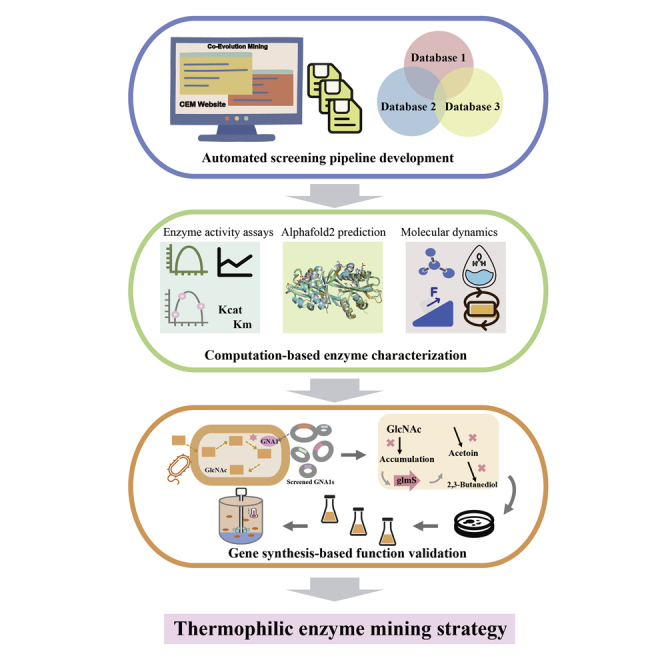
Summary
The host-guest incompatibility between a production host and non-native enzymes has posed an arduous challenge for synthetic biology, particularly between mesophile-derived enzymes and a thermophilic chassis. In the present study, we develop a thermophilic enzyme mining strategy comprising an automated co-evolution-based screening pipeline (http://cem.sjtu.edu.cn), computation-based enzyme characterization, and gene synthesis-based function validation. Using glucosamine-6-phosphate acetyltransferase (GNA1) as an example, we successfully mined four novel GNA1s with excellent thermostabilities and catalytic performances. Calculation and analysis based on AlphaFold2-generated structures were also conducted to uncover the mechanism underlying their excellent properties. Finally, our mined GNA1s were used to enable the high-temperature N-acetylglucosamine (GlcNAc) production with high titers of up to 119.3 g/L, with the aid of systems metabolic engineering and temperature programming. This study demonstrates the effectiveness of the enzyme mining strategy, highlighting the application prospects of mining new enzymes from massive databases and providing an effective solution for tackling host-guest incompatibility.
Subject areas: Enzyme engineering, Synthetic biology
Graphical abstract
Enzyme engineering; Synthetic biology
Introduction
Synthetic biology adopts the bottom-up concept of engineering, integrates different components in biological hosts to create new cell factories, and builds various artificial life systems for specific purposes.1,2 The incompatibility between a host and non-native enzymes is unavoidable in practice. For example, introducing anaerobe-derived enzymes into cyanobacteria with a high-oxygen intracellular environment led to the failure of functional expression.3 Moreover, when non-native enzymes are heterologously expressed into acidophiles, it will cause enzyme instability and poor catalytic efficiency under low pH conditions.4 These common and inevitable situations severely limit the potential and applications of synthetic biology, especially in the development of productive microbial cell factories.
High-temperature fermentation by thermophilic cell factories is considered a promising next-generation biotechnology due to its multiple superiorities in minimizing the cost such as reducing the risk of bacterial contamination, accelerating the production processes, and saving energy consumption in cooling.5,6,7 Savings associated with a 5°C increase in the fermentation temperature are estimated to be more than US$390,000 per year for a 30,000-kL scale ethanol plant.8 Chemical production at elevated temperatures has great potential to significantly outperform traditional processes and helps the growth of the Bioeconomy.9,10 At present, high-temperature fermentation of useful products has made great progress, such as isobutanol,11 bioethanol,12,13 acetoin,14 lactate,15 and 2,3-butanediol.16
However, the host-guest incompatibility is particularly problematical in high-temperature fermentation. High-temperature production of chemicals by thermophiles is greatly limited by using mesophile-derived (unstable) enzymes, which is a primary challenge for promoting the development of high-temperature cell factories.11 Most reported enzymes related to biomanufacture are derived from mesophilic microorganisms, which may be inactivated under high-temperature conditions.17 For example, spCas9, derived from the mesophilic bacterium Streptococcus pyogenes, was reported to lose its catalytic activity at 42°C, which limits the utilization of spCas9 in thermophilic Bacillaceae strains.18,19 Moreover, the production of d-lactic acid has long been limited due to the low thermostabilities of d-LDHs until a thermophilic d-LDH was mined to achieve the highest d-lactic acid titer (226.6 g/L) at 50°C.15,20,21 Thus, to achieve the high-temperature production of biochemicals, there is a great demand for stable and thermophilic enzymes that are compatible with thermophilic hosts.
Advances in genome sequencing technology have led to an explosive growth of genome sequence data in biological databases.22,23 The NCBI statistics show that there are more than 250 million genomic DNA sequences in the GenBank database as of January 2022 (https://www.ncbi.nlm.nih.gov/search/all/?term=Genbank). There is a huge amount of unexplored thermophilic enzyme resources in such an extensive genome database. The mining of thermophilic enzymes presented in the databases will fulfill the great demand for thermophilic enzymes, and give rise to the realization of high-temperature chemical production with thermophilic chassis.
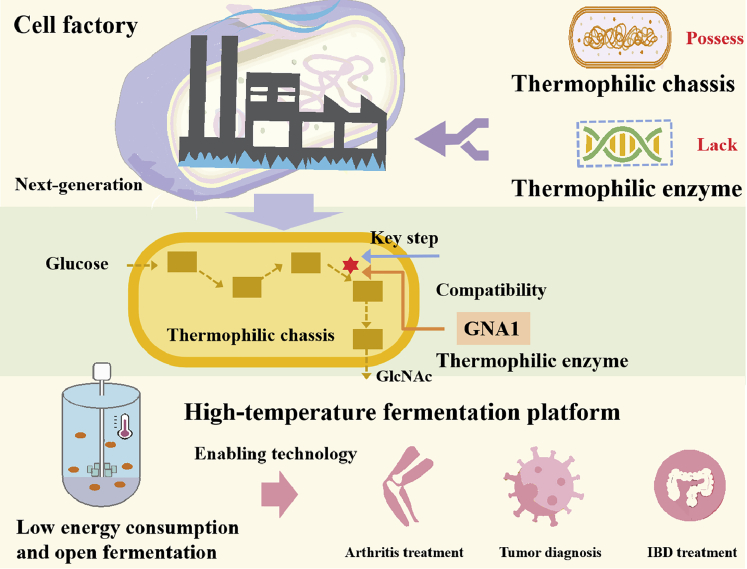
N-Acetylglucosamine (GlcNAc) has been widely used as a medicine and health product to treat the prevalent disease arthritis and improve joint mobility function.24,25,26 As the global aging population grows, the demand for GlcNAc as a joint health dietary supplement will continue to increase.27 In addition, GlcNAc is a potential candidate to treat inflammatory bowel disease (IBD),24,28,29 and plays a certain help on tumor diagnosis.30,31 A recent study provided evidence to support GlcNAc as an anti-inflammatory and adjunctive treatment for COVID-19.32 Currently, microbial fermentation is considered as the most promising process for producing non-shellfish-derived GlcNAc with biosafety in mind,24 compared with chemical acidolysis and the chitinase degradation method.27,33,34,35,36 In the GlcNAc synthesis pathway, the glucosamine-6-phosphate acetyltransferase (GNA1) is the key enzyme for the overproduction of GlcNAc requiring heterologous expression.37 Currently, the reported GNA1s are mostly derived from mesophiles, such as yeasts, which limits the host options for microbial fermentation.38,39,40,41 To date, the strains used for GlcNAc production include Escherichia coli,42 Saccharomyces cerevisiae,43 Bacillus subtilis,44 and Corynebacterium glutamicum.45 Despite great achievements in using these hosts, the mesophilic fermentation conditions (≤37°C) remain the problems of higher energy consumption and cost. Thus, it is highly demanded to search for thermally robust GNA1s that can break the constraints of mesophiles to realize high temperature fermentation of GlcNAc, which in turn effectively lower costs and benefits most people (Figure 1).
Figure 1.
Scheme for GlcNAc high-temperature fermentation with engineered thermophilic chassis and elements
In the present study, we developed a thermophilic enzyme mining strategy to eliminate guest-host incompatibility in the thermophilic chassis. First, we established a pipeline (http://cem.sjtu.edu.cn.) to achieve in silico thermophilic enzyme mining from massive amounts of publicly available genomes. Using GNA1 from yeast as the template, 20 thermophilic candidates GNA1s were mined with the aid of computational biology analysis for preliminary GlcNAc production tests in Bacillus licheniformis that is a potential thermophilic chassis because of its superiorities in growth rate and biosafety.15 We chose four mined GNA1s with higher GlcNAc titers in shake-flask experiments, which were further compared in terms of their thermostabilities and catalytic performances. Besides, calculation and analysis based on AlphaFold2-generated structures were also carried out. The systems metabolic engineering and temperature programming were conducted, which led to a significant increase in the GlcNAc titer. Finally, 50-L fed-batch fermentation of GlcNAc was conducted at elevated temperatures to demonstrate the industrial potential of the developed strains.
