Abstract
Climate change is a defining challenge of the 21st century, and this decade is a critical time for action to mitigate the worst effects on human populations and ecosystems. Plant science can play an important role in developing crops with enhanced resilience to harsh conditions (e.g. heat, drought, salt stress, flooding, disease outbreaks) and engineering efficient carbon-capturing and carbon-sequestering plants. Here, we present examples of research being conducted in these areas and discuss challenges and open questions as a call to action for the plant science community.
We discuss research aimed at improving carbon sequestering capacity and climate resilience in plants to illustrate how plant science can help to mitigate climate change and enhance food security.
Introduction
Climate change is caused by an accumulation of greenhouse gases (GHGs) (e.g. CO2, methane) in the atmosphere leading to increased planetary heat-trapping and global warming. The IPCC Sixth assessment report (IPCC, 2022) strongly suggests that limiting global warming to 1.5ºC above pre-industrial levels will be needed to avoid severe climate change effects. This will require halving global CO2 emissions by 2030 and cutting them to net zero by 2050, as well as removing an additional 2–10 billion metric tons (Gt) of CO2 each year. In some locations, warming may benefit certain crops, and, over time, the optimal growing regions may shift farther away from the equator. However, the effects of climate change are not limited to increasing temperatures and heatwaves in many parts of the world but include changes in rainfall, more severe and frequent storms, increased drought, and increased threat of wildfires. All of these effects are anticipated to adversely affect crop yields and food security worldwide within the next 20 years (Zhao et al., 2017; Li et al. 2019; Jägermeyr et al., 2021). As the impact of climate change on crop systems intensifies, the need to develop stress-resilient crops to combat food insecurity rises.
In this article, we explore several ways in which plant scientists are working on solutions related to carbon sequestration to help achieve net zero CO2 emissions and crop improvements to protect and enhance yields for increased food security. The first section outlines challenges and approaches for enhancing the carbon sequestration capacity of crops (annual and perennial) and seagrasses, followed by a section on improving photosynthesis. A third section addresses engineering climate resilience in crops (resistance or tolerance to abiotic and biotic stresses). The final section describes the vision of a sustainable global bioeconomy rooted in plant biology. We acknowledge that there are other areas, not covered here, in which plant science can play a role in mitigating adverse climate change effects, including bioenergy, forestry, and ecosystem conservation. Solutions in all of these areas are needed in the very near future, and in the longer term. We do not provide an in-depth review of these topics. Rather, the examples provided here illustrate a few of the many avenues of research being conducted by plant scientists around the world. A companion review by Verslues et al. (2023) addresses unresolved questions in plant abiotic stress. We hope that these stories help to inform the plant science community of the possibilities, stimulate further research, and motivate plant scientists at any stage of their careers to become involved in work aimed at mitigating climate change and enhancing food and energy security. Mitigating the climate change crisis will require all hands on deck.
How can more carbon be retained in soil and biomass?
Carbon sequestration in annual cropping systems
(By John K. McKay)
Annual cropping systems present opportunities for carbon sequestration that have yet to be exploited. In addition to the need to reduce GHG emissions, active atmospheric CO2 removal strategies, also called Negative Emissions Technologies (NETs), are needed to attain net CO2 reductions and avoid the most damaging climate change outcomes (National Academies of Sciences, Engineering, and Medicine, 2019). Atmospheric CO2 removal technologies need to be implemented now and increase to levels on the order of 10 Gt CO2 per year by 2050, and 20 Gt CO2 per year by 2100 (National Academies of Sciences, Engineering, and Medicine, 2019).
Among NET for CO2 removal, soil carbon sequestration is the least expensive and most ready to scale in the next decades (National Academies of Sciences, Engineering, and Medicine, 2019). Current US cropping systems use genetics that were not designed to minimize GHG emissions nor to maximize carbon sequestration, yet heritable genetic variation for these traits exists in many crops. In addition, agricultural soils experienced well-documented decreases in soil carbon over the last century (Davidson and Ackerman, 1993) and are capable of sequestering all of the CO2 currently in the atmosphere (Ciais et al., 2013). Here, I review the challenges with attempts to achieve soil carbon sequestration in current annual cropping systems, both with the way in which the maize (Zea mays)–soy (Glycine max) rotation was designed and the science to date on how management might lead to predictable increases in soil carbon. I then focus on genetic changes that are needed to create carbon-negative crops, including optimal combinations of traits that can be addressed in breeding programs.
The major, unaddressed problem for sustainability and GHG emissions in annual cropping systems is excess nitrogen (N) in the form of synthetic fertilizer (Northrup et al., 2021), which leaches into groundwater, rivers, and oceans and into the atmosphere as N2O, a GHG with an effect size ∼300× that of CO2 (Albritton et al., 2001). An obvious example is ethanol production from maize, where N is responsible for >80% of GHG emissions overall (Kim et al., 2014). For the parts of the world where the Green Revolution was successfully deployed (Evenson and Gollin, 2003), a major consequence is the exclusive use of crop genotypes that require high N inputs. To fix this N problem, we need to improve N use efficiency (NUE) and greatly reduce N input. Increasing NUE is feasible (Hirel et al., 2007; Northrup et al., 2021) and can be achieved in part by removing a small number of large-effect mutations that were selected to high frequency in elite germplasm in the Green Revolution (Moyers et al., 2018). Getting farmers to reduce N input is a much greater challenge. First, overfertilizing every other year is a well-established management practice of the maize-soy rotation that encompasses 73 million hectares of farmland in the USA. Although soybean is an N-fixing species, in modern cropping systems high-yielding soy crops require hundreds of kilograms of N per hectare (Salvagiotti et al., 2008). Although fertilizer has recently increased in price, so have crop commodity prices, and thus farmers remain incentivized to maximize N inputs. In the USA, the maize–soy rotation is highly subsidized by federal funds in the form of direct payment to farmers as well as mandates on using ethanol from fermentation of maize grain and biodiesel from transesterification of soy lipids.
Most efforts in using annual cropping systems for soil carbon sequestration have focused on changes in management that were originally designed for soil health (Ogle et al., 2019), such as reduced tillage, greater residue retention, and cover crops that are designed to increase the amount of above-ground plant biomass left in the field per unit area per year (McClelland et al., 2021). Most of the published studies on the effect of management on soil carbon are limited to the top 30 cm of soil, which is where most of the carbon inputs are expected (Ogle et al., 2019). However, this top 30 cm is also the least durable soil carbon and can respire back into CO2 in a few years. Getting soil carbon inputs deeper into the soil is needed to achieve greater and more durable carbon sequestration in agricultural systems (Paustian et al., 2016a, 2016b, 2019) and will require genetic changes in crops.
Genetic changes in annual cropping systems are needed both to reduce inputs (Northrup et al., 2021) and achieve carbon sequestration levels of tons per hectare per year (Paustian et al., 2016a). Some changes can be achieved by selecting against large-effect mutations that went to high frequency during the Green Revolution. Prior to the Green Revolution, putting large amounts of synthetic N on agricultural fields reduced yield, as tall crops heavy with grain were highly prone to lodging. In many cases selection during the Green Revolution was based on recurrent backcrossing to dwarf lines and involved small effective population sizes and low levels of effective recombination (Moyers et al., 2018). For example, in rice (Oryza sativa), breeding during the Green Revolution led to the fixation of mutations that reduce NUE (OsTCP19; Liu et al., 2021b) and root growth (Dro1; Arai-Sanoh et al., 2014) in the elite breeding lines.
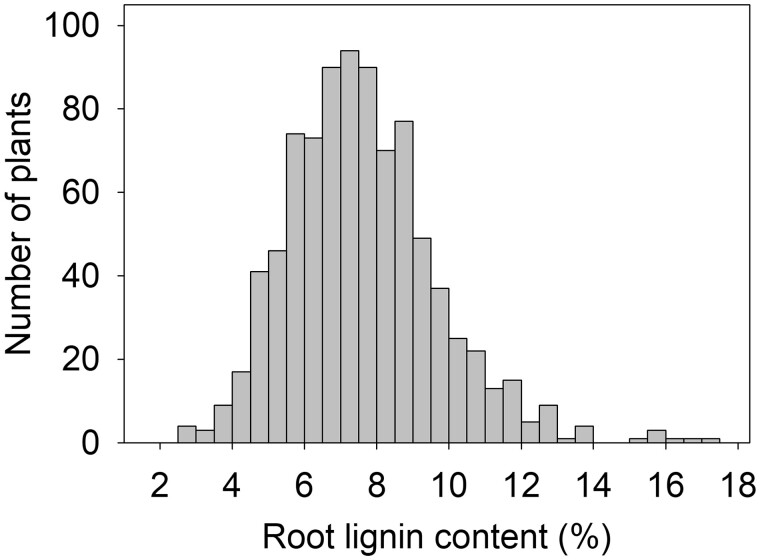
It is worth considering the traits of an ideal annual crop for carbon-negative supply chains for food, feed, fiber, and fuel. As mentioned, genetic changes to lower N requirements and create deeper, more massive root systems can make annual biomass feedstock production carbon negative (Paustian et al., 2016a). Another key trait for carbon sequestration is population density, where increasing the number of individuals per hectare leads to more root systems and greater carbon input. Crop species that were not part of the Green Revolution have promise in this respect (Amaducci et al., 2008). For example, industrial hemp (Cannabis sativa) was never bred for high N inputs, can be grown at population densities of 500,000 plants per hectare, and has greater root biomass below 50 cm than other major crops (Amaducci et al., 2008). Root carbon composition is also a genetic target, as some forms of carbon may be more recalcitrant to degradation and therefore longer lived in soils. The idea of engineering roots to create more recalcitrant forms of carbon, such as suberin, is discussed below by Busch and Chory. Suberin is one example; another is lignin, which is a parameter in models of soil carbon (Parton, 1996). We found large heritable variation in percent lignin in maize roots (Figure 1) and are testing the prediction that genotypes with greater root lignin will lead to greater quantity and durability of soil carbon.
Figure 1.
Quantitative variation in lignin content in maize root systems from a field study of 358 maize inbred lines. A description of the experiment can be found in Woods et al. (2022).
Root exudates, a diverse set of simple carbon molecules that are released passively or actively into the soil, also contribute to soil organic carbon (SOC). Little is known regarding the degree to which root exudates are controlled by genetics versus the environment. Even less is known about the genetic control of the abundance and composition of root exudates, even in model species. This is due in part to the difficulty of measuring the relevant phenotypes in agriculturally relevant environments. On the soil modeling side, recent progress has been made in separating biomass and exudate inputs to soil carbon, where exudates are predicted to lead to increases in mineral-associated organic matter, which in turn is predicted to have a longer residence time than other soil carbon fractions (Zhang et al., 2021b). Finally, the soil and root microbiome, which is influenced by root exudates and plant genotype (Peiffer et al., 2013; Wagner et al., 2020; Favela et al., 2021), influences the carbon retention properties of soils, although data on effect sizes are lacking (Naylor et al., 2020). Ectomycorrhizae are thought to be key drivers of SOC accumulation in forests (Soudzilovskaia et al., 2019) and could be exploited in cropping systems. Manipulating the soil and root microbiome of cropping systems at scale will be much more difficult than obtaining seed from new crop genotypes but is a possible tool for engineering annual cropping systems for enhanced carbon sequestration.
Harnessing plants: A global initiative to enhance plant-based carbon sequestration
(By Wolfgang Busch and Joanne Chory)
We consider solutions for carbon sequestration based on plants’ abilities to draw down CO2 from the atmosphere via photosynthesis and convert it to biomass. Earth’s soils contain a large amount of carbon, estimated at approximately 2,300 Gt carbon to 3-m depth, which constitutes about three times the current atmospheric pool of CO2 (Schlesinger and Bernhardt, 2020). The main source of SOC is plant material (e.g. aboveground plant biomass, roots, and root exudates), which can be stored in the soil or respired back into the atmosphere. It is estimated that cropland and grazing land soils (about 5 billion hectares globally) have an enormous capacity for storing carbon (Sanderman et al., 2017). Combined with existing agricultural infrastructure, this capacity provides an opportunity to leverage genetics to improve traits related to plant-mediated carbon sequestration.
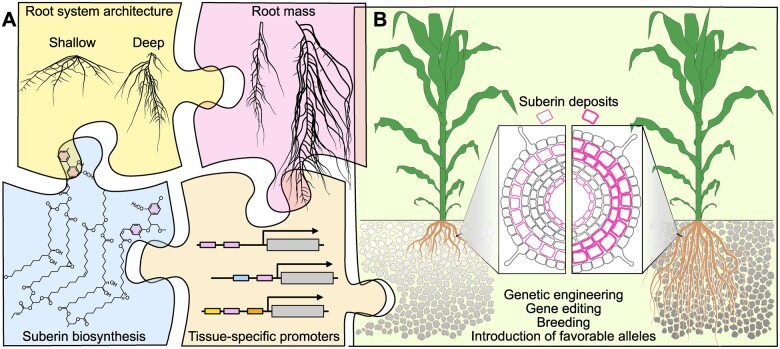
Several plant traits are good candidates for facilitating plant carbon sequestration (Figure 2). Root biomass is one, as it is estimated that a given mass of root inputs contributes about five times more SOC than the equivalent mass of aboveground litter (Jackson et al., 2017). However, traits associated with mechanisms that increase recalcitrance of SOC to breakdown by soil microorganisms (SOC protection) will also be required to increase residence time in soils. Mechanisms of SOC protection include a complex interplay between the chemical makeup of SOC, physical occlusion of SOC within soil aggregates, formation of stable organo-mineral complexes, and water-film connectivity between SOC and microbes (Schmidt et al., 2011; Lehmann et al., 2020). More than half of the global SOC is found in deep soil layers (Jobbágy and Jackson, 2000), and the mean residence time of SOC increases with depth, implying lower decomposability of root-derived carbon in deeper soil layers (Gill et al., 1999; Prieto et al., 2016). Root biochemistry also influences decomposability, and a prime candidate trait is the amount of the natural product suberin in roots. Suberin is a lipophilic complex polyester that is composed of very long-chain fatty acids and polyaromatic compounds. Suberin may be a good source for stable SOC due to its intrinsic biochemical stability (Lorenz et al., 2007) and its interaction with soil minerals and occlusion in topsoil microaggregates (Kell, 2012; Lin and Simpson, 2016). We note that there are numerous other plant traits that promise to be useful for enhancing the capacity of plants to sequester carbon in the soil (see some additional examples in the previous section by McKay).
Figure 2.
Toward an ideal carbon-capturing crop plant. A, The ideal plant should accumulate suberin in the cell wall of its root cells and form a vast and deep root system. To realize this goal, the existing literature and experimental evidence are curated to look for candidate genes affecting root system architecture and root mass. This information is combined with root-specific promoters and suberin biosynthetic genes. B, The ideal plant is created by capitalizing on both classical (breeding) and more recent (genome editing, genetic engineering) approaches to introduce favorable alleles and genes that will increase root biomass and transgenes that will increase the deposition of suberin in the root. In addition to trapping more carbon, these ideal plants will replenish carbon-depleted soils with degradation-recalcitrant carbon polymers (indicated by the darker color of the soil on the right). Figure credit: P. Salomé.
There are several significant challenges to utilizing crops for carbon sequestration. Genetic trait enhancement is a lengthy process and its adoption by the public will be challenging. Establishing a link between root traits and carbon accumulation and permanence in agricultural soils will require substantial experimental efforts. Carbon accumulation and persistence are also dependent on soil type, climate parameters, and agricultural practices such as the use of cover crops and no-till farming (Schmidt et al., 2018). Although there is good potential for plant-based carbon sequestration in the surface soil layer (up to 1.85 Gt C/year in the top 30 cm of global cropland soils alone; Zomer et al., 2017), an enhanced rooting depth and altered biochemical makeup of roots could yield a much larger sequestration capacity. Finally, time is pressing—every year that goes by without significant carbon drawdown will negatively impact billions of humans and decrease the biodiversity of our planet.
The Salk Harnessing Plants Initiative is working to identify genetic and molecular mechanisms to increase root biomass, root depth, and suberin root content. We use examples of this research to highlight considerations for plant-based carbon sequestration that we have identified during this work. Each of the target traits comes with specific challenges and opportunities. For instance, increased root mass will elevate the carbon input into soils and can improve the ability of roots to forage for nutrients and water. However, increasing root mass beyond a certain level might come at the expense of yield. Nevertheless, the relationship between root biomass and yield is not necessarily a zero-sum game as enhanced water and nutrient uptake of a bigger root system can support a larger shoot. This might be particularly relevant under drought or nutrient-limited conditions. An example of the lack of a strict tradeoff of root biomass and yield in major crops is the lack of correlation of yield and root biomass in maize as well as soybean in a multi-location, multi-year study (Ordóñez et al., 2020). Increasing root depth promises to increase the lifetime of the average carbon molecule deposited by roots in the soil, provide roots access to deeper soil layers that can contain more moisture, and facilitate the capture of nitrate that leaches deeper into the soil during the growing season. However, surface roots are still important for foraging immobile nutrients such as phosphorus. Therefore, achieving an optimal balance between shallow roots and deep roots will be important. As an effective apoplastic barrier, suberin in specific areas of the root could provide enhanced flood and drought resilience and might enhance root growth in deeper, more anoxic layers of the soil. Extensive variation for each of these traits between and within species indicates that there are genetic mechanisms that can be leveraged to improve them.
Our work in enhancing these traits is being conducted in parallel with model plants via forward and reverse genetic approaches, as well as in diversity collections of major row crops and cover crops to identify crop-specific targets using genome-wide association studies (GWASs). While we are interested in trait changes that will work in the field and maintain crop productivity, it is not feasible to measure all these root traits in the field at high throughput. We therefore rely on initial screening approaches in the laboratory or the greenhouse to measure and engineer root traits, subsequently moving to in-soil or field-testing with a subset of lines that display distinctive traits. We focus on root mass in relation to depth, as the engineering or breeding goal is to direct as much root mass as possible to a deeper depth, and on enhancing the accumulation of suberin. Suberin is a highly effective apoplastic diffusion barrier and producing it everywhere in the root would be detrimental to plant health. Therefore, we focus on specific root tissues that already produce suberin such as the periderm or the exodermis, which are outer layers in mature root systems. We are targeting such tissues as suberin sinks by using tissue-specific promoters to drive suberin production, as well as utilizing genes involved in the formation of these tissues to produce additional tissue layers.
To quantitatively link these root traits to carbon characteristics in the soil, we are working with soil scientists to better estimate the soil carbon impact of crop varieties that have different root mass, depth, and suberin content. We aim to test the effects of genetic alterations via gene editing or gene engineering approaches in crops over the next few years. Recent advances in high-throughput phenotyping, sequencing, and functional single-cell genomics now provide a way to leverage genes, gene constructs, and genetic variants within and between species. We aim to have the proof of concepts for enhanced crop traits within the next 3 years to then partner with both NGOs and agriculture companies to enhance varieties that are of interest to farmers.
There are numerous other opportunities for plant biologists to contribute to climate change mitigation efforts, ranging from work on traits that will reduce agricultural N2O or methane emissions to creating carbon sequestration-friendly microbiota or mycorrhizal associations. As a community, we should think of and work toward promising plant biology-based solutions.
Rapid de novo domestication of perennial crops
(By Lee R. DeHaan)
Most agricultural soils have lost 50%–70% of the SOC that they had previously accumulated under native plant communities; therefore, raising the carbon levels in historically tilled agricultural soils offers the potential to partially mitigate climate change by capturing 30–60 Gt of organic carbon (Lal, 2003). The restoration of SOC in agricultural soils would not only mitigate climate change through sequestration but would also contribute to adaptation to climate change by developing soils with greater nutrient holding capacity, resistance to erosion from extreme rain events, increased water infiltration, and water storage to stabilize productivity in the face of erratic rainfall (Blanco-Canqui et al., 2013).
Although planting long-lived perennial plants on degraded agricultural soils would be one of the most effective ways to rapidly restore soil carbon levels, this approach is limited because the herbaceous perennials currently available for use in agriculture (mainly forage crops) produce biomass that is unsuitable for direct human consumption (Paustian et al., 2016a). Therefore, efforts are underway to develop new crop plants that would have extensive long-lived root systems and would achieve carbon sequestration levels similar to perennial biofuels (Crews and Rumsey, 2017; Dheri et al., 2022) while simultaneously producing abundant human-edible protein, starch, and oils through mechanically harvestable grain (Glover et al., 2010).
Efforts to develop perennial grain crops began decades ago, but recent advances in genetics and breeding are accelerating the timeline and the first successful perennial grains are now entering fields and markets. A perennial rice breeding program was initiated in 1996, targeting the roughly 19 million hectares of upland rice grown worldwide where forest land is often cleared and degraded (Sacks et al., 2003). Annual rice (Oryza sativa ssp. indica) and the rhizomatous perennial relative Oryza longistaminata were hybridized, and a breeding program has produced lines for flooded paddies that persist through eight harvests with yields and quality traits on par with modern rice cultivars (Huang et al., 2018; Hu et al., 2022). Perennial paddy rice is expected to reduce GHG emissions and water consumption relative to annual rice (Oda et al., 2019). The development of perennial rice for upland conditions also remains possible in the near term.
Perennial grain sorghum is being developed through wide hybridization of annual grain sorghum (Sorghum bicolor) with perennial species (Figure 3). Progress for yield and survival has been made by selecting among progeny of crosses between S. bicolor and the tetraploid perennial Sorghum halepense, and evaluation under tropical conditions suggests no barrier to high-yielding perennial varieties in warmer regions (Cox et al., 2018b). Recently, diploid perennial grain sorghum lines have been derived from diploid × tetraploid crosses (Cox et al., 2018a) and from crosses between S. bicolor and the perennial diploid species Sorghum propinquum (Foster et al., 2020). Working at the diploid level is expected to expedite the development of perennial grain sorghum by simplifying crosses between perennial germplasm and locally adapted S. bicolor varieties. Now, marker-assisted selection is being initiated to accelerate progress in breeding for traits related to perenniality and productivity (Cox et al., 2018b).
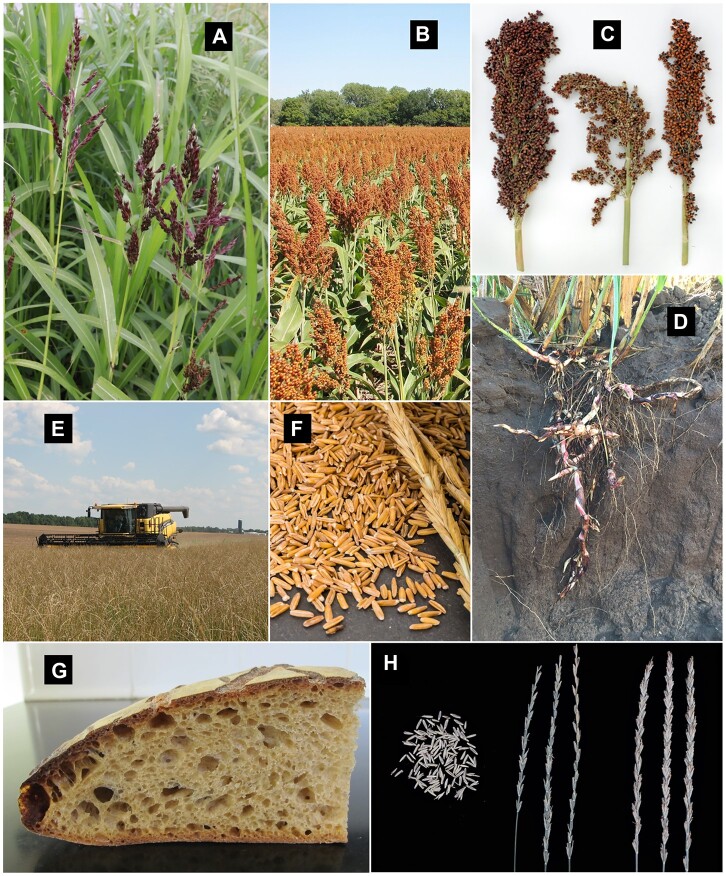
Figure 3.
Examples of wide hybridization and direct domestication to develop perennial grains. The wild perennial Sorghum halepense (A) was hybridized with the domestic species Sorghum bicolor (B) and selective breeding of the progeny produced lines with intermediate head and seed size (C) and the ability to regrow from underground rhizomes (D). In an example of direct domestication, the mostly wild grass Thinopyrum intermedium can be harvested with conventional equipment (E) and cleaned to obtain a human-edible grain (F) that has properties similar to wheat, as seen in this loaf made with an 80/20 blend of wheat and Th. intermedium flour (G). Domesticated Th. intermedium types now possess domestication traits, such as shatter resistance (H, at right).
A wide array of perennial grain crops could likely be developed either by direct domestication of wild perennial species or wide hybridization between crops and related perennials. Perennial wheat with potential to improve soil quality (Audu et al., 2022) is being developed through wide hybridization (Hayes et al., 2018). A direct domestication program is underway to develop the perennial sunflower relative Silphium integrifolium into a dual-purpose forage and grain crop (Van Tassel et al., 2017). Various perennial leguminous species are also being considered for their suitability for use as perennial grains (Schlautman et al., 2018). Perennial flax (Linum) species are being evaluated for direct domestication as perennial oilseeds (Tork et al., 2019).
Direct domestication of the cool season perennial grass species intermediate wheatgrass (Thinopyrum intermedium; Figure 3) was initiated in the 1980s, and now the harvested grain is being produced and sold in North America under the trade name Kernza (DeHaan and Ismail, 2017). With its extensive root system (Sprunger et al., 2019), the crop has potential for carbon storage belowground (De Oliveira et al., 2020) and to accumulate microbial necromass (Peixoto et al., 2020). However, genetic improvement for grain yield is needed, since selected populations still have a yield potential of less than half that of bread wheat (Triticum aestivum) in the same region (Culman et al., 2013). In addition to the currently limited genetic potential for grain yield, the crop faces many challenges. New crops always struggle with the need to coordinate supply chain development in concert with expanding acreage. Novel perennial grains also introduce a new array of challenges for farmers and agronomists, such as controlling pests and diseases and managing for sustained yield over many years. With intermediate wheatgrass, the decline in yield that occurs in aging stands is an ongoing challenge (Pinto et al., 2021).
Recent developments in plant biology, genetics, and breeding have opened the door to breeding new crops with carbon-storing perennial root systems and abundant grain production at a time scale that can proceed at the pace of commercial enterprise development (Runck et al., 2014). Low-cost genome sequencing and innovative genome assembly approaches are allowing the rapid generation of reference sequences even for large-genome perennial species. Genomic information is now leveraged to perform genomic selection (Meuwissen et al., 2001) which can greatly accelerate the breeding of perennial crops. Whereas traditional breeding of a perennial crop might require 5 or more years per generation, involving field evaluation of multiple years followed by intermating of selected individuals, genomic selection uses a genomic prediction model based on the performance of plants grown from multiple generations over many years. Applying genomic models to genetic marker data from seedlings of intermediate wheatgrass has accurately predicted mature plant performance (Crain et al., 2021). Speed breeding (Watson et al., 2018) paired with genomic selection has the potential to further accelerate perennial crop improvement by increasing the number of generations that can be completed per year. Genomic selection with speed breeding is currently being implemented with intermediate wheatgrass to complete two full cycles of selection per year, compared to one cycle every 3 years with classical approaches. Although these methods hold great promise, they remain to be validated across the many repeated cycles of selection necessary to produce a highly productive domestic crop.
The application of genome editing techniques to the domestication of wild species creates exciting possibilities to compress the development timeline for new crops (Zsögön et al., 2018). For instance, by comparing the genome sequence of the perennial Thinopyrum intermedium with related domestic grains, targets for genome editing to obtain domestic phenotypes have been identified and a roadmap for rapid domestication established (DeHaan et al., 2020).
Although new crop development is the primary approach being used to produce new perennial grains, the rediscovery of “orphan” perennial grain species is another approach worthy of investigation. Pigeonpea (Cajanus cajan) is an N-fixing semi-perennial shrub that is grown in Asia and southern Africa. Although types that can be grown for several seasons without replanting have been used in erosion control and are still grown by some farmers, annual pigeonpea is now the dominant form. A recent study in Malawi indicated that farmers are less likely to adopt erratically performing perennial pigeonpea due to social pressures and lack of trust in the technology (Grabowski et al., 2019). Expanded acreage of soil-conserving perennial pigeonpea may depend on the development of improved management techniques spread through peer learning, and new cultivars that enable consistent production.
The basic genetic and physiological control of the perennial growth habit has only recently been the subject of experimentation and remains poorly understood (Park et al., 2017). This lack of understanding has thus far hindered the development of perennial grain crops. With a clear understanding of the pathways involved, rapid conversion of existing annual crops into perennials could be possible. Combined approaches of wide hybridization, genome editing, mutagenesis, and transgenics could be used to achieve perennial growth in high-yielding cultivars. Because the need for carbon sequestration in soils is urgent, these approaches could be implemented in parallel, following the approach used to develop COVID vaccines (Ball, 2020) to develop an array of high-yielding perennial crops in the coming years.
The promise of seagrasses for carbon capture and storage
(By Carlos M. Duarte)
Seagrasses are a group of about 74 angiosperm species that complete their life cycle in the marine environment, where they form lush meadows that rank amongst the world’s most productive ecosystems (Duarte and Chiscano, 1999; Hemminga and Duarte, 2000). Seagrass meadows are strongly autotrophic, producing more organic matter than consumed in the ecosystem (Duarte et al., 2010) and acting, therefore, as sinks for CO2, much of which is buried in seagrass soils (Duarte et al., 2005, 2013a; Fourqurean et al., 2012). The role of seagrasses as intense carbon sinks in the biosphere is supported by their high photosynthetic efficiency, low nutrient requirements, adaptations that minimize carbon losses, and their capacity to cope with anoxic, sulfide-rich sediments. Indeed, whereas seagrasses occupy an estimated 0.08% of the ocean seafloor, they contribute an estimated 12.7% of all organic carbon annually buried in the ocean seafloor (Duarte et al., 2005). Yet at least one-third of the historical global area occupied by seagrasses has been lost, leading to the loss of this carbon sink and the risk of remineralization and subsequent CO2 emission of the carbon stocks accumulated in their soils over millennia. Hence, seagrass meadows represent a key component of the so-called “blue carbon” strategies aimed at avoiding losses and restoring coastal vegetated habitats to contribute to climate change mitigation, through carbon capture and storage, and climate change adaptation through the coastal protection seagrasses offer (Duarte et al., 2013a; Macreadie et al., 2021).
A range of tools within plant sciences, from genomics and metabolomics to microbiome investigations are providing important insights into the underpinnings of the remarkable carbon capture capacity of seagrass. Whereas the role of seagrasses in carbon capture and storage has been addressed largely through the quantification of stocks (Fourqurean et al., 2012) and burial rates (Duarte et al., 2005, 2013a), seagrass traits related to carbon capture and storage have been poorly addressed. Here, I discuss the fundamental plant traits that render seagrasses so efficient in carbon removal and identify a number of promising areas where further research may provide additional insights on their role. Further efforts in resolving carbon concentration mechanisms and the role of the microbiome, specifically the root component, offer promise to contribute to developments in carbon capture technologies and to increase the efficiency of seagrass restoration, respectively.
What do we know?
The high productivity of seagrass meadows even under low light conditions (Duarte and Chiscano, 1999) supplies much of the carbon sequestered in seagrass meadows (Kennedy et al., 2010). The keys to the high productivity of seagrass meadows are efficient light use (Enríquez et al., 1994), low nutrient requirements (Duarte, 1990), and carbon concentrating mechanisms that allow seagrasses to use both CO2 and to support their high photosynthetic rates (Larkum et al., 2006). The analysis of the full genome sequence of the seagrass Zostera marina pointed to a number of evolutionary adaptations required for these species to colonize the ocean from freshwater angiosperm ancestors (Olsen et al., 2016). Some of these adaptations help explain their high carbon removal, including the loss of volatiles, consistent with the loss of stomata through which they are emitted for airborne communication and plant defense, which reduces losses of carbon and the probability of infections, as stomata are a main entry point for pests and pathogens in terrestrial plants (Olsen et al., 2016).
