Abstract
Background
A novel human parechovirus 3 Australian recombinant (HPeV3-AR) strain emerged in 2013 and coincided with biennial outbreaks of sepsis-like illnesses in infants. We evaluated the molecular evolution of the HPeV3-AR strain and its association with severe HPeV infections.
Methods
HPeV3-positive samples collected from hospitalized infants aged 5–252 days in 2 Australian states (2013–2020) and from a community-based birth cohort (2010–2014) were sequenced. Coding regions were used to conduct phylogenetic and evolutionary analyses. A recombinant-specific polymerase chain reaction was designed and utilized to screen all clinical and community HPeV3-positive samples.
Results
Complete coding regions of 54 cases were obtained, which showed the HPeV3-AR strain progressively evolving, particularly in the 3′ end of the nonstructural genes. The HPeV3-AR strain was not detected in the community birth cohort until the initial outbreak in late 2013. High-throughput screening showed that most (>75%) hospitalized HPeV3 cases involved the AR strain in the first 3 clinical outbreaks, with declining prevalence in the 2019–2020 season. The AR strain was not statistically associated with increased clinical severity among hospitalized infants.
Conclusions
HPeV3-AR was the dominant strain during the study period. Increased hospital admissions may have been from a temporary fitness advantage and/or increased virulence.
Keywords: HPeV3, parechovirus, pediatric, viral evolution, Australia, sepsis, infants, picornavirus, community, recombination
The continent-wide emergence, dominance, and decline of a human parechovirus 3 recombinant strain (HPeV3-AR) in Australia mirrored a rise and fall in HPeV infant hospitalizations; however, HPeV3-AR did not correlate with increased disease severity in hospital or community cohorts.
Human parechovirus (HPeV) is a nonenveloped, single-stranded, positive-sense RNA virus within the Picornaviridae family. The parechovirus genome is linear with a continuous coding sequence (CDS) containing both structural and nonstructural genes that is bookended by terminal noncoding sequences. HPeV is currently classified into 18 genotypes based upon the sequence diversity of the VP1 capsid protein [1]. Subclinical HPeV infections are common. Sewage surveillance samples from Scotland and weekly stool samples from healthy, Australian children report continuous HPeV circulation [2, 3]. Hospital-based studies from the United States, Europe, and Asia of young infants and children presenting with febrile illnesses have also found HPeV year-round in sterile (blood, cerebrospinal fluid [CSF]) and nonsterile (respiratory secretions, stools) site specimens [4–6].
HPeV type 3 (HPeV3) was first described in 1999 in a 1-year old Japanese infant with fever and transient flaccid paralysis [7]. Subsequently it has been observed in seasonal outbreaks of meningoencephalitis and sepsis-like illnesses in neonates and infants in multiple countries, including Australia [8–13]. Following its initial recognition in Australia in 2013, there have been 4 biennial HPeV3 outbreaks, each occurring over the southern hemisphere spring/summer months (September–February). The most recent was from December 2019 to March 2020. These outbreaks were experienced across Australia, but most cases were identified in the major eastern seaboard cities of Brisbane, Sydney, and Melbourne.
In 2017, sequencing of 9 HPeV3 genomes from cases with severe infections revealed a novel recombinant strain [14]. This recombinant (henceforth the Australian recombinant [AR] strain or HPeV3-AR) was then detected in stored HPeV3-positive samples from 2015 [15].
It has been hypothesized that HPeV3-AR may have caused the severe HPeV infections observed in Australian infants during the past decade [14]. Of 145 infants hospitalized with HPeV infections across the state of Queensland in 2013–2016 and for whom clinical information was available, 23% were admitted to a pediatric intensive care unit (PICU) and 14% developed neurodevelopmental sequelae [9]. However, the HPeV3 recombination status of the viruses detected in this cohort was not determined. To date, direct comparisons of AR status and clinical presentation in large numbers of hospital, as well as community, cases have not been conducted. We therefore conducted this study to examine the relationship between the emergence of clinically severe HPeV disease in children and the ongoing viral evolution. We did this by documenting the molecular evolution of the HPeV3-AR strain in Australia from the time of its emergence in 2013 until 2020 in 2 hospital cohorts and 1 community cohort. We also describe the associated pediatric clinical phenotype in infected children from the hospital cohort.
METHODS
Study Population, Clinical Data, and Sample Collection
HPeV-positive samples came from the following sources: (1) an Australian hospital-based cohort of infants with an HPeV infection identified by medical record review and admitted over the spring to fall (September–April) period of 2013–2020; and (2) the Observational Research in Childhood Infectious Diseases (ORChID) community-based birth cohort study [3, 16, 17].
Hospital cohort samples came from a centralized pathology laboratory servicing government-funded hospitals and clinics within Queensland (population 5.2 million), along with a similar laboratory in Western Australia (population 2.7 million).
The community cohort samples originated from the ORChID study, which was conducted between September 2010 and October 2014 in Brisbane, Queensland [16]. Participants were healthy term infants, enrolled at birth, following which parents collected stool swabs weekly and recorded a daily symptom diary until their child’s second birthday. HPeV detection and genotyping were reported previously [3]. The Supplementary Material further describes both cohorts, and an overview of study sample workflow is summarized in Supplementary Figure 1.
Clinical data for the hospital cohort (Queensland only) were extracted from medical records. This included sociodemographic characteristics as well as markers of disease severity [9] (PICU admission, duration of hospitalization, apnea, seizures, brain magnetic resonance imaging [MRI] results, and developmental assessment after 6 months of age). A subgroup underwent prospective follow-up 1-year postinfection for formal developmental assessment (Ages and Stages Questionnaire) as part of another study [18, 19], and these data were included where available.
HPeV Screening
All samples were rescreened by a generic HPeV real-time polymerase chain reaction (PCR) assay (Bensch-ParechoF/K) to obtain cycle threshold (Ct) values, followed by identifying HPeV3 using sequencing-based genotyping or a specific HPeV3 real-time PCR (HPeV3-F/R). An HPeV3-AR–specific real-time PCR (HPeV3RecomF/R) was designed and evaluated (Supplementary Materials, Supplementary Tables 6-8) using the study’s generated genomic data (see below), and then employed as a high-throughput screening tool for all HPeV3-positive samples. The HPeV3-AR detection algorithm (Supplementary Figure 2) required the sample to be positive in both the HPeV3 and HPeV3-AR assays, with the assays targeting regions upstream and downstream of the AR recombination site, respectively.
