Abstract
The generation times of our recent ancestors can tell us about both the biology and social organization of prehistoric humans, placing human evolution on an absolute time scale. We present a method for predicting historical male and female generation times based on changes in the mutation spectrum. Our analyses of whole-genome data reveal an average generation time of 26.9 years across the past 250,000 years, with fathers consistently older (30.7 years) than mothers (23.2 years). Shifts in sex-averaged generation times have been driven primarily by changes to the age of paternity, although we report a substantial increase in female generation times in the recent past. We also find a large difference in generation times among populations, reaching back to a time when all humans occupied Africa.
Men have always had children at an older age than women, even among diverse populations, but this age gap has recently shrunk.
INTRODUCTION
Knowledge of the human generation time (or “generation interval”) in the recent past is important for many fields. While genetic data have provided deep insights into human history, population genetic methods typically scale history in terms of generations [e.g., (1, 2)]. This makes knowing the generation time especially important for determining the absolute timing of historic events, including migrations to new continents (3) or gene flow with extinct hominids (4). To transform these population genetic estimates into absolute time, it is commonly assumed that current generation times have persisted across hundreds of thousands of years or that studies of extant hunter-gatherer (forager) societies provide representative generation times across the span of human history (5, 6). However, neither assumption is likely to be correct: The average age at which males and females have children depends on many environmental, demographic, and cultural factors that can change rapidly (7), while contemporary hunter-gatherer societies differ substantially from each other and from past societies (8). It is also clear that generation times have evolved among the great apes (9) and may therefore have evolved along the branch leading to modern humans.
Previous genetic approaches to estimating historical generation times (the average age at which individuals conceive children) have taken advantage of the compounding effects of either recombination (10) or mutation (11) on modern human DNA sequence divergence from ancient samples. While these estimates have provided substantial insight, they are averaged both across the sexes and across the past 40,000 to 45,000 years. Greater resolution through time is possible by examining the mutations that originated at specific times in the past, together with a model that accurately predicts the generation times of individuals producing those mutations. Here, we develop a model that uses the spectrum of de novo mutations as a predictor of parental age. By coupling this model with variants whose ages have been estimated from genome-wide genealogical information, we are able to separately estimate the male and female generation times at many different points across the past 250,000 years.
RESULTS
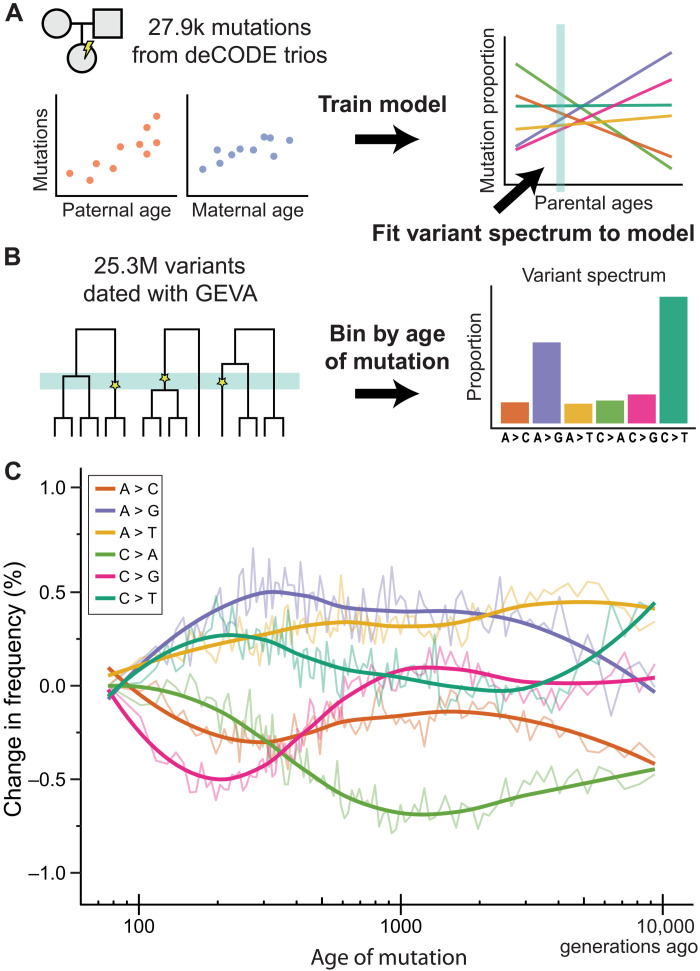
As humans age, the number and type of de novo mutations that they transmit to their offspring change (12, 13). We use information on mutations from a large pedigree study with parents whose ages at conception are known (14) to model the relationship between parental age and the counts of the six different types of single-nucleotide mutations (fig. S1). These mutation counts are regressed on both paternal and maternal age in a Dirichlet-multinomial model (Fig. 1A and fig. S2). To obtain mutation spectra from many different periods in the past, we used the estimated time of origin for current polymorphisms from the genealogical estimation of variant age (GEVA) approach (Fig. 1B) (15). This method estimates when, in the past, each of ~43 million variants from the 1000 Genomes Project arose by mutation. After filtering variants with the same criteria applied to de novo mutations used to train our model, we retained 25.3 million variants for our analysis.
