Abstract
The gut microbiome has been referred to as the “forgotten organ.” Although much about the gut microbiome remains incompletely understood, data on its clinical importance is emerging at rapid speed. Many practicing clinicians may be unaware of the essential role that the microbiome plays in both health and disease. This review aims to improve clinical understanding of the gut microbiome by discussing key terminology and foundational concepts. The role of a healthy microbiome in normal host function is described, as well as the consequences of a disrupted microbiome (i.e., dysbiosis). Management strategies to restore the gut microbiome from a disrupted to a healthy state are also briefly discussed. Lastly, we review emerging areas for therapeutic potential and opportunity to bring determinants of microbiome health from the bench to bedside.
Keywords: Clostridioides difficile infection, dysbiosis, gut microbiome
1. INTRODUCTION
The role of the gut microbiome in human health and disease has been an area of increasing clinical interest in recent decades. Its contributions to overall health are significant but often under‐appreciated, leading it to be called the “forgotten organ.” 1 The gut microbiome is a complex ecosystem comprised of an estimated 100 trillion organisms and their genetic content. 2 , 3 The diverse organisms within the microbiome ecosystem are called microbiota. 2 , 3 Microbiota are bacteria, 2 viruses including phages, 2 fungi, eukarya, and archaea. 2 , 3 , 4 Most research to date has focused on bacterial microbiota and their roles within the microbiome given their predominance relative to other microbes. 2 There are more than 1000 different species of bacteria in a healthy gut 2 , 3 , 4 with the majority residing in the lumen of the large intestine. 5 In a state of “healthy” homeostasis, gut microbiota aid with digestion, metabolism, and immune modulation. Disruption of this homeostatic state, called dysbiosis, is believed to be associated with a variety of potential consequences, including infection and several disease states or medical conditions. Despite its complexities, several core elements of the gut microbiome have been described. This review aims to introduce practicing clinicians to foundational concepts related to the microbiome, including discussion of key commensal bacteria and examples of their functional roles in maintaining host health. We also discuss factors that can result in microbiome disruption, the consequences of disruption, and practical mitigation strategies. Finally, areas for further research are summarized.
2. METHODS
A PubMed search was conducted using “gut microbiome” for inclusion in the title and MeSH terms. Peer‐reviewed articles published in English before April 30, 2022, were eligible for inclusion. Initial searches focused on comprehensive overviews of the gut microbiome to identify key terminology and foundational concepts. Articles describing the gut microbiome in the context of specific diseases (e.g., inflammatory bowel disease) or modifiers (e.g., diet) were omitted for initial review. Studies specifically pertaining to gut microbiome research in animals were also omitted. References from any articles deemed to be particularly informative by the authors of this manuscript were reviewed to find similar summaries of the gut microbiome. References from these comprehensive summary articles were also used to identify additional references that focused on relevant niche concepts, such as specific key bacteria or bacterial groups, the role of the microbiome in specific diseases, dysbiosis including the impact of medications, and specific areas of function of the microbiome (e.g., bile acid homeostasis). This article was not designed as a comprehensive literature review or a systematic review.
3. RESULTS
3.1. Gut microbiome composition
Gut microbiome composition evolves from birth 3 and is affected by numerous factors, including diet, 2 , 5 , 6 environment, 2 , 6 age, 6 and medications taken. Therefore, every individual's gut microbiome is believed to be unique, 2 , 7 , 8 akin to a fingerprint. However, there are compositional characteristics that appear to distinguish a “healthy” microbiome from an “unhealthy” one. These characteristics are largely described in the context of certain patterns among commensal bacteria.
Bacteria within the gut microbiome can be characterized with variable levels of granularity using taxonomy. The kingdom of bacteria is described using taxonomic units of phylum, class, order, family, genus, species, and strain. 9 Although units vary by study, bacterial microbiome analysis to the phylum level is common. The most frequently discussed phyla are Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria, as these four phyla constitute up to 99% of intestinal microbiota in healthy adults. 2 Firmicutes and Bacteroidetes appear to collectively predominate (up to 90%). 3 , 9 Characteristics and examples of species within the four most common phyla are outlined in Table 1. It is important to note that phylum nomenclature was updated in late 2021 by the International Committee of Systematics of Prokaryotes. 10 However, for this review, we opted to use historical nomenclature to maintain consistency with existing literature. Bacterial classification at the phylum level is often performed using 16S rRNA sequencing, which is able to identify bacteria to the genus level. 9 More granular bacterial classification approaches, including to the species and strain level, require shotgun metagenomic methods. 11 Although shotgun metagenomic sequencing strategies are often more accurate and descriptive than 16S rRNA sequencing, they are more resource intensive. 11
TABLE 1.
