Abstract
The presence of senescent cells within tissues has been functionally linked to malignant transformations. Here, using tension-gauge tethers technology, particle-tracking microrheology, and quantitative microscopy, we demonstrate that senescent-associated secretory phenotype (SASP) derived from senescent fibroblasts impose nuclear lobulations and volume shrinkage on malignant cells, which stems from the loss of RhoA/ROCK/myosin II-based cortical tension. This loss in cytoskeletal tension induces decreased cellular contractility, adhesion, and increased mechanical compliance. These SASP-induced morphological changes are, in part, mediated by Lamin A/C. These findings suggest that SASP induces defective outside-in mechanotransduction from actomyosin fibers in the cytoplasm to the nuclear lamina, thereby triggering a cascade of biophysical and biomolecular changes in cells that associate with malignant transformations.
Significance Statement.
Senescent cells promote adverse physiologies and malignant transformations in many organisms. During ageing, senescent cells accumulate and impose mechano-structural alterations on their local microenvironment, mainly through the hyper-secretion of soluble factors that influence the phenotypes of neighboring cells. Here we show that secreted factors from senescent cells induce a defective outside-in signaling cascade to promote nuclear lobulations in cancer cells. These nuclear lobulations result from the loss of cytoskeletal tension through and decreased RhoA/ROCK/Myosin II-based contractility and are capable of triggering a cascade of changes in cells, ranging from cytoskeletal-induced changes in morphologies that associates with the propagation of malignant transformations and phenotype-switching in cancer cells.
Introduction
Cellular senescence is a process of cell-cycle arrest that is characterized by a loss of proliferative capacity in mitotic cells (1–3). Senescence is triggered in numerous cell types in response to a range of stressors, including replicative, genotoxic, or oncogenic stresses. Studies have shown that overexpressing the oncogenes KRAS (Kirsten Rat Sarcoma Oncogene), or the induction of DNA double-strand breaks, either by ionizing radiation or other strong mitogenic signals, can induce senescence (4–8). Under certain conditions, when damage is irreparable, cells halt their progress through the cell cycle to limit the perpetuation of cellular damage (9, 10).
While seemingly beneficial from a tumor-prevention standpoint, senescence can induce critical problems to the overall well-being of organisms. The accumulation of senescent cells within aging tissues results in tissue dysfunctions (11, 12) that in part stem from soluble factors secreted by senescent cells, termed senescence-associated secretory phenotype (SASP)(7). A wide range of transcriptional changes accompanies senescence, including significant increases in the secretion of cytokines, chemokines, proteases, and growth factors (7, 13, 14). Utilizing in vivo and in vitro models, SASP was shown to stimulate the survival and proliferation of tumor cells (15–17), enable tumor cell invasion through ECM-degrading enzymes, e.g., MMPs (Matrix metalloproteinases) (17), and promote tumor-driven angiogenesis through the expression of vascular endothelial growth factor (VEGF) (18). Furthermore, SASP can drive epithelial-to-mesenchymal transition (EMT) (7, 19) and enable the dissemination and migration of cells to distal sites. Recently, we demonstrated that SASP promotes cancer cell migration by reorganizing cytoskeletal proteins through the modulation of RhoA activity (20). Since cytoskeletal proteins are implicated in the shaping of interphase nuclei (21), we asked whether SASP directly influenced nuclear architecture, mechanics, and associated gene expression.
Alterations and defects in nuclear morphology are associated with numerous human pathologies (22), including Hutchison–Gilford progeria syndrome (23), Werner syndrome, and more commonly, cancer (12, 24) and aging (12, 25, 26). Among the morphological changes observed in tumor cells, defects in nuclear morphology and architecture remain a key diagnostic feature of malignant cells (24, 27). These include changes in nuclear size and shape, as well as changes in chromatin organization. The spatial arrangement of the nucleus is maintained by the nuclear matrix, which consists of the nuclear lamina and a network of proteins and RNA (27). The most abundant nuclear matrix proteins are lamin A/C and lamin B, which compose the nuclear lamina. The nuclear lamina aids in organizing chromatin within the nucleus, where lamin and lamina-associated proteins bind directly to DNA sequences called lamina-associating domains (LADs) (28). Because the nuclear lamina directly interacts with chromatin, changes in nuclear shapes have been shown to alter chromatin organization and gene positioning, thereby influencing gene expression patterns and cellular function (29). The nuclear shape is tightly coupled to cytoskeletal elements via the linker of nucleoskeleton and cytoskeleton (LINC) complexes and can therefore be influenced by changes in intracellular (inside-out) and extracellular (outside-in) forces (30). This occurs through a process called mechanotransduction, where adhesion receptors such as integrins and cadherins are physically coupled to cytoskeletal filaments, linking the nuclear membrane to nucleoli, chromatin, and DNA (31).
Here, we show that SASP secreted by senescent stromal fibroblasts substantially alters the nuclear morphology of cancer cells. The nuclei of SASP-exposed cancer cells form deep invaginations and exhibit large-scale volume shrinkage. These changes in nuclear shapes, which are also accompanied by changes in underlying molecular patterns, are highly dependent on intact lamin A/C-mediated connections and the reduction of actomyosin tension. The loss of this cellular tension decreases the lateral pulling forces exerted on the nucleus, thereby leading to membrane relaxation and the formation of nuclear lobulations. We demonstrate that RhoA/ROCK-mediated actomyosin tension is both required and sufficient to induce nuclear-shape changes.
Results
SASP induces changes in nuclear morphology and volume of cancer cells
To assess the impact of SASP on the nuclear properties of carcinoma cells, we induced senescence in human WI-38 lung fibroblasts and aged human dermal fibroblast GT125 via exposure to bleomycin and collected the SASP-containing media (20). This conditioned media (CM) derived from senescent cells was added to human T47D epithelial breast carcinoma cells. Upon exposure, we observed and quantified cellular responses by way of changes in nuclear morphology. Prior to the addition of CM from senescent fibroblasts (denoted as Sen CM), most cancer cells displayed a round morphology, with less than 5% of cells displaying an elongated morphology. However, when exposed to Sen CM (48 h), approximately 70% of the cells developed an elongated cellular morphology, and 30% retained a rounded morphology (32).
To visualize real-time morphological changes in cells exposed to Sen CM, we first utilized phase-contrast microscopy (Figure 1a). Most cells switched from a rounded to an elongated morphology, with increased cell extensions and significant decreases in cell-substrate contact area and circularity (Figure 1b and c and Figure S1a and b). Furthermore, we observed an increase in the integrated intensity measured from phase contrast micrographs within the traced cell region (Figure 1a to c). This apparent optical change indicated that cells underwent significant changes in cell height along the z-direction, leading us to ask whether SASP exposure also influenced nuclear morphology.
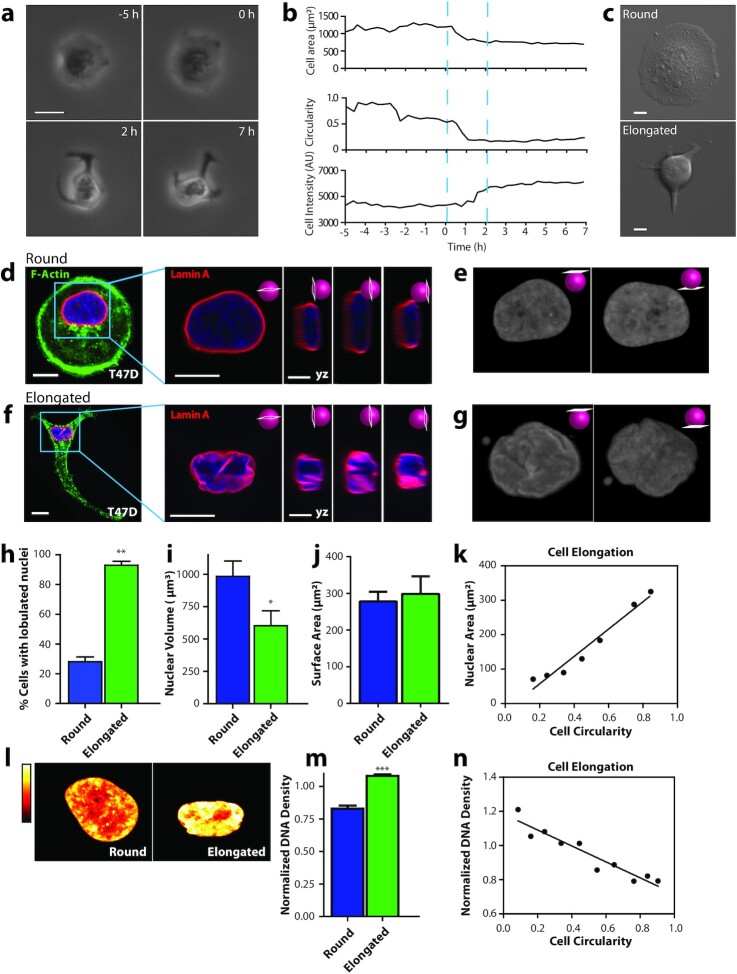
Figure 1.