The seagrass genome also revealed new combinations of structural traits related to the cell wall, enabling the synthesis of cutin-cuticular waxes, suberin–lignin near the plasma membrane, and macroalgal-like sulfated polysaccharides (Olsen et al., 2016), recently confirmed by direct analyses of the seagrass cell walls, which revealed the presence of fucose-containing sulfated polysaccharides, apiogalacturonan and lignin (particularly in roots and rhizomes; Pfeifer et al., 2022). This composition, together with low N and phosphorus content, renders seagrass tissues highly recalcitrant to microbial degradation (Enríquez et al., 1993), helping to explain high seagrass-derived lignin concentrations in seagrass soils (Nakakuni et al., 2021) and the high organic carbon preservation supporting high carbon sequestration rates. The full genome sequence conducted to date excluded endophytic prokaryotes (Olsen et al., 2016), which also have important contributions, as exemplified by the recent discovery of a symbiosis with an N-fixing, root-endophytic bacteria, which helps explain the high productivity of seagrass in oligotrophic environments (Mohr et al., 2021).
Seagrass morphology is a basic underpinning of their role in carbon removal. They are able to form dense canopies, exceeding 15 m2 of leaf surface per m2 of ground covered (Romero et al., 2006), and their rhizomes and roots also form a dense web in the sediments, with 0.18–3 m2 of rhizome per squaremeter and 0.47–1 m2 of roots per squaremeter of soil (Duarte et al., 1998). The dense web of seagrass leaves acts as a filter that retains particles entrained in the flow and dissipates wave and turbulent energy, enhancing the deposition and retention of particles in their soils (Hendriks et al., 2008). Meanwhile, the dense web of rhizomes and roots in the sediments injects a significant fraction of seagrass net production (2.8%–48.6% of total net production; Duarte et al., 1998) into the soil and provides physical cohesion, thereby reinforcing the soils against the erosive force of storms and extreme-energy events, such as tsunamis (Chatenoux and Peduzzi, 2007; Sasa et al., 2012).
Rhizome growth and meristematic dominance are the keys to the exponential clonal growth of seagrasses, which is a major driver of the efficiency of seagrass restoration projects in restoring seagrass carbon removal (Duarte et al., 2013b), as demonstrated in assessments of the carbon removal benefits of seagrass restoration (Marbà et al., 2015; Oreska et al., 2020). Seagrass restoration traditionally was small in scale and relatively expensive and inefficient, largely due to small planting units (van Katwijk et al., 2016). However, observations from hundreds of restoration projects (van Katwijk et al., 2016) have led to major recent successes, such as the cost-effective restoration of 36 km2 of Zostera marina meadows in Virginia’s coastal waters, with major carbon removal benefits (Orth et al., 2020), as well as the long-term success of Posidonia australis restoration in SW Australia, again coupled with important carbon removal benefits (Marbà et al., 2015). Hence, seagrass restoration has a significant scope to contribute to climate action (Macreadie et al., 2021). There is ample scope for plant science to contribute to enhancing the success of seagrass restoration, through, for instance, the use of probiotic applications (Peixoto et al., 2022) or selective breeding of seagrasses used for restoration to enhance their resistance, and thereby restoration success, in areas experiencing marine heat waves (Zabin et al., 2022).
Lack of oxygen in seagrass soils, where oxygen penetration is limited to the top few mm of seagrass soils, slows down microbial degradation and the bioturbation activity of benthic fauna, thereby improving the efficiency of carbon burial. Anoxic sediments support sulfate-reducing bacteria, producing sulfide that is toxic to seagrass. However, seagrasses protect themselves from toxic sulfide intrusions by releasing oxygen through their roots, transported from photosynthetically produced oxygen in their leaves to their roots and rhizomes (Borum et al., 2006), thereby maintaining a protective oxidized layer a few millimeters thick around their roots and rhizomes (Brodersen et al., 2015). Oxygen transport from photosynthetic production sites to roots is enabled by the development of a lacunae system that provides gaseous connectivity between leaves, rhizomes, and roots (Borum et al., 2006). While continuous within organs, they are interrupted between organs by diaphragms one cell thick, perforated by interstitial pores (0.5–1.0 μm), which provide protection from flooding while allowing gas flow (Roberts et al., 1984). In addition, the below-ground tissues of seagrasses exhibit physiological adaptations which allow them to rely temporarily on anaerobic fermentative metabolism (Borum et al., 2006).
Known unknowns
Carbon concentrating mechanisms that allow seagrasses to support their high photosynthetic rates and circumvent boundary-layer rate-limiting effects are not fully resolved (Larkum et al., 2006). Seagrass carbon metabolism remains poorly understood and seems to neither fully conform to C4 nor Crassulacean acid metabolism (Larkum et al., 2006). Genomic analyses conducted to date have focused on the seagrass genome and ignored the rich community of endophytes. There is a growing number of analyses of the seagrass microbiome, including bacteria and fungi (Tarquinio et al., 2019; Garcias-Bonet et al., 2021; Torta et al., 2022), but they remain mostly descriptive and functional analyses are limited, despite evidence that endophytes may play a major role in supporting nutrient metabolism (Mohr et al., 2021) and detoxification (Crump et al., 2018). For instance, recently discovered cable bacteria in seagrass roots could alleviate critical sulfide toxicity and promote nutrient uptake by mobilizing soil iron and phosphorous with acidification associated with electrogenic sulfide oxidation, and by stimulating dissimilatory nitrate reduction to ammonium and even fixing N2 (Scholz et al., 2021).
Opportunities around unknown unknowns
Overall, limited progress has been made in applying modern concepts and tools of plant science to further our understanding of seagrass carbon removal, where an ecological focus prevails. This is not surprising given that seagrasses represent only 0.02% of angiosperm species and have little scope to emerge as model organisms. Yet, the strong selection pressure required for angiosperms to cope with life in the marine environment and anoxic, sulfide-rich sediments is likely to have generated novel mechanisms that can open new pathways in biotechnology. Understanding the carbon concentration mechanism of seagrass can open the door for hybrid photosynthesis technologies for carbon removal (Kornienko et al., 2018), while resolving the functional role of their microbiome can help improve the outcomes of seagrass restoration. The limited effort of plant science on seagrass research to date suggests the existence of “unknown unknowns” and, therefore, a potential for new discoveries that can lead to applications in carbon removal, conservation ecology and, more broadly, plant science.
Can we improve photosynthesis?
Photosynthesis: A key target for improving crop productivity, sustainability, and resilience in the face of climate change
(By Elizabeth A. Ainsworth and Andrew D.B. Leakey)
Photosynthesis heavily influences crop productivity, resource use efficiency, and sensitivity to stresses. Therefore, strategic engineering of photosynthetic metabolism and the morphological features of leaves that control carbon and water fluxes can: (1) increase the food, fuel, fiber, and feed produced by crops; while (2) reducing demand for water and improving agricultural GHG balance; and (3) making crops more resilient to future climatic and atmospheric conditions. Detailed models of photosynthetic metabolism (Zhu et al., 2012; Bellasio, 2019) and crop function can identify engineering strategies (Kromdijk et al., 2016; Leakey et al., 2019; Wu et al., 2019). Synthetic biology is also opening doors for novel photosynthetic systems to be custom designed to new environments (Zhu et al., 2020, and discussed below by Lu and Liao). Here, we discuss engineering for greater photosynthesis under near-future elevated atmospheric CO2 concentrations and temperatures, plus improved photosynthetic water use efficiency (WUE) and NUE.
Despite a general effect of higher atmospheric CO2 enhancing photosynthesis in C3 plants, global warming is expected to have profoundly negative consequences for crop photosynthesis and productivity by the middle to end of this century (Slattery and Ort, 2019). Rising temperatures also increase vapor pressure deficit (Ficklin and Novick, 2017), which may increase irrigation demand in the future and limit the potential yield of current crop genotypes grown under standard management practices (Ort and Long, 2014; DeLucia et al., 2019). Photosynthesis is a temperature-dependent process, with rates increasing to an optimum, then decreasing once that temperature optimum is exceeded (Moore et al., 2021). This temperature dependency reflects the biochemical processes that determine rate limitations, namely Rubisco activity (and the balance between photosynthetic carbon assimilation and photorespiration) and ribulose-1,5-bisphosphate regeneration. While in vitro Rubisco carboxylation rates increase beyond ∼50°C, decreased discrimination by Rubisco for oxygen and increased solubility of oxygen relative to CO2 with rising temperatures inhibit net photosynthetic carbon assimilation in temperate C3 crops at temperatures exceeding ∼30°C, due to increased photorespiration (Moore et al., 2021). Rubisco activase is a key target for improving photosynthesis at elevated temperatures because of the thermolability of the enzyme (Salvucchi and Crafts-Brandner, 2004) and the observation that activases from species or genotypes adapted to warmer climates are more thermostable (Scafaro et al., 2016).
Work in Arabidopsis thaliana suggested that simply overexpressing a thermostable Rubisco activase could improve photosynthesis and growth in high temperature conditions (Kurek et al., 2007), but that result was not translated to crops where overexpression of Rubisco activase resulted in lower Rubisco content (Fukayama et al., 2012, 2018). Studies in rice discovered that over-expression of both Rubisco and Rubisco activase were required for enhanced photosynthesis at both optimal and high temperatures (Qu et al., 2021; Suganami et al., 2021). A highly thermostable Rubisco activase identified in the Crassulacean acid metabolism plant Agave tequilana (Shivhare and Mueller-Cajar, 2017) and greater understanding of the mechanisms of thermostability in different Rubisco activase isoforms (Scafaro et al., 2019; Degen et al., 2020) provide potential guides for further improving thermotolerance in crops.
Another target for improving photosynthesis at elevated temperatures is reducing photorespiration, the process that recycles 2P-glycolate at the expense of ATP and NADH (Walker et al., 2016). A number of genetic engineering strategies have successfully demonstrated that photorespiration can be partially bypassed, resulting in improved photosynthetic carbon assimilation (Kebeish et al., 2007; Carvalho et al., 2012; South et al., 2019). Recently, transgenic tobacco (Nicotiana tabacum) was developed to recycle 2P-glycolate in the chloroplast via overexpression of plant malate synthase and Chlamydomonas (C. reinhardtii) glycolate dehydrogenase and simultaneous RNAi to downregulate a glycolate–glycerate transporter (South et al., 2019). When these plants were grown in the field at elevated temperatures (+ 5°C), they showed greater resilience to heat stress compared to wild-type (Cavanagh et al., 2022), providing strong proof-of-concept for this strategy.
Growth at elevated CO2 (550–600 ppb, which is in the range of predicted average atmospheric CO2 concentrations by 2050) generally enhances yields of C3 crops in major temperate growing regions (Ainsworth and Long, 2021). This primarily results from enhanced photosynthetic CO2 fixation driven by greater Rubisco carboxylation rates combined with inhibition of Rubisco oxygenation rates (Stitt, 1991). Even if C3 plants acclimate to elevated CO2 in the long term by downregulating investment in Rubisco content and electron transport capacity, photosynthesis is generally stimulated along with NUE (Leakey et al., 2009). Field experiments with transgenic plants overexpressing Calvin–Benson–Bassham (CBB) cycle enzymes further enhanced the benefits of elevated CO2 on carbon gain and yield by increasing photosynthetic electron transport capacity (Rosenthal et al., 2011; Köhler et al., 2017). If coupled with breeding or engineering to maintain high sink capacity, which is a prerequisite to maximizing the potential of photosynthetic enhancements in elevated CO2 (Ainsworth and Long, 2021), this provides a widely applicable pathway to a greater CO2-fertilization effect on yield.
Greater atmospheric CO2 also causes stomatal closure, resulting in lower transpiration and greater WUE (Leakey et al., 2009, 2019). This can reduce drought-induced stress and yield loss (Fitzgerald et al., 2016). However, interactions with abscisic acid signaling, canopy micrometeorology, and N fixation can also cause the CO2-fertilization effect on yield to be lost under hot and dry conditions (Gray et al., 2016). There is also significant uncertainty about which of these responses will occur in tropical locations where water availability, high temperatures, and soil fertility might be most limiting (Leakey et al., 2012). A possible target to improve yield in times and places of drought is to reduce the amount of water lost through stomata to the atmosphere relative to photosynthetic CO2 uptake, that is increasing WUE by reducing stomatal density or accelerating stomatal closing speed (Leakey et al., 2019). Modeling suggests that prioritizing reductions in water use over increases in carbon gain when trying to enhance WUE may lead to better yield outcomes in many growing environments for both C3 and C4 species, especially as atmospheric CO2 concentrations continue to rise (Leakey et al., 2019; Wu et al., 2019). Successful pursuit of this strategy would increase productivity while making currently marginal land viable for production, reduce freshwater use for irrigation, and make crops more resilient to climate change.
Crop productivity today is highly dependent on fertilizer application, which has negative environmental effects through nitrate run-off and release of the potent GHG nitrous oxide. The need for N inputs is strongly linked to the high N cost of photosynthetic proteins. However, there may be potential to re-invest N in different photosynthetic components to increase carbon gain and improve NUE (Evans and Clarke, 2019).
Enhancing plant CO2 fixation through synthetic biology
(By Kuan-Jen Lu and James C. Liao)
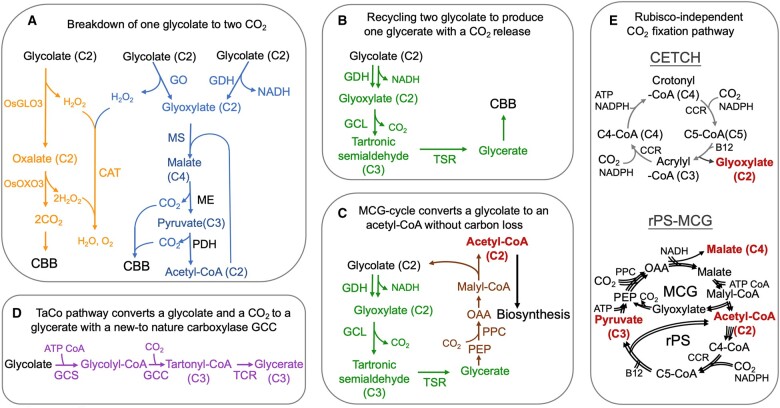
Synthetic biology encompasses engineering natural or non-natural enzymes or pathways into plants to accomplish a designated purpose. In addition to the approaches discussed above, here we discuss attempts using synthetic biology to enhance CO2 fixation, focusing on recycling photorespiration products and CO2-fixation pathways (Figure 4).
Figure 4.
Synthetic biology approaches for recycling photorespiration and CO2-fixation pathways. A and B, Photorespiration engineered for breakdown of one glycolate to two CO2 molecules (A) or conversion of two glycolate to one glycerate plus one CO2 molecule (B). The CO2 released in the chloroplast is recycled back to the CBB cycle for carbon reassimilation (Maier et al., 2012; Shen et al., 2019; South et al., 2019). C, The MCG-cycle engineered to convert glycolate to acetyl-CoA without carbon loss (Yu et al., 2018b). D, Creation of a new enzyme, such as glycolyl-CoA carboxylase, to achieve glycolate recycling to produce glycerate with input of ATP and an additional CO2 molecule (Scheffen et al., 2021). E, Rubisco-independent CETCH and rPS-MCG synthetic CO2-fixation pathways (Luo et al., 2022).
Recycling photorespiration products
Plant photorespiration produces a nonproductive product, 2P-glycolate, through the oxygenase activity of Rubisco. 2P-glycolate is converted to glycerate in peroxisomes and to CO2 in mitochondria in a process requiring ATP and NADPH with CO2 and ammonium released (Walker et al., 2016). Current synthetic pathways for reducing photorespiratory CO2 loss involve the following types:
Breakdown of one glycolate (a C2 compound) to two CO2 in chloroplasts without ATP or NADPH consumption (Figure 4A)
The released CO2 can be reassimilated by Rubisco, and no ammonium would be released. For example, an engineered “GOC” pathway in rice consists of a glycolate oxidase (OsGLO3), an oxalate oxidase (OsOXO3), and a catalase (OsCATC) overexpressed in rice chloroplasts (Shen et al., 2019). Glycolate is converted to oxalate, which is completely oxidized to two CO2 by OsOXO3. OsCATC is required for decomposing H2O2, preventing plants from oxidative stress. Rice plants engineered with the GOC pathway showed a 22% increase in photosynthesis, but increases in yield were inconsistent and dependent on the season in field tests (Shen et al., 2019). An earlier example overexpressed a malate synthase (MS) from pumpkin (Cucurbita pepo), a catalase (CAT) from Escherichia coli, and a peroxisomal glycolate oxidase (GO) in Arabidopsis chloroplasts (Maier et al., 2012). In this manner, glycolate is completely oxidized to CO2 via both the heterologous and endogenous enzymes. The transgenic Arabidopsis had a greater rosette number and size with higher biomass under the ambient CO2, short-day conditions. However, introduction of the above three genes in tobacco did not result in increased biomass in greenhouse studies (South et al., 2019). When GO was replaced with Chlamydomonas glycolate dehydrogenase (GDH), which produces NADH instead of H2O2, transgenic tobacco showed higher carbon assimilation rates, resistance to photorespiration stress, and a significant increase in biomass in the field tests (South et al., 2019). Under high temperatures (+5°C), this pathway decreased yield loss by 11%–21% (Cavanagh et al., 2022).
Conversion of two glycolate (C2) to one glycerate (C3) with CO2 release in chloroplasts (Figure 4B)
The synthetic pathway originated from E. coli, consisting of dehydrogenase (GDH), glyoxylate carboligase (GCL), and tartronic semialdehyde reductase (TSR). Unlike the first approach, this synthetic pathway preserves 75% of carbon from two glyoxylate to produce one glycerate, which is returned to the CBB cycle (Kebeish et al., 2007). The remaining carbon is CO2 produced via GCL. Expressing the above genes in Arabidopsis chloroplasts increased the growth rate and biomass yield. This synthetic pathway was shown to benefit crop plants such as Camelina sativa and potato (Solanum tuberosum) in greenhouse and growth chamber conditions (Nolke et al., 2014; Dalal et al., 2015).
Fixation of an additional CO2 to compensate for the carbon loss by GCL (Figure 4C)
The synthetic malyl-CoA glycerate (MCG) cycle also uses GCL, and TSR to convert two glyoxylates to glycerate, which is then converted to phosphoenolphyruvate (PEP). The oxygen-insensitive PEP carboxylase (PPC) then carboxylates CO2 and PEP to OAA (C4), followed by splitting OAA to acetyl-CoA and glyoxylate (Yu et al., 2018b). The glyoxylate is then recycled in the GCL reaction. The net result is the conversion of glyoxylate (or glycolate) to a productive biosynthetic product, acetyl-CoA, without carbon loss. The MCG cycle has been accomplished in Synechococcus elongatus PCC7942, a photoautotrophic cyanobacterium (Yu et al., 2018b). Compared to the wild-type, the strain expressing the MCG cycle fixed higher amounts of CO2 to produce more acetyl-CoA and its derived compound ketoisocaproate, an intermediate in leucine biosynthesis.
Fixation of an additional CO2 to glycolate after activation (Figure 4D)
An elegant tartronyl-CoA (TaCo) pathway was demonstrated recently, in which glycolate is activated to glycoly-CoA, which is then caboxylated to tartronyl-CoA and then to glycerate. This approach requires a new-to-nature enzyme, glycolyl-CoA carboxylase, which was developed by rational design and high-throughput screening (Scheffen et al., 2021).
Rubisco-independent, synthetic CO2 fixation pathways
Six Rubisco-independent CO2-fixation pathways in microorganisms have been identified in nature (Berg, 2011), and a number of theoretical synthetic pathways have been designed in silico based on reported enzyme activities and thermodynamics (Bar-Even, 2018). The first step in implementing synthetic pathways is to demonstrate the pathway feasibility in a cell-free system. This in vitro demonstration requires in-depth processes in solving problems in co-factor regeneration, enzyme stability, and pathway control. Through these processes, incompatibility of enzyme reactions, kinetic barrier, and thermodynamic limitations can be identified. To date, two Rubisco-independent synthetic CO2-fixing pathways, CETCH and reductive pyruvate synthesis (rPS)–MCG (Figure 4E), have been demonstrated, and achieved similar or increased CO2 fixation rates in vitro compared with the CBB cycle in vivo (Schwander et al., 2016; Luo et al., 2022). The CETCH pathway consists of 17 enzymes from different organisms (Schwander et al., 2016). An oxygen-insensitive carboxylase/reductase (CCR) from Methylorubrum extorqens was chosen as the carboxylase to fix CO2 in the CETCH cycle because of its high carboxylase activity and broad substrate range. The carboxylation substrate acrylyl-CoA and crotonyl-CoA in CETCH were regenerated to complete the cycle for continuous fixation of CO2. The fixed carbon is converted to glyoxylate as the output.
The rPS–MCG cycle consists of two parts (Luo et al., 2022). The first utilizes the MCG cycle described above. In the second part, rPS converts acetyl-CoA to pyruvate through a series of reactions that takes two acetyl-CoA to make a crotonyl-CoA, which is carboxylated by CCR to produce a C5 compound. The C5 compound is split into a C3 (pyruvate) and C2 (acetyl-CoA) through a series of carbon rearrangement reactions that complete the cycle. The rPS–MCG cycle exhibits a self-replenishing feature as it can export any of its intermediates as a product, such as acetyl-CoA (C2), pyruvate (C3), and malate (C4). This self-replenishing characteristic is also seen in almost all naturally evolved cycles. Since the output C2, C3, or C4 intermediates are essential for cell growth, it is potentially malleable for in vivo engineering. Introduction of the CETCH cycle or the rPS–MCG cycle in a plant would require the activity of many heterologous enzymes, along with co-enzyme B12, which is absent in plants. Hence, enzyme design, pathway evaluation in prokaryotes, plant-associated microbiome engineering, and various genome editing strategies have been proposed to facilitate this process (Erb et al., 2017; Gupta et al., 2021; Ke et al., 2021).
Engineering carbon dioxide-responsive C3 crops to sustain higher productivity under a CO2-rich, warmer climate
(By Rajeev N. Bahuguna and S. V. Krishna Jagadish)
C4 plant species are overrepresented in agriculture systems and have substantially higher productivity compared to C3 crops mainly due to higher photosynthetic efficiency (Rao et al., 2012; Sales et al., 2021). Yet a number of C3 crops are important food sources for millions of people globally, including cereals such as wheat, rice, barley (Hordeum vulgare), oats (Avena sativa), and many vegetable and tree crops. Therefore, efforts to increase the photosynthetic efficiency and productivity of C3 crops are underway to help meet the increasing global food demand (Cui 2021). The high CO2 saturation point for photosynthesis of C3 plants (intercellular CO2 levels ∼600 µmol mol−1) makes them more responsive to elevated CO2 than C4 plants, which are saturated for CO2 under current atmospheric CO2 levels (Loladze, 2014; Dingkuhn et al., 2020; Kundu et al., 2022). Thus, C3 crops provide a unique opportunity to harvest more carbon from a CO2-rich environment and convert it to biomass and yield (Broberg et al., 2019; Ainsworth and Long, 2021).
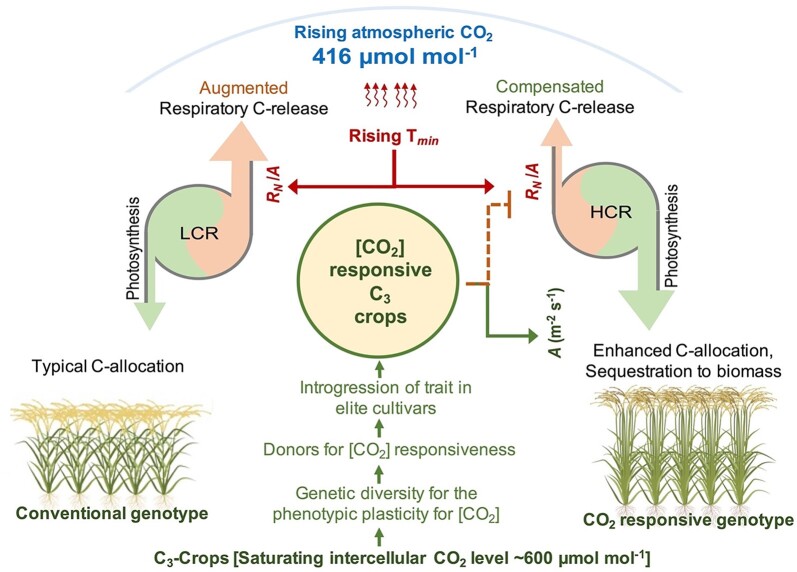
In contrast to the positive effect of CO2 on C3 photosynthesis, the global rise in temperature is a major factor limiting the yield of major cereal crops (Lobell and Gourdji, 2012; Teixeira et al., 2013; Zhao et al., 2017). A rise in night temperature has been shown to have a large impact on the productivity of C3 crops such as rice (Peng et al., 2004; Welch et al., 2010) and wheat (Hein et al., 2020, 2022; Impa et al., 2021). Recent studies suggest that high night temperature (HNT) is related to physiological changes such as an increased rate of night respiration (RN) and a reduced rate of starch accumulation in developing grains in rice (Bahuguna et al., 2017; Shi et al., 2017), wheat (Narayanan et al., 2016a, 2016b; Impa et al., 2020), and barley (García et al., 2015, 2016). Hence, the positive effect of CO2 on C3 photosynthesis and augmented rate of night respiration under HNT have opposing effects on carbon-balance dynamics under CO2-rich, warmer environments (Song et al., 2014; Dusenge et al., 2019). While the sensitivity of RN to a rise in temperature is well documented (Atkin and Tjoelker, 2003), variable effects of the long- and short-term impact of elevated CO2 on RN have been reported, ranging from direct inhibition of respiration to no significant impact or even an increase under long-term exposure to elevated CO2 (Griffin et al., 1996; Gonzalez-Meler et al., 1996, 2004; Ziska and Bunce, 1998; Drake et al., 1999; Baker et al., 2000; Davey et al., 2004; Ayub et al., 2014). However, none of these studies considered the genetic background for CO2 responsiveness, which could be a major determinant of the effect of elevated CO2 on RN, and carbon balance dynamics in C3 crops (Figure 5).
Figure 5.
Schematic diagram showing average annual atmospheric [CO2] level for 2021 and the effect of rising night temperature (Tmin) on rice productivity by enhanced respiration: photosynthesis ratio (RN/A) resulting in augmented release of carbon at the cost of biomass and yield in conventional genotypes. On the contrary, introgression of CO2-responsiveness trait in C3 crops facilitates enhanced carbon sequestration and allocation of additional carbon into biomass, and compensating Tmin-induced carbon losses. LCR, least CO2-responsive; HCR, high CO2-responsive.
Despite the well-documented photosynthetic enhancement of C3 crops under elevated CO2 (Leakey et al., 2009), active selection in C3 crops for CO2 responsiveness has not been given adequate attention (Ziska et al., 2012; Dingkuhn et al., 2020). The complexity of field-based CO2 enrichment facilities and space constraints for screening and characterizing a large number of genotypes remain major bottlenecks for identifying potential CO2-responsive genotypes. Recently, Shimono et al. (2014) and Kikuchi et al. (2017) demonstrated that altering planting density provides a means of assessing phenotypic plasticity in rice genotypes under enhanced resource availability (e.g. space, light, nutrients). Interestingly, genotypes responsive to higher available resources under low planting density responded similarly under an elevated CO2 environment (Shimono et al., 2014). Subsequently, in a series of field experiments, Bahuguna et al. (2022) assessed the variable phenotypic plasticity of 194 diverse rice genotypes by measuring parameters related to photosynthesis, biomass, and yield under different planting densities. A wide genetic variability observed for the phenotypic plasticity under a resource-rich environment showed a strong relationship (R2 = 0.71) with CO2 responsiveness under realistic CO2 conditions using a field-based free air CO2 enrichment facility. Further, the high CO2-responsive (HCR) genotypes showed significantly higher rates of photosynthesis (A) and lower rates of RN resulting in a lower RN/A ratio as compared to the least CO2-responsive (LCR) genotypes. Interestingly, elevated CO2 was identified as the major driver influencing carbon-balance dynamics and the phenotypic response of HCR genotypes resulting in higher biomass and yield under elevated CO2 + HNT conditions, whereas the LCR genotype was severely affected by HNT despite exposure to elevated CO2.
This study demonstrated that the impact of HNT on grain yield, total biomass, and grain weight was compensated by elevated CO2, but this response was mainly confined to the HCR genotypes (Bahuguna et al., 2022). Thus, LCR or conventional genotypes are expected to lose biomass and yield under an elevated CO2, warmer climate due to augmented respiratory carbon losses, whereas HCR genotypes could accumulate more carbon per unit area and maintain their biomass and yield by compensating for carbon losses under HNT (Figure 5). In addition, the ability to fix additional carbon with a lower respiration-to-photosynthesis ratio in HCR genotypes provides an opportunity to sequester a substantial amount of carbon into biomass. There is, however, a need for prediction models for simulated carbon fluxes at temporal and spatial scales to assess the carbon sequestration potential of CO2-responsive C3 crops. In conclusion, the introgression of a ‘CO2-responsiveness’ trait into elite rice varieties and other C3 crops could help sustain and enhance crop yield in a warmer environment.
The C4 rice project
(By Jane Langdale)
In the majority of photosynthetic organisms, both in water and on land, CO2 is fixed by Rubisco into the three-carbon compound 3-phospho-glycerate, the first intermediate of the CBB cycle. The efficiency of this C3 photosynthetic pathway is compromised because Rubisco also reacts with oxygen, forming 2-phospho-glycolate, which has to be detoxified in the energetically costly photorespiratory pathway (Walker et al., 2016). Because of this energetically wasteful competitive reaction, the decrease in atmospheric CO2 levels that occurred during the Oligocene (Pearson et al., 2009) would have been accompanied by photosynthetic inefficiencies at a global scale.
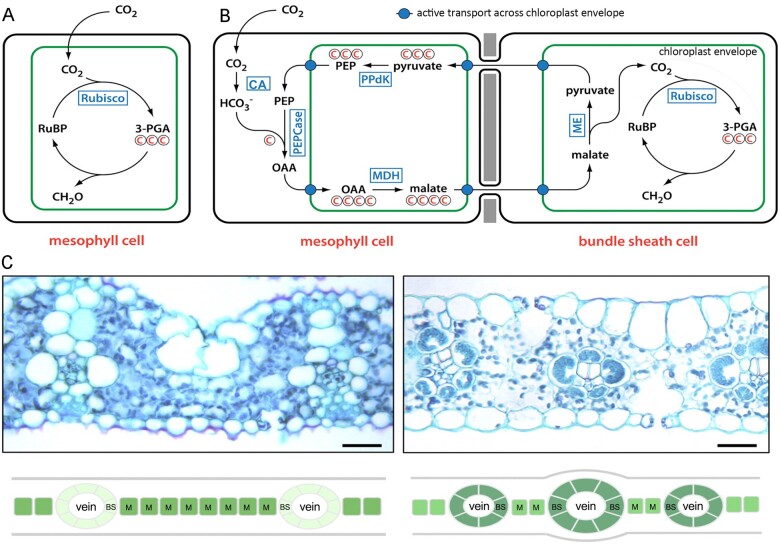
The reported drop from ∼800 to ∼400 ppm atmospheric CO2 during this period is thought to have driven, at least in part, the evolution of the C4 photosynthetic pathway that concentrates CO2 at the site of Rubisco and thus minimizes photorespiration (Sage, 2016). In the C4 pathway, CO2 is initially fixed by phosphoenolpyruvate carboxylase (PEPCase), which is oxygen insensitive. This carboxylation reaction occurs in the outer mesophyll cells of the leaf, with the four-carbon reaction product subsequently transported to inner bundle sheath cells for decarboxylation and re-fixation by Rubisco in the Calvin cycle (Figure 6). Given the specialized leaf anatomy and compartmentalization of metabolic reactions required for C4 function, evolution of the pathway must have involved functional modification of multiple genes, including those encoding enzymes, metabolite transporters, and regulators of cell-type patterning. Despite this apparent complexity, the C4 photosynthetic pathway evolved over 60 times independently and is represented in diverse families of flowering plants (Sage, 2016). The adaptive success of the C4 photosynthetic strategy is demonstrated by the fact that just 2% of plant species utilize the pathway but C4 plants are responsible for ∼25% of terrestrial primary productivity (Still et al., 2003).
Figure 6.
