Abstract
The efficacy and safety of drugs are widely known to be determined by their interactions with multiple molecules of pharmacological importance, and it is therefore essential to systematically depict the molecular atlas and pharma-information of studied drugs. However, our understanding of such information is neither comprehensive nor precise, which necessitates the construction of a new database providing a network containing a large number of drugs and their interacting molecules. Here, a new database describing the molecular atlas and pharma-information of drugs (DrugMAP) was therefore constructed. It provides a comprehensive list of interacting molecules for >30 000 drugs/drug candidates, gives the differential expression patterns for >5000 interacting molecules among different disease sites, ADME (absorption, distribution, metabolism and excretion)-relevant organs and physiological tissues, and weaves a comprehensive and precise network containing >200 000 interactions among drugs and molecules. With the great efforts made to clarify the complex mechanism underlying drug pharmacokinetics and pharmacodynamics and rapidly emerging interests in artificial intelligence (AI)-based network analyses, DrugMAP is expected to become an indispensable supplement to existing databases to facilitate drug discovery. It is now fully and freely accessible at: https://idrblab.org/drugmap/
Graphical Abstract
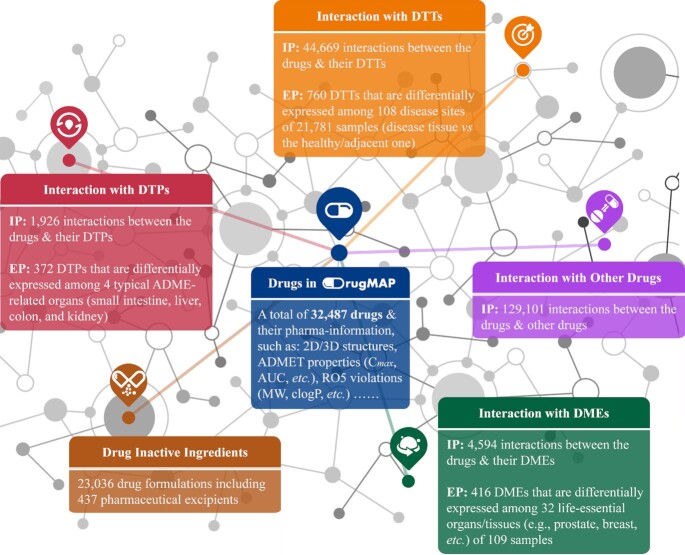
Graphical Abstract.
DrugMAP provides a comprehensive list of interacting molecules for >30 000 drugs/drug candidates, illustrates the differential expression pattern of >5000 interacting molecules among disease sites, ADME-related organs or physiological tissues, and weaves a comprehensive and precise network containing >200 000 interactions among drugs and molecules.
INTRODUCTION
The efficacy and safety of drugs are widely known to be determined by their interactions with some molecules of pharmacological importance (1–3). For example, a drug's therapeutic effect and resistance profile are largely shaped by its interactions with its target (4–6); the absorption, disposition and elimination of drugs are extensively affected by their interactions with channels/transporters (7–9); drug metabolism and prodrug synthesis are substantially regulated by specific enzymatic interactions (10–13); and so on. In other words, the efficacy and safety of a drug derive from the collective regulation by many molecules of an extremely sophisticated mechanism (14,15). Thus, it is essential to systematically depict the molecular atlas of regulation mechanisms for each drug, which is expected to facilitate the design/repurposing of drugs (16,17), the prediction of drug–drug interactions (3,18,19), the reversion of multidrug resistances (20), and so on.
However, our understanding of such a molecular atlas of each drug is neither comprehensive nor precise (21–23). On the one hand, available pharmaceutical studies tend to focus on specific types of interacting molecules (24,25), and thus lack comprehensive descriptions of all molecules of pharmacological importance (26,27). On the other hand, these molecules are found to be differentially expressed among disease sites, ADME (absorption, distribution, metabolism and excretion)-related organs and other physiological sites (15,28), and the differentially expressed pattern of these molecules is not precisely illustrated (3,29). Thus, a large number of studies have been conducted to discover the comprehensive set of drug-centered molecular interactions (30–32) and illustrate the expression pattern of those interacting molecules among various organs/tissues (33–36). With the wide application of artificial intelligence (AI) techniques in current drug discovery, it is crucial to construct a comprehensive and precise ‘network’ (based on the findings of previous publications) to describe a large number of drugs and their interacting molecules of pharmacological importance (30,37,38). In other words, it is key to have a database that provides such valuable network data for facilitating the discovery of efficacious combination therapy (39), the understanding of off-target mechanisms and undesirable side effects (40–42), etc.
So far, a variety of reputable databases related to the topic above have been constructed (43–53). Some of them provide complex networks to describe the interactions among drugs and molecules [such as STITCH (43), DGIdb (44), BindingDB (45) and ChEMBL (46)]. Such networks usually have a huge number of interactions, but the vast majority of them have little reported relation to a drug’s pharmacological effect (54–56). Some others describe the relationship between the interacting molecule and a drug’s pharmacological effect [such as DrugBank (47), TTD (48), UCSF-FDA (49), VARIDT (50), BRENDA (51) and KinaseMD (52)], but these databases only focus on providing specific types of interacting molecules (e.g. targets, channels/transporters or enzymes). In other words, none of them provides a comprehensive and precise ‘network’ containing a large number of drugs and their interacting molecules of pharmacological importance.
In this study, a newly constructed database named ‘Molecular Atlas and Pharma-information of drugs (DrugMAP)’ was therefore introduced. First, a comprehensive literature review on ∼2500 approved drugs, ∼8900 drugs ever tested in clinical trials, ∼6000 patented/pre-clinical drugs and ∼16 000 investigative agents was conducted. Second, interacting molecules of pharmacological importance to the above drugs were collected, and a total of 5067 pharmacologically important molecules were identified as interacting with 32 487 drugs/drug candidates. Third, the expression patterns of 1539 molecules among 108 disease sites of 21 781 patients, 1323 molecules among four typical ADME-related organs (small intestine, liver, colon and kidney) of 236 healthy individuals and 1286 molecules among 32 physiological/life-essential sites/organs (such as prostate, breast, etc.) of 109 samples, were collected. Finally, a comprehensive and precise ‘network’ containing 50 180 drug–molecule interactions was constructed. The subnetwork for each drug and the entire network for all drugs are freely accessible in DrugMAP. All in all, since such a comprehensive and precise ‘network’ is essential for the understanding of the molecular atlas of drugs, this database is expected to have great implications and impact on modern drug discovery.
FACTUAL CONTENT AND DATA RETRIEVAL
Systematic collection of pharma-information for a comprehensive set of drugs
A comprehensive set of drugs was collected from multiple sources. A total of ∼2500 approved drugs were collected from the official website of the US Food and Drug Administration (US FDA), ∼8900 drugs ever tested in clinical trials were collected from the official site of ClinicalTrial.gov and ∼6000 patented/pre-clinical drugs together with ∼16 000 investigative drugs were collected by literature review or from existing databases [such as TTD (57)]. In DrugMAP, a total of ∼33 000 drug entities of clinical importance were stored, and their general information (name, synonyms, indications, therapeutic classes, etc.) was extracted. Approximately 700 diseases corresponding to the indication of drugs were manually checked and mapped to the International Classification of Diseases (ICD-11) (58). Moreover, 2D and 3D structures of ∼26 000 drugs (∼81.0%) were made visualizable and fully downloadable (in an SDF file) from the online database.
Moreover, additional pharma-information of drugs was also collected and provided in DrugMAP. In particular, chemical identifiers (canonical SMILES, formula, InChI, InChIKey, etc.) and RO5 violations of drugs were collected and confirmed from PubChem and the ChEMBL database (46,59), and ADMET properties (volume of distribution, bioavailability, metabolism, clearance, elimination, half-life, etc.) of each drug were collected by additional literature review. Moreover, to cross-reference DrugMAP with external databases, the cross-matching IDs of drugs to other databases (PubChem, ChEBI, CAS, TTD, etc.) was provided. The detailed statistics and description of the general and pharma-information of the drugs in this database are illustrated in Figure 1.
Figure 1.
The primary data components and the corresponding statistics of DrugMAP. The basic pharma-information of drugs (blue), the interactions between drugs and three types of molecules of pharmacological importance, i.e. drug therapeutic target (DTT, orange), drug transporter (DTP, red) and drug-metabolizing enzyme (DME, green), the drug–drug interaction information (purple) and the data of drug inactive ingredients (brown).
