SUMMARY
Asymmetric localization of mRNAs is crucial for cell polarity and cell fate determination. By performing fractionation RNA-seq, we report here that a large number of maternal RNAs are associated with the ER in Xenopus oocytes but are released into the cytosol after oocyte maturation. We provide evidence that the majority of ER-associated RNA-binding proteins (RBPs) remain associated with the ER after oocyte maturation. However, all ER-associated RBPs analyzed exhibit reduced binding to some of their target RNAs after oocyte maturation. Our results further show that the ER is remodeled massively during oocyte maturation, leading to the formation of a widespread tubular ER network in the animal hemisphere that is required for the asymmetric localization of mRNAs in mature eggs. Thus, our findings demonstrate that dynamic regulation of RNA-ER association and remodeling of the ER are important for the asymmetric localization of RNAs during development.
Graphical Abstract
In brief
Hwang et al. report that remodeling of the ER and dynamic regulation of the RNA-ER association during Xenopus oocyte maturation is essential for the asymmetric localization of maternal RNAs and facilitates the transition of oocytes from a dormant state into meiotically mature eggs that are ready for fertilization.
INTRODUCTION
In multicellular organisms, cellular machinery and organelles are organized uniquely in different cell lineages, allowing each type of cell to serve a specific function. Although adult animals have numerous types of cells with distinct structural features, all these cells are derived from fertilized eggs. For decades, much attention has been given to transcriptional regulatory networks and signaling pathways that control lineage specification and differentiation events. It remains largely unclear how the cellular machinery and organelles are remodeled during development and whether the remodeling of cellular machinery and organelles may in turn influence cell fate determination and cellular differentiation.
The endoplasmic reticulum (ER) is the largest organelle and plays many important “housekeeping” functions. As a continuous membrane-enclosed network, the ER consists of cisternae and tubules that extend from the outer nuclear envelope into the peripheral zone of the cytoplasm.1-4 Remodeling of the ER during development is best studied during oocyte maturation, an important developmental process during which the fully grown oocyte is transformed from the meiotic prophase I into the mature metaphase II-arrested egg.5 It is well known that, after germinal vesicle breakdown (GVBD), the ER undergoes massive remodeling. As such, a substantial amount of the ER is placed beneath the cell membrane, near the sperm entry site. This unique distribution of the ER allows rapid release of calcium upon sperm entry, preparing the egg for fertilization.6-14
The ER is the primary compartment of protein translation.15-17 Since the pioneering work by Caro and Palade,18 which demonstrates the translation of secretory proteins by ER-bound ribosomes, the roles of the ER in controlling the translation of secreted factors and integral membrane proteins have been extensively studied. Interestingly, there is growing evidence that mRNAs encoding cytosolic proteins are present and translated on the ER as well.19-30 A few studies suggest that the ER is important for the localization of RNAs in some contexts. In budding yeast, the RNA-binding protein (RBP) She2p anchors ASH and a small group of mRNAs to the ER, allowing these mRNAs to comigrate with ER tubules to the growing bud.31 In Xenopus oocytes, the ER is involved in asymmetric localization of gdf1 (previously known as Vg1), wnt11, nanos1, and dazl, which encode cell fate determinants essential for early embryonic patterning.32-34 It has been shown that Igf2bp3 (previously known as Vera) binds the 3′ UTR of gdf1 and nanos1 and anchors them to the ER.32,34 A similar phenomenon was observed in ascidian eggs, where HrPEM and macho1 mRNAs are associated with the ER in a thin cortical layer and form a gradient along the animal-vegetal axis.35,36 Currently, how mRNA-ER association is regulated during development and to what extent mRNA-ER association may influence asymmetric localization of mRNAs in general remain open questions.
Asymmetric localization of mRNAs, by sequestering mRNAs to specific subcellular locations for translation, results in asymmetric distribution of cytoplasmic proteins. It is fundamentally important for cell polarity and cell fate determination during development.37-44 We previously reported that mRNAs encoding proteasome components undergo vegetal-to-animal translocation during Xenopus oocyte-to-embryo transition (OET).45 In the oocyte, proteasome mRNAs are present in a gradient, with the highest levels in the animal pole and the trailing end extending into the vegetal hemisphere, where it overlaps with vegetally localized germline determinants, such as dnd146-48 and pgat.49 During the OET, proteasome mRNAs undergo vegetal-to-animal translocation and are separated from vegetally localized germline determinants, creating a permissive environment in the vegetal pole for the accumulation of germline determinants and subsequent primordial germ cell development.45 The aim of the present study was to understand the mechanisms governing the dynamic regulation of proteasome mRNAs during the OET.
RESULTS
Sorting of maternal mRNAs during the OET
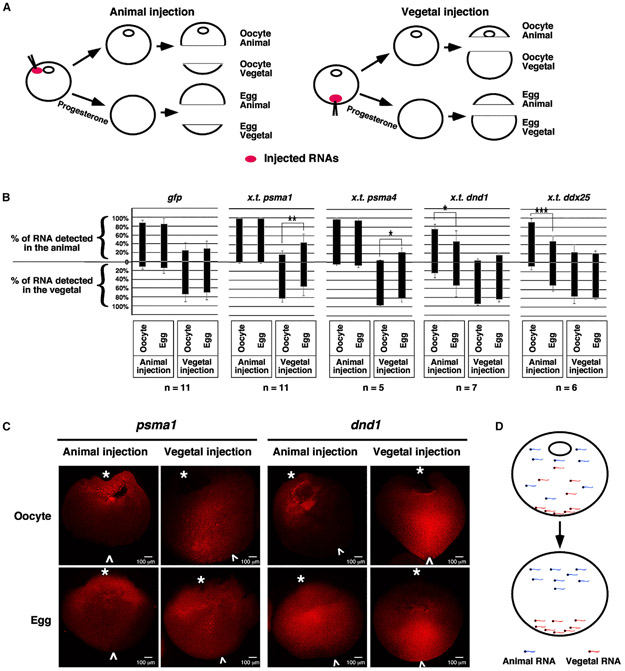
We first analyzed the vegetal-to-animal translocation of proteasome mRNAs (psma1 and psma4) during the OET. Two vegetally localized RNAs, dnd146 and ddx25 (previously known as dead-south),50 and gfp, which is not localized, served as controls. Xenopus tropicalis dnd1, ddx25, psma1, and psma4 were used in this experiment, as polymorphisms in their sequences distinguish them from their orthologous genes in Xenopus laevis oocytes. Oocytes injected with these RNAs were cultured in normal oocyte culture medium (OCM) or OCM with progesterone to induce meiotic maturation. Each oocyte and egg were dissected into animal and vegetal pieces for RNA extraction and subsequent qRT-PCR analysis (Figure 1A). The percentage of injected RNAs detected in the animal and vegetal pieces were calculated.
Figure 1. Sorting of localized maternal transcripts during oocyte maturation.
(A and B) A schematic drawing shows the design of the experiments. RNAs were injected into the animal or vegetal poles of oocytes, respectively. Injected oocytes were cultured in either the OCM or OCM containing progesterone. Oocytes and mature eggs were dissected into animal and vegetal pieces for RNA extraction and subsequent qRT-PCR. The percentages of RNAs detected from each animal and vegetal pair were calculated. In (B), each black bar is 100%. These bars are placed at specific positions along the y axis of the graph to show the percentage distribution of RNAs along the animal/vegetal axis. Two-tailed Student’s t tests were performed. *p < 0.05, **p < 0.01, ***p < 0.001; n, number of animal and vegetal pairs being analyzed.
(C) Confocal images show the distribution of Alexa 546-labeled psma1 and dnd1 RNAs in hemi-sectioned oocytes and mature eggs. White “*” and “^” label the animal and vegetal poles, respectively.
(D) A schematic drawing shows the sorting of localized maternal transcripts during oocyte maturation.
As shown in Figure 1B, the majority of animally injected psma1 and psma4 RNAs were detected in the animal hemisphere in the oocyte and mature egg. When injected into the vegetal pole of the oocyte, we observed a significant increase in the percentages of psma1 and psma4 in the animal hemisphere after oocyte maturation. dnd1 and ddx25, which are vegetally localized, exhibit the opposite pattern. When injected vegetally, dnd1 and ddx25 were mainly detected in the vegetal hemisphere. Their distribution was not significantly changed after oocyte maturation. In contrast, when injected into the animal pole, the percentages of dnd1 and ddx25 detected in the vegetal pole increased significantly after oocyte maturation. The distribution of gfp along the animal/vegetal axis remained unchanged during oocyte maturation (Figure 1B).
Parallel to these studies, we injected fluorescence-labeled psma1 and dnd1 into oocytes and cultured them in normal OCM or treated with progesterone. After progesterone-treated oocytes were arrested at metaphase II, samples were fixed, hemi-sectioned, and analyzed by confocal microscopy. When injected animally, psma1 and dnd1 RNAs were detected in the animal hemisphere in the oocyte. In mature eggs, while psma1 remained in the animal hemisphere, a significant amount of dnd1 RNA was detected in the vegetal hemisphere. When injected vegetally, psma1 and dnd1 RNAs were detected in the vegetal hemisphere in the oocyte. After oocyte maturation, dnd1 RNA remained in the vegetal hemisphere. However, a large amount of psma1 RNA was transported to the animal hemisphere (Figure 1C). Collectively, these observations argue for the existence of a sorting mechanism that acts during oocyte maturation to ensure that RNAs are localized to the correct location (Figure 1D).
ER dynamics during the OET
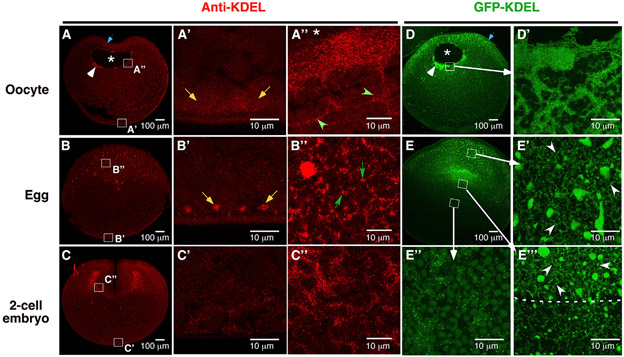
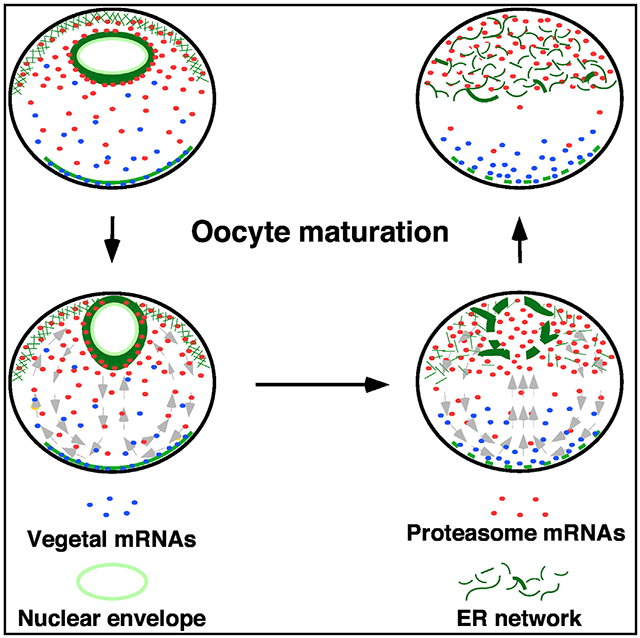
Next, we investigated the mechanism through which proteasome mRNAs are retained in the animal hemisphere of the mature egg. We analyzed oocytes and eggs by transmission electron microscopy (TEM), aiming to identify a structure that is asymmetrically present along the animal-vegetal axis in the egg. Since the proteasome mRNA expression domain matches with the lower edge of the pigmentation on the egg surface (white arrowhead, Figure S1A), and is located above the heavy yolk platelet-rich territory (dotted line, Figure S1C), we carefully analyzed the ultrastructure of the “yolk-loose” cytoplasm in the animal hemisphere (Figures S1G and S1H). Intriguingly, we observed many tubular ER in the animal hemisphere of the egg (arrows, Figure S1G). In some random locations, ER sheets stacked on top of each other and formed dense patches (circle, Figures S1H and S1H′). This unique feature was neither observed in the vegetal hemisphere of the egg (Figure S1I) nor in the oocyte (Figures S1D, S1E, and S1F). To confirm that the ER is indeed asymmetrically distributed along the animal-vegetal axis, we stained oocytes, eggs, and two-cell-stage embryos using antibodies against “KDEL,” the ER localization sequence51 (Figures 2A, 2B, and 2C), pan-Atlastin (Figures S1J, S1K, and S1L), a GTPase located on the ER membrane,52 and GRP78 (Figures S1M, S1N, and S1O), an ER chaperone protein.53 Similar staining patterns were obtained from these experiments. In the oocyte, the ER was detected in the animal subcortical region, around the GV, and in the vegetal cortex (Figures 2A, S1J, and S1M). The ER sheets in the animal hemisphere appear smooth and lack branching (green arrowheads, Figure 2A′). After GVBD, a tubular ER network with abundant three-way junctions forms in the entire animal hemisphere (Figure 2B′). Consistent with the literature,54 we found that the vegetal cortical ER was remodeled into small patches after oocyte maturation (Figures 2A′ and 2B′). The dense ER network in the animal hemisphere persists in two-cell-stage embryos (Figures 2C, 2C″, S1L, and S1O). To further confirm these observations, we overexpressed GFP-KDEL, which specifically labels the ER.12 We detected abundant ER sheets in the perinuclear region in the oocyte (Figures 2D and 2D′). After oocyte maturation, a widespread tubular ER network forms in the entire animal hemisphere (Figures 2E, 2E′, and 2E′). In contrast, the vegetal hemisphere contains only a small amount of ER (Figure 2E″). The distribution of the ER is strikingly similar to the expression pattern of proteasome mRNAs during the OET.
