Abstract
Non-alcoholic steatohepatitis (NASH) is the clinically aggressive variant of non-alcoholic fatty liver disease. Hippo pathway dysregulation can contribute to NASH development and progression. The use of probiotics is effective in NASH management. Our aim is to investigate the efficacy of kefir Milk in NASH management via modulation of hepatic mRNA-miRNA based panel linked to NAFLD/NASH Hippo signaling and gut microbita regulated genes which was identified using bioinformatics tools. Firstly, we analyzed mRNAs (SOX11, SMAD4 and AMOTL2), and their epigenetic regulator (miR-6807) followed by validation of target effector proteins (TGFB1, IL6 and HepPar1). Molecular, biochemical, and histopathological, analyses were used to evaluate the effects of kefir on high sucrose high fat (HSHF) diet -induced NASH in rats. We found that administration of Kefir proved to prevent steatosis and development of the inflammatory component of NASH. Moreover, Kefir improved liver function and lipid panel. At the molecular level, kefir down-regulated the expression of miR 6807-5p with subsequent increase in the expression of SOX 11, AMOTL2 associated with downregulated SMAD4, resulting in reduction in the expression of the inflammatory and fibrotic markers, IL6 and TGF-β1 in the treated and prophylactic groups compared to the untreated rats. In conclusion, Kefir suppressed NASH progression and improved both fibrosis and hepatic inflammation. The produced effect was correlated with modulation of SOX11, SMAD4 and AMOTL2 mRNAs) – (miR-6807-5p) – (TGFB, IL6 and, HepPar1) expression.
Subject terms: Molecular biology, Gastroenterology, Molecular medicine
Introduction
Nonalcoholic fatty liver disease (NAFLD) is a pathological condition with accumulation of fat in the liver with no excessive alcohol intake, varying from simple hepatic steatosis (SS) to nonalcoholic steatohepatitis (NASH) with fibrosis and cirrhosis1. Due to the increasing epidemic of obesity and type 2 diabetes worldwide, a 178% rise in liver deaths has been estimated because of NASH by 20302. NASH could be induced by multiple conditions acting in parallel, including genetic predisposition, oxidative stress, abnormal lipid metabolism, lipotoxicity, mitochondrial dysfunction, endoplasmic reticulum stress, altered production of cytokines and adipokines and gut dysbiosis. Accordingly, hepatic inflammation could precede steatosis in NASH3. At the present time, no FDA-approved medical treatment for NASH or liver fibrosis are confirmed4.
Hippo signaling pathway was previously reported to have a critical role in the regulation of hepatic size, proliferation, apoptosis, and stress response5. Accordingly, its dysregulation could have an effect on NASH pathogenesis via liver dedifferentiation and tumorigenesis6.
Moreover, former data have reported that noncoding RNAs (ncRNA), including microRNAs (miRNAs) and long noncoding RNAs (lncRNAs)7 may play crucial regulatory roles in NASH initiation and progression. The coregulatory links between these ncRNAs may demonstrate the molecular regulation and its involvement in NASH progression and could be used as appropriate biomarkers for assessing severity of the disease8.
Additionally, the use of intestinal microbiota as a potential therapeutic target for NASH have been achievable. Probiotics are live microorganisms, which give health benefits to the host if consumed in appropriate amounts9. Probiotics slow down disease progression and hinder gastrointestinal complications by affecting intestinal flora, intestinal permeability, and inflammatory response10. Currently, the use of probiotics in the management of NAFLD have not been addressed by any guidelines and the molecular mechanisms of their health benefits are not entirely known as well11.
Kefir is an acidic-alcoholic fermented milk, produced by kefir grains. Therefore, it is stable and has a precise balance of lactic acid bacteria and yeast12. It contains around 30 unique species of “good bacteria” that could benefit gut health13 and so used as a potential therapy to treat gastrointestinal diseases and ischemic heart disease14 because of its microflora, and the existence of some metabolites as organic acids. Further, it has several biological effects e.g. antibacterial, antioxidant, immunomodulatory, antidiabetic, and cholesterol lowering actions15. Ho et al. 21 reported that kefir milk has anti-adipogenic effects by suppressing adipocyte differentiation and inhibiting the expression of the SREBP-1, ACC and FAS proteins during cholesterol synthesis16.
Clearly, bioinformatics presents novel clues and initial data for screening potential biomarkers for different diseases and drug monitoring17. Moreover, it can be used to identify genes related to specific biological functions and predict drug-molecular signaling that may lead to a breakthrough in targeted therapy18.
Therefore, the present study aimed at investigating the efficacy of kefir Milk in NASH management via modulation of hepatic mRNA-miRNA based panel linked to NAFLD/NASH Hippo signaling and gut microbiota regulated genes which are identified using bioinformatics tools.
Materials and methods
Chemicals and drugs
Cholesterol and cholic acid were purchased from Ralin BV (Lijinbaan, Netherlands). Ready-made kefir milk was purchased from Heal Pharmaceutical every week and stored at temperature (4 °C), it contains Lactobacillus lactis as the largest bacterial colony representing approximately 80% and Lactobacillus paracasei, Lactobacillus acidophilus, Lactobacillus delbrueckii ssp. bulgaricus, Lactiplantibacillus plantarum, Lactobacillus kefiranofaciens, Leuconostoc mesenteroides, Streptococcus thermophilus and yeast of kefir (Danisco®) representing the other 20%.
The total number of micro-organisms in the fermented milk produced contain at least 107 colony-forming units (CFU)/ml and the yeast number not less than 104 CFU/ml19,20.
Experimental animals and diets
Forty male (6 weeks-old) Wistar rats (140–160 g) were obtained from the Scientific Research Institute, Cairo, Egypt, housed under specific conditions (20 ± 2 °C), 12 h light/dark cycle with free access to water and normal rat chow. All experimental procedures were done according to the guidelines of the Institutional Animal Care and were approved by the Ethics Committee of Ain Shams Faculty of Medicine, Egypt (Ethical Approval Number; FWA000017585).
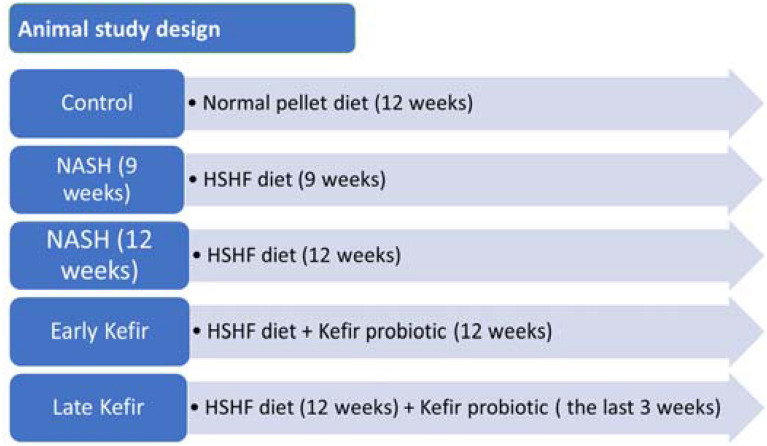
After a one-week acclimatization period, the rats were randomly divided into three groups (n = 8 for each group where treatment was given orally by gavage21: (I) Normal control group (NC) fed a normal pellet diet; (II) NASH model group, this group was further subdivided into IIa: (9-weeks NASH model) and IIb: (12-weeks NASH model)22, Group III (probiotic-treated), this group was further subdivided into IIIa: (NASH/early probiotic-treated), rats were fed HSFD for 12 weeks and received probiotic treatment daily from day one for 12 weeks and IIIb: (NASH/late probiotic-treated), rats were fed HSHF for 12 weeks and received probiotic treatment daily in the last 3 weeks. NASH model was induced by feeding rats high fat/high sucrose diet consisting of 70% standard chow, 20% lard, 10% sucrose, 1% cholesterol, and 0.25% cholic acid22. Kefir milk was mixed and then administered by oral gavage at a dose of 1.8 mL/rat/day. All rats were weighed at the beginning of the study, at the end of each week, and before animal sacrifice (Fig. 1).
Figure 1.
Flow chart showing the experimental design of the animal studies. NASH nonalcoholic steatohepatitis, HSHF high sucrose and high fat.
