Abstract
Fungi are highly diverse organisms, which provide multiple ecosystem services. However, compared with charismatic animals and plants, the distribution patterns and conservation needs of fungi have been little explored. Here, we examined endemicity patterns, global change vulnerability and conservation priority areas for functional groups of soil fungi based on six global surveys using a high‐resolution, long‐read metabarcoding approach. We found that the endemicity of all fungi and most functional groups peaks in tropical habitats, including Amazonia, Yucatan, West‐Central Africa, Sri Lanka, and New Caledonia, with a negligible island effect compared with plants and animals. We also found that fungi are predominantly vulnerable to drought, heat and land‐cover change, particularly in dry tropical regions with high human population density. Fungal conservation areas of highest priority include herbaceous wetlands, tropical forests, and woodlands. We stress that more attention should be focused on the conservation of fungi, especially root symbiotic arbuscular mycorrhizal and ectomycorrhizal fungi in tropical regions as well as unicellular early‐diverging groups and macrofungi in general. Given the low overlap between the endemicity of fungi and macroorganisms, but high conservation needs in both groups, detailed analyses on distribution and conservation requirements are warranted for other microorganisms and soil organisms.
Keywords: biodiversity, biogeography, climate change, conservation priorities, global change vulnerability, global maps, mycorrhizal fungi, pathogens, saprotrophs
Using a long‐read metabarcoding approach, we found that the endemicity of all fungi and most functional groups peaks in tropical habitats, including Amazonia, Yucatan and West‐Central Africa, with a negligible island effect compared with plants and animals. Fungi are predominantly vulnerable to drought, heat and land cover change, particularly in dry tropical regions with high human population density. Fungal conservation areas of highest priority include herbaceous wetlands, tropical forests and woodlands.
![]()
1. INTRODUCTION
Human activities affect nearly all habitats through changes in climate and land‐use, impacting vegetation cover and composition. These changes negatively influence many plant and animal species that have narrow environmental tolerances and limited dispersal capacities across anthropogenic landscapes (Schulte to Bühne et al., 2021). Anthropogenic impacts most strongly affect endemic species—that is, taxa with small distribution ranges and typically narrow ecological niches (Brook et al., 2008). The diversity of endemic plants and animals is higher in areas characterized by long‐term climatic stability, high precipitation, environmental heterogeneity and insularity. Unfortunately, these areas—which are mostly located in the tropics and subtropics—usually coincide with major human degradations of the environment (Kier et al., 2009; Sandel et al., 2020; Stein et al., 2014). Unlike the wealth of information that has accumulated for plants and animals, global patterns of fungal endemism and vulnerability to environmental change remain virtually unknown (Cameron et al., 2019; Guerra, Bardgett, et al., 2021; but see Davison et al., 2015). Only a few Alnus‐associated ectomycorrhizal (EcM) fungi (Põlme et al., 2013) and fewer than 100 soil‐borne saprotrophic and pathogenic fungi (Egidi et al., 2019) can be considered to be cosmopolitan, suggesting that the vast majority of fungal species may be endemic at least at a subcontinental scale (Talbot et al., 2014). This paucity of knowledge about the endemicity and vulnerability of fungi to global change is alarming, given the fundamental roles of fungi in soil carbon and nutrient cycling processes and as devastating crop and forest pathogens (Crowther et al., 2019; Wardle & Lindahl, 2014).
Comparative studies indicate that aboveground and belowground biodiversity is driven by different environmental predictors at local and global scales (Cameron et al., 2019; Le Provost et al., 2021). This suggests differential responses of macro‐ and microorganisms to land use and climate change (Guerra, Delgado‐Baquerizo, et al., 2021). Despite the vast global climatic gradients, soil pH appears to be the main driver of microbial diversity. For example, fungal richness peaks at weakly acidic soils (Tedersoo, Bahram, Põlme, et al., 2014; Tedersoo et al., 2022), whereas bacteria and protists are highest in neutral soils (Aslani et al., 2022; Bahram et al., 2018; Delgado‐Baquerizo, Oliverio, et al., 2018). Another study that did not measure pH found that the fungal diversity peaked at lower temperatures, but dominant fungal species were most‐influenced by mean annual temperature (Větrovský et al., 2019). As for plants and animals (Foden et al., 2019; Pacifici et al., 2015), soil fungal communities are likely vulnerable to global change drivers. For instance, high temperature stress (Barcenas‐Moreno et al., 2009; Malcolm et al., 2008; Misiak et al., 2021) and prolonged drought (de Vries et al., 2018; Schmidt et al., 2018) can alter fungal growth, functionality and community composition. Likewise, changes in land use that result in habitat fragmentation may lead to shifts in the prevalence of pathogenic, mutualistic and free‐living fungal groups (Brinkmann et al., 2019; Le Provost et al., 2021; Makiola et al., 2019; Rodriguez‐Ramos et al., 2021).
Compared with macroorganisms, information about the conservation needs of fungi and other microbes is very limited. While thousands of plant and animal species are listed as threatened on the International Union for Conservation of Nature (IUCN) global Red List, only 262 out of an estimated 2.2–3.8 million fungal species (Hawksworth & Lücking, 2017) have been listed as such. The majority of these are from high‐income countries in temperate regions (IUCN, 2021) and are from fungal groups that form conspicuous macroscopic fruiting bodies (Cao et al., 2021). However, the vast majority of fungi produce no, microscopic or inconspicuous fruiting bodies and are therefore difficult to survey using traditional morphology‐ and culturing‐based identification methods, which have hampered assessments of their distribution (Gonçalves et al., 2021). Consequently, we have no information for which parts of the world fungal conservation needs are the highest.
Here we used the most advanced high‐resolution sequencing technology to globally survey soil fungi and assess their endemicity and vulnerability to global change. We hypothesized that (i) as observed for plants and animals (Rosauer & Jetz, 2015), the endemicity of fungi is relatively higher in the tropics due to greater regional‐scale climatic stability and also coincides with that of plants and animals and (ii) the vulnerability of fungi to global change is highest in habitats experiencing the strongest warming effects (e.g., polar regions) and intensive land use (e.g., dry tropics). We predicted that endemicity and vulnerability patterns are more evident for macrofungi that require abundant resources for building reproductive structures and for obligately biotrophic groups compared with saprotrophic microfungal groups. From these findings, we identify regions where fungal conservation may be the most warranted, and propose global conservation priorities.
2. MATERIALS AND METHODS
2.1. Data sets
To study fungal endemicity and vulnerability to global change, we combined data from the Global Soil Mycobiome consortium (GSMc) open data set (Tedersoo, Mikryukov, et al., 2021) with materials from five other global soil biological surveys (Figure 1)—BIODESERT (Maestre et al., 2022), MUSGONET (including the natural sites in Delgado‐Baquerizo et al., 2021), CLIMIFUN (Bastida et al., 2021), GlobalAM (Davison et al., 2021), GlobalWetlands (Bahram et al., 2022) as well as Sanger sequence data from soil‐inhabiting fungi obtained from the UNITE database (Nilsson et al., 2019) covering GenBank. We obtained soil DNA from these five surveys and performed new DNA metabarcoding analyses following the protocols outlined for the GSMc data set (Tedersoo, Mikryukov, et al., 2021).
FIGURE 1.
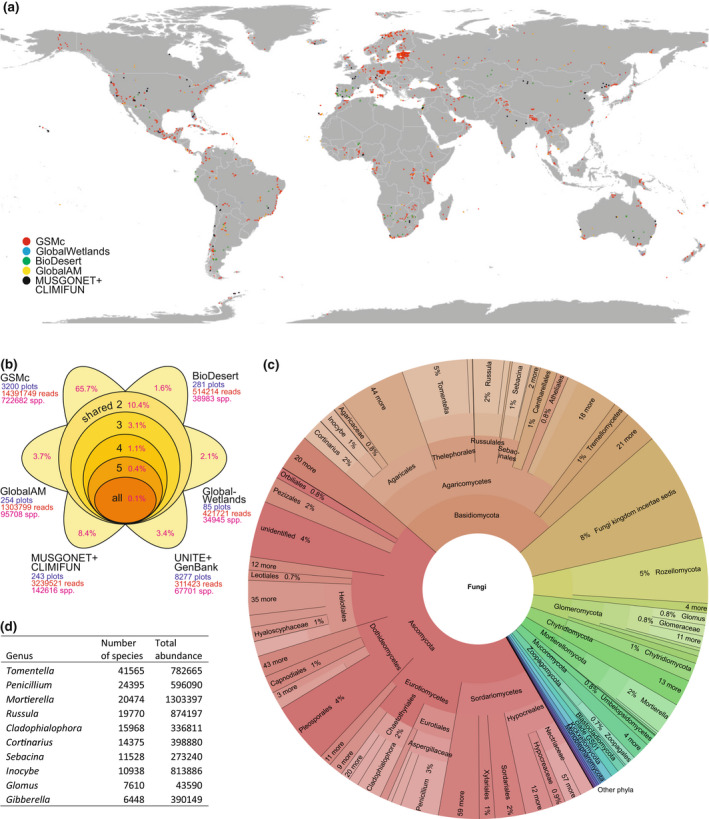
Distribution of samples and fungal species across data sets. (a) Global sampling map, with different colours representing different data sets; (b) species distribution of fungi among data sets, with the proportion of unique and shared species indicated; (c) Krona chart indicating the taxonomic distribution of fungal species (interactive chart can be browsed at https://plutof.ut.ee/#/doi/10.15156/BIO/2483900); and (d) species richness and total read abundance of the top 10 most diverse fungal genera.