Fig. 1. The mutation spectrum changes with human generation time.
(A) Data on de novo mutations from 1247 Icelandic trios (14) were used to train a model that predicts the effect of both maternal and paternal age on the mutation spectrum. (B) Data from 25.3 million segregating variants whose date of origin was estimated using GEVA (15) were used to assess the mutation spectrum at different periods in the past. The mutation spectrum from each time period (bin) was used as input to the model from (A) to estimate the generation interval for males and females. (C) Differences in the frequency of each of the six different mutation types through time, as compared to the most recent time period (smoothed lines from local regression). Figure S15 presents the absolute frequencies of the same mutation data over time.
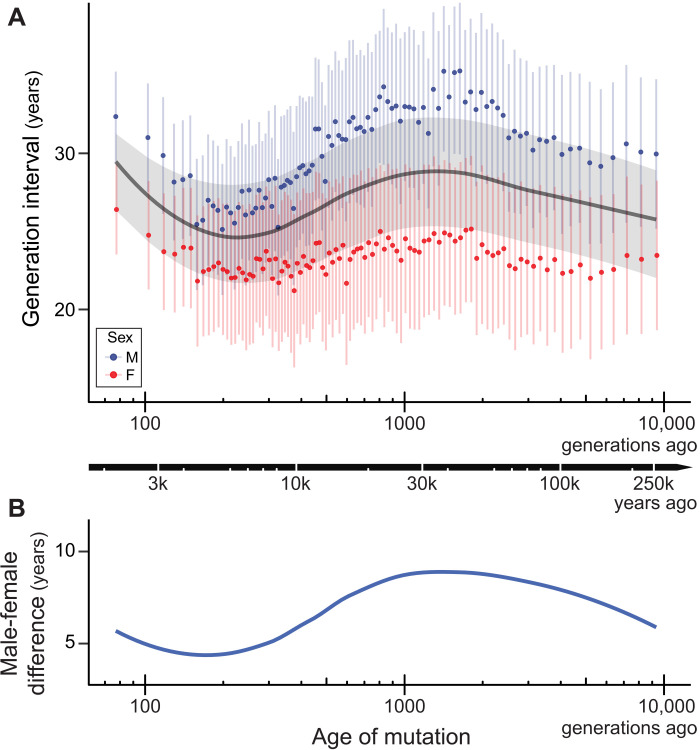
Applying our mutation spectrum model to the polymorphism data allows us to estimate generation times for males and females across the past 250,000 years (Fig. 2A). Within this time frame, we find the average human generation interval to be 26.9 ± 3.4 years (SE) with an average for males of 30.7 ± 4.8 years and an average for females of 23.2 ± 3.0 years. The results show that human generation times have undergone a rapid increase in the recent past after declining for over a thousand generations. The average human generation interval was at a recent minimum of 24.9 ± 3.5 years at ~250 generations ago (6.4 ka ago), roughly concurrent with the historic rise of early civilizations. Before this, it had declined from a peak of 29.8 ± 4.1 years at ~1400 generations ago (38 ka ago), just before the beginning of the Last Glacial Maximum. Note that these estimates are a composite across multiple human populations (see below for separate estimates from different continental populations).