The interaction pattern of the collected drugs with diverse types of molecules
As reported, the pharmacokinetic and pharmacodynamic characteristics of a drug are determined by its interactions with various molecules of pharmacological importance (1–3). The interactions between drugs and these molecules can be grouped into two different types: drug efficacy-related interactions that are dominating in a drug's therapeutic effects and resistant profiles (4–6) and drug ADME-related interactions that are key for drug absorption/disposition/elimination/metabolism and prodrug biosynthesis (7–13). These two types of drug-centric molecular interactions jointly formed the interaction pattern of a drug with its corresponding interacting molecules. In particular, drug efficacy-related interactions are largely shaped by the drug's target, and the ADME-related interactions are greatly affected by channel/transporter/enzyme. Moreover, with the extensive applications of drug combinations (60) and the growing concerns about the safety of drug formulation (61), drug–drug interactions (DDIs) and the biological activities of drug inactive ingredients (DIGs) have gained significantly increased attention due to their essential roles in pharmacovigilance and therapeutic management (62–66). In other words, the information of DDIs and DIGs for drugs is valuable when assessing the risk of adverse DDIs and the safety of new drug formulations (67).
Collection of the interaction data for all drugs in DrugMAP
First, the drug-centric molecular interactions were collected by a comprehensive literature review. The corresponding literature was retrieved by searching PubMed using keyword combinations, including: ‘Drug Name’ + therapeutics, ‘Drug Name’ + targets, ‘Drug Name’ + absorption, ‘Drug Name’ + distribution, ‘Drug Name’ + metabolism, ‘Drug Name’ + transporters, ‘Drug Name’ + metabolizing enzymes, ‘Drug Name’ + ADME and ‘Drug Name’ + interacting molecules.
Second, the newly identified literature was systematically validated, and reliable interactions were identified. For a drug's interactions with its targets, only the primary interactions were considered in DrugMAP as a drug's therapeutic target (DTT) by following a strict ‘target validation procedure’ well established by a previous publication (56). That ‘validation procedure’ stated that the primary target(s) should have (i) experimentally determined drug potencies (such as IC50), (ii) observed effects of drugs against disease models (cell lines, ex vivo and in vivo) linked to the primary target(s) and (iii) observed effects of knockout, knockdown, trans-genetics, RNA interference, antibody or antisense-treated in vivo models for primary targets. For a drug's interactions with its metabolizing enzymes, a protein was considered as a drug-metabolizing enzyme (DME) in DrugMAP only if its metabolized drug was experimentally confirmed by cellular/biochemical assays (68), and the DME(s) which mainly metabolize that drug was retrieved from the official site of the FDA (Drugs@FDA, https://www.accessdata.fda.gov/scripts/cder/daf/) and underlined as the ‘main DME’ in DrugMAP. Taking the anti-arthritic drug apremilast as an example, its ‘Label’ file (https://www.accessdata.fda.gov/drugsatfda_docs/label/2014/205437s000lbl.pdf) that indicates its original approval was identified from Drugs@FDA, and its main DME (CYP3A4, under these circumstances) was identified from the ‘Metabolism’ subsection of the ‘12.3 Pharmacokinetics’ section in its Label file. As a result, among all drugs with reported DME information, 60.0% of them have their ‘main DME’ reported. A total of 90.4, 7.1, 2.3 and 0.2% of these drugs with a reported ‘main DME’ had one, two, three and four main DMEs, respectively. For a drug's interactions with its transporters, a protein was considered to be a drug transporter (DTP) in DrugMAP only if its transported drug was experimentally validated using a cell line/in vivo model (7). As reported, the estimation of the contribution of transporters to total tissue uptake and excretion is necessary for understanding their importance in drug disposition (69,70). However, from the mechanistic point of view, a DTP offers complexities that are distinct from a DME (71), which makes the identification of the main DTP more difficult and complex than that of the main DME (71). In particular, such a difficulty may originate from the following: (i) different from DMEs, the concentration of a drug in a specific tissue is determined by both uptake and efflux DTPs (72); (ii) in contrast to DMEs, that are concentrated in the liver and intestine, DTPs are expressed in different tissues, and their abundance varies considerably among these tissues (73); and (iii) compared with DMEs, it is more difficult to identify probes or inhibitors for DTPs, as they are complex integral membrane proteins lacking a crystal structure (74). Therefore, although some progress has been made, there is a paucity of relevant data for the main DTP of drugs. However, during the construction of DrugMAP, a comprehensive literature review was also conducted to collect the DTPs’ kinetic data (Km) for each drug, which were considered as an indirect indication of the ‘main DTP(s)’ of drugs (75). As a result, a total of 156 drugs were reported in DrugMAP to have such kinetic data; this will also be continually updated.
Third, a total of 44 669 drug efficacy-related interactions between 32 148 drugs/drug candidates and 3095 DTTs, together with 6520 drug ADME-related interactions between 2194 drugs and 1118 DTPs and DMEs were identified and then described in DrugMAP. In total, 5067 molecules were collected, which show a certain pharmacological effect by interacting with 32 487 drugs/drug candidates, and these molecules were linked to 624 disease classes as defined by the WHO International Classification of Diseases (58).
Data of drug–drug interactions and drug inactive ingredients
In addition to the interaction data described above, information on DDIs and DIGs was also reported as essential for pharmacovigilance and therapeutic management (62–64). On the one hand, DDIs were considered as one of the key reasons for insufficient efficacy and adverse drug reactions (76–78). On the other hand, for a drug formulation, the major components by mass were not the active pharmaceutical ingredient (API) but rather the DIGs (also known as excipients). DIGs can usually reach a much higher concentration (up to 100 times in the gastrointestinal tract) than that achieved by APIs, which raised great concerns regarding their clinical toxicity and DIG–drug interactions, and were reported as critical to a drug's ADMET property and treatment outcomes (67). Herein, a systematic literature review on DDIs and DIGs was thus conducted. For example, to have comprehensive information on DIGs, a list of FDA-approved drugs was first collected from TTD (57), which led to >2000 drugs approved by the US FDA. Second, a systematic literature review on these approved drugs was performed, and a total of 23 627 formulations (for a total of 1132 drugs) that consisted of 437 DIGs were identified. Third, a variety of DIG functions in the drug formulation (that were critical to formulation design) were carefully identified and systematically recorded using a well-defined functional classification system (64). Moreover, ∼130 000 clinically validated DDIs that were induced by the co-administrations of therapeutics among 1344 drugs approved or in clinical trial were identified, and the mechanism underlying and the severity of each DDI were explicitly described in the online database. For pharmaceutical scientists, it is of great interest to distinguish the victim and perpetrator within each DDI, and the mechanistic data covered by DrugMAP were therefore very important for understanding the occurrence or avoidance of any studied DDI. The detailed statistics of these interaction patterns were shown in Figure 1.
Description and visualization of the drug's interaction pattern
It is known that the efficacy and safety of a drug are largely determined by its multidimensional interactions with diverse types of molecules, which collectively constitute its interaction pattern (79). In other words, only by considering the interaction patterns in a holistic and comprehensive manner (rather than focusing only on parts of it), can we fully comprehend the principles of how a drug works or fails (80). In DrugMAP, the interaction pattern for each drug was thus described and visualized. On the one hand, all interacting molecules of a drug were explicitly described in a tabular form online, and all corresponding data could be readily accessed and downloaded. On the other hand, a drug-centric graph (shown in Figure 2A), which illustrated its interaction pattern with multiple ‘landmarks’ marking all interacting molecules, was drawn as a vivid visualization. Such an explicit description and vivid visualization were provided online in DrugMAP to clarify the sophisticated interaction pattern of each drug.
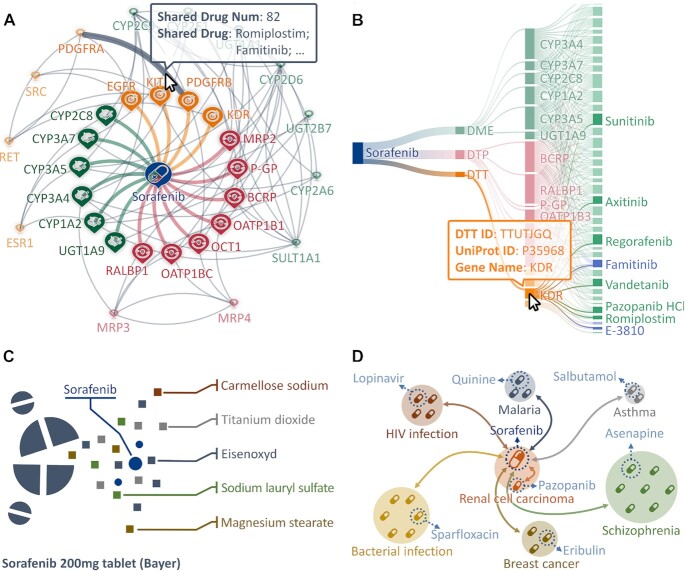
Figure 2.