All data sets included information on geographical coordinates and soil pH. Based on geographical coordinates, we assigned the following climatic and land‐cover metadata to the samples: (i) CHELSA v2.1 bioclimatic variables for the period 1981–2010 (Karger et al., 2017), (ii) CHELSA‐TraCE21k v1.0. for the last glacial maximum (LGM; Karger et al., 2021), and (iii) CHELSA v2.1 climate extrapolations for the year 2070 following the RCP8.5 global warming scenario with SSP5 socioeconomic conditions and the GFDL‐ESM4 global circulation model (Karger et al., 2020); (iv) normalized difference vegetation index (NDVI; Filipponi et al., 2018); (v) SoilGrids v.2 soil pH from 0 to 5 cm depth (Poggio et al., 2021); (vi) land‐cover type using Copernicus classification v.3 (Buchhorn et al., 2020) for the year 2015; and (vii) human footprint index based on the Land‐Use Harmonization (LUH2; Hurtt et al., 2020) for the year 2015 extrapolation. Based on original descriptions of vegetation (age, cover, relative abundance of species, fire history) or remote sensing data (Google Earth Pro; https://earth.google.com/), samples were assigned to biomes (Olson et al., 2001) and land‐cover types. Based on z‐transformed differences in all 19 present and LGM bioclimatic variables, we calculated for each sample an averaged LGM climate change index. Furthermore, for each sample we estimated the human footprint index as the cumulative sum of land‐use state transitions, with the year 1960 used as a baseline.
2.2. Molecular analysis and functional assignments
To infer fungal species and taxonomy, we used a long‐read sequencing approach involving the ribosomal RNA 18S gene V9 subregion, internal transcribed spacer 1 (ITS1), 5.8S gene and ITS2 to enhance taxonomic resolution and accuracy. We used degenerate, universal eukaryotic primers to cover as many divergent taxa within the fungi and micro‐eukaryotes as possible (Tedersoo, Albertsen, et al., 2021). The amplicon samples were prepared in 82 PacBio SMRTbell sequencing libraries and sequenced on 48 PacBio Sequel 8M SMRT cells. The obtained reads were quality‐filtered, demultiplexed to samples, trimmed to include only the full‐length ITS region, and clustered to operational taxonomic units (conditionally termed as species) at 98% sequence similarity, which roughly corresponds to species‐level divergence. Taxonomy was assigned based on information from the 10 best BLASTn matches against the UNITE 9.1 beta data set (https://doi.org/10.15156/BIO/1444285). The resulting species‐by‐sample matrices were manually checked library‐wise for external and cross‐contamination and rates of index switching artefacts. We excluded several samples for which we suspected contamination, and removed rare occurrences of dominant species using the following thresholds: abundances = 1 for species with total abundance of >99 and abundances = 2 for species with total abundance of >999.
Based on FungalTraits 1.3 (Põlme et al., 2020), species belonging to the kingdom Fungi were assigned to functional groups based on ecological or physiological characters: (i) arbuscular mycorrhizal (AM) fungi (including all Glomeromycota but excluding all Endogonomycetes, owing to the paucity of information distinguishing AM species from free‐living species); (ii) EcM fungi (excluding dubious lineages); (iii) non‐EcM Agaricomycetes (mostly saprotrophic fungi with usually macroscopic fruiting bodies); (iv) molds (including Mortierellales, Mucorales, Umbelopsidales and Aspergillaceae and Trichocomaceae of Eurotiales and Trichoderma of Hypocreales); (v) putative pathogens (including plant, animal and fungal pathogens as primary or secondary lifestyles); (vi) opportunistic human parasites (OHPs; excluding Mortierellales); (vii) yeasts (excluding dimorphic yeasts); and (viii) other unicellular (non‐yeast) fungi (including chytrids, aphids, rozellids, and other early‐diverging fungal lineages). Other groups such as lichen‐forming fungi were not considered, owing to their relative infrequency in the soil samples and across ecoregions. Among these groups, mostly non‐EcM Agaricomycetes and EcM fungi include many red‐listed species of macrofungi, with fruiting bodies that are conch‐shaped (polypores), resupinate (corticioids), or stipitate (agarics, boletes, etc.) and are hence considered to be of higher conservation interest because of their charismatic appearance (Cao et al., 2021; IUCN, 2021). Here, we test whether this approach is also justified from the endemicity and vulnerability perspectives.
2.3. Endemicity
To infer endemicity patterns in fungi, samples were assigned to ecological regions (Olson et al., 2001) based on their geographical coordinates, allowing a 10‐km buffer zone between terrestrial ecological regions and water (due to low resolution of the map layers in shore areas). Based on climatic and floristic similarities and data availability (Kier et al., 2009), the ecological regions were further subjectively aggregated into larger areas or split into smaller, geographically distinct units, which we refer to as ecoregions (Figure 2; Table S1). Each of the 174 ecoregions covered 1–45 soil samples, with surplus samples excluded randomly from two intensively sampled ecoregions. The ecoregions were allowed to include unlimited Sanger‐sequenced sites from UNITE. We distinguished the ecoregions located on islands and determined their minimum distance to nearest continents or larger islands using the Google Earth Pro ground distance measurement tool.
FIGURE 2.
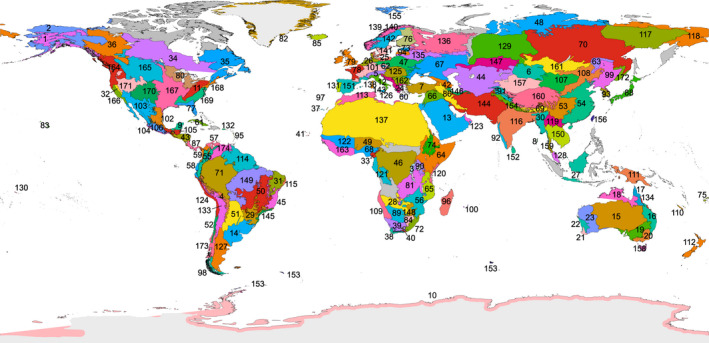
Distribution of 174 ecoregions used in endemicity analyses. Ecoregions excluded from the analyses due to the lack of data are indicated in grey. Their definition and relationship with Olson's ecological regions are given in Table S1.
Using the betapart package v.1.5.4 (Baselga & Orme, 2012) of R v.4.1.10 (R Core Team, 2021), five indices of endemism—the number of endemic species (weight = 16.7%), proportion of endemic species (weight = 16.7%), mean maximum geographical range of taxa (weight = 33.3%), Jaccard index (weight = 16.7%) and beta‐sim index (weight = 16.7%)—were calculated based on the community matrix (Crisp et al., 2001; Villéger & Brosse, 2012; Box S1). The first two and the last two indices reflect similar aspects of endemicity and were therefore downweighed when averaging these indices based on z‐score transformation. To account for differences in sampling intensity, we calculated residuals for the numbers of all species and endemic species by regressing these against the logarithmically transformed number of samples and sequencing depth (Table S2).
Of the indices used, only the number of endemic taxa and proportion of endemic taxa were significantly positively correlated with fungal species richness (r = .707 and r = .212, respectively). Furthermore, species richness was not included among the best predictors of averaged endemicity, indicating that these metrics are independent. Endemicity indices of all fungi and functional groups were subjected to random forest machine learning analysis to pre‐select the 10 most important environmental variables for general linear modelling (GLM). We used the variance (as coefficient of variation) and averaged values of bioclimatic variables, area, latitude, longitude, altitude, and soil pH as well as continents (dummy variables) to explain endemicity. GLMs were fitted using second‐order polynomial terms for continuous variables. Only significant variables (p < .050; r 2 > .020) were kept in the final models. Based on the predictions revealed by GLMs, endemicity maps were constructed using the sf v.1.0.5 package (Pebesma, 2018) of R.