HPeV3 Genome Sequencing and Analyses
Only HPeV3 samples, as determined by specific PCR or sequencing (see above), were selected for sequencing if their Ct value was ≤29 in the generic HPeV screening assay. A genome-walking amplification and Sanger sequencing approach was used to sequence the viral coding region and flanking sequences. Genome assembly, phylogenetic analyses, and recombination were conducted in Geneious (Build 2020-12-23; Biomatters Ltd, New Zealand). Nextstrain [20] was used to analyze AR genome evolution over time. The Tajima test of neutrality [21] was used to assess selective pressure.
Statistical Analyses
Statistical analyses were performed using R statistical software [22]. P values < .05 were considered significant. For the purposes of clinical correlation analyses, a positive case was defined as a subject who had HPeV3 detected in at least 1 sample type during a 1-week period.
Further detailed methodology is described in the Supplementary Materials.
Ethical Approvals
The Children’s Health Queensland Human Research Ethics Committee approved this study (HREC/16/QRCH/223). The Children’s Health Queensland (HREC/10/QRCH/16), the Royal Brisbane and Women’s Hospital (HREC/10/QRBW125), and The University of Queensland (2010000820) Human Research Ethics Committees approved the ORChID study.
RESULTS
HPeV3 Prevalence Between 2010 and 2020
Queensland Hospital Cohort
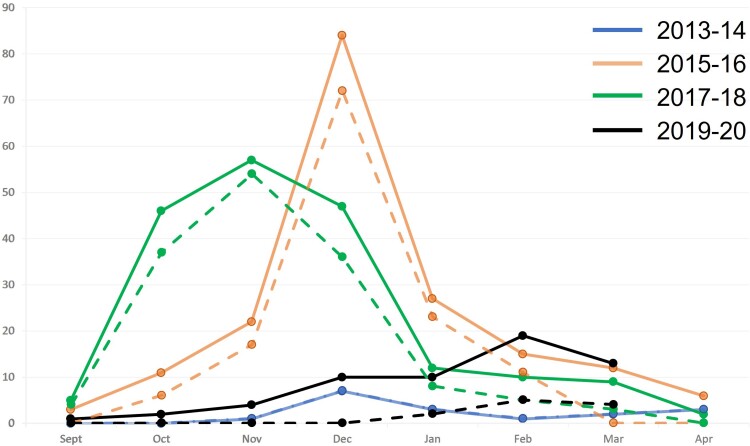
Overall, 559 HPeV-positive samples from blood, CSF, stools, and nasopharyngeal swabs were available from 71.5% (n = 322) of 450 HPeV-diagnosed cases. Multiple samples from individual hospitalized cases represented parallel collections of different sample types. These samples represented 68.0% (17/25), 79.2% (141/178), 84.6% (159/188), and 18.6% (11/59), of the HPeV-infected patients diagnosed in each of the 4 biennial epidemic periods of 2013–2014, 2015–2016, 2017–2018, and 2019–2020, respectively. HPeV3 was detected in 100% (n = 17), 94.3% (n = 133), 92.4% (n = 147), and 100% (n = 11) of HPeV-positive cases in each of these epidemic seasons, respectively (Supplementary Table 1). Most cases occurred during the spring/summer period (September–February). Notably, the epidemic occurred later (January–March) in 2019–2020, with an accompanying decrease in the number of positive cases (Figure 1).
Figure 1.
Seasonality of hospitalized human parechovirus (HPeV) cases across the Queensland, Australia, spring/fall period. The solid lines represent total diagnosed HPeV cases within the state within that outbreak period, and the smaller, dashed lines underneath represent the respective cases captured for the study, which were genotyped as HPeV3. Note all 2013–2014 samples that were diagnosed were also captured for the study. Y-axis shows number of cases.
Western Australian Hospital Cohort
Twenty-two CSF samples collected between 2014 and 2017 were available and met the minimum Ct value criteria. All were HPeV3 positive.
Queensland Community Cohort
As described previously [3], 18.3% (52/289) of HPeV episodes within the community cohort were HPeV3 positive and peaked during the 2011–2012 and 2013–2014 summer seasons of this 4-year study.
Genome Sequencing and Characterization
Of the 383 available HPeV3 samples from all cohorts, 122 (31.8%) met sequencing selection criteria (Supplementary Figure 1). Of these samples, 53 did not have sufficient working volumes, leaving 69 HPeV3 successfully sequenced genomes, with most (n = 54) having at least their whole CDS captured as well as portions of their flanking noncoding regions (Supplementary Table 2). The CDS from the remaining 15 viral genomes could not be sequenced completely due to amplification failure, most likely from low virus concentration and/or RNA degradation. Only 10 of 15 partial genomes yielded sufficient coverage to be included in the study.
