Abstract
Glioblastoma, also referred to as glioblastoma multiforme (GBM), is grade IV astrocytoma characterized by being fast-growing and the most aggressive brain tumor. In adults, it is the most prevalent type of malignant brain tumor. Despite the advancements in both diagnosis tools and therapeutic treatments, GBM is still associated with poor survival rate without any statistically significant improvement in the past three decades. Patient's genome signature is one of the key factors causing the development of this tumor, in addition to previous radiation exposure and other environmental factors. Researchers have identified genomic and subsequent molecular alterations affecting core pathways that trigger the malignant phenotype of this tumor. Targeting intrinsically altered molecules and pathways is seen as a novel avenue in GBM treatment. The present review shed light on signaling pathways and intrinsically altered molecules implicated in GBM development. It discussed the main challenges impeding successful GBM treatment, such as the blood brain barrier and tumor microenvironment (TME), the plasticity and heterogeneity of both GBM and TME and the glioblastoma stem cells. The present review also presented current advancements in GBM molecular targeted therapy in clinical trials. Profound and comprehensive understanding of molecular participants opens doors for innovative, more targeted and personalized GBM therapeutic modalities.
Keywords: glioblastoma multiforme, molecular targeted therapy, metabolism, signal transduction, blood-brain barrier, tumor microenvironment
1. Introduction
Glioblastoma (GBM) is the most aggressive and deadly form of malignant brain cancers. It accounts for ~80% of all primary brain gliomas and ~60% of all adult brain tumors (1). Currently, surgical resection of the tumor, followed by radiotherapy and temozolomide, is the typical treatment used for GBM (2). GBM is associated with poor survival rate, despite the enhancements in diagnosis tools, surgical techniques and therapeutic approaches; the median survival rate is ~14–20 months and fewer than 5% of the patients survive 5 years post-treatment (3). Moreover, the survival rates for patients with GBMs have not shown statistically significant improvements in the last three decades (4). Thus, there is a need of new therapeutic approaches that inhibit GBM growth, impair its migration and invasion ability and sensitize it to therapy.
The use of high throughput technology in the last decade allowed researchers to classify GBM according to their genomic signature. Based on The Cancer Genome Atlas analysis, four different GBM sub-classes are defined: i) The neural subtype that represents 16% of GBM and are characterized by the expression of neuron markers such as NEFL, GABRA1, SYT1, and SLC1A5, ii) the pro-neural subtype that shows an alteration of PGFRA, a point mutation in IDH1and TP3 and an overexpression in development genes such as NKX2-2, OLIG2, iii) the mesenchymal subtype that is distinguished with alterations in neurofibromatosis type 1 (NF1), phosphatase and tensin homolog (PTEN) point mutation and expression of mesenchymal genes such as MET, CD44, iv) the classical subtype that displays EGFR amplification, CDKN2 A deletion, and p53 mutations (5). Subsequently, researchers have detected several molecular and genomic alterations in core pathways that regulate GBM cell viability, growth, metastasis and invasion. It is thus important to target these altered molecules in order to inhibit GBM proliferation and progression. The present review highlights current understanding of the molecular alterations commonly involved in GBM (Fig. 1). Thorough understanding of molecular alterations facilitates the development of current therapeutic targeted therapy, and opens the way to novel therapeutic approaches. The present review also highlight molecular elements that are currently targeted in GBM clinical trials and others that represent promising potential new targets.
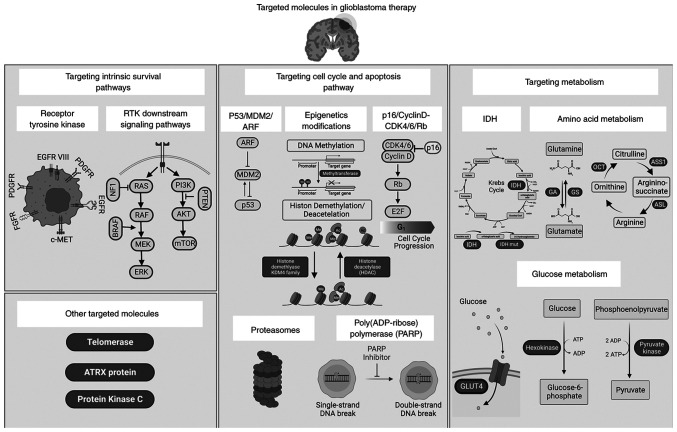
Figure 1.
Overview of signaling pathways and intrinsically altered molecules implicated in GBM development. Several molecules are targeted in clinical trials for glioblastoma cancer therapy. These include molecules implicated in survival pathways (RTKs, BRAF and PI3K), cell cycle pathways (p53, MDM2, CDK4/6, proteasomes and PARP), and metabolism (IDH, hexokinase, glutamine and arginine), in addition to the hTERT and PKC. Increased understanding of glioblastoma biology revealed novel GBM molecular players that can be considered as potential new therapeutic targets for GBM treatment, such as PTEN, GLUT and PKM2. Created with Biorender.com. GBM, glioblastoma; RTKs, receptor tyrosine kinases; BRAF, B-Raf proto-oncogene MDM2, mouse double minute 2 homolog; PARP, Poly(ADP-ribose) polymerase; IDH, isocitrate dehydrogenase; hTERT, human telomerase reverse transcriptase; PKC, protein kinase C; PTEN, phosphatase and tensin homolog; GLUT, glucose transporter; PKM2, pyruvate kinase muscle 2. Created with Biorender.com.
2. Targeting intrinsic survival pathways
Receptor tyrosine kinases (RTKs)
RTKs are transmembrane proteins that consist of an extracellular ligand-binding domain, a single transmembrane helix, and an intracellular catalytic domain (6). RTK superfamily includes epidermal growth factor receptor (EGFR), platelet-derived growth factors (PDGPR), hepatocyte growth factor receptor (c-MET), and fibroblast growth factor receptor (FGFR) (7). Under normal physiological conditions, RTKs are involved in maintaining cellular homeostasis by regulating cell-cell communication, cell survival, migration, proliferation, differentiation, metabolism, and cell cycle. Hence, dysregulation of the RTK pathway is thought to play an important role in GBM initiation, development, and progression (8,9).
EGFR
Genomic analysis revealed that 57% of GBM cells harbor EGFR genetic alterations (10). EGFR amplification and overexpression were identified in 40 and 60% of primary glioblastoma respectively. Other types of genetic alterations were also detected; these include EGFR rearrangement, point mutations, and deletions such as the deletion of exons 2–7 which leads to the truncated mutant variant III (EGFRvIII) (11). EGFR amplification and overexpression lead to constitutive activation of the receptor and enhance GBM cell proliferation, survival, invasion and resistance to treatments (12–15). Talasila et al (16) demonstrated that cells from patients with GBM and with EGFR gene amplification are able to invade the tumor microenvironment (TME) in an angiogenesis independent manner. Additionally, EGFRvIII mutations lacking an extra cellular domain, tend to maintain the EGFR signaling pathway constitutively active in a ligand independent manner. These mutations are detected in 25% of GBM cases and promote survival, tumor growth, migration, invasion and angiogenesis (17–21). Despite the lack of an extracellular domain, EGFRvIII mutation maintains the EGFR signaling It is important to note that 50–60% of GBMs overexpressing wild-type EGFR also express EGFRvIII (22,23). GFRvIII is therefore a potential therapeutic target for GBM.
EGFR is targeted in 77 clinical trials according to clinicaltrials.gov (August 10, 2022). The most common strategy for EGFR targeting is through the use of monoclonal antibodies (Table I). Several anti-EGFR antibodies have been developed since the first chimeric antibody Cetuximab. While Cetuximab and Panitumumab did not show promising results (24), Nimotuzumab and Depatuxizumab-mafodotin (ABT-414), an antibody-drug conjugate, showed survival benefits when combined with radiotherapy and chemotherapeutic Temozolomide (TMZ), respectively (25,26). EGFRs are also targeted by inhibitors of tyrosine kinase activity. Several inhibitors have been evaluated in clinical trials with minimal or no benefits such as Erlotinib, Gefitinib and Dacomitinib. However, using Afatinib, an irreversible pan-inhibitor of the ErbB family resulted in an increase in the progression-free survival (PFS) in patients with overexpressing EGFR or expressing EGFRvIII (27). Notably, EGFR could also be inhibited by anti-tumor vaccines such as ACTIVATe (Phase II NCT00643097), a vaccine against tumor-specific EGFRvIII. ACTIVATe reportedly induces immune responses and an elimination of EGFRvIII-expressing tumor cells (28).
Table I.
Therapeutic agents targeting RTK pathways for the treatment of glioblastoma in interventional clinical trials. Only interventional Phase II, III and IV clinical trials are listed here for therapeutic agents targeting RTKa.
| Therapeutic agent | Specificity | Therapeutic strategy | Cancer condition | Phase | Status | (Refs.) |
|---|---|---|---|---|---|---|
| Cetuximab | EGFR | Monotherapy | Newly diagnosed glioblastoma | Phase I/II NCT02861898 | Recruiting | |
| In combination with Bevacizumab | Recurrent glioblastoma multiforme | Phase II NCT02800486 | Recruiting | |||
| In combination with Irinotecan and Bevacizumab | Recurrent glioblastomas | Phase II NCT00463073 | Completed | |||
| Depatuxizumab-mafodotin (ABT-414) | EGFR or mutant EGFRvIII | ABT-414 to concomitant radiotherapy and TMZ followed by combination of ABT-414 with adjuvant TMZ | Newly diagnosed Glioblastoma with EGFR Amplification | Phase II/III NCT02573324 | Completed | |
| ABT-414 alone or ABT-414 plus | Glioblastoma | Phase II NCT02343406 | Completed | (337) | ||
| TMZ vs. Lomustine or TMZ | ||||||
| Erlotinib (Tarceva) | EGFR | Monotherapy | Glioblastoma | Phase II NCT00337883 | Completed | |
| In combination with Bevacizumab | Glioblastoma | Phase II NCT00671970 | Completed | (338) | ||
| Monotherapy | Recurrent glioblastoma multiforme and anaplastic astrocytoma | Phase I/II NCT00301418 | Completed | (339) | ||
| Monotherapy | Recurrent malignant glioma or recurrent or progressive meningioma | Phase I/II NCT00045110 | Completed | (340) | ||
| In combination with radiotherapy | Newly diagnosed gliomas | Phase I/II NCT00124657 | Completed | (341) | ||
| In combination with TMZ during and following radiotherapy | Malignant glioma (glioblastoma or gliosarcoma) | Phase II NCT00187486 | Completed | (342) | ||
| Monotherapy | Recurrent or progressive glioblastoma multiforme | Phase II NCT00054496 | Unknown | |||
| Gefitinib (Iressa/ZD1839) | EGFR | Monotherapy | Recurrent glioblastoma | Phase II NCT00250887 | Completed | (343) |
| In combination with radiotherapy | Glioblastoma multiforme | Phase I/II NCT00052208 | Completed | |||
| In combination with radiotherapy | Newly diagnosed gliomas | Phase I/II NCT00042991 | Completed | |||
| GC1118 | EGFR | GC1118 with standard concurrent chemoradiation | Recurrent glioblastoma with high EGFR amplification | Phase II NCT03618667 | Unknown | |
| Sym004 | EGFR | Monotherapy | Recurrent malignant glioma (glioblastoma or gliosarcoma) | Phase II NCT02540161 | Completed | |
| Olaratumab (IMC-3G3) | PDGFRα | Compared to Ramucirumab (anti-VEGFR2) | Recurrent glioblastoma multiforme | Phase II NCT00895180 | Completed | |
| MEDI-575 | PDGFRα | Monotherapy | Recurrent glioblastoma multiforme | Phase II NCT01268566 | Completed | |
| APL-101 | c-MET | Monotherapy | Advanced solid tumors including glioblastoma multiforme | Phase I/II NCT03175224 | Recruiting | |
| Onartuzumab (MetMAb) | c-MET | In combination with Bevacizumab compared with Bevacizumab alone or Onartuzumab monotherapy | Recurrent glioblastoma | Phase II NCT01632228 | Completed | |
| Infigratinib (BGJ398) | FGFR1, FGFR2, and FGFR3 | Monotherapy | Recurrent glioblastoma multiforme | Phase II NCT01975701 | Completed | |
| Dacomitinib (PF-299804/Vizimpro) | EGFR/HER1, HER2, and HER4 | Monotherapy | Recurrent Glioblastoma With EGFR Amplification or Presence of EGFRvIII Mutation | Phase II NCT01520870 | Completed | |
| Monotherapy | Recurrent glioblastoma | Phase II NCT01112527 | Completed | |||
| Afatinib (BIBW 2992) | ErbB family | With or without daily TMZ | Recurrent malignant glioma | Phase II NCT00727506 | Completed | |
| AEE788 | ErbB & VEGFR family | Monotherapy | Glioblastoma Multiforme | Phase I/II NCT00116376 | Completed | |
| Tesevatinib | EGF, HER2, and VEGF | Monotherapy | Recurrent glioblastoma | Phase II NCT02844439 | Completed | |
| Anlotinib | VEGFR, FGFR, PDGFR, Kit | Monotherapy | Recurrent glioblastoma | Phase I/II NCT04004975 | Unknown | (344) |
| In combination with dose-dense TMZ | First recurrent or progressive glioblastoma | Phase II NCT04547855 | Recruiting | |||
| Sorafenib | VEGFR, FLT-3, PDGFR-β, Kit | Radiotherapy and TMZ followed by TMZ plus Sorafenib | Glioblastoma Multiforme | Phase II NCT00544817 | Completed | (345) |
| A combination of Sorafenib tosylate, Valproic acid and Sildenafil | Recurrent high-grade glioma | Phase II NCT01817751 | Active, not recruiting | (303) | ||
| Dasatinib | c-KIT, EPHA2, and PDGFR-β | Monotherapy | Recurrent glioblastoma multiforme or gliosarcoma | Phase II NCT00423735 | Completed | (304) |
Data acquired from the U.S. National library of medicine (http://clinicaltrials.gov, accessed on 10 August 2022). Terminated studies are not included. c-MET, hepatocyte growth factor receptor; EGFR, epidermal growth factor receptor; EPHA2, Ephrin type-A receptor 2; ErbB, epidermal growth factor receptor; FGFR, fibroblast growth factor receptor; FLT-3, fms-like tyrosine kinase 3; HER, human epidermal growth factor receptor; PDGFR, platelet-derived growth factors; RTK, receptor tyrosine kinase; TMZ, Temozolomide; VEGFR, vascular endothelial growth factor receptor.
PDGFR
The receptor tyrosine kinase PDGFR is the second therapeutic target in GBM proneural subtype. PDGFR gene amplification is found in 15% of GBM cases (10). Overexpression of PDGFR and its ligands (PDGF-AA/-AB/-BB/-CC/-DD) is observed in gliomas of all grades and associated with poor prognosis (10,29). Once activated, PDGFR triggers intracellular signaling cascades that regulate cancer cell survival, growth and progression (30,31). Therefore, dysregulation of PDGF signaling stimulates malignant transformation of normal neural stem cells into glioblastoma and enhances GBM cell growth and motility through autocrine signaling (32–34).
In clinical trials, PDGFR is either targeted by multikinase inhibitors or specific anti-PDGFR antibodies (Table I). To date, multikinase inhibitors such as Sunitinib, Imatinib and Dasatinib have not shown promising clinical benefits (24). In addition, two Phase II clinical trials assessed the tolerance and efficacy of Ramucirumab (IMC-3G3) and MEDI-575 anti-PDGFR antibodies (NCT00895180 and NCT01268566 respectively) in patients with recurrent glioblastoma multiforme. However, these monotherapies did not show improved survival.
cMET
MET is a transmembrane receptor with a tyrosine kinase activity. Different types of MET genetic alterations are detected in GBM cells. MET gene amplification and overexpression are detected in 5–13% of GBM cases (35), and MET fusion genes are found in pediatric GBM (36). Overexpression of MET and its ligand HGF promotes tumor growth, migration, invasion and drug resistance (37–40). Increased activation of MET/HGF pathway is strongly and selectively associated with highly anaplastic GBM cells. The c-MET pathway is either targeted directly by c-MET antibodies or inhibitors or through antibodies targeting its ligand, HGF (Table I). Treatment with MET inhibitors can potentially be viable from the standpoint of drug selectivity, thus avoiding toxicity to normal cells (41). However, and in order to avoid drug resistance, combination of both PI3K inhibitors along with MET inhibitors can be favorably considered in the upcoming trials to efficiently target patients with GBMs (42). On the other hand, one promising result for c-MET antibodies was shown by a randomized, double-blind, placebo-controlled, multicenter Phase II study (NCT01632228) assessing Onartuzumab (MetMAb, an anti-cMET antibody) with or without Bevacizumab. Survival benefits were reported in patients with high HGF expression (43).
FGFR
Genomic alterations in FGFR are rarely detected in GBM (44); however, the constitutive activation of its downstream pathways stimulate tumor progression, proliferation and resistance to apoptosis (44,45). Infigratinib (BGJ 398) selectively binds to and inhibits FGFRs and was used as a monotherapy in Phase II NCT01975701 clinical trial in patients with recurrent glioblastoma or other glioma subtypes. However, Infigratinib was not licensed and the indication was abandoned (Table I).
Ras/Raf/MEK/ERK pathway
Ras/Raf/MEK/Erk (also known as the Ras/MAPK pathway) is a chain of effectors downstream of RTKs that regulate cell survival and proliferation (46). Alterations in various components of this pathway are detected in GBM.
B-Raf proto-oncogene (BRAF)
BRAF is a serine/threonine kinase that belongs to the RAF family. Multiple BRAF gene alterations are associated with GBM; however, the BRAF V600E mutation is the most relevant. This missense mutation leads to constitutive activation of Ras/Raf/MEK/Erk pathway, promoting tumor cell proliferation, survival and inhibit apoptosis (47).
NF1
The NF1 gene encodes neurofibromin, a GTPase activating protein that controls cell growth and survival. It regulates the conversion of active GTP-bound Ras to its inactive GDP-bound form, thus inhibiting Ras/Raf/MEK/Erk signaling pathway (48). NF1 mutation or deletion is found in 10% of glioblastoma cases, especially in the mesenchymal GBM subtype (10). Despite the fact that genomic alterations of this tumor suppressor gene enhance the epithelial-mesenchymal transition in neurofibromatosis and can induce malignant transformation (49), its role in glioblastoma is not fully understood.
In glioblastoma, MEK inhibitors are common drugs used to target Ras/Raf/MEK/Erk signaling pathway (Table II). For instance, Atorvastatin, a Ras/MAPK inhibitor, showed encouraging results when evaluated in combination with radiotherapy and TMZ in patients with GBMs (Phase II NCT02029573) (50). Additionally, BRAF inhibitors such as Dabrafenib and Encorafenib are evaluated in clinical trials in combination with MEK inhibitors trametinib (Phase II NCT03919071) and Binimetinib, respectively (Phase II NCT03973918).
Table II.
Therapeutic agents targeting the Ras/MAPK pathway for the treatment of glioblastoma in interventional clinical trialsa.
| Therapeutic agents | Therapeutic target | Therapeutic strategy | Cancer condition | Clinical phase | Status | (Refs.) |
|---|---|---|---|---|---|---|
| ABM-1310 and Cobimetinib | BRAF V600E (ABM-1310) and MEK1/2 (Cobimetinib) | ABM-1310 monotherapy or in combination with Cobimetinib | Advanced solid tumors | Phase I NCT04190628 | Recruiting | |
| Dabrafenib and Trametinib | BRAF (Dabrafenib) and MEK1/2 (Trametinib) | Dabrafenib in combination with Trametinib | BRAF V600 mutation positive low grade glioma or relapsed or refractory high grade glioma. | Phase II NCT02684058 | Active, not recruiting | (346) |
| Dabrafenib Combined with Trametinib after radiotherapy Newly-diagnosed high-grade glioma | Phase II (NCT03919071 | Recruiting | (346) | |||
| Encorafenib and Binimetinib | BRAF (Encorafenib) and MEK1/2 (Binimetinib) | Combination treatment with Encorafenib and Binimetinib | Recurrent BRAF V600-Mutated high grade glioma | Phase II NCT03973918 | Active, not recruiting | |
| Atorvastatin | Ras/MAPK | In combination with Radiotherapy and TMZ | Glioblastoma | Phase II NCT02029573 | Completed | (50) |
| Ulixertinib (BVD-523) | ERK1/2 | Monotherapy | Advanced solid tumors | NCT04566393 | Available | |
| LY2228820 | p38 MAPK | With radiotherapy plus concomitant TMZ | Newly diagnosed glioblastoma | Phase I/II NCT02364206 | Completed | |
| RSC-1255 | Ras | Monotherapy | Advanced malignancies including glioblastoma | Phase I NCT04678648 | Recruiting |
Data acquired from the U.S. National library of medicine (http://clinicaltrials.gov, accessed on 10 August 2022). Terminated studies are not included. BRAF, B-Raf proto-oncogene; ERK, extracellular signal-regulated kinase; MAPK, mitogen activated protein kinase; MEK, mitogen-activated protein kinase kinase; TMZ, Temozolomide.
PI3K/AKT/mTOR pathway
Phosphatidylinositol-3-kinase (PI3K) is a family of lipid kinases that regulates cell survival, growth, motility and metabolism (51). It consists of three subclasses depending on their structure and substrate specificities (51). Activated PI3Ks phosphorylate the lipid phosphatidylinositol (4,5)-bisphosphate (PIP2) to generate phosphatidylinositol (3,4,5)-trisphosphate (PIP3) (51). PI3K signaling pathway dysregulation is predominant in glioblastoma and is mainly caused by gain-of-function mutations in PIK3CA gene, PIK3R1 gene and loss of PTEN gene.
PTEN
PTEN mediates the conversion of PIP3 to PIP2 which regulates cell proliferation, apoptosis, metabolism, motility and angiogenesis (52). PTEN deletion or mutation, found in 5–40% of GBM (53), inhibit AKT (protein kinase B) phosphorylation and induce the hyperactivation of PI3K signaling pathway, which in turn accelerates tumor growth, progression, and metastasis (54).
PI3K
Heterodimeric Class IA PI3Ks consist of a catalytic subunit (p110α, p110β, or p110) and a p85-type regulatory subunit (55). PIK3CA and PIK3R genes encode for p110α and p85α respectively (55,56). Several studies show that mutations in PIK3R1, identified in 8–10% of GBM cases, are found to be mutually exclusive with mutations in PIK3CA in primary GBM cases (57,58). Constitutively active PIK3CA and PIK3R1 upregulate the PI3K/AKT signaling pathway and promote tumorigenesis (59,60). Somatic mutations in PIK3CA occurs in 6–17% of GBM (57,58,61) and PIK3CA activating mutations are correlated with poor prognosis, aggressive and more disseminated phenotype and with shorter survival rate (62). Alterations in PIK3CD (p110δ) and PIK3CB (p110β) are also detected in GBM (56).
The PI3K pathway is generally targeted by several PI3K pan-inhibitors in clinical trials (Table III). One example is Pictilisib, a PI3K isoform inhibitor shown to sensitize tumors to radio- and chemotherapy. Effectiveness of Pictilisib is being compared with immunotherapeutic Pembrolizumab (MK-3475) in Phase II clinical trial in patients with glioblastoma (NCT02430363). Paxalisib (GDC-0084) is another small molecule inhibitor of PI3K currently evaluated as an adjuvant therapy after surgical resection and concomitant chemoradiation therapy with TMZ in patients with GBM and unmethylated MGMT promoter status (Phase II NCT03522298). Preliminary results show enhanced overall survival (OS; 17.7 months for treated group compared with 12.7 months for patients treated with TMZ) (24). The PI3K/AKT/mammalian target of rapamycin (mTOR) pathway is also targeted by mTOR inhibitors. While several mTOR inhibitors do not show clinical benefits, AZD2014 is proposed as a promising drug for radiosensitizing glioblastoma stem cells (GSCs) in vitro and in vivo, and is currently being evaluated in clinical trials (Phase I NCT02619864). Moreover, another clinical trial is evaluating the efficiency of ABI-009 (Nab-Rapamycin), a novel albumin-bound mTOR inhibitor, in recurrent and newly diagnosed glioblastoma (Phase II NCT03463265).
Table III.
PI3K inhibitors applied for the treatment of glioblastoma in interventional clinical trialsa.
| Therapeutic agents | Therapeutic strategy | Cancer condition | Clinical phase | Status |
|---|---|---|---|---|
| Paxalisib (GDC-0084) | Adjuvant therapy following surgical resection and initial chemoradiation with TMZ | Newly-diagnosed glioblastoma with unmethylated MGMT promoter status | Phase II NCT03522298 | Active not recruiting |
| With metformin while maintaining a ketogenic diet | Glioblastoma | Phase II NCT05183204 | Not yet recruiting | |
| Multiple drugs and drug combinations (TMZ, Lomustine, Regorafenib, Radiation, Paxalisib, VAL-083, VT1021, Troriluzole) | Newly diagnosed and recurrent glioblastoma | Phase II/III NCT03970447 | Recruiting | |
| Pictilisib | Compared with Pembrolizumab (MK-3475/anti-PD-1 monoclonal antibody) | Glioblastoma | Phase I/II NCT02430363 | Unknown |
Data acquired from the U.S. National library of medicine (http://clinicaltrials.gov, accessed on 10 August 2022). Terminated studies are not included. MGMT, O6-methylguanine DNA methyltransferase; PD-1, programmed cell death protein 1; PI3K, phosphatidylinositol-3-kinase; TMZ, Temozolomide.
