Abstract
Background:
Environmental exposures are implicated in diabetes etiology, but are poorly understood due to disease heterogeneity, complexity of exposures, and analytical challenges. Machine learning and data mining are artificial intelligence methods that can address these limitations. Despite their increasing adoption in etiology and prediction of diabetes research, the types of methods and exposures analyzed have not been thoroughly reviewed.
Objective:
We aimed to review articles that implemented machine learning and data mining methods to understand environmental exposures in diabetes etiology and disease prediction.
Methods:
We queried PubMed and Scopus databases for machine learning and data mining studies that used environmental exposures to understand diabetes etiology on September 19th, 2022. Exposures were classified into specific external, general external, or internal exposures. We reviewed machine learning and data mining methods and characterized the scope of environmental exposures studied in the etiology of general diabetes, type 1 diabetes, type 2 diabetes, and other types of diabetes.
Results:
We identified 44 articles for inclusion. Specific external exposures were the most common exposures studied, and supervised models were the most common methods used. Well-established specific external exposures of low physical activity, high cholesterol, and high triglycerides were predictive of general diabetes, type 2 diabetes, and prediabetes, while novel metabolic and gut microbiome biomarkers were implicated in type 1 diabetes.
Discussion:
The use of machine learning and data mining methods to elucidate environmental triggers of diabetes was largely limited to well-established risk factors identified using easily explainable and interpretable models. Future studies should seek to leverage machine learning and data mining to explore the temporality and co-occurrence of multiple exposures and further evaluate the role of general external and internal exposures in diabetes etiology.
Keywords: environmental exposures, diabetes mellitus, machine learning, data mining
1. INTRODUCTION:
Diabetes mellitus is a group of chronic metabolic disorders characterized by dysfunctional carbohydrate metabolism and glucose dysregulation. With an estimated 34.2 million people affected and 1.5 million new cases per year in the United States, diabetes mellitus is an increasingly prevalent public health crisis (1). Significant morbidity and mortality are attributed to diabetes progression, as diabetes is the leading cause of kidney failure, lower-limb amputation, and adult blindness and the 7th leading cause of death in the United States (1, 2). Diabetes also represents a substantial financial burden for patients and healthcare organizations alike, with an average 2.3 times increase in expenditure in diabetic patients compared to controls (3). Together, these statistics highlight the need for early detection, improved diagnosis, and primary prevention of diabetes and its complications.
Strategies to prevent and predict onset of diabetes are complicated by the heterogeneity of diabetes phenotypes and limited understanding of diabetes etiology. There are several types of diabetes mellitus, type 1 diabetes mellitus (T1DM) and type 2 diabetes mellitus (T2DM) being the most common. T1DM results from autoimmune destruction of insulin-producing pancreatic beta cells and accounts for approximately 10% of diabetes cases worldwide (4, 5). In contrast, T2DM results from a combination of deficient insulin secretion and insulin resistance among insulin-sensitive tissues and affects 462 million individuals globally (4, 6). The etiology of autoimmune destruction in T1DM and insulin malfunction in T2DM are poorly understood but are thought to be due to genetic and environmental factors (4, 7, 8).
Environmental exposures can be holistically defined by the exposome – the total exposures an individual experiences from the prenatal period until death (9, 10). Environmental exposures include general external exposures such as urban environment and climate; specific exposures such as diet and physical activity; and internal exposures such as the gut microbiome and the metabolome (10). Recent findings suggest a role for upper respiratory infections, early dietary exposures, and psychological stress in T1DM (11) and diet quantity and quality, limited physical activity, increased sedentary time, exposure to noise, exposure to fine dust, sleep disturbances, and smoking in T2DM (12). Despite these findings, no individual exposures have been attributed to diabetes etiology. Investigating the role of environmental exposures in diabetes etiology is complex due to the vast number and variety of exposures, variations in exposure collection, and insufficient statistical methods to consider the complex relationships between multiple exposures (13, 14).
Machine learning and data mining approaches are promising analytical approaches that have been used in diabetes research to address these limitations. Previous reviews have detailed the use of data mining and machine learning in diabetes research (15–20). A wide range of algorithms have been implemented, with supervised algorithms (15, 18) being more commonly reviewed than unsupervised and data mining algorithms (20). The most frequently used algorithms ranged from support vector machines (15, 16), neural networks (16, 18), logistic regressions (16), and decision trees (16). Though many studies tested multiple machine learning algorithms (15, 20), external validation with alternate datasets was rarely performed (18). Notably, these studies discuss applications of data mining and machine learning methods across fields of prediction and diagnosis, genetic and environmental components of etiopathology, diabetes complications, and healthcare systems and management. However, few studies systematically review literature applying these approaches to diabetes onset prediction and etiology. Big-data repositories of environmental data in individuals susceptible to diabetes and improvements in electronic medical record collection and storage have enabled the implementation of these advanced computational methods to elucidate diabetes etiopathology.
Despite these advances, literature on the use of machine learning and data mining methods to elucidate environmental factors involved in diabetes etiology remains poorly characterized. Therefore, the objective of this scoping review was to explore machine learning and data mining approaches to understand environmental exposures in diabetes etiology. This review is organized as follows: first, a detailed description of the methodology used to identify articles is presented. Next, the general results of the included articles and pertinent findings relating to each diabetes type are summarized. This review concludes with a summary of the main findings, discussion of the gaps in environmental exposures, machine learning, and data mining methods implemented, and strengths and limitations of the study.
2. MATERIALS AND METHODS:
This scoping review follows the guidelines of the Preferred Reporting Items for Systematic reviews and Meta-Analyses extension for Scoping Reviews (PRISMA-ScR) (21). The search strategy, screening and eligibility criteria, and data extraction and analysis procedures are described below.
2.1. Search Strategy:
To systematically identify articles that applied machine learning and data mining techniques using environmental exposure data for diabetes prediction and etiology, we searched original research articles using the PubMed and Scopus databases on April 9th, 2021, and updated the search on September 19th, 2022. The search terms for both searches were constructed by concatenating “diabetes,” “machine learning,” or “data mining,” and terms related to environmental exposures. The search criteria for environmental exposures were derived from the Exposure Science Ontology (ExO) exposure stressor class and included “exposome”, “environment”, “exposure”, “biological”, “biomechanical”, “chemical”, “ecological”, “transport”, “physical”, “psychological”, and “social” (22). Additional articles were identified by backward reference literature search and duplicated articles were removed. Articles from both searches were merged (Supplementary Methods: Search Strategy).
2.2. Eligibility & Selection:
English articles and articles published in peer-reviewed journals or conference proceedings were considered for screening. Articles were screened on the title and abstract review for relevance to prediction or etiology of diabetes, use of machine learning or data-mining techniques, and inclusion of environmental exposures. Full-text articles were retrieved with institutional licenses and further assessed for eligibility. Articles not relating to diabetes onset prediction or etiology were excluded. Articles that did not use environmental exposures and machine learning or data mining methods were also excluded. Finally, articles conducted in non-human data or samples were excluded.
2.3. Data Extraction & Analysis:
Criteria for data extraction were developed using Guidelines for Developing and Reporting Machine Learning Predictive Models in Biomedical Research (MLP) (23) and the Transparent reporting of multivariable prediction model for individual prognosis or diagnosis (TRIPOD) Statement (24). Extracted information included article information, study characteristics, outcome variables, predictor variables, patient characteristics, methods, results, and discussion. A detailed list of extraction items is presented in Supplementary Table 1.
Exposures were classified as internal exposures, specific external exposures, or general external exposures, as proposed by Wild (10). Articles were grouped by the diabetes type used for the predictor variable as defined by the American Diabetes Association guidelines (4) because the pathogenesis of each type of diabetes is different, and separate environmental exposures may be involved in disease onset. The article groups include general diabetes (GD), T1DM, T2DM, and other. Methods were categorized as data mining if the approaches were used for knowledge discovery and as machine learning if the approaches were used for prediction (25, 26). Machine learning algorithms were separated into supervised machine learning and unsupervised machine learning. Descriptive statistics were calculated for relevant extraction criteria, including the median and interquartile range (IQR) for numeric variables and the percentage and count for categorical variables.
3. RESULTS:
Overall, 2,426 unique articles were retrieved from Scopus, PubMed, and backward reference literature searches. Screening on title and abstract removed 2,286 articles. Full-text review of the remaining 140 articles resulted in excluding an additional 96 articles, leaving 44 articles for inclusion in the final review (Figure 1).
