Meta-analyses across cancers show that common germline risk variants affect not only cancer predisposition but the age of cancer onset and burden of somatic alterations, including total mutations and copy-number alterations.
Abstract
Aggregation of genome-wide common risk variants, such as polygenic risk score (PRS), can measure genetic susceptibility to cancer. A better understanding of how common germline variants associate with somatic alterations and clinical features could facilitate personalized cancer prevention and early detection. We constructed PRSs from 14 genome-wide association studies (median n = 64,905) for 12 cancer types by multiple methods and calibrated them using the UK Biobank resources (n = 335,048). Meta-analyses across cancer types in The Cancer Genome Atlas (n = 7,965) revealed that higher PRS values were associated with earlier cancer onset and lower burden of somatic alterations, including total mutations, chromosome/arm somatic copy-number alterations (SCNA), and focal SCNAs. This contrasts with rare germline pathogenic variants (e.g., BRCA1/2 variants), showing heterogeneous associations with somatic alterations. Our results suggest that common germline cancer risk variants allow early tumor development before the accumulation of many somatic alterations characteristic of later stages of carcinogenesis.
Significance:
Meta-analyses across cancers show that common germline risk variants affect not only cancer predisposition but the age of cancer onset and burden of somatic alterations, including total mutations and copy-number alterations.
Introduction
Cancer is a genomic disease driven by the accumulation of somatic alterations, but germline variants also contribute to the process of carcinogenesis (1). Through comprehensive genomic analyses, such as The Cancer Genome Atlas (TCGA; ref. 2) and Pan-Cancer Analysis of Whole Genomes (PCAWG; ref. 3), numerous somatic driver alterations [including mutations and somatic copy-number alterations (SCNA)] have been identified, and their effects on carcinogenesis have been extensively evaluated in a pan-cancer manner. Similar efforts have been extended to rare germline pathogenic variants, such as BRCA1/2 variants, which are observed in approximately 8% of all patients with cancer and present in high-penetrance cancer-associated genes (1). These rare germline pathogenic variants contribute to carcinogenesis through coordination with somatic alterations, as exemplified by frequent colocalization of rare germline pathogenic variants with somatic mutations or loss of heterozygosity (LOH) of the same predisposition gene (two-hit theory; ref. 1). However, such rare pathogenic variants account for only a small fraction of the familial risk, leaving much heritability unexplained.
Recent genome-wide association studies (GWAS) have identified hundreds of common germline risk variants, which show low-penetrance and have relatively smaller effect sizes. Such common risk variants are estimated to exist in a genome-wide manner as more than thousands in total for a wide range of cancer types (i.e., polygenicity; ref. 4). Although individual risk variants have small effects, aggregation of genome-wide risk variants can explain genetic liability to cancers, affecting a great number of people; common germline risk accounts for a higher proportion of cancer incidence than lifestyle-related risk factors for most cancer types at the population level (5). Polygenic risk score (PRS), a score reflecting the combined effect of genome-wide common risk variants identified by GWAS, is a widely used approach to measure polygenic germline risk. PRS has a clinical potential to promote personalized prevention and early detection of cancer by identifying individuals at substantially elevated risks (5). PRS has been shown to effectively predict the risk of cancer development for most cancer types in the general population (6), and even in carriers of rare germline pathogenic variants for breast and ovarian cancers (7). However, there are only sporadic reports on PRS associations with certain driver mutations and cancer subtypes (8). A comprehensive evaluation of polygenic germline–somatic associations is thus needed.
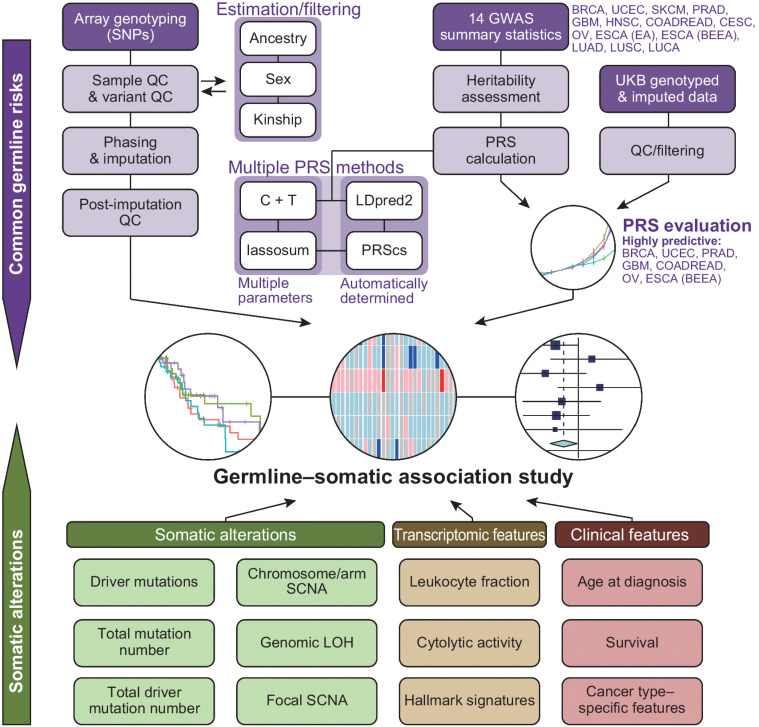
Here, we constructed cancer PRSs with high-prediction capability by applying multiple PRS construction methods in parallel and calibrating them using the UK Biobank (UKB) resources. We examined the PRS associations with a wide range of somatic alterations and clinical features in TCGA by jointly modeling PRSs and rare germline pathogenic variants, to provide a comprehensive portrait of germline–somatic associations (Fig. 1).
