Abstract
DNA sensing through the cGAS-STING pathway plays an important role in cancer immunosurveillance. Pharmaceutical activation of STING in the tumor environment is considered an attractive approach to induce antitumor immunity, but had limited efficacy in the clinic. Several studies have found that STING is epigenetically silenced in many tumors, including colon cancer. This suggests that STING silencing in tumor cells contributes to immune escape and may limit the application of STING agonists. We previously found that inhibition of the KDM5 family histone demethylases restored STING expression in human breast cancer cells and activated the cGAS-STING pathway. In this study, we used MC38 and CT26 syngeneic mouse colorectal cancer models to show that loss of STING in tumor cells accelerates tumor growth. KDM5 inhibitors activate STING expression in mouse colorectal cancer cells and suppress colon cancer growth in immune competent mice in a STING-dependent manner. This study highlights KDM5 inhibitors as novel immune modulators in cancer therapies.
Keywords: Cancer epigenetics, STING, Histone demethylase, KDM5, Cancer Immunology
1. Introduction
Immunogenic tumor cells escape immune restriction through various mechanisms [1,2]. To combat immune escape, restoration of immunosurveillance, e.g. by targeting immune checkpoint molecules, has been used as an effective clinical approach to treat certain types of cancer [3]. However, most cancer patients do not benefit from current immunotherapies and their mechanisms are still largely unknown [4]. Thus, searching for novel approaches to boost anti-tumor immune response is under intense investigation.
Stimulator of interferon genes (STING) is a key signal transductor in the DNA sensing pathway and innate immune response [5]. Cyclic GMP-AMP (cGAMP) synthase (cGAS) recognizes pathogen- or abnormally released self-DNA and produces cGAMP [6]. cGAMP can then be transported from tumor cells into immune cells through gap junctions [7,8] to bind to STING and induce its dimerization, translocation, and activation. Activated STING recruits TANK-binding kinase 1 (TBK1) and triggers a downstream signal cascade, including the activation of the type-I interferon pathway and interferon-stimulated genes (ISGs) [9]. Many ISGs are key regulators of the immune response [10]. Thus, STING and its pathway play a critical role in innate and adaptive immunity in infection and cancer settings.
The genes in the cGAS-STING pathways are largely ubiquitously expressed, but most studies focus on STING’s function in immune cells, e.g. in dendritic cells [11,12]. In contrast, the role of STING in tumor cells is understudied. STING was found to be silenced in many tumors [13-17], suggesting a tumor suppressor function for STING. We reported previously that histone H3K4 demethylases, KDM5B and KDM5C, repress immune response via suppression of STING in cancer cells [18]. Depletion of KDM5B and KDM5C using CRISPR/Cas9 or inhibition of KDM5 with small molecule inhibitors activated STING expression, IFN-β secretion, and ISG induction in cancer cells in vitro [18], providing a potential new approach to boost anti-tumor immunity. In this study, we showed that STING is a key suppressor of tumor progression in syngeneic colorectal tumor models in immune competent mice. Loss of STING in tumor cells significantly accelerated tumor growth. In contrast, KDM5 inhibitor treatment suppresses tumor growth in a tumor cell intrinsic STING-dependent manner. This study nominates KDM5 inhibitors as novel regulators of anti-tumor immunity through STING induction.
2. Materials and methods
2.1. Cell lines and treatment
Murine MC38 colon cancer cells, B16-F10 melanoma cells, 293T cells were cultured in DMEM medium. Murine CT26 colon cancer cells were cultured in RPMI-1640 medium. 10% FBS and 100 U/mL penicillin plus 100 μg/mL streptomycin were supplemented into all cell culture medium. All mediums and cell culture supplements were obtained from Gibco. All cells were cultured at 37 °C in a humid atmosphere containing 5% CO2. KDM5 inhibitor CPI-455 was purchased from Selleck. To induce STING expression, MC38, and CT26 were treated with 25 μM CPI-455 for 10 days.
2.2. Generation of plasmids
Guide sequences were cloned into LentiCRISPRv2 as described previously [19]. The control guide sequences are: Ctrl sg1, GACCGGAACGATCTCGCGTA; Ctrl sg2, ATTGAGAATTCGTTTCAAGG; STING sg1, CCACAGAGGGTTACCTGGAC; STING sg2, TGCGGTTGATCTTACCAGGT; IFRaR1: GATTCGTATGGTGTAACTGA; IRF3: GGCTGGACGAGAGCCGAACG. STING was amplified from cDNA and cloned into pLX304 (Addgene) by Gateway recombination. An 3xHA tag was added into the N-terminal of STING. An N154S was introduced by site-directed mutagenesis to generate mutant STING expression plasmid.
2.3. Generation of knockout and overexpression cell lines
CRISPR/Cas9-mediated gene knockout was carried as described previously [19]. Briefly, lentivirus was produced by transfecting 293T cells with LentiCRISPRv2 plasmid carrying a guide sequence, psPAX2 and pMD2. G (all obtained from Addgene) and Lipofectamine 2000 (Invitrogen). Lentivirus-containing medium was used to infect cells following with selection using puromycin (ThermoFish). Lentivirus-introduced overexpression stable cell lines were generated in a similar way except that the selection was performed with blasticidin (ThermoFish).
2.4. Western blot
Homogenization of cells was carried out in cell lysis buffer containing PMSF and protease inhibitor cocktail (all from Beyotime), then proteins were separated in SDS-PAGE gel and electroblotted onto nitrocellulose membranes. After blocking, the membranes were incubated with STING antibody (CST, D2P2F, 1:1000) and Vinculin antibody (CST, #4650, 1:5000) at 4 °C overnight. After washing, the secondary antibody conjugated to HRP was incubated at room temperature for 2 h. The protein bands were detected with ELC reagent (YEASEN) on Amersham Imager 600 (GE Healthcare Life Sciences).