3. Targeting cell cycle and apoptosis pathways
p53/mouse double minute 2 homolog (MDM2)/ARF and p16/cyclinD-CDK4/6/Rb signaling pathways regulate cell cycle and cell proliferation. According to the Cancer Genome Atlas Research Network, 87 and 77% of the analyzed GBM samples harbor a mutation in p53 and Rb signaling pathway respectively (57).
p53/MDM2/ARF pathway
The p53/MDM2/ARF pathway is one of the major core pathways deregulated in 84% of patients with GBMs. p53, the key protein of this pathway and also known as ‘guardian of the genome’, protects cells from external or internal stress signals by regulating cell processes such as cell cycle, DNA repair, angiogenesis, metabolism, cell death (apoptosis and autophagy) and senescence (63).
p53 genetic alterations were detected in 30% of primary GBM and 65% of secondary GBM (64). Mainly two different types of genomic alteration are detected in GBM cells; point mutation associated with overexpression of the mutant version of p53 and deletion that provokes the loss of function of p53. These alterations result in either gain or loss of p53 function (65). The oncogenic potential of p53 is mediated by the accumulation of p53 mutants in cells. Indeed, mutations in the MDM2 binding domain of p53 affect its regulation by MDM2, leading to the accumulation of p53 inside the cell and the subsequent gain of function phenotype. This enhances tumorigeneses in GBM by promoting inflammation, genomic instability, tumor growth, invasion, metastasis and neo-angiogenesis (66,67). Pedrote et al (68) revealed that accumulation of amyloid-like p53 with p53 gain of function phenotype leads to increased chemo-resistant GBM cells. Thus, p53 gain of function mutations are associated with great oncogenic potential and aggressive GBM phenotype.
Loss of p53 tumor suppression function is not only mediated by p53 mutations. Previous studies demonstrated that different components in the p53/MDM2/ARF pathway, such as MDM2/MDM4 protein, negatively regulate the activity of p53 (69–72). Under normal physiological conditions, MDM2 protein, which is an E3 ubiquitin ligase, binds p53 transcriptional domain and controls its activity by preventing its transcriptional function and promoting its degradation. However, the activity of p53 is positively regulated by the alternative reading frame tumor suppressor (ARF), an upstream molecule that suppresses the MDM2 activity (73). p53 mutations or deletions, MDM2 amplification or overexpression and ARF homozygous deletion inactivate p53 and trigger the p53 loss of function phenotype in glioblastoma, thus contributing to tumor growth, progression and therapy resistance (65,74).
RG7388 (Idasanutlin) is a small molecule antagonist of MDM2 used to target p53/MDM2 pathway. Recent Phase I/II trials are recruiting to test for molecularly matched targeted therapies (APG101, Alectinib, Idasanutlin, Atezolizumab, Vismodegib, Temsirolimus and Palbociclib) in combination with radiotherapy in patients with newly diagnosed glioblastoma without MGMT promoter methylation (NCT03158389). A more recent Phase I clinical trial is studying the uptake and tolerance of BI 907828 (an MDM2 inhibitor) in combination with radiotherapy in newly diagnosed glioblastoma patients (NCT05376800). Markedly, several different strategies are currently employed for targeting p53 using gene therapy in different cancer types. In GBM, only one clinical trial reported using SGT-53, a human wild type p53 DNA sequence encapsulated in nanodelivery liposome. However, this study was terminated due to the very small number of participants (Phase II NCT02340156).
Epigenetic modifications
In addition to genetic alterations, epigenetic modulators affect expression by interacting with drivers of GBM cell proliferation, without causing any changes in the DNA sequence (75). In GBM, DNA methylation is strongly correlated with responses to TMZ treatment that methylates adenine in position N3 and guanine in position O6 and N7 (76). Guanine methylation at O6 position leads to strand breaks, activating p53-mediated apoptosis through Fas/CD95/Apo-1 receptor or by the mitochondrial pathway (77). To decrease the effects of O6-methylguanine DNA methyltransferase (MGMT) methylation, synthetic inhibitors of MGMT entered human trials (78). Nevertheless, several studies revealed that inhibitors such as O6-benzylguanine and PaTrim-2 (Lomeguatrib) did not show survival improvement in response to TMZ (79–81).
Histone demethylases (KDM)4C is often overexpressed in GBM and is associated with epigenetic regulation of both tumor suppressor genes and oncogenes (82). Lee et al (82) showed that KDM4C binds to the promoter of c-Myc oncogene inducing its expression/activation and suppressing the functions of p53 by demethylating p53K372me1, thus inducing apoptosis. KDM4C knockdown significantly suppresses both the proliferation and tumorigenesis of glioblastoma cells in vitro and in vivo, thus suggesting KDM4C inhibition as a promising tool in targeting glioblastoma.
Histone deacetylases (HDAC) also serve an important role in regulating cell growth and survival of cancer cells (83). Inhibition of HDACs leads to cell cycle arrest as well as apoptosis, and, in GBM, causes the rebalance of histones acetylation (84,85). In clinical trials, testing Romidepsin (FR901228, an HDAC inhibitor) exhibited failure in treating patients with GBMs (NCT00085540) (24). Other HDAC inhibitors, such as Vorinostat. did not show improvement in the median OS or PFS both alone or in combination with Bortezomib (NCT00641706) or Bevacuzimab (NCT01738646) (24).
p16/CyclinD-CDK4/6/Rb pathway
The p16/cyclinD-CDK4/6/Rb signaling pathway regulates cell cycle progression and is also disrupted in GBM cells (10). The cyclin D-CDK4/6-Rb axis, which is a crucial cell cycle checkpoint, controls cell transition from G1 to S phase (86). Indeed, cyclin D forms a complex with CDK 4/6 inducing the phosphorylation of Rb and inhibiting Rb-E2F formation (87). Released E2F mediates transcription of genes that facilitate the transition into S phase and consequently cell cycle progression (88). Under carcinogenic conditions, p16 protein inhibits cyclin D-CDK 4/6 complex formation (74). Thus, Rb associates with E2F and triggers cell cycle arrest. It has been demonstrated that genetic alterations such as deletion in CDKN2A/B, amplification of CDK4/6 and mutations of Rb and p16 genes are detected in 79% of patients with GBM (10). Consequently, cells harboring such alterations undergo uncontrolled cell proliferation and tumor growth. Collectively, targeting cell cycle and apoptosis pathways could emerge as promising tool for treating GBM. There are three CDK4/6 inhibitors evaluated in clinical trials for GBM treatment in clinical trials (Table IV). Ribociclib and Abemaciclib are being evaluated in current studies, whereas PD 0332991 (Palbociclib) monotherapy was not effective in treating patients with recurrent glioblastoma (89).
Table IV.
Therapeutic agents targeting CDK4/6, proteasomes and Poly(ADP-ribose) polymerase applied for the treatment of glioblastoma in interventional clinical trialsa.
| A, CDK4/6 inhibitors | |||||
|---|---|---|---|---|---|
|
| |||||
| Therapeutic agents | Therapeutic strategy | Cancer condition | Clinical phase | Status | (Refs.) |
| Abemaciclib (LY2835219) | In combination with TMZ and Irinotecan vs. Abemaciclib in combination with TMZ | Solid tumors | Phase I NCT04238819 | Recruiting | |
| In combination with Bevacizumab | Recurrent glioblastoma patients with loss of CDKN2A/B or gain or amplification of CDK4/6 | Early Phase I NCT04074785 | Active, not recruiting | ||
| With or without surgery | Recurrent glioblastoma | Phase II NCT02981940 | Recruiting | ||
| In combination with LY3214996 (ERK Inhibitor) | Recurrent glioblastoma | Early Phase I NCT04391595 | Recruiting | ||
| Ribociclib (LEE011) | Before surgical tumor resection | Preoperative glioma and meningioma | Early Phase I NCT02933736 | Recruiting | |
| In combination with Everolimus | Preoperative recurrent high-grade glioma | Early Phase I NCT03834740 | Active, not recruiting | ||
| Ribociclib and Everolimus following Radiotherapy | High grade gliomas | Phase I NCT03355794 | Active, not recruiting | ||
| Monotherapy | Recurrent glioblastoma or anaplastic glioma | Phase I NCT02345824 | Unknown | ||
|
| |||||
| B, Proteasome inhibitors | |||||
|
| |||||
| Therapeutic agents | Therapeutic strategy | Cancer condition | Clinical phase | Status | (Refs.) |
|
| |||||
| Bortezomib | In combination with TMZ | Glioblastoma | Phase I/II NCT03643549 | Recruiting | (347) |
| In combination with TMZ | Brain tumors or other solid tumor that have not responded to treatment | Phase I NCT00544284 | Completed | ||
| In combination with bevacizumab and escalating doses of TMZ | Recurrent glioblastoma multiforme | Phase I NCT01435395 | Completed | ||
| In combination with TMZ and radiotherapy | Newly diagnosed glioblastoma multiforme or gliosarcoma | Phase II NCT00998010 | Completed | ||
| In combination with Bevacizumab | Recurrent Malignant Glioma | Phase II NCT00611325 | Completed | ||
| Marizomib | In combination with TMZ and radiotherapy | Newly diagnosed glioblastoma | Phase III NCT03345095 | Active, not recruiting | |
| In combination with Bevacizumab | Malignant glioma and glioblastoma | Phase I/II NCT02330562 | Completed | (348) | |
| In combination with TMZ and radiotherapy | Malignant glioma and glioblastoma | Phase I NCT02903069 | Completed | (348) | |
| Ixazomib (MLN9708) | Monotherapy | Glioblastoma | Early Phase I NCT02630030 | Completed | (95) |
| Disulfiram (DSF) | After radiotherapy with TMZ | Glioblastoma multiforme | Early Phase I NCT01907165 | Completed | |
| DSF-Copper in combination with TMZ | Recurrent glioblastoma | Phase II NCT03034135 | Completed | (97) | |
| DSF-Copper with concurrent radiotherapy and TMZ | Glioblastoma multiforme | Phase I/II NCT02715609 | Active, not recruiting | ||
|
| |||||
| C, PARP inhibitors | |||||
|
| |||||
| Therapeutic agents | Therapeutic strategy | Cancer condition | Clinical phase | Status | (Refs.) |
|
| |||||
| Iniparib (BSI-201) | In combination with TMZ | Newly diagnosed malignant glioma | Phase I/II NCT00687765 | Completed | |
| Olaparib | Cediranib maleate and Olaparib work compared with bevacizumab | Recurrent glioblastoma | Phase II NCT02974621 | Active, not recruiting | |
| In combination with TMZ | Relapsed glioblastoma | Phase I NCT01390571 | Completed | (101) | |
| In combination with radiotherapy compared with Pamiparib with radiotherapy | Newly diagnosed and recurrent glioblastoma | Early Phase I NCT04614909 | Recruiting | ||
| Monotherapy | Advanced glioma, cholangiocarcinoma, or solid tumors with IDH1 or IDH2 mutations | Phase II NCT03212274 | Recruiting | ||
| Combination therapy of Pembrolizumab, Olaparib and TMZ | Glioblastoma and recurrent glioblastoma | Phase II NCT05463848 | Not yet recruiting | ||
| Afatinib, Dasatinib, Palbociclib, Everolimus or Olaparib based on patient's genetic profile | Glioblastoma and recurrent glioblastoma | Early Phase I NCT05432518 | Not yet recruiting | ||
| Veliparib | In combination with TMZ | Young patients with recurrent or refractory CNS tumors | Phase I NCT00946335 | Completed | |
| In combination with TMZ and radiotherapy | Younger patients with newly diagnosed diffuse pontine gliomas | Phase I/II NCT01514201 | Completed | (103) | |
| In combination with TMZ | Recurrent glioblastoma | Phase I/II NCT01026493 | Completed | (102) | |
| In combination with TMZ and radiotherapy | Newly diagnosed glioblastoma multiforme | Phase I NCT00770471 | Completed | ||
| In combination with TMZ and radiotherapy | Newly diagnosed malignant glioma without H3 K27M or BRAFV600 mutations | Phase II NCT03581292 | Active, not recruiting | ||
| TMZ with or without Veliparib | Newly diagnosed glioblastoma multiforme | Phase II/III NCT02152982 | Active, not recruiting | (349) | |
| Pamiparib | In combination with TMZ and radiotherapy | Newly diagnosed or recurrent glioblastoma | Phase I/II NCT03150862 | Completed | (350) |
Data acquired from the U.S. National library of medicine (http://clinicaltrials.gov, accessed on 10 August 2022). Terminated studies are not included. CDK, cyclin-dependent kinase; CNS, central nervous system; DSF, Disulfiram; ERK, extracellular signal-regulated kinase; IDH, Isocitrate dehydrogenase; PARP, poly(ADP-ribose) polymerase; TMZ, Temozolomide.
Proteasomes
Proteasomes are protein complexes that are central for degradation of unneeded or damaged proteins (90). They regulate cell cycle and homeostasis in normal and cancer cells by regulating p53 and endoplasmic reticulum stress. Proteasomes can thus influence drug resistance in tumor cells (91). Due to the complexity of GBM physiology, there is a need for therapeutic options that could target broad systems contributing to the tumorgenicity in GBM. Proteasome inhibition is thus identified as a promising strategy for GBM treatments. Currently, four proteasome inhibitors are tested in clinical trials to target proteasomes in GBM: Bortezomib, Ixazomib, Marizomib and Disulfiram (Table IV). Bortezomib is the first-generation proteasome inhibitor and showed promising results when studied in combination with TMZ and radiotherapy (NCT00998010) (92). Marizomib is a second-generation proteasome inhibitor that has the ability to cross the BBB (93). It is currently being evaluated in ongoing clinical trials combined with Bevacizumab, TMZ, or ABI-009 (Nab-rapamycin, nanoparticle albumin-bound rapamycin). Encouraging observations are reported in Phase III study (NCT03345095) for Marizomib administered in combination with radiotherapy and TMZ to treat patients with newly diagnosed glioblastoma (94). Ixazomib, on the other hand, was evaluated in early Phase I clinical trial (NCT02630030) for its ability to reach brain tumors due to its distinctive permeability to tumor tissues (95). Disulfiram is another interesting proteasome inhibitor that has an improved BBB penetration to employ its anti-tumor action (96). Disulfiram was well tolerated in Phase II clinical trial in combination with TMZ but showed limited activity for unselected population (97).
Poly(ADP-ribose) polymerase (PARP)
Elements of DNA damage repair mechanisms are considered promising radio-sensitizing agents for cancer therapy (98). PARP is an important protein in DNA repair pathways and high PARP-1 mRNA expression is associated with poor survival in classic GBMs (99). A few PARP-1 inhibitors have been evaluated in clinical studies (Table IV). For instance, a Phase I/II clinical trial (NCT00687765) recently completed a study to evaluate the safety and efficiency of Iniparib (BSI-201), a PARP1 inhibitor (results as yet unpublished). Olaparib is another inhibitor of PARP that shows an effective radio-sensitizing result in GBM cell lines and preclinical glioma models (98,100). A Phase I trial OPARATIC (NCT01390571) reported that Olaparib penetrates core and margin regions of GBM at radio-sensitizing concentrations. It was also reported to be safe when used with continuous low-dose of TMZ (101). PARP inhibitor Veliparib was also evaluated in several clinical trials. However, results showed that Veliparib did not show improved clinical survival when combined with TMZ (Phase I/II NCT01026493) (102), nor when added to radiation followed by TMZ (Phase I/II NCT01514201) (103).
4. Targeting metabolism
Isocitrate dehydrogenase (IDH)
WHO classifies GBM depending on IDH mutational status (104). The IDH enzyme plays a pivotal role in major cellular metabolic processes. It is involved in the Krebs cycle, lipid and glutamine metabolism and oxidative stress regulation. During the Krebs cycle, IDH catalyzes the oxidative decarboxylation of isocitrate to α-ketoglutarate [α-KG, also known as 2-oxyglutarate (2OG)] and reduces the cofactor NADP+ into NADPH which is essential to decrease the level of reactive oxygen species (ROS) (105–107).
Three different isoforms of IDH exist in human cells: IDH1, detected in the cytoplasm and in peroxisomes, whereas IDH2 and IDH3 are both present in mitochondria (108). IDH mutations were found in 6% of primary GBM and in 54% of secondary GBM (109). Missense mutations, caused by the substitution of arginine residue in codons 132 and 172 in the enzymatic site of the IDH1 and IDH2, are frequently detected in glioblastoma cells (58,110,111). IDH mutants catalyze the production of 2-hydroxyglutaric acid (2-HG) from isocitrate. 2-HG affects cellular metabolism, enhances the oxidation of NADPH in NADP+ which, in turn, disrupts cellular homeostasis and increases the level of ROS (112). It has been suggested that the accumulation of this oncometabolite inhibits the activity of α-KG dependent dehydrogenase, a family of enzymes involved in methylation of histones and DNA, which includes KDMs and 10–11 translocation (TET, a family of DNA hydroxylases that leads to a hypermethylation state and genetic instability) (113). Moreover, modification of the epigenetic profile is associated with the inhibition of glioma stem cell differentiation (114). Furthermore, overexpression of hypoxia-inducible factor 1-α (HIF-1α), VEGF and platelet-derived growth factor subunit A (PDGF-A) are detected in GBM cells holding IDH mutation (115–117). Hence, these alterations induced by the high levels 2-HG are associated with an aggressive and invasive carcinogenic phenotype.
Patients with GBMs and IDH mutations are often treated with inhibitors such as Olaparib targeting PARP (Phase II NCT03212274). More clinical trials are investigating other PARP inhibitors such as Nivolumab (Phase II NCT03718767, recruiting), Talazoparib (Phase II NCT04740190, recruiting) and BGB-290 (Phase II NCT03914742, active, not recruiting/Phase I NCT03749187) to treat patients with advanced gliomas including glioblastoma. Notably, a recently completed study investigated the safety and clinical activity of Enasidenib (AG-221), a small molecule inhibitor of IDH2 in patients with advanced solid tumors, including glioma (Phase I/II NCT02273739). Patients with similar cancer types were treated in a recently completed study (Phase I/II NCT03684811) with Olutasidenib (FT-2102), another agent that specifically inhibits mutant IDH1 at arginine R132. Currently, a number of other clinical trials are evaluating IDH inhibitors, therefore, more time and research are needed to conclude about their efficiency to treat gliomas (Table V).
Table V.
Therapeutic agents targeting metabolism of gliomas (including glioblastoma) in interventional clinical trialsa.
| A, IDH | |||||
|---|---|---|---|---|---|
|
| |||||
| Therapeutic agents | Therapeutic strategy | Cancer condition | Clinical phase | Status | (Refs.) |
| Enasidenib (AG-221), Inhibitor of mutant IDH2 | Monotherapy | Solid tumor, glioma, angioimmunoblastic T-cell lymphoma, intrahepatic cholangiocarcinoma, chondrosarcoma | Phase I/II NCT02273739 | Completed | |
| Olutasidenib (FT-2102), Inhibitor of R132 mutant IDH1 | In combination with other anti-cancer drugs (Azacitidine, Nivolumab, Gemcitabine and Cisplatin) | Advanced solid tumors and gliomas (glioblastomas) | Phase I/II NCT03684811 | Completed | |
| AG-120, Inhibitor of R132 mutant IDH1 | Monotherapy | Advanced solid tumors and gliomas | Phase I/II NCT02073994 | Active, not recruiting | (351) |
| IDH305, Inhibitor of | Monotherapy | Advanced | |||
| mutant IDH1 | malignancies with IDH1 R132 mutations | Phase I NCT02381886 | Active, not recruiting | (352) | |
| LY3410738, Inhibitor of R132 mutant IDH1, and R140 or R172 mutant IDH2 | Monotherapy or in combination with gemcitabine and Cisplatin, or Durvalumab | Advanced solid tumors with IDH1 or IDH2 mutations | Phase I NCT04521686 | Recruiting | |
|
| |||||
| B, Hexokinase | |||||
|
| |||||
| Therapeutic agents | Therapeutic strategy | Cancer condition | Clinical phase | Status | (Refs.) |
|
| |||||
| Posaconazole | Monotherapy | Glioblastoma | Early Phase I NCT04825275 | Recruiting | |
| Ketoconazole | Monotherapy | Glioblastoma | Early Phase I NCT04869449 | Recruiting | |
Data acquired from the U.S. National library of medicine (http://clinicaltrials.gov, accessed on 10 August 2022). Terminated studies are not included. IDH, Isocitrate dehydrogenase.
Glucose metabolism
Normal cells rely on oxidative phosphorylation as the main energy source. However, cancer cells shift to aerobic glycolysis even in the presence of oxygen. This phenomenon, known as Warburg effect, protects cancer cells from apoptosis, stimulates the production of new precursors and increases invasion capacity (118–121). Therefore, targeting glucose transporters, such as glucose transporter (GLUT), and metabolic enzymes, such as Hexokinase 2 and Pyruvate kinase muscle, may provide a novel avenue in glioblastoma treatment.
GLUT
GLUT is a transmembrane protein that belongs to the major facilitator superfamily (122), Currently, 14 different isoforms are identified in human tissues and divided into three classes (123). Class I, which consists of GLUT 1/2/3/4, is mainly expressed in brain cells. In GBM, GLUT-1 and GLUT-3 are the predominantly expressed isoforms and associated with poor survival rates. These transporters, characterized by their high affinity for glucose, promote glycolysis metabolism by increasing glucose uptake to maintain the survival, proliferation and growth of cancer cells. GLUT-3, also known as neural glucose transporter, is highly expressed by brain tumor initiating cells (BTICs) and has higher affinity for glucose compared with GLUT-1 (124).
Overexpression of GLUT-3 enhances chemoresistance and survival of BTICs in glucose deficient microenvironment (124). Recently, Libby et al (125) proposed that overexpression of GLUT-3 is positively correlated with the invasion phenotype of GBM. They found that the C-terminal tail of GLUT-3 reduces invasion (125). Accordingly, the design of drugs targeting GLUT-3 could be improved to inhibit molecular functions outside its role in metabolism as a means to limit potential brain toxicity. This can be achieved by potentially targeting the C-terminal tail or the protein interactions driving GLUT-3 mediated invasion. While glucose transporters provide promising results in research laboratories, they are not yet targeted in clinical trials.
Hexokinase
Hexokinase 2 (HK2), the driver of the aerobic glycolysis, regulates the first step of glucose metabolism by converting glucose to glucose 6-phosphate. In GBM, HK2 promotes tumor progression by enhancing cancer cell growth, lactate production and chemoresistance (126,127). It also binds to the mitochondrial membrane and controls the release of cytochrome c, protecting cancer cells from apoptosis (128). Notably, depletion of HK2 induces the shift to oxidative glucose metabolism, impairs cancer cell proliferation, reduces angiogenesis and sensitizes GBM cells to chemoradiotherapy (129). Microarray analysis reveals that HK2 expression is negligible in normal cells but predominant in GBM (126). Therefore, targeting HK2 or its activity may be a novel therapeutic strategy to selectively kill cancer cells without damaging normal cells. Ketoconazole and Posaconazole are antifungals known to inhibit tumor metabolism. These drugs also selectively target HK2 in glioblastoma cells (129). Two recent recruiting early Phase I clinical trials are studying the delivery and activity of Ketoconazole and Posaconazole in brain tumors (NCT04869449 and NCT04825275, respectively).
Pyruvate kinase muscle (PKM)2
PKM consists of two isoforms PKM1 and PKM2 (130). PKM1 is mainly expressed in muscle and brain cells while PKM2 is highly expressed in embryonic cells, stem cells and cancer cells (131). PKM converts phosphoenolpyruvate to pyruvate during the last irreversible step of glycolysis (132). Mukherjee et al reported an overexpression of PKM2 and an isoform switching between metabolic enzymes PKM1 and PKM2 in GBM (133).
Three different forms of PKM2 exist in mammalian cells: A tetrameric form, known as the metabolic/glycolytic form and is involved in aerobic glycolysis, and less active monomeric and dimeric forms. The latter two forms promote GBM tumor growth (134,135) and protect it from apoptosis (136,137). This is achieved either by diverting the glycolysis pathway to an anabolic pentose phosphate pathway, or by activating the transcription of a number of oncogenes such as c-Myc and cyclin D (138). The ratio of monomeric/dimeric and tetrameric forms of PKM2 decides whether cells will undergo glycolysis or phosphate pathway (139). In addition, a study conducted by Sizemore et al (140) reveals that the ATM-PKM2-CtIP axis enhances DNA double-stand break repair efficiency and resistance to genotoxic damage caused by radiation. Overexpression of PKM2 is hence associated with a radio-resistant phenotype. Therefore, targeting PKM2 might be a promising therapeutic strategy for glioblastoma treatment.
Amino acid metabolism
Amino acid metabolism is another intriguing metabolism route important for GBM cells. Data suggest that GBM cells produce an increased pool of free amino acids associated with poor disease prognosis (141). Research provided evidence that the hypoxic stress in glioma and GBM cells increases protein catabolism (142,143). Under hypoxic conditions, and due to elevated levels of redox stress, hypoxic tumor cells maintain redox homeostasis that is dependent on increased glutathione synthesis. Thus, GBM cells want to maintain high levels of glutathione, consequently favoring conditions that are resistant to chemoradiotherapy (142). Arginine, asparagine, glutamine and lysine are among the relevant amino acids studied, however, the present review will discuss metabolic pathways of both glutamine and arginine that are targeted to therapeutically control cancer cell proliferation and spreading.
Glutamine metabolism
Glutamine is a non-essential amino acid that is highly concentrated in blood (144) and required for the tricarboxylic acid (TCA) cycle and the synthesis of nucleotide bases, amino acids, fatty acids and proteins. Glutamine metabolism regulates oxidative stress by regulating glutathione synthesis (145). Additionally, healthy neural cells in the brain synthesize glutamate, the main activator neurotransmitter, by the uptake of glutamine from astrocytes in the glutamine-glutamate cycle (146). Glutamine in the extracellular space is transported into neural cells through SNAT1 transporter where it is hydrolyzed by the glutaminase enzyme (GA) to form glutamate. Glutamate is then packed in synaptic vesicles and cleared during neurotransmission and transported into astrocytes via glutamate transporters GLT-1 and GLAST. In astrocytes, glutamine synthetase (GS) catalyzes the generation of glutamine from glutamate. The cycle is completed by the release of glutamine from astrocytes into the synaptic cleft by the SN1 transporter (147).