Blood sample and liver tissue collection
At the end of the study, rats were fasted for 12 h. and anesthetized with one dose of urethane (1.2 g/kg, IP)23 for sacrificing. Blood samples were collected from retro-orbital vein, centrifuged at 3000 rpm for 10 min for serum separation, then aliquoted and stored at − 20 °C for subsequent biochemical analysis. Whole livers were removed promptly, weighed and dissected. Part of the hepatic tissues was immediately stored in − 80 °C for RNAs and protein assessment whereas the remaining parts were rapidly fixed in 10% neutral buffered formalin for histopathological and immunohistochemical analyses.
Assessment of liver function and lipid profile markers
We assessed levels of serum aspartate transaminase (AST), alanine transaminase (ALT), total and direct bilirubin, gamma-glutamyl transferase (GGT), alkaline phosphatase (ALP) and Lipid profile [Total cholesterol (TC), Triglycerides (TG), HDL cholesterol (HDL-C), and LDL cholesterol (LDL-C)] in serum samples24 using commercial kits according to the manufacturer’s instructions and proceeded by a multifunctional biochemistry analyzer (AU680, Beckman Coulter Inc, CA).
Hepatic histopathological evaluation
For light microscopy, the buffered formalin-fixed liver tissues were dehydrated in ethanol, immersed in paraffin wax and sliced into Sections (5-μm thick). Standard hematoxylin–eosin (HE) for assessment of histological features of steatohepatitis and Masson’s Trichrome staining for detecting collagen fibers were performed25.
Each section was scored for the severity of hepatic steatosis, inflammation and fibrosis individually according to the published criteria26, Table 1.
Table 1.
Histological grading of steatosis, inflammation and staging of fibrosis.
| Steatosis | Grades | |
| 0 | ˂ 5% steatosis | |
| 1 | 5–25% steatosis | |
| 2 | 26–50% steatosis | |
| 3 | 51–75% steatosis | |
| 4 | More than 75% steatosis | |
| Inflammation | Grades | |
| 0 | None | |
| 1 | Focal collection of mononuclear cells | |
| 2 | Diffuse infiltrates of mononuclear cells | |
| 3 | Focal collection of polymorphonuclear cells | |
| 4 | Diffuse infiltrates of polymorphonuclear | |
| Fibrosis | Stages | |
| F0 | No evidence of fibrosis | |
| F1 | Mild fibrosis | |
| F2 | Moderate fibrosis | |
| F3 | Advanced fibrosis | |
In-silico filtration of hepatic mRNA-miRNA based panel linked to hippo signaling, gut microbita regulated genes and NAFLD/NASH
Firstly; we have selected AMOTL2, SOX11 and SMAD4 mRNAs from public microarray dataset; QuickGO (https://www.ebi.ac.uk/QuickGO/), Comparative Toxicogenomics Database (http://ctdbase.org/) and Gene atlas database (https://www.ebi.ac.uk/gxa) and by literature reviews because of their strong correlation to NASH pathogenesis (Supplementary Fig. 1A–E). Secondly; we verified the gene ontology of the selected mRNAs and their link to Hippo signaling and hepatocyte stem cell proliferation/differentiation using Harmonizome database (http://amp.pharm.mssm.edu/Harmonizome/) (Supplementary Fig. 2A–E). Thirdly; the selected mRNAs were verified for their relation to gut microbiota related genes by using Encyclopedia of gut microbiota regulated genes (http://microbiota.wall.gu.se/) (Supplementary Fig. 3A–C). Fourthly, AMOTL2, SOX11 and SMAD4 mRNAs were then imported into Protein–Protein Interaction Networks Functional Enrichment Analysis (STRING) database (https://string-db.org/) to ensure their protein protein interaction and their linkage with HIPPO pathway target effectors (Supplementary Fig. 4A). Then, the mRNA was mapped to Hippo signaling using KEGG pathway of hippo (Supplementary Fig. 4B). Fifthly, we used miRWalk 3.0 (http://mirwalk.umm.uni-heidelberg.de/), that combine the prediction results of both TargetScan and MiRBase to retrieve has-miR-6807-5p based on target complementarity and ranking score (Supplementary Fig. 5A–C). We verified the expression of has-miR-6807-5p in liver using miRmine database (Supplementary Fig. 6A). https://guanfiles.dcmb.med.umich.edu/mirmine/index.html). Lastly; pathway enrichment analysis of has-miR-6807-5p by using DIANA database tools: miRpath (http://snf-515788.vm.okeanos.grnet.gr/) that confirmed its relation to Hippo signaling and stem cell proliferation/differentiation (Supplementary Fig. 6B).
Extraction of total RNA (mRNA and miRNA)
Total RNA from the liver tissue was extracted27 using miRNeasy Mini Kit (Cat. No. 217004, Qiagen, Germany) as per instructions from the manufacturer. The concentration and purity of total RNA were assessed using NanoDrop (Thermoscientific, USA); the purity of the isolated RNAs (A260/A280) was 1.8–2. The reverse transcription of the extracted total RNA into complementary DNA (cDNA) was immediately proceeded with miScript II RT (Cat. No. 218161, Qiagen) and RT2 First Strand Kit (Cat.No. 330404, Qiagen) according to the manufacturer’s protocol using Thermo Hybaid PCR express (Thermo Fisher Scientific, Massachusetts, USA).
Real-time quantitative polymerase chain reaction (RT-qPCR)
The expression levels of SMAD4, SOX11 and AMOTL2 mRNAs ((Accession: NM_NM_005359.6, NM_ _003108.4 and NR_002819, _001113490.2, respectively)) in the liver tissues were assessed using a Quantitect SYBR Green ROX qPCR Mastermix Kit (Cat no. 330523) (Qiagen, Germany), hsa-miR-6807-5p (Accession: MIMAT0027514: 5' gugagccaguggaauggagagg 3′) expression in tissue samples was assessed by using miScript SYBR Green PCR Kit (Cat. No. 218073, Qiagen, Germany). The GAPDH and Hs_SNORD72_11 miScript Primer were used as the house keeping gene to normalize the raw data and then compared with a control sample. RT-qPCR amplification was performed in Applied Biosystems 7500 Real Time PCR system (Applied Biosystems, Foster City, USA) thermal cycler. The PCR program for the SYBR Green-based qPCR was as follows: denaturation at 95 °C for 15 min; 40 cycles of denaturation for 10 s at 94 °C; then annealing for 30 s at 55 °C; and lastly, extension for 30 s at 70 °C. Each reaction was performed in duplicate. The threshold cycle (Ct) value of each sample was calculated using The Applied Biosystems™ 7500 Real-Time PCR System version 2.0 software which also calculated the efficiency of the PCR (Applied Biosystems). The melting curves were analyzed to affirm the specificities of the amplicons for the SYBR Green-based PCR amplification. Calculation of the relative quantification of RNA expression was performed by the software according to Livak method, where RQ = 2−ΔΔCt28,29.
Detection of Hepatocyte specific antigen (Hep Par 1) as Protein-based biomarkers for NAFLD/NASH progression by immunohistochemical staining30
Immunohistochemistry was performed on liver sections embedded in paraffin (4 μm) using Benchmark Ventana (GX) automated staining platform (Ventana Medical Systems, USA). Liver sections were put on positively charged slides, dewaxed in xylene, and rehydrated using graded alcohols. Where, heat-induced epitope retrieval was done with Ventana Cell Conditioning Solution 1 for 48 min and the diluted primary antibodies and Hepatocyte specific antigen (Hep Par 1) (EP265), monoclonal rabbit antibody (7 ml prediluted, dilution 1:100. Ref no: 264R-18, Lot no: 0000027465) were applied to sections at 37 °C for 32 min. The presence of antigen was visualized using Ventana ultraView Universal DAB Detection Kit. Slides were counterstained with hematoxylin, dehydrated, cleared, and mounted with a permanent mounting media. The result of immunohistochemistry staining was counted by using the method described by31 to calculate the histoscore (H-score) which involves a semiquantitative assessment of both the percentage of positive cells and the intensity of staining (graded as: 0, non-staining; 1, weak staining; 2, medium staining; and 3, strong staining). Hep Par 1 positive cells were counted using fraction area in Image J software.
