Abstract
As geographical location can impact the gut microbiome, it is important to study region-specific microbiome signatures of various diseases. Therefore, we profiled the gut microbiome of breast cancer (BC) patients of the Midwestern region of the United States. The bacterial component of the gut microbiome was profiled utilizing 16S ribosomal RNA sequencing. Additionally, a gene pathway analysis was performed to assess the functional capabilities of the bacterial microbiome. Alpha diversity was not significantly different between BC and healthy controls (HC), however beta diversity revealed distinct clustering between the two groups at the species and genera level. Wilcoxon Rank Sum test revealed modulation of several gut bacteria in BC specifically reduced abundance of those linked with beneficial effects such as Faecalibacterium prausnitzii. Machine learning analysis confirmed the significance of several of the modulated bacteria found by the univariate analysis. The functional analysis showed a decreased abundance of SCFA (propionate) production in BC compared to HC. In conclusion, we observed gut dysbiosis in BC with the depletion of SCFA-producing gut bacteria suggesting their role in the pathobiology of breast cancer. Mechanistic understanding of gut bacterial dysbiosis in breast cancer could lead to refined prevention and treatment.
Subject terms: Cancer, Biomarkers, Diseases, Oncology, Microbiology, Microbial communities, Microbial genetics
Introduction
The global incidence rate of breast cancer has increased substantially since the 1980s, and this heterogenous disease now represents the most diagnosed cancer worldwide1. In the United States, breast cancer is responsible for nearly one-third of all cancers diagnosed in women2. There are numerous risk factors associated with breast cancer, including both environmental factors (e.g., reproductive history, hormone replacement therapy, obesity, etc.) and familial factors (e.g., family history of genetic mutations in BRCA1 and BRCA2, etc.)3–5. However, up to 50% of breast cancer cases cannot be attributed to these known risk factors6,7, suggesting that other, unknown factors can also lead to the development of breast cancer. Recently, research has focused on the interactions between the host microbiome and cancer, though the nature of these interactions remains elusive. Although specific bacterial species have been linked to some cancers, such as Helicobacter pylori with gastric cancer and Fusobacterium nucleatum with colorectal cancers4, there is no single bacterium linked with the pathobiology of breast cancer.
The human microbiome consists of many species of bacteria, viruses, fungi, and archaea, and estimates suggest that there are at least as many microbes in and on the human body as human cells8. These microbes exist in a complex relationship with the human host and are essential to homeostasis9. Bacteria represent the most abundant microorganism that inhabit the human host and interact with the host through manipulation of signaling pathways, hormone release, DNA double-strand breaks, apoptosis and senescence, and inflammation3,4,10. Dysbiosis, an atypical microbiome composition, has been correlated with many disease states, including cancer11. Evidence suggests that breast cancer patients have bacterial dysbiosis in both the breast microbiome3 and the gut microbiome3,5,10,12–15.
An association between microbial dysbiosis and breast cancer was reported as early as 1990 in a study that identified significantly different fecal microbial compositions of postmenopausal breast cancer patients (n = 11) compared to healthy controls (n = 7)12. More recently, studies have identified significantly different gut microbial compositions in breast cancer patients based on BMI or clinical cancer stage16–18. Goedert et al. demonstrated that postmenopausal breast cancer patients (n = 48) had a significantly lower alpha diversity compared to healthy controls (n = 48)19. In contrast, Zhu et al. observed that postmenopausal breast cancer patients (n = 44) had higher alpha diversity compared to postmenopausal healthy controls (n = 46)20. As alpha diversity observes community richness, these studies display contradicting results of the total types of microbes present. These studies show significant variability, but this heterogeneity is not surprising since many factors such as geographical location, weather conditions, population genetics, dietary habits, and green spaces strongly impact gut microbiome composition. Thus, to better understand the role of the microbiome in breast cancer, we need data from multiple geographical regions. Therefore, this study was undertaken to determine whether there is a gut dysbiosis in breast cancer patients from the Midwest region of the United States.
We recruited patients with breast cancer (BC) through the Breast Molecular Epidemiology Resource (BMER) of the Holden Comprehensive Cancer Center (HCCC) and healthy controls (HC) at the University of Iowa. In this pilot study, we report a significant difference in gut microbial composition in BC when compared to race-, body mass index (BMI)-, and sex-matched HC.
Results
The composition of the gut microbiome differs between patients with breast cancer and healthy controls
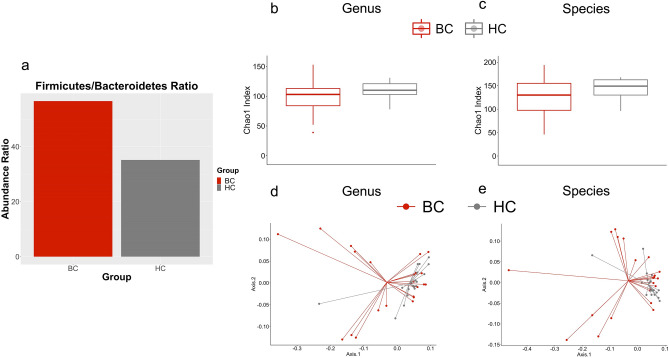
To observe the gut microbiome of the BC (n = 24) and HC (n = 23) cohorts, metagenomic sequencing of the V3-V4 region of 16S rRNA was utilized. After removing subjects with low-quality sequences and a precancerous patient, we had 22 BC patients and 19 HC for further analysis. First, the ratio between Firmicutes/Bacteroidetes was observed as it is considered a marker for dysbiosis21. This comparison was performed before filtering or normalization of feature abundances to observe the raw value differences. This ratio was not significantly different between the two cohorts (p value: 0.06241) (Fig. 1A). Next the differences between the groups at genera and species levels were analyzed.
Figure 1.
The composition of the gut microbiome differs between patients with breast cancer and healthy controls. (a) The Firmicutes/Bacteroidetes Ratio found in patients with BC and HC. The ratios are not significantly different (p = 0.06241). (b) The Chao1 Index was utilized to measure genera richness. This comparison was not significantly different between BC and HC (p = .111). (c) This measurement was also utilized to analyze the species level. There was not a significant difference between the two groups at the species level (p = .129). (d) The Weighted UniFrac distance metric was utilized to analyze beta diversity at the genus level and BC and HC significantly clustered (p = 0.011). (e) This metric was also utilized to analyze the species level and BC and HC again significantly clustered (p = 0.014).
In total, 519 species and 340 genera were identified. Of these 519 species, 61 were exclusively found in HC, and 81 were exclusively found in BC patients. Of these 340 genera, 28 were unique to HC, and 49 were unique to BC patients. After filtering, 114 species and 92 genera remained. Alpha diversity of the pre-normalized data was measured with the Chao1 Index; however, it was not significant at the species (p = 0.129) or genera (p = 0.111) level between BC and HC cohorts (Fig. 1B and C). Shannon diversity was also not significantly different between BC and HC at the species or genera level (p = 0.344, p = 0.414, respectively). Beta diversity was measured utilizing the Weighted UniFrac distance metric, which compares the microbiomes of each sample by assessing the quantity of the features present while also including the phylogenetic relationships between these features. Beta diversity was statistically significant at the species (p = 0.014) and genus (p = 0.011) levels, which can also be seen by the distinct clustering of BC and HC into separate groups at both levels (Fig. 1D and E).