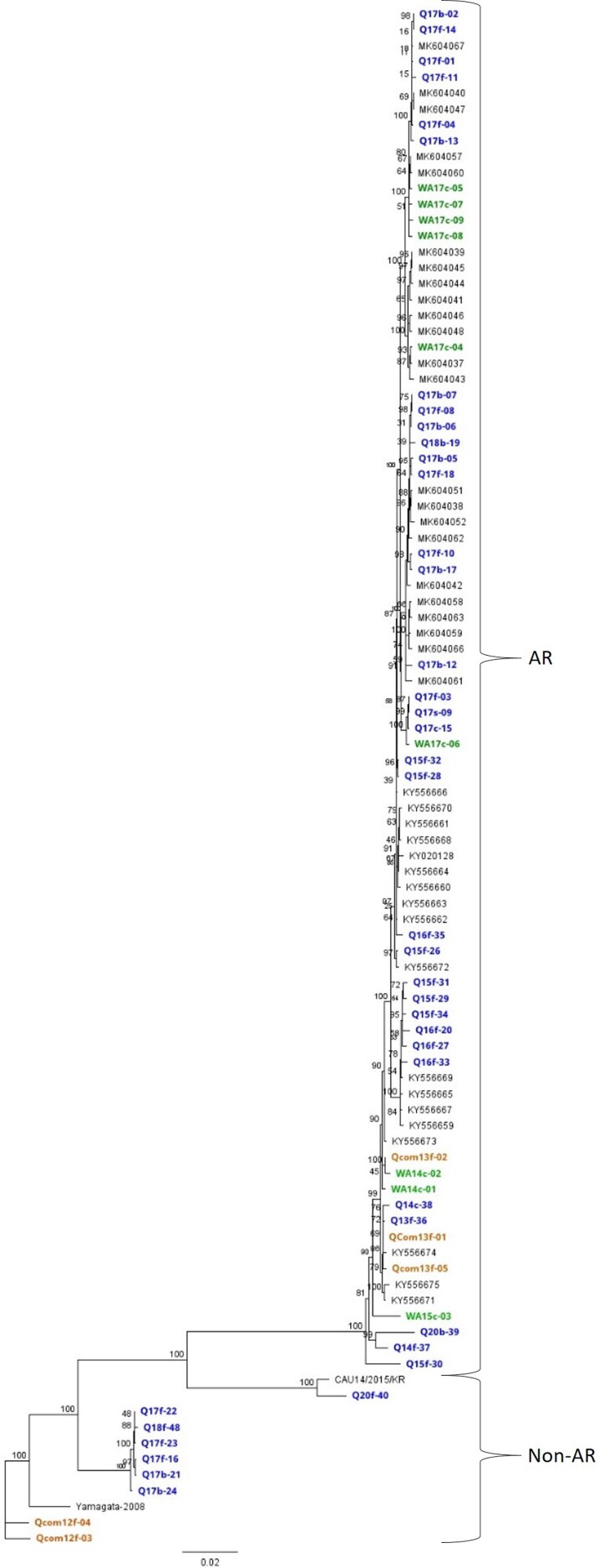
A maximum likelihood phylogenetic analysis of the CDS upstream of the AR breakpoint (nt 3114) generated by this study and previously published Australian sequences [14, 15] showed the expected divergence between the AR and non-AR strains (Figure 2), which was also reflected at the amino acid level (Supplementary Figure 3). Of note, the community-sourced AR viral genomes clustered with hospital viral genomes (Figure 2).
Figure 2.
Maximum likelihood phylogenetic analyses using 1000 bootstraps of all available Australian human parechovirus 3 (HPeV3) genomes’ coding sequence regions upstream of the recombination breakpoint (nt 1–3114), including 2 non-AR, non-Australian (Yamagata-2008 and CAU14/82015/KR) HPeV3 reference strains. Node colors indicate strain genomes generated in this study and are color coded by Australian state where they were collected: blue, Queensland hospital; green, Western Australian hospital; orange, Queensland community; gray, previously published Australian AR genomes collected during 2014–2016 from New South Wales, Victoria, and Tasmania.
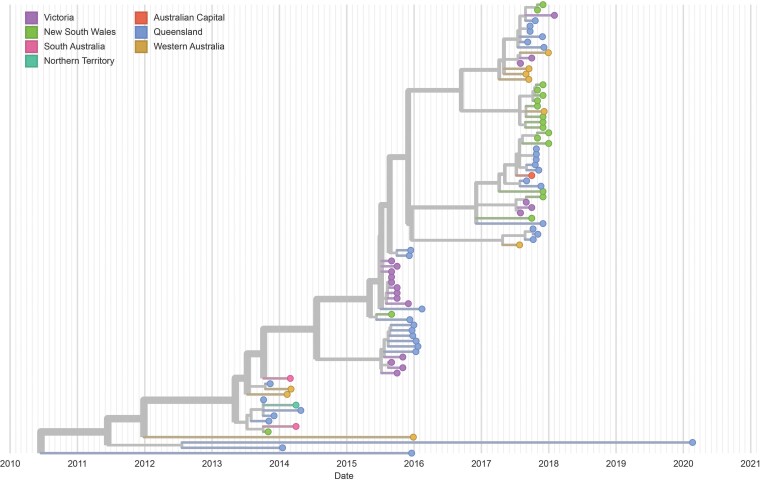
With few exceptions, analysis of the AR genomes showed a progressive evolution of the AR strain, which seemed independent of geographic origin (Figure 3). However, several viral genomes from 2016 and 2020 did not conform to this pattern and instead showed separate lineages that likely shared a recent common ancestor with most AR strains pre-2013. The addition of incomplete genomes in an analysis of the structural gene section of the AR and non-AR genomes showed multiple non-AR HPeV3 lineages clustering by year/outbreak period of collection (Supplementary Figure 4).
Figure 3.
Maximum likelihood phylogenetic analyses of available human parechovirus 3 Australian recombinant (AR) strains coding sequences as a function of time and categorized by geographic area of origin. The Nextstrain plot shows the progressive evolution of the AR strain for most of the disease outbreak periods.
The AR strain CDS was under significant selective pressure (Tajima D = −2.07, P < .05), with a Shannon entropy plot showing increased amino acid alterations accumulating in the recombinant section of the AR strain CDS (Supplementary Figure 5). In addition, 1 novel recombinant between HPeV4 and HPeV3-AR was detected (Supplementary Materials, Supplementary Figure 6).