Figure 1:
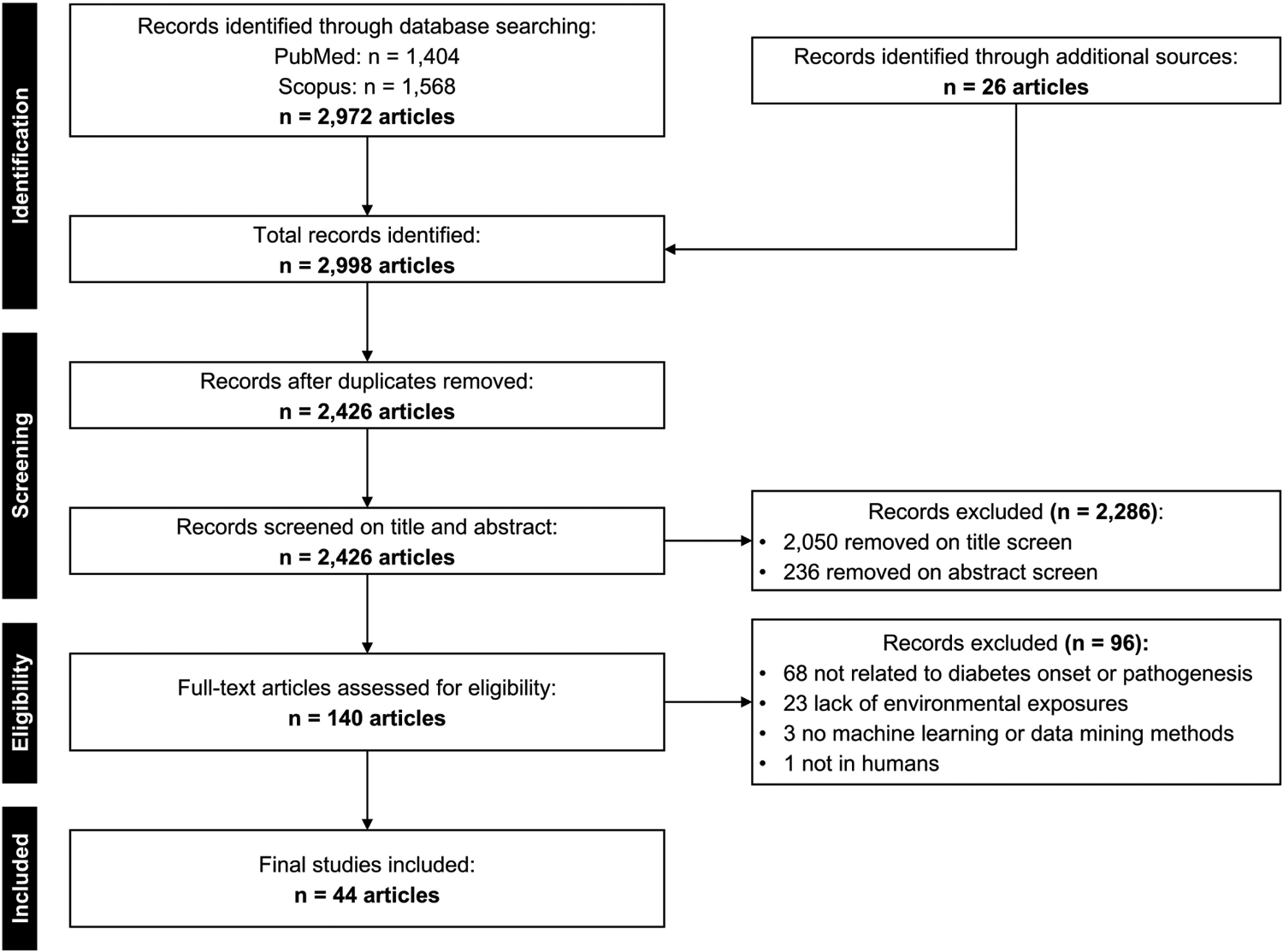
Preferred Reporting Items for Systematic Reviews and Meta-Analyses Extension for Scoping Reviews (PRISMA-ScR) flowchart of article inclusion and exclusion criteria.
3.1. General Study Results:
This section discusses study characteristics, datasets, and machine learning methods implemented in all 44 articles included in the study (Supplementary Table 2).
3.1.1. Article Descriptions:
Articles spanned a 13-year range, with a substantial rise in publications after 2017 (Figure 2A). Data collection occurred in several countries, with most studies being conducted in the United States (n = 13) and China (n = 9) (Figure 2B). Study designs included cohort (n = 22), cross-sectional (n = 16), and case-control (n = 3), with three studies not specifying the design framework. Studies were similarly split between retrospective (n = 22) and prospective (n = 19) methods of data collection. Overall, the median number of subjects included was 3,589 (IQR: 8,132 subjects), and the median number of variables included was 18 (IQR: 30 variables).
Figure 2:
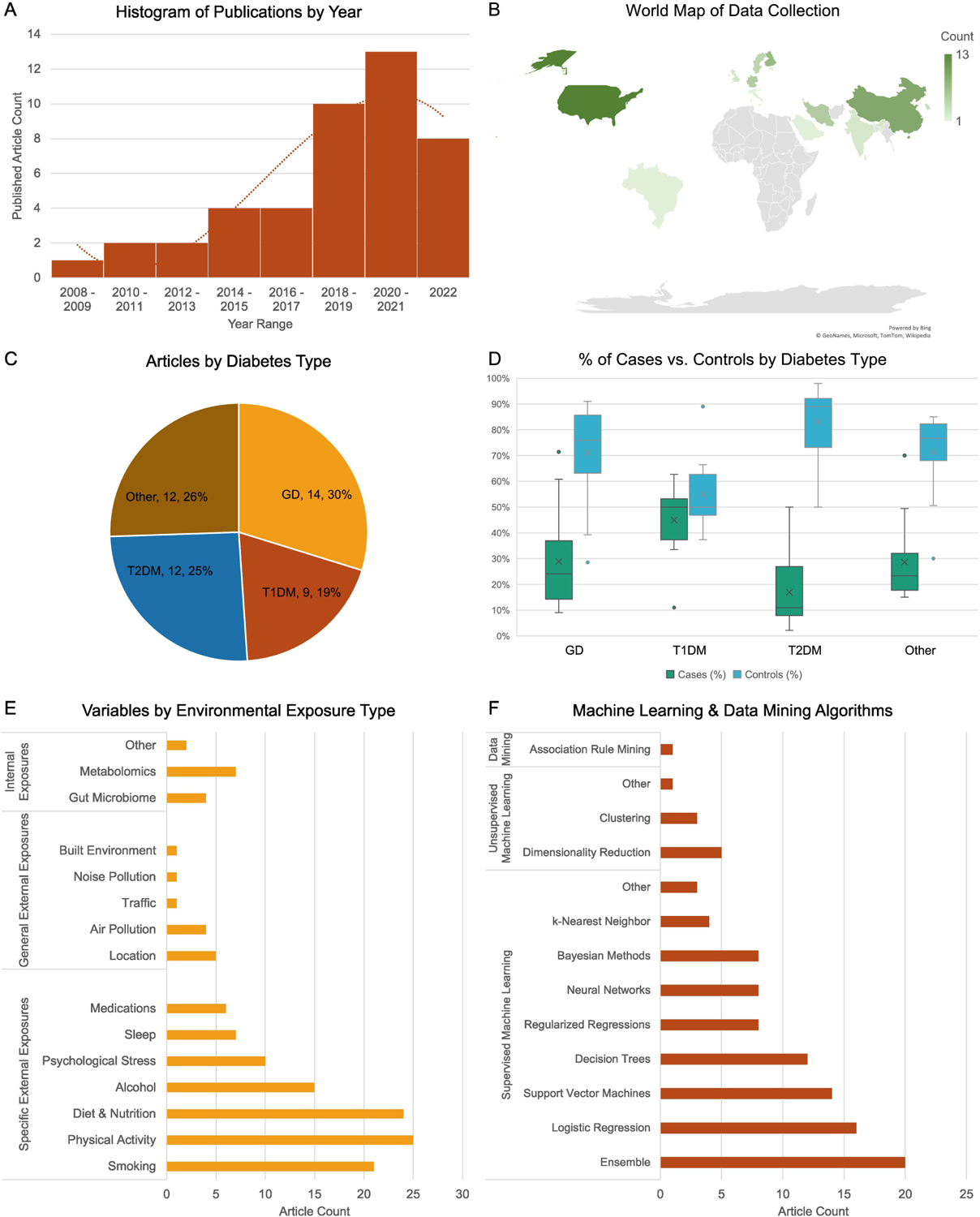
General Study Characteristics. (A) Histogram of the number of publications by year. (B) World map of publications by country. (C) Pie chart of articles by diabetes type including general diabetes (GD), type 1 diabetes mellitus (T1DM), type 2 diabetes mellitus (T2DM), and other diabetes. (D) Percentage of cases and controls by diabetes type. (E) Count of exposure variables analyzed across all articles categorized by the Wild classification system. (F) Count of machine learning and data mining algorithms implemented across all articles.
3.1.2. Diabetes Types:
Diabetes was classified as GD in 30% (n = 14), T1DM in 19% (n = 9), T2DM in 25% (n = 12), and as other in 26% (n = 12) of articles (Figure 2C). The other category included prediabetes alone as well as prediabetes and diabetes. Clinical diagnosis of diabetes was evaluated by fasting plasma glucose (n = 19), oral glucose tolerance testing (n = 14), or islet autoantibody measurements (n = 7), and 10 studies did not specify how diabetes was determined. While the percentage of cases and controls was similar for T1DM (cases median: 50%, controls median: 50%), the GD (cases median: 24%, controls median: 76%), T2DM (cases median: 11%, controls median: 89%), and other (cases median: 23%, controls median: 77%) groups had larger proportions of controls to cases (Figure 2D). Despite this discrepancy, only seven studies employed methods to correct or adjust the analysis for class imbalance.
3.1.3. Exposure Variables:
Exposure data was collected using a variety of techniques, including biological samples (n = 26), survey (n = 24), clinical measurements (n = 9), medical records (n = 6), and sensors (n = 4). Most studies used existing data sources (n = 30), and 14 collected novel data. Exposures were classified as specific external exposures, general external exposures, and internal exposures. Detailed definitions are provided (Supplementary Table 3). Specific external exposures were the most common exposure studied. Variables in articles included smoking (n = 21), physical activity (n = 25), dietary and nutritional factors (n = 24), alcohol (n = 15), psychological stress (n = 10), sleep (n = 7), and medications (n = 6) (Figure 2E). There was significant variability in how various exposures were defined, with many studies not specifying how some exposures were defined (Supplementary Table 3). General external exposures were the least common exposure studies. General exposures evaluated in articles included location (n = 5), air pollution (n = 4), traffic (n = 1), noise pollution (n = 1), and built environment (n = 1) (Figure 2E). Finally, at least one internal exposure was included in 13 articles. Data types included gut microbiome (n = 4), metabolome (n = 7), vaginal samples (n = 1), and analytical measurements from hair and urine (n = 1) (Figure 2E).