SASP induces changes in cell and nuclear morphology. (a) Phase contrast micrographs of a T47D breast cancer cell undergoing a dramatic change in morphology induced by Sen CM. Scale bar, 20 µm. (b) Changes in cell area, cell circularity, and cell intensity for the cell shown in (a). (c) phase contrast micrographs showing examples of rounded and elongated T47D cells after exposure to Sen CM for 48 h (d and f) Fluorescence confocal micrographs of round (d) and elongated (f) T47D cells stained for F-actin (green), Lamin A (red), and nuclear DNA (blue) after exposure to Sen CM for 48 h. Blue squares indicate zoomed regions of T47D cells. Scale bar, 10 µm. Following panels are zoomed in xy and yz cross-sections of the left panel. These include the xy equatorial cross-section, yx left, equatorial, and right cross section respectively. Scale bar, 5 µm. (e and g) Surface rendering of lamin-A-stained nuclei in panels d and f. The left and right panels indicated top and bottom views of the nuclei, respectively. (h) Percentage of cells of round (blue) and elongated cell morphology with lobulated nuclei 48 h after exposure to Sen CM. Numbers of examined cells are n = 24 and n = 43 for round and elongated cells, respectively. (i) Nuclear volume of cells having round and elongated cell morphology using consequential z-slices; n = 8 and n = 10, respectively. (j) Nuclear surface area of round and elongated cells; n = 8 and n = 10, respectively. (k) Binned correlation between cellular circularity and nuclear area. The blue triangle indicates a decrease in cell circularity from left to right (elongated to round). (l) Intensities of DNA staining were color coded from high to low (white, yellow, orange, red, and black). Condensed regions have higher fluorescence intensity compared to the less condensed regions. (m) Normalized nuclear density between round and elongated cells. Normalization was performed among repeats (n = 87 and n = 157 for round and elongated cells, respectively). (n) Correlation between cellular circularity and normalized DNA density. The blue triangle indicates a decrease in cell circularity from left to right (elongated to round).
To better understand this, we collected z-stacks of fluorescent images using confocal microscopy and then reconstructed cells in 3D to visualize rounded versus Sen CM-induced elongated cells (Figure 1d to g). Cellular and nuclear morphologies were visualized based on F-actin and nuclear lamin-A staining. In the presence of Sen CM, the nuclei of rounded cells featured a smooth disk-like morphology, whereas the nuclei of elongated cells displayed large deformations with deep invaginations (Figure 1e and g). These nuclear deformations were readily visualized through y-z cross-sections and top/bottom-surface views of stained nuclei (right panels in Figure 1d and f). In the presence of Sen CM, we observed that ∼25% of rounded T47D cells displayed nuclear lobulations, while ∼90% of elongated cells displayed nuclear lobulations (Figure 1h). The extent of lobulations was determined based on the number of folds present as visualized via lamin A staining; nuclei with ≥ 2 nuclear folds were deemed lobulated (see the "methods" section and Supplementary Material). Similarly, nuclear deformations were observed in tumorigenic, noninvasive human breast epithelial MCF7, melanoma A375, and pancreatic PANC-1 cells exposed to Sen CM (Figure S1a and b and Figure S6a and b, d, and f to h).
In conjunction with lobulated nuclear morphology, Sen CM-induced elongated cells displayed a ∼50% reduction in their projected nuclear area (Figure S1c). To determine whether this reduction in nuclear area corresponded to a similar reduction in nuclear volume, we utilized a scanning-laser confocal and integrated nuclear areas of serially sectioned, GFP-lamin A-labeled nuclei (33). We measured a 40% reduction in nuclear volume in elongated cells compared with control round cells (Figure 1i). This volume decrease occurred with no significant changes in the surface area of the nucleus (Figure 1J), presumably because of the observed extensive folding and wrinkling of the nuclear envelope. Despite this large decrease in nuclear volume, the height of the nuclei of elongated cells significantly increased (Figure S1d), explaining the observed overall bulging observed under phase contrast microscopy when cells were exposed to Sen CM (Figure 1a to c).
As cells transitioned from a rounded to an elongated morphology, they displayed large decreases in cell contact area accompanied by the formation of cell extensions (Figure 1a to c). However, because nuclear shape changes seemed to occur synchronously with cellular shape changes, we next asked whether changes in cell morphology correlated with changes in nuclear morphology. We found that the projected nuclear area decreased as cell circularity decreased. i.e., rounder cells featured larger nuclei (Figure 1k). Similarly, larger cells featured larger nuclei (Figure S1e). This suggests that SASP-induced nuclear morphological changes may in part be driven by cellular morphology changes.
Large-scale changes in cell morphology have been shown to not only influence nuclear shape but also affect chromatin remodeling and compaction (34). To determine whether SASP-induced changes in intranuclear architecture, we examined the organization of chromatin utilizing DNA staining via Hoechst 33342. This DNA fluorochrome stoichiometrically stains DNA, allowing us to determine cell cycle status, as well as chromatin distribution and condensation (35, 36). Brightly stained regions indicate increased compaction, denoting heterochromatic regions, while dimly stained regions indicate euchromatic regions, denoting less compaction. A significant reorganization of chromatin accompanied nuclear deformation. Nuclei of elongated cells had regions with significantly higher DNA staining compared to control cells (Figure 1l). DNA density (total intensity of DAPI-stained DNA divided by nuclear area) was increased ∼25% in elongated cells compared to control cells (Figure 1m), which increased correlated with progressive elongation in cell morphology (Figure 1n).
Together, these results suggest that SASP-induced nuclear deformation and DNA condensation occur in conjunction with changes in cellular morphology.
Coupled mechanical forces generated by F-actin are required for nuclear shaping
The actin cytoskeletal network is a major mediator of nuclear remodeling processes (29, 37) in response to changes in cellular morphology (21). To determine whether Sen CM elicited changes in filamentous actin structures, we compared cells exposed to fresh medium (FM) and Sen CM. Cells exposed to Sen CM displayed reduced F-actin (Figure 2a to c), accompanied by the disappearance of the rich cortical actin structures observed in FM (Figure 2a and b).
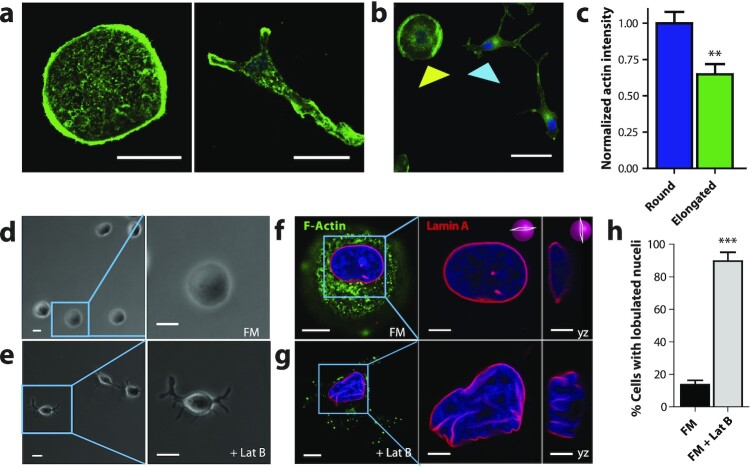
Figure 2.