Recombination Status Between 2013 and 2020
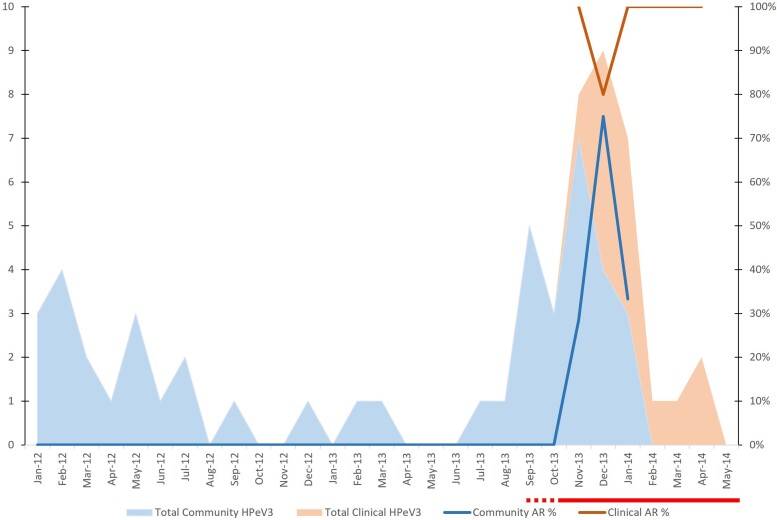
All Queensland community (n = 52) and clinical samples (n = 559) were tested using the HPeV3 AR strain-specific screening PCR and detection algorithm (Supplementary Materials, Supplementary Figure 2). In hospital cohort samples, the AR strain was highly prevalent in each period, in contrast with the community cohort samples where the AR strain was only detected in the final years (2013–2014) of that study (Table 1). The Queensland community and hospital cohort sample collections overlapped during the initial emergence of the AR strain–associated disease outbreak (Figure 4). From 2010 no AR strains were detected in the community cohort until the summer of 2013–2014, coincident with the first observed hospital cohort outbreak and AR strain–positive clinical samples (Figure 4).
Table 1.
Human Parechovirus 3 (HPeV3) Australian Recombinant Strain Screening Results of All Available Hospital and Community HPeV3-Positive Subjects
| Subject Type | Time Period | AR Strain | Non-AR | Equivocal |
|---|---|---|---|---|
| QLD hospital (n = 17) | 2013–2014 | 13 (76.5) | 1 (5.9) | 3 (17.6) |
| ȃH/C overlap (n = 9) | 13 Nov–14 Jan | 8 (88.9) | 1 (11.1) | 0.0 |
| QLD hospital (n = 133) | 2015–2016 | 102 (76.7) | 17 (12.9) | 14 (10.5) |
| QLD hospital (n = 147) | 2017–2018 | 129 (87.7) | 12 (8.1) | 6 (4.1) |
| QLD hospital (n = 11) | 2020 | 2 (18.2) | 8 (72.7) | 1 (9.1) |
| WA hospital (n = 4) | 2013–2014 | 4 (100.0) | 0.0 | 0.0 |
| WA hospital (n = 4) | 2015–2016 | 1 (25.0) | 3 (75.0) | 0.0 |
| WA hospital (n = 12)a | 2017–2018 | 12 (100.0) | 0.0 | 0.0 |
| QLD community (n = 21) | 2011–2012 | 0.0 | 21 (100.0) | 0.0 |
| QLD community (n = 6) | 2012–2013 | 0.0 | 6 (100.0) | 0.0 |
| QLD community (n = 25) | 2013–2014 | 7 (28.0) | 18 (72.0) | 0.0 |
| ȃH/C overlap (n = 14) | 13 Nov–14 Jan | 6 (42.8) | 8 (57.2) | 0.0 |
Data are presented as No. (%) unless otherwise indicated.
Hospital-based sample time periods represent the 3 outbreaks of human parechovirus 3 (HPeV) disease, whereas the longitudinal community sampling timeframes represent a June–July period. A 3-month overlap from November 2013 to January 2014 between the QLD hospital and QLD community cohorts is also shown as “H/C overlap.”
Abbreviations: AR, Australian recombinant; H/C, hospital/community cohorts; QLD, Queensland; WA, Western Australia.
One 2017 sample was not included as it fell outside of the June to July period; however, it was also an HPeV3-AR strain.
Figure 4.
Total human parechovirus 3 (HPeV3) and Australian recombinant (AR) strain prevalence in the Queensland community (blue) and hospital-based (peach) cohorts each month over an interval period spanning the first observed HPeV disease outbreak. Only an overlap of several months from November to January was available for both datasets. Red bar indicates duration of the first HPeV3 disease outbreak; the dashed line portion indicates uncertainty in when the outbreak began. Y-axis shows number of cases.
High-frequency, longitudinal collections in the community cohort allowed us to examine the relative viral load and shedding duration in both AR and non-AR HPeV-positive cases in the first 2 years of life. Ct values were used as a proxy of relative viral RNA quantification; however, no significant difference was observed between AR (mean Ct, 29.2 [standard deviation {SD}, 4.3]) and non-AR (mean Ct, 29.7 [SD, 3.6]) cases (P = .57).
Clinical Associations of AR and Non-AR Strains Within Queensland Hospital and Birth Cohorts
Hospital Cohort
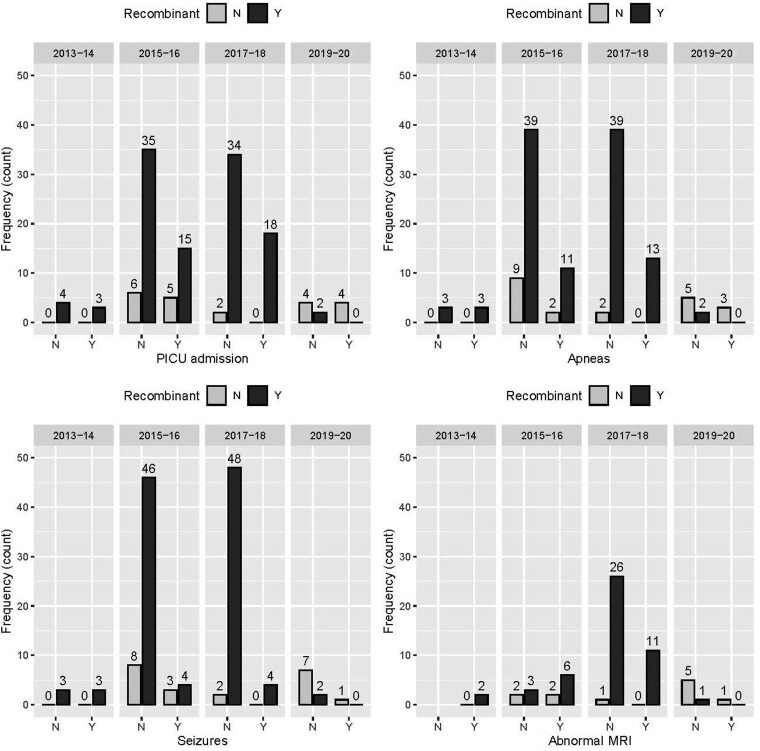
Minimum sociodemographic and clinical data were available in 147 of 320 (45.5%) HPeV3-positive Queensland hospital cohort cases for 2013–2020 (Supplementary Tables 1, 3), of which 136 were also characterized for AR status (Table 2). The median age of hospitalized children was 28 days (interquartile range, 15.5–55 days). Most children were born at >36 weeks’ gestation (n = 119 [80.95%]). No statistically significant relationship between AR status and the selected clinical markers of disease severity was seen in the hospitalized children (Table 2). This remained true for each separate outbreak of HPeV3 (Figure 5). Over time, there were also no significant differences in age, length of stay, or length of PICU stay between the patients in each of the 4 outbreaks (Supplementary Table 4).
