Abstract
Nucleotide excision repair (NER) is a major pathway to deal with bulky adducts induced by various environmental toxins in all cellular organisms. The two sub-pathways of NER, global genome repair (GGR) and transcription-coupled repair (TCR), differ in the damage recognition modes. In this review, we describe the molecular mechanism of NER in mammalian cells, especially the details of damage recognition steps in both sub-pathways. We also introduce new sequencing methods for genome-wide mapping of NER, as well as recent advances about NER in chromatin by these methods. Finally, the roles of NER factors in repairing oxidative damages and resolving R-loops are discussed.
Keywords: nucleotide excision repair, transcription-coupled repair, global genome repair, damage recognition, repair mapping
Introduction
As the carrier of genetic information, DNA is the target of many endogenous and exogenous genetic toxic agents [1]. The former contain metabolic products such as reactive oxygen species (ROS) and aldehydes [2]; while the latter have a long list including ultraviolet (UV) in the sunlight [3], polycyclic aromatic hydrocarbons from air pollutants [4], aflatoxin from contaminated food [5], chemotherapeutic drugs like cisplatin [6], and natural products such as aristolochic acids [7] and illudin S [8].
These agents cause various types of DNA base modifications and adducts, which will affect base pairing and interfere with DNA replication and transcription [9], and finally threaten genomic stability, resulting in cancer and aging [ 10– 12] . To maintain genomic integrity, DNA repair pathways, mainly base excision repair (BER) and nucleotide excision repair (NER), are evolved to deal with these damages.
The BER pathway utilizes specific glycosylases to recognize and excise the corresponding base modifications, generating apurinic/apyrimidinic (AP) sites which are further processed by APE1 and other BER factors [ 13– 15] . However, there are only 11 glycosylases identified from the human genome [16], and each glycosylase can only recognize a couple of lesions sharing similar structures [17]. Thus, a limited range of damages can be repaired by the BER pathway, while an unpredictable number of structurally heterogeneous base modifications and adducts are left unrepaired. Therefore, a piece of versatile repair machinery is strongly required, and NER is such a pathway. To cope with such a diversity of lesions, NER aims for common features of base modifications and adducts instead of unique structures, which is discussed below.
NER exists in all three domains of life [18], albeit there are two pieces of evolutionarily unrelated machinery: bacterial NER and eukaryotic NER. Intriguingly, NER in bacteria and eukaryotes utilizes the same strategy to recognize various lesions, but the core factors are completely different between the two domains [19]. The bacterial type of NER has been identified in certain species of Archaea in vitro, which is likely due to horizontal gene transfer [20]. In Archaea, some proteins homologous to eukaryotic NER factors were also found [21], but a functional eukaryotic type of NER has not been demonstrated yet [22]. In this review, we will focus on NER in eukaryote, especially in mammalian cells.
Defects of NER factors in humans can cause several inherited diseases with far different phenotypes, including xeroderma pigmentosum (XP), Cockayne syndrome (CS), and ultraviolet sensitive syndrome (UV SS) [23]. Patients with XP are identified by an extremely high chance of skin cancer [24], while CS patients suffer from severe growth retardation, progeria, and photosensitivity but without an increased risk of skin cancer [25]. In contrast, patients of UV SS only have a higher sensitivity to UV in sunlight but have neither developmental abnormality nor elevated possibility of skin cancer [ 26– 28] .
The concept of NER was first raised in the 1960s to describe the “dark repair” of UV damage in E. coli [29], in contrast to “light repair” operated by photolyases [ 30, 31] . The basic mode of NER, namely “dual incisions”, was revealed in the 1980s for bacteria [ 32– 34] , and in the 1990s for mammalian cells [ 35– 37] , both through in vitro reconstituted reactions. In recent years, this mode was finally demonstrated in both eukaryotic and prokaryotic cells [ 34, 38– 40] . Dr. Aziz Sancar is one of the main contributors in this field and was awarded the Nobel Prize in Chemistry in 2015 [41]. Although the mechanism of dual incision reaction was described in considerable details, it is still elusive how repair factors seek and recognize damage efficiently across the genome.
In this review, we will summarize the basic mechanism of NER, and introduce recent advances of the above question by various approaches including biochemistry, cell biology, structure biology, and genomics. We will also discuss the roles of NER factors in repairing some non-classical substrates.
Molecular Mechanism of Eukaryotic Nucleotide Excision Repair
NER removes damaged nucleotides by dual incisions bracketing the lesion and releasing a short (22–30 nucleotides in mammalian cells) single-stranded DNA fragment containing the damage [ 34, 37, 39, 42, 43] . Repair synthesis and ligation are then performed to recover intact double-strand DNA [ 44– 47] . In brief, NER can be divided into 3 major steps: (1) damage recognition, including initial recognition and damage verification; (2) dual incisions and release of excision products; and (3) gap filling, which means repair synthesis and DNA ligation ( Figure 1). In contrast to BER which identifies the specific structures of modified bases, NER recognizes damage-induced double-strand distortions or RNA polymerase II stalled by lesions. The former mechanism deals with damage across the whole genome thus named Global Genome Repair (GGR), while the latter mode only repairs damage on the template strands of transcribed regions, which is called Transcription-Coupled Repair (TCR) [ 48– 50] .
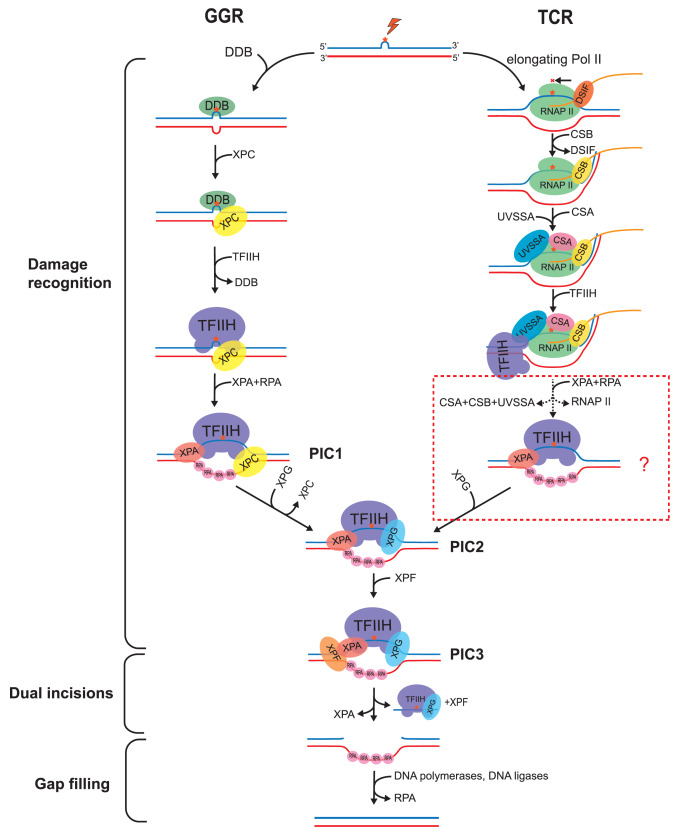
Figure 1 .
Mechanistic model of nucleotide excision repair
In GGR, lesions are first recognized by DDB complex, then XPC, TFIIH, XPA and RPA, XPG, XPF are sequentially recruited while DDB and XPC are released before the dual incisions step. In TCR, blocked Pol II serves as the damage sensor to recruit CSB, CSA and UVSSA, which corporately recruit TFIIH. The details happened between TFIIH loading and dual incisions are unclear. Nonetheless, for both sub-pathways, XPG and XPF make the 3′ and 5′ incisions, respectively, to generate ~26 nt-long ssDNA fragments containing damage which are released in complex with TFIIH and XPG. The resulted gaps are sealed by DNA polymerases and ligases to recover intact dsDNA.
GGR was successfully reconstituted in vitro with 6 purified factors (XPC, TFIIH, XPA, RPA, XPF, and XPG) [ 35, 36] , whereas TCR has not been reconstructed till now. Among these 6 factors, XPC is only involved in GGR, and the other 5 factors are necessary for both sub-pathways [ 51– 53] . In addition, the DDB complex is required for GGR initiation in vivo [ 54, 55] , while CSB, CSA and UVSSA are needed for TCR [ 26, 28, 56, 57] , as summarized in Table 1.
Table 1 Human nucleotide excision repair factors
|
Stage |
Factor |
Component(s) |
Enzymatic activity/function |
Related disease(s) |
|
Initial damage recognitionfor TCR |
CSA |
CSA |
Ubiquitin E3 ligase (in complex withDDB1, CUL4A & RBX1) |
CS or UV SS |
|
CSB |
CSB |
Translocase activity |
CS or UV SS |
|
|
UVSSA |
UVSSA |
Deubiquitinase (in complex with USP7) |
UV SS |
|
|
Initial damage recognitionfor GGR |
DDB |
DDB1, DDB2/XPE |
Damage recognition in chromatin, ubiquitinE3 ligase (in complex with CUL4A & RBX1) |
XP |
|
XPC |
XPC, HR23b, CETN2 |
Damaged DNA binding |
XP |
|
|
Damage verification andPIC assembly |
XPA |
XPA |
Damaged DNA binding |
XP |
|
RPA |
p70, p32, p14 |
ssDNA binding |
– |
|
|
TFIIH |
p89/XPB, p62, p52, p44,p34, p8, p80/XPD, Cdk7*,Cyclin H*, MAT1* |
3′ to 5′ (XPB) and 5′ to 3′ (XPD)helicases |
XP or XP-CS |
|
|
Dual incisions |
XPF |
XPF, ERCC1 |
Structure-specific endonuclease for 5′ incision |
XP or XP-CS |
|
XPG |
XPG |
Structure-specific endonuclease for 3′ incision |
XP or XP-CS |
* indicates the Cdk-activating kinase complex (CAK) of TFIIH which is not essential for NER. Blue, TCR specific factors; yellow, GGR specific factors; green, common factors for both sub-pathways. CS, Cockayne syndrome; UV SS, Ultraviolet-sensitive syndrome; XP, Xeroderma pigmentosum; XP-CS, Xeroderma pigmentosum-Cockayne syndrome complex (the combination of XP and CS).
Damage recognition by GGR
The efficiency of GGR is dependent on the extent of double-strand distortion. For instance, UV can induce two major lesions, i. e., pyrimidine-pyrimidone (6–4) photoproducts [(6–4) PPs] and cyclobutane pyrimidine dimers (CPDs), the former of which causes stronger distortion and is efficiently repaired by GGR [58], while the latter has less impact on DNA structure and is a poor substrate for GGR [59]. Nonetheless, both damages could be excised in vitro by the 6-factor system. In this reaction, the lesion is recognized by the cooperation of XPC (usually in the form of XPC-TFIIH), XPA, and RPA. Although either of the three factors is unable to discriminate damages from normal DNA independently, loading of one factor could facilitate binding of other factors, and eventually achieve specific damage recognition [ 19, 60, 61] .
