Abstract
DNA replication is essential for all living organisms. Several events can disrupt replication, including DNA damage (e.g., pyrimidine dimers, crosslinking) and so-called “roadblocks” (e.g., DNA-binding proteins or transcription). Bacteria have several well-characterized mechanisms for repairing damaged DNA and then restoring functional replication forks. However, little is known about the repair of stalled or arrested replication forks in the absence of chemical alterations to DNA. Using a library of random transposon insertions in Bacillus subtilis, we identified 35 genes that affect the ability of cells to survive exposure to an inhibitor that arrests replication elongation, but does not cause chemical alteration of the DNA. Genes identified include those involved in iron-sulfur homeostasis, cell envelope biogenesis, and DNA repair and recombination. In B. subtilis, and many bacteria, two nucleases (AddAB and RecJ) are involved in early steps in repairing replication forks arrested by chemical damage to DNA and loss of either nuclease causes increased sensitivity to DNA damaging agents. These nucleases resect DNA ends, leading to assembly of the recombinase RecA onto the single-stranded DNA. Notably, we found that disruption of recJ increased survival of cells following replication arrest, indicating that in the absence of chemical damage to DNA, RecJ is detrimental to survival. In contrast, and as expected, disruption of addA decreased survival of cells following replication arrest, indicating that AddA promotes survival. The different phenotypes of addA and recJ mutants appeared to be due to differences in assembly of RecA onto DNA. RecJ appeared to promote too much assembly of RecA filaments. Our results indicate that in the absence of chemical damage to DNA, RecA is dispensable for cells to survive replication arrest and that the stable RecA nucleofilaments favored by the RecJ pathway may lead to cell death by preventing proper processing of the arrested replication fork.
Author summary
DNA replication is essential for life. A variety of events, including chemical damage to DNA (e.g., caused by irradiation with ultraviolet light) and other cellular stresses that do not cause chemical modification of the DNA, can perturb and stop ongoing replication. Cell survival often depends on the ability to resume (restart) replication after such disruptions and many organisms have multiple mechanisms for restarting replication. Many bacterial species have two pathways for restarting DNA replication following DNA damage (e.g., exposure to ultraviolet light), and loss of either of these pathways makes cells more sensitive to UV irradiation. We found that one of these two pathways is detrimental and the other pathway is beneficial for bacterial survival following replication arrest in the absence of chemical damage to DNA. These surprising findings highlight key differences between the two pathways and the fine balance organisms face in maintaining replication restart pathways that are sometimes helpful and other times harmful.
Introduction
DNA replication is essential for all living organisms, and a variety of events can perturb replication, including DNA damage and replication arrest due to “roadblocks” such as DNA-binding proteins or transcription. In any of these cases, some or all the components of the replication machinery (the replisome) may disassemble from the complex, leading to the collapse of the replication fork [1–3]. If a subsequent replisome reaches unrepaired lesions, nicks, or collapsed forks, it can turn these defects into double-strand breaks, which, if not repaired, are lethal.
Proper cell growth and division following replication arrest depend on the restart of replication. A wide range of bacteria use a similar mechanism for replication restart: the DNA is processed to enable the formation of a structure resembling a replication fork, followed by reassembly of the replisome (reviewed in [4]). This processing typically involves homologous recombination mediated by RecA [5, 6]. To initiate homologous recombination, helicases and nucleases process DNA at the arrested fork to generate single-stranded DNA (ssDNA), in a process called end-resection [7, 8].
Bacillus subtilis has two nucleases capable of performing end-resection, RecJ and AddAB. In response to DNA damage, the exonuclease activity of RecJ can expand ssDNA gaps by several kilobases [7, 9]. In contrast, the helicase-nuclease complex AddAB, a functional homolog of RecBC in Escherichia coli, binds blunt or near blunt DNA ends. AddAB degrades both strands until reaching a Chi site [10] where it switches to degrade only a single DNA strand in the 5’ to 3’ direction [8]. Both the RecJ and AddAB end-resection pathways in B. subtilis lead to loading of RecA onto the DNA by the loader protein RecO [11]. RecA forms nucleofilaments that can search and invade complementary double-stranded DNA, creating cross-shaped DNA structures known as Holliday junctions. Finally, Holliday junctions are processed into a substrate for the helicase and replication restart protein PriA. PriA functions downstream of AddAB and RecJ, binds to stalled replication forks and promotes assembly of the replicative helicase and primosome enabling replication restart (reviewed in [12, 13]).
Most of what is known about the bacterial response to replication arrest comes from experiments using DNA damage to disrupt DNA replication. Repair of damaged DNA and replication restart are both required for optimal survival, making it difficult to differentiate between processes related to the damage itself or the arrest of replication forks. We were interested in identifying genes that influence the ability of cells to survive replication arrest, without the confounding effects of chemical damage (e.g., pyrimidine dimers, cross-links) to DNA.
To cause replication fork arrest independent of DNA damage, we used HPUra (6-(p-hydroxyphenylazo)-uracil), an azopyrimidine that reversibly binds to and inhibits the catalytic subunit of DNA polymerase III (PolC) in B. subtilis [14]. We used Tn-seq (transposon insertion mutagenesis with massively parallel sequencing) to identify candidate genes that affect cell survival following the arrest of replication elongation caused by treatment with HPUra.
We found that many processes appeared to be important for survival, including: regulation of oxidative homeostasis, cell wall stability, and DNA repair pathways. Interestingly, we found that one of the DNA end-resection pathways that is important for surviving DNA damage was detrimental to surviving replication arrest. Loss of recJ, recO, and recF was beneficial to cell survival, indicating that the functional genes were detrimental. In contrast, functional addA, recN, and recU were beneficial for cell survival following replication fork arrest. Our findings indicate that although AddAB and RecJ both carry out end-resection, they commit replication fork repair to distinct pathways. We found that these pathways led to different amounts of RecA that formed filaments on the ssDNA. Following replication fork arrest, RecJ or the recombinase loader and regulator (recO and recF, respectively) led to excessive RecA loading, accumulation of unrepaired forks, and ultimately an increase in cell death. Our data indicate that B. subtilis activates at least two pathways to repair arrested replication forks due to replisome malfunction and that one pathway (AddAB) is beneficial and the other (RecJ) is detrimental to cell survival.
Results
Identification of genes involved in cell survival following replication arrest
Using Tn-seq, we identified non-essential genes that significantly affected the ability of cells to survive and recover from replication arrest caused by treatment with HPUra. We used a previously described library of ~1.7 x 105 unique transposon insertions of a modified version of the magellan6 transposon, magellan6x, distributed throughout the B. subtilis genome [15]. This library of cells was split in two and treated or not with HPUra for one hour. The cells were then removed from HPUra, allowing replication and cell growth to resume. Aliquots of cells were harvested 2, 3, and 4 hours later, and the location and relative number of transposon insertions in a given chromosomal site were determined by high-throughput sequencing. In a population of random transposon insertions, mutations that lead to decreased survival would be underrepresented, and the ratio between the frequency of insertions in treated and untreated samples would be <1. Conversely, insertion mutations that improved survival would be overrepresented, resulting in frequency ratios >1. The vast majority of genes had a similar number of insertions (frequency ratios of ~1) with or without HPUra, indicating that these are neutral (S1 Table).
Thirty-five genes were classified as affecting survival and recovery from HPUra (see Material and Methods for analysis and criteria), and 13 of these were validated using targeted gene disruptions (Table 1). Several cellular processes appeared to affect survival, including: (i) iron-sulfur and oxidative homeostasis (rex, nadABC, defB, and nifS), likely related to the induction of oxidative stress and genes regulating NAD/NADH following treatment with HPUra [16]; (ii) cell wall stability (ltaS, rseP, oppAB, ydiL, cwlO and ponA); (iii) induction of lysogenic phage (yoyI, yonH from the SPß prophage and xpf, xkdBC from PBSX); and (iv) DNA repair and recombination. Below, we focus on several of the DNA recombination and repair genes (Fig 1A). We also validated seven of the other genes identified in the screen using targeted gene disruptions (Table 1), but did not further evaluate them as part of this study. The number of insertion sites and read frequencies for all open reading frames are presented (S1 Table).
Table 1. Genes identified in Tn-seq analysis of cells surviving replication arrest in the absence of DNA damage.
| Gene | Function | Log2 FC, Tn-seqa | Survival of null mutantsb | Strainb |
|---|---|---|---|---|
| DNA repair and recombination | ||||
| polA | DNA polymerase I | -6.51 | 0.06 (0.01) | LSF41 |
| recU | Holliday junction-specific endonuclease | -2.46 | 0.22 (0.09) | GP891 |
| recN | recognition of DNA double strand breaks | -1.65 | 0.14 (0.08) | LSF298 |
| addA | ATP-dependent deoxyribonuclease subunit A | -1.55 | 0.10 (0.06) | LSF253 |
| nfo | endonuclease IV | -1.11 | ND | |
| recJ | 5’-3’ exonuclease | 1.04 | 12 (1.7) | LSF200 |
| walJ | metallo-β-lactamase/ 5’-3’ exonuclease | 1.29 | 2.89 (0.07) | BKE40370 |
| Prophages | ||||
| yonH | unknown function (SPß) | 1.09 | 1.6 (0.3)c | LSF203 |
| yoyI | transmembrane protein (SPß) | 1.24 | ||
| xkdC | unknown function (PBSX) | 1.39 | 1.9 (0.1)d | LSF204 |
| xkdB | protein with LexA-type HTH domain (PBSX) | 1.46 | ||
| xpf | positive control sigma-like factor (PBSX) | 1.47 | ||
| Iron-Sulfur and oxidative homeostasis | ||||
| rex | redox-sensing transcriptional repressor | -1.24 | ND | |
| nadC | nicotinate-nucleotide pyrophosphorylase | 1.39 | ND | |
| nadA | quinolinate synthetase | 1.47 | ND | |
| defB | peptide deformylase | 1.64 | ND | |
| nifS | cysteine desulfurase | 1.84 | ND | |
| nadB | L-aspartate oxidase | 2.03 | ND | |
| Cell wall and membrane | ||||
| ltaS | polyglycerolphosphate lipoteichoic acid synthase | -1.79 | ND | |
| rseP | inner membrane zinc metalloprotease | -1.68 | ND | |
| oppA | oligopeptide ABC transporter binding lipoprotein | -1.67 | 0.21 (0.07) | JRL131 |
| ponA | peptidoglycan glycosyltransferase | -1.43 | 0.11 (0.03) | CMJ293 |
| oppB | oligopeptide ABC transporter permease | -1.40 | 0.3 (0.11) | JRL189 |
| ydiL | membrane protease | -1.05 | ND | |
| cwlO | secreted cell wall DL-endopeptidase | 1.10 | 2.2 (0.4) | CMJ374 |
| Other | ||||
| hrcA | heat-inducible transcription repressor | -1.93 | 0.26 (0.09) | LSF233 |
| bkdB | E2 subunit, lipoamide acyltransferase | -1.43 | ND | |
| speE | spermidine synthase | -1.38 | 0.3 (0.15) | LSF20 |
| yutD | unknown function | -1.12 | ND | |
| ytpQ | unknown function | -1.10 | ND | |
| argG | argininosuccinate synthase | 1.07 | ND | |
| ywdH | aldehyde dehydrogenase | 1.08 | ND | |
| yoaQ | unknown function | 1.25 | ND | |
| ycgE | transcriptional regulator | 1.39 | 4 (2) | LSF444 |
| argH | argininosuccinate lyase | 1.42 | ND | |
a Log2 FC: Log2 of the ratio of normalized number of insertions from the Tn-seq analyses in the indicated gene 4 h after removal of HPUra compared to that of cells that had not been treated with HPUra.