Table 2.
Relationship Analyses Between Clinical Variables, Severity Markers, and Australian Recombinant Status in Human Parechovirus 3–Positive Children
| Variable | Non-AR | AR | P Value |
|---|---|---|---|
| Seizures | .26 | ||
| ȃNo | 17 (80.95) | 99 (86.09) | … |
| ȃYes | 4 (19.05) | 11 (9.57) | … |
| ȃNA | 0 | 5 (4.35) | … |
| PICU admission | .45 | ||
| ȃNo | 12 (57.14) | 75 (65.22) | … |
| ȃYes | 9 (42.86) | 36 (31.3) | … |
| ȃNA | 0 | 4 (3.48) | … |
| Apneas | 1 | ||
| ȃNo | 16 (76.19) | 83 (72.17) | … |
| ȃYes | 5 (23.81) | 27 (23.48) | … |
| ȃNA | 0 | 5 (4.35) | … |
| HPeV positive CSF | 1 | ||
| ȃNo | 5 (23.81) | 22 (19.13) | … |
| ȃYes | 15 (71.43) | 73 (63.48) | … |
| ȃNA | 1 (4.76) | 20 (17.39) | … |
| Abnormal MRI | .73 | ||
| ȃNo | 8 (38.1) | 30 (26.09) | … |
| ȃYes | 3 (14.29) | 19 (16.52) | … |
| ȃNA | 10 (47.62) | 66 (57.39) | … |
| Normal development at 6 mo | 1 | ||
| ȃNo | 1 (4.76) | 16 (13.91) | … |
| ȃYes | 7 (33.33) | 58 (50.43) | … |
| ȃNA | 13 (61.9) | 41 (35.65) | … |
| Normal ASQ | .49 | ||
| ȃNo | 2 (9.52) | 18 (15.65) | … |
| ȃYes | 0 | 18 (15.65) | … |
| ȃNA | 19 (90.48) | 79 (68.7) | … |
Data are presented as No. (%) unless otherwise indicated.
Abbreviations: ASQ, ages and stages questionnaire; AR, Australian recombinant; CSF, cerebrospinal fluid; HPeV, human parechovirus; MRI, magnetic resonance imaging; NA, data not available, or test not performed; PICU, pediatric intensive care unit.
Figure 5.
Clinical features relating to Australian recombinant status over the 4 sampled epidemic periods. Abbreviations: MRI, magnetic resonance imaging; N, no; PICU, pediatric intensive care unit; Y, yes.
Community Cohort
The AR strain was only detectable from October 2013 (n = 7). Prior to this, all strains genotyped had been non-AR strain (n = 29). Subjects had a mean AR strain infection age of 1.45 years (SD, 0.27; range, 1.21–1.95 years) compared to 1.20 years (SD, 0.45; range, 5 days–2.08 years) in the non-AR strain group. Of note, however, the longitudinal community cohort was older at the time of AR strain emergence, which would have meant no infants would have been sampled during that key time period. Within the period of AR detection (October 2013–January 2014), the AR strain group (n = 7) was significantly (P = .03, 2-tailed t test) younger (mean, 1.45 years [SD, 0.27 years]; range, 1.21–1.95 years) than the non-AR strain group (n = 10) (mean, 1.74 years [SD, 0.18 years]; range, 1.02–2.10). No HPeV3-positive children required hospitalization, with subclinical infections accounting for 21 of 43 (48.8%) HPeV3 episodes (Supplementary Table 5). The remaining 22 infections were accompanied by respiratory symptoms, although respiratory viruses were co-detected on 17 of 22 (77.2%) such occasions. There was no significant difference in the proportion of symptomatic children with AR or non-AR HPeV (4/7 [42.8%] and 21/39 [46.1%], respectively; P = 1.0).
Comparison of Community and Hospital Cohorts
Parallel data from community and hospitalized cases of HPeV were only available from October 2013 to January 2014. During this period, 14 and 9 HPeV3 infections were detected in the community and hospitalized cohorts, respectively. There was a significantly greater proportion of children with the AR strain in the hospitalized group than the community group (8/9 [88.9%] vs 6/14 [42.8%]; P = .0052, Fisher exact test). However, these groups were not directly comparable due to the difference in ages between the groups. Children in the community cohort were significantly older, with a mean age of 1.50 years (range, 1.22–1.88 years) compared to the hospitalized children (mean age, 30.6 days; range, 7–87 days).
DISCUSSION
This study examined the genetic diversity of HPeV3 across Australia during a 7-year period where a severe disease phenotype emerged in concert with a new recombinant (AR) strain. By thoroughly sampling the clinical HPeV3 cases in the state of Queensland across all of the biennial epidemic periods, we were able to estimate the true prevalence of the AR strain within the hospitalized infant population, as well as investigate the clinical presentations of both the AR and nonrecombinant circulating strains. Within this hospital-based population, we found that the AR strain was the dominant form of HPeV3 across the 4 epidemic periods; however, it was not associated with significant differences in disease severity or nature of clinical presentation when compared with non-HPeV3 AR cases within each cohort.