In GBM, glutamine is an important source for the TCA cycle and nucleotide and fatty acid synthesis, thereby sustaining tumor growth and stimulating glutathione synthesis (148). Concentrations of glutamine and its related metabolites are proportional to decreased cell survival (149). Due to developments in noninvasive image collection and analysis, in vivo glutamine measurement is now clinically feasible. Previous research demonstrated that glutamine imaging may have prognostic significance (150). In the line of the role of glutamine in cancer cell growth and promoting chemo-and radiotherapy resistance, targeting glutamine metabolism constitutes an attractive research area for the treatment of brain tumors.
Research strategies to target glutamine metabolism included targeting enzymes involved in their synthesis (GA and GS), or targeting transporters for glutamine/glutamate uptake (147,151–153). Modulating the GA enzyme was performed using the RNA interference approach or the use of allosteric inhibitors. GLS, a gene encoding GA, was targeted in GBM cell lines, patient derived GBM cells and mouse xenografts (147,154). Several research groups report a decreased cancer cell viability by silencing GLS, whereas GLS2 isoform shows an opposite effect as its overexpression reverses the aggressive GBM phenotype (155–157). These data suggest approached for a combinatorial therapeutic approach of both GLS inhibition and GLS2 overexpression thereby enhancing the antitumor effect. On the other hand, GS enzyme is also targeted, and its overexpression resulted in decreased proliferation and migration in rat glioma cell lines (158). Along with similar supporting data, GS is suggested to have an anti-glioma effect.
To date, targeting glutamine uptake by neural cells through silencing SNAT transporters has not shown significant effects on the proliferation of GBM cells (147). Conversely, targeting glutamate transporters such as GLT-1 and GLAST showed more promising results in GBM cell lines and xenografts [reviewed in details in (147)]. Another promising target is system xc- (SXC), a cysteine-glutamate exchanger that is shown to mediate >50% of glutamate transport in GBM cell lines (159). Notably, SXC is targeted and inhibited by an FDA approved sulfasalazine (SAS) for the treatment of bowl disease (159). While SAS resulted in increased cell death of GBM cells in vitro and in xenografts (160), Phase I/II clinical trials were terminated due to incidents of severe side effects (161). Research studies using SAS in gliomas, in combination with radiotherapy, are still in progress (162) and more studies are needed to reveal the effectiveness of the combined treatment.
There are several pharmacological strategies to inhibit glutamine metabolism (147,163). Telaglenastat (CB-839) is a GLS inhibitor currently being evaluated in a Phase I clinical trial (NCT03528642) combined with radiotherapy and TMZ for treatment of IDH-mutant astrocytomas and anaplastic astrocytomas. Collectively, several combinatorial strategies targeting various facets of glutamine cancer metabolism can be applied and this should lead to more research in this field (148).
Arginine metabolism
Arginine is a semi-essential amino acid synthesized from citrulline via urea cycle enzymes, argininosuccinate synthetase-1 (ASS1) and argininosuccinate lyase (ASL) (164). Cancer cells, which are characterized with a high proliferation rate, depend on exogenous arginine in their microenvironment (165). Therefore, reduced expression of urea cycle enzymes ASS1, ASL and OCT makes cancer cells auxotrophic and completely reliant on extracellular arginine sources (166,167). Arginine deprivation has been shown to promote cell death and impairs cell motility and invasion. Hence, arginine deprivation is a promising potential therapy target for selective destruction of tumor cells.
Arginine deprivation serves a role in enhancing endoplasmic reticulum stress and inducing the expression of genes involved in unfolded protein responses, thus resulting in cancer cell death (168). Furthermore, arginine deprivation reduces the extent of β-actin arginylation which disturbs cell cytoskeletal organization, alters cell adhesion and impairs cell migration and invasion (169). HuArgI (Co)-PEG5000 is human arginase I, characterized by the addition of two cobalt ions and polyethylene glycol that results in improved enzymatic catalytic activity, increased stability, decreased immunogenicity and lower dissociation constant at neutral pH (170,171). Promising effects were shown when using HuArgI (Co)-PEG5000 to target arginine auxotrophy in different tumor types (166,172–175). Our laboratory research found evidence that HuArgI (Co)-PEG5000 induces autophagy cell death in hepatocellular carcinoma (176), pancreatic carcinoma (173,176), acute lymphoblastic leukemia (174), renal cell carcinoma (177), prostate cancer (178), colorectal cancer (CRC) (172), ovarian carcinoma (175) and breast cancer (179). In GBM, we observed that HuArgI (Co)-PEG5000 induces autophagy-dependent cancer cell death. The addition of exogenous citrulline failed to rescue any of the GBM cell lines from arginine depletion-induced cytotoxicity, indicating a high level of arginine dependence (166). A recent recruiting Phase I clinical trial (NCT04587830) is currently assessing the safety and tolerability of ADI-PEG 20, an arginine deprivation agent, in combination with radiotherapy and TMZ in patients newly diagnosed with GBM (180). Taken together, targeting GBM cells using arginine depletion is a potent and selective potential treatment for GBM.
5. Other targeted molecules
Other molecules such as human telomerase reverse transcriptase (hTERT), ATRX and protein kinase C, have also gained attention as potential targets for GBM cancer therapy. This is due to the finding that alterations in these molecules enhance GBM cells survival and progression.
Telomerase
hTERT
Telomerase is a ribonucleoprotein that consists of two subunits: i) the protein catalytic hTERT that mediates reverse transcriptase activity and ii) the human telomerase RNA template. Telomerase is mainly active in embryonic cells, stem cells and precursor cells. Notably, studies show that hTERT expression is upregulated in glioblastoma (181,182). In addition, ~80% of primary glioblastoma cells harbor a mutation or methylation in the hTERT promoter (183). Genomic sequencing revealed a high incidence of mutually exclusive point mutations C228T and C250T in hTERT promotor of glioblastoma cells (183). Arita et al (181) show that hTERT expression is 6.1 times higher in tumors carrying these mutations than in wild-type tumors and therefore leads to the upregulation of TERT. Upregulation of hTERT enhances telomerase activity and promotes an immortal phenotype (183). In addition, hTERT mutations are associated with poor prognosis and a multifocal invasive phenotype in GBM (184,185). Thus, hTERT promotor mutation can be recognized as a novel biomarker and therapeutic target molecule for GBMs (184,186,187). Currently, two immunization strategies are currently targeting hTERT in clinical trials. A Recruiting Phase II/III study (NCT03548571) is investigating the efficiency of immunization with IMP dendritic cells that are transfected with mRNA hTERT, as well as autologous tumor stem cells and survivin, compared with standard adjuvant chemoradiotherapy. Additionally, an active, not recruiting Phase I/II study, (NCT03491683) is evaluating the safety of INO-5401 (a combination of three separate DNA plasmids targeting hTERT, Wilms tumor gene-1 antigen, and prostate-specific membrane antigen) in patients with a glioblastoma tumor with an unmethylated MGMT promote.
Alpha thalassemia/mental retardation syndrome X-linked (ATRX)
ATRX is a DNA helicase/ATPase that belongs to the SWI2/SNF2 family (188). It is involved in chromatin remodeling and telomere maintenance. ATRX mutations occur in 57% of secondary glioblastoma and are associated with IDH1 and p53 mutations (189). ATRX loss of function mutation triggers a telomerase length mechanism also known as ‘alternative lengthening of telomeres’ that maintains telomere length, thus enabling cancer cells to escape cell senescence and promoting tumor growth (190). In addition, ATRX mutations impair non-homologous end joining and pDNA-PKcs recruitment, causing genetic instability and leading to DNA damage (190). Therefore, ATRX is another potential target for GBM treatment, however, it is not yet targeted in clinical trials.
Protein kinase C (PKC)
PKC is a serine threonine kinase that serves a crucial role in GBM growth and progression. PKC consists of 11 isoforms and is divided into three subfamilies: Classical, novel and atypical classes (191). Classical PKCs consist of two members, PKCα and PKCβ (192). Leirdal et al (193) show that PKC enhances GBM survival via MAK-ERK12 pathway. PKCα induces GBM progression via PKC-progesterone pathway (194). Once stimulated with 12-O tetradecanoylphorbol-13-acetate or lysophosphatidic acid, PKCa translocates to the nucleus, enhances progesterone receptor phosphorylation and promotes the migration and invasion capacity of GBM (194,195). PKCb serves an essential role in enhancing angiogenesis, thus facilitating GBM progression (196). Moreover, PKCβ leads to GBM cell death. Conversely, Liu et al (197) show that in an orthotopic mouse xenograft model, PKCβ II expression suppresses GBM tumor growth and extends mouse survival by inhibiting YAP/TAZ. Thus, PKCβ has two opposite roles in GBM.
The novel PKC subfamily contains four members: PKCδ, PKCɛ, PKCη and PKCθ. Inhibition of PKCδ suppresses tumor spheroid formation in vitro and tumor development in vivo (198). In glioblastoma, PKCδ is involved in tumor initiation (199), cancer cell migration and invasion (194,200) and radio-chemoresistance (199). Activation of PKCδ phosphorylates glycerol-3-phosphate dehydrogenase enzyme and serves an essential role in glucose metabolism and phospholipid synthesis (201). PKCɛ, on the other hand, is overexpressed in glioblastoma cell (202) and controls tumor survival (203) cancer cell adhesion and motility via activation of the ERK pathway (204,205). PKCη promotes GBM proliferation (206,207) and reduces cell sensitivity to radiotherapy (208).
Atypical PKCs ɩ and ζ are involved in GBM viability, migration, and invasion (209–211). Baldwin et al (211) show that inhibition of PKCi increases the activity of Rho B that, in turn, improves actin fiber formation and reduces GBM cell migration and invasion. Similarly, Guo et al (210) found that the suppression of PKCz enhances actin polymerization and reduces GBM cell migration and invasion.
Enzastaurin is a kinase inhibitor that particularly inhibits protein kinase C, thus decreasing tumor growth and cell proliferation (212). Results from recent clinical studies (Phase II NCT00586508 and Phase III NCT03776071) did not show improved PFS compared with bevacizumab and other therapeutic agents (212,213).
6. Challenges
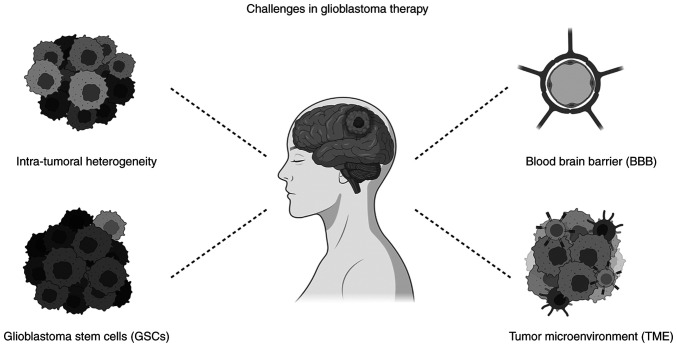
Targeting intrinsically altered molecules that trigger GBM malignant phenotype is an innovative therapeutic strategy that aims to destroy tumor cells without damaging normal cells. However, despite the promising in vitro results, this novel therapy faces various barriers in vivo (Fig. 2).
Figure 2.
Challenges for glioblastoma treatment. Intra-tumoral and tumor microenvironment heterogeneity, the blood brain barrier and the glioblastoma stem cells are barriers against effective therapeutic strategies for glioblastoma patients. They also confer resistance to drugs and may obstruct proper drug deliver. Created with Biorender.com.
Intra-tumoral heterogeneity
One reason behind the complexity of GBM management is that the genetic profiles of recurrent glioma cells and the initial cancer derived from the same patient are different (214). Thus, the plasticity and heterogeneity of GBM cell populations can explain the failure of clinical trials based on monotherapy in vivo. Clones of the genes that are identified by different number of chromosomal sets, exhibit a high level of heterogeneity in the expression of cell membrane markers. This leads to phenotypic heterogeneity at the level of the population providing an advantage for tumor survival (215). Neftel et al (216) found that gliomas rarely have identical proportions of single-cell transcriptomic states and this proportion is skewed by genetic associations that are specific to each patient leading to abundant heterogeneity. Therefore, it is recommended to carefully weigh the genetic, epigenetic, and molecular profile of each patient. Single-cell RNA-sequencing reveals the presence of various molecular and genetic cell subtypes within the same patient biopsy (215,217). These subtypes can change progressively and become more resistant to chemoradiotherapy (214). Neftel et al (216) describe multiple cellular states and determine a correlation between the cellular states, plasticity and genetic signature of tumor cells. They define four cellular states of malignant GBM tumor cells: Neural-progenitor-like, oligodendrocyte-progenitor-like, astrocyte-like and mesenchymal-like states. These states are characterized by the overexpression of CDK4, PDGPRα, epidermal growth factor (EGF) and NF1 alterations, respectively. Neftel et al (216) also showed the co-existence of multiple cellular states and their transition potential within the same patient sample and evidenced the ability of a single cell to generate all cellular states. Amplification of the EGFR and PDGFRA genes that specifically encode for RTKs has been recognized in GBM tumors (218). Snuderl et al (219) established that the amplification of different RTKs was infrequently found in the same region of the tumor; yet, different RTKs (MET, EGFR and PDGFRA) were amplified in diverse subpopulations of the GBM tumoral cells.
Another advancing technology that expands the understanding of the heterogeneity of GBM is the whole genome amplification (WGA) methods. WGA helps to identify different imbalances on the level of the chromosomes (218). In one study, Nobusawa et al (220) applied the WGA method on GBM samples (14 different primary samples) from two to five locations within each tumor. They recognized not only common modifications between all studied locations, but also changes that were specific to a certain region among the studied GBM tumor samples. A later study targeted 33 cancer genes with single molecular inversion probes and reported regional mutational heterogeneity among different patients with GBMs, as well as among the same patient (221).
Recently, Bhaduri et al (222) discovered the existence of ‘cancer stem cell likes’ that are able to give rise to heterogeneous cancer cell populations within the same tumor (discussed in details in the section below). In their study, Pang et al (223) discovered a path characterized by the progressive renovation of the GBM stem cells to reach an invasive state, known as the ‘stem-to-invasion path’. A gradual expression of the invasion signatures linked with GBM, failure in expressing the markers of GBM stem cells and different molecular cascades related to the invasion path were detected, along with different key factors such as transcription factors and long noncoding RNAs (223). Such findings help the understanding of the progression of glioma tumors and thus facilitate screening for efficient methods to therapeutically target GBM.
Efficient treatment options necessitate building a model the mimics a patient's GBM biology. Jacob et al (224) generated a new organoid model from patient-derived primary cancer cells that conserved the original tumor's histological, genetic and molecular signatures in addition to its cellular heterogeneity. In order to generate patient-derived glioblastoma organoids (GBOs), Jacob et al (224) obtained tissues along the tumor margin. These tissues were refined from necrosis and surrounding brain tissues, dissected and cultured in optimized serum-free GBO medium. When allowed to grow larger, GBO created gradients of hypoxia reflecting a hallmark of GBM. Immunohistological analyses reported markers for glia, immature neurons, neural progenitors and glioma stem cells, resembling the cellular composition of parental tumors. Also, single-cell transcriptome analysis confirmed cell-type heterogeneity and suggested that specific elements of the TME were preserved. Upon transplantation into adult rodent brains, GBOs showed reliable engraftment and aggressive infiltration (224). This suggested that a biobank of patient-derived GBO can be used as a prescreening drug model to investigate drug response and to evaluate its effectiveness on patient cells.
Glioblastoma stem cells (GSCs)
‘Cancer stem cell likes’, also referred to as ‘cancer stem cells’ (CSCs) are self-autonomous units that play a significant role in tumor initiation and growth as well as therapeutic resistance. GSCs are derived from the malignant transformation of normal neural stem cells or the dedifferentiation of tumor cells after radiotherapy or chemotherapy (225,226). GSCs exhibit prolonged proliferation and are able to metastasize and suppress anti-inflammatory responses and confer resistance to therapeutic treatments (227). Thus, GSCs are important factors that limit clinical options and treatments of GBM (228,229). Numerous studies that are currently being undertaken to target GSC were made possible by improved understanding of the biology of GCS (24). GSCs represent a hot topic for improved GBM treatment.
Some studies aim to target and kill GSCs directly by targeting stem cell biomarkers. For instance, GSCs that are CD133 positive are important for sustaining and spreading GBM (230). Patients with GBM have a higher survival rate when CD133 positive stem cells are eliminated (231). CD133 stem cells may be eliminated by BMI1 gene suppression, an oncogene involved in the control of stem cell self-renewal and differentiation (232). Inducing apoptosis in GSCs can be also achieved by targeting the signaling pathways that play a role in their self-renewal. For instance, the Wnt signaling pathway has been targeted since 2014 as it is involved in neural stem cell development (233). Targeting glycogen synthase kinase-3β (GSK-3β) and β-catenin is suggested to inhibit the Wnt pathway in vitro (234). AR-A01441, LiCl and SEN461 are examples of GSK-β inhibitors that lead to increased apoptosis of GBMs cells (24). Notably, Celecoxib is a β-catenin inhibitor that is already studied in clinical trials, however, no promising results have been reported yet (Table VI).
Table VI.
Therapeutic agents targeting glioblastoma stem cells in interventional clinical trialsa.
| Therapeutic target | Therapeutic agent | Therapeutic strategy | Cancer condition | Phase | Status | (Refs.) |
|---|---|---|---|---|---|---|
| CD133 positive stem cells | ICT-121 DC vaccine | Four intradermal injections of the autologous ICT-121 dendritic cell vaccine | Recurrent glioblastoma | Phase I NCT02049489 | Completed | |
| Activated T cells | ATC against glioma CSC antigens recruiting administered intravenously at one timepoint | Recurrent glioblastoma | Phase I NCT05341947 | Not yet | ||
| Wnt pathway (b-catenin) | Celecoxib | TMZ alone or in combination with Thalidomide and/or Isotretinoin and/or Celecoxib after radiotherapy | Glioblastoma multiforme | Phase II NCT00112502 | Completed | (353) |
| Combination of Celecoxib, 6-Thioguanine, and Xeloda (Capecitabine), with TMZ or Lomustine (CCNU) | Recurrent anaplastic glioma and glioblastoma multiforme | Phase II NCT00504660 | Completed | |||
| TMZ, Thalidomide, and Celecoxib after radiotherapy | Patients with newly diagnosed glioblastoma multiforme | Phase II NCT00047294 | Completed | (354) | ||
| Coordinated undermining of survival paths by 9 repurposed drugs (aprepitant, auranofin, captopril, celecoxib, disulfiram, itraconazole, minocycline, ritonavir and sertraline) combined with metronomic TMZ | Recurrent glioblastoma | Phase I/II NCT02770378 | Completed | (355) | ||
| Thalidomide, Celecoxib, and combination chemotherapy | Relapsed or refractory malignant glioma | Phase II NCT00047281 | Completed | |||
| Notch pathway (g-secretase) | RO4929097 | RO4929097 in combination with Cediranib Maleate | Advanced solid tumors including glioblastoma | Phase I NCT01131234 | Completed | |
| RO4929097, TMZ, and radiotherapy | Advanced solid tumors including glioblastoma | Phase I NCT01119599 | Completed | |||
| Hedgehog (SHH) pathway (SMO) | Vismodegib (GDC-0449) | Monotherapy | Recurrent glioblastoma multiforme | Phase II NCT00980343 | Completed | |
| Umbrella protocol of molecularly matched targeted therapies (APG101, Alectinib, Idasanutlin, Atezolizumab, Vismodegib, Temsirolimus, Palbociclib) in combination with radiotherapy | Glioblastoma without MGMT promoter methylation | Phase I/II NCT03158389 | Recruiting | |||
| Sonidegib (LDE225) | Oral LDE225 in combination with BKM120 | Advanced solid tumors including recurrent glioblastoma multiforme | Phase I NCT01576666 | Completed | ||
| Molecularly-Driven doublet therapy for Gemcitabine, Ribociclib, Sonidegib, and Trametinib | Children and young adults with recurrent brain tumors | Phase I NCT03434262 | Recruiting | |||
| Glasdegib (PF-04449913) | In combination with TMZ | Newly diagnosed glioblastoma | Phase I/II NCT03466450 | Active, not recruiting | ||
| STAT pathway | Napabucasin (BBI608) | In combination with TMZ | Recurrent or progressed glioblastoma | Phase I/II NCT02315534 | Completed | |
| WP1066 | Monotherapy | Malignant glioma including recurrent glioblastoma | Phase I NCT01904123 | Completed |
Data acquired from the U.S. National library of medicine (http://clinicaltrials.gov, accessed on 10 August 2022). Terminated studies are not included. ATC, activated T cells; CSC, cancer stem cells; DC, dendritic cells; MGMT, O6-methylguanine DNA methyltransferase; STAT, signal transducer and activator of transcription; TMZ, Temozolomide.
The Notch pathway is another targeted pathway for GBM treatment as it is involved in resistance to immunotherapies (235). γ-secretase is an enzyme that mediates Notch's cleavage and translocation to the nucleus. Therefore, γ-secretase is suggested as a plausible target for Notch inhibition (236). In fact, several clinical trials are testing the effect of RO4929097, a γ-secretase inhibitor, as a monotherapy for GBM treatment or in combination with other treatments. In addition to the aforementioned pathways, Hedgehog (SHH) pathway is also targeted to inhibit the renewal of GSCs, as it confers resistance to conventional GBM treatment. SHH pathway is targeted by inhibiting smoothened (SMO), its downstream effector (24). Vismodegib is one of the SMO inhibitors used in several clinical trials and it showed enhanced PFS when used before surgical resection (NCT00980343). Finally, the STAT3 pathway is also targeted to control neural stem development (237). WP1066 and Napabucasin (BBI608) are STAT3 inhibitors currently in Phase I (NCT01904123) and II (NCT02315534) clinical trials, respectively (no results are reported yet).
Notably, GSCs can be targeted by targeting the mitochondria or via specific antibiotics. For instance, oxidative phosphorylation (OXPHOS) is a major source of ATP in a number of types of cancer and plays a significant role in carcinogenesis and tumor growth (238). GSCs are OXPHOS-dependent GBM cells that require mitochondrial translation (239). The bacterial antibiotic quinupristin/dalfopristin (Q/D) blocks mitochondrial translation, which not only inhibits the formation of GSCs but also disrupts the cell cycle and increases apoptosis (239). These results suggest that Q/D may be taken into consideration for the treatment of GBM and that reducing mitochondrial translation may be examined to inhibit GSC growth. On the other hand, Salinomycin, is a K+ ionophore antibiotic that causes lysosomal iron sequestration, and leads to the production of ROS and lysosome membrane permeabilization (240). Salinomycin is shown to favorably kill GSCs and other types of CSCs (240,241). Although clinical trials examining the possible efficiency of Salinomycin in the treatment of glioblastoma have yet to be published, Salinomycin and its derivatives are now being researched intensively as anti-CSCs treatments for a variety of malignancies (241). Collectively, these results imply that focusing on GSCs is a promising approach for treating GBM more effectively.
Blood Brain Barrier (BBB)
The BBB forms an interface between the brain and the systematic blood circulation. It consists of endothelial cells (ECs) that line blood vessels, are surrounded by astrocytic perivascular pseudopodium and pericytes, and interconnected by neuronal ending and microglia (242,243). Under normal physiological conditions, ECs, connected by tight junction proteins, create a compact barrier. In addition, efflux transporters, carried by BBB cells, regulate molecular and cellular transport across the BBB and protect the brain from toxins, xenobiotics, and pathogens (243). The development and progression of GBM impair the integrity and function of the BBB, resulting in a heterogenous resistant barrier known as Blood Brain Tumor Barrier (BBTB). In glioblastoma, the BBTB is characterized by the downregulation of tight junction proteins such as claudin and occludin and overexpression of efflux transporters (244–247). These tight junction proteins increase the permeability of the BBTB, enhance paracellular transport and thus facilitate the penetration of cells and small molecules to the interstitial space of the brain. Accumulation of such molecules might have neuro-toxic effects. In addition, the heterogeneous permeability of the BBTB caused by the ‘leaking’ structure of the new blood vessels as against the compact structure of the local blood vessels leads to unequal distribution of the drug (248–250). Several FDA-approved drugs, including chemotherapy drugs, have affinity for efflux transporters, including P-gp and BCRP (251–256). Hence, overexpression of efflux transporters in glioblastoma restricts drug delivery to the brain.
In the line of BBB heterogeneity and overexpression of efflux transporters, cellular, chemical and physical approaches are being investigated to enhance drug delivery through the BBTB. One of these advanced approaches is the nanomaterial system which is based on different types of nanocarriers including, polymer-based, viral, drug-conjugated, lipid-based and inorganic nanoparticles (NPs) (257). NPs are usually loaded with drugs and are known to have a small size within a range between 1–100 nm (258). Budama-Kilinc et al (259) used a nanoparticle system based on a poly (ε-caprolactone) to deliver a tripeptide, glycyl-L-histidyl-L-lysine (GHK), to the GBM cells. GHK is known as a natural growth modulating tripeptide in vitro; thus, it decreases the viability of GBM cells to ~65%. Another study was based on a nanobubble system that consists of NPs made up of iron and platinum. NPs were either surface-functionalized with transferrin to target the glioma cells or loaded with doxorubicin (DOX) in a mouse model. These NPs resulted in reduced growth of the tumor by ~70% (260). On the other hand, Ambruosi et al (261) used a rat glioma model to test poly (n-butyl cyanoacrylate) (PBCA) as a potential therapy for GBM. PBCA NPs were loaded with DOX chemotherapeutic drug and coated with polysorbate 80 (surfactant). NPs successfully delivered the drug across the BBB and increased the median survival rate by 35% among tested rats living for the whole time of the study period.
