Abstract
Phase separation (PS) proteins form droplets to regulate myriad membraneless organelles (MLOs) and cellular pathways such as transcription, signaling transduction and protein degeneration. PS droplets are usually liquid-like and can convert to hydrogel/solid-like under certain conditions. The PS behavior of proteins is regulated by co-PS partners and mutations, modifications, oligomerizations, repeat regions and alternative splicing of the proteins. With growing interest in PS condensates and associated proteins, we established PhaSepDB 1.0, which provided experimentally verified PS proteins and MLO-related proteins. The past few years witnessed a surge in PS-related research works; thus, we kept updating PhaSepDB. The current PhaSepDB contains 1419 PS entries, 770 low-throughput MLO-related entries and 7303 high-throughput MLO-related entries. We provided more detailed annotations of PS proteins, including PS verification experiments, regions used in experiments, phase diagrams of different experimental conditions, droplet states, co-PS partners and PS regulatory information. We believe that researchers can go further in studying PS proteins with the updated PhaSepDB (http://db.phasep.pro/).
INTRODUCTION
Phase separation (PS) compartmentalizes proteins and nucleic acids in cells and acts as a driving force underlying the organization of many membraneless organelles (MLOs) (1), such as stress granules, nuclear speckles, nucleoli and P-bodies. PS regulates various cellular functions, including transcription activation (2), genome organization (3), signaling transduction (4) and protein degeneration (5). Emerging evidence has shown that aberrant PS is associated with various diseases, including cancer, neurodegeneration and infectious diseases (6). More importantly, PS condensates can concentrate small-molecule drugs and affect their therapeutic effects (7), which implies novel insights for the future development of efficacious therapeutics. Thus, it is important to investigate the components of PS condensates.
Protein is the most well-studied component in PS condensates. Previous research works conclude that proteins form PS condensates via multivalent interactions between disordered regions or modular interacting domains (8). Researchers performed in vitro and in vivo experiments such as droplet formation, droplet fusion, fluorescence recovery after photobleaching (FRAP) and 1,6-hexanediol treatment to identify novel PS proteins and explore their properties (9). Different proteins might form droplets with different material states: proteins such as BRD4 and MED1 form liquid droplets that fuse and recover fast after photobleaching (10), while proteins such as TDP-43 and FUS form hydrogel or solid droplets that set high energy barrier for their components when diffusing with the outside matrix (11,12). When studying the PS behavior of proteins, researchers not only explore how the proteins phase separate in vivo and in vitro, but also care about the macromolecules that regulate the PS process of proteins, and changes in proteins that affect the PS ability of proteins. Some proteins can self-assemble to form condensates, while other proteins form PS condensates with the assistance of partner components such as protein, RNA or DNA (13). In addition, the PS behavior of proteins could be affected by post-translational modifications (PTMs) (14), mutations (15), oligomerizations (16), repeat regions (17) and alternative splicing (18) of proteins.
Several databases have been developed to contain information related to PS proteins, such as PS protein databases LLPSDB (19), PhaSePro (20) and DrLLPS (21), MLO-related protein databases RNAgranuleDB (22) and MSGP (23), and PS- and MLO-related disease database MloDisDB (24). Previously, we built the PS-related protein database PhaSepDB (25). Aside from PS proteins with experimental evidence, PhaSepDB includes many MLO-related proteins, even though their PS ability has not been well studied in biological labs. We reckon that they might locate to MLOs via PS, thus constituting possible PS proteins. Given a large amount of newly identified PS proteins and their regulatory mechanisms, we keep curating new PS proteins and updating the database. In July 2021, we released PhaSepDB 2.0 and provided a newly designed website. This year, we updated the database to version 2.1.
PhaSepDB 2.1 (http://db.phasep.pro/) contains 1419 PS entries for 868 PS proteins, 770 low-throughput MLO-related entries for 590 proteins and 7303 high-throughput MLO-related entries for 5292 proteins. PhaSepDB also provides three classes of annotations:
Proteins’ PS experiments: droplet state, phase diagram, verification experiment, PS region, PS domain, organism and cell line.
PS regulating macromolecules: co-PS protein, RNA and chemicals/DNA.
PS regulating events on proteins: PTM, mutation, oligomerization, repeat region and alternative splicing.
