Abstract
Rheumatoid arthritis (RA) is an inflammatory disease that leads to disability; however, existing therapies are still unsatisfactory. Activated fibroblast-like synoviocytes (FLSs) play an essential role in synovitis formation and joint destruction in RA. The Hedgehog signaling pathway is aberrantly activated and contributes to the aggressive phenotype of RA-FLSs. However, it remains uncertain whether inhibiting Smoothened (SMO), a critical component of the Hedgehog signaling pathway, is an effective treatment for RA. Here, we design a series of small interfering RNAs (siRNAs) that specifically target the SMO gene. With precise chemical modifications, siRNAs’ efficacy and stability are significantly improved, and the off-target effects are minimized. The optimized chemically modified siRNA (si-S1A3-Chol) decreases RA-FLS proliferation and invasiveness without the transfection reagent. Furthermore, si-S1A3-Chol injected intra-articularly effectively alleviates joint destruction and improves motor function in collagen-induced arthritis mouse models. Consequently, our results demonstrate that chemically modified siRNA targeting the Hedgehog signaling pathway may be a potential therapy for RA.
Keywords: MT: Oligonucleotides: therapies and applications, rheumatoid arthritis, Hedgehog signaling, Smoothened, chemical modification, small interfering RNA
Graphical abstract
Introduction
Rheumatoid arthritis (RA) is an autoimmune-mediated chronic inflammatory disease characterized by progressive synovitis and bone erosion.1 Because of the complexity of the pathogenesis and pathophysiology in RA, currently available disease-modifying antirheumatic drugs (DMARDs) are insufficient to prevent progressive joint destruction.2,3,4 Therefore, it is urgently necessary to adopt novel techniques for developing a new therapy that can conquer the persistent inflammation in RA.
Activated fibroblast-like synoviocytes (FLSs) are the predominant cell types of the RA hyperplastic synovial lining layer and play a fundamental role in the synovial pannus formation and joint damage.5 Hyperproliferation, apoptosis resistance, and invasiveness enhancement are characteristics of RA-FLS aggressive phenotype, which results in pannus formation and bone erosion.6 Targeting activated RA-FLSs is an attractive and promising approach for RA therapy.7
The Hedgehog signaling pathway is a conserved pathway that plays a critical role in embryonic development.8 In mammals, Hedgehog ligands interact with the transmembrane receptors Patched 1 and relieve the suppression against Smoothened (SMO). Subsequently, the signaling cascade results in activation and nuclear trans localization of glioma-associated oncogene family zinc finger 1–3 (GLI1–3).9 As the transcriptional factor, GLI drives the expression of target genes involved in pathway feedback regulation (PTCH1, PTCH2, GLI1), proliferation (CCND, CCNE, MYC), survival (BCL2), angiogenesis (ANG1/2, VEGFA), and epithelial-to-mesenchymal transitions (SNAI1, TWIST1).10 In healthy adult tissues, the Hedgehog signaling pathway is usually quiescent.11 Aberrant activation and dysregulation of the Hedgehog signaling pathway in adult tissues were identified in the tumorigenesis and metastasis of various malignancies, including basal cell carcinoma (BCC), medulloblastoma, melanoma, acute myeloid leukemia (AML), and prostate, ovarian, colorectal, and liver cancers.12 Additionally, the Hedgehog signaling pathway was found to be aberrantly activated in rheumatic diseases such as RA, osteoarthritis, ankylosing spondylitis, and systemic sclerosis.13 Considering these findings, inhibiting the Hedgehog signaling pathway may aid in the treatment of various cancers and inflammatory diseases. Nowadays, several small-molecule antagonists of SMO, such as vismodegib, sonidegib, and glasdegib, are approved by the Food and Drug Administration (FDA) to treat BCC and AML, respectively.14,15,16 However, the adverse effects of systemic administration and SMO drug-resistant mutations restrict the therapeutic utility of antagonists, motivating the quest for novel therapies.14,17,18
As small interfering RNA (siRNA) can mediate the targeted mRNA degradation in a sequence-specific manner and may be an alternative approach to interfering with the expression target gene.19 With the FDA approval of patisiran, givosiran, lumasiran, inclisiran, and vutrisiran, siRNA therapeutics are emerging as a potential and popular field for new drug development. However, because drug carriers are easily enriched in the liver and spleen, all approved siRNA drugs only target liver diseases. Successfully stabilizing siRNA and efficiently delivering it to target tissues have become challenges for therapeutic applications of siRNA drugs. Accumulated evidence has suggested that chemical modifications make it possible to improve siRNA performance. Appropriate modification of siRNA can enhance its efficacy, specificity, and stability while minimizing its biotoxicity and immunogenicity.20 Moreover, topical administration contributes to delivering siRNA to intended tissues beyond the liver and reduces unintended organs’ side effects.21
This study aims to develop a series of siRNAs that specifically target SMO mRNA. With the chemical modification and cholesterol conjugation, the optimized chemically modified anti-SMO siRNA regulates RA-FLS proliferation and invasiveness without the transfection reagent. Moreover, we demonstrate that siRNA accumulates in the intended joints by intra-articular injection, ameliorates arthritis progression, and maintains motor function in collagen-induced arthritis (CIA) mouse models. Therefore, chemically modified anti-SMO siRNA injected intra-articularly may be a potential therapeutic approach for RA.
Results
Anti-SMO siRNAs designed against various sites efficiently inhibit SMO expression in RA-FLSs
FLSs were isolated from patients with active RA and used as the cell model for siRNA screening in vitro. Primary RA-FLSs exuded from synovial tissues within 7 days (Figure S1A). The morphological characteristics of RA-FLSs at passage 3 were unique, with a spindle and woven shape (Figure S1B). The surface molecules of RA-FLSs at passage 3 were characterized by flow cytometry. The positive staining rate of CD55 and CD90, the fibroblast markers, were 97.9% and 99.4%; meanwhile, CD14 and CD68, the macrophage markers, were negatively stained, indicating the high purity of RA-FLSs used in the study (Figure S1C).
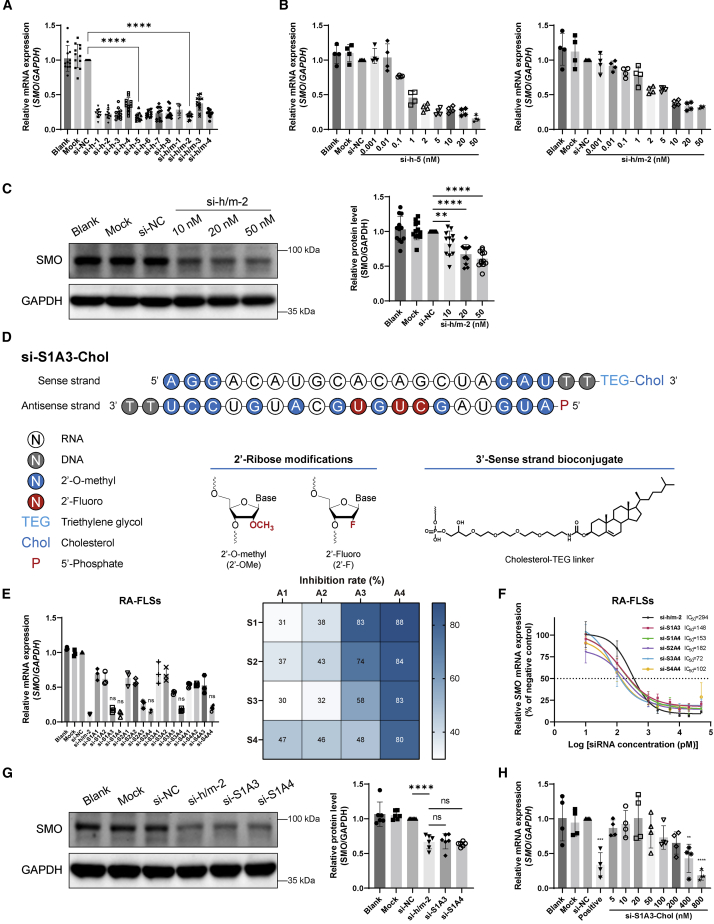
Because the target site accessibility highly impacted the effectiveness of siRNA,22 we designed 12 siRNAs against various target sites of coding sequences in mRNA, including eight siRNAs specifically targeting Homo sapiens SMO mRNA (here called si-h-1 to si-h-8), and four siRNAs targeting the homologous region between Homo sapiens SMO mRNA and Mus musculus Smo mRNA (here called si-h/m-1 to si-h/m-4). The sequences of siRNAs are listed in Table S1, and the target sites of siRNAs are shown in Figure S2.
RA-FLSs isolated from 12 patients with active RA were used as 12 biological repetitions to maximize the applicability of siRNAs. All siRNAs transfected by transfection reagent inhibited SMO mRNA expression in RA-FLSs with high efficiency (Figure 1A). Compared with the negative control siRNA (si-NC), si-h-5 (0.18 ± 0.06 versus 1.00 ± 0, p < 0.0001) and si-h/m-2 (0.19 ± 0.04 versus 1.00 ± 0, p < 0.0001) showed the highest silencing efficiency. The inhibition rates of si-h-5 and si-h/m-2 were concentration dependent (Figure 1B). si-h/m-2 was selected as a candidate for subsequent experiments because it targets the homologous region between humans and mice. Compared with si-NC, si-h/m-2 transfected to RA-FLSs inhibited the expression of SMO protein in a concentration-dependent manner (10 nM, 0.83 ± 0.18 versus 1.00 ± 0, p < 0.01; 20 nM, 0.67 ± 0.14 versus 1.00 ± 0, p < 0.0001; 50 nM, 0.61 ± 0.11 versus 1.00 ± 0, p < 0.0001) (Figure 1C).
Figure 1.
Chemically modified siRNAs inhibit SMO expression with high efficiency
(A) Silencing efficiency screening of anti-SMO siRNAs. Relative SMO mRNA expression was assessed by qPCR after RA-FLSs were transfected with different siRNAs (50 nM) for 48 h, shown as fold change versus si-NC (50 nM) (n = 12). (B) Concentration dependence of siRNAs in the silence of SMO mRNA expression was determined by qPCR after RA-FLSs were transfected with si-h-5 (left) or si-h/m-2 (right) at indicated concentrations (0.001–50 nM) for 48 h. Relative expression is shown as fold change versus si-NC (50 nM) (n = 4). (C) Concentration dependence of si-h/m-2 in inhibiting SMO protein level was determined by western blot after RA-FLSs were transfected with si-h/m-2 at indicated concentrations (10, 20, and 50 nM) for 72 h. GAPDH was used as a loading control. Relative expression was calculated as the ratio of SMO/GAPDH and is presented as fold change versus si-NC (50 nM) (n = 12). (D) Representative schematic diagram of chemical modification pattern of si-S1A3-Chol. (E) Silencing efficiency screening of siRNAs with different chemical modification patterns. Relative SMO mRNA expression was assessed by qPCR after RA-FLSs were transfected with naked siRNA or chemically modified siRNAs (50 nM) for 48 h, shown as fold change versus si-NC (50 nM) (left) (n = 3). Heatmap (right) shows the percentages of the mean inhibition rate of siRNAs with different modifications in RA-FLSs. (F) Half-maximal inhibitory concentration of selected chemical modification anti-SMO siRNAs. Concentration dependence of siRNAs in the silence of SMO mRNA expression was assessed by qPCR after RA-FLSs were transfected with naked siRNA or chemically modified siRNAs at indicated concentrations (0.01, 0.1, 1, 2, 5, 10, 20, and 50 nM) for 48 h, shown as percentages of si-NC (50 nM) (n = 3). (G) Protein silencing efficiency of selected chemically modified siRNA. Representative western blot of SMO protein after RA-FLSs were transfected with si-h/m-2, si-S1A3, or si-S1A4 (50 nM) for 72 h. GAPDH was used as a loading control. Relative expression was calculated as the ratio of SMO/GAPDH and is presented as fold change versus si-NC (50 nM) (n = 6). (H) Concentration dependence of si-S1A3-Chol in the silence of SMO mRNA expression was determined by qPCR after RA-FLSs were transfected with si-S1A3-Chol at indicated concentrations without any transfection reagent for 48 h. si-S1A3 (50 nM) transfected with Lipofectamine 3000 served as the positive control, and relative expression is shown as fold change versus si-NC (50 nM) (n = 4). Data are presented as mean ± SD. ns, p > 0.05; ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 versus si-NC group (in A, C, and H) or versus si-h/m-2 (naked siRNA with transfection reagent) group (E and G) by one-way ANOVA with Dunnett’s test for multiple comparisons.