Schematics of C3 CBB and NADP-ME C4 Cycles. A, CBB C3 cycle. B, NADP-ME C4 cycle. C, Transverse leaf sections and corresponding schematics of C3 rice (left) and C4 maize (right). Bars = 30 μm. Adapted from Langdale (2011), Figures 1 and 3.
Why C4 rice?
In addition to strategies that aim to improve the efficiency of the C3 photosynthetic pathway (discussed above, and see Ort et al., 2015; Johnson, 2022) or to introduce Crassulacean acid metabolism into C3 plants (Schiller and Bräutigam, 2021), the enhanced efficiency of C4 photosynthesis provides a potential engineering opportunity for improved yield and resilience against abiotic stresses in C3 crops. Although the C4 pathway utilizes two extra ATP molecules per CO2 fixed than the C3 pathway, in warm and dry environments where dissolved oxygen conditions are relatively high, these energy costs are offset by those not spent on photorespiration (3.5 ATP per O2 fixed). In general, C4 plants also use less water (Kocacinar et al., 2008) and N (Evans and von Caemmerer, 2000) per CO2 fixed and have substantially faster growth rates (Monteith 1978). Physiological models that incorporate these factors predict that if C4 traits could be introduced into C3 plants, enhanced radiation, N, and WUEs could generate substantial yield increases, particularly in warm environments where crops are rainfed and fertilizer applications are limited (Mitchell and Sheehy, 2006).
Importantly, the level of atmospheric CO2 at which C4 outcompetes C3 is dependent on temperature; C4 is favored below 550 ppm CO2 at 35°C, 450 ppm at 30°C, and 350 ppm at 25°C (Ehleringer et al., 1997). Although future predictions of atmospheric CO2 levels differ depending on fossil fuel usage scenarios, with current levels at 419 ppm and annual increases of 2–3 ppm over the last decade (https://gml.noaa.gov/ccgg/trends/gl_gr.html), the status quo would result in atmospheric CO2 levels of ∼500 ppm by 2050. C4 plants could thus outperform C3 plants where temperatures exceeded ∼33oC, which given climate warming predictions could be much of the global agricultural landscape for at least part of the year. Leaving predictions aside, long-term field experiments at elevated (+180 ppm) CO2 demonstrated that the biomass of C3 but not C4 grasses was enhanced over the first 12 years of the project but then C4 outperformed C3 in the following 8 years (Reich et al., 2018). This switch was correlated with net N mineralization rates in the soil, which were initially enhanced by elevated CO2 in C3 plots but were later depressed. Despite the difficulties in predicting exactly how plants will respond to global change, C4 engineering is thus a plausible strategy, albeit one with significant challenges.
Strategy
The C3 species rice is an obvious target for C4 engineering because it is one of the world’s top three staple crops and in many parts of Asia it is the major source of calorie intake. With predicted population increases, the one hectare of land that provided enough rice to feed 27 people in Asia in 2007 will need to support at least 43 people by 2050—a 60% increase in demand (Zeigler, 2007). Successful conversion of a C3 plant into one that utilizes the C4 pathway requires that leaf anatomy be modified to reduce the number of mesophyll cells between veins to the extent that there is an approximate 1:1 ratio of mesophyll:bundle sheath cells in the leaf; that chloroplast development is activated in the normally achlorophyllous bundle sheath cells; and that C4 pathway enzymes and metabolite transporters are compartmentalized and functional in either the mesophyll or bundle sheath cells.
When the C4 Rice Project (www.c4rice.com) was initiated, genes encoding all of the enzymes of the C4 pathway had been identified in maize and other C4 species, as had some of the genes encoding metabolite transporters (reviewed in Langdale, 2011), but regulators of C4 leaf anatomy had not been identified. The strategy to introduce C4 traits into rice was thus three-pronged: (1) introduce compartmentalized C4 metabolism into existing bundle sheath cells and the mesophyll cells immediately adjacent to them by expressing maize genes in specific cell-types of rice; (2) activate chloroplast development and photosynthesis in existing bundle sheath cells by expressing a known regulator of chloroplast development in maize (the Golden2 [ZmG2] gene; Hall et al., 1998); and (3) identify regulators of C4 leaf anatomy in maize with a view to future manipulation in rice. The ultimate goal was to combine the metabolic prototypes generated in the first two strands with the anatomical prototype.
Much of the first decade of the project was spent developing tools in rice to enable this strategy, for example robust transformation pipelines, cell-type-specific promoters, and modular cloning technology. Ongoing research continues to characterize potential regulators of C4 leaf anatomy and to evaluate whether manipulation in rice can modify cell-type patterning in the leaf (Wang et al., 2013a, 2017a; Schuler et al., 2018; Hughes et al., 2019; Hughes and Langdale, 2020, 2022)—but much more discovery research is needed before an anatomical prototype can be designed and engineered (Sedelnikova et al., 2018). Recent work has, however, made progress toward engineering C4 metabolic prototypes.
Progress
Maize genes encoding C4 pathway enzymes have been expressed in specific cell types of both an elite cultivar of indica rice (IR64) and a model cultivar of japonica rice (Kitaake), and in both cases the pathway is partly functional. Specifically, primary carboxylation by PEPCase is seen in mesophyll cells, but subsequent decarboxylation in bundle sheath cells has yet to be detected (Lin et al., 2020; Ermakova et al., 2021). Creating a fully functional cycle will require a better understanding of metabolite flux within and between the two cell types, which may require the development of more sensitive detection methods. In a second advance, chloroplast development has been activated in the normally achlorophyllous bundle sheath cells of rice, through constitutive expression of ZmG2 (Wang et al., 2017b). No fitness penalty was observed in greenhouse-grown lines expressing ZmG2, in either IR64 or Kitaake backgrounds (Wang et al., 2017b) and although only evaluated in the nonelite Kitaake background, field-grown lines overexpressing ZmG2 exhibited up to 30% yield increases (Li et al., 2020). These examples validate the overall engineering strategy but there is still a long way to go before a full transition to C4 can be achieved in any C3 species.
Can we develop climate-resilient crops?
The trait development pipeline: Bridging the gap between upstream science and breeding for adaptation to climate change
(By J. Damien Platten and Amelia Henry)
Improving the adaptability of crops is a key strategy to mitigate the effects of climate change on productivity (Aggarwal et al., 2019). We focus on rice breeding in this section; however, the pipeline we describe (Figure 7) could easily be extended to other crops, taking into consideration the challenges and parameters unique to each species. For example, the platform is being adopted across the CGIAR partnership for global food security (https://www.cgiar.org/) for other mandate crops. In rice breeding, abiotic stress tolerance was not a selection target during the Green Revolution, and some evidence suggests that stress tolerance was even selected against due to tight linkage between stress tolerance loci and loci conferring unfavorable agronomic traits (Vikram et al., 2015). Subsequently, a range of breeding approaches has been taken to improve stress tolerance, including introgression of quantitative trait loci (QTLs) for stress tolerance traits as well as direct selection for grain yield under stress using traditional varieties as the sources of stress tolerance. Characterization of stress-tolerant varieties has revealed that combinations of physiological traits have been affected by selection for yield under stress (Anantha et al., 2016; Kumar et al., 2021), which may explain some of the difficulty in developing superior varieties through introgression of single genes/QTLs. With a few exceptions (i.e. Sub1 varieties for submergence such as Swarna-sub1 [Mackill et al., 2012] and drought-tolerant DRR dhan 42 [IR64 qDTY2.2 + qDTY4.1; Swamy et al. 2013]) the majority of recently released stress-tolerant varieties were conventionally bred (i.e. by crossing and selecting over several generations).
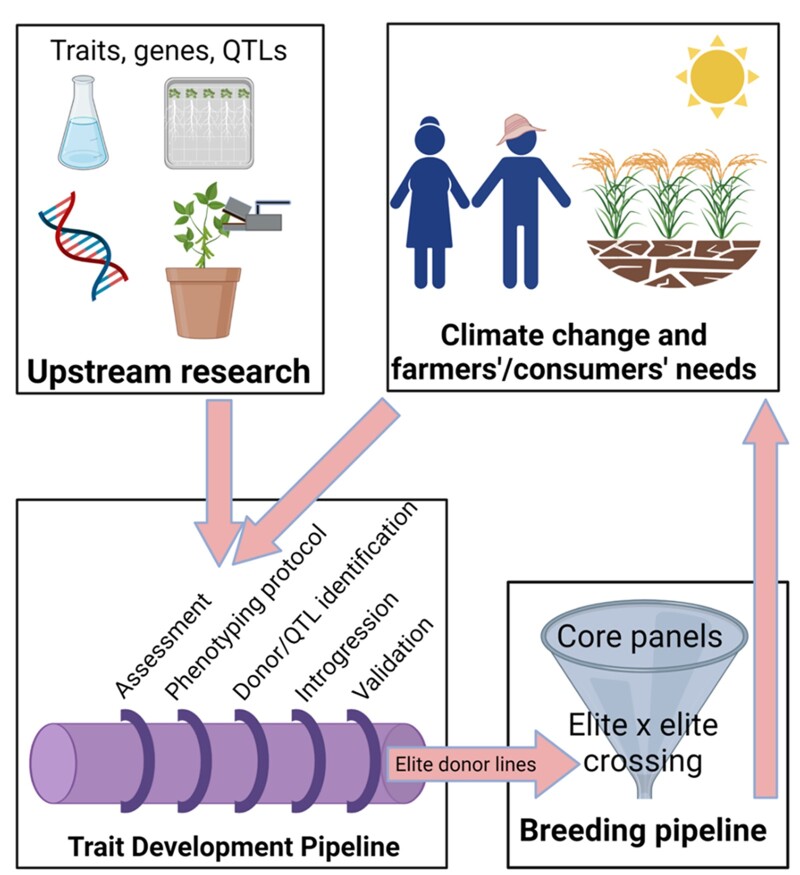
Figure 7.
The Trait Development Pipeline for delivery of valuable stress tolerance traits/genes/QTLs from upstream research into the elite breeding pool for improvement of crop productivity under climate change. The Trait Development Pipeline is organized into six stages: 1) guidelines for prioritizing traits (assessment), 2) defining standards for phenotyping protocols, 3) identifying donors and QTL (including refining marker quality metrics), 4) introgressing and 5) validating traits/genes/QTLs into elite genetic backgrounds to develop the elite donor lines that are 6) handed to the breeding program for crossing. Those elite donor lines will then be systematically crossed and tested in target environments where climate change is increasingly affecting the degree of abiotic stress affecting crop production. Created with BioRender.com
The use of genes and QTLs through marker-assisted selection could shorten the breeding process. Although hundreds of stress-tolerance genes, QTLs, and physiological mechanisms have been identified, only a small number of these research outputs have been used in breeding (Wissuwa et al., 2016; Cobb et al., 2019; Platten et al., 2019) and the frequency of known abiotic stress QTLs in the current elite breeding material remains low (Juma et al., 2021). There is thus a need to bridge the gap between upstream science and breeding for adaptation to climate change so that valuable traits/genes/QTLs are more actively utilized in breeding pipelines.
Modern breeding strategies have shifted to a paradigm of population improvement based on elite x elite crossing (Juma et al., 2021) within core panels, which for stress-prone areas have been selected from the most stress-tolerant genotypes available (i.e. Khanna et al., 2022). This strategy presents significant opportunities for upstream plant biologists to contribute to breeding efforts. With a defined list of genetic backgrounds (many of which have already been sequenced; see, for example, Mansueto et al., 2017) to which potential stress tolerance traits/genes/QTLs can be compared, those that best complement the elite breeding pool can be prioritized. However, although traditional varieties are the most promising source of stress tolerance, they also typically possess detrimental traits that make them unsuitable for use in elite × elite crossing. A defined protocol is needed to deliver useful traits/genes/QTLs from traditional varieties into elite backgrounds and into the breeding pool.
In seeking to bridge the gap between upstream plant science and breeding for stress tolerance, an understanding of breeding program needs is critical. In the briefest terms, reliability is key: a gene/QTL must reliably improve the target trait, in relevant elite genomic backgrounds and in relevant environments (field locations). Therefore, the growth stages, genetic backgrounds, and environmental conditions relevant to breeding programs should be reflected in the study systems used in upstream research. One example is in the validation of candidate genes for stress tolerance: this is frequently done in the background of japonica rice due to the established transformation protocols. However, the rice type preferred in most stress-prone rice-growing regions is indica, which grows better than temperate japonica rice in field trials in the tropics. Use of the relevant genetic background is important because stress tolerance alleles are often absent in japonica genomes and thus their level of stress tolerance is more easily improved, exaggerating apparent effect size. Other recommendations for increasing the likelihood of upstream research outputs being taken up by breeding are to take additional steps such as validation of identified QTLs in relevant elite genomic backgrounds and to link with researchers who can evaluate the material under field conditions.
On the other side of the gap, downstream science must make released varieties and advanced breeding lines more accessible to upstream scientists. Familiarity with this material is important because in some cases, key genes have been identified and advocated as promising breeding targets without the recognition that they are already present in the breeding pool. Furthermore, the availability of improved material to upstream researchers will help to ensure that target loci are effective in those genetic backgrounds. Downstream science also must stay up to date and gain access to the most recently identified traits/genes/QTLs to incorporate into the breeding program, while at the same time effectively connecting with local breeders who can conduct widespread testing and who understand the needs of farmers in stress-prone regions. This “bridge building” among scientific disciplines is critical to the development of more efficient pipelines that bring novel improvements to crops for climate change-affected farmers.
To strengthen linkages between upstream and downstream development efforts, a framework has been developed that organizes and codifies trait development efforts. This “Trait Development Pipeline” developed at the International Rice Research Institute (Figure 7) applies stage gate systems widely used in industry (Covarrubias-Pazaran et al., 2022) to assess trait development progress against defined advancement criteria. The organization by stages enables external review of the progress at each stage and provides decision points on whether to proceed, giving an opportunity to discontinue efforts that are not likely to make an impact in breeding programs.
The current Trait Development Pipeline is organized into six stages that link a variety of research disciplines, and the pipeline provides a structure that gives a framework for teamwork between these areas. The pipeline starts with an assessment of the trait of interest in the context of priority traits needed by farmers and consumers that are not already present in the elite breeding pool (see “Product concepts” and “Market segments”; Covarrubias-Pazaran et al., 2022). A set of criteria regarding the availability and reliability of phenotyping protocols for the trait, potential donor genotypes, mapping populations, QTLs, and markers determine advancement to subsequent stages in the Trait Development Pipeline. The pipeline is dynamic and subject to modification over time based on researcher feedback and as techniques and technologies change. The outputs of the Trait Development Pipeline are validated donor lines containing new traits/genes/QTLs in a fully elite background. These “elite donor lines” can be used in the elite × elite crossing work to improve the most advanced breeding lines which will be evaluated in multilocation trials, evaluated by local researchers, and considered for release as varieties for dissemination to farmers. In this way, the outputs of trait/gene/QTL discovery realize an ongoing impact across the breeding pool rather than improving just a single variety. Such sustained improvement through mainstream breeding programs will facilitate the deployment of new technologies to as many climate-change-affected crop production market segments as possible.
Application of the pipeline to known genes and QTLs helps to identify gaps in knowledge and products available, and addressing these gaps is already enabling the rapid introduction of a wide variety of genes contributing to disease resistance, heat, drought, cold, and salinity tolerance into mainstream rice breeding efforts. Breeding programs are thus able to respond in a far more agile manner to changing climate and market demands. As these genes are deployed into elite backgrounds, it becomes easier for small breeding programs to also leverage their value; the “heavy lifting” of eliminating highly unfavorable genomic backgrounds, breaking linkage drag, and developing coupling-phase linkages has been done, so only minimal or no additional effort is required to move the new genes to other elite breeding programs. Thus, the value of new genes is no longer exclusively available to large, well-resourced programs.
Enhancing climate resilience through the use of crop wild relatives
(By Damaris A. Odeny)
Crop wild relatives (CWRs) are wild species that are closely related to domesticated crops and can be used for crop improvement. Breeders have traditionally used CWRs as sources of superior traits, including key traits for enhancing adaptation to climate change (Dempewolf et al. 2014). The main breeding objectives for climate change adaptation include resilience to abiotic stresses (drought, heat, salinity, and flooding/waterlogging) and biotic stresses brought about as a result of the increase in atmospheric CO2 and elevated average temperatures. Here, we provide examples of recent progress in the use of CWRs in managing these stresses and highlight specific areas where work is needed. Table 1 provides a summary of the use of CWRs in breeding for tolerance to abiotic stresses.
Table 1.
Examples of wild relatives used to enhance abiotic stress tolerance in cultivated crops
| Crop | More resilient wild species | Trait of interest | Reference |
|---|---|---|---|
| Wheat | Aegilops cylindrica | Drought | Pour-Aboughadareh et al. (2017) |
| Ae. crassa | |||
| Ae. caudata | |||
| Triticum urartu | |||
| T. monococcum | Heat | Khanna-Chopra and Viswanathan (1999); Peng et al. (2013); El Haddad et al. (2021) | |
| T. dicoccoides | |||
| Ae. speltoides ssp. liqustica | |||
| T. ararticum | |||
| Ae. speltoides | Salinity | Ahmadi et al. (2018) | |
| Ae. caudata | |||
| Ae. cylindrica | |||
| T. boeoticum | |||
| Sorghum | Sorghum macrospermum | Drought | Cowan et al. (2020) |
| S. brachypodum | Ochieng et al. (2020) | ||
| S. arundinaceum | |||
| S. sudanense | |||
| S. purpureosericeum | |||
| Banana | Musa balbisiana; | Drought | Eyland et al. (2022) |
| M. acuminata ssp. errans | |||
| Rice | Oryza rufipogon | Salinity | Tin et al. (2021) |
| O. nivara; O. coarctata | Zhang and Xie (2014) | ||
| O. nivara; O. rufipogon | Flooding | Niroula et al. (2012) | |
| O. meridionalis Ng. | Heat | Scafaro et al. (2012); Scafaro et al. (2016); Scafaro et al. (2018) | |
| O. australiensis | |||
| Maize | Zea nicaraguensis | Flooding | Mano et al. (2006); |
| Z. luxurians | Mano et al. (2005); | ||
| Z. mays ssp. huehuetenangensis | |||
| Z. diploperennis | Drought | Shaibu et al. (2021) | |
| Tomato | Solanum cheesmaniae; | Salinity | Dehan and Tal (1978); Shalata and Tal (1998); Mittova et al. (2002); Frary et al. (2010); Pailles et al. (2020) |
| S. pennellii | |||
| S. galapagense | |||
| S. pimpinellifolium | Heat | Driedonks (2018) | |
| Tepary bean | Wild Phaseolus acutifolius | Drought | Buitrago-Bitar et al. (2021) |
| Adzuki bean | Vigna nakashimae; V. riukiensis | Salinity | Yoshida et al. (2016) |
| Eggplant | Solanum insanum | Salinity | Brenes et al. 2020) |
| Chickpea | Cicer reticulatum | Drought | Moenga et al. (2020) |
| Sugarcane | Saccharum spontaneum | Salinity | Kasirajan et al. (2021) |
Drought stress
Despite the complexity of drought stress (Ilyas et al., 2021), CWRs have been reported that are more efficient than crop relatives in drought-related physiological processes such as higher WUE, higher CO2 assimilation, deeper root systems, more efficient regulatory networks, leaf curling, and stomatal closure, as well as showing an abundance of allelic diversity within candidate genes. For example, higher WUE, higher carbon assimilation, and greater carboxylation efficiency were reported in wild lettuce (Lactuca serriola; Eriksen et al., 2020), and Moenga et al. (2020) reported novel divergent drought tolerance mechanisms in wild chickpea (Cicer reticulatum) that would be a great resource for improving cultivated chickpea (Cicer arietinum). Drought-related transcription factors of the Asr (abscisic acid, stress, ripening) family have a high level of diversity in CWRs (Cortés et al. 2012) that might be further exploited to improve cultivated crops. Most drought studies to date in CWRs have focused on major crops, and there is tremendous scope to undertake similar studies in minor and under-researched crops.
Heat stress
Heat stress is one of the greatest concerns for crop production considering the increasing effects of climate change. The wild wheat relatives Triticum monococcum, T. dicoccoides, and Aegilops speltoides ssp. liqustica and CWR-derived wheat genotypes were among the most heat tolerant when tested alongside elite wheat genotypes (Peng et al., 2013; El Haddad et al., 2021). Similar observations have been made in wild rice, Oryza meridionalis Ng. and O. australiensis, in which heat tolerance was associated with a more stable activation of Rubisco (Scafaro et al., 2016). Overexpressing a thermostable variant of Rubisco activase from CWR significantly improved yield in domesticated rice (Oryza sativa L.; Scafaro et al., 2018). More studies on the physiological and molecular basis of heat tolerance in wild versus domesticated species are needed to enhance the deployment of novel heat tolerant alleles in crop improvement.
Salinity tolerance
Halophytic plants adapt to salinity through three distinct mechanisms, all of which have been identified in various CWRs: osmotic stress tolerance, Na+ or Cl− exclusion, and tolerance of tissue to accumulated Na+ or Cl−. Wild relatives of adzuki bean, Vigna nakashimae and V. riukiensis, prevented Na+ accumulation in roots and stems, and tolerated accumulated Na+, respectively (Yoshida et al., 2016). A wild relative of tomato, Solanum pennellii, showed greater induction of antioxidant activity than cultivated tomato (Solanum lycopersicum L.) under salt stress (Frary et al., 2010). Salinity tolerance has also been reported in Oryza glaberrima (Platten et al., 2013), Hordeum spontaneum (Kiani-Pouya et al., 2020), and in Aegilops spp. (Zamani Babgohari et al., 2013).
Flooding tolerance
Flooding tolerance has been mainly studied in rice leading to the identification of the SUB1 locus (Mackill et al., 2012). Additional submergence-tolerant alleles (SUB1A-1) were identified from wild rice species O. nivara and O. rufipogon, together with a likely presence of other submergence mechanisms in other wild rice accessions (Niroula et al., 2012). Two anaerobic germination QTLs (qAGP1 and qAGP3) from O. nivara introgression lines (Liu et al., 2021a) potentially can be used to enhance flooding tolerance in elite SUB1 genotypes, which are not always tolerant to anaerobic conditions during germination. Wild species with tolerance to waterlogging/stagnant flooding have been reported to possess unique alleles for aerenchyma formation (Zhang et al., 2017), or to provide a stronger barrier to radial oxygen loss (Pedersen et al., 2021). The availability of these different sources of flooding/waterlogging resistance in CWRs provides an opportunity to introgress the beneficial alleles into elite varieties, especially where genomics-assisted introgression and selection is possible.
CWRs as sources of resistance/tolerance to biotic stress
Introgression of disease resistance genes into cultivated crop species is perhaps the most beneficial use of CWRs in crop improvement to date. Major genes have been introgressed from CWRs for resistance to late blight (Phytophthora infestans) in potato (Solanum tuberosum L.; Ghislain et al., 2019), blast disease (Magnaporthe oryzae) resistance in rice (Yoshida and Miyashita, 2009) and several other key pathogens in wheat (Rani et al., 2020) and tomato (Sharlach et al., 2013), just to mention a few. Pest resistance also benefited from CWRs (Therezan et al., 2021), among the most recent being the introduction of fall armyworm (Spodoptera frugiperda) resistance from wild relatives of maize (Singh et al., 2022). Climate change-related warmer average temperatures and altered weather patterns are contributing to altered patterns in the occurrence of crop pests and pathogens and the emergence of new pests and pathogens around the globe, as explored in more detail in the section by Rim et al. below. More studies will be needed to focus on the introgression of quantitative resistance from wild to cultivated species to improve the durability of resistance to various diseases and pests.
De novo domestication for resilience to climate change
Despite several wild relatives having remarkable tolerance to biotic and abiotic stresses, successful introgression of these traits into elite backgrounds has been difficult due to linkage drag (Nevo and Chen, 2010) and the complexity of most traits. De novo domestication, the incorporation of domesticated genes into the nondomesticated species to develop new crops (Razzaq et al., 2021), presents a novel opportunity for immediate utilization of the novel resilience alleles in CWRs. The availability of vast genomic and phenomic resources allow for machine learning (Niazian and Niedbała, 2020) and more precise genome editing (Hua et al., 2019). An excellent example of de novo domestication has been reported in Solanum pimpinellifolium (Zsögön et al., 2018). There are now several countries that have exempted genome-edited plants from genetically modified organism regulations, making it possible to utilize de novo domesticated plants as soon as they are generated.
Development of disease-resistant crops for a changing climate
(By Ellen Youngsoo Rim, Alexandra M. Shigenaga, Pamela C. Ronald)
Plant reactions to a single stress differs from those of plants exposed to combined abiotic and biotic stresses, with a shift in signaling pathways and transcriptomic responses (Atkinson and Urwin, 2012; Prasch and Sonnewald, 2013; Sharma et al., 2013). Thus, understanding how plants respond to pathogen stress under nonoptimal environmental conditions is essential for the development of resilient crops in a changing climate (Chaloner et al., 2021; Velásquez et al., 2018).
Diverse plant–pathogen interactions have been shown to be affected by adverse environmental conditions, leading to increased host susceptibility, or in some cases, increased host resistance (Velásquez et al., 2018). Various plant species become more susceptible to fungal, viral, or bacterial pathogens in response to elevated temperatures (Cohen and Leach, 2020; Velásquez et al., 2018). For example, exposure to elevated temperature combined with drought stress led to greater susceptibility to Turnip mosaic virus due to downregulated defense response gene expression (Prasch and Sonnewald, 2013). However, there are examples where high temperatures led to enhanced host resistance against pathogen pressure (Venkatesh and Kang, 2019). Similarly, the impact of increased atmospheric CO2 in relation to plant–pathogen interactions remains unsettled. For example, high CO2 concentrations led to increased susceptibility of wheat to fungal infection (Váry et al., 2015), whereas soybean (Glycine max) exhibited either enhanced or reduced susceptibility to infection, depending on the pathogen studied (Eastburn et al., 2010). Deeper insight into the complex interplay among abiotic and biotic stresses will inform ongoing work to mitigate crop damage caused by extreme climate conditions or pathogens.
One mitigation strategy is the application of beneficial microbes that enhance plant health and immunity. For instance, Actinobacteria in the genus Streptomyces are enriched in the root microbiome of plants under drought stress (Naylor et al., 2017). Application of Streptomyces strains to seeds improved wheat growth and yield in drought field conditions (Yandigeri et al., 2012). Beneficial strains of Trichoderma fungus heightened plant immunity and antagonized pathogenic fungi (Tyśkiewicz et al., 2022). Soil application of Trichoderma reduced fungal infection in soybeans, tomatoes, peanuts (Arachis hypogaea), and other crops (Zin and Badaluddin, 2020). Inoculation with Trichoderma has also been shown to enhance tolerance to abiotic stresses such as drought and salinity (Zhang et al., 2016; Scudeletti et al., 2021). Biocontrol strategies present an opportunity to enhance resilience to environmental stress and disease pressure, while reducing the use of chemical pesticides or fertilizers that can further damage the environment.
Another strategy well-aligned with sustainable agriculture practices is the introduction of genetic improvements that protect crops against pathogens and confer resilience to abiotic stress in a heritable manner. There are numerous examples of genetic alterations in crops leading to resistance to specific pathogens, many of which were achieved through the introduction of immune receptors (Ercoli et al., 2022). Immune receptors are activated by direct or indirect interaction with microbial molecules to elicit host defense responses. Advances in genome sequencing and analysis have accelerated the discovery of immune receptors and other beneficial genetic traits in cultivated crop varieties and their wild relatives (Ercoli et al., 2022; Zsögön et al., 2022; and discussed in the section above by Odeny). For instance, a previously unknown variant of the immune receptor FLS2 was identified in the genome of a wild grape species (Vitis riparia; Fürst et al., 2020). The introduction of the new FLS2 variant conferred resistance to Agrobacterium tumefaciens in tobacco, offering a potential strategy to control crown gall disease, which affects many crops including nut trees and grapevines. Once identified, desirable genetic traits can be introduced into crops through methods such as marker-assisted breeding or genetic engineering. The use of gene-stacking to introduce multiple protective genes into a single background will likely be an important consideration in engineering climate resilience (Figure 8).
Figure 8.
Development of crops with enhanced resilience to abiotic and biotic stress. Crops are exposed to a variety of stresses. Abiotic stresses will intensify as the following climate conditions change: water availability, precipitation, temperatures, and atmospheric CO2 levels. Biotic stressors that plants encounter will vary, but may consist of: bacteria, fungi, oomycetes, nematodes, viruses, and insect pests. As climate change alters environmental conditions and plant-pathogen interactions, strategies to develop more climate-ready and disease-resistant crop varieties include breeding or genome engineering approaches with stacking disease resistance genes, stacking climate tolerance and disease resistance genes, and/or addition of beneficial microbes (see text for examples).
Genome engineering tools such as CRISPR–Cas9 and transcription activator-like effector nucleases allow greater control over the sequence and genomic location of these genetic changes. Crop engineering is an especially promising avenue for mitigating vector-borne plant diseases, which are anticipated to rise as higher temperatures expand the geographical distribution and survival of insect pests (Perilla-Henao and Casteel, 2016; Huang et al., 2020; Skendžić et al., 2021). For instance, introduction of proteins that target the insect vector or the pathogen itself can confer host resistance. Expression of antimicrobial proteins that bind the membrane of Xylella fastidiosa, the causative agent of Pierce’s Disease, decreased disease incidence in grapevines (Vitis vinifera; Dandekar et al., 2019). Such genetic strategies may lead to more effective and sustainable management of vector-borne diseases, which have relied heavily on chemical insecticides.
Introduction of beneficial traits, however, have mainly focused on the development of crop varieties with resistance to a single stress. Major hurdles remain in engineering crops with combined stress tolerance (Steinwand and Ronald, 2020). For one, enhanced stress tolerance is often accompanied by fitness costs, such as reduced plant growth and yields (Velásquez et al., 2018; Venkatesh and Kang, 2019). Additional genetic interventions can reduce detrimental effects of overactive defenses. For instance, growth penalties associated with powdery mildew resistance in wheat were reversed upon ectopic activation of genes encoding sugar transporters through a mechanism yet to be elucidated (Li et al., 2022). In another example, necrosis associated with broad spectrum potato virus resistance was eliminated through mutating the regulatory region of the resistance-conferring immune receptor (Harris et al., 2013). Alternatively, protective genes can be designed to be expressed under specific conditions. High temperatures increase susceptibility of the model plant Arabidopsis to the bacterial pathogen Pseudomonas syringae pv. tomato DC3000 (Pst; Wang et al., 2009; Huot et al., 2017). Disease resistance genes expressed under a heat-inducible promoter protected against Pst infection after exposure to high temperature without pleiotropic growth defects (Leng et al., 2021). Another challenge is that pathogens can overcome resistant traits by developing novel virulence strategies or by evolving mechanisms to evade detection by existing immune receptors. Gene stacking might be used to delay or prevent the evolution of resistance-breaking pathogens under diverse climate stresses.
A promising avenue to simultaneously reduce crop loss to pathogen and environmental stress is introducing disease resistance in the context of climate resilience (Rivero et al., 2022; Figure 8). Identification of resistance genes that are more effective under abiotic stress, such as increased temperature, is one approach (Chen et al., 2018; Dossa et al., 2020). For example, stacking the rice disease resistance genes Xa4 and Xa7 provided enhanced resistance to Xanthomonas oryzae pv. oryzae (Xoo), the causal agent of bacterial blight disease, under high temperature conditions (Dossa et al., 2020). Alternatively, disease resistance genes can be introduced into crop varieties that already sustain high resilience to abiotic stress. The introduction of six genes associated with resistance to the fungal pathogen Magnaporthe oryzae, the causal agent of rice blast disease, into a rice variety with elevated drought tolerance resulted in plants that are both resistant to blast infection and tolerant of drought stress in the field (Carrillo et al., 2021). Another approach is stacking genes, through breeding or genome engineering, to confer both abiotic stress tolerance and disease resistance. For example, submergence (Sub1) and salt (Saltol) tolerance genes were stacked with eight pathogen and pest resistance genes in an elite rice line (Das and Rao, 2015); this line showed resistance to M. oryzae, Xoo and gall midge, as well as tolerance of submergence and salinity. In some cases, genes can act as regulatory hubs to control abiotic and biotic signaling pathways and are particularly valuable candidates to target for crop engineering (Husaini, 2022). For example, the rice transcription factor MADS26 orchestrates abiotic and biotic stress responses. Downregulation of MADS26 led to enhanced resistance to M. oryzae and Xoo as well as drought tolerance in the field (Khong et al., 2015). The effectiveness of each of these strategies to develop resilience to multiple types of stress will vary depending on the crop, pathogen, and environmental conditions. Research on various strategies and their evaluation under field conditions will be crucial to combat the negative effects of climate change on agricultural systems.