Quantification of hepatic inflammatory cytokine IL-6 and fibrotic marker TGF-β1 by Enzyme Linked Immunosorbant Assay (ELISA)
Interleukin-6 (IL-6) and transforming growth factor β1 (TGF-β1) in liver tissues were determined32 using sandwich ELISA kits (cat no: E0079r and E0124R, respectively; EIAab, Wuhan, China) and according to manufacturer’s instructions. IL-6 and TGF-β1 were selected as they are either highly involved in cell proliferation or strongly correlated to Hippo signaling pathway as well as strongly interacted with the pre-selected protein markers through string tool.
Statistical analysis
The results were expressed as mean ± SEM. Statistical analysis was carried out using GraphPad Prism, software program, version 6.0. Inc., CA, USA. Statistical difference among groups was determined using one-way ANOVA followed by post hoc test for comparison between more than two groups of parametric data. P values < 0.05 were considered statistically significant.
Ethical approval &Institutional Review Board Statement
The study is reported in accordance with ARRIVE guidelines. All experimental procedures were done according to the guidelines of the Institutional Animal Care and were approved by the Ethics Committee of Ain Shams Faculty of Medicine, Egypt (Ethical Approval Number; FWA000017585).
Results
The effect of Kefir on body weight and relative liver weight
There was a significant increase in the body weight of the rats fed the HSHF diet in NASH groups compared with rats fed with the normal pellet in normal controls (NC), (P < 0.05). As well as Early Kefir group showed a significant decrease in body weight and the ratio of liver weight to body weight than NASH (12 weeks) group (P < 0.05), Table 2.
Table 2.
The effect of kefir milk on body weight and liver mass in different study groups.
| Control (n = 8) | NASH (9 weeks) (n = 8) | NASH (12 weeks) (n = 8) | Early kefir treatment (n = 12) | Late kefir treatment (n = 10) | ||
|---|---|---|---|---|---|---|
| Mean ± SD | ||||||
| Body weight (g) | Week 0 | 144.5 ± 17 | 158.5 ± 16.1 | 153.3 ± 6.79 | 161.2 ± 17.37 | 151.5 ± 15.31 |
| Week 9 | 240 ± 32.95 | 288.8 ± 30.09€ | 277.9 ± 16.2€ | 269.9 ± 27.78€a | 301.7 ± 18.79 | |
| Week12 | 263.8 ± 41.4 | 293.8 ± 16.3 | 299.5 ± 29.57 | 312.7 ± 12 | ||
| Liver weight (g) | 5.62 ± 1.06 | 10.63 ± 1.99* | 12.31 ± 1.03* | 10.67 ± 1.23 | 10.8 ± 1.687 | |
Values are mean ± SD; n: number of animals = 8 rats/each group. One-Way ANOVA test followed by Tukey’s multiple comparison test.
*P < 0.001 compared to control group.
#P < 0.001 compared to NASH (9 weeks) group.
δP < 0.001 compared to NASH (12 weeks) group.
aP < 0.001 compared to Late Kefir.
€P < 0.05 compared to control group.
The previous results were also associated with a significant increase in liver relative weight in NASH groups (P < 0.001) compared with the NC Fig. 2. The liver to body weight ratio analysis revealed that both early and late Kefir treatment may be able to alleviate liver injury, where they showed significant decrease in the liver/body weight ratios (P < 0.001) compared to that of NASH group.
Figure 2.
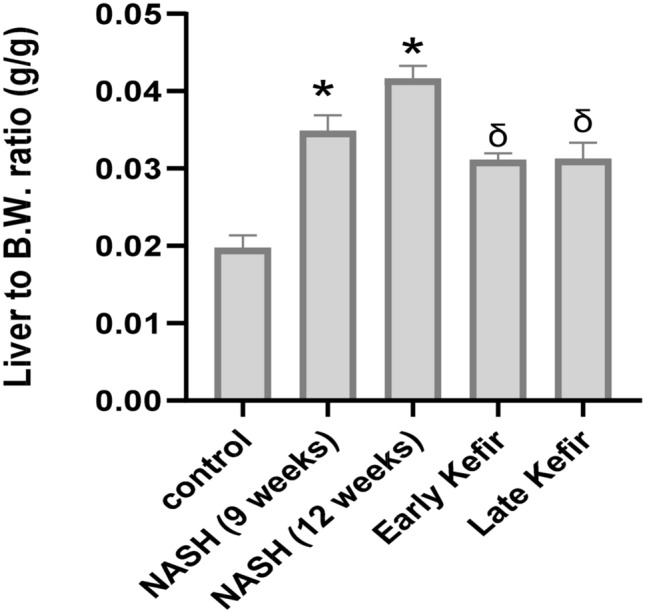
Liver/body ratio in different study groups. Values are mean ± SEM; number of animals = 8 rats/each group. One-Way ANOVA test followed by Tukey’s multiple comparison test. *P < 0.001 compared to control group, δP < 0.001 compared to NASH (12 weeks) group.
Histopathological findings
The liver from NC rats had a visible normal brownish red color with smooth and shiny appearance. Whereas, NASH rats liver, showed the typical fatty liver features and looked enlarged and extensively infiltrated with yellowish spots. Additionally, a netlike pattern on the livers surface from NASH group revealed the existence of fibrosis. Liver tissue of HSHF-fed rats administered with either early or late Kefir treatment looked redder and lighter than those of NASH groups.
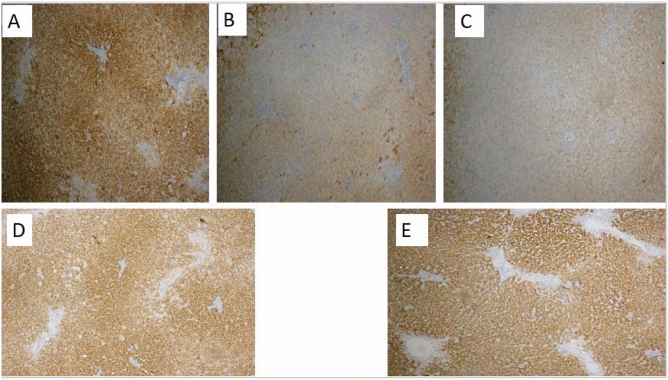
Under the light microscope, liver of NC rats exhibited normal histological structure without inflammation or fibrosis. It was formed of classic hepatic lobules which were nearly hexagonal in shape. On contrary, NASH groups demonstrated development of steatosis with large fat droplets, inflammation and fibrosis were visible as well (Fig. 3). Compared with the NASH groups, the Kefir treated groups showed visible reductions in fat droplets, improvements in steatosis, and inflammation. The effects were more prominent in early treated group.
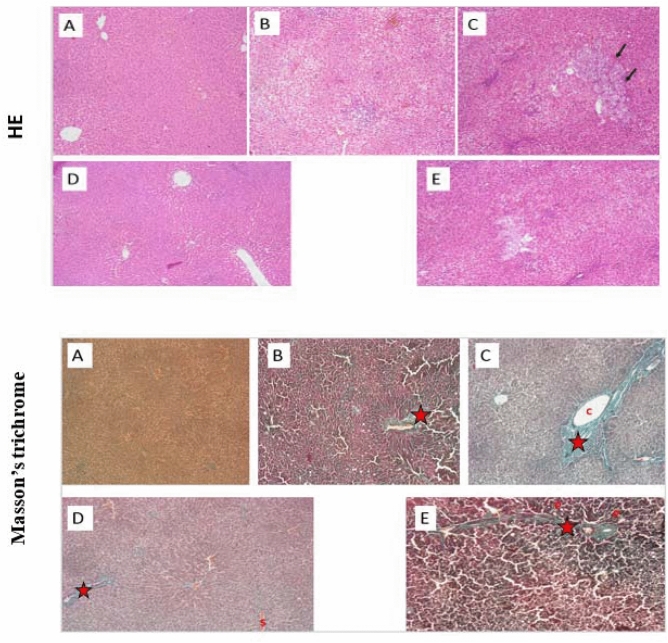
Figure 3.