Fig. 2. Estimating the male and female generation interval across 250,000 years.
(A) Male (blue points), female (red points), and sex-averaged (gray line) generation intervals over the past 10,000 generations. The data were divided into 100 time periods with equal numbers of variants; generation intervals in each were independently estimated using the Dirichlet-multinomial model. Sex-averaged generation intervals are shown here as a line smoothed by local regression. Confidence intervals (±1 SE) displayed for estimates of the mean, and for males and females separately, were obtained by resampling both the de novo mutation data for bootstrapped models and the variants in each time period for bootstrapped spectra. The absolute timeline (black arrow) was calculated by integrating sex-averaged generation-time estimates across generations elapsed since the present (section S3.3). (B) The smoothed difference (loess) between estimates of the male and female generation interval over time.
Our model estimates a longer generation interval for males than females across all analyzed time periods (Fig. 2B). These results are consistent with studies of contemporary cultures, more than 99% of which show a longer male generation interval (5). Overall, there is a high correlation between the average generation interval and the male-female difference (Pearson’s r = 0.88; P < 1 × 10−10), likely because of a relatively constant generation interval in females (σ2 = 0.9 years) and a large amount of variation in males across time (σ2 = 6.8 years). Males and females reach puberty at approximately the same age (16), but the reproductive age in males can extend more than 20 years beyond that in females. Sociocultural factors are likely to have acted in concert with the higher bound on male reproductive age to produce the greater variance observed in male generation interval. The male-female difference follows a similar pattern to that of the average generation time except for the most recent windows, which show a smaller increase in male-female difference than expected during the recent uptick in generation times (compare panels A and B of Fig. 2). This smaller difference appears to be driven by a relatively larger increase in recent female generation intervals: The most recent time period is significantly higher than at any point in the past 250,000 years (P < 0.005, z test).
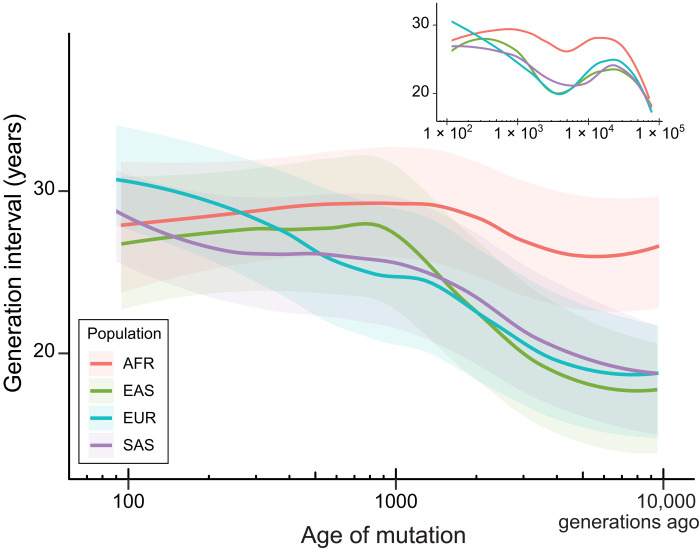
To investigate differences in generation times among human populations, we repeated our analysis using four major continental populations within the 1000 Genomes Project. Variants are counted as part of a continental population as long as they are polymorphic among samples from that population. Private variants from each population suggest that the mutation process in the recent past is consistent between them (fig. S3). While the continental labels for each population are used across the span of the analysis, note that beyond roughly 2000 generations ago, all non-African populations were likely located in Africa and show little differentiation among themselves; coalescence among all ancestral populations living in Africa does not occur until more than 10,000 generations ago (15).
