Abstract
Streptococcus mutans, which causes dental caries in the human oral cavity and occasionally causes infective endocarditis in the heart, withstands adverse environmental stress through diverse alterations in protein synthesis. Differential gene expression in response to environmental stress was analyzed by RNA fingerprinting using arbitrarily primed PCR with a panel of 11mer primers designed for differential display in Enterobacteriaceae. Dot and Northern blot hybridization confirmed that the transcription of several genes was up- or down-regulated following exposure to acid shock from pH 7.5 to 5.5. RNA of a gene designated AP-185 (acid-stress protein) was induced specifically by acid treatment, while RNA of GSP-781 (general-stress protein) was up-regulated significantly when bacteria were exposed to high osmolarity and temperature, as well as low pH. The deduced amino acid sequence of AP-185 shares homology (78% identity) with branched-chain amino acid aminotransferase. Cloning and sequence analysis of GSP-781 revealed a potential secreted protein of a molecular mass of about 43 kDa and with a pI predicted to be 5.5. Transcriptional levels of another gene, designated AR-186 (acid-repressed protein), which encodes putative aconitase, were repressed by acid treatment but were enhanced by plasma or serum components. Analogous results were identified in icd and citZ genes, and repression of these genes, along with AR-186, was also observed when they were exposed to high osmolarity and temperature. These results indicate that differential regulation of specific genes at the transcriptional level is triggered by different stress and that genes responsible for glutamate biosynthesis in the citrate pathway are coordinately regulated during the stress response of S. mutans.
Members of the Streptococcus mutans group are pathogens causing dental caries and can cause infective endocarditis, but at a lower frequency than Streptococcus sanguis and Streptococcus oralis (17, 33). In the oral cavity, S. mutans colonizes tooth surfaces to form dental plaque, a typical biofilm, and survives fluctuating pH, starvation, and oxygen tension (3). A similar situation may be encountered in the biofilm adherent to the heart valves, where plasma components with bacteriocidal activities must be overcome. The ability of S. mutans to survive in these diverse environments, as well as the underlying physiological adjustments, is an interesting model for studying stress responses.
Recent advances in molecular techniques, in conjunction with progress in determining microbial genome sequences, have enabled comprehensive exploration of cellular functions. The genome sequencing project of S. mutans is currently under way in the University of Oklahoma's Advanced Center for Genome Technology (OU-ACGT). DNA microarrays enable a large-scale, systematic approach to the analysis of transcriptional profiles in microorganisms (22). Analogously, differential display has been successfully used for the comparative analysis of gene expression in eukaryotes (21). A simple, modified technology named RNA arbitrarily primed PCR (RAP-PCR) has been used successfully for detecting differentially expressed genes in prokaryotes (23, 34) and was applied recently in the oral streptococcus Streptococcus gordonii (8). Primers have been designed for RAP-PCR in the Enterobacteriaceae group, based on statistical evaluation of species-specific coding regions extracted from 3′ and 5′ regions of the Escherichia coli genes (9). The strategy was to use a combination of 10 10mer and 11mer primers for bacterial mRNA, which exhibits a low level of polyadenylation. The 11mer primers used for reverse transcription (RT) were selected for localization in the 3′ region of the RNA containing a stop codon. The 10mer oligonucleotides, used subsequently for RT-PCR, bound to the 5′ end of the RNA harboring an ATG start codon. Theoretically, these oligonucleotides could be used as arbitrary primers for nonstringent RT-PCR in any species of microorganism. Practically, these primers have been tested successfully in RAP-PCR for identifying stress-inducible genes of another member of Enterobacteriaceae, Salmonella enterica serovar Typhimurium, in addition to E. coli (35). It was interesting to test the efficacy of these primers for gram-positive bacteria, because computer analysis and searching of the available data bank revealed that 10mer or 11mer primers also should bind to numerous coding sequences in these bacteria.
A comprehensive analysis of multiple-stress responses by two-dimensional polyacrylamide electrophoresis confirmed that S. mutans responded to environmental stresses such as oxidation, heat, acidity, high salt, etc., by specific or coordinated regulation at protein levels (30). However, genetic analysis is still limited, except that the transcriptional levels of two genes, dnaK and ffh, encoding heat shock protein Hsp70 and a homologue of the 54-kDa subunit of signal recognition particle, respectively, were enhanced during acid-shock response (15, 18). To investigate transcriptional regulation in response to different stress conditions reconstituted in vitro, we initiated genetic analysis of S. mutans RNA by RAP-PCR with primers designed for prokaryotic differential display. In this small-scale differential screen, we demonstrated that these primers derived from the genome of E. coli can be applied to gram-positive microorganisms as well for fingerprinting of RNA and identified several genes which are induced or repressed by acidic pH. Transcriptional analysis and genetic characterization of an acid-specific and a general stress protein are reported. Our results also provide data to indicate that three genes responsible for glutamate synthesis are coordinately regulated in stress responses of S. mutans.
MATERIALS AND METHODS
Bacteria and cultivation conditions.
E. coli JM109 was used as the plasmid host, and cultures were grown in Luria-Bertani medium supplemented with ampicillin (100 μg/ml) and/or agar (2%) as required. The E. coli XL1-Blue MRF′ strain, used for the phage library, and the XLOLR strain, used for phagemid recovery, were grown and maintained according to the manufacturer's instructions (Stratagene, La Jolla, Calif.). The transformation-competent S. mutans GS-5 and XC (19) laboratory strains were grown and maintained in brain heart infusion broth (BHI; Difco Laboratories Inc., Detroit, Mich.). For stress challenge, bacteria were grown to the early exponential phase in BHI with 20 mM glucose at pH 7.5. The cells were rapidly concentrated by centrifugation (5,000 × g for 15 min) and washed in sterile saline (0.16 M NaCl), and aliquots (∼106 cells/ml) were inoculated into 10 ml of fresh BHI medium with 20 mM glucose buffered with 40 mM phosphate-citrate buffer under the following conditions: (i) at pH 5.5, (ii) in high osmolarity (+0.5 M NaCl), (iii) with a chaotropic agent (2 M urea), (iv) with antimicrobial agents (40 mg of lysozyme/ml or 50 mg of spectinomycin/ml), and (v) with human serum or saliva (1/10 volume [vol]). Unchallenged control and treated cells (except the pH condition) were incubated at pH 7.5 at 37°C for 30 min. To isolate bacteria from different growth phases at a constant pH and oxygen tension, the growth of S. mutans cells was monitored in a bioreactor (Bioflo3000; New Brunswick Scientific, Edison, N.J.) and cells from the early exponential or stationary phase of growth were recovered for analysis.
RNA preparation and analysis.
