Abstract
The self-complementary triplet 5′UGG3′/5′UGG3′ is a particular structural motif containing a noncanonical G–G pair and two U·G wobble pairs. It constitutes a specific structural and electrostatic environment attracting metal ions, particularly Ba2+ ions. Crystallographic research has shown that two Ba2+ cations are located in the major groove of the helix and interact directly with the UGG triplet. A comparison with the unliganded structure has revealed global changes in the RNA structure in the presence of metal ions, whereas thermodynamic measurements have shown increased stability. Moreover, in the structure with Ba2+, an unusual noncanonical G(syn)-G(syn) pair is observed instead of the common G(anti)-G(syn). We further elucidated the metal binding properties of the UGG/UGG triplet by performing crystallographic and thermodynamic studies using DSC and UV melting with other metal ions. The results explain the preferences of the UGG sequence for Ba2+ cations and point to possible applications of this metal-binding propensity.
Keywords: UGG repeats; U·G wobble, G(syn)–G(syn) pair; RNA crystallography; RNA UV melting, RNA DSC measurement
INTRODUCTION
RNA contains many noncanonical base pairs that increase the structural richness of RNA compared to fully complementary DNA. Due to their unsaturated potential for base pairing, these noncanonical pairs have an additional capacity for binding external ligands, including metal cations. These bound cations play a role in the folding of RNA into tertiary structures and can also be involved in catalysis conducted by ribozymes.
The G·U wobble is the most common noncanonical base pair found in RNA structures (Varani and Mcclain 2000). In most cases, the G·U pair is stabilized by two hydrogen bonds: N1···O2 and O6···N3. The guanosine residue is inclined toward the minor groove, while the uridine residue is shifted toward the major groove. This arrangement results in the exposition of functional groups (N2 exo-amino of G, O4 carbonyl of U) and the formation of a specific topological and electrostatic landscape within the RNA helix. The hydrogen bond donors and acceptors not involved in G·U pairing show a high binding potential for ligands (Strobel and Cech 1995, 1996; Ananth et al. 2013). The O4 carbonyl group of uridine together with the N7 and O6 atoms of guanine creates a broad electronegative potential (Xu et al. 2007). This attracts metal ions and other positively charged ligands (Allain and Varani 1995; Burgstaller 1997; Ananth et al. 2013).
X-ray and NMR studies have provided much information about structures containing either single or tandem G·U wobbles and enriched our knowledge about the properties of this noncanonical base pair (Biswas and Sundaralingam 1997; McDowell et al. 1997; Mooers et al. 1997; Mueller et al. 1999; Trikha et al. 1999; Sashital et al. 2007; Gu et al. 2015; Garg and Heinemann 2018; Berger et al. 2019; Ruszkowska et al. 2022). An important feature of the G·U pair is that it is nonisosteric, that is, the superposition of N-glycosic bonds of 5′G·U3′ with the 5′U·G3′ pair results in a large shift between the bases (Westhof et al. 2019). The most pronounced outcome of this nonisostericity is observed in the twist angle between G·U and the neighboring pair. For G·U, the following pair is usually undertwisted while for U·G it is overtwisted (Ananth et al. 2013). Overall, the twist is distinct from a typical value observed for A-RNA but the overall effect on the helix structure is moderate and absorbed by local changes in the sugar–phosphate backbone.
There are several known three-dimensional models of a wobble pair with bound metal ions in the major groove (Allain and Varani 1995; Colmenarejo and Tinoco 1999; Kacer et al. 2003; Keel et al. 2007; Gu et al. 2015; Price et al. 2015; Leonarski et al. 2017, 2019; Rozov et al. 2019; Ruszkowska et al. 2022). Structural studies have confirmed the ability of G·U to interact with metal species but have also indicated that the potential of the wobble motif to bind cations depends on their structural context (Keel et al. 2007). The structural and electrostatic requirements for the metal ions to bind this wobble pair have not yet been fully investigated. Here, we present crystallographic and thermodynamic studies of the self-complementary triplet 5′UGG3′/5′UGG3′ motif consisting of two U·G wobble pairs interrupted by a G–G pair. The presented motif can be found in humans, within the mRNA of certain genes as a repeated sequence. Intronic UGG repeats participate in alternative splicing of the CD44 gene that codes the CD44 antigen involved in cell–cell interactions, adhesion and migration (Galiana-Arnoux et al. 2005). The crystallographic structures reveal that the UGG motif shows particular affinity for Ba2+ forming direct interactions with bound metal ions. The presence of Ba2+ is associated with global changes in RNA structure and with the presence of an unusual noncanonical G(syn)-G(syn) pair instead of the common G(anti)-G(syn). To elucidate the metal binding properties of the UGG/UGG triplet, we performed crystallographic and thermodynamic studies, using DSC and UV-melting, with other metal ions. The results explain the preferences of the UGG sequence for Ba2+ cations. These properties can be useful in crystallographic research, for phasing with anomalous signal, and in biotechnology, whether using RNA molecules as a metal sponge or as a sensor of metal ions.
RESULTS
The RNA models
Seven crystal structures of the RNA duplex (AUGUGGCAU)2 containing the 5′UGG3′/5′UGG3′ motif were determined: in an unliganded form and in the presence of the following metal cations: Cs+, Sr2+, Ca2+, Cd2+, and two models with Ba2+ ions (Supplemental Table S1). All six of the ion-liganded structures were crystallized in similar conditions, containing MPD as the precipitant, while the unliganded structure crystallized in the presence of ammonium sulfate. The Ba2+ ions were identified as strong peaks (approx. 14 r.m.s.d.) on the anomalous electron density map, and confirmed by an examination of their coordination spheres and distances from ligand molecules. Crystals containing Cs+, Sr2+, Ca2+, and Cd2+ were obtained by the cocrystalization of RNA with the appropriate ions, in the absence of Ba2+. In all four structures the RNA molecules showed the same packing in the crystal lattice, resulting in the same space group and unit cell parameters. The asymmetric unit of the monoclinic crystals contained fourteen independent RNA duplexes. A weak anomalous signal was also observed for Cs+ and Sr2+ structures.