Notably, another GGR-specific factor, the DDB complex, was not required in vitro, and the addition of purified DDB did not improve repair efficiency [62]. The DDB complex, composed of DNA damage-binding protein 2 (DDB2/XPE) and DNA damage-binding protein 1 (DDB1), forms a complex with the ubiquitin E3 ligase Cul4A-RBX1 (CRL4 DDB2) [ 63, 64] . Although DDB is dispensable for in vitro repair [ 35, 36] , it plays important physiological roles as defects in DDB2/XPE gene strongly impede the repair of CPD in vivo, and can also cause xeroderma pigmentosum [ 65– 67] . Therefore, DDB is thought to be involved in the repair of lesions with less distortion ( e. g., CPDs) in chromatin [ 67, 68] . However, the exact roles of DDB are more complex and not completely clear. Firstly, DDB2/XPE protein has the highest affinity and selectivity to both (6–4) PP and CPD among all GGR factors [ 69, 70] . Local U V irradiation revealed that DDB was recruited to damaged sites ahead of XPC [71]. Furthermore, structure studies suggested the binding of DDB kink the double-stranded DNA and promote the binding of XPC [72]. It was recently reported that DDB can bind to nucleosomal lesions and shift the DNA to expose the lesions facing the nucleosome core [73]. Thus, DDB can help recruit XPC to damage sites, especially those with minor distortion or hard to access. However, it was reported that DDB prefers lesions located in linkers and nucleosome-free regions rather than nucleosome core regions in vivo [74]. Consistently, a recently published study indicated that DDB can be recruited to linker regions after UV irradiation to stimulate the displacement of linker histones and relax heterochromatin compaction, and finally facilitate repair in heterochromatin domains [75]. Moreover, upon binding to damage, the CRL4 DDB2 ubiquitin ligase can ubiquitinate surrounding proteins including DDB2 itself, XPC [55], and histones [67], which is thought to promote damage handover [54] and decompaction of damaged nucleosomes [76]. Therefore, DDB can stimulate GGR in both direct and indirect ways.
Unlike the in vitro system, XPC is supposed to be the first factor following the recruitment of DDB. XPC itself can bind to DNA and scan for damages both in vitro and in vivo [77], albeit it cannot efficiently distinguish lesions with weaker DNA distortion from undamaged DNA [78]. Structure studies of Rad4 (yeast homolog of XPC) complex and damaged DNA indicated that it binds to the opposite strand of the lesion and the double-stranded DNA at 3′ downstream to the lesion ( Figure 1) [ 58, 78, 79] . Notably, (6–4) PPs that strongly affect DNA structure can be successfully repaired in vivo without DDB though with an apparent delay [80].
Once loaded onto damage sites, XPC can recruit the scaffold factor TFIIH through their interaction [ 81– 83] . TFIIH is a multi-subunit factor involved in both transcription initiation and NER [ 84– 87] . NER requires the TFIIH core complex which consists of 7 subunits, including two DNA helicases (XPB and XPD) and other structural peptides [88], while transcription initiation needs an extra 3-subunit CAK module [89]. As discussed above, initial damage recognition is not a strict process, such that XPC can bind to undamaged DNA or minor base modifications which should not be excised by NER. Therefore, the suitability of NER substrates is verified before dual incisions through cooperative binding and kinetic proofreading, both of which are mediated by TFIIH [ 90– 92] . The loading of TFIIH, together with DDB2 ubiquitination, can stimulate the dissociation of DDB2 and stabilize the binding of XPC [54]. The helicase activity of TFIIH subunits can separate DNA double strands, allowing the binding of XPA and RPA [93]. The structure of the TFIIH-XPA-DNA complex revealed that TFIIH adopt different conformation in transcription and repair, and the binding of XPA can promote and stabilize the conformation change of TFIIH and stimulate the helicase activity of TFIIH on undamaged DNA [ 94, 95] . On the other hand, the presence of bulky adducts ( e. g., cisplatin-damage), but not minor modifications ( e. g., AP sites), can reduce helicase activity of TFIIH, and the inhibition is further enhanced by XPA [90]. Therefore, TFIIH is trapped by appropriate bulky adducts with the help of XPA, achieving specific damage verification.
In the in vitro reaction, XPC-TFIIH, XPA, and RPA can form a stable complex with damaged DNA, called pre-incision complex 1 (PIC1). Then XPG endonuclease is recruited to damage through its interaction with TFIIH, while XPC leaves the complex, forming pre-incision complex 2 (PIC2). This process is driven by the ATP hydrolysis activity of TFIIH, while XPG can stimulate the helicase activity of TFIIH in the absence of damage. The other endonuclease XPF is finally recruited to assemble pre-incision complex 3 (PIC3) [ 96, 97] . Although XPF has a strong interaction with XPA [98], the loading of XPF also depends on the recruitment (but not the incision) of XPG [99].
Damage recognition by TCR
RNA polymerase II (Pol II) is efficiently blocked by bulky adducts and serves as a damage sensor to initiate TCR [ 9, 100] . Thus, in comparison with GGR, TCR can only deal with lesions on the template strands of transcribed regions, but it repairs various substrates with similar efficiency despite their different impacts on DNA structure [101], for Pol II indirectly detects bulky lesions by their transcription-blocking feature.
Although TCR was first identified in mouse cells more than 35 years ago [102], it has not been reconstituted in vitro till now. Thus, the molecular details of this mechanism are not as clear as GGR. The initial clue came from human genetics that CS was connected with defects in TCR [ 103, 104] , and two genes responsible for CS, i. e., CSA [105] and CSB [104], were identified as essential factors of TCR. In 2012, the third factor of TCR, namely UVSSA, was characterized through the study of UV SS [ 26– 28] . Indeed, new TCR players have still been reported even during the last two years [106]. Therefore, the mechanism of TCR is still a hot topic in the DNA repair field.
When an elongating Pol II encounters a lesion and stalls at that site, CSB, a member of the SWI2/SNF2 ATPase family of chromatin remodelers [107], is the first repair factor to be recruited [ 108– 110] . Actually, CSB is required for normal transcription even without bulky adducts [111]. It was reported that Rad26, the yeast ortholog of CSB, can act as an elongation factor to help Pol II to overcome nucleosome barriers in vitro [112]. The structure of the Pol II-Rad26 complex revealed that Rad26 binds to DNA upstream of Pol II, and the binding sites of Rad26 overlap with that of the transcription elongation factor Spt4-Spt5 [ 113, 114] . Therefore, it was speculated that when Pol II temporally stalls during elongation, Spt4-Spt5 should be replaced by CSB/Rad26 which can “push” Pol II to overcome “small” barriers. If it is a “large” obstacle like CPD that cannot be bypassed, a stable complex of Pol II-CSB/Rad26-DNA damage will be formed to recruit downstream repair factors, i. e., CSA and UVSSA [ 111, 115, 116] .
Similar to DDB2, CSA forms a complex with the ubiquitin E3 ligase DDB1-Cul4A-RBX1 (CRL4 CSA) which mediates UV-induced ubiquitination of TCR factors including Pol II, CSB, CSA, and UVSSA, resulting in the instability of this complex [64]. Paused Pol II, CSB, and CSA together recruit UVSSA which is in complex with the deubiquitinase USP7 [56]. CRL4 CSA and UVSSA-USP7 can cooperatively balance the stability of CSB, as the depletion of UVSSA reduces CSB protein level following UV irradiation. Deficiency of UVSSA will lead to an earlier release of Pol II from damaged sites [117]. However, CSB overexpression cannot rescue the UV hypersensitivity caused by UVSSA mutation, indicating other roles of UVSSA in TCR [28]. It was reported that UVSSA can directly interact with the p62 subunit of TFIIH and is essential for the recruitment of TFIIH [ 56, 81] . Cramer et al. [118] reported the structures of human TCR damage recognition complexes (including Pol II, CSB, CRL4 CSA, UVSSA, etc.). In the basic complex, CSB binds to upstream DNA; UVSSA localizes to downstream DNA; CSA sets between them as a bridge. Moreover, their results confirmed that stalled Pol II can induce the replacement of DSIF (Spt4-Spt5) by CSB which “pulls” DNA and facilitates Pol II to move forward. CSA can stimulate the ATPase activity of CSB and help CSB push Pol II. The activity center of CRL4 CSA contacts Pol II (near K1268, see below) and CSB in two different conformations, while in vitro experiments also confirmed that CRL4 CSA can ubiquitinate Pol II K1268 and CSB.
Despite their essential roles in TCR, mutations in CSB, CSA, or UVSSA genes can cause two different diseases, i. e., CS or UV SS, respectively, in most cases. However, a few cases of UV SS were reported to be due to defects in CSB [119] and CSA [ 57, 120] , respectively. Although there are several hypotheses about the relationship between genetic defects and phenotypes, the exact underlying mechanism is unclear.
Besides the dedicated TCR factors mentioned above, Pol II can be regarded as another critical damage recognition factor of TCR. It has long been known that RPB1, the catalytic subunit of Pol II, is ubiquitinated following UV irradiation [ 121– 124] . Even so, the UV-induced ubiquitination site of RPB1 was just identified recently [ 125, 126] . Two groups simultaneously reported that K1268 of RPB1 is the main UV-induced ubiquitinated residue, and the K1268 ubiquitination is important for transcription recovery and cell survival after UV treatment [ 125, 126] . However, the roles of K1268 ubiquitination are, to some extent, controversial in two studies. On one hand, Svejstrup and colleagues reported that K1268 ubiquitination mainly regulates the pool of Pol II through UV-induced proteolysis, which is important for DNA damage response and cell survival [126]. On the other hand, Ogi and colleagues found that loss of K1268 ubiquitination impairs the recruitment of TFIIH, thus strongly inhibits TCR [125]. Further studies are required to unveil the roles of K1268 ubiquitination in TCR and transcriptional response to UV damage.
Recently, a general elongation factor Elof1 emerged from independent screens for damage-sensitivity factors [127]. Elof1 is a conserved small protein (~10 kDa) that exists in the Pol II elongation complex [ 128, 129] . The structure of Pol II elongation complexes revealed that Elf1 (yeast orthologue of Elof1) binds to downstream of Pol II on the DNA and plays a role in elongating through nucleosomes [130]. Two back-to-back studies reported that loss of Elof1 greatly impedes the recruitment of UVSSA to damage sites and abrogates TCR [ 106, 131] . The simulated structure suggested that Elof1 binding site on Pol II is close to K1268, and experimental evidence indicated that Elof1 is involved in UV-induced Pol II ubiquitination at K1268. This may explain the mechanistic role of Elof1 in TCR.
Unlike GGR whose mechanism has been well studied, what happens after TFIIH loading remains elusive in TCR. Common NER factors, i. e., XPA, RPA, XPG, and XPF, should also be recruited to perform dual incisions. However, whether they are recruited in the same way as in GGR or not is unknown. Furthermore, the fate of TCR-specific factors including Pol II, CSB, CSA, and UVSSA is an open question. Although in vitro experiments indicated that paused Pol II would not inhibit dual incision reaction by GGR [116], it was supposed that Pol II stalling at damage sites should either be backtracked or removed during TCR [ 123, 132] . Evidence from recent sequencing-based studies suggested that stalled Pol II should dissociate from damage sites, since nascent transcriptions mainly restart from transcription starting sites after UV irradiation [ 133– 136] .
Dual incisions and release of excision products
In vitro studies indicated that after the assemble of PIC3, two structure-specific endonucleases, XPG and XPF, sequentially carry out incisions on the damaged strand at 4–7 nucleotides downstream and 16–21 nucleotides upstream to the lesion, respectively [ 37, 99] . The primary excised oligomers are released in complex with TFIIH, and then slowly degraded to shorter fragments 15-20-nt in length which are bound by RPA [137]. However, it was not clear whether in vivo repair has the same excision pattern, especially in TCR that cannot be reconstructed in vitro. This question was resolved by the detection of in vivo excised oligomers from UV-irradiated human cells [101]. Analyses of in vivo excision products suggested that both nucleases make incisions at the same positions as in vitro reaction, generating excision products of the same length and bound by the same proteins, i. e., TFIIH and XPG. The in vivo degradation rate of primary products is faster than in vitro, while the 15–20-nt long degraded products are also bound by RPA, and further degraded to fragments that are too short to be detected [101]. However, the nucleases responsible for this degradation process are not clear. More importantly, excision products from XP-C cells which have only TCR show identical properties with those from CS-B cells that have only GGR, indicating that dual incisions and release of excision products are the same for both GGR and TCR in vivo [101]. In addition to human cells and UV damage, the in vivo excision products were identified in different species including lemur cells [138], mice [ 139– 141] , Drosophila [142], Arabidopsis [ 39, 143, 144] , and yeast [145], and for various damage types such as cisplatin [ 139– 141, 146] and BPDE [147], suggesting that “dual incisions” is a universal mechanism for eukaryotes.
