which cleaves the apocarotenoid apo-10-zeaxanthinal (C27) at the C13, C14 double bond, forming the C18-apocarotenoid zaxinone (Wang et al., 2019). Zaxinone is a growth-promoting metabolite required for normal rice growth and a negative regulator of SL biosynthesis and release (Wang et al., 2019, 2020). A rice loss-of-function zas mutant showed reduced root zaxinone level, retarded growth, that is, lower root and shoot biomass, tiller number, and higher SL content (Wang et al., 2019). Confirming its biological function, the exogenous application of zaxinone restored several phenotypes of the zas mutant (Wang et al., 2019). Though all other CCD subfamilies are conserved, nonmycorrhizal species, such as A. thaliana and other members of the Brassicales, lack ZAS orthologs, indicating a role of ZAS in AM symbiosis (Fiorilli et al., 2019; Wang et al., 2019). Indeed, the zas mutant displayed a lower level of AM colonization compared to the wild-type (WT; Wang et al., 2019).
The rice genome contains three OsZAS homologs, previously called OsZAS-L1 (renamed here to OsZAS1b), OsZAS-L2 (renamed here to OsZAS1c), and OsZAS-L3 (renamed here to OsZAS2) with unknown function (Wang et al., 2019). In this study, we investigated the biological function of OsZAS2 by studying its enzymatic activity and by generating and characterizing the corresponding mutant and GUS reporter lines. Obtained data suggest that OsZAS2 is a nonredundant, root- and arbusculated cell-localized zaxinone-forming enzyme required for proper growth and normal SL homeostasis and mycorrhization level.
Results
OsZAS2 represents a separate clade in the ZAS CCD subfamily
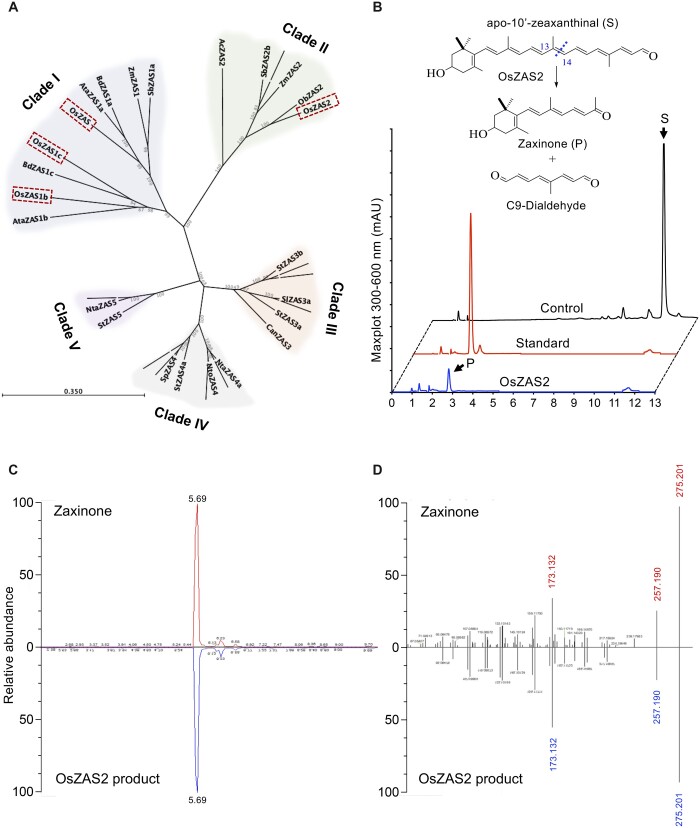
To clarify the phylogenetic relationship of OsZASs (OsZAS, OsZAS-L1, OsZAS-L2, and OsZAS-L3) with other plant ZAS genes, we first constructed a phylogenetic tree, using ZAS sequences from selected monocot and dicot species (Supplemental Table S2; Supplemental File S1). This analysis divided the ZAS proteins into five clades (I–V) (Figure 1A). Three OsZAS enzymes, including OsZAS (Os09g0321200), OsZAS-L1 (Os08g0369800), and OsZAS-L2 (Os08g0371608) grouped in clade I, while OsZAS-L3 (Os06g0162550) clustered in clade II. Based on this analysis we renamed OsZAS-L1, OsZAS-L2, and OsZAS-L3 to OsZAS1b, OsZAS1c, and OsZAS2, respectively (Figure 1A). The clades III, IV, and V contain only enzymes from dicot species, which we called ZAS3, ZAS4, and ZAS5 (Supplemental Table S3).
Figure 1.
Phylogenetic analysis of ZAS enzymes and analysis of OsZAS2 enzymatic activity. A, Phylogenetic tree analysis of ZAS orthologs from selected monocot and dicot plants, showing bootstrap values on nodes of each cluster. Dashed rectangles represent rice ZAS members. The scale bar represents the number of amino acid change per site. B, HPLC chromatogram of in vitro incubation of OsZAS2 with apo-10′-zeaxanthinal (I) yielded zaxinone (II) and a presumed C9-dialdehyde. The maximum absorbance (mAU) peak for substrate and product is shown at 347 and 450 nm (mAU), respectively. The representation of zaxinone production in the Figure B adapted from Wang et al. (2019), which is permitted to adaptation under a Creative Commons Attribution 4.0 International License. C, Verification of OsZAS2 in vitro product, based on retention time, (D) accurate mass and MS/MS pattern and in comparison to zaxinone standard.
OsZAS2 is a zaxinone-forming enzyme
Next, we investigated the enzymatic activity of OsZAS2: we expressed OsZAS2 fused to maltose-binding protein (MBP) in Escherichia coli cells and incubated the soluble fraction of these cells with different apocarotenoids, that is, β-apo-10′-(C27), 9-cis-β-apo-10′-(C27), β-apo-12′-(C25), and apo-8′-zeaxanthinal (3-OH-β-apo-8′-carotenal, C30) (Supplemental Figure S1). In addition, we incubated the MBP–OsZAS2 fusion with carotenoids, that is, β-carotene, zeaxanthin, and lutein (Supplemental Figure S1). Finally, we performed in vivo activity test by expressing a thioredoxin–OsZAS2 fusion in β-carotene, zeaxanthin, and lycopene-accumulating E. coli cells. In all these assays, we only detected a conversion of apo-10′-zeaxanthinal (C27) that was cleaved by OsZAS2 at the C13, C14 double bond, yielding zaxinone (3-OH-β-apo-13-carotenone, C18) and a predicted C9-dialdehyde (Figure 1B). We confirmed the identity of zaxinone by UHPLC (Ultra High Performance Liquid Chromatography) and LC–MS (Liquid chromatography–mass spectrometry) analysis, using a synthetic standard (Figure 1, C and D).
OsZAS2 is predominantly localized in the plastid
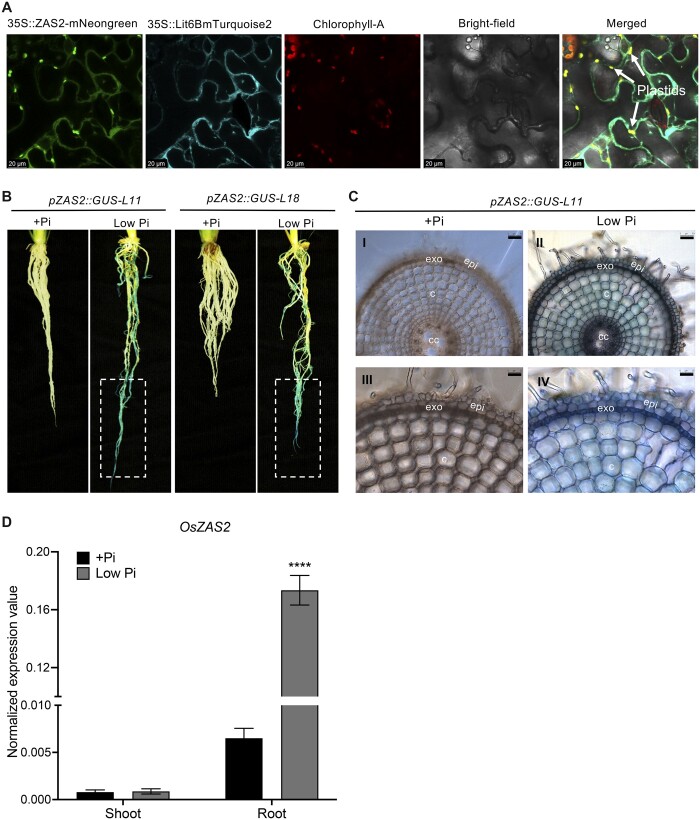
The ChloroP Server program (Emanuelsson et al., 1999) predicts the presence of a plastid transit peptide in the OsZAS2, indicating a plastid localization of this enzyme. To confirm this prediction, we transiently expressed OsZAS2 cDNA fused with the sequence encoding mNeonGreen fluorescence protein under the control of the 35S promoter (35S:OsZAS2:mNeonGreen) in Nicotiana benthamiana leaves epidermal cells, alone or together with the gene encoding the plasma membrane specific Turquoise2 marker protein (35S::Lit6BmTurquoise2) (Cutler et al., 2000). As shown in Figure 2A, the green fluorescent signal of the OsZAS2 fusion clearly overlapped with the red autofluorescence of chlorophyll A. However, it also showed co-localization with the 35S::Lit6B Turquoise marker of the plasma membrane. Overall, we observed a stronger green fluorescent signal of the OsZAS2 fusion in plastids than in plasma membranes, supporting a plastid localization of this enzyme.
Figure 2.
OsZAS2 subcellular localization and expression pattern. A, Subcellular localization of OsZAS2 transiently expressed in N. benthamiana epidermis leave tissue. B, GUS staining of roots of two independent pZAS2:GUS reporter lines (pZAS2::GUS-L11, pZAS2::GUS-L18) under normal (+Pi) and low Pi conditions. Dash rectangle emphasizes the root tip. C, Cross-section of pZAS2:GUS11 line primary root was examined with two different resolutions under microscope: in the first resolution (parts I and II, bars correspond to 50 μm) all types of tissues were observed while the second resolution of the same samples (parts III and IV, bars correspond to 25 μm) showed a close view of epidermal and cortex cells. Exo, exoderms; epi, epidermis; c, cortex; cc, central cylinder. D, Normalized expression value of OsZAS2 under normal (+Pi) and low Pi conditions in root and shoot tissue of 21-day-old rice plants. Values in (D) are means ± sd (n = 4). Student’s t test used for the statistical analysis (****P ≤ 0.0001).
