Abstract
Monoclonal antibodies (MAbs) specific for Plasmodium falciparum rhoptry-associated protein 1 (RAP-1) were generated and tested for inhibition of parasite growth in vitro. The majority of indirect immunofluorescence assay (IFA)-positive MAbs raised against recombinant RAP-1 positions 23 to 711 (rRAP-123–711) recognized epitopes located in the immunodominant N-terminal third of RAP-1. MAbs specific for the building block 35.1 of the synthetic peptide malaria vaccine SPf66 also yielded an IFA staining pattern characteristic for rhoptry-associated proteins and reacted specifically with rRAP-1 and parasite-derived RAP-1 molecules p67 and p82. Cross-reactivity with RAP-1 was blocked by the 35.1 peptide. Epitope mapping with truncated rRAP-1 molecules and overlapping peptides identified the linear RAP-1 sequence Y218KYSL222 as a target of the anti-35.1 MAbs. This sequence lacks primary sequence similarity with the 35.1 peptide (YGGPANKKNAG). Cross-reactivity of the anti-35.1 MAbs thus appears to be associated with conformational rather than sequence homology. While the anti-35.1 MAb SP8.18 exhibited parasite growth-inhibitory activity, none of the tested anti-rRAP-123–711 MAbs inhibited parasite growth, independently of their fine specificity for the RAP-1 sequences at positions 33 to 42, 213 to 222, 243 to 247, 280 to 287, or 405 to 446. The growth-inhibitory activity of MAb SP8.18 was, however, accelerated by noninhibitory anti-RAP-1 MAbs. Results demonstrate that in addition to fine specificity, other binding parameters are also crucial for the inhibitory potential of an antibody.
The protective potential of antibodies against Plasmodium falciparum malaria has been demonstrated by passive transfer studies in which purified immunoglobulin (IgG) from individuals living in regions of hyperendemic malaria had curative effects (4, 7). This has generated much interest in the identification and characterization of parasite structures recognized by protective antibodies. Antibodies to a number of parasite antigens expressed on free merozoites or the surface of infected erythrocytes have been shown to inhibit in vitro growth, reinvasion or development of P. falciparum. However, the mechanisms of immune protection in vivo and the influence of fine specificity and the kinetic and thermodynamic binding parameters of protective antibodies on antibody effector functions in malaria are still incompletely understood.
A variety of malaria blood stage vaccine candidate antigens have been identified primarily by protection studies in animal models and by monoclonal antibodies (MAbs) that have inhibitory activity in vitro (15). Several of these candidate antigens are expressed on the rhoptries, apical organelles involved in erythrocyte invasion (23). Experimental immunization of Saimiri monkeys with purified complexes of rhoptry-associated protein 1 (RAP-1) and RAP-2 have been shown to confer partial protection against P. falciparum infection (40) and inhibitory activities of certain anti-RAP-1 MAbs in vitro (18, 19, 42) suggest that antibodies to this antigen may reduce the replication of the parasite. Furthermore, IgG reactivities to RAP-1 have been found to be inversely correlated with parasite density in Tanzanian children less than 5 years of age, which suggests that immune recognition of RAP-1 is associated with control of parasitemia (26). Unlike many other candidate antigens, RAP-1 exhibits minimal genetic polymorphism. It is synthesized as an 86-kDa precursor, which subsequently is N-terminally cleaved to generate an 82-kDa molecule (p82). In late schizogony a fraction of p82 is further processed at amino acid residue 191 to yield a 67-kDa molecule (p67) (5, 6, 21, 22). As part of their maturation the processed RAP-1 products bind RAP-2 and RAP-3 to form heterooligomeric complexes (22). Two major species of RAP-1, the mature protein p82 and its N-terminally processed product p67, dominate in mature schizonts (21). MAbs with specificity for linear RAP-1 sequences close to the p82→p67 processing site at position 191 (N200TLTPLEELYPT211 and L238VAQKEEFEYDENMEKAKQDKKKAL262, respectively) have been shown to inhibit parasite growth in vitro (18, 19, 42).
In 1987, three peptide sequences derived from proteins isolated from P. falciparum-infected erythrocytes, which conferred partial protection against P. falciparum infection in Aotus monkeys, were described (34). These partial sequences were incorporated into the synthetic peptide vaccine SPf66 (33, 34). Two of the three sequences, 35.1 (YGGPANKKNAG) and 55.1 (DELEAETQNVYAA) were derived from as yet unidentified proteins. In this study we show that two independently derived anti-35.1 MAbs were both cross-reactive with a RAP-1-derived sequence located close to the proteolytic cleavage site at the amino terminus of p67. Parasite growth-inhibitory activities of these antibodies are compared with those of MAbs elicited against recombinantly expressed RAP-1.
MATERIALS AND METHODS
Peptides and RAP-1 His6 fusion proteins.
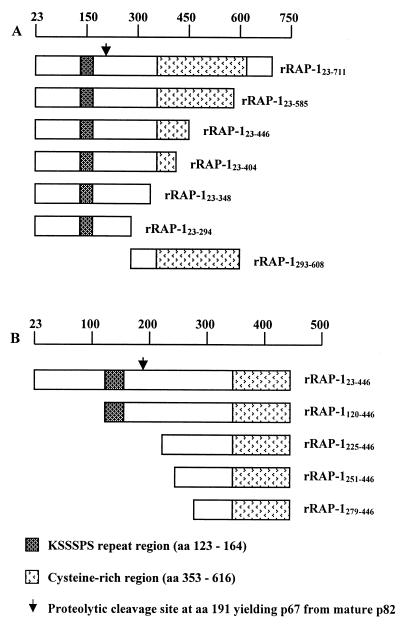
A series of recombinant RAP-1 (rRAP-1) sequences with a C-terminal six-histidine (His6) tag (Fig. 1A) were expressed in Escherichia coli and purified as described (12). Because of the presence of putative alternative initiation sites (internal methionine codons), rRAP-1 preparations contained additional N-terminally truncated molecular species, as represented for rRAP-1 positions 23 to 446 rRAP-123–446 in Fig. 1B. Coding sequences were derived from the RAP-1 allele of P. falciparum clone K1. Polymeric SPf66 (SPf66pol) (CDELEAETQNVYAAPNANPYSLFQKEKMVLPNANPPANKKNAGC), monomeric SPf66 (SPf66mon) without the terminal cysteines of the SPf66pol peptide, and the polymeric SPf66 building blocks 35.1pol (CYGGPANKKNAGC), 55.1pol (CGDELEAETQNVYAAGC), and 83.1pol (CGYSLFQKEKMVLGC) were a kind gift of M. E. Patarroyo. 35.1mon (YGGPANKKNAG) and RAP–147–57 (YWTPINKKEFL) were obtained from Sigma-Genosys.
FIG. 1.
Schematic representation of P. falciparum His6 rRAP-1 proteins (A) and N-terminally truncated forms of rRAP-123–446 (B). The lengths of the rRAP-1 fragments and the locations of structural elements of RAP-1 are indicated.
Generation of hybridomas and production of MAb.
Hybridomas were generated from mice immunized as described (36) with SPf66 or with His6 rRAP-123–711 using PAI mouse myeloma cells as a fusion partner (38). Hybrids were selected in hypoxanthine-aminopterin-thymidine (HAT) medium, and cells that secreted MAbs specific for the 35.1 building block of SPf66 or for His6 rRAP-123–711, respectively, were identified by enzyme-linked immunosorbent assay (ELISA). For large-scale MAb production hybridoma cell lines were cultured in 1-liter spinner bottles or in the Tecnomouse hollow-fiber bioreactor system (Integra Biosciences, Woburn, Mass.) (31). MAbs were purified by affinity chromatography using rabbit anti-mouse-Ig polyclonal antibodies coupled to Sepharose. Purified MAbs were dialyzed against phosphate-buffered saline (PBS), aliquoted, and stored at −80°C.
ELISA.
ELISA plates (Immunolon 4B, Dynatech, Embrach, Switzerland) were coated at 4°C overnight with 100 μl of a 10-μg/ml solution of SPf66pol, 35.1pol, 55.1pol, 83.1pol, or rRAP-1 in PBS (pH 7.2). Wells were then blocked with 5% milk powder in PBS for 1 h at 37°C followed by three washings with PBS containing 0.05% Tween 20. Plates were than incubated with serial dilutions of mouse serum, hybridoma cell culture supernatants, or purified MAbs in PBS containing 0.05% Tween 20 and 0.5% milk powder for 3 h at 37°C. After washing, the plates were incubated with alkaline phosphatase-conjugated goat anti-mouse IgG (γ-chain)-or IgM (μ-chain)-specific antibodies (Sigma, St. Louis, Mo.) for 1 h at 37°C, washed, and rinsed with deionized water. Phosphatase substrate solution (1 mg of p-nitrophenyl phosphate [Sigma] per ml in a pH 9.8 buffer solution containing 10% [vol/vol] diethanolamine and 0.02% MgCl2) was added and incubated at room temperature, and the optical density was read at 405 nm using a Titertek Multiscan MCC/340 reader (Labsystems, Helsinki, Finland). In peptide competition ELISAs, antigen-coated plates were incubated for 2 h at 37°C with MAbs in the presence of increasing concentrations of competitor peptides. MAb concentrations were kept constant in these assays.
