Abstract
Background
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and monkeypox virus (MPXV) severely threaten human health; however, currently, no vaccine can prevent a co-infection with both viruses.
Methods
Five antigens were selected to predict dominant T and B cell epitopes screened for immunogenicity, antigenicity, toxicity, and sensitization. After screening, all antigens joined in the construction of a novel multiepitope vaccine. The physicochemical and immunological characteristics, and secondary and tertiary structures of the vaccine were predicted and analyzed using bio- and immunoinformatics. Finally, codon optimization and cloning in-silico were performed.
Results
A new multiepitope vaccine, named S7M8, was constructed based on four helper T lymphocyte (HTL) epitopes, six cytotoxic T lymphocyte (CTL) epitopes, five B cell epitopes, as well as Toll-like receptor (TLR) agonists. The antigenicity and immunogenicity scores of the S7M8 vaccine were 0.907374 and 0.6552, respectively. The S7M8 vaccine was comprised of 26.96% α-helices, the optimized Z-value of the tertiary structure was −5.92, and the favored area after majorization in the Ramachandran plot was 84.54%. Molecular docking showed that the S7M8 vaccine could tightly bind to TLR2 (−1100.6 kcal/mol) and TLR4 (−950.3 kcal/mol). In addition, the immune stimulation prediction indicated that the S7M8 vaccine could activate T and B lymphocytes to produce high levels of Th1 cytokines and antibodies.
Conclusion
S7M8 is a promising biomarker with good antigenicity, immunogenicity, non-toxicity, and non-sensitization. The S7M8 vaccine can trigger significantly high levels of Th1 cytokines and antibodies and may be a potentially powerful tool in preventing SARS-CoV-2 and MPXV.
Keywords: SARS-CoV-2, Monkeypox Virus (MPXV), Biomarker, Vaccine, Prevention
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) belongs to the beta genus of coronaviruses, which causes novel coronavirus pneumonia with characteristics of rapid mutation, epidemics, and devastating power. As of November 29, 2022, there were 638,175,811 confirmed cases of coronavirus disease 2019 (COVID-19), including 6,612,970 deaths, reported to the World Health Organization (WHO) (https://covid19.who.int/). Currently, 199 vaccines are in preclinical development, 175 are in clinical trials, and more than ten COVID-19 vaccines have been approved for emergency use to prevent COVID-19, including BNT162b2 [1], mRNA-1273 [2], AZD1222 [3], Ad26.COV2-S [4], Sputnik V [5], BBV152/Covaxin [6], CoronaVac [7], Sinopharm BBIBP-CorV [7], NVX-CoV2373 [8], [9], [10], and EpiVacCorona [11]. These vaccines are designed based on surface spike glycoprotein (QHR63290.1) [12], [13], nucleocapsid protein (QHR63298.1) [14], [15], [16], and membrane glycoprotein (QHR63293.1) [17], [18]. However, the emergence of variants of interest (VOIs) and variants of concern (VOCs) affects the characterization of SARS-CoV-2, such as transmission, disease severity, vaccine effectiveness, and therapeutic medicines [19], [20], [21], [22].
Monkeypox is a zoonotic disease caused by the monkeypox virus (MPXV) of the Orthopoxvirus genus [23], [24], [25]. The disease was first identified in monkeys by a laboratory in Denmark in 1958, and the first human infection with the disease was reported in a child in the Democratic Republic of Congo in 1970 [26]. On July 23, 2022, the WHO designated the monkeypox outbreak a Public Health Emergency of International Concern (PHEIC) and as of November 28, 2022, >81,107 confirmed cases of Monkeypox from 110 countries/regions have been reported to the WHO (https://worldhealthorg.shinyapps.io/mpx_global/). The judicious use of vaccines can help countries worldwide respond to monkeypox outbreaks and prevent human-to-human transmission of the disease. Currently, three smallpox vaccines available are useful in preventing MPXV infection: ACAM2000 (Emergent BioSolutions), LC16 (KM Biologics), and MVA-BN (Bavarian Nordic) [27]. Unfortunately, no specific vaccine has been approved to prevent MPXV infection. Therefore, to deal with possible monkeypox epidemics in the future, it is necessary to develop specific MPXV vaccines. Recent studies have designed several multiepitope vaccines for the prevention of MPXV infection using an immunoinformatics approach [28], [29], [30].
International travel has played a significant role in the spread of SARS-CoV-2 and MPXV. It has been reported that travel lockdowns imposed by the Chinese government in the early stages of the COVID-19 outbreak prevented 70.5% of these exportations, with daily exportation rates decreasing by an average of 81.3% in the first three weeks of travel restrictions being employed [31]. Similarly, some studies analyzed the transmission route of MPXV and found that it was transmitted between different countries by international travelers [32], [33]. Although restrictions on international travel may temporarily alleviate the spread of SARS-CoV-2 and MPXV, a lockdown could impact global economic vitality. Therefore, the development of novel vaccines that can prevent a co-infection with SARS-CoV-2 and MPXV is critical for reducing the international spread of both viruses.
In this study, we aimed to develop a novel multiepitope vaccine to fight SARS-CoV-2 and MPXV co-infections using bio and immunoinformatics. This study designed a multiepitope vaccine based on three antigens from SARS-CoV-2 and two antigens from MPXV, including surface spike and membrane glycoprotein, nucleocapsid, myristylated (MPXVgp127), and Crm-B-secreted TNF-α receptor-like proteins (MPXVgp190). The helper T lymphocyte (HTL), cytotoxic T lymphocyte (CTL), and B cell epitopes were predicted from these five antigens and screened using bioinformatics based on their immunogenicity, antigenicity, toxicity, and sensitization. Immunodominant epitopes were then adopted after a cascade of screenings to construct a multiepitope vaccine. Finally, the multiepitope vaccine's physicochemical, structural, and immunological properties were analyzed using bioinformatics and immunoinformatics.
2. Methods
2.1. Identification of target antigens
Surface spike (QHR63290.1) and membrane (QHR63293.1) glycoproteins, and nucleocapsid protein (QHR63298.1), from SARS-CoV-2 were selected as candidate antigens for epitope prediction due to their significant usefulness in COVID-19 vaccines [8], [19], [22]. In addition, MPXVgp127 and MPXVgp190 were selected as preparatory antigens against MPXV because of their superiority for both myricetin and TNF-α [34]. Protective antigens were predicted using VaxiJen, based on auto-cross-covariance (ACC) transformation. Full-length amino acid sequences were obtained from the National Center of Biotechnology Information (NCBI; https://www.ncbi.nlm.nih.gov/gene/) database.
2.2. Screening of immunodominant HTL epitopes
The main histocompatibility complex (MHC) II server (https://tools.iedb.org/mhcii/) in the Immune Epitope Database (IEDB) was used to predict the HTL epitope. In brief, IEDB recommendation 2.22 was selected as the prediction method, and the total human leukocyte antigen (HLA) reference set (HLA-DR, HLA-DP, HLA-DQ) was chosen as the human MHC alleles. HTL epitopes with a percentile rank <0.5 (lower means higher for epitopes bonded to MHC II) were identified as candidates. The antigenicity of these identified epitopes was evaluated using VaxiJen v2.0 (https://www.ddg-pharmfac.net/vaxijen/VaxiJen/VaxiJen.html) with a threshold of 0.4 following a previous study [35]. Finally, the interferon gamma (IFN-γ) inducibility of epitopes was determined using the IFN-γ epitope server (https://crdd.osdd.net/raghava/ifnepitope/index.php), according to a previous study [36]. The final epitopes with immunodominance were identified as eligible HTL epitopes for constructing a multiepitope vaccine.
2.3. Screening of immunodominant CTL epitopes
The IEDB MHC I server (https://tools.iedb.org/mhci/) was used to predict CTL epitopes. IEDB Recommendation 2020.09 (NetMHCpanEL4.1) was the primary qualifier, with epitopes of all lengths of human HLA alleles being the secondary qualifier. Epitopes with a percentile level <0.5 were eligible for the subsequent analysis step. The Class I immunogenicity server (https://tools.iedb.org/immunogenicity/) was then used to analyze the immunogenicity of these CTL epitopes, and those with percentile levels <0.5 and immune scores >0 were selected for further study. Finally, the VaxiJen v2.0 server was used to predict antigenicity with a threshold value of 0.4 [29] referring to a previous study. Finally, CTL epitopes with immunological advantages were used to construct a multiepitope vaccine.
2.4. Screening of the immunodominant linear B cell epitopes
B cells play an essential role in the fight against various viruses [37], [38], [39]. The ABCpred server (https://webs.iiitd.edu.in/raghava/abcpred/ABC_submission.html) was used to predict linear B-cell epitopes because of its high accuracy (65.93%) [40]. The epitope length was limited to 20, and the filtering threshold was maintained at the default value of 0.51 (a higher threshold implies higher specificity but lower sensitivity). Linear B-cell epitopes that remained ultimately eligible for enrollment were included in the construction of subsequent multiepitope vaccines.
2.5. Construction of the multiepitope vaccine
A novel multiepitope vaccine was constructed by linking immunodominant CTL, HTL, and B-cell epitopes using AAY, GPGPG, and KK linkers, respectively. To improve the antigenicity and immunogenicity of this multiepitope vaccine, the toll-like receptor 4 (TLR4) agonist RS-09 [41] and a 13 amino acid nonnatural pan DR epitope (PADRE) were added to the amino terminus of the multiepitope vaccine, and the TLR2 agonist dipalmitoyl-S-glycero-cysteine (Pam2Cys) and a 6 × His tag (HHHHH) were added to its carboxyl terminus [42], [43]. TLR agonists and PADRE were linked to epitopes using an EAAAK linker.
2.6. Assessment of immunogenicity, antigenicity, allergenicity, and toxicity of the multiepitope vaccine
The immunogenicity and antigenicity of the multiepitope vaccine were analyzed using the IEDB Immunogenicity Server, VaxiJen v2.0, and ANTIGENpro server (https://scratch.proteomics.ics.uci.edu/) according to a previous study [44]. The allergenicity of the multiepitope vaccine was predicted using AllerTOP v.2.0 server (https://www.ddg-pharmfac.net/AllerTOP/) and Allergen FP v.1.0. The AllerTOP v2.0 server explored the physicochemical properties of proteins by developing techniques such as k-nearest neighbors (kNN), ACC transformations, and machine learning of amino acid E descriptors [45], [46]. Finally, toxicity of the multiepitope vaccine was determined using the ToxinPred server (https://crdd.osdd.net/raghava/toxinpred/).
2.7. Assessment of physicochemical properties and solubility of multiepitope vaccine
The Expasy Protparam server (https://web.expasy.org/protparam/) was used to predict the physicochemical properties of multiepitope vaccines, such as molecular weight, theoretical isoelectric point (PI), amino acid composition, in vitro and in vivo half-life, instability, lipid index, and Grand average of hydropathicity (GRAVY). The solubility of the multiepitope vaccine was predicted using the Protein-Sol server (https://protein-sol.manchester.ac.uk/). An expected solubility value higher than 0.45 for an antigen means that the antigen has good solubility in the experimental solubility dataset (solubility of E. coli proteins) [47].
2.8. Global population coverage of the 15 candidate epitopes
Recognition of T cells with pathogen-derived epitopes depends on MHC molecules, which are highly polymorphic. The selection of epitopes with different HLA-binding specificities will increase the coverage of multiepitope vaccines in the target population. Hence, the global population coverage of MHC-I and MHC-II epitopes in the multiepitope vaccine was analyzed using the Population Coverage tool in the IEDB database (https://tools.iedb.org/population/) following a previous study [29].