Description and visualization of sorafenib in DrugMAP. (A) The interactable interaction graph showing the interactions between the drug and the molecules of pharmacological importance. (B) Sankey diagram illustrating a list of drugs interacting with one of the interacting molecules of sorafenib. (C) The schematic representation of the DIGs for sorafenib 200 mg tablet formulation; the corresponding data were represented in tabular format in the ‘Drug Inactive Ingredients and Formulations of This Drug’ part of sorafenib's drug page. (D) The schematic representation of the DDI data of sorafenib in the same indication (renal cell carcinoma) and comorbidities (HIV, malaria, asthma, etc.); the corresponding information is also represented in tabular format in the above-mentioned section of sorafenib's drug page.
Taking sorafenib as an example, Figure 2A illustrates a network diagram of its interaction pattern, and all interacting molecules were categorized using different colors (DTTs, DTPs and DMEs were indicated in orange, red and green, respectively). As illustrated, the inner nodes scattered around the drug represented these molecules interacting directly with sorafenib, while those outer nodes indicated the interacting molecules of other drug(s) that also interacted with the molecules in the inner nodes. The number of drugs interacting with two molecules can be viewed by hovering the mouse over the gray line between them, and an exemplar drug is simultaneously provided to give referencing information. Moreover, to gain explicit knowledge of the molecules interacting with multiple drugs, an additional diagram was drawn online in our database. As illustrated in Figure 2B, a comprehensive list of drugs interacting with one of those interacting molecules of sorafenib was provided on the right side of the Sankey diagram, and the detailed information of both nodes and edges could be viewed via hovering the mouse over them. Last, but not least, the information on DIGs and DDIs of sorafenib was also provided in those sections of ‘Drug Inactive Ingredients and Formulations of This Drug’ and ‘Drug–Drug Interaction (DDI) Information of This Drug’ on the ‘Drug’ page sorafenib. As provided, the schematic representations of the relationship between drugs and their corresponding DIGs (Figure 2C) and DDIs (Figure 2D) were fully described. Such relationships were systematically presented in the online database in a tabular format.
The expression pattern of drug-interacting molecules among different organs
The interacting molecules of drugs were frequently reported to be differentially expressed among disease sites, ADME-related organs/tissues and other physiological sites, which can significantly affect the efficacy and safety of drug (15,28). First, the differential expression of drug-interacting molecules among different disease sites is critical for the ADMET properties and potential failure of new chemical entities in clinical trials (81). Second, the differential expression of molecules among various ADME-related organs/tissues is key for explaining the interindividual variability in pharmacokinetics, understanding the mechanisms underlying DDIs and predicting the hepatic clearance of drugs (22,71). Third, the differential expression of the interacting molecules among other physiological sites is also vital for maintaining the delicate balance between a drug’s efficacy and safety (82). Thus, a precise and systematic description of the molecular atlas of the regulation mechanism for a drug calls for an explicit elucidation of the differential expression pattern of its interacting molecules, which has been systematically described in DrugMAP.
Collection of the expression data for drug-interacting molecules
The differential expression data of drug-interacting molecules among disease sites were collected using the following process. First, a total of 5534 series records of human gene expression raw data based on the Affymetrix HGU133 Plus 2.0 platform were collected from the GEO database (83), and the corresponding disease and tissue distribution were identified from the sample annotations. By comparing with the disease indications of drugs, 436 records were finally selected to illustrate the expression atlas of all drug-interacting molecules. For the series records of small sample size, multiple records of the same disease and same tissue were integrated to address the issue of low statistical power (84). To enable data integration, a well-established strategy was applied (85), which has been widely adopted by recent studies (86–89). Second, these newly collected records were further pre-processed using a standardized procedure of normalization, log transformation, data integration, perfect match correction, quantization, powerful multi-array averaging, median polishing, and so on (90–93). Third, the differential expression between distinct groups of samples (disease tissues of patients versus normal tissues of healthy people) were assessed by calculating the fold change, Z-score and P-value of the Student t-test (92,94). Finally, the interactable box plot depicting the differential expression pattern of the interacting molecules under the circumstances of a given disease among sample groups was drawn based on the Apache EChart visualization library.
Moreover, the differential expression data of drug-interacting molecules among various ADME-related organs/tissues and other physiological sites were collected by the following process. First, a benchmark dataset providing gene expression data among human ADME-related organs/tissues and other physiological sites was collected (95). Second, the intensity of the interacting molecules was processed by following the same pre-processing procedure as that described in previous publications (90–93). Finally, the expression atlas of interacting molecules among ADME-related tissues or physiological/life-essential organs was provided based on the Apache EChart visualization library and presented in DrugMAP as a number of interactable bar plots.
As a result, the expression patterns of 1539 molecules among 108 disease sites of 21 781 patients, 1323 molecules among four typical ADME-related organs (liver, colon, kidney and small intestine) of 236 healthy individuals and 1286 molecules among 32 physiological organs (prostate, breast, etc.) of 109 samples were systematically collected and then provided in the DrugMAP database. All these molecules were described in this database as interacting with at least one drug.
Visualization of the expression pattern of drug-interacting molecules
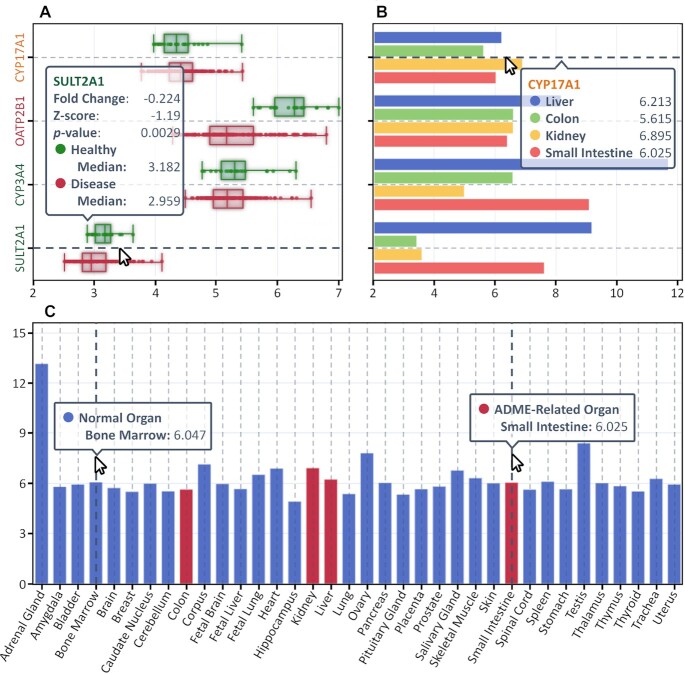
The differential expression data of drug-interacting molecules were visualized in an intuitive and interactable way, which could be freely downloaded from DrugMAP. Taking abiraterone acetate as an example, it is an approved drug for prostate cancer by targeting CYP17A1, and it is reported to be transported by OATP2B1 and metabolized by CYP3A4 and SULT2A1 after administration. As shown in Figure 3A, a box plot illustrating the differential expression of these four molecules between the prostate tissue of patients and that of healthy individuals was provided in DrugMAP, and the detailed data of fold change, Z-score and P-value could be viewed by hovering the mouse over the boxes. Moreover, the differential expression of these interacting molecules among four ADME-related organs is described in Figure 3B, which provides the detailed statistics of the relative expression intensities. To illustrate the differential expression of these molecules in other physiological sites, an additional bar chart was also drawn online. As demonstrated in Figure 3C, the expression intensities of CYP17A1 among 32 physiological/life-essential (colored in blue) and four ADME-related (colored in red) organs are quantitatively provided in DrugMAP.
Figure 3.
Differential expression data of abiraterone acetate-interacting molecules in DrugMAP. (A) A box plot illustrating the differential expression of four molecules between the prostate tissue of patients and that of healthy individuals. (B) A bar chart showing the differential expression of four interacting molecules among different ADME-related organs (liver, colon, kidney and small intestine). (C) A bar chart showing the expression intensity of CYP17A1 among 32 physiological (colored in blue) and four ADME-related (colored in red) organs.
