Abstract
We propose a BlackBox Counterfactual Explainer, designed to explain image classification models for medical applications. Classical approaches (e.g., , saliency maps) that assess feature importance do not explain how imaging features in important anatomical regions are relevant to the classification decision. Such reasoning is crucial for transparent decision-making in healthcare applications. Our framework explains the decision for a target class by gradually exaggerating the semantic effect of the class in a query image. We adopted a Generative Adversarial Network (GAN) to generate a progressive set of perturbations to a query image, such that the classification decision changes from its original class to its negation. Our proposed loss function preserves essential details (e.g., support devices) in the generated images.
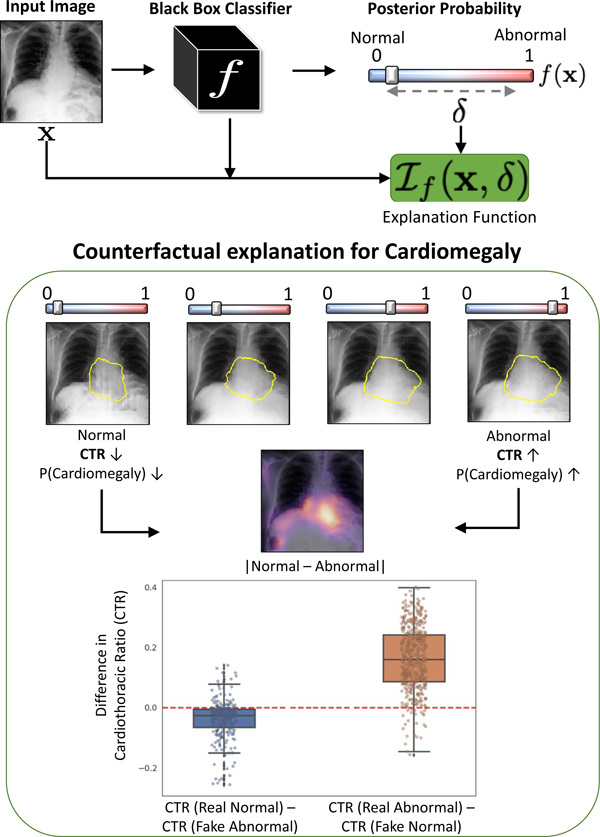
We used counterfactual explanations from our framework to audit a classifier trained on a chest x-ray dataset with multiple labels. Clinical evaluation of model explanations is a challenging task. We proposed clinically-relevant quantitative metrics such as cardiothoracic ratio and the score of a healthy costophrenic recess to evaluate our explanations. We used these metrics to quantify the counterfactual changes between the populations with negative and positive decisions for a diagnosis by the given classifier.
We conducted a human-grounded experiment with diagnostic radiology residents to compare different styles of explanations (no explanation, saliency map, cycleGAN explanation, and our counterfactual explanation) by evaluating different aspects of explanations: (1) understandability, (2) classifier’s decision justification, (3) visual quality, (d) identity preservation, and (5) overall helpfulness of an explanation to the users. Our results show that our counterfactual explanation was the only explanation method that significantly improved the users’ understanding of the classifier’s decision compared to the no-explanation baseline. Our metrics established a benchmark for evaluating model explanation methods in medical images. Our explanations revealed that the classifier relied on clinically relevant radiographic features for its diagnostic decisions, thus making its decision-making process more transparent to the end-user.
Keywords: Explainable AI, Interpretable Machine Learning, Counterfactual Reasoning, Chest X-Ray diagnosis
Graphical Abstract
1. Introduction
Machine learning, specifically Deep Learning (DL), is being increasingly used for sensitive applications such as Computer-Aided Diagnosis (Hosny et al., 2018) and other tasks in the medical imaging domain (Rajpurkar et al., 2018; Rodriguez-Ruiz et al., 2019). However, for real-world deployment (Wang et al., 2020), the decision-making process of these models should be explainable to humans to obtain their trust in the model (Gastounioti and Kontos, 2020; Jiang et al., 2018). Explainability is essential for auditing the model (Winkler et al., 2019), identifying various failure modes (Oakden-Rayner et al., 2020; Eaton-Rosen et al., 2018) or hidden biases in the data or the model (Larrazabal et al., 2020), and for obtaining new insights from large-scale studies (Rubin et al., 2018).
With the advancement of DL methods for medical imaging analysis, deep neural networks (DNNs) have achieved near-radiologist performance in multiple image classification tasks (Seah et al., 2021; Rajpurkar et al., 2017). However, DNNs are criticized for their “black-box” nature, i.e., they fail to provide a simple explanation as to why a given input image produces a corresponding output (Tonekaboni et al., 2019).To address this concern, multiple model explanation techniques have been proposed that aim to explain the decision-making process of DNNs (Selvaraju et al., 2017; Cohen et al., 2021). The most common form of explanation in medical imaging is a class-specific heatmap overlaid on the input image. It highlights the most relevant regions (where) for the classification decision (Rajpurkar et al., 2017; Young et al., 2019). However, the location information alone is insufficient for applications in medical imaging. Different diagnoses may affect the same anatomical regions, resulting in similar explanations for multiple diagnosis, resulting in inconclusive explanations. A thorough explanation should explain what imaging features are present in those important locations, and how changing such features modifies the classification decision.
To address this problem, we propose a novel explanation method to provide a counterfactual explanation. A counterfactual explanation is a perturbation of the input image such that the classification decision is flipped. By comparing, the input image and its corresponding counterfactual image, the end-users can visualize the difference in important image features that leads to a change in classification decision. Fig. 1 shows an example. The input image is predicted as positive for pleural effusion (PE), while the generated counterfactual image is negative for PE. The changes are mostly concentrated in the lower lobe region, which is known to be clinically important for PE (Lababede, 2017). Counterfactual explanation is used to derive a pseudo-heat-map, highlighting the regions that change the most in the transformation (difference map in Fig. 1).
Fig. 1.
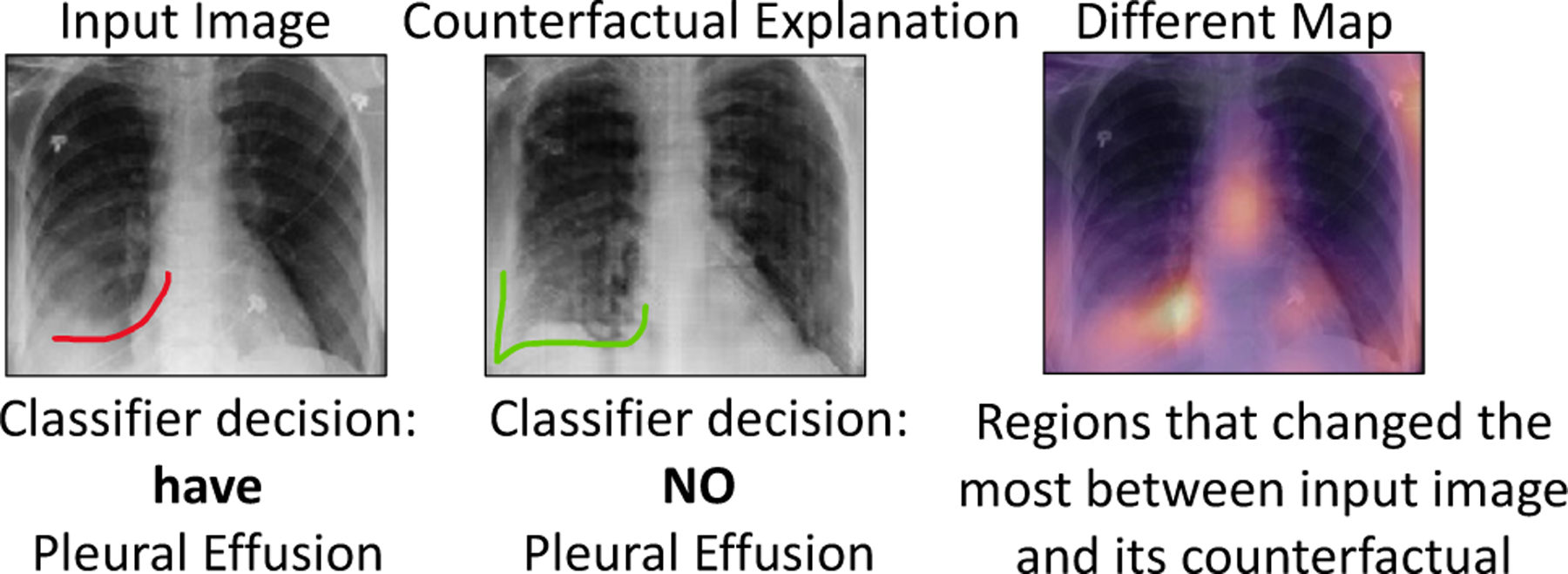
Counterfactual explanation shows “where” + “what” minimum change must be made to the input to flip the classification decision. For Pleural Effusion, we can observe vanishing of the meniscus (red) in counterfactual image as compared to the query image.
We demonstrate the performance of the counterfactual explainer on a chest x-ray (CXR) dataset. Rather than generating just one counterfactual image at the end of the prediction spectrum, our explanation function generates a series of perturbed images that gradually traverse the decision boundary from one extreme (negative decision) to another (positive decision) for a given target class. We adopted a conditional Generative Adversarial Network (cGAN) as our explanation function (Singla et al., 2019). We extend the cGAN to preserve small or uncommon details during image generation (Bau et al., 2019). Preserving such details is particularly important in our application, as the missing information may include support devices that may influence human users’ perceptions. To this end, we incorporated semantic segmentation and object detection into our loss function to preserve the shape of the anatomy and foreign objects during image reconstruction. We evaluated the quality of our explanations using different quantitative metrics, including clinical measures. Further, we performed a clinical study with 12 radiology residents to compare the explanations for the proposed method and the baseline models.
1.1. Related work
Posthoc explanation is a popular approach that aims to improve human understanding of a pre-trained classifier. Our work broadly relates to the following posthoc methods:
Feature Attribution methods provide explanation by producing a saliency map that shows the importance of each input component (e.g., pixel) to the classification decision. Gradient-based approaches (Simonyan et al., 2013; Springenberg et al., 2015; Bach et al., 2015; Shrikumar et al., 2017; Sundararajan et al., 2017; Lundberg et al., 2017; Selvaraju et al., 2017) produce a saliency map by computing the gradient of the classifier’s output with respect to the input components. Such methods are often applied to the medical imaging studies, e.g., CXR (Rajpurkar et al., 2017), skin imaging (Young et al., 2019), brain MRI (Eitel and Ritter, 2019) and retinopathy (Sayres et al., 2019).
Perturbation-based methods identify salient regions by directly manipulating the input image and analyzing the resulting changes in the classifier’s output. Such methods modify specific pixels or regions in an input image, either by masking with constant values (Dabkowski and Gal, 2017) or with random noise, occluding (Zhou et al., 2015), localized blurring (Fong and Vedaldi, 2017), or in-filling (Chang et al., 2019). Especially for medical images, such perturbations may introduce anatomically implausible features or textures. Our proposed method also generates a perturbation of the query image such that classification decision is flipped. But in contrast to the above methods, we enforce consistency between the perturbed data and the real data distribution to ensure that the perturbations are plausible and visually similar to the input.
Counterfactual Explanations are a type of contrastive (Dhurandhar et al., 2018) explanation that provides a useful way to audit the classifier and determine causal attributes that lead to the classification decision (Parafita Martinez and Vitria Marca, 2019; Singla et al., 2021). Similar to our method, generative models like GANs and variational autoencoders (VAE) are used to compute interventions that generate realistic counterfactual explanations (Cohen et al., 2021; Joshi et al., 2019). Much of this work is limited to simpler image datasets like MNIST, celebA (Liu et al., 2019; Van Looveren and Klaise, 2019) or simulated data (Parafita Martinez and Vitria Marca, 2019). For more complex natural images, previous studies (Chang et al., 2019; Agarwal and Nguyen, 2020) focused on finding and infilling salient regions to generate counterfactual images. In contrast, our explanation function doesn’t require any re-training for generating explanations for a new image at inference time. In another line of work (Wang and Vasconcelos, 2020; Goyal et al., 2019) provide counterfactual explanations that explain both the predicted and the counter class. Further, researcher (Narayanaswamy et al., 2020; DeGrave et al., 2020) has used a cycle-GAN (Zhu et al., 2017) model to perform image-to-image translation between normal and abnormal images. Such methods are independent of the classifier. In contrast, our framework uses a classifier consistency loss to enable image perturbation that is consistent with the classifier.
1.2. Contributions
In this paper, we propose a progressive counterfactual explainer, that explains the decision of a pre-trained image classifier. Our contributions are summarized as follows:
We developed a cGAN-based framework to generate progressively changing perturbations of the query image, such that classification decision changes from being negative to being positive for a given target class. We performed a thorough qualitative and quantitative evaluation of our explanation function to audit a classifier trained on a CXR dataset.
Our method preserved the anatomical shape and foreign objects such as support devices across generated images by adding a specialized reconstruction loss. The loss incorporates context from semantic segmentation and foreign object detection networks.
