Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus causing COVID-19, has continued to mutate and spread worldwide despite global vaccination efforts. In particular, the Omicron variant, first identified in South Africa in late November 2021, has become the dominant strain worldwide. Compared to the original strain identified in Wuhan, Omicron features 50 genetic mutations, with 15 mutations in the receptor-binding domain (RBD) of the spike protein, which binds to the human angiotensin-converting enzyme 2 (ACE2) receptor for viral entry. However, it is not completely understood how these mutations alter the interaction and binding strength between the Omicron RBD and ACE2. In this study, we used a combined steered molecular dynamics (SMD) simulation and experimental microscale thermophoresis (MST) approach to quantify the interaction between Omicron RBD and ACE2. We report that the Omicron brings an enhanced RBD-ACE2 interface through N501Y, Q498R, and T478K mutations; the changes further lead to unique interaction patterns, reminiscing the features of previously dominated variants, Alpha (N501Y) and Delta (L452R and T478K). Among the Q493K and Q493R, we report that Q493R shows stronger binding to ACE2 than Q493K due to increased interactions. Our MST data confirmed that the Omicron mutations in RBD are associated with a five-fold higher binding affinity to ACE2 compared to the RBD of the original strain. In conclusion, our results could help explain the Omicron variant’s prevalence in human populations, as higher interaction forces or affinity for ACE2 likely promote greater viral binding and internalization, leading to increased infectivity.
Graphical Abstract
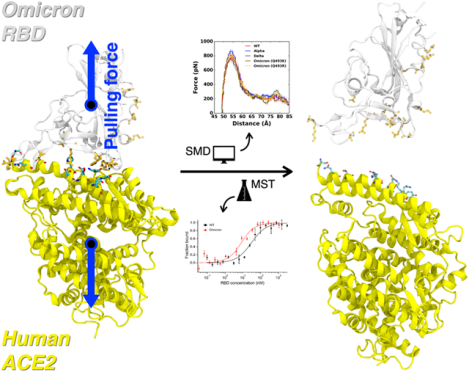
1. INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a positive-sense RNA virus causing the COVID-19 pandemic.1 Although vaccines for the virus have been administered to the public, several variants of concern (VOCs) defined by the World Health Organization (WHO), including the Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), and Delta (B.1.617.2), as well as the recently identified Omicron variant (B.1.1.529), have caused an increase in infectivity, lowering the efficacy of current vaccines.2,3
The Omicron (B.1.1.529) variant was first identified in South Africa in late November 2021. It has overtaken the Delta variant and become the dominant strain circulating in many countries, including the United States, causing record-high new COVID-19 cases in December 2021 and January 2022.3,4 The Omicron variant features 50 genetic mutations compared to the original (i.e., wild-type, WT) strains identified in Wuhan, China, with 36 mutations located in the spike (S) glycoprotein, over three times more than the number of mutations identified in the four other VOCs.5 More specifically, the receptor-binding domain (RBD) of the Omicron spike contains 15 mutations, much more than the 1, 3, 3, and 2 mutations found in RBDs of the Alpha, Beta, Gamma, and Delta variants, respectively.6 The RBD is the protein structure used by SARS-CoV-2 to bind to the human angiotensin-converting enzyme 2 (ACE2) receptor to gain host entry.7 Such a large number of mutations on the Omicron RBD suggests potential changes in both protein structure and binding affinity to hACE2. Indeed, several recent studies have tackled these issues. Han et al. reported the X-ray crystallography structure and biophysical binding kinetics of the Omicron RBD-ACE2 complex and claimed that Omicron RBD interacts with ACE2 at a similar affinity compared to the WT RBD.8 However, another protein crystallography and experimental study by Lan et al.9 and an MD simulation study by Lupala et al.10 reported that mutations in the Omicron RBD resulted in stronger binding toward ACE2. Additionally, recent studies claimed that the Omicron involves either Q493K or Q493R, while other mutations remain the same.8,11–15 With the answer to the critical question of whether the Omicron mutations influence the strength of the RBD-ACE2 interaction remains inconclusive, two new Omicron lineages (BA.4 and BA.5, involving additional L452R and F486V mutations) were reported in South Africa and spread out internationally, exhibiting stronger neutralization evasion than BA.1 and BA.2.16–18
We recently reported a combined all-atom steered molecular dynamics (SMD) simulation and experimental microscale thermophoresis (MST) approach in studying the binding between human ACE2 and the RBDs of SARS-CoV-2 VOCs Alpha, Beta, Gamma, and Delta in addition to Epsilon and Kappa.19 In this study, we applied the same approach to investigate the Omicron (B.1.1.529) RBD-ACE2 interaction. Our study reveals that the Omicron mutations enhance binding between RBD and ACE2 by forming unique interaction patterns caused mainly by N501Y, Q498R, and T478K mutations. We also disclose the effects of Q493K and Q493R in terms of Omicron RBD-ACE2 interaction. This study provides a better understanding of the role of each mutation in terms of RBD-ACE2 interaction at the molecular level.
2. METHODS
2.1. Computational Methods
A fully-glycosylated SARS-CoV-2 RBD and ACE2 complex model was achieved from CHARMM-GUI Archive COVID-19 Protein Library (6vsb_1_1_1_6vw1.pdb).20 The model complex includes five and one N-linked glycans in ACE2 (Asn53, Asn90, Asn103, Asn322, and Asn546) and RBD (Asn343), respectively. CHARMM-GUI Solution Builder21 and Input Generator22 were used for system generation. In each variant, the Alpha includes N501Y mutation, Delta has L452R and T478K mutations, and Omicron contains 15 mutated amino acids, i.e., G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493K/R, G496S, Q498R, N501Y, and Y505H (see also Figures 1B and 2D).8,12 Thus, corresponding residues were mutated from the WT RBD during the system generation, since the Omicron RBD structures were solved recently.23,24 This limitation may not fully capture the conformational and dynamic information from our initial structure, but almost all major interactions were well reproduced in our simulations (see Figure S3,S4). The CHARMM36(m) force field was used for protein and carbohydrates.25,26 The TIP3P water model27 was employed with 0.15 M of K+ and Cl− ions for mimicking a physiological condition. A large enough system size (190 Å ×190 Å × 190 Å) was considered to make both proteins to be sufficiently solvated when they are fully dissociated. The number of atoms in each system is approximately 550,000.
Figure 1.
