Abstract
High-throughput sequencing for B cell receptor (BCR) repertoire provides useful insights for the adaptive immune system. With the continuous development of the BCR-seq technology, many efforts have been made to develop methods for analyzing the ever-increasing BCR repertoire data. In this review, we comprehensively outline different BCR repertoire library preparation protocols and summarize three major steps of BCR-seq data analysis, i. e., V(D)J sequence annotation, clonal phylogenetic inference, and BCR repertoire profiling and mining. Different from other reviews in this field, we emphasize background intuition and the statistical principle of each method to help biologists better understand it. Finally, we discuss data mining problems for BCR-seq data and with a highlight on recently emerging multiple-sample analysis.
Keywords: high-throughput sequencing, BCR repertoire analysis, statistical method, profiling and data mining
Introduction
The antigen receptor on B cells, B cell receptor (BCR), recognizes the antigen and plays key roles in B cell development, survival and activation. The secreted form of BCR is also called antibody, which contains two identical immunoglobulin (Ig) heavy (IgH) chains and two identical Ig light (IgL) chains [1]. In the human genome, the IgH locus is located at 14q32.33, and Igκ and Igλ are located at 2p11.2 and 22q11.2, respectively [2]. Both IgH and IgL chains can be divided into the variable N-terminal Ig domain (IgV) and constant C-terminal Ig domain (IgHC or IgLC). The IgV contains highly diversified sequences and is primarily responsible for antigen recognition, while the sequence constant domain can activate downstream immune responses.
In this review, different BCR repertoire library preparation protocols are outlined and three major steps of BCR-seq data analysis, i. e., V(D)J sequence annotation, clonal phylogenetic inference, and BCR repertoire profiling and mining are summarized.
Two Layers of BCR Diversification Processes
To recognize different antigens, in each B cell, Ig genes undergo two layers of diversification processes to contribute to the total BCR/antibody repertoire, including the antigen-independent and antigen-dependent processes.
V(D)J recombination, occurring during B cell development in the omentum, fetal liver, or adult bone marrow, assembles the IgV exon to shape a primary antibody repertoire. In this antigen-independent process, the germline variable (V), diversity (D), and joining (J) gene segments are assembled in an ordered manner from a panel of gene segments spanning in the Ig locus. The diversity of primary BCR repertoire comes from the numbers of V/D/J gene segments and is also contributed by the insertion and deletion (indel) at the joining junction [3]. In the IgV domain, the sequence can be further divided into complementarity-determining regions (CDRs) and framework regions (FWRs) [4]. The FWRs and CDR1/2 are contributed by the V gene segment, while CDR3 covers a sequence containing the V-D-J junctions. As a result, the CDR3 is the most diversified sequence and usually plays the utmost important role during antigen recognition [ 5, 6] . From the peptide sequence aspect, CDR3 can be identified from several features, i. e., starting with a cysteine (C) and ending with phenylalanine (F) or tryptophan (W), as highlighted in the sequence annotation from IMGT (international ImMunoGeneTics information system, https://www.imgt.org/) [7].
Upon antigen stimulation, BCR undergoes another layer of diversification, including IgH class-switch recombination (CSR) and IgV somatic hypermutation (SHM), both of which are initiated by activation-induced cytidine deaminase (AID) [ 8, 9] . In CSR, BCR can switch from IgM to another isotype including IgG, IgA, or IgE [10].While in SHM, AID can initiate mutation or small indels at IgV exons [11]. B cells expressing mutated variable BCRs undergo affinity-based selection in the secondary lymphoid structure named germinal center, leading to a process named affinity maturation [12]. Along with these antigen-dependent processes, naïve mature B cells further differentiate into either antibody-secreting plasma cells or memory B cells [13]. In this context, the B cell pools express an advanced antibody repertoire containing highly potent antibodies of different classes.
Preparation of BCR Repertoire Library
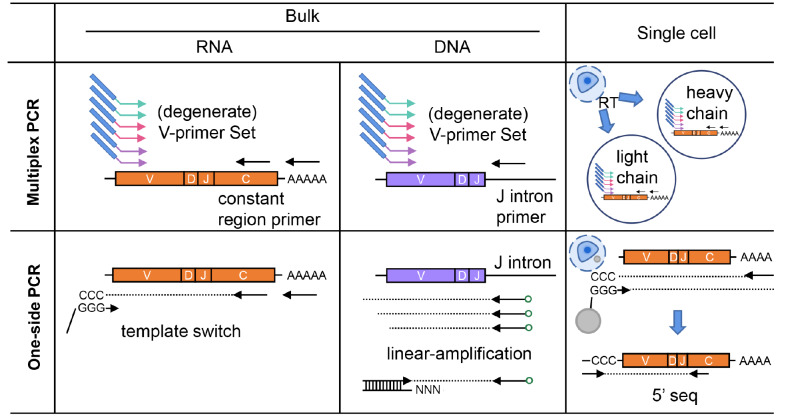
To present a landscape of BCR repertoire, researchers have developed diverse approaches to prepare such a library ( Figure 1), which can be grouped based on the input materials (bulk or single cell, genomic DNA or mRNA) and cloning strategies (multiplex PCR or one-side PCR).
Figure 1 .
Methods for BCR repertoire preparation
Multiplex PCR: regardless of the template types, degenerated primer sets targeting the V gene segments are adopted. Different colors of arrows symbolizes different primer sets targeting various V classes. When mRNA (from bulk or single cell) is used as input, reverse primers annealing to C segments or poly-A sequence are commonly used to amplify the V(D)J. While when genomic DNA is subject to library preparation, the reverse primers usually anneal to the J intron region. One-side PCR methods: In the context of using bulk mRNA as template, after reverse transcription, several cytidines are over-hanged to the 3’ end of synthesized cDNA. Then a “template switch” primer with poly-G sequence will add adaptor on this side. In bulk mRNA samples, 5’ RACE is a popular example. The biotinylated primers annealing to J intron segment are used to select the target ssDNA sequence with streptavidin. In the next step, a dsDNA “bridge primer” is used to mediate the following ligation and synthesize of the other strand. The one-side PCR strategy of single-cell samples, e.g., 5’ seq, is similar to that of bulk mRNA, and the “template switch” primers are attached to a barcoded bead for discrimination between different cell samples.
RNA from a bulk of cells
The mRNA from a pool of B cells can be subject to library preparation with a multiplex PCR strategy. The complementary DNA (cDNA) can be prepared with a random hexamer or primers annealing to the Ig gene constant exon. Degenerated primer sets targeting the V gene segments [ 14, 15] are applied. Along with the development of the method, many efforts have been made to minimize the multiplex PCR bias. On the other way, the one-side PCR method, e.g., 5’ Rapid Amplification of cDNA Ends (5’ RACE), was also applied to amplify the V(D)J sequence in an unbiased way, as exampled in [ 15– 17] . Regardless of the PCR strategy, using RNA as the input complicates the downstream quantification analysis as mRNA levels are not directly correlated with the numbers of B cell clones [18], as BCR mRNA transcripts vary among different B cells. To present a relatively quantitative profile, several strategies can be applied. For example, in one approach, the authors separated the B cells into different aliquots before extracting RNA [ 19, 20] . In another, a unique molecular identifier (UMI) is introduced to tag RNA molecules during cDNA synthesis [21].