We proposed quantitative metrics based on clinical definition of two diseases (cardiomegaly and PE). We are one of the first methods to use such metrics for quantifying DNN model explanation. Specifically, we used these metrics to quantify statistical differences between the real images and their corresponding counterfactual images.
We are one of the first methods to conduct a thorough human-grounded study to evaluate different counterfactual explanations for medical imaging task. Specifically, we collected and compared feedback from diagnostic radiology residents, on different aspects of explanations: (1) understandability, (2) classifier’s decision justification, (3) visual quality, (d) identity preservation, and (5) overall helpfulness of an explanation to the users.
2. Methodology
We consider a black-box image classifier f , with high prediction accuracy. We assume that f is a differentiable function and we have access to its value as well as its gradient with respect to the input . We also assume access to either the training data for f , or an equivalent dataset with competitive prediction accuracy.
Notation:
The classification function is defined as , where d is the dimensionality of the image space and K is the number of classes. The classifier produces point estimates for posterior probability of class k as .
Explanation function:
We aim to explain the decision of function f for a target class k. We consider visual explanation of the black-box as a generative process that produces a plausible and realistic perturbation of the query image x such that the classification decision for class k is changed to a desired value c. This idea allows us to view c as a “knob”. By gradually changing the desired output c in range [0, 1], we generate progressively changing perturbations of the query image x, such that classification decision changes from being negative to being positive for a class k.
To achieve this, we propose an explanation function . This function takes two arguments: a query image x and the desired posterior probability c for the target class k. The explanation function generates a perturbed image xc such that . For simplicity, we will drop k from subsequent notations. Fig. 2 summarizes our framework. We design the explanation function to satisfy the following properties:
Fig. 2.
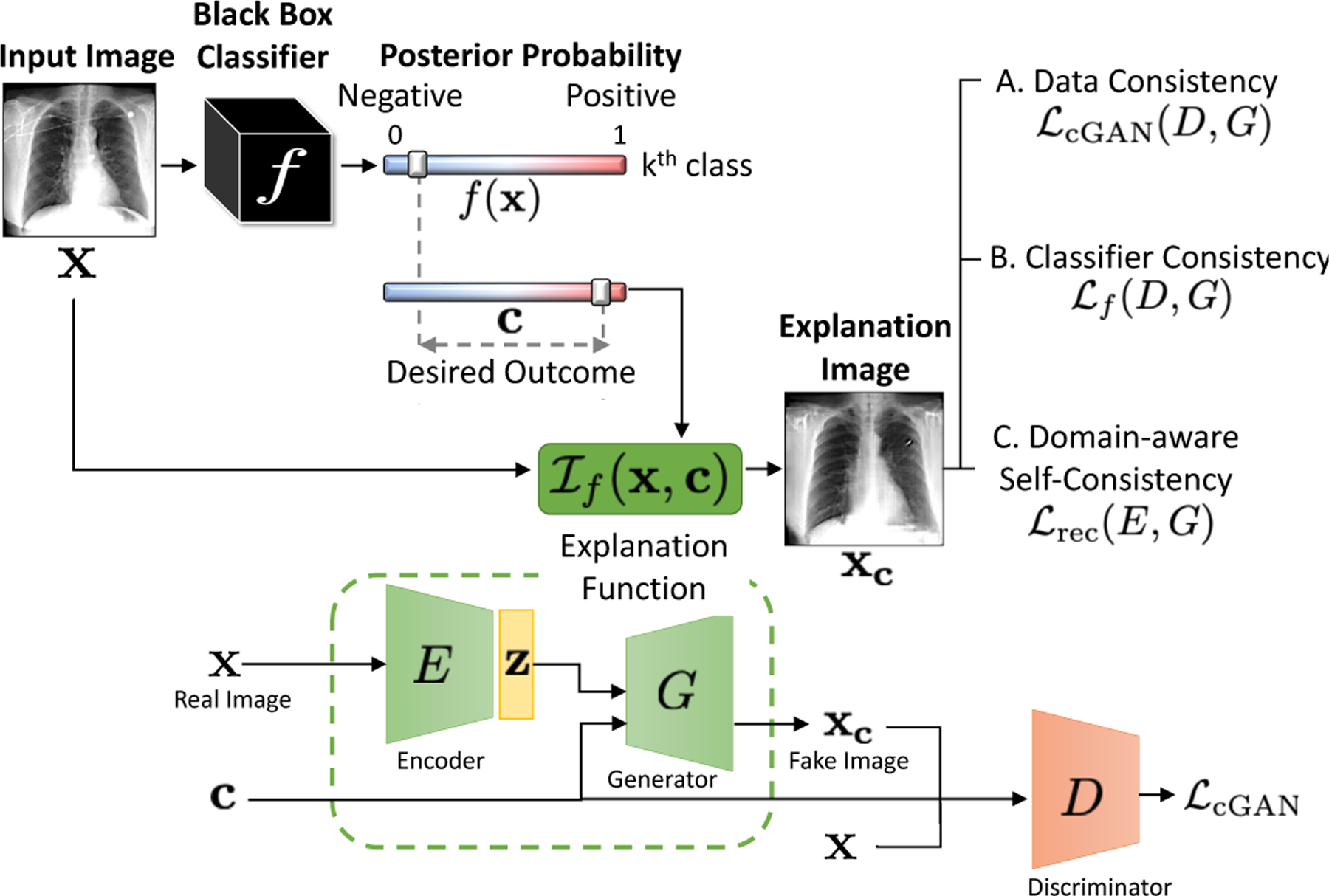
Explanation function for classifier f. Given an input image x, we generates a perturbation of the input, xc as explanation, such that the posterior probability, f , changes from its original value, f (x), to a desired value c while satisfying the three consistency constraints.
(A). Data consistency:
xc should resemble data instance from input space i.e., if input space comprises of CXRs, xc should look like a CXR with minimum artifacts or blurring.
(B). Classifier consistency:
xc should produce the desired output from the classifier f, i.e., .
(C). Context-aware self-consistency:
On using the original decision as the condition, i.e., c = f (x), the explanation function should reconstruct the query image. We forced this condition for self-consistency as and for cyclic-consistency as . Further, we constrained the explanation function to achieve the aforementioned reconstructions while preserving anatomical shape and foreign objects (e.g., pacemaker) in the input image.
Overall objective:
Our explanation function is trained end to end to learn parameters for three networks, an image encoder E(∙), a conditional GAN generator G(∙) and a discriminator D(∙), to satisfy the above three properties while minimizing the following objective function:
| (1) |
where is a conditional GAN-based loss function that enforces data-consistency, enforces classifier consistency through a KullbackLeibler (KL) divergence loss and is a reconstruction loss that enforces self-consistency. Hyperparamerters, λcGAN , λf and λrec controls the balance between the terms. In the following sections, we will discuss each property and the associated loss term in detail.
2.1. Data consistency
We formulated the explanation function, , as an image encoder E(∙) followed by a conditional GAN (cGAN) (Miyato and Koyama, 2018), with c as the condition. The encoder enables the transformation of a given image, while the GAN framework generates realistic-looking transformations as an explanation image. The cGAN is a variant of GAN that allows the conditional generation of the data by incorporating extra information as the context. Like GANs, cGAN is composed of two deep networks, generator G(∙) and discriminator D(∙). The G, D are trained adversarially by optimizing the following objective function,
| (2) |
where c denotes a condition and z is noise sampled from a uniform distribution Pz. In our formulation, z is the latent representation of the input image x, learned by the encoder E(·). Finally, the explanation function is defined as,
| (3) |
For the discriminator in cGAN, we adapted the loss function from the Projection GAN (Miyato and Koyama, 2018). The Projection GAN imposes the following structure on the discriminator loss function:
| (4) |
Here, r(x) is the discriminator logit that evaluates the visual quality of the generated image. It is the discriminator’s attempt to separate real images from the fakes images created by the generator. The second term evaluates the correspondence between the generated image xc and the condition c.
To represent the condition, the discriminator learns an embedding matrix V with N rows, where N is the number of conditions. The condition is encoded as an N-dimensional one-hot vector which is multiplied by the embedding-matrix to extract the condition-embedding. When c = n, the conditional embedding is given as the n-th row of the embedding-matrix (vn). The projection is computed as the dot product of the condition-embedding and the features extracted from the fake image,
| (5) |
where, n is the current class for the conditional generation and ϕ is the feature extractor.
In our use-case, the condition c is the desired posterior probability from the classification function f . c is a continuous variable with values in range [0, 1]. Projection-cGAN requires the condition to be a discrete variable, to be mapped to the embedding matrix V. Hence, we discretize the range [0, 1] into N bins, where each bin is one condition. One can view change from f (x) to c as changing the bin index from the current value C( f (x)) to C(c) where C(·) returns the bin index.
2.2. Classifier consistency
Ideally, cGAN should generate a series of smoothly transformed images as we change condition c in range [0, 1]. These images, when processed by the classifier f should also smoothly change the classification prediction between [0, 1]. To enforce this, rather than considering bin-index C(c) as a scalar, we consider it as an ordinal-categorical variable, i.e., C(c1) < C(c2) when c1 < c2. Specifically, rather than checking one condition that desired bin-index is equal to some value n, C(c) = n, we check n − 1 conditions that desired binindex is greater than all bin-index which are less than n i.e., (Frank and Hall, 2001).
We adapted Eq. 5 to account for a categorical variable as the condition, by modifying the second term to support ordinal multi-class regression. The modified loss function is as follows:
| (6) |
Along with conditional loss for the discriminator, we need additional regularization for the generator to ensure that the actual classifier’s outcome, i.e., f (xc), is very similar to the condition c. To ensure this compatibility with f , we further constrain the generator to minimize the KullbackLeibler (KL) divergence that encourages the classifier’s score for xc to be similar to c. Our final condition-aware loss is as follows,
| (7) |
Here, the first term evaluates a conditional probability associated with the generated image given the condition c and is a function of both G and D. The second term minimize the KL divergence between the posterior probability for new image f (xc) and the desired prediction distribution c. It influences only the G. Please note, the term r(x) is not appearing in Eq. 7 as it is independent of c.
2.3. Context-aware self consistency
A valid explanation image is a small modification of the input image, and should preserve the inputs’ identity i.e., patient-specific information such as the shape of the anatomy or any foreign objects (FO) if present. While images generated by GAN are shown to be realistic looking (Karras et al., 2019), GAN with an encoder may ignore small or uncommon details in the input image (Bau et al., 2019). To preserve such details, we propose a context-aware reconstruction loss (CARL) that exploits extra information from the input domain to refine the reconstruction results. This additional information comes as semantic segmentation and detection of any FO present in the input image. The CARL is defined as,
| (8) |
Here, S (·) is a pre-trained semantic segmentation network that produces a label map for different regions in the input domain. Rather than minimizing a distance such as ℓ1 over the entire image, we minimize the reconstruction loss for each segmentation label ( j). Such a loss heavily penalizes differences in small regions to enforce local consistency.
O(x) is a pre-trained object detector that, given an input image x, outputs a number of bounding boxes called region of interests (ROIs). For each bounding box, it outputs 2-d coordinates in the image where the box is located and an associated probability of presence of an object. Using the input image x, we obtain the ROIs and associated O(x), which is a probability vector, stating probability of finding an object in each ROI. For reconstructed image x′, we reuse the ROIs obtained from image x and computed the associated probabilities for the reconstructed image as O(x′). Next, we used KL divergence to quantify the difference between probability vectors as , in eq 8.
Finally, we used the CAR loss to enforce two essential properties of the explanation function:
If c = f (x), the self-reconstructed image should resemble the input image.
For c ≠ f (x), applying a reverse perturbation on the explanation image x should recover the initial image i.e., .
We enforce these two properties by the following loss,
| (9) |
where is defined in Eq. 8. We minimize this loss only while reconstructing the input image (either by performing self or cyclic reconstruction). Please note, the classifier f and support networks S (·) and O(·) remained fixed during training.
3. Implementation and Evaluation
3.1. Dataset
We performed our experiments on MIMIC-CXR Johnson et al. (2019) dataset consisting of 377K CXR images from 65K patients. The dataset provides image-level labels for the presence of 14 observations, namely, enlarged cardiomediastinum, cardiomegaly, lung-lesion, lung-opacity, edema, consolidation, pneumonia, atelectasis, pneumothorax, pleural effusion, pleural other, fracture, support devices and no-finding.
3.2. Implementation details
Classification model:
We consider classification model that take as input a single-view chest radiograph and output the probability of each of the 14 observations. Following the prior work on diagnosis classification (Irvin et al., 2019), we used DenseNet-121 (Huang et al., 2016) architecture for the classifier. We use the Adam optimizer with default β-parameters of β1 = 0.9, β2 = 0.999 and learning rate 1× 10−4 which is fixed for the duration of the training. We used a batch size of 16 images and train for 3 epochs, saving checkpoints every 4800 iterations.
The classifier is trained on 198K (~80%) frontal view CXR from 51K patients and is test on a held-out set of 50K images from 12K non-overlapping patients. The images are resized to 256 × 256 and are pre-processed using a standard pipeline involving cropping, re-scaling, and intensity normalization. Our classification model achieved an AUC-ROC of 0.87 for Cardiomegaly, 0.95 for pleural effusion, and 0.91 for edema. These results are comparable to performance of the published model (Irvin et al., 2019).