Overview of four most common bacterial phyla within the gut microbiome, listed by generally decreasing relative abundances in healthy individuals a , 2 , 9 , 10
| Phylum (2021 revised nomenclature) | Brief description | Genus examples |
|---|---|---|
| Bacteroidetes (Bacteroidota) | Gram‐negative; typically obligately anaerobic; often abundant | Bacteroides, Prevotella |
| Firmicutes (Bacillota) | Gram‐positive; typically obligately anaerobic; often abundant; highly diverse phyla | Clostridium, Enterococcus, Eubacterium, Faecalibacterium, Lactobacillus, Roseburia, Ruminococcus, Streptococcus |
| Actinobacteria (Actinomycetota) | Gram‐positive; typically obligately anaerobic/requires low oxygen levels | Bifidobacterium, Corynebacterium, Eggerthella |
| Proteobacteria (Pseudomonadota) | Gram‐negative; mostly facultatively anaerobic; includes many pathogenic species | Enterobacter, Escherichia, Klebsiella, Serratia |
In addition to trends in the relative abundances of key bacteria, the degree of bacterial species diversity, or richness, within the phyla and overall gut microbiota community is another important signal of health and homeostasis. This is supported by the 1000 of bacterial species that are often isolated from healthy individuals. Richness within a sample is measured by alpha diversity, often using the Shannon Index. 12 Microbial diversity within the gut microbiome is believed to contribute to resilience against disruption from a “normal” state, so that some losses within the bacterial community will not compromise the health of the larger ecosystem. 8 , 13 Therefore, changes in the abundances of certain phyla and/or less species diversity may indicate that a microbiome has deviated from a healthy or homeostatic state. 7
3.2. Functions of a healthy microbiome
As discussed, the microbiome is comprised of a diverse community of commensal organisms existing in various abundances. These organisms interact in myriad ways that are complex and multifaceted, with microbial and host crosstalk being incompletely understood. 14 However, studies indicate that commensal gut microbiota participate in several healthy and beneficial functions. Bacteria within the gut microbiome ecosystem have been shown to play important roles in maintenance of gut epithelium integrity, digestion, metabolism, and synthesis of beneficial substances including vitamins. 14 The microbiome also helps combat infection and inflammation by interfacing with and modulating our immune systems 4 (Figure 1). Some of these functions can be performed by several bacteria belonging to different phyla, whereas other functions appear to be performed in a complementary or symbiotic fashion by select bacteria. Although our understanding continues to evolve, several critical functions that have been well described among gut microbiota are discussed below.
FIGURE 1.
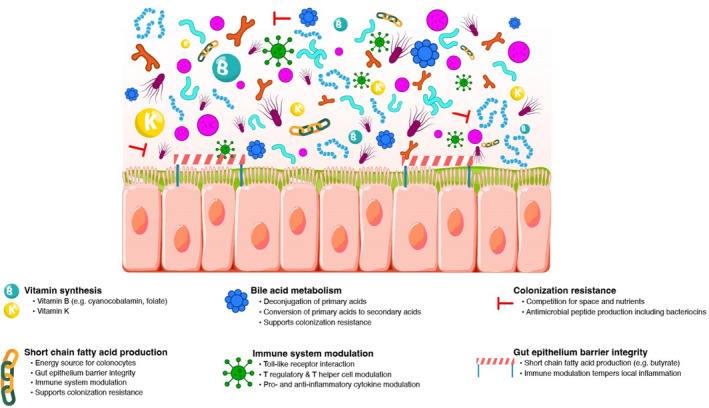
The gut microbiome comprises a diverse community of commensal organisms existing in various abundances that participate in several healthy and beneficial functions
3.2.1. Protective functions
Colonization resistance
One beneficial function offered by a diverse consortium of gut microbes in homeostatic abundances is colonization resistance. 2 , 14 Colonization resistance, while complex, generally refers to strategies that gut microbiota employ to mitigate risk of local colonization with potentially pathogenic bacteria. It also refers to strategies that help maintain “healthy” homeostatic abundances of bacteria in the microbiome community. For example, gut microbes compete with surrounding organisms for nutrients and space, helping prevent overgrowth of niche populations. 2 , 14 Several bacteria also exhibit their own antimicrobial systems to mitigate overgrowth and/or to target cells that are deemed foreign. Some commensals excrete diffusible proteins called bacteriocins, which can be highly toxic to surrounding species and even to different strains within the originating species by inducing pore formation, nucleic acid degradation, and/or interference with cell wall synthesis. 5 Bacteroidetes and Proteobacteria exhibit antibacterial secretion systems that can induce damage to any genetically unidentical surrounding cells. 5 Though mechanisms of colonization resistance vary, all of these functions aim to help maintain gut microbiome homeostasis while simultaneously mitigating risk of local and systemic infection to the host.