OsZAS2 is expressed in roots and induced under low Pi
To determine the expression pattern of OsZAS2, we generated the GUS reporter lines pOsZAS2::GUS11 and pZAS2::GUS18 by fusing a 1.2-kb upstream OsZAS2 fragment to GUS and transforming the resulting pZAS2-MDC162 plasmid into rice. The staining of the two reporter lines showed that OsZAS2 expression significantly increased under low Pi conditions, while the GUS signal was not detectable at all under normal conditions (Figure 2B). Moreover, the GUS signal increased substantially toward the tip of the primary and crown roots. The RT-qPCR (Reverse transcription-quantitative polymerase chain reaction) analysis also showed that the OsZAS2 transcript level increased about 20-fold under low Pi compared to normal (+Pi) conditions in roots (Figure 2D). We further investigated OsZAS2 localization at the cellular level using cross-sectioning. Using a confocal microscope, we detected a strong GUS signal in the exodermis of primary roots, while the cortex, epidermis, and other root tissues showed only mild signals (Figure 2C).
CRISPR/Cas9-generated Oszas2 mutant lines show severe growth defects
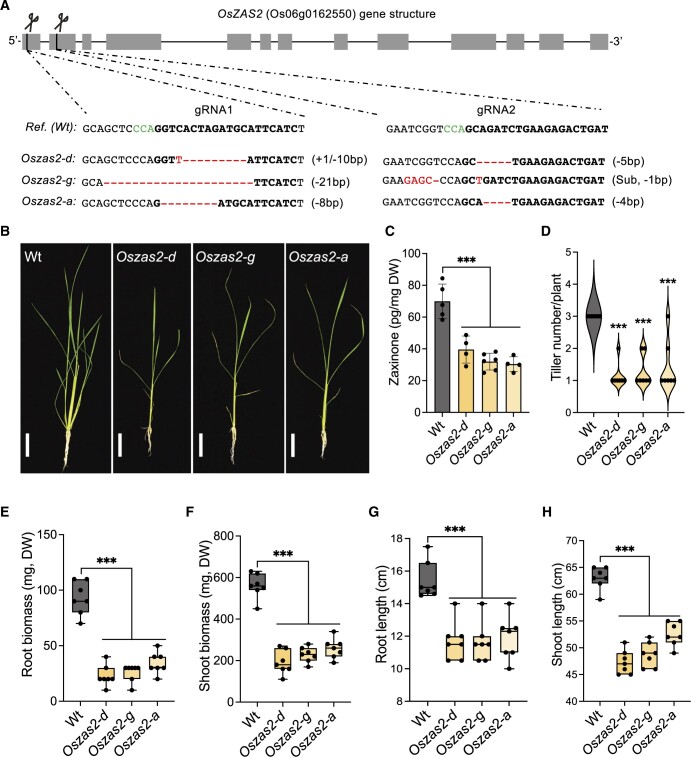
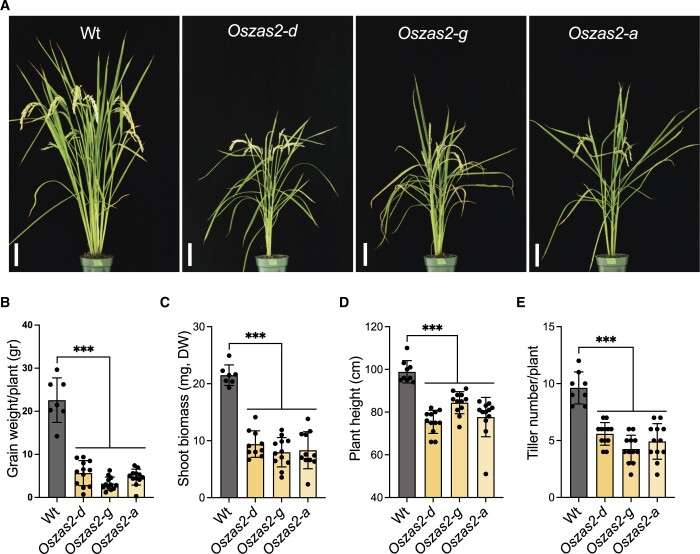
Next, we generated Oszas2 mutant knockout lines in the Dongjin (DJ) variety by employing CRISPR/Cas9. For this purpose, we used two guide RNAs, gRNA1 and gRNA2, which target coding sequence in exons 1 and 2, respectively (Figure 3A). We identified three independent Oszas2 mutants, zas2-d, zas2-g, and zas2-a, with mutations in the first and second exon (Figure 3A), which resulted in premature stop codons (Supplemental Figure S5). To validate the function of OsZAS2 as a zaxinone synthesizing enzyme in planta, we quantified zaxinone in roots of the three mutant lines grown hydroponically under normal and low Pi supply. Compared to WT, zaxinone content was reduced up to 45% in Oszas2 mutant lines under normal conditions (Figure 3C), but was unchanged under low Pi conditions (Supplemental Figure S4A). Next, we grew the mutant lines in soil and characterized them at the seedling and mature stage. At seedling stage, Oszas2 mutants displayed shorter roots and shoots and a striking reduction in root and shoot biomass (Figure 3, B and E–H). Moreover, they produced one tiller on average, while the WT developed three (Figure 3D). The low-tillering and reduced shoot biomass phenotypes remained pronounced after growing the mutants for 3 months in GH and caused a significant reduction in grain weight per plant, compared to the corresponding WT (Figure 4, A–E).
Figure 3.
Characterization of CRISPR/Cas9-mediated Oszas2 mutant lines at the seedling stages. A, Schematic representation of three individual mutations of OsZAS2 gene generated by CRISPR–Cas9. B, The seedling phenotype of WT (DJ) and three independent Oszas2 mutants. The scale bar in the pictures represents 7.5 cm. C, Quantification of zaxinone content in WT and Oszas2 mutants roots. D–H, Root biomass (D), shoot biomass (E), root length (F), shoot length (G), and tiller number (H) of the WT and Oszas2 mutants are shown in (B). Boxes in boxplots represent the median, first and third quartile. The minimum and maximum values are shown with the length of the whiskers. Dots represent the biological replicates. Values in (C–H) are means ± sd (n ≥ 4). Student’s t test used for the statistical analysis (***P ≤ 0.001).
Figure 4.
Characterization of Oszas2 mutant lines at the maturing stage. A, The picture of the 3-month-old WT and Oszas2 mutants grown in the greenhouse. The scale bar in the pictures represents 7.5 cm. B–E, Grain weight per plant (B), shoot biomass (C), plant height (D), and tiller number (E) of the WT and Oszas2 mutants represented in (A). Values in (B–E) are means ± sd (n ≥ 7). Student’s t test was applied for the statistical analysis (***P ≤ 0.001).
OsZAS2 is involved in AM colonization
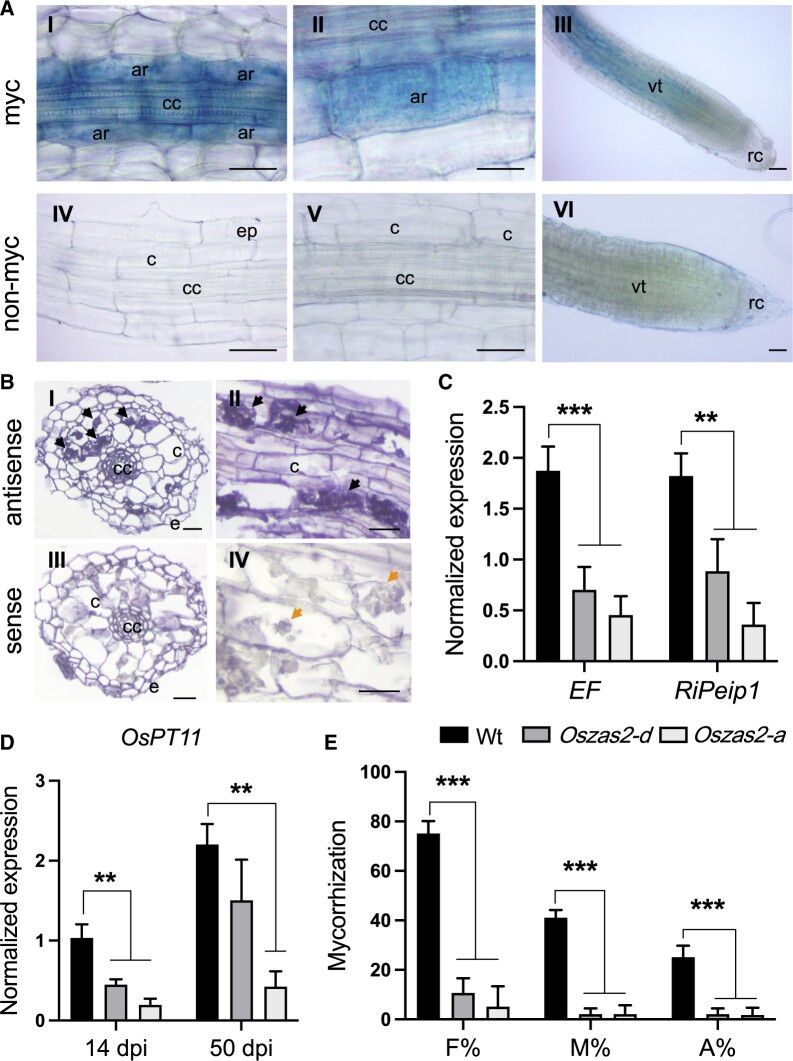
The expression analysis of OsZAS2 in whole roots at early and late stages of AM colonization revealed an upregulation at 21 days postinoculation (dpi) (Supplemental Figure S2A), when arbuscules are present, as witnessed by the abundance of the AM-inducible plant marker OsPT11 transcript (Paszkowski et al., 2002; Supplemental Figure S2B). To obtain deeper insights into the spatial expression of OsZAS2 during mycorrhization, we inoculated pZAS2::GUS reporter lines with AM fungi and monitored the GUS signal. Interestingly, we detected GUS activity only in arbusculated cells (Figure 5A; Supplemental Figure S2C), and did not observe any signal in any other root cells, including cortical cells and cells crossed by fungal hyphae. We further validated this observation by using in situ hybridization assays on mycorrhizal roots of WT plants. OsZAS2 mRNA exclusively accumulated in cells with fully developed arbuscules, in which we detected a strong chromogenic signal (Figure 5B). We did not observe any signal in noncolonized cells or upon using the OsZAS2 sense probe (Figure 5B).