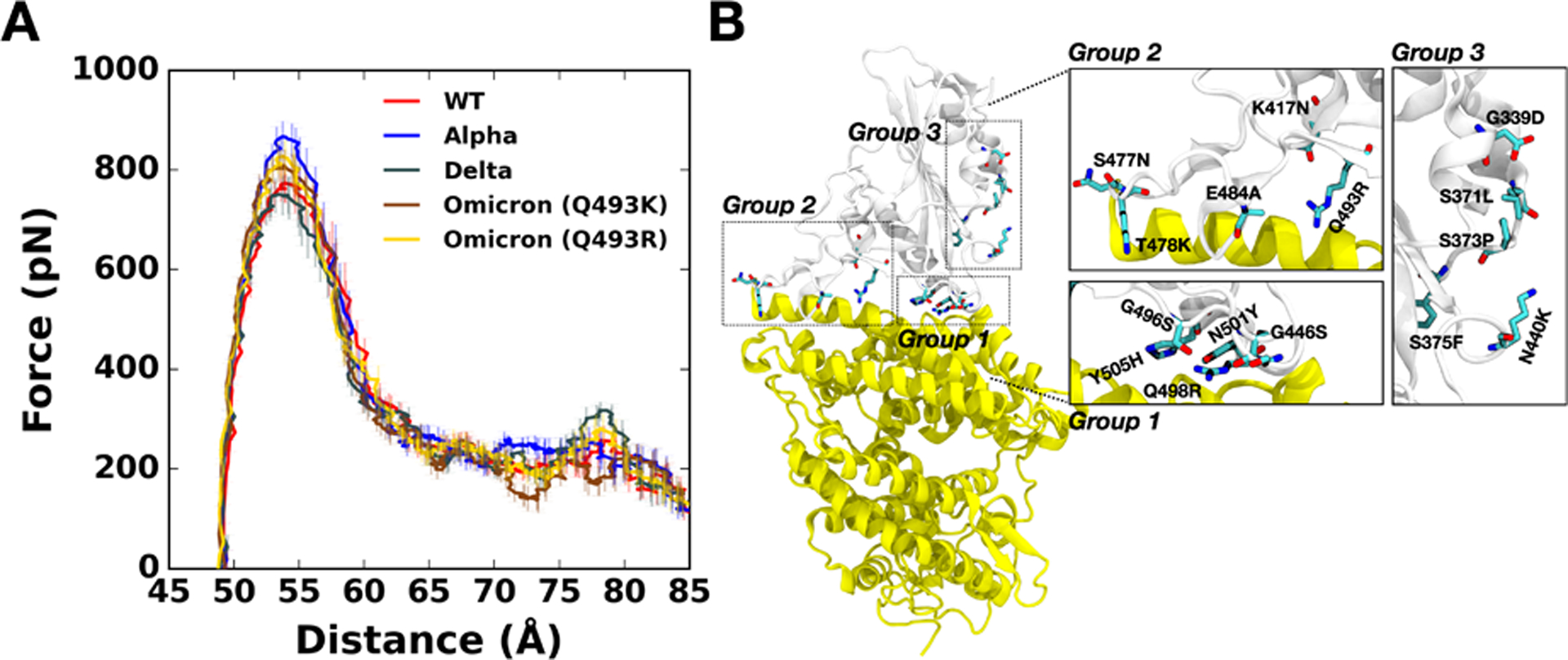
(A) Average force profiles of WT (red), Alpha (blue), Delta (gray), and Omicron variants (brown for Q493K and gold for Q493R) as a function of the distance between the center of mass of ACE2 and RBD. (B) The initial structure of Omicron (Q493R). Mutated residues from the WT are shown as solid sticks that are grouped and magnified into three panels with labels on the right. RBD and ACE2 are respectively colored in light gray (top) and yellow (bottom). All N-glycans, water, and ions are hidden for clarity.
Figure 2.
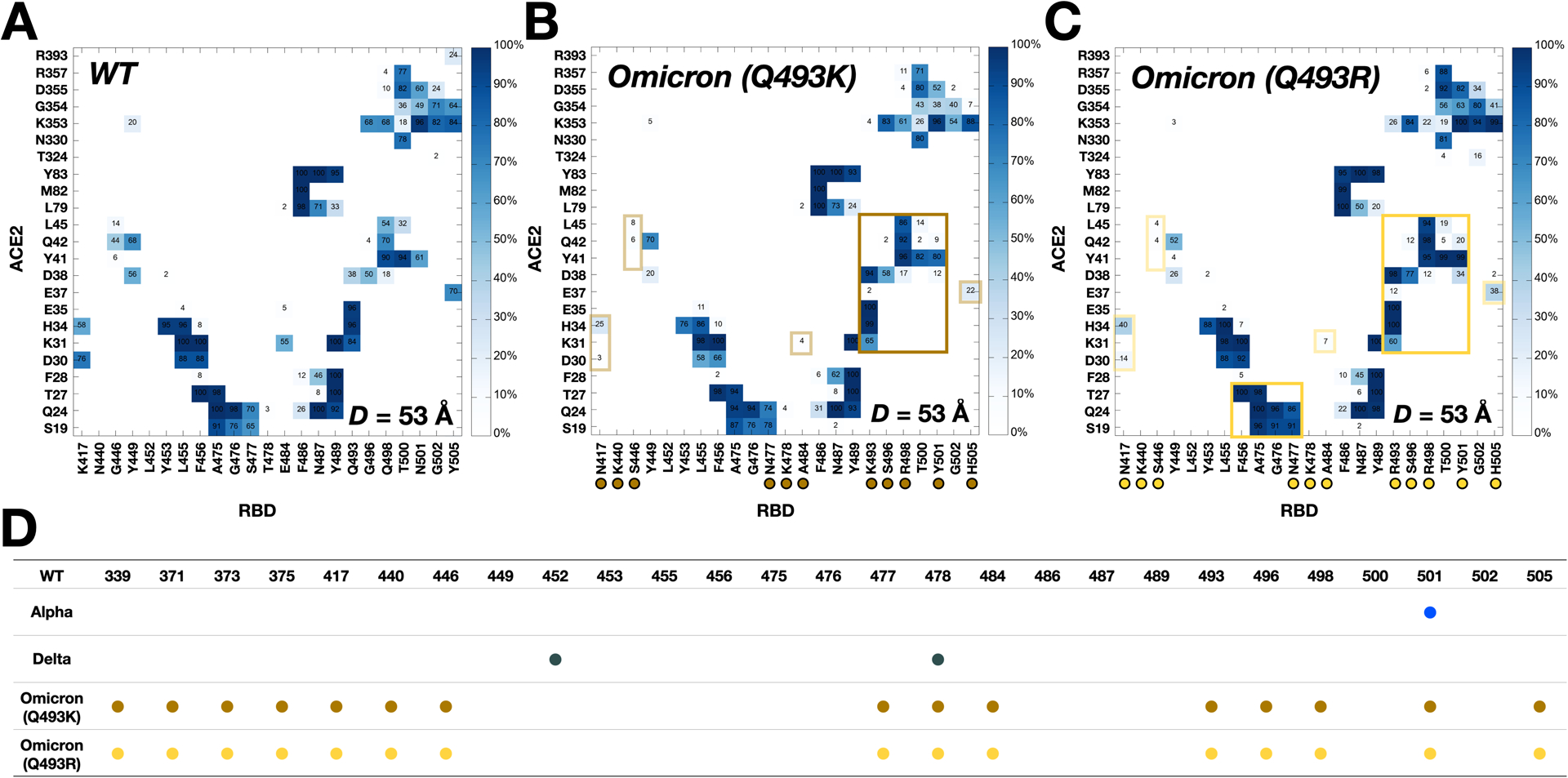
Two-dimensional contact maps of residues in RBD-ACE2 interface at D = 53 Å. (A) Interacting residue pairs between RBDWT and ACE2. RBD residues subjected to mutation are marked with colored circles at the bottom: (B) brown for Omicron Q493K and (C) gold for Omicron Q493R. The contact frequency is displayed with colors from light blue to dark blue. Solid and transparent boxes on the map respectively represent increased and decreased interactions between RBD and ACE2 upon mutations. (D) A summary of RBD mutations in the Omicron variants.