Figure 2. Remodeling of the ER during Xenopus OET.
(A–C) Immunofluorescence using an anti-KDEL antibody to stain the oocyte (A), egg (B), and two-cell-stage embryo (C). (A′, A″, B′, B′, C′, and C″) High-magnification images of the boxed areas in (A), (B), and (C), respectively.
(D and E) Confocal images show the expression of GFP-KDEL in the oocyte (D and D′) and egg (E, E′, E″, and E‴). (D′) High-magnification image of the boxed area in (D). (E′, E″, and E‴) High magnification images of the boxed areas in (E). The position of GV in (A and D) is marked by “*.” White arrowheads in (A) and (D) point to the dense ER patches associated with the GV. Blue arrows point to the animal subcortical region. Yellow arrows point to the ER in the vegetal cortex. Green arrowheads in (A″) highlight the long unbranched ER sheets around GV. Green arrows in (B″) point to three-way ER junctions. White arrowheads in (E′) and (E′) point to the tubular ER that forms a network in the animal hemisphere of the egg.
Dynamic regulation of proteasome mRNA-ER association during oocyte maturation
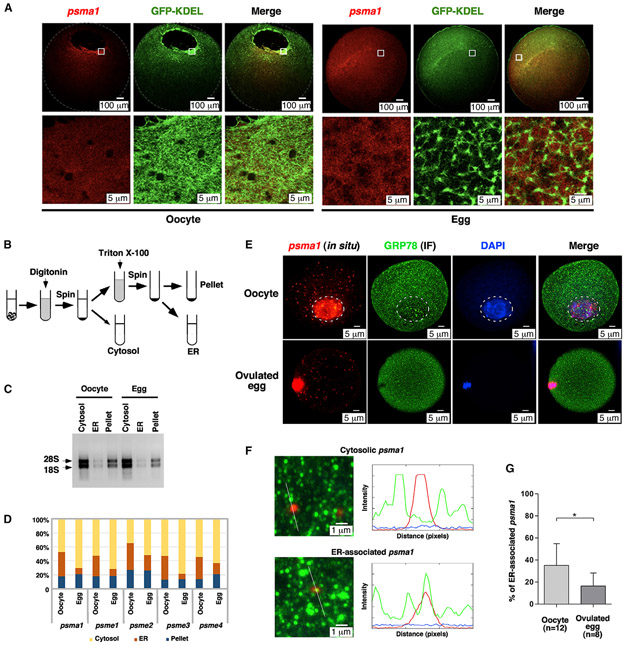
To determine if the ER is important for proteasome mRNA localization, we revisited psma1 and psme3 expression in the oocyte and mature egg. We modified the in situ hybridization protocol by staining samples in BM-purple solution for a short amount of time (3 h), which allowed us to detect mainly the strongest signal inside the cell. Following this protocol, the strongest signals of psma1 and psme3 mRNAs were detected around the GV (Figures S2G and S2H, arrowheads), where large ER sheets are located in the oocyte. After maturation, psma1 and psme3 mRNAs were uniformly distributed in the entire animal hemisphere (Figures S2I and S2J), reminiscent of the ER dynamics during the OET. In parallel, we injected fluorescent psma1 RNA into oocytes expressing GFP-KDEL. We found that psma1 RNA colocalized with the ER in the perinuclear region in the oocyte. After oocyte maturation, psma1 was detected in the entire animal hemisphere, appearing trapped in the “honeybee nest”-shaped ER network (Figure 3A).
Figure 3. Association of proteasome mRNAs with the ER is dynamically regulated during Xenopus and mouse oocyte maturation.
(A) Confocal images show the distribution of Alexa 546-labeled psma1 RNA in GFP-KDEL overexpressed oocytes and eggs. Lower panel images are high-magnification views of the boxed area in the upper panel.
(B) A schematic drawing shows the design of the fractionation experiments.
(C) Agarose gel electrophoresis of RNAs extracted from the cytosolic, ER, and pellet fractions of oocytes and eggs. Arrows point to 18S and 28S rRNAs.
(D) qRT-PCR results show the percentage distribution of psma1, psme1, psme2, psme3, and psme4 across the cytosolic, ER, and pellet fractions. The values of proteasome mRNAs were normalized to that of the spike-in gfp. The normalized values were used to calculate the percentage distribution of individual RNAs across three fractions.
(E) Confocal images show the association of psma1 with the ER during mouse oocyte maturation. Mouse oocytes and ovulated eggs were analyzed by fluorescent in situ hybridization of psma1, in conjunction with immunofluorescence (IF) of GRP78. White circles in the upper panel mark the GV of the oocyte.
(F) Representative images for cytosolic psma1 (upper panel) and ER-associated psma1 (lower panel). The left panels are high-magnification confocal images. The right panels show an analysis of psma1 and ER by ImageJ.
(G) Bar graphs show the percentage of ER-associated psma1 in oocytes and ovulated eggs. Two-tailed Student’s t tests were performed. *p < 0.05; n, number of samples being analyzed.
We further performed fractionation to quantify ER association of proteasome mRNAs during the OET (Figure 3B). As expected, Atlastin and Hsc70 proteins were enriched in the ER and cytosolic fractions, respectively (Figure S3). We observed large amounts of 18S and 28S rRNAs in the cytosolic and pellet fractions. The ER fraction contained a very low level of rRNAs (Figure 3C). qRT-PCR revealed that about 35%–40% of proteasome mRNAs were associated with the ER in the oocyte. After oocyte maturation, the percentages of ER-associated psma1, psme1, psme2, psme3, and psme4 were all decreased (Figure 3D). We conclude that proteasome mRNAs are associated with the ER in the oocyte but are released into the cytosol after oocyte maturation.
We extended our investigation by analyzing the mRNA-ER association during mouse oocyte maturation, using manually isolated oocytes and ovulated eggs. We performed in situ hybridization for psma1 and immunofluorescence for GRP78. We were able to detect psma1 in the cytoplasm of oocytes and ovulated eggs (Figure 3E). The signal inside the germinal vesicle in the oocyte and around the spindle in ovulated eggs appears to be non-specific, as the negative control also showed a similar pattern (data not shown). After GVBD, the ER was more enriched in the cortical and subcortical regions, consistent with previous reports8,11 (Figure 3E). On high-magnification images, some psma1 mRNAs were associated with ER (Figure 3F, lower panel), whereas others were freely distributed in the cytoplasm (Figure 3F, upper panel). We counted psma1 mRNAs in 12 oocytes and 8 ovulated eggs. As shown in Figure 3G, 35% of psma1 transcripts were associated with the ER in oocytes. In ovulated eggs, the percentage of ER-associated psma1 decreased to 16%. These results demonstrate that the release of proteasome mRNAs from the ER into the cytosol during oocyte maturation is evolutionarily conserved.
Transcriptome-scale profiling of the RNA-ER association during the OET
Inspired by the above findings, we performed a fractionation RNA-seq. Immediately after fractionation, a mixture of five spike RNAs ranging from 0.01 to 100 pg (corresponding to 0.0173–325 amol) was added into each fraction. This allowed us to convert relative transcript abundance from RNA-seq (transcripts per a million reads, TPM) to the absolute amount of RNAs (amol) in each sample (Figure S4A). We also scaled the absolute amount of RNAs, assuming that the total RNAs of each sample with the same number of oocytes or eggs should be comparable, so their sum would be identical across samples (Figure S4B). To reduce the noise from the low abundant mRNAs, we filtered out transcripts less than 0.125 amol (Figure S4C) and used the remaining 17,811 transcripts for further analysis. Our analysis reveals that all biological replicates were clustered well based on their fraction and developmental stages, confirming the consistency of our quantification method for absolute mRNA abundance in each fractionation sample (Figures S4D and S4E).
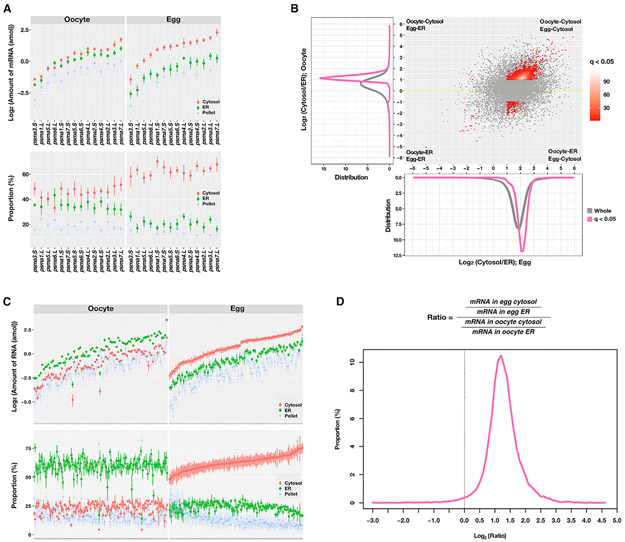
We examined all mRNAs encoding proteasome α subunits (from psma1 to psma7 with homologous L/S genes) in our transcriptome data. Consistent with the above results (Figure 3D), we detected a massive increase in the amount and proportion of all proteasome mRNAs in the cytosolic fraction of the mature egg compared with the oocyte (Figure 4A; Table S1), demonstrating that these mRNAs are released from the ER into the cytosol after oocyte maturation. Next, we determined if RNAs exhibit such an ER-to-cytosol translocation pattern during oocyte maturation at the whole-transcriptome level. Since the proportion of the pellet fractions was similar between the oocyte and egg (Figure S4F), we simplified our analysis by focusing on the ratio of mRNA proportion between the cytosolic and ER fractions. This allowed us to investigate the mRNA localization dynamics during oocyte maturation. Our results revealed that the majority of transcripts skewed toward being enriched in the cytosol both in the oocyte and the egg (Figures 4B and S5). Although many mRNAs were preferentially localized in the cytosol even before oocyte maturation (mean log2(cytosol/ER) in the oocyte is ~0.34; y axis of Figure 4B), this bias toward the cytosol is significantly increased after maturation (mean log2(cytosol/ER) in the egg is 1.62; x axis of Figure 4B), demonstrating that the release of ER-associated transcripts into the cytosol occurs at the whole-transcriptome level during oocyte maturation. Based on their differential enrichment in the cytosolic or ER fractions in the oocyte and egg, we categorized all transcripts into four groups: (1) transcripts staying in the cytosol preferentially (oocyte-cytosol/egg-cytosol), (2) transcripts moved from the cytosol to ER (oocyte-cytosol/egg-ER), (3) transcripts staying in the ER (oocyte-ER/egg-ER), and (4) transcripts moved from ER to cytosol (oocyte-ER/egg-cytosol) during oocyte maturation. Among transcripts present on the ER in the oocyte, only a small amount remained associated with the ER in mature eggs (oocyte-ER/egg-ER) (Figure 4B; Table S2), and the remaining were released to the cytosol in the egg (oocyte-ER/egg-cytosol) (Figures 4B and 4C). Both the absolute amount of RNAs (Figure 4C, top panel) and the relative proportion (Figure 4C, bottom panel) confirmed that those mRNAs were translocated into the cytosol (Table S2 for more details). Besides, we confirmed that the overall ratio of cytosolic/ER transcripts increased during oocyte maturation (mean of log2(egg(cytosol/ER)/oocyte(cytosol/ER)) is ~1.3) (Figure 4D), reflecting the global tendency of the increase of the cytosolic RNAs in the egg. We performed validation experiments using RNAs purified from the detergent-based fractionation (Figure S6, middle panel) and the standard sucrose density gradient fractionation (Figure S6, lower panel). We assessed the distribution of nme3.S and got1.S, which show a similar distribution pattern as proteasome RNAs in the RNA-seq, mprip.S, snph.S, and spata13.S, which are highly enriched in the ER fraction in the oocyte and egg, sptan1.S and capn5.L, which are highly enriched in the cytosolic fraction in the oocyte and egg. Indeed, psma1.S, nme3.S, and got1.S, but not mprip.S, snph.S, spata13.S, sptan1.S, and capn5.L, show a decreased ER association after oocyte maturation. Collectively, results from the fractionation RNA-seq reveal that, at the transcriptome level, RNAs are released from the ER into the cytosol during oocyte maturation.
