Summary
With the widespread vaccinations against coronavirus disease 2019 (COVID-19), we are witnessing gradually waning neutralizing antibodies and increasing cases of breakthrough infections, necessitating the development of drugs aside from vaccines, particularly ones that can be administered outside of hospitals. Here, we present two cross-reactive nanobodies (R14 and S43) and their multivalent derivatives, including decameric ones (fused to the immunoglobulin M [IgM] Fc) that maintain potent neutralizing activity against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) after aerosolization and display not only pan-SARS-CoV-2 but also varied pan-sarbecovirus activities. Through respiratory administration to mice, monovalent and decameric R14 significantly reduce the lung viral RNAs at low dose and display potent pre- and post-exposure protection. Furthermore, structural studies reveal the neutralizing mechanisms of R14 and S43 and the multiple inhibition effects that the multivalent derivatives exert. Our work demonstrates promising convenient drug candidates via respiratory administration against SARS-CoV-2 infection, which can contribute to containing the COVID-19 pandemic.
Keywords: SARS-CoV-2, Omicron, nanobody, IgM antibody, inhalable administration, cross-reactive antibody, sarbecovirus
Graphical abstract
Highlights
-
•
Nanobodies R14 and S43 display not just pan-SARS-CoV-2 but varied pan-sarbecovirus activities
-
•
The multivalent nanobodies show enhanced neutralization, fitting the avidity effect
-
•
Intranasally administered MR14 potently prevents Omicron infections in mice
-
•
Structural studies reveal the neutralizing mechanisms of R14 and S43 as well as MR14
Liu et al. isolate two nanobodies (R14 and S43) and construct their multivalent derivatives with increased potencies and breadth against SARS-CoV-2 variants. Particularly, decameric R14, maintaining similar activities after aerosolization, efficiently prevents BA.2 infections in both pre- and post-exposure prophylactic settings in vivo and deserves further development.
Introduction
Emerging and re-emerging viruses are a threat to global public health.1 The coronavirus disease 2019 (COVID-19) pandemic is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2),2,3 which is continuously evolving into multiple variants. The current SARS-CoV-2 variant of concern (VOC), Omicron, has novel epidemiological and biological characteristics, making it more contagious than previous VOCs, including Alpha, Beta, Gamma, and Delta. Even worse, Omicron has evolved into multiple sub-variants (BA.1, BA.2, BA.2.12.1, BA.2.75, BA.3, BA.4, and BA.5), yielding increased transmissibility, infectivity, and immune evasion.4,5,6 Indeed, the most current sub-variants, BA.4 and BA.5, are reported to have more severe immune evasion than BA.2.7
Together with SARS-CoV, which is another coronavirus that caused an epidemic two decades ago, SARS-CoV-2 belongs to the SARS-like coronaviruses (sarbecoviruses) that contain a cluster of phylogenetically related viruses, such as RaTG13, RshSTT182, RshSTT200, GX/P2V/2017, and GD/1/2019. These viruses show varied capacities to bind to human angiotensin-converting enzyme 2 (ACE2), suggesting their potential to cause new pandemics.8,9,10,11,12 Therefore, cross-reactive antibodies that broadly prevent the infections of multiple SARS-CoV-2 variants and sarbecoviruses are urgently needed.
During the current pandemic, therapeutic antibodies advanced into clinical applications in an extremely short amount of time and play important roles in treating the disease. They prevent SARS-CoV-2 infections by multiple mechanisms, mainly by blocking the binding between the receptor-binding domain (RBD) and the ACE2 receptor.13,14,15,16 However, several authorized therapeutic antibodies display weakened protective efficacy against SARS-CoV-2 variants, especially the emerging Omicron VOC, and were revoked.17 In addition, most therapeutic antibodies are conventional monoclonal antibodies (mAbs; ∼150 kDa) that require intravenous injection, which reduces their concentration in the lungs, where antibodies could make the most significant impact.18,19 However, for respiratory viral infections, no antibodies delivered via respiratory administration have been authorized despite the fact that it would be better to deliver antibody directly to the lungs rather than via intravenous injection.
Compared with conventional antibodies, nanobodies only contain the variable domain of the heavy chain and are consequently much smaller (∼15 kDa), more stable, and easier to manipulate. Due to their small size and stability, nanobodies can penetrate the barrier between the respiratory epithelium and capillary endothelium.20 In response to the current COVID-19 pandemic, several nanobodies against SARS-CoV-2 have been reported that target the RBD, for example Nb21,21 Nb15,22 and MR3.23 However, data concerning the neutralizing activities of these nanobodies against Omicron VOC are limited. Regardless, based on their small size and lack of a light chain, nanobodies can be flexibly constructed into multiple versions, such as dimers, trimers, and multivalent antibodies. Therefore, to develop convenient drugs against SARS-CoV-2 variants and sarbecoviruses, nanobodies and their derivatives administered through respiratory routes are a likely solution.
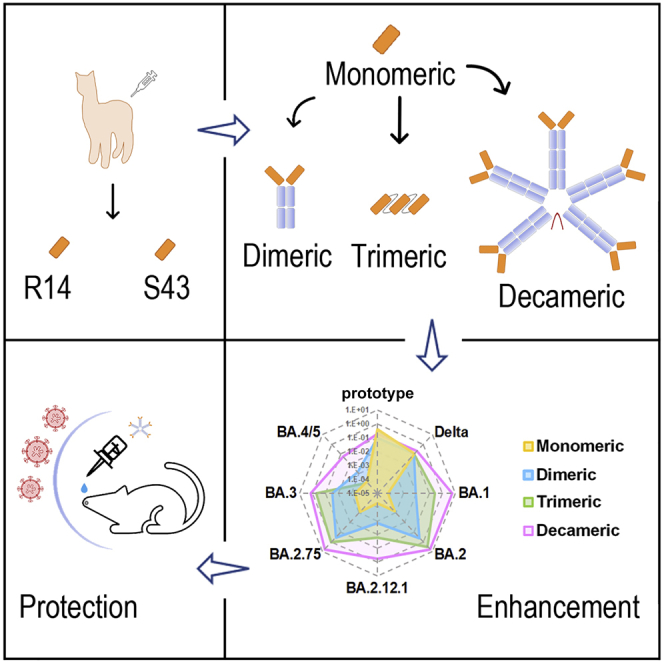
In this study, we present two pan-SARS-CoV-2 and pan-sarbecovirus nanobodies, R14 and S43, that maintained neutralizing activity after being aerosolized. Both nanobodies significantly reduced lung viral RNAs via intranasal (i.n.) administration prior to SARS-CoV-2 infection. Specifically, aerosolized R14 also inhibited SARS-CoV-2 infection in mice in both pre- (PrEP) and post-exposure prophylactic (PEP) settings. To further increase their potencies and broaden their spectrum against Omicron VOC, we engineered the nanobodies into multiple constructs, including dimeric derivatives by tagging them to the Fc of the immunoglobulin G1 (IgG1) (DR14 and DS43), homo-trimeric ones by tandem-repeat constructions (TR14 and TS43), and decameric ones by fusing them to the Fc of IgM (MR14 and MS43). Notably, trimeric and decameric constructs maintained similar activities after aerosolization and exhibited higher antiviral activities against the Omicron sub-variants BA.1, BA.2, BA.2.12.1, BA.2.75, BA.3, and BA.4/5. Compared with the parental nanobodies, the inhalable derivatives also displayed longer residence time in the lung, as well as longer half-life (t1/2) in the blood when intraperitoneally (i.p.) administered. Additionally, using a mouse infection model, MR14 displayed great potency to prevent Omicron BA.2 infections in both PrEP and PEP settings via i.n. and i.p. administration routes. Further structural analysis revealed the neutralizing mechanisms of R14 and S43 and indicated the mechanisms by which the multivalent derivatives substantially increased their inhibition potencies. Thus, these data indicate that R14, S43, and their multivalent derivatives have great potential to be further developed as convenient inhalable therapeutics that are effective and broad spectrum for preventing infections of SARS-CoV-2 variants and potential emerging sarbecoviruses.
Results
Nanobodies prevented SARS-CoV-2 infection in vitro and in vivo
To obtain nanobodies, we immunized two alpacas with the SARS-CoV-2 spike (S) and RBD proteins, respectively. Nanobodies were displayed following the protocols previously described24 (Figure S1A). Approximately 500 clones were identified by phage ELISA assays during panning, and the positive clones were sequenced. In our current study, we concentrated on two nanobodies, R14 and S43.
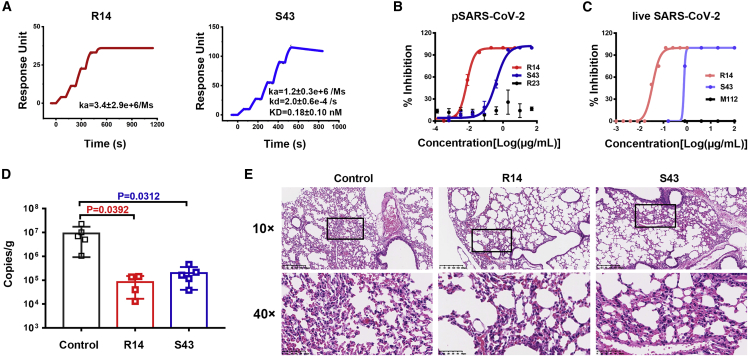
Both R14 and S43 specifically recognized the SARS-CoV-2 RBD, as confirmed by phage ELISA (Figure S1). Moreover, S43 could also bind to SARS-CoV S, as shown by flow cytometry (Figure S2). Both R14 and S43 also blocked the binding between the SARS-CoV-2 RBD and human ACE2 (hACE2) (Figure S3). As determined by surface plasmon resonance (SPR) assays, S43 was bound to the SARS-CoV-2 RBD with high affinity, with an equilibrium dissociation constant (KD) value of 0.18 nM, while R14 displayed no detectable dissociation from the SARS-CoV-2 RBD over 10 min (Figure 1A). These results indicate that R14 has a stronger binding affinity for SARS-CoV-2 than that of S43.
Figure 1.
Anti-SARS-CoV-2 activity of R14 and S43 in vitro and in vivo
(A) The binding kinetics of R14 (left) or S43 (right) to the SARS-CoV-2 RBD obtained using a BIAcore 8K system in single-cycle mode. Values are the mean ± standard deviation (SD) of three independent results.
(B) Neutralization of pseudotyped SARS-CoV-2 (pSARS-CoV-2) by R14 and S43 in Vero cells in vitro; n = 3 per dilution. Experiments were repeated independently three times with similar results, and one representative curve is displayed. Nanobody R23 was used as the negative control.
(C) Neutralization of live SARS-CoV-2 by R14 and S43 in Vero cells in vitro; n = 4 per dilution. Experiments were independently repeated twice with similar results, and one representative curve is displayed. M112 represents the negative control.