Noncanonical pairs in RNA duplexes
The AUGUGGCAU oligomer forms self-complementary duplexes. The six flanking nucleotides are engaged in Watson–Crick base pairing between complementary nucleotides of the two strands (Fig. 1A), with the exception of the RNA-Ba(1) structure, in which the 1A residue of chain C and the 9U residue of chain D form a noncanonical pair (see details in Supplemental Material Fig. S1). The UGG motif is located in the middle of the duplex and is engaged in noncanonical pairing. Two U·G pairs are wobbled, and the G–G pairs are in most cases G(syn)–G(anti), but in the RNA-Ba(2) structure there is also an instance of the G(syn)–G(syn) conformation (Fig. 2A–D). The G(syn)–G(anti) pairs interact via two H-bonds (N1···O6 and N2···N7) between their respective Watson–Crick and Hoogsteen edges, as described earlier (Fig. 2B; Kiliszek et al. 2011). The G(syn)–G(syn) pair has not been described previously. The G(syn) bases interact via two solvent molecules wedged between them. The innermost ligand is 2.2–2.5 Å from the carbonyl O6 atoms and the other ligand. Its distance from the N7 atoms is 2.5–2.8 Å. The other ligand is 2.4–2.6 Å from the two N7 atoms. The closeness of the inner ligand to the carbonyl O2 atoms suggests that it could be Na+ rather than a water molecule, while the outer ligand could be water-donating H-bonds to the imino groups (Fig. 2D; Supplemental Fig. S2). The unusual alignment of the G rings is stabilized by the presence of two Ba2+ ions in the major groove at 3.0–3.1 Å from the carbonyl groups (Figs. 1B, 3A). Indications of G(syn)–G(syn) pairs are also observed in RNA-Cs, RNA-Cd and RNA-Ba(1) structures but the modeling is ambiguous due to static disorder and low resolution (Supplemental Fig. S3).
FIGURE 1.
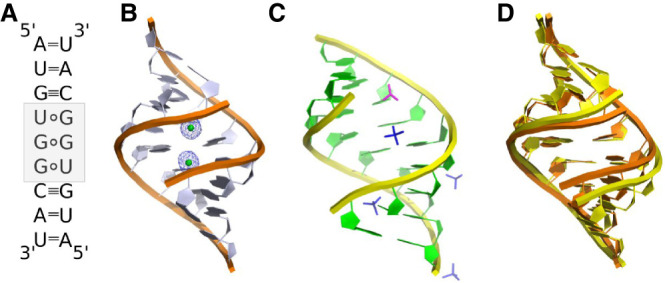
Secondary and tertiary structure models of the AUGUGGCAU duplex. (A) The UGG/UGG motif (gray) forms two U·G pairs and one noncanonical G–G pair. (B) Cartoon representation of the RNA-Ba(2) model with bound Ba2+ ions (green balls) in the major groove of the helix. The 2Fo–Fc electron density map (gray) is contoured at the 1σ level. (C) Cartoon representation of the unliganded model with sulfate (dark blue sticks) and acetate ions (pink sticks) located in the major and minor grooves of the helix. (D) A superposition of RNA-Ba(2) (orange) and unliganded structures (yellow).
FIGURE 2.
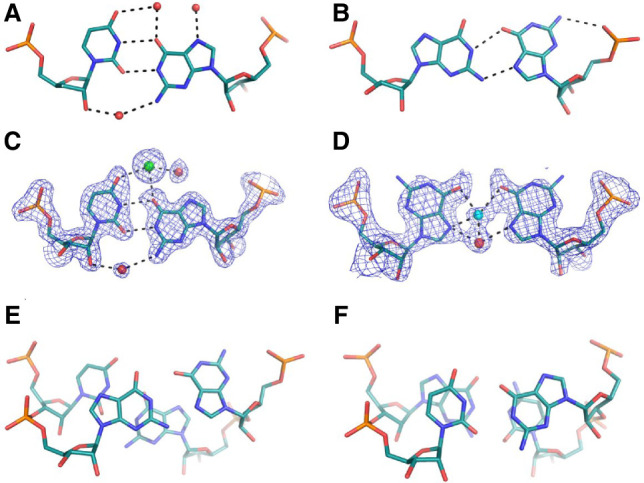
Noncanonical base pairs observed in crystallographic models of the AUGUGGCAU duplex. (A,C) The U·G wobble interacting with solvent molecules. (B) Stick representation of the G(anti)–G(syn) pair and (D) G(syn)–G(syn) base pairs. The stacking interactions observed for G–G (E) and U·G pairs (F). Panels A and B are based on the unliganded structure; C–F are based on the Ba(2) model. Water molecules are red spheres, Ba2+ ions are green, and Na+ is blue. The 2Fo–Fc electron density map (gray) is contoured at the 1σ level. The H-bonds are black dashed lines.
FIGURE 3.
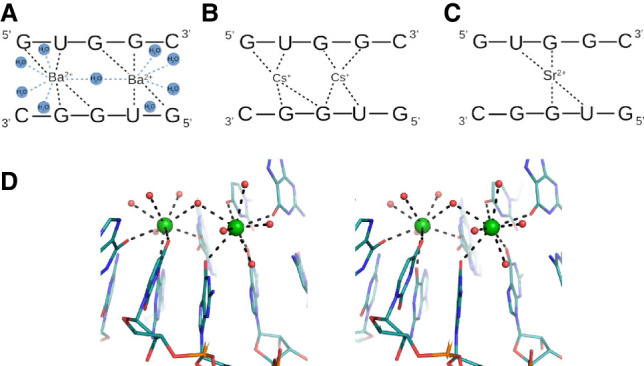
Interactions of RNA residues with metal ions: (A) with Ba2+, (B) Cs+ and (C) Sr2+ ions. (D) Stereo view of RNA interacting with Ba2+ ions (green spheres).
Interactions of RNA with metal ions
Each UGG motif can bind up to two metal ions in the major groove. Complexes with Ba2+, Cs+, and Sr2+ are formed easily, while Ca2+ and Cd2+ are difficult to observe. Ba2+ and Cs+ occupy two binding sites, while Sr2+ shows a mixture of binding modes (Figs. 1B, 3A–C; Supplemental Fig. S4). In seven out of fourteen crystallographically independent duplexes single Sr2+ ions are bound; in three cases there are two ions while in four cases, the occupancy of the second ion is only partial (Supplemental Fig. S4). Single Cd2+ ions are observed in only three duplexes. In the structure crystallized with Ca2+, the solvent structure is indistinct. Three single peaks of the electron density map observed in the major groove have been interpreted as water molecules, not Ca2+ (Supplemental Fig. S4). Their coordination distances are at least 2.7 Å, while for calcium, they should be ∼2.4 Å (Harding et al. 2010).
When two ions (Ba2+, Cs+, or Sr2+) interact with the UGG motif, they are located on each side of the G–G pair, above the U·G wobble (Fig. 3A,B). For Ba2+, the pattern of interactions is the most ordered. Each ion forms four direct interactions with the carbonyl groups of RNA, spanning both strands of the duplex. Two bonds are observed with O6 of 3G and O4 of 4U residues of one chain and with O6 of 5G and O6 of 6G of the second strand. In addition, Ba2+ interacts with up to five water molecules, supplementing its coordination sphere to a maximum value of nine ligands. The range of distances is 2.5–3.1 Å. The ions are ∼4.8 Å apart and share only one water molecule (Fig.3A,D). In the case of Cs+ the pattern of interactions is less ordered. Each Cs+ ion forms four direct contacts with RNA, as in the case of Ba2+, but in some cases, one of the ions is shifted toward the G–G pair engaging the carbonyl groups of both guanosines (Fig. 3B). This is reflected in a wide range of Cs–Cs distances, from 4.8 to 5.7 Å. Similarly, in the Sr-structure, one of the metal ions tends to be shifted toward the G–G pair. When only one ion (Sr2+ or Cd2+) is bound to RNA, it is placed just above the G–G pair interacting with their carbonyl groups. Two additional contacts can be observed with the carbonyl groups of the 4U residues of each strand (Fig. 3C).