Nuclear shape requires intact F-actin structures. (a to e) Fluorescence confocal and phase contrast micrographs of T47D cells after 48 h exposure to FM and FM + 250 µM of actin-depolymerizing drug Latruculin B, respectively, for 48 h. Cells are stained for F-actin (green), Lamin A (red), and nuclear DNA (blue). Scale bar, 10 µm. Panels to the right of f and g show zoomed in xy and yz cross-sections of the left panels. Scale bar 5 µm. (h) Percentage of cells featuring a lobulated nucleus 48 h after exposure to FM (black) or FM + Lat B (gray). Numbers of examined cells are n = 92 andn = 49 for FM and FM + Lat B stimulated cells, respectively.
To determine and probe the role of the actin cytoskeleton in regulating normal nuclear morphology, cells were treated with 250 nM of the small-molecule inhibitor of F-actin assembly, Latrunculin B (LatB) in FM for 1 h. Similar to Sen CM treatment, LatB-treated cells formed long, thick extensions together with significant bulging in the nuclear region (Figure 2e) relative to control cells in FM (Figure 2d). To visualize the effects of cellular morphological changes on nuclear morphology, cells were stained for F-actin and Lamin A (Figure 2f and g). In addition, LatB-treated cells formed gross nuclear lobulations with a similar proportion of cells having nuclear lobulations as cells exposed to Sen CM (Figure 2h). These findings indicate that LatB-dependent F-actin remodeling elicits similar nuclear morphology changes to Sen CM-treated cells.
Lamin A/C mediates SASP-induced nuclear changes
While B-type lamins are required for cell viability, A-type lamins (lamin A/C) are major nuclear scaffolding proteins, which are required for the maintenance of nuclear structure and organization. Lamin A/C plays a pivotal role in mechanotransduction, and the depletion of lamin A/C disrupts the physical connectivity between the cytoplasm and nucleoplasm in normal cells (38). LINC complex proteins (linkers of nucleoskeleton and cytoskeleton) are SUN and KASH-domain-containing proteins, which constitute mechanical tethers that span the nuclear envelope and connect a network of lamins and lamin-associated proteins in the nucleus to the cytoskeleton in the cytoplasm (29, 39). Hence, we asked whether SASP-induced nuclear deformations occurred due to the direct connection between the cytoskeleton and nucleoskeleton through lamin A/C.
We found that the expression and localization of lamin A, the nuclear envelope protein Emerin, and the LINC complex proteins SUN1, Nesprin2giant, and Nesprin3 were largely unchanged following exposure of cells to Sen CM (Figure S2a and b). These results suggest that Sen CM-induced nuclear invagination is not necessarily due to a loss in expression or changes in the localization of these key nuclear-envelope proteins.
Next, we depleted cells of lamin A/C and then exposed to Sen CM (Figure S3a). We visualized nuclear folds via staining of the nuclear-lamina-associated protein Lap2β. Interestingly, lamin A/C-depleted cells exposed to Sen CM failed to alter their nuclear shape (Figure 3a to d and Figure S3b to f). While 80% of control cells featured nuclear lobulations in response to Sen CM stimulation, less than 5% of lamin A/C-depleted cells formed nuclear lobulations in Sen CM (Figure 3e). Although the nuclei of lamin A/C-depleted cells did not form lobulations in FM, an increase in nuclear height was still observed. These results indicate that nuclear lamin proteins—lamin A/C and associated nucleoskeleton−cytoskeleton connections are required for the wrinkling process, i.e., without lamin A/C, the nucleus fails to wrinkle in response to SASP exposure.
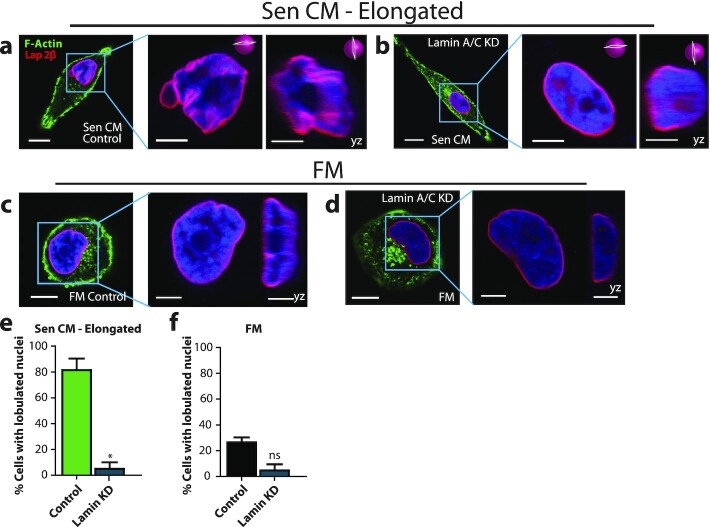
Figure 3.
Lamin A/C mediates SASP-induced nuclear changes. (a to d) Fluorescence confocal micrographs of control and lamin A/C knockdown cells after stimulation with Sen CM and FM for 48 h. Cells were stained with phalloidin, Lap2β, and Hoechst 33342 DNA stain. The following panels are zoomed in xy and yz cross-sections of the left panel. Scale bar 5 µm. These include the xy and yz equatorial cross-sections. (e) and (f) Percentage of control and lamin A/C knockdown cells having lobulated nuclei 48 h after exposure to Sen CM (e) and FM (f). n = 62, n = 33, n = 43, and n = 31 for Sen CM, Sen CM + lamin KD, FM, and FM + lamin KD, respectively.
Because lamin A/C contributes to the maintenance of the structural integrity of the nucleus, we also asked whether lamin A/C deficiency was sufficient to induce nuclear shape changes in FM. We compared the nuclear morphology of control and lamin A/C-depleted cells in FM. In both conditions, nuclei maintained a round morphology (Figure 3c, d, and f and Figure S3c and f). This suggests that, although lamin A/C is a major mediator of nuclear shape changes induced by SASP, the depletion of lamin A/C alone is not sufficient to produce SASP-induced changes in nuclear morphology.
Decreased cytoskeletal tension and cellular contractility drive nuclear shape changes
Cytoplasmic mechanical compliance (deformability) critically depends on nuclear–cytoskeletal connections (40). To measure cytoplasmic compliance, we utilized particle tracking microrheology (41). Carboxylate-modified 100-nm fluorescent polystyrene beads were ballistically injected into the cytoplasm of T47D cells. Since the sizes of the beads are significantly larger than the effective mesh size of cytoskeletal network (20 to 40 nm), the movements of beads reflect the viscoelastic properties of the cells in which they are embedded (42, 43). Cells were then exposed to Sen CM for 24 h. Using the time-dependent coordinates of the bead trajectories, we calculated the ensemble-averaged mean square displacement (MSD) of beads embedded within cells exposed to either FM or Sen CM (Figure 4a to d). We observed that particles lodged in the cytoplasm of Sen CM-exposed cells had significantly larger displacements at all measured time-lags compared to particles in round cells in FM (Figure 4e to h). Together, these results indicate that elongated cells exposed to Sen CM are more deformable than cells exposed to FM, which is consistent with reduced F-actin content in cells exposed to Sen CM (Figure 2a to c).
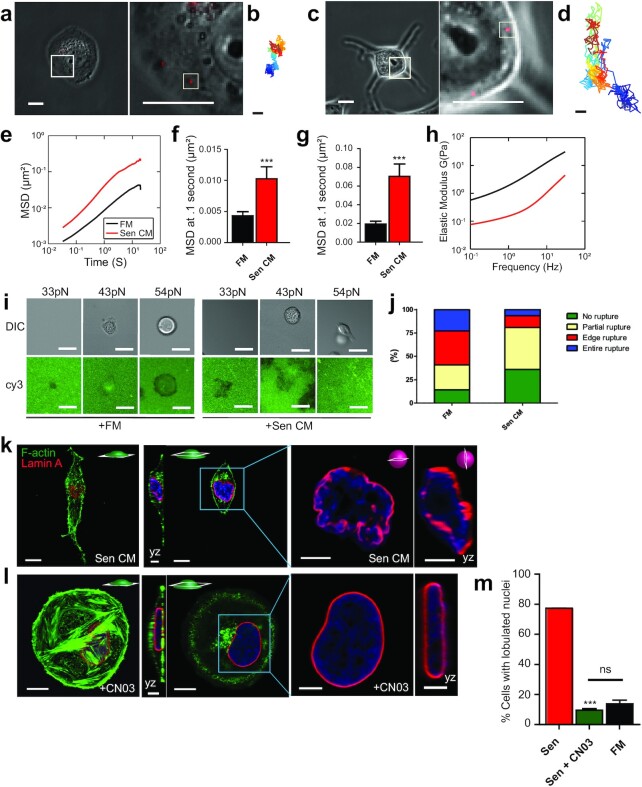
Figure 4.