2.9. Prediction and optimization of secondary/three-dimensional structures of the multiepitope vaccine
The structure of a protein profoundly influences its biological function. PSIPRED is a widely used tool to predict the secondary structure of target proteins and can predict the transmembrane topology, transmembrane helix, folding, and domain recognition of proteins [48]. The Prabi server predicted the secondary structures of candidate proteins using GOR4, and the average prediction accuracy was 64.4% [49]. In this study, we used PSIPRED (https://bioinf.cs.ucl.ac.uk/inspired/) and the Prabi server (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_ gor4.html) to analyze the secondary structure of the multiepitope vaccine. In addition, the three-dimensional (3D) spatial structure and function of the multiepitope vaccine were analyzed using the I-TASSER server (https://zhanggroup.org//I-TASSER/) according to a previous study [50]. The I-TASSER server generated 3D atomic models of the vaccine through multi-threaded comparisons and iterative structural assembly simulations. The estimated accuracy of the predictions is reflected in the modeling confidence score (C-score), which typically ranges from −5 to 2 (higher values indicate higher accuracy) [51]. Then, regarding optimizing the 3D model of the multiepitope vaccine, the GalaxyRefine web server (https://galaxy.seoklab.org/cgi-bin/submit.cgi?type=REFINE) was awarded this optimization work according to a previous study [52].
2.10. Validation and evaluation of 3D structures
The 3D structure of multiepitope vaccines constructed by the GalaxyRefine web server often contains errors. Therefore, the possible errors in the 3D structure of the produced multiepitope vaccine were validated using the ProSA-web (https://prosa.services.came.sbg.ac.at/prosa.php) and the ERRAT-web servers (https://saves.mbi.ucla.edu/), according to previous studies [53], [54]. Then, the SWISS-MODEL server (https://swissmodel.expasy.org/ assess) was used to generate Ramachandran plots to show favorable regions of backbone dihedral angles of amino acid residues in protein structures [55]. The most relevant scores provided by MolProbity were presented on the structure assessment page, and visual presentation of the results helped identify low-quality residue positions in the model or structure [55].
2.11. Prediction of discontinuous B-cell epitopes
It has been found that many B cell epitopes are not continuous in the amino acid sequence of a protein, and these discontinuous B cell epitopes play a vital role in biological function [56]. In the current study, discontinuous B-cell epitopes of the multiepitope vaccine were predicted using the ElliPro server (https://tools.iedb.org/ellipro/).
2.12. Molecular docking between the multiepitope vaccine and TLRs
Molecular docking is one of the most fundamental and valuable methods for assessing the interaction and binding coherence between vaccines and human TLRs. First, the Protein Data Bank (PDB) files of TLR2 (PDB ID:6NIG) and TLR4 (PDB ID:4G8A) were obtained from the NCBI Molecular Modeling Database (MMDB) (https://www.ncbi.nlm.nih.gov/structure/). Ligand-receptor docking analysis was performed using ClusPro 2.0, an online server (https://cluspro.bu.edu/home.php), following a previous study [57]. Finally, the PDB file of the docking results was loaded into LigPlot + to visualize the effects of cross-interface interactions using DIMPLOT plots.
2.13. Prediction of molecular dynamics and immunological responses induced by the multiepitope vaccine
The molecular dynamics and flexibility of the multiepitope vaccine were evaluated using the iMODS web server (https://imods.iqfr.csic.es/) [58]. The coordinated motion of the multiepitope vaccine in internal coordinates was demonstrated by the iMODS server based on Normal Modal Analysis (NMA) [59]. Furthermore, we applied an additional program to iMODS, namely, elementary dynamics simulation (EDM), to understand the macromolecular mobility and stability of the docked complexes. The evaluation parameters consist of special terms, such as B-factor, deformability, covariance, elastic model, and eigenvalues.
Potential immune responses induced by the multiepitope vaccine were predicted using the C-ImmSim server (https://150.146.2.1/C-IMMSIM/index.php). The C-ImmSim server shows excellent performance in predicting cellular and humoral responses generated by the vaccine in the mammalian immune system [60]. This robust server uses a 'site-specific score matrix' and machine-learning methods to assess specific antigen-mediated immune responses. In this study, HLA alleles A0101, A0201, B0702, B0801, DRB10101, and DRB1501 were used as the established parameters for implementing the simulation injections, and the number of immunizations was set to three.
2.14. Codon optimization and further cloning
Codon optimization can lead to superior expression of recombinant proteins. Referring to tools used by other vaccine researchers in the past, the Optimizer server was chosen to accomplish the optimization (https://genomes.urv.es/OPTIMIZER/) [61] which provides an optimized version depending on the codon of the target DNA sequence of the selected organism [62]. E. coli 536 was selected as the expression vector because of its wide range of applications. The results included several indicators, such as the codon adaptation index (CAI) and percentage GC content. The ideal value for CAI is 1 and the GC content is 30–70% [63]. After restriction site screening, the optimized nucleotide sequence of the multiepitope vaccine was inserted between the SacI and NcoI restriction sites in the pET30a (+) plasmid. In some previous studies, pcDNA3.1 expression vector [64] and pET-28a (+) expression vector [65], [66] were selected for cloning candidate vaccines. In this study, we chose the pET-30a (+) expression vector rather than the pET-28a (+) or pcDNA3.1 expression vector based on the following considerations: (1) Both pET-28a (+) and pET-30a (+) plasmids are commonly used prokaryotic expression vectors for high efficiency of fusion proteins; (2) Both plasmids contain kanamycin resistance genes, and their expression is induced by T7 RNA polymerase; (3) The number of restriction sites of pET-30a (+) plasmid was more than that of pET-28a (+) plasmid, which made it more advantageous in the expression of the target protein; (4) The pcDNA3.1 expression vector is used for high-level eukaryotic expression rather than prokaryotic expression in mammalian host cells. Finally, computer cloning was performed using SnapGene 6.0 software (SnapGene Inc., Chicago, USA).
3. Result
3.1. A novel multiepitope vaccine was constructed, based on immunodominant epitopes
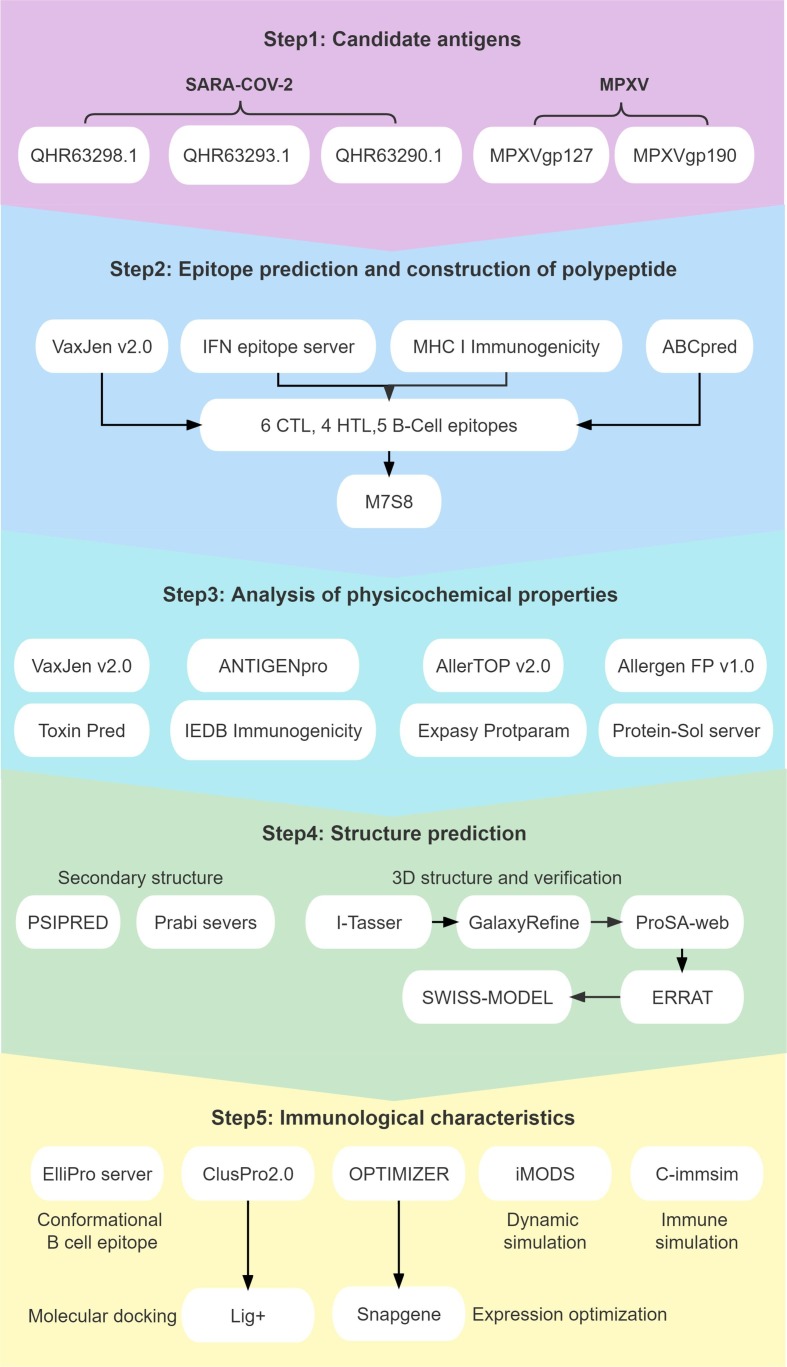
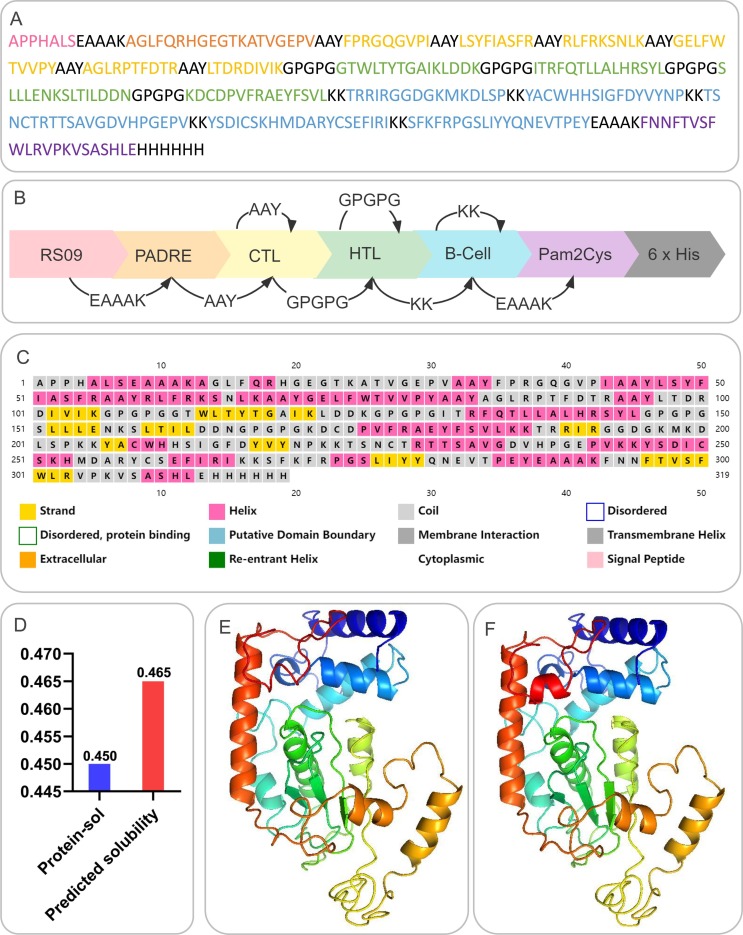
A flow chart for predicting and screening immunodominant epitopes and producing multiepitope vaccines is shown in Fig. 1 . The inclusion criteria for immunodominant epitopes were as follows: (1) HTL epitopes with antigenicity >0.7 and the highest IFN prediction score; (2) CTL epitopes with the highest immunogenicity score; and (3) B-cell epitopes with the highest ABC prediction score (>0.9). Finally, four HTL, six CTL, and five B-cell immunodominant epitopes were selected to construct a new multiepitope vaccine termed “S7M8″ (Table 1 ). GGPPG, AY, and KK linkers were used to link HTL, CTL, and B-cell epitopes, respectively. TLR4 agonist RS-09 (APPHALS), TLR2 agonist Pam2Cys (FNNFTVSFWLRVPKVSASHLE), and helper peptide PADRE (AGLFQRHGEGTKATVGEPV) were linked by the EAAK linker (Fig. 2 A). The final amino acid sequence and secondary structure of the S7M8 vaccine are shown in Fig. 2B and 2C, respectively.