Results
Pipeline establishment for mining thermophilic GNA1s
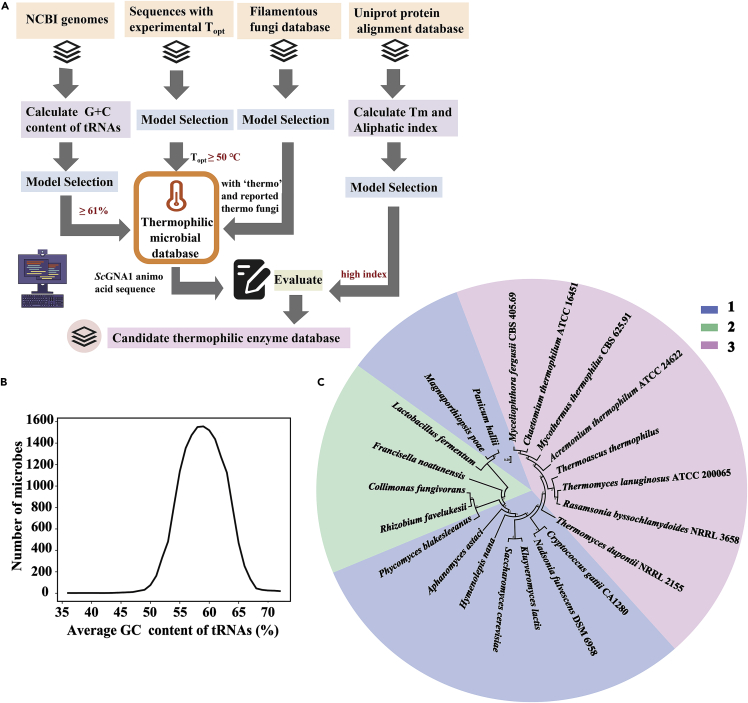
A pipeline for mining thermophilic enzymes was developed based on the positive correlation between the optimal growth temperature (OGT) of microorganisms and the thermostabilities of enzymes encoded in their genomes (Figure 2A).46,47 The co-evolution mining website (http://cem.sjtu.edu.cn) was established according to the pipeline, based on the concept of co-evolution between thermophilic enzymes and their hosts. Since the housekeeping enzymes and the other enzymes in the genomes of thermophilic microorganisms are co-evolving, it is easier to mine thermophilic enzymes in the genomes of thermophilic hosts. We used the amino acid sequence of GNA1 from S. cerevisiae as a template for candidate thermophilic GNA1 enzyme mining. Based on a filamentous fungus database (https://pub.fungalgenomics.ca/), we screened microorganisms with the word “thermo” and the thermophilic fungi reported previously to form a thermophilic fungus database (Figure 2A, Table S1). The scores based on the NCBI PSI blast of the screened candidate thermophilic GNA1s ranged from 83 to 308, and eight candidate GNA1s with higher scores were further screened. We also filtered 224,799 microbial genomes according to the collated OGT data established by Engqvist,48 and selected 1,672 microbial genomes with OGT greater than 50°C to establish a database. In addition, since microbial OGT is positively correlated with the GC content of tRNAs.49,50 We found 15,271 microbial genome data in the NCBI microbial genome database that could conduct tRNA annotations and GC content calculations, and could be further analyzed by the open reading frame (ORF) prediction. Figure 2B shows the distribution of the average GC content of tRNAs in all microbes. The average GC content of the tRNAs of most microorganisms was 58%. We selected the microbial genomes, in which the GC content of tRNAs was higher than 61%, and included these in our database for thermophilic enzyme mining. Finally, we screened for GNA1s both in the two databases with Topt ≥ 50°C and tRNA GC content higher than 61% as thermophilic candidate enzymes. Moreover, we used the enzyme GNA1 from S. cerevisiae (ScGNA1) as the template for alignment and calculated the Tm index and aliphatic indices of the mined GNA1 enzymes using the Uniprot database. Eight candidates thermophilic GNA1s derived based on Tm index >1, and aliphatic indices of enzymes greater than that of ScGNA1 were selected (Figure 2A, Table S2). Finally, 20 selected candidate GNA1s from different databases were used to perform multiple alignments with ScGNA1 as a template for constructing the phylogenetic tree (Figure 2C).
Figure 2.
Model development for mining thermophilic GNA1s with two strategies
(A) Schematic overview of the process to build candidate thermophilic enzyme database. Topt, the optimal growth temperatures of organisms.
(B) Variation curve of microbial numbers with NCBI genomic average GC content of tRNAs (%).
(C) Phylogenetic tree analysis of the 20 selected candidate GNA1 thermophilic enzymes. 1. Based on Uniprot alignment according to ScGNA1 amino acid sequence. 2. Based on the GC content of tRNAs in microbial genomes and reported optimal growth temperatures of organisms. 3. Based on the thermophilic fungus database.
Performances of thermophilic enzymes GNA1s
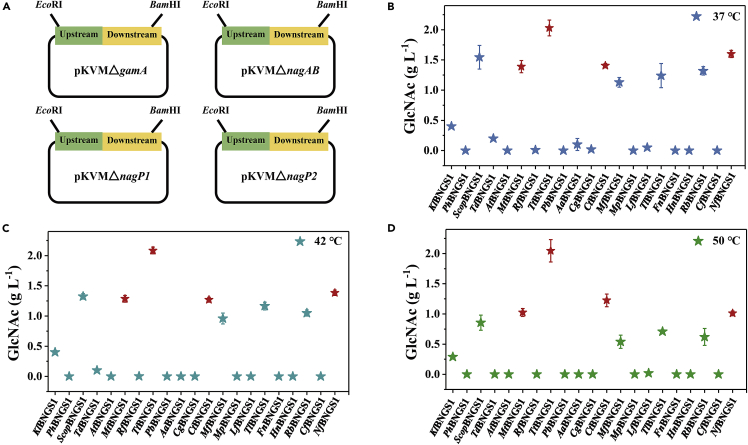
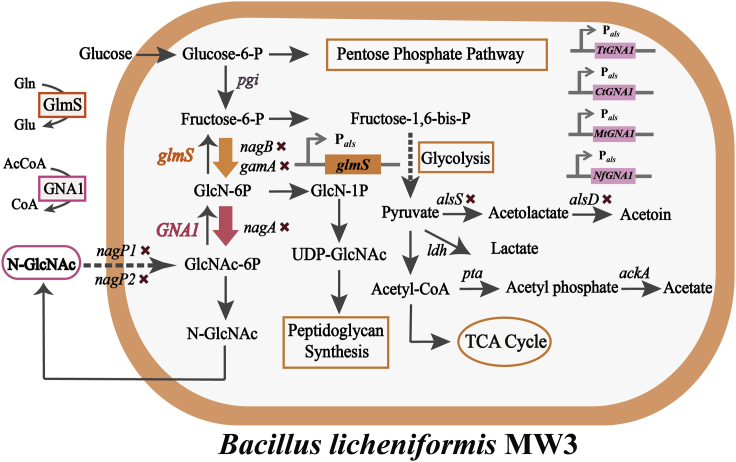
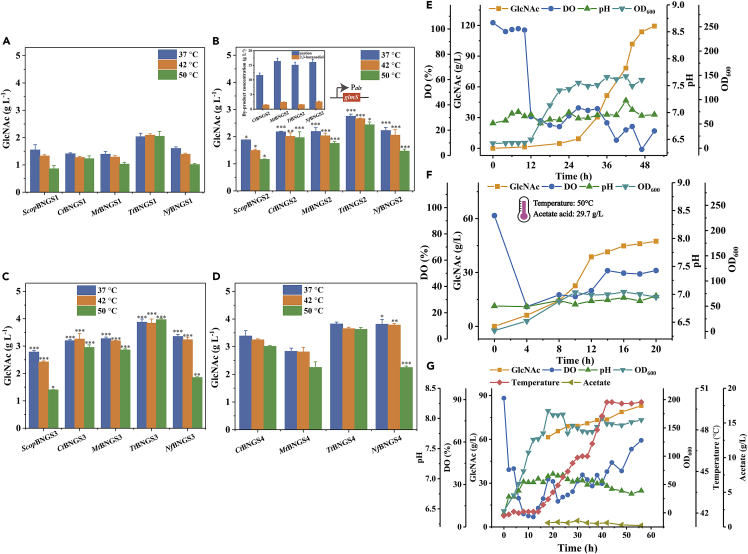
To verify the functions of the above 20 candidate thermophilic GNA1 enzymes, the genes in the catabolic pathway of GlcNAc such as nagP1 and nagP2 (encoding the GlcNAc-specific enzyme of phosphotransferase system), nagA (encoding GlcNAc-6-P deacetylase), and gamA and nagB (encoding GlcN-6-P deaminase) in B. licheniformis MW3 were blocked to obtain the five-gene knockout test strain BNGS1 (Figure 3A and S1). We introduced the Scopgna1 gene (the codon-optimized version of S. cerevisiae GNA1 using Jcat http://www.jcat.de/) into the BNGS1 strain with the B. licheniformis expression vector pHY300PLK to obtain the strain ScopBNGS1 for performance testing. The results showed that GlcNAc was successfully produced (Figure S2). Besides, compared with the control ScBNGS1 (the gna1 gene without codon optimization from S. cerevisiae, introduced into the BNGS1 strain named ScBNGS1), the GlcNAc production of ScopBNGS1 increased from 0.24 g/L to 0.9 g/L at 50°C, which showed that codon optimization was effective and necessary (Figure S2). Thus, the nucleotide sequences of the 20 thermophilic GNA1 enzymes were codon-optimized using Jcat (http://www.jcat.de/). The 20 codon-optimized candidates GNA1s, which were mined according to our established pipeline, were expressed respectively into the strain BNGS1 containing five constitutive promoters for GlcNAc flask tests. Optimal promoter Pals with the highest GlcNAc titers was selected to obtain 20 candidate strains for GlcNAc production (Figure S2). The twenty candidate GlcNAc production strains were cultured in flasks at 37°C (the optimum growth temperature for common microorganisms), 42°C (the critical temperature for mesophiles and thermophiles for microbes), and 50°C (the optimum growth temperature for B. licheniformis MW3) (Figures 3B−3D). The results showed that among the 20 strains, eight strains produced GlcNAc at the temperatures of 37°C, 42°C, and 50°C, respectively, indicating that the eight candidates of thermophilic GNA1 enzymes were successfully expressed heterologously in B. licheniformis. Four candidate strains TtBNGS1, NfBNGS1, MtBNGS1, and CtBNGS1 were selected because they exhibited high titers of GlcNAc at 42 and 50°C (Figures 3B−3D). The four thermophilic GNA1s in the four selected strains, named TtGNA1, NfGNA1, MtGNA1, and CtGNA1, were further analyzed for subsequent enzymatic properties.
Figure 3.
Shake flask fermentation of 20 candidate thermophilic strains for thermophilic enzyme GNA1 performance tests
(A−D) The construction of five gene-knockout plasmids (A). Shake flask fermentation of 20 candidate thermophilic strains at 37°C (B), 42°C (C), and 50°C (D). Triplicate experiments were done for physiological measurements, and error bars represent SD.
Enzymatic properties and AlphaFold2 protein structure prediction
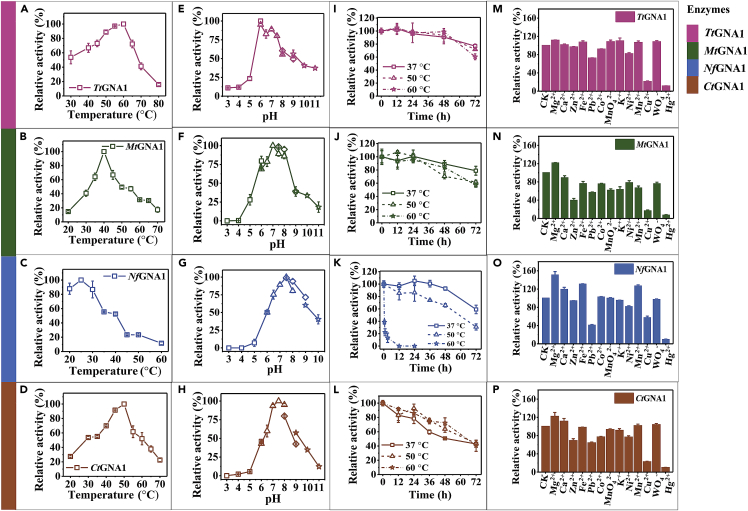
By amino acid sequence alignment, four proteins NfGNA1, TtGNA1, MtGNA1, and CtGNA1 share 47.1%, 45.2%, 44.2% and 43.0% amino acid identity with ScGNA1, respectively (Figure S3). We inserted NfGNA1, TtGNA1, MtGNA1, and CtGNA1 into the pETDuet-1 vector, and transformed them into E. coli BL21 (DE3), respectively, for further protein expression and purification. Each of the four proteins NfGNA1, TtGNA1, MtGNA1, and CtGNA1 showed a single lane after purification that appears at approximately 20 kDa (Figure S4). The four purified proteins were further characterized in terms of its enzymatic properties (Figures S4 and S5). The optimum temperatures for CtGNA1, TtGNA1, MtGNA1, and NfGNA1 were 50°C, 60°C, 40°C, and 25°C, respectively (Figures 4A−4D). Among them, TtGNA1 shows good catalytic activity in a broad temperature range, retaining the relative activities of over 60% maximum activity at 37−60°C (Figure 4A). The optimum pH for CtGNA1, TtGNA1, MtGNA1, and NfGNA1 was 7.5, 6, 7, and 7.5, respectively (Figures 4E−4H), which are required for the growth of B. licheniformis. Moreover, the four GNA1 enzymes were thermally stable, showing more than 60% of the initial activity residue after 48 h of incubation at 37 and 50°C (Figures 4I−4L). Among the four enzymes, TtGNA1 has the highest thermal stability, showing more than 90% of the initial activity residue after 48 h of incubation at 37°C, 50°C, and 60°C, respectively (Figure 4I). Because of the low optimum temperature of NfGNA1, its activity at 60°C rapidly dropped to zero within 12 h, however, its thermal stabilities at 37 and 50°C were excellent, which probably had little effect on the fermentation of B. licheniformis MW3 at 50°C. Compared with the four screened GNA1s, ScGNA1 exhibited much lower thermostability, with only 20% of the initial activity residue after 48 h of incubation at 50°C (Figure S6). In addition, differential scanning calorimeter (DSC) characteristics of the five enzymes showed that the Tm value of ScGNA1 (47.4) was much lower than that of TtGNA1 (60.1), CtGNA1 (59.3), MtGNA1 (54.8), NfGNA1 (58.5), which exhibited excellent protein stabilities of our screened four GNA1s (Figure S7).