The overall simulation details are similar to our previous works.19,28 We used NAMD simulation software for the equilibrium and pulling simulations with the COLVARS method.29 Initially, the SARS-CoV-2 RBD and ACE2 proteins were aligned and maintained along the X-axis by applying the external forces to the center of mass (COM) of each protein. The effective pulling force applied to the COMs of Cα of both proteins was calculated through the following equation:
where k is the spring constant, v is the moving speed of the dummy atoms (i.e., spring potentials), R(t) is the vector between the COMs of RBD and ACE2 proteins, and n is the unit vector of R(t). This force allows the spring-connected proteins to be pulled in the opposite directions. The moving speed of proteins was set to 0.5 Å/ns, and 5 kcal/mol/Å2 of the spring constant was applied to the COM of each protein to let both proteins move along the X direction. To improve statistical results, 20 independent simulation runs of two Omicron variants, Q493K and Q493R, were performed for at least 40 ns to secure that the RBD and ACE2 are entirely dissociated from each other (Figure S1). The initial and final distances between the COMs of RBD and ACE2 are approximately 48 Å and 85 Å, respectively, where elongated loop structures of RBD upon full dissociation could affect the distance calculation (see Figure S2 for the dissociated structure). Note that we used our previous results19,28 for WT, Alpha, and Delta for comparison in this study.
The van der Waals interactions were smoothly switched off over 10–12 Å using a force-based switching function30 and the particle-mesh Ewald method31 with a mesh size of 1 Å was used for the electrostatic interactions. The SHAKE algorithm32 was employed to constrain bond lengths including hydrogen atoms. The Langevin piston method and the Langevin damping control method were respectively used for pressure control (1.01325 bar) and simulation temperature (303.15 K).33 The NVT ensemble with positional and dihedral restrained was applied for the equilibration simulations for 250 ps, and the NPT ensemble was applied for pulling simulations. The 4 fs simulation time-step was used with the hydrogen mass repartitioning method.34,35
2.2. Experimental Methods
Recombinant human ACE2 protein (GenBank accession: AF291820.1, Sino Biological 10108-H08H, Wayne, PA) was labeled with RED-NHS (2nd Generation) dye using the Monolith Protein Labeling Kit (NanoTemper Technologies, MO-L011, München, Germany). Labeled ACE2 (5 nM, final concentration) was mixed with the RBD proteins in a serial 15-step 2-fold dilution starting from 4 μM (for WT) or 1 (for Omicron) μM in PBS buffer supplanted with 0.1 % Pluronic® F-127. Both the WT and Omicron RBD proteins were from ACRObiosystems, Newark, DE (WT: SPD-C52H3, GenBank accession: QHD43416.1; Omicron: SPD-C522e). The mixed RBD+ACE2 samples were separately loaded into 16 premium glass capillaries (NanoTemper Technologies, MO-K025). The 16 capillaries were then placed in the reaction chamber in the order from low to high concentration. MST measurements were conducted on a Monolith NT.115 instrument (NanoTemper Technologies) at 20% excitation power at 24 °C. The measurement was repeated at least three times. Kd calculations were performed using the MO Affinity Analysis software (NanoTemper Technologies).
3. RESULTS AND DISCUSSION
3.1. The Omicron exhibits stronger RBD-ACE2 binding than WT
Pulling force analysis was performed (Figure 1A) as a function of distance, D, between COMs of RBDOmicrons and ACE2 to obtain molecular-level insight into the Omicron variants (Q493K/R). Like our previous study,19,28 we utilized our fully-glycosylated S RBD-ACE2 complex model for the pulling simulation.20 Compared to WT, the Omicron Q493K/R present increased force profiles evidenced by both the first (D = 53 Å) and second maximum (D = 79 Å) peaks (Figure 1A). As shown in Figure 1B, there are many mutations in the RBDOmicron, and notably, most of the mutations are located at the RBD-ACE2 interface, which can affect the enhanced RBD-ACE2 binding.
3.2. N501Y and Q498R mutations of the Omicron increase maximum force
Figure 2A,B,C compare two-dimensional RBD-ACE2 interaction patterns between WT and Omicron variants at D = 53 Å. As shown in Figure 2B,C, Omicron shows increased contact frequency (solid box) compared to WT (Figure 2A), which explains the higher forces than WT at D = 53 Å when both proteins are pulled (Figure 1A). The contact information is accordant with recent RBDOmicrons-ACE2 crystal and cryo-EM structures,8,9 indicating that our model system is well-validated (Figure S3). To analyze the contact frequency, the number of contacts was calculated between RBD residues 501 and 498 (N501 and Q498 for WT; Y501 and R498 for Omicron) and ACE2 (Figure 3A). The contact was counted if the residues in RBD are located within 4.5 Å of heavy atoms of key interacting residues of ACE2. The Omicron variants (Q493K/R) contain more contacts than other variants, indicating that N501Y and Q498R mutations induce a reinforced RBD-ACE2 interface. It is worthwhile to mention that the N501Y mutation also could impact on the populations of RBD-up conformation necessary for its host recognition through ACE2 binding.36 As shown in Figure 3B,C, at D = 55 Å, Omicron Y501 is positioned closer to ACE2 Y41 and K353 than WT N501, and the π-π and π-cation interactions with neighboring ACE2 Y41 and K353 of the Omicron contribute to holding RBD-ACE2 interface more tightly than WT.
Figure 3.