Immune modulation
Microbiota appear to aid in both innate and adaptive immunity via several complex and poorly understood mechanisms. 14 In a broad sense, microbiota interact with the host immune system via a variety of mechanisms to help modulate pro‐ and anti‐inflammatory responses in the gut. In a healthy state, commensals appear to help the host immune system differentiate between microbial “friend” or “foe,” thereby helping to temper or induce an inflammatory response, respectively. For example, Toll‐like receptors in the gut epithelium interact with local bacteria to help the body's immune system distinguish familiar commensals from pathogenic microbes. 2 Commensal bacteria are also proposed to help induce secretion of immunoglobulin A in the gut, 2 as well as interact with gut T lymphocytes to help maintain balanced interactions between immune‐stimulating T helper and immune‐suppressing regulatory T cells. 2 , 14 Firmicutes (e.g., Clostridia species), Bacteroidetes (e.g., B. fragilis), and Bifidobacterium infantis have all been associated with regulatory T‐cell recruitment. 2 , 5 , 14 Other bacteria, such as Faecalibacterium prausnitzii of the Firmicutes phylum, appear to decrease levels of pro‐inflammatory cytokines (e.g., Interleukin [IL]‐12) and increase levels of anti‐inflammatory cytokines (e.g., IL‐10). 14 Therefore, the gut microbiome appears to not only help induce an appropriate inflammatory immune response (e.g., in the setting of infection), but also tempers “unnecessary” inflammation to support maintenance of healthy gut epithelium. 2 , 14
3.2.2. Metabolic functions
Bile acid homeostasis
Bile acids, which are produced in the liver, aid with fat digestion and absorption of fat‐soluble vitamins upon secretion into the small intestine. While primary bile acids are secreted in their conjugated form, several commensals (Firmicutes, e.g., Clostridium, Lactobacillus, and Enterococcus species; Bacteroidetes, for example, Bacteroides species; and Actinobacteria, for example, Bifidobacterium species) 15 express a bile salt hydrolase enzyme that deconjugates the acids back to an unconjugated form. This supports enterohepatic recirculation of most of the bile acid pool. 16 The small percentage of primary bile acids that reach the large intestine are converted by commensals to secondary bile acids. This process is performed by select microbiota (<0.025% of gut microbes) that express alpha dehydroxylase enzymes. 15 The most well‐characterized species in this role is Clostridium scindens. 15 , 17 , 18 Both primary and secondary bile acids are involved in glucose metabolism and have antimicrobial detergent properties that support colonization resistance. 14 Secondary bile acids play a large role in inhibiting Clostridioides difficile growth in particular. 18 Given the essential role that microbiota play in bile acid homeostasis, this is an emerging biomarker to help distinguish between a “healthy” (i.e., predominance of secondary bile acids relative to primary bile acids in the colon) and “unhealthy” microbiome. 17
Short‐chain fatty acid production
Gut microbiota ferment unabsorbed starches and soluble dietary fiber from the host's diet, which results in generation of short‐chain fatty acids (SCFA). 2 , 14 Diets that are rich in fiber (e.g., plant‐based) and fermented foods are therefore considered “gut healthy” because they are sources of SCFA. The most predominant SCFA are butyrate, propionate, and acetate. 2 SCFA production is widespread among microbiota, with generation of specific SCFA varying among specific bacterial groups. 19 Butyrate generation is particularly important as it is the preferred energy source of colonic epithelial cells. 2 Several members of the Firmicutes phylum (e.g., Roseburia and Ruminococcus strains) are key butyrate producers. 7 , 19 Butyrate produced by gut microbiota is estimated to provide 5–10% of caloric requirements for the host by supplying energy to colon cells. 2 , 14 SCFA are also believed to be involved in other functions, including appetite and glucose tolerance. 14 Furthermore, SCFA have been shown to have various immune‐modulating properties, 6 including antagonizing the proinflammatory effects of certain cytokines on the gut epithelium 14 and the ability to interact with neutrophils to either stimulate or suppress local activity. 2 SCFA are therefore important contributors to gut barrier health by both providing energy to epithelial cells and tempering local inflammation. 2 , 14 These functions support the purported health benefits of a high‐fiber diet and, conversely, the potential loss of benefits associated with a traditional Western diet. 8
Vitamin synthesis
Intestinal microbiota are essential for synthesis of select vitamins. Endogenous B vitamins such as cyanocobalamin are synthesized exclusively by gut microbiota (e.g., Firmicutes, Actinobacteria, and Proteobacteria strains), 5 which are essential for a range of healthy metabolic processes including DNA replication and repair. 14 Several commensal bacteria also produce vitamin K, including Bacteroides fragilis, Eubacterium lentum, Enterobacter agglomerans, Serratia marcescens, and Enterococcus faecium. 14 Alteration of the abundances of these bacteria can therefore affect the availabilities of these endogenous vitamins.
3.3. Dysbiosis
Disruption of the gut microbiome that deviates from a healthy or normal state (i.e., eubiosis) is known as dysbiosis. 4 Dysbiosis is characterized by alterations in the composition and/or functions of the microbiome 6 , 8 , 11 (Figure 2). Altered composition can occur in the setting of reduction in species diversity and/or via changes in the relative abundances of “healthy” relative to “less healthy” microbes. 6 , 11 , 14 Although there are consistencies in microbial distribution patterns among healthy individuals, an Anna Karenina principle has been proposed for dysbiosis 20 in that a microbiome can be considered “unhealthy” in myriad individualistic ways 8 , 21 ; therefore, “dysbiosis” is a broadly encompassing term. 22 An example of dysbiosis that is often described in the gut microbiome literature is an increased relative abundance of Proteobacteria, 7 , 11 , 17 , 19 , 21 including Escherichia coli and Klebsiella species. For example, although the relative abundances of Proteobacteria in healthy individuals is generally <10%, 9 , 23 relative abundances of about 20–30% have been observed in patients with dysbiosis. 11 , 19
FIGURE 2.
