Abstract
Liquid chromatography, mass spectrometry, and metal analyses of cytosol and mitochondrial filtrates from healthy copper-replete Saccharomyces cerevisiae cells revealed that metallothionein CUP1 was a notable copper-containing species in both compartments, with its abundance dependent upon the level of copper supplementation in the growth media. Electrospray ionization mass spectrometry of cytosol and soluble mitochondrial filtrates displayed a full isotopologue pattern of CUP1 in which the first eight amino acid residues were truncated and eight copper ions were bound. Neither apo-CUP1 nor intermediate copper-bound forms were detected, but chelator treatment could generate apo-CUP1. Mitoplasting revealed that mitochondrial CUP1 was located in the intermembrane space. Fluorescence microscopy demonstrated that 34 kDa CUP1-GFP entered the organelle, discounting the possibility that 7 kDa CUP1 enters folded and metalated through outer membrane pores. How CUP1 enters mitochondria remains unclear, as does its role within the organelle. Although speculative, mitochondrial CUP1 may limit the concentrations of low-molecular-mass copper complexes in the organelle.
Introduction
Copper has unique catalytic properties which make it essential for living systems. In respiring eukaryotic cells, this d-block transition metal helps catalyze the reduction of O2 by cytochrome c oxidase (COX) in the mitochondrial inner membrane (IM). The active site of this respiratory complex includes a copper ion called CuB which works in conjunction with a heme to bind and reduce O2. Electrons used in reduction pass through the CuA redox site also in COX.
Cytosolic copper ions derived from nutrients are trafficked to mitochondria and installed into the CuA and CuB sites during COX assembly. After copper ions enter the intermembrane space (IMS), the trafficking pathway for delivery to these sites is generally understood. COX17, a soluble 8 kDa copper-containing protein in the IMS homologous to metallothioneins, transfers copper to chaperones SCO1 and COX11.1−5 SCO1 functions along with COA6 and SCO2 to install copper into CuA,6,7 whereas COX11 delivers copper to CuB.5 COX17 may also donate copper to CCS1, the chaperone that installs copper into superoxide dismutase 1 (SOD1), which partially localizes a portion of which localizes to the IMS.8
By contrast, the pathway of copper trafficking from the cytosol to the IMS is uncertain. In the mid 1990s, Tzagoloff and co-workers3−9 discovered that: (a) COX17 has dual localization, with ∼40% in the cytosol and the remainder in the IMS, (b) mutations of COX17 inhibit COX assembly and cause a respiratory defect, (c) ΔCOX17 cells are rescued when grown in copper-supplemented media, and (d) COX17 does not bind copper tightly enough to prevent dissociation during purification. They concluded that COX17 traffics cytosolic copper into the IMS for eventual delivery to COX.
However, COX17 was subsequently found to enter mitochondria through the MIA40/ERV1 pathway10 which requires that all cysteine residues are reduced and that the protein is in the apo-unfolded state.11 Once in the IMS, MIA40 oxidizes the twin Cx9C cysteines, forming two disulfide bonds that prevent the protein from returning to the cytosol.1,2,10 Morgan et al.11 found that Zn2+ ions bound to a Mia40 protein substrate impeded entry into the IMS, suggesting that Cu1+ ions bound to COX17 might do the same.
Also, Winge and co-workers showed that COX17 tethered to the mitochondrial IM (by fusing it with the SCO1 transmembrane domain) in an otherwise COX17-free cell does not result in a respiratory defect or abolish COX activity.12 The COX17 chimera was not detected in the cytosol nor were high copper concentrations in the media required for normal respiratory activity. They concluded that COX17 functions exclusively in the mitochondrial IMS and that it receives copper from an unidentified chaperone.
Soon thereafter, Winge and Cobine examined soluble mitochondrial extracts (SMEs) using liquid chromatography.13,14 They concluded that 85% of total mitochondrial copper is present as a low-molecular-mass (LMM) complex in the matrix called CuL. Although CuL remains unidentified, it has been characterized as a nonproteinaceous and anionic complex that is stable to boiling and organic extractions. Its chromatographic apparent mass was originally reported as 13 kDa, but later investigations imply a lower mass more typical of mononuclear copper coordination complexes with small organic ligands. Ligand L is reportedly fluorescent and detectable in the cytosol.15 IM proteins PIC2 and MRS3 reportedly import CuL into the matrix.15,16 The CuL pool is proposed to dynamically store cellular copper and, once exported back into the IMS by an unknown transporter, provide copper for COX and other copper-containing mitochondrial proteins.17
The presence of labile copper in mitochondria has been independently verified.18,19 Additionally, Cobine et al.14 targeted SOD1 and metallothionein CRS5 to the matrix in cells lacking the high-affinity plasma-membrane-bound copper importer CTR1. This inhibited respiration and diminished the intensity of the CuL chromatography peaks in extracts from those cells. The effect could be reversed by supplementing growth media with copper. These results suggest that matrix-localized SOD1 and CRS5 apo-proteins extracted copper from CuL and that this inhibited COX metalation.
Our group has attempted to identify CuL in mitochondria by isolating the organelle, filtering SMEs through a 10 kDa cutoff membrane, and then passing the flow-through solution (FTS) through size-exclusion chromatography columns.20−22 Unexpectedly, the major detected copper species migrated with an apparent mass of ∼5000 Da. The chromatographic region in which LMM species migrated, defined as having apparent masses between 200 and 2000 Da, exhibited only minor-intensity Cu peaks, albeit with some batch-to-batch variation. Copper species that migrated similarly to Cu5000 were recently observed in FTSs from the cytosol and vacuoles.23,24 The intensity of such peaks in cytosol FTSs increased when cells were grown on copper-supplemented media, and Cu5000 was absent in traces of cytosol FTSs from ΔCUP1 cells. These results, along with the apparent mass of Cu5000, supported speculation that this species was metallothionein CUP1, a well-known cytosolic protein.
In the current study, we used liquid chromatography electrospray ionization mass spectrometry (LC-ESI-MS) and liquid chromatography inductively coupled plasma mass spectrometry (LC-ICP-MS) to demonstrate that Cu5000 indeed arises from CUP1 and that this protein colocalizes to the cytosol and IMS of mitochondria isolated from healthy respiring WT yeast cells. Confocal fluorescence images of green fluorescent protein (GFP)-labeled CUP1 suggest that copper-bound CUP1 does not enter mitochondria through pores on the outer membrane (OM). Additionally, CUP1 concentration within the mitochondria had an inverse relationship with those of LMM Cu complexes, suggesting that CUP1 limits that metal pool. Although the pathway by which CUP1 enters mitochondria and its role within the organelle remain tentative, our results demand a fundamental re-evaluation of mitochondrial copper metabolism.
Materials and Methods
Yeast Strains and Cell Culture
DTY WT (MATα trp1–l gall metl3 can1 CUP1S ura3-50 Ade-His-) and CUP1Δ (MATα trp1-l gall metl3 can1 CUP1Δ ura3-50 Ade-His-) cells25 were received as a gift from Dr. Dennis Thiele. BY4741 CUP1-1-GFP and CUP1-2-GFP clones were acquired from the Yeast GFP Clone Collection (Thermo Scientific). These cells, along with WT strains W303 (MATα ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1) and BY4741 (MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0), were maintained as described.26 Fifty mL precultures were inoculated with single colonies taken from YPAD plates (20 g/L glucose, 10 g/L yeast extract, 20 g/L peptone, 100 mg/L adenine, and 10 g/L bactoagar) at 30 °C. Precultures were transferred to larger volume cultures (1, 1.5, or 2 L) once the optical density at 600 nm (OD600) reached ∼1. Transfers and isolations were conducted using exponentially growing cells.
For cytosol isolations, W303 WT cells were cultured in a 2.8 L baffled flask containing 2 L of MM, including 20 g/L glucose, 5 g/L ammonium sulfate, 1.7 g/L yeast nitrogen base without ammonium sulfate or ferric chloride (MP Bio), 100 mg/L leucine, 50 mg/L adenine, 20 mg/L histidine, 20 mg/L uracil, 48 mg/L tryptophan, 40 μM ferric citrate, and 50 μM copper sulfate. Flasks were swirled at 180 rpm in an incubator (MaxQ 8000, Thermo Scientific) at 30 °C. For isotopic enrichment studies, natural-abundance copper sulfate was replaced with 50 μM copper(II) chloride of the indicated isotope. Baffled flasks were soaked in 5% HNO3 overnight and rinsed with double distilled water. For mitochondrial isolations, W303 WT cells were cultured in either 1.5 or 25 L of MM in which 30 mL/L glycerol and 10 mL/L ethanol replaced glucose. Larger volume growths used a custom-built 24 L glass reactor with a titanium stir paddle and tubing. Pressurized air was bubbled through the media. For a comparison study of the effect of changing the media, mitochondria were also isolated from W303 WT cells cultured in 1.5 L YPEG composed of 30 mL/L glycerol, 10 g/L yeast extract, 20 g/L peptone, 100 mg/L adenine, and 50 μM copper sulfate.
Due to poor growth on MM, DTY cells were cultured in 2.8 L baffled flasks containing 2 L of YPAD media including 20 g/L glucose, 10 g/L yeast extract, 20 g/L peptone, 100 mg/L adenine, and 50 μM copper sulfate. BY4741 cells, which also grew poorly on MM, were cultured in 2.8 L baffled flasks containing 1.5 L of respiring CSM media, including 30 mL/L glycerol, 0.64 g/L CSM-Leu-Trp (Sunrise Science), 1.7 g/L yeast nitrogen base (without ammonium sulfate, ferric chloride, or copper sulfate (MP Bio)), 5 g/L ammonium sulfate, 100 mg/L leucine, 50 mg/L adenine, 48 mg/L tryptophan, 40 μM ferric citrate, and 50 μM copper sulfate. For a comparison study, mitochondria were isolated from DTY005 WT cells cultured in 1.5 L of CSM respiring media.
All cultures were grown aerobically. Initial centrifugation steps to acquire initial whole cell pellets were also conducted aerobically. 20AA6.5 buffer (20 mM ammonium acetate pH 6.5) and SAA2 buffer (0.6 M sorbitol in 20AA6.5 were degassed on a Schenk line and imported into a refrigerated glovebox (Mbraun Labmaster 130, 5 °C, <5 ppm O2). Isolations were conducted in a glovebox, taken out sealed for centrifugation steps, and then brought back into the glovebox.