Chemically modified siRNA efficiently inhibits SMO expression in RA and murine FLSs with high stability
Chemical modifications to the siRNA duplex were implemented to improve the stability and specificity. Chemical modifications including 2′-O-methyl, 2′-fluoro, and 5′-phosphate were introduced to the sense strand (here called S1, S2, S3, and S4) and antisense strand (here called A1, A2, A3, and A4) of si-h/m-2 in four respective patterns (Figure S3). Four patterns of the sense and antisense strand were annealed in pairs to form a 16-siRNA duplex (here called si-S1A1 to si-S4A4). All chemically modified siRNAs were based on si-h/m-2, and their sequences are listed in Table S2. A schematic diagram of chemically modified siRNA (si-S1A3-Chol) (Chol = cholesterol) is shown in Figure 1D.
After 3 h of human serum incubation, 62.28% of the si-h/m-2 (naked siRNA) was degraded (Figure S4). Conversely, most chemically modified siRNA samples could retain 41.25%–88.04% of the intact siRNA after 48 h of exposure to human serum. Chemically modified siRNAs effectively resisted nuclease degradation in human serum. The serum stability of siRNA increased with the level of chemical modification in the ribonucleic acid.
Meanwhile, as the level of chemical modification in the antisense strand was increasing, the inhibition rate of siRNA was elevated (Figure 1E). For example, the siRNA modified with the patterns of S1A1, S1A2, S1A3, and S1A4 showed increasing efficacy. Compared with naked siRNA, several chemically modified siRNAs still maintain similar silencing efficiency in RA-FLSs (Figure 1E) and murine FLSs (Figure S5A). Besides, compared with si-h/m-2, chemically modified siRNAs inhibited SMO mRNA expression in a concentration-dependent manner with a lower half-maximal inhibitory concentration (IC50) value (72–182 pM versus 294 pM) (Figure 1F). Moreover, compared with si-h/m-2, si-S1A3 and si-S1A4 suppressed SMO protein expression in RA and murine FLSs with similar efficiency (Figures 1G and S5B). Therefore, si-S1A3 and si-S1A4 were selected as candidates for further experiments considering the serum stability and silencing efficiency.
Cholesterol-conjugated chemically modified siRNA efficiently inhibits the activation of Hedgehog signaling without transfection reagent
To avoid the influence of transfection reagent and enhance cellular uptake efficiency, si-S1A3 and si-S1A4 were conjugated with cholesterol at the 3′ end of the sense strand (here called si-S1A3-Chol and si-S1A4-Chol; sequences are shown in Table S3). To evaluate the silencing efficiency, RA-FLSs were treated with si-S1A3-Chol and si-S1A4-Chol at different concentrations without any transfection reagent, with si-S1A3 or si-S1A4 transfected with Lipofectamine 3000 serving as the respective positive control. Compared with si-NC, the SMO mRNA expression in groups treated with si-S1A3-Chol at 400 nM (0.43 ± 0.20 versus 1.00 ± 0, p < 0.01) and 800 nM (0.18 ± 0.07 versus 1.00 ± 0, p < 0.0001) were significantly reduced (Figure 1H), and RA-FLSs treated with si-S1A4-Chol achieved similar outcomes (Figure S6). Moreover, cholesterol-conjugated chemically modified siRNAs maintained the integrity stably in synovial fluids from patients with active RA (Figure S7). For instance, si-S1A3-Chol kept 53.41% of intact siRNA in synovial fluids for 2 days and 63.62% of intact si-S1A4-Chol for 7 days. Based on the high efficiency and appropriate stability, si-S1A3-Chol (at 800 nM) was used to assess the effect of siRNAs in follow-up experiments.
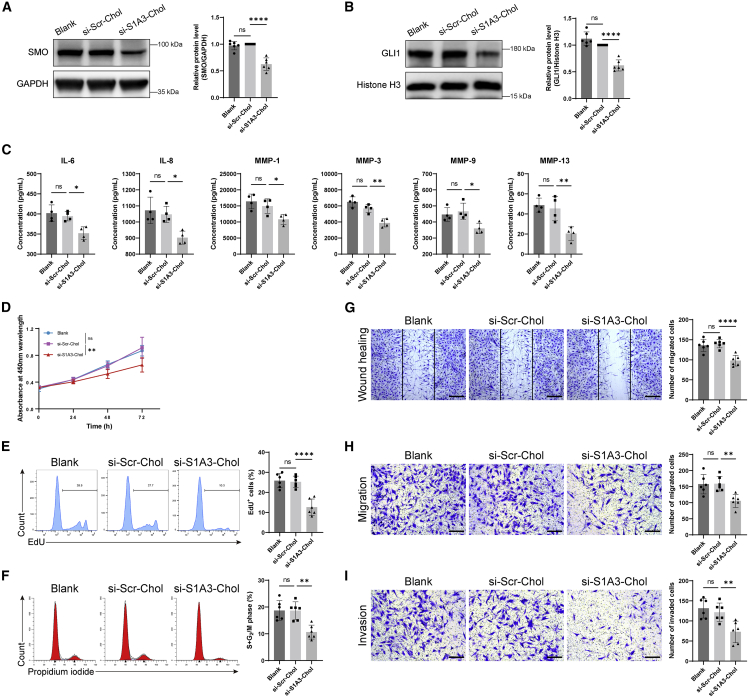
In comparison with the healthy control FLSs, the SMO protein level was highly expressed in RA-FLSs (Figure S8A). Compared with scrambled control siRNA (here called si-Scr-Chol), the expression of SMO protein (0.63 ± 0.12 versus 1.00 ± 0, p < 0.0001) and the nuclear accumulation of GLI1 protein (0.62 ± 0.10 versus 1.00 ± 0, p < 0.0001) in RA-FLSs treated with si-S1A3-Chol was significantly decreased (Figures 2A and 2B). Moreover, as the results showed with the reduction of PTCH1 and PTCH2, the product of the target genes, the activity of the Hedgehog signaling pathway was wholly inhibited through interfering SMO expression with si-S1A3-Chol treatment (Figure S8B), and the expression of target genes such as Cyclin D1 and Cyclin E1 were also downregulated (Figure S8C). The pro-inflammatory cytokines in cell-culture medium supernatants were detected by enzyme-linked immunosorbent assay (ELISA) (Figure 2C). si-S1A3-Chol significantly reduced the production of interleukin-6 (IL-6), IL-8, matrix metalloproteinase 1 (MMP-1), MMP-3, MMP-9, and MMP-13 (all with p < 0.05).
Figure 2.
Cholesterol-conjugated chemically modified siRNA reverses RA-FLSs dysfunction
(A) The inhibiting effect on SMO protein level after si-S1A3-Chol treatment. Representative western blot of SMO protein in total protein after RA-FLSs were treated with si-Scr-Chol or si-S1A3-Chol (800 nM) for 72 h. GAPDH was used as a loading control. Relative expression was calculated as the ratio of SMO/GAPDH and is presented as fold change versus si-Scr-Chol (n = 6). (B) The inhibiting effect on GLI1 protein level after si-S1A3-Chol treatment. Representative western blot of GLI1 protein in nuclear protein after RA-FLSs were treated with si-Scr-Chol or si-S1A3-Chol (800 nM) for 72 h. Histone H3 was used as a loading control. Relative expression was calculated as the ratio of GLI1/Histone H3 and is presented as fold change versus si-Scr-Chol (n = 6). (C) ELISA assay determined the concentrations of cytokines IL-6, IL-8, MMP-1, MMP-3, MMP-9, and MMP-13 in cell-culture medium supernatants after RA-FLSs were treated with si-Scr-Chol or si-S1A3-Chol (800 nM) for 72 h (n = 4). (D) The impact on cell viability was evaluated by CCK-8 assay after RA-FLSs were treated with si-Scr-Chol or si-S1A3-Chol (800 nM) for 0, 24, 48, and 72 h, and the absorbance was detected at indicated time points (n = 6). (E) The cell proliferation was determined by flow cytometry. The percentages of EdU-positive cells were detected after RA-FLSs were treated with si-Scr-Chol or si-S1A3-Chol (800 nM) for 48 h, shown in the bar graph (n = 6). (F) The cell-cycle phase distribution was detected by flow cytometry after RA-FLSs were treated with si-Scr-Chol or si-S1A3-Chol (800 nM) for 48 h, and the percentages of cells in the S and G2/M phases are shown in the bar graph (n = 6). (G) The migration capacity of RA-FLSs after si-S1A3-Chol treatment was examined by scratch wound healing assay. Representative images were captured after RA-FLSs were treated with si-Scr-Chol or si-S1A3-Chol (800 nM) for 24 h, and the number of migrated cells beyond the scratch line is shown in the bar graph (n = 6). Scale bars, 400 μm. (H) The migration ability was detected using chemotaxis assay for 12 h after RA-FLSs were treated with si-Scr-Chol or si-S1A3-Chol (800 nM) for 48 h. Representative images were captured, and the number of migrated cells is shown in the bar graph (n = 6). Scale bars, 200 μm. (I) The invasion ability was assessed using transwell chambers with Matrigel-coated membrane for 24 h after RA-FLSs were treated with si-Scr-Chol or si-S1A3-Chol (800 nM) for 48 h. Representative images were captured, and the number of invaded cells is shown in the bar graph (n = 6). Scale bars, 200 μm. Data are presented as mean ± SD. ns, p > 0.05; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001 versus si-Scr-Chol group by one-way ANOVA with Dunnett’s test for multiple comparisons.
Chemically modified siRNA inhibits RA-FLS proliferation
To determine whether si-S1A3-Chol inhibited RA-FLS proliferation, the cell counting kit-8 (CCK-8) assay and 5-ethynyl-2′-deoxyuridine (EdU) assay were performed. The cell viability was significantly decreased in RA-FLSs treated with si-S1A3-Chol compared with si-Scr-Chol treatment (0.66 ± 0.11 versus 0.91 ± 0.16, p < 0.01) (Figure 2D). In addition, even though RA-FLSs were treated with an exceptionally high concentration (3,200 nM), less than a 25% reduction of cell viability was achieved (Figure S9A). The EdU assay was conducted to elucidate the effects of si-S1A3-Chol on cell proliferation of RA-FLSs. The percentages of EdU-positive cells in RA-FLSs treated with si-S1A3-Chol were diminished under detection using flow cytometry (12.63% ± 4.04% versus 25.25% ± 2.84%, p < 0.0001) and confocal microscopy (13.05% ± 4.15% versus 37.96% ± 4.26%, p < 0.0001) (Figures 2E and S9B). Moreover, to eliminate the toxicity of siRNA, the apoptosis assay was performed, the results of which showed no significant difference in the cell apoptotic rate between groups (Figure S9C).