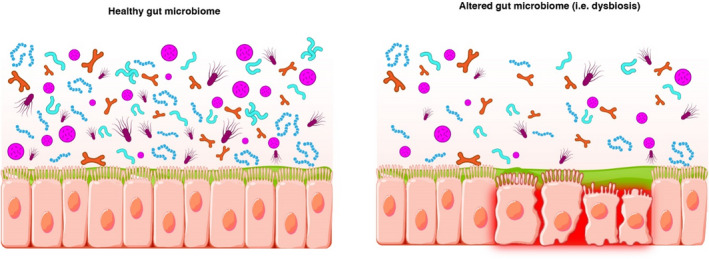
Dysbiosis is associated with alterations in the composition and/or functions of the microbiome
Although there is no gold standard approach to identifying dysbiosis, several indices have been proposed. These are primarily derived from studies of patients with recurrent C. difficile infection (CDI), given its association with microbiome disruption. One proposed dysbiosis index is a ratio of the total number of Proteobacteria strains divided by the total number of overall bacterial strains. 11 Another, called the Microbiome Health Index (MHI), considers bacterial distributions within the Firmicutes, Bacteroidetes, and Proteobacteria phyla. MHI, evaluated after antibiotic exposure, is measured by the relative abundances of bacteria belonging to the Clostridia (Firmicutes phylum) and Bacteroidia (Bacteroidetes phylum) classes, given their associations with healthy functions, compared with the abundances of Bacilli (Firmicutes phylum) and Gammaproteobacteria (Proteobacteria phylum), given their association with pathogenesis. 24
3.3.1. Etiologies of dysbiosis: Focus on medications
Precipitating factors for dysbiosis that are described in the literature include genetic defects, stress, diet, alcohol consumption, infection, and medication exposure including both antibiotics and non‐antibiotics. 2 , 14 , 25 Several articles address the effects of antimicrobials on dysbiosis, including in the context of CDI, in greater detail. 26 , 27 , 28 , 29 , 30 Notably, durations of microbiota perturbations appear to vary among antimicrobials, lasting in some cases for months (e.g., cephalosporins) to years (e.g., fluoroquinolones, clindamycin). Some agents (e.g., tetracyclines, macrolides, and sulfonamides) are associated with a shorter perturbation time, 28 perhaps related to their narrower antimicrobial spectrums. Non‐systemically absorbed antibiotics, including fidaxomicin and orally administered vancomycin, also demonstrate variable effects. Fidaxomicin is more commensal sparing than vancomycin, 17 , 31 in that vancomycin has appreciable activity against a variety of Firmicutes (e.g., Clostridia, Ruminococcus species) and Bacteroidetes. 31 Vancomycin over‐exposure may contribute to selection of pathogenic bacterial strains, including vancomycin‐resistant Enterococci 32 and increased abundances of Proteobacteria. 17 Untoward effects of vancomycin on the microbiome can be observed for a month or more after treatment. 31 , 33
Whereas the potential for antibiotics to induce dysbiosis is well appreciated, several non‐antimicrobial medications appear to have comparably undesirable effects on the gut microbiome. Proton pump inhibitors have been shown to modulate the abundances and densities of commensals throughout the gastrointestinal tract, including increasing Proteobacteria and decreasing Actinobacteria in both the small intestine and colon. 34 Psychiatric and mood disorder medications (e.g., selective serotonin reuptake inhibitors, olanzapine) have also been described as potentially contributing to dysbiosis by exhibiting antimicrobial properties. 35
3.3.2. Consequences of dysbiosis
Dysbiosis is associated with numerous consequences, many of which can be appreciated when considering the numerous beneficial functions performed by the gut microbiome in a healthy state. Decreased abundances of important commensals can lead to loss of colonization resistance that may lead to overgrowth of pathogenic bacteria including C. difficile. 2 There also may be an element of functional immune compromise, given the role that gut microbiota play in immune modulation in a state of homeostasis. 2 , 6 Increased gut inflammation may occur, as has been observed with higher abundances of Proteobacteria, 14 which can increase risk of systemic infection including bacteremia. 19 Inflammation may be further exacerbated with decreased production of SCFAs, such as butyrate, that exert anti‐inflammatory effects and help maintain gut barrier integrity. 14 , 25
CDI after antibiotic exposure is one of the most well‐described consequences of dysbiosis. Various species of gut microbiota may become collateral damage depending on the antibiotic(s) used, resulting in decreased bacterial diversity and altered abundances of communities within the microbiome. Loss of these communities may result in loss of protective functions that would otherwise be provided in the presence of C. difficile. For example, antibiotic exposure may result in a higher amount of primary relative to secondary bile acids in the gut, due to loss of Clostridia species that are known to perform this conversion. If C. difficile spores are present, these primary bile acids will trigger spore germination into toxin‐producing vegetative cells. 15 , 18 Inhibitory effects on vegetative cells, which would typically be provided by secondary bile acids, are diminished in the setting of decreased conversion of primary to secondary acids. 15 , 18 These vegetative C. difficile cells can then proliferate when colonization resistance is compromised, via less competition for nutrients and reduced expression of antibacterial peptides by commensals. 2 , 5 , 14 Local gut inflammation and damage to colonic cells that likely were precipitated by antibiotic‐associated dysbiosis are further exacerbated by toxins secreted by vegetative C. difficile cells. Local immune function that is typically modulated by the presence of commensals may also be compromised, further facilitating growth of C. difficile. To complicate this scenario, there appears to be a bidirectional element of dysbiosis in that it can precipitate CDI, be further exacerbated by certain anti‐CDI antibiotics, and perpetuate a cycle of recurrent infections via the mechanisms described above in the absence of microbiome restoration.
In addition to being well characterized for its role in CDI, dysbiosis has also been linked to several disease states and conditions affecting a variety of organ systems. A non‐inclusive list is featured in Table 2. Notably, dysbiosis as the cause versus consequence of a condition or disease is often not well understood. 8 This bidirectional relationship between dysbiosis and disease has been proposed for inflammatory bowel diseases, 14 as well as neurologic and psychiatric disorders, via disruption of the gut‐brain signaling axis. 36 In some cases, pathogenesis is postulated to involve compromised gut barrier integrity or a “leaky gut.” This is likely a consequence of localized inflammation, which leads to host exposure to gut substances that can act like antigens to induce both local and systemic inflammatory responses. 14 Although proposed underlying mechanisms of dysbiosis and disease are often complex, they are likely multifactorial. 8
TABLE 2.