3.1.4. Machine Learning and Data Mining Methods:
Articles used a variety of machine learning and data mining models, including supervised models (n = 35), unsupervised (n = 5), both supervised and unsupervised (n = 3), and data mining (n = 1) methods. Most studies tested multiple algorithms (n = 29), with a median of 2.5 algorithms tested (IQR: 3). Ensemble algorithms were the most common algorithm overall (n = 20), followed by logistic regressions (n = 16), support vector machines (n = 14), and decision trees (n = 12) (Figure 2F). The most common ensemble method used was random forest (Table 1). Regularized regression models, Bayesian methods, and neural networks were trained in eight articles each, while k-Nearest neighbor models were trained in four articles (Table 1). Unsupervised and data mining algorithms were less common than supervised methods (Figure 2F). Unsupervised algorithms were primarily used for dimensionality reduction (n = 5), while clustering was implemented in three articles (Table 1). Only one article used a data mining method for rule-based learning with the apriori algorithm (Figure 2F).
Table 1: Machine Learning and Data Mining Algorithms Summary:
Counts of machine learning and data mining algorithms and specific models by diabetes type. Types of diabetes include general diabetes (GD), type 1 diabetes mellitus (T1DM), type 2 diabetes mellitus (T2DM), and other diabetes. Algorithms were divided into supervised machine learning, unsupervised machine learning, and data mining. Criterion used for each model category are also presented as relevant.
| Algorithm Type | Algorithm Name | Algorithm Count | Articles | Model Specifications | Model Count |
|---|---|---|---|---|---|
| Supervised | Ensemble | 20 | GD: (27–29, 32, 36, 40) T1DM: (42, 44, 45, 47, 49) T2DM: (52, 56, 57, 61) Other: (36, 52, 63–65, 67, 70) |
Random Forest | 20 |
| AdaBoost | 2 | ||||
| XGBoost | 5 | ||||
| Gradient Boosting | 4 | ||||
| Decision Jungle | 1 | ||||
| Other/not specified | 3 | ||||
| Logistic Regression | 16 | GD: (28, 29, 36–38) T1DM: (44, 45, 47) T2DM: (57–59, 61) Other: (36, 37, 63, 66, 67, 70) |
Other/not specified | 16 | |
| Support Vector Machines | 14 | GD: (28, 32, 34, 36–38) T1DM: (44, 45, 49) T2DM: (59) Other: (36, 37, 64, 67, 68, 70) |
Linear | 5 | |
| Polynomial | 3 | ||||
| Radial Basis Function | 6 | ||||
| Gaussian | 2 | ||||
| Sigmoid | 1 | ||||
| Other/not specified | 5 | ||||
| Decision Trees | 12 | GD: (27, 28, 35) T2DM: (51, 54, 56, 61) Other: (62, 64, 66, 69, 70) |
C4.5 | 3 | |
| C5.0 | 2 | ||||
| CART | 2 | ||||
| QUEST | 1 | ||||
| Other/not specified | 6 | ||||
| Regularized Regressions | 8 | GD: (31, 40) T1DM: (42, 43, 49) T2DM: (53, 55) Other: (65) |
LASSO | 4 | |
| Least Squares | 1 | ||||
| ElasticNet | 3 | ||||
| Neural Networks | 8 | GD: (27–29) T2DM: (58) Other: (63, 66–68) |
Artificial Neural Network | 6 | |
| Other/not specified | 2 | ||||
| Bayesian Methods | 8 | GD: (28, 29, 39) T1DM: (44, 45, 48) Other: (64, 67) |
Naïve Bayes | 7 | |
| Bayesian Networks | 2 | ||||
| Other/not specified | 2 | ||||
| k-Nearest Neighbor | 4 | GD: (29) T1DM: (44, 45) T2DM: (59) |
Other/not specified | 4 | |
| Other | 3 | T1DM: (44, 45) Other: (67) |
Average Perceptron | 1 | |
| Linear Discriminant Analysis | 2 | ||||
| Unsupervised | Dimensionality Reduction | 5 | GD: (30) T1DM: (41, 47) T2DM: (53, 60) |
Principal Component Analysis | 4 |
| Partial Least-Squares | 1 | ||||
| Clustering | 3 | GD: (33) T1DM: (46) T2DM: (60) |
Clustering | 2 | |
| Gaussian-based Model | 1 | ||||
| Other | 1 | T1DM: (42) | Weighted Correlation Network Analysis | 1 | |
| Data Mining | Association Rule Mining | 1 | T2DM: (50) | Apriori Algorithm | 1 |
3.2. General Diabetes (GD):
This section discusses the following 14 articles related to GD (27–40).
3.2.1. Patient Characteristics:
Studies predicting GD were primarily conducted in adults (n = 10). Of the 7 studies that reported the proportions of patients by sex, the median percentage of males was greater than females (males: 55%, females: 45%).
3.2.2. Predictor Variables:
Articles studying GD included a wide array of exposures (Figure 3A). Specific external exposures were the most common exposures studied, with physical activity (n = 9), smoking (n = 8), and diet and nutrition (n = 8) being studied most frequently. General external and internal exposures were less frequently evaluated, with only two articles including air pollution measures and one article including location, built environment, metabolomics, hair samples, and urine samples each. Among non-exposure variables, anthropometric (n = 11) and demographic features (n = 11) were often included while no studies evaluated genetic markers (Figure 3B).
Figure 3:
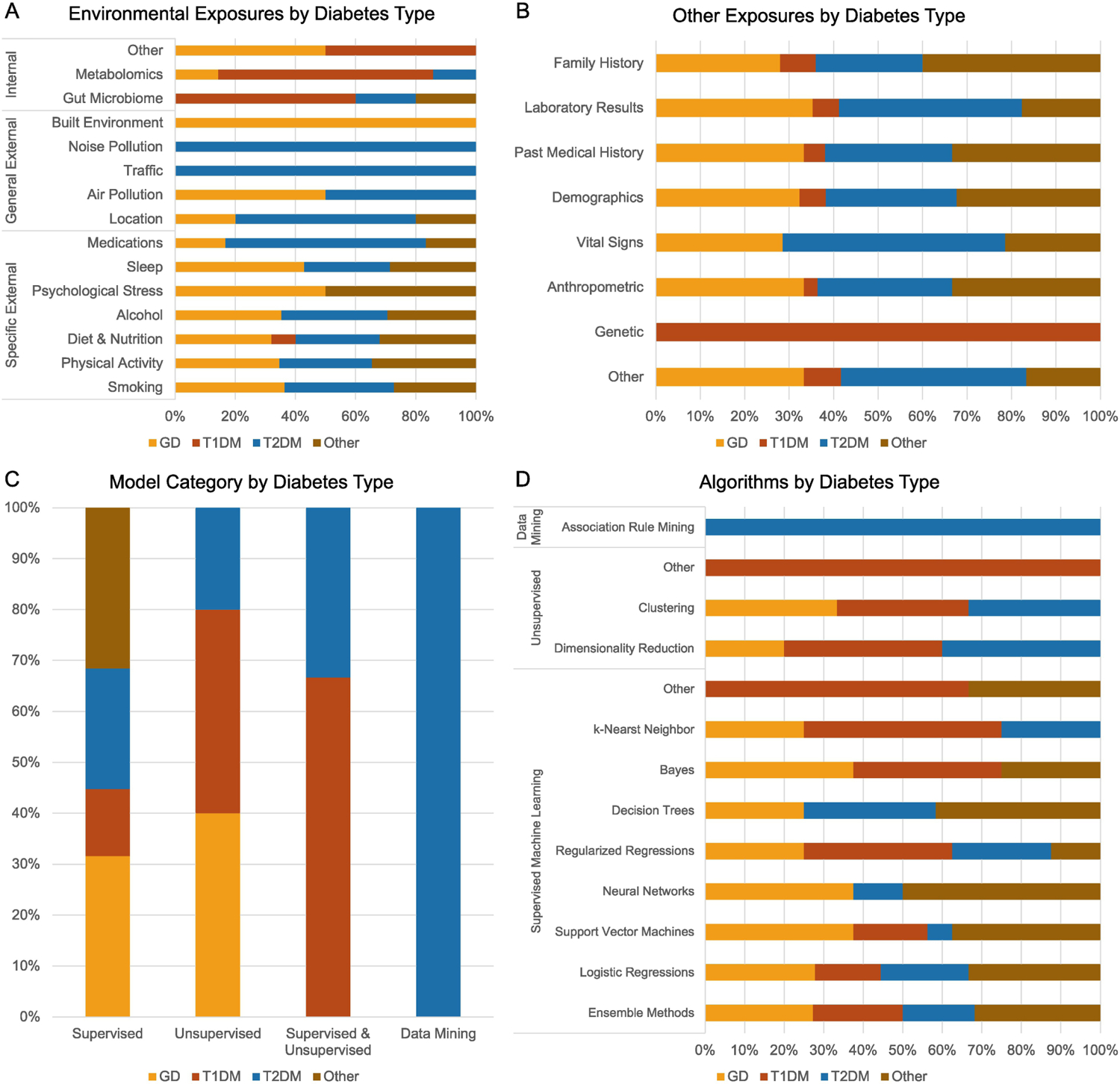
Description of Variables and Machine Learning Models by Diabetes Type. Types of diabetes include general diabetes (GD), type 1 diabetes mellitus (T1DM), type 2 diabetes mellitus (T2DM), and other diabetes. (A) Stacked bar graph of the exposure variables categorized by the Wild classification system and diabetes type. (B) Stacked bar graph of the non-exposure variables by diabetes type. (C) Stacked bar graph of the categories of machine learning and data mining methods used. (D) Stacked bar graph of the types of machine learning and data mining algorithms used.