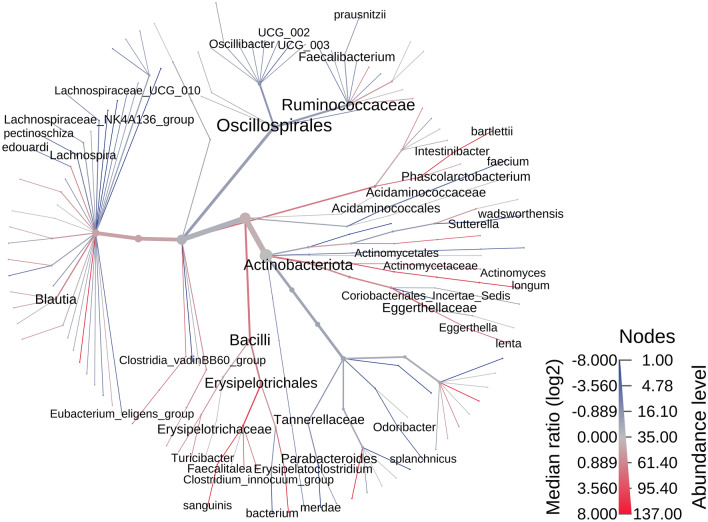
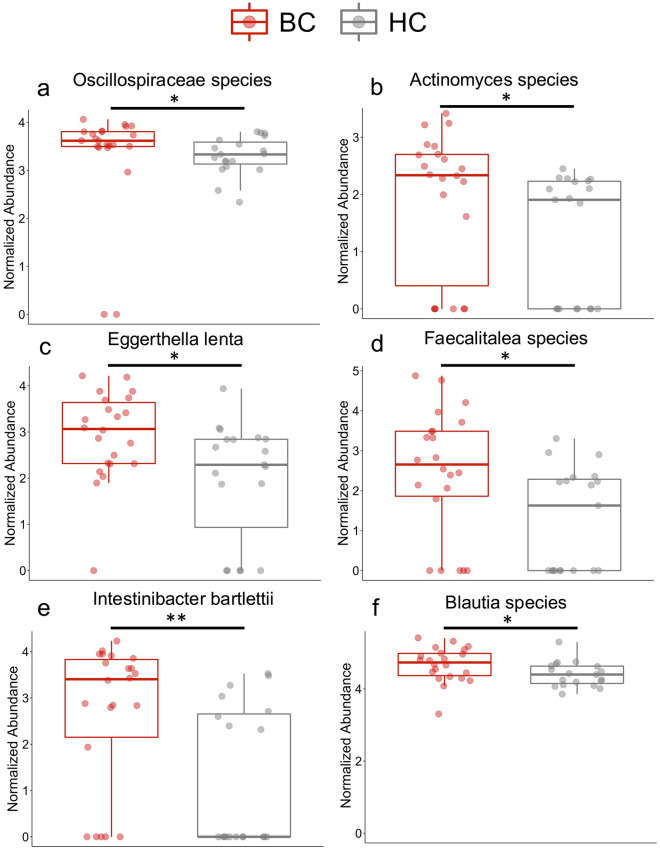
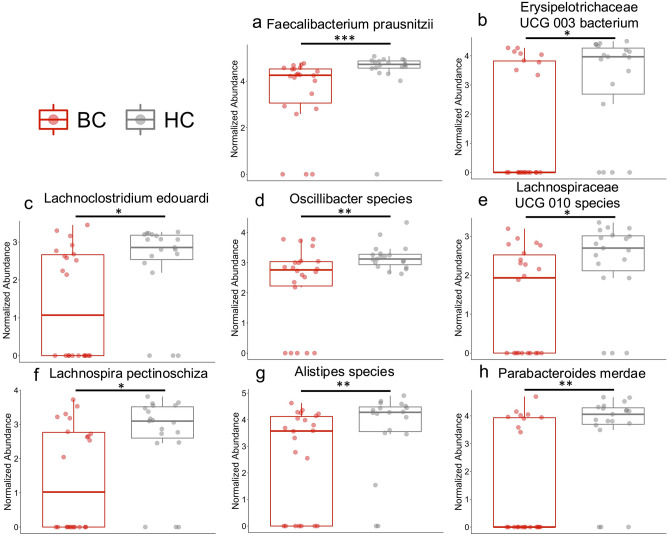
A heat tree was utilized for an overview of the fecal microbiome. This provides a visual representation of the bacterial features enriched or reduced/depleted between groups (Fig. 2). For an overall summary of the most abundant genera, family, and phylum, we included a stacked bar plot at each of these taxa levels in Supplementary Fig. 1. A closer look at these features revealed 16 species that were significantly different between BC and HC based on their normalized log abundance with the Wilcoxon signed rank sum test. Significant features had a p value ≤ 0.05 and an adjusted p value ≤ 0.15. The notable species that were more abundant in the BC cohort compared to the HC cohort include Oscillospiraceae species, Actinomyces species, Eggerthella lenta, Faecalitalea species, Intestinibacter bartlettii, and Blautia species (Fig. 3A–F). The species showing lower abundance in the BC cohort compared to the HC cohort include Faecalibacterium prausnitzii, Erysipelotrichaceae UCG 003 bacterium, Alistipes species, Oscillibacter species, Lachnospiraceae UCG 010 species, Lachnoclostridium edouardi, Lachnospira pectinoshiza, and Parabacteroides merdae (Fig. 4A–H). A full summary of these results can be found in Supplementary Table 1.
Figure 2.
Phylogenetic diversity differences between patients with breast cancer and healthy controls. This heat tree represents the phylogenetic differences between HC and BC. Red indicates a higher abundance in BC compared to HC and blue indicates a lower abundance in BC than HC. This heat tree was created using the MicrobiomeAnalyst web-based interface with updates as of spring 202278,79.
Figure 3.
Bacteria significantly increased in patients with breast cancer compared to healthy controls. (a–f) Based on the Wilcoxon test and the Benjamini–Hochberg procedure, 6 features were significantly higher in abundance in the breast cancer cohort compared to the healthy controls (p ≤ 0.05, q ≤ .15). Significance: * < 0.05 and ** < 0.01.
Figure 4.
Bacteria significantly decreased in patients with breast cancer compared to healthy controls. (a–h) Based on the Wilcoxon test and the Benjamini–Hochberg procedure, 8 features weresignificantly lower in abundance in the breast cancer cohort compared to the healthy controls (p ≤ 0.05, q ≤ 0.15). Significance: * < 0.05, ** < 0.01, and *** < 0.001.