Segmentation network:
Semantic segmentation network S (·) is a 2D U-Net (Ronneberger et al., 2015) that marks the lung and the heart contour in a CXR. In the absence of ground truth lung and heart segmentation on the MIMIC-CXR dataset, we pre-trained the segmentation network trained on 385 CXRs from Japanese Society of Radiological Technology (JSRT) (van Ginneken et al., 2006) and Montgomery (Jaeger et al., 2014) datasets. The pre-trained segmentation network is used in our explanation function to enforce CARL loss and to compute Cardio Thoracic Ratio (CTR). Please refer SM-Sec.6.6 for details on segmentation network.
Object detector:
We trained a Fast Region-based CNN (Ren et al., 2015) network as object detector O(·). We trained three independent detectors for three use-cases: detecting foreign objects (FO) such as pacemakers and hardware, detecting healthy costrophenic (CP) recess and detecting blunt CP recess.
For constructing a training dataset for this object detection, we first collect candidate CXRs for each object by parsing the radiology reports associated with the CXR to find positive mention for “blunting of the costophrenic angle” for blunt CP recess, and “lungs are clear” for healthy CP recess. For each object, we manually collect bounding box annotations for 300 candidate CXRs.
Explanation Function:
Our explanation function is implemented using TensorFlow version 2.0 and is trained on NVIDID P100 GPU. Before training the explanation function, we assume access to the pre-trained classification function, that we aim to explain. We also assume access to pre-trained segmentation and object detection networks, that are used to enforce CARL loss.
In cGAN, we adapted a ResNet (He et al., 2016) architecture for the encoder, generator, and discriminator networks. For optimization, we used Adam optimizer (Kingma and Ba, 2015), with hyper-parameters set to α = 0.0002, β1 = 0, β2 = 0.9 and updated the discriminator five times per one update of the generator, and the encoder.
In our experiments, we train three independent explanation functions, for explaining classifier’s decision for three class labels; cardiomegaly, pleural effusion (PE), and edema. For training, we divide into N = 10 equally-sized bins and trained the cGAN with 10 conditions. To construct the training-set for the explanation function, we randomly sample images from the test-set of the classifier such that each condition (binindex) have 2500 – 3000 images. Similarly, we created a non-overlapping (unique subjects) evaluation dataset, of 20K images for the explanation function. We created one such dataset for each class label.
3.3. Evaluation
For evaluating the explanations, we randomly sample two groups of real images from the test set of the explanation function (1) a real-negative group defined as . It consists of real CXR that are predicted as negative by the classifier f for a target class k. (2) A real-positive group defined as . For , we generated a counterfactual group by setting condition c = 1.0 as, . Similarly for , we derived a counterfactual group as . We create one such dataset for each target class k. Combining the two groups, our set of real images is and corresponding set of counterfactual explanations is . All the results are computed on this evaluation dataset.
We employ several metrics to quantify different aspects of a valid counterfactual explanation.
Frechet Inception Distance (FID) score:
FID score quantifies the visual similarity between the real images and the synthetic counterfactuals. It computes the distance between the activation distributions as follow,
| (10) |
where µ’s and Σ’s are mean and covariance of the activation vectors derived from the penultimate layer of a Inception v3 network Heusel et al. (2017) pre-trained on MIMIC-CXR dataset.
Counterfactual Validity (CV) score:
CV score (Mothilal et al., 2020) is defined as the fraction of counterfactual explanations that successfully flipped the classification decision i.e., if the input image is negative, then the explanation is predicted as positive for the target class. CV score is computed as,
| (11) |
where, τ is the margin between the two prediction distributions. We used τ = 0.8 in our experiments.
Foreign Object Preservation (FOP) score:
FOP score is the fraction of the real images, with successful detection of FO, in which FO was also detected in the corresponding explanation image xc.
| (12) |
where, O(x) is the probability of finding a FO in image x as predicted by a pre-trained object detector. i.e., we consider images with positive detection of FO in set .
Next, we define two clinical metrics to quantify the counterfactual changes that leads to the flipping of the classifier’s decision. Precisely, we translated the clinical definition of cardiomegaly and pleural effusion into metrics that can be computed using a CXR.
Cardio Thoracic Ratio (CTR):
We used CTR as the clinical metric to quantify cardiomegaly. CTR is the ratio of the cardiac diameter to the maximum internal diameter of the thoracic cavity. A CTR ratio greater than 0.5 indicates cardiomegaly (Mensah et al., 2015; Centurión et al., 2017; Dimopoulos et al., 2013). We followed the approach in (Chamveha et al., 2020) to calculate CTR from a CXR. We use the pre-trained segmentation network S (·) to mark the heart and lung region. We calculated heart diameter as the distance between the leftmost and rightmost points from the lung centerline on the heart segmentation. The thoracic diameter is the horizontal distance between the widest points on the lung mask.
Score for detecting a healthy Costophrenic recess (SCP):
We first identify CP recess in a CXR and then classify it as healthy or blunt to quantify pleural effusion. The fluid accumulation in CP recess may lead to the diaphragm’s flattening and the associated blunting of the angle between the chest wall and the diaphragm arc, called costophrenic angle (CPA). The blunt CPA is an indication of pleural effusion (Maduskar et al., 2016; Lababede, 2017). Marking the CPA angle on a CXR requires expert supervision, while annotating the CP region with a bounding box is a much simpler task (see SM-Fig. 15). We learned an object detector to identify healthy or blunt CP recess in the CXRs and used SCP as our evaluation metric.
4. Experiments and Results
We performed four sets of experiments on CXR dataset:
In Section 4.1, we evaluated the validity of our counterfactual explanations and compared them against xGEM (Joshi et al., 2018) and CycleGAN (Narayanaswamy et al., 2020; DeGrave et al., 2020).
In Section 4.2, we compared against the saliency-based methods to provide post-hoc model explanation.
In Section 4.3, we associate the counterfactual changes in our explanation with the clinical definitions of two diagnosis, cardiomegaly and pleural effusion.
In Section 4.4, we present a clinical study that collects subjective feedback from radiology residents on three different explanation approaches, saliency maps, cycleGAN and ours.
4.1. Validity of counterfactual explanations
A valid counterfactual explanation resembles the query image while having perceivable differences that achieves an opposite classification decision as compared to the query image from the classifier. In Fig. 4, we present qualitative examples of counterfactual explanations from our method and compared them against those obtained from xGEM and CycleGAN.
Fig. 4.
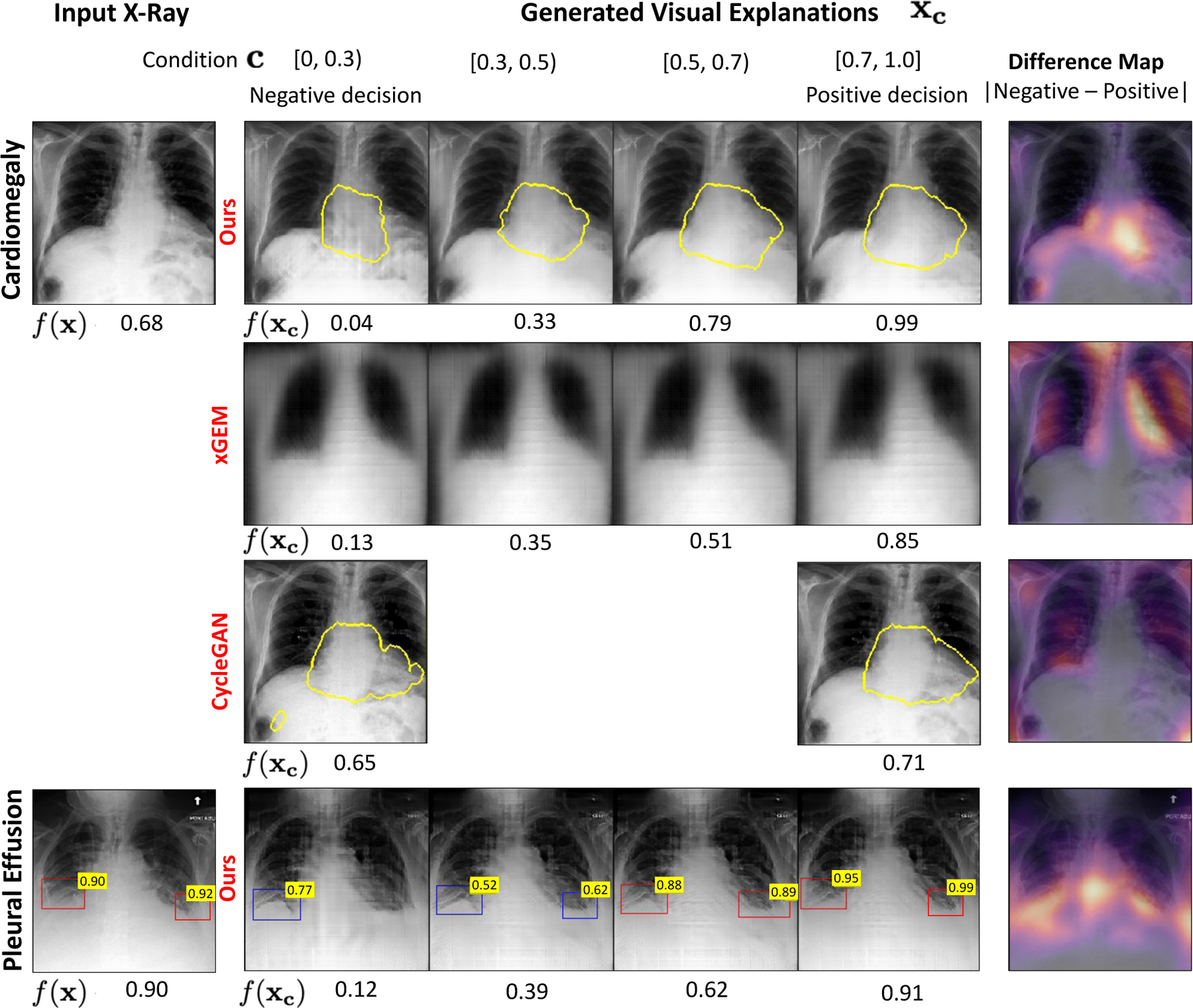
Qualitative comparison of the counterfactual explanations generated for two classification tasks, cardiomegaly (first row) and pleural effusion (PE) (last row). The bottom labels are the classifier’s predictions for the specific task. For the input image in first column, our model generates a series of images xc as explanations by gradually changing c in range [0, 1]. The last column presented a pixel-wise difference map between the explanations at the two extreme ends i.e., with condition c = 0 (negative decision) and with condition c = 1 (positive decision). The heatmap highlights the regions that changed the most during the transformation. For cardiomegaly, we show the heart border in yellow. For PE, we showed the results of an object detector as a bounding-box (BB) over the healthy (blue) and blunt (red) CP recess regions. The number on the top-right of the blue-BB is the Score for detecting a healthy CP recess (SCP). The number on red-BB is 1-SCP.
4.1.1. Classifier consistency
In Fig. 4, we observe that the explanation images gradually flip their decision f (xc) (bottom label) as we go from left to right. Table. 1 summarizes our results on CV score metric. A high CV score for our model confirms that the condition used in cGAN is successfully met and the generated explanations are successfully flipping the classification decision and hence, are consistent with the classifier.
Table 1.
The counterfactual validity (CV) score is the fraction of explanations that have an opposite prediction compared to the input image. The FID score quantifies the visual appearance of the explanations. We have normalized the FID scores with respect to the best method (cycleGAN).
| Cardiomegaly | Pleural Effusion | Edema | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Ours | xGEM | CycleGAN | Ours | xGEM | CycleGAN | Ours | xGEM | CycleGAN | |
|
Counterfactual Validity Score
| |||||||||
| 0.91 | 0.91 | 0.43 | 0.97 | 0.97 | 0.49 | 0.98 | 0.66 | 0.57 | |
|
FID score | |||||||||
| Negative group: | 4.3 | 12.5 | 1 | 3.7 | 9.2 | 1 | 1.9 | 5.0 | 1 |
| Positive group: | 2.4 | 5.6 | 1 | 3.4 | 10.1 | 1 | 1.3 | 3.5 | 1 |
On the other hand, cycleGAN achieves a CV score of about 50%, thus creating valid counterfactual images only half of the times. In a deployment scenario, a counterfactual explanation that fails to flip the classification decision would be rejected as an invalid, and hence half of the explanations provided by cycleGAN would be rejected.
Our formulation constraints the condition c to vary linearly with the actual prediction f (xc) i.e., if we increase c in range [0, 1] then the cGAN should create an image xc such that condition c is met and the actual prediction f (xc) should also increase. Further, consider a scenario when c = 1.0. The expected behaviour is and also, , where different xc are generated using same x but different conditions.
To verify this behaviour, we group images in the test-set of the explanation function, into five non-overlapping groups based on their original prediction f (x). Next, for each image, we created 10 explanation images by discretising the range [0, 1] into 10 bins. In Fig 5, we represented each input group as a line and plotted the average response of the classifier i.e., f (xc) for explanations generated with a same condition c. The positive slope of the line-plot, parallel to y = x line confirms that starting from images with low f (x), our model creates fake images such that f (xc) is high and vice-versa.