These annotations enable us to study the properties and regulatory rules of PS proteins profoundly.
MATERIALS AND METHODS
PS entries’ collection and classification
We rechecked the publications in PhaSepDB 1.0 for more detailed annotations such as droplet states and co-PS partners. Then, we searched PubMed with the same keywords as we used when curating PhaSepDB 1.0: ‘(phase transition [Title/Abstract] OR phase separation [Title/Abstract] OR membraneless organelles [Title/Abstract] OR biomolecular condensates [Title/Abstract]) AND protein AND cell’. Finally, 1812 publications from 1 July 2019 (endpoint of PhaSepDB 1.0) to 1 April 2022 were manually curated and examined. For reviews describing PS proteins, we checked the original publications cited by the reviews for detailed annotations. All collected data in PhaSepDB were at least double checked.
In PhaSepDB, each PS entry represents PS validation experiments of a protein illustrated in one publication. Thus, well-studied PS proteins are described in many entries as they are investigated by many publications, while different PS proteins investigated in one publication are recorded in different entries. PS entries were further classified into two categories: 511 PS-self and 908 PS-other entries (Table 1). Proteins in the PS-self category can undergo PS in vitro by themselves, while proteins falling in the PS-other category require interacting partners to form droplets in vitro, or they can form droplets in vivo. It should be noted that there are some PS-other proteins whose ability to undergo PS in vitro by themselves was not verified in the original publication, yet it could be possible for them to be PS-self proteins.
Table 1.
Overview of the datasets in PhaSepDB v1.0, v2.0 and v2.1
| PhaSepDB datasets | v1.0 | v2.0 | v2.1 |
|---|---|---|---|
| Release date | 1 September 2019 | 1 July 2021 | 1 June 2022 |
| Literature publication deadline | 1 July 2019 | 1 May 2021 | 1 April 2022 |
| PS entries | 565 | 961 | 1419 |
| PS proteins | 352 | 593 | 868 |
| PS-self entries | – | 356 | 511 |
| PS-other entries | – | 605 | 908 |
| Low-throughput MLO entries | 431 | 698 | 770 |
| High-throughput MLO entries | 3182 | 6981 | 7303 |
| MLOs | 16 | 59 | 73 |
| Number of PS entries with following annotations | |||
| Droplet state | 0 | 787 | 1075 |
| Phase diagram | 0 | 177 | 281 |
| Verification experiment | 0 | 961 | 1419 |
| PS region | 0 | 672 | 1015 |
| PS domain | 0 | 511 | 709 |
| Co-PS protein | 0 | 531 | 837 |
| Co-PS RNA | 0 | 147 | 214 |
| Co-PS chemicals/DNA | 0 | 84 | 160 |
| Post-translational modification | 0 | 113 | 146 |
| Mutation | 0 | 266 | 416 |
| Oligomerization | 0 | 80 | 111 |
| Repeat | 0 | 26 | 41 |
| Alternative splicing | 0 | 4 | 11 |
PS entries’ annotation
Proteins in PhaSepDB were mapped to UniProt (26). All PS entries were assigned with one or more of the four PS verification experiments below based on the original publication: (i) in vitro droplet formation; (ii) in vivo droplet formation; (iii) in vitro FRAP; and (iv) in vivo FRAP. Droplet formation indicates a protein’s ability to form or be recruited to PS droplets; the FRAP experiment shows the mobility of proteins within droplets.
Entries were also annotated with the following information:
Organisms of proteins and cell lines used for experiments.
MLO location and nuclear/cytoplasm location of PS proteins.
Phase diagrams that show the formation of droplets in different experimental conditions.
Regions used in PS experiments, and domains that are important for the proteins to undergo PS.
Material states of the PS droplets: liquid, hydrogel and solid.
PS partners for the proteins, including proteins, RNAs and others such as chemicals and DNAs.
Regulation of the PS ability of proteins, including PTMs, mutations, oligomerizations, repeat regions and alternative splicing.
We provided detailed descriptions from the original publications for these annotations.
