Abstract
To understand the RNA expression in response to acid stress of Helicobacter pylori in genomic scale, a microarray membrane containing 1,534 open reading frames (ORFs) from strain 26695 was used. Total RNAs of H. pylori under growth conditions of pH 7.2 and 5.5 were extracted, reverse transcribed into cDNA, and labeled with biotin. Each microarray membrane was hybridized with cDNA probe from the same strain under two different pH conditions and developed by a catalyzed reporter deposition method. Gene expression of all ORFs was measured by densitometry. Among the 1,534 ORFs, 53 ORFs were highly expressed (≧30% of rRNA control in densitometry ratios). There were 445 ORFs which were stably expressed (<30% of rRNA in densitometry) under both pH conditions without significant variation. A total of 80 ORFs had significantly increased expression levels at low pH, while expressions of 4 ORFs were suppressed under acidic condition. The remaining 952 ORFs were not detectable under either pH condition. These data were highly reproducible and comparable to those obtained by the RNA slot blot method. Our results suggest that microarray can be used in monitoring prokaryotic gene expression in genomic scale.
Helicobacter pylori is the causative agent of chronic superficial gastritis in humans, and the presence of this organism increases risk of development of peptic ulcer disease and adenocarcinoma and mucosa-associated lymphoid tissue lymphoma of the distal stomach (2, 3, 10, 19, 21). The mechanism of pathogenesis remains largely unknown. The ability to survive under acidic stomach conditions might be one of the virulence mechanisms. Monitoring the response of H. pylori genes during acid stress may be helpful to understand the pathogenesis.
Expression of many bacterial genes is induced in response to environmental stimuli (17). For technical reasons, only a limited subset of genes could be simultaneously analyzed until recently. Differential display of subsets of mRNA on a sequencing gel allows a broad search for expression differences (11, 15), but the method has been difficult to standardize. An rRNA subtraction approach has been used to identify differentially expressed genes in Mycobacterium avium (23); however, removal of rRNA by subtraction caused loss of mRNA. Chuang et al. (6) have analyzed differentially expressed genes in Escherichia coli by a method based on hybridization to spot overlapping λ clones. This method required subcloning and subsequent sequencing for the identification of relevant genes.
Recently, a system for monitoring of a large number of gene expressions has been developed in eukaryotes using DNA microarray (5, 23, 24) or oligonucleotide microarray (16, 22, 29). Labeled cDNA, or in vitro-transcribed mRNA, was hybridized to the high-density probe microarray. Microarrays are able to analyze the expression of hundreds of genes in a single hybridization experiment. DNA array has also been recently adopted for monitoring gene expressions in bacteria. Unfortunately, since bacterial mRNA could not be separated from rRNA, large amounts of total RNAs are needed for probe labeling, and confocal microscopy is usually necessary for result interpretation (8, 28). A method of “differential expression using customized amplification libraries” that needs only small amounts of total RNA for analysis has been developed (1). However, the optimal restriction enzyme for cosmid library construction, one that would exclude contamination by all the rRNA clones, was difficult to select. We adopted a catalyzed amplification of reporter deposition (CARD) method to explore the gene expression of H. pylori in genomic scale (5).
MATERIALS AND METHODS
Bacterial strains, growth conditions, and total RNA isolation.
An H. pylori strain from a patient with duodenum ulcer at National Taiwan University Hospital, H. pylori NTU-D1, was used for analysis. Columbia agar with 5% sheep blood and antibiotics supplement (GIBCO BRL, Rockville, Md.) were used for culture. The pH value of the medium was titrated by HC1 to pH 7.2 and 5.5, respectively. The bacterial cells were cultured at pH 7.2 and 5.5 at 37°C in a microaerophilic chamber (Don Whitley, West Yorkshire, England) containing 10% CO2, 5% O2, and 85% N2 for 48 h. Cells were harvested after 48 h of incubation for total RNA extraction. Bacterial cells were grown to 48 h on Columbia agar plates, collected, washed with Tris-EDTA (TE) buffer (pH 7.4), and pelleted. Cell pellets then were resuspended and lysed in boiled 1% sodium dodecyl sulfate (SDS)-TES buffer (TES is 50 mM Tris-hydrochloride [pH 8.0] 1 mM EDTA, and 50 mM NaCl) for 5 min; then they were subjected to phenol (pH 4.5) extractions at 65°C for 5 min, extracted twice with chloroform–isoamyl alcohol (24:1) solution, precipitated with an equal volume of isopropanol, and stored at −70°C. These crude RNA fractions typically contained substantial amounts of genomic DNA; therefore, DNA was removed by treating 30 μg of RNA with 2 U of RNase-free DNase I (Roche, Mannheim, Germany) at 37°C for 30 min. RNA samples were subjected to phenol-chloroform extraction and isopropanol precipitation, were washed once in 70% ethanol, and were redissolved in diethyl pyrocarbonate-treated water until use. Total RNAs that showed prominent 23S and 16S ribosomal bands were examined by gel electrophoresis, and the A260/A280 ratio obtained by spectrometry was kept within 1.8 to 1.9.
Microarray preparation.
PCR products that contained 1,534 predicted open reading frames (ORFs) of H. pylori strain 26695 were kindly provided by Invitrogen (San Diego, Calif.). Fifty-six ORFs—which were less than 100 nucleotides, were untranslated, or for which more than one copy of homologue was found—have been excluded from the microarray membrane (Table 1). PCR products were concentrated by evaporation at 95°C to obtain a concentration of 2 to 3 μg/μl before they were spotted onto a positively charged nylon membrane (Roche). A computer-controlled XYZ translation system (PM500; Newport, Inc., Fountain Valley, Calif.) outfitted with Teflon (Teflon-AF; DuPont, Wilmington, Del.)-coated tool steel pins was used for arraying the PCR products (11). Samples were held at the tip of the pins for delivery by the action of surface tension, and the delivery volume was governed by the number of elements in an array; arraying tools consisting of 1 pin were used. It took approximately 6 h to print an array consisting of 1,534 ORFs.
TABLE 1.