On the molecular level, transferrin receptor (TfR) is a protein commonly targeted to enhance the delivery of therapeutical drugs through the BBB (262). Poly (lactic-co-glycolic acid) NPs loaded with TMZ and coated with TfR-monoclonal antibodies showed higher internalization in GBM cells compared with control cells (263). Kuang et al (264) examined the use of polymer-based NPs coupled to a peptide having the ability to target TfR on both the BBB and GBM cells to transport doxorubicin and RNA. Results showed higher efficiency in in vitro tumor targeting and cellular uptake, with a decline in the tumor growth leading to improved median survival time.
Focused ultrasound therapy (FUS) is another method used to facilitate transport through the BBB. It is a non-invasive and image-guided method that enhances the efficacy of drug delivery to GBM cells (265). Wei et al (266) showed an enhancement in the local delivery of TMZ to tumor cells through the FUS-mediated disruption of the BBB. Results reported an increase in the OS of rats with experimentally induced gliomas. Notably, MRI-guided FUS (MRgFUS) was evaluated to attain enhanced tissue delivery of TMZ in mice (267), liposome-encapsulated doxorubicin in rats (266) and cisplatin-conjugated gold NPs in mice (268). This method is advantageous as it reduces the systematic toxic effects of the drugs used (low doses of therapeutic drugs can be used) (269) and leads to temporary opening of the BBB rather than long-term opening associated with BBB disruptions (270). These studies show that the use of different NPs (nanomaterial system) or FUS has the potential to enhance the efficiency of therapeutical drug delivery in GBM management.
TME
The TME comprises fibroblasts, endothelial cells, immune cells, ECM and soluble factors surrounding the tumor. It provides biophysical and biochemical support for the tumor and contributes significantly to tumor initiation, growth, progression, and resistance to therapy (271).
GBM is classified as a hypoxic solid tumor where hypoxia is considered one of the non-cellular TME components. The hypoxic microenvironment is a major inducer of angiogenesis; a process in which ECs branch from preexisting small vessels to form sprouts of capillaries. Angiogenesis thus promotes tumor growth by supplying cancer cells with O2 and nutrients and facilitates their metastasis (272,273). Moreover, hypoxia induces the activation of a number of cellular processes that promote the migration, invasion and radio-chemoresistance of GBM cells (274–280). This is due to the overexpression of VEGF/HIF-1α protein. Therefore, and in order to control the hypoxic environment, researchers developed Bevacizumab, an FDA approved anti-VEGF antibody, for GBM treatment. Bevacizumab showed encouraging results as monotherapy or when combined with traditional treatment, as reviewed in the study by Cruz Da Silva et al (24).
GBM secretes a wide range of chemo-attractants and cytokines that recruit immune cells to the TME. For instance, GBM stem cells recruit macrophages and promote their transition from the antitumor (M1) to the pro-tumor (M2) phenotype (281). Tumor associated macrophages (TAM) are the most abundant cells inflating tumor lesion and accounting for ≤40% of the tumor mass. Indeed, the disrupted structure of BBTB facilitates the infiltration of TAM. In addition, GBM cells inhibit the proliferation of cytotoxic T cells and activate regulatory T cells (282). Occurrence of immune cells in the TME leads to the formation of an extensively immune-suppressive microenvironment and enhances GBM cell migration and invasion.
At present, researchers are investigating new therapeutic approaches that aim to reactivate the immune system against GBM and restore the immune surveillance in GBM. These approaches include reversing the polarization of M2 macrophages to M1, targeting immune checkpoints using checkpoint inhibitors to rebut T cell responses, and implementing cytokine therapy. Yang et al (283), discovered that the dual-targeting of IL-6, which stimulates different macrophages activation in GBM, and CD40 may help in reversing tumor immunosuppression mediated by macrophages thus informing the immunotherapy based on T-cells against GBM. In their study, it was shown that CD40 stimulation and IL-6 inhibition reverse tumor immunosuppression mediated by macrophages and increase the survival rates of the animals in GBM models (283). Similar findings supported the use of macrophages-based therapies as effective therapeutic approaches when targeting GBM.
Cytokine therapy represents a hot topic for immunotherapeutic approaches since cytokines are molecular messengers of innate and adaptive immunity (284). This approach promotes human natural killer (NK) and T cell activity, survival and proliferation thus coordinating immune responses against malignancies (285). TGF-β, IFN-α, IFN-γ and IL-12 and other cytokines have anti-tumor effect in GBM, thus reducing tumor size and inhibiting carcinogenesis at early stages (286). Tumoral injection of Ad-RTS-hIL-12 in combination with veledimex (VDX, an hIL-12 activator) and an FDA approved antibody cemiplimab was tolerated well and exhibited increased circulating cytotoxic T cells in patients with recurrent or progressive GBM (Phase II NCT04006119). IFN-β immunotherapy in combination with TMZ was also evaluated in clinical trials (287). Han et al (288), used cytokine-induced killer therapy with TMZ on patients with GBMs and reported higher PFS rates in treated vs. control untreated group. The therapeutic efficacy of various cytokines in the treatment of gliomas is still being investigated. The cytokines' safety and tolerance are being defined, and some are thought to be a promising supplement to conventional treatments (289).
Glioma cells frequently employ a variety of strategies to avoid immune surveillance. Therefore, several therapeutic approaches have been developed to activate immune responses in the TME. Cytotoxic T lymphocyte-associated protein 4 (CTLA-4), programmed cell death protein 1 (PD-1), and PD ligand 1 (PD-L1) are immune checkpoint proteins targeted for monoclonal antibody inhibition (290,291). Monoclonal antibodies act by freeing T lymphocytes from their negative regulation thus suppressing cancer evasion of the immune system (292,293). Several immune checkpoint inhibitor antibodies have been approved by the FDA for the treatment of several types of cancer (294). Neoadjuvant therapy with pembrolizumab, an anti-PD-1 monoclonal antibody, resulted in improved OS and PFS in patients with recurrent surgically respectable GBM (Phase II NCT02337686) (43). Wang et al (295) demonstrated that genetically engineered multifunctional NK cells (CD73.mCARpNKs) can lead to efficient anti-GBM activity mainly by bypassing the heterogeneity and the immunosuppressive structures of GBM tumors. Immune responses in the TME can be also stimulated by adoptive cell therapy (ACT). This method involves isolating autologous T lymphocytes with anti-tumor activity from cancer patients, expanding them ex vivo, and transferring amplified engineered T cells to the patients (296,297). Among the different ACT methods developed, chimeric antigen receptor (CAR)-T cell gene therapy has attracted the most attention for killing cancer cells. CAR-modified T cells may identify different types of antigens regardless of how they are presented on MHC molecules (298). To date, there are currently 24 clinical trials examining the efficacy of CAR-T cell treatment for GBM according to Clinicaltrials.gov (accessed on 10 August 2022; Table VII). Several CARs have been generated to specifically target GBM such as EGFRvIII, HER2, IL13R2, and CD70 (290). Collectively, studies using CAR-T cell immunotherapy exhibit encouraging results. However, obstacles such as tumor heterogeneity, the heterogenous expression of antigens, and the role of T cells at the tumor's sites make it challenging to efficiently eradicate the tumor (290).
Table VII.
CAR-T cells and cancer vaccines applied in interventional clinical trials for the treatment of glioblastomaa.
| A, CAR-T cells | ||||||
|---|---|---|---|---|---|---|
|
| ||||||
| Therapeutic agent | Specificity | Therapeutic strategy | Cancer condition | Phase | Status | (Refs.) |
| CAR-T Cell Receptor Immunotherapy | EGFRvIII | In combination with Aldesleukin, Fludarabine or Cyclophosphamide | Malignant gliomas expressing EGFRvIII | Phase I/II NCT01454596 | Completed | (356) |
| CAR-T-EGFR-IL13 Ra2 cells | EGFR epitope 806 and IL13Ra2 | CART-EGFR-IL13 Ra2 cells following lymphodepleting chemotherapy | Recurrent glioblastoma | Phase I NCT05168423 | Not yet recruiting | |
| CAR-T-EGFRvIII cells | EGFRvIII | In combination with pembrolizumab (PD-1 Inhibitor) | With newly diagnosed, MGMT-unmethylated glioblastoma | Phase I NCT03726515 | Completed | |
| CD147-CAR-T Cells | CD147 | Monotherapy | Recurrent glioblastoma CD147 Positive | Early Phase I NCT04045847 | Unknown | |
| B7-H3CAR-T | IFN-1 | Monotherapy | Recurrent glioblastoma multiforme | Phase I NCT05474378 | Completed | |
| NK-92/5.28.z Cells | NK-cells | With the anti-PD-1 antibody Ezabenlimab | Recurrent HER2-positive glioblastoma | Phase I NCT03383978 | Recruiting | (308) |
|
| ||||||
| B, Cancer vaccine | ||||||
|
| ||||||
| Therapeutic agent | Specificity | Therapeutic strategy | Cancer condition | Phase | Status | (Refs.) |
|
| ||||||
| DCVax®-L | DC based vaccines | Monotherapy | Newly diagnosed glioblastoma multiforme | Phase III NCT00045968 | Active, not recruiting | (357) |
| ICT-107 | DC based vaccines | After surgery and chemotherapy | Glioblastoma multiforme | Phase II NCT01280552 | Completed | |
| ACTIVATe (PEP-3 vaccine) | EGFRvIII | In combination with sargramostim and TMZ | Newly diagnosed glioblastoma multiforme | Phase II NCT00643097 | Completed | (28) |
| Rindopepimut (CDX-110, ACT IV vaccine) | EGFRvIII | With GM-CSF, TMZ and KLH | Newly diagnosed glioblastoma | Phase III NCT01480479 | Completed | (358) |
| With GM-CSF, Bevacizumab and KLH | Recurrent EGFRvIII-positive glioblastoma | Phase II NCT01498328 | Completed | (359) | ||
| Personalized Neo Antigen Vaccine | Personalized neoantigens | After radiotherapy, with Pembrolizumab | Newly diagnosed glioblastoma multiforme | Phase I NCT02287428 | Recruiting | (318) |
| GAPVAC (APVAC1 and APVAC2) | Personalized mutated and unmutated tumor antigens | With immunomodulators concurrent to TMZ | Newly diagnosed glioblastoma | Phase I NCT02149225 | Completed | (319) |
| AV-GBM-1 | Following primary surgery plus concurrent chemoradiation | Newly diagnosed glioblastoma | Phase II NCT03400917 | Active, not recruiting | ||
Data acquired from the U.S. National library of medicine (http://clinicaltrials.gov, accessed on 10 August 2022). Terminated studies are not included. CAR-T, chimeric antigen receptor-T cell; EGFR, epidermal growth factor receptor; GM-CSF, granulocyte-macrophage colony-stimulating factor; IL, interleukin; INF-1, type I interferon; KLH, Keyhole Limpet Hemocyanin; MGMT, O6-methylguanine DNA methyltransferase; NK, natural killer cells; PD-1, programmed cell death protein 1; TMZ, Temozolomide.
7. Discussion
Glioblastoma is characterized with high cell proliferation and neovascularization leading to tumor infiltration reaching crucial structures in the brain. This increases the possibility of tumor recurrence, scoring low survival rates (299,300). Therefore, more effective treatment procedures are required to extend the lives of patients. Conventional treatments such as radiotherapy and chemotherapy are regarded as a two-edged sword. This is because these treatments are frequently accompanied with normal tissue damage or genetic changes, nonspecific drug distribution and multidrug resistance. However, chemoradiotherapy promises improved survival and quality of life that may outweigh their disadvantages; thus, these modalities remain the primary cancer weapons. Substantial research has been done, and continues to be done, to develop novel therapeutic approaches that are less toxic for normal tissues and endow increased survival and improved quality of the lives of patients. Researchers have discovered genetic and subsequent molecular changes influencing critical pathways that cause aggressive behavior of tumors (24,214). Therefore, targeting these altered molecules and pathways is seen as a promising new approach to GBM treatment.
Over the previous few decades, biomarker-driven methods, such as small molecule inhibitors and monoclonal antibodies, have been developed. These methods aim to target key altered players that are directly correlated with tumor progression and invasion. EGFRs constitute one of the mostly targeted molecules in clinical trials to treat patients with GBMs (Table I). However, targeted therapy has shown little to no promising results due the intra-tumoral heterogeneity of GBM and drug resistance (216,218). Tumor heterogeneity could be a plausible explanation for GBM resistance to EGFR-targeted treatments. Moreover, resistance to EGFR therapy can also be caused by upregulation of redundant RTKs and downregulation of EGFR downstream molecules (218). Consequently, co-targeting of EGFR and other RTKs such as c-MET could offer a solution as seen in vivo (301). Multi-targeted therapeutic approaches offer an advantage where drugs can target numerous important nodes for GBM growth and progression, compensating for the inefficiency and quick acquisition of resistance seen with monotherapies (302). Multi-targeted therapeutic approaches also include the administration of multi-kinase inhibitors (303,304). Moreover, more complicated pathways have been identified and targeted in vitro. In-depth-investigation of these pathways helps to identify novel drugs.
Glioblastomas are intrinsically immunologically ‘cold’ tumors due to presence of few T cells but predominant pro-tumor immune cells in the TME, constituting a common reason for drug resistance (305). Immunotherapies thus aim to use a patient's immune system to re-direct immune cells against a tumor. Researchers have established several other immunotherapeutic strategies to target patients with GBMs with immunostimulatory conditions. These include immune check-point inhibition, autologous stimulated lymphocytes, cytokine therapy and dendritic cell therapy (306). Except for certain cases, immunotherapy in glioblastoma clinical trials has been disappointing overall. CAR-T cell therapy, for instance, faces a significant hurdle since glioblastoma tumors can rapidly adapt via antigen escape (307). CAR-NK cell therapy, on the other hand, offers an advantage as it can be administered to an HLA-mismatched patient, and Phase I clinical trial is showing promising preliminary results (NCT03383978) (308). Yet, NK cell expansion and manufacturing are still time and money consuming (309). Akin to targeted therapy, tumor heterogeneity is also a limiting factor for immunotherapy. Moreover, combing immunotherapy with chemotherapy may also lead to drug resistance (310). Anti-inflammatory corticosteroids, for example, may counteract the therapeutic effects of immunotherapy (311). Additionally, glioblastoma cells can adapt to immune checkpoint inhibition by increasing the expression of alternative checkpoints (312). The immunosuppressive TME is another limitation for immunotherapy, thus, combinatorial immunotherapy approaches may overcome this issue (313,314). In fact, preclinical research on combination immunotherapy yielded promising outcomes [reviewed in (314)]. The most successful approaches are those that influence the cancer-immunity cycle at different times and/or sites, leading to the activation of immune response and the inhibition of immunosuppressive elements (314).
Cancer vaccines are one of the very promising immunotherapies and are being thoroughly researched as a method to reduce tumor development and eliminate tumor cells in cancer patients. Peptide-based, dendritic cell (DC)-based, and personalized vaccines are being explored as potential vaccine therapies in GBM (305,315) (Table VII). Cancer vaccine design depends on selecting an ideal antigen specifically expressed and present on all cancer cells, highly immunogenic and essential for cancer cell survival (316). These antigens are therefore processed by DCs aiding in recruiting and activating CD8+ and CD4+ T cells that will promote tumor eradication (317). DCVax®-L and ICT-107 are DC-based vaccines tested in clinical trials for safety, tolerance and efficacy in patients with GBMs (NCT00045968 and NCT01280552 respectively). Personalized vaccination is now made possible using multi- epitope-based patient-specific vaccine to target neoantigens only (Phase I NCT02287428) (318) or both neoantigens and unmutated tumor-specific antigens (Phase I NCT02149225) (319). Corresponding results reported neoantigen-specific CD4+ and CD8+ T cell responses migrating into the tumor with increased number of infiltrating T cells (318,319). AV-GBM-1 is another personalized cancer vaccine based on autologous dendritic cells full of autologous tumor neoantigens. Phase II clinical study (NCT03400917) reported an enhanced PFS (320) demonstrating a promising positive effect of vaccination in patients with GBM.
Most of the current therapeutic strategies need to overcome the obstacles of drug delivery due to BBB, tumor and TME heterogeneity and abundant GSC niches. Burkhardt et al (321) propose promising results for an intra-arterial brain administration of Bevacizumab after temporary destruction of the BBB by mannitol. Followed by intravenous administration, their technique is able to overcome the poor intracerebral availability of the drug and enhanced PFS in patients with recurrent GBMs. Several studies suggest that molecular subgroups exist within histologically similar GBM tumors (322,323). Research into the dynamics of various glioblastoma subtypes would help understand the survival features of the TME. This highlights that future treatment options should no longer be based exclusively on morphological assessments, but must now take molecular and cellular heterogeneity into account. Furthermore, the plasticity of GBM cells caused by the bidirectional communication between GSCs and their differentiated counterparts complicates heterogeneity (227). These two populations respond differently to radio-, chemo- and targeted therapies. Thus, targeting both components should be well considered during the course of treatment.
Due to genetic heterogeneity and tumor growth, single-target therapy generates recurrence and consequent resistance to initial treatment. Therefore, therapeutic strategies are implementing targeted therapy along with immunotherapy. Notably, immunostimulatory and immunosuppressive effects are also mediated by targeted therapy (324). Inhibition of several molecular targets, such as anti-angiogenic agents, has already been proved to improve immunotherapy clinical results in a variety of types of cancer (325). It is worth noting here that, when it comes to combinatorial therapeutic approaches, greater laboratory and clinical efforts are required to validate their efficiency.
The increased understanding of GBM biology outlined above was made possible in part by the growth of murine preclinical GBM models. These models were key for translatable research from preclinical to clinical studies. Glioblastoma cell-line xenografts, patient-derived xenografts, and genetically engineered mouse models are currently available. However, these models fail to recapitulate tumoral and TME heterogeneity of GBM. Thus, the effects of advanced therapies in animal models rarely replicate clinical data, which is a persistent issue in glioblastoma research. The success of such therapies may rely on the development of reliable GBM models. Several reproducible glioma models in pigs are evolving as preclinical large-animal models (326–329). Therefore, xenograft and genetically engineered porcine models are suggested as reliable prospective models to allow for a transitional step between murine models and human clinical trials (330).
Effective drug delivery systems that can cross the BBB are also attracting attention. This includes non-viral vectors such as nanocarriers and viral vectors such as oncolytic viruses. Due to advanced research and promising outcomes, chemoradiotherapy nanomedicine is expected to be progressively applied in the clinic in the coming years (331). Targeted gene therapy employing nano-based carriers is being explored and its applicability for a number of forms of cancer are currently under Phase I–III development (332). It is thus expected that combing monoclonal antibodies and nano-carriers could also improve the efficiency of immunotherapy. On the other hand, oncolytic viruses are vectors of targeted gene therapy that can cross the BBB and are considered a promising method for treating GBM (333–336). While drug and gene delivery via oncolytic viruses are currently in clinical trials, more research and large randomized controlled Phase II/III trials are needed to assess their beneficial clinical outcome.
8. Conclusion
An efficient GBM treatment starts by understanding the molecular alterations in cells derived from the patient and that are driving the malignant phenotype. This is followed by studying the effect of a selected treatment in vitro using the ‘ideal’ model that can conserve the molecular signature of patient cells. Prior to incorporating the patient in any clinical trial, it is also recommended that researchers or physicians analyze the structure of the BBB to evaluate the ability of a selected drug to pass across the BBB. Finally, the use of a combined therapy strategy that targets tumor cells and cells of the surrounding microenvironment may increase the efficiency of the GBM treatment.
Acknowledgements
Not applicable.
Funding Statement
Funding: No funding was received.
Availability of data and materials
Not applicable.