Fig. 1.
Flow chart of epitope prediction, screening, and construction of the S7M8 vaccine. The S7M8 vaccine was developed in five steps and shown in five colors, including Candidate antigens, Epitope prediction and construction of polypeptide, Analysis of physicochemical properties, Structure prediction, and Immunological characteristics.
Table 1.
The information of the HTL, CTL, and B cell immunodominant epitopes selected to construct the S7M8 vaccine.
| Protein | Peptide sequence | Length | Alleles | Percentile rank | Antigenicity score | IFN-γ Inducer score | Immunogenicity score | ABC pred score |
|---|---|---|---|---|---|---|---|---|
| SARS-Cov-2 | ||||||||
| HTL epitopes | ||||||||
| QHR63293.1 | NRFLYIIKLIFLWLL | 15 | HLA-DRB4*01:01 | 0.12 | 1.0532 | 1 | – | – |
| QHR63290.1 | ITRFQTLLALHRSYL | 15 | HLA-DRB5*01:01 | 0.26 | 1.0412 | 3 | – | – |
| CTL epitopes | ||||||||
| QHR63298.1 | FPRGQGVPI | 9 | – | – | 1.0979 | – | 1.647 | – |
| QHR63293.1 | LSYFIASFR | 9 | – | – | 1.207 | – | 1.4994 | – |
| QHR63290.1 | RLFRKSNLK | 9 | – | – | 1.3419 | – | 1.7563 | – |
| B cellular epitopes | ||||||||
| QHR63293.1 | RSMWSFNPETNILLNV | 20 | – | – | – | – | – | 0.89 |
| QHR63290.1 | YACWHHSIGFDYVYNP | 20 | – | – | – | – | – | 0.96 |
| MPXV | ||||||||
| HTL epitopes | ||||||||
| MPXVgp127 | SLLLENKSLTILDDN | 15 | HLA-DRB1*13:02 | 0.31 | 0.9087 | 0.09220719 | – | – |
| MPXVgp190 | KDCDPVFRAEYFSVL | 15 | HLA-DPA1*01:03/DPB1*04:01 | 0.06 | 0.90993 | 0.1643051 | – | – |
| CTL epitopes | ||||||||
| MPXVgp127 | GELFWTVVPY | 10 | HLA-A*01:01 | 0.01 | 1.0979 | – | 0.20881 | – |
| AGLRPTFDTR | 10 | HLA-B*08:01 | 0.01 | 1.207 | – | 0.15678 | – | |
| LTDRDIVIK | 9 | HLA-A*03:01 | 0.01 | 1.3419 | – | 0.03632 | – | |
| B cellular epitopes | ||||||||
| MPXVgp127 | TSNCTRTTSAVGDVHPGEPV | 20 | – | – | – | – | – | 0.95 |
| YSDICSKHMDARYCSEFIRI | 20 | – | – | – | – | – | 0.89 | |
| SFKFRPGSLIYYQNEVTPEY | 20 | – | – | – | – | – | 0.87 | |
Fig. 2.
Construction, amino acid sequence, solubility, and structure analysis of the S7M8 vaccine. (A) Schematic diagram of the S7M8 vaccine construction. (B) The amino acid sequence of the S7M8 vaccine. (C) Second structure: the S7M8 vaccine contained 26.96% (86/319) of α-helix, 15.99% (51/319) of extended strand, and 57.05% (182/319) of random coil. (D) Solubility analysis predicted by the Protein-Sol server: the solubility of S7M8 vaccine was 0.465, indicating that S7M8 vaccine had good solubility. (E) 3-D structure of an unoptimized polypeptide molecule. (F) 3-D structure of the optimized polypeptide molecule.
3.2. Physicochemical properties and solubility of the S7M8 vaccine
The physical properties of proteins significantly affect their immune functions [67]. Therefore, the physicochemical characteristics of the S7M8 vaccine were analyzed using the Expasy ProtParam server. Our results showed that the S7M8 vaccine consisted of 319 amino acids with a molecular weight of 35463.46 Da, theoretical PI of 9.57, lipid index of 69.50, GRAVY of −0.419, and an instability index of 25.05 (Table 2 ), and these data were consistent with several previous studies [68], [69], [70], [71]. In addition, the estimated half-lives of the S7M8 vaccine in mammalian reticulocytes (in vitro), yeast (in vivo), and Escherichia coli (in vivo) were 4.4 h, >20 h, and >10 h, respectively (Table 2). Interestingly, we found that the half-life of the S7M8 vaccine in mammalian reticulocytes (in vitro) was shorter than that of several previous reported multiepitope vaccines [68], [69], [70], [71], suggesting that the half-life of the S7M8 vaccine in mammalian reticulocytes should be improved in the future. These data indicate that the S7M8 vaccine was stable in yeast and E. coli. Furthermore, the solubility of the S7M8 vaccine predicted by the Protein-Sol server was 0.465 (Fig. 2D), indicating good solubility.
Table 2.
Physical, chemical, and biological immunological characteristics of the S7M8 vaccine.
| No./Sl. | Features | Server | Assessment | Remark (left with blue means negative; red means the contrary) | |
|---|---|---|---|---|---|
| 1 | Antigenicity | Vaxijen | 0.6202 | – | Probable Antigen |
| 2 | ANTIGENpro server | 0.907374 | – | ||
| 3 | immunogenicity | IEDB | 0.6552 | – | Great immunogenicity |
| 4 | Predicted solubility | Protein-sol serve | 0.465 | – | Soluble well |
| 5 | pI | 10.05 | – | – | |
| 6 | Sensitization | AllerTOP v.2.0 | / | Probable Non-Allergen | – |
| 7 | AllerFP v.1.0 | Probable Non-Allergen | – | ||
| 8 | Toxicity | ToxinPred server | Non-Toxin | – | |
| 9 | Formula | Expasy Protparam server | C1617H2461N445O444S7 | – | – |
| 10 | The estimated half-life: mammalian reticulocytes, in vitro | Expasy Protparam server | 4.4 h | – | – |
| 11 | The estimated half-life: yeast, in vivo | >20 h | – | – | |
| 12 | The estimated half-life: Escherichia coli, in vivo | >10 h | – | – | |
| 13 | The instability index (II) | 25.05 | – | Classify the protein as stable | |
| 14 | Aliphatic index | 69.5 | – | Thermostable | |
| 15 | Grand average of hydropathicity (GRAVY) | −0.419 | – | Hydrophilic | |
3.3. The antigenicity, immunogenicity, sensitization, and toxicity of the S7M8 vaccine
An ideal vaccine candidate for preventing SARS-CoV-2 and MPXV co-infection should have good immunogenicity and antigenicity and should not be toxic or sensitizing. In our study, several tools were used to evaluate the properties of the S7M8 vaccine. Our results showed that the immunogenicity predicted by the IEDB database was 0.6552, and the antigenicity indicated by VaxiJenv2.0 and ANTIGENpro were 0.6202 and 0.907374, respectively (Table 2). These data suggest that the S7M8 vaccine is highly antigenic and immunogenic and can induce strong immune responses. The results showed that the S7M8 vaccine was non-allergenic and non-toxic. These data suggest that S7M8 may be a promising vaccine against SARS-CoV-2 and MPXV co-infection.
3.4. Analysis of the secondary and tertiary structure of the S7M8 vaccine
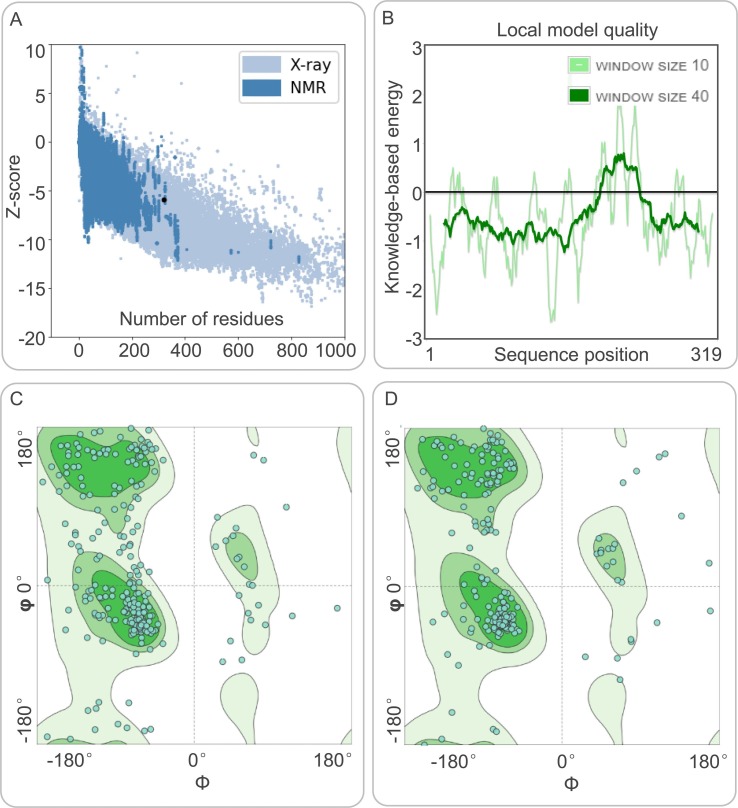
The results of the PSIPRED and Prabi servers showed that the S7M8 vaccine contained 26.96% (86/319) of alpha helices, 15.99% (51/319) of extended chains, and 57.05% (182/319) of random coils (Fig. 2C). The 3D structure of the S7M8 vaccine was subsequently predicted using the I-TASSER server, with the predicted z-scores of the tertiary structure model lying between 0.36 and 5.62. Their confidence scores (C-scores) were −3.26, −4.65, −5, −3.79, and −3.77, respectively. The model with a C-score = -3.26 was selected as the best 3D model with a corresponding TM value of 0.35 ± 0.12, and an expected root mean square deviation (RMSD) of 14.3 ± 3.8 Å (Fig. 2E). The TM value has been used as the recommended solution for the computational scaling of structural similarity and RMSD problems, which are sensitive to local errors. The 3D models of the S7M8 vaccine were refined using the GalaxyRefine web server. The level of consistency and quality of the candidate proteins improved after refinement. As a result, five Galaxy server-optimized 3D models were presented to researchers. After loop refinement and energy minimization analysis, Model 4 was used for subsequent studies, with several advantages: a GDTHA value of 0.9412 (the highest relative value), RMSD value of 0.447, MolProbity value of 2.84, and Clash score of 29.6 (Fig. 2F). The quality and potential errors of the 3D model of the S7M8 vaccine were checked and validated using the ProSA-web and ERRAT-web servers. The Z-value of the optimized S7M8 model was −5.92 (Fig. 3 A), and the energy plot is shown in Fig. 3B. The overall quality factor of the S7M8 model was 73.1392 after optimization using the ERRAT web server. The Ramachandran plot showed that the model had 68.82% favored regions, 11.18% outlier regions, and 17.37% rotamer regions (Fig. 3C). Interestingly, after optimization, the optimized model performed at 84.54%, 6.31%, and 2.32% in the favored, outlier, and rotamer regions of the S7M8 vaccine, respectively (Fig. 3D).
Fig. 3.
Evaluation and validation of the tertiary structure model of the S7M8 vaccine. (A) Z-score predicted by ProSA-web server: −5.92. (B) Energy map of the S7M8 validation. (C) Before optimization, the Ramachandran diagram showed the favored, outlier, and rotamer regions in the S7M9 vaccine were 68.82%, 11.18%, and 17.37%, respectively. (D) After optimization, the Ramachandran diagram showed the favored, outlier, and rotamer regions in the S7M9 vaccine were 84.54%, 6.31%, and 2.32%, respectively.