2.4. Global change vulnerability
Fungal vulnerability was estimated based on high‐throughput sequencing data at the soil sample level. The vulnerability of soil fungal groups was estimated relative to three global change drivers, viz. heat (maximum monthly temperature), drought (negative of inverse hyperbolic sine‐transformed precipitation in the driest quarter) and land cover change, for the year 2070 (relative to a 2015 baseline) using the community‐mean percentile vulnerability index (V 2; Smith, Jovan, et al., 2020). This index is based on averaging percentiles of all species at a given global change driver value.
where is the presence (0 or 1) of species j at site i, F j (x i ) the percentile of species j given site parameter value x i , x i the parameter value of site i and n the total number of species observed. Precipitation in the driest quarter was selected as a proxy for drought because bioclimatic variables cover larger areas (including islands) and offer greater resolution compared with other measures of soil water content and indicators of drought. The vulnerability scores were calculated for each soil sample using the vuln v.0.0.05 (Smith, Jovan, et al., 2020) package of R. We also constructed the average vulnerability score by equally weighing all components based on z‐scores. The vulnerability scores were unrelated to sequencing depth and sample size.
We performed a similar random forest and GLM modelling exercise to determine the main predictors of vulnerability as described above but allowed two‐way interaction terms between categorical and continuous predictors and used a more relaxed threshold for retaining variables in the model (p < .001; R 2 > .01) due to greater sample size. To predict vulnerability scores for each global driver and estimate their prediction uncertainty, thin plate splines (basis dimensionality = 3) were fitted using a generalized additive model with the mgcv v.1.8–38 (Wood, 2011) package of R. To incorporate the spatial autocorrelation signal, we calculated residuals at the sampling sites and used inverse distance weighting to interpolate residuals beyond the sampling sites. To obtain final vulnerability predictions, interpolated residuals were added to the results based on the predicted regression part (Hengl & MacMillan, 2019). By using the relative vulnerability values, we also prepared the map of fungal vulnerability ascribed to each of the three components. Vulnerability maps were visualized using the raster v.3.5–9 (Hijmans et al., 2021) package of R.
The maps for conservation priorities were calculated for all fungi using the same sampling points used for the vulnerability analyses, except for points corresponding to cropland and urban and village land cover types. For each sampling point, the respective average endemicity, γ‐diversity and vulnerability scores were z‐transformed, followed by adding a constant (5, to exclude negative values), multiplied (to downweight areas with any low values), and used in a regression approach (Table S3) as described for vulnerability.
3. RESULTS AND DISCUSSION
3.1. General findings
We used the recently generated Global Soil Mycobiome consortium data set (GSMc; 3200 plots, Tedersoo, Mikryukov, et al., 2021) along with data from five other global soil surveys (Figure 1) and international nucleotide sequence databases to determine the diversity and endemicity of fungal functional groups—viz. AM fungi, ectomycorrhizal (EcM) fungi, non‐EcM Agaricomycetes (mostly saprotrophic macrofungi), molds, pathogens, OHPs (mostly thermophilic saprotrophs), early‐diverging unicellular lineages and yeasts. Compared with previous meta‐analytical approaches (e.g., Větrovský et al., 2019), our cumulative data comprise the largest available globally standardized database based on directly comparable soil sampling and long‐read molecular analysis protocols. Collectively, all data sets yielded 20,182,427 fungal reads composed of 905,841 “species”—operational taxonomic units (OTUs), each defined at <98% sequence similarity of the rRNA ITS barcode from all other OTUs. Tomentella (Basidiomycota), Penicillium (Ascomycota), and Mortierella (Mortierellomycota) were the most species‐rich genera recorded (Figure 1).
3.2. Fungal endemicity
To estimate relative endemism among the world's ecoregions (Figure 2, Table S4), we combined indices of community similarity, uniqueness, and species ranges into an overall endemicity index. We found that the endemicity of all fungi peaked in tropical rainforest and tropical woodland biomes of Amazonia (including cerrado), Yucatan, West‐Central Africa, Sri Lanka, and New Caledonia (Figure 3). Endemicity was positively related to mean annual air temperature (MAT; R 2 adj = .277; Figure 3) and soil acidity (R 2 adj = .108; Table S4).
FIGURE 3.
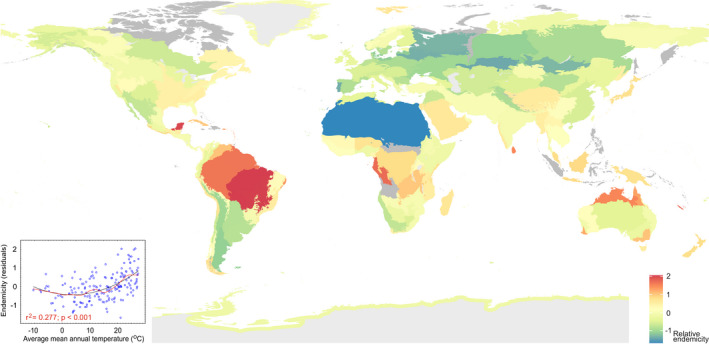
Average endemicity of soil fungi across ecoregions (defined in Figure 2). The inset graph indicates that endemicity has a positive and U‐shaped relationship with mean annual air temperature, the best predictor of endemicity. Grey ecoregions were excluded because of insufficient data.
While endemicity patterns of non‐EcM Agaricomycetes and AM fungi were similar to those shown for all fungi, different patterns were found for other functional groups (Figure S1). Endemicity of EcM fungi was related to high mean annual precipitation (R 2 adj = .147), with peaks in moist and dry tropical forests and temperate rainforests of Patagonia and New Zealand. Molds had relatively high endemicity in Amazonia, whereas pathogens and yeasts showed multiple endemicity hotspots. Molds (R 2 adj = .199) and pathogens (R 2 adj = .105) had relatively greater endemicity in strongly acidic or alkaline soils, indicating that extreme soil conditions may support unique soil biota, with limited effective dispersal across edaphically extreme habitats (Figure S1). Human footprint (see Section 2) had a weak negative effect on endemicity of all fungi (R 2 adj = .018), pathogens (R 2 adj = .015) and OHPs (R 2 adj = .056), suggesting that anthropogenic habitat loss or homogenization may affect endemic species (Finderup Nielsen et al., 2019). European ecoregions had the lowest endemicity for all fungi (R 2 adj = .065), pathogens (R 2 adj = .086) and unicellular fungi (R 2 adj = .035) compared with those of other subcontinents. Averaged current aerial bioclimatic variables better explained endemicity compared with the ranges of those variables or bioclimatic variables of soil and LGM. Climate change since the LGM had a weak positive effect on endemicity of molds (mean diurnal range and overall climate change: R 2 adj = .073) and OHPs (bioclimatic variables climate isothermality and mean diurnal range: R 2 adj = .056; Table S4).
We found that patterns in fungal endemicity were relatively consistent among the five individual endemicity indices (Figure S1) and that, in agreement with our first hypothesis, they resembled endemicity patterns of vascular plants and animals, which exhibit major hotspots in wet tropical habitats (Barlow et al., 2018; Kier et al., 2009). However, in a striking contrast to plants and animals (Kier et al., 2009), fungal endemicity showed no detectable relationship with insularity (Figure 4). This lack of insularity effect may reflect the greater long‐distance dispersal capacity of fungal spores relative to plant propagules and animals (Golan & Pringle, 2017). The negligible effects of soil nutrients on fungal diversity (Tedersoo, Bahram, Põlme, et al., 2014; Tedersoo, Bahram, Ryberg, et al., 2014) and endemicity (this study) suggest that nutrient availability may not limit fungal growth or promote speciation.
FIGURE 4.