Cultures of treated or untreated S. mutans were centrifuged, and the cells were suspended in 200 μl of Tris-EDTA (TE) containing 5 mg of lysozyme per ml. After standing for 20 to 30 min on ice, cell suspensions were treated with 20 μl of 10% sodium dodecyl sulfate and boiled for 2 min (28). Cellular debris was removed by centrifugation, and the supernatants were extracted using the Ultraspec RNA isolation reagent (Biotecx Laboratories, Houston, Tex.). After ethanol precipitation, by addition of 20 μl of 3 M sodium acetate and 500 μl of ethanol, RNA was dissolved in 100 μl of TE. The RNA samples were treated with DNase I (Gibco BRL, Life Technologies, Inc., Gaithersburg, Md.) and subsequently purified by ethanol precipitation. A total of 2.5 to 20 μg of each sample, in a volume of 5 μl, was subjected to dot or Northern (RNA) blot analysis (27). Relative amounts of the transcripts were quantified by densitometric analysis using the Electrophoresis Documentation and Analysis System 120 (Scientific Imaging Systems, Eastman Kodak Co., New York, N.Y.). Two DNA fragments, 719 bp of 16S rRNA (accession no. X58303) and 300 bp of gbp (accession no. M30945, encoding a 74-kDa glucan-binding protein), were used as internal controls for total RNA loaded on dot or Northern blots. The quantity estimated from the intensity of the bands correlated with the scintillation counting of bands cut directly from the membranes.
Fingerprinting of RNA and identification of RAP-PCR products.
Fingerprinting of RNA by RAP-PCR was employed essentially as described by Welsh et al. with modification (34). Ten-microliter samples containing 12 ng of RNA were incubated at 65°C for 10 min and then put on ice. Ten microliters of cDNA synthesis buffer, containing 100 mM Tris-HCl (pH 8.0), 100 mM KCl, 8 mM MgCl2, 20 mM dithiothreitol, 2 mM deoxynucleoside triphosphates, 1 μM primer (see below), and 5 U of Moloney murine reverse transcriptase (Stratagene), was added. The cDNA synthesis was carried out at 37°C for 1 h. After cDNA synthesis, 2 μl of sample was subjected to PCR with the same primer or in combination with other primers at a concentration of 2 μM. The PCR products were labeled with [α-33P]dATP (2,000 Ci/mmol). The thermal cycling parameters were as follows: 94°C (0.5 min), 40°C (2 min), 72°C (1 min) for 40 cycles. The primers selected for initial testing were 10 10mer RT primers (Ea1 to Ea10) and 4 11mer PCR primers (Es1, Es6, Es8, and Es11), originally designed for RAP-PCR in Enterobacteriaceae (9). Subsequent RAP-PCR for differential display of RNA samples from bacteria exposed to different stresses was carried out with two 11mer RT primers, Ea1 (TTTTATCCAGC) and Ea7 (TCTTTTTTACC), and three 10mer PCR primers, Es1 (GCTGGAAAAA), Es3 (GAAGTGCTGG), and Es8 (TGCCGATGAA). The RT-PCR samples were analyzed by standard sequencing gels, and RAP-PCR fingerprints were visualized by autoradiography using Kodak XAR film. Two independent RNA samples prepared from bacteria treated under different stress conditions were fingerprinted, and duplicated signals from at least two RAP-PCRs were isolated and analyzed. In addition, in each reaction, parallel experiments carried out without cDNA synthesis yielded nondetectable signals and, therefore, confirmed that the amplified products were not due to DNA contamination. The bands of RAP-PCR products were cut from the gel, and the DNA was eluted into 50 μl of TE by heating at 65°C for at least 1 h. Five microliters of the eluent was PCR amplified with the same sets of primers, cloned into the pT7Blue(R) vector (Novagen, Inc., Madison, Wis.), and sequenced using Sequenase, version 2 (U.S. Biochemical, Cleveland, Ohio). The deduced nucleotide and amino acid sequences were analyzed by BLASTN and BLASTX (National Center for Biotechnology Information, Los Alamos, N.Mex.), which are available via the Internet.
Genomic library and cloning.
Chromosomal DNA from S. mutans GS-5 was partially digested with Sau3AI and size fractionated by gel electrophoresis. The 4- to 12-kbp size range was eluted from the gel, ligated to the BamHI-cut arms of Lambda ZAP Express (Stratagene), packaged, and amplified according to the manufacturer's instructions. Plaques were transferred to nylon membrane and hybridized with DNA probes labeled with 32P using the random-primer method, as instructed by the manufacturer (Boehringer GmbH, Mannheim, Germany). The pBK-CMV phagemids containing hybridizing inserts were excised from the Lambda ZAP Express vector as described by Stratagene. The partial sequence of the GSP-781 was submitted previously by another group under accession no. U78607. See “Nucleotide sequence accession number” below regarding submission of the complete sequence.
Gene disruption mutagenesis.
AP-185 was disrupted by integration of pEVP3, a lacZ reporter vector for insertion mutagenesis in streptococci, targeted by an internal fragment of the gene, as previously described (26). The S. mutans GS-5 and XC strains were used for mutagenesis. Mutants were selected on BHI agar containing chloramphenicol. The single integration of pEVP3 in the targeted gene was verified by Southern hybridization. To target integration of pEVP3 into the ASP-185 locus, a mutagenic plasmid was made by subcloning a 305-bp EcoRI-PstI fragment of this gene into pEVP3 digested with the same enzymes. The resulting plasmid was transformed into S. mutans GS-5 or XC strain to disrupt the corresponding chromosomal loci by homologous recombination. GS-B13 was constructed as a control strain by duplication of the promoter region of the gtfB gene, leaving a copy of intact gtfB downstream of the integrated pEVP3. The phenotypic characteristics of mutants carrying the desired mutations, GS-85, XC-85, and GS-B13, were tested by biochemical reactions and serological typing and showed no distinct changes compared with wild-type strains.
General genetic manipulations.
Transformation of S. mutans was performed as described by Perry and Kuramitsu (25), and DNA extraction, Southern blots, and PCR were performed as described previously (4, 5). Automated DNA sequencing was performed by the Molecular Biology Facility, College of Medicine, National Taiwan University. Sequencing was carried out with the custom oligonucleotides, and all sequencing was in both directions.
Evaluation of adaptation to environmental stress.
The environmental adaptability of the wild type (GS-5 or XC) and mutants (GS-85, XC-85, or GS-B13) was evaluated by monitoring bacterial growth in Todd-Hewitt broth (Difco) containing 50 mM sodium acetate at appropriate pH values or the indicated concentrations of NaCl. Temperature sensitivity was determined from 37 to 42°C. To initiate the growth experiments, bacteria grown to early log phase were inoculated into 3 ml of Todd-Hewitt broth with or without the indicated modification. Bacterial growth was monitored by measurement of optical density at 550 mm. Adaptation to acid tolerance was measured by the ability to survive a 3-h exposure to a killing pH of 3.0, as previously described (29). The number of surviving cells was determined for each mutant strain tested and was compared with that of the corresponding wild-type strain.