2.5. Quantitative Reverse Transcription PCR (qRT-PCR)
4 ug of 2′,3′-cGAMP (Sigma) was transfected into cells in 6-well plate using Lipofectamine 2000 (ThermoFisher) following product’s user guide. Cells were harvested 6 h after transfection. Total RNA was isolated using RNeasy mini kit (Qiagen). Reverse transcription was conducted using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). qPCR was conducted using SYBR Green Master Mix (Bio-Rad). Primer sequences are available upon request.
2.6. Colony formation assay
Cells were seeded into 6-well plates at a density of 1000 cells/well and incubated for 10–14 days until visible clones appeared. Colonies were stained with 0.5% crystal violet and ImageJ was used to quantify number of colonies from scanned images.
2.7. Flow cytometry analysis
FACS antibody staining were performed as previously described [20]. Briefly, tumors were stripped from subcutaneous transplantation models. For preparation of a single-cell suspension, the tumor dissociation kit (Miltenyi Biotec, #130–096–730) in combination with the gentleMACS dissociators were used. Cells in single-cell suspension were fixed with 2% PFA (Santa-Cruz, #sc-281692). After washing with a Flow Cytometry Staining Buffer (eBioscience, #00–4222–26), cells were stained with antibodies for cell-surface marker for 1 h on ice in the dark. For staining of intracellular proteins, the cells were washed and resuspended in the permeabilization buffer (BD, #554723) and stained by antibodies in the permeabilization buffer for 1 h on ice in the dark. The cells were then pelleted and resuspended in the Flow Cytometry Staining Buffer for flow cytometry analysis. Antibodies used for flow cytometry include: V450 Mouse Anti-Mouse CD45.2 (BD Horizon, #560697, Clone 104), APC anti-mouse NK1.1 Antibody (BioLegend, #108710, clone PK136), FITC anti-human/mouse Granzyme B Antibody (BioLegend, #515403, clone GB11), CD8a Monoclonal Antibody (53–6.7) PE-Cyanine7 (eBioscience, #25–0081-82, clone 53–6.7), Alexa Fluor® 488 anti-mouse CD8a Antibody (BioLegend, #100726, clone 53–6.7), CD4 Monoclonal Antibody (RM4–5)PE (eBioscience, #12–0042-82, clone RM4–5), PE/Cyanine7 anti-mouse CD4 Antibody (BioLegend, #100527, clone RM4–5), PE/Cyanine7 anti-mouse CD45.2 Antibody (BioLegend, # 109829, Clone 104), PE anti-mouse CD69 Antibody (BioLegend, # 104507, Clone H1.2F30), FITC anti-mouse CD8a Antibody (BioLegend, #100705, Clone 53–6.7), Brilliant Violet 421™ anti-mouse IFN-γ Antibody (BioLegend, # 505829, Clone XMG1.2), APC/Cyanine7 anti-mouse CD3 Antibody (BioLegend, # 100221, clone 17A2). The data were acquired on Cytoflex S (Beckman Coulter) and analyzed using FlowJo software.
2.8. Murine tumor models
Studies were conducted in compliance with US guidelines for the care and use of laboratory animals and were approved by the Institutional Animal Care and Use Committee of Rutgers University, Yale University, or Fudan University Shanghai Cancer Center. MC38 (5 ×105), CT26 (5 ×105) and B16-F10 (2 ×105) cells were suspended in 100 μL PBS and injected subcutaneously into the flank of mice. C57BL/6 or BALB/c mice of 6–8 weeks of age were used and purchased from the Jackson laboratory or Chinese Academy of Science Shanghai Laboratory Animal Center. The tumor size was measured by caliper. Tumor volume was calculated by the modified ellipsoidal formula: V = ½ (Length × Width2). For NK and T cell depletion experiment, InVivoMAb anti-mouse NK1.1 (Bio X cell-BE0036), InVivoMAb anti-mouse CD8a (Bio X cell-BE0117), and InVivoMAb anti-mouse CD4 (Bio X cell-BE0003–3) were mixed. InVivoMAb rat IgG2b isotype control, anti-keyhole limpet hemocyanin (Bio X cell-BE0090) was used as a control. 600 mg total antibodies (control or depletion antibodies mix) per animal were injected into mice at day – 2, 1, 4, 7, 10, and 12. KDM5 inhibitor CPI-48 was provided by the Early Translational Branch, National Center for Advancing Translational Sciences, the US National Institutes of Health. Mice were injected daily intraperitoneally with CPI-48 or vehicle from one day after tumor cell injection at 100 mg / kg body weight. The injections contained CPI-48 in a solution of PBS (30%), DMSO (5%), DMA (5%), PEG400 (20%), and PG (40%) to ensure solubility. The vehicle contained the same reagents but without CPI-48.
2.9. Statistical analysis
Statistical analysis of the data was performed using SPSS version 22.0 or GraphPad Prism version 9.0.0. The tumor volume at the end points, gene expression, FACS results and stained cells were compared using Student’s t-test between two groups or one-way ANOVA in all multiple group comparisons. In multiple group comparisons, comparisons were conducted among all groups and the Tukey test was applied for multiple testing correction. Differences were considered significant when P < 0.05.
2.10. Data availability
The data generated in this study are available upon request from the corresponding author.