Figure 1.
Overview of this study. A graphical overview of germline–somatic association analyses. CESC, cervical cancer; COADREAD, colorectal cancer; ESCA (BEEA), esophageal adenocarcinoma and Barrett's esophagus; ESCA (EA), esophageal adenocarcinoma; GBM, glioblastoma multiforme; HNSC, head and neck squamous cell carcinoma; LUAD, lung adenocarcinoma; LUCA, lung cancer; LUSC, lung squamous cell carcinoma; OV, ovarian serous carcinoma; PRAD, prostate cancer; QC, quality control; SKCM, skin cutaneous melanoma; SNP, single-nucleotide polymorphism; UCEC, uterine endometrial carcinoma.
Materials and Methods
PRS
We used four methods to construct PRS, Clumping and Thresholding (C+T), PRScs (released on April 24, 2020, https://github.com/getian107/PRScs; ref. 9), lassosum (v0.4.5, https://github.com/tshmak/lassosum; ref. 10), and LDpred2 (specifically, LDpred2-auto in bigsnpr R package v1.4.4, https://privefl.github.io/bigsnpr/; ref. 11), with the 1000 Genomes Project (phase 3) European samples as the reference panel (Fig. 1). PRScs and LDpred2 are Bayesian approaches using HapMap3 variants (9, 11), whereas lassosum-selected variants from genome-wide variants using penalized regression (10). We used PRScs with automated optimization of the parameter phi (PRScs-auto). We applied these methods to public GWASs and GWASs obtained from the data provider as summarized in Supplementary Table S1. For breast cancer (BRCA), multiple studies with the same case–control definition were available, and we chose the study with the largest sample size. We note that “ESCA (BEEA)” is a GWAS for Barrett's esophagus and esophageal adenocarcinoma, which considered both diseases as a single entity because of a very high genetic correlation between these two diseases. On the other hand, “ESCA (EA)” is a GWAS only for esophageal adenocarcinoma. To guarantee the consistency of PRS between UKB and TCGA, we restricted the variants in the GWAS summary statistics to the variants included in all of UKB, TCGA, and the reference panel. For the GWASs in which the summary statistics were available, we matched the palindrome variants between the summary statistics and the reference panel if the difference of their allele frequencies was less than 0.1. We also evaluated the strand of these palindrome variants and removed them if there was strand inconsistency. All palindrome variants were excluded for the GWASs in which the allele frequencies were not reported in the summary statistics. We used plink1.9 (v1.90b6.16, https://www.cog-genomics.org/plink/1.9/; ref. 12) for clumping with the command “plink–clump–clump-r2 0.1–clump-kb 250,” and examined eight thresholds for GWAS P values, 1×10−1, 1×10−2, …, 1×10−7, 5×10−8. The PRSs were evaluated in UKB data with age, sex, and top 20 genetic principal components as covariates. We defined the control group as the individuals who were not diagnosed with cancer and were not reported as patients with cancer by themselves. For the case group, we included both individuals with relevant International Classification of Diseases (ICD)-10 codes and individuals who self-reported to be diagnosed with relevant cancer types. If the cancer types were defined histologically, we did not include self-reported individuals in the case group. The ICD-10 codes, histology, and cancer types are described in Supplementary Table S2. We picked up the hyperparameters that gained the largest area under the curve (AUC) per PRS method after excluding scores that negatively correlated with disease status, and compared the R2, AUC, and odds ratios of deciles across the methods. We used the Delong's test to evaluate the difference of AUC between the full model and the reduced model including all covariates but PRS, and the P values were subsequently adjusted by the Benjamini–Hochberg method. We note that the skin cutaneous melanoma (SKCM) and uterine endometrial carcinoma (UCEC) studies included UKB participants in their discovery cohorts, which may inflate the PRS performance of those studies.
Evaluation of germline–somatic associations
PRSs were evaluated in cancer types in which the adjusted P value was below 0.05 and R2 was above 0.01 in the UKB cohort. We restricted our subsequent analysis to patients in which primary solid tumor or primary blood-derived cancer samples were available (TCGA sample code 01 or 03). SKCM was excluded because the majority of the samples were from metastatic sites. Because our PRSs were generated from cohorts that mostly included European individuals, we evaluated the association of PRS values with somatic alterations and clinical features only in European samples (see also ancestry analysis section of Supplementary Materials and Methods). Samples with atypical sex chromosomes (see sex inference section of Supplementary Materials and Methods) as well as samples with the do_not_use flag in the TCGA PanCan Atlas Project (merged_sample_quality_annotations.tsv), such as those with a history of unacceptable prior treatment, were also excluded.