Cytosol Isolation
Cytosol was isolated as described23 with minor modifications. When 2 L cell cultures reached 0.6 < OD600 < 0.8, they were spun by centrifugation at 5000 × g for 5 min in a Sorvall Lynx 6000 centrifuge (Thermo Scientific), resuspended in deionized water (5 mL water per g cells, typically 4 g cells), and recentrifuged. Water washing was performed three times. Pelleted cells were brought into the glovebox preceding the third rinse’s removal and resuspended in 20AA6.5, 10 mM DTT (5 mL/g cells). The suspension was agitated at 180 rpm at 30 °C in an incubator shaker and then pelleted at 4000 × g for 6 min. The supernatant was discarded, and the pellet was resuspended in 5 mL of SAA2 buffer per g cells. A 0.2 M stock of PMSF (Sigma) was prepared in ethanol, and an aliquot was added to the suspension (10 mM, final). Zymolyase (Amsbio 120493-1) was added to the suspension (1 mg (100 KU) per g cells). Aliquots were taken before and after zymolyase addition to monitor cell wall digestion using OD600. The suspension was agitated in the 30 °C incubator shaker until OD600 had declined to <30% of its original value. The spheroplasts were spun by centrifugation at 1800 × g for 5 min, and the supernatant was discarded. The spheroplasts were then resuspended in SAA2 buffer, 10 mM PMSF, 6 mL/g according to the original pellet mass (typically 24 mL of SAA2 buffer for 4 g cells). The spheroplast suspension was homogenized in a 40 mL capacity Dounce tissue grinder (Kimble, glass pestle A with clearance 0.0030–0.0060 inches), in 15 mL aliquots and 10 strokes per aliquot. Homogenates were combined and centrifuged at 1000 × g for 15 min. The supernatant was removed and then centrifuged at 15000 × g for 30 min. The resulting supernatant was removed and centrifuged at 112,000 × g for 60 min. Any detectable lipid layer was removed from the top of the supernatant, and the supernatant was then centrifuged at 185,000 × g for 150 min. The resulting supernatant, defined as cytosol, was filtered through either a 10 kDa cutoff filter (Amicon Ultra 2 mL Centrifugal filter) by centrifugation at 3858 × g for 1 h or a 0.2 μm pore syringe filter (Titan3 regenerated cellulose, Thermo Scientific). For the study incubating the cytosol with varying equivalents of BCS, CUP1 concentration was calculated from the integrated chromatographic peak. Mixtures were incubated overnight in the glovebox.
Large-scale lysates were prepared from 25 L bioreactor cultures. W303 WT cells were cultured in glucose-based MM. Cells were centrifuged at 5000 × g for 5 min. The pellet was resuspended in 5 mL of 20 mM ammonium acetate +10 mM DTT per g cells. The suspension was agitated in the incubator shaker and then pelleted at 4000 × g for 6 min. Cells were resuspended in 5 mL of SAA2 buffer and treated with zymolyase as above. The spheroplast pellet was resuspended in 180 mL of 20AA6.5 plus 16 mM PMSF per g whole cells. The spheroplast suspension was homogenized, 40 mL at a time, in the Dounce tissue homogenizer for 5 min (approximately 40 strokes). The homogenates were combined and centrifuged at 4000 × g for 7 min. The supernatant was pelleted at 4000 × g for 20 min, 12,000 × g for 30 min, and 15,000 × g for 30 min, each time discarding the pellet. The final supernatant was then filtered on a 250 mL Amicon Stir Cell with a 10 kDa cutoff regenerated cellulose membrane. The filtrate was then concentrated using the same stir cell with a 3 kDa cutoff membrane and then further with a 3 kDa cutoff Amicon Ultra 2 mL Centrifugal filter (Sigma) centrifuged at 3858 × g for 45 min. One independent preparation of the retentate was frozen and lyophilized. An unfrozen aliquot was incubated in 20 mM BCS overnight in the glovebox. Retentates were analyzed by LC-ESI-MS.
Cytosol, SME, and cell lysate samples were lyophilized (Labconco Freezone 2.5 Plus), resuspended in a minimal volume of double-distilled water, and then subjected to LC-ESI-MS.
Mitochondrial Sample Preparation
Yeast mitochondria were isolated as described27 with minor modifications. Large-scale (25 L) cell cultures were harvested at OD600 = 0.6–0.8; small-scale (1.5 L) isolations were harvested at OD600 = 1.2–1.4. Standard density gradients consisted of two 10 mL layers of 20AA6.5 solutions containing 32% (top) and 60% (bottom) (w/v) sucrose. An alternative gradient used 40% (top) and 50% (bottom); another alternative used 7 mL of 32% (top), 7 mL of 45% (middle), and 7 mL of 60% (bottom) sucrose. Alternative gradients were run simultaneously using aliquots from the same mitochondrial suspension. SH buffer was replaced with SAA2.
Mitochondrial suspensions were pelleted at 12,000 × g for 10 min. SMEs were generated by resuspending mitochondrial pellets in 20× dilution of lysis buffer, consisting of 5 mM dodecylmaltoside and 20AA6.5. Mitoplasts were generated by hypotonic lysis essentially as described.13 Mitochondrial pellets were resuspended in 20× dilution of 10 mM ammonium acetate for 15 min. Mitoplasts and fragments were centrifuged at 25,655 × g for 15 min. The supernatant, which represented the IMS fraction, was removed. Soluble mitoplast extracts (called SmpEs) were generated by resuspending the mitoplast pellet in the same volume of lysis buffer used to generate SMEs. IMS samples were concentrated 10× for Western blot analysis using a 3 kDa cutoff Amicon Ultra 2 mL Centrifugal filter centrifuged at 3858 × g for 45 min. Protein concentration was evaluated using a Pierce BCA Protein Assay Kit (Thermo Scientific).
Proteinase K and NaCl Solution Treatment of Mitochondria
Mitochondria were pelleted at 12,000 × g for 10 min, and mitoplasts were separated from the IMS fraction as indicated above. Proteinase K stock solution was prepared in SAA2 buffer (1 mg/mL). One milliliter of the ProK stock solution was added to ∼90 mg of pelleted mitochondria. After 30 min, the mitochondria were pelleted at 12,000 × g for 10 min. The SME was generated by addition of 1 mL of 5 mM dodecylmaltoside and 20AA6.5 (initial concentrations) and adding 100 μL of 0.2 M PMSF stock solution in ethanol to inhibit any residual ProK. One milliliter of ProK stock solution was also added to 1.8 mL of individual IMS fractions and incubated at 5 °C in the glovebox for 30, 60, and 120 min durations. At the end of each duration, protease activity was halted by adding 100 μL of the PMSF solution.
About 90 mg of mitochondria were resuspended and incubated in a NaCl solution (100 mM NaCl plus 0.6 M sorbitol) at 5 °C in the glovebox for 15 min. Mitochondria were pelleted at 12,000 × g for 10 min, and the supernatant was removed. Lysis buffer was then added to generate the SMEs as already described.
LC-ICP-MS and Elemental Analysis
Samples (100 μL) were injected into an LC-ICP-MS instrument configured as described.28 All samples were filtered using a 0.2 μm pore syringe filter (Titan3, Thermo Scientific) prior to injection on the LC-ICP-MS. Samples were injected into either low-mass (Superdex 30 Peptide or Increase 10/300 GL, Cytiva) or high-mass (Superdex 200 Increase 10/300 GL, Cytiva) at a flow rate of 0.6 mL/min. The low-mass Peptide column was replaced mid-study with the Increase column, resulting in minor shifts of elution volumes. For fractionation, lysate was injected onto two low-mass columns connected in series at a flow rate of 0.25 mL/min. Fractions (1.5 mL) obtained using an Agilent 1260 Infinity Bioinert fraction collector were frozen and lyophilized for LC-ESI-MS analysis. SAA2 and lysis buffers were exchanged with 20AA6.5 preceding lyophilization. This involved concentrating samples, diluting 10× with buffer, and re-concentrating (repeated twice).
For elemental analysis, 150 μL of double-distilled water was added to 100–200 mg of pelleted mitochondria. A portion (175 μL) of the suspension was mixed with 300 μL of trace-metal-grade 70% (w/v) HNO3 (Fisher) in a 15 mL plastic screw-top tube (Corning) which was finally sealed with electrical tape. Samples were incubated at 70 °C for 2 days. Once cooled, samples were treated with 1.5 mL of 35% (w/v) H2O2. Samples were additionally incubated at 80 °C for 1 day and then diluted with double-distilled water to a final volume of 21 mL (0.5% HNO3 and 2.5% H2O2). The metal concentrations were determined by ICP-MS. All samples from three independent batches were run in technical triplicate. A packing efficiency of 0.77 for pelleted mitochondria was assumed.22
Sample Manipulations
The cytosol and SMEs were concentrated for LC-ESI-MS analysis. Prior to concentration, samples were filtered through 10 kDa cutoff Amicon Ultra Centrifugal filters, 2 mL capacity, spun at 3858 × g for 1 h. The 10 kDa filtrates were then concentrated in 3 kDa cutoff Amicon Ultra 2 mL Centrifugal filters spun at 3858 × g for 1 h. Concentrating in this manner typically required four consecutive rounds of centrifugation. After each round, the filtrate was removed, and an additional sample was added to the retentate in the same filter.
Western blots were run as described23 using antibodies at the same concentrations. Anti-COX3 (Thermo Scientific) and anti-SOD1 (Stress Marq) were diluted 2000×.
Microscopy
GFP clones were cultured from single colonies maintained on YPAD plates. Clones were cultured in 50 mL of YPAD supplemented with 75 μM copper sulfate. Cells were pelleted at 5000 × g for 5 min. Whole cells were suspended in 20 mM ammonium bicarbonate, pH 7.5. Isolated mitochondria were suspended in SAA2 buffer and kept in a refrigerated glovebox overnight following isolation. Slides were coated with poly-l-lysine for 1 h on a rocker at RT. Excess was removed and slides were dried at RT overnight. MitoTracker Red CMXRos (Thermo Scientific) was prepared in a dimethyl sulfoxide stock solution of 1 mM from the desiccated solid. MitoTracker was added to the mitochondrial suspension to a final concentration of 100 nM. The suspension was agitated in a 30 °C incubator shaker for 45 min. Mitochondria were pelleted at 12,000 × g for 10 min and resuspended in minimal SAA2 buffer. Samples were imaged using a Zeiss LSM 780 NLO multiphoton microscope and an airy scan detector. MitoTracker was excited at 560 nm, and its emission was measured at 599 nm. GFP was excited at 488 nm, and its emission was measured at 510 nm. GFP emission was measured after filtering >570 nm light. Image brightness and contrast were adjusted using the “despeckle” function in ImageJ.