3. Results
3.1. Tumor cell intrinsic STING hinders tumor growth in immunocompetent host
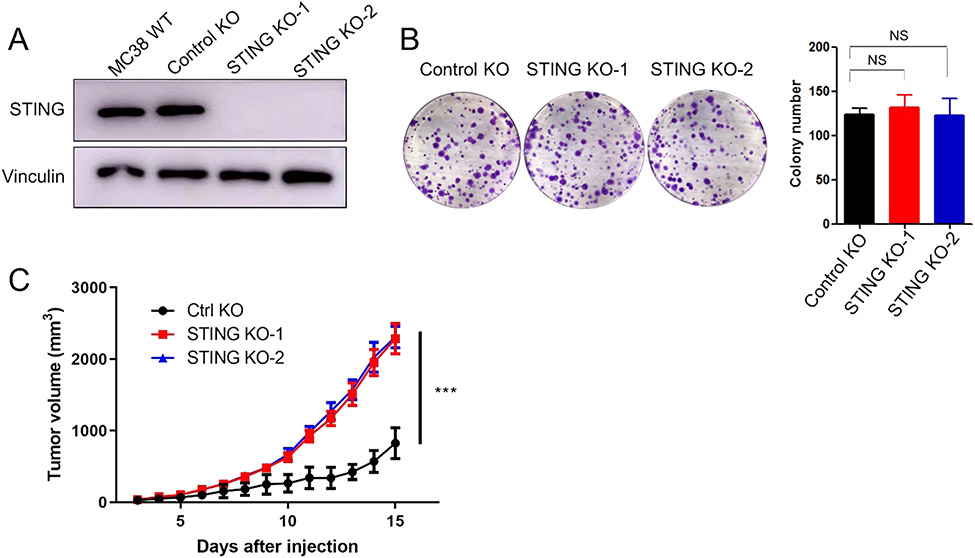
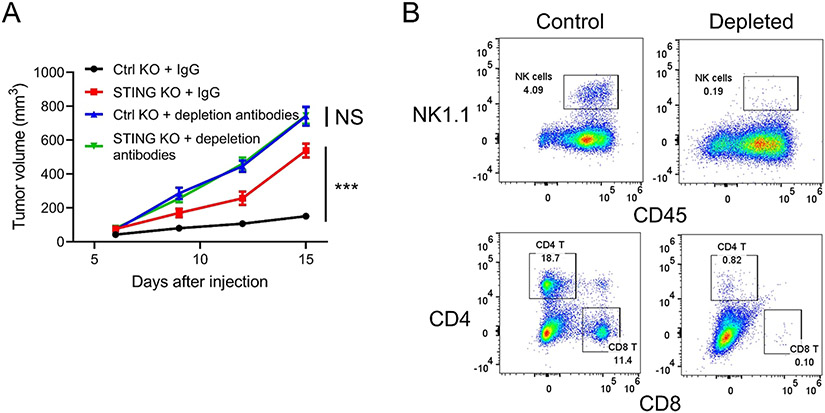
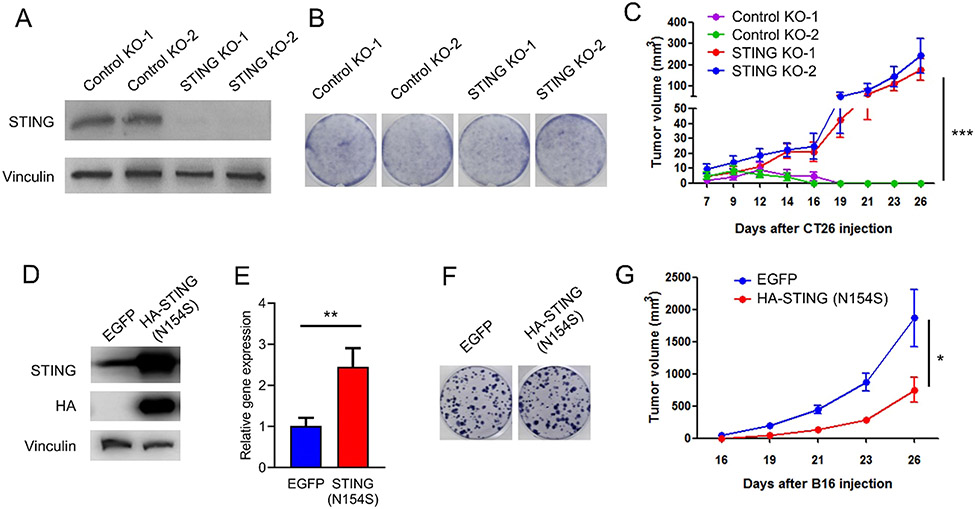
Previous studies have shown loss of STING in some colorectal carcinomas. To explore the function of the STING pathway in colorectal cancer cells, STING knockout (KO) cell lines were generated using CRISPR/Cas9 in MC38, a mouse colon cancer cell line (Fig. 1A). Colony formation assays showed that loss of tumor cell intrinsic STING had no impact on the proliferation of cancer cells in vitro (Fig. 1B). In contrast, the growth of MC38 tumors with STING KO was significantly accelerated in immunocompetent C57BL/6 mice, compared to control tumors (Fig. 1C). We then depleted CD4 T cells, CD8 T cells, and NK cells using antibodies and repeated the tumor growth assay (Fig. 2A and B). We found that the difference between control and STING knockout cells disappeared in T and NK cell depleted hosts. We conducted similar experiments using another murine colon cancer cell line, CT26. Consistent with results from MC38 cells, KO of STING in CT26 cells had no effect on in vitro cell growth, while significantly accelerating tumor growth compared to control tumors in syngeneic immunocompetent BALB/c mice (Fig. 3A, B, and C). These results suggest that tumor cell intrinsic STING is important for immune restriction.
Fig. 1.
Loss of STING accelerates MC38 tumor growth in immunocompetent mice. (A) Western blot analysis of STING expression in wild type, control knockout, and two independent STING knockout MC38 cell lines. (B) Colony formation (left) and its quantification (right) of the controls and STING knockout MC38 cell lines. (C) In vivo tumor growth curves of control knockout and two STING knockout MC38 cell lines. N = 5. * ** P < 0.001. NS: not significant.
Fig. 2.