ITC.
Calorimetric titration experiments were performed using an MSC-isothermal titration calorimetry (ITC) instrument (MicroCal, Northampton, Mass.). The design and operation of the instrument have been previously described (60). MAbs were titrated with peptides essentially as described (37). The sample cell (1.34 ml) was filled with MAb solution (typically 4 μM) in PBS. The injection syringe (nominal volume, 250 μl) was filled with a peptide solution (typically 100 μM SPf66mon or 35.1mon) in PBS. The reference cell contained a solution of 0.01% sodium azide. During equilibration the stirrer was set to rotate at 400 rpm. After baseline stability was reached, 2 μl of solution was preinjected to correct for any exchange of the syringe and cell contents and to ensure that the first experimental injection made was 10 μl. The preinjection was followed by a succession of 24 injections of 10 μl each. Injections were separated by 250 s, and each occurred over 5 s. The instrument was equilibrated with an external circulating bath at least 5°C below the experimental temperature. Experiments were carried out at 25°C. Prior to each experiment sample cell and syringes were rinsed with PBS. After each experiment sample cell and syringes were first rinsed with PBS, then cleaned with 200 ml of 0.1% sodium dodecyl sulfate solution (Merck-Schuchardt, Darmstadt, Germany), and finally rinsed with at least 1 liter of water. The isothermal titration curve was registered and analyzed using ORIGIN software (MicroCal) provided with the MCS-ITC instrument. Values for binding constants (Kb) and binding enthalpies (ΔHb) were determined from a fit of the heat exchanged per injection as a function of the amount of injected titrant as described (60).
Indirect immunofluorescence assay (IFA).
Twelve-well multitest immunofluorescence microscopy slides (Flow Laboratories, Baar, Switzerland) were pretreated with 0.01% poly-l-lysine (Sigma) for 30 min at room temperature and washed five times with RPMI basal salt medium (Gibco BRL). Erythrocytes from in vitro cultures of P. falciparum clones with about 10% parasitemia were washed two times in RPMI at room temperature. Cells were resuspended in RPMI and mixed with 2 volumes of a solution containing 4% paraformaldehyde and 0.1% Triton X-100. Droplets of 30 μl of cell suspension were added to each well, incubated at room temperature for 30 min, and washed five times with PBS. Wells were incubated for 15 min at room temperature with blocking solution containing 10% fatty acid-free bovine serum albumin (BSA) in PBS. Immunostaining was started by incubating the wells with 20 μl of purified MAb (0.1 mg/ml) in blocking solution for 1 h at room temperature in a humid chamber. After five washes with blocking solution, 20 μl of 5-μg/ml cyanine dye (Cy3)-conjugated affinity-pure F(ab′)2 fragment goat anti-mouse IgG (Fcγ-specific) antibodies (Jackson Immuno Research Laboratories, West Grove, Pa.), diluted in blocking solution containing Hoechst stain (catalog no. 33258; 0.01 mg/ml; Sigma), was added to the wells and incubated for 1 h at room temperature. Afterwards slides were washed five times with PBS; mounted with 90% (vol/vol) glycerol containing 0.1 M Tris-Cl, pH 8.0, and o-phenylenediamine (2 mg/ml); and covered with a coverslip. Antibody binding and DNA staining were assessed by fluorescence microscopy.
Western blot analysis.
Total parasite protein preparations were obtained by saponin lysis of erythrocytes from parasite cultures as described (30). Briefly, cultured parasites were washed three times with serum-free RPMI medium. Pelleted infected red blood cells were lysed by mixing with a large volume (adjusted to 5% hematocrit) of 0.015% (wt/vol) saponin in 150 mM NaCl and 15 mM sodium citrate (pH 7.0) and incubated on ice for 20 min. Finally, the pelleted parasites were resuspended in 3 volumes of SSC buffer (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and stored at −80°C before use. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was performed essentially as described (13). Briefly, parasite lysates or rRAP-1 (2 μg per lane) were run on 10% gels. As a molecular weight marker, RPN 800 (Amersham) was used. Separated proteins were transferred electrophoretically to a nitrocellulose filter (Protean Nitrocellulose, BA 85; Schleicher & Schuell). Blots were blocked with 1% BSA and then incubated with anti-35.1 or anti-RAP-1 MAb. After several washing steps, blots were incubated with a 1:1,000 dilution of a goat anti-mouse IgG alkaline phosphatase-conjugated IgG (Sigma) for 2 h. Blots were developed using BCIP (5-bromo-chloro-3-indolylphosphate, Bio-Rad) and nitroblue tetrazolium (Bio-Rad) to visualize bands. In peptide competition analyses, blots were incubated with MAb solution in the presence of 100 μg of 35.1pol, 55.1pol, or 83.1pol per ml.
Peptide epitope mapping.
Binding of MAbs to linear RAP-1 sequences was analyzed using the Geysen technique (14). In brief, a set of 15-mer peptides with an offset of five amino acids covering the RAP-1 sequence from residues 23 to 294 was synthesized on a 96-pin plastic support in an epitope scanning kit (Chiron Technologies Pty Ltd, Clayton, Victoria, Australia). MAb reactivity was assessed by ELISA according to the manufacturer's protocol. Peptides tested were 1, INVNGDNNYGKTIIN; 2, DNNYGKTIINNDFNF; 3, KTIINNDFNFDDYNY; 4, NDFNFDDYNYWTPIN; 5, DDYNYWTPINKKEFL; 6, WTPINKKEFLNSYED; 7, KKEFLNSYEDEFSSE; 8, NSYEDEFSSESFLEN; 9, EFSSESFLENKSSVD; 10, SFLENKSSVDDGNIN; 11, KSSVDDGNINLTDTS; 12, DGNINLTDTSTSNKS; 13, LTDTSTSNKSSKKGH; 14, TSNKSSKKGHGRSRV; 15, SKKGHGRSRVRSASA; 16, GRSRVRSASAAAILE; 17, RSASAAAILEEDDSK; 18, AAILEEDDSKDDMEF; 19, EDDSKDDMEFKASPS; 20, DDMEFKASPSVVKTS; 21, KASPSVVKTSTPSGT; 22, VVKTSTPSGTQTSGL; 23, TPSGTQTSGLKSSSP; 24, QTSGLKSSSPSSTKS; 25, KSSSPSSTKSSSPSN; 26, SSTKSSSPSNVKSAS; 27, SSPSNVKSASPHGES; 28, VKSASPHGESNSSEE; 29, PHGESNSSEESTTKS; 30, NSSEESTTKSSKRSA; 31, STTKSSKRSASVAGI; 32, SKRSASVAGIVGADE; 33, SVAGIVGADEEAPPA; 34, VGADEEAPPAPKNTL; 35, EAPPAPKNTLTPLEE; 36, PKNTLTPLEELYPTN; 37, TPLEELYPTNVNLFN; 38, LYPTNVNLFNYKYSL; 39, VNLFNYKYSLNNMEE; 40, YKYSLNNMEENINIL; 41, NNMEENINILKNEGD; 42, NINILKNEGDLVAQK; 43, KNEGDLVAQKEEFEY; 44, LVAQKEEFEYDENME; 45, EEFEYDENMEKAKQD; 46, DENMEKAKQDKKKAL; 47, KAKQDKKKALEKIGK; 48, KKKALEKEGKESDEA; 49, EKIGKESDEAPFMFS; 50, ESDEAPFMFSENKFL; 51, PFMFSENKFLENQVK; 52, ENKFLENQVKERNVA; and 53, KFLENQVKERNVAGS.
Parasite culture and in vitro growth inhibition assays.
P. falciparum strains FVO (55), NF54 (59), K1 (57), RO33 (50), MAD20 (54), W2 (56), HB3 (53), and RFCR3 were cultured in vitro as described previously (30). Culture medium was supplemented with 0.5% lipid-rich BSA (AlbuMAX; Gibco, Paisley, Scotland) as a substitute for human serum (10). Cultures were synchronized by sorbitol treatment (28). Serogroup A+ erythrocytes for passages were obtained from the Swiss Red Cross (Bern, Switzerland).
For growth inhibition studies P. falciparum clone FVO was used, which expresses a RAP-1 allele (GenBank accession number AF205284), which is identical in all identified linear epitopes with the allele of clone K1 (M32853) used for recombinant expression of RAP-1. Synchronous schizonts were diluted with fresh red blood cells to give 0.5% parasitemia and were mixed with purified MAb. The final hematocrit was 0.5%. Each culture was set up in sextuplicate in 96-well flat-bottom culture plates. After 96 h plates were centrifuged at 180 × g for 5 min and culture supernatants were removed. The erythrocytes were resuspended in 200 μl of 15 μg of hydroethidine fluorescent vital stain (10-mg/ml stock solution in dimethyl sulfoxide; Polysciences, Inc., Warrington, Pa.) per ml in PBS and incubated for 30 min at 37°C, as described (11). The red blood cells were washed twice in PBS, resuspended in PBS, and analyzed in a flow cytometer (FACSscan, Becton-Dickinson, San Jose, Calif.) with CELLQuest 3.2.1fl software. The hydroethidine emission was detected in the FL2 channel by logarithmic amplification, and the erythrocytes were gated on the basis of their forward and side scatters. A total of 10,000 cells per sample were analyzed. Percent inhibition was calculated from the geometric mean parasitemias of sextuplicate test and control wells as 100 × (control − test)/control.