b Survival after HPUra treatment of defined null mutants (strain numbers listed) relative to that of the isogenic parental strain, with one standard deviation of the mean in parenthesis.
c Sensitivity of a strain devoid of SPß was used, rather than individual yonH and yoyI mutants.
d Sensitivity of a strain devoid of PBSX was used, rather than individual xkdC, xkdB, and xpf mutants.
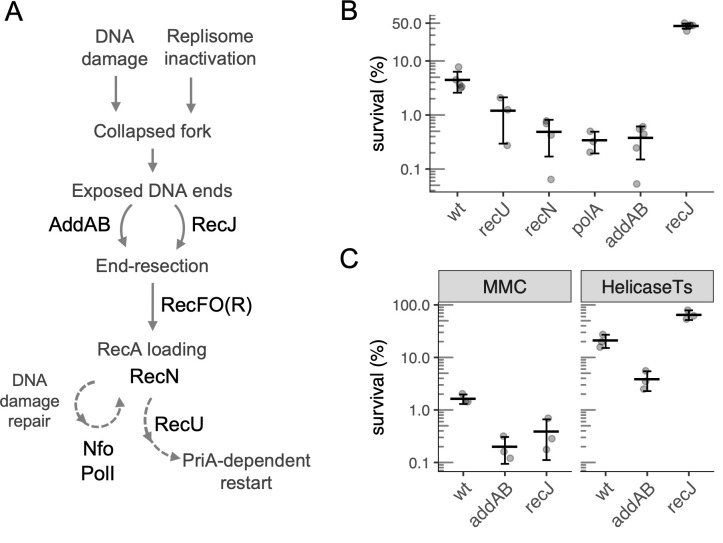
Fig 1. Pathways and genes involved in cell survival after arrest of DNA replication.
A. General view of the roles of the DNA repair and recombination genes identified in the Tn-seq screen. The dashed arrows represent multistep pathways. B. Percent survival of wild type (strain JMA222) and recU (GP891), polA (LSF41), recN (LSF298), addAB (LSF253) and recJ (LSF200) null mutants after 3 h of replication arrest caused by HPUra treatment. Percent survival is relative to cell viability at the time of arrest. All mutants were statistically different from wild type, as assessed by two-sample t-tests (P < 0.025). C. Effects of recJ and addAB null mutations on cell survival following replication arrest due to treatment with the DNA damaging agent mitomycin-C (MMC) or inactivation of the replicative helicase (DnaC). Left panel: survival of wild-type (JMA222), ΔaddAB (LSF253) and ΔrecJ (LSF200) cells after 3 h of treatment with 0.33 μg/ml of MMC. Right panel: wild-type, ΔrecJ and ΔaddAB cells carrying a dnaCts allele (LSF176, LSF274, and LSF326) were grown at permissive temperature (30°C) to early exponential phase and shifted to non-permissive temperature (49°C) for 4 h. Percent survival is relative to the population before treatment (left) or the temperature shift (right). The deletion mutants in both conditions were statistically different from wild type, as assessed by two-sample t-tests (P < 0.025). Throughout this work, error bars correspond to one standard deviation of the mean and each point is from an independent culture (biological replicates).
Effects of null mutations in genes involved in DNA repair and recombination on survival after replication arrest
In order to confirm the importance of the DNA repair and recombination genes identified in the Tn-seq screen, we made null mutations in six of the loci identified (polA, recU, recN, recJ, walJ, and addAB; note that addB and addA comprise an operon and for all experiments except where indicated, both genes were deleted) and tested their effects on survival after HPUra-mediated replication fork arrest. We treated wild-type and mutant cells with HPUra for 3 h and then removed HPUra and measured the number of viable cells remaining. Approximately 4.5% of wild-type cells survived this prolonged replication arrest (Fig 1B). In contrast, only 0.5–1% of polA, recU, recN, and addAB mutant cells survived.
In contrast to the detrimental effects of the loss of polA, recU, recN, and addAB on surviving replication fork arrest, we found that loss of recJ promoted survival. In the recJ null mutant, approximately 45% of cells remained viable after 3 hr of replication arrest (Fig 1B). These results indicate that normally RecJ is detrimental to survival of replication arrest in the absence of DNA lesions. Together, these results confirm the initial Tn-seq data that indicated that these genes are important for promoting survival following replication fork arrest in the absence of DNA damage.
Comparison of recJ and addAB mutants in response to replication arrest
Both RecJ and AddAB are involved in end-resection (Fig 1A), and both improve survival after treatment with DNA-damaging agents [17–20]. Therefore, it was somewhat surprising that a recJ null mutant had increased survival in response to replication fork arrest in the absence of externally induced chemical damage to DNA.
We confirmed that ΔrecJ and ΔaddAB mutants were more sensitive than wild-type cells to treatment with the DNA damaging agent mitomycin C (MMC). Approximately 1.5% of wild-type cells survived treatment with MMC (0.33 μg/ml) for 3 h (Fig 1C). In contrast, addAB and recJ mutants had 7 and 4-fold lower survival, respectively.
To verify that the effects of recJ and addAB on survival and recovery from HPUra were due to replication arrest, rather than some other possible effect, we tested their survival under conditions in which replication was arrested with a temperature-sensitive allele of the replicative DNA helicase (dnaCts) [21]. Inactivation of the replicative helicase mirrored the phenotypes observed during HPUra-mediated replication arrest (Fig 1C). We grew dnaCts (LSF176), dnaCts ΔaddAB (LSF326), and dnaCts ΔrecJ (LSF274) strains at a permissive temperature (30°C). Cells were then shifted to non-permissive temperature (49°C) to arrest replication elongation. After 4 h at 49°C, we measured the number of viable cells from each of the three strains. In the otherwise wild-type cells, prolonged replication arrest at the non-permissive temperature caused a decrease in cell viability to about 20% of the original population (Fig 1C). The loss of addAB exacerbated this effect, and only 4% of cells were viable. In contrast, the loss of recJ largely protected the cells from the outcome of replication arrest due to inactivation of the replicative helicase, and 65% of cells survived (Fig 1C). Together, our results indicate that recJ is detrimental to surviving replication fork arrest in the absence of DNA lesions, and confirm previous work [17, 19] demonstrating that recJ contributes to survival following chemical damage to DNA.
Loss of RecA loader RecO or regulator RecF protects ΔaddAB cells from arrest-induced death
End-resection by AddAB or RecJ produces the substrate for interaction with RecA. RecO is the main protein responsible for loading RecA onto the ssDNA substrate (Fig 1A), and RecF stabilizes the resulting RecA nucleofilaments [11]. Insertions in recO and recF were not recovered in our initial Tn-seq screen because they were depleted from the initial library prior to treatment with HPUra (S1 Table). We suspect that this depletion was because cells were not kept in the dark during library preparation and these mutants are probably sensitive to ambient UV light.
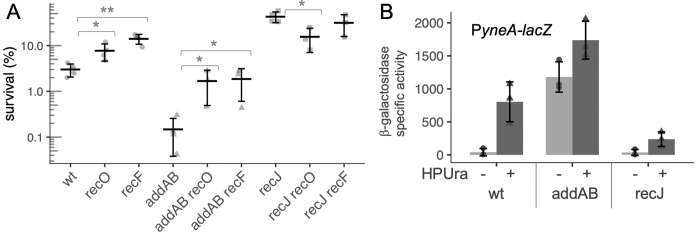
To test if recO and recF contribute to survival following replication arrest, we constructed recO and recF null mutants. We found that deletion of either recO or recF significantly increased cell survival following replication fork arrest (HPUra treatment) in comparison to wild type (Fig 2A). Both deletions suppressed the sensitivity of ΔaddAB mutants to HPUra (Fig 2A, see addAB recO and addAB recF), indicating that loading RecA was at least partially responsible for decreased survival of the ΔaddAB mutant.
Fig 2. RecA loading is detrimental to survival during HPUra-induced replication arrest.