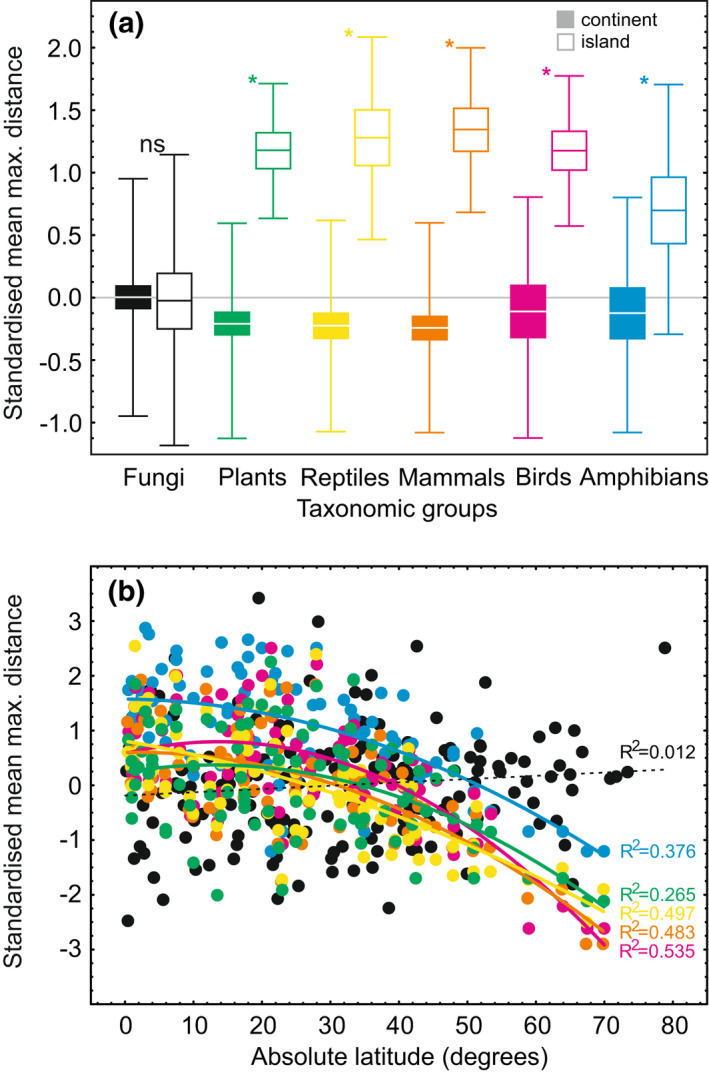
Endemicity patterns of fungi compared with plants and various vertebrate groups: (a) insularity effect and (b) latitudinal distribution. The analyses are based on world's ecoregions that differ for fungi compared with plants and animals (Kier et al., 2009). For all data sets, the mean maximum distance of all species was available and therefore taken as a measure of relative endemicity. For comparative purposes, these values were logarithm‐transformed, corrected for sequencing and sampling depth (only fungi) and z‐transformed. In (a) lines, boxes and error bars represent mean, SE and SD, respectively. In (b) the endemicity values were regressed against latitude using second‐order polynomial function; colours represent different organism groups as indicated in (a). For fungi, n = 156 and n = 28 for continent and island ecoregions, respectively; for other groups, n = 76 and n = 14 for continent and island ecoregions, respectively.
The literature abounds with hypotheses, including narrower niche breadth, more asymmetric biotic interactions (i.e., greater specialization to symbiotic partners), long‐term climatic stability (LGM and earlier epochs), and more rapid evolution due to environmental energy (metabolic hypothesis; Brown, 2014; Vázquez & Stevens, 2004), to explain the greater macroorganism richness and endemicity found in the tropics than elsewhere. The observed negligible effects of the LGM suggest that climatic stability is not an important driver of fungal endemicity, a pattern which contrasts with that of plants and animals (Rosauer & Jetz, 2015). The greater phylogenetic diversity and mean neighbour taxonomic distance of fungi noted for the tropics (Tedersoo et al., 2018) reflect tropical origins for many lineages, as well as extensive radiation and rapid speciation of certain genera in higher latitude areas (Kennedy et al., 2012; Sánchez‐Ramírez et al., 2015; Tedersoo, Bahram, Ryberg, et al., 2014). On a global scale, plant diversity does not appear to be causally related to fungal diversity (Tedersoo, Bahram, Põlme, et al., 2014), but there is some evidence for stronger mutualistic plant–fungal interactions related to high rainfall (Põlme et al., 2018). Pathogenic interactions warrant further research in this respect, given their major importance as regulators of plant diversity (Chen et al., 2019). Tropical soil fungi have relatively greater dispersal limitations (Bahram et al., 2013) and narrower distribution ranges (this study), suggesting that high local diversity may contribute to greater regional‐scale endemicity.
3.3. Vulnerability to global change drivers
We evaluated the relative vulnerability of soil fungal functional groups by estimating the percentage of species occurring at their upper niche limits to three major global change drivers—land use (land cover change), heat (maximum monthly temperature), and drought (lowest quarterly precipitation). For all fungi taken together, predicted vulnerability to heat (best predictor: maximum monthly temperature; R 2 adj = .583) and drought (precipitation seasonality; R 2 adj = .456) were greatest in the drylands of tropical and subtropical latitudes. Vulnerability to land use change (best predictor: climate isothermality; R 2 adj = .145) peaked in the tropics (Figure S2; Table S5). The overall additive global change vulnerability was, thus, the highest in densely populated and drier tropical and subtropical regions, especially in India and the sub‐Saharan Sudanian savanna (Figure 5), and was positively associated with temperature‐related variables (cumulative R 2 adj = .382). Fungal functional groups had similar vulnerability patterns, which were mostly related to temperature (Figures S2 and S3; Table S5). Among fungal groups, average vulnerability scores were highest for AM and EcM symbionts and unicellular fungi, but these scores differed only slightly across the global change drivers (Figure 5c). The actual vulnerability was probably underestimated for biotrophic pathogens and EcM fungi because these groups associate with a limited number of plant species and are sometimes host‐specific (Kennedy et al., 2015). Therefore, the loss of one of only a few key symbiotic partners may greatly reduce the biotic niches of specialist fungi.
FIGURE 5.
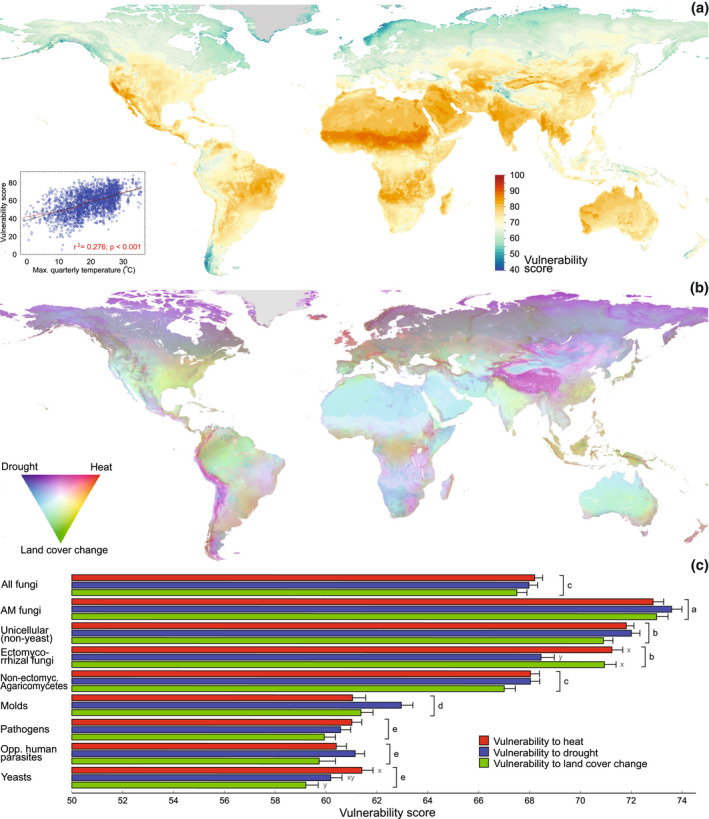
Vulnerability of fungi to global change drivers: (a) average vulnerability of all fungi to drought, heat and land cover change. The inset shows the near‐linear relationship of vulnerability to the air temperature of the warmest quarter. (b) Relative importance of predicted vulnerability of all fungi to drought, heat and land cover change as indicated by colour mixes. For example, the purple colour indicates high vulnerability to both heat and drought but low vulnerability to land cover change. (c) Relative importance of global change drivers for the predicted vulnerability of soil fungi and functional groups. Different letters indicate statistically significant (p < .001) differences among functional groups (a–e) and among global change drivers within functional groups (x–z). Mapping error estimates for panel (a) are given in Figure S4.
Patterns of vulnerability in fungi are somewhat similar to those of terrestrial plants and animals, and in agreement with our second hypothesis, vulnerability peaks in drylands prone to desertification (Warren et al., 2013), Arctic/alpine areas (cold‐adapted species) and regions with dense human populations (Watson et al., 2013). The relatively low vulnerability to heat in tundra‐inhabiting fungi can be explained by their relatively high‐temperature optima (Maynard et al., 2019; but see Misiak et al., 2021), acclimation (Romero‐Olivares et al., 2017) and poleward migration potentials, despite relatively greater predicted warming in Arctic ecosystems. Above certain tolerance thresholds, soil organisms may be physiologically constrained by increasing soil temperature and evaporation, lower soil water potentials and loss of oxygen due to greater respiration and faster decomposition, which result in hampered soil functioning and ecosystem multifunctionality (Delgado‐Baquerizo et al., 2017). Open areas are predicted to increase due to climate change and human activities. This will further expose soil to solar radiation and result in erosion and the loss of plant hosts for fungi. Losses of soil carbon and nutrients, but also anthropogenic nitrogen deposition, may exacerbate fungal vulnerability. These drivers are expected to be more influential locally (e.g., Correia et al., 2021; van der Linde et al., 2018) and are partly accounted for by land cover change in this study. Furthermore, while here we calculated average vulnerabilities by summing the effects of individual drivers, global change impacts tend to be synergistic (Rillig et al., 2019), so actual vulnerabilities may be much higher.