SASP-induced nuclear shape is accompanied with decreased cell contractility and increased cell deformability. (a) and (c) Phase contrast micrographs of T47D cells bombarded with 100nm-diameter fluorescent beads. Post bombardment, cells were exposed to FM (a) and Sen CM (c) for 48 h. Scale bar, 20 µm. White squares indicate the regions of each cell expanded to better display the embedded fluorescent particles in the following panel. The second panel shows a zoomed view of 100-nm-diameter fluorescent beads within the cytoplasmic region of the cell in the previous figure. White squares indicate individual particles within each T47D cell. Scale bar, 5 µm. (b) and (d) Particle trajectories of beads indicated in the second panel of (a) where passage of time is indicated by color change. Beads were tracked with ∼10 nm spatial resolution and 33 ms temporal resolution for 20 s. (e) Population-averaged MSD of particles within the cytoplasm of round and elongated T47D cells as a function of time lag. The numbers of examined cells are n = 85 and n = 48, for FM and Sen CM conditions, respectively. (f) to (g) Population-averaged MSD evaluated at time lags of 0.1 s (f) and 1 s (g). (h) Computed elastic modulus of the cell exposed to FM or Sen CM. (i) FM and Sen CM-exposed T47D cell adhesion on TGT surfaces with different tension tolerances (33, 43 and 54 pN) after 3 h incubation. Images show adherent T47D cells on TGT surfaces and TGT rupture patterns. A loss of Cy3 signal indicates the molecular force larger than tension tolerance of each TGT was exerted by cells. (j) Percentage of rupture patterns of T47D cells exposed to FM or Sen CM. n = 92 for FM, n = 65 for Sen CM. Entire rupture indicates TGT rupture in the entire cell projected area. Edge rupture indicates TGT rupture at the cell periphery. Partial rupture indicates any other small ruptures underneath a cell. (k) Fluorescence confocal micrographs of T47D cells after 48 h exposure to Sen CM and Sen CM + 1.0 µg/ml Rho activator II CNO3. Cells were stained for F-actin (green), Lamin A (red), and nuclear DNA (blue). First and third panels are basel and equatorial xy cross section of imaged cells. Scale bar, 10 µm. Second panels display yz cross sections. Scale bar, 5 µm. Blue squares indicate zoomed regions of T47D cells in the following panels. The last panels are xy and yz equatorial cross sections of nuclei in previous panels. Scale bar, 5 µm. (i) Percentage of cells with lobulated nuclei 48 h after exposure to Sen CM (red; n = 53 tested cells), Sen CM + CN03 (green; n = 54), and FM (black; n = 92).
Cells exposed to Sen CM display decreased number and size of focal adhesions (Figure S4a and b) and decreased traction forces on the underlying matrix compared to control cells (20). For control cells, maximal traction forces, and adhesions are formed at the cellular cortex. Because traction forces exerted by cells are governed by a cell-intrinsic surface balance of contractile forces (44), we sought to determine whether elongated cells were less contractile than their round counterparts (45, 46). To assess cellular cortical tension at molecular levels, we employed the tension gauge tethers (TGT) assay (47) where a ligand for cell membrane receptors (RGD for integrins in this case) is tethered to a surface through a rupturable DNA tether. Polymer-passivated glass substrates coated with RGD-conjugated TGTs having tension tolerance Ttol of 33, 43 or 54pN were used. For Ttol = 33 pN, under exposure to both FM and Sen CM, cells did not adhere stably, and imaging of fluorescence Cy3 labels on the tether showed fluorescence loss, likely because the cells failed to experience > 40 pN forces needed to activate adhesion response through single integrin-ligand bonds due to TGT rupture (47). For Ttol = 43 pN, both FM and Sen CM-exposed cells adhered stably on the TGT surface as expected (47) and retained a round morphology. For Ttol = 54 pN, cells exposed to FM remained round with significant TGT rupture in the cell periphery or cell center) (Figure 4i and j and Figure S4d), indicating that cells can apply tension stronger than 54 pN, likely through large focal adhesions (48). In contrast, cells exposed to Sen CM developed an elongated cell morphology with minor ruptures mostly at the front and back extensions, supporting a reduction in cortical cell tension for cells exposed to Sen CM relative to FM.
Next, we asked whether increased contractility would prevent nuclear-shape changes. In nonmuscle cells, contractility is modulated via myosin II, primarily by phosphorylation of myosin regulatory light chain (49, 50). Myosin light chain (MLC) phosphorylation can occur directly via a myriad of proteins, including MLC kinase (MLCK) and Rho-associated coiled–coiled containing kinase (ROCK), an effector of RhoA GTPase (51). Therefore, stimulation of the Rho-Rock pathway induces myosin II-mediated contractility and tension (52). Hence, to increase cell tension and contractility, we increased RhoA activity by treating cells exposed to Sen CM with CN03 (constitutive activator of RhoA). In the presence of Sen CM and in response to increased RhoA activity, cells retained their rounded morphology, preserved prominent actin stress fibers and a high number of large focal adhesions, and prevented SASP-induced nuclear deformations (Figure 4k to m, Figure S4c).
These results suggest that the reduction in RhoA-based contractility mediates SASP-induced nuclear lobulations.
Reduction in cell contractility induces nuclear deformation
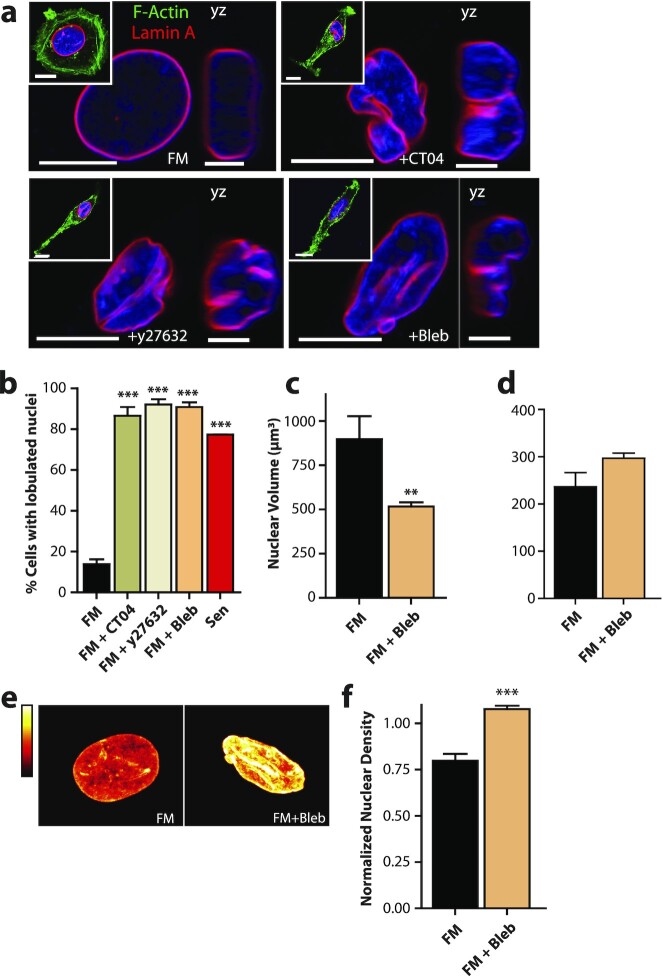
We asked whether the reduction in cellular tension and contractility alone was sufficient to recapitulate SASP-induced nuclear deformation. We treated cells with inhibitors of various proteins of the acto-myosin network, including CT04 (FM + CT04), Y27632 (FM + Y27632), and Blebbistatin (FM + Bleb) to inhibit RhoA, ROCK, and myosin II ATPase activity in FM, respectively. Treated cells not only acquired an elongated morphology (53), but also developed nuclear deformations and invaginations (Figure 5a). Cells with inhibited RhoA, ROCK, and myosin II activity had similar fractions of cells with lobulated nuclei as cells exposed to Sen CM (Figure 5b). This result was also consistent in cells treated with the microtubule destabilizer Nocodozole, with a rescue observed in cells first treated with Sen CM the 1μM Jasplankinolide for 1 h (Figure S6d and e)
Figure 5.