Data standardization, access, retrieval and the similarity-based search tools
To facilitate the access and use of DrugMAP data by users, all collected data were systematically cleaned up and then standardized, which included (i) disease standardization using the latest version of the WHO International Classification of Diseases (ICD-11); (ii) standardization of ADMET properties through literature review and FDA labeling; (iii) the structure standardization of 26 286 drugs (∼81.0% of all drugs in DrugMAP) to SDF format (both 2D and 3D); (iv) the extension of the structure coverage of 4858 proteins (∼95.9% of all interacting molecules in DrugMAP) using both literature reviews and AlphaFold predictions (96); and (v) the extended pharma-information of drugs and their interacting molecules by cross-linking to other existing databases [such as CAS Registry Number (97), Ensembl (98), ClinicalTrial.gov (99), Drugs@FDA (100), DrugBank (47), ChEBI (101), InChl (102), INTEDE (30), KEGG (103), NCBI Gene-Taxonomy (104,105), PDB (106), PubChem (59), TTD (48), VARIDT (50) and UniProt (107)].
Tools for data search based on structure and sequence similarity
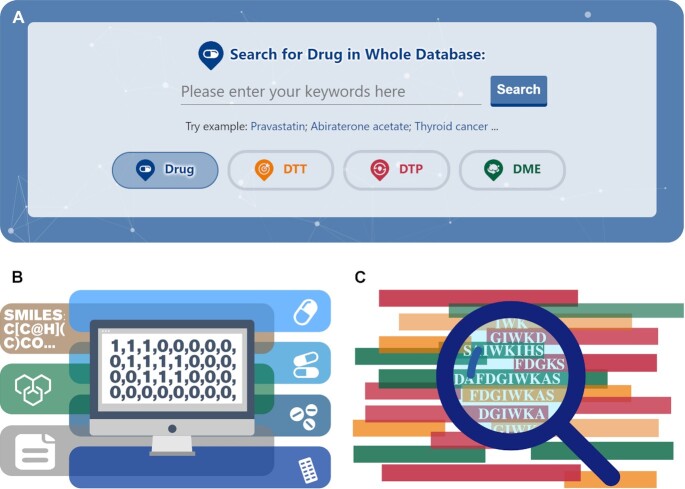
To facilitate an accurate and convenient data search in DrugMAP, diverse retrieval methods were implemented. As shown in Figure 4A, a search engine using keywords was constructed to enable the whole database search of drugs, DTTs, DTPs and DMEs. Users can click on the different buttons at the bottom of the search box to switch among different search objects (drug, DTT, DTP and DME). Moreover, another search engine based on the drug structure similarity was developed to identify drugs of high structural similarity with the studied structure. As illustrated in Figure 4B, users can upload their studied drug structures by inputting the corresponding SMILES strings, manually drawn drug structures or SDF/MOL structural files, and a list of drugs with similar structures to those input will be calculated and provided online. Meanwhile, a search engine based on protein sequence similarity was also developed to identify the proteins of high sequence similarity with those studied. As shown in Figure 4C, users can upload a protein sequence of interest, and a list of proteins of pharmacological importance similar to that input will be predicted and described in DrugMAP.
Figure 4.
Diverse search engines implemented in DrugMAP. (A) A search engine using keywords enabling the whole database search of drugs, drug targets, drug transporters and drug-metabolizing enzymes; (B) another search engine based on drug structural similarity to discover drugs of high structural similarity to the studied chemical structure; (C) another search engine based on the protein sequence similarity to identify proteins of high sequence similarity to those studied.
CONCLUSION AND PERSPECTIVES
DrugMAP provides comprehensive interaction data for each drug and quantitatively shows the differential expression data of all of a drug's interacting molecules, which makes this new database an important supplement to existing databases. With the rapid adoption of AI in drug discovery, it is found that a comprehensive ‘interacting network’ including a large number of drugs and their interacting molecules is highly favored when using AI methods (108–114). For example, some tools were constructed to predict drug–target interaction based on the network of heterogeneous drug-centered interactions (109), and other tools were made available to learn the topology-preserving representation of drugs and targets based on the heterogeneous network data of drug–target, drug–drug and target–target interaction (110). Because of these emerging demands on such interaction-based big data, DrugMAP made the first endeavor to weave a comprehensive network containing >200 000 interactions among >30 000 drugs/drug candidates and >5000 molecules of pharmacological importance. Such a drug-centric ‘interacting network’ for each drug can be freely viewed online and fully downloaded by all users in the popular format of Cytoscape (115), which is expected to have great implications for drug repurposing (57,116), target discovery (117–119) and drug development (120–122). Moreover, an overall interacting network including all interactions is downloadable from DrugMAP to facilitate network analyses (123–126).
To facilitate modern drug development, it is essential to have the explicit pharma-information for not only the drugs but also the molecules of pharmacological importance (127). Therefore, various data were implemented into DrugMAP to significantly enrich such valuable pharma-information, which is expected to be extremely useful for those non-bioinformaticians such as clinicians. In this database, the newly implemented pharma-information for each drug included its chemical identifiers (such as formula, canonical SMILES, InChI and InChIKey), the characteristics of the Lipinski's Rule of Five (i.e. molecular weight, topological polar surface area, rotatable bonds, hydrogen bond donors and hydrogen bond acceptors), the ADME properties (such as volume of distribution, bioavailability, metabolism, clearance, half-life and elimination) and drug structures in 2D and 3D formats. Furthermore, additional information on the molecules of pharmacological importance was implemented into DrugMAP, which were also valuable data, especially for target discovery and ADME-related analysis. Such information included the biochemical classes (such as kinases, GPCRs and transcription factors), physiological or pathological functions, affiliated signaling pathways, endogenous substrates associated with enzyme/channel/transporter, protein structures either resolved by experiments or predicted using AlphaFold, clinically validated DDIs, detailed information on DIGs, etc. To facilitate users’ quick utilization of DrugMAP, a systematic user manual with exemplar demonstrations illustrated by webpage screenshots is also provided in the latest version of DrugMAP.
All in all, with the increasing efforts made to elucidate the complex mechanisms underlying drug pharmacokinetics and pharmacodynamics (1–3) and the rapidly emerging interest in AI-based network studies, the volume and requirement for the interacting data of drugs are both constantly booming (18). As those data collected by DrugMAP are only representative of what is happening at the current time point and is not fully indicative of future trends, this database will be continuously updated using the latest data to catch up with the frontiers of modern drug research. This database is now freely accessible by all users at: https://idrblab.org/drugmap/.
Contributor Information
Fengcheng Li, College of Pharmaceutical Sciences, The Second Affiliated Hospital, Zhejiang University School of Medicine, Zhejiang University, Hangzhou 310058, China; Innovation Institute for Artificial Intelligence in Medicine of Zhejiang University, Alibaba–Zhejiang University Joint Research Center of Future Digital Healthcare, Hangzhou 330110, China.
Jiayi Yin, College of Pharmaceutical Sciences, The Second Affiliated Hospital, Zhejiang University School of Medicine, Zhejiang University, Hangzhou 310058, China.
Mingkun Lu, College of Pharmaceutical Sciences, The Second Affiliated Hospital, Zhejiang University School of Medicine, Zhejiang University, Hangzhou 310058, China.
Minjie Mou, College of Pharmaceutical Sciences, The Second Affiliated Hospital, Zhejiang University School of Medicine, Zhejiang University, Hangzhou 310058, China.
Zhaorong Li, Innovation Institute for Artificial Intelligence in Medicine of Zhejiang University, Alibaba–Zhejiang University Joint Research Center of Future Digital Healthcare, Hangzhou 330110, China.
Zhenyu Zeng, Innovation Institute for Artificial Intelligence in Medicine of Zhejiang University, Alibaba–Zhejiang University Joint Research Center of Future Digital Healthcare, Hangzhou 330110, China.
Ying Tan, State Key Laboratory of Chemical Oncogenomics, Key Laboratory of Chemical Biology, Tsinghua Shenzhen International Graduate School, Tsinghua University, Shenzhen 518055, China.
Shanshan Wang, Qian Xuesen Collaborative Research Center of Astrochemistry and Space Life Sciences, Institute of Drug Discovery Technology, Ningbo University, Ningbo 315211, China.
Xinyi Chu, Qian Xuesen Collaborative Research Center of Astrochemistry and Space Life Sciences, Institute of Drug Discovery Technology, Ningbo University, Ningbo 315211, China.
Haibin Dai, College of Pharmaceutical Sciences, The Second Affiliated Hospital, Zhejiang University School of Medicine, Zhejiang University, Hangzhou 310058, China.
Tingjun Hou, College of Pharmaceutical Sciences, The Second Affiliated Hospital, Zhejiang University School of Medicine, Zhejiang University, Hangzhou 310058, China.