LC-ESI-MS
Samples were analyzed using a Thermo Scientific Q Exactive Focus coupled with an LC unit (ultimate 3000 RS). Samples were separated by injecting 10 μL of sample into a BioBasic-18 (2.1 × 150 mm; 3 μm) C18 column (Thermo Scientific). The mobile phase consisted of 10 mM ammonium acetate (eluent A) and methanol with 10 mM ammonium acetate (eluent B). The flow rate was set at 300 μL/min with the following gradient: 0–5.0 min 5% B, 5–12 min to 5–100% B, 12–15 min hold 100% B, 15.1–20 min hold 5% B. A Q Exactive Focus HESI source was operated in full MS in positive and negative ESI mode. The mass resolution was tuned to 70,000 full width at half maximum at m/z 200. The spray voltage was set to 3.75 kV for positive mode and 3.3 kV for negative mode. The sheath gas and auxiliary gas flow rates were set to 35 and 10 arbitrary units, respectively. The transfer capillary temperature and the auxiliary gas heater temperature were held at 275 and 300 °C, respectively. The S-Lens RF level was set at 50 v. Exactive Series 2.8 SP1/Xcalibur 4.0 software was used for data acquisition and processing.
Results
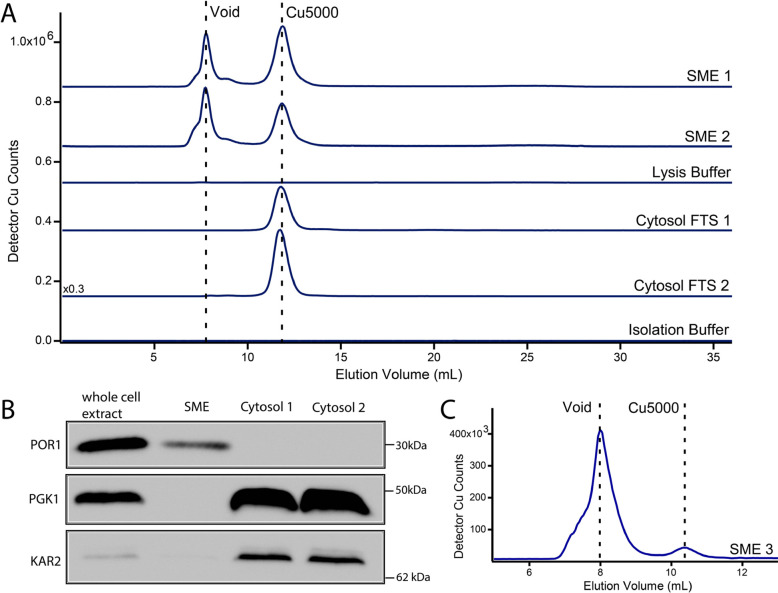
While investigating copper species within isolated mitochondria and the cytosol of Saccharomyces cerevisiae, we noticed that the major copper species detected in isolated mitochondria, previously called Cu5000, co-migrated with the dominant copper species in the 10 kDa-filtered cytosol. To investigate further, yeast strain W303 cells were grown in the respiring minimal medium supplemented with 50 μM Cu, a significant but nontoxic concentration exhibiting no growth deficit. Both cellular compartments were isolated. Mitochondria from two independent batches were solubilized and passed through a 0.2 μm filter to obtain SMEs. The filter functioned only to remove large particles that could have fouled the LC system. Using improved methods,28 LC traces revealed Cu5000 along with some higher-molecular-mass copper-containing species at or near the void volume of the column (Figure 1A). The lysis buffer used was devoid of detectable Cu species.
Figure 1.
Detection of copper species in the cytosol and mitochondria. (A) Copper-detected LC-ICP-MS chromatograms of two independent SMEs and two independent cytosolic FTSs. Control traces of the buffers used to prepare samples were devoid of peaks. Left and right vertical dashed lines indicate the column void volume and elution volume of Cu5000, respectively. Cell cultures were supplemented with 50 μM CuSO4. (B) Western blot of whole cell extracts, unfiltered SME, and cytosol solutions staining for mitochondrial porin (POR1), cytosol localized phosphoglycerate kinase (PGK1), and endoplasmic reticulum localized protein KAR2. Each lane was loaded with 12.5 μg of protein. (C) Copper-detected LC-ICP-MS chromatogram of the SME prepared from a cell culture supplemented with 10 μM CuSO4. Use of a Superdex Increase rather than Peptide column resulted in minor elution volume shifts relative to in A.
Cytosol from two independent batches of fermenting cells were isolated and passed through a 10 kDa cutoff membrane. LC traces of cytosolic FTSs displayed a significant Cu peak that co-migrated with Cu5000 (Figure 1A). No other notable Cu-detected peaks were evident in these traces or in traces from the isolation buffer. A Western blot of mitochondrial porin (POR1) and cytosolic phosphoglycerate kinase (PGK1) (Figure 1B) indicated that these two isolated cellular compartments were nearly free of contamination. This provided strong evidence that mitochondrial Cu5000 was not an artifact of cytosolic contamination and that cytosolic Cu5000 was not an artifact of mitochondrial contamination. Significant levels of this copper-containing protein were present in both cellular compartments.
This was an important point as even modest contamination of cytosol (or ER) in mitochondrial extracts could be responsible for the observed Cu5000 in those extracts. To investigate this further, the density gradients used in mitochondrial isolations were altered to determine whether Cu5000 intensities varied in proportion to the contamination level. We found that Cu5000 peak intensity was correlated with mitochondrial markers POR1 and COX3 and not with markers for other organelles (Figure S1). This verified that Cu5000 observed in SMEs originated from mitochondria, not from cytosol or ER contamination. The average copper concentration of acid-digested isolated mitochondria from cells supplemented with 50 μM CuSO4 was 87 ± 18 μM (n = 3 independent batches). Of this, 32 ± 2 μM was soluble copper that could pass through a 0.2 μm filter. Based on calibrated peak areas, 18 ± 3 μM copper was associated with Cu5000, which corresponded to 21% of total mitochondrial copper and 56% of soluble mitochondrial copper under the condition tested.
We had anticipated that low-mass copper species would constitute the majority of mitochondrial copper in line with the current CuL hypothesis. In contrast, the low-mass region (Figure 1A, 2000–200 Da; 18–35 mL elution volume) in LC traces of SMEs was virtually devoid of copper species.
The level of copper supplementation in the growth media affected the concentration of Cu5000 in the cytosol,23 and we wondered whether a similar effect would be observed in SMEs. To investigate, we supplemented media with less copper (10 μM CuSO4) and found that the Cu5000 peak was present in SME traces at a reduced intensity (Figure 1C) relative to peaks in the void region.
Mitochondrial Cu5000 was observed in all preparations tested, but its peak intensity also varied with the growth medium (Figure S2). Using glycerol/ethanol as a carbon source, the intensity of Cu5000 relative to the copper void peaks declined in the YPEG medium compared to that in CSM or MM. We concluded that Cu5000 is present in the mitochondria of healthy (unstressed) cells but that its concentration depends on the growth medium and the level of copper supplementation in the medium.
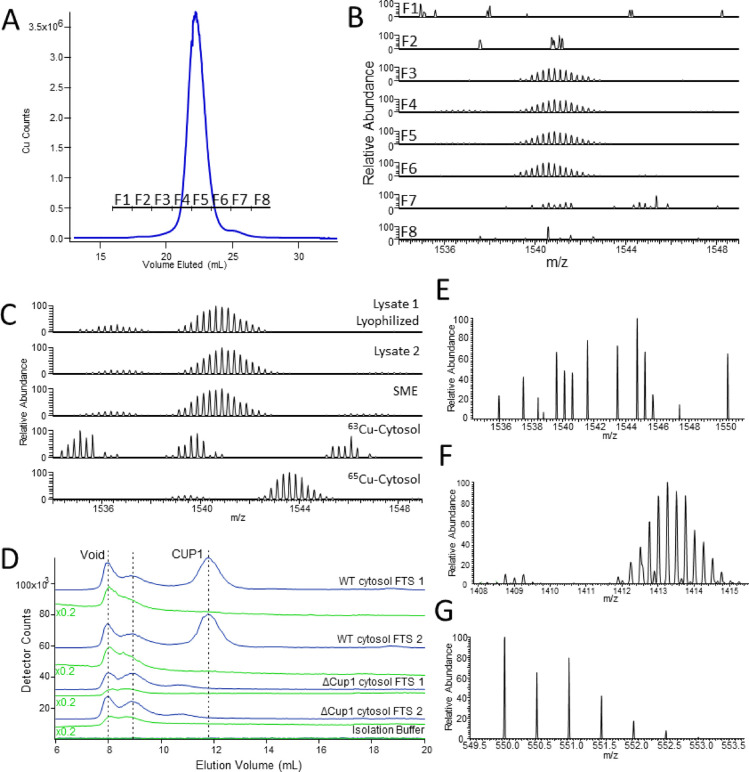
Our next objective was to identify Cu5000. To ensure a sufficient concentration for detection by LC-ESI-MS, we isolated 66 grams of wet cells from a 24 L culture. We lysed the cells and passed the soluble fraction through a 10 kDa cutoff membrane. We then passed the FTS through a 3 kDa cutoff membrane and collected the retentate. This solution was lyophilized, resuspended in buffer, and passed through two size-exclusion columns connected in series. An intense Cu5000 peak was detected (Figure 2A).
Figure 2.
Identification of Cu5000 as metallothionein CUP1. (A) Copper-detected LC-ICP-MS chromatogram of the purified cell lysate with collected fractions indicated. (B) LC-ESI-MS (negative mode) spectra of lyophilized collected chromatographic fractions from A. (C) LC-ESI-MS spectra of filtered samples including (from top) lyophilized cell lysate, matched nonlyophilized cell lysate, SME, cytosol from cells grown on 63Cu media, and the cytosol from cells grown on 65Cu media. (D) Copper-detected (blue) and zinc-detected (green) LC-ICP-MS chromatograms of the isolated cytosol FTS from two independent batches of WT DTY005 cells and two independent batches of ΔCUP1 cells. The left vertical dashed line indicates the void volume, and the middle line indicates the volume of a copper peak that is stronger in the ΔCUP1 cytosol than in the WT cytosol (suspected of being metallothionein CRS5). The right vertical line indicates the position of CUP1. (E) LC-ESI-MS spectra of lysate 2 incubated in 20 mM BCS collected in negative mode. (F) apo-CUP1 detected in the same spectrum as E; (G) [Cu(BCS)2H]−2 detected in the same spectrum as E. CUP1 and apo-CUP1 species were located by screening the LC-ESI-MS chromatogram for masses near those expected.