Figure 5.
OsZAS2 is required for AM establishment. A, GUS staining of roots of pZAS2:GUS-L18 reporter line inoculated (I, II, and III) for 35 days with F. mosseae and noninoculated (IV, V, and VI). B, Localization of OsZAS2 mRNA in sections from differentiated regions of inoculated roots by cold in situ hybridization. Sections of mycorrhizal roots treated with OsZAS2 antisense probe are shown in parts I and II; arrows highlight the strong chromogenic signal, which mirrors the presence of the OsZAS2 transcript in arbuscule-containing cells. Sections of mycorrhizal roots treated with the OsZAS2 sense probe are shown in parts III and IV; arrows in part IV indicate arbusculated cells that are not labeled. C, Relative expression of fungal genes; RiEF and RiPeip1 in WT and Oszas2 mutants at 50 dpi. D, Relative expression of OsPT11 at 14 and 40 dpi in WT and Oszas2 mutants. E, Frequency of mycorrhizal colonization (F%), the intensity of colonization (M%), and a total number of arbuscules (A%) in WT and Oszas2 mutants at 50 dpi. cc, central cylinder; c, noncolonized cortical cells; e, epidermal cells; vt, vascular tissue; ar, arbuscule containing cells; rc, root cap. Bars (A and B) correspond to 50 µm. Values in (C–E) are means ± sd (n ≥ 3). Student’s t test was applied for the statistical analysis (**P ≤ 0.01; ***P ≤ 0.001).
To determine the impact of OsZAS2 on AM symbiosis, we inoculated the Oszas2 mutant lines with AM fungi and assessed the colonization level by morphological analysis and by monitoring the transcript abundance of OsPT11 at two time points (14 and 50 dpi) and of the fungal genes RiEF and RiPeip1 (Fiorilli et al., 2016) at 50 dpi. Both mutant lines showed a lower frequency of mycorrhizal colonization (F%), intensity of colonization (M%), and total number of arbuscules (A%), compared to WT plants (Figure 5E). Molecular analysis confirmed these results: the expression level of fungal and plant genes was significantly lower in the mutant lines (Figure 5, C and D).
SL biosynthesis increased in Oszas2 mutants
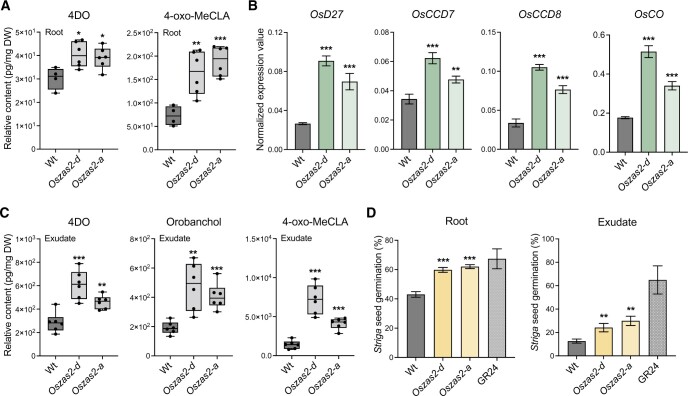
The low-tillering phenotype of Oszas2 mutants (Figures 3, D and 4, E) indicated that they may have higher SL content. To test this hypothesis, we profiled their SLs in both roots and root exudates under low Pi conditions. We also analyzed the expression level of SL biosynthetic genes in roots. In roots, the contents of a noncanonical SL, a tentative 4-oxo-methylcarlactonate (4-oxo-MeCLA) (Yoneyama et al., 2018; Ito et al., 2022), and of the canonical SL 4-deoxyorobanchol (4DO) were significantly increased in Oszas2 lines compared to WT (Figure 6A). As shown in Figure 6B, we also observed an increase in the transcript level of SL biosynthetic genes, that is, D27, CCD7, CCD8, and OsCO. To confirm the increase in SLs, we conducted a Striga seed germination assay with root extracts. Extracts of Oszas2 roots showed a significantly higher germination rate (around 60%), compared to those of WT (around 42%, Figure 6D). In root exudates, both canonical (4DO, orobanchol) and noncanonical (4-oxo-MeCLA) SLs were significantly increased in Oszas2 mutants, compared to WT (Figure 6C). Here again, we performed a Striga seed germination assay, in which Oszas2 mutant lines displayed about15% higher activity compared to WT (Figure 6D), which confirms the LC–MS quantification.
Figure 6.
SL biosynthesis increased in Oszas2 mutants. A, Relative quantification of 4DO and 4-oxo-MeCLA in root tissue of Oszas2 mutants. B, Normalized expression value of SL biosynthetic genes in Oszas2 mutants. C, Relative quantification of canonical (4DO, Orobanchol) and noncanonical (4-oxo-MeCLA) SL in root exudate of Oszas2 mutants. D, Striga seed germination assay conducted with root exudate and tissue of Oszas2 mutants. For both root exudate and tissue bioassay, 1 μM of GR24 was used as control, which showed about 65% and 67% of Striga seed germination, respectively. Boxes in boxplots in (A) and (C) represent the median, first and third quartile. The minimum and maximum values are showed with the length of the whiskers. Dots represent the biological replicates. Values in (A–D) are means ± sd (n ≥ 4) and student’s t test was applied for the statistical analysis (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001).
Exogenous zaxinone application repressed SL biosynthesis in Oszas2 mutant
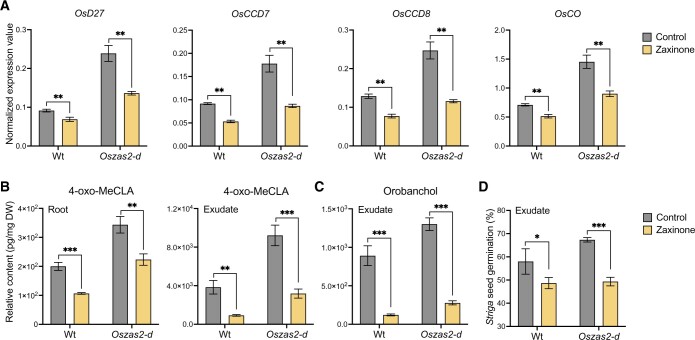
Next, we treated 3 weeks old, hydroponically grown Oszas2 seedlings (grown 1 week in Hoagland agar and 2 weeks in low Pi) with 5 μM zaxinone for 6 h. As shown in Figure 7A, zaxinone treatment repressed transcript levels of the SL biosynthetic genes D27, CCD7, CCD8, and OsCO in Oszas2-d back to the WT level. Furthermore and as shown before (Wang et al., 2019, 2020), exogenous zaxinone application decreased the transcript level of SL biosynthetic genes in WT as well (Figure 7A). Zaxinone application also decreased the content of the noncanonical SL 4-oxo-MeCLA in roots and root exudates of Oszas2-d and WT (Figure 7B). In addition, it caused a reduction in the level of the canonical SL orobanchol in root exudates of both Oszas2-d and WT (Figure 7C). Surprisingly, 4DO content was slightly increased upon zaxinone treatment in root tissues of both Oszas2-d and WT while it was not affected in root exudates (Supplemental Figure S3). Moreover, root exudates of both Oszas2 and WT plants showed, upon zaxinone treatment, decreased Striga seed germination (Figure 7D).
Figure 7.
Zaxinone treatment reduced SL biosynthesis in Oszas2 mutant. A, SL biosynthetic genes; OsD27, OsCCD7, OsCCD8, and OsCO expression in WT and Oszas2 mutant upon zaxinone (5 μM) treatment. B, Relative content of 4-oxo-MeCLA after zaxinone (5 μM) treatment in root tissue and exudate of WT and Oszas2 mutant. C, Relative content of Orobanchol after zaxinone (5 μM) treatment in root exudate of WT and Oszas2 mutant. D, Striga seed germination assay with exudate of WT and Oszas2 mutant upon zaxinone (5 μM) treatment. About 1 μM of GR24 was used as control, which showed about 63% of Striga seed germination. Values in (A–D) are means ± sd (n ≥ 4). Student’s t test was applied for the statistical analysis (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001).
Zaxinone treatment partially rescued the growth defects of Oszas2 mutant
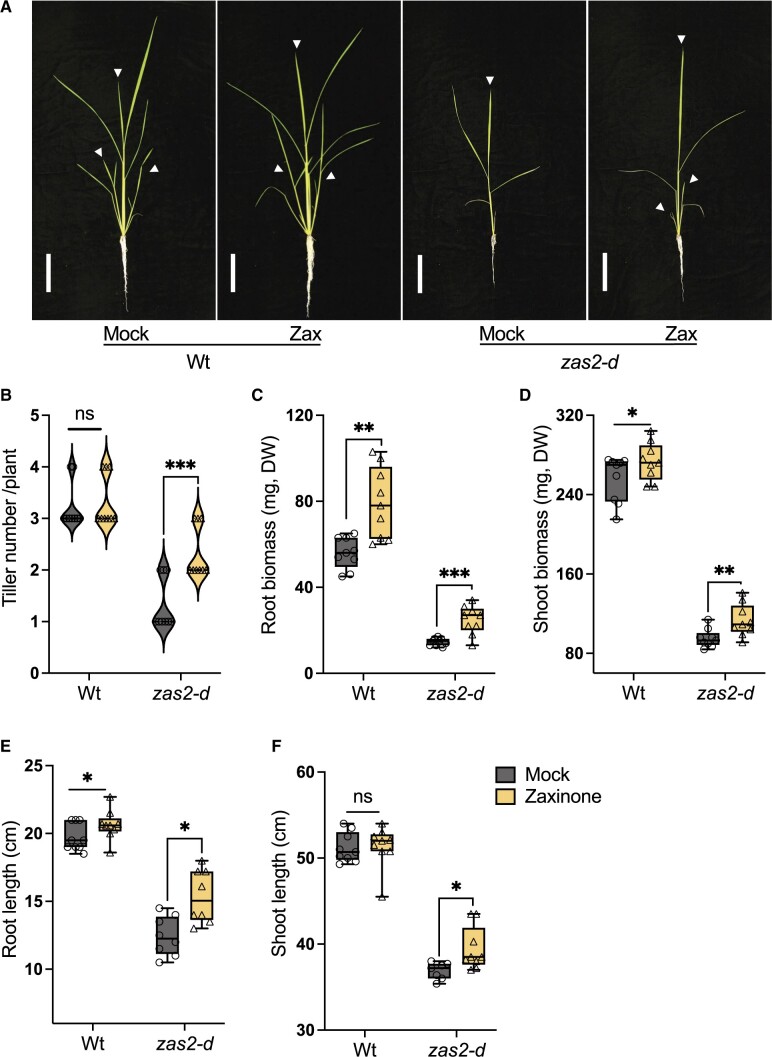
Next, we supplied Oszas2 seedlings, grown in soil, with exogenous zaxinone at a concentration of 10 μM. After 2 weeks, we observed an increase in the tiller number of Oszas2-d mutant, but not of that of the WT (Figure 8, A and B). Moreover, zaxinone treatment significantly increased root and shoot biomass and shoot length of Oszas2-d (Figure 8, C–F). We also observed an increase in root length and root and shoot biomass of treated WT plants upon zaxinone treatment, while their shoot length remained unaffected (Figure 8, C, D, and F).
