SUMMARY
Blood-feeding mosquitoes survive by feeding on nectar for metabolic energy but require a blood meal to develop eggs. Aedes aegypti females must accurately discriminate blood and nectar because each meal promotes mutually exclusive feeding programs with distinct sensory appendages, meal sizes, digestive tract targets, and metabolic fates. We investigated the syringe-like blood-feeding appendage, the stylet, and discovered that sexually dimorphic stylet neurons taste blood. Using pan-neuronal calcium imaging, we found blood is detected by four functionally distinct stylet neuron classes, each tuned to specific blood components associated with diverse taste qualities. Stylet neurons are insensitive to nectar-specific sugars and respond to glucose only in the presence of additional blood components. The distinction between blood and nectar is therefore encoded in specialized neurons at the very first level of sensory detection in mosquitoes. This innate ability to recognize blood is the basis of vector-borne disease transmission to millions of people world-wide.
Keywords: Aedes aegypti, mosquito, stylet, labrum, labium, taste, sexual dimorphism, blood-feeding behavior, nectar-feeding behavior, ionotropic receptors, gustatory receptors, GCaMP calcium imaging, Q-system, chemogenetics
INTRODUCTION
Animals actively obtain energy from protein and carbohydrates in food, which are distinguished by their savory (“umami”) or sweet taste, respectively (Liman et al., 2014; Yarmolinsky et al., 2009). These two taste qualities signal different nutritional values, and animals use diverse strategies to prioritize ingestion of the food source that best matches their current metabolic requirements. For feeding specialists, discrimination between savory and sweet tastes can be hardwired into the animal’s genetic code. Cats are obligate carnivores that have lost the canonical sweet taste receptor but retain a functional umami receptor (Li et al., 2005). Hummingbirds, which are nectar-feeding specialists, have adapted the ancestral umami receptor into a novel sweet taste receptor (Baldwin et al., 2014). For feeding generalists like flies, rodents, and humans, both protein and carbohydrates are useful energy sources and these animals can detect both savory and sweet tastes. Detection of either taste promotes feeding unless an animal becomes deficient in a specific nutrient and develops a nutrient-specific appetite (Deutsch et al., 1989; Leitao-Goncalves et al., 2017; Liu et al., 2017; Murphy et al., 2018; Ribeiro and Dickson, 2010; Simpson et al., 2015; Steck et al., 2018; Vargas et al., 2010). After days of protein deprivation, for example, animals can still detect savory and sweet, but the sensitivity of savory taste circuits is increased to promote a protein-specific appetite (Liu et al., 2017; Steck et al., 2018). Intrinsic indifference to a taste is ideally suited for specialists that utilize only one food source, while acute neuromodulation of taste preference is an effective means for generalists to conditionally prioritize one food source. However, female blood-feeding mosquitoes are specialists with two parallel and specific appetites for protein and carbohydrates that each utilize a different feeding program and fulfil distinct physiological processes. The mechanism that enables mosquitoes to engage mutually exclusive feeding programs for each food source is unknown.
The specialized feeding demands of blood-feeding mosquitoes, including Ae. aegypti, are linked to nutritional value. Although carbohydrates supplied from plant nectar are sufficient for energy metabolism in both females and males, protein obtained from host blood is required for females to develop eggs and successfully reproduce (Briegel, 2003; Duvall et al., 2019). Females sporadically produce a very small number of eggs without a blood meal, but these are rare exceptions (Ariani et al., 2015; Gulia-Nuss et al., 2015; Lea, 1964). Mosquitoes obtain nectar from flowers, which attract insects by producing floral cues (Barredo and DeGennaro, 2020; Lahondere et al., 2020; Van Handel, 1972).Typically, Ae. aegypti mosquitoes obtain a blood meal from a human or other vertebrate animal and integrate sensory cues like gaseous carbon dioxide (CO2), heat, and odor to locate their victim (Dekker et al., 2005; Liu and Vosshall, 2019; McMeniman et al., 2014; Takken and Kline, 1989). To procure necessary nutrients from these distinct food sources, females employ two behaviorally and anatomically distinct feeding programs: nectar feeding and blood feeding. Each feeding program is linked to a distinct feeding appendage, meal size, and digestive tract (Gordon and Lumsden, 1939; Trembley, 1952). Nectar is detected by the labium (Pappas and Larsen, 1978; Sanford et al., 2013). Blood is likely detected by the stylet, which pierces skin and directly contacts blood (Gordon and Lumsden, 1939; Trembley, 1952). The stylet is a needle-like feeding tube and stylet neurons are located on the part of the stylet referred to as the labrum (Lee, 1974). All parts of the stylet, including the labrum, maxillae, mandibles, and hypopharynx pierce the skin and directly contact blood, but the labrum is the only innervated part of the stylet. Females typically take small nectar meals but engorge on blood, consuming a volume that approximately doubles their body weight and provides sufficient protein to allow them to produce 100 – 150 eggs per blood meal. Finally, the nectar meal is routed initially to the crop, whereas ingested blood entirely bypasses the crop and is directed to the midgut (Gordon and Lumsden, 1939; Trembley, 1952), which is lateral to the crop in Diptera (Dow, 1987). Thus, the mosquito has parallel feeding pathways for blood and nectar from the sensory periphery, to visceral organs, to the ultimate metabolic function of the meal. This strict separation in feeding programs may allow the female to take a nectar meal to sustain her metabolism while maintaining her hunger for blood.
The ability of a female to distinguish between blood and nectar is finely tuned but the underlying mechanism remains unclear. In the absence of human sensory cues like heat and CO2, female mosquitoes readily ingest nectar via the nectar-feeding program. In the presence of human sensory cues, females will bite and feed on warm blood delivered in an artificial feeder (Bishop and Gilchrist, 1946; McMeniman et al., 2014). But if the blood meal is replaced with nectar sugars in the presence of the human cues heat and CO2, females reject the meal entirely (Bishop and Gilchrist, 1946). Therefore, the mechanism that distinguishes between blood and nectar must be flexible enough to promote ingestion of nectar only when a mosquito intends to feed on nectar and not when she intends to feed on blood.
The sensory mechanisms of whole blood recognition prior to initiating blood-feeding behavior are unknown. The stylet is the only sensory appendage that directly contacts blood and is therefore likely the primary structure that evaluates blood prior to initiation of blood feeding. Electron microscopy studies have demonstrated the presence of female-specific sensory sensilla at the tip of the stylet (Lee, 1974). Sensilla are specialized, porous cuticle structures that house sensory neuron dendrites. Chemical ligands enter chemosensory sensilla through pores to directly contact these dendrites (Stocker, 1994). Extracellular recordings from one stylet sensillum type documented neuronal activity in response to specific plasma components (Werner-Reiss et al., 1999a; Werner-Reiss et al., 1999b, c). Blood contains components that are traditionally associated with distinct taste qualities including sodium chloride (salty), protein (umami), glucose (sweet), and CO2 (sour/carbonation). It is not known if blood is recognized as a single taste quality, or if multiple taste qualities are integrated to form the perception of blood.
Here we show that Ae. aegypti mosquitoes possess sexually dimorphic stylet neurons specialized to distinguish blood from nectar. Using pan-neuronal GCaMP calcium imaging, we found that stylet neurons robustly respond to blood and its components but are insensitive to nectar-specific sugars. A mixture of four blood components—adenosine triphosphate, glucose, sodium bicarbonate, and sodium chloride— that was previously shown to potently trigger blood-feeding behavior (Galun et al., 1963; Galun et al., 1984; Hosoi, 1959) activated the same population of stylet neurons as blood. By presenting these ligands individually or as mixtures, we show that the taste of blood is combinatorial across multiple taste qualities. We defined functionally distinct subsets of stylet sensory neurons selectively tuned to specific blood components. We performed RNA-seq on the stylet to identify genetic markers that selectively label these neuronal subsets. We identified Ir7a and Ir7f as female stylet-specific transcripts and generated driver lines for both genes using CRISPR-Cas9 genome editing. We found that each driver line labels a functionally distinct subset of blood-sensitive stylet neurons activated by different components of blood. Finally, we discovered polymodal stylet “Integrator” neurons that respond to physiological levels of blood glucose only in the presence of additional blood components. Importantly, all stylet neurons, including Integrator neurons, are not activated by high concentrations of nectar-specific sugars. Since glucose is a redundant cue in blood and nectar, coincident detection of multiple blood components in Integrator neurons confers context-specific information about glucose. These experiments reveal that specialized sensory neurons in the mosquito stylet innately encode the distinction between blood and nectar.
RESULTS
Sensory Detection Prior to Blood and Nectar Feeding
When a female bites a human, she retracts the labium, uncovering the needle-like stylet required to draw blood. During blood feeding, the needle-like stylet pierces the skin to come into direct contact with blood. In contrast, the labium rests on the skin’s surface, which prevents it from contacting blood (Figure 1A,B) (Gordon and Lumsden, 1939; Griffiths and Gordon, 1952; Ramasubramanian et al., 2008). During nectar feeding, the labium directly contacts the nectar source and the stylet remains recessed and ensheathed within the labium (Figure 1C,D). In this configuration the stylet serves as a feeding tube for passing liquids after pumping is initiated. There is a striking difference in the meal volume consumed and how these meals are metabolized by the digestive system after ingestion. The average sugar meal size is 0.87 μL, in stark contrast to the average blood meal size of 3.20 μL (Figure 1E,F). Finally, the blood meal is immediately directed to the midgut for blood protein digestion, whereas the sugar meal is first directed to the crop (Figure 1G).
Figure 1. Sensory Detection Prior to Blood and Nectar Feeding.
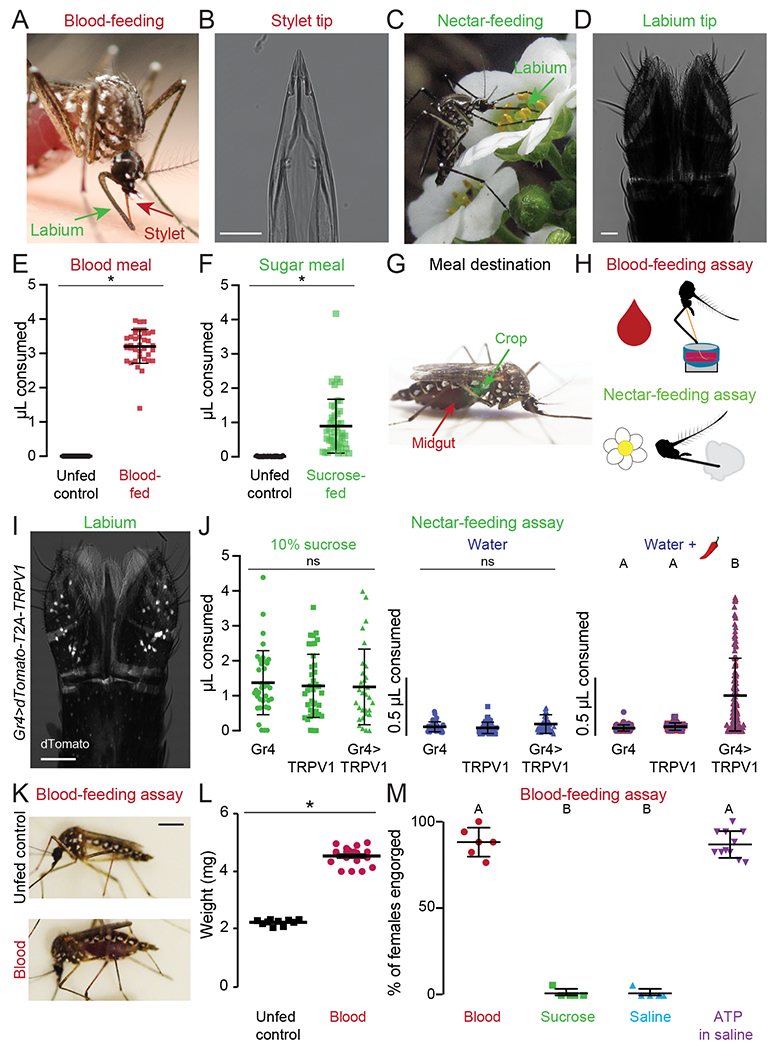
(A,C) An Ae. aegypti female feeding on human skin (A, Photo: Benjamin Matthews) or flower nectar (C, Photo: Eric Eaton).
(B,D) Transmitted light image of the female stylet (B) or labium (D). Scale bars: 25 μm
(E,F) Volume of meal consumed after presenting blood (E) or sugar (F). Unfed controls were not given the option to feed and therefore represent the baseline for the assay. Each data point represents 1 female (mean ± SD, N=37-46; * p < 0.05 Mann-Whitney test).
(G) Ae. aegypti female with a blood meal in the midgut (red) and a 10% sucrose meal in the crop (green). Green food dye added to 10% sucrose to visualize meal location.
(H) Schematic of blood- (top) and nectar-feeding (bottom) behavior assay.
(I) Confocal image of dTomato expression in Gr4>dTomato-T2A-TRPV1 labium with transmitted light overlay. Scale bar: 50 μm.
(J) Volume of meal consumed by the indicated genotypes. Each data point represents 1 female: 10% sucrose N=30-40 females/genotype; water N=41-60 females/genotype; water + 50 μM capsaicin (red chili pepper): Gr4 N=61, TRPV1 N=62, Gr4>TRPV1 N=124 females.
(K) Female mosquitoes following 15 min exposure to different meals. Scale bar, 1 cm.
(L) Sampled weight measurements from data for engorged females offered blood or unfed controls not offered any meal; N=10-19 weight measurements/meal (mean ± SEM; * p < 0.05 unpaired t-test).
(M) Female engorgement on the indicated meal delivered via Glytube. Each data point denotes 1 trial with 15-20 females/trial: N=5-11 trials/meal.
In (J, M) data labeled with different letters are significantly different from each other (mean ± SD; Kruskal-Wallis test with Dunn’s multiple comparison, p < 0.05).
See Figure S1 for chemogenetic and blood-feeding behavioral experiments.
We used blood- and nectar-feeding assays (Figure 1H) (Costa-da-Silva et al., 2013; Liesch et al., 2013) to quantify features of each behavioral program and substituted meal components to determine the requirements for feeding initiation. The blood-feeding assay offers females warmed meals in the presence of CO2 and heat, which attracts them to the artificial feeder (Liu and Vosshall, 2019; McMeniman et al., 2014). Upon landing, a parafilm membrane separates the female from the meal, forcing her to pierce it with the stylet just as she pierces skin to contact blood. In contrast, the nectar-feeding assay offers females room temperature meals on a cotton ball, allowing the labium to directly contact the meal upon landing.
To understand how mosquito nectar feeding is initiated, we searched for mosquito orthologues of Drosophila melanogaster Gustatory Receptor (GR) genes that are selectively tuned to sweet tastants (Scott, 2018). Ae. aegypti Gr4 is the closest orthologue of both D. melanogaster Gr5a and Gr64f (Kent and Robertson, 2009; Matthews et al., 2018). To label and manipulate neurons that express Gr4, we used CRISPR-Cas9 genome editing to insert the QF2 transcriptional activator at the endogenous Gr4 locus (Kistler et al., 2015; Matthews et al., 2019; Potter et al., 2010; Riabinina et al., 2016). We also generated an effector QUAS line to express both the dTomato fluorescent reporter and the rat cation channel TRPV1 in Gr4-expressing neurons (Tobin et al., 2002). In Gr4>dTomato-T2A-TRPV1 mosquitoes, we detected dTomato expression in the labium and legs, the two major taste appendages of insects (Figure 1I, Figure S1A–C).
To ask if activation of Gr4 neurons is sufficient to initiate nectar-feeding behavior, we performed chemogenetic experiments that used capsaicin to activate TRPV1. We confirmed that capsaicin did not alter ingestion of water or sucrose by wild-type animals in the nectar-feeding assay (Figure S1D). Similar to previous observations in D. melanogaster (Marella et al., 2006), addition of 50 μM capsaicin to water promoted ingestion of the otherwise inert water meal only in animals expressing TRPV1 in Gr4 neurons (Figure 1J). Thus, nectar feeding can be initiated by activation of sensory neurons that express sweet taste receptors.
What are the minimal sensory inputs required to initiate blood feeding? When we used the blood-feeding assay to offer females warm sheep blood in the presence of heat and CO2, they engorged on the meal, roughly doubling their initial body weight (Figure 1K–M). To separate meal composition from human cues, we maintained CO2 and heat delivery and exchanged the warm blood meal for warm sucrose or a saline solution that was isotonic with blood. Females consistently rejected both sucrose and saline in the blood-feeding assay, indicating that engorgement requires a separate step of evaluation after the female encounters a meal in the presence of human cues (Figure 1M). Classic work from Hosoi and Galun indicated that the nutritional value of blood as a protein source can be uncoupled from blood-feeding behavior. These studies identified ATP as a phagostimulant that could trigger engorgement only when co-presented with additional plasma components like sodium chloride (NaCl) and sodium bicarbonate (NaHCO3) (Galun et al., 1963; Galun et al., 1984). We replicated these experiments in the Liverpool Aedes aegypti laboratory strain and confirmed that an artificial blood meal sufficient for egg production, which consists of blood proteins, NaCl, and NaHCO3 (Kogan, 1990), did not trigger engorgement unless ATP was added (Figure S1E,F). As previously reported, a protein-free solution of saline and ATP, or its non-hydrolyzable analogues, is sufficient for engorgement. (Figure 1M, Figure S1G,H) (Galun et al., 1963; Galun et al., 1985; Galun et al., 1984). Finally, changing the concentration of ATP altered the probability of initiating engorgement (Figure S1I), but did not affect the meal size (Figure S1J). These behavioral data confirm classic observations by Galun and Hosoi and suggest that females can accurately recognize specific sensory features of blood and nectar to choose the appropriate feeding response.
The Stylet is Poised to Evaluate Meal Quality Prior to Blood Feeding
To track the stylet of individual females in response to different meals presented with heat and CO2, we used the biteOscope assay (Hol et al., 2020). The biteOscope consists of a transparent bite substrate mounted in the wall of a cage for high-resolution imaging of freely behaving mosquitoes. Subsequent manual video analysis enables the characterization of landing, piercing, and feeding dynamics at the individual mosquito level. The biteOscope allowed us to visualize the stylet as it pierces a membrane and to determine whether the female subsequently engorged on warmed meals of water, saline, or ATP in saline (Figure 2A and Video 1). We selected ATP in saline as a proxy for blood since biteOscope meals must be optically clear to enable stylet video tracking. In all three conditions, the females repeatedly landed on the membrane and pierced it, bringing the stylet into direct contact with the meal, but females engorged only on ATP in saline (Figure 2B–E). Once females engorged, they did not return to the membrane (Figure 2B,D). We conclude that human cues like heat and CO2 are sufficient to cause the female to pierce with her stylet and contact the meal, but additional blood-specific cues from the meal itself are required to trigger and sustain engorgement.
Figure 2. The Stylet is Poised to Evaluate Meal Quality Prior to Blood Feeding.