Diseases or conditions proposed to have gut microbiome‐associated pathophysiology 3 , 14 , 25 , 36 , 39
| Type of disease/condition | Specific examples |
|---|---|
| Immune‐mediated/autoimmune diseases |
Inflammatory bowel disease (Crohn's disease and ulcerative colitis) Irritable bowel syndrome Celiac disease Systemic lupus erythematosus Type 1 diabetes Rheumatoid arthritis Atopic disease (e.g., childhood allergic asthma) Necrotizing enterocolitis Autoimmune liver diseases (primary sclerosing cholangitis, primary biliary cirrhosis, autoimmune hepatitis) Immunoglobulin A deficiency Graft‐versus‐host disease |
| Metabolic/cardiovascular disorders |
Obesity Type 2 diabetes Hypertension Atherosclerosis |
| Cancer | Colorectal cancer |
| Neuropsychiatric |
Autism spectrum disorder Alzheimer's disease Depression Parkinson's disease |
| Infectious disease |
Clostridioides difficile infection Human immunodeficiency virus (HIV) |
| Other |
Chronic kidney disease Liver disease (non‐alcoholic fatty liver disease, non‐alcoholic steatohepatitis) Sepsis Small intestinal bacterial overgrowth |
Dysbiosis may also affect the activities of certain medications. For instance, gut microbiota can convert tacrolimus to less potent metabolites, which may help to explain why oral tacrolimus can result in variable drug exposure between patients. 37 Metformin efficacy appears to be due in part to commensals in that its antihyperglycemic effect can be significantly reduced with the concomitant administration of oral vancomycin. 38 Digoxin appears to be inactivated by Eggerthella lenta, 39 and the effect of vitamin K antagonists such as warfarin may be more pronounced in patients who are receiving antibiotics that eliminate microbiota that produce vitamin K. 40 In addition, the efficacy of immune checkpoint inhibitors that inhibit tumor growth can be improved or restored with the supplementation of certain commensal bacteria. 41 Ultimately, the role of the gut microbiome in medication metabolism and the potential clinical implications remain poorly understood. It has been postulated that species within the Clostridia and Bacilli bacterial classes might be involved in P‐glycoprotein expression. 42 Receptors in the gut may interact with both microbiota‐derived compounds such as butyrate and exogenous compounds, including medications. Given that some gut receptors regulate expression of metabolic and transporter systems such as cytochrome P450, 25 it can be postulated that dysbiosis could alter metabolism of drugs that utilize these pathways.
3.4. Microbiome restoration
When dysbiosis occurs, restoration of the microbiome toward a healthy pre‐dysbiosis state can begin either organically upon removal of the inciting factor (e.g., antibiotic) or occur via more deliberate therapeutic approaches. One highly accessible approach to restoration or supplementation of existing healthy bacteria is via administration of pre‐ or probiotics. Prebiotics are intended to serve as a nutritional source for healthy gut bacteria (e.g., those that produce butyrate). 43 Probiotics, defined as “live microorganisms which when administered in adequate amounts confer a health benefit to the host,” are intended to have positive modulatory effects on the gut microbiome, including via pleiotropic effects. Common probiotic formulations include Lactobacillus, Bifidobacterium, and/or Saccharomyces species. 43 Although probiotics may be beneficial in certain scenarios (e.g., mitigation of antibiotic‐associated diarrhea), they are not regulated by the United States Food and Drug Administration (FDA) and can pose safety concerns including risk of bloodstream infection in select high‐risk populations such as the immunocompromised. 43 , 44 Probiotics also do not fully address the extensive bacterial diversity of a healthy microbiome, and in some cases may delay recovery to a pre‐dysbiosis state. 45 Although probiotics are often discussed in the context of CDI‐associated dysbiosis, current guidelines do not recommend their use for primary or secondary infection prevention. 44
A more resource‐intensive approach to microbiome restoration is through microbiota‐based transplantation via the stool of a healthy donor. Fecal microbiota transplantation (FMT) has been shown to restore bacterial diversity in patients with recurrent CDI 46 and is currently recommended by clinical practice guidelines for this indication. 44 , 47 , 48 Investigational FMT has also been used for a variety of dysbiosis‐associated conditions outside of CDI, including graft‐versus‐host disease, malignancies, and decolonization of multidrug‐resistant organisms. 36 Notably, FMT is not standardized in its administration techniques nor is it FDA‐approved for any therapeutic indication. Some safety concerns are associated with FMT, including risk of transmission of multidrug‐resistant organisms from donor to recipient, as outlined in recent case reports. 49 However, several novel and standardized microbiota‐based restoration therapies that range from more narrow (e.g., Firmicutes only; few deliberately selected bacterial strains) to broad consortium (e.g., the collective microbiome community) products are being studied in late‐stage clinical trials to reduce the risk of recurrent CDI. Though various means to evaluate the effectiveness of microbiome restoration are of interest, there is no gold‐standard approach to determining “engraftment” of a product in a recipient. However, identification of key commensals in both the recipient and originating product, confirmation of appropriate microbiome‐associated functions, and resolution or mitigation of the condition of interest appear to be highly relevant indicators of successful restoration.