STING regulates tumor growth in a host immune response-dependent manner. (A) In vivo tumor growth curves of control or STING knockout MC38 cells injected into mice receiving control IgG or NK and T cell depletion antibodies. N = 10. (B) FACS analyses of NK, CD4, and CD8 T cell markers of splenocytes isolated from mice receiving control IgG or NK and T cell depletion antibodies. * ** P < 0.001. NS: not significant.
Fig. 3.
STING regulates tumor growth in immunocompetent mice. (A) Western blot analysis of STING expression in two independent control knockout, and two independent STING knockout CT26 cell lines. (B) Colony formation of the controls and STING knockout CT26 cell lines. (C) In vivo tumor growth curves of controls and STING knockout CT26 cell lines. N = 10. (D) Western blot analysis of STING expression in EGFP or STING (N154S)-overexpressed B16F10 cell lines. (E) RT-qPCR analysis of IFNβ expression in EGFP or STING (N154S)-overexpressed B16F10 cell lines. (F) Colony formation of EGFP or STING(N154S) overexpressed B16F10 cells. (G) In vivo tumor growth curves of control and STING (N154S)-overexpressed B16F10 cell lines. N = 6. * P < 0.05, * * P < 0.01, * ** P < 0.001.
To further substantiate the above conclusion, we examined the effects of STING activation. An asparagine-to-serine substitution at amino acid residue 154 (N154S) of STING causes its constitutive activation and triggers the interferon pathway without cGAS activation [21]. Overexpression of STING (N154S) mutant in B16-F10 cells, a non-immunogenic murine melanoma cell line, significantly increased the expression of IFN-β (Fig. 3D and E). There was no in vitro growth difference between control and STING (N154S)-overexpressed cells (Fig. 3F). However, STING (N154S) overexpression significantly suppressed tumor growth in immunocompetent C57BL/6 mice, compared to control cells (Fig. 3G). This suggests that activation of STING in tumor cells alone suppresses tumor growth in immunocompetent host.
3.2. The function of cancer-intrinsic STING is independent of IFN-sensing by tumor cells
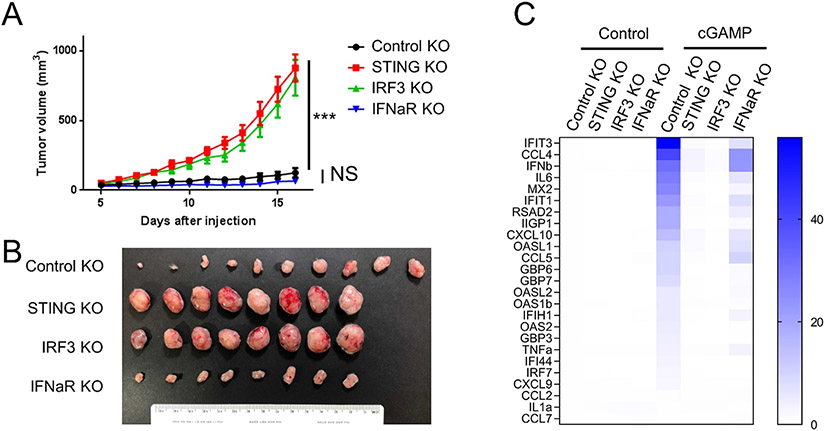
In tumors, cytosolic dsDNA can be induced by IFN-γ, a common tumor feature, and then activates the STING pathway in cancer cells [22]. The activation of STING leads to phosphorylation of IRF3 and secretion of type-I interferons, e.g. IFN-β. Both cancer cells and immune cells express IFN-α/β receptor (IFNAR). The binding of type-I interferons to IFNAR on tumor cells activates transcription of a large group of ISGs [18], which may have tumor cell intrinsic effects. Some ISGs are cytokines and chemokines, thus activation of IFN pathway in tumor cells can also indirectly regulate immune cells. On the other hand, type-I interferons can bind to IFNAR on immune cells and directly regulate their function. To determine whether the tumor cell intrinsic STING regulates immune cells indirectly through IFNAR on tumor cells, we knockouted IFNAR1 in MC38 cells. IRF3 is downstream of STING and upstream of IFNAR in the cGAS/STING/IFN pathway. We also included an IRF3 KO line. As expected, STING and IRF3 KO significantly accelerated tumor growth, compared to control knockout. In contrast, IFNAR1 KO had no impact on tumor growth (Fig. 4A and B). We also measured the induction of a group of genes regulated by STING pathway in the presence of cGAMP, a STING ligand. These genes include direct targets of IRF3, e.g. IFN-β, and genes downstream of IFNAR1, e.g. IFIT3. We found that STING and IRF3 knockouts blocked the induction of both groups of genes and IFNAR1 knockout only blocked some of these genes (Fig. 4C). Notably, IFN-β induction was not affected in IFNAR1 knockout cells. This result suggests that although the tumor cell intrinsic STING pathway suppresses tumor growth, it is independent of IFN-sensing in cancer cells.
Fig. 4.
The tumor suppression function of cancer-intrinsic STING is independent of IFN-sensing by tumor cells. (A and B) In vivo tumor growth curves (A) and tumor images at end point (B) of control knockout MC38 cells, and MC38 cells with knockout of STING, IRF3, or IFNAR1. N = 8 or 10. (C) RT-qPCR analysis of selected genes regulated by STING pathway in control, STING, IRF3, or IFNAR1 knockout MC38 cells at the presence or absence of cGAMP. * ** P < 0.001. NS: not significant.