ORFs which are not included in the H. pylori microarray membranea
| ORF | ORF | |
|---|---|---|
| HP0007 | HP0533 | |
| HP0009 | HP0548 | |
| HP0023 | HP0610 | |
| HP0058 | HP0691 | |
| HP0143 | HP0698 | |
| HP0208 | HP0705 | |
| HP0225 | HP0713 | |
| HP0269 | HP0725 | |
| HP0289 | HP0738 | |
| HP0336 | HP0744 | |
| HP0341 | HP0857 | |
| HP0342 | HP0903 | |
| HP0353 | HP0904 | |
| HP0359 | HP0922 | |
| HP0360 | HP1007 | |
| HP0361 | HP1194 | |
| HP0381 | HP1198 | |
| HP0400 | HP1280 | |
| HP0402 | HP1353 | |
| HP0429 | HP1369 | |
| HP0450 | HP1417 | |
| HP0451 | HP1460 | |
| HP0453 | HP1500 | |
| HP0456 | HP1522 | |
| HP0458 | Hp1528 | |
| HP0461 | HP1536 | |
| HP0521 | HP1538 | |
| HP0527 | HP1589 |
ORFs that were less than 100 nucleotide in size, that were untranslated, or for which more than one copy of the homologous putative ORF was found were excluded from the microarray.
Additional 23S rRNA and 16S rRNA genes were amplified and spotted on the membrane for visual detection before scanning.
cDNA probe preparation and microarray hybridization.
Total RNAs from two culture conditions, pH 7.2 and 5.5, were isolated separately according to the method described above. A total of 10 μg of total RNAs from each condition was used for one labeling reaction during reverse transcription (RT) in the presence of a 12 μM concentration of random primers; a 1 mM concentration (each) of dCTP, dATP, and dGTP; 80 μM dTTP; 80 μM biotin-16-dUTP; ribonuclease inhibitor (0.5 U/μl; Roche); 10 mM DDT; and 400 U of moloney murine leukemia virus reverse transcriptase (GIBCO BRL) in 62.5 μl of solution. The reaction mixture was incubated at 42°C for 90 min, and the reaction was stopped by heating the mixture to 95°C for 5 min. The RNA was degraded by addition of 6.9 μl of 3 M NaOH followed by a 30-min incubation at 50°C. The labeled samples were neutralized by addition of 6.9 μl of 3 M acetic acid and then precipitated by addition of 75 μl of 3 M sodium acetate (pH 5.2), 20 μg of tRNA as carrier (Roche), 562.5 μl of isopropanol, and water to make a total of 784.55 μl.
The membrane carrying the PCR products targets was prehybridized in 2 ml of hybridization buffer (0.1% N-lauroylsarcosine, 0.1% SDS, 1% blocking reagent [Roche], and 40 μg of herring sperm DNA per μl) at 65°C for 2 h before hybridization. cDNA probe was mixed with 50 μl of hybridization buffer, and the reaction mixture was sealed with a membrane. The sealed bag was incubated at 95°C for 5 min and then hybridization was performed at 63°C for up to 14 h. The membrane was then washed twice with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) containing 0.1% SDS for 5 min at room temperature followed by three washes with 0.1× SSC containing 0.1% SDS at 65°C for 15 min each.
Colorimetric detection and data analysis.
After hybridization and washing, the membrane was blocked by 2 ml of blocking buffer, 1× phosphate-buffered saline (PBS) (GIBCO BRL) containing 0.05% Tween 20 and 7% casein (Sigma, St. Louis, Mo.). Horseradish peroxidase-conjugated streptavidin (GIBCO BRL) was used to detect the spots on the membrane. The membrane was incubated with 2 ml of reaction mixture containing 350-fold horseradish peroxidase-conjugated streptavidin 4% polyethylene glycol 8000 (Sigma), 0.1-fold blocking buffer in bovine serum albumin (BSA) buffer (1× PBS containing 0.05% Tween 20 [GIBCO BRL] and 1% BSA [Sigma]) at room temperature for 1 h. The membrane was then washed 5 min with a washing buffer (1× PBS containing 0.05% Tween 20) four times. The membrane was incubated in a 0.1 M borate buffer (pH 8.5; Serva, Ingelheim, Germany) containing 0.0035% hydroperoxidase and biotinyl-tyramide (15 μg/ml) for 15 min without shaking and washed 5 min with a washing buffer four times after incubation. The membrane was then incubated with 2 ml of reaction mixture containing β-galactosidase-conjugated streptavidin (2.76 U/ml) (GIBCO BRL), 4% polyethylene glycol 8000 (Sigma), and 0.1× blocking buffer in BSA buffer at room temperature for 1 h and was washed with a washing buffer four times (5 min each). The chromogens were generated by treating the membrane at 37°C for approximately 10 min with 2 ml of X-Gal substrate, which contained 1.2 mM X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) 1 mM MgCl2, 3 mM K3Fe(CN)6, and 3 mM K4Fe(CN)6 in 1× TBS buffer solution (10 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.3% BSA) The membrane was then briefly rinsed with deionized water, and the color development reactions were stopped by adding 1× PBS containing 20 mM EDTA.
To determine the results from arrays of different density, the membrane was scanned by a PowerLook 3000 scanner (Umax, Hsinchu, Taiwan) with an optical resolution of 3,048 by 3,048 dpi. Densitometry was performed with imaging software (version 1.62; National Institutes of Health, Bethesda, Md.) after magnification (0.46 by 0.46 in. to 12.3 by 12.3 in.) of the scanned images, using 23S or 16S rRNA genes as internal standard.
Slot blots.
Ten micrograms of total RNA was analyzed on 1% denaturing agarose gel. The total RNAs were transferred onto a nylon membrane by passive vacuum pressure. Membrane was prehybridized with a hybridization buffer at 55°C for 18 h and hybridized with digoxigenin (DIG)-labeled antisense RNA at 55°C for 18 h. Detection was performed with a DIG Luminescent Detection kit (Roche) according to the manufacturer's instructions.
Semiquantitative PCR.
One microgram of total RNA was isolated and reverse transcribed by using the specific antisense primer and moloney murine leukemia virus reverse transcriptase (Ambion, Austin, Tex.) at 37°C for 2 h. Tenfold serial dilutions of cDNA were made, and the end point was determined by negativity of PCR results. PCR was performed with Taq polymerase and a buffer containing 1.75 mM MgCl2 for 30 cycles of annealing at 55°C for 15 s, denaturing at 94°C for 30 s, and extension at 72°C for 2 min. Primer pair sequences were as follows: for HP1037-f, 5′-ATGAAAGGATTAGAAAGAGAATCGC-3′; for HP1037-r, 5′-CAAAAGCTCAGACCTAGAATTTTTG-3′.