3.2.3. Pre-processing Methods:
Of the 14 articles in this group, five completely removed missing variables, one applied mean imputation, one applied k-nearest neighbor imputation, and the remaining articles did not specify how missing data was handled. The most common train-test split was 80% training and 20% testing (n = 3), with two articles using a 70%−30% split, one using a 66%−33% split, and the remaining not specifying the train-test split. Class imbalance was addressed using Synthetic Minority Oversampling Technique (SMOTE) in two articles.
3.2.4. Machine Learning and Data Mining Methods:
The most common machine learning methods used were supervised (n = 12), while only two studies used unsupervised methods (Figure 3C). Of the 14 articles in this group, 9 tested and compared the performance of multiple machine learning models. Overall, studies assessing GD evaluated the widest range of machine learning algorithms (Table 1). The most common methods were ensemble methods (n = 6) and support vector machines (n = 6), followed by logistic regressions (n = 5) (Figure 3D). Cross-validation was performed in 10 articles, with 10-fold cross-validation being the most common (n = 7). Model performance was evaluated using accuracy (n = 6), precision (n = 2), recall (n = 6), specificity (n = 5), F-score (n = 3), and area under the receiver operating curve (AUROC) (n = 9). Feature importance was assessed in 7 articles.
3.2.5. Results:
Two studies comparing multiple models reported that neural networks achieved the best predictive performance. Esmaeily et al. (38) found that their artificial neural network model had improved accuracy and that the most important exposure variables were total cholesterol and triglyceride levels. Xie et al. (28) found that while their neural network model outperformed eight other models, the decision tree model demonstrated superior sensitivity and found that sleeping fewer than six hours per day increased the risk for diabetes. Olivera et al. (29) also compared the performance of several machine learning methods and found that both artificial neural networks and logistic regressions achieved the best AUROC; however, they did not detail variable importance.
Tree-based algorithms and ensemble methods demonstrated superior performance compared to other models. Esmaily et al. (27) compared the performance of decision trees and random forest algorithms. Their random forest model demonstrated improved specificity, accuracy, and AUROC and found that triglyceride, low-density lipoprotein, and total cholesterol levels were the most important features for predicting diabetes status (27). Cuesta et al. (35) also exclusively trained decision trees and reported body mass index and age as the overall most important features. Environmental exposures of the number of hours outside, psychological distress, physical activity, and lot trash were also important for diabetes prediction. In comparison, Dinh et al. (36) evaluated the performance of several algorithms, including logistic regressions, support vector machines, random forest, and gradient boosting. They found that XGBoost has the best performance, with sodium intake being the only exposure predictive of diabetes onset.
Non-tree-based ensemble methods were also used to predict GD. Deberneh et al. (32) found that ensemble methods achieved better accuracy than non-ensemble methods. The most predictive exposures included triglycerides, smoking status, drinking status, and physical activity (32). Interestingly, this set of features demonstrated superior prediction than the traditional 5-predictor model of plasma glucose, HbA1c, body mass index, age, and sex. Chen et al. (34) used principal component analysis on elemental concentrations derived from hair samples and found that age-dependent concentrations of zinc and iron distinguished GD cases from controls.
Two articles used chemical exposure data to predict the onset of GD (39, 40). Oh et al. (39) found that Bayesian network classifiers incorporating environment-polluting chemicals such as aryl hydrocarbon receptor ligands, mitochondria-inhabiting substances determined by ATP contents, and mitochondria-inhabiting substances determined by reactive oxygen species levels substantially improved predictive performance of impaired glucose tolerance and GD. In contrast, Wei et al. (40) used a LASSO regression model to predict GD using a combination of chemical exposures and other risk factors. Among the chemical exposures, lead, mercury, arsenic, and cadmium were the most influential features. However, they also found that environmental chemical exposures were less predictive of GD than traditional risk factors such as age, body mass index, and nutritional intake.
The remaining studies used a variety of machine learning techniques to predict GD. Yu et al. (37) found similar performance between support vector machine and logistic regression models; however, the feature importance of environmental exposure variables was low. Similarly, Chandrakar et al. (33) found that k-means clustering with the Euclidean distance identified three clusters that distinguished diabetic and non-diabetic patients but did not evaluate feature importance. Kumarage et al. (31) used the least absolute shrinkage and selection operator and found that the number of physical activity hours and number of sedentary life hours were important exposure predictors for diabetes. Li et al. (30) utilized metabolomics data and identified five potential diabetes biomarkers that were all implicated in hyperglycemia or deregulation of fatty acid metabolism.
3.3. Type 1 Diabetes Mellitus (T1DM):
This section discusses the following nine articles related to T1DM (41–49).
3.3.1. Patient Characteristics:
Studies predicting T1DM were only conducted in children (n = 9). The median percentage of females was equal to the median number of males (females: 50%, males: 50%) as reported in 5 studies.
3.3.2. Predictor Variables:
Internal exposures were the most common exposures in studies evaluating T1DM (Figure 3A), with three studies using gut microbiome data, five using metabolomics, and one using maternal vaginal microbiome data. In contrast to studies evaluating GD, no studies evaluating T1DM incorporated general external exposures, and only two incorporated diet and nutrition exposures. Among non-exposure variables, genetic markers were the most studied covariate (n = 3, Figure 3B).
3.3.3. Pre-processing Methods:
Missing data was imputed using GmSimpute, a label-free metabolomic imputation tool (n = 2), random forest (n = 1), or minimum value imputation (n = 1), while two articles completely removed missing data. Train-test split was not indicated in most articles, and class imbalance methods were not applied.
3.3.4. Machine Learning and Data Mining Methods:
Similar to articles studying GD, articles studying T1DM primarily used supervised algorithms (n = 5), though several used unsupervised (n = 2) or a combination of supervised and unsupervised algorithms (n = 2) (Figure 3C). Of the nine articles in this group, five compared the performance of multiple machine learning algorithms. Similar to articles studying GD, ensemble methods were the most common model trained (n = 5) (Figure 3D). Unlike other groups, decision trees and neural networks were not assessed by articles evaluating T1DM (Table 1). Cross-validation was performed in five articles and the most common performance metric was AUROC (n = 2). Feature importance was assessed in five articles.
3.3.5. Results:
Three studies applied a combination of unsupervised and supervised methods to evaluate gut microbiome data. Brown et al. (41) found that T1DM patients had fewer butyrate-producing and mucin-degrading bacteria and greater short-chain fatty acid-producing bacteria than controls. Biassoni et al. (42) found that patients with newly diagnosed T1DM demonstrated differential abundance of several Bacteroides, Bifidobacterium, and Gammaproteobacteria. Using random forest, unsupervised analysis did not identify any exposures related to gut microbiome clusters (42). However, other Bacteroides species implicated in the microbiota of T1DM individuals were identified as important features in a random forest algorithm developed by Fernandez-Edreira et al. (49). Unlike other studies, Ruotsalainen et al. (47) evaluated the role of maternal vaginal microbiomes to T1DM risk in infants. They found that an increase in bacteriome diversity and a decrease in mycobiome diversity were more common in mothers of children with T1DM compared to controls using principal components analysis. Using random forest models, they further found that bacteria and fungal data significantly improved the prediction of T1DM status compared to models generated with bacteria or fungi alone (47).
Two studies conducted by Li et al. (43, 46) utilized metabolomics and lipidomics data on an elastic net conditional logistic regression model. They found that islet autoimmunity was preceded by reduced serum proline and branched-chain amino acids (43). They further found that individuals with reduced serum ascorbic acid and cholesterol experienced islet autoimmunity at earlier ages and that serum Vitamin D levels were lower in individuals who progressed to T1DM using a gaussian clustering approach (46).
Three additional studies combined multivariate datasets to predict the development of islet autoantibodies and T1DM. Webb-Robertson et al. (44) used feature selection to identify a subset of environmental, genetic, and metabolomic variables and found that 22 metabolites and lipids, exposure to prebiotic formula, and 18 genetic markers were predictive of persistent islet autoantibody development. The following year, Webb-Robertson et al. (48) developed a model to predict the development of T1DM by six years of age using temporal environmental, genetic, and metabolomic measurements collected at 3, 6, and 9 months of life. Exposure to cow’s milk before age 6 months, three metabolites at three months, five metabolites at 6 months, and three metabolites at nine months were among the top predictive features (48). Frohnert et al. (45) also utilized genetic, immunologic, metabolomic, and proteomic biomarkers to predict the development of islet autoantibodies and progression to T1DM. They identified a novel set of 16 metabolomic markers that predicted islet autoantibody development and further found that serum glucose, adenosine diphosphate (ADP) fibrinogen, and mannose predicted progression to T1DM (45).
3.4. Type 2 Diabetes Mellitus (T2DM):
This section discusses the following 12 articles related to T2DM (50–61).
3.4.1. Patient Characteristics:
All studies predicting T2DM were conducted in adults (n = 12). Of the seven studies that reported the proportions of patients by sex, the median percentage of males was greater than females (females: 45%, males: 55%). One study stratified its predictive model by sex (54).
3.4.2. Predictor Variables:
Similar to studies evaluating GD, studies evaluating T2DM demonstrated a wide array of exposure variables included in training models (Figure 3A). Specific external exposures were frequently studied, specifically smoking (n = 8), physical activity (n = 8), and diet and nutrition (n = 7). General external exposure variables included were noise pollution (n = 1), traffic (n = 1), air pollution (n = 2), and urbanicity (n = 3). Few studies evaluated internal exposure variables. Studies evaluating T2DM also examined several non-exposure variables in predictive models, including anthropometric (n = 10) and demographic (n = 10) variables (Figure 3B).
3.4.3. Pre-processing Methods:
Most articles completely removed any instances of missing data (n = 8), and the remaining articles applied Classification and Regression Trees (CART) (n = 1), k-nearest neighbor (n = 1), and most frequent value imputation (n = 1). Train-test split strategies included 70–30 or 80–20 in three articles each. Two articles used SMOTE, and one used random under-sampling to correct for class imbalance.