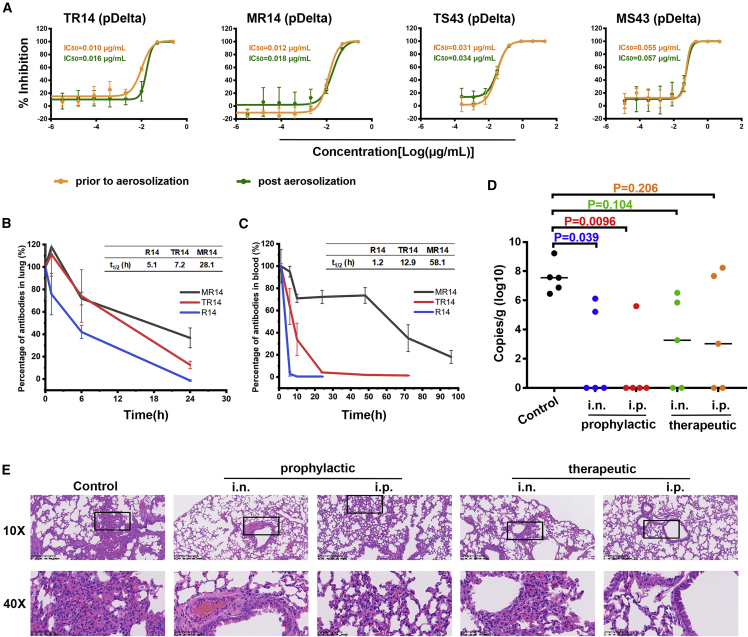
(D) Viral RNA loads in the lungs of mice treated with 5 mg/kg R14 or S43 via the i.n. route prior to SARS-CoV-2 infection. Two-tailed ordinary one-way ANOVA with Sidak’s multiple comparisons was used for statistical analyses.
(E) Representative histopathology of lung tissues of SARS-CoV-2-infected mice (5 days post-infection [dpi]). The images and areas of interest (black boxes) are magnified 10× and 40×, respectively.
We next assessed the neutralizing capabilities of both nanobodies against pseudotyped and live SARS-CoV-2 in vitro and found that R14 and S43 have potent neutralizing activities against pseudotyped SARS-CoV-2, with half maximal inhibitory concentration (IC50) values of 0.16 (0.0024 μg/mL) and 11 nM (0.16 μg/mL), respectively (Figure 1B). They also neutralized live SARS-CoV-2 with IC50 values of 1.3 (0.02 μg/mL) and 40.7 nM (0.61 μg/mL), respectively (Figure 1C).
We further evaluated the in vivo prophylactic efficacy of the two nanobodies against SARS-CoV-2 in an Ad5-hACE2 mouse model. We found that 5 mg/kg R14 and S43 significantly reduced lung viral RNAs when administered via the i.n. route prior to SARS-CoV-2 infection (Figure 1D). In particular, R14 displayed an approximately 100-fold reduction of lung viral RNAs compared with the control mice. Lung pathology analyses revealed that SARS-CoV-2 can cause severe interstitial pneumonia, characterized by inflammatory cell infiltration and alveolar septal thickening in control mice, while only very mild lesions were observed when the mice received R14 or S43 by the i.n. route prior to challenge with SARS-CoV-2 (Figure 1E).
Aerosolized R14 maintained neutralizing activity and prevented infection
We then tried to evaluate the effectiveness of the nanobody via inhalation (i.h.) delivery. Because R14 was superior to S43 in terms of neutralization activity in vitro and preventive efficacy in vivo via i.n. administration, we chose to assess the effects of R14 by aerosol delivery with a whole-body exposure platform (Figure 2A).
Figure 2.
Aerosolization and the prophylactic and therapeutic efficacy of R14 against SARS-CoV-2
(A) Schematic layout of the whole-body exposure platform.
(B) Aerosol size distributions of six monitoring points in the exposure chamber (red triangles in A).
(C) Aerosol mass concentration as a function of time when the flowrate of the carrier air was 5, 10, or 20 LPM.
(D) Neutralization of SARS-CoV-2 pseudotyped virus by R14 and S43 post-aerosolization in Vero cells; n = 3 per nanobody concentration points. Experiments were independently repeated twice with similar results, and one representative curve is displayed.
(E) Viral RNA loads in the lungs of mice treated with a single dose of 0.25 or 5 mg/kg R14 by the i.n. route and i.h. route prior to SARS-CoV-2 infection, respectively.
(F) Viral RNA loads in the lungs of mice treated with 0.75 mg/kg of R14 at 10 min, 1.5 h, and 7 h by the i.h. route post-SARS-CoV-2 infection.
Two-tailed ordinary one-way ANOVA with Sidak’s multiple comparisons was used in the statistical analyses for (E) and (F).
The whole-body exposure system designed in this study mainly contained an Aerogen Solo vibrating mesh nebulizer (Aerogen, Chicago, IL, USA) and an exposure chamber. Aerosol size distributions were measured at six monitoring sites (red triangles in Figure 2A) on the middle cross-section of the exposure chamber and showed no significant differences among the monitoring sites (Figure 2B), indicating that each mouse was exposed to similar conditions. To select the proper flowrate of the carrier air, the average mass concentration was monitored and suggested to be higher when the carrier air flowrate was maintained at 10 and 5 L per minute (LPM) compared with 20 LPM (Figure 2C). Finally, the flowrate of the carrier air was maintained at 10 LPM during exposure experiments due to remarkably deposited aerosol at the bottom of the chamber when the flowrate was 5 LPM.
Notably, the aerosolization process, a large environmental change, did not influence the neutralization activities of the two nanobodies against SARS-CoV-2 pseudotyped viruses (Figure 2D), providing the basis for further trials of i.h. administration. Thus, mice were administrated a single 0.25 mg/kg dose of R14 via the i.h. route prior to SARS-CoV-2 infection, while a single 5 mg/kg dose administrated via the i.n. route was used as a control. We found that the lung viral RNAs in the i.h. group decreased to a similar level to those mice in the i.n. group, though the administrated dose via the i.h. route was 20 times lower than that via the i.n. route (Figure 2E).
We further evaluated the therapeutic effect of R14 against SARS-CoV-2 via the i.h. route at different times. After SARS-CoV-2 infection, mice were treated with a single 0.75 mg/kg dose of R14 at 10 min, 1.5 h, or 7 h, while the control mice were administrated PBS (Figure 2F). We found that the viral RNAs in the lung were significantly reduced when the nanobody was administrated at 10 min or 1.5 h post-infection. Although no significant difference was observed in the 7 h group, the viral titer decreased compared with the control group, suggesting that the preventive effect is related to the time when the nanobody is delivered post-infection. Indeed, at 10 min, the viral copies decreased by three orders of magnitude, which is similar to the efficacy of R14 administered prior to infection by the i.h. route with a dose of 0.25 mg/kg (Figure 2E).
Both R14 and S43 displayed pan-SARS-CoV-2 and pan-sarbecovirus activities
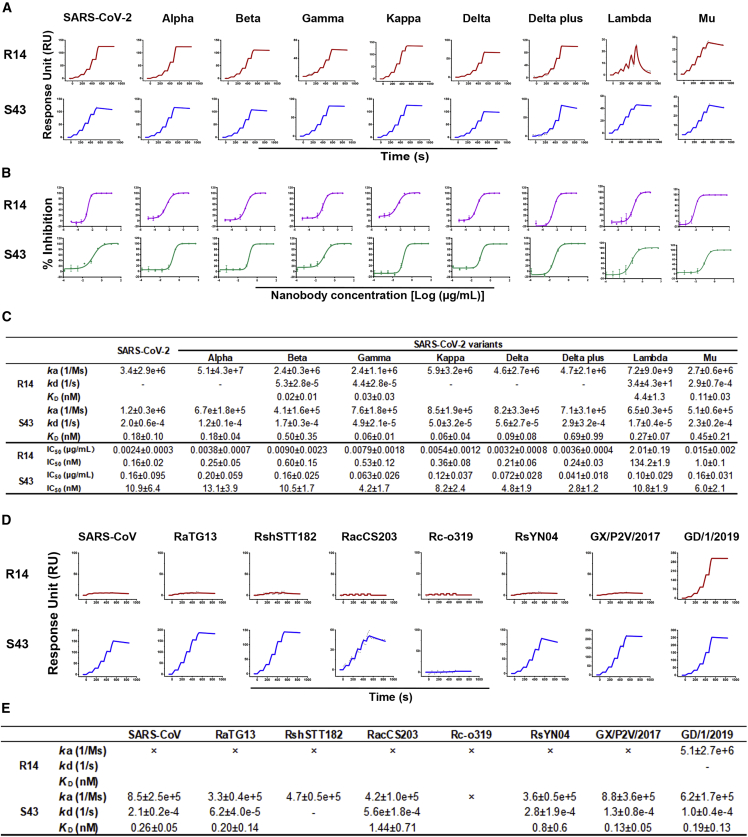
The effectiveness of R14 and S43 against the prototype SARS-CoV-2 prompted us to further evaluate their efficacy against SARS-CoV-2 variants. We measured the binding affinities and neutralizing activities of R14 and S43 (Figures 3A and 3B). Notably, both R14 and S43 could bind to all of the tested RBDs from Alpha, Beta, Gamma, Kappa, Delta, Delta plus, Lambda, and Mu (Figures 3A and 3C). Additionally, both R14 and S43 broadly neutralized multiple SARS-CoV-2 variants (Figures 3B and 3C). R14 displayed similar neutralizing ability against the SARS-CoV-2 prototype and its variants, with IC50 values ranging from 0.16 to 1 nM (0.0024–0.015 μg/mL), except for the Lambda variant. The neutralizing activity of R14 against Lambda was 134 nM (2.01 μg/mL), which is ∼800-fold lower than that against the prototype strain. S43 displayed weaker neutralizing ability against these pseudotyped viruses than R14, with IC50 values ranging from 2.75–13.05 nM (0.041–0.20 μg/mL). The results of the two nanobodies against the Omicron variant are covered in the next section.
Figure 3.
The cross-binding and cross-neutralization of R14 and S43 to SARS-CoV-2 and its variants
(A) The binding kinetics of R14 and S43 to RBDs from SARS-CoV-2 and its variants obtained using a BIAcore 8K system in single-cycle mode. Values summarized in (C) are the mean ± SD of three independent experiments. “-” represents no detectable dissociation from the RBD.
(B and C) (B) Neutralization of R14 and S43 against SARS-CoV-2 and its variant pseudotyped viruses in Vero cells. n = 3 per nanobody concentration points. IC50 values summarized in (C) are the mean ± SD of three independent results.
(D) The binding between R14 or S43 and multiple sarbecoviruses. The binding kinetics of R14 and S43 with RBDs from several sarbecoviruses were obtained using a BIAcore 8K system in single-cycle mode. Human Fc-tagged RBD proteins were captured on the chip surface, and serial dilutions of His-tagged R14 or S43 were then injected over the chip surface. Values summarized in (E) are the mean ± SD of three independent experiments. “-” represents no detectable dissociation from the RBD. “×” represents no detectable binding to the RBD.
To further explore the cross-reactivity of R14 and S43, we also tested the binding affinities of the two nanobodies to several sarbecoviruses, including SARS-CoV, RaTG13, RshSTT182, RacCS203, Rco319, RsYN04, GX/P2V/2017, and GD/1/2019.11 We found that R14 bound to the GD/1/2019 RBD with high affinity, but no binding was observed to the other sarbecoviruses (Figures 3D and 3E) at the highest tested concentration of 100 nM. Nevertheless, S43 bound to the RBDs from SARS-CoV, RaTG13, RshSTT182, RacC203, RsYN04, GX/P2V/2017, and GD/1/2019 (Figure 3D and 3E). These data indicate that S43 has broader cross-reactivity to sarbecoviruses than R14.