3.3. Tumor-derived STING expression affects the activation of tumor infiltrating lymphocytes
To investigate the role of tumor cell intrinsic STING on host immune cells, we analyzed tumor infiltrating lymphocytes isolated from control or STING knockout MC38 tumors using flow cytometry. We found that STING deficiency in MC38 cells had no significant impact on the number of tumor infiltrating lymphocytes (CD45 +), NK cells (NK1.1 +) and CD8 + T cells (CD8a +) (Fig. 5). However, when we examined the cytotoxic markers, such as Granzyme B, CD69, and IFNγ, we found significant decreases of activated NK cells in the STING knockout tumors, compared to control tumors (Fig. 6). CD8 + T cells also showed significant decrease of cell surface CD69 in the STING knockout tumors (Fig. 6). These data suggest that tumor intrinsic STING suppresses tumor growth by promoting the activation of cytotoxic NK and CD8 + T cells. Similarly, CD69 was significantly reduced in both CD8 + T cells and NK cells isolated from IRF3 knockout tumors but was not affected in IFNAR1 knockout tumors (Fig. 7).
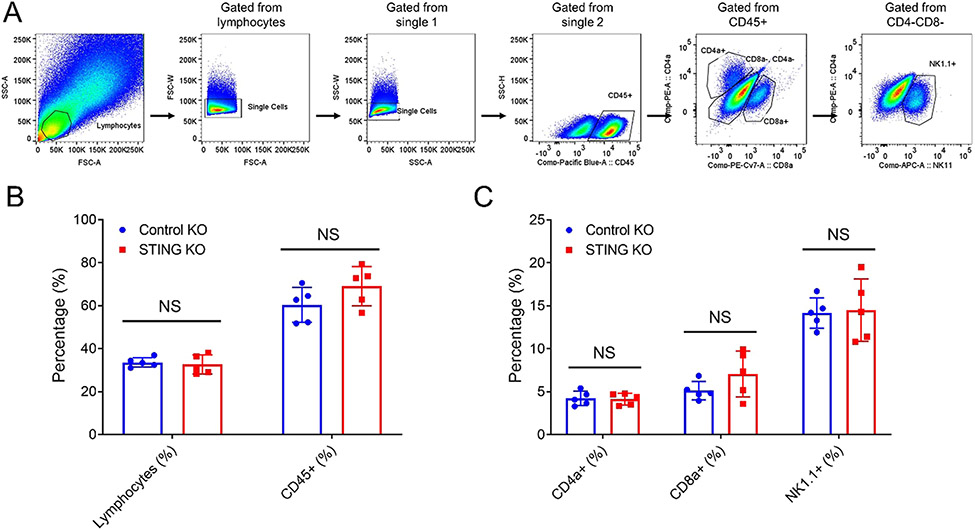
Fig. 5.
Tumor cell intrinsic STING has no effect on tumor infiltrating immune cells. (A) The gating strategy for the FACS analysis. (B) The quantification of FACS analysis of tumor infiltrating immune cells (CD45 +), NK cells (NK1.1 +), and cytotoxic T cells (CD8a +) isolated from control KO or STING KO tumors. N = 5. NS: not significant.
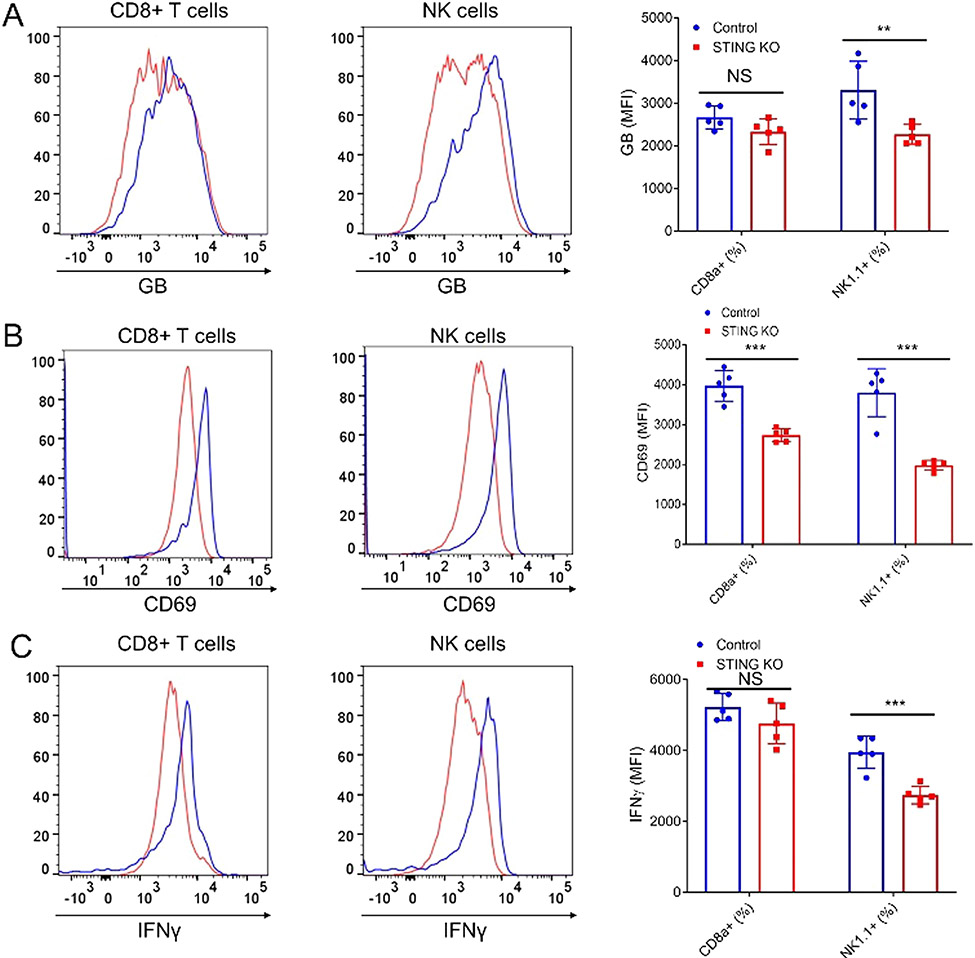
Fig. 6.