We find subtle changes to the average human generation interval among populations in the last 1000 generations (Fig. 3 and fig. S4). Average generation times in European and South Asian populations have increased slightly, while generation times in African and East Asian populations have changed little. Similar results in the recent past were observed when using only private alleles (fig. S5). We estimate a shorter sex-averaged generation interval for Europeans (26.1 years) than East Asians (27.1 years) over the past 40,000 years, supporting a recent estimate derived from divergence to archaic DNA (11). Beyond this most recent time frame, the average generation interval in each of the ancestral non-African populations grows progressively shorter into the past. The dominating pattern across the past 10,000 generations is a significantly shorter sex-averaged generation interval for East Asian, European, and South Asian populations—20.1 ± 3.9, 20.6 ± 3.8, and 21.0 ± 3.7 years—compared to the African population, 26.9 ± 3.5 years (P < 1 × 10−10, t test). The estimated generation times do not converge between populations until we expand our analysis to include periods older than 10,000 generations ago (Fig. 3, inset).
Fig. 3. Change in generation interval across different human populations.
Generation intervals were estimated in ancestors of four major continental human populations included in the 1000 Genomes Project; sex-averaged generation intervals are shown here as smoothed by loess (see fig. S6 for full results). Confidence intervals for each population were obtained by bootstrapping, as in Fig. 2. The inset shows results from including polymorphisms that date back to 78,000 generations ago; note that age estimates of mutations in the very distant past have decreased accuracy (15). AFR, Africa; EAS, East Asia; EUR, Europe; SAS, South Asia.
DISCUSSION
The large difference in generation times between populations suggests that different time scales are needed to estimate events outside of Africa (20 to 21 years per generation) versus those in Africa (27 years per generation). These results are consistent with the prediction of a shorter generation time in non-Africans, based on the observation of a slightly elevated per-year mutation rate in these populations (17). Estimates of the error in our model fit do not show increasing error with either genetic or geographic distance from Iceland (figs. S6 and S7), the origin of the pedigreed mutation data used to train our model. Such a trend may have been expected if differences in mutational spectra were driven by genetic or environmental differences among populations. Note that the difference among populations beyond 2000 generations ago reflects population structure in humans before their dispersal out of Africa, a structure that is not fully captured by the 1000 Genomes AFR sample (3, 15, 17, 18). This implies that the simple labels of “African” and “non-African” for these populations conceal differences in generation times that existed on our ancestral continent.
Our study builds upon advances in understanding the characteristics of de novo mutations (14) and in estimating genome-wide genealogies (15) to create a model for generation times that can be applied to ancient populations. While it is clear that the frequency of individual mutation types can evolve rapidly (19–21), even small changes to the generation interval can reshape the overall mutation spectrum (22, 23). Our results are consistent with previous estimates of the average generation time over the past 40,000 to 45,000 years (10, 11) but offer unprecedented resolution of sex-specific generation times across 250,000 years of human history. While information on the mutation spectrum far into the past (>10,000 generations ago) is limited by the coalescent process (and subsequent lack of ancient polymorphisms), fine-scale estimates of generation times from the most recent 100 generations will be possible with larger population samples [cf. (24)]. Large-enough samples will bring estimates from population genetic data close enough in time to overlap with historical birth records [e.g., (25)]. As it stands, our results offer a unique look into the biology of our ancestors and provide a more detailed picture of human demographic history.
MATERIALS AND METHODS
We developed a parental age model for the mutation spectrum based on data from a large study of de novo mutations in an Icelandic population (14). Mutation count data from each proband was modeled as coming from a Dirichlet-multinomial distribution with parental ages treated as covariates (section S1). We excluded variants from mutation classes prone to homoplasy (CpG → TpG mutations), as well as TCC → TTC and related triplet transitions, which have been inferred to be the result of a recent mutation pulse (19, 20). After filtering mutations, our model was trained on 27,902 phased mutations from 1247 trios.