Repair synthesis and ligation
In most cases, the excision gaps are directly filled by DNA polymerase ε in proliferating cells or polymerase δ/κ in non-proliferating cells in the presence of proliferating cell nuclear antigen (PCNA) [ 40, 45, 148] . The size of repair patches is about 30 nt [ 149, 150] , consistent with the length of excised fragments. However, a small portion of excision gaps are enlarged by exonuclease I (Exo I) to generate a long stretch of ssDNA which is occupied by RPA and serves as the initial signal for ATR-mediated DNA damage response [ 151– 153] . In dividing cells, the final nick is mainly sealed by DNA ligase I [ 40, 45] , while the XRCC1-ligase3 complex performs ligation in non-dividing cells. Although the gap-filling process is not essential for dual incisions, inhibition of repair synthesis and ligation can hinder the degradation of RPA-bound fragments and reduce the repair rate of UV damages [154].
Genome-wide Maps of Nucleotide Excision Repair
In eukaryotic cells, genomic DNA is packaged with histones. Thus, NER is performed in chromatin rather than on naked DNA [155]. Meanwhile, complicated events occurring in chromatin, including transcription and DNA replication, also have impacts on NER ( Figure 2A). In order to investigate how chromatin environment affects NER, many efforts have been made to acquire genome-wide maps of NER and unveil the correlations between NER and chromatin compaction, transcription, etc.
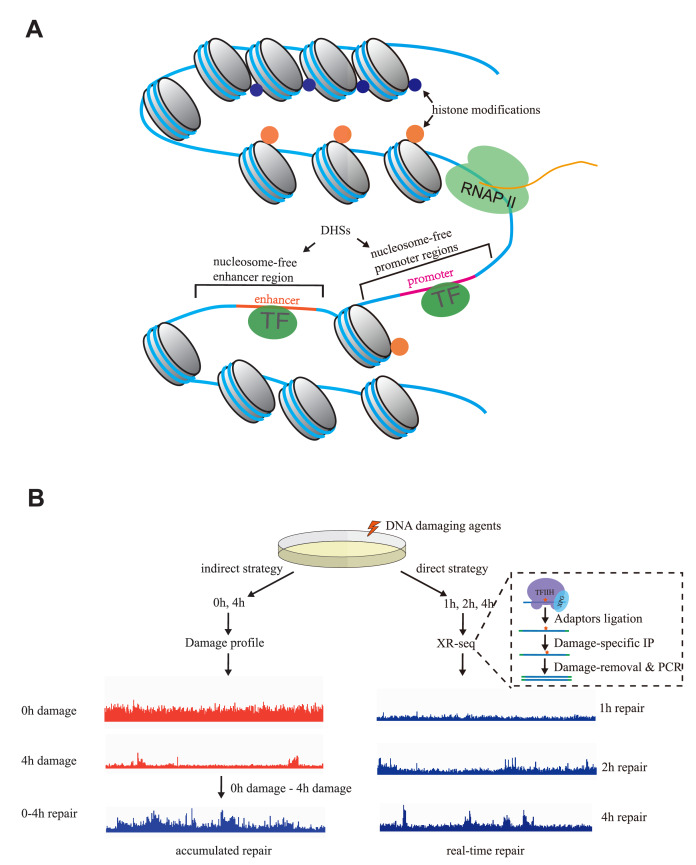
Figure 2 .
Nucleotide excision repair in chromatin environment
(A) Chromatin factors that may affect NER, including histone modifications, transcription, transcription factor binding, nucleosome positioning and genome accessibility. TF is short for transcription factor. (B) The methodology to map NER across the genome. The indirect strategy measures genomic profiles of damage at different time points and acquires the pattern of accumulated repair during this time course by comparing damage profiles. The direct strategy (XR-seq) maps NER by capturing and sequencing excised oligonucleotides and obtains the snapshot of repair at a specific time point.
Methodology for mapping nucleotide excision repair
There are two strategies to profile NER across the whole genome ( Figure 2B). The first one is achieved by assessing the genome-wide distribution of bulky adducts (the substrates of NER) in a time course and calculating the rate of disappearance at different loci throughout the genome. Accordingly, a couple of methods were developed in recent years to map adducts at base resolution. One type of these methods, including CPD-seq [156], Adduct-seq [157], and Circle-damage-seq [158], took advantage of T4 Endonuclease V to cut the damaged DNA strand at CPD sites and captured these DNA ends for sequencing. Other methods such as Damage-seq [ 146, 159] and cisplatin-seq [160] utilized specific antibodies [for CPD, (6–4) PP, cisplatin adducts, etc.] or a damage-binding protein (engineered HMGB1 for cisplatin adducts) to capture DNA strands containing lesions, respectively, and then detected the exact positions by high-fidelity DNA polymerases which can be blocked by the lesions. However, it is not a good choice to measure repair by comparing damage distribution in a time course, especially when only a small portion of damage is repaired, e. g., at early time points or in cells partially deficient in repair, since it would be inaccurate to determine a small value by subtracting a big number from another big number.
The second strategy is directly profiling NER by isolating and sequencing in vivo excision products. The method, named XR-seq (eXcision-Repair sequencing), captured primary excision products by co-immunoprecipitation with anti-TFIIH (XPB or p62) or anti-XPG antibodies, and then added adaptors to both ends of excision products. Afterwards ligation products were purified by immunoprecipitation with damage-specific antibodies, and the lesions were directly reversed by photolyases (for UV-induced damages) or chemical treatment (for cisplatin-adduct) to enable PCR-amplification and sequencing [ 146, 161, 162] . Comparing with indirect assays that assess repair by subtraction, XR-seq possesses much higher sensitivity and provides snapshots of repair instead of cumulative changes of damage. XR-seq can detect repair from 1 min to 48 h after UV irradiation [163], and profile GGR and TCR in TCR-deficient and GGR-deficient cells, respectively [162]. Notably, the genome-wide distribution of damage is also valuable for the exploration of repair, as the repair events captured by XR-seq are determined by both relative repair capacity and local damage density, and the cumulative repair maps are complementary to the snapshots.
Patterns of nucleotide excision repair throughout the genome
The repair maps by XR-seq revealed intriguing patterns of TCR and GGR. In XPC mutant cells that have only TCR, repair of both UV-induced damages occurs exclusively on the template strands of transcribed regions. Clear repair signals on the coding strands at the upstream regions of TSSs and on both strands around enhancers suggested that the bidirectional transcription by Pol II in mammalian cells is also able to trigger TCR [162]. In contrast, rDNA regions transcribed by Pol I showed no preferential repair of templates strands, indicating that Pol I cannot cause TCR [164].
In addition to TCR, GGR is also promoted by transcription. In CSB mutant cells that lack TCR, elevated repairs on both strands around active TSSs were observed, probably due to the relaxed chromatin within these regions [162]. Furthermore, GGR is also related to other factors including histone modifications, DNase I hypersensitive sites (DHSs), super-enhancers, nucleosome occupation and transcription factor binding [ 163, 165– 168] . In general, “open” regions with active transcription and high accessibility are more accessible for the repair factors and thus repaired faster.
However, in the TSS surrounding regions where damages are repaired faster in general, the impaired repair was observed at specific loci. For instance, time-course XR-seq identified a valley at early time points at downstream region (less than 1kb from TSSs) which turned to be a peak at late time points [165]. The location of the valley and peak coincided with the H3K4me3 peak, which is thought to reflect the first nucleosome downstream of TSSs [169]. Damage-seq verified that the repressed repair at early time points resulted in the accumulation of damage at these loci, which caused the late repair peak [165]. Hindered repair was also observed at transcription factor binding sites in the upstream regions of TSSs, which is related to increased mutation frequencies in cancer genomes [ 167, 168] . This phenomenon was attributed to transcription factor binding which inhibited the access of repair factors. The heterogeneity of repair was generally more obvious at early time points, e.g. repair hotspots were identified at super-enhancers as early as 1 min, and disappeared with time, likely due to the change of damage distribution (as described above) and UV-induced alteration of chromatin compaction [163].
An interesting question is the contributions of TCR and GGR in the repair of different damages, e. g., UV-induced (6–4) PP and CPD. In mutant cell lines which have only one sub-pathway of NER, both lesions have similar repair patterns. In repair-proficient cells, (6–4) PP repair showed virtually no strand bias, like that in TCR-deficient cells, indicating that this lesion is mainly eliminated by GGR [162]. On the other hand, CPDs on template strands are preferentially repaired, although repair signals on the non-template strands can also be detected [162]. As discussed above, (6–4) PP induces stronger double-strand distortion than CPD, thus is more readily to be eliminated by GGR. The strand difference of CPD repair decreases over time, due to the disappearance of damages on template strands [159]. Surprisingly, at a very early time point (12 min), no asymmetric repair of CPD on two strands was observed in NHF1 human fibroblasts and HeLa cells, implying a delay of TCR after UV [ 163, and our unpublished data]. This phenomenon could only be detected by direct measurement of repair ( e. g., XR-seq), and the underlying mechanism is unknown.
XR-seq was also used to detect the repair of cisplatin-induced damage in mice [ 139– 141] . Repair maps at different time points of one day revealed the impact of circadian rhythm in two ways. Firstly, the template strands of circadian-controlled genes are preferably repaired when they are being actively transcribed, which is driven by TCR. In contrast, the repair of non-template of all genes and intergenic regions peaks at Zeitgeber time ZT08 when the expression of XPA gene is upregulated by circadian rhythm, indicating the influence of circadian on GGR [ 139, 140] . Moreover, repair in different organs (kidney, liver, lung, and spleen) of mice are shown to be related to tissue-specific transcription patterns and epigenomic profiles [141]. Therefore, NER in living animals is much more complicated and regulated by many factors not existing in cultured cells.
The Roles of NER Proteins other than Bulky Adducts Repair
It is well known that TFIIH is an essential factor for transcription initiation [170], while RPA is involved in many DNA-related events like replication [171]. The rest of NER factors also possess other functions, since they are not restricted to specific lesions. The recognition factors identify damage by the double-strand distortion or blocked Pol II, no matter they are proper substrates of NER or not. The nucleases, XPF and XPG, just recognize DNA with flap-structure and cut at the single strand-double strand junctions [172]. Therefore, they can operate on DNA with similar property or structure, and perform other functions. Indeed, XPF is also an essential factor in the Fanconi Anemia pathway for the repair of inter-strand crosslink damage [173]. Here we will discuss the roles of NER factors in the repair of oxidative damage, and the important physiological functions of the NER nucleases in resolving R-loops.
NER proteins and oxidative DNA damage
In general, oxidative damage is eliminated by BER in mammalian cells [174]. However, some “bulky” oxidative lesions, e. g., 8,5′-cyclo-2′-deoxyadenosine and 8,5′-cyclo-2′-deoxyguanosine, are thought to be repaired by NER [175]. In addition, further oxidation products of the most common oxidative lesion 7,8-dihydro-8-oxo-2′-deoxyguanosine (8-oxo-dG), i. e., spiroiminodihydantoin and guanidinohydantoin, were found to be excised by both BER and NER in vitro by cell extracts that have both BER and NER systems, while 8-oxo-dG was preferred to be repaired by BER under the same condition [ 175, 176] . Although NER excision products cannot be detected under that condition, both 8-oxo-dG and its repair intermediate abasic sites can be recognized by in vitro reconstituted NER system [177], albeit they are efficiently removed by OGG1 and APE1 via BER pathway in vivo [178]. Whether NER can repair these lesions in the absence of BER in vivo is unclear.