3.5. Role of the clinician
Clinicians can promote gut health among their patients through endorsement of a healthy diet and via appropriate use of medications. Clinicians can promote a diet rich in fiber to patients with or at risk for dysbiosis, given the beneficial role it can play in restoring and maintaining a healthy gut microbiome (e.g., as a source of SCFA). For medications, clinicians can steward use of agents that may precipitate or contribute to dysbiosis (e.g., antimicrobials, proton pump inhibitors). In accordance with best antimicrobial stewardship practices, clinicians should prioritize use of the narrowest spectrum antimicrobial possible, used for the shortest possible duration to minimize collateral damage on the microbiome. Similar consideration for collateral damage should be made when evaluating risk versus benefit of oral vancomycin, particularly in the setting of CDI prophylaxis. For patients who will receive microbiome restoration therapy such as FMT, clinicians should help ensure that anti‐CDI therapy has a sufficient washout period (e.g., 24–72 h or as recommended by the treating specialist) leading up to FMT administration. It can also help to be mindful of the potential effects of the microbiome on certain medications (e.g., warfarin, tacrolimus, and checkpoint inhibitors) and any potential role that dysbiosis may play in their effectiveness or safety.
4. FUTURE DIRECTIONS
Microbiome research continues to evolve at rapid speed. Several areas of interest are in various “omics” (e.g., metagenomics, metabolomics) relating to the microbiome. Research findings can lead to increased understanding of the microbiome that can then be translated for clinical purposes. For example, studies in metabolomics can help identify biomarkers associated with specific functions of the gut microbiome. Characterization of bile acid and/or SCFA profiles have been proposed to help distinguish a “healthy” versus “unhealthy” microbiome. These analyses can supplement bacterial identification studies to confirm that key bacteria are not only present but also functioning appropriately. Furthermore, these analyses can be particularly helpful in identifying patients who may benefit from microbiome restoration, be it in a broad or microbiota‐specific manner. Microbiota‐specific findings, particularly as they relate to specific diseases on the acute or chronic disease continuum, can be used to develop more targeted therapeutics. Functional analyses can help inform successfulness of “engraftment” for microbiota‐based therapeutics, paired with clinical success (e.g., absence of recurrent CDI). Given the extensive therapeutic potential of the gut microbiome, the hope is that technology will evolve to be convenient, accurate, precise, and rapid enough to bring microbiome‐based medicine from bench to bedside.
5. CONCLUSIONS
The gut microbiome is often considered the forgotten organ due to its role in both health and disease. In a healthy state of homeostasis, gut microbiota aid with digestion, metabolism, and immune modulation, among other functions. The most prominent bacteria aiding in these functions are Firmicutes and Bacteroidetes, followed by Actinobacteria and Proteobacteria. Dysbiosis can occur via numerous means, including upon antibiotic exposure. Dysbiosis is associated with deviation from a “healthy” microbiome profile and has been linked to various conditions and diseases. Even though cause versus consequence is poorly understood in many cases of dysbiosis and disease, research in this space is emerging rapidly. Increased availability of microbiota‐based therapeutics and clinically feasible testing methodologies may help accelerate the gut microbiome's place in restoring health and mitigating disease.
AUTHOR CONTRIBUTIONS
TPL and MRB contributed to study conceptualization and design. All authors contributed to drafting of the manuscript and critical revision of the manuscript for important intellectual content.
CONFLICT OF INTEREST
MRB is an employee of Ferring Pharmaceuticals. TPL is a consultant for Ferring Pharmaceuticals. ALVH reports no conflicts. Authors did not receive specific funding or compensation for this work.
ACKNOWLEDGMENT
Administrative support in the form of copy‐editing, fact‐checking, referencing, and figure creation was funded by Ferring Pharmaceuticals.
Bidell MR, Hobbs ALV, Lodise TP. Gut microbiome health and dysbiosis: A clinical primer. Pharmacotherapy. 2022;42:849‐857. doi: 10.1002/phar.2731
REFERENCES
- 1. O'Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7(7):688‐693. doi: 10.1038/sj.embor.7400731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lawley TD, Walker AW. Intestinal colonization resistance. Immunology. 2013;138(1):1‐11. doi: 10.1111/j.1365-2567.2012.03616.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Amon P, Sanderson I. What is the microbiome? Arch Dis Child Educ Pract Ed. 2017;102(5):257‐260. doi: 10.1136/archdischild-2016-311643 [DOI] [PubMed] [Google Scholar]