Nucleotide sequence accession number.
The complete sequence of GSP-781 was submitted to GenBank on 19 January 2001 and was assigned accession no. AF338445 (BankIt 381076).
RESULTS
Differential display of stress-responsive genes.
S. mutans RNA was submitted to differential display by RAP-PCR by using sets of primers of different G+C content, 10 11mer RT and 4 10mer PCR primers (9), in various combinations. After initial evaluation of various reaction conditions and concentrations, we found that PCRs were optimal at an annealing temperature of 40°C. We used [α-33P]dATP incorporation instead of [α-32P]dATP, which had been used in previously reported RAP-PCR with E. coli or Salmonella serovar Typhimurium (9, 35). The PCR products with a size ranging from 150 to 450 bp gave the most reliable reproducibility (Fig. 1). Although all 10 11mer RT primers (Ea1 to Ea10), in combination with any of the 4 10mer PCR primers (Es1, Es3, Es8, and Es11), could amplify cDNA, 2 RT primers, Ea1 and Ea7, in combination with any of the 4 PCR primers, gave better amplification than the other primers in terms of the number of bands generated. We also found that pairs of RT primers, instead of combinations with PCR primers (for example, Ea 1 with Ea 1, Ea 7, or Ea 8), could successfully amplify a significant number of bands with high reproducibility. Therefore, these primers, Ea1, Ea7, Es1, Es3, and Es8, were used for RAP-PCR of RNA samples from S. mutans submitted to various stresses.
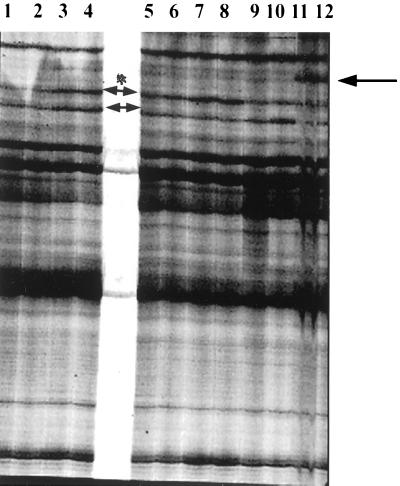
FIG. 1.
Representative RAP-PCR fingerprinting of RNAs. Total RNA from S. mutans GS-5 cells in the early exponential phase, after various stress treatments for 30 min, were RAP-PCR fingerprinted with primers Ea7 and Es8 in two independent reactions. A portion of the gel is shown with fragment lengths ranging from 180 to 360 bp. Lanes 1 and 2, control RNA from cells growing in pH 7.4 BHI broth; lanes 3 and 4, cells exposed in pH 5.5; lanes 5 and 6, cells treated with spectinomycin (50 mg/ml); lanes 7 and 8, cells treated with 500 mM NaCl; lanes 9 and 10, cells treated with whole saliva; lanes 11 and 12, cells treated with serum. Arrowheads indicate differentially amplified products. ∗, AP-781 gene fragment which was stimulated by low pH and high osmotic pressure as confirmed by Northern blot.
Identifying and characterizing acid-induced or -repressed genes.
Distinct bands in response to different stresses, such as an acidic pH of 5.5, elevated temperature, high osmolarity, and lysozyme, were obtained, and identification of their sequences is currently under way in this laboratory. In this paper, we report six genes that were activated (AP-11, AP-712, AP-711, GSP-781, AP-785, and AP-185) and three (AR-186, -731, and -732) that were repressed by exposure to acid stress (Table 1). All were identified from signals duplicated in at least two independent RAP-PCRs and were subsequently confirmed by dot blot analysis of RNA, with independent RNA preparations for estimating the ratios of induction or repression. Computer analysis (BLASTX) mapped three of the genes (AP-731, AP-732, and GSP-781) to S. mutans isocitrate synthase (icd) (6), citrate synthase (citZ) (6), and the partial sequence of a secreted protein (unpublished data). By using the S. mutans genome sequence provided by OU-ACGT, the sequence of AR-186 was mapped to a putative aconitase gene (acn). The positions as well as the sequence that the RAP-PCR primers annealed in these genes are listed in Table 2.
TABLE 1.
Homology analyses of nucleotide sequences of DNA fragments derived from differential display RT-PCR
| Gene | RT/PCR primer | Predicted protein (organism) | Ratio of induction or repressiona | No. of residues compared (% identity) | Accession no. (reference) |
|---|---|---|---|---|---|
| AP-11 | Ea7/Es1 | Hippurate hydrolase hipO (Bacillus subtilis) | 2.3 | 148/293 (50) | E69640 (20) |
| AP-712 | Ea7/Es1 | Acetolactate synthase large subunit (L. lactis) | 3.0 | 17/35 (48) | Q02137 (13) |
| AP-711 | Ea7/Es3 | Pyruvate carboxylase subunit B (Methanococcus jannaschii) | 2.5 | 196/333 (58) | Q58628 (2) |
| GSP-781 | Ea7/Es8 | Putative secreted protein (S. mutans) | 3.0 | 160/160 (100) | AAD00288 |
| AP-785 | Ea7/Es8 | Conserved hypothetical protein (Thermotoga maritima) | 2.0 | 50/162 (30) | AAD35257 (24) |
| AP-185 | Ea1/Es8 | Branched-chain amino acid aminotransferase (Haemophilus influenzae) | 3.0 | 98/125 (78) | P54689 (10) |
| AR-186 | Ea1/Es8 | Aconitate hydratase (B. subtilis) | 0.1 | 90/119 (75) | G69599 (7) |
| AR-731b | Ea7/Ea7 | Isocitrate dehydrogenase (S. mutans) | 0.1 | 126/131 (96) | Q59940 (6) |
| AR-732b | Ea7/Ea7 | Citrate synthase (S. mutans) | 0.1 | 38/38 (100) | Q59939 (6) |
The ratio of induction or repression was estimated by the intensity of the bands of RNA from samples treated with pH 5.5 for 30 min over the intensity of the bands from untreated samples detected on dot or Northern blots.
AR-731 and -732 were identical PCR products from independent RAP-PCRs but were cloned in opposite orientation in pT7Blue(R).
TABLE 2.