Figure 8.
Exogenous zaxinone application rescued the growth defects of Oszas2 mutant. A, The images of the WT (DJ) and Oszas2-d mutant grown for 2 weeks in soil supplemented with 10 μM of zaxinone and tap water (0.01% [v/v] acetone) as mock. The white bar represents 10 cm of scale. The white arrows represent main tillers. B–F, Tiller number (B), root biomass (C), shoot biomass (D), root length (E), and shoot length (F) of the WT and Oszas2 mutants are shown in (A). Boxes in boxplots represent the median, first and third quartile. The minimum and maximum values are showed with the length of the whiskers. Dots represent the biological replicates. Values in (B–F) are means ± sd (n ≥ 7). Student’s t test used for the statistical analysis (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ns, not significant).
Discussion
The identification of a zaxinone synthase, the rice ZAS, revealed the presence of a widely distributed plant CCD subfamily (Wang et al., 2019). Functional studies and characterization of a corresponding T-DNA insertion mutant demonstrated the importance of ZAS for plant growth, interaction with AM fungi, and SL homeostasis. Furthermore, exogenous treatment with zaxinone revealed the growth-promoting effect of this apocarotenoid and its impact on hormone homeostasis and sugar metabolism, indicating that it may be a candidate for a novel plant hormone (Wang et al., 2019, 2021a, 2021b). Rice contains three OsZAS homologs (Wang et al., 2019), called here ZAS1b, ZAS1c, and ZAS2, with unknown functions. The severe phenotypes of zas mutant indicate the importance of this gene and suggest that its activity cannot be compensated by that of its homolog(s), and that the latter may exert different function(s). Therefore, we were interested in investigating the biological function of these enzymes.
Phylogenetic analysis placed OsZAS2 in a clade different from that of ZAS, ZAS1b, and 1c, indicating a different biological role and maybe enzymatic activity (Figure 1A). Therefore, we focused in this study on OsZAS2. However, the enzymatic studies performed here demonstrate that OsZAS2 catalyzes the same reaction as ZAS, that is, it converts apo-10′-zeaxanthinal into zaxinone (Figure 1B). Indeed, the enzyme did not convert other substrates tested, for example, β-carotene, zeaxanthin, or different apocarotenoids, or produced other products, pointing to zaxinone formation as its enzymatic function. The same activity was reported for OsZAS. However, OsZAS cleaved, in addition to apo-10′-zeaxanthinal, apo-12′- and apo-8′-zeaxanthinal, but with lower activity (Wang et al., 2019). This difference might be caused by a wider substrate specificity. However, it is also possible that the heterologously expressed OsZAS is generally more active than ZAS2.
To explore the biological function of OsZAS2 in planta, we generated Oszas2 knockout lines using the CRISPR/Cas9 technology. Disrupting OsZAS2 led to around 40% decrease in roots zaxinone content. This decrease supports the in vitro enzymatic activity of OsZAS2 and suggests that OsZAS2 is an enzyme, besides OsZAS, responsible for zaxinone biosynthesis in rice. Oszas2 mutants still contained a substantial amount of zaxinone in roots. This could be due to the activity of OsZAS, which might compensate for the zaxinone production in rice roots. Nevertheless, zaxinone is common at higher levels in green tissues than in roots and is also present in plant species, such as Arabidopsis, which lacks ZAS genes, indicating that it can be synthesized via alternative route(s) independent of ZAS enzymes (Mi et al., 2019; Wang et al., 2019; Ablazov et al., 2020). However, the phenotypes observed with Oszas and Oszas2 mutants suggest the importance of these enzymes and indicate the zaxinone content in roots is crucial for normal growth and development. We are currently generating Oszas/Oszas2 double mutants, which could give us a hint about the involvement of other route(s) in zaxinone biosynthesis.
Zaxinone is a negative regulator of SL biosynthesis in rice (Wang et al., 2019). Indeed, the Oszas mutant contained and released higher amounts of SLs (Wang et al., 2019) under Pi starvation, and this increase could be suppressed by exogenous zaxinone application. Based on its zaxinone-forming activity and the low-tillering phenotype of the Oszas2 mutants, we assumed that the loss of OsZAS2 may also cause an increase in SL content. Therefore, we quantified the SL and zaxinone content in Oszas2 mutants under low Pi conditions. Indeed, transcript levels of the SL biosynthetic genes OsD27, OsCCD7, OsCCD8, and OsCO transcript levels were upregulated in Oszas2 compared to WT (Figure 6A). In parallel, both canonical and noncanonical SLs were significantly increased in roots and root exudates of Oszas2 mutants (Figure 6, A and C), as confirmed by LC–MS analysis and by using the Striga seed germination bioassay. Interestingly, zaxinone content was not changed in Oszas2 mutants under low Pi conditions (Supplemental Figure S4A), compared to the WT, albeit an increase in SL biosynthesis that is assumed to be caused by a decrease in zaxinone content. Since OsZAS is still functional in the Oszas2 mutant, we hypothesized that it might compensate for the loss of Oszas2 activity. In fact, OsZAS expression was upregulated, but not that of its homologs OsZAS1b and OsZAS1c, in Oszas2 mutant under low Pi conditions (Supplemental Figure S4B). It might be speculated that changes in zaxinone content in certain root cells are crucial for regulating SL biosynthesis and that the increase in OsZAS activity may lead to a generally higher zaxinone content but cannot replace OsZAS2 in cells expressing this enzyme. Clarifying this point requires precise localization of both enzymes under low Pi conditions.
Oszas2 mutants showed severe reduction in root and shoot biomass (Figure 3, E and F) and developed fewer tillers compared to WT (Figure 3D). The retarded growth of Oszas2 mutants demonstrates that OsZAS2 is necessary for normal rice growth and development. In general, the phenotypes of the Oszas2 mutants, reduced zaxinone content, retarded growth, and reduced tiller number, are similar to that of the Oszas mutant under normal conditions (Wang et al., 2019). Hence, it can be concluded that rice requires both ZAS and ZAS2 genes to keep the root zaxinone concentration at a certain level, as well as for normal rice growth and development under normal conditions.
We checked the expression pattern of OsZAS2 at tissue and cellular level using RT-qPCR and promoter–GUS–reporter lines. Similar to OsZAS (Wang et al., 2019), OsZAS2 is expressed in roots and induced upon Pi starvation. A robust upregulation of both OZAS and OsZAS2 in rice roots in response to Pi starvation indicates their involvement in the plant’s response to Pi deficiency. Interestingly, analysis of the GUS reporter lines (pOsZAS2::GUS11 and pZAS2::GUS18) demonstrated that the OsZAS2 expression level was higher in root tips (Figure 2A). The cross-sectioning of the primary roots of the pOsZAS2::GUS11 further showed that OsZAS2 is highly expressed in exoderms. Here again, it would be very interesting to monitor OsZAS expression patterns at the cellular level to get insights into the function of OsZAS and OsZAS2 and understand why both of them are important for proper rice growth.
In a previous work, we demonstrated that zas mutant showed a lower AM colonization level, compared to WT plants (Wang et al., 2019). Moreover, we revealed that the ZAS gene family is absent in genomes of nonmycorrhizal species, such as Arabidopsis (Fiorilli et al., 2019; Wang et al., 2019), suggesting a strong link between ZAS and AM symbiosis. To investigate the role of OsZAS2 in the different steps of AM colonization, we monitored its expression level during a time course experiment in mycorrhizal and nonmycorrhizal roots. Contrarily to OsZAS which was upregulated during both early and later stages, OsZAS2 was only upregulated at the maximum of arbuscules formation (21 dpi), suggesting an involvement in arbuscules development/formation. This assumption is supported by in situ hybridization and using the pZAS2::GUS reporter lines: indeed both assays revealed that OsZAS2 expression is localized in arbusculated cells. To further clarify the OsZAS2 involvement during the AM symbiosis, we assessed the Oszas2 mutant lines (Oszas2-d and Oszas2-a) colonization level at the morphological level and using molecular analyses. Although Oszas2 mutants displayed in nonmycorrhizal condition a higher level of SLs in roots and root exudates, they showed a severe reduction of AM colonization level; however, no defects in arbuscules morphology were detected. A similar phenotype was also observed in the Oszas mutant (Wang et al., 2019). We recently demonstrated that the lower AM colonization rate of the Oszas mutant is due to SL deficiency at the early stage of the AM interaction (Votta et al., 2022): during this phase OsZAS activity is required to induce SL production possibly through the Dwarf14-Like (D14L) signaling pathway which was shown to regulate AM colonization in rice (Gutjahr et al., 2015). We can hypothesize that also OsZAS2 acts as a component of a regulatory network that involves SL and D14L pathways. Although further experiments are needed to prove this hypothesis, all the above data demonstrate that OsZAS2, in analogy to OsZAS, is required to reach a correct level of AM colonization.
We assume that the growth retardation of the Oszas2 mutants is more likely due to decreased root zaxinone levels under normal conditions. Therefore, we applied zaxinone exogenously in to the Oszas2-d seedlings grown in soil. The exogenous application of zaxinone partially rescued the low-tillering, reduced root and shoot biomass, and shorter root and shoot length phenotypes of Oszas2-d mutant (Figure 8, C–F). This result indicates that a certain level of zaxinone is required to keep normal SL homeostasis and, hence, maintain normal tillering degree. Moreover, the effects of zaxinone treatment highlight again the importance of appropriate zaxinone concentrations of this apocarotenoid for regular growth and development of rice and support the idea of its function as a growth-promoting metabolite.