Su Zeng, College of Pharmaceutical Sciences, The Second Affiliated Hospital, Zhejiang University School of Medicine, Zhejiang University, Hangzhou 310058, China.
Yuzong Chen, State Key Laboratory of Chemical Oncogenomics, Key Laboratory of Chemical Biology, Tsinghua Shenzhen International Graduate School, Tsinghua University, Shenzhen 518055, China; Qian Xuesen Collaborative Research Center of Astrochemistry and Space Life Sciences, Institute of Drug Discovery Technology, Ningbo University, Ningbo 315211, China.
Feng Zhu, College of Pharmaceutical Sciences, The Second Affiliated Hospital, Zhejiang University School of Medicine, Zhejiang University, Hangzhou 310058, China; Innovation Institute for Artificial Intelligence in Medicine of Zhejiang University, Alibaba–Zhejiang University Joint Research Center of Future Digital Healthcare, Hangzhou 330110, China.
FUNDING
Funded by the Natural Science Foundation of Zhejiang Province [LR21H300001]; National Natural Science Foundation of China [81872798 and U1909208]; Leading Talent of the ‘Ten Thousand Plan’—National High-Level Talents Special Support Plan of China; Fundamental Research Fund of Central University [2018QNA7023]; Key R&D Program of Zhejiang Province [2020C03010]; National Key R&D Program of China Synthetic Biology Research [2019YFA0905900]; ‘Double Top-Class’ University [181201*194232101]; Space Exploration Breeding Grant of Qian Xuesen Lab [TKTSPY-2020-04-03]; Scientific Research Grant of Ningbo University [215–432000282]; Ningbo Top Talent Proj [215–432094250]; Shenzhen Municipal Government grant [NO.2019156, JCYJ20170413113448742, NO.201901]; Department of Science & Technology of Guangdong Province [2017B030314083]; Westlake Laboratory (Westlake Laboratory of Life Sciences and Biomedicine); Alibaba–Zhejiang University Joint Research Center of Future Digital Healthcare; Alibaba Cloud; and Information Technology Center of Zhejiang University.
Conflict of interest statement. None declared.
REFERENCES
- 1. Santos M., Niemi M., Hiratsuka M., Kumondai M., Ingelman-Sundberg M., Lauschke V.M., Rodriguez-Antona C.. Novel copy-number variations in pharmacogenes contribute to interindividual differences in drug pharmacokinetics. Genet. Med. 2018; 20:622–629. [DOI] [PubMed] [Google Scholar]
- 2. Roden D.M., McLeod H.L., Relling M.V., Williams M.S., Mensah G.A., Peterson J.F., Van Driest S.L.. Pharmacogenomics. Lancet. 2019; 394:521–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fuhr U., Hsin C.H., Li X., Jabrane W., Sorgel F.. Assessment of pharmacokinetic drug–drug interactions in humans: in vivo probe substrates for drug metabolism and drug transport revisited. Annu. Rev. Pharmacol. Toxicol. 2019; 59:507–536. [DOI] [PubMed] [Google Scholar]
- 4. Haley B., Roudnicky F.. Functional genomics for cancer drug target discovery. Cancer Cell. 2020; 38:31–43. [DOI] [PubMed] [Google Scholar]
- 5. Passirani C., Vessieres A., La Regina G., Link W., Silvestri R.. Modulating undruggable targets to overcome cancer therapy resistance. Drug Resist. Updat. 2022; 60:100788. [DOI] [PubMed] [Google Scholar]
- 6. Yu A.M., Choi Y.H., Tu M.J.. RNA drugs and RNA targets for small molecules: principles, progress, and challenges. Pharmacol. Rev. 2020; 72:862–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nigam S.K. What do drug transporters really do?. Nat. Rev. Drug Discov. 2015; 14:29–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhou F.F., Zhu L., Wang K., Murray M.. Recent advance in the pharmacogenomics of human solute carrier transporters (SLCs) in drug disposition. Adv. Drug. Deliv. Rev. 2017; 116:21–36. [DOI] [PubMed] [Google Scholar]
- 9. Ning B., Yu D., Yu A.M. Advances and challenges in studying noncoding RNA regulation of drug metabolism and development of RNA therapeutics. Biochem. Pharmacol. 2019; 169:113638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dalvie D., Di L.. Aldehyde oxidase and its role as a drug metabolizing enzyme. Pharmacol. Ther. 2019; 201:137–180. [DOI] [PubMed] [Google Scholar]
- 11. Lai Y., Chu X., Di L., Gao W., Guo Y., Liu X., Lu C., Mao J., Shen H., Tang H.et al.. Recent advances in the translation of drug metabolism and pharmacokinetics science for drug discovery and development. Acta Pharm. Sin. B. 2022; 12:2751–2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kovalchuk N., Zhang Q.Y., Van Winkle L., Ding X.X.. Contribution of pulmonary CYP-mediated bioactivation of naphthalene to airway epithelial injury in the lung. Toxicol. Sci. 2020; 177:334–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ding L., Li L., Liu S., Bao X., Dickman K.G., Sell S.S., Mei C., Zhang Q.Y., Gu J., Ding X.X.. Proximal tubular vacuolization and hypersensitivity to drug-induced nephrotoxicity in male mice with decreased expression of the NADPH-cytochrome P450 reductase. Toxicol. Sci. 2020; 173:362–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Neul C., Schaeffeler E., Sparreboom A., Laufer S., Schwab M., Nies A.T.. Impact of membrane drug transporters on resistance to small-molecule tyrosine kinase inhibitors. Trends Pharmacol. Sci. 2016; 37:904–932. [DOI] [PubMed] [Google Scholar]
- 15. Li X., Tian Y., Tu M.J., Ho P.Y., Batra N., Yu A.M.. Bioengineered miR-27b-3p and miR-328-3p modulate drug metabolism and disposition via the regulation of target ADME gene expression. Acta Pharm. Sin. B. 2019; 9:639–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Peng J., Wang Y., Guan J., Li J., Han R., Hao J., Wei Z., Shang X.. An end-to-end heterogeneous graph representation learning-based framework for drug–target interaction prediction. Brief. Bioinform. 2021; 22:bbaa430. [DOI] [PubMed] [Google Scholar]
- 17. Perez D.R., Sklar L.A., Chigaev A., Matlawska-Wasowska K.. Drug repurposing for targeting cyclic nucleotide transporters in acute leukemias—a missed opportunity. Semin. Cancer Biol. 2021; 68:199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang S., Amahong K., Zhang C., Li F., Gao J., Qiu Y., Zhu F.. RNA–RNA interactions between SARS-CoV-2 and host benefit viral development and evolution during COVID-19 infection. Brief. Bioinform. 2021; 23:bbab397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huo X., Meng Q., Wang C., Wu J., Wang C., Zhu Y., Ma X., Sun H., Liu K.X.. Protective effect of cilastatin against diclofenac-induced nephrotoxicity through interaction with diclofenac acyl glucuronide via organic anion transporters. Br. J. Pharmacol. 2020; 177:1933–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu Y., Zheng X., Yu Q., Wang H., Tan F., Zhu Q., Yuan L., Jiang H., Yu L., Zeng S.. Epigenetic activation of the drug transporter OCT2 sensitizes renal cell carcinoma to oxaliplatin. Sci. Transl. Med. 2016; 8:348ra397. [DOI] [PubMed] [Google Scholar]
- 21. Maeda K., Sugiyama Y.. Transporter biology in drug approval: regulatory aspects. Mol. Aspects Med. 2013; 34:711–718. [DOI] [PubMed] [Google Scholar]
- 22. Schlessinger A., Welch M.A., van Vlijmen H., Korzekwa K., Swaan P.W., Matsson P.. Molecular modeling of drug–transporter interactions—an international transporter consortium perspective. Clin. Pharmacol. Ther. 2018; 104:818–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xue W., Fu T., Deng S., Yang F., Yang J., Zhu F.. Molecular mechanism for the allosteric inhibition of the human serotonin transporter by antidepressant escitalopram. ACS Chem. Neurosci. 2022; 13:340–351. [DOI] [PubMed] [Google Scholar]
- 24. Fang H., De Wolf H., Knezevic B., Burnham K.L., Osgood J., Sanniti A., Lledo Lara A., Kasela S., De Cesco S., Wegner J.K.et al.. A genetics-led approach defines the drug target landscape of 30 immune-related traits. Nat. Genet. 2019; 51:1082–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bicker J., Alves G., Falcao A., Fortuna A.. Timing in drug absorption and disposition: the past, present, and future of chronopharmacokinetics. Br. J. Pharmacol. 2020; 177:2215–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yalcin-Ozkat G. Molecular modeling strategies of cancer multidrug resistance. Drug Resist. Updat. 2021; 59:100789. [DOI] [PubMed] [Google Scholar]