Besides the potential involvement of the whole NER pathway, individual repair factors may participate in the repair of oxidative damage in collaboration with BER pathway, as reviewed by Kumar et al. [16]. Among NER factors, XPA, XPG, CSA, CSB, UVSSA, XPC, and DDB were all reported to stimulate the repair of oxidative damage like 8-oxo-dG in different studies [ 175, 179– 181] . However, the conclusions are to some extent conflicting. For instance, XPC was reported to be able to stimulate the activity of OGG1 directly [182]. However, genetic experiments indicated that XPC and XPA are involved in the same 8-oxo-dG repair pathway which may be different from that of CSB and OGG1 [181]. The roles of XPG and XPA in oxidative damage repair are also discordant [ 177, 183, 184] . Meanwhile, Guo et al. [185] identified transcription-coupled repair of 8-oxo-dG in a CSB-dependent manner, and the recruitment of CSB to oxidative damage sites was also verified by other studies [ 181, 186, 187] . However, as 8-oxo-dG is unable to block Pol II [188], the underlying mechanism is unknown. Finally, a recent study reported the role of DDB in BER [189]. The existence of nucleosomes can greatly inhibit the activity of DNA glycosylates, while DDB was shown to play a role in repairing nucleosomal oxidative damages, just as it did in NER [189]. Further work is needed to clarify the functions and underlying mechanisms of NER factors in the repair of oxidative damage.
The roles of NER nucleases in resolving R-loops
R-loop is a specific 3-strand structure consisting of a DNA-RNA hybrid and displaced single-stranded DNA [190]. It can be physiologically formed during transcription and is involved in multiple cellular processes, including transcription regulation and termination [191], class switch recombination of immunoglobulin genes [ 192, 193] , etc. However, R-loop can also be induced accidentally and cause genome instability. The flap structure of R-loop makes it a potential substrate of the two NER endonucleases XPG and XPF [194]. It was reported that the absence of the RNA/DNA helicases Aquarius causes the accumulation of R-loops which are further digested by XPG and XPF to generate DSBs. This process depends on the TCR factor CSB and common NER factors TFIIH and XPA, thus is thought to be a TCR-like reaction [194]. However, since R-loops are behind the elongating RNA polymerases, how they can trigger a TCR-like reaction is unclear.
Another study reported that R-loops can stimulate high-fidelity DSB repair by a Rad52 and XPG-dependent mechanism [195]. DSBs in actively transcribing regions can induce R-loops which help recruit Rad52 to facilitate the high-fidelity homologous recombination repair (HR) and suppress the error-prone non-homologous end-joining (NHEJ). In this process, XPG but not XPF is recruited by Rad52 to resolve R-loops and initiate homologous recombination repair. This study revealed the role of XPG in DSB repair via its activity on R-loops.
A more recent study revealed another mechanism for XPG and XPF to be enrolled in resolving R-loops [196]. When R-loops are induced by RNA polymerase stalling, e. g., in the case of transcription-blocking damage, the splicing factor XAB2 can interact with XPG and XPF-ERCC1 independent of other NER factors to stimulate the processing of R-loops and play a role in maintaining genome integrity. These studies suggested that the NER nucleases, especially XPG, are involved in R-loop processing in multiple ways.
Conclusions and Perspectives
Although the basic mechanism of NER was unveiled more than 20 years ago, the molecular details of TCR, as well as that of the initial damage recognition by DDB in GGR, remained unclear for a long time due to the lack of the in vitro system. Significant progress has been made in the past few years based on the advancement of methodologies in structural biology, in vivo imaging, genomics, high-throughput screen, etc. However, there are still a couple of remaining questions: (1) Does DDB play other roles in GGR in addition to its reported functions? How do different functions of DDB coordinate in response to UV? (2) How do local chromatin compaction and histone modifications change following UV irradiation across the genome? How do they affect GGR? (3) What determines the phenotype of TCR-deficient patients, i. e., CS or UV SS? And why do patients possessing some TFIIH, XPF and XPG mutations have CS-like phenotypes? (4) What are the endogenous substrates of NER? Are they related to CS, especially neuro-associated phenotypes? (5) How is a lesion transferred from damage recognition factors to the pre-incision complex during TCR? What is the fate of damage-blocked Pol II and associated TCR factors? (6) Does NER serve as a backup of BER in oxidative damage repair? How important are NER factors for the repair of oxidative damage? (7) Does XPG participate in the processing of all R-loops? Is there any general mechanism for XPG (and XPF) to involve in R-loop resolving?
New answers to the above questions will certainly emerge in next few years, which can help to reveal the molecular details underlying NER and uncover the link between NER and human health diseases.
Acknowledgments
The authors would like to thank Dr. Wentao Li of the University of Georgia (Athens, USA) for his careful reading of the manuscript and valuable suggestions.
COMPETING INTERESTS
The authors declare that they have no conflict of interest.
Funding Statement
This work was supported by the grants from the National Natural Science Foundation of China (NSFC) (No. 31870804), Shanghai Municipal Natural Science Foundation (No. 22ZR1413900), Program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institutions of Higher Learning (2019), Shanghai Outstanding Young Talent Program (2019), and Innovative Research Team of High-level Local University in Shanghai (to J.H.).
References
- 1.Lindahl T. Instability and decay of the primary structure of DNA. Nature. . 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 2.De Bont R, van Larebeke N. Endogenous DNA damage in humans: a review of quantitative data. Mutagenesis. . 2004;19:169–185. doi: 10.1093/mutage/geh025. [DOI] [PubMed] [Google Scholar]
- 3.Kciuk M, Marciniak B, Mojzych M, Kontek R. Focus on UV-induced DNA damage and repair—disease relevance and protective strategies. Int J Mol Sci. . 2020;21:7264. doi: 10.3390/ijms21197264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ewa B, Danuta M-Š. Polycyclic aromatic hydrocarbons and PAH-related DNA adducts. J Appl Genet. 2017, 58: 321–330 . [DOI] [PMC free article] [PubMed]
- 5.Rushing BR, Selim MI. Aflatoxin B1: a review on metabolism, toxicity, occurrence in food, occupational exposure, and detoxification methods. Food Chem Toxicol. . 2019;124:81–100. doi: 10.1016/j.fct.2018.11.047. [DOI] [PubMed] [Google Scholar]
- 6.Ghosh S. Cisplatin: the first metal based anticancer drug. BioOrg Chem. . 2019;88:102925. doi: 10.1016/j.bioorg.2019.102925. [DOI] [PubMed] [Google Scholar]
- 7.Schmeiser HH, Schoepe KB, Wiessler M. DNA adduct formation of aristolochic acid I and II in vitro and in vivo . Carcinogenesis. . 1988;9:297–303. doi: 10.1093/carcin/9.2.297. [DOI] [PubMed] [Google Scholar]
- 8.Jaspers NGJ, Raams A, Kelner MJ, Ng JM, Yamashita YM, Takeda S, McMorris TC, et al. Anti-tumour compounds illudin S and irofulven induce DNA lesions ignored by global repair and exclusively processed by transcription- and replication-coupled repair pathways. DNA Repair. . 2002;1:1027–1038. doi: 10.1016/S1568-7864(02)00166-0. [DOI] [PubMed] [Google Scholar]
- 9.Lans H, Hoeijmakers JH, Vermeulen W, Marteijn JA. The DNA damage response to transcription stress. Nat Rev Mol Cell Biol. . 2019;20:766–784. doi: 10.1038/s41580-019-0169-4. [DOI] [PubMed] [Google Scholar]
- 10.Maslov AY, Vijg J. Genome instability, cancer and aging. Biochim Biophys Acta (BBA) - Gen Subj. . 2009;1790:963–969. doi: 10.1016/j.bbagen.2009.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoeijmakers JH. DNA damage, aging, and cancer. N Engl J Med. . 2009;361:1475–1485. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- 12.Marteijn JA, Lans H, Vermeulen W, Hoeijmakers JH. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat Rev Mol Cell Biol. . 2014;15:465–481. doi: 10.1038/nrm3822. [DOI] [PubMed] [Google Scholar]
- 13.Fortini P, Parlanti E, Sidorkina OM, Laval J, Dogliotti E. The type of DNA glycosylase determines the base excision repair pathway in mammalian cells. J Biol Chem. . 1999;274:15230–15236. doi: 10.1074/jbc.274.21.15230. [DOI] [PubMed] [Google Scholar]
- 14.Drohat AC, Maiti A. DNA glycosylases: mechanisms. in: lennarz WJ, lane MD eds. Encyclopedia of biological chemistry (second edition) waltham: Academic Press. 2013: 16–20
- 15.Lindahl T. The intrinsic fragility of DNA (nobel lecture) Angew Chem Int Ed. . 2016;55:8528–8534. doi: 10.1002/anie.201602159. [DOI] [PubMed] [Google Scholar]
- 16.Kumar N, Raja S, Van Houten B. The involvement of nucleotide excision repair proteins in the removal of oxidative DNA damage. Nucleic Acids Res. . 2020;48:11227–11243. doi: 10.1093/nar/gkaa777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zharkov DO, Grollman AP. The DNA trackwalkers: principles of lesion search and recognition by DNA glycosylases. Mutat Res Fundamental Mol Mech Mutagenesis. . 2005;577:24–54. doi: 10.1016/j.mrfmmm.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 18.Farnell DA. Nucleotide excision repair in the three domains of life. WURJ:HNS. . 2011;2:1. doi: 10.5206/wurjhns.2010-11.1. [DOI] [Google Scholar]
- 19.Reardon JT, Sancar A. Nucleotide excision repair. Prog Nucleic Acid Res Mol Biol. 2005: 183–235 . [DOI] [PubMed]
- 20.Ögrünç M̈, Becker DF, Ragsdale SW, Sancar A. Nucleotide excision repair in the third kingdom. J Bacteriol. . 1998;180:5796–5798. doi: 10.1128/JB.180.21.5796-5798.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.White MF. Archaeal DNA repair: paradigms and puzzles. Biochem Soc Trans. . 2003;31:690–693. doi: 10.1042/bst0310690. [DOI] [PubMed] [Google Scholar]
- 22.Marshall CJ, Santangelo TJ. Archaeal DNA repair mechanisms. Biomolecules. . 2020;10:1472. doi: 10.3390/biom10111472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cleaver JE, Lam ET, Revet I. Disorders of nucleotide excision repair: the genetic and molecular basis of heterogeneity. Nat Rev Genet. . 2009;10:756–768. doi: 10.1038/nrg2663. [DOI] [PubMed] [Google Scholar]
- 24.Boulikas T. Xeroderma pigmentosum and molecular cloning of DNA repair genes. Anticancer Res. 1996, 16: 693–708 . [PubMed]
- 25.Hanawalt PC. The bases for cockayne syndrome. Nature. . 2000;405:415. doi: 10.1038/35013197. [DOI] [PubMed] [Google Scholar]
- 26.Zhang X, Horibata K, Saijo M, Ishigami C, Ukai A, Kanno S, Tahara H, et al. Mutations in UVSSA cause UV-sensitive syndrome and destabilize ERCC6 in transcription-coupled DNA Repair. Nat Genet. . 2012;44:593–597. doi: 10.1038/ng.2228. [DOI] [PubMed] [Google Scholar]