In conclusion, we revealed the function of OsZAS2, a member of the CCD gene family, which is crucial for normal rice growth and development. Besides OsZAS, OsZAS2 contributes to zaxinone production in rice roots and is a further determinant of SL content. Moreover, it is involved in regulating the levels of mycorrhizal colonization. Thus, manipulation of OsZAS2 expression level could be a tool to modulate rice architecture and improve AM symbiosis.
Materials and methods
Plant material and phenotyping
Rice seedlings (O. sativa L. cv DJ) were grown in a Biochamber under the following conditions: a 12-h photoperiod, 500 µmol photons m−2 s−1 and day/night temperature of 27/25°C. Briefly, rice seeds were surface sterilized in a 50% (v/v) household bleach for 15 min and rinsed 5 times with distilled water. Then, sterilized seeds were germinated in the dark for 2 days in the magenta boxes containing 50 mL of 0.4% (w/v) agarose half-strength Hoagland medium with pH 5.8 at 30°C. The pregerminated seeds were transferred to the biochamber and kept for 5 days.
For metabolite quantification, gene expression analysis, and Striga seed germination assay, 1-week-old rice seedlings were transferred into 50-mL black falcon tubes filled with half-strength modified Hoagland nutrient solution with adjusted pH to 5.8. The nutrient solution consisted of 5.6-mM NH4NO3, 0.8-mM MgSO4.7H2O, 0.8-mM K2SO4, 0.18-mM FeSO4.7H2O, 0.18-mM Na2EDTA.2H2O, 1.6-mM CaCl2.2H2O, 0.8-mM KNO3, 0.023-mM H3BO3, 0.0045-mM MnCl2.4H2O, 0.0003-mM CuSO4.5H2O, 0.0015-mM ZnCl2, 0.0001-mM Na2MoO4.2H2O, and 0.4-mM K2HPO4.2H2O. For normal conditions (+Pi), the 1-week-old seedlings were grown in the Hoagland nutrient solutions (+Pi) for another 2 weeks. For phosphate starvation, the seedlings were grown for 2 weeks in lower phosphate (4 µM, K2HPO4⋅2H2O) nutrient solution. The nutrient solution was replaced every 3 days. For zaxinone treatment, 3-week-old seedlings were treated with 5 µM of zaxinone for 6 h: tissues were collected and immediately frozen into liquid N2.
For phenotyping, 1-week-old Oszas2 seedlings were transferred into pots filled with soil and grown in growth chamber under the above-mentioned conditions. Tap water was supplied when needed. After 18 days, roots were cleaned off from the soil. Then, the seedlings were photographed with a digital camera, and root and shoot lengths were measured. To analyze the dry weight (DW) of roots and shoots biomass, samples were kept for 3 days in a 65°C oven. For phenotyping of Oszas2 mutants at the mature stage, 1-week-old seedlings were transferred into greenhouse and grown until the mature stage with a day/night temperature of 28°C/25°C. One-time tap water and a one-time half-strength nutrient solution were supplied when necessary. After 3 months, yield-related traits were recorded. This experiment was repeated twice.
For rescue experiments, 7-days-old seedlings were transferred into 1-L pots filled with soil and grown in Biochamber. Initially, for zaxinone treatment 200 mL of 10-µM zaxione solution (pH 5.8) was added per pot. The same volume of tap water containing 0.01% (v/v) acetone was used as control. Every 3 days, 50 mL of 10 µM of zaxione solution and an equivalent amount of water were added to the treatment and control groups, respectively. After 2 weeks of treatment, seedlings were phenotyped as above.
Generation of transgenic lines
The CRISPR/Cas9 genome-editing technique was used to knock out OsZAS2 (Os06g0162550) in O. sativa ssp. japonica variety DJ. The gRNA sequences were designed using the CRISPR-PLANT webserver (www.genome.arizona.edu/crispr/). Two different gRNA sites targeted OsZAS2 in exon regions; at exon 1 which encodes (5′-GGTCACTAGATGCATTCATC-3′) and exon 2 which encodes (5′-GCAGATCTGAAGAGACTGAT-3′). The gRNA spacers were fused to a tRNA sequence as described by Xie et al. (2015) (Supplemental Figure S6) and synthesized from GENEWIZ (South Plainfield, NJ, USA). Then, the corresponding fragment was cloned into pRGEB32 (Kanamycin) using 5′-BsaI and 3′-BsaI sites. The pRGEB32-OsZAS2 construct was further introduced into Agrobacterium tumefaciens strain EHA105 competent cells via electroporation. To construct the pOsZAS2::GUS reporter plasmid, the 1.2-kb promoter region of OsZAS2 was amplified with the Phusion enzyme from the genomic DNA of rice using promoter-specific primers (Supplemental Table S1). The PCR product was ligated into pJet1.2 intermediate plasmid following the instruction of CloneJET PCR Cloning Kit (K1232; Thermo Scientific). The pOsZAS2 sequence was amplified from the pJet1.2 plasmid with specific primers (Supplemental Table S1) and cloned into the pENTR/D-TOPO plasmid. Then, OsZAS2:pENTR/D-TOPO were inserted into pMDC162 (Curtis and Grossniklaus, 2003) by Gateway cloning.
Rice transformation was conducted according to Hiei and Komari (2008). The mutations of transformed lines were analyzed by PCR using a Thermo Scientific Phire Plant Direct PCR Master Mix Kit. Gene-specific primers (Supplemental Table S1) were used for PCR amplification to detect the mutation sites. Then, PCR products were cleaned up using ExoSAP-IT PCR Product Cleanup Reagent and submitted to the Sanger sequencing core lab team, at KAUST. The Sanger sequencing data (abi file) were analyzed following the instruction of DSDecode (Degenerate Sequence Decode; http://skl.scau.edu.cn/dsdecode/). Three independent homozygote mutant lines were identified and grown until T3 generation.
Metabolite quantification
For quantification, plant material was lyophilized with freeze-dryer and ground with Geno Grinder 2010. D3-zaxinone (customized synthesis; Buchem B.V., Apeldoorn, the Netherlands) was used as an internal standard. Zaxinone was extracted according to Wang et al. (2019). SLs were extracted from the root tissues as described by Mi et al. (2018). SL extraction from the root exudates was performed according to Wang et al. (2022). In the final step, the dried extract was dissolved in 110 μL of acetonitrile: water (90:10, v:v) and filtered through a 0.22-μm filter for LC–MS/MS analysis. The samples were run on UHPLC- Triple-Stage Quadrupole Mass Spectrometer (TSQ-Altis) with parameters as described in Wang et al. (2022).
Striga seed germination bioassays
Striga seeds were preconditioned as described by Jamil et al. (2019). After 10 days, the preconditioned Striga seeds were treated with rice root extracts and exudates, using 50 μL per disc (n = 3–6). Root extracts were prepared following the above described SL extraction method. For treatment, 5 μL of root extracts were diluted in 400 μL of MilliQ water before application. Root exudates were collected following the above described protocol. For treatment, 200 μL of SL enriched solution was diluted in 1,800 μL of MilliQ water before application. The discs were also treated with water and GR24 (1 μM) as negative and positive control, respectively. The plates were sealed with parafilm and incubated at 30°C for 24 h. The discs were scanned in a microscope and germinated and nongerminated seeds were counted from these scanned images by using the software SeedQuant (Braguy et al., 2021) to calculate the percentage of germination.
RT-qPCR analysis
Rice tissues were ground and homogenized in liquid nitrogen, and total RNA was isolated using a Direct-zol RNA Miniprep Plus Kit following the manufacturer’s instructions (ZYMO RESEARCH, Irvine, CA, USA). Briefly, a 1-μg RNA sample was reverse transcribed using an iScript cDNA Synthesis Kit (BIO-RAD Laboratories Inc., 2000 Alfred Nobel Drive, Hercules, CA, USA). The RT-qPCR was performed using SYBR Green Master Mix (Applied Biosystems, Waltham, MA, USA; http://www.lifetechnologies.com) in a CFX384 Touch Real-Time PCR Detection System (BIO-RAD Laboratories, Inc., 2000 Alfred Nobel Drive, Hercules, CA, USA). Primer-BLAST webserver (Ye et al., 2012) was used to design the gene-specific RT-qPCR primers (Supplemental Table S1) and Ubiquitin was used as an internal control. The relative gene expression level was calculated according to 2ΔCT method.
In vitro assays
The OsZAS2 cDNA was amplified using primers listed in Supplemental Table S1 and cloned into the pET-His6 MBP N10 TEV LIC cloning vector (2C-T vector; http://www.addgene.org/29706/) with MBP tags at the N-terminus. OsZAS2-MBP construct transformed into the BL21 Rosetta E. coli cells. A single colony of the transformed E. coli was cultured overnight and 0.5 mL of this culture was inoculated into 50-mL liquid media and grown at 37°C to OD (optical density) 0.6 at 600 nm. Then, bacteria were induced by IPTG (150 μM final concentration) and kept shaking at 28°C for 4 h. Cells were harvested by centrifugation and resuspended in lysis buffer (sodium phosphate buffer pH 8 containing 1% (v/v) Triton X-100 and 10 mM of dithiothreitol, lysozyme (1 mg mL−1)) and incubated on ice for 30 min. Next, the crude lysate was sonicated and centrifuged at 12,000 rpm and 4°C for 10 min, and the supernatant containing the protein was collected for in vitro incubation with the substrate. Synthetic substrates were purchased from Buchem B. V. (Apeldoorn, the Netherlands). Substrates were prepared according to Wang et al. (2019). Dried substrates were resuspended in 0.4% (v/v) Triton X-100 dissolved in ethanol. The mixture was then dried using a vacuum centrifuge to produce an apocarotenoid-containing gel. The gel was resuspended in incubation buffer (2-mM tris 2-carboxyethylphosphine, 0.4-mM FeSO4, and 2-mg/mL catalase in 200-mM Hepes/NaOH, pH 8). OsZAS2 crude cell lysate, that is, 50 μL of the soluble fraction of overexpressing cells, was added to the assay. The assay was incubated for 4 h under shaking at 140 rpm at 28°C in dark. The reaction was stopped by adding two volumes of acetone and the lipophilic compounds were separated by partition extraction with petroleum ether: diethyl ether 1: 4 (v/v), dried, and resuspended in methanol for HPLC (High Performance Liquid Chromatography) analysis. For this purpose, we used an Ultimate 3000 UHPLC system and a YMC Carotenoid C30 column (150 × 3.0 mm, 5 μm) following the parameters described in Wang et al. (2019). This experiment was repeated at least 3 times.