Multivalent derivatives presented improved efficacy against Omicron VOC
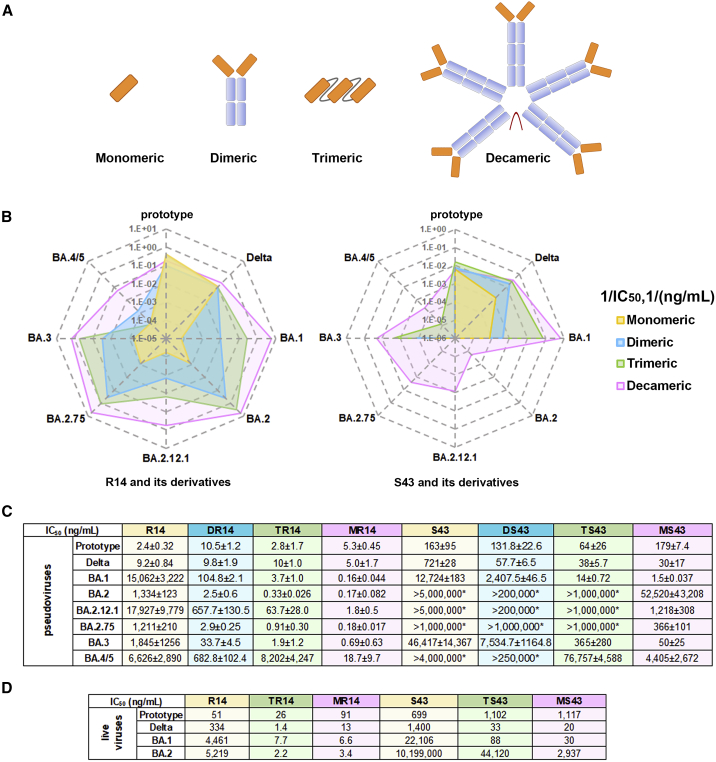
To increase their neutralization and broaden their activities against the Omicron VOC, we constructed multivalent forms of the two nanobodies, including dimeric derivatives by tagging them to the Fc of the IgG1 (DR14 and DS43), homo-trimeric ones by tandem-repeat constructions (TR14 and TS43), and decameric ones by fusing them to the Fc of IgM (MR14 and MS43) (Figure 4A), as it is known that multivalent mAbs can display improved efficacy.21,25,26 Indeed, compared with the monomeric R14, both DR14 and TR14 greatly improved the neutralizing activities, with 27- to 500- and 280- to 4,000-fold enhancement, respectively, against pseudotyped Omicron sub-variants BA.1, BA.2, BA.2.12.1, BA.2.75, and BA.3. However, their activities against pseudotyped BA.4/5 were only slightly higher than that of R14. When R14 was constructed to the decameric MR14, the activity against BA.4/5 was substantially increased, with 350-fold change. Additionally, compared with DR14 and TR14, MR14 displayed further enhancement of the activities against sub-variants BA.1, BA.2, BA.2.12.1, BA.2.75, and BA.3, with 94,000-, 7,800-, 9,900-, 6,700-, and 2,600-fold changes, respectively, although, as with TR14, MR14 displayed similar binding affinities to the sub-variant RBDs compared with those of R14 (Figure S5).
Figure 4.
Enhanced neutralization by dimer, homo-trimer, and IgM versions of R14 or S43
(A) The dimers were constructed via nanobody fusing with human IgG1 Fc, homo-trimers were constructed with three nanobodies by head-to-tail fusion, and the IgM versions (decameric constructs) were constructed via nanobody fusing to the Fc of IgM.
(B) Radar plot demonstrating the neutralization profile of antibodies to prototype and Delta and Omicron sub-variants. For the antibody with IC50 of <100% inhibition at the highest tested concentration, 1/IC50 is thought of as zero.
(C) IC50 values of multivalent constructs of R14 and S43 against pSARS-CoV-2 prototype, Delta, and Omicron (BA.1, BA.2, BA.2.12.1, BA.2.75, BA.3, and BA.4/5) in Vero cells. IC50 values for pseudotyped viruses are the mean ± SD of three independent results. Experiments were independently repeated three times with similar results, and one representative curve is displayed in the Figure S4. “∗” indicates that the antibody showed <100% inhibition at the highest tested concentration.
(D) IC50 values for live viruses are the mean of two independent experiments.
However, S43 displayed weak potency, with IC50 >12 μg/mL against pseudotyped Omicron sub-variants, but enhanced neutralizing activity was also observed with DS43, TS43, and MS43. In addition, the multivalent forms of R14 and S43 demonstrated similar neutralizing activities as their respective monomeric forms against the SARS-CoV-2 prototype and Delta variants (Figures 4B and 4C).
We then chose the homo-trimeric and decameric derivatives for evaluation using live viruses since both strategies have been used to develop therapeutic antibodies that entered clinical trials as nasal administration drugs27,28 (https://clinicaltrials.gov/). Both forms of constructs also greatly increased the neutralizing activities against sub-variants BA.1 and BA.2 compared with the monomeric nanobodies (Figure 4D).
MR14 displayed effective protection in both PrEP and PEP settings via i.n. and i.p. in vivo
As with the parental nanobodies, the aerosolization process did not influence the neutralization activities of both homo-trimeric and decameric versions of R14 and S43 (Figure 5A), which provides the basis for their applications as inhalable drugs. We further focused on R14, TR14, and MR14 to evaluate the decay rate, due to their superiorities in terms of neutralization activities and their broad potencies against Omicron sub-variants. We found that both aerosolized TR14 and MR14 displayed longer t1/2 (7.2 and 28.1 h, respectively) in lungs compared with the aerosolized R14 (5.1 h) (Figure 5B), indicating an extended residence time and thereby higher protective efficacy by the two R14 multimers in the respiratory system when administered via the i.h. route. In addition, we also assessed the t1/2 of TR14 and MR14 in blood after i.p. injection and found that they had substantially longer t1/2 (12.9 and 58.1 h, respectively) compared with monovalent R14 (1.2 h) (Figure 5C).
Figure 5.
The prophylactic and therapeutic efficacy of MR14 against Omicron BA.2 in vivo
(A) Neutralization of pseudotyped virus by homo-trimeric and IgM versions of nanobodies post-aerosolization in Vero cells; n = 3 per nanobody concentration point. Experiments were independently repeated twice with similar results, and one representative curve is displayed.
(B) The percentage of remaining nanobodies in the lungs of BALB/c mice post-aerosolization (n = 3).
(C) The percentage of remaining nanobodies in the blood after a single 20 mg/kg dose in BALB/c mice via intraperitoneal injection (n = 3).
(D) Lung viral sgRNA_E loads in the BA.2 variant-infected mice with the indicated treatments; n = 5 biologically independent mice in all groups. The solid lines indicate the median lung viral sgRNA_E loads. Ordinary one-way ANOVA with Sidak’s multiple comparisons was used in the statistical analyses.
(E) Representative histopathology of lung tissues in BA.2-infected mice (3 dpi). The images and areas of interest (black boxes) are magnified 10× and 40×, respectively.
We next evaluated the prophylactic and therapeutic efficacy of MR14 against Omicron sub-variant BA.2 using a K18-hACE2 mouse model. A single 5 mg/kg dose of MR14 was administered 6 h prior to BA.2 infection by the i.n. or i.p. route to evaluate prophylactic efficacy. Mice receiving PBS were used as a control. For therapeutic evaluation, MR14 was administered in three i.n. doses at 6, 30, and 54 h post-BA.2 infection, but only a single dose of MR14 was administered via the i.p. route 6 h post-infection. Subgenomic RNA (sgRNA) encoding for the SARS-CoV-2 E genes in lung tissues was analyzed 3 days post-infection. We found that MR14 demonstrated significant prophylactic efficacy when administered by the i.n. or i.p. route (Figure 5D). Notably, three mice (n = 5) in the i.n. group and four mice (n = 5) in the i.p. group showed undetectable sgRNA levels. MR14 also yielded decreased sgRNA loads in therapeutic groups, with two mice (n = 5) displaying undetectable sgRNA in the i.n. and i.p. groups. Pathological analyses of lung tissue revealed that MR14 alleviated lung injury, especially in prophylactic treatment via i.n. delivery (Figure 5E). These results suggest that MR14 has potent efficacy for the prevention and treatment of COVID-19.
Structural insight into the binding between SARS-CoV-2 RBD and R14 or S43
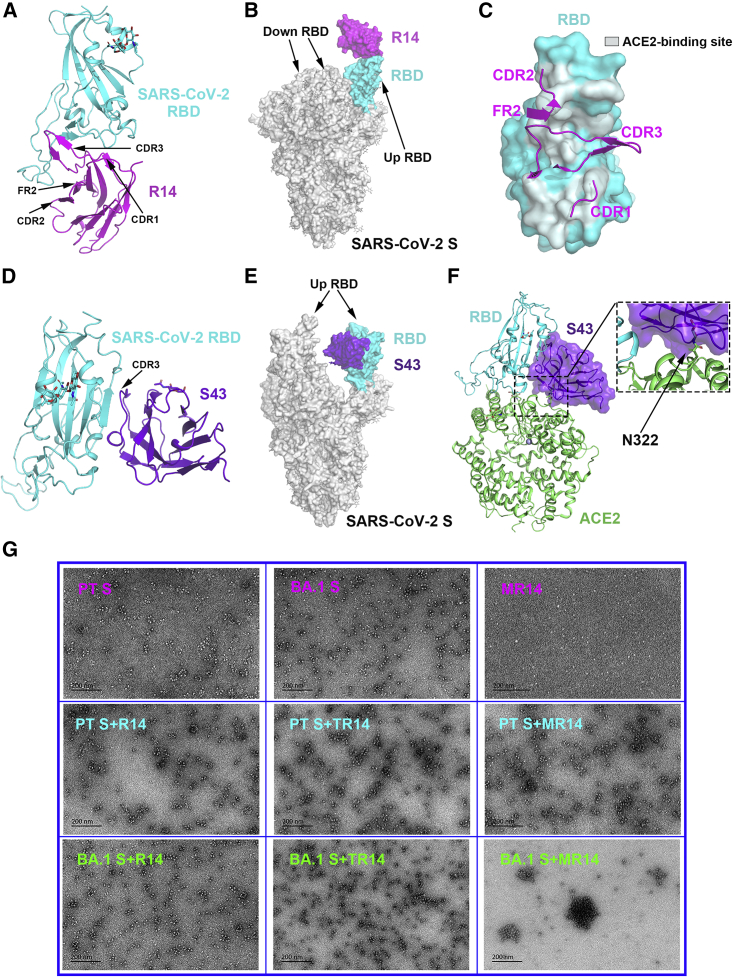
To illustrate the molecular basis of R14 and S43 neutralizing virus infection, we prepared SARS-CoV-2 prototype RBD/R14 complex and SARS-CoV-2 prototype RBD/S43 complex proteins (Figure S6) and solved their crystal structures at resolutions of 2.5 and 2.69 Å, respectively (Table S1). Overall, the two nanobodies recognized distinct conformational epitopes on the RBD.
R14 bound to the receptor-binding motif (RBM) with all three CDRs (CDR1-3) and one framework region (FR, FR2) involved in the interaction (Figure 6A), and it could span the epitopes of RBD-1, RBD-2, RBD-3, and RBD-4 antibodies due to its long CDR3 and large binding area (∼1,015 Å2).29 We superimposed the prototype RBD/R14 complex onto the SARS-CoV-2 prototype S trimer (PDB: 6VYB), revealing that R14 preferentially bound to the RBD in the up conformation. However, when R14 interacted with the down RBD, it would sterically clash with the adjacent RBD in both up and down conformations (Figure 6B). Additionally, the R14 epitope would clash with the hACE2-binding site, as shown by superimposing the prototype RBD/R14 complex onto the prototype RBD/hACE2 complex (PDB: 6LZG) (Figure 6C). Therefore, R14 likely neutralizes SARS-CoV-2 by preventing viral attachment to host cells via competition with the ACE2 entry receptor.