In each cancer type, generalized linear model was fitted using genetic or clinical feature as dependent variables [after rank-based inverse normal transformation (INT) to quantitative variables] and PRS values of the corresponding cancer type (after INT), subtypes, and the presence of germline pathogenic variants (a binary variable) as independent variables. Samples with missing values were excluded from the analysis. Although we cannot exclude the possibility that the germline pathogenic variants were in linkage disequilibrium (LD) with common germline variants used for PRS calculation, such LD should be weak, if at all. Furthermore, we calculated PRSs from a large number of variants (Supplementary Table S3), and therefore the confounding effects of LD for individual variants should be minimized. We assumed the normal distribution for quantitative phenotypes and the binomial distribution for the mutations of individual driver genes. Then, the effect sizes and standard errors were combined through fixed-effect meta-analysis to derive a final P value. Meta-analysis was conducted using the R package “meta.”
Data availability
Our findings are supported by data that are available from public online repositories, or data that are publicly available upon request from the data provider. Specifically, GWAS summary statistics were downloaded from the Breast Cancer Association Consortium (https://bcac.ccge.medschl.cam.ac.uk/bcacdata/; BRCA), the Prostate Cancer Association Group to Investigate Cancer Associated Alterations in the Genome consortium (http://practical.icr.ac.uk/blog/; prostate cancer, PRAD), the database of Genotypes and Phenotypes (dbGaP) accession phs001868.v1.p1 (SKCM), GWAS catalog (https://www.ebi.ac.uk/gwas/home; UCEC and ovarian serous carcinoma, OV), and the Harvard Dataverse (https://doi.org/10.7910/DVN/2VBLLP; cervical cancer, CESC). The GWAS summary statistics of colorectal cancer (COADREAD), lung cancer (LUCA), lung adenocarcinoma (LUAD), and lung squamous cell carcinoma (LUSC) were obtained from Supplementary Tables of the original articles. The GWAS summary statistics of ESCA (BEEA), ESCA (EA), glioblastoma multiforme (GBM), and head and neck squamous cell carcinoma (HNSC) were provided by the authors. The UKB resource (https://www.ukbiobank.ac.uk/) was accessed through application number 47821. For TCGA, birdseed files were downloaded from GDC Portal (dbGaP accession phs000178.v10.p8). Somatic and germline mutational data in Mutation Annotation Format, RNA-seq expression data, In Silico Admixture Removal (ISAR)–corrected copy-number segment data, and clinical information were downloaded from GDC (https://gdc.cancer.gov/about-data/publications/pancanatlas and https://gdc.cancer.gov/about-data/publications/PanCanAtlas-Germline-AWG).
Further details of the methods are provided in the Supplementary Materials and Methods.
Results
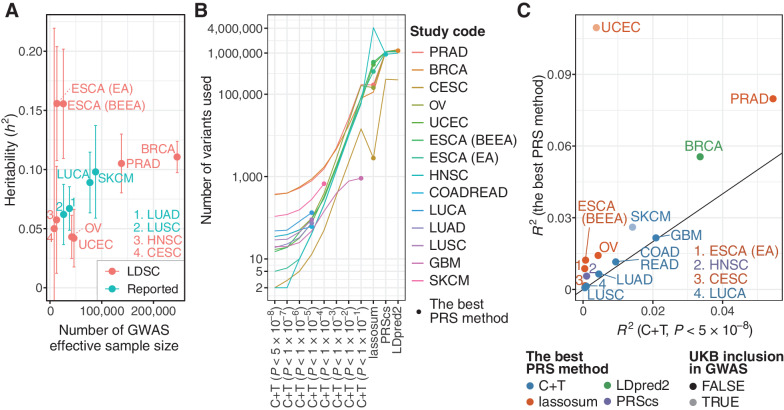
We first widely collected GWAS summary statistics and obtained them from 14 studies (n = 9,347–247,173; median 64,905) of 12 cancer types (Supplementary Table S1). GWAS effective sample sizes, calculated as a doubled harmonic mean of the number of cases and controls, were between 7,949 and 245,620 (median 39,658; Fig. 2A). All GWASs used in this study were predominantly from populations with European ancestry. Single-nucleotide polymorphism (SNP)-based heritability estimates were different across cancer types, ranging from h2 = 0.042 [95% confidence interval (CI), 0.018–0.066] for UCEC to h2 = 0.16 (95% CI, 0.11–0.20) for ESCA (EA). We then applied multiple PRS construction methods to available variants in these GWASs to obtain the best predictive PRSs. C+T, a simple PRS construction method, was conducted for all studies, with different P value thresholds (minimum 5×10−8). In addition, we adopted three LD-aware methods [PRScs (9), lassosum (10), and LDpred2 (11)] for 8 (57%) studies (from 7 cancer types) with complete GWAS summary statistics. Compared with C+T, these methods used a larger number of variants, which may contain potentially informative variants (Fig. 2B; Supplementary Table S3; refs. 9–11). Although LDpred2 and PRScs produced one PRS, C+T and lassosum produced 8 and 80 PRSs according to their hyperparameters, respectively, thus generating a total of 90 PRSs per individual for each study.
Figure 2.