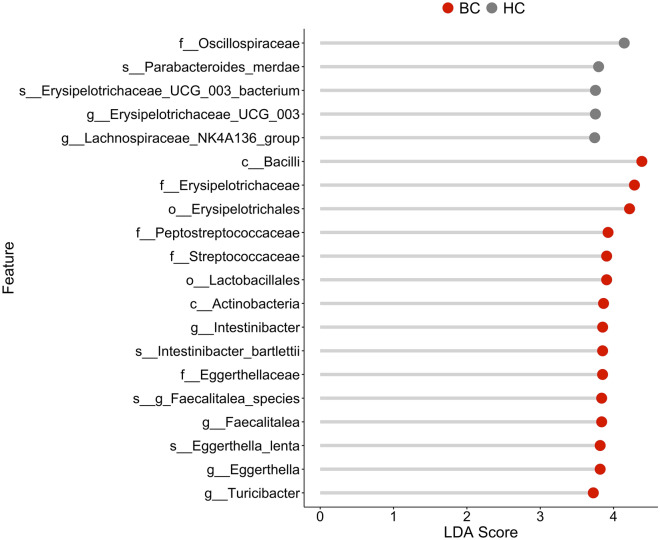
Lastly, we performed Linear discriminant analysis Effect Size (LEfSe) which identifies features distinguishing the two groups by combining statistical test with biological consistency and significance22. Using LEfSe, we observed five features with an LDA score of at least three in HC and 15 features with an LDA score of at least three in BC. All the species identified by LEfSe were also identified in the univariate test with the same relationship between HC and BC (Fig. 5).
Figure 5.
Distinguishing taxa between patients with breast cancer and healthy controls. Top 20 significant features selected by LEfSe analysis. The LDA score indicates the effect size of each feature.
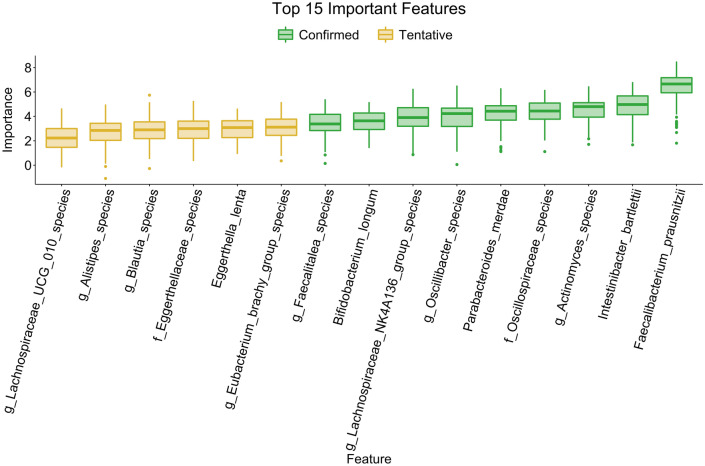
Random Forest identifies key species in differentiating between the microbiome of patients with breast cancer and healthy controls
The random forest machine learning algorithm was applied to assess the ability of the fecal microbiome to predict the BC phenotype. Specifically, a bootstrapped random forest of 500 trees was utilized to produce a predictive model based on BC and HC samples. The top 15 features important in identifying which cohort the sample is from are shown in Fig. 6. We then utilized the Boruta23 function in R with a significance level of 0.01 to identify the bacterial species important in differentiating between the BC and HC samples. Nine of these 15 features were significant, as seen in green. Seven of these nine significant species were also identified in our univariate analysis as significantly different. The species found to be lower in BC compared to HC in the univariate analysis and were also considered significant in the random forest analysis include Faecalibacterium prausnitzii, Parabacteroides merdae, and Oscillibacter species. The species that were higher in BC compared to HC in the univariate analysis that were also considered significant in the random forest analysis include Intestinibacter bartelli, Actinomyces species, Faecalitalea species, and Oscillospiraceae species. Two species were found to be significant based on the random forest analysis but not in the univariate test, Bifidobacterium longum and Lachnospiraceae NK4A136 group species.
Figure 6.
Random Forest identifies key species in differentiating between the microbiome of patients with breast cancer and healthy controls. The random forest machine learning algorithm was utilized to see if the fecal microbiome could differentiate between BC and HC. We utilized a bootstrapped random forest algorithm of 500 trees. Significance was based on the Boruta algorithm with a significance level of 0.01 highlighted in green.
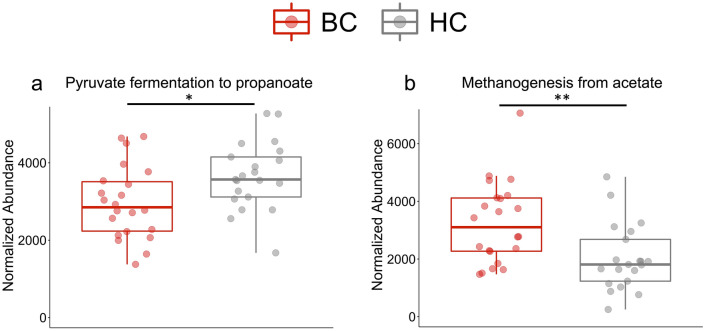
The functional profile of the gut microbiome differs between patients with breast cancer and healthy controls
To estimate the functional profile of the microbiome (metagenome) of our samples, PICRUSt2 (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States) was utilized24. This bioinformatics tool analyzed the metagenome of the bacteria in our fecal samples by using the 16S rRNA sequences. In brief, PICRUSt2 estimates the abundance of the gene families in the sample to determine the composition of the metagenome. Through this analysis, 43 statistically significant pathways were identified (Supplementary Table 2). Two of these pathways are involved in short chain fatty acid (SCFA) metabolism: Pyruvate fermentation to propanoate and Methanogenesis from acetate (Fig. 7A and B). Thus, our marker-based functional profiling suggests that BC have distinct functional pathways compared to HC with reduced abundance of pathways involved in the production of SCFA.
Figure 7.
The functional profile of the gut microbiome differs between patients with breast cancer and healthy controls. Based on the Wilcoxon test and the Benjamini–Hochberg procedure, 43 pathways were significantly different between BC and HC. Two pathways were highlighted as they are involved in short chain fatty acid metabolism, (a) Pyruvate fermentation to propanoate and (b) Methanogenesis from acetate. Significance: * < 0.05 and ** < 0.01.
Discussion
The gut microbiome has emerged as a potential factor in the pathobiology of breast cancer. As geographical region and environment play an important role in shaping the individual microbiome, a new region-specific study is required to profile the fecal microbiome in patients with breast cancer. This study is the first to analyze the gut microbiome of breast cancer patients and healthy controls from the Midwest region of the United States for taxonomic composition and predictive functional profiling. Our results demonstrate an altered gut microbiome with reduced SCFA-producing gut bacteria in BC patients.
Our data showing gut dysbiosis in BC is in accordance with prior studies, suggesting a role of the gut microbiome in breast cancer3,5,10. Of the eight species showing reduced abundance in our BC cohort, six either produce SCFAs (i.e., F. prausnitzii, L. pectinoshiza, and P. merdae) or are members of SCFA-producing genera (i.e., Lachnoclostridium, Alistipes, and Oscillibacter). F. prausnitzii embodies 5% of human fecal bacteria25,26 and is a major butyrate (C4) producing species in the gut27–29. L. pectinoschiza, which ferments polygalacturonic acid to formate (C1) and acetate (C2)30, also showed reduced abundance in the BC cohort. Finally, P. merdae, which produces acetate (C2)31, was reduced in the BC cohort.