Authors' contributions
OEA and RN equally designed the review and wrote most of the article. MA researched references and contributed to the writing of the manuscript. RAH and MES (PIs) wrote the final draft and edited the manuscript. Data authentication is not applicable. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Hanif F, Muzaffar K, Perveen K, Malhi SM, Simjee SHU. Glioblastoma multiforme: A review of its epidemiology and pathogenesis through clinical presentation and treatment. Asian Pac J Cancer Prev. 2017;18:3–9. doi: 10.22034/APJCP.2017.18.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karachi A, Dastmalchi F, Mitchell DA, Rahman M. Temozolomide for immunomodulation in the treatment of glioblastoma. Neuro Oncol. 2018;20:1566–1572. doi: 10.1093/neuonc/noy072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Delgado-López PD, Corrales-García EM. Survival in glioblastoma: A review on the impact of treatment modalities. Clin Transl Oncol. 2016;18:1062–1071. doi: 10.1007/s12094-016-1497-x. [DOI] [PubMed] [Google Scholar]
- 4.Tamimi AF, Juweid M. Epidemiology and outcome of glioblastoma. In: De Vleeschouwer S, editor. Glioblastoma. Codon Publications; Brisbane, AU: 2017. Available from: [DOI] [PubMed] [Google Scholar]
- 5.Verhaak RGW, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hubbard SR. Structural analysis of receptor tyrosine kinases. Prog Biophys Mol Biol. 1999;71:343–358. doi: 10.1016/S0079-6107(98)00047-9. [DOI] [PubMed] [Google Scholar]
- 7.Montor WR, Salas AROSE, Melo FHM. Receptor tyrosine kinases and downstream pathways as druggable targets for cancer treatment: The current arsenal of inhibitors. Mol Cancer. 2018;17:55. doi: 10.1186/s12943-018-0792-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- 9.Hunter T. Tyrosine phosphorylation: Thirty years and counting. Curr Opin Cell Biol. 2009;21:140–146. doi: 10.1016/j.ceb.2009.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brennan CW, Verhaak RGW, McKenna A, Campos B, Noushmehr H, Salama SR, Zheng S, Chakravarty D, Sanborn JZ, Berman SH, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2018;359:492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 12.Chakravarti A, Chakladar A, Delaney MA, Latham DE, Loeffler JS. The epidermal growth factor receptor pathway mediates resistance to sequential administration of radiation and chemotherapy in primary human glioblastoma cells in a RAS-dependent manner. Cancer Res. 2002;62:4307–4315. [PubMed] [Google Scholar]
- 13.Mazzoleni S, Politi LS, Pala M, Cominelli M, Franzin A, Sergi Sergi L, Falini A, De Palma M, Bulfone A, Poliani PL, Galli R. Epidermal growth factor receptor expression identifies functionally and molecularly distinct tumor-initiating cells in human glioblastoma multiforme and is required for gliomagenesis. Cancer Res. 2010;70:7500–7513. doi: 10.1158/0008-5472.CAN-10-2353. [DOI] [PubMed] [Google Scholar]
- 14.Li L, Dutra A, Pak E, Labrie JE, III, Gerstein RM, Pandolfi PP, Recht LD, Ross AH. EGFRvIII expression and PTEN loss synergistically induce chromosomal instability and glial tumors. Neuro Oncol. 2009;11:9–21. doi: 10.1215/15228517-2008-081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hatanpaa KJ, Burma S, Zhao D, Habib AA. Epidermal growth factor receptor in glioma: Signal transduction, neuropathology, imaging, and radioresistance. Neoplasia. 2010;12:675–684. doi: 10.1593/neo.10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Talasila KM, Soentgerath A, Euskirchen P, Rosland GV, Wang J, Huszthy PC, Prestegarden L, Skaftnesmo KO, Sakariassen PØ, Eskilsson E, et al. EGFR wild-type amplification and activation promote invasion and development of glioblastoma independent of angiogenesis. Acta Neuropathol. 2013;125:683–698. doi: 10.1007/s00401-013-1101-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonavia R, Inda MM, Vandenberg S, Cheng SY, Nagane M, Hadwiger P, Tan P, Sah DW, Cavenee WK, Furnari FB. EGFRvIII promotes glioma angiogenesis and growth through the NF-κB, interleukin-8 pathway. Oncogene. 2012;31:4054–4066. doi: 10.1038/onc.2011.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katanasaka Y, Kodera Y, Kitamura Y, Morimoto T, Tamura T, Koizumi F. Epidermal growth factor receptor variant type III markedly accelerates angiogenesis and tumor growth via inducing c-myc mediated angiopoietin-like 4 expression in malignant glioma. Mol Cancer. 2013;12:31. doi: 10.1186/1476-4598-12-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng H, Hu B, Vuori K, Sarkaria JN, Furnari FB, Cavenee WK, Cheng SY. EGFRvIII stimulates glioma growth and invasion through PKA-dependent serine phosphorylation of Dock180. Oncogene. 2014;33:2504–2512. doi: 10.1038/onc.2013.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eskilsson E, Rosland GV, Talasila KM, Knappskog S, Keunen O, Sottoriva A, Foerster S, Solecki G, Taxt T, Jirik R, et al. EGFRvIII mutations can emerge as late and heterogenous events in glioblastoma development and promote angiogenesis through Src activation. Neuro Oncol. 2016;18:1644–1655. doi: 10.1093/neuonc/now113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roos A, Dhruv HD, Peng S, Inge LJ, Tuncali S, Pineda M, Millard N, Mayo Z, Eschbacher JM, Loftus JC, et al. EGFRvIII-Stat5 signaling enhances glioblastoma cell migration and survival. Mol Cancer Res. 2018;16:1185–1195. doi: 10.1158/1541-7786.MCR-18-0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sugawa N, Ekstrand AJ, James CD, Collins VP. Identical splicing of aberrant epidermal growth factor receptor transcripts from amplified rearranged genes in human glioblastomas. Proc Natl Acad Sci USA. 1990;87:8602–8606. doi: 10.1073/pnas.87.21.8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frederick L, Wang XY, Eley G, James CD. Diversity and frequency of epidermal growth factor receptor mutations in human glioblastomas. Cancer Res. 2000;60:1383–1387. [PubMed] [Google Scholar]
- 24.Cruz Da Silva E, Mercier MC, Etienne-Selloum N, Dontenwill M, Choulier L. A systematic review of glioblastoma-targeted therapies in phases II, III, IV clinical trials. Cancers (Basel) 2021;13:1795. doi: 10.3390/cancers13081795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Den Bent M, Eoli M, Sepulveda JM, Smits M, Walenkamp A, Frenel JS, Franceschi E, Clement PM, Chinot O, De Vos F, et al. INTELLANCE 2/EORTC 1410 randomized Phase II study of Depatux-M alone and with temozolomide vs temozolomide or lomustine in recurrent EGFR amplified glioblastoma. Neuro Oncol. 2020;22:684–693. doi: 10.1093/neuonc/noz222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Solomon MT, Miranda N, Jorrín E, Chon I, Marinello JJ, Alert J, Lorenzo-Luaces P, Crombet T. Nimotuzumab in combination with radiotherapy in high grade glioma patients: A single institution experience. Cancer Biol Ther. 2014;15:504–509. doi: 10.4161/cbt.28021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reardon DA, Nabors LB, Mason WP, Perry JR, Shapiro W, Kavan P, Mathieu D, Phuphanich S, Cseh A, Fu Y, et al. Phase I/randomized Phase II study of afatinib, an irreversible ErbB family blocker, with or without protracted temozolomide in adults with recurrent glioblastoma. Neuro Oncol. 2015;17:430–439. doi: 10.1093/neuonc/nou160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sampson JH, Aldape KD, Archer GE, Coan A, Desjardins A, Friedman AH, Friedman HS, Gilbert MR, Herndon JE, McLendon RE, et al. Greater chemotherapy-induced lymphopenia enhances tumor-specific immune responses that eliminate EGFRvIII-expressing tumor cells in patients with glioblastoma. Neuro Oncol. 2011;13:324–333. doi: 10.1093/neuonc/noq157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paulsson J, Lindh MB, Jarvius M, Puputti M, Nistér M, Nupponen NN, Paulus W, Söderberg O, Dresemann G, von Deimling A, et al. Prognostic but not predictive role of platelet-derived growth factor receptors in patients with recurrent glioblastoma. Int J Cancer. 2011;128:1981–1988. doi: 10.1002/ijc.25528. [DOI] [PubMed] [Google Scholar]
- 30.Plate KH, Breier G, Farrell CL, Risau W. Platelet-derived growth factor receptor-beta is induced during tumor development and upregulated during tumor progression in endothelial cells in human gliomas. Lab Invest. 1992;67:529–534. [PubMed] [Google Scholar]
- 31.Camorani S, Esposito CL, Rienzo A, Catuogno S, Iaboni M, Condorelli G, de Franciscis V, Cerchia L. Inhibition of receptor signaling and of glioblastoma-derived tumor growth by a novel PDGFRβ aptamer. Mol Ther. 2014;22:828–841. doi: 10.1038/mt.2013.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vassbotn FS, Ostman A, Langeland N, Holmsen H, Westermark B, Heldin CH, Nistér M. Activated platelet-derived growth factor autocrine pathway drives the transformed phenotype of a human glioblastoma cell line. J Cell Physiol. 1994;158:381–389. doi: 10.1002/jcp.1041580221. [DOI] [PubMed] [Google Scholar]
- 33.Lokker NA, Sullivan CM, Hollenbach SJ, Israel MA, Giese NA. Platelet-derived growth factor (PDGF) autocrine signaling regulates survival and mitogenic pathways in glioblastoma cells: Evidence that the novel PDGF-C and PDGF-D ligands may play a role in the development of brain tumors. Cancer Res. 2002;62:3729–3735. [PubMed] [Google Scholar]
- 34.Cattaneo MG, Gentilini D, Vicentini LM. Deregulated human glioma cell motility: Inhibitory effect of somatostatin. Mol Cell Endocrinol. 2006;256:34–39. doi: 10.1016/j.mce.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 35.Kwak Y, Kim SI, Park CK, Paek SH, Lee ST, Park SH. C-MET overexpression and amplification in gliomas. Int J Clin Exp Pathol. 2015;8:14932–14938. [PMC free article] [PubMed] [Google Scholar]
- 36.Bender S, Gronych J, Warnatz HJ, Hutter B, Gröbner S, Ryzhova M, Pfaff E, Hovestadt V, Weinberg F, Halbach S, et al. Recurrent MET fusion genes represent a drug target in pediatric glioblastoma. Nat Med. 2016;22:1314–1320. doi: 10.1038/nm.4204. [DOI] [PubMed] [Google Scholar]
- 37.Wen PY, Schiff D, Cloughesy TF, Raizer JJ, Laterra J, Smitt M, Wolf M, Oliner KS, Anderson A, Zhu M, et al. A Phase II study evaluating the efficacy and safety of AMG 102 (rilotumumab) in patients with recurrent glioblastoma. Neuro Oncol. 2011;13:437–446. doi: 10.1093/neuonc/noq198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Y, Li A, Glas M, Lal B, Ying M, Sang Y, Xia S, Trageser D, Guerrero-Cázares H, Eberhart CG, et al. c-Met signaling induces a reprogramming network and supports the glioblastoma stem-like phenotype. Proc Natl Acad Sci USA. 2011;108:9951–9956. doi: 10.1073/pnas.1016912108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jahangiri A, De Lay M, Miller LM, Carbonell WS, Hu YL, Lu K, Tom MW, Paquette J, Tokuyasu TA, Tsao S, et al. Gene expression profile identifies tyrosine kinase c-Met as a targetable mediator of antiangiogenic therapy resistance. Clin Cancer Res. 2013;19:1773–1783. doi: 10.1158/1078-0432.CCR-12-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson J, Ascierto ML, Mittal S, Newsome D, Kang L, Briggs M, Tanner K, Marincola FM, Berens ME, Vande Woude GF, Xie Q. Genomic profiling of a hepatocyte growth factor-dependent signature for MET-targeted therapy in glioblastoma. J Transl Med. 2015;13:306. doi: 10.1186/s12967-015-0667-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cruickshanks N, Zhang Y, Yuan F, Pahuski M, Gibert M, Abounader R. Role and therapeutic targeting of the HGF/MET pathway in glioblastoma. Cancers (Basel) 2017;9:87. doi: 10.3390/cancers9070087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang K, Wu Z, Zhang H, Zhang N, Wu W, Wang Z, Dai Z, Zhang X, Zhang L, Peng Y, et al. Glioma targeted therapy: Insight into future of molecular approaches. Mol Cancer. 2022;21:39. doi: 10.1186/s12943-022-01513-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cloughesy TF, Mochizuki AY, Orpilla JR, Hugo W, Lee AH, Davidson TB, Wang AC, Ellingson BM, Rytlewski JA, Sanders CM, et al. Neoadjuvant anti-PD-1 immunotherapy promotes a survival benefit with intratumoral and systemic immune responses in recurrent glioblastoma. Nat Med. 2019;25:477–486. doi: 10.1038/s41591-018-0337-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Torre M, Vasudevaraja V, Serrano J, DeLorenzo M, Malinowski S, Blandin AF, Pages M, Ligon AH, Dong F, Meredith DM, et al. Molecular and clinicopathologic features of gliomas harboring NTRK fusions. Acta Neuropathol Commun. 2020;8:107. doi: 10.1186/s40478-020-00980-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jimenez-Pascual A, Mitchell K, Siebzehnrubl FA, Lathia JD. FGF2: A novel druggable target for glioblastoma? Expert Opin Ther Targets. 2020;24:311–318. doi: 10.1080/14728222.2020.1736558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feldkamp MM, Lau N, Guha A. The farnesyltransferase inhibitor L-744,832 inhibits the growth of astrocytomas through a combination of antiproliferative, antiangiogenic, and proapoptotic activities. Ann N Y Acad Sci. 1999;886:257–260. doi: 10.1111/j.1749-6632.1999.tb09430.x. [DOI] [PubMed] [Google Scholar]
- 47.Kowalewski A, Durślewicz J, Zdrenka M, Grzanka D, Szylberg Ł. Clinical relevance of BRAF V600E mutation status in brain tumors with a focus on a novel management algorithm. Target Oncol. 2020;15:531–540. doi: 10.1007/s11523-020-00735-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dischinger PS, Tovar EA, Essenburg CJ, Madaj ZB, Gardner EE, Callaghan ME, Turner AN, Challa AK, Kempston T, Eagleson B, et al. NF1 deficiency correlates with estrogen receptor signaling and diminished survival in breast cancer. NPJ Breast Cancer. 2018;4:29. doi: 10.1038/s41523-018-0080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arima Y, Hayashi H, Kamata K, Goto TM, Sasaki M, Kuramochi A, Saya H. Decreased expression of neurofibromin contributes to epithelial-mesenchymal transition in neurofibromatosis type 1. Exp Dermatol. 2010;19:e136–e141. doi: 10.1111/j.1600-0625.2009.01017.x. [DOI] [PubMed] [Google Scholar]
- 50.Altwairgi AK, Alghareeb WA, AlNajjar FH, Alhussain H, Alsaeed E, Balbaid AAO, Aldanan S, Orz Y, Alsharm AA. Atorvastatin in combination with radiotherapy and temozolomide for glioblastoma: A prospective Phase II study. Invest New Drugs. 2021;39:226–231. doi: 10.1007/s10637-020-00992-5. [DOI] [PubMed] [Google Scholar]
- 51.Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7:606–519. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- 52.Mondin VE, Ben El Kadhi K, Cauvin C, Jackson-Crawford A, Bélanger E, Decelle B, Salomon R, Lowe M, Echard A, Carréno S. PTEN reduces endosomal PtdIns(4,5)P2 in a phosphatase-independent manner via a PLC pathway. J Cell Biol. 2019;218:2198–2214. doi: 10.1083/jcb.201805155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Han F, Hu R, Yang H, Liu J, Sui J, Xiang X, Wang F, Chu L, Song S. PTEN gene mutations correlate to poor prognosis in glioma patients: A meta-analysis. Onco Targets Ther. 2016;9:3485–3492. doi: 10.2147/OTT.S99942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen C, Zhu S, Zhang X, Zhou T, Gu J, Xu Y, Wan Q, Qi X, Chai Y, Liu X, et al. Targeting the synthetic vulnerability of PTEN-deficient glioblastoma cells with MCL1 inhibitors. Mol Cancer Ther. 2020;19:2001–2011. doi: 10.1158/1535-7163.MCT-20-0099. [DOI] [PubMed] [Google Scholar]
- 55.Zhao JJ, Liu Z, Wang L, Shin E, Loda MF, Roberts TM. The oncogenic properties of mutant p110alpha and p110beta phosphatidylinositol 3-kinases in human mammary epithelial cells. Proc Natl Acad Sci USA. 2005;102:18443–18448. doi: 10.1073/pnas.0508988102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thorpe LM, Yuzugullu H, Zhao JJ. PI3K in cancer: Divergent roles of isoforms, modes of activation and therapeutic targeting. Nat Rev Cancer. 2015;15:7–24. doi: 10.1038/nrc3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McLendon R, Friedman A, Bigner D, Van Meir EG, Brat DJ, Mastrogianakis GM, Olson JJ, Mikkelsen T, Lehman N, Aldape K, et al. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Parsons DW, Jones S, Zhang X, Lin JCH, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weber GL, Parat MO, Binder ZA, Gallia GL, Riggins GJ. Abrogation of PIK3CA or PIK3R1 reduces proliferation, migration, and invasion in glioblastoma multiforme cells. Oncotarget. 2011;2:833–849. doi: 10.18632/oncotarget.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Quayle SN, Lee JY, Cheung LWT, Ding L, Wiedemeyer R, Dewan RW, Huang-Hobbs E, Zhuang L, Wilson RK, Ligon KL, et al. Somatic mutations of PIK3R1 promote gliomagenesis. PLoS One. 2012;7:e49466. doi: 10.1371/journal.pone.0049466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gallia GL, Rand V, Siu IM, Eberhart CG, James CD, Marie SKN, Oba-Shinjo SM, Carlotti CG, Caballero OL, Simpson AJ, et al. PIK3CA gene mutations in pediatric and adult glioblastoma multiforme. Mol Cancer Res. 2006;4:709–714. doi: 10.1158/1541-7786.MCR-06-0172. [DOI] [PubMed] [Google Scholar]
- 62.Tanaka S, Batchelor TT, Iafrate AJ, Dias-Santagata D, Borger DR, Ellisen LW, Yang D, Louis DN, Cahill DP, Chi AS. PIK3CA activating mutations are associated with more disseminated disease at presentation and earlier recurrence in glioblastoma. Acta Neuropathol Commun. 2019;7:66. doi: 10.1186/s40478-019-0720-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.England B, Huang T, Karsy M. Current understanding of the role and targeting of tumor suppressor p53 in glioblastoma multiforme. Tumour Biol. 2013;34:2063–2074. doi: 10.1007/s13277-013-0871-3. [DOI] [PubMed] [Google Scholar]
- 64.Ohgaki H. Genetic pathways to glioblastomas. Neuropathology. 2005;25:1–7. doi: 10.1111/j.1440-1789.2004.00600.x. [DOI] [PubMed] [Google Scholar]
- 65.Olafson LR, Gunawardena M, Nixdorf S, McDonald KL, Rapkin RW. The role of TP53 gain-of-function mutation in multifocal glioblastoma. J Neurooncol. 2020;147:37–47. doi: 10.1007/s11060-019-03318-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fontemaggi G, Dell'Orso S, Trisciuoglio D, Shay T, Melucci E, Fazi F, Terrenato I, Mottolese M, Muti P, Domany E, et al. The execution of the transcriptional axis mutant p53, E2F1 and ID4 promotes tumor neo-angiogenesis. Nat Struct Mol Biol. 2009;16:1086–1093. doi: 10.1038/nsmb.1669. [DOI] [PubMed] [Google Scholar]
- 67.Ham SW, Jeon HY, Jin X, Kim EJ, Kim JK, Shin YJ, Lee Y, Kim SH, Lee SY, Seo S, et al. TP53 gain-of-function mutation promotes inflammation in glioblastoma. Cell Death Differ. 2019;26:409–425. doi: 10.1038/s41418-018-0126-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pedrote MM, Motta MF, Ferretti GDS, Norberto DR, Spohr TCLS, Lima FRS, Gratton E, Silva JL, de Oliveira GAP. Oncogenic gain of function in glioblastoma is linked to mutant p53 amyloid oligomers. iScience. 2020;23:100820. doi: 10.1016/j.isci.2020.100820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tao W, Levine AJ. P19(ARF) stabilizes p53 by blocking nucleo-cytoplasmic shuttling of Mdm2. Proc Natl Acad Sci USA. 1999;96:6937–6941. doi: 10.1073/pnas.96.12.6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nakamura M, Watanabe T, Klangby U, Asker C, Wiman K, Yonekawa Y, Kleihues P, Ohgaki H. p14ARF deletion and methylation in genetic pathways to glioblastomas. Brain Pathol. 2001;11:159–168. doi: 10.1111/j.1750-3639.2001.tb00388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Reifenberger G, Liu L, Ichimura K, Schmidt EE, Collins VP. Amplification and overexpression of the MDM2 gene in a subset of human malignant gliomas without p53 mutations. Cancer Res. 1993;53:2736–2739. [PubMed] [Google Scholar]
- 72.Riemenschneider MJ, Büschges R, Wolter M, Reifenberger J, Boström J, Kraus JA, Schlegel U, Reifenberger G. Amplification and overexpression of the MDM4 (MDMX) gene from 1q32 in a subset of malignant gliomas without TP53 mutation or MDM2 amplification1. Cancer Res. 1999;59:6091–6096. [PubMed] [Google Scholar]
- 73.Xiong Y, Zhang Y, Xiong S, Williams-Villalobo AE. A glance of p53 functions in brain development, neural stem cells, and brain cancer. Biology (Basel) 2020;9:285. doi: 10.3390/biology9090285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sherr CJ, McCormick F. The RB and p53 pathways in cancer. Cancer Cell. 2002;2:103–112. doi: 10.1016/S1535-6108(02)00102-2. [DOI] [PubMed] [Google Scholar]
- 75.Allen BK, Stathias V, Maloof ME, Vidovic D, Winterbottom EF, Capobianco AJ, Clarke J, Schurer S, Robbins DJ, Ayad NG. Epigenetic pathways and glioblastoma treatment: Insights from signaling cascades. J Cell Biochem. 2015;116:351–363. doi: 10.1002/jcb.24990. [DOI] [PubMed] [Google Scholar]
- 76.Romani M, Pistillo MP, Banelli B. Epigenetic targeting of glioblastoma. Front Oncol. 2018;8:448. doi: 10.3389/fonc.2018.00448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Roos WP, Batista LFZ, Naumann SC, Wick W, Weller M, Menck CFM, Kaina B. Apoptosis in malignant glioma cells triggered by the temozolomide-induced DNA lesion O6-methylguanine. Oncogene. 2007;26:186–197. doi: 10.1038/sj.onc.1209785. [DOI] [PubMed] [Google Scholar]
- 78.Yu W, Zhang L, Wei Q, Shao A. O6-methylguanine-DNA methyltransferase (MGMT): Challenges and new opportunities in glioma chemotherapy. Front Oncol. 2020;9:1547. doi: 10.3389/fonc.2019.01547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang J, Chen L, Han L, Shi Z, Zhang J, Pu P, Kang C. EZH2 is a negative prognostic factor and exhibits pro-oncogenic activity in glioblastoma. Cancer Lett. 2015;356:929–936. doi: 10.1016/j.canlet.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 80.Sharma V, Malgulwar PB, Purkait S, Patil V, Pathak P, Agrawal R, Kulshreshtha R, Mallick S, Julka PK, Suri A, et al. Genome-wide ChIP-seq analysis of EZH2-mediated H3K27me3 target gene profile highlights differences between low- and high-grade astrocytic tumors. Carcinogenesis. 2017;38:152–161. doi: 10.1093/carcin/bgw126. [DOI] [PubMed] [Google Scholar]
- 81.Mohammad F, Weissmann S, Leblanc B, Pandey DP, Højfeldt JW, Comet I, Zheng C, Johansen JV, Rapin N, Porse BT, et al. EZH2 is a potential therapeutic target for H3K27M-mutant pediatric gliomas. Nat Med. 2017;23:483–492. doi: 10.1038/nm.4293. [DOI] [PubMed] [Google Scholar]
- 82.Lee DH, Kim GW, Yoo J, Lee SW, Jeon YH, Kim SY, Kang HG, Kim DH, Chun KH, Choi J, Kwon SH. Histone demethylase KDM4C controls tumorigenesis of glioblastoma by epigenetically regulating p53 and c-Myc. Cell Death Dis. 2021;12:89. doi: 10.1038/s41419-020-03380-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ceccacci E, Minucci S. Inhibition of histone deacetylases in cancer therapy: Lessons from leukaemia. Br J Cancer. 2016;114:605–611. doi: 10.1038/bjc.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lee P, Murphy B, Miller R, Menon V, Banik NL, Giglio P, Lindhorst SM, Varma AK, Vandergrift WA, III, Patel SJ, Das A. Mechanisms and clinical significance of histone deacetylase inhibitors: Epigenetic glioblastoma therapy. Anticancer Res. 2015;35:615–625. [PMC free article] [PubMed] [Google Scholar]
- 85.Chen R, Zhang M, Zhou Y, Guo W, Yi M, Zhang Z, Ding Y, Wang Y. The application of histone deacetylases inhibitors in glioblastoma. J Exp Clin Cancer Res. 2020;39:138. doi: 10.1186/s13046-020-01643-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Harbour JW, Luo RX, Santi AD, Postigo AA, Dean DC. Cdk phosphorylation triggers sequential intramolecular interactions that progressively block Rb functions as cells move through G1. Cell. 1999;98:859–869. doi: 10.1016/S0092-8674(00)81519-6. [DOI] [PubMed] [Google Scholar]
- 87.Matsushime H, Ewen ME, Strom DK, Kato JY, Hanks SK, Roussel MF, Sherr CJ. Identification and properties of an atypical catalytic subunit (p34PSK-J3/cdk4) for mammalian D type G1 cyclins. Cell. 1992;71:323–334. doi: 10.1016/0092-8674(92)90360-O. [DOI] [PubMed] [Google Scholar]
- 88.Cooper S, Shayman JA. Revisiting retinoblastoma protein phosphorylation during the mammalian cell cycle. Cell Mol Life Sci. 2001;58:580–595. doi: 10.1007/PL00000883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Taylor JW, Parikh M, Phillips JJ, James CD, Molinaro AM, Butowski NA, Clarke JL, Oberheim-Bush NA, Chang SM, Berger MS, Prados M. Phase-2 trial of palbociclib in adult patients with recurrent RB1-positive glioblastoma. J Neurooncol. 2018;140:477–483. doi: 10.1007/s11060-018-2977-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426:895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- 91.Narayanan S, Cai CY, Assaraf YG, Guo HQ, Cui Q, Wei L, Huang JJ, Ashby CR, Jr, Chen ZS. Targeting the ubiquitin-proteasome pathway to overcome anti-cancer drug resistance. Drug Resist Updat. 2020;48:100663. doi: 10.1016/j.drup.2019.100663. [DOI] [PubMed] [Google Scholar]