Figure 6.
The complex structure of R14 or S43 bound to the SARS-CoV-2 RBD
(A) The overall complex structure of R14 bound to the SARS-CoV-2 RBD.
(B) The overall features of R14 bound to the SARS-CoV-2 S trimer. The RBD/R14 complex was superimposed onto the S trimer (PDB: 6VYB), which contains one up RBD and two down RBDs.
(C) Comparison between R14 and ACE2 bound to the RBD, respectively. The CDRs and FR of R14 involved in the interaction with the RBD are displayed as cartoons, and the footprint of ACE2 on the RBD is shown in pale cyan.
(D) The overall complex structure of S43 bound to the RBD.
(E) The overall features of S43 bound to the SARS-CoV-2 S trimer. The RBD/S43 complex was superimposed onto the S trimer (PDB: 7LYK), which contains two up RBDs and one down RBD.
(F) Comparison between S43 bound to the RBD and ACE2 bound to the RBD. The glycosylated residue N322 of ACE2 is framed and magnified.
(G) Representative negatively stained images of protein particles. The morphology of protein particles was determined using TEM (JEM-1400). Experiments were independently repeated twice with similar results, and representative images are displayed.
Distinct from R14, S43 recognized a cryptic epitope on one side of the RBD (RBD-7)29 using only CDR3 (Figure 6D). When the prototype RBD/S43 complex was superimposed onto the SARS-CoV-2 prototype S trimer (PDB: 7LYK), S43 was observed to interact with the up RBD-containing S, which possesses at least two up RBDs (Figure 6E). Although the S43 epitope is away from the RBM, and no overlap with the ACE2-binding site was observed, we found that this nanobody displays steric hindrance with the ACE2 N322 glycan by superimposing the prototype RBD/S43 complex onto the prototype RBD/hACE2 complex (PDB: 6LZG) (Figure 6F). Therefore, S43 likely neutralizes SARS-CoV-2 infection by partially interfering with S binding to ACE2 caused by ACE2 glycosylation. This probable mechanism was also observed with SARS-CoV-2 neutralizing antibodies S30430 and EY6A,31 both of which also belong to RBD-7 and apply their constant regions to prevent the association between the receptor and the neighboring RBDs.
The detailed interactions, containing van der Waals (vdw), hydrogen bond (H-bond), and salt bridge interactions, between R14 and RBD are displayed (Table S2). However, we found 10 mutations (K417N, G446S, L452R/Q/M, T478K, F486V, Q493R, G496S, Q498R, N501Y, and Y505H) in the Omicron RBD that belong to R14-binding sites (Figure S7A; Table S2). They break partial H-bond (G446S, Q493R, Q498R, N501Y, and Y505H) and hydrophobic (G446S, L452R, F486V, and G496S) interactions, and some showed potential clashes with R14 due to their large side chain (such as Q498R) (Figures S7B and S7D). On the contrary, T478K added a salt bridge interaction with D62, and N501Y added a π-π interaction with Y119 and a hydrophobic interaction with F27, L29, and P100. Nevertheless, R14 ultimately displayed decreased binding and neutralizing activities to Omicron sub-variants due to the combination of several mutations. Notably, because the BA.2 RBD has no G446S and G496S mutations, R14 demonstrated higher binding and neutralizing abilities to BA.2 than to BA.1.
S43 recognized a cryptic epitope, defined by previous studies,32 and its footprint did not overlap with the ACE2-binding site (Figure 6F). The detailed interactions between S43 and the RBD are shown in Table S3. Notably, all of the interactions were contributed by CDR3, which adopted a pan-like conformation parallel to the RBD. Notably, only the S375F mutation in the BA.1 RBD is included in the S43-binding site, which leads to the loss of two H-bonds formed by S375 with G110 and T112 in S43 (Figure S7E). This may reveal why S43 showed slightly reduced binding to the BA.1 RBD compared with the prototype RBD. However, due to the preferential conformation of BA.1 S in one-RBD-up,33 S43 displayed significantly reduced neutralization against BA.1. Aside from the S375F mutation, the T376A, D405N, and R408S mutations in the BA.2, BA.2.12.1, and BA.4/5 RBDs also belong to the S43-binding site, which further impaired the interaction with S43 (Figure S7F). Therefore, S43 displayed poor neutralization against other Omicron sub-variants besides BA.1.
Both TR14 and MR14 bind to the BA.1 RBD at similar levels to R14 but display extremely higher neutralizing activity against pseudotyped and authentic BA.1 than R14. To explain this phenomenon, we negatively stained samples of S protein incubated with antibodies and imaged them with transmission electron microscopy (TEM) (Figure 6G), which suggested that particles are formed between BA.1 S protein with TR14 or MR14 but not with R14. Indeed, large particles without free S protein were observed in samples of BA.1 S protein with the MR14 antibody, explaining its high neutralizing activity. The clustering effect of TR14 or MR14 to SARS-CoV-2 prototype S protein was not obvious, consistent with their similar potencies against SARS-CoV-2 prototype infections compared with the parental R14.
Discussion
When facing Omicron variants, countermeasures, such as vaccines and therapeutic drugs, display weaker or even lost efficacy.6,34 Multiple strategies have been tried to increase the efficacies of vaccines, including the optimization of immunization protocols,35 sequential immunization with heterologous vaccines,36 and updated immunogens that stimulate cross-protection effects.37 Nevertheless, breakthrough infections are still a great challenge to contain SARS-CoV-2 infection, suggesting an urgent need to develop drugs with cross-reactivities. Aside from small molecules and traditional Chinese medicine, therapeutic antibodies, such as human monoclonal neutralizing antibodies, have made great contributions to antiviral treatment. However, due to the mutations in the Omicron RBDs, the majority of previously identified RBM-targeting antibodies (RBD-1, RBD-2, and RBD-3) and RBD-4 antibodies have lost binding and neutralizing activities. Although RBD-5, RBD-6, and RBD-7 antibodies maintain interactions, their neutralizing potencies are relatively low, with IC50 >1 μg/mL for most members.6 These results highlight the need to develop new antibody drugs by integrating more mechanisms.
Compared with conventional IgG antibodies, nanobodies have advantage for the development of inhalable drugs and benefit for the treatment of respiratory pathogens (such as coronavirus and influenza virus). Although several nanobodies have been reported to be effective against SARS-CoV-2 infection, such as the Nb21, Nb15, and MR3 nanobodies targeting the RBD,21,22,23 no one nanobody has yet been approved for COVID-19 treatment. Here, we isolated two nanobodies, R14 and S43, targeting different epitopes on the RBD. They both showed pan-SARS-CoV-2 but varied pan-sarbecovirus activities. Notably, in the mice infection model, R14 administrated through the inhaling route can prevent SARS-CoV-2 infection in both PrEP and PEP settings.
However, R14 and S43 nanobodies showed reduced neutralizing activities against the emerging Omicron variant. To address this concern, the two nanobodies were flexibly engineered into different multivalent versions, including IgG1 Fc-fusion dimer, head-to-tail homo-trimer, and IgM Fc-fusion decamer, to enhance antiviral efficacy and overcome emerging SARS-CoV-2 variants. Previous data reported the construction of a homo-trimeric nanobody, exemplified by ALX-0171, that was developed against respiratory syncytial virus infection38 and displayed safety and good tolerance in the clinical trials, suggesting a homo-trimer is a potential solution. Notably, at mucosal surfaces, IgM antibody is thought to play a critical role in promoting mucosal tolerance and shaping the microbiota together with IgA antibodies. Typically, IgM antibodies have 10 (in the pentamer) binding sites that yield their amplified ability to eliminate foreign antigens. Several IgM antibodies have entered into clinical trials (https://clinicaltrials.gov) as the injective reagents, which also verifies their safety and good tolerance in humans. However, most of these IgM antibodies are of natural origin, without extensive somatic hypermutation, and thus exhibited low affinity and specificity,39 which leads to their failures in the clinical trials. Engineered IgM antibodies, with the variable domains of an IgG1 antibody, have been tried to increase the efficacies. Currently, Ku et al. reported such an engineered IgM antibody (IgM-14), which showed broad protection against SARS-CoV-2 variants27 and has recently been approved for a phase I clinical trial as a nasally delivered drug candidate, suggesting that engineered IgM is also a potential strategy to combat SARS-CoV-2 (https://clinicaltrials.gov).
Thus, we focused on both of the above strategies to engineer R14 and S43 in this study. The homo-trimer of R14 or S43 was linked by a (G4S)5 linker, and IgM derivatives were constructed by fusing the nanobodies with the Fc of IgM. Especially, as the single variable domain of the nanobody without the light chain, the complexities for the assembly as well as the manufacturing of these IgM derivatives decreased from the traditional 21 chains (10 heavy chains, 10 light chain, and 1 J chain) to the current 11 chains (10 heavy chains and 1 J chain). Since the manufacturing is a challenge for the development of IgM antibodies, the nanobody-based IgM derivatives reported here provide a potential design that would solve this problem and advance their further applications.
The data suggest that these multivalences, including the dimer, homo-trimer, and decamer, enable them to have pan-SARS-CoV-2 neutralizing activities, especially against the latest Omicron VOC. As expected, dimeric (DR14 and DS43), homo-trimeric (TR14 and TS43), and decameric derivatives (MR14 and MS43) displayed significant improvement against Omicron sub-variants compared with monomer. Particularly, MR14 exhibited more than 300-fold enhancement and also had potent in vivo prophylactic and therapeutic efficacy against the BA.2 sub-variant when delivered by i.n. and i.p. administration in mice.
As indicated by the complex structures, R14 has large binding area on the RBD and could overlap with RBD-1, RBD-2, RBD-3, and RBD-4 antibodies. It binds to the RBD in the up conformation and blocks the interaction between the virus and the receptor. S43 belongs to the RBD-7 and needs to function when at least two RBDs are in the up conformations. Thus, R14 would be greatly influenced by the residue mutations on the RBM region, such as L452Q/R/M, F486V, and Q493R. Although S43 targets the relatively conserved region and shows broader spectrum against sarbecoviruses than R14, the conformational shifts of the S protein would influence its activity. When constructed to the multivalent derivatives, their remarkably increased efficacy for preventing viral infection was observed in Omicron sub-variants but not with the prototype or Delta. Negatively stained TEM images showed that multivalent R14, especially the decameric MR14, engages more S proteins and causes the aggregation of the BA.1 S proteins. It is reported that, compared with the prototype S protein, the S trimer of BA.1 predominantly has one RBD in the up position, allowing the stable binding of the R14 protomer in the multimers. This also provides a reasonable explanation for why the activity of MS43 did not increase a lot.
With the global spread of SARS-CoV-2 and its variants, the demand for therapeutic drugs that exert antiviral effects through different mechanisms has become increasingly prominent, as demonstrated by the excitement after the approval of the oral antiviral drugs molnupiravir and Paxlovid.40 Our study provides two cross-reactive nanobodies and their multivalent descendants as promising candidates against current SARS-CoV-2 variants and many sarbecoviruses. The most potent one in this study, MR14, had improved prophylactic and therapeutic activities in vivo, serving as a potential drug candidate for respiratory administration for COVID-19 prevention and treatment.