DNA from a bulk of cells
When genomic DNA was used as an input, both multiplex and one-side PCR can be applied. In multiplexed PCR, forward primer sets annealing to the V segments and reverse primers annealing to J segments are usually used [14]. Linear amplification-mediated one-side PCR (LAM-PCR) is used to amplify the V(D)J fragments [ 22, 23] . In this context, DNA-input repertoire allows analysis of both productive and non-productive V(D)J rearranged products, while the mRNA of no-productive allele is degraded through a nonsense-mediated mRNA decay pathway [24] and less-covered in RNA-input library. Furthermore, the BCR sequence number is correlated with the B cell numbers, allowing a more precise evaluation of B cell clonal expansion. However, the DNA-input repertoire losses the IgH class information, as the constant exon is several kilo-base pairs from the V(D)J exon [25].
Single cell
The emerging single-cell methods enable the analysis of BCR at the single-cell level. Multiplex PCR can be apply to a single B cell to amplify the IgH and IgL chains [26], as exampled by the cloning of anti-viral neutralizing antibodies [27]. On the other hand, 5’ single-cell RNA sequencing (5′ scRNA-seq), as a one-side PCR-based method, utilizes microfluidic devices to profile the antibody V(D)J sequence together with the transcriptome. Compared to the repertoire library of bulk-input, single-cell BCR repertoire retains the IgH and IgL paring information but is low-throughput and costly.
Analyses of BCR Repertoire Sequencing Data
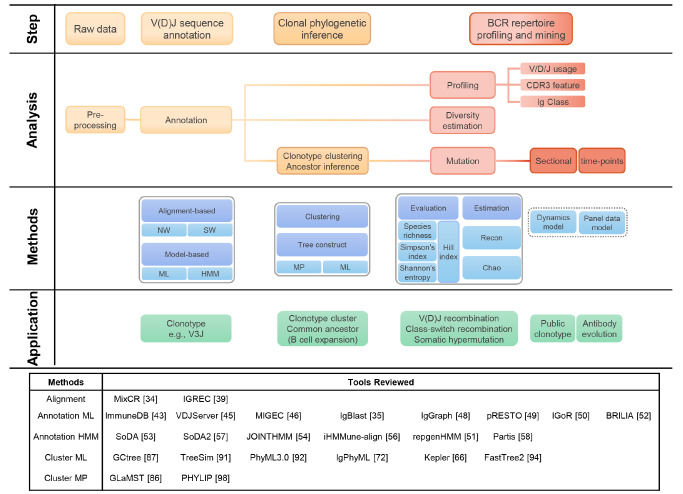
The BCR repertoire sequencing (BCR-seq) libraries are usually sequenced through high-throughput sequencing (HTS) to generate data in FASTQ format. Different from other types of HTS data, the BCR-seq data analysis can be summarized into the following three major steps [ 28– 30] : (1) V(D)J sequence annotation; (2) clonal phylogenetic inference; and (3) BCR repertoire profiling and mining ( Figure 2). In the following section, we will review the key ideas and their statistical principles in each step.
Figure 2 .
A reference analysis pipeline for BCR-seq data
V(D)J sequence annotation
As the first step, V(D)J sequence annotation infers the V(D)J gene segments and CDR3 nucleotide/amino acid sequences from the preprocessed BCR-seq data. Generally, there are two ways to annotate the sequence: alignment-based and model-based algorithms, both of which use germline Ig sequences as reference. In this task, IMGT [7] is the most frequently used database for Ig reference.
Alignment-based algorithms
Sequence alignment, an algorithm to compare two or more biological sequences, is probably the most fundamental procedure in data analyses [ 31– 33] . It was intensively used in many applications such as functional annotation of an unknown protein/DNA sequence, phylogenetic analysis, and database searching [ 32, 34, 35] . According to different optimization objectives, sequence alignment can be divided into the following two types: global algorithm (Needleman-Wunsch, NW [36]) and local algorithm (Smith-Waterman, SW [37]).
Needleman-Wunsch alignment algorithm aims to calculate the best overall similarity score between the query sequence and a target sequence by a dynamic programming algorithm. On the contrary, the Smith-Waterman algorithm does not devote to comparing the entire sequence, but to finding the fragments with high similarity in two sequences. Both algorithms are successfully applied to DNA and protein sequence analyses, while the local algorithm is more frequently used in BCR-seq analysis. For example, Bolotin et al. [34] proposed MixCR, a comprehensive framework of adaptive immunity analysis with features of low-quality sequence rescue and cluster-based clonotype inference. It uses Subread [38], a local alignment algorithm to annotate the V(D)J gene segment of each core sequence. Another annotation tool in the same category is IGREC [39], which performs a two-step alignment to find the longest subsequence of k-mers between reads and Ig segments to infer the Ig segment, e.g., Ighutil [40]. The annotation step yields the antibody clonotypes. For example, the cAb-Rep database [41] summarized a total of 267.9 million IgH and 72.9 million IgL clonotypes annotated by the SONAR pipeline [42].
Model-based algorithms
Due to the complexity of the BCR repertoire, researchers further introduced statistic-based methods to precisely annotate V(D)J sequences. Two types of models are frequently used in this category, i. e., classical probabilistic model and Hidden Markov Model (HMM).
Classical probabilistic model
Classical probabilistic model describes the biological process by a series of dependent or independent probabilistic events, and parameters can be estimated by maximizing the likelihood function of the observed data. This kind of model is widely used in many tools including ImmuneDB, VDJServer, and MIGEC. Among them, ImmuneDB [43] is a tool for adaptive immune repertoire analysis and repertoire data storage. Besides the alignment-based method, ImmuneDB also provides the likelihood of closely related V genes [44]. Two other probabilistic models VDJServer [45] and MIGEC [46] use a model-based searching tool called IgBlast [ 35, 47] to perform the annotation, and IgGraph [48] uses colored de Bruijn graphs to help the annotation. pRESTO [49] is also a model-based annotation scheme that labels individual reads by extending the sequence descriptions.
We take IGoR as an example to illustrate the classical probabilistic model in detail. IGoR [50] models the antibody recombination as three basic types of recombination events, i. e., germline Ig segments choice, insertion, and deletion (without consideration of mutation events). These interconnected events were described as a conditional probability density function, which is also applied by other tools [51].
For each V(D)J sequence x, denote the objective function of it as P rcomb( x, θ), where θ represents the set of parameters to be determined. When obtaining the value of θ, each query sequence can be easily annotated as the V(D)J gene segment that gives the highest observation probability. In detail, it first performs a local or global alignment between the query sequence x i and the germline gene sequence g j . Second, it marks each nucleotide as one of the basic types (germline Ig segments choice, insertion, and deletion) of recombination. Third, IGoR substitutes the parameters ( θ) and observed values from the last step into the objective function so that we would get the probability p i·j (the query sequence x i is expanded from the germline gene sequence g j ). Fourth, IGoR repeats the above steps until all the germline gene sequences are traversed. Then the query sequence x i will be annotated as the germline gene sequence g j* with the highest probability p i·j* among all j germline gene sequences.
The key step of this method is how to optimize the parameter θ. Parameter θ can be obtained from previous experiments or estimated with the following procedure (for most cases). Based on available observations, one can calculate the likelihood function based on the probabilistic model and maximize the likelihood function to find the best parameters, and the difference between probabilistic and likelihood models is described in Box 1 ( Supplementary Data). IGoR utilized Expectation Maximum (EM) algorithm to calculate the MLE estimation of parameter. The EM algorithm is a powerful statistical method in biological data analysis with latent variables, which is shown as a “coin” example in Box 2 ( Supplementary Data). Briefly, the recombination event (latent variable) is an analogy to the side of “coins”, and the probability of head-side up (the observable variable) is affected by parameter θ. After repeating the E and M steps iteratively, both parameter θ and recombination events will be obtained. Once we have the parameter θ, recombination events of other homogeneous samples can be inferred with ML- or Bayesian-based method directly.