Fig. 5.
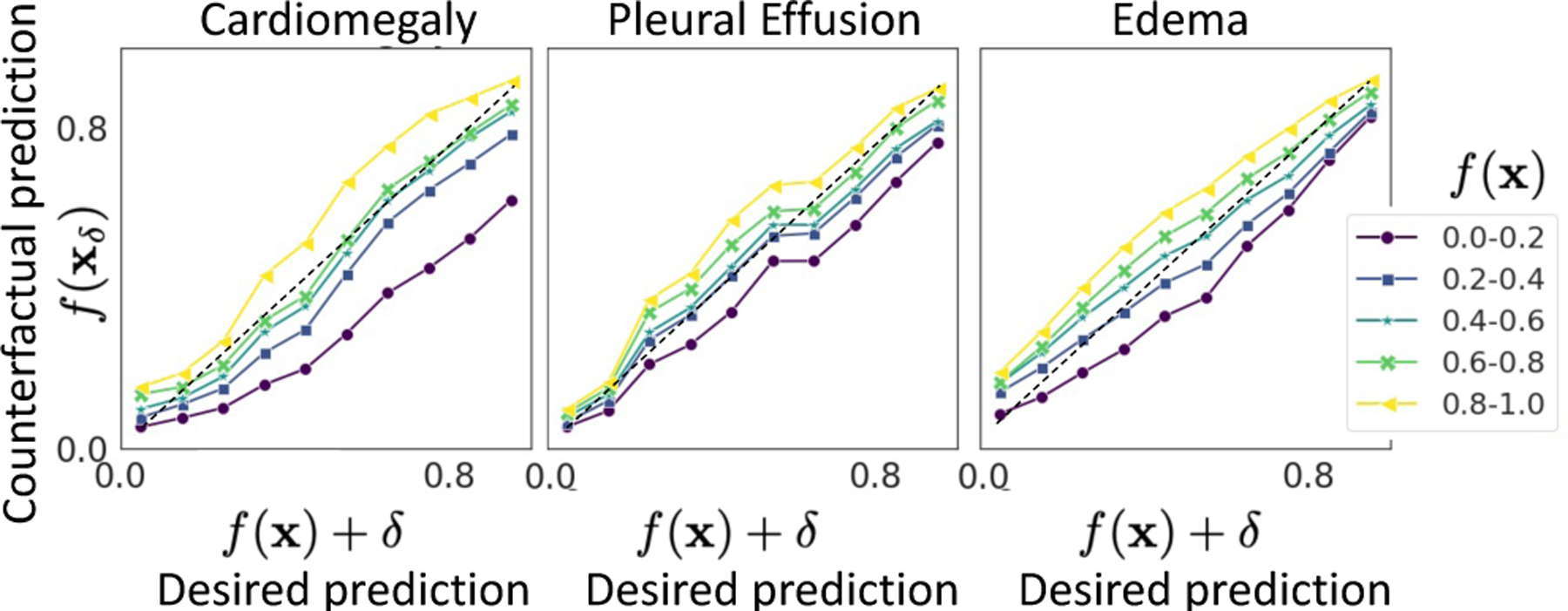
The plot of condition, c (desired prediction), against actual response of the classifier on generated explanations, f (xc). Each line represents a set of input images with prediction f (x) in a given range. Plots for xGEM and cycleGAN are shown in SM-Fig. 18.
4.1.2. Visual quality
Qualitatively, the counterfactual explanations generated by our method look visually similar to the query image (see Fig. 4). Table. 1 reports the FID score for our method and compares them against xGEM and cycleGAN. Our approach achieved a lower FID score as compared to xGEM. xGEM has very high FID score, thus creating counterfactual images that are very different from the query image and hence are unsuitable for deployment. CycleGAN achieved the least FID score, thus generating the most realistic images as explanations.
4.1.3. Identity preservation
Ideally, a counterfactual explanation should differ in semantic features associated with the target task while retaining unique properties of the input, such as foreign objects (FOs). FO provide critical information to identify the patient in an x-ray. The disappearance of FO in explanation images may create confusion that explanation images show a different patient.
In this experiment, we quantify the strength of our revised CARL loss in preserving FO in explanation images compared to an image-level ℓ1 reconstruction loss. In Table 2, we report the results on the FOP score metric. Our model with CARL obtained a higher FOP score. The FO detector network has an accuracy of 80%. Fig. 6 presents examples of counterfactual explanations generated by our model with and without the CARL. Our results confirm that CARL is an improvement over ℓ1 reconstruction loss. We further provide a detailed ablation study over different components of our loss in SM-Sec.6.13.
Table 2.
The foreign object preservation (FOP) score for our model with and without the context-aware reconstruction loss (CARL). FOP score depends on the performance of FO detector.
| Foreign Object (FO) | FOP score | |
|---|---|---|
| Ours with CARL | Ours with ℓ1 | |
| Pacemaker | 0.52 | 0.40 |
| Hardware | 0.63 | 0.32 |
Fig. 6.
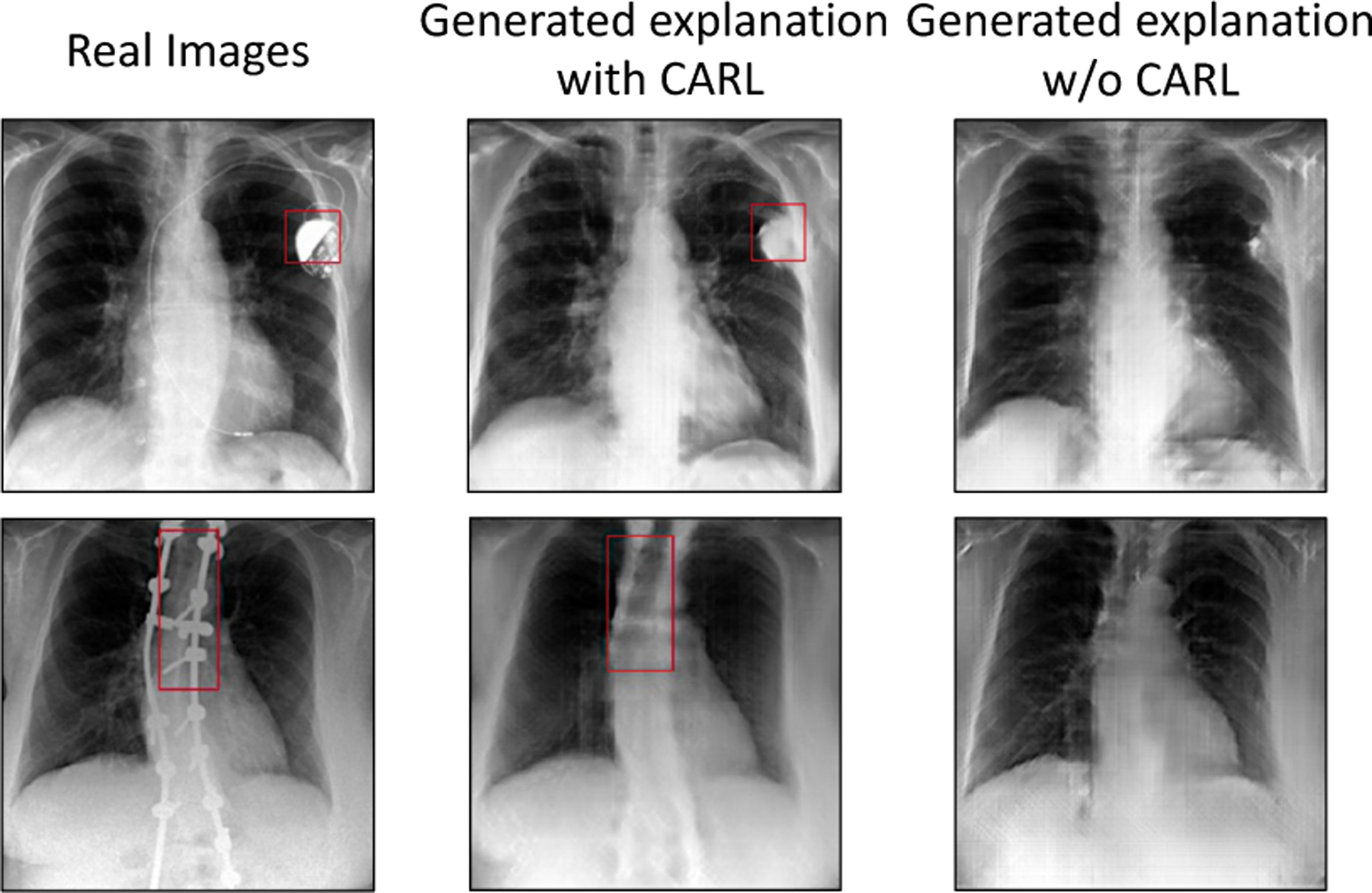
Fidelity of generated images with respect to preserving FO.
4.2. Comparison with saliency maps
Popular existing approaches for post-hoc model explanation includes explaining using a saliency-map (Pasa et al., 2019; Irvin et al., 2019). To compare against such methods, we approximated a saliency map as a pixel-wise difference map between the explanations at the two extreme ends i.e., with condition c = 0 (negative decision) and with condition c = 1 (positive decision). For proper comparison, we normalized the absolute values of the saliency maps in the range [0, 1].
In clinical setting, multiple diagnosis may affect the same anatomical region. In this case, the saliency map may highlight same regions as important for multiple target tasks. Fig. 8 is showing one such example. Our method not only provides a saliency map, but also counterfactual images to demonstrate how image features in those relevant regions should be modify to change the classification decision.
Fig. 8.
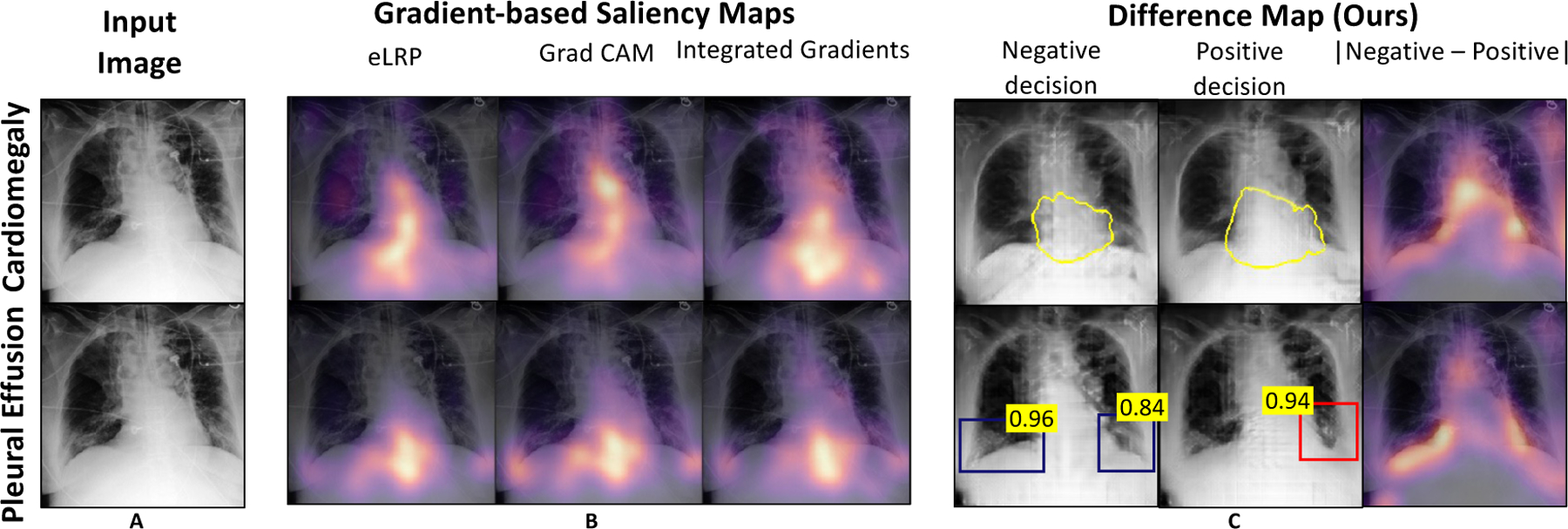
Comparison of our method against different gradient-based methods. A: Input image; B: Saliency maps from existing works; C: Our simulation of saliency map as difference map between the normal and abnormal explanation images. More examples are shown in SM-Fig. 21.
Quantitatively evaluation:
In this experiment, we quantitatively compare different methods for generating saliency maps, to show that important regions identified by these methods are actually relevant for classification decision. Specifically, we used the deletion evaluation metric (Petsiuk et al., 2018; Samek et al., 2017). For each image in set , we derived saliency maps using different methods. We used the saliency information to sort the pixels based on their importance. Next, we gradually removed top x% of important pixels by selectively impainting the removed region based on its surroundings. We processed the resulting image through the classifier and measure the output probability. We repeated this process while gradually increasing the fraction of removed pixels.