Genomic Variation Over Time
The HPeV genome is capable of recombination; however, HPeV3 is unusual as its genome is more stable than that observed with other HPeV genotypes [23–25]. Thus, the emergence of a new recombinant HPeV3 strain offers an opportunity to examine at the population genetics level the emergence of a novel variant. While HPeV3 episodes were frequently detected in the community cohort between May 2011 and January 2014 [3], detection of the AR strain (October 2013–January 2014) was temporally associated with increased hospital admissions in infants with a severe phenotype of HPeV infection in September–December 2013 [9]. Seventy-four percent of these infections were HPeV3-AR. Two subsequent Australian epidemics also had a predominance (>75%) of HPeV3-AR. Of note, the most recent outbreak period of 2019–2020 showed a marked reduction in overall HPeV cases (n = 59), as well as the proportion of infections due to the AR strain (from >75% to 18%).
These data support the predicted major AR strain recombination event occurring between March 2012 and November 2013 [14]. The AR genomes circulating in the community were nearly identical to contemporaneous clinical AR genomes, despite some of those viral genomes being collected from the eastern and western seaboards of Australia, separated in distance by as much as 4030 km (2500 miles). These findings add further evidence to support a previous report [26] of a rapid expansion of the AR strain across the Australian continent after the major recombination event.
A gradual evolution of the AR genome was observed in viruses collected from multiple Australian regions. These changes were primarily driven by mutations in the recombined nonstructural portion of the genome, a finding which supports the previous analyses of more limited AR datasets [14, 26]. The progressive evolution of the AR strain over the epidemic periods indicates that the strain maintained a presence within the community between epidemic peaks.
Surprisingly, a small number of archaic AR genomes also appeared to be co-circulating during this time. This resulted in the AR strains circulating during the 2019–2020 season being more closely related to the archaic AR strains rather than those of the previous season. This is unusual, as in general each new season contained AR strain viruses progressively evolved from the previous season’s viruses. Non-AR genomes tended to cluster by collection period; however, the 2020 genome was notable for having a greater phylogenetic (nucleotide) distance compared to previous years' clusters. Taken together, the small peak of the clinical 2019–2020 epidemic season, the dramatically decreased representation of the AR strain in clinical cases and the emergence of a large genetic shift in the non-AR virus suggests that a transition event was occurring and a new non-AR variant was supplanting AR as the dominant strain. An alternative hypothesis could be that maternal antibody levels increased over time to impact the number of hospitalized HPeV3-AR cases, as maternal HPeV strain–specific antibody levels have been previously correlated with hospitalization rates [27, 28]. Increasing maternal antibody levels against HPeV3-AR are unlikely, however, to explain the observed decline in hospitalized HPeV3-AR cases given that the immune-interacting HPeV3 structural capsid proteins are conserved and HPeV3 antibodies have been shown to cross-neutralize HPeV3-AR [28]. Thus, the strain supplantation event offers the most likely explanation for the observed reduction of hospitalized cases in 2019–2020.
Relationship Between AR Strain and Clinical Severity
Overall, HPeV3 disease severity in hospitalized infants during the study period was substantial, with nearly a third of cases requiring PICU admission, and 15% showing abnormal MRI results in the clinical cohorts up to 2020. Indeed, it has been suggested that the AR strain might cause more severe clinical disease than other HPeV3 variants [14]. Interestingly, among hospitalized infants, we found no statistical difference in clinical markers of disease severity with either AR or non-AR infections, suggesting a common pathogenesis of severe disease between the 2 strains. Despite these findings, the AR strain may be capable of increased likelihood of clinical disease requiring hospitalization in young infants; however, there were insufficient data in this study to fully address this possibility. While data from 2013 to 2014 suggested a significantly greater proportion of HPeV3-AR detections among hospitalized cases than community cases, direct comparison of these groups is confounded by the significant differences in cohort ages. As the AR strain emerged toward the end of the longitudinal community study, only older children approaching their second birthday were being sampled. The lack of significant correlation between symptoms and AR status in community-based children may also have been mitigated by their older age. Overall, these data highlight the vulnerability of younger infants to hospitalization with HPeV3 infections, although the reasons for this vulnerability are yet to be elucidated.
Apart from correlative links, there is still a poor understanding of the pathogenesis of severe HPeV3 infection. Immune-mediated mechanisms have been hypothesized [29], similar to those seen in enterovirus 71 [30]. Data from the infants in this study are similar to those from previous cohorts that show no correlation between detectable virus in CSF and poor long-term outcome [31, 32], arguing against a direct neuropathic effect of the virus. Similarities in MRI brain findings in HPeV encephalitis and hypoxic-ischemic encephalopathy suggests that potentially vasculitis-mediated changes are responsible for the clinical presentation rather than direct cytopathic effects in the central nervous system from HPeV3 [33].
An alternative, but not mutually exclusive, hypothesis is that the AR recombination increased the infectivity or overall fitness of the virus rather than its virulence, such as by being able to infect younger children more easily. This fitness advantage consequently would have allowed the virus to become more prevalent within the community during 2012–2018, thereby leading to a proportionally larger number of infants presenting with the severe disease phenotype. Such changes in fitness would most likely be due to the nonstructural portion of the virus CDS, given the relative conservation of the structural protein genes in comparison.
The strengths of this study lie in its comprehensive screening and sequencing of HPeV3 viruses collected from hospitalized infants across a 7-year period and incorporation of data from geographically separate parts of Australia, as well as the screening and sequencing of HPeV3 isolates detected in a community cohort, which overlapped in timing with the first clinical epidemic. It also combines both genomic and clinical data to help resolve HPeV3 population dynamics during a novel strain ingression in relation to hospitalizations. However, there are important limitations to consider. The collected clinical data were limited to half the hospitalized cases within the Queensland state cohort, alongside absent clinical data from Western Australia and small numbers of AR cases in the community cohort, all of which could have decreased the precision around the clinical association estimates. The clinical data available from the Queensland Hospital cohort was primarily from the major metropolitan hospitals, which may have resulted in a bias toward more severe cases, given that severe cases are often referred to the major hospitals from outlying regions. Despite genomic sequencing and validation of the AR-specific screening PCR algorithm, it is still possible some isolates could have additional cryptic recombination events that could not be detected by the algorithm. As remarked on previously, the small sample sizes and divergent populations between the hospital and community cohorts in the temporal overlap period also hindered direct comparisons. Finally, the community AR prevalence may be skewed compared to the hospital-based prevalence estimates due to potentially different demographic profiles. The community cohort was from a high socioeconomic background, which may not represent the government-funded hospital patient populations used in this study.