RESULTS
Generation of monoclonal antibodies specific for the 35.1 building block of SPf66.
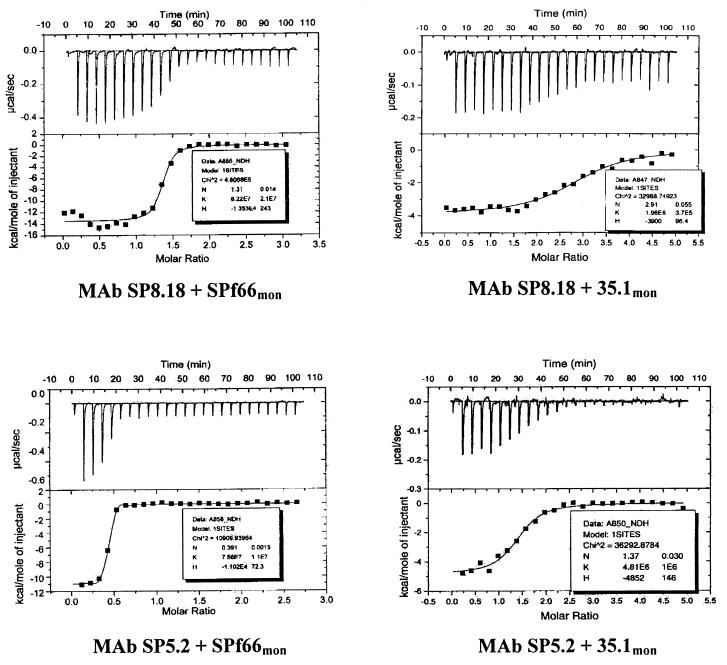
In an attempt to generate MAbs against all building blocks of SPf66, 64 B-cell hybridoma clones that secreted SPf66-specific IgG were generated from spleen cells of a mouse immunized with SPf66. When the fine specificity of the antibodies was assessed by peptide building block-specific ELISA, 18 of the MAbs turned out to be 83.1-specific and 14 were 55.1 specific. The remaining 32 MAbs bound to the SPf66 polymer but to none of the three SPf66 building blocks. These results indicated that the 35.1 sequence has limited immunogenicity in the context of the SPf66 peptide. To obtain 35.1-specific MAbs, we therefore subsequently screened B-cell hybridoma culture supernatants in a 35.1-specific peptide ELISA directly for the presence of anti-35.1 antibodies. From three additional fusion experiments with spleen cells from SPf66 immunized mice, direct screening yielded two anti-35.1 MAbs, designated SP5.2 (IgG1:κm) and SP8.18 (IgG1:κh). MAb SP8.18 was derived from a mouse which carried Ig constant region gene replacement mutations (36) and therefore has a human kappa light chain constant region. In ELISA, MAbs SP5.2 and SP8.18 both specifically bound to SPf66 and to the 35.1 peptide but not to the other two SPf66 building blocks 55.1 and 83.1 (data not shown). Binding to the 35.1 peptide sequence and to SPf66 was reconfirmed by ITC experiments (Fig. 2). ITC measures the heat of reaction and allows simultaneous determination of Kb, ΔHb, entropy, and the number of bindings sites (60). The values for Kb and ΔHb calculated from calorimetric titrations of MAbs SP5.2 and SP8.18 are presented in Table 1. All binding reactions were exothermic. Both MAbs showed comparable Kb values for binding to the monomeric peptides 35.1mon (>105 M−1) and SPf66mon (>106 M−1), respectively.
FIG. 2.
ITC profiles of the binding of anti-35.1 MAbs SP8.18 and SP5.2 to 35.1mon and SPf66mon peptides (upper curves) and integrated heats of binding of injected peptide (lower curves). Aliquots (2 μl) of 100 μM titrant in PBS were injected into a solution of 1.34 ml of MAb at 4 μM. The measuring temperature was 25°C.
TABLE 1.
Thermodynamic parameters of peptide-MAb binding interactions as determined by ITCa
| MAb | Peptide | Kb (107 mol−1) | ΔHb (kJ mol−1) |
|---|---|---|---|
| SP5.2 | SPf66mon | 7.5 (1.1) | −46.1 (0.3) |
| 35.1mon | 0.5 (0.1) | −20.3 (0.6) | |
| SP8.18 | SPf66mon | 6.2 (2.1) | −62.2 (1) |
| 35.1mon | 0.2 (0.04) | −16.3 (0.4) |
Mean values for ΔHb and Kb (standard deviations shown in parentheses) were determined from a nonlinear least-squares fit of the heat exchanged per injection as a function of total injected peptide using Microcal ORIGIN software.
Binding of 35.1-specific MAbs to the RAP-1 protein of P. falciparum.
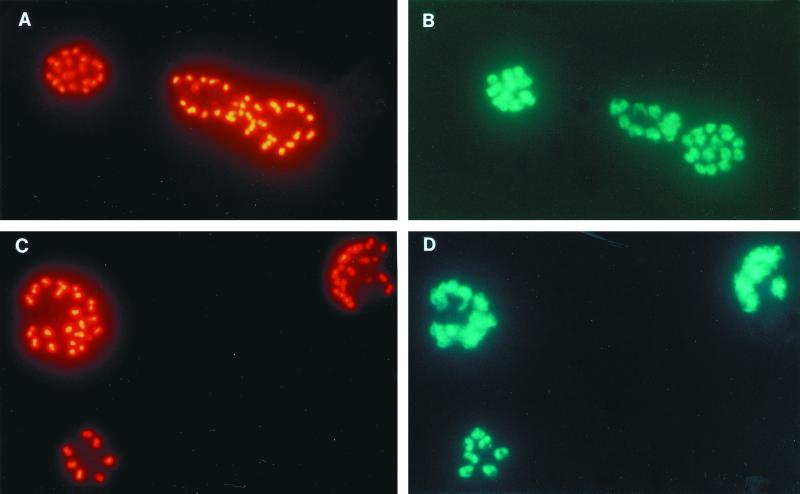
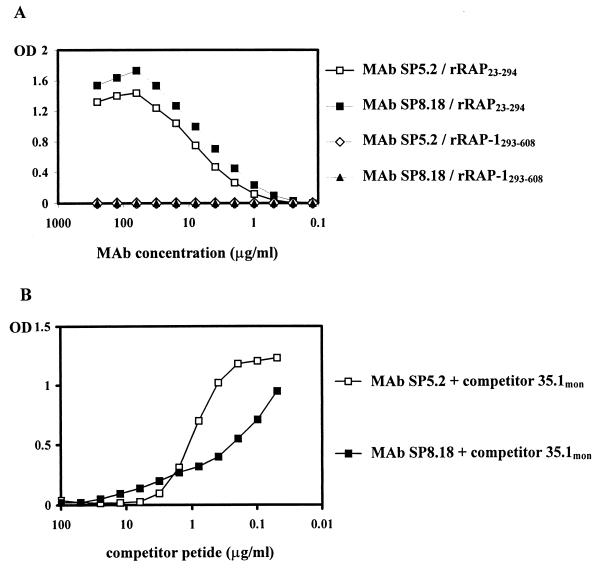
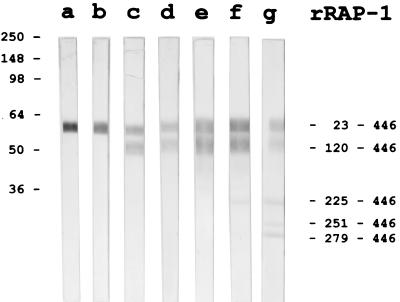
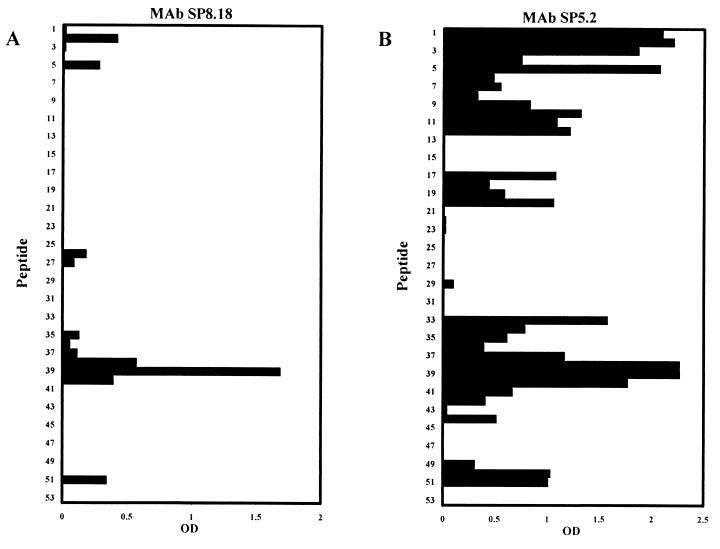
Both 35.1-specific MAbs exhibited a double-dot staining pattern characteristic for rhoptry-associated proteins in IFA with blood stage P. falciparum parasites (Fig. 3). In Western blots using parasite blood stage lysates, two major proteins bands of an approximate molecular mass of 67 and 82 kDa were stained (Fig. 4). Similar staining patterns were observed with all parasite clones (HB3, K1, MAD20, NF54, RFCR3, RO33, and W2) tested (not shown). Staining of the two bands was inhibited by the 35.1 peptide, but not by the 55.1 or the 83.1 peptides (Fig. 4). MAbs raised against recombinant RAP-1 yielded identical recognition patterns in both the IFA and immunoblotting (Fig. 3 and 4). Since no sequence stretch identical to the 35.1 sequence was found within the RAP-1 sequence, we have experimentally characterized the portion of RAP-1 that is recognized by the anti-35.1 MAbs. No binding to RAP-147–57 (YWTPINKKEFL), which has the most significant linear sequence homology with 35.1 (YGGPANKKNAG), was observed (data not shown). Both the SP5.2 and the SP8.18 MAb bound, however, to the N-terminal rRAP-123–294 fragment, but not to the C-terminal rRAP-1293–608 fragment of RAP-1 (Fig. 5A). Binding to rRAP-123–294 was specifically inhibited by the 35.1 peptide in competition ELISA (Fig. 5B). The portion of RAP-1 recognized by the anti-35.1 MAbs was narrowed down further by Western blot analysis using an rRAP-1 preparation which contained in addition to rRAP-123–446 the N-terminally truncated molecules rRAP-1120–446, rRAP-1225–446, rRAP-1251–446, and rRAP-1279–446. Both MAbs stained rRAP-123–446 and rRAP-1120–446, but not rRAP-1225–446, rRAP-1251–446, and rRAP-1279–446 (Fig. 6), which indicates that they bind to a sequence located between residues 120 and 224. Recognition of the 67-kDa fragment of RAP-1 in Western blots with parasite lysates had indicated that the anti-35.1 MAbs bind C-terminally of the proteolytic cleavage site at residue 191. To reconfirm the deduced mapping of the epitope between residues 191 and 224, we performed a peptide epitope mapping analysis using the Geysen technique (14). In this analysis the RAP-1 sequence from residues 23 to 294 was covered by a set of 15-mer peptides with an offset of five amino acids. MAb SP8.18 exhibited a significantly higher binding to the three overlapping peptides L208YPTNVNLFNYKYSL222, V213NLFNYKYSLNNMEE227, and Y218KYSLNNMEENINIL232, which share the motif Y218KYSL222, than to any of the other peptides (Fig. 7A). MAb SP5.2 recognized the same three peptides (Fig. 7B) but exhibited a far more scattered binding pattern than MAb SP8.18. The 35.1 sequence YGGPANKKNAG has no obvious sequence similarity with the RAP-1 sequence Y218KYSL222.