We used results from GEVA (15) on the time at which new mutations arose in human history (section S2). GEVA dates each variant independently by inferring when on the local underlying genealogy it occurred. Using the local genealogy (“gene tree”), GEVA avoids problems associated with hemiplasy (26), although it does assume that every mutation only occurred once at a site. This assumption means that mutation classes with very high mutation rates (e.g., CpG → CpT) can be mismapped; to be consistent with the de novo mutation data used to train our model above, these mutations were filtered from both datasets (section S2.2). Human variants dated by the GEVA approach were subject to several additional filters to ensure their appropriateness for estimating generation time. We considered only biallelic single-nucleotide sites and discarded singletons and variants with derived allele frequencies higher than 98%. More than 80% of the sampled variants arose in the last 10,000 generations, but very few are from the last 100 generations (fig. S8). Because the sampled variants are unevenly distributed through time, we divided the data from the past 10,000 generations into bins with equal numbers of variants. Maternal and paternal ages were then estimated by fitting the variant spectrum in each of the 100 historical bins to our Dirichlet-multinomial model by minimizing compositional (Aitchison) distance between the observed spectrum and the model (section S3). We found that the mutation spectrum from the large pedigree study (14) consistently differed from the variant spectrum inferred from the 1000 Genomes Project data, possibly because we removed singletons from the polymorphism dataset to reduce errors. Therefore, to obtain absolute generation times for historical periods, we centered the observed spectra on the most recent bin, subtracting its difference with the average mutation spectrum estimated in (14) from each historical spectrum. This has the effect of assuming that parental ages in the pedigreed mutation dataset reflect generation times in the most recent historical bin. We find this assumption to be robust for both the relative difference in generation time between the sexes and the overall pattern of historical generation times (see section S4.3).
We carried out multiple analyses to ensure the accuracy and robustness of our results. Error in our model fit did not increase with increasing time since the present (section S3.5 and fig. S6), as might be expected if multiple aspects of the mutation spectrum had evolved over time. We also find that our method estimates mean generation times with very low error on simulated data, although increasing variance in parental ages slightly increased error rates (fig. S9). In general, we expect that multiple sources of variation within populations are likely to contribute to error in our estimates, including the aforementioned variation in parental ages or genetic variation in the mutation spectrum among individuals. Intragenomic differences in the mutation spectrum due to variation in recombination rate (section S4.1) and replication timing (section S4.2) (27) are likely to contribute to variance in estimates from our genome-wide model. The mutation spectrum differs significantly across genomic regions where recombination rates differ (fig. S10A), and if we estimate generation times from only subsets of the genome, then these estimates would be consistently higher when inferred from genomic regions with higher recombination rates (fig. S10C). To ensure that the genomic regions used to build the model are the same as those used to estimate historical mutation spectra, we resampled multiple datasets matched for recombination rate and replication timing; there was no effect on our estimates after controlling for the slight differences in the distribution of mutations and polymorphisms with respect to these variables (fig. S10, G and H). Systemic effects of recombination on generation time estimates due to biased gene conversion or linked selection may be more difficult to rule out. However, we do not find any significant increase in the frequency of mutations toward G or C with increasing variant age, as might be expected from the effects of GC-biased gene conversion over time (fig. S11). Lastly, the trends found for human generation time were not substantially affected by the stringency of mutation filters on the training set (fig. S12A), the masking of regions introgressed from Neanderthals (fig. S13A), nor the precision of allele ages in the GEVA dataset (fig. S14).
Acknowledgments
We thank J. Raff and A. Bentley for helpful input early in this project, as well as D. Schrider and two anonymous reviewers for comments on the manuscript.
Funding: This work was supported by grants to M.W.H. from the Precision Health Initiative of Indiana University and the NSF (DEB-1936187).
Author contributions: Conceptualization: R.J.W., J.R., and M.W.H. Methodology: R.J.W., S.I.A.-S., and M.W.H. Investigation: R.J.W. and S.I.A.-S.. Funding acquisition: M.W.H. Supervision: J.R. and M.W.H. Writing (original draft): R.J.W. and M.W.H. Writing (review and editing): R.J.W., J.R., and M.W.H.
Competing interests: In addition to authors’ listed affiliations, J.R. holds a position as a Core Scientist at the Wisconsin National Primate Research Center. The authors declare that they have no other competing interests.