A. Survival after HPUra treatment is greater in the absence of RecA loader and regulator (RecO, RecF). Fold change in survival after 3 h of arrest for wild type (JMA222), ΔrecO (LSF814), ΔrecF (LSF270), ΔaddAB (LSF253), ΔaddAB ΔrecO (LSF817), ΔaddAB ΔrecF (LSF709), ΔrecJ (LSF200), ΔrecJ ΔrecO (LSF815), ΔrecJ ΔrecF (LSF282). Significant difference in means (two-sample T-test): * P < 0.05; ** P < 0.01. B. RecA activity is increased in the absence of addAB and decreased in the absence of recJ. Transcription from the promoter of the SOS-inducible gene yneA is derepressed when RecA is active and serves as an indicator of RecA activity. Cultures of wild-type, ΔrecJ, and ΔaddAB strains containing amyE::PyneA-lacZ (LSF633, LSF634, and LSF635, respectively) were treated or not with HPUra. After 30 min, aliquots were collected for measuring ß-galactosidase activity. Results from three independent experiments are presented.
In contrast to effects on the addAB mutant, deletion of recF had little if any effect on the sensitivity of the ΔrecJ mutant to HPUra, indicating that RecF and RecJ likely affect the same process–formation of stable RecA filaments (Fig 2A).
The SOS response is reduced in the absence of recJ and increased in the absence of addAB
We postulated that if the increased survival of ΔrecJ is a consequence of lower stability of RecA filaments, then ΔrecJ might have lower RecA activity. Once assembled onto ssDNA, RecA induces autocleavage of the SOS repressor LexA, allowing expression of many genes involved in the response to genotoxic stress [22–24]. Consequently, expression of genes repressed by LexA can be used as a proxy for the levels of activated RecA. yneA encodes an inhibitor of cell division [25], and is repressed by LexA and de-repressed in response to DNA damage and replication fork arrest [26].
We used a fusion of the promoter region of yneA to lacZ (PyneA-lacZ) and measured expression in the mutants in the presence and absence of HPUra. For wild-type cells in defined minimal medium, ß-galactosidase activity from the PyneA-lacZ fusion increased approximately 20-fold after 30 min of replication arrest (Fig 2B). In the ΔrecJ mutant, there was a basal level of activity similar to that in wild type, but only an approximately 6-fold increase after addition of HPUra, indicating that recJ plays an important role in activation of RecA. The absence of addAB led to very high PyneA-lacZ activity during exponential growth (Fig 2B), indicating that there is an abundance of active RecA in these cells.
Lysogenic phages are not responsible for the effects of recJ and addAB on survival after replication arrest
It seemed plausible that the impact of recJ and addAB on survival following replication stress could be due to different levels of phage induction, as both mutations affect the activity of RecA. The B. subtilis strains used in the experiments described above have two resident lysogenic phages, both of which undergo RecA-dependent activation after genotoxic stress: SPß and the defective phage PBSX [16, 26–28]. Both encode toxins and autolysins which, when expressed, can cause cell death. We found that insertions in two genes from SPß (yoyI, yonH) and three genes involved in the early steps of PBSX induction (xpf, xkdBC) were overrepresented in the screen, indicating that loss of these genes helped cells survive arrest of replication elongation (Table 1).
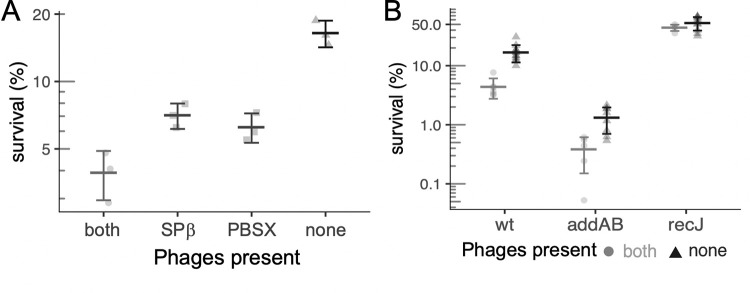
We measured survival of otherwise wild-type cells missing either PBSX, SPß, or both following replication arrest with HPUra. In these experiments, the survival of wild-type cells (containing both lysogenic phages) was 3.5% (Fig 3A). Loss of either phage increased survival to approximately 7%, and loss of both phages increased survival to 15–20% (Fig 3A).
Fig 3. Effect of the resident lysogenic phages SPß and PBSX on survival after replication arrest.
A. The lysogenic phages SPß and PBSX (defective) negatively affect cell survival after replication arrest (3 h HPUra treatment). Percent survival of strains JMA222 (which has both phages), LSF204 (only SPß; ΔPBSX), LSF203 (only PBSX; ΔSPß), and LSF225 (no phage; ΔPBSX, ΔSPß). B. recJ and addAB affect survival following replication arrest, even in cells without SPß and PBSX. Survival after 3 h of replication arrest for strains lacking both phage is plotted as dark gray triangles with black error bars; wt (LSF225), ΔaddAB (LSF254), and ΔrecJ (LSF231). Data from corresponding strains with both phages (JMA222, LSF253, and LSF200, respectively), plotted as light gray circles with gray error bars, are shown for comparison, and are the same as presented in Fig 1B. Statistically different means, by two-tailed t-test: * P < 0.05; ** P < 0.001; *** P < 0.0001).
Although the presence of SPß and PBSX contributed to cell death following replication arrest, we found that addAB and recJ affected survival following replication arrest even in cells missing both PBSX and SPß. In strains cured of phages, loss of addAB caused a 10-fold drop in survival, indicating that this mutant was still more sensitive to replication arrest than wild-type cells (Fig 3B). This effect was comparable to that caused by loss of addAB in strains with both phages (~8-fold). The absence of recJ significantly increased the survival of cells after treatment with HPUra to ~50%, regardless of the presence or absence of PBSX and SPß. The increase in survival of the phage null mutant caused by loss of recJ was approximately 3-fold (Fig 3B). Together, these results indicate that the effects of addAB and recJ on survival are largely independent of killing by SPß or PBSX.
End-resection is needed for survival following replication fork arrest, but RecA is not
We sought to determine how strains lacking recJ and addAB-encoded nucleases involved in end resection or lacking recA behaved following arrest of replication forks with HPUra. RecA is essential for survival following treatment with DNA damaging agents. Due to the inability to efficiently load RecA in the absence of the nucleases AddAB and RecJ, a double ΔaddAB ΔrecJ mutant has a similar extreme sensitivity to DNA damaging agents as ΔrecA [17, 19, 29].
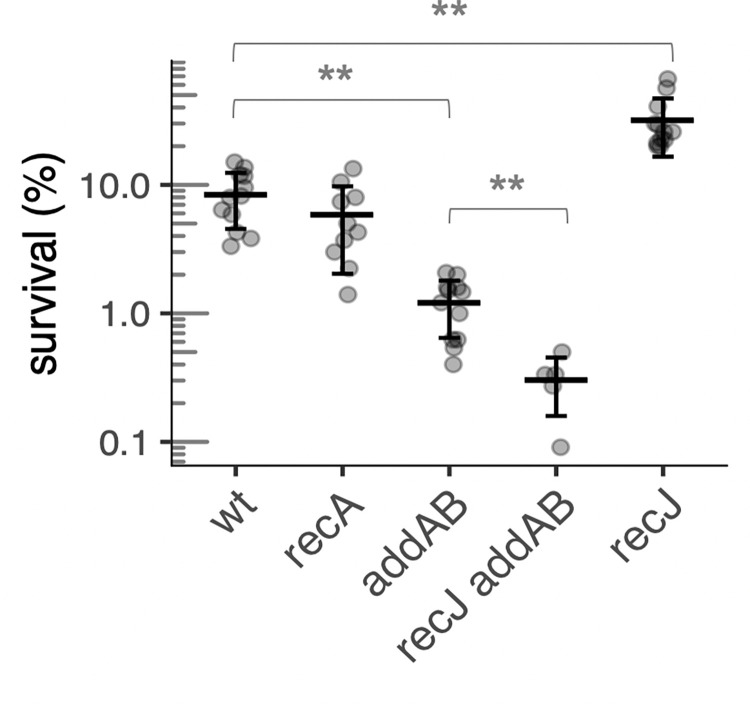
The survival following replication arrest of a strain missing both addAB and recJ was 0.3% (Fig 4). This is significantly worse than the effect caused by ΔaddAB and indicates that the loss of both end-resection pathways is synergistic and that end-resection is needed for survival in response to replication arrest in the absence of chemical damage to the DNA.
Fig 4. Effect of RecA and end-resection on survival of HPUra-induced replication arrest, in the absence of phages.
Percent survival after 3 h of replication arrest for wild-type (LSF225), and recA (LSF658), addAB (LSF254), recJ addAB (LSF648), and recJ (LSF231) null mutants. ** Statistically different means, by two-tailed t-test (P < 0.005).
In contrast, recA did not detectably affect survival following replication arrest. Survival of the recA null mutant was similar to that of otherwise wild-type cells, but lower than the recJ mutant (Fig 4). Based on these results, we conclude that end-resection mediated by either AddAB or RecJ is essential for recovery from replication fork arrest, but that RecA and RecA-mediated homologous recombination is not required.
It is formally possible that RecA is required in the absence of RecJ. For example, survival in the absence of RecJ depends on AddAB and this could be due to end processing by AddAB and subsequent assembly of RecA onto the ssDNA. If true, then survival of a recJ recA double mutant should be less than that of a recJ single mutant. We found that the increased survival conferred by the loss of recJ was not dependent on the presence of recA (Table 2), indicating that the AddAB pathway that is required for surviving replication arrest in the absence of RecJ was not dependent on RecA.
Table 2. recA is not needed for increased survival to replication stress in the absence of recJ.
| Strain1 | Survival relative to wild type2 |
|---|---|
| Wild type (LSF225) | 1.0 ± 0.5 |
| recJ (LSF231) | 3.8 ± 1.7 |
| recA (LSF658) | 1.0 ± 0.5 |
| recJ recA (LSF659) | 3.2 ± 1.4 |
1The indicated strains were grown to mid-exponential phase in defined minimal medium, treated with HPUra for 3 h, and the fraction of cells that survived were measured.
2Survival of each strain was determined and is presented relative to that of the wild type grown and treated in parallel. Data for each strain are averages and standard error of the mean from four independent matched experiments.