3.5. The S7M8 vaccine showed a high population coverage
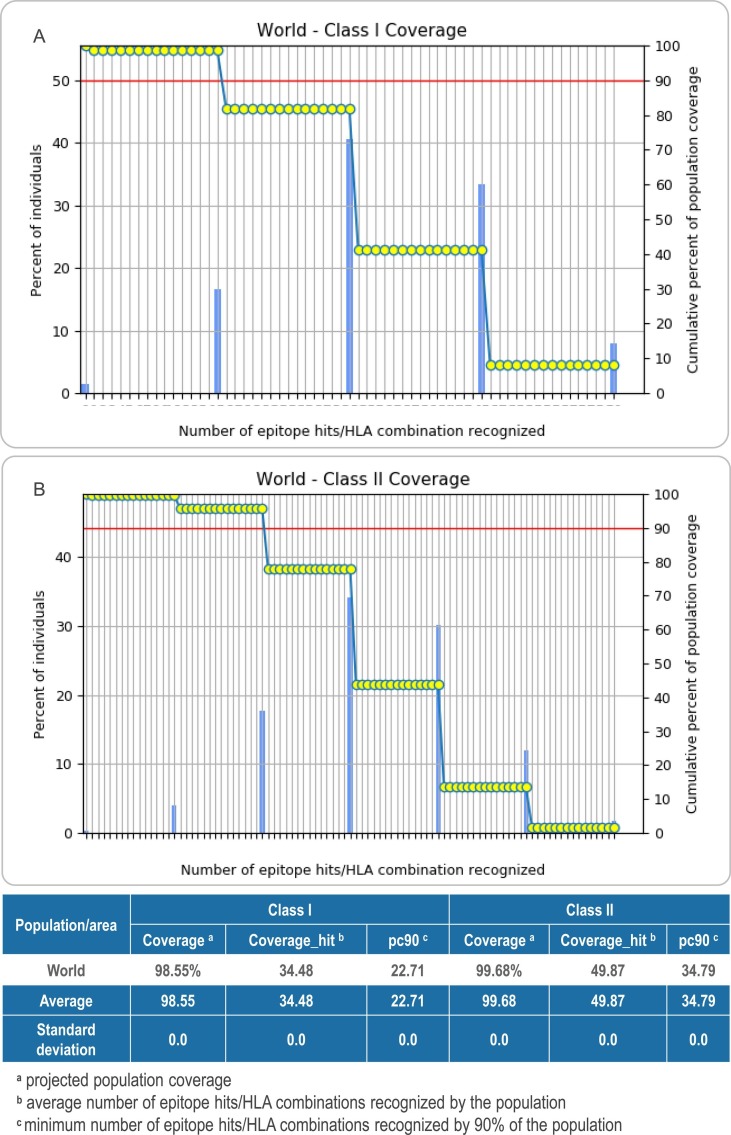
The results showed that the global population coverage of HLA I and HLA II of the multiepitope vaccine S7M8 was 98.55% and 99.68, respectively. These data suggest that the S7M8 vaccine has a comprehensive population coverage and can be recognized by most people worldwide. Detailed information is shown in Fig. 4 .
Fig. 4.
The population coverage rate of the C7M8 vaccine. (A) Class I Coverage: 98.55% worldwide. (B) Class II Coverage: 99.68% worldwide.
3.6. Conformational B-cell epitopes of the S7M8 vaccine
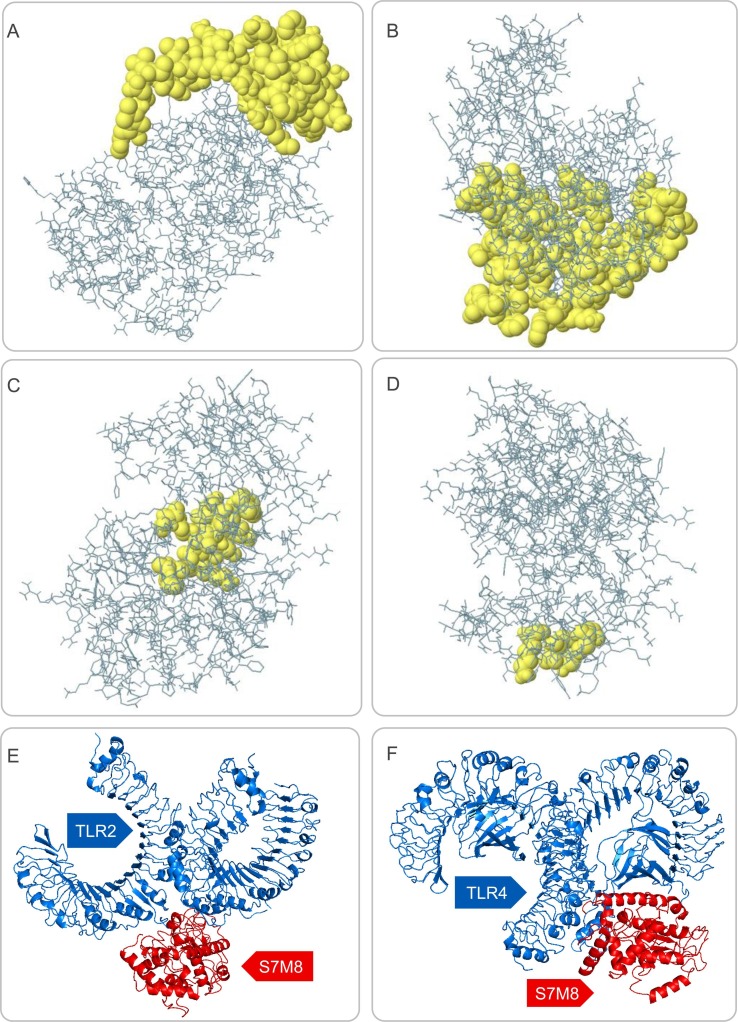
Based on data obtained from the ElliPro server, 193 residues were distributed across the four putative B-cell epitopes, with values ranging from 0.567 to 0.762. Among the predicted discontinuous epitopes, those with scores >0.66 were selected for further analysis. The conformations of the four B-cell epitopes of the S7M8 vaccine contained 128 residues, with scores ranging from 0.661 to 0.762 (Fig. 5 A–D and Table 3 ).
Fig. 5.
The discontinuous B cell epitopes of the S7M8 vaccine and molecular docking between the S7M8 vaccine and TLR2/4. (A–D) The conformation or discontinuous B cell epitopes of the S7M8 vaccine. The yellow balls in panels (A–D) represented four conformations or discontinuous B cell epitopes, respectively, and the rest were shown in gray. (E-F) 3-D structure diagram of molecular docking between S7M8 and TLR2/4. Ligand–receptor interactions: S7M8 was displayed in red, and TLR2 and TLR4 were shown in blue. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Table 3.
The conformational B cell epitope residues of the S7M8 vaccine predicted by the ElliPro.
| No. | Residues | Number of residues | Score |
|---|---|---|---|
| 1 | A:R192, A:G193, A:G194, A:D195, A:G196, A:K197, A:M198, A:K199, A:D200, A:L201, A:S202, A:P203, A:K204, A:K205, A:Y206, A:A207, A:C208, A:W209, A:H210, A:H211, A:S212, A:I213, A:G214, A:F215, A:D216, A:Y217, A:V218, A:Y219, A:N220, A:P221, A:K222, A:K223, A:T224, A:S225, A:N226, A:C227, A:T228, A:R229, A:T230, A:T231, A:S232, A:A233, A:V234, A:G235, A:D236, A:V237, A:H238, A:R264 | 48 | 0.762 |
| 2 | A:A1, A:P2, A:P3, A:H4, A:A5, A:L6, A:S7, A:E8, A:A9, A:A10, A:A11, A:K12, A:A13, A:F16, A:Q17, A:H19, A:G20, A:E21, A:G22, A:T23, A:K24, A:A25, A:T26, A:V27, A:G28, A:E29, A:P30, A:V31, A:A32, A:A33, A:F35, A:P36, A:Q39, A:Y58, A:R62, A:F74, A:A289, A:A290, A:K292, A:F293, A:N294, A:N295, A:F296, A:T297, A:V298, A:S299, A:F300, A:W301, A:L302, A:R303, A:V304, A:P305, A:K306, A:V307, A:S308, A:H311, A:H316, A:H317, A:H318, A:H319 | 60 | 0.737 |
| 3 | A:K121, A:L122, A:D123, A:D124, A:K125, A:G126, A:P127, A:G128, A:I161, A:L162, A:D163, A:D164, A:N165, A:G166, A:P167, A:G168 | 16 | 0.667 |
| 4 | A:K245, A:D248, A:S251, A:K252 | 4 | 0.661 |
3.7. Molecular docking of the S7M8 vaccine with human TLR-2 and TLR-4
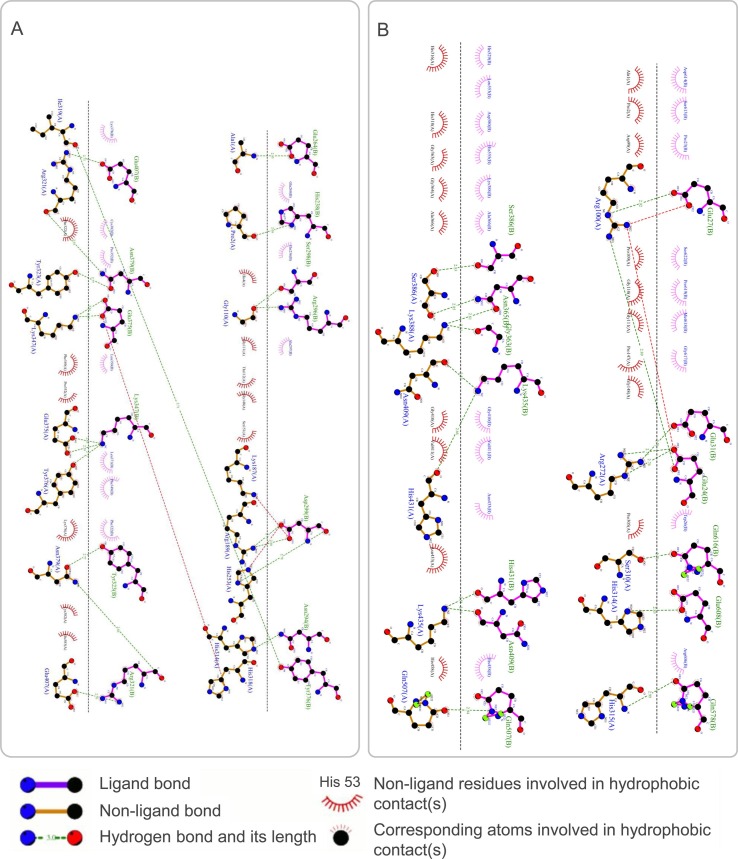
The results showed that the S7M8 vaccine formed 30 models after docking with TLR2 or TLR4. Among these models, the TLR2-S7M8 model (central weighted score: −768.9; lowest energy weighted score: −1100.6) and TLR4-S7M8 model (central weighted score: −950.3; lowest energy weighted score: −950.3) with lower complex binding energies were selected for further analysis. The docking effects of both models were presented in Fig. 5E and F, respectively. In addition, the hydrogen bonding and non-adhesive interactions in TLR2-S7M8 (Fig. 6 A) and TLR4-S7M8 (Fig. 6B) were visualized using LigPlot+ via a DIMPLOT plot. Our results showed 23 and 19 interactions in the TLR2-S7M8 and TLR4-S7M8 models, respectively. These results indicated that the S7M8 vaccine had excellent performance in tightly binding to TLR2/4 to trigger a strong immune response.
Fig. 6.
The DIMPLOT of the S7M8-TLR2 (A) and S7M8-TLR4 (B) complexes. There were 23 interaction sites in TLR2-S7M8 and 19 interaction sites in TLR4-S7M8. These results showed that the interaction between TLR2/4 and S7M8 was strong, and the docking effect was good.