Detection limit of the CARD method.
An 800-bp fragment of the 23S rRNA gene was first amplified by PCR. After isopropanol precipitation to concentrate the PCR products, the PCR products were spotted onto the nylon membrane (Roche). The PCR products were also subcloned into a pCR2.1 vector by a TA Cloning kit (Invitrogen). Ten micrograms of 23S rRNA was synthesized according to the standard protocols provided in the Maxi in vitro transcription kit (Ambion). The biotin-labeled cDNA probe was obtained through reverse transcription. After quantification of the cDNA probe, 10-fold dilutions were made by following the microarray hybridization protocols mentioned above. The end point was determined by negativity of colorimetric development. Primer pair sequences were as follows: for 23SR, 5′-GCGTTGAATTGAAGCCCGAG-3′; for 23SF, 5′-TGTGTGCTACCCAGCGATGC-3′.
RESULTS
Expression of H. pylori genes on microarray.
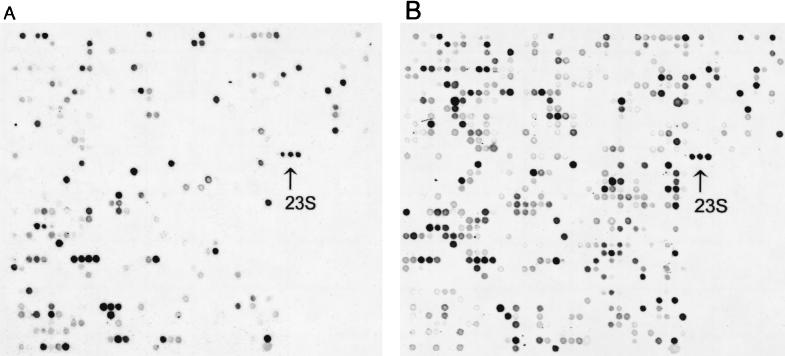
To identify genes of H. pylori that were expressed in responses to acid stress, array hybridization signals were compared between RNA from pH 7.2 and 5.5 conditions after standardization with internal rRNA controls (Fig. 1). Among 1,534 ORFs, 53 ORFs were highly expressed under both growth conditions (defined by a densitometry ratio of ≧0.3 compared with the 23S rRNA internal standard, including 22 ORFs with 23S rRNA ratios of >1.0 and 31 ORFs with ratios between 0.3 and 1.0) (Table 2). There were 445 ORFs that were detectable under both pH conditions (with 23S rRNA ratios of <0.3), and the variations of those ORFs between the two growth conditions were not significant (data not shown). There were 80 ORFs that had significantly increased (defined as greater-than-fivefold increase of mean densitometry) expression levels at pH 5.5 (Table 3). These 80 ORFs were divided into three groups: (i) 16 ORFs that were reported to be involved in acidic stress response, (ii) 21 ORFs that had no database match, and (iii) 43 ORFs homologous to previously reported sequences but not reported to be related to acidic stress response. In contrast, only four ORFs had decreased (defined as greater-than-fivefold decrease of mean densitometry) expression levels during acidic stress (Table 3). The remaining 952 genes were not detectable under either pH condition.
FIG. 1.
Colorimetric detection of H. pylori total RNA expression on a microarray. Comparison of acidic stress responses by culture at pH 7.2 (A) and pH 5.5 (B). The expression level of 23S rRNA at pH 5.5 was slightly higher than those at pH 7.2; however, each mRNA expression was standardized by using 23S rRNA as an internal control. 23S: additional 23S rRNA gene. The figures are amplified from 0.4 by 0.4 in. to 4 by 4 in.
TABLE 2.
H. pylori ORFs highly expressed in microarraya
| ORF (expressionb) | Putative gene product (gene) |
|---|---|
| HP0011 (1.2) | Cochaperone (groES) |
| HP0056 (0.7) | Delta-1-pyrroline-5-carboxylate dehydrogenase |
| HP0072 (1.5) | Urease beta subunit (urea amidohydrolase) (ureB) |
| HP0097 (1.0) | Hypothetical protein |
| HP0242 (0.3) | Hypothetical protein |
| HP0294 (1.1) | Aliphatic amidase (aimE) |
| HP0306 (0.8) | Glutamate-1-semialdehyde 2,1-aminomutase (hemL) |
| HP0311 (0.9) | Hypothetical protein |
| HP0389 (1.0) | Superoxide dismutase (sodB) |
| HP0390 (0.4) | Adhesin-thiol peroxidase (tagD) |
| HP0411 (0.6) | Hypothetical protein |
| HP0452 (1.1) | Hypothetical protein |
| HP0486 (1.2) | Hypothetical protein |
| HP0515 (0.6) | Heat shock protein (hslV) |
| HP0540 (0.6) | cag pathogenicity island protein (cag19) |
| HP0547 (1.2) | cag pathogenicity island protein (cag26) |
| HP0570 (0.9) | Aminopeptidase a/i (pepA) |
| HP0558 (1.1) | Beta ketoacyl-acyl carrier protein synthase II (fabF) |
| HP0561 (0.7) | 3-Ketoacyl-acyl carrier protein reductase (fabG) |
| HP0591 (0.6) | Ferredoxin oxidoreductase, gamma subunit |
| HP0593 (0.9) | Adenine specific DNA methyltransferase (mod) |
| HP0596 (1.3) | Hypothetical protein |
| HP0601 (1.2) | Flagellin A (flaA) |
| HP0627 (0.7) | Hypothetical protein |
| HP0648 (1.7) | UDP-N-acetylglucosamine enolpyruvyl transferase (murZ) |
| HP0682 (0.9) | Hypothetical protein |
| HP0695 (1.4) | Hydantoin utilization protein A (hyuA) |
| HP0696 (1.4) | N-Methylhydantoinase |
| HP0697 (0.6) | Hypothetical protein |
| HP0719 (0.9) | Hypothetical protein |
| HP0784 (1.4) | Hypothetical protein |
| HP0865 (0.7) | Deoxyuridine 5′-triphosphate nucleotidohydrolase (dut) |
| HP0866 (0.4) | Transcription elongation factor GreA (greA) |