Mycorrhizal and rhizobial symbioses under climate change challenges
(By Xiaowei Zhang, Ertao Wang)
N cycling strongly influences climate change as it is closely correlated to the production of CO2, N2O, and CH4. Currently, crop productivity is highly dependent on fertilizer application, particularly N, which has negative environmental effects through nitrate run-off and release of the potent GHG nitrous oxide. The development of high-yielding, disease-resistant crops can be aided significantly by improving associations with symbiotic microorganisms that enhance nutrient assimilation in the host plant. Here, we summarize the potential application of engineered mycorrhizal and rhizobial symbioses in developing self-fertilizing crops and maintaining sustainable agriculture in the era of global climate change (Figure 9).
Figure 9.
Mycorrhizal symbiosis and N self-fertilizing crops. A, Positive effects of mycorrhizal symbiosis. The mycorrhizal hyphal network forms a mycorrhizosphere (light brown) which can enlarge the plant nutrient absorption area and supply a convenient zone for root-related microbes. Benefits from mycorrhizal symbiosis include increased tolerance or resistance to abiotic or biotic stresses. B, Three steps to develop N self-fertilizing cereal crops to enhance climate change resilience. (1) Increasing associative N fixation. The mucilage (light green) is rich in carbohydrates and harbors abundant diazotrophic microbiota (pink). Engineered cereal plants (such as maize) have the ability to produce rhizophine, which can be perceived by engineered diazotrophs (orange). (2) Transferring symbiotic N fixation to cereal plants. Cereal crops are engineered for symbiotic N fixation by expressing the chimeric receptors perceiving rhizobia signals and overexpressing key symbiotic regulators (CSSP genes, CRE1, etc.) and nodule development genes (SCR-SHR, LBD16, etc.) to form nodule-like structures. (3) Autonomous N fixation in cereal crops. The ideal plant which could assimilate N2 into ammonium is created by overexpressing rhizobial N fixation genes in plant cells.
Mycorrhizal symbiosis
Plant roots are associated with diverse microbes, including bacteria, fungi, and viruses collectively called the rhizosphere microbiome. Among them, mycorrhizal fungi are known to improve plant access to nutrients, particularly phosphorus and N. Two major groups are arbuscular mycorrhizal fungi (AMF), which colonize host roots and are widely distributed in plants, and ectomycorrhizae, mainly associated with trees and shrubs (Genre et al., 2020). The soil region influenced by mycorrhizal roots is called the mycorrhizosphere (Priyadharsini et al., 2016), where mycorrhizal fungi sequester carbon and form aggregate particles in soil that have a major impact on the composition of microbial and plant communities (Priyadharsini et al., 2016; Wang et al., 2021). Under a warmer climate, mycorrhizal fungi can increase carbon sequestration by influencing the root/shoot ratio (Zhou et al., 2022). Ectomycorrhizal fungi can significantly affect the carbon sequestration capacity of certain soils, for example in boreal forests (Clemmensen et al., 2015; Genre et al., 2020). Colonization by AMF can mitigate adverse effects of drought and salt stress by improving nutrient uptake, minimizing oxidative damage, and increasing osmotic adjustment (Hameed et al., 2014; Klinsukon et al., 2021).
Several challenges restrict the application of AMF in agriculture as a biofertilizer. Plants vary widely in response to individual mycorrhizal fungi (Klironomos, 2003), and results of research focusing on one or few AMF species under controlled conditions may not translate to field conditions. In addition, many aspects of the signaling and nutrient exchange pathways are shared between mycorrhizal fungi and biotrophic pathogens (Wang et al., 2012; Zhang et al., 2015; Jacott et al., 2017; Jiang et al., 2017; Zhang et al., 2021a). Thus, it is critical to dissect how plants distinguish between AMF- and pathogen-influenced agronomic traits and engage productively with symbiotic microorganisms while simultaneously restricting pathogens.
Mycorrhizal fungi strongly influence host plant phosphorus acquisition. Interestingly, the phosphate starvation response was found to be a core regulator in both a direct phosphate uptake pathway via root hairs and epidermis and an indirect phosphate uptake pathway via mycorrhizal symbiosis (Shi et al., 2021), suggesting the possibility of developing crops that use phosphorus and N more efficiently by coordination of the direct phosphate uptake pathway and mycorrhizal pathway in future.
N fixation in legumes and nonlegumes
Biological N fixation is the process by which nitrogenase (an enzyme found only in certain prokaryotes known as diazotrophs) converts dinitrogen gas from the atmosphere into ammonia and is the main path for the formation of combined N in nature. Three forms of N fixation are found in nature: free-living, associative, and symbiotic (Soumare et al., 2020). In associative N fixation, N-fixing microorganisms living on the surfaces or in the interstitial spaces of the plant host use photosynthetic products from the plant as carbon sources to fix N for their own use and provide the excess fixed N to the host (Soumare et al., 2020). In symbiotic N fixation, N-fixing bacteria colonize the cells of plant organs such as root nodules and supply N to support host growth and development, in systems such as Rhizobium/legume, Frankia/alder, and Cyanobacteria/Australian cycads (Pankievicz et al., 2019; Soumare et al., 2020). The rhizobium–legume symbiosis is the most important N fixation system in terrestrial communities.
Three mechanisms have been proposed to develop N self-fertilizing cereal crops to enhance climate change resilience (Figure 9B):
1. Increasing associative N fixation
Diazotrophs are present in the carbon-enriched mucilage in maize aerial roots and were found to contribute 29%–82% of the N nutrition of Sierra Mixe maize in a 5-year field experiment (Van Deynze et al., 2018). Engineering the cereal host and/or the diazotrophs to enhance this association is therefore a promising avenue to increase biologically fixed N in crops.
It has been shown that the engineered expression in Medicago truncatula and barley (Hordeum vulgare) of rhizopine, a small molecule compound synthesized by a few rhizobia, could be sensed by engineered bacteria Azorhizobium caulinodans ORS571 with a 103-fold increase in perception sensitivity (Geddes et al., 2019; Haskett et al., 2022). This provides the possibility of increasing N fixation from endophytic and free-living bacteria associated with crop plants, although the in situ nitrogenase activity was suboptimal. Further experiments should explore optimizing the expression levels of rhizopine biosynthetic genes to reduce fitness costs in host plants due to excessive gene expression.
2. Transferring symbiotic N fixation to cereal plants
The association of legumes with N-fixing bacteria requires several molecular processes common to the mycorrhizal associations that are more widespread in plants, showing that the evolution of the Rhizobium–legume symbiosis utilized many existing processes that facilitate mycorrhizal interactions (Roy et al., 2020; Wang et al., 2022). This close relationship provides a possibility to engineer symbiotic N fixation into non-legume cereal crops by synthetic biology (Mus et al., 2016). Some progress has been made toward this goal: (i) The overexpression of chimeric receptors, for which the extracellular domains of the rice Myc factor receptors MYC FACTOR RECEPTOR1 (OsMYR1) and CHITIN ELICITOR RECEPTOR KINASE1 (OsCERK1) were replaced with those from the M. truncatula Nod factor receptors NOD FACTOR PERCEPTION (MtNFP) and RECEPTOR-LIKE KINASE3 (MtLYK3), respectively, triggers calcium spiking in response to a low concentration Nod factor treatment in rice (He et al., 2019). (ii) The overexpression of several symbiotic regulators induces spontaneous root-nodule-like structures (Soyano et al., 2013; Tirichine et al., 2007; Yang et al., 2022). (iii) The key development genes in M. truncatula SHORT ROOT (MtSHR), SCARECROW (MtSCR), and LLOB-DOMAIN PROTEIN16 (MtLBD16) specify cortical cell fate with the ability to de-differentiate to form nodule primordia in response to symbiotic signals (Schiessl et al., 2019; Soyano et al., 2019; Dong et al., 2021). This constitutes a genetic toolkit to generate nodule-like structures to accommodate N-fixing rhizobia, that is by engineering the expression of these key regulators of nodule organogenesis in cereal crops. However, creating the micro-aerobic conditions necessary for rhizobia in the nodule organs of cereal crops to perform N fixation is still a black box.
3. Autonomous N fixation in cereal crops
An ideal approach for self-fertilizing cereal crops would be to make them fix N autonomously. A detailed study showed that the smallest N fixation operon consists of 9 genes, nifB, nifH, nifD, nifK, nifE, nifN, nifX, hesA, and nifV in Paenibacillus WLY78 (Wang et al., 2013b). Transgenic Arabidopsis expressing a nine-nif gene cassette (nifBHDKENXhesAnifV) showed moderate nitrogenase activity and resulted in higher biomass and chlorophyll compared to control plants grown in low-N or N-free medium (Yao et al., 2021). If the results of this study can be validated, this will provide the possibility to construct cereal crops capable of autonomous N fixation in the future.
Enhancing climate resilience in the hardy staple crop cassava
(By Marnin Wolfe, Eder Jorge de Oliveira, and Ismail Rabbi)
Cassava (Manihot esculenta) is a staple root crop grown on more than 28 million hectares and crucial to the food security of almost half a billion people. Cassava is uniquely positioned as one of the most climate change resilient crops due to its ability to tolerate prolonged droughts, often exceeding 5 months. The cultivation of cassava has continued to increase in tropical regions, where climate change impacts will be particularly adverse (El-Sharkawy, 1993; Parry and Rosenzweig, 1993; de Oliveira Aparecido et al., 2020). In this section, we overview the innovations that have recently accelerated cassava genetic improvement, the challenges that drought and heat are expected to pose in coming decades, and address prospects to improve climate resilience through interdisciplinary innovations.
The NextGen cassava breeding project: A decade of innovation
Cassava is a clonally propagated crop domesticated in South America that continues to radiate throughout the tropics. Phenotypic recurrent selection has been the mainstay of cassava breeding in much of its history. As a result of its 12- to 24-month growth cycle, low multiplication rate and low-seed set, phenotypic selection requires 4–6 years between crosses, a major bottleneck for genetic improvement (Ceballos et al., 2015). Cassava has emerged as a model for the adoption of new breeding technologies among root and tuber crops, including the incorporation of improved experimental designs and phenotyping, as well marker-assisted selection (Mbanjo et al., 2021).
In 2012, the Next-Generation Cassava (NGC) Breeding Project initiated a multi-disciplinary effort to accelerate genetic improvement, notably using genomic selection at breeding programs in Africa and Latin America. NGC partners in Africa include the International Institute of Tropical Agriculture and the National Root Crops Research Institute in Nigeria; the West Africa Center for Crop Improvement in Ghana; the National Crops Resources Research Institute, Uganda, and Makerere University in Uganda; and the Tanzania Agricultural Research Institute in Tanzania. In South America, collaborators include EMBRAPA in Brazil and the International Center for Tropical Agriculture in Colombia. In the USA, collaborators are Cornell University, the Boyce Thompson Institute at Cornell, the University of Hawaii, and the USDA-ARS in Ithaca, NY. The details of partners and funding found at https://www.nextgencassava.org.
Instead of requiring phenotyping breeding lines over many years before selecting new parents, genomic selection enables breeders to predict performance based on genome-wide genetic markers, even at the seedling stage (Figure 10A). Genotyping of all germplasm and targeted phenotyping of representative subsets make breeding value prediction in early stages possible, increasing selection intensity and reducing selection cycle time (Heffner et al., 2009). The transition from phenotypic to genomic selection has gained momentum in cassava through the NGC, and while breeding cycle times are 50% shorter, selection intensity and accuracy are higher (Wolfe et al., 2017), and the rate of improvement is demonstrably increased relative to previous decades (Figure 10B).
Figure 10.
Genomic selection (GS) in a cassava breeding program. A, Each breeding cycle begins with a crossing block trial where seeds are generated. The first evaluation, a seedling nursery (SDN) usually involves >10K plants, but cassava does not produce storage roots when planted from seed and no yield data is collected. After 12 months, seedlings are cloned (5–10 cuttings/plant) into their first single-row, unreplicated clonal evaluation trial (CET) followed by at least three stages of yield trials (preliminary [PYT], advanced [AYT], and uniform [UYT]). All lines entering CET are genotyped genome-wide; sometimes this is done during the seedling nursery. As a result, genomic prediction enables selection of new parents for crossing even as early as the SDN (dashed red arrow). B, GS has resulted in demonstrable acceleration in the rate of genetic improvement since initiation in 2012. Results shown are from the IITA GS population. The genomically predicted performance of GS-era (purple) and historical (yellow) clones relative to a multi-trait selection index (y-axis) is plotted against the year when each clone was first generated (x-axis). C, Field trial showing variability for one of the major future challenges to cassava: drought. The top image shows plants 3 months after planting, under irrigation at Petrolina (Pernambuco, Brazil). The bottom image shows plants 3 months later under water deficit.
Several additional innovations came to cassava under NGC, including: (1) GWAS enabling the cataloging and validation of trait-linked single-nucleotide polymorphisms used for marker-assisted selection (Wolfe et al., 2017; Zhang et al., 2018; Rabbi et al., 2020); (2) Cassavabase.org, an open-access, breeding database for efficient management of phenotype and genotype data (Morales et al., 2022); and (3) use of plant growth regulators for improved flowering and seed set (Hyde et al., 2020). Genomic resources developed during the last decade, including reference genomes (Lyons et al., 2021) and HapMap (Ramu et al., 2017; Kuon et al., 2019) have laid a foundation for trait-discovery research. These technologies collectively will enable breeders worldwide to tackle the food-security challenges posed by climate change.
Climate resilient cassava breeding: Innovations for the next decade
Although cassava is considered a drought-tolerant species (Okogbenin et al., 2013), there is still a large gap between the yield obtained by farmers in semi-arid regions (9.5 t.ha−1) and yield observed under experimental water deficit (23.6 t.ha−1) with improved genotypes (de Oliveira et al., 2015). Fortunately, there is enormous genetic variability to tap for drought tolerance for future genetic improvement (de Oliveira et al., 2017; Figure 10C). Most cassava field testing by breeders is done in both high-rainfall and drought-prone environments. Over the annual cropping cycle, genotypes are routinely exposed to 3–5 months of drought and higher temperatures during which they are evaluated for leaf retention, greenness, and damage by dry season pests such as green mites (Ezenwaka et al., 2018). Advanced testing is usually done in multienvironment trials, including low rainfall, heat-stressed environments (Hershey, 1984). Although genetic control of drought tolerance in cassava, as measured by yield under drought, is complex with strong genotype-by-environment interaction (de Oliveira et al., 2015), a recent GWAS identified candidate genes with known association to drought tolerance and markers useful for breeding (dos Santos Silva et al., 2021).
The complexity of drought-tolerance genetic architecture suggests that genomic selection, augmented by genome editing and cutting-edge phenomics, will be necessary for the rapid development of climate-resilient cassava varieties. Currently, final yield is the basis for selection for drought tolerance (Khadka et al., 2020). However, yield is affected by many factors into drought and is only measurable after 10–12 months. Earlier stage, nondestructive evaluation of physiological drought responses and root bulking is needed. Remote sensing of photosynthetic performance using drones with hyperspectral imaging (Verma et al., 1993; Banerjee et al., 2020) and root yield using ground penetrating radar is now possible (Agbona et al., 2021). High-throughput phenotyping plus genomic prediction and GWAS-based discovery are powerful tools for climate-resilience breeding (Juliana et al., 2019; Jha et al., 2020). Pilot tests have been conducted on cassava for association with above and below-ground traits (Selvaraj et al., 2020).
Genome editing and metabolic engineering are promising supplements to exploiting existing natural diversity. Transformation of cassava to express isopentenyl transferase resulted in increased water retention and leaf retention under water stress (Zhang et al., 2010). The overexpression of transcription factors like DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN (DREB), ABA-RESPONSIVE ELEMENT BINDING PROTEIN1 (AREB1), and ABA-RESPONSIVE ELEMENT BINDING FACTOR2 (ABF2) were also shown to increase drought tolerance in some species (Rivero et al., 2007). In Arabidopsis, CRISPR/Cas9 was used to modify the OPEN STOMATA2 (OST2) gene resulting in greater drought tolerance through enhanced stomatal response (Osakabe et al., 2016). Engineering multiple traits such as improving light reaction efficiency, reducing photorespiration, improving sucrose synthesis to increase sucrose loading and stimulate cambium activity could improve starch synthesis and metabolite transport into storage roots and increase sink capacity (Obata et al., 2020; Sonnewald et al., 2020). In cassava, there are no published studies on the use of genome editing to mitigate drought response, but several studies have demonstrated the feasibility of using CRISPR/Cas9 for virus resistance (Gomez et al., 2019) and reducing cyanogenic compounds (Gomez et al., 2021).
Climate change will disproportionately impact already food insecure regions of the world (Easterling and Apps, 2005). Cassava, already a hardy crop, can help to attenuate some of those negative impacts. For example, cassava planted under free-air CO2 enrichment has been shown to positively respond with increased yield and higher WUE (Rosenthal et al., 2012; Ruiz-Vera et al., 2020). Given sufficient investment, the role of cassava as a food-security and industrial crop will continue to expand and serve as a buffer to future climate change-related food insecurity. We have described ways in which cassava is a “climate-smart” crop and an important staple for millions in the tropics. Now is the time to continue the modernization in cassava breeding and biotechnology to benefit the most vulnerable populations.
The carbon nutrient penalty: Will it matter?
(By Gabriel Castrillo, Martin R. Broadley, and David E. Salt)
Hidden hunger, the lack of sufficient dietary micronutrients including iron (Fe) and zinc (Zn), is a major problem for a significant portion of the world’s human population (Kumssa et al., 2015; Lenaerts and Demont, 2021). Experiments with plants cultivated in growth chambers have suggested that elevated atmospheric CO2 is associated with a decline in mineral nutrients in a number of crops, for example, decreased Fe and Zn concentrations in wheat, barley, and rice (Manderscheid et al., 1995; Fangmeier et al., 1997; Seneweera and Conroy, 1997; la Puente de et al., 2000; Pleijel et al., 2000). Free-air CO2 enrichment (FACE) experiments with plants grown under standard field management practices, with various crops including soybean, sorghum (Sorghum bicolor), potatoes, wheat, barley, and rice (Prior et al., 2008; Högy and Fangmeier, 2009; Högy et al., 2009; Erbs et al., 2010; Fernando et al., 2014a, 2014b; Ujiie et al., 2019), showed similar decreases in mineral nutrients. A more comprehensive set of FACE experiments were reported across three countries, with multiple sites and crops (Myers et al., 2014, Dietterich et al., 2015), which confirmed decreases in Zn and Fe concentration of 5%–10% for C3 grains and legumes at the elevated CO2 concentrations predicted for 2050 (546–586 ppm). A large meta-analysis representing numerous FACE and non-FACE experiments also identified similar reductions in Zn, and in other dietary mineral nutrients such as calcium (Ca) and magnesium (Mg; Loladze, 2014). This carbon nutrient penalty was projected to cause a decrease in the global availability of dietary Fe and Zn of between 2.5% and 3.6% by 2050 (Beach et al., 2019); producing the forecast that many countries that currently have high levels of hidden hunger will continue to do so.
A better understanding of the impact of elevated CO2 on mineral nutrient concentrations in crops requires concomitant consideration of elevated temperature, as they go hand in hand. Combined FACE and temperature (T-FACE) experiments have begun to address the possible impact of elevated temperatures on the carbon nutrient penalty. In soybean, elevated CO2 caused a decrease in seed Fe and Zn concentrations (as previously observed), while elevated temperature had the opposite effect; but the combined effect of elevated temperature and CO2 restored seed Fe and Zn concentrations (Köhler et al., 2019). A similar compensating effect of elevated temperature on the carbon nutrient penalty was also observed in rice and wheat (Guo et al., 2022). Under uniform global temperature increases, the carbon nutrient penalty may therefore be expected to disappear. However, rising global temperatures will not be uniform across the globe, with different regions experiencing different levels of warming. Predicting if elevated temperatures will balance nutrient loss due to elevated CO2 may be more complex and uncertain.
Improved access to diverse diets, comprising more nutrient-dense foods, can play a role in alleviating hidden hunger. However, access to micronutrient-adequate diets is unlikely for many people in the coming decades, for socioeconomic reasons (Nelson et al., 2018). Geographical constraints to micronutrient availability in many food systems, reported from recent GeoNutrition surveys, further compound this challenge (Gashu et al., 2021). Interventions to alleviate hidden hunger include supplements, food fortification, and biofortification of staple crops through breeding and agronomy. Zn-biofortified wheat varieties released in India and Pakistan (Zia et al., 2020; Govindan et al., 2022), and Zn-biofortified hybrid maize varieties in Guatemala and Colombia (Maqbool and Beshir, 2019) can increase grain Zn concentration by more than the anticipated decreases due to elevated CO2. The continued development of crops that can reliably accumulate sufficient quantities of mineral nutrients against a backdrop of climate change is an important part of this solution. The use of micronutrient fertilizers (Joy et al., 2017) and “regenerative” agricultural interventions (Manzeke-Kangara et al., 2021) can also play a role in reducing hidden hunger.
Our understanding of mineral nutrient homeostasis in plants is extensive, with over 176 genes identified to date (Whitt et al., 2020), but far from complete. Of these known genes, over 80 are characterized as ion transporters, many of which were investigated based on their predicted function as transmembrane proteins. High-throughput elemental analysis of plant material, also known as ionomics (Salt et al., 2008), has proved to be a powerful forward genetic screening tool that allows the discovery of genes involved in mineral nutrient homeostasis and the study of natural genetic variation in the system (Huang and Salt, 2016). This approach highlights the critical importance of the Casparian strip in the endodermal cell wall in controlling mineral nutrient homeostasis (Hosmani et al., 2013; Pfister et al., 2014; Kamiya et al., 2015; Reyt et al., 2020, 2021; Alcock et al., 2021). Ionomics has also revealed a global pattern of natural variation in the leaf and seed ionome of Arabidopsis (Campos et al., 2021), and in rice (Pinson et al., 2015), barley (Houston et al., 2020), soybean (Ziegler et al., 2018), common bean (Phaseolus vulgaris; Nazir et al., 2022), peanut (Zhang et al., 2019), and wheat (Gardiner et al., 2018). The application of genome-wide association mapping to this variation has led to the identification of genes controlling variation in numerous elements (Baxter et al., 2010; Chao et al., 2012, 2014; Forsberg et al., 2015; Yang et al., 2018). Ecological studies are starting to reveal the adaptive benefit of this variation for coastal populations (Busoms et al., 2018, 2021).
Soil microbiota contributes to the biogeochemical cycling of elements, soil regeneration, and plant and animal growth and productivity (Custódio et al., 2022). In experiments with Arabidopsis, the root microbiome was shown to control differentiation of the endodermis, a diffusion barrier that affects mineral nutrient homeostasis, through the repression of responses to the phytohormone abscisic acid in the root (Salas-González et al., 2021). However, these mechanisms have not been evaluated under future elevated CO2 scenarios. Elevated CO2 in the short term increases metabolic activity and microbial biomass in the soil, with a concomitant promotion of plant growth and root exudation, conditions that reduce soil N content (Chen et al., 2014; Xiong et al., 2015; Yu et al., 2018a). Thus, in the long-term, elevated CO2 is predicted to have a negative impact on the soil carbon cycle, promoting the depletion of easily decomposed carbon and increasing the degradation of mineralized SOC with a net increase in atmospheric CO2 (Yang et al., 2019a). Elevated CO2 influences microbial enzymatic activities for phosphorus and N cycling but this effect changes depending on the ecosystem (Naylor et al., 2020). We need to understand microbiome stability in diverse ecological contexts, considering spatial resolution, microbial connectivity, and multi-kingdom composition. This will allow us to feed current models with realistic experimental data to predict the impact of climate changes on soil microbial populations and their interactions with plants, helping us to develop microbial-based strategies to alleviate climate change impacts on soil and food production.
Can we achieve a biomass-based bioeconomy?
(By Maureen C. McCann and Nicholas C. Carpita)
Gross domestic product, a measure of economic prosperity, is tightly correlated with energy consumption. Fossil fuels accounted for 80% of global energy resources in 2020. Coal and gas can eventually be displaced by renewable energy from wind and solar, geothermal and hydroelectric, and nuclear energy (U.S. Department of Energy, 2015). Oil, however, provides both liquid transportation fuels and raw materials for the petrochemical industry. As addressing climate change becomes increasingly urgent, we now need to shift from oil derived from long-dead organisms to living organisms that can provide chemicals, fuels, and materials (Carpita and McCann, 2020). In this section, we imagine a biomass-based, circular bioeconomy, enabled by recombinant DNA technologies, with the potential to decouple our prosperity from fossil fuel consumption (National Academies of Science, Engineering, and Medicine, 2020). To succeed, this bioeconomy must be fully rooted in plant biology.
Natural and engineered oil-accumulating plants and microalgae, such as cyanobacteria, are an important source of liquid hydrocarbons for use as fuel components. To address how plants can displace a significant proportion of oil consumption also requires use of the sugars and aromatics derived from plant cell walls (McCann and Carpita, 2015). Electric and hybrid vehicles powered by renewable energy sources are becoming viable long-term options for light ground transportation (U.S. Department of Energy, 2015). However, air, marine, and heavy-duty modes of transportation, which contribute one-third of US transportation GHG emissions, will remain dependent upon energy-dense, liquid-hydrocarbon fuels for decades because of slow fleet turnover: aircraft, for example, have a service lifetime of 25–30 years. Advanced biofuels, fully compatible with existing engines and transportation infrastructure, can include liquid hydrocarbons produced by chemical or enzymatic catalytic conversion of biomass-derived sugars and aromatics (Huber et al., 2003; Wang et al., 2014).
Plant-based biofuels also offer the potential for the production of valuable chemical co-products. Decades of research have overcome the technological barriers to the production of cellulose-derived glucose and, more recently, lignin-derived aromatics. As a result of new deconstruction technologies that preserve aromatic ring structures (Bozell et al., 2011; Labbé et al., 2012; Parsell et al., 2013; Socha et al., 2014), lignin is no longer a major source of biomass recalcitrance. Catalytic depolymerization of lignin has been achieved without decomposition of cellulose or xylan, enabling the concept of the “lignin-first” biorefinery, where aromatic fuel substrates are removed before cellulose and other carbohydrates are processed (Ragauskas et al., 2014; Schutyser et al., 2015; Key and Bozell, 2016; Yang et al., 2019b). To produce hydrocarbon fuels, deoxygenation reactions must proceed to full chemical reduction, but for chemical products, reactions must necessarily be highly selective to preserve desirable functional chemical groups. The petroleum industry produces a handful of platform chemicals from oil, including ethylene, propylene, C4-olefins, benzene, toluene, and xylene, that are oxygenated to make tens of thousands of chemicals (Wang et al., 2014; Parsell et al., 2015). Plants synthesize highly oxygenated polymers, and the chemical moieties in these structures hold tremendous value as useful building blocks for chemical co-products. Pathways that employ lignin-derived aromatics as substrates to replace commodity chemicals have been envisioned using either enzymatic or chemical catalysis (Wellisch et al., 2010; Zakzeski et al., 2010). Controlled fractionation of biomass with downstream catalytic upgrading provides several value-added streams for the major biomass components: xylans to furfural (Vinueza et al., 2015), lignin to aromatics and dicarboxylic acids (Zeng et al., 2015), and cellulose to hydroxymethylfurfural (Hewetson et al., 2016).
The diversity of plant metabolism, natural and engineered, also provides a foundation for engineering biology to create economic value. Living plant cells synthesize between 100,000 and 1 million kinds of molecules (Fang et al., 2019). Making natural or synthetic products directly in plants can take advantage of orders-of-magnitude greater metabolic complexity and potential product yields than can be achieved in microbial chassis organisms. Efficient production of target compounds in plants will require a systems-level understanding of metabolism and constraints, including tradeoffs between carbon fluxes and cellular energy balances.
The structural complexity of plant cell wall components can provide oligomeric and polymeric substrates for materials such as thermosets, thermoplastics, composites, cellulose nanocrystals, and nanofibers. Thermoset materials include epoxy, silicone, and polyurethane. Lignin- and carbohydrate-derived monomers have been incorporated into polymers to create new bio-based materials with improved performance characteristics compared to fossil fuel-derived thermoset materials (Zhao and Abu-Omar, 2015; Jiang et al., 2018; Chen et al., 2019). Poplar fibers have also been directly incorporated into composites with polylactic acid as a replacement for conventional carbon nanofibers that reinforce polymers for large-scale 3D printing applications (Zhao et al., 2019). In contrast to thermosets, thermoplastics can be melted, and some of their monomers may be recycled. The entire pathway to polyhydroxybutyrate was engineered in cotton over 25 years ago (John and Keller, 1996), and more recently in the bioenergy crop switchgrass (Panicum virgatum) (Somleva et al., 2008). Routes for the biological synthesis of polyhydroxyurethane have been envisioned (Nohra et al., 2013). When pulped wood particles are treated with acids, cellulose nanocrystals, and cellulose nanofibers are recovered, derivatives of which are used for several kinds of synthetic materials as replacements for plastics (Moon et al., 2011; Zhu et al., 2016).
Maximizing the recovery of biomass carbon into fuels and co-products requires flexible design capabilities to produce cell wall architectures that can be easily and completely deconstructed for current and future conversion processes (McCann and Carpita, 2015). As robust cell wall architectures are integral to plant growth and development, genetic variants that are tailored with regard to biomass quality for conversion processes must not be compromised for yield or sustainability traits in field performance. Major knowledge gaps include how biosynthetic products are integrated into composite structures, how their individual structural complexities contribute to molecular- to macro-scale architectures, and how cell wall architectures might be redesigned for production of high-value products (Carpita and McCann, 2020).
Material use is tightly coupled to energy use, GHG emissions, land and water use, and waste flows. About one-third of global GHG emissions comes from industrial manufacturing. To decarbonize this economic sector will require changing the means of manufacture as well as the nature of material inputs (Chui et al., 2020). We might imagine how to build simplified production systems with the components of plant cells or make biohybrid materials outside an intact organism. We might imagine designing plants to synthesize homopolymers, heteropolymers and composite materials, displacing structural concrete and steel with new materials like superwood (Chen et al., 2020), or developing new-to-nature materials with advantageous properties.
Food security is a paramount concern and there are land use issues to consider in raising crops for nonfood versus food production. Nonfood cash crops, such as—traditionally—cotton (Gossypium hirsutum), tobacco, hemp, hops (Humulus lupulus), and biofuel feedstocks, to name just a few, can be of considerable value to growers and mitigate financial risk. The diversification of food and nonfood plant products within a single cropping system, or a single crop, coupled with principles of sustainability and climate change resilience, could thus be an advantage toward achieving both food and energy security. The decarbonization of agriculture could include the use of bioenergy crops to displace fossil fuels as a source of hydrogen for ammonia production (Gencer et al., 2020) as well as displacement of fossil fuels for harvesting and drying.
In the USA, an annual sustainable resource of over 1.6 billion tons of lignocellulosic biomass could be considered a strategic carbon reserve (U.S. Department of Energy, 2016). This quantity of biomass represents double the entire annual output of the US agricultural system—grains, fruits, vegetables, hay, and pasture grasses. To double or triple the capacity of the current agricultural system for a biomass-based bioeconomy, additional acres must be brought into production, all crops must be high-yielding, and growers must benefit from diversification of plant products.
Conclusions
We presented examples of some plant biology-based solutions that we believe show promise toward enhancing terrestrial carbon sequestration and engineering climate resilient crops. Although we addressed several disparate topics, a few overarching conclusions emerge.
Innovation
Some of the ideas described here may seem far-fetched to today’s readers, but we believe that for our planet to remain inhabitable and sustainable, many of the ideas proposed here—or others like them—will need to be realized, and we will need plant scientists to help achieve them.