In conclusion, this study leveraged both clinical and community cohorts to investigate the role of the HPeV3-AR strain in disease, while more than doubling the amount of available AR genomes. In doing so, the population dynamics of an emergent HPeV3 strain could be tracked from its emergence to its decline. The emergence of the AR strain was associated with markedly increased hospitalizations, but not specifically with more severe illness among hospitalized infants with HPeV. The nature of this observational study, however, precluded drawing definitive inferences over whether the increased numbers of hospitalizations associated with the AR strain were due to increased fitness and/or virulence compared with that of other HPeV3 strains.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Contributor Information
Seweryn Bialasiewicz, School of Chemistry and Molecular Biosciences, The University of Queensland, Australian Centre for Ecogenomics, St Lucia, Australia; Children’s Health Queensland Hospital and Health Service, Centre for Children’s Health Research, South Brisbane, Australia.
Meryta May, Children’s Health Queensland Hospital and Health Service, Centre for Children’s Health Research, South Brisbane, Australia; Sullivan Nicolaides Pathology, Brisbane, Australia.
Sarah Tozer, Children’s Health Queensland Hospital and Health Service, Centre for Children’s Health Research, South Brisbane, Australia.
Rebecca Day, Children’s Health Queensland Hospital and Health Service, Centre for Children’s Health Research, South Brisbane, Australia.
Anne Bernard, QCIF Bioinformatics, Institute for Molecular Bioscience, The University of Queensland, St Lucia, Australia.
Julian Zaugg, School of Chemistry and Molecular Biosciences, The University of Queensland, Australian Centre for Ecogenomics, St Lucia, Australia.
Kyana Gartrell, Children’s Health Queensland Hospital and Health Service, Centre for Children’s Health Research, South Brisbane, Australia.
Soren Alexandersen, School of Medicine, Deakin University, Geelong, Australia; Statens Serum Institut, Copenhagen, Denmark.
Anthony Chamings, School of Medicine, Deakin University, Geelong, Australia.
Claire Y T Wang, Children’s Health Queensland Hospital and Health Service, Centre for Children’s Health Research, South Brisbane, Australia.
Julia Clark, Children’s Health Queensland Hospital and Health Service, South Brisbane, Australia.
Keith Grimwood, School of Medicine and Dentistry, Menzies Health Institute Queensland, Griffith University, Southport, Australia; Departments of Infectious Diseases and Paediatrics, Gold Coast Health, Southport, Australia.
Claire Heney, Department of Microbiology, Pathology Queensland, Herston, Australia.
Luregn J Schlapbach, Children’s Health Queensland Hospital and Health Service, South Brisbane, Australia; Department of Intensive Care and Neonatology, Children’s Research Center, University Children’s Hospital Zürich, Zürich, Switzerland.
Robert S Ware, School of Medicine and Dentistry, Menzies Health Institute Queensland, Griffith University, Southport, Australia.
David Speers, Department of Microbiology, PathWest Laboratory Medicine Western Australia, Queen Elizabeth II Medical Centre, Nedlands, Australia; School of Medicine, University of Western Australia, Crawley, Australia.
Ross M Andrews, National Centre for Epidemiology and Population Health, Australian National University, Canberra, Australia.
Stephen Lambert, Child Health Research Centre, The University of Queensland, South Brisbane, Australia; National Centre for Immunisation Research and Surveillance, Westmead, Australia.
Notes
Acknowledgments. The authors thank the Pathology Queensland staff for assistance in obtaining clinical samples, and the Queensland Children’s Hospital clinical research nurses for collecting clinical data. We acknowledge the assistance provided by Tania Duarte with our early attempts of nanopore sequencing.
Financial support. This work was supported by the National Health and Medical Research Council (NHMRC) Centre of Research Excellence; the Australian Partnership for Preparedness Research on Infectious Disease Emergencies (AppID 1116530); and the Pathology Queensland Study Education and Research Committee (grant number 5793). The ORChID study was supported by a project grant from the NHMRC (grant number GNT615700) and a program grant from the Children’s Hospital Foundation Queensland (grant number 5006). Funding to pay the Open Access publication charges for this article was covered by the Read & Publish agreement between the Council of Australian University Librarians and the publisher.