- 27. Mazerska Z., Mroz A., Pawlowska M., Augustin E.. The role of glucuronidation in drug resistance. Pharmacol. Ther. 2016; 159:35–55. [DOI] [PubMed] [Google Scholar]
- 28. Foretz M., Guigas B., Viollet B.. Understanding the glucoregulatory mechanisms of metformin in type 2 diabetes mellitus. Nat. Rev. Endocrinol. 2019; 15:569–589. [DOI] [PubMed] [Google Scholar]
- 29. Mao Q., Lai Y., Wang J.. Drug transporters in xenobiotic disposition and pharmacokinetic prediction. Drug Metab. Dispos. 2018; 46:561–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yin J., Li F., Zhou Y., Mou M., Lu Y., Chen K., Xue J., Luo Y., Fu J., He X.et al.. INTEDE: interactome of drug-metabolizing enzymes. Nucleic Acids Res. 2021; 49:D1233–D1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Guney E., Menche J., Vidal M., Barabasi A.L.. Network-based in silico drug efficacy screening. Nat. Commun. 2016; 7:10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Carona A., Bicker J., Silva R., Fonseca C., Falcao A., Fortuna A.. Pharmacology of lacosamide: from its molecular mechanisms and pharmacokinetics to future therapeutic applications. Life Sci. 2021; 275:119342. [DOI] [PubMed] [Google Scholar]
- 33. Leandro K., Bicker J., Alves G., Falcao A., Fortuna A.. ABC transporters in drug-resistant epilepsy: mechanisms of upregulation and therapeutic approaches. Pharmacol. Res. 2019; 144:357–376. [DOI] [PubMed] [Google Scholar]
- 34. Ali Y., Shams T., Wang K., Cheng Z., Li Y., Shu W., Bao X., Zhu L., Murray M., Zhou F.F.. The involvement of human organic anion transporting polypeptides (OATPs) in drug–herb/food interactions. Chin. Med. 2020; 15:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhu H. Big data and artificial intelligence modeling for drug discovery. Annu. Rev. Pharmacol. Toxicol. 2020; 60:573–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tang J., Fu J., Wang Y., Li B., Li Y., Yang Q., Cui X., Hong J., Li X., Chen Y.et al.. ANPELA: analysis and performance assessment of the label-free quantification workflow for metaproteomic studies. Brief. Bioinform. 2020; 21:621–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mendez-Lucio O., Baillif B., Clevert D.A., Rouquie D., Wichard J.. De novo generation of hit-like molecules from gene expression signatures using artificial intelligence. Nat. Commun. 2020; 11:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hu D.G., Mackenzie P.I., Nair P.C., McKinnon R.A., Meech R.. The expression profiles of ADME genes in human cancers and their associations with clinical outcomes. Cancers. 2020; 12:3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cheng F., Kovacs I.A., Barabasi A.L.. Network-based prediction of drug combinations. Nat. Commun. 2019; 10:1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu K., Ding R.F., Xu H., Qin Y.M., He Q.S., Du F., Zhang Y., Yao L.X., You P., Xiang Y.P.et al.. Broad-spectrum profiling of drug safety via learning complex network. Clin. Pharmacol. Ther. 2020; 107:1373–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shaked I., Oberhardt M.A., Atias N., Sharan R., Ruppin E.. Metabolic network prediction of drug side effects. Cell Syst. 2016; 2:209–213. [DOI] [PubMed] [Google Scholar]
- 42. Ali Y., Shams T., Cheng Z., Li Y., Chun C.S., Shu W., Bao X., Zhu L., Murray M., Zhou F.F.. Impaired transport activity of human organic anion transporters (OATs) and organic anion transporting polypeptides (OATPs) by wnt inhibitors. J. Pharm. Sci. 2021; 110:914–924. [DOI] [PubMed] [Google Scholar]
- 43. Szklarczyk D., Santos A., von Mering C., Jensen L.J., Bork P., Kuhn M.. STITCH 5: augmenting protein–chemical interaction networks with tissue and affinity data. Nucleic Acids Res. 2016; 44:D380–D384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Freshour S.L., Kiwala S., Cotto K.C., Coffman A.C., McMichael J.F., Song J.J., Griffith M., Griffith O.L., Wagner A.H.. Integration of the drug–gene interaction database (DGIdb 4.0) with open crowdsource efforts. Nucleic Acids Res. 2021; 49:D1144–D1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gilson M.K., Liu T., Baitaluk M., Nicola G., Hwang L., Chong J.. BindingDB in 2015: a public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res. 2016; 44:D1045–D1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mendez D., Gaulton A., Bento A.P., Chambers J., De Veij M., Felix E., Magarinos M.P., Mosquera J.F., Mutowo P., Nowotka M.et al.. ChEMBL: towards direct deposition of bioassay data. Nucleic Acids Res. 2019; 47:D930–D940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Sajed T., Johnson D., Li C., Sayeeda Z.et al.. DrugBank 5.0: a major update to the drugbank database for 2018. Nucleic Acids Res. 2018; 46:D1074–D1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhou Y., Zhang Y., Lian X., Li F., Wang C., Zhu F., Qiu Y., Chen Y.. Therapeutic target database update 2022: facilitating drug discovery with enriched comparative data of targeted agents. Nucleic Acids Res. 2022; 50:D1398–D1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Morrissey K.M., Wen C.C., Johns S.J., Zhang L., Huang S.M., Giacomini K.M.. The UCSF-FDA transportal: a public drug transporter database. Clin. Pharmacol. Ther. 2012; 92:545–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fu T., Li F., Zhang Y., Yin J., Qiu W., Li X., Liu X., Xin W., Wang C., Yu L.et al.. VARIDT 2.0: structural variability of drug transporter. Nucleic Acids Res. 2022; 50:D1417–D1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chang A., Jeske L., Ulbrich S., Hofmann J., Koblitz J., Schomburg I., Neumann-Schaal M., Jahn D., Schomburg D. BRENDA, the ELIXIR core data resource in 2021: new developments and updates. Nucleic Acids Res. (2021); 49:D498–D508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hu R., Xu H., Jia P., Zhao Z.. KinaseMD: kinase mutations and drug response database. Nucleic Acids Res. 2021; 49:D552–D561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Huang L.H., He Q.S., Liu K., Cheng J., Zhong M.D., Chen L.S., Yao L.X., Ji Z.L.. ADReCS-Target: target profiles for aiding drug safety research and application. Nucleic Acids Res. 2018; 46:D911–D917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. IJzerman A.P., Guo D. Drug–target association kinetics in drug discovery. Trends Biochem. Sci. 2019; 44:861–871. [DOI] [PubMed] [Google Scholar]
- 55. Ye Q., Hsieh C.Y., Yang Z., Kang Y., Chen J., Cao D., He S., Hou T.. A unified drug–target interaction prediction framework based on knowledge graph and recommendation system. Nat. Commun. 2021; 12:6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhu F., Shi Z., Qin C., Tao L., Liu X., Xu F., Zhang L., Song Y., Liu X., Zhang J.et al.. Therapeutic target database update 2012: a resource for facilitating target-oriented drug discovery. Nucleic Acids Res. 2012; 40:D1128–D1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wang Y., Zhang S., Li F., Zhou Y., Zhang Y., Wang Z., Zhang R., Zhu J., Ren Y., Tan Y.et al.. Therapeutic target database 2020: enriched resource for facilitating research and early development of targeted therapeutics. Nucleic Acids Res. 2020; 48:D1031–D1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. The Lancet ICD-11. Lancet. 2019; 393:2275. [DOI] [PubMed] [Google Scholar]
- 59. Kim S., Chen J., Cheng T., Gindulyte A., He J., He S., Li Q., Shoemaker B.A., Thiessen P.A., Yu B.et al.. PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res. 2021; 49:D1388–D1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Onder G., Marengoni A.. Polypharmacy. JAMA. 2017; 318:1728. [DOI] [PubMed] [Google Scholar]