Figure 4. Transcriptome profiling analysis on the RNA localization associated with ER during oocyte maturation.
(A) A plot shows the absolute amount of RNAs(amol; top panels) and the proportion (percentage; bottom) of 14 20S proteasome α subunit mRNAs in each fraction (red, cytosol; green, ER; blue, pellet). The mean and standard errors of three biological replicates are presented.
(B) Scatterplot shows the cytosol to ER ratio in the oocyte (y axis) and egg (xaxis). Distribution plots (bottom, egg; left, oocyte) show the distribution of cytosol/ER ratios for all transcripts. Differentially localized transcripts were defined as the adjusted p value (q value) less than 0.05 and a greater than 2-fold difference between cytosol and egg (see also Figure S5). The yellow line represents an expected value for a transcript with no RNA localization change during oocyte maturation. Along both axes, the gray line shows the distribution of whole transcripts, and the pink line and red dots show the distribution of differentially localized transcripts.
(C) Plots show the absolute amount and distribution of the top 100 differentially localized transcripts belonging to the oocyte-ER/egg-cytosol group in each fraction.
(D) Distribution plot shows many transcripts were altered their localization during oocyte maturation, represented as a shift of the cytosol/ER ratio (dotted line, expected ratio for a transcript with no localization change). All-transcriptome profile was performed on biologically independent triplicated samples.
Since the ER is enriched in the animal hemisphere (Figure 2), we determined if the mRNA-ER association is important for the asymmetric localization of RNAs along the animal-vegetal axis. We compared our fractionation RNA-seq data with the subcellular transcriptomic analysis by Sindelka et al.,55 in which localization of RNAs was analyzed by dissecting the egg along the animal-vegetal axis, followed by RNA-seq. Among 10,871 transcripts that were robustly detected in both studies, 10,720 were enriched in the animal hemisphere, with 87.67% in the oocyte-cytosol/egg-cytosol category and 11.33% in the oocyte-ER/egg-cytosol category. A small number of transcripts (151) were vegetally localized. Strikingly, these RNAs have a higher tendency to be associated with the ER in the oocyte, with 37.75% of them falling into the oocyte-ER/egg-cytosol category (Table S3). This result is reminiscent of ER-dependent localization of gdf1, wnt11, nanos1, and dazl,32-34 and suggests that mRNA-ER association may play a more general role in regulating vegetal localization of RNAs.
We further extended our investigation by asking if releasing mRNAs from the ER may influence the chance of mRNA translation. mRNAs that are actively translating are enriched in the polysome fraction. Yang et al.56 recently investigated mRNA translation during Xenopus oocyte maturation by sequencing RNAs enriched in the polysome fractions. The results show that 368 transcripts exhibit a significant increase in the polysome fraction after oocyte maturation. Among these, 200 transcripts (Table S4) are abundantly expressed (more than 0.125 amol). Intriguingly, 94% of these transcripts are released from the ER into the cytosol significantly (Figure S7, black dots), whereas the other 6% of transcripts show no significant change in their ER association (Figure S7, red dots). Of note, while 81% of these transcripts are relatively evenly distributed across the cytosol and ER fractions in the oocyte (left panel; between blue dotted lines), 83% are enriched in the cytosol after oocyte maturation (right panel; above the upper blue dotted line). It appears that the majority of transcripts that become more actively translated in the egg are those being released from the ER during oocyte maturation, suggesting that releasing mRNAs into the cytosol may increase their chance for translation after oocyte maturation.
Contribution of RBPs to mRNA-ER association during oocyte maturation
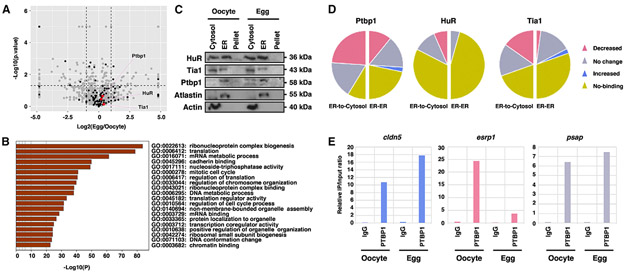
RNAs are sequestered to the ER by either ER-bound ribosomes or ER-associated RBPs. Since the amount of ER-associated rRNAs remains unchanged after oocyte maturation (Figure 3C), we went on to determine if the amount of ER-associated RBPs is reduced after oocyte maturation and if ER association of RBPs bind to their target RNAs with a lower affinity after oocyte maturation. We performed a comparative proteomic analysis using microsomes purified from oocytes and mature eggs. We detected 1,720 proteins in the oocyte microsomes and 1,721 proteins in the egg microsomes (Figure 5A; Table S5). Many of these proteins are involved in ribonucleoprotein complex biogenesis, translation, and RNA binding (Figure 5B). Among 106 ER-associated RBPs, only Khsrp, Alyref, Lsm1, and Ilf3 show a statistically significant increase in their ER association, and Xpo5, Elavl2, Supt5h, Cpeb1, and Pum3 show a statistically significant decrease in their ER association (Table S5). The vast majority of microsome-associated proteins, including most of the identified ER-associated RBPs, remain unchanged during oocyte maturation (Figure 5A; Table S5). To validate this finding, we examined the expression of several widely expressed RBPs, including HuR (also known as Elval1), Ptbp1, and Tia1. Indeed, the subcellular distribution of these RBPs remains unchanged during oocyte maturation (Figure 5C). Thus, it seems unlikely that the decrease in mRNA-ER association after oocyte maturation is caused by a reduction in the amount of ER-associated RBPs.
Figure 5. Roles of ER-associated RBPs in regulating mRNA-ER association during oocyte maturation.
(A) Volcano plot shows the proteomic profiling ofmicrosomes purified from oocytes and mature eggs. Differentially expressed proteinswere defined asap value lessthan 0.05 (dashed horizontal line) and greater than a 2-fold difference between oocyte and egg (dashed vertical lines) (gray dots, whole 1,976 proteins; black dots, RBPs). All-proteomic profile was performed on biologically independent triplicated samples.
(B) Bar graph shows gene ontology (GO) enrichment analysis of ER-associated proteins that remain unchanged during oocyte maturation. This analysis was performed by Metascape.
(C) Western blot shows the expression of RBPs (Ptbp1, HuR, and Tia1) in cytosolic, ER, and pellet fractions in the oocyte and mature egg. Atlastin and Actin were enriched in the ER and cytosolic fractions, respectively.
(D) Pie charts show the summary of RIP assays. In all three RIP assays, 23 transcripts in the oocyte-ER/egg-ER groups and 23 transcripts in the oocyte-ER/egg-cytosol groups were analyzed (pink, the proportion of mRNAs with decreased binding; gray, the proportion of mRNAs with no change; blue, the proportion of mRNAs with increased binding; yellow, the proportion of mRNAs not bound by RBPs).
(E) Bar graphs are Ptbp1-RIP of cldn5, esrp1, and psap, which are shown as representative results of the increased, decreased, and unchanged categories, respectively.
To determine if ER-associated RBPs may bind to their target RNAs with a lower affinity after oocyte maturation, we performed ribonucleoprotein immunoprecipitation (RIP) using the ER fraction purified from oocytes and eggs. We chose to analyze 46 transcripts that are statistically significantly enriched in the ER fraction in the oocyte (Table S6). Among these, 23 remain associated with the ER in the egg (ER-ER), whereas the other 23 are released from the ER into the cytosolic fraction (ER-to-cytosol). Our results reveal that 32 RNAs were coimmunoprecipitated with ER-associated Ptbp1 in the oocyte. These include 19 ER-to-cytosol RNAs and 13 ER-ER RNAs. After oocyte maturation, only one ER-ER RNA (cldn5) exhibited an increase in Ptbp1 binding. Reduced Ptbp1 binding was detected in 38.5% ER-ER RNAs and 57.9% ER-to-cytosol RNAs. The binding between Ptbp1 and the remaining RNAs remained unchanged (Figure 5D; Table S6). Ptbp1-RIP results for cldn5, esrp1, and psap are shown to represent RNAs exhibiting increased, decreased, and unchanged Ptbp1 binding after oocyte maturation, respectively (Figure 5E). Our results further reveal that HuR bound eight ER-to-cytosol RNAs and two ER-ER RNAs. After oocyte maturation, the binding between HuR and ER-ER RNAs remained unchanged. In the case of ER-to-cytosol RNAs, reduced HuR binding was detected in 37.5% of RNAs. The remaining 62.5% of RNAs showed no change (Figure 5D; Table S6). We also identified 14 ER-to-cytosol RNAs and 9 ER-ER RNAs as targets of Tia1 in the oocyte. Among these, only ccdc50, which belongs to the ER-ER group, showed an increase in Tia1 binding after oocyte maturation (Table 6). Other Tia1 targets showed either reduced Tia1 binding (34.8%) or no change (60.9%) after oocyte maturation (Figure 5D; Table S6). As all RBPs analyzed show a decrease in their binding to a subset of their target RNAs after oocyte maturation, it seems likely that decreased mRNA-ER association after oocyte maturation is caused by a weakened interaction between ER-associated RBPs and their target RNAs.