RecA activity correlates with the accumulation of repair centers formed during HPUra-induced replication arrest
Since RecA itself was not required to survive HPUra-induced replication arrest, but mutants with lower RecA activity like ΔrecJ and ΔrecF had increased survival, we considered the possibility that the activity of RecJ and the formation of RecA filaments were interfering with the proper processing of the arrested replication forks. If true, then there should be greater accumulation of recombination intermediates and exposed DNA ends in strains with more loading of RecA. Upon formation of a double-strand break, the DNA ends are processed by end-resection allowing RecA to be loaded onto the ssDNA. It was previously believed that following chemical damage to DNA, RecN was recruited directly to dsDNA breaks before RecA loading [19, 30, 31]. However, recent evidence indicates that RecN is recruited to the already-processed DNA in a manner dependent on RecA [17]. Nonetheless, the formation of foci of RecN can be used as a proxy for DNA repair centers, likely as a result of a dsDNA break or gaps that have been processed to allow RecA to assemble into a filament [17, 19, 30, 31].
RecN with a C-terminal tag has been shown to be functional in DNA repair and was used previously to monitor repair centers formed following DNA damage [17, 19, 30]. We constructed strains in which the native recN was replaced by recN-mNeongreen, and monitored formation of foci to determine the frequency of repair centers arising from DNA damage-independent replication arrest. Different mutants containing the fusion were treated with HPUra and scored by the presence of foci after 30 min of replication arrest (Fig 5A). In wild-type cells growing exponentially, cells with RecN-mNeongreen foci were uncommon (< 2% cells). Thirty minutes after replication arrest, approximately 20% of wild-type cells had at least one focus of RecN-mNeongreen (Fig 5A).
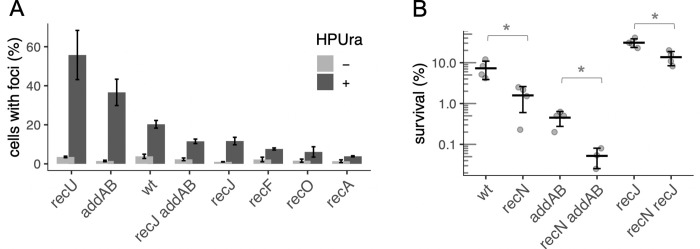
Fig 5. Accumulation of exposed DNA ends during replication arrest by HPUra.
A. Repair centers in response to exposed DNA ends were visualized by fluorescence microscopy of cells expressing the RecN-mNeongreen fusion. Cell membranes were stained with FM 4–64 and DNA was stained with DAPI. Wild-type (LSF708), ΔrecU (LSF740), ΔaddA (LSF650), ΔrecO (LSF818), ΔrecJ (LSF649), ΔrecF (LSF816), ΔrecJ ΔaddA (LSF706), and ΔrecA (LSF756) cells were analyzed without (gray) or 30 min after treatment with HPUra (black). At least 800 cells from two biological replicates were analyzed from each strain for the presence of RecN-mNeongreen foci. The means and standard errors are presented. B. Loss of recN causes a phenotype in cells with or without addAB or recJ. Percent survival of wild type (LSF225), ΔrecN (LSF536), ΔaddAB (LSF254), ΔrecN ΔaddA (LSF652), ΔrecJ (LSF231), and ΔrecN ΔrecJ (LSF651) are plotted. *Statistically different means according to two-tailed t-test: * P < 0.05.
Effects of mutations on the formation of RecN-mNeongreen foci were significant. In the absence of recU or addA, 56% and 37% of cells, respectively, had at least one focus of RecN-mNeongreen (Fig 5A). In contrast, only ~12% of ΔrecJ cells contained foci and a double ΔrecJ ΔaddA mutant behaved similarly to ΔrecJ (Fig 5A). Finally, deleting recA or recO or recF, the genes encoding the RecA-loader and stabilizer, greatly reduced the number of foci of RecN-mNeongreen (Fig 5A). This indicates that association of RecN to the DNA damage centers is dependent on RecA during damage-independent replication arrest and is consistent with similar findings after treatment with agents that induce DNA damage [17].
These results support the model that RecJ and the recombinase loader and regulator impede the proper processing of the replication fork via excessive formation of RecA filaments in the absence of chemical damage to DNA, and indicate that the RecJ pathway is the major source of RecA assembly following HPUra-induced replication arrest. The neutral effect of recA in survival (Fig 4) may be a combination of increased survival due to lack of loading on DNA processed by RecJ and a decrease in RecA-mediated processes for restart. In addition, there are likely to be RecA-independent processes for replication restart (see Discussion).
RecN is important for survival if RecA is assembled onto DNA
Our results indicate that there are breaks or gaps in dsDNA that lead to DNA processing and RecA loading in wild-type cells following replication fork arrest. Furthermore, we found that the loss of recN caused a decrease in survival in otherwise wild-type cells following replication fork arrest (Table 1 and Fig 5B). In the absence of addAB, there was a further decrease in survival upon loss of recN (Fig 5B), consistent with the results indicating that RecA is more active and there are more dsDNA breaks in the absence of addAB than in otherwise wild-type cells. In addition, loss of recN also decreased survival of the recJ mutant (Fig 5B), indicating that RecN is beneficial regardless of which pathway (AddAB or RecJ) is used to process DNA ends.
Discussion
The replisome pauses or collapses not only when it encounters damaged DNA, but also when roadblocks, such as DNA binding proteins, are encountered, or when the replisome itself is inhibited or damaged. We specifically focused on how cells recover from replication arrest by replisome inhibition, in the absence of chemical damage to DNA. It is well known that RecA activity and both the RecJ and AddAB end-resection pathways are critically important for surviving exposure to DNA damage-inducing agents [32–34]. In contrast, we found a different scenario when there is no DNA damage and a replisome component is inhibited (chemically or using a thermosensitive allele). In this situation, end-resection was necessary, but the RecJ pathway and high RecA activity were detrimental, and the AddAB pathway was beneficial. Our results show that different types of genotoxic stresses require different levels of RecA activity, which are obtained by the AddAB vs. RecJ pathways, and that the choice of pathway affects survival.
The consequences of using each end-resection pathway depend on the kind of replication arrest
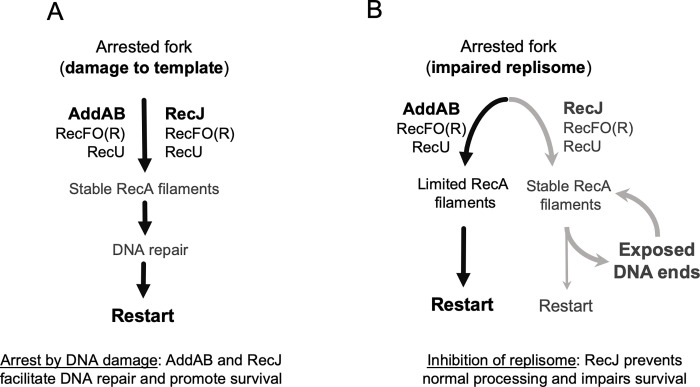
As depicted in Fig 6A, when replication is blocked, both end-resection pathways work together to process the collapsed replication fork. After fork regression, which involves reannealing of the template DNA and annealing of the two daughter strands, AddAB can unwind and degrade the daughter strands until it reaches a Chi site, after which it begins generating a 3’ ssDNA tail [10]. RecJ can target the collapsed fork directly, or an already resected fork [35], probably extending the ssDNA substrate generated by AddAB and leading to longer RecA nucleofilaments. RecFO(R) then ensures RecA nucleofilament loading and stability [11]. In this case, the role of RecA is not to promote recombination, but rather to stabilize the DNA at the fork until the DNA damage has been repaired [32, 33]. Our results indicate that when replication stops because a replisome component is inhibited, AddAB activity supports optimal recovery (black arrows in the left side of Fig 6B) and after fork regression, AddAB generates less substrate for RecA loading than does RecJ.
Fig 6. The role of end-resection pathways AddAB and RecJ on survival outcomes after replication arrest.
A. When replication arrest is caused by lesions on the DNA template or other genotoxic stresses requiring RecA activity, both end-resection pathways work together, generating stable RecA filaments that delay restart and support DNA repair. B. If replication arrest is caused by the inhibition of replisome components or other stresses that do not require RecA, AddAB favors a pathway (black arrows) with limited RecA filaments that promotes survival. In this situation, RecJ and the RecA loader RecO and regulator RecF impair survival by promoting a pathway (gray arrows) with high RecA activity. This could prevent proper processing of the fork, exposing DNA ends and creating a futile circle.
Our results indicate that RecJ and the loader RecO (and regulator RecF) negatively affect the process of fork repair by enhancing RecA activity and preventing fork regression (gray arrows by the right side of Fig 6B). RecJ can target a range of DNA substrates [9, 36]. In addition to competing with AddAB for access to an already regressed fork, it can degrade a daughter strand in the collapsed fork, which would prevent fork regression and the generation of the blunt ends AddAB requires. Also, RecJ and RecO localize to the replication fork via interaction with SSB [37], and therefore have immediate access to the replication fork, whereas AddAB does not. In the absence of DNA damage, the resulting stable RecA filaments that help promote survival in other conditions may expose DNA ends that unnecessarily delay fork processing. Any DNA breaks or exposed ends arising during this step would need to go through an additional round of end-resection and RecA loading, feeding the detrimental pathway.
Different types of genotoxic stresses require different levels and duration of RecA activity
The different effects that RecA has on survival, depending on the kind of replication arrest, could be due to the complexity of the resulting collapsed fork and duration of RecA activity necessary for restart. Treatment with MMC, for example, impedes replication due to template lesions and “dirty” DNA breaks, i.e. ones that cannot be ligated because they lack a 3’ hydroxyl or 5’ phosphate. The recovery from this kind of arrest can take up to 3 h in B. subtilis and relies on high levels of RecA activity [29, 30]. The repair of a “clean”, ligatable single break in the chromosome, however, requires very transient RecA filaments and takes less than 20 min, at least in E. coli [38].