Limitations of the study
MR14 retained stability before and after aerosolization, while we did not administrate MR14 via inhalable route on the mice model because of the limited resources of the animal biosafety level 3 facility. Additionally, due to the continuous evolution of SARS-CoV-2, novel Omicron sub-variants are emerging. The efficacies of the monovalent nanobodies and their multivalent derivatives against the currently circulating Omicron sub-variants, including BF.7, BQ.1.1, and XBB, need further evaluation.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| DMEM medium | Invitrogen | Cat# C11995500BT |
| Gibco fetal bovine sera | Gibco | Cat# 10099141C |
| SMM 293-TII Expression Medium | SinoBiological | Cat# M293TII-1L |
| Sinofection Transfection Reagent | SinoBiological | Cat# STF02-5ML |
| SMS 293-SUPI cell culture supplement | SinoBiological | Cat# M293-SUPI-100mL |
| Polyethylenimine (PEI) | Polysciences | Cat# 24885-2 |
| Antibodies | ||
| Mouse anti-his mAb | ZSGB-BIO | Cat# TA-02; RRID:AB_2801388 |
| Goat anti-Mouse IgG (H&L)-HRP conjugated | Easybio | Cat# BE0102; RRID:AB_2923205 |
| Goat anti-Llama IgG H&L (HRP) | Abcam | Cat# ab112786; RRID:AB_10858603 |
| 410,712 APC anti-human IgG Fc | BioLegend | Cat# 410712; RRID:AB_2565790 |
| Anti-HIS-APC, GG11-8F3.5.1, 100 tests | Miltenyi Biotec | Cat# 130-119-782; RRID:AB_2751840 |
| M13 Bacteriophage Antibody (HRP) | SinoBiological | Cat# 11973-MM05T-H; RRID:AB_2857928 |
| Anti-Human IgM antibody [IM260] | Abcam | Cat# ab200541 |
| Critical commercial assays | ||
| HisTrap HP 5 mL column | GE Healthcare | Cat# 17524802 |
| Protein A HP 5 mL column | GE Healthcare | Cat#17040303 |
| HiLoad 16/600 Superdex 200 pg | GE Healthcare | Cat# 28989335 |
| Hitrap IgM purification 5×1ML | GE Healthcare | Cat# 17511001 |
| Superose 6 Increase 10/300 GL | GE Healthcare | Cat# 29091596 |
| NuPAGE 3–8% native PAGE | Invitrogen | Cat# EA03752BOX |
| Series S Sensor Chip SA | GE Healthcare | Cat# 29104992 |
| Series S Sensor Chip Protein A | GE Healthcare | Cat#29127556 |
| Deposited data | ||
| R14/SARS-CoV-2 RBD complex | This paper | PDB: 7WD1 |
| S43/SARS-CoV-2 RBD complex | This paper | PDB: 7WD2 |
| Experimental models: Cell lines | ||
| HEK293T cells | ATCC | Cat# CRL-3216; RRID:CVCL_0063 |
| HEK293F cells | Gibco | Cat# 11625-019 |
| BHK-21 cells | ATCC | Cat# CCL-10; RRID:CVCL_1915 |
| Sf9 Cells | Invitrogen | Cat# 11496015 |
| Hi5 cells | Invitrogen | Cat# B85502 |
| Vero cells | ATCC | Cat# CCL-81; RRID:CVCL_0059 |
| Bacterial and virus strains | ||
| Escherichia coli (E. coli) strain DH5α | Vazyme | Cat# C502-02 |
| Live SARS-CoV-2 prototype strain hCoV-19/China/CAS-B001/2020 | This paper | GISAID: EPI_ISL_514256-7 |
| Live SARS-CoV-2 prototype BetaCoV/Wuhan/IVDC-HB-envF13/2020 | This paper | GISAID: EPI_ISL_408511 |
| Live SARS-CoV-2 Beta strain | This paper | NPRC 2.192100004 |
| Live SARS-CoV-2 BA.1 strain | This paper | NPRC 2.192100007 |
| Live SARS-CoV-2 BA.2 strain | This paper | NPRC 2.192100010 |
| Live SARS-CoV-2 BA.4 strain | This paper | NPRC 2.192100012 |
| Live SARS-CoV-2 BA.5 strain | This paper | NPRC 2.192100014 |
| Experimental models: Organisms/strains | ||
| SPF BALB/c mice, female, 6–8 weeks | Vital River (Beijing, China) | Cat# 01011 |
| SPF K18-hACE2 mice, female, 6–8 weeks | GemPharmatech Co., Ltd (Jiangsu, China) | Strain NO. T037657 |
| Recombinant DNA | ||
| pCAGGS | MiaoLingPlasmid | Cat# P0165 |
| pMES4 | Addgene | Cat# 154837 |
| pEGFP-N1 | MiaoLingPlasmid | Cat# P0133 |
| pFastbac1 | Invitrogen | Cat# 10360014 |
| Prototype RBD, residues 319-541 | This paper | GenBank: MN908947 |
| Prototype RBD with N501Y mutation, residues 319–541 (Alpha) | This paper | N/A |
| Prototype RBD with K417N, E484K, N501Y mutation, residues 319–541 (Beta) | This paper | N/A |
| Prototype RBD with K417T, E484K, N501Y mutation, residues 319–541 (Gamma) | This paper | N/A |
| Prototype RBD with L452R, T478K mutation, residues 319–541 (Delta) | This paper | N/A |
| Prototype RBD with K417N, L452R, T478K mutation, residues 319–541 (Delta plus) | This paper | N/A |
| Prototype RBD with L452R, E484Q mutation, residues 319–541 (Kappa) | This paper | N/A |
| Prototype RBD with L452Q, F490S mutation, residues 319–541 (Lambda) | This paper | N/A |
| prototype RBD with R346K, E484K, N501Y mutation, residues 319–541 (Mu) | This paper | N/A |
| Omicron BA.1 RBD, residues 319-541 | This paper | GISAID: EPI_ISL_6640916 |
| Omicron BA.2 RBD, residues 319-541 | This paper | GISAID: EPI_ISL_9652748 |
| Omicron BA.2 RBD with L452Q, residues 319–541 (BA.2.12.1) | This paper | N/A |
| Omicron BA.2 RBD with D339H, G446S, N460K, R493Q, residues 319–541 (BA.2.75) | This paper | N/A |
| Omicron BA.3 RBD, residues 319-541 | This paper | GISAID: EPI_ISL_7605589 |
| Omicron BA.4/5 RBD, residues 319-541 | This paper | GISAID: EPI_ISL_12029894 |
| SARS-CoV RBD, residues 306-527 | This paper | GenBank: NC_004718 |
| RaTG13 RBD, residues 319-541 | This paper | GenBank: QHR63300.2 |
| RshSTT182 RBD, residues 319-541 | This paper | GISAID: EPI_ISL_852604 |
| RacCS203 RBD, residues 319-541 | This paper | GenBank: QQM18864.1 |
| Rc-o319 RBD, residues 319-541 | This paper | GenBank: BCG66627.1 |
| RsYN04 RBD, residues 309-527 | This paper | GISAID: EPI_ISL_1699444 |
| GX/P2V/2017 RBD, residues 319-541 | This paper | GISAID: EPI_ISL_410542 |
| GD/1/2019 RBD, residues 319-541 | This paper | GISAID: EPI_ISL_410721 |
| Software and algorithms | ||
| FlowJo V10 | FLOWJO | https://www.flowjo.com/solutions/flowjo/downloads |
| Graphpad Prism 7 | GraphPad Software | https://www.graphpad.com/ |
| BIAcore® 8K Evaluation software | GE Healthcare | N/A |
| PKSolver | Zhang et al.41 | N/A |
| HKL2000 | Otwinowski et al.42 | https://www.hkl-xray.com/hkl-2000 |
| Phaser | Read R.J. et al.43 | N/A |
| COOT | Emsley et al.44 | http://www.mrc-lmb.cam.ac.uk/personal/peemsley/coot/ |
| Phenix | Adams et al.45 | http://www.phenix-online.org/ |
| MolProbity | Williams et al.46 | http://molprobity.biochem.duke.edu/index.php |
| PyMOL software | Molecular Graphics System, Version 1.8 Schrö dinger | https://pymol.org/2/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Qihui Wang (wangqihui@im.ac.cn).
Materials availability
All unique/stable reagents generated in this study are available from the lead contact with a completed Materials Transfer Agreement.
Experimental model and subject details
Cells
HEK293T cells (ATCC CRL-3216), Vero cells (ATCC CCL81), and BHK-21 cells (ATCC CCL-10) were maintained in Dulbecco’s Modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FBS) and cultured at 37°C in a humidified incubator with 5% CO2. Freestyle 293F cells were cultured in SMM 293-TII medium at 37°C in a shaker with 5% CO2. Sf9 (Invitrogen) and Hi5 (Invitrogen) insect cells were cultured in Insect-XPRESS medium (Lonza, USA) at 28°C.
Viruses
The SARS-CoV-2 strain hCoV-19/China/CAS-B001/2020 (GISAID: EPI_ISL_514256-7) was from the Institute of Microbiology, Chinese Academy of Sciences (IMCAS). The SARS-CoV-2 strains BetaCoV/Wuhan/IVDC-HB-envF13/2020 (GISAID: EPI_ISL_408511), Delta (NPRC 2.192100004), BA.1 (NPRC 2.192100007), BA.2 (NPRC 2.192100010), BA.4 (NPRC 2.192100012), and BA.5 (NPRC 2.192100014) were isolated by the National Institute for Virus Disease Control and Prevention (Chinese Center for Disease Control and Prevention, China CDC).
Animals
Specific pathogen-free (SPF) female BALB/c mice were purchased from Vital River (Beijing, China). SPF female K18-hACE2 mice (6–8 weeks) were purchased from GemPharmatech Co., Ltd (Jiangsu, China). Animal experiments were approved by the Animal Ethics Committee [Ethical approval No. 2017(55)] at Shanxi Agricultural University (Taigu, Shanxi, China), the Ethics Committee of IMCAS (Ethical approval No. APIMCAS2021091) and the Ethics Committee of China CDC (Ethical approval No. 20211124088 and No. 202205160054). The animal experiments with SARS-CoV-2 challenge were conducted in an Animal Biosafety Level 3 (ABSL3) facility at IMCAS or China CDC.
Method details
Protein expression
The coding sequences of the SARS-CoV-2 RBD (including prototype, Omicron sub-variant BA.2.12.1, BA.3 and BA.4/5) and S, as well as nanobodies with a C-terminal His-tag were cloned into the pCAGGS vector, respectively. The coding sequences of RBDs with human IgG Fc-tag (hFc) were also cloned into the pCAGGS vector, including prototype, Alpha, Beta, Gamma, Kappa, Delta, Delta plus, Lambda, and Mu, as well as eight sarbecoviruses (SARS-CoV, RaTG13, RshSTT182, RacCS203, Rc-o319, RsYN04, GX/P2V/2017, and GD/1/2019). The recombinant plasmids were transfected into Freestyle 293F cells to express the SARS-CoV-2 RBD, S, or nanobody proteins. After 5 days, the supernatants were collected, and soluble protein was purified by Ni affinity chromatography using a HisTrap EXCEL 5-mL column (GE Healthcare). The protein was further purified via gel filtration chromatography with a Superdex 200 column (GE Healthcare) or Superose 6 Increase 10/300 GL column (GE Healthcare) in a buffer composed of 20 mM Tris-HCl (pH 8.0) and 150 mM NaCl. The hFc-tagged SARS-CoV-2 RBD was used in blocking or SPR assays.