- 27.Schwertman P, Lagarou A, Dekkers DHW, Raams A, van der Hoek AC, Laffeber C, Hoeijmakers JHJ, et al. UV-sensitive syndrome protein UVSSA recruits USP7 to regulate transcription-coupled repair. Nat Genet. . 2012;44:598–602. doi: 10.1038/ng.2230. [DOI] [PubMed] [Google Scholar]
- 28.Nakazawa Y, Sasaki K, Mitsutake N, Matsuse M, Shimada M, Nardo T, Takahashi Y, et al. Mutations in UVSSA cause UV-sensitive syndrome and impair RNA polymerase IIo processing in transcription-coupled nucleotide-excision repair. Nat Genet. . 2012;44:586–592. doi: 10.1038/ng.2229. [DOI] [PubMed] [Google Scholar]
- 29.Harm W. Dark repair of photorepairable UV lesions in Escherichia coli . Mutat Res Fundamental Mol Mech Mutagenesis. . 1968;6:25–35. doi: 10.1016/0027-5107(68)90100-0. [DOI] [PubMed] [Google Scholar]
- 30.Rupert CS. Photoreactivation of transforming DNA by an enzyme from bakers’ yeast. J Gen Physiol. . 1960;43:573–595. doi: 10.1085/jgp.43.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rupert CS, Goodgal SH, Herriott RM. Photoreactivation in vitro of ultraviolet-inactivated hemophilus influenzae transforming factor . J Gen Physiol. . 1958;41:451–471. doi: 10.1085/jgp.41.3.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Selby CP, Sancar A. ABC excinuclease incises both 5′ and 3′ to the CC-1065-DNA adduct and its incision activity is stimulated by DNA helicase II and DNA polymerase I. Biochemistry. . 1988;27:7184–7188. doi: 10.1021/bi00419a004. [DOI] [PubMed] [Google Scholar]
- 33.Orren DK, Sancar A. The (A)BC excinuclease of Escherichia coli has only the UvrB and UvrC subunits in the incision complex . Proc Natl Acad Sci USA. . 1989;86:5237–5241. doi: 10.1073/pnas.86.14.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sancar A, Rupp WD. A novel repair enzyme: UVRABC excision nuclease of Escherichia coli cuts a DNA strand on both sides of the damaged region . Cell. . 1983;33:249–260. doi: 10.1016/0092-8674(83)90354-9. [DOI] [PubMed] [Google Scholar]
- 35.Mu D, Park CH, Matsunaga T, Hsu DS, Reardon JT, Sancar A. Reconstitution of human DNA repair excision nuclease in a highly defined system. J Biol Chem. . 1995;270:2415–2418. doi: 10.1074/jbc.270.6.2415. [DOI] [PubMed] [Google Scholar]
- 36.Aboussekhra A, Biggerstaff M, Shivji MKK, Vilpo JA, Moncollin V, Podust VN, Protić M, et al. Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell. . 1995;80:859–868. doi: 10.1016/0092-8674(95)90289-9. [DOI] [PubMed] [Google Scholar]
- 37.Huang JC, Svoboda DL, Reardon JT, Sancar A. Human nucleotide excision nuclease removes thymine dimers from DNA by incising the 22nd phosphodiester bond 5′ and the 6th phosphodiester bond 3′ to the photodimer. Proc Natl Acad Sci USA. . 1992;89:3664–3668. doi: 10.1073/pnas.89.8.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu J, Selby CP, Adar S, Adebali O, Sancar A. Molecular mechanisms and genomic maps of DNA excision repair in Escherichia coli and humans . J Biol Chem. . 2017;292:15588–15597. doi: 10.1074/jbc.R117.807453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Canturk F, Karaman M, Selby CP, Kemp MG, Kulaksiz-Erkmen G, Hu J, Li W, et al. Nucleotide excision repair by dual incisions in plants. Proc Natl Acad Sci USA. . 2016;113:4706–4710. doi: 10.1073/pnas.1604097113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wood RD. Nucleotide excision repair in mammalian cells. J Biol Chem. . 1997;272:23465–23468. doi: 10.1074/jbc.272.38.23465. [DOI] [PubMed] [Google Scholar]
- 41.Sancar A. Mechanisms of DNA repair by photolyase and excision nuclease (Nobel Lecture) Angew Chem Int Ed. . 2016;55:8502–8527. doi: 10.1002/anie.201601524. [DOI] [PubMed] [Google Scholar]
- 42.Svoboda DL, Taylor JS, Hearst JE, Sancar A. DNA repair by eukaryotic nucleotide excision nuclease. Removal of thymine dimer and psoralen monoadduct by HeLa cell-free extract and of thymine dimer by Xenopus laevis oocytes . J Biol Chem. . 1993;268:1931–1936. doi: 10.1016/S0021-9258(18)53943-0. [DOI] [PubMed] [Google Scholar]
- 43.Guzder SN, Habraken Y, Sung P, Prakash L, Prakash S. Reconstitution of yeast nucleotide excision repair with purified rad proteins, replication protein A, and transcription factor TFIIH. J Biol Chem. . 1995;270:12973–12976. doi: 10.1074/jbc.270.22.12973. [DOI] [PubMed] [Google Scholar]
- 44.Kemp MG. Damage removal and gap filling in nucleotide excision repair. Enzymes. 2019, 45: 59–97 . [DOI] [PubMed]
- 45.Sancar A. DNA excision repair. Annu Rev Biochem. . 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 46.Husain I, Van Houten B, Thomas DC, Abdel-Monem M, Sancar A. Effect of DNA polymerase I and DNA helicase II on the turnover rate of UvrABC excision nuclease. Proc Natl Acad Sci USA. . 1985;82:6774–6778. doi: 10.1073/pnas.82.20.6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lehmann AR. DNA polymerases and repair synthesis in NER in human cells. DNA Repair. . 2011;10:730–733. doi: 10.1016/j.dnarep.2011.04.023. [DOI] [PubMed] [Google Scholar]
- 48.Atanassov B, Velkova A, Mladenov E, Anachkova B, Russev G. Comparison of the global genomic and transcription-coupled repair rates of different lesions in human cells. Z für Naturforschung C. . 2004;59:445–453. doi: 10.1515/znc-2004-5-628. [DOI] [PubMed] [Google Scholar]
- 49.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol. . 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 50.Lans H, Marteijn JA, Schumacher B, Hoeijmakers JHJ, Jansen G, Vermeulen W. Involvement of global genome repair, transcription coupled repair, and chromatin remodeling in UV DNA damage response changes during development. PLoS Genet. . 2010;6:e1000941. doi: 10.1371/journal.pgen.1000941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fousteri M, Mullenders LH. Transcription-coupled nucleotide excision repair in mammalian cells: molecular mechanisms and biological effects. Cell Res. . 2008;18:73–84. doi: 10.1038/cr.2008.6. [DOI] [PubMed] [Google Scholar]
- 52.Sugasawa K, Ng JM, Masutani C, Iwai S, van der Spek PJ, Eker AP, Hanaoka F, et al. Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol Cell. . 1998;2:223–232. doi: 10.1016/S1097-2765(00)80132-X. [DOI] [PubMed] [Google Scholar]
- 53.van Hoffen A, Venema J, Meschini R, van Zeeland AA, Mullenders LH. Transcription-coupled repair removes both cyclobutane pyrimidine dimers and 6-4 photoproducts with equal efficiency and in a sequential way from transcribed DNA in xeroderma pigmentosum group C fibroblasts. EMBO J. . 1995;14:360–367. doi: 10.1002/j.1460-2075.1995.tb07010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ribeiro-Silva C, Sabatella M, Helfricht A, Marteijn JA, Theil AF, Vermeulen W, Lans H. Ubiquitin and TFIIH-stimulated DDB2 dissociation drives DNA damage handover in nucleotide excision repair. Nat Commun. . 2020;11:4868. doi: 10.1038/s41467-020-18705-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sugasawa K, Okuda Y, Saijo M, Nishi R, Matsuda N, Chu G, Mori T, et al. UV-induced ubiquitylation of XPC protein mediated by UV-DDB-Ubiquitin ligase complex. Cell. . 2005;121:387–400. doi: 10.1016/j.cell.2005.02.035. [DOI] [PubMed] [Google Scholar]
- 56.van der Weegen Y, Golan-Berman H, Mevissen TET, Apelt K, González-Prieto R, Goedhart J, Heilbrun EE, et al. The cooperative action of CSB, CSA, and UVSSA target TFIIH to DNA damage-stalled RNA polymerase II. Nat Commun. . 2020;11:2104. doi: 10.1038/s41467-020-15903-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fei J, Chen J. KIAA1530 protein is recruited by Cockayne syndrome complementation group protein A (CSA) to participate in transcription-coupled repair (TCR) J Biol Chem. . 2012;287:35118–35126. doi: 10.1074/jbc.M112.398131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Paul D, Mu H, Zhao H, Ouerfelli O, Jeffrey PD, Broyde S, Min JH. Structure and mechanism of pyrimidine–pyrimidone (6-4) photoproduct recognition by the Rad4/XPC nucleotide excision repair complex. Nucleic Acids Res. . 2019;47:6015–6028. doi: 10.1093/nar/gkz359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sugasawa K. Regulation of damage recognition in mammalian global genomic nucleotide excision repair. Mutat Res Fundamental Mol Mech Mutagenesis. . 2010;685:29–37. doi: 10.1016/j.mrfmmm.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 60.Volker M, Moné MJ, Karmakar P, van Hoffen A, Schul W, Vermeulen W, Hoeijmakers JH, et al. Sequential assembly of the nucleotide excision repair factors in vivo . Mol Cell. . 2001;8:213–224. doi: 10.1016/S1097-2765(01)00281-7. [DOI] [PubMed] [Google Scholar]
- 61.Riedl T, Hanaoka F, Egly JM. The comings and goings of nucleotide excision repair factors on damaged DNA. EMBO J. . 2003;22:5293–5303. doi: 10.1093/emboj/cdg489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kazantsev A, Mu D, Nichols AF, Zhao X, Linn S, Sancar A. Functional complementation of xeroderma pigmentosum complementation group E by replication protein A in an in vitro system . Proc Natl Acad Sci USA. . 1996;93:5014–5018. doi: 10.1073/pnas.93.10.5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Scrima A, Fischer ES, Lingaraju GM, Böhm K, Cavadini S, Thomä NH. Detecting UV-lesions in the genome: the modular CRL4 ubiquitin ligase does it best! FEBS Lett. . 2011;585:2818–2825. doi: 10.1016/j.febslet.2011.04.064. [DOI] [PubMed] [Google Scholar]
- 64.Fischer ES, Scrima A, Böhm K, Matsumoto S, Lingaraju GM, Faty M, Yasuda T, et al. The molecular basis of CRL4DDB2/CSA ubiquitin ligase architecture, targeting, and activation. Cell. . 2011;147:1024–1039. doi: 10.1016/j.cell.2011.10.035. [DOI] [PubMed] [Google Scholar]
- 65.Nichols AF, Ong P, Linn S. Mutations specific to the xeroderma pigmentosum group E Ddb − phenotype . J Biol Chem. . 1996;271:24317–24320. doi: 10.1074/jbc.271.40.24317. [DOI] [PubMed] [Google Scholar]
- 66.Tang J, Chu G. Xeroderma pigmentosum complementation group E and UV-damaged DNA-binding protein. DNA Repair. . 2002;1:601–616. doi: 10.1016/S1568-7864(02)00052-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kapetanaki MG, Guerrero-Santoro J, Bisi DC, Hsieh CL, Rapić-Otrin V, Levine AS. The DDB1-CUL4A DDB2 ubiquitin ligase is deficient in xeroderma pigmentosum group E and targets histone H2A at UV-damaged DNA sites . Proc Natl Acad Sci USA. . 2006;103:2588–2593. doi: 10.1073/pnas.0511160103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.RapićOtrin V, Kuraoka I, Nardo T, McLenigan M, Eker AP, Stefanini M, Levine AS, et al. Relationship of the xeroderma pigmentosum group E DNA repair defect to the chromatin and DNA binding proteins UV-DDB and replication protein A. Mol Cell Biol. . 1998;18:3182–3190. doi: 10.1128/MCB.18.6.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reardon JT, Nichols AF, Keeney S, Smith CA, Taylor JS, Linn S, Sancar A. Comparative analysis of binding of human damaged DNA-binding protein (XPE) and Escherichia coli damage recognition protein (UvrA) to the major ultraviolet photoproducts: T[c,s]T, T[t,s]T, T[6-4]T, and T[Dewar]T . J Biol Chem. . 1993;268:21301–21308. doi: 10.1016/S0021-9258(19)36924-8. [DOI] [PubMed] [Google Scholar]