For each image, we plotted the updated classification probability as a function of the fraction of removed pixels, to obtain the deletion curve and measure its area under the deletion curve (AUDC). A sharp decline in classification probability shows that the removed pixels were actually important for classification decision. A sharp decline results in smaller AUDC, and demonstrates the high sensitivity of the classifier in the salient regions. In Fig. 7, we reported the mean and standard deviation of the AUDC metric over the set . Our method achieved the lowest AUDC, confirming the high sensitivity of the classifier in the salient regions identified by our method.
Fig. 7.
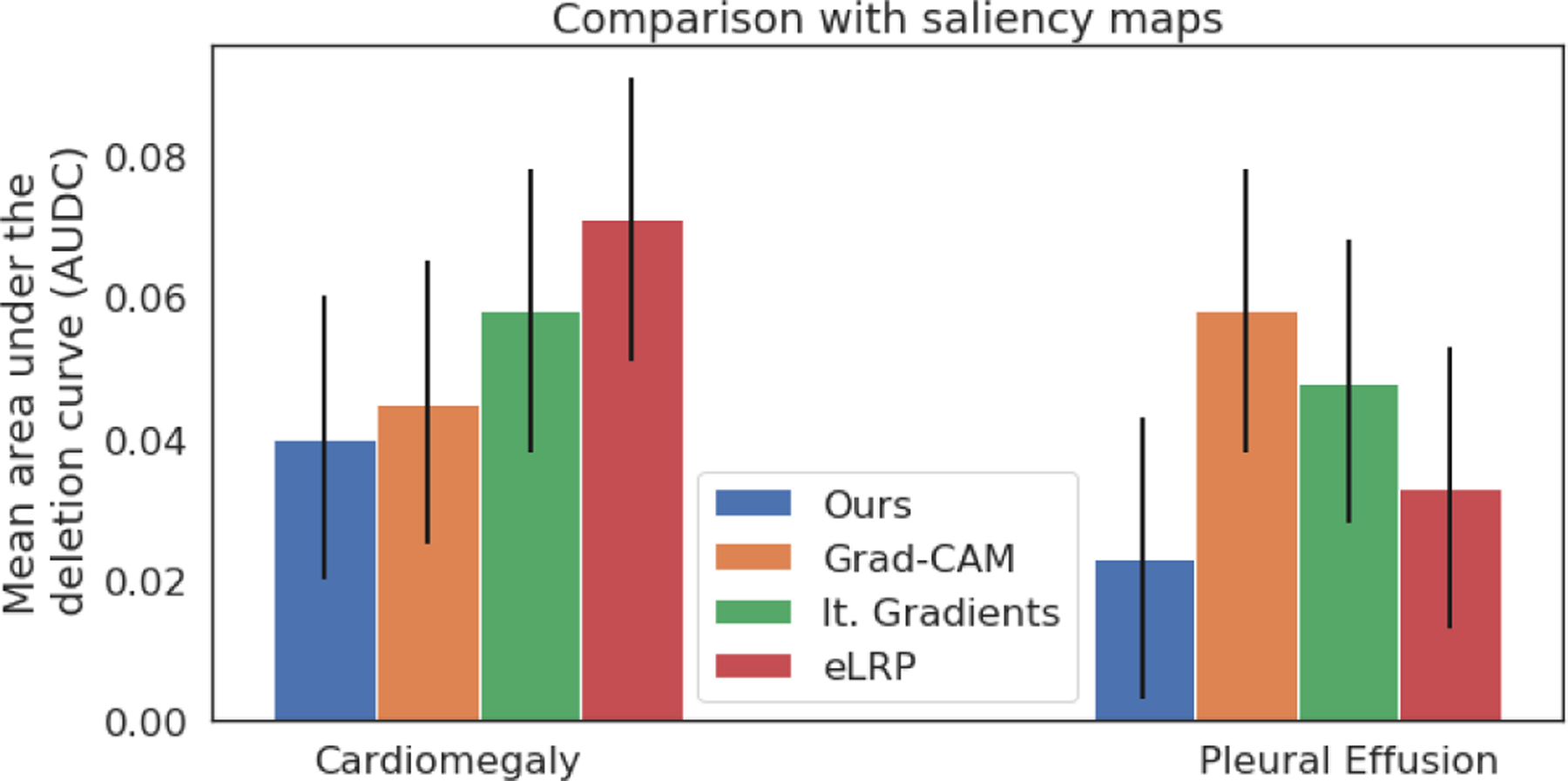
Quantitative comparison of our method against gradient-based methods. Mean area under the deletion curve (AUDC), plotted as a function of the fraction of removed pixels. A low AUDC shows a sharp drop in prediction accuracy as more pixels are deleted.
4.3. Disease-specific evaluation
In this experiment, we demonstrated the clinical relevance of our explanations. We defined two clinical metrics, cardiothoracic ratio (CTR) for cardiomegaly and score of the normal costophrenic recess (SCP) for pleural effusion. We used these metrics to quantify the counterfactual changes between normal (negative diagnosis) and abnormal (positive diagnosis) populations, as identified by the given classifier. If the change in classification decision is associated with the corresponding change in clinical-metric, we can conclude that the classifier considers clinically relevant information in its diagnosis prediction.
We conducted a paired t-test to determine the effect of counterfactual perturbation on the clinical metric for the respective diagnosis. To perform the test, we considered the two groups of real images and and their corresponding counterfactual groups and respectively. In Fig. 9, we showed the distribution of differences in CTR for cardiomegaly and SCP for PE in a pair-wise comparison between real images and their respective counterfactuals. Patients with cardiomegaly have higher CTR as compared to normal subjects. Consistent with clinical knowledge, in Fig. 9, we observe a negative mean difference for (a p-value of < 0.0001) and a positive mean difference for (with a p-value of ≪ 0.0001). The low p-value in the dependent t-test statistics supports the alternate hypothesis that the difference in the two groups is statistically significant, and this difference is unlikely to be caused by sampling error or by chance.
Fig. 9.
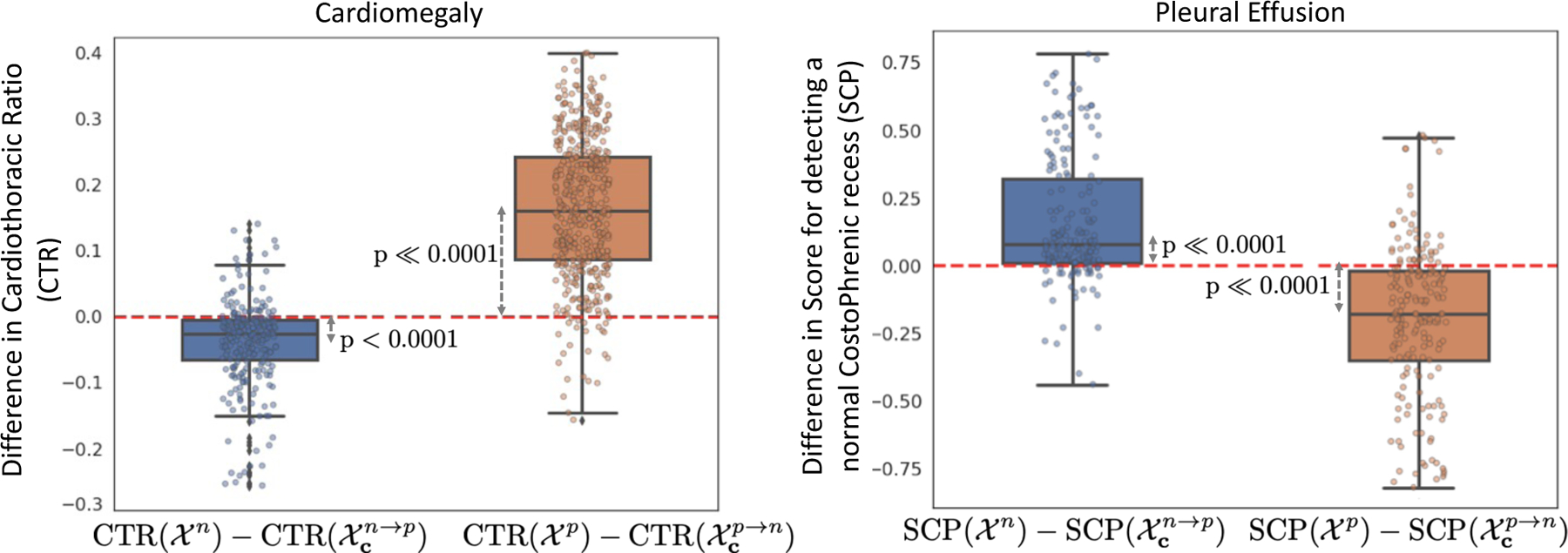
Box plots to show distributions of pairwise differences in clinical metrics, CTR for cardiomegaly and the Score of normal CP recess (SCP) for pleural effusion, before (real) and after (counterfactual) our generative counterfactual creation process. The mean value corresponds to the average causal effect of the clinical-metric on the target task. The low p-values for the dependent t-test statistics confirm the statistically significant difference in the distributions of metrics for real and counterfactual images. The mean and standard deviation for the statistic tests are summarized in SM-Table 8.
By design, the object detector assigns a high SCP to a healthy CP recess with no evidence of blunting CPW. Consistent with our expectation, we observe a positive mean difference for (with a p-value of ≪ 0.0001) and a negative mean difference for (with a p-value of ≪ 0.0001). A low p-value confirmed the statistically significant difference in SCP for real images and their corresponding counterfactuals.
4.4. Human evaluation
We conducted a human-grounded experiment with diagnostic radiology residents to compare different styles of explanations (no explanation, saliency map, cycleGAN explanation, and our counterfactual explanation) by evaluating different aspects of explanations: (1) understandability, (2) classifier’s decision justification, (3) visual quality, (d) identity preservation, and (5) overall helpfulness of an explanation to the users.
Our results show that our counterfactual explanation was the only explanation method that significantly improved the users’ understanding of the classifier’s decision compared to the no-explanation baseline. In addition, our counterfactual explanation had a significantly higher classifier’s decision justification than the cycleGAN explanation, indicating that the participants found a good evidence for the classifier’s decision more frequently in our counterfactual explanation as compared to cycleGAN explanation.
Further, cycleGan explanation performed better in terms of visual quality and identity preservation. However, at times the cycleGAN explanations were identical to the query image, thus providing inconclusive explanations. Overall the participants found our explanation method the most helpful method in understanding the assessment made by the AI system in comparison to other explanation methods. Below, we describe the design of the study, the data analysis methods, along with the results of the experiment in detail.
4.4.1. Experiment Design
We conducted an online survey experiment with 12 diagnostic radiology residents. Participants first reviewed an instruction script, which described the AI system developed to provide an autonomous diagnosis for CXR findings such as cardiomegaly. The study comprised of the radiologists evaluating six CXR images which were presented in random order to them. For selecting these siz CXR, we first, divided the test-set of the explanation function for cardiomegaly into three groups, positive ( f (x) ∈ [0.8, 1.0]), mild ( f (x) ∈ [0.4, 0.6]) and negative ( f (x) ∈ [0.0, 0.2]). Next, we randomly selected two CXR images from each group. The six CXR images were anonymized as part of the MIMIC-CXR dataset protocol.
For each image, we had the same procedure consisted of a diagnosis tasks, followed by four explanation conditions, and ended by a final evaluation question between the explanation conditions. Further details of the study design are includes in SM-Section 6.1.
Diagnosis:
For each CXR image, we first asked a participant to provide their diagnosis for cardiomegaly. This question ensures that the participants carefully consider the imaging features that helped them diagnose. Subsequently, the participants were presented with the classifier’s decision and were asked to provide feedback on whether they agreed.
Explanation Conditions:
Next, the study provides the classifier’s decision with the following explanation conditions:
No explanation (Baseline): This condition simply provides the classifier decision without any explanation, and is used as the control condition.
Saliency map: A heat map overlaid on the query CXR, highlighting essential regions for the classifier’s decision.
CycleGAN explanation: A video loop over two CXR images, corresponding to the query CXR transformation with a negative and a positive decision for cardiomegaly.
Our counterfactual explanation: A video showing a series of CXR images gradually changing the classifier’s decision from negative to positive.
Please note that after showing the baseline condition, we provided the other explanation conditions in random order to avoid any learning or biasing effects.
Evaluation metrics:
Given the classifier’s decision and corresponding explanation, we consider the following metrics to compare different explanation conditions:
Understandability: For each explanation condition, the study included a question to measure whether the end-user understood the classifier’s decision, when explanation was provided. The participants were asked to rate agreement with “I understand how the AI system made the above assessment for Cardiomegaly”.
Classifier’s decision justification: Human user’s may perceive explanations as the reason for the classifier’s decision. For the cycleGAN and our counterfactual explanation conditions, we quantify whether the provided explanation were actually related to the classification task by measuring the participants’ agreement with “The changes in the video are related to Cardiomegaly”.
Visual quality: The study quantifies the proximity between the explanation images and the query CXR by measuring the participants’ agreement with ”Images in the video look like a chest x-ray.”.
Identity preservation: The study also measures the extent to which participants think the explanation images correspond to the same subject as the query CXR by measuring the participants’ agreement with “Images in the video look like the chest x-ray from a given subject”.