Due to the huge amount of reads in a typical BCR experiment, a classical probabilistic model usually takes substantial computation time. In order to save the computation time and reduce the inference error, other researchers claimed to cluster the sequences first and annotated sequences in the same cluster as one clonal type [52].
Hidden Markov Model
Hidden Markov Model (HMM) is another popular annotation algorithm implemented in SoDA [53], JOINTHMM [54] (used in [55]), iHMMune-align [56] and SoDA2 [57]. According to HMM, each reference germline sequence corresponds to a hidden state, which could not be directly observed in BCR-seq data. In contrast, the observed BCR-seq sequences, which undergo recombination events, correspond to the observed series (at each position, the observation value correlates with the hidden state ). The entire modeling process is named as Markov process. Different from classical probabilistic models, HMM is built up with two probability matrices: transition matrix and emission matrix.
For one germline sequence, each nucleotide of it corresponds to a hidden stage, which will ‘emit’ an observation among A, T, G, and C. In particular, if the hidden stage of a nucleotide is C, it will mostly emit a C, but it will also emit to other nucleotides such as T with a small probability, indicating a C>T mutation event. In the transition matrix, each entry quantifies the transition probability of the i-th hidden stage to the ( i+1)-th hidden stage, which can be understood as a simple Markov property. In BCR-seq analysis, the indel events further complicate the model from classical HMM. For deletion events, each hidden stage can either be transited from the initial state or skip all stages behind it to the next gene segment. Naturally, the closer to the start (or end) of gene segment (J or V) that stage position is, the more probably the deletion event would happen. For insertion events, an N-region topology is inserted at the junction site, which is made up of four stages (A/G/C/T) and forms a self-transition structure. Thus, the transition matrix defines the pair-wise transition probabilities between two nucleotides and a loop-break probability at each position.
There are several ways to improve the classical HMM model, e.g., one can use a blank stage between gene segments to block the dependence of cross-segment probabilities (cross V, D, J) and decrease the computing complexity [ 51, 58] ). Alternatively, one can first use the alignment-based method to initialize the parameters of HMM, then apply the model with current parameters to other samples [59].
Clonal phylogenetic inference
Upon antigen stimulation, B cells undergo clonal expansion in germinal center reaction and are selected by their affinities [60], leading to antibody affinity maturation or antibody evolution (in the long run). Thus, methods for constructing B-cell lineage (phylogenetic tree) and inferring the common ancestor (the root of the tree) are needed. The methods for phylogenetic inference [ 29, 61– 64] mainly solve two issues: (1) clustering, to determine the clonotype scope of a certain phylogenetic tree by a given distance measurement; (2) inferring the ancestral sequence as the root node of the tree, to convert the undirected tree to a directed one. These two issues can be solved separately or integrated [ 64– 66] .
Clustering
Clustering is an unsupervised learning method to divide the unlabeled data into several classes and thus can show the unknown data structure and topology. In BCR-seq analysis, clustering is the key step to define a clonotype cluster that potentially represents B cell clonal expansion from one ancestor. As previously demonstrated, the highest abundance antibody usually is not a high-affinity antibody in a BCR repertoire [67]. That’s why we prefer to do clustering for additional valuable information rather than only considering the abundance.
Two steps of clustering
Clustering is usually performed in BCR-seq analysis in both annotation and phylogenetic inference steps. In BCR-seq, both PCR-amplification and sequencing steps could introduce errors [ 39, 48] . In order to reduce the systematical error, in the above “V(D)J sequence annotation” step, an extremely strict cluster strategy is frequently applied to infer antibody clonotype. A clonotype is represented by the core sequence to produce more confident results [ 34, 48] , which also helps to reduce the computational complexity as millions or billions of antibody sequences are produced in a BCR-seq experiment [ 45, 68] . Meanwhile, in the clonal phylogenetic inference step, a relatively loose cluster strategy is applied to infer the clonotype cluster. In addition, different from other data, BCR-seq data are characterized by a huge number of categories with small sizes. In graph theory, these data could be described as a large graph with many dense sub-graphs [69], which requires customized clustering methods.
Clustering strategy
Clustering algorithms can be roughly divided into the following five groups, i. e., partition-, model-, density-, hierarchical-, and spectral-based. Basically, all methods aim to find a partition of samples in a way that samples from the same clusters are as similar as possible, while those from different clusters are as different as possible. Many classical algorithms were developed in each group. For example, in single-cell transcriptome data analysis, hundreds of clustering algorithms were developed, most of which require explicit specification of the cluster number k [70]. In a typical single-cell transcriptome analysis, the cluster number k, which indicates the number of cell types, is usually less than 100 [70]. But in BCR-seq analysis, a fixed kis often infeasible for B-cell repertoire clustering due to the huge number of clonal types [69].
Most clustering algorithms require a distance matrix as the input. Distance between two sequences is calculated based on their similarity or alignment score of either nucleotide or amino acid sequences. Nucleotide sequences could contain mutation hotspot information, while amino acid sequences have a much clear biological significance [ 71, 72] . At the nucleotide level, 90% to 95% similarity threshold of CDR3 was mainly used to define a cluster [ 73– 78] . For the whole V(D)J sequence, the 97% similarity threshold is preferred [ 79, 80] . Moreover, other researchers proposed a hybrid threshold of 97% similarity for the whole sequence and 90% similarity for CDR3 [75]. At the amino acid level, most algorithms used identical sequences or with only 1-2 mismatch as a threshold to define clusters in CDR3 [ 19, 79, 81– 83] . An exception was adopted by Meng et al. [84] who used a low similarity threshold of 85% in CDR3, i. e., 2.4 mismatch in a CDR3 of 16 aa.
Hamming distance and Minimum edit distance (MED) are usually used to define the distance between two sequences. Hamming distance is the simplest algorithm that only counts the number of mismatches in the alignment [ 73– 75, 77, 78] . MED is also called the substitution model, integrating mutation, insertion, and deletion events through a quantitative model [ 85, 86] . MED method has been frequently used by GLaMST, TraCeR, and other tools [ 83, 85, 87, 88] . There are also some heuristic distance metrics based on sequence alignment. For example, BRILIA [52] adopted a penalty algorithm to increase the distance accumulation for consecutive mismatches.
Strategies to accelerate clustering
The distance matrix requires a great deal of computing resources especially for the high-throughput BCR-seq data. In practice, this problem can be alleviated by either using an approximate algorithm or further dividing the dataset into subgroups. For sub-grouping methods, clonotypes can be grouped based on the combination of V and J gene segments or sequence length. Other methods in this line include IgRepertoireConstructor, which constructs a Hamming Graph with many sub-graphs that are defined by a threshold of hamming distance [69]. A few methods apply approximate algorithms, e.g., IGREC [39] uses a fast minimizers algorithm to find the longest subsequence of k- mers between reads and germline sequences as a new filtration strategy to cluster sequences.
Inference of common ancestor
Maximum likelihood (ML) and maximum parsimony (MP) are the most popular methods to construct a phylogenetic tree inside a clonotype cluster and infer the common ancestor, which was originally developed in evolutionary biology [89]. ML method attempts to construct a tree with the highest probability [64], while the MP method puts its effort into minimizing the number of mutation events (sum of all edge weights).
Maximum likelihood method
The ML method is used in both classical probabilistic model-based annotation and phylogenetic tree construction. We take GCtree [86] as an example to show the basic framework of the ML-based method for phylogenetic tree construction. GCtree uses both distance and abundance information to construct a phylogenetic tree. Two key parameters p and q are introduced, where p defines whether the node bifurcates, and q defines whether the descendant contains a mutant. These two parameters were assumed as independent in calculating the likelihood of a GCtree, which is defined by multiplying the likelihood function of each node (clonotype) [86]. At last, the infinite type of assumption is raised to make sure each node can be identified with one sub-tree in the original lineage tree. An EM algorithm is applied to estimate the parameters, while the tree topology is treated as a latent variable , and the parameter =( p, q) as observable variables . Using the initial tree topology and the observed abundance, we can get the estimated parameter . Based on , the user can reconstruct the tree and perform another iteration of estimation.