The tubular ER network in the egg is required for the asymmetric localization of proteasome mRNAs
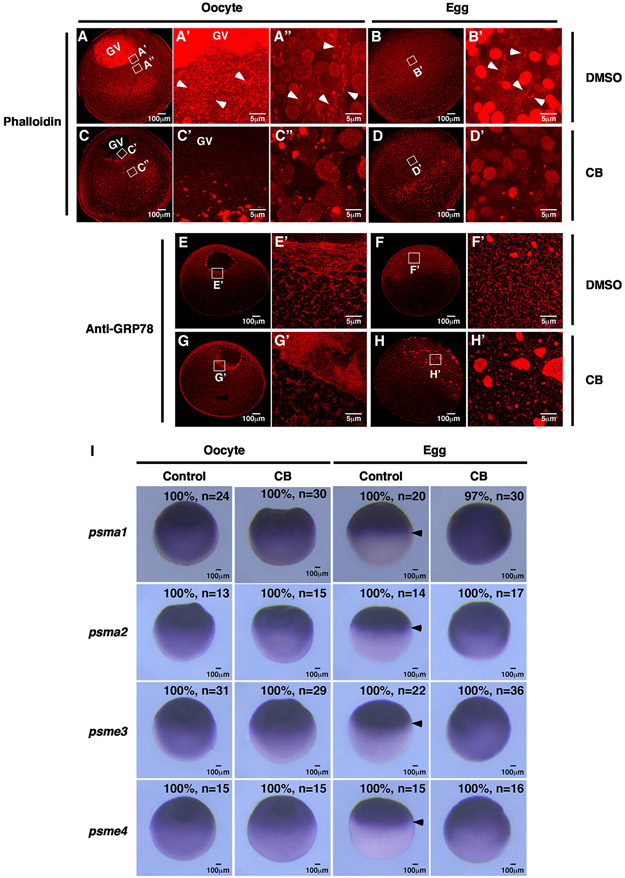
We further extended our investigation by asking if the tubular ER network in the animal hemisphere of the egg is important for the asymmetric localization of proteasome mRNAs. To interfere with the formation of the tubular ER network, we first investigated the roles of the cytoskeleton during ER remodeling. We performed phalloidin staining to assess F-actin in the oocyte (Figure 6A) and mature egg (Figure 6B). In agreement with the literature,57 we observed a strong staining signal inside the GV in the oocyte (Figure 6A) and a dense F-actin meshwork around the GV in the animal hemisphere (Figures 6A' and 6A″, arrowheads). In mature eggs, F-actin was detected in a broad area in the animal hemisphere (Figure 6B'). These staining signals were not detected in F-actin inhibitor cytochalasin B (CB)-treated oocytes (Figures 6C, 6C′, and 6C″) and eggs (Figures 6D and 6D′). Our results further reveal that CB severely affects ER morphology. In control oocytes, large ER patches were tightly associated with the GV. Some ER was detected in the cortical region (Figures 6E and 6E′). In CB-treated oocytes, while the ER was still present in the perinuclear and cortical regions, they were loosened, forming a wide and diffuse band around the GV (Figures 6G and 6G′). After oocyte maturation, a fine tubular ER network was detected in the entire animal hemisphere in the egg (Figures 6F and 6F′). CB-treated eggs failed to form such a tubular ER network and often had many large ER patches that were randomly distributed in the animal hemisphere (Figures 6H and 6H’). These results reveal that F-actin plays a critical role in supporting the ER network during the OET.
Figure 6. Interfering with the tubular ER network via dissociation of F-actin impairs the localization of proteasome mRNAs during oocyte maturation.
(A–D) Confocal images of phalloidin-stained oocytes and eggs. White boxes indicate the areas where high-magnification images were taken from. Arrowheads point to F-actin in control oocytes and eggs.
(E–H) Confocal images of GRP78 staining show the morphology of the ER network in control and F-actin inhibitor-treated samples.
(I) In situ hybridization results show the effects of F-actin inhibitors on proteasome mRNA distribution in oocytes and eggs. Arrows mark the lower edge of the proteasome mRNA expression domain in the equator of mature eggs. CB, cytochalasin B; n, number of samples being analyzed.
We then assessed the distribution of proteasome mRNAs in CB-treated oocytes and eggs. Consistent with our previous findings, psma1, psma2, psme3, and psme4 mRNAs formed an animal-to-vegetal gradient in oocytes. After oocyte maturation, these mRNAs were uniformly distributed in the animal hemisphere (arrows, Figure 6I). Disruption of F-actin by CB had minimal effect on the subcellular distribution of psma1, psma2, psme3, and psme4 mRNAs in the oocyte. In the egg, however, psma1, psma2, psme3, and psme4 mRNAs were detected in the entire egg (Figure 6I). These results support the idea that the tubular ER network in the egg is important for the asymmetric localization of proteasome mRNAs.
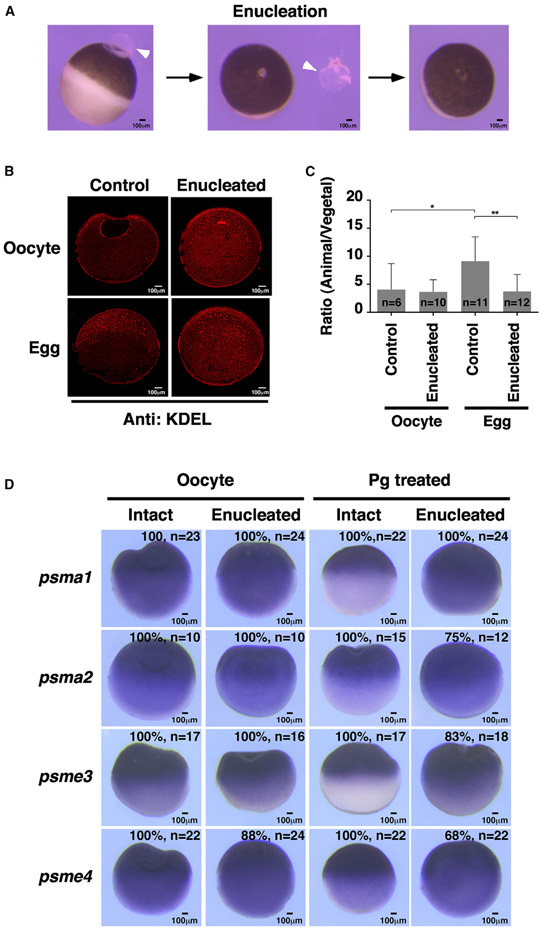
To strengthen the above conclusion, we performed an enucleation experiment.58 Xenopus oocytes can be enucleated (Figure 7A) and still undergo meiotic maturation upon progesterone treatment.59-62 We reasoned that, as a result of enucleation, the nuclear envelope of the GV and some ER associated with the GV would be removed. This would partially deplete the membrane needed for the formation of a tubular ER network in the animal hemisphere of the egg, offering a unique opportunity to further determine if the ER dynamics during oocyte maturation is essential for the localization of proteasome mRNAs.
Figure 7. Enucleation disrupts ER remodeling and proteasome mRNA localization during oocyte maturation.
(A) Images show the process of enucleation. Arrowheads point to the GV.
(B) Confocal images of anti-KDEL antibody staining, showing that enucleated oocytes failed to develop an ER network in the animal hemisphere after progesterone treatment.
(C) Anti-KDEL-positive signals in images of (B) were quantified using ImageJ. Two-tailed Student’s t tests were performed. *p < 0.05, **p < 0.01; n, number of samples being analyzed.
(D) In situ hybridization shows that enucleation disrupted animal localization of proteasome mRNAs after oocyte maturation. The number of samples exhibiting the phenotype and the sample size is provided at the lower right corner of each figure.
We thus compared ER dynamics and translocation of proteasome mRNAs in control and enucleated oocytes. As shown in Figures 7B and 7C, the ER was detected mainly in the animal subcortical region and around GV in intact oocytes. After removal of the GV, large ER patches, presumably some of those associated with the GV before enucleation, were scattered in the animal hemisphere. When intact oocytes were treated with progesterone, a fine ER network formed in the animal hemisphere. In contrast, in 70% of enucleated oocytes, the ER became randomly distributed and was detected in both animal and vegetal hemispheres (Figures 7B and 7C). We further analyzed the localization of proteasome mRNAs. While psma1, psma2, psme3, and psme4 were transported to the animal hemisphere in progesterone-treated control oocytes, they were detected in the entire cell in progesterone-treated enucleated samples (Figure 7D). These results provide additional support for our hypothesis that the tubular ER network in the animal hemisphere is essential for the asymmetric localization of the proteasome mRNAs in the mature egg.
DISCUSSION
The roles of the ER for the translation of secreted factors and integral membrane proteins have been studied extensively.15-17 Recent works, especially a series of elegant studies by Nicchitta and co-workers,19-30 have demonstrated that many mRNAs encoding cytosolic proteins are abundantly present and translated on the ER as well, highlighting the fundamental function of the ER as a primary compartment for protein translation. The mRNA-ER association can be dynamically regulated. It has been reported that, in response to protein folding stress, mRNAs encoding signal sequences are released from the ER to the cytosol, offering a mechanism to quickly reduce protein synthesis and protein flux into the ER.63 Here, we report that the mRNA-ER association is dynamically regulated during Xenopus oocyte maturation. What is the significance of differentially regulated mRNA-ER association?
During early oogenesis, oocytes accumulate large amounts of maternal products important for early embryonic development. Once these processes are completed, fully grown oocytes enter into a dormant state and can be stored in the ovary for a long time until meiotic resumption in response to hormone stimulation.5,64 We observed that, starting from early oogenesis, a large fraction of proteasome mRNAs are present on the ER in the oocyte (Figure S2), a compartment with only a small amount of rRNAs. After oocyte maturation, mRNAs are released into the cytosol where rRNAs are most abundant (Figure 3C). It is well known that the protein synthesis rate is low in fully grown Xenopus oocytes, but increases abruptly after oocyte maturation.65-68 We speculate that the relatively high ratio of RNA-ER association in the oocyte may serve as a mechanism for the long-term storage of maternal mRNAs. Releasing RNAs from the ER into the cytosol during oocyte maturation likely increases the chance for maternal RNAs to be translated. Indeed, the majority of transcripts becoming more actively translated after oocyte maturation exhibit a decrease in their ER association in the egg (Figure S7; Table S4). It seems likely that the dynamic regulation of mRNA-ER association is an important tuning mechanism that contributes to the overall increase in protein synthesis rate after oocyte maturation.
Our results further argue that dynamic regulation of mRNA-ER association, together with ER remodeling during oocyte maturation, provides an important mechanism for the control of asymmetric localization of RNAs. We show that the subcellular localization of proteasome RNAs is heavily influenced by the ER. We found that a large amount of proteasome RNAs are associated with the ER in the oocyte. The distribution of proteasome RNAs is a near-mirror image of the ER. This pattern is disrupted during oocyte maturation when proteasome RNAs, together with many other RNAs are released into the cytosol. Since these RNAs are no longer tethered to the ER, they can move freely in the cytosol. Likely due to cytoplasmic streaming and local retention, a mechanism controlling the polarity establishment in Drosophila oocytes,69 proteasome RNAs, and germline determinants are “transported” to the animal and vegetal hemispheres, respectively. Meanwhile, the newly formed tubular ER network plays a key role in retaining proteasome RNAs in the animal hemisphere. Interfering with the ER network by dissociation of F-actin or enucleation resulted in the widespread distribution of proteasome RNAs in the entire egg. It is worth noting that one-third of RNAs localized to the vegetal hemisphere in early embryos are enriched in the ER fraction in the oocyte (Table S3). This observation is consistent with the findings that the ER is essential for the vegetal localization of gdf1, wnt11, nanos1, and dazl.32-34 It raises the striking possibility that the ER may play a more general role in the vegetal localization of maternal RNAs in Xenopus oocytes. Collectively, our results demonstrate a fundamentally important function of the ER in controlling the asymmetric localization of RNAs.
In many species, females are born with a finite number of oocytes. Some oocytes are stored in the ovary throughout the reproductive lifespan. Oocytes contain a large amount of maternal factors important for early embryonic development. Many of these are synthesized early during oogenesis. It is currently unclear how these maternal products are preserved in the oocyte for such a long time. We propose that the association of RNAs with the ER may be an important mechanism for the long-term storage of maternal mRNAs in the oocyte. Since a large fraction of maternal mRNAs is sequestered on the ER, the oocyte cytoplasm is less crowded. More importantly, compartmentalization of maternal mRNAs would reduce their accessibility to the RNA degradation machinery, hence increasing the stability of maternal mRNAs. During oocyte maturation, as many mRNAs previously “packed” on the ER are released into the cytosol, they regain the ability to move in the oocyte. In animals, such as Xenopus, zebrafish, and Drosophila, early embryonic patterning relies on asymmetrically localized maternal cell fate determinants. Remodeling of the ER and releasing RNAs into the cytosol during oocyte maturation thus offers an important mechanism to sort maternal RNAs and correct localization errors that may have occurred during early oogenesis. In agreement with this view, we found that mislocalized proteasome and germline RNAs can be relocated to the correct location after oocyte maturation in Xenopus (Figure 1). We speculate that precisely regulated RNA-ER association is an important mechanism that allows the oocyte to leave the dormant state and get ready for fertilization.
In summary, our work reported here demonstrates that the ER plays a multifaced role in controlling the asymmetric localization of RNAs during the OET. We speculate that dynamic regulation of mRNA-ER association and ER remodeling may be involved in many processes during early development and adult tissue homeostasis.