Subcellular localization
The 35S::OsZAS2:mNeongreen was constructed by amplifying the coding sequence of OsZAS2 using specific primers (Supplemental Table S1). The PCR product was sub-cloned into the pDONR221 entry vector by BP recombination reaction. Then, the OsZAS2::pDONR221 fragment was fused into pB7FWG2,0 by Gateway cloning. The construct was introduced into A. tumefaciens strain GV3101 by electroporation. Nicotiana benthamiana infiltration was performed as described by Aljedaani et al. (2021). 35S::OsZAS2-mNeongreen, membrane protein marker (35S::Lit6BmTurquoise2) and p19 helper plasmid were co-infiltrated into the abaxial leave side of N. benthamiana. The fluorescence expression was checked 3-day postinfiltration by the confocal microscope. Leaf tissues of the infiltrated N. benthamiana was mounted with water on microscope slides and visualized by using a high-resolution laser confocal microscope (STELLARIS 8 FALCON, Leica). Images were acquired using an HC PL APO CS2 63x/1.20 WATER immersion objective, with 512 × 512 pixel resolution with a line average of 8. The mNeongreeen was excited with a 488 laser, and emission was collected at 500–558 nm. The laser was with 40% intensity, the gain was 180 gain and the pinhole was 1 air unit (AU) pinhole. The mTurquoise2 was excited with a 440 laser and the emission was collected at 445–479. The laser power was 12% intensity. The gain was 74 and the pinhole was 1 AU. This experiment repeated at least 3 times.
Mycorrhization
Rice seeds of WT cv. DJ, Oszas2 (Oszas2-d and Oszas2-a) were germinated in pots containing a sand and incubated for 10 days in a growth chamber under a 14-h light (23°C)/10-h dark (21°C). All genotypes were colonized with approximately 1,000 sterile spores of Rhizophagus irregularis DAOM 197198 (Agronutrition, Labège, France). Mycorrhizal plants were grown in sand and watered with a modified Long-Ashton solution containing 3.2-μM Na2HPO4·12H2O and kept in a growth chamber as described before. WT and Oszas2 mutant plants were sampled at 14 and 50 dpi. To analyze OsZAS2 gene expression profiles WT plants were inoculated with a fungal inoculum of Funneliformis mosseae (BEG 12, MycAgroLab, France) mixed (25% [w/v]) with sterile quartz. Nonmycorrhizal and mycorrhizal plants were sampled at 7 and 21 dpi. For all experiments, mycorrhizal roots were stained with cotton blue, and the level of mycorrhizal colonization was assessed according to Trouvelot et al. (1986) using MYCOCALC (http://www2.dijon.inra.fr/mychintec/Mycocalc-prg/download.html). For molecular analyses, roots were immediately frozen in liquid nitrogen and stored at −80°C. This experiment repeated at least 2 times.
In situ hybridization and GUS staining
For sample preparation and embedding, rice roots were fixed in 4% (v/v) paraformaldehyde in phosphate-buffered saline (PBS; 130-mM NaCl; 7-mM Na2HPO4; 3-mM NaH2PO4, pH 7.4) overnight at 4°C. To facilitate the fixation, samples were placed under vacuum for the first 15–30 min. Then the tissues were dehydrated in successive steps, each of 30–60 min duration, in 30%, 50%, 70%, 80%, 95%, and 100% (v/v) ethanol and 100% (v/v) Neo-Clear (Xylene substitute; Sigma-Aldrich, St Louis, MO, USA). Finally, samples were embedded in paraffin wax (Paraplast plus; Sigma) at 60°C. Sections of 7–8 μm were then transferred to slides treated with 100 mg mL−1 poly-L-Lys (Sigma) and dried on a warm plate at 40°C overnight. In parallel, DIG-labeled RNA probes were synthesized starting with 1 μg of PCR-obtained template (Langdale, 1993). DIG-labeled riboprobes (antisense and sense probes) were produced with DIG-UTP by in vitro transcription using the Sp6 and T7 promoters, according to the manufacturer’s protocol (RNA-labeling kit; Roche, Basel, Switzerland). The sections were treated as follows: they were de-paraffinized in Neo-Clear, rehydrated through an ethanol series, treated with 0.2-M HCl for 20 min, washed in sterile water for 5 min, incubated in 2× SSC for 10 min, washed in sterile water for 5 min, incubated with proteinase K (1 mg mL−1 in 100-mM Tris–HCl, pH 8.0, 50-mM EDTA; Roche) at 37°C for 30 min, washed briefly in PBS, and then treated with 0.2% (w/v) Glycine in PBS for 5 min. After two rinses in PBS, slides were incubated in 4% paraformaldehyde in PBS for 20 min, washed in PBS (2, 3, and 5 min), and then dehydrated in an ethanol series from 30% to 100% (v/v). Hybridizations were carried out overnight at 55°C with denatured DIG-labeled RNA probes in 50% (v/v) formamide, 20×SSC, 20% (w/v) SDS (Sodium dodecyl sulfate), 50-mg mL−1 tRNA, 40- μg mL−1 Salmon Sperm DNA. Slides were then washed twice in 1× SSC, 0.1% (w/v) SDS at room temperature, and rinsed with 0.2× SSC, 0.1% (w/v) SDS at 55°C (2, 3, and 10 min). After rinsing with 2× SSC for 5 min at room temperature, the nonspecifically bound DIG-labeled probe was removed by incubating in 10-mg mL−1 RNase A in 2× SSC at 37°C for 30 min. Slides were then rinsed twice in 2% (v/v) SSC before proceeding to the next stage. The hybridized probe was detected using an alkaline phosphatase antibody conjugate (Roche). After rinsing in TBS (100-mM Tris–HCl, pH 7.5, 400-mM NaCl) for 5 min, slides were treated with 0.5% (w/v) blocking reagent in TBS (Tris Buffered Saline) for 1 h, incubated for 2 h with the anti-DIG alkaline phosphatase conjugate diluted 1:500 in 0.5% (v/v) Bovine Serum Albumin Fraction V in TBS, and then washed in TBS (3 × 5 min). Color development was carried out according to Torres et al. (1995). The color reaction was stopped by washing in distilled water, and the sections were then dehydrated through an ethanol series, deparaffinized in Neo-Clear, and mounted in Neomount (Merck, Kenilworth, NJ, USA) (Balestrini et al., 1997).
The GUS assay was performed on roots of pOsZAS2:GUS-L11 and L18 colonized by F. mosseae and sampled at 35 dpi. Rice mycorrhizal root segments were cut and placed in single wells of a Multiwell plate and covered with freshly prepared GUS buffer (0.1-M sodium phosphate buffer pH 7.0, 5-mM K4Fe(CN)6, 5-mM K3Fe(CN)6, 0.3% (v/v) Triton X, 0.3% (w/v) x-Glc). To improve buffer penetration into the root segments, they were placed under vacuum for 10 min. Finally, samples were incubated at 37°C for 16 h in the dark, de-stained with 70% (v/v) ethanol, and observed under an optical microscope (Nikon Eclipse E300).
Accession numbers
The cDNA and promoter sequence of rice ZAS2 is available in NCBI under the accession number LOC107275952. Accessions of SL biosynthetic genes; LOC_Os11g37650 (OsD27), LOC_Os04g46470 (OsCCD7), LOC_Os01g54270 (OsCCD8), and LOC_Os01g50520 (OsCO).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Structures of carotenoids and apocarotenoids used as substrates in ZAS2 in vitro and vivo assays.
Supplemental Figure S2. OsZAS2 expression during AM establishment.
Supplemental Figure S3. Relative content of 4DO after zaxinone (5 μM) treatment in root tissue and exudate of Wt and Oszas2 mutant.
Supplemental Figure S4. Zaxinone quantification and OsZAS genes expression analysis in Oszas2 mutants under low Pi conditions.
Supplemental Figure S5. Truncated amino acid sequences of Oszas2 mutant lines after CRISPR-Cas9 genome editing.
Supplemental Figure S6. gRNA targets of OsZAS2 were fused to tRNA sequences (Xie et al., 2015).
Supplemental File S1. Clustal alignment of ZAS members.
Supplemental Table S1. Primer sequences used in this study.
Supplemental Table S2. Distribution of ZAS members across monocot and dicot plants.
Supplemental Table S3. Protein accessions of ZAS members in different organisms.
Supplementary Material
Acknowledgments
We are thankful to Dr. Imran Haider for helping with rice transformation, and Mr. Saad Hammad and Ms. Vijayalakshmi Ponnakanti for their help with greenhouse work.
Funding
This work was supported by baseline funding and Competitive Research Grants (CRG 2017 and CRG 2020) given to Salim Al-Babili from King Abdullah University of Science and Technology.
Conflict of interest statement. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Contributor Information
Abdugaffor Ablazov, Biological and Environmental Sciences and Engineering Division, Center for Desert Agriculture (CDA), King Abdullah University of Science and Technology (KAUST), The BioActives Lab, Thuwal, 23955-15 6900, Saudi Arabia; The Plant Science Program, Biological and Environmental Science and Engineering Division, King Abdullah University of Science and Technology (KAUST), Thuwal, Saudi Arabia.
Cristina Votta, Department of Life Sciences and Systems Biology, University of Torino, Torino 10125, Italy.
Valentina Fiorilli, Department of Life Sciences and Systems Biology, University of Torino, Torino 10125, Italy.
Jian You Wang, Biological and Environmental Sciences and Engineering Division, Center for Desert Agriculture (CDA), King Abdullah University of Science and Technology (KAUST), The BioActives Lab, Thuwal, 23955-15 6900, Saudi Arabia.
Fatimah Aljedaani, The Plant Science Program, Biological and Environmental Science and Engineering Division, King Abdullah University of Science and Technology (KAUST), Thuwal, Saudi Arabia; Plant Cell and Developmental Biology, Biological and Environmental Sciences and Engineering (BESE), King Abdullah University of Science and Technology (KAUST), Thuwal 23955-6900, Saudi Arabia.
Muhammad Jamil, Biological and Environmental Sciences and Engineering Division, Center for Desert Agriculture (CDA), King Abdullah University of Science and Technology (KAUST), The BioActives Lab, Thuwal, 23955-15 6900, Saudi Arabia.