Tools for inferring phylogenetic tree by the ML framework based on different model assumptions. For example, TreeSim [90] generates a lineage tree by modeling the extant species evolution process as a dynamic method––episodic birth-death process (EBDP). PhyML3.0 [91] utilizes the nearest neighbor interchanges (NNIs) to get a fast approximate MLE. Bonsignori et al. [92] used PhyML3.0 to construct a phylogenetic tree and infer the common ancestor of the VRC01 anti-HIV-1 antibody lineage. Paschold et al. [87] used a fast approximate MLE method, FastTree2 [93], to infer the phylogenetic tree of SARS-CoV-2-specific antibody. Though not as accurate as ML-based methods, it is 100-1000 times faster. IgPhyML [71] estimates a transition rate matrix by considering the hot-spot and cold-spot motifs and other situations. Kepler’s group developed a computational framework that iteratively infers a phylogenetic tree by taking the unmutated ancestral sequence with the highest posterior probability and re-annotate the sequence set [66]. It can integrate the VDJ recombination and phylogenetics through the iterative algorithm.
Maximum parsimony method
Maximum parsimony (MP), also called the minimum spanning method, is an approximate algorithm for phylogenetic tree construction. The main procedure of MP usually starts with inferring an undirected tree by a certain distance metric and is followed by an iterative step of trimming and rewiring the phylogenetic tree. The undirected tree can be constructed by the Kruskal’s algorithm [94] or Prim’s algorithm [ 95, 96] , which are based on edges and nodes, respectively. They sort the edges by their weights (or nodes’ distance in Prim) and continuously choose the edge (node) that keeps the graph acyclic in order to construct a minimum spanning tree (MST).
Some existing MP-based tools, such as GLaMST [85], use a heuristic method to literally trim and rewire the lineage tree and complete the tree by generating the intermediate sequences that were not observed. Liberman et al. [97] used PHYLIP, an MP-based method to produce a lineage tree and detect population selection [98]. Li et al. [76] constructed an undirected tree and used it to analyze lymph node IgA-expressing.
These two types of methods have their advantage and disadvantage. ML methods can bring the best result under its model assumption, while MP methods have a significant advantage in computation time. For both types of methods, we can use the conservative tree estimation process to obtain a more confident result of both the structure of the tree and other analyses such as substitution model. For instance, we can exclude nodes without descendants or just remove trees with only a two-tier structure.
BCR repertoire profiling and mining
The BCR-seq data contain multiple layers of information, which can be displayed in various ways. Here, we review the popular single-sample profiling approaches and also introduce the emerging profiling and data-mining approaches for multiple-sample analysis.
Single-sample analysis
Outcomes of BCR-seq analysis mainly include the following categories: profiling, diversity estimation, mutation frequencies (substitution model), similarity analysis to known antibodies, public (shared) clonotypes, convergent among samples, and so on [ 29, 64, 99, 100] .
Data display
BCR repertoire can be profiled in the following ways. First, the V, D, and J gene segment usage (distribution) can be visualized by using a simple bar plot or scatter plot as well as the Ig class [ 101, 102] . The sequence features, like nucleotide or amino acid motifs, can be shown with a logo plot [103]. Meanwhile, the distribution of CDR3 length, clonotype abundance, or amino acid charge can be displayed [ 68, 77, 104– 107] . In a clustered tree, the substitution model can be applied to estimate the frequency of mutation and indel generated in V(D)J recombination and somatic hypermutation (SHM) processes, which is an important problem in BCR repertoire analysis [ 71, 72] .
Diversity evaluation and estimation
Diversity of a BCR repertoire is an important feature of antibody diversification. Traditionally, diversity can be evaluated with three commonly used indexes: species richness, Simpson′s index, and Shannon′s entropy [108], which provide different aspects of abundance partition. For example, species richness is the total number of species, which can be simply an analogy to clonotype richness. Simpson′s index reflects the abundance of dominant species in a certain sample, and is regarded as an index named “dominance concentration” [109]. The Shannon index quantifies the uncertainty in the species identity of an individual that is randomly picked from the dataset [110]. Those indexes reflect incomplete information of diversity. Hill [108] integrated them and gave a unified index–Hill index, which covers all of the above three indexes as special cases. The Hill index is suggested as the “true diversity” index by many researchers [ 110– 115] . To compare the diversity among multiple samples ( evaluation problem), the parameter q in the Hill index can be set as 2 or +∞. To infer the diversity of the whole repertoire ( estimate problem), i. e., the total number of clonotypes, the parameter q in Hill index should be set as 0. For example, Bashford-Rogers et al. [101] used the Gini index to evaluate the unevenness of the number of RNA molecules, while Galson et al. [116] evaluated the repertoire diversity by Shannon index. For estimation problem, published studies have explored the relationship between sequencing depth and observed clonotype number, and used Recon [117] or Chao [ 118– 120] estimator to estimate the missing number of unique clonotypes [ 14, 68, 76, 121] .
Multi-sample analysis
Currently, the BCR-seq technology is widely used in profiling the different BCR repertoires from multiple individuals or samples collected at different time points. However, tumor-infiltrate BCR repertoires are found to be heterogeneous and convergent among different cancer types, healthy degrees, and pathological stages [ 87, 122] . Discovering the shared clonotypes among different individuals is a useful way to further analyze BCR repertoire [ 68, 123] . For example, after comparing the numbers of common clonotypes in different samples with the random simulation samples, Soto et al. [121] found that the overlapping clonotypes in the human BCR repertoire are at a higher level than the simulation one. Computing the public clonotypes among samples from different individuals [68] is a useful way to further analyze BCR repertoire.
In clinical scenarios, multiple samples can be either time-point samples or sectional samples. The former corresponds to samples of an individual in different time points, presenting a dynamic profile. The latter refers to samples from different individuals at one time point. For example, time-point samples can be obtained in vaccine research, and the BCR dynamic profiles can be applied to predict vaccine response. For sectional samples, public clonotypes could reveal a convergent response, which was reported in several studies [ 61, 124, 125] .
Perspective
In the past few decades, many efforts have been made to the annotation and phylogenetic inference for BCR repertoire analysis. With the continuous development of the BCR-seq technology, methods for extra-large data set are urgently needed. Compared to other types of biological big-data, BCR-seq data need more stable and accurate statistical models to describe the BCR repertoire in a quantitative way. Due to the large sample size (number of BCRs) in a typical experiment, deep learning models are promising to participant in different tasks of BCR-seq data analyses and mining. For example, Chen et al. [126] used T cell receptor diversity data to train a multimodal recurrent neural network to predict the likelihood of antigen presentation. Tang et al. [127] developed DeepSHM, a deep convolution neural network (CNN) model to identify extended motifs of SHM beyond hotspot targeting. Since deep learning can solve the problems of classification, clustering, regression, and pattern recognition in other fields, making it is suitable to tackle the problems in immune profiling. We could anticipate more comprehensive applications of deep learning in BCR-seq data analysis in the near future.
COMPETING INTERESTS
The authors declare that they have no conflict of interest.
Funding Statement
This work was supported by the grants from the National Key R & D Program of China (No. 2018YFA0900600), the National Natural Science Foundation of China (No. 61972257), the Key Laboratory of Data Science and Intelligence Education (Hainan Normal University), the Ministry of Education (No. DSIE202002), and the Research Base of Online Education for Shanghai Middle and Primary Schools.