When a DNA-binding protein acting as a roadblock arrests replication, E. coli cells are able to process these forks and recover normally, regardless of the presence of recA [39]. In this case, there are no chemical lesions to be repaired or complex structures to be disassembled at the fork before processing. Likewise, our data indicate that the collapsed forks resulting from the inhibition of a replisome component (PolC by HPUra treatment or arrest of the replicative helicase DnaC using at temperature sensitive mutant) readily undergo fork regression and processing. In this scenario, we found that the excess RecA activity that results from RecJ and the recombinase loader and regulator is not only unnecessary but also detrimental to survival.
Replication-transcription conflicts also cause replication arrest (reviewed in [3, 40]). The importance of RecA in replication restart at these sites is unclear. One report found that RecA was not required in E. coli [41], but another found that RecA was needed in B. subtilis [42]. The fact that RecA is required in B. subtilis for resolving head-on replication-transcription conflicts, but not for recovery from replication arrest caused by HPUra may be due to the different structures that are formed after each of these treatments. HPUra treatment has been shown to leave many of the replisome components largely intact, at least initially [43–46], whereas replication-transcription conflicts likely cause replisome disassembly [47], and this may predispose arrested forks to different restart pathways.
RecJ leads to increased RecA loading and DNA repair centers and increased cell death when fork repair does not require RecA activity
We hypothesize that during HPUra-induced arrest in a wild-type cell, the stable RecA nucleofilaments favored by RecJ prevent optimal repair of the collapsed fork by unnecessarily delaying restart, perhaps by inhibiting the ability of PriA to function or increasing the frequency of double-strand breaks, or both. We noted a positive correlation between the predicted levels of RecA activity in different mutants following HPUra treatment and the accumulation of DNA repair centers (increased RecN-mNeongreen foci; Fig 5A). These are likely a result of increased DNA breaks or gaps that have undergone end-resection and RecA loading, followed by RecN recruitment [17, 48]. Stable RecJ-dependent RecA filaments may factor into the propensity to cause breaks in these long stretches of coated ssDNA, leading to more DNA repair centers. Similar to what is observed for repair in the context of DNA damage, we found that deletion of recN decreased survival of HPUra treatment in wild type, ΔaddAB, and ΔrecJ mutants, indicating that under all of these conditions, recruitment of RecN helps to repair the DNA, regardless of the level of activity of RecA. These two mechanisms–increased DNA repair centers (likely from increased dsDNA breaks or gaps) and unnecessarily stalled restart–presumably synergistically contribute to cell death during HPUra treatment.
Few studies have focused on the survival outcome of recombination mutants to replisome inhibition in the absence of external DNA damage. Nevertheless, there are some hints of a disadvantageous role of RecJ in situations where RecA is not required in E. coli, which could be explained by excessive loading of the recombinase. A strain lacking recB (analogous to ΔaddAB in B. subtilis) can still process a fork after collapse due to a roadblock, but survival was low, while a strain lacking recA was indistinguishable from wild type [39]. Although a mechanism for these findings was not proposed, the results indicate that a pathway parallel to RecBCD (presumably RecJ/gap repair) may be processing the collapsed forks in a way that decreases viability. Toxicity caused by RecJ, RecFOR, and RecA also occurs in the absence of the helicases (DNA translocases) encoded by rep and uvrD in E. coli and pcrA in B. subtilis. This toxicity is apparently due to excess or improper loading of RecA at blocked replication forks and led to the understanding that the helicases normally help remove or clear RecA to ensure proper replication restart [49–51]. These examples highlight the importance of controlling RecA loading and how improper or excessive loading can be detrimental to the cell.
RecJ homologs are widespread from bacteria to eukaryotes [52]. It is not known if the detrimental nature of RecJ and excessive loading of RecA in response to replication arrest is widespread, but the results from E. coli discussed above, and the conserved nature of RecJ and RecA indicate that this might be the case.
Possible mechanisms of surviving replication fork arrest in the absence of RecA
Our data indicate that although RecA is dispensable, and at times detrimental, to the repair of replication arrest in absence of external DNA damage, end-resection is still required for survival. This indicates that formation of RecA-dependent Holliday junctions is not required for survival, and that the end-resection has a purpose other than loading of RecA. In E. coli, replication fork reversal can be accomplished independently of RecA, using RecBCD (analogous to AddAB in B. subitilis) and RuvAB, and has been observed in situations of stalled forks in the absence of chemical damage to DNA, including fork stalling caused by mutations in the DNA polymerase holoenzyme (reviewed in [2]), conflicts caused by DNA binding proteins [39], or head-on collisions with RNA polymerase [41]. A similar mechanism may occur in the absence of RecA after HPUra treatment, however it is also possible there are other RecA-independent mechanisms in B. subtilis, perhaps including non-homologous end-joining [53] or a role for DNA polymerase I [54, 55]. Additionally, breaks that occur at the replication fork resulting in damage to only one copy of the chromosome would yield one viable cell, rather than two, but would not be lethal nor require recombination. Indeed, in E. coli, in the absence of RecA, damaged chromosomes are degraded by RecBCD and can result in cells containing an odd number of chromosomes due to selective degradation of the damaged chromosomes [56].
Bacteria use all pathways for repair, even when it hinders proper recovery
Each end-resection mechanism seems adapted to a kind of genotoxic stress in B. subtilis: AddAB for double-strand breaks from damage or fork collapse, and RecJ for gaps from DNA repair. However, our data highlighted that bacteria may use an unfavorable (“wrong”) pathway for repair. RecJ commits repair of some DNA damage-independent collapsed forks to a pathway that decreases viability, and it probably competes with AddAB for blunt DNA ends. RecJ and stable RecA loading only promote survival during DNA repair, but our data indicate that they are also used during replication stresses that do not require RecA. Ultimately, this indicates that cells do not have an effective mechanism to discriminate between these different types of broken forks, potentially leading to increased cell death in some, but enabling robust repair and restart in other circumstances.
Materials and methods
Bacterial strains
Unless otherwise indicated, all B. subtilis strains used in this work derive from JMA222 [57], a version of JH642 [58] cured of the integrative and conjugative element ICEBs1. Strain genotypes and references for previously described strains are listed in Table 3. Previously described alleles were introduced into JMA222 by transforming naturally competent cells with genomic DNA from the relevant strain. These alleles include: dnaC30(ts)-mls [21], recU::cat [59], recA260 [60], walJ::erm [61], recF::spc [62], amyE::Pyne-lacZ cat [63], ponA::spc [64], and addAB::kan [65].
Table 3. B. subtilis strains used.
| Strain | Relevant genotypea [reference] |
|---|---|
| AG174 (a.k.a., JH642) | pheA1 trpC2 [58, 66] |
| BKE40370 | walJ::erm (168 background) [61] |
| CMJ293 | ponA::spc |
| CMJ374 | cwlO::spc |
| GP891 (a.k.a., LSF625) | recU::cat trpC2 ICEBs1 (168 background) [59] |
| JMA222 | pheA1 trpC2 ICEBs10 [57] |
| JRL131 | ΔoppA (AG174 background) [67] |
| JRL189 | ΔoppB (AG174 background) [67] |
| LSF20 | speE::spc |
| LSF41 | polA::cat (a.k.a., AB41) |
| LSF176 | dnaC30(ts)-mls |
| LSF197 | ΔSPß::lox-kan-lox |
| LSF198 | ΔPBSX::lox-kan-lox |
| LSF200 | recJ::spc |
| LSF201 | ΔPBSX::lox-kan-lox (PY79 strain background) [68] |
| LSF203 | ΔSPß::lox |
| LSF204 | ΔPBSX::lox |
| LSF225 | ΔSPß::lox ΔPBSX::lox |
| LSF231 | ΔSPß::lox ΔPBSX::lox recJ::spc |
| LSF233 | ICEBs10 hrcA::cat |
| LSF253 | addBA::kan |
| LSF254 | ΔSPß::lox ΔPBSX::lox addBA::kan |
| LSF270 | recF::spc (a.k.a., CAL1614) |
| LSF274 | dnaC30(ts) recJ::spc |
| LSF282 | recJ::cat recF::spc |
| LSF284 | recN::[recN-9aa-mNeongreen kan]; referred to as recN-mng kan |
| LSF298 | recN::kan |
| LSF326 | dnaC30(ts) addBA::kan |
| LSF444 | SPß::lox PBSX::lox ycgE::spc |
| LSF536 | ΔSPß::lox ΔPBSX::lox recN::kan |
| LSF633 | amyE::[PyneA-lacZ cat] |
| LSF634 | amyE::[PyneA-lacZ cat] recJ::spc |
| LSF635 | amyE::[PyneA-lacZ cat] addBA::kan |
| LSF648 | ΔSPß::lox ΔPBSX::lox recJ::spc addBA::kan |
| LSF649 | ΔSPß::lox ΔPBSX::lox recN-mng kan recJ::spc |
| LSF650 | ΔSPß::lox ΔPBSX::lox recN-mng kan addA::spc |
| LSF651 | ΔSPß::lox ΔPBSX::lox recN::kan recJ::spc |
| LSF652 | ΔSPß::lox ΔPBSX::lox recN::kan addA::spc |
| LSF658 | ΔSPß::lox ΔPBSX::lox recA260 (mls, cat) |
| LSF659 | ΔSPẞ::lox ΔPBSX::lox recJ::spc recA260 (mls, cat) |
| LSF706 | ΔSPß::lox ΔPBSX::lox recN-mng kan recJ::cat addA::spc |
| LSF708 | ΔSPß::lox ΔPBSX::lox recN-mng kan |
| LSF709 | addBA::kan recF::spc |
| LSF740 | ΔSPß::lox ΔPBSX::lox recN-mng kan recU::cat |
| LSF756 | ΔSPß::lox ΔPBSX::lox recN-mng kan recA260 (mls, cat) |
| LSF814 | recO::cat |
| LSF815 | recO::cat recJ::spc |
| LSF816 | ΔSPß::lox ΔPBSX::lox recN-mng kan recF::spc |
| LSF817 | recO::cat addBA::kan |
| LSF818 | ΔSPß::lox ΔPBSX::lox recN-mng kan recO::cat |
a Unless otherwise indicated, strains are derived from JMA222, and the trpC2, pheA1, and ICEBs10 alleles are not shown.