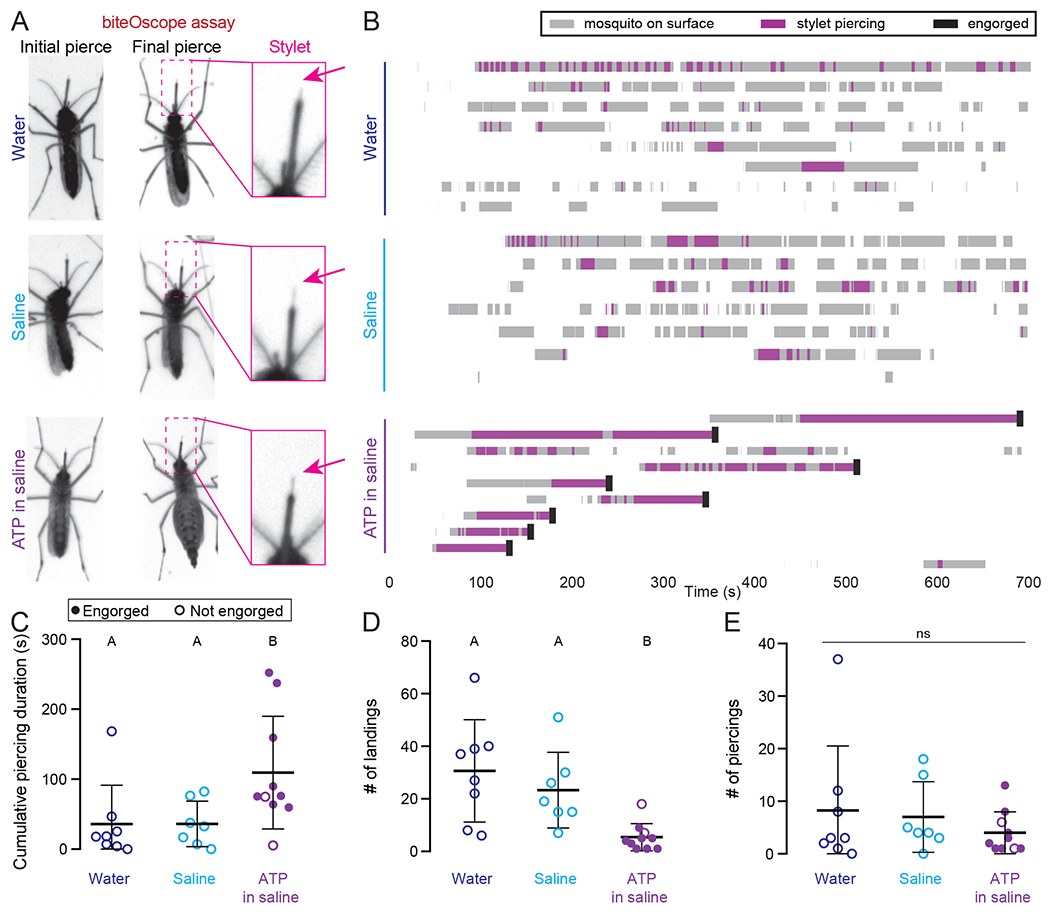
(A) Still video frames of female in biteOscope assay when stylet contacted meal for the first (left panel) or last (middle panel) time during the trial. Inset at right is from middle panel.
(B) biteOscope ethogram of landing events (gray boxes), stylet piercing events (pink boxes), and engorgement events (black boxes) for individual females provided water (N=8 females), saline (N=7 females), or 1mM ATP in saline (N=10 females) over 700 sec trial. Each row is an ethogram from 1 female.
(C-E) Summary statistics from individual female ethograms in (A) for cumulative piercing duration during trial (C), # of landings (D), and # of piercings (E) for indicated meal. Each dot denotes 1 female, filled dot represents an engorged female. In C,E, data labeled with different letters are significantly different from each other (mean ± SD; Kruskal-Wallis test with Dunn’s multiple comparison, p < 0.05). In D, data labeled with different letters are significantly different from each other (mean ± SD; one-way ANOVA with Tukey’s multiple comparisons test).
See Video 1 representative biteOscope movies.
Video 1.
The Stylet Repeatedly Pierces and Directly Contacts the Meal, Even When Females do not Eventually Engorge, Related to Figure 2
Representative biteOscope movies of individual females presented with a meal of warmed water, saline, or ATP in saline (1x speed).
The Stylet is Sexually Dimorphic
Previous electron microscopy studies showed that females have three bilaterally symmetric sets of sensory sensilla, all of which are likely to directly contact blood underneath the skin (Lee, 1974). The first two sets are putative chemosensory sensilla, located at the distal tip and found only in the female stylet (Jung et al., 2015; Lee, 1974) (Figure S2A, pink arrows). The third set comprises mechanosensory sensilla and is found in both the female and male stylet (Jung et al., 2015; Lee, 1974) (Figure S2A, white arrows). Beyond this early description of the external morphology of stylet sensilla, there has been limited investigation of its neuroanatomy.
To reveal the organization of the stylet, we used reagents to stain cell nuclei and actin filaments, and visualized dTomato-labeled neurons in a Brp>dTomato-T2A-GCaMP6s pan-neuronal reporter strain (Zhao et al., 2020) (Figure 3A–F). Nuclear staining indicated that there is a concentration of rounded nuclei within the first 300 μm from the distal tip of the stylet, with more proximal nuclei showing a flatter elongated morphology (Figure S2B). When we examined dTomato expression in Brp>dTomato-T2A-GCaMP6s animals, we found that all stylet neurons are located within the distal region (Figure S2C). Moreover, this section of the stylet is dramatically sexually dimorphic. When compared to males, females have a greater number of nuclei (Figure 3A,C), neurons (Figure 3B,D), and dendritic processes that innervate the distal tip (Figure 3E,F, Figure S2D–G). In agreement with previous electron microscopy data, we found that sensory dendrites innervated mechanosensory sensilla in both males and females (Figure 3E,F).
Figure 3. Sensory Neurons in the Female Stylet are Sexually Dimorphic and Project to a Unique Subesophageal Zone Region.
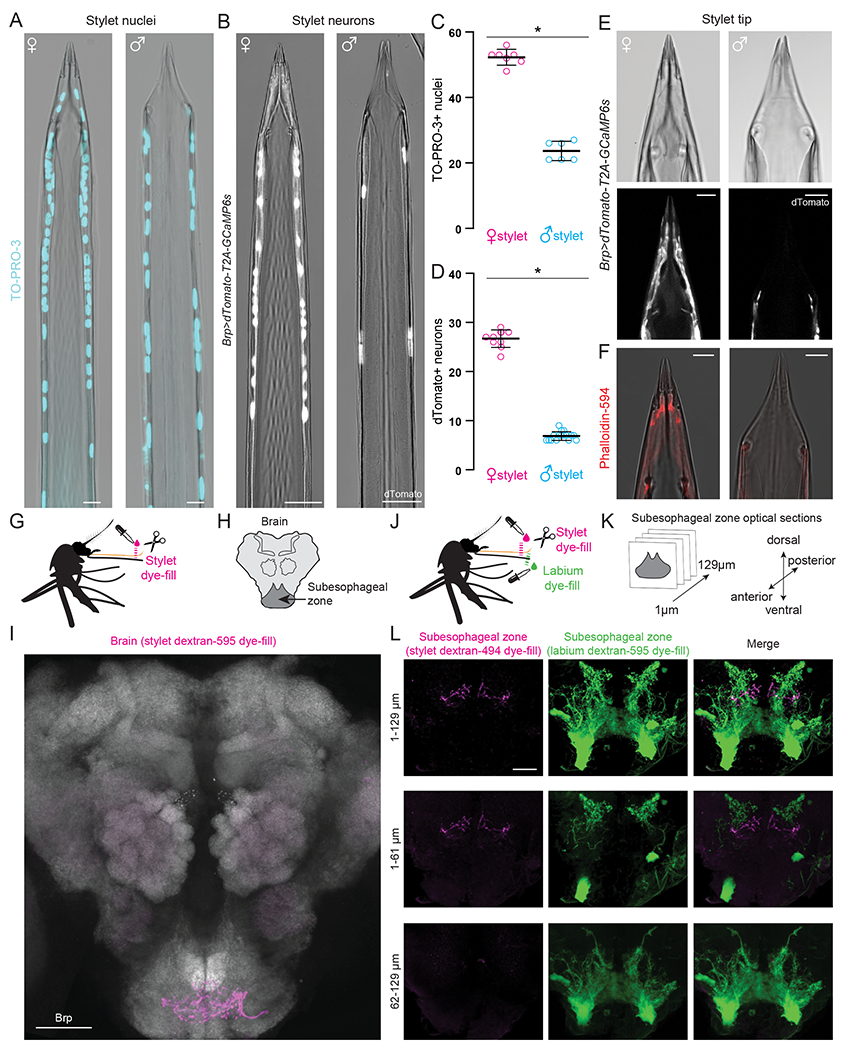
(A,B) Confocal image with transmitted light overlay of TO-PRO-3 nuclear staining (cyan) in wild-type female (A, left) and male (A, right) stylets, and dTomato expression (gray) in Brp>dTomato-T2A-GCaMP6s female (B, left) and male (B, right) stylets.
(C,D) Average # of TO-PRO-3 nuclei/stylet for most distal 300 μm (C, N=7 females, N=6 males), and dTomato neurons/stylet (D, N=10 females, N=16 males). Each dot denotes 1 animal (mean ± SD, * p < 0.05 Mann-Whitney test).
(E) Confocal image of transmitted light (top) and dTomato (gray, bottom) in Brp>dTomato-T2A-GCaMP6s female (left) and male (right) stylet tip.
(F) Confocal image with transmitted light overlay of phalloidin-594 (red) staining in wild-type female (left) and male (right) stylets.
(G, J) Schematic of stylet (G) and double (J) dye-fill experiment set-up performed in (I) and (L), respectively.
(H, K) Schematic of mosquito brain region captured in (I), and subesophageal zone optical sections captured in (L).
(I) Stylet neuron projection pattern (magenta) revealed by dextran-595 dye-fill. Neuropil stained with anti-Drosophila Brp (gray).
(L) Optical subesophageal zone sections from most anterior (top row) to most posterior (bottom row) of stylet (left, magenta) and labium (middle, green) projection pattern revealed by dual dextran-494 and dextran-595 dye-fill.
Scale bar: 50 μm (I), 25 μm (A,B,L), 10 μm (E,F).
See Figure S2 for additional analysis of stylet sexual dimorphism and Video 2 for confocal Z-stacks of dual dye-fills.
We next asked where these female stylet neurons project in the mosquito brain. If the stylet detects the taste of blood, we would expect innervation of the subesophageal zone, the first processing center in the insect taste system (Ito et al., 2014; Scott, 2018). We performed dye-fill experiments to label axon terminals from all stylet neurons (Figure 3G) and found that stylet innervation was restricted to a discrete anterior and ventral region in the subesophageal zone (Ignell and Hansson, 2005) (Figure 3H, I). A previous study reported additional innervation of the antennal lobe, the primary olfactory processing center, upon dye-filling the stylet (Jung et al., 2015), but we could not replicate this result (Figure 3I and Figure S2H).
To ask how female stylet neuron projections relate to projections from female labium neurons in the subesophageal zone, we performed a dual dye-fill experiment in which we labelled stylet and labium neurons with different dye colors in the same animal (Figure 3J). The female labium projects to the posterior region of the subesophageal zone and there is no overlap with stylet neuron projections (Figure 3K–L, Figure S2I, and Video 2). Therefore, inputs from the stylet and labium are segregated at the first synapse in the subesophageal zone.
Video 2.
Projections from Female Stylet and Labium Sensory Neurons do not Overlap at the First Synapse in the Brain, Related to Figure 3
Representative confocal Z-stack of stylet (magenta) and labium (green) sensory neuron projection pattern in the subesophageal zone upon dual dextran-494 and dextran-595 dye fill. Scale bar 25 μm.
Stylet Neurons Detect Blood
To test if stylet neurons can directly detect blood, we developed an ex vivo calcium imaging preparation with the pan-neuronal Brp>dTomato-T2A-GCaMP6s mosquito (Zhao et al., 2020) (Figure 4A,B). Because all stylet neurons are located in one optical plane, we were able to image responses from all neurons simultaneously. When we applied 500 mM potassium chloride (KCl) as a depolarizing stimulus, we observed strong responses in all stylet neurons (Figure 4C). Since whole blood is opaque, it was necessary to restrict blood to the stylet tip so that it did not interfere with GCaMP6s signal in the cell bodies. To solve this problem, we used the BioPen microfluidic device to deliver blood to the chemosensory pores that are innervated by sexually dimorphic distal processes (Figure 4D and Video 3). We developed an analysis pipeline to calculate peak ΔF/F responses to individual ligands for each stylet neuron (Figure S3A–D). Stylet neurons consistently responded to 3 presentations of blood (denoted as 1st, 2nd, and 3rd blood) separated by 60 sec intervals, and not to water (Figure 4E–G and Video 4). Within a given female, the peak ΔF/F response to multiple presentations of blood was stable, but the exact number and position of blood-sensitive neurons was not stereotyped across individuals (Figure 4G and Figure S3E,F). Across individuals approximately 50% of stylet neurons responded to blood (Figure S3E). Different neurons within an individual had unique GCaMP6s response waveforms that were stable across every blood presentation for a given neuron (Figure S3G). These results demonstrate that a large population of stylet chemosensory neurons responds directly to whole blood.
Figure 4. Sexually Dimorphic Stylet Neurons Directly Sense Blood.
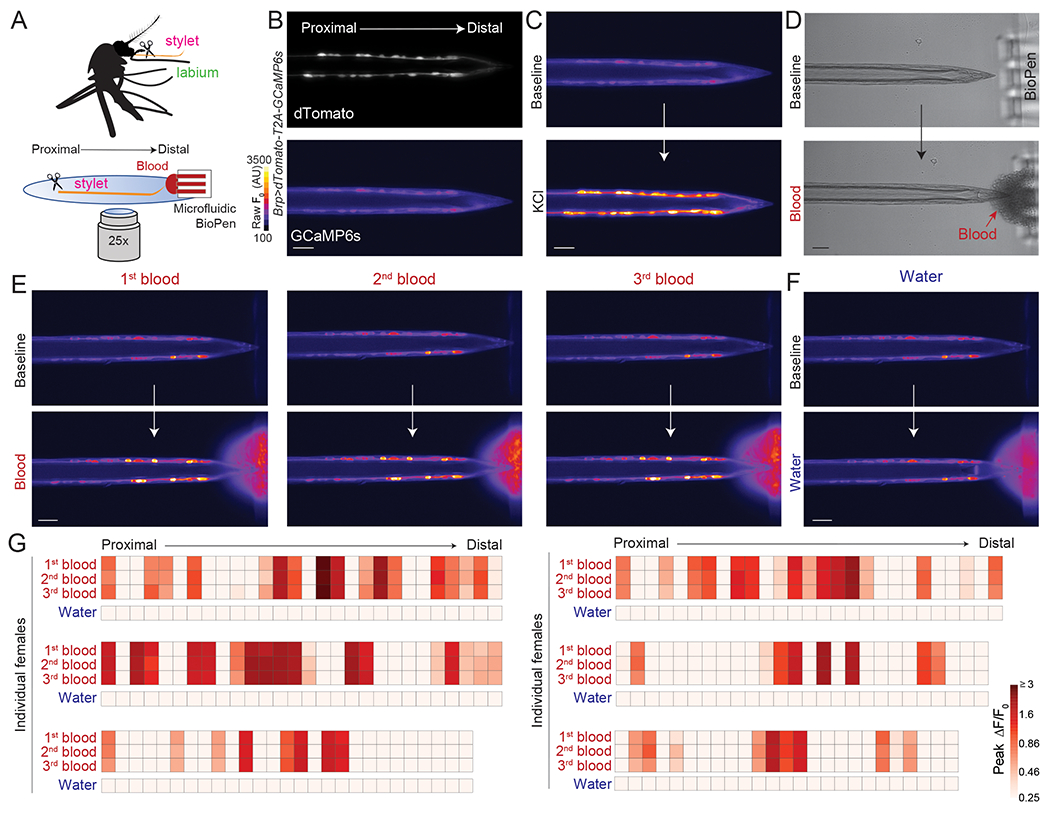
(A) Schematic of ex vivo stylet imaging preparation.
(B) Wide-field image of dTomato (top) and baseline GCaMP6s (bottom, scale: arbitrary units) for a representative stylet, oriented proximal to distal.
(C) Representative image of GCaMP6s fluorescence increase to bulk neuronal depolarization with 500 mM KCl (bottom) compared to baseline (top).
(D) Representative bright-field image before (top) and during (bottom) delivery of sheep blood to the stylet tip via the BioPen.
(E,F) Representative image of GCaMP6s fluorescence increase to indicated blood presentation (bottom, E) or water control (bottom, F), compared to baseline (top).
(G) Heat maps of peak ΔF/F0 response to the indicated ligand. Each square is the average of the peak ΔF/F0 measured in 3 separate trials. Each column represents 1 neuron and each row represents the response to indicated ligand for all neurons from 1 individual female, with neurons ordered from proximal to distal. N=6 individual females. In (B-F) scale bar: 25 μm. 0.0002% fluorescein was added to blood and water stimuli to visualize ligand delivery zone.
See Video 3, 4 for representative movies of BioPen stimulus delivery and stylet responses to blood or water, and Figure S3 for details on calcium imaging analysis.
Video 3.
The Stylet Responds to Blood, Related to Figure 4
Representative movie of the BioPen delivering a drop of blood to chemosensory sensilla located at the stylet’s tip. The stylet is from a Brp>dTomato-T2A-GCaMP6s female, images captured with transmitted light (left) and 488 nm wavelength (right), scale bar 25 μm.
Video 4.
The Stylet Responds to Consecutive Blood Presentations and not Water, Related to Figure 4
Representative movie of stylet neuron responses to presentations of blood or water (5x speed). The stylet is from a Brp>dTomato-T2A-GCaMP6s female, scale bar 25 μm.
Blood Detection is Combinatorial Across Taste Qualities
How is the complex mixture of blood represented by stylet neurons? We used a reductionist approach to understand how the taste of blood is encoded in stylet neurons. We selected 4 blood components [ATP, glucose, NaHCO3, and NaCl] that have been individually shown to contribute to the probability of engorgement (Galun et al., 1984; Gonzales et al., 2018). ATP and unbuffered NaHCO3 (pH = 8 - 9) are not associated with canonical taste qualities, but glucose and NaCl are sweet and salty, respectively. We selected concentrations of glucose, NaHCO3, and NaCl within range of standard blood values for vertebrate species. The in vivo concentration of ATP present when the female bites a human is unknown because ATP is derived from multiple sources and is rapidly hydrolyzed. For example, up to millimolar-range ATP can be released from the deformation or lysis of red blood cells or from epithelial cells lining the blood vessel as a damage response to being pierced by the stylet (Born and Kratzer, 1984; Forsyth et al., 2011). We selected 1 mM because it elicited robust behavioral responses (Figure S1I). Using the blood-feeding assay, we found that the combination of these 4 ligands (hereafter referred to as Mix+ATP) was sufficient to trigger engorgement (Figure 5A,B).