The coding sequences of the Omicron sub-variant BA.1 and BA.2 RBD with a C-terminal His-tag were cloned into the baculovirus transfer pFastbac1 vector (Invitrogen) and subsequently transformed into Escherichia coli DH10Bac competent cells to generate the recombinant bacmids. Transfection of bacmids and virus amplification were conducted in Sf9 cells, while Hi5 cells were used for protein expression. The supernatants of Hi5 cells were collected 48 h post-infection, and soluble proteins were purified by metal affinity chromatography using a HisTrap HP 5 mL column (GE Healthcare) and Superdex 200 column (GE Healthcare) in a buffer composed of 20 mM Tris-HCl (pH 8.0) and 150 mM NaCl. The His-tagged RBDs from Omicron BA.1 and BA.2 were used in SPR assays.
Alpaca immunization and generation of nanobodies
Alpaca immunization was handled according to the alpaca guidelines approved by the Animal Ethics Committee [2017(55)] at Shanxi Agricultural University (Taigu, Shanxi, China). One alpaca (female, 3 years old) was subcutaneously immunized with 200 μg of SARS-CoV-2 RBD on days 0, 14, 28, and 42, and another (female, 3 years old) was immunized with 200 μg of SARS-CoV-2 S on days 0, 7, 28, 42, and 56. Blood was collected 1 week after the last immunization for the preparation of lymphocytes. Nanobodies were generated by phage display technology following a general protocol as described previously.47 Different single colonies were randomly selected from each round of panning and tested by phage ELISA. The positive clones (OD450nm > 0.2) were sequenced to obtain the sequences of the nanobodies. Then, the coding sequence of the nanobodies with a C-terminal His-tag were cloned into the pCAGGS vector. The recombinant nanobodies were expressed in Freestyle 293F cells and purified as previously described.
Flow cytometry assay
The binding between nanobodies and the SARS-CoV-2 S was first evaluated by flow cytometry. Briefly, pCAGGS vectors containing the SARS-CoV-2 S or SARS-CoV S fused to GFP were transfected into BHK-21 cells. 72 h later, 2×105 cells were harvested, resuspended in PBS, and incubated with the supernatants containing His-tagged R14 or S43 protein at 37°C for 30 min. Cells were then washed twice and stained with anti-His/APC antibodies (Miltenyi) for 30 min at 37°C. The cells incubated with a nanobody (named R67 that was determined in our lab previously) cross-binding to SARS-CoV-2 and SARS-CoV were used as positive controls, and the cells only incubated with the secondary antibody were used as the negative control (NC).
The ability of nanobodies to block the binding between the SARS-CoV-2 RBD and human ACE2 (hACE2) was also evaluated by flow cytometry. Briefly, pEGFP-N1 vectors containing hACE2 were transfected into BHK-21 cells and incubated for 48 h. Then, 0.5 μg of hFc-tagged SARS-CoV-2 RBD protein was mixed with 2.5 μg of nanobodies at 4°C for 30 min. The mixtures were added to BHK-21 cells (2×105) and incubated at 37°C for 1 h. The cells were washed with PBS three times and then stained with anti-hFc/APC antibodies (BioLegend). The cells incubated with only the secondary antibody were used as blank control, and the cells incubated with the SARS-CoV-2 RBD with an irrelevant nanobody (R23) were used as negative control.
Engineering and generation of dimer, homo-trimer, and IgM versions of antibodies
The dimer constructs (DR14 and DS43) were engineered via fusing nanobody to hFc. The homo-trimer versions of R14 and S43 (TR14 and TS43) were constructed via head-to-tail with (GGGGS)3 linkers and a C-terminal His-tag. The coding sequence of R14 or S43 fused to the Fc of human IgM was cloned into the pCAGGS vector to generate IgM versions of the antibodies (MR14 and MS43). The coding sequence of TR14 was synthesized by Nanjing GenScript Biotech Co., Ltd., and TS43 was synthesized by Tsingke Biotechnology Co., Ltd. DR14 and DS43 were expressed in Freestyle 293F cells and were purified using a HiTrap Protein A 5-mL column (GE Healthcare) and Superdex 200 column (GE Healthcare). TR14 and TS43 were expressed in Freestyle 293F cells and were purified using a HisTrap EXCEL 5-mL column (GE Healthcare) and Superdex 200 column (GE Healthcare). The recombinant plasmid for MR14 or MS43 and a human J-chain expressing vector were co-transfected into Freestyle 293F cells to express the MR14 or MS43 proteins, which were further purified by HiTrap™ IgM Purification HP (GE Healthcare) and Superose 6 Increase 10/300 GL (GE Healthcare) chromatography.
Surface plasmon resonance (SPR) assay
The binding affinities of R14 or S43 with RBDs from SARS-CoV-2 and its variants (Alpha, Beta, Gamma, Kappa, Delta, Delta plus, Lambda, and Mu), as well as eight sarbecoviruses (SARS-CoV, RaTG13, RshSTT182, RacCS203, Rc-o319, RsYN04, GX/P2V/2017, and GD/1/2019) were evaluated by SPR analysis. The experiments were performed using a BIAcore8000 system (GE Healthcare) with protein A chips (Cytiva Life Sciences) at 25°C in single-cycle mode. All proteins used for kinetic analysis were exchanged into the PBST buffer (2.7 mM KCl, 137 mM NaCl, 4.3 mM Na2HPO4, 1.4 mM KH2PO4, and 0.005% (v/v) Tween 20). The supernatants containing hFc-tagged RBD proteins were captured by the protein A chip at more than 400 response units. Then, serial dilutions of His-tagged R14 or S43 were injected over the chip surface to test binding. After each cycle, regeneration of the sensor chips was performed using Glycine (pH 1.7).
To test the binding between multivalent nanobodies and RBD, biotinylated monomer, homo-trimer and IgM versions of R14 or S43 were loaded onto a preconditioned Series S Sensor Chip SA chip (Cytiva Life Sciences), and 2-fold serial dilutions of His-tagged RBDs of SASR-CoV-2 prototype, Omicron BA.1, BA.2, BA.2.12.1, BA.3, and BA.4/5 strains were flowed over the chip surface to test binding. The ka, kd, KD, and the parameters (Chi2 value and Rmax) values for each pair of interactions was calculated with BIAcore 8K evaluation software (GE Healthcare). Figures were prepared using OriginPro 9.1.
Pseudotyped virus neutralization
SARS-CoV-2 pseudotyped virus was prepared as previously described.48 In brief, HEK293T cells expressing the S proteins of SARS-CoV-2 prototype or its variants were infected with the rVSV-ΔG virus. 30 h later, the supernatants containing the indicated pseudotyped viruses were sequentially collected, centrifuged, and filtered through 0.45 μm sterilized membranes. For the neutralization assay, 1×104 of Vero cells were plated into each well of a 96-well plate 24 h before infection. 5-fold serial dilutions of nanobodies were incubated with an equal volume of supernatant containing 1,000 fluorescence focus units (FFU) of pseudotyped virus for 1 h at 37°C. Then, the mixture was added to the Vero cells in triplicate. Fifteen hours later, the infected cells were measured using CQ1 Confocal Quantitative Image Cytometer (Yokogawa). Nanobody R23 represents the negative control. The half maximal inhibitory concentration (IC50) was calculated using GraphPad Prism 7.0.
Live SARS-CoV-2 virus neutralization
Live virus neutralization assays were performed based on the cytopathic effect (CPE). The monomer, homo-trimer, and IgM versions of the nanobody were serially diluted to 50 μL and incubated with an equal volume of 100 TCID50 of SARS-CoV-2 for 1 h at 37°C. The mixtures were then added to Vero cells in quadruplicate and incubated for 3 days at 37°C. CPE was observed and recorded on day 4 after infection. The IC50 values of the nanobody was calculated using GraphPad Prism 7.0. All experiments were performed in the BSL3 facility of China CDC.
Negative-stain assay by transmission electron microscopy (TEM)
S protein and nanobody were incubated on ice for 48 h. The protein solution was then diluted to an appropriate concentration with PBS, and 10 μL of the sample was placed onto a glow discharged grid. After 1 min, excess sample was blotted away by touching the edge of the grid with a small wedge of filter paper. Then, the grid was quickly touched to the surface of the drop of ddH2O and one edge of the grid was rapidly touched to a wedge of filter paper again to wick away excess ddH2O. Finally, the grid was stained with 1% uranyl acetate, and TEM was performed with a JEM-1400 instrument (JEDL, Japan) at an acceleration voltage of 80 kV.
Nanobody aerosolization
The whole-body exposure platform was designed by Dr. Jingkun Jiang’s group from Tsinghua University. This whole-body exposure system mainly contains an Aerogen Solo vibrating mesh nebulizer (Aerogen Inc., Chicago, USA), an exposure chamber, and a filter (HEPA). The size distribution at six monitoring points (red triangles in Figure 3A) on the middle cross section were measured using a scanning mobility particle spectrometer (SMPS) that contains an electrostatic classifier (Model 3081; TSI Inc., Shoreview, USA), a soft X-ray neutralizer (Model 3088; TSI Inc.), a Differential Mobility Analyzer (Model 3081A; TSI Inc.), and a condensation particle counter (Model 3772; TSI Inc.). To determine the proper flowrate of the carrier air, the aerosol mass concentration at the central point of the exposure chamber was monitored using a Dusttrak (Model 8533, TSI Inc.). To test the neutralizing activity of post nebulization-nanobodies, an all-glass SKC (Eighty Four, PA, USA) containing 20 mL of PBS was used to collect the post-nebulized nanobodies, and neutralization assays were performed as described previously.
The inhaled dose was determined via analysis of bronchoalveolar lavage fluid of mice. An ELISA assay was performed to determine the concentrations of nanobodies in the lavage fluid. SARS-CoV-2 RBD was coated on the plate with 500 ng/well at 4°C overnight. After blocking with 5% skim milk at room temperature (RT) for 1 h, 50 μL of 1% diluted lavage fluid was added to the plates and incubated for 1 h at RT; R14 or S43 dissolved in the 1% diluted lavage fluid from a mouse without any administration was used to generate a standard curve. After washing, HRP-conjugated antibody was added and incubated at RT for 1 h. After washing, the TMB substrate was added, and 2 M H2SO4 was used to stop the reaction. The plate was read on a TECAN multimode plate reader (Infinite 200 PRO), and data were analyzed.
Animal experiments with SARS-CoV-2 challenge
For the prototyped SARS-CoV-2 challenge, BALB/c mice model transduced intranasally with a recombinant adenovirus 5 expressing human ACE2 (Ad5-hACE2) was used. Female SPF BALB/c mice (6–8 weeks) were purchased from Vital River (Beijing, China). Mice were intranasally (i.n.) transduced with 8×109 vp of Ad5-hACE2 for the generation of a mouse model of SARS-CoV-2 infection as previously reported.49 After 5 days, the mice were infected with 5× 105 TCID50 of SARS-CoV-2 via the i.n. route.
Intranasal administration of R14 and S43 at 5 mg/kg performed in the ABSL-3 Laboratory of IMCAS. The mice were given R14 or S43 nanobody via the i.n. route 6 h prior to SARS-CoV-2 infection. Then, the mice were euthanized and necropsied 5 days after SARS-CoV-2 challenge.