Collaboration
By definition, efforts to mitigate global climate change must be large scale. Plant scientists contributing meaningfully in this arena most likely will be those who seek out effective collaboration—not only with other plant scientists but also with those in other disciplines, including for example agronomy, bioinformatics, data science, engineering, forestry, and soil science. Improved communication and collaboration across disciplines and between academia and industry can also be viewed as a low-tech effort that can have a strong impact. Identifying and seeking out potential collaborators who can link the research to impactful pathways should be a primary goal early in the planning stages of research projects for maximum benefit. Socio-economic and political perspectives will also be crucial in determining which approaches will be adopted and how quickly they will be implemented. Networking, discussion, and collaboration in the socio-political arena and with industry, governmental, and nongovernmental organizations may also be crucial.
Implementation
Many current practices have substantial potential for mitigating CO2 emissions, including reducing food and agricultural waste, shifting to plant-based diets, reducing deforestation coupled with afforestation/reforestation, and restoring coastal wetlands. Some technological solutions also have potential in the shorter term, including direct air capture, biochar, enhanced rock weathering, and bioenergy combined with carbon capture and storage. To date, none of these options has been implemented globally due to cost, timeline, inefficiency, lack of scalability, or an uncertain and evolving carbon price and market. Estimates for the potential of available technologies vary widely and will depend on the ability of nations to realize effective measures (Roe et al., 2019). We have explored ways that plant science can help to tip the balance toward enhanced climate change mitigation and crop resilience. The section on enhancing carbon capture and sequestration in annual cropping systems speaks to our immediate needs for carbon capture on a massive scale and the possibility that plant scientists can achieve a meaningful impact in this arena.
Some of the goals of examples discussed may require years to realize fully, such as engineering C4 photosynthesis into rice, symbiotic N fixation into cereals, and crops that produce a variety of synthetic products. Although time is pressing, this does not make them unworthy of attention. First, aspects of these longer-term goals may provide significant benefits in the short term, and second, the need for carbon capture and enhancing crop resilience and food security will continue in the future. The need is urgent for every plant biologist to consider today how their research can contribute to addressing climate change, ensuring food security, and achieving a sustainable biomass-based bioeconomy.
Acknowledgments
We thank two anonymous reviewers and the editors for their helpful comments. Wolfgang Busch and Joanne Chory are grateful for the work of colleagues in the Harnessing Plants Initiative (HPI): Julie Law, Joe Noel, Todd Michael, and all other HPI team members.
Funding
Collaborative research in the J.K. McKay (Colorado State University) and C. Topp (Donald Danforth Plant Science Center) labs is funded by U.S. Department of Energy Advanced Research Projects Agency-Energy award DE-AR0000826 (variation in NUE and root growth responses to N in rice breeding lines) and by funding from Wells Fargo IN2 (carbon sequestration potential of hemp crops grown for grain and fiber). The work of HPI is funded by gifts from the TED Audacious Program, the Bezos Earth Fund, the Hess Corporation, SEMPRA Energy and others. The NextGen Cassava Breeding project is funded by the UK’s Foreign, Commonwealth & Development Office (FCDO) and the Bill and Melinda Gates Foundation (Grant INV-007637). Research on improving photosynthetic water use efficiency by the A.D.B. Leakey group is funded by the Office of Biological and Environmental Research in the U.S. Department of Energy, Office of Science (DE-SC0018277 and DE-SC0018420). The C4 Rice project is funded by a grant from the Bill & Melinda Gates Foundation to the University of Oxford (INV-002970). Work in the P.C. Ronald lab is funded by gifts from the CHAN ZUCKERBERG INITIATIVE and grants from the National Science Foundation (2027795 to P.C.R.), the United States Department of Agriculture, the National Institutes of Health (GM122968 and GM55962 to P.C.R.), and the Joint BioEnergy Institute funded by the US Department of Energy (No. DE-AC02-05CH11231 to J.C.M and P.C.R.).
Conflict of interest statement. None declared.
Contributor Information
Nancy A Eckardt, Senior Features Editor, The Plant Cell, American Society of Plant Biologists, USA.
Elizabeth A Ainsworth, USDA ARS Global Change and Photosynthesis Research Unit, Urbana, Illinois 61801, USA.
Rajeev N Bahuguna, Centre for Advanced Studies on Climate Change, Dr Rajendra Prasad Central Agricultural University, Samastipur 848125, Bihar, India.
Martin R Broadley, School of Biosciences, University of Nottingham, Nottingham, NG7 2RD, UK; Rothamsted Research, West Common, Harpenden, Hertfordshire, AL5 2JQ, UK.
Wolfgang Busch, Plant Molecular and Cellular Biology Laboratory, Salk Institute for Biological Studies, La Jolla, California 92037, USA.
Nicholas C Carpita, Biosciences Center, National Renewable Energy Laboratory, Golden, Colorado 80401, USA.
Gabriel Castrillo, School of Biosciences, University of Nottingham, Nottingham, NG7 2RD, UK; Future Food Beacon of Excellence, University of Nottingham, Nottingham, NG7 2RD, UK.
Joanne Chory, Plant Molecular and Cellular Biology Laboratory, Salk Institute for Biological Studies, La Jolla, California 92037, USA; Howard Hughes Medical Institute, Salk Institute for Biological Studies, La Jolla, California 92037, USA.
Lee R DeHaan, The Land Institute, Salina, Kansas, USA.
Carlos M Duarte, Red Sea Research Center (RSRC) and Computational Bioscience Research Center, King Abdullah University of Science and Technology (KAUST), Thuwal, 23955-6900, Saudi Arabia.
Amelia Henry, International Rice Research Institute, Rice Breeding Innovations Platform, Los Baños, Laguna 4031, Philippines.
S V Krishna Jagadish, Department of Plant and Soil Science, Texas Tech University, Lubbock, Texas 79410, USA.
Jane A Langdale, Department of Biology, University of Oxford, Oxford, OX1 3RB, UK.
Andrew D B Leakey, Department of Plant Biology, Department of Crop Sciences, and Institute for Genomic Biology, University of Illinois at Urbana-Champaign, Illinois 61801, USA.
James C Liao, Institute of Biological Chemistry, Academia Sinica, Taipei 11528, Taiwan.
Kuan-Jen Lu, Institute of Biological Chemistry, Academia Sinica, Taipei 11528, Taiwan.
Maureen C McCann, Biosciences Center, National Renewable Energy Laboratory, Golden, Colorado 80401, USA.
John K McKay, Department of Agricultural Biology, Colorado State University, Fort Collins, Colorado 80523, USA.
Damaris A Odeny, The International Crops Research Institute for the Semi-Arid Tropics–Eastern and Southern Africa, Gigiri 39063-00623, Nairobi, Kenya.
Eder Jorge de Oliveira, Embrapa Mandioca e Fruticultura, Rua da Embrapa, Cruz das Almas, BA, Brazil.
J Damien Platten, International Rice Research Institute, Rice Breeding Innovations Platform, Los Baños, Laguna 4031, Philippines.
Ismail Rabbi, International Institute of Tropical Agriculture (IITA), PMB 5320 Ibadan, Oyo, Nigeria.
Ellen Youngsoo Rim, Department of Plant Pathology and the Genome Center, University of California, Davis, California 95616, USA.
Pamela C Ronald, Department of Plant Pathology and the Genome Center, University of California, Davis, California 95616, USA; Innovative Genomics Institute, Berkeley, California 94704, USA.
David E Salt, School of Biosciences, University of Nottingham, Nottingham, NG7 2RD, UK; Future Food Beacon of Excellence, University of Nottingham, Nottingham, NG7 2RD, UK.
Alexandra M Shigenaga, Department of Plant Pathology and the Genome Center, University of California, Davis, California 95616, USA.
Ertao Wang, National Key Laboratory of Plant Molecular Genetics, Center for Excellence in Molecular Plant Sciences, Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai 200032, China.
Marnin Wolfe, Auburn University, Dept. of Crop Soil and Environmental Sciences, College of Agriculture, Auburn, Alabama 36849, USA.
Xiaowei Zhang, National Key Laboratory of Plant Molecular Genetics, Center for Excellence in Molecular Plant Sciences, Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai 200032, China.
Authors are listed alphabetically (with the exception of the lead author/coordinating editor). All authors contributed to writing and revising the article.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Nancy A. Eckardt (neckardt@aspb.org).
References
- Agbona A, Teare B, Ruiz-Guzman H, Dobreva ID, Everett ME, Adams T, Montesinos-Lopez OA, Kulakow PA, Hays DB (2021) Prediction of root biomass in cassava based on ground penetrating radar phenomics. Remote Sens 13: 4908 [Google Scholar]
- Aggarwal P, Vyas S, Thornton P, Campbell BM (2019) How much does climate change add to the challenge of feeding the planet this century? Environ Res Lett 14: 043001 [Google Scholar]
- Ahmadi J, Pour-Aboughadareh A, Fabriki-Ourang S, Mehrabi AA, Siddique KH (2018) Screening wild progenitors of wheat for salinity stress at early stages of plant growth: insight into potential sources of variability for salinity adaptation in wheat. Crop Pasture Sci 69: 649–658 [Google Scholar]
- Ainsworth EA, Long SP (2021) 30 years of free-air carbon dioxide enrichment (FACE): What have we learned about future crop productivity and its potential for adaptation? Global Change Biol 27: 27–49 [DOI] [PubMed] [Google Scholar]
- Albritton DL, Meira Filho LG, Cubasch U, Dai X, Ding Y, Griggs DJ, Hewitson B, Houghton JT, Isaksen I, Karl T. et al. (2001) Technical Summary. In Houghton JT, Ding Y, Griggs DJ, Noguer M, van der Linden PJ, Dai X, Maskell K, Johnson CA, eds, Climate Change 2001: The scientific basis. Contribution of working group I to the third assessment report of the intergovernmental panel on climate change, Cambridge University Press, Cambridge, United Kingdom and New York, NY, USA, pp. 881 [Google Scholar]
- Alcock TD, Thomas CL, Ó Lochlainn S, Pongrac P, Wilson M, Moore C, Reyt G, Vogel-Mikuš K, Kelemen M, Hayden R, et al. (2021) Magnesium and calcium overaccumulate in the leaves of a schengen3 mutant of Brassica rapa. Plant Physiol 186: 1616–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaducci S, Zatta A, Raffanini M, Venturi G (2008) Characterisation of hemp (Cannabis sativa L.) roots under different growing conditions. Plant Soil 313: 227 [Google Scholar]
- Anantha MS, Patel D, Quintana M, Swain P, Dwivedi JL, Torres RO, Verulkar SB, Variar M, Mandal NP, Kumar A, et al. (2016) Trait combinations that improve rice yield under drought: Sahbhagi Dhan and new drought-tolerant varieties in South Asia. Crop Sci 56: 408–421 [Google Scholar]
- Arai-Sanoh Y, Takai T, Yoshinaga S, Nakano H, Kojima M, Sakakibara H, Kondo M, Uga Y (2014) Deep rooting conferred by DEEPER ROOTING 1 enhances rice yield in paddy fields. Scient Rep 4: 5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkin OK, Tjoelker MG (2003) Thermal acclimation and the dynamic response of plant respiration to temperature. Trends Plant Sci 8: 343–351 [DOI] [PubMed] [Google Scholar]
- Atkinson NJ, Urwin PE (2012) The interaction of plant biotic and abiotic stresses: from genes to the field. J Exp Bot 63: 3523–3543 [DOI] [PubMed] [Google Scholar]
- Audu V, Ruf T, Vogt-Kaute W, Emmerling C (2022) Changes in microbial biomass and activity support ecological intensification of marginal land through cultivation of perennial wheat in organic agriculture. Biol Agric Hortic. doi: 10.1080/01448765.2022.2040589 [Google Scholar]
- Ayub G, Zaragoza-Castells J, Griffin KL, Atkin OK (2014) Leaf respiration in darkness and in the light under pre-industrial, current and elevated atmospheric CO2 concentrations. Plant Sci 226: 120–130 [DOI] [PubMed] [Google Scholar]
- Bahuguna RN, Chaturvedi AK, Pal M, Viswanathan C, Jagadish SK, Pareek A (2022) Carbon dioxide responsiveness mitigates rice yield loss under high night temperature. Plant Physiol 188: 285–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahuguna RN, Solis CA, Shi W, Jagadish SVK (2017) Post-flowering night respiration and altered sink activity account for high night temperature-induced grain yield and quality loss in rice (Oryza sat- iva L.). Physiol Plant 159: 59–73 [DOI] [PubMed] [Google Scholar]
- Baker JFT, Allen LRA Jr, Boote KNJ, Pickering NB (2000) Direct effects of atmospheric carbon dioxide concentration on whole canopy dark respiration of rice. Global Change Biol 6: 275–286 [Google Scholar]
- Banerjee BP, Joshi S, Thoday-Kennedy E, Pasam RK, Tibbits J, Hayden M, Spangenberg G, Kant S (2020) High-throughput phenotyping using digital and hyperspectral imaging-derived biomarkers for genotypic nitrogen response. J Exp Bot 71: 4604–4615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball P (2020) The lightning-fast quest for COVID vaccines—and what it means for other diseases. Nature 589: 16–18 [DOI] [PubMed] [Google Scholar]
- Bar-Even A (2018) Daring metabolic designs for enhanced plant carbon fixation. Plant Sci 273: 71–83 [DOI] [PubMed] [Google Scholar]
- Baxter I, Brazelton JN, Yu D, Huang YS, Lahner B, Yakubova E, Li Y, Bergelson J, Borevitz JO, Nordborg M, et al. (2010) A coastal cline in sodium accumulation in Arabidopsis thaliana is driven by natural variation of the sodium transporter AtHKT1;1. PLoS Genet 6: e1001193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach RH, Sulser TB, Crimmins A, Cenacchi N, Cole J, Fukagawa NK, Mason-D'Croz D, Myers S, Sarofim MC, Smith M, et al. (2019) Combining the effects of increased atmospheric carbon dioxide on protein, iron, and zinc availability and projected climate change on global diets: a modelling study. Lancet Planet Health 3: e307–e317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellasio C (2019) A generalised dynamic model of leaf-level C3 photosynthesis combining light and dark reactions with stomatal behaviour. Photosyn Res 141: 99–118 [DOI] [PubMed] [Google Scholar]
- Berg IA (2011) Ecological aspects of the distribution of different autotrophic CO2 fixation pathways. Appl Environ Microbiol 77: 1925–1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Canqui H, Shapiro CA, Wortmann CS, Drijber RA, Mamo M, Shaver TM, Ferguson RB (2013) Soil organic carbon: the value to soil properties. J Soil Water Conserv 68: 129A–134A [Google Scholar]
- Borum J, Sand-Jensen K, Binzer T, Pedersen O, Greve TM (2006) Oxygen movement in seagrasses. InSeagrasses: Biology, Ecology and Conservation. Springer, Dordrecht, pp 255–270 [Google Scholar]
- Bozell JJ, Black SK, Myers M, Cahill D, Miller WP, Park S (2011) Solvent fractionation of renewable woody feedstocks: Organosolv generation of biorefinery process streams for the production of biobased chemicals. Biomass Bioenerg 35: 4197–4208 [Google Scholar]
- Brenes M, Solana A, Boscaiu M, Fita A, Vicente O, Calatayud Á, Prohens J, Plazas M (2020) Physiological and biochemical responses to salt stress in cultivated eggplant (Solanum melongena L.) and in S. insanum L., a close wild relative. Agronomy 10: 651 [Google Scholar]
- Broberg MC, Högy P, Fen Z, Pleijel H (2019) Effects of elevated CO2 on wheat yield: non-linear response and relation to site productivity. Agronomy 9: 243 [Google Scholar]
- Brodersen KE, Nielsen DA, Ralph PJ, Kühl M (2015) Oxic microshield and local p H enhancement protects Z ostera muelleri from sediment derived hydrogen sulphide. New Phytol 205: 1264–1276 [DOI] [PubMed] [Google Scholar]
- Buitrago-Bitar MA, Cortés AJ, López-Hernández F, Londoño-Caicedo JM, Muñoz-Florez JE, Muñoz LC, Blair MW (2021) Allelic diversity at abiotic stress responsive genes in relationship to ecological drought indices for cultivated tepary bean, Phaseolus acutifolius A. Gray, and its wild relatives. Genes 12: 556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busoms S, Paajanen P, Marburger S, Bray S, Huang XY, Poschenrieder C, Yant L, Salt DE (2018) Fluctuating selection on migrant adaptive sodium transporter alleles in coastal Arabidopsis thaliana. Proc Natl Acad Sci USA 115: E12443–E12452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busoms S, Terés J, Yant L, Poschenrieder C, Salt DE (2021) Adaptation to coastal soils through pleiotropic boosting of ion and stress hormone concentrations in wild Arabidopsis thaliana. New Phytol 232: 208–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos ACAL, van Dijk WFA, Ramakrishna P, Giles T, Korte P, Douglas A, Smith P, Salt DE (2021) 1,135 ionomes reveal the global pattern of leaf and seed mineral nutrient and trace element diversity in Arabidopsis thaliana. Plant J 106: 536–554 [DOI] [PubMed] [Google Scholar]
- Carpita NC, McCann MC (2020) Redesigning plant cell walls for the biomass-based bioeconomy. J Biol Chem 295: 15144–15157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo MGC, Martin F, Variar M, Bhatt JC, Perez-Quintero AL, Leung H, Leach JE, Vera Cruz CM (2021) Accumulating candidate genes for broad-spectrum resistance to rice blast in a drought-tolerant rice cultivar. Sci Rep 11: 21502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho J, Madgwick P, Powers S, Keys A, Lea P, Parry M (2012) An engineered pathway for glyoxylate metabolism in tobacco plants aimed to avoid the release of ammonia in photorespiration. BMC Biotech 11: 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh AP, South PF, Bernacchi CJ, Ort DR (2022) Alternative pathway to photorespiration protects growth and productivity at elevated temperatures in a model crop. Plant Biotech J 20: 711–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceballos H, Kawuki RS, Gracen VE, Yencho GC, Hershey CH (2015) Conventional breeding, marker-assisted selection, genomic selection and inbreeding in clonally propagated crops: a case study for cassava. Theor Appl Genet 128: 1647–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaloner TM, Gurr SJ, Bebber DP (2021) Plant pathogen infection risk tracks global crop yields under climate change. Nat Clim Change 11: 710–715 [Google Scholar]
- Chao DY, Chen Y, Chen J, Shi S, Chen Z, Wang C, Danku JM, Zhao FJ, Salt DE (2014) Genome-wide association mapping identifies a new arsenate reductase enzyme critical for limiting arsenic accumulation in plants. PLoS Biol 12: e1002009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao DY, Silva A, Baxter I, Huang YS, Nordborg M, Danku J, Lahner B, Yakubova E, Salt DE (2012) Genome-wide association studies identify heavy metal ATPase3 as the primary determinant of natural variation in leaf cadmium in Arabidopsis thaliana. PLoS Genet 8: e1002923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatenoux B, Peduzzi P (2007) Impacts from the 2004 Indian Ocean Tsunami: analysing the potential protecting role of environmental features. Natural Hazards 40: 289–304 [Google Scholar]
- Chen YP, Liu Q, Liu YJ, Jia FA, He XH (2014) Responses of soil microbial activity to cadmium pollution and elevated CO2. Sci Rep 4: 4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Kuang Y, Zhu S, Burgert I, Keplinger T, Gong A, Li T, Berglung L, Eichorn SJ, Hu L (2020) Structure-property–function relationships of natural and engineered wood. Nat Rev Mater 5: 642–666 [Google Scholar]
- Chen C-H, Tung S-H, Jeng R-J, Abu-Omar MA, Lin C-H (2019) A facile strategy to achieve fully bio-based epoxy thermosets from eugenol. Green Chem 21: 4475–4488 [Google Scholar]
- Chen S, Zhang W, Bolus S, Rouse MN, Dubcovsky J (2018) Identification and characterization of wheat stem rust resistance gene Sr21 effective against the Ug99 race group at high temperature. PLoS Genet 14: e1007287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chui M, Evers M, Manyika J, Zheng A, Nisbet T (2020) The Bio Revolution: innovations transforming economies, societies, and our lives. Report of the McKinsey Global Institute. McKinsey & Co., San Francisco. www.mckinsey.com/mgi
- Ciais P, Sabine C, Bala G, Bopp L, Brovkin V, Canadell J, Chhabra A, DeFries R, Galloway J, Heimann M, et al. (2013) Carbon and other biogeochemical cycles. InStocker TF, Qin D, Plattner G-K, Tignor M, Allen SK, Boschung J, Nauels A, Xia Y, Bex V, Midgley PM (eds) Climate Change 2013: The Physical Science Basis. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge University Press, Cambridge, UK and New York, NY, pp 465–470 [Google Scholar]
- Clemmensen KE, Finlay RD, Dahlberg A, Stenlid J, Wardle DA, Lindahl BD (2015) Carbon sequestration is related to mycorrhizal fungal community shifts during long-term succession in boreal forests. New Phytol 205: 1525–1536 [DOI] [PubMed] [Google Scholar]
- Cobb JN, Biswas PS, Platten JD (2019) Back to the future: revisiting MAS as a tool for modern plant breeding. Theor Appl Genet 132: 647–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SP, Leach JE (2020) High temperature-induced plant disease susceptibility: more than the sum of its parts. Curr Opin Plant Biol 56: 235–241 [DOI] [PubMed] [Google Scholar]
- Cortés AJ, Chavarro MC, Madriñán S, This D, Blair MW (2012) Molecular ecology and selection in the drought-related Asr gene polymorphisms in wild and cultivated common bean (Phaseolus vulgaris L.). BMC Genet 13: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covarrubias-Pazaran G, Gebeyehu Z, Gemenet D, Werner C, Labroo M, Sirak S, Coaldrake P, Rabbi I, Kayondo SI, Parkes E, et al. (2022) Breeding schemes: what are they, how to formalize them, and how to improve them? Front Plant Sci 12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan MF, Blomstedt CK, Norton SL, Henry RJ, Møller BL, Gleadow R (2020) Crop wild relatives as a genetic resource for generating low-cyanide, drought-tolerant Sorghum. Environ Exp Bot 169: 103884 [Google Scholar]
- Cox S, Nabukalu P, Paterson AH, Kong W, Auckland S, Rainville L, Cox S, Wang S (2018a) High proportion of diploid hybrids produced by interspecific diploid× tetraploid Sorghum hybridization. Genet Resour Crop Evol 65: 387–390 [Google Scholar]
- Cox S, Nabukalu P, Paterson AH, Kong W, Nakasagga S (2018b) Development of perennial grain sorghum. Sustainability 10: 172 [Google Scholar]
- Crain J, DeHaan L, Poland J (2021) Genomic prediction enables rapid selection of high‐performing genets in an intermediate wheatgrass breeding program. Plant Genome 14: e20080. [DOI] [PubMed] [Google Scholar]
- Crews TE, Rumsey BE (2017) What agriculture can learn from native ecosystems in building soil organic matter: a review. Sustainability 9: 578 [Google Scholar]
- Crump BC, Wojahn JM, Tomas F, Mueller RS (2018) Metatranscriptomics and amplicon sequencing reveal mutualisms in seagrass microbiomes. Front Microbiol 9: 388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H (2021) Challenges and approaches to crop improvement through C3-to-C4 engineering. Front Plant Sci. doi: 10.3389/fpls.2021.715391 (First published September 14, 2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culman SW, Snapp SS, Ollenburger M, Basso B, DeHaan LR (2013) Soil and water quality rapidly responds to the perennial grain Kernza wheatgrass. Agron J 105: 735–744 [Google Scholar]
- Custódio V, Gonin M, Stabl G, Bakhoum N, Oliveira MM, Gutjahr C, Castrillo G (2022) Sculpting the soil microbiota. Plant J 109: 508–522 [DOI] [PubMed] [Google Scholar]
- Dalal J, Lopez H, Vasani NB, Hu Z, Swift JE, Yalamanchili R, Dvora M, Lin X, Xie D, Qu R, et al. (2015) A photorespiratory bypass increases plant growth and seed yield in biofuel crop Camelina sativa. Biotechnol Biofuels 8: 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandekar AM, Jacobson A, Ibáñez AM, Gouran H, Dolan DL, Agüero CB, Uratsu SL, Just R, Zaini PA (2019) Trans-graft protection against Pierce’s disease mediated by transgenic grapevine rootstocks. Front Plant Sci 10: 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das G, Rao GJN (2015) Molecular marker assisted gene stacking for biotic and abiotic stress resistance genes in an elite rice cultivar. Front Plant Sci 6: 698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey PA, Hunt S, Hymus GJ, DeLucia EH, Drake BG, Karnosky DF, Long SP (2004) Respiratory oxygen uptake is not decreased by an instantaneous elevation of [CO2], but is increased with long-term growth in the field at elevated [CO2]. Plant Physiol 134: 520–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EA, Ackerman IL (1993) Changes in soil carbon inventories following cultivation of previously untilled soils. Biogeochemistry 20: 161–193 [Google Scholar]
- de Oliveira Aparecido LE, da Silva Cabral de Moraes JR, de Meneses KC, Lorençone PA, Lorençone JA, de Olanda Souza GH, Torsoni GB (2020) Agricultural zoning as tool for expansion of cassava in climate change scenarios. Theor Appl Climatol 142: 1085–1095 [Google Scholar]
- de Oliveira EJ, de Tarso Aidar T, Morgante CV, Chaves de Melo Chaves AR, Cruz JL, Coelho Filho MA (2015) Genetic parameters for drought-tolerance in cassava. Pesqui Agropecu Bras 50: 233–241 [Google Scholar]
- de Oliveira EJ, Morgante CV, de Tarso Aidar S, de Melo Chaves AR, Antonio RP, Cruz JL, Filho MAC (2017) Evaluation of cassava germplasm for drought tolerance under field conditions. Euphytica 213: 188 [Google Scholar]
- De Oliveira G, Brunsell NA, Crews TE, DeHaan LR, Vico G (2020) Carbon and water relations in perennial Kernza (Thinopyrum intermedium): an overview. Plant Sci 295: 110279. [DOI] [PubMed] [Google Scholar]
- Degen GE, Worrall D, Carmo-Silva E (2020) An isoleucine residue acts as a thermal and regulatory switch in wheat Rubisco activase. Plant J 103: 742–751 [DOI] [PubMed] [Google Scholar]
- DeHaan L, Larson S, López-Marqués RL, Wenkel S, Gao C, Palmgren M (2020) Roadmap for accelerated domestication of an emerging perennial grain crop. Trends Plant Sci 25: 525–537 [DOI] [PubMed] [Google Scholar]
- DeHaan LR, Ismail BP (2017) Perennial cereals provide ecosystem benefits. Cereal Foods World 62: 278–281 [Google Scholar]
- Dehan K, Tal M (1978) Salt tolerance in the wild relatives of the cultivated tomato: responses of Solanum pennellii to high salinity. Irrig Sci 1: 71–76 [Google Scholar]
- DeLucia EH, Chen S, Guan K, Peng B, Li Y, Gomez-Casanovas N, Kantola IB, Bernacchi CJ, Huang Y, Long SP, et al. (2019) Are we approaching a water ceiling to maize yields in the United States? Ecosphere 10: e02773 [Google Scholar]
- Dempewolf H, Eastwood RJ, Guarino L, Khoury CK, Müller JV, Toll J (2014) Adapting agriculture to climate change: a global initiative to collect, conserve, and use crop wild relatives. Agroecol Sustain Food Syst 38: 369–377 [Google Scholar]
- Dheri GS, Lal R, Moonilall NI (2022) Soil carbon stocks and water stable aggregates under annual and perennial biofuel crops in central Ohio. Agric Ecosyst Environ 324: 107715 [Google Scholar]
- Dietterich LH, Zanobetti A, Kloog I, Huybers P, Leakey AD, Bloom AJ, Carlisle E, Fernando N, Fitzgerald G, Hasegawa T, et al. (2015) Impacts of elevated atmospheric CO2 on nutrient content of important food crops. Sci Data 2: 150036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingkuhn M, Luquet D, Fabre D, Muller B, Yin X, Paul MJ (2020) The case for improving crop carbon sink strength or plasticity for a CO2-rich future. Curr Opin Plant Biol 56: 259–272 [DOI] [PubMed] [Google Scholar]
- Dong WT, Zhu YY, Chang HZ, Wang CH, Yang J, Shi J, Gao J, Yang W, Lan L, Wang Y, et al. (2021) An SHR-SCR module specifies legume cortical cell fate to enable nodulation. Nature 589: 586. [DOI] [PubMed] [Google Scholar]
- dos Santos Silva PP, Sousa MBe, de Oliveira EJ, Morgante CV, de Oliveira CRS, Vieira SL, Borel JC (2021) Genome-wide association study of drought tolerance in cassava. Euphytica 217: 60 [Google Scholar]
- Dossa GS, Quibod I, Atienza-Grande G, Oliva R, Maiss E, Vera Cruz C, Wydra K (2020) Rice pyramided line IRBB67 (Xa4/Xa7) homeostasis under combined stress of high temperature and bacterial blight. Sci Rep 10: 683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake BG, Azcon‐Bieto J, Berry J, Bunce J, Dijkstra P, Farrar J, Gifford RM, Gonzalez‐Meler MA, Koch G, Lambers H, et al. (1999) Does elevated atmospheric CO2 concentration inhibit mitochondrial respiration in green plants? Plant Cell Environ 22: 649–657 [Google Scholar]
- Driedonks NJ (2018) From flower to fruit in the heat-Reproductive thermotolerance in tomato and its wild relatives. Doctoral dissertation, Radboud University Nijmegen, Netherlands.