Figure 5. Stylet Neurons Integrate Across Taste Modalities to Detect Blood.
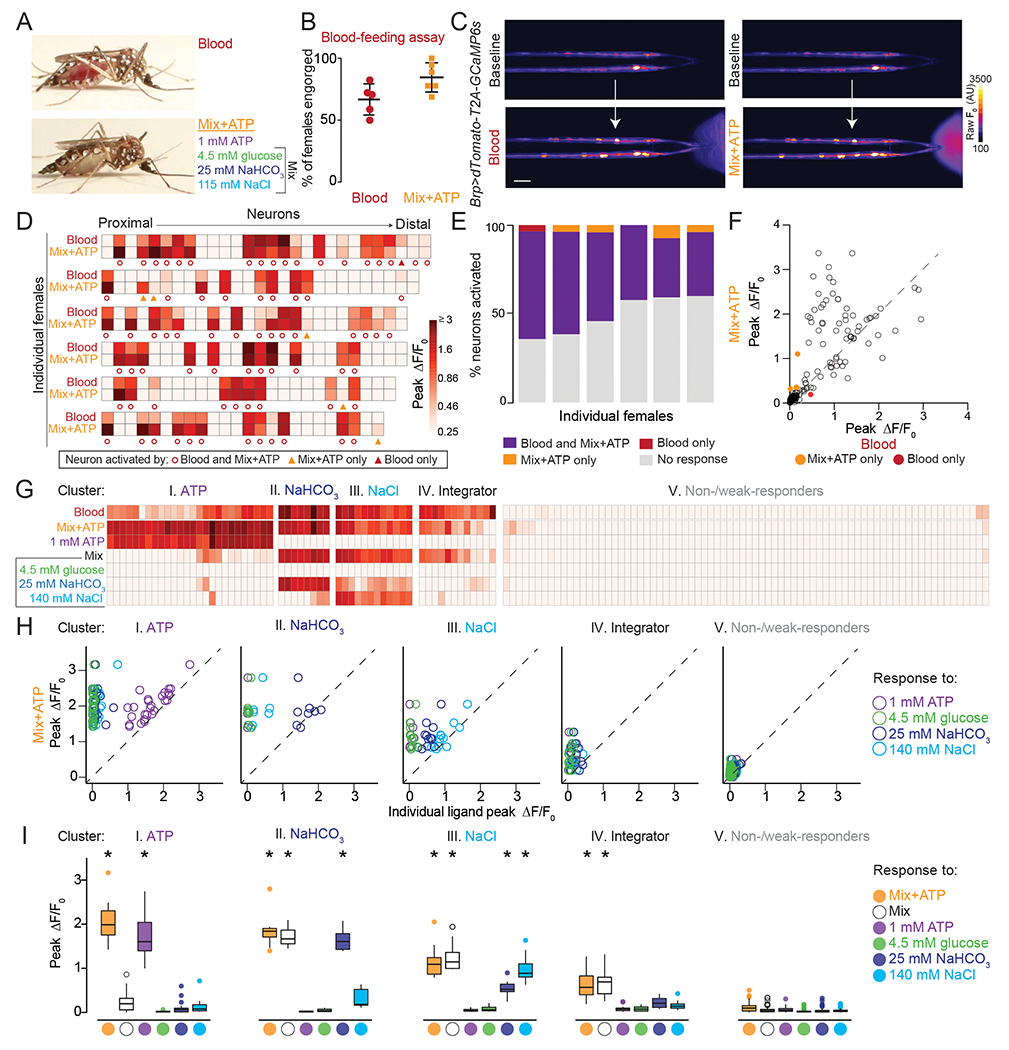
A) Representative engorged Ae. aegypti female following 15-min exposure to blood (top) or Mix+ATP (bottom) via Glytube assay.
(B) Female engorgement on blood (N=5 trials) and Mix+ATP (N=6 trials) delivered via Glytube (lines denote mean ± SD, 15–20 females/trial, p = 0.0714, Mann-Whitney test).
(C) Representative image of GCaMP6s fluorescence increase (scale: arbitrary units) to blood (bottom, left) or Mix+ATP (bottom, right), compared to baseline (top). Scale bar: 25 μm.
(D) Heat maps of peak ΔF/F0 response to the indicated ligand. Each square is the average of 3 ligand exposures and each column represents one neuron. Each row represents the response to indicated ligand for all neurons from 1 individual female, with neurons ordered from proximal to distal. N=6 individual females.
(E) Summary of % neurons with ≥ 0.25 peak ΔF/F0 to the indicated ligand from (D), each column represents 1 female.
(F) Scatter plot comparing peak ΔF/F0 in response to Mix+ATP (y-axis) and blood (x-axis) summarized across N=6 females from (D,E). Each dot represents 1 neuron, dots that fall on the dashed line have the same peak ΔF/F0 in response to blood and Mix+ATP. Dots that fall above the line respond more to Mix+ATP than to blood and dots that fall below the line respond more to blood than to Mix+ATP.
(G-I) 5 clusters of blood-sensitive neurons identified by unsupervised hierarchical clustering of peak ΔF/F responses to the ligands indicated in (G). Clustering removes proximal-distal ordering and female identity. N=5 females (* p < 0.05, one-sample Wilcoxon signed-rank test).
In (A-I) and all subsequent experiments “Mix” is 4.5 mM glucose, 25 mM NaHCO3, 115 mM NaCl and “Mix+ATP” is Mix supplemented with 1 mM ATP. To visualize ligand delivery zone, 0.0002% and 0.00002% fluorescein was added to blood and Mix+ATP, respectively, in BioPen experiments.
See Video 5 for representative movies of stylets responding to blood and Mix+ATP, Data File 1 for raw imaging data and p values for Figure 5I, Figure S4 for responses of individual females to blood components, and Figure S5 for details of the hierarchical clustering method.
Since both blood and Mix+ATP trigger engorgement we asked if there are differences in how stylet neurons respond to these taste stimuli. When we delivered blood or Mix+ATP to Brp>dTomato-T2A-GCaMP6s animals, we found that blood and Mix+ATP activated the same population of stylet neurons (Figure 5C–F and Video 5). Although the magnitude of response can vary within a given neuron (Figure 5D,F), Mix+ATP-responsive neurons track with blood-responsive neurons across individuals, irrespective of variability in the position of the neuronal cell body along the proximal-distal axis of the stylet (Figure 5D,E).
Video 5.
Blood and Mix+ATP Activate the Same Subsets of Stylet Neurons, Related to Figure 5
Representative movie of stylet neuron responses to presentations of blood or Mix+ATP (5x speed). The stylet is from a Brp>dTomato-T2A-GCaMP6s female, scale bar 25 μm.
To understand how blood components contribute to the perception of whole blood, we used Mix+ATP as a chemically-defined mixture that activates blood-responsive neurons. When we presented each component of Mix+ATP individually, we found that blood-sensitive neurons are a heterogenous population and that different neuronal subsets within each female can respond to distinct blood components (Figure S4). Moreover, all components except 4.5 mM glucose reliably activated subpopulations of stylet neurons when presented individually (Figure S4A). Unsupervised hierarchical clustering of this dataset was performed to group neurons into 5 clusters according to their functional response profile (Figure 5G and Figure S5). We performed several analyses to validate this clustering method and found that the dataset is highly clusterable (Hopkins statistic, Figure S5A), the optimal number of clusters is 5 (Silhouette analysis, Figure S5B), cluster identity is stable (Jaccard bootstrap mean, Figure S5C), and that cluster membership is not correlated with female identity (PCA analysis, Figure S5D–F). For each neuron in a cluster, we calculated a ratio of peak ΔF/F response to Mix+ATP compared to the peak ΔF/F response to any individual ligand (Figure 5H). The first 3 clusters represent neurons activated by an individual component: ATP, NaHCO3, and NaCl, respectively (Figure 5I). Although Cluster 4 was not reliably activated by any individual ligand, it was activated by a mixture of NaHCO3, NaCl, and glucose (hereafter referred to as “Mix”) (Figure 5I). We define these as “Integrator” neurons and explore their function in subsequent experiments. Cluster 5 neurons were non-responsive or showed weak responses (Figure 5I). Neurons from the 5 clusters were found across different females, but the exact number of neurons per cluster was not stereotyped across individual females (Figure S4A,B). Together these experiments demonstrate that subsets of blood-sensitive neurons are selectively tuned to specific blood components that span multiple canonical and noncanonical taste qualities.
Ir7a and Ir7f Mark Functionally Distinct Populations of Blood-Sensitive Neurons
We next asked if these functionally distinct blood-sensitive subsets are transcriptionally-defined populations. To identify candidate genetic markers for neuronal subsets, we profiled transcript abundance in the female stylet using RNA-seq and compared it to the male stylet and the female labium and selected transcripts significantly enriched in the female stylet compared to both the labium and male stylet (Figure S6A,B, fuchsia data points). We further filtered the data to select transcripts that were expressed at very low levels (< 0.5 transcripts per million, TPM) in a comprehensive transcriptome dataset that included other sensory appendages, brain, and ovary (Figure S6C) (Matthews et al., 2018; Matthews et al., 2016). Of the four transcripts that met these criteria for female stylet-specific expression, two were members of the ionotropic receptor (IR) superfamily, Ir7a and Ir7f (Figure S6D,E).
We generated QF2 driver lines for Ir7a and Ir7f and crossed these to reporter lines to reveal sparse expression in subsets of chemosensory neurons in the female stylet (Figure 6A,B). Ir7a and Ir7f are expressed in approximately 1 - 2 neurons and 3 - 4 neurons, respectively. No expression of either gene was detected in male stylets. The sparse nature of these drivers revealed dendritic innervation of the bilaterally symmetric set of two chemosensory sensilla at the stylet tip (Figure 6A,B). Both populations of neurons innervate the same ventral subesophageal zone region identified in our stylet dye-fills (Figure 6C–F and Video 6). Importantly, no regions in the male brain or additional regions in the female brain were labeled in these strains, highlighting the exquisite selectivity of Ir7a and Ir7f gene expression to the female stylet.
Figure 6. Ir7a and Ir7f Mark the NaHCO3 and Integrator Neurons.
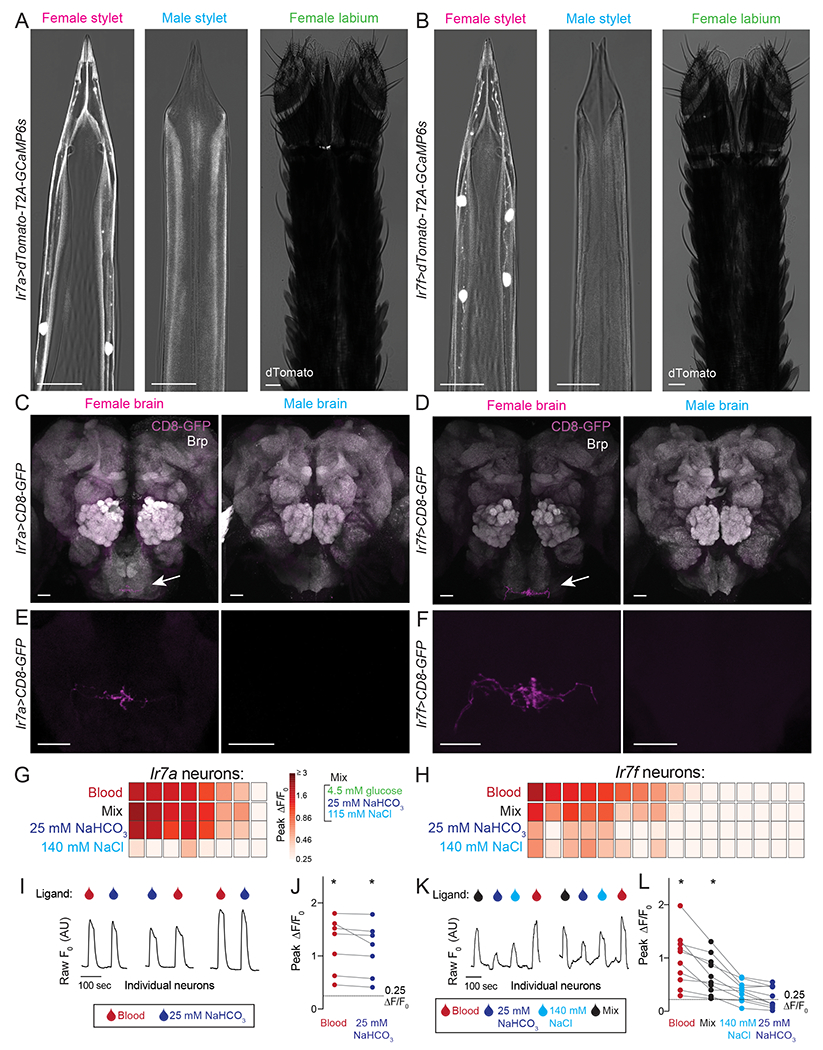
(A,B) Confocal image with transmitted light overlay of dTomato expression (gray) in the female stylet (left panel), male stylet (middle panel), and female labium (right panel) of Ir7a>dTomato-T2A-GCaMP6s (A) and Ir7f>dTomato-T2A-GCaMP6s (B) animals. Ir7a expression: 10/13 females = 2 neurons, 2/13 females = 1 neuron, 1/13 females = 0 neurons. Ir7f expression: 6/11 females = 4 neurons, 5/11 females = 3 neurons.
(C-F) mCD8:GFP expression (magenta, white arrow) of Ir7a>mCD8:GFP (C,E) and Ir7f>mCD8:GFP (D,F) in female (left) and male (right) brain (top) and subesophageal zone (bottom). Neuropil in C and D is labeled with anti-Drosophila Brp (gray). The brain and subesophageal zone images in C-F were acquired from different individuals.
(G,H) Heat maps of peak ΔF/F0 response to the indicated ligand in Ir7a>dTomato-T2A-GCaMP6s (G) and Ir7f>dTomato-T2A-GCaMP6s (H) neurons across N=5 females. Each square is the average of 3 ligand exposures and each column represents one neuron. Columns are sorted by largest to smallest peak ΔF/F0 in response to blood.
(I,K) Raw F0 traces from individual neurons in response to indicated ligand.
(J,L) For blood-sensitive neurons, peak ΔF/F0 to indicated ligand. Each data point denotes the response from 1 neuron and responses from the same neuron are connected by a line (* p < 0.05, one-sample Wilcoxon signed-rank test).
In (A-F) scale bar: 25 μm. 0.0002% fluorescein was added to blood and 140 mM NaCl, and 0.00002% was added to Mix and 25 mM NaHCO3 in the BioPen to visualize ligand delivery zone.
See Video 6 for confocal Z-stack movies of Ir7a- and Ir7f- labeled neurons, Figure S6 for RNA-seq data and behavioral analysis of Ir7a and Ir7a mutants and chemogenetic manipulation, and Data File 1 for p values for Figure 6J,L.
Video 6.
Projections from Ir7a>CD8-GFP or Ir7f>CD8-GFP Neurons Innervate the Same Subesophageal Zone Region Identified in Stylet Dye-Fills, Related to Figure 6
Representative confocal Z-stack of Ir7a>CD8-GFP or Ir7f>CD8-GFP projections (red) in the mosquito brain. Neuropil labelled with anti-Drosophila Brp (gray), scale bar 50 μm.
To determine the functional properties of Ir7a and Ir7f neurons, we performed cell-type specific calcium imaging experiments and found that almost all Ir7a neurons and a subpopulation of Ir7f neurons responded to blood (Figure 6G,H). Ir7a- and Ir7f-expressing blood-responsive populations respond to Mix (glucose, NaHCO3, and NaCl) but not ATP (Figure S6F,G). Ir7a blood-sensitive neurons were robustly activated by NaHCO3 (Figure 6I,J), sharing a profile with NaHCO3 neurons identified in Cluster II (Figure 5G–I). In contrast, Ir7f blood-sensitive neurons were consistently activated by Mix and had variable responses to 140 mM NaCl and/or 25 mM NaHCO3 (Figure 6K,L), sharing a profile most similar to Integrator neurons in Cluster IV (Figure 5G–I). Thus these two female stylet-specific driver lines define the molecular and functional identity of two non-overlapping blood-sensitive neuron populations in the female stylet.
Specialization in Stylet Neurons Enables Discrimination Between Blood and Nectar
Glucose is a redundant cue in blood and nectar (Figure 7A). Since stylet neurons are the only sensory neurons that directly contact the meal during blood feeding, do they have a specialized taste coding strategy to selectively distinguish blood components from nectar components? To address this question, we first measured the behavioral response to 298 mM nectar sugars in the context of the nectar- and blood-feeding assay. This concentration is approximately equivalent to the female’s normal sugar meal that is sufficient for energy metabolism (Van Handel, 1972, 1984). Females readily ingested all three sugars when the labium directly contacted the meal in the nectar-feeding assay, where no host cues are present (Figure 7B and Figure S7A). In contrast, they rejected these same sugars in the blood-feeding assay when the stylet directly contacted the meal in the presence of heat and CO2 (Figure 7C and Figure S7B). In control experiments we showed that blood stimulated robust consumption in the blood-feeding assay (Figure 7C).
Figure 7. The Stylet is Specialized to Detect Blood over Nectar.
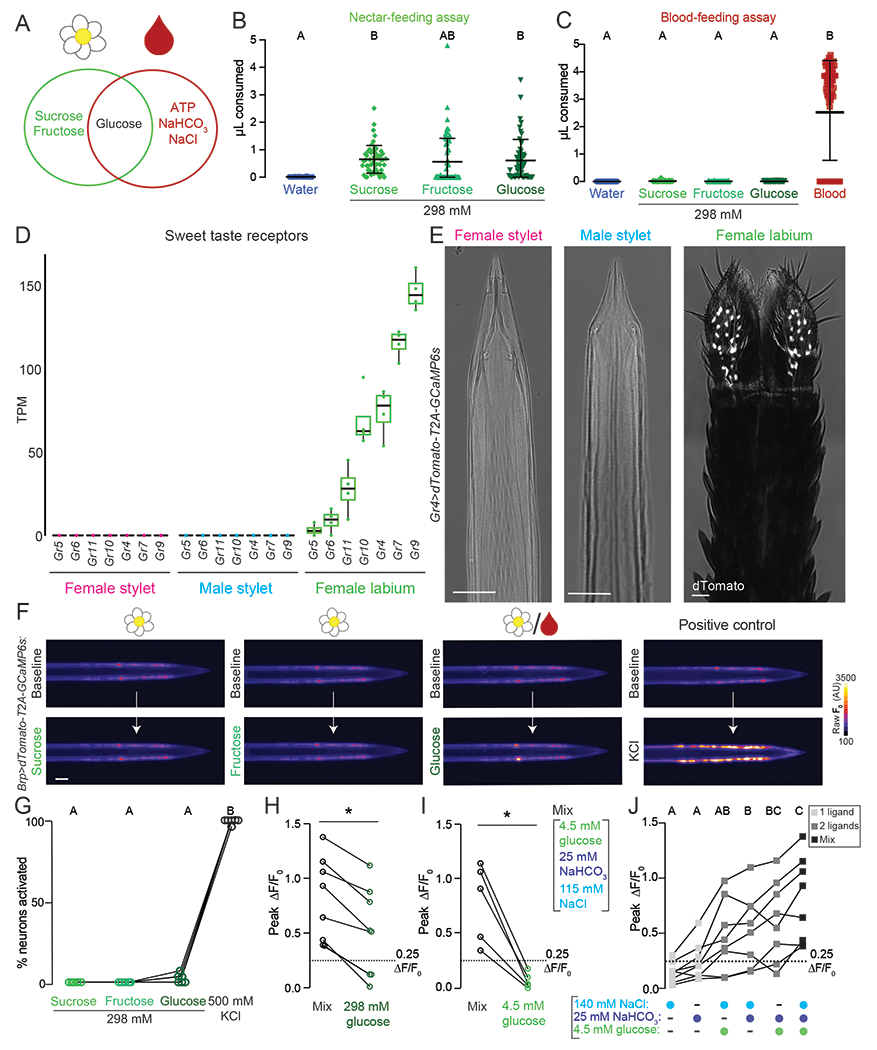
(A) Venn diagram schematizing the similarity and differences between nectar (left circle) and blood (right circle) components.