Limitations of the study
Currently, we do not fully understand how the ER-to-cytosol translocation of RNA is regulated during oocyte maturation. In principle, the mRNA-ER association may be mediated by ER-bound ribosomes or ER-associated RBPs. Since rRNAs associated with the ER remain unchanged after oocyte maturation, we speculated that the RNA-ER association in the oocyte might be mediated by ER-associated RBPs. Our results reveal that the amount of ER-associated RBPs is not significantly changed after oocyte maturation (Figure 5A). However, ER-associated RBPs exhibit reduced binding to a subset of their target RNAs (Figure 5D). This suggests that ER-associated RBPs may bind to their target RNAs less efficiently after oocyte maturation. Interestingly, all three RBPs tested showed reduced binding to only a subset of their target RNAs, suggesting that the accessibility of RNAs to ER-associated RBPs is dynamically regulated during oocyte maturation. Recently, Shi et al.70 reported that dynamic structural changes in the 3′ UTR of maternal mRNAs heavily influence their accessibility to RBPs and are critical for the stability of maternal RNAs during zebrafish maternal to zygotic transition. It will be of great interest to determine if the accessibility of RNAs is dynamically regulated during oocyte maturation.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Jing Yang (yangj@illinois.edu).
Materials availability
This study did not generate any new unique reagents.
Data and code availability
RNA sequencing data generated in this study have been deposited to the Gene Expression Omnibus (GEO) with the dataset identifier GEO:GSE199254. Proteomic profile data generated in this study have been deposited to the ProteomeXchange Consortium via PRIDE partner repository with the dataset identifier PRIDE: PXD033018
This paper does not report the original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Xenopus laevis
Xenopus procedures were approved by the University of Illinois at Urbana-Champaign Institutional Animal Care and Use Committee (IACUC), under animal protocol #20125, and performed in accordance with the recommendations of the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. Oocytes were collected from ovarian tissues by manual defolliculation74 or collagenase treatment, and cultured in the oocyte culture medium (OCM).75 To induce meiotic maturation, oocytes were cultured in the OCM with 2 μM progesterone and incubated at 18°C overnight. Mature eggs with white spots, indicating GVBD, were collected for further experiments. To assess the distribution of injected RNAs along the animal-vegetal axis, oocytes and eggs were dissected into the animal and vegetal pieces. For animally injected RNAs, we measured how much RNAs could reached the vegetal pole. The dissection plan thus was closer to the vegetal pole. For vegetally injected RNAs, the dissection plan was closer to the animal pole. Xenopus embryos were obtained as described.76 Embryos were collected by in vitro fertilization, cultured in 0.2x Marc’s Modified Ringer’s Solution (MMR; 100 mM NaCl, 2 mM KCl, 1 mM MgSO4, 2 mM CaCl2, 5 mM HEPES) and harvested at the desired stages.
Mouse
All mouse work was approved by the University of Illinois at Urbana-Champaign Institutional Animal Care and Use Committee (IACUC), under animal protocol #21002. Female CD1 mice aged 35 days were obtained from Charles River Laboratories and housed at the University of Illinois at Urbana-Champaign at the Carl R. Woese Institute for Genomic Biology Animal Facility. Animals were provided food and water ad libitum and housed in a controlled animal room environment, maintained at a temperature of 22 ± 1°C and light-dark cycles of 12 h. Oocytes were manually isolated from antral follicles freshly collected from the ovaries of mice in DMEM. Mice used for ovulated eggs collection were allowed to acclimate for at least one week before undergoing the superovulation procedure. To obtain the ovulated eggs, female mice were submitted to a superovulation protocol receiving 6 IU of PMSG (Prospec Bio) at 4:00 PM and 6 IU of HCG (Millipore Sigma) 45 h later, at 1:00 PM. Then, females were euthanized and their oviducts collected in holding medium (MOPS buffer supplemented with 5% FBS). Using a stereomicroscope warmed at 37°C, the ampullas of the oviducts were perforated with a needle, and ovulated eggs surrounded by cumulus cells were transferred to a dish containing fresh holding medium. Subsequently, hyaluronidase (500 mg/mL) was added to the dish to dissociate the eggs from the cumulus cells for approximately one minute. Eggs were washed twice in holding medium and then fixed in 4% PFA overnight.
METHOD DETAILS
Enucleation
The protocol was as described.58,77 In brief, defolliculated stage VI oocytes were incubated in the OCM for an hour and pricked at the animal pole with a 26G gauge 5/8 length syringe needle. The oocyte was slightly squeezed with forceps to facilitate the germinal vesicle (GV) extrusion. To minimize the extrusion of cytoplasm, enucleated oocytes were immediately transferred to the healing solution (90 mM potassium phosphate pH7.2, 10 mM NaCl, 1 mM MgSO4) and incubated for 2 h. Then, healed oocytes were transferred to the OCM containing 2 drops of 50 mg/mL gentamicin and 0.1% polyvinyl pyrrolidone and incubated for 2 h to ensure complete wound closure. All manipulations were performed on the concave-convex agar plate to immobilize the oocyte.
Treatment of inhibitors
To inhibit F-actin, defolliculated stage VI oocytes were exposed to 25 μg/mL Cytochalasin B (CB, an F-actin inhibitor). Dimethyl sulfoxide (DMSO) treated samples served as controls. Some CB-treated oocytes were cultured in OCM with 2 μM progesterone overnight at 18°C to induce oocyte maturation. Control and CB-treated samples were harvested for immunofluorescence and in situ hybridization.
Immunofluorescence
For sample preparation, defolliculated oocytes and mature eggs were fixed in MEMFA (0.1 M MOPS pH 7.4, 2 mM EGTA, 1 mM MgSO4, and 3.7% formaldehyde solution), embedded into paraffin wax, and sectioned at 4 μm thickness in the microtome. In experiments where stage I or II oocytes were analyzed, the ovary tissue was fixed and sectioned. For immunofluorescence, sections were deparaffinized with xylene, rehydrated in serial dilution of ethanol, and performed antigen retrieval with sodium citrate buffer (10 mM sodium citrate pH 6.0 containing 0.05% Tween 20) for 20 min in the pressure cooker. Then, sections were incubated in blocking buffer (1% BSA, 0.1% Triton X-100, and 0.05% Tween 20 in PBS with 2% normal serum from the same host as the secondary antibody) for an hour at room temperature and stained with the diluted primary antibody in antibody buffer (1% BSA, 0.5% Triton X-100 in PBS) overnight at 4°C. Sections were washed with PBST (PBS containing 0.1% Tween 20) three times for 5 min each, stained with the secondary fluorescence antibody in PBST for an hour at room temperature, and washed again in PBST three times for 10 min each. Stained sections were mounted in a mounting medium (Fluoromount-G). Images were acquired using Nikon A1Rsi confocal microscope.
Quantification of Figure 7B was done as follows. All images were divided into half and were regarded as the animal and vegetal hemispheres. Using the ImageJ Otsu method, the threshold was set to detect the ER patches (the threshold value is the same for all sample analyses), then the number of ER patches was obtained through the particle analysis tool. The graph of Figure 7C was obtained through an analysis of the animal-to-vegetal ratio with the number of ER patches collected from each animal and vegetal hemisphere using the above method.
Imaging of fluorescent RNAs in Xenopus oocytes and eggs
Fluorescent psma1 and dnd1 RNAs were labeled by Alexa Fluor™ 546-14-UTP. After being injected with fluorescent RNAs, oocytes were cultured in the OCM or OCM with 2 mM progesterone to induce meiotic maturation overnight at 18°C. Then, samples were fixed with MEMFA (0.1 M MOPS pH 7.4,2 mM EGTA, 1 mM MgSO4, and 3.7% formaldehyde solution) for 90 min at room temperature and washed with PBS twice for 5 min each. Fixed oocytes and mature eggs were hemi-sectioned using a razor blade and mounted with TBS pH 8.0 containing 100mM NaN3 in depression slides. Images were acquired within 12 h after mounting using Nikon A1Rsi confocal microscope.
Phalloidin staining
The protocol was as described.78 Briefly, defolliculated stage VI oocyte and mature eggs were fixed with Fixation buffer (80 mM K-PIPES pH 6.8, 1 mM MgCl2, 5 mM EGTA, and 0.2% Triton X-100) containing 3.7% formaldehyde for 12 h at room temperature and washed with TBSN (Tris-buffered saline containing 0.1% NP-40) for 24 h. Then, samples were bisected using a razor blade. Next, hemi-sectioned stage VI oocyte and mature eggs were stained with 4 units/mL phalloidin in TBSN containing 5% DMSO overnight at room temperature. After staining, samples were washed with TBSN for 48 h at room temperature and mounted with TBS pH 8.0 containing 100 mM NaN3 in depression slides. Images were acquired using Nikon A1Rsi confocal microscope.
Transmission electron microscopy
Oocytes and eggs were fixed overnight at 4°C in 2% paraformaldehyde and 2.5% glutaraldehyde in 0.1 M Na-Cacodylate buffer, pH 7.4. Fixed samples were submitted to the University of Illinois Materials Research Lab Core Facility for Toluidine blue basic Fusin staining and TEM analysis.
Whole-Mount fluorescence in situ hybridization-immunofluorescence (Immuno-wmFISH)
Briefly, mouse oocytes and ovulated eggs were fixed with 4% PFA solution overnight at 4°C. After washing twice 5 min each with PBST (PBS containing 0.1% Tween 20), the samples were dehydrated through 25%, 50%, 75%, and 100% methanol for 5 min each step at room temperature. Dehydrated samples were stored in 100% methanol at −20°C until the in situ hybridization was initiated. The wmFISH was performed using the RNAscope Multiplex Fluorescent Reagent Kit v.2 according to the manufacturer’s instructions for RNAscope assay on whole zebrafish embryos with slight modification (Modification: the samples were incubated in the target retrieval buffer for 20 min and the protease plus reagent for 25 min. Next, fluorophore Opal reagent was added with a dilution of 1:100. After probe hybridization, all wash steps were performed twice for 5 min each at room temperature). DapB, the Bacillus subtilis gene, was used as a negative control probe.
For immunofluorescence, after wmFISH, oocytes and ovulated eggs were washed with washing buffer (PBS containing 0.1% Triton X-100, 0.1% polyvinyl pyrrolidone) for 10 min followed by incubation in blocking buffer (PBS containing 0.1% Triton X-100, 1% BSA, 0.1 M glycine, and 10% donkey serum) for 2 h at room temperature. Then, samples were washed with washing buffer for 25 min, incubated with an anti-GRP78 antibody (1:100) in antibody buffer (PBS containing 0.1% Triton X-100 and 1% BSA) overnight at 4°C. After staining, samples were washed with washing buffer three times for 10 min each, incubated with diluted secondary antibody in antibody buffer for 2 h at room temperature, and washed with antibody buffer twice for 20 min each, followed by washing again with washing buffer three times for 10 min each. Finally, samples were incubated with DAPI for 2 min at room temperature and mounted with a mounting medium (Vectashield) on glass plates. All steps were performed on the depression glass spots plates.
To quantify ER-associated psma1, all red signals (psma1) were counted from each sample. Then, the intensity of the red signals was measured using ImageJ and only those greater than 100 were selected. Those selected red signals were taken as the total number of psma1. If the intensity of the green signal (ER) within the distance range of the selected red signal peak was detected as greater than 100, it was taken as ER-associated psma1. Finally, the percentage of ER-associated psma1 of each sample was analyzed with the total number of psma1 and the number of ER-associated psma1 obtained by the above analysis.
Cell fractionation
Fractionation was performed as described.30 In brief, stage VI oocytes and mature eggs were treated with 50 μg/mL cycloheximide for 30 min at room temperature, washed with ice-cold PBS quickly, and then washed again with ice-cold washing buffer (150 mM KOAc, 2.5 mM MgOAc2, and 20 mM K-HEPES pH 7.4). To extract cytosolic fractions, four stage-VI oocytes or four mature eggs were homogenized in 160 μL of extraction buffer (150 mM KOAc, 2.5 mM MgOAc2, 20 mM K-HEPES pH 7.4, 2 mM DTT, 1 mM PSMF, 50 μg/mL cycloheximide, and 200 units/mL RNase inhibitor) containing 0.1% digitonin and centrifugated at 800 x g for 5 min to obtain a supernatant (cytosolic fraction) and pellet (ER and insoluble fractions). After centrifuging, to extract the ER from the insoluble fraction, the pellet was resuspended in cold extraction buffer containing 0.5% Triton X-100. The resuspended solution was centrifugated at 800 x g for 5 min to obtain the ER fraction (supernatant) and pellet fractions. Next, the cytosolic and ER fractions were centrifugated at 10,000 x g for 10 min to remove contaminating organelles such as mitochondria and cell debris. All centrifugations were performed in a refrigerated centrifuge at 4°C.