RNA sequencing (RNA-seq) was performed to further clarify the regulation mechanism of si-S1A3-Chol treatment. As the volcano plot and heatmap plot showed, 423 downregulated genes and 247 upregulated genes were found in RA-FLSs treated with si-S1A3-Chol (Figures S10A and S10B). Because of the interfering effect of siRNA, the downregulated genes might involve the key mechanism of si-S1A3-Chol. Gene Ontology (GO) enrichment analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis, and gene set enrichment analysis (GSEA) demonstrated downregulated genes enriched in the homologous chromosome segregation process, and regulation of the cell division process, cell-cycle pathway, and DNA replication pathway (Figures S10C–S10E).
The effect of si-S1A3-Chol on cell-cycle distribution was then evaluated by flow cytometry, which showed that the percentages of RA-FLSs in the S and G2/M phases were decreased compared with si-Scr-Chol treatment (10.66% ± 2.61% versus 18.67% ± 3.28%, p < 0.01%) (Figure 2F).
Off-target effect analysis
To determine the off-target effect of si-S1A3-Chol on RA-FLS treatment, RNA-seq was performed to recognize the expression of the transcripts. As microRNA-like off-target effects were sequence dependent, sequencing of si-S1A3-Chol was performed by nucleotide BLAST in NCBI databases to find the top 100 similar transcripts (listed in Table S4). Subsequently, the expression of these transcripts was evaluated within RNA-seq data. As shown in the volcano plot, among all similar transcripts, only four unintended transcripts (CCN4, UBE2G1, HELLS, and PLXNC1) were downregulated (Figure S10F).
Chemically modified siRNA inhibits RA-FLS migration and invasion
To examine whether si-S1A3-Chol regulates the migration of RA-FLS, scratch wound healing and chemotaxis assays were performed. The number of migrated cells treated with si-S1A3-Chol was fewer than those treated with si-Scr-Chol in the wound healing assay (97.83 ± 13.38 versus 140.33 ± 10.42, p < 0.0001) (Figure 2G) and the chemotaxis assay (105.56 ± 20.30 versus 159.25 ± 23.03, p < 0.01) (Figure 2H).
To determine whether si-S1A3-Chol modulates the invasiveness of RA-FLSs, transwell chambers with Matrigel coating were used. The number of invaded cells treated with si-S1A3-Chol was fewer than cells treated with si-Scr-Chol (72.93 ± 26.81 versus 121.79 ± 22.94, p < 0.01) (Figure 2I).
Intra-articular injection reduces the amount of siRNA in blood circulation
The concentration of sense and antisense strands was detected by quantitative real-time PCR (qPCR) to test whether intra-articular injection could reduce siRNA entering the blood circulation in comparison with intravenous injection (schematic diagram shown in Figure S11A). Compared with intravenous injection, si-S1A3-Chol injected intra-articularly displayed lower Cmax (the maximum measured concentrations), delayed tmax (the time at which Cmax was measured), and reduced AUC (area under the concentration-time profile from time zero to 168 h) (Figure S11B and Table S5).
Chemically modified siRNA ameliorates arthritis progression and maintains motor function in CIA mice
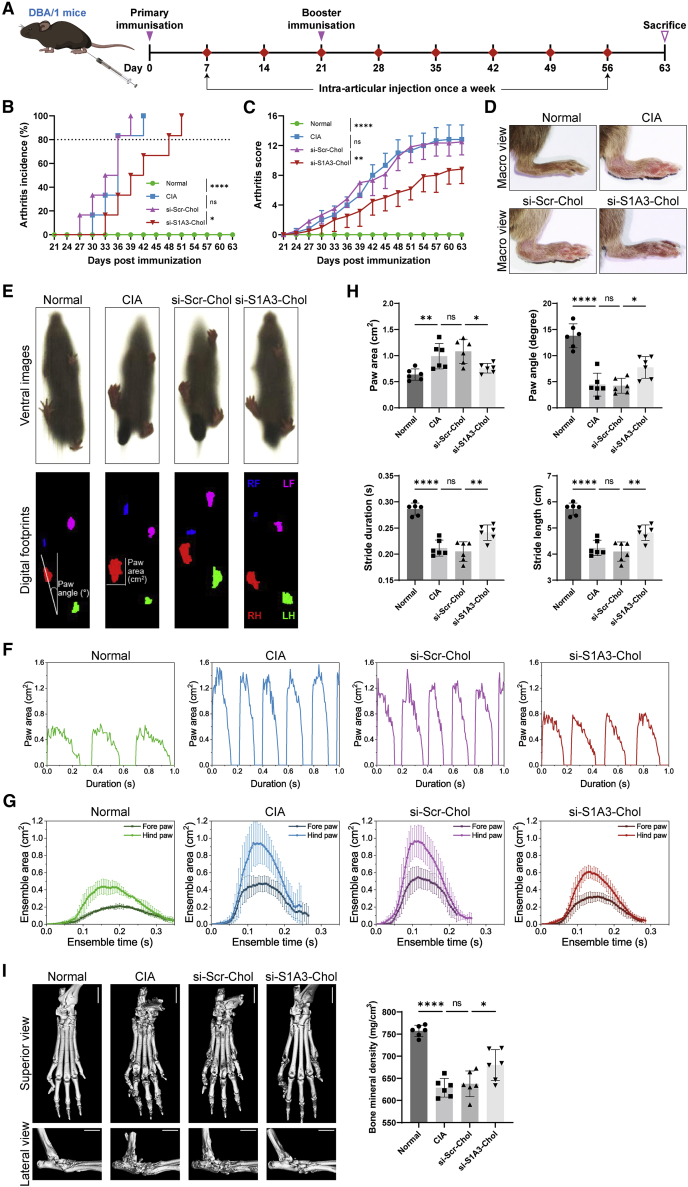
To evaluate the in vivo therapeutic effect of si-S1A3-Chol, CIA mice were established and administered with saline, si-Scr-Chol, or si-S1A3-Chol (250 nmol/kg body weight) (diagram shown in Figure 3A). More than 80% of mice in the CIA group and the si-Scr-Chol group developed arthritis within 36 days after primary immunization, but this was delayed 12 days in the si-S1A3-Chol group (Figure 3B). The results thus indicated that si-S1A3-Chol slowed down arthritis occurrence. Moreover, si-S1A3-Chol ameliorated arthritis progression and alleviated the severity of arthritis compared with the si-Scr-Chol group (8.83 ± 1.94 versus 12.50 ± 1.76, p < 0.01) (Figures 3C and 3D).
Figure 3.
Cholesterol-conjugated chemically modified siRNA ameliorates arthritis progression and maintains gait function in CIA mice
(A) Schematic diagram of siRNA treatment in CIA mice. DBA/1 mice were immunized on days zero and 21 and were injected with siRNA intra-articularly once a week from day 7 to day 56. Respectively, si-S1A3-Chol (250 nmol/kg body weight), si-Scr-Chol (250 nmol/kg body weight), or an equal volume of saline was injected into the bilateral tibiotalar joint. All mice were sacrificed, and specimens were harvested on day 63. (B) The arthritis score was conducted from day 21 to day 63, with an arthritis score greater than 4 indicating successful induction of the CIA model. The effect of si-S1A3-Chol on arthritis occurrence was determined by the percentages of arthritis incidence, and the si-Scr-Chol group served as the negative control group (n = 6). (C) The mean (SD) arthritis scores from day 21 to day 63 showed the effect of si-S1A3-Chol on arthritis progression in CIA mice (n = 6). (D) Representative macro view of right hind paws showed the effect of si-S1A3-Chol on alleviating arthritis severity in CIA mice. (E) The motor function of CIA mice was assessed by gait analysis. Representative ventral images were captured by a high-speed camera, and the digital footprints were generated from those images. (F) The effect of si-S1A3-Chol on CIA mice’s gait function was evaluated by gait parameters. The representative profiles show the dynamic change of the hind paw area with the stride duration in different treatment groups. (G) The mean (SD) ensemble area of the fore paw and the hind paw changed with the ensemble time, showing the effect of si-S1A3-Chol on reversing motor dysfunction in CIA mice (n = 6). (H) The effect of si-S1A3-Chol on paw area, paw angle, stride duration, and stride length of CIA mice were measured by gait analysis (n = 6). (I) Representative micro-CT superior view and lateral view display the protective effect of si-S1A3-Chol on bone erosion in the hind paws of CIA mice. The bone mineral density was calculated as the indicator of bone erosion and is shown in the bar graph (n = 6). Scale bars, 2 mm. Data are presented as mean ± SD. ns, p > 0.05; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001 versus between groups by one-way ANOVA with Dunnett’s test for multiple comparisons.
The ventral motor images of CIA mice were transformed into digital footprints (Figure 3E and Video S1). CIA mice in the si-S1A3-Chol treatment group showed a reduction in stride frequency (Figures 3F and S12B) and ensemble paw area (Figure 3G). On account of the swollen paws, the paw area of the CIA group was increased, whereby there was no difference between the si-Scr-Chol group and the CIA group. However, compared with the si-Scr-Chol group, CIA mice with si-S1A3-Chol treatment had reduced paw area (0.76 ± 0.09 cm2 versus 1.08 ± 0.23 cm2, p < 0.05) (Figure 3H). The paw angle of mice in the CIA group was lowered due to ankylosis of the limbs, and there was no amelioration between the si-Scr-Chol group and the CIA group. Nevertheless, si-S1A3-Chol improved the ankylosis of hind limbs with an increased paw angle (7.74° ± 2.08° versus 4.23° ± 1.42°, p < 0.05) (Figure 3H). Moreover, with increasing arthritis severity, the stride duration of the CIA group was decreased, and there was no improvement in the si-Scr-Chol group. Nonetheless, si-S1A3-Chol ameliorated the dynamic gait with an increased stride duration (0.24 ± 0.01 s versus 0.20 ± 0.02 s, p < 0.01) (Figure 3H). Because of the fixed walking speed, stride length was also improved (Figure 3H). A full stride comprised braking, propulsion, stance, and swing phases (Figure S12A). Consistent with the increasing stride duration in the si-S1A3-Chol group, the swing duration was also increased in contrast to braking duration, propulsion duration, and stance duration (Figure S12B).
The motor function of CIA mice was assessed by gait analysis. Representative ventral images were captured, and the digital footprints were generated. The paw area and stride frequency were increased in CIA mice. si-S1A3-Chol reduced the paw area and stride frequency, showing the effect on reversing gait dysfunction.
A micro-computed tomography (micro-CT) scan was employed to investigate the effect of si-S1A3-Chol in inhibiting bone erosion in CIA mouse models. The bone structures of ankles in the CIA group and si-Scr-Chol group displayed rough bone surfaces and serious bone erosion (Figure 3I). However, the si-S1A3-Chol group showed improved bone structure and bone erosion, with an elevated bone mineral density compared with the si-Scr-Chol control group (680.00 ± 34.85 mg/cm3 versus 637.58 ± 29.08 mg/cm3, p < 0.05).
si-S1A3-Chol alleviates the inflammation level and pathological damage in CIA mice
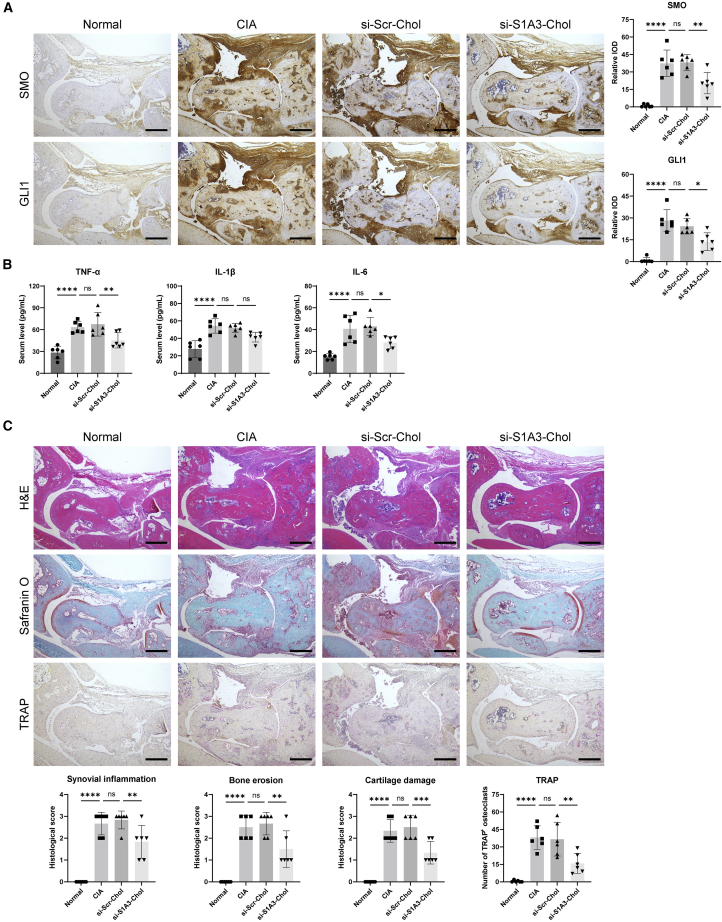
As the immunohistochemical results show, SMO and GLI1 were highly expressed in the ankle joint sections, indicating activation of the Hedgehog signaling pathway (Figure 4A). si-S1A3-Chol treatment markedly reduced the relative expression levels of SMO (20.45 ± 9.24 versus 38.07 ± 6.84, p < 0.01) and GLI1 (13.68 ± 6.13 versus 24.26 ± 5.24, p < 0.05) compared with the si-Scr-Chol group.