3.4. Implications for conservation
Most fungi and soil organisms do not enjoy the protection and conservation measures that are afforded to more charismatic animals and plants (Ducarme et al., 2013). Nonetheless, fungi and other soil biota are pivotal to soil and plant health, carbon and nutrient cycling, water storage, food security and many other ecosystem services. Their biodiversity should, hence, be brought to the focus of global sustainability thinking and conservation planning. For example, these organisms should be factored in when selecting protected areas, which are otherwise based on the distributions of plants or animals (Guerra, Bardgett, et al., 2021). The fact that many EcM and plant pathogenic fungal species are associated with specific host plants indicates that on the local scale it is not only the narrowly distributed species, but also those with unique or specific biotic associations, which require focused conservation measures. From the fungal perspective, it is particularly important to protect plant species that act as hubs in modules of biotic interaction networks, because these hub species typically associate with multiple, distinct fungal partners (Põlme et al., 2018). In other cases, certain unique plant species or higher taxonomic groups should be prioritized. For example, in southern South America, the drought‐sensitive tree family Nothofagaceae is the only group known to support EcM fungi that are mostly endemic to the region (Godoy & Marin, 2019).
Although the vulnerability scores to environmental change differed among fungal groups, their overall global patterns were similar. This suggests that broad habitat conservation measures may work for most fungal groups, including the macroscopic non‐EcM Agaricomycetes and EcM fungi as well as more cryptic pathogens and other groups. To accomplish this, fungi need to be incorporated into conservation frameworks (Gonçalves et al., 2021). Actions to fill existing information gaps at the local and global levels must also be taken, and global‐scale surveys should account for soil biodiversity assessments, complementing traditional collections‐based assessment with metabarcoding of environmental DNA. For fungi—and many other soil organisms—analyses of distribution and conservation requirements are now feasible based on large‐scale molecular surveys that can be highly standardized and rapidly performed. Such studies can distinguish cryptic species and ameliorate the enormous data coverage biases in GBIF and other global databases. Data produced by such studies can then be used in national conservation programs and global policy‐making initiatives, such as the System of Environmental Economic Accounting of the United Nations, World Biodiversity Forum and Post‐2020 Global Biodiversity Framework. Furthermore, promoting the red‐listing of endangered fungal species at the national and global levels is critical (FAO, 2020; IUCN, 2021). Fungi need active and specific inclusion in national and global conservation policies and strategies, not just passive and implicit protection.
Our study provides evidence that soil fungi may be highly vulnerable to global change, which needs to be considered when planning how to preserve these ecologically pivotal organisms in a changing world. As with plants and animals, fungi appear to be sensitive to the strong impacts of land cover change, low moisture and high temperatures on the taxonomic and functional composition of communities (Brinkmann et al., 2019; Makiola et al., 2019; this study). The endemicity of fungi is highest in tropical forest biomes, so conservation measures advocated for tropical plants and animals (Barlow et al., 2018; Brooks et al., 2006) are likely to conserve fungi. Tropical forests are under continued threat from deforestation and degradation driven by expanding agriculture, extractive industries and infrastructural projects (Bebbington et al., 2018). Conservation of herbaceous wetlands, tropical rainforests and tropical woodlands is supported by our global fungal conservation priority map that accounts for endemicity, vulnerability and γ‐diversity (Figure 6). Additionally, given the importance of soil pH for soil microbial diversity and composition (Aslani et al., 2022; Bahram et al., 2018; Delgado‐Baquerizo, Oliverio, et al., 2018; Tedersoo, Bahram, Põlme, et al., 2014; Tedersoo, Bahram, Ryberg, et al., 2014; Tedersoo et al., 2022), it is essential to prioritize areas with high pedodiversity or mixed landscapes including bogs, various forest types and grasslands. As a crucial measure, desertification and loss of soil organic matter need to be controlled by reducing the conversion of primary forest to crops and pasture (Smith, Calvin, et al., 2020). This is important, not only to prevent land degradation processes from impairing the diversity of fungi and other soil biota (Bach et al., 2020; Guerra, Bardgett, et al., 2021), but also to sustain the capacity of drylands to provide essential ecological functions and ecosystem services, such as soil fertility, carbon storage and food production for more than one billion people (Delgado‐Baquerizo, Eldridge, et al., 2018; Sivakumar, 2007).
FIGURE 6.
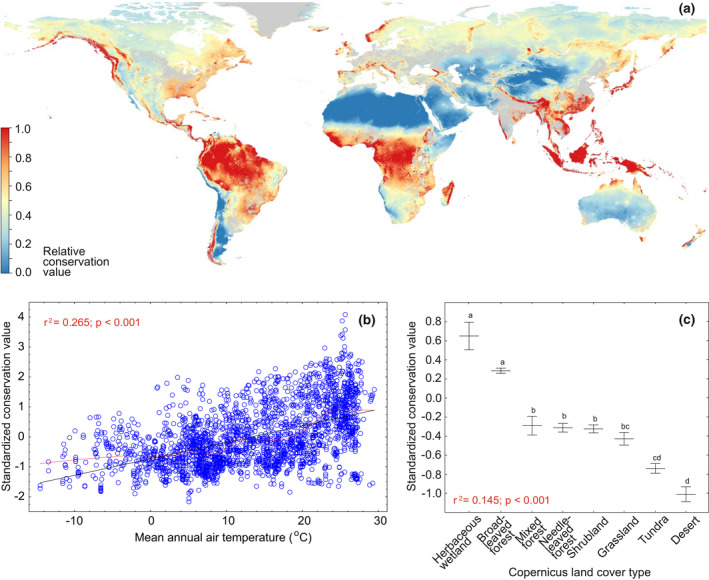
Conservation priority areas for all fungi (a) and their relationships with mean annual temperature (b) and Copernicus land cover types (c). In (b), black and red lines indicate best‐fitting linear and lowess functions, respectively. In (c), central lines and whiskers indicate mean and standard errors, respectively; letters above whiskers indicate statistically significant differences among land cover types.
4. CONCLUSIONS
Soil fungi show strong endemicity patterns, which differ between functional groups and are driven by both climatic and edaphic factors. Fungal groups also differ strongly in their relative vulnerability scores to global change, which peak in heavily populated dryland areas. Unfortunately, these are the very areas most prone to further land degradation and desertification, with the potential loss of many species. Fungal endemicity and vulnerability patterns only partly mirror those of vascular plants and animals, with virtually no insularity effects detected among fungi—a pattern that may be ascribed to their more efficient dispersal mechanisms. Global conservation efforts should include fungal biodiversity surveys alongside assessments of soil health, below‐ and aboveground feedbacks and areas of highest conservation priority, to secure the protection of keystone host species and natural habitats. Furthermore, they should include the monitoring of regional fungal communities over time, to detect relevant changes and provide early warning signals of impending change. The analysis for soil fungi presented here strongly suggests that microorganisms, and soil organisms in general, also deserve detailed assessments of their geographic distributions and conservation needs.
AUTHOR CONTRIBUTIONS
Leho Tedersoo, Vladimir Mikryukov, Alexander Zizka, Mohammad Bahram, Nadejda A. Soudzilovskaia, Alexandre Antonelli, Urmas Kõljalg, and Kessy Abarenkov designed the study; Vladimir Mikryukov, Leho Tedersoo, Alexander Zizka, Marijn Bauters, Niloufar Hagh‐Doust, Sten Anslan and Oleh Prylutskyi analysed data; Leho Tedersoo, Manuel Delgado‐Baquerizo, Fernando T. Maestre, Jaan Pärn, Maarja Öpik, Mari Moora, Martin Zobel, Mikk Espenberg, and Ülo Mander contributed DNA extracts from global surveys; other authors contributed materials, data and/or chemical analyses; Leho Tedersoo wrote the first draft, and all authors contributed to the writing of the article.
CONFLICT OF INTEREST
The authors declare no competing interests.
Supporting information
Appendix S1
Figure S1
Table S1
Table S2
Table S3
ACKNOWLEDGMENTS
The full acknowledgements are provided in Table S6. The bulk of the funding is derived from the Estonian Science Foundation (grants PRG632, PRG1170, PRG1615, MOBTP198), EEA Financial Mechanism Baltic Research Programme (EMP442), and Novo Nordisk Fonden (NNF20OC0059948). All collected soil samples are preserved in collection of DNA and environmental samples of University of Tartu Natural History Museum.