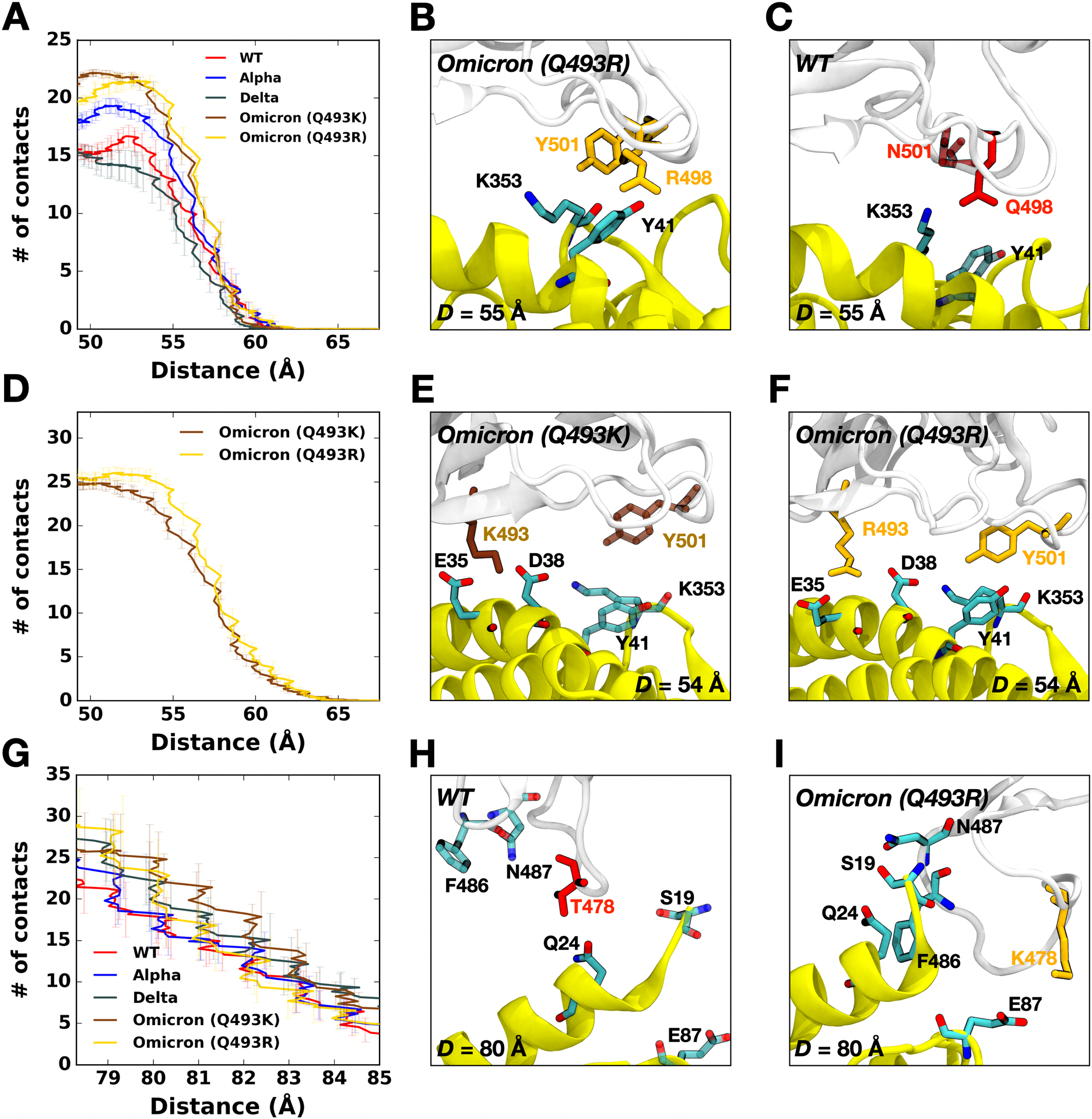
(A) The average number of contacts between RBD residues 498 and 501 and ACE2. (B and C) Representative snapshots at D = 55 Å of (B) Omicron Q493R and (C) WT. (D) The average number of contacts of Omicron variants between residues in RBD and ACE2 and (E and F) their interacting snapshots of (E) Q493K and (F) Q493R at D = 53 Å. (G) The average number of contacts between RBD residues and ACE2 and (H and I) key interaction pairs at D = 80 Å of (H) WT and (I) Omicron Q493R. The overall color scheme is the same as in Figure 1 (i.e., red for WT, brown for Omicron Q493K, and gold for Omicron Q493R). Interacting residues are presented as solid sticks, and residues losing their interactions are depicted as transparent sticks.
3.3. Q493K/R: Q493R shows stronger binding to ACE2 than Q493K
Recent studies report that the Omicron contains either Q493K or Q493R, while other mutations remain the same.8,11–15 Therefore, we built up two independent Omicron model systems retaining Q493K or Q493R to address both cases (Figure 2D). The force profiles of Omicron Q493K at D = 53 Å (Figure 1A) present weaker maximum forces than Omicron Q493R, although both show higher forces than WT at the same distance. Figure 2B,C show the two-dimensional contact maps of Omicron Q493K and Q493R. The overall interaction patterns are similar, but a few differences result in the gap in the force profile. Figure 3D shows the number of contact analysis between RBD Omicron Q493K/R and selected residues in ACE2. From the initial state (about D = 50 Å), Q493R displays more contacts with ACE2 than Q493K up to D = 58 Å. As shown in Figure 3E, a nitrogen atom positioned at the end of K493 holds two ACE2 residues, E35 and E38, by forming hydrogen bonds with four oxygen atoms of both E35 and E38. However, R493, which contains two nitrogen atoms, interacts with both D35 and D38 through hydrogen bonds via four oxygen atoms, showing increased contact frequency and maintaining RBD Y501 interactions with ACE2 Y41 and K353. This difference induces the gap in the number of contact analysis (Figure 3D) and makes the RBDOmicron-Q493R-ACE2 interface stronger than the RBDOmicron-Q493K-ACE2 interface. The stronger R493 interaction of Omicron Q493R further brings about increased contacts between RBD (F456, A475, G476, and N477) and ACE2 (S19, Q24, and T27) compared to WT or Omicron Q493K (Figure 2C). During the pulling process, Group 1 in Figure 1B is dissociated first, followed by Group 2 (Figure S2). Thus, each group plays a role of hinges between RBD and ACE2 and is responsible for having the first and second maximum forces in the force profile (Figure 1A). This indicates that the Omicron Q493R mutation contributes to having a reinforced RBD-ACE2 interface. The Q493R mutation has been reported to neutralize antibody,37–39 yet, compared to Q493K, it still shows better binding to ACE2 than Q493K, which is shown by other studies as well.9,13,40 It is worthwhile to mention that the mutations in Group 3 (Figure 1B) play a role in antibody neutralization,37,39 but they show insufficient abilities for the neutralization by counter effects of huge mutations in the RBD-ACE2 interface.