The effect of early and late Kefir treatment on hepatic steatosis, inflammation and fibrosis using HE and Masson’s trichrome staining (Magnifications: × 100). (A) Control group showed no pathological changes, (B) NASH (9 weeks) group developed mainly micro vesicular steatosis, mild hepatic lobular inflammation and mild fibrous septa(star) (C) NASH (12 weeks) group showed diffuse and extensive micro and macro-vesicular steatosis, severe lobular inflammation, focal necrosis, hepatocellular ballooning, marked expansion of facultative hepatic progenitor cells (arrows), dense fibrous septa and collagen fibers (star) (D) Early Kefir and (E) Late Kefir groups showed a lower degree of steatosis, inflammation and fibrosis, where Early Kefir showed more remarkable histological improvements.
The effect of Kefir treatment on liver function and lipid profile
As shown in Table 3, upon feeding the experimental rats with HSHF diet in NASH groups, the serum levels of AST, ALT, GGT, ALP, total bilirubin, and direct bilirubin have been increased significantly (P < 0.001), compared to the NC group. On contrary, oral administration of either early or late Kefir treatment , along with HSHF feeding, significantly reduced (P < 0.001) the elevated levels of these variables. Noticeably, this ameliorative effect was more remarkable when Kefir was administered prophylactically (Early Kefir).
Table 3.
The effect of Kefir treatment on liver function and lipid profile.
| Laboratory parameters | Control | NASH (9 weeks) | NASH (12 weeks) | Early kefir | Late kefir |
|---|---|---|---|---|---|
| Mean ± SD | |||||
| AST (U/L) | 20.38 ± 3.5 | 71.1 ± 4.6* | 80.5 ± 5.451* # | 33.67 ± 5.48# δ a | 60.8 ± 4.367#δ |
| ALT (U/L) | 27 ± 4.5 | 81.88 ± 3.94* | 126.6 ± 20.03*# | 41 ± 4.936#δa | 59.6 ± 3.34δ# |
| GGT (U/L) | 14.38 ± 2.2 | 64 ± 7.03* | 79.38 ± 13.39*# | 32.83 ± 5.02#δa€ | 43.3 ± 9.62δ# |
| ALP (U/L) | 29.13 ± 7.7 | 90.75 ± 3.15* | 105.9 ± 7.2*# | 51.5 ± 6.11#δa | 68.4 ± 7.57#δ |
| Total bilirubin (mg/dL) | 0.308 ± 0.05 | 1.06 ± 0.17* | 1.844 ± 0.505*# | 0.463 ± 0.05#δ | 0.73 ± 0.067δ#€ |
| Direct bilirubin (mg/dL) | 0.122 ± 0.02 | 0.57 ± 0.08* | 0.806 ± 0.14*# | 0.223 ± 0.05#δ | 0.297 ± 0.0194#δ |
| Total cholesterol (mg/dL) | 88.25 ± 7.7 | 121.1 ± 5.51* | 181.9 ± 3.48*# | 99 ± 9.323#δa | 126.7 ± 1.63δ |
| Triglyceride (mg/dL) | 45.38 ± 4.5 | 115.6 ± 5.9* | 141.5 ± 10.06*# | 73.5 ± 3.7#δa | 94.5 ± 4.882#δ |
| HDL-C (mg/dL) | 64.50 ± 5.8 | 32 ± 3.928* | 30.13 ± 2.35* | 50.17 ± 3.04#δa | 60.5 ± 2.321#δ |
| LDL-C (mg/dL) | 14.68 ± 4 | 65.9 ± 6.35* | 123.5 ± 1.46*# | 34.13 ± 11.3#δa | 47.3 ± 4.187#δ |
Values are mean ± SD; number of animals = 8 rats/each group. One-Way ANOVA test followed by Tukey’s multiple comparison test.
LDL-C low-density lipoproteins, HDL-C high-density lipoprotein, AST Aspartate aminotransferase, ALT Alanine aminotransferase, ALP Alkaline phosphatase, GGT Gamma-glutamyl transferase.
*P < 0.001 compared to control group.
#P < 0.001 compared to NASH (9 weeks) group.
δP < 0.001 compared to NASH (12 weeks) group.
aP < 0.001 compared to Late Kefir.
€P < 0.05 compared to control group.
Regarding lipid profile, there was a significant (P < 0.001) upsurge in the serum levels of TC, TG, and LDL-C coupled with a significant (P < 0.001) decrease of serum HDL-C level in animals of NASH groups compared to NC group. NASH (12 weeks) group exhibited statistically significant increase than NASH (9 weeks) group regarding TC, Triglyceride, LDL, Liver function tests and TGFB1(P < 0.001).
When Kefir was given therapeutically simultaneously with HSHF diet, serum TC, TG and LDL-C levels decreased significantly (P < 0.001) while serum HDL-C increased significantly (P < 0.001), compared to NASH groups. These meliorative effects were augmented by prophylactic supplementation with Kefir. The achieved results showed that the use of gut microbiota-based treatments could decrease hepatocyte injury and improve serum lipid profile in NASH animal model.
The effect of Kefir treatment on the expression of hepatic SOX11, SMAD4 and AMOTL2 mRNAs
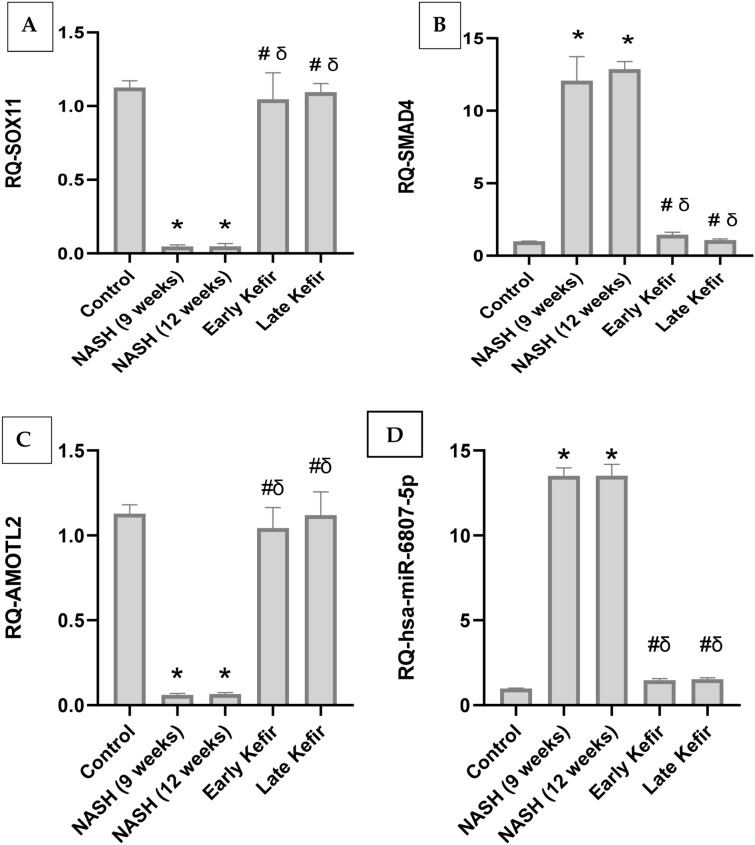
SOX11, SMAD4 and AMOTL2 expression levels were assessed in the liver tissue of the experimental animals (Fig. 4A − C). Results revealed that feeding rats with HSHF diet resulted in a sharp significant (P < 0.001) decrease in SOX11 expression level in NASH group animals. Kefir administration has normalized the significant decreases in hepatic SOX11 expression level shown in untreated NASH group rats. Additionally, no significant change in hepatic SOX11 expression level was observed between early and late treated groups. Meanwhile, NASH group recorded a remarkable significant increase in level of SMAD4(P < 0.001), compared to NC which was significantly corrected with Kefir treatment, compared to NASH group animals. Moreover, data manifested that hepatic AMOTL2 expression level in NASH group animals was significantly decreased (P < 0.001), in comparison to NC and was significantly corrected by Kefir administration.
Figure 4.