- 70.Batty D, Rapic’-Otrin V, Levine AS, Wood RD. Stable binding of human XPC complex to irradiated DNA confers strong discrimination for damaged sites. J Mol Biol. . 2000;300:275–290. doi: 10.1006/jmbi.2000.3857. [DOI] [PubMed] [Google Scholar]
- 71.Wakasugi M, Kawashima A, Morioka H, Linn S, Sancar A, Mori T, Nikaido O, et al. DDB accumulates at DNA damage sites immediately after UV irradiation and directly stimulates nucleotide excision repair. J Biol Chem. . 2002;277:1637–1640. doi: 10.1074/jbc.C100610200. [DOI] [PubMed] [Google Scholar]
- 72.Scrima A, Konícková R, Czyzewski BK, Kawasaki Y, Jeffrey PD, Groisman R, Nakatani Y, et al. Structural basis of UV DNA-damage recognition by the DDB1–DDB2 complex. Cell. . 2008;135:1213–1223. doi: 10.1016/j.cell.2008.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Matsumoto S, Cavadini S, Bunker RD, Grand RS, Potenza A, Rabl J, Yamamoto J, et al. DNA damage detection in nucleosomes involves DNA register shifting. Nature. . 2019;571:79–84. doi: 10.1038/s41586-019-1259-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fei J, Kaczmarek N, Luch A, Glas A, Carell T, Naegeli H. Regulation of nucleotide excision repair by UV-DDB: prioritization of damage recognition to internucleosomal DNA. PLoS Biol. . 2011;9:e1001183. doi: 10.1371/journal.pbio.1001183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fortuny A, Chansard A, Caron P, Chevallier O, Leroy O, Renaud O, Polo SE. Imaging the response to DNA damage in heterochromatin domains reveals core principles of heterochromatin maintenance. Nat Commun. . 2021;12:2428. doi: 10.1038/s41467-021-22575-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Luijsterburg MS, Lindh M, Acs K, Vrouwe MG, Pines A, van Attikum H, Mullenders LH, et al. DDB2 promotes chromatin decondensation at UV-induced DNA damage. J Cell Biol. . 2012;197:267–281. doi: 10.1083/jcb.201106074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hoogstraten D, Bergink S, Ng JM, Verbiest VH, Luijsterburg MS, Geverts B, Raams A, et al. Versatile DNA damage detection by the global genome nucleotide excision repair protein XPC. J Cell Sci. . 2008;121:2850–2859. doi: 10.1242/jcs.031708. [DOI] [PubMed] [Google Scholar]
- 78.Chen X, Velmurugu Y, Zheng G, Park B, Shim Y, Kim Y, Liu L, et al. Kinetic gating mechanism of DNA damage recognition by Rad4/XPC. Nat Commun. . 2015;6:5849. doi: 10.1038/ncomms6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Min JH, Pavletich NP. Recognition of DNA damage by the Rad4 nucleotide excision repair protein. Nature. . 2007;449:570–575. doi: 10.1038/nature06155. [DOI] [PubMed] [Google Scholar]
- 80.Moser J, Volker M, Kool H, Alekseev S, Vrieling H, Yasui A, van Zeeland AA, et al. The UV-damaged DNA binding protein mediates efficient targeting of the nucleotide excision repair complex to UV-induced photo lesions. DNA Repair. . 2005;4:571–582. doi: 10.1016/j.dnarep.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 81.Okuda M, Nakazawa Y, Guo C, Ogi T, Nishimura Y. Common TFIIH recruitment mechanism in global genome and transcription-coupled repair subpathways. Nucleic Acids Res. . 2017;45:13043–13055. doi: 10.1093/nar/gkx970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yokoi M, Masutani C, Maekawa T, Sugasawa K, Ohkuma Y, Hanaoka F. The xeroderma pigmentosum group C protein complex XPC-HR23B plays an important role in the recruitment of transcription factor IIH to damaged DNA. J Biol Chem. . 2000;275:9870–9875. doi: 10.1074/jbc.275.13.9870. [DOI] [PubMed] [Google Scholar]
- 83.Bernardes de Jesus BM, Bjørås M, Coin F́́, Egly JM. Dissection of the molecular defects caused by pathogenic mutations in the DNA repair factor XPC. Mol Cell Biol. . 2008;28:7225–7235. doi: 10.1128/MCB.00781-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Svejstrup J. The multiple roles of transcription/repair factor TFIIH. Trends Biochem Sci. . 1996;21:346–350. doi: 10.1016/0968-0004(96)10046-3. [DOI] [PubMed] [Google Scholar]
- 85.Iben S, Tschochner H, Bier M, Hoogstraten D, Hozák P, Egly JM, Grummt I. TFIIH plays an essential role in RNA polymerase I transcription. Cell. . 2002;109:297–306. doi: 10.1016/S0092-8674(02)00729-8. [DOI] [PubMed] [Google Scholar]
- 86.Holstege FC, Fiedler U, Timmers HT. Three transitions in the RNA polymerase II transcription complex during initiation. EMBO J. . 1997;16:7468–7480. doi: 10.1093/emboj/16.24.7468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kumar KP, Akoulitchev S, Reinberg D. Promoter-proximal stalling results from the inability to recruit transcription factor IIH to the transcription complex and is a regulated event. Proc Natl Acad Sci USA. . 1998;95:9767–9772. doi: 10.1073/pnas.95.17.9767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Coin F, Bergmann E, Tremeau-Bravard A, Egly JM. Mutations in XPB and XPD helicases found in xeroderma pigmentosum patients impair the transcription function of TFIIH. EMBO J. . 1999;18:1357–1366. doi: 10.1093/emboj/18.5.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Compe E, Egly JM. Nucleotide excision repair and transcriptional regulation: TFIIH and beyond. Annu Rev Biochem. . 2016;85:265–290. doi: 10.1146/annurev-biochem-060815-014857. [DOI] [PubMed] [Google Scholar]
- 90.Li CL, Golebiowski FM, Onishi Y, Samara NL, Sugasawa K, Yang W. Tripartite DNA lesion recognition and verification by XPC, TFIIH, and XPA in nucleotide excision repair. Mol Cell. . 2015;59:1025–1034. doi: 10.1016/j.molcel.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lainé JP, Egly JM. Initiation of DNA repair mediated by a stalled RNA polymerase IIO. EMBO J. . 2006;25:387–397. doi: 10.1038/sj.emboj.7600933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Reardon JT, Sancar A. Thermodynamic cooperativity and kinetic proofreading in DNA damage recognition and repair. Cell Cycle. . 2004;3:139–142. doi: 10.4161/cc.3.2.645. [DOI] [PubMed] [Google Scholar]
- 93.Coin F, Oksenych V, Egly JM. Distinct roles for the XPB/p52 and XPD/p44 subcomplexes of TFIIH in damaged DNA opening during nucleotide excision repair. Mol Cell. . 2007;26:245–256. doi: 10.1016/j.molcel.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 94.Kokic G, Chernev A, Tegunov D, Dienemann C, Urlaub H, Cramer P. Structural basis of TFIIH activation for nucleotide excision repair. Nat Commun. . 2019;10:2885. doi: 10.1038/s41467-019-10745-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Coin F, Oksenych V, Mocquet V, Groh S, Blattner C, Egly JM. Nucleotide excision repair driven by the dissociation of CAK from TFIIH. Mol Cell. . 2008;31:9–20. doi: 10.1016/j.molcel.2008.04.024. [DOI] [PubMed] [Google Scholar]
- 96.Wakasugi M, Sancar A. Order of assembly of human DNA repair excision nuclease. J Biol Chem. . 1999;274:18759–18768. doi: 10.1074/jbc.274.26.18759. [DOI] [PubMed] [Google Scholar]
- 97.Wakasugi M, Sancar A. Assembly, subunit composition, and footprint of human DNA repair excision nuclease. Proc Natl Acad Sci USA. . 1998;95:6669–6674. doi: 10.1073/pnas.95.12.6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Park CH, Sancar A. Formation of a ternary complex by human XPA, ERCC1, and ERCC4(XPF) excision repair proteins. Proc Natl Acad Sci USA. . 1994;91:5017–5021. doi: 10.1073/pnas.91.11.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mu D, Hsu DS, Sancar A. Reaction mechanism of human DNA repair excision nuclease. J Biol Chem. . 1996;271:8285–8294. doi: 10.1074/jbc.271.14.8285. [DOI] [PubMed] [Google Scholar]
- 100.Mellon I. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. . 1987;51:241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- 101.Hu J, Choi JH, Gaddameedhi S, Kemp MG, Reardon JT, Sancar A. Nucleotide excision repair in human cells. J Biol Chem. . 2013;288:20918–20926. doi: 10.1074/jbc.M113.482257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bohr V. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. . 1985;40:359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- 103.Andrews AD, Barrett SF, Yoder FW, Robbins JH. Cockayne’s syndrome fibroblasts have increased sensitivity to ultraviolet light but normal rates of unscheduled DNA synthesis. J Investig Dermatol. . 1978;70:237–239. doi: 10.1111/1523-1747.ep12541383. [DOI] [PubMed] [Google Scholar]
- 104.Troelstra C, van Gool A, de Wit J, Vermeulen W, Bootsma D, Hoeijmakers JH. ERCC6, a member of a subfamily of putative helicases, is involved in Cockayne’s syndrome and preferential repair of active genes. Cell. . 1992;71:939–953. doi: 10.1016/0092-8674(92)90390-X. [DOI] [PubMed] [Google Scholar]
- 105.Henning KA, Li L, Iyer N, McDaniel LD, Reagan MS, Legerski R, Schultz RA, et al. The Cockayne syndrome group A gene encodes a WD repeat protein that interacts with CSB protein and a subunit of RNA polymerase II TFIIH. Cell. . 1995;82:555–564. doi: 10.1016/0092-8674(95)90028-4. [DOI] [PubMed] [Google Scholar]
- 106.Geijer ME, Zhou D, Selvam K, Steurer B, Mukherjee C, Evers B, Cugusi S, et al. Elongation factor ELOF1 drives transcription-coupled repair and prevents genome instability. Nat Cell Biol. . 2021;23:608–619. doi: 10.1038/s41556-021-00692-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Boetefuer EL, Lake RJ, Fan HY. Mechanistic insights into the regulation of transcription and transcription-coupled DNA repair by Cockayne syndrome protein B. Nucleic Acids Res. . 2018;46:7471–7479. doi: 10.1093/nar/gky660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tantin D, Kansal A, Carey M. Recruitment of the putative transcription-repair coupling factor CSB/ERCC6 to RNA polymerase II elongation complexes. Mol Cell Biol. . 1997;17:6803–6814. doi: 10.1128/MCB.17.12.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.van den Boom V, Citterio E, Hoogstraten D, Zotter A, Egly JM, van Cappellen WA, Hoeijmakers JH, et al. DNA damage stabilizes interaction of CSB with the transcription elongation machinery. J Cell Biol. . 2004;166:27–36. doi: 10.1083/jcb.200401056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.van Gool AJ, Citterio E, Rademakers S, van Os R, Vermeulen W, Constantinou A, Egly JM, et al. The Cockayne syndrome B protein, involved in transcription-coupled DNA repair, resides in an RNA polymerase II-containing complex. EMBO J. . 1997;16:5955–5965. doi: 10.1093/emboj/16.19.5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Selby CP, Sancar A. Cockayne syndrome group B protein enhances elongation by RNA polymerase II. Proc Natl Acad Sci USA. . 1997;94:11205–11209. doi: 10.1073/pnas.94.21.11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Duan M, Selvam K, Wyrick JJ, Mao P. Genome-wide role of Rad26 in promoting transcription-coupled nucleotide excision repair in yeast chromatin. Proc Natl Acad Sci USA. . 2020;117:18608–18616. doi: 10.1073/pnas.2003868117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wang W, Xu J, Chong J, Wang D. Structural basis of DNA lesion recognition for eukaryotic transcription-coupled nucleotide excision repair. DNA Repair. . 2018;71:43–55. doi: 10.1016/j.dnarep.2018.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Xu J, Lahiri I, Wang W, Wier A, Cianfrocco MA, Chong J, Hare AA, et al. Structural basis for the initiation of eukaryotic transcription-coupled DNA repair. Nature. . 2017;551:653–657. doi: 10.1038/nature24658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Selby CP, Sancar A. Human transcription-repair coupling factor CSB/ERCC6 is a DNA-stimulated ATPase but is not a helicase and does not disrupt the ternary transcription complex of stalled RNA polymerase II. J Biol Chem. . 1997;272:1885–1890. doi: 10.1074/jbc.272.3.1885. [DOI] [PubMed] [Google Scholar]