Matching of primer sequences to known S. mutans templates
| Locus | RT primer (position) and sequence | PCR primer (position) and sequence | Coding sequencea | Length of PCR product (bp) |
|---|---|---|---|---|
| AR-186 | Ea1 (855) | Es8 (455) | ||
| 5′-TTTTATCCAGC | 5′-TGCCGATGAA | |||
| IIIIII | IIIII | 1∼2667 | 401 | |
| 5′-GAAAGTCCAGG | 5′-GCGTTATGAA | |||
| AR-731(icd)/AR-732(citZ) | Ea7 (2110) | Ea7 (1771) | ||
| 5′-TCTTTTTTACC | 5′-TCTTTTTTACC | 1798∼1916 (citZ) | ||
| IIIIIIIIII | IIIIIIII | 1918∼3099 (icd) | 340 | |
| U62799b | 5′-CTTTTTTTACC | 5′-CTATTTTTACC | ||
| GSP-781 | Ea7 (1244) | Es8 (896) | ||
| 5′-TCTTTTTTACC | 5′-TGCCGATGAA | |||
| I IIIII | I I III II | 1∼1295 | 349 | |
| 5′-CGGTAGTTACC | 5′-TTCTGATCAA |
The sequence of AR-186 was derived from the S. mutans genome sequence provided by the University of Oklahoma's Advanced Center for Genome Technology.
EMBL accession number.
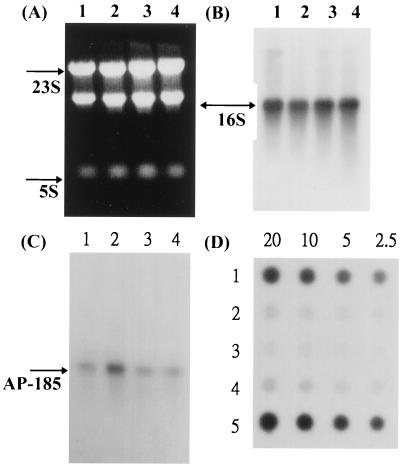
Northern blot analysis further confirmed that acid stress induced around a threefold increase in the transcription of AP-185 compared to that of the untreated control (Fig. 2C, lane 2). The changes were not due to the difference in total RNA loaded, as confirmed by the signals of 16S tRNA (Fig. 2A and B). On the other hand, the transcript of AR-186 was significantly reduced by acid stress, to about 10% of the untreated level (Fig. 2D, lane 2). The significant repression in the transcription of AR-186 was also detectable when S. mutans was exposed to high osmolarity or temperature, in addition to acid stress (lanes 3 and 4). The transcripts of both AP-185 and GSP-781 appeared to be monocistronic, with a size of around 1.5 kb on Northern blots. Interestingly, transcription of AR-186 was enhanced significantly, about twofold, by the addition of 1/10 vol of plasma in the BHI medium (Fig. 2D, lane 5). These results confirm our initial hypothesis that adjustment of S. mutans to various environmental stresses, such as low pH and serum components, involves differential regulation at the transcriptional level.
FIG. 2.
Northern and dot blot analysis of RNA from cells in the early exponential phase and after exposure to low pH (5.5) and other stress. (A, B, and C) Total RNAs were prepared from cultures of S. mutans GS-5 without treatment (control, lane 1) or treated for 30 min with pH 5.5 (lane 2), 1/10 vol of plasma (lane 3), and serum (lane 4). Ten micrograms of each RNA was electrophoresed through 1% agarose–2.2 M formaldehyde gels and transferred to nylon membrane. (A) Total RNA on gels stained with ethidium bromide. (B) The membrane was hybridized with a 719-bp 16S rRNA fragment used to control the amount of RNA loaded. (C) The same membrane was hybridized with radiolabeled DNA fragments from AP-185. (D) Dot blot analysis of different amounts of RNA as indicated (20, 10, 5, and 2.5 μg). The membrane was hybridized with AR-186 for untreated RNA (lane 1) or RNA treated for 30 min with pH 5.5 (lane 2) or 400 mM NaCl (lane 3) and incubated at 42°C (lane 4) and with 1/10 vol of plasma (lane 5).
Induction time course and growing phase dependence of acid-specific AP-185 and general-stress AP-781.
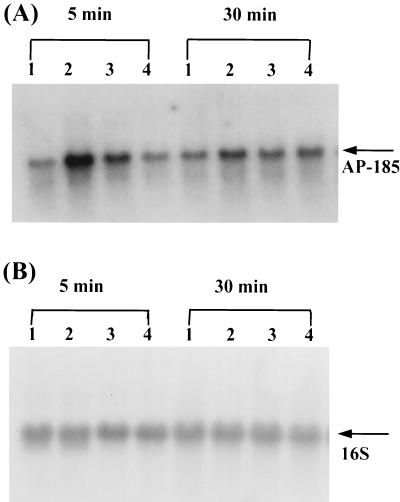
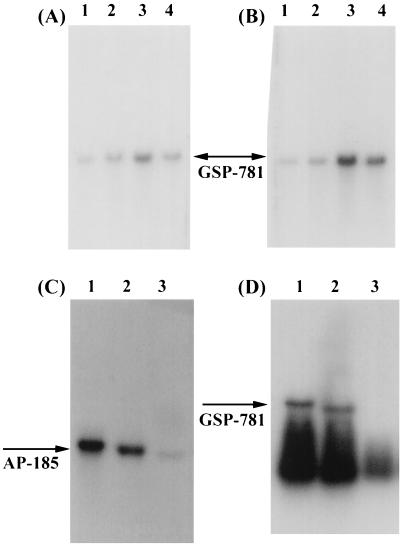
In order to investigate further the specificity and the time required for induction of AP-185 and GSP-781, in response to different stresses, RNA was extracted from S. mutans exposed to high osmolarity or high temperature for different time intervals for Northern blot analysis. Induction of both AP-185 and GSP-781 was detectable only 5 min after exposure to acidic medium. The increase in transcription of AP-185, in response to acid stress, was higher (around eightfold) at 5 min than at 30 min (Fig. 3A) and persisted for at least 1 h. Enhanced expression of AP-185 was restricted to acid stress, although a slight increase was observed initially when cells were exposed to high osmotic pressure for 5 min (Fig. 3A, lane 3). The transcription of GSP-781 increased slightly after exposure to acidic medium for 5 min and increased continuously to 2.5-fold at 30 min (Fig. 4A and B, lane 2). Therefore, acid stress induced a rapid expression of AP-185 as a function of pH. Distinct from AP-185, the induction of GSP-781 was more pronounced when cells were exposed to high osmolarity and temperature, with a stimulation fold higher (fivefold versus threefold) by the former than the latter (Fig. 4A and B, lanes 3 and 4). To determine whether the induction of AP-185 and GSP-781 is dependent on growth phase, S. mutans cells were monitored growing at a constant pH of 7.5 under anaerobic conditions in a bioreactor and cells from the early, exponential, and stationary phases of growth were recovered. The expression of both AP-185 and GSP-781 appeared to be growth phase dependent, with significantly higher levels of transcription in the exponential phase than in the stationary phase (Fig. 4C and D).