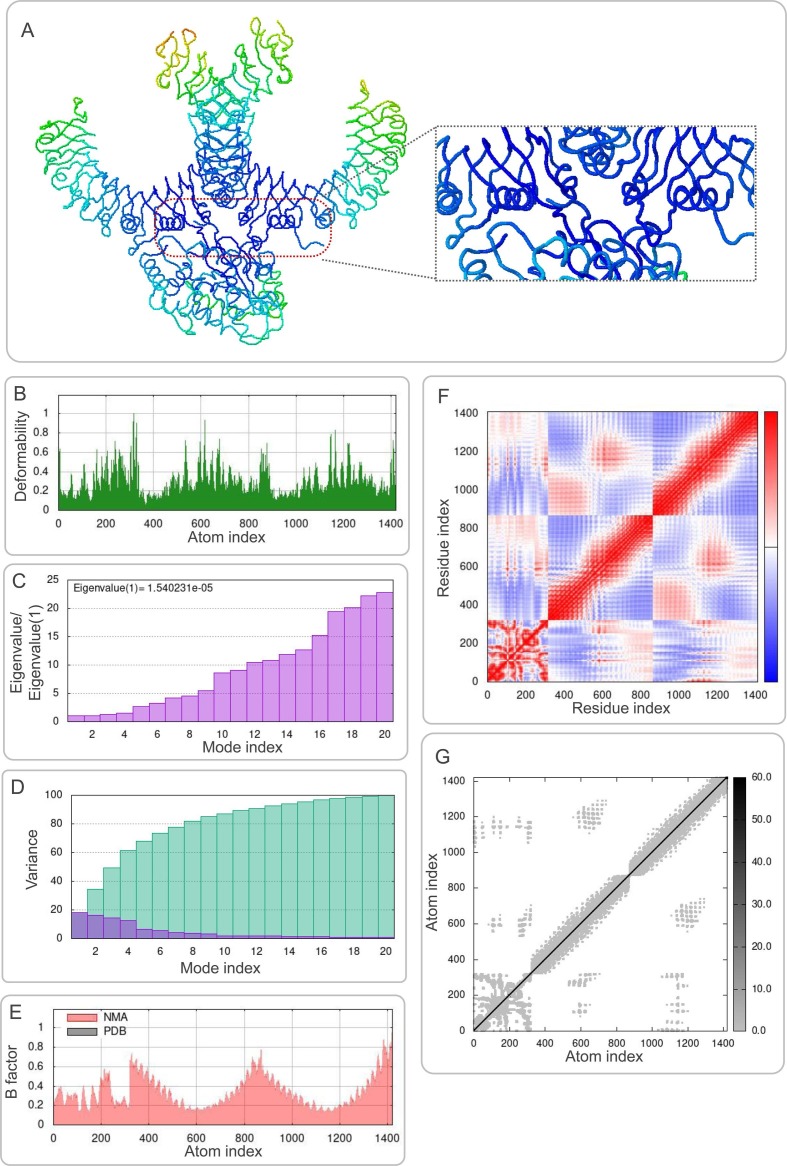
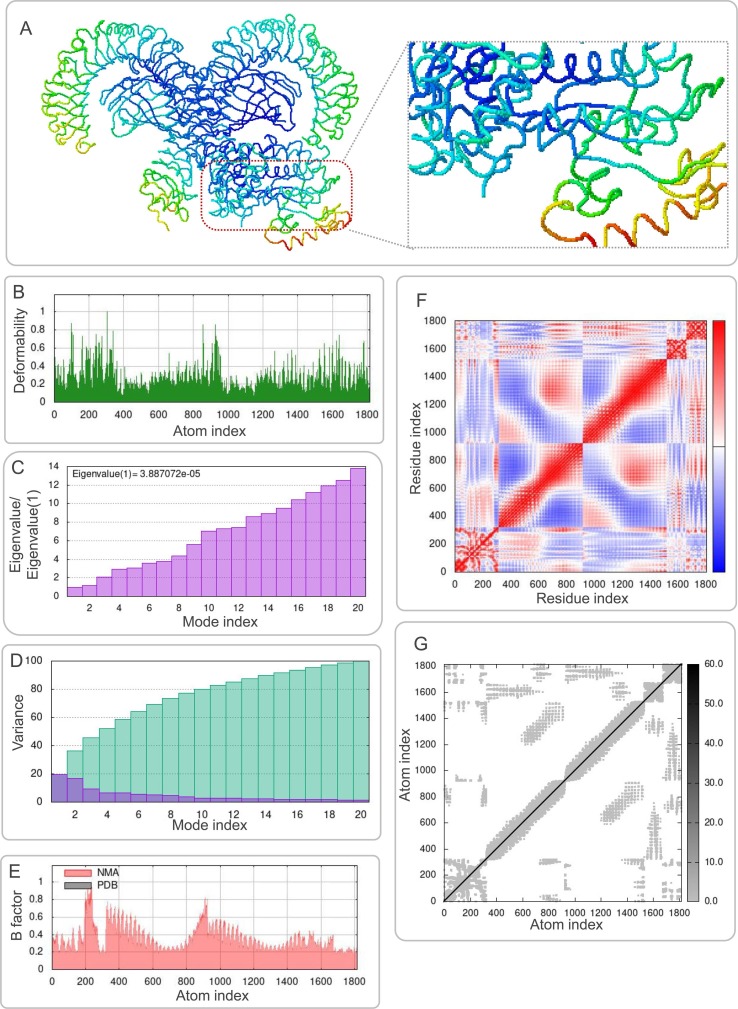
3.8. Molecular dynamics simulations between the S7M8 vaccine and TLRs
The results of molecular dynamics simulations and NMA of the docking complexes of the S7M8 vaccine with TLR2 and TLR4 were presented in Figs. 7 A and 8A, respectively. Simulations predicted the ability to determine the movement of molecules and atoms in the antigen structure. The deformability of TLR2-S7M8 (Fig. 7B) and TLR4-S7M8 (Fig. 8 B) complexes showed an isolated peak in the deformable region of the S7M8 vaccine. The eigenvalues for–TLR2-S7M8 and TLR4-S7M8 were 1.540231e-05 (Fig. 7C) and 3.887072e−05 (Fig. 8C), respectively. Covariograms showed cumulative or individual variance in green and purple colors, respectively (Figs. 7D and 8D). B-factor plots showed the relationship between the docked complexes in the NMA and PDB sectors (Figs. 7E and 8E). In the covariogram of the complexes, the motion between pairs of residues was indicated in different colors based on the mode of correlation: correlated in red, uncorrelated in white, and anti-correlated in blue (Figs. 7F and 8F). According to the elastic network model, the atoms of the docked protein molecules (C-α) were interconnected in a somewhat 'spring-like' manner (stiffer springs were shown in dark gray) (Figs. 7G and 8G).
Fig. 7.
Molecular dynamics simulation of the S7M8 vaccine and TLR2. (A) NMA mobility. The direction of the arrow indicates the direction of molecular motion. Blue represents the S7M8 molecule, and green expresses the TLR2 receptor. (B) variability; (C) eigenvalues; (D) variance (Purple for individual variance, green for cumulative variance); (E) B-factor; (F) covariance plot (red for correlated motion, white for uncorrelated motion, the blue color indicates anti-correlated motion); (G) elastic network (dark gray areas indicate harder areas). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 8.
Molecular dynamics simulation of the S7M8 vaccine and TLR4. (A) NMA mobility. The direction of the arrow indicates the direction of molecular motion. Blue represents the S7M8 molecule, and green expresses the TLR4 receptor. (B) variability; (C) eigenvalues; (D) variance (Purple for individual variance, green for cumulative variance); (E) B-factor; (F) covariance plot (red for correlated motion, white for uncorrelated motion, the blue color indicates anti-correlated motion); (G) elastic network (dark gray areas indicate harder areas). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.9. Immune responses induced by the S7M8 vaccine
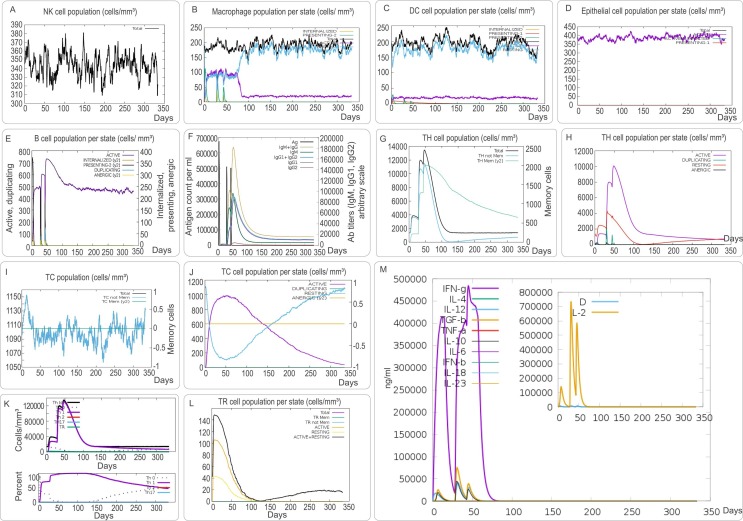
As an intracellular pathogen, cellular and humoral immunity are essential for killing and eliminating SARS-CoV-2 and MPXV. We found that the S7M8 vaccine effectively stimulated the innate immune responses predicted by immune mimics. The results showed that after S7M8 stimulation, (1) the number of natural killer (NK) cells in each state increased in a phased and fluctuating manner, reaching a peak on day 100/150 at 380 cells/mm3 and then fluctuated around 350 cells/mm3 (Fig. 9 A); (2) the number of macrophages formed three peaks at 5, 30, and 40 d, which were 120, 90, and 50 cells/mm3, respectively (Fig. 9B); (3) the overall condition of dendritic cells (DCs) was mainly determined by resting DCs (180/200) (Fig. 9C), and the number of active DCs peaked and fluctuated around 20 cells/mm3 (Fig. 9C); and (4) the number of active epithelial cells remained significantly high over time (Fig. 9D).
Fig. 9.
The innate and adaptive immune responses induced by the S7M8 vaccine in the C-ImmSim server. (A) Expression of NK cells after antigen stimulation. (B) Expression in Macrophages (black) after antigen stimulation. (C) Expression of DC cells (black) after antigen stimulation. (D) Secretion from active epithelial cells (purple) after antigen stimulation. The adaptive immune responses induced by S7M8 in the C-ImmSim server (E-M). (E) Active B cell (purple) secretion after antigen stimulation. (F) The primary B cell antibody produced after antigen stimulation is IgM+ IgG (yellow) changes over time. (G, H) Changes in the secretion level of helper T lymphocytes and the secretion levels of helper T lymphocytes of different memory types. (I, J) Changes in the level of CD8+ T secretion after antigen stimulation and the secretion of different types of CD8+ T cells. (K) The number and proportion of helper T cell subtypes. (L) Changes in the number of regulatory T cell subtypes. (G) Changes in secretion levels of cytokines, mainly IFN-γ (purple) and IL-2 (yellow). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Innate immune cells play an essential role in the early stages of viral infection and are the first line of defense against viral invasion. However, innate immune cells alone cannot completely eliminate or kill the virus, and specific immune cells are indispensable [20], [72], [73]. The results showed that the S7M8 vaccine induced three peaks in the number of active B lymphocytes (Fig. 9E) and three peaks in the antibody levels after three vaccine doses (Fig. 9F). Furthermore, the S7M8 vaccine induced the formation of three gradually rising peaks in the T helper (TH) cell population after three injections (Fig. 9G), and the active TH cell population reached a peak of 10,000 cells/mm3 on day 50 after the first injection (Fig. 9H).
In addition, CTLs have been reported to play an essential role in killing pathogen-infected host cells by producing granzyme and nitric oxide [74], [75]. Here, we found that the number of non-memory cytotoxic T cells per state gradually increased and peaked on day 15 after the first injection (1,152 cells/mm3), while the number of memory cytotoxic T cells remained at 1,105 cells/mm3 (Fig. 9I). Furthermore, we found that the number of activated toxic T lymphocytes in each state increased gradually and peaked at 900 cells/mm3 on day 50 after the first injection; however, duplication of CTLs showed the opposite trend (Fig. 9J). We found that immunization with the S7M8 vaccine induced three peaks in Th1 cell numbers (25,000 cells/mm3, 110,000 cells/mm3, and 130,000 cells/mm3) on days 10, 35, and 50 after the first immunization (Fig. 9K). Furthermore, the number of active and resting regulatory T cells peaked on day ten after the first injection (Fig. 9L). Interestingly, our results showed that the S7M8 vaccine induced significantly higher levels of IFN-γ (480,000 ng/ml) and interleukin 2 (IL-2) (730,000 ng/ml) (Fig. 9M). These data are consistent with a previous report [69].