References
- 1.Alt FW, Zhang Y, Meng FL, Guo C, Schwer B. Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell. . 2013;152:417–429. doi: 10.1016/j.cell.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lefranc MP, Lefranc G. The Immunoglobulin FactsBook. 2001
- 3.Jung D, Giallourakis C, Mostoslavsky R, Alt FW. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu Rev Immunol. . 2006;24:541–570. doi: 10.1146/annurev.immunol.23.021704.115830. [DOI] [PubMed] [Google Scholar]
- 4.Tonegawa S. Somatic generation of antibody diversity. Nature. . 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 5.Xu JL, Davis MM. Diversity in the CDR3 region of VH is sufficient for most antibody specificities. Immunity. . 2000;13:37–45. doi: 10.1016/S1074-7613(00)00006-6. [DOI] [PubMed] [Google Scholar]
- 6.Ippolito GC, Schelonka RL, Zemlin M, Ivanov II, Kobayashi R, Zemlin C, Gartland GL, et al. Forced usage of positively charged amino acids in immunoglobulin CDR-H3 impairs B cell development and antibody production. J Exp Med. . 2006;203:1567–1578. doi: 10.1084/jem.20052217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lefranc MP, Giudicelli V, Duroux P, Jabado-Michaloud J, Folch G, Aouinti S, Carillon E, et al. IMGT®, the international ImMunoGeneTics information system® 25 years on. Nucleic Acids Res. . 2015;43:D413–D422. doi: 10.1093/nar/gku1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. . 2000;102:553–563. doi: 10.1016/S0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 9.Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. . 2000;102:565–575. doi: 10.1016/S0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 10.Stavnezer J. Immunoglobulin class switching. Current Opinion in Immunology 1996, 8: 199-205. [DOI] [PubMed]
- 11.Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem. . 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- 12.Mesin L, Ersching J, Victora GD. Germinal center B cell dynamics. Immunity. . 2016;45:471–482. doi: 10.1016/j.immuni.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cyster JG, Allen CDC. B cell responses: cell interaction dynamics and decisions. Cell. . 2019;177:524–540. doi: 10.1016/j.cell.2019.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roskin KM, Simchoni N, Liu Y, Lee JY, Seo K, Hoh RA, Pham T, et al. IgH sequences in common variable immune deficiency reveal altered B cell development and selection. Sci Transl Med. . 2015;7:302ra135. doi: 10.1126/scitranslmed.aab1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vázquez Bernat N, Corcoran M, Hardt U, Kaduk M, Phad GE, Martin M, Karlsson Hedestam GB. High-quality library preparation for NGS-based immunoglobulin germline gene inference and repertoire expression analysis. Front Immunol. . 2019;10:660. doi: 10.3389/fimmu.2019.00660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramsköld D, Luo S, Wang YC, Li R, Deng Q, Faridani OR, Daniels GA, et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol. . 2012;30:777–782. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frohman MA, Dush MK, Martin GR. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci USA. . 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boyd SD, Joshi SA. High-throughput DNA sequencing analysis of antibody repertoires. Microbiol Spectr. . 2014;2 doi: 10.1128/microbiolspec.aid-0017-2014. [DOI] [PubMed] [Google Scholar]
- 19.Hu Q, Hong Y, Qi P, Lu G, Mai X, Xu S, He X, et al. Atlas of breast cancer infiltrated B-lymphocytes revealed by paired single-cell RNA-sequencing and antigen receptor profiling. Nat Commun. . 2021;12:2186. doi: 10.1038/s41467-021-22300-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang C, Liu Y, Xu LT, Jackson KJL, Roskin KM, Pham TD, Laserson J, et al. Effects of aging, cytomegalovirus infection, and EBV infection on human B cell repertoires. J Immunol. . 2014;192:603–611. doi: 10.4049/jimmunol.1301384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turchaninova MA, Davydov A, Britanova OV, Shugay M, Bikos V, Egorov ES, Kirgizova VI, et al. High-quality full-length immunoglobulin profiling with unique molecular barcoding. Nat Protoc. . 2016;11:1599–1616. doi: 10.1038/nprot.2016.093. [DOI] [PubMed] [Google Scholar]
- 22.Chen H, Zhang Y, Ye AY, Du Z, Xu M, Lee CS, Hwang JK, et al. BCR selection and affinity maturation in Peyer’s patch germinal centres. Nature. . 2020;582:421–425. doi: 10.1038/s41586-020-2262-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu J, Meyers RM, Dong J, Panchakshari RA, Alt FW, Frock RL. Detecting DNA double-stranded breaks in mammalian genomes by linear amplification–mediated high-throughput genome-wide translocation sequencing. Nat Protoc. . 2016;11:853–871. doi: 10.1038/nprot.2016.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lykke-Andersen S, Jensen TH. Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nat Rev Mol Cell Biol. . 2015;16:665–677. doi: 10.1038/nrm4063. [DOI] [PubMed] [Google Scholar]
- 25.Stavnezer J, Guikema JEJ, Schrader CE. Mechanism and regulation of class switch recombination. Annu Rev Immunol. . 2008;26:261–292. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, Nussenzweig MC. Predominant autoantibody production by early human B cell precursors. Science. . 2003;301:1374–1377. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- 27.von Boehmer L, Liu C, Ackerman S, Gitlin AD, Wang Q, Gazumyan A, Nussenzweig MC. Sequencing and cloning of antigen-specific antibodies from mouse memory B cells. Nat Protoc. . 2016;11:1908–1923. doi: 10.1038/nprot.2016.102. [DOI] [PubMed] [Google Scholar]
- 28.Rubelt F, Busse CE, Bukhari SAC, Bürckert JP, Mariotti-Ferrandiz E, Cowell LG, Watson CT, et al. Adaptive Immune Receptor Repertoire Community recommendations for sharing immune-repertoire sequencing data. Nat Immunol. . 2017;18:1274–1278. doi: 10.1038/ni.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lees WD. Tools for adaptive immune receptor repertoire sequencing. Curr Opin Syst Biol. . 2020;24:86–92. doi: 10.1016/j.coisb.2020.10.003. [DOI] [Google Scholar]
- 30.Liu H, Pan W, Tang C, Tang Y, Wu H, Yoshimura A, Deng Y, et al. The methods and advances of adaptive immune receptors repertoire sequencing. Theranostics. . 2021;11:8945–8963. doi: 10.7150/thno.61390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Löytynoja A, Goldman N. Phylogeny-aware gap placement prevents errors in sequence alignment and evolutionary analysis. Science. . 2008;320:1632–1635. doi: 10.1126/science.1158395. [DOI] [PubMed] [Google Scholar]
- 32.Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH, Weng Z, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. . 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gibbs RA, Rogers J, Katze MG, Bumgarner R, Weinstock GM, Mardis ER, Remington KA, et al. Evolutionary and biomedical insights from the rhesus macaque genome. Science. . 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- 34.Bolotin DA, Poslavsky S, Mitrophanov I, Shugay M, Mamedov IZ, Putintseva EV, Chudakov DM. MiXCR: software for comprehensive adaptive immunity profiling. Nat Methods. . 2015;12:380–381. doi: 10.1038/nmeth.3364. [DOI] [PubMed] [Google Scholar]
