Abstract
Human PRPF39 is a homolog of the yeast Prp39 and Prp42 paralogs. We have previously shown that human PRPF39 forms a homodimer that interacts with the CTD of U1C, mirroring the yeast Prp39/Prp42 heterodimer. We demonstrate here that PRPF39 knockdown in HEK293 cells affects many alternative splicing events primarily by reducing the usage of weak 5′ ss. Additionally, PRPF39 preferentially binds to a GC-rich RNA, likely at the interface between its NTD and CTD. These data indicate that PRPF39 potentially recruits U1 snRNP to a weak 5′ ss, serving as a previously unrecognized alternative splicing factor. We further demonstrate that human TIA1 binds to U1C through its RRM1 and RRM3 + Q domains and has weak binding to PRPF39. Finally, all three human LUC7L isoforms directly interact with U1C and with PRPF39. These results reveal significant parallels to the yeast U1 snRNP structure and support the use of yeast U1 snRNP as a model for understanding the mechanism of human alternative splicing.
Keywords: human PRPF39, alternative splicing, 5′ ss, U1 snRNP
INTRODUCTION
Splicing is an essential process required for eukaryotic gene expression and it is catalyzed by the spliceosome (Berget et al. 1977; Chow et al. 1977; Wahl et al. 2009). The spliceosome is a multimegadalton RNA–protein complex composed of U1, U2, U4, U5, and U6 small nuclear ribonucleoproteins (snRNPs) and other non-snRNP associated proteins (Zhou et al. 2002). The spliceosome assembles on each pre-mRNA, forming at least 10 distinct complexes through the assembly, activation, catalysis, and disassembly stages of the splicing cycle, to remove introns and ligate exons (Wahl et al. 2009).
The vast majority of human genes contain introns and undergo alternative splicing (Pan et al. 2008). Alternative splicing increases proteome diversity through the expression of various RNA transcripts from a single gene and is a fundamental approach of gene regulation in eukaryotes (Graveley 2001). A major tactic of alternative splicing regulation is through protein factors which target U1 snRNP (Eperon et al. 2000; Forch et al. 2002; Daniels et al. 2021). U1 snRNP recognizes the 5′ splice site (ss) of the pre-mRNA through base-pairing between the 5′ end of U1 snRNA and the 5′ ss (Lerner et al. 1980; Zhuang and Weiner 1986; Seraphin et al. 1988; Siliciano and Guthrie 1988). Alternative splicing factors can facilitate or prevent the binding of U1 snRNP to the pre-mRNA 5′ ss (Eperon et al. 2000; Forch et al. 2000; Busch and Hertel 2012; Daniels et al. 2021). Due to the transient nature of the interaction between these alternative splicing factors and human U1 snRNP, it has been difficult to capture these interactions structurally and the molecular details of how an alternative splicing factor interacts with U1 snRNP are often lacking.
On the other hand, the yeast homologs of many human alternative splicing factors stably associate with the yeast U1 snRNP (Gottschalk et al. 1998). While purified human U1 snRNP contains only 10 proteins (seven Sm proteins, U1-70K, U1A, and U1C), yeast U1 snRNP has homologs to these core proteins as well as seven additional stably associated proteins (Luc7, Nam8, Prp39, Prp40, Prp42, Snu71, and Snu56). Several of these yeast auxiliary proteins (Luc7, Nam8, Prp40, and Snu71) have homologs that are known alternative splicing factors (LUC7L/LUC7L2/LUC7L3, TIA1, PRPF40A/B, and RBM25, respectively) (Forch et al. 2000; Puig et al. 2007; Zhou et al. 2008; Becerra et al. 2015; Carlson et al. 2017; Lorenzini et al. 2019). The cryo-EM structures of the yeast U1 snRNP alone (Li et al. 2017) or in complex with the Act1 or Ubc4 pre-mRNA (Li et al. 2019) determined by our laboratory lay the foundation for using yeast U1 snRNP as a model in understanding the mechanism of human alternative splicing.
The structure of the yeast U1 snRNP core is similar to the human U1 snRNP with auxiliary proteins interacting with the yeast U1 snRNP core (Fig. 1A; Kondo et al. 2015; Li et al. 2017, 2019). Among the auxiliary proteins, the Prp39/Prp42 paralogs form a heterodimer-like structure that interacts extensively with the carboxyl terminus of U1C (Fig. 1A). We demonstrated biochemically that human PRPF39 forms a homodimer (Li et al. 2017), supported by the crystal structure of the murine PRPF39 (De Bortoli et al. 2019). The human PRPF39 homodimer directly interacts with the carboxy-terminal domain of human U1C, mirroring yeast Prp39/Prp42 (Li et al. 2017). These observations led us to hypothesize that human PRPF39 is a previously unrecognized alternative splicing factor that helps recruit U1 snRNP to weak 5′ ss. Below we will present evidence supporting this hypothesis. We will also demonstrate how human TIA1 (homolog of yeast Nam8) and LUC7Ls (homologs of yeast Luc7) interact with U1 snRNP as well as the similarities and differences between these interactions and those observed in yeast U1 snRNP.
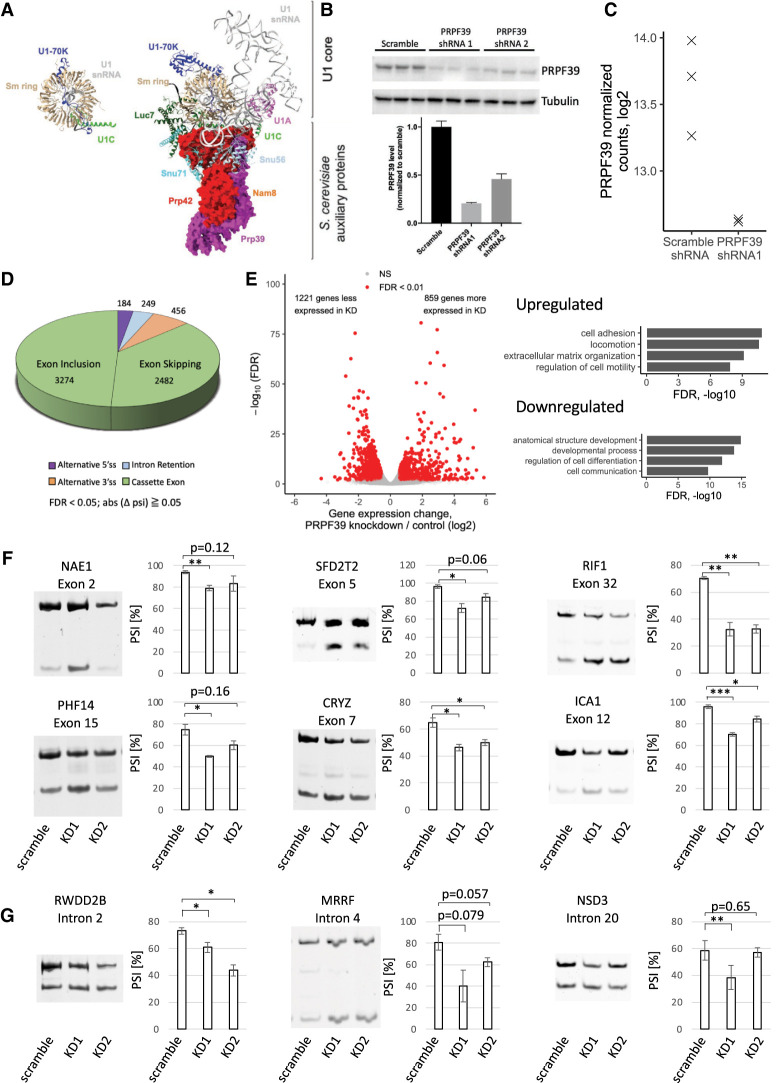
FIGURE 1.