A reduction in cell contractility alone is sufficient to induce nuclear deformation following SASP stimulation. Fluorescence confocal micrographs of T47D cells after 48 h exposure to FM, FM + 2.0 µg/ml Rho Inhibitor I CT04, FM + 15 µM ROCK inhibitor Y27632, and FM + 25 µM myosin II inhibitor Blebbistatin (Bleb). Cells are stained for F-actin (green), Lamin A (red), and nuclear DNA (blue). Images display xy and yz equatorial cross-sections, respectively. Scale bar, 5 µm. Whole cell mid-plane images of are displayed as insets. Scale bar, 10 µm. (b) Percentage of single cells with lobulations when exposed to Sen CM (red; n = 53), FM (black; n = 92), FM + CT04 olive; n = 74), FM + Y27632 (beige; n = 38) and FM + Bleb (orange; n = 49) after 48 h. (c) Nuclear volume of T47D cells after 48 h exposure to FM and FM + 25 µM Blebbistatin—using consequential z-slices; n = 13 and n = 15, respectively. (d) Nuclear surface area of FM and FM + Blebbistatin stimulated cells; n = 13 and n = 15, respectively. (e) Intensities of DNA staining per condition were color coded from high to low (white, yellow, orange, red, and black). Condensed regions have higher fluorescence intensity compared to the less condensed regions. (f) Normalized nuclear density between FM and FM + blebbistatin stimulated cells. Normalization was performed among repeats (n = 43 and n = 112 for FM and FM + Blebbistatin stimulated cells, respectively). (g) and (h) Bar plots showing the transcriptional programs for overlapping upregulated (g) and downregulated (h) genes between Sen CM and FM + Bleb treated cells.
Since myosin II is a direct regulator of cellular tension, we asked whether Blebbistatin treatment could recapitulate the phenotypic effects of Sen CM. In line with previous reports that cells with pronounced myosin contractility typically harbor large focal adhesion complexes (50), we observed decreased staining of focal adhesion protein vinculin after blebbistatin treatment (Figure S5a and b). Furthermore, we found that blebbistatin-treated cells displayed a similar decrease in cell spreading and nuclear area (Figure S5c and d) as Sen CM-stimulated cells (20).
Next, measuring nuclear volume and nuclear surface area, we observed that myosin II inhibition induced a similar reduction in nuclear volume without a significant change in nuclear surface area, with a compensating increase in nuclear height, indicating similar nuclear wrinkling and bulging phenotypes (Figure 5c and d and Figure S5e) to Sen CM-exposed cells. Furthermore, measuring chromatin compaction after myosin II inhibition, we observed the characteristic increase in nuclear density via H33342 staining (Figure 5e and f). Lastly, we wondered whether external modulation of cytoskeletal tension via substrate stiffness also induced nuclear deformations upon treatment with Sen CM. To test this, we seeded T47D cells onto either glass or 25 kPa polyacrylamide gels, and exposed them to the FM of Sen CM. We observed that even on softer substrates, Sen CM also induced nuclear deformations and invaginations (Figure S6c and d).
Together these results point to a notion that the inhibition of intracellular tension and contractility are necessary (Figure 4) and sufficient (Figure 5,and Figure S6) to recapitulate the nuclear phenotypes induced by Sen CM.
Discussion
SASP produced by senescent fibroblasts in the stromal space and nearby connective tissues of tumors can alter the cellular phenotypes of neighboring cancer cells. These phenotypes include enhanced invasion and proliferation, as well as EMT (7, 15, 16, 54, 55). Recently, we have demonstrated that SASP stimulation facilitates the reorganization of cytoskeletal proteins, which induces onset of migration in natively nonmotile cancer cells (20). Here, we observe that the nuclei of Sen CM-stimulated cells display large-scale lobulations and deep nuclear invaginations. The nuclei of Sen CM-stimulated cells also display a large decrease in volume accompanied by an increased height with unchanged surface area (33). This indicates a wrinkled nuclear phenotype with contraction in the lateral direction but an expansion in the normal direction, thereby suggesting a loss of lateral and apical mechanical support. Along with nuclear shape changes, SASP stimulation promotes chromatin reorganization and condensation. These SASP-stimulated alterations in nuclear shape and chromatin compaction correlate well with global cell morphology changes, indicating that these events occur concurrently.
SASP-mediated nuclear lobulations and invagination, and the associated decreased in nuclear volume, critically depend on F-actin assembly and actomyosin contractility. Numerous studies have demonstrated that cell shape information is transduced into nuclear shape changes via the physical transmission of mechanical forces through the actin cytoskeleton (21, 37, 56). External forces distort specific adhesion molecules such as integrins, which link to actin microfilaments through focal adhesions (57). In fibroblasts and several other types of normal cells, nuclear shape modulation via actin is mediated in large part by a dome-like contractile actomyosin structure called the perinuclear actin cap (21, 30, 38). Dorsal actin fibers associate with LINC complex proteins at the apical surface and sides of the nucleus and promote dynamic anchoring between the cytoskeleton and the nuclear interior through lamin A/C (21, 58, 59). However, apical cross sections of both round and elongated cancer cells displayed punctate actin structures on the top and sides of nuclei, with no apparent nucleus-associated fiber structures resembling the perinuclear actin cap. However, nuclear shape is also regulated by central actin filaments pulling laterally on the nuclear membrane via anchor proteins and terminating with adhesion proteins (37, 45). To assess the role of actin cytoskeleton, we first determined the role of nucleus-cytoskeletal connection in regulating SASP-stimulated nuclear deformation, we depleted cells of lamin A/C and assessed the response. While Sen CM exposure leads to a reorganization of the cytoskeleton in both control and lamin-depleted cells, lamin A/C-depleted cancer cells in FM failed to modulate their nuclear shape, unlike normal cells. This suggests that lamin A/C and intact nucleus-cytoskeletal connections are required for the formation of nuclear invaginations in cancer cells in response to SASP stimulation. This also suggests that cell shapes and cytoskeletal forces are transmitted into nuclear deformations via lamin A/C, further strengthening the paradigm that transmitted extracellular forces influence nuclear morphology and chromatin organization.
While intact nucleus-cytoskeletal connections are required for the transmission of forces from the cytoskeleton to the nuclear interior, nuclear shape changes occur through the fluctuations and changes in intracellular and extracellular forces (29, 56, 60). Because extracellular forces remained unchanged between FM and Sen CM-stimulated conditions, we measured the intracellular mechanics of the cells. Utilizing particle-tracking microrheology, we determined that Sen CM stimulated cells were considerably more deformable than control cells. We also observed reduced number and size of focal adhesions (20) and a significant reduction in traction forces in Sen CM (20). Together, these results indicate that Sen SC stimulation facilitates the loss in cellular tension and contractility. Therefore, we speculated that this decreased actomyosin force-profiles acting on the nucleus may result in the nuclear wrinkling and formation of invaginations. To evaluate this, we stimulated cellular contractility/tension by the constitutive activation of upstream regulator of myosin II, Rho A. With RhoA stimulation, nuclei remained round and disk-shaped in the presence of Sen CM. This result suggests that the loss of tension/contractility is required for SASP induced nuclear deformation.
We asked whether a loss in cell tension/contractility was sufficient to promote a nuclear phenotype similar to SASP stimulation. To evaluate this, we inhibited Rho/ROCK/myosin II activity and assessed nuclear morphology. Inhibition of cell tension was sufficient to recapitulate nuclear phenotypes similar to SASP stimulation, where nuclei developed a similar wrinkling and chromatin condensation. Together, these suggests that the decrease in cell tension/contractility are both necessary and sufficient to induce SASP-induced nuclear deformation.