MLO entries and MLOs
Besides the entries with clear PS evidence, PhaSepDB 2.1 contains 770 low-throughput MLO-related entries as well. These entries refer to proteins localized in MLOs as confirmed by in vivo experiments, but no clear PS evidence has been provided. A part of these entries includes database review entries in PhaSepDB 1.0. Furthermore, we collected 7303 high-throughput MLO-related entries from 20 papers; these papers were listed at http://db.phasep.pro/data_summary/. Seventy-three PS-related MLOs were gathered from dispersive literature when curating PS and MLO entries, including both classic and novel bodies.
Implementation of web services
To provide a user-friendly interface and a more stable application of the database, we improved the web implementation of PhaSepDB. The datasets were organized through SQLite. HTML, JavaScript, Django, uwsgi and Nginx were used for interface design and website background management.
RESULTS
Database summary
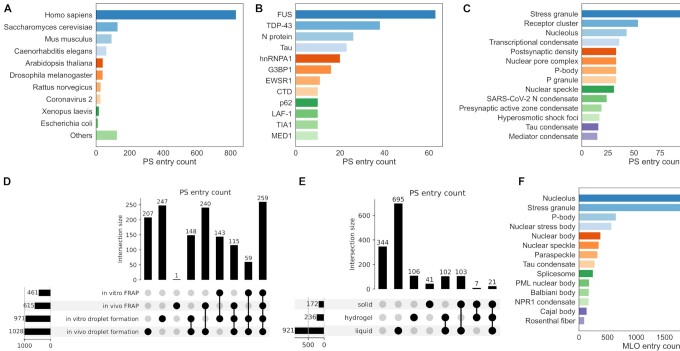
Since the first release of PhaSepDB 1.0 in September 2019, we have released PhaSepDB 2.0 in July 2021 and PhaSepDB 2.1 in June 2022. Overview of the datasets in PhaSepDB v1.0, v2.0 and v2.1 is shown in Table 1. PhaSepDB 2.1 contains 1419 PS entries for 868 PS proteins, among which 566 proteins are not included in other three phase-separating protein databases (19–21) (Supplementary Figure 1). In PhaSepDB 2.1, 833 entries describe human proteins, 129 entries describe yeast proteins and 93 entries describe mouse proteins (Figure 1A). Well-known PS proteins such as RNA-binding protein FUS, stress granule proteins G3BP1 and TIA1, RNA polymerase II, autophagic adaptor p62, mediator complex subunit MED1 and heterogeneous nuclear ribonucleoprotein hnRNPA1 own many entries in PhaSepDB (Figure 1B). Besides, some PS proteins with many entries are famous disease-related proteins, such as virus nucleocapsid (N) protein, TDP-43 and Tau, which are key proteins in neurodegenerative diseases, and Ewing sarcoma driver EWS protein (Figure 1B). PS proteins in stress granule, receptor cluster, nucleolus, transcriptional condensate, postsynaptic density and nuclear pore complex are exhaustively annotated in our database (Figure 1C).
Figure 1.
Statistical analysis of PS and MLO entries in PhaSepDB. (A) Organism distribution of PS entries. (B) Proteins with many PS entries. (C) MLOs with many PS entries. (D) Counts and overlaps of PS entries with different PS verification experiments. (E) Counts and overlaps of PS entries with different droplet states. (F) MLOs with many MLO entries.
We recorded the organism of PS proteins and cell lines used for PS experiments. PS verification experiments including in vitro droplet formation, in vivo droplet formation, in vitro FRAP and in vivo FRAP were recorded for all PS entries. There are 259 entries where the PS ability of proteins has been verified by four kinds of experiments and 964 entries where the PS ability of proteins has been verified by at least two kinds of experiments (Figure 1D). We classified PS entries into PS-self and PS-other classes based on whether the protein can undergo PS in vitro by itself. PS proteins form droplets with different material states. Liquid droplets are formed via liquid–liquid PS, while hydrogel/solid droplets are formed directly by proteins or converted from liquid droplets, which are common signs in neurodegenerative diseases (27). We extracted the droplet states of PS proteins from the original literature and found that most PS droplets are liquid-like. There are also some PS proteins that can form droplets with more than one material state (Figure 1E). These proteins usually undergo state transitions or their droplet states are affected by PTM, mutations, etc. As many researchers conducted PS experiments with crafted protein fragments rather than full-length proteins, we extracted the detailed regions and key domains used in PS experiments (Table 1), so that bioinformatics researchers can build PS predictors to give residue-level predictions and identify key regions for the proteins to undergo PS.