PRPF39 KD affects alternative splicing events. (A) The human U1 snRNP (PDB 4PJO) with coloring to match the yeast U1 snRNP structure (PDB 6N7R). The yeast U1 snRNP shows the interaction between Prp39/Prp42 with U1C and other auxiliary proteins. Prp39/Prp42 are shown in surface representation, and the other proteins and RNAs are shown in ribbon representation. All structural representations in this paper are prepared using Chimera X (Pettersen et al. 2021). (B) Western blot (top) and quantification (bottom) demonstrate PRPF39 KD in HEK293 cells with two shRNAs. (C) Normalized sequencing read counts from RNA-seq analysis show dramatic reduction of PRPF39 mRNA in KD sample. (D) Type and number of alternative splicing events affected by PRPF39 KD. (E) Volcano plot demonstrating the expression changes caused by PRPF39 KD (left) and gene ontology analyses of these changes (right). (F) RT-PCR analyses of selected skipped exon events identified thorough RNA-seq indicate increased short isoform after PRPF39 KD. Standard deviation and statistical significance were derived from three biological replicates of KDs. (*), (**), and (***) denote P < 0.05, 0.01, and 0.001, respectively. (G) RT-PCR analyses of alternative 5′ ss events identified through RNA-seq indicate changed 5′ splice site selection after PRPF39 KD. Standard deviation and statistical significance were derived from three biological replicates of KDs. (*) and (**) denote P < 0.05 and 0.01, respectively.
RESULTS
PRPF39 knockdown affects alternative splicing events
To test the hypothesis that PRPF39 is a previously unrecognized alternative splicing factor, we knocked down PRPF39 in HEK293 cells with a short hairpin RNA (shRNA1) and confirmed that PRPF39 was knocked down by ∼80% using western blot analysis (Fig. 1B). We next performed RNA-seq experiments using HEK293 cells transfected with a scrambled control or shRNA1 targeting PRPF39, each in three biological replicates. Cells transfected with the PRPF39-targeting shRNA1 have a significant decrease of normalized counts of PRPF39 compared to the scrambled control, confirming PRPF39 knockdown (KD) at the RNA level (Fig. 1C). Analyses of the RNA-seq data show that PRPF39 affected a wide range of alternative splicing events, including cassette exon (5756 events), alternative 3′ ss (456), intron retention (249), and alternative 5′ ss (184) (Fig. 1D). The PRPF39 KD cells exhibit a number of expression changes (that could be indirect consequences of the splicing changes), including 1221 genes expressed significantly less and 859 genes significantly more compared to the scramble control (Fig. 1E, left). Gene ontology analyses reveal enrichment of genes in processes important during development (Fig. 1E, right), indicating that PRPF39 could potentially play a role in the differentiation of HEK293 cells.
To validate the RNA-seq results, we carried out PRPF39 KD using a second shRNA which has ∼54% KD efficiency based on western blot analyses (Fig. 1B). We performed RT-PCR on total RNA extracted from scramble or both KD cells for a number of affected alternative splicing events. The skipped exon events we evaluated all have an increase in the short isoform in KD compared to the scramble control demonstrated by a decrease in the PSI (percentage spliced in) values (Fig. 1F). The alternative 5′ ss RT-PCR results also show a decrease of the long isoform with PRPF39 KD, consistent with the RNA-seq results (Fig. 1G).
PRPF39 KD reduces the usage of weak 5′ splice sites
Given that PRPF39 directly interacts with U1 snRNP (Li et al. 2017), we hypothesized that PRPF39 potentially recruits U1 snRNP to weak 5′ ss. Alternative 5′ ss events provide the best resource to test this hypothesis since the two alternative 5′ ss directly compete with each other, and their relative 5′ ss strength is the main variable determining the 5′ ss usage. We therefore analyzed the relationship between alternative 5′ ss usage and 5′ ss strength (Yeo and Burge 2004) upon PRPF39 KD.
Alternative 5′ splice sites result in the production of one of two isoforms: either a longer isoform that contains a longer upstream exon due to the use of a downstream 5′ ss, or a shorter isoform that contains a shorter upstream exon due to the use of an upstream 5′ ss (Fig. 2A, top). We classified all expressed alternative 5′ splice site events into three categories: those in which the longer isoform decreased in abundance following PRPF39 KD (Fig. 2A, orange), those in which the shorter isoform decreased in abundance following KD (Fig. 2A, purple), and those in which the relative abundance of the two isoforms did not change following KD (Fig. 2A, gray). We then compared the difference in 5′ splice site strength (Yeo and Burge 2004) between the two isoforms for the events in each category. Transcriptome-wide, we found that for the events in which the long isoform decreased in abundance following KD, the 5′ splice site associated with the long isoform was weaker than the 5′ splice site of its short isoform counterpart, as indicated by the orange curve shifting to the left of the gray curve (Fig. 2A, bottom). Similarly, when the short isoform decreased in abundance following KD, the 5′ splice site of the short isoform was weaker than the 5′ splice site of its long isoform counterpart, as indicated by the purple curve shifting to the right of the gray curve (Fig. 2A, bottom). These findings suggest that PRPF39 promotes the use of relatively weaker 5′ splice sites as its removal reduces the use of these weaker 5′ splice sites.
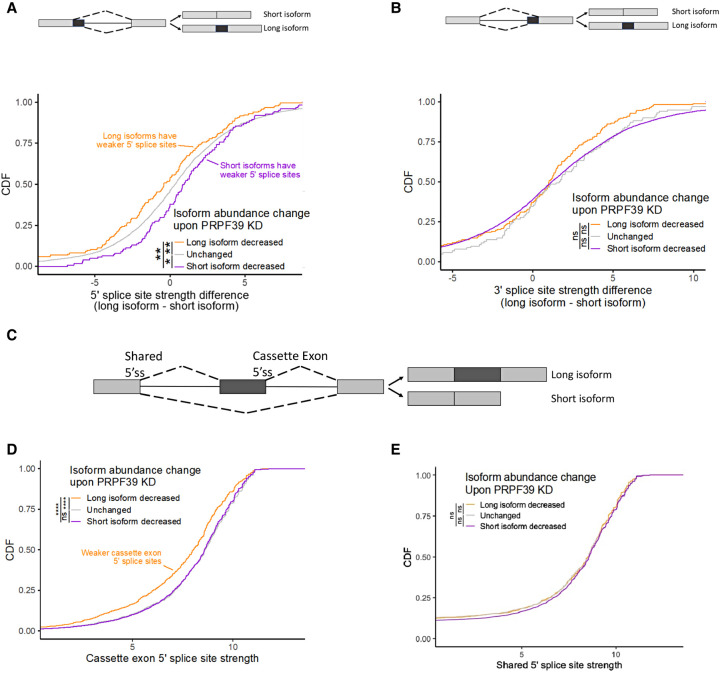
FIGURE 2.
PRPF39 KD reduces the usage of weak 5′ splice sites. (A) Cumulative distribution function (CDF) of different groups of alterative 5′ ss events versus the 5′ ss strength difference between the long and short isoform. (*) P < 0.05; (**) P < 0.01. A schematic representation of the long and short isoform generated from alternative 5′ ss is shown on the top. (B) CDF of alternative 3′ ss events versus the 3′ ss strength difference between the long and short isoform. A schematic representation of the long and short isoform generated from alternative 3′ ss is shown at the top. (C) A schematic representation of the long and short isoform generated from cassette exon events. (D) CDF of cassette exon events versus the strength of the cassette exon 5′ ss. (****) P < 0.0001. (E) CDF of cassette exon events versus the strength of the shared 5′ ss.
If the action of PRPF39 primarily occurred through 5′ splice sites, we would not expect there to be a relationship between alternative 3′ splice site usage and the strength of their corresponding 3′ splice sites as both alternative isoforms share a common 5′ splice site. In fact, we see no such relationship (Fig. 2B), further suggesting that PRPF39 acts primarily through sequences associated with 5′ splice sites.
We then turned to a similar analysis of cassette exon isoforms. Cassette exon events can result in a long isoform in which the cassette exon is included or a short isoform in which it is skipped (Fig. 2C). These two isoforms share a common 5′ splice site while the use of the cassette exon 5′ splice site is specific to and required for the long isoform. As before, we binned cassette exon events into those in which the long isoform decreased in relative abundance upon PRPF39 KD (Fig. 2D, orange), those in which the short isoform decreased in relative abundance upon KD (purple), and those in which relative isoform abundance was unchanged (gray).
We observed that for the events in which the long isoform decreased in abundance (i.e., skipped exon events), the cassette exon-specific 5′ splice site was much weaker than expected (Fig. 2D). This is again consistent with PRPF39 promoting the use of these weaker 5′ splice sites as PRPF39 KD decreases their usage, resulting in increased prevalence of the shorter isoform. Note that PRPF39 KD results in 2482 skipped exon events, significantly more than the other types of alternative splicing events affected (alternative 5′ ss, alternative 3′ ss, and intron retention) (Fig. 1D). This is consistent with many previous observations that the predominant phenotype caused by a weakened 5′ ss in mammalian cells is exon skipping based on the exon definition model (Berget 1995).