Eight fractions (F1–F8) were collected, corresponding to before, during, and after elution of Cu5000. Each fraction was subjected to negative-mode LC-ESI-MS. Fractions F3–F7 displayed a complex isotopologue pattern with a base peak at m/z = 1540.87 and parent peak at m/z = 1539.12 (z = 4) (Figure 2B). This indicated species with 6160.48 Da. The intensity of the pattern increased, maximized, and decreased harmoniously with the Cu5000 LC intensity, confirming that the detected pattern arose from Cu5000.
Related satellite spectral patterns were evident on both sides of the main pattern, with Δm ± 16–17 amu. The same main spectral pattern was evident in SMEs (Figure 2C, third trace), confirming that Cu5000 was present in both cellular compartments. Lyophilization had no effect on the pattern (Figure 2C, first vs second trace). The same species was detected in positive mode with a base peak m/z = 1542.89 (z = 4) and was similarly unaffected by lyophilization (Figure S3).
A species of similar mass, with base peak m/z = 1542.2 (z = 4), has been observed using positive-mode CE-MS for metallothionein CUP1 from S. cerevisiae.29 This protein, with a mass of 6156 Da, corresponds to CUP1 in which the first eight amino acid residues are truncated, eight copper ions are bound, and all coordinating cysteines are deprotonated. The mass of our species of interest corresponded well in both positive and negative modes, and so we tentatively attributed it to CUP1.
In their mass spectral study of CUP1, Knudsen et al.29 also observed a satellite pattern at −16 amu, which they attributed to cyclization of the N-terminal glutamate of CUP1 to form pyroglutamate. The additional satellite pattern that we observed at +17 amu is most likely due to an ammonium adduct (−H and +NH4+), due to the elevated buffer concentration after lyophilization.
The mass obtained using the base peak of the detected isotopologue pattern matched that of metallothionein CUP1 with a statistical mixture of the two dominant copper isotopes according to their natural abundances (69.2% 63Cu; 30.8% 65Cu). To verify this assignment, we isolated the cytosol from a batch of yeast grown on medium supplemented exclusively with 63Cu and from another batch exclusively grown with 65Cu. The resulting ESI-MS patterns shifted according to the isotope used if eight copper ions were bound, verifying the assignment of Cu5000 as CUP1 (Figure 2C). The observed deviation of 1 amu was likely the result of minor isotopic impurities in reagents and/or growth media.
To evaluate whether Cu5000 arose from CUP1 using a genetics-based approach, we examined LC traces of cytosol from ΔCUP1 cells. As a positive control, Cu5000 was detected in LC traces of two independent batches of cytosolic FTSs isolated from the parent strain, DTY005 (Figure 2D). As anticipated, LC traces of FTS from two batches of ΔCUP1 cells did not exhibit Cu5000, confirming results and conclusions from an earlier study.23 The higher mass copper species that coeluted with zinc in that study was proposed to be the other metallothionein in S. cerevisiae, CRS5.
We verified the number of copper ions bound to CUP1 by adding an excess of the copper-specific chelator BCS to cell lysate. Negative-mode ESI-MS spectra of the resulting solutions lacked the original isotopologue pattern and exhibited a new pattern (Figure 2E) with a base peak at m/z = 1413.26 and parent peak at m/z = 1412.01, both z = 4 (Figure 2F). The resultant molecular mass of 5652.04 amu was assigned to apo-CUP1. The parent mass was 508.44 amu lower than that of holo-CUP1 prior to incubation with BCS. A loss of eight 63Cu ions would contribute 504 amu of this loss; the remaining 4 amu was likely due to protonation events. The expected Cu(BCS)2 complex also formed post-incubation, detected as [Cu(BCS)2H]−2 (Figure 2G). We concluded that the detected CUP1 species contains eight Cu ions that could be removed by the BCS chelator. Unless BCS was added, apo-CUP1 was not detected in any cell lysates or in any isolated batch of cytosol over the course of the entire study (Figure S4). Similarly, no form of CUP1 containing intermediate numbers of copper ions bound was detected. Increasing equivalents of BCS were incubated with WT cytosol (Figure S5), but still no intermediates were observed. Collectively, these results suggest cooperative Cu binding to CUP1.
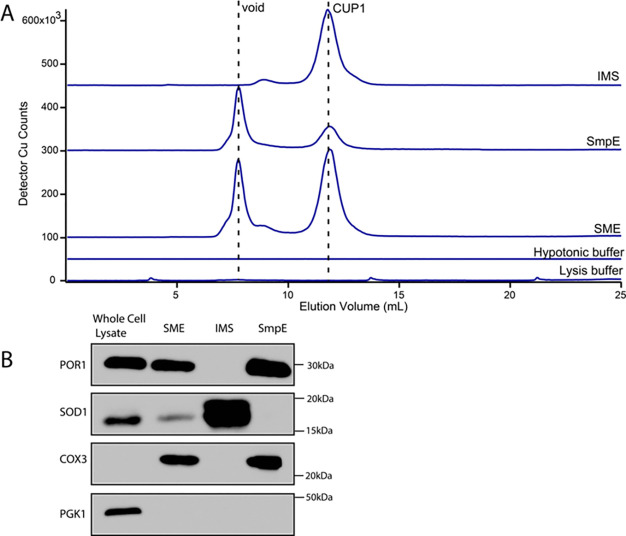
To determine the submitochondrial location of CUP1, we generated mitoplasts from isolated mitochondria by means of hypotonic lysis.13,30,31 After selective rupture of the OM, the buffer should contain IMS proteins and perhaps proteins that are weakly associated with the OM. Mitoplast lysates should contain soluble proteins from the matrix and perhaps proteins that are weakly associated with the IM. After separating the IMS fraction, mitoplasts were lysed, and the resulting SmpE was passed through a 0.2 μm filter. The LC trace of the resulting IMS FTS exhibited an intense CUP1 peak, as well as a minor second peak with an apparent mass of ∼11 kDa (Figure 3A, top trace). We hypothesize that this minor peak arose from COX17.
Figure 3.
Mitoplasting indicates that mitochondrial CUP1 is localized to the IMS. (A) Copper-detected LC-ICP-MS chromatograms of IMS and SmpE subfractions of isolated mitochondria. Control traces of hypotonic and lysis buffers were essentially devoid of copper peaks. (B) Western blot of respective unfiltered samples used in A. The IMS sample was concentrated before blotting. Each lane was loaded with 14 μg of protein.
The LC trace of the SmpE exhibited a diminished CUP1 peak (Figure 3A, second trace). It also exhibited intense high-mass (>10 kDa) unresolved Cu species near or at the void, which likely included other Cu-binding proteins, e.g., COX. The minor peak hypothesized to be COX17 was absent, consistent with that assignment. In principle, the traces of the SME (Figure 3A, third trace) should be the sum of the traces from the IMS and SmpE, and this was qualitatively the case apart from a slight mixing of IMS CUP1 with the mitoplast lysate. Thus, the procedure successfully divided the mitochondrial copper content into the two indicated subfractions. LC traces of the hypotonic and lysis buffers used in the experiment were devoid of significant copper features (Figure 3 bottom two traces). Since >80% of the CUP1 peak intensity was found in the IMS fraction, we concluded that CUP1 localizes to the IMS in intact mitochondria.
A Western blot was obtained in conjunction with this experiment to confirm separation of the IMS fraction and mitoplasts. The presence of POR1, SOD1, COX3, and PGK1 was investigated in the whole cell lysate, the SME, the IMS fraction after mitoplasting, and the SmpE, which should contain IM and matrix fractions. The forementioned proteins are found in the OM, IMS, IM, and cytosol, respectively. The SME served as a positive control for mitochondrial proteins and contained POR1, SOD1, and COX3, as expected. The IMS fraction contained SOD1 as expected. SOD1 colocalizes to both the cytosol and IMS;32 however, the lack of an observable PGK1 band in all mitochondrial samples suggested that the detected SOD1 arose from the population within the IMS and was not due to cytosolic contamination. Finally, the SmpE exhibited a strong COX3 band, as expected.
We considered that CUP1, which we tentatively concluded to be in the IMS, might be cytosolic CUP1 that had adhered to the exterior of mitochondria during isolation. In that case, CUP1 could have potentially been released into SMEs and the IMS fraction during OM rupture. To investigate, we incubated the IMS fraction and intact mitochondria separately with the protease proteinase K. As a positive control, the IMS fraction was treated with the protease for 30, 60, and 120 min. The protease inhibitor PMSF was added at the end of each timepoint to inhibit further hydrolytic activity. The corresponding CUP1 LC peaks shifted with incubation time to lower apparent masses, indicating time-dependent hydrolysis (Figure 4A, top traces). In contrast, the CUP1 peak was unaffected in an SME solution when intact mitochondria were treated with proteinase K (Figure 4A, third trace); this suggested that CUP1 in the IMS was protected from the protease by the OM. Neither the PMSF nor the ethanol used to dissolve the reagent affected Cu binding or the apparent mass of the CUP1 peak (Figure 4A, second trace). Parenthetically, the peak tentatively assigned to COX17 declined in intensity upon exposure to proteinase K but was unaffected in intact mitochondria. Since COX17 is an IMS protein, this result supported our assignment.
Figure 4.
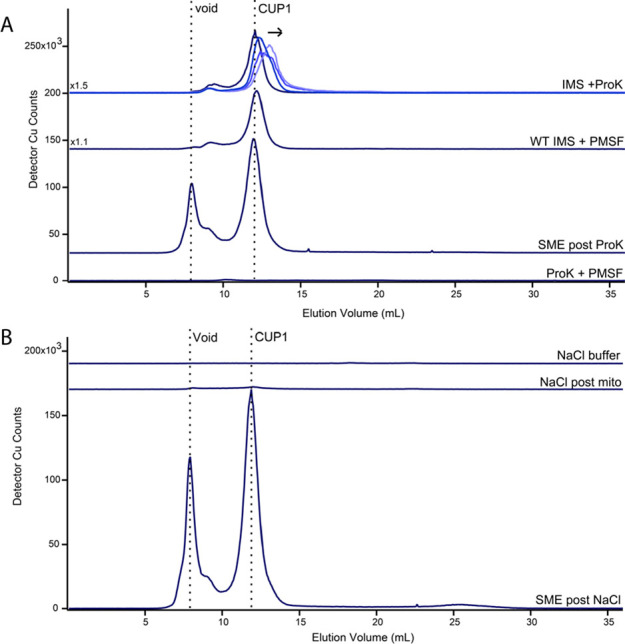
CUP1 does not adhere to the mitochondrial exterior. (A) Copper-detected LC-ICP-MS chromatograms of the IMS fraction treated with proteinase K, an IMS fraction not treated with ProK, and the SME fraction after treatment with ProK. Incubation durations in the top chromatograms were 30, 60, and 120 min, indicated by darker to lighter blue traces and the arrow direction. PMSF was added to inhibit protease activity at the end of the duration. The control trace of ProK + PMSF solution was devoid of copper peaks. (B) Copper-detected LC-ICP-MS chromatograms of the mitochondrial supernatant after incubation with NaCl buffer and centrifugation. Bottom trace is a positive control demonstrating that CUP1 was in the mitochondrial sample used. Top trace is a negative control showing the absence of copper in the NaCl buffer.