Introduction of new alleles was done by transforming cells with linear DNA products generated either by traditional cloning or by isothermal assembly [69]. Insertion-deletion constructs contained antibiotic resistance cassettes typically flanked by 800–1000 bp of genomic sequences upstream and downstream of region to be deleted. These alleles include addA::spc, recJ::spc, recN::kan, recO::cat, recF::spc, ycgE::spc, hrcA::cat, polA::cat and cwlO::spc.
To generate deletions of SPß (LSF203) and PBSX (LSF204), each was substituted by a kanamycin resistance gene flanked by loxP sites, generating strains LSF197 (ΔSPß::lox-kan-lox) and LSF198 (ΔPBSX::lox-kan-lox). The cassettes were removed by the Cre recombinase (from bacteriophage P1) using the cre expression vector pDR244 as previously described [70], leaving a 75 bp insertion (’scar’ containing a single lox site). A strain devoid of both phages (LSF225) resulted from transforming LSF203 (ΔSPß::lox) with genomic DNA from LSF201 (a ΔPBSX::lox-kan-lox strain), and recombining the antibiotic cassette out. The ΔPBSX allele also removes spoIISABC (toxin-antitoxin-antitoxin) which is just downstream of PBSX but is not normally thought to be part of it [71].
To visualize DNA repair centers, we constructed a strain with a recN-mNeongreen (recN-mng) fusion, inserted in the native locus such that the fusion is the only recN allele. The full linear DNA product contained the 800 bp at the 3’ end of recN, an in-frame linker (5’- CTCGAGGGATCTGGCCAAGGAAGCGGC-3’; encoding 9 amino acids), the mNeongreen coding sequence (a gift from Ethan Garner), a kanamycin resistance cassette, and 800 bp genomic sequence downstream of the recN stop codon. Transformation of this construct into JMA222 generated strain LSF284 that contains recN::[recN-9aa-mNeongreen kan]; referred to as recN-mng kan. The recN-mng kan allele from LSF284 was then introduced into various strains, including LSF225 (generating LSF708).
Media and antibiotics
All experiments were performed in defined minimal medium containing 50 mM MOPS (S750) and supplemented with 1% glucose, 0.1% glutamate, 40 μg/ml phenylalanine and 40 μg/ml tryptophan [72]. When required, the following concentration of antibiotics were used: 5 mg/ml chloramphenicol, 100 μg/ml spectinomycin, 5 μg/ml kanamycin, and 0.5μg/ml erthyromycin plus 12.5 μg/ml lincomycin to select for macrolide-lincosamide-streptogramin B (MLS) resistance. Serial dilutions of cultures for spot-plating were made in Spizizen minimal salts medium [73].
Viability assays
B. subtilis strains were streaked from -80°C freezer stocks on LB plates and grown overnight at 37°C. Single colonies were transferred to S750, and dilutions of those were grown overnight with vigorous shaking at 37°C. Starter cultures between OD600 0.05–0.5 were diluted to OD600 0.025, grown for at least three generations, and adjusted to OD600 0.1. recA mutants were grown in the dark (ΔrecA mutants, and many other mutants altered in the DNA damage response, are sensitive to ambient UV light). HPUra (6-(p-hydroxyphenylazo)-uracil; [11]), 38 μg/ml, or mitomycin C (Sigma-Aldrich), 0.33 μg/ml, was added as indicated, and cultures were kept at 37°C with shaking for 3 h. Viability was assessed before and at various times after addition of HPUra or MMC by making 10-fold serial dilutions from 100 μl of cultures in a 96-well plate and spotting 10 μl on LB plates, in triplicate. Results are in percentage of colony forming units present after 3 h treatment in relation to immediately before addition of HPUra or MMC.
Temperature sensitive mutants with the dnaCts (replicative helicase) allele [21] were grown at 30°C. At OD600 0.1, the cultures were shifted to 49°C, the non-permissive temperature, to arrest DNA replication. Dilutions of the cultures were plated before and every hour after the shift to assess survival. Results are percentages of colony forming units (at 30°C) present after 4 h in relation to immediately before the temperature shift.
Statistical comparison between different groups was performed in R, using the varequal and t.test functions. Two-tailed, paired t-tests were computed using a pooled estimate of variances for similarly distributed samples or were estimated separately for both groups and the Welch modification to the degrees of freedom was used.
Tn-seq screen
The transposon insertion library used in this work has been described in detail [15]. The library contains ~1–2 x 105 unique transposon insertions in strain JMA222.
An aliquot of the transposon insertion library was diluted in minimal medium, and grown from an OD600 of 0.02 to 0.3 in defined minimal medium at 37°C. The culture was then split and HPUra (38 μg/ml) was added to half to arrest replication, and the other half was untreated. After 1 h at 37°C, cells from both cultures were harvested by centrifugation, washed, and resuspended in fresh minimal medium, diluting the cultures back to OD600 0.15. They were allowed to recover for 4 h, and aliquots were harvested every hour during this time. DNA was prepared for sequencing as described previously [15], and sequencing was done using an Illumina HiSeq by the MIT BioMicro Center.
Tn-seq data analysis
Tn-seq data were initially processed as described [15]. Sequences adjacent to the transposon were mapped to JMA222 genome using Bowtie2 [74]. The resulting files contained the number of reads per genomic coordinate in each sample. Our resulting mapped libraries had approximately 105 independent insertions each, with an average of one insertion per 37 base pairs, and 19 insertions per non-essential gene.
Subsequent analyses and file manipulations were performed using custom-made R scripts. Any genomic position with less than 3 reads was discarded, to avoid potential noise due to rare insertions or misaligned reads. Then, we used inter-sample quantile normalization from the preprocessCore package (available at www.bioconductor.org) to ensure that different samples were comparable. Finally, we calculated the number of reads interrupting each gene in every sample–only insertions mapping to 5–95% internal sequence were considered since insertions in the extremities of essential genes are sometimes tolerated.
To assess enrichment or depletion of insertion mutants, we calculated the ratio between the number of reads in the treated and control samples. For proper analysis of Tn-seq, it is important that libraries being compared were expanded roughly the same number of generations before sequencing. Since replication and cell division in the treated library was arrested during 1 h (leaving them one generation “behind” the control libraries), we compared the HPUra-treated libraries with the controls harvested an hour earlier: the HPUra sample harvested after 4 h of recovery was compared to the control harvested after 3 h, and HPUra 2 h with control cells harvested after 1 h of recovery.
Finally, for a gene to be considered to affect survival in HPUra in relation to the control, it had to satisfy the following requisites: (i) be longer than 200 bp; (ii) have more than 5 insertions in the corresponding control library; (iii) have log2 fold change > 1 at 4 h; (iv) have an amplified change in read frequency over time. These criteria aimed to restrict the number of candidate genes and decrease false-positives. The importance of additional candidate genes of interest (e.g., recO, recF, and recA) that did not meet all of these criteria were evaluated using targeted gene disruptions.
ß-Galactosidase activity assay
Cultures in mid-exponential phase growing in minimal medium were diluted to OD600 0.1. One-milliliter aliquots were permeabilized with 15 μl toluene and stored at -20°C. For cells undergoing replication arrest, HPUra (38 μg/ml) was added for 30 min, 1.5 ml aliquots were centrifuged at 3000 x g for 2 min and cells were resuspended in 1.5 ml of fresh medium. One milliliter aliquots were permeabilized and frozen, and the remainder was used to measure OD600 to correct for cell recovery. ß-galactosidase specific activity was determined as previously described [75, 76].
Fluorescence microscopy
We used RecN-mNeongreen as a marker to measure DNA repair centers. Cells containing this construct were grown in minimal media until OD600 ~ 0.1. Cells were either untreated or treated with HPUra (38 μg/ml) for 20 min. Aliquots were then added to a tube containing 4’,6’-diamidino-2-phenylindole (DAPI, 1 μg/ml final concentration) and FM4-64 (2.5 μg/ml), to stain nucleoids and membranes, respectively, incubated for 10 min at 37°C, and then prepared for imaging.
For imaging, cultures were concentrated roughly four-fold by centrifugation (2 min at 2000x g) and 2 μl cells were placed on a slice of 1.5% UltraPure agarose (Invitrogen) made with minimal medium. The agarose slice was placed, cells down, on standard coverslips and imaged on a Nikon Ti-E inverted microscope. Fluorescence was generated using excitation/emission of 500/535 nm for RecN-mNeongreen (1 ms exposure), 350/460 nm for DAPI-stained nucleoids (100 ms) and 510/630 for FM4-64-stained membranes (100 ms). Image processing was performed using Fiji [77], where foci were detected with the Laplacian of Gaussians (LoG) detector in the TrackMate plugin [78], with a 0.7 nm blob diameter and threshold between 10 and 20 (determined by the control samples).
Supporting information
Samples were collected and analyzed 2, 3, and 4 hours after treatment, or from parallel untreated cultures. The sequencing data used for these analyses have been deposited in the NCBI Gene Expression Omnibus [79] and are accessible through GEO Series accession number GSE221151.
(XLSX)
The excel spreadsheet contains the underlying data for the experiments presented in each of the figures and Table 2.