3.4.4. Machine Learning and Data Mining Methods:
Supervised methods were the most used (n = 9), while two studies used unsupervised methods, one study used a combination of supervised and unsupervised methods, and one used data mining (Figure 3C). Seven articles compared the performance of multiple machine learning algorithms. Ensemble methods (n = 4), decision trees (n = 4) and ensemble methods (n = 4) were the most common algorithms trained (Figure 3D). Similar to the GD articles, T2DM articles implemented a wide array of supervised machine learning algorithms, though no studies implemented Bayesian methods (Table 1). Cross-validation was performed in six articles, with two studies using 5-fold cross-validation and four using 10-fold cross-validation. The most common performance metrics were AUROC (n = 6) and sensitivity (n = 5). Feature importance was assessed in seven articles.
3.4.5. Results:
Three studies were conducted using the Tehran Lipid and Glucose study data set by Ramezankhani et al. (50, 51, 54). The first study using association rule mining stratified results by sex and found that length of stay in the city greater than 40 years, total cholesterol to high-density lipoprotein ratio greater than 5.3, and low physical activity levels were exposures that increased the risk for diabetes occurrence in men only (50). The second study utilized decision trees and identified triglycerides as important environmental exposures for predicting T2DM (51). The third study also exclusively trained decision trees and found that the Quick Unbiased Efficient Statistical Tree (QUEST) algorithm had the highest sensitivity among models trained on male and female data (54).
Two studies evaluated the predictive performance of internal exposures in predicting T2DM. Peddiniti et al. (55) performed regularized least-square regression modeling and found nine metabolites that were negatively associated and 25 that were positively associated with progression to T2DM, including alpha-tocopherol and bradykinin hydroxyproline. Reitmeier et al. (52) identified 13 taxa with abnormal signatures in gut microbiome membership for patients with T2DM. These taxa all shared a common metabolic function with diurnal oscillations of gut bacteria, suggesting a link between the circadian rhythm and the gut microbiome in T2DM etiology (52).
Two studies identified dietary risk factors for T2DM. He et al. (53) compared the performance of a polyexposure risk model alone, a polygenic risk model alone, and a combination of polyexposure and polygenic risk model in predicting T2DM. The combination of polyexposure and polygenic risk models improved T2DM classification accuracy according to the C-statistic (53). Of the 12 most important variables, the exposure variables included alcohol intake, dietary changes in the past five years, milk type used by fat percentage, dietary restrictions, spread type used (butter or other), tea intake per day, and past tobacco use (53). Xue et al. compared the performance of several decision-tree-based models (56). They found that XGBoost has the best performance, and important exposures included smoking amount, physical activity, drinking status, the ratio of meat to vegetables in their diet, the amount of alcohol consumed, smoking status, and an oil-loving diet (56).
Two studies found that ensemble methods had the best performance metrics (59, 61). Though Ganie et al. (59) used lifestyle data in their model, they did not assess feature importance. In contrast, Liu et al. (61) used a combination of lifestyle data and laboratory measurements and found that exercise status was among the most important environmental exposures in predicting T2DM.
The remaining studies compared multiple machine learning approaches. Riches et al. (60) assessed the cooperative effects of multiple air pollution measurements on the incidence of T2DM using principal component analysis and k-means clustering. Clusters with the highest incidences of T2DM were associated with the highest mean concentrations of CO, NO2, PM10, PM2.5, and SO2 in one model and CO, NO2, Ni, NO3, Zn, and Zr in the second model. Wang et al. (58) evaluated T2DM using artificial neural networks and multivariate logistic regression models and found that the artificial neural network model yielded higher accuracy, sensitivity, specificity, and AUROC. However, the authors did not discuss what environmental exposures were important for prediction. Lam et al. (57) sought to determine if accelerometer data could distinguish individuals with T2DM from normoglycemic controls from UK Biobank participants. They trained random forest and hidden Markov models to classify temporal physical activity phenotypes derived from raw accelerometer data. They then trained XGBoost, random forest, and logistic regression models to classify T2DM patients from controls using these phenotypes, anthropometric, and environmental variables (57). All three classifiers demonstrated high AUROC and F1 scores.
3.5. Other (Prediabetes or Prediabetes and Diabetes):
This section discusses the following 12 articles related to other types of diabetes (36, 37, 52, 62–70).
3.5.1. Patient Characteristics:
Studies predicting prediabetes alone or prediabetes and diabetes together were primarily conducted in adults (n = 10), with one study in both children and adults and another in adolescents. Similar to the T2DM studies, the median percentage of males was greater than females in the seven studies that reported patient sex (females: 44%, males: 56%).
3.5.2. Predictor Variables:
Exposures studied in articles predicting prediabetes alone or prediabetes and diabetes together focused on specific external exposures similar to GD and T2DM studies (Figure 3A), and the most commonly studied variables were physical activity (n = 9), diet and nutrition (n = 8), and smoking (n = 6). Only one article included general external and internal exposures. Non-exposure variables were also studied, with anthropometric (n =11), demographics (n = 11), and family history (n = 10) being the most common (Figure 3B).
3.5.3. Pre-processing Methods:
Five articles completely removed missing variables, with two performing Multiple Imputation by Chained Equation (MICE) method, one using mean imputation, and the remaining did not specify how missing variables were handled. Train-test split was 70–30 or 80–20 in four articles each. One article used SMOTE analysis to correct for class imbalance, and another compared the performance of under-sampling, over-sampling, random over-sampling (ROSE), and SMOTE.
3.5.4. Machine Learning Methods:
Supervised algorithms were the most common methods used (n = 12) (Figure 3C). Nine articles compared the performance of multiple algorithms. Similar to GD and T2DM articles, ensemble methods were the most common algorithms trained (n = 7), followed by logistic regressions (n = 6), and support vector machines (n = 6), and decision trees (n = 5) (Figure 3D). Similar to the GD and T2DM articles, other articles implemented a wide array of supervised machine learning algorithms, though no studies implemented k-nearest neighbor methods (Table 1). Cross-validation was performed in eight articles, including 10-fold (n = 5) and 5-fold (n = 3). Model performance was most evaluated using AUROC (n = 8) and sensitivity (n = 7). Feature importance was assessed in 10 articles, and external validation was performed in three articles.
3.5.5. Results:
Three studies focused on the prediction of prediabetes only. Silva et al. (63) trained four machine learning algorithms and compared the performance to the Centers for Disease Control and Prevention (CDC) prediabetes screening tool. All four models outperformed the CDC tool, and 20 novel predictors were identified by the logistic regression model and five novel predictors by the ensemble models, of which environmental predictors included serum triglyceride, serum potassium, vigorous activity status, and serum calcium (63). Dihn et al. (36) found that ensemble models achieved higher AUROC scores in predicting prediabetes and found that sodium intake was predictive of prediabetes. Choi et al. (68) compared the performance of artificial neural networks and support vector machines using an internal and external validation dataset and found that the support vector machine model produced superior performance.
Nine studies grouped prediabetes and diabetes into one predictive outcome. Decision trees were identified as the most accurate models in five articles. Three articles conducted by Pei et al. (62, 64, 69) found that the J48 decision tree algorithm was most predictive of prediabetes or diabetes according to accuracy and AUROC. Together, these articles identified that less than six hours of sleep, work-related stress, salty food preference, and low physical activity levels were predictive of prediabetes or diabetes (62, 64, 69). Three additional studies compared the performance of multiple machine learning models. Meng et al. (66) found that decision trees demonstrated superior classification accuracy. This model identified environmental exposures of preference for salt food, coffee drinking status, and sleep duration as important predictors for prediabetes or diabetes (66). Syed et al. (67) found that decision trees performed better than other supervised machine learning according to the F1-score; however, the model-specific predictors were not evaluated. In contrast to the previous studies, Hu et al. (70) focused on the detection of adolescents with prediabetes and diabetes. They found that weighted voting classifiers yielded the highest AUROC and accuracy, and implicated exposures included dietary information such as water, protein, and sodium intake.
Only one article used internal exposures. Reitmeier et al. (52) performed Ward hierarchical clustering on gut microbiome data of patients with prediabetes and diabetes and identified three fecal microbial clusters. Cluster 1 had the lowest microbial richness and elevated relative abundance of Bacteroides, Cluster 2 had an elevated abundance of Ruminococcus, and Cluster 3 had an elevated abundance of Prevotella (52). One additional article sought to predict fasting plasma glucose levels for the diagnosis of patients with prediabetes or diabetes. Kopitar et al. (65) used gradient boosting machines, random forest, and generalized linear models with regularizations and identified smoking status, depression, and physical activity levels as important predictors of glucose levels in the diagnostic range for prediabetes and diabetes.
4. DISCUSSION:
This scoping review provides an overview of machine learning and data mining approaches utilizing environmental exposures to elucidate diabetes etiology. Across the 44 included articles, GD was the most common topic of the studies, specific external exposures were the most common exposures, and supervised machine learning models were the most common methods studied (Figure 2). Well-established specific external exposures of low physical activity, high cholesterol, and high triglycerides were predictive of GD, T2DM, and other diabetes, while novel metabolic and gut microbiome biomarkers were implicated in T1DM. Easily interpretable and explainable machine learning models, such as decision trees and support vector machines, were highly represented across studies investigating all types of diabetes.
Overall, this review summarized machine learning and data mining methods to understand environmental exposures in diabetes etiology, characterized the scope of exposures, and categorized the types of machine learning and data mining methods tested. Future studies should seek to leverage the full potential of machine learning by 1.) exploring a wider array of environmental exposures, 2.) investigating the temporality, sequences, and co-occurrence of exposures in diabetes etiology, 3.) discussing qualities of the big-data used in analysis and incorporating data from more-diverse datasets, and 4.) targeting novel disease-specific applications of machine learning and data mining methods.