- Duarte CM (1990) Seagrass nutrient content. Marine Ecol Prog Ser 6: 201–207 [Google Scholar]
- Duarte CM, Chiscano CL (1999) Seagrass biomass and production: a reassessment. Aquatic Bot 65: 159–174 [Google Scholar]
- Duarte CM, Losada IJ, Hendriks IE, Mazarrasa I, Marbà N (2013a) The role of coastal plant communities for climate change mitigation and adaptation. Nat Climate Change 3: 961–968 [Google Scholar]
- Duarte CM, Marbà N, Gacia E, Fourqurean JW, Beggins J, Barrón C, Apostolaki ET (2010) Seagrass community metabolism: assessing the carbon sink capacity of seagrass meadows. Global Biogeochem Cycles 24: GB4032 [Google Scholar]
- Duarte CM, Merino M, Agawin NS, Uri J, Fortes MD, Gallegos ME, Marbà N, Hemminga MA (1998) Root production and belowground seagrass biomass. Marine Ecol Prog Ser 171: 97–108 [Google Scholar]
- Duarte CM, Middelburg JJ, Caraco N (2005) Major role of marine vegetation on the oceanic carbon cycle. Biogeosciences 2: 1–8 [Google Scholar]
- Duarte CM, Sintes T, Marbà N (2013b) Assessing the CO 2 capture potential of seagrass restoration projects. J Appl Ecol 50: 1341–1349 [Google Scholar]
- Dusenge ME, Duarte AG, Way DA (2019) Plant carbon metabolism and climate change: elevated CO 2 and temperature impacts on photosynthesis, photorespiration and respiration. New Phytol 221: 32–49 [DOI] [PubMed] [Google Scholar]
- Eastburn DM, Degennaro MM, Delucia EH, Dermody O, Mcelrone AJ (2010) Elevated atmospheric carbon dioxide and ozone alter soybean diseases at SoyFACE. Global Change Biol 16: 320–330 [Google Scholar]
- Easterling W, Apps M (2005) Assessing the consequences of climate change for food and forest resources: a view from the IPCC. InSalinger J, Sivakumar MVK, Motha RP, eds,Increasing Climate Variability and Change: Reducing the Vulnerability of Agriculture and Forestry. Springer Netherlands, Dordrecht, pp 165–189 [Google Scholar]
- Ehleringer JR, Cerling TE, Helliker BR (1997) C4 photosynthesis, atmospheric CO2 and climate. Oecologia 112: 285–299 [DOI] [PubMed] [Google Scholar]
- El Haddad N, Kabbaj H, Zaïm M, El Hassouni K, Sall AT, Azouz M, Ortiz R, Baum M, Amri A, Gamba F, et al. (2021) Crop wild relatives in durum wheat breeding: drift or thrift? Crop Sci 61: 37–54 [Google Scholar]
- El-Sharkawy MA (1993) Drought-tolerant cassava for Africa, Asia, and Latin America. Bioscience 43: 441–451 [Google Scholar]
- Enríquez S, Agustí S, Duarte CM (1994) Light absorption by marine macrophytes. Oecologia 98: 121–129 [DOI] [PubMed] [Google Scholar]
- Enríquez S, Duarte CM, Sand-Jensen K (1993) Patterns in decomposition rates among photosynthetic organisms: the importance of detritus C: N: P content. Oecologia 94: 457–471 [DOI] [PubMed] [Google Scholar]
- Erb TJ, Jones PR, Bar-Even A (2017) Synthetic metabolism: metabolic engineering meets enzyme design. Curr Opin Chem Biol 37: 56–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erbs M, Manderscheid R, Jansen G, Seddig S, Pacholski A, Hans-Joachim Weigel H-J (2010) Effects of free-air CO2 enrichment and nitrogen supply on grain quality parameters and elemental composition of wheat and barley grown in a crop rotation. Agric Ecosyst Environ 136: 59–68 [Google Scholar]
- Ercoli MF, Luu DD, Rim EY, Shigenaga A, Teixeira de Araujo A, Chern M, Jain R, Joe A, Ruan R, Stewart V, et al. (2022) Plant immunity: rice XA21-mediated resistance to bacterial infection. Proc Natl Acad Sci USA 119: e2121568119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksen RL, Adhikari ND, Mou B (2020) Comparative photosynthesis physiology of cultivated and wild lettuce under control and low-water stress. Crop Sci 60: 2511–2526 [Google Scholar]
- Ermakova M, Arrivault S, Giuliani R, Danila F, Alonso-Cantabrana H, Vlad D, Ishihara H, Feil R, Guenther M, Borghi GL, et al. (2021) Installation of C4 photosynthetic pathway enzymes in rice using a single construct. Plant Biotechnol J 19: 575–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JR, Clarke VC (2019) The nitrogen cost of photosynthesis. J Exp Bot 70: 7–15 [DOI] [PubMed] [Google Scholar]
- Evans JR, von Caemmerer S (2000) Would C4 rice produce more biomass than C3 rice? In JE Sheehy, PL Mitchell, B Hardy, eds, Redesigning Rice Photosynthesis to Increase Yield (IRRI), Elsevier Science BV, Amsterdam, pp 53–71 [Google Scholar]
- Evenson RE, Gollin D (2003) Assessing the impact of the Green Revolution, 1960 to 2000. Science 300: 758–762 [DOI] [PubMed] [Google Scholar]
- Eyland D, Luchaire N, Cabrera-Bosquet L, Parent B, Janssens SB, Swennen R, Welcker C, Tardieu F, Carpentier SC (2022) High-throughput phenotyping reveals differential transpiration behaviour within the banana wild relatives highlighting diversity in drought tolerance. Plant Cell Environ. doi: 10.1111/pce.14310 (First published March 16, 2022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezenwaka L, Carpio DP, Jannink J-L, Rabbi I, Danquah E, Asante I, Danquah A, Blay E, Egesi C (2018) Genome‐wide association study of resistance to cassava green mite pest and related traits in cassava. Crop Sci 58: 1907–1918 [Google Scholar]
- Fang C, Fernie AR, Luo J (2019) Exploring the diversity of plant metabolism. Trends Plant Sci 24: 83–98 [DOI] [PubMed] [Google Scholar]
- Fangmeier A, Grüters U, Högy P, Vermehren B, Jäger H-J (1997) Effects of elevated CO2, nitrogen supply and tropospheric ozone on spring wheat-II. Nutrients (N, P, K, S, Ca, Mg, Fe, Mn, Zn). Environ Pollut 96: 43–59 [DOI] [PubMed] [Google Scholar]
- Favela A, Bohn MO, Kent AD (2021) Maize germplasm chronosequence shows crop breeding history impacts recruitment of the rhizosphere microbiome. ISME J 15: 2454–2464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando N, Panozzo J, Tausz M, Norton RM, Neumann N, Fitzgerald GJ, Myers S, Nicolas ME, Seneweer S (2014a) Intra-specific variation of wheat grain quality in response to elevated [CO2] at two sowing times under rain-fed and irrigation treatments. J Cereal Sci 59: 137–144 [Google Scholar]
- Fernando N, Panozzo J, Tausz M, Norton RM, Neumann N, Fitzgerald GJ, Seneweer S (2014b) Elevated CO2 alters grain quality of two bread wheat cultivars grown under different environmental conditions. Agric Ecosyst Environ 185: 24–33 [Google Scholar]
- Ficklin DL, Novick KA (2017) Historic and projected changes in vapor pressure deficit suggest a continental-scale drying of the United States atmosphere. J Geophys Res Atmos 122: 2061–2079 [Google Scholar]
- Fitzgerald GJ, Tausz M, O’Leary G, Mollah MR, Tausz-Posch S, Seneweera S, Mock I, Low M, Partington DL, McNeill D, et al. (2016) Elevated atmospheric [CO2] can dramatically increase wheat yields in semi-arid environments and buffer against heat waves. Global Change Biol 22: 2269–2284 [DOI] [PubMed] [Google Scholar]
- Forsberg SK, Andreatta ME, Huang XY, Danku J, Salt DE, Carlborg Ö (2015) The multi-allelic genetic architecture of a variance-heterogeneity locus for molybdenum concentration in leaves acts as a source of unexplained additive genetic variance. PLoS Genet 11: e1005648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster TL, Baldi HD, Shen X, Burson BL, Klein RR, Murray SC, Jessup RW (2020) Development of novel perennial Sorghum bicolor × S. propinquum hybrids. Crop Sci 60: 863–872 [Google Scholar]
- Fourqurean JW, Duarte CM, Kennedy H, Marbà N, Holmer M, Mateo MA, Apostolaki ET, Kendrick GA, Krause-Jensen D, McGlathery KJ, et al. (2012) Seagrass ecosystems as a globally significant carbon stock. Nat Geosci 5: 505–509 [Google Scholar]
- Frary A, Göl D, Keleş D, Ökmen B, Pınar H, Şığva HÖ, Yemenicioğlu A, Doğanlar S (2010) Salt tolerance in Solanum pennellii: antioxidant response and related QTL. BMC Plant Biol 10: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukayama H, Ueguchi C, Nishikawa K, Katoh N, Ishikawa C, Masumoto C, Hatanaka T, Misoo S (2012) Overexpression of Rubisco activase decreases the photosynthetic CO2 assimilation rate by reducing Rubisco content in rice leaves. Plant Cell Physiol 53: 976–986 [DOI] [PubMed] [Google Scholar]
- Fukayama H, Mizumoto A, Ueguchi C, Katsunuma J, Morita R, Sasayama D, Hatanaka T, Azuma T (2018) Expression level of Rubisco activase negatively correlates with Rubisco content in transgenic rice. Photosyn Res 137: 465–474 [DOI] [PubMed] [Google Scholar]
- Fürst U, Zeng Y, Albert M, Witte AK, Fliegmann J, Felix G (2020) Perception of Agrobacterium tumefaciens flagellin by FLS2XL confers resistance to crown gall disease. Nat Plants 6: 22–27 [DOI] [PubMed] [Google Scholar]
- García GA, Dreccer MF, Miralles DJ, Serrago RA (2015) High night temperatures during grain number determination reduce wheat and barley grain yield: a field study. Global Change Biol 21: 4153–4164 [DOI] [PubMed] [Google Scholar]
- García GA, Serrago RA, Dreccer MF, Miralles DJ (2016) Post-anthesis warm nights reduce grain weight in field-grown wheat and barley. Field Crops Res 195: 50–59 [Google Scholar]
- Garcias‐Bonet N, Eguíluz VM, Díaz‐Rúa R, Duarte CM (2021) Host‐association as major driver of microbiome structure and composition in Red Sea seagrass ecosystems. Environ Microbiol 23: 2021–2034 [DOI] [PubMed] [Google Scholar]
- Gardiner LJ, Joynson R, Omony J, Rusholme-Pilcher R, Olohan L, Lang D, Bai C, Hawkesford M, Salt D, Spannagl M, et al. (2018) Hidden variation in polyploid wheat drives local adaptation. Genome Res 28: 1319–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gashu D, Nalivata PC, Amede T, Ander EL, Bailey EH, Botoman L, Chagumaira C, Gameda S, Haefele SM, Hailu K, et al. (2021) The nutritional quality of cereals varies geospatially in Ethiopia and Malawi. Nature 594: 71–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geddes BA, Paramasivan P, Joffrin A, Thompson AL, Christensen K, Jorrin B, Brett P, Conway SJ, Oldroyd GED, Poole PS (2019) Engineering transkingdom signalling in plants to control gene expression in rhizosphere bacteria. Nat Commun 10: 3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gencer E, Burniske GR, Doering OC, Tyner W, Agrawal R, Delgass WN, Ejeta G, McCann MC, Carpita NC (2020) Sustainable production of ammonia fertilizers from biomass. Biofuels Bioprod Biorefining. doi: 10.1002/bbb.2101 (First published May 10, 2020) [DOI] [Google Scholar]
- Genre A, Lanfranco L, Perotto S, Bonfante P (2020) Unique and common traits in mycorrhizal symbioses. Nat Rev Microbiol 18: 649–660 [DOI] [PubMed] [Google Scholar]
- Ghislain M, Byarugaba AA, Magembe E, Njoroge A, Rivera C, Román ML, Tovar JC, Gamboa S, Forbes GA, Kreuze JF, et al. (2019) Stacking three late blight resistance genes from wild species directly into African highland potato varieties confers complete field resistance to local blight races. Plant Biotechnol J 17: 1119–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill R, Burke IC, Milchunas DG, Lauenroth WK (1999) Relationship between root biomass and soil organic matter pools in the shortgrass steppe of eastern Colorado. Ecosystems 2: 226–236 [Google Scholar]
- Glover JD, Reganold JP, Bell LW, Borevitz J, Brummer EC, Buckler ES, Cox CM, Cox TS, Crews TE, Culman SW, et al. (2010) Increased food and ecosystem security via perennial grains. Science 328: 1638–1639 [DOI] [PubMed] [Google Scholar]
- Gomez MA, Berkoff KC, Gill BK, Iavarone AT, Lieberman SE, Ma JM, Schultink A, Wyman SK, Chauhan RD, Taylor NJ, et al. (2021) CRISPR-Cas9-mediated knockout of CYP79D1 and CYP79D2 in cassava attenuates toxic cyanogen production. bioRxiv: 2021.10.08.462827 [DOI] [PMC free article] [PubMed]
- Gomez MA, Lin ZD, Moll T, Chauhan RD, Hayden L, Renninger K, Beyene G, Taylor NJ, Carrington JC, Staskawicz BJ, et al. (2019) Simultaneous CRISPR/Cas9-mediated editing of cassava eIF4E isoforms nCBP-1 and nCBP-2 reduces cassava brown streak disease symptom severity and incidence. Plant Biotechnol J 17: 421–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Meler MA, Ribas-Carbo M, Siedow JN, Drake BG (1996) Direct inhibition of plant mitochondrial respiration by elevated CO2. Plant Physiol 112: 1349–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Meler MA, Taneva LINA, Trueman RJ (2004) Plant respiration and elevated atmospheric CO2 concentration: cellular responses and global significance. Annals Bot 94: 647–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindan V, Singh RP, Juliana P, Mondal S, Bentley AR (2022) Mainstreaming grain zinc and iron concentrations in CIMMYT wheat germplasm. J Cereal Sci 105: 103473 [Google Scholar]
- Grabowski P, Olabisi LS, Adebiyi J, Waldman K, Richardson R, Rusinamhodzi L, Snapp S (2019) Assessing adoption potential in a risky environment: the case of perennial pigeonpea. Agric Syst 171: 89–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray SB, Siebers M, Locke AM, Rosenthal D, Strellner R, Paul RE, Klein SP, McGrath JM, Dermody O, Ainsworth EA, et al. (2016) Intensifying drought eliminates the expected benefits of elevated [CO2] for soybean. Nat Plants 2: 16132. [DOI] [PubMed] [Google Scholar]
- Griffin KL, Ball JT, Strain BR (1996) Direct and indirect effects of elevated CO2 on whole-shoot respiration in ponderosa pine seedlings. Tree Physiol 16: 33–41 [DOI] [PubMed] [Google Scholar]
- Guo X, Huang B, Zhang H, Cai C, Li G, Li H, Zhang Y, Struik PC, Liu Z, Dong M, et al. (2022) T-FACE studies reveal that increased temperature exerts an effect opposite to that of elevated CO2 on nutrient concentration and bioavailability in rice and wheat grains. Food Energy Secur 11: e336 [Google Scholar]
- Gupta D, Sharma G, Saraswat P, Ranjan R (2021) Synthetic biology in plants, a boon for coming decades. Mol Biotechnol 63: 1138–1154 [DOI] [PubMed] [Google Scholar]
- Hall LN, Rossini L, Cribb L, Langdale JA (1998) GOLDEN 2: a novel transcriptional regulator of cellular differentiation in the maize leaf. Plant Cell 10: 925–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hameed A, Wu Q-S, Abd-Allah EF, Hashem A, Kumar A, Ahmad Lone H, Ahmad P (2014) Role of AM fungi in alleviating drought stress in plants. InMiransari M (ed) Use of Microbes for the Alleviation of Soil Stresses. Springer, New York, NY, pp 55–75 [Google Scholar]
- Harris CJ, Slootweg EJ, Goverse A, Baulcombe DC (2013) Stepwise artificial evolution of a plant disease resistance gene. Proc Natl Acad Sci USA 110: 21189–21194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haskett TL, Paramasivan P, Mendes MD, Green P, Geddes BA, Knights HE, Jorrin B, Ryu MH, Brett P, Voigt CA, et al. (2022) Engineered plant control of associative nitrogen fixation. Proc Natl Acad Sci USA 119: e2117465119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes RC, Wang S, Newell MT, Turner K, Larsen J, Gazza L, Anderson JA, Bell LW, Cattani DJ, Frels K, et al. (2018) The performance of early-generation perennial winter cereals at 21 sites across four continents. Sustainability 10: 1124 [Google Scholar]
- He JM, Zhang C, Dai HL, Liu H, Zhang XW, Yang J, Chen X, Zhu Y, Wang DP, Qi XF, et al. (2019) A LysM receptor heteromer mediates perception of arbuscular mycorrhizal symbiotic signal in rice. Mol Plant 12: 1561–1576 [DOI] [PubMed] [Google Scholar]
- Heffner EL, Sorrells ME, Jannink J-L (2009) Genomic selection for crop improvement. Crop Sci 49: 1–12 [Google Scholar]
- Hein NT, Bheemanahalli R, Wagner D, Vennapusa AR, Bustamante C, Ostmeyer T, Pokharel M, Chiluwal A, Fu J, Srikanthan DS, et al. (2020) Improved cyber-physical system captured post-flowering high night temperature impact on yield and quality of field grown wheat. Scient Rep 10: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein NT, Impa SM, Wagner D, Bheemanahalli R, Kumar R, Tiwari M, Prasad PVV, Tilley M, Wu X, Neilsen M, et al. (2022) Grain micronutrient composition and yield components in field-grown wheat are negatively impacted by high night-time temperature. Cereal Chem. doi: 10.1002/cche.10523 [DOI] [Google Scholar]
- Hemminga MA, Duarte CM (2000) Seagrass Ecology. Cambridge University Press, Cambridge, UK [Google Scholar]
- Hendriks IE, Sintes T, Bouma TJ, Duarte CM (2008) Experimental assessment and modeling evaluation of the effects of the seagrass Posidonia oceanica on flow and particle trapping. Marine Ecol Prog Ser 356: 163–173 [Google Scholar]
- Hershey CH (1984) Breeding cassava for adaptation to stress conditions: development of a methodology. In Symposium of the International Society for Tropical Root Crops, Lima, Peru
- Hewetson B, Zhang X, Mosier NS (2016) Enhanced acid-catalyzed biomass conversion to hydroxymethylfurfural following cellulose solvent-and organic solvent-based lignocellulosic fractionation pretreatment. Energy Fuels 30: 9975–9977 [Google Scholar]
- Hirel B, Le Gouis J, Ney B, Gallais A (2007) The challenge of improving nitrogen use efficiency in crop plants: towards a more central role for genetic variability and quantitative genetics within integrated approaches. J Exp Bot 58: 2369–2387 [DOI] [PubMed] [Google Scholar]
- Högy P, Fangmeier A (2009) Atmospheric CO2 enrichment affects potatoes: 2. Tuber quality traits. Eur J Agron 30: 85–94 [Google Scholar]
- Högy P, Wieser H, Köhler P, Schwadorf K, Breuer J, Franzaring J, Muntifering R, Fangmeier A (2009) Effects of elevated CO2 on grain yield and quality of wheat: results from a 3-year free-air CO2 enrichment experiment. Plant Biol 11: 60–69. [DOI] [PubMed] [Google Scholar]
- Hosmani PS, Kamiya T, Danku J, Naseer S, Geldner N, Guerinot ML, Salt DE (2013) Dirigent domain-containing protein is part of the machinery required for formation of the lignin-based Casparian strip in the root. Proc Natl Acad Sci USA 110: 14498–14503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston K, Qiu J, Wege S, Hrmova M, Oakey H, Qu Y, Smith P, Situmorang A, Macaulay M, Flis P, et al. (2020) Barley sodium content is regulated by natural variants of the Na+ transporter HvHKT1;5. Commun Biol 3: 258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu F, Zhang S, Huang G, Zhang Y, Lv X, Wan K, Liang J, Dao J, Wu S, Zhang L, et al. (2022) Perennial rice improves farmer livelihood and ecosystem security. Preprint posted to Research Square February 2, 2022, doi: 10.21203/rs.3.rs-1302277/v1 [DOI]
- Hua K, Zhang J, Botella JR, Ma C, Kong F, Liu B, Zhu J-K (2019) Perspectives on the application of genome-editing technologies in crop breeding. Mol Plant 12: 1047–1059 [DOI] [PubMed] [Google Scholar]
- Huang W, Reyes-Caldas P, Mann M, Seifbarghi S, Kahn A, Almeida RPP, Béven L, Heck M, Hogenhout SA, Coaker G (2020) Bacterial vector-borne plant diseases: unanswered questions and future directions. Mol Plant 13: 1379–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XY, Salt DE (2016) Plant ionomics: from elemental profiling to environmental adaptation. Mol Plant 9: 787–797 [DOI] [PubMed] [Google Scholar]
- Huang G, Qin S, Zhang S, Cai X, Wu S, Dao J, Zhang J, Huang L, Harnpichitvitaya D, Wade LJ, et al. (2018) Performance, economics and potential impact of perennial rice PR23 relative to annual rice cultivars at multiple locations in Yunnan Province of China. Sustainability 10: 1086 [Google Scholar]
- Huber GW, Shabaker JW, Dumesic JA (2003) Raney Ni–Sn catalyst for H production from biomass-derived hydrocarbons. Science 300: 2075–2077 [DOI] [PubMed] [Google Scholar]
- Hughes TE, Langdale JA (2020) SCARECROW gene function is required for photosynthetic development in maize. Plant Direct 4: e00264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes TE, Langdale JA (2022) SCARECROW is deployed in distinct contexts during rice and maize leaf development. Development 149: e200410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes TE, Sedelnikova OV, Wu H, Becraft PW, Langdale JA (2019) Redundant SCARECROW genes pattern distinct cell layers in roots and leaves of maize. Development 146: e177543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huot B, Castroverde CDM, Velásquez AC, Hubbard E, Pulman JA, Yao J, Childs KL, Tsuda K, Montgomery BL, He SY (2017) Dual impact of elevated temperature on plant defence and bacterial virulence in Arabidopsis. Nature Commun 8: 1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husaini AM (2022) High-value pleiotropic genes for developing multiple stress-tolerant biofortified crops for 21st-century challenges. Heredity. doi: 10.1038/s41437-022-00500-w (First published February 16, 2022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde PT, Guan X, Abreu V, Setter TL (2020) The anti-ethylene growth regulator silver thiosulfate (STS) increases flower production and longevity in cassava (Manihot esculenta Crantz). Plant Growth Regul 90: 441–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilyas M, Nisar M, Khan N, Hazrat A, Khan AH, Hayat K, Fahad S, Khan A, Ullah A (2021) Drought tolerance strategies in plants: a mechanistic approach. J Plant Growth Regul 40: 926–944 [Google Scholar]
- Impa SM, Raju B, Hein NT, Sandhu J, Prasad PV, Walia H, Jagadish SK (2021) High night temperature effects on wheat and rice: current status and way forward. Plant Cell Environ 44: 2049–2065 [DOI] [PubMed] [Google Scholar]
- Impa SM, Vennapusa AR, Bheemanahalli R, Sabela D, Boyle D, Walia H, Jagadish SK (2020) High night temperature induced changes in grain starch metabolism alters starch, protein, and lipid accumulation in winter wheat. Plant Cell Environ 43: 431–447 [DOI] [PubMed] [Google Scholar]
- IPCC (2022) Climate Change 2022: impacts, adaptation, and vulnerability. InPörtner H-O, Roberts DC, Tignor M, Poloczanska ES, Mintenbeck K, Alegría A, Craig M, Langsdorf S., Löschke S., Möller V., et al. , eds, Contribution of Working Group II to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge University Press, Cambridge, UK and New York, NY, USA, pp 3056 [Google Scholar]
- Jackson RB, Lajtha K, Crow SE, Hugelius G, Kramer MG, Piñeiro G (2017) The ecology of soil carbon: pools, vulnerabilities, and biotic and abiotic controls. Annu Rev Ecol Evol Syst 48: 419–445 [Google Scholar]
- Jacott CN, Murray JD, Ridout CJ (2017) Trade-offs in arbuscular mycorrhizal symbiosis: disease resistance, growth responses and perspectives for crop breeding. Agronomy 7: 75 [Google Scholar]
- Jägermeyr J, Müller C, Ruane AC, Elliott J, Balkovic J, Castillo O, Faye B, Foster I, Folberth C, Franke JA, et al. (2021) Climate impacts on global agriculture emerge earlier in new generation of climate and crop models. Nat Food 2: 873–885 [DOI] [PubMed] [Google Scholar]
- Jha UC, Bohra A, Nayyar H (2020) Advances in “omics” approaches to tackle drought stress in grain legumes. Plant Breed 139: 1–27 [Google Scholar]
- Jiang YN, Wang WX, Xie QJ, Liu N, Liu LX, Wang D, Zhang X, Yang C, Chen X, Tang D, et al. (2017) Plants transfer lipids to sustain colonization by mutualistic mycorrhizal and parasitic fungi. Science 356: 1172–1175. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Ding D, Zhao S, Zhu H, Kenttämaa HI, Abu-Omar MM (2018) Renewable thermosets based on lignin and carbohydrate derived monomers. Green Chem 20: 1131–1138 [Google Scholar]
- Jobbágy EG, Jackson RB (2000) The vertical distribution of soil organic carbon and its relation to climate and vegetation. Ecol Appl 10: 423–436 [Google Scholar]
- John ME, Keller G (1996) Metabolic pathway engineering in cotton: biosynthesis of polydroxybutryate in fiber cells. Proc Natl Acad Sci USA 93: 12768–12773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SL (2022) A year at the forefront of engineering photosynthesis. Biol Open 11: bio059335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joy EJM, Ahmad W, Zia MH, Kumssa DB, Young SD, Ander EL, Watts MJ, Stein AJ, Broadley MR (2017) Valuing increased zinc (Zn) fertiliser-use in Pakistan. Plant Soil 411: 139–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliana P, Montesinos-López OA, Crossa J, Mondal S, González Pérez L, Poland J, Huerta-Espino J, Crespo-Herrera L, Govindan V, Dreisigacker S, et al. (2019) Integrating genomic-enabled prediction and high-throughput phenotyping in breeding for climate-resilient bread wheat. Theor Appl Genet 132: 177–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juma RU, Bartholomé J, Thathapalli Prakash P, Hussain W, Platten JD, Lopena V, Verdeprado H, Murori R, Ndayiragije A, Katiyar SK, et al. (2021) Identification of an elite core panel as a key breeding resource to accelerate the rate of genetic improvement for irrigated rice. Rice 14: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya T, Borghi M, Wang P, Danku JM, Kalmbach L, Hosmani PS, Naseer S, Fujiwara T, Geldner N, Salt DE (2015) The MYB36 transcription factor orchestrates Casparian strip formation. Proc Natl Acad Sci USA 112: 10533–10538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasirajan L, Valiyaparambth R, Velu J, Hari H, Srinivasavedantham V, Athaiappan S (2021) Gene expression studies of Saccharum spontaneum, a wild relative of sugarcane in response to salinity stress. Biotechnol Appl Biochem 68: 288–296 [DOI] [PubMed] [Google Scholar]
- Ke J, Wang B, Yoshikuni Y (2021) Microbiome engineering: synthetic biology of plant-associated microbiomes in sustainable agriculture. Trends Biotechnol 39: 244–261 [DOI] [PubMed] [Google Scholar]
- Kebeish R, Niessen M, Thiruveedhi K, Bari R, Hirsch H-J, Rosenkranz R, Stäbler N, Schönfeld B, Kreuzaler F, Peterhansel C (2007) Chloroplastic photorespiratory bypass increases photosynthesis and biomass production in Arabidopsis thaliana. Nat Biotechnol 25: 593–599 [DOI] [PubMed] [Google Scholar]
- Kell DB (2012) Large-scale sequestration of atmospheric carbon via plant roots in natural and agricultural systems: why and how. Phil Trans R Soc B 367: 1589–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy H, Beggins J, Duarte CM, Fourqurean JW, Holmer M, Marbà N, Middelburg JJ (2010) Seagrass sediments as a global carbon sink: isotopic constraints. Global Biogeochem Cycles 24: GB4026 [Google Scholar]
- Key RE, Bozell JJ (2016) Progress toward lignin valorization via selective catalytic technologies and the tailoring of biosynthetic pathways. ACS Sustain Chem Eng 4: 5123–5135 [Google Scholar]
- Khadka K, Earl HJ, Raizada MN, Navabi A (2020) A physio-morphological trait-based approach for breeding drought tolerant wheat. Front Plant Sci 11: 715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna A, Anumalla M, Catolos M, Bartholome J, Fritsche-Neto R, Platten JD, Pisano DJ, Gulles A, Cruz MTS, Ramos J, et al. (2022) Genetic trends estimation in IRRIs rice drought breeding program and identification of high yielding drought-tolerant lines. Rice 15: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna-Chopra R, Viswanathan C (1999) Evaluation of heat stress tolerance in irrigated environment of T. aestivum and related species. I. Stability in yield and yield components. Euphytica 106: 169–180 [Google Scholar]
- Khong GN, Pati PK, Richaud F, Parizot B, Bidzinski P, Mai CD, Bes M, Bourrie I, Meynard D, Beeckman T, et al. (2015) OsMADS26 negatively regulates resistance to pathogens and drought tolerance in rice. Plant Physiol 169: 2935–2949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiani-Pouya A, Rasouli F, Rabbi B, Falakboland Z, Yong M, Chen ZH, Zhou M, Shabala S (2020) Stomatal traits as a determinant of superior salinity tolerance in wild barley. J Plant Physiol 245: 153108. [DOI] [PubMed] [Google Scholar]
- Kikuchi S, Bheemanahalli R, Jagadish SVK, Kumagai E, Masuya Y, Kuroda E, Raghavan C, Dingkuhn M, Abe A, Shimono H (2017) Genome-wide association mapping for phenotypic plasticity in rice. Plant Cell Environ 40: 1565–1575 [DOI] [PubMed] [Google Scholar]
- Kim S, Dale BE, Keck P (2014) Energy requirements and greenhouse gas emissions of maize production in the USA. Bioenerg Res 7: 753–764 [Google Scholar]
- Klinsukon C, Lumyong S, Kuyper TW, Boonlue S (2021) Colonization by arbuscular mycorrhizal fungi improves salinity tolerance of eucalyptus (Eucalyptus camaldulensis) seedlings. Sci Rep-Uk 11: 4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klironomos JN (2003) Variation in plant response to native and exotic arbuscular mycorrhizal fungi. Ecology 84: 2292–2301 [Google Scholar]
- Kocacinar F, Mckown AD, Sage TL, Sage RF (2008) Photosynthetic pathway influences xylem structure and function in Flaveria (Asteraceae). Plant Cell Environ 31: 1363–1376 [DOI] [PubMed] [Google Scholar]
- Köhler IH, Huber SC, Bernacchi CJ, Baxter IR (2019) Increased temperatures may safeguard the nutritional quality of crops under future elevated CO2 concentrations. Plant J 97: 872–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler IH, Ruiz-Vera UM, VanLoocke A, Thomey ML, Clemente T, Long SP, Ort DR, Bernacchi CJ (2017) Expression of cyanobacterial FBP/SBPase in soybean prevents yield depression under future climate conditions. J Exp Bot 68: 715–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornienko N, Zhang JZ, Sakimoto KK, Yang P, Reisner E (2018) Interfacing nature’s catalytic machinery with synthetic materials for semi-artificial photosynthesis. Nat Nanotechnol 13: 890–899 [DOI] [PubMed] [Google Scholar]
- Kromdijk J, Glowacka K, Leonelli L, Gabilly ST, Iwai M, Niyogi KK, Long SP (2016) Improving photosynthesis and crop productivity by accelerating recovery from photoprotection. Science 354: 857–861 [DOI] [PubMed] [Google Scholar]
- Kumar S, Tripathi S, Singh SP, Prasad A, Akter F, Syed MA, Badri J, Das SP, Bhattarai R, Natividad MA, et al. (2021) Rice breeding for yield under drought has selected for longer flag leaves and lower stomatal density. J Exp Bot 72: 4981–4992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumssa DB, Joy EJ, Ander EL, Watts MJ, Young SD, Walker S, Broadley MR (2015) Dietary calcium and zinc deficiency risks are decreasing but remain prevalent. Sci Rep 5: 10974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu P, Goel K, Zinta G (2022) Nutritional imbalance in plants under rising atmospheric CO2. In V Kumar, A Kumar Srivastava, P Suprasanna, eds, Plant Nutrition and Food Security in the Era of Climate Change. Elsevier Academic Press, Amsterdam, pp 513–536 [Google Scholar]