FIG. 3.
Time course and stress-specific induction of AP-185. Total RNA from S. mutans GS-5 cells without treatment (control, lane 1) or treated with pH 5.5 (lane 2), with 400 mM NaCl (lane 3), and at 42°C (lane 4) for 5 min or 30 min was hybridized with radiolabeled DNA fragments from AP-815 (A) or 16S rRNA (B).
FIG. 4.
Northern blot analysis of gene expression in response to different stresses and growth phases. (A and B) Total RNA from S. mutans GS-5 cells without treatment (control, lane 1) or treated with pH 5.5 (lane 2), with 400 mM NaCl (lane 3), and at 42°C for 5 min (A) or 30 min (B) was hybridized with radiolabeled GSP-781. (C and D) S. mutans GS-5 was grown in a bioreactor (Bioflo3000; New Brunswick Scientific) under constant oxygen tension and a pH of 7.2 ± 0.2. Total RNA from cells grown to early exponential (lane 1), middle exponential (lane 2), and early stationary (lane 3) phase was probed with AP-185 (C) or GSP-781 (D).
Adaptation to stress conditions of the AP-185 mutant.
To confirm the specific role of AP-185 in response to acid stress, the gene was inactivated by insertion of plasmid pEVP3. Strains XC-85 and GS-85, with AP-185 inactivated, showed retarded growth when cultured in acidic medium (pH = 5.5) but not in high osmolarity (400 mM NaCl) nor at elevated temperature (42°C). In addition, strains XC-85 and GS-85 also exhibited decreased resistance to the killing effect of pH 3.0 compared to the wild-type XC strain (55% reduction in survival). Both XC-85 and GS-85 grew at the same rate as the wild-type strains at pH 7.5. These results indicated that AP-185 might be an acid-stress-inducible gene.
Cloning and sequence analysis of general-stress AP-781.
To analyze further the genetic characteristics of GSP-781, genetic cloning was performed and the complete sequence of GSP-781 was determined. The predicted length of the transcript, around 1.5 kb, was consistent with that observed on the Northern blot (Fig. 3 and 4). The sequence of 349 bp, obtained from RAP-PCR, mapped to the C terminus of AP-781 from nucleotides 896 to 1244 and confirmed the identity of AP-781 (Table 2). AP-781 encoded a putative protein with a molecular mass of around 45 kDa and with a pI of 4.8. The deduced polypeptide of this gene showed 75% homology with a secreted protein, Usp45, from Lactococcus lactis with no previously known biological significance (32).
Transcription and regulation of genes in citrate pathway after stress exposure.
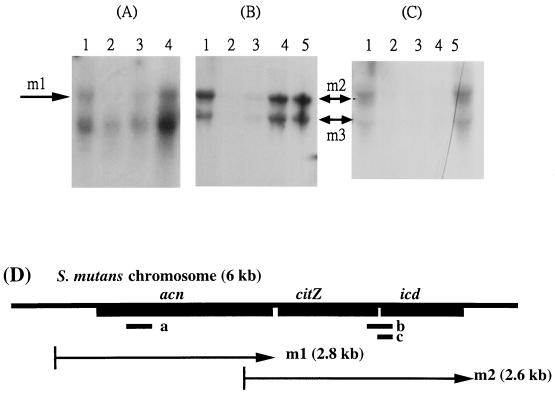
The AR-186 (acn, putative aconitase), AR-731 (icd), and AR-732 (citZ) genes are clustered together on the S. mutans UA159 chromosome and encode metabolic enzymes responsible for the synthesis of glutamate and constitute the citrate pathway (6). The repression of the AR-186 transcriptional level by acid stress warranted the investigation of the response of these functionally coupled genes to other stress treatments. Northern blots prepared with RNA isolated from S. mutans after different stress exposures were hybridized with three different probes, as indicated in Fig. 5. Figure 5A shows the results obtained with a probe prepared from AR-186 whose sequence is complementary to acn. A band named m1, whose size (2.8 kb) corresponds to the predicted open reading frame of aconitase, was detected in lane 1. This 2.8-kb transcript was almost abolished after 30 min of exposure to acid or high osmolarity (Fig. 5A, lanes 2 and 3), but was enhanced about twofold when 1/10 vol of plasma was added in the challenge medium (lane 4), correlating with the results detected earlier on dot blot (Fig. 1D). A transcript of approximately 1.4 kb was also present when hybridized with AR-186, and the identity of this band awaits further investigation.
FIG. 5.
Northern blot analysis of acn, citZ, and icd. (A) Samples of total RNA were prepared from cells in early exponential growth phase (lane 1, untreated), after 30 min in pH 5.5 (lane 2), in 400 mM NaCl (lane 3), and with 1/10 vol of plasma (lane 4). The hybridization was achieved with the 32P-labeled probe deduced from AR-186 (probe a), with the sequence complementary to the 5′ end of acn. (B) Samples of total RNA were prepared from cells in early exponential growth phase (lane 1, untreated), after 30 min in pH 5.5 (lane 2), in 400 mM NaCl (lane 3), and after incubation at 42°C and in 1/10 vol of plasma (lane 4) or serum (lane 5). The hybridization was achieved with the 32P-labeled probe deduced from AR-731 (probe b), with a sequence complementary to both citZ and icd. (C) Total RNA from S. mutans GS-5 cells without treatment (control, lane 1) or treated with pH 5.5 (lane 2), with 400 mM NaCl (lane 3) and at 42°C (lane 4), and with 1/10 vol of plasma (lane 5) for 30 min was hybridized with radiolabeled probe c, with a sequence complementary to the 5′ end of icd. (D) Schematic representation of the S. mutans chromosome containing the genes acn, citZ, and icd, involved in the citrate pathway. The small black bars under the cluster indicate positions of the different probes used for Northern blots. Arrows indicate positions of the predicted transcripts, m1 (acn) and m2 (citZ-icd operon). The m3 might correspond to the transcript of icd only or might be a processed product of m2.
Northern blots hybridized with AR-732, complementary to a sequence between citZ and icd, showed two bands, m2 and m3, with sizes of 2.6 and 1.4 kb, respectively (Fig. 5B). An identical pattern of hybridization was detected on a Northern blot hybridized with a probe specific to the 5′ end of icd (Fig. 5C). Based on these results, the transcript of m2 corresponds to the operon of citZ and icd, while m3 might correspond to the transcript of icd only or might be a processed product of m2. These results also suggested that transcription initiation and the promoter sequence of m2 may locate in the coding regions of acn.