As a control, we also compared the strengths of the shared 5′ splice sites among our three categories (long isoform decreased, short isoform decreased, and unchanged) of cassette exon events. Since these 5′ splice sites are used in production of both the short and long isoforms, we would not expect a difference in the 5′ splice site strengths among the three categories of cassette exon events. In fact, we do not observe a difference (Fig. 2E), further indicating that the mode of action for PRPF39 is through differentially used 5′ splice sites.
PRPF39 interacts with GC-rich RNA
If PRPF39 acts to promote weaker 5′ splice site usage, we reasoned that it may then be binding near those sites through some sequence-specific interactions. To identify candidate RNA sequences that interact with PRPF39, we set out to identify 6-mer sequences that were enriched near 5′ splice sites whose usage was sensitive to PRPF39 KD. To do this, we used the cassette exon data and pulled sequences that flanked the cassette exon 5′ splice site, including 100 nt of the cassette exon and 150 nt of the downstream intron excluding the 5′ splice site itself. We compared these sequences from cassette exon events that were sensitive to PRPF39 KD (Fig. 2D, orange) to sequences from cassette exon events that were insensitive to PRPF39 KD (Fig. 2D, purple + gray).
We found that the sequence content surrounding sensitive 5′ splice sites was very GC-rich and A-poor, illustrated by several different formats including CDF plots, volcano plots, and sequence logos for exonic and intronic regions (Fig. 3A). We then experimentally tested if PRPF39 preferentially binds to GC-rich RNA by evaluating the direct binding between PRPF39 and an arbitrary GC-rich RNA oligo (GGCCCCCCGG) or a 10 nt poly(A) RNA oligo using electrophoretic mobility shift assay (EMSA) experiments. These experiments demonstrate that PRPF39 binds to the GC-rich RNA with a Kd of 516 ± 256 nM (Fig. 3B; Supplemental Fig. S1), while PRPF39 only achieved ∼25% binding to the poly(A) oligo at the highest protein concentration (Fig. 3C).
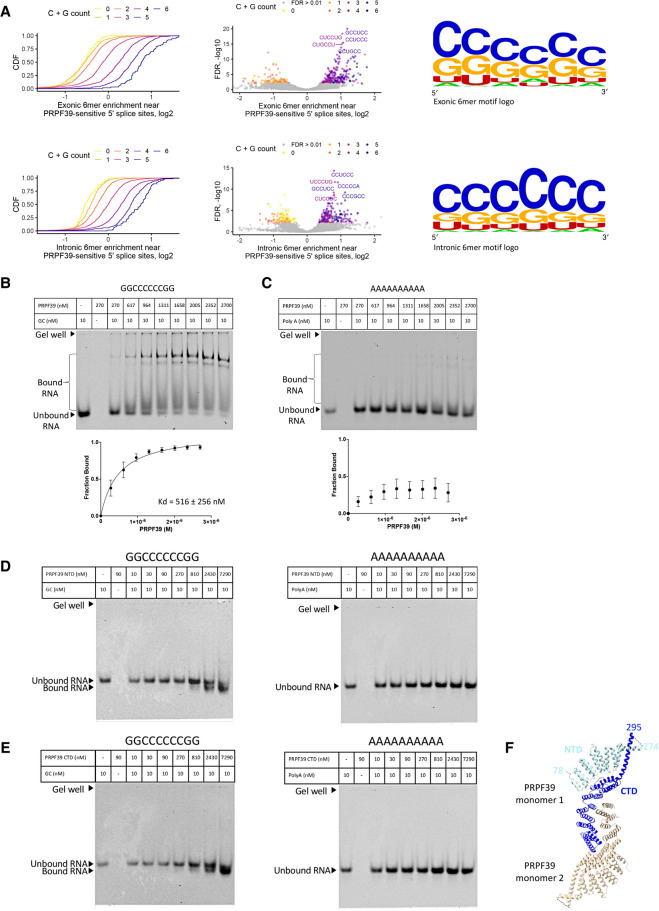
FIGURE 3.
PRPF39 interacts with RNA. (A) Intronic and exonic 6mer enrichment near PRPF39-sensitive 5′ splice sites demonstrated using CDF (left), volcano plot (middle), and logo (right, derived from WebLogo [Schneider and Stephens 1990; Crooks et al. 2004]). (B) EMSA of full-length PRPF39 with a GC-rich oligo (GGCCCCCCGG) demonstrates that PRPF39 binds the GC oligo with a Kd of 516 ± 256 nM. Identity of the protein bound RNA was analyzed using western blot analyses (Supplemental Fig. S1). (C) EMSA of full-length PRPF39 with a 10-mer poly(A) control oligo. (D) EMSA of PRPF39 NTD with the GC and poly(A) oligo. (E) EMSA of PRPF39 CTD with the GC and poly(A) oligo. Note that the migration of a molecule in native gel is determined by the combined effect of its size, charge, and shape, and the substantial negative charge of NTD and CTD (pI of 4.62 and 6.15, respectively) likely have led to a faster migrating band despite the increased mass when bound to the RNA. (F) The dimeric human PRPF39 homologous model based on the murine PRPF39 structure (PDB: 6G70) showing the boundary of the NTD (cyan) and CTD (blue) in one monomer. Dashed line indicates the disordered region separating the NTD and CTD with the flanking residue numbers labeled.
To further probe this interaction, we tested PRPF39 NTD or CTD (separated between residues 296 and 297 based on disorder prediction, Supplemental Fig. S2) with the GC-rich and poly(A) oligo using EMSA. Our results demonstrate that both constructs bind the GC-rich oligo but not the poly(A) oligo (Fig. 3D,E), suggesting that the RNA may bind to the interface between the two domains (Fig. 3F).
TIA1 interacts mainly with U1C and weakly with PRPF39
Given that the above results demonstrate the successful use of yeast U1 snRNP as a model to understand how human PRPF39 functions, we next ask whether it can guide our studies of other human alternative splicing factors such as TIA1 and LUC7Ls. TIA1, the human homolog of yeast Nam8, affects the alternative splicing of select pre-mRNAs in a U1 snRNP-dependent manner by binding to U-rich RNA regions adjacent to the 5′ ss (Dember et al. 1996; Kedersha et al. 1999; Forch et al. 2000). In yeast U1 snRNP, Nam8 interacts with both U1C CTD and Prp42 (Fig. 4A). We asked whether human TIA1 also interacts with both U1C and PRPF39. Using pull-down experiments with recombinant proteins purified from E. coli, we demonstrate that TIA1 interacts with U1C, consistent with a previous report (Forch et al. 2002). This interaction is mostly with the U1C NTD (residues 1–61), different from the yeast structure (Fig. 4B). Additionally, we show that RRM1 as well as RRM3 with the Q-rich tail of TIA1 directly bind U1C (Fig. 4C,D). Finally, we show that TIA1 binds to PRPF39 weakly when compared to the GST control (Fig. 4E).
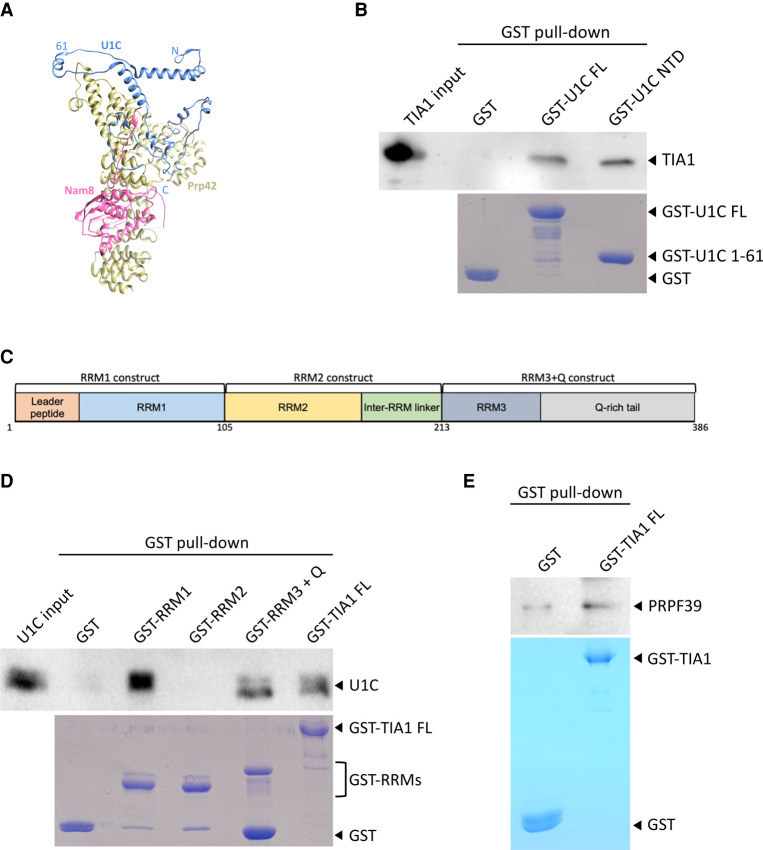
FIGURE 4.