| HP0871 (0.8) | CDP-diglyceride hydrolase (cdh) |
| HP0873 (1.2) | Hypothetical protein |
| HP0897 (0.5) | Hypothetical protein |
| HP0900 (0.6) | Hydrogenase expression-formation protein (hypB) |
| HP0950 (1.1) | Acetyl coenzyme A carboxylase beta subunit (accD) |
| HP0951 (0.9) | Fumarate reductase, flavoprotein subunit (frdA) |
| HP0978 (0.9) | Cell division protein (ftsA) |
| HP0979 (0.9) | Cell division protein (ftsZ) |
| HP0981 (1.1) | Exonuclease VII-like protein (xseA) |
| HP1153-2 (1.3) | Yalyl-tRNA synthetase (valS) |
| HP1241-2 (0.5) | Alanyl-tRNA synthetase (alaS) |
| HP1243-2 (0.7) | Outer membrane protein (omp28) |
| HP1390 (1.4) | Hypothetical protein |
| HP1398 (1.3) | Alanine dehydrogenase (ald) |
| HP1445 (0.7) | Biopolymer transport protein (exbB) |
| HP1454 (0.8) | Hypothetical protein |
| HP1468 (0.9) | Branched-chain amino acid aminotransferase (ilvE) |
| HP1469 (0.5) | Outer membrane protein (omp31) |
| HP1558 (1.3) | Conserved hypothetical protein |
| HP1527 (0.8) | Hypothetical protein |
| 23S (1.0)c | 23S rRNA |
Among of 1,534 ORFs, 53 ORFs were highly expressed (i.e., had a densitometry ratio of >0.3 compared to the internal standard [23S rRNA]), including 22 ORFs that were very highly expressed (ratio > 1.0) and 31 ORFs with densitometry ratios between 0.3 and 1.0.
The expression level is shown as the mean densitometry ratio of each ORF to 23S rRNA.
The 23S rRNA gene was used as an internal control.
TABLE 3.
H. pylori ORFs that were up- or down-regulated under acidic culture conditionsa
| Group | Level of expression at ptt:
|
ORF no. | Putative gene product (gene) | |
|---|---|---|---|---|
| 5.5 | 7.2 | |||
| i | 49,466 | 0 | HP0110 | Cochaperone and heat shock protein (grpE) |
| 46,604 | 4,073 | HP0634 | Guinone-reactive Ni-Fe hydrogenase (hydD) | |
| 96,464 | 4,680 | HP0632 | Quinone-reactive Ni-Fe hydrogenase, large subunit (hydB) | |
| 113,952 | 0 | HP1131 | ATP synthase F1, subunit epsilon (atpC) | |
| 68,410 | 0 | HP1374 | ATP-dependent protease, ATPase subunit (clpX) | |
| 75,581 | 0 | HP1134 | ATP synthase F1, subunit alpha (atpA) | |
| 49,090 | 0 | HP1174 | Glucose-Galactose transporter (gluP) | |
| 77,173 | 0 | HP1263 | NADH-ubiquinone oxidoreductase, NQO4 subunit (NQO4) | |
| 51,034 | 2,264 | HP0010-2 | Chaperone and heat shock protein (groEL) | |
| 19,024 | 0 | HP1562 | Iron(III) ABC transporter, periplasmic iron-binding protein (ceuE) | |
| 54,576 | 0 | HP1072 | Copper-transporting ATPase, P type (copA) | |
| 40,839 | 879 | HP0853 | ABC transporter, ATP-binding protein (yheS) | |
| 72,552 | 3,597 | HP0300 | Dipeptide ABC transporter, permease protein (dppC) | |
| 22,505 | 1,879 | HP1011 | Dihydroorotate dehydrogenase (pyrD) | |
| 40,494 | 0 | HP0194 | Triosephosphate isomerase (tpi) | |
| 80,230 | 0 | HP0140 | l-Lactate permease (lctP) | |
| ii | 66,557 | 0 | HP0150 | Hypothetical protein |
| 18,029 | 0 | HP0831 | Conserved hypothetical ATP binding protein | |
| 91,934 | 4,092 | HP0487 | Hypothetical protein | |
| 52,218 | 3,854 | HP0406 | Hypothetical protein | |
| 29,790 | 1,756 | HP1440 | Hypothetical protein | |
| 41,783 | 1,735 | HP1363 | Conserved hypothetical integral membrane protein | |
| 58,274 | 3,196 | HP1335 | Conserved hypothetical protein | |
| 39,315 | 0 | HP1175 | Conserved hypothetical integral membrane protein | |
| 18,625 | 988 | HP0595 | Hypothetical protein | |
| 64,383 | 0 | HP0156 | Hypothetical protein | |
| 34,019 | 5,719 | HP0746 | Hypothetical protein | |
| 103,799 | 19,796 | HP1037 | Conserved hypothetical protein | |
| 109,248 | 0 | HP1162 | Conserved hypothetical integral membrane protein | |
| 61,191 | 654 | HP1163 | Hypothetical protein | |
| 39,204 | 1,668 | HP1211 | Hypothetical protein | |
| 62,761 | 0 | HP0270 | Hypothetical protein | |
| 68,324 | 0 | HP0880 | Hypothetical protein | |
| 104,157 | 0 | HP1214 | Conserved hypothetical protein | |
| 50,423 | 887 | HP0222 | Hypothetical protein | |
| 92,485 | 5,194 | HP0983 | Conserved hypothetical integral membrane protein | |
| 58,392 | 2,075 | HP0232 | Secreted protein involved in flagellar motility | |
| iii | 52,051 | 0 | HP1201 | Ribosomal protein L1 (rpl1) |
| 92,256 | 0 | HP1300 | Preprotein translocase subunit (secY) | |
| 54,264 | 0 | HP0588 | Ferrodoxin-like protein | |
| 40,818 | 0 | HP0911 | rep helicase, single-stranded DNA-dependent ATPase (rep) | |
| 49,973 | 0 | HP1304 | Ribosomal protein L6 (rpl6) | |
| 94,154 | 3,953 | HP1104 | Cinnamyl-alcohol dehydrogenase ELI3-2 (cad) | |
| 75,662 | 898 | HP1470 | DNA polymerase I (polA) | |