3.4. The Omicron presents enhanced second maximum force by T478K mutation
Omicron variants (Q493K/R) have shown unprecedented transmissibility than any other SARS-CoV-2 variants,4,41 and the force profile in Figure 1A confirms that the Omicron Q493K/R, compared to WT, present increased force profiles around D = 80 Å as well as D = 53 Å. To quantify what brings the increased second maximum peak of the Omicron, the number of contacts was calculated (Figure 3G), where all residues in both RBD and ACE2 interacting with each other were considered, and the residues located within 4.5 Å were considered to be in contact. As shown in Figure 3H,I, the WT T478 shows contacts with ACE2 Q24 without having interactions with ACE2 E87 (Figure 3H), but the Omicron K478 makes contacts with ACE2 E87 (Figure 3I) in addition to having more RBDOmicron-ACE2 interactions (N487-S19 and F486-Q24). Recently published studies claimed that the Omicron may9–11,13,14,40 or may not8,38 show enhanced binding compared to WT. Our results showing enhanced RBDOmicrons-ACE2 binding are consistent with the previous experimental and computational studies,9–11,13,14,40 and explain the differential interactions of Omicron Q493K/R, although we only considered single RBD out of trimeric SARS-CoV-2 S protein.
3.5. The Omicron RBD manifests enhanced binding affinity toward ACE2
To further validate our simulation results, we conducted an experimental protein binding assay using MST.42 As a reliable liquid-phase affinity measurement technique, MST has been used to detect viral protein-receptor interactions.43 We have recently used MST to quantify the interaction between human ACE2 and the S protein RBDs of some VOCs and the WT virus.19 Herein, we measured the binding affinity between ACE2 and the Omicron RBD bearing the Q493R mutation (Figure 4A). Fitting the MST saturation curve to a first-order 1:1 binding kinetics model yielded a binding affinity of 5.5 ± 1.4 nM, indicating that the RBDOmicron-ACE2 interaction has a five-fold higher affinity compared to the 27.5 ± 4.8 nM affinity of the RBDWT-ACE2 interaction that we reported recently (Figure 4B). Consistent with our simulation results, the RBDOmicron-ACE2 interaction also has a higher affinity than all the other VOCs that we measured earlier, including the Alpha, Beta, and Delta VOCs (Figure 4B). Moreover, we compared our experimental results with the published affinity data of variant RBD-ACE2 interactions from ten recent publications.8,9,38,44–52 As shown in Supplementary Table S1, the affinities measured by MST are within the range of the reported affinities of each variant. We also noticed a considerable variation among the published results, with up to one order of magnitude difference in some variants such as the WT, Alpha, and Omicron. Since most of the reported data were measured by surface plasmon resonance (SPR) or bio-layer interferometry (BLI), we attribute such variation to the quality of the samples, such as purity and protein aggregations, which tend to influence affinities detected in solid/liquid interfaces, such as the SPR and BLI, significantly. Finally, to see whether there is a correlation between our SMD simulations and MST experiments, we performed a linear regression analysis (Figure S5). Compared to other variants, Omicron shows a less correlation due to its relatively high variation in our MST study, but the result shows a favorable correlation.
Figure 4.
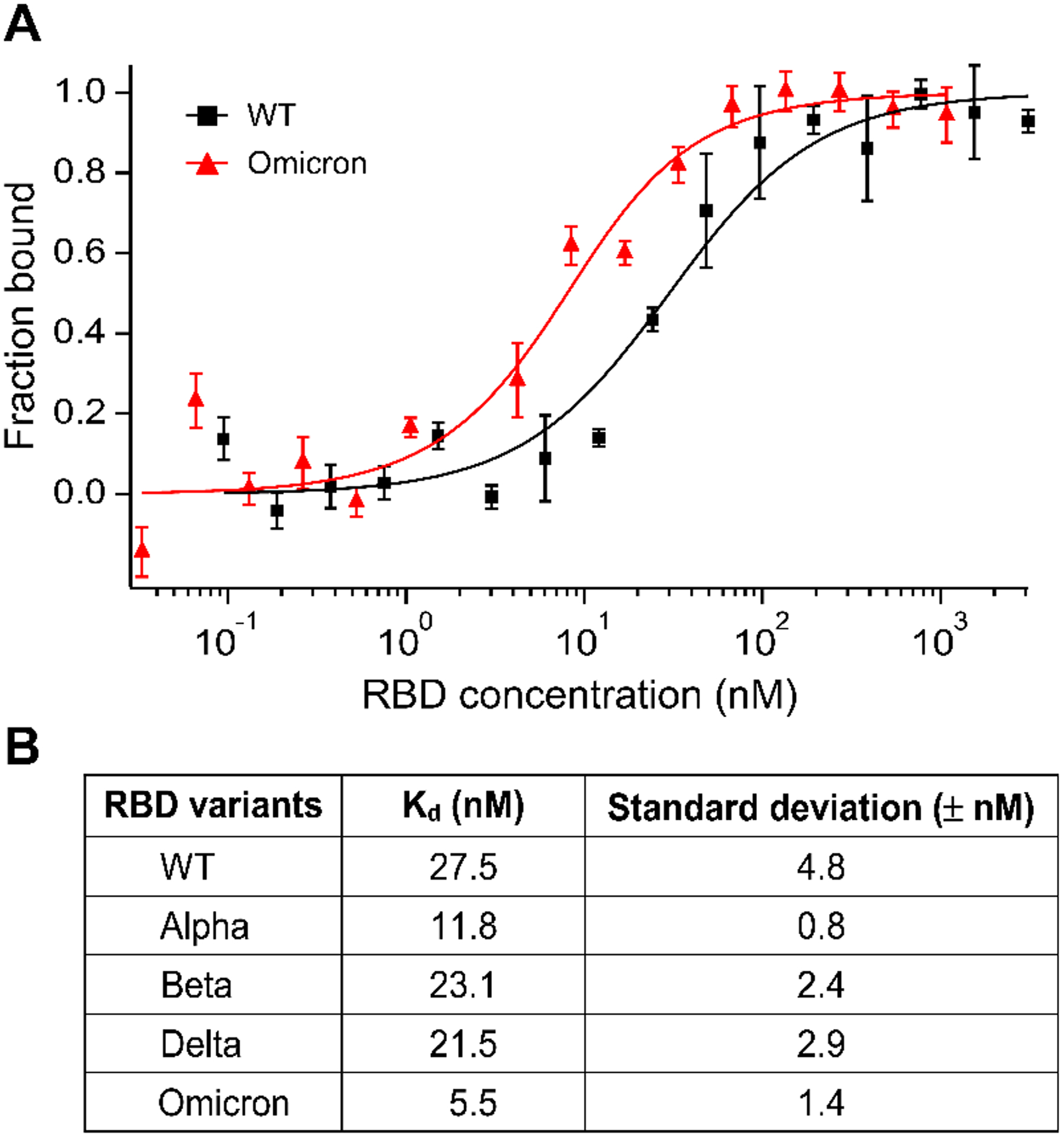
(A) Microscale thermophoresis (MST) analysis of the interaction between ACE2 and RBD of the WT or Omicron variants. Error bars represent standard deviations from five individual repeat measurements. The binding affinities were determined by fitting the data with the ‘Kd’ model of the MO Affinity software. (B) Affinities of ACE2 binding to RBD variants detected by MST. The MST responses were fitted to the 1:1 binding model. The Kd rates are shown as fit ± one standard deviation. Data of the WT, Alpha, Beta, and Delta variants were adapted from our prior study.19
4. CONCLUSIONS
This study characterizes interactions between ACE2 and the Omicron B.1.1.529 RBDs (Q493K or Q493R). Both SMD simulation and MST experiment confirm that the Omicron variant RBD displays stronger binding toward ACE2 than the WT RBD. Our analysis shows that the Omicron variants exhibit unique interaction patterns reminiscing the features of previously dominated Alpha (N501Y) and Delta (L452R and T478K) variants. Compared to the WT virus, the N501Y, Q498R, and T478K mutations are responsible for the higher force for RBDOmicrons-ACE2 dissociation. Among the Q493K and Q493R, Q493R shows stronger binding to ACE2 than Q493K due to increased interactions.