Alternative splicing factor TIA1 directly interacts with U1C and PRPF39. (A) Nam8 (pink) contacts U1C (blue) and Prp42 (yellow) in the yeast U1 snRNP structure (PDB 6N7R). N, C, and 61 indicate the amino- and carboxy-terminal ends and residue 61 (which separates the NTD and CTD) of U1C. (B) Purified GST-U1C FL and NTD (bottom, Coomassie stained SDS PAGE) can pull down TIA1 (top, western blot with anti-TIA1 antibody). (C) A schematic diagram of TIA1 domains and the three TIA1 truncation constructs used in subsequent experiments. (D) Purified GST-RRM1 and RRM3 + Q, but not RRM2 (bottom, Coomassie stained SDS PAGE), can pull down FL U1C (top, western blot with anti-U1C antibody). (E) GST pull-down experiment using GST-TIA1 as a bait (bottom, Coomassie stained SDS PAGE) and PRPF39 as prey (top, western blot with anti-PRPF39 antibody) shows weak binding when compared to the GST control.
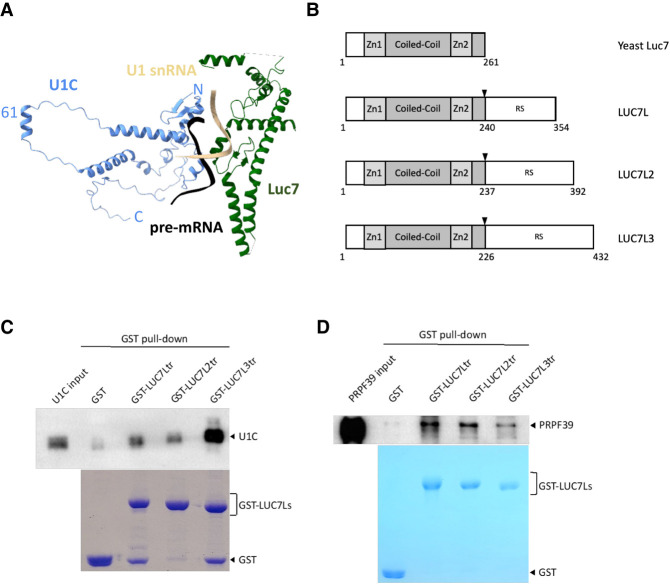
LUC7L isoforms directly interact with U1C and PRPF39
Our cryo-EM structures of the yeast U1 snRNP in complex with either the Act1 or Ubc4 pre-mRNA reveal that Luc7 interacts with U1C and the U1 snRNA–5′ ss RNA duplex (Fig. 5A; Li et al. 2019), which is also observed in the yeast A complex structure (Plaschka et al. 2018). Luc7 has three mammalian homologs (LUC7L, LUC7L2, and LUC7L3) which have significant sequence homology with Luc7 at their amino-terminal region but contain a carboxy-terminal arginine-serine (RS) domain absent in yeast Luc7 (Fig. 5B). Since we have difficulty expressing and purifying full-length proteins of LUC7L isoforms with RS domains in E. coli and their yeast homolog Luc7 has no RS domain, we decided to express and purify truncated LUC7L isoforms without the RS domain. These truncated isoforms are used to evaluate if they mimic the interaction between yeast Luc7 and U1C. Pull-down experiments using purified GST-LUC7Ls show that all truncated isoforms directly interact with U1C, with LUC7L3 having the strongest interaction (Fig. 5C). Although Luc7 does not directly interact with PRP39/PRP42 in the yeast U1 snRNP structure (Li et al. 2017), we showed that human LUC7s interact with PRPF39 in pull-down experiments (Fig. 5D).
FIGURE 5.
LUC7L isoforms interact with U1C and PRPF39. (A) In the yeast U1 snRNP structure (PDB ID 6N7R), Luc7 (green) directly interacts with both U1C (blue) and the U1 snRNA-5′ ss duplex (beige/black). (B) A schematic diagram of yeast Luc7 and human LUC7L isoforms. Black arrowheads indicate positions for the truncation constructs used for subsequent experiments. (C) Purified GST-LUC7L truncations of all three isoforms (bottom, Coomassie stained SDS PAGE) can pull down FL U1C (top, western blot with anti-U1C antibody). (D) Purified GST-LUC7L truncations (bottom, Coomassie stained SDS PAGE) can pull down purified PRPF39 protein (top, western blot with anti-PRPF39 antibody).
DISCUSSION
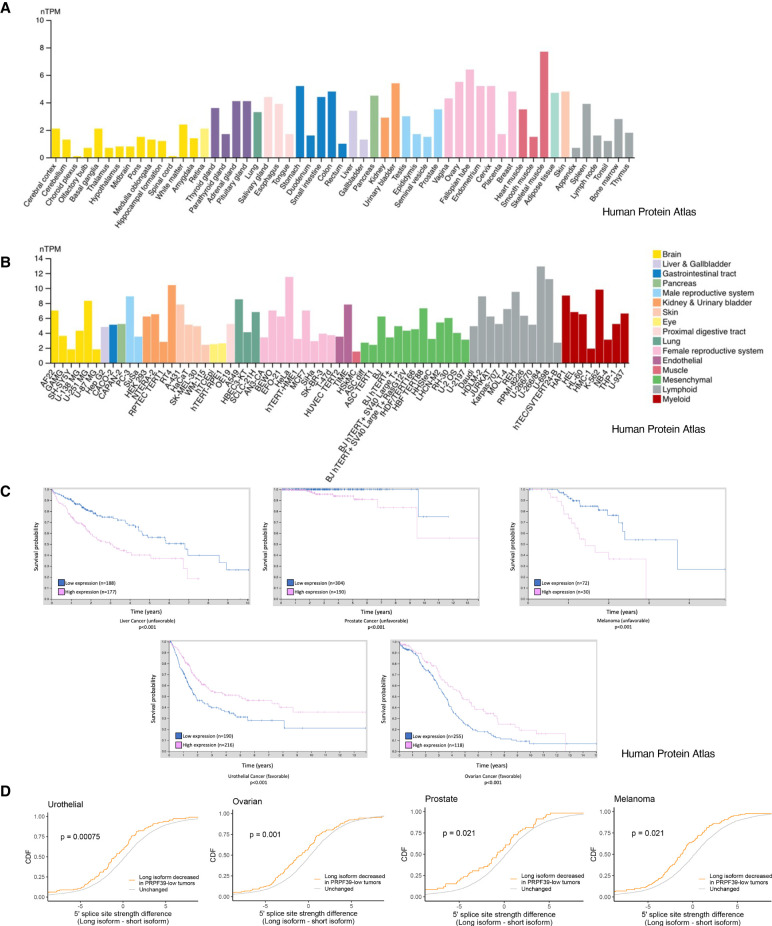
Our study identifies a previously unrecognized alternative splicing factor, PRPF39, which recruits U1 snRNP to weak 5′ splice sites, likely by binding to GC-rich RNA in proximity of the weak 5′ ss. We demonstrate that PRPF39 KD in HEK293 cells affects a wide range of alternative splicing events by reducing the usage of weak 5′ ss. A search of the Human Protein Atlas (Uhlen et al. 2015; Karlsson et al. 2021) shows that PRPF39 mRNA is ubiquitously expressed in various tissues and cell lines (Fig. 6A,B). However, the level of expression fluctuates among these tissues and cell lines, and this may contribute to the regulation of alternative splicing. One way to regulate the level of PRPF39 is through the inclusion of a “poison” exon (Lareau et al. 2007; Ni et al. 2007) conserved between the human and murine PRPF39 which leads to nonsense-mediated decay (NMD) of PRPF39 mRNA (De Bortoli et al. 2019). The inclusion of this “poison” exon varies in different murine tissues, with high inclusion in testis and low inclusion in lymph nodes (De Bortoli et al. 2019). Additionally, the expression levels of PRPF39 may be temporally regulated or in response to other signaling events. For example, the aforementioned “poison” exon in murine PRPF39 is preferentially included in memory T cells compared to naïve T cells, leading to NMD and reduced level of PRPF39, which potentially regulates the naïve to memory T cell differentiation (De Bortoli et al. 2019). Furthermore, PRPF39 is observed to overexpress in different cancers (Uhlen et al. 2017), and its overexpression correlates significantly with the prognosis of several types of cancers including liver (unfavorable), urothelial (favorable), prostate (unfavorable), melanoma (unfavorable), and ovarian (favorable) cancer (Fig. 6C). We further analyzed data sets of these cancers in The Cancer Genome Atlas (TCGA) and demonstrated that lower PRPF39 expression levels correlate with reduced usage of 5′ ss in multiple cancer types (Fig. 6D), consistent with our in vitro results in cell lines. These results suggest that the expression level of PRPF39 may change alternative splicing, consequently affecting disease prognosis in these cancers.