Microsome purification and mass spectrophotometry
Microsome purification protocol was adapted from Ezure et al.79 with slight modification. Briefly, oocytes were obtained from Xenopus ovary by a Collagenase treatment. To obtain mature eggs, oocytes were treated with 2 μM progesterone overnight. The oocytes and eggs were homogenized using a Dounce homogenizer in buffer A (40 mM HEPES-KOH (pH 7.9), 250 mM Sucrose and 1 mM DTT). The suspension was centrifuged for 10 min at 10,000 x g at 4°C. The supernatant was layered over buffer B (40 mM HEPES-KOH (pH 7.9), 1.3M Sucrose and 1 mM DTT). An approximately 3:1 ratio of Supernatant to buffer B was taken and centrifuged for 150 min at 140,000 x g at 4°C. The supernatant from this centrifugation was discarded and the pellets were submitted for Mass Spectrophotometry analysis.
Microsome samples purified from stage VI oocytes and mature eggs were digested by trypsin, separated on a nano-scale reverse-phase HPLC capillary column. Eluted peptides were ionized by electrospray and analyzed in the LTQ Orbitrap Velos Elite mass spectrometer (Thermo Fisher). The peptide-spectrum matching was performed using the SEQUEST program with the Xenopus laevis proteome database from Uniprot (Proteome ID: UP000186698). The error rate of a peptide FDR was controlled to 2% with the linear discrimination analysis. Only peptides detected twice or more in either stage VI oocytes or mature eggs were selected for further analysis. The sum of the total number of peptides detected in three replicates was used to compare oocyte and egg. Generation of raw data and LC-MS/MS analysis was performed by the Harvard Medical School Taplin Mass Spectrometry Facility. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE80 partner repository with the dataset identifier PXD033018 and 10.6019/PXD033018.
Ribonucleoprotein-immunoprecipitation (RIP) assay
To prepare the ER fraction for RIP, Xenopus stage VI oocytes and mature eggs were treated with 50 μg/mL cycloheximide for 30 min at room temperature and washed with ice-cold washing buffer (150 mM KOAc, 2.5 mM MgOAc2, and 20 mM K-HEPES pH 7.4). To remove the cytosolic fraction, fifty Xenopus stage VI oocytes or mature eggs were homogenized in 1 mL of extraction buffer (150 mM KOAc, 2.5 mM MgOAc2, 20 mM K-HEPES pH 7.4, 2 mM DTT, 1 mM PSMF, 50 μg/mL cycloheximide, and 200 units/mL RNase inhibitor) containing 0.1% digitonin and centrifugated at 800 x g for 5 min. After discarding the supernatant (cytosolic fraction), the pellet was resuspended with RIP buffer (50 mM Tris pH 7.6, 125 mM NaCl, 1 mM EDTA, 0.5% NP-40, 0.2% glycerol, 0.1 mM DTT and 100 U/mL RNasin) and centrifugated at 10,000 x g for 10 min. After collecting and combining supernatant (5 mL of ER fraction lysate harvested from 250 stage VI oocytes or mature eggs, respectively), 50 μL of the lysate was saved as ‘1% input’ and the remaining lysates were used for RIP. RIP assays were performed using a protocol modified from our previous work.48 A typical RIP experiment contained 800 μL of the ER fraction lysate, 10 μL of antibody (anti- Ptbp1, HuR, and Tia1 antibodies) or control IgG. Lysates were incubated at 4°C for 4 h with gentle rotation. After 4 h incubation, samples were centrifuged at 13,000 x g at 4°C for 10 min. The supernatants were transferred to the clean 1.5 mL tube. Meanwhile, 250 μL of Dynabeads Protein G (Life Technologies) was washed 3 times with washing buffer (50 mM Tris pH 7.6, 125 mM NaCl, 1 mM EDTA, 0.5% NP-40) and resuspended in 1 mL RIP buffer. 100 μL Dynabeads were added into the ER fraction followed by incubation at 4°C for an hour with gentle rotation. Afterward, samples were washed with washing buffer five times. RNAs associated with the beads were recovered using TRIzol reagent for cDNAs synthesis and subsequent qPCR. All primers used are listed in the table (Table S7).
In vitro transcription
All mRNAs used in this study were synthesized from 2 μg of plasmid templates using the mMESSAGE mMACHINE Kit for SP6 (Ambion). All plasmids used are listed in the table. To incorporate fluorescence UTPs into the newly synthesized mRNAs, 1 μL of ChromaTide Alexa fluor 546-14-UTP (Life Technologies) were added in the in vitro transcription reaction solution.
Xenopus In situ hybridization
Stage I, II, III, VI oocytes and mature eggs were fixed with MEMFA for an hour at room temperature, washed with PBS twice, and dehydrated in methanol. Dehydrated samples were stored in 100% methanol at −20°C until the in situ hybridization was initiated. For in situ hybridization, samples were rehydrated through 100%, 75%, 50%, 25%, and 0% methanol in PBSW (PBS with 0.1% Tween 20) for 5 min each step at room temperature, then all samples were bisected using a razor blade. After obtaining hemi-sectioned samples, in situ hybridization were performed as described.76
RNA extraction and quantitative RT-PCR
RNAs were extracted from fractionated samples or bisected samples using TRIzol reagent or RNA Mini Kit according to the manufacturer’s instructions. To obtain cDNAs, reverse transcription was performed using M-MLV Reverse Transcriptase, and then real-time PCR was performed using the Bimake 2x SYBR Green qPCR Master Mix. Ct values were acquired using Applied Biosystems QuantStudio 3 Real-Time PCR System. All primers used are listed in the table (Table S7).
Western blot
For western blots, lysates were made from fractionated samples, mixed with 2 x SDS sample buffer, boiled for 5 min at 100°C, and loaded on SDS-PAGE. After transferring to PVDF membranes, the membranes were blocked in TBST (Tris-buffered saline containing 0.1% Tween 20) containing 5% nonfat dry milk, then incubated sequentially with the primary and secondary antibodies. After extensive washing, the membranes were developed using ECL™ Prime Western Blotting Detection Reagent.
Sample preparation for RNA-Sequencing
For RNA-seq analysis, each sample contained three biological replicates. Oocytes and eggs were fractionated as described the above. Before RNA extraction, a mixture of spike-in RNAs (egfp – 100 pg, mCherry – 10 pg, mRuby2 – 1 pg, firefly – 0.1 pg, renilla – 0.01 pg) were added into each 100 μL of the cytosolic, ER, and pellet fraction samples. Total RNA was extracted using TRIzol reagent according to the manufacturer’s instructions. Before library construction, ribosomal RNA was removed with the Ribozero HMR kit (Illumina), and the RNA libraries were prepared using the TruSeq Stranded mRNA-seq Sample Prep kit (Illumina). The libraries were quantitated by qPCR and sequenced on one lane for 101 cycles from each end of the fragments on a NovaSeq 6000 using a NovaSeq S1 reagent kit. FASTQ files were generated and demultiplexed with the bcl2fastq Conversion Software v2.20 (Illumina). Ribosomal RNA depletion, library construction, and generation of raw data were performed by the University of Illinois Roy J. Carver Biotechnology Center Sequencing Core Facility.
Absolute quantification of RNA-seq data
Adaptor sequences and low-quality bases were trimmed using Trimmomatic (0.39v81), then the abundance of transcripts (Transcripts Per Kilobase Million; TPM) was quantified by Kallisto v0.46.2,82 based on the Xenopus laevis transcriptome (v 9.2) from the XenBase (http://www.xenbase.org; RRID:SCR_00328083) as the reference. TPM values for relative transcript abundance of each RNA-seq data were converted to the absolute atto-moles (amol) with the linear regression model trained by five spike-in RNAs (Figure S4A), and scaled for normalization (Figure 4B). To reduce the noise from the low abundant mRNAs, all transcripts with an abundance less than 0.125 amol, were filtered out. The remaining 17,811 transcripts were used for further analysis (Figure S4C). Three replicates of each sample were validated through PCA and hierarchical clustering analysis.
Since RNAs in the pellet fraction were showed a similar distribution in both stage VI oocytes and mature eggs (Figure S4F), differentially localized RNAs were analyzed using the cytosol and ER ratio, where the absolute value of the log2-transformed ratio is greater than two-fold, and q-value is less than 0.05. We used Student’s T test for comparison, and the p values were adjusted to q-value for multiple testing with an R package (http://github.com/jdstorey/qvalue). The RNA-seq data have been deposited to the GEO with the dataset identifier GSE199254.
Comparison to RNA localization data in Xenopus laevis eggs
The distribution of RNAs along the animal-vegetal axis in the Xenopus laevis eggs was examined previously.55 In total, 10,871 transcripts were detected in our fractionation RNA-seq and the Sindelka study. The results from the Sindelka study were simplified by categorizing all transcripts into animally (combined ‘extremely animal RNAs’ and ‘animal RNAs’) and vegetally (combined ‘extremely vegetal RNAs’ and ‘vegetal RNAs’) enriched groups. The difference in RNA distribution of the two datasets was tested by Fisher’s exact test.
QUANTIFICATION AND STATISTICAL ANALYSIS
All information about the statistical details is reported in the figure legends. Statistical tests and visualization for RNA-seq and proteomic profile-related results were performed by R studio. All image analysis and statistical tests were performed by ImageJ and GraphPad Prism 9, respectively.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-KDEL | EMD Millipore | Cat. 420400; RRID:AB_212090 |
| Rabbit anti-GRP78 | Abcam | Cat. Ab32618; RRID:AB_732737 |
| Rabbit anti-Pan Atlastin | Rismanchi et al., 200852 | N/A |
| Goat anti-Tia1 | Santa Cruz | Cat. sc-1751; RRID:AB_2201433 |
| Mouse anti-HuR/ELAV1 | Santa Cruz | Cat. sc-5261; RRID:AB_627770 |
| Mouse anti-PTB (for RIP) | EMD Millipore | Cat. MABE986 |
| Mouse anti-PTBP1 (for western blot) | Invitrogen | Cat. 32–4800; RRID:AB_2533082 |
| Mouse anti-HSC70 | Santa Cruz | Cat. sc-7298; RRID:AB_627761 |
| Rabbit anti-Actin | Sigma-Aldrich | Cat. A2066; RRID:AB_476693 |
| Secondary anti-mouse-HRP | GE Healthcare | Cat. NA931V |
| Secondary anti-rabbit-HRP | GE Healthcare | Cat. NA934V |
| Secondary anti-goat-HRP | Santa Cruz | Cat. sc-2020 |
| Alexa Fluor™ goat anti-rabbit-594 | Thermo Fisher | Cat. A11012; RRID:AB_2534079 |
| Alexa Fluor™ goat anti rabbit-488 | Thermo Fisher | Cat. A11008; RRID:AB_143165 |
| Alexa Fluor™ goat anti-mouse-594 | Thermo Fisher | Cat. A11005; RRID:AB_2534073 |
| Alexa Fluor™ donkey anti-rabbit-488 | Thermo Fisher | Cat. A21206; RRID:AB_2535792 |
| Chemicals, peptides, and recombinant proteins | ||
| Cytochalasin B | Sigma-Aldrich | Cat. C6762 |
| Dynabeads Protein G | Thermo Fisher | Cat. 10004D |
| RiboLock RNase Inhibitor | Thermo Fisher | Cat. EO0381 |
| TRIzol reagent | Ambion | Cat. 15596018 |
| Digitonin | EMD Millipore | Cat. 300410 |
| Fluoromount-G Mounting medium | SouthernBiotech | Cat. 0100-01 |
| Mounting medium | Vectatstain | Cat. H-1200 |
| 2x SYBR Green qPCR Master Mix | Bimake | Cat. B21203 |
| Alexa Fluor™ 594 phalloidin | Thermo Fisher | Cat. A12381 |
| ChromaTide Alexa fluor 546-14-UTP | Life Technologies | Cat. C11404 |
| PMSG | Prospec Bio | Cat. HOR-272 |
| HCG | Millipore Sigma | Cat. 230734 |
| BM Purple AP substrate | Roche | Cat. 11442074001 |
| Critical commercial assays | ||
| Opal™ 570 Reagent Pack | Akoya Biosciences | FP1488001KT |
| mMESSAGE mMACHINE Kit for SP6 | Ambion | Cat. AM1340 |
| RNAscope Multiplex Fluorescent Reagent Kit v.2 | ACD | Cat. 323100 |
| RNAscope Probe -Mm-psma1 | ACD | Cat. 557901 |
| RNAscope 3-plex Negative Control Probe | ACD | Cat. 320871 |
| M-MLV Reverse Transcriptase | Promega | Cat. M1701 |
| PureLink™ RNA Mini Kit | Ambion | Cat. 12183025 |
| ECL™ Prime Western Blotting Detection Reagent | Amersham | Cat. RPN2236 |
| Deposited data | ||
| RNA-sequencing raw and analyzed data | This paper | GEO: GSE199254 |
| Mass spectrophotometry raw and analyzed data | This paper | PRIDE: PXD033018 |
| RNA-seq raw and analyzed data for A-V axis | Sindelka et al., 201855 | GEO: GSE104848 |
| Sequencing data for polysome-associated transcripts | Yang et al., 202056 | GEO: GSE134537 |
| Experimental models: Organisms/strains | ||
| Xenopus laevis: Wild type | Nasco | LM00715 |
| Mouse: CD1 | Charles River Laboratories | 022 |
| Oligonucleotides | ||
| See Table S7 for a list of oligonucleotides | ||
| Recombinant DNA | ||
| PSMA1 | Francis Crick Institute | TGas125b12 |
| PSMA2 | Francis Crick Institute | TEgg045c15 |
| PSMA4 | Francis Crick Institute | TGas091k05 |
| DDX25 | Francis Crick Institute | TEgg065i20 |
| DND1 | Francis Crick Institute | TEgg041p23 |
| pCS2-PSME1 | Hwang et al., 201945 | N/A |
| pCS2-PSME2 | Hwang et al., 201945 | N/A |
| pCS2-PSME3 | Hwang et al., 201945 | N/A |
| pCMV-SPORT6-PSME4 | Hwang et al., 201945 | N/A |
| pSP64-GFP-KDEL | Terasaki et al., 199671 | N/A |
| Software and algorithms | ||
| ImageJ | Schneider et al., 201272 | https://imagej.nih.gov/ij/ |
| GraphPad Prism | GraphPad | https://www.graphpad.com/ |
| Kallisto | Bray et al., 201682 | https://pachterlab.github.io/kallisto/ |
| Metascape | Zhou et al., 201973 | https://metascape.org/gp/index.html#/main/step1 |
Highlights.