Aparna Balakrishna, Biological and Environmental Sciences and Engineering Division, Center for Desert Agriculture (CDA), King Abdullah University of Science and Technology (KAUST), The BioActives Lab, Thuwal, 23955-15 6900, Saudi Arabia.
Raffaella Balestrini, National Research Council, Institute for Sustainable Plant Protection, Turin 10135, Italy.
Kit Xi Liew, Biological and Environmental Sciences and Engineering Division, Center for Desert Agriculture (CDA), King Abdullah University of Science and Technology (KAUST), The BioActives Lab, Thuwal, 23955-15 6900, Saudi Arabia.
Chakravarthy Rajan, Biological and Environmental Sciences and Engineering Division, Center for Desert Agriculture (CDA), King Abdullah University of Science and Technology (KAUST), The BioActives Lab, Thuwal, 23955-15 6900, Saudi Arabia.
Lamis Berqdar, Biological and Environmental Sciences and Engineering Division, Center for Desert Agriculture (CDA), King Abdullah University of Science and Technology (KAUST), The BioActives Lab, Thuwal, 23955-15 6900, Saudi Arabia.
Ikram Blilou, The Plant Science Program, Biological and Environmental Science and Engineering Division, King Abdullah University of Science and Technology (KAUST), Thuwal, Saudi Arabia; Plant Cell and Developmental Biology, Biological and Environmental Sciences and Engineering (BESE), King Abdullah University of Science and Technology (KAUST), Thuwal 23955-6900, Saudi Arabia.
Luisa Lanfranco, Department of Life Sciences and Systems Biology, University of Torino, Torino 10125, Italy.
Salim Al-Babili, Biological and Environmental Sciences and Engineering Division, Center for Desert Agriculture (CDA), King Abdullah University of Science and Technology (KAUST), The BioActives Lab, Thuwal, 23955-15 6900, Saudi Arabia; The Plant Science Program, Biological and Environmental Science and Engineering Division, King Abdullah University of Science and Technology (KAUST), Thuwal, Saudi Arabia.
S.A.B. and A.A. conceived and designed the research. S.A.B. supervised the experiments. A.A. generated transgenic lines with the help of CR. C.V., V.F., R.B., and L.L. planned and performed the mycorrhization experiments. M.J. and A.A. conducted the Striga seed germination assays. A.B. performed the in vitro assays. A.A. characterized the transgenic lines with help of M.J. and L.B. J.Y.W., K.X.L., and A.A. performed the metabolite analysis. F.A. performed the subcellular localization experiment. I.B. supervised the subcellular localization and microscopy experiments. A.A. analyzed the data and generated the figures and wrote the manuscript with the help of J.Y.W., V.F., C.V., L.L., and F.A. S.A.B. edited and approved the article.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is Salim Al-Babili (salim.babili@kaust.edu.sa).
References
- Abe S, Sado A, Tanaka K, Kisugi T, Asami K, Ota S, Kim HI, Yoneyama K, Xie X, Ohnishi T (2014) Carlactone is converted to carlactonoic acid by MAX1 in Arabidopsis and its methyl ester can directly interact with AtD14 in vitro. Proc Natl Acad Sci USA 111: 18084–18089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ablazov A, Mi J, Jamil M, Jia KP, Wang JY, Feng Q, Al-Babili S (2020) The apocarotenoid zaxinone is a positive regulator of strigolactone and abscisic acid biosynthesis in Arabidopsis roots. Front Plant Sci 11: 578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahrazem O, Gómez-Gómez L, Rodrigo MJ, Avalos J, Limón MC (2016) Carotenoid cleavage oxygenases from microbes and photosynthetic organisms: features and functions. Int J Mol Sci 17: 1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama K, Matsuzaki KI, Hayashi H (2005) Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435: 824–827 [DOI] [PubMed] [Google Scholar]
- Al-Babili S, Bouwmeester HJ (2015) Strigolactones, a novel carotenoid-derived plant hormone. Ann Rev Plant Biol 66: 161–186 [DOI] [PubMed] [Google Scholar]
- Alder A, Jamil M, Marzorati M, Bruno M, Vermathen M, Bigler P, Ghisla S, Bouwmeester H, Beyer P, Al-Babili S (2012) The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science 335: 1348–1351 [DOI] [PubMed] [Google Scholar]
- Aljedaani F, Rayapuram N, Blilou I (2021) A semi-in vivo transcriptional assay to dissect plant defense regulatory modules. In S Mukhtar, ed, Modeling Transcriptional Regulation. Salmon Tower Building, NY, Springer US, pp. 203–214. [DOI] [PubMed] [Google Scholar]
- Auldridge ME, Block A, Vogel JT, Dabney-Smith C, Mila I, Bouzayen M, Magallanes‐Lundback M, DellaPenna D, McCarty DR, Klee HJ (2006a) Characterization of three members of the Arabidopsis carotenoid cleavage dioxygenase family demonstrates the divergent roles of this multifunctional enzyme family. Plant J 45: 982–993 [DOI] [PubMed] [Google Scholar]
- Auldridge ME, McCarty DR, Klee HJ (2006b) Plant carotenoid cleavage oxygenases and their apocarotenoid products. Curr Opin Plant Biol 9: 315–321 [DOI] [PubMed] [Google Scholar]
- Balestrini R,, Jose-Estanyol M,, Puigdomenech P,, Bonfante P (1997) Hydroxyproline-rich glycoprotein mRNA accumulation in maize root cells colonized by an arbuscular mycorrhizal fungus as revealed by in situ hybridization. Protoplasma 198: 36–42 [Google Scholar]
- Ballottari M, Alcocer MJ, D’Andrea C, Viola D, Ahn TK, Petrozza A, Polli D, Fleming GR, Cerullo G, Bassi R (2014) Regulation of photosystem I light harvesting by zeaxanthin. Proc Natl Acad Sci USA 111: E2431–E2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier F, Dogbo O, Camara B (2003) Biosynthesis of the food and cosmetic plant pigment bixin (annatto). Science 300: 2089–2091 [DOI] [PubMed] [Google Scholar]
- Braguy J, Ramazanova M, Giancola S, Jamil M, Kountche BA, Zarban R, Felemban A, Wang JY, Lin PY, Haider I, et al (2021) SeedQuant: a deep learning-based tool for assessing stimulant and inhibitor activity on root parasitic seeds. Plant Physiol 186: 1632–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno M, Beyer P, Al-Babili S (2015) The potato carotenoid cleavage dioxygenase 4 catalyzes a single cleavage of β-ionone ring-containing carotenes and non-epoxidated xanthophylls. Arch Biochem Biophys 572: 126–133 [DOI] [PubMed] [Google Scholar]
- Bruno M, Hofmann M, Vermathen M, Alder A, Beyer P, Al-Babili S (2014) On the substrate-and stereospecificity of the plant carotenoid cleavage dioxygenase 7. FEBS Lett 588: 1802–1807 [DOI] [PubMed] [Google Scholar]
- Bruno M, Koschmieder J, Wuest F, Schaub P, Fehling-Kaschek M, Timmer J, Beyer P, Al-Babili S (2016) Enzymatic study on AtCCD4 and AtCCD7 and their potential to form acyclic regulatory metabolites. J Exp Bot 67: 5993–6005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernys JT, Zeevaart JA (2000) Characterization of the 9-cis-epoxycarotenoid dioxygenase gene family and the regulation of abscisic acid biosynthesis in avocado. Plant Physiol 124: 343–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler SR, Ehrhardt DW, Griffitts JS, Somerville CR (2000) Random GFP::cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc Natl Acad Sci USA 97: 3718–3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker EL, Alder A, Hunn S, Ferguson J, Lehtonen MT, Scheler B, Kerres KL, Wiedemann G, Safavi-Rizi V, Nordzieke S (2017) Strigolactone biosynthesis is evolutionarily conserved, regulated by phosphate starvation and contributes to resistance against phytopathogenic fungi in a moss, Physcomitrella patens. New Phytologist 216: 455–468 [DOI] [PubMed] [Google Scholar]
- Dhar MK, Mishra S, Bhat A, Chib S, Kaul S (2020) Plant carotenoid cleavage oxygenases: structure–function relationships and role in development and metabolism. Brief Funct Genom 19: 1–9 [DOI] [PubMed] [Google Scholar]
- Dickinson AJ,, Lehner K,, Mi J,, Jia KP,, Mijar M,, Dinneny J,, Al-Babili S,, Benfey PN (2019) β-Cyclocitral is a conserved root growth regulator. Proc Natl Acad Sci USA 116: 10563–10567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Von Heijne G (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8: 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felemban A, Braguy J, Zurbriggen MD, Al-Babili S (2019) Apocarotenoids involved in plant development and stress response. Front Plant Sci 10: 1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorilli V,, Belmondo S,, Khouja HR,, Abbà S,, Faccio A,, Daghino S,, Lanfranco L (2016) RiPEIP1, a gene from the arbuscular mycorrhizal fungus Rhizophagus irregularis, is preferentially expressed in planta and may be involved in root colonization. Mycorrhiza 26: 609–621 [DOI] [PubMed] [Google Scholar]
- Fiorilli V, Wang JY, Bonfante P, Lanfranco L, Al-Babili S (2019) Apocarotenoids: old and new mediators of the arbuscular mycorrhizal symbiosis. Front Plant Sci 10: 1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser PD, Bramley PM (2004) The biosynthesis and nutritional uses of carotenoids. Prog Lipid Res 43: 228–265 [DOI] [PubMed] [Google Scholar]
- Giuliano G, Al-Babili S, Von Lintig J (2003) Carotenoid oxygenases: cleave it or leave it. Trends Plant Sci 8: 145–149 [DOI] [PubMed] [Google Scholar]
- Gomez-Roldan V, Fermas S, Brewer PB, Puech-Pagès V, Dun EA, Pillot JP, Letisse F, Matusova R, Danoun S, Portais JC (2008) Strigolactone inhibition of shoot branching. Nature 455: 189–194 [DOI] [PubMed] [Google Scholar]
- Gutjahr C, Gobbato E, Choi J, Riemann M, Johnston MG, Summers W, Carbonnel S, Mansfield C, Yang SY, Nadal M, et al (2015) Rice perception of symbiotic arbuscular mycorrhizal fungi requires the karrikin receptor complex. Science 350: 1521–1524 [DOI] [PubMed] [Google Scholar]