FIGURE 6.
PRPF39 mRNA is expressed in most or all tissues and cell lines at varying levels and its expression levels correlate with disease prognosis in several cancer types. All data are from the Human Protein Atlas (Uhlen et al. 2015, proteinatlas.org). (A) Normalized transcripts per million (nTPM) of PRPF39 in different tissues (https://www.proteinatlas.org/ENSG00000185246-PRPF39/tissue from v21.proteinatlas.org). (B) Normalized transcripts per million (nTPM) of PRPF39 in different cell lines separated by organ type (https://www.proteinatlas.org/ENSG00000185246-PRPF39/cell+line from v21.proteinatlas.org). (C) Kaplan–Meier plots demonstrate that high PRPF39 levels significantly correlate (P < 0.001) with patient survival in several cancer types with either favorable (urothelial and ovarian cancer) or unfavorable (melanoma, liver, and prostate cancer) outcomes (adapted from https://www.proteinatlas.org/ENSG00000185246-PRPF39/pathology from v21.proteinatlas.org). (D) Cumulative distribution function (CDF) of alternative 5′ ss usage versus the 5′ ss strength difference between the long and short isoform in tumors where PRPF39 levels are low.
Most alternative splicing events affected by PRPF39 KD are cassette exon incidents. Nearly 2500 events are exon skipping, which is consistent with the exon definition model and previous observations that the presence of a weak 5′ss mostly leads to exon skipping (Berget 1995). Interestingly, PRPF39 KD also leads to a large number of (3274) cassette exon inclusion events. This can potentially be an indirect consequence of PRPF39 KD which affected the splicing of proteins that regulate exon inclusion. For example, PRPF39 KD led to splicing pattern changes in several hnRNPs and SRSF proteins, including alternative 5′ ss choices in hnRNPH3 and hnRNPU; exon skipping in hnRNPH1, hnRNPD, hnRNPA2/B1, SRSF2, and SRSF6; as well as exon inclusion in hnRNPC, hnRNPD, SRSF4, and SRSF10. Typically, hnRNPs act as splicing repressors and promote exon skipping (Geuens et al. 2016), while SRSF proteins enhance splicing and promote exon inclusion (Zhou and Fu 2013), although their response can be dependent on the location to which these proteins bind along the pre-mRNA (Sanford et al. 2009; Han et al. 2011; Busch and Hertel 2012; Zhou and Fu 2013). The changing of splicing patterns of hnRNPs or SRSFs can potentially change their functions, leading to exon inclusion in their target genes. Alternatively, the large number of exon inclusion events induced by PRPF39 KD can be a consequence of the competition between PRPF39 and SRSF proteins in RNA binding. For example, SRSF2 is known to bind to GC-rich RNA and promote inclusion of exons with weak splice sites (Liu et al. 2000). It is possible that the KD of PRPF39 reduces its occupancy on GC-rich regions and allows SRSF2 to bind and promote the inclusion of weak 5′ ss containing exons.
Our motif analysis showed an enrichment of GC-rich 6-mer that is devoid of A nucleotides within 100–150 nt flanking the weak 5′ ss of a skipped exon upon PRPF39 KD. We demonstrated using EMSA that PRPF39 preferentially binds the GC oligo while there is only very weak interaction with a poly(A) oligo (Fig. 3B,C). Future eCLIP experiments will help determine the specific motif to which PRPF39 binds. Unlike the other alterative splicing factors we discuss here (TIA1 and LUC7Ls), PRPF39 does not contain a common RNA-binding motif such as the RRM or zinc finger (ZnF) domain. PRPF39 mainly consists of half-a-tetratricopeptide (HAT) repeats. HAT repeats are typically associated with protein–protein interactions (Blatch and Lässle 1999). On the other hand, HCF107 is a sequence-specific RNA binding protein with 11 HAT repeats which interacts with a short (11 nt) single stranded RNA (Hammani et al. 2012), although details of this interaction are unknown. In PRPF39, the interaction with RNA likely occurs at the interface between the amino- and carboxy-terminal domains of PRPF39, given that both the NTD and CTD of PRPF39 bind the GC oligo in EMSA (Fig. 3D,E).
In yeast, the Prp42/Prp39 paralogs interact with multiple stably associated auxiliary factors, mediating their interaction with the U1 snRNP core (Li et al. 2017). For example, the yeast Nam8 directly contacts Prp42 and U1C CTD (Li et al. 2017). We speculated that in addition to PRPF39's role as an alternative splicing factor by binding to the pre-mRNA and recruiting U1 snRNP to particular 5′ss, it may serve as a mediator for the function of the other alternative splicing factors by interacting and bringing them to the U1 snRNP. We showed that TIA1, the human homolog of Nam8, interacts mostly with the U1C NTD. Additionally, both TIA1 and LUC7Ls interact with PRPF39, albeit weakly compared to U1C (Figs. 4, 5). These data suggest that PRPF39 mainly serves as an alternative splicing factor on its own and whether it does play a role as a mediator of TIA1 or LUC7Ls’ function remains to be tested experimentally. Moreover, we find that both RRM1 and RRM3 + Q domains of TIA1 interact with U1C. This has both similarity and differences with previous reports that the RRM3 + Q region but not RRM1 interacts with U1C (Fig. 4D; Forch et al. 2002). However, our result is consistent with the crystal structure of the TIA1 RRM1 with the NTD of U1C determined by Dr. Michael Sattler's laboratory (PDB ID 6ELD) (unpubl.).
Finally, in the yeast U1 snRNP + pre-mRNA structure, Luc7 contacts U1C and interacts with the yeast U1-5′ ss RNA snRNA duplex through its ZnF domain (Li et al. 2019). Human homologs of yeast Luc7 (LUC7L, LUC7L2, and LUC7L3) all contain an amino-terminal domain homologous to yeast Luc7, with an additional carboxy-terminal RS domain unique to the human isoforms. We demonstrated that all three isoforms without the RS domain can pull down U1C (Fig. 5C). U1C seems to be a major hub for alternative splicing factor binding, and it has also been shown to directly bind PRPF39 (Li et al. 2017), TIA1 (Fig. 4B,D; Forch et al. 2002), as well as the LUC7L paralogs (Fig. 5C). Another U1 snRNP protein that interacts with alternative splicing factors is U1-70K that binds SR proteins (Cho et al. 2011). Furthermore, LUC7Ls are shown to bind U1 snRNA while LUC7L2 and LUC7L3 bind to an AAGAAG sequence of exonic splicing enhancer motifs near weak 5′ splice sites in eCLIP experiments (Daniels et al. 2021), suggesting that LUC7Ls may also help stabilize the U1-5′ ss RNA duplex, similar to its yeast counterpart. The significant parallel between these observations and the yeast U1 snRNP structure support the use of yeast U1 snRNP as a model for understanding the mechanism of human alternative splicing.