In the metal-free UGG structure, water molecules are observed in the major groove of the U·G wobble. Each water molecule interacts with U·G and the neighboring G(syn)–G(anti) pair (Fig. 2A). In addition to water, four sulfate and one acetate ions have been identified, either in the major or the minor groove, interacting mostly with positively charged amino groups of RNA (Fig. 1C; Supplemental Fig. S5). Sulfate binding to the exo-amino groups of U·G pairs has been described previously (Masquida et al. 1999; Auffinger et al. 2004).
Impact of metal ions on the RNA structure
In all the described models the RNA has the A-form (Fig. 1B,C; Supplemental Fig. S4). The presence of the noncanonical base pairs seems to have a local rather than global effect on the structure. The conformation of the U·G and G–G pairs is reflected in the shear and shift parameters. Both have distinct values from the neighboring pairs (Supplemental Tables S2, S3). Additionally, the arrangement of the two bulky G–G residues is reflected in the stretch parameter. Other parameters, such as helical twist, groove width or propeller, are associated with the presence of metal ions. A comparison of the unliganded structure with Ba-models revealed that in the presence of Ba2+, the major groove is narrowed, and access to it is limited (Fig. 1D). The major groove width for the native model is 14.2 Å, while for Ba-RNA the values are in the range of 9.7–10.8 Å. In the other complexes with metal ions, groove narrowing is not observed. Rather, they exhibit a range of groove widths (10.1–15.7 Å). The twist parameter is correlated with the groove width. For the Ba-models, the twist has the highest value (34.5° ± 5.2°), while for other metal-structures, it is 33.3° ± 3.0°, and for the unliganded form, it is 32.1° ± 3.7° (Supplemental Table S4). The other parameter associated with Ba2+ ions is the propeller twist, which seems to be elevated (16.9°–23.0°) in comparison to the native structure (14.0°) and other metal-structures (<17°) (Supplemental Table S5).
Stacking interactions and electrostatic profile distribution
The distribution of stacking interactions in all the models is similar. Since each duplex possesses self-symmetry, the stacking is symmetric. Intrastrand overlaps are predominant. Only for the 2U3G/7C8A steps is an interstack observed between the guanosine and adenosine residues. Most extensive stacking interactions are observed between the neighboring 3G4U and 6G7C residues. Measuring only the overlap of base rings (excluding exocyclic atoms) the area is on average 4 Å2, which covers almost all of the pyrimidine plane (Fig. 2F). The observed distribution of stacking indicates that the 4U and 6G residues involved in wobble pairs interact extensively, while guanosines forming the G–G pairs are engaged least of all the nucleotides in the duplex (Fig. 2E,F).
All the helices exhibit similar electrostatic potential and surface profile. The major groove is mostly electronegative due to the presence of carbonyl groups. Patches of positive potential come from the exposed Watson–Crick 5G(syn) edge and the exo-amino group of 8C residues (Fig. 4A,C). The G–G pairs are located in the center of the helix and form a platform of positive and negative potential, while the neighboring wobble pairs are strictly electronegative. The guanosine from the U·G pair is pushed toward the minor groove and forms a niche, but it does not seem to be occupied, and the metal ions occupy the central part of the electronegative area of the wobble pairs (Fig. 4C). When only one ion is bound to the RNA, it is located in the center of the helix above the negatively charged region of the G–G pair. The minor groove shows a characteristic RNA electrostatic profile with alternating stripes of positive and negative potential. The exo-amino group of the wobbled guanosines seems to be less exposed in comparison to other residues (Fig. 4B,D).
FIGURE 4.
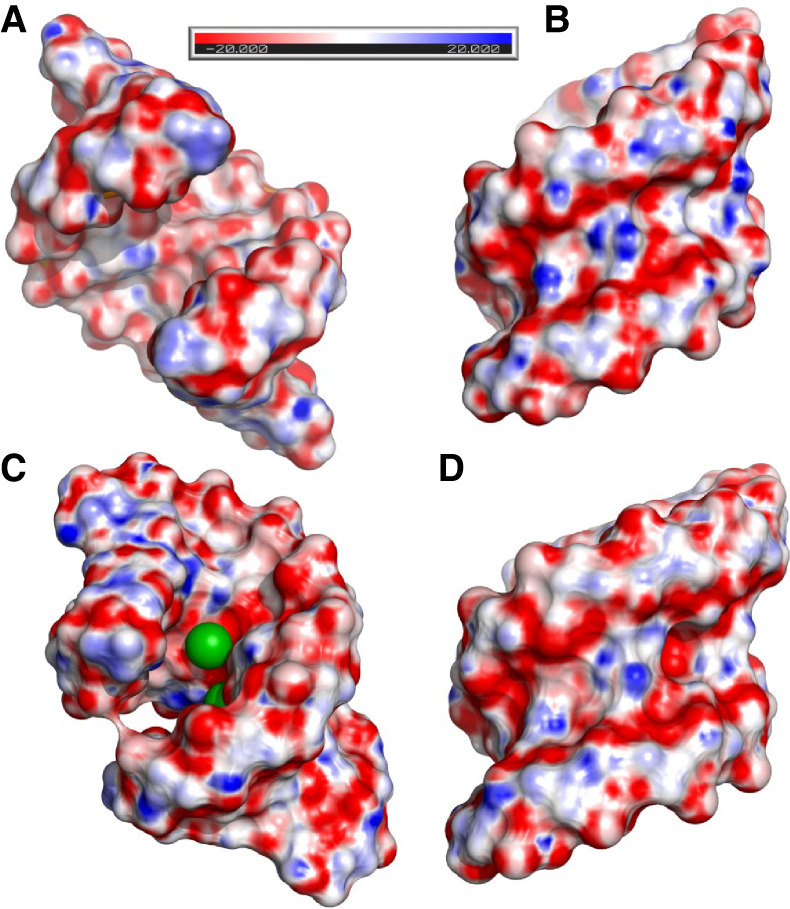
The electrostatic potential distribution calculated for (A,B) the unliganded structure and (C,D) the RNA-Ba(2) model. The Ba2+ ions are green spheres.