Effect of Kefir on the expression level of hepatic m-RNAs SOX11, SMAD4 and AMOTL2 (A − C), hepatic miR 6807-5p (D). Values are mean ± SEM; number of animals = 8 rats/each group. *P < 0.001 compared to control group, #P < 0.001 compared to NASH (9 weeks) group, δP < 0.001 compared to NASH (12 weeks) group. One-way ANOVA followed by Tukey’s multiple comparison test. RQ relative quantification, SOX11 SRY-Related HMG-box gene 11, SMAD4 SMAD family member 4, AMOTL2 Angiomotin-like 2, hsa-miR 6807-5p homo sapiens miR 6807-5p.
The effect of kefir treatment on the expression of hepatic miR-6807-5p
As shown in Fig. 4D, the RQ of miR-6807-5p in the liver tissue of NASH group animals was drastically increased (P < 0.001) compared to NC. Furthermore, both early and late Kefir treatment normalized the expression of hepatic miR-6807-5p, compared with that of NASH groups.
The effect of kefir on hepatic cell differentiation and proliferation
The state of liver progenitor cell proliferation and differentiation were evaluated in the liver by immunohistochemical analysis and assessment of Hepatocyte specific antigen (HepPar1). The increased in HepPar1 expression seen in NC (Fig. 6) compared to NASH groups results from Granular cytoplasmic staining due to mitochondrial binding. Hepatic progenitor cells (HPCs) gradually lose their biliary features, including markers as (HepPar1) and these transitional cells in the hepatocytic lineage become negative for the marker (Fig. 5B,C). Kefir administration showed up-regulation of HepPar1-positive cells in the liver tissues and the effect was more prominent in group received early treatment (Fig. 5D,E).
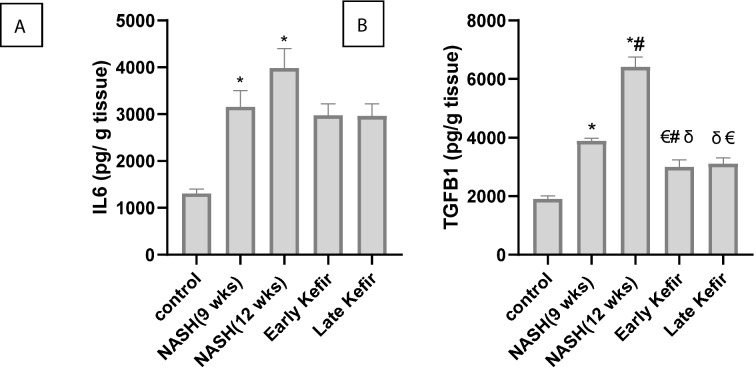
Figure 6.
The effect of Kefir treatment on (A) Level of IL-6 (B) Level of TGF-β1 in liver tissues from various groups. Values are mean ± SEM, number of animals = 8 rats/each group. *P < 0.001 compared to control group, #P < 0.001 compared to NASH (9 weeks) group, δP < 0.001 compared to NASH (12 weeks) group, €P < 0.05 compared to control group. One-way ANOVA followed by Tukey’s multiple comparison test. IL6 Interleukin 6, TGFB1 transforming growth factor beta 1.
Figure 5.
Immunohistochemical staining for hepatocyte specific antigen (HepPar1). (A) Controls showed a strong signal with Hep Par 1 (hepatocyte marker) producing distinct granular, cytoplasmic staining of hepatocytes, whereas in both (B) NASH (9 weeks) and (C) NASH (12 weeks) groups were mostly negative with HepPar-1. By comparison, (96%) hepatocytes in (D) Early Kefir group were immunoreactive for HepPar-1 and (E) Late Kefir group tended to show weaker, patchy positivity (HepPar1 × 100) indicated to hepatocyte differentiation.
The effect of Kefir on hepatic level of IL-6 and TGF-β1
The hepatic contents of IL-6 and TGF-β1 proteins were used as markers of inflammation and fibrogenesis, respectively. In the NASH groups, IL-6 and TGF-β1 proteins were significantly increased (P < 0.001, Fig. 6), compared to NC group. Kefir administration significantly decreased TGF-β1 compared with NASH groups (P < 0.001). These findings implied that both of early and late Kefir treatment effectively diminished the level of inflammation and fibrosis observed in NASH rats.
Discussion
Non-alcoholic fatty liver disease (NAFLD) has become a very common disease because of the prevailing increase in obesity worldwide. Currently, several NAFLD therapies are being targeted to improve insulin resistance (IR), but there is no effective treatment33. Probiotic treatment was shown to improve NASH through modulating insulin resistance, the key factor which plays a major role in the development of a serious liver condition34. Due to correlation between small intestinal bacterial overgrowth (SIBO) and NAFLD observed in experimental and clinical studies35,36, probiotics could also delay disease progression and prevent complications by modulating intestinal flora, intestinal permeability, and inflammatory response37.
In this context, a high fat/high sucrose diet (HFD) induced NASH model experiment was conducted to assess the beneficial effects of the Kefir formula on the degree of hepatic fibrosis and steatosis, inflammation, and body composition via modulation at both epigenetic and genetic level. Previous studies have showed that kefir improved NAFLD regarding to BW, energy expenditure and basal metabolic rate through inhibition of the lipogenesis pathway38. This may explain our findings where the supplementation with Kefir prophylactically or therapeutically along with HFD, decreased the percentage and stage of steatosis, inflammation and fibrosis seen in NASH rats. Moreover, the serum levels of ALT and AST, and the hepatic content of fat, TGFB 1 and IL 6 proteins were significantly reduced with the treatment of the Kefir formula compared to NASH groups. In agreement with the previous finding that Kefir can diminish the inflammatory response in association with a decrease in tumor necrosis factor alpha (TNF-α), interleukin-1 beta (IL-1β) and transforming growth factor beta (TGF-β) cytokines39.
Hepatic stem/progenitor cells (HPCs or HpSCs, in humans) or oval cells (in rodents) are bipotential stem cells that can differentiate into mature hepatocytes and cholangiocytes40. They represent a reserve compartment that can be activated to reactive ductulus (Or ductular reaction: DR) only when the mature epithelial cells of the liver are continuously damaged or blocked in their replication or in cases of serious cell loss41.
Hippo pathway could promote cell death and differentiation and inhibit cell proliferation, however, the regulatory mechanisms for this signaling pathway are not clearly understood42. The downstream effector YAP1 is shown to be involved in hepatic cell proliferation, survival, development and differentiation43. Activated YAP1 leads to activation of hepatic stellate cells (HSC), prolonged activation of these cells causes liver fibrogenesis44. Therefore, a key objective is to understand the mechanisms that stimulate the switch of quiescent HSCs in a healthy liver to activated, myofibroblastic HSCs in NASH.
In this sense, variety of public microarray databases and computation algorithms have been investigated in the current work, for the selection of hepatic mRNA-miRNA panel linked to NAFLD/NASH Hippo signaling pathway and gut microbita regulated genes . We identified 3 mRNAs (SOX11, SMAD4 and AMOTL2), their epigenetic regulator (miR-6807) and their target effector proteins (TGFB, IL6 and HepPar1). Protein -protein interaction between the selected mRNAs protein products and HIPPO target effectors was obvious by STRING database as shown in (Supplementary Fig. 4A).
Cytosolic AMOTL2 proteins can attach YAP1 and TAZ in their unphosphorylated states, providing a Hippo independent mechanism to down-regulate the activities of these proteins45. AMOTL2 mRNA is downregulated in the current study in the rat liver tissue of NASH compared to NC or treated groups (P < 0.001), which indicated the freeing of YAP1 with subsequent activation of hepatic stellate cells and fibrogenesis participating in the pathogenesis of NASH. On the other hand, AMOTL2 is up regulated by the intake of kefir in rats which possibly ameliorates the features of NASH in the treated rat group by bounding YAP1.
SMAD4 interacts with SMAD2/3 and participates in the intracellular TGF-β signaling pathway. Knockout of SMAD4 from mesangial cells resulted in inhibition of TGF-β1-produced ECM synthesis46.These previous published results confirmed that SMAD4 has a significant role in the pathogenesis of fibrosis by controlling the ability of SMAD3 to activate transcription of a number of fibrogenic genes (collagens) , markers (α-SMA and E-cadherin)47 and explain the significant overexpression of SMAD4 in NASH group than NC and treated groups observed in the current study (P < 0.001).