Figure 4.
Activity assays of TtGNA1, MtGNA1, NfGNA1, and CtGNA1
(A−D) Effects of temperature on the activities of TtGNA1, MtGNA1, NfGNA1, and CtGNA1. Error bars indicate standard deviations.
(E−H) Effects of pH on activities of TtGNA1, MtGNA1, NfGNA1, and CtGNA1. Square, citric acid-sodium citrate; Triangle, NaH2PO4-Na2HPO4; Rhombus, Tris-HCl; Star, NaHCO3-Na2CO3. Error bars indicate standard deviations.
(I−L) Thermal stabilities of TtGNA1, MtGNA1, NfGNA1, and CtGNA1 at 37°C, 50°C, and 60°C. Thermal inactivation of each enzyme was evaluated after incubation at 37°C, 50°C, or 60°C for 72 h with timing sampling. The residual activity of each enzyme was determined as a percentage of the initial activity. Error bars indicate standard deviations.
(M−P) Effects of metal ions on the activities of TtGNA1, MtGNA1, NfGNA1, and CtGNA1. Error bars indicate standard deviations. Triplicate experiments were carried out for physiological measurements, and error bars represent SD.
In addition, we measured the specific kinetic parameters of the four GNA1 enzymes. Compared with the catalytic efficiencies (kcat/Km) of ScGNA1, those of TtGNA1, NfGNA1, CtGNA1, and MtGNA1 for GlcN-6P were 1,867.56, 322.52, 1,817.43, and 1,764.88 s−1 mM−1, respectively, which were 11.12, 1.92, 10.82, and 10.51 times that of ScGNA1, respectively (Table S3). The kcat/Km values of TtGNA1, NfGNA1, CtGNA1, and MtGNA1 for Ac-CoA were 1,671.41, 238.99, 1,580.29, and 763.78 s−1 mM−1, respectively, which were 10.52, 1.54, 9.95, and 4.81 times that of ScGNA1, respectively (Table S3). These results indicate that TtGNA1 exhibited higher catalytic efficiencies for GlcN-6P and Ac-CoA than the other three GNA1 enzymes. Figures 4M−4P show the enzyme activity effects of different metal ions on TtGNA1, NfGNA1, CtGNA1, and MtGNA1. Only Mg2+ improved the activity of the four enzymes, Cu2+ and Hg2+ had a significant inhibitory effect on the four GNA1 enzymes, and the remaining metal ions had no obvious effect on the activity of the four GNA1s.
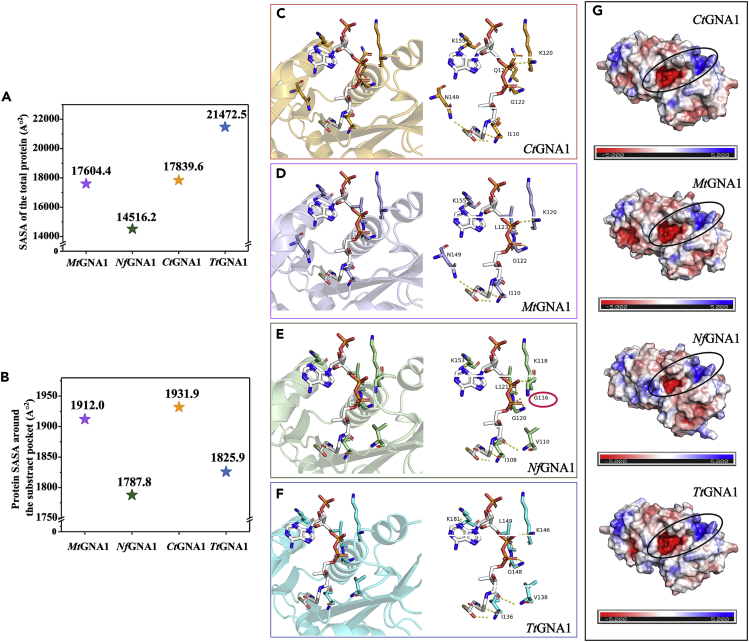
AlphaFold2 was used to predict the protein structures of the four GNA1 enzymes,51 and FoldX was used to calculate the stabilities of the four proteins.52,53 The crystal structure of ScGNA1 was downloaded from PDB, with PDB number 1i21, and we used the RepairPDB module in FoldX to minimize energy before final stabilities of the five proteins (ΔG) were calculated.54,55 The structures of the four predicted proteins (TtGNA1, NfGNA1, CtGNA1, and MtGNA1) were relatively similar, and the substrate pockets tended to be conserved. The free energy of unfolding (ΔG) calculated by FoldX for TtGNA1, NfGNA1, CtGNA1, MtGNA1, and ScGNA1 was −62.69, −76.32, −42.29, −45.74, and 10.23 kcal/mol, respectively, indicating that our screened enzymes have higher stabilities than ScGNA1 (Figure S8). Figures 5A−5B show the solvent-accessible surface area (SASA) of each GNA1 enzyme around the substrate pocket and for the entire protein. The SASA values for the entire proteins of TtGNA1, CtGNA1, MtGNA1, and NfGNA1 were 21,472.5, 17,839.6, 17,604.4, and 14,516.2 Å2, respectively. The SASA values around the substrate pocket (acetyl-CoA) for TtGNA1, CtGNA1, MtGNA1, and NfGNA1 were 1,825.9, 1,931.9, 1,912.0, and 1,787.8 Å2, respectively. These results suggested that TtGNA1, CtGNA1, and MtGNA1 were more hydrophilic than that NfGNA1, which is consistent with their Km values. In addition, the number of hydrogen bonds around the substrate acetyl-CoA differed slightly among the four GNA1 enzymes (Figures 5C−5F). Compared with TtGNA1, CtGNA1, and MtGNA1, in NfGNA1, additional hydrogen bonds were formed between the backbone NH of Gly116 with O4A of acetyl-CoA and CoA-GlcNAc-6P, indicating the stronger stability of the substrate Ac-CoA and the product CoA-GlcNAc-6P in NfGNA1, which facilitates the establishment of a catalytic conformation (Figures S9−S12). We also displayed the surface electrostatic potential of the four proteins using Advanced Poisson-Boltzmann Solver (APBS). The results showed considerable differences in the distribution of electrostatic potential around the substrate pocket of NfGNA1 compared with the other three GNA1 enzymes (Figure 5G), which might potentially affect the binding ability of the substrate.
Figure 5.
Protein structure prediction of TtGNA1, MtGNA1, NfGNA1, and CtGNA1
(A) Calculated solvent-accessible surface area (SASA) for the entire protein of TtGNA1, MtGNA1, NfGNA1, and CtGNA1.
(B) Calculated solvent-accessible surface area (SASA) around the substrate pocket of TtGNA1, MtGNA1, NfGNA1, and CtGNA1.
(C−F) Calculated the number of hydrogen bonds bound to the substrate acetyl-CoA for TtGNA1, MtGNA1, NfGNA1, and CtGNA1.
(G) Predicted Advanced Poisson-Boltzmann Solver (APBS) results in the distribution of electrostatic potential around the substrate pocket for TtGNA1, MtGNA1, NfGNA1, and CtGNA1.
In addition, thermophilic enzymes tend to form more alpha-helices than mesophilic enzymes to make the protein structure more stable, and β-branched residues (Val, Ile, and Thr) can affect helix stability. TtGNA1, MtGNA1, and CtGNA1 showed less β-branched residue content in the helices and the whole sequence, whereas TtGNA1 had more alpha helices, which increased the stability of the protein (Table S4). Flexibility indices were calculated to analyze the thermostabilities of the four proteins. The value of TtGNA1 (0.9703) was lower than those of MtGNA1 (0.9866), NfGNA1 (0.9839), and CtGNA1(0.9862), indicating that TtGNA1 was relatively more thermostable (Table S5). This result is in concordance with the enzymatic properties of TtGNA1, which exhibited increased stability at 60°C.
Metabolic flux remodeling in the GlcNAc production
To further increase the GlcNAc production, we used systematic metabolic engineering to reprogram the metabolic flow (Figure 6). First, we inserted a stronger promoter Pals before glmS in the genome of B. licheniformis BNGS1 (Figure S13), and transformed pHY300PLK-TtGNA1, pHY300PLK-NfGNA1, pHY300PLK-CtGNA1, and pHY300PLK-MtGNA1 to obtain four strains TtBNGS2, NfBNGS2, CtBNGS2, and MtBNGS2, respectively. Flask fermentation was performed for 24 h at 37°C, 42°C, and 50°C (Figures 7A and 7B). The GlcNAc production increased by 35.5%, 39.1%, 54.8%, and 57.6% at 37°C in TtBNGS2, NfBNGS2, CtBNGS2, and MtBNGS2, respectively, compared with TtBNGS1, NfBNGS1, CtBNGS1, and MtBNGS1 (Figures 7A and 7B). At 42°C, the GlcNAc titers of TtBNGS2, NfBNGS2, CtBNGS2, and MtBNGS2 increased by 27.6%, 48.4%, 58.3%, and 58.0%, respectively (Figures 7A and 7B). At 50°C, the GlcNAc titers of TtBNGS2, NfBNGS2, CtBNGS2, and MtBNGS2 increased by 19.1%, 45.1%, 60.0%, and 70.7%, respectively (Figures 7A and 7B). Figure 7B also shows the detection of byproduct concentrations after 24 h of flask fermentation at 50°C. The acetoin concentrations of TtBNGS2, NfBNGS2, MtBNGS2, and CtBNGS2 reached 15.0, 16.0, 16.3, and 11.6 g/L, respectively, and that of the 2,3-butanediol concentrations reached 1.51, 2.49, 2.31, and 1.41 g/L, respectively, which was considered as the main byproducts.
Figure 6.