- 61. Pottel J., Armstrong D., Zou L., Fekete A., Huang X.P., Torosyan H., Bednarczyk D., Whitebread S., Bhhatarai B., Liang G.et al.. The activities of drug inactive ingredients on biological targets. Science. 2020; 369:403–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Niu J., Straubinger R.M., Mager D.E.. Pharmacodynamic drug–drug interactions. Clin. Pharmacol. Ther. 2019; 105:1395–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yin J., Sun W., Li F., Hong J., Li X., Zhou Y., Lu Y., Liu M., Zhang X., Chen N.et al.. VARIDT 1.0: variability of drug transporter database. Nucleic Acids Res. 2020; 48:D1042–D1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhang C., Mou M., Zhou Y., Zhang W., Lian X., Shi S., Lu M., Sun H., Li F., Wang Y.et al.. Biological activities of drug inactive ingredients. Brief. Bioinform. 2022; 23:bbac160. [DOI] [PubMed] [Google Scholar]
- 65. Huo X., Meng Q., Wang C., Zhu Y., Liu Z., Ma X., Ma X., Peng J., Sun H., Liu K.X.. Cilastatin protects against imipenem-induced nephrotoxicity via inhibition of renal organic anion transporters (OATs). Acta Pharm. Sin. B. 2019; 9:986–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Li Y.H., Li X.X., Hong J.J., Wang Y.X., Fu J.B., Yang H., Yu C.Y., Li F.C., Hu J., Xue W.W.et al.. Clinical trials, progression-speed differentiating features and swiftness rule of the innovative targets of first-in-class drugs. Brief. Bioinform. 2020; 21:649–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Reker D., Shi Y., Kirtane A.R., Hess K., Zhong G.J., Crane E., Lin C.H., Langer R., Traverso G.. Machine learning uncovers food– and excipient–drug interactions. Cell Rep. 2020; 30:3710–3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hitchings R., Kelly L.. Drug metabolism as a community effort. Cell Metab. 2019; 30:235–237. [DOI] [PubMed] [Google Scholar]
- 69. Cantrill C., Houston J.B.. Understanding the interplay between uptake and efflux transporters within in vitro systems in defining hepatocellular drug concentrations. J. Pharm. Sci. 2017; 106:2815–2825. [DOI] [PubMed] [Google Scholar]
- 70. Storelli F., Yin M., Kumar A.R., Ladumor M.K., Evers R., Chothe P.P., Enogieru O.J., Liang X., Lai Y., Unadkat J.D.. The next frontier in ADME science: predicting transporter-based drug disposition, tissue concentrations and drug–drug interactions in humans. Pharmacol. Ther. 2022; 24:108271. [DOI] [PubMed] [Google Scholar]
- 71. Giacomini K.M., Huang S.M., Tweedie D.J., Benet L.Z., Brouwer K.L., Chu X., Dahlin A., Evers R., Fischer V., Hillgren K.M.et al.. Membrane transporters in drug development. Nat. Rev. Drug Discov. 2010; 9:215–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zhang D., Hop C., Patilea-Vrana G., Gampa G., Seneviratne H.K., Unadkat J.D., Kenny J.R., Nagapudi K., Di L., Zhou L.et al.. Drug concentration asymmetry in tissues and plasma for small molecule-related therapeutic modalities. Drug Metab. Dispos. 2019; 47:1122–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Mandal A., Agrahari V., Khurana V., Pal D., Mitra A.K.. Transporter effects on cell permeability in drug delivery. Expert Opin. Drug Deliv. 2017; 14:385–401. [DOI] [PubMed] [Google Scholar]
- 74. Cesar-Razquin A., Snijder B., Frappier-Brinton T., Isserlin R., Gyimesi G., Bai X., Reithmeier R.A., Hepworth D., Hediger M.A., Edwards A.M.et al.. A call for systematic research on solute carriers. Cell. 2015; 162:478–487. [DOI] [PubMed] [Google Scholar]
- 75. Severance A.C., Sandoval P.J., Wright S.H.. Correlation between apparent substrate affinity and OCT2 transport turnover. J. Pharmacol. Exp. Ther. 2017; 362:405–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zhu Y., Huo X., Wang C., Meng Q., Liu Z., Sun H., Tan A., Ma X., Peng J., Liu K.X.. Organic anion transporters also mediate the drug–drug interaction between imipenem and cilastatin. Asian J. Pharm. Sci. 2020; 15:252–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Tornio A., Filppula A.M., Niemi M., Backman J.T.. Clinical studies on drug–drug interactions involving metabolism and transport: methodology, pitfalls, and interpretation. Clin. Pharmacol. Ther. 2019; 105:1345–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Quinney S.K. Opportunities and challenges of using big data to detect drug–drug interaction risk. Clin. Pharmacol. Ther. 2019; 106:72–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Casas A.I., Hassan A.A., Larsen S.J., Gomez-Rangel V., Elbatreek M., Kleikers P.W.M., Guney E., Egea J., Lopez M.G., Baumbach J.et al.. From single drug targets to synergistic network pharmacology in ischemic stroke. Proc. Natl Acad. Sci. USA. 2019; 116:7129–7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Muzio G., O’Bray L., Borgwardt K.. Biological network analysis with deep learning. Brief. Bioinform. 2021; 22:1515–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Foti R.S., Tyndale R.F., Garcia K.L., Sweet D.H., Nagar S., Sharan S., Rock D.A.. Target-site drug metabolism and transport. Drug Metab. Dispos. 2015; 43:1156–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Nixon M., Mackenzie S.D., Taylor A.I., Homer N.Z., Livingstone D.E., Mouras R., Morgan R.A., Mole D.J., Stimson R.H., Reynolds R.M.et al.. ABCC1 confers tissue-specific sensitivity to cortisol versus corticosterone: a rationale for safer glucocorticoid replacement therapy. Sci. Transl. Med. 2016; 8:352ra109. [DOI] [PubMed] [Google Scholar]
- 83. Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M.et al.. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res. 2013; 41:D991–D995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Xiao F., Luo X., Hao N., Niu Y.S., Xiao X., Cai G., Amos C.I., Zhang H.. An accurate and powerful method for copy number variation detection. Bioinformatics. 2019; 35:2891–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K.. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015; 43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Seoane J.A., Kirkland J.G., Caswell-Jin J.L., Crabtree G.R., Curtis C.. Chromatin regulators mediate anthracycline sensitivity in breast cancer. Nat. Med. 2019; 25:1721–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Yang Q., Li B., Tang J., Cui X., Wang Y., Li X., Hu J., Chen Y., Xue W., Lou Y.et al.. Consistent gene signature of schizophrenia identified by a novel feature selection strategy from comprehensive sets of transcriptomic data. Brief. Bioinform. 2020; 21:1058–1068. [DOI] [PubMed] [Google Scholar]
- 88. Khaliq A.M., Erdogan C., Kurt Z., Turgut S.S., Grunvald M.W., Rand T., Khare S., Borgia J.A., Hayden D.M., Pappas S.G.et al.. Refining colorectal cancer classification and clinical stratification through a single-cell atlas. Genome Biol. 2022; 23:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Moosavi S.H., Eide P.W., Eilertsen I.A., Brunsell T.H., Berg K.C.G., Rosok B.I., Brudvik K.W., Bjornbeth B.A., Guren M.G., Nesbakken A.et al.. De novo transcriptomic subtyping of colorectal cancer liver metastases in the context of tumor heterogeneity. Genome Med. 2021; 13:143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Gautier L., Cope L., Bolstad B.M., Irizarry R.A.. affy—analysis of Affymetrix genechip data at the probe level. Bioinformatics. 2004; 20:307–315. [DOI] [PubMed] [Google Scholar]
- 91. Fu J., Zhang Y., Wang Y., Zhang H., Liu J., Tang J., Yang Q., Sun H., Qiu W., Ma Y.et al.. Optimization of metabolomic data processing using NOREVA. Nat. Protoc. 2022; 17:129–151. [DOI] [PubMed] [Google Scholar]
- 92. Yang Q., Wang Y., Zhang Y., Li F., Xia W., Zhou Y., Qiu Y., Li H., Zhu F.. NOREVA: enhanced normalization and evaluation of time-course and multi-class metabolomic data. Nucleic Acids Res. 2020; 48:W436–W448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Li B., Tang J., Yang Q., Li S., Cui X., Li Y., Chen Y., Xue W., Li X., Zhu F.. NOREVA: normalization and evaluation of MS-based metabolomics data. Nucleic Acids Res. 2017; 45:W162–W170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Tang J., Mou M., Wang Y., Luo Y., Zhu F.. MetaFS: performance assessment of biomarker discovery in metaproteomics. Brief. Bioinform. 2021; 22:bbaa105. [DOI] [PubMed] [Google Scholar]
- 95. Ge X., Yamamoto S., Tsutsumi S., Midorikawa Y., Ihara S., Wang S.M., Aburatani H.. Interpreting expression profiles of cancers by genome-wide survey of breadth of expression in normal tissues. Genomics. 2005; 86:127–141. [DOI] [PubMed] [Google Scholar]