SOX-11(SRY-related HMG-box) is a member of the group C SOX transcription factor family involved in the regulation of embryonic development and in the determination of the cell fate48. Sox 11 participates in positive regulation of hippo pathway leading to phosphorylation of YAP/TAZ, resulting in their cytoplasmic retention (Supplementary Fig. 2E). This may explain the decreased expression of its mRNA in NASH versus other study groups included in the current study (P < 0.001), with subsequent increase in nuclear translocation of YAP/TAZ forming functional transcriptional complexes with TEA domain proteins 1–4 (TEAD1–4)49. YAP/TAZ–TEADs promote the expression of Hippo-responsive genes that have a role in the production of proinflammatory cytokines (including interleukin 6 and TGF-β) and development of nonalcoholic steatohepatitis (NASH)50.
This agrees with another study that validated hepatic expression of a group of mRNAs and reported SOX11 downregulation in Obesity-Related Nonalcoholic Steatohepatitis compared to normal tissue51.
MiRNAs are shown to play important role in the development of diseases, including NASH5. In patients and animal models with NASH, circulating miRNAs represent significant differences compared to healthy controls, moreover, NASH shows significantly distinct miRNA expression profile compared to NAFLD52.
MiR 6807-5p is shown to be a novel microRNA biomarker for detecting gastric cancer (GC) but no previous reports have correlated its expression with the incidence or progression of NASH or with other liver diseases53. Our bioinformatics data analysis revealed its expression in liver (Supplementary Fig. 6A) and its correlation to Hippo signaling pathway (Supplementary Fig. 6B). Moreover, SOX11, SMAD4 and AMOTL2 are shown to be direct mRNA targets of miRNA-6807-5p as was retrieved from miRwalk database (Supplementary Fig. 5A–C). Our results revealed that there was a significant increase in miR 6807-5p concomitant with marked increase in SMAD4 and significant down regulation of SOX11, and AMOTL2 mRNA levels in the liver tissue of NASH group compared to NC or treated groups. This deregulated molecular profile reverts to normal after treatment with kefir milk.
TGF‐β was significantly increased in NASH as compared with other groups (P < 0.001) which is explained by previous work reporting that activation of TGF‐β signaling pathway was involved in the occurrence and development of NASH through motivating HSCs and the formation of extracellular matrix (ECM)54,55.
Continuous increased triacylglycerols buildup produces reactive oxygen species (ROS) and proinflammatory cytokines e.g. IL6, which induces NASH. IL-6 expression was significantly increased in the hepatic biopsy of patients with nonalcoholic steatohepatitis (NASH) than patients having simple steatosis or normal biopsies. Morever, there was a positive correlation between fibrosis staging and inflammation degree confirming the presence of hepatic IL-6 expression in human NASH56. Our results revealed a marked increase in IL-6 level in rat NASH liver when compared to NC (P < 0.001), but it was returned back to normal levels after kefir treatment.
In the present study, proliferation of oval cells was enhanced in NASH groups and decreased in Kefir groups. Previous studies found that HPC activation and the expansion of DR have been correlated with progressive fibrosis in adult and pediatric NASH and in HCV associated cirrhosis57.
In addition, upon differentiation towards hepatocytes, HPCs were previously shown to gradually lose their biliary features, including markers such as keratin 19 (K19) and keratin 7 (K7)58 as well as Hepatocyte Paraffin 1(Hep Par1). For the best of our knowledge, Hep Par1 was used for the first time as a marker of hepatocellular differentiation in NASH where it is shown to be mostly negative in the liver of NASH group while 96% of hepatocytes were immunoreactive for HepPar-1. with kefir treatment (Fig. 5).
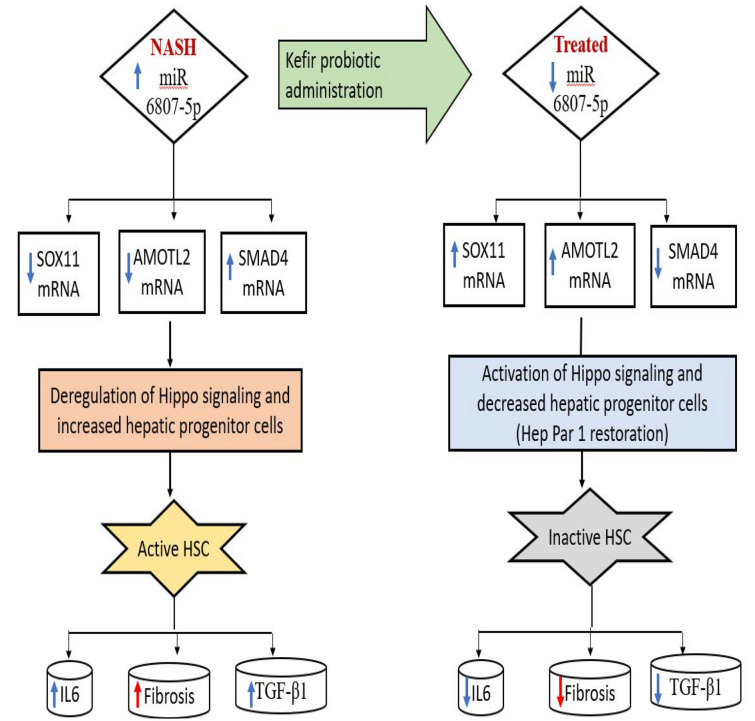
Taken all together, our experimental model hypothesized that treatment with probiotic Kefir down-regulated miR 6807-5p with subsequent upregulation of SOX 11, and AMOTL2 and downregulation of SMAD4. Accordingly, YAP1 is negatively regulated with its subsequent sequestration in the cytosol. This cytoplasmic translocation of YAP1 inhibited cell proliferation and suppressed the expression of inflammatory IL6 and fibrotic TGF-β1 (Fig. 7).
Figure 7.
Proof of concept map of the study hypothesis.
Conclusion
Kefir Milk was effective to improve the pathological and biochemical disturbances induced in NASH animal model through modulation of Hippo signaling/gut regulated genes linked RNA panel. This treatment down-regulated the expression of miR 6807-5p and SMAD4 as well as up-regulated the expression of SOX11& AMOTL2 mRNAs in rat hepatic tissue. The protein products of these genes interact with YAP1 in the Hippo signaling pathway controlling inflammation, and fibrosis progression in NASH.
Limitations
Limitations of the present study includes the lack of Control rat group receiving only Kefir to show the intervention effect on miRNA-mRNA signature in a healthy model of rats. In addition, investigating the effect of kefir on the gut microbiota composition should be done in further studies.
Supplementary Information
Abbreviations
- HSHF diet
High sucrose high fat diet
- NAFLD
Nonalcoholic fatty liver disease
- NASH
Non-alcoholic steatohepatitis
- H-score
Histoscore
- IL6
Interleukin 6
- TGFb1
Transforming growth factor beta
- Hep Par 1
Hepatocyte specific antigen
- lncRNA
Long noncoding rna
- miRNA
Micro-RNA
Author contributions
N.S. carried out nucleic acid extraction, participated in real time PCR experiments, participated in statistical analysis. S.E. supervised the study work, acquired fund, revisied and edited the first draft, submitted the manuscript. A.M. participated in statistical analysis. N.A. prepared probiotic. A.H. designed and conducted the animal model. M.H. participated in real time PCR experiments and immunohistochemistry. M.E. participated in the immunohistochemistry experiments. M.M. carried out the study design, bioinformatics analysis, and participated in real time PCR experiments and statistical analysis. All authors read and approve the final manuscript.
Funding
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB). This work was supported by STDF- Science and Technology Development fund for accredited centers of scientific excellence (JESOR 2017-2961), Cairo, Egypt (Grant no. 411851).
Data availability
Please contact corresponding author for data requests.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Sanaa Eissa, Email: Drsanaa_mohamed@med.asu.edu.eg, Email: Dr_sanaa_eissa@yahoo.com.
Marwa Matboli, Email: marwasayed472@yahoo.com, Email: DrMarwa_Matboly@med.asu.edu.eg.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-27353-x.