(B,C) Volume of indicated meal consumed in the nectar-feeding (B) and blood-feeding (C) assay. Each data point represents 1 female: water N=36-40; sucrose N=53–60; fructose N=40-74; glucose N=55-59. Blood in (C) is a positive control for blood-feeding assay, N=76 females.
(D) Sweet taste receptor expression from RNA-seq analysis of the indicated tissues. N=4 replicates/tissue. Median indicated by black line, bounds of box represent first and third quartile, whiskers are 1.5 times the inter-quartile range, and dots represent TPM value from each biological replicate. The outlier is denoted by a dot without whisker.
(E) Confocal image with transmitted light overlay of dTomato expression (gray) in the female stylet (left panel), male stylet (middle panel), and female labium (right panel) of Gr4>dTomato-T2A-GCaMP6s animals. Scale bar: 25 μm.
(F) Representative image of GCaMP6s fluorescence increase to indicated 298 mM sugar presentation (bottom) compared to baseline (top). Flower/blood symbol (3rd from left) indicates that sugar is found in nectar and blood.
(G) Quantification of % neurons with ≥ 0.25 peak ΔF/F0 to the indicated ligand, each data point denotes the response from 1 female, responses from the same female are connected by a line, N=6 females.
(H,I) For Integrator neurons, peak ΔF/F0 to 298 mM glucose (H, N=8 neurons) and 4.5 mM glucose (I, N=5 neurons). Each dot represents 1 neuron (mean ± SD, * p < 0.05 Mann-Whitney test).
(J) For Integrator neurons, peak ΔF/F0 to indicated ligand(s). Each data point denotes the response from 1 neuron, N=8 neurons. Data labeled with different letters are significantly different from each other (one-way repeated measures ANOVA, with the Geisser-Greenhouse correction and Tukey’s multiple comparisons test, p < 0.05).
In (B,C,G) data labeled with different letters are significantly different from each other (mean ± SD; Kruskal-Wallis test with Dunn’s multiple comparison, p < 0.05) and in (H, I, J) responses from the same neuron are connected by a line.
See Figure S7 for behavioral and imaging data with nectar sugars.
These results lead to the question of whether stylet neurons can detect these nectar sugars at all. While the female labium expresses all predicted orthologues of D. melanogaster canonical sugar receptors (Figure 7D), no sweet GR transcripts were detected in the female or male stylet (Figure 7D). The Gr4 reporter line showed no expression in the female and male stylet but was expressed in a large group of labial neurons (Figure 7E), which project axons to the posterior region of the subesophageal zone that we identified in our labium dye-fill experiments, but not to the anterior, ventral region occupied by stylet neuron projections (Figure S7C,D). Thus stylet neurons do not express canonical sweet taste gustatory receptors.
We next examined stylet neuron responses to nectar sugars using calcium imaging in stylets from Brp>dTomato-T2A-GCaMP6s animals. If stylet neurons lack a canonical sweet taste pathway, we expect that they would not respond to sucrose, fructose, and glucose. Indeed, no stylet neurons responded to 298 mM nectar-specific sugars (Figure 7F,G and Figure S7E) (Werner-Reiss et al., 1999c). In half of the females, we observed occasional responses to 298 mM glucose, which is the only sugar found in both blood and nectar (Figure 7F,G and Figure S7E). In positive control experiments, we confirmed that all stylet neurons responded to KCl (Figure 7F,G and Figure S7E).
Although responses to 298 mM glucose were rare, 298 mM glucose-sensitive neurons were blood-sensitive and shared a functional profile with Integrator neurons (Figure S7F,G). Integrator neurons consistently responded more to Mix than this high concentration of glucose (Figure 7H). We therefore asked if physiological levels of blood glucose directly contribute to Mix responses observed in Integrator neurons. Since Integrator neurons do not respond to 4.5 mM glucose alone (Figure 5G–I and Figure 7I), we tested if the addition of 4.5 mM glucose to other Mix components increased the total neuronal response. Integrator neurons responded to 4.5 mM glucose when co-presented with NaCl or NaHCO3, and by co-presentation of all three (Figure 7J). These results demonstrate that individual sensory neurons can directly integrate glucose (sweet), NaCl (salty), and NaHCO3. Taken together, our results demonstrate that the stylet is specialized to detect blood over nectar.
DISCUSSION
Anatomical, Molecular, and Functional Properties of the Stylet
The female stylet is an unconventional sensory organ whose functional properties are poorly understood. The microneedle-like biophysical properties needed to efficiently pierce skin (Choumet et al., 2012; Ramasubramanian et al., 2008) may influence its unique anatomical organization into two single-file rows of cells along each side. Consistent with its role in female-specific blood-feeding behavior, we identified dramatic sexual dimorphism in neuron number and innervation of chemosensory sensilla. The sparse, stylet-specific Ir7a and Ir7f driver lines allowed us to show that individual neurons send ipsilateral dendrites into one of the two chemosensory sensilla found on each side of the stylet tip. Interestingly, we observed inter-individual differences in blood-sensitive neuron number and cell body position. We do not yet understand the mechanism of developmental patterning that produces variable cell body position along the proximal-distal axis of the stylet. Variability in the exact distance of the cell to the stylet tip may be tolerated because all stylet neuron dendrites terminate at the tip, irrespective of cell body position.
By generating two female stylet-specific driver lines, we identified non-overlapping blood-sensitive neurons belonging to two functionally distinct subsets: Ir7f blood mixture-sensitive neurons and Ir7a NaHCO3-sensitive neurons. Together, these driver lines mark approximately one quarter of total stylet neurons. Future work will allow us to determine if Ir7a and Ir7f, along with additional putative chemosensory receptors identified in our stylet RNA-seq dataset, contribute to blood ligand detection. We were unable to identify orthologues to P2X ATP receptors in the Ae. aegypti genome (Matthews et al., 2018). Although P2X receptors have been identified in diverse species such as vertebrates (Khakh et al., 2001) and ticks (Bavan et al., 2011), they are absent in D. melanogaster and Caenorhabditis elegans (Burnstock and Verkhratsky, 2009; Lima and Miesenbock, 2005). Therefore, the receptor used by Ae. aegypti mosquitoes to detect ATP remains to be identified.
A major finding of this work is that four ligands previously shown to increase the probability of initiating blood-feeding behavior do indeed directly activate the stylet. When presented as a mixture, these four blood components—ATP, glucose, NaHCO3, and NaCl—are sufficient to activate the same neurons as blood and initiate blood-feeding behavior. Our functional imaging shows that roughly half of the 40 stylet neurons can be activated by blood. The remaining stylet neurons may respond to a variety of different ligands, including those found only when the stylet contacts an intact capillary microenvironment. These unidentified ligands may be detected in an in vivo context, but none are required for blood-feeding behavior or egg development. There may also be circulating factors released from surrounding cells as a damage response to the piercing stylet or ligands specific to human blood. Some of the remaining stylet neurons may respond to additional taste qualities observed in other feeding appendages. For example, responses to osmolarity, high salt, CO2, and bitters have been observed in D. melanogaster labellar neurons (Liman et al., 2014; Yarmolinsky et al., 2009). Bitters are of particular interest because specific bitters added to blood prevent feeding (Dennis et al., 2019). Finally, the stylet could be capable of thermosensation or mechanosensation related to sensing blood flow or tissue penetration. The pan-neuronal stylet imaging preparation we have developed will facilitate future systematic analyses of stylet responses to diverse sensory stimuli.
Stylet Neurons Integrate Across Taste Qualities to Detect Blood
Blood-sensitive neurons can be divided into functionally distinct subtypes, each activated by a behaviorally-relevant concentration of a ligand, or mix of ligands, found in blood. Glucose and NaCl are associated with the distinct taste qualities of sweet and salty, but it is unclear if NaHCO3 or ATP overlap with a canonical taste quality. In blood, NaHCO3 is buffered at pH 7.4 and predominately present as HCO3- (Centor, 1990). While CO2 contributes to sour taste and encodes the taste of carbonation (Chandrashekar et al., 2009; Fischler et al., 2007), HCO3- has not yet been assigned to a defined taste quality. Similarly, there is no description of the taste of ATP. Our work shows that the taste of blood is multidimensional and that multiple taste qualities, both canonical and noncanonical, are integrated across subsets of blood-sensitive neurons and for the particular subset of Integrator neurons, within individual neurons.
We directly observed integration by a subset of stylet neurons maximally activated by co-presentation of glucose, NaCl, and NaHCO3. Simultaneous detection of sweet, salty, and NaHCO3 in one neuron is unexpected because distinct taste qualities are thought to activate non-overlapping sensory neuron populations in both mammals and insects (Yarmolinsky et al., 2009). Yet here we only detect responses to physiological levels of blood glucose (4.5 mM) in the presence of NaCl or NaHCO3. We speculate that polymodal Integrator neurons act as coincidence detectors and that 4.5 mM glucose alone produces subthreshold responses without the co-presentation of NaCl and/or NaHCO3. Since glucose is a redundant cue in blood and nectar, this unconventional taste coding mechanism confers an important distinction between glucose present in blood versus nectar.
We speculate that taste quality integration occurs across the distinct blood-sensitive neuronal subsets to form the neural representation of blood. We found that behaviorally-relevant concentrations of ATP, NaHCO3, and NaCl were individually sufficient to activate a subset of stylet neurons. However, any individual component was unable to trigger blood-feeding behavior or activate all blood-sensitive stylet neurons. Consistent with these observations, we found that activation of either Ir7a- and Ir7f-expressing subpopulations alone using the TRPV1 chemogenetic system did not promote engorgement (Figure S6J–L). Moreover, mutant mosquitoes lacking either Ir7a or Ir7f still engorged on blood or ATP in saline (Figure S6H,I). These results are consistent with the importance of integration of sensory information from multiple stylet neuronal subtypes, and the hypothesis that multiple chemosensory receptor genes can be expressed in a given sensory neuron (Abuin et al., 2011; Slone et al., 2007). We propose that activation of multiple stylet neuron subsets is required to initiate blood feeding to decrease the possibility that a female accidentally engorges on nectar instead of blood. For instance, 298 mM glucose occasionally activated blood-sensitive neurons, but females still rejected this meal in the blood-feeding assay. Determining which receptors and neuronal subsets are sufficient and necessary for blood feeding remains an important area for future work.
The Stylet is Specialized to Detect Blood Over Nectar
The needle-like anatomy of the stylet is ideally adapted to blood feeding (Choumet et al., 2012; Ramasubramanian et al., 2008) and we discovered that its functional properties directly encode a distinction between blood and nectar. We propose that specialization of peripheral sensory neurons in the stylet may explain why sugars do not promote nectar feeding in the context of blood feeding. This mechanism is distinct from previously described examples of food source valence changes upon nutrient deprivation or mating in D. melanogaster, which typically involve a state-change that modulates the sensitivity of sensory neurons, and/or their downstream processing, to a given ligand (Devineni et al., 2019; Inagaki et al., 2012; Steck et al., 2018; Walker et al., 2015). One key difference between D. melanogaster and Ae. aegypti feeding is that Ae. aegypti have two distinct feeding appendages. We speculate that feeding appendage segregation and specialization is a mechanism to ensure that the female ingests blood and not nectar in the context of blood feeding. Furthermore, female-specific stylet sensilla are conserved across blood-feeding mosquito species and are absent in non-blood-feeding Toxorhynchites species (Lee and Craig, 1983). Although mosquito species differ in the minimum blood components required to initiate blood feeding (Galun, 1987), blood detection via stylet neurons may be a conserved mechanism across blood-feeding mosquito species. Blood detection is an important step for Ae. aegypti mosquitoes to transmit diseases like Zika and dengue because they acquire these flaviviruses by ingesting a blood meal from an infected person (Ruckert and Ebel, 2018). An understanding of blood detection is fundamental to prevent mosquito blood-feeding behavior, which is responsible for transmission of vector-borne diseases to hundreds of millions of people world-wide each year.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for reagents should be directed to and will be fulfilled by the lead contact, Leslie Vosshall (leslie.vosshall@rockefeller.edu).
Materials Availability
All plasmids described in this paper are available at Addgene. Genetically modified mosquitoes are available upon request.
Data and Code Availability
All data in the paper (with the exception of raw video files) are available on Github at https://github.com/VosshallLab/Jove_Vosshall_2020. Sequencing reads have been deposited at the NCBI Sequence Read Archive (SRA) under BioProject PRJNA605870. Custom Python code for biteOscope data analysis is available on Github at https://github.com/felixhol/biteOscope. Custom R scripts for merged genome annotation and calcium imaging analysis are available on Github at https://github.com/VosshallLab/Jove_Vosshall_2020.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human and Animal Ethics Statement
Blood-feeding procedures with live mouse and human hosts were approved and monitored by The Rockefeller University Institutional Animal Care and Use Committee (IACUC protocol 17108) and Institutional Review Board (IRB protocol LV-0652), respectively. Human subjects gave their written informed consent to participate.
Mosquito Rearing and Maintenance
Ae. aegypti wild-type and genetically-modified strains were maintained and reared at 25 - 28°C, 70–80% relative humidity with a photoperiod of 14 hours light: 10 hours dark (lights on at 7 a.m.) as previously described (DeGennaro et al., 2013). Adult females were blood-fed on mice for stock maintenance, and occasionally on human subjects in the early stages of generating genetically modified strains. Approximately the same number of female and male pupae were placed in one cage prior to eclosion. Adults were allowed to mate freely for at least 7 days prior to performing experiments. Adult mosquitoes were provided constant access to 10% sucrose. 14 – 24 hours prior to behavioral experiments, mosquitoes were briefly anesthetized at 4°C and females were sorted into groups of 15-20 females and were placed into a 32 oz. HDPE plastic cup (VWR #89009-668). Upon returning to the insectary, females were fasted by replacing 10% sucrose with a water source. All behavior experiments were carried between ZT6 and ZT13 and ended before the lights off time based on the photoperiod.
METHOD DETAILS
Generation of Genetically-Modified Mosquito Strains
All CRISPR-Cas9 and transgene injections followed previously established methods (Kistler et al., 2015; Matthews et al., 2019) and were carried out at the Insect Transformation Facility (ITF) at the University of Maryland Institute for Bioscience & Biotechnology Research.
All new strains generated in this paper were generated in the Vosshall Lab using the wild-type Liverpool strain of Aedes aegypti. Brp-QF2w was generated in the McBride Lab using the wild-type Orlando strain of Aedes aegypti (Zhao et al., 2020). We back-crossed Brp-QF2w to wild-type Liverpool for at least 4 generations before crossing to QUAS-dTomato-T2A-GCaMP6s, which was generated in the Liverpool background.
For instances where a transgene was integrated into the genome using homologous recombination, proper payload integration was confirmed using polymerase chain reaction (PCR). Animals were then back-crossed to wild-type Liverpool for at least three generations before crossing to corresponding QF2 or QUAS for experimental use. Details of plasmid construction are below. All homology arms for homology-directed integration were isolated by PCR from genomic DNA isolated from the Liverpool strain, except for Brp-QF2w, which was derived from the Orlando strain. When Gibson assembly was utilized in plasmid construction, oligonucleotide sequences are displayed in lower case to indicate homology to the adjacent fragment and upper case to indicate the target sequence.
For instances of a gene-disrupting insertion/deletion at a specific locus, a frame-shift mutation was confirmed using PCR and Sanger DNA sequencing (Genewiz). Mutants were then back-crossed to wild-type Liverpool for 3 total generations before inbred to generate a stable homozygous mutant line.
3xP3-eYFP-SV40-15xQUAS-dTomato-T2A-TRPV1-SV40 (Addgene plasmid#140945)
This plasmid was generated using NEBuilder HiFi DNA Assembly (New England Biolabs #E5520S), using the following fragments generated by PCR from the indicated template with the indicated primers:
Plasmid backbone with pBAC arms from 15xQUAS-dTomato-T2A-GCaMP6s (Addgene plasmid #130666) (Matthews et al., 2019) (Primers: Forward, 5’-GATCTTTGTGAAGGAACCTTACTTCTGTGGTGTG-3’; Reverse, 5’-ATCCCCCGGGCTGCAGGA-3’)
QUAS-dTomato-T2A from 15xQUAS-dTomato-T2A-GCaMP6s (Primers: Forward, 5’-tcaatgtatcttaACTAGAGCGGCCGCCACC-3’; Reverse, 5’-cccgttgttccatAGGGCCGGGATTCTCCTC-3’)
3xP3-eYFP-SV40 with YFP open reading frame from Addgene plasmid #62291 (Primers: Forward, 5’-atcgaattcctgcagcccgggggatGTTCCCACAATGGTTAATTC-3’; Reverse, 5’-ggccgctctagtTAAGATACATTGATGAGTTTGG-3’).
Rattus norvegicus TRPV1 (Genbank accession NM_031982.1) from ASH:TRPV1 (Bargmann Lab plasmid #10.33.42, with permission from Dr. David Julius of UCSF) (Tobin et al., 2002) (Primers: Forward, 5’-aatcccggccctATGGAACAACGGGCTAGC-3’; Reverse, 5’-gaagtaaggttccttcacaaagatcACCCAGATAACGTCAACC-3’).
200 embryos were injected with 200 ng/μL plasmid and 200 ng/μL pBAC mRNA. Two independent transgenic lines were recovered, one of which was sex-linked. In pilot experiments, both lines showed qualitatively similar behavioral effects in the Gr4>TRPV1 capsaicin experiments. All subsequent behavior and expression pattern experiments were performed using the non-sex-linked line.