Exposomics approaches offer new potential for investigating diabetes etiology and progression but remain largely untapped. Our analysis revealed that the use of machine learning and data mining methods to elucidate environmental triggers of diabetes were primarily limited to well-established risk factors (Figure 2E). General external exposures, including built environment, air pollution, climate, and socioeconomic factors, have been shown to play a role in diabetes etiology (71–73), but were only considered in a few studies included in this review. Therefore, future studies should seek to utilize machine learning and data mining methods to further evaluate the role of general external in combination with well-established specific external risk factors in diabetes etiology.
Temporality and co-occurrence of multiple exposures are important analytical considerations for assessing risk of environmental exposures in health and designing effective prevention strategies. Our findings indicated that no articles examined the temporality of exposures, and few explicitly examined co-occurrence of multiple exposures. Novel algorithms, including sequential pattern mining (74), long short term memory networks (75), and trajectory clustering (76–78), are well-equipped to investigate complex temporal, sequential, and co-occurrence relationships, but have thus far not been used to understand environmental components of diabetes etiology. Future studies should seek to utilize machine learning and data mining to explore the temporality and co-occurrence of multiple exposures to improve the understanding of environmental exposures in diabetes etiology. With respect to T1DM, using these methods to understand the relationship between exposures and immune markers is an open area of investigation.
Robust implementation of machine learning and data mining approaches requires big data that is high in volume, velocity, variety, value, and veracity (79). In this study, we found diversity in the volume and variety, but minimal discussion of velocity, value, and veracity. Drawing clinically-meaningful conclusions about the role of environmental exposures in diabetes etiology was difficult because there was significant variability in how environmental exposures were defined (Supplementary Table 3). These definitions reflect the value and veracity of big data and are important in assessing the external validity and reliability of machine learning and data mining approaches (80). Thus far, access to high-quality data in diabetes has been limited. Repositories containing open or limited access to a variety of longitudinal exposures, such as the Environmental Determinants of Diabetes in the Young (TEDDY) (81), Human Early Life Exposome (HELIX) (82), and the United Kingdom Biobank (83), will enable rigorous investigation of environmental exposures in diabetes etiology.
We identified disease-specific implementations of machine learning and data mining algorithms to elucidate environmental contributors to disease (Table 1). Among all types of diabetes included in this analysis, few studies evaluated unsupervised machine learning and data mining methods. Unsupervised machine learning and data mining methods are essential for characterizing patterns and identifying data-driven subgroups (84, 85) and these algorithms may serve as important tools in conducting analyzes of co-exposures. We also identified disease-specific gaps in applications of machine learning and data mining methods. Decision trees and neural networks were not assessed in studies evaluating T1DM, Bayesian models were not assessed in studies evaluating T2DM, and k-nearest neighbor models were not assessed in studies evaluating other diabetes. Future studies should seek to apply these methods to the disease-specific prediction of environmental exposures in diabetes subtypes.
Though there have been several reviews on machine learning and data mining methods in diabetes research, this is the first systematic approach to review machine learning and data mining methods used to understand environmental exposures in diabetes etiology and progression. Strengths of this review include the comprehensive search strategy and rigorous data extraction and analysis based on multiple validated guidelines for computational research. We used the Exposure Science Ontology to query environmental exposures; however, these criteria may not encompass all environmental exposures comprehensively. Additionally, we stratified our results by GD, T1DM, T2DM, and other diabetes. There may be exposures involved in diabetes etiology that overlap across these subtypes or other types of diabetes, such as monogenic diabetes of youth onset and gestational diabetes, but were not explored in this analysis.
5. CONCLUSION:
Environmental exposures play an important role in diabetes etiology and machine learning and data mining methods are poised to help identify novel environmental triggers. The objective of this work was to review articles that implemented machine learning and data mining methods to understand environmental exposures in diabetes etiology, characterize the scope of exposures analyzed, and categorize the types of machine learning and data mining methods tested. Our findings suggest that the use of artificial intelligence methods to elucidate environmental triggers of diabetes were largely limited to well-established risk factors identified using easily explainable and interpretable models. Future studies should seek to leverage the full potential of machine learning by exploring a wider array of environmental exposures, investigating the temporality and co-occurrence of exposures in diabetes etiology, utilizing more diverse datasets, and targeting novel disease-specific applications.
Supplementary Material
HIGHLIGHTS:
AI is poised to elucidate complex environmental exposures in diabetes etiology
Current approaches were limited to well-established risk factors using traditional models
Future work should leverage AI’s full potential while maintaining explainability
Studies exploring temporality, sequences & co-occurrence of environmental exposures are needed
ACKNOWLEDGEMENTS:
Funding Statement:
This work was partially supported by the National Library of Medicine (T15LM007124), the National Institute of Diabetes and Digestive and Kidney Diseases (F30DK134113), and the National Center for Advancing Translational Sciences (UL1TR002538).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest Statement: No competing financial interests exist.
REFERENCES:
- 1.Prevention CfDCa. National Diabetes Statistics Report, 2020. Atlanta, GA: Centers for Disease Control and Prevention, U.S. Dept of Health and Human Services, 2020. [Google Scholar]
- 2.Bertoni AG, Krop JS, Anderson GF, Brancati FL. Diabetes-Related Morbidity and Mortality in a National Sample of U.S. Elders. Diabetes Care. 2002;25(3):471–5. doi: 10.2337/diacare.25.3.471. [DOI] [PubMed] [Google Scholar]
- 3.Economic Costs of Diabetes in the U.S. in 2017. Diabetes Care. 2018;41(5):917–28. Epub 2018/03/24. doi: 10.2337/dci18-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Association AD. 2. Classification and Diagnosis of Diabetes: Standards of Medical Care in Diabetes—2021. Diabetes Care. 2020;44(Supplement_1):S15–S33. doi: 10.2337/dc21-S002. [DOI] [PubMed] [Google Scholar]
- 5.Mobasseri M, Shirmohammadi M, Amiri T, Vahed N, Hosseini Fard H, Ghojazadeh M. Prevalence and incidence of type 1 diabetes in the world: a systematic review and meta-analysis. Health Promot Perspect. 2020;10(2):98–115. Epub 2020/04/17. doi: 10.34172/hpp.2020.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khan MAB, Hashim MJ, King JK, Govender RD, Mustafa H, Al Kaabi J. Epidemiology of Type 2 Diabetes - Global Burden of Disease and Forecasted Trends. J Epidemiol Glob Health. 2020;10(1):107–11. Epub 2020/03/17. doi: 10.2991/jegh.k.191028.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knip M, Veijola R, Virtanen SM, Hyöty H, Vaarala O, Akerblom HK. Environmental triggers and determinants of type 1 diabetes. Diabetes. 2005;54 Suppl 2:S125–36. Epub 2005/11/25. doi: 10.2337/diabetes.54.suppl_2.s125 [DOI] [PubMed] [Google Scholar]
- 8.Murea M, Ma L, Freedman BI. Genetic and environmental factors associated with type 2 diabetes and diabetic vascular complications. Rev Diabet Stud. 2012;9(1):6–22. Epub 2012/09/14. doi: 10.1900/rds.2012.9.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vrijheid M. The exposome: a new paradigm to study the impact of environment on health. Thorax. 2014;69(9):876–8. Epub 2014/06/08. doi: 10.1136/thoraxjnl-2013-204949 [DOI] [PubMed] [Google Scholar]
- 10.Wild CP. The exposome: from concept to utility. Int J Epidemiol. 2012;41(1):24–32. Epub 2012/02/03. doi: 10.1093/ije/dyr236 [DOI] [PubMed] [Google Scholar]