Data and materials availability: All data used in this analysis are publicly available from references (14, 15). All code used for this analysis has been made publicly available at https://github.com/Wang-RJ/generationtimes/ and has also been archived, accessible through doi:10.5281/zenodo.6545159. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Supplementary Methods
Figs. S1 to S15
Table S1
References
Other Supplementary Material for this : manuscript includes the following:
Data S1
REFERENCES AND NOTES
- 1.Li H., Durbin R., Inference of human population history from individual whole-genome sequences. Nature 475, 493–496 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Speidel L., Forest M., Shi S., Myers S. R., A method for genome-wide genealogy estimation for thousands of samples. Nat. Genet. 51, 1321–1329 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malaspinas A.-S., Westaway M. C., Muller C., Sousa V. C., Lao O., Alves I., Bergström A., Athanasiadis G., Cheng J. Y., Crawford J. E., Heupink T. H., Macholdt E., Peischl S., Rasmussen S., Schiffels S., Subramanian S., Wright J. L., Albrechtsen A., Barbieri C., Dupanloup I., Eriksson A., Margaryan A., Moltke I., Pugach I., Korneliussen T. S., Levkivskyi I. P., Moreno-Mayar J. V., Ni S., Racimo F., Sikora M., Xue Y., Aghakhanian F. A., Brucato N., Brunak S., Campos P. F., Clark W., Ellingvåg S., Fourmile G., Gerbault P., Injie D., Koki G., Leavesley M., Logan B., Lynch A., Matisoo-Smith E. A., McAllister P. J., Mentzer A. J., Metspalu M., Migliano A. B., Murgha L., Phipps M. E., Pomat W., Reynolds D., Ricaut F.-X., Siba P., Thomas M. G., Wales T., Wall C. M., Oppenheimer S. J., Tyler-Smith C., Durbin R., Dortch J., Manica A., Schierup M. H., Foley R. A., Lahr M. M., Bowern C., Wall J. D., Mailund T., Stoneking M., Nielsen R., Sandhu M. S., Excoffier L., Lambert D. M., Willerslev E., A genomic history of Aboriginal Australia. Nature 538, 207–214 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Huerta-Sánchez E., Jin X., Asan, Bianba Z., Peter B. M., Vinckenbosch N., Liang Y., Yi X., He M., Somel M., Ni P., Wang B., Ou X., Huasang, Luosang J., Cuo Z. X. P., Li K., Gao G., Yin Y., Wang W., Zhang X., Xu X., Yang H., Li Y., Wang J., Wang J., Nielsen R., Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA. Nature 512, 194–197 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fenner J. N., Cross-cultural estimation of the human generation interval for use in genetics-based population divergence studies. Am. J. Phys. Anthropol. 128, 415–423 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Matsumura S., Forster P., Generation time and effective population size in Polar Eskimos. Proc. Biol. Sci. 275, 1501–1508 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bocquet-Appel J. P., When the world's population took off: The springboard of the Neolithic Demographic Transition. Science 333, 560–561 (2011). [DOI] [PubMed] [Google Scholar]
- 8.P. Jordan, in The Oxford Handbook of the Archaeology and Anthropology of Hunter-Gatherers, V. Cummings, P. Jordan, M. Zvelebil, Eds. (Oxford Univ. Press, 2014). [Google Scholar]
- 9.Langergraber K. E., Prüfer K., Rowney C., Boesch C., Crockford C., Fawcett K., Inoue E., Inoue-Muruyama M., Mitani J. C., Muller M. N., Robbins M. M., Schubert G., Stoinski T. S., Viola B., Watts D., Wittig R. M., Wrangham R. W., Zuberbühler K., Pääbo S., Vigilant L., Generation times in wild chimpanzees and gorillas suggest earlier divergence times in great ape and human evolution. Proc. Natl. Acad. Sci. U.S.A. 109, 15716–15721 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moorjani P., Sankararaman S., Fu Q., Przeworski M., Patterson N., Reich D., A genetic method for dating ancient genomes provides a direct estimate of human generation interval in the last 45,000 years. Proc. Natl. Acad. Sci. U.S.A. 113, 5652–5657 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coll Macià M., Skov L., Peter B. M., Schierup M. H., Different historical generation intervals in human populations inferred from Neanderthal fragment lengths and mutation signatures. Nat. Commun. 