Construction and evaluation of PRSs. A, A scatter plot showing heritability (h2) and effective sample size of the GWAS summary statistics. Heritability was estimated by LD score regression (LDSC) for GWASs, in which genome-wide variants were available. For others, heritability estimated by LDSC was obtained from the original articles if reported (Reported). GBM and COADREAD were excluded because heritability estimated by LDSC was not reported for these cancer types. Error bars, 95% confidence interval. B, The number of variants used for calculating PRSs. For six GWAS summary statistics in which genome-wide variants were unavailable, only C+T was conducted for PRS calculation. C, The difference of Nagelkerke's R2 for the C+T PRS with genome-wide significant variants (i.e., P < 5×10−8; x-axis) and the best PRS (y-axis). It was calculated in UKB as a difference between the full model and the reduced model including all covariates but PRS. UCEC and SKCM are shown in light colors because UKB was included in the GWAS cohort for these two cancer types.
We evaluated the predictive performance of PRSs, including Nagelkerke's R2and AUC for the case–control status in individuals of European ancestry in the UKB cohort (n = 335,048) and selected the PRS with the highest performance for each study (see Materials and Methods, Figs. 1 and 2C; Supplementary Fig. S1–S3, and Supplementary Tables S2–S4). For 8 studies with complete GWAS summary statistics, the LD-aware methods consistently produced PRSs with higher predictive performance than C+T. For the remaining 6 studies, of which, GWAS summary statistics were available for a limited set of variants, the best PRS was achieved by C+T with the most lenient P value threshold (Supplementary Table S3). Although even the best PRS showed limited improvement in these studies (Fig. 2C), the best PRS was able to stratify individuals more successfully than the C+T PRS with genome-wide significant variants (i.e., P < 5×10−8 as used in previous studies; refs. 5, 8) for all studies (Supplementary Fig. S3), suggesting that the substantial heritability attributed to sub-GWAS significant variants. The best PRS tended to show higher predictive performance for the studies that had a larger effective sample size and a higher heritability, including BRCA, PRAD, and SKCM (Fig. 2A and C). Taken together, we constructed highly predictive PRS for each study by applying the LD-aware methods to genome-wide variants.
We next applied the best PRSs selected in each study to the TCGA cohort. Because PRSs were calculated using European GWASs, we performed ancestry admixture analysis (Supplementary Fig. S4 and Supplementary Table S5) and restricted our subsequent analyses to the samples (n = 7,965) of European ancestry without sex aneuploidy to prevent confounding by low PRS transferability between populations (13). After excluding SKCM, mainly consisting of metastatic samples, we focused on the PRSs with high predictive performance (i.e., R2 > 0.01 and adjusted P < 0.05; see Materials and Methods) calculated from 7 studies (for 7 cancer types), for 5 of which complete GWAS summary statistics were available (Fig. 1; Supplementary Table S4). The predictability of the PRSs was confirmed by the highest PRS values in the corresponding cancer type (Supplementary Fig. S5A and S5B). We evaluated the pair-wise correlation of PRS values among the 7 cancer types in the TCGA cohort (n = 2,924), which revealed several positive and negative correlations (Supplementary Fig. S5C). The most prominent was the positive correlation between UCEC and BRCA, which was validated in the UKB cohort (n = 269,544). This relationship was further supported by an additional genetic correlation analysis using LD score regression, showing a significant positive correlation (rg = 0.20; 95% CI, 0.08–0.32) between UCEC and BRCA. These results suggest common germline susceptibility between these biologically related cancer types. Pathway enrichment analysis demonstrated the overrepresentation of cell-cycle pathway genes in both cancer types (FDR = 2.4×10−2 and 6.9×10−6 for UCEC and BRCA, respectively), among which, CCND1, CDKN2A, and CDKN2B were associated with both cancer types (P < 0.05, Supplementary Table S6). The subset of the PRS values only using the variants within the three genes were more highly correlated to each other [r = 0.33 (TCGA) and 0.32 (UKB)] than the entire PRS values [r = 0.11 (TCGA) and 0.07 (UKB)], suggesting the contribution of cell-cycle susceptibility to both cancer types.