FIG. 3.
Immunofluorescence staining of mature schizonts of P. falciparum K1 by anti-35.1 MAb SP8.18 (A) and anti-rRAP-123–711 MAb RAP1-14 (C). Both MAbs exhibited a double-dot staining pattern characteristic for rhoptry-associated proteins. Parasite DNA was stained using Hoechst 33258 dye (B and D).
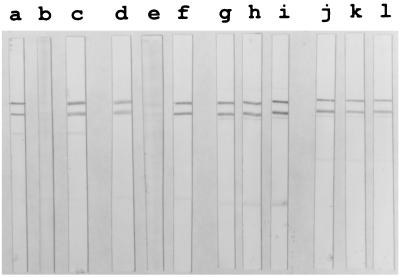
FIG. 4.
MAb binding to RAP-1 molecular species in Western blots using P. falciparum K1 blood stage lysates as antigen. The anti-35.1 MAbs SP5.2 (lane a) and SP8.18 (lane d) and the anti-rRAP-123–711 MAbs RAP5-2 (lane g) and RAP1-14 (lane j) stained two bands of approximately 82 and 67 kDa. Binding of MAbs SP5.2 and SP8.18 was inhibited by peptide 35.1pol (lanes b and e, respectively) but not by peptide 55.1pol (lanes c and f, respectively). Binding of MAbs RAP5-2 and RAP1-14 was inhibited neither by peptide 35.1pol (lane h and k), nor by 55.1pol (lanes i and l).
FIG. 5.
35.1-inhibitable binding of anti-35.1 MAbs to rRAP-1. Binding of MAbs SP5.2 and SP8.18 to rRAP-123–294 and rRAP-1293–608 was analyzed by ELISA (A). Binding to rRAP-123–294 was inhibited by peptide 35.1mon (B).
FIG. 6.
MAb binding to rRAP-123–446 and its N-terminally truncated variants rRAP-1120–446, rRAP-1225–446, rRAP-1251–446, rRAP- 1279–446. Binding of MAbs RAP1-8 (lane a), RAP1-22 (lane b), SP5.2 (lane c), SP8.18 (lane d), RAP1-14 (lane e), RAP2-25 (lane f), and RAP5-2 (lane g) to the different molecular species of RAP-1 was analyzed by Western blotting.
FIG. 7.
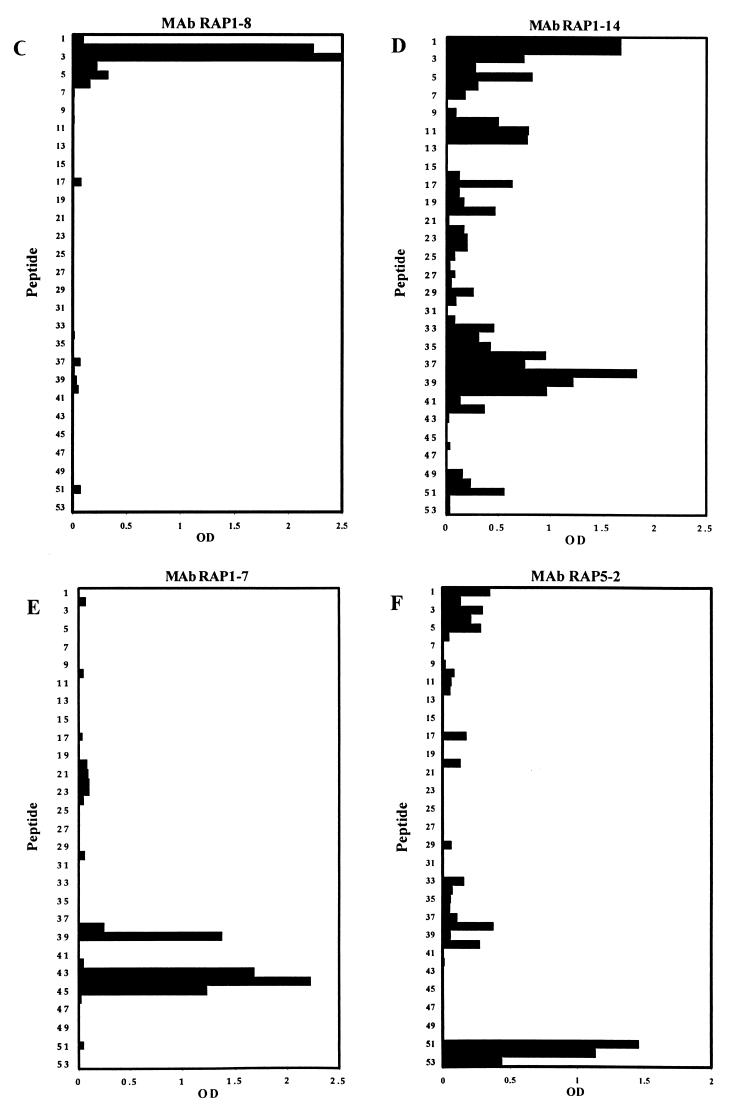
Binding of MAbs to a set of 15-mer peptides with an offset of five amino acids that cover the RAP-1 sequence from residues 23 to 294. Every second peptide sequence is shown. Binding of MAb SP8.18 (A), MAb SP5.2 (B), MAb RAP1-8 (C), MAb RAP1-14 (D), MAb RAP1-7 (E), and MAb RAP5-2 (F) was tested by ELISA.
Generation and characterization of MAbs raised against rRAP-1.