- 4. Thursby E, Juge N. Introduction to the human gut microbiota. Biochem J. 2017;474(11):1823‐1836. doi: 10.1042/bcj20160510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wexler AG, Goodman AL. An insider's perspective: Bacteroides as a window into the microbiome. Nat Microbiol. 2017;2:17026. doi: 10.1038/nmicrobiol.2017.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Buford TW. (Dis)Trust your gut: the gut microbiome in age‐related inflammation, health, and disease. Microbiome. 2017;5(1):80. doi: 10.1186/s40168-017-0296-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shahinas D, Silverman M, Sittler T, et al. Toward an understanding of changes in diversity associated with fecal microbiome transplantation based on 16S rRNA gene deep sequencing. mBio. 2012;3(5):1‐10. doi: 10.1128/mBio.00338-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. McBurney MI, Davis C, Fraser CM, et al. Establishing what constitutes a healthy human gut microbiome: state of the science, regulatory considerations, and future directions. J Nutr. 2019;149(11):1882‐1895. doi: 10.1093/jn/nxz154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rinninella E, Raoul P, Cintoni M, et al. What is the healthy gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases. Microorganisms. 2019;7(1):1‐22. doi: 10.3390/microorganisms7010014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Oren A, Garrity GM. Valid publication of the names of forty‐two phyla of prokaryotes. Int J Syst Evol Microbiol. 2021;71(10):1‐7. doi: 10.1099/ijsem.0.005056 [DOI] [PubMed] [Google Scholar]
- 11. Verma S, Dutta SK, Firnberg E, Phillips L, Vinayek R, Nair PP. Identification and engraftment of new bacterial strains by shotgun metagenomic sequence analysis in patients with recurrent Clostridioides difficile infection before and after fecal microbiota transplantation and in healthy human subjects. PLoS One. 2021;16(7):e0251590. doi: 10.1371/journal.pone.0251590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Spellerberg IF, Fedor PJ. A tribute to Claude Shannon (1916–2001) and a plea for more rigorous use of species richness, species diversity and the ‘Shannon–Wiener’ Index. Glob Ecol Biogeogr. 2003;12(3):177‐179. doi: 10.1046/j.1466-822X.2003.00015.x [DOI] [Google Scholar]
- 13. Wilson BC, Vatanen T, Jayasinghe TN, et al. Strain engraftment competition and functional augmentation in a multi‐donor fecal microbiota transplantation trial for obesity. Microbiome. 2021;9(1):107. doi: 10.1186/s40168-021-01060-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kho ZY, Lal SK. The human gut microbiome – a potential controller of wellness and disease. Front Microbiol. 2018;9:1835. doi: 10.3389/fmicb.2018.01835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Winston JA, Theriot CM. Impact of microbial derived secondary bile acids on colonization resistance against Clostridium difficile in the gastrointestinal tract. Anaerobe. 2016;41:44‐50. doi: 10.1016/j.anaerobe.2016.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Urdaneta V, Casadesús J. Interactions between bacteria and bile salts in the gastrointestinal and hepatobiliary tracts. Front Med. 2017;4:1‐13. doi: 10.3389/fmed.2017.00163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Qian X, Yanagi K, Kane AV, et al. Ridinilazole, a narrow spectrum antibiotic for treatment of Clostridioides difficile infection, enhances preservation of microbiota‐dependent bile acids. Am J Physiol Gastrointest Liver Physiol. 2020;319(2):G227‐g37. doi: 10.1152/ajpgi.00046.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shen A. A gut odyssey: the impact of the microbiota on Clostridium difficile spore formation and germination. PLoS Pathog. 2015;11(10):e1005157. doi: 10.1371/journal.ppat.1005157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sorbara MT, Pamer EG. Microbiome‐based therapeutics. Nat Rev Microbiol. 2022;20(6):365‐380. doi: 10.1038/s41579-021-00667-9 [DOI] [PubMed] [Google Scholar]
- 20. Ke S, Pollock NR, Wang XW, et al. Integrating gut microbiome and host immune markers to understand the pathogenesis of Clostridioides difficile infection. Gut microbes. 2021;13(1):1‐18. doi: 10.1080/19490976.2021.1935186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chang JY, Antonopoulos DA, Kalra A, et al. Decreased diversity of the fecal Microbiome in recurrent Clostridium difficile‐associated diarrhea. J Infect Dis. 2008;197(3):435‐438. doi: 10.1086/525047 [DOI] [PubMed] [Google Scholar]
- 22. Hooks KB, O'Malley MA. Dysbiosis and its discontents. mBio. 2017;8(5):1‐11. doi: 10.1128/mBio.01492-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular‐phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104(34):13780‐13785. doi: 10.1073/pnas.0706625104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Blount K, Jones C, Walsh D, Gonzalez C, Shannon WD. Development and Validation of a Novel Microbiome‐Based Biomarker of Post‐antibiotic Dysbiosis and Subsequent Restoration. Front MicroBiol. 2021;12:781275. doi: 10.3389/fmicb.2021.781275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stolfi C, Maresca C, Monteleone G, Laudisi F. Implication of intestinal barrier dysfunction in gut dysbiosis and diseases. Biomedicines. 2022;10(2):1‐27. doi: 10.3390/biomedicines10020289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Langdon A, Crook N, Dantas G. The effects of antibiotics on the microbiome throughout development and alternative approaches for therapeutic modulation. Genome Med. 2016;8(1):39. doi: 10.1186/s13073-016-0294-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Slimings C, Riley TV. Antibiotics and healthcare facility‐associated Clostridioides difficile infection: systematic review and meta‐analysis 2020 update. J Antimicrob Chemother. 2021;76(7):1676‐1688. doi: 10.1093/jac/dkab091 [DOI] [PubMed] [Google Scholar]
- 28. Zimmermann P, Curtis N. The effect of antibiotics on the composition of the intestinal microbiota – a systematic review. J Infect. 2019;79(6):471‐489. doi: 10.1016/j.jinf.2019.10.008 [DOI] [PubMed] [Google Scholar]