| 80,292 | 0 | HP0196 | UDP-3-0-(3-hydroxymyristoyl) glucosamine N-acyltransferase (lpxD) | |
| 84,545 | 1,021 | HP0214 | Sodium-dependent transporter (huNaDC-1) | |
| 28,505 | 2,112 | HP0743 | Rod shape-determining protein (mreB) | |
| 89,336 | 1,948 | HP0830 | Amidase | |
| 58,542 | 0 | HP0331 | Cell division inhibitor (minD) | |
| 42,286 | 0 | HP0237 | Porphobilinogen deaminase (hemC) | |
| 63,728 | 1,477 | HP0265 | Cytochrome c biogenesis protein (ccdA) | |
| 38,511 | 0 | HP0295 | Flagellin B homolog (fla) | |
| 88,959 | 865 | HP1209 | Ulcer-associated gene restriction endonuclease (iceA) | |
| 15,816 | 0 | HP1050 | Homoserine kinase (thrB) | |
| 21,697 | 0 | HP1539 | Ubiquinol cytochrome c oxidoreductase, cytochrome b subunit (fbcH) | |
| 53,777 | 1,371 | HP1540 | Ubiquinol cytochrome c oxidoreductase, Rieske 2Fe-2S subunit (fbcF) | |
| 30,078 | 0 | HP1035 | Flagellar biosynthesis protein (flhF) | |
| 42,540 | 0 | HP0680 | Ribonucleoside-diphosphate reductase 1, alpha subunit (nrdA) | |
| 79,427 | 1,565 | HP0786 | Preprotein translocase subunit (secA) | |
| 57,499 | 1,007 | HP0103 | methyl-accepting chemotaxis protein (tlpB) | |
| 109,974 | 3,501 | HP1069 | Cell division protein (ftsH) | |
| 19,721 | 0 | HP1543 | toxR-activated gene (tagE) | |
| 108,001 | 1,634 | HP0825 | Thioredoxin reductase (trxB) | |
| 72,762 | 1,842 | HP1190 | Histidyl-tRNA synthetase (hisS) | |
| 59,353 | 1,438 | HP0348 | Single-stranded-DNA-specific exonuclease (recJ) | |
| 5.5 | 7.2 | |||
| 21,081 | 0 | HP0286 | Cell division protein (ftsH) | |
| 50,433 | 0 | HP0370 | Biotin carboxylase (accC) | |
| 72,258 | 0 | HP0480 | GTP-binding protein, fusA product homolog (yihK) | |
| 26,381 | 0 | HP0123 | Threonyl-tRNA synthetase (thrS) | |
| 21,951 | 0 | HP0099 | methyl-accepting chemotaxis protein (tlpA) | |
| 71,210 | 1,754 | HP0183 | Serine hydroxymethyltransferase (glyA) | |
| 115,474 | 0 | HP0407 | Biotin sulfoxide reductase (bisC) | |
| 12,266 | 0 | HP1139 | SpoOJ regulator (soj) | |
| 84,907 | 11,995 | HP0634 | Quinone-reactive Ni-Fe hydrogenase (hydD) | |
| 53,468 | 5,219 | HP0572-2 | Adenine phosphoribosyltransferase (apt) | |
| 166,46 | 0 | HP1372 | Rod shape-determining protein (mreC) | |
| 107,016 | 14,166 | HP0632 | Quinone-reactive Ni-Fe hydrogenase, large subunit (hydB) | |
| 104,705 | 0 | HP1290 | Nicotinamide mononucleotide transporter (pnuC) | |
| 21,740 | 4,465 | HP1448 | Ribonuclease P, protein component (rnpA) | |
| Down-regulated | 4,016 | 29,484 | HP0947 | Hypothetical protein |
| 1,308 | 11,868 | HP0918 | Hypothetical protein | |
| 3,712 | 23,231 | HP1271 | NADH-ubiquinone oxidoreductase, NQO12 subunit (NQO12) | |
| 2,779 | 31,518 | HP1272 | NADH-ubiquinone oxidoreductase, NQO13 subunit (NQO13) | |
A total of 80 ORFs had increased expression levels during acid stress: group i, ORFs that were known corresponded to acid stress; group ii, those without database match; and group iii, ORFs that were not reported to be related to acid response. Four ORFs had decreased expression levels under acid conditions (Down-regulated).
Comparison of expression levels in slot blotting and microarray.
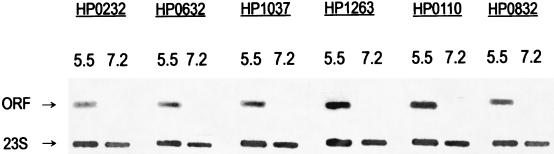
To confirm the expression level in microarray, six ORFs were randomly selected for fluorescence-labeled slot hybridization. Those ORFs included (i) HP0632, which encodes the enzyme quinone-reactive Ni-Fe hydrogenase; (ii) HP0110, which encodes a cochaperone and heat shock protein; (iii) HP1263, which encodes a homologue of the NADH-ubiquinone oxidoreductase NQO4 subunit, (iv) HP0830, which encodes the Glu-tRNA amidotransferase A subunit; (v) HP0232, which encodes a secreted protein involved in flagellar motility; and (vi) HP1037, which encodes a conserved hypothetical protein. The results of slot blotting were quantified by using NIH Image 1.62 software to evaluate rRNA levels, and the ratios are shown in Table 4. Expression levels of the six ORFs obtained in the slot blot assay correlated well with those obtained by microarray (Fig. 2 and Table 4).
TABLE 4.
Comparison of expression levels by microarray at different pHsa
| Method | pH | Expression levelb
|
|||||
|---|---|---|---|---|---|---|---|
| HP0232 | HP0632 | HP1037 | HP1263 | HP0110 | HP0832 | ||
| Slot blot | 7.2 | 0 | 0 | 0 | 0 | 0 | 0 |
| Slot blot | 5.5 | 0.7 | 0.7 | 0.6 | 0.7 | 0.8 | 0.5 |
| Microarray | 5.5 | 0.6 | 1.1 | 1 | 0.8 | 0.7 | 0.8 |
The amounts of each spot of microarray and slot blot hybridization were calculated by densitometry with NIH Image 1.62 software.