For comparative analyses, a second set of parasite-binding anti-RAP-1 MAbs was generated after immunization of mice with rRAP-123–711. Thirteen anti-rRAP-123–711 MAbs that stained blood stage parasites in IFA were obtained. All of them yielded the characteristic rhoptry-associated double-dot staining pattern (shown for RAP1-14 in Fig. 3). Western blot analyses with C- and N-terminally truncated rRAP-1 preparations identified the portions of RAP-1 recognized by the different MAbs (Table 2). Except for MAbs RAP2-5 and RAP2-16 (see below), all anti-rRAP23–711 MAbs bound to all six C-terminally truncated rRAP-1 proteins (rRAP-123–294, rRAP-123–348, rRAP-123–404, rRAP-123–446, rRAP-123–585, and rRAP-123–711) tested. These 11 MAbs thus recognize sequences located between amino acids 23 and 294. Reactivity patterns with the N-terminally truncated rRAP-1 in Western blot analyses narrowed the presumed location of epitopes further (Table 2; Fig. 6 shows selected results). Two MAbs (RAP1-8 and RAP1-22) thus seem to bind between residues 23 and 119, three MAbs (RAP1-14, RAP1-15, and RAP8-2) seem to bind between residues 121 and 224, five MAbs (RAP1-7, RAP1-25, RAP2-21, RAP2-25, and RAP4-9) seem to bind between residues 225 and 250, and one MAb (RAP5-2) seems to bind between residues 279 and 294. Since MAb RAP2-5 bound to rRAP-123–348, but not to rRAP-123–294, the epitope recognized by this MAb appears to be located between residues 294 and 348. MAb RAP2-16 did not bind to rRAP-123–294, rRAP-123–348, and rRAP-123–404, but bound to rRAP-123–446 and thus seems to recognize a sequence between residues 405 and 446. Epitope mapping results with the set of 15-mer peptides covering residues 23 to 294 of RAP-1 are shown in Fig. 7 and are summarized in Table 2. MAb RAP1-8 recognized the peptides D28NNYGKTIINNDFNF42 and K33TIINNDFNFDDYNY47 (Fig. 7C), which share the sequence K33TIINNDFNF42. MAb RAP1-14 showed at a broad range of antibody concentrations a scattered binding pattern, giving the highest reactivity to the two overlapping peptide pairs I23NVNGDNNYGKTIIN37-D28NNYGKTIINNDFNF42 and L208YPTNVNLFNYKYSL222-V213NLFNYKYSLNNMEE227 (Fig. 7D). Based on the Western blot results, which localized the epitope between residues 120 and 225, it is likely that the sequence most relevant for binding is located within V213NLFNYKYSL222. The binding pattern of MAb RAP8-2 was comparable to that of MAb RAP1-14 (data not shown). MAbs RAP1-14 and RAP8-2 yielded the highest reactivity to peptide L208YPTNVNLFNYKYSL222, while the 35.1-specific MAb SP8.18 showed the highest reactivity to peptide V213NLFNYKYSLNNMEE227, which overlaps with the former in the 10 amino acids V213NLFNYKYSL222. In ELISA, MAbs RAP1-14 and RAP8-2 showed a very weak cross-reactivity to the 35.1 building block (not shown). MAb RAP1-7 recognized the overlapping peptides K233NEGDLVAQKEEFEY247, L238VAQKEEFEYDENME252, and E243EFEYDNMEKAKQD257 (Fig. 7E), which share the sequence E243EFEY247. In addition, significant binding to the peptide V213NLFNYKYSLNNMEE227 was found. MAbs RAP1-25, RAP2-21, and RAP2-25 yielded binding patterns comparable to that of MAb RAP1-7 (not shown). MAb RAP5-2 bound to the three overlapping peptides PFMFSENKFLENQVK, ENKFLENQVKERNVA, and KFLENQVKERNVAGS (Fig 7F), which share the sequence K280FLENQVK287.
TABLE 2.
Epitope analysis of RAP-1 binding MAbs by Western blotting and peptide mappingg
| MAb | Immunogen | Isotype | Epitope assignment by:
|
|
|---|---|---|---|---|
| Western blotting | Peptide mapping | |||
| RAP1-8 | rRAP-123–711 | IgM: κm | 23–119a | K33TIINNDFNF42 |
| RAP1-22 | rRAP-123–711 | IgG1: κm | 23–119a | NDh |
| RAP1-14 | rRAP-123–711 | IgG1: κm | 120–224b | V213NLFNYKYSL222 |
| RAP1-15 | rRAP-123–711 | IgG1: κm | 120–224b | ND |
| RAP8-2 | rRAP-123–711 | IgG2a: κm | 120–224b | V213NLFNYKYSL222 |
| RAP1-7 | rRAP-123–711 | IgG1: κm | 225–250c | E243EFEY247 |
| RAP1-25 | rRAP-123–711 | IgG1: κm | 225–250c | E243EFEY247 |
| RAP2-21 | rRAP-123–711 | IgG1: κm | 225–250c | E243EFEY247 |
| RAP2-25 | rRAP-123–711 | IgG1: κm | 225–250c | E243EFEY247 |
| RAP4-9 | rRAP-123–711 | IgG1: κm | 225–250c | ND |
| RAP5-2 | rRAP-123–711 | IgG1: κm | 279–294d | K280FLENQVK287 |
| RAP2-5 | rRAP-123–711 | IgM: κm | 295–348e | ND |
| RAP2-16 | rRAP-123–711 | IgM: κm | 405–446f | ND |
| SP8.18 | SPf66 | IgG1: κh | 120–224b | Y218KYSL222 |
| SP5.2 | SPf66 | IgG1: κm | 120–224b | Y218KYSL222 |
Binding to rRAP-123–446 but not to rRAP-1120–446.
binding to rRAP-1120–446 but not to rRAP-1225–446.
Binding to rRAP-1225–446 but not to rRAP-1251–446.
Binding both to rRAP-123–294 and to rRAP-1279–446.
Binding to rRAP-123–348 but not to rRAP-123–294.
Binding to rRAP-123–446 but not to rRAP-123–404.
Western blot analyses using parasite lysates indicated that RAP2-5 and RAP1-8 cross-react with RAP-2 (preliminary results). Furthermore MAbs RAP1-8 and RAP1-22 stain p67, which is inconsistent with the deduced location of their primary linear target epitope.
ND, not done.
Epitope assignments by peptide mapping were consistent with results obtained by Western blotting with the sets of N- and C-terminally truncated rRAP-1 molecules (Table 2). All 13 MAbs generated against rRAP-123–711 recognized epitopes located in the N-terminal portion between residues 33 and 446. Only two MAbs (RAP1-18 and RAP1-22) bound to a sequence upstream of the N terminus of p67 at residue 191. Like the anti-35.1 MAbs, three of the anti-rRAP-123–711 MAbs (RAP1-14, RAP1-15, and RAP8-2) recognized a sequence close to the RAP-1 processing site at position 191.
P. falciparum in vitro growth inhibition assays with anti-RAP-1 MAbs elicited against SPf66 or rRAP-1.
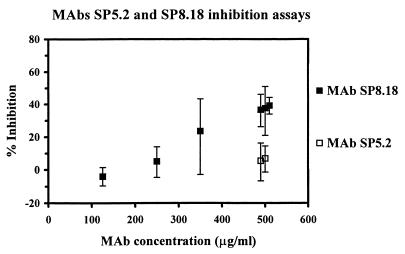
Selected RAP-1 binding antibodies were used for P. falciparum in vitro growth inhibition analyses. Assays were conducted for two cycles of merozoite invasion in order to enhance the sensitivity. The statistical significance for inhibition was ensured by setting up sample cultures in sextuplicate. At 500 μg/ml the anti-35.1 MAb SP8.18 consistently showed growth inhibitory effects (37.9% average inhibition) in independent experiments (Fig. 8). This inhibition was statistically significant, as judged by a two-sided t test. At lower concentrations the inhibitory effect of MAb SP8.18 soon became insignificant. In contrast to MAb SP8.18, the second anti-35.1 MAb SP5.2 exerted no inhibitory activity at 500 μg/ml (Fig. 8). A range of anti-rRAP-123–711 MAbs—RAP1-8, RAP1-14, RAP1-7, RAP5-2, and RAP2-16—which mapped to different regions of RAP-1 (residues 33 to 42, 213 to 222, 243 to 247, 280 to 287, and 405 to 446, respectively) were also tested in the growth inhibition assay. At high concentrations, all anti-rRAP-123–711 MAbs tested showed a parasite growth-accelerating effect, rather than a growth-inhibitory effect (Table 3). However, an inhibition potentiating effect (49 to 57% inhibition) was consistently observed when MAb SP8.18 was tested in combination with the noninhibitory MAb SP5.2, RAP1-7, or RAP1-14 (Table 3 and 4). This effect was specific since no potentiation of the inhibitory activity was observed when MAb SP8.18 was tested in combination with a noninhibitory isotype-matched control MAb (not shown). All binary combinations of the three noninhibitory MAbs—SP5.2, RAP1-7, and RAP1-14—still exhibited growth accelerating activity (data not shown).
FIG. 8.
Parasite growth-inhibitory activity of anti-35.1 MAbs SP5.2 and SP8.18. The vertical bars indicate the 95% confidence intervals.
TABLE 3.
P. falciparum in vitro growth-accelerating activity of anti-RAP-123–711 MAbsa
| MAb(s) (concn [μg/ml]) | % Inhibitionb | 95% CIc |
|---|---|---|
| RAP1-7 (350) | −47.1 | −106–−4.9 |
| RAP1-7 (500) | −34.3 | −52.9–−17.9 |
| RAP1-8 (500) | −22.9 | −54.4–2.2 |
| RAP1-14 (350) | −15.5 | −78–25 |
| RAP1-14 (500) | 1.1 | −7.4–8.9 |
| RAP1-7 + RAP1-14 (350)d | −64.7 | −123.1–−21.6 |
| RAP2-16 (500) | −44.5 | −77.9–−17.4 |
| RAP5-2 (100) | −6.3 | −41.4–20.1 |
| RAP5-2 (200) | −19.3 | −59.5–10.7 |
| RAP5-2 (300) | −26.3 | −51.9–−4.6 |
| RAP5-2 (400) | −35.9 | −66.1–−11.2 |
| RAP5-2 (500) | −21.2 | −46.9–0.2 |
Schizont stage parasites were plated in sextuplicate at initial parasitemia values of 0.5%. After 96 h the final parasitemia was determined by flow cytometry.