Tumor cell intrinsic STING promotes the activation of NK and CD8+ T cells. FACS analysis (left) of activation markers, Granzyme b (A), CD69 (B), and IFN-γ (C) of tumor infiltrating NK cells and CD8 T cells isolated from control KO or STING KO tumors and their quantification (right). N = 5. * * P < 0.01, * ** P < 0.001. NS: not significant.
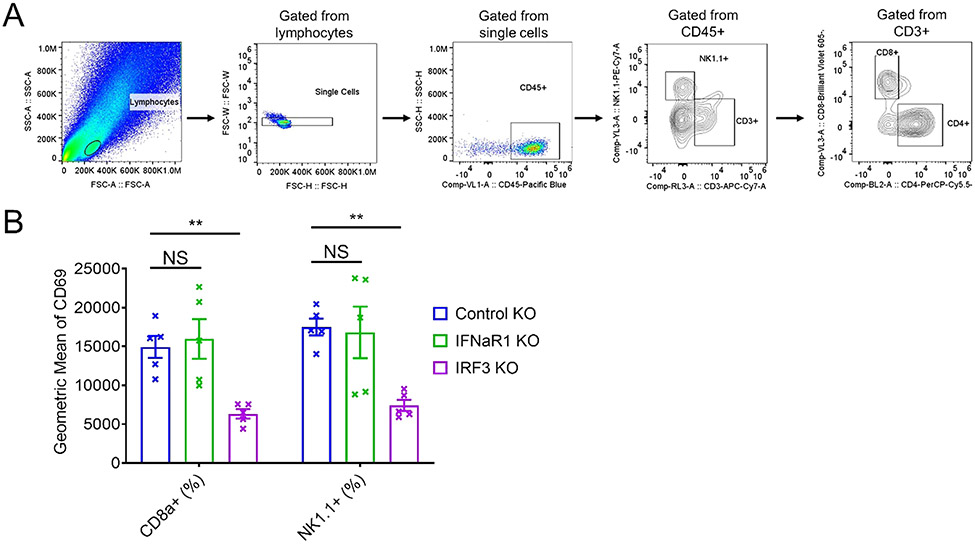
Fig. 7.
The function of cancer-intrinsic STING on activating T and NK cells is independent of IFN-sensing by tumor cells(A) The gating strategy for the FACS analysis. (B) The quantification of FACS analysis of activation markers, CD69 of tumor infiltrating NK cells and CD8 T cells isolated from control, IFNaR1, or IRF3 KO tumors. N = 5. * * P < 0.01. NS: not significant.
3.4. KDM5 inhibitor activates tumor cell intrinsic STING and suppresses tumor growth in immunocompetent mice
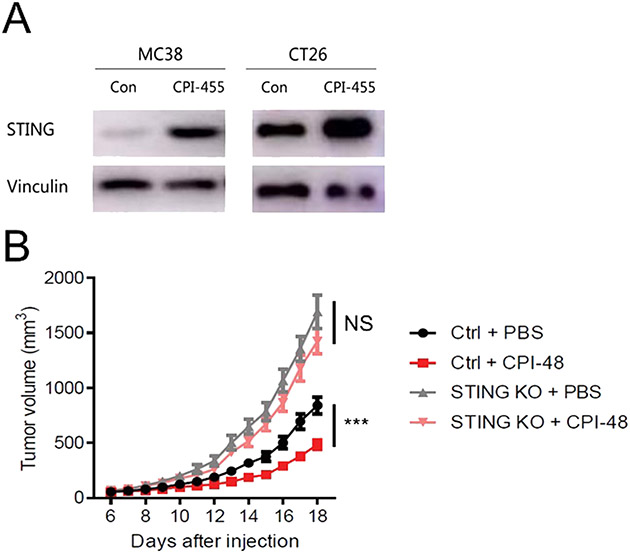
STING is commonly silenced in colon cancer cells [23]. Our previous study showed that the expression of STING was epigenetically suppressed by KDM5 demethylases in breast cancer cell lines [18]. Thus, KDM5 inhibitors are potential immune modulating agents in colon cancer. Treatment of mouse colon cancer cell lines, MC38 and CT26, with CPI-455, a KDM5 inhibitor, significantly increased STING expression (Fig. 8A). We then performed animal experiments with CPI-48, a KDM5 inhibitor amenable for in vivo studies [24]. Allografts were generated with MC38 cells in immunocompetent mice. Tumor-carrying mice were then treated with CPI-48 or a vehicle control. STING knockout MC38 cells were also included as controls. Consistent with previous results, STING knockout in tumor cells significantly accelerated tumor growth. CPI-48 treatment suppressed the growth of wild type tumors but had no effects on STING KO tumors (Fig. 8B). These results suggest that KDM5 inhibitors induce STING expression to suppress immune evasion.
Fig. 8.
KDM5 inhibitors activate tumor cell intrinsic STING and suppress tumor growth. (A) Western blot showing the induction of STING expression after CPI-455 treatment in MC38, and CT26 cells. (B) In vivo tumor growth curves of control or STING knockout MC38 cells with or without CPI-48 treatment. N = 10. * ** P < 0.001. NS: not significant.
4. Discussion
STING is a central player in the cytosolic DNA-sensing pathway that drives activation of type-I IFN in the host immune response. Since its role in cancer immunology was discovered, pharmaceutical activation of the STING pathway to induce anti-tumor immunity has been heavily investigated. For example, intratumoral injection of DMXAA, a direct ligand for mouse STING, induced profound regression of established tumors in mice [25]. Intratumoral injection of a low-dose of ADU-S100, another STING agonist, activated tumor-specific CD8 + T cells, induced durable anti-tumor immunity, and enhanced the efficiency of immune checkpoint inhibitors [26]. Indeed, more than a dozen STING activation agents are at various stages in clinical studies [27]. However, the majority of the antitumor effects caused by STING activation are considered to be dependent on IFN-β production by antigen-presenting cells (APCs), which promotes CD8 T cell priming against tumor-associated antigens [28]. The fact that the endogenous STING ligand, cGAMP, can be transported from tumor cells into immune cells through cell-cell junctions further highlights immune cells as a major contributor of the STING function [7,8].