PS ability of proteins is affected by co-PS partners and their own properties. Thus, we recorded 837 entries with protein partners, 214 entries with RNA partners and 160 entries with chemicals or DNA partners (Table 1). Besides, we obtained 416 entries where the PS ability of proteins is affected by mutations, 146 entries where they are affected by PTMs, 111 entries where they are affected by oligomerization, 41 entries where repeat regions play such important roles and 11 entries where alternative splicing influences proteins’ PS ability (Table 1). This regulatory information enables us to have a thorough understanding of proteins’ PS behavior.
More and more MLOs were shown to be formed or regulated by PS; thus, we collected 73 PS-related MLOs from dispersive literature and provided their synonyms, locations and descriptions on the website (Table 1). As PS has only been widely studied in recent years while MLOs have been investigated for a long time, there are many MLO-related proteins that are potential PS proteins. Thus, we collected 770 low-throughput MLO-related entries where the proteins have been experimentally proved to locate in MLOs, and 7303 high-throughput MLO-related entries identified by organelle purification, proximity labeling and immunofluorescence image-based screening. Many proteins in MLO-related entries locate in the nucleolus, stress granule, P-body, nuclear stress body, nuclear body and nuclear speckle (Figure 1F). These proteins constitute a valuable candidate PS protein dataset, and their PS abilities deserve further studies.
The web application
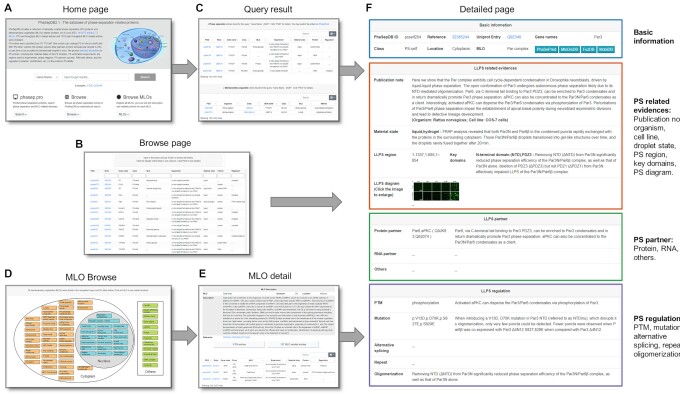
A freely available and fully functional website has been developed to access the collected datasets (Figure 2). The website comprises five sections: ‘Home’, ‘Browse’, ‘MLO Browse’, ‘About’ and ‘Download’. Users can search or browse all data in PhaSepDB according to gene name, UniProt entry, MLO location and other annotations on the ‘Home’ (Figure 2A) or ‘Browse’ (Figure 2B) page. The query results are presented in two tables (Figure 2C): the PS entry table and the MLO entry table. The PS entry table contains information on entry class, MLO location, PS verification experiment, droplet state, co-PS partner and regulation summary, while the MLO entry table contains information on entry class, MLO location and identification method. Both the PS entry table and the MLO entry table support customized searches; users can search all columns via the search box at the top right of each table. The ‘MLO Browse’ (Figure 2D) page provides user-friendly graphical navigation and a table that enables users to browse 73 MLOs based on their locations and descriptions; users can click each MLO to go to the ‘MLO detail’ (Figure 2E) page to see the description of the MLO and related PS and MLO entries.
Figure 2.
The interfaces of PhaSepDB. (A) Home page: users can search according to gene name, UniProt entry and MLOs. (B) Browse page: users can browse all PS entries via combinatorial search. (C) Query result page: two tables of PS entries and MLO entries. (D) MLO Browse page: all MLOs are displayed in graphical navigation and a table. (E) MLO detail page: basic information and related entries of the MLO. (F) Detailed page: detailed information for each entry.
Users can click on the unique PhaSepDB ID (PSID) on the ‘Browse’ page, query result page and ‘MLO detail’ page to navigate to the detailed page (Figure 2F). The detailed page for PS entries includes four sections:
Basic information: protein name, UniProt entry, reference (PubMed ID), entry class and MLO location.