Gr4, Ir7a, and Ir7f QF2 strains
These knock-in/knock-out strains were generated through CRISPR-mediated homologous recombination of the QF2 transcription factor (Potter et al., 2010; Riabinina et al., 2015) into the endogenous locus of the Ae. aegypti genome. In vitro transcription was performed using HiScribe Quick T7 kit (New England Biolabs #E2050S) following the manufacturer’s directions and incubating for 3 hr at 37°C. Following transcription and DNAse treatment for 15 min at 37°C, sgRNA was purified using RNAse-free SPRI beads (Ampure RNAclean, Beckman-Coulter #A63987), and eluted in Ultrapure water (Invitrogen #10977–015). For each line, 2000 embryos were injected with 600 ng/μL plasmid, 300 ng/μL Cas9 protein, and 40 ng/μL sgRNA. sgRNA DNA template was prepared by annealing oligonucleotides as previously described (Kistler et al., 2015). For all plasmids, fragments were generated by PCR from the indicated template with the indicated primers and assembled using NEBuilder HiFi DNA Assembly as detailed below.
Gr4-T2A-QF2 -SV40-3xP3-dsRed (Addgene plasmid#140944)
Plasmid backbone from pUC19 (Primers: Forward, 5’- CTAGAGTCGACCTGCAGGC −3’; Reverse, 5’- CCCGGGTACCGAGCTCGA −3’).
Gr4 left homology arm (NCBI LOC5563657) (Primers: Forward, 5’- agtgaattcgagctcggtacccgggACTCTCCTAAAATCTCAAGTATAC-3’; Reverse, 5’- tctgccctctccTGCACGTTTGGGATACTTG-3’).
Gr4 right homology arm (NCBI LOC5563657) (Primers: Forward, 5’- caatgtatcttaCAGGGAAAACTGGATCCATG-3’; Reverse, 5’- ttgcatgcctgcaggtcgactctagGTGTATTTGGAGCCTCAG-3’).
T2A- QF2-SV40-3xP3-dsRed with QF2 and dsRed open reading frame from ppk301-T2A-QF2 (Addgene plasmid #130667) (Matthews et al., 2019) (Primers: Forward, 5’- tcccaaacgtgcaGGAGAGGGCAGAGGAAGTC-3’; Reverse, 5’- ccagttttccctgTAAGATACATTGATGAGTTTGGACAAAC-3). The sgRNA targeted exon 2 of the Gr4 locus, target sequence with protospacer adjacent motif (PAM) underlined: GTATCCCAAACGTGCAACCAGGG.
Ir7a-T2A-QF2 -SV40-3xP3-dsRed (Addgene plasmid#140943)
Plasmid backbone from pUC19 (Primers: Forward, 5’-cgatcaactataaCTAGAGTCGACCTGCAGGC −3’; Reverse, 5’- aatttgctttttaCCCGGGTACCGAGCTCGA-3’.
Ir7a left homology arm (Primers: Forward, 5’-cggtacccgggTAAAAAGCAAATTTCACCATG-3’; Reverse, 5’- tctgccctctccATATACGTGACCCCAAATATC-3’).
Ir7a right homology arm (Primers: Forward, 5’- caatgtatcttaATCCAGAACGGGTGCGGTAG-3’; Reverse, 5’- ggtcgactctagTTATAGTTGATCGAGGAATTTCCGAATCC-3’).
T2A- QF2-SV40-3xP3-dsRed with QF2 and dsRed open reading frame from ppk301-T2A-QF2 (Addgene plasmid #130667) (Matthews et al., 2019) (Primers: Forward, 5’- gggtcacgtatatGGAGAGGGCAGAGGAAGTC-3’; Reverse, 5’- acccgttctggatTAAGATACATTGATGAGTTTGGACAAAC-3’). The sgRNA targeted exon 1 of the Ir7a locus, target sequence with PAM underlined: TGGGGTCACGTATATCCAAATGG.
Ir7a was not annotated in the AaegL5 NCBI RefSeq Annotation version 101 (Matthews et al., 2018). Genomic coordinates (NC_035107.1:37734383-37736188; FASTA file available in Data File 1) were identified using the manual chemoreceptor annotation (Matthews et al., 2018). See the “Transcript abundance and differential expression analysis” section below for additional annotation information.
Ir7f-T2A-QF2 -SV40-3xP3-dsRed (Addgene plasmid#140942)
Plasmid backbone from pUC19 (Primers: Forward, 5’-attttgaggcgggCTAGAGTCGACCTGCAGGC-3’; Reverse, 5’-aatcagccagtcaCCCGGGTACCGAGCTCGA-3’).
Ir7f left homology arm (NCBI LOC5565007) (Primers: Forward, 5’-ctcggtacccgggTGACTGGCTGATTAGCTCATCCTATATAAGAA-3’; Reverse, 5’- ctctgccctctccACGCTCGCCACGCATCGAGAAACACCCGG-3’).
Ir7f right homology arm (NCBI LOC5565007) Primers: Forward, 5’-tcaatgtatcttaTGTCGGTGATGAGGTCCAG −3’; Reverse, 5’-aggtcgactctagCCCGCCTCAAAATGTGCAC-3’).
T2A- QF2-SV40-3xP3-dsRed with QF2 and dsRed open reading frame from ppk301-T2A-QF2 (Addgene plasmid #130667) (Matthews et al., 2019) (Primers: Forward, 5’-gcgtggcgagcgtGGAGAGGGCAGAGGAAGTC-3’; Reverse, 5’-ctcatcaccgacaTAAGATACATTGATGAGTTTGGACAAAC-3’). The sgRNA targeted exon 1 of the Ir7f locus, target sequence with PAM underlined: GATGCGCGGTGAACGCATGTCGG.
Brp-QF2w strain
This knock-in strain was generated in the McBride Lab (Zhao et al., 2020) in the wild-type Orlando strain background using CRISPR-mediated homologous recombination of the QF2w transcription factor (Potter et al., 2010; Riabinina et al., 2015) into the endogenous bruchpilot locus (NCBI LOC5570381) of the Ae. aegypti genome.
Ir7a−/− and Ir7f−/− loss-of-function strains
These mutant strains were generated using CRISPR-Cas9 as described previously (Kistler et al., 2015) except that 4 sgRNA (instead of 2) were targeted to Exon 1 in Ir7a or Ir7f, respectively, to increase the probability of cutting. In vitro transcription was performed using HiScribe Quick T7 kit (New England Biolabs #E2050S) following the manufacturer’s directions and incubating for 3 hr at 37°C. Following transcription and DNAse treatment for 15 min at 37°C, sgRNA was purified using RNAse-free SPRI beads (Ampure RNAclean, Beckman-Coulter #A63987), and eluted in Ultrapure water (Invitrogen #10977–015). For each line, 400 embryos were injected with 200 ng/μL ssODN, 4x 40 ng/μL sgRNA, and 300 ng/μL Cas9. Although a ssODN was injected into both strains, the recovered loss-of-function strains did not have successful integration. Both strains contain a frame-shift deletion.
Ir7a−/− 70 base pair frame-shift
ssODN: TTGAAGACAGAAAAAGGCGGCTGGTTCTTCGGGGAGTCGAAGGAGATATGCTGACGATCATTTCAAGAAGAATGAACTTCTCGATTTAGGTTAGAGTTCCCACGGGTGAAGATATTTGGGGTCACGTATATCCAGTCATGGCTAATTAATTAAGCTGTTGTAGCGGTGGTTGTCGGTACGGCATACTTAGTTATACTCATATTCAGTTGTCCGCTAATGGGGTACTTCAACCATTCTCCAGCATTGACTCTGTATCGGACAACTATTGGGGATTCCCTCCCATCACTTCCAACAGGAAACTT
-
All sgRNA targeted exon 1 of the Ir7a locus, target sequences with PAM underlined:
CACCCGTGGGAACTCTAACCTGG
TGGGGTCACGTATATCCAAATGG
GATTTGGATAGGCATGGCGGTGG
ACTCATATTCAGTTGTCCGCTGG
-
[3] PCR primers for Sanger DNA sequencing and genotyping:
Forward, 5’- GAGATATGCTGACGATCATTTCAAG-3’; Reverse, 5’-TAGAACATTTGTAGCTCTCCCTTAT-3’.
To control for genetic background, Ir7a+/− females were mated to Ir7a+/− males to generate animals for the behavior experiments in Figure S6H. This allowed Ir7a+/− females to be directly compared to Ir7a−/− females. All animals were genotyped after behavior experiments so that the experimenter was blind to genotype during the experiment.
Ir7f−/− 260 base pair frame-shift mutation
ssODN: CACTCCAGCGCCAGCCAACGTGTACAATTTCACCATCATCCAGGTGACAGCACTAAACGGTCGGAACATCTTCTCGAACGCCGTGTAGGGCCTTCCCTAATAAGGATCCATAACCTAAGGTACGTGAAGTTCAGCTCCGAGGAAATCATGTTCAGCATGTCGCCTTCTATTTTACGTAGTCTTCGGCGACCTCCAATCCA
-
All sgRNA targeted exon 1 of the Ir7f locus, target sequences with PAM underlined:
AGCGCCAGCCAACGTGTACAAGG
GCCGTGTAGGGCCTTCCCGGTGG
GGAGCTGAACTTCACGTACGAGG
GGAGGTCGCCGAAGACTACGTGG
-
PCR primers for Sanger DNA sequencing and genotyping:
Forward, 5’-ATA CGT TGA ACA TCA CTG TGA ACA T-3’; Reverse, 5’-AGCCAACGTGTACAAGGTC-3’
To control for genetic background, Ir7f+/− females were mated to Ir7f+/− males to generate animals for the behavior experiments in Figure S6I. This allowed Ir7f+/− females to be directly compared to Ir7f−/− females. All animals were genotyped after behavior experiments so that the experimenter was blind to genotype during the experiment.
Ligands for Feeding Experiments
Sheep blood:
(Hemostat Laboratories #DSB100) was used within 1 week of arrival.
Nucleotides:
ATP (Adenosine 5′-triphosphate disodium salt hydrate, Sigma #A6419), AMP-PNP (β,γ-imidoadenosine 5’-triphosphate lithium salt hydrate, Millipore Sigma #10102547001), AMP-CPP (α,β-methyleneadenosine 5’-triphosphate lithium salt, Jena Bioscience #NU-421-25), AMP-PCP (β,γ-Methyleneadenosine 5′-triphosphate disodium salt, Millipore Sigma #M7510). ATP and non-hydrolyzable analogues were reconstituted and aliquoted in 25 mM NaHCO3.
Sugars:
sucrose (Fisher Scientific #S5-3), cellobiose [D-(+)-cellobiose, Millipore Sigma #22150], fructose [D-(−)-Fructose, Millipore Sigma #F0127], glucose [D-(+)-Glucose, Millipore Sigma #G7528].
Additional blood components:
NaCl (Millipore Sigma #S6546), NaHCO3 (Fisher Scientific #S233), albumin (human serum, Millipore Sigma #A9511), hemoglobin (human, Millipore Sigma #G4386), gamma-globulin (human blood, Millipore Sigma, #H7379).
Capsaicin:
(E)-capsaicin (Tocris #0462)
Blood-Feeding Assay (Glytube)
7 to 21 day-old female mosquitoes were anesthetized at 4°C and sorted into groups of 15-20 females, and placed into a 32 oz. HDPE plastic cup (VWR #89009-668). The cup was prepared by cutting a 10 cm hole in the lid with a razor blade, covering the cup with a 20 cm x 20 cm piece of white 0.8 mm polyester mosquito netting (American Home & Habit Inc. #F03A-PONO-MOSQ-M008-ZS) and securing the mesh to the cup by snapping on the modified lid. Animals recovered overnight at 25 - 28°C, 70–80% relative humidity with access to water. The assay chamber was a modification of previously published methods (McMeniman et al., 2014) and used a translucent polypropylene storage box 36 cm L x 31 cm W x 32 cm H with a removable lid. One 1.5 cm hole was made on the chamber wall and was used to introduce silicone tubing for CO2 delivery. The CO2 diffusion pad (8.9 cm x 12.7 cm; Tritech Research) was affixed to the inner center of the lid to allow delivery of purified air and CO2 to condition the chamber atmosphere during the trial. Up to 4 cups were placed in the chamber per trial and feeding positions were randomized according to meal during assays. Females were fed sheep blood or test ligands using Glytube membrane feeders exactly as described (Costa-da-Silva et al., 2013), except the Parafilm feeding surface was not rubbed on human skin prior to offering the Glytube to mosquitoes to avoid introducing contact chemosensory cues as secondary stimuli in our experiments. In Figure 1, Figure 2 and Figure S1, the saline meal contained 110 mM NaCl and 20 mM NaHCO3. All meals and Glytubes were preheated for at least 15 min in a 45°C water bath and, if required, ATP or non-hydrolyzable ATP analogues were added to meals immediately before feeding and mixed by vortexing. At the start of each trial, cups were placed in the assay chamber and allowed to acclimate for 5 min before 1 Glytube containing 1.5 mL of a given meal was placed on top each cup and CO2 was turned on for 15 min. In Figure 1M, Figure S1E,G,I, and Figure 5A–B, fed females were scored by eye for engorgement of the abdomen. In the rare cases that females partially fed they were counted as non-fed and discarded. To sample the weights of these females (Figure 1K,L and Figure S1F,H,J), a selection of engorged individuals was weighed in groups of 5 females and the resulting weight in mg was divided by 5 to report the average weight per female.
In Figure 1E and Figure 7C, Glytube feeding was performed as described, except that fluorescein (Amresco #0681) was added as a fluorescent tracer to each meal (blood, sucrose, fructose, glucose, or water) at a final concentration of 0.002%. After feeding, females were frozen at −20°C until they were processed for fluorescence reading. A 96-well PCR plate was prepared with one 3 mm diameter borosilicate solid-glass bead (Millipore Sigma #Z143928) and 100 μl PBS in each well. 8 wells were used to generate a reference standard curve. These wells contained a single unfed mosquito and the following volumes of the same fluorescent meal fed to test mosquitoes: 5, 2.5, 1.25, 0.625, 0.3125, 0.15625, 0.078125, or 0 μL. One test group mosquito was added to each of the remaining wells. Tissue was disrupted using TissueLyser II (Qiagen) and briefly centrifuged at 2000 rpm for 1 – 2 min. 20 μL of tissue lysate from each well was added to 180 μL PBS in a well of a black 96-well plate (ThermoFisher #12-566-09). Fluorescent intensity for each well was measured using the 485/520 excitation/emission channel of a Varioskan Lux (ThermoFisher #VL0000D0) plate reader. Using the reference dilution curve, fluorescent measurements were converted to volume (μL) of solution ingested. Measurements below the level of detection were quantified as 0 for plotting and statistical analysis.
Nectar-Feeding Assay
Animals were prepared exactly as described for the Glytube assay. Consumption of nectar was quantified by supplementing the meal with 0.002% fluorescein. A cotton ball (Fisher Scientific #22456880) was soaked in each test meal, the cotton ball was briefly dabbed on a Kimwipe to prevent excess liquid from dripping through the mesh, and placed on top of the mesh covering the cup. Animals were allowed to feed for 4 hours. After feeding, animals were frozen at −20°C and fluorescence reading was performed as described.
Meal Size Quantification
In Figure 1E,F, we analyzed the average meal size of mosquitoes that fed on blood or sugar respectively. Mosquitoes that did not feed were excluded from meal size analysis. To set a cut-off for whether or not a mosquito fed, we included unfed control groups that were not offered a meal and therefore reflected a true 0. We detected fluctuations in baseline from 0 – 0.0304 μL. We therefore set a cut-off at 0.05 μL and excluded animals in the blood or sugar experimental group that measured < 0.05 μL. We then applied this 0.05 μL cut-off for statistical analysis in subsequent meal size quantification experiments in Figure 1J, Figure S1D, and Figure 7B,C: all values < 0.051 were replaced with 0.05. This cut-off was also applied to determine whether or not a female fed in Figure S7A,B.
Chemogenetic Capsaicin Feeding Assays
Chemogenetic experiments using capsaicin to activate Gr4>TRPV1 sensory neurons were carried out exactly as the nectar-feeding experiments except that 50 μM capsaicin in 0.1% DMSO or 0.1% DMSO only-control was added to the meals. Chemogenetic experiments using capsaicin to activate Ir7a>TRPV1 and Ir7f>TRPV1 sensory neurons were carried out exactly as the blood-feeding (Glytube) experiments except that 50 μM capsaicin in 0.1% DMSO or 0.1% DMSO only-control was added to the meals.
biteOscope Assay
Stylet piercing behavior was characterized using the biteOscope (Hol et al., 2020). Briefly, all meals were prepared exactly as for the Glytube experiments. The meal was applied on the rectangular section on the outside of a 70 mL Falcon cell culture flask and covered with parafilm. To maintain meal temperature, the flask was filled with warm water maintained at 37°C using a Raspberry Pi controlled Peltier element. The flask was mounted in the floor of a 10 cm x 10 cm x 10 cm acrylic cage. A camera (Basler #acA2040-90um) and two white LED arrays for illumination (Vidpro #LED-312) were mounted outside the cage to image mosquitoes interacting with the bite substrate. At least 12 hours prior to the experiment, females were fasted by replacing 10% sucrose with a water source. At the start of each trial, an individual female was introduced into the cage and the experimenter (F.J.J.H.) blew on the cage 2 times 10 sec to provide human cues. Images were acquired at 10 frames/sec using Basler Pylon 5 software running on Ubuntu 18.04. Each female was recorded for 700 sec regardless of engorgement status. Images were processed using custom code written in Python (available from Github: https://github.com/felixhol/biteOscope) using SciPy (Virtanen et al., 2019), TrackPy (Allan et al., 2019), and OpenCV (Bradski, 2000) packages to determine the presence and location of a mosquito. Engorgement status of a mosquito was determined by measuring abdominal size by fitting an active contour model to its abdomen. Stylet piercing events were scored by manual visual analysis of the images.
Tissue Fixation Protocol
Tissue fixation followed modification of previously published methods (Matthews et al., 2019) as follows. Heads were carefully removed from the body by pinching at the neck with sharp forceps. Heads were placed in a 1.5 mL tube for fixation with 4% paraformaldehyde, 0.1 M Millonig’s Phosphate Buffer (pH 7.4), 0.25% Triton X-100, and nutated for 3 hour at 4°C. Samples were dissected and samples of the same tissue were grouped into a cell strainer cap (Fisher Scientific #08-771-23) that was cut to fit into 1 well of a 24-well plate containing PBS with 0.25% Triton X-100 (PBT). All subsequent steps were performed on a low-speed orbital shaker at room temperature. Samples were washed at least 5 times 20 min and transferred to PBT. All dissections were performed using this protocol unless otherwise noted.
TO-PRO-3 Staining
7 to 14 day-old animals were anesthetized on ice. Heads were removed and fixed prior to tissue dissection according to the tissue fixation protocol. Samples were transferred to a well of PBT with 1:400 TO-PRO-3 (ThermoFisher #T3605) for 2 days. Samples were washed at least 5 times 20 min in 0.25% PBT. After washing, tissues were briefly transferred to a well of SlowFade diamond (ThermoFisher #S36972) to eliminate excess PBT. Samples were then mounted in SlowFade. Within each experiment, all image acquisition parameters were maintained across both sexes.
dTomato Visualization
7 to 14 day-old mosquitoes were anesthetized on ice. Heads were removed and fixed prior to tissue dissection according to the tissue fixation protocol. Samples were briefly transferred to a well of Vectashield (Vector Laboratories #H-1000) to remove excess PBT. Samples were then mounted in Vectashield. Within each genotype, all image acquisition parameters were maintained across tissue types. At higher laser power, we observed very faint cells in Ir7f>dTomato-T2A-GCaMP6s female labiums (Figure 6B, right panel) but we suspect that they are not neurons because we did not observe nerve fibers exiting the labium or projecting to the posterior subesophageal zone where labium neurons normally terminate (Figure 3L, Figure S2I).