Thermal stability of RNA
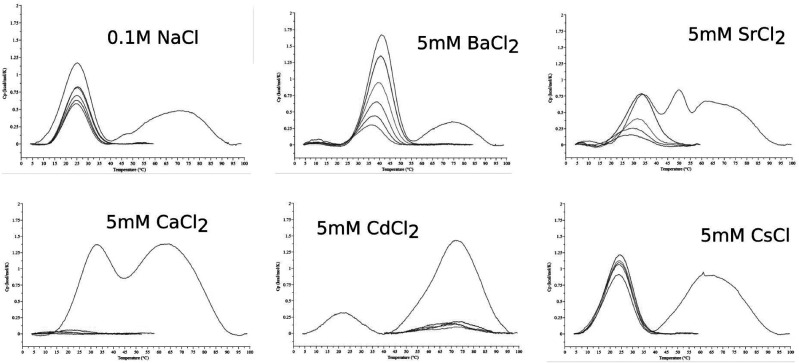
Thermodynamic and thermal studies were performed for the AUGUGGCAU duplex in the presence of Cs+, Sr2+, Ca2+, Cd2+, or Ba2+ ions using the UV melting method and differential scanning calorimetry (DSC). The measurements were performed in 100 mM NaCl and 5 mM of metal ions, since the same conditions were present in the crystallization solutions. The reference RNA sample was melted in the buffer containing only 100 mM NaCl. Additionally, the thermal stability of the duplex was determined in high salt concentrations: (1) 1 M NaCl and (2) 1 M NaCl with 5 mM BaCl2. The UV-melting data were used to calculate thermodynamic parameters such as ΔH, ΔG, and ΔS and to determine the melting temperature TM of the duplex at 100 µM RNA concentration (Table 1; Supplemental Fig. S6). Each DSC measurement was performed in five runs of the denaturation–renaturation cycle to define the melting point of the duplex Tm (as distinct from TM which was calculated for the UV-melting experiments), the degree of degradation or refolding of RNA and the presence of other forms of RNA aggregation (Table 1; Supplemental Table S6).
TABLE 1.
The thermodynamic and thermal parameters of the AUGUGGCAU oligomer in different buffer conditions
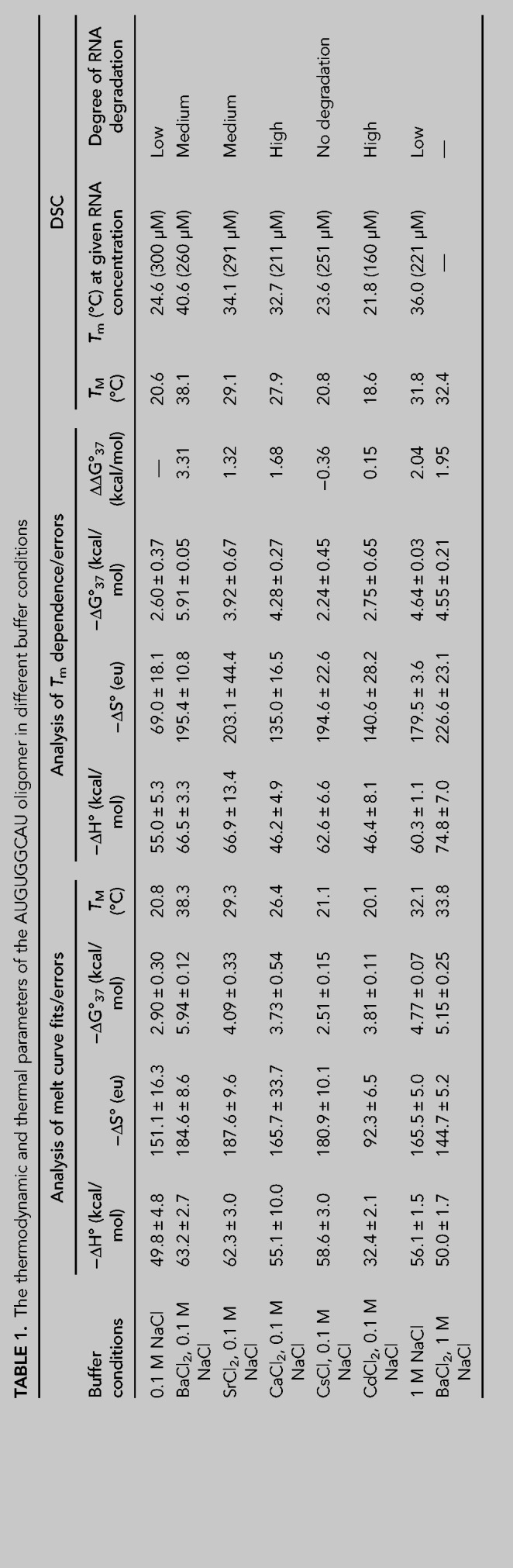
Both methods show similar effects on the thermal stability of the investigated samples. The RNA is stabilized the most in the presence of Ba2+ (Table 1). The ΔG is 3.3 kcal/mol lower than that of the reference sample. This amounts to a higher melting temperature by 17.6°C for UV melting and by 16°C for DSC measurement. Sr2+ and Ca2+ seem to have a similar stabilizing effect. For Sr2+, it is 1.3 kcal/mol and 8.5°C or 9.5°C (UV melting and DSC, respectively), and in the presence of Ca2+, it is 1.7 kcal/mol and 7.3°C or 8.1°C (UV melting, DSC). Cs+ and Cd2+ do not stabilize the duplex or show only a slight destabilizing effect. In the presence of Cd2+, the RNA has a lower melting temperature by ∼2°C and ΔΔG = −0.2 kcal/mol. In the presence of Cs+, the melting temperature is similar to that of the control sample, while ΔG is higher by 0.4 kcal/mol. In 1 M NaCl the Tm of the duplex is higher than that in 0.1 M NaCl by 11°C and ΔG is lower by 2 kcal/mol. These parameters indicate that a high concentration of NaCl stabilizes the RNA substantially, but not as much as Ba2+ ions. When the duplex is melted in 1 M NaCl and 5 mM Ba2+, the stabilization effect of the metal ions is not observed (the same ΔG for both conditions: 1 M NaCl and 5 mM Ba2+ in 1 M NaCl).
In all cases, the DSC spectra show an additional broad peak at higher temperatures (45°C–90°C) (Fig. 5). It is present only in the first denaturation–renaturation cycle. The height and width of the peak were different for each RNA preparation. In Ba2+, the high-temperature peak is the smallest compared to the first peak, while for Cd2+, it is the highest peak. The peaks observed in the initial melting event could indicate some higher-order aggregates that disaggregate into a duplex form when heated, and do not reform within the time frame of the heating-cooling cycles. In the presence of Sr2+, a third peak can be distinguished at 49°C. The DSC experiments show RNA degradation with each cycle, indicated by diminishing heights of the main peak. The fastest loss of the signal is observed for Ca2+ and Cd2+ ions, while the highest protection of RNA is observed for Cs+. In NaCl, RNA degrades relatively slowly while, in the presence of Ba2+ and Sr2+, the RNA is hydrolyzed at an intermediate rate (Fig. 5).
FIGURE 5.
DSC spectra obtained for the AUGUGGCAU oligomer in different buffer conditions.
DISCUSSION
UGG is interesting from the perspective of structural biology as a motif containing noncanonical pairs G·U and G–G or as a component of microsatellite repeats, but it also plays a biological role. UGG repeats are found in the transcripts of certain genes, mostly in the 3′UTRs (Kozlowski et al. 2010). Intronic UGG repeats participate in alternative splicing of the CD44 gene which encodes the CD44 antigen involved in cell–cell interactions, adhesion and migration (Galiana-Arnoux et al. 2005).