Maternal RNAs are uniquely stored on the ER in Xenopus oocyte
mRNAs are released from the ER into the cytosol after oocyte maturation
The ER is remodeled into a tubular network after oocyte maturation in Xenopus
The tubular ER network is essential for the asymmetric localization of RNA
ACKNOWLEDGMENTS
We thank Mike Gilchrist for the suggestion of using spike RNA mixture in fractionation RNA-seq experiments, Mark Terasaki for providing pSP64-GFP-KDEL plasmid, Craig Blackstone for providing anti-Atlastin antibody, Jia Fu for technical support, Jodi Flaws and Liying Gao for helping with mouse oocytes collection, Lou Ann Miller for TEM analysis of Xenopus oocytes and eggs, Alvaro Hernandez and Christy Wright for ribosomal RNA depletion/library construction/RNA sequencing, and Ross Tomaino for proteomic analysis of microsome proteins. J.Y. is supported by a grant from NIH (R35 GM131810). W.M. is supported by NIH grants (R03AI146900 and R01GM140306). T.K. is supported by the Basic Science Research Program funded by the Ministry of Education (2018R1A6A1A03025810), and the Future-leading Project Research Fund of UNIST (1.220023.01).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111802.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Shibata Y, Hu J, Kozlov MM, and Rapoport TA (2009). Mechanisms shaping the membranes of cellular organelles. Annu. Rev. Cell Dev. Biol 25, 329–354. 10.1146/annurev.cellbio.042308.113324. [DOI] [PubMed] [Google Scholar]
- 2.Shibata Y, Shemesh T, Prinz WA, Palazzo AF, Kozlov MM, and Rapoport TA (2010). Mechanisms determining the morphology of the peripheral ER. Cell 143, 774–788. 10.1016/j.cell.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Friedman JR, and Voeltz GK (2011). The ER in 3D: a multifunctional dynamic membrane network. Trends Cell Biol. 21, 709–717. 10.1016/j.tcb.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwarz DS, and Blower MD (2016). The endoplasmic reticulum: structure, function and response to cellular signaling. Cell. Mol. Life Sci 73, 79–94. 10.1007/s00018-015-2052-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schultz RM, Stein P, and Svoboda P (2018). The oocyte-to-embryo transition in mouse: past, present, and future. Biol. Reprod 99, 160–174. 10.1093/biolre/ioy013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jaffe LA, and Terasaki M (1994). Structural changes in the endoplasmic reticulum of starfish oocytes during meiotic maturation and fertilization. Dev. Biol 164, 579–587. 10.1006/dbio.1994.1225. [DOI] [PubMed] [Google Scholar]
- 7.Terasaki M, and Jaffe LA (1991). Organization of the sea urchin egg endoplasmic reticulum and its reorganization at fertilization. J. Cell Biol 114, 929–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mehlmann LM, Terasaki M, Jaffe LA, and Kline D (1995). Reorganization of the endoplasmic reticulum during meiotic maturation of the mouse oocyte. Dev. Biol 170, 607–615. 10.1006/dbio.1995.1240. [DOI] [PubMed] [Google Scholar]
- 9.Shiraishi K, Okada A, Shirakawa H, Nakanishi S, Mikoshiba K, and Miyazaki S (1995). Developmental changes in the distribution of the endoplasmic reticulum and inositol 1, 4, 5-trisphosphate receptors and the spatial pattern of Ca2+ release during maturation of hamster oocytes. Dev. Biol 170, 594–606. 10.1006/dbio.1995.1239. [DOI] [PubMed] [Google Scholar]
- 10.Kume S, Yamamoto A, Inoue T, Muto A, Okano H, and Mikoshiba K (1997). Developmental expression of the inositol 1, 4, 5-trisphosphate receptor and structural changes in the endoplasmic reticulum during oogenesis and meiotic maturation of Xenopus laevis. Dev. Biol 182, 228–239. 10.1006/dbio.1996.8479. [DOI] [PubMed] [Google Scholar]
- 11.Kline D (2000). Attributes and dynamics of the endoplasmic reticulum in mammalian eggs. Curr. Top. Dev. Biol 50, 125–154. [DOI] [PubMed] [Google Scholar]
- 12.Terasaki M, Runft LL, and Hand AR (2001). Changes in organization of the endoplasmic reticulum during Xenopus oocyte maturation and activation. Mol. Biol. Cell 12, 1103–1116. 10.1091/mbc.12.4.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.FitzHarris G, Marangos P, and Carroll J (2007). Changes in endoplasmic reticulum structure during mouse oocyte maturation are controlled by the cytoskeleton and cytoplasmic dynein. Dev. Biol 305, 133–144. 10.1016/j.ydbio.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 14.Stitzel ML, and Seydoux G (2007). Regulation of the oocyte-to-zygote transition. Science 316, 407–408. 10.1126/science.1138236. [DOI] [PubMed] [Google Scholar]
- 15.Walter P, and Lingappa VR (1986). Mechanism of protein translocation across the endoplasmic reticulum membrane. Annu. Rev. Cell Biol 2, 499–516. 10.1146/annurev.cb.02.110186.002435. [DOI] [PubMed] [Google Scholar]
- 16.Rapoport TA (2007). Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature 450, 663–669. 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- 17.Reid DW, and Nicchitta CV (2015). Diversity and selectivity in mRNA translation on the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol 16, 221–231. 10.1038/nrm3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caro LG, and Palade GE (1964). Protein synthesis, storage, and discharge in the pancreatic exocrine cell. An autoradiographic study. J. Cell Biol 20, 473–495. 10.1083/jcb.20.3.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoffman AM, Chen Q, Zheng T, and Nicchitta CV (2019). Heterogeneous translational landscape of the endoplasmic reticulum revealed by ribosome proximity labeling and transcriptome analysis. J. Biol. Chem 294, 8942–8958. 10.1074/jbc.RA119.007996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsu JCC, Reid DW, Hoffman AM, Sarkar D, and Nicchitta CV (2018). Oncoprotein AEG-1 is an endoplasmic reticulum RNA-binding protein whose interactome is enriched in organelle resident protein-encoding mRNAs. RNA 24, 688–703. 10.1261/rna.063313.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Voigt F, Zhang H, Cui XA, Triebold D, Liu AX, Eglinger J, Lee ES, Chao JA, and Palazzo AF (2017). Single-molecule quantification of translation-dependent association of mRNAs with the endoplasmic reticulum. Cell Rep. 21, 3740–3753. 10.1016/j.celrep.2017.12.008. [DOI] [PubMed] [Google Scholar]
- 22.Jagannathan S, Reid DW, Cox AH, and Nicchitta CV (2014). De novo translation initiation on membrane-bound ribosomes as a mechanism for localization of cytosolic protein mRNAs to the endoplasmic reticulum. RNA 20, 1489–1498. 10.1261/rna.045526.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jagannathan S, Hsu JCC, Reid DW, Chen Q, Thompson WJ, Moseley AM, and Nicchitta CV (2014). Multifunctional roles for the protein translocation machinery in RNA anchoring to the endoplasmic reticulum. J. Biol. Chem 289, 25907–25924. 10.1074/jbc.M114.580688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kraut-Cohen J, Afanasieva E, Haim-Vilmovsky L, Slobodin B, Yosef I, Bibi E, and Gerst JE (2013). Translation-and SRP-independent mRNA targeting to the endoplasmic reticulum in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 24, 3069–3084. 10.1091/mbc.E13-01-0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cui XA, Zhang H, and Palazzo AF (2012). p180 promotes the ribosome-independent localization of a subset of mRNA to the endoplasmic reticulum. PLoS Biol. 10, e1001336. 10.1371/journal.pbio.1001336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reid DW, and Nicchitta CV (2012). Primary role for endoplasmic reticulum-bound ribosomes in cellular translation identified by ribosome profiling. J. Biol. Chem 287, 5518–5527. 10.1074/jbc.M111.312280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Q, Jagannathan S, Reid DW, Zheng T, and Nicchitta CV (2011). Hierarchical regulation of mRNA partitioning between the cytoplasm and the endoplasmic reticulum of mammalian cells. Mol. Biol. Cell 22, 2646–2658. 10.1091/mbc.E11-03-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pyhtila B, Zheng T, Lager PJ, Keene JD, Reedy MC, and Nicchitta CV (2008). Signal sequence- and translation-independent mRNA localization to the endoplasmic reticulum. RNA 14, 445–453. 10.1261/rna.721108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stephens SB, and Nicchitta CV (2008). Divergent regulation of protein synthesis in the cytosol and endoplasmic reticulum compartments of mammalian cells. Mol. Biol. Cell 19, 623–632. 10.1091/mbc.e07-07-0677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lerner RS, Seiser RM, Zheng T, Lager PJ, Reedy MC, Keene JD, and Nicchitta CV (2003). Partitioning and translation of mRNAs encoding soluble proteins on membrane-bound ribosomes. RNA 9, 1123–1137. 10.1261/rna.5610403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmid M, Jaedicke A, Du TG, and Jansen RP (2006). Coordination of endoplasmic reticulum and mRNA localization to the yeast bud. Curr. Biol 16, 1538–1543. 10.1016/j.cub.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 32.Deshler JO, Highett MI, and Schnapp BJ (1997). Localization of Xenopus Vg1 mRNA by Vera protein and the endoplasmic reticulum. Science 276, 1128–1131. 10.1126/science.276.5315.1128. [DOI] [PubMed] [Google Scholar]
- 33.Alarcón VB, and Elinson RP (2001). RNA anchoring in the vegetal cortex of the Xenopus oocyte. J. Cell Sci 114, 1731–1741. [DOI] [PubMed] [Google Scholar]
- 34.Chang P, Torres J, Lewis RA, Mowry KL, Houliston E, and King ML (2004). Localization of RNAs to the mitochondrial cloud in Xenopus oocytes through entrapment and association with endoplasmic reticulum. Mol. Biol. Cell 15, 4669–4681. 10.1091/mbc.e04-03-0265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prodon F, Dru P, Roegiers F, and Sardet C (2005). Polarity of the ascidian egg cortex and relocalization of cER and mRNAs in the early embryo. J. Cell Sci 118, 2393–2404. 10.1242/jcs.02366. [DOI] [PubMed] [Google Scholar]
- 36.Sardet C, Nishida H, Prodon F, and Sawada K (2003). Maternal mRNAs of PEM and macho 1, the ascidian muscle determinant, associate and move with a rough endoplasmic reticulum network in the egg cortex. Development 130, 5839–5849. 10.1242/dev.00805. [DOI] [PubMed] [Google Scholar]
- 37.Bashirullah A, Cooperstock RL, and Lipshitz HD (1998). RNA localization in development. Annu. Rev. Biochem 67, 335–394. 10.1146/annurev.biochem.67.1.335. [DOI] [PubMed] [Google Scholar]