Helpfulness: For each CXR image, we asked the participants to select the most helpful explanation condition in understanding the classifier’s decision, “Which explanation helped you the most in understanding the assessment made by the AI system?”. This evaluation metric directly compares the different explanation conditions. All metrics, but the helpfulness metric were evaluated for agreement on a 5-point Likert scale, where one means “strongly disagree” and five means “strongly agree”.
Free-form Response:
After each question, we also asked the participants a free-form question: “Please explain your selection in a few words.” We used answers to these questions to triangulate our findings and complement our quantitative metrics by understanding our participants’ thought-processes and reasoning.
Participants.
Our participants include 12 diagnostic radiology residents who have completed medical school and have been in the residency program for one or more years. On average, the participants finished the survey in 40 minutes and were paid $100 for their participation in the study.
4.4.2. Data analysis
For each evaluation metric, the study asked the same question to the participants while showing them different explanations. For each question, we gather 72 responses (6 - number of CXR images × 12 - number of participants).
For the understandability and helpfulness metrics, we conducted a one-way ANOVA test to determine if there is a statistically significant difference between the mean metric scores for the four explanation conditions. Specifically, we built a one-way ANOVA with the metric as our dependent variable and analyzed agreement rating as the independent variable. If we found a significant difference in the ANOVA method, we ran Tukey’s Honestly Significant Difference (HSD) posthoc test to perform a pair-wise comparison between different explanation conditions.
We measured the classifier’s decision justification, visual quality and identity preservation metrics only for the cycleGAN and our counterfactual explanations. We conducted paired t-tests to compare these evaluation metrics between these two explanation conditions. We also qualitatively analyzed the participants’ free-form responses to find themes and patterns in their responses.
4.4.3. Results
Fig. 10 shows the mean score for the evaluation metrics of understandability, classifier’s decision justification, visual quality, and identity preservation among the different explanation conditions. Below, we report the statistical analysis for these results, followed by analysis of the participants’ free-form responses to understand the reasons behind these results.
Fig. 10.
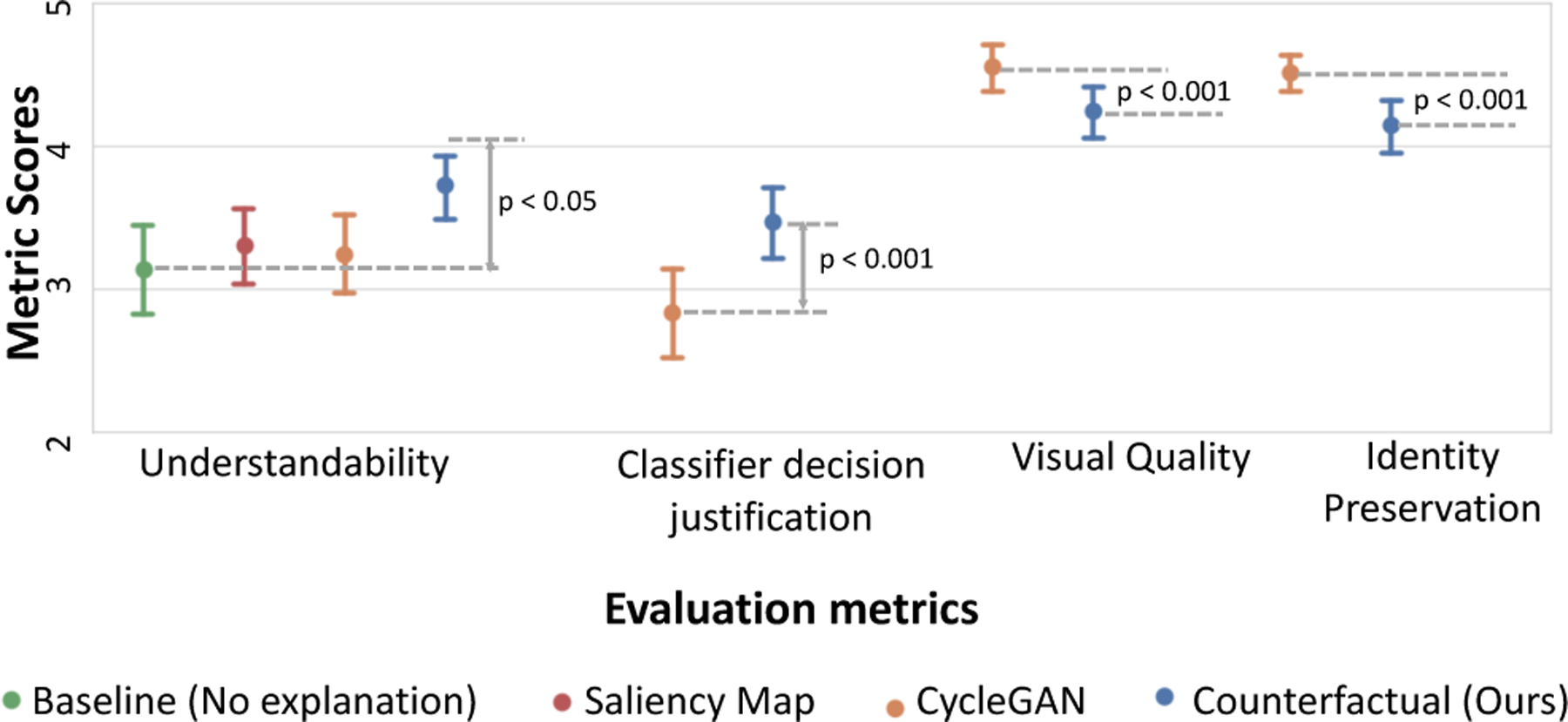
Comparing the evaluation metrics of understandability, classifier’s decision justification, visual quality, and identity preservation across the different explanation conditions.
Understandability:
The results show that our counterfactual explanation was the most understandable explanation to the participants. A one-way ANOVA revealed that there was a statistically significant difference in the understandability metric between at least two explanation conditions (F(3, 284) = [3.39], p=0.019). The Tukey post-hoc test showed that the understandability metric for our counterfactual explanation was significantly higher than the no-explanation baseline (p = 0.018). However, there was no statistically significant difference in mean scores between other pairs of explanations (refer to Table 3, “Understandability” column). This finding indicates that providing our counterfactual explanations along with the classifier’s decision made the algorithm most understandable to our clinical participants, while other explanation conditions, saliency map and cycleGAN failed to achieve significant difference from no-explanation baseline on the understandability metric. Next, we use responses from free-text question to supplement our findings.
Table 3.
Results for one-way ANOVA for understandability metric, followed by Tukey’s HSD post-hoc test between different levels of agreement. Note that the mean value for E4 (our counterfactual explanation) is the highest, indicating that our explanations helped users the most in understanding the classifier’s decision.
| Understandability F(3, 284) = 3.39 p < 0.05 |
Helpfulness F(3, 284) = 21.5 p < 0.001 |
||||
|---|---|---|---|---|---|
| Explanation method | p | Explanation method | p | ||
| E1 (No explanation) M=3.14 SD=1.39 |
E2 E3 E4 |
* |
E1 M=0.05 SD=0.23 |
E2 E3 E4 |
*** |
| E2 (Saliency Map) M=3.31 SD=1.13 |
E1 E3 E4 |
E2 M=0.18 SD=0.39 |
E1 E3 E4 |
*** |
|
| E3 (CycleGAN) M=3.24 SD=1.19 |
E1 E2 E4 |
E3 M=0.16 SD=0.37 |
E1 E2 E4 |
*** |
|
| E4 (Our counterfactual explanation) M=3.72 SD=0.97 |
E1 E2 E3 |
* | E4 M=0.24 SD=0.42 |
E1 E2 E3 |
***
*** *** |
p < 0.05
p < 0.0001
For the no-explanation baseline, the primary reason for poor understanding was the absence of explanation (n=30), (e.g., they stated that “there is no indication as to how the AI made this decision”). Interestingly, many responses (n=23) either associated their high understanding with the correct classification decision i.e., participants understood the decision as the decision is correct (“I agree, it is small and normal”) or they assumed the AI-system is using similar reasoning as them to arrive at its decision (“I assume the AI is just measuring the width of the heart compared to the thorax”, “Assume the AI measured the CT ratio and diagnosed accordingly.”).
Participants’ mostly found saliency maps to be correct but incomplete (n=23), (“Unclear how assessment can be made without including additional regions”). Specifically, for cardiomegaly, the saliency maps were highlighting parts of the heart and not its border (“Not sure how it gauges not looking at the border”) or thoracic diameter (“thoracic diameter cannot be assessed using highlighted regions of heat map”). We observe a similar result in Fig. 8, where the heatmap focuses on the heart but not its border. Further, some participants expressed a concern that they didn’t understand how relevant regions were used to derive the decision (“i understand where it examined but not how that means definite cardiomegaly”).
For cycleGAN explanation, the primary reason for poor understanding was the minimal perceptible changes between the negative and positive images (n=3), (“There is no change in the video.”). In contrast, many participant’s explicitly reported an improved understanding of the classifier’s decision in the presence of our counterfactual explanations (n=33), (“I think the AI looking at the borders makes sense.”, “i can better understand what the AI is picking up on with the progression video”).
Classifier’s decision justification:
Our counterfactual explanation (M=3.46; SD=1.12) achieved a positive mean difference of 0.63 on this metric as compared to cycleGAN (M=2.83; SD=1.33), with t(71)=3.55 and p < 0.001. This result indicates that the participants found a good evidence for the predicted class (cardiomegaly), much frequently in our counterfactual explanations as compared to cycleGAN.
Most responses (n=25) explicitly mentioned visualizing changes related to cardiomegaly such as an enlarged heart in our explanation video as compared to cycleGAN (n=17). In cycleGAN, many reported that changes in the explanation video was not perceptible (n=23). Further, the participants reported changes in density, windowing level or other attributes which were not related to cardiomegaly (“Decreasing the density does not impact how I assess for cardiomegaly.”, “they could be or just secondary to windowing the radiograph”). Such responses were observed in both cycleGAN (n=17) and our explanation (n=17). This indicates that the classifier may have associated such secondary information (short-cuts) with cardiomegaly diagnosis. A more in-depth analysis is required to quantify the classifiers’ behaviour.
Visual quality and identity preservation:
We observe a negative mean difference of 0.31 and 0.37 between our and cycleGAN explanation methods in visual quality and identity preservation metrics, respectively. The mean score for visual quality was higher for cycleGAN (M=4.55; SD=0.71) as compared to our method (M=4.24; SD=0.80) with t(71)=3.49 and p < 0.001. Similarly, the mean score for identity preservation was also higher for cycleGAN (M=4.51; SD=0.56) as compared to our method (M=4.14; SD=0.78) with t(71)=3.96 and p < 0.001.
Most of the responses (n=69) agreed that the CycleGAN explanation were marked as highly similar to the query CXR image. These results are consistent with our earlier results, that cycleGAN has better visual quality with a lower FID score (see Table. 1). However, in some responses, the participants pointed out that the explanation images were almost identical to the query image (“There’s virtually no differences. This is within the spectrum of a repeat chest x-ray for instance.”). An explanation image identical to the query image provides no information about the classifier’s decision. Further, similar looking CXR will also result in similar classification decision, and hence will fail to flip the classification decision. As a result, we also observed a lower agreement in the classifier consistency metric and a lower counterfactual validity score in Table. 1 for cycleGAN.
Helpfulness:
In our concluding question, “Which explanation helped you the most in understanding the assessment made by the AI system?”, 57% of the responses selected our counterfactual explanation as the most helpful method. A one-way ANOVA revealed that there was a statistically significant difference in the helpfulness metric between at least two explanation conditions (F(3, 284) = [21.5], p < 0.0001). In pair-wise Tukey’s HSD posthoc test, we found that the mean usefulness metric for our counterfactual explanations was significantly different from all the rest explanation conditions(p < 0.0001). Table 3 ( “Helpfulness” column) summarizes these results.
These results indicates that the participant’s selected our counterfactual explanations as the most helpful form of explanation for understanding the classifier’s decision.
5. Discussion And Conclusion
We provided a BlackBox Counterfactual Explainer, designed to explain image classification models for medical applications. Our framework explains the decision by gradually transforming the input image to its counterfactual, such that the classifier’s prediction is flipped. We have formulated and evaluated our framework on three properties of a valid counterfactual transformation: data consistency, classifier consistency, and self-consistency. Our results in Section 4.1 showed that our framework adheres to all three properties.
Comparison with xGEM and cycleGAN:
Our model creates visually appealing explanations that produce a desired outcome from the classification model while retaining maximum patient-specific information. In comparison, both xGEM and cycleGAN failed on at least one essential property. xGEM model fails to create realistic images with a high FID score. Furthermore, the cycleGAN model fails to flip the classifier’s decision with a low CV score (∼ 50%).
Further, we present a thorough comparison between cycleGAN and our explanation in a human evaluation study. The clinical experts’ expressed high agreement that explanation images from cycleGAN were of high quality and they resembles the query CXR. But at the same time, users found the explanation images to be too similar to query CXR, and the cycleGAN explanations failed to provide the counterfactual reasoning for the decision.