- 92.Kong XT, Nguyen NT, Choi YJ, Zhang G, Nguyen HN, Filka E, Green S, Yong WH, Liau LM, Green RM, et al. Phase 2 study of bortezomib combined with temozolomide and regional radiation therapy for upfront treatment of patients with newly diagnosed glioblastoma multiforme: safety and efficacy assessment. Int J Radiat Oncol Biol Phys. 2018;100:1195–1203. doi: 10.1016/j.ijrobp.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Di K, Lloyd GK, Abraham V, MacLaren A, Burrows FJ, Desjardins A, Trikha M, Bota DA. Marizomib activity as a single agent in malignant gliomas: Ability to cross the blood-brain barrier. Neuro Oncol. 2016;18:840–848. doi: 10.1093/neuonc/nov299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Roth P, Mason WP, Richardson PG, Weller M. Proteasome inhibition for the treatment of glioblastoma. Expert Opin Investig Drugs. 2020;29:1133–1141. doi: 10.1080/13543784.2020.1803827. [DOI] [PubMed] [Google Scholar]
- 95.Quillin J, Patel R, Herzberg E, Alton D, Bikzhanova G, Geisler L, Olson J. A phase 0 analysis of ixazomib in patients with glioblastoma. Mol Clin Oncol. 2020;13:43. doi: 10.3892/mco.2020.2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Huang J, Campian JL, Gujar AD, Tsien C, Ansstas G, Tran DD, DeWees TA, Lockhart AC, Kim AH. Final results of a Phase I dose-escalation, dose-expansion study of adding disulfiram with or without copper to adjuvant temozolomide for newly diagnosed glioblastoma. J Neurooncol. 2018;138:105–111. doi: 10.1007/s11060-018-2775-y. [DOI] [PubMed] [Google Scholar]
- 97.Huang J, Chaudhary R, Cohen AL, Fink K, Goldlust S, Boockvar J, Chinnaiyan P, Wan L, Marcus S, Campian JL. A multicenter Phase II study of temozolomide plus disulfiram and copper for recurrent temozolomide-resistant glioblastoma. J Neurooncol. 2019;142:537–544. doi: 10.1007/s11060-019-03125-y. [DOI] [PubMed] [Google Scholar]
- 98.Carruthers R, Chalmers AJ. Improving the therapeutic ratio of radiotherapy by targeting the DNA damage response. In: Tofilon PJ, Camphausen K, editors. Increasing the Therapeutic Ratio of Radiotherapy. Series: Cancer Drug Discovery and Development. Humana Press; Cham: 2017. pp. 1–34. [DOI] [Google Scholar]
- 99.Murnyák B, Kouhsari MC, Hershkovitch R, Kálmán B, Marko-Varga G, Klekner Á, Hortobágyi T. PARP1 expression and its correlation with survival is tumour molecular subtype dependent in glioblastoma. Oncotarget. 2017;8:46348–46362. doi: 10.18632/oncotarget.18013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chalmers AJ. Overcoming resistance of glioblastoma to conventional cytotoxic therapies by the addition of PARP inhibitors. Anticancer Agents Med Chem. 2010;10:520–533. doi: 10.2174/187152010793498627. [DOI] [PubMed] [Google Scholar]
- 101.Hanna C, Kurian KM, Williams K, Watts C, Jackson A, Carruthers R, Strathdee K, Cruickshank G, Dunn L, Erridge S, et al. Pharmacokinetics, safety, and tolerability of olaparib and temozolomide for recurrent glioblastoma: Results of the Phase I OPARATIC trial. Neuro Oncol. 2020;22:1840–1850. doi: 10.1093/neuonc/noaa104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Robins HI, Zhang P, Gilbert MR, Chakravarti A, de Groot JF, Grimm SA, Wang F, Lieberman FS, Krauze A, Trotti AM, et al. A randomized Phase I/II study of ABT-888 in combination with temozoflomide in recurrent temozolomide resistant glioblastoma: An NRG oncology RTOG group study. J Neurooncol. 2016;126:309–316. doi: 10.1007/s11060-015-1966-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Baxter PA, Su JM, Onar-Thomas A, Billups CA, Li XN, Poussaint TY, Smith ER, Thompson P, Adesina A, Ansell P, et al. A Phase I/II study of veliparib (ABT-888) with radiation and temozolomide in newly diagnosed diffuse pontine glioma: A pediatric brain tumor consortium study. Neuro Oncol. 2020;22:875–885. doi: 10.1093/neuonc/noaa016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 World Health Organi-zation classification of tumors of the central nervous system: A summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 105.Jo SH, Son MK, Koh HJ, Lee SM, Song IH, Kim YO, Lee YS, Jeong KS, Kim WB, Park JW, et al. Control of mitochondrial redox balance and cellular defense against oxidative damage by mitochondrial NADP+-dependent isocitrate dehydrogenase. J Biol Chem. 2001;276:16168–16176. doi: 10.1016/S0021-9258(19)30242-X. [DOI] [PubMed] [Google Scholar]
- 106.Lee SM, Koh HJ, Park DC, Song BJ, Huh TL, Park JW. Cytosolic NADP(+)-dependent isocitrate dehydrogenase status modulates oxidative damage to cells. Free Radic Biol Med. 2002;32:1185–1196. doi: 10.1016/S0891-5849(02)00815-8. [DOI] [PubMed] [Google Scholar]
- 107.Kim SY, Park JW. Cellular defense against singlet oxygen-induced oxidative damage by cytosolic NADP+-dependent isocitrate dehydrogenase. Free Radic Res. 2003;37:309–316. doi: 10.1080/1071576021000050429. [DOI] [PubMed] [Google Scholar]
- 108.Krell D, Assoku M, Galloway M, Mulholland P, Tomlinson I, Bardella C. Screen for IDH1, IDH2, IDH3, D2HGDH and L2HGDH mutations in glioblastoma. PLoS One. 2011;6:e19868. doi: 10.1371/journal.pone.0019868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Arita H, Narita Y, Yoshida A, Hashimoto N, Yoshimine T, Ichimura K. IDH1/2 mutation detection in gliomas. Brain Tumor Pathol. 2015;32:79–89. doi: 10.1007/s10014-014-0197-x. [DOI] [PubMed] [Google Scholar]
- 110.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Capper D, Weissert S, Balss J, Habel A, Meyer J, Jäger D, Ackermann U, Tessmer C, Korshunov A, Zentgraf H, et al. Characterization of R132H mutation-specific IDH1 antibody binding in brain tumors. Brain Pathol. 2010;20:245–254. doi: 10.1111/j.1750-3639.2009.00352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Wang P, Xiao MT, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, Edwards CR, Khanin R, Figueroa ME, Melnick A, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yalaza C, Ak H, Cagli MS, Ozgiray E, Atay S, Aydin HH. R132H mutation in IDH1 gene is associated with increased tumor HIF1-alpha and serum VEGF levels in primary glioblastoma multiforme. Ann Clin Lab Sci. 2017;47:362–364. [PubMed] [Google Scholar]
- 116.Flavahan WA, Drier Y, Liau BB, Gillespie SM, Venteicher AS, Stemmer-Rachamimov AO, Suvà ML, Bernstein BE. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature. 2016;529:110–114. doi: 10.1038/nature16490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fu Y, Zheng S, Zheng Y, Huang R, An N, Liang A, Hu C. Glioma derived isocitrate dehydrogenase-2 mutations induced up-regulation of HIF-1α and β-catenin signaling: Possible impact on glioma cell metastasis and chemo-resistance. Int J Biochem Cell Biol. 2012;44:770–775. doi: 10.1016/j.biocel.2012.01.017. [DOI] [PubMed] [Google Scholar]
- 118.Warburg O. On respiratory impairment in cancer cells. Science. 1956;124:269–270. doi: 10.1126/science.124.3215.269. [DOI] [PubMed] [Google Scholar]
- 119.Stern R, Shuster S, Neudecker BA, Formby B. Lactate stimulates fibroblast expression of hyaluronan and CD44: The Warburg effect revisited. Exp Cell Res. 2002;276:24–31. doi: 10.1006/excr.2002.5508. [DOI] [PubMed] [Google Scholar]
- 120.Kroemer G, Pouyssegur J. Tumor cell metabolism: Cancer's Achilles' heel. Cancer Cell. 2008;13:472–482. doi: 10.1016/j.ccr.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 121.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Saier MH., Jr Families of transmembrane sugar transport proteins. Mol Microbiol. 2000;35:699–710. doi: 10.1046/j.1365-2958.2000.01759.x. [DOI] [PubMed] [Google Scholar]
- 123.Scheepers A, Joost H, Schürmann A. The glucose transporter families SGLT and GLUT: Molecular basis of normal and aberrant function. JPEN J Parenter Enteral Nutr. 2004;28:364–371. doi: 10.1177/0148607104028005364. [DOI] [PubMed] [Google Scholar]
- 124.Flavahan WA, Wu Q, Hitomi M, Rahim N, Kim Y, Sloan AE, Weil RJ, Nakano I, Sarkaria JN, Stringer BW, et al. Brain tumor initiating cells adapt to restricted nutrition through preferential glucose uptake. Nat Neurosci. 2013;16:1373–1382. doi: 10.1038/nn.3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Libby CJ, Gc S, Benavides GA, Fisher JL, Williford SE, Zhang S, Tran AN, Gordon ER, Jones AB, Tuy K, et al. A role for GLUT3 in glioblastoma cell invasion that is not recapitulated by GLUT1. Cell Adh Migr. 2021;15:101–115. doi: 10.1080/19336918.2021.1903684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wolf A, Agnihotri S, Micallef J, Mukherjee J, Sabha N, Cairns R, Hawkins C, Guha A. Hexokinase 2 is a key mediator of aerobic glycolysis and promotes tumor growth in human glioblastoma multiforme. J Exp Med. 2011;208:313–326. doi: 10.1084/jem.20101470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Vartanian A, Agnihotri S, Wilson MR, Burrell KE, Tonge PD, Alamsahebpour A, Jalali S, Taccone MS, Mansouri S, Golbourn B, et al. Targeting hexokinase 2 enhances response to radio-chemotherapy in glioblastoma. Oncotarget. 2016;7:69518–69535. doi: 10.18632/oncotarget.11680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Pastorino JG, Shulga N, Hoek JB. Mitochondrial binding of hexokinase II inhibits Bax-induced cytochrome c release and apoptosis. J Biol Chem. 2002;277:7610–7618. doi: 10.1074/jbc.M109950200. [DOI] [PubMed] [Google Scholar]
- 129.Agnihotri S, Mansouri S, Burrell K, Li M, Mamatjan Y, Liu J, Nejad R, Kumar S, Jalali S, Singh SK, et al. Ketoconazole and posaconazole selectively target HK2-expressing glioblastoma cells. Clin Cancer Res. 2019;25:844–855. doi: 10.1158/1078-0432.CCR-18-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Harada K, Saheki S, Wada K, Tanaka T. Purification of four pyruvate kinase isozymes of rats by affinity elution chromatography. Biochim Biophys Acta. 1978;524:327–339. doi: 10.1016/0005-2744(78)90169-9. [DOI] [PubMed] [Google Scholar]
- 131.Verma H, Cholia RP, Kaur S, Dhiman M, Mantha AK. A short review on cross-link between pyruvate kinase (PKM2) and glioblastoma multiforme. Metab Brain Dis. 2021;36:751–765. doi: 10.1007/s11011-021-00690-y. [DOI] [PubMed] [Google Scholar]
- 132.Uyeda K. Pyruvate kinase. In: Lennarz WJ, Lane MD, editors. Encyclopedia of Biological Chemistry. (2nd edition) Academic Press; Waltham: 2013. pp. 719–721. Available from: [DOI] [Google Scholar]
- 133.Mukherjee J, Phillips JJ, Zheng S, Wiencke J, Ronen SM, Pieper RO. Pyruvate kinase M2 expression, but not pyruvate kinase activity, is up-regulated in a grade-specific manner in human glioma. PLoS One. 2013;8:e57610. doi: 10.1371/journal.pone.0057610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yang W, Xia Y, Hawke D, Li X, Liang J, Xing D, Aldape K, Hunter T, Alfred Yung WK, Lu Z. PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell. 2012;150:685–696. doi: 10.1016/j.cell.2012.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Jiang Y, Li X, Yang W, Hawke DH, Zheng Y, Xia Y, Aldape K, Wei C, Guo F, Chen Y, Lu Z. PKM2 regulates chromosome segregation and mitosis progression of tumor cells. Mol Cell. 2014;53:75–87. doi: 10.1016/j.molcel.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Cholia RP, Dhiman M, Kumar R, Mantha AK. Oxidative stress stimulates invasive potential in rat C6 and human U-87 MG glioblastoma cells via activation and cross-talk between PKM2, ENPP2 and APE1 enzymes. Metab Brain Dis. 2018;33:1307–1326. doi: 10.1007/s11011-018-0233-3. [DOI] [PubMed] [Google Scholar]
- 137.Liang J, Cao R, Wang X, Zhang Y, Wang P, Gao H, Li C, Yang F, Zeng R, Wei P, et al. Mitochondrial PKM2 regulates oxidative stress-induced apoptosis by stabilizing Bcl2. Cell Res. 2017;27:329–351. doi: 10.1038/cr.2016.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yang W, Xia Y, Ji H, Zheng Y, Liang J, Huang W, Gao X, Aldape K, Lu Z. Nuclear PKM2 regulates β-catenin transactivation upon EGFR activation. Nature. 2011;480:118–122. doi: 10.1038/nature10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Chaneton B, Gottlieb E. Rocking cell metabolism: Revised functions of the key glycolytic regulator PKM2 in cancer. Trends Biochem Sci. 2012;37:309–316. doi: 10.1016/j.tibs.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 140.Sizemore ST, Zhang M, Cho JH, Sizemore GM, Hurwitz B, Kaur B, Lehman NL, Ostrowski MC, Robe PA, Miao W, et al. Pyruvate kinase M2 regulates homologous recombination-mediated DNA double-strand break repair. Cell Res. 2018;28:1090–1102. doi: 10.1038/s41422-018-0086-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Rieger J, Bähr O, Maurer GD, Hattingen E, Franz K, Brucker D, Walenta S, Kämmerer U, Coy JF, Weller M, Steinbach JP. ERGO: A pilot study of ketogenic diet in recurrent glioblastoma. Int J Oncol. 2014;44:1843–1852. doi: 10.3892/ijo.2014.2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kucharzewska P, Christianson HC, Belting M. Global profiling of metabolic adaptation to hypoxic stress in human glioblastoma cells. PLoS One. 2015;10:e0116740. doi: 10.1371/journal.pone.0116740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ogunrinu TA, Sontheimer H. Hypoxia increases the dependence of glioma cells on glutathione. J Biol Chem. 2010;285:37716–37724. doi: 10.1074/jbc.M110.161190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Robertson DS. The physical chemistry of brain and neural cell membranes: An overview. Neurochem Res. 2010;35:681–687. doi: 10.1007/s11064-010-0121-7. [DOI] [PubMed] [Google Scholar]
- 145.Newsholme P, Lima MMR, Procopio J, Pithon-Curi TC, Doi SQ, Bazotte RB, Curi R. Glutamine and glutamate as vital metabolites. Braz J Med Biol Res. 2003;36:153–163. doi: 10.1590/S0100-879X2003000200002. [DOI] [PubMed] [Google Scholar]
- 146.Bak LK, Schousboe A, Waagepetersen HS. The glutamate/GABA-glutamine cycle: Aspects of transport, neurotransmitter homeostasis and ammonia transfer. J Neurochem. 2006;98:641–653. doi: 10.1111/j.1471-4159.2006.03913.x. [DOI] [PubMed] [Google Scholar]
- 147.Obara-Michlewska M, Szeliga M. Targeting glutamine addiction in gliomas. Cancers (Basel) 2020;12:310. doi: 10.3390/cancers12020310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Michalak KP, Maćkowska-Kędziora A, Sobolewski B, Woźniak P. Key roles of glutamine pathways in reprogramming the cancer metabolism. Oxid Med Cell Longev. 2015;2015:964321. doi: 10.1155/2015/964321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Rubin H. Deprivation of glutamine in cell culture reveals its potential for treating cancer. Proc Natl Acad Sci USA. 2019;116:6964–6968. doi: 10.1073/pnas.1815968116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ekici S, Nye JA, Neill SG, Allen JW, Shu HK, Fleischer CC. Glutamine imaging: A new avenue for glioma management. AJNR Am J Neuroradiol. 2022;43:11–18. doi: 10.3174/ajnr.A7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.de Groot JF, Liu TJ, Fuller G, Yung WKA. The excitatory amino acid transporter-2 induces apoptosis and decreases glioma growth in vitro and in vivo. Cancer Res. 2005;65:1934–1940. doi: 10.1158/0008-5472.CAN-04-3626. [DOI] [PubMed] [Google Scholar]
- 152.Lyons SA, Chung WJ, Weaver AK, Ogunrinu T, Sontheimer H. Autocrine glutamate signaling promotes glioma cell invasion. Cancer Res. 2007;67:9463–9471. doi: 10.1158/0008-5472.CAN-07-2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Dranoff G, Elion GB, Friedman HS, Bigner DD. Combination chemotherapy in vitro exploiting glutamine metabolism of human glioma and medulloblastoma. Cancer Res. 1985;45:4082–4086. [PubMed] [Google Scholar]
- 154.Seltzer MJ, Bennett BD, Joshi AD, Gao P, Thomas AG, Ferraris DV, Tsukamoto T, Rojas CJ, Slusher BS, Rabinowitz JD, et al. Inhibition of glutaminase preferentially slows growth of glioma cells with mutant IDH1. Cancer Res. 2010;70:8981–8987. doi: 10.1158/0008-5472.CAN-10-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Majewska E, Márquez J, Albrecht J, Szeliga M. Transfection with GLS2 glutaminase (GAB) sensitizes human glioblastoma cell lines to oxidative stress by a common mechanism involving suppression of the PI3K/AKT pathway. Cancers (Basel) 2019;11:115. doi: 10.3390/cancers11010115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Szeliga M, Obara-Michlewska M, Matyja E, Łazarczyk M, Lobo C, Hilgier W, Alonso FJ, Márquez J, Albrecht J. Transfection with liver-type glutaminase cDNA alters gene expression and reduces survival, migration and proliferation of T98G glioma cells. Glia. 2009;57:1014–1023. doi: 10.1002/glia.20825. [DOI] [PubMed] [Google Scholar]
- 157.Szeliga M, Zgrzywa A, Obara-Michlewska M, Albrecht J. Transfection of a human glioblastoma cell line with liver-type glutaminase (LGA) down-regulates the expression of DNA-repair gene MGMT and sensitizes the cells to alkylating agents. J Neurochem. 2012;123:428–436. doi: 10.1111/j.1471-4159.2012.07917.x. [DOI] [PubMed] [Google Scholar]
- 158.Yin Y, Sun W, Xiang J, Deng L, Zhang B, Xie P, Qiao W, Zou J, Liu C. Glutamine synthetase functions as a negative growth regulator in glioma. J Neurooncol. 2013;114:59–69. doi: 10.1007/s11060-013-1168-5. [DOI] [PubMed] [Google Scholar]
- 159.Ye ZC, Rothstein JD, Sontheimer H. Compromised glutamate transport in human glioma cells: Reduction-mislocalization of sodium-dependent glutamate transporters and enhanced activity of cystine-glutamate exchange. J Neurosci. 1999;19:10767–10777. doi: 10.1523/JNEUROSCI.19-24-10767.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Chung WJ, Lyons SA, Nelson GM, Hamza H, Gladson CL, Gillespie GY, Sontheimer H. Inhibition of cystine uptake disrupts the growth of primary brain tumors. J Neurosci. 2005;25:7101–7110. doi: 10.1523/JNEUROSCI.5258-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Robe PA, Martin DH, Nguyen-Khac MT, Artesi M, Deprez M, Albert A, Vanbelle S, Califice S, Bredel M, Bours V. Early termination of ISRCTN45828668, a phase 1/2 prospective, randomized study of sulfasalazine for the treatment of progressing malignant gliomas in adults. BMC Cancer. 2009;9:372. doi: 10.1186/1471-2407-9-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Sleire L, Skeie BS, Netland IA, Førde HE, Dodoo E, Selheim F, Leiss L, Heggdal JI, Pedersen PH, Wang J, Enger PØ. Drug repurposing: Sulfasalazine sensitizes gliomas to gamma knife radiosurgery by blocking cystine uptake through system Xc-, leading to glutathione depletion. Oncogene. 2015;34:5951–5959. doi: 10.1038/onc.2015.60. [DOI] [PubMed] [Google Scholar]
- 163.Choi YK, Park KG. Targeting glutamine metabolism for cancer treatment. Biomol Ther (Seoul) 2018;26:19–28. doi: 10.4062/biomolther.2017.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Morris SM., Jr Enzymes of arginine metabolism. J Nutr. 2004;134((Suppl 10)):2743S–2747S. 2765S–2767S. doi: 10.1093/jn/134.10.2743S. [DOI] [PubMed] [Google Scholar]
- 165.Kuo MT, Savaraj N, Feun LG. Targeted cellular metabolism for cancer chemotherapy with recombinant arginine-degrading enzymes. Oncotarget. 2010;1:246–251. doi: 10.18632/oncotarget.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Khoury O, Ghazale N, Stone E, El-Sibai M, Frankel AE, Abi-Habib RJ. Human recombinant arginase I (Co)-PEG5000 [HuArgI (Co)-PEG5000]-induced arginine depletion is selectively cytotoxic to human glioblastoma cells. J Neurooncol. 2015;122:75–85. doi: 10.1007/s11060-014-1698-5. [DOI] [PubMed] [Google Scholar]
- 167.Choy CT, Wong CH, Loong HHF. Low expressions of ASS1 and OTC in glioblastoma suggest the potential clinical use of recombinant human arginase (rhArg) J Neurooncol. 2016;129:579–581. doi: 10.1007/s11060-016-2209-7. [DOI] [PubMed] [Google Scholar]
- 168.Bobak Y, Kurlishchuk Y, Vynnytska-Myronovska B, Grydzuk O, Shuvayeva G, Redowicz MJ, Kunz-Schughart LA, Stasyk O. Arginine deprivation induces endoplasmic reticulum stress in human solid cancer cells. Int J Biochem Cell Biol. 2016;70:29–38. doi: 10.1016/j.biocel.2015.10.027. [DOI] [PubMed] [Google Scholar]
- 169.Pavlyk I, Rzhepetskyy Y, Jagielski AK, Drozak J, Wasik A, Pereverzieva G, Olchowik M, Kunz-Schugart LA, Stasyk O, Redowicz MJ. Arginine deprivation affects glioblastoma cell adhesion, invasiveness and actin cytoskeleton organization by impairment of β-actin arginylation. Amino Acids. 2015;47:199–212. doi: 10.1007/s00726-014-1857-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Stasyk OV, Boretsky YR, Gonchar MV, Sibirny AA. Recombinant arginine-degrading enzymes in metabolic anticancer therapy and bioanalytics. Cell Biol Int. 2015;39:246–252. doi: 10.1002/cbin.10383. [DOI] [PubMed] [Google Scholar]
- 171.Stone EM, Chantranupong L, Georgiou G. The second-shell metal ligands of human arginase affect coordination of the nucleophile and substrate. Biochemistry. 2010;49:10582–10588. doi: 10.1021/bi101542t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Al-Koussa H, Al-Haddad M, Abi-Habib R, El-Sibai M. Human recombinant arginase I [HuArgI (Co)-PEG5000]-induced arginine depletion inhibits colorectal cancer cell migration and invasion. Int J Mol Sci. 2019;20:6018. doi: 10.3390/ijms20236018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Khalil N, Abi-Habib RJ. [HuArgI (co)-PEG5000]-induced arginine deprivation leads to autophagy dependent cell death in pancreatic cancer cells. Invest New Drugs. 2020;38:1236–1246. doi: 10.1007/s10637-019-00883-4. [DOI] [PubMed] [Google Scholar]
- 174.Tanios R, Bekdash A, Kassab E, Stone E, Georgiou G, Frankel AE, Abi-Habib RJ. Human recombinant arginase I(Co)-PEG5000 [HuArgI(Co)-PEG5000]-induced arginine depletion is selectively cytotoxic to human acute myeloid leukemia cells. Leuk Res. 2013;37:1565–1571. doi: 10.1016/j.leukres.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 175.Nasreddine G, El-Sibai M, Abi-Habib RJ. Cytotoxicity of [HuArgI (co)-PEG5000]-induced arginine deprivation to ovarian cancer cells is autophagy dependent. Invest New Drugs. 2020;38:10–19. doi: 10.1007/s10637-019-00756-w. [DOI] [PubMed] [Google Scholar]
- 176.Glazer ES, Stone EM, Zhu C, Massey KL, Hamir AN, Curley SA. Bioengineered human arginase I with enhanced activity and stability controls hepatocellular and pancreatic carcinoma xenografts. Transl Oncol. 2011;4:138–146. doi: 10.1593/tlo.10265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Yoon CY, Sung DJ, Lee JH, Kim AR, Oh CW, Je JH, Weon BM, Seol SK, Pyun A, Hwu Y, et al. Imaging of renal and prostate carcinoma with refractive index radiology. Int J Urol. 2007;14:96–103. doi: 10.1111/j.1442-2042.2007.01614.x. [DOI] [PubMed] [Google Scholar]
- 178.Hsueh EC, Knebel SM, Lo WH, Leung YC, Cheng PNM, Hsueh CT. Deprivation of arginine by recombinant human arginase in prostate cancer cells. J Hematol Oncol. 2012;5:17. doi: 10.1186/1756-8722-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Wang Z, Shi X, Li Y, Fan J, Zeng X, Xian Z, Wang Z, Sun Y, Wang S, Song P, et al. Blocking autophagy enhanced cytotoxicity induced by recombinant human arginase in triple-negative breast cancer cells. Cell Death Dis. 2014;5:e1563. doi: 10.1038/cddis.2014.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Hou X, Chen S, Zhang P, Guo D, Wang B. Targeted arginine metabolism therapy: A dilemma in glioma treatment. Front Oncol. 2022;12:938847. doi: 10.3389/fonc.2022.938847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Arita H, Narita Y, Fukushima S, Tateishi K, Matsushita Y, Yoshida A, Miyakita Y, Ohno M, Collins VP, Kawahara N, et al. Upregulating mutations in the TERT promoter commonly occur in adult malignant gliomas and are strongly associated with total 1p19q loss. Acta Neuropathol. 2013;126:267–276. doi: 10.1007/s00401-013-1141-6. [DOI] [PubMed] [Google Scholar]
- 182.Nonoguchi N, Ohta T, Oh JE, Kim YH, Kleihues P, Ohgaki H. TERT promoter mutations in primary and secondary glioblastomas. Acta Neuropathol. 2013;126:931–937. doi: 10.1007/s00401-013-1163-0. [DOI] [PubMed] [Google Scholar]
- 183.Killela PJ, Reitman ZJ, Jiao Y, Bettegowda C, Agrawal N, Diaz LA, Jr, Friedman AH, Friedman H, Gallia GL, Giovanella BC, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci USA. 2013;110:6021–6066. doi: 10.1073/pnas.1303607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Arita H, Yamasaki K, Matsushita Y, Nakamura T, Shimokawa A, Takami H, Tanaka S, Mukasa A, Shirahata M, Shimizu S, et al. A combination of TERT promoter mutation and MGMT methylation status predicts clinically relevant subgroups of newly diagnosed glioblastomas. Acta Neuropathol Commun. 2016;4:79. doi: 10.1186/s40478-016-0351-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Kikuchi Z, Shibahara I, Yamaki T, Yoshioka E, Shofuda T, Ohe R, Matsuda KI, Saito R, Kanamori M, Kanemura Y, et al. TERT promoter mutation associated with multifocal phenotype and poor prognosis in patients with IDH wild-type glioblastoma. Neurooncol Adv. 2020;2:vdaa114. doi: 10.1093/noajnl/vdaa114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Vuong HG, Nguyen TQ, Ngo TNM, Nguyen HC, Fung KM, Dunn IF. The interaction between TERT promoter mutation and MGMT promoter methylation on overall survival of glioma patients: A meta-analysis. BMC Cancer. 2020;20:897. doi: 10.1186/s12885-020-07364-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Patel B, Taiwo R, Kim AH, Dunn GP. TERT, a promoter of CNS malignancies. Neurooncol Adv. 2020;2:vdaa025. doi: 10.1093/noajnl/vdaa025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Picketts DJ, Higgs DR, Bachoo S, Blake DJ, Quarrell OW, Gibbons RJ. ATRX encodes a novel member of the SNF2 family of proteins: Mutations point to a common mechanism underlying the ATR-X syndrome. Hum Mol Genet. 1996;5:1899–1907. doi: 10.1093/hmg/5.12.1899. [DOI] [PubMed] [Google Scholar]