Expression levels were calculated as mean ORF/23S rRNA ratios.
FIG. 2.
Slot blot hybridization of RNA isolated from H. pylori cells grown at pH 7.2 and 5.5. Probes were either the cloned 23S H. pylori rRNA gene or the pCRII insert (H. pylori ORFs). The amount of RNA used for all conditions was standardized at 10 μg per slot. The expressions of these ORFs were much greater at acidic pHs than at pH 7.2. 23S, H. pylori 23S rRNA, 5.5, total RNA isolated from growth condition at pH 5.5, 7.2, total RNA isolated from growth condition at pH 7.2; ORF, probes that were used for hybridization.
Detection and evaluation of HP1037 mRNA expression by semiquantitative RT-PCR.
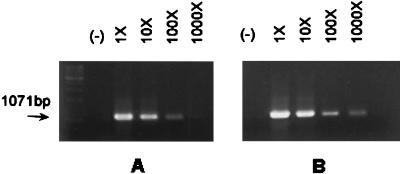
Because HP1037 was detectable by microarray at pH 7.2 but not by slot blotting (Table 4), a semiquantitative RT-PCR assay was done by 10-fold serial dilution of cDNA. At pH 7.2, there were 5 × 109 copies of HP1037 mRNA and there were 1011 copies of HP1037 mRNA at pH 5.5 as estimated by this method (Fig. 3). Therefore, the sensitivity of the CARD method should be less than 5 × 109 copies.
FIG. 3.
Semiquantification of HP1037 ORF mRNA by PCR. Total RNAs were isolated at pH 7.2 (A) and pH 5.5 (B) separately. One microgram from each was reverse transcribed with antisense primer HP1037-r. Tenfold serial dilutions of cDNA were made, and the end point was determined by negativity of PCR. (−), RNA templates not subjected to the RT reaction served as negative control to exclude the possibility of contamination of genomic DNA; 1×, 1 μg of total RNA used as template; 10×, 100×, and 1000×, sample was at a 10-, 100-, or 1,000-fold serial dilution, respectively.
Detection limit of CARD method.
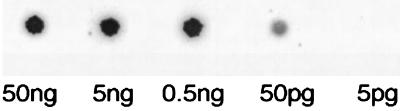
To determine the sensitivity of the CARD method, 23S rRNA was transcribed in vitro and then biotin-labeled cDNA probe was obtained through RT. CARD was done by 10-fold dilution of cDNA probe. The 23S rRNA gene could be detected by probes from 50 pg but not by those from 5 pg of rRNA. Therefore, it was estimated that the limitation of the CARD method was 108 copies of mRNA/10 μg of rRNA.
Variations of each ORF in microarray results.
To estimate the reproducibility of the RNA expression detected on microarray, the variations of densitometry value of each gene under the same pH condition and on different membranes were compared. In four experiments at pH 7.2, 554 ORFs were detected by microarray. Of these 554 ORFs, the variations of densitometry values of 227 ORFs (50%) were less than 10%, those of 185 ORFs (33%) were less than 20%, and those of the remaining 92 ORFs were less than 40%. In three experiments at pH 5.5, 582 ORFs were detected by microarray. The variations of the densitometry values of 360 ORFs (62%) were less than 10%, and those of 149 ORFs (25%) were less than 20%. The variations of densitometry value of the remaining 73 ORFs were less than 40%.
DISCUSSION
The densitometry values obtained by the microarray in up to four experiments at pH 7.2 and three experiments at pH 5.5 were evaluated. All the expression levels varied less than 40% by densitometry, and for 80% of them the variations were within 20% by densitometry. In our data, slot blot hybridization using a DIG labeling detection method correlated well with results by the CARD method on microarray (Fig. 2 and Table 4). However, the CARD method seemed to be slightly more sensitive than the slot blot method, since we observed one ORF (HP1037) detected by microarray but undetectable by slot blotting. Semiquantification by cDNA dilution and PCR confirmed that the sensitivity of microarray should be less than 109 copies of mRNA in each hybridization. The limitation of the CARD method by in vitro-transcribed 23S rRNA reached approximately 108 copies/10 μg of RNA, which was in agreement with the semiquantitative RT-PCR (Fig. 4). The CARD method was slightly less sensitive, but its sensitivity was close to that of the laser-induced fluorescence method, which reached approximated 6 × 107 copies in each hybridization (4, 7). However, in colorimetric methods, the patterns of gene expression can be visually observed, and the DNA on the membrane can be identified by this method. The colorimetric method took 24 h in each hybridization. Laser-induced fluorescence detection is faster than the colorimetric method since no enzyme incubation period is needed. However, the costs of an imaging system based on either laser-induced fluorescence emission or phosphorescence from β-particle emission is higher than those of the colorimetric method.
FIG. 4.
Determination of detection limit of CARD method. To determine the detection limit of the CARD method, spotted 23S rRNA genes in the array membrane were hybridized with in an vitro-transcribed 23S rRNA fragment. Biotin-cDNA probe was synthesized during the RT reaction. Tenfold dilutions of cDNA probes from 1 μg of rRNA were made according to CARD protocols. The limit of CARD was determined to be 50 pg, which corresponds to approximately 108 copies of 23S rRNA.
There were 53 ORFs that were highly expressed under both neutral and acidic conditions (Table 2). This implicated the fact that these genes have important or essential functions in bacteriological physiology. For example, with HP0601, flaA, which encodes a major flagellin, was found to be highly expressed at either pH 7.2 or 5.5, and this gene has been shown to be essential for H. pylori motility and colonization (9, 13). It allows the bacteria to spread through the viscous mucus covering the epithelial cells of the gastric mucosa (9, 13).