Dot and Northern blots confirmed that in addition to acid, high osmolarity and elevated temperature repressed the expression of m1, m2, and m3 simultaneously with comparable ratios of reduction, around 85 to 92% (Fig. 2D and Fig. 5). By contrast, exposure of the bacteria to either plasma or serum induced enhanced expression of m1, m2, and m3 with a similar fold increase (1.5- to 2-fold on Northern blot). These results indicated that citZ and icd without acn constitute an operon and that acn and the citZ-icd operon of S. mutans are coordinately regulated in the multiple-stress response.
DISCUSSION
Although considerable information about gram-negative bacteria, such as E. coli and Salmonella, has been accumulated recently, little is known about the stress responses in gram-positive bacteria. In S. mutans, two genes essential for stress resistance, dgk and sgp, were identified by transposon mutagenesis (1, 36). In this study we identified acid-stress-responsive genes in S. mutans using RAP-PCR. Although the application of differential display to prokaryotes has been well documented, studies of gram-positive microorganisms are limited. We demonstrated that the primers originally designed for the Enterobacteriaceae (9) can be used for gram-positive microorganisms also, despite the fact that G+C content (36 to 38% for S. mutans) is an important parameter in primer design. The rationale to test these primers was that none of the 11mer RT or 10mer PCR primers show homology to rRNA or other abundant small RNA species, but can bind to numerous coding sequences in gram-positive bacteria. In contrast to previous RAP-PCR studies performed with E. coli or Salmonella serovar Typhumurium (6, 31), our differential display RT-PCR was labeled with [α-33P]dATP instead of [α-32P]dATP. The 33P has a radiation energy and half-life between that of 32P and 35S. We found that direct incorporation of [α-33P]dATP in PCRs resulted in satisfactory band intensity for products ranging from 200 to 500 bp. DNA fragments of these sizes were amenable to subsequent isolation and cloning. Smaller molecular bands, less than 150 bp, were difficult to visualize by autoradiography, a problem which could be solved by using 32P-end-labeled primers as reported previously (31).
To identify conditionally induced or repressed genes, both the number of the bands and their intensity on autoradiography were key factors to be controlled. We found that the G+C contents of the microorganisms should be taken into consideration when optimizing the RAP-PCR fingerprinting. As shown in the present study, the primers that gave better results were found to have a low G+C content, as is the case for S. mutans chromosomal DNA (12). Pairs of RT primers, instead of combinations with PCR primers, could successfully amplify a significant number of bands with high reproducibility (Table 2; Ea7 alone could serve as both RT and PCR primers). Therefore, these sequence-based designs of primers indeed could be efficiently adopted for functional screening of conditionally controlled genes in gram-positive microorganisms even though the genome sequences of S. mutans are not yet completed. It should be noted that the RAP-PCR products must be verified subsequently by dot or Northern blots, even though the signals could be reproduced from independent RT-PCRs. As shown in Table 1, all of the nine genes were confirmed by dot blots, whereas false-positive RAP-PCR products were occasionally identified during differential display. Such drawbacks could be minimized if identical signals or RAP-PCR products could be obtained by using different sets of primers.
The results of the present study also confirm the hypothesis that in order to survive in different tissue compartments and adjust to the environmental stresses encountered, S. mutans could respond to external stimuli through the transcriptional regulation of multiple genes. Previous analysis of stress-responsive elements in S. mutans by two-dimensional polyacrylamide electrophoresis had identified more than 69 proteins which were up-regulated and 49 proteins which were down-regulated in the exponential phase when the bacteria were treated with different stresses, including low pH (pH = 5.0), salt, heat, oxidation, and starvation (30). The proteins that exhibited increased synthesis were classified as either stress-specific or general-stress proteins, according to their responses to single or multiple stimuli. Genetic evidence and characteristics of site-specific mutants defective at the AP-185 locus indicated that AP-185 might code for an acid-stress protein. Sequence alignment of a portion of AP-185 revealed homology with amino acid aminotransferases from several microorganisms, including the gram-positive B. subtilis (20). Interestingly, a computer-based sequence analysis of the B. subtilis complete genome has found that several putative regulatory proteins display significant similarity to aminotransferases (and these enzymes have been reported to show similarity to repressors) (20). The genetic information obtained from this study also indicated that GSP-781 might encode a protein which is responsive to multiple stress stimuli, including low pH, high osmotic pressure, and temperature. The molecular mass and pI derived from the nucleotide sequence are 43 kDa and 5.5, respectively. Sequence comparison of this protein indicated that it shares 75% homology with a secreted protein, Usp45, of unknown function from L. lactis (32). In addition, the predicted signal sequences of both proteins share nearly 95% identity. Preliminary data from this laboratory indicated that the GSP-781-encoded protein is a cell wall-associated protein, which can be extracted from S. mutans cells with 8 M urea and might be essential for the maintenance of the stability of cell membrane or cell wall.
As far as pathogenesis is concerned, genes regulated by acidic pressure as well as serum components are of particular interest because S. mutans induces dental caries and causes infective endocarditis in vivo. We found that transcription of AR-186 was induced specifically within 30 min by exposure to plasma or serum but was depressed significantly by acid stress. This result led us to speculate that the AR-186-encoded protein might be important for the survival of S. mutans in the blood circulation, and experiments are currently in progress in this laboratory to test this hypothesis. The partial nucleotide sequence of AR-186 shared 75% homology with aconitate hydrase (aconitase) from B. subtilis, which belongs to the superfamily of iron-responsive element-binding proteins (7). By using the S. mutans genome sequence provided by OU-ACGT, the putative aconitase gene (acn) was located immediately upstream of citZ. The sequence of AR-186 completely mapped without alterations to the 5′ end of this gene (Table 2). The function of aconitase is to convert citrate to isocitrate in the citrate pathway of glutamate biosynthesis. The citrate pathway has recently been reported for S. mutans, although the identity and biological activities of the aconitase have not yet been characterized (6). The tandemly arranged icd gene, coding for isocitrate dehydrogenase, and citZ gene, encoding citrate synthase, were proposed to constitute the pathway for the synthesis of glutamic acid in collaboration with aconitase (6). Insertional inactivation of icd resulted in an auxotrophic mutant which required the addition of glutamate for growth (14). Transcriptional analyses of icd and citZ have not yet been reported, but a twofold increase in the enzymatic activities of both enzymes was detected when S. mutans was grown in minimal medium, as compared with those of cells grown in rich medium (6). Dot and Northern blot analysis conducted in the present study confirmed a differential regulation of acn, icd, and citZ when exposed to acid stress or serum components. Our results also indicated that the transcriptions of all three genes, icd, citZ, and AR-186, are coordinately regulated in response to different environmental stresses, such as acid, osmotic pressure, and high temperature.