Experimental evidence: a publication note describing the PS behavior of the protein, the organisms and cell lines used in the experiment, material state of the PS droplet, regions used in PS experiments, key domains regulating the PS behavior of the protein, and phase diagrams showing the formation of droplets in different experimental conditions.
Co-PS partner: protein, RNA and other (chemicals or DNA) PS partners.
PS regulation: mutations, PTMs, oligomerizations, repeat regions and alternative splicing that regulate the PS behavior of the protein.
For example, Figure 2F summarizes the PS experiments of rat Par3 (UniProt entry: Q9Z340, PhaSepDB ID: psself204) conducted by Liu et al. (28). Par3 is a component of Par complex. Three regions (1–1337, 1–685, 1–854) of Par3 were shown to be able to form droplets in vitro or in vivo (COS-7 cells), and the droplets transitioned from liquid-like into gel-like over time. Both the N-terminal domain (NTD) and the PDZ3 domain are important for the protein’s PS ability. NTD mediates oligomerization of Par3, and PDZ3 interacts with Par6β. Par6β and aPKC can co-phase separate with Par3. Par6β promotes Par3 PS, while activated aPKC can disperse the Par3/Par6 condensates via phosphorylation of Par3. p.V13D and p.D70K mutations in NTD disrupt the oligomerization process, and aPKC-mediated phosphomimetic p.S827E and p.S829E mutations impaired PS of Par3. Overall, users can learn about protein’s PS properties and regulatory rules with this page.
The detailed page for MLO entries includes protein name, UniProt entry, reference (PubMed ID), entry class, MLO location, the organisms and cell lines used in the experiment, and publication note describing the relation of the protein and the MLO.
The user guide and data summary have been illustrated on the ‘About’ page. Users can download all data in PhaSepDB freely on the ‘Download’ page.
DISCUSSION
Phase separation provides a means to concentrate and segregate proteins and nucleic acids in a spatiotemporally defined manner and sheds new light on how cellular condensates fulfil their diverse functions. However, we still do not know how many proteins can undergo PS at the proteome level. We also know little about the molecular grammar underlying PS. With detailed annotations in the updated PhaSepDB, researchers can go further in studying the properties of PS proteins, like classifying self-assembling and partner-dependent phase-separating proteins (13), investigating the differences between liquid and hydrogel/solid condensates, identifying the mutations or PTMs that influence proteins’ PS ability, and predicting PS promoting regions and domains.
These are also limitations of our work. First, in some publications, researchers did not state the protein isoforms used for PS experiments clearly. Thus, it is hard to specify which isoforms were used in these publications. Second, some of the mutation annotations do not specify mutation sites due to lack of clear descriptions in the original publications.
We believe that it will be of great interest for both experimental and computational researchers to work with the data in the new PhaSepDB, and our database will benefit future PS-related studies. We will keep updating PhaSepDB with more entries, more MLOs and new annotations in future.
DATA AVAILABILITY
All data described in this manuscript are freely accessible at http://db.phasep.pro/download/.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to acknowledge the contributions of all of the other PhaSepDB version 1.0 authors. We are grateful to all scientific researchers in the field of phase separation.
Contributor Information
Chao Hou, Department of Biomedical Informatics, School of Basic Medical Sciences, Peking University Health Science Center, Beijing 100191, China; Key Laboratory for Neuroscience, Ministry of Education/National Health Commission of China, Peking University, Beijing 100191, China.
Xinxin Wang, Department of Biomedical Informatics, School of Basic Medical Sciences, Peking University Health Science Center, Beijing 100191, China; Key Laboratory for Neuroscience, Ministry of Education/National Health Commission of China, Peking University, Beijing 100191, China.
Haotai Xie, Department of Cardiology, Peking University First Hospital, Beijing 100034, China.
Taoyu Chen, Department of Biomedical Informatics, School of Basic Medical Sciences, Peking University Health Science Center, Beijing 100191, China; Key Laboratory for Neuroscience, Ministry of Education/National Health Commission of China, Peking University, Beijing 100191, China.
Peiyu Zhu, Department of Biomedical Informatics, School of Basic Medical Sciences, Peking University Health Science Center, Beijing 100191, China; Key Laboratory for Neuroscience, Ministry of Education/National Health Commission of China, Peking University, Beijing 100191, China.