Tedersoo, L. , Mikryukov, V. , Zizka, A. , Bahram, M. , Hagh‐Doust, N. , Anslan, S. , Prylutskyi, O. , Delgado‐Baquerizo, M. , Maestre, F. T. , Pärn, J. , Öpik, M. , Moora, M. , Zobel, M. , Espenberg, M. , Mander, Ü. , Khalid, A. N. , Corrales, A. , Agan, A. , Vasco‐Palacios, A.‐M. … Abarenkov, K. (2022). Global patterns in endemicity and vulnerability of soil fungi. Global Change Biology, 28, 6696–6710. 10.1111/gcb.16398
DATA AVAILABILITY STATEMENT
The data supporting the results is archived in Zenodo (https://doi.org/10.5281/zenodo.6983158 and https://doi.org/10.5281/zenodo.7027315) and PlutoF (https://doi.org/10.15156/BIO/2263453). The scripts for analyses are available via GitHub (https://github.com/Mycology‐Microbiology‐Center/Fungal_Endemicity_and_Vulnerability).
REFERENCES
- Aslani, F. , Geisen, S. , Ning, D. , Tedersoo, L. , & Bahram, M. (2022). Towards revealing the global diversity and community assembly of soil eukaryotes. Ecology Letters, 25, 65–76. [DOI] [PubMed] [Google Scholar]
- Bach, E. M. , Ramirez, K. S. , Fraser, T. D. , & Wall, D. H. (2020). Soil biodiversity integrates solutions for a sustainable future. Sustainability, 12, 2662. [Google Scholar]
- Bahram, M. , Hildebrand, F. , Forslund, S. K. , Anderson, J. L. , Soudzilovskaia, N. A. , Bodegom, P. M. , & Bork, P. (2018). Structure and function of the global topsoil microbiome. Nature, 560, 233–237. [DOI] [PubMed] [Google Scholar]
- Bahram, M. , Koljalg, U. , Courty, P. E. , Diedhiou, A. G. , Kjøller, R. , Polme, S. , Ryberg, M. , Veldre, V. , & Tedersoo, L. (2013). The distance decay of similarity in communities of ectomycorrhizal fungi in different ecosystems and scales. Journal of Ecology, 101, 1335–1344. [Google Scholar]
- Bahram, M. , Espenberg, M. , Pärn, J. , Lehtovirta‐Morley, L. , Anslan, S. , Kasak, K. , & Mander, Ü. (2022). Structure and function of the soil microbiome underlying N2O emissions from global wetlands. Nature Communications, 13, 1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcenas‐Moreno, G. E. , Gomez‐Brandon, M. A. , Rousk, J. , & Bååth, E. (2009). Adaptation of soil microbial communities to temperature: Comparison of fungi and bacteria in a laboratory experiment. Global Change Biology, 15, 2950–2957. [Google Scholar]
- Barlow, J. , França, F. , Gardner, T. A. , Hicks, C. C. , Lennox, G. D. , Berenguer, E. , & Graham, N. A. (2018). The future of hyperdiverse tropical ecosystems. Nature, 559, 517–526. [DOI] [PubMed] [Google Scholar]
- Baselga, A. , & Orme, C. D. L. (2012). Betapart: An R package for the study of beta diversity. Methods in Ecology and Evolution, 3, 808–812. [Google Scholar]
- Bastida, F. , Eldridge, D. J. , García, C. , Kenny Png, G. , Bardgett, R. D. , & Delgado‐Baquerizo, M. (2021). Soil microbial diversity–biomass relationships are driven by soil carbon content across global biomes. The ISME Journal, 15, 2081–2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bebbington, A. J. , Humphreys Bebbington, D. , Sauls, L. A. , Rogan, J. , Agrawal, S. , Gamboa, C. , & Verdum, R. (2018). Resource extraction and infrastructure threaten forest cover and community rights. Proceedings of the National Academy of Sciences of the USA, 115, 13164–13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmann, N. , Schneider, D. , Sahner, J. , Ballauff, J. , Edy, N. , Barus, H. , & Polle, A. (2019). Intensive tropical land use massively shifts soil fungal communities. Scientific Reports, 9, 3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brook, B. W. , Sodhi, N. S. , & Bradshaw, C. J. (2008). Synergies among extinction drivers under global change. Trends in Ecology & Evolution, 23, 453–460. [DOI] [PubMed] [Google Scholar]
- Brooks, T. M. , Mittermeier, R. A. , Da Fonseca, G. A. , Gerlach, J. , Hoffmann, M. , Lamoreux, J. F. , & Rodrigues, A. S. (2006). Global biodiversity conservation priorities. Science, 313, 58–61. [DOI] [PubMed] [Google Scholar]
- Brown, J. H. (2014). Why are there so many species in the tropics? Journal of Biogeography, 41, 8–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchhorn, M. , Lesiv, M. , Tsendbazar, N. E. , Herold, M. , Bertels, L. , & Smets, B. (2020). Copernicus global land cover layers—Collection 2. Remote Sensing, 12, 1044. [Google Scholar]
- Cameron, E. K. , Martins, I. S. , Lavelle, P. , Mathieu, J. , Tedersoo, L. , Bahram, M. , & Eisenhauer, N. (2019). Global mismatches in aboveground and belowground biodiversity. Conservation Biology, 33, 1187–1192. [DOI] [PubMed] [Google Scholar]
- Cao, Y. , Wu, G. , & Yu, D. (2021). Include macrofungi in biodiversity targets. Science, 372, 1160. [DOI] [PubMed] [Google Scholar]
- Chen, L. , Swenson, N. G. , Ji, N. , Mi, X. , Ren, H. , Guo, L. , & Ma, K. (2019). Differential soil fungus accumulation and density dependence of trees in a subtropical forest. Science, 366, 124–128. [DOI] [PubMed] [Google Scholar]
- Correia, M. , Espelta, J. M. , Morillo, J. A. , Pino, J. , & Rodríguez‐Echeverría, S. (2021). Land‐use history alters the diversity, community composition and interaction networks of ectomycorrhizal fungi in beech forests. Journal of Ecology, 109, 2856–2870. [Google Scholar]
- Crisp, M. D. , Laffan, S. , Linder, H. P. , & Monro, A. (2001). Endemism in the Australian flora. Journal of Biogeography, 28, 183–198. [Google Scholar]
- Crowther, T. W. , Van den Hoogen, J. , Wan, J. , Mayes, M. A. , Keiser, A. D. , Mo, L. , Averill, C. , & Maynard, D. S. (2019). The global soil community and its influence on biogeochemistry. Science, 365, eaav0550. [DOI] [PubMed] [Google Scholar]
- Davison, J. , Moora, M. , Semchenko, M. , Adenan, S. B. , Ahmed, T. , Akhmetzhanova, A. A. , & Öpik, M. (2021). Temperature and pH define the realised niche space of arbuscular mycorrhizal fungi. New Phytologist, 231, 763–776. [DOI] [PubMed] [Google Scholar]
- Davison, J. , Moora, M. , Öpik, M. , Adholeya, A. , Ainsaar, L. , Bâ, A. , & Zobel, M. (2015). Global assessment of arbuscular mycorrhizal fungus diversity reveals very low endemism. Science, 349, 970–973. [DOI] [PubMed] [Google Scholar]
- de Vries, F. T. , Griffiths, R. I. , Bailey, M. , Craig, H. , Girlanda, M. , Gweon, H. S. , & Bardgett, R. D. (2018). Soil bacterial networks are less stable under drought than fungal networks. Nature Communications, 9, 3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado‐Baquerizo, M. , Eldridge, D. J. , Liu, Y. R. , Sokoya, B. , Wang, J. T. , Hu, H. W. , He, J. Z. , Bastida, F. , Moreno, J. L. , Bamigboye, A. R. , & Blanco‐Pastor, J. L. (2021). Global homogenization of the structure and function in the soil microbiome of urban greenspaces. Science Advances, 7, eabg5809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado‐Baquerizo, M. , Eldridge, D. J. , Ochoa, V. , Gozalo, B. , Singh, B. K. , & Maestre, F. T. (2017). Soil microbial communities drive the resistance of ecosystem multifunctionality to global change in drylands across the globe. Ecology Letters, 20, 1295–1305. [DOI] [PubMed] [Google Scholar]
- Delgado‐Baquerizo, M. , Eldridge, D. J. , Travers, S. K. , Val, J. , Oliver, I. , & Bissett, A. (2018). Effects of climate legacies on above‐ and belowground community assembly. Global Change Biology, 24, 4330–4339. [DOI] [PubMed] [Google Scholar]
- Delgado‐Baquerizo, M. , Oliverio, A. M. , Brewer, T. E. , Benavent‐González, A. , Eldridge, D. J. , Bardgett, R. D. , & Fierer, N. (2018). A global atlas of the dominant bacteria found in soil. Science, 359, 320–325. [DOI] [PubMed] [Google Scholar]
- Ducarme, F. , Luque, G. M. , & Courchamp, F. (2013). What are “charismatic species” for conservation biologists. BioSciences Master Reviews, 10, 1–8. [Google Scholar]