Figure 4.
Cholesterol-conjugated chemically modified siRNA alleviates the inflammation level and pathological damage in CIA mice
(A) Representative immunohistochemical images of SMO and GLI1 protein in hind paws of CIA mice show the effect of si-S1A3-Chol on Hedgehog signaling pathway activity. The relative IOD is presented as fold change versus the normal group (n = 6). Scale bars, 400 μm. (B) ELISA assay determined the serum level of TNF-α, IL-1β, and IL-6 in CIA mice treated with si-Scr-Chol or si-S1A3-Chol (n = 6). (C) Representative H&E staining, Safranin O staining, and TRAP staining in CIA mice show pathological damage prevention of si-S1A3-Chol. Histopathologic scores of synovial inflammation, bone erosion, and cartilage damage were scored from 0 to 3, and the number of TRAP-stained positive osteoclasts was counted manually (n = 6). Scale bars, 400 μm. Data are presented as mean ± SD. ns, p > 0.05; ∗p < 0.05, ∗∗p < 0.01 versus between groups by one-way ANOVA with Dunnett’s test for multiple comparisons.
The ELISA assay was used to determine the serum levels of pro-inflammatory cytokines after si-S1A3-Chol treatment (Figure 4B). The results indicated that the levels of cytokines increased with arthritis in CIA mice. si-S1A3-Chol treatment suppressed the production of cytokines including tumor necrosis factor α (TNF-α) (45.22 ± 10.49 pg/mL versus 67.36 ± 16.22 pg/mL, p < 0.01), IL-1β (41.47 ± 5.57 pg/mL versus 51.92 ± 5.18 pg/mL, p = 0.07), and IL-6 (28.16 ± 5.41 pg/mL versus 42.94 ± 8.11 pg/mL, p < 0.05) compared with the si-Scr-Chol group.
Paw sections were stained with hematoxylin & eosin (H&E) and safranin O to assess the therapeutic effect on pathological damage in CIA mice. Compared with the si-Scr-Chol treatment, mice in the si-S1A3-Chol group showed a marked reduction in pathological scores in synovial inflammation (1.83 ± 0.75 versus 2.83 ± 0.41, p < 0.05), bone erosion (1.50 ± 0.84 versus 2.67 ± 0.52, p < 0.05), and cartilage damage (1.33 ± 0.52 versus 2.50 ± 0.55, p < 0.01) (Figure 4C). The number of mature osteoclasts was also reduced (15.83 ± 8.54 versus 36.50 ± 14.29, p < 0.05).
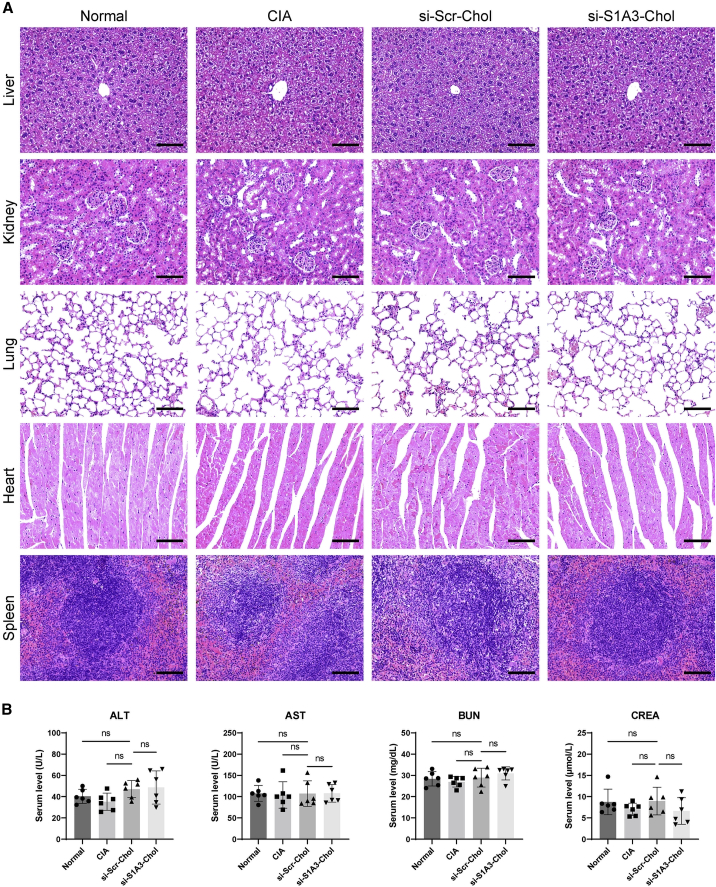
In vivo toxicity of si-S1A3-Chol in CIA mice
Histopathology and enzymology of major organs were performed to evaluate whether siRNA treatment by intra-articular injection could cause toxicity in vivo. None of the CIA mice treated with si-Scr-Chol and si-S1A3-Chol appeared to have any clinical discomfort until sacrifice during the whole experimental period. The major organs, including the liver, kidney, lung, heart, and spleen, of all groups were harvested at sacrifice and showed a normal appearance. Additionally, histopathological examinations indicated that all organ sections maintained typical physiological structure and cell morphology (Figure 5A). Furthermore, the serum levels of alanine aminotransferase (ALT), aspartate aminotransferase (AST), blood urea nitrogen (BUN), and creatinine (CREA) were detected at sacrifice and used as liver and kidney function indicators. All indicators were within the normal ranges, and there was no significant difference among CIA mice in the different treatment groups (Figure 5B).
Figure 5.
In vivo toxicity of cholesterol-conjugated chemically modified siRNA in CIA mice
(A) Images show H&E-stained sections of CIA mice’s organs (liver, kidney, lung, heart, and spleen) in different treatment groups harvested on day 63. Scale bars, 100 μm. (B) The serum levels of ALT, AST, BUN, and CREA were used as indicators of liver function and kidney function in CIA mice (n = 6). Data are presented as mean ± SD. ns, p > 0.05 versus the si-Scr-Chol group by one-way ANOVA with Dunnett’s test for multiple comparisons.
Discussion
Our study demonstrated that si-S1A3-Chol, a chemically modified siRNA, reversed RA-FLS dysfunction and ameliorated arthritis progression by targeting the Hedgehog signaling pathway.
While the development of DMARDs has improved disease prognosis, there has been no cure for RA.4 Moreover, long-term use of DMARDs has been limited by their adverse effects, including hepatotoxicity and bone marrow suppression for conventional synthetic DMARDs (methotrexate and leflunomide), and increased risk of severe infections for biologic DMARDs (infliximab and tocilizumab).3 Therefore, it is necessary to discover new targets and apply novel technologies to treat RA more efficaciously and safely.
Recently, we reported on the upregulation of Hedgehog signaling in RA synovium and RA-FLSs.23,24 The Hedgehog signaling pathway was associated with the aggressive phenotype of RA-FLSs, such as excessive proliferation and invasiveness.25,26 Suppressing the Hedgehog signaling pathway may be a potential alternative for RA treatment.27 However, there are several limitations to the FDA-approved small-molecule antagonists targeting SMO in clinical practice.28 On the one hand, because of the lack of selective target ability of small-molecule antagonists, adverse effects such as alopecia, muscle spasms, and gastrointestinal distress are common in patients with orally administered SMO antagonists.29 As a result of the cumulative toxicity, antagonists are inappropriate for the long-term treatment of chronic inflammatory diseases such as RA. On the other hand, SMO mutations are common and confer drug resistance to inhibitors.30 Owing to the complexity of mutant structural changes, further optimization of efficient antagonists is challenging and time consuming.31,32 siRNA can mediate RNA-induced silencing complex to specifically cleave target mRNA before protein translation.19 Anti-SMO siRNA has the potential to overcome the drawbacks of small-molecule antagonists due to its high specificity and ease of customization.
This study used FLSs isolated from 12 patients with active RA as in vitro cell experimental models to simulate an actual clinical condition. The silencing efficiency screening showed that all anti-SMO siRNAs, precisely designed to target various sites, efficiently interfered with RA-FLS SMO mRNA expression (Figure 1A). However, despite the high silencing efficiency in vitro, naked siRNA was promptly degraded in vivo because of the abundant nuclease in body fluids.33 Procedures for improving the stability and specificity of siRNA were essential for further clinical applications.
To enhance the performances of si-h/m-2, we applied various extents of chemical modifications to its sense and antisense strands (Figure S3). A large number of siRNA duplex candidates are readily available by pair annealing. Our findings indicated that the stability of siRNA increased in direct proportion to the degree of chemical modification (Figure S4). Previous research has demonstrated that the chemical modification of nucleotides effectively enhances their resistance to degradation by endogenous nucleases.34 The present study demonstrated that the stability of siRNA could be tailored by varying degrees of chemical modifications (Figure S4).
Additionally, the influence of chemical modifications on siRNA interfering efficiency was investigated. Although siRNA silencing efficiency was somewhat affected by the modification of the sense strand, the increasing extent of antisense strand modifications overcame this obstacle (Figure 1E). Our results indicated that with the increasing level of antisense strand chemical modification, higher inhibition rate and lower IC50 were achieved (Figure 1F). si-S1A3 was conjugated with cholesterol to improve cellular uptake efficiency.35 The silencing efficiency of si-S1A3-Chol (800 nM) was better than that of the naked siRNA, which needs transfection reagent assistance to enter targeted cells (Figure 1H). These results were consistent with the previous study indicating that modestly modified siRNAs displayed highly efficient silencing activity in comparison with naked siRNAs.36 Thus, our study suggested that optimized chemical modification could enhance the stability and efficiency of siRNAs.
Furthermore, optimized chemical modification successfully mitigates the off-target effects of si-S1A3-Chol. It is well known that naked siRNA can trigger the off-target silencing of unintended mRNA.37 With precise design and optimized modification, the silencing efficiency was enhanced and the specificity was improved. These improvements were beneficial in minimizing the occurrence of off-target effects.38 As the RNA-seq results demonstrated, the off-target rate of si-S1A3-Chol was less than that of naked siRNA, which usually ranges from 5% to 80%.39 Hence, chemical modification may be a reliable technique to minimize off-target effects and to facilitate siRNA drug development.