- 35.Ye J, Ma N, Madden TL, Ostell JM. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. . 2013;41:W34–W40. doi: 10.1093/nar/gkt382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. . 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 37.Smith TF, Waterman MS. Identification of common molecular subsequences. J Mol Biol. . 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 38.Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. . 2013;41:e108. doi: 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shlemov A, Bankevich S, Bzikadze A, Turchaninova MA, Safonova Y, Pevzner PA. Reconstructing antibody repertoires from error-prone immunosequencing reads. J Immunol. . 2017;199:3369–3380. doi: 10.4049/jimmunol.1700485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCoy CO, Bedford T, Minin VN, Bradley P, Robins H, Matsen Iv FA. Quantifying evolutionary constraints on B-cell affinity maturation. Phil Trans R Soc B. . 2015;370:20140244. doi: 10.1098/rstb.2014.0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo Y, Chen K, Kwong PD, Shapiro L, Sheng Z. cAb-Rep: a database of curated antibody repertoires for exploring antibody diversity and predicting antibody prevalence. Front Immunol. . 2019;10:2365. doi: 10.3389/fimmu.2019.02365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schramm CA, Sheng Z, Zhang Z, Mascola JR, Kwong PD, Shapiro L. SONAR: A high-throughput pipeline for inferring antibody ontogenies from longitudinal sequencing of B cell transcripts. Front Immunol. . 2016;7:372. doi: 10.3389/fimmu.2016.00372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rosenfeld AM, Meng W, Luning Prak ET, Hershberg U. ImmuneDB, a novel tool for the analysis, storage, and dissemination of immune repertoire sequencing data. Front Immunol. . 2018;9:2107. doi: 10.3389/fimmu.2018.02107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang B, Meng W, Luning Prak ET, Hershberg U. Discrimination of germline V genes at different sequencing lengths and mutational burdens: A new tool for identifying and evaluating the reliability of V gene assignment. J Immunological Methods. . 2015;427:105–116. doi: 10.1016/j.jim.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Christley S, Scarborough W, Salinas E, Rounds WH, Toby IT, Fonner JM, Levin MK, et al. VDJServer: a cloud-based analysis portal and data commons for immune repertoire sequences and rearrangements. Front Immunol. . 2018;9:976. doi: 10.3389/fimmu.2018.00976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shugay M, Britanova OV, Merzlyak EM, Turchaninova MA, Mamedov IZ, Tuganbaev TR, Bolotin DA, et al. Towards error-free profiling of immune repertoires. Nat Methods. . 2014;11:653–655. doi: 10.1038/nmeth.2960. [DOI] [PubMed] [Google Scholar]
- 47.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. . 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bonissone SR, Pevzner PA. Immunoglobulin classification using the colored antibody graph. J Comput Biol. . 2016;23:483–494. doi: 10.1089/cmb.2016.0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vander Heiden JA, Yaari G, Uduman M, Stern JNH, O′Connor KC, Hafler DA, Vigneault F, et al. pRESTO: a toolkit for processing high-throughput sequencing raw reads of lymphocyte receptor repertoires. Bioinformatics. . 2014;30:1930–1932. doi: 10.1093/bioinformatics/btu138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marcou Q, Mora T, Walczak AM. High-throughput immune repertoire analysis with IGoR. Nat Commun. . 2018;9:561. doi: 10.1038/s41467-018-02832-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Elhanati Y, Marcou Q, Mora T, Walczak AM. repgenHMM: a dynamic programming tool to infer the rules of immune receptor generation from sequence data. Bioinformatics. . 2016;32:1943–1951. doi: 10.1093/bioinformatics/btw112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee DW, Khavrutskii IV, Wallqvist A, Bavari S, Cooper CL, Chaudhury S. BRILIA: integrated tool for high-throughput annotation and lineage tree assembly of B-cell repertoires. Front Immunol. . 2017;7:1. doi: 10.3389/fimmu.2016.00681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Volpe JM, Cowell LG, Kepler TB. SoDA: implementation of a 3D alignment algorithm for inference of antigen receptor recombinations. Bioinformatics. . 2006;22:438–444. doi: 10.1093/bioinformatics/btk004. [DOI] [PubMed] [Google Scholar]
- 54.Krogh A, Brown M, Mian IS, Sjölander K, Haussler D. Hidden Markov models in computational biology. J Mol Biol. . 1994;235:1501–1531. doi: 10.1006/jmbi.1994.1104. [DOI] [PubMed] [Google Scholar]
- 55.Ohm-Laursen L, Nielsen M, Larsen SR, Barington T. No evidence for the use of DIR, D?D fusions, chromosome 15 open reading frames or V H replacement in the peripheral repertoire was found on application of an improved algorithm, JointML, to 6329 human immunoglobulin H rearrangements . Immunology. . 2006;119:265–277. doi: 10.1111/j.1365-2567.2006.02431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gaëta BA, Malming HR, Jackson KJL, Bain ME, Wilson P, Collins AM. iHMMune-align: hidden Markov model-based alignment and identification of germline genes in rearranged immunoglobulin gene sequences. Bioinformatics. . 2007;23:1580–1587. doi: 10.1093/bioinformatics/btm147. [DOI] [PubMed] [Google Scholar]
- 57.Munshaw S, Kepler TB. SoDA2: a Hidden Markov Model approach for identification of immunoglobulin rearrangements. Bioinformatics. . 2010;26:867–872. doi: 10.1093/bioinformatics/btq056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ralph DK, Matsen FA. Consistency of VDJ rearrangement and substitution parameters enables accurate B cell receptor sequence annotation. PLoS Comput Biol. . 2016;12:e1004409. doi: 10.1371/journal.pcbi.1004409.1503.04224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.R D, SR E, A K, G M. Biological Sequence Analysis: Probabilistic Models of Proteins and Nucleic Acids. Cambridge University Press 1998
- 60.Victora GD, Nussenzweig MC. Germinal centers. Annu Rev Immunol. . 2012;30:429–457. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- 61.Davidsen K, Matsen Iv FA. Benchmarking tree and ancestral sequence inference for B Cell receptor sequences. Front Immunol. . 2018;9:2451. doi: 10.3389/fimmu.2018.02451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gascuel O, Steel M. Neighbor-joining revealed. Mol Biol Evol. . 2006;23:1997–2000. doi: 10.1093/molbev/msl072. [DOI] [PubMed] [Google Scholar]
- 63.Breda J, Zavolan M, van Nimwegen E. Bayesian inference of gene expression states from single-cell RNA-seq data. Nat Biotechnol. . 2021;39:1008–1016. doi: 10.1038/s41587-021-00875-x. [DOI] [PubMed] [Google Scholar]