In comparison, our explanation were most helpful in understanding the classification decision. Though the users reported inconsistencies in the visual appearance, but the overall sentiment looks positive and they selected our method as their preferred explanation method for improved understandability.
Clinical relevance of the explanations:
From a clinical perspective, we demonstrated that the counterfactual changes associated with normal (negative) or abnormal (positive) classification decisions are also associated with corresponding changes in disease-specific metrics such as CTR and SCP. In our clinical study, multiple radiologist reported using CTR as the metric to diagnose cardiomegaly. As radiologist annotations are expensive, and it is not efficient to perform human evaluation on a large test set, our results with CTR calculations provides a quantitative way t evaluate difference in real and counterfactual populations.
We acknowledge that our GAN-generated counterfactual explanations may have missing details such as small wires. In our extended experiments, we found that the foreign objects such as pacemaker have minimal importance in the classification decision (see SM-Sec. 6.10.1). We attempted to improve the preservation of such information through our revised context-aware reconstruction loss (CARL). However, even with CARL, the FO preservation score is not perfect. A possible reason for this gap is the limited capacity of the object detector used to calculate the FOP score. Training a highly accurate FO detector is outside the scope of this study.
Further, a resolution of 256 × 256 for counterfactually generated images is smaller than a standard CXR. Small resolution limits the evaluation for fine details by both the algorithm and the interpreter. Our formulation of cGAN uses conditional-batch normalization (cBN) to encapsulate condition information while generating images. For efficient cBN, the mini-batches should be class-balanced. To accommodate high-resolution images with smaller batch sizes, we must decrease the number of conditions to ensure class-balanced batches. Fewer conditions resulted in a coarse transformation with abrupt changes across explanation images. In our experiments, we selected the largest N, which created a class-balanced batch that fits in GPU memory and resulted in stable cGAN training. However, with the advent of larger-memory GPUs, we intend to apply our methods to higher resolution images in future work; and assess how that impacts interpretation by clinicians.
To conclude, this study uses counterfactual explanations as a way to audit a given black-box classifier and evaluate whether the radio-graphic features used by that classifier have any clinical relevance. In particular, the proposed model did well in explaining the decision for cardiomegaly and pleural effusions and was corroborated by an experienced radiology resident physician. By providing visual explanations for deep learning decisions, radiologists better understand the causes of its decision-making. This is essential to lessen physicians’ concerns regarding the “BlackBox” nature by an algorithm and build needed trust for incorporation into everyday clinical workflow. As an increasing amount of artificial intelligence algorithms offer the promise of everyday utility, counterfactually generated images are a promising conduit to building trust among diagnostic radiologists.
By providing counterfactual explanations, our work opens up many ideas for future work. Our framework showed that valid counterfactuals can be learned using an adversarial generative process that is regularized by the classification model. However, counterfactual reasoning is incomplete without a causal structure and explicitly modeling of the interventions. An interesting next step should explore incorporating or discovering plausible causal structures and creating explanations grounded with them.
Supplementary Material
Fig. 3.
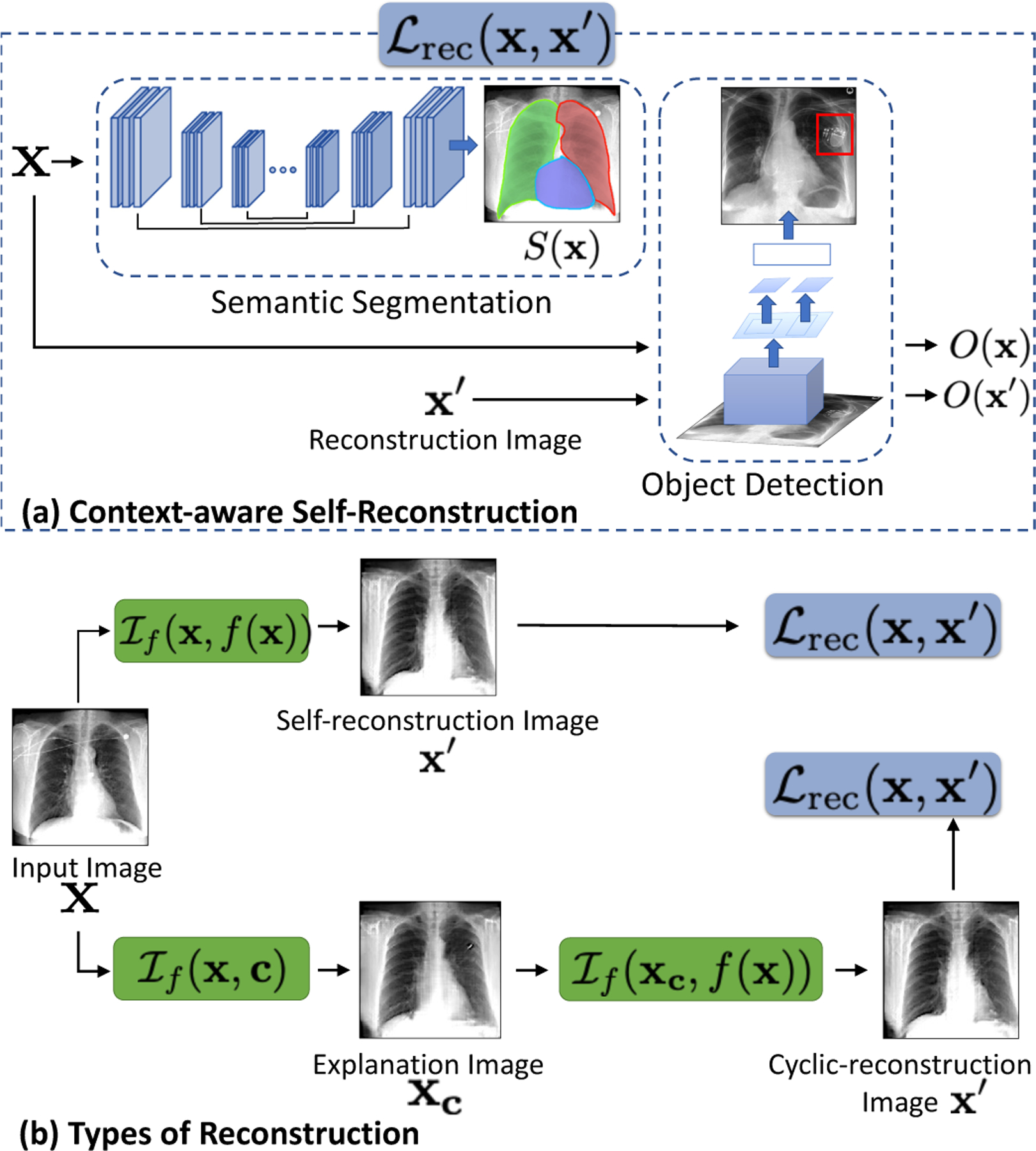
(a) A domain-aware self-reconstruction loss with pre-trained semantic segmentation S (x) and object detection O(x) networks. (b) The self and cyclic reconstruction should retain maximum information from x.
We proposed a BlackBox Counterfactual Explainer designed to explain image classification models for medical applications.
We fill the gap in existing deep-learning model explanation methods by explaining what image features are important for classification decisions and how those features should be modified to flip the decision.
We developed a cGAN-based framework to generate progressively changing perturbations of the query image, such that classification decision changes from being negative to being positive for a given target class.
Our method preserved the anatomical shape and foreign objects such as support devices across generated images by adding a specialized reconstruction loss. The loss incorporates context from semantic segmentation and foreign object detection networks.
We developed quantitative metrics based on the clinical definition of two diseases (cardiomegaly and pleural effusion). We used these metrics to quantify counterfactual differences in normal and abnormal populations, as identified by the classifier.
We are one of the first methods to conduct a thorough human-grounded study to evaluate different counterfactual explanations for medical imaging task. Specifically, we collected and compared feedback from diagnostic radiology residents, on different aspects of explanations: (1) understandability, (2) classifier’s decision justification, (3) visual quality, (d) identity preservation, and (5) overall helpfulness of an explanation to the users.
Our explanations revealed that the classifier relied on clinically relevant radiographic features for its diagnostic decisions.
Acknowledgments
This work was partially supported by NIH Award Number 1R01HL141813-01, NSF 1839332 Tripod+X, SAP SE, and Pennsylvania’s Department of Health. We are grateful for the computational resources provided by Pittsburgh SuperComputing grant number TG-ASC170024.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
Dr Kayhan Batmanghelich reports financial support was provided by National Institutes of Health. Dr Kayhan Batmanghelich reports financial support was provided by National Science Foundation.
References
- Agarwal C, Nguyen A, 2020. Explaining image classifiers by removing input features using generative models Asian Conference on Computer Vision (ACCV) . [Google Scholar]
- Bach S, Binder A, Montavon G, Klauschen F, Müller KR, Samek W, 2015. On pixel-wise explanations for non-linear classifier decisions by layer-wise relevance propagation. PLOS ONE 10, 1–46. doi: 10.1371/journal.pone.0130140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bau D, Zhu JY, Wulff J, Peebles W, Strobelt H, Zhou B, Torralba A, 2019. Seeing what a gan cannot generate IEEE International Conference on Computer Vision (ICCV) , 4502–4511. [Google Scholar]
- Centurión OA, Scavenius K, Miño L, Sequeira OR, 2017. Evaluating Cardiomegaly by Radiological Cardiothoracic Ratio as Compared to Conventional Echocardiography. Journal of Cardiology & Current Research (JCCR) 9 doi: 10.15406/jccr.2017.09.00319. [DOI] [Google Scholar]
- Chamveha I, Promwiset T, Tongdee T, Saiviroonporn P, Chaisang-mongkon W, 2020. Automated Cardiothoracic Ratio Calculation and Cardiomegaly Detection using Deep Learning Approach. arXiv e-prints , arXiv:2002.07468. [Google Scholar]
- Chang CH, Creager E, Goldenberg A, Duvenaud D, 2019. Explaining Image Classifiers by Counterfactual Generation International Conference on Learning Representations (ICLR) . [Google Scholar]
- Cohen JP, Brooks R, En S, Zucker E, Pareek A, Lungren MP, Chaudhari A, 2021. Gifsplanation via latent shift: A simple autoencoder approach to counterfactual generation for chest x-rays. Medical Imaging with Deep Learning (MIDL) [Google Scholar]
- Dabkowski P, Gal Y, 2017. Real time image saliency for black box classifiers 31st International Conference on Advances in Neural Information Processing Systems (NeurIPS) , 6970–6979. [Google Scholar]
- DeGrave AJ, Janizek JD, Lee SI, 2020. AI for radiographic COVID-19 detection selects shortcuts over signal. medRxiv , 10.1101/2020.09.13.20193565. [DOI] [Google Scholar]
- Dhurandhar A, Chen PY, Luss R, Tu CC, Ting P, Shanmugam K, Das P, 2018. Explanations based on the missing: Towards contrastive explanations with pertinent negatives International Conference on Advances in Neural Information Processing Systems (NeurIPS) . [Google Scholar]
- Dimopoulos K, Giannakoulas G, Bendayan I, Liodakis E, Petraco R, Diller GP, Piepoli MF, Swan L, Mullen M, Best N, Poole-Wilson PA, Francis DP, Rubens MB, Gatzoulis MA, 2013. Cardiothoracic ratio from postero-anterior chest radiographs: A simple, reproducible and independent marker of disease severity and outcome in adults with congenital heart disease. International Journal of Cardiology 166. [DOI] [PubMed] [Google Scholar]
- Eaton-Rosen Z, Bragman F, Bisdas S, Ourselin S, Cardoso MJ, 2018. Towards safe deep learning: Accurately quantifying biomarker uncertainty in neural network predictions International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI) , 691–699. [Google Scholar]
- Eitel F, Ritter K, 2019. Testing the robustness of attribution methods for convolutional neural networks in mri-based alzheimer’s disease classification. International Workshop on Interpretability of Machine Intelligence in Medical Image Computing (IMIMIC) 11797 LNCS, 3–11. [Google Scholar]
- Fong RC, Vedaldi A, 2017. Interpretable Explanations of Black Boxes by Meaningful Perturbation IEEE International Conference on Computer Vision (ICCV) doi: 10.1109/ICCV.2017.371. [DOI] [Google Scholar]
- Frank E, Hall M, 2001. A simple approach to ordinal classification. European Conference on Machine Learning , 145–156. [Google Scholar]
- Gastounioti A, Kontos D, 2020. Is It Time to Get Rid of Black Boxes and Cultivate Trust in AI? Radiology: Artificial Intelligence 2, e200088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ginneken B, Stegmann MB, Loog M, 2006. Segmentation of anatomical structures in chest radiographs using supervised methods: a comparative study on a public database. Medical Image Analysis 10, 19–40. doi: 10.1016/j.media.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Goyal Y, Wu Z, Ernst J, Batra D, Parikh D, Lee S, 2019. Counterfactual Visual Explanations 36th International Conference on Machine Learning (ICML) 97, 2376–2384. [Google Scholar]
- Hansell DM, Bankier AA, MacMahon H, et al. , 2008. Fleischner Society: Glossary of terms for thoracic imaging. Radiology [DOI] [PubMed] [Google Scholar]
- He K, Zhang X, Ren S, Sun J, 2016. Deep residual learning for image recognition IEEE Conference on Computer Vision Pattern Recoginition (CVPR) . [Google Scholar]
- Heusel M, Ramsauer H, Unterthiner T, Nessler B, Hochreiter S, 2017. Gans trained by a two time-scale update rule converge to a local nash equilibrium International Conference on Advances in Neural Information Processing Systems (NeurIPS) . [Google Scholar]
- Hosny A, Parmar C, Quackenbush J, Schwartz LH, Aerts HJ, 2018. Artificial intelligence in radiology. Nature Reviews Cancer 18, 500–510. doi: 10.1038/s41568-018-0016-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Liu Z, van der Maaten L, Weinberger KQ, 2016. Densely connected convolutional networks 30th IEEE Conference on Computer Vision Pattern Recoginition (CVPR) , 2261–2269. [Google Scholar]