MATERIALS AND METHODS
PRPF39 knockdown in HEK293 cells
The human embryonic kidney 293FT cell line, HEK293FT, was cultured in DMEM supplemented with 10% fetal bovine serum (FBS) in 5% CO2 at 37°C. shRNA constructs on TRC2 pLKO.5 lentivirus vector targeting human PRPF39 (TRCN0000424447 as shRNA1 for RNA-seq and TRCN0000422853 as shRNA2 for RT-PCR validation) and nonmammalian control shRNA (SHC202) from the MISSION TRC lentiviral shRNA library were purchased through the Functional Genomics Core Facility at the University of Colorado. To produce lentiviruses, HEK293FT cells at 30%–50% confluency were cotransfected with 8 µg of the corresponding shRNA pLKO.5-puro plasmid plus 3.3 µg of each of the packaging plasmids (Sigma) using calcium phosphate (Han et al. 2019). The medium was changed 24 h post-transfection and cells were cultured for an additional 48 h. The resulting virus-containing supernatants (10 mL) were used to infect the corresponding cells (10-cm dish) in medium supplemented with 8 µg/mL of polybrene (Sigma) overnight. Cells were then selected with 2 µg/mL of puromycin for at least two passages or until all cells in a negative control transfection were killed. Selected cells were harvested and either lysed in lysis buffer (40 mM HEPES pH 7.5, 150 mM NaCl, 0.5% Triton X-100, 10% glycerol, 2 mM EDTA) for western blot analysis or pelleted and flash frozen for subsequent RNA extraction.
RNA-seq analyses
RNA was extracted from three biological replicates of HEK293 cells treated with scramble or shRNA targeting PRPF39. All RNA samples were treated with DNase I prior to submission to the Genomics and Microarray Core at the University of Colorado Anschutz Medical Campus. The core prepared RNA-seq libraries after poly(A) selection and obtained ∼100 million pair-end 150 bp sequencing reads using the NovaSEQ 6000 system (Illumina).
Sequence reads were aligned to the human genome (hg38 GENCODE 28) using STAR (Dobin et al. 2013). Differences in alternative splicing between control and PRPF39 knockdown samples were determined using rMATS (Shen et al. 2014). Splice site score strengths were calculated using MaxEntScore (Yeo and Burge 2004). Differentially used splice sites were defined as those covered by at least 50 reads and with PSI (percent spliced in) value differences between control and PRPF39 knockdown samples of at least 0.05 and an FDR (false discovery rate) of 0.05 or less. We also tested using a PSI difference of 0.1 as the cutoff which generated essentially the same results.
Kmer analyses for sequences surrounding 5′ splice sites were performed by first extracting sequences surrounding the splice site. These included the first 150 nt of the intron or the last 100 nt of the upstream exon. Sequence contents of these regions drawn from affected and unaffected 5′ splice sites were then compared by counting the abundances of 6-mers present within each sequence set.
For expression analyses, transcript abundances were calculated using Salmon (Patro et al. 2017), collapsed to gene level abundances using tximport (Soneson et al. 2015), and differentially expressed genes were identified using DESeq2 (Love et al. 2014) using an FDR threshold of 0.05. GO analyses were done using GOrilla (Eden et al. 2009).
To validate RNA-seq results by RT-PCR, 1 µg of total RNA was used in a reverse-transcription reaction with random primers (NEB) to produce cDNA. PCR product generated from the cDNA using gene-specific primers were separated on 8% denaturing urea polyacrylamide gels, stained with SYBR Gold, and quantified with Chemidoc Image Laboratory (BioRad). Gene-specific primers are located in the two exons flanking the cassette exon or the alternative 5′ ss.
RNA-seq data sets from primary tumor samples were downloaded from TCGA (https://www.cancer.gov/tcga). Within a tumor, samples were ranked based on the PRPF39 RNA expression levels. RNA-seq data from PRPF39-high samples (90th percentile and above) and PRPF39-low samples (10th percentile and below) was compared to identify alternative splicing events that were differentially regulated between these two sets of samples. This analysis was completed using rMATS in an identical manner to the HEK293 PRPF39 shRNA and control samples.
Protein expression and purification
PRPF39 (FL, NTD, CTD), LUC7L, LUC7L2, LUC7L3, TIA1 (FL, RRM1, RRM2, RRM3 + Q), U1C (FL, NTD) were subcloned into pGEX-6p1 as a GST-fusion protein. U1C FL was also subcloned into pET28a as a His-tagged protein (all U1C proteins were codon optimized for E. coli expression).
Most GST-tagged fusion proteins were expressed in Escherichia coli XA90 cells in 2xYT media overnight at 16°C (PRPF39 FL) or 3 h at 37°C (GST, PRPF39 NTD/CTD, LUC7Ls, U1C 1-61, and all TIA1). Cells were induced at OD600 of 0.8 with 0.3 mM (PRPF39 FL) or 1 mM (GST, PRPF39 NTD/CTD, LUC7Ls, U1C 1-61, and all TIA1) IPTG. All the following steps were performed at 4°C. Cells were resuspended in lysis buffer (50 mM Tris/HCl pH 7.5, 500 mM NaCl, 0.5% v/v Triton X-100, 5% v/v glycerol, 1 mM DTT) with protease inhibitors and lysed by sonication. PEI treatment (0.4%) with a high salt lysis buffer (50 mM Tris/HCl pH 7.5, 600 mM NaCl, 0.5% v/v Triton X-100, 5% v/v glycerol, 1 mM DTT) was used to precipitate contaminating nucleic acids for PRPF39 FL, LUC7Ls, and TIA1 FL. Soluble cell lysate was centrifuged for 60 min at 10,000 rpm in a GSA rotor (Sorvall), and the supernatant was bound to glutathione resin (GE). GST-tag was cleavage overnight at 4°C with 1:50 PreScission or eluted using 30 mM reduced glutathione. The elution was further purified on a Superose 6 increase (GE Healthcare) size exclusion column (SEC). Peak fractions were visualized by SDS-PAGE. Target protein fractions were pooled, concentrated, and snap-frozen in liquid nitrogen for storage at −80°C.
GST-tagged full length U1C fusion protein was expressed in Escherichia coli BL21 (DE3) cells in 2xYT media overnight at 16°C, induced at OD600 of 2 with 0.5 mM IPTG, resuspended in FL lysis buffer (50 mM HEPES pH 7.5, 500 mM NaCl, 0.5% v/v Triton X-100, 5% v/v Glycerol, and 1 mM DTT) with protease inhibitor cocktail, and lysed by sonication. All remaining steps of purification are the same as detailed above.
His-tagged full length U1C fusion protein was expressed in Escherichia coli BL21 (DE3) cells in 2xYT media overnight at 16°C, induced at OD600 of 2 with 0.5 mM IPTG, resuspended in FL lysis buffer (20 mM HEPES pH 7.5, 1 M NaCl, 1 M Urea, 1 mM TCEP) with protease inhibitor cocktail tablets, and lysed by sonication. All the following steps were performed at 4°C. Lysate was cleared by centrifugation, and lysate bound to nickel resin (ABT). Nickel resin was washed with increasing imidazole (20 and 40 mM) and eluted with 500 mM imidazole. The elution was further purified on a Superdex 75 column (GE Healthcare) in SEC buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 1 mM TCEP). Peak fractions were visualized by SDS-PAGE. Target protein was then snap-frozen in liquid nitrogen for storage at −80°C.
Pull-down
Pull-downs were performed as previously described (Li et al. 2017). Briefly, purified GST-tagged proteins were bound on glutathione resin in high salt buffer (50 mM Tris pH 7.5, 500 mM NaCl, 5% glycerol, 0.5 mM DTT), washed three times with high salt buffer and then incubated with 6 µg of prey protein for 1.5 h at 4°C with gentle rotation. Resin was then washed three times each with low salt buffer (50 mM Tris pH 7.5, 150 mM NaCl, 5% glycerol, 0.5 mM DTT) and medium salt buffer (50 mM Tris pH 7.5, 250 mM NaCl, 5% glycerol, 0.5 mM DTT), followed by a high salt wash, transfer of resin to a new tube, and a final high salt wash. After the last wash, SDS-sample buffer was added, samples were boiled, and analyzed by western blot.