The clustered organization and coordinate regulation of acn, citZ, and icd merit further investigation of the transcriptional units of these genes. acn was separated from the downstream citZ by 5 bp, and citZ was also separated from icd by 2 bp only. No putative transcriptional terminator or putative ςA consensus sequence was present between acn and citZ or between citZ and icd. In the 5′-end untranscribed region, about 300 bp of acn shared 51% homology with the promoter region in the citB gene of B. subtilis (7). These data initially suggested that acn, citZ, and icd might constitute an operon but that was found not to be the case. Northern blot made with AR-186 complementary to acn hybridized to a transcript of 2.8 kb, while AR-732 with a sequence complementary to both citZ and icd, or a specific probe mapped to the 5′ end of icd, hybridized to signals of approximately 2.6 and 1.4 kb (Fig. 5). Although none of these probes detected signals equivalent to the expected operon size of a transcript (more than 5 kb) containing all three genes, the possible acn-citZ-icd operon could not be neglected. Instead of the long transcript, citZ and icd without acn constitute an operon, as confirmed by the 2.6-kb transcript on Northern blot and the existence of the RT-PCR product of AR-731 and -732 which contained the sequences of both citZ and icd. These results indicated that although acn, citZ, and icd were coordinately regulated, acn or citZ-icd had their own regulatory cis element. In addition, icd might also have its own promoter and regulatory cis element, located in the coding regions of citZ, or citZ-icd might be processed to another shorter transcript and be regulated by stress such as acid, high osmolarity, or temperature. The existence of multiple transcription units in these genes strongly suggested the importance of maintaining regulation in the citrate pathway. Work is in progress to understand the molecular basis responsible for this regulation and to identify the candidate regulator(s) of these genes.
It was interesting to find that most genes retrieved from RAP-PCR encode metabolic enzymes which were regulated in response to stress conditions. In Salmonella serovar Typhimurium, the synthesis of glutamate was assumed to maintain the steady-state potassium pool and K+ glutamate was required for optimal growth of this gram-negative bacterium (37). Obviously, this may not be the case for S. mutans growing in the presence of low pH or high salt concentrations or temperature, because all genes specifically responsible for the glutamate synthesis were almost shut down at the transcriptional level. Therefore, it is likely that the synthesis of glutamate must be tightly regulated for the optimal growth of S. mutans under stress conditions. This is the first report that the transcription of genes encoding enzymes responsible for glutamate synthesis in S. mutans is significantly repressed (below 10 to 15% of the level in neutral pH) in acidic and other stress conditions. On the other hand, several genes encoding metabolic enzymes were up-regulated by acid stress, for example, AP-711, which putatively encodes pyruvate carboxylase, a member of the pyruvate dehydrogenase complex (16). Giard et al. recently reported similar results for another gram-positive microorganism, Enterococcus faecalis; the enzymes involved in pyruvate metabolism, such as l-lactate dehydrogenase, lipoamide dehydrogenase, and pyruvate decarboxylase, were up-regulated during stress conditions in order to compensate for the defects caused by inactivation of a general-stress protein, Gls24 (11). These data suggested that the enzymes in pyruvate metabolism may function as general-stress proteins and may respond to regulations triggered by different stress treatments.
ACKNOWLEDGMENTS
We thank H. K. Kuramitsu, T. Koga, and S. Hamada for bacterial strains and D. A. Morrison for plasmid pEVP3. We thank Chia-Wei Chang for technical assistance in differential display. The sequence of acn was obtained from the University of Oklahoma's Advanced Center for Genome Technology (URI: www.genome.ou.edu). We thank Tim J. Harrison, Reader in Molecular Virology, Royal Free and University College Medical School, for his kind review and help in the preparation of this paper.
This work was supported in part by the National Science Council (grants NSC-862314-B002-113, 872314-B-002-263, and 89-2314-B-002-184) and National Health Research Institute (grant NHRI-GT-ZX89B814C).
REFERENCES
- 1.Baev D, England R, Kuramitsu H K. Stress-induced membrane association of the Streptococcus mutansGTP-binding protein, an essential G protein, and investigation of its physiological role by utilizing an antisense RNA strategy. Infect Immun. 1999;67:4510–4516. doi: 10.1128/iai.67.9.4510-4516.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 3.Carlsson J. Bacterial metabolism in dental biofilms. Adv Dent Res. 1997;11:75–80. doi: 10.1177/08959374970110012001. [DOI] [PubMed] [Google Scholar]
- 4.Chia J S, Hsu T Y, Teng L J, Chen J Y, Hahn L J, Yang C S. Glucosyltransferase gene polymorphism among Streptococcus mutansstrains. Infect Immun. 1991;59:1656–1660. doi: 10.1128/iai.59.5.1656-1660.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chia J S, Lin S W, Hsu T Y, Chen J Y, Kwan H W, Yang C S. Analysis of a DNA polymorphic region in the gtfB and gtfC genes of Streptococcus mutansstrains. Infect Immun. 1993;61:1563–1566. doi: 10.1128/iai.61.4.1563-1566.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cvitkovitch D G, Gutierrez J A, Bleiweis A S. Role of the citrate pathway in glutamate biosynthesis by Streptococcus mutans. J Bacteriol. 1997;179:650–655. doi: 10.1128/jb.179.3.650-655.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dingman D W, Sonenshein A L. Purification of aconitase from Bacillus subtilis and correlation of its N-terminal amino acid sequence with the sequence of the citBgene. J Bacteriol. 1987;169:3062–3067. doi: 10.1128/jb.169.7.3062-3067.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dû L D, Kolenbrander P E. Identification of saliva-regulated genes of Streptococcus gordoniiDL1 by differential display using random arbitrarily primed PCR. Infect Immun. 2000;68:4834–4837. doi: 10.1128/iai.68.8.4834-4837.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fislage R, Berceanu M, Humboldt Y, Wendt M, Oberender H. Primer design for a prokaryotic differential display RT-PCR. Nucleic Acids Res. 1997;25:1830–1835. doi: 10.1093/nar/25.9.1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J F, Dougherty B A, Merrick J M, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 11.Giard J-C, Rince A, Capiaux H, Auffray Y, Hartke A. Inactivation of the stress- and starvation-inducible gls24 operon has a pleiotrophic effect on cell morphology, stress sensitivity, and gene expression in Enterococcus faecalis. J Bacteriol. 