- 64.Yaari G, Kleinstein SH. Practical guidelines for B-cell receptor repertoire sequencing analysis. Genome Med. . 2015;7:121. doi: 10.1186/s13073-015-0243-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Olson BJ, Matsen Iv FA. The Bayesian optimist′s guide to adaptive immune receptor repertoire analysis. Immunol Rev. . 2018;284:148–166. doi: 10.1111/imr.12664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kepler TB. Reconstructing a B-cell clonal lineage. I. Statistical inference of unobserved ancestors. F1000Res. . 2013;2:103. doi: 10.12688/f1000research.2-103.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Khodadoust MS, Olsson N, Chen B, Sworder B, Shree T, Liu CL, Zhang L, et al. B-cell lymphomas present immunoglobulin neoantigens. Blood. . 2019;133:878–881. doi: 10.1182/blood-2018-06-845156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Briney B, Inderbitzin A, Joyce C, Burton DR. Commonality despite exceptional diversity in the baseline human antibody repertoire. Nature. . 2019;566:393–397. doi: 10.1038/s41586-019-0879-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Safonova Y, Bonissone S, Kurpilyansky E, Starostina E, Lapidus A, Stinson J, DePalatis L, et al. IgRepertoireConstructor: a novel algorithm for antibody repertoire construction and immunoproteogenomics analysis. Bioinformatics. . 2015;31:i53–i61. doi: 10.1093/bioinformatics/btv238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Duò A, Robinson MD, Soneson C. A systematic performance evaluation of clustering methods for single-cell RNA-seq data. F1000Res. . 2018;7:1141. doi: 10.12688/f1000research.15666.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hoehn KB, Lunter G, Pybus OG. A phylogenetic codon substitution model for antibody lineages. Genetics. . 2017;206:417–427. doi: 10.1534/genetics.116.196303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yaari G, Vander Heiden JA, Uduman M, Gadala-Maria D, Gupta N, Stern JNH, O′Connor KC, et al. Models of somatic hypermutation targeting and substitution based on synonymous mutations from high-throughput immunoglobulin sequencing data. Front Immunol. . 2013;4:358. doi: 10.3389/fimmu.2013.00358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Helmink BA, Reddy SM, Gao J, Zhang S, Basar R, Thakur R, Yizhak K, et al. B cells and tertiary lymphoid structures promote immunotherapy response. Nature. . 2020;577:549–555. doi: 10.1038/s41586-019-1922-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Petitprez F, de Reyniès A, Keung EZ, Chen TWW, Sun CM, Calderaro J, Jeng YM, et al. B cells are associated with survival and immunotherapy response in sarcoma. Nature. . 2020;577:556–560. doi: 10.1038/s41586-019-1906-8. [DOI] [PubMed] [Google Scholar]
- 75.Cabrita R, Lauss M, Sanna A, Donia M, Skaarup Larsen M, Mitra S, Johansson I, et al. Tertiary lymphoid structures improve immunotherapy and survival in melanoma. Nature. . 2020;577:561–565. doi: 10.1038/s41586-019-1914-8. [DOI] [PubMed] [Google Scholar]
- 76.Li H, Limenitakis JP, Greiff V, Yilmaz B, Schären O, Urbaniak C, Zünd M, et al. Mucosal or systemic microbiota exposures shape the B cell repertoire. Nature. . 2020;584:274–278. doi: 10.1038/s41586-020-2564-6. [DOI] [PubMed] [Google Scholar]
- 77.Lee J, Boutz DR, Chromikova V, Joyce MG, Vollmers C, Leung K, Horton AP, et al. Molecular-level analysis of the serum antibody repertoire in young adults before and after seasonal influenza vaccination. Nat Med. . 2016;22:1456–1464. doi: 10.1038/nm.4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lavinder JJ, Wine Y, Giesecke C, Ippolito GC, Horton AP, Lungu OI, Hoi KH, et al. Identification and characterization of the constituent human serum antibodies elicited by vaccination. Proc Natl Acad Sci USA. . 2014;111:2259–2264. doi: 10.1073/pnas.1317793111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.McDaniel JR, Pero SC, Voss WN, Shukla GS, Sun Y, Schaetzle S, Lee CH, et al. Identification of tumor-reactive B cells and systemic IgG in breast cancer based on clonal frequency in the sentinel lymph node. Cancer Immunol Immunother. . 2018;67:729–738. doi: 10.1007/s00262-018-2123-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.DeKosky BJ, Kojima T, Rodin A, Charab W, Ippolito GC, Ellington AD, Georgiou G. In-depth determination and analysis of the human paired heavy- and light-chain antibody repertoire. Nat Med. . 2015;21:86–91. doi: 10.1038/nm.3743. [DOI] [PubMed] [Google Scholar]
- 81.Li X, Zhang W, Huang M, Ren Z, Nie C, Liu X, Yang S, et al. Selection of potential cytokeratin-18 monoclonal antibodies following IGH repertoire evaluation in mice. J Immunological Methods. . 2019;474:112647. doi: 10.1016/j.jim.2019.112647. [DOI] [PubMed] [Google Scholar]
- 82.Hu X, Zhang J, Wang J, Fu J, Li T, Zheng X, Wang B, et al. Landscape of B cell immunity and related immune evasion in human cancers. Nat Genet. . 2019;51:560–567. doi: 10.1038/s41588-018-0339-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mandric I, Rotman J, Yang HT, Strauli N, Montoya DJ, Van Der Wey W, Ronas JR, et al. Profiling immunoglobulin repertoires across multiple human tissues using RNA sequencing. Nat Commun. . 2020;11:3126. doi: 10.1038/s41467-020-16857-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Meng W, Zhang B, Schwartz GW, Rosenfeld AM, Ren D, Thome JJC, Carpenter DJ, et al. An atlas of B-cell clonal distribution in the human body. Nat Biotechnol. . 2017;35:879–884. doi: 10.1038/nbt.3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang X, Tipton CM, Woodruff MC, Zhou E, Lee FEH, Sanz I, Qiu P. GLaMST: grow lineages along minimum spanning tree for b cell receptor sequencing data. BMC Genomics. . 2020;21:583. doi: 10.1186/s12864-020-06936-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.DeWitt WS, Mesin L, Victora GD, Minin VN, Matsen FA. Using genotype abundance to improve phylogenetic inference. Mol Biol Evol. . 2018;35:1253–1265. doi: 10.1093/molbev/msy020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Paschold L, Simnica D, Willscher E, Vehreschild MJGT, Dutzmann J, Sedding DG, Schultheiß C, et al. SARS-CoV-2–specific antibody rearrangements in prepandemic immune repertoires of risk cohorts and patients with COVID-19. J Clin Investigation. . 2021;131:1. doi: 10.1172/JCI142966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stubbington MJT, Lönnberg T, Proserpio V, Clare S, Speak AO, Dougan G, Teichmann SA. T cell fate and clonality inference from single-cell transcriptomes. Nat Methods. . 2016;13:329–332. doi: 10.1038/nmeth.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rokas A, Charlesworth D. Molecular Evolution and Phylogenetics . By M. Nei and S. Kumar. Oxford University Press. 2000. ISBN: 0-19-513584-9 (hbk); 0-19-513585-7 (pbk). xiv+333 pages. Price: £65 (hbk); £32.50 (pbk). . Genet Res. . 2001;77:117–120. doi: 10.1017/S0016672301219405. [DOI] [Google Scholar]
- 90.Stadler T. Simulating trees with a fixed number of extant species. Systatic Biol. . 2011;60:676–684. doi: 10.1093/sysbio/syr029. [DOI] [PubMed] [Google Scholar]
- 91.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systatic Biol. . 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 92.Bonsignori M, Scott E, Wiehe K, Easterhoff D, Alam SM, Hwang KK, Cooper M, et al. Inference of the HIV-1 VRC01 antibody lineage unmutated common ancestor reveals alternative pathways to overcome a key glycan barrier. Immunity. . 2018;49:1162–1174.e8. doi: 10.1016/j.immuni.2018.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Price MN, Dehal PS, Arkin AP. Fasttree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE. . 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kruskal JB. On the Shortest Spanning Subtree of a Graph and the Traveling Salesman Problem. Proceedings of the American Mathematical Society 1956, 7
- 95.Prim RC. Shortest connection networks and some generalizations. Bell Syst Technical J. . 1957;36:1389–1401. doi: 10.1002/j.1538-7305.1957.tb01515.x. [DOI] [Google Scholar]
- 96.Martel C. The expected complexity of Prim′s minimum spanning tree algorithm. Inf Processing Lett. . 2002;81:197–201. doi: 10.1016/S0020-0190(01)00220-4. [DOI] [Google Scholar]