The newly reported Omicron lineages such as BA.4 and BA.5 are showing stronger antibody escape than existing variants, leading to making the current vaccines be less effective. In other word, the Omicron could be continuously evolving to survive under the immune pressure. Thus, understanding the effect of each mutation is inevitably important to prevent its continuous prevalence through the mutation. We hope this study provides valuable information on the mechanism behind the significantly increased transmissibility of heavily mutated Omicron variants.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported in part by NIH AI133634 and NSF CBET-1804117 (to X.F.Z.), NIH R21 AI163708 (to X.F.Z. and W.I.), NIH GM138472, and MCB-1810695 (to W.I.), and an internal grant from Lehigh University (to X.F.Z, and W.I.). This work was also supported by the National Supercomputing Center with supercomputing resources including technical support (KSC-2021-CRE-0274).
Footnotes
CONFLICT OF INTEREST
The authors declare no competing financial interest.
DATA AVAILABILITY STATEMENT
All data is available upon request.
REFERENCES
- (1).Holmes EC; Goldstein SA; Rasmussen AL; Robertson DL; Crits-Christoph A; Wertheim JO; Anthony SJ; Barclay WS; Boni MF; Doherty PC; Farrar J; Geoghegan JL; Jiang X; Leibowitz JL; Neil SJD; Skern T; Weiss SR; Worobey M; Andersen KG; Garry RF; Rambaut A The origins of SARS-CoV-2: A critical review. Cell 2021, 184, 4848–4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Cascella M; Rajnik M; Aleem A; Dulebohn SC; Napoli RD Features, Evaluation, and Treatment of Coronavirus (COVID-19). Treasure Island (FL): StatPearls Publishing, 2022. https://www.ncbi.nlm.nih.gov/books/NBK554776/ [PubMed] [Google Scholar]
- (3).COVID Data Tracker. Centers for Disease Control and Prevention. https://covid.cdc.gov/covid-data-tracker/#datatracker-home (accessed 2022-01-24).
- (4).Karim SSA; Karim QA Omicron SARS-CoV-2 variant: a new chapter in the COVID-19 pandemic. The Lancet 2021, 398, 2126–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Flemming A Omicron, the great escape artist. Nat. Rev. Immunol 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Mehra R; Kepp KP Structure and Mutations of SARS-CoV-2 Spike Protein: A Focused Overview. ACS Infect. Dis 2022, 8, 29–58. [DOI] [PubMed] [Google Scholar]
- (7).Jackson CB; Farzan M; Chen B; Choe H Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol 2022, 23, 3–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Han P; Li L; Liu S; Wang Q; Zhang D; Xu Z; Han P; Li X; Peng Q; Su C; Huang B; Li D; Zhang R; Tian M; Fu L; Gao Y; Zhao X; Liu K; Qi J; Gao GF; Wang P Receptor binding and complex structures of human ACE2 to spike RBD from omicron and delta SARS-CoV-2. Cell 2022, 185, 630–640 e610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Lan J; He X; Ren Y; Wang Z; Zhou H; Fan S; Zhu C; Liu D; Shao B; Liu T-Y; Wang Q; Zhang L; Ge J; Wang T; Wang X bioRxiv 2022.
- (10).Lupala CS; Ye Y; Chen H; Su XD; Liu H Mutations on RBD of SARS-CoV-2 Omicron variant result in stronger binding to human ACE2 receptor. Biochem. Biophys. Res. Commun 2021, 590, 34–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Shah M; Woo HG Omicron: A heavily mutated SARS-CoV-2 variant exhibits stronger binding to ACE2 and potently escape approved COVID-19 therapeutic antibodies. bioRxiv 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Wu L; Zhou L; Mo M; Liu T; Wu C; Gong C; Lu K; Gong L; Zhu W; Xu Z SARS-CoV-2 Omicron RBD shows weaker binding affinity than the currently dominant Delta variant to human ACE2. Signal Transduct. Targeted Ther 2022, 7, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Cameroni E; Saliba C; Bowen JE; Rosen LE; Culap K; Pinto D; De Marco A; Zepeda SK; di Iulio J; Zatta F; Kaiser H; Noack J; Farhat N; Czudnochowski N; Havenar-Daughton C; Sprouse KR; Dillen JR; Powell AE; Chen A; Maher C; Yin L; Sun D; Soriaga L; Gustafsson C; Franko NM; Logue J; Iqbal NT; Mazzitelli I; Geffner J; Grifantini R; Chu H; Gori A; Riva A; Giannini O; Ceschi A; Ferrari P; Franzetti-Pellanda A; Garzoni C; Hebner C; Purcell LA; Piccoli L; Pizzuto MS; Walls AC; Telenti A; Virgin HW; Lanzavecchia A; Veesler D; Snell G; Corti D Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. bioRxiv 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Ortega JT; Jastrzebska B; Rangel HR Omicron SARS-CoV-2 Variant Spike Protein Shows an Increased Affinity to the Human ACE2 Receptor: An In Silico Analysis. Pathogens 2021, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Schmidt F; Muecksch F; Weisblum Y; Da Silva J; Bednarski E; Cho A; Wang Z; Gaebler C; Caskey M; Nussenzweig MC; Hatziioannou T; Bieniasz PD Plasma neutralization properties of the SARS-CoV-2 Omicron variant. medRxiv 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Cao Y; Yisimayi A; Jian F; Song W; Xiao T; Wang L; Du S; Wang J; Li Q; Chen X; Yu Y; Wang P; Zhang Z; Liu P; An R; Hao X; Wang Y; Wang J; Feng R; Sun H; Zhao L; Zhang W; Zhao D; Zheng J; Yu L; Li C; Zhang N; Wang R; Niu X; Yang S; Song X; Chai Y; Hu Y; Shi Y; Zheng L; Li Z; Gu Q; Shao F; Huang W; Jin R; Shen Z; Wang Y; Wang X; Xiao J; Xie XS BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Tuekprakhon A; Nutalai R; Dijokaite-Guraliuc A; Zhou D; Ginn HM; Selvaraj M; Liu C; Mentzer AJ; Supasa P; Duyvesteyn HME; Das R; Skelly D; Ritter TG; Amini A; Bibi S; Adele