Systemically delivering siRNA to target tissues beyond the liver or other filtering organs was challenging.40,41 In the current study, the pharmacokinetic analysis demonstrated that intra-articular injection effectively reduced si-S1A3-Chol entering the blood circulation (Figure S11). Considering the relative enveloped property of the articular capsule and the approachability of FLSs, this indicates that siRNA could accumulate in intended joints and interfere with a relatively high concentration.42,43 Our results showed that the silencing efficiency of chemically modified siRNAs was concentration dependent (Figure 1), which would be conducive to improving the efficacy of synovial tissue treatment and reducing the toxicity of unintended organs. Moreover, si-S1A3-Chol stably retained integrity in synovial fluid for a long time while it was readily degraded in serum (Figure S7). It was also beneficial in reducing the side effects on the unintended tissues even if si-S1A3-Chol entered the blood system when it was locally injected into an articular cavity. Therefore, intra-articular injection was suitable for si-S1A3-Chol administration for RA treatment.
The therapeutic effect of si-S1A3-Chol was investigated in CIA mice, considered the classic animal model for RA.44 Our findings revealed that si-S1A3-Chol treatment delayed arthritis onset, reduced arthritic severity, alleviated synovial inflammation, ameliorated cartilage damage, and prevented bone erosion (Figures 3 and 4). These results were consistent with a previous study in which intraperitoneally injected cyclopamine, a Hedgehog pathway antagonist, effectively reduced synovitis and cartilage destruction in rats with adjuvant-induced arthritis.27 Moreover, the efficacy of si-S1A3-Chol on CIA mouse motor function was objectively assessed by gait analysis. In previous studies, common assessment methods were usually subjective and semi-quantitative.45 Here, gait analysis results demonstrated that si-S1A3-Chol significantly reduced paw area, elevated paw angle, increased stride duration, and increased stride length (Figure 3H). This indicates that si-S1A3-Chol treatment improved paw swelling, limb ankylosis, and motor function of CIA mice. Most research has evaluated the symptom, radiology, and histopathology of the CIA mouse model. Few studies have assessed functional impairments during arthritis progression.46 However, in clinical practice, whether therapies improve joint function or not is quite significant for the prognosis of RA patients.47,48 Our results indicated that si-S1A3-Chol treatment alleviated functional impairment in CIA mice. Besides, si-S1A3-Chol treatment was relatively safe in CIA mice because no histopathological toxicity was observed in vivo (Figure 5).
The therapeutic effect of si-S1A3-Chol on CIA mice might be attributed to suppression of the Hedgehog signaling pathway. Although Hedgehog signaling is modulated finely on the basis of the negative feedback regulators (PTCH1 and PTCH2) and the positive feedback regulator (GLI1), the abnormal regulation of pathway feedback contributes to carcinogenesis.49 Because the target genes were involved in cell proliferation, survival, and metastasis, aberrant activation of the Hedgehog signaling pathway led to RA-FLSs’ tumor-like behavior.50,51 Here, si-S1A3-Chol reversed RA-FLS dysfunction by inhibiting cell proliferation and invasiveness (Figure 2). RNA-seq and enrichment analysis demonstrated that downregulated genes were involved in the DNA replication pathway and cell division process. Combined with the molecular and phenotypic change, the results confirmed the hypothesis of si-S1A3-Chol. Specifically, they indicated that si-S1A3-Chol inhibited cell proliferation of RA-FLSs through inhibiting the activity of the Hedgehog signaling pathway, interfering with the expression of the cyclin family, and arresting the transit of cells from G1 to S phase. All these findings strongly support the assumption that targeting SMO effectively suppressed the tumor-like character of RA-FLSs. Previous studies suggested that RA-FLSs mediate direct tissue damage and joint destruction.52 Our results indicated that si-S1A3-Chol ameliorated arthritis progression by suppressing the hyperactivation of the Hedgehog signaling pathway in FLSs.
Nonetheless, several limitations need to be addressed with regard to this study. First, while chemical modifications significantly enhanced siRNA stability, it is unclear how they alter the interference effect. Illuminating the mechanisms by which chemical modification patterns contribute to siRNA efficiency will aid in improving siRNA treatment efficacy and dose efficiency. Second, although anti-SMO siRNA ameliorated the progression of arthritis in CIA mice, larger animal models, such as non-human primates, are required to validate its efficacy. Third, despite the fact that intra-articular injection of si-S1A3-Chol increases the concentration in the targeted joint and eliminates the effects on other organs, arthritis frequently presents in more than one joint in clinical practice. The development of siRNA drug-delivery carriers for inflamed joints will have significant clinical utility in the treatment of RA.
In summary, we designed and modified anti-SMO siRNAs to improve their effectiveness and stability. The results demonstrated that si-S1A3-Chol reversed the tumor-like character of RA-FLSs in vitro and alleviated the progression of arthritis in CIA mice in vivo. These findings indicate that chemically modified siRNA injected intra-articularly could be a promising therapy for RA patients.
Materials and methods
Ethics and samples collection
The study was approved by the Medical Ethics Committee of the Sixth Affiliated Hospital of Sun Yat-sen University. All patients provided written informed consent to participate in this study under the Declaration of Helsinki. Animal experimental procedures were performed with the approval of the Institutional Animal Care and Use Committee of Guangzhou Institutes of Biomedicine and Health and conducted following the Guide for the Care and Use of Laboratory Animals of the National Research Council.
Fifteen patients with active RA, including 4 males and 11 females (from 55 to 78 years of age), were recruited. All patients were classified according to the 2010 American College of Rheumatology/European League Against Rheumatism classification criteria53 and exhibited moderate to severe disease activity (Disease Activity Score of 28 joint counts >3.2). Six traumatic patients (three males and three females, from 18 to 46 years of age) without arthritis were recruited as healthy controls. Synovial tissue samples were obtained from 12 RA patients undergoing knee arthroscopy or synovectomy and six traumatic patients undergoing knee arthroscopy or surgery. Synovial fluid samples were collected by needle aspiration from the knees of three RA patients. Synovial fluids were centrifuged immediately, and the supernatant was stored at −80°C until use.
Cell isolation, culture, and characterization
Synovial tissue specimens were minced into small pieces, transferred into a tissue flask, and cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, C11995500BT) supplemented with 10% fetal bovine serum (FBS; Gibco, 10099141C) at 37°C in a humidified atmosphere containing 5% CO2. RA-FLSs migrated out of the tissue explant and formed an adherent cell monolayer. After growing to approximately 90% confluence, cells were trypsinized and reseeded for cell subculture. RA-FLSs between passages 3 and 5 were used for the in vitro experiments. The morphological characteristics of RA-FLSs were confirmed under the optical microscope. The purity analysis of surface markers was detected by flow cytometry. RA-FLSs from passages 3 were stained with antibodies including anti-CD55 (BioLegend, 311311), anti-CD90 (BioLegend, 328107), anti-CD14 (BioLegend, 325603), and anti-CD68 (BioLegend, 333805). Stained cells were detected using a BD LSRFortessa Cell Analyzer (BD Biosciences). The data were analyzed using FlowJo software (BD Biosciences). The murine FLSs (Procell, CP-M083) were cultured in murine synoviocytes’ complete culture medium (Procell, CM-M083). To ensure mycoplasma-free conditions throughout this study, mycoplasma detection was performed routinely.
Design and transfection of siRNA
Eight siRNAs targeted Homo sapiens SMO mRNA (NCBI: NM_005631.5), and four siRNAs targeted the homologous region between Homo sapiens SMO mRNA and Mus musculus Smo mRNA (NCBI: NM_176996.4) were designed according to design parameters.22,54,55 The naked siRNAs were designed to target different sites of coding sequences in mRNA, and an siRNA with no homology to any genes was designed as si-NC. The sequences of 13 naked siRNAs are listed in Table S1. Chemical modifications were introduced to different sites of siRNA sequences according to modification principles to improve the performance of siRNA.36,56 The sequences of 16 chemically modified siRNAs are listed in Table S2. Moreover, to avoid the influence of transfection reagent, two optimized chemically modified siRNAs were chosen to conjugate with cholesterol at the 3′ end of the sense strand (si-S1A3-Chol and si-S1A4-Chol). Besides, a scrambled control siRNA conjugated with cholesterol was designed (si-Scr-Chol). The sequences of three cholesterol-conjugated chemically modified siRNAs are listed in Table S3. All siRNAs were synthesized and purified by Ribobio (China).
The siRNAs were dissolved in RNase-free water (Takara Bio, 9012) and diluted to the final concentration of 50 nM unless otherwise noted. RA-FLSs were seeded onto 6-well plates at a density of 1 × 105 cells per well. When the confluence was around 50%–60%, RA-FLSs were transiently transfected with naked siRNAs or chemically modified siRNAs using Lipofectamine 3000 transfection reagent (Invitrogen, L3000015). Blank control group (treated without transfection reagent), mock control group (treated with transfection reagent in the absence of siRNA), and negative control group (treated with transfection reagent containing si-NC) served as control groups. Furthermore, cholesterol-conjugated chemically modified siRNAs were treated to RA-FLSs without transfection reagent.
Total RNA extraction and qPCR analysis
The expression of SMO mRNA was determined by qPCR to compare the silencing efficiency of each siRNA. RA-FLSs were seeded at a density of 5 × 104 cells per well in 12-well plates for 24 h and then transfected with si-NC or anti-SMO siRNAs using the transfection reagent for 48 h. To examine the silencing efficiency of cholesterol-modified siRNA without transfection reagent, RA-FLSs were treated with cholesterol-conjugated chemically modified siRNAs for 48 h. Total RNAs were extracted from RA-FLSs or murine FLSs using TRIzol reagent (Invitrogen, 15596018) followed by the addition of chloroform, and precipitated with isopropanol in the supernatant after centrifugation. cDNA was synthesized from equal amounts of total RNA using iScript cDNA Synthesis Kit (Bio-Rad, 1708891) on an S1000 Thermal Cycler (Bio-Rad). qPCR was performed using iQ SYBR Green Supermix (Bio-Rad, 1708882AP) on a CFX Connect Real-Time System (Bio-Rad). The sequences of the primers used for qPCR amplification are listed in Table S6. Targeted mRNA expression was normalized toward the internal control gene (glyceraldehyde-3-phosphate dehydrogenase, GAPDH for humans, Gapdh for mice). The comparative delta cycle threshold (Ct) method was used to calculate the relative expression level against the negative control sample. Dose-response curves for siRNAs at gradient concentrations were fitted using Prism9 software (GraphPad). The IC50 of each siRNA was calculated using a log(inhibitor) versus response − variable slope (four parameters) equation.