Linear parasitemia values were transformed into logarithmic scale, and the mean was determined. Percent inhibition was calculated as follows: 100 × [1 − (geometric sample mean)/(geometric control mean)].
P < 0.05 by two-sided t test. Confidence intervals (CI) were calculated by antilogging the confidence limits calculated on the log scale.
Both MAbs were at the same concentration.
TABLE 4.
Enhancement of in vitro growth-inhibitory activity of MAb SP8.18 by noninhibitory MAbs SP5.2, RAP1-7, and RAP1-14a
| MAb or MAb combination | Concn (μg/ml) | % Inhibition | 95% CI |
|---|---|---|---|
| SP8.18 | 500 | 39.3 | 34–44.2 |
| SP8.18 + SP5.2 | 500 | 56.7 | 47.7–64.1 |
| SP8.18 + RAP1-14 | 500 | 56.3 | 49.5–62.1 |
| SP8.18 | 350 | 23.7 | −2.9–43.4 |
| SP8.18 + RAP1-7 | 350 | 50.9 | 34–63.5 |
| SP8.18 + RAP1-14 | 350 | 49.2 | 31.2–62.5 |
MAb SP8.18 was mixed with a second MAb and added to the parasite culture. Experiments were performed in sextuplicate, and inhibition was calculated as explained in the footnotes of Table 3. CI, confidence interval.
DISCUSSION
Aotus monkeys, which are susceptible to infection with P. falciparum (8, 9, 17, 27, 34, 35, 39, 41, 44–47, 58) without prior splenectomy (48), are a World Health Organization-recommended model to test the efficacy of malaria blood stage vaccine candidates. An evaluation of the protective potential of a series of peptide sequences derived from proteins isolated from P. falciparum-infected erythrocytes in the Aotus infection model identified the partially protective peptides 35.1, 55.1, and 83.1 (34). These three sequences were then incorporated as linked building blocks into the synthetic peptide vaccine SPf66 (33). While the 83.1 peptide turned out to correspond to amino acids 43 to 53 of merozoite surface protein-1 (MSP-1) of P. falciparum, proteins containing the 35.1 or 55.1 sequences have not yet been identified. Our attempts to clone parasite proteins that contain either the 35.1 or the 55.1 sequence by using degenerated oligonucleotides deduced from these peptide sequences as PCR primers or as DNA probes have failed (unpublished data). Therefore, we have generated MAbs specific for the SPf66 building blocks and identified the P. falciparum proteins recognized by these MAbs. While 83.1- and 55.1-specific MAbs bound to MSP-1 (38) and P. falciparum heat shock proteins (preliminary results), respectively, both 35.1-specific MAbs that we succeeded in obtaining cross-reacted with RAP-1. No binding to RAP-147–57 (YWTPINKKEFL), which has the most significant linear sequence homology with 35.1 (YGGPANKKNAG), was observed. Epitope mapping studies with C- and N-terminally truncated rRAP-1 molecules showed that both anti-35.1 MAbs bind to a sequence within RAP-1 residues 191 and 224, and epitope mapping using sets of overlapping peptides (14) indicated that the RAP-1 sequence Y218KYSL222, which has no significant homology with the 35.1 peptide, is recognized. Conversely, the anti-RAP-1 MAbs RAP1-14 and RAP8-2, which mapped to the sequence V213NLFNYKYSL222, cross-reacted weakly but significantly with the 35.1 peptide. Cross-reactivities between malaria antigens in the absence of apparent primary sequence homology have been described before and may reflect the existence of cross-reactive conformational epitopes (2). Although RAP-1 and RAP-2, for example, have no apparent homology in primary structure, a majority of mice immunized with the RAP-2-derived peptide E25TEFSKLY32 raised antibodies against native RAP-1 (52). Reactivity of antisera raised against MSP-2 derived peptide sequences with MSP-1 (29) is another example of a cross-reactivity that appears to be primarily associated with conformational rather than sequence homology (49, 54).
Eleven out of the 13 IFA-positive anti-rRAP-123–711 MAbs isolated here recognized epitopes located within the N-terminal 294 amino acids of RAP-1, suggesting that the N-terminal third of this protein is immunodominant. This agrees with previous immunogenicity studies in mice (51) and in humans (12, 20, 24, 25).
Although there is mounting evidence that a number of P. falciparum antigens have a potential as targets for inhibitory antibodies, the mechanisms of antibody-mediated parasite growth inhibition and of in vivo immune protection are incompletely understood. The function(s) of the processing product p67 are not known, but it has been hypothesized that binding of antibodies to epitopes close to the N terminus of p67 can inhibit parasite growth by interfering with the processing of p82 to p67 (18). Similarly, certain anti-MSP-1 MAbs, which inhibit merozoite invasion, prevent the processing of MSP-1 (16). Parasite growth inhibition described here for the anti-35.1 MAb SP8.18, which binds to the RAP-1 sequence Y218KYSL222, supports the concept that binding of antibodies to epitopes of RAP-1 that are located between position 200 and 260 may inhibit parasite growth. The concentration of MAb SP8.18 required for significant in vitro growth inhibition was >250 μg/ml, which is comparable to MICs of previously described inhibitory antibodies against anti-RAP-1 (18), the receptor binding domain of EBA-175 (32), and the N terminus (R. Moreno et al., unpublished data) or the C terminus of MSP-1 (3). In addition to fine specificity, other binding parameters seem to be crucial for the parasite growth inhibitory potential of a MAb. This is demonstrated by the fact that SP5.2, the second anti-35.1 MAb described here, exhibited essentially the same fine specificity as MAb SP8.18 but did not inhibit parasite growth. Furthermore, the anti-rRAP-123–711 MAbs RAP1-14 and RAP1-7, which bind to sequences between position 200 and 260 (V213NLFNYKYSL222 and E243EFEY247 respectively), also had no growth inhibitory activity. Kinetic (on and off rates) as well as thermodynamic parameters (avidity) of antibody binding may be crucial for anti-infectious activity (1). In fact, all five anti-rRAP-123–711 MAbs that were tested (RAP1-8, RAP1-14, RAP1-7, RAP5-2, and RAP2-16) had an in vitro growth-accelerating rather than a growth-inhibitory effect, irrespective of their fine specificity for positions 33 to 42, 213 to 222, 243 to 247, 280 to 287, or 405 to 446, respectively. It has been suggested that certain anti-RAP-1 antibodies can increase invasion by promoting binding of merozoites to red blood cells or by increasing adventitious protein-protein interactions (18, 19, 42). Interestingly, a potentiation of growth inhibition by MAb SP8.18 was found, when noninhibitory MAbs (SP5.2, RAP1-7, or RAP1-14) were added. Conversely, MSP-1 specific antibodies have been described that compete with processing-inhibitory anti-MSP-119 antibodies for binding to the merozoite surface and neutralize the growth-inhibitory activity of the latter (16). Identification of protective epitopes of vaccine candidate antigens may therefore be crucial for the optimal design of an effective malaria vaccine.
It is remarkable, that screening of sequences for immunoprotective structures in the Aotus malaria infection model led to the identification of the 35.1 peptide (34), which has, as demonstrated here, the potential to elicit inhibitory antibodies that cross-react with RAP-1. This emphasizes the fact that this primate infection model is a suitable tool to identify building blocks for an epitope-focused multivalent, multistage malaria vaccine. Such a vaccine should contain both suitable B- and T-cell epitopes (43). The results presented here reemphasize the fact that in the case of B-cell epitopes, conformational properties are of crucial importance.
ACKNOWLEDGMENTS
This work was conducted within the framework of the Swiss National Science Foundation project 31-52068.97.
We thank Alberto Moreno and Martha Torres for introduction to parasite growth inhibition assays and Manuel E. Patarroyo for providing us with SPf66 and its building blocks. We gratefully acknowledge Tom Smith for support in statistical analysis and are grateful to Jutta Marfurt and Erika Flury for assistance with hybridoma fusions and cultures. We thank Christian Scheurer for providing parasite cultures and logistical support and Till Voss and Tobias Spielmann for helpful comments.