(XLSX)
Acknowledgments
We thank Ethan Garner and Alexandre Bisson-Filho for providing the mNeongreen allele and initial help with microscopy, Juan Alonso for helpful comments on the manuscript, and Lyle Simmons for providing HPUra.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was supported, in part, by grants from the National Institute of General Medical Sciences of the National Institutes of Health under award numbers R37 GM041934 and R35 GM122538 to ADG. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Cox MM, Goodman MF, Kreuzer KN, Sherratt DJ, Sandler SJ, Marians KJ. The importance of repairing stalled replication forks. Nature. 2000;404: 37–41. doi: 10.1038/35003501 [DOI] [PubMed] [Google Scholar]
- 2.Michel B, Boubakri H, Baharoglu Z, LeMasson M, Lestini R. Recombination proteins and rescue of arrested replication forks. DNA Repair. 2007;6: 967–980. doi: 10.1016/j.dnarep.2007.02.016 [DOI] [PubMed] [Google Scholar]
- 3.Lang KS, Merrikh H. The clash of macromolecular titans: replication-transcription conflicts in bacteria. Annu Rev Microbiol. 2018;72: 71–88. doi: 10.1146/annurev-micro-090817-062514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sinha AK, Possoz C, Leach DRF. The roles of bacterial DNA double-strand break repair proteins in chromosomal DNA Replication. Fems Microbiol Rev. 2020;44: 351–368. doi: 10.1093/femsre/fuaa009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kowalczykowski SC. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem Sci. 2000;25: 156–165. doi: 10.1016/s0968-0004(00)01569-3 [DOI] [PubMed] [Google Scholar]
- 6.Lusetti SL, Cox MM. The bacterial RecA protein and the recombinational DNA repair of stalled replication forks. Annu Rev Biochem. 2002;71: 71–100. doi: 10.1146/annurev.biochem.71.083101.133940 [DOI] [PubMed] [Google Scholar]
- 7.Lovett ST, Kolodner RD. Identification and purification of a single-stranded-DNA-specific exonuclease encoded by the recJ gene of Escherichia coli. Proc Natl Acad Sci U S A. 1989;86: 2627–2631. doi: 10.1073/pnas.86.8.2627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chédin F, Ehrlich SD, Kowalczykowski SC. The Bacillus subtilis AddAB helicase/nuclease is regulated by its cognate Chi sequence in vitro. J Mol Biol. 2000;298: 7–20. doi: 10.1006/jmbi.2000.3556 [DOI] [PubMed] [Google Scholar]
- 9.Dianov G, Sedgwick B, Daly G, Olsson M, Lovett S, Lindahl T. Release of 5′-terminal deoxyribose-phosphate residues from incised abasic sites in DNA by the Escherichia coli RecJ protein. Nucleic Acids Res. 1994;22: 993–998. doi: 10.1093/nar/22.6.993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chédin F, Noirot P, Biaudet V, Ehrlich SD. A five-nucleotide sequence protects DNA from exonucleolytic degradation by AddAB, the RecBCD analogue of Bacillus subtilis. Mol Microbiol. 1998;29: 1369–1377. doi: 10.1046/j.1365-2958.1998.01018.x [DOI] [PubMed] [Google Scholar]
- 11.Lenhart JS, Brandes ER, Schroeder JW, Sorenson RJ, Showalter HD, Simmons LA. RecO and RecR are necessary for RecA loading in response to DNA damage and replication fork stress. J Bacteriol. 2014;196: 2851–2860. doi: 10.1128/JB.01494-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bénédicte M, Sandler SJ. Replication Restart in Bacteria. Journal of Bacteriology. 2017;199: e00102–17. doi: 10.1128/JB.00102-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Windgassen TA, Wessel SR, Bhattacharyya B, Keck JL. Mechanisms of bacterial DNA replication restart. Nucleic Acids Research. 2018;46: 504–519. doi: 10.1093/nar/gkx1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown NC. 6-(p-Hydroxyphenylazo)-uracil: a selective inhibitor of host DNA replication in phage-infected Bacillus subtilis. Proc Natl Acad Sci U S A. 1970;67: 1454–1461. doi: 10.1073/pnas.67.3.1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson CM, Grossman AD. Identification of host genes that affect acquisition of an integrative and conjugative element in Bacillus subtilis. Mol Microbiol. 2014;93: 1284–1301. doi: 10.1111/mmi.12736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goranov AI, Katz L, Breier AM, Burge CB, Grossman AD. A transcriptional response to replication status mediated by the conserved bacterial replication protein DnaA. Proceedings of the Nationall Academy of Science U S A. 2005;102: 12932–7. doi: 10.1073/pnas.0506174102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McLean EK, Lenhart JS, Simmons LA. RecA Is Required for the Assembly of RecN into DNA Repair Complexes on the Nucleoid. Journal of Bacteriology. 2021;203: e00240–21. doi: 10.1128/JB.00240-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanchez H, Kidane D, Reed P, Curtis FA, Cozar MC, Graumann PL, et al. The RuvAB branch migration translocase and RecU Holliday junction resolvase are required for double-stranded DNA break repair in Bacillus subtilis. Genetics. 2005;171: 873–883. doi: 10.1534/genetics.105.045906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanchez H, Kidane D, Cozar MC, Graumann PL, Alonso JC. Recruitment of Bacillus subtilis RecN to DNA double-strand breaks in the absence of DNA end processing. J Bacteriol. 2006;188: 353–360. doi: 10.1128/JB.188.2.353-360.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carrasco B, Yadav T, Serrano E, Alonso JC. Bacillus subtilis RecO and SsbA are crucial for RecA-mediated recombinational DNA repair. Nucleic Acids Res. 2015;43: 5984–5997. doi: 10.1093/nar/gkv545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karamata D, Gross JD. Isolation and genetic analysis of temperature-sensitive mutants of B. subtilis defective in DNA synthesis. Mol Gen Genet. 1970;108: 277–287. doi: 10.1007/BF00283358 [DOI] [PubMed] [Google Scholar]
- 22.Little JW, Edmiston SH, Pacelli LZ, Mount DW. Cleavage of the Escherichia coli lexA protein by the recA protease. Proc Natl Acad Sci U S A. 1980;77: 3225–3229. doi: 10.1073/pnas.77.6.3225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wojciechowski MF, Peterson KR, Love PE. Regulation of the SOS response in Bacillus subtilis: evidence for a LexA repressor homolog. J Bacteriol. 1991;173: 6489–6498. doi: 10.1128/jb.173.20.6489-6498.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Au N, Kuester-Schoeck E, Mandava V, Bothwell LE, Canny SP, Chachu K, et al. Genetic composition of the Bacillus subtilis SOS system. J Bacteriol. 2005;187: 7655–7666. doi: 10.1128/JB.187.22.7655-7666.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawai Y, Moriya S, Ogasawara N. Identification of a protein, YneA, responsible for cell division suppression during the SOS response in Bacillus subtilis. Molecular Microbiology. 2003;47: 1113–1122. doi: 10.1046/j.1365-2958.2003.03360.x [DOI] [PubMed] [Google Scholar]
- 26.Goranov AI, Kuester-Schoeck E, Wang JD, Grossman AD. Characterization of the global transcriptional responses to different types of DNA damage and disruption of replication in Bacillus subtilis. Journal of Bacteriology. 2006;188: 5595–605. doi: 10.1128/JB.00342-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Craig NL, Roberts JW. E. coli recA protein-directed cleavage of phage λ repressor requires polynucleotide. Nature. 1980;283: 26–30. doi: 10.1038/283026a0 [DOI] [PubMed] [Google Scholar]
- 28.Phizicky EM, Roberts JW. Kinetics of recA protein-directed inactivation of repressors of phage λ and phage P22. J Mol Biol. 1980;139: 319–328. doi: 10.1016/0022-2836(80)90133-3 [DOI] [PubMed] [Google Scholar]
- 29.Kidane D, Graumann PL. Dynamic formation of RecA filaments at DNA double strand break repair centers in live cells. J Cell Biol. 2005;170: 357–366. doi: 10.1083/jcb.200412090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kidane D, Sanchez H, Alonso JC, Graumann PL. Visualization of DNA double-strand break repair in live bacteria reveals dynamic recruitment of Bacillus subtilis RecF, RecO and RecN proteins to distinct sites on the nucleoids. Mol Microbiol. 2004;52: 1627–1639. doi: 10.1111/j.1365-2958.2004.04102.x [DOI] [PubMed] [Google Scholar]
- 31.Rösch TC, Altenburger S, Oviedo-Bocanegra L, Pediaditakis M, Najjar NE, Fritz G, et al. Single molecule tracking reveals spatio-temporal dynamics of bacterial DNA repair centres. Scientific Reports. 2018;8: 16450. doi: 10.1038/s41598-018-34572-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Courcelle J, Hanawalt PC. RecQ and RecJ process blocked replication forks prior to the resumption of replication in UV-irradiated Escherichia coli. Molecular & general genetics: MGG. 1999;262: 543–551. [DOI] [PubMed] [Google Scholar]
- 33.Courcelle J, Hanawalt PC. RecA-Dependent Recovery of Arrested DNA Replication Forks. Annu Rev Genet. 2003;37: 611. doi: 10.1146/annurev.genet.37.110801.142616 [DOI] [PubMed] [Google Scholar]