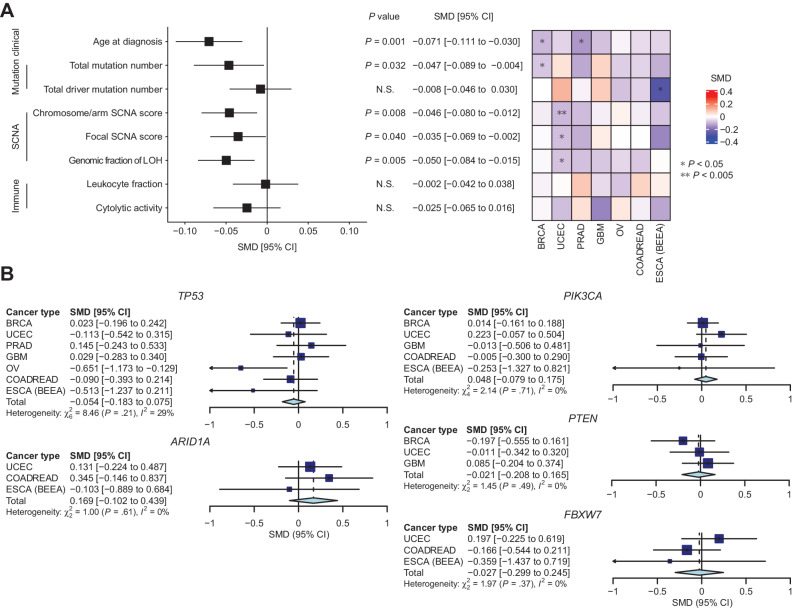
We next evaluated associations of PRS values for the corresponding cancer types with somatic alterations and clinical features in the 7 cancer types of the TCGA cohort (n = 82–759; median 399) using generalized linear regression (Fig. 3A; Supplementary Figs. S5 and S6). As PRS has been reported as an independent risk of cancer development from rare germline variants in breast and ovarian cancers (7), and their presence did not affect the distribution of PRS values in our analysis (Supplementary Fig. S5D), we incorporated the presence of rare germline pathogenic variants (1) as covariates (Supplementary Fig. S5E). Because PRS values were slightly different between subtypes in certain cancer types, we also adjusted for subtype (Supplementary Fig. S5F). First, higher PRS values were associated with younger age at diagnosis in BRCA and PRAD (P = 0.014 and 0.041, respectively; Fig. 3A; Supplementary Fig. S6A). Similar trends were observed across all evaluated cancer types, and this association was significant in the meta-analysis (P = 0.001), suggesting that individuals with higher common germline risk develop malignancy at a younger age, generalizing the previous finding in certain cancer types (14). Second, after the removal of hypermutator samples, higher PRS values were associated with a smaller number of total mutations per tumor (P = 0.032; Fig. 3A; Supplementary Fig. S6C). Given tumor mutation burden increases with age, this observation reflects early tumor onset in individuals with increased common germline risk. Intriguingly, PRS values also showed a negative association with chromosome/arm SCNA scores, which reflect the numbers and extents of SCNAs, without apparent heterogeneity across cancer types. This result suggests a limited role of aneuploidy in the carcinogenic process for individuals with increased common germline risk (P = 0.008; Fig. 3A; Supplementary Fig. S6G). Consistently, the genomic fraction of LOH, loss of one parental allele causing allelic imbalance, showed a negative association with PRS values (P = 0.005; Fig. 3A; Supplementary Fig. S6K). The scores of focal SCNAs, another class of SCNAs caused by different biological mechanisms from chromosome/arm SCNAs, were also negatively associated with PRS values in the meta-analysis (P = 0.040; Fig. 3A; Supplementary Fig. S6I). These negative associations were consistent even when amplification and deletion were considered separately (P = 0.147 and 0.030 for chromosome/arm SCNAs and 0.350 and 0.090 for focal SCNAs, respectively; Supplementary Fig. S7). As increased genomic instability is characteristic of later stages of carcinogenesis (15), these results suggest that common germline risk enables early tumor development before many mutations and SCNAs accumulate.
Figure 3.
Germline–somatic association analyses in TCGA. A, The associations between PRS values and somatic/clinical features of cancer. Left, pooled effect size (standardized mean difference, SMD) of PRS values for evaluated features. Right, heatmap indicating the SMD of PRS values for each feature in each cancer type. B, Forest plots of cancer type–specific and pooled SMD of PRS values for driver mutations. Genes listed as drivers in three or more cancer types were evaluated.
In contrast, the associations with PRS values were not significant for the number of driver mutations across cancer types (P = 0.677; Fig. 3A; Supplementary Fig. S6E). We also examined the associations of PRS values with mutations of individual driver genes, but all associations were not significant except for TP53 mutations in OV (Supplementary Fig. S8). The associations remained insignificant after meta-analyzing the genes that were considered as drivers in three or more cancer types (Fig. 3B). These observations suggest that common germline risk does not necessarily affect the necessity of driver mutations during cancer development.
For other genetic and clinical features, we found no significant association with PRS values as well, including tumor immunity (leukocyte fraction and cytolytic activity; Fig. 3A; Supplementary Fig. S6M and S6O) and transcriptomic signatures (50 hallmark signatures; Supplementary Fig. S9). Patient survival (overall survival and progression-free survival) was evaluated by univariate Cox proportional hazards model, which revealed no difference between PRS value-high and -low samples (Supplementary Fig. S10A and S10B). Clinical features specific to cancer type (histological grade, including Gleason score of PRAD, and sidedness of COADREAD; Supplementary Fig. S10C–S10E) were not significantly associated with PRS values. Taken together, our results suggest that the common germline risk have no or minimal effect on the transcriptomic and clinical phenotype in cancer.
Next, we assessed the associations of rare germline pathogenic variants with somatic alterations and clinical features. Younger age at diagnosis was associated with the presence of rare germline pathogenic variants (P = 1.89×10−6; Supplementary Fig. S6B), especially in OV, BRCA, and COADREAD, confirming the early onset of familial cancers. We also detected associations of rare germline pathogenic variants with total mutation number and genomic fraction of LOH in BRCA and OV (P = 2.96×10−4 and 2.43×10−4 for total mutation number and 0.017 and 0.033 for genomic fraction of LOH, respectively; Supplementary Fig. S6D and S6L), likely reflecting the mutagenic effect of germline BRCA1 and BRCA2 variants in these cancer types (16). However, the associations with genetic features were heterogeneous across cancer types and not significant in the meta-analysis. These findings suggest the functional heterogeneity of rare germline pathogenic variants across responsible genes and cancer types, demonstrating their differences from common germline variants.