References
- 1.Siddiqui MS, Van Natta ML, Connelly MA, Vuppalanchi R, Neuschwander-Tetri BA, Tonascia J, Chalasani N. Impact of obeticholic acid on the lipoprotein profile in patients with non-alcoholic steatohepatitis. J. Hepatol. 2020;72(1):25–33. doi: 10.1016/j.jhep.2019.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Estes C, Razavi H, Loomba R, Younossi Z, Sanyal AJ. Modeling the epidemic of nonalcoholic fatty liver disease demonstrates an exponential increase in burden of disease. Hepatology. 2018;67(1):123–133. doi: 10.1002/hep.29466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kutlu O, Kaleli HN, Ozer E. Molecular pathogenesis of nonalcoholic steatohepatitis-(NASH-) related hepatocellular carcinoma. Can. J. Gastroenterol. Hepatol. 2018;2018:1–9. doi: 10.1155/2018/8543763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.EASL, Easd, EASO EASL-EASD-EASO Clinical Practice Guidelines for the Management of Non-Alcoholic Fatty Liver Disease. Obes. Facts. 2016;9:65–90. doi: 10.1159/000443344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gadallah SH, Eissa S, Ghanem HM, Ahmed EK, Hasanin AH, El Mahdy MM, Matboli M. Probiotic-prebiotic-synbiotic modulation of (YAP1, LATS1 and NF2 mRNAs/miR-1205/lncRNA SRD5A3-AS1) panel in NASH animal model. Biomed. Pharmacother. 2021;140:111781. doi: 10.1016/j.biopha.2021.111781. [DOI] [PubMed] [Google Scholar]
- 6.Ardestani A, Lupse B, Maedler K. Hippo signaling: Key emerging pathway in cellular and whole-body metabolism. Trends Endocrinol. Metab. 2018;29(7):492–509. doi: 10.1016/j.tem.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Z, Wen H, Peng B, Weng J, Zeng F. Downregulated microrna-129-5p by long non-coding RNA NEAT1 upregulates PEG3 expression to aggravate non-alcoholic steatohepatitis. Front. Genet. 2021;11:1407. doi: 10.3389/fgene.2020.563265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang R, Duan X, Fan J, Li G, Wang B. Role of noncoding RNA in development of nonalcoholic fatty liver disease. Biomed. Res. Int. 2019;26:2019. doi: 10.1155/2019/8690592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, Morelli L, et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014 doi: 10.1038/nrgastro.2014.66. [DOI] [PubMed] [Google Scholar]
- 10.Suez J, Zmora N, Zilberman-Schapira G, Mor U, Dori-Bachash M, Bashiardes S, Horn M. Post-antibiotic gut mucosal microbiome reconstitution is impaired by probiotics and improved by autologous FMT. Cell. 2018;174(6):1406–1423. doi: 10.1016/j.cell.2018.08.047. [DOI] [PubMed] [Google Scholar]
- 11.Nobili V, Putignani L, Mosca A, Del Chierico F, Vernocchi P, Alisi A, Drago L. Bifidobacteria and lactobacilli in the gut microbiome of children with non-alcoholic fatty liver disease: Which strains act as health players? Arch. Med. Sci. 2018;14(1):81. doi: 10.5114/aoms.2016.62150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang SY, Chen KN, Lo YM, Chiang ML, Chen HC, Liu JR, Chen MJ. Investigation of microorganisms involved in biosynthesis of the kefir grain. Food Microbiol. 2012;32(2):274–285. doi: 10.1016/j.fm.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 13.Prado MR, Blandón LM, Vandenberghe LP, Rodrigues C, Castro GR, Thomaz-Soccol V, Soccol CR. Milk kefir: Composition, microbial cultures, biological activities, and related products. Front. Microbiol. 2015;6:1177. doi: 10.3389/fmicb.2015.01177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farnworth ER, Mainville I. Kefir: A fermented milk product. Handb. Ferment. Funct. Foods. 2003;2:89–127. [Google Scholar]
- 15.Vujičić IF, Vulić M, Könyves T. Assimilation of cholesterol in milk by kefir cultures. Biotech. Lett. 1992;14(9):847–850. doi: 10.1007/BF01029151. [DOI] [Google Scholar]
- 16.Ho JN, Choi JW, Lim WC, Kim MK, Lee IY, Cho HY. Kefir inhibits 3T3-L1 adipocyte differentiation through down-regulation of adipogenic transcription factor expression. J. Sci. Food Agric. 2013;93(3):485–490. doi: 10.1002/jsfa.5792. [DOI] [PubMed] [Google Scholar]
- 17.Ye Z, Wang F, Yan F, Wang L, Li B, Liu T, Fu Z. Bioinformatic identification of candidate biomarkers and related transcription factors in nasopharyngeal carcinoma. World J. Surg. Oncol. 2019;17(1):1–10. doi: 10.1186/s12957-019-1605-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matboli M, Gadallah SH, Rashed WM, Hasanin AH, Essawy N, Ghanem HM, Eissa S. mRNA-miRNA-lncRNA regulatory network in nonalcoholic fatty liver disease. Int. J. Mol. Sci. 2021;22(13):6770. doi: 10.3390/ijms22136770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Codex Alimentarius Commission. Milk and Milk Products (CODEX STAN 243–2003) 6–16 (2011).
- 20.Punaro GR, Maciel FR, Rodrigues AM, Rogero MM, Bogsan CSB, Oliveira MN, Ihara SSM, et al. Kefir administration reduced progression of renal injury in STZ-diabetic rats by lowering oxidative stress. Nitric Oxide. 2014;37:53–60. doi: 10.1016/j.niox.2013.12.012. [DOI] [PubMed] [Google Scholar]
- 21.Monteiro BL, Dias AT, Wanderkoke SC, Yokota R, Casarini DE, Leal MAS, Nogueira BV, et al. Protective effects of kefir in the angiotensin II-dependent hypertension. J. Funct. Foods. 2020;75:104260. doi: 10.1016/j.jff.2020.104260. [DOI] [Google Scholar]
- 22.Guo JH, Han DW, Li XQ, Zhang Y, Zhao YC. The impact of small doses of LPS on NASH in high sucrose and high fat diet induced rats. Eur. Rev. Med. Pharmacol. Sci. 2014;18(18):2742–2747. [PubMed] [Google Scholar]
- 23.Fisher CD, Lickteig AJ, Augustine LM, Oude Elferink RPJ, Besselsen DG, Erickson RP, Cherrington NJ. Experimental non-alcoholic fatty liver disease results in decreased hepatic uptake transporter expression and function in rats. Eur. J. Pharmacol. 2009;613(1–3):119–127. doi: 10.1016/j.ejphar.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meng Y, Liu Y, Fang N, Guo Y. Hepatoprotective effects of Cassia semen ethanol extract on non-alcoholic fatty liver disease in experimental rat. Pharm. Biol. 2019;57(1):98–104. doi: 10.1080/13880209.2019.1568509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen SW, Chen YX, Shi J, Lin Y, Xie WF. The restorative effect of taurine on experimental nonalcoholic steatohepatitis. Dig. Dis. Sci. 2006;51(12):2225–2234. doi: 10.1007/s10620-006-9359-y. [DOI] [PubMed] [Google Scholar]