Phalloidin, DAPI, and FITC Staining
7 to14 day-old mosquitoes were anesthetized on ice. Stylets were dissected and placed directly into a 24 well-plate containing 4% paraformaldehyde, 0.1 M Millonig’s Phosphate Buffer (pH 7.4) and 0.25% Triton X-100. All subsequent steps were performed on a low-speed orbital shaker at room temperature. Samples were washed at least 4 times 15 min in PTx.2 (for 1L: 100 mL PBS 10x, 2 mL TritonX-100) before placed overnight in iDISCO permeabilization solution (for 500 mL: 400 mL PTx.2, 11.5 g glycine, 100 mL DMSO) (Renier et al., 2014). Samples were then incubated in iDISCO PTwH solution (for 1L: 100 mL 10x PBS, 2mL Tween-20, 1 mL of 10mg/mL Heparin stock solution) with 5% DMSO for at least 2 days at room temperature with the following reagents: (1) 1:20 AlexaFluor 594 phalloidin (ThermoFisher #A12381) (Figure 3F) or (2) 1:20 AlexaFluor 488 phalloidin (ThermoFisher #A12379) and 1:500 DAPI (Millipore Sigma #D9542) (Figure S2D) or (3) 1:20 AlexaFluor 647 phalloidin (ThermoFisher #A22287) (Figure S2G) or (4) 2 mg/mL FITC (Millipore Sigma #1.24546) (Figure S2A). Samples were then washed at least 4 times 15 min with PTx.2 solution and mounted in Vectashield. If a sample contained AlexaFluor 647, it was mounted in SlowFade instead of Vectashield because this fluorophore was better preserved in this mounting medium.
Dextran Dye-Fills
7 to 14 day-old mosquitoes were anesthetized on ice. The labium was separated from the stylet using forceps. Mosquitoes were affixed on their side to a plastic dish (Falcon #353001) using UV-curable glue (Bondic, Amazon #B0181BEHQU) or double-sided tape so that the stylet and labium were flat on the dish and distal tips were separated. For stylet dye-fills, a scalpel was used to cut approximately 300-750 μm away from the distal tip and 1 μL of Dextran, Texas Red, 3000 MW, Lysine Fixable (ThermoFisher #D3328) diluted to 1 mg/10 μL in External Saline was added immediately. The External Saline recipe (Matthews et al., 2019) is based on D. melanogaster imaging saline: 103 mM NaCl, 3 mM KCl, 5 mM 2-[Tris(hydroxymethyl)methyl]-2-aminoethanesulfonic acid (TES), 1.5 mM CaCl2, 4 mM MgCl2, 26 mM NaHCO3, 1 mM NaH2PO4, 10 mM trehalose, 10 mM glucose, pH 7.3, osmolality adjusted to 275 mOsm/kg. The mosquito was left on ice and covered for approximately 3-5 min before excess dye was removed by pipette. Mosquitoes were left at 4°C overnight in a closed Petri dish with a moist Kimwipe placed in the corner to prevent desiccation. Heads were then removed and fixed prior to tissue dissection according to the tissue fixation protocol.
For double dye-fills of stylet and labium, the mosquito was prepared as described for single dye-fills above. The labium was cut at the base of the labellar lobes using a scalpel and 1 μL of Dextran, Texas Red diluted to 1 mg/10 μL in External Saline was added immediately. The mosquito was left on ice and covered for approximately 3-5 min before excess dye was removed by pipette. The stylet was cut approximately 300 – 750 μm away from the distal tip and 1 μL of Dextran, Fluorescein and Biotin, 3000 MW, Lysine Fixable (ThermoFisher #D7156) diluted to 1 mg/10 μL in External Saline was immediately added. The mosquito was left on ice and covered for approximately 3-5 min before excess dye was pipetted up. Mosquitoes were left at 4°C overnight with a moist Kimwipe to prevent desiccation. Heads were then removed and fixed prior to tissue dissection according to the tissue fixation protocol.
Fixed heads of both single and double dye-fill preparations were then dissected and brains were placed in cell-strainer caps (Falcon #352235) in a 24 well-plate. Brains were stained using a modification of previously published methods (Matthews et al., 2019). All subsequent steps were performed on a low-speed orbital shaker. Brains were washed at room temperature in PBT for at least 4 times 15 min. Brains were permeabilized with 4% Triton X-100 with 2% normal goat serum (Jackson ImmunoResearch #005-000-121) in PBS at 4°C for 2 days. Brains were washed at least 5 times 15 min with PBT at room temperature before being incubated in PBT plus 2% normal goat serum for 3 days at 4°C degrees. The following primary antibodies at the following dilutions were used: rabbit anti-fluorescein (ThermoFisher #A889) 1:500 and mouse anti-Drosophila Brp (nc82) 1:50. The nc82 hybridoma developed by Erich Buchner of Universitätsklinikum Würzburg was obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at The University of Iowa, Department of Biology, Iowa City, IA 52242. Following primary antibody incubations, brains were washed at least 5 times 15 min with PBT at room temperature. Brains were incubated with secondary antibody for 3 days at 4°C with secondary antibodies at 1:500: goat anti-rabbit Alexa Fluor 488 (ThermoFisher #A-11008) and goat anti-mouse Alexa Fluor 647 (ThermoFisher #A-21236). Brains were then washed PBT and mounted in Vectashield.
Brain Immunostaining
8 to 9 day-old mosquitoes were anaesthetized on ice. Heads were then removed and fixed prior to tissue dissection according to the tissue fixation protocol. Primary antibodies were used at the following dilutions: rat anti-mCD8 (Invitrogen #14008185) 1:100, and a concentrated aliquot of mouse anti-Drosophila Brp 1:5000 generated in-house with the nc82 hybridoma obtained from DHSB. Brains were then washed 5x for at least 30 min at room temperature. Brains were then incubated with secondary antibodies in PBT with 2% normal goat serum for 2 days at 4˚C. The following secondary antibodies were used at 1:500 dilutions: goat anti-rat Alexa Fluor 647 (Invitrogen #A21247) and goat anti-mouse Alexa Fluor 555 (Invitrogen #A32727). Brains were then washed 6 times in PBT at room temperature for at least 30 min then mounted in SlowFade diamond. 3xP3 was used as a promoter to mark transgene insertion as previously described (Matthews et al., 2019). To avoid any interference from possible 3xP3 signal, we used a different laser excitation/secondary antibody for monitoring Ir7a, Ir7f, and Gr4 expression. Within each genotype, all image acquisition parameters were maintained across both sexes. Ir7a and Ir7f are expressed in a maximum of 2 and 4 neurons, respectively (Figure 6A,B), which is far fewer neurons than Gr4 (Figure 1I and Figure S1A). We also noted that it was easier to detect processes in the subesophageal zone of Gr4 > CD8-GFP animals (Figure S7C,D) compared to Ir7a > CD8-GFP or Ir7f > CD8-GFP animals (Figure 6C,D), leading us to use a higher laser power to acquire these images. Upon generating the max projections for Figure 6C,D, we noted that the background signal from tissue autofluorescence is higher in Ir7a > CD8-GFP and Ir7f > CD8-GFP animals. However, this background signal is not correlated with innervation from stylet neurons since females and males from the same genotype were both imaged at the same settings and both have similar background, but only females have subesophageal zone innervation (Figure 6C–F). The complete image stacks used to generate the max projections are available in Movie 6 (Ir7a) and Movie 7 (Ir7f) and show the clear difference between neuronal innervation and background tissue autofluorescence.
Confocal Image Acquisition
Images were acquired with a Zeiss Axio Observer Z1 Inverted LSM 880 NLO laser scanning confocal microscope (Zeiss) with a 25x/0.8 NA immersion-corrected objective at a resolution of 2048 x 2048 or 1024 x 1024 pixels. When necessary, tiled images were stitched with 10% overlap. Confocal images were processed in ImageJ (NIH).
Ex-Vivo Stylet Prep for Calcium Imaging
Calcium imaging was performed on an inverted Ti-2E wide-field microscope (Nikon) with a dual FITC/TRITC bandpass cube and alternating emission wheel with 520/40 GFP and 628/40 RFP bandpass filters. A nd2 filter was added with the 628/40 RFP bandpass filter to attenuate dTomato signal. Images were acquired with a 25x/0.9 N.A. water-immersion objective (Nikon) and Zyla 4.2 Plus camera. Calcium imaging experiments were performed on female mosquitoes that were 7–14 days post-eclosion.
Prior to dissection, the imaging chamber was prepared by affixing a Gold Seal Cover Glass, No. 1 22 x 40 mm coverslip (Ted Pella #260353) to a recording chamber using silicone lubricant (Dow Molykote 111 O-Ring Silicone Lubricant). A fast exchange recording chamber (Warner Instruments #64-0230) was used for perfusion-only experiments and a low-profile large bath recording chamber (Warner Instruments #640236) was used to accommodate the BioPen apparatus. A drop of silicone lubricant approximately 100-200 μm in diameter was placed slightly off-center on the coverslip.
After preparing the chamber, females were anesthetized briefly at 4°C for dissection. The labium was removed to expose the stylet, and then the stylet was detached at the proximal end using a scalpel (Feather disposable scalpel, No. 11, Fisher Scientific #FH/CX7281A). The severed end was immediately placed in the drop of silicone lubricant with the stylet tip facing the center of the coverslip. Great care was taken to place the stylet flat along the coverslip so that all stylet neurons could be imaged in one plane. This process often involved carefully removing the maxillae and mandibles without damaging the stylet. However, if the stylet was already flat, it was not necessary to remove additional appendages as they did not interfere with image acquisition. The most distal 300 μm of the stylet tip remained free of silicone lubricant to prevent interference with ligand delivery. Once the stylet was secured to the coverslip, the chamber was filled with MilliQ water and the perfusion and/or BioPen fluidics were inserted into the chamber.
dTomato fluorescence was examined before and throughout imaging to verify that the stylet nerves were intact. The sample remained stable during the duration of the imaging session in all animals that were included in this study. Each image acquisition captured one GCaMP image and one dTomato image separated by less than the 100 ms required to switch the filter wheel. Image acquisition was triggered at a rate of approximately 2 frames per sec for each channel (2 sets of GCaMP/dTomato images per sec).
Perfusion Ligand Delivery
Two independent ValveBank8 Pinch Valve perfusion systems (Automate Scientific #13-pp-54) with BubbleStop8 60 mL Syringe Heater (Automate Scientific #10-8-60-G) were automatically controlled by NIS-Elements software (Nikon). To ensure full perfusion chamber exchange, ligands were perfused for 30 sec followed by a 45 sec recovery period before the next ligand. Ligand delivery switched from water (baseline) to ligand of interest with the following exceptions. Since ATP is rapidly hydrolyzed in water, ATP was always delivered in a buffer of 25 mM NaHCO3. 25 mM NaHCO3 was delivered for 30 sec to establish a baseline, after which ATP dissolved in 25 mM NaHCO3 was applied. Responses above the baseline were considered ATP responses. In control experiments, we demonstrated that ATP dissolved in PBS activated these same neurons after pre-equilibration in PBS. In Figure 7F,G and Figure S7E, stylets were pre-equilibrated in 298 mM cellobiose for 30 sec prior to the isomolar sugar of interest to control for osmotic effects. 298 mM cellobiose was behaviorally inactive in both the blood- and nectar-feeding assays (Matthews et al., 2019) (see raw data at https://github.com/VosshallLab/Jove_Vosshall_2020).
Ligands were delivered in the following order for the indicated experiment:
(“>” indicates water recovery before adding next ligand)Figure 4G The stimulus order alternated between the following options so that each animal experienced at least one of each: 1. water > 1st blood > 2nd blood > 3rd blood and 2. 1st blood >2nd blood > 3rd blood > water
Figure 5D The stimulus order alternated between the following options so that each animal experienced at least one of each: 1. blood > mix+ATP and 2. mix+ATP > blood
Figure 5G The stimulus order alternated between the following options: 1. Blood > NaCl > glucose > NaHCO3 > NaHCO3, ATP > Mix+ ATP > Mix and 2. Blood > Mix > NaCl > glucose > NaHCO3 > NaHCO3, ATP > Mix+ATP
Figure 7G The stimulus order alternated between the following options so that each animal experienced one of each: 1. cellobiose, glucose > cellobiose, sucrose > cellobiose, fructose and 2. cellobiose, fructose > cellobiose, sucrose > cellobiose, glucose and 3. cellobiose, sucrose > cellobiose, fructose > cellobiose, glucose
Microfluidic Ligand Delivery Using the BioPen
The BioPen tip holder (Fluicell) was secured using a MP-285 micromanipulator (Sutter #SU-MP-285). Each BioPen tip was prepared according to the manufacturer’s instructions with the following exceptions. First, the initial “New Tip” protocol was run with MilliQ water in each well to prime the microfluidic channels. Once the protocol was completed, water was removed from each BioPen well and replaced with test ligands. 0.0002% fluorescein was added to each test ligand to visualize the size and location of ligand delivery in each trial. For solutions containing NaHCO3, the fluorescein signal was much brighter, so 0.00002% fluorescein was used instead. For each ligand, the BioPen stimulus was ON for 20 sec with a 60 sec recovery before the next stimulus.
Analysis of GCaMP6s Data
All calcium imaging data were processed with Nikon Elements software. Regions of interest (ROIs) were selected based on the dTomato fluorescence intensity and used for analysis of GCaMP6s signal. Great care was taken to draw ROIs on the cell body of interest and not on en passant processes or slightly overlapping cell bodies. To exclude background noise, a cut-off of 0.25 peak ΔF/F0 was set as the minimum threshold for activation. This cut-off intentionally filters for clear activation and does not distinguish between background noise and weak activation. Occasionally (less than 1 cell body per animal) it was difficult to avoid the halo, especially if baseline GCaMP fluorescence was very low in a given cell body. In these rare cases, the cell body was not considered to be activated. All traces with sample motion, as determined by dTomato fluorescence instability, were discarded.
Once raw fluorescence values were extracted for each neuron/stimulus (ligand) pair, ΔF/F0 calculations were performed using a custom R script (R version 3.6.0) where ΔF/F0 = (F – F0)/F0. To determine the baseline fluorescence (F0) 5 frames (~2 fps) were averaged before stimulus presentation. To determine peak F to a given stimulus, the average of 3 frames at the peak during stimulus delivery was determined for each stimulus. This process was repeated twice for each stimulus so that the peak ΔF/F0 value represented in all plots is the average peak ΔF/F0 for 3 independent stimulus presentations. Stimulus trains were delivered so that each stimulus was only presented once per trial. Therefore, the final value represents the average peak stimulus response collected from three trials. Once all averages had been calculated, the dataset from individual females were analyzed and represented in multiple ways. Heat maps in Figure 4G, Figure 5D, Figure 6G,H, Figure S4A, and Figure S7E were generated using a custom R script available at https://github.com/VosshallLab/Jove_Vosshall_2020. Each box represents average peak ΔF/F0 to a given stimulus as described above. The heat map color scale is log2 to increase dynamic range and the minimum and maximum color value was set to 0.25 and 3 respectively. Plots in Figure 5E, Figure 7G, Figure S3E, Figure S4D, and Figure S7G were plotted using Prism 8 (GraphPad) and a neuron was considered activated if peak ΔF/F0 > 0.25. All peak ΔF/F0 scatter plots were generated using Prism 8 (GraphPad) except the scatter plots (Figure 5H) and box plots (Figure 5I) of peak ΔF/F0 for neurons within a given cluster were plotted in base R.
Hierarchical Clustering
In Figure 5G–I 134 individual neurons from the 5 females in Figure S4A were pooled and subjected to hierarchical clustering using Euclidean distance with complete linkage and visualized with the pheatmap R package v 1.0.12 (https://CRAN.R-project.org/package=pheatmap). Clustering was based on each neuron’s response profile to 7 ligands: blood, mix+ATP, mix, ATP, NaCl, NaHCO3, glucose. The peak ΔF/F0 of each neuron in response to each ligand was recorded 3 times to calculate an average peak ΔF/F0 per ligand per neuron, similar to the protocol described in Figure S3. Raw peak ΔF/F0 values for biological (“Fig 5G_input_to_clustering”) and technical replicates (“FigS4C_raw_values_technical_rep”) can be found in Data File 1. In Figure S5A suitability of the normalized response measurements to clustering was assessed by the Hopkins statistic (h) (Lawson and Jurs, 1990) using the factoextra R package v 1.0.7 and the get_clust_tendency function (https://CRAN.R-project.org/package=factoextra). The derived Hopkins statistic of 0.9046932 showed the dataset contains suitable information for clustering. In datasets which are not amenable to clustering, the distances between neighboring closest points will be close to the random dataset and the Hopkins statistic will approach 0. In a dataset with clusters present, the distances between neighboring closest points will be low compared to the random dataset and the Hopkins statistic will approach 1 (Lawson and Jurs, 1990). To show the significance of this clustering tendency, the p-value for the Hopkins statistic, 4.0126e-39, was calculated using the beta distribution in base R. In Figure S5B the optimal number of clusters to be drawn for the data was established by the Silhouette method (Rousseeuw, 1987) using the NbClust R package v 3.0 (Charrad et al., 2014) with potential cluster numbers in the range of 2 to 10. 5 was the optimal cluster number with the highest mean silhouette value 0.769 across clusters. The factoextra package was used to visualize the silhouette analysis results and show the distribution of silhouette widths for all members of each cluster (Kassambara and Mundt, 2020). From the visualization it can be seen that one single neuron in Cluster IV can be considered mis-clustered with a silhouette width less than 0. To evaluate the stability of the 5 clusters in Figure S5C, we assessed the bootstrap distribution of the Jaccard coefficient of resampled versus original data (Hennig, 2007, 2008). Clusters showing a Jaccard bootstrap mean of less than 0.5 can be considered unstable and unreliable, and an average Jaccard bootstrap mean across clusters above 0.85 shows a highly stable clustering (Hennig, 2007). To calculate the Jaccard bootstrap mean for all clusters, we used the fpc v 2.2-5 R package’s clusterboot function following the recommendations of 100 bootstraps (https://CRAN.R-project.org/package=fpc). All clusters identified have Jaccard bootstrap mean values above 0.7 indicating a set of stable of clusters and an average Jaccard bootstrap mean across clusters of 0.8727142. The hierarchical clustering approach used in this study is agnostic to female identity and therefore was assessed to ensure no biases in clustering are associated with specific individuals. In Figure S5D,E, principal component analysis (PCA) was applied to neuronal responses using base R and the FactoMineR package (Lê et al., 2008) to visualize the contribution of female or cluster to derived principal components. Comparison of PCA plots demonstrates that the differing clusters are well distributed and defined by the major principal components in the data (Figure S5D), whereas the animals are distributed throughout principal components (Figure S5E). Cluster membership can be seen to highly significantly correlated with all principal components, but female showed low correlation and no significant association with any principal components (Figure S5F). To determine which ligand(s) robustly activate the neuronal subpopulation belonging to each cluster, we performed the one-sample Wilcoxon signed rank test (wilcox.test in R) on each subpopulation’s average response to each ligand in Figure 5I. P values for this statistical analysis can be found in Data File 1 (“Fig 5I_p_values”). A cluster was considered activated by a given ligand if p < 0.05 when compared to the hypothetical value 0.25: wilcox.test(GroupA_Values, mu = 0.25, alternative = “greater”) because a neuron was considered activated if peak ΔF/F0 > 0.25. Scatter plots (Figure 5H) and Box plots (Figure 5I) of peak ΔF/F0 for neurons within a given cluster were plotted in base R.