- Huang H, Li Z, He R, Sun Z, Tan T, 2018. IntroVAE: Introspective Variational Autoencoders for Photographic Image Synthesis International Conference on Advances in Neural Information Processing Systems (NeurIPS) , 10236–10245. [Google Scholar]
- Irvin J, Rajpurkar P, Ko M, Yu Y, Ciurea-Ilcus S, Chute C, Marklund H, Haghgoo B, et al. , 2019. Chexpert: A large chest radiograph dataset with uncertainty labels and expert comparison 33rd AAAI Conference on Artificial Intelligence , 590–597. [Google Scholar]
- Jaeger S, Candemir S, Antani S, Wáng YXJ, Lu PX, Thoma G, 2014. Two public chest x-ray datasets for computer-aided screening of pulmonary diseases. Quantitative imaging in medicine and surgery 4, 475–477. doi: 10.3978/j.issn.2223-4292.2014.11.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Kim B, Guan M, Gupta M, 2018. To trust or not to trust a classifier International Conference on Advances in Neural Information Processing Systems (NeurIPS) 31. [Google Scholar]
- Johnson AE, Pollard TJ, Berkowitz SJ, Greenbaum NR, Lungren MP, Deng CY, Mark RG, Horng S, 2019. MIMIC-CXR, a de-identified publicly available database of chest radiographs with free-text reports. Scientific data 6, 317. doi: 10.1038/s41597-019-0322-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi S, Koyejo O, Kim B, Ghosh J, 2018. xGEMs: Generating Examplars to Explain Black-Box Models. arXiv e-prints , arXiv:1806.08867 [Google Scholar]
- Joshi S, Koyejo O, Vijitbenjaronk W, Kim B, Ghosh J, 2019. Towards Realistic Individual Recourse and Actionable Explanations in Black-Box Decision Making Systems. arXiv e-prints , arXiv:1907.09615 [Google Scholar]
- Karras T, Laine S, Aila T, 2019. A style-based generator architecture for generative adversarial networks IEEE Conference on Computer Vision Pattern Recoginition (CVPR) , 4401–4410. [DOI] [PubMed] [Google Scholar]
- Kingma DP, Ba J, 2015. Adam: A Method for Stochastic Optimization International Conference on Learning Representation (ICLR) . [Google Scholar]
- Lababede O, 2017. Pleural effusion imaging: Overview, radiography, computed tomography URL: https://emedicine.medscape.com/article/355524-overview. [Google Scholar]
- Larrazabal A, Nieto N, Peterson V, Milone D, Ferrante E, 2020. Gender imbalance in medical imaging datasets produces biased classifiers for computer-aided diagnosis. Proceedings of the National Academy of Sciences 117, 201919012. doi: 10.1073/pnas.1919012117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Kailkhura B, Loveland D, Han Y, 2019. Generative Counterfactual Introspection for Explainable Deep Learning IEEE Global Conference on Signal and Information Processing (GlobalSIP) , 1–5 doi: 10.1109/GlobalSIP45357.2019.8969491. [DOI] [Google Scholar]
- Lundberg SM, Allen PG, Lee SI, 2017. A unified approach to interpreting model predictions 31st International Conference on Advances in Neural Information Processing Systems (NeurIPS) , 4768–4777. [Google Scholar]
- Maduskar P, Hogeweg L, Philipsen R, van Ginneken B, 2013. Automated localization of costophrenic recesses and costophrenic angle measurement on frontal chest radiographs. Medical Imaging: Computer-Aided Diagnosis [Google Scholar]
- Maduskar P, Philipsen RH, Melendez J, Scholten E, Chanda D, Ayles H, Sánchez CI, van Ginneken B, 2016. Automatic detection of pleural effusion in chest radiographs. Medical Image Analysis [DOI] [PubMed] [Google Scholar]
- Mensah Y, Mensah K, Asiamah S, Gbadamosi H, Idun E, Brakohiapa W, Oddoye A, 2015. Establishing the Cardiothoracic Ratio Using Chest Radiographs in an Indigenous Ghanaian Population: A Simple Tool for Cardiomegaly Screening. Ghana medical journal [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyato T, Kataoka T, Koyama M, Yoshida Y, 2018. Spectral normalization for generative adversarial networks International Conference on Learning Representations (ICLR) . [Google Scholar]
- Miyato T, Koyama M, 2018. cGANs with Projection Discriminator International Conference on Learning Representations (ICLR) . [Google Scholar]
- Mothilal RK, Sharma A, Tan C, 2020. Explaining Machine Learning Classifiers through Diverse Counterfactual Explanations. Conference on Fairness, Accountability, and Transparency (FAT) , 607–617. [Google Scholar]
- Narayanaswamy A, Venugopalan S, Webster DR, Peng L, Corrado GS, Ruamviboonsuk P, Bavishi P, Brenner M, Nelson PC, Varadarajan AV, 2020. Scientific Discovery by Generating Counterfactuals using Image Translation International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI) , 273–283. [Google Scholar]
- Oakden-Rayner L, Dunnmon J, Carneiro G, Ré C, 2020. Hidden stratification causes clinically meaningful failures in machine learning for medical imaging ACM Conference on Health, Inference, and Learning 2020, 151–159. doi: 10.1145/3368555.3384468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parafita Martinez A, Vitria Marca J, 2019. Explaining visual models by causal attribution IEEE International Conference on Computer Vision Workshop (ICCVW) , 4167–4175 doi: 10.1109/ICCVW.2019.00512. [DOI] [Google Scholar]
- Pasa F, Golkov V, Pfeiffer F, Cremers D, Pfeiffer D, 2019. Efficient Deep Network Architectures for Fast Chest X-Ray Tuberculosis Screening and Visualization. Scientific Reports 9, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petsiuk V, Das A, Saenko K, 2018. Rise: Randomized input sampling for explanation of black-box models British Machine Vision Conference (BMVC) . [Google Scholar]
- Rajpurkar P, Irvin J, Ball RL, Zhu K, Yang B, Mehta H, Duan T, Ding D, Bagul A, et al. , 2018. Deep learning for chest radiograph diagnosis: A retrospective comparison of the CheXNeXt algorithm to practicing radiologists. PLOS Medicine 15, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajpurkar P, Irvin JA, Zhu K, Yang B, Mehta H, Duan T, Ding D, Bagul A, Langlotz C, Shpanskaya K, Lungren M, Ng A, 2017. Chexnet: Radiologist-level pneumonia detection on chest x-rays with deep learning. arXiv e-prints , arXiv:1711.05225 [Google Scholar]
- Ren S, He K, Girshick R, Sun J, 2015. Faster r-cnn: Towards real-time object detection with region proposal networks International Conference on Advances in Neural Information Processing Systems (NeurIPS) 28. [Google Scholar]
- Rodriguez-Ruiz A, Lång K, Gubern-Mérida A, Broeders M, Gennaro G, Clauser P, Helbich T, Chevalier M, Tan T, Mertelmeier T, Wallis M, Andersson I, Zackrisson S, Mann R, Sechopoulos I, 2019. Stand-alone artificial intelligence for breast cancer detection in mammography: Comparison with 101 radiologists. Journal of the National Cancer Institute 111. doi: 10.1093/jnci/djy222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronneberger O, Fischer P, Brox T, 2015. U-net convolutional networks for biomedical image segmentation International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI) 9351, 234–241. [Google Scholar]
- Rubin J, Sanghavi D, Zhao C, Lee K, Qadir A, Xu-Wilson M, 2018. Large Scale Automated Reading of Frontal and Lateral Chest X-Rays using Dual Convolutional Neural Networks. arXiv e-prints , arXiv:1804.07839arXiv:1804.07839 [Google Scholar]
- Samek W, Binder A, Montavon G, Lapuschkin S, Müller KR, 2017. Evaluating the visualization of what a deep neural network has learned. IEEE Transactions on Neural Networks and Learning Systems 28, 2660–2673. doi: 10.1109/TNNLS.2016.2599820. [DOI] [PubMed] [Google Scholar]
- Sayres R, Taly A, Rahimy E, Blumer K, Coz D, Hammel N, Krause J, Narayanaswamy A, Rastegar Z, Wu D, Xu S, Barb S, Joseph A, Shumski M, Smith J, Sood AB, Corrado GS, Peng L, Webster DR, 2019. Using a Deep Learning Algorithm and Integrated Gradients Explanation to Assist Grading for Diabetic Retinopathy. Ophthalmology 126, 552–564. [DOI] [PubMed] [Google Scholar]
- Seah JC, Tang CH, Buchlak QD, Holt XG, Wardman JB, Aimoldin A, Esmaili N, Ahmad H, Pham H, Lambert JF, et al. , 2021. Effect of a comprehensive deep-learning model on the accuracy of chest x-ray interpretation by radiologists: a retrospective, multireader multicase study. The Lancet Digital Health 3, e496–e506. [DOI] [PubMed] [Google Scholar]
- Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D, 2017. Grad-cam: Visual explanations from deep networks via gradient-based localization IEEE International Conference on Computer Vision (ICCV) , 618–626. [Google Scholar]
- Shrikumar A, Greenside P, Kundaje A, 2017. Learning important features through propagating activation differences 34th International Conference on Machine Learning (ICML) 70, 3145–3153. [Google Scholar]
- Simonyan K, Vedaldi A, Zisserman A, 2013. Deep inside convolutional networks: Visualising image classification models and saliency maps. Computing Research Repository abs/1312.6034 [Google Scholar]
- Simonyan K, Zisserman A, 2014. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv e-prints , arXiv 1409.1556. [Google Scholar]
- Singla S, Pollack B, Chen J, Batmanghelich K, 2019. Explanation by Progressive Exaggeration International Conference on Learning Representations (ICLR) . [Google Scholar]
- Singla S, Wallace S, Triantafillou S, Batmanghelich K, 2021. Using causal analysis for conceptual deep learning explanation International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI) , 519–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springenberg JT, Dosovitskiy A, Brox T, Riedmiller MA, 2015. Striving for Simplicity: The All Convolutional Net International Conference on Learning Representations (ICLR-workshop track) . [Google Scholar]
- Sundararajan M, Taly A, Yan Q, 2017. Axiomatic Attribution for Deep Networks 34th International Conference on Machine Learning (ICML) 70, 3319–3328. [Google Scholar]
- Tonekaboni S, Joshi S, McCradden MD, Goldenberg A, 2019. What Clinicians Want: Contextualizing Explainable Machine Learning for Clinical End Use URL: http://arxiv.org/abs/1905.05134. [Google Scholar]
- Van Looveren A, Klaise J, 2019. Interpretable Counterfactual Explanations Guided by Prototypes. arXiv e-prints , arXiv:1907.02584 [Google Scholar]
- Wada K, 2016. labelme Image Polygonal Annotation with Python https://github.com/wkentaro/labelme. [Google Scholar]
- Wang F, Kaushal R, Khullar D, 2020. Should health care demand interpretable artificial intelligence or accept “black Box” Medicine? Annals of Internal Medicine 172, 59–61. doi: 10.7326/M19-2548. [DOI] [PubMed] [Google Scholar]
- Wang P, Vasconcelos N, 2020. SCOUT: Self-Aware Discriminant Counterfactual Explanations IEEE Conference on Computer Vision and Pattern Recognition (CVPR) . [Google Scholar]
- Winkler J, Fink C, Toberer F, Enk A, Deinlein T, Hofmann-Wellenhof R, Thomas L, Lallas A, Blum A, Stolz W, Haenssle H, 2019. Association between surgical skin markings in dermoscopic images and diagnostic performance of a deep learning convolutional neural network for melanoma recognition. JAMA Dermatology 155. doi: 10.1001/jamadermatol.2019.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young K, Booth G, Simpson B, Dutton R, Shrapnel S, 2019. Deep neural network or dermatologist? International Workshop on Interpretability of Machine Intelligence in Medical Image Computing (IMIMIC) 11797 LNCS, 48–55. [Google Scholar]
- Zhou B, Khosla A, Lapedriza Ágata, Oliva A, Torralba A, 2015. Object detectors emerge in deep scene cnns International Conference on Learning Representations (ICLR) . [Google Scholar]
- Zhu JY, Park T, Isola P, Efros AA, 2017. Unpaired Image-to-Image Translation using Cycle-Consistent Adversarial Networks IEEE International Conference on Computer Vision (ICCV) . [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