S; Johnson SA; Constantinides B; Webster H; Temperton N; Klenerman P; Barnes E; Dunachie SJ; Crook D; Pollard AJ; Lambe T; Goulder P; Paterson NG; Williams MA; Hall DR; Consortium O; Consortium IC; Fry EE; Huo J; Mongkolsapaya J; Ren J; Stuart DI; Screaton GR Antibody escape of SARS-CoV-2 Omicron BA.4 and BA.5 from vaccine and BA.1 serum. Cell 2022, 185, 2422–2433 e2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Qu P; Faraone J; Evans JP; Zou X; Zheng YM; Carlin C; Bednash JS; Lozanski G; Mallampalli RK; Saif LJ; Oltz EM; Mohler PJ; Gumina RJ; Liu SL Neutralization of the SARS-CoV-2 Omicron BA.4/5 and BA.2.12.1 Subvariants. N. Engl. J. Med 2022, 386, 2526–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Kim S; Liu Y; Lei Z; Dicker J; Cao Y; Zhang XF; Im W Differential Interactions between Human ACE2 and Spike RBD of SARS-CoV-2 Variants of Concern. J. Chem. Theory Comput 2021, 17, 7972–7979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Woo H; Park S-J; Choi YK; Park T; Tanveer M; Cao Y; Kern NR; Lee J; Yeom MS; Croll TI; Seok C; Im W Developing a Fully Glycosylated Full-Length SARS-CoV-2 Spike Protein Model in a Viral Membrane. J. Phys. Chem. B 2020, 124, 7128–7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Jo S; Kim T; Iyer VG; Im W CHARMM‐GUI: a web‐based graphical user interface for CHARMM. J. Comput. Chem 2008, 29, 1859–1865. [DOI] [PubMed] [Google Scholar]
- (22).Lee J; Cheng X; Swails JM; Yeom MS; Eastman PK; Lemkul JA; Wei S; Buckner J; Jeong JC; Qi Y; Jo S; Pande VS; Case DA; Brooks CL; MacKerell AD; Klauda JB; Im W CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field. J. Chem. Theory Comput 2016, 12, 405–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Mannar D; Saville JW; Zhu X; Srivastava SS; Berezuk AM; Tuttle KS; Marquez AC; Sekirov I; Subramaniam S SARS-CoV-2 Omicron variant: Antibody evasion and cryo-EM structure of spike protein-ACE2 complex. Science 2022, 375, 760–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Han P; Li L; Liu S; Wang Q; Zhang D; Xu Z; Han P; Li X; Peng Q; Su C; Huang B; Li D; Zhang R; Tian M; Fu L; Gao Y; Zhao X; Liu K; Qi J; Gao GF; Wang P Receptor binding and complex structures of human ACE2 to spike RBD from Omicron and Delta SARS-CoV-2. Cell 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Hatcher E; Guvench O; MacKerell AD CHARMM Additive All-Atom Force Field for Aldopentofuranoses, Methyl-aldopentofuranosides, and Fructofuranose. J. Phys. Chem. B 2009, 113, 12466–12476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Huang J; Rauscher S; Nawrocki G; Ran T; Feig M; de Groot BL; Grubmüller H; MacKerell AD CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods 2017, 14, 71–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Jorgensen WL; Chandrasekhar J; Madura JD; Impey RW; Klein ML Comparison of simple potential functions for simulating liquid water. J. Chem. Phys 1983, 79, 926–935. [Google Scholar]
- (28).Cao W; Dong C; Kim S; Hou D; Tai W; Du L; Im W; Zhang XF Biomechanical characterization of SARS-CoV-2 spike RBD and human ACE2 protein-protein interaction. Biophys. J 2021, 120, 1011–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Phillips JC; Braun R; Wang W; Gumbart J; Tajkhorshid E; Villa E; Chipot C; Skeel RD; Kale L; Schulten K Scalable molecular dynamics with NAMD. J. Comput. Chem 2005, 26, 1781–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Dion M; Rydberg H; Schröder E; Langreth DC; Lundqvist BI Van der Waals density functional for general geometries. Phys. Rev. Lett 2004, 92, 246401. [DOI] [PubMed] [Google Scholar]
- (31).Essmann U; Perera L; Berkowitz ML; Darden T; Lee H; Pedersen LG A smooth particle mesh Ewald method. J. Chem. Phys 1995, 103, 8577–8593. [Google Scholar]
- (32).Ryckaert J-P; Ciccotti G; Berendsen HJ Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys 1977, 23, 327–341. [Google Scholar]
- (33).Feller SE; Zhang Y; Pastor RW; Brooks BR Constant pressure molecular dynamics simulation: the Langevin piston method. J. Chem. Phys 1995, 103, 4613–4621. [Google Scholar]
- (34).Gao Y; Lee J; Smith IPS; Lee H; Kim S; Qi Y; Klauda JB; Widmalm G. r.; Khalid S; Im W CHARMM-GUI Supports Hydrogen Mass Repartitioning and Different Protonation States of Phosphates in Lipopolysaccharides. J. Chem. Inf. Model 2021, 61, 831–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Hopkins CW; Le Grand S; Walker RC; Roitberg AE Long-time-step molecular dynamics through hydrogen mass repartitioning. J. Chem. Theory Comput 2015, 11, 1864–1874. [DOI] [PubMed] [Google Scholar]
- (36).Yang TJ; Yu PY; Chang YC; Liang KH; Tso HC; Ho MR; Chen WY; Lin HT; Wu HC; Hsu SD Effect of SARS-CoV-2 B.1.1.7 mutations on spike protein structure and function. Nat. Struct. Mol. Biol 2021, 28, 731–739. [DOI] [PubMed] [Google Scholar]