Finally, to independently validate the germline–somatic associations, we analyzed additional three PRAD cohorts in PCAWG (n = 32, 40, and 116) because the associations between PRS values and somatic alterations were most prominent in PRAD in our analysis. These cohorts showed similar results on associations between PRS values and clinical and genetic features to that from TCGA (Supplementary Fig. S11). Meta-analysis of the four PRAD cohorts showed significant associations of PRS values with earlier age at diagnosis and a lower number of total mutations and SCNAs. In contrast, the associations of rare germline pathogenic variants were not significant for any of those features.
Discussion
Through cross-cancer meta-analyses of TCGA resources, we found that elevated PRS values were associated with earlier tumor onset and a lower number of somatic mutations and SCNAs, which was confirmed in independent PRAD cohorts. Although the association between PRS and early tumor onset has been reported for breast and prostate cancers (14), our analysis is the first to generalize such observations to a wide range of cancer types. Recent cancer evolution studies revealed the temporal ordering of somatic alterations, which showed genomic instability as later-stage events of carcinogenesis (15). Our findings suggest that individuals with higher common germline risk require relatively lower somatic alterations characteristic of later stages of carcinogenesis for tumor development. This is in contrast with germline pathogenic variants, which showed heterogeneous associations with somatic features across cancer types and responsible genes.
The mechanisms of high incidence and early onset of cancer in individuals with germline pathogenic variants may include (i) promoting the selection of certain somatic driver alterations, especially in the same gene or pathway, (ii) promoting the accumulation of somatic alterations, as in the cases of germline pathogenic mismatch repair gene variants (such as MLH1 and MSH2), and (iii) enhancing the oncogenic process; for example, by modulating the expression or function of certain genes (17). Our results suggest that common germline variants may not promote the accumulation of somatic alterations and/or certain driver alterations, but enhance the oncogenic processes in conjunction with somatic driver mutations. Given PRS values are not associated with the number of driver mutations, the effects of somatic driver mutations on carcinogenesis may be different according to common germline risk. Indeed, differential effects of driver mutations among individuals have already been reported in clonal hematopoiesis of indeterminate potential, which has somatic driver mutations similar to hematologic malignancies but not meeting its criteria (18). The heterogeneous consequences of driver mutations may be affected by common germline risk, which is an interesting direction of future research.
Although several germline–somatic associations have been reported for common germline variants (8, 19), such associations were limited to individual cancer types and specific somatic alterations and clinical features. Furthermore, although some common germline variants have been reported to affect somatic features in locus-specific manners, such as rs8051518, an RBFOX1 intronic variant that enhances the effect of SF3B1 mutation on RNA splicing (20), their global effects are yet to be known. To the best of our knowledge, this is the first study showing the universal effect of PRS on somatic alterations maintained across cancers. As the PRS was constructed independently and included different common germline variants between cancer types, our analysis demonstrates the general effect of common germline variants rather than the effect of a specific variant.
There may be some possible limitations in this study. First, although we constructed highly predictive PRSs with the LD-aware methods, variance explained by PRSs was still smaller than heritability predicted by common germline variants. In the future, generation of large GWASs and methodological advances for PRS will improve the predictive performance of PRS. Second, our analysis was restricted to European samples because both GWASs (Supplementary Table S1) and large-scale cancer genetic studies (2) have been conducted mainly for Europeans. Thus, replication studies will be required to generalize our results to other populations. Finally, our power to detect the associations between germline risks and various genetic and clinical features is limited. However, we have provided the best available evidence using some of the largest datasets and multiple PRS construction methods, although further validation of our findings using much larger-scale datasets should be warranted.
In conclusion, our comprehensive assessment of germline–somatic associations reveals the overall effects of common germline variants and their difference from rare pathogenic variants. Particularly, increased common germline risk enables accelerated tumor development without many somatic alterations frequently observed in later stages of oncogenesis. Such understanding helps to improve the predictive capacity for early-onset cancers, contributing to the refinement of their management strategy.