Immunoblotting
The protein levels were evaluated by western blot to verify the effects of anti-SMO siRNAs. RA-FLSs were treated with siRNAs for 72 h. In brief, for the detection of protein levels, the total protein was extracted using RIPA Lysis Buffer (Beyotime, P0013B) supplemented with a cocktail of protease inhibitor (Solarbio, P6730), phosphatase inhibitor (Solarbio, P1260), and phenylmethylsulfonyl fluoride (Solarbio, P0100). The nuclear protein was isolated using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Scientific, 78833) to detect GLI1 expression. Protein lysate was loaded and separated on SDS-PAGE using 4%–20% SurePAGE gels (Genscript, M00657). The proteins were blotted onto a 0.22-μm pore size polyvinylidene fluoride (PVDF) membrane (Millipore, ISEQ00010) for the detection of Histone H3, or transferred onto a 0.45-μm pore size PVDF membrane (Millipore, IPVH00010) for the detection of other proteins. After blocking with 5% skim milk for 2 h, the PVDF membranes were incubated in primary antibodies at 4°C overnight. Primary antibodies included anti-SMO (1:100; Santa Cruz Biotechnology, sc-166685), anti-GAPDH (1:2,000; Cell Signaling Technology, 5174), anti-GLI1 (1:1,000; Cell Signaling, 3538), Anti-Histone H3 (1:2,000; Abcam, ab1791), anti-PTCH1 (1:1,000; Cell Signaling, 2468), anti-GLI2 (1:100; Santa Cruz, sc-271786), anti-PTCH2 (1:1,000; Cell Signaling, 2470), anti-SUFU (1:1,000; Cell Signaling, 2520), anti-SHH (1:1,000; Cell Signaling, 2207), anti-Cyclin E1 (1:1,000; Cell Signaling, 4129), anti-Cyclin D1 (1:1,000; Cell Signaling, 2978), anti-p27 (1:1,000; Cell Signaling, 3686), and anti-p21 (1:1,000; Cell Signaling, 2947). Subsequently, membranes were incubated in horseradish peroxidase (HRP)-conjugated secondary antibodies, including anti-rabbit IgG (1:5,000; Cell Signaling, 7074) or anti-mouse IgG (1:5,000; Cell Signaling, 7076) for 1 h. Immobilon Western Chemiluminescent HRP Substrate (Millipore, WBKLS0500) was added to illuminate the bands. The signals of protein bands were detected using a Sapphire biomolecular imager (Azure Biosystems). The gray values of bands were quantified using ImageJ software (National Institutes of Health [NIH]). The protein expression was normalized to GAPDH or Histone H3, which served as an internal reference. Data were presented as the expression relative to that of the negative control group.
siRNA stability measurement
The stabilities of siRNA duplex against human serum (Sigma-Aldrich, H4522) or RA patients’ synovial fluids were investigated using gel electrophoresis. In brief, naked siRNAs or chemically modified siRNAs were incubated with 50% human serum or synovial fluids at 37°C in water baths for indicated time points. Samples were loaded on 2.5% agarose gel containing SYBR Gold Nucleic Acid Gel Stain (Invitrogen, S11494), followed by gel electrophoresis at 130 V for 15 min. The siRNA bands on the gels were visualized under UV illumination using a Tanon 1600 Gel Imaging System (Tanon). The integrity of siRNA was quantitated using ImageJ software (NIH).
ELISA
RA-FLSs were treated with si-Scr-Chol or si-S1A3-Chol for 72 h, and the levels of cytokines, such as interleukins and MMPs, in cell-culture supernatants were measured by ELISA assays. ELISA kits used were Human IL-6 Valukine ELISA Kit (Novus, VAL102), Human IL-8 Valukine ELISA Kit (Novus, VAL103), Human MMP-1 ELISA (Raybiotech, ELH-MMP1-1), Human MMP-3 ELISA (Raybiotech, ELH-MMP3-1), Human MMP-9 Valukine ELISA Kit (Novus, VAL113), and Human MMP-13 ELISA (Raybiotech, ELH-MMP13-1).
Cell viability assay
RA-FLSs were seeded at a density of 2.5 × 103 cells per well in 96-well plates for 24 h and treated with si-Scr-Chol or si-S1A3-Chol at 800 nM or gradient concentration from 50 to 3,200 nM for indicated times. After 0, 24, 48, and 72 h, cell viability was assessed using CCK-8 (Bimake, B34304). In brief, 10 μL of CCK-8 solution was added to each well and incubated in darkness for 2 h. The absorbance value at 450 nm wavelength was measured using a Varioskan LUX multimode microplate reader (Thermo Scientific).
EdU assay
EdU is a nucleoside analog of thymidine, which can be incorporated into the DNA duplex during DNA synthesis. RA-FLSs were seeded at a density of 1 × 104 cells per well in μ-Slide 8 Well (Ibidi, 80826) and treated with si-Scr-Chol or si-S1A3-Chol for 48 h. Cell proliferation assay was performed using the Cell-Light EdU Apollo567 In Vitro Kit (Ribobio, C10310-1). After treatment, RA-FLSs were incubated with 50 μM EdU at 37°C for 8 h. After being fixed with 4% paraformaldehyde for 30 min, RA-FLSs were treated with 0.5% Triton X-100 for 10 min. Thereafter, RA-FLSs were exposed to 100 μL of stain reaction cocktail for 30 min and incubated with Hoechst 33342 (Beyotime, C1028) to stain nuclei. Images were captured using a confocal microscope (TCS SP8 STED; Leica Microsystems).
Cell proliferation assay
RA-FLSs were seeded at a density of 1 × 105 cells per well in 6-well plates for 24 h and treated with si-Scr-Chol or si-S1A3-Chol for 48 h. According to the manufacturer’s instructions, the percentages of proliferating cells were assessed using the Cell-Light EdU Apollo567 In Vitro Flow Cytometry Kit (Ribobio, C10338-1). The ratio of proliferating cells was measured using a BD LSRFortessa Cell Analyzer (BD Biosciences). The data were analyzed using FlowJo software (BD Biosciences).
Cell-cycle analysis
RA-FLSs were seeded at a density of 1 × 105 cells per well in 6-well plates for 24 h. To synchronize the cell cycle, cells were serum starved for 24 h before treatment. After being treated with si-Scr-Chol or si-S1A3-Chol for 48 h, cells were harvested and fixed in 70% ethanol at −20°C overnight. Cells were subsequently stained using PI/RNase staining buffer (BD Biosciences, 550825). The cell-cycle phase distribution of RA-FLSs was detected using a BD LSRFortessa Cell Analyzer (BD Biosciences). The data were evaluated using ModFit LT software (Verity Software House).
Apoptosis detection
RA-FLSs were seeded at a density of 1 × 105 cells per well into 6-well plates and treated with si-Scr-Chol or si-S1A3-Chol for 48 h to detect cell apoptosis. According to the manufacturer’s instructions, cell apoptosis detection was performed using the FITC-Annexin V Apoptosis Detection Kit I (BD Biosciences, 556547). In brief, RA-FLSs were collected, resuspended, and incubated with FITC-Annexin V in darkness at room temperature for 15 min. Subsequently, stained cells were measured by a BD LSRFortessa Cell Analyzer (BD Biosciences) within 1 h. Annexin V-positive staining was regarded as apoptosis, and the percentages of apoptotic cells were calculated using FlowJo software.
Scratch wound healing assay
RA-FLSs were cultured to confluence in 24-well plates and scratched with micropipette tips. The cells were then incubated with DMEM containing 1% FBS in the presence of si-Scr-Chol or si-S1A3-Chol. After 24 h of incubation, cells were fixed and stained with crystal violet staining solution (Beyotime, C0121). The number of RA-FLSs that migrated beyond the scratching line was counted.
Cell migration and invasion assay
The migration ability of RA-FLSs was measured using Transwell Permeable Supports with an 8.0-μm pore size membrane (Corning, 3422). In brief, RA-FLSs were treated with si-Scr-Chol or si-S1A3-Chol for 48 h, then resuspended with the serum-free medium at a final density of 1.5 × 105 cells/mL and 100 μL loaded in the upper chambers. DMEM containing 10% FBS (600 μL) was placed as a chemoattractant in the lower wells. After 12 h of incubation, the non-migrating cells were wiped away with a cotton swab. The cells that passed through the membrane were fixed and stained with crystal violet staining solution. The migration ability of RA-FLSs was quantified by calculating the mean number of migrated cells in at least five random fields. A Matrigel-coated membrane was used in the invasion assay. The Matrigel Basement Membrane Matrix (Corning, 356234) was diluted with serum-free DMEM and then coated onto the upper side of the chambers. RA-FLSs were resuspended at a 3 × 105 cells/mL density and 100 μL loaded in the upper chambers. After 24 h of incubation, the subsequent experiments were similar to the chemotaxis assay.
RNA-seq and data analysis
RA-FLSs were treated with si-S1A3-Chol at 800 nM for 48 h. Total RNAs were isolated from RA-FLSs using TRIzol reagent according to the manufacturer’s protocol. The library preparation, cluster generation, and sequencing were performed by Novogene Bioinformatic Technology. In brief, the cDNA libraries were generated using the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (NEB, E7760L) following the manufacturer’s instructions. Subsequently, clustering of samples was performed on a cBot Cluster Generation System using TreSeq PE Cluster Kit v3-cBot-HS (Illumina, PE-401-3001). After cluster generation, the library preparations were sequenced by synthesis on an Illumina Novaseq platform and 150-bp paired-end reads were generated. After quality control and reads mapping to the hg38 reference genome, gene expression levels were quantified by the FPKM method. Differential expression analysis was assessed via the DESeq2 R package. Genes with an adjusted p value of less than 0.05 and absolute fold change of 2 were considered as differentially expressed. GO enrichment analysis and KEGG enrichment analysis of differentially expressed genes was assessed by the clusterProfiler R package. GSEA was conducted using version 4.2.3 with GO data and KEGG data. The sequence of si-S1A3-Chol was performed by nucleotide BLAST in NCBI databases to find the top 100 similar transcripts, and the off-target effects were evaluated within RNA-seq data.
siRNA pharmacokinetic studies
Sprague-Dawley rats were purchased from Beijing Vital River Laboratory Animal Technology. All rats were group-housed in a specific pathogen-free facility under a 12:12-h light/dark cycle at 22°C–24°C. All rats were males aged 8–10 weeks, weighing 401 ± 17 g. Six rats were randomly assigned to two groups to evaluate the pharmacokinetic (PK) feature of anti-SMO siRNAs: the intravenous and intra-articular injection groups. Each group contained three rats. The rats were administered a single dose of si-S1A3-Chol intravenously or intra-articularly at a 25 nmol/kg dose of body weight. The blood samples were harvested using the retro-orbital eye bleed procedure at 0, 0.08, 0.25, 0.5, 1, 2, 4, 8, 24, 48, 96, and 168 h after administration. Heparin served as an anticoagulant, and blood samples were centrifuged to harvest plasma. The concentrations of the sense and antisense strand of si-S1A3-Chol in plasma were quantified by stem-loop qPCR as described previously.57 In brief, plasma samples were heated in 0.25% Triton X-100 for 10 min. The siRNA-containing supernatants then underwent stem-loop reversed transcription followed by qPCR using the Bulge-Loop miRNA qRT-PCR Starter Kit (Ribobio, C10211). The sequences of the primers are listed in Table S6. The siRNA concentrations were determined using a standard curve. Mean siRNA plasma concentration-time profiles were used to estimate PK parameters, including Cmax, tmax, and AUC.
Induction and assessment of CIA mouse models
DBA/1 mice were purchased from Shanghai Slaccas Experimental Animal Company. All mice were group-housed in a specific pathogen-free facility under a 12:12-h light/dark cycle at 22°C–24°C. All mice were males aged 7–8 weeks, weighing between 20 and 22 g. Twenty-four mice were randomly assigned to four experimental groups to evaluate the therapeutic efficacy of anti-SMO siRNA: the normal control group, CIA control group, si-Scr-Chol-treated group, and si-S1A3-Chol-treated group. Each group contained six mice. For the establishment of arthritis models in the last three groups, DBA/1 mice were injected intradermally at the base of the tail with 100 μg of bovine type II collagen (Chondrex, 20022) emulsified in 50 μL of complete Freund’s adjuvant (Chondrex, 7001) as primary immunization on day zero. On day 21, mice received a booster immunization of bovine type II collagen emulsified in incomplete Freund’s adjuvant (Chondrex, 7002). Starting on day 7, the CIA mice were respectively administered an equal volume of either saline (vehicle), si-Scr-Chol (250 nmol/kg body weight), or si-S1A3-Chol (250 nmol/kg body weight) by intra-articular injection into the bilateral tibiotalar joint once a week for 8 weeks. For assessment of arthritic severity, clinical signs were assessed every 3 days and scored as follows: 0, normal joint; 1, mild swelling restricted to the ankle or tarsals; 2, mild swelling involving the ankle and tarsals; 3, moderate swelling spread from the ankle to metatarsals; and 4, severe swelling encompassing the ankle, metatarsals, and digits, or ankylosis in the limb.45 The arthritis scores of individual limbs were summed up, giving a maximum score for each mouse of 16. An arthritis score greater than 4 indicated the CIA model was induced successfully. On day 63, all mice were sacrificed and specimens harvested.