UGG repeats are noted for forming tetraplexes (Sobczak et al. 2010; Malgowska et al. 2014). A question then arises: why do all the structures in this study have the duplex form? It seems that the crucial variables are the nature of the cations and ionic strength. Tetraplexes have been observed to form mainly in the presence of K+, whereas the duplexes presented here contain divalent cations (Ba2+, Sr2+, Ca2+, Cd2+) or the monovalent Cs+, which stands out, however, as the largest ionic species under study. In the absence of the metal cations, the duplex could be crystallized under the conditions of high ionic strength (2.5 M ammonium sulfate). Another factor favoring the duplex could be the presence of flanking sequences. Although all the examined crystal structures show this oligomer in the duplex form, it cannot be excluded that in solution tetraplexes could present. Further studies, by other techniques might be needed to resolve this.
Several RNA crystal structures show Ba2+ binding at sites consisting of a single U·G pair and carbonyl oxygen atoms from neighboring bases, preferably on the 5′-sides (Supplemental Table S7; Kacer et al. 2003; Hancock et al. 2004; Serganov et al. 2009; Huang et al. 2010, 2016; Schroeder et al. 2012; Daldrop and Lilley 2013; Price et al. 2015; Bachas and Ferré-D'Amaré 2018; Knappenberger et al. 2018; Heber et al. 2019; Ruszkowska et al. 2022). The structure presented in this paper can bind up to two metal ions. The site consists of the 5′UGG/3′GGU motif and two carbonyl oxygen atoms from guanosine rings at the 5′-side of each strand (Fig. 3A,D). The two cation binding sites are asymmetric, due to the asymmetry of the central G(syn)–G(anti) pair, and they seem to have different affinities for the ions. The exception is the previously unobserved instance of G(syn)–G(syn), which is symmetric and binds the ligands symmetrically. In the strong complexes, both cation sites are occupied, whereas in the weaker complexes, there can be only one metal ion per UGG motif. The complexes with Ba2+ stand out as the best ordered, showing detailed coordination spheres of the metal ions (Fig. 3A). Each ion has four RNA ligands and up to five coordinating water molecules. The observed coordination number of nine is the most common number observed for barium complexes in the CCDC structural database (Hancock et al. 2004). In the complex with Cs+, there are also two cations in each UUG. The structure with Sr2+ shows a tendency to bind only one metal ion per UGG, and the site is shifted toward the central G–G pair. Cd2+ is barely seen, with single sites located in three out of 14 duplexes, while in the structure soaked in Ca2+, the presence of the cations is unclear. The apparent tendency to weaker binding and shift toward the G–G pair can be correlated with the decreasing radii of the doubly charged metal ions (Ba2+ > Sr2+ > Ca2+ > Cd2+) (Shannon 1976). This series is reproduced in calorimetric measurements in terms of decreasing thermal stability of the complexes.
Temperature-melting curves and the thermodynamic parameters derived from them were used to determine the amount of stabilization bestowed by the ions on the RNA in solution (Table 1). Ba2+ stabilized the most, followed by Sr2+ and Ca2+, while Cs+ and Cd2+ had no stabilization effect. The thermodynamic results corroborate the crystallographic observations, in terms of their ability to form well-defined complexes, with the exception of Cs+ ions, which are clearly visible in the crystal structure but have no impact on the thermal stability of the RNA duplex. They do, however, protect the RNA from hydrolysis, compared to all the other examined cations.
How can Cs+ shield the chemical integrity of RNA without enhancing its thermal stability? The lack of stabilizing effect could mean that the addition of 5 mM Cs+ does not make a significant contribution to stability in comparison to the already present 100 mM Na+. Both cations belong to the same group in the periodic table of elements and could play similar roles, balancing the negative charge of the RNA. However, Cs+ is much larger, which makes it easier to observe in the crystal structure, and it could fit the major groove more snugly. On the other hand, the protection by Cs+ against RNA hydrolysis could mean that this large, singly charged cation does not polarize water efficiently for nucleophilic attack.
Another observation from the thermodynamic study is that the stabilizing effect of Ba2+ (ΔΔG = 3.0 kcal/mol) is seen only at low ionic strength (0.1 M NaCl), whereas in the presence of 1 M NaCl the amount of stabilization due to Ba2+ is much lower (ΔΔG = 0.4 kcal/mol) (Table 1). This can be compared with the results of crystallization experiments in which all the complexes with metal ions were crystallized only under low salt conditions (80 mM NaCl), whereas the unliganded RNA could only be crystallized in the presence of high salt (2.5 M ammonium sulfate). The bulk effect of high salt can stabilize the duplex to a comparable degree to the more specifically acting divalent cations. This apparent effect of ionic strength could be relevant to studies in which high salt conditions are used as a point of reference.
Two common physiological cations, Mg2+ and K+, were included in some crystallization solutions but have not been observed in the crystal structures. Mg2+ was present, but unidentified, in the structure designated as unliganded. In terms of electron density, Mg2+ is similar to water molecules but it can often be recognized by its characteristic coordination sphere. One possibility is that the resolution of the X-ray data was insufficient to identify the cations or perhaps Mg2+ did not bind in a sufficiently ordered manner. As for K+, no useful crystals were obtained in its presence. When considering the potential of the UGG motif to bind different metal cations, crystallography and calorimetric measurements show that Ba2+ is optimal for binding the UGG motif. The weaker binding of other cations is related to both their size and valence, or more precisely the way their charge is distributed. K+ has a radius similar to Ba2+ but it is monovalent. On the other hand Mg2+ is divalent and very directional in the way it forms its coordination sphere, but it is much smaller and may not align as well with the binding potential of the UGG motif.
Among the RNA motifs containing U·G pairs, the structure presented here seems to be an enhanced version of related motifs in terms of its ability to bind metal cations, especially Ba2+. It seems that the central G–G pair increases the number of bound Ba2+ to two. The affinity for Ba2+ is indicated by the well–ordered crystal structures and the amount of thermodynamic stabilization. A distinguishing feature of the RNA complex with Ba2+, compared to other cations, is the closing of the major groove around Ba2+. This could reflect a good site complementarity of Ba2+ in terms of its size and coordination number.
Possible points of interest of this RNA sequence could be an application as an ionic “sponge” or even a detector, because the closing of the major groove could be used as a means to activate suitable fluorophores attached to the opposite ends of the motif. A more obvious application could be the use of bound Ba2+ in crystallographic phasing. Last, it should be cautioned that the ability of Ba2+ to bind, with high affinity, to some biological molecules could be a risk factor in the use of barium salts in medical imaging.