Supplementary Material
Figures S1–S11, Tables S1–S6, Supplementary Materials and Methods, Supplementary Notes, and Supplementary References
Acknowledgments
The acknowledgments and the details of the participating consortia are included in the Supplementary Notes. S. Namba was supported by Takeda Science Foundation. Y. Saito was supported by JSPS KAKENHI (22K20808). S. MacGregor, D.C. Whiteman, and P. Gharahkhani were supported by Australian National Health and Medical Research Council Fellowships/Investigator grants. Y. Okada was supported by JSPS KAKENHI (22H00476), AMED (JP21gm4010006, JP22km0405211, JP22ek0410075, JP22km0405217, and JP22ek0109594), JST Moonshot R&D (JPMJMS2021 and JPMJMS2024), Takeda Science Foundation, and Bioinformatics Initiative of Osaka University Graduate School of Medicine, Osaka University. K. Kataoka was supported by AMED (JP21cm0106575 and JP22ama221510), JST Moonshot R&D (JPMJMS2022), the Uehara Memorial Foundation, and Keio University Academic Development Funds.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Disclosures
S. Namba reports other support from Takeda Science Foundation during the conduct of the study. Y. Saito reports grants from JSPS KAKENHI during the conduct of the study. Y. Kogure reports personal fees from Takeda Pharmaceutical Co., Ltd., Daiichi Sankyo Co., Ltd., Nippon Shinyaku Co., Ltd., and Kyowa Kirin Co., Ltd. outside the submitted work. P. Gharahkhani reports grants from NHMRC (Investigator Grant) during the conduct of the study. A. Hillmer reports grants from Dracen Pharmaceuticals outside the submitted work. S. MacGregor reports grants from Australian National Health and Medical Research Council during the conduct of the study. D.C. Whiteman reports grants from National Health and Medical Research Council of Australia during the conduct of the study and personal fees from Pierre Fabre outside the submitted work. Y. Okada reports grants from AMED, JSPS, JST Moonshot R&D, and Takeda Science Foundation, and nonfinancial support from Bioinformatics Initiative of Osaka University Graduate School of Medicine, Osaka University during the conduct of the study, as well as grants from JST CREST, Ono Pharmaceutical, Boehringer Ingelheim, Teijin Pharma, Otsuka Pharmaceutical, and The Nippon Foundation, grants and personal fees from Chugai Pharmaceutical, and personal fees from Daiichi Sankyo, Kyowa Kirin, MEDICAL and BIOLOGICAL LABORATORIES, Taisho Pharmaceutical, Astellas Foundation for Research on Metabolic Disorders, Novo Nordisk, and Astrazeneca outside the submitted work. K. Kataoka reports grants from AMED, JST Moonshot R&D, Uehara Memorial Foundation, and Keio University Academic Development Funds during the conduct of the study; as well as grants, personal fees, and nonfinancial support from Otsuka Pharmaceutical, grants from Chordia Therapeutics, Asahi Kasei Pharma, Shionogi, Teijin Pharma, Japan Blood Products Organization, Mochida Pharmaceutical, JCR Pharmaceuticals, and Nippon Shinyaku, grants and personal fees from Chugai Pharmaceutical, Takeda Pharmaceutical, Eisai, Ono Pharmaceutical, Kyowa Kirin, and Sumitomo Dainippon Pharma, and personal fees from Celgene, Astellas Pharma, Novartis, AstraZeneca, Janssen Pharmaceutical, SymBio Pharmaceuticals, Bristol Myers Squibb, Pfizer, Nippon Shinyaku, Daiichi Sankyo, Alexion Pharmaceuticals, AbbVie, Meiji Seika Pharma, and Sanofi outside the submitted work; as well as reports a patent for Genetic alterations as a biomarker in T-cell lymphomas licensed to Kyoto University and PD-L1 abnormalities as a predictive biomarker for immune checkpoint blockade therapy licensed to Kyoto University. No disclosures were reported by the other authors.
Authors' Contributions
S. Namba: Conceptualization, data curation, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. Y. Saito: Conceptualization, data curation, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. Y. Kogure: Data curation, formal analysis, investigation, writing–review and editing. T. Masuda: Data curation, writing–review and editing. M.L. Bondy: Resources, writing–review and editing. P. Gharahkhani: Resources, writing–review and editing. I. Gockel: Resources, writing–review and editing. D. Heider: Resources, writing–review and editing. A. Hillmer: Resources, writing–review and editing. J. Jankowski: Resources, writing–review and editing. S. MacGregor: Resources, writing–review and editing. C. Maj: Resources, writing–review and editing. B. Melin: Resources, writing–review and editing. Q.T. Ostrom: Resources, writing–review and editing. C. Palles: Resources, writing–review and editing. J. Schumacher: Resources, writing–review and editing. I. Tomlinson: Resources, writing–review and editing. D.C. Whiteman: Resources, writing–review and editing. Y. Okada: Conceptualization, supervision, funding acquisition, methodology, writing–original draft, project administration, writing–review and editing. K. Kataoka: Conceptualization, supervision, funding acquisition, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Huang K-L, Mashl RJ, Wu Y, Ritter DI, Wang J, Oh C, et al. Pathogenic germline variants in 10,389 adult cancers. Cell 2018;173:355–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bailey MH, Tokheim C, Porta-Pardo E, Sengupta S, Bertrand D, Weerasinghe A, et al. Comprehensive characterization of cancer driver genes and mutations. Cell 2018;173:371–85.