To further evaluate whether CUP1 was in the IMS or adventitiously associated with the OM, we washed isolated mitochondria with a NaCl solution and examined the resulting wash and SME by LC-ICP-MS. A similar procedure had been used to remove proteins adhering to the mitochondrial OM.31,32 The LC trace of the wash solution was devoid of the CUP1 peak both before and after incubation with intact mitochondria (Figure 4, A and B). This again supported our conclusion that CUP1 was located within the organelle, not bound on the surface. The SME obtained from the NaCl-treated mitochondria exhibited an intense CUP1 peak. These trends, observed using the W303 strain, were also observed in analogous studies using BY4741 cells (Figure S6). The results of both the NaCl solution and proteinase K experiments indicated that CUP1 was contained within the mitochondrial IMS rather than adhered to the exterior OM surface.
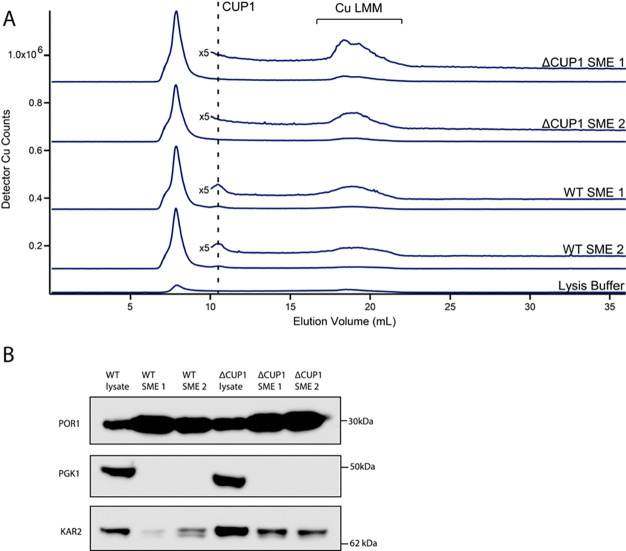
To provide insight as to the potential function of CUP1 within the mitochondria, we prepared SMEs from WT and ΔCUP1 cells in a DTY005 background. Mirroring observations previously made from the isolated cytosol, the trace of the ΔCUP1 SME (Figure 5A, top traces) lacked the CUP1 peak which was observed in the WT control (Figure 5A, bottom traces). The presence of CUP1 within DTY005 mitochondria excluded the possibility that the colocalization of CUP1 was unique to the W303 strain.
Figure 5.
SME of isolated WT and ΔCUP1 mitochondria. (A) LC-ICP-MS copper-detected chromatogram of filtered SMEs generated from independent replicates of isolated ΔCUP1 (top traces) and WT (bottom traces). The insets above traces are magnified regions depicting CuLMM, multiplied by the indicated factor. Use of an Increase column resulted in minor elution volume shifts relative to that in Figure 2. (B) Western blot of unfiltered samples from A. Each lane was loaded with 8.7 μg of protein.
We additionally observed LMM copper complexes in the DTY005 background SME (Figure 5, bracketed regions). Deletion of CUP1 resulted in an approximately twofold increase in the LMM copper complex intensity.
This suggested that the mitochondrial concentration of these complexes may be inversely related to the abundance of CUP1 in the IMS. Supporting this, LMM copper complexes were observed in SMEs from W303 cells cultured under a condition where CUP1 abundance was reduced (Figure S2).
Since CUP1 is encoded in the nucleus and present in the cytosol, we considered that cytosolic CUP1 might enter mitochondria by passing through OM pores. The limit for protein passage through these pores has been estimated at 4–12 kDa.33 To investigate this, we obtained a strain of yeast in which CUP1 was fused with the GFP at the C-terminus.
The molecular mass of the CUP1-GFP chimera (∼ 34 kDa) was sufficiently high to exclude the possibility that it could enter mitochondria through OM pores. One complication was that S. cerevisiae contains two identical genetic copies of CUP1, called CUP1-1 and CUP1-2. Separate strains in which each gene was fused with GFP were obtained, but a strain in which both were GFP tagged was unavailable. CUP1 chromatographic peaks were observed in SMEs obtained from the BY4741 background strain (Figure 6A). This further confirmed that the localization of CUP1 in mitochondria was not unique to a single strain; it was detected independently within W303, DTY005, and BY4741 mitochondria. SMEs from CUP1-1-GFP and CUP1-2-GFP cells exhibited CUP1 peaks that were less intense relative to the high-mass unresolved void peak as the WT SME isolated from BY4741 parent cells (Figure 6A). The observed CUP1 peaks arose from expression of the CUP1 gene that was not fused with GFP, i.e., CUP1-2 in the CUP1-1-GFP strain and CUP1-1 in the CUP1-2-GFP strain. Also evident are low-intensity peaks in the LMM region which might reflect nonproteinaceous copper coordination complexes (Figure 6A, bracketed regions). Interestingly, the fusion of GFP to CUP1 also perturbed these LMM complexes relative to the WT control, though the effect was less prominent than observed with the DTY005 background.
Figure 6.
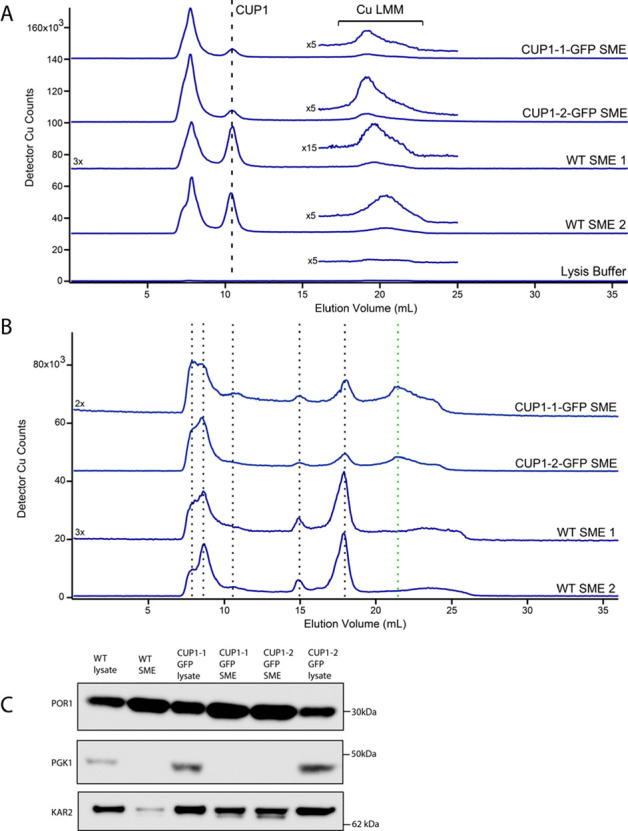
LC-ICP-MS chromatograms of BY4741 WT and CUP1-GFP chimera SMEs. (A) Copper-detected LC-ICP-MS chromatograms of SMEs generated from CUP1-1-GFP, CUP1-2-GFP, and two batches of WT BY4741 background cells. The low-mass Increase column was used. The vertical line indicates the position of CUP1, whereas the bracket indicates a range of elution volumes in which LMM copper complexes are detected. The insets above traces show magnified regions depicting CuLMM, multiplied by the indicated factor. (B) Copper-detected LC-ICP-MS chromatograms of the SME (same samples as in A). The high-mass column was used. (C) Western blot of unfiltered samples used in parts A and B. Each lane was loaded with 5.9 μg of protein.
The same solutions, when chromatographed on a column capable of resolving higher masses, exhibited a half-dozen copper-detected peaks (Figure 6B). The peaks were generally reproducible for SMEs obtained from WT and CUP1-GFP mutant cells. They arose from unassigned high-molecular-mass copper proteins or complexes in the mitochondria. The only qualitative difference was the presence of a Cu-detected peak in the CUP1-GFP SMEs that could potentially be assigned to the chimera (green vertical dashed line). A Western blot suggested that the detected mitochondrial CUP1 was not due to cytosolic contamination, as a PGK1 band was not observed in SMEs used in the study (Figure 6C).
To investigate whether CUP1-GFP was present in SMEs, we collected fluorescence confocal microscopic images of isolated intact mitochondria from CUP1-GFP cells that had been labeled with MitoTracker. The two fluorescent labels overlapped in merged images in both clones (Figure 7A,B), providing further evidence that CUP1-GFP was indeed able to enter mitochondria. Given the large size of the CUP1-GFP chimera, our experiments suggest that CUP1-GFP (and by extension CUP1 itself) may enter mitochondria by a mechanism that does not involve passage through OM pores. We also examined whole CUP1-GFP cells using the same imaging method. Fluorescence from CUP1-GFP was evident throughout the cells, including the cytosol and mitochondria, but vacuoles lacked fluorescence, suggesting that the concentration of intact CUP1-GFP in these organelles was very low, if not absent (Figure 7C).
Figure 7.
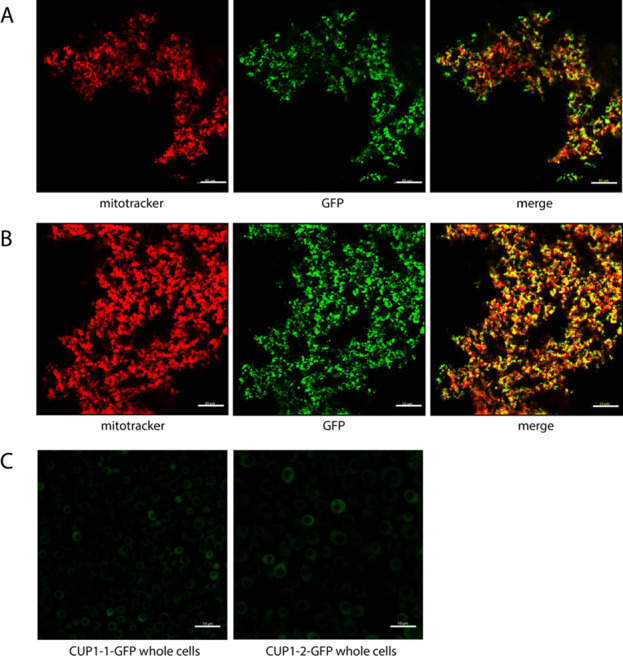
Confocal microscopic images of mitochondria and whole cells from CUP1-GFP clones. (A) Isolated CUP1-1-GFP and (B) CUP1-2-GFP mitochondria incubated with MitoTracker Red. The left image is MitoTracker fluorescence; the middle image is GFP fluorescence, and the right image is the merge. (C) GFP fluorescence of CUP1-1-GFP (left) and CUP1-2-GFP (right) whole cells. Scale bars for all images: 10 μm.