3.10. Result of codon optimization and further cloning by computer simulation
We used the Optimizer server to predict the molecular cloning and expression levels of the S7M8 vaccine in vitro to determine the maximum protein expression in Escherichia coli 536 (E. coli). The data showed that the CAI value of the S7M8 vaccine was 1.000 and the average GC content of the adaptation sequence was 55.90%, indicating that the S7M8 vaccine could be highly expressed in E. coli. Finally, the gene sequence of the optimized S7M8 vaccine was inserted into the pET30a (+) plasmid to construct a recombinant plasmid (Fig. 10 ).
Fig. 10.
The polypeptide molecular sequence was electronically cloned into the pET30a expression vector by SnapGene software.
4. Discussion
With the rapid development of bioinformatics, structural biology, and computing tools, biomarker selection and construction methods for vaccine development are diversified and information-based [76], [77], [78]. In this study, we developed a promising multiepitope vaccine, S7M8, to prevent SARS-CoV-2 and MPXV co-infection, based on immunodominant epitopes identified from three SARS-CoV-2 antigens and two MPXV antigens. To significantly improve the immunogenicity and antigenicity of the S7M8 vaccine, PADRE helper peptide, TLR2 agonist Pam2Cys and TLR4 agonist RS-09 were incorporated into the vaccine design. PADRE, an epitope peptide derived from HLA-DR and tetanus toxin, is commonly used as a helper peptide to induce Th1 cell polarization [79]. In addition, the PADRE peptide can bind to various types of MHC-II alleles to enhance the immune response to vaccines [80]. TLR proteins are pattern recognition receptors for multiple pathogens, including SARS-CoV-2 and MPXV [22], [34]. In this study, the TLR2 agonist Pam2Cys and TLR4 agonist RS-09 were included in the S7M8 vaccine to trigger activation of TLR2 and TLR4 signaling [81], [82], [83], [84], [85], [86]. Furthermore, our results suggested that the S7M8 vaccine could stably bind to TLR2 and TLR4, which laid the foundation for the S7M8 vaccine to recognize and activate TLR pathways effectively.
An ideal peptide-based vaccine designed using bio- and immunoinformatics methods should have good immunogenicity and antigenicity, be non-toxic, and non-allergenic. Furthermore, it can induce a robust immune response without causing side effects [87], [88]. Our results demonstrate that S7M8 is a promising vaccine with good antigenicity, immunogenicity, non-toxicity, and non-sensitization. The secondary structure of the S7M8 vaccine contained 26.96% α-helices, 15.99% extended strand, and 57.05% random coils. Naturally unfolding protein regions and α-helical coiled coil motifs are considered as the basic types of “structural antigens,” and the increase in both structures would facilitate the recognition of antibodies induced after infection [89]. Our results showed that 84.54% of the amino acid residues of the S7M8 vaccine were located in the preferred region, indicating that the prediction quality of the entire model was acceptable.
SARS-CoV-2 first recognizes the ACE2 receptor on the surface of the pulmonary epithelial cells [90]. After shelling and other steps, RNA is contained in the vesicles in the interior of cells. TLRs on the surface of vesicles act on NF-kB and IRFs with the cooperation of MAVS [91]. These two molecules then enter the nucleus to transcribe, translate, and release type 1 and 3 interferons from the nucleus of pulmonary epithelial cells [92]. Interferons can bind to receptors on the surface of dendritic cells or CD8+ T cells. Dendritic cells can cooperate with CD8+T cells to kill infected cells and present antigen molecules to B cells to produce corresponding antibodies to assist the innate immune system in killing or participating in virus neutralization [20], [72], [93], [94]. Furthermore, the MPXV can attach to the cell surface, the virion binds and fuses with the host cell membrane, and the virus core is released into the cytoplasm of the host cells. Virions are assembled into intracellular mature viruses (MVs), remain in the cytoplasm in the form of intracellular mature virions and are released into the extracellular envelope of viruses during cell lysis [95], [96], [97]. MVs can also wrap an extra capsule and attach to the cell membrane, which are then released by exocytosis and continue to invade other normal cells of the human body [24], [98].
Previous studies have shown that TLRs mediate pro-inflammatory signals induced by SARS-CoV-2 [20]. Moreover, DCs, the most robust antigen-presenting cells in the body, express TLRs. With the help of TLRs to recognize pathogen-associated molecular pattern (PAMP) molecules, the vaccine can activate DCs to trigger a non-specific, albeit rapid, innate immune response characterized by inflammation, cytokine production, immune cell recruitment, and activation of phagocytes that neutralize pathogens and infected cells [99]. Therefore, TLRs are bridges for the activation of DCs caused by microbial components such as SARS-CoV-2, MPXV, and Mycobacterium tuberculosis [100]. Thus, the addition of TLR agonists to the design of a multiepitope vaccine can enhance the preventive effect. In our study, agonists of TLR2/4 were added, which enabled TLRs to play a recognition role in acquired immunity.
In contrast, TLRs activate post-transcriptional molecules that play an essential role in viral resistance. For example, TLR2 preferentially activates the transcription of P19, a recently proven protein that forms heterodimers with P40, similar to that of IL-12 [101], [102]. TLR4 agonists can stimulate the production of interferon-inducible protein-10 (IP-10) in large quantities [103], [104]. IP-10 is a chemical activator of CXC produced by different cells in response to IFN-γ and microbial components, which can chemisorb monocytes and NK cells [105]. More importantly, Th1 cells prioritize the expression of IP-10 receptor CXCR3 [106], which may explain why Th1 cells were the main component of the simulated immune results in our study (Fig. 9K). At the same time, macrophages in non-specific immunity can maintain a high level of activated macrophages (Fig. 9B) after each injection, which means that when pathogens invade, macrophages can respond quickly and participate in the process of pathogen resistance.
In addition, inflammation attracts antigen-presenting cells (APCs) to the infection site, where they absorb antigens and transport them to secondary lymphoid organs, such as lymph nodes and the spleen, to guide the activation of T and B cells, which constitutes an adaptive immune response [21], [107], [108]. Although this depends on innate immune activation, the acquired immune system is a typical pathogen-specific response. In secondary lymphoid tissues, APCs drive the expansion of different T cell subsets [109], [110]. Notably, they trigger CD8+ cytotoxic T cells (Fig. 9I and J), which can recognize and kill cells infected by specific pathogens. APCs can also enhance the role of CD4+ helper T cells (Fig. 9G), which contributes to the differentiation of B cells (Fig. 9E), and finally produces pathogen-specific antibodies (Fig. 9F). These antibodies are essential for clearing infections by binding to microorganisms to prevent cell entry or labeling pathogens for complement or innate immune cell destruction. Eventually, these T and B cells adopt a memory phenotype (Fig. 9G, I, and Supplementary file 1), which prepares the cells to expand and reactivate to cope with possible future pathogens. Notably, the next time the body comes into contact with the same pathogen, this memory can respond more quickly and effectively. Interestingly, our results suggested that the S7M8 vaccine could induce high levels of IFN-γ and IL-2 (Fig. 9M). IFN-γ plays a vital role in activating T cells (mainly Th1 and CTL), promoting NK cell activity, presenting antigens, and activating macrophage lysosomes [111], [112], [113]. IL-2 plays a critical role in the immune response against viral infections. It is capable of being stimulated by specific antigens or mitogenic factors and then activates T cell proliferation, activates CD4 + T cells to exert cytotoxic effects, promotes other cytokine production, stimulates NK cell proliferation, enhances NK cell killing activity and cytokine production, induces LAK cell production, promotes B cell proliferation to secrete antibodies, and activates macrophages [114], [115], [116].
These results suggest that the S7M8 vaccine is a promising vaccine for SARS-CoV-2 and MPXV prevention. This vaccine has several advantages: (1) the S7M8 vaccine has excellent antigenicity, immunogenicity, non-toxicity, non-sensitization, stability, and solubility; (2) the S7M8 vaccine contained the PADRE helper peptide, TLR2 agonist Pam2Cys, and TLR4 agonist RS-09, which could target APCs such as macrophages to initiate innate immune responses, induce B lymphocytes to secrete high levels of specific antibodies, and activate CD4+ and CD8+ T lymphocytes to produce high levels of cytokines such as IFN-γ and IL-2; (3) in contrast to the original peptide molecules for the prevention of SARS-CoV-2 or MPXV alone, the new peptide molecule S7M8 constructed herein was selected with due reference to the immunodominant antigens selected during the construction of the existing vaccines in the selection of the preparative antigens [88]. This study confirmed that S7M8 provided a protective effect for the host in the case of either SARS-CoV-2 or MPXV infection alone or in the case of co-infection. This study also has some limitations: (1) the antigens for predicting and screening immunodominant epitopes were limited to five antigens, which may reduce the efficacy of the S7M8 vaccine to fight against the co-infection of SARS-CoV-2 and MPXV; (2) the physicochemical and immunological properties of the S7M8 vaccine were based on bioinformatics analysis and not validated with real-world data.
In the next step, we will carry out research from the following aspects: (1) the sequences of CTL, HTL and B cell epitopes will be further optimized to improve the solubility and expression of the S7M8 vaccine; (2) the spatial three-dimensional structure of the vaccine will be analyzed by transmission electron microscopy, scanning electron microscopy and freezing electron microscopy, providing information for understanding the targets of the S7M8 vaccine; (3) the characteristics of innate and adaptive immune responses induced by the S7M8 vaccine will be evaluated by in vitro and in vivo experiments, and the protective efficacy of the S7M8 vaccine will be determined by animal models.
5. Conclusions
In this study, we manufactured a novel multiepitope vaccine, named S7M8, using bioinformatics and immunoinformatics. The S7M8 vaccine consists of four HTL epitopes, six CTL epitopes, five B cell epitopes, TLR agonists RS-09 and Pam2Cys, and the helper peptide PADRE. Our results suggest that the S7M8 vaccine has good immunogenicity, antigenicity, non-toxicity, and non-allergy properties, and can effectively induce robust immune responses without causing side effects. This study provides a new vaccine candidate for preventing SARS-CoV-2 and MPXV co-infection and highlights new strategies for interrupting the international spread of both viruses.
Declarations
Ethics approval and consent to participate
This paper does not include humans or animals involved in the experiment, so there is no need for a permit from Ethics Committee.
Consent for publication
Not applicable.
Competing interests
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Funding.
This study was funded by the Beijing Municipal Science & Technology Commission (Grant No. L192065 and 7212103) and the National key research and development program of China (Grant No. 2022YFA1303500-003).
Availability of data and materials.
All data generated or analyzed during this study are included in this published article and its supplementary information files.
CRediT authorship contribution statement
Fan Jiang: Software, Methodology, Writing – original draft. Yinping Liu: Methodology. Yong Xue: Methodology. Peng Cheng: Methodology. Jie Wang: Methodology. Jianqi Lian: Conceptualization, Funding acquisition. Wenping Gong: Conceptualization, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Not applicable.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.intimp.2023.109728.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
Data availability
All data are from public databases.