- 116.Selby CP, Drapkin R, Reinberg D, Sancar A. RNA polymerase II stalled at a thymine dimer: footprint and effect on excision repair. Nucleic Acids Res. . 1997;25:787–793. doi: 10.1093/nar/25.4.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.van den Heuvel D, van der Weegen Y, Boer DE, Ogi T, Luijsterburg MS. Transcription-coupled DNA repair: from mechanism to human disorder. Trends Cell Biol. . 2021;31:359–371. doi: 10.1016/j.tcb.2021.02.007. [DOI] [PubMed] [Google Scholar]
- 118.Kokic G, Wagner FR, Chernev A, Urlaub H, Cramer P. Structural basis of human transcription–DNA repair coupling. Nature. . 2021;598:368–372. doi: 10.1038/s41586-021-03906-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Miyauchi-Hashimoto H, Akaeda T, Maihara T, Ikenaga M, Horio T. Cockayne syndrome without typical clinical manifestations including neurologic abnormalities. J Am Acad Dermatol. . 1998;39:565–570. doi: 10.1016/S0190-9622(98)70005-2. [DOI] [PubMed] [Google Scholar]
- 120.Li Y, Zheng L, Chen F, Yao Z, Li M. Two novel mutations in the ERCC8 gene in a patient with ultraviolet-sensitive syndrome. Acta Derm Venerol. . 2018:0. doi: 10.2340/00015555-3032. [DOI] [PubMed] [Google Scholar]
- 121.Huibregtse JM, Yang JC, Beaudenon SL. The large subunit of RNA polymerase II is a substrate of the Rsp5 ubiquitin-protein ligase. Proc Natl Acad Sci USA. . 1997;94:3656–3661. doi: 10.1073/pnas.94.8.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gregersen LH, Svejstrup JQ. The cellular response to transcription-blocking DNA damage. Trends Biochem Sci. . 2018;43:327–341. doi: 10.1016/j.tibs.2018.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Geijer ME, Marteijn JA. What happens at the lesion does not stay at the lesion: transcription-coupled nucleotide excision repair and the effects of DNA damage on transcription in cis and trans. DNA Repair. . 2018;71:56–68. doi: 10.1016/j.dnarep.2018.08.007. [DOI] [PubMed] [Google Scholar]
- 124.Elia AE, Boardman AP, Wang DC, Huttlin EL, Everley RA, Dephoure N, Zhou C, et al. Quantitative proteomic atlas of ubiquitination and acetylation in the DNA damage response. Mol Cell. . 2015;59:867–881. doi: 10.1016/j.molcel.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Nakazawa Y, Hara Y, Oka Y, Komine O, van den Heuvel D, Guo C, Daigaku Y, et al. Ubiquitination of DNA damage-stalled RNAPII promotes transcription-coupled repair. Cell. . 2020;180:1228–1244.e24. doi: 10.1016/j.cell.2020.02.010. [DOI] [PubMed] [Google Scholar]
- 126.Tufegdžić VA, Mitter R, Kelly GP, Neumann M, Harreman M, Rodríguez-Martínez M, Herlihy A, et al. Regulation of the RNAPII pool is integral to the DNA damage response. Cell. . 2020;180:1245–1261.e21. doi: 10.1016/j.cell.2020.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Olivieri M, Cho T, Álvarez-Quilón A, Li K, Schellenberg MJ, Zimmermann M, Hustedt N, et al. A genetic map of the response to DNA damage in human cells. Cell. . 2020;182:481–496.e21. doi: 10.1016/j.cell.2020.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Daniels JP, Kelly S, Wickstead B, Gull K. Identification of a crenarchaeal orthologue of Elf1: implications for chromatin and transcription in Archaea. Biol Direct. . 2009;4:24. doi: 10.1186/1745-6150-4-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Prather D, Krogan NJ, Emili A, Greenblatt JF, Winston F. Identification and characterization of Elf1, a conserved transcription elongation factor in Saccharomyces cerevisiae . Mol Cell Biol. . 2005;25:10122–10135. doi: 10.1128/MCB.25.22.10122-10135.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ehara H, Kujirai T, Fujino Y, Shirouzu M, Kurumizaka H, Sekine SI. Structural insight into nucleosome transcription by RNA polymerase II with elongation factors. Science. . 2019;363:744–747. doi: 10.1126/science.aav8912. [DOI] [PubMed] [Google Scholar]
- 131.van der Weegen Y, de Lint K, van den Heuvel D, Nakazawa Y, Mevissen TET, van Schie JJ, San Martin Alonso M, et al. ELOF1 is a transcription-coupled DNA repair factor that directs RNA polymerase II ubiquitylation. Nat Cell Biol. . 2021;23:595–607. doi: 10.1038/s41556-021-00688-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Noe Gonzalez M, Blears D, Svejstrup JQ. Causes and consequences of RNA polymerase II stalling during transcript elongation. Nat Rev Mol Cell Biol. . 2021;22:3–21. doi: 10.1038/s41580-020-00308-8. [DOI] [PubMed] [Google Scholar]
- 133.Lavigne MD, Konstantopoulos D, Ntakou-Zamplara KZ, Liakos A, Fousteri M. Global unleashing of transcription elongation waves in response to genotoxic stress restricts somatic mutation rate. Nat Commun. . 2017;8:2076. doi: 10.1038/s41467-017-02145-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Liakos A, Konstantopoulos D, Lavigne MD, Fousteri M. Continuous transcription initiation guarantees robust repair of all transcribed genes and regulatory regions. Nat Commun. . 2020;11:916. doi: 10.1038/s41467-020-14566-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chiou YY, Hu J, Sancar A, Selby CP. RNA polymerase II is released from the DNA template during transcription-coupled repair in mammalian cells. J Biol Chem. . 2018;293:2476–2486. doi: 10.1074/jbc.RA117.000971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Andrade-Lima LC, Veloso A, Paulsen MT, Menck CM, Ljungman M. DNA repair and recovery of RNA synthesis following exposure to ultraviolet light are delayed in long genes. Nucleic Acids Res. . 2015;43:2744–2756. doi: 10.1093/nar/gkv148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kemp MG, Reardon JT, Lindsey-Boltz LA, Sancar A. Mechanism of release and fate of excised oligonucleotides during nucleotide excision repair. J Biol Chem. . 2012;287:22889–22899. doi: 10.1074/jbc.M112.374447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Akkose U, Kaya VO, Lindsey-Boltz L, Karagoz Z, Brown AD, Larsen PA, Yoder AD, et al. Comparative analyses of two primate species diverged by more than 60 million years show different rates but similar distribution of genome-wide UV repair events. BMC Genomics. . 2021;22:600. doi: 10.1186/s12864-021-07898-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yang Y, Adebali O, Wu G, Selby CP, Chiou YY, Rashid N, Hu J, et al. Cisplatin-DNA adduct repair of transcribed genes is controlled by two circadian programs in mouse tissues. Proc Natl Acad Sci USA. . 2018;115 doi: 10.1073/pnas.1804493115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yang Y, Liu Z, Selby CP, Sancar A. Long-term, genome-wide kinetic analysis of the effect of the circadian clock and transcription on the repair of cisplatin-DNA adducts in the mouse liver. J Biol Chem. . 2019;294:11960–11968. doi: 10.1074/jbc.RA119.009579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yimit A, Adebali O, Sancar A, Jiang Y. Differential damage and repair of DNA-adducts induced by anti-cancer drug cisplatin across mouse organs. Nat Commun. . 2019;10:309. doi: 10.1038/s41467-019-08290-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Deger N, Yang Y, Lindsey-Boltz LA, Sancar A, Selby CP. Drosophila, which lacks canonical transcription-coupled repair proteins, performs transcription-coupled repair . J Biol Chem. . 2019;294:18092–18098. doi: 10.1074/jbc.AC119.011448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kaya S, Adebali O, Oztas O, Sancar A. Genome‐wide excision repair map of cyclobutane pyrimidine dimers in Arabidopsis and the roles of CSA1 and CSA2 proteins in transcription‐coupled repair . Photochem Photobiol. . 2021, doi: 10.1111/php.13519 doi: 10.1111/php.13519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Oztas O, Selby CP, Sancar A, Adebali O. Genome-wide excision repair in arabidopsis is coupled to transcription and reflects circadian gene expression patterns. Nat Commun. . 2018;9:1503. doi: 10.1038/s41467-018-03922-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Li W, Adebali O, Yang Y, Selby CP, Sancar A. Single-nucleotide resolution dynamic repair maps of UV damage in Saccharomyces cerevisiae genome . Proc Natl Acad Sci USA. . 2018;115:E3408. doi: 10.1073/pnas.1801687115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hu J, Lieb JD, Sancar A, Adar S. Cisplatin DNA damage and repair maps of the human genome at single-nucleotide resolution. Proc Natl Acad Sci USA. . 2016;113:11507–11512. doi: 10.1073/pnas.1614430113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Li W, Hu J, Adebali O, Adar S, Yang Y, Chiou YY, Sancar A. Human genome-wide repair map of DNA damage caused by the cigarette smoke carcinogen benzo[a]pyrene. Proc Natl Acad Sci USA. . 2017;114:6752–6757. doi: 10.1073/pnas.1706021114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ogi T, Limsirichaikul S, Overmeer RM, Volker M, Takenaka K, Cloney R, Nakazawa Y, et al. Three DNA polymerases, recruited by different mechanisms, carry out NER repair synthesis in human cells. Mol Cell. . 2010;37:714–727. doi: 10.1016/j.molcel.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 149.Edenberg H, Hanawalt P. Size of repair patches in the DNA of ultraviolet-irradiated HeLa cells. Biochim Biophys Acta (BBA) - Nucleic Acids Protein Synthesis. . 1972;272:361–372. doi: 10.1016/0005-2787(72)90389-9. [DOI] [PubMed] [Google Scholar]
- 150.Reardon JT, Thompson LH, Sancar A. Rodent UV-sensitive mutant cell lines in complementation groups 6–10 have normal general excision repair activity. Nucleic Acids Res. . 1997;25:1015–1021. doi: 10.1093/nar/25.5.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Sertic S, Pizzi S, Cloney R, Lehmann AR, Marini F, Plevani P, Muzi-Falconi M. Human exonuclease 1 connects nucleotide excision repair (NER) processing with checkpoint activation in response to UV irradiation. Proc Natl Acad Sci USA. . 2011;108:13647–13652. doi: 10.1073/pnas.1108547108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lindsey-Boltz LA, Kemp MG, Reardon JT, DeRocco V, Iyer RR, Modrich P, Sancar A. Coupling of human DNA excision repair and the DNA damage checkpoint in a defined in vitro system . J Biol Chem. . 2014;289:5074–5082. doi: 10.1074/jbc.M113.542787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. . 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 154.Kemp MG, Gaddameedhi S, Choi JH, Hu J, Sancar A. DNA repair synthesis and ligation affect the processing of excised oligonucleotides generated by human nucleotide excision repair. J Biol Chem. . 2014;289:26574–26583. doi: 10.1074/jbc.M114.597088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Mao P, Wyrick JJ, Roberts SA, Smerdon MJ. UV-induced DNA damage and mutagenesis in chromatin. Photochem Photobiol. . 2017;93:216–228. doi: 10.1111/php.12646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Mao P, Wyrick JJ. Genome-wide mapping of UV-induced DNA damage with CPD-seq. Methods Mol Biol. 2020, 2175: 79–94 . [DOI] [PubMed]