Metabolic engineering scheme for efficient production of GlcNAc
×, gene knockout in B. licheniformis MW3; Thick downward arrow, gene overexpression; AcCoA, acetyl-coenzyme A; GlcN-6P, glucosamine-6-phosphate; GlcNAc-6P, N-acetylglucosamine-6-phophate; GlmS, glucosamine synthase; Gln, glutamine; Glu, glutamate; GNA1, GlcN-6P N-acetyltransferase.
Figure 7.
Shake-flask fermentation and 50-L bioreactor fed-batch fermentation
(A) Shake-flask fermentation of ScopBNGS1, TtBNGS1, NfBNGS1, CtBNGS1, and MtBNGS1 at 37°C, 42°C, and 50°C, respectively. Triplicate experiments were carried out for physiological measurements, and error bars represent SD.
(B) Shake-flask fermentation of ScopBNGS2, TtBNGS2, NfBNGS2, CtBNGS2, and MtBNGS2 at 37°C, 42°C, and 50°C, respectively and by-product detection of TtBNGS2, NfBNGS2, CtBNGS2, and MtBNGS2 at 50°C. Triplicate experiments were carried out for physiological measurements, and error bars represent SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 as determined by t test compared with ScopBNGS1, TtBNGS1, NfBNGS1, CtBNGS1, and MtBNGS1 at 37°C, 42°C, and 50°C, respectively.
(C) Shake-flask fermentation of ScopBNGS3, TtBNGS3, NfBNGS3, CtBNGS3, and MtBNGS3 at 37°C, 42°C, and 50°C, respectively. Triplicate experiments were carried out for physiological measurements, and error bars represent SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 as determined by t test compared with ScopBNGS2, TtBNGS2, NfBNGS2, CtBNGS2, and MtBNGS2 at 37°C, 42°C, and 50°C, respectively.
(D) Shake-flask fermentation of TtBNGS4, NfBNGS4, CtBNGS4, and MtBNGS4 at 37°C, 42°C, and 50°C, respectively. Triplicate experiments were carried out for physiological measurements, and error bars represent SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 as determined by t test compared with TtBNGS3, NfBNGS3, CtBNGS3, and MtBNGS3 at 37°C, 42°C, and 50°C, respectively.
(E) Fed-batch fermentation of NfBNGS4 in a 50-L bioreactor at 42°C.
(F) Fed-batch fermentation of TtBNGS4 in a 50-L bioreactor at 50°C.
(G) Fed-batch fermentation of TtBNGS4 in a 50-L bioreactor with temperature programming. OD600, optical density at 600 nm. 0−15 h, 42°C, 15−33 h, the temperature rises 0.5°C every 2h from 42°C to 46°C. 33−41 h, the temperature rises 1°C every 2 h to 50°C.
To further improve the production of GlcNAc, acetoin and 2,3-butanediol biosynthetic genes encoding acetolactate synthase (AlsS) and acetolactate decarboxylase (AlsD) were knocked out on the basis of BNGS2 to block the synthesis pathways. The four plasmids pHY300PLK-TtGNA1, pHY300PLK-NfGNA1, pHY300PLK-CtGNA1, and pHY300PLK-MtGNA1 were transformed into BNGS2 to obtain TtBNGS3, NfBNGS3, CtBNGS3, and MtBNGS3, respectively. The four strains TtBNGS3, NfBNGS3, CtBNGS3, and MtBNGS3 were tested in shake flasks (Figures 7C, S14, and S15). After eliminating the acetoin and 2,3-butanediol pathways, the accumulation of the two by-products was not detected. The highest GlcNAc concentrations in TtBNGS3, NfBNGS3, CtBNGS3, and MtBNGS3 reached 3.85, 3.35, 3.20, and 3.47 g/L, which were 40.0%, 50.6%, 46.9%, and 58.5% higher than that of TtBNGS2, NfBNGS2, CtBNGS2, and MtBNGS2, respectively, at 37°C (Figure 7C). At 42°C, the GlcNAc titers of TtBNGS3, NfBNGS3, CtBNGS3, and MtBNGS3 reached 3.39, 3.23, 3.26, and 3.39 g/L, which was 44.2%, 57.2%, 61.9%, and 67.0% higher than that of TtBNGS2, NfBNGS2, CtBNGS2, and MtBNGS2, respectively (Figure 7C). At 50°C, the GlcNAc concentration of TtBNGS3, NfBNGS3, CtBNGS3, and MtBNGS3 reached 3.97, 1.85, 2.95, and 2.86 g/L, which was 62.7%, 25.9%, 50.5%, and 63.4% higher than that of TtBNGS2, NfBNGS2. CtBNGS3, and MtBNGS3, respectively (Figure 7C).
Although plasmids are easy to insert into cells and allow strong gene expression, they are prone to genetic instability, which is unfavorable for the industrial production of chemicals.56 Therefore, the four GNA1s, TtGNA1, NfGNA1, MtGNA1, and CtGNA1 with constitutive promoter Pals were integrated into the BNGS2 chromosome separately, to obtain four plasmid-free strains TtBNGS4, NfBNGS4, CtBNGS4, and MtBNGS4. As shown in Figure 7D, among the four plasmid-free strains, only NfBNGS4 improved the concentrations of GlcNAc from 3.23 g/L to 3.78 g/L at 42°C, and 1.85 g/L to 2.25 g/L at 50°C, which was 17.0 and 21.6% higher, respectively, than that of NfBNGS3. The GlcNAc concentration of the other three strains showed little change or a slight decrease with no statistically significant difference, which could be owing to the low copy number of GNA1 after integration into the chromosome.
Scale-up production of GlcNAc in a 50 L fermenter
To test the stability and robustness of the four GNA1s integration systems in a large-scale bioreactor, the four strains TtBNGS4, NfBNGS4, CtBNGS4, and MtBNGS4 were used to scale up the production of GlcNAc in a 50 L fermenter. NfBNGS4 showed the highest GlcNAc production of 119.3 g/L with 0.22 g/g glucose after purification and 2.39 g/(L⋅h) high productivity at 42°C in 50 h (Figures 7E, S16, and S17), which was even better than the 101.2 g/L of TtBNGS4 (Figure S18). At 50°C, TtBNGS4 showed the highest GlcNAc production of 47.4 g/L with 2.37 g/(L⋅h), compared with NfBNGS4, CtBNGS4, and MtBNGS4, which is in concordance with the excellent enzymatic characteristics of TtGNA1 (Figures 7F, S19, and S20). However, fermentation of the four strains showed an overflow of acetic acid at 50°C, leading to the inhibition of cell growth and limited GlcNAc production (Figure 7F). This may be caused by insufficient ATP and NAD+ under the conditions of high temperatures and more vigorous metabolism, leading to an imbalance between cellular substance metabolism and energy demand. In addition, NAD+ is unstable at high temperature conditions.57 Therefore, we speculated that an appropriate temperature reduction may achieve a perfect balance to mitigate the acetic acid flow. The temperature reduction gradient was set from 50°C to 42°C (Figures S21 and S22). When the temperature dropped to 47°C, the GlcNAc titers increased by 82.3% from 47.4 g/L to 86.4 g/L with acetate acid decreasing by 257.8% from 29.7 g/L to 8.3 g/L, showing little effect on GlcNAc fermentation (Figure S22), which was considered as the critical temperature to balance substance metabolism and energy demand for avoiding acetic acid overflow. To further increase fermentation temperature and reduce acetic acid overflow, the temperature programming was proposed by slowly increasing the fermentation temperature from 42°C to 50°C to gradually adapt the GlcNAc production host to rising temperatures (Figure 7G). As shown in Figure 7G, the fermentation temperature was mainly maintained from 42°C to 47°C during the fermentation cycle. Finally, the GlcNAc production reached 83.0 g/L with almost no acetate produced, indicating the effectiveness of our proposed temperature programming to reduce acetate overflow.
Multi-omics analysis of transcriptome and metabolome
To identify key genes and metabolic pathways to further increase GlcNAc production, we collected bacteria in the logarithmic stage from BNGS3, NfBNGS4, TtBNGS4, NfBNGS3, and TtBNGS3 at 50°C in a 5 L fermenter, and analyzed the transcriptome and metabolome data of the fermentation samples.
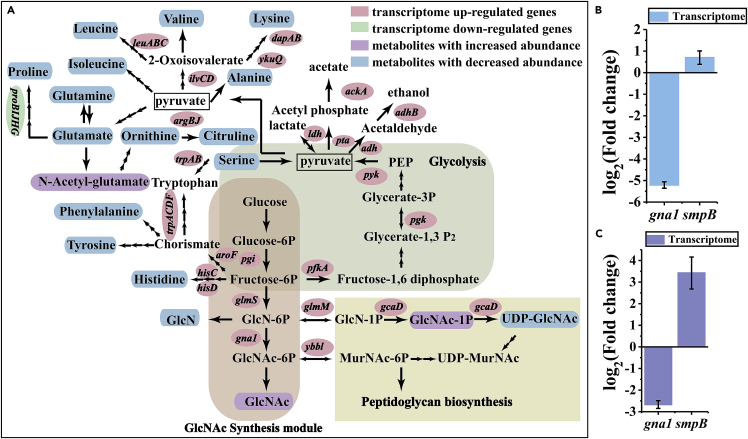
Comparing BNGS3 and NfBNGS4, the transcriptome results showed that the differentially expressed genes were classified into 83 categories according to the Kyoto Encyclopedia of Genes and Genomes (KEGG). Significant differences in KEGG pathways were found in “biosynthesis of amino acids,” “oxidative phosphorylation,” and “glycolysis and gluconeogenesis” (Figure S23). Especially in metabolic engineering of oxidative phosphorylation, the genes related to F-type ATPase were all up-regulated. In addition, in the respiratory streptoquinol oxidase pathway, the cytochrome aa3 menoquinol oxidase-related coding gene (qoxC) and cytochrome bd complex oxidase coding gene (cydA) were up-regulated (Figure S24). As the production of GlcNAc requires the consumption of ATP, the expression of GNA1 would promote the up-regulation of ATP synthesis-related genes and the respiratory chain cytochrome P450 oxidase coding genes to rapidly synthesize and provide ATP for GlcNAc production in cells. The related genes in glycolysis were also up-regulated, revealing that the glycolysis took on the task of supplying energy for cell growth and metabolism (Figure 8A). In addition, the insertion of NfGNA1 not only promoted the up-regulation of related genes in the metabolic pathway of GlcNAc production, such as pfkA, pgi, and glmS, but also the genes related to the competitive metabolic pathway, such as ybbl (from GlcNAc-6P to MurNAc), and glmM (from GlcN-6P to GlcN-1P) (Figure 8A), which affected the yield of GlcNAc production and needed to be suppressed. In addition, the decreased abundance of UDP-GlcNAc in NfBNGS4 also demonstrated that the GlcNAc synthesis module competes with the peptidoglycan synthesis pathway. In addition, the abundance of almost all amino acids was reduced, including Leu, Ile, Ala, His, Glu, Lys, and Orn. However, the 34 genes related to the synthesis of amino acids were up-regulated, suggesting that the insertion of NfGNA1 opened up the production route of GlcNAc, thus promoting the consumption of glutamic acid to produce glutamine to provide amino donors. Increased demand for glutamic acid production causes cells to be more inclined to synthesize glutamate, which significantly reduces the synthesis of other amino acids, resulting in the up-regulation of genes related to amino acid synthesis. In addition, owing to the rapid growth of cells, even if the cells were more inclined to synthesize glutamate, the demand for glutamate was high and the consumption was rapid, resulting in a general decrease in the abundance of glutamate in general. Furthermore, the insertion of NfGNA1 also promoted the acetylation of glutamate, and metabolomic data revealed an increase in the abundance of N-acetyl glutamic acid (Figure 8A).