- 11.Rewers M, Ludvigsson J. Environmental risk factors for type 1 diabetes. Lancet. 2016;387(10035):2340–8. Epub 2016/06/16. doi: 10.1016/s0140-6736(16)30507-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kolb H, Martin S. Environmental/lifestyle factors in the pathogenesis and prevention of type 2 diabetes. BMC Medicine. 2017;15(1):131. doi: 10.1186/s12916-017-0901-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rønningen KS. Environmental Trigger(s) of Type 1 Diabetes: Why So Difficult to Identify? BioMed Research International. 2015;2015:321656. doi: 10.1155/2015/321656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dendup T, Feng X, Clingan S, Astell-Burt T. Environmental Risk Factors for Developing Type 2 Diabetes Mellitus: A Systematic Review. Int J Environ Res Public Health. 2018;15(1). Epub 2018/01/06. doi: 10.3390/ijerph15010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kavakiotis I, Tsave O, Salifoglou A, Maglaveras N, Vlahavas I, Chouvarda I. Machine Learning and Data Mining Methods in Diabetes Research. Comput Struct Biotechnol J. 2017;15:104–16. doi: 10.1016/j.csbj.2016.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharma T, Shah M. A comprehensive review of machine learning techniques on diabetes detection. Visual Computing for Industry, Biomedicine, and Art. 2021;4(1):30. doi: 10.1186/s42492-021-00097-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woldaregay AZ, Årsand E, Walderhaug S, Albers D, Mamykina L, Botsis T, et al. Data-driven modeling and prediction of blood glucose dynamics: Machine learning applications in type 1 diabetes. Artif Intell Med. 2019;98:109–34. Epub 2019/08/07. doi: 10.1016/j.artmed.2019.07.007 [DOI] [PubMed] [Google Scholar]
- 18.Jaiswal V, Negi A, Pal T. A review on current advances in machine learning based diabetes prediction. Prim Care Diabetes. 2021;15(3):435–43. Epub 2021/03/02. doi: 10.1016/j.pcd.2021.02.005 [DOI] [PubMed] [Google Scholar]
- 19.Saputro SA, Pattanaprateep O, Pattanateepapon A, Karmacharya S, Thakkinstian A. Prognostic models of diabetic microvascular complications: a systematic review and meta-analysis. Syst Rev. 2021;10(1):288. Epub 20211101. doi: 10.1186/s13643-021-01841-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choubey DK, Kumar M, Shukla V, Tripathi S, Dhandhania VK. Comparative Analysis of Classification Methods with PCA and LDA for Diabetes. Curr Diabetes Rev. 2020;16(8):833–50. doi: 10.2174/1573399816666200123124008 [DOI] [PubMed] [Google Scholar]
- 21.Tricco AC, Lillie E, Zarin W, O’Brien KK, Colquhoun H, Levac D, et al. PRISMA Extension for Scoping Reviews (PRISMA-ScR): Checklist and Explanation. Ann Intern Med. 2018;169(7):467–73. Epub 2018/09/05. doi: 10.7326/m18-0850 [DOI] [PubMed] [Google Scholar]
- 22.Mattingly CJ, McKone TE, Callahan MA, Blake JA, Hubal EA. Providing the missing link: the exposure science ontology ExO. Environ Sci Technol. 2012;46(6):3046–53. Epub 2012/02/14. doi: 10.1021/es2033857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo W, Phung D, Tran T, Gupta S, Rana S, Karmakar C, et al. Guidelines for Developing and Reporting Machine Learning Predictive Models in Biomedical Research: A Multidisciplinary View. J Med Internet Res. 2016;18(12):e323. Epub 2016/12/18. doi: 10.2196/jmir.5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moons KG, Altman DG, Reitsma JB, Ioannidis JP, Macaskill P, Steyerberg EW, et al. Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD): explanation and elaboration. Ann Intern Med. 2015;162(1):W1–73. Epub 2015/01/07. doi: 10.7326/m14-0698 [DOI] [PubMed] [Google Scholar]
- 25.Fayyad U, Piatetsky-Shapiro G, Smyth P. From Data Mining to Knowledge Discovery in Databases. AI Magazine. 1996;17(3):37. doi: 10.1609/aimag.v17i3.1230. [DOI] [Google Scholar]
- 26.Janga SC, Zhu D, Chen JY, Zaki MJ. Knowledge Discovery Using Big Data in Biomedical Systems [Guest Editorial]. IEEE/ACM Transactions on Computational Biology and Bioinformatics. 2015;12(4):726–8. doi: 10.1109/TCBB.2015.2454551. [DOI] [PubMed] [Google Scholar]
- 27.Esmaily H, Tayefi M, Doosti H, Ghayour-Mobarhan M, Nezami H, Amirabadizadeh A. A Comparison between Decision Tree and Random Forest in Determining the Risk Factors Associated with Type 2 Diabetes. J Res Health Sci. 2018;18(2):e00412. Epub 2018/05/23 [PubMed] [Google Scholar]
- 28.Xie Z, Nikolayeva O, Luo J, Li D. Building Risk Prediction Models for Type 2 Diabetes Using Machine Learning Techniques. Prev Chronic Dis. 2019;16:E130. Epub 2019/09/21. doi: 10.5888/pcd16.190109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olivera AR, Roesler V, Iochpe C, Schmidt MI, Vigo A, Barreto SM, et al. Comparison of machine-learning algorithms to build a predictive model for detecting undiagnosed diabetes - ELSA-Brasil: accuracy study. Sao Paulo Med J. 2017;135(3):234–46. Epub 2017/07/27. doi: 10.1590/1516-3180.2016.0309010217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li X, Xu Z, Lu X, Yang X, Yin P, Kong H, et al. Comprehensive two-dimensional gas chromatography/time-of-flight mass spectrometry for metabonomics: Biomarker discovery for diabetes mellitus. Anal Chim Acta. 2009;633(2):257–62. Epub 2009/01/27. doi: 10.1016/j.aca.2008.11.058 [DOI] [PubMed] [Google Scholar]
- 31.Kumarage PM, Yogarajah B, Ratnarajah N, editors. Efficient Feature Selection for Prediction of Diabetic Using LASSO. 2019 19th International Conference on Advances in ICT for Emerging Regions (ICTer); 2019. 2–5 Sept. 2019. [Google Scholar]
- 32.Deberneh HM, Kim I. Prediction of Type 2 Diabetes Based on Machine Learning Algorithm. Int J Environ Res Public Health. 2021;18(6). Epub 2021/04/04. doi: 10.3390/ijerph18063317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chandrakar O, Saini JR. Development of Indian Weighted Diabetic Risk Score (IWDRS) using Machine Learning Techniques for Type-2 Diabetes. Proceedings of the 9th Annual ACM India Conference; Gandhinagar, India: Association for Computing Machinery; 2016. p. 125–8. [Google Scholar]
- 34.Chen H, Tan C, Lin Z, Wu T. The diagnostics of diabetes mellitus based on ensemble modeling and hair/urine element level analysis. Comput Biol Med. 2014;50:70–5. Epub 2014/05/20. doi: 10.1016/j.compbiomed.2014.04.012 [DOI] [PubMed] [Google Scholar]
- 35.Cuesta HA, Coffman DL, Branas C, Murphy HM. Using decision trees to understand the influence of individual- and neighborhood-level factors on urban diabetes and asthma. Health Place. 2019;58:102119. Epub 2019/06/17. doi: 10.1016/j.healthplace.2019.04.009 [DOI] [PubMed] [Google Scholar]
- 36.Dinh A, Miertschin S, Young A, Mohanty SD. A data-driven approach to predicting diabetes and cardiovascular disease with machine learning. BMC Med Inform Decis Mak. 2019;19(1):211. Epub 2019/11/07. doi: 10.1186/s12911-019-0918-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu W, Liu T, Valdez R, Gwinn M, Khoury MJ. Application of support vector machine modeling for prediction of common diseases: the case of diabetes and pre-diabetes. BMC Med Inform Decis Mak. 2010;10:16. Epub 2010/03/24. doi: 10.1186/1472-6947-10-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Esmaeily H, Tayefi M, Ghayour-Mobarhan M, Amirabadizadeh A. Comparing Three Data Mining Algorithms for Identifying the Associated Risk Factors of Type 2 Diabetes. Iran Biomed J. 2018;22(5):303–11. Epub 2018/01/28. doi: 10.29252/ibj.22.5.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oh R, Lee HK, Pak YK, Oh MS. An Interactive Online App for Predicting Diabetes via Machine Learning from Environment-Polluting Chemical Exposure Data. Int J Environ Res Public Health. 2022;19(10). Epub 20220510. doi: 10.3390/ijerph19105800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wei H, Sun J, Shan W, Xiao W, Wang B, Ma X, et al. Environmental chemical exposure dynamics and machine learning-based prediction of diabetes mellitus. Sci Total Environ. 2022;806(Pt 2):150674. Epub 20210929. doi: 10.1016/j.scitotenv.2021.150674 [DOI] [PubMed] [Google Scholar]
- 41.Brown CT, Davis-Richardson AG, Giongo A, Gano KA, Crabb DB, Mukherjee N, et al. Gut microbiome metagenomics analysis suggests a functional model for the development of autoimmunity for type 1 diabetes. PLoS One. 2011;6(10):e25792. Epub 2011/11/02. doi: 10.1371/journal.pone.0025792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Biassoni R, Di Marco E, Squillario M, Barla A, Piccolo G, Ugolotti E, et al. Gut Microbiota in T1DM-Onset Pediatric Patients: Machine-Learning Algorithms to Classify Microorganisms as Disease Linked. The Journal of Clinical Endocrinology & Metabolism. 2020;105(9):e3114–e26. doi: 10.1210/clinem/dgaa407. [DOI] [PubMed] [Google Scholar]
- 43.Li Q, Parikh H, Butterworth MD, Lernmark A, Hagopian W, Rewers M, et al. Longitudinal Metabolome-Wide Signals Prior to the Appearance of a First Islet Autoantibody in Children Participating in the TEDDY Study. Diabetes. 2020;69(3):465–76. Epub 2020/02/08. doi: 10.2337/db19-0756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Webb-Robertson BM, Bramer LM, Stanfill BA, Reehl SM, Nakayasu ES, Metz TO, et al. Prediction of the development of islet autoantibodies through integration of environmental, genetic, and metabolic markers. J Diabetes. 2021;13(2):143–53. Epub 2020/10/31. doi: 10.1111/1753-0407.13093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frohnert BI, Webb-Robertson BJ, Bramer LM, Reehl SM, Waugh K, Steck AK, et al. Predictive Modeling of Type 1 Diabetes Stages Using Disparate Data Sources. Diabetes. 2020;69(2):238–48. Epub 2019/11/20. doi: 10.2337/db18-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Q, Liu X, Yang J, Erlund I, Lernmark A, Hagopian W, et al. Plasma Metabolome and Circulating Vitamins Stratified Onset Age of an Initial Islet Autoantibody and Progression to Type 1 Diabetes: The TEDDY Study. Diabetes. 2021;70(1):282–92. Epub 2020/10/28. doi: 10.2337/db20-0696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ruotsalainen AL, Tejesvi MV, Vanni P, Suokas M, Tossavainen P, Pirttila AM, et al. Child type 1 diabetes associated with mother vaginal bacteriome and mycobiome. Med Microbiol Immunol. 2022;211(4):185–94. Epub 20220614. doi: 10.1007/s00430-022-00741-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Webb-Robertson BM, Nakayasu ES, Frohnert BI, Bramer LM, Akers SM, Norris JM, et al. Integration of Infant Metabolite, Genetic, and Islet Autoimmunity Signatures to Predict Type 1 Diabetes by Age 6 Years. J Clin Endocrinol Metab. 2022;107(8):2329–38. doi: 10.1210/clinem/dgac225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fernández-Edreira D, Liñares-Blanco J, Fernandez-Lozano C. Machine Learning analysis of the human infant gut microbiome identifies influential species in type 1 diabetes. Expert Systems with Applications. 2021;185:115648. doi: 10.1016/j.eswa.2021.115648. [DOI] [Google Scholar]