- Gutjahr C, Parniske M (2013) Cell and developmental biology of arbuscular mycorrhiza symbiosis. Ann Rev Cell Dev Biol 29: 593–617 [DOI] [PubMed] [Google Scholar]
- Ha CV, Leyva-González MA, Osakabe Y, Tran UT, Nishiyama R, Watanabe Y, Tanaka M, Seki M, Yamaguchi S, Dong NV (2014) Positive regulatory role of strigolactone in plant responses to drought and salt stress. Proc Natl Acad Sci USA 111: 851–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Uragami C, Cogdell RJ (2016) Carotenoids and photosynthesis. Carotenoids in Nature. Springer, Berlin, Germany, pp. 111–139 [DOI] [PubMed] [Google Scholar]
- Hiei Y, Komari T (2008) Agrobacterium-mediated transformation of rice using immature embryos or calli induced from mature seed. Nat Protocol 3: 824–834 [DOI] [PubMed] [Google Scholar]
- Ilg A, Beyer P, Al-Babili S (2009) Characterization of the rice carotenoid cleavage dioxygenase 1 reveals a novel route for geranial biosynthesis. FEBS J 276: 736–747 [DOI] [PubMed] [Google Scholar]
- Ilg A, Bruno M, Beyer P, Al-Babili S (2014) Tomato carotenoid cleavage dioxygenases 1A and 1B: relaxed double bond specificity leads to a plenitude of dialdehydes, mono-apocarotenoids and isoprenoid volatiles. FEBS Open Biol 4: 584–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Braguy J, Wang JY, Yoda A, Fiorilli V, Takahashi I, Jamil M, Felemban A, Miyazaki S, Mazzarella T (2022) Canonical strigolactones are not the tillering-inhibitory hormone but rhizospheric signals in rice. BioRxiv 10.1101/2022.04.05.487102 [DOI] [PMC free article] [PubMed]
- Jamil M, Kountche BA, Haider I, Wang JY, Aldossary F, Zarban RA, Jia KP, Yonli D, Shahul Hameed UF, Takahashi I (2019) Methylation at the C-3′ in D-ring of strigolactone analogs reduces biological activity in root parasitic plants and rice. Front Plant Sci 10: 353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia KP, Baz L, Al-Babili S (2018) From carotenoids to strigolactones. J Exp Bot 69: 2189–2204 [DOI] [PubMed] [Google Scholar]
- Jia KP, Dickinson AJ, Mi J, Cui G, Xiao TT, Kharbatia NM, Guo X, Sugiono E, Aranda M, Blilou I (2019) Anchorene is a carotenoid-derived regulatory metabolite required for anchor root formation in Arabidopsis. Sci Adv 5: eaaw6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia KP, Mi J, Ablazov A, Ali S, Yang Y, Balakrishna A, Berqdar L, Feng Q, Blilou I, Al-Babili S (2021) Iso‐anchorene is an endogenous metabolite that inhibits primary root growth in Arabidopsis. Plant J 107: 54–66 [DOI] [PubMed] [Google Scholar]
- Langdale JA (1993) In situ hybridization. In M Freeling, V Walbot, eds, The Maize Handbook. Springer-Verlag, New York, pp 165–180 [Google Scholar]
- Liang MH, He YJ, Liu DM, Jiang JG (2021) Regulation of carotenoid degradation and production of apocarotenoids in natural and engineered organisms. Critic Rev Biotechnol 41: 513–534 [DOI] [PubMed] [Google Scholar]
- Mi J, Jia KP, Wang JY, Al-Babili S (2018) A rapid LC-MS method for qualitative and quantitative profiling of plant apocarotenoids. Anal Chim Acta 1035: 87–95 [DOI] [PubMed] [Google Scholar]
- Mi J, Jia KP, Balakrishna A, Wang JY, Al-Babili S (2019) An LC-MS profiling method reveals a route for apocarotene glycosylation and shows its induction by high light stress in Arabidopsis. Analyst 144: 1197–1204 [DOI] [PubMed] [Google Scholar]
- Moise AR, Al-Babili S, Wurtzel ET (2014) Mechanistic aspects of carotenoid biosynthesis. Chem Rev 114: 164–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno JC, Mi J, Alagoz Y, Al‐Babili S (2021) Plant apocarotenoids: from retrograde signaling to interspecific communication. Plant J 105: 351–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara E, Marion-Poll A (2005) Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol 56: 165–185 [DOI] [PubMed] [Google Scholar]
- Nisar N, Li L, Lu S, Khin NC, Pogson BJ (2015) Carotenoid metabolism in plants. Mol Plant 8: 68–82 [DOI] [PubMed] [Google Scholar]
- Paszkowski U,, KrokenS, , Roux C,, Briggs SP (2002) Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci USA 99: 13324–13329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg Z, Blumwald E (2011) Hormone balance and abiotic stress tolerance in crop plants. Curr Opin Plant Biol 14: 290–295 [DOI] [PubMed] [Google Scholar]
- Ramel F, Birtic S, Ginies C, Soubigou-Taconnat L, Triantaphylidès C, Havaux M (2012) Carotenoid oxidation products are stress signals that mediate gene responses to singlet oxygen in plants. Proc Natl Acad Sci USA 109: 5535–5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Concepcion M, Avalos J, Bonet ML, Boronat A, Gomez-Gomez L, Hornero-Mendez D, Limon MC, Meléndez-Martínez AJ, Olmedilla-Alonso B (2018) A global perspective on carotenoids: metabolism, biotechnology, and benefits for nutrition and health. Prog Lipid Res 70: 62–93 [DOI] [PubMed] [Google Scholar]
- Schwartz SH, Tan BC, Gage DA, Zeevaart JA, McCarty DR (1997) Specific oxidative cleavage of carotenoids by VP14 of maize. Science 276: 1872–1874 [DOI] [PubMed] [Google Scholar]
- Torres MA,, Rigau J,, Puigdomenech P,, Stiefel V (1995) Specific distribution of mRNAs in maize growing pollen tubes observed by whole-mount in situ hybridization with non-radioactive probes. Plant J 8: 317–321 [Google Scholar]
- Trouvelot A, Kough JL, Gianinazzi-Pearson V (1986) Estimation of vesicular arbuscular mycorrhizal infection levels. Research for methods having a functional significance. Physiological and Genetical Aspects of Mycorrhizae = Aspects Physiologiques et Genetiques des Mycorhizes: Proceedings of the 1st European Symposium on Mycorrhizae, Dijon, 1–5 July 1985. Institut National de le Recherche Agronomique, Paris [Google Scholar]
- Umehara M, Hanada A, Yoshida S, Akiyama K, Arite T, Takeda-Kamiya N, Magome H, Kamiya Y, Shirasu K (2008) Inhibition of shoot branching by new terpenoid plant hormones. Nature 455: 195–200 [DOI] [PubMed] [Google Scholar]
- Vogel JT, Tan BC, McCarty DR, Klee HJ (2008) The carotenoid cleavage dioxygenase 1 enzyme has broad substrate specificity, cleaving multiple carotenoids at two different bond positions. J Biol Chem 283: 11364–11373 [DOI] [PubMed] [Google Scholar]
- Votta C, Fiorilli V, Haider I, Wang JY, Balestrini R, Petřík I, Tarkowská D, Novák O, Serikbayeva A, Bonfante P, et al. (2022). Zaxinone synthase controls arbuscular mycorrhizal colonization level in rice. Plant J 111: 1688–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JY, Alseekh S, Xiao T, Ablazov A, Perez de Souza L, Fiorilli V, Anggarani M, Lin PY (2021a) Multi-omics approaches explain the growth-promoting effect of the apocarotenoid growth regulator zaxinone in rice. Commun Biol 4: 1–1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JY, Chen GT, Jamil M, Braguy J, Sioud S, Liew KX, Balakrishna A, Al-Babili S (2022) Protocol for characterizing strigolactones released by plant roots. STAR Protocol 3: 101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JY, Haider I, Jamil M, Fiorilli V, Saito Y, Mi J, Baz L, Kountche BA, Jia KP, Guo X (2019) The apocarotenoid metabolite zaxinone regulates growth and strigolactone biosynthesis in rice. Nat Commun 10: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JY, Jamil M, Lin PY, Ota T, Fiorilli V, Novero M, Zarban RA, Kountche BA (2020) Efficient mimics for elucidating zaxinone biology and promoting agricultural applications. Mol Plant 13: 1654–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JY, Lin PY, Al-Babili S (2021b) On the biosynthesis and evolution of apocarotenoid plant growth regulators. Semin Cell Dev Biol 109: 3–11 [DOI] [PubMed] [Google Scholar]
- Wang W, Shi J, Xie Q, Jiang Y, Yu N, Wang E (2017) Nutrient exchange and regulation in arbuscular mycorrhizal symbiosis. Mol Plant 10: 1147–1158 [DOI] [PubMed] [Google Scholar]
- Waters MT, Gutjahr C, Bennett T, Nelson DC (2017) Strigolactone signaling and evolution. Ann Rev Plant Biol 68: 291–322 [DOI] [PubMed] [Google Scholar]
- Xie K, Minkenberg B, Yang Y (2015) Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc Natl Acad Sci USA 112: 3570–3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Yoneyama K, Yoneyama K (2010) The strigolactone story. Ann Rev Phytopathol 48: 93–117 [DOI] [PubMed] [Google Scholar]
- Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL (2012) Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinform 13: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama K, Xie X, Yoneyama K, Kisugi T, Nomura T, Nakatani Y, Akiyama K, McErlean CS (2018) Which are the major players, canonical or non-canonical strigolactones? J Exp Bot 69: 2231–2239 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Van Dijk AD, Scaffidi A, Flematti GR, Hofmann M, Charnikhova T, Verstappen F, Hepworth J, Van Der Krol S (2014) Rice cytochrome P450 MAX1 homologs catalyze distinct steps in strigolactone biosynthesis. Nat Chem Biol 10: 1028–1033 [DOI] [PubMed] [Google Scholar]
- Zheng X, Giuliano G, Al-Babili S (2020) Carotenoid biofortification in crop plants: citius, altius, fortius. Biochim Biophys Acta Mol Cell Biol Lipids 1865: 158664. [DOI] [PubMed] [Google Scholar]
- Zheng X, Yang Y, Al-Babili S (2021) Exploring the diversity and regulation of apocarotenoid metabolic pathways in plants. Front Plant Sci 12: 787049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.