Lachnoclostridium, Alistipes, and Oscillibacter are all genera associated with SCFA production. Lachnoclostridium symbiosum produces butyrate (C4)32, Alistipes produces minor amounts of acetate (C2), valerate (C5), propionate (C3), and butyrate (C4)33, and Oscillibacter valericigenes and Oscillibacter ruminantium produce valerate (C5)34 and butyrate (C4)35, respectively. The species of Alistipes and Oscillibacter reduced in our BC cohort were unclassified, though they are potential SCFA-producing species due to the properties of phylogenetically similar species. L. edouardi is phylogenetically related to L. symbiosum (with a 16S rRNA gene sequence identity of 94.26%)36, thus suggesting reduction of an additional SCFA producer in our BC cohort.
In the large intestines, SCFAs are the primary bacterial fermentation metabolites of non-digestible carbohydrates. In the human gut microbiome, SCFAs are predominantly acetate (C2), propionate (C3), and butyrate (C4)37, but also include formate (C1) and valerate (C5). A change in SCFAs may be associated with various inflammatory conditions (e.g., multiple sclerosis, inflammatory bowel disease, and obesity)38–40. In addition, evidence suggests that SCFAs are important for homeostasis through modulation of colonic epithelium integrity, adipocyte lipolysis, and regulation of the immune system41. Many of the effects of SCFAs are likely mediated through G-protein coupled receptors GPR43 and GPR4142. Specific to breast cancer, SCFAs activate GPR41 and GPR43-mediated signaling pathways in the MCF-7 human breast cancer cell line43, and these receptors have demonstrated reduced expression in invasive breast carcinoma and aggressive triple-negative breast tumors when compared to healthy breast tissue44.
In contrast, two species associated with SCFA production were significantly enriched in the BC cohort (Intestinibacter bartletti and Faecalitalea species). I. bartletti produces acetate (C2), valerate (C5), and butyrate (C4)45, and Faecalitalea produces butyrate (C4)46. The unclassified species of Faecalitalea that was enriched in our BC cohort is a potential SCFA-producing species. It is possible that these SCFA-producing bacteria are dependent on bacteria enriched in the BC cohort and/or SCFA metabolites produced by them feed into inflammatory pathways. Overall, there are more SCFA-producing bacteria significantly depleted in our BC cohort than those enriched. These results suggests that dysbiosis of SCFA-producing bacteria in our BC cohort could be influential to the pathobiology of breast cancer.
We identified 43 significant differentially abundant functional pathways. Interestingly, the pyruvate-fermentation-to-propanoate-I pathway was significantly decreased in the BC cohort, and the methanogenesis-from-acetate pathway was significantly increased in the BC cohort. These changes result in reduction of SCFAs and increased gut methane. Previous studies have associated an increase in gut methane with inflammatory disorders, such as multiple sclerosis and irritable bowel syndrome47,48. Microbial SCFAs in the gut increase colonic serotonin production, promoting gastrointestinal motility49. A decrease in SCFAs would therefore decrease gut motility, and increased gut methane also slows intestinal transit50,51. This decrease in intestinal transit is postulated to increase nutrient absorption, leading to weight gain and obesity52. Obesity contributes to systemic inflammation53 and increases the risk of developing breast cancer in postmenopausal women54,55. Thus, reduced abundance of SCFA-producing bacteria can contribute to breast cancer through induction of inflammation by modulating serotonin.
In addition to SCFA-producing bacteria, Eggerthella lenta and a species of the genus Blautia were significantly enriched in BC patients compared to HC. Enrichment of E. lenta is associated with various inflammatory states, including colitis56 and multiple sclerosis57. The role of Blautia in disease is more controversial, as both depletion and enrichment of this genus has been associated with inflammatory states (e.g.—depletion of Blautia in patients with Crohn’s disease58, colorectal cancer59, and multiple sclerosis60; enrichment of Blautia in patients with multiple sclerosis48 and inflammatory bowel syndrome61).
In this study, we aimed to identify the region-specific microbial composition of breast cancer patients in the midwestern United States. It is well established that geography can impact the gut microbiome, as highlighted by a study showing a distinct microbiome composition in individuals from the United States compared to other countries62. Relative to our study, Yatsunenko et al. identified that adults from metropolitan areas of the United States have increased abundance of an unidentified species of Alistipes when compared to adults from Malawi and Venezuela62. In this study, we identified a decreased abundance of an unidentified species of Alistipes in our BC cohort. Due to the increased abundance of an unidentified species of Alistipes in healthy adults from the United States, this could represent a United States-specific dysbiosis.
Few studies have investigated the intracontinental region-specific differences of gut microbial compositions at the genus- or species-level. Previously, Chen et al. analyzed the gut microbial composition of 118 midwestern subjects that varied by sex, race, BMI, age, alcohol use, and tobacco use to establish a midwestern reference population for gut microbiome research63. Relevant to our study, they found that the genera Faecalibacterium, Parabacteroides, Lachnospira, and Blautia represented some of the most prevalent genera in this midwestern healthy population63. In our study, we identified that midwestern BC patients had significantly differential abundance of species within these genera. Specifically, our BC cohort displayed decreased abundance of Faecalibacterium prausnitzii, Parabacteroides merdae, and Lachnospira pectinoschiza, and increased abundance of an unidentified Blautia species. Importantly, this region-specific reference population did not show significant difference in alpha- or beta-diversity based on age63, suggesting that adult age may play less of a role in microbial diversity in the Midwest region of the United States.
In conclusion, this pilot study displayed dysbiosis in our BC cohort with decreased abundance of SCFA-producing bacteria, decreased production of propionate, and increased conversion of acetate to methane. Our findings support the hypothesis that decreased abundance of SCFA-producing fecal bacteria can contribute to breast cancer pathobiology64. We were unable to measure fecal SCFA due to technical difficulties including sample storage. A prior study has shown that SCFAs evaporate from stool at contact with the atmosphere and thus when measuring SCFA levels, samples must be properly stored immediately65. Future studies should measure the abundance of SCFAs in stool samples of breast cancer patients to confirm these findings, as previously done in similar studies15. A better understanding of the role of gut dysbiosis in breast cancer could lead to refined prevention, treatment, and prognosis.
Limitations to this study include a small sample size, a lack of information on the menopausal status of the healthy controls, a significant difference in cohort ages, and the possible effects of different treatments on the microbiome. Due to the small sample sizes, we were unable to stratify by breast cancer subtypes, but future studies should aim to recruit large enough cohorts to allow for this analysis.
Our BC cohort was significantly older than our HC cohort. Although age could also play a role in the differences in gut microbial composition between our two cohorts, studies are conflicting on the role of age in microbial composition after middle-age. Ghosh et al. studied a cohort of 2500 individuals and found that elderly individuals had differential abundances of specific taxa in relationship to disease state (i.e., inflammatory bowel disease, colorectal cancer, cirrhosis, type II diabetes, and polyps) when compared to either young adults or middle-aged adults66. In contrast, de la Cuesta-Zuluaga et al. demonstrated that the microbial diversity of healthy women in cohorts from the United States, United Kingdom, and Columbia increases with age and plateaus around 40 years old67. In the same study, a cohort from China showed no effect on microbial diversity when stratified by age67. These studies suggest that after age 40, age may play a role in the microbiota of some disease states but does not appear to have a significant role in the microbiota of healthy controls. More research in this area is required to better understand the interplay of age, disease, and the microbiome.