- 189.Jiao Y, Killela PJ, Reitman ZJ, Rasheed BA, Heaphy CM, de Wilde RF, Rodriguez FJ, Rosemberg S, Oba-Shinjo SM, Nagahashi Marie SK, et al. Frequent ATRX, CIC, FUBP1 and IDH1 mutations refine the classification of malignant gliomas. Oncotarget. 2012;3:709–722. doi: 10.18632/oncotarget.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Koschmann C, Calinescu AA, Nunez FJ, Mackay A, Fazal-Salom J, Thomas D, Mendez F, Kamran N, Dzaman M, Mulpuri L, et al. ATRX loss promotes tumor growth and impairs nonhomologous end joining DNA repair in glioma. Sci Transl Med. 2016;8:328ra28. doi: 10.1126/scitranslmed.aac8228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.do Carmo A, Balça-Silva J, Matias D, Lopes MC. PKC signaling in glioblastoma. Cancer Biol Ther. 2013;14:287–294. doi: 10.4161/cbt.23615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Steinberg SF. Structural basis of protein kinase C isoform function. Physiol Rev. 2008;88:1341–1378. doi: 10.1152/physrev.00034.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Leirdal M, Sioud M. Protein kinase Calpha isoform regulates the activation of the MAP kinase ERK1/2 in human glioma cells: Involvement in cell survival and gene expression. Mol Cell Biol Res Commun. 2000;4:106–110. doi: 10.1006/mcbr.2000.0259. [DOI] [PubMed] [Google Scholar]
- 194.Marquina-Sánchez B, González-Jorge J, Hansberg-Pastor V, Wegman-Ostrosky T, Baranda-Ávila N, Mejía-Pérez S, Camacho-Arroyo I, González-Arenas A. The interplay between intracellular progesterone receptor and PKC plays a key role in migration and invasion of human glioblastoma cells. J Steroid Biochem Mol Biol. 2017;172:198–206. doi: 10.1016/j.jsbmb.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 195.Valdés-Rives SA, Arcos-Montoya D, de la Fuente-Granada M, Zamora-Sánchez CJ, Arias-Romero LE, Villamar-Cruz O, Camacho-Arroyo I, Pérez-Tapia SM, González-Arenas A. LPA1 receptor promotes progesterone receptor phosphorylation through PKCα in human glioblastoma cells. Cells. 2021;10:807. doi: 10.3390/cells10040807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Yoshiji H, Kuriyama S, Ways DK, Yoshii J, Miyamoto Y, Kawata M, Ikenaka Y, Tsujinoue H, Nakatani T, Shibuya M, Fukui H. Protein kinase C lies on the signaling pathway for vascular endothelial growth factor-mediated tumor development and angiogenesis. Cancer Res. 1999;59:4413–4418. [PubMed] [Google Scholar]
- 197.Liu Z, Wei Y, Zhang L, Yee PP, Johnson M, Zhang X, Gulley M, Atkinson JM, Trebak M, Wang HG, Li W. Induction of store-operated calcium entry (SOCE) suppresses glioblastoma growth by inhibiting the Hippo pathway transcriptional coactivators YAP/TAZ. Oncogene. 2019;38:120–139. doi: 10.1038/s41388-018-0425-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Chen Z, Forman LW, Williams RM, Faller DV. Protein kinase C-δ inactivation inhibits the proliferation and survival of cancer stem cells in culture and in vivo. BMC Cancer. 2014;14:90. doi: 10.1186/1471-2407-14-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Kim MJ, Kim RK, Yoon CH, An S, Hwang SG, Suh Y, Park MJ, Chung HY, Kim IG, Lee SJ. Importance of PKCδ signaling in fractionated-radiation-induced expansion of glioma-initiating cells and resistance to cancer treatment. J Cell Sci. 2011;124:3084–3094. doi: 10.1242/jcs.080119. [DOI] [PubMed] [Google Scholar]
- 200.Sarkar S, Yong VW. Reduction of protein kinase C delta attenuates tenascin-C stimulated glioma invasion in three-dimensional matrix. Carcinogenesis. 2010;31:311–317. doi: 10.1093/carcin/bgp297. [DOI] [PubMed] [Google Scholar]
- 201.Lu J, Xu Z, Duan H, Ji H, Zhen Z, Li B, Wang H, Tang H, Zhou J, Guo T, et al. Tumor-associated macrophage interleukin-β promotes glycerol-3-phosphate dehydrogenase activation, glycolysis and tumorigenesis in glioma cells. Cancer Sci. 2020;111:1979–1990. doi: 10.1111/cas.14408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Sharif TR, Sharif M. Overexpression of protein kinase C epsilon in astroglial brain tumor derived cell lines and primary tumor samples. Int J Oncol. 1999;15:237–243. [PubMed] [Google Scholar]
- 203.Okhrimenko H, Lu W, Xiang C, Hamburger N, Kazimirsky G, Brodie C. Protein kinase C-epsilon regulates the apoptosis and survival of glioma cells. Cancer Res. 2005;65:7301–7309. doi: 10.1158/0008-5472.CAN-05-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Besson A, Davy A, Robbins SM, Yong VW. Differential activation of ERKs to focal adhesions by PKC epsilon is required for PMA-induced adhesion and migration of human glioma cells. Oncogene. 2001;20:7398–7407. doi: 10.1038/sj.onc.1204899. [DOI] [PubMed] [Google Scholar]
- 205.Besson A, Wilson TL, Yong VW. The anchoring protein RACK1 links protein kinase cepsilon to integrin beta chains. Requirements for adhesion and motility. J Biol Chem. 2002;277:22073–22084. doi: 10.1074/jbc.M111644200. [DOI] [PubMed] [Google Scholar]
- 206.Aeder SE, Martin PM, Soh JW, Hussaini IM. PKC-eta mediates glioblastoma cell proliferation through the Akt and mTOR signaling pathways. Oncogene. 2004;23:9062–9069. doi: 10.1038/sj.onc.1208093. [DOI] [PubMed] [Google Scholar]
- 207.Uht RM, Amos S, Martin PM, Riggan AE, Hussaini IM. The protein kinase C-eta isoform induces proliferation in glioblastoma cell lines through an ERK/Elk-1 pathway. Oncogene. 2007;26:2885–2893. doi: 10.1038/sj.onc.1210090. [DOI] [PubMed] [Google Scholar]
- 208.Hussaini IM, Carpenter JE, Redpath GT, Sando JJ, Shaffrey ME, Vandenberg SR. Protein kinase C-eta regulates resistance to UV- and gamma-irradiation-induced apoptosis in glioblastoma cells by preventing caspase-9 activation. Neuro Oncol. 2002;4:9–21. doi: 10.1215/S1522851701000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Phillips E, Lang V, Bohlen J, Bethke F, Puccio L, Tichy D, Herold-Mende C, Hielscher T, Lichter P, Goidts V. Targeting atypical protein kinase C iota reduces viability in glioblastoma stem-like cells via a notch signaling mechanism. Int J Cancer. 2016;139:1776–1787. doi: 10.1002/ijc.30234. [DOI] [PubMed] [Google Scholar]
- 210.Guo H, Gu F, Li W, Zhang B, Niu R, Fu L, Zhang N, Ma Y. Reduction of protein kinase C zeta inhibits migration and invasion of human glioblastoma cells. J Neurochem. 2009;109:203–213. doi: 10.1111/j.1471-4159.2009.05946.x. [DOI] [PubMed] [Google Scholar]
- 211.Baldwin RM, Parolin DA, Lorimer IA. Regulation of glioblastoma cell invasion by PKC iota and RhoB. Oncogene. 2008;27:3587–3595. doi: 10.1038/sj.onc.1211027. [DOI] [PubMed] [Google Scholar]
- 212.Odia Y, Iwamoto FM, Moustakas A, Fraum TJ, Salgado CA, Li A, Kreisl TN, Sul J, Butman JA, Fine HA. A Phase II trial of enzastaurin (LY317615) in combination with bevacizumab in adults with recurrent malignant gliomas. J Neurooncol. 2016;127:127–135. doi: 10.1007/s11060-015-2020-x. [DOI] [PubMed] [Google Scholar]
- 213.Sevastre AS, Costachi A, Tataranu LG, Brandusa C, Artene SA, Stovicek O, Alexandru O, Danoiu S, Sfredel V, Dricu A. Glioblastoma pharmacotherapy: A multifaceted perspective of conventional and emerging treatments (review) Exp Ther Med. 2021;22:1408. doi: 10.3892/etm.2021.10844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Johnson BE, Mazor T, Hong C, Barnes M, Aihara K, McLean CY, Fouse SD, Yamamoto S, Ueda H, Tatsuno K, et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343:189–193. doi: 10.1126/science.1239947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Stieber D, Golebiewska A, Evers L, Lenkiewicz E, Brons NHC, Nicot N, Oudin A, Bougnaud S, Hertel F, Bjerkvig R, et al. Glioblastomas are composed of genetically divergent clones with distinct tumourigenic potential and variable stem cell-associated phenotypes. Acta Neuropathol. 2014;127:203–219. doi: 10.1007/s00401-013-1196-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Neftel C, Laffy J, Filbin MG, Hara T, Shore ME, Rahme GJ, Richman AR, Silverbush D, Shaw ML, Hebert CM, et al. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell. 2019;178:835–849.e21. doi: 10.1016/j.cell.2019.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Lauko A, Lo A, Ahluwalia MS, Lathia JD. Cancer cell heterogeneity and plasticity in glioblastoma and brain tumors. Semin Cancer Biol. 2022;82:162–175. doi: 10.1016/j.semcancer.2021.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Snuderl M, Fazlollahi L, Le LP, Nitta M, Zhelyazkova BH, Davidson CJ, Akhavanfard S, Cahill DP, Aldape KD, Betensky RA, et al. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell. 2011;20:810–817. doi: 10.1016/j.ccr.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 220.Nobusawa S, Lachuer J, Wierinckx A, Kim YH, Huang J, Legras C, Kleihues P, Ohgaki H. Intratumoral patterns of genomic imbalance in glioblastomas. Brain Pathol. 2010;20:936–944. doi: 10.1111/j.1750-3639.2010.00395.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Kumar A, Boyle EA, Tokita M, Mikheev AM, Sanger MC, Girard E, Silber JR, Gonzalez-Cuyar LF, Hiatt JB, Adey A, et al. Deep sequencing of multiple regions of glial tumors reveals spatial heterogeneity for mutations in clinically relevant genes. Genome Biol. 2014;15:530. doi: 10.1186/s13059-014-0530-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Bhaduri A, Di Lullo E, Jung D, Müller S, Crouch EE, Espinosa CS, Ozawa T, Alvarado B, Spatazza J, Cadwell CR, et al. Outer radial glia-like cancer stem cells contribute to heterogeneity of glioblastoma. Cell Stem Cell. 2020;26:48–63.e6. doi: 10.1016/j.stem.2019.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Pang B, Xu J, Hu J, Guo F, Wan L, Cheng M, Pang L. Single-cell RNA-seq reveals the invasive trajectory and molecular cascades underlying glioblastoma progression. Mol Oncol. 2019;13:2588–2603. doi: 10.1002/1878-0261.12569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Jacob F, Salinas RD, Zhang DY, Nguyen PTT, Schnoll JG, Wong SZH, Thokala R, Sheikh S, Saxena D, Prokop S, et al. A patient-derived glioblastoma organoid model and biobank recapitulates inter- and intra-tumoral heterogeneity. Cell. 2020;180:188–204.e22. doi: 10.1016/j.cell.2019.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Iwadate Y. Plasticity in glioma stem cell phenotype and its therapeutic implication. Neurol Med Chir (Tokyo) 2018;58:61–70. doi: 10.2176/nmc.ra.2017-0089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Koso H, Takeda H, Yew CCK, Ward JM, Nariai N, Ueno K, Nagasaki M, Watanabe S, Rust AG, Adams DJ, et al. Transposon mutagenesis identifies genes that transform neural stem cells into glioma-initiating cells. Proc Natl Acad Sci USA. 2012;109:E2998–E3007. doi: 10.1073/pnas.1215899109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Wang X, Prager BC, Wu Q, Kim LJY, Gimple RC, Shi Y, Yang K, Morton AR, Zhou W, Zhu Z, et al. Reciprocal signaling between glioblastoma stem cells and differentiated tumor cells promotes malignant progression. Cell Stem Cell. 2018;22:514–528.e5. doi: 10.1016/j.stem.2018.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Hsu JF, Chu SM, Liao CC, Wang CJ, Wang YS, Lai MY, Wang HC, Huang HR, Tsai MH. Nanotechnology and nanocarrier-based drug delivery as the potential therapeutic strategy for glioblastoma multiforme: An update. Cancers (Basel) 2021;13:195. doi: 10.3390/cancers13020195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Lathia JD, Heddleston JM, Venere M, Rich JN. Deadly teamwork: Neural cancer stem cells and the tumor microenvironment. Cell Stem Cell. 2011;8:482–485. doi: 10.1016/j.stem.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Yuan X, Curtin J, Xiong Y, Liu G, Waschsmann-Hogiu S, Farkas DL, Black KL, Yu JS. Isolation of cancer stem cells from adult glioblastoma multiforme. Oncogene. 2004;23:9392–9400. doi: 10.1038/sj.onc.1208311. [DOI] [PubMed] [Google Scholar]
- 231.Brescia P, Ortensi B, Fornasari L, Levi D, Broggi G, Pelicci G. CD133 is essential for glioblastoma stem cell maintenance. Stem Cells. 2013;31:857–869. doi: 10.1002/stem.1317. [DOI] [PubMed] [Google Scholar]
- 232.Carlsson SK, Brothers SP, Wahlestedt C. Emerging treatment strategies for glioblastoma multiforme. EMBO Mol Med. 2014;6:1359–1370. doi: 10.15252/emmm.201302627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Nusse R. Wnt signaling and stem cell control. Cell Res. 2008;18:523–527. doi: 10.1038/cr.2008.47. [DOI] [PubMed] [Google Scholar]
- 234.Sareddy GR, Kesanakurti D, Kirti PB, Babu PP. Nonsteroidal anti-inflammatory drugs diclofenac and celecoxib attenuates Wnt/β-catenin/Tcf signaling pathway in human glioblastoma cells. Neurochem Res. 2013;38:2313–2322. doi: 10.1007/s11064-013-1142-9. [DOI] [PubMed] [Google Scholar]
- 235.Yu JB, Jiang H, Zhan RY. Aberrant Notch signaling in glioblastoma stem cells contributes to tumor recurrence and invasion. Mol Med Rep. 2016;14:1263–1268. doi: 10.3892/mmr.2016.5391. [DOI] [PubMed] [Google Scholar]
- 236.Shih IM, Wang TL. Notch signaling, gamma-secretase inhibitors, and cancer therapy. Cancer Res. 2007;67:1879–1882. doi: 10.1158/0008-5472.CAN-06-3958. [DOI] [PubMed] [Google Scholar]
- 237.de la Iglesia N, Puram SV, Bonni A. STAT3 regulation of glioblastoma pathogenesis. Curr Mol Med. 2009;9:580–590. doi: 10.2174/156652409788488739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Porporato PE, Filigheddu N, Pedro JMBS, Kroemer G, Galluzzi L. Mitochondrial metabolism and cancer. Cell Res. 2018;28:265–280. doi: 10.1038/cr.2017.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Sighel D, Notarangelo M, Aibara S, Re A, Ricci G, Guida M, Soldano A, Adami V, Ambrosini C, Broso F, et al. Inhibition of mitochondrial translation suppresses glioblastoma stem cell growth. Cell Rep. 2021;35:109024. doi: 10.1016/j.celrep.2021.109024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Mai TT, Hamaï A, Hienzsch A, Cañeque T, Müller S, Wicinski J, Cabaud O, Leroy C, David A, Acevedo V, et al. Salinomycin kills cancer stem cells by sequestering iron in lysosomes. Nat Chem. 2017;9:1025–1033. doi: 10.1038/nchem.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Sharifzad F, Ghavami S, Verdi J, Mardpour S, Mollapour Sisakht M, Azizi Z, Taghikhani A, Łos MJ, Fakharian E, Ebrahimi M, Hamidieh AA. Glioblastoma cancer stem cell biology: Potential theranostic targets. Drug Resist Updat. 2019;42:35–45. doi: 10.1016/j.drup.2018.03.003. [DOI] [PubMed] [Google Scholar]
- 242.Saunders NR, Dreifuss JJ, Dziegielewska KM, Johansson PA, Habgood MD, Møllgård K, Bauer HC. The rights and wrongs of blood-brain barrier permeability studies: A walk through 100 years of history. Front Neurosci. 2014;8:404. doi: 10.3389/fnins.2014.00404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Daneman R, Prat A. The blood-brain barrier. Cold Spring Harb Perspect Biol. 2015;7:a020412. doi: 10.1101/cshperspect.a020412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Liebner S, Fischmann A, Rascher G, Duffner F, Grote EH, Kalbacher H, Wolburg H. Claudin-1 and claudin-5 expression and tight junction morphology are altered in blood vessels of human glioblastoma multiforme. Acta Neuropathol. 2000;100:323–331. doi: 10.1007/s004010000180. [DOI] [PubMed] [Google Scholar]
- 245.Wolburg H, Wolburg-Buchholz K, Kraus J, Rascher-Eggstein G, Liebner S, Hamm S, Duffner F, Grote EH, Risau W, Engelhardt B. Localization of claudin-3 in tight junctions of the blood-brain barrier is selectively lost during experimental autoimmune encephalomyelitis and human glioblastoma multiforme. Acta Neuropathol. 2003;105:586–592. doi: 10.1007/s00401-003-0688-z. [DOI] [PubMed] [Google Scholar]
- 246.Ishihara H, Kubota H, Lindberg RLP, Leppert D, Gloor SM, Errede M, Virgintino D, Fontana A, Yonekawa Y, Frei K. Endothelial cell barrier impairment induced by glioblastomas and transforming growth factor beta2 involves matrix metalloproteinases and tight junction proteins. J Neuropathol Exp Neurol. 2008;67:435–448. doi: 10.1097/NEN.0b013e31816fd622. [DOI] [PubMed] [Google Scholar]
- 247.Rama AR, Alvarez PJ, Madeddu R, Aranega A. ABC transporters as differentiation markers in glioblastoma cells. Mol Biol Rep. 2014;41:4847–4851. doi: 10.1007/s11033-014-3423-z. [DOI] [PubMed] [Google Scholar]
- 248.Lockman PR, Mittapalli RK, Taskar KS, Rudraraju V, Gril B, Bohn KA, Adkins CE, Roberts A, Thorsheim HR, Gaasch JA, et al. Heterogeneous blood-tumor barrier permeability determines drug efficacy in experimental brain metastases of breast cancer. Clin Cancer Res. 2010;16:5664–5678. doi: 10.1158/1078-0432.CCR-10-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Taskar KS, Rudraraju V, Mittapalli RK, Samala R, Thorsheim HR, Lockman J, Gril B, Hua E, Palmieri D, Polli JW, et al. Lapatinib distribution in HER2 overexpressing experimental brain metastases of breast cancer. Pharm Res. 2012;29:770–781. doi: 10.1007/s11095-011-0601-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Tiwary S, Morales JE, Kwiatkowski SC, Lang FF, Rao G, McCarty JH. Metastatic brain tumors disrupt the blood-brain barrier and alter lipid metabolism by inhibiting expression of the endothelial cell fatty acid transporter Mfsd2a. Sci Rep. 2018;8:8267. doi: 10.1038/s41598-018-26636-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Elmeliegy MA, Carcaboso AM, Tagen M, Bai F, Stewart CF. Role of ATP-binding cassette and solute carrier transporters in erlotinib CNS penetration and intracellular accumulation. Clin Cancer Res. 2011;17:89–99. doi: 10.1158/1078-0432.CCR-10-1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.de Vries NA, Buckle T, Zhao J, Beijnen JH, Schellens JHM, van Tellingen O. Restricted brain penetration of the tyrosine kinase inhibitor erlotinib due to the drug transporters P-gp and BCRP. Invest New Drugs. 2012;30:443–449. doi: 10.1007/s10637-010-9569-1. [DOI] [PubMed] [Google Scholar]
- 253.Oberoi RK, Mittapalli RK, Elmquist WF. Pharmacokinetic assessment of efflux transport in sunitinib distribution to the brain. J Pharmacol Exp Ther. 2013;347:755–764. doi: 10.1124/jpet.113.208959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Lin F, de Gooijer MC, Roig EM, Buil LCM, Christner SM, Beumer JH, Würdinger T, Beijnen JH, van Tellingen O. ABCB1, ABCG2, and PTEN determine the response of glioblastoma to temozolomide and ABT-888 therapy. Clin Cancer Res. 2014;20:2703–2713. doi: 10.1158/1078-0432.CCR-14-0084. [DOI] [PubMed] [Google Scholar]
- 255.de Gooijer MC, de Vries NA, Buckle T, Buil LCM, Beijnen JH, Boogerd W, van Tellingen O. Improved brain penetration and antitumor efficacy of temozolomide by inhibition of ABCB1 and ABCG2. Neoplasia. 2018;20:710–720. doi: 10.1016/j.neo.2018.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Choi YH, Yu AM. ABC transporters in multidrug resistance and pharmacokinetics, and strategies for drug development. Curr Pharm Des. 2014;20:793–807. doi: 10.2174/138161282005140214165212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Tran S, DeGiovanni PJ, Piel B, Rai P. Cancer nanomedicine: A review of recent success in drug delivery. Clin Transl Med. 2017;6:44. doi: 10.1186/s40169-017-0175-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Sun T, Zhang YS, Pang B, Hyun DC, Yang M, Xia Y. Engineered nanoparticles for drug delivery in cancer therapy. Angew Chem Int Ed Engl. 2014;53:12320–12364. doi: 10.1002/anie.201403036. [DOI] [PubMed] [Google Scholar]
- 259.Budama-Kilinc Y, Kecel-Gunduz S, Cakir-Koc R, Aslan B, Bicak B, Kokcu Y, Ozel AE, Akyuz S. Structural characterization and drug delivery system of natural growth-modulating peptide against glioblastoma cancer. Int J Pept Res Ther. 2021;27:2015–2028. doi: 10.1007/s10989-021-10229-5. [DOI] [Google Scholar]
- 260.Pasut G. Grand challenges in nano-based drug delivery. Front Med Technol. 2019;1:1. doi: 10.3389/fmedt.2019.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Ambruosi A, Gelperina S, Khalansky A, Tanski S, Theisen A, Kreuter J. Influence of surfactants, polymer and doxorubicin loading on the anti-tumour effect of poly(butyl cyanoacrylate) nanoparticles in a rat glioma model. J Microencapsul. 2006;23:582–592. doi: 10.1080/02652040600788080. [DOI] [PubMed] [Google Scholar]
- 262.Caraway CA, Gaitsch H, Wicks EE, Kalluri A, Kunadi N, Tyler BM. Polymeric nanoparticles in brain cancer therapy: A review of current approaches. Polymers (Basel) 2022;14:2963. doi: 10.3390/polym14142963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Ramalho MJ, Sevin E, Gosselet F, Lima J, Coelho MAN, Loureiro JA, Pereira MC. Receptor-mediated PLGA nanoparticles for glioblastoma multiforme treatment. Int J Pharm. 2018;545:84–92. doi: 10.1016/j.ijpharm.2018.04.062. [DOI] [PubMed] [Google Scholar]
- 264.Kuang Y, Jiang X, Zhang Y, Lu Y, Ma H, Guo Y, Zhang Y, An S, Li J, Liu L, et al. Dual functional peptide-driven nanoparticles for highly efficient glioma-targeting and drug codelivery. Mol Pharm. 2016;13:1599–1607. doi: 10.1021/acs.molpharmaceut.6b00051. [DOI] [PubMed] [Google Scholar]
- 265.Mathew EN, Berry BC, Yang HW, Carroll RS, Johnson MD. Delivering therapeutics to glioblastoma: Overcoming biological constraints. Int J Mol Sci. 2022;23:1711. doi: 10.3390/ijms23031711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Wei KC, Chu PC, Wang HYJ, Huang CY, Chen PY, Tsai HC, Lu YJ, Lee PY, Tseng IC, Feng LY, et al. Focused ultrasound-induced blood-brain barrier opening to enhance temozolomide delivery for glioblastoma treatment: A preclinical study. PLoS One. 2013;8:e58995. doi: 10.1371/journal.pone.0058995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Treat LH, McDannold N, Vykhodtseva N, Zhang Y, Tam K, Hynynen K. Targeted delivery of doxorubicin to the rat brain at therapeutic levels using MRI-guided focused ultrasound. Int J Cancer. 2007;121:901–907. doi: 10.1002/ijc.22732. [DOI] [PubMed] [Google Scholar]
- 268.Coluccia D, Figueiredo CA, Wu MY, Riemenschneider AN, Diaz R, Luck A, Smith C, Das S, Ackerley C, O'Reilly M, et al. Enhancing glioblastoma treatment using cisplatin-gold-nanoparticle conjugates and targeted delivery with magnetic resonance-guided focused ultrasound. Nanomedicine. 2018;14:1137–1148. doi: 10.1016/j.nano.2018.01.021. [DOI] [PubMed] [Google Scholar]
- 269.Timbie KF, Mead BP, Price RJ. Drug and gene delivery across the blood-brain barrier with focused ultrasound. J Control Release. 2015;219:61–75. doi: 10.1016/j.jconrel.2015.08.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Burgess A, Shah K, Hough O, Hynynen K. Focused ultrasound-mediated drug delivery through the blood-brain barrier. Expert Rev Neurother. 2015;15:477–491. doi: 10.1586/14737175.2015.1028369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Dutta D, Heo I, Clevers H. Disease modeling in stem cell-derived 3D organoid systems. Trends Mol Med. 2017;23:393–410. doi: 10.1016/j.molmed.2017.02.007. [DOI] [PubMed] [Google Scholar]
- 272.Nishida N, Yano H, Nishida T, Kamura T, Kojiro M. Angiogenesis in cancer. Vasc Health Risk Manag. 2006;2:213–219. doi: 10.2147/vhrm.2006.2.3.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Hillen F, Griffioen AW. Tumour vascularization: Sprouting angiogenesis and beyond. Cancer Metastasis Rev. 2007;26:489–502. doi: 10.1007/s10555-007-9094-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Joseph JV, Conroy S, Pavlov K, Sontakke P, Tomar T, Eggens-Meijer E, Balasubramaniyan V, Wagemakers M, den Dunnen WF, Kruyt FA. Hypoxia enhances migration and invasion in glioblastoma by promoting a mesenchymal shift mediated by the HIF1α-ZEB1 axis. Cancer Lett. 2015;359:107–116. doi: 10.1016/j.canlet.2015.01.010. [DOI] [PubMed] [Google Scholar]
- 275.Huang W, Ding X, Ye H, Wang J, Shao J, Huang T. Hypoxia enhances the migration and invasion of human glioblastoma U87 cells through PI3K/Akt/mTOR/HIF-1α pathway. Neuroreport. 2018;29:1578–1585. doi: 10.1097/WNR.0000000000001156. [DOI] [PubMed] [Google Scholar]