Custom R scripts for all analyses are available at https://github.com/VosshallLab/Jove_Vosshall_2020. A comprehensive supplementary document on clustering methods is available at https://github.com/VosshallLab/Jove_Vosshall_2020/tree/master/Clustering_validation
Tissue Dissection and RNA Extraction
7 to 11 day-old mosquitoes were cold-anesthetized and kept on ice for up to 30 min or until dissections were complete. For labium samples, the labium was removed by forceps and immediately flash-frozen in DNA Lo-bind nuclease-free tubes (Fisher Scientific #13-698-790) contained in a CoolRack (Biocision #BCS0137) in dry ice for snap-freezing tissue. For female and male stylet samples, the labium was removed first. The stylet was detached half-way from the tip using a scalpel and immediately flash-frozen as described above. Extreme caution was taken during the tissue dissection and RNA extraction process to ensure that there was no contamination from other mosquito tissues or RNases. Each dish, forcep, and scalpel was carefully cleaned with 70% ethanol and RNase-away (ThermoFisher #7003) after every dissection or dissection attempt. Once the labium was removed, the stylet was discarded if there was any contact between the stylet and any surface other than the cleaned dish, forceps, or scalpel. A dedicated pair of stylet-only forceps was used to place the detached stylet into the collection tube. The following number of mosquitoes was used for each female library: female stylet, 25; male stylet, 25; female labium, 4. Each sample group was dissected in parallel to avoid batch effects. Dissected tissue was stored at −80°C until RNA extraction.
RNA extraction was performed using the PicoPure Kit (ThermoFisher #KIT0204) with the following exception for homogenizing tissue: instead of lysis buffer, 240 μL of TRIzol (ThermoFisher #15596018) was added to the collection tube on ice. Custom-order molecular biology grade, low-binding zirconium beads in 100 μm, 200 μm and 800 μm were used to disrupt tissue (OPS diagnostics). An RNase free spatula (Corning #CLS3013) was used to add 1 scoop each of 100 μm and 200 μm beads and ~100 μL of 800 μm beads to collection tube. Tubes were briefly spun down in a tabletop centrifuge before disruption in a TissueLyser II (Qiagen #85300) for 2 min 30 sec at 30 Hz. Tubes were briefly spun down again in tabletop centrifuge and returned to the TissueLyser II for an additional 2 min at 30 Hz. The remaining TRIzol extraction steps were performed in a chemical fume hood according to manufacturer’s instructions: tubes stood at room temperature for 5 min before 48 μL of chloroform:isoamyl alcohol 24:1 was added (Sigma #C0549). Tubes were hand-shaken for 30 sec and left to stand for 2 min before centrifuging at 12,000 x g for 15 min at 4°C. The aqueous Trizol layer was then removed and added into the PicoPure column, up to 180 μL at one time. Subsequent steps were performed according to PicoPure manufacturer’s instructions, including DNase treatment.
RNA-seq Library Preparation and Sequencing
Labium samples were run on Bioanalyzer RNA Pico Chip (Agilent #5067-1513) to determine RNA quantity and quality and were used as a proxy for overall sample integrity because female and male stylet samples fell below the level of detection. Labium samples were diluted 1:10 before cDNA amplification to more closely approximate stylet samples. cDNA synthesis was performed using SMART-Seq v4 Ultra Low Input RNA Kit for Sequencing (Takara #634894) according to the manufacturer’s instructions except that 10 μL instead of 9 μL was used to optimize for low RNA input. The number of PCR amplification cycles was adjusted for each sample group based on the number of cycles needed to detect RNA in the lowest input sample as determined by the Bioanalyzer High Sensitivity DNA Kit (Agilent #5067-4627). Negative controls for each group were run in parallel to ensure that additional cycles did not result in unspecific background product. All samples within one group were subjected to the same number of PCR amplification cycles. The female labium and female stylet samples underwent 20 cycles and male stylet 22 cycles. The full-length cDNA output was processed with Nextera XT DNA library preparation kit (Illumina #FC-131-1024) according to manufacturer’s instructions. Library quantity and quality were evaluated using High Sensitivity DNA ScreenTape Analysis (Agilent #5067-5585) prior to pooling. Bar-coded samples from all tissues were pooled in an equal ratio before distributing the pool across 3 sequencing lanes. Sequencing was performed at The Rockefeller University Genomics Resource Center on a NextSeq 500 sequencer (Illumina). All reads were 1 × 75 bp. Data were de-multiplexed and delivered as fastq files for each library. Sequencing reads have been deposited at the NCBI Sequence Read Archive (SRA) under BioProject PRJNA605870.
Transcript Abundance and Differential Expression Analysis
All reads were trimmed using TrimGalore version 0.4.2 (https://github.com/FelixKrueger/TrimGalore) with minimum read length of 35 base pairs. Reads from individual libraries were mapped to the AaegL5 genome (Matthews et al., 2018) using STAR version 2.5.2a (Dobin et al., 2013). All raw data are available in Data File 1 and use gene names with the LOCXXX naming format derived from the most recent NCBI RefSeq annotation of the Aedes aegypti genome (https://www.ncbi.nlm.nih.gov/assembly/GCF_002204515.2/)(Matthews et al., 2018). Gene names with the legacy AAELXXX naming format are easily cross-referenced to the new gene names by searching Vectorbase.
A custom gene annotation was generated by merging AaegL5 with the more recent manual chemoreceptor annotation for ORs, GRs and IRs (Matthews et al., 2018). This merged annotation and the R script used to generate it is available at https://github.com/VosshallLab/Jove_Vosshall_2020. For each of these chemoreceptors, the manual annotation replaced the AaegL5 RefSeq annotation. If the chemoreceptor did not previously exist in AaegL5 RefSeq, it was added. Reads mapping to each were mapped to transcript coding regions (UTRs and multi-mappers were excluded) using featureCounts version 1.5.0-p3 (Liao et al., 2014). For abundance visualization, raw counts were converted to TPM (Data File 1). RNA-seq TPM plots were generated using ggplot2 version 3.2.0 (R Development Core Team, 2017) in RStudio R 3.6.0. Raw counts were used for differential expression analysis in R using DESeq2 version 1.24.0 (Love et al., 2014). Sweet GRs analyzed in Figure 7D were derived from the Ae. aegypti genome reannotation (Matthews et al., 2018). TPM data from the stylet RNA-seq experiment are available for all predicted coding transcripts in Data File 1.
A previous study reported orco, Or8, and Or49 expression in the female stylet (Jung et al., 2015), but we found no strong evidence for orco and Or8 transcripts in our stylet RNA-seq data (Data File 1). We did not examine Or49 because it was annotated as a predicted pseudogene (Matthews et al., 2018). We also could not detect orco in the stylet by RNA in-situ hybridization (data not shown). We did detect orco in the female labium RNA-seq dataset (Data File 1), which is in agreement with previous experiments that detected orco in the labium of Anopheles gambiae (Kwon et al., 2006; Riabinina et al., 2016). We did not pursue further experiments related to odorant receptor (OR) expression in the female stylet since the presence or absence of ORs does not affect the interpretation of the data presented here.
Filtering for Stylet-Specific Transcripts
To obtain the 53 transcripts enriched in the female stylet compared to the female labium and male stylet (Figure S6C), we examined TPM values for non-mouthpart tissues that were previously profiled in a comprehensive dataset (Matthews et al., 2018; Matthews et al., 2016). A transcript was considered female stylet-specific if the average TPM expression across a given tissue was < 0.5 TPM for all tissues profiled by Matthews and colleagues, except for the Proboscis and Rostrum samples because these samples included mouthparts. To calculate average TPM, we used the most recent dataset aligned to the L5 genome and quantified using NCBI RefSeq Annotation version 101 (Matthews et al., 2018). If a transcript was present in the NCBI RefSeq annotation and the manual chemoreceptor annotation published alongside (Matthews et al., 2018), we used the TPM value quantified using the manual chemoreceptor annotation because the NCBI RefSeq annotation is missing a handful of chemoreceptors, including Ir7a. A DESeq2 results table and a TPM table filtered for these 53 transcripts are provided in Data File 1.
Quantification and Statistical Analysis
All statistical analysis was performed using Graphpad Prism Version 8 and RStudio R 3.6.0. For experiments where data were quantified as percent of females engorged, non-parametric tests were performed. For all other analyses, we first tested whether the values were normally distributed using D’Agostino–Pearson omnibus and Shapiro–Wilk normality tests. When data were normally distributed, we used parametric tests and when data were not normally distributed, we used non-parametric tests. Data collected as raw values are shown as mean ± SEM or mean ± SD. Details of statistical methods are reported in the figure legends.
Supplementary Material
Data File 1. RNA-seq Data, Related to Figure 6 and Figure S6; Imaging Data for Hierarchical Clustering, Related to Figure 5 and Figure S4,5; and Ir7a and Ir7f Data, Related to Figure 6 and Figure S6
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-fluorescein | ThermoFisher | #A889 |
| Mouse anti-Drosophila Brp (nc82) | Developmental Studies Hybridoma Bank | RRID: AB_2314866 |
| Rat anti-mCD8 | Invitrogen | #14008185 |
| Goat anti-rat Alexa Fluor 647 | Invitrogen | #A21247 |
| Goat anti-rabbit Alexa Fluor 488 | ThermoFisher | #A-11008 |
| Goat anti-mouse Alexa Fluor 647 | ThermoFisher | #A-21236 |
| Goat anti-mouse Alexa Fluor 555 | Invitrogen | #A32727 |
| Critical Commercial Assays | ||
| SMART-Seq v4 Ultra Low Input RNA Kit for Sequencing | Takara | #634894 |
| PicoPure Kit | ThermoFisher | #KIT0204 |
| Deposited Data | ||
| Stylet RNA-seq Dataset | NCBI Sequence Read Archive | BioProject PRJNA605870 |
| Experimental Models: Organisms/Strains | ||
| Aedes aegypti Liverpool strain | Vosshall Lab | N/A |
| Aedes aegypti 15x-QUAS-dTomato-T2A-GCaMP6s strain | This paper | N/A |
| Aedes aegypti Ir7f-T2A-QF2 strain | This paper | N/A |
| Aedes aegypti 15x-QUAS-mCD8-GFP strain | PMID: 31112133 | N/A |
| Aedes aegypti Brp-T2A-QF2 strain | This paper | N/A |
| Aedes aegypti Ir7a-T2A-QF2 strain | This paper | N/A |
| Aedes aegypti Gr4-T2A-QF2 strain | This paper | N/A |
| Aedes aegypti 15x-QUAS-dTomato-T2A-TRPV1 strain | This paper | N/A |
| Oligonucleotides | ||
| Gr4 sgRNA (GTATCCCAAACGTGCAACCAGGG) | This paper | N/A |
| Ir7a sgRNA (TGGGGTCACGTATATCCAAATGG) | This paper | N/A |
| Ir7f sgRNA (GATGCGCGGTGAACGCATGTCGG) | This paper | N/A |
| Brp sgRNA (GCAACTGGTACAGATGACACAGG) | This paper | N/A |
| Gr4 left homology arm (Forward, 5’- caatgtatcttaCAGGGAAAACTGGATCCATG-3’) | This paper | N/A |
| Gr4 left homology arm (Reverse, 5’- tctgccctctccTGCACGTTTGGGATACTTG-3’) | This paper | N/A |
| Gr4 right homology arm (Forward, 5’- caatgtatcttaCAGGGAAAACTGGATCCATG-3’) | This paper | N/A |
| Gr4 right homology arm (Reverse, 5’- ttgcatgcctgcaggtcgactctagGTGTATTTGGAGCCTCAG-3’) | This paper | N/A |
| Ir7a left homology arm (Forward, 5’-cggtacccgggTAAAAAGCAAATTTCACCATG-3’) | This paper | N/A |
| Ir7a left homology arm (Reverse, 5’- tctgccctctccATATACGTGACCCCAAATATC-3’) | This paper | N/A |
| Ir7a right homology arm (Forward, 5’- caatgtatcttaATCCAGAACGGGTGCGGTAG-3’) | This paper | N/A |
| Ir7a right homology arm (Reverse, 5’- ggtcgactctagTTATAGTTGATCGAGGAATTTCCGAATCC-3’) | This paper | N/A |
| Ir7f left homology arm (Forward, 5’-ctcggtacccgggTGACTGGCTGATTAGCTCATCCTATATAAGAA-3’) | This paper | N/A |
| Ir7f left homology arm (Reverse, 5’- ctctgccctctccACGCTCGCCACGCATCGAGAAACACCCGG-3’) | This paper | N/A |
| Ir7f right homology arm (Forward, 5’-tcaatgtatcttaTGTCGGTGATGAGGTCCAG -3’) | This paper | N/A |
| Ir7f right homology arm (Reverse, 5’-aggtcgactctagCCCGCCTCAAAATGTGCAC-3’) | This paper | N/A |
| Recombinant DNA | ||
| Plasmid: Ir7f-T2A-QF2 HDR | This paper | Addgene #140942 |
| Plasmid: Ir7a-T2A-QF2 HDR | This paper | Addgene #140943 |
| Plasmid: Gr4-T2A-QF2 HDR | This paper | Addgene #140944 |
| Plasmid: 15x-QUAS-dTomato-T2A-TRPV1 | This paper | Addgene #140945 |
| Plasmid: TRPV1 Bargmann lab | PMID: 12160748 | Bargmann Lab Plasmid #10.33.42 |
| Plasmid: pQF2wWB | PMID: 25581800 | Addgene #61313 |
| Plasmid: ppk301-T2A-QF2 HDR | PMID: 31112133 | Addgene #130667 |
| Plasmid: 3xP3-eYFP-SV40 | PMID: 25869647 | Addgene #62291 |
| Plasmid: 15xQUAS-dTomato-T2A-GCaMP6s | PMID: 31112133 | Addgene #130666 |
| Plasmid: Brp-T2A-QF2 HDR | This paper | Addgene #141094 |
| Software and Algorithms | ||
| RStudio | https://www.r-project.org | N/A |
| MacVector | MacVector | N/A |
| GraphPad Prism | GraphPad | N/A |
| ImageJ (FIJI) | https://imagej.net/Fiji | N/A |
| TrimGalore | https://github.com/FelixKrueger/TrimGalore | N/A |
| STAR | PMID: 23104886 | N/A |
| featureCounts | PMID: 24227677 | N/A |
| Basler Pylon 5 | https://www.baslerweb.com/en/sales-support/downloads/software-downloads/ | N/A |
| Python | https://www.python.org/ | N/A |
| Other | ||
| Additional resources for RNA-seq data and calcium imaging data analysis | This paper | https://github.com/VosshallLab/Jove_Vosshall_2020 |
| Additional resources for biteOscope data analysis | This paper | https://github.com/felixhol/biteOscope |
ACKNOWLEDGMENTS
We thank J. Clowney, M. DeGennaro, E.J. Dennis, M. Dietrich, L. Duvall, A. Handler, R. Jové, K.J. Lee, M.Z. Liu, B. J. Matthews, P. Muller, N. Yapici, and members of the Vosshall Lab for comments on the manuscript; T. Morita for guidance and technical assistance with stylet GCaMP calcium imaging; B. Matthews for advice on RNA-seq and transgenic strain design; L. Duvall for advice on Glytube assays; G. Gordon, L. Mejia, and A. Pandey for mosquito rearing; W. Freiwald, P. Muller, and V. Ruta for discussions; D. Gross and J. Petrillo for advice on imaging hardware; G. Jeffries of Fluicell for optimizing the BioPen; C. Zhao and B. Zhang for help with RNA-seq library preparation; A. North and C. Pyrgaki for help with confocal and structured illumination image acquisition; R. Harrell II at ITF for embryo injections; T. Siskoglou and A. Gerson of Morrell Instruments for help with imaging hardware; B. Matthews, M. Younger, and the Aedes Toolkit Group for early access to unpublished strains.