On the other hand, STING was found to be widely suppressed in human tumor cells and cancer cell lines [14,18,23,29-31]. For example, more than 20% of late stage colon cancers lost expression of STING [23]. Several studies have shown that suppression of the STING pathway in tumor cells contributed to immune evasion [30,32-36]. In addition, tumor intrinsic STING and its induced immune response are required for the efficacy of 5-Fluorouracil, a chemotherapeutic drug [37], immune checkpoint blockade [22], and oncolytic viruses [14]. Mechanistically, the STING pathway is found to be silenced by major oncogenic drivers, e.g. HER2 [36], LKB1 [30], and SOX2 [38]. STING can be degraded through the ubiquitin-proteasome pathway [35] or by autophagy [38]. The recurrent silencing of STING in tumor cells suggests an anti-tumor function of tumor intrinsic STING.
The silencing of the STING pathway is predominantly mediated by epigenetic regulators, including DNMT1 [23,30,34,39], EZH2 [15], and KDM5 [18]. An earlier study found that cGAS and STING were absent in some colon cancer and melanoma cell lines. Treating cells with 5-aza, a DNMT inhibitor, rescued the expression of cGAS in some cell lines but without significantly inducing STING [23]. Similarly, 5-aza treatment was able to reactivate cGAS but not STING in most melanoma cell lines [14]. We previously found that KDM5 family histone demethylases bind to the STING promotor, catalyze the removal of H3K4me3 transcriptional active marks, and suppress STING transcription in human cancer cell lines [18]. In these cell lines, in vitro KDM5 pan-inhibitor treatment enhanced STING expression, promoted IFN-β secretion, and activated ISG [18], providing a new approach to boost anti-tumor immunity.
In this study, we found that the silencing of tumor intrinsic STING promotes colorectal tumor growth in immunocompetent mice. KDM5 inhibitors activated STING expression in mouse colon cancer cell lines and suppressed tumor growth in a tumor intrinsic STING-dependent manner. Our findings suggest that tumoral STING contributes significantly to STING-induced anti-tumor immunity. Thus, an intact STING pathway in tumor cells may be important to maximize the efficacy of STING agonists in cancer therapies. When STING or other components downstream is epigenetically silenced in tumor cells, especially in late-stage tumors, combining epigenetic drugs, e.g. KDM5 inhibitors, to reinstall the STING pathway, and STING agonists may achieve better clinical response.
Supplementary Material
Acknowledgements
We thank Drs. Daniel Jansen, Ganesha Rai, Stephen Kales, Anton Simeonov, and Matthew Hall from the Early Translational Branch of National Center for Advancing Translational Sciences for providing KDM5 inhibitor CPI-48. We thank Dr. Hao Liu from the Rutgers Cancer Institute of New Jersey for advice on statistical analyses. This study was supported by a start-up fund and the 2021 Center of Excellence in Cancer Immunology and Metabolism (CoECIM) Seed Grant (both to J.C.) provided by Rutgers Cancer Institute of New Jersey and Cancer Center Support Grant P30CA072720, National Institutes of Health Awards R01CA237586 and P50CA121974 (to Q.Y.), National Natural Science Foundation of China NSFC-82072876 (to R.L.), and Science and Technology Commission of Shanghai Municipality 19ZR1410300 (to H.Z.).
Footnotes
CRediT authorship contribution statement
Hui Zheng: Investigation, Writing – original draft. Lizhen Wu: Conceptualization, Investigation. Qian Xiao: Investigation, Visualization. Xin Meng: Investigation. Alex Hafiz: Writing – review & editing. Qin Yan: Supervision, Writing – review & editing, Funding acquisition. Renquan Lu: Supervision, Funding acquisition. Jian Cao: Conceptualization, Investigation, Writing – original draft, Writing – review & editing, Supervision, Funding acquisition.
Conflict of interest
The authors declare no conflict of Interest. The authors declare no potential conflicts of interest.
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.biopha.2022.114033.
Data Availability
No data was used for the research described in the article.