References
- 1. Romero JR, Selvarangan R. The human parechoviruses: an overview. Adv Pediatr 2011; 58:65–85. [DOI] [PubMed] [Google Scholar]
- 2. Harvala H, Calvert J, Van Nguyen D, et al. Comparison of diagnostic clinical samples and environmental sampling for enterovirus and parechovirus surveillance in Scotland, 2010–2012. Euro Surveill 2014; 19:pii20772. [DOI] [PubMed] [Google Scholar]
- 3. Wang CYT, Ware RS, Lambert SB, et al. Parechovirus A infections in healthy Australian children during the first 2 years of life: a community-based longitudinal birth cohort study. Clin Infect Dis 2020; 71:116–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yamamoto SP, Kaida A, Naito T, et al. Human parechovirus infections and child myositis cases associated with genotype 3 in Osaka City, Japan, 2014. J Med Microbiol 2015; 64:1415–24. [DOI] [PubMed] [Google Scholar]
- 5. Benschop KS, Schinkel J, Minnaar RP, et al. Human parechovirus infections in Dutch children and the association between serotype and disease severity. Clin Infect Dis 2006; 42:204–10. [DOI] [PubMed] [Google Scholar]
- 6. Souverbielle CT, Wang H, Feister J, et al. Year-round, routine testing of multiple body site specimens for human parechovirus in young febrile infants. J Pediatr 2021; 229:216–22.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ito M, Yamashita T, Tsuzuki H, et al. Isolation and identification of a novel human parechovirus. J Gen Virol 2004; 85:391–98. [DOI] [PubMed] [Google Scholar]
- 8. Piralla A, Furione M, Rovida F, Marchi A, Stronati M, Gerna G. Human parechovirus infections in patients admitted to hospital in northern Italy, 2008–2010. J Med Virol 2012;84:686–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Joseph L, May M, Thomas M, et al. Human parechovirus 3 in infants: expanding our knowledge of adverse outcomes. Pediatr Infect Dis J 2019; 38:1–5. [DOI] [PubMed] [Google Scholar]
- 10. Strenger V, Diedrich S, Boettcher S, et al. Nosocomial outbreak of parechovirus 3 infection among newborns, Austria, 2014. Emerg Infect Dis 2016; 22:1631–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Selvarangan R, Nzabi M, Selvaraju SB, Ketter P, Carpenter C, Harrison CJ. Human parechovirus 3 causing sepsis-like illness in children from midwestern United States. Pediatr Infect Dis J 2011; 30:238–42. [DOI] [PubMed] [Google Scholar]
- 12. Renaud C, Harrison CJ. Human parechovirus 3: the most common viral cause of meningoencephalitis in young infants. Infect Dis Clin North Am 2015; 29:415–28. [DOI] [PubMed] [Google Scholar]
- 13. Piñeiro L, Vicente D, Montes M, Hernández-Dorronsoro U, Cilla G. Human parechoviruses in infants with systemic infection. J Med Virol 2010; 82:1790–6. [DOI] [PubMed] [Google Scholar]
- 14. Nelson TM, Vuillermin P, Hodge J, et al. An outbreak of severe infections among Australian infants caused by a novel recombinant strain of human parechovirus type 3. Sci Rep 2017; 7:44423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Alexandersen S, Nelson TM, Hodge J, Druce J. Evolutionary and network analysis of virus sequences from infants infected with an Australian recombinant strain of human parechovirus type 3. Sci Rep 2017; 7:3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lambert SB, Ware RS, Cook AL, et al. Observational Research in Childhood Infectious Diseases (ORChID): a dynamic birth cohort study. BMJ Open 2012; 2:e002134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sarna M, Lambert SB, Sloots TP, et al. Viruses causing lower respiratory symptoms in young children: findings from the ORChID birth cohort. Thorax 2018; 73:969–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Silcock R, Doyle R, Clark J, Kynaston A, Thomas M, May M. HPeV infection in infants—evidence-based parental counselling for paediatricians. J Paediatr Child Health 2022; 58:856–62. [DOI] [PubMed] [Google Scholar]
- 19. Doyle R, Clark J, Philips L, Bernard A, Kynaston A, Thomas May M, Monitoring outcomes of human parechovirus admissions to Queensland Children’s Hospital, on behalf of the CHQ HPeV Working Group . In: Australasian Epidemiological Association Annual Scientific Meeting, Brisbane, Australia, 23–25 October 2019.
- 20. Tajima F. Statistical methods to test for nucleotide mutation hypothesis by DNA polymorphism. Genetics 1989; 123:585–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hadfield J, Megill C, Bell SM, et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics 2018; 34:4121–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. R Core Team . R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing, 2020. [Google Scholar]
- 23. Benschop KS, Williams CH, Wolthers KC, Stanway G, Simmonds P. Widespread recombination within human parechoviruses: analysis of temporal dynamics and constraints. J Gen Virol 2008; 89:1030–35. [DOI] [PubMed] [Google Scholar]
- 24. Benschop KS, deVries M, Minnaar RP, et al. Comprehensive full-length sequence analyses of human parechoviruses: diversity and recombination. J Gen Virol 2010; 91:145–54. [DOI] [PubMed] [Google Scholar]
- 25. Calvert J, Chieochansin T, Benschop KS, et al. Recombination dynamics of human parechoviruses: investigation of type-specific differences in frequency and epidemiological correlates. J Gen Virol 2010; 91:1229–38. [DOI] [PubMed] [Google Scholar]
- 26. Aizawa Y, Watanabe K, Oishi T, Hirano H, Hasegawa I, Saitoh A. Role of maternal antibodies in infants with severe diseases related to human parechovirus type 3. Emerg Infect Dis 2015; 21:1966–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Karelehto E, Brouwer L, Benschop K, Kok J, Basile K, McMullan B, et al. Seroepidemiology of parechovirus A3 neutralising antibodies, Australia, the Netherlands and United States. Emerg Infect Dis 2019; 25:148–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chamings A, Druce J, Caly L, et al. Evolutionary analysis of human parechovirus type 3 and clinical outcomes of infection during the 2017–18 Australian epidemic. Sci Rep 2019; 9:8906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fortuna D, Cárdenas AM, Graf EH, et al. Human parechovirus and enterovirus initiate distinct CNS innate immune responses: pathogenic and diagnostic implications. J Clin Virol 2017; 86:39–45. [DOI] [PubMed] [Google Scholar]
- 30. Zhang W, Huang Z, Huang M, Zeng J. Predicting severe enterovirus 71-infected hand, foot, and mouth disease: cytokines and chemokines. Mediators Inflamm 2020; 2020:9273241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kadambari S, Braccio S, Ribeiro S, et al. Enterovirus and parechovirus meningitis in infants younger than 90 days old in the UK and Republic of Ireland: a British Paediatric Surveillance Unit study. Arch Dis Child 2019; 104:552–7. [DOI] [PubMed] [Google Scholar]
- 32. Antolín L F, Kadambari S, Braccio S, et al. Increased detection of human parechovirus infection in infants in England during 2016: epidemiology and clinical characteristics. Arch Dis Child 2018; 103:1061–6. [DOI] [PubMed] [Google Scholar]
- 33. Amarnath C, Helen Mary T, Periakarupan A, Gopinathan K, Philson J. Neonatal parechovirus leucoencephalitis—radiological pattern mimicking hypoxic-ischemic encephalopathy. Eur J Radiol 2016; 85:428–34. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.