Xiaofeng Xu, Department of Biomedical Informatics, School of Basic Medical Sciences, Peking University Health Science Center, Beijing 100191, China; Key Laboratory for Neuroscience, Ministry of Education/National Health Commission of China, Peking University, Beijing 100191, China.
Kaiqiang You, Department of Biomedical Informatics, School of Basic Medical Sciences, Peking University Health Science Center, Beijing 100191, China; Key Laboratory for Neuroscience, Ministry of Education/National Health Commission of China, Peking University, Beijing 100191, China.
Tingting Li, Department of Biomedical Informatics, School of Basic Medical Sciences, Peking University Health Science Center, Beijing 100191, China; Key Laboratory for Neuroscience, Ministry of Education/National Health Commission of China, Peking University, Beijing 100191, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key Research and Development Program of China [2021YFF1200900, 2018YFA0507504]; National Natural Science Foundation of China [32070666]. Funding for open access charge: National Key Research and Development Program of China [2021YFF1200900].
Conflict of interest statement. None declared.
REFERENCES
- 1. Zhang H., Ji X., Li P., Liu C., Lou J., Wang Z., Wen W., Xiao Y., Zhang M., Zhu X.. Liquid–liquid phase separation in biology: mechanisms, physiological functions and human diseases. Sci. China Life Sci. 2020; 63:953–985. [DOI] [PubMed] [Google Scholar]
- 2. Hnisz D., Shrinivas K., Young R.A., Chakraborty A.K., Sharp P.A.. A phase separation model for transcriptional control. Cell. 2017; 169:13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Feric M., Misteli T.. Phase separation in genome organization across evolution. Trends Cell Biol. 2021; 31:671–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Su X., Ditlev J.A., Hui E., Xing W., Banjade S., Okrut J., King D.S., Taunton J., Rosen M.K., Vale R.D.. Phase separation of signaling molecules promotes T cell receptor signal transduction. Science. 2016; 352:595–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fu A., Cohen-Kaplan V., Avni N., Livneh I., Ciechanover A.. p62-containing, proteolytically active nuclear condensates, increase the efficiency of the ubiquitin-proteasome system. Proc. Natl Acad. Sci. U.S.A. 2021; 118:e2107321118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Alberti S., Dormann D. Liquid–liquid phase separation in disease. Annu. Rev. Genet. 2019; 53:171–194. [DOI] [PubMed] [Google Scholar]
- 7. Klein I.A., Boija A., Afeyan L.K., Hawken S.W., Fan M., Dall’Agnese A., Oksuz O., Henninger J.E., Shrinivas K., Sabari B.R.et al.. Partitioning of cancer therapeutics in nuclear condensates. Science. 2020; 368:1386–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Boeynaems S., Alberti S., Fawzi N.L., Mittag T., Polymenidou M., Rousseau F., Schymkowitz J., Shorter J., Wolozin B., Van Den Bosch Let al.. Protein phase separation: a new phase in cell biology. Trends Cell Biol. 2018; 28:420–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Alberti S., Gladfelter A., Mittag T.. Considerations and challenges in studying liquid–liquid phase separation and biomolecular condensates. Cell. 2019; 176:419–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sabari B.R., Dall’Agnese A., Boija A., Klein I.A., Coffey E.L., Shrinivas K., Abraham B.J., Hannett N.M., Zamudio A.V., Manteiga J.C.et al.. Coactivator condensation at super-enhancers links phase separation and gene control. Science. 2018; 361:eaar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen A.K., Lin R.Y., Hsieh E.Z., Tu P.H., Chen R.P., Liao T.Y., Chen W., Wang C.H., Huang J.J.. Induction of amyloid fibrils by the C-terminal fragments of TDP-43 in amyotrophic lateral sclerosis. J. Am. Chem. Soc. 2010; 132:1186–1187. [DOI] [PubMed] [Google Scholar]