Inhalational administration of R14 for prophylactic and therapeutic experiments was performed in the ABSL-3 Laboratory of IMCAS and China CDC, respectively. For the prophylactic experiments, mice were given R14 or S43 nanobody via the i.n. or i.h. route 6 h prior to SARS-CoV-2 infection. PBS was used as a control. Nanobodies dissolved in 50 μL of PBS were administered via the i.n. route, whereas for i.h. delivery, mice were exposed in the whole-body exposure system for 10 min to inhale aerosols containing nanobodies. For the therapeutic experiments, mice were given R14 nanobody via the i.h. route as previously described. Mice were exposed in the whole-body exposure system for 30 min to inhale aerosols containing R14 at the indicated time post SARS-CoV-2 infection (10 min, 1.5 h, and 7 h). PBS was used as the control. Mice were euthanized and necropsied 5 days after SARS-CoV-2 challenge.
IN and i.p. administration of MR14 was performed in the ABSL-3 Laboratory of China CDC. Female SPF K18-hACE2 mice (6–8 weeks) were purchased from GemPharmatech Co., Ltd (Jiangsu, China). For the prophylactic experiments, mice were given 5 mg/kg of MR14 via the IN or i.p. route 6 h prior to BA.2 infection. For the therapeutic experiments, mice were given 5 mg/kg of MR14 via the IN route at indicated time post BA.2 infection (6, 30, and 54 h), but only a single dose of MR14 was administrated via i.p. 6 h post infection. PBS was used as a control. The mice were euthanized and necropsied 3 days after BA.2 challenge.
Lung tissues were harvested for virus titer and pathological examination. Viral RNAs were extracted using a Qiagen 52,906 QIAamp Viral RNA Mini Kit (Qiagen) following the manufacturer’s instructions. Virus titers were determined using a One Step Prime Script RT-PCR kit (TaKaRa). Graphs were generated using GraphPad Prism version 7.
Histopathology and immunohistochemistry
The lungs were fixed in 4% (v/v) paraformaldehyde solution for 72 h, and paraffin sections (3–4 μm) were prepared routinely. Hematoxylin and eosin staining was used to identify histopathological changes in the lungs. The histopathology of the lung tissue was observed by light microscopy.
Pharmacokinetic studies
Female SPF BALB/c mice (6–8 weeks) were purchased from Vital River (Beijing, China). For pharmacokinetic studies by i.p. injection, four groups of mice (three mice per group) were used. Briefly, mice were anesthetized with isoflurane, and 200 μL of R14, TR14, or MR14 at 2 mg/mL (total dose 20 mg/kg) was slowly injected. Serum samples were collected by retro-orbital puncture at 1, 6, 10, 24, 48, 72, and 96 h post injection. The concentrations of antibodies in serum were determined by ELISA assays. The half-time (t1/2) of the nanobodies was calculated using PKSolver.41
For pharmacokinetic studies by IH delivery, mice were anesthetized with tribromoethanol via i.p. injection and then received aerosolized R14, TR14, or MR14 continuously for 10 min. Bronchoalveolar lavage fluids were collected at 0, 1, 6, and 24 h after aerosol inhalation. The antibody concentration in the bronchoalveolar lavage fluid was also determined by ELISA assays, and the half-time (t1/2) of the nanobodies in the lungs was calculated using PKSolver.41
For ELISA assays to test his-tagged R14 and TR14, 96-well ELISA plates (Corning Costar, Cat# 3590) were coated with 500 ng of SARS-CoV-2 RBD-hFc overnight. After blocking with 5% skim milk at room temperature for 1 h and washing in PBS + Tween 20, 2% mouse plasma (or bronchoalveolar lavage fluids) diluted in PBS was added and incubated for 1h at room temperature. After washing, mouse anti-his mAb (ZSGB-BIO, Cat# TA-02, 1:2,000 diluted) was added and the plates were incubated for 1 h at room temperature. After washing, goat anti-mouse IgG (H&L)-HRP conjugated (Easybio, Cat# BE0102-100, 1:3,000 diluted) was added and the plates were incubated for 1 h at room temperature. After washing, 100 μL of TMB Substrate Solutions (Beyotime, Cat# P0209-500 mL) were added and incubated for around 20 min. Following addition of 50 μL of 2M H2SO4, plates were read on the Infinite 200 PRO microplate plate reader (Tecan). Sample concentrations were calculated according to the standard curve.
For ELISA assays to test MR14, 96-well ELISA plates (Corning Costar, Cat# 3590) were coated with 500 ng of his-tagged SARS-CoV-2 RBD overnight. After blocking with 5% skim milk at room temperature for 1 h and washing in PBS + Tween 20, 2% mouse plasma (or bronchoalveolar lavage fluids) diluted in PBS was added and incubated for 1h at room temperature. After washing, Anti-Human IgM antibody (Abcam, Cat# ab200541, 1:200 diluted) was added and the plates were incubated for 1 h at room temperature. After washing, goat anti-mouse IgG (H&L)-HRP conjugated (Easybio, Cat# BE0102-100, 1:3,000 diluted) was added and the plates were incubated for 1 h at room temperature. After washing, 100 μL of TMB Substrate Solutions (Beyotime, Cat# P0209-500 mL) were added and incubated for around 20 min. Following addition of 50 μL of 2M H2SO4, plates were read on the Infinite 200 PRO microplate plate reader (Tecan). Sample concentrations were calculated according to the standard curve.
Crystal screening and structure determination
The crystallization of the nanobody/SARS-CoV-2 RBD complex was performed using the vapor-diffusion sitting-drop method with 0.8 μL of protein and 0.8 μL of reservoir solution at 18°C. High-resolution crystals of the SARS-CoV-2 RBD/R14 complex were obtained in a solution consisting of 0.10% (w/v) n-Octyl-b-D-glucoside, 0.1 M sodium citrate tribasic dihydrate (pH 5.5), and 22% (w/v) polyethylene glycol 3,350 at a protein concentration of 5 mg/mL. The crystals of the SARS-CoV-2 RBD/S43 complex were obtained in a solution consisting of 0.2 M lithium sulfate, 0.1 M Tris (pH 8.5), and 30% (w/v) PEG 4000 at a protein concentration of 10 mg/mL. Diffraction data were collected at the Shanghai Synchrotron Radiation Facility (SSRF) BL17U. The data were processed with HKL2000 software.42 The complex structures of the SARS-CoV-2 RBD with the nanobodies were determined by the molecular replacement method using Phaser43 with the previously reported SARS-CoV-2 RBD complex structure (PDB: 2AJF). The atomic models were built with Coot44 and refined with Phenix,45 and the stereochemical qualities of the final models were assessed with MolProbity.46 Data collection, processing, and refinement statistics are summarized in Table S1. All structural figures were generated using Pymol software (https://pymol.org/2/).
Quantification and statistical analysis
Binding analysis
KD values of SPR experiments were obtained with BIAcore 8K Evaluation software (GE Healthcare), using a 1:1 binding model. The values indicate the mean ± SD of three independent experiments.
Neutralization analysis
IC50 values of neutralization experiments were obtained using GraphPad Prism 7 software.
Acknowledgments
We thank Dr. Linhui Wang for generous support of pMES4, phage VSCM13, and E. coli TG1. We are grateful for the help from the staff of BSL-3 at the Institute of Microbiology, Chinese Academy of Sciences. We thank Dr. Xin Zhou from Yangzhou University for supporting our project. We thank Kun Xu (Beijing Institutes of Life Science, Chinese Academy of Sciences) for generous support of Ad5-hACE2. We thank Zheng Fan (Institutional Center for Shared Technologies and Facilities, Institute of Microbiology, Chinese Academy of Sciences) and Yuanyuan Chen (Institute of Biophysics, Chinese Academy of Sciences) for technical help with Biacore experiments. We thank Jingnan Liang (Institute of Microbiology, Chinese Academy of Sciences) for technical help with negative-stain experiments by TEM. We also thank Qingwen He, Xinxin Yan, Xiaoyun Wang, Anqi Zheng, Chunli Wu, Shihua Li, and Yunfei Jia (Institute of Microbiology, Chinese Academy of Sciences) for supporting our project. This work was supported by CAS Project for Young Scientists in Basic Research (YSBR-010); National Natural Science Foundation of China (81922044, 52091541, 82041047, and 82225021); Ministry of Science and Technology of the People’s Republic of China (2021YFC0863300 and 2022YFC2303403); Strategic Priority Research Program of the Chinese Academy of Sciences (XDB29040203); and Youth Innovation Promotion Association CAS (2018119).
Author contributions
Q.W., G.F.G., and H.L. designed this study. S.Q. and R.F. conducted the alpaca immunization. H.L. selected the two nanobodies R14 and S43 by phage display technology. H.L., B.L., and X.R. performed protein expression. H.L. and B.L. performed flow cytometry, SPR, and crystallization assays. H.L. and X.R. performed neutralization assay of pseudotyped virus. K.X. and W.L. performed neutralization assay of live virus in BSL-3 facility. J.J., J.D., D.W., and L. Wang designed the whole-body exposure system and detected aerosol distribution and mass concentration. H.L. and B.L. performed the bronchoalveolar lavage of murine lungs. With the help of W.J.L. and G.W., W.L. and K.X. performed animal experiments using live virus in the BSL-3 Laboratory of the Chinese Center for Disease Control and Prevention. With the help of Y.B., L. Wu and P.D. performed animal experiments using live virus in the BSL-3 Laboratory of the Institute of Microbiology, Chinese Academy of Sciences. J.Q. and C.S. collected the diffraction data and built structural model. Q.W. and L. Wu analyzed the complex structure. H.C. and X.Z. analyzed the mutations of multiple variants. H.L., L. Wu, and P.D. wrote the manuscript. Q.W., G.F.G., J. J., and Q.C. revised the manuscript with inputs from other co-authors.
Declaration of interests
Q.W., G.F.G., H.L., and L. Wu are listed as inventors on patent applications for R14 and S43 based antiviral treatment. Q.W., G.F.G., H.L., B.L., and L. Wu are listed as inventors on patent applications for MR14 and MS43 based antiviral treatment.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: January 12, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2023.100918.
Contributor Information
Ruiwen Fan, Email: ruiwenfan@163.com.
Jingkun Jiang, Email: jiangjk@tsinghua.edu.cn.
Guizhen Wu, Email: wugz@ivdc.chinacdc.cn.
George F. Gao, Email: gaof@im.ac.cn.
Qihui Wang, Email: wangqihui@im.ac.cn.