- 97.Felsenstein J. PHYLIP: phylogeny inference package. Cladistics 1993
- 98.Liberman G, Benichou JIC, Maman Y, Glanville J, Alter I, Louzoun Y. Estimate of within population incremental selection through branch imbalance in lineage trees. Nucleic Acids Res. . 2016;44:e46. doi: 10.1093/nar/gkv1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Davis MM, Boyd SD. Recent progress in the analysis of αβ T cell and B cell receptor repertoires. Curr Opin Immunol. . 2019;59:109–114. doi: 10.1016/j.coi.2019.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nielsen SCA, Boyd SD. Human adaptive immune receptor repertoire analysis-Past, present, and future. Immunol Rev. . 2018;284:9–23. doi: 10.1111/imr.12667. [DOI] [PubMed] [Google Scholar]
- 101.Bashford-Rogers RJM, Bergamaschi L, McKinney EF, Pombal DC, Mescia F, Lee JC, Thomas DC, et al. Analysis of the B cell receptor repertoire in six immune-mediated diseases. Nature. . 2019;574:122–126. doi: 10.1038/s41586-019-1595-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tipton CM, Hom JR, Fucile CF, Rosenberg AF, Sanz I. Understanding B-cell activation and autoantibody repertoire selection in systemic lupus erythematosus: A B-cell immunomics approach. Immunol Rev. . 2018;284:120–131. doi: 10.1111/imr.12660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Adler AS, Mizrahi RA, Spindler MJ, Adams MS, Asensio MA, Edgar RC, Leong J, et al. Rare, high-affinity anti-pathogen antibodies from human repertoires, discovered using microfluidics and molecular genomics. mAbs. . 2017;9:1282–1296. doi: 10.1080/19420862.2017.1371383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rosenfeld AM, Meng W, Chen DY, Zhang B, Granot T, Farber DL, Hershberg U, et al. Computational evaluation of B-cell clone sizes in bulk populations. Front Immunol. . 2018;9:1472. doi: 10.3389/fimmu.2018.01472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dunn-Walters D, Townsend C, Sinclair E, Stewart A. Immunoglobulin gene analysis as a tool for investigating human immune responses. Immunol Rev. . 2018;284:132–147. doi: 10.1111/imr.12659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Khass M, Vale AM, Burrows PD, Schroeder Jr HW. The sequences encoded by immunoglobulin diversity (D H ) gene segments play key roles in controlling B-cell development, antigen-binding site diversity, and antibody production . Immunol Rev. . 2018;284:106–119. doi: 10.1111/imr.12669. [DOI] [PubMed] [Google Scholar]
- 107.DeKosky BJ, Lungu OI, Park D, Johnson EL, Charab W, Chrysostomou C, Kuroda D, et al. Large-scale sequence and structural comparisons of human naive and antigen-experienced antibody repertoires. Proc Natl Acad Sci USA. . 2016;113:E2636–2645. doi: 10.1073/pnas.1525510113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hill MO. Diversity and evenness: a unifying notation and its consequences. Ecology. . 1973;54:427–432. doi: 10.2307/1934352. [DOI] [Google Scholar]
- 109.Whittaker RH. Dominance and diversity in land plant communities: numerical relations of species express the importance of competition in community function and evolution. Science. . 1965;147:250–260. doi: 10.1126/science.147.3655.250. [DOI] [PubMed] [Google Scholar]
- 110.Tuomisto H. A consistent terminology for quantifying species diversity? Yes, it does exist. Oecologia. . 2010;164:853–860. doi: 10.1007/s00442-010-1812-0. [DOI] [PubMed] [Google Scholar]
- 111.Jost L. Entropy and diversity . Oikos. . 2006;113:363–375. doi: 10.1111/j.2006.0030-1299.14714.x. [DOI] [Google Scholar]
- 112.Jost L. Mismeasuring biological diversity: Response to Hoffmann and Hoffmann (2008) Ecol Economics. . 2009;68:925–928. doi: 10.1016/j.ecolecon.2008.10.015. [DOI] [Google Scholar]
- 113.Jost L. Partitioning diversity into independent alpha and beta components. Ecology. . 2007;88:2427–2439. doi: 10.1890/06-1736.1. [DOI] [PubMed] [Google Scholar]
- 114.Tuomisto H. A diversity of beta diversities: straightening up a concept gone awry. Part 1. Defining beta diversity as a function of alpha and gamma diversity. Ecography. . 2010;33:2–22. doi: 10.1111/j.1600-0587.2009.05880.x. [DOI] [Google Scholar]
- 115.Tuomisto H. A diversity of beta diversities: straightening up a concept gone awry. Part 2. Quantifying beta diversity and related phenomena. Ecography. . 2010;33:23–45. doi: 10.1111/j.1600-0587.2009.06148.x. [DOI] [Google Scholar]
- 116.Galson JD, Schaetzle S, Bashford-Rogers RJM, Raybould MIJ, Kovaltsuk A, Kilpatrick GJ, Minter R, et al. Deep sequencing of B cell receptor repertoires From COVID-19 patients reveals strong convergent immune signatures. Front Immunol. . 2020;11:605170. doi: 10.3389/fimmu.2020.605170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kaplinsky J, Arnaout R. Robust estimates of overall immune-repertoire diversity from high-throughput measurements on samples. Nat Commun. . 2016;7:11881. doi: 10.1038/ncomms11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ganne P, Najeeb S, Chaitanya G, Sharma A, Krishnappa NC. Digital eye strain epidemic amid COVID-19 pandemic–a cross-sectional survey. Ophthalmic Epidemiol. . 2021;28:285–292. doi: 10.1080/09286586.2020.1862243. [DOI] [PubMed] [Google Scholar]
- 119.A C. Estimating the population size for capture-recapture data with unequal catchability. %J Biometrics. 1987, 43: 783-791. [PubMed]
- 120.Eren MI, Chao A, Hwang WH, Colwell RK. Estimating the richness of a population when the maximum number of classes is fixed: a nonparametric solution to an archaeological problem. PLoS ONE. . 2012;7:e34179. doi: 10.1371/journal.pone.0034179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Soto C, Bombardi RG, Branchizio A, Kose N, Matta P, Sevy AM, Sinkovits RS, et al. High frequency of shared clonotypes in human B cell receptor repertoires. Nature. . 2019;566:398–402. doi: 10.1038/s41586-019-0934-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Davis MM, Brodin P. Rebooting human immunology. Annu Rev Immunol. . 2018;36:843–864. doi: 10.1146/annurev-immunol-042617-053206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lin K, Zhou Y, Ai J, Wang YA, Zhang S, Qiu C, Lian C, et al. B cell receptor signatures associated with strong and poor SARS-CoV-2 vaccine responses. Emerging Microbes Infects. . 2022;11:452–464. doi: 10.1080/22221751.2022.2030197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wang Z, Schmidt F, Weisblum Y, Muecksch F, Barnes CO, Finkin S, Schaefer-Babajew D, et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. . 2021;592:616–622. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Stamatopoulos K, Agathangelidis A, Rosenquist R, Ghia P. Antigen receptor stereotypy in chronic lymphocytic leukemia. Leukemia. . 2017;31:282–291. doi: 10.1038/leu.2016.322. [DOI] [PubMed] [Google Scholar]
- 126.Chen B, Khodadoust MS, Olsson N, Wagar LE, Fast E, Liu CL, Muftuoglu Y, et al. Predicting HLA class II antigen presentation through integrated deep learning. Nat Biotechnol. . 2019;37:1332–1343. doi: 10.1038/s41587-019-0280-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tang C, Krantsevich A, MacCarthy T. Deep learning model of somatic hypermutation reveals importance of sequence context beyond hotspot targeting. iScience. . 2022;25:103668. doi: 10.1016/j.isci.2021.103668. [DOI] [PMC free article] [PubMed] [Google Scholar]