Gait analysis
Gait dynamics analysis was performed using a DigiGait imaging system (Mouse Specifics). In brief, mice ran on a motor-driven transparent treadmill belt, whose speed was adapted to 20 cm/s. A high-speed video camera captured the ventral images at 200 frames per second. The digital paw prints and dynamic gait signals were generated automatically. Temporal and spatial gait parameters of each paw movement were measured using DigiGait Analysis software (Mouse Specifics) according to stride phases.
Micro-CT imaging
The hind paws of CIA mice were used to determine the degree of bone erosion. The claws were scanned using a Latheta LCT-200 Micro-CT Scanner (Hitachi) with a voxel size of 24 μm at 50 kV. Subsequently, the three-dimensional images were reconstructed from the dataset using VGStudio MAX software (Volume Graphics). Bone mineral density of hind paws was measured as the comparative indicator of bone destruction using Latheta software (Hitachi).
Immunohistochemistry
On day 63, hind paws were dissected from the CIA mice, fixed in 4% paraformaldehyde for 24 h, and decalcified in 14% EDTA decalcification solution (Servicebio, G1105) for at least 28 days. Decalcified paws were embedded in paraffin blocks and serially sectioned at 4-μm thickness. Paraffin sections were subjected to antigen retrieval using citrate antigen retrieval solution (Servicebio, G1202). The slides were treated with 3% hydrogen peroxide to quench endogenous peroxidase activity, followed by blocking with 5% BSA. The sections were incubated in primary antibodies, including anti-SMO (1:40; Santa Cruz, sc-166685) and anti-GLI1 (1:200; Cell Signaling, 2643) in a moist chamber at 4°C overnight and then incubated in anti-mouse IgG HRP-conjugated secondary antibody (Servicebio, G1214) for 1 h. HRP staining was developed using the DAB (3,3′-diaminobenzidine) Chromogenic Kit (Servicebio, G1212), followed by counterstaining with hematoxylin. The integrated optical density (IOD) value was calculated by assessing the area and density of the dyed region of the immunohistochemistry section using ImageJ software (NIH). The relative IOD was presented as fold change relative to the normal control group.
Cytokine profile in CIA mice
Blood was collected using the retro-orbital eye bleed procedure on day 63 to investigate the cytokine levels of CIA mice. The cytokine profile of serum samples, including TNF-α, IL-1β, and IL-6, were analyzed using commercial kits, including QuantiCyto Mouse TNF-α ELISA kit (Neobioscience, EMC102a), QuantiCyto Mouse IL-1β ELISA kit (Neobioscience, EMC001b), and QuantiCyto Mouse IL-6 ELISA kit (Neobioscience, EMC004).
Histopathologic evaluation
To evaluate the synovial inflammation, bone erosion, and cartilage damage, paraffin sections of murine paws were stained using H&E Stain Solution (Servicebio, G1005), Modified Safranin O-Fast Green Cartilage Stain Kit (Solarbio, G1371), and Tartrate-Resistant Acid Phosphatase (TRAP) Staining Kit (Servicebio, G1050). The semi-quantitative assessment was graded from 0 (healthy) to 3 (severe) based on the severity of joint pathology as described previously.58 Numbers of TRAP-stained positive synovial osteoclasts were counted manually. Scoring and measurements for every histopathologic feature were performed on the same consistent region of the right hind paw for each mouse.
Toxicological evaluation
For histopathological assessment, the major organ samples from the liver, kidney, lung, heart, and spleen were embedded, sectioned, and stained with H&E. Serum samples were examined using the ALT assay kit (Rayto, S03030), AST assay kit (Rayto, S03040), BUN assay kit (Rayto, S03036), and CREA assay kit (Rayto, S03076) according to the manufacturer’s instructions to assess the toxic effect of anti-SMO siRNA on the liver function and renal function.
Statistical analysis
Statistical analysis was performed using Prism9 software (GraphPad). Data are presented as mean ± standard deviation (SD). The normality of data was assessed by the Shapiro-Wilk test, and the homogeneity of variances was examined by Levene’s test. Statistical differences among multiple groups were tested by one-way ANOVA. Dunnett’s test was applied for multiple comparisons where appropriate. Across all comparisons, the difference with a p value of <0.05 was regarded as statistically significant. The numbers of observations, statistical tests, and p values are reported in the figure panels or legends.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81871270), the International Science and Technology Cooperation Program of Guangdong Province (2019A050510028), and the Guangdong Pearl River Talents Program (2017GC010411). The authors are grateful to Kun Wang (The Third Affiliated Hospital, Sun Yat-sen University) for synovium biopsy, Yuyun Mai (Stomatological Hospital, Southern Medical University) for schematic illustration, and all Wu lab members for experimental assistance and valuable suggestion.
Author contributions
L.-P.W. and J.H. conceived the idea; L.L., S.Z., and H.H. conducted the experiments; all authors analyzed and discussed data; L.L. and L.-P.W. wrote the paper with contributions from all co-authors.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2022.12.008.
Contributor Information
Lin-Ping Wu, Email: wu_linping@gibh.ac.cn.
Jianlin Huang, Email: hjianl@mail.sysu.edu.cn.
Supplemental information
Data availability
All raw data supporting the findings are available from the corresponding authors upon reasonable request.
References
- 1.Smolen J.S., Aletaha D., McInnes I.B. Rheumatoid arthritis. Lancet. 2016;388:2023–2038. doi: 10.1016/S0140-6736(16)30173-8. [DOI] [PubMed] [Google Scholar]
- 2.You S., Koh J.H., Leng L., Kim W.U., Bucala R. The tumor-like phenotype of rheumatoid synovium: molecular profiling and prospects for precision medicine. Arthritis Rheumatol. 2018;70:637–652. doi: 10.1002/art.40406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aletaha D., Smolen J.S. Diagnosis and management of rheumatoid arthritis: a review. JAMA. 2018;320:1360–1372. doi: 10.1001/jama.2018.13103. [DOI] [PubMed] [Google Scholar]
- 4.Fraenkel L., Bathon J.M., England B.R., St Clair E.W., Arayssi T., Carandang K., Deane K.D., Genovese M., Huston K.K., Kerr G., et al. 2021 American College of Rheumatology guideline for the treatment of rheumatoid arthritis. Arthritis Care Res. 2021;73:924–939. doi: 10.1002/acr.24596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bottini N., Firestein G.S. Duality of fibroblast-like synoviocytes in RA: passive responders and imprinted aggressors. Nat. Rev. Rheumatol. 2013;9:24–33. doi: 10.1038/nrrheum.2012.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoshitomi H. Regulation of immune responses and chronic inflammation by fibroblast-like synoviocytes. Front. Immunol. 2019;10:1395. doi: 10.3389/fimmu.2019.01395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nygaard G., Firestein G.S. Restoring synovial homeostasis in rheumatoid arthritis by targeting fibroblast-like synoviocytes. Nat. Rev. Rheumatol. 2020;16:316–333. doi: 10.1038/s41584-020-0413-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Varjosalo M., Taipale J. Hedgehog: functions and mechanisms. Genes Dev. 2008;22:2454–2472. doi: 10.1101/gad.1693608. [DOI] [PubMed] [Google Scholar]
- 9.Briscoe J., Thérond P.P. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat. Rev. Mol. Cell Biol. 2013;14:416–429. doi: 10.1038/nrm3598. [DOI] [PubMed] [Google Scholar]
- 10.Salaritabar A., Berindan-Neagoe I., Darvish B., Hadjiakhoondi F., Manayi A., Devi K.P., Barreca D., Orhan I.E., Süntar I., Farooqi A.A., et al. Targeting Hedgehog signaling pathway: paving the road for cancer therapy. Pharmacol. Res. 2019;141:466–480. doi: 10.1016/j.phrs.2019.01.014. [DOI] [PubMed] [Google Scholar]
- 11.Scales S.J., de Sauvage F.J. Mechanisms of Hedgehog pathway activation in cancer and implications for therapy. Trends Pharmacol. Sci. 2009;30:303–312. doi: 10.1016/j.tips.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 12.Clara J.A., Monge C., Yang Y., Takebe N. Targeting signalling pathways and the immune microenvironment of cancer stem cells - a clinical update. Nat. Rev. Clin. Oncol. 2020;17:204–232. doi: 10.1038/s41571-019-0293-2. [DOI] [PubMed] [Google Scholar]
- 13.Su Y., Xing H., Kang J., Bai L., Zhang L. Role of the Hedgehog signaling pathway in rheumatic diseases: an overview. Front. Immunol. 2022;13:940455. doi: 10.3389/fimmu.2022.940455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sekulic A., Migden M.R., Oro A.E., Dirix L., Lewis K.D., Hainsworth J.D., Solomon J.A., Yoo S., Arron S.T., Friedlander P.A., et al. Efficacy and safety of vismodegib in advanced basal-cell carcinoma. N. Engl. J. Med. 2012;366:2171–2179. doi: 10.1056/NEJMoa1113713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Migden M.R., Guminski A., Gutzmer R., Dirix L., Lewis K.D., Combemale P., Herd R.M., Kudchadkar R., Trefzer U., Gogov S., et al. Treatment with two different doses of sonidegib in patients with locally advanced or metastatic basal cell carcinoma (BOLT): a multicentre, randomised, double-blind phase 2 trial. Lancet Oncol. 2015;16:716–728. doi: 10.1016/S1470-2045(15)70100-2. [DOI] [PubMed] [Google Scholar]
- 16.Norsworthy K.J., By K., Subramaniam S., Zhuang L., Del Valle P.L., Przepiorka D., Shen Y.L., Sheth C.M., Liu C., Leong R., et al. FDA approval summary: glasdegib for newly diagnosed acute myeloid leukemia. Clin. Cancer Res. 2019;25:6021–6025. doi: 10.1158/1078-0432.CCR-19-0365. [DOI] [PubMed] [Google Scholar]
- 17.Sharpe H.J., Pau G., Dijkgraaf G.J., Basset-Seguin N., Modrusan Z., Januario T., Tsui V., Durham A.B., Dlugosz A.A., Haverty P.M., et al. Genomic analysis of smoothened inhibitor resistance in basal cell carcinoma. Cancer Cell. 2015;27:327–341. doi: 10.1016/j.ccell.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Atwood S.X., Sarin K.Y., Whitson R.J., Li J.R., Kim G., Rezaee M., Ally M.S., Kim J., Yao C., Chang A.L.S., et al. Smoothened variants explain the majority of drug resistance in basal cell carcinoma. Cancer Cell. 2015;27:342–353. doi: 10.1016/j.ccell.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Setten R.L., Rossi J.J., Han S.P. The current state and future directions of RNAi-based therapeutics. Nat. Rev. Drug Discov. 2019;18:421–446. doi: 10.1038/s41573-019-0017-4. [DOI] [PubMed] [Google Scholar]
- 20.Hu B., Zhong L., Weng Y., Peng L., Huang Y., Zhao Y., Liang X.J. Therapeutic siRNA: state of the art. Signal Transduct. Target. Ther. 2020;5:101. doi: 10.1038/s41392-020-0207-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roberts T.C., Langer R., Wood M.J.A. Advances in oligonucleotide drug delivery. Nat. Rev. Drug Discov. 2020;19:673–694. doi: 10.1038/s41573-020-0075-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tafer H., Ameres S.L., Obernosterer G., Gebeshuber C.A., Schroeder R., Martinez J., Hofacker I.L. The impact of target site accessibility on the design of effective siRNAs. Nat. Biotechnol. 2008;26:578–583. doi: 10.1038/nbt1404. [DOI] [PubMed] [Google Scholar]