We acknowledge chemotherapy and radiation affect the microbiome. In this study, chemotherapy treatment ended at least 145 days prior to sample collection for all BC patients. Currently, it is unknown if chemotherapy has long-lasting effects on the microbiome. Radiation therapy ended at least 31 days prior to sample collection and was targeted to tumors within the breast, which would have spared the gut microbiome. Additionally, we compared the microbiome of BC on chemotherapy (n = 4) vs no chemotherapy (n = 18) and BC on radiation therapy (n = 9) vs no radiation therapy (n = 13) prior to our BC vs HC analysis and found that alpha and beta diversity were not significantly different nor were any species or genera identified as significantly different between these therapies within the BC cohort. Thus, our preliminary investigation suggested that if chemotherapy or radiation had effects on the gut microbiome, those changes had subsided by the time of this analysis and thus BC samples did not need to be further split based on these therapies when compared to HC.
We also acknowledge that hormonal therapy could impact the microbiome. The hormonal therapies that our BC patients were on in this study were selective estrogen receptor modulators (SERMs; i.e. Nolvadex) and aromatase inhibitors (i.e., Arimidex, Aromasin, and Femara). The gut microbiome modulates circulating estrogen levels68, and SERMs are toxic to specific gut bacterial species10. To our knowledge, the bacteria found to be significant in our study have not been shown to be affected by SERMs. However, the specific effects of aromatase inhibitors on the gut microbiome have not been well established10. Future studies should address these shortcomings to strengthen our understanding of the gut microbiome’s relation to breast cancer.
Methods
Patient recruitment and demographics
This study was approved by the University of Iowa Institutional Review Board (Iowa City, IA, USA). Patients with BC (n = 24) were recruited from the BMER at the HCCC. Inclusion criteria were a diagnosis of invasive breast cancer of any stage and age 18–90 years old. Exclusion criteria were antibiotic use during sample collection and premalignant or in situ breast disease without concurrent invasive cancer. For BC patients, data was collected on body mass index (BMI), race, age, lymph node status, menopausal status, types and dates of treatments received, and cancer stage. For all BC patients, chemotherapy treatment had ended 145 days or more before sample collection, and the most recent radiation treatment was 31 days or more before sample collection. Of the BC patients, 22 were on hormonal therapies at the time of sample collection.
HC (n = 23) were recruited through the University of Iowa College of Nursing. Inclusion criteria were females ages 18–90. Exclusion criteria were antibiotic or laxative use within four weeks of sample collection and colonoscopy within three months. For healthy controls, data was collected on BMI, race, and age.
One BC patient and four HC were excluded from analysis due to poor sequence quality, and one BC patient was excluded due to a premalignant lesion. This resulted in 22 BC patients and 19 HC. Subject characteristics are described in Table 1.
Table 1.
Patient and healthy control demographics.
| HC (n = 19)* | BC (n = 22)* | p value | ||
|---|---|---|---|---|
| Age (mean, SD, in years) | 56.10 ± 9.04 | 67.82 ± 9.56 | 0.0048 | |
| BMI (mean, SD, in kg/m2) | 25.36 ± 3.92 | 27.26 ± 4.68 | 0.1645 | |
| Sex | F = 19 | F = 22 | ||
| M = 0 | M = 0 | |||
| Race | White = 19 | White = 22 | ||
| Menopausal status | Unknown |
Post = 20 Pre = 1 Unknown = 1 |
||
| Breast Cancer Subtype | HR + | 18 | ||
| HER2+ | 1 | |||
| TNBC | 4 | |||
| Clinical Stage | 0 | 0 | ||
| 1 | 15 | |||
| 2A | 6 | |||
| 2B | 1 | |||
| Hormonal Therapy | Nolvadex | 8 | ||
| Arimidex | 5 | |||
| Aromasin | 1 | |||
| Femara | 3 | |||
*Sample demographics utilized in analysis. HR = hormone receptor, HER2 = human epidermal growth factor receptor 2, TNBC = triple negative breast cancer.
Sample collection, DNA extraction and 16S sequencing
Stool samples were collected by patients in Commode Specimen Collection kits (Fisher PA, USA) provided by our laboratory. Stool samples were shipped on ice and received within 24 h of collection. The stool was aliquoted and stored at − 80 °C within 24 h of receipt. For fecal DNA extraction from the samples, we utilized Qiagen DNeasy PowerLyser PowerSoil Kit (Qiagen, Germantown, MD). We followed the manufacturer’s instructions by performing the bead-beating step (PowerLyzer 24 Homogenizer, Omni International, USA). Sequencing of the V3-V4 region of the 16S rRNA was performed as previously described by our laboratory69.
Metagenomic profiling
We processed the raw sequence data of fecal samples utilizing the V3-V4 region of the bacterial 16S rRNA and the DADA2 pipeline70. Briefly, we removed the primers, truncated the rest of the sequence based on a Phred quality score of 25, and then denoised the reads. Denoising was used to eliminate inaccurate base calling. Next, paired reads were merged, and chimeras were removed. The remaining sequences produced our amplicon sequence variants (ASVs). To assign taxonomy to these ASVs, the Silva database was utilized (Version 138.1, released March 2021)71. After taxonomy assignment, one BC sample and four HC samples were removed due to having a low read depth (i.e., less than 27,000 reads), resulting in 22 BC patients and 19 HC. The remaining samples had 27,000 to 86,874 counts, averaging 64,265 counts per sample.
Functional profiling
To identify the possible functions of the microbiome, we utilized tools from DADA2 that converted our cleaned sequence data to an ASV table with a corresponding FASTA sequence file. Then, with the use of PICRUSt224, we predicted potential functional pathways.
Statistical analysis and visualization
For analyses and figure creation, we utilized R (Version 4.0.3)72. The alpha diversity, beta diversity, and differential abundance analyses of the present features were performed with in-house scripts that utilized phyloseq73, microbiomeMarker74, vegan75, and ggpubr76. Data were normalized by sum scaling to one million reads at the sample level and log (base 10) transforming at the bacteria level. Features with a prevalence of less than 20 and a relative abundance of less than 1e-4 were also filtered out. These cut-offs were chosen to eliminate inaccurate claims of significance due to the absence of the feature in one group and a small presence in the other. In total, 519 species and 340 genera were identified. After filtering, 114 species and 92 genera remained. For our pathway analysis, pathways with a relative abundance threshold less than 0.0001 (percent composition) were filtered out. Alpha diversity was measured utilizing the Chao1 index and Shannon Diversity. For the differential abundance analyses, the Wilcoxon signed-rank test measured significance, and adjusted p values were calculated by the Benjamini–Hochberg algorithm. For beta diversity, the Weighted UniFrac distance metric was utilized, and significance of sample clustering was identified by PERMANOVA. LEFSe was performed utilizing the function run_lefse from the microbiomeMarker R package. Random forest was performed with the randomForest77 and Boruta23 functions in R. More details about the Random Forest analysis can be found in the "Random Forest identifies key species in differentiating between the microbiome of patients with breast cancer and healthy controls" section. LEFSe is a commonly used differential analysis method while Random Forest is a machine learning-based approach. LEFSe identifies the taxa that are significantly increased in abundance in one group compared to the other while also calculating each feature’s effect size. Random Forest, on the other hand, utilizes many decision trees and bagging (majority vote of decision trees) to decide which features help most in differentiating the groups. Thus, applying both very different approaches, and finding the same features in both, allows us to have more confidence in the features identified as significant.