2000;182:4512–4520. doi: 10.1128/jb.182.16.4512-4520.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilmour M N, Whittam T S, Kilian M, Selander R K. Genetic relationships among the oral streptococci. J Bacteriol. 1987;169:5247–5257. doi: 10.1128/jb.169.11.5247-5257.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Godon J J, Chopin M C, Ehrlich S D. Branched-chain amino acid biosynthesis genes in Lactococcus lactis subsp. lactis. J Bacteriol. 1992;174:6580–6589. doi: 10.1128/jb.174.20.6580-6589.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gutierrez J A, Crowley P J, Brown D P, Hamilton J D, Yangman P, Bleiweis A. Insertional mutagenesis and recovery of interrupted genes of Streptococcus mutans by using transposon Tn917: preliminary characterization of mutants displaying acid sensitivity and nutritional requirements. J Bacteriol. 1996;178:4166–4175. doi: 10.1128/jb.178.14.4166-4175.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gutierrez J A, Crowley P J, Cvitkovitch D G, Brady L J, Hamilton I R, Hillman J D, Bleiweis A S. Streptococcus mutans ffh, a gene encoding a homologue of the 54 kDa subunit of the signal recognition particle, is involved in resistance to acid stress. Microbiology. 1999;145:357–366. doi: 10.1099/13500872-145-2-357. [DOI] [PubMed] [Google Scholar]
- 16.Hemila H, Palva A, Paulin L, Arvidson S, Palva I. Secretory S complex of Bacillus subtilis: sequence analysis and identity to pyruvate dehydrogenase. J Bacteriol. 1990;172:5052–5063. doi: 10.1128/jb.172.9.5052-5063.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horaud T, Delbos F. Viridans streptococci in infective endocarditis: species distribution and susceptibility to antibiotics. Eur Heart J. 1984;5(Suppl. C):39–44. doi: 10.1093/eurheartj/5.suppl_c.39. [DOI] [PubMed] [Google Scholar]
- 18.Jayaraman G C, Penders J E, Burne R A. Transcriptional analysis of the Streptococcus mutans hrcA, grpE, and dnaKgenes and regulation of expression in response to heat shock and environmental acidification. Mol Microbiol. 1997;25:329–341. doi: 10.1046/j.1365-2958.1997.4671835.x. [DOI] [PubMed] [Google Scholar]
- 19.Koga T, Asakawa H, Okahashi N, Takahashi I. Effect of subculturing on expression of a cell-surface protein antigen by Streptococcus mutans. J Gen Microbiol. 1989;135:3199–3207. doi: 10.1099/00221287-135-12-3199. [DOI] [PubMed] [Google Scholar]
- 20.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell S C, Bron S, Brouillet S, Bruschi C V, Caldwell B, Capuano V, Carter N M, Choi S K, Codani J J, Connerton I F, Danchin A. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 21.Liang P, Pardee A B. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 22.Liang P, Pardee A B. Recent advances in differential display. Curr Opin Immunol. 1995;7:274–280. doi: 10.1016/0952-7915(95)80015-8. [DOI] [PubMed] [Google Scholar]
- 23.Mathieu-Daude F, Trenkle T, Welsh J, Jung B, Vogt T, McClelland M. Identification of differentially expressed genes using RNA fingerprinting by arbitrarily primed polymerase chain reaction. Methods Enzymol. 1999;303:309–324. doi: 10.1016/s0076-6879(99)03020-7. [DOI] [PubMed] [Google Scholar]
- 24.Nelson K E, Clayton R A, Gill S R, Gwinn M L, Dodson R J, Haft D H, Hickey E K, Peterson J D, Nelson W C, Ketchum K A, McDonald L, Utterback T R, Malek J A, Linher K D, Garrett M W, Stewart A M, Cotton M D, Pratt M S, Phillips C A, Richardson D, Heidelberg J, Sutton G G, Fleischmann R D, Eisen J A, Fraser C M, et al. Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima. Nature. 1999;399:323–329. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- 25.Perry D, Kuramitsu H K. Genetic transformation of Streptococcus mutans. Infect Immun. 1981;32:1295–1297. doi: 10.1128/iai.32.3.1295-1297.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pestova E V, Morrison D A. Isolation and characterization of three Streptococcus pneumoniae transformation-specific loci by use of a lacZreporter insertion vector. J Bacteriol. 1998;180:2701–2710. doi: 10.1128/jb.180.10.2701-2710.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 28.Shiroza T, Kuramitsu H K. Construction of a model secretion system for oral streptococci. Infect Immun. 1993;61:3745–3755. doi: 10.1128/iai.61.9.3745-3755.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Svensater G, Larsson U-B, Greif E C G, Cvitkovitch D G, Hamilton I R. Acid tolerance response and survival by oral bacteria. Oral Microbiol Immunol. 1997;12:266–273. doi: 10.1111/j.1399-302x.1997.tb00390.x. [DOI] [PubMed] [Google Scholar]
- 30.Svensater G, Sjogreen B, Hamilton I R. Multiple stress responses in Streptococcus mutansand the induction of general and stress-specific proteins. Microbiology. 2000;146:107–117. doi: 10.1099/00221287-146-1-107. [DOI] [PubMed] [Google Scholar]
- 31.Trentmann S M, van der Knaap E, Kende H. Alternatives to 35S as a label for the differential display of eukaryotic messenger RNA. Science. 1995;267:1186–1187. doi: 10.1126/science.7855603. [DOI] [PubMed] [Google Scholar]
- 32.Van Asseldonk M, Rutten G, Oteman M, Siezen R J, de Vos M W, Simons G. Cloning of usp45, a gene encoding a secreted protein from Lactococcus lactis subsp. lactisMG1363. Gene. 1990;95:155–160. doi: 10.1016/0378-1119(90)90428-t. [DOI] [PubMed] [Google Scholar]
- 33.Van de Rijn I, George M, Bouvet A, Roberts R B. Enzyme-linked immunosorbent assay for the detection of antibodies to nutritionally variant streptococci in patients with endocarditis. J Infect Dis. 1986;153:116–121. doi: 10.1093/infdis/153.1.116. [DOI] [PubMed] [Google Scholar]
- 34.Welsh J, Chada K, Dalal S S, Cheng R, Ralph D, McClelland M. Arbitrarily primed PCR fingerprinting of RNA. Nucleic Acids Res. 1992;11:4965–4970. doi: 10.1093/nar/20.19.4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wong K K, McClelland M. Stress-inducible gene of Salmonella typhimuriumidentified by arbitrary primed PCR of RNA. Proc Natl Acad Sci USA. 1994;91:639–643. doi: 10.1073/pnas.91.2.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamashita Y, Takehara T, Kuramitsu H K. Molecular characterization of a Streptococcus mutansmutant altered in environmental stress responses. J Bacteriol. 1993;175:6220–6228. doi: 10.1128/jb.175.19.6220-6228.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yan D, Ikeda T P, Shauger A E, Kustu S. Glutamate is required to maintain the steady-state potassium pool in Salmonella typhimurium. Proc Natl Acad Sci USA. 1996;93:6527–6531. doi: 10.1073/pnas.93.13.6527. [DOI] [PMC free article] [PubMed] [Google Scholar]