There were 445 ORFs that were stably expressed (the densitometry ratios were less than 0.3 compared to 23S rRNA as an internal control, and the variations of these genes under two pH conditions were not significant). These genes were probably also needed to maintain the basic physiology in H. pylori. However, one of them, the ureI gene, has been reported to be involved in an H+-gated urea channel regulating cytoplasmic urease activity (27). The gene is essential for gastric survival and colonization and will increase its protein level approximately 100-fold during acid stress (27). The fact that the mRNA level of ureI did not significantly increase as shown by our microarray detection suggested that the regulation of this gene could be at the posttranscriptional level. CagA protein expression has been reported to increase twofold under low pH conditions, indicating that cagA plays a role in acidic adaptation of H. pylori under particular circumstances (14). In our results, the mRNA expression of cagA was increased approximately 1.4-fold under acidic conditions. However, it did not increase fivefold, which was our cutoff point. There were 952 ORFs that remained undetectable for any signal in microarray. Why these ORFs were undetectable could be due to one of the following reasons. (i) The expression levels of these genes might be lower than the detection limit of the CARD method, or these ORFs might be expressed only under specific conditions. (ii) Some of these ORFs might not represent true genes with transcripts.
There are 80 ORFs that increased their expression levels significantly during acid stress (Table 3). We divided them into three groups. (i) Those reported to be involved in acidic stress response constituted the first group. For example, Omp11, encoded by HP0472, is a member of the proton-translocating ATPase family. Omp11 plays a role in pH regulation by extruding protons from the cytoplasm. This is in agreement with its functions, which were involved in importing divalent cation and eliminating toxic metals (18). Furthermore, arginase, encoded by HP1399, is orthologous to the product of Baccillus subtilis rocF gene. It is involved in urea cycle regulation and crucial for acid protection in vitro since the rocF mutant was 1,000-fold more sensitive to acid exposure (20). Regulation of the iron uptake system is another important feature related to the survival of the bacteria under extremely acidic conditions (18). HP1562, which encodes an iron(III) ATP-binding cassette (ABC) transporter, an important protein for the ferric siderophore enterochelin uptake. (ii) Those without database match constituted the second group. (iii) Those homologous to reported sequences not related to acidic stress response constituted the third group. For example, HP0632, which encodes quinone-reactive Ni-Fe dehydrogenase, was an important protein in respiratory electron-generating dehygrogenase for metabolic energy generation. Bacterial respiratory chains have a modular character, comprising dehydrogenase complexes, quinone pools, and terminal oxidoreductases. This enzyme's bioenergic functions include generation of proton motive force, maintenance of intracellular redox balance, and control of dioxygen concentration. Thus, this regulation system could help bacteria respond to and survive acidic environmental changes. Although the functions of these ORFs and their roles in acid regulation of H. pylori remain unknown, our results could give a direction for future studies.
Four ORFs had decreased expression levels during acid stress (Table 3). Two of them, HP1271 and HP1272, encode NADH-ubiquinone oxidoreductase, which is involved in NADH-quinone oxidoreductase respiratory chain complex-1 (NDH-1) synthesis (26). In bacteria, this complex is made up of 14 protein subunits (30). The H. pylori NDH-1 operon has genes encoding 12 subunits (18), which are HP1260 to HP1263 and HP1266 to HP1273. This complex functions as a proton pump in H. pylori. Expression of HP1271 and HP1272 was profoundly suppressed during acid stress, indicating that the translocation of protons across the membrane could have been suppressed during acid response (10). In others related genes of NDH-1 synthesis, such as HP1260 to HP1263 and HP1266 to HP1270 the fact that suppression cannot be detected under acidic conditions may be due to the tiny amount of its expression levels. The remaining two ORFs without database match, HP0918 and HP0947, await further studies.
In summary, we have monitored H. pylori gene expression at the genomic level by a microarray and a CARD method. Expressions of 952 ORFs are undetectable at both pH 7.2 and 5.5. There are 498 ORFs stably expressed under both pH conditions without significant variation, 80 ORFs whose expression levels significantly increase under acidic conditions, and four ORFs that are suppressed by acidic conditions. Under the same pH conditions, expression levels of all detectable ORFs varied less than 40% in different hybridization experiments. The expression levels obtained by microarray correlate well with those obtained by slot blotting. The detection limit of the CARD method reached 108 copies in each hybridization. Therefore, this method can be used for genomic-scale detection of prokaryotic gene expressions.
ACKNOWLEDGMENTS
This study was supported by grants from the Taiwan Department of Health (DOH88-TD-1118J and DOH89-TD-1145).
We thank Uday Matrubutham, Michelle Sindici, and Martin Gleeson (Invirogen Corporation) for providing the PCR products of each ORF of H. pylori and Douglas Berg, Washington University, St. Louis, Mo., for providing the chromosomal DNA for H. pylori strain 26695.