To explain these findings, we propose a model by which actomyosin tension exerted on the nucleus mediated by lamin A/C together with alterations in nuclear-cytoplasmic force balance creates an outside-in physical signaling cascade. With control cells (FM), the actomyosin forces pull laterally on the nucleus and flattens it into a disk shape (Figure S6g). Here, actomyosin forces are balanced by mature focal adhesions, enabling a rounded cellular morphology. However, SASP stimulation reduces RhoA activity (20). This in turn reduced myosin II-mediated contractility, as observed by the reduction of cell traction forces (20) and cell adhesion. Because, RhoA mediates the maturation and turnover of focal adhesions through myosin II (61), the reduction of cytoskeletal tension and reduction in cellular adhesions promote a decrease in the actomyosin forces laterally pulling on the nucleus. Moreover, reduction in cytoskeletal tension also alters the nucleoplasm-cytoplasm mass transport and pressure difference, which results in the loss of water and protein content in the nucleus and the formation of folds and invaginations within the nucleus walls (33). This reduction in nuclear mass helps to propagate the observed alterations in cellular and nuclear mechanics. These factors together trigger a cascade that leads to the force-dependent changes in projected gene expression profiles.
Lastly, these results paint a mechanistic picture for the role of SASP in driving phenotypic changes and nuclear deformations in cancer cells via cytoskeletal tension. However, there are still many unanswered questions that require attention. For instance, it is unclear whether and how these results translate to cells embedded within 3D matrices, or what specific factors within the SASP drive these effects. Based on published data on SASP compositions, and data generated within this paper, we have some clues, pointing to a potential role of proinflammatory interleukins (Figure S6i). For instance, comparing the secretions of nonsenescent and senescent dermal fibroblasts indicates a heightened expression of IL6, IL9, and IL13. However, IL8 and IL5 show a differential expression with IL8 having a more than 2-fold increase, and IL5 having a ∼0.5-fold increase in Sen CM compared to nonsenescent media. While interesting, mechanistic studies are needed to further decipher what molecules drive this SASP-associated nuclear deformations.
Materials and Methods
Cell culture
T47D, MCF-7, A375, and PANC-1 cells (ATCC) were cultured in RPMI 1640 medium (ATCC) or DMEM, respectively, supplemented with 10% (v/v) fetal bovine serum (Hyclone Standard) and 1% (v/v) Penicillin/streptomycin (Sigma). Wi-38 (ATCC) cell were cultured in Eagle's minimal essential medium (EMEM; ATCC) supplemented with 10% (v/v) fetal bovine serum (Hyclone Standard) and 1% (v/v) penicillin/streptomycin (Sigma). GT125 primary human dermal fibroblast (89-year-old Caucasian male) were cultured in DMEM supplemented with 15% (v/v) fetal bovine serum (Hyclone Standard) and 1% (v/v) penicillin/streptomycin (Sigma). All cells were maintained at 37°C and 5% CO2 in a humidified environment during culture and imaging. For experiments, cancer cells were seeded onto collagen-I (Corning) coated glass-bottom dishes (50 µg/ml). For experiments on softer substrates, cells were seeded onto commercial 25 kPa polyacrylamide hydrogels on coverslips (Matrigen).
Senescence induction of human fibroblasts to generate senescence CM
Human fibroblasts lines (WI-38 and GT125) were exposed to 50 µM Bleomycin for 4 h in cell culture medium supplemented with FBS and P/S. After 4 h, cells were washed three times with 1× PBS to remove Bleomycin and fresh media was added to each dish. After 7 additional days of culture, senescence was verified based on ki67 staining, cell counting to determine proliferation rates over a 4-day period, the presence of gamma H2AX foci indicating DNA damage, and in some cases the expression of senescence associate beta galactosidase. To generate CM, 100,000 senescent/nonsenescent cells were seeded onto 10 cm dishes in 10 ml of fresh media. Cell were cultures for 48 h, after which media was collected, spun to remove large debris, then frozen at -80 degrees. For experiments, frozen media was thawed, diluted 50:50 with fresh media, then used for cell exposures per condition.
Drug treatments
The microtubule destabilizer nocodazole (Sigma), the F-actin disassembly drug latrunculin B (Sigma), the nonmuscle myosin II inhibitor Blebbistatin (Sigma), the ROCK inhibitor Y27632 (Sigma), the Rho inhibitor CT04 (Cytoskeleton Inc.), and the Rho activator CN03 (Cytoskeleton Inc.), the actin polymerization drug Jasplakinolide (Sigma) were diluted from stock using culture medium. Latrunculin B was used final concentration of 250 nM; Blebbistatin was used at a final concentration of 25 mM; Y27632 was used at a final concentration of 15 mM; CT04 was used at a final concentration of 2 µg/ml; CN03 was used at a final concentration of 1 µg/ml; Nocodazole was used at a final concentration of 1 uM; Jasplakinolide was used at a final concentration of 1 µM. For Nocodazole and Jasplakinolide studies, cells were first incubated 48 h with FM or CM, and then treated for 1 h with Nocodazole or Jasplakinolide.
Immunofluorescence
To visualize Lamin A, microtubules, and F-actin, cells were fixed in 4% paraformaldehyde (Electronic Microscopy Systems) for 10 min and permeabilized with 0.1%Triton X-100 (Fisher Chemicals) for 10 min. Cells were blocked at room temperature for 1 h in 1% BSA (Sigma). Subsequently, cells were incubated in primary antibodies for 1 h at room temperature. Antibodies used included: Rb. anti-lamin A (1:50; ab16667 Abcam), sMs. Anti-vinculin (1:400; Sigma). Cells were then incubated with either Alexa 568 conjugated donkey anti-mouse or anti-rabbit secondary antibodies, Hoechst 33342 and phalloidin for 1 h at room temperature, after which cells were washed with PBS. Fluorescent images were collected after 48 h exposure to FM or CM conditions. Cells were imaged using a Nikon A1 or a Leica Stellaris-5 confocal microscope with a 63X oil-immersion lens.
Tension gauge tether (TGT) assay
The TGT assay was utilized to estimate molecular tension on a single integrin-ligand bond during cell adhesion and spreading of T47D as described previously (47). In brief, a cyclic peptide RGDfK (cRGDfK) ligand and a Cy3 dye were conjugated to each end of an 18-nucleotide single-stranded DNA (5-/Cy3/GGC CCG CAG CGA CCA CCC/cRGDfK/-3). The ssDNA was hybridized with a biotin-tagged complementary ssDNA (5-/GGG TGG TCG CTG CGG GCC/-3) at distinct locations, resulting in a double-stranded DNA with different tension tolerances (Ttol = 33, 43, and 54 pN). Then, 1 µM of each TGTs was immobilized on a PEGylated glass slide via a neutravidin-biotin interaction. Next, T47D cells at the density of 105 cells/ml were seeded on the TGT surfaces with either the FM or the conditioned medium from the senescent cells (Sen CM). After 3 h incubation at 37°C, the cells were fixed with 4% para-formaldehyde. The fixed cells and TGT rupture patterns induced by the cells were imaged by an epi-fluorescence microscope (Nikon Ti-E, Nikon Inc.). The number of adherent cells on each TGT surface and the rupture patterns were further analyzed using ImageJ.
Nuclear volume
The nuclear volume for T47D cells under the various conditions was measured from randomly selected cells stained with Hoechst 33342. Confocal microscopy was used to image nuclei by 0.3 µm z-stacks. Nuclear area per each cell was traced using NIS-Elements image analysis software (Nikon) and nuclear volume was then calculated by summing the nuclear area per 0.3 µm z-stack.
Image analysis
Image analysis was performed using a customized MATLAB program to segment cell and nuclear boundaries using phalloidin actin stain and Hoechst 33342 DNA stain, respectively. Fluorescence intensity was quantified per pixel within the segmented region. Nuclear density was calculated by summing the total DNA intensity within the traced nuclear region divided by the nuclear area. Cellular circularity was calculated as 4πA/P2. Where A is the area and P is the perimeter of the measured cell. A Fraction of cells with lobulated nuclei were manually scored based on 3D z-stacks of single cells. Cells having two or more nuclear invaginations were deemed lobulated.