- 34.Lenhart JS, Schroeder JW, Walsh BW, Simmons LA. DNA repair and genome maintenance in Bacillus subtilis. Microbiol Mol Biol Rev. 2012;76: 530–564. doi: 10.1128/MMBR.05020-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Viswanathan M, Lovett ST. Single-strand DNA-specific exonucleases in Escherichia coli: roles in repair and mutation avoidance. Genetics. 1998;149: 7–16. doi: 10.1093/genetics/149.1.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morimatsu K, Kowalczykowski SC. RecQ helicase and RecJ nuclease provide complementary functions to resect DNA for homologous recombination. Proc Natl Acad Sci U S A. 2014;111: E5133–E5142. doi: 10.1073/pnas.1420009111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Costes A, Lecointe F, McGovern S, Quevillon-Cheruel S, Polard P. The C-terminal domain of the bacterial SSB protein acts as a DNA maintenance hub at active chromosome replication forks. Matic Ivan, editor. PLoS Genet. 2010;6: e1001238. doi: 10.1371/journal.pgen.1001238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Amarh V, White MA, Leach DRF. Dynamics of RecA-mediated repair of replication-dependent DNA breaks. J Cell Biol. 2018;217: 2299–2307. doi: 10.1083/jcb.201803020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Possoz C, Filipe SR, Grainge I, Sherratt DJ. Tracking of controlled Escherichia coli replication fork stalling and restart at repressor-bound DNA in vivo. EMBO J. 2006;25: 2596–2604. doi: 10.1038/sj.emboj.7601155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Merrikh H, Zhang Y, Grossman AD, Wang JD. Replication–transcription conflicts in bacteria. Nat Rev Microbiol. 2012;10: 449–458. doi: 10.1038/nrmicro2800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.De Septenville AL, Duigou S, Boubakri H, Michel B. Replication Fork Reversal after Replication–Transcription Collision. PLoS Genet. 2012;8: e1002622. doi: 10.1371/journal.pgen.1002622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Million-Weaver S, Samadpour AN, Merrikh H. Replication restart after replication-transcription conflicts requires RecA in Bacillus subtilis. Henkin T M, editor. J Bacteriol. 2015;197: 2374–2382. doi: 10.1128/JB.00237-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liao Y, Li Y, Schroeder JW, Simmons LA, Biteen JS. Single-Molecule DNA Polymerase Dynamics at a Bacterial Replisome in Live Cells. Biophys J. 2016;111: 2562–2569. doi: 10.1016/j.bpj.2016.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liao Y, Schroeder JW, Gao B, Simmons LA, Biteen JS. Single-molecule motions and interactions in live cells reveal target search dynamics in mismatch repair. Proc Natl Acad Sci U S A. 2015;112: E6898–E6906. doi: 10.1073/pnas.1507386112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Y, Chen Z, Matthews LA, Simmons LA, Biteen JS. Dynamic exchange of two essential DNA polymerases during replication and after fork arrest. Biophys J. 2019;116: 684–693. doi: 10.1016/j.bpj.2019.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goranov AI, Breier AM, Merrikh H, Grossman AD. YabA of Bacillus subtilis controls DnaA-mediated replication initiation but not the transcriptional response to replication stress. Molecular Microbiology. 2009;74: 454–466. doi: 10.1111/j.1365-2958.2009.06876.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mangiameli SM, Merrikh CN, Wiggins PA, Merrikh H. Transcription leads to pervasive replisome instability in bacteria. van Oijen AM, editor. eLife. 2017;6: e19848. doi: 10.7554/eLife.19848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Keyamura K, Sakaguchi C, Kubota Y, Niki H, Hishida T. RecA Protein Recruits Structural Maintenance of Chromosomes (SMC)-like RecN Protein to DNA Double-strand Breaks *. Journal of Biological Chemistry. 2013;288: 29229–29237. doi: 10.1074/jbc.M113.485474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Florés M-J, Sanchez N, Michel B. A fork-clearing role for UvrD. Molecular Microbiology. 2005;57: 1664–1675. doi: 10.1111/j.1365-2958.2005.04753.x [DOI] [PubMed] [Google Scholar]
- 50.Lestini R, Michel B. UvrD and UvrD252 Counteract RecQ, RecJ, and RecFOR in a rep Mutant of Escherichia coli. Journal of Bacteriology. 2008;190: 5995–6001. doi: 10.1128/JB.00620-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Petit M-A, Ehrlich D. Essential bacterial helicases that counteract the toxicity of recombination proteins. The EMBO Journal. 2002;21: 3137–3147. doi: 10.1093/emboj/cdf317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sanchez-Pulido L, Ponting CP. Cdc45: the missing RecJ ortholog in eukaryotes? Bioinformatics. 2011;27: 1885–1888. doi: 10.1093/bioinformatics/btr332 [DOI] [PubMed] [Google Scholar]
- 53.Bertrand C, Thibessard A, Bruand C, Lecointe F, Leblond P. Bacterial NHEJ: a never ending story. Mol Microbiol. 2019;111: 1139–1151. doi: 10.1111/mmi.14218 [DOI] [PubMed] [Google Scholar]
- 54.Cooper PK, Hanawalt PC. Role of DNA Polymerase I and the rec System in Excision-Repair in Escherichia coli. Proceedings of the National Academy of Sciences. 1972;69: 1156–1160. doi: 10.1073/pnas.69.5.1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Patel PH, Suzuki M, Adman E, Shinkai A, Loeb LA. Prokaryotic DNA polymerase I: evolution, structure, and “base flipping” mechanism for nucleotide selection11Edited by J. H. Miller. Journal of Molecular Biology. 2001;308: 823–837. doi: 10.1006/jmbi.2001.4619 [DOI] [PubMed] [Google Scholar]
- 56.Skarstad K, Boye E. Degradation of individual chromosomes in recA mutants of Escherichia coli. Journal of Bacteriology. 1993;175: 5505–5509. doi: 10.1128/jb.175.17.5505-5509.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Auchtung JM, Lee CA, Monson RE, Lehman AP, Grossman AD. Regulation of a Bacillus subtilis mobile genetic element by intercellular signaling and the global DNA damage response. Proc Natl Acad Sci U S A. 2005;102: 12554–12559. doi: 10.1073/pnas.0505835102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Perego M, Spiegelman GB, Hoch JA. Structure of the gene for the transition state regulator, abrB: regulator synthesis is controlled by the spo0A sporulation gene in Bacillus subtilis. Molecular Microbiology. 1988;2: 689–699. [DOI] [PubMed] [Google Scholar]
- 59.Gunka K, Tholen S, Gerwig J, Herzberg C, Stülke J, Commichau FM. A high-frequency mutation in Bacillus subtilis: requirements for the decryptification of the gudB glutamate dehydrogenase gene. J Bacteriol. 2012;194: 1036–1044. doi: 10.1128/JB.06470-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cheo DL, Bayles KW, Yasbin RE. Molecular characterization of regulatory elements controlling expression of the Bacillus subtilis recA+ gene. Biochimie. 1992;74: 755–762. doi: 10.1016/0300-9084(92)90148-8 [DOI] [PubMed] [Google Scholar]
- 61.Koo B-M, Kritikos G, Farelli JD, Todor H, Tong K, Kimsey H, et al. Construction and analysis of two genome-scale deletion libraries for Bacillus subtilis. Cell Syst. 2017;4: 291–305.e7. doi: 10.1016/j.cels.2016.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sciochetti SA, Blakely GW, Piggot PJ. Growth phase variation in cell and nucleoid morphology in a Bacillus subtilis recA mutant. J Bacteriol. 2001;183: 2963–2968. doi: 10.1128/JB.183.9.2963-2968.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Biller SJ, Wayne KJ, Winkler ME, Burkholder WF. The putative hydrolase YycJ (WalJ) affects the coordination of cell division with DNA replication in Bacillus subtilis and may play a conserved role in cell wall metabolism. J Bacteriol. 2011;193: 896–908. doi: 10.1128/JB.00594-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Popham D L, Setlow P. Cloning, nucleotide sequence, and mutagenesis of the Bacillus subtilis ponA operon, which codes for penicillin-binding protein (PBP) 1 and a PBP-related factor. Journal of Bacteriology. 1995;177: 326–335. doi: 10.1128/jb.177.2.326-335.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Meima R, Haijema BJ, Venema G, Bron S. Overproduction of the ATP-dependent nuclease AddAB improves the structural stability of a model plasmid system inBacillus subtilis. Molecular and General Genetics MGG. 1995;248: 391–398. doi: 10.1007/BF02191638 [DOI] [PubMed] [Google Scholar]
- 66.Smith JL, Goldberg JM, Grossman AD. Complete genome sequences of Bacillus subtilis subsp. subtilis laboratory strains JH642 (AG174) and AG1839. Genome announcements. 2014;2: e00663–14. doi: 10.1128/genomeA.00663-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.LeDeaux JR, Solomon JM, Grossman AD. Analysis of non-polar deletion mutations in the genes of the spo0K (opp) operon of Bacillus subtilis. Fems Microbiol Lett. 1997;153: 63–69. doi: 10.1111/j.1574-6968.1997.tb10464.x [DOI] [PubMed] [Google Scholar]
- 68.Schroeder JW, Simmons LA. Complete Genome Sequence of Bacillus subtilis Strain PY79. Genome Announcements. 2013;1: e01085–13. doi: 10.1128/genomeA.01085-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA III, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature Methods. 2009;6: 343. doi: 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- 70.Meisner J, Llopis PM, Sham L, Garner E, Bernhardt TG, Rudner DZ. FtsEX is required for CwlO peptidoglycan hydrolase activity during cell wall elongation in Bacillus subtilis. Mol Microbiol. 2013;89: 1069–1083. doi: 10.1111/mmi.12330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gabriško M, Barák I. Evolution of the SpoIISABC Toxin-Antitoxin-Antitoxin System in Bacilli. Toxins. 2016;8: 180. doi: 10.3390/toxins8060180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jaacks KJ, Healy J, Losick R, Grossman AD. Identification and characterization of genes controlled by the sporulation regulatory gene SpooH in Bacillus subtilis. J Bacteriol. 1989;171: 4121–4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Harwood CR, Cutting SM. Molecular biological methods for Bacillus. Chichester; New York: Wiley; 1990. [Google Scholar]
- 74.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9: 357–359. doi: 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY; 1972. [Google Scholar]
- 76.Jones JM, Grinberg I, Eldar A, Grossman AD. A mobile genetic element increases bacterial host fitness by manipulating development. Levin PA, Storz G, Dubnau D, Dunny G, editors. eLife. 2021;10: e65924. doi: 10.7554/eLife.65924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9: 676–682. doi: 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tinevez J-Y, Perry N, Schindelin J, Hoopes GM, Reynolds GD, Laplantine E, et al. TrackMate: An open and extensible platform for single-particle tracking. Methods. 2017;115: 80–90. doi: 10.1016/j.ymeth.2016.09.016 [DOI] [PubMed] [Google Scholar]
- 79.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002; 30:207–10. doi: 10.1093/nar/30.1.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Samples were collected and analyzed 2, 3, and 4 hours after treatment, or from parallel untreated cultures. The sequencing data used for these analyses have been deposited in the NCBI Gene Expression Omnibus [79] and are accessible through GEO Series accession number GSE221151.
(XLSX)
The excel spreadsheet contains the underlying data for the experiments presented in each of the figures and Table 2.
(XLSX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.