Figure 8.
Multi-omics analysis of transcriptomes and metabolomes
(A) Multi-omics analysis of transcriptomes and metabolomes in BNGS3 and NfBNGS4. Transcriptome, N = 3 biological replicates; Metabolome, N = 6 biological replicates.
(B) The screen of differently expressed genes in transcriptome analysis of TtBNGS3 and TtBNGS4. N = 3 biological replicates; the SD is shown as black error bars.
(C) The screen of differently expressed genes in transcriptome analysis of NfBNGS3 and NfBNGS4. N = 6 biological replicates; the SD is shown as black error bars.
In the case of TtBNGS4 and NfBNGS4, the transcription level of genes in NfBNGS4 was generally up-regulated compared with that in TtBNGS4, including “biosynthesis of amino acids,” “glycolysis and gluconeogenesis,” and “amino sugar and nucleotide sugar metabolism,” indicating that the enzymes NfGNA1 and TtGNA1 exposed different effects on the metabolism (Figure S24). As the enzyme activity of TtGNA1 was higher than that of NfGNA1, in TtBNGS4, more carbon metabolism flowed to GlcNAc and less to energy metabolism and growth, resulting in an overall down-regulation of the transcriptional levels of genes related to bypassing metabolic pathways.
As shown in Figures 8B and 8C, the transcription level of NfGNA1 in NfBNGS3 was up-regulated compared to that in NfBNGS4, and a similar result was observed for TtGNA1 expression in TtBNGS3 compared to that in TtBNGS4. It can be speculated that the gna1 mRNA accumulation in the cells of strain TtBNGS3 or NfBNGS3 is much higher than that in the cells of strain TtBNGS4 or NfBNGS4. In addition, smpB transcription in NfBNGS4 was significantly up-regulated compared with that in NfBNGS3. This result suggests that NfBNGS4 had more tmRNAs to free the ribosomes trapped on the mRNAs, thus allowing the correct expression and translation of the protein and further improving GlcNAc production.
Discussion
The continued advancement of synthetic biology has facilitated the introduction of a growing number of genes into cells to build versatile cell factories.1 Hence, the problem of host-guest incompatibility between the chassis and non-native enzymes has arisen, the solving of which is highly desired, such as the incompatibility between oxygen-generating cyanobacteria and anaerobic enzymes,3 acidophiles and acid-labile enzymes,58 halophiles and salt-sensitive enzymes,59 and mesophile-derived enzymes and thermophilies.15 This problem is particularly prominent in the development of cell factories for high-temperature fermentation. The lack of stable and thermophilic enzymes significantly affects the development and application of a thermophilic chassis, even though they possess many crucial superiorities.60 Considering that it is difficult and unpredictable to modify enzymes to overcome host-guest incompatibility through protein engineering,61 mining diverse enzymes from massive genomic data has great potential as an alternative method. In this study, a thermophilic enzyme mining strategy was established with co-evolution in mind, which can provide excellent elements for building thermophilic cell factories. We used GlcNAc production as an example, successfully mined four thermophilic GNA1s, and achieved GlcNAc titers of 119.3 g/L at 42°C, which is ready for industrial application. To our knowledge, this is the first demonstration of GlcNAc production at elevated temperatures, representing a significant breakthrough in the GlcNAc production process (Table S6). The excellent in vivo and in vitro performances of our mined thermophilic GNA1s suggest that our mining strategy is feasible and effective for tackling the unsuitability of critical enzymes in thermophilic hosts. Our established pipeline contained abundant microbial genomic information with broad OGT ranges, suggesting that our mining strategy is universally applicable.
The NfBNGS4 strain achieved the highest GlcNAc titer at 42°C, even though the optimum temperature for NfGNA1 was 25°C. NfGNA1 could remain at 60% activities after incubation for 48 h at 37 and 50°C, respectively (Figure 4). It is reasonable to suggest that high thermostability contributes to good fermentation titers. These results also suggest that a comprehensive characterization is required to fully measure the advantage of an enzyme, rather than merely determining the optimal temperature.
Although the advantage of TtGNA1 with higher catalytic efficiency, and higher thermostability stands out when the fermentation temperature rises to 50°C, the GlcNAc production decreased to 47.4 g/L, which showed lower titers than GlcNAc production at 42°C. We observed the rapid cell growth and overflow of acetic acid when fermented at 50°C, which may greatly limit GlcNAc production. Thus, to further increase the GlcNAc production at 50°C, acetic acid overflow must be resolved. We exhausted ineffectual attempts to engineer acetate metabolism pathways. We hypothesize that the acetate production genes, such as pta and ackA are crucial for the metabolism of B. licheniformis; hence, a complete gene knockout of acetic acid may be lethal. When cells grow too fast in a glucose-rich medium, the transcription levels of genes related to acetic acid synthetic pathways were up-regulated, leading to more acetic acid being converted as a storage carbon source for utilization, when necessary, in terms of substance metabolism and energy metabolism.62,63,64 As the OGT of B. licheniformis MW3 is 50°C, appropriately lowering the initial fermentation temperature, might reduce growth and resolve the problem of acetic acid overflow. According to our results, the decreased temperature was able to slow down cell growth when the temperature was reduced to 47°C (Figure S22). The acetic acid concentration decreased from 29.7 g/L to 8.3 g/L, and the GlcNAc titers increased by 82.3% from 47.4 g/L to 86.4 g/L. When the fermentation temperature was slowly increased from 42°C to 50°C, there was almost no acetic acid overflow (Figure 7G), indicating the feasibility of our proposed temperature programming. The temperature programming can avoid tedious gene knockout and metabolic engineering operations, which is easy to implement for large-scale applications. In addition, the temperature programming further improved the applicability of the enzymes obtained using our mining strategy.
After NfGNA1 was integrated into the chromosome, the transcriptional level of NfGNA1 in NfBNGS4 was down-regulated because of lower gene copy numbers than in NfBNGS3 (vector) (Figure 8B). However, GlcNAc production by NfBNGS4 was much higher, indicating that the copies of nfgna1 may not always correspond to GlcNAc titers. The transcriptome data analysis showed that the smpB was greatly up-regulated in NfBNGS4 compared to that in NfBNGS3 (Figure 8B). The tmRNA-SmpB complex system could rescue the ribosomes stalled on defective mRNAs to ensure quality control of protein synthesis and correct translation in prokaryotes.65,66 It is reasonable to suggest that compared with NfBNGS3, there are more freed ribosomes to help increase protein translation efficiency in NfBNGS4, which in turn compensates for the low nfgna1 copy numbers on the chromosomes. In addition, compared to NfBNGS4, NfBNGS3 showed poor growth with the addition of tetracycline (Figure S14), which may further affect GlcNAc production when nfgna1 is expressed in the vector. These results suggest that the mined enzymes can achieve high GlcNAc titers despite their low copy numbers, which further demonstrates their excellent adaptability in a thermophilic host, indicating the effectiveness of our strategy.
In conclusion, we develop a systematic thermophilic enzyme mining strategy and successfully demonstrate the high-temperature fermentation of GlcNAc with high titers, which is a major breakthrough in the production process of GlcNAc. The in vitro and in vivo enzymatic performance tests of the four enzymes reveal high thermostabilities, proving the feasibility of our mining pipeline. Combining the thermophilic enzyme mining pipeline with a systems biology strategy may improve the production of GlcNAc. To make the enzyme mining strategy available for worldwide users, we have established a website that is able to automatically realize thermophilic enzyme mining (http://cem.sjtu.edu.cn). Our enzyme mining strategy provides a direction and paves the way for eliminating the host-guest incompatibility challenge faced by synthetic biology.
Limitation of the study
Our engineered strains for high-temperature fermentation of GlcNAc with high titers have strong potentials of high robustness and low energy consumption, being ready for industrialization. However, in this study, we have only been successful in 50-L bioreactors at most. Future work is that larger-scale fermentations for industrialization will be carried out, such as in a large scale of 200-ton bioreactors in our collaborative factories, benefiting from the high robustness and low energy consumption.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Clone-express Vltra One Step Cloning kit | Vazyme | Cat#C115-01 |
| Isopropyl-β-d-thiogalactoside (IPTG), | Glpbio | Cat#GC30002-5 |
| phenylmethanesulfonyl fluoride (PMSF) | Glpbio | Cat#GC10477-10000 |
| Acetic acid | Sigma-Aldrich | Cat#V900798-500 mL |
| Acetoin | Merck | Cat#40127-U |
| GlcNAc standards | Glpbio | Cat#GC41283-1 |
| 2,3-Butanediol | Macklin | Cat#B822252-500 mL |
| Tetracycline hydrochloride | Amresco | Cat#0422-25g |
| Acetyl CoA | Sigma-Aldrich | Cat#A2056-5 mg |
| DTNB | Sigma-Aldrich | Cat#D218200-1g |
| d-Glucosamine 6-phosphate | Sigma | Cat#G5509-10 mg |
| Deposited data | ||
| Metabolomics data | This study | Accession number MTBLS5385 |
| Transcriptome data | This study | GEO accession number PRJNA869503 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Fei Tao (taofei@sjtu.edu.cn).
Materials availability
All the requests for the generated plasmids and strains should be directed to the lead contact and will be made available on request after completion of a Materials Transfer Agreement.
Experimental model and subject details
Strains and growth media
Wild type B. licheniformis MW3 strain and its derivatives were used in this study. The double mutant MW3 (ΔhadR1, ΔhsdR2) of B. licheniformis ATCC 14580 was a kindly gift from Professor Friedhelm Meinhardt.67 E. coli strains DH5α and BL21 (DE3) were used for the vector construction and protein expression, respectively. E. coli S17-1 was used as the donor strain for conjugation. The pETDuet-1 vector was used for protein expression. The conjugative shuttle vector pKVM1 carries ampicillin and erythromycin-resistance genes was used as knock-out vector plasmid pHY300PLK was a general gift from Professor Shouwen Chen for protein expression in B. licheniformis.68 For all related experiments, we always prepared fresh LB plates of B.licheniformis cells (from the glycerol stock of −80°C freezer) as the starting material. The strains and plasmids are listed in Tables S7 and S8. Primers are listed in Table S9.
Method details
Website establishment
The CEM website (Co-Evolution Mining) started running from February 2022. It contained more than 200,000 microbial genome data, which can be downloaded from the NCBI database, the microbial whole genome data according to the collated OGT data from Engqvist48 and a filamentous fungus database (https://pub.fungalgenomics.ca/). The website can provide users with three methods for thermophilic enzyme mining, such as the microbial optimum growth temperatures, the thermophilic fungus genomes, and the structural RNA GC content. It can achieve rapid mining of thermophilic enzymes in a short time of no more than 10 min. Users only need to input the amino acid sequence of an enzyme in fasta. format and choose an enzyme mining method, the rapid mining of thermophilic enzymes can be realized. The information about mined candidate thermophilic enzymes will be sent to users in the form of reports with tsv. format, which can be downloaded directly from the website.