References
- 1.Polack F.P., et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;383(27):2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baden L.R., et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2021;384(5):403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Voysey M., et al. Single-dose administration and the influence of the timing of the booster dose on immunogenicity and efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccine: a pooled analysis of four randomised trials. Lancet. 2021;397(10277):881–891. doi: 10.1016/S0140-6736(21)00432-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ledford H. J&J's one-shot COVID vaccine offers hope for faster protection. Nature. 2021 doi: 10.1038/d41586-021-00119-7. [DOI] [PubMed] [Google Scholar]
- 5.Logunov D.Y., et al. Safety and efficacy of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine: an interim analysis of a randomised controlled phase 3 trial in Russia. Lancet. 2021;397(10275):671–681. doi: 10.1016/S0140-6736(21)00234-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sapkal G.N., et al. Inactivated COVID-19 vaccine BBV152/COVAXIN effectively neutralizes recently emerged B.1.1.7 variant of SARS-CoV-2. J. Travel Med. 2021;28(4) doi: 10.1093/jtm/taab051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim J.H., Marks F., Clemens J.D. Looking beyond COVID-19 vaccine phase 3 trials. Nat. Med. 2021;27(2):205–211. doi: 10.1038/s41591-021-01230-y. [DOI] [PubMed] [Google Scholar]
- 8.Dunkle L.M., et al. Efficacy and safety of NVX-CoV2373 in adults in the United States and Mexico. N. Engl. J. Med. 2022;386(6):531–543. doi: 10.1056/NEJMoa2116185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heath P.T., et al. Safety and Efficacy of NVX-CoV2373 Covid-19 Vaccine. N. Engl. J. Med. 2021;385(13):1172–1183. doi: 10.1056/NEJMoa2107659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shinde V., et al. Efficacy of NVX-CoV2373 Covid-19 Vaccine against the B.1.351 Variant. N. Engl. J. Med. 2021;384(20):1899–1909. doi: 10.1056/NEJMoa2103055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.A. Barchuk et al., Gam-COVID-Vac, EpiVacCorona, and CoviVac effectiveness against lung injury during Delta and Omicron variant surges in St. Petersburg, Russia: a test-negative case-control study, Respir. Res. 23(1) (2022) 276. [DOI] [PMC free article] [PubMed]
- 12.Amraei R., Rahimi N. Covid-19, Renin-angiotensin system and endothelial dysfunction. Cells. 2020;9(7) doi: 10.3390/cells9071652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumari P., et al. Potential diagnostics and therapeutic approaches in COVID-19. Clin. Chim. Acta. 2020;510:488–497. doi: 10.1016/j.cca.2020.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bai Z., et al. The SARS-CoV-2 nucleocapsid protein and its role in viral structure, biological functions, and a potential target for drug or vaccine mitigation. Viruses. 2021;13(6) doi: 10.3390/v13061115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaur H., et al. Ivermectin as a potential drug for treatment of COVID-19: an in-sync review with clinical and computational attributes. Pharmacol. Rep. 2021;73(3):736–749. doi: 10.1007/s43440-020-00195-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li D., Li J. Immunologic testing for SARS-CoV-2 infection from the antigen perspective. J. Clin. Microbiol. 2021;59(5) doi: 10.1128/JCM.02160-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang Y., et al. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020;41(9):1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xia X. Domains and functions of spike protein in Sars-Cov-2 in the context of vaccine design. Viruses. 2021;13(1) doi: 10.3390/v13010109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jia Z., Gong W. Will mutations in the spike protein of SARS-CoV-2 lead to the failure of COVID-19 vaccines? J. Korean Med. Sci. 2021;36(18):e124. doi: 10.3346/jkms.2021.36.e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson C.B., et al. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022;23(1):3–20. doi: 10.1038/s41580-021-00418-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merad M., et al. The immunology and immunopathology of COVID-19. Science. 2022;375(6585):1122–1127. doi: 10.1126/science.abm8108. [DOI] [PubMed] [Google Scholar]
- 22.Sanami S., et al. Exploring SARS-COV-2 structural proteins to design a multi-epitope vaccine using immunoinformatics approach: an in silico study. Comput. Biol. Med. 2021;133 doi: 10.1016/j.compbiomed.2021.104390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adler H., et al. Clinical features and management of human monkeypox: a retrospective observational study in the UK. Lancet Infect. Dis. 2022;22(8):1153–1162. doi: 10.1016/S1473-3099(22)00228-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kabuga A.I., El Zowalaty M.E. A review of the monkeypox virus and a recent outbreak of skin rash disease in Nigeria. J. Med. Virol. 2019;91(4):533–540. doi: 10.1002/jmv.25348. [DOI] [PubMed] [Google Scholar]
- 25.Thornhill J.P., et al. Monkeypox virus infection in humans across 16 countries - April-June 2022. N. Engl. J. Med. 2022;387(8):679–691. doi: 10.1056/NEJMoa2207323. [DOI] [PubMed] [Google Scholar]
- 26.Bhattacharya M., Dhama K., Chakraborty C. Recently spreading human monkeypox virus infection and its transmission during COVID-19 pandemic period: a travelers' prospective. Travel Med. Infect. Dis. 2022;49 doi: 10.1016/j.tmaid.2022.102398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.WHO, Vaccines and immunization for monkeypox Interim guidance, W.H. Organization, Editor. 2022, World Health Organization: Geneva, p. 20.
- 28.Yousaf M., et al. Immuno-informatics profiling of monkeypox virus cell surface binding protein for designing a next generation multi-valent peptide-based vaccine. Front. Immunol. 2022;13:1035924. doi: 10.3389/fimmu.2022.1035924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bhattacharya M., et al. Designing, characterization, and immune stimulation of a novel multi-epitopic peptide-based potential vaccine candidate against monkeypox virus through screening its whole genome encoded proteins: an immunoinformatics approach. Travel Med. Infect. Dis. 2022;50 doi: 10.1016/j.tmaid.2022.102481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Akhtar N., et al. Immunoinformatics-aided design of a peptide based multiepitope vaccine targeting glycoproteins and membrane proteins against Monkeypox virus. Viruses. 2022;14(11) doi: 10.3390/v14112374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wells C.R., et al. Impact of international travel and border control measures on the global spread of the novel 2019 coronavirus outbreak. PNAS. 2020;117(13):7504–7509. doi: 10.1073/pnas.2002616117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adegboye O.A., et al. Travel-related monkeypox outbreaks in the era of COVID-19 pandemic: are we prepared? Viruses. 2022;14(6) doi: 10.3390/v14061283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farahat R.A., et al. Monkeypox and human transmission: Are we on the verge of another pandemic? Travel Med. Infect. Dis. 2022;49 doi: 10.1016/j.tmaid.2022.102387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swetha R.G., et al. Multi-epitope vaccine for monkeypox using pan-genome and reverse vaccinology approaches. Viruses. 2022;14(11) doi: 10.3390/v14112504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doytchinova I.A., Flower D.R. VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinf. 2007;8:4. doi: 10.1186/1471-2105-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dhanda S.K., Vir P., Raghava G.P. Designing of interferon-gamma inducing MHC class-II binders. Biol. Direct. 2013;8:30. doi: 10.1186/1745-6150-8-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ju B., et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584(7819):115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- 38.Quast I., Tarlinton D. B cell memory: understanding COVID-19. Immunity. 2021;54(2):205–210. doi: 10.1016/j.immuni.2021.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Song W., et al. Development of Tbet- and CD11c-expressing B cells in a viral infection requires T follicular helper cells outside of germinal centers. Immunity. 2022;55(2):290–307.e5. doi: 10.1016/j.immuni.2022.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saha S., Raghava G.P. Prediction of continuous B-cell epitopes in an antigen using recurrent neural network. Proteins. 2006;65(1):40–48. doi: 10.1002/prot.21078. [DOI] [PubMed] [Google Scholar]
- 41.Albutti A. An integrated computational framework to design a multi-epitopes vaccine against Mycobacterium tuberculosis. Sci. Rep. 2021;11(1):21929. doi: 10.1038/s41598-021-01283-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meza B., et al. A novel design of a multi-antigenic, multistage and multi-epitope vaccine against Helicobacter pylori: an in silico approach. Infect. Genet. Evol. 2017;49:309–317. doi: 10.1016/j.meegid.2017.02.007. [DOI] [PubMed] [Google Scholar]
- 43.Jackson D.C., et al. A totally synthetic vaccine of generic structure that targets Toll-like receptor 2 on dendritic cells and promotes antibody or cytotoxic T cell responses. PNAS. 2004;101(43):15440–15445. doi: 10.1073/pnas.0406740101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Magnan C.N., et al. High-throughput prediction of protein antigenicity using protein microarray data. Bioinformatics. 2010;26(23):2936–2943. doi: 10.1093/bioinformatics/btq551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dimitrov I., et al. AllerTOP vol 2–a server for in silico prediction of allergens. J. Mol. Model. 2014;20(6):2278. doi: 10.1007/s00894-014-2278-5. [DOI] [PubMed] [Google Scholar]
- 46.Dimitrov I., et al. AllergenFP: allergenicity prediction by descriptor fingerprints. Bioinformatics. 2014;30(6):846–851. doi: 10.1093/bioinformatics/btt619. [DOI] [PubMed] [Google Scholar]
- 47.Hebditch M., et al. Protein-Sol: a web tool for predicting protein solubility from sequence. Bioinformatics. 2017;33(19):3098–3100. doi: 10.1093/bioinformatics/btx345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McGuffin L.J., Bryson K., Jones D.T. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16(4):404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 49.Garnier J., Gibrat J.F., Robson B. GOR method for predicting protein secondary structure from amino acid sequence. Methods Enzymol. 1996;266:540–553. doi: 10.1016/s0076-6879(96)66034-0. [DOI] [PubMed] [Google Scholar]
- 50.Yang J., et al. The I-TASSER Suite: protein structure and function prediction. Nat. Methods. 2015;12(1):7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roy A., Kucukural A., Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 2010;5(4):725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.L. Heo, H. Park, C. Seok, GalaxyRefine: protein structure refinement driven by side-chain repacking, Nucl. Acids Res. 41(Web Server issue) (2013) W384-8. [DOI] [PMC free article] [PubMed]
- 53.M. Wiederstein, M.J. Sippl, ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins, Nucl. Acids Res. 35(Web Server issue) (2007) W407-10. [DOI] [PMC free article] [PubMed]
- 54.Colovos C., Yeates T.O. Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 1993;2(9):1511–1519. doi: 10.1002/pro.5560020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Waterhouse A., et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucl. Acids Res. 2018;46(W1):W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ponomarenko J., et al. ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinf. 2008;9:514. doi: 10.1186/1471-2105-9-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kozakov D., et al. The ClusPro web server for protein-protein docking. Nat. Protoc. 2017;12(2):255–278. doi: 10.1038/nprot.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lopéz-Blanco J.R., Garzón J.I., Chacón P. iMod: multipurpose normal mode analysis in internal coordinates. Bioinformatics. 2011;27(20):2843–2850. doi: 10.1093/bioinformatics/btr497. [DOI] [PubMed] [Google Scholar]
- 59.J.R. López-Blanco et al., iMODS: internal coordinates normal mode analysis server, Nucl. Acids Res. 42(Web Server issue) (2014) W271-6. [DOI] [PMC free article] [PubMed]