- 50.Ramezankhani A, Pournik O, Shahrabi J, Azizi F, Hadaegh F. An application of association rule mining to extract risk pattern for type 2 diabetes using tehran lipid and glucose study database. Int J Endocrinol Metab. 2015;13(2):e25389. Epub 2015/05/01. doi: 10.5812/ijem.25389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramezankhani A, Pournik O, Shahrabi J, Khalili D, Azizi F, Hadaegh F. Applying decision tree for identification of a low risk population for type 2 diabetes. Tehran Lipid and Glucose Study. Diabetes Res Clin Pract. 2014;105(3):391–8. Epub 2014/08/03. doi: 10.1016/j.diabres.2014.07.003 [DOI] [PubMed] [Google Scholar]
- 52.Reitmeier S, Kiessling S, Clavel T, List M, Almeida EL, Ghosh TS, et al. Arrhythmic Gut Microbiome Signatures Predict Risk of Type 2 Diabetes. Cell Host Microbe. 2020;28(2):258–72 e6. Epub 2020/07/04. doi: 10.1016/j.chom.2020.06.004 [DOI] [PubMed] [Google Scholar]
- 53.He Y, Lakhani CM, Rasooly D, Manrai AK, Tzoulaki I, Patel CJ. Comparisons of Polyexposure, Polygenic, and Clinical Risk Scores in Risk Prediction of Type 2 Diabetes. Diabetes Care. 2021;44(4):935–43. Epub 2021/02/11. doi: 10.2337/dc20-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ramezankhani A, Hadavandi E, Pournik O, Shahrabi J, Azizi F, Hadaegh F. Decision tree-based modelling for identification of potential interactions between type 2 diabetes risk factors: a decade follow-up in a Middle East prospective cohort study. BMJ Open. 2016;6(12):e013336. Epub 2016/12/03. doi: 10.1136/bmjopen-2016-013336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peddinti G, Cobb J, Yengo L, Froguel P, Kravic J, Balkau B, et al. Early metabolic markers identify potential targets for the prevention of type 2 diabetes. Diabetologia. 2017;60(9):1740–50. Epub 2017/06/10. doi: 10.1007/s00125-017-4325-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xue M, Su Y, Li C, Wang S, Yao H. Identification of Potential Type II Diabetes in a Large-Scale Chinese Population Using a Systematic Machine Learning Framework. J Diabetes Res. 2020;2020:6873891. Epub 2020/10/09. doi: 10.1155/2020/6873891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lam B, Catt M, Cassidy S, Bacardit J, Darke P, Butterfield S, et al. Using Wearable Activity Trackers to Predict Type 2 Diabetes: Machine Learning-Based Cross-sectional Study of the UK Biobank Accelerometer Cohort. JMIR Diabetes. 2021;6(1):e23364. Epub 2021/03/20. doi: 10.2196/23364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang C, Li L, Wang L, Ping Z, Flory MT, Wang G, et al. Evaluating the risk of type 2 diabetes mellitus using artificial neural network: an effective classification approach. Diabetes Res Clin Pract. 2013;100(1):111–8. Epub 2013/03/05. doi: 10.1016/j.diabres.2013.01.023 [DOI] [PubMed] [Google Scholar]
- 59.Ganie SM, Malik MB, Arif T. Performance analysis and prediction of type 2 diabetes mellitus based on lifestyle data using machine learning approaches. J Diabetes Metab Disord. 2022;21(1):339–52. Epub 20220314. doi: 10.1007/s40200-022-00981-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Riches NO, Gouripeddi R, Payan-Medina A, Facelli JC. K-means cluster analysis of cooperative effects of CO, NO2, O3, PM2.5, PM10, and SO2 on incidence of type 2 diabetes mellitus in the US. Environ Res. 2022;212(Pt B):113259. Epub 20220420. doi: 10.1016/j.envres.2022.113259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu Q, Zhang M, He Y, Zhang L, Zou J, Yan Y, et al. Predicting the Risk of Incident Type 2 Diabetes Mellitus in Chinese Elderly Using Machine Learning Techniques. J Pers Med. 2022;12(6). Epub 20220531. doi: 10.3390/jpm12060905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pei D, Yang T, Zhang C. Estimation of Diabetes in a High-Risk Adult Chinese Population Using J48 Decision Tree Model. Diabetes Metab Syndr Obes. 2020;13:4621–30. Epub 2020/12/05. doi: 10.2147/DMSO.S279329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.De Silva K, Jonsson D, Demmer RT. A combined strategy of feature selection and machine learning to identify predictors of prediabetes. J Am Med Inform Assoc. 2020;27(3):396–406. Epub 2020/01/01. doi: 10.1093/jamia/ocz204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pei D, Gong Y, Kang H, Zhang C, Guo Q. Accurate and rapid screening model for potential diabetes mellitus. BMC Med Inform Decis Mak. 2019;19(1):41. Epub 2019/03/15. doi: 10.1186/s12911-019-0790-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kopitar L, Cilar L, Kocbek P, Stiglic G, editors. Local vs. Global Interpretability of Machine Learning Models in Type 2 Diabetes Mellitus Screening 2019; Cham: Springer International Publishing. [Google Scholar]
- 66.Meng XH, Huang YX, Rao DP, Zhang Q, Liu Q. Comparison of three data mining models for predicting diabetes or prediabetes by risk factors. Kaohsiung J Med Sci. 2013;29(2):93–9. Epub 2013/01/26. doi: 10.1016/j.kjms.2012.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Syed AH, Khan T. Machine Learning-Based Application for Predicting Risk of Type 2 Diabetes Mellitus (T2DM) in Saudi Arabia: A Retrospective Cross-Sectional Study. IEEE Access. 2020;8:199539–61. doi: 10.1109/ACCESS.2020.3035026. [DOI] [Google Scholar]
- 68.Choi SB, Kim WJ, Yoo TK, Park JS, Chung JW, Lee YH, et al. Screening for prediabetes using machine learning models. Comput Math Methods Med. 2014;2014:618976. Epub 2014/08/29. doi: 10.1155/2014/618976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pei D, Zhang C, Quan Y, Guo Q. Identification of Potential Type II Diabetes in a Chinese Population with a Sensitive Decision Tree Approach. J Diabetes Res. 2019;2019:4248218. Epub 2019/02/26. doi: 10.1155/2019/4248218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hu H, Lai T, Farid F. Feasibility Study of Constructing a Screening Tool for Adolescent Diabetes Detection Applying Machine Learning Methods. Sensors (Basel). 2022;22(16). Epub 20220817. doi: 10.3390/s22166155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pasala SK, Rao AA, Sridhar GR. Built environment and diabetes. Int J Diabetes Dev Ctries. 2010;30(2):63–8. doi: 10.4103/0973-3930.62594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thiering E, Heinrich J. Epidemiology of air pollution and diabetes. Trends in Endocrinology & Metabolism. 2015;26(7):384–94. [DOI] [PubMed] [Google Scholar]
- 73.Everson SA, Maty SC, Lynch JW, Kaplan GA. Epidemiologic evidence for the relation between socioeconomic status and depression, obesity, and diabetes. Journal of psychosomatic research. 2002;53(4):891–5. [DOI] [PubMed] [Google Scholar]
- 74.Fournier-Viger P, Lin JC-W, Kiran RU, Koh YS, Thomas R. A survey of sequential pattern mining. Data Science and Pattern Recognition. 2017;1(1):54–77. [Google Scholar]
- 75.Malhotra P, Vig L, Shroff G, Agarwal P, editors. Long short term memory networks for anomaly detection in time series. Proceedings; 2015. [Google Scholar]
- 76.Lee J-G, Han J, Whang K-Y, editors. Trajectory clustering: a partition-and-group framework. Proceedings of the 2007 ACM SIGMOD international conference on Management of data; 2007. [Google Scholar]
- 77.Genolini C, Falissard B. KmL: k-means for longitudinal data. Computational Statistics. 2010;25(2):317–28. [Google Scholar]
- 78.Genolini C, Pingault J-B, Driss T, Côté S, Tremblay RE, Vitaro F, et al. KmL3D: a nonparametric algorithm for clustering joint trajectories. Computer methods and programs in biomedicine. 2013;109(1):104–11. [DOI] [PubMed] [Google Scholar]
- 79.Demchenko Y, Grosso P, Laat Cd, Membrey P, editors. Addressing big data issues in Scientific Data Infrastructure. 2013 International Conference on Collaboration Technologies and Systems (CTS); 2013. 20–24 May 2013. [Google Scholar]
- 80.Rajkomar A, Dean J, Kohane I. Machine learning in medicine. New England Journal of Medicine. 2019;380(14):1347–58. [DOI] [PubMed] [Google Scholar]
- 81.Group TS. The Environmental Determinants of Diabetes in the Young (TEDDY) Study. Ann N Y Acad Sci. 2008;1150:1–13. doi: 10.1196/annals.1447.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Maitre L, de Bont J, Casas M, Robinson O, Aasvang GM, Agier L, et al. Human Early Life Exposome (HELIX) study: a European population-based exposome cohort. BMJ Open. 2018;8(9):e021311. Epub 20180910. doi: 10.1136/bmjopen-2017-021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, et al. UK Biobank: An Open Access Resource for Identifying the Causes of a Wide Range of Complex Diseases of Middle and Old Age. PLOS Medicine. 2015;12(3):e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Koh HC, Tan G. Data mining applications in healthcare. J Healthc Inf Manag. 2005;19(2):64–72 [PubMed] [Google Scholar]
- 85.Deo RC. Machine Learning in Medicine. Circulation. 2015;132(20):1920–30. doi: 10.1161/circulationaha.115.001593. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