- Kuon JE, Qi W, Schläpfer P, Hirsch-Hoffmann M, von Bieberstein PR, Patrignani A, Poveda L, Grob S, Keller M, Shimizu-Inatsugi R, et al. (2019) Haplotype-resolved genomes of geminivirus-resistant and geminivirus-susceptible African cassava cultivars. BMC Biol 17: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurek I, Chang TK, Bertain SM, Madrigal A, Liu L, Lassner MW, Zhu G (2007) Enhanced thermostability of Arabidopsis Rubisco activase improves photosynthesis and growth rates under moderate heat stress. Plant Cell 19: 3230–3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- la Puente de LS, Perez PP, Martinez-Carrasco R, Morcuende RM, del Molino IMM (2000) Action of elevated CO2 and high temperatures on the mineral chemical composition of two varieties of wheat. Agrochimica 44: 221–230 [Google Scholar]
- Labbé N, Kline LM, Moens L, Kim K, Kim PC, Hayes DG (2012) Activation of lignocellulosic biomass by ionic liquid for biorefinery fractionation. Bioresour Technol 104: 701–707 [DOI] [PubMed] [Google Scholar]
- Lal R (2003) Global potential of soil carbon sequestration to mitigate the greenhouse effect. Crit Rev Plant Sci 22: 151–184 [Google Scholar]
- Langdale JA (2011) C4 cycles: past, present, and future research on C4 photosynthesis. Plant Cell 23: 3879–3892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkum WD, Orth RJ, Duarte CM (2006) Seagrasses: biology, Ecology and Conservation. Springer, Dordrecht, The Netherlands [Google Scholar]
- Leakey ADB, Ainsworth EA, Bernacchi CJ, Rogers A, Long SP, Ort DR (2009) Elevated CO2 effects on plant carbon, nitrogen, and water relations: six important lessons from FACE. J Exp Bot 60: 2859–2976 [DOI] [PubMed] [Google Scholar]
- Leakey ADB, Bishop KA, Ainsworth EA (2012) A multi-biome gap in understanding of crop and ecosystem responses to elevated CO2. Curr Opin Plant Biol 15: 228–236 [DOI] [PubMed] [Google Scholar]
- Leakey ADB, Ferguson JN, Pignon CP, Wu A, Jin Z, Hammer GL, Lobell DB (2019) Water use efficiency as a constraint and target for improving the resilience and productivity of C3 and C4 crops. Ann Rev Plant Biol 70: 781–808 [DOI] [PubMed] [Google Scholar]
- Lehmann J, Hansel CM, Kaiser C, Kleber M, Maher K, Manzoni S, Nunan N, Reichstein M, Schimel JP, Torn MS, et al. (2020) Persistence of soil organic carbon caused by functional complexity. Nat Geosci 13: 529–534 [Google Scholar]
- Lenaerts B, Demont M (2021) The global burden of chronic and hidden hunger revisited: new panel data evidence spanning 1990-2017. Glob Food Sec 28: 100480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng J, Tu W, Hou Y, Cui H (2021) Temperature-inducible transgenic EDS1 and PAD4 in Arabidopsis confer an enhanced disease resistance at elevated temperature. Plants 10: 1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Lin D, Zhang Y, Deng M, Chen Y, Lv B, Li B, Lei Y, Wang Y, Zhao L, et al. (2022) Genome-edited powdery mildew resistance in wheat without growth penalties. Nature 602: 455–460 [DOI] [PubMed] [Google Scholar]
- Li X, Wang P, Li J, Wei SB, Yan YY, Yang J, Zhao M, Langdale JA, Zhou WB (2020) Maize GOLDEN2-LIKE genes enhance biomass and grain yields in rice by improving photosynthesis and reducing photoinhibition. Commun Biol 3: 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Guan K, Schnitkey GD, DeLucia E, Peng B (2019) Excessive rainfall leads to maize yield loss of a comparable magnitude to extreme drought in the United States. Global Change Biol 25: 2325–2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Arrivault S, Coe RA, Karki S, Covshoff S, Bagunu E, Lunn JE, Stitt M, Furbank RT, Hibberd JM, et al. (2020) A partial C4 photosynthetic biochemical pathway in rice. Front Plant Sci 11: e564463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin LH, Simpson MJ (2016) Enhanced extractability of cutin- and suberin-derived organic matter with demineralization implies physical protection over chemical recalcitrance in soil. Organic Geochem 97: 111–121 [Google Scholar]
- Liu L, Li X, Liu S, Min J, Liu W, Pan X, Fang B, Hu M, Liu Z, Li Y, et al. (2021a) Identification of QTLs associated with the anaerobic germination potential using a set of Oryza nivara introgression lines. Genes Genom 43:399–406. 10.1007/s13258-021-01063-6 [DOI] [PubMed] [Google Scholar]
- Liu Y, Wang H, Jiang Z, Wang W, Xun R, Wang Q, Zhang Z, Li A, Liang Y, Ou S, et al. (2021b) Genomic basis of geographical adaptation to soil nitrogen in rice. Nature 590: 600–605 [DOI] [PubMed] [Google Scholar]
- Lobell DB, Gourdji SM (2012) The influence of climate change on global crop productivity. Plant Physiol 160: 1686–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loladze I (2014) Hidden shift of the ionome of plants exposed to elevated CO2 depletes minerals at the base of human nutrition. eLife 3: e02245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz K, Lal R, Preston CM, Nierop KGJ (2007) Strengthening the soil organic carbon pool by increasing contributions from recalcitrant aliphatic bio(macro)molecules. Geoderma 142: 1–10 [Google Scholar]
- Luo S, Lin PP, Nieh L-Y, Liao G-B, Tang P-W, Chen C, Liao JC (2022) A cell-free self-replenishing CO2-fixing system. Nat Catalysis 5: 154–162 [Google Scholar]
- Lyons JB, Bredeson JV, Mansfeld BN, Bauchet GJ, Berry J, Boyher A, Mueller LA, Rokhsar DS, Bart RS (2021) Current status and impending progress for cassava structural genomics. Plant Mol Biol 109: 177–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackill DJ, Ismail AM, Singh US, Labios RV, Paris TR (2012) Development and rapid adoption of submergence-tolerant (Sub1) rice varieties. Adv Agron 115: 299–352 [Google Scholar]
- Macreadie PI, Costa MD, Atwood TB, Friess DA, Kelleway JJ, Kennedy H, Lovelock CE, Serrano O, Duarte CM (2021) Blue carbon as a natural climate solution. Nat Rev Earth Environ 2: 826–839 [Google Scholar]
- Maier A, Fahnenstich H, von Caemmerer S, Engqvist MK, Weber AP, Flugge UI, Maurino VG (2012) Transgenic Introduction of a glycolate oxidative cycle into A. thaliana chloroplasts leads to growth improvement. Front Plant Sci 3: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manderscheid R, Bender J, Jäger H-J, Weigel HJ (1995) Effects of season long CO2 enrichment on cereals. II. Nutrient concentrations and grain quality. Agric Ecosyst Environ 54: 175–185 [Google Scholar]
- Mano YO, Muraki M, Fujimori M, Takamizo T, Kindiger B (2005) Identification of QTL controlling adventitious root formation during flooding conditions in teosinte (Zea mays ssp. huehuetenangensis) seedlings. Euphytica 142: 33–42 [Google Scholar]
- Mano YO, Omori FU, Takamizo TA, Kindiger B, Bird RM, Loaisiga CH (2006) Variation for root aerenchyma formation in flooded and non-flooded maize and teosinte seedlings. Plant Soil 281: 269–279 [Google Scholar]
- Mansueto L, Fuentes RR, Borja FN, Detras J, Abriol-Santos JM, Chebotarov D, Sanciangco M, Palis K, Copetti D, Poliakov A, et al. (2017) Rice SNP-seek database update: new SNPs, indels, and queries. Nucleic Acids Res 45: D1075–D1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzeke-Kangara MG, Joy EJM, Mtambanengwe F, Chopera P, Watts MJ, Broadley MR, Mapfumo P (2021) Good soil management can reduce dietary zinc deficiency in Zimbabwe. CAB Agric Biosci 2: 36 [Google Scholar]
- Maqbool AM, Beshir A (2019) Zinc biofortification of maize (Zea mays L.): status and challenges. Plant Breed 138: 1–28 [Google Scholar]
- Marbà N, Arias‐Ortiz A, Masqué P, Kendrick GA, Mazarrasa I, Bastyan GR, Garcia‐Orellana J, Duarte CM (2015) Impact of seagrass loss and subsequent revegetation on carbon sequestration and stocks. J Ecol 103: 296–302 [Google Scholar]
- Mbanjo EGN, Rabbi IY, Ferguson ME, Kayondo SI, Eng NH, Tripathi L, Kulakow P, Egesi C (2021) Technological innovations for improving cassava production in sub-saharan Africa. Front Genet 11: 623736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann MC, Carpita NC (2015) Biomass recalcitrance: a multi-scale, multi-factor and conversion-specific property. J Exp Bot 66: 4109–4118 [DOI] [PubMed] [Google Scholar]
- McClelland SC, Paustian K, Schipanksi ME (2021) Management of cover crops in temperate climates influences soil organic carbon stocks - a meta-analysis. Ecol Appl 31: e02278. [DOI] [PubMed] [Google Scholar]
- Meuwissen TH, Hayes BJ, Goddard M (2001) Prediction of total genetic value using genome-wide dense marker maps. Genetics 157: 1819–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell PL, Sheehy JE (2006) Supercharging rice photosynthesis to increase yield. New Phytol 171: 688–693 [DOI] [PubMed] [Google Scholar]
- Mittova V, Guy M, Tal M, Volokita M (2002) Response of the cultivated tomato and its wild salt-tolerant relative Lycopersicon pennellii to salt-dependent oxidative stress: increased activities of antioxidant enzymes in root plastids. Free Radic Res 36: 195–202 [DOI] [PubMed] [Google Scholar]
- Moenga SM, Gai Y, Carrasquilla-Garcia N, Perilla-Henao LM, Cook DR (2020) Gene co-expression analysis reveals transcriptome divergence between wild and cultivated chickpea under drought stress. Plant J 104: 1195–1214 [DOI] [PubMed] [Google Scholar]
- Mohr W, Lehnen N, Ahmerkamp S, Marchant HK, Graf JS, Tschitschko B, Yilmaz P, Littmann S, Gruber-Vodicka H, Leisch N, et al. (2021) Terrestrial-type nitrogen-fixing symbiosis between seagrass and a marine bacterium. Nature 600: 105–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteith JL (1978) Reassessment of maximum growth-rates for C3 and C4 crops. Exp Agric 14: 1–5 [Google Scholar]
- Moon RJ, Martini A, Nairn J, Simonsen J, Youngblood J (2011) Cellulose nanomaterials review: structure, properties and nanocomposites. Chem Soc Rev 40: 3941–3994 [DOI] [PubMed] [Google Scholar]
- Moore CE, Meacham-Hensold K, Lemonnier P, Slattery RA, Benjamin C, Bernacchi CJ, Lawson T, Cavanagh AP (2021) The effect of increasing temperature on crop photosynthesis: from enzymes to ecosystems. J Exp Bot 72: 2822–2844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales N, Ogbonna AC, Ellerbrock BJ, Bauchet GJ, Tantikanjana T, Tecle IY, Powell AF, Lyon D, Menda N, Simoes CC, et al. (2022) Breedbase: a digital ecosystem for modern plant breeding G3 12: kac078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyers BT, Morell PL, McKay JK (2018) Genetic costs of domestication and improvement. J Heredity 109: 103–116 [DOI] [PubMed] [Google Scholar]
- Mus F, Crook MB, Garcia K, Garcia Costas A, Geddes BA, Kouri ED, Paramasivan P, Ryu MH, Oldroyd GED, Poole PS, et al. (2016) Symbiotic nitrogen fixation and the challenges to its extension to nonlegumes. Appl Environ Microbiol 82: 3698–3710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers SS, Zanobetti A, Kloog I, Huybers P, Leakey AD, Bloom AJ, Carlisle E, Dietterich LH, Fitzgerald G, Hasegawa T, et al. (2014) Increasing CO2 threatens human nutrition. Nature 510: 139–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakakuni M, Watanabe K, Kaminaka K, Mizuno Y, Takehara K, Kuwae T, Yamamoto S (2021) Seagrass contributes substantially to the sedimentary lignin pool in an estuarine seagrass meadow. Sci Total Environ 793: 148488. [DOI] [PubMed] [Google Scholar]
- Narayanan S, Prasad PV, Welti R (2016a) Wheat leaf lipids during heat stress: II. Lipids experiencing coordinated metabolism are detected by analysis of lipid co‐occurrence. Plant Cell Environ 39: 608–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan S, Tamura PJ, Roth MR, Prasad PV, Welti R (2016b) Wheat leaf lipids during heat stress: I. High day and night temperatures result in major lipid alterations. Plant Cell Environ 39: 787–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Academies of Sciences, Engineering, and Medicine (2019) Negative Emissions Technologies and Reliable Sequestration: A Research Agenda. The National Academies Press, Washington, DC: [PubMed] [Google Scholar]
- National Academies of Science, Engineering, and Medicine (2020) Safeguarding the Bioeconomy. The National Academies Press, Washington, DC: [PubMed] [Google Scholar]
- Naylor D, Sadler N, Bhattacharjee A, Graham EB, Anderton CR, McClure R, Lipton M, Hofmockel KS, Jansson JK (2020) Soil microbiomes under climate change and implications for carbon cycling. Annu Rev Environ Resour 45: 29–59 [Google Scholar]
- Naylor D, DeGraaf S, Purdom E, Coleman-Derr D (2017) Drought and host selection influence bacterial community dynamics in the grass root microbiome. ISME J 11: 2691–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazir M, Mahajan R, Mansoor S, Rasool S, Mir RA, Singh R, Thakral V, Kumar V, Sofi PA, El-Serehy HA, et al. (2022) Identification of QTLs/candidate genes for seed mineral contents in common bean Phaseolus vulgaris L through genotyping-by-sequencing. Front Genet 13: 750814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson G, Bogard J, Lividini K, Arsenault J, Riley M, Sulser TB, Mason-D’Croz D, Power B, Gustafson D, Herrero M, et al. (2018) Income growth and climate change effects on global nutrition security to mid-century. Nat Sustain 1: 773–781 [Google Scholar]
- Nevo E, Chen G (2010) Drought and salt tolerances in wild relatives for wheat and barley improvement. Plant Cell Environ 33: 670–685 [DOI] [PubMed] [Google Scholar]
- Niazian M, Niedbała G (2020) Machine learning for plant breeding and biotechnology. Agriculture 10: 436 [Google Scholar]
- Niroula RK, Pucciariello C, Ho VT, Novi G, Fukao T, Perata P (2012) SUB1A-dependent and -independent mechanisms are involved in the flooding tolerance of wild rice species. Plant J 72: 282–293 [DOI] [PubMed] [Google Scholar]
- Nohra B, Candy L, Blanco J-F, Guerin C, Raoul Y, Mouloungui Z (2013) From petrochemical polyurethanes to biobased polyhydroxyurethanes. Macromolecules 46: 3771−3792 [Google Scholar]
- Nolke G, Houdelet M, Kreuzaler F, Peterhansel C, Schillberg S (2014) The expression of a recombinant glycolate dehydrogenase polyprotein in potato (Solanum tuberosum) plastids strongly enhances photosynthesis and tuber yield. Plant Biotechnol J 12: 734–742 [DOI] [PubMed] [Google Scholar]
- Northrup DL, Basso B, Wang MQ, Morgan CLS, Benfey PN (2021) Novel technologies for emission reduction complement conservation agriculture to achieve negative emissions from row-crop production. Proc Natl Acad Sci USA 118: e2022666118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obata T, Klemens PAW, Rosado-Souza L, Schlereth A, Gisel A, Stavolone L, Zierer W, Morales N, Mueller LA, Zeeman SC, et al. (2020) Metabolic profiles of six African cultivars of cassava (Manihot esculenta Crantz) highlight bottlenecks of root yield. Plant J 102: 1202–1219 [DOI] [PubMed] [Google Scholar]
- Ochieng G, Ngugi K, Wamalwa LN, Manyasa E, Muchira N, Nyamongo D, Odeny DA (2020) Novel sources of drought tolerance from landraces and wild sorghum relatives. Crop Sci 61: 104–118 [Google Scholar]
- Oda M, Nguyen HC, Huynh VT (2019) Evaluation of cropping method for perennial ratoon rice: Adaptation of SALIBU to triple-cropping in Vietnam. F1000Res 8: 1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogle SM, Alsaker C, Baldock J, Bernoux M, Jay Breidt F, McConkey B, Regina K, Vazquez-Amabile GG (2019) Climate and soil characteristics determine where no-till management can store carbon in soils and mitigate greenhouse gas emissions. Sci Rep 9: 11665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okogbenin E, Setter TL, Ferguson M, Mutegi R, Ceballos H, Olasanmi B, Fregene M (2013) Phenotypic approaches to drought in cassava: review. Front Physiol 4: 93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JL, Rouzé P, Verhelst B, Lin YC, Bayer T, Collen J, Dattolo E, De Paoli E, Dittami S, Maumus F, et al. (2016) The genome of the seagrass Zostera marina reveals angiosperm adaptation to the sea. Nature 530: 331–335 [DOI] [PubMed] [Google Scholar]
- Ordóñez RA, Archontoulis SV, Martinez-Feria R, Hatfield JL, Wright EE, Castellano MJ (2020) Root to shoot and carbon to nitrogen ratios of maize and soybean crops in the US Midwest. Eur J Agron 120: 126130 [Google Scholar]
- Oreska MP, McGlathery KJ, Aoki LR, Berger AC, Berg P, Mullins L (2020) The greenhouse gas offset potential from seagrass restoration. Scient Rep 10: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ort DR, Long SP (2014) Limits on yields in the corn belt. Science 344: 483–484 [DOI] [PubMed] [Google Scholar]
- Ort DR, Merchant SS, Alric J, Barkan A, Blankenship RE, Bock R, Croce R, Hanson MR, Hibberd JM, Long SP, et al. (2015) Redesigning photosynthesis to sustainably meet global food and bioenergy demand. Proc Natl Acad Sci USA 112: 8529–8536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth RJ, Lefcheck JS, McGlathery KS, Aoki L, Luckenbach MW, Moore KA, Oreska MP, Snyder R, Wilcox DJ, Lusk B (2020) Restoration of seagrass habitat leads to rapid recovery of coastal ecosystem services. Sci Adv 6: eabc6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe Y, Watanabe T, Sugano SS, Ueta R, Ishihara R, Shinozaki K, Osakabe K (2016) Optimization of CRISPR/Cas9 genome editing to modify abiotic stress responses in plants. Scient Rep 6: 26685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pailles Y, Awlia M, Julkowska M, Passone L, Zemmouri K, Negrão S, Schmöckel SM, Tester M (2020) Diverse traits contribute to salinity tolerance of wild tomato seedlings from the Galapagos islands. Plant Physiol 182: 534–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankievicz VCS, Irving TB, Maia LGS, Ane JM (2019) Are we there yet? The long walk towards the development of efficient symbiotic associations between nitrogen-fixing bacteria and non-leguminous crops. BMC Biol 17: 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JY, Kim H, Lee I (2017) Comparative analysis of molecular and physiological traits between perennial Arabis alpina Pajares and annual Arabidopsis thaliana Sy-0. Scient Rep 7: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry M, Rosenzweig C (1993) The potential effects of climate change on world food supply. In MB Jackson, CR Black, eds, Interacting Stresses on Plants in a Changing Climate. Springer; Berlin Heidelberg, Berlin Heidelberg, pp 1–26 [Google Scholar]
- Parsell TH, Owen BC, Klein I, Jarell TM, Marcum CL, Haupert LJ, Amundson LM, Kenttämaa HI, Riberio F, Miller JT, et al. (2013) Cleavage and hydrodeoxygenation (HDO) of C–O bonds relevant to lignin conversion using Pd/Zn synergistic catalysis. Chem Sci 4: 806–813 [Google Scholar]
- Parsell T, Yohe S, Degenstein J, Jarrell T, Klein I, Gençer E, Hewetson B, Hurt M, Kim JI, Choudari H, et al. (2015) A synergistic biorefinery based on catalytic conversion of lignin prior to cellulose starting from lignocellulosic biomass. Green Chem 17: 1492–1499 [Google Scholar]
- Parton W (1996) The CENTURY model. InPowlson DS, Smith P, Smith JU. (eds) Evaluation of Soil Organic Matter Models. Springer, Berlin, pp 283–291 [Google Scholar]
- Paustian K, Campbell N, Dorich C, Marx E, Swan A (2016a) Assessment of potential greenhouse gas mitigation from changes to crop root mass and architecture. Final report to ARPA-E, 34. https://arpa-e.energy.gov/sites/default/files/documents/files/Revised_Final_Report_to_ARPA_Bounding_Analysis.pdf (April 18, 2022)
- Paustian K, Larson E, Kent J, Marx E, Swan A (2019) Soil C sequestration as a biological negative emission strategy. Front Climate 1: 8 [Google Scholar]
- Paustian K, Lehmann J, Ogle S, Reay D, Robertson GP, Smith P (2016b) Climate-smart soils. Nature 532: 49–57 [DOI] [PubMed] [Google Scholar]
- Pearson PN, Foster GL, Wade BS (2009) Atmospheric carbon dioxide through the Eocene–Oligocene climate transition. Nature 461: 1110–1113 [DOI] [PubMed] [Google Scholar]
- Pedersen O, Nakayama Y, Yasue H, Kurokawa Y, Takahashi H, Heidi Floytrup A, Omori F, Mano Y, David Colmer T, Nakazono M (2021) Lateral roots, in addition to adventitious roots, form a barrier to radial oxygen loss in Zea nicaraguensis and a chromosome segment introgression line in maize. New Phytol 229: 94–105 [DOI] [PubMed] [Google Scholar]
- Peiffer JA, Spor A, Koren O, Jin Z, Tringe SG, Dangl JL, Buckler ES, Ley RE (2013) Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc Natl Acad Sci USA 110: 6548–6553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peixoto L, Elsgaard L, Rasmussen J, Kuzyakov Y, Banfield CC, Dippold MA, Olesen JE (2020) Decreased rhizodeposition, but increased microbial carbon stabilization with soil depth down to 3.6 m. Soil Biol Biochem 150: 108008 [Google Scholar]
- Peixoto RS, Voolstra CR, Sweet M, Duarte CM, Carvalho S, Villela H, Lunshof JE, Gram L, Woodhams DC, Walter J, et al. (2022) Harnessing the microbiome to prevent global biodiversity loss. Nat Microbiol 7: 1726–1735 [DOI] [PubMed] [Google Scholar]
- Peng J, Sun D, Peng Y, Nevo E (2013) Gene discovery in triticum dicoccoides, the direct progenitor of cultivated wheats. Cereal Res Commun 41: 1–22 [Google Scholar]
- Peng S, Huang J, Sheehy JE, Laza RC, Visperas RM, Zhong X, Centeno GS, Khush GS, Cassman KG (2004) Rice yields decline with higher night temperature from global warming. Proc Nat Acad Sci USA 101: 9971–9975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perilla-Henao LM, Casteel CL (2016) Vector-borne bacterial plant pathogens: interactions with hemipteran insects and plants. Front Plant Sci 7: 1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer L, van Erven G, Sinclair EA, Duarte CM, Kabel MA, Classen B (2022) Profiling the cell walls of seagrasses from A (Amphibolis) to Z (Zostera). BMC Plant Biol 22: 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister A, Barberon M, Alassimone J, Kalmbach L, Lee Y, Vermeer JE, Yamazaki M, Li G, Maurel C, Takano J, et al. (2014) A receptor-like kinase mutant with absent endodermal diffusion barrier displays selective nutrient homeostasis defects. eLife 3: e03115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinson SRM, Tarpley L, Yan W, Yeater K, Lahner B, Yakubova E, Huang X-Y, Zhang M, ML Guerinot, Salt DE (2015) Worldwide genetic diversity for mineral element concentrations in rice grain. Crop Sci 55: 1–18 [Google Scholar]
- Pinto P, DeHaan L, Picasso V (2021) Post-harvest management practices impact on light penetration and Kernza intermediate wheatgrass yield components. Agronomy 11: 442 [Google Scholar]
- Platten JD, Cobb JN, Zantua RE (2019) Criteria for evaluating molecular markers: comprehensive quality metrics to improve marker-assisted selection. PLoS One 14: e0210529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platten JD, Egdane JA, Ismail AM (2013) Salinity tolerance, Na+ exclusion and allele mining of HKT1;5 in Oryza sativa and O. glaberrima: many sources, many genes, one mechanism? BMC Plant Biol 13: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleijel H, Gelang J, Sild E, Danielsson H, Younis S, Karlsson P-K, Wallin G, Skärby L, Selldén G (2000) Effects of elevated carbon dioxide, ozone and water availability on spring wheat growth and yield. Physiol Planta 108: 61–70 [Google Scholar]
- Pour-Aboughadareh A, Ahmadi J, Mehrabi AA, Etminan A, Moghaddam M, Siddique KHM (2017) Physiological responses to drought stress in wild relatives of wheat: implications for wheat improvement. Acta Physiol Plant 39: 106 [Google Scholar]
- Prasch CM, Sonnewald U (2013) Simultaneous application of heat, drought, and virus to arabidopsis plants reveals significant shifts in signaling networks. Plant Physiol 162: 1849–1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto I, Stokes A, Roumet C (2016) Root functional parameters predict fine root decomposability at the community level. J Ecol 104: 725–733. [Google Scholar]
- Prior SA, Runion GB, Rogers HH, Torbert HA (2008) Effects of atmospheric CO2 enrichment on crop nutrient dynamics under no-till conditions. J Plant Nutr 31: 758–773 [Google Scholar]
- Priyadharsini P, Rojamala K, Koshila Ravi R, Muthuraja R, Nagaraj K, Muthukumar T (2016) Mycorrhizosphere: the extended rhizosphere and its significance. In DK Choudhary, A Varma, N Tuteja, eds, Plant-Microbe Interaction: An Approach to Sustainable Agriculture. Springer, New York, pp 97–124 [Google Scholar]
- Qu Y, Sakoda K, Fukayama H, Kondo E, Suzuki Y, Makino A, Terashima I, Yamori W (2021) Overexpression of both Rubisco and Rubisco activase rescues rice photosynthesis and biomass under heat stress. Plant Cell Environ 44: 2308–2320 [DOI] [PubMed] [Google Scholar]
- Rabbi IY, Kayondo SI, Bauchet G, Yusuf M, Aghogho CI, Ogunpaimo K, Uwugiaren R, Smith IA, Peteti P, Agbona A, et al. (2020) Genome-wide association analysis reveals new insights into the genetic architecture of defensive, agro-morphological and quality-related traits in cassava. Plant Mol Biol 109: 195–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragauskas AJ, Beckham GT, Biddy MJ, Chandra R, Chen F, Davis MF, Davison BH, Dixon RA, Gilna P, Keller M, et al. (2014) Lignin valorization: improving lignin processing in the biorefinery. Science 344: 1246843. [DOI] [PubMed] [Google Scholar]
- Ramu P, Esuma W, Kawuki R, Rabbi IY, Egesi C, Bredeson JV, Bart RS, Verma J, Buckler ES, Lu F (2017) Cassava haplotype map highlights fixation of deleterious mutations during clonal propagation. Nat Genet 49: 959–963 [DOI] [PubMed] [Google Scholar]
- Rani K, Raghu BR, Jha SK, Agarwal P, Mallick N, Niranjana M, Sharma JB, Singh AK, Sharma NK, Rajkumar S, et al. (2020) A novel leaf rust resistance gene introgressed from Aegilops markgrafii maps on chromosome arm 2AS of wheat. Theoret Appl Genet 133: 2685–2694 [DOI] [PubMed] [Google Scholar]
- Rao Z, Chen F, Zhang X, Xu Y, Xue Q, Zhang P (2012) Spatial and temporal variations of C3/C4 relative abundance in global terrestrial ecosystem since the Last Glacial and its possible driving mechanisms. Chinese Sci Bull 57: 4024–4035 [Google Scholar]
- Razzaq A, Saleem F, Wani SH, Abdelmohsen SA, Alyousef HA, Abdelbacki AM, Alkallas FH, Tamam N, Elansary HO (2021) De-novo domestication for improving salt tolerance in crops. Frontiers Plant Sci 12: 1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich PB, Hobbie SE, Lee TD, Pastore MA (2018) Unexpected reversal of C3 versus C4 grass response to elevated CO2 during a 20-year field experiment. Science 360: 317–320 [DOI] [PubMed] [Google Scholar]
- Reyt G, Chao Z, Flis P, Salas-González I, Castrillo G, Chao DY, Salt DE (2020) Uclacyanin proteins are required for lignified nanodomain formation within casparian strips. Curr Biol 30: 4103–4111.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyt G, Ramakrishna P, Salas-González I, Fujita S, Love A, Tiemessen D, Lapierre C, Morreel K, Calvo-Polanco M, Flis P, et al. (2021) Two chemically distinct root lignin barriers control solute and water balance. Nat Commun 12: 2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivero RM, Mittler R, Blumwald E, Zandalinas SI (2022) Developing climate‐resilient crops: improving plant tolerance to stress combination. Plant J 109: 373–389 [DOI] [PubMed] [Google Scholar]
- Rivero RM, Kojima M, Gepstein A, Sakakibara H, Mittler R, Gepstein S, Blumwald E (2007) Delayed leaf senescence induces extreme drought tolerance in a flowering plant. Proc Natl Acad Sci USA 104: 19631–19636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts DG, McComb AJ, Kuo J (1984) The structure and continuity of the lacunar system of the seagrass Halophila ovalis (R. Br.) Hook f.(Hydrocharitaceae). Aquatic Bot 18: 377–388 [Google Scholar]
- Roe S, Streck C, Obersteiner M, Frank S, Grisom B, Drouet L, Fricko O, Gusti M, Harris N, Hasegawa T, et al. (2019) Contribution of the land sector to a 1.5°C world. Nat Climate Change 9: 817–828 [Google Scholar]
- Romero J, Lee KS, Pérez M, Mateo MA, Alcoverro T (2006) Nutrient dynamics in seagrass ecosystems. InSeagrasses: Biology, Ecology and Conservation. Springer, Dordrecht, pp 227–254 [Google Scholar]
- Rosenthal DM, Locke AM, Khozaei M, Raines CA, Long SP, Ort DR (2011) Over-expressing the C3 photosynthesis cycle enzyme sedoheptulose-1-7 bisphosphatase improves photosynthetic carbon gain and yield under fully open air CO2 fumigation (FACE). BMC Plant Biol 11: 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal DM, Slattery RA, Miller RE, Grennan AK, Cavagnaro TR, Fauquet CM, Gleadow RM, Ort DR (2012) Cassava about-FACE: greater than expected yield stimulation of cassava (Manihot esculenta) by future CO2levels. Global Change Biol 18: 2661–2675 [Google Scholar]
- Roy S, Liu W, Nandety RS, Crook A, Mysore KS, Pislariu CI, Frugoli J, Dickstein R, Udvardi MK (2020) Celebrating 20 years of genetic discoveries in legume nodulation and symbiotic nitrogen fixation. Plant Cell 32: 15–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Vera UM, De Souza AP, Ament MR, Gleadow RM, Ort DR (2020) High sink-strength prevents photosynthetic down-regulation in cassava grown at elevated CO2 concentration. J Exp Bot 72: 542–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runck BC, Kantar MB, Jordan NR, Anderson JA, Wyse DL, Eckberg JO, Barnes RJ, Lehman CL, DeHaan LR, Stupar RM, et al. (2014) The reflective plant breeding paradigm: a robust system of germplasm development to support strategic diversification of agroecosystems. Crop Sci 54: 1939–1948 [Google Scholar]
- Sacks EJ, Roxas JP, Cruz MTS (2003) Developing perennial upland rice II: field performance of S1 families from an intermated Oryza sativa/O. longistaminata population. Crop Sci 43: 129–134 [Google Scholar]
- Sage RF (2016) A portrait of the C4 photosynthetic family on the 50th anniversary of its discovery: species number, evolutionary lineages, and Hall of Fame. J Exp Bot 67: 4039–4056 [DOI] [PubMed] [Google Scholar]
- Salas-González I, Reyt G, Flis P, Custódio V, Gopaulchan D, Bakhoum N, Dew TP, Suresh K, Franke RB, Dangl JL, et al. (2021) Coordination between microbiota and root endodermis supports plant mineral nutrient homeostasis. Science 371: eabd0695. [DOI] [PubMed] [Google Scholar]
- Sales CR, Wang Y, Evers JB, Kromdijk J (2021) Improving C4 photosynthesis to increase productivity under optimal and suboptimal conditions. J Exp Bot 72: 5942–5960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salt DE, Baxter I, Lahner B (2008) Ionomics and the study of the plant ionome. Annu Rev Plant Biol 59: 709–733 [DOI] [PubMed] [Google Scholar]
- Salvagiotti F, Cassman KG, Specht JE, Walters DT, Weiss A, Dobermann A (2008) Nitrogen uptake, fixation and response to fertilizer N in soybeans: a review. Field Crops Res 108: 1–13 [Google Scholar]