Supplemental information
Data and code availability
The atomic coordinates for the complex of the SARS-CoV-2 RBD with R14 or S43 have been deposited in the PDB (www.rcsb.org). The PDB IDs are 7WD1 and 7WD2. This paper does not report the original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Gao G.F. From "A"IV to "Z"IKV: attacks from emerging and re-emerging pathogens. Cell. 2018;172:1157–1159. doi: 10.1016/j.cell.2018.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu Z., Liu K., Gao G.F. Omicron variant of SARS-CoV-2 imposes a new challenge for the global public health. Biosaf. Health. 2022;4:147–149. doi: 10.1016/j.bsheal.2022.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Du P., Gao G.F., Wang Q. The mysterious origins of the Omicron variant of SARS-CoV-2. Innovation. 2022;3:100206. doi: 10.1016/j.xinn.2022.100206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang M., Wu L., Zheng A., Xie Y., He Q., Rong X., Han P., Du P., Han P., Zhang Z., et al. Atlas of currently available human neutralizing antibodies against SARS-CoV-2 and escape by Omicron sub-variants BA.1/BA.1.1/BA.2/BA.3. Immunity. 2022;55:1501–1514.e3. doi: 10.1016/j.immuni.2022.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao Y., Yisimayi A., Jian F., Song W., Xiao T., Wang L., Du S., Wang J., Li Q., Chen X., et al. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature. 2022;608:593–602. doi: 10.1038/s41586-022-04980-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Niu S., Wang J., Bai B., Wu L., Zheng A., Chen Q., Du P., Han P., Zhang Y., Jia Y., et al. Molecular basis of cross-species ACE2 interactions with SARS-CoV-2-like viruses of pangolin origin. EMBO J. 2021;40:e107786. doi: 10.15252/embj.2021107786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu K., Pan X., Li L., Yu F., Zheng A., Du P., Han P., Meng Y., Zhang Y., Wu L., et al. Binding and molecular basis of the bat coronavirus RaTG13 virus to ACE2 in humans and other species. Cell. 2021;184:3438–3451.e10. doi: 10.1016/j.cell.2021.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao K., Zhai J., Feng Y., Zhou N., Zhang X., Zou J.J., Li N., Guo Y., Li X., Shen X., et al. Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature. 2020;583:286–289. doi: 10.1038/s41586-020-2313-x. [DOI] [PubMed] [Google Scholar]
- 11.Wang Q., Chen H., Shi Y., Hughes A.C., Liu W.J., Jiang J., Gao G.F., Xue Y., Tong Y. Tracing the origins of SARS-CoV-2: lessons learned from the past. Cell Res. 2021;31:1139–1141. doi: 10.1038/s41422-021-00575-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lam T.T.Y., Jia N., Zhang Y.W., Shum M.H.H., Jiang J.F., Zhu H.C., Tong Y.G., Shi Y.X., Ni X.B., Liao Y.S., et al. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature. 2020;583:282–285. doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- 13.Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.Y., et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904.e9. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- 15.Wu Y., Wang F., Shen C., Peng W., Li D., Zhao C., et al. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science. 2020;368:1274–1278. doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Earle K.A., Ambrosino D.M., Fiore-Gartland A., Goldblatt D., Gilbert P.B., Siber G.R., Dull P., Plotkin S.A. Evidence for antibody as a protective correlate for COVID-19 vaccines. Vaccine. 2021;39:4423–4428. doi: 10.1016/j.vaccine.2021.05.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forchette L., Sebastian W., Liu T. A comprehensive review of COVID-19 virology, vaccines, variants, and therapeutics. Curr. Med. Sci. 2021;41:1037–1051. doi: 10.1007/s11596-021-2395-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guilleminault L., Azzopardi N., Arnoult C., Sobilo J., Hervé V., Montharu J., Guillon A., Andres C., Herault O., Le Pape A., et al. Fate of inhaled monoclonal antibodies after the deposition of aerosolized particles in the respiratory system. J. Control. Release. 2014;196:344–354. doi: 10.1016/j.jconrel.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 19.Respaud R., Vecellio L., Diot P., Heuzé-Vourc'h N. Nebulization as a delivery method for mAbs in respiratory diseases. Expert Opin. Drug Deliv. 2015;12:1027–1039. doi: 10.1517/17425247.2015.999039. [DOI] [PubMed] [Google Scholar]
- 20.Jovčevska I., Muyldermans S. The therapeutic potential of nanobodies. BioDrugs. 2020;34:11–26. doi: 10.1007/s40259-019-00392-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiang Y., Nambulli S., Xiao Z., Liu H., Sang Z., Duprex W.P., Schneidman-Duhovny D., Zhang C., Shi Y. Versatile and multivalent nanobodies efficiently neutralize SARS-CoV-2. Science. 2020;370:1479–1484. doi: 10.1126/science.abe4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu X., Cheng L., Fu M., Huang B., Zhu L., Xu S., Shi H., Zhang D., Yuan H., Nawaz W., et al. A potent bispecific nanobody protects hACE2 mice against SARS-CoV-2 infection via intranasal administration. Cell Rep. 2021;37:109869. doi: 10.1016/j.celrep.2021.109869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li T., Cai H., Yao H., Zhou B., Zhang N., van Vlissingen M.F., Kuiken T., Han W., GeurtsvanKessel C.H., Gong Y., et al. A synthetic nanobody targeting RBD protects hamsters from SARS-CoV-2 infection. Nat. Commun. 2021;12:4635. doi: 10.1038/s41467-021-24905-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wrapp D., De Vlieger D., Corbett K.S., Torres G.M., Wang N., Van Breedam W., Roose K., van Schie L., VIB-CMB COVID-19 Response Team. Hoffmann M., et al. Structural basis for potent neutralization of betacoronaviruses by single-domain camelid antibodies. Cell. 2020;181:1436–1441. doi: 10.1016/j.cell.2020.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schoof M., Faust B., Saunders R.A., Sangwan S., Rezelj V., Hoppe N., Boone M., Billesbølle C.B., Puchades C., Azumaya C.M., et al. An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike. Science. 2020;370:1473–1479. doi: 10.1126/science.abe3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Z., Wang Y., Jin Y., Zhu Y., Wu Y., Li C., Kong Y., Song W., Tian X., Zhan W., et al. A non-ACE2 competing human single-domain antibody confers broad neutralization against SARS-CoV-2 and circulating variants. Signal Transduct. Target. Ther. 2021;6:378. doi: 10.1038/s41392-021-00810-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ku Z., Xie X., Hinton P.R., Liu X., Ye X., Muruato A.E., Ng D.C., Biswas S., Zou J., Liu Y., et al. Nasal delivery of an IgM offers broad protection from SARS-CoV-2 variants. Nature. 2021;595:718–723. doi: 10.1038/s41586-021-03673-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.RESPIRE study group. Piedra P.A., Martinon-Torres F., Szymanski H., Brackeva B., Dombrecht E., Detalle L., Fleurinck C., Cunningham S., Piedra P.A., et al. Nebulised ALX-0171 for respiratory syncytial virus lower respiratory tract infection in hospitalised children: a double-blind, randomised, placebo-controlled, phase 2b trial. Lancet Respir. Med. 2021;9:21–32. doi: 10.1016/S2213-2600(20)30320-9. [DOI] [PubMed] [Google Scholar]
- 29.Hastie K.M., Li H., Bedinger D., Schendel S.L., Dennison S.M., Li K., Rayaprolu V., Yu X., Mann C., Zandonatti M., et al. Defining variant-resistant epitopes targeted by SARS-CoV-2 antibodies: a global consortium study. Science. 2021;374:472–478. doi: 10.1126/science.abh2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piccoli L., Park Y.J., Tortorici M.A., Czudnochowski N., Walls A.C., Beltramello M., Silacci-Fregni C., Pinto D., Rosen L.E., Bowen J.E., et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020;183:1024–1042.e21. doi: 10.1016/j.cell.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou D., Duyvesteyn H.M.E., Chen C.P., Huang C.G., Chen T.H., Shih S.R., Lin Y.C., Cheng C.Y., Cheng S.H., Huang Y.C., et al. Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient. Nat. Struct. Mol. Biol. 2020;27:950–958. doi: 10.1038/s41594-020-0480-y. [DOI] [PubMed] [Google Scholar]
- 32.Yuan M., Wu N.C., Zhu X., Lee C.-C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cui Z., Liu P., Wang N., Wang L., Fan K., Zhu Q., Wang K., Chen R., Feng R., Jia Z., et al. Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron. Cell. 2022;185:860–871.e13. doi: 10.1016/j.cell.2022.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ren S.Y., Wang W.B., Gao R.D., Zhou A.M. Omicron variant (B.1.1.529) of SARS-CoV-2: mutation, infectivity, transmission, and vaccine resistance. World J. Clin. Cases. 2022;10:1–11. doi: 10.12998/wjcc.v10.i1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao X., Li D., Ruan W., Chen Z., Zhang R., Zheng A., Qiao S., Zheng X., Zhao Y., Dai L., et al. Effects of a prolonged booster interval on neutralization of Omicron variant. N. Engl. J. Med. 2022;386:894–896. doi: 10.1056/NEJMc2119426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ai J., Zhang H., Zhang Y., Lin K., Zhang Y., Wu J., Wan Y., Huang Y., Song J., Fu Z., et al. Omicron variant showed lower neutralizing sensitivity than other SARS-CoV-2 variants to immune sera elicited by vaccines after boost. Emerg. Microbes Infect. 2022;11:337–343. doi: 10.1080/22221751.2021.2022440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu K., Gao P., Liu S., Lu S., Lei W., Zheng T., Liu X., Xie Y., Zhao Z., Guo S., et al. Protective prototype-Beta and Delta-Omicron chimeric RBD-dimer vaccines against SARS-CoV-2. Cell. 2022;185:2265–2278.e14. doi: 10.1016/j.cell.2022.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Detalle L., Stohr T., Palomo C., Piedra P.A., Gilbert B.E., Mas V., Millar A., Power U.F., Stortelers C., Allosery K., et al. Generation and characterization of ALX-0171, a potent novel therapeutic nanobody for the treatment of respiratory syncytial virus infection. Antimicrob. Agents Chemother. 2016;60:6–13. doi: 10.1128/AAC.01802-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vollmers H.P., Brändlein S. Nature’s best weapons to fight cancer. Revival of human monoclonal IgM antibodies. Hum. Antibodies. 2002;11:131–142. [PubMed] [Google Scholar]
- 40.Willyard C. How antiviral pill molnupiravir shot ahead in the COVID drug hunt. Nature. 2021 doi: 10.1038/d41586-021-02783-1. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Y., Huo M., Zhou J., Xie S. PKSolver: an add-in program for pharmacokinetic and pharmacodynamic data analysis in Microsoft Excel. Comput. Methods Programs Biomed. 2010;99:306–314. doi: 10.1016/j.cmpb.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 42.Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 43.Read R.J. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr. D Biol. Crystallogr. 2001;57:1373–1382. doi: 10.1107/s0907444901012471. [DOI] [PubMed] [Google Scholar]
- 44.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 45.Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Williams C.J., Headd J.J., Moriarty N.W., Prisant M.G., Videau L.L., Deis L.N., Verma V., Keedy D.A., Hintze B.J., Chen V.B., et al. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 2018;27:293–315. doi: 10.1002/pro.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pardon E., Laeremans T., Triest S., Rasmussen S.G.F., Wohlkönig A., Ruf A., Muyldermans S., Hol W.G.J., Kobilka B.K., Steyaert J. A general protocol for the generation of nanobodies for structural biology. Nat. Protoc. 2014;9:674–693. doi: 10.1038/nprot.2014.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nie J., Li Q., Wu J., Zhao C., Hao H., Liu H., Zhang L., Nie L., Qin H., Wang M., et al. Quantification of SARS-CoV-2 neutralizing antibody by a pseudotyped virus-based assay. Nat. Protoc. 2020;15:3699–3715. doi: 10.1038/s41596-020-0394-5. [DOI] [PubMed] [Google Scholar]
- 49.Sun J., Zhuang Z., Zheng J., Li K., Wong R.L.Y., Liu D., Huang J., He J., Zhu A., Zhao J., et al. Generation of a broadly useful model for COVID-19 pathogenesis, vaccination, and treatment. Cell. 2020;182:734–743.e5. doi: 10.1016/j.cell.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates for the complex of the SARS-CoV-2 RBD with R14 or S43 have been deposited in the PDB (www.rcsb.org). The PDB IDs are 7WD1 and 7WD2. This paper does not report the original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.