- 157.Premi S, Han L, Mehta S, Knight J, Zhao D, Palmatier MA, Kornacker K, et al. Genomic sites hypersensitive to ultraviolet radiation. Proc Natl Acad Sci USA. . 2019;116:24196–24205. doi: 10.1073/pnas.1907860116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Jin SG, Pettinga D, Johnson J, Li P, Pfeifer GP. The major mechanism of melanoma mutations is based on deamination of cytosine in pyrimidine dimers as determined by circle damage sequencing. Sci Adv. . 2021;7:eabi6508. doi: 10.1126/sciadv.abi6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Hu J, Adebali O, Adar S, Sancar A. Dynamic maps of UV damage formation and repair for the human genome. Proc Natl Acad Sci USA. . 2017;114:6758–6763. doi: 10.1073/pnas.1706522114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Shu X, Xiong X, Song J, He C, Yi C. Base-resolution analysis of cisplatin-DNA adducts at the genome scale. Angew Chem Int Ed. . 2016;55:14246–14249. doi: 10.1002/anie.201607380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hu J, Li W, Adebali O, Yang Y, Oztas O, Selby CP, Sancar A. Genome-wide mapping of nucleotide excision repair with XR-seq. Nat Protoc. . 2018;14:248–282. doi: 10.1038/s41596-018-0093-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Hu J, Adar S, Selby CP, Lieb JD, Sancar A. Genome-wide analysis of human global and transcription-coupled excision repair of UV damage at single-nucleotide resolution. Genes Dev. . 2015;29:948–960. doi: 10.1101/gad.261271.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Jiang Y, Li W, Lindsey-Boltz LA, Yang Y, Li Y, Sancar A. Super hotspots and super coldspots in the repair of UV-induced DNA damage in the human genome. J Biol Chem. . 2021;296:100581. doi: 10.1016/j.jbc.2021.100581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Yang Y, Hu J, Selby CP, Li W, Yimit A, Jiang Y, Sancar A. Single-nucleotide resolution analysis of nucleotide excision repair of ribosomal DNA in humans and mice. J Biol Chem. . 2019;294:210–217. doi: 10.1074/jbc.RA118.006121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Adar S, Hu J, Lieb JD, Sancar A. Genome-wide kinetics of DNA excision repair in relation to chromatin state and mutagenesis. Proc Natl Acad Sci USA. . 2016;113:E2124–E2133. doi: 10.1073/pnas.1603388113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Li W, Sancar A. Methodologies for detecting environmentally induced DNA damage and repair. Environ Mol Mutagen. 2020, 61: 664–679 . [DOI] [PMC free article] [PubMed]
- 167.Sabarinathan R, Mularoni L, Deu-Pons J, Gonzalez-Perez A, López-Bigas N. Nucleotide excision repair is impaired by binding of transcription factors to DNA. Nature. . 2016;532:264–267. doi: 10.1038/nature17661. [DOI] [PubMed] [Google Scholar]
- 168.Plaschka C, Hantsche M, Dienemann C, Burzinski C, Plitzko J, Cramer P. Transcription initiation complex structures elucidate DNA opening. Nature. . 2016;533:353–358. doi: 10.1038/nature17990. [DOI] [PubMed] [Google Scholar]
- 169.Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet. . 2011;12:7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 170.Compe E, Egly JM. TFIIH: when transcription met DNA repair. Nat Rev Mol Cell Biol. . 2012;13:343–354. doi: 10.1038/nrm3350. [DOI] [PubMed] [Google Scholar]
- 171.Zou Y, Liu Y, Wu X, Shell SM. Functions of human replication protein A (RPA): from DNA replication to DNA damage and stress responses. J Cell Physiol. . 2006;208:267–273. doi: 10.1002/jcp.20622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Nishino T, Ishino Y, Morikawa K. Structure-specific DNA nucleases: structural basis for 3D-scissors. Curr Opin Struct Biol. . 2006;16:60–67. doi: 10.1016/j.sbi.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 173.Bhagwat N, Olsen AL, Wang AT, Hanada K, Stuckert P, Kanaar R, D’Andrea A, et al. XPF-ERCC1 participates in the fanconi anemia pathway of cross-link repair. Mol Cell Biol. . 2009;29:6427–6437. doi: 10.1128/MCB.00086-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Whitaker AM, Schaich MA, Smith MR, Flynn TS, Freudenthal BD. Base excision repair of oxidative DNA damage from mechanism to disease. Front Biosci. . 2017;22:1493–1522. doi: 10.2741/4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Shafirovich V, Geacintov NE. Removal of oxidatively generated DNA damage by overlapping repair pathways. Free Radical Biol Med. . 2017;107:53–61. doi: 10.1016/j.freeradbiomed.2016.10.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.McKibbin PL, Fleming AM, Towheed MA, Van Houten B, Burrows CJ, David SS. Repair of hydantoin lesions and their amine adducts in DNA by base and nucleotide excision repair. J Am Chem Soc. . 2013;135:13851–13861. doi: 10.1021/ja4059469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Reardon JT, Bessho T, Kung HC, Bolton PH, Sancar A. In vitro repair of oxidative DNA damage by human nucleotide excision repair system: possible explanation for neurodegeneration in Xeroderma pigmentosum patients . Proc Natl Acad Sci USA. . 1997;94:9463–9468. doi: 10.1073/pnas.94.17.9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Gorini F, Scala G, Cooke MS, Majello B, Amente S. Towards a comprehensive view of 8-oxo-7,8-dihydro-2’-deoxyguanosine: highlighting the intertwined roles of DNA damage and epigenetics in genomic instability. DNA Repair. . 2021;97:103027. doi: 10.1016/j.dnarep.2020.103027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Melis JPM, van Steeg H, Luijten M. Oxidative DNA damage and nucleotide excision repair. Antioxid Redox Signal. . 2013;18:2409–2419. doi: 10.1089/ars.2012.5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Kumar N, Moreno NC, Feltes BC, Menck CF, Houten BV. Cooperation and interplay between base and nucleotide excision repair pathways: from DNA lesions to proteins. Genet Mol Biol. . 2020;43:e20190104. doi: 10.1590/1678-4685-gmb-2019-0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Parlanti E, D’Errico M, Degan P, Calcagnile A, Zijno A, van der Pluijm I, van der Horst GT, et al. The cross talk between pathways in the repair of 8-oxo-7,8-dihydroguanine in mouse and human cells. Free Radical Biol Med. . 2012;53:2171–2177. doi: 10.1016/j.freeradbiomed.2012.08.593. [DOI] [PubMed] [Google Scholar]
- 182.D’Errico M, Parlanti E, Teson M, de Jesus BMB, Degan P, Calcagnile A, Jaruga P, et al. New functions of XPC in the protection of human skin cells from oxidative damage. EMBO J. . 2006;25:4305–4315. doi: 10.1038/sj.emboj.7601277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Klungland A, Höss M, Gunz D, Constantinou A, Clarkson SG, Doetsch PW, Bolton PH, et al. Base excision repair of oxidative DNA damage activated by XPG protein. Mol Cell. . 1999;3:33–42. doi: 10.1016/S1097-2765(00)80172-0. [DOI] [PubMed] [Google Scholar]
- 184.Wang HT, Choi B, Tang M. Melanocytes are deficient in repair of oxidative DNA damage and UV-induced photoproducts. Proc Natl Acad Sci USA. . 2010;107:12180–12185. doi: 10.1073/pnas.1005244107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Guo J, Hanawalt PC, Spivak G. Comet-FISH with strand-specific probes reveals transcription-coupled repair of 8-oxoGuanine in human cells. Nucleic Acids Res. . 2013;41:7700–7712. doi: 10.1093/nar/gkt524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Menoni H, Wienholz F, Theil AF, Janssens RC, Lans H, Campalans A, Radicella JP, et al. The transcription-coupled DNA repair-initiating protein CSB promotes XRCC1 recruitment to oxidative DNA damage. Nucleic Acids Res. . 2018;46:7747–7756. doi: 10.1093/nar/gky579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Wong HK, Muftuoglu M, Beck G, Imam SZ, Bohr VA, Wilson Iii DM. Cockayne syndrome B protein stimulates apurinic endonuclease 1 activity and protects against agents that introduce base excision repair intermediates. Nucleic Acids Res. . 2007;35:4103–4113. doi: 10.1093/nar/gkm404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Konovalov KA, Pardo-Avila F, Tse CKM, Oh J, Wang D, Huang X. 8-Oxo-guanine DNA damage induces transcription errors by escaping two distinct fidelity control checkpoints of RNA polymerase II. J Biol Chem. . 2019;294:4924–4933. doi: 10.1074/jbc.RA118.007333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Jang S, Kumar N, Beckwitt EC, Kong M, Fouquerel E, Rapić-Otrin V, Prasad R, et al. Damage sensor role of UV-DDB during base excision repair. Nat Struct Mol Biol. . 2019;26:695–703. doi: 10.1038/s41594-019-0261-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Thomas M, White RL, Davis RW. Hybridization of RNA to double-stranded DNA: formation of R-loops. Proc Natl Acad Sci USA. . 1976;73:2294–2298. doi: 10.1073/pnas.73.7.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Niehrs C, Luke B. Regulatory R-loops as facilitators of gene expression and genome stability. Nat Rev Mol Cell Biol. . 2020;21:167–178. doi: 10.1038/s41580-019-0206-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Pavri R. R Loops in the regulation of antibody gene diversification. Genes. . 2017;8:154. doi: 10.3390/genes8060154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Roy D, Yu K, Lieber MR. Mechanism of R-loop formation at immunoglobulin class switch sequences. Mol Cell Biol. . 2008;28:50–60. doi: 10.1128/MCB.01251-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Sollier J, Stork CT, García-Rubio ML, Paulsen RD, Aguilera A, Cimprich KA. Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol Cell. . 2014;56:777–785. doi: 10.1016/j.molcel.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Yasuhara T, Kato R, Hagiwara Y, Shiotani B, Yamauchi M, Nakada S, Shibata A, et al. Human rad52 promotes XPG-mediated R-loop processing to initiate transcription-associated homologous recombination repair. Cell. . 2018;175:558–570.e11. doi: 10.1016/j.cell.2018.08.056. [DOI] [PubMed] [Google Scholar]
- 196.Goulielmaki E, Tsekrekou M, Batsiotos N, Ascensão-Ferreira M, Ledaki E, Stratigi K, Chatzinikolaou G, et al. The splicing factor XAB2 interacts with ERCC1-XPF and XPG for R-loop processing. Nat Commun. . 2021;12:3153. doi: 10.1038/s41467-021-23505-1. [DOI] [PMC free article] [PubMed] [Google Scholar]