- 26.Schnabl B, Farshchi-Heydari S, Loomba R, Mattrey RF, Hoh CK, Sirlin CB, Vera DR. Staging of fibrosis in experimental non-alcoholic steatohepatitis by quantitative molecular imaging in rat models. Nucl. Med. Biol. 2016;43(2):179–187. doi: 10.1016/j.nucmedbio.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 27.Matsunami T, Sato Y, Ariga S, Sato T, Kashimura H, Hasegawa Y, Yukawa M. Regulation of oxidative stress and inflammation by hepatic adiponectin receptor 2 in an animal model of nonalcoholic steatohepatitis. Int. J. Clin. Exp. Pathol. 2010;3(5):472. [PMC free article] [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using realtime quantitative PCR and the 2(Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 29.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, et al. The MIQE Guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 30.Garcia-Martinez I, Alen R, Rada P, Valverde AM. Insights into extracellular vesicles as biomarker of NAFLD pathogenesis. Front. Med. 2020;7:395. doi: 10.3389/fmed.2020.00395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khatun S, Fujimoto J, Toyoki H, Tamaya T. Clinical implications of expression of ETS-1 in relation to angiogenesis in ovarian cancers. Cancer Sci. 2003;94(9):769–773. doi: 10.1111/j.1349-7006.2003.tb01517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng Ze, Xiaohua Xu, Zhang X, Wang A, Zhang C, Hüttemann M, Grossman LI, et al. Exposure to ambient particulate matter induces a NASH-like phenotype and impairs hepatic glucose metabolism in an animal model. J. Hepatol. 2013;58(1):148–154. doi: 10.1016/j.jhep.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smeuninx B, Boslem E, Febbraio MA. Current and future treatments in the fight against non-alcoholic fatty liver disease. Cancers. 2020;12(7):1714. doi: 10.3390/cancers12071714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Silva-Sperb AS, Moraes HA, de Moura BC, Alves BC, Bruch-Bertani JP, Azevedo VZ, Dall’Alba V. Effect of probiotic supplementation in nonalcoholic steatohepatitis patients: Probiliver trial protocol. Trials. 2019;20(1):1–8. doi: 10.1186/s13063-019-3679-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takaki A, Kawai D, Yamamoto K. Multiple hits, including oxidative stress, as pathogenesis and treatment target in non-alcoholic steatohepatitis (NASH) Int. J. Mol. Sci. 2013;14(10):20704–20728. doi: 10.3390/ijms141020704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caligiuri A, Gentilini A, Marra F. Molecular pathogenesis of NASH. Int. J. Mol. Sci. 2016;17(9):1575. doi: 10.3390/ijms17091575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feldman M, Friedman LS, Brandt LJ. Sleisenger and Fordtran’s Gastrointestinal and Liver Disease E-book: Pathophysiology, Diagnosis, Management. Elsevier; 2020. [Google Scholar]
- 38.Chen HL, Tsai TC, Tsai YC, Liao JW, Yen CC, Chen CM. Kefir peptides prevent high-fructose corn syrup-induced non-alcoholic fatty liver disease in a murine model by modulation of inflammation and the JAK2 signaling pathway. Nutr. Diabetes. 2016;6(12):e237–e237. doi: 10.1038/nutd.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peluzio MD, e Dias MD, Martinez JA, Milagro FI. Kefir and intestinal microbiota modulation: Implications in human health. Front. Nutr. 2021 doi: 10.3389/fnut.2021.638740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beaufrère A, Calderaro J, Paradis V. Combined hepatocellular-cholangiocarcinoma: An update. J. Hepatol. 2021 doi: 10.1016/j.jhep.2021.01.035. [DOI] [PubMed] [Google Scholar]
- 41.Van Haele M, Roskams T. Hepatic progenitor cells: An update. Gastroenterol. Clin. 2017;46(2):409–420. doi: 10.1016/j.gtc.2017.01.011. [DOI] [PubMed] [Google Scholar]
- 42.Meng Z, Moroishi T, Guan KL. Mechanisms of Hippo pathway regulation. Genes Dev. 2016;30(1):1–17. doi: 10.1101/gad.274027.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Manmadhan S, Ehmer U. Hippo signaling in the liver–a long and ever-expanding story. Front. Cell Dev. Boil. 2019;7:33. doi: 10.3389/fcell.2019.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu HX, Yao Y, Bu FT, Chen Y, Wu YT, Yang Y, Meng XM. Blockade of YAP alleviates hepatic fibrosis through accelerating apoptosis and reversion of activated hepatic stellate cells. Mol. Immunol. 2019;107:29–40. doi: 10.1016/j.molimm.2019.01.004. [DOI] [PubMed] [Google Scholar]
- 45.Rojek KO, Krzemień J, Doleżyczek H, Boguszewski PM, Kaczmarek L, Konopka W, Prószyński TJ. Amot and Yap1 regulate neuronal dendritic tree complexity and locomotor coordination in mice. PLoS Biol. 2019;17(5):e3000253. doi: 10.1371/journal.pbio.3000253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsuchida KI, Zhu Y, Siva S, Dunn SR, Sharma K. Role of Smad4 on TGF-β–induced extracellular matrix stimulation in mesangial cells. Kidney Int. 2003;63(6):2000–2009. doi: 10.1046/j.1523-1755.2003.00009.x. [DOI] [PubMed] [Google Scholar]
- 47.Xu F, Liu C, Zhou D, Zhang L. TGF-β/SMAD pathway and its regulation in hepatic fibrosis. J. Histochem. Cytochem. 2016;64(3):157–167. doi: 10.1369/0022155415627681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiao Y, Zhao J, Zhang Z, Li M, Yu X, Yang Y, Li X. SRY-Box containing gene 4 promotes liver steatosis by upregulation of SREBP-1c. Diabetes. 2018;67(11):2227–2238. doi: 10.2337/db18-0184. [DOI] [PubMed] [Google Scholar]
- 49.Bae SJ, Luo X. 2018. Activation mechanisms of the Hippo kinase signaling cascade. Biosci. Rep. [DOI] [PMC free article] [PubMed]
- 50.Song K, Kwon H, Han C, Chen W, Zhang J, Ma W, Dash S, Gandhi CR, Wu T. Yes-associated protein in kupffer cells enhances the production of proinflammatory cytokines and promotes the development of nonalcoholic steatohepatitis. Hepatology. 2020;72(1):72–87. doi: 10.1002/hep.30990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gerhard GS, Legendre C, Still CD, Chu X, Petrick A, DiStefano JK. Transcriptomic profiling of obesity-related nonalcoholic steatohepatitis reveals a core set of fibrosis-specific genes. J. Endocr. Soc. 2018;2(7):710–726. doi: 10.1210/js.2018-00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim TH, Lee Y, Lee Y-S, Gim J-A, Ko E, Yim SY, Jung YK, et al. Circulating miRNA is a useful diagnostic biomarker for nonalcoholic steatohepatitis in nonalcoholic fatty liver disease. Sci. Rep. 2021;11(1):1–9. doi: 10.1038/s41598-021-94115-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Iwasaki H, Shimura T, Yamada T, Okuda Y, Natsume M, Kitagawa M, Kataoka H. A novel urinary microRNA biomarker panel for detecting gastric cancer. J. Gastroenterol. 2019;54(12):1061–1069. doi: 10.1007/s00535-019-01601-w. [DOI] [PubMed] [Google Scholar]
- 54.Richardson N, Ng ST, Wraith DC. Antigen-specific immunotherapy for treatment of autoimmune liver diseases. Front. Immunol. 2020;11:1586. doi: 10.3389/fimmu.2020.01586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qin G, Wang GZ, Guo DD, Bai RX, Wang M, Du SY. Deletion of Smad4 reduces hepatic inflammation and fibrogenesis during nonalcoholic steatohepatitis progression. J. Dig. Dis. 2018;19(5):301–313. doi: 10.1111/1751-2980.12599. [DOI] [PubMed] [Google Scholar]
- 56.Wieckowska A, Papouchado BG, Li Z, Lopez R, Zein NN, Feldstein AE. Increased hepatic and circulating interleukin-6 levels in human nonalcoholic steatohepatitis. Off. J. Am. Coll. Gastroenterol. ACG. 2008;103(6):1372–1379. doi: 10.1111/j.1572-0241.2007.01774.x. [DOI] [PubMed] [Google Scholar]
- 57.Nobili V, Carpino G, Alisi A, Franchitto A, Alpini G, De Vito R, Gaudio E. Hepatic progenitor cells activation, fibrosis and adipokines production in pediatric nonalcoholic fatty liver disease. Hepatology. 2012;56(6):2142–2153. doi: 10.1002/hep.25742. [DOI] [PubMed] [Google Scholar]
- 58.Ko S, Russell JO, Molina LM, Monga SP. Liver progenitors and adult cell plasticity in hepatic injury and repair: Knowns and unknowns. Annu. Rev. Pathol. 2020;15:23–50. doi: 10.1146/annurev-pathmechdis-012419-032824. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Bae SJ, Luo X. 2018. Activation mechanisms of the Hippo kinase signaling cascade. Biosci. Rep. [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
Please contact corresponding author for data requests.