- Egidi, E. , Delgado‐Baquerizo, M. , Plett, J. M. , Wang, J. , Eldridge, D. J. , Bardgett, R. D. , & Singh, B. K. (2019). A few Ascomycota taxa dominate soil fungal communities worldwide. Nature Communications, 10, 2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO , ITPS , GSBI , SCBD , EC . (2020). State of knowledge of soil biodiversity – Status, challenges and potentialities . FAO. 10.4060/cb1928en. [DOI]
- Filipponi, F. , Valentini, E. , Nguyen Xuan, A. , Guerra, C. A. , Wolf, F. , Andrzejak, M. , & Taramelli, A. (2018). Global MODIS fraction of green vegetation cover for monitoring abrupt and gradual vegetation changes. Remote Sensing, 10, 653. [Google Scholar]
- Finderup Nielsen, T. , Sand‐Jensen, K. , Dornelas, M. , & Bruun, H. H. (2019). More is less: Net gain in species richness, but biotic homogenization over 140 years. Ecology Letters, 22, 1650–1657. [DOI] [PubMed] [Google Scholar]
- Foden, W. B. , Young, B. E. , Akçakaya, H. R. , Garcia, R. A. , Hoffmann, A. A. , Stein, B. A. , & Huntley, B. (2019). Climate change vulnerability assessment of species. Wiley Interdisciplinary Reviews: Climate Change, 10, e551. [Google Scholar]
- Godoy, R. , & Marin, C. (2019). Mycorrhizal studies in temperate rainforests of southern Chile. In Pagano M. C. & Lugo M. A. (Eds.), Mycorrhizal fungi in South America (pp. 315–341). Springer. [Google Scholar]
- Golan, J. J. , & Pringle, A. (2017). Long‐distance dispersal of fungi. Microbiology Spectrum, 5, 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves, S. C. , Haelewaters, D. , Furci, G. , & Mueller, G. M. (2021). Include all fungi in biodiversity goals. Science, 373, 403. [DOI] [PubMed] [Google Scholar]
- Guerra, C. A. , Bardgett, R. D. , Caon, L. , Crowther, T. W. , Delgado‐Baquerizo, M. , Montanarella, L. , & Eisenhauer, N. (2021). Tracking, targeting, and conserving soil biodiversity. Science, 371, 239–241. [DOI] [PubMed] [Google Scholar]
- Guerra, C. A. , Delgado‐Baquerizo, M. , Duarte, E. , Marigliano, O. , Görgen, C. , Maestre, F. T. , & Eisenhauer, N. (2021). Global projections of the soil microbiome in the Anthropocene. Global Ecology and Biogeography, 30, 987–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawksworth, D. L. , & Lücking, R. (2017). Fungal diversity revisited: 2.2 to 3.8 million species. Microbiology Spectrum, 5, 79–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengl, T. , & MacMillan, R. A. (2019). Predictive soil mapping with R . www.soilmapper.org (OpenGeoHub foundation, 2019).
- Hijmans, R. J. , Van Etten, J. , Mattiuzzi, M. , Cheng, J. , Sumner, M. , & Greenberg, J. A. (2021). Raster: Geographic data analysis and modeling. R package version 3.5–9. https://CRAN.R‐project.org/package=raster
- Hurtt, G. C. , Chini, L. , Sahajpal, R. , Frolking, S. , Bodirsky, B. L. , Calvin, K. , & Zhang, X. (2020). Harmonization of global land use change and management for the period 850–2100 (LUH2) for CMIP6. Geoscientific Model Development, 13, 5425–5464. [Google Scholar]
- IUCN . (2021). IUCN red list of threatened species . version 2021–2. IUCN, Gland. https://www.iucnredlist.org/
- Karger, D. N. , Conrad, O. , Böhner, J. , Kawohl, T. , Kreft, H. , Soria‐Auza, R. W. , Zimmermann, N. E. , Linder, H. P. , & Kessler, M. (2017). Climatologies at high resolution for the earth's land surface areas. Scientific Data, 4, 170122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karger, D. N. , Nobis, M. P. , Normand, S. , Graham, C. H. , & Zimmermann, N. E. (2021) CHELSA‐TraCE21k v1. 0. Downscaled transient temperature and precipitation data since the last glacial maximum. Climate Past Discussions, 2021, 30. [Google Scholar]
- Karger, D. N. , Schmatz, D. R. , Dettling, G. , & Zimmermann, N. E. (2020). High‐resolution monthly precipitation and temperature time series from 2006 to 2100. Scientific Data, 7, 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy, P. G. , Matheny, P. B. , Ryberg, K. M. , Henkel, T. W. , Uehling, J. K. , & Smith, M. E. (2012). Scaling up: Examining the macroecology of ectomycorrhizal fungi. Molecular Ecology, 21, 4151–4154. [DOI] [PubMed] [Google Scholar]
- Kennedy, P. G. , Walker, J. K. M. , & Bogar, L. M. (2015). Interspecific mycorrhizal networks and non‐networking hosts: Exploring the ecology of host genus Alnus . Ecologial Studies, 224, 227–254. [Google Scholar]
- Kier, G. , Kreft, H. , Lee, T. M. , Jetz, W. , Ibisch, P. L. , Nowicki, C. , & Barthlott, W. (2009). A global assessment of endemism and species richness across Island and mainland regions. Proceedings of the National Academy of Sciences of the USA, 106, 9322–9327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Provost, G. , Thiele, J. , Westphal, C. , Penone, C. , Allan, E. , Neyret, M. , Van Der Plas, F. , Ayasse, M. , Bardgett, R. D. , Birkhofer, K. , & Boch, S. (2021). Contrasting responses of above‐and belowground diversity to multiple components of land‐use intensity. Nature Communications, 12, 3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makiola, A. , Dickie, I. A. , Holdaway, R. J. , Wood, J. R. , Orwin, K. H. , & Glare, T. R. (2019). Land use is a determinant of plant pathogen alpha‐but not beta‐diversity. Molecular Ecology, 28, 3786–3798. [DOI] [PubMed] [Google Scholar]
- Maestre, F. T. , Eldridge, D. J. , Gross, N. , Le Bagousse‐Pinguet, Y. , Saiz, H. , Gozalo, B. , Ochoa, V. , & Gaitán, J. J. (2022). The BIODESERT survey: Assessing the impacts of grazing on the structure and functioning of global drylands. Web Ecology. In press. 10.5194/we-21-1-2021 [DOI] [Google Scholar]
- Malcolm, G. M. , Lopez‐Gutierrez, J. C. , Koide, R. T. , & Eissenstat, D. M. (2008). Acclimation to temperature and temperature sensitivity of metabolism by ectomycorrhizal fungi. Global Change Biology, 14, 1169–1180. [Google Scholar]
- Maynard, D. S. , Bradford, M. A. , Covey, K. R. , Lindner, D. , Glaeser, J. , Talbert, D. A. , & Crowther, T. W. (2019). Consistent trade‐offs in fungal trait expression across broad spatial scales. Nature Microbiology, 4, 846–853. [DOI] [PubMed] [Google Scholar]
- Misiak, M. , Goodall‐Copestake, W. P. , Sparks, T. H. , Worland, M. R. , Boddy, L. , Magan, N. , & Newsham, K. K. (2021). Inhibitory effects of climate change on the growth and extracellular enzyme activities of a widespread Antarctic soil fungus. Global Change Biology, 27, 1111–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson, R. H. , Larsson, K. H. , Taylor, A. F. S. , Bengtsson‐Palme, J. , Jeppesen, T. S. , Schigel, D. , & Abarenkov, K. (2019). The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Research, 47(D1), D259–D264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson, D. M. , Dinerstein, E. , Wikramanayake, E. D. , Burgess, N. D. , Powell, G. V. N. , Underwood, E. C. , & Kassem, K. R. (2001). Terrestrial ecoregions of the world: A new map of life on earth. Bioscience, 51, 933–938. [Google Scholar]
- Pacifici, M. , Foden, W. B. , Visconti, P. , Watson, J. E. , Butchart, S. H. , Kovacs, K. M. , & Rondinini, C. (2015). Assessing species vulnerability to climate change. Nature Climate Change, 5, 215–224. [Google Scholar]
- Pebesma, E. (2018). Simple features for R: Standardized support for spatial vector data. R Journal, 10, 439–446. [Google Scholar]
- Poggio, L. , De Sousa, L. M. , Batjes, N. H. , Heuvelink, G. , Kempen, B. , Ribeiro, E. , & Rossiter, D. (2021). SoilGrids 2.0: Producing soil information for the globe with quantified spatial uncertainty. The Soil, 7, 217–240. [Google Scholar]
- Põlme, S. , Abarenkov, K. , Henrik Nilsson, R. , Lindahl, B. D. , Clemmensen, K. E. , Kauserud, H. , & Tedersoo, L. (2020). FungalTraits: A user‐friendly traits database of fungi and fungus‐like stramenopiles. Fungal Diversity, 105, 1–16. [Google Scholar]