Bacterial culture conditions
For seed cultivation, the B. licheniformis MW3 and its derivates were grown in Luria-Bertani broth (LB) medium at 50 °C at 200 rpm.
Gene cloning and synthesis, expression, and purification
The coding gene of codon-optimized 21 GNA1 genes was synthesized by GeneWiz (Table S10). The resulting fragment was fusion with Pals, P43, PaprE, Pst or Pbl9 by PCR, and the fusion fragment were digested with BamHI/EcoRI and inserted into the expression vector pHY300PLK. The several constructed vectors were electro-transformed into BNGS1, separately. The Ttgna1, Nfgna1, Ctgna1, Mtgna1 and Scopgna1 genes were amplified and inserted into pETDuet-1 to generate plasmids pETDuet-Ttgna1, pETDuet-Nfgna1, pETDuet-Ctgna1, pETDuet-Mtgna1, and pETDuet-Scopgna1, respectively. All the enzymes (TtGNA1, NfGNA1, MtGNA1, CtGNA1, and ScGNA1) were codon-optimized and expressed in E. coli BL21 (DE3) and purified using the method of Zheng et al.69
Enzyme performance testing, characterization, and AlphaFold2 protein structure prediction
The specific determination method of GNA1 enzyme activity was as follows: The 5 μL of sample protein was incubated at different temperatures or pH buffers in 200 μL reaction system which including 200 μM GlcN-6-P, 5 mM MgCl2, 50 mM pH buffer, 10% (v/v) glycerol, 200 μM Ac-CoA and incubated for 4 min, then adding the 100 μL termination system including 50 mM Tris-HCl (pH 7.5) and 6.4 M guanidine hydrochloride, finally adding the 100 μL measurement system including 50 mM Tris-HCl (pH 7.5), 1 mM EDTA and DTNB. The product was detected at 412 nm to assess the GNA1 activity. The Michaelis-Menten equation was used to determine the kinetic parameters. Superdex 200 (ÄTKA purifier) was used for analysis of size exclusion chromatography. Protein thermostability tests for CtGNA1, MtGNA1, TtGNA1, NfGNA1, and ScGNA1 were used with nano differential scanning calorimeter (Nano DSC, the TA instruments, USA). FoldX was used for protein stability prediction of TtGNA1, MtGNA1, NfGNA1, CtGNA1, and ScGNA1. Putative stabilizing factors and flexibility indices of TtGNA1, MtGNA1, NfGNA1, and CtGNA1 were calculated based on amino acid sequences and AlphaFold2 predicted structures of the four proteins.
Genetic manipulation in B. licheniformis
B. licheniformis MW3 transformation with pHY300PLK was performed by electrotransformation.68 The detailed gene-knockout operation method was performed by Li et al.15 with pKVM1 plasmid. E. coli S17-1 harboring pKVM plasmids were used as the donors in conjugation with B. licheniformis MW3. All the deletion and insertion mutations were verified by PCR amplification of the genomic DNA with appropriate primers (Table S9) and sequencing of the amplified products.
Shake-flask fermentation and fed-batch fermentation
The constructed B. licheniformis grown on LB agar was transferred to 5 mL seed medium (LB), then the seed strain was transferred into a 500 mL shake flask with 50 mL shake-flask fermentation medium including 12 g/L yeast extract, 6 g/L tryptone, 6 g/L (NH4)2SO4, 18.75 g/L K2HPO4•3H2O, 5.75 g/L KH2PO4, and 30 g/L glucose, then cultivated in 37°C, 42°C, or 50°C for 24 h, respectively.
The initial fed-batch fermentation culture included 40 g/L glucose, 5 g/L yeast extract, 30 g/L corn steep liquor, 12.5 g/L K2HPO4, 2.5 g/L KH2PO4, 3 g/L MgSO4, 6 g/L (NH4)2SO4, and 10 g/L groundnut meal. The glucose concentration was set at 40 g/L for fed-batch fermentation. The pH and DO were set at 7.0 and 20–40%, respectively, in 3 L or 50 L bioreactors.
Analytical methods
GlcNAc and other by-product were determined by HPLC (Agilent 1200 series, USA) equipped with a Bio-Rad Aminex HPX-87H column (300∗7.8 mm) and a refractive index detector. Analysis was performed with a mobile phase of 5 mM H2SO4 in 0.5 mL/min with 60°C column temperature and 55°C RID detector temperature. Fermentation supernatant was filtered with a 0.22 μL membrane filter. The glucose and l-lactic acid concentration were measured with SBA.
Extraction and measurement of metabolomes
Based on the previous studies,70,71 the extraction of the metabolites was carried out for the bacteria cells with minor modifications. Briefly, the cells were quenched and extracted rapidly with 1 mL of 80:20 MeOH/H2O (containing 0.1% formic acid, −80°C) and then frozen in liquid nitrogen. Then, the supernatants of the samples were collected for LC-MS analysis after being frozen-thawed three times to extract the metabolites.
The five kinds of samples, BNGS1, TtBNGS4, NfBNGS4, TtBNGS3, and NfBNGS3 were subjected to the Thermo UPLC Q-Extractive (QE) coupled with ESI ion source for metabolomics analysis according to the previous study.72 The chromatographic separation was achieved with the Acquity UPLC BEH C18 column (1.7 μm, 2.1 × 100 mm). Mobile phase A is aqueous of 0.1% formic acid in diluted water, and mobile phase B is the organic phase of 0.1% formic acid in pure acetonitrile. The LC gradient elution program was as follows: t = 0.0 min, 99% A; t = 5.0 min, 99% A; t = 5.5 min, 70% A; t = 9 min, 100% B; t = 11 min, 100% B; and t = 12.1 min, 99% A. The mass parameters of the C18-ESIMS are as follows: full mass spectrometry scanning range 80−1,000 m/z, the capillary voltage of 4,500 V, the gas flow rate was 1.6 bar, the velocity of dry gas at 220°C was 6.0 L/min. According to the previous studies,72 the network tool of MetaboAnalyst73 was used to conduct the metabolome data analysis. And the pathway enrichment was used the OmicsBean Multi-omics function.74
In the metabolomics analysis, six replicates were used. The dysregulated metabolites were identified with p value of <0.05 and fold change of >1.2. The experimental data were analyzed using two-tailed Student’s t-tests. Metabolite annotation was performed using the commercial software Compound Discoverer (ThermoFisher Scintific, USA). The data of metabolomics were available in the MetaboLights database with an access number of MTBLS5385.
Transcriptome analysis
Total RNA was extracted from the tissue using TRIzol Reagent according the manufacturer’s instructions (Invitrogen) and genomic DNA was removed using DNase I (TaKara). Then RNA quality was determined by 2100 Bioanalyser (Agilent) and quantified using the ND-2000 (NanoDrop Technologies). Only high-quality RNA sample (OD260/280 = 1.8–2.0, OD260/230 ≥ 2.0, RIN ≥6.5, 28S:18S ≥ 1.0, concentration ≥100 ng/μL, total amount ≥2 μg) was used to construct sequencing library.
RNA-seq transcriptome library was prepared following TruSeqTM RNA sample preparation Kit from Illumina (San Diego, CA) using 2 μg of total RNA. Shortly, rRNA depletion instead of poly(A) purification was performed by Ribo-Zero Magnetic kit (epicenter) and then all mRNAs were broken into short (200 nt) fragments by adding fragmentation buffer firstly. Secondly double-stranded cDNA was synthesized using a Super-Script double-stranded cDNA synthesis kit (Invitrogen, CA) with random hexamer primers (Illumina). When the second strand cDNA was synthesized, dUTP was incorporated in place of dTTP. Then the synthesized cDNA was subjected to end-repair, phosphorylation, and ‘A’ base addition according to Illumina’s library construction protocol. The second strand cDNA with dUTP was recognized and degraded by UNG enzyme. Libraries were size selected for cDNA target fragments of 200 bp on 2% Low Range Ultra Agarose followed by PCR amplified using Phusion DNA polymerase (NEB) for 15 PCR cycles. After quantified by TBS380, paired-end RNA-seq sequencing library was sequenced with the Illumina HiSeq × TEN (2 × 150 bp read length). The processing of original images to sequences, base-calling, and quality value calculations were performed using the Illumina GA Pipeline (version 1.6), in which 150 bp paired-end reads were obtained. A Perl program was written to select clean reads by removing low-quality sequences, reads with more than 5% of N bases (unknown bases) and reads containing adaptor sequences.
The data generated from Illumina platform were used for bioinformatics analysis. All of the analyses were performed using the free online platform of Majorbio Cloud Platform (www.majorbio.com) from Shanghai Majorbio Bio-pharm Technology Co., Ltd. The differentially expressed genes were identified through overlapping gene analysis of three biological duplicate experiments using a 2-fold change as an empirical criterion. The experimental data were analyzed using two-tailed Student’s t-tests. P-values less than 0.05 were considered as significant. The transcriptome data discussed in this article were deposited into NCBI’s Gene Expression Omnibus (GEO) Series with an accession number (PRJNA869503).
Quantification and statistical analysis
At least three biological replicates were performed for each experiment. Data are given as the means ± SD. All comparisons to determine differences were performed by applying Student’s t test. SPPS 18.0 software (SPSS Inc., Chicago, IL, USA) was used for data processing and analysis. Mean values were regarded as significantly different at p < 0.05.
Acknowledgments
This work was supported by the grants (22138007, 32170105, and 31870088) of National Natural Science Foundation of China. We highly thank Xiaoling Mu, Hao Tang, Sihong Chen, Weili Li, Chunlin Tan, Jiahui Zhi, Yuanyuan Feng, and Hang Xu for their assistance in the experiments regarding this study. We thank Prof. Shouwen Chen for supplying the pHY300PLK plasmid. We acknowledge T&J Bio-engineering (Shanghai) Co., LTD and Shanghai Bailun Biological Technology Co., LTD for their supplying the bioreactors to preliminary experiments.
Author contributions
Fei Tao, Yutong Wu, and Ping Xu designed the experiments. Yutong Wu, Jiongqin Liu, Xuanlin Meng, Mengke Li, Hongsong Xue, and Jing Wang performed the experiments. Yutong Wu, Xiao Han, and Yuhan Yang prepared the materials and reagents. Yutong Wu wrote the article. Yutong Wu, Fei Tao, and Ping Xu revised the article. Yutong Wu and Fei Tao analyzed the data. Ping Xu and Fei Tao conceived the projects.
Declaration of interests
The authors declare that they have no competing interests.
Published: January 20, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105774.