- (37).Cao Y; Wang J; Jian F; Xiao T; Song W; Yisimayi A; Huang W; Li Q; Wang P; An R; Wang J; Wang Y; Niu X; Yang S; Liang H; Sun H; Li T; Yu Y; Cui Q; Liu S; Yang X; Du S; Zhang Z; Hao X; Shao F; Jin R; Wang X; Xiao J; Wang Y; Xie XS Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature 2022, 602, 657–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Dejnirattisai W; Huo J; Zhou D; Zahradnik J; Supasa P; Liu C; Duyvesteyn HME; Ginn HM; Mentzer AJ; Tuekprakhon A; Nutalai R; Wang B; Dijokaite A; Khan S; Avinoam O; Bahar M; Skelly D; Adele S; Johnson SA; Amini A; Ritter TG; Mason C; Dold C; Pan D; Assadi S; Bellass A; Omo-Dare N; Koeckerling D; Flaxman A; Jenkin D; Aley PK; Voysey M; Costa Clemens SA; Naveca FG; Nascimento V; Nascimento F; Fernandes da Costa C; Resende PC; Pauvolid-Correa A; Siqueira MM; Baillie V; Serafin N; Kwatra G; Da Silva K; Madhi SA; Nunes MC; Malik T; Openshaw PJM; Baillie JK; Semple MG; Townsend AR; Huang KA; Tan TK; Carroll MW; Klenerman P; Barnes E; Dunachie SJ; Constantinides B; Webster H; Crook D; Pollard AJ; Lambe T; Consortium O; Consortium IC; Paterson NG; Williams MA; Hall DR; Fry EE; Mongkolsapaya J; Ren J; Schreiber G; Stuart DI; Screaton GR SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell 2022, 185, 467–484 e415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Yin W; Xu Y; Xu P; Cao X; Wu C; Gu C; He X; Wang X; Huang S; Yuan Q; Wu K; Hu W; Huang Z; Liu J; Wang Z; Jia F; Xia K; Liu P; Wang X; Song B; Zheng J; Jiang H; Cheng X; Jiang Y; Deng SJ; Xu HE Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody. Science 2022, 375, 1048–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Jawad B; Adhikari P; Podgornik R; Ching WY Binding Interactions between Receptor-Binding Domain of Spike Protein and Human Angiotensin Converting Enzyme-2 in Omicron Variant. J Phys. Chem. Lett 2022, 3915–3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Torjesen I Covid-19: Omicron may be more transmissible than other variants and partly resistant to existing vaccines, scientists fear. BMJ 2021, 375, n2943. [DOI] [PubMed] [Google Scholar]
- (42).Plach MG; Grasser K; Schubert T MicroScale Thermophoresis as a tool to study protein-peptide interactions in the context of large eukaryotic protein complexes. Bio-Protoc. 2017, 7, e2632–e2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Walls A; Tortorici MA; Bosch BJ; Frenz B; Rottier PJ; DiMaio F; Rey FA; Veesler D Crucial steps in the structure determination of a coronavirus spike glycoprotein using cryo‐electron microscopy. Protein Sci. 2017, 26, 113–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Barton MI; MacGowan SA; Kutuzov MA; Dushek O; Barton GJ; van der Merwe PA Effects of common mutations in the SARS-CoV-2 Spike RBD and its ligand, the human ACE2 receptor on binding affinity and kinetics. Elife 2021, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Cui Z; Liu P; Wang N; Wang L; Fan K; Zhu Q; Wang K; Chen R; Feng R; Jia Z; Yang M; Xu G; Zhu B; Fu W; Chu T; Feng L; Wang Y; Pei X; Yang P; Xie XS; Cao L; Cao Y; Wang X Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron. Cell 2022, 185, 860–871 e813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Han P; Su C; Zhang Y; Bai C; Zheng A; Qiao C; Wang Q; Niu S; Chen Q; Zhang Y; Li W; Liao H; Li J; Zhang Z; Cho H; Yang M; Rong X; Hu Y; Huang N; Yan J; Wang Q; Zhao X; Gao GF; Qi J Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants. Nat. Commun 2021, 12, 6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Lee JH; Lee Y; Lee SK; Kim J; Lee CS; Kim NH; Kim HG Versatile role of ACE2-based biosensors for detection of SARS-CoV-2 variants and neutralizing antibodies. Biosens. Bioelectron 2022, 203, 114034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Shang J; Ye G; Shi K; Wan Y; Luo C; Aihara H; Geng Q; Auerbach A; Li F Structural basis of receptor recognition by SARS-CoV-2. Nature 2020, 581, 221–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Supasa P; Zhou D; Dejnirattisai W; Liu C; Mentzer AJ; Ginn HM; Zhao Y; Duyvesteyn HME; Nutalai R; Tuekprakhon A; Wang B; Paesen GC; Slon-Campos J; Lopez-Camacho C; Hallis B; Coombes N; Bewley KR; Charlton S; Walter TS; Barnes E; Dunachie SJ; Skelly D; Lumley SF; Baker N; Shaik I; Humphries HE; Godwin K; Gent N; Sienkiewicz A; Dold C; Levin R; Dong T; Pollard AJ; Knight JC; Klenerman P; Crook D; Lambe T; Clutterbuck E; Bibi S; Flaxman A; Bittaye M; Belij-Rammerstorfer S; Gilbert S; Hall DR; Williams MA; Paterson NG; James W; Carroll MW; Fry EE; Mongkolsapaya J; Ren J; Stuart DI; Screaton GR Reduced neutralization of SARS-CoV-2 B.1.1.7 variant by convalescent and vaccine sera. Cell 2021, 184, 2201–2211 e2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Tian F; Tong B; Sun L; Shi S; Zheng B; Wang Z; Dong X; Zheng P N501Y mutation of spike protein in SARS-CoV-2 strengthens its binding to receptor ACE2. Elife 2021, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Zhang X; Wu S; Wu B; Yang Q; Chen A; Li Y; Zhang Y; Pan T; Zhang H; He X SARS-CoV-2 Omicron strain exhibits potent capabilities for immune evasion and viral entrance. Signal Transduct. Target Thers 2021, 6, 430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Cameroni E; Bowen JE; Rosen LE; Saliba C; Zepeda SK; Culap K; Pinto D; VanBlargan LA; De Marco A; di Iulio J; Zatta F; Kaiser H; Noack J; Farhat N; Czudnochowski N; Havenar-Daughton C; Sprouse KR; Dillen JR; Powell AE; Chen A; Maher C; Yin L; Sun D; Soriaga L; Bassi J; Silacci-Fregni C; Gustafsson C; Franko NM; Logue J; Iqbal NT; Mazzitelli I; Geffner J; Grifantini R; Chu H; Gori A; Riva A; Giannini O; Ceschi A; Ferrari P; Cippa PE; Franzetti-Pellanda A; Garzoni C; Halfmann PJ; Kawaoka Y; Hebner C; Purcell LA; Piccoli L; Pizzuto MS; Walls AC; Diamond MS; Telenti A; Virgin HW; Lanzavecchia A; Snell G; Veesler D; Corti D Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature 2022, 602, 664–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data is available upon request.