REFERENCES
- 1.Alland D, Kramnik I, Weisbrod T R, Otsubo L, Cerny R, Miller L P, Jacobs W R, Jr, Bloom B R. Identification of differentially expressed mRNA in prokaryotic organisms by customized amplification libraries (DECAL): the effect of isoniazid on gene expression in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 1998;95:13227–13232. doi: 10.1073/pnas.95.22.13227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bayerdorffer E, Neubauer A, Rudolph B, Thiede C, Lehn N, Eidt S, Stolte M. Regression of primary gastric lymphoma of mucosa-associated lymphoid tissue type after cure of Helicobacter pylori infection. MALT Lymphoma Study Group. Lancet. 1995;345:1591–1594. doi: 10.1016/s0140-6736(95)90113-2. [DOI] [PubMed] [Google Scholar]
- 3.Blaser M J. The bacteria behind ulcers. Sci Am. 1996;274:104–107. doi: 10.1038/scientificamerican0296-104. [DOI] [PubMed] [Google Scholar]
- 4.Chen F T, Tusak A, Pentoney S, Jr, Konrad K, Lew C, Koh E, Sternberg J. Semiconductor laser-induced fluorescence detection in capillary electrophoresis using a cyanine dye. J Chromatogr A. 1993;652:355–360. doi: 10.1016/0021-9673(93)83253-o. [DOI] [PubMed] [Google Scholar]
- 5.Chen J J, Wu R, Yang P C, Huang J Y, Sher Y P, Han M H, Kao W C, Lee P J, Chiu T F, Chang F, Chu Y W, Wu C W, Peck K. Profiling expression patterns and isolating differentially expressed genes by cDNA microarray system with colorimetry detection. Genomics. 1998;51:313–324. doi: 10.1006/geno.1998.5354. [DOI] [PubMed] [Google Scholar]
- 6.Chuang S E, Daniels D L, Blatner F R. Global regulation of gene expression in Escherichia coli. J Bacteriol. 1993;175:2026–2036. doi: 10.1128/jb.175.7.2026-2036.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeRisi J L, Iyer V R, Brown P O. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 8.De Saizieu A, Certa U, Warrington J, Gray C, Keck W, Mous J. Bacterial transcript imaging by hybridization of total RNA to oligonucleotide arrays. Nat Biotechnol. 1998;16:45–48. doi: 10.1038/nbt0198-45. [DOI] [PubMed] [Google Scholar]
- 9.Eaton K A. Motility as a factor in the colonization of gnotobiotic piglets by Helicobacter pylori. J Med Microbiol. 1992;37:123–127. doi: 10.1099/00222615-37-2-123. [DOI] [PubMed] [Google Scholar]
- 10.Finel M. Does NADH play a central role in energy metabolism in Helicobacter pylori? Trends Biochem Sci. 1998;23:412–414. doi: 10.1016/s0968-0004(98)01276-6. [DOI] [PubMed] [Google Scholar]
- 11.Fislage R, Berceanu M, Humboldt Y, Wendt M, Oberender H. Primer design for a prokaryotic differential display PT-POP. Nucleic Acids Res. 1997;25:1830–1835. doi: 10.1093/nar/25.9.1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graham D Y, Lew G M, Klein P D, Evans D G, Evans D J, Jr, Saeed Z A, Malaty H M. Effect of treatment of Helicobacter pylori infection on the long-term recurrence of gastric or duodenal ulcer. A randomized, controlled study. Ann Intern Med. 1992;116:705–708. doi: 10.7326/0003-4819-116-9-705. [DOI] [PubMed] [Google Scholar]
- 13.Josenhans C, Labigne A, Suerbaum S. Comparative ultrastructural and functional studies of Helicobacter pylori and Helicobacter mustelae flagellin mutants: both flagellin subunits, FlaA and FlaB, are necessary for full motility in Helicobacter species. J Bacteriol. 1995;11:3010–3020. doi: 10.1128/jb.177.11.3010-3020.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karita M, Blaser M J. Acid-tolerance response in Helicobacter pylori and differences between cagA+ and cagA− strains. J Infect Dis. 1998;178:213–219. doi: 10.1086/515606. [DOI] [PubMed] [Google Scholar]
- 15.Liang P, Pardee A B. Differential display eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 16.Lockhart D J, Dong H, Byme M C, Follettie M T, Gallo M V, Chee M S. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- 17.Mahan M J, Slauch J M, Hanna P C, Camilli A, Tobias J W, Waldor M K, Mekalanos J J. Selection for bacterial genes that are specifically induced in host tissues: the hunt for virulence factors. Infect Agents Dis. 1993;2:263–268. [PubMed] [Google Scholar]
- 18.Marais A, Mendz G L, Hazell S L, Megraud F. Metabolism and genetics of Helicobacter pylori: the genome era. Microbiol Mol Biol Rev. 1999;63:642–674. doi: 10.1128/mmbr.63.3.642-674.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marshall B J, Warren J R. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1984;i:1311–1315. doi: 10.1016/s0140-6736(84)91816-6. [DOI] [PubMed] [Google Scholar]
- 20.McGee D J, Radcliff F J, Mendz G L, Ferrero R L, Mobley H L. Helicobacter pylori rocF is required for arginase activity and acid protection in vitro but is not essential for colonization of mice or for urease activity. J Bacteriol. 1999;181:7314–7322. doi: 10.1128/jb.181.23.7314-7322.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parsonnet J, Hansen S, Radriguez L, Gelb A B, Warnke R A, Jellum E, Orentreich N. Helicobacter pylori infection and gastric lymphoma. N Engl J Med. 1994;330:1267–1271. doi: 10.1056/NEJM199405053301803. [DOI] [PubMed] [Google Scholar]
- 22.Pease A C, Solas D, Sullivan E J, Cronin M T, Holmes C P, Fodor S P A. Sight-directed oligonucleotide arrays for rapid DNA sequence analysis. Proc Natl Acad Sci USA. 1994;91:5022–5026. doi: 10.1073/pnas.91.11.5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Plum G, Clark-Curtiss J E. Induction of Mycobacterium avium gene expression following phagocytosis by human macrophages. Infect Immun. 1994;62:476–483. doi: 10.1128/iai.62.2.476-483.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schena M, Shalon D, Davis R W, Brown P O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 25.Shalon D, Smith S J, Brown P O. A DNA microarray system for analyzing complex DNA samples using two-color fluorescent probe hybridization. Genome Res. 1996;6:639–645. doi: 10.1101/gr.6.7.639. [DOI] [PubMed] [Google Scholar]
- 26.Tomb J F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, Nelson K, Quackenbush J, Zhou L, Kirkness E F, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H G, Glodek A, McKenney K, Fitzegerald L M, Lee N, Adams M D, Venter J C. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 27.Weeks D L, Eskandari S, Scott D R, Sachs G. A H+-gated urea channel: the link between Helicobacter pylori urease and gastric colonization. Science. 2000;287:482–485. doi: 10.1126/science.287.5452.482. [DOI] [PubMed] [Google Scholar]
- 28.Wilson M, DeRisi J, Kristensen H H, Imboden P, Rane S, Brown P O, Schoolnik G K. Exploring drug-induced alterations in gene expression in Mycobacterium tuberculosis by microarray hybridization. Proc Natl Acad Sci USA. 1999;96:12833–12838. doi: 10.1073/pnas.96.22.12833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wodicka L, Dong H, Mittmann M, Ho M-H, Lockhart D. genome-wide expression monitoring in Saccharomyces cerevisiae. Nat Biotechnol. 1997;15:1359–1367. doi: 10.1038/nbt1297-1359. [DOI] [PubMed] [Google Scholar]
- 30.Yagi T, Yano T, Di Bernardo S, Matsuno-Yagi A. Procaryotic complex I (NDH-I), an overview. Biochim Biophys Acta. 1998;1364:125–133. doi: 10.1016/s0005-2728(98)00023-1. [DOI] [PubMed] [Google Scholar]