12, 5317 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kong A., Frigge M. L., Masson G., Besenbacher S., Sulem P., Magnusson G., Gudjonsson S. A., Sigurdsson A., Jonasdottir A., Jonasdottir A., Wong W. S. W., Sigurdsson G., Walters G. B., Steinberg S., Helgason H., Thorleifsson G., Gudbjartsson D. F., Helgason A., Magnusson O. T., Thorsteinsdottir U., Stefansson K., Rate of de novo mutations and the importance of father’s age to disease risk. Nature 488, 471–475 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rahbari R., Wuster A., Lindsay S. J., Hardwick R. J., Alexandrov L. B., Turki S. A., Dominiczak A., Morris A., Porteous D., Smith B., Stratton M. R.; UK10K Consortium, Hurles M. E., Timing, rates and spectra of human germline mutation. Nat. Genet. 48, 126–133 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jónsson H., Sulem P., Kehr B., Kristmundsdottir S., Zink F., Hjartarson E., Hardarson M. T., Hjorleifsson K. E., Eggertsson H. P., Gudjonsson S. A., Ward L. D., Arnadottir G. A., Helgason E. A., Helgason H., Gylfason A., Jonasdottir A., Jonasdottir A., Rafnar T., Frigge M., Stacey S. N., Th. Magnusson O., Thorsteinsdottir U., Masson G., Kong A., Halldorsson B. V., Helgason A., Gudbjartsson D. F., Stefansson K., Parental influence on human germline de novo mutations in 1,548 trios from Iceland. Nature 549, 519–522 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Albers P. K., McVean G., Dating genomic variants and shared ancestry in population-scale sequencing data. PLoS Biol. 18, e3000586 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parent A.-S., Teilmann G., Juul A., Skakkebaek N. E., Toppari J., Bourguignon J.-P., The timing of normal puberty and the age limits of sexual precocity: Variations around the world, secular trends, and changes after migration. Endocr. Rev. 24, 668–693 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Mallick S., Li H., Lipson M., Mathieson I., Gymrek M., Racimo F., Zhao M., Chennagiri N., Nordenfelt S., Tandon A., Skoglund P., Lazaridis I., Sankararaman S., Fu Q., Rohland N., Renaud G., Erlich Y., Willems T., Gallo C., Spence J. P., Song Y. S., Poletti G., Balloux F., van Driem G., de Knijff P., Romero I. G., Jha A. R., Behar D. M., Bravi C. M., Capelli C., Hervig T., Moreno-Estrada A., Posukh O. L., Balanovska E., Balanovsky O., Karachanak-Yankova S., Sahakyan H., Toncheva D., Yepiskoposyan L., Tyler-Smith C., Xue Y., Abdullah M. S., Ruiz-Linares A., Beall C. M., di Rienzo A., Jeong C., Starikovskaya E. B., Metspalu E., Parik J., Villems R., Henn B. M., Hodoglugil U., Mahley R., Sajantila A., Stamatoyannopoulos G., Wee J. T. S., Khusainova R., Khusnutdinova E., Litvinov S., Ayodo G., Comas D., Hammer M. F., Kivisild T., Klitz W., Winkler C. A., Labuda D., Bamshad M., Jorde L. B., Tishkoff S. A., Watkins W. S., Metspalu M., Dryomov S., Sukernik R., Singh L., Thangaraj K., Pääbo S., Kelso J., Patterson N., Reich D., The Simons Genome Diversity Project: 300 genomes from 142 diverse populations. Nature 538, 201–206 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scerri E. M. L., Thomas M. G., Manica A., Gunz P., Stock J. T., Stringer C., Grove M., Groucutt H. S., Timmermann A., Rightmire G. P., d’Errico F., Tryon C. A., Drake N. A., Brooks A. S., Dennell R. W., Durbin R., Henn B. M., Lee-Thorp J., deMenocal P., Petraglia M. D., Thompson J. C., Scally A., Chikhi L., Did our species evolve in subdivided populations across Africa, and why does it matter? Trends Ecol. Evol. 33, 582–594 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harris K., Evidence for recent, population-specific evolution of the human mutation rate. Proc. Natl. Acad. Sci. U.S.A. 112, 3439–3444 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harris K., Pritchard J. K., Rapid evolution of the human mutation spectrum. eLife 6, e24284 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mathieson I., Reich D., Differences in the rare variant spectrum among human populations. PLOS Genet. 