Electrophoretic mobility shift assay (EMSA)
EMSA was performed using IRDye-700 labeled 10 nt poly(A) and GC (GGCCCCCCGG) RNA oligos (Integrated DNA Technologies). PRPF39 was added to the RNA sample (10 nM) in binding buffer (50 mM Tris pH 7, 150 mM NaCl, 20% glycerol, 3 mM MgCl2, 1 mM DTT) to final concentrations of 270, 617, 964, 1311, 1658, 2005, 2352, and 2700 nM. The concentration range for PRPF39 NTD and CTD were 10, 30, 90, 270, 810, 2430, and 7290 nM. All reactions had a final volume of 5 µL including murine RNA inhibitor (New England Biolabs) and were incubated at 30°C for 15 min. The EMSA samples were analyzed on a nondenaturing PAGE (5% acrylamide; 37.5:1 acrylamide: N,N-methylene-bis-acrylamide [BioRad]) with 0.2× TBE buffer (26 mM Tris pH 7.5, 9 mM boric acid, 0.5 mM EDTA) and 2.5% glycerol (Liu et al. 2016). The gels were prerun at 100 V for 40 min before loading sample and subsequently electrophoresed at 100 V for 65 min. The RNA was visualized using an Odyssey infrared imaging system (LiCor). The band intensities were measured using AzureSpot (Azure Biosystems) analysis software. After background subtraction, the fraction of RNA bound was calculated, fitted with the Hill equation, and graphed using Prism software (GraphPad).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank members of the Zhao laboratory for insightful discussion. We also would like to thank Remix Therapeutics for the codon optimized human U1C plasmid and Dr. Juan Valcarcel for the human TIA1 plasmid. Research reported in this paper was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award numbers R01GM126157 (R.Z.), R35GM145289 (R.Z.), R35GM133385 (J.M.T.), F32GM145067 (F.D.B.), and T32GM008730 (H.Y.G.L.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. S.E. is a Howard Hughes Medical Institute Gilliam Fellow. Molecular graphics and analyses were performed with UCSF ChimeraX, developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from National Institutes of Health R01-GM129325 and the Office of Cyber Infrastructure and Computational Biology, National Institute of Allergy and Infectious Diseases.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.079320.122.
MEET THE FIRST AUTHOR
Sara Espinosa.

Meet the First Author(s) is a new editorial feature within RNA, in which the first author(s) of research-based papers in each issue have the opportunity to introduce themselves and their work to readers of RNA and the RNA research community. Sara Espinosa is the first author of this paper, “Human PRPF39 is an alternative splicing factor recruiting U1 snRNP to weak 5′ splice sites.” Sara did this work as a PhD student in molecular biology in the Zhao laboratory at the University of Colorado Anschutz Medical Campus. The main focus of this work is to determine whether yeast U1 snRNP can be used as a viable model to understand and make testable predictions about how alternative splicing factors function in relationship to human U1 snRNP.
What are the major results described in your paper and how do they impact this branch of the field?
We learned that yeast U1 snRNP can provide valuable insights into how alternative splicing factors interact with human U1 snRNP and each other. We used the yeast U1 snRNP structure as a model to identify a new alternative splicing factor, PRPF39, which binds to GC-rich RNA motif and recruits U1 snRNP to weak 5′ splice sites. We further found that yeast U1C seems to be a major hub for interactions with alternative splicing factors (PRPF39, TIA1, and LUC7Ls) and that these alternative splicing factors interact with each other to varying degrees. I believe that the major impact of this work on the field is the demonstration of yeast U1 snRNP as a valuable model for understanding the mechanism of human alternative splicing.
What led you to study RNA or this aspect of RNA science?
When I initially applied for graduate school, I thought I wanted to study cancer biology due to family history. I met a graduate student while interviewing, who inspired me to look more into structural biology. Ultimately, I spoke with Dr. Rui Zhao, who is a crystallographer by training. It was August 2015 and the structure of the tri-snRNP had just been published. Rui's passion for splicing, RNA biology, and structural biology could not be denied. I completed my first rotation in the Zhao laboratory and with each new rotation I could not deny being invariably drawn back to RNA and structural biology. It was just a matter of time before I decided to join the laboratory and focus my dissertation on the spliceosome.
During the course of these experiments, were there any surprising results or particular difficulties that altered your thinking and subsequent focus?
The main issues I encountered with this project was the large number of individual proteins that required separate expression conditions, buffers, and tags, and ultimately not all these proteins did well with long-term storage or with our experimental setups. We tried to obtain biophysical information from these protein pairs and found that due to the weak interactions, a large amount of protein was needed. I have done a lot of protein expression and purification and tried many biophysical approaches including microscale thermophoresis (MST), isothermal calorimetry (ITC), and surface plasmon resonance (SPR). In more than one instance, my proteins crashed while running these experiments. Although results from these experiments were not used in the end, they provided me with extensive experience in protein expression, purification, and protein interaction characterization.
What are some of the landmark moments that provoked your interest in science or your development as a scientist?
My landmark moments primarily include the people who I have met along my journey, and not all are scientists. My mother and grandmother are huge influences in persistence and a drive to further my education. My grandmother was denied a higher education and she always pushed me to fight for what I want. I encountered my first scientific mentors at 17 years old while volunteering at Rush University Medical Center, and they enforced my love for science. Dr. Judith Luborsky and Dr. Seerin Shatavi allowed me into the laboratory, trained me to be a productive member, and showed me what it was like to work on a team.
What are your subsequent near- or long-term career plans?
I am currently looking for opportunities as a science writer/editor and industry scientist. I am enjoying having a wide variety of options to pursue as a future career and learning about these fields through informational interviews. I find that throughout my education I have been driven by clear-cut goals and having this freedom is a uniquely liberating experience.
REFERENCES
- Becerra S, Montes M, Hernández-Munain C, Suñé C. 2015. Prp40 pre-mRNA processing factor 40 homolog B (PRPF40B) associates with SF1 and U2AF65 and modulates alternative pre-mRNA splicing in vivo. RNA 21: 438–457. 10.1261/rna.047258.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berget SM. 1995. Exon recognition in vertebrate splicing. J Biol Chem 270: 2411–2414. 10.1074/jbc.270.6.2411 [DOI] [PubMed] [Google Scholar]
- Berget SM, Moore C, Sharp PA. 1977. Spliced segments at the 5′ terminus of adenovirus 2 late mRNA. Proc Natl Acad Sci 74: 3171–3175. 10.1073/pnas.74.8.3171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatch GL, Lässle M. 1999. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays 21: 932–939. [DOI] [PubMed] [Google Scholar]
- Busch A, Hertel KJ. 2012. Evolution of SR protein and hnRNP splicing regulatory factors. Wiley Interdiscip Rev RNA 3: 1–12. 10.1002/wrna.100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson SM, Soulette CM, Yang Z, Elias JE, Brooks AN, Gozani O. 2017. RBM25 is a global splicing factor promoting inclusion of alternatively spliced exons and is itself regulated by lysine mono-methylation. J Biol Chem 292: 13381–13390. 10.1074/jbc.M117.784371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S, Hoang A, Sinha R, Zhong XY, Fu XD, Krainer AR, Ghosh G. 2011. Interaction between the RNA binding domains of Ser-Arg splicing factor 1 and U1-70K snRNP protein determines early spliceosome assembly. Proc Natl Acad Sci 108: 8233–8238. 10.1073/pnas.1017700108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow LT, Gelinas RE, Broker TR, Roberts RJ. 1977. An amazing sequence arrangement at the 5′ ends of adenovirus 2 messenger RNA. Cell 12: 1–8. 10.1016/0092-8674(77)90180-5 [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14: 1188–1190. 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels NJ, Hershberger CE, Gu X, Schueger C, DiPasquale WM, Brick J, Saunthararajah Y, Maciejewski JP, Padgett RA. 2021. Functional analyses of human LUC7-like proteins involved in splicing regulation and myeloid neoplasms. Cell Rep 35: 108989. 10.1016/j.celrep.2021.108989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bortoli F, Neumann A, Kotte A, Timmermann B, Schüler T, Wahl MC, Loll B, Heyd F. 2019. Increased versatility despite reduced molecular complexity: evolution, structure and function of metazoan splicing factor PRPF39. Nucleic Acids Res 47: 5867–5879. 10.1093/nar/gkz243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dember LM, Kim ND, Liu KQ, Anderson P. 1996. Individual RNA recognition motifs of TIA-1 and TIAR have different RNA binding specificities. J Biol Chem 271: 2783–2788. 10.1074/jbc.271.5.2783 [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. 2009. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10: 48. 10.1186/1471-2105-10-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eperon IC, Makarova OV, Mayeda A, Munroe SH, Cáceres JF, Hayward DG, Krainer AR. 2000. Selection of alternative 5′ splice sites: role of U1 snRNP and models for the antagonistic effects of SF2/ASF and hnRNP A1. Mol Cell Biol 20: 8303–8318. 10.1128/MCB.20.22.8303-8318.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forch P, Puig O, Kedersha N, Martinez C, Granneman S, Seraphin B, Anderson P, Valcarcel J. 2000. The apoptosis-promoting factor TIA-1 is a regulator of alternative pre-mRNA splicing. Mol Cell 6: 1089–1098. 10.1016/S1097-2765(00)00107-6 [DOI] [PubMed] [Google Scholar]