Microrheology
T47D cells were plated overnight and allowed to adhere. Particle-tracking measurements are detailed in Wu et al (41).
Statistical analysis
The number of cells and biological repeats (n) for all experiments are indicated in the figure captions. Mean values, SE and all statistical analyses were calculated and plotted using Graphpad Prism 9 (Graphpad Software, San Diego, CA, USA). One-way ANOVA and unpaired t tests were conducted to determine significance, depending on the number of variables assessed. Statistical significance (p) was indicated within the figures using the following scale: *** for P < 0.001, ** for P < 0.01, and * for P < 0.05. All experiments were performed in duplicate of triplicates, with an average of 20 to 25 cells imaged per replicate, unless otherwise stated. The numbers of cells per condition are denoted in the figure captions.
Supplementary Material
ACKNOWLEDGEMENTS
T.H. is an investigator of the Howard Hughes Medical Institute. We thank Dr. Denis Wirtz for the critical discussions and feedback on the results that helped shape the final manuscript. This manuscript was posted as a preprint on BioRxiv: https://doi.org/10.1101/2021.11.17.468902
Notes
Competing Interest: The authors declare no competing interests.
Contributor Information
Ivie Aifuwa, Johns Hopkins Physical Sciences - Oncology Center, Institute for Nanobiotechnology, Johns Hopkins University, Baltimore, MD 21218, USA; Department of Chemical and Biomolecular Engineering, Johns Hopkins University, Baltimore, MD 21218, USA.
Byoung Choul Kim, Department of Biomedical Engineering, Johns Hopkins University, Baltimore, MD 21218, USA; Department of Biophysics and Biophysical Chemistry, Johns Hopkins School of Medicine, Baltimore, MD 21205, USA; Division of Nano-Bioengineering, Incheon National University, Incheon 22012, South Korea.
Pratik Kamat, Johns Hopkins Physical Sciences - Oncology Center, Institute for Nanobiotechnology, Johns Hopkins University, Baltimore, MD 21218, USA; Department of Chemical and Biomolecular Engineering, Johns Hopkins University, Baltimore, MD 21218, USA.
Bartholomew Starich, Johns Hopkins Physical Sciences - Oncology Center, Institute for Nanobiotechnology, Johns Hopkins University, Baltimore, MD 21218, USA; Department of Chemical and Biomolecular Engineering, Johns Hopkins University, Baltimore, MD 21218, USA.
Anshika Agrawal, Johns Hopkins Physical Sciences - Oncology Center, Institute for Nanobiotechnology, Johns Hopkins University, Baltimore, MD 21218, USA; Department of Chemical and Biomolecular Engineering, Johns Hopkins University, Baltimore, MD 21218, USA.
Derin Tanrioven, Johns Hopkins Physical Sciences - Oncology Center, Institute for Nanobiotechnology, Johns Hopkins University, Baltimore, MD 21218, USA.
Teresa R Luperchio, Department of Biological Chemistry, Johns Hopkins University, Baltimore, MD 21218, USA.
Angela M Jimenez Valencia, Johns Hopkins Physical Sciences - Oncology Center, Institute for Nanobiotechnology, Johns Hopkins University, Baltimore, MD 21218, USA; Department of Chemical and Biomolecular Engineering, Johns Hopkins University, Baltimore, MD 21218, USA.
Tania Perestrelo, Johns Hopkins Physical Sciences - Oncology Center, Institute for Nanobiotechnology, Johns Hopkins University, Baltimore, MD 21218, USA; Department of Chemical and Biomolecular Engineering, Johns Hopkins University, Baltimore, MD 21218, USA.
Karen Reddy, Department of Biological Chemistry, Johns Hopkins University, Baltimore, MD 21218, USA.
Taekjip Ha, Department of Biomedical Engineering, Johns Hopkins University, Baltimore, MD 21218, USA; Department of Biophysics and Biophysical Chemistry, Johns Hopkins School of Medicine, Baltimore, MD 21205, USA; Howard Hughes Medical Institute, Baltimore, MD 21205, USA.
Jude M Philip, Johns Hopkins Physical Sciences - Oncology Center, Institute for Nanobiotechnology, Johns Hopkins University, Baltimore, MD 21218, USA; Department of Chemical and Biomolecular Engineering, Johns Hopkins University, Baltimore, MD 21218, USA; Department of Biomedical Engineering, Johns Hopkins University, Baltimore, MD 21218, USA; Department of Oncology, Sidney Kimmel Cancer Center, Johns Hopkins School of Medicine, Baltimore, MD 21205, USA.
Funding
This work was supported in part by the National Institutes of Health Grants U01AG060903 (JMP), a National Science Foundation grant PHY1430124 (to T.H.), and by the Johns Hopkins University Older Americans Independence Center of the National Institute on Aging (NIA) under award number P30AG021334 (JMP).
Authors' Contributions
I.A. and J.M.P. conceived and designed the study. I.A., B.C.K., P.K., B.S., A.A., D.T., T.L., A.J., and T.P. performed the experiments. I.A, J.M.P., P.K., B.S., B.C.K., T.L., and K.R. analyzed the results. J.M.P., K.R., and T.H. supervised the study. I.A., J.M.P., B.C.K., and T.H. wrote and edited the manuscript.
Data Availability
The authors declare that all the data supporting the findings of this study are available within the paper and its Supplementary Material.
References
- 1. Hayflick L. 1965. The limited in vitro lifetime of human diploid cell strains. Exp Cell Res. 37:614–636. [DOI] [PubMed] [Google Scholar]
- 2. van Deursen JM. 2014. The role of senescent cells in ageing. Nature. 509:439–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Di Micco R, Krizhanovsky V, Baker D, d'Adda di Fagagna F. 2021. Cellular senescence in ageing: from mechanisms to therapeutic opportunities. Nat Rev Mol Cell Biol. 22:75–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ben-Porath I, Weinberg RA. 2005. The signals and pathways activating cellular senescence. Int J Biochem Cell Biol. 37:961–976. [DOI] [PubMed] [Google Scholar]
- 5. Braig M, Schmitt CA. 2006. Oncogene-induced senescence: putting the brakes on tumor development. Cancer Res. 66:2881–2884. [DOI] [PubMed] [Google Scholar]
- 6. Campisi J. 2005. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell. 120:513–522. [DOI] [PubMed] [Google Scholar]
- 7. Coppe JP, et al. 2008. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 6:2853–2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lombard DB, et al. 2005. DNA repair, genome stability, and aging. Cell. 120:497–512. [DOI] [PubMed] [Google Scholar]
- 9. Campisi J. 2001. Cellular senescence as a tumor-suppressor mechanism. Trends Cell Biol. 11:S27–S31. [DOI] [PubMed] [Google Scholar]
- 10. Coppe JP, et al. 2008. Senescence-associated secretory phenotypes reveal nell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. Plos Biol. 6:2853–2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Baker DJ, et al. 2011. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature. 479:232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Phillip JM, Aifuwa I, Walston J, Wirtz D.. 2015. The Mechanobiology of Aging. Annu Rev Biomed Eng. 17:113–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Freund A, Orjalo AV, Desprez PY, Campisi J. 2010. Inflammatory networks during cellular senescence: causes and consequences. Trends Mol Med. 16: 238–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Basisty N, et al. 2020. A proteomic atlas of senescence-associated secretomes for aging biomarker development. PLoS Biol. 18:e3000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Krtolica A, Parrinello S, Lockett S, Desprez PY, Campisi J.. 2001. Senescent fibroblasts promote epithelial cell growth and tumorigenesis: a link between cancer and aging. Proc Nat Acad Sci USA. 98:12072–12077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu D, Hornsby PJ. 2007. Senescent human fibroblasts increase the early growth of xenograft tumors via matrix metalloproteinase secretion. Cancer Res. 67:3117–3126. [DOI] [PubMed] [Google Scholar]
- 17. Parrinello S, Coppe JP, Krtolica A, Campisi J.. 2005. Stromal-epithelial interactions in aging and cancer: senescent fibroblasts alter epithelial cell differentiation. J Cell Sci. 118:485–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Coppe JP, Kauser K, Campisi J, Beausejour CM. 2006. Secretion of vascular endothelial growth factor by primary human fibroblasts at senescence. J Biol Chem. 281:29568–29574. [DOI] [PubMed] [Google Scholar]