The heat tree was created in MicrobiomeAnalyst78,79. The minimum feature count was set to 20, the percent prevalence in each sample was set to 20, and 10% of features were removed based on their inter-quartile range. Total sum scaling was used to normalize the feature data to create the heat tree.
Ethics approval
This study was performed in accordance with the ethical standards as laid down in the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards. Research specimens and/or clinical data were obtained through the University of Iowa Holden Comprehensive Cancer Center's 'Breast Molecular Epidemiology Resource' (BMER), an Institutional Review Board-approved biospecimen repository and data registry (IRB 201003791).
Consent to participate
Informed consent was obtained from all individual participants included in the study.
Supplementary Information
Acknowledgements
Research reported in this publication was supported by the Patients Enhancing Research Collaboration at Holden Program (IRB 201708847) facilitated by the Biospecimen Procurement and Molecular Epidemiology Resource Core (BioMER) at Holden Comprehensive Cancer Center, National Cancer Institute/Nation Institute of Health under Award Number P30CA086862. We acknowledge funding from the National Institutes of Health/NIAID 1RO1AI137075 (A.K.M), Veteran Affairs Merit Award 1I01CX002212 (A.K.M), University of Iowa Environmental Health Sciences Research Center, NIEHS/NIH P30 ES005605 (A.K.M), Gift from P. Heppelmann and M. Wacek (A.K.M), Carver Trust Pilot Grant (A.K.M) and pilot award from Center for Biocatalysis and Bioprocessing (CBB) (A.K.M.), R.L.S. was supported by the Informatics Fellowship from the University of Iowa. J.E.K. was supported on a National Institutes of Health grant (T32 GM139776 to University of Iowa Medical Scientist Training Program).
Author contributions
A.K.M., M.Y., C.C., M.C., PV., S.S., S.P., E.F., and R.G. contributed to the study conception and design. Material preparation and data collection were performed by N.C., M.Y., and J.H.. Data analysis was performed by R.L.S. The manuscript was written by R.L.S. and J.K. All authors read and approved the final manuscript.
Data availability
The 16S microbiome data has been uploaded to the Sequence Read Archive (SRA) under the BioProject ID: PRJNA872152 for free public access. The rest of the data can be made available through contacting the corresponding author.
Competing interests
A.K.M. is one of the inventors of a technology claiming the use of Prevotella histicola to treat autoimmune diseases. A.K.M. received royalties from Mayo Clinic (paid by Evelo Biosciences). However, no fund or product from the patent were used in the present study. All other authors declare no commercial or financial relationships that could be a potential conflict of interest.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Rachel L. Shrode, Jessica E. Knobbe and Nicole Cady.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-27436-3.
References
- 1.Sung H, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J. Clin. 2021;71:7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 3.Eslami-S Z, Majidzadeh-A K, Halvaei S, Babapirali F, Esmaeili R. Microbiome and breast cancer: New role for an ancient population. Front. Oncol. 2020;10:120. doi: 10.3389/fonc.2020.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen J, et al. The microbiome and breast cancer: A review. Breast Cancer Res. Treat. 2019;178:493–496. doi: 10.1007/s10549-019-05407-5. [DOI] [PubMed] [Google Scholar]
- 5.Laborda-Illanes A, et al. Breast and gut microbiota action mechanisms in breast cancer pathogenesis and treatment. Cancers (Basel) 2020;12:1–27. doi: 10.3390/cancers12092465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lacey JV, Jr, et al. Breast cancer epidemiology according to recognized breast cancer risk factors in the Prostate Lung Colorectal and Ovarian (PLCO) Cancer Screening Trial Cohort. BMC Cancer. 2009;9:84. doi: 10.1186/1471-2407-9-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Madigan MP, Ziegler RG, Benichou J, Byrne C, Hoover RN. Proportion of breast cancer cases in the United States explained by well-established risk factors. JNCI J. Natl. Cancer Inst. 1995;87:1681–1685. doi: 10.1093/jnci/87.22.1681. [DOI] [PubMed] [Google Scholar]
- 8.Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016;14:e1002533. doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Honda K, Littman DR. The microbiota in adaptive immune homeostasis and disease. Nature. 2016;535:75–84. doi: 10.1038/nature18848. [DOI] [PubMed] [Google Scholar]
- 10.Mikó E, et al. Microbiome—microbial metabolome—cancer cell interactions in breast cancer—familiar, but unexplored. Cells. 2019;8:293. doi: 10.3390/cells8040293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Armstrong H, Bording-Jorgensen M, Dijk S, Wine E. The complex interplay between chronic inflammation, the microbiome, and cancer: Understanding disease progression and what we can do to prevent it. Cancers (Basel) 2018;10:83. doi: 10.3390/cancers10030083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Minelli EB, et al. Intestinal microflora as an alternative metabolic source of estrogens in women with uterine leiomyoma and breast cancer. Ann. N Y Acad. Sci. 1990;595:473–479. doi: 10.1111/j.1749-6632.1990.tb34337.x. [DOI] [Google Scholar]
- 13.Ma J, et al. Alter between gut bacteria and blood metabolites and the anti-tumor effects of Faecalibacterium prausnitzii in breast cancer. BMC Microbiol. 2020;20:82. doi: 10.1186/s12866-020-01739-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Attraplsi S, Abbasi R, Mohammed Abdul M, Salih M, Mutlu E. Fecal microbiota composition in women in relation to factors that may impact breast cancer development. Am. J. Gastroenterol. 2013;108:S183. doi: 10.14309/00000434-201310001-00625. [DOI] [Google Scholar]
- 15.He C, Liu Y, Ye S, Yin S, Gu J. Changes of intestinal microflora of breast cancer in premenopausal women. Eur. J. Clin. Microbiol. Infect. Dis. 2020;40:503–513. doi: 10.1007/s10096-020-04036-x. [DOI] [PubMed] [Google Scholar]
- 16.Bard J-M, et al. Relationship between intestinal microbiota and clinical characteristics of patients with early stage breast cancer. FASEB J. 2015;29:914.2. doi: 10.1096/fasebj.29.1_supplement.914.2. [DOI] [Google Scholar]
- 17.Luu T, et al. Intestinal proportion of Blautia sp. is associated with clinical stage and histoprognostic grade in patients with early-stage breast cancer. Nutr. Cancer. 2017;69:267–275. doi: 10.1080/01635581.2017.1263750. [DOI] [PubMed] [Google Scholar]