MATERIALS AND METHODS
Synthesis, purification, and crystallization of the AUGUGGCAU oligomer
The oligomer was synthesized by the solid phase method in an Applied Biosystems DNA/RNA synthesizer using TOM protected phosphoramidites. The synthesis was carried out in DMT-ON mode resulting in the oligomer carrying a trityl group at the 5′ end. The RNA was purified according to the protocol suitable for Glen-Pak cartridges (Glen Research) and then desalted and lyophilized under vacuum using a Speed-Vac. The purity of the oligomer was monitored using analytical HPLC separation (Lang and Micura 2008). Usually, the purity was at least 90%, which is suitable for crystallization. Before crystallization the RNA was dissolved in 10 mM sodium cacodylate pH 7.0 to a final concentration of 1 mM. The oligomer was denatured for 5 min at 95°C and cooled to ambient temperature within 2 h. Crystals were grown by the sitting drop method at 19°C. Most of the crystals grew under similar conditions: 0.08 M NaCl, 0.04 M sodium cacodylate pH 6.0, 45% MPD and 12 mM spermine. Depending on the metal ions the solution was supplemented with 20 mM BaCl2 or CdCl2 or SrCl2 or CaCl2 or CsCl. Only the unliganded structure was crystallized in 10 mM magnesium acetate, 50 mM MES pH 5.6 and 2.5 M ammonium sulfate. The RNA was mixed with the crystallization solution at a ratio of 3:1.
X-ray data collection, structure solution, and refinement
X-ray diffraction data were collected on the BL 14.1, 14.2, and 14.3 beam lines at the BESSY II electron storage ring operated by Helmholtz-Zentrum Berlin (Mueller et al. 2015). Only the unliganded crystals were cryoprotected by 20% glycerol (v/v) in the mother liquor. The data were integrated and scaled using XDSAPP or XDSGUI software implementing the XDS package (Kabsch 2010; Krug et al. 2012). The x-ray data are summarized in Supplemental Table S1. First, the structure of RNA-Ba(1) was solved by anomalous phasing. Although the data were collected at λ = 0.895 Å wavelength, far from the absorption edge of barium, the anomalous signal was observed up to a resolution of 3.4 Å. The initial phases were calculated using the Auto-Rickshaw web server (Panjikar et al. 2009). An inspection of the electron density map indicated that it corresponded well with the determined location of heavy atoms. Moreover, the location of the RNA chain could be resolved. The atomic model was manually built using Coot (Emsley and Cowtan 2004). The remaining structures were solved by molecular replacement with PHASER using RNA-Ba(1) as a search model (Read et al. 2013). Early stages of the refinement were carried out using Refmac5 from the CCP4 program suite (Vagin et al. 2004; Winn et al. 2011), and then refinement was continued with PHENIX (Liebschner et al. 2019). The models are summarized in Supplemental Table S1. The helical parameters were calculated with 3DNA (Zheng et al. 2009) using a sequence-independent method based on vectors connecting the C1′ atoms of the paired residues to avoid computational artifacts arising from noncanonical base-pairing. The program PBEQ-Solver (Jo et al. 2008) was used to calculate the electrostatic potential map. All pictures were drawn using PyMOL v0.99rc6 (Schrödinger and DeLano 2020). Atomic coordinates of the crystallographic models have been deposited with the Protein Data Bank (accession codes 8AMG, 8AMI, 8AMJ, 8AMK, 8AML, 8AMM, 8AMN). X-ray diffraction images are deposited in the MX-RDR database (Macromolecular Xtallography Raw Data Repository) (https://mxrdr.icm.edu.pl/) with appropriate DOI (Supplemental Table S1).
UV melting of oligonucleotides
UV thermal melting studies were performed according to a previous report (Rypniewski et al. 2016) on a Jasco V-700 spectrometer with a thermoprogrammer. The RNA oligomers were dissolved in a buffer containing 0.1 or 1 M sodium chloride, 20 mM sodium cacodylate pH 7.0 and appropriately 5 mM of BaCl2 or CdCl2 or SrCl2 or CaCl2 or CsCl. Each duplex was prepared in nine different concentrations in the range of 10−5 to 10−6 M. The concentrations of single-strand oligomers were calculated from a high temperature (>80°C) absorbance and single strand extinction coefficients approximated by the nearest-neighbor model. The UV absorption versus temperature was measured at 260 nm at a heating rate of 0.5°C/min in the range 5°C–90°C. The melting curves were analyzed, and the thermodynamic parameters were calculated using MeltWin 3.5.
DSC melting of oligonucleotides
The RNA oligomers were dissolved in buffer I containing 0.1 or 1 M sodium chloride, 20 mM sodium cacodylate pH 7.0 and appropriately 5 mM of BaCl2, CdCl2, SrCl2, CaCl2, or CsCl. The samples were dialyzed overnight at 4°C against buffer I to equilibrate RNA and the reference solutions. DSC experiments were carried out on a MicroCal PEAQ-DSC calorimeter (Malvern Instruments Ltd.). Each measurement was performed in five cycles of heating and cooling in the range of 2°C–110°C and a scan rate of 1°C/min. First, reference scans of buffer I were carried out to establish the instrument thermal history and to reach a near perfect baseline repeatability. Then, RNA samples were measured at concentrations ranging from 160–298 µM. The results were analyzed using dedicated software implemented by Malvern Instruments. The melting temperature Tm was calculated by applying a two-state model fitting.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Institute of Bioorganic Chemistry, Polish Academy of Sciences.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.079414.122.
MEET THE FIRST AUTHOR
Agnieszka Kiliszek.

Meet the First Author(s) is a new editorial feature within RNA, in which the first author(s) of research-based papers in each issue have the opportunity to introduce themselves and their work to readers of RNA and the RNA research community. Agnieszka Kiliszek is the first author of this paper, “Structure and thermodynamics of a UGG motif interacting with Ba2+ and other metal ions: accommodating changes in the RNA structure and the presence of a G(syn)–G(syn) pair.” Agnieszka is an Associate Professor at the Institute of Bioorganic Chemistry, Polish Academy of Sciences. She is a crystallographer working in the Department of Structure and Function Relationship of Biomolecules.
What are the major results described in your paper and how do they impact this branch of the field?
We have shown that the self-complementary UGG/UGG motif shows affinity for divalent cations, particularly for Ba2+. Crystallographic observations were supported by thermal studies using DSC and UV-melting. We have explained that Ba2+ ions are optimal for binding to the UGG motif since their size and valence suits the RNA landform and electrostatic distribution. We have pointed to a possible application of this metal-binding propensity.
What led you to study RNA or this aspect of RNA science?
RNA molecules exhibit spectacular ability to form three-dimensional structures. This structural richness drew my scientific interest into RNA crystallography. This method allows determination of atomic models of molecules which help in understanding the function of RNA from a structural perspective.
During the course of these experiments, were there any surprising results or particular difficulties that altered your thinking and subsequent focus?
I was surprised that the UGG motif binds two Ba2+ ions and that binding is associated with closing the major groove of the RNA helix. The interaction of Ba2+ seemed to be specific, which is not obvious for RNA molecules. Therefore it was challenging to provide evidence explaining this specificity in terms of the structural and chemical properties of RNA.
What are some of the landmark moments that provoked your interest in science or your development as a scientist?