- 19. Laberge RM, Awad P, Campisi J, Desprez PY.. 2012. Epithelial-mesenchymal transition induced by senescent fibroblasts. Cancer Microenviron. 5:39–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Aifuwa I, et al. 2015. Senescent stromal cells induce cancer cell migration via inhibition of RhoA/ROCK/myosin-based cell contractility. Oncotarget. 6:30516–30531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Khatau SB, et al. 2009. A perinuclear actin cap regulates nuclear shape. Proc Natl Acad Sci U S A. 106:19017–19022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wu PH, et al. 2015. Evolution of cellular morpho-phenotypes in cancer metastasis. Sci Rep. 5:18437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Scaffidi P, Misteli T. 2005. Reversal of the cellular phenotype in the premature aging disease Hutchinson-Gilford progeria syndrome. Nat Med. 11:440–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Webster M, Witkin KL, Cohen-Fix O.. 2009. Sizing up the nucleus: nuclear shape, size and nuclear-envelope assembly. J Cell Sci. 122:1477–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Phillip JM, et al. 2017. Biophysical and biomolecular determination of cellular age in humans. Nat Biomed Eng. 1:0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Phillip JM, Han K-S, Chen W-C, Wirtz D, Wu P-H. 2021. A robust unsupervised machine-learning method to quantify the morphological heterogeneity of cells and nuclei. Nat Protoc. 16:754–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zink D, Fischer AH, Nickerson JA. 2004. Nuclear structure in cancer cells. Nat rev Cancer. 4:677–687. [DOI] [PubMed] [Google Scholar]
- 28. Luperchio TR, Wong XR, Reddy KL. 2014. Genome regulation at the peripheral zone: lamina associated domains in development and disease. Curr Opin Genet Dev. 25:50–61. [DOI] [PubMed] [Google Scholar]
- 29. Dahl KN, Ribeiro AJS, Lammerding J. 2008. Nuclear shape, mechanics, and mechanotransduction. Circ Res. 102:1307–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim JK, et al. 2017. Nuclear lamin A/C harnesses the perinuclear apical actin cables to protect nuclear morphology. Nat Commun. 8:2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang N, Tytell JD, Ingber DE. 2009. Mechanotransduction at a distance: mechanically coupling the extracellular matrix with the nucleus. Nat rev Mole cell biol. 10:75–82. [DOI] [PubMed] [Google Scholar]
- 32. Aifuwa I, et al. 2015. Senescent stromal cells induce cancer cell migration via inhibition of RhoA/ROCK/myosin-based cell contractility. Oncotarget. 6:30516–30531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim DH, et al. 2015. Volume regulation and shape bifurcation in the cell nucleus. J Cell Sci. 128:3375–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Webster M, Witkin KL, Cohen-Fix O. 2009. Sizing up the nucleus: nuclear shape, size and nuclear-envelope assembly. J Cell Sci. 122:1477–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Darzynkiewicz Z. 2010. Critical aspects in analysis of cellular DNA content. Curr Protoc Cytom. Chapter 7, Unit7 2, doi:10.1002/0471142956.cy0702s52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vergani L, Grattarola M, Nicolini C. 2004. Modifications of chromatin structure and gene expression following induced alterations of cellular shape. Int J Biochem Cell Biol. 36: 1447–1461. [DOI] [PubMed] [Google Scholar]
- 37. Jean RP, Gray DS, Spector AA, Chen CS. 2004. Characterization of the nuclear deformation caused by changes in endothelial cell shape. J Biomech Eng. 126:552–558. [DOI] [PubMed] [Google Scholar]
- 38. Kim DH, Wirtz D. 2015. Cytoskeletal tension induces the polarized architecture of the nucleus. Biomaterials. 48:161–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Alam S, Lovett DB, Dickinson RB, Roux KJ, Lele TP. 2014. Nuclear forces and cell mechanosensing. Prog Mol Biol Transl. 126:205–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stewart-Hutchinson PJ, Hale CM, Wirtz D, Hodzic D. 2008. Structural requirements for the assembly of LINC complexes and their function in cellular mechanical stiffness. Exp Cell Res. 314:1892–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wu PH, et al. 2012. High-throughput ballistic injection nanorheology to measure cell mechanics. Nat Protoc. 7:155–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hale CM, Sun SX, Wirtz D. 2009. Resolving the role of actoymyosin contractility in cell microrheology. PLoS One. 4:e7054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wirtz D 2009. Particle-tracking microrheology of living cells: principles and applications. Annu Rev Biophys. 38:301–326. [DOI] [PubMed] [Google Scholar]
- 44. Chicurel ME, Chen CS, Ingber DE. 1998. Cellular control lies in the balance of forces. Curr Opin Cell Biol. 10:232–239. [DOI] [PubMed] [Google Scholar]
- 45. Chancellor TJ, Lee J, Thodeti CK, Lele T. 2010. Actomyosin tension exerted on the nucleus through nesprin-1 connections influences endothelial cell adhesion, migration, and cyclic strain-induced reorientation (vol 99, pg 115, 2010). Biophys J. 99: 1329–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sims JR, Karp S, Ingber DF. 1992. Altering the cellular mechanical force balance results in integrated changes in cell, cytoskeletal and nuclear shape. J Cell Sci. 103:1215–1222. [DOI] [PubMed] [Google Scholar]
- 47. Wang X, Ha T. 2013. Defining single molecular forces required to activate integrin and notch signaling. Science. 340:991–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang X, et al. 2015. Integrin molecular tension within motile focal adhesions. Biophys J. 109:2259–2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ikebe M, Hartshorne DJ. 1985. Phosphorylation of smooth muscle myosin at two distinct sites by myosin light chain kinase. J Biol Chem. 260:10027–10031. [PubMed] [Google Scholar]
- 50. Riento K, Ridley AJ.. 2003. Rocks: multifunctional kinases in cell behaviour. Nat Rev Mol Cell Biol. 4:446–456. [DOI] [PubMed] [Google Scholar]
- 51. Vicente-Manzanares M, Ma X, Adelstein RS, Horwitz AR. 2009. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat Rev Mol Cell Biol. 10:778–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chrzanowska-Wodnicka M, Burridge K. 1996. Rho-stimulated contractility drives the formation of stress fibers and focal adhesions. J Cell Biol. 133:1403–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Stroka KM, et al. 2017. Loss of giant obscurins alters breast epithelial cell mechanosensing of matrix stiffness. Oncotarget. 8:54004–54020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Coppe JP, et al. 2010. A human-like senescence-associated secretory phenotype is conserved in mouse cells dependent on physiological oxygen. PLoS One. 5:e9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ohuchida K, et al. 2004. Radiation to stromal fibroblasts increases invasiveness of pancreatic cancer cells through tumor-stromal interactions. Cancer Res. 64:3215–3222. [DOI] [PubMed] [Google Scholar]
- 56. Versaevel M, Grevesse T, Gabriele S. 2012. Spatial coordination between cell and nuclear shape within micropatterned endothelial cells. Nat Commun. 3:671. [DOI] [PubMed] [Google Scholar]
- 57. Zaidel-Bar R, Itzkovitz S, Ma'ayan A, Iyengar R, Geiger B. 2007. Functional atlas of the integrin adhesome. Nat Cell Biol. 9:858–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Luxton GW, Gomes ER, Folker ES, Vintinner E, Gundersen GG. 2010. Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science. 329:956–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Luxton GW, Gomes ER, Folker ES, Worman HJ, Gundersen GG. 2011. TAN lines: a novel nuclear envelope structure involved in nuclear positioning. Nucleus. 2:173–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Neelam S, et al. 2015. Direct force probe reveals the mechanics of nuclear homeostasis in the mammalian cell. Proc Natl Acad Sci USA. 112: 5720–5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Geiger B, Bershadsky A, Pankov R, Yamada KM. 2001. Transmembrane crosstalk between the extracellular matrix—cytoskeleton crosstalk. Nat rev Mol cell biol. 2:793–805. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all the data supporting the findings of this study are available within the paper and its Supplementary Material.