- 18.Frugé AD, et al. Fecal Akkermansia muciniphila is associated with body composition and microbiota diversity in overweight and obese women with breast cancer participating in a presurgical weight loss trial. J. Acad. Nutr. Diet. 2020;120:650–659. doi: 10.1016/j.jand.2018.08.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goedert JJ, et al. Investigation of the association between the fecal microbiota and breast cancer in postmenopausal women: a population-based case-control pilot study. JNCI J. Natl. Cancer Inst. 2015;107:147. doi: 10.1093/jnci/djv147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu J, et al. Breast cancer in postmenopausal women is associated with an altered gut metagenome. Microbiome. 2018;6:136. doi: 10.1186/s40168-018-0515-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Magne F, et al. The firmicutes/bacteroidetes ratio: A relevant marker of gut dysbiosis in obese patients? Nutrients. 2020;12:1474. doi: 10.3390/nu12051474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Segata N, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kursa MB, Rudnicki WR. Feature selection with the Boruta Package. J. Stat. Softw. 2010;36:1–13. doi: 10.18637/jss.v036.i11. [DOI] [Google Scholar]
- 24.Langille M, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suau A, et al. Fusobacterium prausnitzii and related species represent a dominant group within the human fecal flora. Syst. Appl. Microbiol. 2001;24:139–145. doi: 10.1078/0723-2020-00015. [DOI] [PubMed] [Google Scholar]
- 26.Hold GL, Schwiertz A, Aminov RI, Blaut M, Flint HJ. Oligonucleotide probes that detect quantitatively significant groups of butyrate-producing bacteria in human feces. Appl. Environ. Microbiol. 2003;69:4320–4324. doi: 10.1128/AEM.69.7.4320-4324.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferreira-Halder CV, de Sousa Faria AV, Andrade SS. Action and function of Faecalibacterium prausnitzii in health and disease. Best Pract. Res. Clin. Gastroenterol. 2017;31:643–648. doi: 10.1016/j.bpg.2017.09.011. [DOI] [PubMed] [Google Scholar]
- 28.Lopez-Siles M, Duncan SH, Garcia-Gil LJ, Martinez-Medina M. Faecalibacterium prausnitzii: From microbiology to diagnostics and prognostics. ISME J. 2017;11:841–852. doi: 10.1038/ismej.2016.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martín R, et al. Functional characterization of novel Faecalibacterium prausnitzii strains isolated from healthy volunteers: A step forward in the use of F. prausnitzii as a next-generation probiotic. Front. Microbiol. 2017;8:1226. doi: 10.3389/fmicb.2017.01226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cornick N, Jensen N, Stahl D, Hartman P, Allison M. Lachnospira pectinoschiza sp. nov., an anaerobic pectinophile from the pig intestine. Int. J. Syst. Evol. Microbiol. 1994;44:87–93. doi: 10.1099/00207713-44-1-87. [DOI] [PubMed] [Google Scholar]
- 31.Sakamoto M, Benno Y. Reclassification of Bacteroides distasonis, Bacteroides goldsteinii and Bacteroides merdae as Parabacteroides distasonis gen. nov., comb. nov., Parabacteroides goldsteinii comb. nov. and Parabacteroides merdae comb. nov. Int. J. Syst. Evol. Microbiol. 2006;56:1599–1605. doi: 10.1099/ijs.0.64192-0. [DOI] [PubMed] [Google Scholar]
- 32.Gutiérrez N, Garrido D. Species deletions from microbiome consortia reveal key metabolic interactions between gut microbes. mSystems. 2019;4:e00185–e219. doi: 10.1128/mSystems.00185-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rautio M, et al. Reclassification of Bacteroides putredinis (Weinberg et al., 1937) in a new genus Alistipes gen. nov., as Alistipes putredinis comb. nov., and description of Alistipes finegoldii sp. nov., from human sources. Syst. Appl. Microbiol. 2003;26:182–188. doi: 10.1078/072320203322346029. [DOI] [PubMed] [Google Scholar]
- 34.Iino T, Mori K, Tanaka K, Suzuki K, Harayama S. Oscillibacter valericigenes gen nov., sp. nov., a valerate-producing anaerobic bacterium isolated from the alimentary canal of a Japanese corbicula clam. Int. J. Syst. Evol. Microbiol. 2007;57:1840–1845. doi: 10.1099/ijs.0.64717-0. [DOI] [PubMed] [Google Scholar]
- 35.Lee G-H, et al. Oscillibacter ruminantium sp. nov., isolated from the rumen of Korean native cattle. Int. J. Syst. Evol. Microbiol. 2013;63:1942–1946. doi: 10.1099/ijs.0.041749-0. [DOI] [PubMed] [Google Scholar]
- 36.Traore SI, et al. Description of ‘Blautia phocaeensis’ sp. nov. and ‘Lachnoclostridium edouardi’ sp. nov., isolated from healthy fresh stools of Saudi Arabia Bedouins by culturomics. New Microbes New Infect. 2017;19:129–131. doi: 10.1016/j.nmni.2017.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Macfarlane GT, Gibson GR, Cummings JH. Comparison of fermentation reactions in different regions of the human colon. J. Appl. Bacteriol. 1992;72:57–64. doi: 10.1111/j.1365-2672.1992.tb04882.x. [DOI] [PubMed] [Google Scholar]
- 38.Machiels K, et al. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut. 2014;63:1275–1283. doi: 10.1136/gutjnl-2013-304833. [DOI] [PubMed] [Google Scholar]
- 39.Melbye P, Olsson A, Hansen T, Søndergaard H, BangOturai A. Short-chain fatty acids and gut microbiota in multiple sclerosis. Acta Neurol. Scand. 2019;139:208–219. doi: 10.1111/ane.13045. [DOI] [PubMed] [Google Scholar]
- 40.den Besten G, et al. Short-chain fatty acids protect against high-fat diet-induced obesity via a PPARγ-dependent switch From lipogenesis to fat oxidation. Diabetes. 2015;64:2398–2408. doi: 10.2337/db14-1213. [DOI] [PubMed] [Google Scholar]
- 41.Morrison DJ, Preston T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes. 2016;7:189–200. doi: 10.1080/19490976.2015.1134082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brown AJ, et al. The orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J. Biol. Chem. 2003;278:11312–11319. doi: 10.1074/jbc.M211609200. [DOI] [PubMed] [Google Scholar]
- 43.Yonezawa T, Kobayashi Y, Obara Y. Short-chain fatty acids induce acute phosphorylation of the p38 mitogen-activated protein kinase/heat shock protein 27 pathway via GPR43 in the MCF-7 human breast cancer cell line. Cell Signal. 2007;19:185–193. doi: 10.1016/j.cellsig.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 44.Thirunavukkarasan M, et al. Short-chain fatty acid receptors inhibit invasive phenotypes in breast cancer cells. PLoS ONE. 2017;12:e0186334. doi: 10.1371/journal.pone.0186334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gerritsen, J. et al. Characterization of Romboutsia ilealis gen. nov., sp. nov., isolated from the gastro-intestinal tract of a rat, and proposal for the reclassification of five closely related members of the genus Clostridium into the genera Romboutsia gen. nov., Intestinibacter gen. nov, Terrisporobacter gen. nov. and Asaccharospora gen. nov. Int J Syst Evol Microbiol64, 1600–1616 (2014). [DOI] [PubMed]
- 46.Maesschalck, C. de et al.Faecalicoccus acidiformans gen. nov., sp. nov., isolated from the chicken caecum, and reclassification of Streptococcus pleomorphus (Barnes et al. 1977), Eubacterium biforme (Eggerth 1935) and Eubacterium cylindroides (Cato et al. 1974) as Faecalicoccus pleomorphus comb. nov., Holdemanella biformis gen. nov., comb. nov. and Faecalitalea cylindroides gen. nov., comb. nov., respectively, within the family Erysipelotrichaceae. Int J Syst Evol Microbiol64, 3877–3884 (2014). [DOI] [PubMed]