I discovered that I want to be a scientist when I was in high school. I decided to study chemistry and I had the opportunity to conduct my Msc project in the field of RNA biochemistry. This was the moment that began my interest in the RNA world.
What are your subsequent near- or long-term career plans?
I would like to continue my research on RNA structures. I am planning to implement, next to crystallography, cryoEM as an alternative method for determination of three-dimensional structures of RNA molecules.
REFERENCES
- Allain FH-T, Varani G. 1995. Divalent metal ion binding to a conserved wobble pair defining the upstream site of cleavage of group I self-splicing introns. Nucleic Acids Res 23: 341–350. 10.1093/nar/23.3.341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananth P, Goldsmith G, Yathindra N. 2013. An innate twist between Crick's wobble and Watson-Crick base pairs. RNA 19: 1038–1053. 10.1261/rna.036905.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auffinger P, Bielecki L, Westhof E. 2004. Anion binding to nucleic acids. Structure 12: 379–388. 10.1016/j.str.2004.02.015 [DOI] [PubMed] [Google Scholar]
- Bachas ST, Ferré-D'Amaré AR. 2018. Convergent use of heptacoordination for cation selectivity by RNA and protein metalloregulators. Cell Chem Biol 25: 962–973.e5. 10.1016/j.chembiol.2018.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger KD, Kennedy SD, Turner DH. 2019. Nuclear magnetic resonance reveals that GU base pairs flanking internal loops can adopt diverse structures. Biochemistry 58: 1094–1108. 10.1021/acs.biochem.8b01027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas R, Sundaralingam M. 1997. Crystal structure of r(GUGUGUA)dC with tandem G·U/U·G wobble pairs with strand slippage. J Mol Biol 270: 511–519. 10.1006/jmbi.1997.1118 [DOI] [PubMed] [Google Scholar]
- Burgstaller P. 1997. Isoalloxazine derivatives promote photocleavage of natural RNAs at G.U base pairs embedded within helices. Nucleic Acids Res 25: 4018–4027. 10.1093/nar/25.20.4018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colmenarejo G, Tinoco I. 1999. Structure and thermodynamics of metal binding in the P5 helix of a group I intron ribozyme. J Mol Biol 290: 119–135. 10.1006/jmbi.1999.2867 [DOI] [PubMed] [Google Scholar]
- Daldrop P, Lilley DMJ. 2013. The plasticity of a structural motif in RNA: structural polymorphism of a kink turn as a function of its environment. RNA 19: 357–364. 10.1261/rna.036657.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132. 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- Galiana-Arnoux D, Del Gatto-Konczak F, Gesnel M-C, Breathnach R. 2005. Intronic UGG repeats coordinate splicing of CD44 alternative exons v8 and v9. Biochem Biophys Res Commun 336: 667–673. 10.1016/j.bbrc.2005.08.153 [DOI] [PubMed] [Google Scholar]
- Garg A, Heinemann U. 2018. A novel form of RNA double helix based on G·U and C·A+ wobble base pairing. RNA 24: 209–218. 10.1261/rna.064048.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Mooers BHM, Thomas LM, Malone J, Harris S, Schroeder SJ. 2015. Structures and energetics of four adjacent G·U pairs that stabilize an RNA helix. J Phys Chem B 119: 13252–13261. 10.1021/acs.jpcb.5b06970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock RD, Siddons CJ, Oscarson KA, Reibenspies JM. 2004. The structure of the 11-coordinate barium complex of the pendant-donor macrocycle 1,4,7,10-tetrakis(carbamoylmethyl)-1,4,7,10-tetraazacyclododecane: an analysis of the coordination numbers of barium(II) in its complexes. Inorg Chim Acta 357: 723–727. 10.1016/j.ica.2003.06.016 [DOI] [Google Scholar]
- Harding MM, Nowicki MW, Walkinshaw MD. 2010. Metals in protein structures: a review of their principal features. Crystallogr Rev 16: 247–302. 10.1080/0889311X.2010.485616 [DOI] [Google Scholar]
- Heber S, Gáspár I, Tants J-N, Günther J, Moya SMF, Janowski R, Ephrussi A, Sattler M, Niessing D. 2019. Staufen2-mediated RNA recognition and localization requires combinatorial action of multiple domains. Nat Commun 10: 1659. 10.1038/s41467-019-09655-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Serganov A, Patel DJ. 2010. Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch. Mol Cell 40: 774–786. 10.1016/j.molcel.2010.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Wang J, Lilley DMJ. 2016. A critical base pair in k-turns determines the conformational class adopted, and correlates with biological function. Nucleic Acids Res 44: 5390–5398. 10.1093/nar/gkw201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S, Vargyas M, Vasko-Szedlar J, Roux B, Im W. 2008. PBEQ-Solver for online visualization of electrostatic potential of biomolecules. Nucleic Acids Res 36: W270–W275. 10.1093/nar/gkn314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. 2010. XDS. Acta Crystallogr D Biol Crystallogr 66: 125–132. 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kacer V, Scaringe SA, Scarsdale JN, Rife JP. 2003. Crystal structures of r(GGUCACAGCCC)2. Acta Crystallogr D Biol Crystallogr 59: 423–432. 10.1107/S0907444902021911 [DOI] [PubMed] [Google Scholar]
- Keel AY, Rambo RP, Batey RT, Kieft JS. 2007. A general strategy to solve the phase problem in RNA crystallography. Structure 15: 761–772. 10.1016/j.str.2007.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiliszek A, Kierzek R, Krzyzosiak WJ, Rypniewski W. 2011. Crystal structures of CGG RNA repeats with implications for fragile X-associated tremor ataxia syndrome. Nucleic Acids Res 39: 7308–7315. 10.1093/nar/gkr368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knappenberger AJ, Reiss CW, Strobel SA. 2018. Structures of two aptamers with differing ligand specificity reveal ruggedness in the functional landscape of RNA. Elife 7: e36381. 10.7554/eLife.36381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlowski P, de Mezer M, Krzyzosiak WJ. 2010. Trinucleotide repeats in human genome and exome. Nucleic Acids Res 38: 4027–4039. 10.1093/nar/gkq127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug M, Weiss MS, Heinemann U, Mueller U. 2012. XDSAPP: a graphical user interface for the convenient processing of diffraction data using XDS. J Appl Crystallogr 45: 568–572. 10.1107/S0021889812011715 [DOI] [Google Scholar]
- Lang K, Micura R. 2008. The preparation of site-specifically modified riboswitch domains as an example for enzymatic ligation of chemically synthesized RNA fragments. Nat Protoc 3: 1457–1466. 10.1038/nprot.2008.135 [DOI] [PubMed] [Google Scholar]