References
- [1].Dunn GP, et al. , Cancer immunoediting: from immunosurveillance to tumor escape, Nat. Immunol 3 (11) (2002) 991–998. [DOI] [PubMed] [Google Scholar]
- [2].Cao J, Yan Q, Cancer Epigenetics, Tumor Immunity, and Immunotherapy, Trends Cancer 6 (7) (2020) 580–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Waldman AD, Fritz JM, Lenardo MJ, A guide to cancer immunotherapy: from T cell basic science to clinical practice, Nat. Rev. Immunol 20 (11) (2020) 651–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Haslam A, Prasad V, Estimation of the percentage of US patients with cancer who are eligible for and respond to checkpoint inhibitor immunotherapy drugs, JAMA Netw. Open 2 (5) (2019), e192535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ishikawa H, Barber GN, STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling, Nature 455 (7213) (2008) 674–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Sun L, et al. , Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway, Science 339 (6121) (2013) 786–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Chen Q, et al. , Carcinoma-astrocyte gap junctions promote brain metastasis by cGAMP transfer, Nature 533 (7604) (2016) 493–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Schadt L, et al. , Cancer-Cell-Intrinsic cGAS Expression Mediates Tumor Immunogenicity, Cell Rep. 29 (5) (2019), p. 1236–1248.e7. [DOI] [PubMed] [Google Scholar]
- [9].Burdette DL, et al. , STING is a direct innate immune sensor of cyclic di-GMP, Nature 478 (7370) (2011) 515–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].McNab F, et al. , Type I interferons in infectious disease, Nat. Rev. Immunol 15 (2) (2015) 87–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Deng L, et al. , STING-dependent cytosolic DNA sensing promotes radiation-induced type I interferon-dependent antitumor immunity in immunogenic tumors, Immunity 41 (5) (2014) 843–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Marcus A, et al. , Tumor-derived cGAMP triggers a STING-mediated interferon response in non-tumor cells to activate the NK cell response, Immunity 49 (4) (2018), p. 754–763.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Xia T, et al. , Deregulation of STING signaling in colorectal carcinoma constrains DNA damage responses and correlates with tumorigenesis, Cell Rep. 14 (2) (2016) 282–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Xia T, Konno H, Barber GN, Recurrent loss of STING signaling in melanoma correlates with susceptibility to viral oncolysis, Cancer Res. 76 (22) (2016) 6747–6759. [DOI] [PubMed] [Google Scholar]
- [15].Konno H, et al. , Suppression of STING signaling through epigenetic silencing and missense mutation impedes DNA damage mediated cytokine production, Oncogene 37 (15) (2018) 2037–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Chen YA, et al. , Extrachromosomal telomere repeat DNA is linked to ALT development via cGAS-STING DNA sensing pathway, Nat. Struct. Mol. Biol 24 (12) (2017) 1124–1131. [DOI] [PubMed] [Google Scholar]
- [17].Low JT, et al. , Epigenetic STING silencing is developmentally conserved in gliomas and can be rescued by methyltransferase inhibition, Cancer Cell 40 (5) (2022) 439–440. [DOI] [PubMed] [Google Scholar]
- [18].Wu L, et al. , KDM5 histone demethylases repress immune response via suppression of STING, PLoS Biol. 16 (8) (2018), e2006134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Cao J, et al. , An easy and efficient inducible CRISPR/Cas9 platform with improved specificity for multiple gene targeting, Nucleic Acids Res. 44 (19) (2016) e149–e149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Shen T, et al. , Neutralizing monoclonal antibody against Dickkopf2 impairs lung cancer progression via activating NK cells, Cell Death Disco 5 (2019) 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Liu Y, et al. , Activated STING in a vascular and pulmonary syndrome, New Engl. J. Med 371 (6) (2014) 507–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Xiong H, et al. , IFN-γ activates the tumor cell-intrinsic STING pathway through the induction of DNA damage and cytosolic dsDNA formation, Oncoimmunology 11 (1) (2022), 2044103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Xia T, et al. , Deregulation of STING signaling in colorectal carcinoma constrains DNA damage responses and correlates with tumorigenesis, Cell Rep. 14 (2) (2016) 282–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Liang J, et al. , Lead optimization of a pyrazolo[1,5-a]pyrimidin-7(4H)-one scaffold to identify potent, selective and orally bioavailable KDM5 inhibitors suitable for in vivo biological studies, Bioorg. Med. Chem. Lett 26 (16) (2016) 4036–4041. [DOI] [PubMed] [Google Scholar]
- [25].Corrales L, et al. , Direct activation of STING in the tumor microenvironment leads to potent and systemic tumor regression and immunity, Cell Rep. 11 (7) (2015) 1018–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Sivick KE, et al. , Magnitude of therapeutic STING activation determines CD8(+) T cell-mediated anti-tumor immunity, Cell Rep. 25 (11) (2018), p. 3074–3085.e5. [DOI] [PubMed] [Google Scholar]
- [27].Amouzegar A, et al. , STING agonists as cancer therapeutics, Cancers 13 (11) (2021) 2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Sokolowska O, Nowis D, STING signaling in cancer cells: important or not? Arch. Immunol. Ther. Exp 66 (2) (2018) 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Sunthamala N, et al. , E2 proteins of high risk human papillomaviruses down-modulate STING and IFN-κ transcription in keratinocytes, PLoS One 9 (3) (2014), e91473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kitajima S, et al. , Suppression of STING associated with LKB1 loss in KRAS-driven lung cancer, Cancer Discov. 9 (1) (2019) 34–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Falahat R, et al. , Epigenetic reprogramming of tumor cell-intrinsic STING function sculpts antigenicity and T cell recognition of melanoma. 2021. 118(15): p. e2013598118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Takahashi M, et al. , The tumor suppressor kinase DAPK3 drives tumor-intrinsic immunity through the STING-IFN-β pathway, Nat. Immunol 22 (4) (2021) 485–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Shu C, et al. , Pharmacological rescue of tumor intrinsic STING expression and immune response in LKB1-mutant lung cancer via the IAP-JAK regulatory axis. 2021: p. 2021.09.17.460294. [Google Scholar]
- [34].Falahat R, et al. , Epigenetic reprogramming of tumor cell-intrinsic STING function sculpts antigenicity and T cell recognition of melanoma, Proc. Natl. Acad. Sci. USA 118 (15) (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Zhang C.-x, et al. , Galectin-9 promotes a suppressive microenvironment in human cancer by enhancing STING degradation, Oncogenesis 9 (7) (2020) 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Wu S, et al. , HER2 recruits AKT1 to disrupt STING signalling and suppress antiviral defence and antitumour immunity, Nat. Cell Biol 21 (8) (2019) 1027–1040. [DOI] [PubMed] [Google Scholar]
- [37].Tian J, et al. , 5-Fluorouracil efficacy requires anti-tumor immunity triggered by cancer-cell-intrinsic STING, EMBO J. 40 (7) (2021) p. e106065–e106065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Tan YS, et al. , Mitigating SOX2-potentiated immune escape of head and neck squamous cell carcinoma with a STING-inducing nanosatellite vaccine, Clin. Cancer Res 24 (17) (2018) 4242–4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Ma F, et al. , LncRNA NEAT1 interacted with DNMT1 to regulate malignant phenotype of cancer cell and cytotoxic T cell infiltration via epigenetic inhibition of p53, cGAS, and STING in lung cancer, Front Genet. 11 (2020) 250. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are available upon request from the corresponding author.
No data was used for the research described in the article.