- 60.Rapin N., et al. Computational immunology meets bioinformatics: the use of prediction tools for molecular binding in the simulation of the immune system. PLoS One. 2010;5(4):e9862. doi: 10.1371/journal.pone.0009862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Puigbò P., et al. OPTIMIZER: a web server for optimizing the codon usage of DNA sequences. Nucl. Acids Res. 2007;35(Web Server issue):W126–W131. doi: 10.1093/nar/gkm219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gould N., Hendy O., Papamichail D. Computational tools and algorithms for designing customized synthetic genes. Front. Bioeng. Biotechnol. 2014;2:41. doi: 10.3389/fbioe.2014.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Puigbò P., Romeu A., Garcia-Vallvé S. HEG-DB: a database of predicted highly expressed genes in prokaryotic complete genomes under translational selection. Nucl. Acids Res. 2008;36(Database issue):D524–D527. doi: 10.1093/nar/gkm831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Safavi A., et al. Efficacy of co-immunization with the DNA and peptide vaccines containing SYCP1 and ACRBP epitopes in a murine triple-negative breast cancer model. Hum. Vaccin. Immunother. 2021;17(1):22–34. doi: 10.1080/21645515.2020.1763693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Safavi A., et al. Production, purification, and in vivo evaluation of a novel multiepitope peptide vaccine consisted of immunodominant epitopes of SYCP1 and ACRBP antigens as a prophylactic melanoma vaccine. Int. Immunopharmacol. 2019;76 doi: 10.1016/j.intimp.2019.105872. [DOI] [PubMed] [Google Scholar]
- 66.Mahdevar E., et al. Immunoprotective effect of an in silico designed multiepitope cancer vaccine with BORIS cancer-testis antigen target in a murine mammary carcinoma model. Sci. Rep. 2021;11(1):23121. doi: 10.1038/s41598-021-01770-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ikai A. Thermostability and aliphatic index of globular proteins. J. Biochem. 1980;88(6):1895–1898. [PubMed] [Google Scholar]
- 68.Mahdevar E., et al. Exploring the cancer-testis antigen BORIS to design a novel multi-epitope vaccine against breast cancer based on immunoinformatics approaches. J. Biomol. Struct. Dyn. 2022;40(14):6363–6380. doi: 10.1080/07391102.2021.1883111. [DOI] [PubMed] [Google Scholar]
- 69.Safavi A., et al. Exploring the out of sight antigens of SARS-CoV-2 to design a candidate multi-epitope vaccine by utilizing immunoinformatics approaches. Vaccine. 2020;38(48):7612–7628. doi: 10.1016/j.vaccine.2020.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Safavi A., et al. In silico analysis of transmembrane protein 31 (TMEM31) antigen to design novel multiepitope peptide and DNA cancer vaccines against melanoma. Mol. Immunol. 2019;112:93–102. doi: 10.1016/j.molimm.2019.04.030. [DOI] [PubMed] [Google Scholar]
- 71.Safavi A., et al. In silico analysis of synaptonemal complex Protein 1 (SYCP1) and Acrosin Binding Protein (ACRBP) antigens to design novel multiepitope peptide cancer vaccine against breast cancer. Int. J. Pept. Res. Ther. 2019;25(4):1343–1359. [Google Scholar]
- 72.Hu B., et al. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021;19(3):141–154. doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tregoning J.S., et al. Progress of the COVID-19 vaccine effort: viruses, vaccines and variants versus efficacy, effectiveness and escape. Nat. Rev. Immunol. 2021;21(10):626–636. doi: 10.1038/s41577-021-00592-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gong W., et al. Peptide-based vaccines for tuberculosis. Front Immunol. 2022;13 doi: 10.3389/fimmu.2022.830497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gong W., et al. Peptides-based vaccine MP3RT induced protective immunity against mycobacterium tuberculosis infection in a humanized mouse model. Front. Immunol. 2021;12(1393) doi: 10.3389/fimmu.2021.666290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cortés-Ciriano I., et al. Computational analysis of cancer genome sequencing data. Nat. Rev. Genet. 2022;23(5):298–314. doi: 10.1038/s41576-021-00431-y. [DOI] [PubMed] [Google Scholar]
- 77.Leman J.K., et al. Macromolecular modeling and design in Rosetta: recent methods and frameworks. Nat. Methods. 2020;17(7):665–680. doi: 10.1038/s41592-020-0848-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Magana M., et al. The value of antimicrobial peptides in the age of resistance. Lancet Infect. Dis. 2020;20(9):e216–e230. doi: 10.1016/S1473-3099(20)30327-3. [DOI] [PubMed] [Google Scholar]
- 79.Kuprianov V.V., et al. Combination of three adjuvants enhances the immunogenicity of a recombinant protein containing the CTL epitopes of non-structural proteins of hepatitis C virus. Virus Res. 2020;284 doi: 10.1016/j.virusres.2020.197984. [DOI] [PubMed] [Google Scholar]
- 80.Tran T.A., et al. Peptide vaccine combined adjuvants modulate anti-tumor effects of radiation in glioblastoma mouse model. Front. Immunol. 2020;11:1165. doi: 10.3389/fimmu.2020.01165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Asghari A., et al. Development of a chimeric vaccine candidate based on Toxoplasma gondii major surface antigen 1 and apicoplast proteins using comprehensive immunoinformatics approaches. Eur. J. Pharm. Sci. 2021;162 doi: 10.1016/j.ejps.2021.105837. [DOI] [PubMed] [Google Scholar]
- 82.Li D., et al. Lupeol protects against cardiac hypertrophy via TLR4-PI3K-Akt-NF-κB pathways. Acta Pharmacol. Sin. 2022;43(8):1989–2002. doi: 10.1038/s41401-021-00820-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Khanmohammadi S., Rezaei N. Role of Toll-like receptors in the pathogenesis of COVID-19. J. Med. Virol. 2021;93(5):2735–2739. doi: 10.1002/jmv.26826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pino M., et al. A yeast expressed RBD-based SARS-CoV-2 vaccine formulated with 3M–052-alum adjuvant promotes protective efficacy in non-human primates. Sci. Immunol. 2021;6(61) doi: 10.1126/sciimmunol.abh3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.M. Silva et al., A particulate saponin/TLR agonist vaccine adjuvant alters lymph flow and modulates adaptive immunity, Sci. Immunol. 6(66) (2021) eabf1152. [DOI] [PMC free article] [PubMed]
- 86.Simpson D.S., et al. Interferon-γ primes macrophages for pathogen ligand-induced killing via a caspase-8 and mitochondrial cell death pathway. Immunity. 2022;55(3):423–441.e9. doi: 10.1016/j.immuni.2022.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Qamar T.U., M.,, et al. Multiepitope-based subunit vaccine design and evaluation against respiratory syncytial virus using reverse vaccinology approach. Vaccines (Basel) 2020;8(2) doi: 10.3390/vaccines8020288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shantier S.W., et al. Novel multi epitope-based vaccine against monkeypox virus: vaccinomic approach. Sci. Rep. 2022;12(1):15983. doi: 10.1038/s41598-022-20397-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Corradin G., Villard V., Kajava A.V. Protein structure based strategies for antigen discovery and vaccine development against malaria and other pathogens. Endocr. Metab. Immune Disord. Drug Targets. 2007;7(4):259–265. doi: 10.2174/187153007782794371. [DOI] [PubMed] [Google Scholar]
- 90.Araf Y., et al. Omicron variant of SARS-CoV-2: Genomics, transmissibility, and responses to current COVID-19 vaccines. J. Med. Virol. 2022;94(5):1825–1832. doi: 10.1002/jmv.27588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zotta A., Hooftman A., O'Neill L.A.J. SARS-CoV-2 targets MAVS for immune evasion. Nat. Cell Biol. 2021;23(7):682–683. doi: 10.1038/s41556-021-00712-y. [DOI] [PubMed] [Google Scholar]
- 92.D.M.G. Halpin et al., Global initiative for the diagnosis, management, and prevention of chronic obstructive lung disease. The 2020 GOLD Science Committee Report on COVID-19 and chronic obstructive pulmonary disease, Am. J. Respir. Crit. Care Med. 203(1) (2021) 24–36. [DOI] [PMC free article] [PubMed]
- 93.Hodgson S.H., et al. What defines an efficacious COVID-19 vaccine? A review of the challenges assessing the clinical efficacy of vaccines against SARS-CoV-2. Lancet Infect. Dis. 2021;21(2):e26–e35. doi: 10.1016/S1473-3099(20)30773-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.V'Kovski P., et al. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021;19(3):155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Altindis M., Puca E., Shapo L. Diagnosis of monkeypox virus – an overview. Travel Med. Infect. Dis. 2022;50 doi: 10.1016/j.tmaid.2022.102459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Billioux B.J., et al. Neurologic complications of smallpox and monkeypox: a review. JAMA Neurol. 2022;79(11):1180–1186. doi: 10.1001/jamaneurol.2022.3491. [DOI] [PubMed] [Google Scholar]
- 97.Huang Y., Mu L., Wang W. Monkeypox: epidemiology, pathogenesis, treatment and prevention. Signal Transduct. Target. Ther. 2022;7(1):373. doi: 10.1038/s41392-022-01215-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mileto D., et al. New challenges in human monkeypox outside Africa: A review and case report from Italy. Travel Med. Infect. Dis. 2022;49 doi: 10.1016/j.tmaid.2022.102386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zheng C., Tang Y.D. The emerging roles of the CDK/cyclin complexes in antiviral innate immunity. J. Med. Virol. 2022;94(6):2384–2387. doi: 10.1002/jmv.27554. [DOI] [PubMed] [Google Scholar]
- 100.Fitzgerald K.A., Kagan J.C. Toll-like receptors and the control of immunity. Cell. 2020;180(6):1044–1066. doi: 10.1016/j.cell.2020.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Re F., Strominger J.L. Toll-like receptor 2 (TLR2) and TLR4 differentially activate human dendritic cells. J. Biol. Chem. 2001;276(40):37692–37699. doi: 10.1074/jbc.M105927200. [DOI] [PubMed] [Google Scholar]
- 102.Vanden Eijnden S., et al. Preferential production of the IL-12(p40)/IL-23(p19) heterodimer by dendritic cells from human newborns. Eur. J. Immunol. 2006;36(1):21–26. doi: 10.1002/eji.200535467. [DOI] [PubMed] [Google Scholar]
- 103.Fußbroich D., et al. Impact of soyasaponin I on TLR2 and TLR4 induced inflammation in the MUTZ-3-cell model. Food Funct. 2015;6(3):1001–1010. doi: 10.1039/c4fo01065e. [DOI] [PubMed] [Google Scholar]
- 104.Stark R., et al. Monophosphoryl lipid A inhibits the cytokine response of endothelial cells challenged with LPS. Innate Immun. 2015;21(6):565–574. doi: 10.1177/1753425914564172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.van Tongeren J., et al. Expression profiling and functional analysis of Toll-like receptors in primary healthy human nasal epithelial cells shows no correlation and a refractory LPS response. Clin. Transl. Allergy. 2015;5:42. doi: 10.1186/s13601-015-0086-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhao Y., et al. A novel PPARα agonist propane-2-sulfonic acid octadec-9-enyl-amide inhibits inflammation in THP-1 cells. Eur. J. Pharmacol. 2016;788:104–112. doi: 10.1016/j.ejphar.2016.06.026. [DOI] [PubMed] [Google Scholar]
- 107.Moss P. The T cell immune response against SARS-CoV-2. Nat. Immunol. 2022;23(2):186–193. doi: 10.1038/s41590-021-01122-w. [DOI] [PubMed] [Google Scholar]
- 108.T.N. Schumacher, D.S. Thommen, Tertiary lymphoid structures in cancer, Science 375(6576) (2022) eabf9419. [DOI] [PubMed]
- 109.Mantel I., Sadiq B.A., Blander J.M. Spotlight on TAP and its vital role in antigen presentation and cross-presentation. Mol. Immunol. 2022;142:105–119. doi: 10.1016/j.molimm.2021.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhou Y., et al. CD4(+) T cell activation and inflammation in NASH-related fibrosis. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.967410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Casanova J.L., Abel L. From rare disorders of immunity to common determinants of infection: following the mechanistic thread. Cell. 2022;185(17):3086–3103. doi: 10.1016/j.cell.2022.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Manik M., Singh R.K. Role of toll-like receptors in modulation of cytokine storm signaling in SARS-CoV-2-induced COVID-19. J. Med. Virol. 2022;94(3):869–877. doi: 10.1002/jmv.27405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sen S., et al. Metal-based anticancer agents as immunogenic cell death inducers: the past, present, and future. Chem. Soc. Rev. 2022;51(4):1212–1233. doi: 10.1039/d1cs00417d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hernandez R., et al. Engineering IL-2 for immunotherapy of autoimmunity and cancer. Nat. Rev. Immunol. 2022;22(10):614–628. doi: 10.1038/s41577-022-00680-w. [DOI] [PubMed] [Google Scholar]
- 115.Kolios A.G.A., Tsokos G.C., Klatzmann D. Interleukin-2 and regulatory T cells in rheumatic diseases. Nat. Rev. Rheumatol. 2021;17(12):749–766. doi: 10.1038/s41584-021-00707-x. [DOI] [PubMed] [Google Scholar]
- 116.Propper D.J., Balkwill F.R. Harnessing cytokines and chemokines for cancer therapy. Nat. Rev. Clin. Oncol. 2022;19(4):237–253. doi: 10.1038/s41571-021-00588-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are from public databases.