REFERENCES
- 1.Bachmann M F, Kalinke U, Althage A, Freer G, Burkhart C, Roost H-P, Aguet M, Hengartner H, Zinkernagel R M. The role of antibody concentration and avidity in antiviral protection. Science. 1997;276:2024–2027. doi: 10.1126/science.276.5321.2024. [DOI] [PubMed] [Google Scholar]
- 2.Berzins K, Anders R. The malaria antigens. In: Wahlgren M, Perlmann P, editors. Malaria. Amsterdam, The Netherlands: Harwood Academic Publishers; 1999. pp. 181–216. [Google Scholar]
- 3.Blackman M J, Heidrich H G, Donachie S, McBride J S, Holder A A. A single fragment of a malaria merozoite surface protein remains on the parasite during red cell invasion and is the target of invasion-inhibiting antibodies. J Exp Med. 1990;172:379–382. doi: 10.1084/jem.172.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouharoun-Tayoun H, Attanath P, Sabchareon A, Chongsuphajaisiddhi T, Druilhe P. Antibodies that protect humans against Plasmodium falciparum blood stages do not on their own inhibit parasite growth and invasion in vitro, but act in cooperation with monocytes. J Exp Med. 1990;172:1633–1641. doi: 10.1084/jem.172.6.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bushell G R, Ingram L T, Fardoulys C A, Cooper J A. An antigenic complex in the rhoptries of Plasmodium falciparum. Mol Biochem Parasitol. 1988;28:105–112. doi: 10.1016/0166-6851(88)90057-6. [DOI] [PubMed] [Google Scholar]
- 6.Clark J T, Anand R, Akoglu T, McBride J S. Identification and characterisation of proteins associated with the rhoptry organelles of Plasmodium falciparum merozoites. Parasitol Res. 1987;73:425–434. doi: 10.1007/BF00538200. [DOI] [PubMed] [Google Scholar]
- 7.Cohen S, McGregor I A, Carrington S. Gamma globulin and acquired immunity to human malaria. Nature. 1961;192:733–737. doi: 10.1038/192733a0. [DOI] [PubMed] [Google Scholar]
- 8.Collins W E, Anders R F, Pappaioanou M, Campbell G H, Brown G V, Kemp D J, Coppel R L, Skinner J C, Andrysiak P M, Favaloro J M. Immunization of Aotus monkeys with recombinant proteins of an erythrocyte surface antigen of Plasmodium falciparum. Nature. 1986;323:259–262. doi: 10.1038/323259a0. [DOI] [PubMed] [Google Scholar]
- 9.Collins W E, Pappaioanou M, Anders R F, Campbell G H, Brown G V, Kemp D J, Broderson J R, Coppel R L, Skinner J C, Procell P M. Immunization trials with the ring-infected erythrocyte surface antigen of Plasmodium falciparum in owl monkeys (Aotus vociferans) Am J Trop Med Hyg. 1988;38:268–282. doi: 10.4269/ajtmh.1988.38.268. [DOI] [PubMed] [Google Scholar]
- 10.Dorn A, Stoffel R, Matile H, Bubendorf A, Ridley R G. Malarial haemozoin/beta-haematin supports haem polymerization in the absence of protein. Nature. 1995;374:269–271. doi: 10.1038/374269a0. [DOI] [PubMed] [Google Scholar]
- 11.Elloso M M, van der Heyde H C, vande Waa J A, Manning D D, Weidanz W P. Inhibition of Plasmodium falciparum in vitro by human γδ T cells. J Immunol. 1994;153:1187–1194. [PubMed] [Google Scholar]
- 12.Fonjungo P N, Stuber D, McBride J S. Antigenicity of recombinant proteins derived from rhoptry-associated protein 1 of Plasmodium falciparum. Infect Immun. 1998;66:1037–1044. doi: 10.1128/iai.66.3.1037-1044.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gentz R, Certa U, Takacs B, Matile H, Dobeli H, Pink R, Mackay M, Bone N, Scaife J G. Major surface antigen p190 of Plasmodium falciparum: detection of common epitopes present in a variety of plasmodia isolates. EMBO J. 1988;7:225–230. doi: 10.1002/j.1460-2075.1988.tb02803.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geysen H M, Meloen R H, Barteling S J. Use of peptide synthesis to probe viral antigens for epitopes to a resolution of a single amino acid. Proc Natl Acad Sci USA. 1984;81:3998–4002. doi: 10.1073/pnas.81.13.3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Good M F, Kaslow D C, Miller L H. Pathways and strategies for developing a malaria blood stage vaccine. Annu Rev Immunol. 1998;16:57–87. doi: 10.1146/annurev.immunol.16.1.57. [DOI] [PubMed] [Google Scholar]
- 16.Guevara Patiño J A, Holder A A, McBride J S, Blackman M J. Antibodies that inhibit malaria merozoite surface protein-1 processing and erythrocyte invasion are blocked by naturally acquired antibodies. J Exp Med. 1997;186:1689–1699. doi: 10.1084/jem.186.10.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall R, Hyde J E, Goman M, Simmons D L, Hope I A, Mackay M, Scaife J, Merkli B, Richle R, Stocker J. Major surface antigen gene of a human malaria parasite cloned and expressed in bacteria. Nature. 1984;311:379–382. doi: 10.1038/311379a0. [DOI] [PubMed] [Google Scholar]
- 18.Harnyuttanakorn P, McBride J S, Donachie S, Heidrich H G, Ridley R G. Inhibitory monoclonal antibodies recognise epitopes adjacent to a proteolytic cleavage site on the RAP-1 protein of Plasmodium falciparum. Mol Biochem Parasitol. 1992;55:177–186. doi: 10.1016/0166-6851(92)90138-a. [DOI] [PubMed] [Google Scholar]
- 19.Howard R F, Jacobson K C, Rickel E, Thurman J. Analysis of inhibitory epitopes in the Plasmodium falciparum rhoptry protein RAP-1 including identification of a second inhibitory epitope. Infect Immun. 1998;66:380–386. doi: 10.1128/iai.66.1.380-386.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howard R F, Jensen J B, Franklin H L. Reactivity profile of human anti-82-kilodalton rhoptry protein antibodies generated during natural infection with Plasmodium falciparum. Infect Immun. 1993;61:2960–2965. doi: 10.1128/iai.61.7.2960-2965.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Howard R F, Narum D L, Blackman M, Thurman J. Analysis of the processing of Plasmodium falciparum rhoptry-associated protein 1 and localization of Pr86 to schizont rhoptries and p67 to free merozoites. Mol Biochem Parasitol. 1998;92:111–122. doi: 10.1016/s0166-6851(97)00238-7. [DOI] [PubMed] [Google Scholar]
- 22.Howard R F, Reese R T. Plasmodium falciparum: hetero-oligomeric complexes of rhoptry polypeptides. Exp Parasitol. 1990;71:330–342. doi: 10.1016/0014-4894(90)90038-e. [DOI] [PubMed] [Google Scholar]
- 23.Howard R J, Pasloske B. Target antigens for asexual malaria vaccine development. Parasitol Today. 1993;9:369–372. doi: 10.1016/0169-4758(93)90085-t. [DOI] [PubMed] [Google Scholar]
- 24.Jacobson K C, Thurman J, Schmidt C M, Rickel E, Oliviera D F, Ferreira-da-Cruz M F, Daniel-Ribeiro C T, Howard R F. A study of antibody and T cell recognition of rhoptry-associated protein-1 (RAP-1) and RAP-2 recombinant proteins and peptides of Plasmodium falciparum in migrants and residents of the state of Rondonia, Brazil. Am J Trop Med Hyg. 1998;59:208–216. doi: 10.4269/ajtmh.1998.59.208. [DOI] [PubMed] [Google Scholar]
- 25.Jakobsen P H, Hviid L, Theander T G, Afare E A, Ridley R G, Heegaard P M, Stuber D, Dalsgaard K, Nkrumah F K. Specific T-cell recognition of the merozoite proteins rhoptry-associated protein 1 and erythrocyte-binding antigen 1 of Plasmodium falciparum. Infect Immun. 1993;61:268–273. doi: 10.1128/iai.61.1.268-273.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jakobsen P H, Lemnge M M, Abu-Zeid Y A, Msangeni H A, Salum F M, Mhina J I, Akida J A, Ruta A S, Ronn A M, Heegaard P M, Ridley R G, Bygbjerg I C. Immunoglobulin G reactivities to rhoptry-associated protein-1 associated with decreased levels of Plasmodium falciparum parasitemia in Tanzanian children. Am J Trop Med Hyg. 1996;55:642–646. doi: 10.4269/ajtmh.1996.55.642. [DOI] [PubMed] [Google Scholar]
- 27.Kumar S, Collins W, Egan A, Yadava A, Garraud O, Blackman M J, Patino J A, Diggs C, Kaslow D C. Immunogenicity and efficacy in Aotus monkeys of four recombinant Plasmodium falciparum vaccines in multiple adjuvant formulations based on the 19-kilodalton C terminus of merozoite surface protein 1. Infect Immun. 2000;68:2215–2223. doi: 10.1128/iai.68.4.2215-2223.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lambros C, Vanderberg J P. Synchronization of Plasmodium falciparum erythrocytic stages in culture. J Parasitol. 1979;65:418–420. [PubMed] [Google Scholar]
- 29.Lord R, Jones G, Saul A. Two immunogenic peptide conjugates derived from P. falciparum MSA2 give rise to antibodies that crossreact with a nonhomologous 195-kDa malarial protein. Peptide Res. 1993;6:191–194. [PubMed] [Google Scholar]
- 30.Matile H, Pink J. Plasmodium falciparum malaria parasite cultures and their use in immunology. In: Lefkovits I, Benvenuto P, editors. Immunological methods. San Diego, Calif: Academic Press, Inc.; 1990. pp. 221–234. [Google Scholar]