- Forch P, Puig O, Martinez C, Seraphin B, Valcarcel J. 2002. The splicing regulator TIA-1 interacts with U1-C to promote U1 snRNP recruitment to 5′ splice sites. EMBO J 21: 6882–6892. 10.1093/emboj/cdf668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuens T, Bouhy D, Timmerman V. 2016. The hnRNP family: insights into their role in health and disease. Hum Genet 135: 851–867. 10.1007/s00439-016-1683-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk A, Tang J, Puig O, Salgado J, Neubauer G, Colot HV, Mann M, Séraphin B, Rosbash M, Lührmann R, et al. 1998. A comprehensive biochemical and genetic analysis of the yeast U1 snRNP reveals five novel proteins. RNA 4: 374–393. [PMC free article] [PubMed] [Google Scholar]
- Graveley BR. 2001. Alternative splicing: increasing diversity in the proteomic world. Trends Genet 17: 100–107. 10.1016/S0168-9525(00)02176-4 [DOI] [PubMed] [Google Scholar]
- Hammani K, Cook WB, Barkan A. 2012. RNA binding and RNA remodeling activities of the half-a-tetratricopeptide (HAT) protein HCF107 underlie its effects on gene expression. Proc Natl Acad Sci 109: 5651–5656. 10.1073/pnas.1200318109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Ding JH, Byeon CW, Kim JH, Hertel KJ, Jeong S, Fu XD. 2011. SR proteins induce alternative exon skipping through their activities on the flanking constitutive exons. Mol Cell Biol 31: 793–802. 10.1128/MCB.01117-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han KJ, Wu Z, Pearson CG, Peng J, Song K, Liu CW. 2019. Deubiquitylase USP9X maintains centriolar satellite integrity by stabilizing pericentriolar material 1 protein. J Cell Sci 132: jcs221663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson M, Zhang C, Mear L, Zhong W, Digre A, Katona B, Sjostedt E, Butler L, Odeberg J, Dusart P, et al. 2021. A single-cell type transcriptomics map of human tissues. Sci Adv 7: eabh2169. 10.1126/sciadv.abh2169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha NL, Gupta M, Li W, Miller I, Anderson P. 1999. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2α to the assembly of mammalian stress granules. J Cell Biol 147: 1431–1442. 10.1083/jcb.147.7.1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo Y, Oubridge C, van Roon AM, Nagai K. 2015. Crystal structure of human U1 snRNP, a small nuclear ribonucleoprotein particle, reveals the mechanism of 5′ splice site recognition. Elife 4: e04986. 10.7554/eLife.04986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE. 2007. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature 446: 926–929. 10.1038/nature05676 [DOI] [PubMed] [Google Scholar]
- Lerner MR, Boyle JA, Mount SM, Wolin SL, Steitz JA. 1980. Are snRNPs involved in splicing? Nature 283: 220–224. 10.1038/283220a0 [DOI] [PubMed] [Google Scholar]
- Li X, Liu S, Jiang J, Zhang L, Espinosa S, Hill RC, Hansen KC, Zhou ZH, Zhao R. 2017. CryoEM structure of Saccharomyces cerevisiae U1 snRNP offers insight into alternative splicing. Nat Commun 8: 1035. 10.1038/s41467-017-01241-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Liu S, Zhang L, Issaian A, Hill RC, Espinosa S, Shi S, Cui Y, Kappel K, Das R, et al. 2019. A unified mechanism for intron and exon definition and back-splicing. Nature 573: 375–380. 10.1038/s41586-019-1523-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HX, Chew SL, Cartegni L, Zhang MQ, Krainer AR. 2000. Exonic splicing enhancer motif recognized by human SC35 under splicing conditions. Mol Cell Biol 20: 1063–1071. 10.1128/MCB.20.3.1063-1071.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WH, Roemer SC, Zhou Y, Shen ZJ, Dennehey BK, Balsbaugh JL, Liddle JC, Nemkov T, Ahn NG, Hansen KC, et al. 2016. The Cac1 subunit of histone chaperone CAF-1 organizes CAF-1-H3/H4 architecture and tetramerizes histones. Elife 5: e18023. 10.7554/eLife.18023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzini PA, Chew RSE, Tan CW, Yong JY, Zhang F, Zheng J, Roca X. 2019. Human PRPF40B regulates hundreds of alternative splicing targets and represses a hypoxia expression signature. RNA 25: 905–920. 10.1261/rna.069534.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, O'Brien G, Shiue L, Clark TA, Blume JE, Ares M Jr. 2007. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev 21: 708–718. 10.1101/gad.1525507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. 2008. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40: 1413–1415. 10.1038/ng.259 [DOI] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. 2017. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14: 417–419. 10.1038/nmeth.4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, Morris JH, Ferrin TE. 2021. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci 30: 70–82. 10.1002/pro.3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaschka C, Lin PC, Charenton C, Nagai K. 2018. Prespliceosome structure provides insights into spliceosome assembly and regulation. Nature 559: 419–422. 10.1038/s41586-018-0323-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig O, Bragado-Nilsson E, Koski T, Seraphin B. 2007. The U1 snRNP-associated factor Luc7p affects 5′ splice site selection in yeast and human. Nucleic Acids Res 35: 5874–5885. 10.1093/nar/gkm505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford JR, Wang X, Mort M, Vanduyn N, Cooper DN, Mooney SD, Edenberg HJ, Liu Y. 2009. Splicing factor SFRS1 recognizes a functionally diverse landscape of RNA transcripts. Genome Res 19: 381–394. 10.1101/gr.082503.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider TD, Stephens RM. 1990. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res 18: 6097–6100. 10.1093/nar/18.20.6097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seraphin B, Kretzner L, Rosbash M. 1988. A U1 snRNA:pre-mRNA base pairing interaction is required early in yeast spliceosome assembly but does not uniquely define the 5′ cleavage site. EMBO J 7: 2533–2538. 10.1002/j.1460-2075.1988.tb03101.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Park JW, Lu ZX, Lin L, Henry MD, Wu YN, Zhou Q, Xing Y. 2014. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc Natl Acad Sci 111: E5593–E5601. 10.1073/pnas.1419161111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siliciano PG, Guthrie C. 1988. 5′ splice site selection in yeast: genetic alterations in base-pairing with U1 reveal additional requirements. Genes Dev 2: 1258–1267. 10.1101/gad.2.10.1258 [DOI] [PubMed] [Google Scholar]
- Soneson C, Love MI, Robinson MD. 2015. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res 4: 1521. 10.12688/f1000research.7563.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, et al. 2015. Proteomics. Tissue-based map of the human proteome. Science 347: 1260419. 10.1126/science.1260419 [DOI] [PubMed] [Google Scholar]
- Uhlen M, Zhang C, Lee S, Sjostedt E, Fagerberg L, Bidkhori G, Benfeitas R, Arif M, Liu Z, Edfors F, et al. 2017. A pathology atlas of the human cancer transcriptome. Science 357: eaan2507. 10.1126/science.aan2507 [DOI] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Luhrmann R. 2009. The spliceosome: design principles of a dynamic RNP machine. Cell 136: 701–718. 10.1016/j.cell.2009.02.009 [DOI] [PubMed] [Google Scholar]
- Yeo G, Burge CB. 2004. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol 11: 377–394. 10.1089/1066527041410418 [DOI] [PubMed] [Google Scholar]
- Zhou Z, Fu XD. 2013. Regulation of splicing by SR proteins and SR protein-specific kinases. Chromosoma 122: 191–207. 10.1007/s00412-013-0407-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Licklider LJ, Gygi SP, Reed R. 2002. Comprehensive proteomic analysis of the human spliceosome. Nature 419: 182–185. 10.1038/nature01031 [DOI] [PubMed] [Google Scholar]
- Zhou A, Ou AC, Cho A, Benz EJ Jr, Huang SC. 2008. Novel splicing factor RBM25 modulates Bcl-x pre-mRNA 5′ splice site selection. Mol Cell Biol 28: 5924–5936. 10.1128/MCB.00560-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y, Weiner AM. 1986. A compensatory base change in U1 snRNA suppresses a 5′ splice site mutation. Cell 46: 827–835. 10.1016/0092-8674(86)90064-4 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.