- 276.Chen JWE, Lumibao J, Blazek A, Gaskins HR, Harley B. Hypoxia activates enhanced invasive potential and endogenous hyaluronic acid production by glioblastoma cells. Biomater Sci. 2018;6:854–862. doi: 10.1039/C7BM01195D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Rosa P, Catacuzzeno L, Sforna L, Mangino G, Carlomagno S, Mincione G, Petrozza V, Ragona G, Franciolini F, Calogero A. BK channels blockage inhibits hypoxia-induced migration and chemoresistance to cisplatin in human glioblastoma cells. J Cell Physiol. 2018;233:6866–6877. doi: 10.1002/jcp.26448. [DOI] [PubMed] [Google Scholar]
- 278.Velásquez C, Mansouri S, Gutiérrez O, Mamatjan Y, Mollinedo P, Karimi S, Singh O, Terán N, Martino J, Zadeh G, Fernández-Luna JL. Hypoxia can induce migration of glioblastoma cells through a methylation-dependent control of ODZ1 gene expression. Front Oncol. 2019;9:1036. doi: 10.3389/fonc.2019.01036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Huang S, Michalek JE, Reardon DA, Wen PY, Floyd JR, Fox PT, Clarke GD, Jerabek PA, Schmainda KM, Muzi M, et al. Assessment of tumor hypoxia and perfusion in recurrent glioblastoma following bevacizumab failure using MRI and 18F-FMISO PET. Sci Rep. 2021;11:7632. doi: 10.1038/s41598-021-84331-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Chédeville AL, Madureira PA. The role of hypoxia in glioblastoma radiotherapy resistance. Cancers (Basel) 2021;13:542. doi: 10.3390/cancers13030542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Zhou W, Ke SQ, Huang Z, Flavahan W, Fang X, Paul J, Wu L, Sloan AE, McLendon RE, Li X, et al. Periostin secreted by glioblastoma stem cells recruits M2 tumour-associated macrophages and promotes malignant growth. Nat Cell Biol. 2015;17:170–182. doi: 10.1038/ncb3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Wei J, Barr J, Kong LY, Wang Y, Wu A, Sharma AK, Gumin J, Henry V, Colman H, Priebe W, et al. Glioblastoma cancer-initiating cells inhibit T-cell proliferation and effector responses by the signal transducers and activators of transcription 3 pathway. Mol Cancer Ther. 2010;9:67–78. doi: 10.1158/1535-7163.MCT-09-0734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Yang F, He Z, Duan H, Zhang D, Li J, Yang H, Dorsey JF, Zou W, Nabavizadeh SA, Bagley SJ, et al. Synergistic immunotherapy of glioblastoma by dual targeting of IL-6 and CD40. Nat Commun. 2021;12:3424. doi: 10.1038/s41467-021-23832-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Banyer JL, Hamilton NH, Ramshaw IA, Ramsay AJ. Cytokines in innate and adaptive immunity. Rev Immunogenet. 2000;2:359–373. [PubMed] [Google Scholar]
- 285.Conlon KC, Miljkovic MD, Waldmann TA. Cytokines in the treatment of cancer. J Interferon Cytokine Res. 2019;39:6–21. doi: 10.1089/jir.2018.0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Zhu VF, Yang J, Lebrun DG, Li M. Understanding the role of cytokines in glioblastoma multiforme pathogenesis. Cancer Lett. 2012;316:139–150. doi: 10.1016/j.canlet.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 287.Wakabayashi T, Kayama T, Nishikawa R, Takahashi H, Hashimoto N, Takahashi J, Aoki T, Sugiyama K, Ogura M, Natsume A, Yoshida J. A multicenter Phase I trial of combination therapy with interferon-β and temozolomide for high-grade gliomas (INTEGRA study): The final report. J Neurooncol. 2011;104:573–577. doi: 10.1007/s11060-011-0529-1. [DOI] [PubMed] [Google Scholar]
- 288.Han MH, Kim JM, Cheong JH, Ryu JI, Won YD, Nam GH, Kim CH. Efficacy of cytokine-induced killer cell immunotherapy for patients with pathologically pure glioblastoma. Front Oncol. 2022;12:851628. doi: 10.3389/fonc.2022.851628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Iwami K, Natsume A, Wakabayashi T. Cytokine therapy of gliomas. Prog Neurol Surg. 2018;32:79–89. doi: 10.1159/000469682. [DOI] [PubMed] [Google Scholar]
- 290.Xu S, Tang L, Li X, Fan F, Liu Z. Immunotherapy for glioma: Current management and future application. Cancer Lett. 2020;476:1–12. doi: 10.1016/j.canlet.2020.02.002. [DOI] [PubMed] [Google Scholar]
- 291.Zhang Y, Zhang Z. The history and advances in cancer immunotherapy: Understanding the characteristics of tumor-infiltrating immune cells and their therapeutic implications. Cell Mol Immunol. 2020;17:807–821. doi: 10.1038/s41423-020-0488-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.de Mello RA, Veloso AF, Esrom Catarina P, Nadine S, Antoniou G. Potential role of immunotherapy in advanced non-small-cell lung cancer. Onco Targets Ther. 2016;10:21–30. doi: 10.2147/OTT.S90459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Dine J, Gordon R, Shames Y, Kasler MK, Barton-Burke M. Immune checkpoint inhibitors: An innovation in immunotherapy for the treatment and management of patients with cancer. Asia Pac J Oncol Nurs. 2017;4:127–135. doi: 10.4103/apjon.apjon_4_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Mpakali A, Stratikos E. The role of antigen processing and presentation in cancer and the efficacy of immune checkpoint inhibitor immunotherapy. Cancers (Basel) 2021;13:134. doi: 10.3390/cancers13010134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Wang J, Toregrosa-Allen S, Elzey BD, Utturkar S, Lanman NA, Bernal-Crespo V, Behymer MM, Knipp GT, Yun Y, Veronesi MC, et al. Multispecific targeting of glioblastoma with tumor microenvironment-responsive multifunctional engineered NK cells. Proc Natl Acad Sci USA. 2021;118:e2107507118. doi: 10.1073/pnas.2107507118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Rosenberg SA, Restifo NP, Yang JC, Morgan RA, Dudley ME. Adoptive cell transfer: A clinical path to effective cancer immunotherapy. Nat Rev Cancer. 2008;8:299–308. doi: 10.1038/nrc2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Redeker A, Arens R. Improving adoptive T cell therapy: The particular role of T cell costimulation, cytokines, and post-transfer vaccination. Front Immunol. 2016;7:345. doi: 10.3389/fimmu.2016.00345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Chruściel E, Urban-Wójciuk Z, Arcimowicz Ł, Kurkowiak M, Kowalski J, Gliwiński M, Marjański T, Rzyman W, Biernat W, Dziadziuszko R, et al. Adoptive cell therapy-harnessing antigen-specific T cells to target solid tumours. Cancers (Basel) 2020;12:683. doi: 10.3390/cancers12030683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Staquicini FI, Smith TL, Tang FHF, Gelovani JG, Giordano RJ, Libutti SK, Sidman RL, Cavenee WK, Arap W, Pasqualini R. Targeted AAVP-based therapy in a mouse model of human glioblastoma: A comparison of cytotoxic versus suicide gene delivery strategies. Cancer Gene Ther. 2020;27:301–310. doi: 10.1038/s41417-019-0101-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Vigneswaran K, Neill S, Hadjipanayis CG. Beyond the World Health Organization grading of infiltrating gliomas: Advances in the molecular genetics of glioma classification. Ann Transl Med. 2015;3:95. doi: 10.3978/j.issn.2305-5839.2015.03.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Goodwin CR, Rath P, Oyinlade O, Lopez H, Mughal S, Xia S, Xia S, Li Y, Kaur H, Zhou X, et al. Crizotinib and erlotinib inhibits growth of c-Met+/EGFRvIII+ primary human glioblastoma xenografts. Clin Neurol Neurosurg. 2018;171:26–33. doi: 10.1016/j.clineuro.2018.02.041. [DOI] [PubMed] [Google Scholar]
- 302.Sestito S, Runfola M, Tonelli M, Chiellini G, Rapposelli S. New multitarget approaches in the War against glioblastoma: A mini-perspective. Front Pharmacol. 2018;9:874. doi: 10.3389/fphar.2018.00874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Lang F, Liu Y, Chou FJ, Yang C. Genotoxic therapy and resistance mechanism in gliomas. Pharmacol Ther. 2021;228:107922. doi: 10.1016/j.pharmthera.2021.107922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Lassman AB, Pugh SL, Gilbert MR, Aldape KD, Geinoz S, Beumer JH, Christner SM, Komaki R, DeAngelis LM, Gaur R, et al. Phase 2 trial of dasatinib in target-selected patients with recurrent glioblastoma (RTOG 0627) Neuro Oncol. 2015;17:992–998. doi: 10.1093/neuonc/nov011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Baratta MG. Glioblastoma is ‘hot’ for personalized vaccines. Nat Rev Cancer. 2019;19:129. doi: 10.1038/s41568-019-0118-8. [DOI] [PubMed] [Google Scholar]
- 306.Wiwatchaitawee K, Quarterman JC, Geary SM, Salem AK. Enhancement of therapies for glioblastoma (GBM) using nanoparticle-based delivery systems. AAPS PharmSciTech. 2021;22:71. doi: 10.1208/s12249-021-01928-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Pearson JRD, Cuzzubbo S, McArthur S, Durrant LG, Adhikaree J, Tinsley CJ, Pockley AG, McArdle SEB. Immune escape in glioblastoma multiforme and the adaptation of immunotherapies for treatment. Front Immunol. 2020;11:582106. doi: 10.3389/fimmu.2020.582106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Burger MC, Zhang C, Harter PN, Romanski A, Strassheimer F, Senft C, Tonn T, Steinbach JP, Wels WS. CAR-engineered NK cells for the treatment of glioblastoma: Turning innate effectors into precision tools for cancer immunotherapy. Front Immunol. 2019;10:2683. doi: 10.3389/fimmu.2019.02683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Granzin M, Wagner J, Köhl U, Cerwenka A, Huppert V, Ullrich E. Shaping of natural killer cell antitumor activity by ex vivo cultivation. Front Immunol. 2017;8:458. doi: 10.3389/fimmu.2017.00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Yu MW, Quail DF. Immunotherapy for glioblastoma: Current progress and challenges. Front Immunol. 2021;12:676301. doi: 10.3389/fimmu.2021.676301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Dietrich J, Rao K, Pastorino S, Kesari S. Corticosteroids in brain cancer patients: Benefits and pitfalls. Expert Rev Clin Pharmacol. 2011;4:233–242. doi: 10.1586/ecp.11.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Koyama S, Akbay EA, Li YY, Herter-Sprie GS, Buczkowski KA, Richards WG, Gandhi L, Redig AJ, Rodig SJ, Asahina H, et al. Adaptive resistance to therapeutic PD-1 blockade is associated with upregulation of alternative immune checkpoints. Nat Commun. 2016;7:10501. doi: 10.1038/ncomms10501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Chan HY, Choi J, Jackson C, Lim M. Combination immunotherapy strategies for glioblastoma. J Neurooncol. 2021;151:375–391. doi: 10.1007/s11060-020-03481-0. [DOI] [PubMed] [Google Scholar]
- 314.Bausart M, Préat V, Malfanti A. Immunotherapy for glioblastoma: The promise of combination strategies. J Exp Clin Cancer Res. 2022;41:35. doi: 10.1186/s13046-022-02251-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Oh T, Sayegh ET, Fakurnejad S, Oyon D, Lamano JB, DiDomenico JD, Bloch O, Parsa AT. Vaccine therapies in malignant glioma. Curr Neurol Neurosci Rep. 2015;15:508. doi: 10.1007/s11910-014-0508-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Hollingsworth RE, Jansen K. Turning the corner on therapeutic cancer vaccines. NPJ Vaccines. 2019;4:7. doi: 10.1038/s41541-019-0103-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Liu J, Fu M, Wang M, Wan D, Wei Y, Wei X. Cancer vaccines as promising immuno-therapeutics: Platforms and current progress. J Hematol Oncol. 2022;15:28. doi: 10.1186/s13045-022-01247-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Keskin DB, Anandappa AJ, Sun J, Tirosh I, Mathewson ND, Li S, Oliveira G, Giobbie-Hurder A, Felt K, Gjini E, et al. Neoantigen vaccine generates intratumoral T cell responses in Phase Ib glioblastoma trial. Nature. 2019;565:234–239. doi: 10.1038/s41586-018-0792-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Hilf N, Kuttruff-Coqui S, Frenzel K, Bukur V, Stevanović S, Gouttefangeas C, Platten M, Tabatabai G, Dutoit V, van der Burg SH, et al. Actively personalized vaccination trial for newly diagnosed glioblastoma. Nature. 2019;565:240–245. doi: 10.1038/s41586-018-0810-y. [DOI] [PubMed] [Google Scholar]
- 320.Inc. AB, corp-author. AIVITA biomedical's phase 2 glioblastoma trial shows improved progression free survival. https://www.prnewswire.com/news-releases/aivita-biomedicals-phase-2-glioblastoma-trial-shows-improved-progression-free-survival-301307757.html [cited 2022 Aug 5]. Available from: [Google Scholar]
- 321.Burkhardt JK, Riina H, Shin BJ, Christos P, Kesavabhotla K, Hofstetter CP, Tsiouris AJ, Boockvar JA. Intra-arterial delivery of bevacizumab after blood-brain barrier disruption for the treatment of recurrent glioblastoma: Progression-free survival and overall survival. World Neurosurg. 2012;77:130–134. doi: 10.1016/j.wneu.2011.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed BV, Curry WT, Martuza RL, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Wang Q, Hu B, Hu X, Kim H, Squatrito M, Scarpace L, deCarvalho AC, Lyu S, Li P, Li Y, et al. Tumor evolution of glioma-intrinsic gene expression subtypes associates with immunological changes in the microenvironment. Cancer Cell. 2017;32:42–56.e6. doi: 10.1016/j.ccell.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Petroni G, Buqué A, Zitvogel L, Kroemer G, Galluzzi L. Immunomodulation by targeted anticancer agents. Cancer Cell. 2021;39:310–345. doi: 10.1016/j.ccell.2020.11.009. [DOI] [PubMed] [Google Scholar]
- 325.Song Y, Fu Y, Xie Q, Zhu B, Wang J, Zhang B. Anti-angiogenic agents in combination with immune checkpoint inhibitors: A promising strategy for cancer treatment. Front Immunol. 2020;11:1956. doi: 10.3389/fimmu.2020.01956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Selek L, Seigneuret E, Nugue G, Wion D, Nissou MF, Salon C, Seurin MJ, Carozzo C, Ponce F, Roger T, Berger F. Imaging and histological characterization of a human brain xenograft in pig: The first induced glioma model in a large animal. J Neurosci Methods. 2014;221:159–165. doi: 10.1016/j.jneumeth.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 327.Khoshnevis M, Carozzo C, Bonnefont-Rebeix C, Belluco S, Leveneur O, Chuzel T, Pillet-Michelland E, Dreyfus M, Roger T, Berger F, Ponce F. Development of induced glioblastoma by implantation of a human xenograft in Yucatan minipig as a large animal model. J Neurosci Methods. 2017;282:61–68. doi: 10.1016/j.jneumeth.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 328.Tora MS, Texakalidis P, Neill S, Wetzel J, Rindler RS, Hardcastle N, Nagarajan PP, Krasnopeyev A, Roach C, James R, et al. Lentiviral vector induced modeling of high-grade spinal cord glioma in minipigs. Sci Rep. 2020;10:5291. doi: 10.1038/s41598-020-62167-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Khoshnevis M, Carozzo C, Brown R, Bardiès M, Bonnefont-Rebeix C, Belluco S, Nennig C, Marcon L, Tillement O, Gehan H, et al. Feasibility of intratumoral 165Holmium siloxane delivery to induced U87 glioblastoma in a large animal model, the Yucatan minipig. PLoS One. 2020;15:e0234772. doi: 10.1371/journal.pone.0234772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Hicks WH, Bird CE, Pernik MN, Haider AS, Dobariya A, Abdullah KG, Aoun SG, Bentley RT, Cohen-Gadol AA, Bachoo RM, et al. Large animal models of glioma: Current status and future prospects. Anticancer Res. 2021;41:5343–5353. doi: 10.21873/anticanres.15347. [DOI] [PubMed] [Google Scholar]
- 331.Zhao CY, Cheng R, Yang Z, Tian ZM. Nanotechnology for cancer therapy based on chemotherapy. Molecules. 2018;23:826. doi: 10.3390/molecules23040826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Zaimy MA, Saffarzadeh N, Mohammadi A, Pourghadamyari H, Izadi P, Sarli A, Moghaddam LK, Paschepari SR, Azizi H, Torkamandi S, Tavakkoly-Bazzaz J. New methods in the diagnosis of cancer and gene therapy of cancer based on nanoparticles. Cancer Gene Ther. 2017;24:233–243. doi: 10.1038/cgt.2017.16. [DOI] [PubMed] [Google Scholar]
- 333.Geletneky K, Hajda J, Angelova AL, Leuchs B, Capper D, Bartsch AJ, Neumann JO, Schöning T, Hüsing J, Beelte B, et al. Oncolytic H-1 parvovirus shows safety and signs of immunogenic activity in a first Phase I/IIa glioblastoma trial. Mol Ther. 2017;25:2620–2634. doi: 10.1016/j.ymthe.2017.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Angelova AL, Barf M, Geletneky K, Unterberg A, Rommelaere J. Immunotherapeutic potential of oncolytic H-1 parvovirus: Hints of glioblastoma microenvironment conversion towards immunogenicity. Viruses. 2017;9:382. doi: 10.3390/v9120382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Suryawanshi YR, Schulze AJ. Oncolytic viruses for malignant glioma: On the verge of success? Viruses. 2021;13:1294. doi: 10.3390/v13071294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Li J, Wang W, Wang J, Cao Y, Wang S, Zhao J. Viral gene therapy for glioblastoma multiforme: A promising hope for the current dilemma. Front Oncol. 2021;11:678226. doi: 10.3389/fonc.2021.678226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Clement PMJ, Dirven L, Eoli M, Sepulveda-Sanchez JM, Walenkamp AME, Frenel JS, Franceschi E, Weller M, Chinot O, De Vos FYFL, et al. Impact of depatuxizumab mafodotin on health-related quality of life and neurological functioning in the Phase II EORTC 1410/INTELLANCE 2 trial for EGFR-amplified recurrent glioblastoma. Eur J Cancer. 2021;147:1–12. doi: 10.1016/j.ejca.2021.01.010. [DOI] [PubMed] [Google Scholar]
- 338.Sathornsumetee S, Desjardins A, Vredenburgh JJ, McLendon RE, Marcello J, Herndon JE, Mathe A, Hamilton M, Rich JN, Norfleet JA, et al. Phase II trial of bevacizumab and erlotinib in patients with recurrent malignant glioma. Neuro Oncol. 2010;12:1300–1310. doi: 10.1093/neuonc/noq099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Mellinghoff IK, Wang MY, Vivanco I, Haas-Kogan DA, Zhu S, Dia EQ, Lu KV, Yoshimoto K, Huang JH, Chute DJ, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med. 2005;353:2012–2024. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- 340.Raizer JJ, Abrey LE, Lassman AB, Chang SM, Lamborn KR, Kuhn JG, Yung WK, Gilbert MR, Aldape KD, Wen PY, et al. A Phase I trial of erlotinib in patients with nonprogressive glioblastoma multiforme postradiation therapy, and recurrent malignant gliomas and meningiomas. Neuro Oncol. 2010;12:87–94. doi: 10.1093/neuonc/nop017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Lucas JT, Jr, Knapp BJ, Uh J, Hua CH, Merchant TE, Hwang SN, Patay Z, Broniscer A. Posttreatment DSC-MRI is predictive of early treatment failure in children with supratentorial high-grade glioma treated with erlotinib. Clin Neuroradiol. 2018;28:393–400. doi: 10.1007/s00062-017-0580-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Prados MD, Chang SM, Butowski N, DeBoer R, Parvataneni R, Carliner H, Kabuubi P, Ayers-Ringler J, Rabbitt J, Page M, et al. Phase II study of erlotinib plus temozolomide during and after radiation therapy in patients with newly diagnosed glioblastoma multiforme or gliosarcoma. J Clin Oncol. 2009;27:579–54. doi: 10.1200/JCO.2008.18.9639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Hegi ME, Diserens AC, Bady P, Kamoshima Y, Kouwenhoven MCM, Delorenzi M, Lambiv WL, Hamou MF, Matter MS, Koch A, et al. Pathway analysis of glioblastoma tissue after preoperative treatment with the EGFR tyrosine kinase inhibitor gefitinib-a Phase II trial. Mol Cancer Ther. 2011;10:1102–1112. doi: 10.1158/1535-7163.MCT-11-0048. [DOI] [PubMed] [Google Scholar]
- 344.Wang Y, Liang D, Chen J, Chen H, Fan R, Gao Y, Gao Y, Tao R, Zhang H. Targeted therapy with anlotinib for a patient with an oncogenic FGFR3-TACC3 fusion and recurrent glioblastoma. Oncologist. 2021;26:173–177. doi: 10.1002/onco.13530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Hainsworth JD, Ervin T, Friedman E, Priego V, Murphy PB, Clark BL, Lamar RE. Concurrent radiotherapy and temozolomide followed by temozolomide and sorafenib in the first-line treatment of patients with glioblastoma multiforme. Cancer. 2010;116:3663–3669. doi: 10.1002/cncr.25275. [DOI] [PubMed] [Google Scholar]
- 346.Shahid S, Kushner BH, Modak S, Basu EM, Rubin EM, Gundem G, Papaemmanuil E, Roberts SS. Association of BRAF V600E mutations with vasoactive intestinal peptide syndrome in MYCN-amplified neuroblastoma. Pediatr Blood Cancer. 2021;68:e29265. doi: 10.1002/pbc.29265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Rahman MA, Brekke J, Arnesen V, Hannisdal MH, Navarro AG, Waha A, Herfindal L, Rygh CB, Bratland E, Brandal P, et al. Sequential bortezomib and temozolomide treatment promotes immunological responses in glioblastoma patients with positive clinical outcomes: A phase 1B study. Immun Inflamm Dis. 2020;8:342–359. doi: 10.1002/iid3.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Morin A, Soane C, Pierce A, Sanford B, Jones KL, Crespo M, Zahedi S, Vibhakar R, Mulcahy Levy JM. Proteasome inhibition as a therapeutic approach in atypical teratoid/rhabdoid tumors. Neurooncol Adv. 2020;2:vdaa051. doi: 10.1093/noajnl/vdaa051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Gupta SK, Kizilbash SH, Carlson BL, Mladek AC, Boakye-Agyeman F, Bakken KK, Pokorny JL, Schroeder MA, Decker PA, Cen L, et al. Delineation of MGMT hypermethylation as a biomarker for veliparib-mediated temozolomide-sensitizing therapy of glioblastoma. J Natl Cancer Inst. 2016;108:djv369. doi: 10.1093/jnci/djv369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Xiong Y, Guo Y, Liu Y, Wang H, Gong W, Liu Y, Wang X, Gao Y, Yu F, Su D, et al. Pamiparib is a potent and selective PARP inhibitor with unique potential for the treatment of brain tumor. Neoplasia. 2020;22:431–440. doi: 10.1016/j.neo.2020.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Lowery MA, Burris HA, III, Janku F, Shroff RT, Cleary JM, Azad NS, Goyal L, Maher EA, Gore L, Hollebecque A, et al. Safety and activity of ivosidenib in patients with IDH1-mutant advanced cholangiocarcinoma: A phase 1 study. Lancet Gastroenterol Hepatol. 2019;4:711–720. doi: 10.1016/S2468-1253(19)30189-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.DiNardo CD, Hochhaus A, Frattini MG, Yee K, Zander T, Krämer A, Chen X, Ji Y, Parikh N, Choi J, Wei AH. A phase 1 study of IDH305 in patients with IDH1R132-mutant acute myeloid leukemia or myelodysplastic syndrome. J Cancer Res Clin Oncol. 2022 Mar 30; doi: 10.1007/s00432-022-03983-6. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Penas-Prado M, Hess KR, Fisch MJ, Lagrone LW, Groves MD, Levin VA, De Groot JF, Puduvalli VK, Colman H, Volas-Redd G, et al. Randomized Phase II adjuvant factorial study of dose-dense temozolomide alone and in combination with isotretinoin, celecoxib, and/or thalidomide for glioblastoma. Neuro Oncol. 2015;17:266–273. doi: 10.1093/neuonc/nou155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Kesari S, Schiff D, Henson JW, Muzikansky A, Gigas DC, Doherty L, Batchelor TT, Longtine JA, Ligon KL, Weaver S, et al. Phase II study of temozolomide, thalidomide, and celecoxib for newly diagnosed glioblastoma in adults. Neuro Oncol. 2008;10:300–308. doi: 10.1215/15228517-2008-005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Halatsch ME, Dwucet A, Schmidt CJ, Mühlnickel J, Heiland T, Zeiler K, Siegelin MD, Kast RE, Karpel-Massler G. In vitro and clinical compassionate use experiences with the drug-repurposing approach CUSP9v3 in glioblastoma. Pharmaceuticals (Basel) 2021;14:1241. doi: 10.3390/ph14121241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Morgan RA, Johnson LA, Davis JL, Zheng Z, Woolard KD, Reap EA, Feldman SA, Chinnasamy N, Kuan CT, Song H, et al. Recognition of glioma stem cells by genetically modified T cells targeting EGFRvIII and development of adoptive cell therapy for glioma. Hum Gene Ther. 2012;23:1043–1053. doi: 10.1089/hum.2012.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Liau LM, Ashkan K, Tran DD, Campian JL, Trusheim JE, Cobbs CS, Heth JA, Salacz M, Taylor S, D'Andre SD, et al. First results on survival from a large Phase 3 clinical trial of an autologous dendritic cell vaccine in newly diagnosed glioblastoma. J Transl Med. 2018;16:142. doi: 10.1186/s12967-018-1507-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Weller M, Butowski N, Tran DD, Recht LD, Lim M, Hirte H, Ashby L, Mechtler L, Goldlust SA, Iwamoto F, et al. Rindopepimut with temozolomide for patients with newly diagnosed, EGFRvIII-expressing glioblastoma (ACT IV): A randomised, double-blind, international phase 3 trial. Lancet Oncol. 2017;18:1373–1385. doi: 10.1016/S1470-2045(17)30517-X. [DOI] [PubMed] [Google Scholar]
- 359.Reardon DA, Desjardins A, Vredenburgh JJ, O'Rourke DM, Tran DD, Fink KL, Nabors LB, Li G, Bota DA, Lukas RV, et al. Rindopepimut with bevacizumab for patients with relapsed EGFRvIII-expressing glioblastoma (ReACT): Results of a double-blind randomized Phase II trial. Clin Cancer Res. 2020;26:1586–1594. doi: 10.1158/1078-0432.CCR-18-1140. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.