Supplemental information
Data and code availability
All data reported in this study will be shared by the lead contact upon request. The raw data of metabolomics were available in the MetaboLights database with an accession number of MTBLS5385. The transcriptome data discussed in this article were deposited into NCBI’s Gene Expression Omnibus (GEO) Series with an accession number of PRJNA869503. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Meng F., Ellis T. The second decade of synthetic biology: 2010–2020. Nat. Commun. 2020;11:5174. doi: 10.1038/s41467-020-19092-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jia H., Schwille P. Bottom-up synthetic biology: reconstitution in space and time. Curr. Opin. Biotechnol. 2019;60:179–187. doi: 10.1016/j.copbio.2019.05.008. [DOI] [PubMed] [Google Scholar]
- 3.Liu H., Ni J., Xu P., Tao F. Enhancing light-driven 1, 3-propanediol production by using natural compartmentalization of differentiated cells. ACS Synth. Biol. 2018;7:2436–2446. doi: 10.1021/acssynbio.8b00239. [DOI] [PubMed] [Google Scholar]
- 4.Sharma A., Kawarabayasi Y., Satyanarayana T. Acidophilic bacteria and archaea: acid stable biocatalysts and their potential applications. Extremophiles. 2012;16:1–19. doi: 10.1007/s00792-011-0402-3. [DOI] [PubMed] [Google Scholar]
- 5.Chen G.-Q., Jiang X.-R. Next generation industrial biotechnology based on extremophilic bacteria. Curr. Opin. Biotechnol. 2018;50:94–100. doi: 10.1016/j.copbio.2017.11.016. [DOI] [PubMed] [Google Scholar]
- 6.Bhandiwad A., Shaw A.J., Guss A., Guseva A., Bahl H., Lynd L.R. Metabolic engineering of Thermoanaerobacterium saccharolyticum for N-butanol production. Metab. Eng. 2014;21:17–25. doi: 10.1016/j.ymben.2013.10.012. [DOI] [PubMed] [Google Scholar]
- 7.Barnard D., Casanueva A., Tuffin M., Cowan D. Extremophiles in biofuel synthesis. Environ. Technol. 2010;31:871–888. doi: 10.1080/09593331003710236. [DOI] [PubMed] [Google Scholar]
- 8.Abdel-Banat B.M.A., Hoshida H., Ano A., Nonklang S., Akada R. High-temperature fermentation: how can processes for ethanol production at high temperatures become superior to the traditional process using mesophilic yeast? Appl. Microbiol. Biotechnol. 2010;85:861–867. doi: 10.1007/s00253-009-2248-5. [DOI] [PubMed] [Google Scholar]
- 9.Straub C.T., Zeldes B.M., Schut G.J., Adams M.W., Kelly R.M. Extremely thermophilic energy metabolisms: biotechnological prospects. Curr. Opin. Biotechnol. 2017;45:104–112. doi: 10.1016/j.copbio.2017.02.016. [DOI] [PubMed] [Google Scholar]
- 10.Arnolds K.L., Dahlin L.R., Ding L., Wu C., Yu J., Xiong W., Zuniga C., Suzuki Y., Zengler K., Linger J.G., Guarnieri M.T. Biotechnology for secure biocontainment designs in an emerging bioeconomy. Curr. Opin. Biotechnol. 2021;71:25–31. doi: 10.1016/j.copbio.2021.05.004. [DOI] [PubMed] [Google Scholar]
- 11.Lin P.P., Rabe K.S., Takasumi J.L., Kadisch M., Arnold F.H., Liao J.C. Isobutanol production at elevated temperatures in thermophilic Geobacillus thermoglucosidasius. Metab. Eng. 2014;24:1–8. doi: 10.1016/j.ymben.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 12.Cripps R.E., Eley K., Leak D.J., Rudd B., Taylor M., Todd M., Boakes S., Martin S., Atkinson T. Metabolic engineering of Geobacillus thermoglucosidasius for high yield ethanol production. Metab. Eng. 2009;11:398–408. doi: 10.1016/j.ymben.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 13.Zhou J., Wu K., Rao C.V. Evolutionary engineering of Geobacillus thermoglucosidasius for improved ethanol production. Biotechnol. Bioeng. 2016;113:2156–2167. doi: 10.1002/bit.25983. [DOI] [PubMed] [Google Scholar]
- 14.Lu C.J., Ge Y., Cao M., Guo X., Liu P., Gao C., Xu P., Ma C. Metabolic engineering of Bacillus licheniformis for production of acetoin. Front. Bioeng. Biotechnol. 2020;8:125. doi: 10.3389/fbioe.2020.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li C., Tao F., Xu P. Carbon flux trapping: highly efficient production of polymer-grade d-lactic acid with a thermophilic D-lactate dehydrogenase. Chembiochem. 2016;17:1491–1494. doi: 10.1002/cbic.201600288. [DOI] [PubMed] [Google Scholar]
- 16.Wang Q., Chen T., Zhao X., Chamu J. Metabolic engineering of thermophilic Bacillus licheniformis for chiral pure d-2, 3-butanediol production. Biotechnol. Bioeng. 2012;109:1610–1621. doi: 10.1002/bit.24427. [DOI] [PubMed] [Google Scholar]
- 17.Adapa V., Ramya L.N., Pulicherla K.K., Rao K.R.S.S. Cold active pectinases: advancing the food industry to the next generation. Appl. Biochem. Biotechnol. 2014;172:2324–2337. doi: 10.1007/s12010-013-0685-1. [DOI] [PubMed] [Google Scholar]
- 18.Mougiakos I., Bosma E.F., Weenink K., Vossen E., Goijvaerts K., van der Oost J., van Kranenburg R. Efficient genome editing of a facultative thermophile using mesophilic spCas9. ACS Synth. Biol. 2017;6:849–861. doi: 10.1021/acssynbio.6b00339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deltcheva E., Chylinski K., Sharma C.M., Gonzales K., Chao Y., Pirzada Z.A., Eckert M.R., Vogel J., Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong C., Drueckhammer D.G., Sweers H.M. Enzymatic vs. fermentative synthesis: thermostable glucose dehydrogenase catalyzed regeneration of NAD(P)H for use in enzymatic synthesis. J. Am. Chem. Soc. 1985;107:4028–4031. doi: 10.1021/ja00299a044. [DOI] [Google Scholar]
- 21.Wang Q., Ingram L.O., Shanmugam K.T. Evolution of d-lactate dehydrogenase activity from glycerol dehydrogenase and its utility for d-lactate production from lignocellulose. Proc. Natl. Acad. Sci. USA. 2011;108:18920–18925. doi: 10.1073/pnas.1111085108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Dijk E.L., Auger H., Jaszczyszyn Y., Thermes C. Ten years of next-generation sequencing technology. Trends Genet. 2014;30:418–426. doi: 10.1016/j.tig.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 23.Prokop J.W., May T., Strong K., Bilinovich S.M., Bupp C., Rajasekaran S., Worthey E.A., Lazar J. Genome sequencing in the clinic: the past, present, and future of genomic medicine. Physiol. Genom. 2018;50:563–579. doi: 10.1152/physiolgenomics.00046.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen J.K., Shen C.R., Liu C.L. N-acetylglucosamine: production and applications. Mar. Drugs. 2010;8:2493–2516. doi: 10.3390/md8092493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang N.J., Lin Y.T., Lin C.C., Wang H.C., Hsu H.C., Yeh M.L. The repair of full-thickness articular cartilage defect using intra-articular administration of N-acetyl-d-glucosamine in the rabbit knee: randomized controlled trial. Biomed. Eng. Online. 2015;14:105. doi: 10.1186/s12938-015-0100-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eriksen P., Bartels E.M., Altman R.D., Bliddal H., Juhl C., Christensen R. Risk of bias and brand explain the observed inconsistency in trials on glucosamine for symptomatic relief of osteoarthritis: a meta-analysis of placebo-controlled trials. Arthritis Care Res. 2014;66:1844–1855. doi: 10.1002/acr.22376. [DOI] [PubMed] [Google Scholar]
- 27.Zhu Y., Liu Y., Li J., Shin H.D., Du G., Liu L., Chen J. An optimal glucose feeding strategy integrated with step-wise regulation of the dissolved oxygen level improves N-acetylglucosamine production in recombinant Bacillus subtilis. Bioresour. Technol. 2015;177:387–392. doi: 10.1016/j.biortech.2014.11.055. [DOI] [PubMed] [Google Scholar]
- 28.Liu Y., Li Z., Liu G., Jia J., Li S., Yu C. Liquid chromatography-tandem mass spectrometry method for determination of N-acetylglucosamine concentration in human plasma. J. Chromatogr., B: Anal. Technol. Biomed. Life Sci. 2008;862:150–154. doi: 10.1016/j.jchromb.2007.11.043. [DOI] [PubMed] [Google Scholar]
- 29.Salvatore S., Heuschkel R., Tomlin S., Davies S.E., Edwards S., Walker-Smith J.A., French I., Murch S.H. A pilot study of N-acetylglucosamine, a nutritional substrate for glycosaminoglycan synthesis, in paediatric chronic inflammatory bowel disease. Aliment. Pharmacol. Ther. 2000;14:1567–1579. doi: 10.1046/j.1365-2036.2000.00883.x. [DOI] [PubMed] [Google Scholar]
- 30.Longo D.L., Moustaghfir F.Z., Zerbo A., Consolino L., Anemone A., Bracesco M., Aime S. EXCI-CEST: exploiting pharmaceutical excipients as MRI-CEST contrast agents for tumor imaging. Int. J. Pharm. 2017;525:275–281. doi: 10.1016/j.ijpharm.2017.04.040. [DOI] [PubMed] [Google Scholar]
- 31.Kumar P., Tambe P., Paknikar K.M., Gajbhiye V. Folate/N-acetyl glucosamine conjugated mesoporous silica nanoparticles for targeting breast cancer cells: a comparative study. Colloids Surf. B Biointerfaces. 2017;156:203–212. doi: 10.1016/j.colsurfb.2017.05.032. [DOI] [PubMed] [Google Scholar]
- 32.Hassan A.E. An observational cohort study to assess N-acetylglucosamine for COVID-19 treatment in the inpatient setting. Ann. Med. Surg. (Lond) 2021;68:102574. doi: 10.1016/j.amsu.2021.102574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu L., Liu Y., Shin H.D., Chen R., Li J., Du G., Chen J. Microbial production of glucosamine and N-acetylglucosamine: advances and perspectives. Appl. Microbiol. Biotechnol. 2013;97:6149–6158. doi: 10.1007/s00253-013-4995-6. [DOI] [PubMed] [Google Scholar]
- 34.Sashiwa H., Fujishima S., Yamano N., Kawasaki N., Nakayama A., Muraki E., et al. Production of N-acetyl-d-glucosamine from alpha-chitin by crude enzymes from Aeromonas hydrophila H-2330. Carbohydr. Res. 2002;337:761–763. doi: 10.1016/s0008-6215(02)00034-4. [DOI] [PubMed] [Google Scholar]
- 35.Zhu W., Wang D., Liu T., Yang Q. Production of N-acetyl-d-glucosamine from mycelial waste by a combination of bacterial chitinases and an insect N-acetyl-d-glucosaminidase. J. Agric. Food Chem. 2016;64:6738–6744. doi: 10.1021/acs.jafc.6b03713. [DOI] [PubMed] [Google Scholar]
- 36.Chen X., Gao C., Guo L., Hu G., Luo Q., Liu J., Nielsen J., Chen J., Liu L. DCEO biotechnology: tools to design, construct, evaluate, and optimize the metabolic pathway for biosynthesis of chemicals. Chem. Rev. 2018;118:4–72. doi: 10.1021/acs.chemrev.6b00804. [DOI] [PubMed] [Google Scholar]