This work was supported in part by NCATS NIH CTSA grant UL1 TR000043, NIH T32-MH095246 (V.J.), HHMI Gilliam Fellowship (V.J.), NSF DGE-1325261 (V.J.). CASI / Burroughs Wellcome Fund (F.J.J.H.), Marie Skłodowska-Curie grant agreement No 841893 — PiQiMosqBite (F.J.J.H.), Jane Coffin Childs Postdoctoral Fellowship (T.R.S.), Kavli Neural Systems Institute postdoctoral fellowship (T.R.S.), NIDCD R00-DC012069 (C.S.M.), NIH DP2-AI124336 (M.P.), USAID Grand Challenges: Zika and Future Threats Award (M.P.). M.P. is an HHMI-Gates Faculty Scholar of the Howard Hughes Medical Institute. C.S.M. is a New York Stem Cell Foundation – Robertson Investigator. L.B.V. is an investigator of the Howard Hughes Medical Institute.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Abuin L, Bargeton B, Ulbrich MH, Isacoff EY, Kellenberger S, and Benton R (2011). Functional architecture of olfactory ionotropic glutamate receptors. Neuron 69, 44–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan D, van der Wel C, Keim N, Caswell TA, Wieker D, Verweij R, Reid C, Thierry, Grueter L, Ramos K, et al. (2019). soft-matter/trackpy: Trackpy v0.4.2 (DOI: 10.5281/zenodo.3492186). [DOI] [Google Scholar]
- Ariani CV, Smith SC, Osei-Poku J, Short K, Juneja P, and Jiggins FM (2015). Environmental and genetic factors determine whether the mosquito Aedes aegypti lays eggs without a blood meal. Am J Trop Med Hyg 92, 715–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin MW, Toda Y, Nakagita T, O’Connell MJ, Klasing KC, Misaka T, Edwards SV, and Liberles SD (2014). Evolution of sweet taste perception in hummingbirds by transformation of the ancestral umami receptor. Science 345, 929–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barredo E, and DeGennaro M (2020). Not just from blood: Mosquito nutrient acquisition from nectar sources. Trends Parasitol 36, 473–484. [DOI] [PubMed] [Google Scholar]
- Bavan S, Farmer L, Singh SK, Straub VA, Guerrero FD, and Ennion SJ (2011). The penultimate arginine of the carboxyl terminus determines slow desensitization in a P2X receptor from the cattle tick Boophilus microplus. Mol Pharmacol 79, 776–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop A, and Gilchrist BM (1946). Experiments upon the feeding of Aedes aegypti through animal membranes with a view to applying this method to the chemotherapy of malaria. Parasitology 37, 85–100. [DOI] [PubMed] [Google Scholar]
- Born GV, and Kratzer MA (1984). Source and concentration of extracellular adenosine triphosphate during haemostasis in rats, rabbits and man. J Physiol 354, 419–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradski G (2000). The OpenCV Library. Dr Dobb’s Journal of Software Tools. [Google Scholar]
- Briegel H (2003). Physiological bases of mosquito ecology. J Vector Ecol 28, 1–11. [PubMed] [Google Scholar]
- Burnstock G, and Verkhratsky A (2009). Evolutionary origins of the purinergic signalling system. Acta Physiol (Oxf) 195, 415–447. [DOI] [PubMed] [Google Scholar]
- Centor RM (1990). Serum Total Carbon Dioxide. In Clinical Methods: The History, Physical, and Laboratory Examinations, Walker H, Hall W, and Hurst J, eds. (Boston: Butterworths; ), pp. 888–889. [PubMed] [Google Scholar]
- Chandrashekar J, Yarmolinsky D, von Buchholtz L, Oka Y, Sly W, Ryba NJ, and Zuker CS (2009). The taste of carbonation. Science 326, 443–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrad M, Ghazzali N, Boiteau V, and Niknafs A (2014). NbClust: An R Package for Determining the Relevant Number of Clusters in a Data Set. J Stat Soft 61, 1–36. [Google Scholar]
- Choumet V, Attout T, Chartier L, Khun H, Sautereau J, Robbe-Vincent A, Brey P, Huerre M, and Bain O (2012). Visualizing non infectious and infectious Anopheles gambiae blood feedings in naive and saliva-immunized mice. PLoS One 7, e50464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-da-Silva AL, Navarrete FR, Salvador FS, Karina-Costa M, Ioshino RS, Azevedo DS, Rocha DR, Romano CM, and Capurro ML (2013). Glytube: a conical tube and parafilm M-based method as a simplified device to artificially blood-feed the dengue vector mosquito, Aedes aegypti. PLoS One 8, e53816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGennaro M, McBride CS, Seeholzer L, Nakagawa T, Dennis EJ, Goldman C, Jasinskiene N, James AA, and Vosshall LB (2013). orco mutant mosquitoes lose strong preference for humans and are not repelled by volatile DEET. Nature 498, 487–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker T, Geier M, and Carde RT (2005). Carbon dioxide instantly sensitizes female yellow fever mosquitoes to human skin odours. J Exp Biol 208, 2963–2972. [DOI] [PubMed] [Google Scholar]
- Dennis EJ, Goldman OV, and Vosshall LB (2019). Aedes aegypti mosquitoes use their legs to sense DEET on contact. Curr Biol 29, 1551–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutsch JA, Moore BO, and Heinrichs SC (1989). Unlearned specific appetite for protein. Physiol Behav 46, 619–624. [DOI] [PubMed] [Google Scholar]
- Devineni AV, Sun B, Zhukovskaya A, and Axel R (2019). Acetic acid activates distinct taste pathways in Drosophila to elicit opposing, state-dependent feeding responses. Elife 8, e47677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow JAT (1987). Insect Midgut Function. In, pp. 187–328. [Google Scholar]
- Duvall LB, Ramos-Espiritu L, Barsoum KE, Glickman JF, and Vosshall LB (2019). Small-molecule agonists of Ae. aegypti neuropeptide Y receptor block mosquito biting. Cell 176, 687–701 e685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischler W, Kong P, Marella S, and Scott K (2007). The detection of carbonation by the Drosophila gustatory system. Nature 448, 1054–1057. [DOI] [PubMed] [Google Scholar]
- Forsyth AM, Wan J, Owrutsky PD, Abkarian M, and Stone HA (2011). Multiscale approach to link red blood cell dynamics, shear viscosity, and ATP release. Proc Natl Acad Sci U S A 108, 10986–10991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galun R (1987). The evolution of purinergic receptors involved in recognition of a blood meal by hematophagous insects. Mem Inst Oswaldo Cruz 82 Suppl 3, 5–9. [DOI] [PubMed] [Google Scholar]
- Galun R, Avi-Dor Y, and Bar-Zeev M (1963). Feeding response in Aedes aegypti: Stimulation by adenosine triphosphate. Science 142, 1674–1675. [DOI] [PubMed] [Google Scholar]
- Galun R, Koontz LC, Gwadz RW, and Ribeiro JMC (1985). Effect of ATP analogues on the gorging response of Aedes aegypti. Physiol Entomol 10, 275–281. [Google Scholar]
- Galun R, Oren N, and Zecharia M (1984). Effect of plasma components on the feeding response of the mosquito Aedes aegypti L. to adenine nucleotides. Physiol Entomol 9, 403–408. [Google Scholar]
- Gonzales KK, Rodriguez SD, Chung HN, Kowalski M, Vulcan J, Moore EL, Li Y, Willette SM, Kandel Y, Van Voorhies WA, et al. (2018). The effect of SkitoSnack, an artificial blood meal replacement, on Aedes aegypti life history traits and gut microbiota. Sci Rep 8, 11023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon RM, and Lumsden WHR (1939). A study of the behaviour of the mouth-parts of mosquitoes when taking up blood from living tissue; together with some observations on the ingestion of microfilariae. Ann Trop Med Parasitol 33, 259–278. [Google Scholar]
- Griffiths RB, and Gordon RM (1952). An apparatus which enables the process of feeding by mosquitoes to be observed in the tissues of a live rodent; together with an account of the ejection of saliva and its significance in Malaria. Ann Trop Med Parasitol 46, 311–319. [DOI] [PubMed] [Google Scholar]
- Gulia-Nuss M, Elliot A, Brown MR, and Strand MR (2015). Multiple factors contribute to anautogenous reproduction by the mosquito Aedes aegypti. J Insect Physiol 82, 8–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig C (2007). Cluster-wise assessment of cluster stability. Comput Stat Data An 52, 258–271. [Google Scholar]
- Hennig C (2008). Dissolution point and isolation robustness: Robustness criteria for general cluster analysis methods. J Multivariate Anal 99, 1154–1176. [Google Scholar]
- Hol FJ, Lambrechts L, and Prakash M (2020). BiteOscope: an open platform to study mosquito blood-feeding behavior. bioRxiv, 10.1101/2020.1102.1119.955641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoi T (1959). The identification of blood components which induce gorging of the mosquito. J Insect Physiol 3, 191–218. [Google Scholar]
- Ignell R, and Hansson BS (2005). Projection patterns of gustatory neurons in the suboesophageal ganglion and tritocerebrum of mosquitoes. J Comp Neurol 492, 214–233. [DOI] [PubMed] [Google Scholar]
- Inagaki HK, Ben-Tabou de-Leon S, Wong AM, Jagadish S, Ishimoto H, Barnea G, Kitamoto T, Axel R, and Anderson DJ (2012). Visualizing neuromodulation in vivo: TANGO-mapping of dopamine signaling reveals appetite control of sugar sensing. Cell 148, 583–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Shinomiya K, Ito M, Armstrong JD, Boyan G, Hartenstein V, Harzsch S, Heisenberg M, Homberg U, Jenett A, et al. (2014). A systematic nomenclature for the insect brain. Neuron 81, 755–765. [DOI] [PubMed] [Google Scholar]
- Jung JW, Baeck SJ, Perumalsamy H, Hansson BS, Ahn YJ, and Kwon HW (2015). A novel olfactory pathway is essential for fast and efficient blood-feeding in mosquitoes. Sci Rep 5, 13444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassambara A, and Mundt F (2020). factoextra: Extract and Visualize the Results of Multivariate Data Analyses. [Google Scholar]
- Kent LB, and Robertson HM (2009). Evolution of the sugar receptors in insects. BMC Evol Biol 9, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakh RS, Burnstock G, Kennedy C, King BF, North RA, Seguela P, Voigt M, and Humphrey PPA (2001). International Union of Pharmacology. XXIV. Current Status of the Nomenclature and Properties of P2X Receptors and Their Subunits. Pharmacological Reviews 53, 107–118. [PubMed] [Google Scholar]
- Kistler KE, Vosshall LB, and Matthews BJ (2015). Genome engineering with CRISPR-Cas9 in the mosquito Aedes aegypti. Cell Rep 11, 51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogan PH (1990). Substitute blood meal for investigating and maintaining Aedes aegypti (Diptera: Culicidae). J Med Entomol 27, 709–712. [DOI] [PubMed] [Google Scholar]
- Lahondere C, Vinauger C, Okubo RP, Wolff GH, Chan JK, Akbari OS, and Riffell JA (2020). The olfactory basis of orchid pollination by mosquitoes. Proc Natl Acad Sci U S A 117, 708–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson RG, and Jurs PC (1990). New index for clustering tendency and its application to chemical problems. J Chem Inf Comput Sci 30, 36–41. [Google Scholar]
- Lê S, Josse J, and Husson F (2008). FactoMineR: An R Package for Multivariate Analysis. J Stat Softw 25. [Google Scholar]
- Lea AO (1964). Selection for autogeny in Aedes aegypti (Diptera: Culicidae). Ann Entomol Soc Am 57, 656–657. [Google Scholar]
- Lee R (1974). Structure and function of the fascicular stylets, and the labral and cibarial sense organs of male and female Aedes aegypti (L.)(Diptera, Culicidae). Quaestiones entomologicae 10, 187–215. [Google Scholar]
- Lee RMKW, and Craig DA (1983). The labrum and labral sensilla of mosquitoes (Diptera: Culicidae): a scanning electron microscope study. Can J Zool 61, 1568–1579. [Google Scholar]
- Leitao-Goncalves R, Carvalho-Santos Z, Francisco AP, Fioreze GT, Anjos M, Baltazar C, Elias AP, Itskov PM, Piper MDW, and Ribeiro C (2017). Commensal bacteria and essential amino acids control food choice behavior and reproduction. PLoS Biol 15, e2000862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Li W, Wang H, Cao J, Maehashi K, Huang L, Bachmanov AA, Reed DR, Legrand-Defretin V, Beauchamp GK, et al. (2005). Pseudogenization of a sweet-receptor gene accounts for cats’ indifference toward sugar. PLoS Genet 1, 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. [DOI] [PubMed] [Google Scholar]
- Liesch J, Bellani LL, and Vosshall LB (2013). Functional and genetic characterization of neuropeptide Y-like receptors in Aedes aegypti. PLoS Negl Trop Dis 7, e2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima SQ, and Miesenbock G (2005). Remote control of behavior through genetically targeted photostimulation of neurons. Cell 121, 141–152. [DOI] [PubMed] [Google Scholar]
- Liman ER, Zhang YV, and Montell C (2014). Peripheral coding of taste. Neuron 81, 984–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu MZ, and Vosshall LB (2019). General visual and contingent thermal cues interact to elicit attraction in female Aedes aegypti mosquitoes. Curr Biol 29, 2250–2257 e2254. [DOI] [PubMed] [Google Scholar]
- Liu Q, Tabuchi M, Liu S, Kodama L, Horiuchi W, Daniels J, Chiu L, Baldoni D, and Wu MN (2017). Branch-specific plasticity of a bifunctional dopamine circuit encodes protein hunger. Science 356, 534–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marella S, Fischler W, Kong P, Asgarian S, Rueckert E, and Scott K (2006). Imaging taste responses in the fly brain reveals a functional map of taste category and behavior. Neuron 49, 285–295. [DOI] [PubMed] [Google Scholar]
- Matthews BJ, Dudchenko O, Kingan SB, Koren S, Antoshechkin I, Crawford JE, Glassford WJ, Herre M, Redmond SN, Rose NH, et al. (2018). Improved reference genome of Aedes aegypti informs arbovirus vector control. Nature 563, 501–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews BJ, McBride CS, DeGennaro M, Despo O, and Vosshall LB (2016). The neurotranscriptome of the Aedes aegypti mosquito. BMC Genomics 17, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews BJ, Younger MA, and Vosshall LB (2019). The ion channel ppk301 controls freshwater egg-laying in the mosquito Aedes aegypti. Elife 8, e43963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMeniman CJ, Corfas RA, Matthews BJ, Ritchie SA, and Vosshall LB (2014). Multimodal integration of carbon dioxide and other sensory cues drives mosquito attraction to humans. Cell 156, 1060–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy M, Peters KZ, Denton BS, Lee KA, Chadchankar H, and McCutcheon JE (2018). Restriction of dietary protein leads to conditioned protein preference and elevated palatability of protein-containing food in rats. Physiol Behav 184, 235–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappas LG, and Larsen JR (1978). Gustatory mechanisms and sugar-feeding in the mosquito, Culiseta inornata. Physiol Entomol 3, 115–119. [Google Scholar]
- Potter CJ, Tasic B, Russler EV, Liang L, and Luo L (2010). The Q system: a repressible binary system for transgene expression, lineage tracing, and mosaic analysis. Cell 141, 536–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasubramanian MK, Barham OM, and Swaminathan V (2008). Mechanics of a mosquito bite with applications to microneedle design. Bioinspir Biomim 3, 046001. [DOI] [PubMed] [Google Scholar]
- Renier N, Wu Z, Simon DJ, Yang J, Ariel P, and Tessier-Lavigne M (2014). iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell 159, 896–910. [DOI] [PubMed] [Google Scholar]
- Riabinina O, Luginbuhl D, Marr E, Liu S, Wu MN, Luo L, and Potter CJ (2015). Improved and expanded Q-system reagents for genetic manipulations. Nat Methods 12, 219–222, 215 p following 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riabinina O, Task D, Marr E, Lin CC, Alford R, O’Brochta DA, and Potter CJ (2016). Organization of olfactory centres in the malaria mosquito Anopheles gambiae. Nat Commun 7, 13010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro C, and Dickson BJ (2010). Sex peptide receptor and neuronal TOR/S6K signaling modulate nutrient balancing in Drosophila. Curr Biol 20, 1000–1005. [DOI] [PubMed] [Google Scholar]
- Rousseeuw PJ (1987). Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J Comput Applied Math 20, 53–65. [Google Scholar]
- Ruckert C, and Ebel GD (2018). How do mosquito-virus interactions lead to viral emergence? Trends Parasitol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford JL, Shields VD, and Dickens JC (2013). Gustatory receptor neuron responds to DEET and other insect repellents in the yellow-fever mosquito, Aedes aegypti. Naturwissenschaften 100, 269–273. [DOI] [PubMed] [Google Scholar]
- Scott K (2018). Gustatory processing in Drosophila melanogaster. Annu Rev Entomol 63, 15–30. [DOI] [PubMed] [Google Scholar]
- Simpson SJ, Le Couteur DG, and Raubenheimer D (2015). Putting the balance back in diet. Cell 161, 18–23. [DOI] [PubMed] [Google Scholar]
- Slone J, Daniels J, and Amrein H (2007). Sugar receptors in Drosophila. Curr Biol 17, 1809–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steck K, Walker SJ, Itskov PM, Baltazar C, Moreira JM, and Ribeiro C (2018). Internal amino acid state modulates yeast taste neurons to support protein homeostasis in Drosophila. Elife 7, e31625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker RF (1994). The organization of the chemosensory system in Drosophila melanogaster: a review. Cell Tissue Res 275, 3–26. [DOI] [PubMed] [Google Scholar]
- Takken W, and Kline DL (1989). Carbon dioxide and 1-octen-3-ol as mosquito attractants. J Am Mosq Control Assoc 5, 311–316. [PubMed] [Google Scholar]
- Tobin D, Madsen D, Kahn-Kirby A, Peckol E, Moulder G, Barstead R, Maricq A, and Bargmann C (2002). Combinatorial expression of TRPV channel proteins defines their sensory functions and subcellular localization in C. elegans neurons. Neuron 35, 307–318. [DOI] [PubMed] [Google Scholar]
- Trembley HL (1952). The distribution of certain liquids in the esophageal diverticula and stomach of mosquitoes. Am J Trop Med Hyg 1, 693–710. [DOI] [PubMed] [Google Scholar]
- Van Handel E (1972). The detection of nectar in mosquitoes. Mosquito News 32, 458. [Google Scholar]
- Van Handel E (1984). Metabolism of nutrients in the adult mosquito. Mosquito News 44, 573–579. [Google Scholar]
- Vargas MA, Luo N, Yamaguchi A, and Kapahi P (2010). A role for S6 kinase and serotonin in postmating dietary switch and balance of nutrients in D. melanogaster. Curr Biol 20, 1006–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virtanen P, Gommers R, Oliphant TE, Haberland M, Reddy T, Cournapeau D, Burovski E, Peterson P, Weckesser W, Bright J, et al. (2019). SciPy 1.0--Fundamental Algorithms for Scientific Computing in Python. arXiv https://arxiv.org/abs/1907.10121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker SJ, Corrales-Carvajal VM, and Ribeiro C (2015). Postmating circuitry modulates salt taste processing to increase reproductive output in Drosophila. Curr Biol 25, 2621–2630. [DOI] [PubMed] [Google Scholar]
- Werner-Reiss U, Galun R, Crnjar R, and Liscia A (1999a). Factors modulating the blood feeding behavior and the electrophysiological responses of labral apical chemoreceptors to adenine nucleotides in the mosquito Aedes aegypti (Culicidae). J Insect Physiol 45, 801–808. [DOI] [PubMed] [Google Scholar]
- Werner-Reiss U, Galun R, Crnjar R, and Liscia A (1999b). Sensitivity of the mosquito Aedes aegypti (Culicidae) labral apical chemoreceptors to blood plasma components. J Insect Physiol 45, 485–491. [DOI] [PubMed] [Google Scholar]
- Werner-Reiss U, Galun R, Crnjar R, and Liscia A (1999c). Sensitivity of the mosquito Aedes aegypti (Culicidae) labral apical chemoreceptors to phagostimulants. J Insect Physiol 45, 629–636. [DOI] [PubMed] [Google Scholar]
- Yarmolinsky DA, Zuker CS, and Ryba NJ (2009). Common sense about taste: from mammals to insects. Cell 139, 234–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Tian D, and McBride CS (2020). Development of a pan-neuronal genetic driver in Aedes aegypti mosquitoes. bioRxiv, 10.1101/2020.1108.1122.262527v262521. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data File 1. RNA-seq Data, Related to Figure 6 and Figure S6; Imaging Data for Hierarchical Clustering, Related to Figure 5 and Figure S4,5; and Ir7a and Ir7f Data, Related to Figure 6 and Figure S6
Data Availability Statement
All data in the paper (with the exception of raw video files) are available on Github at https://github.com/VosshallLab/Jove_Vosshall_2020. Sequencing reads have been deposited at the NCBI Sequence Read Archive (SRA) under BioProject PRJNA605870. Custom Python code for biteOscope data analysis is available on Github at https://github.com/felixhol/biteOscope. Custom R scripts for merged genome annotation and calcium imaging analysis are available on Github at https://github.com/VosshallLab/Jove_Vosshall_2020.