- Salvucchi ME, Crafts-Brandner SJ (2004) Relationship between the heat tolerance of photosynthesis and the thermal stability of Rubisco activase in plants from contrasting thermal environments. Plant Physiol 134: 1460–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderman J, Hengl T, Fiske GJ (2017) Soil carbon debt of 12,000 years of human land use. Proc Natl Acad Sci USA 114: 9575–9580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasa S, Sawayama S, Sakamoto S, Tsujimoto R, Terauchi G, Yagi H, Komatsu T (2012) Did huge tsunami on 11 March 2011 impact seagrass bed distributions in Shizugawa Bay, Sanriku Coast, Japan? InRemote Sensing of the Marine Environment II (Vol. 8525, p. 85250X). International Society for Optics and Photonics [Google Scholar]
- Scafaro AP, Atwell BJ, Muylaert S, Reusel BV, Ruiz GA, Rie JV, Gallé A (2018) A thermotolerant variant of Rubisco activase from a wild relative improves growth and seed yield in rice under heat stress. Front Plant Sci 20: 1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scafaro AP, Bautsoens N, den Boer B, Van Rie J, Gallé A (2019) A conserved sequence from heat-adapted species improves Rubisco activase thermostability in wheat. Plant Physiol 181: 43–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scafaro AP, Gallé A, Van Rie J, Carmo-Silva E, Salvucci ME, Atwell BJ (2016) Heat tolerance in a wild Oryza species is attributed to maintenance of Rubisco activation by a thermally stable Rubisco activase ortholog. New Phytol 211: 899–911 [DOI] [PubMed] [Google Scholar]
- Scafaro AP, Yamori W, Carmo-Silva AE, Salvucci ME, von Caemmerer S, Atwell BJ (2012) Rubisco activity is associated with photosynthetic thermotolerance in a wild rice (Oryza meridionalis). Physiol Planta 146: 99–109 [DOI] [PubMed] [Google Scholar]
- Scheffen M, Marchal DG, Beneyton T, Schuller SK, Klose M, Diehl C, Lehmann J, Pfister P, Carrillo M, He H, et al. (2021) A new-to-nature carboxylation module to improve natural and synthetic CO2 fixation. Nat Catalysis 4: 105–115 [Google Scholar]
- Schiessl K, Lilley JLS, Lee T, Tamvakis I, Kohlen W, Bailey PC, Thomas A, Luptak J, Ramakrishnan K, Carpenter MD, et al. (2019) NODULE INCEPTION recruits the lateral root developmental program for symbiotic nodule organogenesis in Medicago truncatula. Curr Biol 29: 3657–3668.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller K, Bräutigam A (2021) Engineering of crassulacean acid metabolism. Ann Rev Plant Biol 72: 77–103 [DOI] [PubMed] [Google Scholar]
- Schlautman B, Barriball S, Ciotir C, Herron S, Miller AJ (2018) Perennial grain legume domestication phase I: criteria for candidate species selection. Sustainability 10: 730 [Google Scholar]
- Schlesinger WH, Bernhardt ES (2020) Biogeochemistry: An Analysis of Global Change. Elsevier Science, Amsterdam [Google Scholar]
- Schmidt MWI, Torn MS, Abiven S, Dittmar T, Guggenberger G, Janssens IA, Kleber M, Kögel-Knabner I, Lehmann J, Manning DAC, et al. (2011) Persistence of soil organic matter as an ecosystem property. Nature 478: 49–56 [DOI] [PubMed] [Google Scholar]
- Schmidt R, Gravuer K, Bossange AV, Mitchell J, Scow K (2018) Long-term use of cover crops and no-till shift soil microbial community life strategies in agricultural soil. PLoS One 13: e0192953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz VV, Martin BC, Meyer R, Schramm A, Fraser MW, Nielsen LP, Kendrick GA, Risgaard-Petersen N, Burdorf LD, Marshall IP (2021) Cable bacteria at oxygen-releasing roots of aquatic plants: a widespread and diverse plant-microbe association. New Phytol 232: 2138–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler ML, Sedelnikova OV, Walker BJ, Westhoff P, Langdale JA (2018) SHORTROOT-mediated increase in stomatal density has no impact on photosynthetic efficiency. Plant Physiol 176: 757–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutyser W, Van den Bosch S, Renders T, De Boe T, Koelewijn SF, Dewaele A, Ennaert T, Verkindern O, Goderis B, Courtin CM, et al. (2015) Influence of bio-based solvents on the catalytic reductive fractionation of birch wood. Green Chem 17: 5035–5045 [Google Scholar]
- Schwander T, Schada von Borzyskowski L, Burgener S, Cortina NS, Erb TJ (2016) A synthetic pathway for the fixation of carbon dioxide in vitro. Science 354: 900–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scudeletti D, Crusciol CAC, Bossolani JW, Moretti LG, Momesso L, Servaz Tubaña B, de Castro SGQ, De Oliveira EF, Hungria M, et al. (2021) Trichoderma asperellum inoculation as a tool for attenuating drought stress in sugarcane. Front Plant Sci 12: 645542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedelnikova OV, Hughes TE, Langdale JA (2018) Understanding the genetic basis of C4 Kranz anatomy with a view to engineering C3 crops. Annu Rev Genet 52: 249–270 [DOI] [PubMed] [Google Scholar]
- Selvaraj MG, Valderrama M, Guzman D, Valencia M, Ruiz H, Acharjee A (2020) Machine learning for high-throughput field phenotyping and image processing provides insight into the association of above and below-ground traits in cassava (Manihot esculenta Crantz). Plant Methods 16: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seneweera SP, Conroy JP (1997) Growth, grain yield and quality of rice (Oryza sativa L.) in response to elevated CO2 and phosphorus nutrition. Soil Sci Plant Nutr 43: 1131–1136 [Google Scholar]
- Shaibu AS, Badu-Apraku B, Ayo-Vaughan MA (2021) Enhancing drought tolerance and Striga hermonthica resistance in maize using newly derived inbred lines from the wild maize relative, Zea diploperennis. Agronomy 11: 177 [Google Scholar]
- Shalata A, Tal M (1998) The effect of salt stress on lipid peroxidation and antioxidants in the cultivated tomato and its wild salt-tolerant relative Lycopersicon pennellii. Physiol Plant 104: 169–174 [DOI] [PubMed] [Google Scholar]
- Sharlach M, Dahlbeck D, Liu L, Chiu J, Jiménez-Gómez JM, Kimura S, Koenig D, Maloof JN, Sinha N, Minsavage GV, et al. (2013) Fine genetic mapping of RXopJ4, a bacterial spot disease resistance locus from Solanum pennellii LA716. Theor Appl Genet 126: 601–609 [DOI] [PubMed] [Google Scholar]
- Sharma R, De Vleesschauwer D, Sharma MK, Ronald PC (2013) Recent advances in dissecting stress-regulatory crosstalk in rice. Mol Plant 6: 250–260 [DOI] [PubMed] [Google Scholar]
- Shen BR, Wang LM, Lin XL, Yao Z, Xu HW, Zhu CH, Teng HY, Cui LL, Liu EE, Zhang JJ, et al. (2019) Engineering a new chloroplastic photorespiratory bypass to increase photosynthetic efficiency and productivity in rice. Mol Plant 12: 199–214 [DOI] [PubMed] [Google Scholar]
- Shi W, Yin X, Struik PC, Solis C, Xie F, Schmidt RC, Huang M, Zou Y, Ye C, Jagadish SVK (2017) High day- and night-time temperatures affect grain growth dynamics in contrasting rice genotypes. J Exp Bot 68: 5233–5245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi JC, Zhao BY, Zheng S, Zhang XW, Wang XL, Dong W, Xie Q, Wang G, Xiao Y, Chen F, et al. (2021) A phosphate starvation response-centered network regulates mycorrhizal symbiosis. Cell 184: 5527–5540.18. [DOI] [PubMed] [Google Scholar]
- Shimono H, Ozaki Y, Jagadish SVK, Sakai H, Usui Y, Hasegawa T, Kumagai E, Nakano H, Yoshinaga S (2014) Planting geometry as a pre-screening technique for identifying CO2 responsive rice geno- types: a case study of panicle number. Physiol Plant 152: 520–528 [DOI] [PubMed] [Google Scholar]
- Shivhare D, Mueller-Cajar O (2017) In vitro characterization of thermostable CAM Rubisco activase reveals a Rubisco interacting surface loop. Plant Physiol 174: 1505–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh GM, Xu J, Schaefer D, Day R, Wang Z, Zhang F (2022) Maize diversity for fall armyworm resistance in a warming world. Crop Sci 62: 1–9 [Google Scholar]
- Skendžić S, Zovko M, Živković IP, Lešić V, Lemić D (2021) The impact of climate change on agricultural insect pests. Insects 12: 440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery RA, Ort DR (2019) Carbon assimilation in crops at high temperatures. Plant Cell Environ 42: 2750–2758 [DOI] [PubMed] [Google Scholar]
- Socha AM, Parthasarathi P, Shi J, Pattathil S, Whyte D, Bergeron M, George A, Tran K, Stavila V, Venkatachalam S, et al. (2014) Efficient biomass pretreatment using ionic liquids derived from lignin and hemicellulose. Proc Natl Acad Sci USA 111: 12582–12587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somleva MN, Snell KD, Beaulieu JJ, Peoples OP, Garrison BR, Patterson NA (2008) Production of polyhydroxybutyrate in switchgrass, a value-added co-product in an important lignocellulosic biomass crop. Plant Biotech J 6: 663–678 [DOI] [PubMed] [Google Scholar]
- Song Y, Yu J, Huang B (2014) Elevated CO2-mitigation of high temperature stress associated with maintenance of positive carbon balance and carbohydrate accumulation in Kentucky bluegrass. PLoS One 9: e89725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnewald U, Fernie AR, Gruissem W, Schläpfer P, Anjanappa RB, Chang S, Ludewig F, Rascher U, Muller O, van Doorn AM, et al. (2020) The Cassava Source‐Sink project: opportunities and challenges for crop improvement by metabolic engineering. Plant J 103: 1655–1665 [DOI] [PubMed] [Google Scholar]
- Soudzilovskaia NA, van Bodegom PM, Terrer C, van’t Zelfde M, McCallum I, Luke McCormack M, Fisher JB, Brundrett MC, César de Sá N, Tedersoo L (2019) Global mycorrhizal plant distribution linked to terrestrial carbon stocks. Nat Commun 10: 5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soumare A, Diedhiou AG, Thuita M, Hafidi M, Ouhdouch Y, Gopalakrishnan S, Kouisni L (2020) Exploiting biological nitrogen fixation: a route towards a sustainable agriculture. Plants (Basel) 9: 1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- South PF, Cavanagh AP, Liu HW, Ort DR (2019) Synthetic glycolate metabolism pathways stimulate crop growth and productivity in the field. Science 363: 6422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyano T, Kouchi H, Hirota A, Hayashi M (2013) NODULE INCEPTION directly targets NF-Y subunit genes to regulate essential processes of root nodule development in Lotus japonicus. PloS Genet 9: e1003352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyano T, Shimoda Y, Kawaguchi M, Hayashi M (2019) A shared gene drives lateral root development and root nodule symbiosis pathways in Lotus. Science 366: 1021–1023 [DOI] [PubMed] [Google Scholar]
- Sprunger CD, Culman SW, Peralta AL, DuPont ST, Lennon JT, Snapp SS (2019) Perennial grain crop roots and nitrogen management shape soil food webs and soil carbon dynamics. Soil Biol Biochem 137: 107573 [Google Scholar]
- Steinwand MA, Ronald PC (2020) Crop biotechnology and the future of food. Nat Food 1: 273–283 [Google Scholar]
- Still CJ, Berry JA, Collatz GJ, DeFries RS (2003) Global distribution of C3 and C4 vegetation: carbon cycle implications. Global Biogeochem Cycles 17: 1–6 [Google Scholar]
- Stitt M (1991) Rising CO2 levels and their potential significance for carbon flow in photosynthetic cells. Plant Cell Environ 14: 741–762 [Google Scholar]
- Suganami M, Suzuki Y, Tazoe Y, Yamori W, Makino A (2021) Co-overproducing Rubisco and Rubisco activase enhances photosynthesis in the optimal temperature range in rice. Plant Physiol 185: 108–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swamy BPM, Ahmed HU, Henry A, Mauleon R, Dixit S, Vikram P, Tilatto R, Verulkar SB, Perraju P, Mandal NP, et al. (2013) Genetic, physiological, and gene expression analyses reveal that multiple QTL enhance yield of rice mega-variety IR64 under drought. PLoS One 8: e62795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarquinio F, Hyndes GA, Laverock B, Koenders A, Säwström C (2019) The seagrass holobiont: understanding seagrass-bacteria interactions and their role in seagrass ecosystem functioning. FEMS Microbiol Lett 366: p.fnz057. [DOI] [PubMed] [Google Scholar]
- Teixeira EI, Fischer G, Van Velthuizen H, Walter C, Ewert F (2013) Global hot-spots of heat stress on agricultural crops due to climate change. Agric Forest Meteorol 170: 206–215 [Google Scholar]
- Therezan R, Kortbeek R, Vendemiatti E, Legarrea S, de Alencar SM, Schuurink RC, Bleeker P, Peres LEP (2021) Introgression of the sesquiterpene biosynthesis from Solanum habrochaites to cultivated tomato offers insights into trichome morphology and arthropod resistance. Planta 254: 11. 10.1007/s00425-021-03651-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tin HQ, Loi NH, Bjornstad Å, Kilian B (2021) Participatory selection of CWR-derived salt-tolerant rice lines adapted to the coastal zone of the Mekong Delta. Crop Sci 61: 277–288 [Google Scholar]
- Tirichine L, Sandal N, Madsen LH, Radutoiu S, Albrektsen AS, Sato S, Asamizu E, Tabata S, Stougaard J (2007) A gain-of-function mutation in a cytokinin receptor triggers spontaneous root nodule organogenesis. Science 315: 104–107 [DOI] [PubMed] [Google Scholar]
- Tork DG, Anderson NO, Wyse DL, Betts KJ (2019) Domestication of perennial flax using an ideotype approach for oilseed, cut flower, and garden performance. Agronomy 9: 707 [Google Scholar]
- Torta L, Burruano S, Giambra S, Conigliaro G, Piazza G, Mirabile G, Pirrotta M, Calvo R, Bellissimo G, Calvo S, et al. (2022) Cultivable fungal endophytes in roots, rhizomes and leaves of Posidonia oceanica (L.) Delile along the coast of Sicily, Italy. Plants 11: 1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyśkiewicz R, Nowak A, Ozimek E, Jaroszuk-Ściseł J (2022) Trichoderma: the current status of its application in agriculture for the biocontrol of fungal phytopathogens and stimulation of plant growth. Int J Mol Sci 23: 2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. Department of Energy (2015) Quadrennial technology review. An assessment of energy technologies and research opportunities. Office of Efficiency and Renewable Energy
- U.S. Department of Energy (2016) Billion-ton report: advancing domestic resources for a thriving bioeconomy. Office of Efficiency and Renewable Energy.
- Ujiie K, Ishimaru K, Hirotsu N, Nagasaka S, Miyakoshi Y, Ota M, Tokida T, Sakai H, Usui Y, Ono K, et al. (2019) How elevated CO2 affects our nutrition in rice, and how we can deal with it. PLoS One 14: e0212840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Deynze A, Zamora P, Delaux PM, Heitmann C, Jayaraman D, Rajasekar S, Graham D, Maeda J, Gibson D, Schwartz KD, et al. (2018) Nitrogen fixation in a landrace of maize is supported by a mucilage-associated diazotrophic microbiota. PLoS Biol 16: e2006352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Katwijk MM, Thorhaug A, Marbà N, Orth RJ, Duarte CM, Kendrick GA, Althuizen IH, Balestri E, Bernard G, Cambridge ML, et al. (2016) Global analysis of seagrass restoration: the importance of large‐scale planting. J Appl Ecol 53: 567–578. [Google Scholar]
- Van Tassel DL, Albrecht KA, Bever JD, Boe AA, Brandvain Y, Crews TE, Gansberger M, Gerstberger P, González-Paleo L, Hulke BS, et al. (2017) Accelerating Silphium domestication: an opportunity to develop new crop ideotypes and breeding strategies informed by multiple disciplines. Crop Sci 57: 1274–1284 [Google Scholar]
- Váry Z, Mullins E, McElwain JC, Doohan FM (2015) The severity of wheat diseases increases when plants and pathogens are acclimatized to elevated carbon dioxide. Global Change Biol 21: 2661–2669 [DOI] [PubMed] [Google Scholar]
- Velásquez AC, Castroverde CDM, He SY (2018) Plant–pathogen warfare under changing climate conditions. Curr Biol 28: R619–R634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh J, Kang B-C (2019) Current views on temperature-modulated R gene-mediated plant defense responses and tradeoffs between plant growth and immunity. Curr Opin Plant Biol 50: 9–17 [DOI] [PubMed] [Google Scholar]
- Verma SB, Sellers PJ, Walthall CL, Hall FG, Kim J, Goetz SJ (1993) Photosynthesis and stomatal conductance related to reflectance on the canopy scale. Remote Sens Environ 44: 103–116 [Google Scholar]
- Verslues PE, Bailey-Serres J, Brodersen C, Buckley TN, Conti L, Christmann A, Dinneny JR, Grill E, Hayes S, Heckman RW (2023) Burning questions for a warming and changing world: 15 unknowns in plant abiotic stress. Plant Cell 35: 67–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vikram P, Swamy BPM, Dixit S, Singh R, Singh BP, Miro B, Kohli A, Henry A, Singh NK, Kumar A (2015) Drought susceptibility of modern rice varieties: an effect of linkage of drought tolerance with undesirable traits. Scient Rep 5: 14799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinueza NR, Kim ES, Gallardo VA, Mosier NS, Abu-Omar MM, Carpita NC, Kenttäma HI (2015) Tandem mass spectrometric characterization of the conversion of xylose to furfural. Biomass Bioenerg 74: 1–5 [Google Scholar]
- Wagner MR, Roberts JH, Balint-Kurti P, Holland JB (2020) Heterosis of leaf and rhizosphere microbiomes in field-grown maize. New Phytol 228: 1055–1069 [DOI] [PubMed] [Google Scholar]
- Walker BJ, VanLoocke A, Bernacchi CJ, Ort DR (2016) The costs of photorespiration to food production now and in the future. Annu Rev Plant Biol 67: 107–129 [DOI] [PubMed] [Google Scholar]
- Wang D, Dong W, Murray J, Wang E (2022) Innovation and appropriation in mycorrhizal and rhizobial symbioses. Plant Cell 34:1573–1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Schornack S, Marsh JF, Gobbato E, Schwessinger B, Eastmond P, Schultze M, Kamoun S, Oldroyd GE (2012) A common signaling process that promotes mycorrhizal and oomycete colonization of plants. Curr Biol 22: 2242–2246 [DOI] [PubMed] [Google Scholar]
- Wang L, Zhang L, Liu Z, Zhao D, Liu X, Zhang B, Xie J, Hong Y, Li P, Chen S, et al. (2013b) A minimal nitrogen fixation gene cluster from Paenibacillus sp. WLY78 enables expression of active nitrogenase in Escherichia coli. PLoS Genet 9: e1003865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Karki S, Biswal AK, Lin H-C, Dionora MJ, Rizal G, Yin X, Schuler ML, Hughes T, Fouracre JP, et al. (2017a) Candidate regulators of early leaf development in maize perturb hormone signalling and secondary cell wall formation when constitutively expressed in rice. Scient Rep 7: 4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Kelly S, Fouracre JP, Langdale JA (2013a) Genome-wide transcript analysis of early maize leaf development reveals gene cohorts associated with the differentiation of C4 Kranz anatomy. Plant J 75: 656–670 [DOI] [PubMed] [Google Scholar]
- Wang P, Khoshravesh R, Karki S, Tapia R, Balahadia CP, Bandyopadhyay A, Quick WP, Furbank RT, Sage TL, Langdale JA (2017b) Re-creation of a key step in the evolutionary switch from C3 to C4 leaf anatomy. Curr Biol 27: 3278–3287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Nolte MW, Shanks BH (2014) Catalytic dehydration of C6 carbohydrates for the production of hydroxymethylfurfural (HMF) as a versatile platform chemical. Green Chem 16: 548–572 [Google Scholar]
- Wang XL, Feng H, Wang YY, Wang MX, Xie XG, Chang H, Wang L, Qu J, Sun K, He W, et al. (2021) Mycorrhizal symbiosis modulates the rhizosphere microbiota to promote rhizobia-legume symbiosis. Mol Plant 14: 503–516 [DOI] [PubMed] [Google Scholar]
- Wang Y, Bao Z, Zhu Y, Hua J (2009) Analysis of temperature modulation of plant defense against biotrophic microbes. Mol Plant Microbe Interact 22: 498–506 [DOI] [PubMed] [Google Scholar]
- Watson A, Ghosh S, Williams MJ, Cuddy WS, Simmonds J, Rey MD, Asyraf Md Hatta M, Hinchliffe A, Steed A, Reynolds D, et al. (2018) Speed breeding is a powerful tool to accelerate crop research and breeding. Nat Plants 4: 23–29 [DOI] [PubMed] [Google Scholar]
- Welch JR, Vincent JR, Auffhammer M, Moya PF, Dobermann A, Dawe D (2010) Rice yields in tropical/subtropical Asia exhibit large but opposing sensitivities to minimum and maximum temperatures. Proc Natl Acad Sci USA 107: 14562–14567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellisch M, Jungmeier G, Karbowski A, Patel MK, Rogulska M (2010) Biorefinery systems - potential contributors to sustainable innovation. Biofuels Bioprod Biorefin 4: 275–286 [Google Scholar]
- Whitt L, Ricachenevsky FK, Ziegler GZ, Clemens S, Walker E, Maathuis FJM, Kear P, Baxter I (2020) A curated list of genes that affect the plant ionome. Plant Direct 4: e00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissuwa M, Kretzschmar T, Rose TJ (2016) From promise to application: root traits for enhanced nutrient capture in rice breeding. J Exp Bot 67: 3605–3615 [DOI] [PubMed] [Google Scholar]
- Wolfe MD, Del Carpio DP, Alabi O, Ezenwaka LC, Ikeogu UN, Kayondo IS, Lozano R, Okeke UG, Ozimati AA, Williams E, et al. (2017) Prospects for genomic selection in cassava breeding. Plant Genome 10: 1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods P, Lehner K, Hein K, Mullen JL, McKay JK (2022) Root pulling force across drought in maize reveals genotype by environment interactions and candidate genes. Front Plant Sci 13: 883209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A, Hammer GL, Doherty A, von Caemmerer S, Farquhar GD (2019) Quantifying impacts of enhancing photosynthesis on crop yield. Nat Plants 5: 380–388 [DOI] [PubMed] [Google Scholar]
- Xiong J, He Z, Shi S, Kent A, Deng Y, Wu L, Van Nostrand JD, Zhou J (2015) Elevated CO2 shifts the functional structure and metabolic potentials of soil microbial communities in a C4 agroecosystem. Sci Rep 5: 9316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yandigeri MS, Meena KK, Singh D, Malviya N, Singh DP, Solanki MK, Yadav AK, Arora DK (2012) Drought-tolerant endophytic actinobacteria promote growth of wheat (Triticum aestivum) under water stress conditions. Plant Growth Regul 68: 411–420. [Google Scholar]
- Yang J, Lan LLY, Jin Y, Yu N, Wang D, Wang ET (2022) Mechanisms underlying legume-rhizobium symbioses. J Integr Plant Biol 64: 244–267 [DOI] [PubMed] [Google Scholar]
- Yang M, Lu K, Zhao FJ, Xie W, Ramakrishna P, Wang G, Du Q, Liang L, Sun C, Zhao H, et al. (2018) Genome-wide association studies reveal the genetic basis of ionomic variation in rice. Plant Cell 30: 2720–2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Zheng Q, Yuan M, Shi Z, Chiariello NR, Docherty KM, Dong S, Field CB, Gu Y, Gutknecht J, et al. (2019a) Long-term elevated CO2 shifts composition of soil microbial communities in a Californian annual grassland, reducing growth and N utilization potentials. Sci Total Environ 652: 1474–1481 [DOI] [PubMed] [Google Scholar]
- Yang H, Zhang X, Luo H, Liu B, Shiga TM, Li X, Kim JI, Rubinelli P, Overton JC, Subramanyam V, et al. (2019b) Overcoming cellulose recalcitrance in woody biomass for the lignin-first biorefinery. Biotechnol Biofuels 12: 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao QH, Peng RH, Wang B, Tian YS, Zhu YM, Gao JJ, Liu ZJ, Fu XY, Xu J, Han HJ, et al. (2021) Endowing Plants with the Capacity for Autogenic Nitrogen Fixation. Preprint posted to Research Square on May 7, 2021, 10.21203/rs.3.rs-436726/v1 [DOI] [Google Scholar]
- Yoshida K, Miyashita NT (2009) DNA polymorphism in the blast disease resistance gene Pita of the wild rice Oryza rufipogon and its related species. Genes Genet Syst 84: 121–136 [DOI] [PubMed] [Google Scholar]
- Yoshida Y, Marubodee R, Ogiso-Tanaka E, Iseki K, Isemura T, Takahashi Y, Muto C, Naito K, Kaga A, Okuno K, et al. (2016) Salt tolerance in wild relatives of adzuki bean, Vigna angularis (Willd.) Ohwi et Ohashi. Genet Resour Crop Evol 63: 627–637 [Google Scholar]
- Yu H, Deng Y, He Z, Van Nostrand JD, Wang S, Jin D, Wang A, Wu L, Wang D, Tai X, Zhou J (2018a) Elevated CO2 and warming altered grassland microbial communities in soil top-layers. Front Microbiol 9: 1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Li X, Duchoud F, Chuang DS, Liao JC (2018b) Augmenting the Calvin-Benson-Bassham cycle by a synthetic malyl-CoA-glycerate carbon fixation pathway. Nat Commun 9: 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabin CJ, Jurgens LJ, Bible JM, Patten MV, Chang AL, Grosholz ED, Boyer KE (2022) Increasing the resilience of ecological restoration to extreme climatic events. Front Ecol Environ 5: 310–318 [Google Scholar]
- Zakzeski J, Bruijnincx PCA, Jongerius AL, Weckhuysen BM (2010) The catalytic valorization of lignin for the production of renewable chemicals. Chem Rev 110: 3552–3599 [DOI] [PubMed] [Google Scholar]
- Zamani Babgohari M, Niazi A, Moghadam AA, Deihimi T, Ebrahimie E (2013) Genome-wide analysis of key salinity-tolerance transporter (HKT1; 5) in wheat and wild wheat relatives (A and D genomes). In Vitro Cellular Dev Biol-Plant 49: 97–106 [Google Scholar]
- Zeng J, Yoo CG, Wang F, Pan X, Vermerris W, Tong Z (2015) Biomimetic Fenton‐catalyzed lignin depolymerization to high‐value aromatics and dicarboxylic acids. ChemSusChem 8: 861–871 [DOI] [PubMed] [Google Scholar]
- Zhang C, He J, Dai H, Wang G, Zhang X, Wang C, Shi J, Chen X, Wang D, Wang E (2021a) Discriminating symbiosis and immunity signals by receptor competition in rice. Proc Natl Acad Sci USA 118: e2023738118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Xie J (2014) Genes and QTLs resistant to biotic and abiotic stresses from wild rice and their applications in cultivar improvements. In W Yan, ed, Rice – Germplasm, Genetics and Improvement, Vol 23. Intech Open, London, pp 59–78 [Google Scholar]
- Zhang H, Mittal N, Leamy LJ, Barazani O, Song B-H (2017) Back into the wild—Apply untapped genetic diversity of wild relatives for crop improvement. Evol Appl 10: 5–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Wang ML, Schaefer R, Dang P, Jiang T, Chen C (2019) GWAS and coexpression network reveal ionomic variation in cultivated peanut. J Agric Food Chem 67: 12026–12036 [DOI] [PubMed] [Google Scholar]
- Zhang XW, Dong WT, Sun JH, Feng F, Deng YW, He ZH, Oldroyd GED, Wang E (2015) The receptor kinase CERK1 has dual functions in symbiosis and immunity signalling. Plant J 81: 258–267 [DOI] [PubMed] [Google Scholar]
- Zhang P, Wang W-Q, Zhang G-L, Kaminek M, Dobrev P, Xu J, Gruissem W (2010) Senescence-inducible expression of isopentenyl transferase extends leaf life, increases drought stress resistance and alters cytokinin metabolism in cassava. J Integr Plant Biol 52: 653–669 [DOI] [PubMed] [Google Scholar]
- Zhang S, Chen X, Lu C, Ye J, Zou M, Lu K, Feng S, Pei J, Liu C, Zhou X, et al. (2018) Genome-wide association studies of 11 agronomic traits in Cassava (Manihot esculenta Crantz). Front Plant Sci 9: 503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Gan Y, Xu B (2016) Application of plant-growth-promoting fungi Trichoderma longibrachiatum T6 enhances tolerance of wheat to salt stress through improvement of antioxidative defense system and gene expression. Front Plant Sci 7: 1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Lavallee JM, Robertson AD, Even R, Ogle SM, Paustian K, Cotrufo MF (2021b) Simulating measurable ecosystem carbon and nitrogen dynamics with the mechanistically defined MEMS 2.0 model. Biogeosciences 18: 3147–3171 [Google Scholar]
- Zhao C, Liu B, Piao S, Wang X, Lobell DB, Huang Y, Huang M, Yao Y, Bassu S, Ciais P, et al. (2017) Temperature increase reduces global yields of major crops in four independent estimates. Proc Natl Acad Sci USA 114: 9326–9331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Abu-Omar MM (2015) Biobased epoxy nanocomposites derived from lignin-based monomers. Biomacromolecules 16: 2025–2031 [DOI] [PubMed] [Google Scholar]
- Zhao X, Tekinalp H, Meng X, Ker D, Benson B, Yunqiao P, Ragauskas AJ, Wang Y, Li K, Webb E, et al. (2019) Poplar as biofiber reinforcement in composites for large-scale 3D printing. ACS Appl Bio Mater 2: 4557–4570 [DOI] [PubMed] [Google Scholar]
- Zhou LY, Zhou XH, He YH, Fu YL, Du ZG, Lu M, Sun XY, Li CH, Lu CY, Liu RQ, et al. (2022) Global systematic review with meta-analysis shows that warming effects on terrestrial plant biomass allocation are influenced by precipitation and mycorrhizal association. Nat Commun 13: 4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Luo W, Ciesielski PN, Fang Z, Zhu JY, Hendriksson G, Himmel ME, Hu L (2016) Wood-derived materials for green electronics, biological devices, and energy applications. Chem Rev 116: 9305–9374 [DOI] [PubMed] [Google Scholar]
- Zhu X-G, Ort DR, Parry MAJ, von Caemmerer S (2020) A wish list for synthetic biology in photosynthesis research. J Exp Bot 71: 2219–2225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X-G, Wang Y, Ort DR, Long SP (2012) e-photosynthesis: a comprehensive dynamic mechanistic model of C3 photosynthesis: from light capture to sucrose synthesis. Plant Cell Environ 36: 1711–1727 [DOI] [PubMed] [Google Scholar]
- Zia MH, Ahmed I, Bailey EH, Lark RM, Young SD, Lowe NM, Joy EJM, Wilson L, Zaman M, Broadley MR (2020) Site-specific factors influence the field performance of a Zn-biofortified wheat variety. Front Sustain Food Syst 4: 135 [Google Scholar]
- Ziegler RS (2007) Foreward. In JE Sheehy, PL Mitchell, B Hardy, eds, Charting new pathways to C4 rice, International Rice Research Institute, Los Baños, Phillipines, pp. 422 [Google Scholar]
- Ziegler G, Nelson R, Granada S, Krishnan HB, Gillman JD, Baxter I (2018) Genomewide association study of ionomic traits on diverse soybean populations from germplasm collections. Plant Direct 2: e00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zin NA, Badaluddin NA (2020) Biological functions of Trichoderma spp. for agriculture applications. Ann Agric Sci 65: 168–178 [Google Scholar]
- Ziska LH, Bunce JA (1998) The influence of increasing growth temperature and CO2 concentration on the ratio of respiration to photosynthesis in soybean seedlings. Global Change Biol 4: 637–643 [Google Scholar]
- Ziska LH, Bunce JA, Shimono H, Gealy DR, Baker JT, Newton PC, Reynolds MP, Jagadish KS, Zhu C, Howden M, et al. (2012) Food security and climate change: on the potential to adapt global crop production by active selection to rising atmospheric carbon dioxide. Proc R Soc B: Biol Sci 279: 4097–4105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zomer RJ, Bossio DA, Sommer R, Verchot LV (2017) Global sequestration potential of increased organic carbon in cropland soils. Sci Rep 7: 15554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zsögön A, Čermák T, Naves E, Notini MM, Edel KH, Weinl S, Freschi L, Voytas DF, Kudla J, Peres LEP (2018) De novo domestication of wild tomato using genome editing. Nat Biotechnol 36: 1211–1216 [DOI] [PubMed] [Google Scholar]
- Zsögön A, Peres LEP, Xiao Y, Yan J, Fernie AR (2022) Enhancing crop diversity for food security in the face of climate uncertainty. Plant J 109: 402–414 [DOI] [PubMed] [Google Scholar]