- 37.Ma W., Liu Y., Lv X., Li J., Du G., Liu L. Combinatorial pathway enzyme engineering and host engineering overcomes pyruvate overflow and enhances overproduction of N-acetylglucosamine in Bacillus subtilis. Microb. Cell Factories. 2019;18:1. doi: 10.1186/s12934-018-1049-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Riegler H., Herter T., Grishkovskaya I., Lude A., Ryngajllo M., Bolger M.E., Essigmann B., Usadel B. Crystal structure and functional characterization of a glucosamine-6-phosphate N-acetyltransferase from Arabidopsis thaliana. Biochem. J. 2012;443:427–437. doi: 10.1042/bj20112071. [DOI] [PubMed] [Google Scholar]
- 39.Peneff C., Mengin-Lecreulx D., Bourne Y. The crystal structures of Apo and complexed Saccharomyces cerevisiae GNA1 shed light on the catalytic mechanism of an amino-sugar N-acetyltransferase. J. Biol. Chem. 2001;276:16328–16334. doi: 10.1074/jbc.M009988200. [DOI] [PubMed] [Google Scholar]
- 40.Wang J., Liu X., Liang Y.H., Li L.F., Su X.D. Acceptor substrate binding revealed by crystal structure of human glucosamine-6-phosphate N-acetyltransferase 1. FEBS Lett. 2008;582:2973–2978. doi: 10.1016/j.febslet.2008.07.040. [DOI] [PubMed] [Google Scholar]
- 41.Dorfmueller H.C., Fang W., Rao F.V., Blair D.E., Attrill H., van Aalten D.M.F. Structural and biochemical characterization of a trapped coenzyme A adduct of Caenorhabditis elegans glucosamine-6-phosphate N-acetyltransferase 1. Acta Crystallogr. D Biol. Crystallogr. 2012;68:1019–1029. doi: 10.1107/s0907444912019592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma Q., Sun Q., Tan M., Xia L., Zhang Y., Yang M., Zhuo M., Zhao K., Li Y., Xu Q., et al. Highly efficient production of N-acetyl-glucosamine in Escherichia coli by appropriate catabolic division of labor in the utilization of mixed Glycerol/Glucose carbon sources. J. Agric. Food Chem. 2021;69:5966–5975. doi: 10.1021/acs.jafc.1c01513. [DOI] [PubMed] [Google Scholar]
- 43.Lee S.W., Lee B.Y., Oh M.K. Combination of three methods to reduce Glucose metabolic rate for improving N-acetylglucosamine production in Saccharomyces cerevisiae. J. Agric. Food Chem. 2018;66:13191–13198. doi: 10.1021/acs.jafc.8b04291. [DOI] [PubMed] [Google Scholar]
- 44.Gu Y., Lv X., Liu Y., Li J., Du G., Chen J., Rodrigo L.A., Liu L. Synthetic redesign of central carbon and redox metabolism for high yield production of N-acetylglucosamine in Bacillus subtilis. Metab. Eng. 2019;51:59–69. doi: 10.1016/j.ymben.2018.10.002. [DOI] [PubMed] [Google Scholar]
- 45.Deng C., Lv X., Li J., Zhang H., Liu Y., Du G., Amaro R.L., Liu L. Synergistic improvement of N-acetylglucosamine production by engineering transcription factors and balancing redox cofactors. Metab. Eng. 2021;67:330–346. doi: 10.1016/j.ymben.2021.07.012. [DOI] [PubMed] [Google Scholar]
- 46.Li G., Rabe K.S., Nielsen J., Engqvist M.K.M. Machine learning applied to predicting microorganism growth temperatures and enzyme catalytic optima. ACS Synth. Biol. 2019;8:1411–1420. doi: 10.1021/acssynbio.9b00099. [DOI] [PubMed] [Google Scholar]
- 47.Lieph R., Veloso F.A., Holmes D.S. Thermophiles like hot T. Trends Microbiol. 2006;14:423–426. doi: 10.1016/j.tim.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 48.Engqvist M.K.M. Correlating enzyme annotations with a large set of microbial growth temperatures reveals metabolic adaptations to growth at diverse temperatures. BMC Microbiol. 2018;18:177. doi: 10.1186/s12866-018-1320-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cimen E., Jensen S.E., Buckler E.S. Building a tRNA thermometer to estimate microbial adaptation to temperature. Nucleic Acids Res. 2020;48:12004–12015. doi: 10.1093/nar/gkaa1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jegousse C., Yang Y., Zhan J., Wang J., Zhou Y. Structural signatures of thermal adaptation of bacterial ribosomal RNA, transfer RNA, and messenger RNA. PLoS One. 2017;12:e0184722. doi: 10.1371/journal.pone.0184722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cramer P. AlphaFold2 and the future of structural biology. Nat. Struct. Mol. Biol. 2021;28:704–705. doi: 10.1038/s41594-021-00650-1. [DOI] [PubMed] [Google Scholar]
- 52.Schymkowitz J., Borg J., Stricher F., Nys R., Rousseau F., Serrano L. The FoldX web server: an online force field. Nucleic Acids Res. 2005;33:W382–W388. doi: 10.1093/nar/gki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guerois R., Nielsen J.E., Serrano L. Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J. Mol. Biol. 2002;320:369–387. doi: 10.1016/S0022-2836(02)00442-4. [DOI] [PubMed] [Google Scholar]
- 54.Studer R.A., Christin P.A., Williams M.A., Orengo C.A. Stability-activity tradeoffs constrain the adaptive evolution of RubisCO. Proc. Natl. Acad. Sci. USA. 2014;111:2223–2228. doi: 10.1073/pnas.1310811111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu B., Wijma H.J., Song L., Rozeboom H.J., Poloni C., Tian Y., Arif M.I., Nuijens T., Quaedflieg P.J.L.M., Szymanski W., et al. Versatile peptide C-terminal functionalization via a computationally engineered peptide amidase. ACS Catal. 2016;6:5405–5414. doi: 10.1021/acscatal.6b01062. [DOI] [Google Scholar]
- 56.Tyo K.E.J., Ajikumar P.K., Stephanopoulos G. Stabilized gene duplication enables long-term selection-free heterologous pathway expression. Nat. Biotechnol. 2009;27:760–765. doi: 10.1038/nbt.1555. [DOI] [PubMed] [Google Scholar]
- 57.Daniel R.M., Cowan D.A. Biomolecular stability and life at high temperatures. Cell. Mol. Life Sci. 2000;57:250–264. doi: 10.1007/PL00000688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang H., Luo H., Bai Y., Wang Y., Yang P., Shi P., Zhang W., Fan Y., Yao B. An acidophilic β-galactosidase from Bispora sp. MEY-1 with high lactose hydrolytic activity under simulated gastric conditions. J. Agric. Food Chem. 2009;57:5535–5541. doi: 10.1021/jf900369e. [DOI] [PubMed] [Google Scholar]
- 59.Cao L., Zhang R., Zhou J., Huang Z. Biotechnological aspects of salt-tolerant xylanases: a review. J. Agric. Food Chem. 2021;69:8610–8624. doi: 10.1021/acs.jafc.1c03192. [DOI] [PubMed] [Google Scholar]
- 60.Jiang Y., Jiang W., Xin F., Zhang W., Jiang M. Thermophiles: potential chassis for lignocellulosic biorefinery. Trends Biotechnol. 2022;40:643–646. doi: 10.1016/j.tibtech.2021.12.008. [DOI] [PubMed] [Google Scholar]
- 61.Sinha R., Shukla P. Current trends in protein engineering: updates and progress. Curr. Protein Pept. Sci. 2019;20:398–407. doi: 10.2174/1389203720666181119120120. [DOI] [PubMed] [Google Scholar]
- 62.Wolfe A.J. The acetate switch. Microbiol. Mol. Biol. Rev. 2005;69:12–50. doi: 10.1128/mmbr.69.1.12-50.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fujita Y. Carbon catabolite control of the metabolic network in Bacillus subtilis. Biosci. Biotechnol. Biochem. 2009;73:245–259. doi: 10.1271/bbb.80479. [DOI] [PubMed] [Google Scholar]
- 64.van Hoek M.J.A., Merks R.M.H. Redox balance is key to explaining full vs. partial switching to low-yield metabolism. BMC Syst. Biol. 2012;6:22. doi: 10.1186/1752-0509-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ramrath D.J.F., Yamamoto H., Rother K., Wittek D., Pech M., Mielke T., Loerke J., Scheerer P., Ivanov P., Teraoka Y., et al. The complex of tmRNA-SmpB and EF-G on translocating ribosomes. Nature. 2012;485:526–529. doi: 10.1038/nature11006. [DOI] [PubMed] [Google Scholar]
- 66.Guyomar C., D'Urso G., Chat S., Giudice E., Gillet R. Structures of tmRNA and SmpB as they transit through the ribosome. Nat. Commun. 2021;12:4909. doi: 10.1038/s41467-021-24881-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Waschkau B., Waldeck J., Wieland S., Eichstädt R., Meinhardt F. Generation of readily transformable Bacillus licheniformis mutants. Appl. Microbiol. Biotechnol. 2008;78:181–188. doi: 10.1007/s00253-007-1278-0. [DOI] [PubMed] [Google Scholar]
- 68.Cai D., Chen Y., He P., Wang S., Mo F., Li X., Wang Q., Nomura C.T., Wen Z., Ma X., Chen S. Enhanced production of poly-γ-glutamic acid by improving ATP supply in metabolically engineered Bacillus licheniformis. Biotechnol. Bioeng. 2018;115:2541–2553. doi: 10.1002/bit.26774. [DOI] [PubMed] [Google Scholar]
- 69.Zheng Z., Sheng B., Ma C., Zhang H., Gao C., Su F., Xu P. Relative catalytic efficiency of ldhL- and ldhD-encoded products is crucial for optical purity of lactic acid produced by lactobacillus strains. Appl. Environ. Microbiol. 2012;78:3480–3483. doi: 10.1128/aem.00058-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cui J., Sun T., Li S., Xie Y., Song X., Wang F., Chen L., Zhang W. Improved salt tolerance and metabolomics analysis of Synechococcus elongatus UTEX 2973 by overexpressing Mrp antiporters. Front. Bioeng. Biotechnol. 2020;8:500. doi: 10.3389/fbioe.2020.00500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang Y., Shi M., Niu X., Zhang X., Gao L., Chen L., Wang J., Zhang W. Metabolomic basis of laboratory evolution of butanol tolerance in photosynthetic Synechocystis sp. PCC 6803. Microb. Cell Factories. 2014;13:151. doi: 10.1186/s12934-014-0151-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Meng X., Zhao X., Ding X., Li Y., Cao G., Chu Z., Su X., Liu Y., Chen X., Guo J., et al. Integrated functional omics analysis of flavonoid-related metabolism in AtMYB12 transcript factor overexpressed tomato. J. Agric. Food Chem. 2020;68:6776–6787. doi: 10.1021/acs.jafc.0c01894. [DOI] [PubMed] [Google Scholar]
- 73.Pang Z., Zhou G., Chong J., Xia J. Comprehensive meta-analysis of COVID-19 global metabolomics datasets. Metabolites. 2021;11:44. doi: 10.3390/metabo11010044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bao X., Guo X., Yin M., Tariq M., Lai Y., Kanwal S., Zhou J., Li N., Lv Y., Pulido-Quetglas C., et al. Capturing the interactome of newly transcribed RNA. Nat. Methods. 2018;15:213–220. doi: 10.1038/nmeth.4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this study will be shared by the lead contact upon request. The raw data of metabolomics were available in the MetaboLights database with an accession number of MTBLS5385. The transcriptome data discussed in this article were deposited into NCBI’s Gene Expression Omnibus (GEO) Series with an accession number of PRJNA869503. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.