- Leonarski F, D'Ascenzo L, Auffinger P. 2017. Mg2+ ions: Do they bind to nucleobase nitrogens? Nucleic Acids Res 45: 987–1004. 10.1093/nar/gkw1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonarski F, D'Ascenzo L, Auffinger P. 2019. Nucleobase carbonyl groups are poor Mg2+ inner-sphere binders but excellent monovalent ion binders—a critical PDB survey. RNA 25: 173–192. 10.1261/rna.068437.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebschner D, Afonine P V, Baker ML, Bunkoczi G, Chen VB, Croll TI, Hintze B, Hung LW, Jain S, McCoy AJ, et al. 2019. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr D Struct Biol 75: 861–877. 10.1107/S2059798319011471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malgowska M, Gudanis D, Kierzek R, Wyszko E, Gabelica V, Gdaniec Z. 2014. Distinctive structural motifs of RNA G-quadruplexes composed of AGG, CGG and UGG trinucleotide repeats. Nucleic Acids Res 42: 10196–10207. 10.1093/nar/gku710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masquida B, Sauter C, Westhof E. 1999. A sulfate pocket formed by three GoU pairs in the 0.97 Å resolution X- ray structure of a nonameric RNA. RNA 5: 1384–1395. 10.1017/S1355838299991173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell JA, He L, Chen X, Turner DH. 1997. Investigation of the structural basis for thermodynamic stabilities of tandem GU Wobble pairs: NMR structures of (rGGA u>GU/u>UCC)2 and (rGGA u>UG/u>UCC)2. Biochemistry 36: 8030–8038. 10.1021/bi970122c [DOI] [PubMed] [Google Scholar]
- Mooers BHM, Eichman BF, Shing Ho P. 1997. The structures and relative stabilities of d(G.G) reverse Hoogsteen, d(G.T) reverse wobble, and d(G.C) reverse Watson-Crick base-pairs in DNA crystals. J Mol Biol 269: 796–810. 10.1006/jmbi.1997.1100 [DOI] [PubMed] [Google Scholar]
- Mueller U, Schübel H, Sprinzl M, Heinemann U. 1999. Crystal structure of acceptor stem of tRNAAla from Escherichia coli shows unique G·U wobble base pair at 1.16 Å resolution. RNA 5: 670–677. 10.1017/S1355838299982304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller U, Förster R, Hellmig M, Huschmann FU, Kastner A, Malecki P, Pühringer S, Röwer M, Sparta K, Steffien M, et al. 2015. The macromolecular crystallography beamlines at BESSY II of the Helmholtz-Zentrum Berlin: current status and perspectives. Eur Phys J Plus 130: 141. 10.1140/epjp/i2015-15141-2 [DOI] [Google Scholar]
- Panjikar S, Parthasarathy V, Lamzin VS, Weiss MS, Tucker PA. 2009. On the combination of molecular replacement and single-wavelength anomalous diffraction phasing for automated structure determination. Acta Crystallogr D Biol Crystallogr 65: 1089–1097. 10.1107/S0907444909029643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price IR, Gaballa A, Ding F, Helmann JD, Ke A. 2015. Mn2+-sensing mechanisms of yybP-ykoY orphan riboswitches. Mol Cell 57: 1110–1123. 10.1016/j.molcel.2015.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read RJ, Adams PD, McCoy AJ. 2013. Intensity statistics in the presence of translational noncrystallographic symmetry. Acta Crystallogr D Biol Crystallogr 69: 176–183. 10.1107/S0907444912045374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozov A, Khusainov I, El Omari K, Duman R, Mykhaylyk V, Yusupov M, Westhof E, Wagner A, Yusupova G. 2019. Importance of potassium ions for ribosome structure and function revealed by long-wavelength X-ray diffraction. Nat Commun 10: 2519. 10.1038/s41467-019-10409-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruszkowska A, Zheng YY, Mao S, Ruszkowski M, Sheng J. 2022. Structural insights into the 5′UG/3′GU Wobble tandem in complex with Ba2+ cation. Front Mol Biosci 8: 762786. 10.3389/fmolb.2021.762786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rypniewski W, Banaszak K, Kuliński T, Kiliszek A. 2016. Watson-Crick-like pairs in CCUG repeats: evidence for tautomeric shifts or protonation. RNA 22: 22–31. 10.1261/rna.052399.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sashital DG, Venditti V, Angers CG, Cornilescu G, Butcher SE. 2007. Structure and thermodynamics of a conserved U2 snRNA domain from yeast and human. RNA 13: 328–338. 10.1261/rna.418407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrödinger L, DeLano W. 2020. PyMOL. https://www.pymol.org/pymol.html? (Accessed July 19, 2022).
- Schroeder KT, Daldrop P, McPhee SA, Lilley DMJ. 2012. Structure and folding of a rare, natural kink turn in RNA with an A•A pair at the 2b•2n position. RNA 18: 1257–1266. 10.1261/rna.032409.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Huang L, Patel DJ. 2009. Coenzyme recognition and gene regulation by a flavin mononucleotide riboswitch. Nature 458: 233–237. 10.1038/nature07642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobczak K, Michlewski G, De Mezer M, Kierzek E, Krol J, Olejniczak M, Kierzek R, Krzyzosiak WJ. 2010. Structural diversity of triplet repeat RNAs. J Biol Chem 285: 12755–12764. 10.1074/jbc.M109.078790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel SA, Cech TR. 1995. Minor groove recognition of the conserved G·U pair at the Tetrahymena ribozyme reaction site. Science 267: 675–679. 10.1126/science.7839142 [DOI] [PubMed] [Google Scholar]
- Strobel SA, Cech TR. 1996. Exocyclic amine of the conserved G·U pair at the cleavage site of the Tetrahymena ribozyme contributes to 5′-splice site selection and transition state stabilization. Biochemistry 35: 1201–1211. 10.1021/bi952244f [DOI] [PubMed] [Google Scholar]
- Trikha J, Filman DJ, Hogle JM. 1999. Crystal structure of a 14 bp RNA duplex with non-symmetrical tandem G·U wobble base pairs. Nucleic Acids Res 27: 1728–1739. 10.1093/nar/27.7.1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin AA, Steiner RA, Lebedev AA, Potterton L, McNicholas S, Long F, Murshudov GN. 2004. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr D Biol Crystallogr 60: 2184–2195. 10.1107/S0907444904023510 [DOI] [PubMed] [Google Scholar]
- Varani G, Mcclain WH. 2000. The G-U wobble base pair diverse biological systems. EMBO Rep 1: 18–23. 10.1093/embo-reports/kvd001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhof E, Yusupov M, Yusupova G. 2019. The multiple flavors of GoU pairs in RNA. J Mol Recognit 32: e2782. 10.1002/jmr.2782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, et al. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67: 235–242. 10.1107/S0907444910045749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Landon T, Greenbaum NL, Fenley MO. 2007. The electrostatic characteristics of G·U wobble base pairs. Nucleic Acids Res 35: 3836–3847. 10.1093/nar/gkm274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G, Lu X, Olson WK. 2009. Web 3DNA–a web server for the analysis, reconstruction, and visualization of three-dimensional nucleic-acid structures. Nucleic Acids Res 37: W240–W246. 10.1093/nar/gkp358 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.