- 23.Wang M., Zhu S., Peng W., Li Q., Li Z., Luo M., Feng X., Lin Z., Huang J. Sonic Hedgehog signaling drives proliferation of synoviocytes in rheumatoid arthritis: a possible novel therapeutic target. J. Immunol. Res. 2014;2014:401903. doi: 10.1155/2014/401903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu S.L., Huang J.L., Peng W.X., Wu D.C., Luo M.Q., Li Q.X., Li Z.X., Feng X.X., Liu F., Wang M.X., et al. Inhibition of smoothened decreases proliferation of synoviocytes in rheumatoid arthritis. Cell. Mol. Immunol. 2017;14:214–222. doi: 10.1038/cmi.2015.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu F., Feng X.X., Zhu S.L., Huang H.Y., Chen Y.D., Pan Y.F., June R.R., Zheng S.G., Huang J.L. Sonic Hedgehog signaling pathway mediates proliferation and migration of fibroblast-like synoviocytes in rheumatoid arthritis via MAPK/ERK signaling pathway. Front. Immunol. 2018;9:2847. doi: 10.3389/fimmu.2018.02847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu S., Ye Y., Shi Y., Dang J., Feng X., Chen Y., Liu F., Olsen N., Huang J., Zheng S.G. Sonic Hedgehog regulates proliferation, migration and invasion of synoviocytes in rheumatoid arthritis via JNK signaling. Front. Immunol. 2020;11:1300. doi: 10.3389/fimmu.2020.01300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li R., Cai L., Ding J., Hu C.M., Wu T.N., Hu X.Y. Inhibition of Hedgehog signal pathway by cyclopamine attenuates inflammation and articular cartilage damage in rats with adjuvant-induced arthritis. J. Pharm. Pharmacol. 2015;67:963–971. doi: 10.1111/jphp.12379. [DOI] [PubMed] [Google Scholar]
- 28.Quaglio D., Infante P., Di Marcotullio L., Botta B., Mori M. Hedgehog signaling pathway inhibitors: an updated patent review (2015-present) Expert Opin. Ther. Pat. 2020;30:235–250. doi: 10.1080/13543776.2020.1730327. [DOI] [PubMed] [Google Scholar]
- 29.Xie P., Lefrançois P. Efficacy, safety, and comparison of sonic Hedgehog inhibitors in basal cell carcinomas: a systematic review and meta-analysis. J. Am. Acad. Dermatol. 2018;79:1089–1100.e17. doi: 10.1016/j.jaad.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 30.Liao S., Floyd C., Verratti N., Leung L., Wu C. Analysis of vismodegib resistance in D473G and W535L mutants of SMO receptor and design of novel drug derivatives using molecular dynamics simulations. Life Sci. 2020;244:117302. doi: 10.1016/j.lfs.2020.117302. [DOI] [PubMed] [Google Scholar]
- 31.Pricl S., Cortelazzi B., Dal Col V., Marson D., Laurini E., Fermeglia M., Licitra L., Pilotti S., Bossi P., Perrone F. Smoothened (SMO) receptor mutations dictate resistance to vismodegib in basal cell carcinoma. Mol. Oncol. 2015;9:389–397. doi: 10.1016/j.molonc.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dijkgraaf G.J.P., Alicke B., Weinmann L., Januario T., West K., Modrusan Z., Burdick D., Goldsmith R., Robarge K., Sutherlin D., et al. Small molecule inhibition of GDC-0449 refractory smoothened mutants and downstream mechanisms of drug resistance. Cancer Res. 2011;71:435–444. doi: 10.1158/0008-5472.CAN-10-2876. [DOI] [PubMed] [Google Scholar]
- 33.Elmén J., Thonberg H., Ljungberg K., Frieden M., Westergaard M., Xu Y., Wahren B., Liang Z., Ørum H., Koch T., Wahlestedt C. Locked nucleic acid (LNA) mediated improvements in siRNA stability and functionality. Nucleic Acids Res. 2005;33:439–447. doi: 10.1093/nar/gki193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choung S., Kim Y.J., Kim S., Park H.O., Choi Y.C. Chemical modification of siRNAs to improve serum stability without loss of efficacy. Biochem. Biophys. Res. Commun. 2006;342:919–927. doi: 10.1016/j.bbrc.2006.02.049. [DOI] [PubMed] [Google Scholar]
- 35.Shmushkovich T., Monopoli K.R., Homsy D., Leyfer D., Betancur-Boissel M., Khvorova A., Wolfson A.D. Functional features defining the efficacy of cholesterol-conjugated, self-deliverable, chemically modified siRNAs. Nucleic Acids Res. 2018;46:10905–10916. doi: 10.1093/nar/gky745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bramsen J.B., Laursen M.B., Nielsen A.F., Hansen T.B., Bus C., Langkjaer N., Babu B.R., Højland T., Abramov M., Van Aerschot A., et al. A large-scale chemical modification screen identifies design rules to generate siRNAs with high activity, high stability and low toxicity. Nucleic Acids Res. 2009;37:2867–2881. doi: 10.1093/nar/gkp106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seok H., Lee H., Jang E.S., Chi S.W. Evaluation and control of miRNA-like off-target repression for RNA interference. Cell. Mol. Life Sci. 2018;75:797–814. doi: 10.1007/s00018-017-2656-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bartoszewski R., Sikorski A.F. Editorial focus: understanding off-target effects as the key to successful RNAi therapy. Cell. Mol. Biol. Lett. 2019;24:69. doi: 10.1186/s11658-019-0196-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qiu S., Adema C.M., Lane T. A computational study of off-target effects of RNA interference. Nucleic Acids Res. 2005;33:1834–1847. doi: 10.1093/nar/gki324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wittrup A., Lieberman J. Knocking down disease: a progress report on siRNA therapeutics. Nat. Rev. Genet. 2015;16:543–552. doi: 10.1038/nrg3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zuckerman J.E., Davis M.E. Clinical experiences with systemically administered siRNA-based therapeutics in cancer. Nat. Rev. Drug Discov. 2015;14:843–856. doi: 10.1038/nrd4685. [DOI] [PubMed] [Google Scholar]
- 42.Ralphs J.R., Benjamin M. The joint capsule: structure, composition, ageing and disease. J. Anat. 1994;184(Pt 3):503–509. [PMC free article] [PubMed] [Google Scholar]
- 43.Uson J., Rodriguez-García S.C., Castellanos-Moreira R., O'Neill T.W., Doherty M., Boesen M., Pandit H., Möller Parera I., Vardanyan V., Terslev L., et al. EULAR recommendations for intra-articular therapies. Ann. Rheum. Dis. 2021;80:1299–1305. doi: 10.1136/annrheumdis-2021-220266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dekkers J.S., Schoones J.W., Huizinga T.W., Toes R.E., van der Helm-van Mil A.H. Possibilities for preventive treatment in rheumatoid arthritis? Lessons from experimental animal models of arthritis: a systematic literature review and meta-analysis. Ann. Rheum. Dis. 2017;76:458–467. doi: 10.1136/annrheumdis-2016-209830. [DOI] [PubMed] [Google Scholar]
- 45.Brand D.D., Latham K.A., Rosloniec E.F. Collagen-induced arthritis. Nat. Protoc. 2007;2:1269–1275. doi: 10.1038/nprot.2007.173. [DOI] [PubMed] [Google Scholar]
- 46.Vincelette J., Xu Y., Zhang L.N., Schaefer C.J., Vergona R., Sullivan M.E., Hampton T.G., Wang Y.X.J. Gait analysis in a murine model of collagen-induced arthritis. Arthritis Res. Ther. 2007;9:R123. doi: 10.1186/ar2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith T.O., Clarke C., Dainty J.R., Watts L., Yates M., Pomeroy V.M., Stanmore E., O'Neill T.W., Macgregor A.J. Clinical and biomechanical factors associated with falls and rheumatoid arthritis: baseline cohort with longitudinal nested case-control study. Rheumatology. 2022;61:679–687. doi: 10.1093/rheumatology/keab388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bombardier C., Barbieri M., Parthan A., Zack D.J., Walker V., Macarios D., Smolen J.S. The relationship between joint damage and functional disability in rheumatoid arthritis: a systematic review. Ann. Rheum. Dis. 2012;71:836–844. doi: 10.1136/annrheumdis-2011-200343. [DOI] [PubMed] [Google Scholar]
- 49.Katoh Y., Katoh M. Hedgehog target genes: mechanisms of carcinogenesis induced by aberrant Hedgehog signaling activation. Curr. Mol. Med. 2009;9:873–886. doi: 10.2174/156652409789105570. [DOI] [PubMed] [Google Scholar]
- 50.Amakye D., Jagani Z., Dorsch M. Unraveling the therapeutic potential of the Hedgehog pathway in cancer. Nat. Med. 2013;19:1410–1422. doi: 10.1038/nm.3389. [DOI] [PubMed] [Google Scholar]
- 51.Fang Q., Zhou C., Nandakumar K.S. Molecular and cellular pathways contributing to joint damage in rheumatoid arthritis. Mediators Inflamm. 2020;2020:3830212. doi: 10.1155/2020/3830212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Turner J.D., Filer A. The role of the synovial fibroblast in rheumatoid arthritis pathogenesis. Curr. Opin. Rheumatol. 2015;27:175–182. doi: 10.1097/BOR.0000000000000148. [DOI] [PubMed] [Google Scholar]
- 53.Aletaha D., Neogi T., Silman A.J., Funovits J., Felson D.T., Bingham C.O., 3rd, Birnbaum N.S., Burmester G.R., Bykerk V.P., Cohen M.D., et al. 2010 Rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis Rheum. 2010;62:2569–2581. doi: 10.1002/art.27584. [DOI] [PubMed] [Google Scholar]
- 54.Fakhr E., Zare F., Teimoori-Toolabi L. Precise and efficient siRNA design: a key point in competent gene silencing. Cancer Gene Ther. 2016;23:73–82. doi: 10.1038/cgt.2016.4. [DOI] [PubMed] [Google Scholar]
- 55.Reynolds A., Leake D., Boese Q., Scaringe S., Marshall W.S., Khvorova A. Rational siRNA design for RNA interference. Nat. Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 56.Birmingham A., Anderson E., Sullivan K., Reynolds A., Boese Q., Leake D., Karpilow J., Khvorova A. A protocol for designing siRNAs with high functionality and specificity. Nat. Protoc. 2007;2:2068–2078. doi: 10.1038/nprot.2007.278. [DOI] [PubMed] [Google Scholar]
- 57.Landesman Y., Svrzikapa N., Cognetta A., 3rd, Zhang X., Bettencourt B.R., Kuchimanchi S., Dufault K., Shaikh S., Gioia M., Akinc A., et al. In vivo quantification of formulated and chemically modified small interfering RNA by heating-in-Triton quantitative reverse transcription polymerase chain reaction (HIT qRT-PCR) Silence. 2010;1:16. doi: 10.1186/1758-907X-1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hayer S., Vervoordeldonk M.J., Denis M.C., Armaka M., Hoffmann M., Bäcklund J., Nandakumar K.S., Niederreiter B., Geka C., Fischer A., et al. 'SMASH' recommendations for standardised microscopic arthritis scoring of histological sections from inflammatory arthritis animal models. Ann. Rheum. Dis. 2021;80:714–726. doi: 10.1136/annrheumdis-2020-219247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The motor function of CIA mice was assessed by gait analysis. Representative ventral images were captured, and the digital footprints were generated. The paw area and stride frequency were increased in CIA mice. si-S1A3-Chol reduced the paw area and stride frequency, showing the effect on reversing gait dysfunction.
Data Availability Statement
All raw data supporting the findings are available from the corresponding authors upon reasonable request.