- 29. Ferrer M, Méndez‐García C, Rojo D, Barbas C, Moya A. Antibiotic use and microbiome function. Biochem Pharmacol. 2017;134:114‐126. doi: 10.1016/j.bcp.2016.09.007 [DOI] [PubMed] [Google Scholar]
- 30. Schwartz DJ, Langdon AE, Dantas G. Understanding the impact of antibiotic perturbation on the human microbiome. Genome Med. 2020;12(1):82. doi: 10.1186/s13073-020-00782-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Louie TJ, Cannon K, Byrne B, et al. Fidaxomicin preserves the intestinal microbiome during and after treatment of Clostridium difficile infection (CDI) and reduces both toxin reexpression and recurrence of CDI. Clin Infect Dis. 2012;55(Suppl 2):S132‐S142. doi: 10.1093/cid/cis338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Garey KW. Perils, pitfalls, and promise of primary prophylaxis for clostridioides difficile infection. Clin Infect Dis. 2020;71(5):1140‐1141. doi: 10.1093/cid/ciz970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Abujamel T, Cadnum JL, Jury LA, et al. Defining the vulnerable period for re‐establishment of Clostridium difficile colonization after treatment of C. difficile infection with oral vancomycin or metronidazole. PLoS One. 2013;8(10):e76269. doi: 10.1371/journal.pone.0076269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bruno G, Zaccari P, Rocco G, et al. Proton pump inhibitors and dysbiosis: Current knowledge and aspects to be clarified. World J Gastroenterol. 2019;25(22):2706‐2719. doi: 10.3748/wjg.v25.i22.2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sjöstedt P, Enander J, Isung J. Serotonin reuptake inhibitors and the gut microbiome: Significance of the gut microbiome in relation to mechanism of action, treatment response, side effects, and tachyphylaxis. Frontiers Psychiatry. 2021;12:682868. doi: 10.3389/fpsyt.2021.682868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Halaweish HF, Boatman S, Staley C. Encapsulated fecal microbiota transplantation: Development, efficacy, and clinical application. Front Cellular Infect Microbiol. 2022;12:826114. doi: 10.3389/fcimb.2022.826114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Guo Y, Crnkovic CM, Won KJ, et al. Commensal gut bacteria convert the immunosuppressant tacrolimus to less potent metabolites. Drug Metab Dispos. 2019;47(3):194‐202. doi: 10.1124/dmd.118.084772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kim E, Kim AH, Lee Y, et al. Effects of vancomycin‐induced gut microbiome alteration on the pharmacodynamics of metformin in healthy male subjects. Clin Transl Sci. 2021;14(5):1955‐1966. doi: 10.1111/cts.13051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Marchesi JR, Adams DH, Fava F, et al. The gut microbiota and host health: a new clinical frontier. Gut. 2016;65(2):330‐339. doi: 10.1136/gutjnl-2015-309990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Camelo‐Castillo A, Rivera‐Caravaca JM, Orenes‐Piñero E, et al. Gut microbiota and the quality of oral anticoagulation in vitamin K antagonists users: A review of potential implications. J Clin Med. 2021;10(4):1‐15. doi: 10.3390/jcm10040715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rani V, Singhal S, Sharma K, et al. Human gut microbiome: A new frontier in cancer diagnostics & therapeutics. Curr Pharm Des. 2021;27(45):4578‐4592. doi: 10.2174/1381612827666211006152112 [DOI] [PubMed] [Google Scholar]
- 42. Foley SE, Tuohy C, Dunford M, et al. Gut microbiota regulation of P‐glycoprotein in the intestinal epithelium in maintenance of homeostasis. Microbiome. 2021;9(1):183. doi: 10.1186/s40168-021-01137-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Su GL, Ko CW, Bercik P, et al. AGA clinical practice guidelines on the role of probiotics in the management of gastrointestinal disorders. Gastroenterology. 2020;159(2):697‐705. doi: 10.1053/j.gastro.2020.05.059 [DOI] [PubMed] [Google Scholar]
- 44. Kelly CR, Fischer M, Allegretti JR, et al. ACG clinical guidelines: Prevention, diagnosis, and treatment of clostridioides difficile infections. Am J Gastroenterol. 2021;116(6):1124‐1147. doi: 10.14309/ajg.0000000000001278 [DOI] [PubMed] [Google Scholar]
- 45. Khoruts A, Staley C, Sadowsky MJ. Faecal microbiota transplantation for Clostridioides difficile: mechanisms and pharmacology. Nat Rev Gastroenterol Hepatol. 2021;18(1):67‐80. doi: 10.1038/s41575-020-0350-4 [DOI] [PubMed] [Google Scholar]
- 46. van Nood E, Vrieze A, Nieuwdorp M, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med. 2013;368(5):407‐415. doi: 10.1056/NEJMoa1205037 [DOI] [PubMed] [Google Scholar]
- 47. McDonald LC, Gerding DN, Johnson S, et al. Clinical Practice Guidelines for Clostridium difficile Infection in Adults and Children: 2017 Update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clin Infect Dis. 2018;66(7):e1‐e48. doi: 10.1093/cid/cix1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Johnson S, Lavergne V, Skinner AM, et al. Clinical Practice Guideline by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA): 2021 Focused Update Guidelines on Management of Clostridioides difficile Infection in Adults. Clin Infect Dis. 2021;73(5):e1029‐e1044. doi: 10.1093/cid/ciab549 [DOI] [PubMed] [Google Scholar]
- 49. DeFilipp Z, Bloom PP, Torres Soto M, et al. Drug‐resistant E. coli bacteremia transmitted by fecal microbiota transplant. N Engl J Med. 2019;381(21):2043‐2050. doi: 10.1056/NEJMoa1910437 [DOI] [PubMed] [Google Scholar]