- 96. Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Zidek A., Potapenko A.et al.. Highly accurate protein structure prediction with alphafold. Nature. 2021; 596:583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Stobaugh R.E. Chemical abstracts service chemical registry system. 11. Substance-related statistics: update and additions. J. Chem. Inf. Comput. Sci. 1988; 28:180–187. [DOI] [PubMed] [Google Scholar]
- 98. Cunningham F., Allen J.E., Allen J., Alvarez-Jarreta J., Amode M.R., Armean I.M., Austine-Orimoloye O., Azov A.G., Barnes I., Bennett R.et al.. Ensembl 2022. Nucleic Acids Res. 2022; 50:D988–D995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Tse T., Fain K.M., Zarin D.A.. How to avoid common problems when using clinicaltrials.gov in research: 10 issues to consider. BMJ. 2018; 361:k1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Schwartz L.M., Woloshin S., Zheng E., Tse T., Zarin D.A.. ClinicalTrials.gov and drugs@fda: a comparison of results reporting for new drug approval trials. Ann. Intern. Med. 2016; 165:421–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Hastings J., Owen G., Dekker A., Ennis M., Kale N., Muthukrishnan V., Turner S., Swainston N., Mendes P., Steinbeck C.. ChEBI in 2016: improved services and an expanding collection of metabolites. Nucleic Acids Res. 2016; 44:D1214–D1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Goodman J.M., Pletnev I., Thiessen P., Bolton E., Heller S.R.. InChI version 1.06: now more than 99.99% reliable. J Cheminform. 2021; 13:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Kanehisa M., Furumichi M., Sato Y., Ishiguro-Watanabe M., Tanabe M.. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 2021; 49:D545–D551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Sayers E.W., Bolton E.E., Brister J.R., Canese K., Chan J., Comeau D.C., Connor R., Funk K., Kelly C., Kim S.et al.. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2022; 50:D20–D26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Federhen S. Type material in the NCBI taxonomy database. Nucleic Acids Res. 2015; 43:D1086–D1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Berman H.M. Synergies between the protein data bank and the community. Nat. Struct. Mol. Biol. 2021; 28:400–401. [DOI] [PubMed] [Google Scholar]
- 107. UniProt Consortium UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021; 49:D480–D489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Thafar M.A., Olayan R.S., Ashoor H., Albaradei S., Bajic V.B., Gao X., Gojobori T., Essack M.. DTiGEMS+: drug–target interaction prediction using graph embedding, graph mining, and similarity-based techniques. J. Cheminform. 2020; 12:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Luo Y., Zhao X., Zhou J., Yang J., Zhang Y., Kuang W., Peng J., Chen L., Zeng J.. A network integration approach for drug–target interaction prediction and computational drug repositioning from heterogeneous information. Nat. Commun. 2017; 8:573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Wan F., Hong L., Xiao A., Jiang T., Zeng J.. NeoDTI: neural integration of neighbor information from a heterogeneous network for discovering new drug–target interactions. Bioinformatics. 2019; 35:104–111. [DOI] [PubMed] [Google Scholar]
- 111. Himmelstein D.S., Lizee A., Hessler C., Brueggeman L., Chen S.L., Hadley D., Green A., Khankhanian P., Baranzini S.E.. Systematic integration of biomedical knowledge prioritizes drugs for repurposing. Elife. 2017; 6:e26726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Xu Q., Liu K., Lin X., Qin Y., Chen L., Cheng J., Zhong M., He Q., Li Y., Wang T.et al.. ADMETNet: the knowledge base of pharmacokinetics and toxicology network. J. Genet. Genomics. 2017; 44:273–276. [DOI] [PubMed] [Google Scholar]
- 113. Xia W., Zheng L., Fang J., Li F., Zhou Y., Zeng Z., Zhang B., Li Z., Li H., Zhu F.. PFmulDL: a novel strategy enabling multi-class and multi-label protein function annotation by integrating diverse deep learning methods. Comput. Biol. Med. 2022; 145:105465. [DOI] [PubMed] [Google Scholar]
- 114. Yang Q., Hong J., Li Y., Xue W., Li S., Yang H., Zhu F.. A novel bioinformatics approach to identify the consistently well-performing normalization strategy for current metabolomic studies. Brief. Bioinform. 2020; 21:2142–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Otasek D., Morris J.H., Boucas J., Pico A.R., Demchak B.. Cytoscape automation: empowering workflow-based network analysis. Genome Biol. 2019; 20:185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Zhang Y., Ying J.B., Hong J.J., Li F.C., Fu T.T., Yang F.Y., Zheng G.X., Yao X.J., Lou Y., Qiu Y.et al.. How does chirality determine the selective inhibition of histone deacetylase 6? A lesson from trichostatin A enantiomers based on molecular dynamics. ACS Chem. Neurosci. 2019; 10:2467–2480. [DOI] [PubMed] [Google Scholar]
- 117. Li F., Zhou Y., Zhang Y., Yin J., Qiu Y., Gao J., Zhu F.. POSREG: proteomic signature discovered by simultaneously optimizing its reproducibility and generalizability. Brief. Bioinform. (2022); 23:bbac040. [DOI] [PubMed] [Google Scholar]
- 118. Fu T., Zheng G., Tu G., Yang F., Chen Y., Yao X., Li X., Xue W., Zhu F.. Exploring the binding mechanism of metabotropic glutamate receptor 5 negative allosteric modulators in clinical trials by molecular dynamics simulations. ACS Chem. Neurosci. 2018; 9:1492–1502. [DOI] [PubMed] [Google Scholar]
- 119. Yang Q., Li B., Chen S., Tang J., Li Y., Li Y., Zhang S., Shi C., Zhang Y., Mou M.et al.. MMEASE: online meta-analysis of metabolomic data by enhanced metabolite annotation, marker selection and enrichment analysis. J. Proteomics. 2021; 232:104023. [DOI] [PubMed] [Google Scholar]
- 120. Wang X., Li F., Qiu W., Xu B., Li Y., Lian X., Yu H., Zhang Z., Wang J., Li Z.et al.. SYNBIP: synthetic binding proteins for research, diagnosis and therapy. Nucleic Acids Res. 2022; 50:D560–D570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Hong J., Luo Y., Zhang Y., Ying J., Xue W., Xie T., Tao L., Zhu F.. Protein functional annotation of simultaneously improved stability, accuracy and false discovery rate achieved by a sequence-based deep learning. Brief. Bioinform. 2020; 21:1437–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Li F., Yin J., Lu M., Yang Q., Zeng Z., Zhang B., Li Z., Qiu Y., Dai H., Chen Y.et al.. ConSIG: consistent discovery of molecular signature from OMIC data. Brief. Bioinform. 2022; 23:bbac253. [DOI] [PubMed] [Google Scholar]
- 123. Hong J., Luo Y., Mou M., Fu J., Zhang Y., Xue W., Xie T., Tao L., Lou Y., Zhu F.. Convolutional neural network-based annotation of bacterial type IV secretion system effectors with enhanced accuracy and reduced false discovery. Brief. Bioinform. 2020; 21:1825–1836. [DOI] [PubMed] [Google Scholar]
- 124. Xue W., Wang P., Tu G., Yang F., Zheng G., Li X., Li X., Chen Y., Yao X., Zhu F.. Computational identification of the binding mechanism of a triple reuptake inhibitor amitifadine for the treatment of major depressive disorder. Phys. Chem. Chem. Phys. 2018; 20:6606–6616. [DOI] [PubMed] [Google Scholar]
- 125. Zhang S., Sun X., Mou M., Amahong K., Sun H., Zhang W., Shi S., Li Z., Gao J., Zhu F.. REGLIV: molecular regulation data of diverse living systems facilitating current multiomics research. Comput. Biol. Med. 2022; 148:105825. [DOI] [PubMed] [Google Scholar]
- 126. Xue W., Yang F., Wang P., Zheng G., Chen Y., Yao X., Zhu F.. What contributes to serotonin–norepinephrine reuptake inhibitors' dual-targeting mechanism? The key role of transmembrane domain 6 in human serotonin and norepinephrine transporters revealed by molecular dynamics simulation. ACS Chem. Neurosci. 2018; 9:1128–1140. [DOI] [PubMed] [Google Scholar]
- 127. Fu J., Zhang Y., Liu J., Lian X., Tang J., Zhu F.. Pharmacometabonomics: data processing and statistical analysis. Brief. Bioinform. 2021; 22:bbab138. [DOI] [PubMed] [Google Scholar]