- 31.Nagel A, Koch S, Valley U, Emmrich F, Marx U. Membrane-based cell culture systems—an alternative to in vivo production of monoclonal antibodies. Dev Biol Stand. 1999;101:57–64. [PubMed] [Google Scholar]
- 32.Narum D L, Haynes J D, Fuhrmann S, Moch K, Liang H, Hoffman S L, Sim B K. Antibodies against the Plasmodium falciparum receptor binding domain of EBA-175 block invasion pathways that do not involve sialic acids. Infect Immun. 2000;68:1964–1966. doi: 10.1128/iai.68.4.1964-1966.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patarroyo M E, Amador R, Clavijo P, Moreno A, Guzmán F, Romero P, Tascon R, Franco A, Murillo L A, Ponton G, Trujillo G. A synthetic vaccine protects humans against challenge with asexual blood stages of Plasmodium falciparum malaria. Nature. 1988;332:158–161. doi: 10.1038/332158a0. [DOI] [PubMed] [Google Scholar]
- 34.Patarroyo M E, Romero P, Torres M L, Clavijo P, Moreno A, Martinez A, Rodriguez R, Guzmán F, Cabezas E. Induction of protective immunity against experimental infection with malaria using synthetic peptides. Nature. 1987;328:629. doi: 10.1038/328629a0. [DOI] [PubMed] [Google Scholar]
- 35.Peltola H O, Case S E, Perri S F, Siddiqui W A. Plasmodium falciparum merozoite vaccination in Aotus monkeys recovered spontaneously from P. falciparum infection: a clinical study. Scand J Infect Dis. 1982;14:217–224. doi: 10.3109/inf.1982.14.issue-3.11. [DOI] [PubMed] [Google Scholar]
- 36.Pluschke G, Joss A, Marfurt J, Daubenberger C, Kashala O, Zwickl M, Stief A, Sansig G, Schläpfer B, Linkert S, van der Putten H, Hardman N, Schröder M. Generation of chimeric monoclonal antibodies from mice that carry human immunoglobulin Cγ1 heavy or Cκ light chain gene segments. J Immunol Methods. 1998;215:27–37. doi: 10.1016/s0022-1759(98)00041-6. [DOI] [PubMed] [Google Scholar]
- 37.Pluschke G, Mutz M. Use of isothermal titration calorimetry in the development of molecularly defined vaccines. J Therm Anal Cal. 1999;57:377–388. [Google Scholar]
- 38.Pöltl-Frank F, Zurbriggen R, Helg A, Stuart F, Robinson J, Gluck R, Pluschke G. Use of reconstituted influenza virus virosomes as an immunopotentiating delivery system for a peptide-based vaccine. Clin Exp Immunol. 1999;117:496–503. doi: 10.1046/j.1365-2249.1999.00989.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reese R T, Trager W, Jensen J B, Miller D A, Tantravahi R. Immunization against malaria with antigen from Plasmodium falciparum cultivated in vitro. Proc Natl Acad Sci USA. 1978;75:5665–5668. doi: 10.1073/pnas.75.11.5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ridley R G, Takacs B, Etlinger H, Scaife J G. A rhoptry antigen of Plasmodium falciparum is protective in Saimiri monkeys. Parasitology. 1990;101:187–192. doi: 10.1017/s0031182000063228. [DOI] [PubMed] [Google Scholar]
- 41.Ruebush T K, Campbell G H, Moreno A, Patarroyo M E, Collins W E. Immunization of owl monkeys with a combination of Plasmodium falciparum asexual blood-stage synthetic peptide antigens. Am J Trop Med Hyg. 1990;43:355–366. doi: 10.4269/ajtmh.1990.43.355. [DOI] [PubMed] [Google Scholar]
- 42.Schofield L, Bushell G R, Cooper J A, Saul A J, Upcroft J A, Kidson C. A rhoptry antigen of Plasmodium falciparum contains conserved and variable epitopes recognized by inhibitory monoclonal antibodies. Mol Biochem Parasitol. 1986;18:183–195. doi: 10.1016/0166-6851(86)90037-x. [DOI] [PubMed] [Google Scholar]
- 43.Shi Y P, Hasnain S E, Sacci J B, Holloway B P, Fujioka H, Kumar N, Wohlhueter R, Hoffman S L, Collins W E, Lal A A. Immunogenicity and in vitro protective efficacy of a recombinant multistage Plasmodium falciparum candidate vaccine. Proc Natl Acad Sci USA. 1999;96:1615–1620. doi: 10.1073/pnas.96.4.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Siddiqui W A. An effective immunization of experimental monkeys against a human malaria parasite, Plasmodium falciparum. Science. 1977;197:388–389. doi: 10.1126/science.406671. [DOI] [PubMed] [Google Scholar]
- 45.Siddiqui W A, Kan S C, Kramer K, Case S, Palmer K, Niblack J F. Use of a synthetic adjuvant in an effective vaccination of monkeys against malaria. Nature. 1981;289:64–66. doi: 10.1038/289064a0. [DOI] [PubMed] [Google Scholar]
- 46.Siddiqui W A, Tam L Q, Kan S C, Kramer K J, Case S E, Palmer K L, Yamaga K M, Hui G S. Induction of protective immunity to monoclonal-antibody-defined Plasmodium falciparum antigens requires strong adjuvant in Aotus monkeys. Infect Immun. 1986;52:314–318. doi: 10.1128/iai.52.1.314-318.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Siddiqui W A, Tam L Q, Kramer K J, Hui G S, Case S E, Yamaga K M, Chang S P, Chan E B, Kan S C. Merozoite surface coat precursor protein completely protects Aotus monkeys against Plasmodium falciparum malaria. Proc Natl Acad Sci USA. 1987;84:3014–3018. doi: 10.1073/pnas.84.9.3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Siddiqui W A, Taylor D W, Kan S C, Kramer K, Richmond-Crum S M, Kotani S, Shiba T, Kusumoto S. Vaccination of experimental monkeys against Plasmodium falciparum: a possible safe adjuvant. Science. 1978;201:1237–1239. doi: 10.1126/science.99814. [DOI] [PubMed] [Google Scholar]
- 49.Smythe J A, Coppel R L, Brown G V, Ramasamy R, Kemp D J, Anders R F. Identification of two integral membrane proteins of Plasmodium falciparum. Proc Natl Acad Sci USA. 1988;85:5195–5199. doi: 10.1073/pnas.85.14.5195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Snewin V A, Herrera M, Sanchez G, Scherf A, Langsley G, Herrera S. Polymorphism of the alleles of the merozoite surface antigens MSA1 and MSA2 in Plasmodium falciparum wild isolates from Colombia. Mol Biochem Parasitol. 1991;49:265–275. doi: 10.1016/0166-6851(91)90070-m. [DOI] [PubMed] [Google Scholar]
- 51.Stowers A, Prescott N, Cooper J, Takacs B, Stueber D, Kennedy P, Saul A. Immunogenicity of recombinant Plasmodium falciparum rhoptry associated proteins 1 and 2. Parasite Immunol. 1995;17:631–642. doi: 10.1111/j.1365-3024.1995.tb01009.x. [DOI] [PubMed] [Google Scholar]
- 52.Stowers A W, Cooper J A, Ehrhardt T, Saul A. A peptide derived from a B cell epitope of Plasmodium falciparum rhoptry associated protein 2 specifically raises antibodies to rhoptry associated protein 1. Mol Biochem Parasitol. 1996;82:167–180. doi: 10.1016/0166-6851(96)02730-2. [DOI] [PubMed] [Google Scholar]
- 53.Szarfman A, Walliker D, McBride J S, Lyon J A, Quakyi I A, Carter R. Allelic forms of gp195, a major blood-stage antigen of Plasmodium falciparum, are expressed in liver stages. J Exp Med. 1988;167:231–236. doi: 10.1084/jem.167.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tanabe K, Mackay M, Goman M, Scaife J G. Allelic dimorphism in a surface antigen gene of the malaria parasite Plasmodium falciparum. J Mol Biol. 1987;195:273–287. doi: 10.1016/0022-2836(87)90649-8. [DOI] [PubMed] [Google Scholar]
- 55.Taylor D W, Siddiqui W A. Effect of falciparum malaria infection on the in vitro mitogen responses of spleen and peripheral blood lymphocytes from owl monkeys. Am J Trop Med Hyg. 1978;27:738–742. doi: 10.4269/ajtmh.1978.27.738. [DOI] [PubMed] [Google Scholar]
- 56.Teklehaimanot A, Collins W E, Nguyen-Dinh P, Campbell C C, Bhasin V K. Characterization of Plasmodium falciparum cloned lines with respect to gametocyte production in vitro, infectivity to Anopheles mosquitoes, and transmission to Aotus monkeys. Trans R Soc Trop Med Hyg. 1987;81:885–887. doi: 10.1016/0035-9203(87)90336-1. [DOI] [PubMed] [Google Scholar]
- 57.Thaithong S, Beale G H. Resistance of ten Thai isolates of Plasmodium falciparum to chloroquine and pyrimethamine by in vitro tests. Trans R Soc Trop Med Hyg. 1981;75:271–273. doi: 10.1016/0035-9203(81)90333-3. [DOI] [PubMed] [Google Scholar]
- 58.Voller A, Richards W H. An attempt to vaccinate owl monkeys (Aotus trivirgatus) against falciparum malaria. Lancet. 1968;ii:1172–1174. doi: 10.1016/s0140-6736(68)91644-9. [DOI] [PubMed] [Google Scholar]
- 59.Walliker D, Quakyi I A, Wellems T E, McCutchan T F, Szarfman A, London W T, Corcoran L M, Burkot T R, Carter R. Genetic analysis of the human malaria parasite Plasmodium falciparum. Science. 1987;236:1661–1666. doi: 10.1126/science.3299700. [DOI] [PubMed] [Google Scholar]
- 60.Wiseman T, Williston S, Brandts J F, Lin L N. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal Biochem. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]