13, e1006581 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carlson J., DeWitt W. S., Harris K., Inferring evolutionary dynamics of mutation rates through the lens of mutation spectrum variation. Curr. Opin. Genet. Dev. 62, 50–57 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang R. J., Raveendran M., Harris R. A., Murphy W. J., Lyons L. A., Rogers J., Hahn M. W., De novo mutations in domestic cat are consistent with an effect of reproductive longevity on both the rate and spectrum of mutations. Mol. Biol. Evol. 39, msac147 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keinan A., Clark A. G., Recent explosive human population growth has resulted in an excess of rare genetic variants. Science 336, 740–743 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Helgason A., Hrafnkelsson B., Gulcher J. R., Ward R., Stefánsson K., A populationwide coalescent analysis of Icelandic matrilineal and patrilineal genealogies: Evidence for a faster evolutionary rate of mtDNA lineages than Y chromosomes. Am. J. Hum. Genet. 72, 1370–1388 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mendes F. K., Hahn M. W., Gene tree discordance causes apparent substitution rate variation. Syst. Biol. 65, 711–721 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Koren A., Handsaker R. E., Kamitaki N., Karlić R., Ghosh S., Polak P., Eggan K., McCarroll S. A., Genetic variation in human DNA replication timing. Cell 159, 1015–1026 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim J., Zhang Y., Day J., Zhou H., MGLM: An R package for multivariate categorical data analysis. R J. 10, 73–90 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.J. Aitchison, The Statistical Analysis of Compositional Data (Monographs on Statistics and Applied Probability, Chapman and Hall, 1986). [Google Scholar]
- 30.J. Nocedal, S. J. Wright, Numerical Optimization (Springer Series in Operations Research and Financial Engineering, Springer, 1999), vol. 35. [Google Scholar]
- 31.Barton N. H., The effect of hitch-hiking on neutral genealogies. Genet. Res. 72, 123–133 (1998). [Google Scholar]
- 32.Glémin S., Arndt P. F., Messer P. W., Petrov D., Galtier N., Duret L., Quantification of GC-biased gene conversion in the human genome. Genome Res. 25, 1215–1228 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lachance J., Tishkoff S. A., Biased gene conversion skews allele frequencies in human populations, increasing the disease burden of recessive alleles. Am. J. Hum. Genet. 95, 408–420 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steinrücken M., Spence J. P., Kamm J. A., Wieczorek E., Song Y. S., Model-based detection and analysis of introgressed Neanderthal ancestry in modern humans. Mol. Ecol. 27, 3873–3888 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anderson-Trocmé L., Farouni R., Bourgey M., Kamatani Y., Higasa K., Seo J.-S., Kim C., Matsuda F., Gravel S., Legacy data confound genomics studies. Mol. Biol. Evol. 37, 2–10 (2020). [DOI] [PubMed] [Google Scholar]
- 36.Kessler M. D., Loesch D. P., Perry J. A., Heard-Costa N. L., Taliun D., Cade B. E., Wang H., Daya M., Ziniti J., Datta S., Celedón J. C., Soto-Quiros M. E., Avila L., Weiss S. T., Barnes K., Redline S. S., Vasan R. S., Johnson A. D., Mathias R. A., Hernandez R., Wilson J. G., Nickerson D. A., Abecasis G., Browning S. R., Zöllner S., O’Connell J. R., Mitchell B. D.; National Heart, Lung, and Blood Institute Trans-Omics for Precision Medicine (TOPMed) Consortium; TOPMed Population Genetics Working Group, O’Connor T. D., De novo mutations across 1,465 diverse genomes reveal mutational insights and reductions in the Amish founder population. Proc. Natl. Acad. Sci. 117, 2560–2569 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Methods
Figs. S1 to S15
Table S1
References
Data S1