- Põlme, S. , Bahram, M. , Jacquemyn, H. , Kennedy, P. , Kohout, P. , Moora, M. , & Tedersoo, L. (2018). Host preference and network properties in biotrophic plant–fungal associations. New Phytologist, 217, 1230–1239. [DOI] [PubMed] [Google Scholar]
- Põlme, S. , Bahram, M. , Yamanaka, T. , Nara, K. , Dai, Y. C. , Grebenc, T. , & Tedersoo, L. (2013). Biogeography of ectomycorrhizal fungi associated with alders (Alnus spp.) in relation to biotic and abiotic variables at the global scale. New Phytologist, 198, 1239–1249. [DOI] [PubMed] [Google Scholar]
- R Core Team . (2021). R: A language and environment for statistical computing . R Foundation for Statistical Computing, Vienna. https://www.R‐project.org/
- Rillig, M. C. , Ryo, M. , Lehmann, A. , Aguilar‐Trigueros, C. A. , Buchert, S. , Wulf, A. , & Yang, G. (2019). The role of multiple global change factors in driving soil functions and microbial biodiversity. Science, 366, 886–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez‐Ramos, J. C. , Cale, J. A. , Cahill, J. F., Jr. , Simard, S. W. , Karst, J. , & Erbilgin, N. (2021). Changes in soil fungal community composition depend on functional group and forest disturbance type. New Phytologist, 229, 1105–1117. [DOI] [PubMed] [Google Scholar]
- Romero‐Olivares, A. L. , Allison, S. D. , & Treseder, K. K. (2017). Soil microbes and their response to experimental warming over time: A meta‐analysis of field studies. Soil Biology and Biochemistry, 107, 32–40. [Google Scholar]
- Rosauer, D. F. , & Jetz, W. (2015). Phylogenetic endemism in terrestrial mammals. Global Ecology and Biogeography, 24, 168–179. [Google Scholar]
- Sánchez‐Ramírez, S. , Etienne, R. S. , & Moncalvo, J. M. (2015). High speciation rate at temperate latitudes explains unusual diversity gradients in a clade of ectomycorrhizal fungi. Evolution, 69, 2196–2209. [DOI] [PubMed] [Google Scholar]
- Sandel, B. , Weigelt, P. , Kreft, H. , Keppel, G. , van der Sande, M. T. , Levin, S. , & Knight, T. M. (2020). Current climate, isolation and history drive global patterns of tree phylogenetic endemism. Global Ecology and Biogeography, 29, 4–15. [Google Scholar]
- Schmidt, P. A. , Schmitt, I. , Otte, J. , Bandow, C. , Römbke, J. , Bálint, M. , & Rolshausen, G. (2018). Season‐long experimental drought alters fungal community composition but not diversity in a grassland soil. Microbial Ecology, 75, 468–478. [DOI] [PubMed] [Google Scholar]
- Schulte to Bühne, H. , Tobias, J. A. , Durant, S. M. , & Pettorelli, N. (2021). Improving predictions of climate change–land use change interactions. Trends in Ecology & Evolution, 36, 29–38. [DOI] [PubMed] [Google Scholar]
- Sivakumar, M. V. K. (2007). Interactions between climate and desertification. Agricultural and Forest Meteorology, 142, 143–155. [Google Scholar]
- Smith, P. , Calvin, K. , Nkem, J. , Campbell, D. , Cherubini, F. , Grassi, G. , Korotkov, V. , Le Hoang, A. , Lwasa, S. , McElwee, P. , & Nkonya, E. (2020). Which practices co‐deliver food security, climate change mitigation and adaptation, and combat land degradation and desertification? Global Change Biology, 26, 1532–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, R. J. , Jovan, S. , & McCune, B. (2020). Climatic niche limits and community‐level vulnerability of obligate symbioses. Journal of Biogeography, 47, 382–395. [Google Scholar]
- Stein, A. , Gerstner, K. , & Kreft, H. (2014). Environmental heterogeneity as a universal driver of species richness across taxa, biomes and spatial scales. Ecology Letters, 17, 866–880. [DOI] [PubMed] [Google Scholar]
- Talbot, J. M. , Bruns, T. D. , Taylor, J. W. , Smith, D. P. , Branco, S. , Glassman, S. I. , & Peay, K. G. (2014). Endemism and functional convergence across the north American soil mycobiome. Proceedings of the National Academy of Sciences of the USA, 111, 6341–6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedersoo, L. , Albertsen, M. , Anslan, S. , & Callahan, B. (2021). Perspectives and benefits of high‐throughput long‐read sequencing in microbial ecology. Applied and Environmental Microbiology, 87, e00626‐21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedersoo, L. , Bahram, M. , Põlme, S. , Kõljalg, U. , Yorou, N. S. , Wijesundera, R. , & Abarenkov, K. (2014). Global diversity and geography of soil fungi. Science, 346, 1256688. [DOI] [PubMed] [Google Scholar]
- Tedersoo, L. , Bahram, M. , Ryberg, M. , Otsing, E. , Kõljalg, U. , & Abarenkov, K. (2014). Global biogeography of the ectomycorrhizal/sebacina lineage (fungi, Sebacinales) as revealed from comparative phylogenetic analyses. Molecular Ecology, 23, 4168–4183. [DOI] [PubMed] [Google Scholar]
- Tedersoo, L. , Mikryukov, V. , Anslan, S. , Bahram, M. , Khalid, A. N. , Corrales, A. , & Abarenkov, K. (2021). The global soil mycobiome consortium dataset for boosting fungal diversity research. Fungal Diversity, 111, 573–588. [Google Scholar]
- Tedersoo, L. , Mikryukov, V. , Zizka, A. , Bahram, M. , Hagh‐Doust, N. , Anslan, S. , & Abarenkov, K. (2022). Towards understanding diversity, endemicity and global change vulnerability of soil fungi. bioRxiv, 2022, 484796. [Google Scholar]
- Tedersoo, L. , Sánchez‐Ramírez, S. , Kõljalg, U. , Bahram, M. , Döring, M. , Schigel, D. , & Abarenkov, K. (2018). High‐level classification of the fungi and a tool for evolutionary ecological analyses. Fungal Diversity, 90, 135–159. [Google Scholar]
- van der Linde, S. , Suz, L. M. , Orme, C. D. L. , Cox, F. , Andreae, H. , Asi, E. , & Bidartondo, M. I. (2018). Environment and host as large‐scale controls of ectomycorrhizal fungi. Nature, 558, 243–248. [DOI] [PubMed] [Google Scholar]
- Vázquez, D. P. , & Stevens, R. D. (2004). The latitudinal gradient in niche breadth: Concepts and evidence. The American Naturalist, 164, E1–E19. [DOI] [PubMed] [Google Scholar]
- Větrovský, T. , Kohout, P. , Kopecký, M. , Machac, A. , Man, M. , Bahnmann, B. D. , & Baldrian, P. (2019). A meta‐analysis of global fungal distribution reveals climate‐driven patterns. Nature Communications, 10, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villéger, S. , & Brosse, S. (2012). Measuring changes in taxonomic dissimilarity following species introductions and extirpations. Ecological Indicators, 18, 552–558. [Google Scholar]
- Wardle, D. A. , & Lindahl, B. D. (2014). Disentangling global soil fungal diversity. Science, 346, 1052–1053. [DOI] [PubMed] [Google Scholar]
- Warren, R. , VanDerWal, J. , Price, J. , Welbergen, J. A. , Atkinson, I. , Ramirez‐Villegas, J. , & Lowe, J. (2013). Quantifying the benefit of early climate change mitigation in avoiding biodiversity loss. Nature Climate Change, 3, 678–682. [Google Scholar]
- Watson, J. E. , Iwamura, T. , & Butt, N. (2013). Mapping vulnerability and conservation adaptation strategies under climate change. Nature Climate Change, 3, 989–994. [Google Scholar]
- Wood, S. N. (2011). Fast stable restricted maximum likelihood and marginal likelihood estimation of semiparametric generalized linear models. Journal of the Royal Statistical Society: Series B (Statistical Methodology), 73, 3–36. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1
Figure S1
Table S1
Table S2
Table S3
Data Availability Statement
The data supporting the results is archived in Zenodo (https://doi.org/10.5281/zenodo.6983158 and https://doi.org/10.5281/zenodo.7027315) and PlutoF (https://doi.org/10.15156/BIO/2263453). The scripts for analyses are available via GitHub (https://github.com/Mycology‐Microbiology‐Center/Fungal_Endemicity_and_Vulnerability).