- 47.Pimentel M, et al. Methane production during lactulose breath test is associated with gastrointestinal disease presentation. Dig. Dis. Sci. 2003;48:86–92. doi: 10.1023/A:1021738515885. [DOI] [PubMed] [Google Scholar]
- 48.Jangi S, et al. Alterations of the human gut microbiome in multiple sclerosis. Nat. Commun. 2016;7:12015. doi: 10.1038/ncomms12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reigstad CS, et al. Gut microbes promote colonic serotonin production through an effect of short-chain fatty acids on enterochromaffin cells. FASEB J. 2015;29:1395–1403. doi: 10.1096/fj.14-259598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pimentel M, et al. Methane, a gas produced by enteric bacteria, slows intestinal transit and augments small intestinal contractile activity. Am. J. Physiol. Gastrointest. Liver Physiol. 2006;290:G1089–1095. doi: 10.1152/ajpgi.00574.2004. [DOI] [PubMed] [Google Scholar]
- 51.Jahng J, Jung IS, Choi EJ, Conklin JL, Park H. The effects of methane and hydrogen gases produced by enteric bacteria on ileal motility and colonic transit time. Neurogastroenterol. Motil. 2012;24:185–90, e92. doi: 10.1111/j.1365-2982.2011.01819.x. [DOI] [PubMed] [Google Scholar]
- 52.Basseri RJ, et al. Intestinal methane production in obese individuals is associated with a higher body mass index. Gastroenterol. Hepatol. (N Y) 2012;8:22–28. [PMC free article] [PubMed] [Google Scholar]
- 53.Ellulu MS, Khaza’ai H, Rahmat A, Patimah I, Abed Y. Obesity can predict and promote systemic inflammation in healthy adults. Int. J. Cardiol. 2016;215:318–324. doi: 10.1016/j.ijcard.2016.04.089. [DOI] [PubMed] [Google Scholar]
- 54.Rosenberg LU, et al. Risk factors for hormone receptor-defined breast cancer in postmenopausal women. Cancer Epidemiol. Biomarkers Prev. 2006;15:2482–2488. doi: 10.1158/1055-9965.EPI-06-0489. [DOI] [PubMed] [Google Scholar]
- 55.Reeves GK, et al. Cancer incidence and mortality in relation to body mass index in the Million Women Study: Cohort study. BMJ. 2007;335:1134–1139. doi: 10.1136/bmj.39367.495995.AE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alexander M, et al. Human gut bacterial metabolism drives Th17 activation and colitis. Cell Host Microbe. 2022;30:17–30.e9. doi: 10.1016/j.chom.2021.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cekanaviciute E, et al. Gut bacteria from multiple sclerosis patients modulate human T cells and exacerbate symptoms in mouse models. Proc. Natl. Acad. Sci. U S A. 2017;114:10713–10718. doi: 10.1073/pnas.1711235114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen L, et al. Characteristics of fecal and mucosa-associated microbiota in Chinese patients with inflammatory bowel disease. Medicine (Baltimore) 2014;93:e51. doi: 10.1097/MD.0000000000000051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang X, Yang Y, Huycke MM. Microbiome-driven carcinogenesis in colorectal cancer: Models and mechanisms. Free Radic. Biol. Med. 2017;105:3–15. doi: 10.1016/j.freeradbiomed.2016.10.504. [DOI] [PubMed] [Google Scholar]
- 60.Zhou X, et al. Gut microbiome of multiple sclerosis patients and paired household healthy controls reveal associations with disease risk and course. Cell. 2022;185:3467–3486.e16. doi: 10.1016/j.cell.2022.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rajilić-Stojanović M, et al. Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology. 2011;141:1792–1801. doi: 10.1053/j.gastro.2011.07.043. [DOI] [PubMed] [Google Scholar]
- 62.Yatsunenko T, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen J, et al. Impact of demographics on human gut microbial diversity in a US Midwest population. PeerJ. 2016;4:e1514. doi: 10.7717/peerj.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mirzaei R, et al. Role of microbiota-derived short-chain fatty acids in cancer development and prevention. Biomed. Pharmacother. 2021;139:111619. doi: 10.1016/j.biopha.2021.111619. [DOI] [PubMed] [Google Scholar]
- 65.Hoving LR, Heijink M, van Harmelen V, van Dijk KW, Giera M. GC-MS analysis of short-chain fatty acids in feces, cecum content, and blood samples. Methods Mol. Biol. 2018;1730:247–256. doi: 10.1007/978-1-4939-7592-1_17. [DOI] [PubMed] [Google Scholar]
- 66.Ghosh TS, Das M, Jeffery IB, O’Toole PW. Adjusting for age improves identification of gut microbiome alterations in multiple diseases. Elife. 2020;9:e50240. doi: 10.7554/eLife.50240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.de la Cuesta-Zuluaga J, et al. Age- and sex-dependent patterns of gut microbial diversity in human adults. mSystems. 2019;4:e00261–e319. doi: 10.1128/mSystems.00261-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kwa M, Plottel CS, Blaser MJ, Adams S. The Intestinal Microbiome and Estrogen Receptor-Positive Female Breast Cancer. J Natl Cancer Inst. 2016;108(8):djw029. doi: 10.1093/jnci/djw029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shahi S, Zarei K, Guseva N, Mangalam A. Microbiota analysis using two-step PCR and next-generation 16S rRNA gene sequencing. J. Vis. Exp. 2019 doi: 10.3791/59980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Callahan BJ, et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Quast C, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.R Core Team. R: A language and environment for statistical computing. Preprint at (2020).
- 73.McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cao Y, et al. microbiomeMarker: an R/Bioconductor package for microbiome marker identification and visualization. Bioinformatics. 2022;38:4027–4029. doi: 10.1093/bioinformatics/btac438. [DOI] [PubMed] [Google Scholar]
- 75.Oksanen, J. et al. Vegan: Community Ecology Package. Preprint at https://cran.r-project.org/package=vegan (2020).
- 76.Kassambara, A. ggpubr: ‘ggplot2’ Based Publication Ready Plots. Preprint at https://cran.r-project.org/package=ggpubr (2020).
- 77.Liaw A, Wiener M. Classification and regression by randomForest. Rnews. 2002;2:18–22. [Google Scholar]
- 78.Chong J, Liu P, Zhou G, Xia J. Using MicrobiomeAnalyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat. Protoc. 2020;15:799–821. doi: 10.1038/s41596-019-0264-1. [DOI] [PubMed] [Google Scholar]
- 79.Dhariwal A, et al. MicrobiomeAnalyst: a web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. 2017;45:W180–W188. doi: 10.1093/nar/gkx295. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 16S microbiome data has been uploaded to the Sequence Read Archive (SRA) under the BioProject ID: PRJNA872152 for free public access. The rest of the data can be made available through contacting the corresponding author.