- 38.Palacios IM, and St Johnston D (2001). Getting the message across: the intracellular localization ofmRNAs in higher eukaryotes. Annu. Rev. Cell Dev. Biol 17, 569–614. 10.1146/annurev.cellbio.17.1.569. [DOI] [PubMed] [Google Scholar]
- 39.Shav-Tal Y, and Singer RH (2005). RNA localization. J. Cell Sci 118, 4077–4081. 10.1242/jcs.02543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martin KC, and Ephrussi A (2009). mRNA localization: gene expression in the spatial dimension. Cell 136, 719–730. 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holt CE, and Bullock SL (2009). Subcellular mRNA localization in animal cells and why it matters. Science 326, 1212–1216. 10.1126/science.1176488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Medioni C, Mowry K, and Besse F (2012). Principles and roles of mRNA localization in animal development. Development 139, 3263–3276. 10.1242/dev.078626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Houston DW (2013). Regulation of cell polarity and RNA localization in vertebrate oocytes. Int. Rev. Cell Mol. Biol 306, 127–185. 10.1016/B978-0-12-407694-5.00004-3. [DOI] [PubMed] [Google Scholar]
- 44.Oh D, and Houston DW (2017). RNA localization in the vertebrate oocyte: establishment of oocyte polarity and localized mRNA assemblages. Results Probl. Cell Differ 63, 189–208. 10.1007/978-3-319-60855-6_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hwang H, Jin Z, Krishnamurthy VV, Saha A, Klein PS, Garcia B, Mei W, King ML, Zhang K, and Yang J (2019). Novel functions of the ubiquitin-independent proteasome system in regulating Xenopus germline development. Development 146. 10.1242/dev.172700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Horvay K, Claussen M, Katzer M, Landgrebe J, and Pieler T (2006). Xenopus Dead end mRNA is a localized maternal determinant that serves a conserved function in germ cell development. Dev. Biol 291, 1–11. 10.1016/j.ydbio.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 47.Mei W, Jin Z, Lai F, Schwend T, Houston DW, King ML, and Yang J (2013). Maternal Dead-End1 is required for vegetal cortical microtubule assembly during Xenopus axis specification. Development 140, 2334–2344. 10.1242/dev.094748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aguero T, Jin Z, Chorghade S, Kalsotra A, King ML, and Yang J (2017). Maternal Dead-end 1 promotes translation of nanos1 by binding the eIF3 complex. Development 144, 3755–3765. 10.1242/dev.152611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hudson C, and Woodland HR (1998). Xpat, a gene expressed specifically in germ plasm and primordial germ cells of Xenopus laevis. Mech. Dev 73, 159–168. [DOI] [PubMed] [Google Scholar]
- 50.MacArthur H, Houston DW, Bubunenko M, Mosquera L, and King ML (2000). DEADSouth is a germ plasm specific DEAD-box RNA helicase in Xenopus related to eIF4A. Mech. Dev 95, 291–295. [DOI] [PubMed] [Google Scholar]
- 51.Munro S, and Pelham HR (1987). A C-terminal signal prevents secretion of luminal ER proteins. Cell 48, 899–907. [DOI] [PubMed] [Google Scholar]
- 52.Rismanchi N, Soderblom C, Stadler J, Zhu PP, and Blackstone C (2008). Atlastin GTPases are required for Golgi apparatus and ER morphogenesis. Hum. Mol. Genet 17, 1591–1604. 10.1093/hmg/ddn046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shiu RP, and Pastan IH (1979). Properties and purification of a glucose-regulated protein from chick embryo fibroblasts. Biochim. Biophys. Acta 576, 141–150. 10.1016/0005-2795(79)90493-8. [DOI] [PubMed] [Google Scholar]
- 54.Larabell CA, and Chandler DE (1988). Freeze-fracture analysis of structural reorganization during meiotic maturation in oocytes of Xenopus laevis. Cell Tissue Res. 251, 129–136. 10.1007/BF00215457. [DOI] [PubMed] [Google Scholar]
- 55.Sindelka R, Abaffy P, Qu Y, Tomankova S, Sidova M, Naraine R, Kolar M, Peuchen E, Sun L, Dovichi N, and Kubista M (2018). Asymmetric distribution of biomolecules of maternal origin in the Xenopus laevis egg and their impact on the developmental plan. Sci. Rep 8, 8315. 10.1038/s41598-018-26592-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang F, Wang W, Cetinbas M, Sadreyev RI, and Blower MD (2020). Genome-wide analysis identifies cis-acting elements regulating mRNA polyadenylation and translation during vertebrate oocyte maturation. RNA 26, 324–344. 10.1261/rna.073247.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gard DL (1999). Confocal microscopy and 3-D reconstruction of the cytoskeleton of Xenopus oocytes. Microsc. Res. Tech 44, 388–414. . [DOI] [PubMed] [Google Scholar]
- 58.Liu XS, and Liu XJ (2006). Oocyte isolation and enucleation. Methods Mol. Biol 322, 31–41. 10.1007/978-1-59745-000-3_3. [DOI] [PubMed] [Google Scholar]
- 59.Schorderet-Slatkine S, and Drury KC (1973). Progesterone induced maturation in oocytes of Xenopus laevis. Appearance of a ’maturation promoting factor’ in enucleated oocytes. Cell Differ. 2, 247–254. 10.1016/0045-6039(73)90013-4. [DOI] [PubMed] [Google Scholar]
- 60.Fox CA, Sheets MD, and Wickens MP (1989). Poly(A) addition during maturation of frog oocytes: distinct nuclear and cytoplasmic activities and regulation by the sequence UUUUUAU. Genes Dev. 3, 2151–2162. [DOI] [PubMed] [Google Scholar]
- 61.Oe T, Nakajo N, Katsuragi Y, Okazaki K, and Sagata N (2001). Cytoplasmic occurrence of the Chk1/Cdc25 pathway and regulation of Chk1 in Xenopus oocytes. Dev. Biol 229, 250–261. 10.1006/dbio.2000.9968. [DOI] [PubMed] [Google Scholar]
- 62.Bayaa M, Booth RA, Sheng Y, and Liu XJ (2000). The classical progesterone receptor mediates Xenopus oocyte maturation through a non-genomic mechanism. Proc. Natl. Acad. Sci. USA 97, 12607–12612. 10.1073/pnas.220302597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reid DW, Chen Q, Tay ASL, Shenolikar S, and Nicchitta CV (2014). The unfolded protein response triggers selective mRNA release from the endoplasmic reticulum. Cell 158, 1362–1374. 10.1016/j.cell.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Conti M, and Franciosi F (2018). Acquisition of oocyte competence to develop as an embryo: integrated nuclear and cytoplasmic events. Hum. Reprod. Update 24, 245–266. 10.1093/humupd/dmx040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ecker RE, and Smith LD (1966). The kinetics of protein synthesis in early amphibian development. Biochim. Biophys. Acta 129, 186–192. 10.1016/0005-2787(66)90020-7. [DOI] [PubMed] [Google Scholar]
- 66.Smith LD, Ecker RE, and Subtelny S (1966). The initiation of protein synthesis in eggs of rana pipiens. Proc. Natl. Acad. Sci. USA 56, 1724–1728. 10.1073/pnas.56.6.1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ecker RE, Smith LD, and Subtelny S (1968). Kinetics of protein synthesis in enucleate frog oocytes. Science 160, 1115–1117. 10.1126/science.160.3832.1115. [DOI] [PubMed] [Google Scholar]
- 68.Ecker RE, and Smith LD (1968). Protein synthesis in amphibian oocytes and early embryos. Dev. Biol 18, 232–249. 10.1016/0012-1606(68)90034-1. [DOI] [PubMed] [Google Scholar]
- 69.Quinlan ME (2016). Cytoplasmic streaming in the Drosophila oocyte. Annu. Rev. Cell Dev. Biol 32, 173–195. 10.1146/annurev-cellbio-111315-125416. [DOI] [PubMed] [Google Scholar]
- 70.Shi B, Zhang J, Heng J, Gong J, Zhang T, Li P, Sun BF, Yang Y, Zhang N, Zhao YL, et al. (2020). RNA structural dynamics regulate early embryogenesis through controlling transcriptome fate and function. Genome Biol. 21, 120. 10.1186/s13059-020-02022-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Terasaki M, Jaffe LA, Hunnicutt GR, and Hammer JA 3rd. (1996). Structural change of the endoplasmic reticulum during fertilization: evidence for loss of membrane continuity using the green fluorescent protein. Dev. Biol 179, 320–328. [DOI] [PubMed] [Google Scholar]
- 72.Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, and Chanda SK (2019). Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun 10, 1523. 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Houston DW (2018). Oocyte host-Transfer and maternal mRNA depletion experiments in Xenopus. Cold Spring Harb. Protoc 2018, pdb. prot096982. 10.1101/pdb.prot096982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Heasman J, Holwill S, and Wylie CC (1991). Fertilization of cultured Xenopus oocytes and use in studies of maternally inherited molecules. Methods Cell Biol. 36, 213–230. [DOI] [PubMed] [Google Scholar]
- 76.Sive H, Grainger R, and Harland R (2000). Early development of Xenopus laevis. A Laboratory Manual (Cold Spring Harbor Press). [Google Scholar]
- 77.Ford CC, and Gurdon JB (1977). A method for enucleating oocytes of Xenopus laevis. J. Embryol. Exp. Morphol 37, 203–209. [PubMed] [Google Scholar]
- 78.Roeder AD, and Gard DL (1994). Confocal microscopy of F-actin distribution in Xenopus oocytes. Zygote 2, 111–124. [DOI] [PubMed] [Google Scholar]
- 79.Ezure T, Nanatani K, Sato Y, Suzuki S, Aizawa K, Souma S, Ito M, Hohsaka T, von Heijine G, Utsumi T, et al. (2014).A cell-freetranslocation system using extracts of cultured insect cells to yield functional membrane proteins. PLoS One 9, e112874. 10.1371/journal.pone.0112874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Perez-Riverol Y, Bai J, Bandla C, García-Seisdedos D, Hewapathirana S, Kamatchinathan S, Kundu DJ, Prakash A, Frericks-Zipper A, Eisenacher M, et al. (2022). The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 50, D543–D552. 10.1093/nar/gkab1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bolger AM, Lohse M, and Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bray NL, Pimentel H, Melsted P, and Pachter L (2016). Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol 34, 525–527. 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 83.Fortriede JD, Pells TJ, Chu S, Chaturvedi P, Wang D, Fisher ME, James-Zorn C, Wang Y, Nenni MJ, Burns KA, et al. (2020).Xenbase: deep integration of GEO & SRA RNA-seq and ChIP-seq data in a model organism database. Nucleic Acids Res. 48, D776–D782. 10.1093/nar/gkz933. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA sequencing data generated in this study have been deposited to the Gene Expression Omnibus (GEO) with the dataset identifier GEO:GSE199254. Proteomic profile data generated in this study have been deposited to the ProteomeXchange Consortium via PRIDE partner repository with the dataset identifier PRIDE: PXD033018
This paper does not report the original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.