- 12. Boczek E.E., Fürsch J., Niedermeier M.L., Jawerth L., Jahnel M., Ruer-Gruß M., Kammer K.M., Heid P., Mediani L., Wang J.et al.. HspB8 prevents aberrant phase transitions of FUS by chaperoning its folded RNA-binding domain. eLife. 2021; 10:e69377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen Z., Hou C., Wang L., Yu C., Chen T., Shen B., Hou Y., Li P., Li T.. Screening membraneless organelle participants with machine-learning models that integrate multimodal features. Proc. Natl Acad. Sci. U.S.A. 2022; 119:e2115369119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Singh H.R., Ostwal Y.B.. Post-translational modification, phase separation, and robust gene transcription. Trends Genet. 2019; 35:89–92. [DOI] [PubMed] [Google Scholar]
- 15. Banani S.F., Afeyan L.K., Hawken S.W., Henninger J.E., Dall’Agnese A., Clark V.E., Platt J.M., Oksuz O., Hannett N.M., Sagi I.et al.. Genetic variation associated with condensate dysregulation in disease. Dev. Cell. 2022; 57:1776–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Case L.B., Ditlev J.A., Rosen M.K.. Regulation of transmembrane signaling by phase separation. Annu. Rev. Biophys. 2019; 48:465–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Boeynaems S., Bogaert E., Kovacs D., Konijnenberg A., Timmerman E., Volkov A., Guharoy M., De Decker M., Jaspers T., Ryan V.H.et al.. Phase separation of C9orf72 dipeptide repeats perturbs stress granule dynamics. Mol. Cell. 2017; 65:1044–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Batlle C., Yang P., Coughlin M., Messing J., Pesarrodona M., Szulc E., Salvatella X., Kim H.J., Taylor J.P., Ventura S.. hnRNPDL phase separation is regulated by alternative splicing and disease-causing mutations accelerate its aggregation. Cell Rep. 2020; 30:1117–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang X., Zhou X., Yan Q., Liao S., Tang W., Xu P., Gao Y., Li Q., Dou Z., Yang W.et al.. LLPSDB v2.0: an updated database of proteins undergoing liquid–liquid phase separation in vitro. Bioinformatics. 2022; 38:2010–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mészáros B., Erdős G., Szabó B., Schád É., Tantos Á., Abukhairan R., Horváth T., Murvai N., Kovács O.P., Kovács M.et al.. PhaSePro: the database of proteins driving liquid–liquid phase separation. Nucleic Acids Res. 2020; 48:D360–D367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ning W., Guo Y., Lin S., Mei B., Wu Y., Jiang P., Tan X., Zhang W., Chen G., Peng D.et al.. DrLLPS: a data resource of liquid–liquid phase separation in eukaryotes. Nucleic Acids Res. 2020; 48:D288–D295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Youn J.Y., Dyakov B.J.A., Zhang J., Knight J.D.R., Vernon R.M., Forman-Kay J.D., Gingras A.C.. Properties of stress granule and P-body proteomes. Mol. Cell. 2019; 76:286–294. [DOI] [PubMed] [Google Scholar]
- 23. Nunes C., Mestre I., Marcelo A., Koppenol R., Matos C.A., Nobrega C.. MSGP: the first database of the protein components of the mammalian stress granules. Database (Oxford). 2019; 2019:baz031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hou C., Xie H., Fu Y., Ma Y., Li T.. MloDisDB: a manually curated database of the relations between membraneless organelles and diseases. Brief. Bioinform. 2021; 22:bbaa271. [DOI] [PubMed] [Google Scholar]
- 25. You K., Huang Q., Yu C., Shen B., Sevilla C., Shi M., Hermjakob H., Chen Y., Li T.. PhaSepDB: a database of liquid–liquid phase separation related proteins. Nucleic Acids Res. 2020; 48:D354–D359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. UniProt Consortium UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019; 47:D506–D515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zbinden A., Pérez-Berlanga M., De Rossi P., Polymenidou M.. Phase separation and neurodegenerative diseases: a disturbance in the force. Dev. Cell. 2020; 55:45–68. [DOI] [PubMed] [Google Scholar]
- 28. Liu Z., Yang Y., Gu A., Xu J., Mao Y., Lu H., Hu W., Lei Q.Y., Li Z., Zhang M.et al.. Par complex cluster formation mediated by phase separation. Nat. Commun. 2020; 11:2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data described in this manuscript are freely accessible at http://db.phasep.pro/download/.