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Campbell PJ, Getz G, Korbel JO, Stuart JM, Jennings JL, Stein LD, et al. Pan-cancer analysis of whole genomes. Nature 2020;578:82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhang YD, Hurson AN, Zhang H, Choudhury PP, Easton DF, Milne RL, et al. Assessment of polygenic architecture and risk prediction based on common variants across fourteen cancers. Nat Commun 2020;11:3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kachuri L, Graff RE, Smith-Byrne K, Meyers TJ, Rashkin SR, Ziv E, et al. Pan-cancer analysis demonstrates that integrating polygenic risk scores with modifiable risk factors improves risk prediction. Nat Commun 2020;11:6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fritsche L, Patil S, Beesley L, VandeHaar P, Salvatore M, Peng R, et al. Cancer PRSweb—an online repository with polygenic risk scores (PRS) for major cancer traits and their Phenome-wide exploration in two independent Biobanks. Am J Hum Genet 2020;1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kuchenbaecker KB, McGuffog L, Barrowdale D, Lee A, Soucy P, Dennis J, et al. Evaluation of polygenic risk scores for breast and ovarian cancer risk prediction in BRCA1 and BRCA2 mutation carriers. J Natl Cancer Inst 2017;109:djw302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Porta-Pardo E, Sayaman R, Ziv E, Valencia A. The landscape of interactions between cancer polygenic risk scores and somatic alterations in cancer cells. bioRxiv 2020. doi: 10.1101/2020.09.28.316851. [DOI] [Google Scholar]
- 9. Ge T, Chen CY, Ni Y, Feng YCA, Smoller JW. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat Commun 2019;10:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mak TSH, Porsch RM, Choi SW, Zhou X, Sham PC. Polygenic scores via penalized regression on summary statistics. Genet Epidemiol 2017;41:469–80. [DOI] [PubMed] [Google Scholar]
- 11. Privé F, Arbel J, Vilhjálmsson BJ. LDpred2: better, faster, stronger. Bioinformatics 2021;36:5424–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chang CC, Chow CC, Tellier LCAM, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 2015;4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Martin AR, Kanai M, Kamatani Y, Okada Y, Neale BM, Daly MJ. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat Genet 2019;51:584–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mars N, Koskela JT, Ripatti P, Kiiskinen TTJ, Havulinna AS, Lindbohm JV, et al. Polygenic and clinical risk scores and their impact on age at onset and prediction of cardiometabolic diseases and common cancers. Nat Med 2020;26:549–57. [DOI] [PubMed] [Google Scholar]
- 15. Gerstung M, Jolly C, Leshchiner I, Dentro SC, Gonzalez S, Rosebrock D, et al. The evolutionary history of 2,658 cancers. Nature 2020;578:122–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sokol ES, Pavlick D, Khiabanian H, Frampton GM, Ross JS, Gregg JP, et al. Pan-cancer analysis of BRCA1 and BRCA2 genomic alterations and their association with genomic instability as measured by Genome-wide loss of heterozygosity. JCO Precis Oncol 2020;4:442–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chatrath A, Ratan A, Dutta A. Germline variants that affect tumor progression. Trends Genet 2021;37:433–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Steensma DP, Bejar R, Jaiswal S, Lindsley RC, Sekeres MA, Hasserjian RP, et al. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood 2015;126:9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhu B, Mukherjee A, Machiela MJ, Song L, Hua X, Shi J, et al. An investigation of the association of genetic susceptibility risk with somatic mutation burden in breast cancer. Br J Cancer 2016;115:752–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Carter H, Marty R, Hofree M, Gross AM, Jensen J, Fisch KM, et al. Interaction landscape of inherited polymorphisms with somatic events in cancer. Cancer Discov 2017;7:410–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1–S11, Tables S1–S6, Supplementary Materials and Methods, Supplementary Notes, and Supplementary References
Data Availability Statement
Our findings are supported by data that are available from public online repositories, or data that are publicly available upon request from the data provider. Specifically, GWAS summary statistics were downloaded from the Breast Cancer Association Consortium (https://bcac.ccge.medschl.cam.ac.uk/bcacdata/; BRCA), the Prostate Cancer Association Group to Investigate Cancer Associated Alterations in the Genome consortium (http://practical.icr.ac.uk/blog/; prostate cancer, PRAD), the database of Genotypes and Phenotypes (dbGaP) accession phs001868.v1.p1 (SKCM), GWAS catalog (https://www.ebi.ac.uk/gwas/home; UCEC and ovarian serous carcinoma, OV), and the Harvard Dataverse (https://doi.org/10.7910/DVN/2VBLLP; cervical cancer, CESC). The GWAS summary statistics of colorectal cancer (COADREAD), lung cancer (LUCA), lung adenocarcinoma (LUAD), and lung squamous cell carcinoma (LUSC) were obtained from Supplementary Tables of the original articles. The GWAS summary statistics of ESCA (BEEA), ESCA (EA), glioblastoma multiforme (GBM), and head and neck squamous cell carcinoma (HNSC) were provided by the authors. The UKB resource (https://www.ukbiobank.ac.uk/) was accessed through application number 47821. For TCGA, birdseed files were downloaded from GDC Portal (dbGaP accession phs000178.v10.p8). Somatic and germline mutational data in Mutation Annotation Format, RNA-seq expression data, In Silico Admixture Removal (ISAR)–corrected copy-number segment data, and clinical information were downloaded from GDC (https://gdc.cancer.gov/about-data/publications/pancanatlas and https://gdc.cancer.gov/about-data/publications/PanCanAtlas-Germline-AWG).
Further details of the methods are provided in the Supplementary Materials and Methods.