Discussion
Dual Localization of Cu8-CUP1
The main conclusion of this paper is that holo-metallothionein CUP1 is the major copper species in the cytosol and is the dominant soluble copper species in mitochondria of healthy WT respiring S. cerevisiae cells grown under copper-replete but nontoxic conditions (minimal media, with glycerol/ethanol as a carbon source and 10 or 50 μM CuSO4).
The first inkling of mitochondrial metallothioneins was in 1961 when “mitochondrocuprein”, a copper-bound sulfur-rich protein, was reported in the mitochondria of newborn livers.34 This protein was soon associated with metallothioneins (MTs).35,36 Sakurai et al.37 used immunohistochemical staining to demonstrate that MTs were present in the cytosol, mitochondria, and nuclei of hepatocytes from rats with hepatitis. Ye et al.33 demonstrated that zinc ions bound to mammalian MTs38 could be imported into isolated liver mitochondria under in vitro conditions. Reinecke et al.39 found that MT expression in HeLa cells was induced when cells were treated with the mitochondrial-acting pesticide rotenone. They concluded that the MT protects mitochondria against ROS and provides a response to various diseased states.
Despite these earlier reports, Cu-bound MTs are commonly thought to be exclusively cytosolic. Wright et al.40 reported no evidence for mitochondrial localization of CUP1. Cobine et al.13 concluded that CUP1 and CRS5 do not contribute to the mitochondrial copper pool. Copper-bound MTs in mitochondria are not described in any major review of copper metabolism in this organelle,7,17,41−44 and proteomics studies do not include MTs as a mitochondrial protein.45,46
The discord regarding MTs and mitochondrial localization has been compounded by uncertainties regarding how cytosolic metal-bound MTs could enter this organelle and what it would do there. MTs play protective cellular roles in metal toxicity and ROS damage, and they also sequester metals as an essential component of homeostatic regulatory mechanisms. However, this functional stereotype may limit consideration of other cellular roles. Lindeque et al.36 lamented that “perhaps the uncertainty of their function or (mode of) import into the mitochondrion makes it hard to finally conclude that MTs are also mitochondrial proteins”. For these reasons, a major focus of the current study was to demonstrate unequivocally the presence of CUP1 in both the cytosol and the IMS of mitochondria of healthy nondistressed (diseased or heavy-metal intoxicated) WT S. cerevisiae cells.
How Does Cytosolic CUP1 Enter Mitochondria?
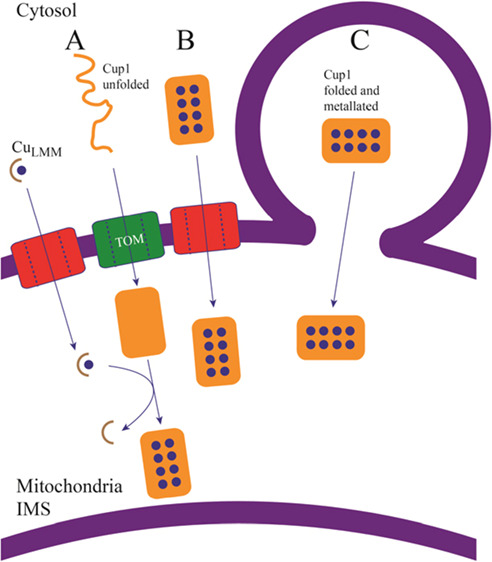
We have considered four scenarios. The first is that CUP1 passes folded and metalated through a channel/pore on the OM (Figure 8B). Ye et al.33 suggested that Zn-MT in its native conformation is translocated through the OM while retaining the bound metal. Small proteins like CUP1 or COX17 could conceivably pass through open-state pores, especially given their dynamic structural flexibility.47,48 However, our results and those of Bragina et al.49 disfavor this import pathway. Given the large size of the CUP1-GFP chimera and its ability to enter mitochondria, it seems unlikely that this protein enters through an OM pore, though it is feasible that CUP1 and CUP1-GFP could enter the mitochondria via differing mechanisms. Perhaps in support of this, the detected CUP1 in WT SMEs is more than twofold that of each of the respective chimera strains (Figure 6A).
Figure 8.
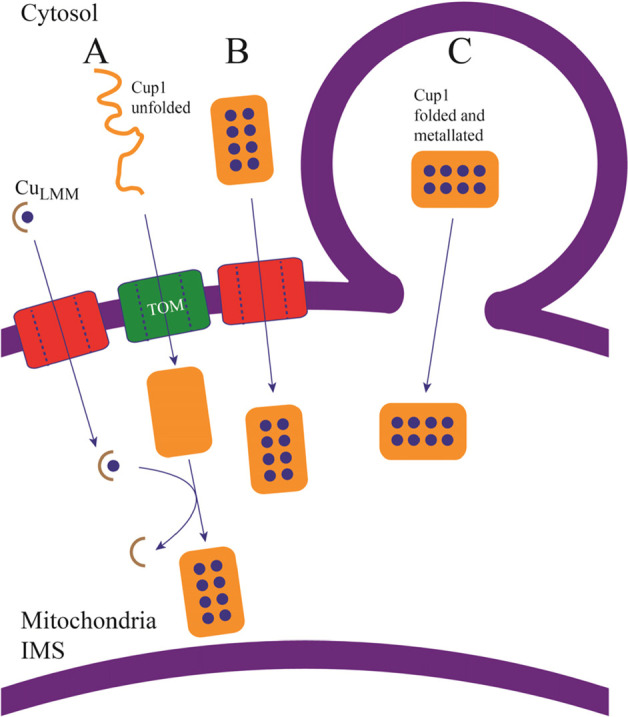
Possible scenarios of CUP1 import into mitochondria. (A) apo-CUP1 may enter unfolded and become folded and metalated once in the IMS. CUP1 would then be metalated within the mitochondria by an unidentified Cu complex, suggested here to be a low-mass CuLMM complex. (B) CUP1 may enter folded and metalated via VDAC pores in the outer mitochondrial membrane. Current evidence disfavors this mode of entry. (C) CUP1 may enter folded and metalated via the secretory pathway, by membrane fusion and direct organelle contact.
The second possibility is that cytosolic CUP1 enters mitochondria via the MIA40/ERV1 pathway10,50−52 unfolded, apo, and with all cysteine residues reduced (Figure 8A). Once in the IMS, apo-CUP1 would fold and become metalated by unidentified IMS copper species. CUP1 contains one of the two major cysteine motifs of MIA40 client protein, but they are contained within other cysteine residues that are not MIA40-related. Moreover, cysteines in the canonical motifs of MIA40 client proteins become oxidized to disulfides after entering the IMS, whereas for CUP1, they remain reduced and coordinated collectively to eight Cu(I) ions. For these reasons, as well as the lack of detected apo-CUP1 in both cellular lysates and the isolated cytosol, we disfavor an MIA40-dependent import mechanism for CUP1.
The third possibility is that CUP1 enters mitochondria via a MIA40-independent pathway such as appears to be used by cytochrome c. CUP1 is encoded with eight additional residues at the N-terminus, but we only detected truncated CUP1. No effect upon cellular copper resistance or its structural integrity was observed when CUP1 was modified with an uncleavable N-terminal extension,40 suggesting an alternative purpose for the extension. Although the reason for cleaving the residues remains unknown, the truncated residues bear resemblance to the presequence of cytochrome c, and the possibility that this is associated with trafficking into mitochondria should be considered further.
The fourth and final considered possibility is that CUP1 enters mitochondria folded and copper bound via the secretory pathway (Figure 8C), though the absence of vacuolar CUP1-GFP in our studies does not support this mechanism.
What Is the Function of CUP1 in the IMS of Mitochondria?
We were initially intrigued by the possibility that CUP1 might deliver cytosolic copper into mitochondria for installation into apo-COX; however, this now seems unlikely. Dual localization is not unusual for mitochondrial proteins; about a third have this property.53,54 If CUP1 were the sought-after copper trafficking species, we would expect a respiration defect in ΔCUP1 cells, but this has not been reported. In our hands, ΔCUP1 cells grew like WT cells on respiring media, though the possibility of a compensatory role for the other MT CRS5 has not been excluded. Moreover, MTs bind and sequester Cu ions extremely tightly.55−57 Such ultratight binding seems inconsistent with a trafficking function. A conformational change or other means of releasing the Cu bound to CUP1 would be required to consider it for a trafficking role.
CUP1 may function to maintain low levels of LMM copper complexes in mitochondria, to lessen copper toxicity, oxidative damage, or metalloprotein mismetalation, consistent with the rescue due to expressing CUP1 in ΔSOD1 mutants.58,59 Our results suggest an inverse relationship between the level of mitochondrial CUP1 and the size of the mitochondrial LMM copper pool. Future studies are needed to evaluate these possibilities. The possibility that LMM Cu complexes provide a source of copper for COX also merits further investigation.
Modification of the CuL Hypothesis
Another major conclusion of this study is that most mitochondrial copper is not in a nonproteinacous LMM form within the matrix. Besides detecting CUP1 in SMEs, we detected numerous soluble copper proteins and low concentrations of LMM nonproteinaceous copper species (which we call CuLMM). These results support some but not all aspects of the CuL hypothesis, and they suggest some modification.
For example, CuLMM might enter the IMS through OM pores and pass copper (directly or indirectly) to COX17. In this scenario, CUP1 would serve to limit the levels of CuLMM in both the cytosol and IMS, such that these LMM complexes would only be present when the CUP1 binding capacity is at or near saturation. This would maintain the levels of CuLMM low enough to prevent copper toxicity in both compartments. LMM Cu complexes have the potential to damage iron–sulfur cluster assembly,60 an essential process in the mitochondrial matrix. Stewart et al.61 has pointed out that, except for cyanobacteria, “cuproproteins are not known to exist inside the bacterial cytoplasm”. In fact, many bacteria contain Cu-transporting P-type ATPases which pump copper out of the cytoplasm62 and into the periplasm, the evolutionary relative of the mitochondrial IMS. This might reflect a strategy of nature to balance the necessity of using copper to catalyze COX-based respiratory reactions while minimizing its toxicity to sensitive processes occurring in the mitochondrial matrix. By scavenging Cu from CuLMM complexes, CUP1 would minimize the import of such complexes into the matrix and the subsequent effect upon the essential processes within. More work is required to determine the localization of the CuLMM species in mitochondria; regardless of whether CuLMM is(are) located in the IMS or matrix, our data indicate that the concentration of such species in mitochondria is low.
In summary, we have determined that yeast MT CUP1 localizes in both the cytosol and the IMS of mitochondria. Although the mechanism by which CUP1 enters mitochondria, its metalation state at entry, and its cellular role once in the IMS remain uncertain, what is certain is that the presence of this protein in mitochondria, coupled with the low levels of LMM labile copper complexes in the organelle previously thought to dominate the copper content of mitochondria, suggests that we re-evaluate and restructure our overall understanding of copper metabolism in this critically important organelle.
Acknowledgments
We thank Dr. Dennis Thiele for providing the DTY005 background WT and ΔCUP1 strains, Dr. Yohannes Rezenom for assistance with LC-ESI-MS, Dr. Robert Burghardt for assistance with microscopy, and Dr. Beiyan Nan for assistance with image processing.
Glossary
Abbreviations
- 20AA6.5
20 mM ammonium acetate pH 6.5
- COX
cytochrome c oxidase
- Cu5000
dominant copper species detected in mitochondria by liquid chromatography
- ESI-MS
electrospray ionization mass spectrometry
- FTS
flow-through solution
- GFP
green fluorescent protein
- ICP-MS
inductively coupled plasma mass spectrometry
- IM
inner mitochondrial membrane
- IMS
intermembrane space
- LMM
low molecular mass
- MT
metallothionein
- OM
outer membrane
- SAA2
buffer consisting of 0.6 M sorbitol and 20 mM ammonium acetate pH 6.5
- SME
soluble mitochondrial extract
- SmpE
soluble mitoplast extract
- WT
wild type
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.biochem.2c00481.
(Figure S1) Use of alternative gradients in mitochondrial isolation; (Figure S2) effect of the media type on mitochondrial Cu5000 abundance; (Figure S3) LC-ESI-MS spectra of cell lysates with and without lyophilization; (Figure S4) Apo-CUP1 not detected in the cytosol or cell lysates; (Figure S5) addition of BCS to the WT cytosol; and (Figure S6) mitoplasting of washed or unwashed BY4741 mitochondria (PDF)
Accession Codes
Author Contributions
J.E.K. performed all of the experiments, analyzed the data, prepared most figures, and wrote some of the paper. P.A.L. directed the project, helped analyze the data, and wrote the first draft of the paper. Both authors approved the final version of the manuscript.
This work was funded by the National Institutes of Health (GM127021) and the Robert A. Welch Foundation (A1170). The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or the Welch Foundation.
The authors declare no competing financial interest.
Supplementary Material
References
- Heaton D. N.; George G. N.; Garrison G.; Winge D. R. The mitochondrial copper metallochaperone Cox17 exists as an oligomeric, polycopper complex. Biochemistry 2001, 40, 743–751. 10.1021/bi002315x. [DOI] [PubMed] [Google Scholar]
- Banci L.; Bertini I.; Ciofi-Baffoni S.; Janicka A.; Martinelli M.; Kozlowski H.; Palumaa P. A structural-dynamical characterization of human Cox17. J. Biol. Chem. 2008, 283, 7912–7920. 10.1074/jbc.M708016200. [DOI] [PubMed] [Google Scholar]
- Glerum D. M.; Shtanko A.; Tzagoloff A. Characterization of Cox17, a yeast gene involved in copper metabolism and assembly of cytochrome oxidase A. J. Biol. Chem. 1996, 271, 14504–14509. 10.1074/jbc.271.24.14504. [DOI] [PubMed] [Google Scholar]
- Glerum D. M.; Shtanko A.; Tzagoloff A. SCO1 and SCO2 act as high copy suppressors of a mitochondrial copper recruitment defect in Saccharomyces cerevisiae. J. Biol. Chem. 1996, 271, 20531–20535. 10.1074/jbc.271.34.20531. [DOI] [PubMed] [Google Scholar]
- Punter F. A.; Glerum D. M. Mutagenesis reveals a specific role for Cox17p in copper transport to cytochrome oxidase. J. Biol. Chem. 2003, 278, 30875–30880. 10.1074/jbc.M302358200. [DOI] [PubMed] [Google Scholar]
- Soma S.; Morgada M. N.; Naik M. T.; Boulet A.; Roesler A. A.; Dziuba N.; Ghosh A.; Yu Q.; Lindahl P. A.; Ames J. B.; Leary S. C.; Vila A. J.; Gohil V. M. COA6 Is Structurally Tuned to Function as a Thiol-Disulfide Oxidoreductase in Copper Delivery to Mitochondrial Cytochrome c Oxidase. Cell Rep. 2019, 29, 4114.e5–4126.e5. 10.1016/j.celrep.2019.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leary S. C. Redox Regulation of SCO Protein Function: Controlling Copper at a Mitochondrial Crossroads. Antioxid. Redox Signaling 2010, 13, 1403–1416. 10.1089/ars.2010.3116. [DOI] [PubMed] [Google Scholar]
- Field L. S.; Furukawa Y.; O’Halloran T. V.; Culotta V. C. Factors controlling the uptake of yeast copper/zinc superoxide dismutase into mitochondria. J. Biol. Chem. 2003, 278, 28052–28059. 10.1074/jbc.M304296200. [DOI] [PubMed] [Google Scholar]
- Beers J.; Glerum D. M.; Tzagoloff A. Purification, characterization, and localization of yeast Cox17p, a mitochondrial copper shuttle. J. Biol. Chem. 1997, 272, 33191–33196. 10.1074/jbc.272.52.33191. [DOI] [PubMed] [Google Scholar]
- Koch J. R.; Schmid F. X. Mia40 targets cysteines in a hydrophobic environment to direct oxidative protein folding in the mitochondria. Nat. Commun. 2014, 5, 3041. 10.1038/ncomms4041. [DOI] [PubMed] [Google Scholar]
- Morgan B.; Ang S. K.; Yan G.; Lu H. Import of Mitochondrial Small Tim Proteins. J. Biol. Chem. 2009, 284, 6818–6825. 10.1074/jbc.M808691200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxfield A. B.; Heaton D. N.; Winge D. R. Cox17 is functional when tethered to the mitochondrial inner membrane. J. Biol. Chem. 2004, 279, 5072–5080. 10.1074/jbc.M311772200. [DOI] [PubMed] [Google Scholar]
- Cobine P. A.; Ojeda L. D.; Rigby K. M.; Winge D. R. Yeast contain a non-proteinaceous pool of copper in the mitochondrial matrix. J. Biol. Chem. 2004, 279, 14447–14455. 10.1074/jbc.M312693200. [DOI] [PubMed] [Google Scholar]
- Cobine P. A.; Pierrel F.; Bestwick M. L.; Winge D. R. Mitochondrial Matrix Copper Complex Used in Metallation of Cytochrome Oxidase and Superoxide Dismutase. J. Biol. Chem. 2006, 281, 36552–36559. 10.1074/jbc.M606839200. [DOI] [PubMed] [Google Scholar]
- Vest K. E.; Wang J.; Gammon M. G.; Maynard M. K.; White O. L.; Cobine J. A.; Mahone W. K.; Cobine P. A. Overlap of copper and iron uptake systems in mitochondria in Saccharomyces cerevisiae. Open Biol. 2016, 6, 150223 10.1098/rsob.150223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vest K. E.; Leary S. C.; Winge D. R.; Cobine P. A. Copper Import into the Mitochondrial Matrix in Saccharomyces cerevisiae is Mediated by Pic2, a Mitochondrial Carrier Family Protein. J. Biol. Chem. 2013, 288, 23884–23892. 10.1074/jbc.M113.470674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leary S. C.; Winge D. R.; Cobine P. A. “Pulling the plug” on cellular copper: The role of mitochondria in copper export. Biochim. Biophys. Acta, Mol. Cell Res. 2009, 1793, 146–153. 10.1016/j.bbamcr.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodani S. C.; Leary S. C.; Cobine P. A.; Winge D. R.; Chang C. J. A targetable fluorescent sensor reveals that copper-deficient SCO1 and SCO2 patient cells prioritize mitochondrial copper homeostasis. J. Am. Chem. Soc. 2001, 133, 8606–8616. 10.1021/ja2004158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L.; McRae R.; Henry M. M.; Patel R.; Lai B.; Vogt S.; Fahrni C. J. Imaging of the intracellular topography of copper with a fluorescent sensor and by synchrotron X-ray fluorescence microscopy. Proc. Natl. Acad. Sci. U. S. A. 2005, 102, 11179–11184. 10.1073/pnas.0406547102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick S. P.; Moore M. J.; Lindahl P. A. Detection of labile low-molecular-mass transition metal complexes in mitochondria. Biochemistry 2015, 54, 3442–3453. 10.1021/bi5015437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl P. A.; Moore M. J. Labile Low-Molecular-Mass metal complexes in mitochondria: Trials and tribulations of a burgeoning field. Biochemistry 2016, 55, 4140–4153. 10.1021/acs.biochem.6b00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M. J.; Wofford J. D.; Dancis A.; Lindahl P. A. Recovery of mrs3Δmrs4Δ Saccharomyces cerevisiae Cells under Iron-Sufficient Conditions and the Role of Fe580. Biochemistry 2018, 57, 672–683. 10.1021/acs.biochem.7b01034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T. Q.; Kim J. E.; Brawley H. N.; Lindahl P. A. Chromatographic detection of low-molecular-mass metal complexes in the cytosol of Saccharomyces cerevisiae. Metallomics 2020, 12, 1094–1105. 10.1039/c9mt00312f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T. Q.; Dziuba N.; Lindahl P. A. Isolated Saccharomyces cerevisiae vacuoles contain low-molecular-mass transition-metal polyphosphate complexes. Metallomics 2019, 11, 2298–1309. 10.1039/c9mt00104b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer D. H.; Thiele D. J.; Lemontt J. E. Function and auto-regulation of yeast copperthionein. Science 1985, 228, 685–690. 10.1126/science.3887570. [DOI] [PubMed] [Google Scholar]
- Kim J. E.; Vali S. W.; Nguyen T. Q.; Dancis A.; Lindahl P. A. Mössbauer and LC-ICP-MS investigation of iron trafficking between vacuoles and mitochondria in Vma2Δ Saccharomyces cerevisiae. J. Biol. Chem. 2021, 296, 100141. 10.1074/jbc.RA120.015907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl P.A.; Morales J.G.; Miao R.; Holmes-Hampton G.. Isolation of Saccharomyces cerevisiae mitochondria for Mössbauer, EPR, and electronic absorption spectroscopic analyses. In Methods of Enzymology, Allison W. S., Ed.; 2009; Vol. 456, pp 267–285. [DOI] [PubMed]
- Brawley H. N.; Lindahl P. A. Low-molecular-mass labile metal pools in Escherichia coli: advances using chromatography and mass spectrometry. J. Biol. Inorg. Chem. 2021, 26, 479–494. 10.1007/s00775-021-01864-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen C.; Bjørnsdottir I.; Jøns O.; Hansen S. Detection of Metallothionein Isoforms from Three Different Species Using On-Line Capillary Electrophoresis-Mass Spectrometry. Anal. Biochem. 1998, 265, 167–175. 10.1006/abio.1998.2874. [DOI] [PubMed] [Google Scholar]
- Glick B. Pathways and Energetics of Mitochondrial Protein Import in Saccharomyces cerevisiae. Methods Enzymol. 1995, 260, 224–231. 10.1016/0076-6879(95)60140-6. [DOI] [PubMed] [Google Scholar]
- Diekert K.; de Kroon A.; Kispal G.; Lill R. Isolation and Subfractionation of Mitochondria from the Yeast Saccharomyces cerevisiae. Methods Cell Biol. 2001, 65, 37–51. 10.1016/S0091-679X(01)65003-9. [DOI] [PubMed] [Google Scholar]
- Sturtz L. A.; Diekert K.; Jensen L. T.; Lill R.; Culotta V. C. A fraction of yeast Cu,Zn-super-oxide dismutase and its metallochaperone, CCS, localize to the intermembrane space of mitochondria. A physiological role for sod1 in guarding against mitochondrial oxidative damage. J. Biol. Chem. 2001, 276, 38084–38089. 10.1074/jbc.M105296200. [DOI] [PubMed] [Google Scholar]
- Ye B.; Maret W.; Vallee B. L. Zinc metallothionein imported into liver mitochondria modulates respiration. Proc. Natl. Acad. Sci. U. S. A. 2001, 98, 2317–2322. 10.1073/pnas.041619198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter H.; Wiener W.; Barker M. Intracellular distribution of copper in immature liver. Biochim. Biophys. Acta 1961, 52, 419–423. 10.1016/0006-3002(61)90398-5. [DOI] [PubMed] [Google Scholar]
- Porter H.; Johnston J.; Porter E. M. Neonatal hepatic mitochondrocuprein 1. Isolation of a protein fraction containing more than 4 percent copper from mitochondria of immature bovine liver. Biochim. Biophys. Acta 1962, 65, 66–73. 10.1016/0006-3002(62)90149-X. [DOI] [PubMed] [Google Scholar]
- Lindeque J. Z.; Levanets O.; Louw R.; van der Westhuizen F. H. The Involvement of Metallothioneins in Mitochondrial Function and Disease. Curr. Protein Pept. Sci. 2010, 11, 292–309. 10.2174/138920310791233378. [DOI] [PubMed] [Google Scholar]
- Sakurai H.; Nakajima K.; Kamada H.; Satoh H.; Otaki N.; Kimura M.; Kawano K.; Hagino T. Copper-metallothionein distribution in the liver of Long-Evans cinnamon rats – studies on immunohistochemical staining, metal determination, gel-filtration and electron spin resonance spectroscopy. Biochem. Biophys. Res. Commun. 1993, 192, 893–898. 10.1006/bbrc.1993.1499. [DOI] [PubMed] [Google Scholar]
- Skutkova H.; Babula P.; Stiborova M.; Eckschlager T.; Trnkova L.; Provaznik I.; Hubalek J.; Kizek R.; Adam V. Structure, Polymorphisms and Electrochemistry of Mammalian Metallothioneins – A Review. Intl. J. Electrochem. Sci. 2012, 7, 12415–12431. [Google Scholar]
- Reinecke F.; Levanets O.; Olivier Y.; Louw R.; Semete B.; Grobler A.; Hidalgo J.; Smeitink J.; Olckers A.; Van der Westhuizen F. H. Metallothionein isoform 2A expression is inducible and protects against ROS-mediated cell death in rotenone treated HeLa cells. Biochem. J. 2006, 395, 405–415. 10.1042/BJ20051253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright C. F.; McKenney K.; Hamer D. H.; Byrd J.; Winge D. R. Structural and functional studies of the amino terminus of yeast metallothionein. J. Biol. Chem. 1987, 262, 12912–12919. 10.1016/S0021-9258(18)45144-7. [DOI] [PubMed] [Google Scholar]
- Baker Z. N.; Cobine P. A.; Leary S. C. The mitochondrion: a central architect of copper homeostasis. Metallomics 2017, 9, 1501–1512. 10.1039/C7MT00221A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson N. J.; Winge D. R. Copper metallochaperones. Annu. Rev. Biochem. 2010, 79, 537–562. 10.1146/annurev-biochem-030409-143539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H.; Jiang Y. H.; Yang Y.; Peng Y. G.; Li C. Copper metabolism in Saccharomyces cerevisiae: an update. BioMetals 2021, 34, 3–14. 10.1007/s10534-020-00264-y. [DOI] [PubMed] [Google Scholar]
- Cobine P. A.; Moore S. A.; Leary S. C. Getting out what you put in: Copper in mitochondria and its impacts on human disease. Biochim. Biophys. Acta, Mol. Cell Res. 2021, 1868, 118867 10.1016/j.bbamcr.2020.118867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sickmann A.; Reinders J.; Wagner Y.; Joppich C.; Zahedi R.; Meyer H. E.; Schönfisch B.; Perschil I.; Chacinska A.; Guiard B.; Rehling P.; Pfanner N.; Meisinger C. The proteome of Saccharomyces cerevisiae Mitochondria. Proc. Natl. Acad. Sci. U. S. A. 2003, 100, 13207–13212. 10.1073/pnas.2135385100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenstern M.; Stiller S. B.; Lubbert P.; Warscheid B. Definition of a high-confidence mitochondrial proteome at quantitative scale. Cell Rep. 2017, 19, 2836–2852. 10.1016/j.celrep.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Isart N.; Vasak M. Advances in the structure and chemistry of metallothioneins. J. Inorg. Biochem. 2002, 88, 388–396. 10.1016/S0162-0134(01)00347-6. [DOI] [PubMed] [Google Scholar]
- Irvine G. W.; Stillman M. J. Residue Modification and Mass Spectrometry for the Investigation of Structural and Metalation Properties of Metallothionein and Cysteine-Rich Proteins. Int. J. Mol. Sci. 2017, 18, 913. 10.3390/ijms18050913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragina O.; Gurjanova K.; Krishtal J.; Kulp M.; Karro N.; Tougu V.; Palumaa P. Metallothionein 2A affects the cell respiration by suppressing the expression of mitochondrial protein cytochrome c oxidase subunit II. J. Bioenergy Biomembr. 2015, 47, 209–216. 10.1007/s10863-015-9609-9. [DOI] [PubMed] [Google Scholar]
- Fraga H.; Ventura S. Protein Oxidative Folding in the Intermembrane Mitochondrial Space: More than Protein Trafficking. Curr. Protein Pept. Sci. 2012, 13, 224–231. 10.2174/138920312800785012. [DOI] [PubMed] [Google Scholar]
- Pfanner N.; Chacinska A. The mitochondrial import machinery: preprotein-conducting channels with binding sites for presequences. Biochim. Biophys. Acta, Mol. Cell Res. 2002, 1592, 15–24. 10.1016/S0167-4889(02)00260-4. [DOI] [PubMed] [Google Scholar]
- Neupert W.; Herrmann J. M. Translocation of proteins into mitochondria. Annu. Rev. Biochem. 2007, 76, 723–749. 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- Ben-Menachem R.; Tal M.; Shadur T.; Pines O. A third of the yeast mitochondrial proteome is dual localized: a question of evolution. Proteomics 2011, 11, 4468–4476. 10.1002/pmic.201100199. [DOI] [PubMed] [Google Scholar]
- Kisslov I.; Naamati A.; Shakarchy N.; Pines O. Dual-targeted proteins tend to be more evolutionarily conserved. Mol. Biol. Evol. 2014, 31, 2770–2779. 10.1093/molbev/msu221. [DOI] [PubMed] [Google Scholar]
- Festa R.; Thiele D. Copper An Essential Metal in Biology. Curr. Biol. 2011, 21, R877–R883. 10.1016/j.cub.2011.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S.; Kumari S.; Rath S.; Priyadarshanee M.; Das S. Diversity, Structure and Regulation of Microbial Metallothionein: Metal Resistance and Possible Applications in Sequestration of Toxic Metals. Metallomics 2020, 12, 1637–1655. 10.1039/d0mt00140f. [DOI] [PubMed] [Google Scholar]
- Mehlenbacher M.; Elsiesy R.; Lakha R.; Villones R.; Orman M.; Vizcarra C.; Meloni G.; Wilcox D.; Austin R. Metal binding and interdomain thermodynamics of mammalian metallalothionein-3: enthalpically favoured Cu+ supplants entropically favoured Zn2+ to form Cu4+ clusters under physiological conditions. Chem. Sci. 2022, 13, 5289–5304. 10.1039/D2SC00676F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.; Thiele D. Oxidative Stress Induces Heat Shock Factor Phosphorylation and HSF-Dependent Activation of Yeast Metallothionein Gene Transcription. Genes Dev. 1996, 10, 592–603. 10.1101/gad.10.5.592. [DOI] [PubMed] [Google Scholar]
- Tamai K.; Gralla E.; Ellerby L.; Valentine J.; Thiele D. Yeast and Mammalian Metallothioneins Functionally Substitute for Yeast Copper-Zinc Superoxide Dismutase. Proc. Natl. Acad. Sci. U. S. A. 1993, 90, 8013–8017. 10.1073/pnas.90.17.8013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brancaccio D.; Gallo A.; Piccioli M.; Novellino E.; Ciofi-Baffoni S.; Banci L. [4Fe-4S] Cluster Assembly in Mitochondria and Its Impairment by Copper. J. Am. Chem. Soc. 2017, 139, 719–730. 10.1021/jacs.6b09567. [DOI] [PubMed] [Google Scholar]
- Stewart L. J.; Thaqi D.; Kobe B.; McEwan A. G.; Waldron K. J.; Djoko K. Y. Handling of nutrient copper in the bacterial envelope. Metallomics 2019, 11, 50–63. 10.1039/C8MT00218E. [DOI] [PubMed] [Google Scholar]
- Abeyrathna N.; Abeyrathna S.; Morgan M. T.; Fahrni C. J.; Meloni G. Transmembrane Cu(I) P-type ATPase pumps are electrogenic uniporters. Dalton Trans. 2020, 49, 16082–16094. 10.1039/D0DT01380C. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.